- 1Frontiers Science Center for Molecular Design Breeding/Key Laboratory of Crop Heterosis and Utilization (Ministry of Education), Beijing Key Laboratory of Crop Genetic Improvement, China Agricultural University, Beijing, China
- 2Institute of Crop Sciences, Xinjiang Academy of Agricultural Sciences, Urumuqi, China
- 3Hebei Key Laboratory of Crop Salt-alkali Stress Tolerance Evaluation and Genetic Improvement, Cangzhou Academy of Agriculture and Forestry Science, Cangzhou, China
Common wheat (Triticum aestivum, BBAADD) is an allohexaploid species combines the D genome from Ae. tauschii and with the AB genomes from tetraploid wheat (Triticum turgidum). Compared with tetraploid wheat, hexaploid wheat has wide-ranging adaptability to environmental adversity such as salt stress. However, little is known about the molecular basis underlying this trait. The plasma membrane Na+/H+ transporter Salt Overly Sensitive 1 (SOS1) is a key determinant of salt tolerance in plants. Here we show that the upregulation of TaSOS1 expression is positively correlated with salt tolerance variation in polyploid wheat. Furthermore, both transcriptional analysis and GUS staining on transgenic plants indicated TaSOS1-A and TaSOS1-B exhibited higher basal expression in roots and leaves in normal conditions and further up-regulated under salt stress; while TaSOS1-D showed markedly lower expression in roots and leaves under normal conditions, but significant up-regulated in roots but not leaves under salt stress. Moreover, transgenic studies in Arabidopsis demonstrate that three TaSOS1 homoeologs display different contribution to salt tolerance and TaSOS1-D plays the prominent role in salt stress. Our findings provide insights into the subgenomic homoeologs variation potential to broad adaptability of natural polyploidy wheat, which might effective for genetic improvement of salinity tolerance in wheat and other crops.
Introduction
Allopolyploidization played a significant role in the speciation and evolution of both wild and cultivated plants (Jiao et al., 2011; Hegarty et al., 2013). Allopolyploid organisms often outperform their ancestral species in several aspects, such as increased vigor and potential adaptation to a wider range of environmental conditions (Otto, 2007; Madlung, 2013; Wu et al., 2020). As an allohexaploid species, common wheat (Triticum aestivum, BBAADD) is originated through hybridization and chromosome doubling of three ancestral diploid species, related to nowadays existing Triticum urartu (AA), Aegilops speltoides (SS), and Ae. tauschii (DD) (Goncharov, 2011). Thus, most of the genes present in common wheat as triplicate homoeologs derived from the ancestral species (Feldman and Levy, 2005; IWGSC et al., 2018). Hexaploid wheat presents broader adaptability to different environments and increasing the tolerance to both biotic and abiotic stresses compared with tetraploid wheat (Dubcovsky and Dvorak, 2007; Feldman et al., 2012; Pallotta et al., 2014; Yang et al., 2014; Yang et al., 2018). Research increasingly suggests that the differential expression and functional diversification of homoeologous genes plays an important role in the evolutionary success of polyploidy wheat species (Shitsukawa et al., 2007; Hu et al., 2013; Li et al., 2014; Yang et al., 2014; Uauy, 2017; Wang et al., 2018). Nevertheless, our understanding of the molecular basis underlying of these traits or the relative contributions of allohexaploidization on the trait development are still limited in hexaploid wheat. Exploring the mechanisms of evolutionary genetic changes or that modulating the expression of triplicate homoeologs may provide clues regarding the origin of important traits that have arisen during the evolution and domestication of allopolyploid wheat (Borrill et al., 2015; Ding and Chen, 2018).
Salinity is considered as one of the most severe environmental hazards for wheat growth and grain yield worldwide (Munns and Tester, 2008). Salinity stress can decrease wheat growth and productivity by reducing water uptake and causing nutrient disorders and ion toxicity in many regions (Huang et al., 2006; Byrt et al., 2014; Munns and Gilliham, 2015). Therefore, understanding the mechanisms of response and adaptation to salt stress and then improving the salinity tolerance of crops are main targets for future researches. It is well known that hexaploid bread wheat is generally more salt tolerant than its tetraploid wheat progenitor (Dvorak et al., 1994; Munns et al., 2012; Yang et al., 2014). In saline environments, hexaploid wheat has a low rate of Na+ transport to the shoot and maintains a high ratio of K+ to Na+ in leaves than durum wheat, this trait is conferred, at least partially, by the Kna1 locus that mapped to the distal region of chromosome 4DL (Gorham et al., 1987; Dubcovsky et al., 1996). Further studies indicated that gene TaHKT1;5-D could be as the candidate gene of Kna1 locus, encodes a Na+-selective transporter located on the plasma membrane of stelar cells in roots of wheat, confers the essential salinity tolerance in bread wheat via shoot Na+ exclusion (Byrt et al., 2014). The tetraploid wheat (genome BBAA) does not exclude Na+ to the same extent as bread wheat, since the A subgenome of tetraploid and hexaploid wheats, which was contributed by Triticum urartu (AuAu), lacks the HKT1;5 gene; the HKT1;5-B, albeit present, may have lost its function for salt tolerance (James et al., 2006).
Besides the HKT-type transporters confer salinity tolerance by promoting sodium exclusion (Ren et al., 2005; Huang et al., 2008), the salt-overly-sensitive (SOS) signal transduction pathway is to be widely regarded as important mechanism for maintaining intracellular ion homeostasis and salt tolerance in plants (Zhu, 2003; Du et al., 2011). This pathway including three key components (SOS1, SOS2 and SOS3). SOS3 encodes a myristoylated Ca2+ -binding protein that capable of sensing Ca2+ oscillations elicited by salt stress and is able to activate and recruit the serine/threonine protein kinase SOS2 to the plasma membrane to achieve phosphorylation and activation of SOS1, a Na+/H+ antiporter, thus causes efflux of excessive Na+ from the cell (Shi et al., 2000; Qiu et al., 2002; Guo et al., 2004; Quintero et al., 2011; Ji et al., 2013). Compared with sos2 and sos3 mutant plants, sos1 mutant plants are even more sensitive to NaCl treatments (Wu et al., 1996). In Arabidopsis, SOS1 preferentially express in epidermal cells at the root tip and in parenchyma cells at the xylem/symplast boundary of roots, stems, and leaves (Shi et al., 2002; Mahi et al., 2019). Under salt condition, the xylem sap of sos1 mutant plants contain more Na+ suggested that SOS1 is crucial for controlling long-distance Na+ transport from root to shoot (Shi et al., 2002). Increased SOS1 activity would also lead to apoplastic alkalinization and cytoplasmic acidification which might serve as a signal to mediate gene regulation (Chung et al., 2008). Recently, the importance of SOS1-facilitated Na+ flux in the salt tolerance of rice, soybean, tomato and cotton were analyzed, suggesting that SOS1 is conserved in higher plants including both monocots and dicots (Olías et al., 2009; Che et al., 2019; Mahi et al., 2019). In wheat, previous studies also indicated expression of TaSOS1 can enhance the salt tolerance of a yeast strain lacking the major Na+ efflux systems and improves the growth and salt tolerance of transgenic Arabidopsis and tobacco (Xu et al., 2008; Feki et al., 2014; Zhou et al., 2016). However, little is known about the relative contributions of TaSOS1 homoeologs to salt tolerance in polyploid wheat. In addition, there are currently no comparative analysis focusing on assessment of genome-specific expression and functional variances among TaSOS1 homoeologous genes in allopolyploid wheat. Except that Feki et al. found the promoter of TaSOS1-AB and TaSOS1-D are age-dependent and TaSOS1-AB is abiotic stress-inducible at different developmental stages (Feki et al., 2015).
In this study, we show that three Na+/H+ antiporter TaSOS1 homoeologs of wheat exhibited subgenome-biased expression and functional diversification in response to salt stress and among three TaSOS1 homoeologs, TaSOS1-D displays the prominent contribution to salt tolerance in transgenic Arabidopsis. Our findings provide insights into the subgenomic homoeologs variation potential to facilitate the wide-ranging adaptability of natural hexaploid wheat and other polyploidy species.
Materials and methods
Plant materials and growth conditions
Two set of synthetic allohexaploid wheat (SCAUP/SQ523, DM4/Y199) and their tetraploid (SCAUP, DM4) and diploid (SQ523, Y199) progenitors, 48 natural wheat accessions with different ploidy levels (Table S1) were used to examine the difference of salt tolerance between different ploidy wheats and to explore the expression pattern of TaSOS1 before and after salt treatment. wheat plants were grown under 16/8 h, light/dark conditions at a temperature of 22°C. wheat variety Nongda3338 was used to clone the three CDS and promoter sequences of TaSOS1 homoeologous genes and analyze the expression pattern of three TaSOS1 homoeologous genes under normal or salt condition.
Arabidopsis (Arabidopsis thaliana) wild-type Columbia was used as the recipient of the overexpression and promoter analysis of pSOS1:uidA reporters transformation. Sterilized seeds were incubated at 4°C for 3 d then sowed on MS plates containing 1% (w/v) Suc and 0.8% (w/v) agar. Seedlings were grown under a 16-h-light/8-h-dark cycle at 22°C in a growth room.
Salt tolerance assays
For salt tolerance comparisons, seedlings of the synthetic allohexaploid wheat genotype (SCAUP/SQ523), and their respective tetraploid and diploid parents, 48 natural wheat accessions with different ploidy levels were grown in vermiculite and watered with half strength Hoagland’s liquid medium (pH 6.0). The seedlings were treated with 0 or 200 mM NaCl at three-leaf stage for 10 d. Then the chlorophyll content, shoot fresh weight, root length and root fresh weight were quantified and photos were taken. Root length was quantified by measurement with a ruler and the fresh mass of the shoots and roots were measured using an automated electronic scale. Chlorophyll were measured by aqueous acetone (80%) described earlier.
For Arabidopsis germination rate analysis, Seeds were sown on MS medium supplemented with 0, 100, 200 mM NaCl, and germination rates were scored every 24 hours. For the salt tolerance assay of plants grown in soil, 5-d-old seedlings were transferred from MS medium to soil. The seedling of wild type and transgenic lines plants grown in soil pots were irrigated with 250 mM NaCl every 5 d, then the chlorophyll content, shoot fresh weight and the leaves electrolyte leakage of each genotype were evaluated after 15 d. For whole growth duration salt tolerance assay, seedlings plants were irrigated every 5 d with or without the addition of 200 mM NaCl until seed maturation.
