- School of Biosciences, Cardiff University, Cardiff, United Kingdom
Recombinant proteins are of paramount importance for research, industrial and medical use. Numerous expression chassis are available for recombinant protein production, and while bacterial and mammalian cell cultures are the most widely used, recent developments have positioned transgenic plant chassis as viable and often preferential options. Plant chassis are easily maintained at low cost, are hugely scalable, and capable of producing large quantities of protein bearing complex post-translational modification. Several protein targets, including antibodies and vaccines against human disease, have been successfully produced in plants, highlighting the significant potential of plant chassis. The aim of this review is to act as a guide to producing recombinant protein in plants, discussing recent progress in the field and summarising the factors that must be considered when utilising plants as recombinant protein expression systems, with a focus on optimising recombinant protein expression at the genetic level, and the subsequent extraction and purification of target proteins, which can lead to substantial improvements in protein stability, yield and purity.
Introduction
Recombinant proteins (RPs) are proteins produced in heterologous systems, usually used to increase yields relative to the native system from which the protein is derived. Frequently, these proteins are pharmaceuticals, including vaccines and antibodies, which are produced as recombinant proteins in such scales that the biopharmaceutical sector has previously been the largest sector of the pharmaceutical market (Owczarek et al., 2019). Furthermore, protein drugs contributed to approximately 10% of the drug market in 2017 (Usmani et al., 2017). For biopharmaceuticals specifically, plants have already been shown to be a viable expression platform, and have been recently reviewed (Shanmugaraj et al., 2020), especially for the production of antibodies (Malaquias et al., 2021). Traditionally, mammalian cell cultures are the most favourable expression chassis, and between 2014 and 2018 they were used to produce 84% of novel recombinant proteins, while E. coli cell cultures were used for production of 8% and S. cerevisiae for 6.5% (Walsh, 2018). Broadly speaking, proteins for vaccines are not needed in substantial yields, but monoclonal antibody production and proteins produced for structural studies need high yields, requiring cost-efficient production platforms (Legastelois et al., 2016). However, it is unlikely that one optimal expression chassis exists for all RPs, as they each have different capabilities, and different RPs have various levels of complexity. For example, some proteins have eukaryotic-specific post-translational modifications (PTMs), and some are membranous meaning they have lower abundance relative to cytosolic proteins due to the reduced area that plasma membranes make up in most cells compared to the cytoplasm. Where possible, recombinant membrane proteins are produced in engineered E. coli strains (Schlegel et al., 2014). However, for proteins with PTMs, eukaryotic cell cultures including insect and mammalian cell lines are usually used (Hunter et al., 2018).
There is however an emerging potential for plants to be used as RP expression systems. Plant-made antibodies, or plantibodies, have been investigated as early as 1989 (Hiatt et al., 1989), and have had dramatic success in recent years (Malaquias et al., 2021), for example in producing a successful vaccine for the Ebola outbreak (Zhang et al., 2014), demonstrating that the system is suitable for expression of complex proteins with high yields. Moreover, technologies are developing that allow dramatically improved protein yields in plants, such as hyper-translatable viral elements for transgenes (Thuenemann et al., 2013) and commercially viable plant expression tools (Sainsbury et al., 2009). Due to these improvements, plant expression chassis have become a viable alternative to the conventional mammalian and bacterial systems, though further optimisation is still necessary.
The field of plant recombinant protein production has been heavily reviewed in recent years (Schillberg et al., 2019; Schillberg and Finnern, 2021; Schillberg and Spiegel, 2022), with reviews focussing on the advantages of plant RP expression systems, including the low upstream costs (Avesani et al., 2013; Chen and Davis, 2016), high capacity for scalability (Burnett and Burnett, 2019), potential for edible vaccines (Merlin et al., 2014; Kurup and Thomas, 2019), lack of endotoxin production (Fischer and Emans, 2000; Merlin et al., 2014), rapid protein production times that make them particularly suitable for vaccine production (Zhang et al., 2014; Chen and Davis, 2016; Rattanapisit et al., 2020; Makatsa et al., 2021), and the ability to produce complex post-translational modifications (Gomord and Faye, 2004; Millar et al., 2019), summarized in Table 1. Glycosylation is particularly important for biopharmaceutical proteins, with engineering efforts made to humanize plant glycosylation and ensure safe immunogenicity (Sethuraman & Stadheim, 2006; Castilho et al., 2010; Montero-Morales & Steinkellner, 2018).
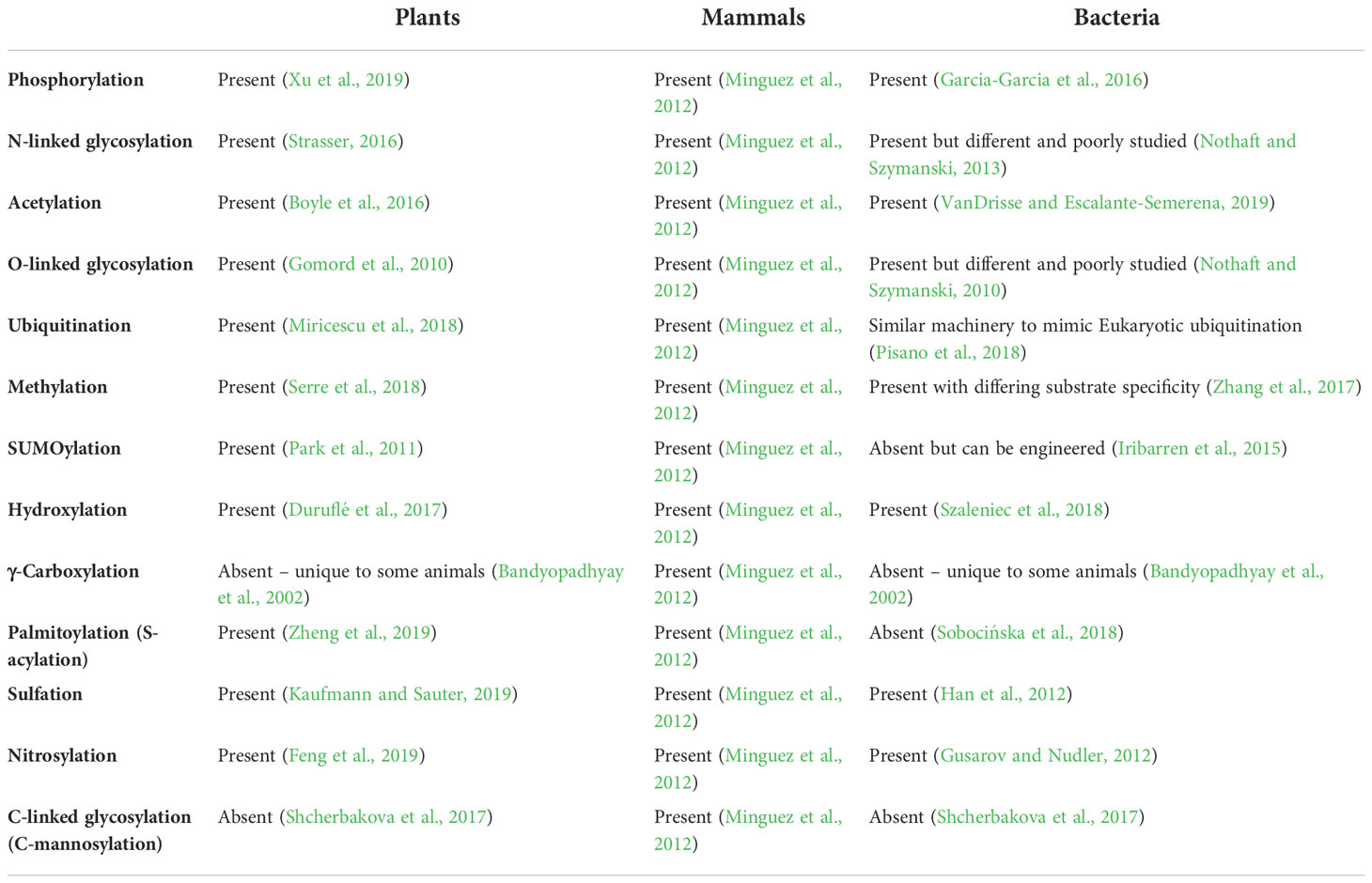
Table 1 Comparison of the capabilities of plants, mammals, and bacteria the carry out the 13 most common Eukaryotic PTMs, according to Minguez et al. (2012).
There are several available plant chassis that each have advantages and disadvantages, reviewed in detail by Sabalza et al. (2013) and more recently by (Gerszberg and Hnatuszko-Konka, 2022). Tobacco (Nicotiana spp.) is often used for RP production due to its rapid growth time, scalability, large number of seeds, and well-established transgenesis methods (Ma et al., 2003; Chen and Lai, 2015; Burnett and Burnett, 2019; Moon et al., 2019). Furthermore, the most commonly used plant cell culture is the tobacco Bright-Yellow 2 (BY-2) cell line, however, plant cell cultures rarely present any significant advantages over other eukaryotic cell cultures (Schillberg et al., 2019; Karki et al., 2021; Sethi et al., 2021). Legumes have high protein content (Burnett and Burnett, 2019), rapid growth (Abd-Aziz et al., 2020) and several transformable species such as Mucuna bracteate, but often show unfavourable heterogenous glycosylation (Abd-Aziz et al., 2020; Debler et al., 2021). Fruit and vegetable crops, including Lactuca and Solanacea species have been made to produce RPs, with the main advantage that they can be eaten, negating downstream purification costs (Merlin et al., 2014; Kurup and Thomas, 2019; Moon et al., 2019; Shanmugaraj et al., 2021). For these, several tissue-specific promoters are available (Lim et al., 2012; Ding et al., 2022), and researchers have found that growth conditions can dramatically affect protein content (Matsuda et al., 2009). Aquatic plants can be used, in particular Lemnoideae or Wolffia species, due to their fast growth rates and high yields, but transformation methods are poorly optimised (Khvatkov et al., 2018; Chanroj et al., 2021). Cereals present another promising expression system, recently reviewed (Mirzaee et al., 2022), with the advantage that their seeds have a high protein content and enable RPs to be stably stored (Stöger et al., 2000). Commonly used cereals include maize (Naqvi et al., 2011; Sabalza et al., 2013), barley (Ritala et al., 2008; Magnusdottir et al., 2013; Shanmugaraj et al., 2021), and rice, the latter particularly as cell-cultures (Huang et al., 2002; Kuo et al., 2013; Huang et al., 2015; Liu et al., 2017). Mosses such as Physcomitrella patens have had success for RP production (Baur et al., 2005; Gitzinger et al., 2009; Niederkrüger et al., 2014; Reski et al., 2018), particularly for glycosylated biopharmaceuticals (Huether et al., 2005; Decker & Reski, 2007; Reski et al., 2015), where RPs can be secreted into culture medium which dramatically reduces downstream purification costs (Schaaf et al., 2005). In fact, α-galactosidase A produced in moss has been shown to be more effective in treating enzymatic deficiencies in mice than the more conventional agalsidase alfa (Shen et al., 2015), demonstrating the potential superiority of the system. Finally, some newer expression systems are available, including cell-free protein synthesis systems such as the tobacco BY-2 cell-line lysate-derived system ALiCE, and several wheat germ systems, which have had success in producing cytoplasmic and microsomal proteins (Buntru et al., 2014; Harbers, 2014; Buntru et al., 2015). Similarly, plant cell packs utilise porous plant cell aggregates without cultivation media (Rademacher et al., 2019), compatible with well-plates which enable high-throughput rapid construct screening and can be automated to minimise batch variation (Gengenbach et al., 2020).
