- 1Chongqing Key Laboratory of Molecular Biology of Plant Environmental Adaptations, College of Life Science, Chongqing Normal University, Chongqing, China
- 2Wuxi Fisheries College, Nanjing Agricultural University, Jiangsu, China
- 3Department of Botany, University of British Columbia, Vancouver, BC, Canada
Calcium-dependent protein kinase (CPK) is a class of Ser/Thr protein kinase that exists in plants and some protozoa, possessing Ca2+ sensing functions and kinase activity. To better reveal the roles that Brassica CPKs played during plant response to stresses, five Brassica species, namely Brassica rapa (B. rapa), Brassica nigra (B. nigra), Brassica oleracea (B. oleracea), Brassica juncea (B. juncea), and Brassica napus (B. napus) were selected and analyzed. In total, 51 BraCPK, 56 BniCPK, 56 BolCPK, 88 BjuCPK, and 107 BnaCPK genes were identified genome wide and phylogenetics, chromosomal mapping, collinearity, promoter analysis, and biological stress analysis were conducted. The results showed that a typical CPK gene was constituted by a long exon and tandem short exons. They were unevenly distributed on most chromosomes except chromosome A08 in B. napus and B. rapa, and almost all CPK genes were located on regions of high gene density as non-tandem form. The promoter regions of BraCPKs, BolCPKs, and BnaCPKs possessed at least three types of cis-elements, among which the abscisic acid responsive-related accounted for the largest proportion. In the phylogenetic tree, CPKs were clustered into four primary groups, among which group I contained the most CPK genes while group IV contained the fewest. Some clades, like AT5G23580.1(CPK12) and AT2G31500.1 (CPK24) contained much more gene members than others, indicating a possibility that gene expansion occurred during evolution. Furthermore, 4 BraCPKs, 14 BolCPKs, and 31 BnaCPKs involved in the Plasmodiophora brassicae (P. brassicae) defense response in resistant (R) or susceptible (S) materials were derived from online databases, leading to the discovery that some R-specific induced CPKs, such as BnaC02g08720D, BnaA03g03800D, and BolC04g018270.2J.m1 might be ideal candidate genes for P. brassicae resistant research. Overall, these results provide valuable information for research on the function and evolution of CDK genes.
Introduction
Ca2+ is a generic form of the second messenger to participate in numerous signaling pathways (Hetherington and Brownlee, 2004; Zhang et al., 2014b). The change of Ca2+ concentration in cells can be captured by calcium sensors. Based on whether or not they are adjusted by the combination with Ca2+, calcium sensors are divided into four classes in plants, namely calmodulin (CaM) or CaM-like proteins (CMLs), calcineurin B-like proteins (CBLs), CBL interacting protein kinases (CIPKs), and the calcium-dependent protein kinases (CPKs) and their relatives, CPK-related kinases (CRKs) (Luan et al., 2002; Yang et al., 2021). Among them, CPKs exist in plants and some protozoa, especially in plant extracts. Additionally, rich calcium-stimulated protein kinase activity is found to be related to CPKs (Harmon et al., 2000). Typical CPK proteins contain an N-terminal variable domain, a protein kinase domain, an autoinhibitory domain, and a calmodulin-like domain (Cheng et al., 2002). The protein kinase domain is the catalytic domain containing an adenosine triphosphate binding site adjacent to the inhibitory domain, and the successive calmodulin domains always contain a protein kinase domain and EF-hands to determine whether the CPK is calmodulin dependent (Cheng et al., 2002; Hrabak et al., 2003).
Numerous studies have reported that CPKs are involved in plant responses to abiotic stresses. For example, Arabidopsis CPK1 and CPK2 were rapidly expressed under drought and high salinity stress, and CPKs played a significant role in enhancing abiotic stress tolerance (Urao et al., 1994). In maize, 12 of 40 CPK genes responded to low temperature, drought, salt, ABA, and hydrogen peroxide (Kong et al., 2013). In wheat, 10 of 14 CPK genes were responsive to abiotic stress, including drought, NaCl, and ABA. In potatoes, StCPKs were responsive to biotic and abiotic stresses as well as hormone stimulation (Bi et al., 2021). Among them, 20 StCPKs showed differential expression patterns in drought-tolerant and drought-sensitive potato varieties under drought stress, which allowed for recognition of abiotic stress of potatoes. In grapes, the response of VpCPKs to NaCl treatment and transcriptional responses at low and high temperatures has been studied (Zhang et al., 2015). Almost all 19 CPKs were up or down-regulated under at least one abiotic stress, but some genes had a rather mild response, and some grape CPK genes, like VpCPK6, VpCPK9, VpCPK14, VpCPK16, and VpCPK19 had positive effects in fighting against powdery mildew. Overall, most CPK-related studies focused on the role in abiotic stress and rarely mentioned biotic stressors.
Brassica is a member of the Brassicaceae family, which contains many important vegetables, oil crops, and forage crops and has high economic value. CPK gene is a candidate gene for modifying plant stress resistance cations. Although there are some studies on CPKs of Brassica plants, such as turnip and canola (Zhang et al., 2014a; Wang et al., 2017), there are few studies on Brassica CPKs as a whole. Additionally, most of the studies on CPKs are conducted in one type of plant, which cannot be accurately used for comparison between plants. In this study, five Brassica Species (B. rapa, B. nigra, B. oleracea, B. juncea, and B. napus) CPKs (BraCPKs, BniBCPKs, BolCPKs, BjuCPKs, BnaCPKs) were identified, for phylogenetic, chromosomal mapping, collinearity, promoter analysis, and biological stress analysis. This study specifically studied the Brassica CPK induced by clubroot, which is caused by the obligate biotrophic pathogen P. brassicae, one of the most devastating diseases affecting Brassicae (Zhang et al., 2016). Our results provide important information on the evolutionary history and biological function of the Brassica Species CPKs in biotic stress.
Materials and methods
Identification of CPK genes
To identify CPK genes in Brassica, the whole-genomes of five Brassica species were downloaded from the Brassica Database (BRAD) (Chen et al., 2022) (http://brassicadb.cn/). The HMMER profiles of the protein kinase domains (PF00069) and EF-hand domains (PF00036, PF13499, or PF13833) derived from Pfam (www.ebi.ac.uk/interpro) were used as queries to search for CPKs in the Brassica protein database, thereby obtaining the first part of the candidate CPK proteins. Furthermore, all Arabidopsis CPK proteins were obtained from TAIR (https://www.arabidopsis.org/) and used as queries to search orthologs in five Brassica species using the BLAST search analysis. Subsequently, the two sets of candidates were merged and subjected to NCBI-BLASTP using the UniProtKB/SwissoPrpt database, and their annotations were downloaded for further screening. Then, we excluded the sequences that were CPK-related proteins, calcium/calmodulin-dependent proteins, and calcium and calcium/calmodulin-dependent protein kinases. The remaining proteins were regarded as CPKs. The conserved domains and EF-hand quantity were predicted by SMART (Letunic and Bork, 2018; Letunic et al., 2021) (http://smart.embl-heidelberg.de/). N-terminal myristoylation sites, N-terminal palmitoylation sites, and N-terminal acetylation sites were predicted using NMT (https://mendel.imp.ac.at/myristate/SUPLpredictor.htm), CSS-Palm4.0 (Ren et al., 2008; Chica, 2020) (http://csspalm.biocuckoo.org/index.php), and NetAcet-1.0 (Kiemer et al., 2005) (https://services.healthtech.dtu.dk/service.php?NetAcet-1.0), respectively.
Gene structure, conserved motif, promoter element, and phylogenetic analysis
TBtools (Chen et al., 2020) was used to visualize the exon-intron organization of CPK genes according to the annotation information of the Brassica genomes in BRAD. MEME suit (https://meme-suite.org/meme/index.html) was used to search for conserved motifs in CPK proteins (Bailey et al., 2015). Approximately 2,000-bp upstream flanking fragments of the CPK genes were derived from the genome, and PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to predict promoter cis-elements (Lescot et al., 2002). Full-length Brassica species CPK protein sequences were aligned via the MUSCLE program and then used to construct a phylogenic tree via the neighbor-joining method of MEGA11.0 software (Tamura et al., 2021). Bootstrap values were estimated to assess the relative support for each branch with 2,000 replicates.
Genomic distribution and synteny analysis
Brassica CPKs were mapped on chromosomes according to genome annotation files from BRAD. The segmental and tandem duplication regions were obtained using the MCscanX software (Wang et al., 2012). For synteny analysis, synteny blocks of the CPK genes were visualized using TBtools (Chen et al., 2020). Nonsynonymous (Ka)/synonymous (Ks) calculation analysis was performed by KaKs_calculator 3.0 (Zhang, 2022).
Expression characteristics of CPK genes induced by Plasmodiophora brassicae
To study the CPKs induced by P. brassicae infection, three RNA-seq datasets of clubroot-resistant (R) and clubroot-susceptible (S) lines of B.rapa (PRJNA298858), B.oleracea (PRJNA298858), and B.napus (PRJNA345072) were downloaded from NCBI (Chen et al., 2015; Zhang et al., 2016; Li et al., 2019). The time points for B. rapa dataset were 0, 12, 72, and 96 hours post-inoculation (hpi). The time points for B.olerascea were 0, 7, and 14 days post-inoculation (dpi), and for B.napus were 0, 12, 24, 60, and 96 hpi. The TPM (Transcripts Per Kilobase Million) value was used to display the gene expression levels, and the heat map was drawn by TBtools (Chen et al., 2020). Correlation analysis was carried out according to their expression levels in the relevant time after P.brassicae infection, so as to judge the influence of P. brassicae in Brassica species.