Genomic DNA and total RNA extraction
The genomic DNA of wheat genotype Nongda3338 was extracted using the CTAB protocol. Total RNA was extracted with TRIzol reagent (Invitrogen), and purified RNA was treated with DNase I. Subsequently, First-strand cDNA synthesis was performed using HiScipt II One Step RT-PCR Kit (Vazyme Biotech, Nanjing, China).
Quantitative RT-PCR
Real-time qRT-PCR analysis was conducted using SYBR Premix EX TagTM (TaKaRa, Dalian, China) in a volume of 20μl in a Bio-Rad CFX96 real-time PCR detection system3. The PCR parameters were as follows: 94°C for 3 min, followed by 40 cycles of 94°C for 15 s, 60°C for 20 s, and 72°C for 20 s. Each sample was quantified at least in triplicate and normalized using Actin as an internal control.
Gene cloning and vector construction
Specific primer sets were designed to obtain the CDS and promoter sequences of TaSOS1-A, TaSOS1-B and TaSOS1-D. The PCR was performed using the Tks Gflex DNA Polymerase (TaRaKa, Beijing, China) and the PCR products were recovered by TIANgel Midi Purification Kit (TIANGEN BIOTECH, Beijing, China) then cloned into the pEASY-Blunt Simple Cloning vector (TransGen Biotech, Beijing, China) and sequenced. Amplified fragments of the full-length coding sequence of TaSOS1-A, TaSOS1-B and TaSOS1-D were cloned into the binary expression vector pCAMBIA1300 (driven by the CaMV 35S promoter). The promoter region of target genes that contains about 2-kb fragment upstream of the coding sequence was fused to the reporter gene encoding GUS and was then cloned into the binary expression vector pCAMBIA1300 to generate reporters. All the constructs were constructed by In-Fusion PCR Cloning Kits (TaRaKa, Beijing, China) and were confirmed by sequencing. These vectors were transferred into Agrobacterium tumefaciens strain GV3101 and transgenic plants were identified on Murashige and Skoog medium containing hygromycin. The primers employed for plasmid construction are listed in Table S2.
GUS histochemical staining
Two-week-old transgenic Arabidopsis seedlings were transferred to MS medium (1% Suc) with 0 or 200 mM NaCl, and after 24h whole seedlings or different tissues expressing the GUS gene were immersed in staining buffer ((100 mM NaPO4, 0.5 mM K3[Fe(CN)6, 0.5 mM K4[Fe(CN)6], 10 mM EDTA, 1 mg/ml 5-bromo-4-chloro-3-indolyl-b-D-glucuronide (X-gluc)) at 37°C in the dark for overnight. Subsequently, samples were washed in 75% ethanol and the intensity of color development in different tissues was monitored and photographed.
Ion measurements
For measurement of Na+ and K+ contents in the seedling leaves, 10-day-old uniform seedlings were transferred to MS plates with 100 mM NaCl solution. 2 days later, the samples were harvested and dried, then dissolved in HNO3 and analyzed by inductively coupled plasma optical emission spectrometry (FP6410, INESA).
Results
Sequence analysis of TaSOS1 homoeologs in hexaploid wheat
In order to identify TaSOS1 homoeologous genes in wheat, we conducted a BLAST search against the wheat genome database (IWGSC RefSeq v1.0) using rice and barley SOS1 sequences as queries. After the redundancy elimination and domain confirmation, the three sets of wheat TaSOS1 homoeologs on chromosome 3AS, 3BS and 3DS were obtained and these homologous alleles were designated as TaSOS1-A, TaSOS1-B, or TaSOS1-D according to their chromosome location (Figure 1A). A sequence comparison between cDNA and genomic DNA revealed that TaSOS1 has 23 exons and 22 introns (Figure S1). The coding sequences (CDS) of the three TaSOS1 homoeologs were comparatively conserved, and can be distinguished from one another by series of single nucleotide polymorphisms (SNPs). In contrast with the CDS, differences in molecular size were found in introns and in upstream/downstream regions. Deduced amino acid sequence alignments showed that TaSOS1-A, TaSOS1-B and TaSOS1-D encode 1142 amino acid residues and three TaSOS1 homoeologs shared 98.16% sequence similarity. The amino acid sequences differed from one another at 49 positions (Figure 1B). In addition, similar to its orthologues from Arabidopsis, the predicted proteins of TaSOS1 homoeologs contain 12 transmembrane domains (TM) in N-terminus and all of them are conserved in three TaSOS1 homoeologs, except that a single amino acid substitution in the first predicted membrane-spanning region of TaSOS1-D (Ala-Gly). Previously analysis in Arabidopsis showed that AtSOS1 protein contains three pivotal and highly conserved consensus motifs, including CNBD activation domain in N-terminus, auto-inhibitory domain and phosphorylation site in C-terminus. The alignments revealed the phosphorylation site and the auto-inhibitory domain highly conserved at the C-terminus of three TaSOS1 homoeologs (Figure 1B). However, the sequence of CNBD activation domain, a conserved domain interacts with auto-inhibitory domain to keep the SOS1 protein with basal activity under control condition, show differences between TaSOS1-D and TaSOS1-A, TaSOS1-B (Figure 1B).

Figure 1 Sequence analysis of three TaSOS1 homoeologous genes. (A) Phylogenetic relationship analysis of three TaSOS1 homoeologs with other members in multiple species. The accession numbers of selected species and gene IDs are listed as Table S3. (B) Comparison of amino acids in three TaSOS1 homoeologs. Identical amino acids are indicated by asterisks, gray boxes indicate conserved domains among the three proteins, and the red arrows indicate amino acid difference within the conserved domains among three TaSOS1 homoeologs.
Expression patterns of wheat TaSOS1 in different tissues under salt stress
The spatial and temporal expression patterns of TaSOS1 under normal and salt stress condition were first analyzed in a wheat variety Nongda3338 by quantitative real-time PCR (qRT-PCR). The results indicated the transcription abundance of TaSOS1 were improved after salt treatment but the transcription patterns were different in seedling shoot and root. In root, the expression of TaSOS1 was induced quickly and peaked at 9 h after salt treatment then the level of mRNA accumulation gradually declined (Figure 2A). However, the mRNA accumulation remains almost unchanged in shoot until 48h after salt condition (Figure 2B).
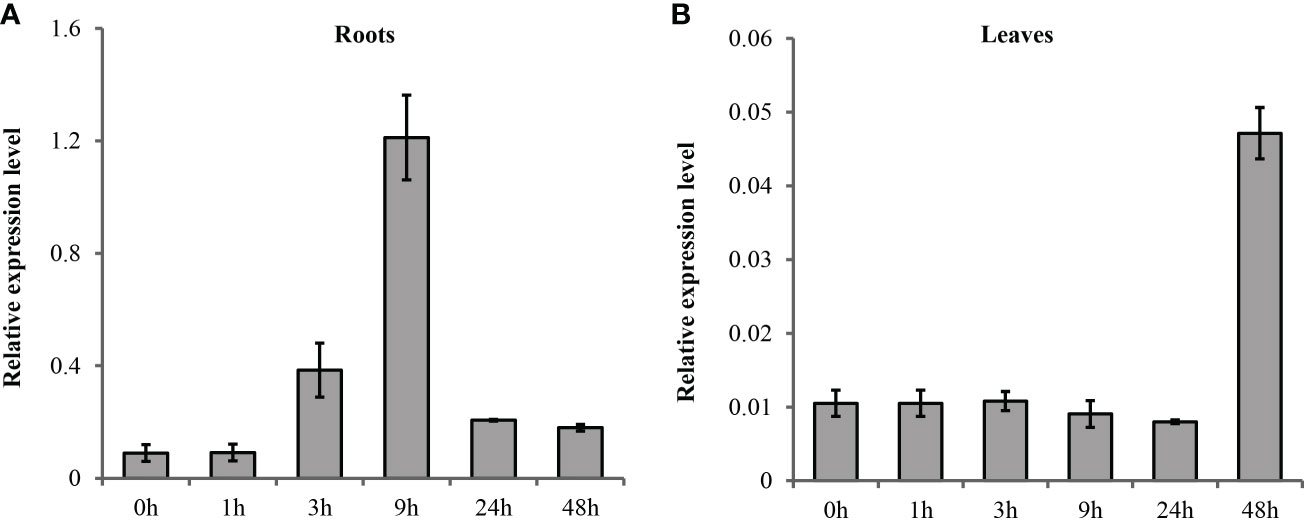
Figure 2 Quantitative RT-PCR analysis of the expression of TaSOS1 genes in response to salt stress in wheat. Transcript levels of TaSOS1 genes detected in 7-day-old wheat seedling roots (A) and leaves (B) were subjected to 0 mM or 200 mM NaCl treatment with indicated time course. The expression of β-Actin was used as an endogenous control. Error bars represent the standard deviations of three technical replicates, and the results were consistent in three biological replicates.
Expression of wheat TaSOS1 gene was correlated with salt tolerance variation in polyploidy wheat
Hexaploid bread wheat has a greater discriminating capacity between Na+ and K+ than durum wheat under salt stress and is generally more salt tolerant. In order to assess the salt tolerance variation after the formation of allopolyploid wheat, two set of synthetic hexaploids (SCAUP/SQ523 and DM4/Y199, BBAADD) with their tetraploid (SCAUP and DM4, BBAA) and diploid Ae. tauschii (SQ523 and Y199, DD) parents were used for salt tolerance assessment. The root length, root and shoot fresh weight for each of the three genotypes under normal and salt stress conditions were recorded and quantified. Consistent with previous study (Zheng et al., 2021), the results indicated the synthetic hexaploid wheats and diploid Ae. tauschii exhibited superior growth vigour relative to the tetraploid lines (Figures 3A-E; Figure S2). We further analyzed the expression patterns of TaSOS1 gene in two synthetic hexaploids with their tetraploid and diploid parents using qRT-PCR. Consistent with our expectation, the TaSOS1 genes were up-regulated in synthetic hexaploid wheats (SCAUP/SQ523, DM4/Y199) compared with their tetraploid parents (SCAUP, DM4) both in seedling roots and shoots upon NaCl treatment. Notably, the expression of TaSOS1 was upregulated more significantly in diploid (SQ523, Y199) parents (Figures 3F-I). Thus, we examined whether the expression pattern of TaSOS1 is correlative to the variation in the salt tolerance (scored root length as a growth indicator and compared the changes before and after salt treatment) in 48 natural wheat accessions with varying ploidy, including 16 allohexaploid wheats, 16 allotetraploid and 16 diploid species Ae. tauschiis (Figures 4A, B; Table S1). Across the three different ploidy levels, the fold change of TaSOS1 upregulation expression was positively correlated with the root length under salt treatment of the natural wheat accessions (BBAADD, BBAA and DD; R2 = 0.613, P < 0.01; Figure 4C). In addition, we also performed the correlation analysis between the TaSOS1 expression pattern with the change of root and shoot fresh weight before and after salt treatment in 48 natural wheat accessions, these results also indicated the fold change of TaSOS1 up-regulation expression was positively correlated with salt tolerance in wheat accessions of different ploidy (Figures S2A-E).