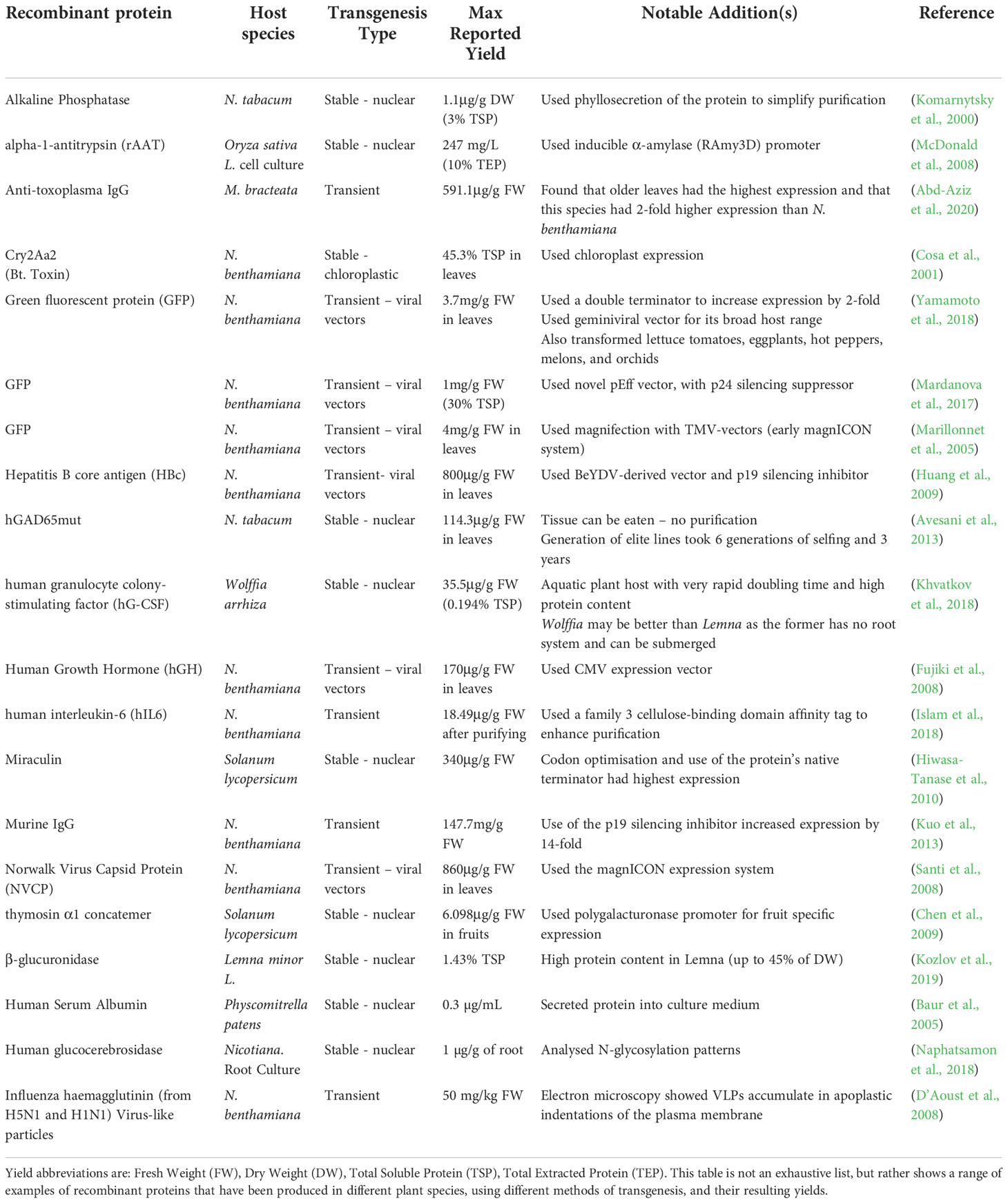
Plants can also be transformed using a variety of different methods, depending on a researcher’s needs. Stable transformation requires a single transformation event which is stably inherited in progeny of the primary transformant. This can be nuclear transformation, which typically results in moderate expression levels, but can be subject to transcriptional gene silencing through DNA methylation (Gallego‐Bartolomé, 2020). The site of transgene integration in the genome is generally random (Jaganathan et al., 2018), though some targeted transgenesis methods have been developed (Sanagala et al., 2017). Alternatively, plastids can be stably transformed, enabling very high expression levels due to the high plastid copy number within cells (Oey et al., 2009) and desirable biocontainment through primarily maternal inheritance (Ruf et al., 2007; Gerszberg and Hnatuszko-Konka, 2022). However, plastids are unable to produce complex PTMs. Lastly, transient expression is very rapid, taking days, with well-developed methods and high-expression levels and the ability to produce complex PTMs (Chen and Lai, 2015; Chen and Davis, 2016), with the main drawback that transformation needs to be repeated for each batch of RP. Table 2 provides a range of examples of recombinant proteins that have been expressed in plants.
While there is extensive literature about RP production in plants, little information is available about optimising expression through genetic manipulation and subsequent protein extraction. Consequently, this review focusses on the optimization of RP expression at the level of transcriptional and translational control, and the subsequent extraction methods employed for RP purification, with the aim of providing a comprehensive summary of the necessary considerations for producing RPs in plants.
Optimising expression
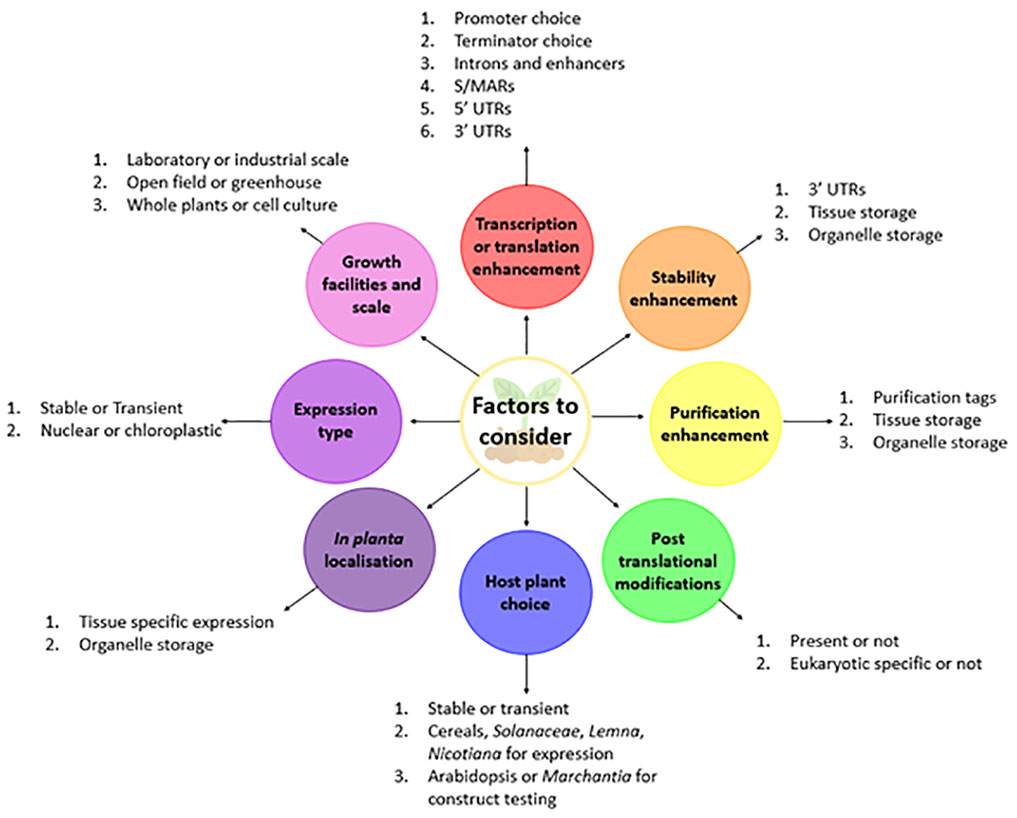
Recombinant protein expression can be optimised through increasing transcription and translation and decreasing mRNA and protein degradation. This section is dedicated to the transgene design considerations that can help improve in planta RP production (summarised in Figure 1).
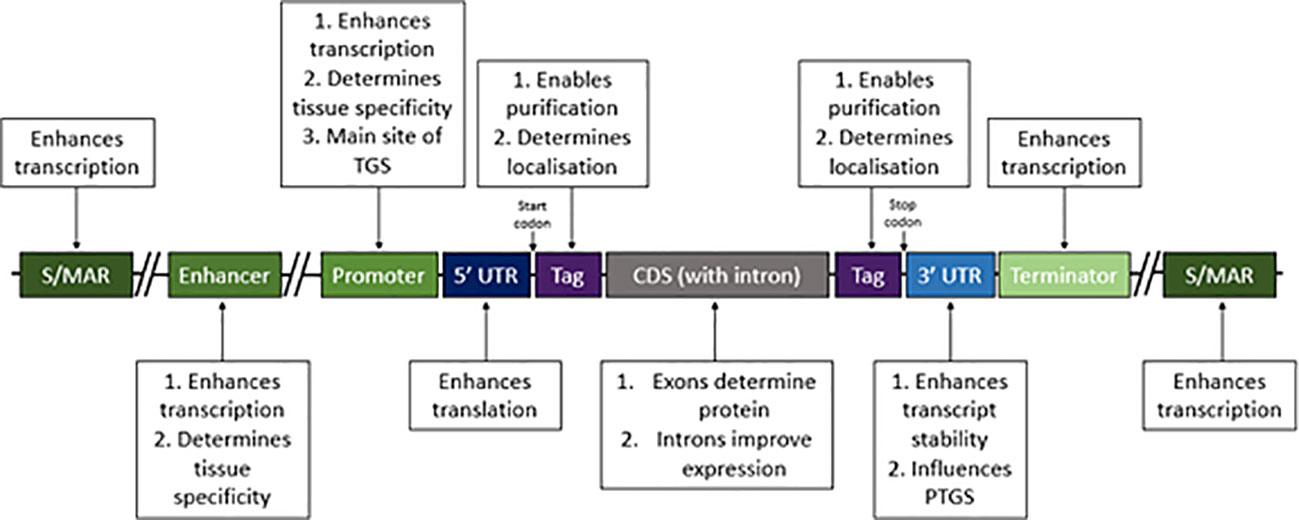
Figure 1 Regulatory DNA elements to consider when making plant RP expression constructs. Some DNA elements are essential in expression constructs, including a promoter, the coding sequence (CDS) and terminator; however many components in expression constructs are non-essential, including scaffold/matrix attachment regions (S/MARs), enhancers, introns, tags and to some extent untranslated regions (UTRs). Each of these elements has implications in recombinant protein expression, by affecting either transcription, translation, stability or efficacy of purification. The main roles of each element are summarised in this figure and should be considered when designing RP expression constructs.
Improving transient transformation efficiency
Agroinfiltration with regular expression vectors only leads to localised RP production in tissues that have been directly infiltrated by the Agrobacterium, leading to patchy and variable expression. Transient transformation efficiency can be improved by using agroinfiltration with deconstructed vectors containing viral elements that allow the spread of the transgene throughout the plant via a process called magnifection, transiently transforming plant cells that usually would not be directly affected by agroinfiltration. These vectors have been extensively reviewed by Peyret and Lomonossoff (2015), but the most commonly used are the TMV-based vectors, using the magnICON system (Chen and Lai, 2015). Despite their frequent use, other viral vectors pose advantages, and should be used in two specific cases. The first is when non-tobacco plants are used, as magnICON vectors, being based on TMV, are only very efficient at transforming tobacco plants. In such cases, other vectors such as geminiviral vectors, which have a broad host range, may be more suitable for other plant species (Yamamoto et al., 2018). Secondly, the magICON vectors are unsuitable for producing proteins with heterosubunits, as multiple vectors have competing replicons, resulting in competing origins of replication, meaning cells will usually only amplify one of the vectors, producing only one of the subunits. Other vectors, including geminiviral vectors, with large copy numbers or without competing replicons are more suitable for co-expression of RPs, or expression of proteins with heterosubunits (Chen and Lai, 2015). Thus, the use of deconstructed viral vectors is often favourable to improve RP expression within transiently transformed plants, but these should be chosen on a case-by-case basis depending on the plant species being transformed and the properties of the RP. Improvements in transient transformation efficiency have also been reported in N. benthamiana plants by Norkunas et al. (2018) who used a modified infiltration buffer, a heat-shock of transformed plants, and co-expression of several different proteins to increase recombinant protein yields. By combining different improvements, these researchers reported a 3.5-fold increase in GUS expression when compared to the already efficacious pEAQ-HT system.
