
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 14 December 2022
Sec. Crop and Product Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1066820
This article is part of the Research Topic Flowering Time Control in Agricultural and Horticultural Crops, Volume II View all 8 articles
 Zhao Wan1,2
Zhao Wan1,2 Yingxiang Liu3
Yingxiang Liu3 Dandan Guo1
Dandan Guo1 Rong Fan3
Rong Fan3 Yang Liu3
Yang Liu3 Kun Xu1
Kun Xu1 Jinlong Zhu1
Jinlong Zhu1 Le Quan3
Le Quan3 Wentian Lu3
Wentian Lu3 Xi Bai3
Xi Bai3 Hong Zhai1*
Hong Zhai1*The distribution of elite soybean (Glycine max) cultivars is limited due to their highly sensitive to photoperiod, which affects the flowering time and plant architecture. The recent emergence of CRISPR/Cas9 technology has uncovered new opportunities for genetic manipulation of soybean. The major maturity gene E1 of soybean plays a critical role in soybean photoperiod response. Here, we performed CRISPR/Cas9-mediated targeted mutation of E1 gene in soybean cultivar Tianlong1 carrying the dominant E1 to investigate its precise function in photoperiod regulation, especially in plant architecture regulation. Four types of mutations in the E1 coding region were generated. No off-target effects were observed, and homozygous trans-clean mutants without T-DNA were obtained. The photoperiod sensitivity of e1 mutants decreased relative to the wild type plants; however, e1 mutants still responded to photoperiod. Further analysis revealed that the homologs of E1, E1-La, and E1-Lb, were up-regulated in the e1 mutants, indicating a genetic compensation response of E1 and its homologs. The e1 mutants exhibited significant changes in the architecture, including initiation of terminal flowering, formation of determinate stems, and decreased branch numbers. To identify E1-regulated genes related to plant architecture, transcriptome deep sequencing (RNA-seq) was used to compare the gene expression profiles in the stem tip of the wild-type soybean cultivar and the e1 mutants. The expression of shoot identity gene Dt1 was significantly decreased, while Dt2 was significantly upregulated. Also, a set of MADS-box genes was up-regulated in the stem tip of e1 mutants which might contribute to the determinate stem growth habit.
Soybean (Glycine max) is highly sensitive to photoperiod; each soybean cultivar has to be planted in a narrow latitude range to obtain maximum yield, which limits the wide distribution of elite soybean varieties. Soybean has successfully adapted to a wide range of latitudes, attributed to natural variations in several major genes that control flowering and the presence of various allelic combinations of a series of major maturity loci. So far, 14 maturity loci have been identified in soybean, including E1-E11, J, Tof5, Tof11, and Tof12 (Bernard, 1971; Buzzell, 1971; McBlain and Bernard, 1987; Ray et al., 1995; Bonato and Vello, 1999; Cober and Voldeng, 2001; Watanabe et al., 2009; Cober et al., 2010; Xia et al., 2012; Samanfar et al., 2017; Wang et al., 2019; Li et al., 2020; Lu et al., 2020).
Maturity locus E1 was cloned using a map-based approach. It is assumed to be a legume-specific transcription factor with a putative nuclear localization signal and a region distantly related to the B3 domain (Xia et al., 2012). E1 demonstrates a key role in repressing flowering and delaying maturity by repressing the expression of GmMDEs, GmFT2a, and GmFT5a in soybean. The E1 locus is largely responsible for the variation in flowering time among soybean cultivars, and has the most prominent effect on flowering time and photoperiod sensitivity (Bernard, 1971; McBlain and Bernard, 1987; Upadhyay et al., 1994; Abe et al., 2003; Tsubokura et al., 2014). The expression level of the functional E1 gene is strongly associated with the flowering time of soybean cultivars (Zhai et al., 2014b). E3 and E4 have been identified as homologues of the photoreceptor phytochrome A (Liu et al., 2008; Watanabe et al., 2009). The E1 expression is modulated by E3 and E4 loci (Xia et al., 2012). The expansion of soybean cultivation in tropical regions can be attributed to the J locus, which controls the long-juvenile trait. J protein physically associates with the E1 promoter to downregulate its transcription, relieving repression of GmFT2a/5a and promoting flowering under short days (Lu et al., 2017). Time of Flowering 11 (Tof11) and Time of Flowering 12 (Tof12) contributed to changes in flowering and early maturity in soybean crop evolution, demonstrating that their effects on flowering are genetically dependent on E1 (Lu et al., 2020). Time of Flowering 5 (Tof5), which promotes soybean flowering and adaptation to high latitudes, also acts downstream of E1 (Dong et al., 2022). E6 is a novel allele of J (Fang et al., 2021). GmFT2a was identified to be responsible for E9 (Zhao et al., 2016). The FLOWERING LOCUS T orthologue, GmFT4 is a strong candidate causal gene for maturity locus E10, and has been used in a breeding program (Samanfar et al., 2017). GmFT4 functions as a flowering repressor, and is induced by E1 (Zhai et al., 2014a). Another FT-like gene GmFT1a was proven to function as a flowering repressor, which is also induced by E1 (Liu et al., 2018). Collectively, the E1 gene serves as the hub of the photoperiodic responses of soybean, which is like a switch that controls control the photoperiod-dependent flowering.
Recent emergence of clustered regularly interspaced short palindromic repeats/CRISPR associated protein 9 (CRISPR/Cas9) technology has brought new opportunities to plant genetic engineering programs. CRISPR/Cas9 can be employed to make precise modification of genes controlling important agronomic traits (Shan et al., 2013; Xu et al., 2020). Cai et al. (2018) successfully edited GmFT2a and GmFT5a in soybean using the CRISPR/Cas9 system and generated late flowering mutants with high yield potential for the tropics (Cai et al., 2018). CRISPR/Cas9-mediated mutation of both LUX1 and LUX2 genes successfully obtained lux1 lux2 double mutant, showing an extreme delay in flowering time and an insensitive response to day-length (Bu et al., 2021). Herein, we hypothesized that CRISPR/Cas9-mediated mutation of E1 could create photoperiod insensitive germplasm, which can be used to expand the soybean genetic variations for breeding. CRISPR/Cas9-mediated mutation of E1 has been conducted in soybean cultivar Jack (Han et al., 2019). However, CRISPR/Cas9 based genome editing technique relies on an efficient genetic transformation protocol, which is highly dependent on genotype. Currently, o nly a few soybean cultivars are amenable to genetic transformation (Donaldson and Simmonds, 2000). Jack is a model soybean cultivar for genetic transformation, but it carries a recessive e1-as allele, a leaky allele that retains partial E1 function (Xia et al., 2012). The function of the e1-as allele is significantly weaker than the dominant E1 allele. In this study, Agrobacterium-mediated transformation was used to introduce the CRISPR/Cas9 expression vector into soybean cultivar Tianlong1 carrying the dominant E1 to analyze the precise effect of E1 mutation, and to create novel germplasm in an elite background that can be used in soybean breeding. In addition, some new roles of E1 were found, including the regulation of stem growth habit and branch number, the genetic compensation response of E1 to its homolog E1-La and E1-Lb, and E1-regulated genes in stem tip. Our findings provides solid evidence that E1 regulate s photoperiod by controlling the flowering time, stem growth habit and brunch number, and will inform future efforts in molecular breeding of photoperiod insensitive soybean cultivars.