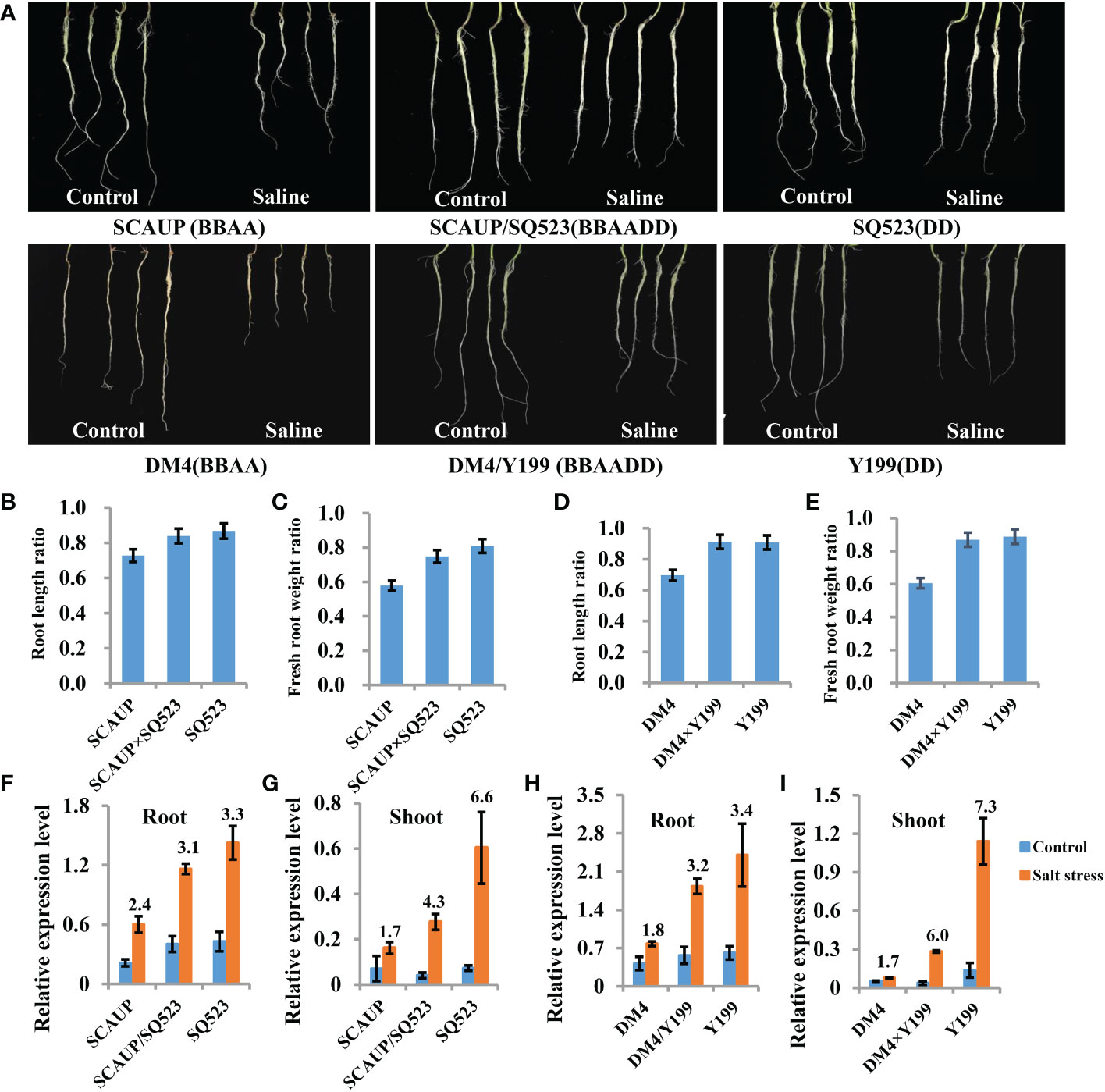
Figure 3 Salt stress phenotypes and transcript level of SOS1 genes in synthetic allopolyploid wheats and their parents. Seedlings of the two set of synthetic hexaploids with their tetraploid and diploid parents were first grown for 10 d without NaCl treatment. The plants were then grown under control conditions (Control) or treated with 200 mM NaCl solution (Saline); phenotypes were measured and photographed after 7 d. SCAUP/SQ523 and DM4/Y199, synthetic allohexaploid wheats, BBAADD; SCAUP and DM4, tetraploid parents, BBAA; SQ523 and Y199, diploid parents, DD. (A) Root phenotypes of with tetraploid and diploid parents upon NaCl treatment. (B-E) Quantification of root length (B, D) and fresh weight (C, E) of two synthetic allotetraploid wheats and their tetraploid and diploid parents. Mean and SD values were derived from measurements of at least 20 seedlings from three independent assays. (F-I) qRT‐PCR analysis of SOS1 genes in roots (F, H) and shoots (G, I) of two set of synthetic hexaploids with their tetraploid and diploid parents in response to salt stress. Seven-day-old seedling plants were subjected to 0 mM or 9h after 200 mM NaCl treatment for root or after 2 days of 200 mM NaCl treatment for shoot. The expression of β-Actin was used to normalize mRNA levels. The values are means (± SE) of three biological replicates.
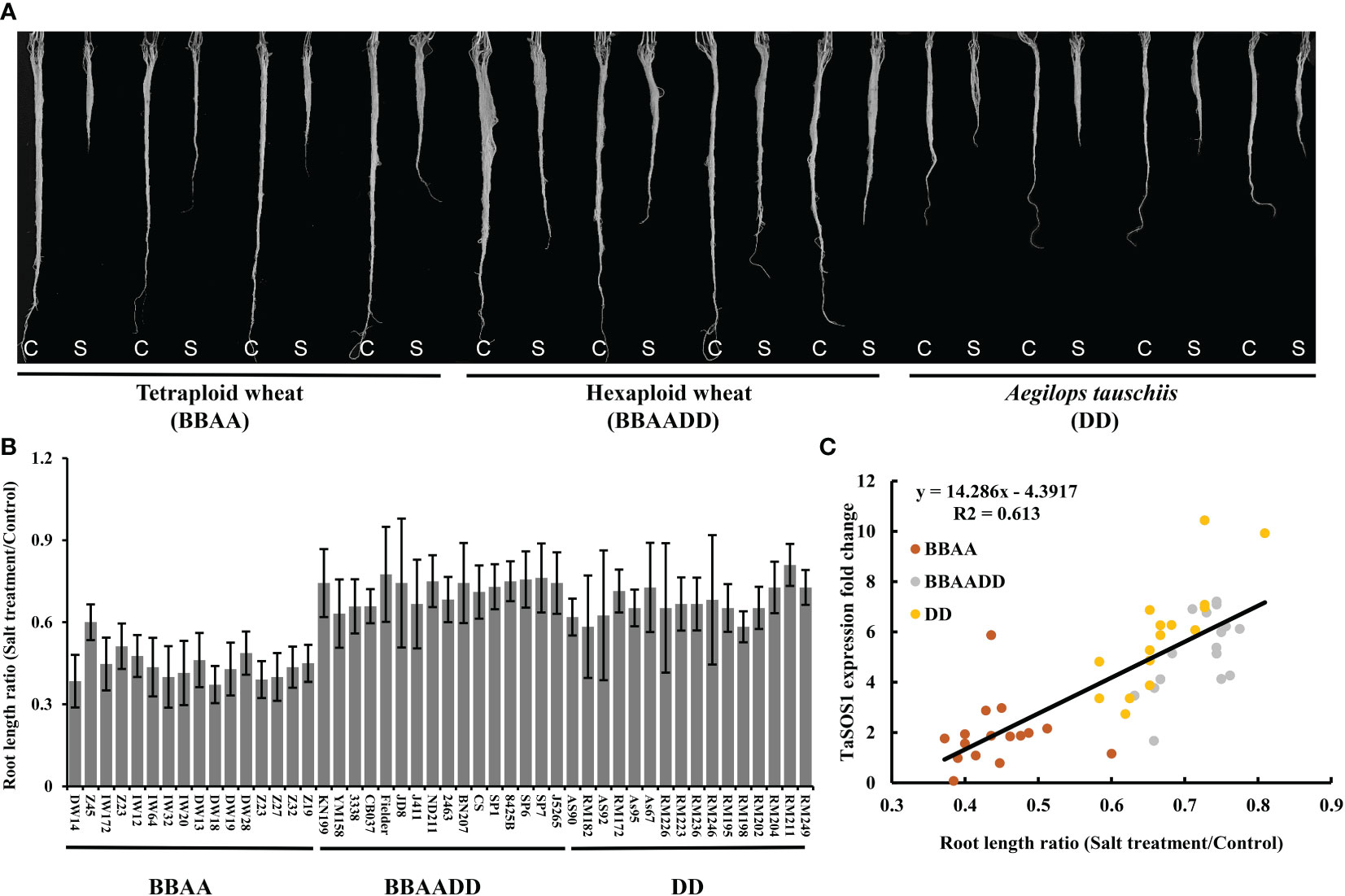
Figure 4 Expression of SOS1 gene was correlated with salt tolerance variation in wheat accessions of different ploidy. (A) Example of root length phenotype of natural wheat accessions before and after 200 mM NaCl treatment. C: Control; S: Salt treatment. (B) A quantitative assessment of the roots length for 48 natural wheat accessions. The y-axis denotes a ratio of NaCl-treated seedlings relative to the same genotype grown on control condition. Mean and SD values were derived from measurements of at least 20 seedlings from three independent assays. (C) Correlation coefficients between the roots length ratio (Salt treatment/Control) and the expression levels of TaSOS1 among different ploidy wheat accessions. The x-axis denotes a ratio of NaCl-treated seedlings relative to the same genotype grown on control condition. The y-axis denotes the fold change of TaSOS1 up-regulation expression in root of wheat accessions with different ploidy before and after 200 mM NaCl treatment.
