- 1College of Life Sciences, Nanjing Agricultural University, Nanjing, China
- 2College of Agro-Grassland Science, Nanjing Agricultural University, Nanjing, China
- 3National experimental Teaching Center for Plant Production, Nanjing Agricultural University, Nanjing, China
- 4Institute of Industrial Crops, Jiangsu Academy of Agricultural Sciences, Nanjing, Jiangsu, China
- 5Jiangsu Collaborative Innovation Center for Solid Organic Waste Resource Utilization, Nanjing Agricultural University, Nanjing, China
It has been shown that multidrug and toxic compound extrusion/detoxification (MATE/DTX) family transporters are involved in the regulation of plant development and stress response. Here, we characterized the mini body1 (mib1) mutants in mungbean, which gave rise to increased branches, pentafoliate compound leaves, and shortened pods. Map-based cloning revealed that MIB1 encoded a MATE/DTX family protein in mungbean. qRT-PCR analysis showed that MIB1 was expressed in all tissues of mungbean, with the highest expression level in the young inflorescence. Complementation assays in Escherichia coli revealed that MIB1 potentially acted as a MATE/DTX transporter in mungbean. It was found that overexpression of the MIB1 gene partially rescued the shortened pod phenotype of the Arabidopsis dtx54 mutant. Transcriptomic analysis of the shoot buds and young pods revealed that the expression levels of several genes involved in the phytohormone pathway and developmental regulators were altered in the mib1 mutants. Our results suggested that MIB1 plays a key role in the control of plant architecture establishment in mungbean.
Introduction
Plant architecture refers to the three-dimensional organization of plant organs, including the branching pattern and the shape and size of lateral organs, which affects plant growth and productivity (Reinhardt and Kuhlemeier, 2002; Wang and Li, 2008). During the last decades, multiple regulators in the control of plant architecture have been identified in model plants, such as rice (Oryza sativa) and Arabidopsis thaliana, which form complex regulatory networks including microRNA, key transcription factors, and phytohormones (Wang and Li, 2008; Guo et al., 2020).
The multidrug and toxic compound extrusion/detoxification (MATE/DTX) family was one of the important groups of multidrug transporters, which plays diverse roles in stress responses including detoxification, iron homeostasis, and drought stress (Diener et al., 2001; Li et al., 2002; Nawrath et al., 2002; Rogers and Guerinot, 2002; Magalhaes et al., 2007; Ishihara et al., 2008; Lu et al., 2019; Upadhyay et al., 2019; Duan et al., 2022; Nimmy et al., 2022). MATE/DTX family proteins also participate in plant development and growth (Thompson et al., 2010; Burko et al., 2011; Li et al., 2014; Suzuki et al., 2015; Jia et al., 2019; Upadhyay et al., 2020; Gani et al., 2022). For example, Arabidopsis ADP1/DTX51, a putative MATE/DTX family transporter, affects plant architecture. Elevated expression of ADP1/DTX51 in Arabidopsis leads to an increase in plant growth rate and branch number by modulating the auxin level (Li et al., 2014). Another MATE/DTX transporter, BIG EMBRYO1 (BIGE1) in maize, regulates embryo development, initiation, and the size of lateral organs (Suzuki et al., 2015). The mutation of the maize BIGE1 gene results in increased leaf number and larger embryo size. Similarly, the mutant of DTX54/BIGE1A (ortholog of BIGE1 in Arabidopsis) exhibits increased leaf number and shortened pods with smaller seeds (Suzuki et al., 2015).
Legume is the third largest plant family, with more than 600 genus and 18,000 species (Graham and Vance, 2003). The plant architecture significantly affects the seed yield of grain legume such as pea (Pisum sativa), soybean (Glycine max), and mungbean (Vigna radiata). In pea, the TCP family gene PsBRC1 integrates phytohormones including auxin, cytokinin (CK) and strigolactones (SL) to regulate shoot branching (Rameau et al., 2015; Kerr et al., 2021). It has been shown that the soybean gene INCREASED LEAF PETIOLE ANGLE 1 (GmILPA1), encoding a subunit of the anaphase-promoting complex, controls the angle of leaf petiole (Gao et al., 2017). Notably, the MicroRNA156 (miR156)-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) module has important roles in controlling plant architecture and agronomic traits in soybean (Bao et al., 2019; Sun et al., 2019). Overexpression of the GmmiR156b in soybean significantly alters plant architecture and improves seed yield (Sun et al., 2019). Consistently, knockout GmmiR156b targeted gene GmSPL9 by gene editing alters plant architecture with improved performance and productivity in soybean (Bao et al., 2019). Recently, it has been shown that an MYB family transcription factor GmMYB14 in soybean regulates plant architecture through the brassinosteroid pathway. GmMYB14-overexpressing soybean plants display the compact plant architecture and improved seed yield (Chen et al., 2021). However, up to now, only a few key factors regulating plant architecture has been identified in legume and the underlying molecular mechanism is still poorly understood (Liu et al., 2020).
In this study, we characterized the mini body1 (mib1) mutant in mungbean, which affected plant growth rate, branch number, and lateral organ size. It was found that MIB1 encoded a member of MATE/DTX family proteins, potentially acting as a transporter in mungbean. Transcriptomic analysis revealed that expression levels of phytohormone pathway genes and developmental regulators were altered in the mib1 mutants. Our results indicated that MIB1 plays a pivotal role in regulating plant architecture in mungbean.
Materials and methods
Plant materials
Three mutants, namely, mib1-1 (A001), mib1-2 (A006), and mib1-3 (I007), were identified from M2 generation of the gamma ray mutagenized cultivar Sulu (Li et al., 2022). For phenotype analysis of wild-type (WT) plants, mutants were grown in the greenhouse at 28 ± 2°C, with a 16-h/8-h day/night photoperiod. The allelic tests for three mutants were carried out by crossing the mib1-1 mutant with the mib1-2 and mib1-3 mutants, respectively. All plants of F1 generation showed the mutated phenotype.
Scanning electron microscopy analysis
The terminal leaflets of the fifth compound leaves were fixed in FAA solution and then the samples were dehydrated in the ethanol/tert-butanol series. Field emission scanning electron microscopic (SU8010, Hitachi, Tokyo, Japan) analysis was conducted as previously described (Jiao et al., 2019).
Map-based cloning of MIB1 gene
The mib1-3 mutants were crossed with cultivar AL127 to generate a population for genetic mapping. A total of 150 plants with mutant phenotype isolated from 642 plants in the F2 population were used to map the MIB1 gene. The primers of the molecular markers used in present study are listed in Supplementary Table 1. The DNA were extracted via a plant Genomic DNA Kit DP305 (Tiangen, Beijing, China). The polymerase chain reaction (PCR) was carried out and the polymorphisms of the markers were analyzed as previously described (Jiao et al., 2016).
The PCR of the MIB1 genomic region was conducted by the primers in Supplementary Table 1. The PCR products were cloned into the pMD18-T (TaKaRa, Dalian, China) and sequenced.
RNA-sequencing analysis and quantitative reverse transcription PCR analysis
Shoot buds (2 weeks after germination) and the young pods (2 days after pollination) of WT and mib1-3 mutants were collected with three biological replicates. RNA was extracted by the RNA Kit R6827-01 (Omega, Shanghai, China). We performed RNA-seq using the Illumina HiSeq X Ten platform (Illumina, San Diego, California, USA). The raw sequences were submitted to the NCBI SRA database with accession numbers SRR16944233–SRR16944244. Number of reads per kilobase of exon region in a gene per million mapped reads (RPKM) was used to value expression levels (Mortazavi et al., 2008), and VC1973A version 1.0 was used as the reference genome (Kang et al., 2014). Based on the methods described by Audic and Claverie (1997), DEGs were identified. Heat maps were generated by the pheatmap package (https://cran.r-project.org).
For qRT-PCR, the first strand cDNA was synthesized via Takara PrimeScript™ RT reagent Kit RR047A (TaKaRa, Dalian, China). qRT-PCR analysis was conducted using TB Green™ Premix Ex™ RR420A (TaKaRa) and the ABI StepOnePlus machine (Applied Biosystems, Foster City, CA, USA). Three biological replicates with three technical repeats were conducted.
Arabidopsis transformation
The WT (Col-0) and dtx54 mutant (WiscDsLoxHs046_04F) were used in the present study. The CDS of the MIB1 gene was cloned into pCAMBIA1304 using primers in Supplementary Table 1. The construct was transformed into the dtx54 mutants through floral dip transformation as previously described (Clough and Bent, 1998). T3 progeny lines of 35S::MIB1/dtx54 (L04 and L06) were used for phenotype analysis in this study.
Complementation assays in Escherichia coli
WT strain K12 and ΔacrB mutant strain of E. coli were obtained from Professor Chuanzhen Jiang (South China Agricultural University). The CDS of the mungbean MIB1 gene was cloned into the pET32a vector using primers in Supplementary Table 1, and the vectors were transformed into K12 and mutant strain. Transformants were selected on Luria-Bertani (LB) plate medium with 100 μg/ml ampicillin. The positive clones were then grown in liquid medium containing ampicillin and 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) to induce the expression of MIB1. The cells were diluted and spotted on medium plates with or without tetrabutyl ammonium (TBA) at 37°C for 24 h. Cell growth curves were determined by the absorbance at 600 nm of the cultures grown at 37°C for 24 h.
