
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 14 December 2022
Sec. Plant Bioinformatics
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1064556
This article is part of the Research Topic Advances and Applications of Cost-Effective, High-Throughput Genotyping Technologies for Sustainable Agriculture View all 14 articles
 Shilpa Devi1†
Shilpa Devi1† Parva Kumar Sharma2
Parva Kumar Sharma2 Tusar Kanti Behera1,3*
Tusar Kanti Behera1,3* Sarika Jaiswal2
Sarika Jaiswal2 G. Boopalakrishnan1
G. Boopalakrishnan1 Khushboo Kumari1
Khushboo Kumari1 Neha Kumari Mandal1
Neha Kumari Mandal1 Mir Asif Iquebal2
Mir Asif Iquebal2 S. Gopala Krishnan4
S. Gopala Krishnan4 Bharti5
Bharti5 Chandrika Ghosal1
Chandrika Ghosal1 Anilabha Das Munshi1*
Anilabha Das Munshi1* Shyam Sundar Dey1*
Shyam Sundar Dey1*Parthenocarpy is an extremely important trait that revolutionized the worldwide cultivation of cucumber under protected conditions. Pusa Parthenocarpic Cucumber-6 (PPC-6) is one of the important commercially cultivated varieties under protected conditions in India. Understanding the genetics of parthenocarpy, molecular mapping and the development of molecular markers closely associated with the trait will facilitate the introgression of parthenocarpic traits into non-conventional germplasm and elite varieties. The F1, F2 and back-crosses progenies with a non-parthenocarpic genotype, Pusa Uday indicated a single incomplete dominant gene controlling parthenocarpy in PPC-6. QTL-seq comprising of the early parthenocarpy and non-parthenocarpic bulks along with the parental lines identified two major genomic regions, one each in chromosome 3 and chromosome 6 spanning over a region of 2.7 Mb and 7.8 Mb, respectively. Conventional mapping using F2:3 population also identified two QTLs, Parth6.1 and Parth6.2 in chromosome 6 which indicated the presence of a major effect QTL in chromosome 6 determining parthenocarpy in PPC-6. The flanking markers, SSR01148 and SSR 01012 for Parth6.1 locus and SSR10476 and SSR 19174 for Parth6.2 locus were identified and can be used for introgression of parthenocarpy through the marker-assisted back-crossing programme. Functional annotation of the QTL-region identified two major genes, Csa_6G396640 and Csa_6G405890 designated as probable indole-3-pyruvate monooxygenase YUCCA11 and Auxin response factor 16, respectively associated with auxin biosynthesis as potential candidate genes. Csa_6G396640 showed only one insertion at position 2179 in the non-parthenocarpic parent. In the case of Csa_6G405890, more variations were observed between the two parents in the form of SNPs and InDels. The study provides insight about genomic regions, closely associated markers and possible candidate genes associated with parthenocarpy in PPC-6 which will be instrumental for functional genomics study and better understanding of parthenocarpy in cucumber.
Cucumber (Cucumis sativus L.) is grown commercially in tropical and subtropical climates around the world (Pradeepkumara et al., 2022). In the Indian sub-continent, cucumber is grown from the highlands to the plains under open fields and protected conditions, including riverbeds. India is considered as the home of cucumber and has a wide range of genetic diversity and variation depending on growth habits, fruit size, fruit composition, and skin color besides several other agronomically important traits (Staub et al., 1997), but this variation has never been fully utilized in the crop improvement. The cultivated cucumber has a narrow genetic base with only 3-8% polymorphism within the cultivated genotypes, and 10-25% between plant species (Behera et al., 2011). The small, diploid genome (367 Mb), annual growth pattern, autogamous mating system, and relatively short life cycle (~ 3 months from generation to offspring) provide important genetic benefits (Wang et al., 2020) and detailed genomics-based studies in cucumber.
In most of the Angiosperms, fruit formation usually occurs after successful pollination followed by fertilization of eggs, which results in ovary growth, however, fruit development without pollination and fertilization is referred to as parthenocarpy (Li et al., 2014). Parthenocarpic fruits are seedless as ovules fertilization is disrupted due to changes in the basic genetic makeup involved in fertilization processes. Sources of genetic parthenocarpy are either obligate or facultative by nature. In sexually transmitted species, parthenocarpic genotypes need to be facultative in nature for successful development of fruits when pollinated. Alternatively, the obligate parthenocarpy can be found in asexually propagated plants (Gorguet et al., 2005). From a consumer perspective, parthenocarpy is a possible way to improve fruit quality and total productivity in several fruit crops. If seed setting fails, flower mortality is a common way to avoid wasting of resources. However, parthenocarpic genotypes are also found in wild or non-fruit species, indicating that there may be a variety of factors that cause the formation of seedless fruit in higher plants. The viability and permanence of parthenocarpy in a variety of plants is mainly the result of human selection (Varoquaux et al., 2000). Parthenocarpy in cucumber is determined by a complex interaction between several genetic factors and phytohormones (Sharif et al., 2022; Gou et al., 2022). Various phytohormones, especially gibberellins, cytokinins and auxins are involved in the processes that follow pollination and fertilization and these are essential factors for fruit and seed development (Fos et al., 2001). Growing seeds are major source of phytohormones that stimulate fruit growth and development (Ozga et al., 2002). The use of gynoecy in combination with parthenocarpy is necessary as cucumber exhibits facultative parthenocarpy as seeded fruit set can occur in parthenocarpic varieties when fertilised with viable pollen source. Gynoecious varieties are advantageous because of increased numbers of pistillate flowers, and thus greater opportunities for higher fruit set and per unit production. Parthenocarpic cucumber varieties offer several advantages over conventional seeded varieties. Parthenocarpic varieties are able to set fruits sequentially without suffering from first-fruit inhibition (Denna, 1973; Sun et al., 2006a). Parthenocarpy should be combined with stable gynoecious habit, because the fruits formed after fertilization of parthenocarpic plants become misshapen, have no economic value and lead to loss of productivity in case the female flowers received viable pollen. Selection of diverse genotypes to be used as a parent in the development of the F1 hybrid to achieve higher yield, uniformity and suitability for protected cultivation (2016; 2017; Jat et al., 2015) is necessary in crop improvement programme. Therefore, the development of molecular markers closely associated with parthenocarpy and its marker-assisted introgression into diverse back-grounds is necessary to facilitate hybrid breeding programme.
Marker assisted selection (MAS) can enhance the efficiency of traditional breeding. In cucumber, breeding of parthenocarpic lines based on molecular markers provides a faster and more efficient way as selection can be based on genotypes itself rather than the phenotypes. There are two key requirements in successful of MAS, i.e. markers should be closely linked to target genes and a moderately saturated or high density genetic linkage map (Miao et al., 2011). The development of genetic linkage maps in cucumber have made possible for molecular characterization of important economic traits which includes fruit quality (Wenzel et al., 1995), resistance to diseases (Park et al., 2000; Zhang et al., 2010), yield (Serquen et al., 1997b; Fazio et al., 2003a), gynoecious sex and fruit colour (Miao et al., 2011) and yellow fruit flesh (Lu et al., 2015). Genetic studies have been largely inconsistent on the mode of inheritance for parthenocarpy in cucumber and have ranged from proposals of a single gene to complex multigenic inheritance (Pike and Peterson, 1969; De Ponti and Garretsen, 1976; El-Shawaf and Baker, 1981; Kim et al., 1992b). In the past, parthenocarpy has been studied by several workers to unravel the genetic and physiological basis of this extremely important trait (Fu et al., 2008; Li et al., 2014; Su et al., 2021; Gou et al., 2022; Mandal et al., 2022). The parthenocarpic genotypes of cucumber can set fruit without pollination however normal seed formation happens with successful pollination with viable pollen grains. This typical phenomenon in cucumber is attributed to the facultative parthenocarpic nature. The majority of the studies in the last two decades suggested that multiple QTLs across the genome are responsible for parthenocarpic fruit development in cucumbers. In a European greenhouse-slicing cucumber genotype, EC-1 parthenocarpy was found to be determined by one major and stable QTL in chromosome 2 (Parth 2.1) revealed through a F2:3 population (Wu et al., 2015). In north American pickling type cucumber 2A, seven QTL were detected for parthenocarpy and one QTL each on chromosomes 5 and 7 (parth5.1 and parth7.1) and two on chromosome 6 (parth6.1 and parth6.2) were found govern parthenocarpy (Lietzow et al., 2016). Besides, in a south China ecotype cucumber, 4 novel QTLs associated with parthenocarpy were detected (Niu et al., 2020).
There is a broad consensus based on the available reports that parthenocarpic fruit set is complex in nature and genomic regions in different chromosomes are responsible for induction of parthenocarpic fruit development in cucumbers. The present study was conducted to identify and map the genomic regions associated with parthenocarpy in one of the commercially cultivated gynoecious parthenocarpic genotype, Pusa Parthenocarpic Cucumber-6 (PPC-6) through QTL-seq approach. Identification of closely linked PCR-based markers and possible identification of candidate genes associated with parthenocarpy in PPC-6 would facilitate the marker-assisted back-cross breeding and characterization of parthenocarpic trait in cucumber.