Subgenome expression divergence of three TaSOS1 homoeologs in hexaploid wheat
To further investigate how the expressions of TaSOS1 homoeologous genes were regulated in allopolyploid wheat under salt stress, gene-specific PCR primers were designed and the amplification efficiency of the primers was confirmed by homoeolog-specific genomic PCR (Figure S3A). Considering the expression of TaSOS1 peaked at 9 h in root and 2 d after salt stress in shoot, we coupled quantitative RT-PCR to quantity the TaSOS1-A, TaSOS1-B and TaSOS1-D subgenome specific expression at these specific time points after salt stress in root and shoot, respectively. Interestingly, different expression patterns were found between the three TaSOS1 homoeologous genes. In root, NaCl treatment increased the transcription level by more than 12 times of TaSOS1-D and 2.7-, 6.4-fold enhancements were observed of the amount of TaSOS1-A and TaSOS1-B (Figure 5A). However, in shoot, the amount of TaSOS1-B mRNA was much higher than TaSOS1-A and TaSOS1-D, while the transcription level of TaSOS1-D was the lowest in both control and salt condition (Figure 5B). Taken together, these results showed that these homoeologs may play versatile roles in different organs under salt stress condition.
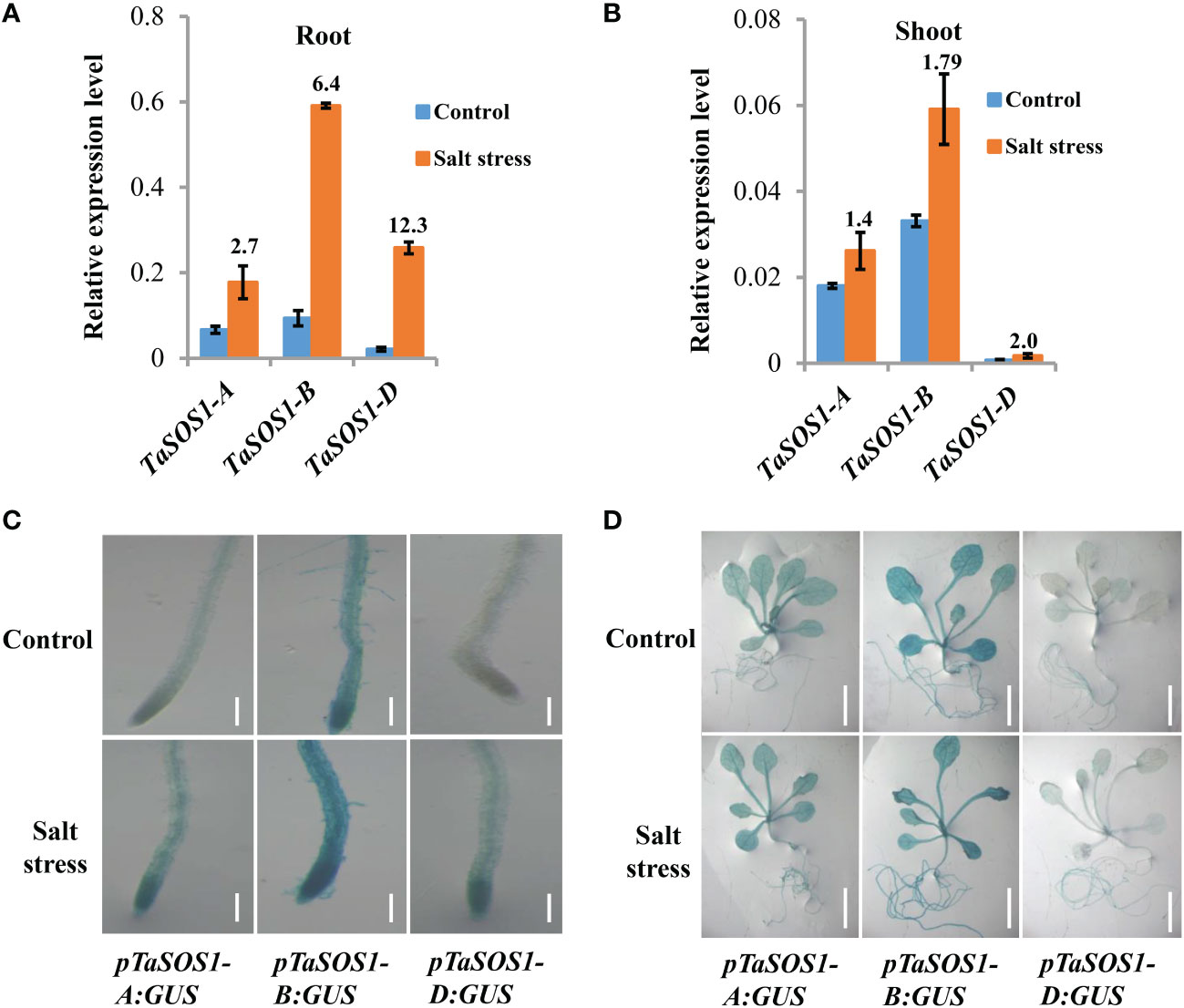
Figure 5 Expression patterns of three TaSOS1 homoeologous genes in response to salt stress in allohexaploid wheat. (A, B) Transcript levels of three TaSOS1 homoeologous genes in roots and shoots of allohexaploid wheat upon NaCl treatment. qRT-PCR analysis the expression of TaSOS1-A, TaSOS1-B and TaSOS1-D genes with gene-specific primers in wheat seedling roots that subjected to 9 h treatment with 200 mM NaCl or in wheat seedling leaves subjected to 2 d treatment with 200 mM NaCl. Fold change ratio was labeled above the bars. The expression of β-Actin was used to normalize mRNA levels. The values are means (± SE) of three biological replicates. (C, D) Spatial expression patterns of the TaSOS1-A, TaSOS1-B and TaSOS1-D genes in transgenic Arabidopsis plants harboring their promoters fused to the GUS gene, respectively. Promoter activity was visualized through histochemical GUS staining in roots (C) and seedlings plants (D) transferred to normal medium (control) or to medium containing 200 mM NaCl (salt stress) for 1 days. Scale bars: root, 200 µm; whole seedling plants, 1 cm.
To gain further insight into the transcriptional divergence of three TaSOS1 homoeologous genes in normal and salt treatment conditions, we generated transgenic Arabidopsis plants expressing the β-glucuronidase (GUS) gene (uidA) under the control of the TaSOS1-A, TaSOS1-B and TaSOS1-D promoter, respectively. About 2 kb genomic DNA fragments which included the sequence upstream of transcription initiation site of three TaSOS1 homoeologs were amplified, respectively. The reporter constructs using uidA fused to the promoter sequence of TaSOS1-A, TaSOS1-B and TaSOS1-D (pTaSOS1-A:GUS, pTaSOS1-B:GUS, pTaSOS1-D:GUS) were transformed into the wild type Arabidopsis (Columbia ecotype) (Figure S3B). The construct was introduced via Agrobacterium-mediated transformation by the floral-dip method. For each transgenic assay, three independent T3 homozygous lines carrying the construct for a single insertion were selected and used for further characterization (Figure S3C). The GUS activity of transgenic Arabidopsis plants carrying pTaSOS1-A:GUS, pTaSOS1-B:GUS and pTaSOS1-D:GUS were examined under normal or salt conditions by histochemical staining (Figures 5C, D). In normal conditions, the GUS activity can be detected in roots, stems and leaves of pTaSOS1-A:GUS and pTaSOS1-B:GUS seedling plants, but undetectable in leaves and slightly in root of pTaSOS1-D:GUS lines (Figures 5C, D). These differential expression pattern between the three promoters are consistent with our qRT-PCR results in wheat. To assess the expression of TaSOS1s under salt stress conditions, transgenic Arabidopsis seedlings were subjected to 200 mM NaCl treatment for 1 d, after which the seedlings were stained. The results showed that NaCl induced deeper blue staining in root of pTaSOS1-B:GUS than pTaSOS1-A:GUS and pTaSOS1-D:GUS lines, but the improvement ratio of GUS activity of TaSOS1-D in root was more than TaSOS1-A and TaSOS1-B (Figure 5C). Meanwhile, the histochemical staining showed that the activities of the three wheat TaSOS1 promoters in leaves under salt stress almost unchanged or a bit higher than untreated controls (Figure 5D). Collectively, our results, on the one hand, were consistent with our qRT-PCR results showing that the transcription patterns of TaSOS1-A, TaSOS1-B and TaSOS1-D existed discrepancies under normal and salt treatment. On the other hand, the results indicated that TaSOS1-A and TaSOS1-B showed high basal expression in roots and leaves in normal condition and they can further up-regulation in roots under salt stress. While TaSOS1-D showed markedly lower expression in both roots and leaves under normal conditions, but showed significant up-regulation in roots but not leaves under salt stress.
The cis-acting element analysis for promoters of three TaSOS1 homoeologs
To explore the molecular basis of the differential expression pattern of three TaSOS1 homoeologous genes, cis-acting elements analysis for promoters of three TaSOS1 homoeologs was carried out through web-based PLANTCARE and PLACE databases (Lescot et al., 2002). Results showed that there are 23 cis-acting elements owned by all the three TaSOS1 promoters and several of them involved in the activation of defense-related genes, including predicted ABREs, which are known to be involved in abscisic acid responsiveness. TGACG-motif, CGTCA-motif, which are involved in jasmonic acid methyl ester (MeJA) responses. AuxRR-core, which is associate with auxin responsiveness. Further, several elements involved in light responsiveness such as G-box, ATC-motif, and CAT-box, which is related to meristem expression were also present in the promoters of three TaSOS1 homoeologs (Figure S4). Moreover, there are several types of cis-acting elements were detected only in one or two promoter regions of three TaSOS1 genes, such as EIRE and O2-site motif only presented in TaSOS1-A; while ACE, ATCT-motif, CATT-motif, HSE and TC-rich repeats are presented both in promoters of TaSOS1-A and TaSOS1-B, but absent in TaSOS1-D. Interestingly, we also found the P-box, AP-2-like and WUN-motif are specifically exhibited in promoter of TaSOS1-D. These results suggest there are genetic variations in cis-regulatory elements or regulatory regions in the three TaSOS1 homoeologs and may be involved in gene expression changes that impact phenotype.
