- College of Life Sciences, Qingdao University, Qingdao, Shandong, China
Wheat and barley are widely distributed cereal crops whose yields are adversely affected by environmental stresses such as drought, salinity, extreme temperatures, and attacks of pathogens and pests. As the interphase between aerial plant organs and their environments, hydrophobic cuticle largely consists of a cutin matrix impregnated and sealed with cuticular waxes. Increasing evidence supports that the cuticle plays a key role in plant adaptation to abiotic and biotic stresses, which could be harnessed for wheat and barley improvement. In this review, we highlighted recent advances in cuticle biosynthesis and its multifaceted roles in abiotic and biotic stress tolerance of wheat and barley. Current strategies, challenges, and future perspectives on manipulating cuticle biosynthesis for abiotic and biotic stress tolerance in wheat and barley are discussed.
Introduction
As the first plants domesticated about 10,000 years ago, wheat and barley are important cereal crops used extensively for human food and animal feed (Haas et al., 2019; Levy and Feldman, 2022). The global population is projected to reach 9.7 billion by 2050 and rise further to 11.2 billion in 2100, which drives the demand for wheat and barley grains (Lee, 2011). However, yields and quality of wheat and barley are adversely affected by numerous environmental stresses such as water deficit (drought), high salinity, extreme temperatures (heat and cold), and attacks of pathogens and pests (P&Ps) (Hura, 2020). For instance, drought stress was documented to reduce yields of 50%-90% and 49%-87% in drought-susceptible cultivars of wheat and barley respectively (Samarah, 2005; Daryanto et al., 2016). Soil salinity affects about 20% of global cultivated land, and seriously threatens the growth and production of glycophytes wheat and barley (Zörb et al., 2019). Temperature stresses such as chilling, freezing, and heat have become more frequent due to climate change and reduced grain yields and quality of wheat and barley (Jacott and Boden, 2020). In addition, a plethora of P&Ps, including pathogenic fungi, oomycetes, bacteria, viruses, nematodes, and herbivorous insects, were responsible for above 20% yield loss in wheat and barley (Savary et al., 2019). Developing and cultivating resistant varieties of wheat and barley are, therefore, essential for ensuring food security under environmental challenges.
As the outmost surface of terrestrial plants, lipophilic cuticle predominantly covers plant aerial organs like non-woody stems, leaves, flowers, and fruits, and protects plant tissues from abiotic and biotic stresses such as drought, salinity, heat, cold, ultraviolet (UV) radiation, mechanical damages, and P&Ps attacks (Domínguez et al., 2017; Kong et al., 2020a; Li and Chang, 2021). In addition to these protective roles, the cuticle also regulates plant developmental processes by inhibiting organ fusion and promoting lateral root formation (Kurdyukov et al., 2006; Ingram and Nawrath, 2017; Berhin et al., 2019). It has been demonstrated that the expression of cuticle biosynthesis genes is governed by DNA-binding transcription factors (TFs), mediators, and epigenetic regulators (Lee and Suh, 2015; Lee and Suh, 2022). At the same time, there is increasing evidence that cuticle biosynthesis mechanisms could be exploited for crop improvement (Wang et al., 2020; Liu et al., 2022b). Although past decades have seen a great advance in the understanding mechanisms of plant cuticle biosynthesis, most of this progress was achieved in model plants. Herein, we focus on recent studies exploring the mechanism of cuticle biosynthesis and its roles in the tolerance of wheat and barley to biotic and abiotic stresses. Potentials, strategies, challenges, as well as future perspectives on harnessing cuticle biosynthesis to improve abiotic and biotic stress tolerance in wheat and barley are discussed.