Analysis of indole-3-acetic acid and abscisic acid contents
Plant hormone levels of indole-3-acetic acid (IAA) and abscisic acid (ABA) in young pods of the WT plant and mutants were determined by high-performance liquid chromatography–mass spectrum/mass spectrum (HPLC/MS/MS) by Agilent 1290 HPLC (Agilent, Santa Clara, CA, USA) and SCIEX-6500 Qtrap (AB Sciex, Foster, CA, USA), as described previously (Pan et al., 2010).
Phylogenetic analysis
In this study, the MIB1 protein sequence was used to search against the mungbean database (Kang et al., 2014). The phylogenetic analysis was conducted by MEGA (version 7.0) using the neighbor-joining method with 1,000 replications (Kumar et al., 2016). The tree was displayed by the Interactive Tree of Life (iTOL; Letunic and Bork, 2016). Protein sequences from this study are listed in Supplementary Table 2.
Results
Isolation and characterization of the mib1 mutants in mungbean
To investigate key components regulating plant architecture in mungbean, we screened mutants with altered branch number and shape and size of lateral organs from the mutagenesis population (Li et al., 2022). Three allelic mutants affecting plant architecture were isolated in mungbean (Figure 1). We named these mutants mini body1-1 (mib1-1), mib1-2, and mib1-3, respectively.
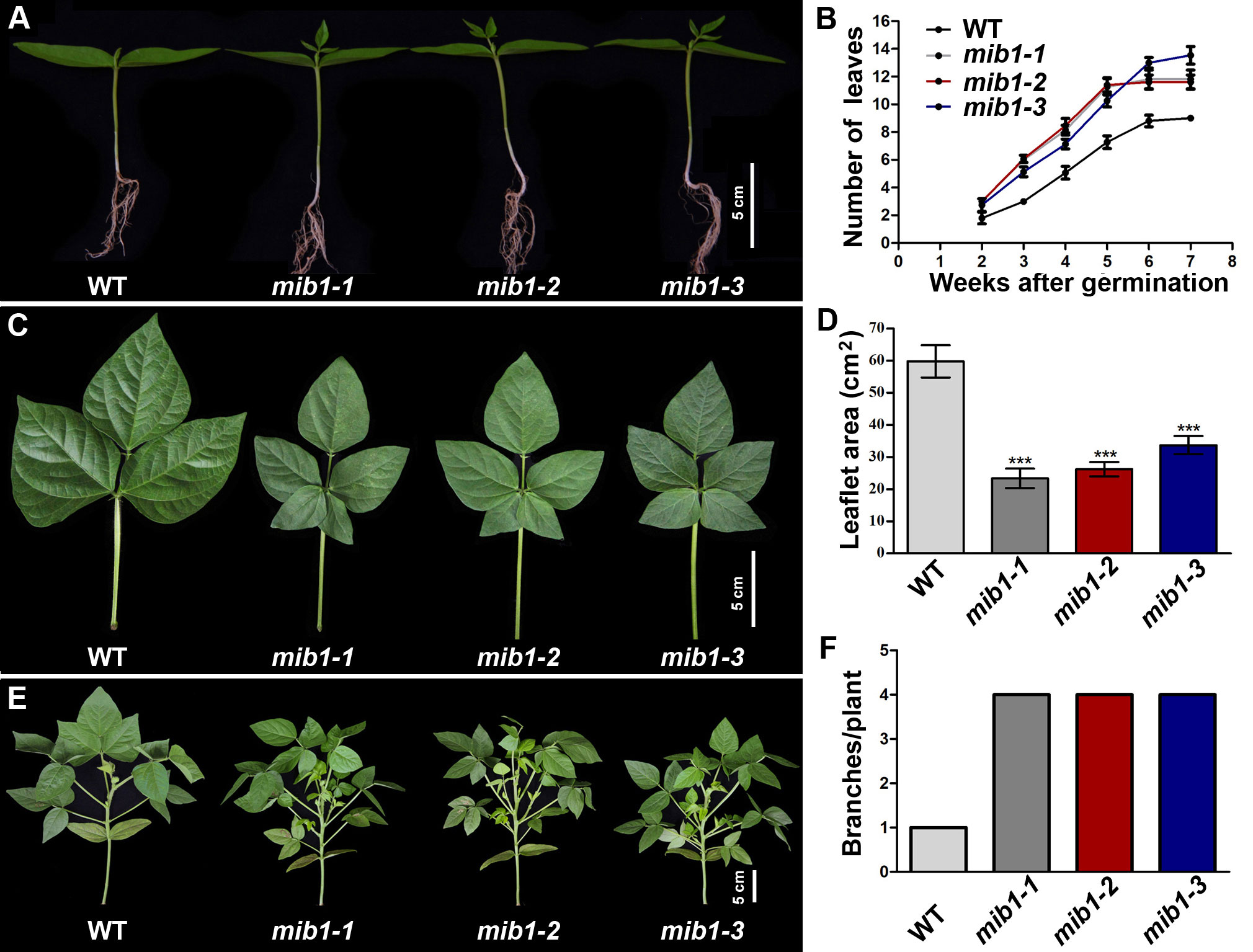
Figure 1 Growth rate characterization of WT and mib1 mutants. (A) Two-week-old seedlings of the wild-type (WT) plant and mib1 mutants. (B) The number of compound leaves of WT and mib1 mutants (n = 10). (C) The fifth compound leaves of WT and mib1 mutants. (D) The size of the terminal leaflets of the fifth compound leaves of WT and mib1 mutants (n = 10). (E) Plant architecture of WT and mib1 mutants at the 4 weeks after germination. (F) The number of branches of WT and mib1 mutants at 4 weeks after germination (n = 10). The data were means ± SD. The Tukey’s multiple comparison test was used. *** p < 0.001.
The leaf production rate in the mib1 mutants was accelerated, compared with that of WT (Figures 1A, B). The juvenile leaves of the mutants were normal, but the adult leaves displayed pentafoliate form, compared to those of WT with trifoliate compound leaves (Figure 1C). In the mib1 mutants, the size of the leaflets was severely reduced by 43.61%–60.93% (Figure 1D). The outgrowth of axillary buds in the mib1 mutants was faster than those in WT (Figure 1E). The number of branches in the mib1 mutants increased significantly (Figure 1F). At 4 weeks after germination, there was only one branch in each WT plant, while each mib1 mutant had four branches (Figure 1F). At 8 weeks after germination, there was no difference in the number of primary branches between WT and mutants of mib1-2 and mib1-3 (the mib1-1 mutant has about two more primary branches than WT; Supplementary Figure 1). However, the secondary branches in the three mib1 alleles increased significantly (Supplementary Figure 1). Thus, the increased branch number in the mutants was caused by accelerated bud outgrowth and sustained branching capacity among early developed primary branches. Additionally, the mib1 mutants had a compact plant architecture, compared with WT (Figure 1E and Supplementary Figure 1A).
The flowers and young pods of the mib1 mutants were smaller than those of WT (Supplementary Figure 2). The matured pods of the mutants were shorter, with decreased seed number and size (Figures 2A–E). The pod length of the mib1 mutants (6.6± 0.03, 6.7± 0.05, and 7.3 ± 0.09 cm, respectively) was decreased, compared to that of WT (9.8 ± 0.11 cm). The seed number per pod of three mib1 mutants (8.5 ± 0.11, 8.7 ± 0.20, and 9.9 ± 0.09, respectively) was much lower than that of WT (11.3 ± 0.65). Compared with the WT, mature seeds of mib1 mutants were rounder and showed significantly decreased length, width, and thickness (Figures 2D, F). Therefore, the 100-seed weight was decreased by 26.82%, 18.63%, and 27.42% in mib1-1, mib1-2, and mib1-3 mutants, compared with that of WT, respectively (Figure 2E).