Three TaSOS1 homoeologous genes show a different contribution to salt tolerance
To further understand the functions of three TaSOS1 homoeologs in wheat, ectopic expression of TaSOS1-A, TaSOS1-B and TaSOS1-D were performed using Arabidopsis transgenic system. For each transgenic line, three independent T3 homozygous overexpression lines carrying the construct for a single insertion were selected and used for further characterization. RT-PCR analysis showed that the TaSOS1 were expressed in all the transgenic Arabidopsis plants, respectively, but was absent in the wild-type (Figure S5A). We investigated the effects of three TaSOS1 homoeologs overexpression on the morphology and growth of transgenic plants under normal and salt conditions. Germination rates of TaSOS1-overexpressing and wild type Arabidopsis seeds were first compared. In normal conditions, the transgenic lines of 35S:TaSOS1-A, 35S:TaSOS1-B and 35S:TaSOS1-D and wild type plants showed similar germination rates (Figure 6A). In MS medium with 100 mM NaCl, three TaSOS1 homoeologs transgenic lines exhibited higher germination rates than wild type plants (Figure 6A). Notably, the transgenic lines of 35S:TaSOS1-D displayed more salt tolerant phenotype than 35S:TaSOS1-A and 35S:TaSOS1-B plants. The germination rate for 35S:TaSOS1-D lines were nearly 60% in 48h under salt stress treatment, but only 20% for 35S:TaSOS1-A and 41% for 35S:TaSOS1-B lines during the same time period (Figure 6B). More significantly, when sowed in MS medium with 200 mM NaCl, the seed germination of 35S:TaSOS1-D transgenic lines were significantly higher than that of the 35S:TaSOS1-A, 35S:TaSOS1-B and wild type plants, with more than 30% of the seeds of the 35S:TaSOS1-D transgenic lines germinating in the 84h after sowing, while less 6% of the 35S:TaSOS1-A, 35S:TaSOS1-B transgenic lines and wild type seeds germinated at the same treatment (Figures S5B, C). We further determined the seedlings growth of the wild type and the three TaSOS1 homoeologs transgenic plants and found that the growth performance of transgenic lines was similar to that of wild type plants when grown in soil pots in non-saline conditions (Figure 6C). When irrigated with 200 mM NaCl for 15 d, three TaSOS1 homoeologs transgenic lines exhibited a salt tolerant phenotype compared with wild type plants (Figure 6C). We scored the shoot fresh weight and compared the changes between transgenic and wild type plants before and after salt treatment. Overall, NaCl had a lower inhibitory effect on TaSOS1 overexpression lines than it had on the wild type plants. The shoot fresh weight of wild type was 38% of the control weight, while the shoot fresh weight of TaSOS1-B and TaSOS1-D overexpression lines was more than 70% of the control weight (Figure 6D). We also determined the relative electrolyte leakage (REL) of wild type, 35S:TaSOS1-A, 35S:TaSOS1-B and 35S:TaSOS1-D plants after salt stress and found that the REL of the TaSOS1-overexpression lines was significantly lower than that of the wild type (Figure 6E). Since low Na+ content and high K+/Na+ ratio in cells are crucial for salt tolerance, we further compared the Na+, K+ content and K+/Na+ ratio in leaves of three TaSOS1 homoeologs transgenic lines and wild type under salt stress conditions. As shown in Figures 6F-H, generally, the TaSOS1 overexpression lines keep less Na+, more K+ content and higher K+/Na+ ratio than wild type and among them, 35S:TaSOS1-D showed lower Na+ content and higher K+/Na+ than the 35S:TaSOS1-A, 35S:TaSOS1-B transgenic lines. Furthermore, all TaSOS1 homoeologs transgenic lines exhibited a salt tolerant phenotype compared with wild type plants in the reproductive growth stage. Among them, 35S:TaSOS1-B and 35S:TaSOS1-D showed much less growth retardation than the 35S:TaSOS1-A transgenic lines and wild type plants, including fresh weight, silique number and plant height (Figures 7A-C; Figure S5D). Thus, the results demonstrate that three TaSOS1 homoeologous genes display different degree of contribution to salt tolerance at different developmental stages and among of them, TaSOS1-A contributes the least to salt tolerance in all developmental stages, while TaSOS1-D plays the prominent role in germination and seedling stage and both TaSOS1-B and TaSOS1-D exhibit a higher contribution in adult stage under salt stress.
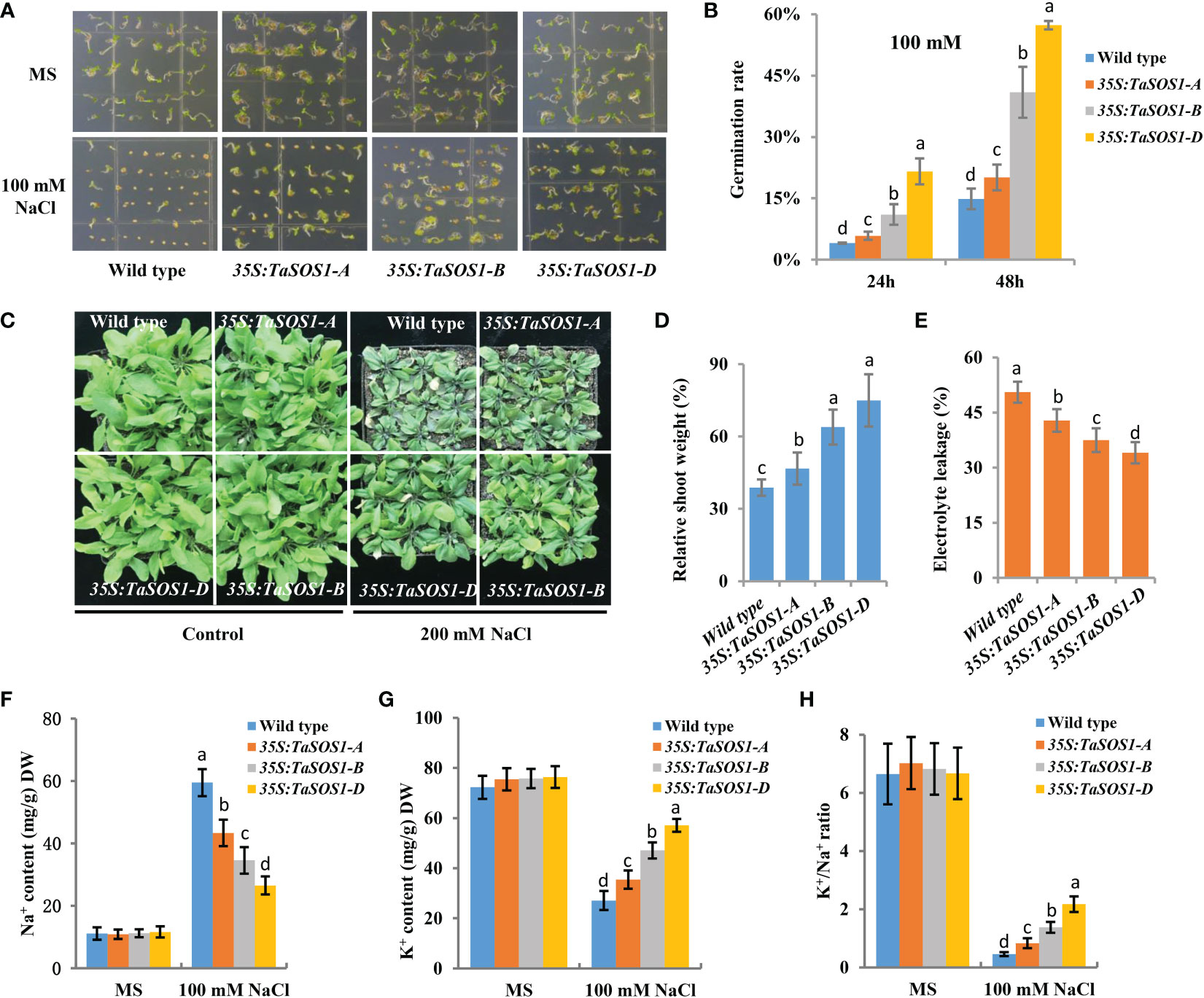
Figure 6 Phenotypic analysis of the 35S:TaSOS1-A, 35S:TaSOS1-B and 35S:TaSOS1-D plants exposed to salt stress. (A, B) Comparison of the seed germination of the wild type, the 35S:TaSOS1-A, 35S:TaSOS1-B and 35S:TaSOS1-D plants. Seeds harvested on the same day were planted on MS medium containing 0, or 100 mM NaCl. Plates were transferred to a growth chamber after stratification at 4°C for 3 d. Photographs were taken, and germination rate was determined 24 h and 48 h after the transfer, respectively. (C, D) Comparison of seedlings growth of the wild type, the 35S:TaSOS1-A, 35S:TaSOS1-B and 35S:TaSOS1-D plants with 200 mM NaCl. Five-day-old seedlings were transferred from MS medium to soil. The plants were then irrigated under well-watered conditions (Control) or irrigated every 5 d with 200 mM NaCl solution; 15 d later, phenotypes were measured and photographed. Shoots fresh weight was measured relative to the same genotype grown on control condition. More than 30 plants were measured for each genotype. (E) Relative electrolyte leakage of leaves from wild type, 35S:TaSOS1-A, 35S:TaSOS1-B and 35S:TaSOS1-D plants after exposure to 200 mM NaCl. (F-H) Comparison of Na+ contents (F), K+ contents (G), and K+ to Na+ ratios (H) of leaves from 35S:TaSOS1-A, 35S:TaSOS1-B and 35S:TaSOS1-D, and wild type plants under salt stress conditions. The values are means ( ± SE) of three biological replicates. The data were analyzed by ANOVA one-way comparison followed by LSD test. Different letters above the bars indicate a significant difference at P < 0.05.