Cuticle composition and biosynthetic machinery in wheat and barley
As a hydrophobic layer covering the plant aerial epidermis, the cuticle is generally composed of lipid, phenolic, and polysaccharide compounds, and its hydrophobic property is mainly conferred by the lipid components cutin and wax (Reynoud et al., 2021). Cutin largely consists of cross-linked polyester of oxygenated C16 and C18 fatty acids, as well as their derivatives, whereas cuticular wax mixtures contain very-long-chain (VLC, >C20) fatty acids, alkanes, aldehydes, alcohols, esters, and ketones (Bhanot et al., 2021). Although lipophilic wax and cutin constitute the major components of plant cuticle, the composition of wax and cutin varies among plant species, organs, developmental stages, and environmental conditions. For instance, VLC alkanes are the major wax constituents of seedling leaves and stems in Arabidopsis, whereas VLC alcohols dominate the wax compositions of seedling leaves in wheat and barley (Rowland et al., 2006; Wang et al., 2015; Li et al., 2018). Notably, Arabidopsis does not produce β-diketones that are abundant in the cuticles covering spikes, flag leaves and stems at flowering wheat and barley plants (Hen-Avivi et al., 2016; Schneider et al., 2016). In addition, C18:0 18-OH acids are identified as the major cutin monomers in the seedling leaves of wheat and barley, whereas C18:2 diacids dominate the cutin composition of Arabidopsis seedling leaves (Hong et al., 2017; Kong and Chang, 2018; Li et al., 2018).
In plant epidermal cells, biosyntheses of cutin and wax occur in the endoplasmic reticulum (ER) through the modification of C16 and C18 fatty acids trafficked from the plastid (Yeats and Rose, 2013). For the cutin biosynthesis, C16 and C18 fatty acids sequentially undergo esterification, aliphatic chain elongation, hydroxylation, and acyltransferation to synthesize the cutin precursor sn-2 monoacylglycerols (2-MAGs) (Fich et al., 2016; Philippe et al., 2020). Long-chain acyl-coenzyme A synthases (LACS) catalyze the esterification of C16 and C18 fatty acids with coenzyme A (CoA) (Fich et al., 2016; Philippe et al., 2020). Cytochrome P450 enzymes (CYP77 and CYP86) and epoxide hydrolases (EH) mediate the hydroxylation of acyl-CoAs, and then glycerol-3-phosphate acyltransferase (GPAT) enzymes convert acyl-CoAs to 2-MAGs precursors (Lee et al., 2020). Cutin precursors are then exported out of plant cell via plasma membrane (PM) localized ATP binding cassette transporter subfamily G (ABCG) proteins and deposited into the cuticle, where cutin synthase (CUS) enzymes catalyze the cutin polymerization (Hong et al., 2017; Elejalde-Palmett et al., 2021; Philippe et al., 2022).
For the wax biosynthesis, fatty acid elongase (FAE) enzyme complexes comprising ketoacyl-CoA synthases (KCS), ketoacyl-CoA reductases (KCR), hydroxyacyl-CoA dehydratases (HCD), and enoyl-CoA reductases (ECR) function together with ECERIFERUM2-LIKE (CER2-LIKE) proteins to catalyze the aliphatic chain elongation of C16 and C18 acyl-CoAs, leading to the formation of VLC acyl-CoAs (Haslam et al., 2015; Haslam and Kunst, 2021; Kim J. et al., 2022). VLC acyl-CoAs could be converted to VLC alkanes by a VLC alkane-forming complex consisting of ECERIFERUM1 (CER1), CER1-LIKE1, CER3, and cytochrome B5 (CYTB5) proteins (Pascal et al., 2019). VLC alkanes then undergo hydroxylation mediated by the CYP95A family cytochrome P450 enzyme midchain alkane hydroxylase 1 (MAH1) to form VLC secondary alcohols and ketones in the alkane-forming pathway (Greer et al., 2007). As an alternative direction, VLC acyl-CoAs could enter the alcohol-forming pathway and are converted to VLC primary alcohols by fatty acyl-coenzyme A reductase (FAR) CER4 and acyl-CoA desaturase LIKE4 (ADS4/CER17) (Rowland et al., 2006; Yang et al., 2017). VLC primary alcohols and acyl-CoAs serve as precursors in the subsequent biosynthesis of wax esters catalyzed by a bifunctional wax ester synthase/diacylglycerol acyltransferase WSD1 (Li et al., 2008; Patwari et al., 2019). These wax constituents such as VLC fatty acids, aldehydes, alkanes, alcohols, ketones, and esters are transported through the Golgi and trans-Golgi network (TGN)-trafficking pathways to the PM, and then exported to the cuticle via ABCG subfamily half transporters and the lipid transfer proteins (LTPs) (Pighin et al., 2004; Debono et al., 2009; Ichino and Yazaki, 2022).