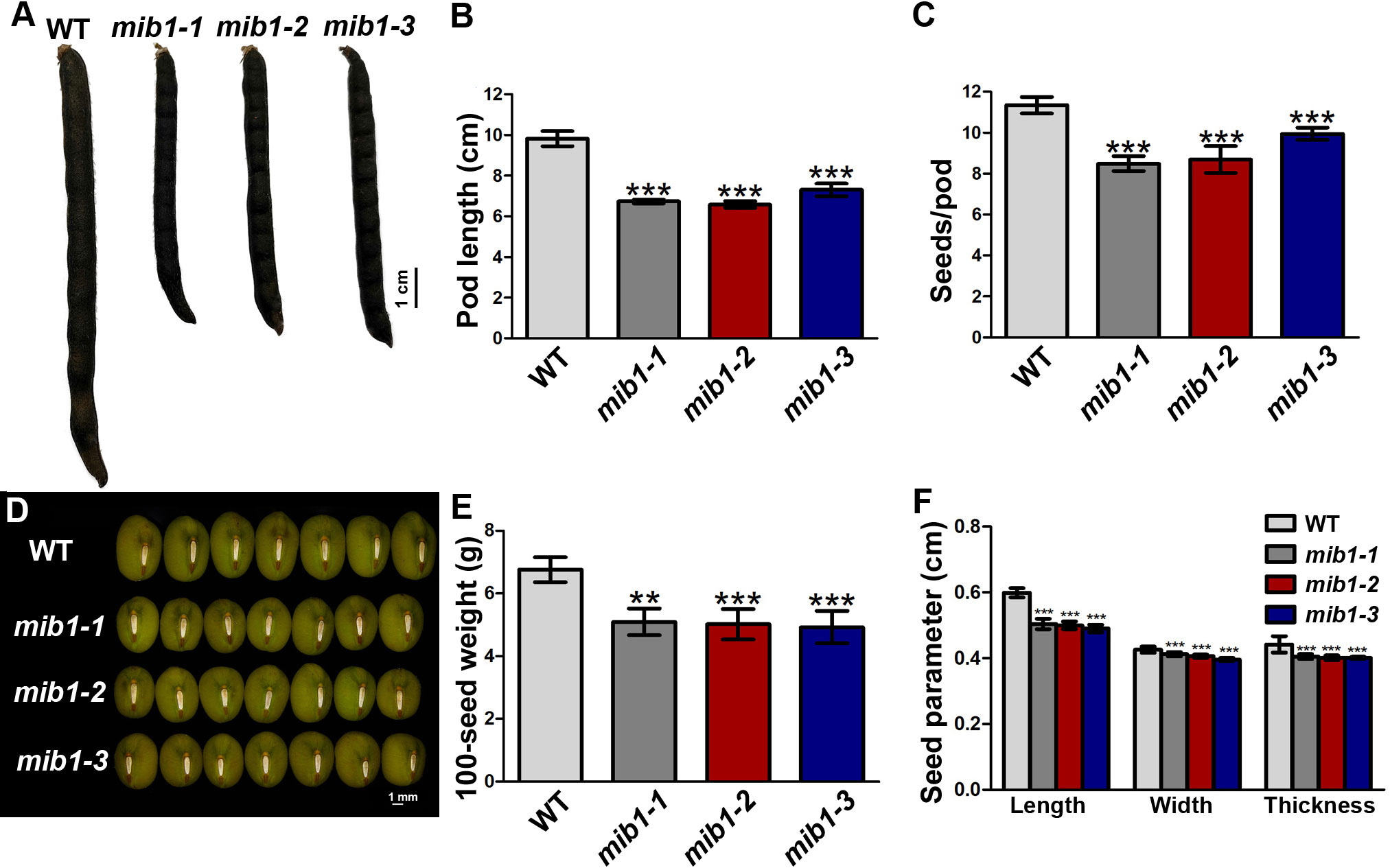
Figure 2 Characterization of pods and seeds of WT and mib1 mutants. (A) Pods of WT and mib1 mutants at matured stage. (B) Pod length of WT and mib1 mutants (n = 150). (C) Seed number per pod of WT and mib1 mutants (n = 150). (D) Seeds of WT and mib1 mutants. (E) The 100-seed weights of WT and mib1 mutants (n = 5). (F) Seed parameters of WT and mib1 mutants (n = 200). The data were means ± SD. The Tukey’s multiple comparison test was used. ** p < 0.01, *** p < 0.001.
The plant organ size is regulated by the coordination of two connected processes, cell division and expansion (Gonzalez et al., 2012). Microscopic examination of leaflet epidermal cells showed that the cell size decreased significantly in the mib1-3 mutants in comparison with that of WT (Supplementary Figure 3A). The area of epidermal cells in the mib1-3 mutants was only about half that in the WT plants (Supplementary Figure 3B), suggesting that MIB1 augments organ size mainly by increasing the cell size.
Molecular characterization of the MIB1 gene in mungbean
Genetic analysis of the mib1 mutants was conducted by backcrossing mib1-3 mutants with the WT plants. All F1 plants were similar to WT. In the F2 population, the WT plants and mutant plants segregated with a 3:1 ratio (87 WT plants and 25 mutants, χ2 = 0.42 < χ2 0.05 = 3.84), indicating that mib1 was a single recessive locus.
We conducted map-based cloning to identify the MIB1 gene (Jiao et al., 2016). The MIB1 gene was preliminarily mapped on chromosome 1 of the VC1973A genome (Kang et al., 2014), linked with the markers ID244 and ID171 (Figure 3A). By developing new markers, the mib1 mapping region was narrowed down to a 1.71-Mb region flanked by the markers ID218 and ID201 (Figure 3A). Based on the functional annotation (Kang et al., 2014) and the mutant phenotype, Vradi01g10280 (LOC106766026) in the mapping region was identified as the candidate (Figure 3B). Sequencing of the PCR products of Vradi01g10280 from WT and mib1 mutants displayed mutations (Figure 3B), showing that three alleles, mib1-1, mib1-2, and mib1-3, carried different deletions (1-bp deletion, 1-bp deletion, and 21-bp deletion, respectively). qRT-PCR analysis of shoot buds (2 weeks after germination) revealed that there were decreased expression of the Vradi01g10280 gene in the mib1 mutants (Figure 3C).
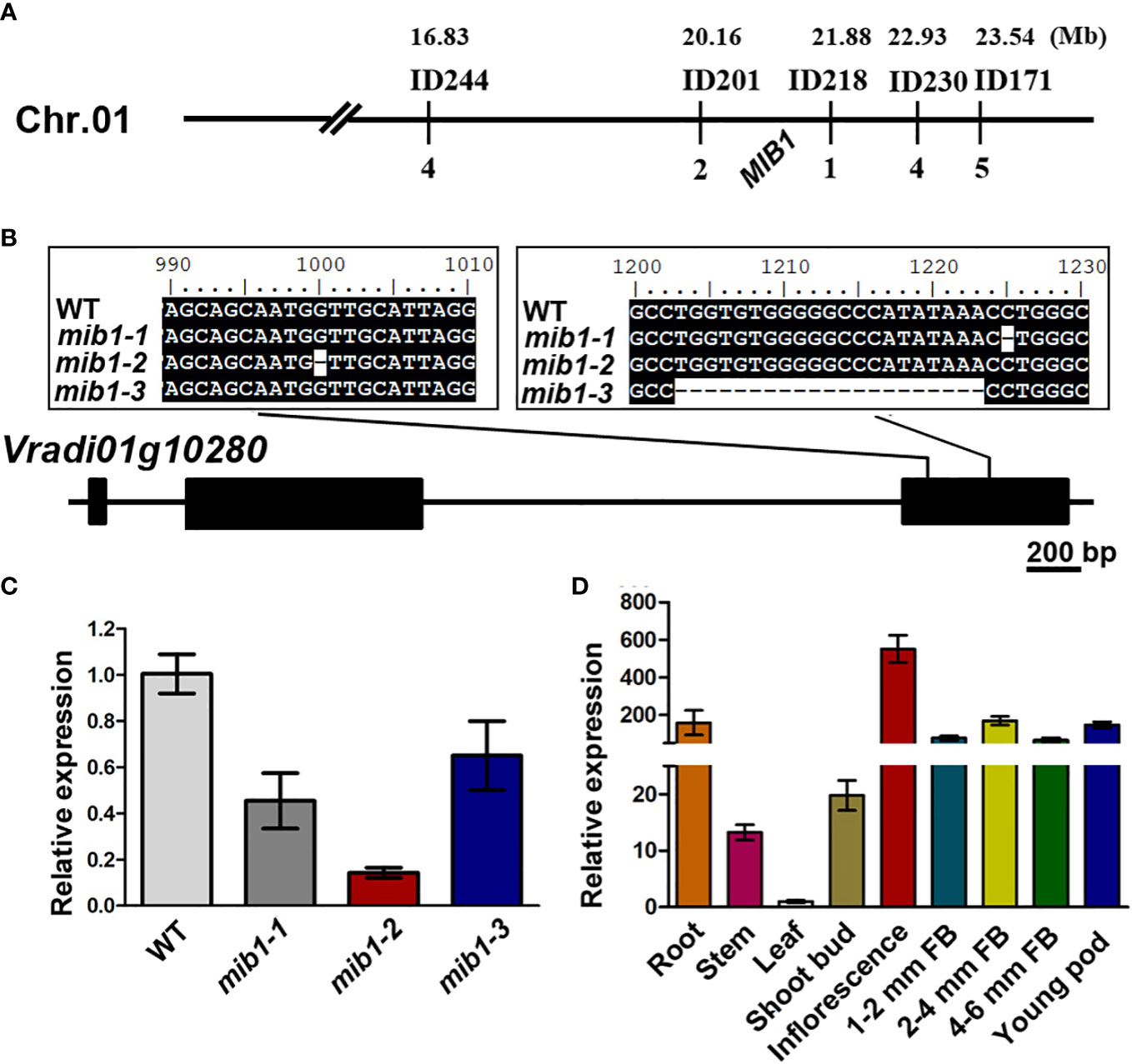
Figure 3 Map-based cloning of MIB1. (A) Genetic map of MIB1 in mungbean. (B) Mutations in the open reading frame of Vradi01g10280. Numbers up the sequence indicate the position on the open reading frame. (C) Analysis of MIB1 expression in shoot buds of WT and mib1 mutant by qRT-PCR. (D) Relative expression level of MIB1 in different tissues of WT.
Segregation analysis showed that 150 mutated plants out of a total of 642 individuals from the F2 mapping population were homozygous for the 21-bp deletion in Vradi02g10020, indicating that the deletion co-segregates with the mutant phenotype. Therefore, MIB1 (Vradi01g10280) encoded a member of MATE/DTX proteins (Figure 4), which was closely related to DTX54/BIGE1A in Arabidopsis and BIGE in maize (Suzuki et al., 2015), affecting plant architecture in mungbean.
