- 1Citrus Research Institute, Southwest University/Chinese Academy of Agricultural Sciences, Chongqing, China
- 2National Citrus Engineering Research Center, Chongqing, China
Morphogenesis of root is a vital factor to determine the root system architecture. Cytokinin response regulators (RRs) are the key transcription factors in cytokinin signaling, which play important roles in regulating the root morphogenesis. In this study, 29 RR proteins, including 21 RRs and 8 pseudo RRs, were identified from the genome of citrus, and termed as CcRR1-21 and CcPRR1-8, respectively. Phylogenetic analysis revealed that the 29 CcRRs could be classified into four types according to their representative domains. Analysis of cis-elements of CcRRs indicated that they were possibly involved in the regulation of growth and abiotic stress resistance in citrus. Within the type A and type B CcRRs, CcRR4, CcRR5, CcRR6 and CcRR16 highly expressed in roots and leaves, and dramatically responded to the treatments of hormones and abiotic stresses. CcRR2, CcRR10, CcRR14 and CcRR19 also highly expressed in roots under different treatments. Characteristic analysis revealed that the above 8 CcRRs significantly and differentially expressed in the three zones of root, suggesting their functional differences in regulating root growth and development. Further investigation of the 3 highly and differentially expressed CcRRs, CcRR5, CcRR10 and CcRR14, in 9 citrus rootstocks showed that the expression of CcRR5, CcRR10 and CcRR14 was significantly correlated to the length of primary root, the number of lateral roots, and both primary root and the number of lateral roots, respectively. Results of this study indicated that CcRRs were involved in regulating the growth and development of the root in citrus with different functions among the members.
Introduction
Root system architecture is a critical characteristic to determine the distribution of roots in soil, the capacity of absorption, anchorage and stress resistance (Lavenus et al., 2013; Singh et al., 2020; Jeon et al., 2021). Formation of root system architecture is a complex biological process regulated by multiple genes, of which the expression greatly affected by environmental conditions. Plant root can sense signals of any environmental changes and modify its primary root length and lateral root density through cell proliferation and cell differentiation to adapt to the changes of environmental factors (Street et al., 2016; Liu and Wirén, 2022). Phytohormones such as auxin, cytokinin, abscisic acid etc., play essential roles in root morphogenesis and growth (de Bang et al., 2020; Pandey et al., 2021; Deng et al., 2022; Ji et al., 2022; Li C. et al., 2022; Timilsina et al., 2022).
Cytokinin, a plant-specific chemical messenger, plays an important role in many aspects of plant growth and development, such as cell division and differentiation of the root meristem, vascular development, stress response and plant immunity (Choi et al., 2010; Ni et al., 2017; Li L. et al., 2022; Papon and Courdavault, 2022). It acts as a potent negative regulator of root formation and elongation, including primary root, lateral root and crown root (Li L. et al., 2022). The cytokinin signaling system is a two-component system, mainly consisting of three important parts: receptor protein histidine kinase (HK), histidine-phosphotransfer protein (HP), and response regulator (RR) (Muller and Sheen, 2007). Among them, RRs play crucial roles in cytokinin signal transduction.
In Arabidopsis, the RR family contains 24 members, which can be divided into four types, type A, type B, type C and pseudo ARR (Makino et al., 2000; Horák et al., 2008; Schaller et al., 2008). Type A ARRs have a conserved aspartic acid-aspartic acid-lysine (D-D-K) region and a short C-terminal with unknown functions, and their expression could be induced by exogenous cytokinin, which occurs in the absence of de novo protein synthesis (To et al., 2004). In addition to the D-D-K domain, type B ARRs contain a long C-terminal sequence encoding a MYB-like DNA binding domain, called ARRM, which is the binding site to downstream target genes for cytokinin signaling (Ramireddy et al., 2013; Sakai et al., 2001). Type B ARRs serve as transcriptional regulators for certain target genes such as the type A ARR genes (Tajima et al., 2004). Type C ARRs have a domain similar to that of the type A ARRs in structure. However, their expression could not be induced by cytokinin (Yang et al., 2016). Pseudo ARRs lack conserved residues for phosphorylation and some of them contain a CCT (for CONSTANS, CONSTANS-like and TOC1) motif in C-termini (Strayer et al., 2000; To et al., 2007; Li et al., 2017).
RR family members were reported to be involved in root development. Previous study showed that ARR1 and ARR12 could bind to the promoter of LAX2, an auxin influx carrier, and inhibited its expression, thus regulating the distribution of auxin in the root apical meristem (Zhang et al., 2013). ARR1 mediated arsenate-induced root growth inhibition by directly binding to the promoters of ASA1 (ANTHRANILATE SYNTHASE ALPHA SUBUNIT) and ASB1 (BETA SUBUNIT 1), and positively regulating their expressions (Tu et al., 2021). Similarly, ARR2 and ARR12 could affect the formation of lateral root primordia by directly and negatively regulating the localization of PIN-FORMED (PIN) proteins, the key factors in regulating cell division and differentiation of root meristem by maintaining adequate auxin concentration (Hwang et al., 2012). The up-regulated ARR16 and ARR17 at the lower water potential side of root were involved in root hydrotropism through promoting the division of cells in the meristem zone (Chang et al., 2019). RcRR1, a Rosa canina type A response regulator gene, homology to ARR8 and ARR9, was reported to be involved in cytokinin-modulated rhizoid organogenesis, and overexpression of RcRR1 resulted in increased primary root length and lateral root density (Gao et al., 2013). Citrus is the most important fruit plant worldwide. However, little attention has been paid to RRs in citrus research. Previous study in our group indicated that RRs probably play essential roles in citrus rootstock roots by RNA-seq analysis (unpublished data).
In this study, the RR genes were comprehensively identified and characterized in the whole citrus genome. The expression profiles of type A and type B CcRRs were analyzed in different plant tissues and organs, under different hormones and stresses treatments as well. Expressions of CcRRs in different zones of root and varieties were also analyzed to mine the functional genes related to root morphogenesis in citrus rootstocks. It is a crucial basis for further study on the regulation of RR genes in citrus root morphogenesis, and provides guidance for genetic modification of rootstocks to solve the issues faced in citriculture.
Materials and methods
Plant materials
The nine citrus rootstock varieties, including Poncirus trifoliata (Pt026, Pt030, Pt034, Pt038), Citrus wilsonii Tanaka ‘Zhique’, C. limonia Pasquale ‘Volkamer’, C. limonia Osbeck ‘Canton Lemon’, C. reshni Hort. Ex Tan ‘Cleopetra’, and C. reticulata ‘Zhuju’, were used in this study. The plant materials were collected from National Citrus Germplasm Repository (Chongqing, China).
The seedlings of ‘Volkamer’ were used for analysis of genes expression in different tissues and organs, and under different hormone treatments and stresses. The seedlings of all varieties were used to study the correlation between root indexes and gene expression.
Genome-wide identification of RR gene family in citrus
The genome and protein information of citrus plant (C. clementina) was downloaded from the Phytozome database (http://phytozome.jgi.doe.gov/pz/portal.html). Twenty-four response regulators’ protein sequences of Arabidopsis thaliana (http://www.arabidopsis.org/) (ARRs) were used as reference sequences to identify the members of the RR family in C. clementina (CcRRs) with protein basic local alignment search tool (BLASTP) (Mahram and Herbordt, 2015). The top E-value was less than 1 × 10−10. All the retrieved sequences were submitted to the HMMER database (https://www.ebi.ac.uk/Tools/hmmer/; Potter et al., 2018), Pfam database (http://pfam.xfam.org/; Punta et al., 2011) and NCBI CDD database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi; Marchler-Bauer et al., 2015) to confirm whether they were the members of RR family. The redundant members were removed manually. ExPASy website (http://web.expasy.org/) was used to predict molecular weight (MW) and isoelectric points (pI) of all proteins of the identified RR family members. CELLO website (http://cello.life.nctu.edu.tw/) was used to predict the subcellular localization of RR family members (Yu et al., 2006).
Chromosomal distribution
The chromosomal location of each RR gene was derived from the genome annotation information. Chromosomal distributions of these genes were generated using MapChart software (Voorrips, 2002).
Phylogenetic analysis
MEGA7.0 software (Kumar et al., 2016) was used to construct the phylogenetic tree with protein sequences of RRs from different plant species based on the Maximum Likelihood Method (MJ). The bootstrap value was repeated 1000 times.
Gene structure, motif and promoter analysis
TBtools (Chen et al., 2020) was used to display the distribution of gene structure of RRs. The analysis of the protein conserved motif was conducted through the online MEME website (https://meme-suite.org/meme/tools/meme; Bailey and Elkan, 1995) to study the differences among RR family members. The maximum number of motifs was set to 5. 2000 bp upstream sequence of each RR gene was extracted from the citrus genome. The cis-elements in the promoter region were predicted using PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002).
Treatments
After removal of seed coats, the embryos were cultured in petri dish at 28°C under moisturized conditions for 3-5 days for germination. The seedlings of Volkamer were grown in hydroponics for 25 days. The roots, stems and leaves were collected for gene expression analysis of CcRRs. Meanwhile, the remaining seedlings were treated with hormones and abiotic stresses. The root samples were collected at different time (0, 1, 3, 6, 12 h) after 50 μM 6-BA, 100 μM ABA, and 20% polyethylene glycol (PEG6000) treatments for RNA extraction, respectively. The roots were also sampled from the seedlings treated at 4°C and 200 mM NaCl for 0, 3, 6, 12 and 24 h, respectively. In addition, the seedlings with lateral root primordium just emerged (as shown in Supplementary Figure 1) were used to explore the expression of type A and type B RRs in different zones of root (RT: the meristematic/elongation zone; RM: the root elongation/differentiation and lateral root initiation zone; RC: lateral root growth zone). This division was based on the reports of Hwang et al. (2012), Goh (2019), Santos Teixeira and ten Tusscher (2019).
The seeds of nine citrus rootstocks under same state were selected and sown in the pots (30 × 30 cm) filled with the mix of vermiculite and perlite. Different zones of root were sampled at 25, 50 and 75 days after sowing. The samples were immediately frozen with liquid nitrogen and stored at -80°C for RNA extraction to explore the expression of the three RR genes. At the same time, the length of primary root and the number of lateral roots in RC zone of the root were counted. All the seedlings were grown at 28°C in a greenhouse under 16 h day length.