Results
Identification and characterization of CPKs in Brassica
To identify CPKs in Brassica, we performed a genome-wide analysis of the CPK gene family in the five Brassica species through a BLAST search and HMMER analysis. Proteins with protein kinase domain and EF-hand domain were identified, and those that might possess similar domains, such as CPK-related proteins, calcium/calmodulin-dependent proteins, and calcium and calcium/calmodulin-dependent protein kinases were carefully excluded. Altogether, we identified 51 BraCPKs, 56 BniCPKs, 56 BolCPKs, 88 BjuCPKs, and 107 BnaCPKs from B. rapa, B. nigra, B. oleracea, B. juncea, and B. napus, respectively. The typical CPK genes were constituted by a long exon and tandem short exons (Table S1; Figure S1). Three quarters of CPK genes in five species possessed six to eight exons, with detailed proportions of each species possessing this number of exons at 80.39% (B. rapa), 78.57% (B. nigra), 78.57% (B. oleracea), 80.68% (B. juncea), and 73.83% (B. napus). The exon number ranged from 2 to 21, such as BjuO012843 having only two short exons and BraA02g042460.3C exhibiting 21 exons of varying lengths.
In general, CPK proteins contained one Protein kinase and four EF-hand domains, with some CPKs varying in the number of EF-hand domains, ranging from one to six. (Table S1; Figure S2). Furthermore, the majority of CPK proteins possessed palmitoylation sites at their N-termini (B. rapa, 75.44%; B. nigra, 76.79%; B. oleracea, 73.21%; B. juncea, 69.32%; B. napus, 70.09%). This is crucial for regulation, as palmitoylation plays important roles in modulating proteins’ trafficking, stability and sorting (Martín and Busconi, 2000; Draper et al., 2007; Greaves and Chamberlain, 2007; Linder and Deschenes, 2007). Moreover, half of CPK proteins contained myristoylation sites, and several contained N-terminal acetylation sites.
Chromosomal localization and gene expansion
To understand the genomic distribution of CPKs, gene positions were acquired from the genome database of Brassica species (Figure 1). BraCPK genes were located on 9 out of 10 chromosomes with an exception of Chromosome A08 (Figure 1A). Chromosome A03 carried 11 evenly distributed BraCPK genes, which was the maximum number on one chromosome. Meanwhile, chromosomes A01 and A04 possessed only three BraCPK genes. 55 BniCPK genes were spread out across eight chromosomes, and among them, 12 BniCPK genes were located on Chromosome B08, mostly in cluster manner (Figure 1B). The BniCPK gene number on other chromosomes varied, ranging from four to nine. As seen in Figure 1C, BolCPK genes distributed on all chromosomes, and most genes clustered on chromosome C03 which was followed by chromosome C07, C09, and C02. As shown in Figure 1D, BjuCPK genes were unevenly dispersed on chromosomes excluding A08 and B04 which carried no BjuCPK genes. The gene number on different chromosomes varied greatly. For example, there are nine genes on chromosome A03 and only 2 genes on chromosome A01. Similar to the distribution of BraCPKs and BolCPKs, BnaCPK genes were unevenly dispersed on chromosomes. This uneven distribution excluded ChrA08 which was the only one that carried no BnaCPK genes (Figure 1E). The quantity of BnaCPK genes on different chromosomes ranged from 2 to 10. ChrC09 had 10 BnaCPK genes which was the largest number found on a single chromosome, while ChrC06 contained only 2 BnaCPKs.
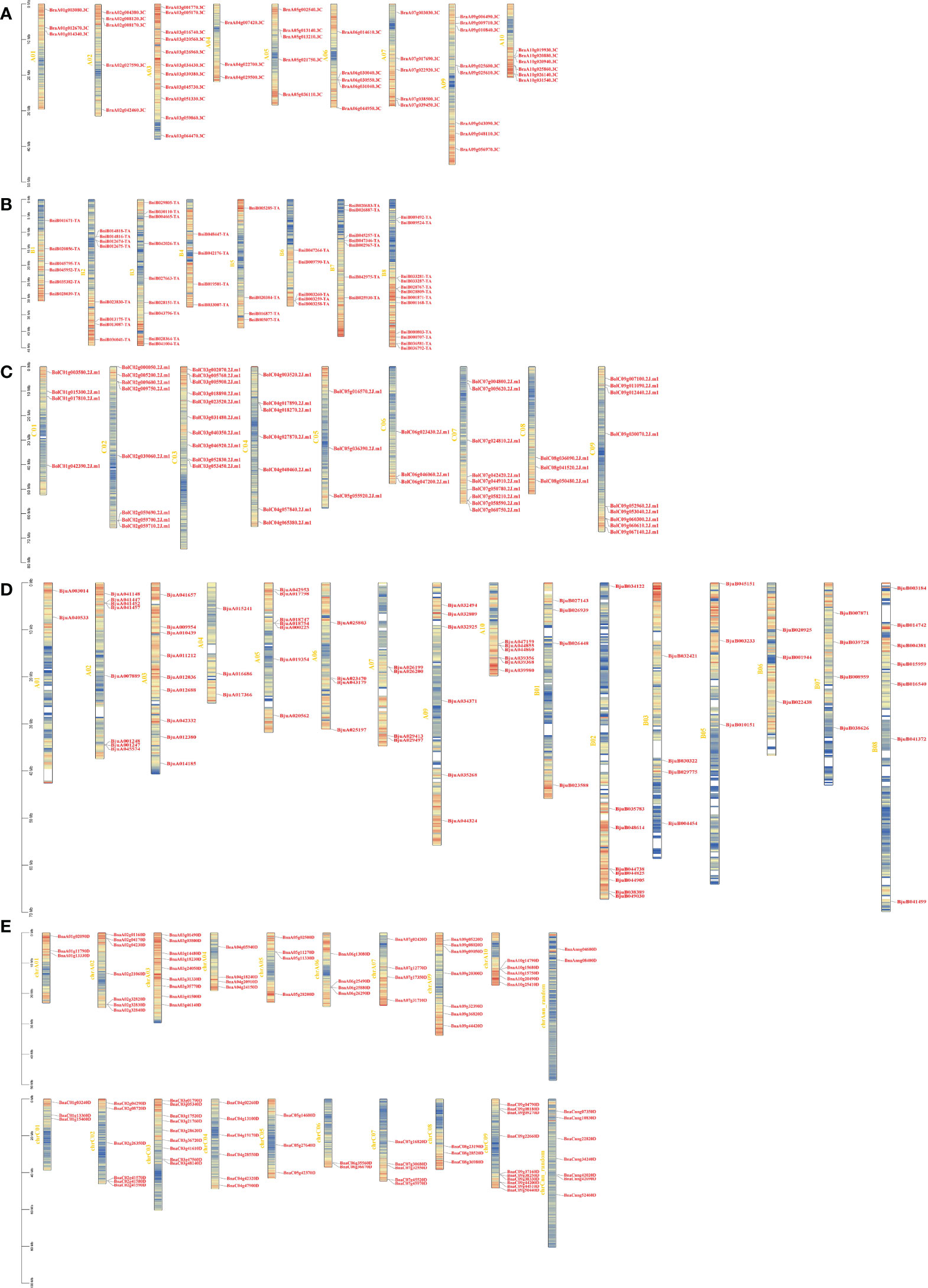
Figure 1 Locations of CPK genes on chromosomes in five Brassica species. Locations of CPK genes in B rapa (A), B. nigra. (B), B. oleracea (C), B. juncea (D), and B. napus (E), respectively. A01-A10, B1-B8, C01-C09, A01-B08, and chrA01-chrCnn represent the chromosomes in B rapa, B. nigra, B. oleracea, B. juncea, and B. napus, respectively. Red indicates high gene density, while blue indicates low.
Considering that B. napus is derived from the recombination of B. rapa and B. oleracea, B. juncea is derived from B. rapa and B. nigra, the positions of BnaCPK genes on chromosomes ChrAs or ChrCs and BjuCPK genes on chromosomes ChrAs or ChrBs showed highly consistent with those of corresponding homologous BraCPK, BolCPK or BniCPK genes. Additionally, almost all CPK genes were located on regions of high gene density as non-tandem form. In total, only seven CPK gene clusters were found in five species, including one in B. rapa (BraA09g325600.3C and BraA09g325610.3C), two in B. nigra (BniB012674-TA and BniB012675-TA; BniB003258-TA, BniB003259-TA and BniB003260-TA), one in B. oleracea (BolC02g059690.2J.m1, BolC02g059700.2J.m1 and BolC02g059710.2J.m1), two in B. juncea (BjuA001247 and BjuA001248, BjuA026199 and BjuA026200), and one in B. napus (BnaC02g41570, BnaC02g41580 and BnaC02g415790).
To better reveal the expansion of CPK genes in five Brassica genomes, the duplication patterns of CPK genes were predicted within and between each genome with the A. thaliana genome added for comparison (Figure 2). The result showed that there were 15 CPK gene pairs in A. thaliana, and the gene pair number of Brassica species was tripled or even tenfold, namely, 47 pairs in B. rapa, 51 pairs in B. nigra, 52 pairs in B. oleracea, 154 pairs in B. juncea, and 200 pairs in B. napus (Figure 2A). Moreover, the possible syntenic relationship of CPK genes between Brassica genomes was also investigated. Subsequently, we obtained 234 orthologous gene pairs between B. rapa and B. napus, 177 between B. rapa and B. juncea, 258 between B. oleracea and B. napus, and 182 between B. nigra and B. juncea, which are shown in Figure 2B. Chromosome A03/A09, B03/B08, and C03/C07/C09 possessed the higher orthologous regions in B. rapa, B. nigra and B. oleracea, respectively. Correspondingly, chrA03, chrA09, chrC03, and chrC09 subgenomes were classified as higher orthologous regions in B.napus. These higher orthologous regions were not present in chromosome A03, A09, B03, and B08 subgenomes in B.juncea, which may be due to the imperfect assembly of the B.juncea genome.