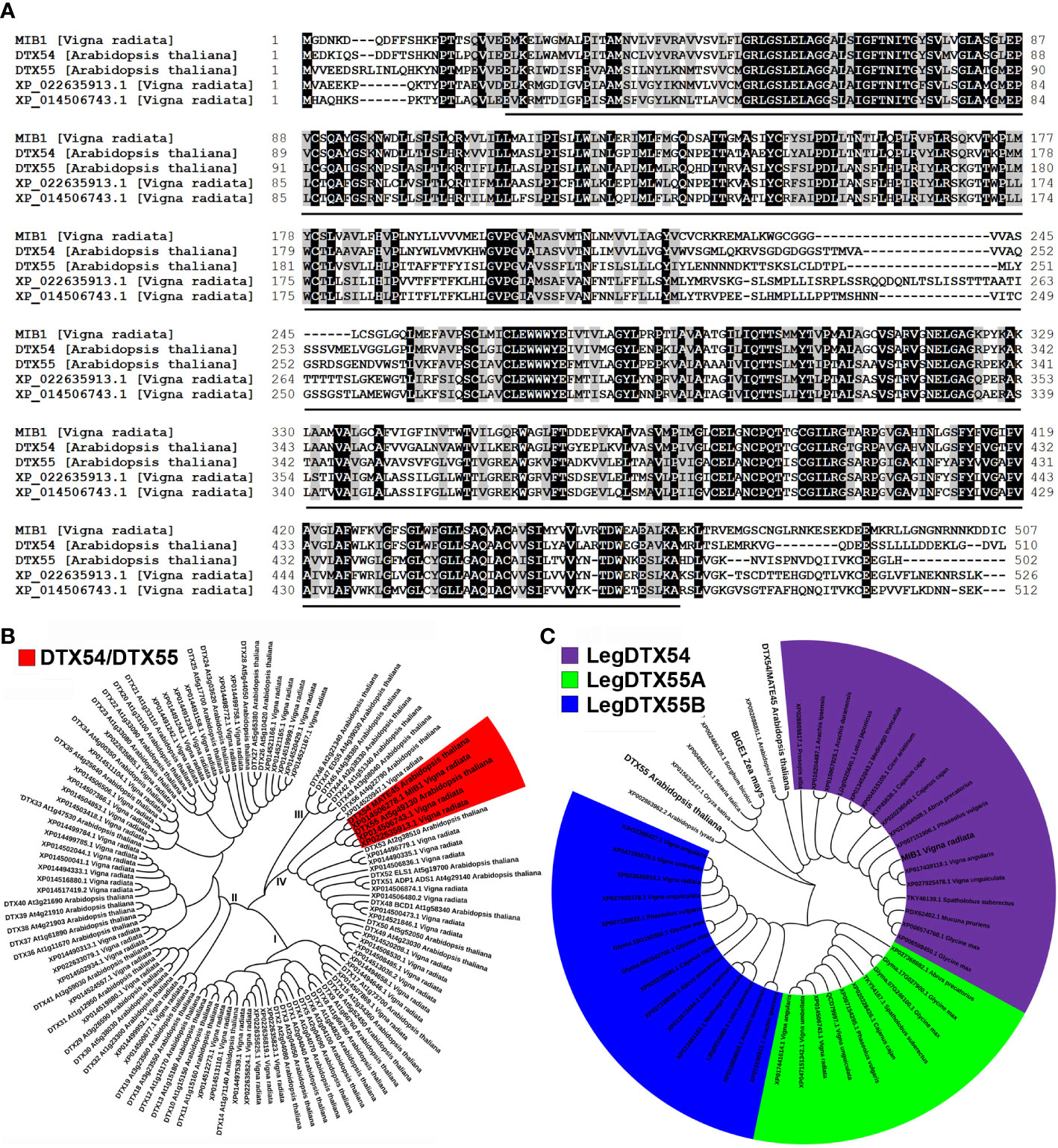
Figure 4 Analysis of the MIB1 protein isolated from mungbean. (A) Alignment of amino acid sequences of MIB1, DTX54, DTX55, XP_022635913.1, and XP_014506743.1. The conserved MatE domain was indicated by the black line. (B) Phylogenetic tree of MATE family proteins in mungbean and Arabidopsis. I, II, III, and IV represent four different groups. (C) Phylogenetic analysis of LegDTX54 clade and LegDTX55 clade proteins in legume.
The relative expression of the MIB1 gene in different tissues of mungbean was analyzed by qRT-PCR. It was found that the MIB1 gene was expressed in all tissues, with the highest expression level in the young inflorescence (Figure 3D).
MIB1 was a member of the MATE/DTX family proteins in mungbean
Multiple amino acid sequence alignments of the MIB1 protein (XP_014506278.1) with its homologs indicated that it shared a conserved MatE domain (Figure 4A). The MIB1 protein was predicted to have 12 transmembrane domains with N-termini towards the inside of the cell (Supplementary Figure 4).
We conducted a BLASTP search for sequences with homology to MIB1 to characterize the MATE/DTX family in the mungbean database (Kang et al., 2014) and found 56 MATE/DTX proteins in the mungbean genome (Supplementary Table 2). These mungbean MATE/DTX proteins were classified into four groups by phylogenetic analysis with Arabidopsis MATE/DTX proteins (Figure 4B; Wang et al., 2016). It was found that MIB1 had two other closely related homologs in mungbean, XP_022635913.1 (Vradi05g00900) and XP_014506743.1 (Vradi07g25110, Figure 4B).
In order to investigate the origin of MIB1 in legume plants, we identified MIB1 closed homologs from a number of public databases (Supplementary Table 2). The phylogenetic tree of aligned legume DTX54 and DTX55 orthologs was constructed (Figure 4B). It was found that one copy encoding the ortholog to DTX54 in legume formed the LegDTX54 clade, which was distinct from the LegDTX55 clade (Figure 4C). In contrast, within the LegDTX55 clade, there were different copies in legume, such as two copies in adzuki bean (V. angularis) and mungbean, and one copy in Medicago truncatula and Lotus japonicus (Figure 4C). The best phylogeny places the legume DTX55A (LegDTX55A) subclade and the legume DTX55B (LegDTX55B) subclade sister together, forming the LegDTX55 clade in legume (Figure 4C).
Heterologous expression of mungbean MIB1 gene increased TBA tolerance in the mutant Escherichia coli
To investigate the functional character of the MIB1 protein, the expression vector containing the MIB1 gene or empty vector was transformed into the WT strain (K12) and mutant strain (ΔacrB) in E. coli. The ΔacrB mutant strain lacks the functional AcrAB complex, the multidrug efflux carrier (Seo et al., 2012), and cannot grow under toxic conditions. The transformed cells were grown on the medium with and without different concentrations of TBA. The ΔacrB mutant cells with empty expressing vector (pET32a) did not grow on an LB plate with 10 and 15 mg/ml TBA (Figure 5A), while the MIB1-expressing ΔacrB cells continued their growth on the LB medium with 10 and 15 mg/ml TBA (Figure 5A), suggesting that MIB1 in mungbean potentially acts as a MATE/DTX family transporter.
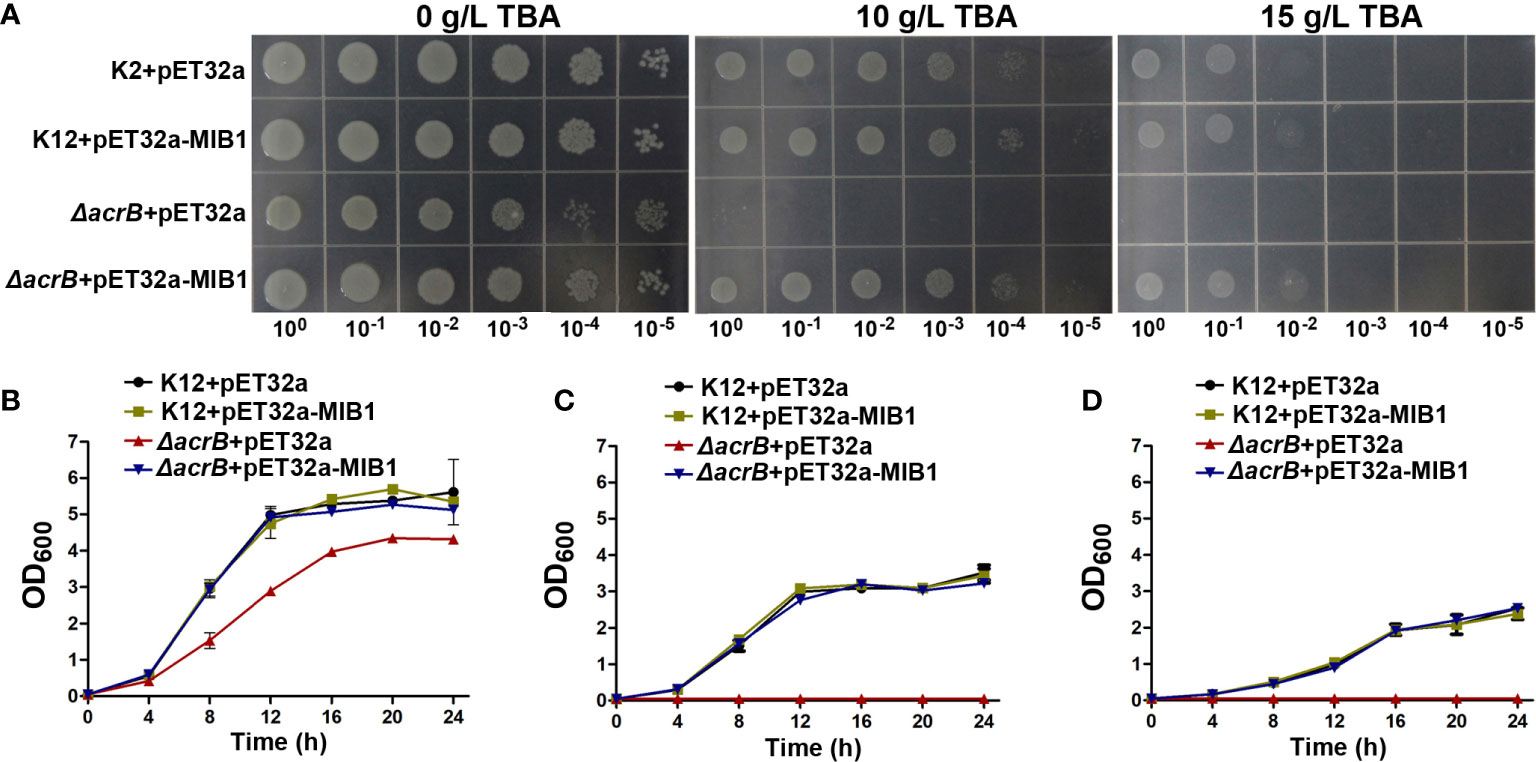
Figure 5 Analysis of MIB1 transport function in Escherichia coli. (A) The effect of MIB1 expression on the growth of (E) coli cells under TBA treatment on the LB plate. (E) coli cells were spotted on the LB plate with 0, 10, and 15 g/L TBA for 24 h. 100, 10−1, 10−2, 10−3, 10−4, and 10−5 represented dilution series. (B–D) The effect of MIB1 expression on the growth curve of (E) coli cells under TBA treatment. (E) coli cells were inoculated in liquid LB medium with 0 (B), 10 (C), and 15 (D) g/L TBA for 24 h. The data were means ± SD (n = 3).