Transcription
Optimising transcription is an easy way to improve expression levels because it requires alteration of the genetic construct without editing the coding sequence, so the resulting RP is not compromised. Cis-acting DNA elements can be included in the constructs to improve transcriptional rates. Broadly, every expression construct needs a promoter and a terminator, both of which influence the efficacy of transcription. Promoter sequences have different ‘strengths’, which ultimately means that strong promoters recruit RNA polymerase II more efficiently than weaker ones and create more transcripts. Some promoters are inducible, which means transcription factors bind the DNA only in response to stimuli, and others are tissue specific, meaning specific transcription factors only bind in certain tissues of multicellular organisms (Beringer et al., 2017). Generally, RPs are aimed to be produced in the largest quantities possible, so typically constitutive, global promoters (ie. expressed in all tissues) are used, such as the 35S Cauliflower Mosaic Virus (CaMV) promoter. Promoters should be chosen based on the need for in planta tissue-specific expression of the RP (if using stable transformants), and only constitutive if the RP will not affect plant growth. Finally, an important note to mention when using promoters is transcriptional gene silencing. Plants methylate homologous promoters, causing transcriptional transgene silencing in cells with two copies of the same promoter (Rajeevkumar et al., 2015). This means that it is beneficial to select for plant transformants with a single transgene integration event, or use different promoters when expressing two transgenes in the same plant line. Similarly, expressing the same RP with multiple constructs using different promoters, known as promoter stacking, can dramatically improve yields by avoiding or reducing transcriptional gene silencing. Damaj et al. (2020) managed to stack several promoters in stably transformed sugarcane plants, increasing expression by 147-fold when using stacked promoters compared to a singular promoter. Finally, there has been some progress made in the development of synthetic promoters, reviewed recently by Ali and Kim (2019), where promoters are engineered by combining different cis-acting DNA elements, sometimes combining multiple promoters in tandem, for novel or enhanced functions. Some synthetic promoters boasted an improvement in protein expression of 25-fold when compared to the commonly used CaMV 35S promoter (Kumar et al., 2011). Thus, a promoter should be chosen based on desired strength and cell-type-specific expression, with synthetic promoters, dual promoters, and promoter stacking available to further improve RP expression.
Choosing a terminator is also important as they terminate transcription and determine transcript stability, influencing transcript copy number. Commonly used terminators are the CaMV 35S, nopaline synthase (NOS), octopine synthase (OCS) and Heat Shock Protein (HSP) terminators (Nagaya et al., 2010; Limkul et al., 2015; Diamos and Mason, 2018). A factor to consider is the use of tandem units of promoters and terminators, as both double promoters and double terminators have been shown to improve protein expression (Khvatkov et al., 2018; Yamamoto et al., 2018). Diamos and Mason (2018) extensively characterised a series of terminators, and matrix attachment regions (MARs) in both tobacco and lettuce plants, showing that the use of different double terminator combinations and the employment of a MAR increased GFP production by over 60-fold compared to constructs using a single NOS terminator and no MAR. Interestingly, this is one of few studies to show that MARs improve RP expression in transient transformants.
Other commonly used but non-essential elements are enhancers, introns, and matrix-attachment regions. Enhancers increase transcript numbers through binding other trans-acting proteins, creating an active-chromatin hub (Tippens et al., 2018). As such, enhancers also contribute to tissue-specific gene expression, determined by the expression of these trans-acting proteins. In plants, introns are often used to improve transgene stability (Bourdon et al., 2001), and many of these contain enhancers, increasing mRNA production by up to 10-fold through intron-mediated enhancement (Rose, 2008). When designing the transgene, cryptic splice sites of AU-rich regions should be avoided to prevent the plant from recognising an unintended intron, and any intended introns should have their 3D-structure analysed before construct creation to avoid hairpins that could create siRNAs to silence the transgene (Smith et al., 2000). Large scale analysis of moss genes and subsequent codon optimization to reduce mRNA splicing resulted in a dramatic 12-fold increase in recombinant protein accumulation and correct intracellular trafficking (Top et al., 2021). Matrix attachment regions bind to the nuclear matrix and consequently improve transcription of nuclear-transformants, so should be considered for stably transformed plants (Allen, 2008; Diamos and Mason, 2018). MARs have been used in stable rice transformants to improve expression 3-fold (Vain et al., 1999)
Post-transcriptional gene silencing (PTGS), also termed RNA interference (RNAi), involves siRNA targeting of mRNA transcripts and subsequent transcript degradation, which is thought to have evolved to protect plants from viral attacks (Zhang et al., 2015). Fortunately, viruses have also evolved a defence against siRNAs, by producing proteins that bind to the siRNAs, preventing the degradation of the target mRNAs. These PTGS inhibitory proteins can be co-expressed in plants to increase expression dramatically. Most often, the p19 PTGS inhibitor from tomato bush stunt virus is used (Kuo et al., 2013; Merlin et al., 2014; Dong et al., 2019; Buyel et al., 2021), but others such as the P24 PTGS inhibitor from grapevine leafroll-associated virus-2 (Mardanova et al., 2017), P0 from Polerovirus, P17 from Aureusvirus, or NSs from Tospovirus are also available, though Peyret et al. (2019) recently established that P19 is the most effective of these. Ideally, these should always be used when high expression of a RP is desired, in both stable and transient transformants.
Finally, transcription is affected by position effects in stable transformants (Chen and Lai, 2015). This is because certain regions of chromatin are more active and euchromatic than others, and lengthy screening processes of many transformants are needed to isolate the best integrant lines. Fortunately, there are gene-targeting tools available for use in plants, including ZFNs, TALENs, and CRISPR variants (Sanagala et al., 2017).
Translation
Translation efficiency can be improved by incorporating certain 5’-UTRs into the transformation construct, including the 5’-UTRs from alfalfa mosaic virus (AMV), which is reported to increase the production of the target protein by up to 4-fold (Mardanova et al., 2017). In fact, one system has developed into a commercial biotechnology product known as HyperTrans. This uses a modified 5’-UTR from cowpea mosaic virus (CPMV), with the deletion of an endogenous in-frame start codon in the UTR, enabling protein accumulation up to 20% of TSP (Sainsbury and Lomonossoff, 2008). These have been developed into commercially viable expression vectors that allow the expression of different transgenes at high levels (Sainsbury et al., 2009). The technology is used commercially for RP production. In fact, engineering efforts have recently been made to optimise both 5’ and 3’ UTRs through rational design; whilst the native CPMV 3’ UTR was not able to be improved, a synthetic 5’ UTR was developed that, when used in conjunction with the 3’ UTR, demonstrated double the protein expression of the HyperTrans system (Peyret et al., 2019).
Additionally, codon optimisation can increase translation efficiency by utilising the codons that are more commonly used in the host plants endogenous proteins (Webster et al., 2016; Buyel et al., 2021). This influences translation as tRNAs and their corresponding codons are in proportionate use (Plotkin and Kudla, 2010). In other words, codons that are rarely used by the host have fewer tRNAs in the cell, which can be a rate limiting step for protein production. One study found that codon optimisation of a Bt toxin, cryIA, increased protein accumulation by 100-fold through only changing codons by 21% (Perlak et al., 1991). Another study found that codon optimisation of miraculin increased protein accumulation by nearly 1.5-fold (Hiwasa-Tanase et al., 2010). A third study found that codon optimisation of the S. cerevisiae phytase enzyme, when expressed in rice, improved expression by nearly 120-fold, though a signal sequence was also added (Hamada et al., 2004). Thus, codon optimisation should be performed to improve translation efficiency for highly expressed proteins, however the secondary structure of the resulting mRNA should be computationally analysed for hairpin structures in case the modified codon would create siRNAs against the transgene and cause PTGS as mentioned above.
Intracellular localisation, degradation and storage
Varying the eventual subcellular localisation of the RP can have implications on yield, stability, and purification. It is possible to sequester proteins into the nucleus, ER, vacuoles, mitochondria, plastids, or secrete them for the cell entirely (Drakakaki et al., 2006). For some proteins, such as Norwalk virus-like particles (NVLP) in N. benthamiana plants, cytosolic targeting (without the addition of any motifs) had the highest yield (Merlin et al., 2014). For others, such as hIL-6, endoplasmic reticulum (ER) targeted RP had expression up to 10-fold higher than apoplast and vacuole-targeted RP (Merlin et al., 2014). Importantly, intracellular localisation can be tissue dependent (Drakakaki et al., 2006), with reported differences between seeds and leaves (Hood et al., 2007), so this should be carefully investigated when using stable nuclear transformants in conjunction with tissue-specific promoter elements. The most common compartment to direct RPs is the ER, using KDEL/HDEL retention signals, typically chosen as the ER allows RPs to be stored in a less harsh environment with few proteases, and can result in antibody levels up to 100-fold higher than those expressed in the apoplast or cytosol (Fischer et al., 1999). However, ER retention causes RPs to avoid the Golgi, which means it is unsuitable for proteins which need downstream glycosylation (Villarejo et al., 2005; Egelkrout et al., 2012). Fortunately, multiple constructs can be created and co-expressed with different localisation sequences which can enhance RP production. Recombinant xylanase targeted to both the peroxisome and chloroplasts improved expression by 1.6-fold and 2.4-fold relative to only the chloroplast or peroxisomes respectively, despite similar levels of transcription (Hyunjong et al., 2005). Finally, some tags such as oleosin can be used to influence localisation and purification; these will be discussed more extensively in section 3.0.
Recombinant protein expression can also be optimised by reducing degradation, which can be done by protecting the protein, inhibiting proteases, or increase the protein’s stability. Tissue-specific or intracellular localisation influences protein stability. Some tissues, such as seeds, have naturally lower protease activity than other tissues and have intracellular storage vesicles (Stöger et al., 2000). This makes them ideal for RP expression when the RP needs to be stored. Alternatively, if the RP is not produced in tissue with naturally low protease activity, and is cytoplasmic, protease inhibitors can be co-expressed (Clemente et al., 2019), or the expression of endogenous proteases can be knocked-down (Kuo et al., 2013). Additionally, intracellular compartmentalisation of RPs can reduce degradation and improve stability.
Importantly, researchers studying roots and cell cultures have found that protein degradation is largely dependent on the expression system, with rhizosecreted proteins experiencing more peptidase activity than proteins secreted into culture medium; further investigation determined that these are largely due to serine and metallo-peptidases (Lallemand et al., 2015). Thus, knocking down expression of these may be considered to reduce recombinant protein degradation. Furthermore, Niemer et al. (2014) found that different recombinant monoclonal antibodies have different susceptibilities to serine- and cysteine-protease degradation, dependent on internal cleavage sites within a recombinant protein, demonstrating the importance of the amino acid sequence of expressed proteins.
Strategies to reduce endogenous plant protease activity have been reviewed by Mandal et al. (2016). The addition of stabilizing agents such as gelatin or polyvinylpyrrolidone (Wongsamuth and Doran, 1997; Häkkinen et al., 2014) can improve stability. Alternatively, co-expression of protease inhibitors such as tomato SlCDI (Goulet et al., 2012) or SlCYS8 (Robert et al., 2013) can reduce degradation, though this has been shown to be dependent on leaf age in transgenic tobacco (Robert et al., 2013). A more recent study by Grosse-Holz et al. (2018) identified three novel protease inhibitors, PR4 and Pot1 from N. benthamiana, and human TIMP, inhibitors of cysteine, serine, and metalloproteases, respectively. The researchers expressed recombinant α-Galactosidase, erythropoietin, and IgG antibody VRC01, and co-expression of each protease inhibitor showed similar improvements in each recombinant protein, between 2- and 27-fold, demonstrating the power of protease inhibition. Similarly, in moss, Hoernstein et al. (2018) identified four serpin proteins that irreversibly inhibit proteases, which may present promising targets to further improve RP stability. Targeted knockdown of genes can reduce degradation as demonstrated by RNAi-mediated knockdown of cysteine protease Rep-1 (Kim et al., 2008) in rice suspension cultures when producing recombinant human proteins. More broadly, aspartic-, cysteine-, metallo- and serine-proteases have been simultaneously knocked down by RNAi to improve RP production in tobacco BY-2 cell cultures (Mandal et al., 2014). Similarly, in moss, a subtilisin family protease was knocked out to improve early production of recombinant human α-Galactosidase A (Hoernstein et al., 2018). Finally, some fusion proteins can increase RP stability in planta, including elastin-like peptides (Conley et al., 2009; Kaldis et al., 2013), zein-derived peptides (Torrent et al., 2009; Joseph et al., 2012), and hydrophobins (Reuter et al., 2014), which largely stabilise the protein through directing the RP to storage structures. The expression of GFP fused to hydrophobin improved protein levels by 2-fold in both stably and transiently transformed Nicotiana plants, and by 3-fold in tobacco BY-2 cell cultures (Joensuu et al., 2010; Gutiérrez et al., 2013; Reuter et al., 2014). However, some proteins, such as β-glucuronidase, are naturally more stable in plant cells than others (Kozlov et al., 2019), so whether a stabilising tag is necessary should be decided based on the specific stability of the expressed RP. Tags should be more carefully considered for purification purposes, discussed in the following section.