Although most of these advances in the understanding of cuticle biosynthesis were derived from studies in the model plants like Arabidopsis and tomato, evolutionarily conserved functions are widely displayed by cuticle biosynthesis genes of wheat and barley. On one hand, ectopic expression of cuticle biosynthesis genes derived from wheat and barley could significantly enhance cuticle formation in Arabidopsis. Indeed, overexpression of wheat TaCER1-1A in Arabidopsis could enhance stem and leaf accumulation of cuticular wax (Li et al., 2019). Heterologous expression of wheat TaFAR2, TaFAR3, and TaFAR4 in Arabidopsis cer4-3 mutant defective in the production of C24 and C26 primary alcohols results in the increased accumulation of primary alcohols (Wang et al., 2016). On the other hand, knockdown or knockout of wheat and barley genes orthologous to Arabidopsis cuticle biosynthesis genes usually attenuated plant cuticle biosynthesis. Silencing of wheat TaECR and TaKCS6 via virus-induced gene silencing (VIGS) led to reduced wax accumulation in wheat leaves (Wang et al., 2019; Kong et al., 2020b). Likewise, barley mutant enhanced Magnaporthe resistance gene1 (emr1) carrying a mutation in HvKCS6 is depleted of leaf wax (Weidenbach et al., 2014). Although some cuticle biosynthesis genes exhibited functional conservation among Arabidopsis, wheat, and barley, functional divergence is also observed in some cuticle biosynthesis genes. For instance, Arabidopsis T-DNA tagged mutant kcs1-1 displayed a marginal change in the total wax load compared with the wild-type plants. In contrast, the barley eceriferum-zh (cer-zh) mutant carrying mutation in the KCS1 gene exhibited significantly reduced wax accumulation (Todd et al., 1999; Li et al., 2018). These studies support the idea that, although cuticle biosynthetic machinery is highly conserved among model and crop plants, the functional divergence has been acquired by some cuticle biosynthesis genes in wheat and barley.
As oxidized hydrocarbons, β-diketones are cuticular wax components of wheat and barley rather than the model plant Arabidopsis (von Wettstein-Knowles, 1972; Kosma and Rowland, 2016; von Wettstein-Knowles, 2017). Characterization of barley eceriferum-c (cer-c), cer-q, and cer-u mutants with altered glaucousness traits shed novel light into the biosynthesis of β-diketone (Kosma and Rowland, 2016; von Wettstein-Knowles, 2017). Map based cloning revealed that barley Cer-c, Cer-q and Cer-u genes reside in the 101 kb Cer-cqu gene cluster as the order Cer-c, Cer-u, Cer-q, and encode a chalcone-synthase-like diketone synthase (DKS), a putative lipase/carboxyl transferase, and a cytochrome P450 hydroxylase, respectively (Schneider et al., 2016). As extensively discussed by von Wettstein-Knowles, β-diketones are proposed to be synthesized from C12, C14, C16 fatty acid and C16-CoA via a polyketide-like pathway involving CER-C, CER-Q, CER-U together with components of FAE complex (von Wettstein-Knowles, 2017). Transcriptomic analysis using chromosomearm substitution lines (CASLs) of wild emmer together with the subsequent gene silencing assays revealed that wheat W1 locus contains a similar gene cluster harboring Diketone Metabolism-PKS (DMP), -Hydrolase (DMH), and -CYP450 (DMC) genes essential for β-diketone biosynthesis, suggesting that the conserved metabolic gene cluster mediates β-diketone biosynthesis in wheat and barley (Hen-Avivi et al., 2016).