In order to further verify the results of the plate experiment, we determined the growth curve of the strains under 0, 10, and 15 mg/ml TBA treatment in liquid LB medium (Figures 5B–D). Compared to those of expressing MIB1 cells and the WT strain, TBA treatment significantly inhibited the growth of the mutant strain (Figures 5B–D). Under 10 and 15 mg/ml TBA treatments for 24 h, the growth curve of the mutant strain expressing MIB1 was similar to those of the WT strain with and without expressing MIB1 (Figures 5C, D). The above results showed that heterologous expression of mungbean MIB1 increased TBA tolerance of the ΔAcrB mutant strain.
Heterologous expression of mungbean MIB1 gene partially rescued the pod phenotype of dtx54 mutant in Arabidopsis
It has been reported that loss of function of DTX54/BIGE1A, MIB1 ortholog in Arabidopsis, leads to the shortened pods (Suzuki et al., 2015). To test whether MIB1 performs a similar function to DTX54/BIGE1A in the control of pod development, the coding sequence of MIB1 driven by the cauliflower mosaic virus 35S (CaMV35S) promoter was transformed into the Arabidopsis dtx54 mutant (35S::MIB1/dtx54, two representative lines L04 and L06). It was found that the shortened pods were partially rescued in 35S::MIB1/dtx54 transgenic lines (Figures 6A, B). The expression of MIB1 was detected in 35S::MIB1/dtx54 transgenic lines (Figure 6C). The above results indicated that the mungbean MIB1 gene plays a conserved role in the pod development.
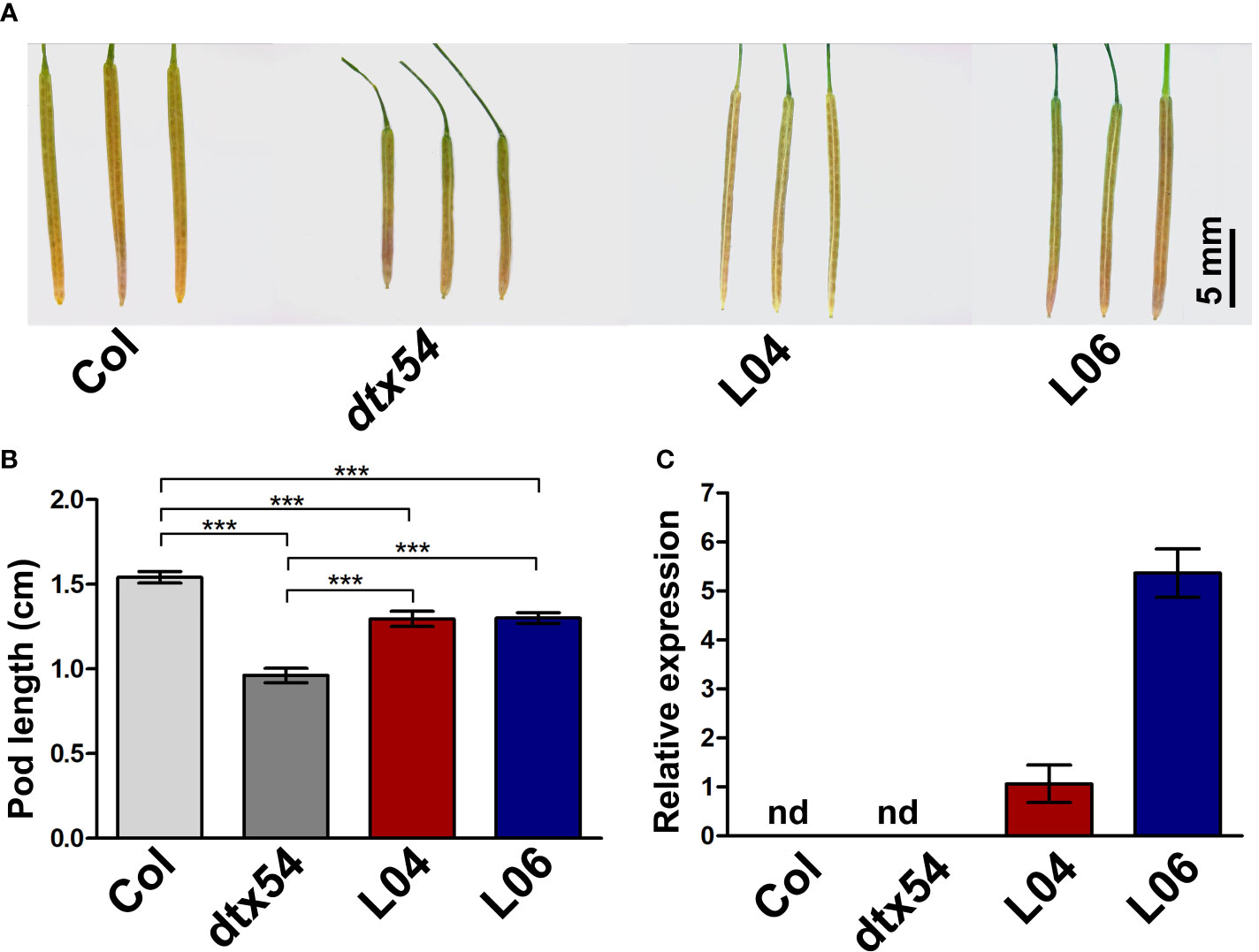
Figure 6 MIB1 partially rescued the shortened pod phenotype of Arabidopsis dtx54 mutant. (A) The pod phenotype of the wild-type plant (Col), dtx54 mutant, and 35S::MIB1 transgenic lines of dtx54 (L04 and L06). (B) The pod length of Col, dtx54 mutants, and two transgenic lines (n = 100). (C) qRT-PCR analysis of MIB1 expression from Col, dtx54 mutant, and two transgenic lines. nd, not detected. The data were means ± SD. One-way ANOVA was used. ***p < 0.001.
RNA-seq analysis of the wild-type plants and mib1 mutants
To investigate the potential genes whose expression was altered in the mutants, RNA-sequencing (RNA-seq) analysis was conducted to compare the gene expression levels in shoot buds and young pods between WT and mib1-3 mutants. A total of 3,173 and 875 differentially expressed genes (DEGs) were identified at the two developmental stages, respectively (Figure 7, Supplementary Tables 3 and 4). The qRT-PCR analysis confirmed the results of the RNA-Seq (Figures 8A, B).