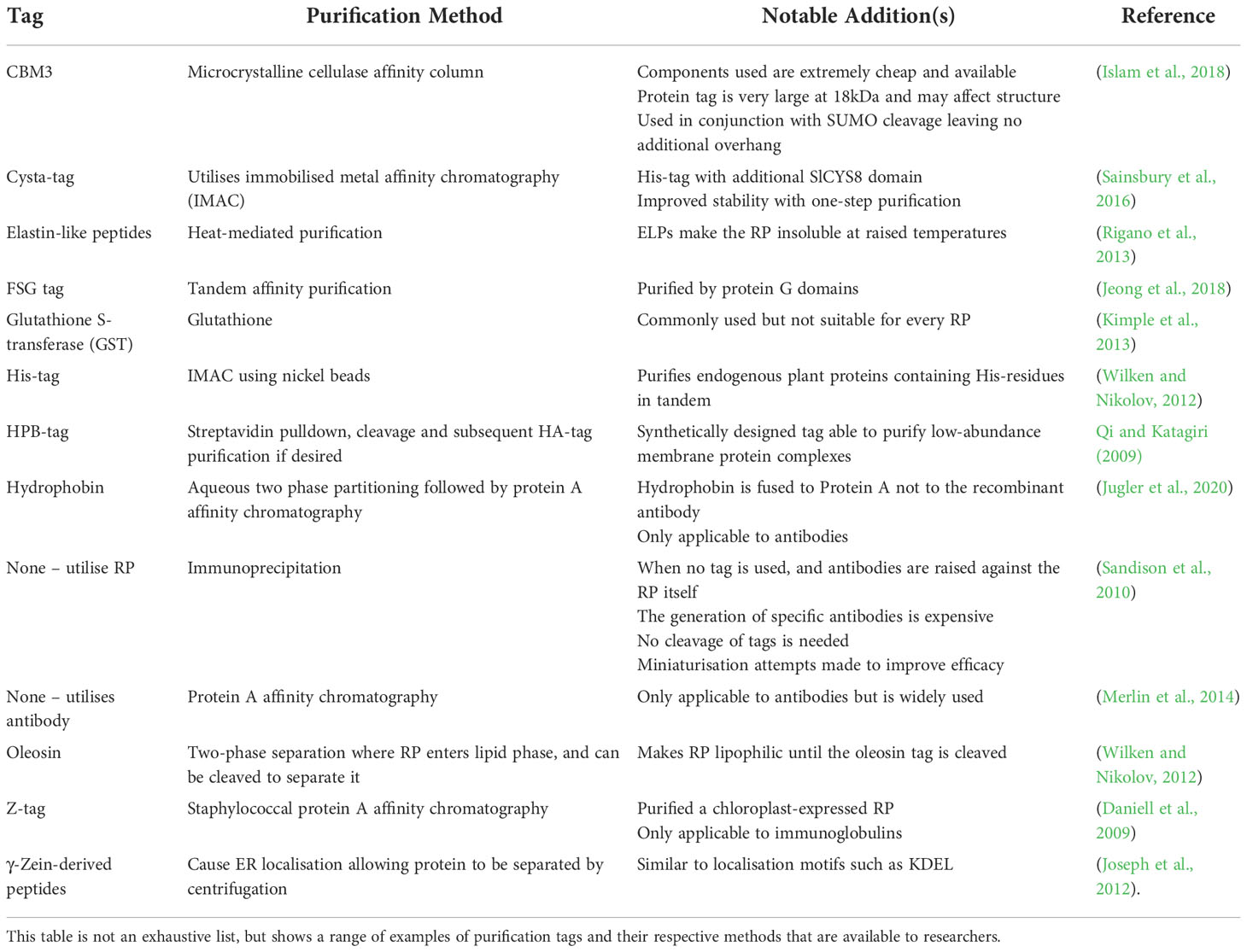
Tags and purification
Recombinant protein purification is considered the costliest stage in plant RP production, with comparable costs to other expression systems (Kuo et al., 2013; Merlin et al., 2014; Chen and Davis, 2016; 2016; Dong et al., 2019). Extraction of plant proteins has been reviewed by Wilken and Nikolov (2012) with some further developments in physical (Buyel and Fischer, 2015) and chemical extraction (Dong et al., 2019). Plant protein purification often relies on chromatography techniques, although non-chromatographic techniques also exist and have shown success in plants. These include flocculation (Elmer et al., 2009; Buyel and Fischer, 2013; Buyel and Fischer, 2014), ultrafiltration and diafiltration (Buyel and Fischer, 2013; Opdensteinen et al., 2019), heat-precipitation (Menzel et al., 2016; Opdensteinen et al., 2020), and pH-precipitation (Menzel et al., 2016; Opdensteinen et al., 2020), with several methods reviewed by Buyel (2022). These will not be discussed in detail in this section, which will instead focus on the use of purification tags in plants as they are encoded in DNA and are often designed with the expression construct.
The use of affinity tags in RP production has been recently reviewed (Mahmoodi et al., 2019), with a diverse array of tags to choose from. There are many existing examples of affinity tag usage for plant RP purification. Antibody production usually utilises protein A affinity chromatography and does not require an additional fusion tag and is an efficient but expensive process that is only applicable to antibodies (Merlin et al., 2014). As a result, a dual-purification technique was developed by Jugler et al. (2020), which utilised a hydrophobin fusion to protein A (HPA). When the HPA fusion protein was mixed with the monoclonal antibody (mAb), two-phase separation could be performed to isolate the mAb from the bulk of endogenous proteins, which are retained in the aqueous phase. Following this, the addition of a low pH buffer can separate the mAb from the HPA fusion protein, allowing high purity and yield mAb recovery. This method is particularly interesting as it utilises a fusion-protein that does not involve the RP (mAb) of interest. However, despite this methods success, due to the use of protein A affinity purification, this protocol is still only applicable to antibodies.
Consequently, most affinity tags used in plant RP purification use fusions to the RP itself. One study utilised Arabidopsis to produce a recombinant membrane protein, STT3a, and used tandem affinity purification of a FSG tag (comprised of three FLAG–streptavidin binding peptide, a TEV protease cleavage site, and two protein G domains) and retrieved the RP following TEV-mediated cleavage with high enough yield and purity for rough structural studies at resolutions of 30Å (Jeong et al., 2018). A cheaper method is the use of IgG-affinity columns to purify proteins with a Staphylococcus aureus Z-tag. This has been used to produce chloroplast-expressed hIGF, at a cleavage efficiency of only 40%, but using hydroxylamine to cleave the tag (Daniell et al., 2009). Histidine residues have an affinity to nickel beads, so His-tags or proteins containing histidine residues in tandem are also used for purification using immobilised metal affinity chromatography (Wilken and Nikolov, 2012). However, despite their common use, His-tags are not always specific, with some endogenous plant proteins containing natural histidine motifs, so one group developed a hybrid tag, termed ‘Cysta-tag’, comprised of the His-tag with an additional multicystatin domain SlCYS8, which enhances RP levels in tobacco, which enabled a tagged RP to have 25% yield recovery and 90% purity (Sainsbury et al., 2016). Other researchers have created hybrid tags containing several different protein domains. Qi and Katagiri (2009) created a purification tag, called the HPB tag, which utilises a biotin carboxyl carrier protein domain (BCCD) derived from the Arabidopsis 3-methylcrotonal CoA carboxylase, and enables purification of low abundance membrane protein complexes using streptavidin beads, and subsequent cleavage of the tag using PreScission protease, leaving only a single short copy of the hemagglutinin (HA) tag for further purification if desired.
Alternatively, tags have been developed that do not rely on chromatography methods, including oleosin, elastin-like proteins (ELPs) and γ-Zein-derived peptides. Oleosin is similar to hydrophobin proteins, as when fused to a RP oleosin causes it to become hydrophobic and localise to the lipid-rich layer of a cell, allowing separation from the bulk of intracellular proteins through centrifugation (Wilken and Nikolov, 2012; Gerszberg and Hnatuszko-Konka, 2022). Parmenter et al. (1995) managed to purify recombinant aprotinin from maize seeds using an oleosin tag, with yield recovery of 49% but relatively low purity of 79%. ELPs allow similar purification, by causing the RP to become insoluble at raised temperatures, allowing easy separation from endogenous proteins (Rigano et al., 2013). Finally, γ-Zein-derived peptides cause localisation into the ER, and allow purification through differential centrifugation (Torrent et al., 2009; Joseph et al., 2012).
The disadvantage of affinity tags is that they sometimes must be cleaved to ensure normal RP function, a step that is usually inefficient and reduces RP yields further. Cleavage mechanisms are also highly varied, with many options available. Often, protease-mediated cleavage is used, where the protein coding sequence incorporates a protease recognition site between the RP and the tag. TEV protease is a widely used and well characterised protease, but several others are available including endoproteases such as factor Xa, enteropeptidase, thrombin and 3C protease, and exoproteases such as aminopeptidases and carboxypeptidases, though exoproteases are rarely used for RP production (Waugh, 2011). Frey & Görlich (2014a) discuss a series of alternative proteases with reportedly improved functions, including SUMO-specific SENP1 and NEDD8-specific proteases from Brachypodium distachyon, NEDP1 protease from Salmo salar, Atg4p from Saccharomyces cerevisiae and Usp2 from Xenopus laevis. Of these, bdSENP1 showed 10,000-fold higher activity than TEV protease at 0-4°C. It also showed increased salt tolerance which is useful as extraction buffers often contain high salt-concentrations. In an accompanying paper, the researchers tested these proteases on several recombinant proteins (Frey & Görlich, 2014b). Similarly, inteins can be used to remove tags, and have been reviewed by Lahiry et al. (2017). These are protein sequences that, under the correct environmental conditions, can ligate flanking peptides called exteins, excising themselves in a process termed protein splicing. Intein activity can be regulated by pH, temperature, the presence of reducing agents, salts, small molecules, and light (Lahiry et al., 2017). However, limitations of these include premature cleavage, product losses, the potential need for unfavourable reducing agents, and varying suitability depending on the amino acid sequence surrounding the exteins. A successful example comes from Islam et al. (2018) who developed a plant-based affinity tag with a family-3 cellulose-binding domain (CBD), CBM3, which binds microcrystalline cellulose (MCC). This was used in conjunction with bdSENP1 which enabled efficient tag cleavage following purification. This method is cheap due to the low cost and high availability of cellulose for purification. One potential disadvantage that the researchers acknowledged was the large size of the CBD (18kDa), which may affect the function and folding of the fused RP. The researchers demonstrated that the CBD tightly bound MCC, allowing RPs to be purified at over 95% purity in a single step. The researchers also noted that traditionally used proteases, including TEV, are expensive, and intein-based cleavage was inefficient and time consuming, requiring high concentrations of potentially damaging reducing agents to activate. Islam et al. (2018) utilised the SUMO-based cleavage as there is no additional overhanging sequence left following cleavage, and the process is highly efficient.
To summarise, protein tags substantially aid purification efficiency, but at the cost of yield due to multi-step purification processes. Thus, purification without the use of tags is often desirable. mAbs can be produced in plants rapidly, cost-effectively, and with high yields (Diamos et al., 2020). For non-antibody proteins, tags are often directly fused to the RP, and must be cleaved. Fortunately, cheaper binding partners are now available and more efficient tags have recently been developed allowing some flexibility when deciding on purification methods. Ultimately, tag usage should also be chosen on a case-by-case basis, paying careful attention to the properties of the RP in different conditions. Table 3 summarises the purification tags discussed. A non-plant-specific table is available in a review by Kimple et al. (2013).
Conclusion
Overall, plants constitute a promising recombinant protein production platform, with several unique advantages that may make them specialists at producing recombinant proteins. When producing RPs, plants give researchers large amounts of flexibility, in that several different modes of transgenesis can be used, whether this is stable or transient, nuclear or chloroplastic, each with their own unique advantages that can cover the breadth of requirements for any recombinant protein. This flexibility results in a series of co-dependent decisions that a researcher can make when expressing RPs in plants, summarised in Figure 2. It is possible that plant-based protein production platforms could become cheaper than prokaryotic systems, as downstream processing methods begin to rely on cheaper reagents which have been developed in recent years. While plants are able to produce nearly any protein, it is most likely they will only be the dominant production system for proteins that have complex post-translational modifications and are required rapidly. To some degree, this is already realised as they are emerging to be the dominant antibody production platform, with pharmaceuticals not far behind, due to their unique advantage that RP-containing tissue can be consumed. Fortunately, recent and seemingly continuous developments in plant RP production and purification are rapidly advancing the field, with researchers now able to dramatically improve transcriptional and translational efficacy, maximise the number of transformed plant cells, and extract RPs with higher yields and purities than before. Consequently, due to these advancements, extremely cheap upstream costs, unparalleled scalability, and relative lack of specialist equipment, it would be unsurprising if plants eventually became the dominant protein production platform worldwide.