The commercially cultivated parthenocarpic genotype, PPC-6 is cultivated widely in India under protected condition and was used as one of the parents for studying the inheritance and development of mapping population for parthenocarpic trait. In contrast, the non-parthenocarpic parent, Pusa Uday (PU), an Indian type cultivar suitable for cultivation under open field conditions was taken for the study. The F1 progeny was developed by crossing the PU with PPC-6 under protected conditions. Development of F1, F2 and back-cross progenies were undertaken under protected conditions. The plants of inbreds and developed progenies were grown under protected conditions using the standard agronomic practices developed by the Division of Vegetable Science, ICAR-Indian Agricultural Research Institute, New Delhi.
The parthenocarpic line, PPC-6 was crossed with the non-parthenocarpic cultivar, Pusa Uday (PU). The resulting F1 generation was selfed to obtain a sufficient number of F2 population. The female flowers were covered with a butter paper bag, one day prior to anthesis to avoid cross-pollination, and pollens collected from the freshly opened male flowers were used for pollination. The observations were recorded for development of parthenocarpic fruits up to the 20th node. If a plant produced parthenocarpic fruits up to the 1st to 5th nodes, it was considered an early parthenocarpic plant while if the fruit set occurred beyond the 10th node, plants were categorized as late parthenocarpic. A total of 498 F2 progenies were grown for recording observation on parthenocarpy and observation was recorded from 400 plants. The observations were recorded separately for early parthenocarpy, late parthenocarpy, and non-parthenocarpy. The goodness of fit of the observed values to the expected segregation ratio for parthenocarpic and non-parthenocarpic plants was tested using the classical Chi-square (χ2) test as expressed below (Panse and Sukhatme, 1985):
Approximately 15g of leaf samples were collected for DNA isolation from 30-35 day-old seedlings at the active vegetative stage during early morning hours. The collected leaf samples were packed in aluminum foil and labeled properly, then frozen into liquid nitrogen and stored at -80°C for further use. Total DNA was isolated from the individual parental lines, F1 hybrids, and mapping populations using the modified cetyl trimethyl ammonium bromide (CTAB) method (Saghai-Maroof et al., 1984). The genomic DNA samples were adjusted to 50ng DNA/µl and stored at 4°C until used as the templates for PCR amplification and sampling for sequencing. The quality and quantity of the extracted DNA were estimated with an Eppendrof Biospectrometer confirmed by running on 0.8% w/v agarose gel.
For QTL-seq analysis, 498 F2 progenies derived from the crossing of the PU × PPC-6, were grown under polyhouse with a partially controlled environment along with their parents. Observation on parthenocarpy was recorded 45 days after sowing when the plants were in the full reproductive stage. One plant from each of the parents, PU and PPC-6 along with two extreme bulks constituting twenty plants each from the parthenocarpic and non-parthenocarpic types were used for sampling (Figure 1). Young leaves from each selected plant were used for the isolation of genomic DNA. After DNA isolation and purification, quantification was done using a Qubit (Thermofisher Scientific, USA). An equal quantity of DNA from each plant taken for bulking to constitute the final bulks.
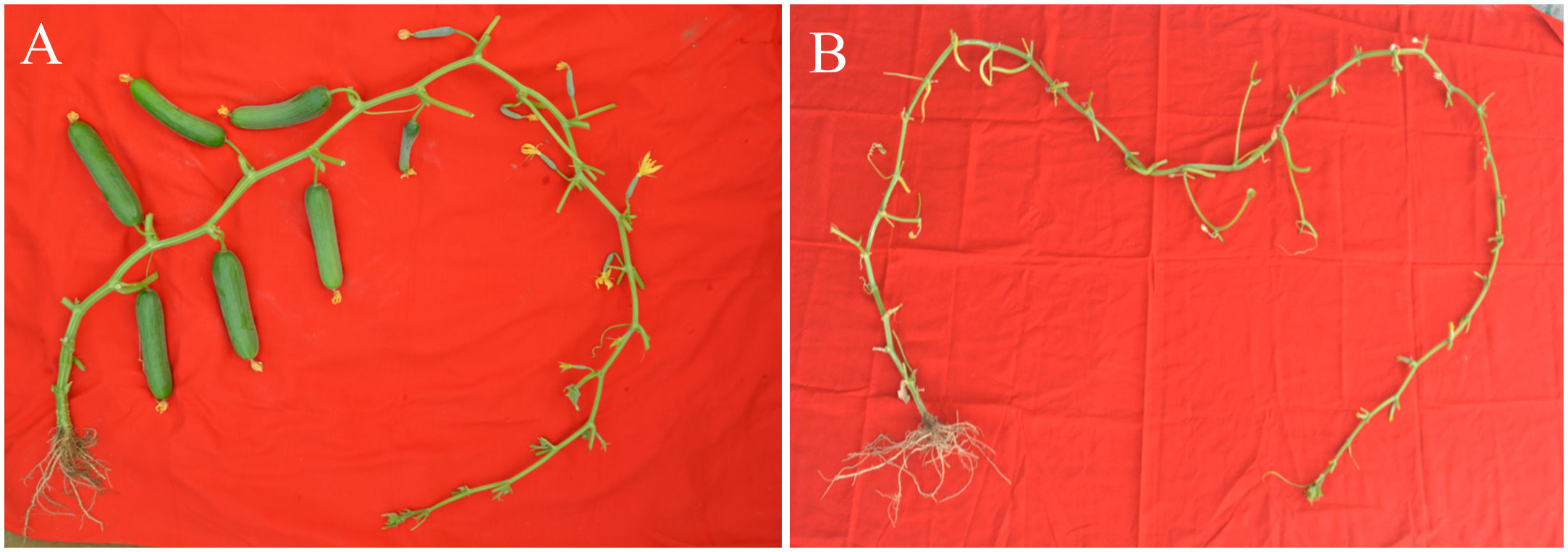
Figure 1 Fruit set in the contrasting parental lines (A) Pusa Parthenocarpic Cucumber-6 with parthenocarpic fruit development (B) Pusa Uday with no parthenocarpic fruit set under protected condition.
Paired-end Illumina reads were obtained for both the parents and the bulks in duplicates. All the reads were 2*150 in length. FastQC version 0.11.8 (Andrews, 2010) was used for visualization of various read parameters and the presence of low-quality bases and of Illumina adapters. Based on the FastQC report the reads were cleaned and trimmed using Trimmomatic v0.39 (Bolger et al., 2014) with command parameters ‘ILLUMINACLIP:TruSeq3-PE.fa:2:30:10:2:keepBothReads SLIDINGWINDOW:4:15 MINLEN:50’. Here, TruSeq3-PE.fa is a fasta file containing Illumina adapter sequences. The obtained high-quality reads were used further for the identification of QTL regions.
QTL-Seq is a fast and efficient method to identify loci related to agronomically important traits (Takagi et al., 2013) in plants. Bulked Segregation Analysis (BSA) is the root of QTL-Seq that detects genomic location(s) showing significant variations in contrasting parents and progenies, produced by the contrasting parents. BSA relies on two main parameters, namely, SNP-index and Δ (SNP-index) (Abe et al., 2012; Takagi et al., 2013). SNP-index is the ratio of a number of reads having a variation, to the total number of reads at a particular position. While Δ (SNP-index) is the difference in the SNP indices of contrasting bulks. The range of SNP index varies from 0 to 1 depending on the parent chosen as a reference. For instance, if parent 1 is used as a reference, and the reads aligned at a particular locus do not have any variation this means that all the loci have been contributed from parent 1 and hence SNP-index = 0, but if all the reads aligned at a particular locus show variation, then it means that the loci have been contributed by another parent and therefore SNP-index = 1. For calculating the SNP-index, a sliding window approach was used with window size of 2000 kb and 100 kb increment followed by their averaging. Graphs were plotted for the Δ (SNP-index) against the chromosomal location. A Δ (SNP-index) value close to zero, indicates that no significant QTL is present at that locus for the studied trait. To obtain significant results, statistical confidence intervals of Δ (SNP-index) were also plotted for all SNP positions with read depth assuming that there are no QTLs as null hypothesis at 95% level of significance (Takagi et al., 2013). We used QTL-seq version 2.2.2 for the detection of significant QTL regions for parthenocarpy in cucumber (Sugihara et al., 2020). The parameters used were [-n1 20 -n2 20 -o qtlseq_results -F 2 -e Cucumis sativus] where n1 and n2 are the numbers of individuals in each bulk, -o is the output directory, F is the filial population (here we had F2) and -e is the dataset of reference genome for identification of the effects due to SNPs. All the other parameters were kept as default. This software uses BWA for the read alignment, SAMtools for filtering and BCF tools for variant calling (Figure 2).
Phenotyping was conducted in F2:3 population developed through selfing of the individual F2 progenies for conventional molecular mapping of the parthenocarpy. An single F1 plant was selfed to obtain the F2 population. An F2:3 population comprising of 94 progenies derived from the cross between parthenocarpic and non-parthenocarpic parents was used to construct linkage map of cucumber. The F2:3 progeny rows along with parental lines were raised under an insect-proof net house during Kharif, July-October, 2021. The F2:3 progenies were grown in two replications with 10 plants in each replication for recording the observation on parthenocarpy. Eight female flowers were bagged one day prior to anthesis from the fifth node onwards on the main stem and eight more from the laterals. At 10 days after anthesis, well-developed and malformed fruits were counted as parthenocarpic fruit, whereas aborted ones were recorded as non-parthenocarpic (Figure 3) as suggested by Wu et al. (2016).