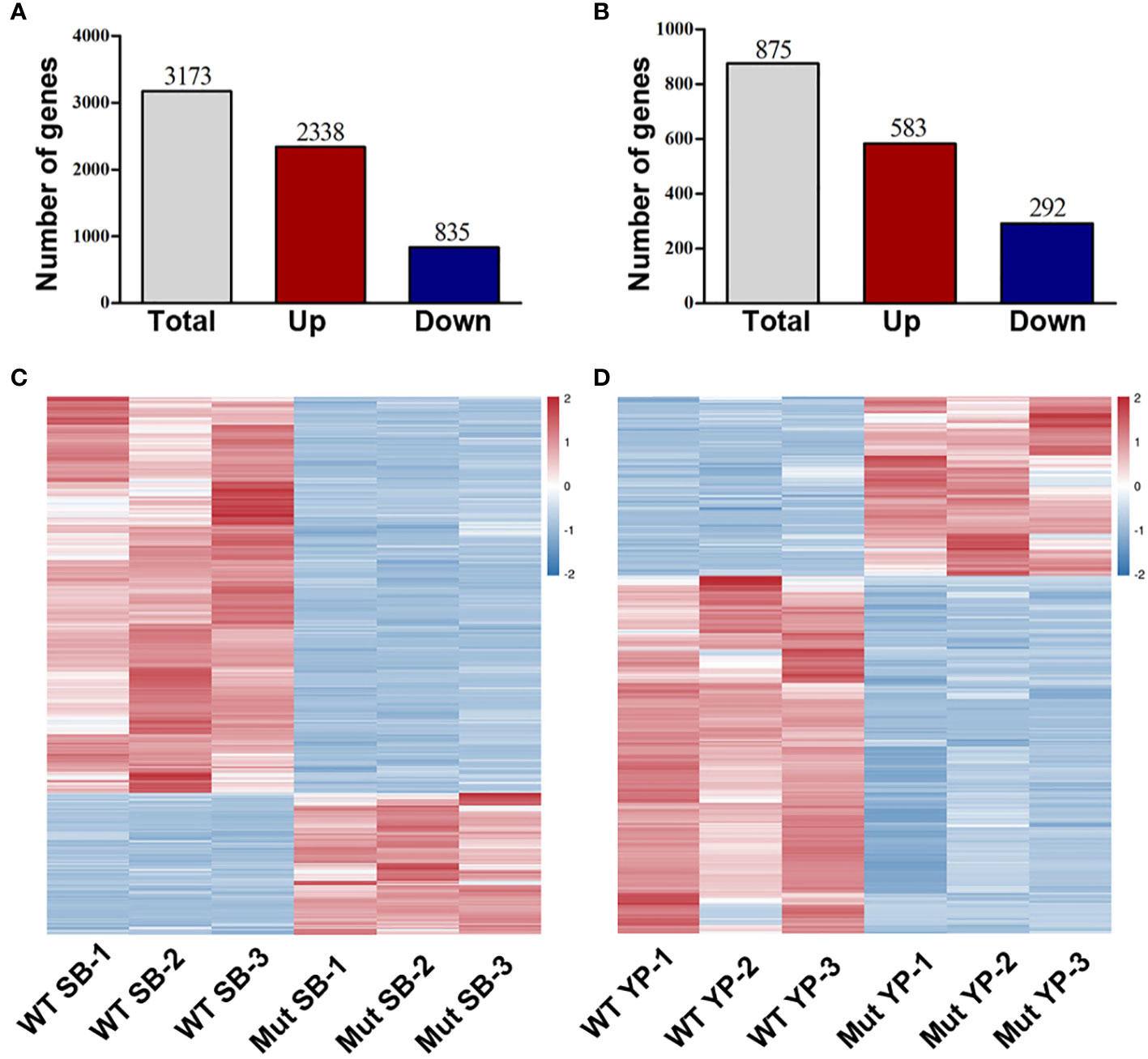
Figure 7 RNA-seq analysis of DEGs between WT and mib1 mutants. (A) Number of DEGs of the shoot buds between WT and mib1 mutants. (B) Number of DEGs of the young pods between WT and mib1 mutants. (C) Heat map of the DEGs of the shoot buds between WT and mib1 mutants. (D) Heat map of the DEGs of the young pods between WT and mib1 mutants.
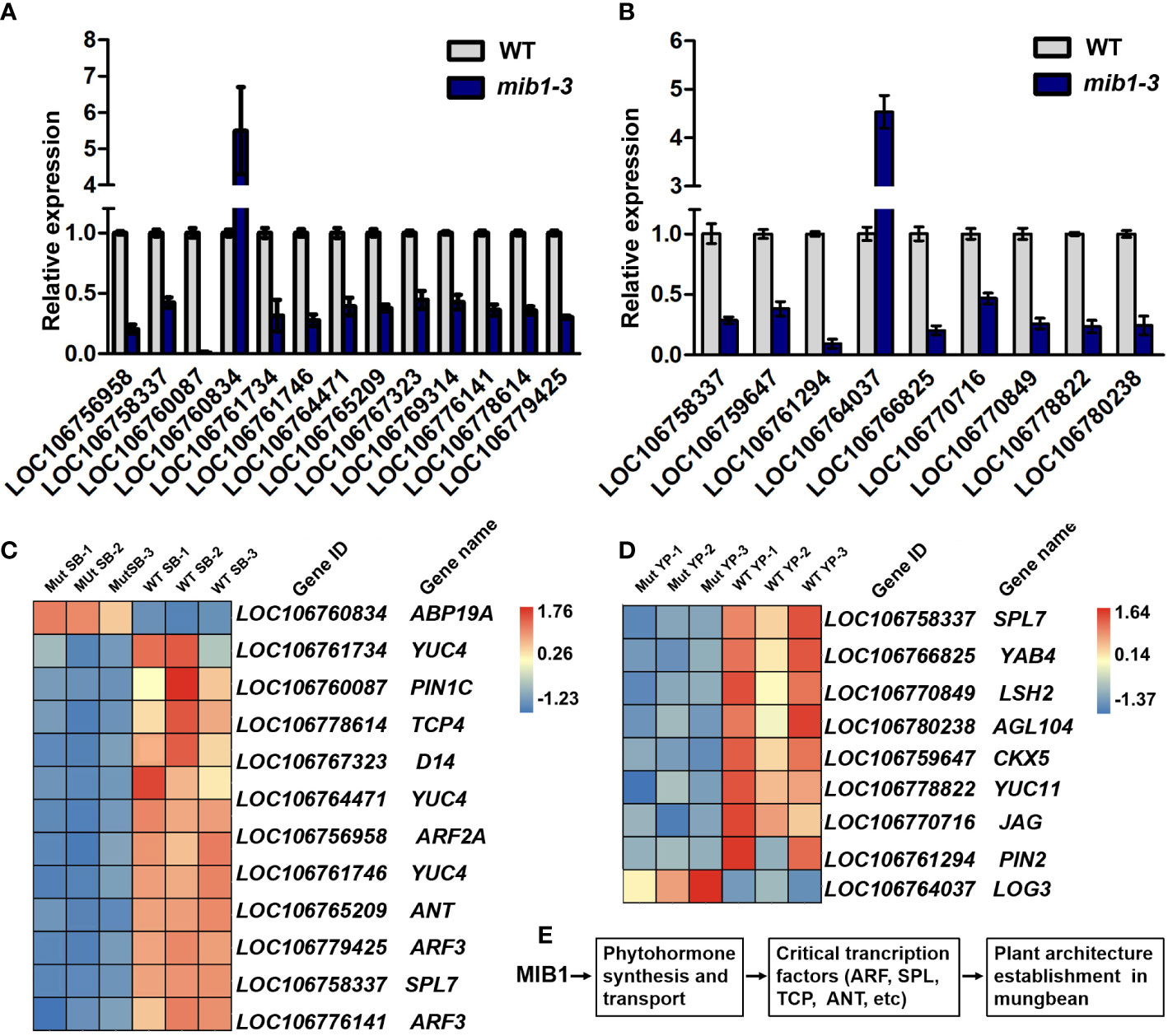
Figure 8 Critical DEGs involved in the control of plant architecture in mungbean. (A) qRT-PCR analysis of 13 DEGs in the shoot buds between WT and mib1 mutants. (B) qRT-PCR analysis of eight DEGs in young pods between WT and mib1 mutants. (C) Heat map showing critical DEGs involved in the control of the rate of leaf production, branch number, and organ size. (D) Heat map showing critical DEGs in involved young pod development. (E) A hypothetical model of MIB1 affecting plant architecture in mungbean.
Consistent with the mutant phenotype of plant architecture, the expression levels of key components of the plant hormone pathway and transcription factors related to plant development and growth were changed in the mib1 mutants (Figures 8C, D). Among DEGs of the shoot buds between WT and mib1 mutants, three auxin biosynthesis genes (LOC106761734, LOC106761746, and LOC106764471, YUCCA 4, YUC4) and a gene encoding auxin transporter (LOC106760087, PIN-FORMED 1C, PIN1C) were downregulated (Figure 8C). Several transcription factor encoding genes involved in plant development, such as LOC06756958 (Auxin Response Factor 2, ARF2), LOC106778614 (TCP family gene, TCP4), LOC106769314 and LOC106758337 (SPL family genes, SPL7 and SPL8), and LOC06765209 (AP2/ERF family gene AINTEGUMENTA, ANT), were downregulated (Figure 8C). Additionally, LOC106767323 (DWARF 14, D14), encoding a key component of the SL signaling pathway (Zhou et al., 2013), was downregulated in the shoot buds of the mib1 mutants (Figure 8C).
It has been shown that auxin and cytokinin pathways play a key role in the control of pod development and seed number per pod (Liu et al., 2021; Qadir et al., 2021; Yu et al., 2022). We found that the auxin biosynthesis gene (LOC106778822, YUC11) and the auxin transporter encoding gene (LOC106761294, PIN2) were downregulated in the young pods of the mib1 mutants (Figure 8D). Consistently, there was a significant reduction in IAA level in young pods of the mib1 mutants, compared to that of WT (Supplementary Figure 5). In addition, the expression levels of LOC106759647 (Cytokinin dehydrogenase 3, CKX3) and LOC106764037 (LONELY GUY 3, LOG3), related to the cytokinin pathway, were also significantly changed in the young pods of the mib1 mutants (Figure 8D).