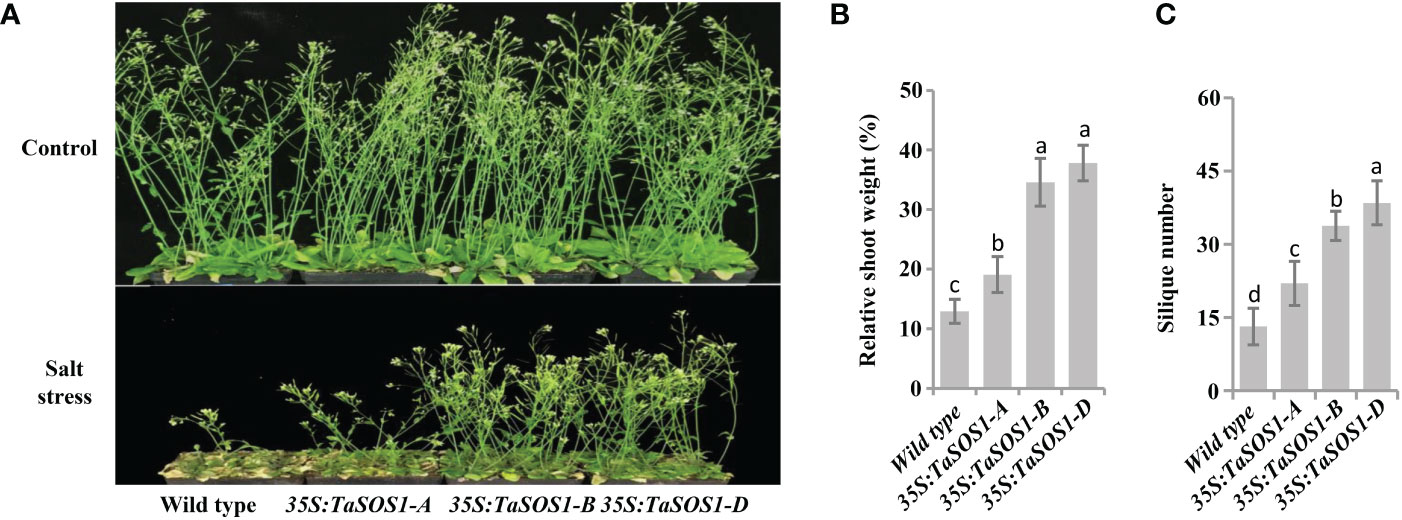
Figure 7 Comparison of 5-week-old of the wild type, the 35S:TaSOS1-A, 35S:TaSOS1-B and 35S:TaSOS1-D plants treated with 200 mM NaCl. (A) Five-day-old seedlings were transferred from MS medium to soil. The plants were then irrigated under well-watered conditions (Control) or irrigated every 5 d with 200 mM NaCl solution; 30 d later, phenotypes were measured and photographed. (B) and (C) Shoots fresh weight and silique number were measured relative to the same genotype grown on control condition. More than 24 plants were measured for each genotype. The values are means ( ± SE) of three biological replicates. The data were analyzed by ANOVA one-way comparison followed by LSD test. Different letters above the bars indicate a significant difference at P < 0.05.
Discussion
The majority of polyploid crops, including wheat, cotton and rape, originated by hybridization between different species. Allopolyploidy results in the convergence of different genomes previously adapted to different environments, thus often exhibit increased vigor and tend to have broader geographic distributions than those of their close diploid relatives (Dubcovsky and Dvorak, 2007; Feldman et al., 2012; Pallotta et al., 2014; Bird et al., 2018). In this study, we determined the phenotypes and measured physiological indexes of two set of synthetic allohexaploid wheat with their parents and 48 natural wheat accessions with different ploidy levels under normal and salt condition. We show that the both the natural and synthetic hexaploid wheat and diploid Ae. tauschii exhibit greater salinity tolerance than tetraploid wheat. This was supported by our observations that for most of the tested phenotype and physiological indexes directly related to salt tolerance, such as growth vigour, relative shoot and root fresh weight, root length, were highly similar between the synthetic hexaploid wheats and diploid parents Ae. tauschii, but substantially different from their tetraploid progenitor species (Figures 3, 4 and Figure S2). These results indicated that tetraploid wheats are relatively sensitive to high salinity and allohexaploid wheats and diploid Ae. tauschii have a higher fitness under salt stress, which was in line with the reports before (Yang et al., 2014; Zheng et al., 2021). The mechanisms behind this trait has always been a topic of great interest to scientists. Previous studies indicated that one of the major contributors behind this difference is that hexaploid wheats maintain a higher K+/Na+ ratios in the leaves (Munns and Tester, 2008; Wang and Xia, 2018), and genetic analysis demonstrated this trait was controlled by Kna1 locus on chromosome 4D (Dvorak et al., 1994; Dubcovsky et al., 1996). Further studies indicated TaHKT1;5-D should be the candidate gene of Kna1 (Huang et al., 2008; Byrt et al., 2014). In the present study, we found there is significant positive correlation between the transcriptional alteration of TaSOS1 and the variation in the salt tolerance of different wheat species, suggesting its important role in the regulation of polyploidy wheat salt tolerance (Figures 3, 4 and Figure S2). Furthermore, both qRT-PCR and histochemical GUS staining results showed that there exist subgenome bias of SOS1 expression in hexaploid wheat. Among of three TaSOS1 homoeologs, TaSOS1-A and TaSOS1-B showed high basal expression in roots and leaves in normal conditions and further up-regulated in roots under salt stress. While TaSOS1-D showed markedly lower expression both in roots and leaves under normal conditions, but significant up-regulated in roots but not leaves under salt stress (Figure 5). It appears to be the rule that allopolyploidization induces rapid and dynamic changes of genome and chromatin structures, as well as reorchestration of homeologous gene expression, which may represent a major advantage of allopolyploidy over its progenitors (Ding and Chen, 2018). Yang et al. found that TaHKT1;5-D exhibited a transcriptional reprogramming from constitutive high basal expression in diploid Ae. tauschii to salt-induced expression in a newly synthetic allohexaploid wheat and natural hexaploid bread wheat (Yang et al., 2014). In this respect, our results are consistent with the previous findings that suggested condition-dependent gene expression reorchestration of the subgenomes might have contributed to the wide-ranging adaptability of natural allopolyploidy plants (Roulin et al., 2013; Powell et al., 2017). It is generally believed the genetic variations in regulatory regions or cis-regulatory elements may lead to gene expression changes that impact phenotype and these variations can be a potential driving force of evolution, creating genetic diversity that can be used for domestication and breeding innovation (Swinnen et al., 2016; Springer et al., 2019). We also found that several types of cis-acting elements were detected only in one or two promoter regions of three TaSOS1 homoeologs (Figure S4). Further research on these three TaSOS1 homoeologs expression divergence, such as identifying the other key element in promoters will be meaningful to reveal the regulation mechanisms. Moreover, a growing body of research suggests that epigenetic modifications contribute to the expression modulation of homoeologs in allopolyploid species (Shitsukawa et al., 2007; Hu et al., 2013; Bird et al., 2018; Ding and Chen, 2018), so it is worth to dissect the possible epigenetic mechanisms of the expression divergence in TaSOS1 homoeologs in wheat.
It has been shown that subgenomes of a given allohexaploid species have redundant but also distinct functions, which may represent a major advantage of allopolyploidy over its diploid progenitors (Shitsukawa et al., 2007; Yang et al., 2014; Uauy, 2017). Notably, we found that three TaSOS1 homoeologous genes showed a differential contribution to salt tolerance at different developmental stages. This was supported by our observations that for most of the measured morphophysiological traits directly related to salt tolerance, including germination rate, growth vigor, osmotic regulation and Na+, K+ content and K+/Na+ ratio, were greater in 35S:TaSOS1-D transgenic lines than that of the 35S:TaSOS1-A, 35S:TaSOS1-B and wild type plants in germination and seedling stage, while both TaSOS1-B and TaSOS1-D exhibit more prominent contribution than TaSOS1-A and wild type plants in adult stage under salt stress condition. Therefore, we speculated the enhancement in salt tolerance of hexaploid wheat maybe partly due to the of TaSOS1-D contribution. This is consistent with the findings of previous study, which demonstrated that to a great extent the salt-tolerant trait of hexaploid wheat can be attributed to the D genome by allopolyploidization (Byrt et al., 2014; Zheng et al., 2021). Nevertheless, in allohexaploid wheat, we found the transcriptional level of TaSOS1-D was suppressed in shoot and mainly induced in root when treated with salt stress. One possible explanation is that the enhanced expression of TaSOS1-D in the root is adequate for extruding Na+ to the external soil environment and provides the first barrier to Na+ uptake in hexaploid wheat (Katschnig et al., 2015; Assaha et al., 2017).
It has been reported that SOS1 encode an over 120-kDa protein with 12 transmembrane domains in the N-terminal part and a long hydrophilic cytoplasmic tail in the C-terminal part (Shi et al., 2000). SOS1 is maintained in resting state through the intra-molecularly interaction of the auto-inhibitory domain with an adjacent domain of SOS1 in the absence of stress (Quintero et al., 2011). Previous study showed that deletion of the auto-inhibitory domains of TdSOS1 conveyed much greater salt tolerance to the transgenic Arabidopsis (Feki et al., 2014). In our study, amino acid sequence alignment showed that both the auto-inhibitory domains and adjacent motif of three TaSOS1 homoeologs were highly conserved (Figure 1B), suggesting these domains maybe not involved in the functional divergence of TaSOS1 homoeologs under salt stress. Research findings in Arabidopsis indicated the activation domain CNBD of SOS1 can interact with its auto-inhibitory domain to keep the transporter in a resting state with basal activity (Shi et al., 2000). The putative CNBDs of the TaSOS1-D is different with TaSOS1-A and TaSOS1-B proteins. There is a leucine (L) at the residue 513 in the TaSOS1-A and TaSOS1-B proteins but an isoleucine (I) in the TaSOS1-D at the same site (Figure 1B). The amino acid difference in the TaSOS1-D protein may create a critical turn in the polypeptide secondary structure and change the interaction between the putative CNBDs and auto-inhibitory domain, thereby alleviating TaSOS1-D auto-inhibition. Moreover, the transmembrane domains in the N-terminal part of SOS1 are critical for SOS1 function in plant salt tolerance. Arabidopsis mutants sos1-3 (R-C), sos1-12 (G-E), which contain single amino acid substitutions in the membrane-spanning region, probably abolish SOS1 antiporter activity (Shi et al., 2000). Protein structure prediction by TMHMM showed that all the three TaSOS1 proteins have the 12 transmembrane domains but there is a single amino acid difference in the first transmembrane domain between TaSOS1-D with TaSOS1-A and TaSOS1-B (Figure 1B). Collectively, we speculated that the amino acid variety maybe have an influence on the antiporter activity, hence TaSOS1-D may have an enhanced capacity in Na+ efflux and salt tolerance in transgenic lines. In addition, there are several amino acid differences at the other sites, such as at the first six amino acids in the N-terminus, but the effects of these differences are not clear, which need further investigation. Moreover, it must be noted that the heterologous expression of TaSOS1 in Arabidopsis may not fully state the function of TaSOS1 in wheat, further studies are required to explore the condition-dependent subfunctionalization of three TaSOS1 homoeologs in wheat and assess specificity as well as generality of the phenomenon in response to abiotic stress. Taken together, our studies suggest that re-orchestration of homeologous genes expression and subfunctionalization of the plasma membrane Na+/H+ transporter TaSOS1s plays a key role of salt tolerance in hexaploid wheat. These results have provided insights into the subgenomic homoeologs variation potential to broad adaptability of natural polyploidy plant in wheat, which might effective for genetic improvement of salinity tolerance in wheat and other crops.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
ZH and QS conceived the original screening and research plans. MZ, JPL, CZ, XL, WC, JCL, FW, WW, WG, MX, YY, HP and ZN performed the experiments and interpreted the data. ZN contributed to materials and methods. MZ and ZH conceived the project and wrote the article with contributions of all the authors. ZH agrees to serve as the author responsible for contact and ensures communication. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the National Key Research and Development Program of China (2021YFD1200603), the Major Program of the National Natural Science Foundation of China (31991210), the National Natural Science Foundation of China (32072001), Frontiers Science Center for Molecular Design Breeding (2022TC145) and the 2115 Talent Development Program of China Agricultural University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1072009/full#supplementary-material
Supplementary Figure 1 | Alignment of genomic and their corresponding cDNA sequences of three TaSOS1 homoeologous genes.