Genotyping was done using a large set of PCR-based markers uniformly distributed all throughout the cucumber genome. For the parental polymorphic survey, 1285 SSRs, Indels, CAPS markers were selected from the Cucumis sativus genome representing 7 linkage groups. In the present experiment, previously reported markers were used for the polymorphic study between the two genotypes, PPC-6 and PU (Miao et al., 2011; Zhu et al., 2016). Linkage analysis was performed using identified 123 polymorphic SSRs, Indels, CAPS markers for the construction of linkage map by IciMapping 4.1.0.0 at LOD threshold of 3.0 (Lander et al., 1987). Segregation of 123 SSRs, Indels, CAPS markers, and parthenocarpy was analyzed and genetic distance between markers was calculated using the Haldane and Kosambi map function (Kosambi, 1944). Parthenocarpy locus was mapped using Inclusive Composite Interval Mapping (ICIM) in ICIMapping 4.1.0.0 software (Wang et al., 2016).
In the present study, the inheritance pattern of parthenocarpy was studied based on the classical dominant-recessive Mendelian model by grouping the cucumber plants into three categories of their fruit development i.e. early parthenocarpic, late parthenocarpic, and non-parthenocarpic fruit development. This information would facilitate the adoption of appropriate breeding strategies for the development of stable parthenocarpic cucumber lines and will improve the efficiency of selection procedures. The genotype, PU produced non-parthenocarpic fruits and it was considered to be homozygous for non-parthenocarpic fruit development. The development of parthenocarpic fruits in PPC-6 is characterised by early parthenocarpy with fruit setting from the beginning or from the base of the plant. Therefore, PPC-6 was used as a homozygous genotype for parthenocarpic fruit development. The F1 hybrid derived from the cross of PU × PPC-6 with heterozygous conditions produced some parthenocarpic fruits on the lower nodes, i.e. 10th node and above (Supplementary Figure 1). In segregating F2 individuals, early, late and non-parthenocarpic fruits were recorded. Out of 400 plants, 307 produced either early parthenocarpic or late parthenocarpic fruits and 93 plants were recorded as non-parthenocarpic plants. The χ2 value indicated a good fit for segregation of parthenocarpy (early, late and non-parthenocarpy) in the F2 population populations confirmed with the expected ratio of 1:2:1 for early parthenocarpy, later parthenocarpy and non-parthenocarpy, respectively (Table 1). In the back cross progeny with the non-parthenocarpic genotype, PU the segregation for late parthenocarpy and early parthenocarpy were in the ratio of 1:1. Similarly, segregation of the plants for early parthenocarpy and late parthenocarpy was in the ratio of 1:1 for early and late parthenocarpy in the back-cross progenies with the parthenocarpic parent (Table 2).
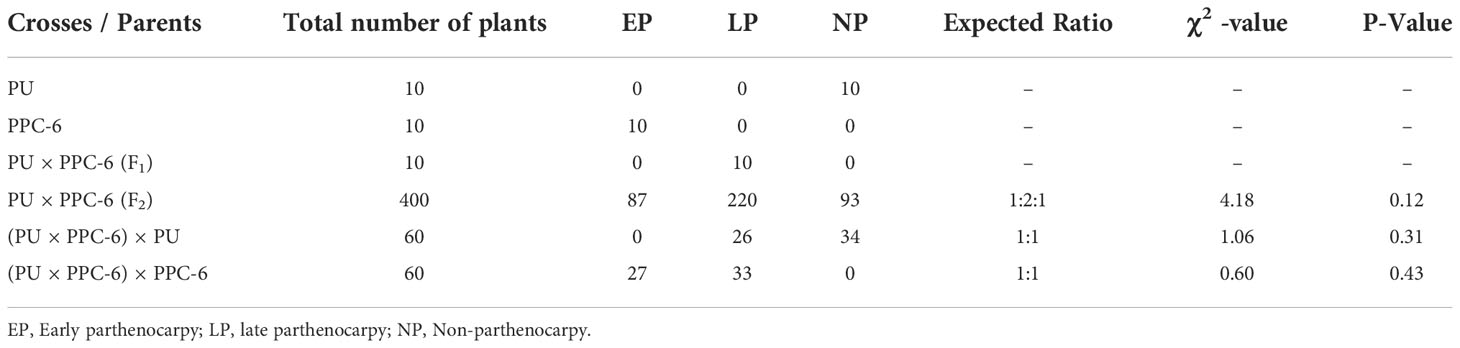
Table 1 Evaluation of the parents along with F1 and F2 and back-cross progenies for studying the inheritance of parthenocarpy.
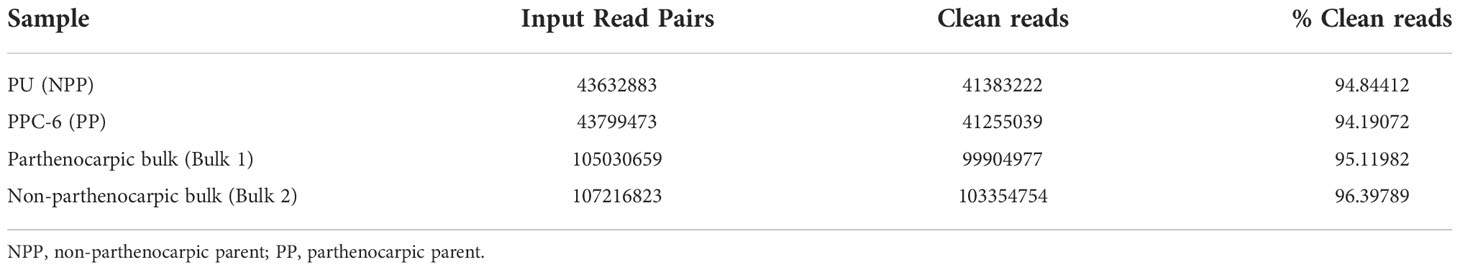
Table 2 Summary of reads’ statistics of the re-sequenced samples of the parents along with the contrasting bulks.
The raw reads for both the parents and bulks were subjected to quality check and removal of adapter sequences. After pre-processing, the non-parthenocarpic parent (NPP), PU retained 41383222 clean reads while parthenocarpic parent (PP), PCC-6 retained 41255039 clean reads. In case of both the bulks, i.e., Parthenocarpic bulk (PB) and Non-Parthenocarpic bulk (NPB), 99904977 and 103354754 cleans reads were obtained, respectively (Table 1).
After alignment and filtering of clean reads followed by variant calling using BWA, SAM tools and BCF tools, two QTL regions related to parthenocarpy were detected. The major QTL was detected on chromosome 6 while a minor QTL region was detected on chromosome 3 (Figure 4; Supplementary Table 1). SNP index of the parthenocarpic and non-parthenocarpic bulks is presented in Supplementary Figure 2. For both the regions, 99% confidence interval was considered. The region covered under the QTL region of chromosome 6 expanded from 13,500,000 till 21,300,000 and 7,000,000 till 9,700,000 for chromosome 3 (Figure 5; Table 3). A total of 5714 variants (SNP and Indels) were identified in chromosome 6 while we found 1129 variants on chromosome 3 (Supplementary Table 2). To view the effect of these variations on the protein sequence, we used the SNPeff software using Cucumis sativus database. All the identified variations were divided into several categories based on the impact on the protein. These categories are High, Moderate, Low and Modifier. Chromosome 3 has 4, 67, 110 and 948 SNPs in each category, respectively while chromosome 6 has 12, 183, 348 and 5171 SNPs, respectively (Figure 6; Supplementary Table 2).