Discussion
MIB1 encoded a MATE/DTX family transporter, affecting plant architecture in mungbean
The plant architecture significantly affects the seed yield of grain legume. However, the underlying molecular mechanism is still poorly understood (Liu et al., 2020). In this study, The mutations of the MIB1 gene in mungbean resulted in bushy and compact plant architecture (Figure 1) and shortened pods with smaller and rounder seeds (Figure 2). Map-based cloning showed that the MIB1 gene encoded a MATE/DTX family protein in mungbean, which was an ortholog of DTX54/BIGE1A in Arabidopsis and BIGE in maize (Figure 4). It has been reported that loss of function of the DTX54/BIGA1A gene, MIB1 ortholog in Arabidopsis, gives rise to increased branch numbers and shortened pods (Suzuki et al., 2015). We found that heterologous expression of the MIB1 gene partially rescued the phenotype of dtx54/bige1a mutant in Arabidopsis, suggesting that MIB1 plays a conserved role in the control of pod development.
MIB1 belonged to group IV of the MATE/DTX family (Figure 4A). Complementation assays in E. coli showed that MIB1 potentially acted as a MATE/DTX transporter in mungbean. Meanwhile, there was a significant reduction in IAA levels in young pods of the mib1 mutants (Supplementary Figure 5). Consistently, transcriptome analysis revealed that expression levels of the genes related to auxin synthesis and transport were decreased (Figure 8). Thus, our results suggested that auxin plays a key role in regulating plant architecture in mungbean. The alteration of plant architecture in the mib1 mutants was probably due to the modulated levels of auxin and other plant hormones and then the altered expression of the downstream genes related to plant growth and development (Figure 8E).
It has been reported that the group IV MATE/DTX transporters are able to modulate plant hormone levels such as auxin and ABA in Arabidopsis and rice (Li et al., 2014; Zhang et al., 2014; Qin et al., 2021). Thus, how plant hormone level is modulated by the MIB1 protein should be investigated in more detail in the future.
Phylogenetic analysis of DTX54 and DTX55 orthologs in legume
It has been shown that the DTX54/BIGE1A and DTX55/BIGE1B in Arabidopsis, two paralogs, have partial functional redundancy and diversity (Suzuki et al., 2015). The mutant of the Arabidopsis DTX54/BIGE1A gene shows increased number of leaves (Suzuki et al., 2015). By contrast, the dtx55 mutant exhibits a slight increase in leaf number, suggesting that Arabidopsis DTX54 has a greater role in the control of leaf initiation, while the leaf number of the double mutants of DTX54/BIGE1A and DTX55/BIGE1B is enhanced compared to the single mutants, indicating that there is an additive interaction between the two genes (Suzuki et al., 2015).
It was found that there were 56 MATE/DTX family proteins in mungbean genome (Figure 4), among which two other MATE/DTX proteins are closely related to MIB1 and might redundantly affect plant development and growth in mungbean. Moreover, based on the public sequences, we identified DTX54 and DTX55 orthologs in legume. We found that there was a single copy encoding the DTX54 orthologs in legume (Figure 4). In contrast, the legDTX55 clade in legume could be further divided into two subclades, LegDTX55A and LegDTX55B (Figure 4B).
At present, mutant libraries for several legume species such as M. truncatula, L. japonicus, and G. max are available (Tadege et al., 2008; Małolepszy et al., 2016; Li et al., 2017), and it would be worth identifying the mutant lines of the LegDTX54 and LegDTX55 clade genes in these species to dissect their function in plant architecture establishment. Moreover, it is also interesting to study the interactions between the LegDTX54 clade and LegDTX55 clade genes during plant development and growth in legume.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
XL wrote the manuscript. XL, YJ, MS, ZJ, HZ, DQ, QC, YX, and XY performed the experiments. XC and ZS supervised the research. XL and ZS analyzed the data and prepared the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Science Foundation of Jiangsu Province, China (BE2021718), Jiangsu Seed Industry Revitalization Project (JBGS[2021]004), the Jiangsu Agricultural Science and Technology Innovation Fund of China (CX(20)3030), and the Students' innovation and entrepreneurship training program of National experimental Teaching Center for Plant Production (ZKF202212).
Acknowledgments
We would like to thank Professor Chuanzhen Jiang (South China Agricultural University) for providing the K12 and ΔacrB mutant strains.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1064685/full#supplementary-material
Supplementary Figure 1 | The number of primary and secondary branches of WT and mib1 mutants at the matured stage. (A) Plant architecture of WT and mib1 mutants at the 8 weeks after germination; (B) The number of primary and secondary branches of WT and mib1 mutants. The data were means ± SD (n=10). The Tukey’s multiple comparison test was used. **p < 0.01, ***p < 0.001.
Supplementary Figure 2 | Characterization of the flowers (A) and young pods (B) of WT and mib1 mutants.
Supplementary Figure 3 | SEM analysis of the terminal leaflets of WT and mib1 mutants. (A) The epidermal cells of the terminal leaflets of the fifth compound leaves in WT and mib1-3 mutants; (B) The size of epidermal cells from the terminal leaflets of WT and mib1-3 mutants. The data were means ± SD (n=100). The Student’s test was used. ***p < 0.001.
Supplementary Figure 4 | The 12 transmembrane domains of the MIB1 protein was predicted using the DeepTMHMM. (A) Most likely topology of MIB1; (B) Posterior probabilities of MIB1.
Supplementary Figure 5 | The hormone levels of IAA and ABA in young pods of WT and mib1 mutants. The data were means ± SD (n=5). The Student’s test was used. **p < 0.01.
Supplementary Table 1 | Primer list used in this study.
Supplementary Table 2 | MATE family proteins used for phylogenetic analysis in this study.
Supplementary Table 3 | DEGs in shoot buds between WT and mib1 mutants.
Supplementary Table 4 | DEGs in young pods between WT and mib1 mutants.
References
Audic, S., Claverie, J. M. (1997). The significance of digital gene expression profiles. Genome Res. 7, 986–995. doi: 10.1101/gr.7.10.986
Bao, A., Chen, H., Chen, L., Chen, S., Hao, Q., Guo, W., et al. (2019). CRISPR/Cas9-mediated targeted mutagenesis of GmSPL9 genes alters plant architecture in soybean. BMC Plant Biol. 19, 131. doi: 10.1186/s12870-019-1746-6
Burko, Y., Geva, Y., Refael-Cohen, A., Shleizer-Burko, S., Shani, E., Berger, Y., et al. (2011). From organelle to organ: ZRIZI MATE-type transporter is an organelle transporter that enhances organ initiation. Plant Cell Physiol. 52, 518–527. doi: 10.1093/pcp/pcr007
Chen, L., Yang, H., Fang, Y., Guo, W., Chen, H., Zhang, X., et al. (2021). Overexpression of GmMYB14 improves high-density yield and drought tolerance of soybean through regulating plant architecture mediated by the brassinosteroid pathway. Plant Biotechnol. J. 19, 702–716. doi: 10.1111/pbi.13496
Clough, S. J., Bent, A. F. (1998). Floral dip: A simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Diener, A. C., Fink, G. R., Gaxiola, R. A. (2001). Arabidopsis ALF5, a multidrug efflux transporter gene family member, confers resistance to toxins. Plant Cell 13, 1625–1637. doi: 10.1105/tpc.010035
Duan, W., Lu, F., Cui, Y., Zhang, J., Du, X., Hu, Y., et al. (2022). Genome-wide identification and characterisation of wheat MATE genes reveals their roles in aluminium tolerance. Int. J. Mol. Sci. 23, 4418. doi: 10.3390/ijms23084418
Gani, U., Nautiyal, A. K., Kundan, M., Rout, B., Pandey, A., Misra, P. (2022). Two homeologous MATE transporter genes, NtMATE21 and NtMATE22, are involved in the modulation of plant growth and flavonol transport in Nicotiana tabacum. J. Exp. Bot. 73 (18), 6186–6206. doi: 10.1093/jxb/erac249
Gao, J., Yang, S., Cheng, W., Fu, Y., Leng, J., Yuan, X., et al. (2017). GmILPA1, encoding an APC8-like protein, controls leaf petiole angle in soybean. Plant Physiol. 174, 1167–1176. doi: 10.1104/pp.16.00074
Gonzalez, N., Vanhaeren, H., Inzé, D. (2012). Leaf size control: complex coordination of cell division and expansion. Trends Plant Sci. 17, 332–340. doi: 10.1016/j.tplants.2012.02.003
Graham, P. H., Vance, C. P. (2003). Legumes: importance and constraints to greater use. Plant Physiol. 131, 872–877. doi: 10.1104/pp.017004