RNA extraction and qRT-PCR
Total RNA was isolated from samples using an EASYspin Plus Plant RNA Rapid Extraction Kit RN38 (Aidlab, Beijing, China) following the manufacturer’s instructions. Subsequently, the RNA was reverse transcribed using the Thermo Scientific RevertAid MM (Thermo Fisher, Lithuania). qRT-PCR was performed with 10 μL reaction mixture containing 2 μL cDNA sample, 5 μL universal SYBR® Green Supermix (2 × iTaq™, Bio-Rad, CA, USA), 0.2 μL of each primer (10 μM) and 2.6 μL sterilized water. The qRT-PCR program was as follows: 95°C for 60 s, followed by 39 cycles of 95°C for 20 s, 60°C for 30s. The primers used in this study are listed in Supplementary Table 1. Each treatment included three replications. The expression levels of the genes were normalized with citrus Actin gene, and fold changes were calculated using 2-ΔΔCt method (Zhang et al., 2020).
Results
Identification of CcRR family members in citrus
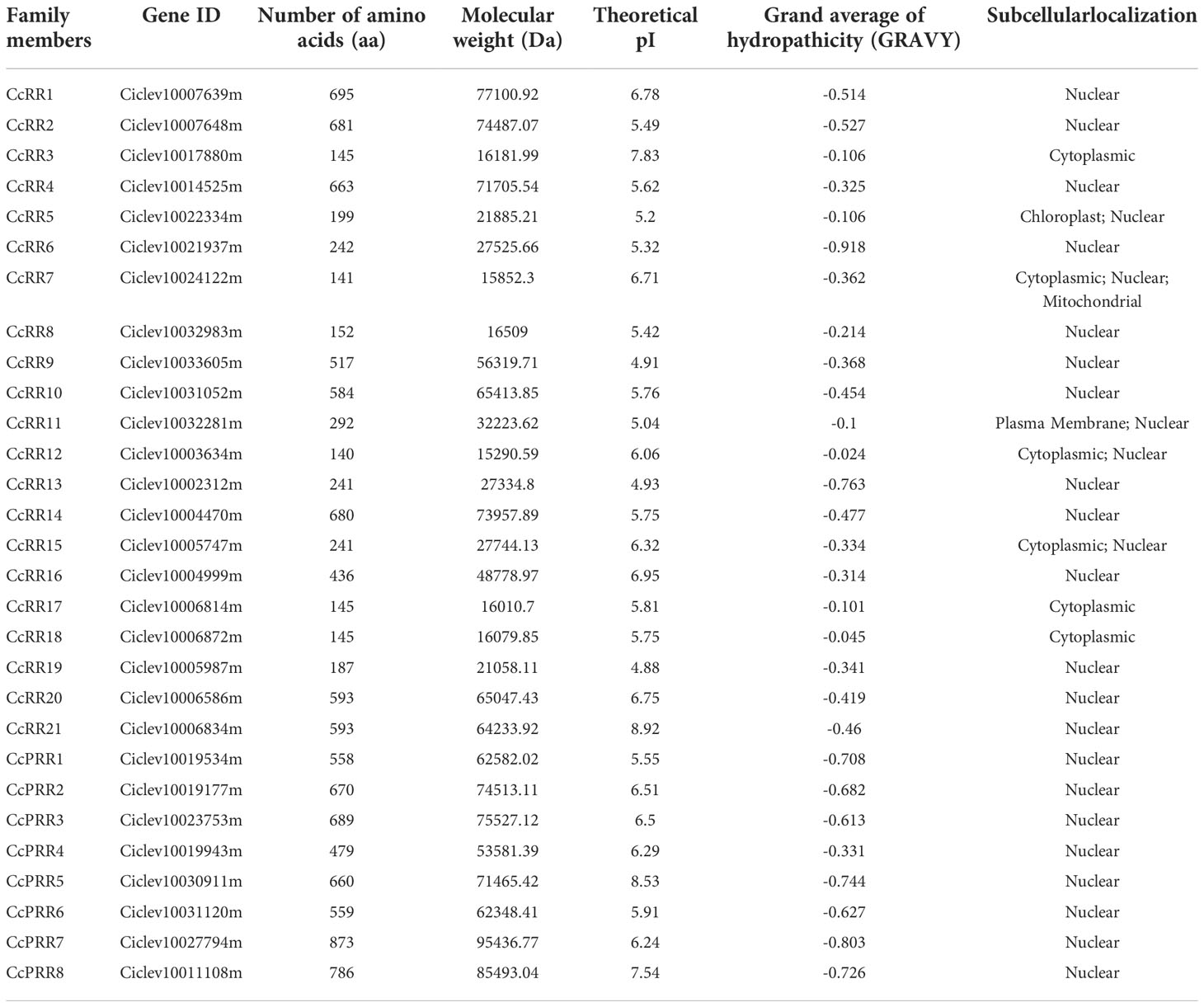
In this study, more than 100 RRs and RR like proteins were searched out from the C. clementina genome. The RRs with sequence redundancy or alternative splicing forms were removed. Furthermore, the RRs containing other domains, or lacking conserved motifs were also eliminated. Finally, a total of 29 potential RR proteins (including 8 pseudo RRs) were identified, and named as CcRR1-21 and CcPRR1-8 according to their positions on the scaffolds. The details of CcRRs, including their names, accession numbers, number of amino acids, molecular weight, isoelectric point (pI), grand average of hydropathicity (GRAVY) and putative subcellular localization were listed in Table 1. The number of amino acids of CcRR proteins varied from 140 (CcRR12) to 873 (CcPRR7), with molecular weight (MW) from 15.29 (CcRR12) to 95.44 kDa (CcPRR7) and pI from 4.88 (CcRR19) to 8.92 (CcRR21). The prediction of subcellular localization showed that most of CcRRs were located in the nucleus, and a few of them were located in the plasma membrane, mitochondria or chloroplast.
Phylogenetic analysis and chromosomal distribution of CcRRs
To uncover the phylogenetic relationship of CcRRs, a phylogenetic tree was generated with 95 RR proteins, of which 33 were from Arabidopsis (24 ARR and 9 APRR), 33 from poplars (22 PtRR and 11 PtPRR) (Ramírez-Carvajal et al., 2008), and 29 from citrus (21 CcRR and 8 CcPRR) (Figure 1). It showed that the 95 RRs were divided into 9 clusters. Based on the feature of protein structure, the 9 clusters could be classified into 4 types. The group in green represented type A CcRRs with only one D-D-K domain, including CcRR5-6, CcRR8, CcRR11-13 and CcRR19. Type C CcRR (pink) was mostly close to type A with a similar D-D-K domain. It is a small group with only four members in citrus (CcRR3, CcRR7 and CcRR17-18). CcRR1-2, CcRR4, CcRR9-10, CcRR14-16 and CcRR20-21 were placed in type B CcRRs (blue) with D-D-K domain and MYB-like DNA-binding domain. The group marked in orange color contained pseudo RRs with D-D-K domain and CCT domain. Chromosome mapping results showed that most of CcRR members were clustered on scaffold 3, 4 and 9, and a few of them were on scaffold 1, 2, 5, 7 and 8 (Figure 2).
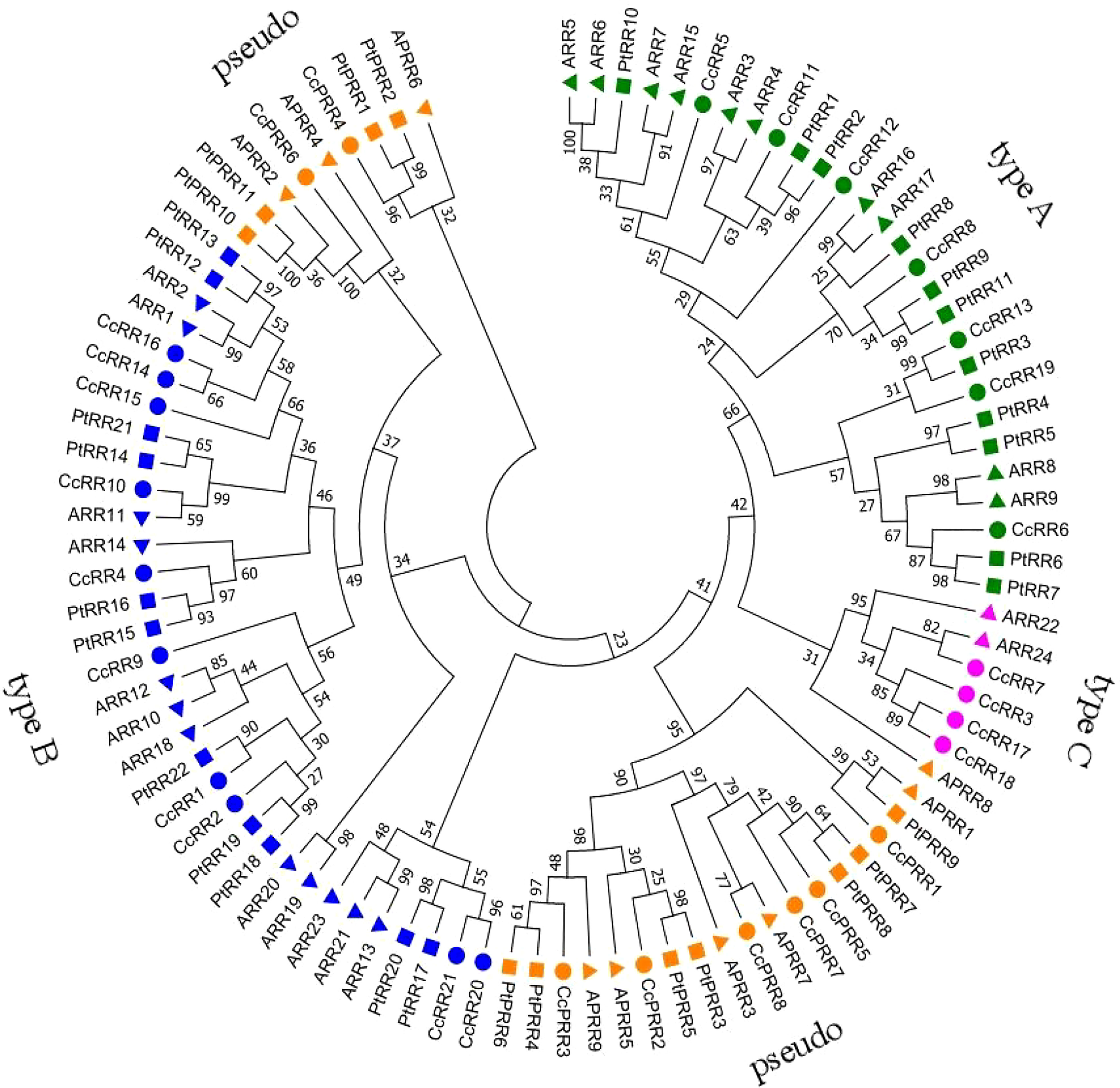
Figure 1 Phylogenetic tree of RR proteins from Citrus clementina, Arabidopsis thaliana and Populus trichocarpa. Groups with different colors represent different types of RRs (Green: type A RRs, with only one D-D-K domain; Blue: type B RRs, with D-D-K domain and MYB-like DNA-binding domain; Pink: type C RRs, with a D-D-K domain similar to type-A CcRRs; Orange: pseudo RRs, with D-D-K domain and CCT domain, except for CcPRR4 and CcPRR6).