Author contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication. RC generated the figures, tables and led the writing.
Funding
This work was funded by the BBSRC GCRF grant BB/S011501/1 and BBSRC SWBIO DTP studentship awarded RC. For the purpose of open access, the author has applied a CC BY public copyright licence (where permitted by UKRI, ‘Open Government Licence’ or ‘CC BY-ND public copyright licence’ may be stated instead) to any Author Accepted Manuscript version arising.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abd-Aziz, N., Tan, B., Rejab, N., Othman, R., Khalid, N. (2020). A new plant expression system for producing pharmaceutical proteins. Mol. Biotechnol. 62, 240–251. doi: 10.1007/s12033-020-00242-2
Ali, S., Kim, W. (2019). A fruitful decade using synthetic promoters in the improvement of transgenic plants. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01433
Allen, G. C. (2008). “The role of nuclear matrix attachment regions in plants,” in Plant cell monographs (Berlin, Heidelberg: Springer).
Avesani, L., Merlin, M., Gecchele, E., Capaldi, S., Brozzetti, A., Falorni, A., et al. (2013). Comparative analysis of different biofactories for the production of a major diabetes autoantigen. Transgenic Res. 23, 281–291. doi: 10.1007/s11248-013-9749-9
Bandyopadhyay, P., Garrett, J., Shetty, R., Keate, T., Walker, C., Olivera, B. (2002). γ-glutamyl carboxylation: An extracellular posttranslational modification that antedates the divergence of molluscs, arthropods, and chordates. Proc. Natl. Acad. Sci. 99, 1264–1269. doi: 10.1073/pnas.022637099
Baur, A., Reski, R., Gorr, G. (2005). Enhanced recovery of a secreted recombinant human growth factor using stabilizing additives and by co-expression of human serum albumin in the moss physcomitrella patens. Plant Biotechnol. J. 3, 331–340. doi: 10.1111/j.1467-7652.2005.00127.x
Beringer, J., Chen, W., Garton, R., Sardesai, N., Wang, P., Zhou, N., et al. (2017). Comparison of the impact of viral and plant-derived promoters regulating selectable marker gene on maize transformation and transgene expression. Plant Cell Rep. 36, 519–528. doi: 10.1007/s00299-017-2099-y
Bourdon, V., Harvey, A., Lonsdale, D. M. (2001). Introns and their positions affect the translational activity of mRNA in plant cells. EMBO Rep. 2, 394–398. doi: 10.1093/embo-reports/kve090
Boyle, P., Schwizer, S., Hind, S., Kraus, C., de la Torre Diaz, S., He, B., et al. (2016). Detecting n-myristoylation and s-acylation of host and pathogen proteins in plants using click chemistry. Plant Methods 12, 38. doi: 10.1186/s13007-016-0138-2
Buntru, M., Vogel, S., Spiegel, H., Schillberg, S. (2014). Tobacco BY-2 cell-free lysate: an alternative and highly-productive plant-based in vitro translation system. BMC Biotechnol. 14, 37. doi: 10.1186/1472-6750-14-37
Buntru, M., Vogel, S., Stoff, K., Spiegel, H., Schillberg, S. (2015). A versatile coupled cell-free transcription-translation system based on tobacco BY-2 cell lysates. Biotechnol. Bioengineering 112, 867–878. doi: 10.1002/bit.25502
Burnett, M., Burnett, A. (2019). Therapeutic recombinant protein production in plants: Challenges and opportunities. PLANTS PEOPLE PLANET 2, 121–132. doi: 10.1002/ppp3.10073
Buyel, J. F. (2022). “Strategies for efficient and sustainable protein extraction and purification from plant tissues,” in Recombinant proteins in plants. methods in molecular biology, vol. 2480 . Eds. Schillberg, S., Spiegel, H. (New York, NY: Humana).
Buyel, J., Fischer, R. (2013). Flocculation increases the efficacy of depth filtration during the downstream processing of recombinant pharmaceutical proteins produced in tobacco. Plant Biotechnol. J. 12, 240–252. doi: 10.1111/pbi.12132
Buyel, J., Fischer, R. (2014). Downstream processing of biopharmaceutical proteins produced in plants. Bioengineered 5, 138–142. doi: 10.4161/bioe.28061
Buyel, J. F., Fischer, R. (2015). A juice extractor can simplify the downstream processing of plant-derived biopharmaceutical proteins compared to blade-based homogenizers. Process Biochem. 50, 859–866. doi: 10.1016/j.procbio.2015.02.017
Buyel, J. F., Stöger, E., Bortesi, L. (2021). Targeted genome editing of plants and plant cells for biomanufacturing. Transgenic Res. 30, 401–426. doi: 10.1007/s11248-021-00236-z
Castilho, A., Strasser, R., Stadlmann, J., Grass, J., Jez, J., Gattinger, P., et al. (2010). In PlantaProtein sialylation through overexpression of the respective mammalian pathway. J. Biol. Chem. 285, 15923–15930. doi: 10.1074/jbc.M109.088401
Chanroj, S., Jaiprasert, A., Issaro, N. (2021). A novel technique for recombinant protein expression in duckweed (Spirodela polyrhiza) turions. J. Plant Biotechnol. 48, 156–164. doi: 10.5010/JPB.2021.48.3.156
Chen, Q., Davis, K. (2016). The potential of plants as a system for the development and production of human biologics. F1000Research 5, 912. doi: 10.12688/f1000research.8010.1
Chen, Q., Lai, H. (2015). Gene delivery into plant cells for recombinant protein production. BioMed. Res. Int. 2015, 932161. doi: 10.1155/2015/932161
Chen, Y., Wang, A., Zhao, L., Shen, G., Cui, L., Tang, K. (2009). Expression of thymosin α1 concatemer in transgenic tomato (Solanum lycopersicum) fruits. Biotechnol. Appl. Biochem. 52, 303. doi: 10.1042/BA20080054
Clemente, M., Corigliano, M., Pariani, S., Sánchez-López, E., Sander, V., Ramos-Duarte, V. (2019). Plant serine protease inhibitors: Biotechnology application in agriculture and molecular farming. Int. J. Mol. Sci. 20, 1345. doi: 10.3390/ijms20061345
Conley, A. J., Joensuu, J. J., Menassa, R., Brandle, J. E. (2009). Induction of protein body formation in plant leaves by elastin-like polypeptide fusions. BMC Biol. 7, 48. doi: 10.1186/1741-7007-7-48
Cosa, B., Moar, W., Lee, S., Miller, M., Daniell, H. (2001). Overexpression of the bt cry2Aa2 operon in chloroplasts leads to formation of insecticidal crystals. Nat. Biotechnol. 19, 71–74. doi: 10.1038/83559
Damaj, M., Jifon, J., Woodard, S., Vargas-Bautista, C., Barros, G., Molina, J., et al. (2020). Unprecedented enhancement of recombinant protein production in sugarcane culms using a combinatorial promoter stacking system. Sci. Rep. 10, 1. doi: 10.1038/s41598-020-70530-z
Daniell, H., Ruiz, G., Denes, B., Sandberg, L., Langridge, W. (2009). Optimization of codon composition and regulatory elements for expression of human insulin like growth factor-1 in transgenic chloroplasts and evaluation of structural identity and function. BMC Biotechnol. 9, 33. doi: 10.1186/1472-6750-9-33
D’Aoust, M.-A., Lavoie, P.-O., Couture, M. M. J., Trépanier, S., Guay, J.-M., Dargis, M., et al. (2008). Influenza virus-like particles produced by transient expression in nicotiana benthamiana induce a protective immune response against a lethal viral challenge in mice. Plant Biotechnol. J. 6, 930–940. doi: 10.1111/j.1467-7652.2008.00384.x
Debler, J., Henares, B., Lee, R. (2021). Agroinfiltration for transient gene expression and characterisation of fungal pathogen effectors in cool-season grain legume hosts. Plant Cell Rep. 40, 805–818. doi: 10.1007/s00299-021-02671-y
Decker, E. L., Reski, R. (2007). Moss bioreactors producing improved biopharmaceuticals. Curr. Opin. Biotechnol. 18, 393–398. doi: 10.1016/j.copbio.2007.07.012
Diamos, A., Hunter, J., Pardhe, M., Rosenthal, S., Sun, H., Foster, B., et al. (2020). High level production of monoclonal antibodies using an optimized plant expression system. Front. Bioengineering Biotechnol. 7. doi: 10.3389/fbioe.2019.00472
Diamos, A., Mason, H. (2018). Chimeric 3’ flanking regions strongly enhance gene expression in plants. Plant Biotechnol. J. 16, 1971–1982. doi: 10.1111/pbi.12931
Ding, X., Yin, Z., Wang, S., Liu, H., Chu, X., Liu, J., et al. (2022). Different fruit-specific promoters drive AtMYB12 expression to improve phenylpropanoid accumulation in tomato. Molecules 27, 317. doi: 10.3390/molecules27010317
Dong, J., Ding, X., Wang, S. (2019). Purification of the recombinant green fluorescent protein from tobacco plants using alcohol/salt aqueous two-phase system and hydrophobic interaction chromatography. BMC Biotechnol. 19, 1–8. doi: 10.1186/s12896-019-0590-y
Drakakaki, G., Marcel, S., Arcalis, E., Altmann, F., Gonzalez-Melendi, P., Fischer, R., et al. (2006). The intracellular fate of a recombinant protein is tissue dependent. Plant Physiol. 141, 578–586. doi: 10.1104/pp.106.076661
Duruflé, H., Hervé, V., Balliau, T., Zivy, M., Dunand, C., Jamet, E. (2017). Proline hydroxylation in cell wall proteins: Is it yet possible to define rules? Front. Plant Sci. 8, 1802. doi: 10.3389/fpls.2017.01802
Egelkrout, E., Rajan, V., Howard, J. (2012). Overproduction of recombinant proteins in plants. Plant Sci. 184, 83–101. doi: 10.1016/j.plantsci.2011.12.005
Elmer, J., Harris, D., Sun, G., Palmer, A. (2009). Purification of hemoglobin by tangential flow filtration with diafiltration. Biotechnol. Prog. 25, 1402–1410. doi: 10.1002/btpr.217
Feng, J., Chen, L., Zuo, J. (2019). Protein s -nitrosylation in plants: Current progresses and challenges. J. Integr. Plant Biol. 61, 1206–1223. doi: 10.1111/jipb.12780
Fischer, R., Emans, N. (2000). Molecular farming of pharmaceutical proteins. Transgenic Res. 9, 279–299. doi: 10.1023/A:1008975123362
Fischer, R., Schumann, D., Zimmermann, S., Drossard, J., Sack, M., Schillberg, S. (1999). Expression and characterization of bispecific single-chain fv fragments produced in transgenic plants. Eur. J. Biochem. 262, 810–816. doi: 10.1046/j.1432-1327.1999.00435.x
Frey, S., Görlich, D. (2014a). A new set of highly efficient, tag-cleaving proteases for purifying recombinant proteins. J. Chromatogr. A 1337, 95–105. doi: 10.1016/j.chroma.2014.02.029
Frey, S., Görlich, D. (2014b). Purification of protein complexes of defined subunit stoichiometry using a set of orthogonal, tag-cleaving proteases. J. Chromatogr. A 1337, 106–115. doi: 10.1016/j.chroma.2014.02.030
Fujiki, M., Kaczmarczyk, J., Yusibov, V., Rabindran, S. (2008). Development of a new cucumber mosaic virus-based plant expression vector with truncated 3a movement protein. Virology 381, 136–142. doi: 10.1016/j.virol.2008.08.022
Gallego-Bartolomé, J. (2020). DNA Methylation in plants: mechanisms and tools for targeted manipulation. New Phytol. 227, 38–44. doi: 10.1111/nph.16529
Garcia-Garcia, T., Poncet, S., Derouiche, A., Shi, L., Mijakovic, I., Noirot-Gros, M. (2016). Role of protein phosphorylation in the regulation of cell cycle and DNA-related processes in bacteria. Front. Microbiol. 7, 184. doi: 10.3389/fmicb.2016.00184
Gengenbach, B. B., Opdensteinen, P., Buyel, J. F. (2020). Robot cookies – plant cell packs as an automated high-throughput screening platform based on transient expression. Front. Bioengineering Biotechnol. 8. doi: 10.3389/fbioe.2020.00393