Regulatory mechanisms of cuticle biosynthesis in wheat and barley
Accumulating evidence support that the expression of cuticle biosynthesis genes in wheat and barley is tightly governed by TFs, mediators, and epigenetic regulators. SHINE (SHN) clade of AP2 domain TF AtSHN1 and its close homologs AtSHN2 and AtSHN3 were firstly identified as transcriptional activators of cuticle lipid biosynthesis in Arabidopsis (Aharoni et al., 2004; Broun et al., 2004; Kannangara et al., 2007). Barley TF WAX INDUCER1 (HvSHN1/WIN1) and wheat TF TaSHN1/WIN1 are homologs of Arabidopsis SHN1. Knockdown of HvSHN1/WIN1 by VIGS resulted in the reduced accumulation of cuticular lipid in barley spikelets, whereas ectopic expression of HvSHN1/WIN1 in tobacco could activate the expression of wax biosynthesis gene NtCER1 and resulted in the altered cuticle property (Kumar et al., 2016; Djemal and Khoudi, 2021). Likewise, Knockout or knockdown of TaSHN1/WIN1 expression in bread wheat attenuated wax and cutin biosynthesis, whereas overexpression of TaSHN1/WIN1 led to enhanced wax accumulation in transgenic wheat plants (Bi et al., 2018; Kong and Chang, 2018). This evidence supports that TFs TaSHN1/WIN1 and HvSHN1/WIN1, resembling their counterparts in Arabidopsis, positively regulate cuticle biosynthesis in wheat and barley. PpWIN1, an SHN1 homolog in Physcomitrium patens, was recently revealed to stimulate cuticle formation in P. patens and Arabidopsis by activating cutin and wax biosynthesis genes, suggesting that the transcriptional activation of cuticle biosynthesis by TF SHN1/WIN1 might be conserved from moss to higher land plants including wheat and barley (Kim R. J. et al., 2022).
In addition to SHN/WINs, myeloblastosis (MYB) and basic helix-loop-helix protein (bHLH) type TFs are identified as key regulators of wheat cuticle biosynthesis. For instance, the wheat MYB TF TaMYB74 was revealed to transactivate wheat cuticle biosynthesis-related gene TaSHN1/WIN1 and respond to drought stress (Bi et al., 2016). Another wheat MYB TF TaEPBM1 could directly bind to the promoter region of the wax biosynthesis gene TaECR and activate TaECR expression (Kong et al., 2020b). TaMYB96, allelic to TaEPBM1, was recently demonstrated to target TaCER1-6A, TaCER1-1A, and TaFAR4, and positively regulate wax biosynthesis as well, supporting that TaMYB96/TaEPBM1 potentiates wheat cuticle biosynthesis via directly activating wax biosynthesis genes (He et al., 2022). In Arabidopsis, bHLH TFs CFLAP1 and CFLAP2 were demonstrated to negatively regulate wax biosynthesis and cuticle formation (Li et al., 2016). The wheat bHLH TF TaKPAB1 was demonstrated to recognize the E-box cis-element in the promoter of wax biosynthesis gene TaKCS6 and activate the transcription of TaKCS6 (Wang et al., 2019). Knockdown of TaKPAB1 and TaKCS6 expression by VIGS results in reduced wax accumulation, suggesting that transactivation of TaKCS6 by TaKPAB1 positively regulates wheat cuticular wax biosynthesis (Wang et al., 2019).
As an essential component of a highly conserved mediator complex, CYCLIN-DEPENDENT KINASE8 (CDK8) functions as a transcriptional co-regulator to activate or repress transcription of target genes (Mao et al., 2019; Agrawal et al., 2021; Chen J. et al., 2022). In Arabidopsis, CDK8 is physically associated with the TF AtSHN1 and positively regulates plant cuticle formation (Zhu et al., 2014). TaCDK8, the wheat homolog of Arabidopsis AtCDK8, also interacts with TaSHN1/WIN1 and activates wax and cutin biosynthesis in bread wheat, suggesting that positive regulation of cuticle biosynthesis by CDK8-WIN1 module might be conserved among dicots and monocots (Kong and Chang, 2018). Notably, TaCDK8 could directly phosphorylate TaSHN1/WIN1, which is essential to the transcriptional activation mediated by TaSHN1/WIN1 (Kong and Chang, 2018). Silencing of TaCDK8 and TaWIN1 by VIGS attenuated wheat wax biosynthesis, further supporting the idea that mediator subunit TaCDK8 phosphorylates the TF TaSHN1/WIN1 to stimulate the expression of wheat biosynthesis genes (Kong and Chang, 2018).