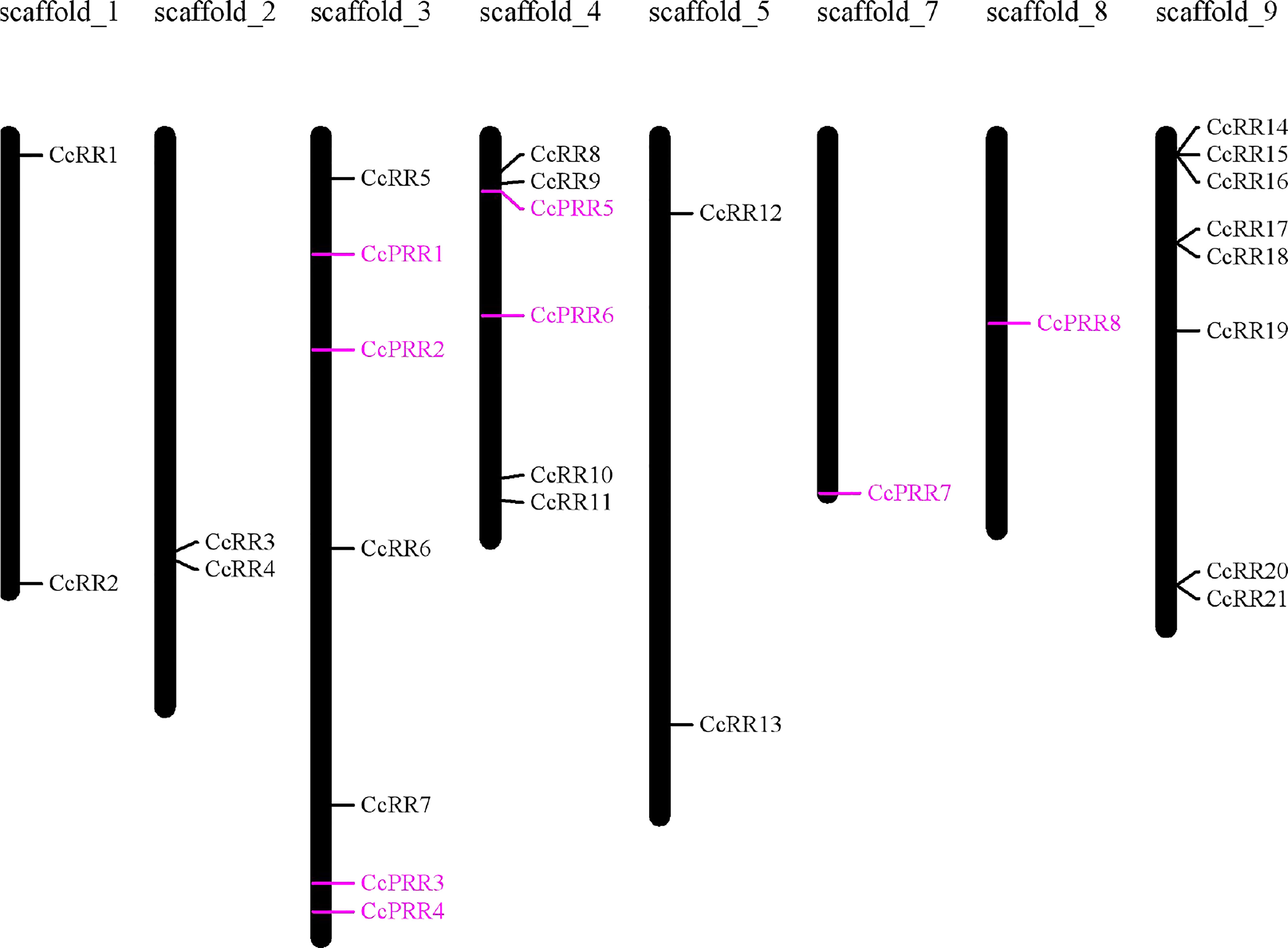
Figure 2 Chromosome location of CcRR gene family members on the C. clementina scaffolds. Pseudo RRs were indicated in purple colour.
Gene structure and motifs of CcRRs
The diversity of exon-intron structure and motifs may reveal the evolution of gene family (Wang et al., 2017; Sun et al., 2022). Intron-exon analysis of CcRR genes (Figure 3) indicated that the CcRRs classified into type A and B possessed 3 to 7 intronic regions, while there was only one intron in the type C CcRRs. The numbers of introns for CcPRRs varied more, ranging from 5 to 10. Five potential conserved motifs were identified from 29 CcRRs by online MEME program. The details of each motif were shown in Supplementary Figure 2. As shown in Figure 3, motifs 1, 2 and 4 encoding the D-D-K domain presented in most of CcRRs, but the sequences of D-D-K domain in some of CcRRs were incomplete. CcRR12 of type A, CcRR9 and CcRR21 of type B, CcPRR5 and CcPRR6 of pseudo RRs, and all type C CcRRs were missing part of the D-D-K domain sequence. All the type B CcRRs and CcPRR8 also possessed motif 3, which was identified as a conserved MYB-binding domain. Motif 5 was deemed as a conserved CCT motif and it existed in almost all CcPRRs except CcPRR4 and CcPRR6. These results suggested that the four types of CcRRs may play different roles in citrus.
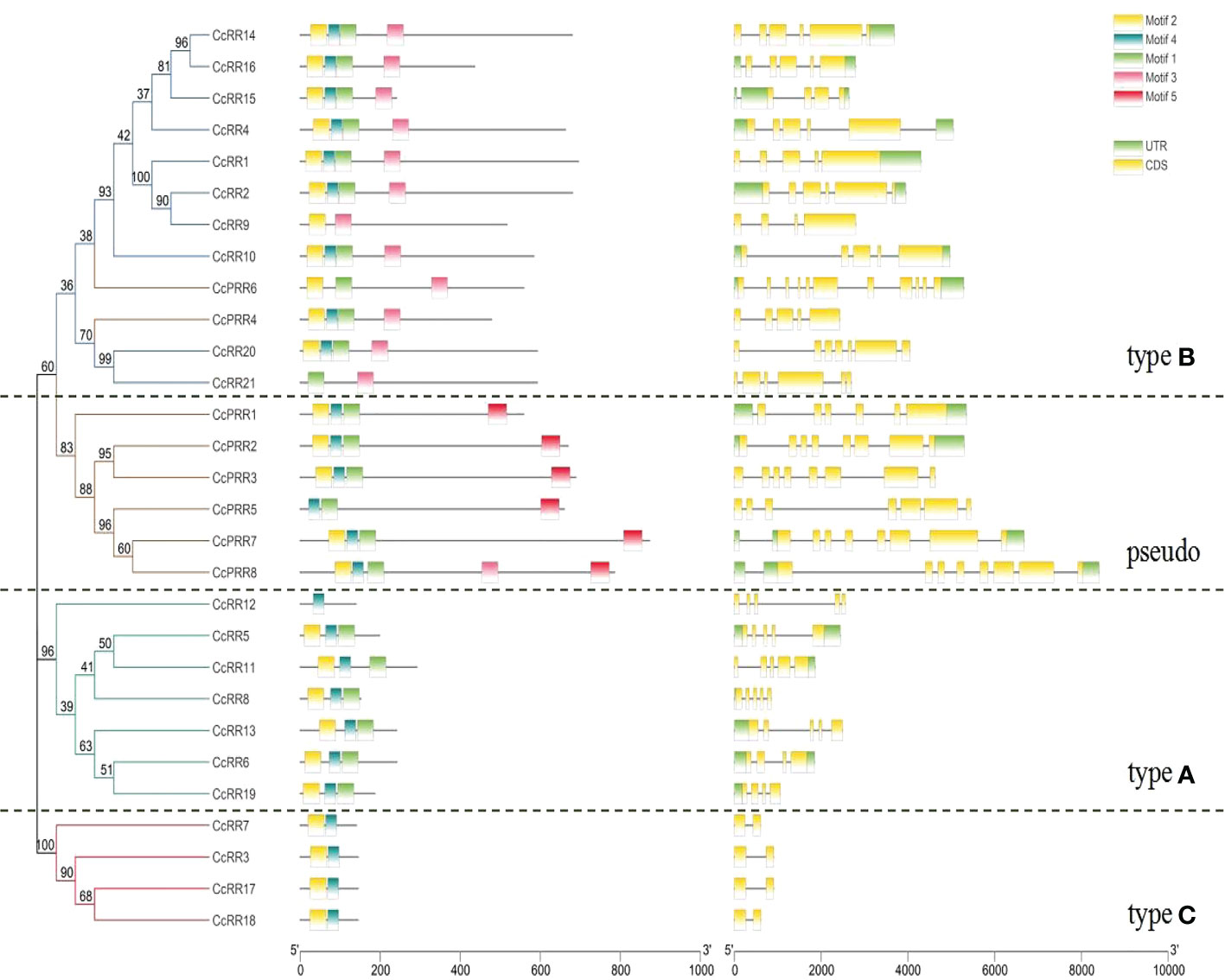
Figure 3 Phylogenetic relationship, conserved domains and exon-intron structures of CcRRs. Left: A phylogenetic tree constructed with the N-J method with MEGA 7. The proteins can be divided into four types, which are indicated with different colored lines (Green: type A; Blue: type B; Red: type C; Yellow: pseudo). Middle: Distribution of conserved domains of CcRRs. The relative positions of each domain are shown by colored bars (Motif 1, 2 and 4: D-D-K domain; Motif 3: MYB-binding domain; Motif 5: CCT domain). Right: The exons, introns and untranslated regions (UTRs) are represented by yellow rectangles, black lines and green rectangles, respectively.