Supplementary Figure 2 | Expression of SOS1 gene was correlated with salt tolerance variation in wheat accessions of different ploidy. (A) Example of aboveground phenotype of natural wheat accessions before and after 200 mM NaCl treatment. A quantitative assessment of shoots fresh weight (B) or roots fresh weight (D) for 48 natural wheat accessions. The y-axis denotes a ratio of NaCl-treated seedlings relative to the same genotype grown on control condition. Mean and SD values were derived from measurements of at least 15 seedlings from three independent assays. Correlation coefficients between the shoots fresh weight ratio (C) or roots fresh weight ratio (E) and the expression levels of TaSOS1 among different ploidy wheat accessions. The x-axis denotes a ratio of NaCl-treated seedlings relative to the same genotype grown on control condition. The y-axis denotes the fold change of TaSOS1 upregulation expression in shoot (C) or root (E) of wheat accessions with different ploidy before and after 200 mM NaCl treatment.
Supplementary Figure 3 | (A) The amplification efficiencies of the gene-specific primers were tested by amplification of wheat genotype Nongda3338 genomic DNA. (B) Schematic of the pTaSOS1-A:uidA, pTaSOS1-B:uidA and pTaSOS1-D:uidA reporters. (C) The pTaSOS1-A:uidA, pTaSOS1-B:uidA and pTaSOS1-D:uidA transgenic lines confirmed by genomic PCR.
Supplementary Figure 4 | The cis-acting regulatory elements analysis for promoter sequences of TaSOS1-A, TaSOS1-B, TaSOS1-D genes from hexaploid wheat genotype Nongda3338 using PlantCARE database. Red boxes: presented both in promoters of TaSOS1-A and TaSOS1-B, but absent in TaSOS1-D. Black boxes: presented specifically in promoter of TaSOS1-A. Blue boxes: presented specifically in promoter of TaSOS1-D.
Supplementary Figure 5 | (A) The 35S:TaSOS1-A, 35S:TaSOS1-B and 35S:TaSOS1-D transgenic lines confirmed by RT-PCR. (B) and (C) Comparison of the seed germination of the wild type, the 35S:TaSOS1-A, 35S:TaSOS1-B and 35S:TaSOS1-D plants on MS medium containing 0, or 200 mM NaCl. Seeds harvested on the same day were planted on MS medium. Plates were transferred to a growth chamber after stratification at 4°C for 3 d. Germination rate was determined every 24 h and photographs were taken at 5 d after the transfer. (D) Comparison the shoots height of 5-week-old of the wild type, the 35S:TaSOS1-A, 35S:TaSOS1-B and 35S:TaSOS1-D plants growth with 200 mM NaCl. Shoots height were measured relative to the same genotype grown on control condition. More than 24 plants were measured for each genotype. The values are means ( ± SE) of three biological replicates. The data were analyzed by ANOVA one-way comparison followed by LSD test. Different letters above the bars indicate a significant difference at P < 0.05.
References
Assaha, D. V. M., Ueda, A., Saneoka, H., AI-Yahyai, R., Yaish, M. W. (2017). The role of na+ and k+ transporters in salt stress adaptation in glycophytes. Front. Physiol. 8, 509. doi: 10.3389/fphys.2017.00509
Bird, K. A., VanBuren, R., Puzey, J. R., Edger, P. P. (2018). The causes and consequences of subgenome dominance in hybrids and recent polyploids. New Phytol. 220 (1), 87–93. doi: 10.1111/nph.15256
Borrill, P., Adamski, N., Uauy, C. (2015). Genomics as the key to unlocking the polyploid potential of wheat. New Phytol. 208 (4), 1008–1022. doi: 10.1111/nph.13533
Byrt, C. S., Xu, B., Krishnan, M., Lightfoot, D. J., Athman, A., Jacobs, A. K., et al. (2014). The na+ transporter, TaHKT1;5-d, limits shoot na+ accumulation in bread wheat. Plant J. 80 (3), 516–526. doi: 10.1111/tpj.12651
Che, B., Cheng, C., Fang, J., Liu, Y., Jiang, L., Yu, B. (2019). The recretohalophyte tamarix TrSOS1 gene confers enhanced salt tolerance to transgenic hairy root composite cotton seedlings exhibiting virus-induced gene silencing of GhSOS1. Int. J. Mol. Sci. 20 (12), 2930. doi: 10.3390/ijms20122930
Chung, J. S., Zhu, J. K., Bressan, R. A., Hasegawa, P. M., Shi, H. (2008). Reactive oxygen species mediate na+-induced SOS1 mRNA stability in arabidopsis. Plant J. 53 (3), 554–565. doi: 10.1111/j.1365-313X.2007.03364.x
Ding, M., Chen, Z. J. (2018). Epigenetic perspectives on the evolution and domestication of polyploid plant and crops. Curr. Opin. Plant Biol. 42, 37–48. doi: 10.1016/j.pbi.2018.02.003
Dubcovsky, J., Dvorak, J. (2007). Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316 (5833), 1862–1866. doi: 10.1126/science.1143986
Dubcovsky, J., María, G. S., Epstein, E., Luo, M. C., Dvořák, J. (1996). Mapping of the K+/Na+ discrimination locus Kna1 in wheat. Theor. Appl. Genet. 92 (3-4), 448–454. doi: 10.1007/BF00223692
Du, W., Lin, H., Chen, S., Wu, Y., Zhang, J., Fuglsang, A. T., et al. (2011). Phosphorylation of SOS3-like calcium-binding proteins by their interacting SOS2-like protein kinases is a common regulatory mechanism in arabidopsis. Plant Physiol. 156, 2235–2243. doi: 10.1104/pp.111.173377
Dvorak, J., Noaman, M. M., Goyal, S., Gorham, J. (1994). Enhancement of the salt tolerance of triticum turgidum l. by the Kna1 locus transferred from the triticum aestivum l. chromosome 4D by homoeologous recombination. Theor. Appl. Genet. 87, 872–877. doi: 10.1007/BF00221141
Feki, K., Brini, F., Ben Amar, S., Saibi, W., Masmoudi, K. (2015). Comparative functional analysis of two wheat Na+/H+ antiporter SOS1 promoters in arabidopsis thaliana under various stress conditions. J. Appl. Genet. 56 (1), 15–26. doi: 10.1007/s13353-014-0228-7
Feki, K., Quintero, F. J., Khoudi, H., Leidi, E. O., Masmoudi, K., Pardo, J. M., et al. (2014). A constitutively active form of a durum wheat Na+/H+ antiporter SOS1 confers high salt tolerance to transgenic arabidopsis. Plant Cell Rep. 33 (2), 277–288. doi: 10.1007/s00299-013-1528-9
Feldman, M., Levy, A. A. (2005). Allopolyploidy–a shaping force in the evolution of wheat genomes. Cytogenet. Genome Res. 109 (1-3), 250–258. doi: 10.1159/000082407
Feldman, M., Levy, A. A., Fahima, T., Korol, A. (2012). Genomic asymmetry in allopolyploid plants: Wheat as a model. J. Exp. Bot. 63 (14), 5045–5059. doi: 10.1093/jxb/ers192
Goncharov, N. P. (2011). Genus triticum l. taxonomy: The present and the future. Plant Syst. Evol. 295, 1–11. doi: 10.1007/s00606-011-0480-9
Gorham, J., Hardy, C., Wyn Jones, R. G., Joppa, L. R., Law, C. N. (1987). Chromosomal location of a K/Na discrimination character in the d genome of wheat. Theor. Appl. Genet. 74 (5), 584–588. doi: 10.1007/BF00288856
Guo, Y., Qiu, Q. S., Quintero, F. J., Pardo, J. M., Ohta, M., Zhang, C., et al. (2004). Transgenic evaluation of activated mutant alleles of SOS2 reveals a critical requirement for its kinase activity and c-terminal regulatory domain for salt tolerance in Arabidopsis thaliana. Plant Cell 16, 435–449. doi: 10.1105/tpc.019174
Hegarty, M., Coate, J., Sherman-Broyles, S., Abbott, R., Hiscock, S., Doyle, J. (2013). Lessons from natural and artificial polyploids in higher plants. Cytogenet. Genome Res. 140 (2-4), 204–225. doi: 10.1159/000353361
Huang, S., Spielmeyer, W., Lagudah, E. S., James, R. A., Platten, J. D., Dennis, E. S., et al. (2006). A sodium transporter (HKT7) is a candidate for Nax1, a gene for salt tolerance in durum wheat. Plant Physiol. 142, 1718–1727. doi: 10.1104/pp.106.088864
Huang, S., Spielmeyer, W., Lagudah, E. S., Munns, R. (2008). Comparative mapping of HKT genes in wheat, barley and rice, key determinants of na+ transport and salt tolerance. J. Exp. Bot. 59, 927–937. doi: 10.1093/jxb/ern033
Hu, Z., Han, Z., Song, N., Chai, L., Yao, Y., Peng, H., et al. (2013). Epigenetic modification contributes to the expression divergence of three TaEXPA1 homoeologs in hexaploid wheat (Triticum aestivum). New Phytol. 197 (4), 1344–1352. doi: 10.1111/nph.12131
IWGSC, Appels, R., Eversole, K., Feuillet, C., Keller, B., Rogers, J, et al. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, 6403. doi: 10.1126/science.aar7191