Histone modification and chromatin remodeling are considered important epigenetic mechanisms in the regulation of plant development and environmental adaptation (Chang et al., 2020; Kumar et al., 2021; Zhi and Chang, 2021). Arabidopsis histone methyl transferases SET DOMAIN GROUP8 (AtSDG8), AtSDG25, histone E3 ligases HISTONE MONOUBIQUITINATION 1 (AtHUB1), AtHUB2, and histone acetyltransferase GENERAL CONTROL NON-REPRESSED PROTEIN5 (AtGCN5) play important roles in the regulation of cuticle biosynthesis (Ménard et al., 2014; Lee et al., 2016; Wang et al., 2018). It was demonstrated that wheat TF TaEPBM1 could directly bind to the transcriptional coactivator TaADA2, an interacting partner of TaGCN5 in the histone acetyltransferase module of Spt-Ada-Gcn5 Acetyltransferase (SAGA) complex (Kong et al., 2020b). Via association with TaEPBM1, the TaGCN5-TaADA2 module is recruited to the TaECR promoter region to mediate histone acetylation (Kong et al., 2020b). Silencing of TaGCN5 and TaADA2 by VIGS resulted in reduced TaECR expression and decreased wax accumulation, suggesting that epigenetic activation of TaECR by histone acetyltransferase complex TaGCN5-TaADA2 triggers wheat wax biosynthesis (Kong et al., 2020b). Likewise, the CHD3-type chromatin remodeling factor TaCHR729 interacts with TF TaKPAB1 and is recruited to the promoter region of TaKCS6 (Wang et al., 2019). Through mediating deposition of permissive epigenetic mark H3K4me3, TaCHR729 promotes TaKCS6 expression and positively regulates wheat wax biosynthesis (Wang et al., 2019). These studies provide evidence that the expression of the wheat cuticle biosynthesis gene is epigenetically governed by multiple epigenetic regulators, including histone modifying enzymes and chromatin remodeling factors.
Interplays of cuticle biosynthesis with other epidermal developmental processes like trichome formation have been extensively discussed by prior reviews (Ingram and Nawrath, 2017; Berhin et al., 2022). Cuticle functions in concert with stomata to tightly control water and gas exchange essential for plant photosynthesis and environmental adaptation. Molecular characterization of barley cer-g.10 and cer-s.31 mutants exhibiting wax-deficiency and stomatal misarrangement demonstrated that Cer-g and Cer-s genes respectively encode a YODA-like (YDA) MAPKKK HvYDA1 and a BREVIS-RADIX (BRX) domain protein HvBRX-Solo, two signaling components in stomatal development (Liu et al., 2022a). Epidermal phenotype analysis of cer-g.10, cer-s.31 and cer-g.10 cer-s.31 double mutants revealed that HvYDA1 and HvBRX function in a common pathway to control wax deposition and epidermal patterning (Liu et al., 2022a). RNA sequencing (RNA-seq) analysis showed that HvYDA1 and HvBRX-Solo coregulate downstream genes associated with cuticle development, epidermal differentiation and patterning, further confirming that HvYDA1-HvBRX-Solo signaling module orchestrates cuticle biosynthesis and epidermal patterning in barley (Liu et al., 2022a).
Regulation of plant abiotic stress tolerance by cuticle biosynthesis in wheat and barley
By limiting non-stomatal water loss, cuticle contributes to plant adaptation to drought conditions, which has been extensively discussed in previous reviews (Xue et al., 2017; Lewandowska et al., 2020; Liu et al., 2022b). Upregulation of cuticle biosynthesis-related genes such as CER1s, FARs, SHN1, MYB74, WXPLs in response to drought stress has been reported in wheat and barley (Wang et al., 2015; Bi et al., 2016; Wang M. et al., 2016; Bi et al., 2017; Bi et al., 2018; Chai et al., 2018; Li et al., 2019; He et al., 2022). As summarized in Table 1 and Figure 1, over-expression of these cuticle biosynthesis genes usually results in reinforced cuticle formation and enhanced plant drought tolerance. In contrast, knockout or knockdown of these cuticle biosynthesis genes expression could attenuate cuticle formation and compromise plant drought resilience. For instance, over-expression of TaSHN1/WIN1 and TaCER1-6A in transgenic wheat plants resulted in enhanced wax accumulation and increase drought tolerance (Table 1, Figure 1) (Bi et al., 2018; He et al., 2022). Ectopic expression of TaCER1-1A and HvSHN1/WIN1 in rice and tobacco could also alter the cuticle property and lead to enhanced plant drought resistance (Table 1, Figure 1) (Li et al., 2019; Djemal and Khoudi, 2021). Consistent with this, wheat TaCER1-6A knockout lines generated by CRISPR/Cas9 genome editing system displayed enhanced cuticle permeability and attenuated plant drought resilience (Table 1, Figure 1) (He et al., 2022). Reduced water retention capacity was also observed in the leaves of barley cuticle mutant eceriferum-ym (cer-ym), cer-zv and eibi1 (Chen et al., 2004; Chen et al., 2011; Li et al., 2015; Li et al., 2017).