Prediction of cis-element in promoters of CcRR genes
Cis-element in promoter region of the gene plays an important role in the regulation of gene transcription and usually combines with TFs to determine the level of gene expression (Wang et al., 2017; Dai et al., 2022). Results of cis-element prediction showed that the promoters of CcRR family members contain a large number of light responsiveness elements (Figure 4). Drought-induced, low-temperature response, defense and stress response elements were also found in promoter regions of CcRRs. In addition, there are many hormone response elements, such as abscisic acid, auxin, methyl jasmonate, salicylic acid, gibberellin response elements and zein metabolism regulation element. Some specific cis-acting elements were also identified, such as anaerobic induction, meristem development, and endosperm expression elements. The analysis of the predicted cis-elements in the promoter indicated that the transcriptional regulation of CcRR gene family members was probably related to the processes of growth and development of citrus plants, and the responses to various stresses.
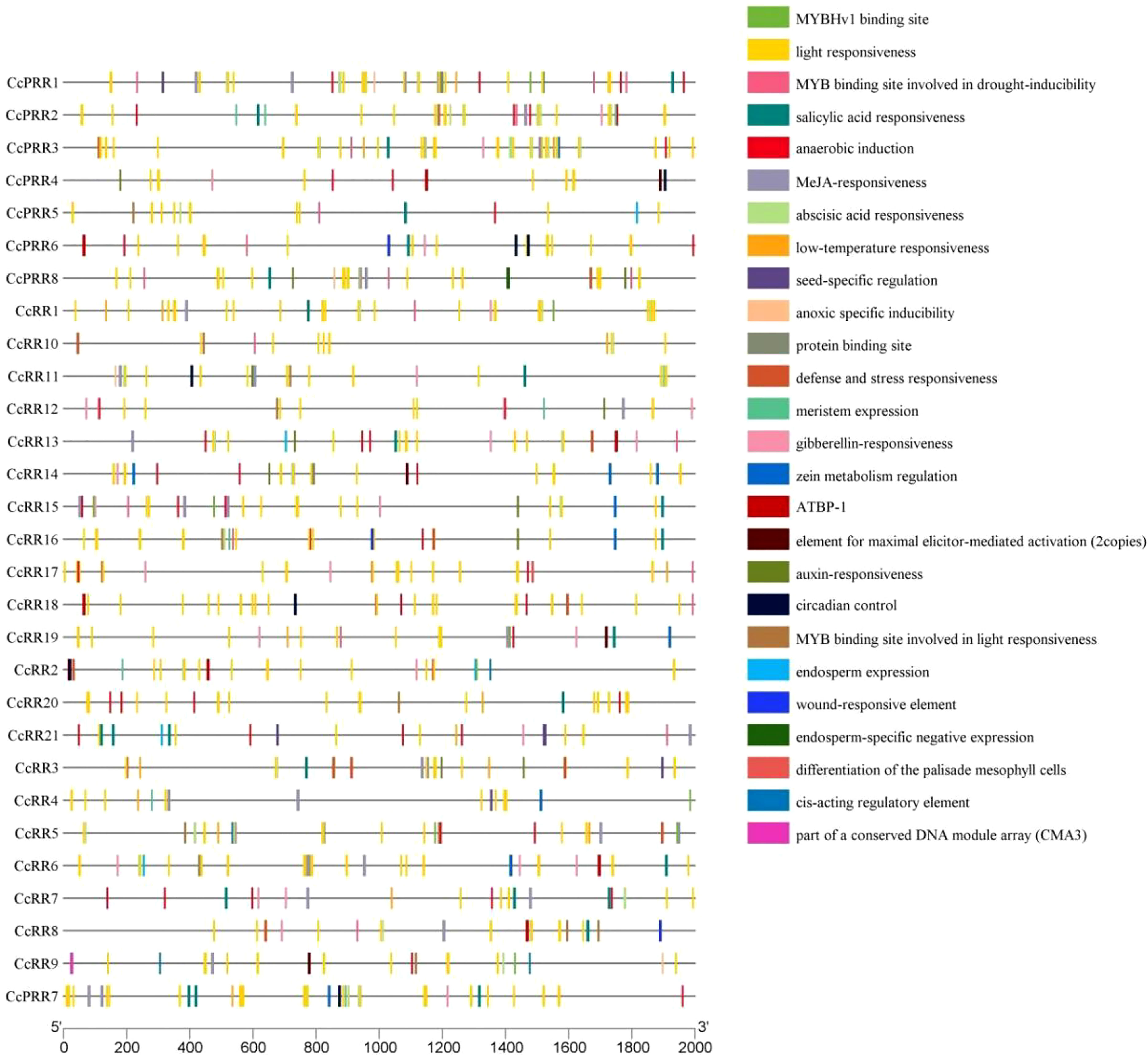
Figure 4 Predicted cis-acting elements in the promoter regions (2000 bp upstream of ATG start code) of CcRRs.
Tissue-specific expression profiles of CcRR genes
Levels of gene expression in the roots, stems and leaves were analyzed by qRT-PCR to characterize the expression of type A and type B CcRR genes in different tissues of citrus (Figure 5). CcRR12, CcRR13, CcRR20 and CcRR21 expressed at very low level in roots, suggesting that they are not active in this tissue, therefore, they were excluded from further work of this study. The remaining 13 CcRR genes showed various expression patterns in different tissues, and most of them had the highest expression in leaves, such as CcRR4-6, CcRR10, CcRR11, CcRR15 and CcRR16. Among them, CcRR4, CcRR5, CcRR6 and CcRR16 also highly expressed in roots.
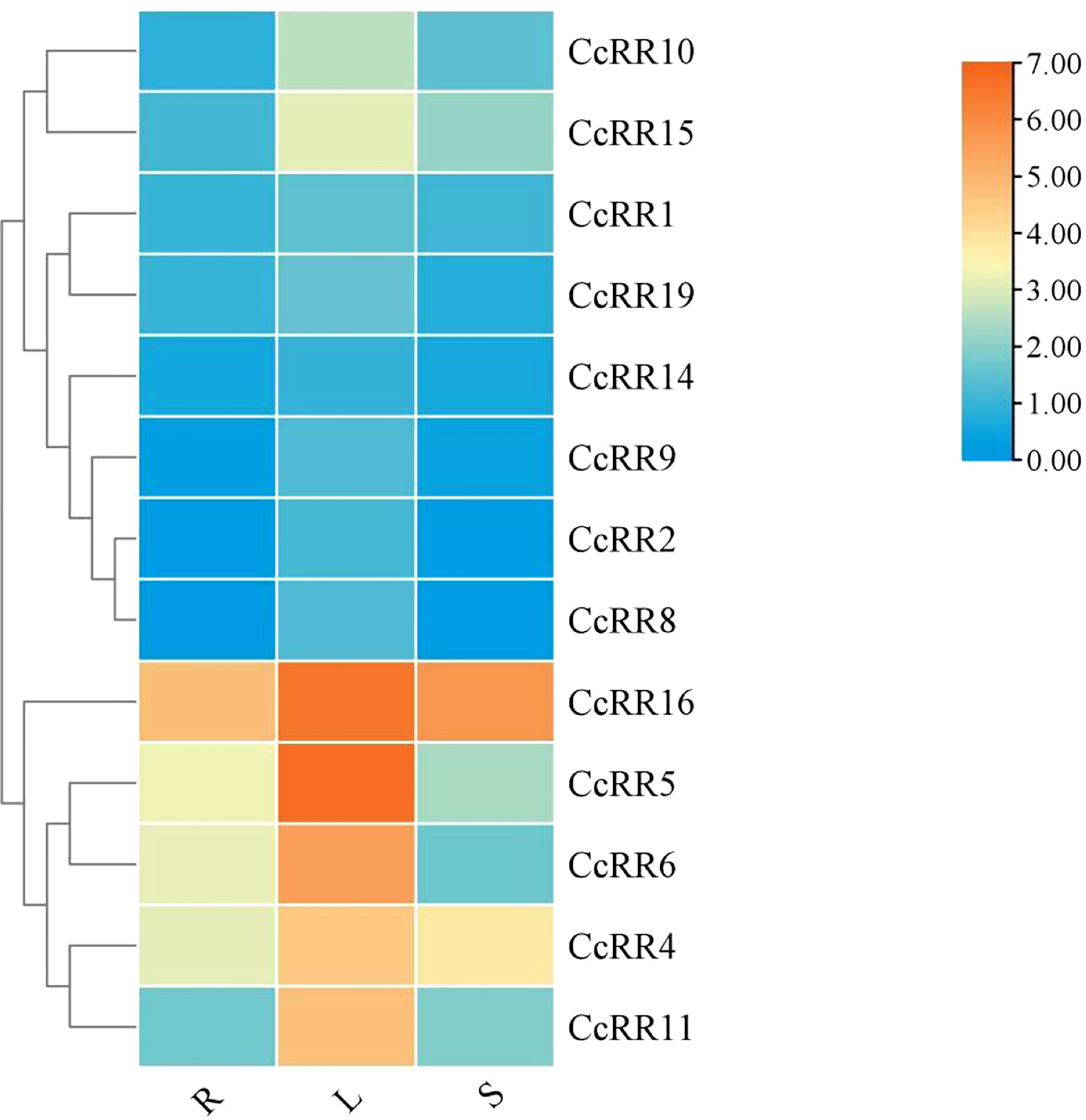
Figure 5 Heat-map of the expression profiles of CcRR genes in different tissues. R: root; L: leaf; S: stem. The vertical color scale shown to the right of the image represents log2 expression values.
Expression of CcRR genes in response to phytohormones and abiotic stresses
Phytohormone and abiotic stress treatments could induce the expression of CcRR genes. Analysis of RR genes’ expression under hormone treatments and abiotic stresses showed that the transcriptional levels of CcRR4, CcRR5, CcRR6, CcRR16 were high and changed significantly under the conditions of PEG, NaCl, cold, ABA or 6-BA treatment, indicating that those CcRRs dynamically responded to the multiple stresses (Figure 6).