Gerszberg, A., Hnatuszko-Konka, K. (2022). Compendium on food crop plants as a platform for pharmaceutical protein production. Int. J. Mol. Sci. 23, 3236. doi: 10.3390/ijms23063236
Gitzinger, M., Parsons, J., Reski, R., Fussenegger, M. (2009). Functional cross-kingdom conservation of mammalian and moss (Physcomitrella patens) transcription, translation and secretion machineries. Plant Biotechnol. J. 7, 73–86. doi: 10.1111/j.1467-7652.2008.00376.x
Gomord, V., Faye, L. (2004). Posttranslational modification of therapeutic proteins in plants. Curr. Opin. Plant Biol. 7, 171–181. doi: 10.1016/j.pbi.2004.01.015
Gomord, V., Fitchette, A., Menu-Bouaouiche, L., Saint-Jore-Dupas, C., Plasson, C., Michaud, D., et al. (2010). Plant-specific glycosylation patterns in the context of therapeutic protein production. Plant Biotechnol. J. 8, 564–587. doi: 10.1111/j.1467-7652.2009.00497.x
Goulet, C., Khalf, M., Sainsbury, F., D'Aoust, M.-A., Michaud, D. (2012). A protease activity-depleted environment for heterologous proteins migrating towards the leaf cell apoplast. Plant Biotechnol. J. 10, 83–94. doi: 10.1111/j.1467-7652.2011.00643.x
Gusarov, I., Nudler, E. (2012). S-nitrosylation signaling in Escherichia coli. Sci. Signaling 5, pe26. doi: 10.1126/scisignal.2003181
Gutiérrez, S. P., Saberianfar, R., Kohalmi, S. E., Menassa, R. (2013). Protein body formation in stable transgenic tobacco expressing elastin-like polypeptide and hydrophobin fusion proteins. BMC Biotechnol. 13, 40. doi: 10.1186/1472-6750-13-40
Grosse-Holz, F., Madeira, L., Zahid, M. A., Songer, M., Kourelis, J., Fesenko, M., et al. (2018). Three unrelated protease inhibitors enhance accumulation of pharmaceutical recombinant proteins in nicotiana benthamiana. Plant Biotechnol. J. 16, 1797–1810. doi: 10.1111/pbi.12916
Häkkinen, S. T., Raven, N., Henquet, M., Laukkanen, M.-L., Anderlei, T., Pitkänen, J.-P., et al. (2014). Molecular farming in tobacco hairy roots by triggering the secretion of a pharmaceutical antibody. Biotechnol. Bioengineering 111, 336–346. doi: 10.1002/bit.25113
Hamada, A., Yamaguchi, K., Ohnishi, N., Harada, M., Nikumaru, S., Honda, H. (2004). High-level production of yeast (Schwanniomyces occidentalis) phytase in transgenic rice plants by a combination of signal sequence and codon modification of the phytase gene. Plant Biotechnol. J. 3, 43–55. doi: 10.1111/j.1467-7652.2004.00098.x
Han, S., Lee, S., Bahar, O., Schwessinger, B., Robinson, M., Shaw, J., et al. (2012). Tyrosine sulfation in a gram-negative bacterium. Nat. Commun. 3, 1153. doi: 10.1038/ncomms2157
Harbers, M. (2014). Wheat germ systems for cell-free protein expression. FEBS Lett. 588, 2762–2773. doi: 10.1016/j.febslet.2014.05.061
Hiatt, A., Caffferkey, R., Bowdish, K. (1989). Production of antibodies in transgenic plants. Nature 342, 76–78. doi: 10.1038/342076a0
Hiwasa-Tanase, K., Nyarubona, M., Hirai, T., Kato, K., Ichikawa, T., Ezura, H. (2010). High-level accumulation of recombinant miraculin protein in transgenic tomatoes expressing a synthetic miraculin gene with optimized codon usage terminated by the native miraculin terminator. Plant Cell Rep. 30, 113–124. doi: 10.1007/s00299-010-0949-y
Hoernstein, S. N. W., Fode, B., Wiedemann, G., Lang, D., Niederkrüger, H., Berg, B., et al. (2018). Host cell proteome of physcomitrella patens harbors proteases and protease inhibitors under bioproduction conditions. J. Proteome Res. 17, 3749–3760. doi: 10.1021/acs.jproteome.8b00423
Hood, E. E., Love, R., Lane, J., Bray, J., Clough, R., Pappu, K., et al. (2007). Subcellular targeting is a key condition for high-level accumulation of cellulase protein in transgenic maize seed. Plant Biotechnol. J. 5, 709–719. doi: 10.1111/j.1467-7652.2007.00275.x
Huang, Z., Chen, Q., Hjelm, B., Arntzen, C., Mason, H. (2009). A DNA replicon system for rapid high-level production of virus-like particles in plants. Biotechnol. Bioengineering 103, 706–714. doi: 10.1002/bit.22299
Huang, J., Nandi, S., Wu, L., Yalda, D., Bartley, G., Rodriguez, R., et al. (2002). Expression of natural antimicrobial human lysozyme in rice grains. Mol. Breed. 10, 83–94. doi: 10.1023/A:1020355511981
Huang, L., Tan, C., Yeh, J., Liu, H., Liu, Y., Ho, S., et al. (2015). Efficient secretion of recombinant proteins from rice suspension-cultured cells modulated by the choice of signal peptide. PloS One 10, e0140812. doi: 10.1371/journal.pone.0140812
Huether, C. M., Lienhart, O., Baur, A., Stemmer, C., Gorr, G., Reski, R., et al. (2005). Glyco-engineering of moss lacking plant-specific sugar residues. Plant Biol. 7, 292–299. doi: 10.1055/s-2005-837653
Hunter, M., Yuan, P., Vavilala, D., Fox, M. (2018). Optimization of protein expression in mammalian cells. Curr. Protoc. Protein Sci. 95, e77. doi: 10.1002/cpps.77
Hyunjong, B., Lee, D.-S., Hwang, I. (2005). Dual targeting of xylanase to chloroplasts and peroxisomes as a means to increase protein accumulation in plant cells. J. Exp. Bot. 57, 161–169. doi: 10.1093/jxb/erj019
Iribarren, P., Berazategui, M., Cazzulo, J., Alvarez, V. (2015). Biosynthesis of SUMOylated proteins in bacteria using the trypanosoma brucei enzymatic system. PloS One 10, e0134950. doi: 10.1371/journal.pone.0134950
Islam, M., Kwak, J., Lee, J., Hong, S., Khan, M., Lee, Y., et al. (2018). Cost-effective production of tag-less recombinant protein in nicotiana benthamiana. Plant Biotechnol. J. 17, 1094–1105. doi: 10.1111/pbi.13040
Jaganathan, D., Ramasamy, K., Sellamuthu, G., Jayabalan, S., Venkataraman, G. (2018). CRISPR for crop improvement: An update review. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00985
Jeong, I., Lee, S., Bonkhofer, F., Tolley, J., Fukudome, A., Nagashima, Y., et al. (2018). Purification and characterization of arabidopsis thaliana oligosaccharyltransferase complexes from the native host: a protein super-expression system for structural studies. Plant J. 94, 131–145. doi: 10.1111/tpj.13847
Joensuu, J. J., Conley, A. J., Lienemann, M., Brandle, J. E., Linder, M. B., Menassa, R. (2010). Hydrophobin fusions for high-level transient protein expression and purification in nicotiana benthamiana. Plant Physiol. 152, 622–633. doi: 10.1104/pp.109.149021
Joseph, M., Ludevid, M. D., Torrent, M., Rofidal, V., Tauzin, M., Rossignol, M., et al. (2012). Proteomic characterisation of endoplasmic reticulum-derived protein bodies in tobacco leaves. BMC Plant Biol. 12, 36. doi: 10.1186/1471-2229-12-36
Jugler, C., Joensuu, J., Chen, Q. (2020). Hydrophobin-protein a fusion protein produced in plants efficiently purified an anti-West Nile virus monoclonal antibody from plant extracts via aqueous two-phase separation. Int. J. Mol. Sci. 21, 2140. doi: 10.3390/ijms21062140
Kaldis, A., Ahmad, A., Reid, A., McGarvey, B., Brandle, J., Ma, S., et al. (2013). High-level production of human interleukin-10 fusions in tobacco cell suspension cultures. Plant Biotechnol. J. 11, 535–545. doi: 10.1111/pbi.12041
Karki, U., Fang, H., Guo, W., Unnold-Cofre, C., Xu, J. (2021). Cellular engineering of plant cells for improved therapeutic protein production. Plant Cell Rep. 40, 1087–1099. doi: 10.1007/s00299-021-02693-6
Kaufmann, C., Sauter, M. (2019). Sulfated plant peptide hormones. J. Exp. Bot. 70, 4267–4277. doi: 10.1093/jxb/erz292
Khvatkov, P., Firsov, A., Shvedova, A., Shaloiko, L., Kozlov, O., Chernobrovkina, M., et al. (2018). Development of wolffia arrhiza as a producer for recombinant human granulocyte colony-stimulating factor. Front. Chem. 6, 304. doi: 10.3389/fchem.2018.00304
Kim, N. S., Kim, T. G., Kim, O. H., Ko, E. M., Jang, Y. S., Jung, E. S., et al. (2008). Improvement of recombinant hGM-CSF production by suppression of cysteine proteinase gene expression using RNA interference in a transgenic rice culture. Plant Mol. Biol. 68, 263–275. doi: 10.1007/s11103-008-9367-8
Kimple, M., Brill, A., Pasker, R. (2013). Overview of affinity tags for protein purification. Curr. Protoc. Protein Sci. 73, 9.9.1–9.9.23. doi: 10.1002/0471140864.ps0909s73
Komarnytsky, S., Borisjuk, N., Borisjuk, L., Alam, M., Raskin, I. (2000). Production of recombinant proteins in tobacco guttation fluid. Plant Physiol. 124, 927–934. doi: 10.1104/pp.124.3.927
Kozlov, O., Mitiouchkina, T., Tarasenko, I., Shaloiko, L., Firsov, A., Dolgov, S. (2019). Agrobacterium-mediated transformation of lemna minor l. with hirudin and β-glucuronidase genes. Appl. Biochem. Microbiol. 55, 805–815. doi: 10.1134/S0003683819080076
Kumar, D., Patro, S., Ranjan, R., Sahoo, D., Maiti, I., Dey, N. (2011). Development of useful recombinant promoter and its expression analysis in different plant cells using confocal laser scanning microscopy. PloS One 6, 24627. doi: 10.1371/journal.pone.0024627
Kuo, Y., Tan, C., Ku, J., Hsu, W., Su, S., Lu, C., et al. (2013). Improving pharmaceutical protein production in oryza sativa. Int. J. Mol. Sci. 14, 8719–8739. doi: 10.3390/ijms14058719
Kurup, V., Thomas, J. (2019). Edible vaccines: Promises and challenges. Mol. Biotechnol. 62, 79–90. doi: 10.1007/s12033-019-00222-1
Lahiry, A., Fan, Y., Stimple, S. D., Raith, M., Wood, D. W. (2017). Inteins as tools for tagless and traceless protein purification. J. Chem. Technol. Biotechnol. 93, 1827–1835. doi: 10.1002/jctb.5415
Lallemand, J., Bouché, F., Desiron, C., Stautemas, J., de Lemos Esteves, F., Périlleux, C., et al. (2015). Extracellular peptidase hunting for improvement of protein production in plant cells and roots. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00037
Legastelois, I., Buffin, S., Peubez, I., Mignon, C., Sodoyer, R., Werle, B. (2016). Non-conventional expression systems for the production of vaccine proteins and immunotherapeutic molecules. Hum. Vaccines Immunotherapeutics 13, 947–961. doi: 10.1080/21645515.2016.1260795
Limkul, J., Misaki, R., Kato, K., Fujiyama, K. (2015). The combination of plant translational enhancers and terminator increase the expression of human glucocerebrosidase in nicotiana benthamiana plants. Plant Sci. 240, 41–49. doi: 10.1016/j.plantsci.2015.08.018
Lim, C. J., Lee, H. Y., Kim, W. B., Lee, B.-S., Kim, J., Ahmad, R., et al. (2012). Screening of tissue-specific genes and promoters in tomato by comparing genome wide expression profiles of arabidopsis orthologues. Molecules Cells 34, 53–59. doi: 10.1007/s10059-012-0068-4
Liu, Y., Lu, C., Chang, J., Lu, C., Tan, C., Huang, L. (2017). Optimization of the culture medium for recombinant protein production under the control of the αAmy3 promoter in a rice suspension-cultured cell expression system. Plant Cell Tissue Organ Culture (PCTOC) 132, 383–391. doi: 10.1007/s11240-017-1337-x
Ma, J. C., Drake, P., Christou, P. (2003). The production of recombinant pharmaceutical proteins in plants. Nat. Rev. Genet. 4, 794–805. doi: 10.1038/nrg1177