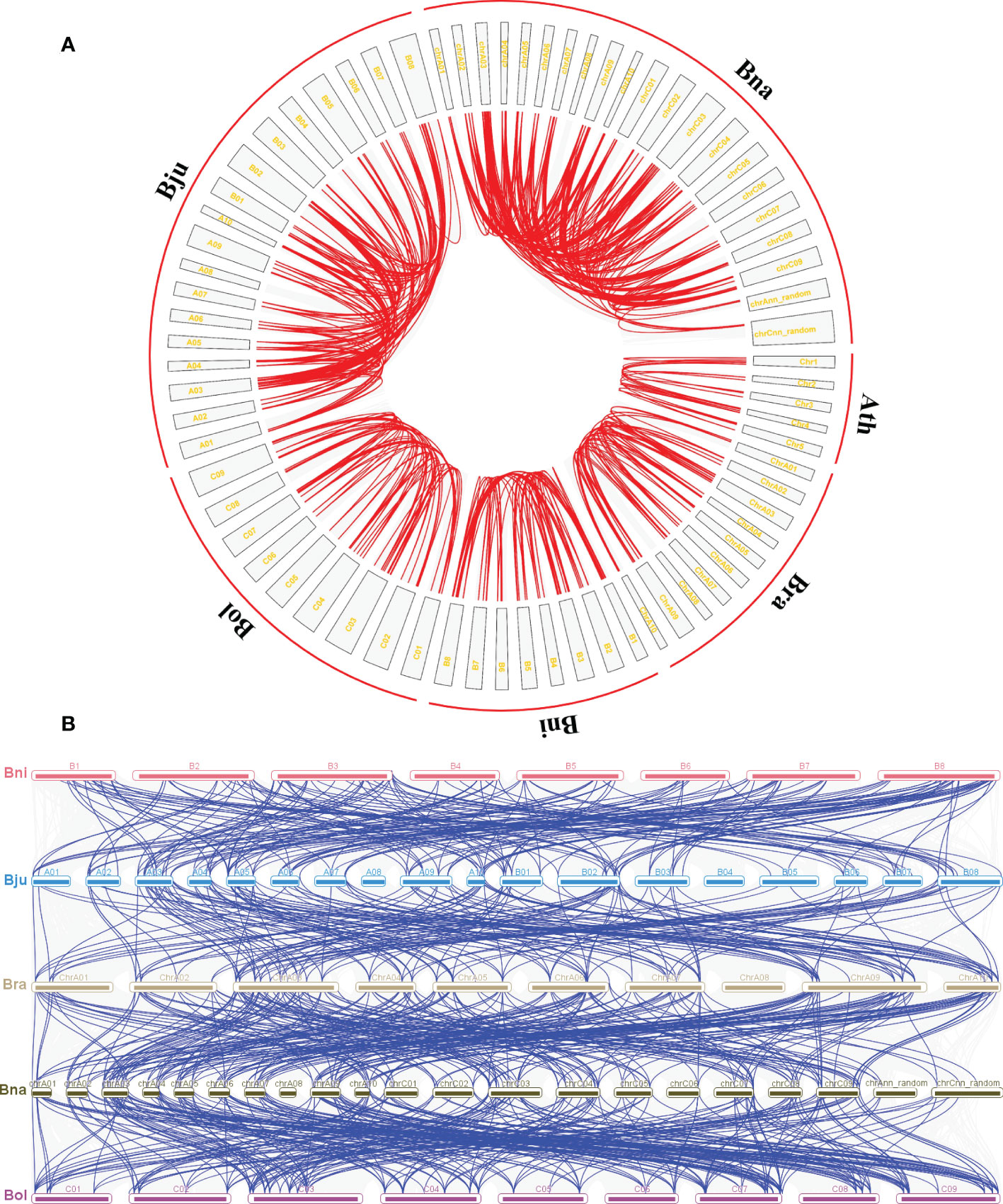
Figure 2 Duplication and syntenic relationship of CPK genes in five Brassica species. (A) Duplication of RLP genes in A. thaliana, B. rapa, B. nigra, B. oleracea, B. juncea, and B. napus, respectively. The red line represents the gene pairs and the gray box represents the chromosome. (B) Syntenic relationship of CPK-encoding genes between B.rapa, B.nigra, B.oleracea, B.juncea, and B.napus. The blue line represents the gene pairs and the colored box represents the chromosome.
Additionally, the nonsynonymous/synonymous mutations (Ka/Ks) analysis was performed on orthologous CPK genes between A. thaliana and the other five Brassica species (Table S2). The Ka/Ks value of the predicted CPK paralogs that had average values of 0.087 (B. rapa), 0.091 (B. nigra), 0.083 (B. oleracea), 0.088 (B. juncea) and 0.083 (B. napus) were less than one, suggesting that all pairs of CPK proteins were under strong purifying selection pressure.
Phylogenetic relationships of CPKs
To explore the phylogenetic relationship of CPK genes, neighbor-joining trees were constructed by CPK protein sequence from five Brassica species (Figure 3). CPKs were clustered into four major groups based on tree topology (group I, group II, group III, group IV), which agreed with the classification of Arabidopsis CPKs (Cheng et al., 2002). In the phylogenetic tree of five species, the CPK genes of five Brassica species and their orthologs in Arabidopsis formed distinct clades. Among four groups, group I contained the most CPK genes, including 15 BraCPKs, 19 BniCPKs, 19 BolCPKs, 31 BjuCPKs, and 37 BnaCPKs. A clade including AT5G23580.1(CPK12) in group I contributed the largest number of orthologs from Brassica species, reaching up to 36. The second largest group was group III, including 105 CPK genes, in which the top two clades were AT2G31500.1 (CPK24) and AT3G51850.1 (CPK13), possessing 20 and 19 members from five species respectively. There were 99 CPK genes in group II, and four clades, namely AT5G12180.1 (CPK17), AT5G19360.1 (CPK34), and AT4G04710.1 (CPK22), AT4G23650.1 (CPK3) occupied about 60% of orthologs from Brassica species in this group, reaching up to 59 genes. The group with a minimum number of members was group IV, including only three clades with a total of 32 genes. In group IV, clades AT5g66210.2 (CPK28), AT2G17890.1 (CPK16), and AT4G36070.2 (CPK18) possessed 14, 11, and 7 members respectively. In addition, some clades had very few members, such as AT4G04700.1 (CPK27), AT4G04695.1 (CPK31), AT2G35890.1 (CPK25), and AT1G50700.1 (CPK33) merely having one or two orthologs from Brassica species.
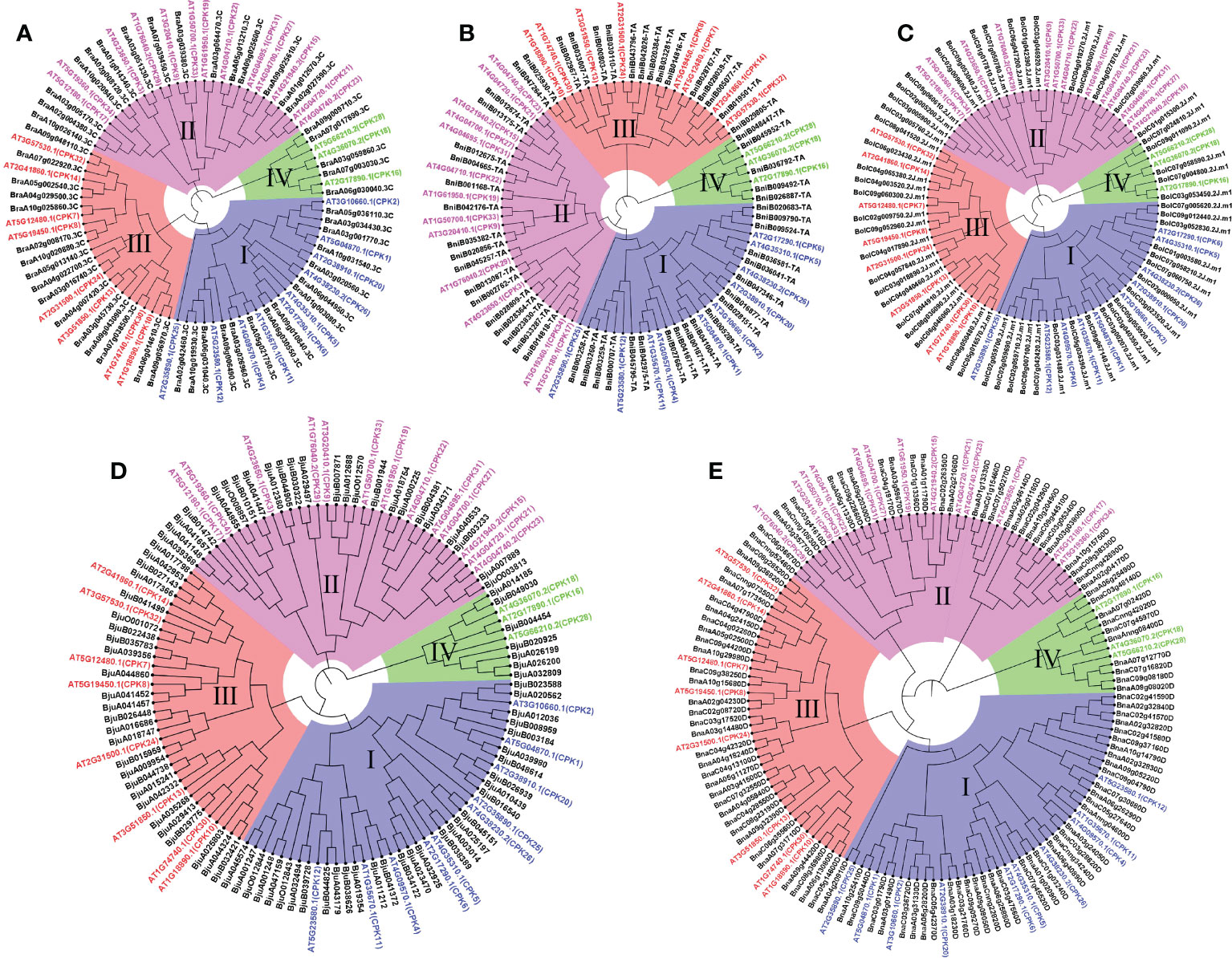
Figure 3 Phylogenetic trees of CPK proteins in five Brassica species. Phylogenetic trees of CPK proteins in A. thaliana and B. rapa (A), B. nigra (B), B. oleracea (C), B. juncea (D), B. napus (E), respectively. The phylogenetic trees were generated by the NJ method with bootstrap analysis (2,000 bootstrap replicates) using an amino acid sequence alignment of CPK proteins from A. thaliana and Brassica species by the MEGA 11.0 program. The CPK proteins in each Brassica species were clustered into four groups (group I, group I, group III, and group IV).