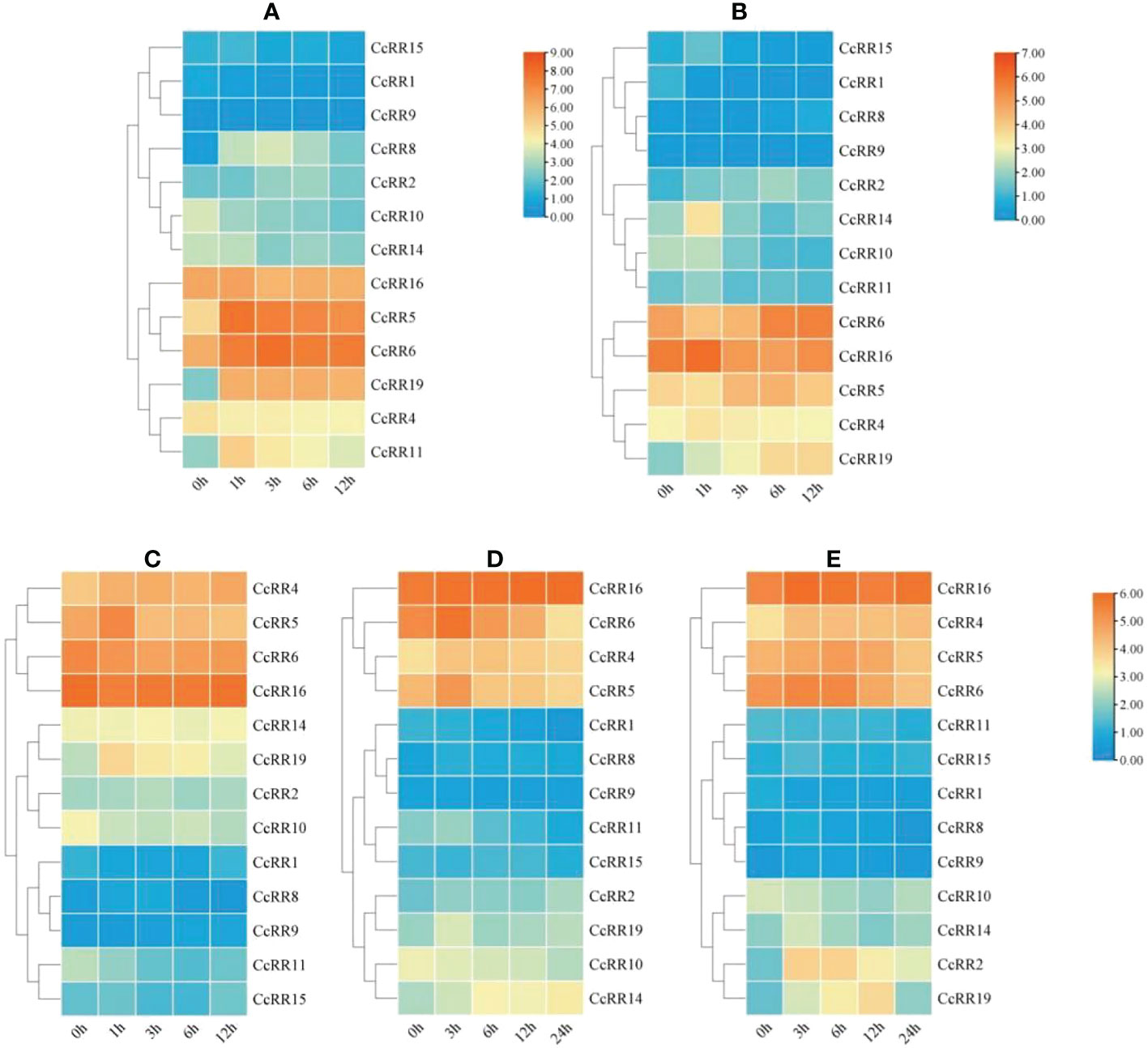
Figure 6 Heat-map of the expression pattern of CcRR genes in roots response to the treatments of hormone and abiotic stresses. (A) 6-BA treatment; (B) ABA treatment; (C) PEG treatment; (D) Cold treatment; (E) NaCl treatment. The figures (C-E) use the same ruler. The 0, 1, 3, 6, 12 and 24 h labels indicate the time after treatment. The vertical color scales shown to the right of the image represent log2 expression values.
Under 6-BA treatment (Figure 6A), the expressions of all the type A CcRRs (CcRR5, CcRR6, CcRR8, CcRR11, CcRR19) and some of type B CcRRs (CcRR2, CcRR15, CcRR16) were up-regulated at the early stage of the treatment and then declined, while the expressions of CcRR1, CcRR4, CcRR9, CcRR10 and CcRR14 were generally suppressed by the treatment. For ABA treatment (Figure 6B), the expression patterns of seven CcRRs (CcRR2, CcRR4, CcRR5, CcRR11, CcRR14, CcRR15, CcRR16) were basically the same. Their expressions up-regulated at the beginning of treatment and then declined, while the expression of CcRR19 was increasing throughout the treatment. However, CcRR1 and CcRR10 showed the opposite trend of decreased expression, while the expressions of CcRR6, CcRR8 and CcRR9 were only down-regulated at the early stage, but increased afterward.
Under PEG treatment (Figure 6C), the expressions of four genes (CcRR2, CcRR5, CcRR8 and CcRR19) were all up-regulated at 1h or 3 h after treatment, but down-regulated from 3h or 6h after treatment, while the expressions of CcRR4 and CcRR9 were up-regulated nearly in the whole period of treatment. On the opposite, the expression of CcRR10 was gradually decreased during the treatment, while the expressions of five genes (CcRR1, CcRR6, CcRR11, CcRR15, CcRR16) were down-regulated at the early stage of treatment and then recovered at late stage. Under low temperature condition (Figure 6D), the expressions of CcRR2, CcRR14 and CcRR16 increased in the entire period of treatment, while the expression level of type A CcRRs (CcRR5, CcRR6, CcRR8, CcRR11, CcRR19) and CcRR4 showed a trend of increasing first and then decreasing. However, the expressions of CcRR1 and CcRR10 were consistently decreasing, while the expressions of CcRR9 and CcRR15 were down-regulated at the early stage of treatment and then slight increased. For NaCl treatment (Figure 6E), the similar expression patterns were observed in the most of genes (CcRR2, CcRR4, CcRR5, CcRR6, CcRR8, CcRR9, CcRR14, CcRR15, CcRR16 and CcRR19). Their expressions were also up-regulated at the beginning of treatment and then declined at varying degree. The expressions of CcRR1 and CcRR11 declined during the whole period of treatment, while the expression of CcRR10 was down-regulated at the early stage of treatment and then up-expressed. The results of above observation indicated that most of CcRR genes involved in the processes responding to the various abiotic stresses and exogenous hormone treatments.
Expression of CcRR genes in different zones of root and citrus varieties
The eight CcRRs highly expressed in roots, also drastically respond to the abiotic stresses and exogenous phytohormone treatments in above study were selected to explore their functions involved in root development with Volkamer seedlings. The levels of gene expression were investigated in RT, RM, and RC zones of the roots as shown in Figure 7. CcRR2, CcRR5, CcRR6 and CcRR19 predominately expressed in RM, among which the expression of CcRR5 was the most prominent. The expressions of CcRR4, CcRR16, especially CcRR10, were higher in RC than that in RT. However, expression of CcRR14 was significantly higher in both RT and RC than in RM. The results suggested that those CcRRs probably play different roles in root morphogenesis. Therefore, the three CcRRs (CcRR5, CcRR10 and CcRR14) that distinguishingly expressed in different zones of root were further investigated to determine their roles in root morphogenesis of 9 citrus rootstocks, which varied in the length of primary roots and the numbers of lateral roots. Results of analysis indicated that the expression of CcRR5 in RT zone was significantly correlated to the length of primary root, while the expression of CcRR10 in RC zone was significantly correlated to the number of lateral roots, and the expressions of CcRR14 were significantly correlated to the two root indexes (Figure 8; Supplementary Table 2).
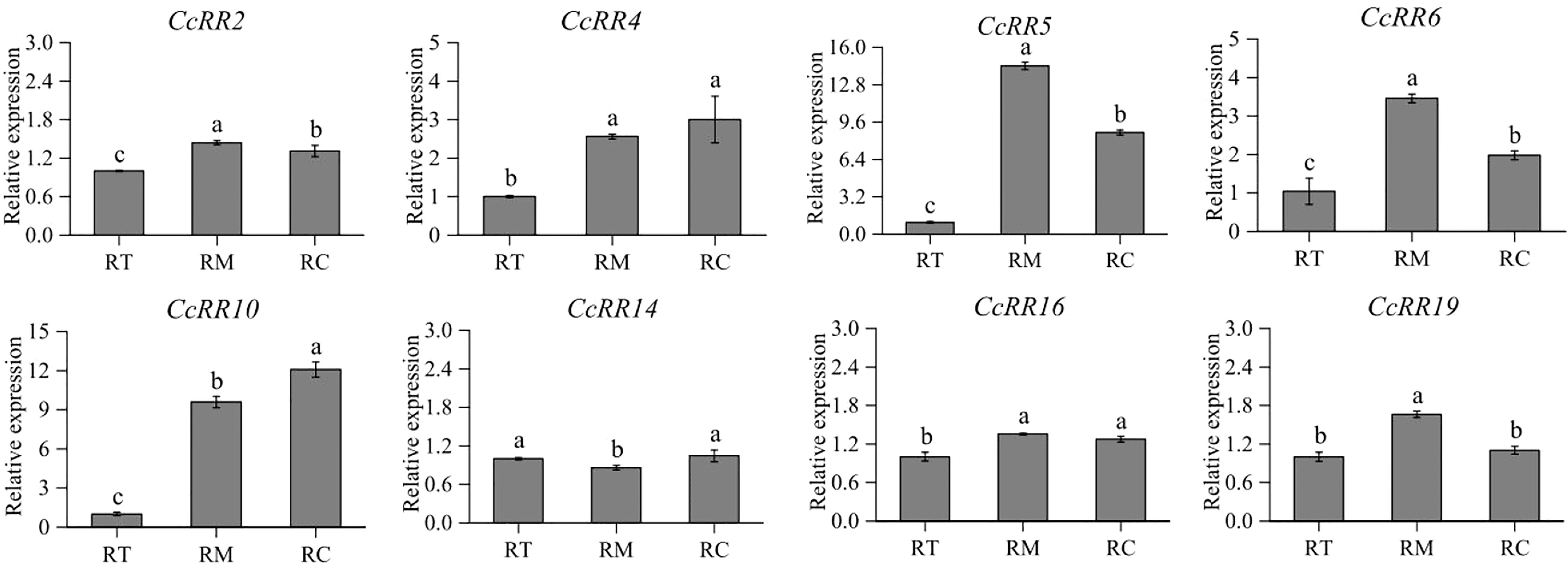
Figure 7 The relative expression of CcRR genes in different zones of citrus root. RT: the meristematic/elongation zone; RM: the root elongation/differentiation and lateral root initiation zone; RC: lateral root growth zone. The values are means ± SD of three independent biological replicates for qRT-PCR.