Magnusdottir, A., Vidarsson, H., Björnsson, J. M., Örvar, B. L.. (2013). Barley grains for the production of endotoxin-free growth factors. Trends Biotechnol. 31, 572–580. doi: 10.1016/j.tibtech.2013.06.002
Mahmoodi, S., Pourhassan-Moghaddam, M., Wood, D., Majdi, H., Zarghami, N. (2019). Current affinity approaches for purification of recombinant proteins. Cogent Biol. 5, 1665406. doi: 10.1080/23312025.2019.1665406
Makatsa, M., Tincho, M., Wendoh, J., Ismail, S., Nesamari, R., Pera, F., et al. (2021). SARS-CoV-2 antigens expressed in plants detect antibody responses in COVID-19 patients. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.589940
Malaquias, A. D. M., Marques, L. E. C., Pereira, S. S., de Freitas Fernandes, C., Maranhão, A. Q., Stabeli, R. G., et al. (2021). A review of plant-based expression systems as a platform for single-domain recombinant antibody production. Int. J. Biol. Macromolecules 193, 1130–1137. doi: 10.1016/j.ijbiomac.2021.10.126
Mandal, M. K., Ahvari, H., Schillberg, S., Schiermeyer, A. (2016). Tackling unwanted proteolysis in plant production hosts used for molecular farming. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00267
Mandal, M. K., Fischer, R., Schillberg, S., Schiermeyer, A. (2014). Inhibition of protease activity by antisense RNA improves recombinant protein production in nicotiana tabacum cv bright yellow 2 (BY-2) suspension cells. Biotechnol. J. 9, 1065–1073. doi: 10.1002/biot.201300424
Mardanova, E., Blokhina, E., Tsybalova, L., Peyret, H., Lomonossoff, G., Ravin, N. (2017). Efficient transient expression of recombinant proteins in plants by the novel pEff vector based on the genome of potato virus X. Front. Plant Sci. 8, 247. doi: 10.3389/fpls.2017.00247
Marillonnet, S., Thoeringer, C., Kandzia, R., Klimyuk, V., Gleba, Y. (2005). Systemic agrobacterium tumefaciens–mediated transfection of viral replicons for efficient transient expression in plants. Nat. Biotechnol. 23, 718–723. doi: 10.1038/nbt1094
Matsuda, R., Kubota, C., Alvarez, M., Cardineau, G. (2009). Biopharmaceutical protein production under controlled environments: Growth, development, and vaccine productivity of transgenic tomato plants grown hydroponically in a greenhouse. HortScience 44, 1594–1599. doi: 10.21273/HORTSCI.44.6.1594
McDonald, K., Hong, L., Trombly, D., Xie, Q., Jackman, A. (2008). Production of human α-1-Antitrypsin from transgenic rice cell culture in a membrane bioreactor. Biotechnol. Prog. 21, 728–734. doi: 10.1021/bp0496676
Menzel, S., Holland, T., Boes, A., Spiegel, H., Bolzenius, J., Fischer, R., et al. (2016). Optimized blanching reduces the host cell protein content and substantially enhances the recovery and stability of two plant-derived malaria vaccine candidates. Front. Plant Sci. 17, 159. doi: 10.3389/fpls.2016.00159
Merlin, M., Gecchele, E., Capaldi, S., Pezzotti, M., Avesani, L. (2014). Comparative evaluation of recombinant protein production in different biofactories: The green perspective. BioMed. Res. Int. 2014, 136419. doi: 10.1155/2014/136419
Millar, A., Heazlewood, J., Giglione, C., Holdsworth, M., Bachmair, A., Schulze, W. (2019). The scope, functions, and dynamics of posttranslational protein modifications. Annu. Rev. Plant Biol. 70, 119–151. doi: 10.1146/annurev-arplant-050718-100211
Minguez, P., Parca, L., Diella, F., Mende, D., Kumar, R., Helmer-Citterich, M., et al. (2012). Deciphering a global network of functionally associated post-translational modifications. Mol. Syst. Biol. 8, 599. doi: 10.1038/msb.2012.31
Miricescu, A., Goslin, K., Graciet, E. (2018). Ubiquitylation in plants: signaling hub for the integration of environmental signals. J. Exp. Bot. 69, 4511–4527. doi: 10.1093/jxb/ery165
Mirzaee, M., Osmani, Z., Frébortová, J., Frébort, I. (2022). Recent advances in molecular farming using monocot plants. Biotechnol. Adv. 58, 107913. doi: 10.1016/j.biotechadv.2022.107913
Montero-Morales, L., Steinkellner, H. (2018). Advanced plant-based glycan engineering. Front. Bioengineering Biotechnol. 6, 81. doi: 10.3389/fbioe.2018.00081
Moon, K., Park, J., Park, Y., Song, I., Lee, H., Cho, H., et al. (2019). Development of systems for the production of plant-derived biopharmaceuticals. Plants 9, 30. doi: 10.3390/plants9010030
Nagaya, S., Kawamura, K., Shinmyo, A., Kato, K. (2010). The HSP terminator of arabidopsis thaliana increases gene expression in plant cells. Plant Cell Physiol. 51, 328–332. doi: 10.1093/pcp/pcp188
Naphatsamon, U., Ohashi, T., Misaki, R., Fujiyama, K. (2018). The production of human β-glucocerebrosidase in nicotiana benthamiana root culture. Int. J. Mol. Sci. 19, 1972. doi: 10.3390/ijms19071972
Naqvi, S., Ramessar, K., Farré, G., Sabalza, M., Miralpeix, B., Twyman, R. M., et al. (2011). High-value products from transgenic maize. Biotechnol. Adv. 29, 40–53. doi: 10.1016/j.biotechadv.2010.08.009
Niederkrüger, H., Dabrowska-Schlepp, P., Schaaf, A. (2014). “Suspension culture of plant cells under phototrophic conditions,” in Industrial scale suspension culture of living cells. Eds. Meyer, H.-P., Schmidhalter, D. R. (Oxford: Wiley), 259–292.
Niemer, M., Mehofer, U., Torres Acosta, J. A., Verdianz, M., Henkel, T., Loos, A., et al. (2014). The human anti-HIV antibodies 2F5, 2G12, and PG9 differ in their susceptibility to proteolytic degradation: Down-regulation of endogenous serine and cysteine proteinase activities could improve antibody production in plant-based expression platforms. Biotechnol. J. 9, 493–500. doi: 10.1002/biot.201300207
Norkunas, K., Harding, R., Dale, J., Dugdale, B. (2018). Improving agroinfiltration-based transient gene expression in nicotiana benthamiana. Plant Methods 14, 71. doi: 10.1186/s13007-018-0343-2
Nothaft, H., Szymanski, C. (2010). Protein glycosylation in bacteria: sweeter than ever. Nat. Rev. Microbiol. 8, 765–778. doi: 10.1038/nrmicro2383
Nothaft, H., Szymanski, C. (2013). Bacterial ProteinN-glycosylation: New perspectives and applications. J. Biol. Chem. 288, 6912–6920. doi: 10.1074/jbc.R112.417857
Oey, M., Lohse, M., Kreikemeyer, B., Bock, R. (2009). Exhaustion of the chloroplast protein synthesis capacity by massive expression of a highly stable protein antibiotic. Plant J. 57, 436–445. doi: 10.1111/j.1365-313X.2008.03702.x
Opdensteinen, P., Clodt, J. I., Müschen, C. R., Filiz, V., Buyel, J. F. (2019). A combined Ultrafiltration/Diafiltration step facilitates the purification of cyanovirin-n from transgenic tobacco extracts. Front. Bioengineering Biotechnol. 6. doi: 10.3389/fbioe.2018.00206
Opdensteinen, P., Lobanov, A., Buyel, J. F. (2020). A combined pH and temperature precipitation step facilitates the purification of tobacco-derived recombinant proteins that are sensitive to extremes of either parameter. Biotechnol. J. 16, 2000340. doi: 10.1002/biot.202000340
Owczarek, B., Gerszberg, A., Hnatuszko-Konka, K. (2019). A brief reminder of systems of production and chromatography-based recovery of recombinant protein biopharmaceuticals. BioMed. Res. Int. 2019, 4216060. doi: 10.1155/2019/4216060
Park, H., Kim, W., Park, H., Lee, S., Bohnert, H., Yun, D. (2011). SUMO and SUMOylation in plants. Molecules Cells 32, 305–316. doi: 10.1007/s10059-011-0122-7
Parmenter, D. L., Boothe, J. G., van Rooijen, G. J. H., Yeung, E. C., Moloney, M. M. (1995). Production of biologically active hirudin in plant seeds using oleosin partitioning. Plant Mol. Biol. 29, 1167–1180. doi: 10.1007/BF00020460
Perlak, F., Fuchs, R., Dean, D., McPherson, S., Fischhoff, D. (1991). Modification of the coding sequence enhances plant expression of insect control protein genes. Proc. Natl. Acad. Sci. 88, 3324–3328. doi: 10.1073/pnas.88.8.3324
Peyret, H., Brown, J., Lomonossoff, G. (2019). Improving plant transient expression through the rational design of synthetic 5′ and 3′ untranslated regions. Plant Methods 15, 108. doi: 10.1186/s13007-019-0494-9
Peyret, H., Lomonossoff, G. (2015). When plant virology metAgrobacterium: the rise of the deconstructed clones. Plant Biotechnol. J. 13, 1121–1135. doi: 10.1111/pbi.12412
Pisano, A., Albano, F., Vecchio, E., Renna, M., Scala, G., Quinto, I., et al. (2018). Revisiting bacterial ubiquitin ligase effectors: Weapons for host exploitation. Int. J. Mol. Sci. 19, 3576. doi: 10.3390/ijms19113576
Plotkin, J., Kudla, G. (2010). Synonymous but not the same: the causes and consequences of codon bias. Nat. Rev. Genet. 12, 32–42. doi: 10.1038/nrg2899
Qi, Y., Katagiri, F. (2009). Purification of low-abundance arabidopsis plasma-membrane protein complexes and identification of candidate components. Plant J. 57 (5), 932–944. doi: 10.1111/j.1365-313X.2008.03736.x
Rademacher, T., Sack, M., Blessing, D., Fischer, R., Holland, T., Buyel, J. (2019). Plant cell packs: a scalable platform for recombinant protein production and metabolic engineering. Plant Biotechnol. J. 17, 1560–1566. doi: 10.1111/pbi.13081
Rajeevkumar, S., Anunanthini, P., Sathishkumar, R. (2015). Epigenetic silencing in transgenic plants. Front. Plant Sci. 6, 693. doi: 10.3389/fpls.2015.00693
Rattanapisit, K., Shanmugaraj, B., Manopwisedjaroen, S., Purwono, P., Siriwattananon, K., Khorattanakulchai, N., et al. (2020). Rapid production of SARS-CoV-2 receptor binding domain (RBD) and spike specific monoclonal antibody CR3022 in nicotiana benthamiana. Sci. Rep. 10, 17698. doi: 10.1038/s41598-020-74904-1
Reski, R., Bae, H., Simonsen, H. T. (2018). Physcomitrella patens, a versatile synthetic biology chassis. Plant Cell Rep. 37, 1409–1417. doi: 10.1007/s00299-018-2293-6
Reski, R., Parsons, J., Decker, E. L. (2015). Moss-made pharmaceuticals: from bench to bedside. Plant Biotechnol. J. 13, 1191–1198. doi: 10.1111/pbi.12401
Reuter, L. J., Bailey, M. J., Joensuu, J. J., Ritala, A. (2014). Scale-up of hydrophobin-assisted recombinant protein production in tobacco by-2 suspension cells. Plant Biotechnol. J. 12, 402–410. doi: 10.1111/pbi.12147
Rigano, M., De Guzman, G., Walmsley, A., Frusciante, L., Barone, A. (2013). Production of pharmaceutical proteins in solanaceae food crops. Int. J. Mol. Sci. 14, 2753–2773. doi: 10.3390/ijms14022753
Ritala, A., Wahlström, E. H., Holkeri, H., Hafren, A., Mäkeläinen, K., Baez, J. (2008). Production of a recombinant industrial protein using barley cell cultures. Protein Expression Purification 59, 274–281. doi: 10.1016/j.pep.2008.02.013
Robert, S., Khalf, M., Goulet, M.-C., D’Aoust, M.-A., Sainsbury, F., Michaud, D. (2013). Protection of recombinant mammalian antibodies from development-dependent proteolysis in leaves of nicotiana benthamiana. PloS One 8, e70203. doi: 10.1371/journal.pone.0070203
Rose, A. B. (2008). “Intron-mediated regulation of gene expression,” in Nuclear pre-mRNA processing in plants. current topics in microbiology and immunology, vol. 326 . Eds. Reddy, A. S. N., Golovkin, M. (Berlin, Heidelberg: Springer).