Analysis of conserved motifs in BraCPK, BolCPK, and BnaCPK
It was found that some CPKs duplicated or lacked certain types of motifs. Most CPKs consisted of 9 to 11 motifs, and covered all motif types (Table S1; Figures S3, S4). As shown in Figure 4, 39 of 51 CPKs in B. rapa, 45 of 56 in B. oleracea, and 78 of 107 in B. napus include ten types of motifs, the proportions of which were up to 76.5%, 80.4%, and 72.90%, respectively. The top three types of motifs were motif 4/1/7 in B. rapa, motif 8/6/4 in B. oleracea, and motif 8/4/1 in B. napus. Further analysis revealed that the composition and the number of conserved motifs or domains varied to a large extent in some CPKs (Figure S3). For example, BnaA02g32830D, BnaA03g46140D, BnaA05g11330D, and BraA09g025610 lacked three to five types of motifs,while BnaC09g22660D had the highest motif number of 15. In addition, some motif types in BraA02g042460 were tripled. Compared to BraCPKs and BolCPKs, BnaCPK proteins showed more diverse combinations of the motifs.
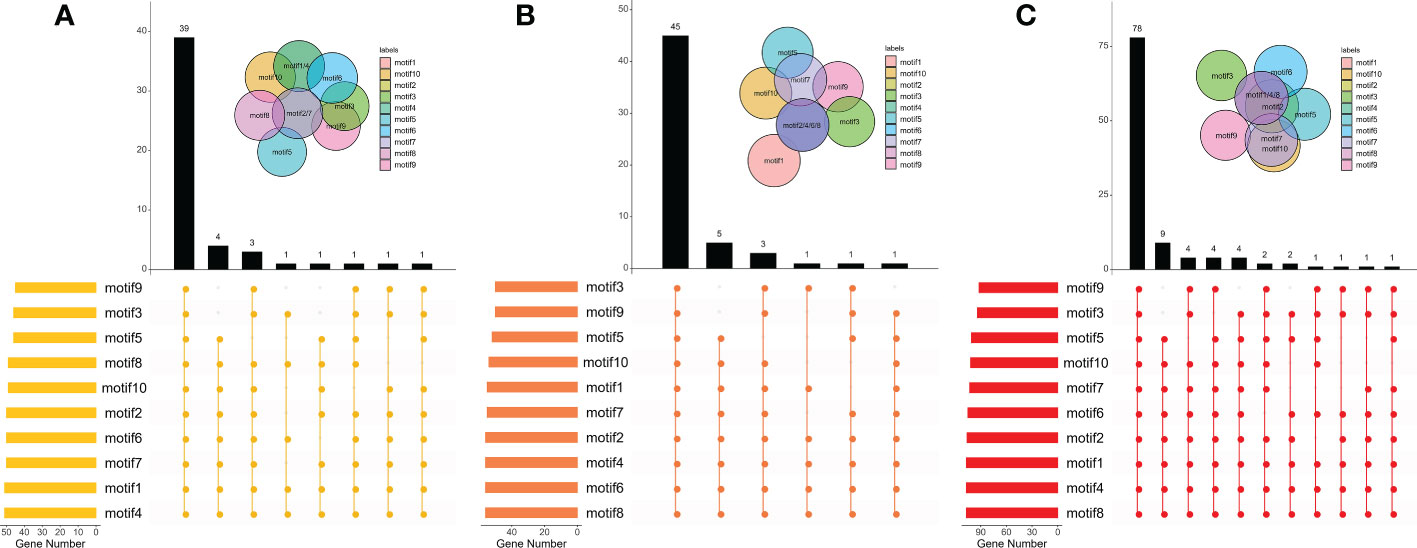
Figure 4 Distributions of motifs in BraCPKs (A), BolCPKs (B), and BnaCPKs (C). The Upset plot shows the distribution of motifs in the CPKs. The number chart above represents the number of genes contained in each type of CPK. The bar chart at the bottom left represents the number of genes included in each type of motif. The dotted line shows the types of motifs contained in the group.
Analysis of cis-acting elements in the promoter region of BraCPKs, BolCPKs, and BnaCPKs
Cis-acting elements distributed in the promoter region can help predict the function of candidate genes. As most CPK genes are related to stress response, we further analyzed the cis-acting elements involved in stress response. Specifically, cis-acting elements are classified as MeJA responsive, gibberellin responsive, abscisic acid responsive, drought inducibility, salicylic acid responsive, and defense and stress responsive related (Table S1; Figure 5 and Figure S5). Among them, BnaCPKs, BraCPKs, and BolCPKs contained the largest number of cis-acting elements related to abscisic acid responsivity, followed by MeJA or gibberellin responsivity (Figure 5). Most genes had three or more promoter elements, but a few genes possessed only one type of cis-acting element, such as six BnaCPK genes (BnaA02g32840D, BnaA03g31330D, BnaA05g11330D, BnaA05g28200D, BnaC05g42370D and BnaC09g04790D), two BraCPK genes (BraA06g030040 and BraA09g009710), and three BolCPK genes (BolC03g031480, BolC06g047200 and BolC07g044910) (Figure S5), and this presence of single type may give them corresponding functional specificities. In addition, some CPK genes contained no promoter elements, which may have a certain impact on gene expressions, such as BnaA04g24150D, BnaC02g41590D, and BolC02g059710, which may not be involved in plants responding to stress.
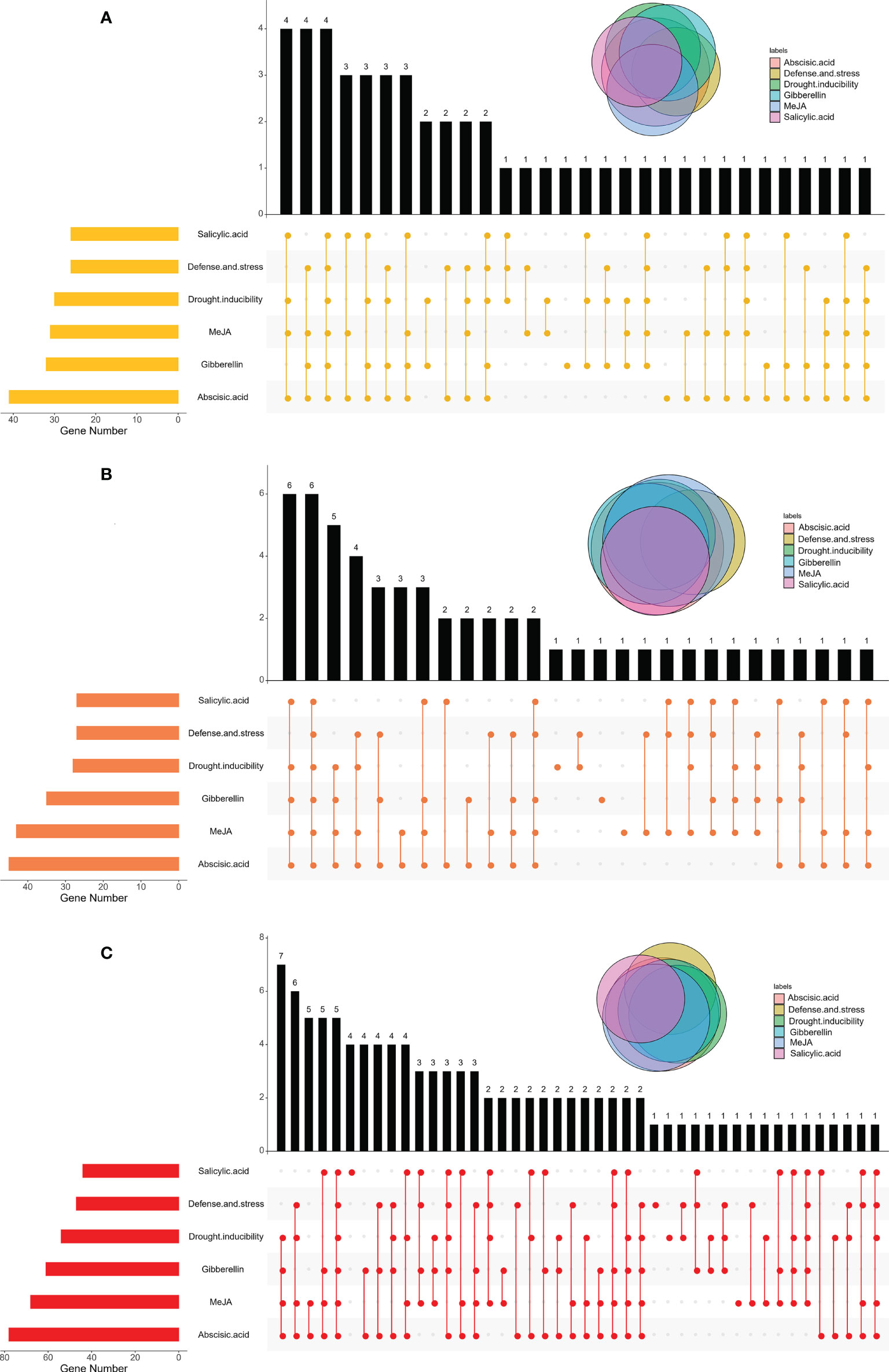
Figure 5 Distributions of cis-acting elements in BraCPKs (A), BolCPKs (B), and BnaCPKs (C) promoter. The Upset plot shows the distribution of cis-acting elements in CPKs promoter. The bar chart above represents the number of genes contained in each type of CPK. The bar chart at the bottom left represents the number of genes included in each type of cis-acting element. The dotted line shows the types of cis-acting elements contained in the group.