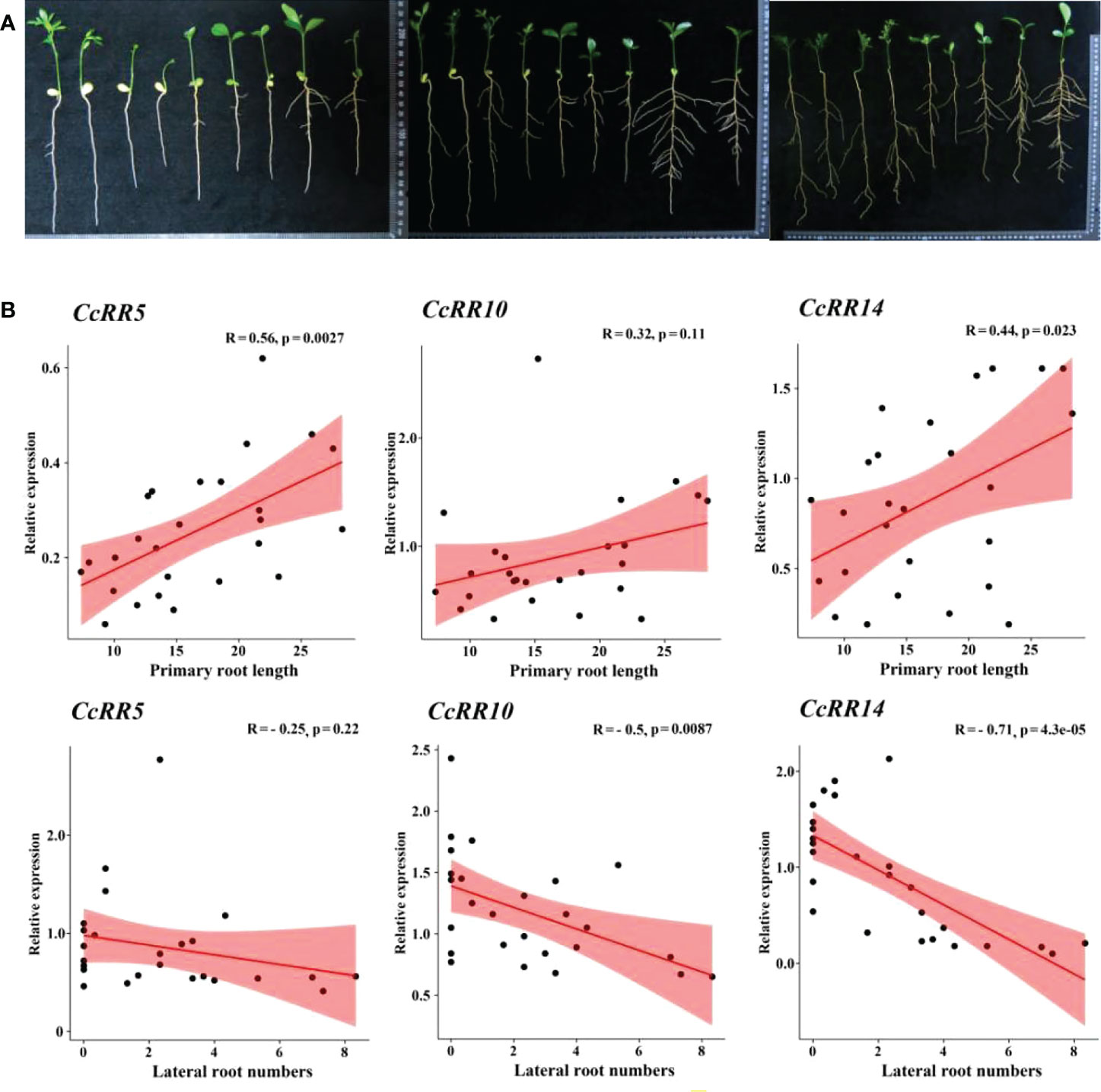
Figure 8 Root indexes and the correlation analysis of CcRR5, 10 and 14 at different stages. (A) Citrus root at 25, 50 and 75 days after sowning. From left to right are Poncirus trifoliata (Pt030, Pt038, Pt034, Pt026), Citrus wilsonii Tanaka ‘Zhique’, C. reticulata ‘Zhuju’, C. reshni Hort. Ex Tan ‘Cleopetra’, C. limonia Pasquale ‘Volkamer’, C. limonia Osbeck ‘Canton Lemon’, respectively; (B) Relationship between root indexes and their expressions.
Discussion
Transcription factor RRs play crucial roles in the cytokinin signaling circuitry, due to their participation in multiple biological functions such as DNA-binding transcriptional regulation (Imamura et al., 1999; Sakai et al., 2001; Zeng et al., 2017). Members of the RR gene family have been identified in several woody plants, such as apple, pear, peach and poplar, and found to be involved in fruit development, adventitious root formation and primary root elongation (Singh and Kumar, 2012; Li et al., 2017; Ni et al., 2017; Zeng et al., 2017). However, in citrus, knowledge of RRs is very limited and the identification of CcRR gene family have not been reported. In this study, 29 RR members were bio-informatically identified from citrus genome. Phylogenetic analysis with Arabidopsis and poplar homologs classified those CcRRs into four types distinguished by their representative domains as reported in other species (Singh and Kumar, 2012; Li et al., 2017). Interestingly, each member of the CcRR family contains a D-D-K domain, suggesting that CcRRs may share a highly conserved function. However, the specific motifs and gene structures presented in different types of CcRR genes implied a diversified and physiological function of CcRRs.
Previous studies indicated that the expressions of the members of RR gene family were tissue specific in relating to their function (Ferreira and Kieber, 2005; Huang et al., 2015; Li et al., 2015). In the four types of RRs, type A and type B RRs play vital roles in aspects of plant growth and development (Jain et al., 2006; Bertheau et al., 2015). Type B ARRs can mediate lateral root formation under cold stress (Jeon et al., 2016). Type A and type B ARRs were reported to be involved in primary root growth, and type-B ARRs negatively modulate primary root growth via AUX-1 mediated auxin translocation (Sharma et al., 2021). Therefore, the type A and type B CcRRs were targeted in this study. The expression patterns of type A and type B CcRRs were explored in root, stem and leaf. Among them, four genes (CcRR4, CcRR5, CcRR6 and CcRR16) were highly expressed in roots and leaves. CcRR12, CcRR13, CcRR20 and CcRR21 were excluded from further work of this study because of their low activities in the root. Promoters of CcRRs contained the certain elements related to hormone responses and abiotic stresses, indicating their function may related to the hormone signaling and abiotic stress response same as in other plants (Jain et al., 2006; Ramírez-Carvajal et al., 2008; Tsai et al., 2012; Yang et al., 2016). This was proved by the results that 8 type A and type B CcRRs drastically responded to the treatments of exogenous hormones and abiotic stresses, partially as reported by Huang et al. (2018) in Arabidopsis. It was reported that root system architecture, including root growth and lateral root branching, was closely related to abiotic stresses resistance (Bielach et al., 2012). Functional analysis of homologous genes indicated that CcRR14 and CcRR16 might play roles in abiotic stress resistance in the root of citrus as they were grouped into the same clade with ARR1, ARR2 and PtRR13. In this group, ARR1 plays leading roles in many stress response processes, such as cold stress, salinity and ABA induction (Zhu et al., 2015; Li et al., 2015; Jeon et al., 2016; Yan et al., 2021), and PtRR13 is a negative regulator of adventitious root development in Populus (Ramírez-Carvajal et al., 2009). In addition, ARR1 and ARR2 are response factors involved in primary root and lateral root meristem initiation, pathogen resistance, and leaf senescence (Hwang et al., 2012).
The specific expression of RR genes may relate to their function in root system architecture (Mason et al., 2004; Ferreira and Kieber, 2005; Garay-Arroyo et al., 2012). In this study, the tissue specific expression of CcRR5, CcRR10 and CcRR14 were significantly correlated to the characteristic indexes of root (primary root length and lateral root numbers) of nine citrus rootstocks. CcRR5, a type A RR gene highly expressed in RM, which is the elongation/differentiation and lateral root initiation zone of the root. Four RRs (ARR5/6/7/15) from Arabidopsis and one RR (PtRR10) from Populus were classified into the same subfamily with CcRR5. PtRR10 is an ortholog of ARR5 (Ramírez-Carvajal et al., 2008; Immanen et al., 2013). Those ARRs are as negative regulators of cytokinin signaling, participating in a negative feedback loop to reduce the sensitivity to cytokinin (Kiba et al., 2003; To et al., 2004; Leibfried et al., 2005; To et al., 2007). Cytokinin plays an inhibitory role on primary root growth arising from effects on cell division in the root meristem and on cell expansion in the root elongation zone (Kieber and Schaller, 2014). Therefore, it could be speculated that CcRR5 should be involved in regulating root growth in citrus. CcRR14 was classified into the same group as ARR1, which was reported to activate the expression of auxin signal inhibitor SHY2 to regulate root meristem size in Arabidopsis (Li et al., 2020). Its expression significantly correlated to the length of primary root in nine citrus rootstocks, suggesting that it would be related to root elongation. Bielach et al. (2012) reported that the mutant of arr1 arr11 in Arabidopsis (orthologues of CcRR14 and CcRR10, respectively) enhanced cytokinin activity in pericycle cells, preventing lateral root initiation in close proximity to existing lateral root primordia. CcRR10 predominantly expressed in the RC of root in citrus. The expressions of CcRR10 and CcRR14 were significantly correlated to the number of lateral roots. This clearly suggested that they are probably involved in the development of lateral roots. Results of this study indicated that CcRR5, CcRR10 and CcRR14 may play important roles in the process of root morphogenesis.