Soybean cultivar Tanlong1 carrying the dominant functional E1 allele was used for genetic transformation. T0, T1 and T2 gene editing plants were planted in a growth chamber under long-day conditions (LD, 16 h/8 h, light/dark) and short-day conditions (SD, 12 h/12 h, light/dark) at 70% relative humidity and 200-300 μmol m−2 S−1 light fluency. Phenotypes of the T2 plants was recorded during the cultivation season (May to October) under a naturally LD conditions (LD, 16 h/8 h, light/dark) and artificially controlled short-day conditions (SD, 12 h/12 h, light/dark) in Harbin. The plant height and flowering time of the R1 stage (the first flower to appear) were recorded according to Fehr’s system (Fehr, 1971). Thirty plants of each genotype were measured and the data collected were subjected to statistical analysis.
CRISPR/Cas9 expression vector was constructed as described previously (Zhai et al., 2022). The CRISPR/Cas9 expression vector was transformed into Agrobacterium strain EHA105 by electroporation. Soybean transformation was performed using the cotyledonary node transformation system described previously (Flores et al., 2008). To identify the e1 mutants, we extracted total genomic DNA from leaf samples of putative mutants using EasyPure® Plant Genomic DNA Kit (TransGen, Beijing, China) according to the manufacturer’s instructions. Subsequently, PCR analysis was performed using E1 sequence-specific primer sets (Supplementary Table 1). PCR products were detected using 1.5% agarose gel electrophoresis and then sequenced. The sequences of T0, T1, and T2 generation plants were analyzed using DSDecodeM to characterize the mutations induced by CRISPR/Cas9. Successfully edited lines were identified via sequence peaks and alignment to the reference sequences. The heterozygous mutants exhibited overlapping peaks near the target site, and the homozygous mutations showed single peaks at the target. The homozygous mutants were then identified by sequence alignment with the WT sequence. To screen and obtain E1 targeted mutants without transgenic elements, we performed PCR analysis using Bar sequence-specific primer sets (Supplementary Table 1) to determine sgRNA/Cas9 on T-DNA elements. Potential off-target sites were predicted with the online tool: CRISPR-P (http://crispr.hzau.edu.cn/CRISPR2/) (Lei et al., 2014). Two potential off-target sites with the highest sequence identities to E1 targets were analyzed. PCR was performed to amply the genome fragment containing the potential off-target sites, using primer pairs listed in Supplementary Table 1. PCR products were detected by 1.5% agarose gel electrophoresis and then sequenced.
Short-day promotion rate (SDHR) was calculated to determine the effect of photoperiod on flowering time, maturity and plant height as follows (Yan et al., 2009):
where VLD represents the phenotype value under LD, and VSD represents the phenotype value under SD.
Wild-type soybean cultivar ‘Tianlong1’ and e1 mutant were used for RNA-seq assay. Stem tips were collected 4 h after dawn from 28-day-old seedlings grown under LD conditions. Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Sequencing cDNA libraries were generated according to the protocol of VAHTS ® Universal V6 RNA-seq Library Prep Kit for Illumina (Vazyme, Nanjing, China). cDNA libraries were then constructed for sequencing on the Illumina NovaSeq 6000 S4 Seq system with 150-bp paired-end read lengths. Tophat2 was used to map clean reads to the soybean reference genome, Glycine max Wm82.a2.v1, from Phytozome (https://phytozome-next.jgi.doe.gov/). Differentially expressed genes (DEGs) were detected using DEGseq under the following parameters: fold change 2.00 and adjusted P-value (Q-value) 0.05. DEGs are listed in Supplemental Data Set S1. Sequence data of RNA-seq from this study were deposited in the National Genomics Data Center (SRA) database under accession number PRJCA010529.
Total RNA was extracted from the leaves of 28-DAE-old plants of wild-type soybean TianLong1 and E1 mutant grown under LD conditions (16 h/8 h, light/dark). Three independent replicates of the total RNA samples were prepared for each analysis.
Total RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA, USA) method according to the manufacturer’s instructions. Subsequently, the RNA was treated with RNase-free recombinant DNase I (Takara, Dalian, China). The integrity of the RNA was determined using NanoDrop™ ND-2000c Spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Equal amounts of isolated RNA were then reverse transcribed to cDNA using the SuperScript™ III Reverse Transcriptase kit. The quality of the cDNA samples was assessed by PCR using GmTubulin A (TUA5) specific primers.
Each cDNA sample was subjected to qRT-PCR analysis using the SYBR Green Master Mix (TransStart Top Green qPCR SuperMix, Beijing, China). The qRT-PCR analysis was performed on LightCycler® 96 real-time PCR detection system (Roche, Switzerland) according to the manufacturer’s protocol. The measured Ct values were converted to relative copy-numbers using the 2-ΔΔCt method. TUA5 gene was used as an internal control to normalize all gene expression data. The qRT-PCR was performed using three fully independent biological replicates and each sample was run in triplicate. Raw data were standardized as described previously (Willems et al., 2008). Primers used for the qRT-PCR and semi-quantitative RT-PCR analyses are listed in Supplementary Table 1.
In our previous study, to investigate the regulation of GmMDE genes by E1, we used CRISPR/Cas9-mediated mutation to inactivate E1 in the soybean cultivar Tianlong1, which carries a functional E1 allele (Zhai et al., 2022). In this study, we obtained four types of homozygous mutations. The e1-1 mutant line harbored a 244-bp deletion in the E1 coding region, whereas the e1-2 mutant line harbored a 247-bp deletion in the E1 coding region. The e1-3 mutant line harbored a 243-bp deletion in the E1 coding region was obtained, whereas the e1-4 mutant line harbored a 209-bp deletion in the E1 coding region. (Supplemental Figure 1). Notably, the e1-1 and e1-2 were frameshift mutations, causing premature termination of translation. The e1-1 and e1-2 produced a truncated protein encoding 98 and 97 amino acids, respectively, resulting in the deletion of all B3 domains and retaining part of the nuclear localization signal. The e1-3 and e1-4 produced a truncated protein encoding 153 and 165 amino acids, respectively, which retained part of the B3 domains and nuclear localization signal (Supplementary Figure 2). Potential off-target sites were predicted and the top 2 genomic regions of homology were selected as most likely off-target sites. Subsequently, each of these regions was amplified by PCR using genomic DNA from the mutant lines as template. The PCR products were further analyzed by Sanger Sequencing. Sequencing analysis did not reveal any potential off-target variants in the T1 mutants (Supplementary Figure 3; Supplementary Table 2). Thus CRISPR/Cas9 expression vector had specific edits at two targets. To obtain trans-clean mutants without T-DNA elements, we performed PCR to examine check whether there were traces of T-DNA in the mutants using Bar gene specific primers (Supplementary Table 1). Among the four T1 mutant lines, two were free of T-DNA in T1 generation derived from e1-1, e1-2, e1-3 and e1-4 (Supplementary Table 3). All the T1 mutant lines was flowering earlier than wild-type soybean cultivar Tianlong1 under LD conditions (Supplementary Figure 4).