Guo, W., Chen, L., Herrera-Estrella, L., Cao, D., Tran, L. P. (2020). Altering plant architecture to improve performance and resistance. Trends Plant Sci. 25, 1154–1170. doi: 10.1016/j.tplants.2020.05.009
Ishihara, T., Sekine, K. T., Hase, S., Kanayama, Y., Seo, S., Ohashi, Y., et al. (2008). Overexpression of the Arabidopsis thaliana EDS5 gene enhances resistance to viruses. Plant Biol. (Stuttg) 10, 451–461. doi: 10.1111/j.1438-8677.2008.00050.x
Jia, M., Liu, X., Xue, H., Wu, Y., Shi, L., Wang, R., et al. (2019). Noncanonical ATG8-ABS3 interaction controls senescence in plants. Nat. Plants 5, 212–224. doi: 10.1038/s41477-018-0348-x
Jiao, K. Y., Li, X., Guo, W. X., Yuan, X. X., Cui, X. Y., Chen, X. (2016). Genome re-sequencing of two accessions and fine mapping the locus of lobed leaflet margins in mungbean. Mol. Breed 36, 128. doi: 10.1007/s11032-016-0552-1
Jiao, K., Li, X., Su, S., Guo, W., Guo, Y., Guan, Y., et al. (2019). Genetic control of compound leaf development in mungbean (Vigna radiata l.). Hortic. Res. 6, 23. doi: 10.1038/s41438-018-0088-0
Kang, Y. J., Kim, S. K., Kim, M. Y., Lestari, P., Kim, K. H., Ha, B. K., et al. (2014). Genome sequence of mungbean and insights into evolution within Vigna species. Nat. Commun. 5, 5443. doi: 10.1038/ncomms6443
Kerr, S. C., Patil, S. B., de Saint Germain, A., Pillot, J. P., Saffar, J., Ligerot, Y., et al. (2021). Integration of the SMXL/D53 strigolactone signalling repressors in the model of shoot branching regulation in Pisum sativum. Plant J. 107, 1756–1770. doi: 10.1111/tpj.15415
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Letunic, I., Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290
Li, L., He, Z., Pandey, G. K., Tsuchiya, T., Luan, S. (2002). Functional cloning and characterization of a plant efflux carrier for multidrug and heavy metal detoxification. J. Biol. Chem. 277, 5360–5368. doi: 10.1074/jbc.M108777200
Li, Z., Jiang, L., Ma, Y., Wei, Z., Hong, H., Liu, Z., et al. (2017). Development and utilization of a new chemically-induced soybean library with a high mutation density. J. Integr. Plant Biol. 59, 60–74. doi: 10.1111/jipb.12505
Li, R., Li, J., Li, S., Qin, G., Novák, O., Pěnčík, A., et al. (2014). ADP1 affects plant architecture by regulating local auxin biosynthesis. PloS Genet. 10, e1003954. doi: 10.1371/journal.pgen.1003954
Li, X., Sun, M., Jia, Y., Qiu, D., Peng, Q., Zhuang, L. (2022). Genetic control of the lateral petal shape and identity of asymmetric flowers in mungbean (Vigna radiata l.). Front. Plant Sci. 13. doi: 10.3389/fpls.2022.996239
Liu, L. M., Zhang, H. Q., Cheng, K., Zhang, Y. M. (2021). Integrated bioinformatics analyses of PIN1, CKX, and yield-related genes reveals the molecular mechanisms for the difference of seed number per pod between soybean and cowpea. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.749902
Liu, S., Zhang, M., Feng, F., Tian, Z. (2020). Toward a "Green revolution" for soybean. Mol. Plant 13, 688–697. doi: 10.1016/j.molp.2020.03.002
Lu, P., Magwanga, R. O., Kirungu, J. N., Hu, Y., Dong, Q., Cai, X., et al. (2019). Overexpression of cotton a DTX/MATE gene enhances drought, salt, and cold stress tolerance in transgenic Arabidopsis. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00299
Magalhaes, J. V., Liu, J., Guimarães, C. T., Lana, U. G., Alves, V. M., Wang, Y. H., et al. (2007). A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat. Genet. 39, 1156–1161. doi: 10.1038/ng2074
Małolepszy, A., Mun, T., Sandal, N., Gupta, V., Dubin, M., Urbański, D., et al. (2016). The LORE1 insertion mutant resource. Plant J. 88, 306–317. doi: 10.1111/tpj.13243
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Nawrath, C., Heck, S., Parinthawong, N., Métraux, J. P. (2002). EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell. 14, 275–286. doi: 10.1105/tpc.010376
Nimmy, M. S., Kumar, V., Suthanthiram, B., Subbaraya, U., Nagar, R., Bharadwaj, C., et al. (2022). A systematic phylogenomic classification of the multidrug and toxic compound extrusion transporter gene family in plants. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.774885
Pan, X., Welti, R., Wang, X. (2010). Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat. Protoc. 5, 986–992. doi: 10.1038/nprot.2010.37
Qadir, M., Wang, X., Shah, S., Zhou, X. R., Shi, J., Wang, H. (2021). Molecular network for regulation of ovule number in plants. Int. J. Mol. Sci. 22, 12965. doi: 10.3390/ijms222312965
Qin, P., Zhang, G., Hu, B., Wu, J., Chen, W., Ren, Z., et al. (2021). Leaf-derived ABA regulates rice seed development via a transporter-mediated and temperature-sensitive mechanism. Sci. Adv. 7, eabc8873. doi: 10.1126/sciadv.abc8873
Rameau, C., Bertheloot, J., Leduc, N., Andrieu, B., Foucher, F., Sakr, S. (2015). Multiple pathways regulate shoot branching. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00741
Reinhardt, D., Kuhlemeier, C. (2002). Plant architecture. EMBO Rep. 3, 846–851. doi: 10.1093/embo-reports/kvf177
Rogers, E. E., Guerinot, M. L. (2002). FRD3, a member of the multidrug and toxin efflux family, controls iron deficiency responses in Arabidopsis. Plant Cell 14, 1787–1799. doi: 10.1105/tpc.001495
Seo, P. J., Park, J., Park, M. J., Kim, Y. S., Kim, S. G., Jung, J. H., et al. (2012). A golgi-localized MATE transporter mediates iron homoeostasis under osmotic stress in Arabidopsis. Biochem. J. 442, 551–561. doi: 10.1042/BJ20111311
Sun, Z., Su, C., Yun, J., Jiang, Q., Wang, L., Wang, Y., et al. (2019). Genetic improvement of the shoot architecture and yield in soya bean plants via the manipulation of GmmiR156b. Plant Biotechnol. J. 17, 50–62. doi: 10.1111/pbi.12946
Suzuki, M., Sato, Y., Wu, S., Kang, B. H., McCarty, D. R. (2015). Conserved functions of the MATE transporter BIG EMBRYO1 in regulation of lateral organ size and initiation rate. Plant Cell. 27, 2288–2300. doi: 10.1105/tpc.15.00290
Tadege, M., Wen, J., He, J., Tu, H., Kwak, Y., Eschstruth, A., et al. (2008). Large-Scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 54, 335–347. doi: 10.1111/j.1365-313X.2008.03418.x
Thompson, E. P., Wilkins, C., Demidchik, V., Davies, J. M., Glover, B. J. (2010). An arabidopsis flavonoid transporter is required for anther dehiscence and pollen development. J. Exp. Bot. 61, 439–451. doi: 10.1093/jxb/erp312
Upadhyay, N., Kar, D., Datta, S. A. (2020). Multidrug and toxic compound extrusion (MATE) transporter modulates auxin levels in root to regulate root development and promotes aluminium tolerance. Plant Cell Environ. 43, 745–759. doi: 10.1111/pce.13658
Upadhyay, N., Kar, D., Deepak Mahajan, B., Nanda, S., Rahiman, R., Panchakshari, N., et al. (2019). The multitasking abilities of MATE transporters in plants. J. Exp. Bot. 70, 4643–4656. doi: 10.1093/jxb/erz246
Wang, L., Bei, X., Gao, J., Li, Y., Yan, Y., Hu, Y. (2016). The similar and different evolutionary trends of MATE family occurred between rice and Arabidopsis thaliana. BMC Plant Biol. 16, 207. doi: 10.1186/s12870-016-0895-0
Wang, Y., Li, J. (2008). Molecular basis of plant architecture. Annu. Rev. Plant Biol. 59, 253–279. doi: 10.1146/annurev.arplant.59.032607.092902
Yu, S. X., Jiang, Y. T., Lin, W. H. (2022). Ovule initiation: the essential step controlling offspring number in Arabidopsis. J. Integr. Plant Biol. 64, 1469–1486. doi: 10.1111/jipb.13314
Zhang, H., Zhu, H., Pan, Y., Yu, Y., Luan, S., Li, L. (2014). A DTX/MATE-type transporter facilitates abscisic acid efflux and modulates ABA sensitivity and drought tolerance in Arabidopsis. Mol. Plant 7, 1522–1532. doi: 10.1093/mp/ssu063
Keywords: legume, mungbean, plant architecture, MIB1, MATE/DTX family, RNA-Seq
Citation: Li X, Jia Y, Sun M, Ji Z, Zhang H, Qiu D, Cai Q, Xia Y, Yuan X, Chen X and Shen Z (2022) MINI BODY1, encoding a MATE/DTX family transporter, affects plant architecture in mungbean (Vigna radiata L.). Front. Plant Sci. 13:1064685. doi: 10.3389/fpls.2022.1064685
Received: 08 October 2022; Accepted: 01 November 2022;
Published: 17 November 2022.
Edited by:
Zhengjun Xia, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Zhenfeng Jiang, Northeast Agricultural University, ChinaZhijuan Wang, Huazhong Agricultural University, China
Akito Kaga, Institute of Crop Science (NARO), Japan
Copyright © 2022 Li, Jia, Sun, Ji, Zhang, Qiu, Cai, Xia, Yuan, Chen and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenguo Shen, emdzaGVuQG5qYXUuZWR1LmNu; Xin Chen, Y3hAamFhcy5hYy5jbg==
 Xin Li
Xin Li Yahui Jia1
Yahui Jia1 Mingzhu Sun
Mingzhu Sun Yan Xia
Yan Xia Xingxing Yuan
Xingxing Yuan Xin Chen
Xin Chen Zhenguo Shen
Zhenguo Shen