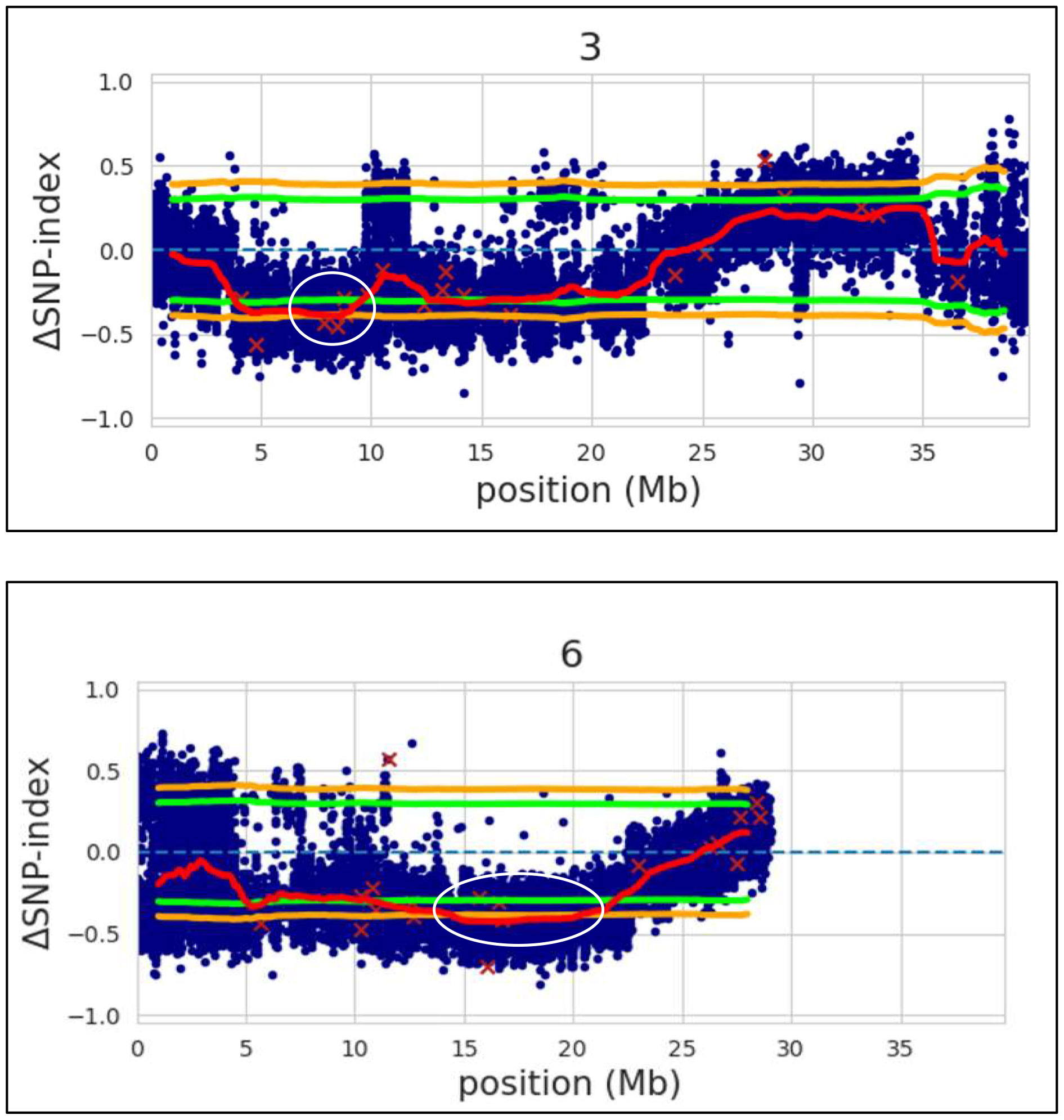
Figure 4 Δ(SNP-index) with statistical confidence intervals (orange, 99%; green, 95%). The major QTL identified on chromosome 6 and a minor QTL region on chromosome 3 of cucumber by QTL-seq (shown in white outline).
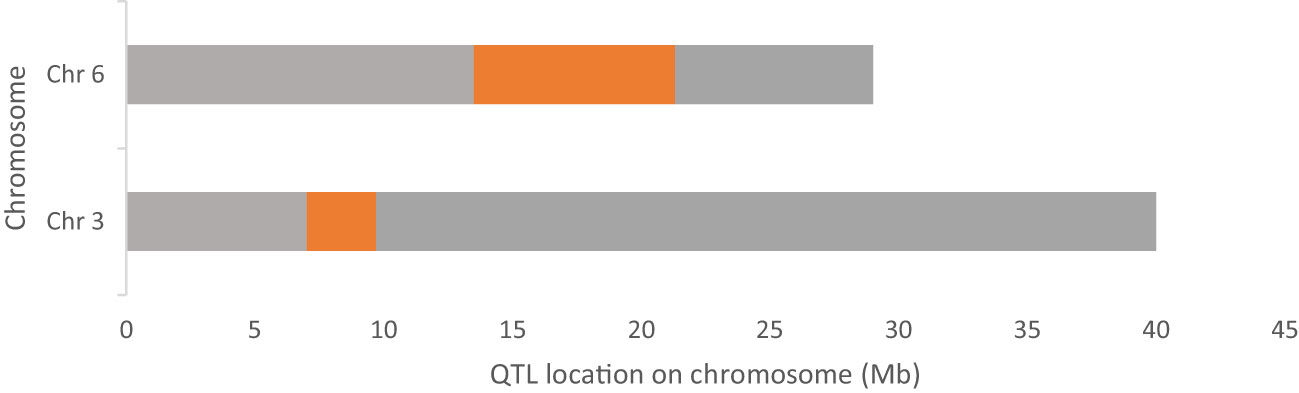
Figure 5 Chromosome 3 and 6 of cucumber showing the identified QTL region for parthenocarpy trait (in orange-coloured regions).

Table 3 Identified QTL regions in cucumber for parthenocarpy and their distribution in chromosome 3 and 6.
A total of 1285 SSR, Indel and CAPS markers were screened for parthenocarpy in polymorphic survey between PU and PPC-6 parental lines to identify polymorphic markers with ability to distinguish the parental lines. The markers were selected for all linkage groups of cucumber. A total of 1285 markers were used for parental polymorphic survey and among them 123 (11.28%) were polymorphic among the parental lines and produced clear and easily identifiable amplicons (Supplementary Table 3). The F2 mapping population comprising of 94 individuals, were genotyped using selected polymorphic markers and respective F2:3 progenies were evaluated for parthenocarpy. A total of 123 polymorphic markers were used for linkage analysis and others were rejected, due to non-amplification, missing data and difficulty in scoring. To confirm the parthenocarpic locus, specificity of the markers genotyping data of these 123 polymorphic markers were used for the construction of linkage map of parthenocarpic locus using Inclusive Composite Interval Mapping (ICIM) method of Ici Mapping (4.1.0.0) software at LOD threshold of 3.0 (Lander et al., 1987) (Figure 7). The linkage map of these polymorphic 123 SSRs, Indels, CAPS markers is presented in Supplementary Figure 3. Out of total 123 markers,10 markers were mapped on 1st linkage group, 20 on 2nd linkage group, 15 on 3rd linkage group, 12 on 4th linkage group, 17 on 5th linkage group, 30 on 6th linkage group and 19 on 7th linkage group (Supplementary Figure 4). Two major effect QTLs associated with parthenocarpy (Parth6.1 and Parth6.2) were mapped to chromosome 6 (Figure 7). These two QTLs had LOD scores of 5.06, 4.59 and phenotypic variance of 16.69% and 12.93%, respectively. The additive effects of Parth6.1 and Parth6.2 were -13.71 and -12.49, respectively indicating the contribution of the parthenocarpy trait from male parent PPC-6. The markers flanking Parth6.1 locus were, SSR 01148 and SSR 01012, spanning a distance of 5.0 cM. The markers flanking Parth6.2 locus were SSR10476 and SSR 19174, spanning a distance of 5.0 cM. The results from this experiment depicted that markers SSR 01148, SSR 01012, SSR10476, and SSR 19174 on chromosome 6 are closely associated with parthenocarpic traits in cucumber.
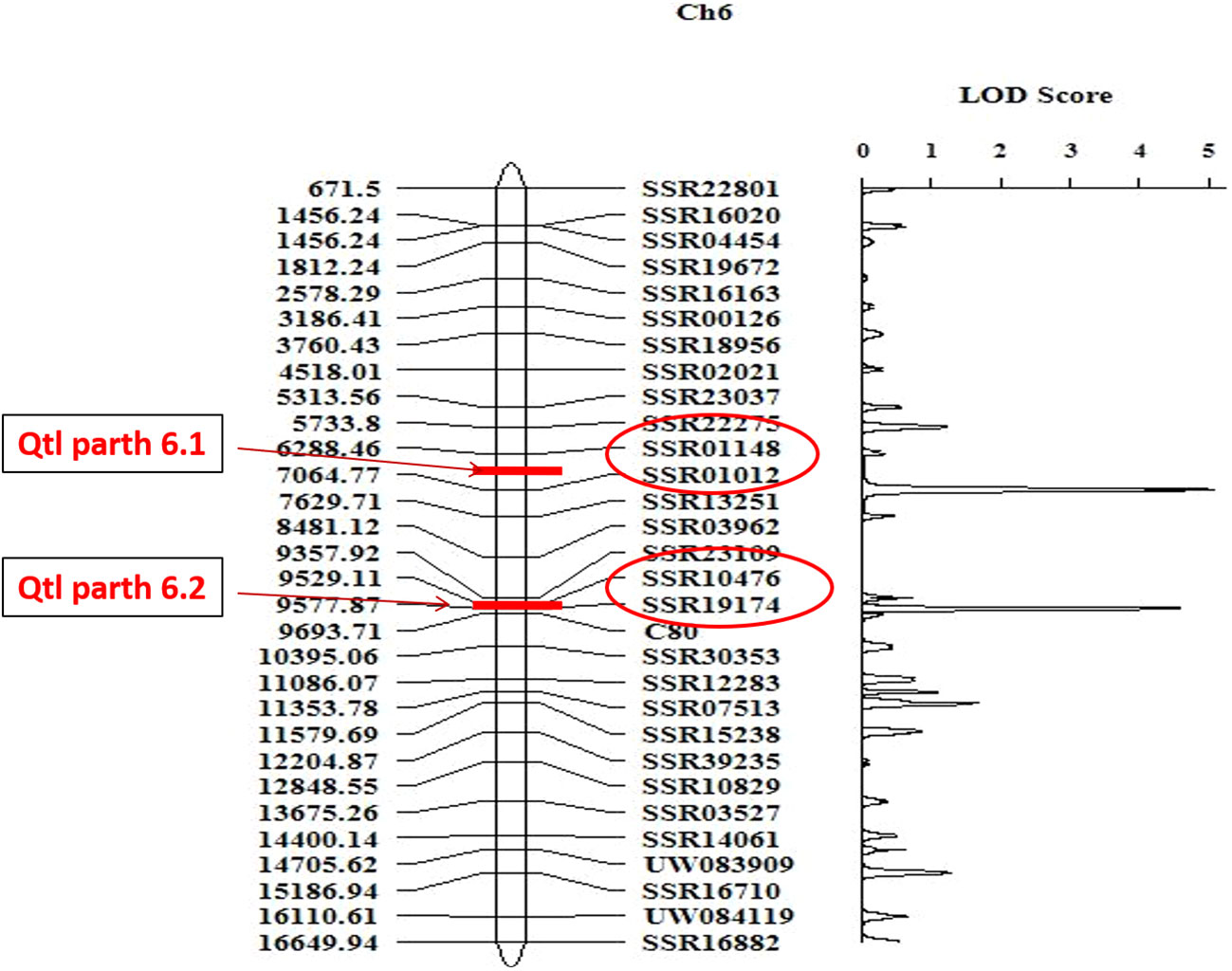
Figure 7 Linkage map of parthenocarpic locus on chromosome 6, constructed using SSR markers. Marker names, LOD score are depicted on the right side of the estimated map and the genetic distances shown in cM.