To analyze the effect of E1 mutation on soybean photoperiod sensitivity, we planted T2 generation seeds of the homozygous mutant e1-1 and e1-2 without T-DNA elements under LD and SD conditions, respectively. The wild-type soybean cultivar Tianlong1 is extremely sensitive to photoperiod. The flowering time, maturation and plant height of Tianlong1 plants grown under LD conditions significantly differed from those under SD conditions (Figures 1A-D). Furthermore, the photoperiod sensitivity of the two E1 mutants decreased greatly. Although the flowering time, maturation and plant height of two E1 mutants under LD were different from those under SD conditions, differences in maturation and plant height between plants grown under LD and SD conditions decreased. The reproductive period R8 of the two E1 mutants under SD conditions was about 25 days earlier than that under LD conditions, while the reproductive period R8 of Tianlong1 under SD conditions was more than 53 days earlier than that under LD conditions. The plant height of Tianlong1 under SD conditions was 60 cm shorter than that under LD conditions, while the plant height of the two E1 mutants under SD conditions was about 25cm shorter than that under LD conditions. Collectively, these results indicate that loss of E1 function reduces the photoperiod sensitivity of soybeans. Short-day hastening rate of flowering time, maturity, and plant height of the two E1 mutants were significantly lower than that of wild-type Tianlong1 (Figure 1E).
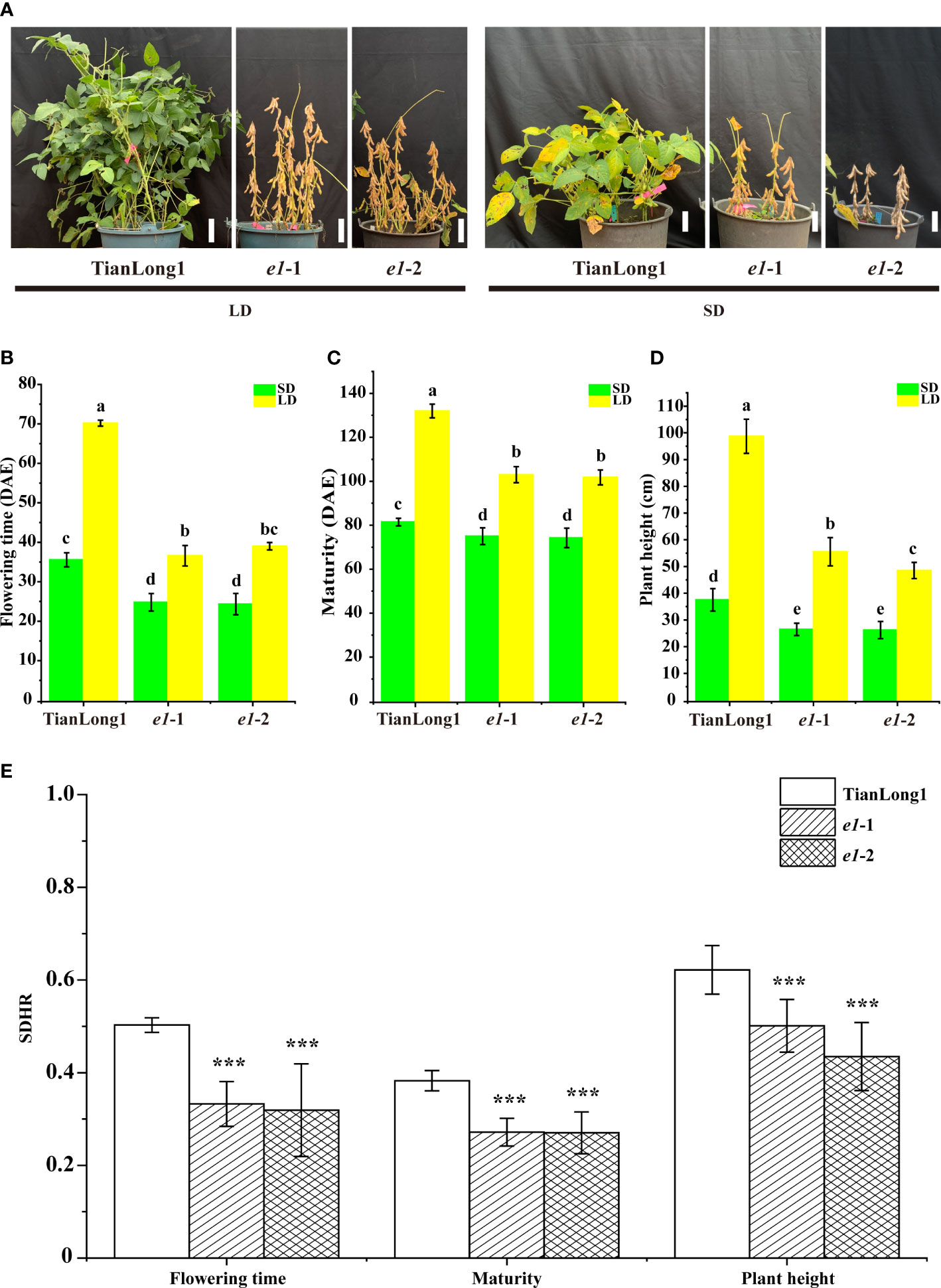
Figure 1 CRISPR/Cas9-induced E1 mutant phenotypes under both LD and SD conditions. (A) Maturity of E1 mutants and wild-type TianLong1 under LD and SD conditions. Scale bar, 10 cm. (B) Flowering time. (C) Time to maturity. (D) Plant height. (E) Short-day hastening rate. All data is shown as the mean values ± standard deviation (n = 20 plants). The diverse lowercase letter above the histogram bars in (B–D) by Two-Way ANOVA, suggests significant differences between the two panels (P > 0.05). The error bars in (E) indicates standard deviation. ***P<0.001, as determined by one-tailed Student’s t-test. DAE, day after emergence.
Since the two E1 mutants were sensitive to photoperiod, we examined whether genetic compensation response of E1 and its homologs exists. The soybean genome harbors two E1 homologs, E1-La and E1-Lb (Xia et al., 2012). The expression of E1-La and E1-Lb in leaves of the e1-2 mutant and its wild-type soybean cultivar Tianlong1 were analyzed. In e1-2 mutant plants, the expression levels of two E1-Ls were significantly higher than those in the wild type (Figures 2A, B), indicating that a genetic compensation response of E1 and its homologs exist.

Figure 2 Expression analyses of E1-Ls/GmFT2a/GmFT5a in WT plants and mutants under LD conditions. (A) Expression analysis of E1-La under LD conditions. (B) Expression analysis of E1-Lb under LD conditions. (C) Expression analysis of GmFT2a under LD conditions. (D) Expression analysis of GmFT5a under LD conditions. Transcript levels were normalized to TUA5. Values represent the means of three biological replicates (n = 3 plants); error bars indicate standard deviation. **P<0.01; ***P<0.001, as determined by one-tailed Student’s t-test.