Conclusions
Twenty-nine RR family members were identified from citrus genome and classified into four types based on their amino acid sequences and conserved domains. Expressions of type A and type B CcRRs varied in different tissues of plant and highly responded to phytohormones and abiotic stresses in the root. Three CcRRs (CcRR5, CcRR10 and CcRR14) were identified to express at high levels in different zones of root, and their expression significantly correlated to the root characteristic indexes in nine citrus rootstocks. Functional analysis of their orthologs in Arabidopsis suggested that they may participate in regulating root morphogenesis.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repositories can be found in the article/Supplementary Material.
Author contributions
MZ, SZ and XZ conceived and designed the experiments. MZ, FW, XW, JF and QY carried out the experiments. MZ, FW, SZ and XZ contributed to the data analysis. MZ, SZ and XZ contributed to paper writing and paper revising. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program of China (2018YFD1000101) and China Agriculture Research System (CARS-Citrus) and the “Double World-classes” Development Plan of Southwest University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1068961/full#supplementary-material
References
Bailey, T. L., Elkan, C. (1995). The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 3, 21–29.
Bertheau, L., Djeghdir, I., Foureau, E., Chefdor, F., Glevarec, G., Oudin, A., et al. (2015). Insights into b-type RR members as signaling partners acting downstream of HPt partners of HK1 in the osmotic stress response in Populus. Plant Physiol. Bioch. 94, 244–252. doi: 10.1016/j.plaphy.2015.06.006
Bielach, A., Podlesáková, K., Marhavy, P., Duclercq, J., Cuesta, C., Müller, B., et al. (2012). Spatiotemporal regulation of lateral root organogenesis in Arabidopsis by cytokinin. Plant Cell. 24 (10), 3967–3981. doi: 10.1105/tpc.112.103044
Chang, J., Li, X., Fu, W., Wang, J., Yong, Y., Shi, H., et al. (2019). Asymmetric distribution of cytokinins determines root hydrotropism in Arabidopsis thaliana. Cell Res. 29 (12), 984–993. doi: 10.1038/s41422-019-0239-3
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Choi, J., Huh, S. U., Kojima, M., Sakakibara, H., Paek, K.-H., Hwang, I. (2010). The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev. Cell. 19 (2), 284–295. doi: 10.1016/j.devcel.2010.07.011
Dai, Z., Dong, S., Miao, H., Liu, X., Han, J., Li, C., et al. (2022). Genome-wide identification of TIFY genes and their response to various pathogen infections in cucumber (Cucumis sativus l.). Scientia Horticult. 295, 110814. doi: 10.1016/j.scienta.2021.110814
de Bang, L., Paez-Garcia, A., Cannon, A. E., Chin, S., Kolape, J., Liao, F., et al. (2020). Brassinosteroids inhibit autotropic root straightening by modifying filamentous-actin organization and dynamics. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00005
Deng, J., Kong, L., Zhu, Y., Pei, D., Chen, X., Wang, Y., et al. (2022). BAK1 plays contrasting roles in regulating abscisic acid-induced stomatal closure and abscisic acid-inhibited primary root growth in Arabidopsis. J. Integr. Plant Biol. 64 (6), 1264–1280. doi: 10.1111/jipb.13257
Ferreira, F. J., Kieber, J. J. (2005). Cytokinin signaling. Curr. Opin. Plant Biol. 8 (5), 518–525. doi: 10.1016/j.pbi.2005.07.013
Gao, B., Fan, L., Li, X., Yang, H., Liu, F., Wang, L., et al. (2013). RcRR1, a Rosa canina type-a response regulator gene, is involved in cytokinin-modulated rhizoid organogenesis. PloS One 8 (8), e72914. doi: 10.1371/journal.pone.0072914
Garay-Arroyo, A., de la Paz Sánchez, M., García-Ponce, B., Azpeitia, E., Alvarez-Buylla, E. R. (2012). Hormone symphony during root growth and development. Dev. Dynam. 241 (12), 1867–1885. doi: 10.1002/dvdy.23878
Goh, T. (2019). Long-term live-cell imaging approaches to study lateral root formation in Arabidopsis thaliana. Microscopy 68 (1), 4–12. doi: 10.1093/jmicro/dfy135
Horák, J., Grefen, C., Berendzen, K. W., Hahn, A., Stierhof, Y.-D., Stadelhofer, B., et al. (2008). The Arabidopsis thaliana response regulator ARR22 is a putative AHP phospho-histidine phosphatase expressed in the chalaza of developing seeds. BMC Plant Biol. 8 (1), 77. doi: 10.1186/1471-2229-8-77
Huang, X., Hou, L., Meng, J., You, H., Li, Z., Gong, Z., et al. (2018). The antagonistic action of abscisic acid and cytokinin signaling mediates drought stress response in Arabidopsis. Mol. Plant 11 (7), 970–982. doi: 10.1016/j.molp.2018.05.001
Huang, W., Xian, Z., Kang, X., Tang, N., Li, Z. (2015). Genome-wide identification, phylogeny and expression analysis of GRAS gene family in tomato. BMC Plant Biol. 15 (1), 209–226. doi: 10.1186/s12870-015-0590-6
Hwang, I., Sheen, J., Müller, B. (2012). Cytokinin signaling networks. Annu. Rev. Plant Biol. 63 (1), 353–380. doi: 10.1146/annurev-arplant-042811-105503
Imamura, A., Hanaki, N., Nakamura, A., Suzuki, T., Taniguchi, M., Kiba, T., et al. (1999). Compilation and characterization of Arabidopsis thaliana response regulators implicated in his-asp phosphorelay signal transduction. Plant Cell Physiol. 40 (7), 733–742. doi: 10.1093/oxfordjournals.pcp.a029600
Immanen, J., Nieminen, K., Silva, H. D., Rojas, F. R., Meisel, L. A., Silva, H., et al. (2013). Characterization of cytokinin signaling and homeostasis gene families in two hardwood tree species: Populus trichocarpa and Prunus persica. BMC Genomics 14, 885. doi: 10.1186/1471-2164-14-885
Jain, M., Tyagi, A. K., Khurana, J. P. (2006). Molecular characterization and differential expression of cytokinin-responsive type-a response regulators in rice (Oryza sativa). BMC Plant Biol. 6 (1), 1–11. doi: 10.1186/1471-2229-6-1
Jeon, J., Cho, C., Lee, M. R., Van Binh, N., Kim, J. (2016). CYTOKININ RESPONSE FACTOR2 (CRF2) and CRF3 regulate lateral root development in response to cold stress in Arabidopsis. Plant Cell. 28 (8), 1828–1843. doi: 10.1105/tpc.15.00909
Jeon, B. W., Kim, M.-J., Pandey, S. K., Oh, E., Seo, P. J., Kim, J. (2021). Recent advances in peptide signaling during arabidopsis root development. J. Exp. Bot. 72 (8), 2889–2902. doi: 10.1093/jxb/erab050
Ji, X. L., Li, H. L., Qiao, Z. W., Zhang, J. C., Sun, W. J., You, C. X., et al. (2022). The BTB protein MdBT2 recruits auxin signaling components to regulate adventitious root formation in apple. Plant Physiol. 189 (2), 1005–1020. doi: 10.1093/plphys/kiac084
Kiba, T., Yamada, H., Sato, S., Kato, T., Tabata, S., Yamashino, T., et al. (2003). The type-a response regulator, ARR15, acts as a negative regulator in the cytokinin-mediated signal transduction in arabidopsis thaliana. Plant Cell Physiol. 44 (8), 868–874. doi: 10.1093/pcp/pcg108
Kieber, J. J., Schaller, G. E. (2014). Cytokinins. Arabidopsis Book 12, e0168. doi: 10.1199/tab.0168
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi: 10.1093/molbev/msw054
Lavenus, J., Goh, T., Roberts, I., Guyomarc’h, S., Lucas, M., De Smet, I., et al. (2013). Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci. 18 (8), 450–458. doi: 10.1016/j.tplants.2013.04.006
Leibfried, A., To, J. P. C., Busch, W., Stehling, S., Kehle, A., Demar, M., et al. (2005). WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438 (7071), 1172–1175. doi: 10.1038/nature04270
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., de Peer, Y. V., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30 (1), 325–327. doi: 10.1093/nar/30.1.325
Li, C., Hou, X., Mou, K., Liu, H., Zhao, Z., Liao, W. (2022). The involvement of abscisic acid in glucose promoted adventitious root development in cucumber. Scientia Horticult. 295, 110816. doi: 10.1016/j.scienta.2021.110816
Li, T., Kang, X., Lei, W., Yao, X., Zou, L., Zhang, D., et al. (2020). SHY2 as a node in the regulation of root meristem development by auxin, brassinosteroids, and cytokinin. J. Integr. Plant Biol. 62 (10), 1500–1517. doi: 10.1111/jipb.12931
Li, S.-B., Ou Yang, W.-Z., Hou, X.-J., Xie, L.-L., Hu, C.-G., Zhang, J.-Z. (2015). Genome-wide identification, isolation and expression analysis of auxin response factor (ARF) gene family in sweet orange (Citrus sinensis). Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00119
Liu, Y., Wirén, N. (2022). Integration of nutrient and water availabilities via auxin into the root developmental program. Curr. Opin. Plant Biol. 65, 102117. doi: 10.1016/j.pbi.2021.102117
Li, Y., Zhang, D., Zhang, L., Zuo, X., Fan, S., Zhang, X., et al. (2017). Identification and expression analysis of cytokinin response-regulator genes during floral induction in apple (Malus domestica borkh). Plant Growth Regul. 83 (3), 455–464. doi: 10.1007/s10725-017-0311-2