To identify the major genes, present in these regions, a reference genome annotation file of cucumber (http://ftp.ebi.ac.uk/ensemblgenomes/pub/release-53/plants/gff3/cucumis_sativus) was used. A total of 998 genes were present in chromosome 6, while 485 genes were present in chromosome 3 of the QTL region (Supplementary Sheet 4). Among the identified genes, majority of them were under the category of hypothetical protein. Two genes, Csa_6G396640 and Csa_6G405890 designated as probable indole-3-pyruvate monooxygenase YUCCA11 and Auxin response factor 16, respectively were the potential candidate genes associated with auxin biosynthesis in plants which is crucial in parthenocarpic fruit development in cucumber. Csa_6G396640 gene showed only one variation (insertion) at position 2179 in PU while in case of Csa_6G405890, more variations were observed between the two parents which includes both SNPs and few INDELs (Figure 8).
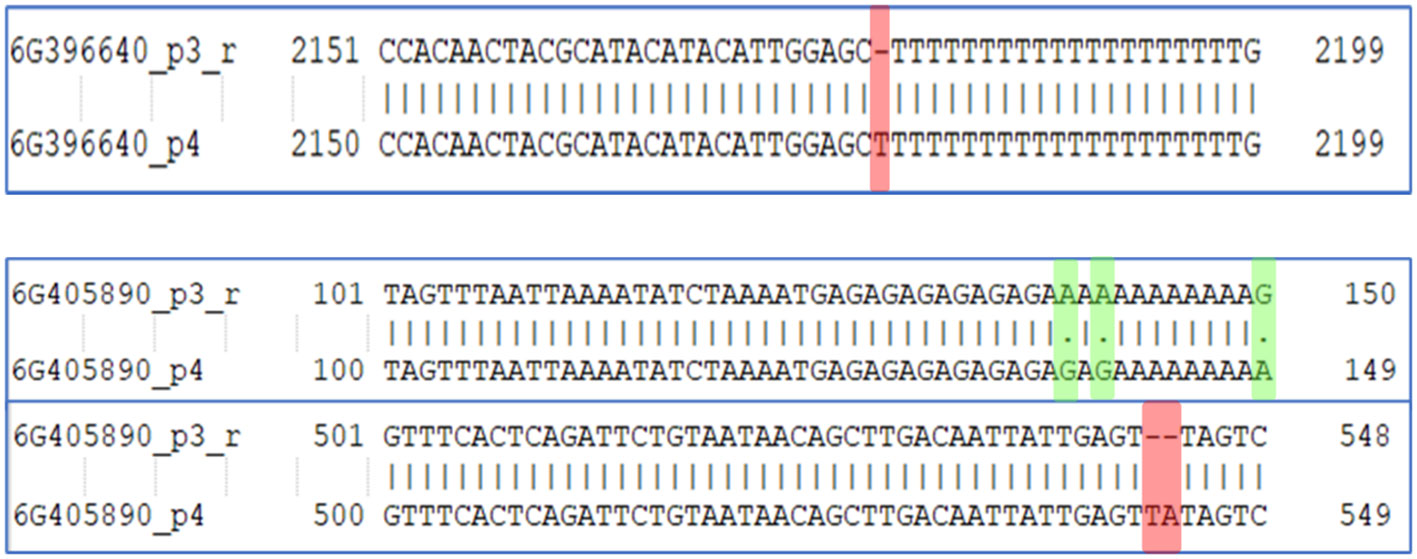
Figure 8 Variation in the Csa_6G396640 and Csa_6G405890 genes associated with indole-3-pyruvate monooxygenase YUCCA11 and Auxin response factor 16, respectively in the parents PU (p3_r) and PPC-6 (p4).
Parthenocarpic fruit development in cucumber is extremely important for its cultivation under protected condition. Studies on inheritance of parthenocarpy by different workers depicted its complex genetics and number of genes/QTLs associated with resistance to parthenocarpy. In cucumber, parthenocarpy is facultative in nature and extent of parthenocarpy varies across different developmental stages of the plants (Joldersma and Liu, 2018). Pike and Peterson (1969) have reported that an incomplete dominant gene, P determines parthenocarpy in cucumber. They have postulated that development of early parthenocarpic fruits in the lower nodes is controlled by the dominant homozygous state, PP while late parthenocarpy and lower extent of parthenocarpy are represented by the heterozygous state, Pp. Whereas, homozygous recessive state, pp is responsible for non-parthenocarpic fruit development. In the homozygous condition, PP produces parthenocarpic fruits early, with the first developing generally by the fifth node. Heterozygous Pp plants produce parthenocarpic fruits later than homozygous plants and are fewer in number. The homozygous recessive pp produces no parthenocarpic fruits. Besides, several other studies have also reported monogenic control of parthenocarpy in cucumber (Hawthorn and Wellington, 1930; Kvasnikov et al., 1970; Juldasheva, 1973; Meshcherov and Juldasheva, 1974). Later on, most of the studies reported polygenic control of parthenocarpy (El-Shawaf and Baker, 1981; Sun et al., 2006; Yan et al., 2009; Lietzow et al., 2016; Wu et al., 2016). In the present study, we have recorded that there was varied types of parthenocarpy in the F2 progenies although the parthenocarpic parent, PPC-6 was early parthenocarpic type and started fruiting from 3-5 nodes onwards. We have recorded the plants as early parthenocarpic when fruiting started from 5th node onwards and late parthenocarpy when parthenocarpic fruit set was recorded after 10th node and considered all the early and late-type plants as parthenocarpic. Based on these observations, the parthenocarpy was found to be controlled by single recessive gene. The present information supported the earlier observation by Pike and Peterson (1969) which postulated that homozygous dominant, heterozygous and homozygous recessive forms are responsible early parthenocarpy, late parthenocarpy and non-parthenocarpy, respectively. However, it was evident there was a significant contribution of the back-ground evidenced from the extent of parthenocarpy and therefore, it is possible to introgress parthenocarpy traits in different elite and non-conventional genotypes of cucumber through marker-assisted back-cross breeding with identification and development of molecular markers closely associated with parthenocarpic trait.
In the recent times, discovery of molecular markers and physical map construction is greatly facilitated by advancement in next-generation sequencing technology (Cao et al., 2021). QTL-seq combines the next-generation sequencing technology with BSA for rapid detection of QTLs for any particular trait and facilitate development of closely associated molecular markers and identification of candidate genes. Thereafter, QTL-seq has been widely used for the detection of QTLs, identification of closely linked molecular markers and identification of candidate genes for number of traits in different crops (Singh et al., 2016; Wang et al., 2016; Wei et al., 2016; Chen et al., 2017; Wen et al., 2019; Arikit et al., 2019; Li et al., 2020). In cucumber, QTL-seq has been used successfully for the identification of QTL for early flowering traits (Lu et al., 2014), flesh thickness (Xu et al., 2015), sub-gynoecy sex expression (Win et al., 2019), pre-harvest sprouting of the seeds (Cao et al., 2021) and resistance to powdery mildew (Zhang et al., 2021). Based on the QTL-seq results, two major QTLs, one each in chromosomes 3 and 6 were identified based on the Δ(SNP-index). The QTL, Parth3.1 was Parth6.1 were spanned 2.7 Mb on chromosome 3 and 7.8 Mb on chromosome 6, respectively. Besides, a large number of variants were detected in both the genomic regions in the form of SNPs and InDels. The identified SNPs and InDels in the genomic regions detected through QTL-seq will be extremely useful in the development of molecular markers and fine mapping of the genomic region associated with parthenocarpy in cucumbers. However, the QTL region identified through QTL-Seq are often not very precise and needs further validation and authentication through additional method of molecular mapping (Xu et al., 2017). Therefore, we have employed mapping of parthenocarpy through conventional F2:3 population. Based on the QTL-seq results, two major QTLs, one each in chromosomes 3 and 6 were identified.
In cucumber, systematic efforts have been made for molecular mapping of number of qualitative traits but for quantitative traits like parthenocarpy progress is slow and hence very few public sector parthenocarpic varieties/hybrids are available in market. Now-a-days, role of marker-assisted selection (MAS) is increasing in conventional plant breeding (Miao et al., 2011). Due to narrow genetic base and low polymorphism, around 30 linkage maps have been constructed (Kennard et al., 1994). These linkage maps involved use of RAPDs (Randomly Amplified Polymorphic DNA) or AFLP (Amplified Fragment Length Polymorphism) (Fukino et al., 2008 and Yuan et al., 2008) that are not breeder friendly. Hence, co-dominant markers like SSR/InDel are best suited for marker-assisted breeding and are breeder friendly.