In our previous study, E1 overexpression repressed the expression of GmFTs (GmFT2a and GmFT5a), two homologs of Arabidopsis FT (Xia et al., 2012). To clarify the correlation between the expression of E1 and GmFT2a/5a in flowering time regulation, we analyzed the transcript levels of GmFT2a and GmFT5a in the leaves of e1-2 mutant and wild-type (Figures 2C, D). GmFT2a and GmFT5a expression increased in e1-2 mutant compared to the wild-type Tianlong 1.
The plant architecture of two E1 mutants was investigated. Mutation of E1 altered the stem growth habit of the mutants. The stem growth habit of the wild-type TianLong1 tended to be indeterminate under natural LD conditions and determinate under natural SD conditions (Figure 3F). Under a longer light period (16h:8 h light/dark), TianLong1 entirely exhibited indeterminate stem growth habit (Figure 3A). Notably, the two E1 mutants exhibited determinate stem growth habits under both LD and SD conditions, characterized by early terminal flowering (Figures 3B, F), reduced plant height and decreased node number along the main stems (Figures 1D, 3C).
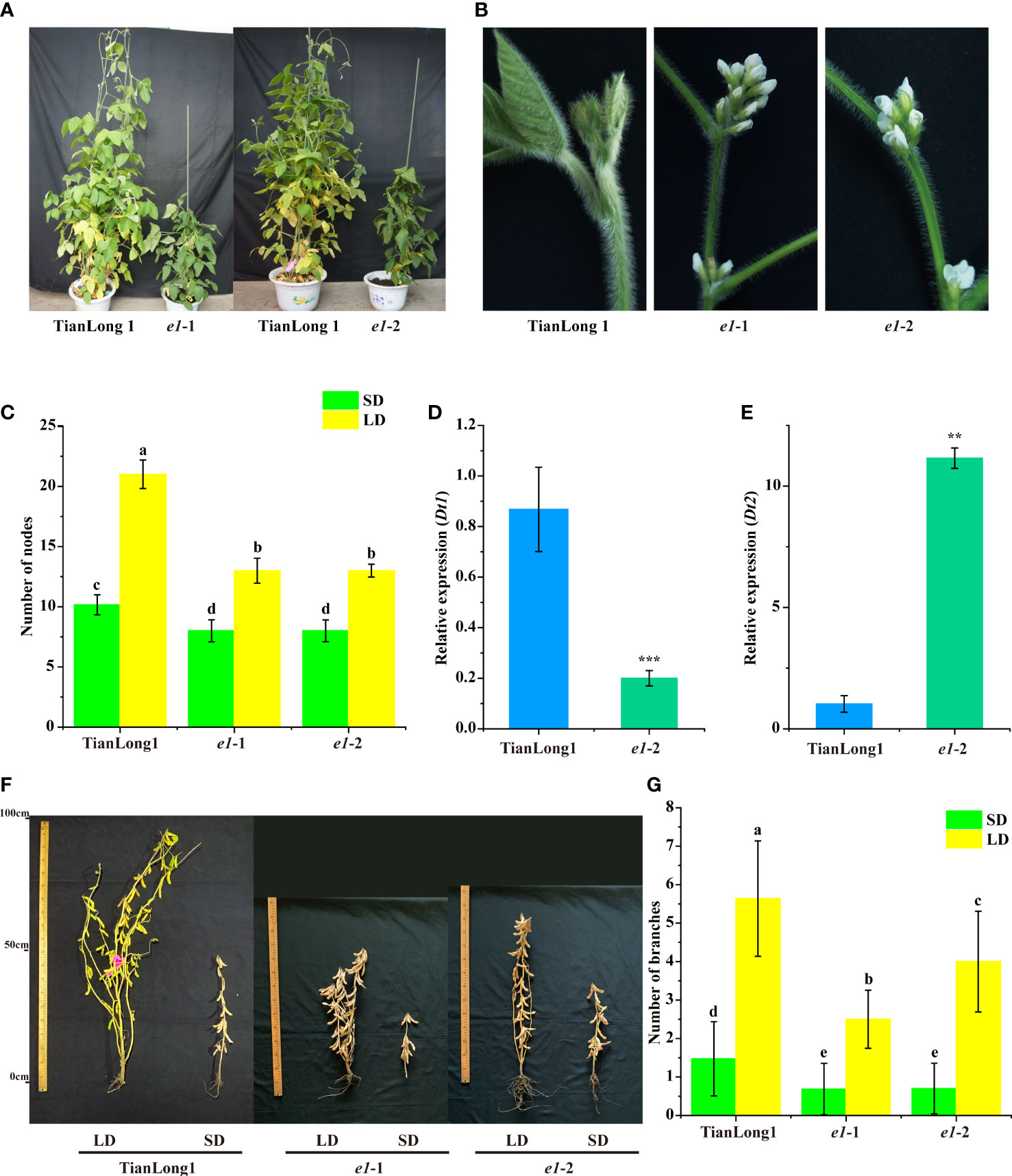
Figure 3 Loss of E1 function changes the Stem Growth Habits of soybean. (A) Comparison of stem growth habit. (B) Morphology of the top of stems in determinate and indeterminate soybean. (C) Number of nodes. (D) Expression analysis of Dt1 under LD conditions. (E) Expression analysis of Dt2 under LD conditions. (F) Phenotypes of E1 mutants and wild-type TianLong1 under LD and SD conditions. (G) Number of branches. Transcript levels were normalized to TUA5. Values represent the means of three biological replicates (n = 3 plants); error bars indicate standard deviation. **P<0.01; ***P<0.001, as determined by one-tailed Student’s t-test. All data is shown as the mean values ± standard deviation (n = 20 plants). The diverse lowercase letter above the histogram bars in (C and G) by Two-Way ANOVA, suggests significant differences between the two panels (P > 0.05).
Soybean stem growth habit is regulated by two major genes, Dt1 and Dt2. Dt1 specifies the indeterminate growth habit, which prevents terminal flowering, leading to taller plants (Liu et al., 2010; Tian et al., 2010; Liu et al., 2016). Dt2 functions as a direct repressor of Dt1, promoting terminal flowering and leading to shorter plants (Liu et al., 2016). Here, we hypothesized that E1 might regulate stem growth habit by modulating Dt1 and Dt2 genes. We thus measured the expression levels of Dt1 and Dt2 in the e1-2 mutant and wild-type Tianlong1 plants. Dt1 expression was higher in the stem tips of wild type plants than in e1-2 mutant, while Dt2 expression was significantly higher in the stem tips of e1-2 mutant than in wild-type stem tips (Figures 3D, E). These results indicate that E1 regulates the stem growth habit via the Dt2- Dt1 signaling pathway.
The branch number of wild type plants and two E1 mutants were investigated. Mutation of E1 altered the branch number. Wild type plants produced much more branches than the two E1 mutants (Figures 3F, G). TianLong1 produced significantly more branch numbers under LD conditions than under SD conditions; however, the branch number of E1 mutants was less different between SD and LD conditions (Figures 3F, G), indicating that mutation of E1 reduces photoperiod sensitivity in branch number.