Expression pattern of CPK genes induced by Plasmodiophora brassicae
Plasmodiophora brassicae Wor., an obligate and biotrophic pathogen Rhizaria (Schwelm et al., 2015), can infect over 3,700 species in Brassicaceae (Hwang et al., 2012), and lead to club-root, causing significant economic losses every year (Dixon, 2009). To further reveal the roles of CPKs in P. brassicae defense responses, we analyzed the corresponding expression profile data available online. Three RNA-seq datasets were downloaded and analyzed (Chen et al., 2015; Zhang et al., 2016; Li et al., 2019). In total, 43 of 51 BraCPK, 49 of 56 BolCPK, and 100 of 107 BnaCPK genes showed expressional fluctuation after inoculation in resistant (R) and susceptible (S) plants (Figure S6). The expression of most CPK genes were down-regulated, and only 4 BraCPK, 14 BolCPK and 31 BnaCPK genes were induced by P. brassicae in resistant (R) or susceptible (S) materials at a certain time point (Figure 6). Among them, most CPKs presented expression specificity between R and S, such as 18 CPKs that were induced merely in R, including 13 BnaCPKs (BnaAnng08400D, BnaC03g05340D, BnaA03g03800D, BnaC04g19170D, BnaC03g41610D, BnaCnng52460D, BnaC03g47900D, BnaC02g08720D, BnaA09g32390D, BnaA06g13080D, BnaA02g32830D, BnaC07g45520D, BnaC09g5044D), one BraCPK (BraA05g013140.3C), and four BolCPKs (BolC02g005200.2J.m1, BolC04g018270.2J.m1, BolC03g018890.2J.m1, BolC08g036090.2J.m1). Additionally, 17 CPKs were only induced in S, including eight BnaCPKs (BnaA10g20490D, BnaA02g01160D, BnaA01g11790D, BnaA03g59670D, BnaA09g20300D, BnaC02g41580D, BnaA06g40890D, BnaA03g18230D), one BraCPK (BraA03g020560.3C), and eight BolCPKs (BolC07g024810.2J.m1, BolC09g060610.2J.m1, BolC03g005900.2J.m1, BolC03g005760.2J.m1, BolC01g015300.2J.m1, BolC06g047200.2J.m1, BolC08g041520.2J.m1, BolC05g036390.2J.m1). Some CPKs were highly induced reaching a level of 14.82 (BnaA03g18230D, 96 hpi in S), 11.02/10.31 (BnaC02g08720D, 12 hpi/60 hpi in R), and 10.31 (BolC09g060610.2J.m1, 14 dpi in S) times comparing to their initial concentrations. Apart from BnaC02g08720D, other R-specific induced genes, like BnaA03g03800D (6.06 folds in 12 hpi, 5.26 folds in 96 hpi) and BolC04g018270.2J.m1 (5.4 folds in 7 dpi, 5.9 folds in 14 dpi) might also be good candidates for P. brassicae resistance research. Moreover, in the phylogenetic tree built by CPKs from Arabidopsis and five Brassica species, up-regulated genes were distributed in all groups (Figure 7). It is remarkable that the majority of BraCPK, BolCPK, and BnaCPK genes in two families, orthologs of AT5G66210.2 (CPK28) and AT5G12180.1 (CPK17), were up-regulated after P. brassicae inoculation, underlining their functional conservation during evolution.
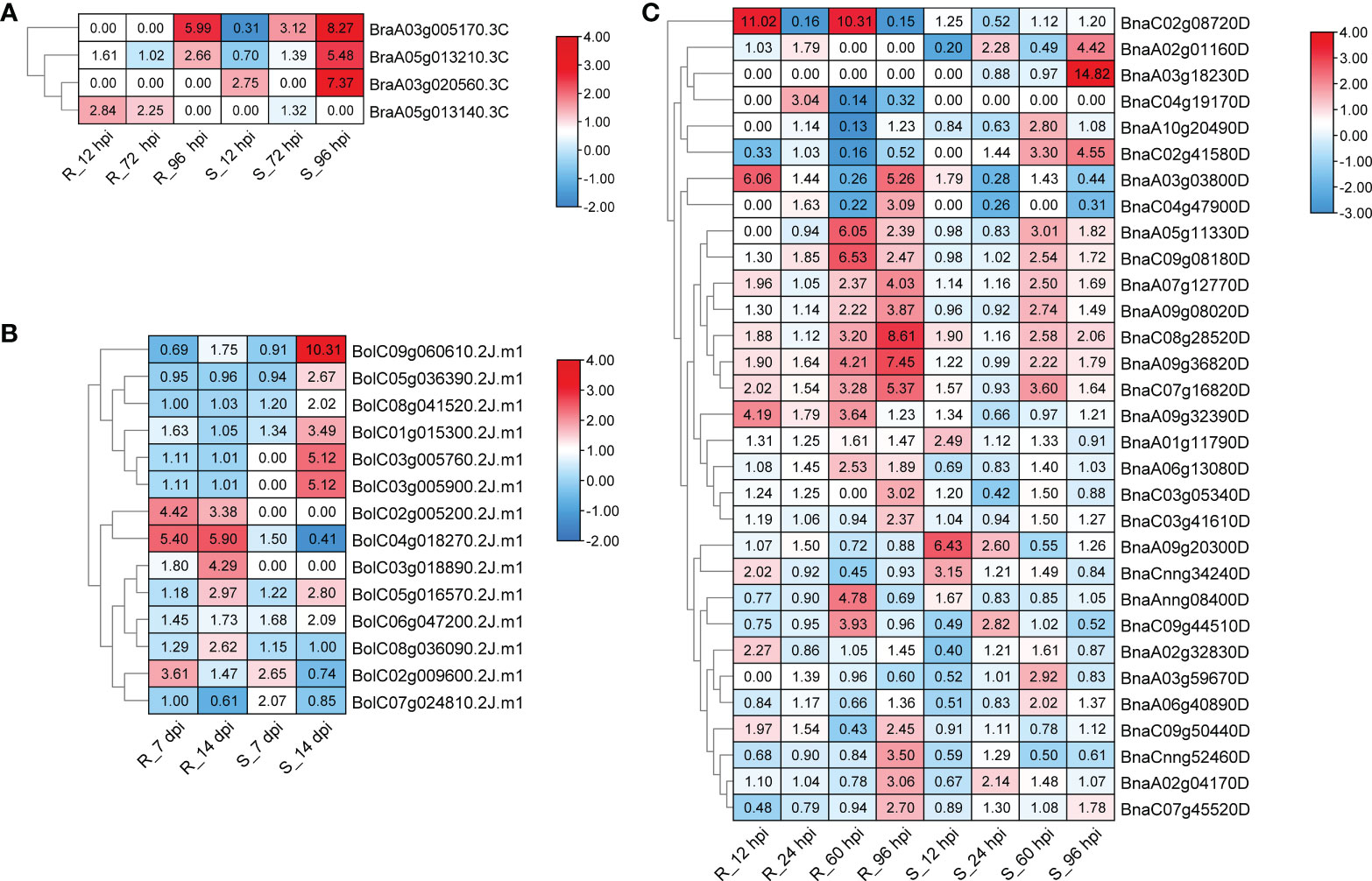
Figure 6 Heat maps of upregulated BraCPKs (A) , BolCPKs (B), and BnaCPKs (C) elicited by P. brassicae. R and S, resistant and susceptible genotype, respectively. hpi: hours post-inoculation, dpi: days post-inoculation. The red color indicates up-regulated, and numbers in boxes indicate fold change. The legends show log2 fold-change. TPM value was used for heat map construction.