Two major QTLs for parthenocarpy at chromosome 6 (Parth6.1 & Parth 6.2) were identified using the F2:3 mapping population. Locus Parth6.1 was flanked by SSR01148 and SSR01012 with a LOD score of 5.06 and 16.69% of PVE (Phenotypic Variance Explained) reflecting that this locus is a major effect QTL. Second QTL Parth 6.2 was flanked by SSR10476 and SSR19174 primers and LOD value was 4.59 with 12.93% of PVE explaining another major effect QTL. Previously, Lietzow et al. (2016) identified seven QTLs associated with parthenocarpic fruit set, one on each chromosomes 5 and 7 (parth5.1 and parth7.1) and two on chromosome 6 (parth 6.1 and parth 6.2) were consistently identified in all experiments. Wu et al. (2016) identified seven novel QTLs on chromosomes 1, 2, 3, 5 and 7. The identification of QTLs is a valuable resource for cucumber breeders for the development of parthenocarpic cultivars (Dey et al., 2022). Molecular markers flanking major effect parthenocarpy QTLs can prove useful in the Marker Assisted Breeding (MAB) programme. However, there is need to further saturate linkage map to narrow-down genetic distance between flanking molecular markers to get markers better suited for foreground selection in endeavour of higher/quality production of cucumber.
Parthenocarpy is a complex trait and determined by interaction of large number of metabolic pathways interlinked with each other. Among the different metabolism, auxin, gibberellins and cytokinins are reported to play key role determining parthenocarpic fruit set in cucumber (Li et al., 2014; Su et al., 2021; Sharif et al., 2022; Gou et al., 2022). Cross-talk between the important phytohomones in determining parthenocarpy in PPC-6 was recently reported by Mandal et al. (2022). The QTL region identified through QTL-seq had two important genes with a possible association with parthenocarpic phenomenon. Indole-3-pyruvate monooxygenase YUCCA11 (Csa_6G396640) was found to be have one SNP in the non-parthenocarpic parent, PU when compared with the parthenocarpic reference genotype. Besides, the auxin response factor 16 (Csa_6G405890) present in the QTL region also showed variation in terms of Indels and SNPs in the parental lines. These two identified genes with key role in auxin biosynthesis could be possible candidate genes for induction of parthenocarpy in cucumber. Auxin, through its influence in cell division and expansion is key determinant in development of fleshy fruits and reported to be integral part in the initial signal for fertilisation and increased fruit (Godoy et al., 2021). After parthenocarpic fruit set their further development is influenced by auxins which was evidenced by the upregulation of the auxin biosynthesis-related genes in the later stages of fruit development in parthenocarpic genotypes in our earlier study (Mandal et al., 2022). Indole-3-pyruvate is one of the important routes for tryptophan-dependent auxin biosynthesis which is believed to be common in all plants (De Smet et al., 2011). Auxin is the key phytohormone besides gibberellins and cytokinin reported to play important role in the induction of parthenocarpy (Sharif et al., 2022). Among the different auxin biosynthesis pathways, the role of Trp-IPyA (tryptophan-indole-3-pyruvic acid) in parthenocarpic fruit development has been reported by several workers. The role of the YUCCA10, PavYUCCA10, SlTAR1, ToFZY2, ToFZY3 and PARENTAL ADVICE-1 (PAD-1) genes in parthenocarpic fruit development of loquat, tomato and eggplants have been reviewed in details by Sharif et al. (2022). However, narrow down of the QTL region through fine mapping is required for the precise identification of candidate genes associated with parthenocarpy in cucumber.
In cucumber parthenocarpic fruit set is extremely important trait facilitated large scale protected cultivation worldwide. In one commercially cultivated parthenocarpic genotype, PPC-6, it was found that, single incomplete dominant gene control this trait in spite of significant effect of genetic back-ground in expression of parthenocarpy. QTL-seq analysis in combination with conventional mapping using F2:3 population identified one major effect QTLs, Parth6.1. The flanking markers, SSR01148 and SSR 01012 for Parth6.1 locus were identified for their use in marker-assisted back-crossing programme. Two major genes, Csa_6G396640 and Csa_6G405890 designated as probable indole-3-pyruvate monooxygenase YUCCA11 and Auxin response factor 16, respectively associated with auxin biosynthesis as potential candidate genes. The study provides insight about the genetics and genomic regions, closely associated markers and possible candidate genes associated with parthenocarpy in PPC-6 for functional genomics studies and future fine mapping.
The data presented in the study are deposited in the NCBI repository, accession number PRJNA885599, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA885599.
Conceived theme of the study and designed experiment: ShyD. Data curation: PS, ShyD, MI, SJ, GS. Investigation: ShiD, ShyD, KK, BG. Resources: ShyD, TB, AM. Supervision: ShyD, AM, TB, GS. Visualization: ShyD, TB, AM. Writing original draft: ShiD, ShyD, SJ. Review and editing: ShyD, TB, GS, MI. All authors contributed to the article and approved the submitted version.
Authors are thankful to the ICAR-Indian Agricultural Research Institute, New Delhi for providing financial support and conduct of the research program of the PhD student, Ms. Shilpa Devi. Authors are thankful to the CRP Molecular Breeding (Cucumber; 12-143 H) programme for funding the research. The research work was partially funded by the NAHEP-CAAST programme of Indian Council of Agricultural Research (ICAR).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1064556/full#supplementary-material
Supplementary Figure 1 | Fruit setting pattern in the F1 hybrid involving Pusa Uday × Pusa Parthenocarpic Cucumber-6.
Supplementary Figure 2 | SNP indices of (A) Parthenocarpic bulk (PB) and (B) Non-partyhenocarpic bulk (NPB) for each chromosomes.
Supplementary Figure 3 | Linkage map of parth locus on chromosome 6, constructed using SSR markers. Marker names, LOD score are depicted on the right side of the estimated map and the genetic distances shown in cM on the left are calculated using software IciMapping ver. 4.1.0.
Supplementary Figure 4 | Distribution of primers across the seven linkage groups and frequency of the amplified and polymorphic markers used for the study.
Supplementary Figure 5 | Amplification pattern of the selected markers closely associated with parthenocarpy in cucumber genotype, PPC-6.
Supplementary File 1 | Identified QTL regions in chromosome 3 and 6 through QTL-seq.
Supplementary File 2 | Identified SNPs and InDels in the QTL regions of chromosome 3 and 6, their impact and associated genes.
Supplementary Table 3 | List of the polymorphic markers used for construction of linkage map and molecular mapping of parthenocarpy using F2:3 population.
Supplementary File 4 | Functional annotation of the genes present in the QTL regions in chromosome 3 and 6.