Transcriptome sequencing (RNA-seq) and expression analysis of differentially expressed genes (DEGs) were performed further to understand the molecular differences between TianLong1 and e1-2. Similar gene expression levels were observed among three biological replicates (Figure 4A), WT and e1-2 were distinguished via principal component analysis (PCA). A total of 1161 DEGs were obtained by comparing the RNA-seq datasets (P< 0.05). Volcano plots were used to visualize the significant DEGs in e1-2 (Figure 4B). The red points in the graphic represent significantly upregulated genes, while the blue points in the graphic represent downregulated genes. Furthermore, we analyzed the gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment of the differentially expressed mRNAs between E1 mutants and controls. The most enriched terms GO terms included cell part (GO: cellular component), metabolic process (GO: biological process), binding (GO: molecular function), cellular process (GO: biological process), catalytic activity (GO: molecular function) and organelle (GO: cellular component) (Figure 4C). The most significantly enriched KEGG pathway was phenylpropanoid biosynthesis, Cutin, suberin and wax biosynthesis (Figure 4D).
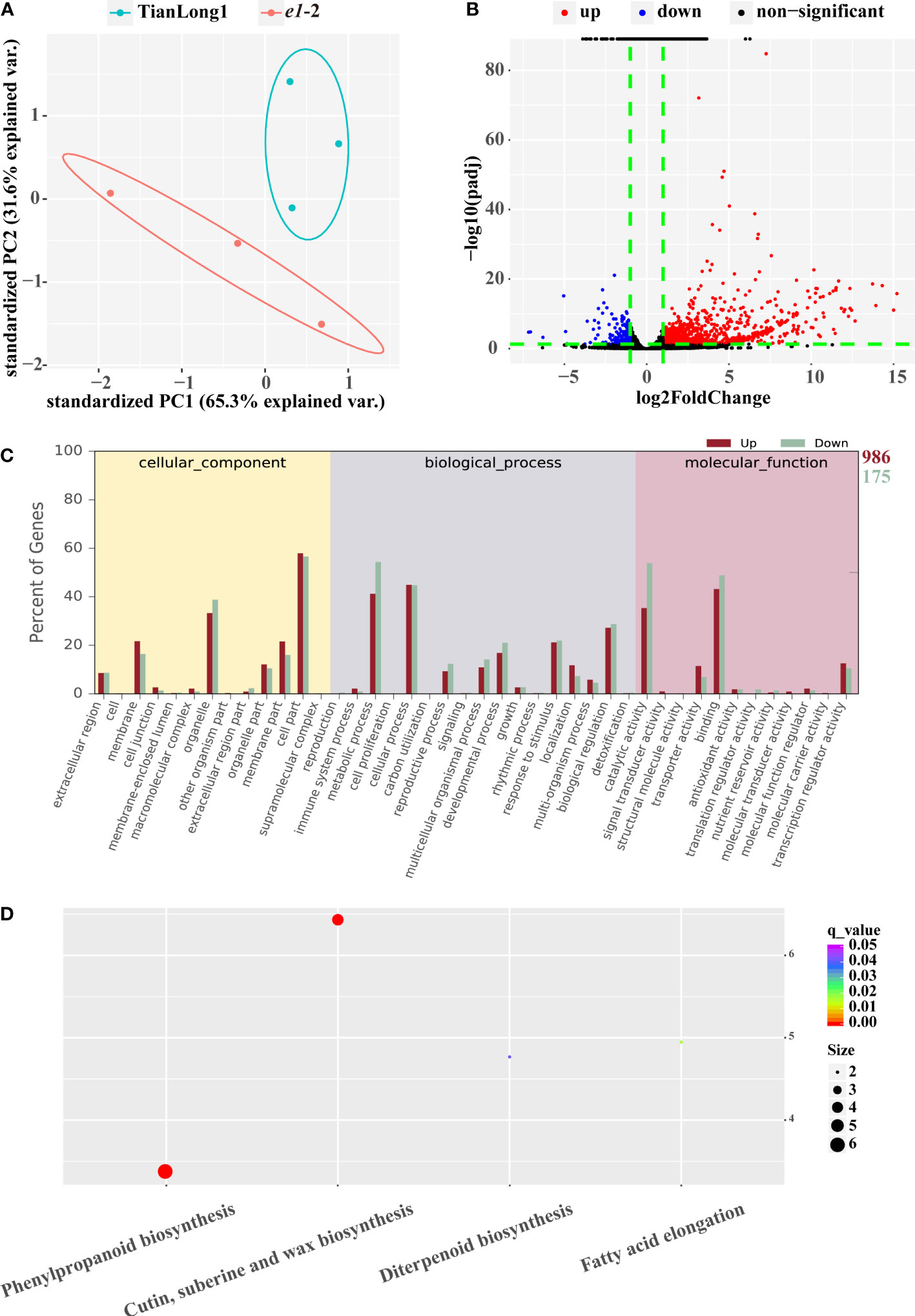
Figure 4 RNA Profiles in e1-2 and TianLong1. (A) PCA between e1-2 and TianLong1. (B) The volcano plot of mRNA expression signals in e1-2 and TianLong1. (C, D) GO (gene ontology) and KEGG (Kyoto encyclopedia of genes and genomes) enrichment analysis of total of 1161 DEGs.
The expressions of several flowering related genes were altered in the E1 knockdown lines, consistent with the e1-2 early flowering phenotype, with most of the genes up-regulated. Among the 32 MADS-box genes, 28 were significantly up-regulated and 4 were down-regulated. Specifically, E1 knockdown significantly up-regulated floral meristem identity genes, LEAFY (LFY) and APETALA1 (AP1), and most floral organ identity genes. Furthermore, the expression of SEPALLATA(SEP), CAULIFLOWER(CAL) and WUSCHEL(WUS) genes were up-regulated. E1 knockdown also affected the expression of multiple key factors in auxin and gibberellin signaling pathways. In particular, the expression of PIN, encoding auxin efflux carrier protein and GA2OX, the key enzymes in Gibberellin (GA) synthesis were upregulated. Also, 12 differentially expressed NAC transcription factors that mediate SAM formation were up-regulated (Figure 5 and Supplementary Table 4).
Studies have shown that E1 confers the most prominent effect on photoperiod sensitivity in plants (Bernard, 1971; Cober et al., 1996; Watanabe et al., 2012; Xia et al., 2012). In soybean, genes that contribute to photoperiodic flowering and domestication, such as E3, E4, J, Tof5 Tof11, Tof12, LUX1, LUX2, and Tof16 all function by modulating E1 expression (Xia et al., 2012; Xu et al., 2015; Lu et al., 2017; Li et al., 2020; Lu et al., 2020; Bu et al., 2021; Dong et al., 2021), indicating that E1 is the core regulator of soybean photoperiodic responses. E1 regulates the photoperiod of soybean like a switch. CRISPR/Cas9 system has recently emerged as an effective method for targeted genome editing and gene function research. Here, we mutated the E1 gene in soybean cultivar Tianlong1 carrying the dominant E1 via CRISPR/Cas9 system to study its function on the photoperiod response of soybean. Short-day hastening rate of flowering time, maturity, and plant height of the two E1 mutants indicated that mutation of the E1 gene reduces photoperiod sensitivity in soybean (Figure 1E).