James, R. A., Davenport, R. J., Munns, R. (2006). Physiological characterization of two genes for na+ exclusion in durum wheat, Nax1 and Nax2. Plant Physiol. 142 (4), 1537–1547. doi: 10.1104/pp.106.086538
Jiao, Y., Wickett, N. J., Ayyampalayam, S., Chanderbali, A. S., Landherr, L., Ralph, P. E., et al. (2011). Ancestral polyploidy in seed plants and angiosperms. Nature 473 (7345), 97–100. doi: 10.1038/nature09916
Ji, H., Pardo, J. M., Batelli, G., Van Oosten, M. J., Bressan, R. A., Li, X. (2013). The salt overly sensitive (SOS) pathway: Established and emerging roles. Mol. Plant 6 (2), 275–286. doi: 10.1093/mp/sst017
Katschnig, D., Bliek, T., Rozema, J., Schat, H. (2015). Constitutive high-level SOS1 expression and absence of HKT1;1 expression in the salt-accumulating halophyte salicornia dolichostachya. Plant Sci. 234, 144–154. doi: 10.1016/j.plantsci.2015.02.011
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30 (1), 325–327. doi: 10.1093/nar/30.1.325
Li, A., Liu, D., Wu, J., Zhao, X., Hao, M., Geng, S., et al. (2014). mRNA and small RNA transcriptomes reveal insights into dynamic homoeolog regulation of allopolyploid heterosis in nascent hexaploid wheat. Plant Cell 26 (5), 1878–1900. doi: 10.1105/tpc.114.124388
Madlung, A. (2013). Polyploidy and its effect on evolutionary success: Old questions revisited with new tools. Hered. (Edinb) 110 (2), 99–104. doi: 10.1038/hdy.2012.79
Mahi, H., Pérez-Hormaeche, J., De Luca, A., Villalta, I., Espartero, J., Gámez-Arjona, F., et al. (2019). A critical role of sodium flux via the plasma membrane Na+/H+ exchanger SOS1 in the salt tolerance of rice. Plant Physiol. 180 (2), 1046–1065. doi: 10.1104/pp.19.00324
Munns, R., Gilliham, M. (2015). Salinity tolerance of crops - what is the cost? New Phytol. 208 (3), 668–673. doi: 10.1111/nph.13519
Munns, R., James, R. A., Xu, B., Athman, A., Conn, S. J., Jordans, C., et al. (2012). Wheat grain yield on saline soils is improved by an ancestral na+ transporter gene. Nat. Biotechnol. 30 (4), 360–364. doi: 10.1038/nbt.2120
Munns, R., Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Olías, R., Eljakaoui, Z., Li, J., De Morales, P. A. Z. A., Marín-Manzano, M. C., Pardo, J. M., et al. (2009). The plasmamembrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of na+ between plant organs. Plant Cell Environ. 32, 904–916. doi: 10.1111/j.1365-3040.2009.01971.x
Otto, S. P. (2007). The evolutionary consequences of polyploidy. Cell 131 (3), 452–462. doi: 10.1016/j.cell.2007.10.022
Pallotta, M., Schnurbusch, T., Hayes, J., Hay, A., Baumann, U., Paull, J., et al. (2014). Molecular basis of adaptation to high soil boron in wheat landraces and elite cultivars. Nature 514 (7520), 88–91. doi: 10.1038/nature13538
Powell, J. J., Fitzgerald, T. L., Stiller, J., Berkman, P. J., Gardiner, D. M., Manners, J. M., et al. (2017). The defence-associated transcriptome of hexaploid wheat displays homoeolog expression and induction bias. Plant Biotechnol. J. 15 (4), 533–543. doi: 10.1111/pbi.12651
Qiu, Q. S., Guo, Y., Dietrich, M. A., Schumaker, K. S., Zhu, J. K. (2002). Regulation of SOS1, a plasmamembrane Na+/H+ exchanger in arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. U.S.A. 99, 8436. doi: 10.1073/pnas.122224699
Quintero, F. J., Martinez-Atienza, J., Villalta, I., Jiang, X., Kim, W. Y., Ali, Z., et al. (2011). Activation of the plasma membrane Na/H antiporter salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory c-terminal domain. Proc. Natl. Acad. Sci. U.S.A. 108 (6), 2611–2616. doi: 10.1073/pnas.1018921108
Ren, Z. H., Gao, J. P., Li, L. G., Cai, X. L., Huang, W., Chao, D. Y., et al. (2005). A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 37, 1141–1146. doi: 10.1038/ng1643
Roulin, A., Auer, P. L., Libault, M., Schlueter, J., Farmer, A., May, G., et al. (2013). The fate of duplicated genes in a polyploid plant genome. Plant J. 73 (1), 143–153. doi: 10.1111/tpj.12026
Shi, H., Ishitani, M., Kim, C., Zhu, J. K. (2000). The arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. U.S.A. 97, 6896–6901. doi: 10.1073/pnas.120170197
Shi, H., Quintero, F. J., Pardo, J. M., Zhu, J. K. (2002). The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance na+ transport in plants. Plant Cell 14 (2), 465–477. doi: 10.1105/tpc.010371
Shitsukawa, N., Tahira, C., Kassai, K., Hirabayashi, C., Shimizu, T., Takumi, S., et al. (2007). Genetic and epigenetic alteration among three homoeologous genes of a class e MADS box gene in hexaploid wheat. Plant Cell 19 (6), 1723–1737. doi: 10.1105/tpc.107.051813
Springer, N., de León, N., Grotewold, E. (2019). Challenges of translating gene regulatory information into agronomic improvements. Trends Plant Sci. 24 (12), 1075–1082. doi: 10.1016/j.tplants.2019.07.004
Swinnen, G., Goossens, A., Pauwels, L. (2016). Lessons from domestication: Targeting cis-regulatory elements for crop improvement. Trends Plant Sci. 21 (6), 506–515. doi: 10.1016/j.tplants.2016.01.014
Uauy, C. (2017). Plant genomics: Unlocking the genome of wheat's progenitor. Curr. Biol. 27 (20), R1122–R1124. doi: 10.1016/j.cub.2017.08.051
Wang, H., Hu, Z., Huang, K., Han, Y., Zhao, A., Han, H., et al. (2018). Three genomes differentially contribute to the seedling lateral root number in allohexaploid wheat: evidence from phenotype evolution and gene expression. Plant J. 95 (6), 976–987. doi: 10.1111/tpj.14005
Wang, M., Xia, G. (2018). The landscape of molecular mechanisms for salt tolerance in wheat. Crop J. 6, 42–47. doi: 10.1016/j.cj.2017.09.002
Wu, S. J., Ding, L., Zhu, J. K. (1996). SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8 (4), 617–627. doi: 10.1105/tpc.8.4.617
Wu, S., Han, B., Jiao, Y. (2020). Genetic contribution of paleopolyploidy to adaptive evolution in angiosperms. Mol. Plant 13 (1), 59–71. doi: 10.1016/j.molp.2019.10.012
Xu, H., Jiang, X., Zhan, K., Cheng, X., Chen, X., Pardo, J. M., et al. (2008). Functional characterization of a wheat plasma membrane Na+/H+ antiporter in yeast. Arch. Biochem. Biophys. 473 (1), 8–15. doi: 10.1016/j.abb.2008.02.018
Yang, C., Yang, Z., Zhao, L., Sun, F., Liu, B. (2018). A newly formed hexaploid wheat exhibits immediate higher tolerance to nitrogen-deficiency than its parental lines. BMC Plant Biol. 18 (1), 113. doi: 10.1186/s12870-018-1334-1
Yang, C., Zhao, L., Zhang, H., Yang, Z., Wang, H., Wen, S., et al. (2014). Evolution of physiological responses to salt stress in hexaploid wheat. Proc. Natl. Acad. Sci. U.S.A. 111 (32), 11882–11887. doi: 10.1073/pnas.1412839111
Zheng, M., Lin, J., Liu, X., Chu, W., Li, J., Gao, Y., et al. (2021). Histone acetyltransferase TaHAG1 acts as a crucial regulator to strengthen salt tolerance of hexaploid wheat. Plant Physiol. 186 (4), 1951–1969. doi: 10.1093/plphys/kiab187
Zhou, Y., Lai, Z., Yin, X., Yu, S., Xu, Y., Wang, X., et al. (2016). Hyperactive mutant of a wheat plasma membrane Na+/H+ antiporter improves the growth and salt tolerance of transgenic tobacco. Plant Sci. 253, 176–186. doi: 10.1016/j.plantsci.2016.09.016
Keywords: adaptability, polyploid, wheat, salt tolerace, homoeolog, expression divergence
Citation: Zheng M, Li J, Zeng C, Liu X, Chu W, Lin J, Wang F, Wang W, Guo W, Xin M, Yao Y, Peng H, Ni Z, Sun Q and Hu Z (2022) Subgenome-biased expression and functional diversification of a Na+/H+ antiporter homoeologs in salt tolerance of polyploid wheat. Front. Plant Sci. 13:1072009. doi: 10.3389/fpls.2022.1072009
Received: 17 October 2022; Accepted: 21 November 2022;
Published: 07 December 2022.
Edited by:
Teruaki Taji, Tokyo University of Agriculture, JapanCopyright © 2022 Zheng, Li, Zeng, Liu, Chu, Lin, Wang, Wang, Guo, Xin, Yao, Peng, Ni, Sun and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhaorong Hu, enJodUBjYXUuZWR1LmNu
†These authors have contributed equally to this work
 Mei Zheng1†
Mei Zheng1† Jingchen Lin
Jingchen Lin Weilong Guo
Weilong Guo Mingming Xin
Mingming Xin Yingyin Yao
Yingyin Yao Zhongfu Ni
Zhongfu Ni Qixin Sun
Qixin Sun Zhaorong Hu
Zhaorong Hu