Li, L., Zheng, Q., Jiang, W., Xiao, N., Zeng, F., Chen, G., et al. (2022). Molecular regulation and evolution of cytokinin signaling in plant abiotic stresses. Plant Cell Physiol., 00(0):1–19 pcac071. doi: 10.1093/pcp/pcac071
Mahram, A., Herbordt, M. C. (2015). NCBI BLASTP on high-performance reconfigurable computing systems. ACM T. Reconfig. Techn. 7, 1–20. doi: 10.1145/2629691
Makino, S., Kiba, T., Imamura, A., Hanaki, N., Nakamura, A., Suzuki, T., et al. (2000). Genes encoding pseudo-response regulators: insight into his-to-Asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol. 41 (6), 791–803. doi: 10.1023/A:1026609112342
Marchler-Bauer, A., Derbyshire, M. K., Gonzales, N. R., Lu, S., Chitsaz, F., Geer, L. Y., et al. (2015). CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43, D222–D226. doi: 10.1093/nar/gku1221
Mason, M. G., Li, J., Mathews, D. E., Kieber, J. J., Schaller, G. E. (2004). Type-b response regulators display overlapping expression patterns in Arabidopsis. Plant Physiol. 135 (2), 927–937. doi: 10.1104/pp.103.038109
Muller, B., Sheen, J. (2007). Advances in cytokinin signaling. Science 318 (5847), 68–69. doi: 10.1126/science.1145461
Ni, J., Bai, S., Gao, L., Qian, M., Zhong, L., Teng, Y. (2017). Identification, classification, and transcription profiles of the b-type response regulator family in pear. PloS One 12 (2), e0171523. doi: 10.1371/journal.pone.0171523
Pandey, B. K., Huang, G., Bhosale, R., Hartman, S., Sturrock, C. J., Jose, L., et al. (2021). Plant roots sense soil compaction through restricted ethylene diffusion. Science 371 (6526), 276–280. doi: 10.1126/science.abf3013
Papon, N., Courdavault, V. (2022). Arresting cytokinin signaling for salt-stress tolerance. Plant Sci. 314, 111116. doi: 10.1016/j.plantsci.2021.111116
Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R., Finn, R. D. (2018). HMMER web server: 2018 update. Nucleic Acids Res. 46, W200–W204. doi: 10.1093/nar/gky448
Punta, M., Coggill, P. C., Eberhardt, R. Y., Mistry, J., Tate, J., Boursnell, C., et al. (2011). The pfam protein families database. Nucleic Acids Res. 40, D290–D301. doi: 10.1093/nar/gkr1065
Ramireddy, E., Brenner, W. G., Pfeifer, A., Heyl, A., Schmülling, T. (2013). In planta analysis of a cis-regulatory cytokinin response motif in Arabidopsis and identification of a novel enhancer sequence. Plant Cell Physiol. 54 (7), 1079–1092. doi: 10.1093/pcp/pct060
Ramírez-Carvajal, G. A., Morse, A. M., Davis, J. M. (2008). Transcript profiles of the cytokinin response regulator gene family in Populus imply diverse roles in plant development. New Phytol. 177 (1), 77–89. doi: 10.1111/j.1469-8137.2007.02240.x
Ramírez-Carvajal, G. A., Morse, A. M., Dervinis, C., Davis, J. M. (2009). The cytokinin type-b response regulator PtRR13 is a negative regulator of adventitious root development in Populus. Plant Physiol. 150 (2), 759–771. doi: 10.1104/pp.109.137505
Sakai, H., Honma, T., Aoyama, T., Sato, S., Kato, T., Tabata, S., et al. (2001). ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294 (5546), 1519–1521. doi: 10.1126/science.1065201
Santos Teixeira, J. A., ten Tusscher, K. H. (2019). The systems biology of lateral root formation: Connecting the dots. Mol. Plant 12, 784–803. doi: 10.1016/j.molp.2019.03.015
Schaller, G. E., Kieber, J. J., Shiu, S.-H. (2008). Two-component signaling elements and histidyl-aspartyl phosphorelays. Arabidopsis Book 6, e0112. doi: 10.1199/tab.0112
Sharma, M., Singh, D., Saksena, H. B., Sharma, M., Tiwari, A., Awasthi, P., et al. (2021). Understanding the intricate web of phytohormone signalling in modulating root system architecture. Int. J. Mol. Sci. 22 (11), 5508. doi: 10.3390/ijms22115508
Singh, G., Kumar, R. (2012). Genome-wide insilico analysis of plant two component signaling system in woody model plant Populus trichocarpa. Res. Plant Biol. 2 (2), 13–23.
Singh, S., Yadav, S., Singh, A., Mahima, M., Singh, A., Gautam, V., et al. (2020). Auxin signaling modulates LATERAL ROOT PRIMORDIUM1 (LRP1) expression during lateral root development in Arabidopsis. Plant J. 101 (1), 87–100. doi: 10.1111/tpj.14520
Strayer, C., Oyama, T., Schultz, T. F., Raman, R., Somers, D. E., Mas, P., et al. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289 (5480), 768–771. doi: 10.1126/science.289.5480.768
Street, I. H., Mathews, D. E., Yamburkenko, M. V., Sorooshzadeh, A., John, R. T., Swarup, R., et al. (2016). Cytokinin acts through the auxin influx carrier AUX1 to regulate cell elongation in the root. Development 143 (21), 3982–3993. doi: 10.1242/dev.132035
Sun, L., Nasrullah, K., Nie, Z., Xu, J., Huang, X., Sun, J., et al. (2022). Genome-wide identification and transcript analysis during fruit ripening of ACS gene family in sweet orange (Citrus sinensis). Scientia Horticult. 294, 110786. doi: 10.1016/j.scienta.2021.110786
Tajima, Y., Imamura, A., Kiba, T., Amano, Y., Yamashino, T., Mizuno, T. (2004). Comparative studies on the type-b response regulators revealing their distinctive properties in the his-to-Asp phosphorelay signal transduction of Arabidopsis thaliana. Plant Cell Physiol. 45 (1), 28–39. doi: 10.1093/pcp/pcg154
Timilsina, R., Kim, Y., Park, S., Park, H., Park, S., Kim, J., et al. (2022). ORESARA 15, a PLATZ transcription factor, controls root meristem size through auxin and cytokinin signalling-related pathways. J. Exp. Bot. 73 (8), 2511–2524. doi: 10.1093/jxb/erac050
To, J., Deruere, J., Maxwell, B., Morris, V., Hutchison, C., Ferreira, F., et al. (2007). Cytokinin regulates type-a Arabidopsis response regulator activity and protein stability via two-component phosphorelay. Plant Cell. 19 (12), 3901–3914. doi: 10.1105/tpc.107.052662
To, J., Haberer, G., Ferreira, F., Deruère, J., Mason, M., Schaller, G., et al. (2004). Type-a Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell. 16 (3), 658–671. doi: 10.1105/tpc.018978
Tsai, Y.-C., Weir, N. R., Hill, K., Zhang, W., Kim, H. J., Shiu, S.-H., et al. (2012). Characterization of genes involved in cytokinin signaling and metabolism from rice. Plant Physiol. 158 (4), 1666–1684. doi: 10.1104/pp.111.192765
Tu, T., Zheng, S., Ren, P., Meng, X., Zhao, J., Chen, Q., et al. (2021). Coordinated cytokinin signaling and auxin biosynthesis mediates arsenate-induced root growth inhibition. Plant Physiol. 185 (3), 1166–1181. doi: 10.1093/plphys/kiaa072
Voorrips, R. E. (2002). MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 93 (1), 77–78. doi: 10.1093/jhered/93.1.77
Wang, Y., Pan, F., Chen, D., Chu, W., Liu, H., Xiang, Y. (2017). Genome-wide identification and analysis of the Populus trichocarpa TIFY gene family. Plant Physiol. Bioch. 115, 360–371. doi: 10.1016/j.plaphy.2017.04.015
Yang, Y., Jiang, Y., Mi, X., Gan, L., Gu, T., Ding, J., et al. (2016). Identification and expression analysis of cytokinin response regulators in Fragaria vesca. Acta Physiol. Plant 38 (8), 198–210. doi: 10.1007/s11738-016-2213-8
Yan, Z., Wang, J., Wang, F., Xie, C., Lv, B., Yu, Z., et al. (2021). MPK3/6-induced degradation of ARR1/10/12 promotes salt tolerance in Arabidopsis. EMBO Rep. 22 (10), e52457. doi: 10.15252/embr.202152457
Yu, C., Chen, Y., Lu, C., Hwang, J. (2006). Prediction of protein subcellular localization. Proteins 64, 643–651. doi: 10.1002/prot.21018
Zeng, J., Zhu, X., Haider, M., Wang, X., Zhang, C., Chen, W. (2017). Genome-wide identification and analysis of the type-b authentic response regulator gene family in peach (Prunus persica). Cytogenet. Genome Res. 151 (1), 41–49. doi: 10.1159/000458170
Zhang, P., Liu, X., Yu, X., Wang, F., Long, J., Shen, W., et al. (2020). The MYB transcription factor CiMYB42 regulates limonoids biosynthesis in citrus. BMC Plant Bio. 20 (1), 254. doi: 10.1186/s12870-020-02475-4
Zhang, W., Swarup, R., Bennett, M., Schaller, G. E., Kieber, J. J. (2013). Cytokinin induces cell division in the quiescent center of the Arabidopsis root apical meristem. Curr. Biol. 23 (20), 1979–1989. doi: 10.1016/j.cub.2013.08.008
Keywords: citrus, root system architecture, response regulators, gene family, abiotic stress
Citation: Zhang M, Wang F, Wang X, Feng J, Yi Q, Zhu S and Zhao X (2022) Mining key genes related to root morphogenesis through genome-wide identification and expression analysis of RR gene family in citrus. Front. Plant Sci. 13:1068961. doi: 10.3389/fpls.2022.1068961
Received: 13 October 2022; Accepted: 07 November 2022;
Published: 22 November 2022.
Edited by:
Jianfu Jiang, Chinese Academy of Agricultural Sciences (CAAS), ChinaReviewed by:
Qiang-Sheng Wu, Yangtze University, ChinaJi-Hong Liu, Huazhong Agricultural University, China
Copyright © 2022 Zhang, Wang, Wang, Feng, Yi, Zhu and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shiping Zhu, emh1c2hpcGluZ0BjcmljLmNu; Xiaochun Zhao, emhhb3hpYW9jaHVuQGNyaWMuY24=
 Manman Zhang
Manman Zhang Fusheng Wang1,2
Fusheng Wang1,2 Shiping Zhu
Shiping Zhu Xiaochun Zhao
Xiaochun Zhao