However, the two E1 mutants were still sensitive to photoperiod. Therefore, we examined whether the genetic compensation response of E1 and its homologs exists. The results showed that E1-Ls were significantly up-regulated in e1-2 mutant plants relative to the wild type plants (Figures 2A, B), suggesting the existence of a genetic compensation response of E1 and its homologs. These results also indicate that simultaneous mutation of E1 and two homologs, E1-la and E1-lb might reduce photoperiod sensitivity more effectively.
In this study, the e1 mutant bloomed and matured earlier than the wild type under both LD and SD conditions (Figures 1B, C). Previously, CRISPR/Cas9 was used to mutate E1 in soybean cultivar Jack carrying the recessive e1-as allele to generate e1 mutant (Han et al., 2019). The flowering time of mutants was significantly earlier than that of wild type plants under LD conditions. However, under SD condition, no significant difference in flowering time was observed between the wild type plants and mutants. The e1-as allele is a leaky allele retaining partial E1 function, which as a flowering suppressor is significantly weaker than the E1 allele (Xia et al., 2012; Zhai et al., 2015). In most soybean cultivars, E1 is highly induced under LD conditions and repressed under SD conditions (Xia et al., 2012; Zhai et al., 2022). The weak function and low expression level of e1-as allele reduce the effect of E1 in soybean cultivar Jack under SD conditions; therefore, E1 knockdown in the cultivar does not change its flowering time under SD conditions. Thus, soybean cultivars carrying functional dominant E1 alleles should be used to determine the precise function of E1.
Soybean is a typical photoperiod-sensitive short-day flowering plant. Its flowering, maturity and plant architecture, including plant height, branch number, node numbers of main stem and pods per plant are mainly regulated by photoperiod (Yan et al., 2009). Studies on the role of E1 have primarily focused on flowering time, maturity and plant height (Xia et al., 2012; Zhang et al., 2016; Han et al., 2019), however, the effect of E1 on the other agronomic traits related to photoperiodic sensitivity remains poorly understood. In this study, mutation of E1 caused phenotypic changes in stem growth habits. The E1 mutants exhibited determinate stem growth habits under both LD and SD conditions (Figures 3A, B, F). Also, E1 knockdown decreased Dt1 expression and increased Dt2 expression in stem tips relative to the wild-type, consistent with the phenotypic changes (Figures 3D, E). Dt1 and Dt2 are the main genes regulating soybean stem growth habits (Liu et al., 2010; Tian et al., 2010; Ping et al., 2014; Liu et al., 2016). Specifically, Dt2 functions as a direct repressor of Dt1, promoting terminal flowering, thus producing shorter plants (Liu et al., 2016). Dt2 encodes a dominant MADS domain factor belonging to the APETALA1/SQUAMOSA (AP1/SQUA) subfamily (Ping et al., 2014). Besides Dt2, a set of genes encoding MADS domain factor were up-regulated (Figure 5). These MADS domain factor potentially regulate the stem growth habit of soybean and might contribute to downregulation of Dt1 in e1-2 mutant.
In our previous study, we mapped the major QTL for branch number to the proximate to the E1 gene, inferring that E1 gene or neighboring genetic factor significantly contributes to the branch number (Yang et al., 2017). Recently, it was proved that Dt2 reduces the branch number in soybean by activating the transcription of the GmAP1 gene family (Liang et al., 2022). In this study, the two E1 mutants produced fewer branches compared with wild-type Tianlong1 under both SD and LD conditions (Figure 3G). Meanwhile, we found that the expression of Dt2 and AP1 were up-regulated in the e1-2 mutants (Figures 3E and 5). This study provides solid evidence that E1 regulates branch number. The branch number is an important agronomic trait related to photoperiodic sensitivity. The branch number of soybean is genotype dependent, with some cultivars showing more sensitivity to photoperiod than others (Yan et al., 2009). This can be attributed to the diverse genetic variation of E1 among soybean cultivars (Xu et al., 2013; Zhai et al., 2015; Li et al., 2017).
Gibberellin promotes shoot branching or tillering in plants, whereas mutation of the gene encoding GA synthesis enzyme decreases branching or tillering (Lo et al., 2008; Ni et al., 2015; Wu et al., 2020; Huang et al., 2021). Physiological observations and molecular studies suggest crosstalk between the GA and auxin, as well as with auxin transport. Auxin transport is reduced in GA mutants (Willige et al., 2011). In this study, RNA-seq analysis indicated that key enzymes in GA synthesis and PIN, encoding auxin efflux carrier protein were upregulated in e1 mutant (Figure 5). This suggests that E1 potentially regulates the branching type by modulating the genes involved in GA synthesis and auxin transport. Notably, hormonal crosstalk of GA and auxin might contribute to the decreased branching phenotype of e1 mutants.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
HZ designed and supervised this research. ZW and YL conducted the experiments and analyzed the data. DG, RF, YL, LQ, and WL conducted the field trial. KX, JZ, and XB provided advice on experimental implementation. ZW and HZ prepared the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by National Natural Science Foundation of China (31971970), Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDA28070302), Natural Science Foundation of Heilongjiang Province of China (YQ2021C034), and Young Scientists Group Project (2022QNXZ05) of Northeast Institute of Geography and Agroecology, Chinese Academy of Sciences and Hainan Yazhou Bay Seed lab (B21HJ0101).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1066820/full#supplementary-material
Abe, J., Xu, D., Miyano, A., Komatsu, K., Kanazawa, A., Shimamoto, Y. (2003). Photoperiod-insensitive Japanese soybean landraces differ at two maturity loci. Crop Sci. 43 (4), 1300–1304. doi: 10.2135/cropsci2003.1300
Bernard, R. L. (1971). Two major genes for time of flowering and maturity in Soybeans1. Crop Sci. 11 (2), cropsci1971.0011183X001100020022x. doi: 10.2135/cropsci1971.0011183X001100020022x