Abe, A., Kosugi, S., Yoshida, K., Natsume, S., Takagi, H., Kanzaki, H., et al. (2012). Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 30 (2), 174–178. doi: 10.1038/nbt.2095
Andrews, S. (2010). FastQC: a quality control tool for high throughput sequence data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
Arikit, S., Wanchana, S., Khanthong, S., Saensuk, C., Thianthavon, T., Vanavichit, A., et al. (2019). QTL-seq identifies cooked grain elongation QTLs near soluble starch synthase and starch branching enzymes in rice (Oryza sativa l.). Sci. Rep. 9 (1), 1–10. doi: 10.1038/s41598-019-44856-2
Behera, T. K., Staub, J. E., Behera, S., Delannay, I. Y., Chen, J. F. (2011). Marker-assisted backcross selection in an interspecific cucumis population broadens the genetic base of cucumber (Cucumis sativus l.). Euphytica 178, 261–272. doi: 10.1007/s10681-010-0315-8
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Cao, M., Li, S., Deng, Q., Wang, H., Yang, R. (2021). Identification of a major-effect QTL associated with pre-harvest sprouting in cucumber (Cucumis sativus l.) using the QTL-seq method. BMC Genomics 22, 249. doi: 10.1186/s12864-021-07548-8
Chen, F., Fu, B., Pan, Y., Zhang, C., Wen, H., Weng, Y., et al. (2017). Fine mapping identifies CsGCN5 encoding a histone acetyltransferase as putative candidate gene for tendril-less1 mutation (td-1) in cucumber. Theor. Appl. Genet. 130 (8), 1549–1558. doi: 10.1007/s00122-017-2909-1
Denna, D. W. (1973). Effect of genetic parthenocarpy and gynoecious flowering habit on fruit production and growth of cucumber (Cucumis sativus l.). J. Am. Soc. Hortic. Sci. 98, 602–604. doi: 10.21273/JASHS.98.6.602
De Ponti, O. M. B., Garretsen, F. (1976). Inheritance of parthenocarpy in pickling cucumbers (Cucumis sativus l.) and linkage with other characters. Euphytica 25 (1), 633–642. doi: 10.1007/BF00041600
De Smet, I., Voß, U., Lau, S., Wilson, M., Shao, N., Timme, R. E., et al. (2011). Unraveling the evolution of auxin signaling. Plant Physiol. 155 (1), 209–221. doi: 10.1104/pp.110.168161
Dey, S. S., Singh, S., Munshi, A. D., Behera, T. K. (2022). “Classical Genetics and Traditional Breeding. In: The Cucumber Genome. Compendium of Plant Genomes. Eds. Pandey, S., Weng, Y., Behera, T. K., Bo, K. Cham: Springer. doi: 10.1007/978-3-030-88647-9_12
El-Shawaf, I. I. S., Baker, L. R. (1981). Inheritance of parthenocarpic yield in gynoecious pickling cucumber for once-over mechanical harvest by diallel analysis of six gynoecious lines. J. Am. Soc Hortic. Sci. 106, 359–364. doi: 10.21273/JASHS.106.3.359
Fazio, G., Staub, J. E., Stevens, M. R. (2003a). Genetic mapping and QTL analysis of horticultural traits in cucumber (Cucumis sativus l.) using recombinant inbred lines. Theor. Appl. Genet. 107, 864–874. doi: 10.1007/s00122-003-1277-1
Fos, M., Proaño, K., Nuez, F., García-Martínez, J. L. (2001). Role of gibberellins in parthenocarpic fruit development induced by the genetic system pat-3/pat-4 in tomato. Physiol. Plant. 111 (4), 545–550. doi: 10.1034/j.1399-3054.2001.1110416.x
Fukino, N., Ohara, T., Monforte, A. J., Sugiyama, M., Sakata, Y., Kunihisa, M., et al. (2008). Identification of QTLs for resistance to powdery mildew and SSR markers diagnostic for powdery mildew resistance genes in melon (Cucumis melo l.). Theor. Appl. Genet. 118 (1), 165–175. doi: 10.1007/s00122-008-0885-1
Fu, F. Q., Mao, W. H., Shi, K., Zhou, Y. H., Asami, T., Yu, J. Q. (2008). A role of brassinosteroids in early fruit development in cucumber. J. Exp. Bot. 59, 2299–2308. doi: 10.1093/jxb/ern093
Godoy, F., Kühn, N., Muñoz, M., Marchandon, G., Gouthu, S., Deluc, L., et al. (2021). The role of auxin during early berry development in grapevine as revealed by transcript profiling from pollination to fruit set. Hortic. Res. 8, 140. doi: 10.1038/s41438-021-00568-1
Gorguet, B., Van Heusden, A. W., Lindhout, P. (2005). Parthenocarpic fruit development in tomato. Plant Biol. 7 (2), 131–139. doi: 10.1055/s-2005-837494
Gou, C., Zhu, P., Meng, Y., Yang, F., Xu, Y., Xia, P., et al. (2022). Evaluation and genetic analysis of parthenocarpic germplasms in cucumber. Genes (Basel) 13 (2), 225. doi: 10.3390/genes13020225
Hawthorn, L. R., Wellington, R. (1930). Geneva, A greenhouse cucumber that develops fruit without pollination. Agr. Exp. Stat. Bull. 580, 1–11. Available at: https://hdl.handle.net/1813/4556.
Jat, G. S., Munshi, A. D., Behera, T. K., Bharadwaj, C. (2017). Inheritance of parthenocarpy in gynoecious cucumber (Cucumis sativus l.) cultivar PPC-2. J. Hortic. Sci. 12 (2), 193–197. Available at: https://jhs.iihr.res.in/index.php/jhs/article/view/23.
Jat, G. S., Munshi, A. D., Behera, T. K., Choudhary, H., Dev, B. (2015). Exploitation of heterosis in cucumber for earliness, yield and yield components utilizing gynoecious lines. Indian J. Hortic. 72 (4), 494–499. doi: 10.5958/0974-0112.2015.00112.7
Jat, G. S., Munshi, A. D., Behera, T. K., Tomar, B. S. (2016). Combining ability estimation of gynoecious and monoecious hybrids for yield and earliness in cucumber (Cucumis sativus). Indian J. Agric. Sci. 86 (3), 399–403. Available at: https://epubs.icar.org.in/index.php/IJAgS/article/view/57033.
Joldersma, D., Liu, Z. (2018). The making of virgin fruit: the molecular and genetic basis of parthenocarpy. J. Exp. Bot. 69 (5), 955–962. doi: 10.1093/jxb/erx446
Juldasheva, L. M. (1973). Inheritance of the tendency towards parthenocarpy in cucumbers. Byull. Vseso yuznogo ordena Lenina Inst. Rastwni evodstva Imeni N.I. Vavilova 32, 58–59. doi: 10.1007/BF00041600
Kennard, W. C., Slocum, M. K., Figdore, S. S., Osborn, T. C. (1994). Genetic analysis of morphological variation in brassica oleracea using molecular markers. Theor. Appl. Genet. 87, 721–732. doi: 10.1007/BF00222898
Kim, S., Okubo, H., Fujieda, K. (1992b). Endogenous levels of IAA in relation to parthenocarpy in cucumber (Cucumis sativus l.). Scientia Hortic. 52, 1–8. doi: 10.1016/0304-4238(92)90002-T
Kosambi, D. D. (1944). The estimation of map distance from recombination values. Ann. Eugen 12, 172–175. doi: 10.1111/j.1469-1809.1943.tb02321.x
Kvasnikov, B. V., Rogova, N. T., Tarakanova, S. I., Ignatova, S. I. (1970). Methods of breeding vegetable crops under the covered ground. Tmtdyprikl. Bot. Genet. Selek. 42, 45–57. Available at: https://agris.fao.org/agris-search/search.do?recordID=US201302372125.
Lander, E. S., Green, P., Abrahamson, J., Barlow, A., Daly, M. J., Lincoln, S. E. (1987). MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1, 174–181. doi: 10.1016/0888-7543(87)90010-3
Lietzow, C. D., Zhu, H., Pandey, S., Havey, M. J., Weng, Y. (2016). QTL mapping of parthenocarpic fruit set in north American processing cucumber. Theor. Appl. Genet. 129 (12), 2387–2401. doi: 10.1007/s00122-016-2778-z
Li, Z., Han, Y., Niu, H., Wang, Y., Jiang, B., Weng, Y. (2020). Gynoecy instability in cucumber (Cucumis sativus l.) is due to unequal crossover at the copy number variation-dependent femaleness (F) locus. Hortic. Res. 7, 32. doi: 10.1038/s41438-020-0251-2
Li, J., Wu, Z., Cui, L., Zhang, T., Guo, Q., Xu, J., et al. (2014). Transcriptome comparison of global distinctive features between pollination and parthenocarpic fruit set reveals transcriptional phytohormone crosstalk in cucumber (Cucumis sativus l.). Plant Cell Physiol. 55, 1325–1342. doi: 10.1093/pcp/pcu051
Lu, H., Lin, T., Klein, J., Wang, S., Qi, J., Zhou, Q., et al (2014). QTL-seq identifies an early flowering QTL located near Flowering Locus T in cucumber. Theor. Appl. Genet. 127 (7), 1491–1499. doi: 10.1007/s00122-014-2313-z
Lu, H., Miao, H., Tian, G., Wehner, T., Gu, X., Zhang, S. (2015). Molecular mapping and candidate gene analysis for yellow fruit flesh in cucumber. Molecular Breeding 35, 1–8. doi: 10.1007/s11032-015-0263-z
Mandal, N. K., Kumari, K., Kundu, A., Arora, A., Bhowmick, P. K., Iquebal, M. A., et al. (2022). Cross-talk between the cytokinin, auxin, and gibberellin regulatory networks in determining parthenocarpy in cucumber. Front. Genet 13, 957360. doi: 10.3389/fgene.2022.957360
Meshcherov, E. T., Juldasheva, L. W. (1974). Parthenocarpy in cucumber. Proc. Appl. Bot. Plant Breed. 51, 204–213.
Miao, H., Zhang, S., Wang, X., Zhang, Z., Li, M., Mu, S., et al. (2011). A linkage map of cultivated cucumber (Cucumis sativus l.) with 248 microsatellite marker loci and seven genes for horticulturally important traits. Euphytica 182, 167–176. doi: 10.1007/s10681-011-0410-5
Niu, Z. H., Song, X. F., Li, X. L., Guo, X. Y., He, S. Q., He, L. J. Z., et al. (2020). Inheritance and QTL mapping for parthenocarpy in cucumber. Sci. Agric. Sin. 53, 160–171. doi: 10.3864/j.issn.0578-1752.2020.01.015
Ozga, J. A., Van, H. R., Reinecke, D. M. (2002). Hormone and seed-specific regulation of pea fruit growth. Plant Physiol. 128 (4), 1379–1389. doi: 10.1104/pp.010800
Panse, V. G., Sukhatme, P. V. (1985). Statistical methods for agricultural workers (New Delhi: ICAR).