Ruf, S., Karcher, D., Bock, R. (2007). Determining the transgene containment level provided by chloroplast transformation. Proc. Natl. Acad. Sci. 104, 6998–7002. doi: 10.1073/pnas.0700008104
Sabalza, M., Vamvaka, E., Christou, P., Capell, T. (2013). Seeds as a production system for molecular pharming applications: status and prospects. Curr. Pharm. Des. 19, 5543–5552. doi: 10.2174/1381612811319310009
Sainsbury, F., Jutras, P., Vorster, J., Goulet, M., Michaud, D. (2016). A chimeric affinity tag for efficient expression and chromatographic purification of heterologous proteins from plants. Front. Plant Sci. 7, 141. doi: 10.3389/fpls.2016.00141
Sainsbury, F., Lomonossoff, G. (2008). Extremely high-level and rapid transient protein production in plants without the use of viral replication. Plant Physiol. 148, 1212–1218. doi: 10.1104/pp.108.126284
Sainsbury, F., Thuenemann, E., Lomonossoff, G. (2009). pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 7, 682–693. doi: 10.1111/j.1467-7652.2009.00434.x
Sanagala, R., Moola, A., Bollipo Diana, R. (2017). A review on advanced methods in plant gene targeting. J. Genet. Eng. Biotechnol. 15, 317–321. doi: 10.1016/j.jgeb.2017.07.004
Sandison, M., Cumming, S., Kolch, W., Pitt, A. (2010). On-chip immunoprecipitation for protein purification. Lab. Chip 10, 2805. doi: 10.1039/c005295g
Santi, L., Batchelor, L., Huang, Z., Hjelm, B., Kilbourne, J., Arntzen, C., et al. (2008). An efficient plant viral expression system generating orally immunogenic Norwalk virus-like particles. Vaccine 26, 1846–1854. doi: 10.1016/j.vaccine.2008.01.053
Schaaf, A., Tintelnot, S., Baur, A., Reski, R., Gorr, G., Decker, E. L. (2005). Use of endogenous signal sequences for transient production and efficient secretion by moss (Physcomitrella patens) cells. BMC Biotechnol. 5, 30. doi: 10.1007/s11240-017-1337-x
Schillberg, S., Finnern, R. (2021). Plant molecular farming for the production of valuable proteins – critical evaluation of achievements and future challenges. J. Plant Physiol. 258-259, 153359. doi: 10.1016/j.jplph.2020.153359
Schillberg, S., Raven, N., Spiegel, H., Rasche, S., Buntru, M. (2019). Critical analysis of the commercial potential of plants for the production of recombinant proteins. Front. Plant Sci. 10, 720. doi: 10.3389/fpls.2019.00720
Schillberg, S., Spiegel, H. (2022). Recombinant protein production in plants: A brief overview of strengths and challenges. Methods Mol. Biol. 2480, 1–13. doi: 10.1007/978-1-0716-2241-4_1
Schlegel, S., Hjelm, A., Baumgarten, T., Vikström, D., de Gier, J. (2014). Bacterial-based membrane protein production. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1843, 1739–1749. doi: 10.1016/j.bbamcr.2013.10.023
Serre, N., Alban, C., Bourguignon, J., Ravanel, S. (2018). An outlook on lysine methylation of non-histone proteins in plants. J. Exp. Bot. 69, 4569–4581. doi: 10.1093/jxb/ery231
Sethi, L., Kumari, K., Dey, N. (2021). Engineering of plants for efficient production of therapeutics. Mol. Biotechnol. 63, 1125–1137. doi: 10.1007/s12033-021-00381-0
Sethuraman, N., Stadheim, T. A. (2006). Challenges in therapeutic glycoprotein production. Curr. Opin. Biotechnol. 17, 341–346. doi: 10.1016/j.copbio.2006.06.010
Shanmugaraj, B., Bulaon, I., C. J., Phoolcharoen, W. (2020). Plant molecular farming: A viable platform for recombinant biopharmaceutical production. Plants 9, 842. doi: 10.3390/plants9070842
Shanmugaraj, B., Bulaon, C. J. I., Malla, A., Phoolcharoen, W. (2021). Biotechnological insights on the expression and production of antimicrobial peptides in plants. Molecules 26, 4032. doi: 10.3390/molecules26134032
Shcherbakova, A., Tiemann, B., Buettner, F., Bakker, H. (2017). DistinctC-mannosylation of netrin receptor thrombospondin type 1 repeats by mammalian DPY19L1 and DPY19L3. Proc. Natl. Acad. Sci. 114, 2574–2579. doi: 10.1073/pnas.1613165114
Shen, J.-S., Busch, A., Day, T. S., Meng, X.-L., Yu, C. I., Dabrowska-Schlepp, P., et al. (2015). Mannose receptor-mediated delivery of moss-made α-galactosidase a efficiently corrects enzyme deficiency in fabry mice. J. Inherited Metab. Dis. 39, 293–303. doi: 10.1007/s10545-015-9886-9
Smith, N. A., Singh, S. P., Wang, M.-B., Stoutjesdijk, P. A., Green, A. G., Waterhouse, P. M. (2000). Total silencing by intron-spliced hairpin RNAs. Nature 407, 319–320. doi: 10.1038/35030305
Sobocińska, J., Roszczenko-Jasińska, P., Ciesielska, A., Kwiatkowska, K. (2018). Protein palmitoylation and its role in bacterial and viral infections. Front. Immunol. 8, 2003. doi: 10.3389/fimmu.2017.02003
Stöger, E., Vaquero, C., Torres, E., Sack, M., Nicholson, L., Drossard, J., et al. (2000). Cereal crops as viable production and storage systems for pharmaceutical scFv antibodies. Plant Mol. Biol. 42, 583–590. doi: 10.1023/A:1006301519427
Strasser, R. (2016). Plant protein glycosylation. Glycobiology 26, 926–939. doi: 10.1093/glycob/cww023
Szaleniec, M., Wojtkiewicz, A., Bernhardt, R., Borowski, T., Donova, M. (2018). Bacterial steroid hydroxylases: enzyme classes, their functions and comparison of their catalytic mechanisms. Appl. Microbiol. Biotechnol. 102, 8153–8171. doi: 10.1007/s00253-018-9239-3
Thuenemann, E., Meyers, A., Verwey, J., Rybicki, E., Lomonossoff, G. (2013). A method for rapid production of heteromultimeric protein complexes in plants: assembly of protective bluetongue virus-like particles. Plant Biotechnol. J. 11, 839–846. doi: 10.1111/pbi.12076
Tippens, N., Vihervaara, A., Lis, J. (2018). Enhancer transcription: what, where, when, and why? Genes Dev. 32, 1–3. doi: 10.1101/gad.311605.118
Top, O., Milferstaedt, S. W. L., van Gessel, N., Hoernstein, S. N. W., Özdemir, B., Decker, E. L., et al. (2021). Expression of a human cDNA in moss results in spliced mRNAs and fragmentary protein isoforms. Commun. Biol. 4, 964. doi: 10.1038/s42003-021-02486-3
Torrent, M., Llop-Tous, I., Ludevid, M. D. (2009). “Recombinant proteins from plants,” in Protein body induction: A new tool to produce and recover recombinant proteins in plants (Berlin, Germany: Springer), 193–208.
Usmani, S., Bedi, G., Samuel, J., Singh, S., Kalra, S., Kumar, P., et al. (2017). THPdb: Database of FDA-approved peptide and protein therapeutics. PloS One 12, 0181748. doi: 10.1371/journal.pone.0181748
Vain, P., Worland, B., Kohli, A., Snape, J. W., Christou, P., Allen, G. C., et al. (1999). Matrix attachment regions increase transgene expression levels and stability in transgenic rice plants and their progeny. Plant J. 18, 233–242. doi: 10.1046/j.1365-313X.1999.00446.x
VanDrisse, C., Escalante-Semerena, J. (2019). Protein acetylation in bacteria. Annu. Rev. Microbiol. 73, 111–132. doi: 10.1146/annurev-micro-020518-115526
Villarejo, A., Burén, S., Larsson, S., Déjardin, A., Monné, M., Rudhe, C., et al. (2005). Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat. Cell Biol. 7, 1224–1231. doi: 10.1038/ncb1330
Walsh, G. (2018). Biopharmaceutical benchmarks 2018. Nat. Biotechnol. 36, 1136–1145. doi: 10.1038/nbt.4305
Waugh, D. S. (2011). An overview of enzymatic reagents for the removal of affinity tags. Protein Expression Purification 80, 283–293. doi: 10.1016/j.pep.2011.08.005
Webster, G. R., Teh, A. Y. H., Ma, J. K. C. (2016). Synthetic gene design-the rationale for codon optimization and implications for molecular pharming in plants. Biotechnol. Bioengineering 114, 492–502. doi: 10.1002/bit.26183
Wilken, L., Nikolov, Z. (2012). Recovery and purification of plant-made recombinant proteins. Biotechnol. Adv. 30, 419–433. doi: 10.1016/j.biotechadv.2011.07.020
Wongsamuth, R., Doran, P. M. (1997). Production of monoclonal antibodies by tobacco hairy roots. Biotechnol. Bioengineering 141–150, 401–415. doi: 10.1002/(SICI)1097-0290(19970605)54:5<401::AID-BIT1>3.0.CO;2-I
Xu, X., Zhu, T., Nikonorova, N., De Smet, I. (2019). Phosphorylation-mediated signalling in plants. Annu. Plant Rev. Online 2, 909–932. doi: 10.1002/9781119312994.apr0702
Yamamoto, T., Hoshikawa, K., Ezura, K., Okazawa, R., Fujita, S., Takaoka, M., et al. (2018). Improvement of the transient expression system for production of recombinant proteins in plants. Sci. Rep. 8, 4755. doi: 10.1038/s41598-018-23024-y
Zhang, Y., Li, D., Jin, X., Huang, Z. (2014). Fighting Ebola with ZMapp: spotlight on plant-made antibody. Sci. China Life Sci. 57, 987–988. doi: 10.1007/s11427-014-4746-7
Zhang, M., Xu, J., Hu, H., Ye, B., Tan, M. (2017). Systematic proteomic analysis of protein methylation in prokaryotes and eukaryotes revealed distinct substrate specificity. PROTEOMICS 18, 300.
Zhang, X., Zhu, Y., Wu, H., Guo, H. (2015). Post-transcriptional gene silencing in plants: a double-edged sword. Sci. China Life Sci. 59, 271–276. doi: 10.1007/s11427-015-4972-7
Keywords: recombinant protein expression, transgenic plants, plant chassis, transformation, protein purification
Citation: Coates RJ, Young MT and Scofield S (2022) Optimising expression and extraction of recombinant proteins in plants. Front. Plant Sci. 13:1074531. doi: 10.3389/fpls.2022.1074531
Received: 19 October 2022; Accepted: 22 November 2022;
Published: 08 December 2022.
Edited by:
Rima Menassa, Agriculture and Agri-Food Canada (AAFC), CanadaReviewed by:
Johannes Felix Buyel, University of Natural Resources and Life Sciences, AustriaAndreas Schiermeyer, Fraunhofer Society (FHG), Germany
Copyright © 2022 Coates, Young and Scofield. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Simon Scofield, U2NvZmllbGRTQGNhcmRpZmYuYWMudWs=
 Ryan J. Coates
Ryan J. Coates Mark T. Young
Mark T. Young Simon Scofield
Simon Scofield