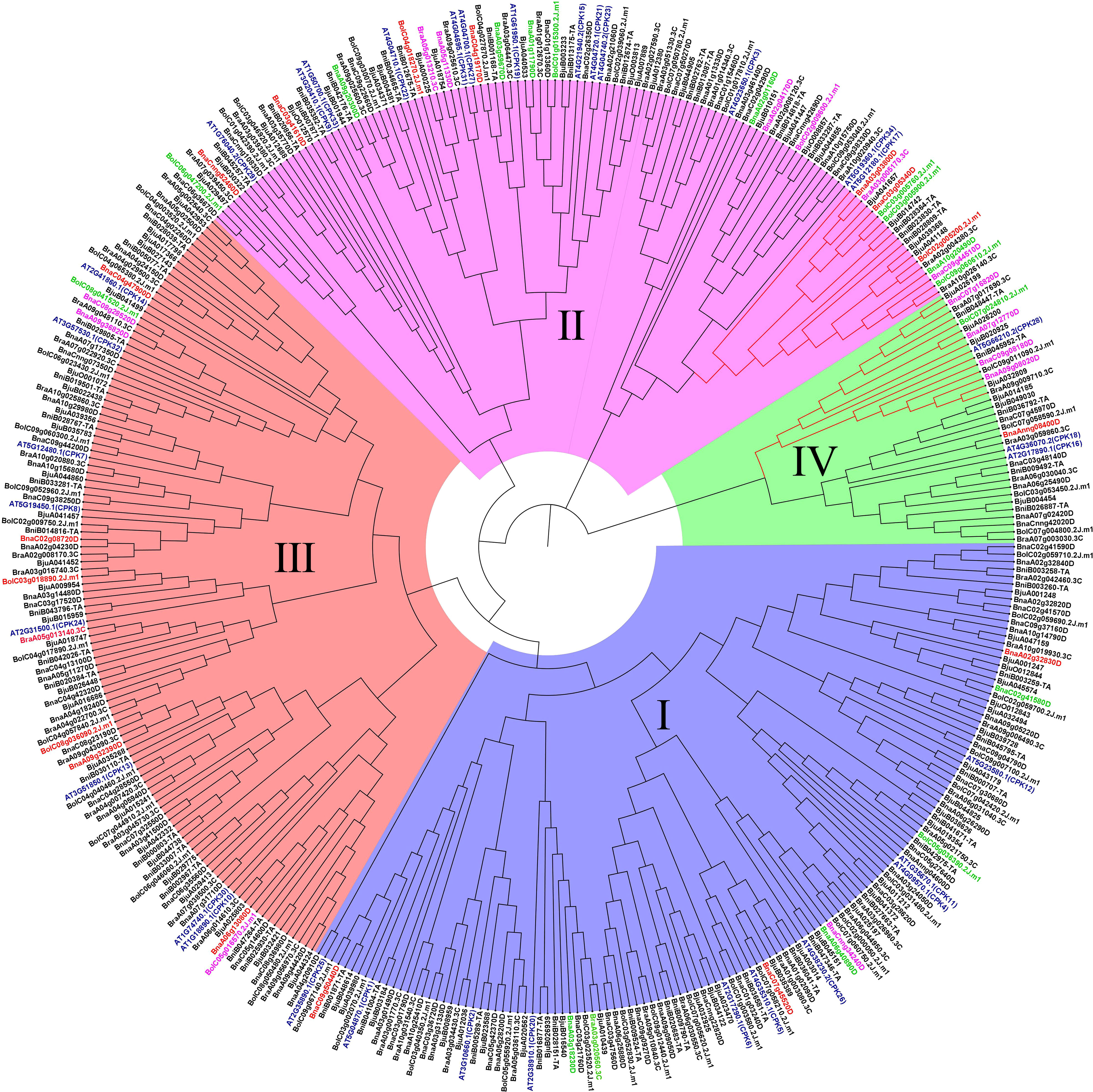
Figure 7 Phylogenetic tree of CPK proteins of A. thaliana and five Brassica species. The phylogenetic tree was generated by the NJ method with bootstrap analysis (2,000 bootstrap replicates) by the MEGA 11.0. The CPK proteins were clustered into four groups (group I, group I, group III, and group IV). Blue mark AthCPK genes, red mark upregulated-CPKs in resistance (R) genotype only, green mark upregulated-CPKs in susceptible (S) genotype only, pink mark upregulated-CPKs both resistance (R) and susceptible (S) genotypes. Red lines mark At5G12180.1 (CPK17) and AT5G66210.3 (CPK28) clades.
Discussion
B. napus (AACC, 2n = 38) and B. juncea (AABB, 2n = 36) are both allopolypoid species formed by the hybridization of two diploid species. B.napus is formed through the hybridrization of B.rapa (AA = 20) and B.oleracea (CC = 18), while the formation of B. juncea occurs through the hybridization of B.rapa (AA = 20) and B. nigra (BB = 16) (Chalhoub et al., 2014; Lu et al., 2019). In our phylogenetic tree, the division of groups was consistent with that of A. thaliana (Cheng et al., 2002). The evolution of CPK genes was relatively conserved among the five Brassica species. In general, the gene number of BraCPKs was almost equal to that of BolCPKs and BniCPKs in the same clade, with duplicated number for BnaCPKs and BjuCPKs. Nevertheless, in terms of individual branches, the quantity of BjuCPK genes was less than two times of that of BraCPKs and BniCPKs, which might be due to inaccurate assembly occurring during genetic recombination or imperfect sequencing. When selecting Arabidopsis CPKs as a reference, there was no doubt that the gene expansion occurred among five species during evolution, especially in some clades, such as Clade AT5G23580.1(CPK12) and AT2G31500.1(CPK24). Of course, the possibility that certain Arabidopsis CPK genes have been lost during evolution cannot be ruled out. As the structure of exons/introns and the type and number of introns can account for the evolutionary history of organisms, we analyzed the exon/introns substructure of all CPKs in this study. Genes in group IV had the most exons, from eleven to thirteen, not including BjuA026200. Most of the CPK genes in other groups contained less than ten exons. In general, CPK genes clustered in the same subfamily showed similar exon/intron structures, indicating a close evolutionary relationship between them.
At present, CPK genes have been identified in Arabidopsis (Cheng et al., 2002), rice (Asano et al., 2005), soybean (Liu et al., 2016), maize (Kong et al., 2013; Ma et al., 2013), cucumber (Xu et al., 2015), tomato (Hu et al., 2016) and many other plants (Yang et al., 2017; Wen et al., 2020; Zhang et al., 2020; Zhao et al., 2021), and their roles in biotic and abiotic stresses have been understood to some extent (Asano et al., 2012; Boudsocq and Sheen, 2013; Ranty et al., 2016; Bredow and Monaghan, 2019). Previous studies found that plant CPKs are involved in abscisic acid signaling pathways (Liu et al., 2006, 1; Yu et al., 2006; Zhu et al., 2007; Jiang et al., 2013; Chen et al., 2019), jasmonic acid signaling pathway (Ulloa et al., 2002; Hettenhausen et al., 2013; Matschi et al., 2015), gibberellin signaling pathway (Abo-el-Saad and Wu, 1995; Abbasi et al., 2004; Ishida et al., 2008; Nakata et al., 2009), and also response to drought stress (Ma and Wu, 2007; Cieśla et al., 2016, 2; Bundó and Coca, 2017; Huang et al., 2018). In line with these results, a large number of cis-acting elements related to abscisic acid, jasmonic acid, gibberellin, and drought stress response were found in the promoter region of Brassica CPK genes in this study.
Plasmodiophora, as one class of obligate biotrophic pathogens (Ludwig-Müller et al., 2009), is highly destructive to many Brassica plants and exerts negative effect on their quality and yield (Dixon, 2009), which seriously hinders the development of agricultural industry. In this study, utilizing the expression data from the resistant (R) and susceptible (S) lines in three Brassica species (B. rapa, B. oleracea, and B. napus), we found 49 up-regulated genes induced by P. brassicae. These up-regulated genes would be helpful to identify positive regulators in the process of response to P. brassicae (Figure 6). Interestingly, the distribution of these 49 genes was extremely uneven among the three Brassica species. Among them, 31 genes were found in B. napus, accounting for 29% of BnaCPKs, while only 4 genes were found in B. rapa, with a proportion of 8%, suggesting that BnaCPKs played an important role in B. napus taking charge in response to P. brassicae infection, while BraCPKs might be mainly involved in other stresses. Moreover, of the 49 up-regulated genes, 18 genes were specifically up-regulated in R, and those genes identified from incompatible interactions might be good candidates for further functional assignment of CPK genes against P.brassicae infection. The up-regulated genes of the three species were distributed in the four groups in varying amounts (Figure 7). There were 9 CPK genes in group I, 23 in group I, 11 in group III, and only 6 in group IV, suggesting that CPK genes involved in P.brassicae response evolved independently in four groups along with the functional divergence in Brassica species. However, the genes in group I may play a broader role in response to P.brassicae infection. It is worth noting that more genes in clade AT5G12180.1 (CPK17) and clade AT5G6621.2 (CPK28) were up-regulated by P.brassicae induction. Evolutionarily, these two clades of genes were functionally conserved, and most likely to be active gene clusters during response to P.brassicae infection in Brassica species.
Conclusion
The gene and protein characterization, localization, evolution, and expression analysis of highly conserved CPK genes in Brassica showed that most of the CPK gene families in this study were relatively conserved during evolution. Genome-wide identification and expression analysis of CPK family members can provide an alternative strategy for enhancing resistance to P. brassicae in Brassica species. These identified genes may be good candidates for further identification of CPK genes of P. brassicae infection and provide theoretical basis and preliminary guidance for prevention of P. brassicae pathogen damage to Brassica plants.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
JL: formal analysis, writing the original draft, data curation, and validation. NY: investigation, resources, and visualization. YZ: data curation. ZC: data validation. TZ: writing-review and editing, and supervision. WL: conceptualization, writing-review and editing, and supervision. All authors contributed to the article and approved the submitted version. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the Chongqing Municipal Education Commission (KJQN201900533) to JL, the National Natural Science Foundation of China (31171588) and the Chongqing Municipal Education Commission (KJZD-K202200508) to TZ, and Chongqing Bureau of Human Resources and Social Security Postdoctoral Funding (0019) to WL. JL and WL were partly supported by fellowships from China Scholarship Council (CSC).