Park, Y., Sensoy, S., Wye, C., Antonise, R., Peleman, J., Havey, M. J. (2000). A genetic map of cucumber composed of RAPDs, RFLPs, AFLPs, and loci conditioning resistance to papaya ringspot and zucchini yellow mosaic viruses. Genome 43, 1003–1010. doi: 10.1139/g00-075
Pike, L. M., Peterson, C. E. (1969). Inheritance of parthenocarpy in the cucumber (Cucumis sativus l.). Euphytica 18, 101–105. doi: 10.1007/BF00021987
Pradeepkumara, N., Sharma, P. K., Munshi, A. D., Behera, T. K., Bhatia, R., Kumari, K., et al. (2022). Fruit transcriptional profiling of the contrasting genotypes for shelf life reveals the key candidate genes and molecular pathways regulating post-harvest biology in cucumber. Genomics 114 (2), 110273. doi: 10.1016/j.ygeno.2022.110273
Saghai-Maroof, M. A., Solimann, K. M., Jorgensen, R. A., Allard, R. W. (1984). Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. 81, 8014–8018. doi: 10.1073/pnas.81.24.8014
Serquen, F. C., Bacher, J., Staub, J. E. (1997b). Mapping and QTL analysis of a narrow cross in cucumber (Cucumis sativus l.) using random amplified polymorphic DNA markers. Mol. Breed. 3, 257–268. doi: 10.1023/A:1009689002015
Sharif, R., Su, L., Chen, X., Qi, X. (2022). Hormonal interactions underlying parthenocarpic fruit formation in horticultural crops. Hortic. Res. 9, uhab024. doi: 10.1093/hr/uhab024
Singh, V. K., Khan, A. W., Jaganathan, D., Thudi, M., Roorkiwal, M., Takagi, H., et al. (2016). QTL-seq for rapid identification of candidate genes for 100-seed weight and root/total plant dry weight ratio under rainfed conditions in chickpea. Plant Biotechnol. J. 14 (11), 2110–2119. doi: 10.1111/pbi.12567
Staub, J. E., Serquen, F. C., Mccreight, J. D. (1997). Genetic diversity in cucumber (Cucumis sativus l.): III. an evaluation of Indian germplasm. Genet. Resour. Crop Evol. 44 (4), 315–326. doi: 10.1023/A:1008639103328
Sugihara, Y., Young, L., Yaegashi, H., Natsume, S., Shea, D. J., Takagi, H., et al. (2020). High performance pipeline for MutMap and QTL seq. PeerJ 10, e13170. doi: 10.7717/peerj.13170
Sun, Z., Lower, R. L., Staub, J. E. (2006a). Analysis of generation means and components of variance for parthenocarpy in cucumber (Cucumis sativus l.). Plant Breed. 123 (3), 277–280. doi: 10.1111/j.1439-0523.2006.01224.x
Sun, Z., Staub, J. E., Chung, S. M., Lower, R. L. (2006). Identification and comparative analysis of quantitative trait loci associated with parthenocarpy in processing cucumber. Plant Breed. 125, 281–287. doi: 10.1111/j.1439-0523.2006.01225
Su, L., Rahat, S., Ren, N., Kojima, M., Takebayashi, Y., Sakakibara, H., et al. (2021). Cytokinin and auxin modulate cucumber parthenocarpy fruit development. Sci. Hortic. 282, 110026. doi: 10.1016/j.scienta.2021.110026
Takagi, H., Abe, A., Yoshida, K., Kosugi, S., Natsume, S., Mitsuoka, C., et al. (2013). QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 74 (1), 174–183. doi: 10.1111/tpj.12105
Varoquaux, F., Blanvillain, R., Delseny, M., Gallois, P. (2000). Less is better: new approaches for seedless fruit production. Trends Biotechnol. 18 (6), 233–242. doi: 10.1016/S0167-7799(00)01448-7
Wang, J., Li, H., Zhang, L., Meng, L. (2016). Users’ Manual of QTL IciMapping. The Quantitative Genetics Group, Institute of Crop Science, Chinese Academy of Agricultural Sciences (CAAS), Beijing 100081, China and Genetic Resources Program, International Maize and Wheat Improvement Center (CIMMYT). (Mexico, D.F., Mexico) Apdo. Postal 6-641, 06600.
Wang, Y., Bo, K., Gu, X., Pan, J., Li, Y., Chen, J., et al. (2020). Molecularly tagged genes and quantitative trait loci in cucumber with recommendations for QTL nomenclature. Hortic. Res. 7, 3. doi: 10.1038/s41438-019-0226-3
Wei, Q. Z., Fu, W. Y., Wang, Y. Z., Qin, X. D., Wang, J., Li, J., et al. (2016). Rapid identification of fruit length loci in cucumber (Cucumis sativus l.) using next-generation sequencing (NGS)-based QTL analysis. Sci. Rep. 6 (1), 1–11. doi: 10.1038/srep27496
Wenzel, G., Kennard, W. C., Havey, M. J. (1995). Quantitative trait analysis of fruit quality in cucumber: QTL detection, confirmation, and comparison with mating-design variation. Theor. Appl. Genet. 91, 53–61. doi: 10.1007/BF00220858
Wen, C., Zhao, W., Liu, W., Yang, L., Wang, Y., Liu, X., et al. (2019). CsTFL1 inhibits determinate growth and terminal flower formation through interaction with CsNOT2a in cucumber. Development 146 (14), 180166. doi: 10.1242/dev.180166
Win, K. T., Zhang, C., Silva, R. R., Lee, J. H., Kim, Y. C., Lee, S. (2019). Identification of quantitative trait loci governing subgynoecy in cucumber. Theor. Appl. Genet. 132 (5), 1505–1521. doi: 10.1007/s00122-019-03295-3
Wu, Z., Li, L., Zhang, T., Zhang, T. L., Li, J., Lou, Q. F., et al. (2015). QTL mapping for parthenocarpy in cucumber. Sci. Agri. Sinica 48, 112–119.
Wu, Z., Zhang, T., Li, L., Xu, J., Qin, X., Zhang, T., et al. (2016). Identification of a stable major-effect QTL (Parth 2.1) controlling parthenocarpy in cucumber and associated candidate gene analysis via whole genome re-sequencing. BMC Plant Biol., 16–182. doi: 10.1186/s12870-016-0873-6
Xu, X., Lu, L., Zhu, B., Xu, Q., Qi, X., Chen, X. (2015). QTL mapping of cucumber fruit flesh thickness by SLAF-seq. Sci. Rep. 5 (1), 1–9. doi: 10.1038/srep15829
Xu, Q., Xu, X., Shi, Y., Qi, X., Chen, X. (2017). Elucidation of the molecular responses of a cucumber segment substitution line carrying Pm5.1 and its recurrent parent triggered by powdery mildew by comparative transcriptome profiling. BMC Genomics 18, 21. doi: 10.1186/s12864-016-3438-z
Yan, L., Luo, L., Feng, Z., Li, X., Lou, Q., Chen, J. (2009). Analysis on mixed major gene and polygene inheritance of parthenocarpy in monoecious cucumber (Cucumis sativus l). Acta Botanica Boreali Occidentalia Sin. 29 (6), 1122–1126.
Yuan, X. J., Li, X. Z., Pan, J. S., Wang, G., Jiang, S., Li, X. H., et al. (2008). Genetic linkage map construction and location of QTLs for fruit-related traits in cucumber. Plant Breed. 127 (2), 180–188. doi: 10.1111/j.1439-0523.2007.01426.x
Zhang, S. P., Miao, H., Gu, X. F., Yang, Y. H., Xie, B. Y., Wang, X. W., et al. (2010). Genetic mapping of the scab resistance gene Ccu in cucumber. J. Am. Soc Hortic. Sci. 135, 53–58. doi: 10.21273/JASHS.135.1.53
Zhang, C., Badri Anarjan, M., Win, K. T., Begum, S., Lee, S. (2021). QTL-seq analysis of powdery mildew resistance in a Korean cucumber inbred line. Theor. Appl. Genet. 134 (2), 435–451. doi: 10.1007/s00122-020-03705-x
Zhu, W. Y., Huang, L., Chen, L., Yang, J. T., Wu, J. N., Qu, M. L., et al. (2016). A high-density genetic linkage map for cucumber (Cucumis sativus l.): based on specific length amplified fragment (SLAF) sequencing and QTL analysis of fruit traits in cucumber. Front. Plant Sci. 7, 437. doi: 10.3389/fpls.2016.00437
Keywords: cucumber, parthenocarpy, inheritance, QTL-seq, molecular mapping, candidate genes
Citation: Devi S, Sharma PK, Behera TK, Jaiswal S, Boopalakrishnan G, Kumari K, Mandal NK, Iquebal MA, Gopala Krishnan S, Bharti, Ghosal C, Munshi AD and Dey SS (2022) Identification of a major QTL, Parth6.1 associated with parthenocarpic fruit development in slicing cucumber genotype, Pusa Parthenocarpic Cucumber-6. Front. Plant Sci. 13:1064556. doi: 10.3389/fpls.2022.1064556
Received: 08 October 2022; Accepted: 17 November 2022;
Published: 14 December 2022.
Edited by:
Nisha Singh, Gujarat Biotechnology University, IndiaReviewed by:
Abhinandan Surgonda Patil, Agharkar Research Institute, IndiaCopyright © 2022 Devi, Sharma, Behera, Jaiswal, Boopalakrishnan, Kumari, Mandal, Iquebal, Gopala Krishnan, Bharti, Ghosal, Munshi and Dey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shyam Sundar Dey, c2h5YW0uaWFyaUBnbWFpbC5jb20=; c2h5YW0uZGV5QGljYXIuZ292Lmlu; Anilabha Das Munshi, YW5pbGFiaG1AeWFob28uY28uaW4=; Tusar Kanti Behera, dHVzYXJAcmVkaWZmbWFpbC5jb20=
†ORCID: Shilpa Devi, orcid.org/0000-0001-9211-8820
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.