Bonato, E. R., Vello, N. A. (1999). E6, a dominant gene conditioning early flowering and maturity in soybeans. Genet. Mol. Biol. 22 (2), 229–232. doi: 10.1590/s1415-47571999000200016
Bu, T., Lu, S., Wang, K., Dong, L., Li, S., Xie, Q., et al. (2021). A critical role of the soybean evening complex in the control of photoperiod sensitivity and adaptation. Proc. Natl. Acad. Sci. 118 (8), e2010241118. doi: 10.1073/pnas.2010241118
Buzzell, R. I. (1971). Inheritance of a soybean flowering response to fluorescent-daylength conditions. Can. J. Genet. Cytol. 13 (4), 703–707. doi: 10.1139/g71-100
Cai, Y., Chen, L., Liu, X., Guo, C., Sun, S., Wu, C., et al. (2018). CRISPR/Cas9-mediated targeted mutagenesis of GmFT2a delays flowering time in soya bean. Plant Biotechnol. J. 16 (1), 176–185. doi: 10.1111/pbi.12758
Cober, E. R., Molnar, S. J., Charette, M., Voldeng, H. D. (2010). A new locus for early maturity in soybean. Crop Sci. 50 (2), 524–527. doi: 10.2135/cropsci2009.04.0174
Cober, E. R., Tanner, J. W., Voldeng, H. D. (1996). Genetic control of photoperiod response in early-maturing, near-isogenic soybean lines. Crop Sci. 36 (3), 601–605. doi: 10.2135/cropsci1996.0011183X003600030013x
Cober, E. R., Voldeng, H. D. (2001). A new soybean maturity and photoperiod-sensitivity locus linked to E1 and T. Crop Sci. 41 (3), 698–701. doi: 10.2135/cropsci2001.413698x
Donaldson, P. A., Simmonds, D. H. (2000). Susceptibility to agrobacterium tumefaciens and cotyledonary node transformation in short-season soybean. Plant Cell Rep. 19 (5), 478–484. doi: 10.1007/s002990050759
Dong, L., Cheng, Q., Fang, C., Kong, L., Yang, H., Hou, Z., et al. (2022). Parallel selection of distinct Tof5 alleles drove the adaptation of cultivated and wild soybean to high latitudes. Mol. Plant 15 (2), 308–321. doi: 10.1016/j.molp.2021.10.004
Dong, L., Fang, C., Cheng, Q., Su, T., Kou, K., Kong, L., et al. (2021). Genetic basis and adaptation trajectory of soybean from its temperate origin to tropics. Nat. Commun. 12 (1), 5445. doi: 10.1038/s41467-021-25800-3
Fang, C., Liu, J., Zhang, T., Su, T., Li, S., Cheng, Q., et al. (2021). A recent retrotransposon insertion of J caused E6 locus facilitating soybean adaptation into low latitude. J. Integr. Plant Biol. 63 (6), 995–1003. doi: 10.1111/jipb.13034
Fehr, W. R., Caviness, C. E., Burmood, D. T., Pennington, J. S. (1971). Stage of development descriptions for soybeans, glycine max (L.) Merrill1. Crop Science 11, 929–931. doi: 10.2135/cropsci1971.0011183X001100060051x
Flores, T., Karpova, O., Su, X., Zeng, P., Bilyeu, K., Sleper, D. A., et al. (2008). Silencing of GmFAD3 gene by siRNA leads to low alpha-linolenic acids (18:3) of fad3-mutant phenotype in soybean [Glycine max (Merr.)]. Transgenic Res. 17 (5), 839–850. doi: 10.1007/s11248-008-9167-6
Han, J., Guo, B., Guo, Y., Zhang, B., Wang, X., Qiu, L. J. (2019). Creation of early flowering germplasm of soybean by CRISPR/Cas9 technology. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01446
Huang, L. J., Luo, J., Wang, Y., Li, N. (2021). From green revolution to green balance: The nitrogen and gibberellin mediated rice tiller growth. Plant Signal Behav. 16 (7), 1917838. doi: 10.1080/15592324.2021.1917838
Jiang, Y., Leng, J. T., Fei, Z. H., Feng, T., Wu, C. X. (2009). Photoperiod responses of a widely-adapted soybean cultivar of zhonghuang 13. Soybean Sci.
Lei, Y., Lu, L., Liu, H.-Y., Li, S., Xing, F., Chen, L.-L. (2014). CRISPR-p: A web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant 7 (9), 1494–1496. doi: 10.1093/mp/ssu044
Liang, Q., Chen, L., Yang, X., Yang, H., Liu, S., Kou, K., et al. (2022). Natural variation of Dt2 determines branching in soybean. Nat. Commun. 13 (1), 6429. doi: 10.1038/s41467-022-34153-4
Li, C., Li, Y. H., Li, Y., Lu, H., Hong, H., Tian, Y., et al. (2020). A domestication-associated gene GmPRR3b regulates the circadian clock and flowering time in soybean. Mol. Plant 13 (5), 745–759. doi: 10.1016/j.molp.2020.01.014
Liu, W., Jiang, B., Ma, L., Zhang, S., Zhai, H., Xu, X., et al. (2018). Functional diversification of flowering locus T homologs in soybean: GmFT1a and GmFT2a/5a have opposite roles in controlling flowering and maturation. New Phytol. 217 (3), 1335–1345. doi: 10.1111/nph.14884
Liu, B., Kanazawa, A., Matsumura, H., Takahashi, R., Harada, K., Abe, J. (2008). Genetic redundancy in soybean photoresponses associated with duplication of the phytochrome a gene. Genetics 180 (2), 995–1007. doi: 10.1534/genetics.108.092742
Liu, B., Watanabe, S., Uchiyama, T., Kong, F., Kanazawa, A., Xia, Z., et al. (2010). The soybean stem growth habit gene Dt1 is an ortholog of arabidopsis TERMINAL FLOWER1. Plant Physiol. 153 (1), 198–210. doi: 10.1104/pp.109.150607
Liu, Y., Zhang, D., Ping, J., Li, S., Chen, Z., Ma, J. (2016). Innovation of a regulatory mechanism modulating semi-determinate stem growth through artificial selection in soybean. PloS Genet. 12 (1), e1005818. doi: 10.1371/journal.pgen.1005818
Li, J., Wang, X., Song, W., Huang, X., Zhou, J., Zeng, H., et al. (2017). Genetic variation of maturity groups and four e genes in the Chinese soybean mini core collection. PloS One 12 (2), e0172106. doi: 10.1371/journal.pone.0172106
Lo, S. F., Yang, S. Y., Chen, K. T., Hsing, Y. I., Zeevaart, J. A., Chen, L. J., et al. (2008). A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20 (10), 2603–2618. doi: 10.1105/tpc.108.060913
Lu, S., Dong, L., Fang, C., Liu, S., Kong, L., Cheng, Q., et al. (2020). Stepwise selection on homeologous PRR genes controlling flowering and maturity during soybean domestication. Nat. Genet. 52 (4), 428–436. doi: 10.1038/s41588-020-0604-7
Lu, S., Zhao, X., Hu, Y., Liu, S., Nan, H., Li, X., et al. (2017). Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat. Genet. 49 (5), 773–779. doi: 10.1038/ng.3819
McBlain, B. A., Bernard, R. L. (1987). A new gene affecting the time of flowering and maturity in soybeans. J. Hered. 78 (3), 160–162. doi: 10.1093/oxfordjournals.jhered.a110349
Ni, J., Gao, C., Chen, M. S., Pan, B. Z., Ye, K., Xu, Z. F. (2015). Gibberellin promotes shoot branching in the perennial woody plant jatropha curcas. Plant Cell Physiol. 56 (8), 1655–1666. doi: 10.1093/pcp/pcv089