Acknowledgments
We sincerely thank Athena Li from the University of British Columbia for the critical reading of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1067723/full#supplementary-material
Supplementary Figure 1 | Gene structures of CPK genes in five Brassica species: (A) BraCPK, (B) BniCPK, (C)BolCPK, (D)BjuCPK, (E)BnaCPK genes. The green bars represent CDS, the black lines represent intron, and the yellow bar represents UTR.
Supplementary Figure 2 | The conserved domains of CPK proteins in five Brassica species. (A)BraCPKs, (B)BniCPKs, (C) BolCPKs, (D)BjuCPKs, (E)BnaCPKs. The green bar represents protein kinase domain (Pkinase), and the yellow bar represents EF-hand domain (EFh).
Supplementary Figure 3 | The motifs in CPK proteins of B. rapa (A), B. oleracea (B), and B. napus (C). Motif labeled as motif 1 - motif 10.
Supplementary Figure 4 | The sequence of motif 1 - motif 10 of B. rapa, B. oleracea, and B. napus.
Supplementary Figure 5 | The cis-acting elements in CPKs promoter of B. rapa (A), B. oleracea (B), and B. napus (C). Approximately 2,000-bp upstream flanking fragments of the CPK genes were derived, and PlantCARE was used to predict promoter cis-elements.
Supplementary Figure 6 | The expression heat map of CPK genes in B. rapa (A), B. oleracea (B), and B. napus (C) elicited by P. brassicae. R and S, resistant and susceptible genotypes, respectively. hpi: hours post-inoculation, dpi: days post-inoculation. The red color indicates up-regulated, and blue color indicates down-regulated. The legends show log2 fold-change. TPM value was used for heat map construction.
References
Abbasi, F., Onodera, H., Toki, S., Tanaka, H., Komatsu, S. (2004). OsCDPK13, a calcium-dependent protein kinase gene from rice, is induced by cold and gibberellin in rice leaf sheath. Plant Mol. Biol. 55, 541–552. doi: 10.1007/s11103-004-1178-y
Abo-el-Saad, M., Wu, R. (1995). A rice membrane calcium-dependent protein kinase is induced by gibberellin. Plant Physiol. 108, 787–793. doi: 10.1104/pp.108.2.787
Asano, T., Hayashi, N., Kikuchi, S., Ohsugi, R. (2012). CDPK-mediated abiotic stress signaling. Plant Signal Behav. 7, 817–821. doi: 10.4161/psb.20351
Asano, T., Tanaka, N., Yang, G., Hayashi, N., Komatsu, S. (2005). Genome-wide identification of the rice calcium-dependent protein kinase and its closely related kinase gene families: Comprehensive analysis of the CDPKs gene family in rice. Plant Cell Physiol. 46, 356–366. doi: 10.1093/pcp/pci035
Bailey, T. L., Johnson, J., Grant, C. E., Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Bi, Z., Wang, Y., Li, P., Sun, C., Qin, T., Bai, J. (2021). Evolution and expression analysis of CDPK genes under drought stress in two varieties of potato. Biotechnol. Lett. 43, 511–521. doi: 10.1007/s10529-020-03037-2
Boudsocq, M., Sheen, J. (2013). CDPKs in immune and stress signaling. Trends Plant Sci. 18, 30–40. doi: 10.1016/j.tplants.2012.08.008
Bredow, M., Monaghan, J. (2019). Regulation of plant immune signaling by calcium-dependent protein kinases. Mol. Plant Microbe Interact. 32, 6–19. doi: 10.1094/MPMI-09-18-0267-FI
Bundó, M., Coca, M. (2017). Calcium-dependent protein kinase OsCPK10 mediates both drought tolerance and blast disease resistance in rice plants. J. Exp. Bot. 68, 2963–2975. doi: 10.1093/jxb/erx145
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A. P., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-neolithic brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cheng, S.-H., Willmann, M. R., Chen, H.-C., Sheen, J. (2002). Calcium signaling through protein kinases. the arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 129, 469–485. doi: 10.1104/pp.005645
Chen, D.-H., Liu, H.-P., Li, C.-L. (2019). Calcium-dependent protein kinase CPK9 negatively functions in stomatal abscisic acid signaling by regulating ion channel activity in arabidopsis. Plant Mol. Biol. 99, 113–122. doi: 10.1007/s11103-018-0805-y
Chen, J., Pang, W., Chen, B., Zhang, C., Piao, Z. (2015). Transcriptome analysis of brassica rapa near-isogenic lines carrying clubroot-resistant and -susceptible alleles in response to plasmodiophora brassicae during early infection. Front. Plant Sci. 6 1183. doi: 10.3389/fpls.2015.01183
Chen, H., Wang, T., He, X., Cai, X., Lin, R., Liang, J., et al. (2022). BRAD V3.0: an upgraded brassicaceae database. Nucleic Acids Res. 50, D1432–D1441. doi: 10.1093/nar/gkab1057
Chica, R. A. (2020). Protein engineering, design and selection. Protein Engineering Design Selection 33, gzaa024. doi: 10.1093/protein/gzaa024
Cieśla, A., Mituła, F., Misztal, L., Fedorowicz-Strońska, O., Janicka, S., Tajdel-Zielińska, M., et al. (2016). A role for barley calcium-dependent protein kinase CPK2a in the response to drought. Front. Plant Sci. 7 1550. doi: 10.3389/fpls.2016.01550
Dixon, G. R. (2009). The occurrence and economic impact of plasmodiophora brassicae and clubroot disease. J. Plant Growth Regul. 28, 194–202. doi: 10.1007/s00344-009-9090-y
Draper, J. M., Xia, Z., Smith, C. D. (2007). Cellular palmitoylation and trafficking of lipidated peptides. J. Lipid Res. 48, 1873–1884. doi: 10.1194/jlr.M700179-JLR200
Greaves, J., Chamberlain, L. H. (2007). Palmitoylation-dependent protein sorting. J. Cell Biol. 176, 249–254. doi: 10.1083/jcb.200610151
Harmon, A. C., Gribskov, M., Harper, J. F. (2000). CDPKs – a kinase for every Ca2+ signal? Trends Plant Sci. 5, 154–159. doi: 10.1016/S1360-1385(00)01577-6
Hetherington, A. M., Brownlee, C. (2004). THE GENERATION OF Ca2+ SIGNALS IN PLANTS. Annu. Rev. Plant Biol. 55, 401–427. doi: 10.1146/annurev.arplant.55.031903.141624
Hettenhausen, C., Yang, D.-H., Baldwin, I. T., Wu, J. (2013). Calcium-dependent protein kinases, CDPK4 and CDPK5, affect early steps of jasmonic acid biosynthesis in nicotiana attenuata. Plant Signal Behav. 8, e22784. doi: 10.4161/psb.22784
Hrabak, E. M., Chan, C. W. M., Gribskov, M., Harper, J. F., Choi, J. H., Halford, N., et al. (2003). The arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 132, 666–680. doi: 10.1104/pp.102.011999
Huang, K., Peng, L., Liu, Y., Yao, R., Liu, Z., Li, X., et al. (2018). Arabidopsis calcium-dependent protein kinase AtCPK1 plays a positive role in salt/drought-stress response. Biochem. Biophys. Res. Commun. 498, 92–98. doi: 10.1016/j.bbrc.2017.11.175
Hu, Z., Lv, X., Xia, X., Zhou, J., Shi, K., Yu, J., et al. (2016). Genome-wide identification and expression analysis of calcium-dependent protein kinase in tomato. Front. Plant Sci. 7, 469. doi: 10.3389/fpls.2016.00469
Hwang, S.-F., Strelkov, S. E., Feng, J., Gossen, B. D., Howard, R. J. (2012). Plasmodiophora brassicae: A review of an emerging pathogen of the Canadian canola (Brassica napus) crop. Mol. Plant Pathol. 13, 105–113. doi: 10.1111/j.1364-3703.2011.00729.x
Ishida, S., Yuasa, T., Nakata, M., Takahashi, Y. (2008). A tobacco calcium-dependent protein kinase, CDPK1, regulates the transcription factor REPRESSION OF SHOOT GROWTH in response to gibberellins. Plant Cell 20, 3273–3288. doi: 10.1105/tpc.107.057489
Jiang, S., Zhang, D., Wang, L., Pan, J., Liu, Y., Kong, X., et al. (2013). A maize calcium-dependent protein kinase gene, ZmCPK4, positively regulated abscisic acid signaling and enhanced drought stress tolerance in transgenic arabidopsis. Plant Physiol. Biochem. 71, 112–120. doi: 10.1016/j.plaphy.2013.07.004
Kiemer, L., Bendtsen, J. D., Blom, N. (2005). NetAcet: Prediction of n-terminal acetylation sites. Bioinformatics 21, 1269–1270. doi: 10.1093/bioinformatics/bti130
Kong, X., Lv, W., Jiang, S., Zhang, D., Cai, G., Pan, J., et al. (2013). Genome-wide identification and expression analysis of calcium-dependent protein kinase in maize. BMC Genomics 14, 433. doi: 10.1186/1471-2164-14-433
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Bork, P. (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496. doi: 10.1093/nar/gkx922
Letunic, I., Khedkar, S., Bork, P. (2021). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460. doi: 10.1093/nar/gkaa937
Li, L., Long, Y., Li, H., Wu, X. (2019). Comparative transcriptome analysis reveals key pathways and hub genes in rapeseed during the early stage of plasmodiophora brassicae infection. Front. Genet. 10, 1075. doi: 10.3389/fgene.2019.01275
Linder, M. E., Deschenes, R. J. (2007). Palmitoylation: policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 8, 74–84. doi: 10.1038/nrm2084
Liu, G., Chen, J., Wang, X. (2006). VfCPK1, a gene encoding calcium-dependent protein kinase from vicia faba, is induced by drought and abscisic acid. Plant Cell Environ. 29, 2091–2099. doi: 10.1111/j.1365-3040.2006.01582.x
Liu, H., Che, Z., Zeng, X., Zhou, X., Sitoe, H. M., Wang, H., et al. (2016). Genome-wide analysis of calcium-dependent protein kinases and their expression patterns in response to herbivore and wounding stresses in soybean. Funct. Integr. Genomics 16, 481–493. doi: 10.1007/s10142-016-0498-8
Luan, S., Kudla, J., Rodriguez-Concepcion, M., Yalovsky, S., Gruissem, W. (2002). Calmodulins and calcineurin b-like proteins: calcium sensors for specific signal response coupling in plants. Plant Cell 14 Suppl, S389–S400. doi: 10.1105/tpc.001115
Ludwig-Müller, J., Prinsen, E., Rolfe, S. A., Scholes, J. (2009). Metabolism and plant hormone action during clubroot disease. D. J. Plant Growth Regul. 28, 229–244. doi: 10.1007/s00344-009-9089-4
Lu, K., Wei, L., Li, X., Wang, Y., Wu, J., Liu, M., et al. (2019). Whole-genome resequencing reveals brassica napus origin and genetic loci involved in its improvement. Nat. Commun. 10, 1154. doi: 10.1038/s41467-019-09134-9
Ma, P., Liu, J., Yang, X., Ma, R. (2013). Genome-wide identification of the maize calcium-dependent protein kinase gene family. Appl. Biochem. Biotechnol. 169, 2111–2125. doi: 10.1007/s12010-013-0125-2
Martín, M. L., Busconi, L. (2000). Membrane localization of a rice calcium-dependent protein kinase (CDPK) is mediated by myristoylation and palmitoylation. Plant J. 24, 429–435. doi: 10.1046/j.1365-313x.2000.00889.x
Matschi, S., Hake, K., Herde, M., Hause, B., Romeis, T. (2015). The calcium-dependent protein kinase CPK28 regulates development by inducing growth phase-specific, spatially restricted alterations in jasmonic acid levels independent of defense responses in arabidopsis. Plant Cell 27, 591–606. doi: 10.1105/tpc.15.00024
Ma, S.-Y., Wu, W.-H. (2007). AtCPK23 functions in arabidopsis responses to drought and salt stresses. Plant Mol. Biol. 65, 511–518. doi: 10.1007/s11103-007-9187-2
Nakata, M., Yuasa, T., Takahashi, Y., Ishida, S. (2009). CDPK1, a calcium-dependent protein kinase, regulates transcriptional activator RSG in response to gibberellins. Plant Signal Behav. 4, 372–374. doi: 10.4161/psb.4.5.8229
Ranty, B., Aldon, D., Cotelle, V., Galaud, J.-P., Thuleau, P., Mazars, C. (2016). Calcium sensors as key hubs in plant responses to biotic and abiotic stresses. Front. Plant Sci. 7, 327. doi: 10.3389/fpls.2016.00327
Ren, J., Wen, L., Gao, X., Jin, C., Xue, Y., Yao, X. (2008). CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng. Des. Sel 21, 639–644. doi: 10.1093/protein/gzn039
Schwelm, A., Fogelqvist, J., Knaust, A., Jülke, S., Lilja, T., Bonilla-Rosso, G., et al. (2015). The plasmodiophora brassicae genome reveals insights in its life cycle and ancestry of chitin synthases. Sci. Rep. 5, 11153. doi: 10.1038/srep11153
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Ulloa, R. M., Raíces, M., MacIntosh, G. C., Maldonado, S., Téllez-Iñón, M. T. (2002). Jasmonic acid affects plant morphology and calcium-dependent protein kinase expression and activity in solanum tuberosum. Physiol. Plant 115, 417–427. doi: 10.1034/j.1399-3054.2002.1150312.x
Urao, T., Katagiri, T., Mizoguchi, T., Yamaguchi-Shinozaki, K., Hayashida, N., Shinozaki, K. (1994). Two genes that encode Ca2+-dependent protein kinases are induced by drought and high-salt stresses in arabidopsis thaliana. Molec. Gen. Genet. 244, 331–340. doi: 10.1007/BF00286684
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Wang, Q., Yin, X., Chen, Q., Xiang, N., Sun, X., Yang, Y., et al. (2017). Genome-wide survey indicates diverse physiological roles of the turnip (Brassica rapa var. rapa) calcium-dependent protein kinase genes. Sci. Rep. 7, 15803. doi: 10.1038/s41598-017-16102-0
Wen, F., Ye, F., Xiao, Z., Liao, L., Li, T., Jia, M., et al. (2020). Genome-wide survey and expression analysis of calcium-dependent protein kinase (CDPK) in grass brachypodium distachyon. BMC Genomics 21, 53. doi: 10.1186/s12864-020-6475-6
Xu, X., Liu, M., Lu, L., He, M., Qu, W., Xu, Q., et al. (2015). Genome-wide analysis and expression of the calcium-dependent protein kinase gene family in cucumber. Mol. Genet. Genomics 290, 1403–1414. doi: 10.1007/s00438-015-1002-1
Yang, Y., Wang, Q., Chen, Q., Yin, X., Qian, M., Sun, X., et al. (2017). Genome-wide survey indicates diverse physiological roles of the barley (Hordeum vulgare l.) calcium-dependent protein kinase genes. Sci. Rep. 7, 5306. doi: 10.1038/s41598-017-05646-w
Yang, H., You, C., Yang, S., Zhang, Y., Yang, F., Li, X., et al. (2021). The role of Calcium/Calcium-dependent protein kinases signal pathway in pollen tube growth. Front. Plant Sci. 12, 633293. doi: 10.3389/fpls.2021.633293
Yu, X.-C., Li, M.-J., Gao, G.-F., Feng, H.-Z., Geng, X.-Q., Peng, C.-C., et al. (2006). Abscisic acid stimulates a calcium-dependent protein kinase in grape berry. Plant Physiol. 140, 558–579. doi: 10.1104/pp.105.074971
Zhang, Z. (2022). KaKs_calculator 3.0: Calculating selective pressure on coding and non-coding sequences. Genomics Proteomics Bioinf. S1672-0229 (21), 00259–0025X. doi: 10.1016/j.gpb.2021.12.002
Zhang, L., Du, L., Poovaiah, B. W. (2014b). Calcium signaling and biotic defense responses in plants. Plant Signal Behav. 9, e973818. doi: 10.4161/15592324.2014.973818
Zhang, K., Han, Y.-T., Zhao, F.-L., Hu, Y., Gao, Y.-R., Ma, Y.-F., et al. (2015). Genome-wide identification and expression analysis of the CDPK gene family in grape, vitis spp. BMC Plant Biol. 15, 164. doi: 10.1186/s12870-015-0552-z
Zhang, X., Liu, Y., Fang, Z., Li, Z., Yang, L., Zhuang, M., et al. (2016). Comparative transcriptome analysis between broccoli (Brassica oleracea var. italica) and wild cabbage (Brassica macrocarpa guss.) in response to plasmodiophora brassicae during different infection stages. Front. Plant Sci. 7, 1929. doi: 10.3389/fpls.2016.01929
Zhang, M., Liu, Y., He, Q., Chai, M., Huang, Y., Chen, F., et al. (2020). Genome-wide investigation of calcium-dependent protein kinase gene family in pineapple: evolution and expression profiles during development and stress. BMC Genomics 21, 72. doi: 10.1186/s12864-020-6501-8
Zhang, H., Liu, W.-Z., Zhang, Y., Deng, M., Niu, F., Yang, B., et al. (2014a). Identification, expression and interaction analyses of calcium-dependent protein kinase (CPK) genes in canola (Brassica napus l.). BMC Genomics 15, 211. doi: 10.1186/1471-2164-15-211
Zhao, P., Liu, Y., Kong, W., Ji, J., Cai, T., Guo, Z. (2021). Genome-wide identification and characterization of calcium-dependent protein kinase (CDPK) and CDPK-related kinase (CRK) gene families in medicago truncatula. Int. J. Mol. Sci. 22, 1044 doi: 10.3390/ijms22031044
Keywords: Brassica species, calcium-dependent protein kinase, evolution, expression pattern, Plasmodiophora brassicae
Citation: Lu J, Yang N, Zhu Y, Chai Z, Zhang T and Li W (2022) Genome-wide survey of Calcium-Dependent Protein Kinases (CPKs) in five Brassica species and identification of CPKs induced by Plasmodiophora brassicae in B. rapa, B. oleracea, and B. napus. Front. Plant Sci. 13:1067723. doi: 10.3389/fpls.2022.1067723
Received: 12 October 2022; Accepted: 03 November 2022;
Published: 21 November 2022.
Edited by:
Cunmin Qu, Southwest University, ChinaReviewed by:
Bingxian Yang, Zhejiang Sci-Tech University, ChinaLunwen Qian, Hunan Agricultural University, China
Copyright © 2022 Lu, Yang, Zhu, Chai, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Li, d2VpbGlAY3FudS5lZHUuY24=; Tao Zhang, emh0MjE4OEAxMjYuY29t
†These authors have contributed equally to this work
 Junxing Lu
Junxing Lu Nan Yang
Nan Yang Yangyi Zhu1
Yangyi Zhu1