Ping, J., Liu, Y., Sun, L., Zhao, M., Li, Y., She, M., et al. (2014). Dt2 is a gain-of-function MADS-domain factor gene that specifies semideterminacy in soybean. Plant Cell 26 (7), 2831–2842. doi: 10.1105/tpc.114.126938
Ray, J. D., Hinson, K., Mankono, J. E. B., Malo, M. F. (1995). Genetic control of a long-juvenile trait in soybean. Crop Sci. 35 (4), cropsci1995.0011183X003500040012x. doi: 10.2135/cropsci1995.0011183X003500040012x
Samanfar, B., Molnar, S. J., Charette, M., Schoenrock, A., Dehne, F., Golshani, A., et al. (2017). Mapping and identification of a potential candidate gene for a novel maturity locus, E10, in soybean. Theor. Appl. Genet. 130 (2), 377–390. doi: 10.1007/s00122-016-2819-7
Shan, Q., Wang, Y., Li, J., Zhang, Y., Chen, K., Liang, Z., et al. (2013). Targeted genome modification of crop plants using a CRISPR-cas system. Nat. Biotechnol. 31 (8), 686–688. doi: 10.1038/nbt.2650
Tian, Z., Wang, X., Lee, R., Li, Y., Specht, J. E., Nelson, R. L., et al. (2010). Artificial selection for determinate growth habit in soybean. Proc. Natl. Acad. Sci. U.S.A. 107 (19), 8563–8568. doi: 10.1073/pnas.1000088107
Tsubokura, Y., Watanabe, S., Xia, Z., Kanamori, H., Yamagata, H., Kaga, A., et al. (2014). Natural variation in the genes responsible for maturity loci E1, E2, E3 and E4 in soybean. Ann. Bot. 113 (3), 429–441. doi: 10.1093/aob/mct269
Upadhyay, A. P., Ellis, R. H., Summerfield, R. J., Roberts, E. H., Qi, A. (1994). Characterization of photothermal flowering responses in maturity isolines of soyabean [Glycine max (L.) Merrill] cv. Clark. Ann. Bot. 74 (1), 87–96. doi: 10.1006/anbo.1994.1097
Wang, F., Nan, H., Chen, L., Fang, C., Zhang, H., Su, T., et al. (2019). A new dominant locus, E11, controls early flowering time and maturity in soybean. Mol. Breed. 39 (5), 70. doi: 10.1007/s11032-019-0978-3
Watanabe, S., Harada, K., Abe, J. (2012). Genetic and molecular bases of photoperiod responses of flowering in soybean. Breed Sci. 61 (5), 531–543. doi: 10.1270/jsbbs.61.531
Watanabe, S., Hideshima, R., Xia, Z., Tsubokura, Y., Sato, S., Nakamoto, Y., et al. (2009). Map-based cloning of the gene associated with the soybean maturity locus E3. Genetics 182 (4), 1251–1262. doi: 10.1534/genetics.108.098772
Willems, E., Leyns, L., Vandesompele, J. (2008). Standardization of real-time PCR gene expression data from independent biological replicates. Anal. Biochem. 379 (1), 127–129. doi: 10.1016/j.ab.2008.04.036
Willige, B. C., Isono, E., Richter, R., Zourelidou, M., Schwechheimer, C. (2011). Gibberellin regulates PIN-FORMED abundance and is required for auxin transport-dependent growth and development in arabidopsis thaliana. Plant Cell 23 (6), 2184–2195. doi: 10.1105/tpc.111.086355
Wu, K., Wang, S., Song, W., Zhang, J., Wang, Y., Liu, Q., et al. (2020). Enhanced sustainable green revolution yield via nitrogen-responsive chromatin modulation in rice. Science 367 (6478). doi: 10.1126/science.aaz2046
Xia, Z., Watanabe, S., Yamada, T., Tsubokura, Y., Nakashima, H., Zhai, H., et al. (2012). Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc. Natl. Acad. Sci. U.S.A. 109 (32), E2155–E2164. doi: 10.1073/pnas.1117982109
Xu, M., Xu, Z., Liu, B., Kong, F., Tsubokura, Y., Watanabe, S., et al. (2013). Genetic variation in four maturity genes affects photoperiod insensitivity and PHYA-regulated post-flowering responses of soybean. BMC Plant Biol. 13, 91. doi: 10.1186/1471-2229-13-91
Xu, M., Yamagishi, N., Zhao, C., Takeshima, R., Kasai, M., Watanabe, S., et al. (2015). The soybean-specific maturity gene E1 family of floral repressors controls night-break responses through down-regulation of FLOWERING LOCUS T orthologs. Plant Physiol. 168 (4), 1735–1746. doi: 10.1104/pp.15.00763
Xu, H., Zhang, L., Zhang, K., Ran, Y. (2020). Progresses, challenges, and prospects of genome editing in soybean (Glycine max). Front. Plant Sci. 11. doi: 10.3389/fpls.2020.571138
Yang, G., Zhai, H., Wu, H.-Y., Zhang, X.-Z., Lü, S.-X., Wang, Y.-Y., et al. (2017). QTL effects and epistatic interaction for flowering time and branch number in a soybean mapping population of Japanese×Chinese cultivars. J. Integr. Agric. 16 (9), 1900–1912. doi: 10.1016/S2095-3119(16)61539-3
Zhai, H., Lü, S., Liang, S., Wu, H., Zhang, X., Liu, B., et al. (2014a). GmFT4, a homolog of FLOWERING LOCUS T, is positively regulated by E1 and functions as a flowering repressor in soybean. PloS One 9 (2), e89030. doi: 10.1371/journal.pone.0089030
Zhai, H., Lü, S., Wang, Y., Chen, X., Ren, H., Yang, J., et al. (2014b). Allelic variations at four major maturity e genes and transcriptional abundance of the E1 gene are associated with flowering time and maturity of soybean cultivars. PloS One 9 (5), e97636. doi: 10.1371/journal.pone.0097636
Zhai, H., Lü, S., Wu, H., Zhang, Y., Zhang, X., Yang, J., et al. (2015). Diurnal expression pattern, allelic variation, and association analysis reveal functional features of the E1 gene in control of photoperiodic flowering in soybean. PloS One 10 (8), e0135909. doi: 10.1371/journal.pone.0135909
Zhai, H., Wan, Z., Jiao, S., Zhou, J., Xu, K., Nan, H., et al. (2022). GmMDE genes bridge the maturity gene E1 and florigens in photoperiodic regulation of flowering in soybean. Plant Physiol. 189 (2), 1021–1036. doi: 10.1093/plphys/kiac092
Zhang, X., Zhai, H., Wang, Y., Tian, X., Zhang, Y., Wu, H., et al. (2016). Functional conservation and diversification of the soybean maturity gene E1 and its homologs in legumes. Sci. Rep. 6, 29548. doi: 10.1038/srep29548
Keywords: E1, CRISPR/Cas9, flowering time, photoperiod, soybean
Citation: Wan Z, Liu Y, Guo D, Fan R, Liu Y, Xu K, Zhu J, Quan L, Lu W, Bai X and Zhai H (2022) CRISPR/Cas9-mediated targeted mutation of the E1 decreases photoperiod sensitivity, alters stem growth habits, and decreases branch number in soybean. Front. Plant Sci. 13:1066820. doi: 10.3389/fpls.2022.1066820
Received: 11 October 2022; Accepted: 29 November 2022;
Published: 14 December 2022.
Edited by:
Fanjiang Kong, Guangzhou University, ChinaReviewed by:
Qing Li, State Key Laboratory for Rice Biology, Chinese Academy of Agricultural Sciences, ChinaCopyright © 2022 Wan, Liu, Guo, Fan, Liu, Xu, Zhu, Quan, Lu, Bai and Zhai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Zhai, emhhaWhAaWdhLmFjLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.