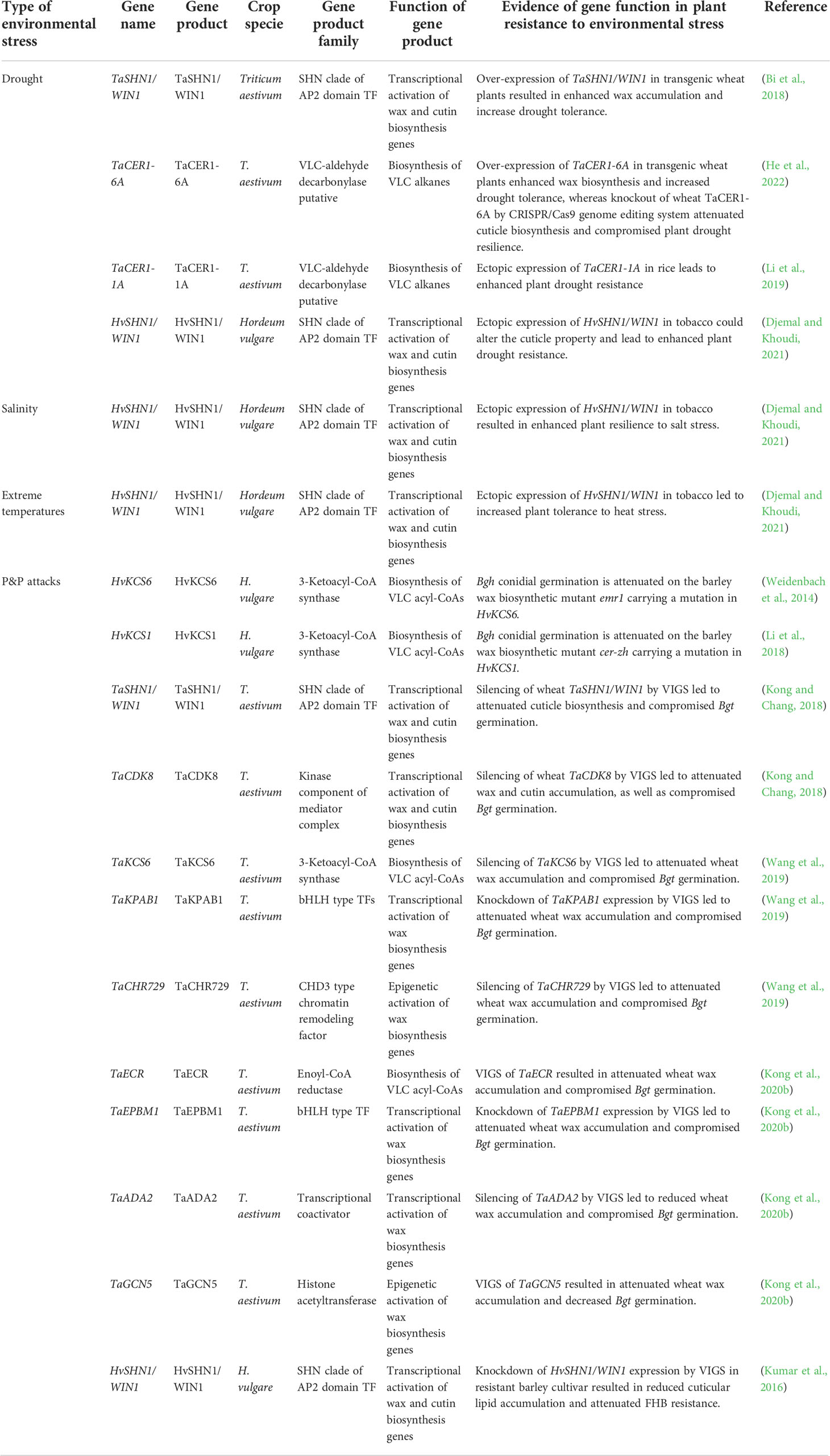
Table 1 Summary of cuticle biosynthesis genes contributing to abiotic and biotic stress tolerance in wheat and barley.
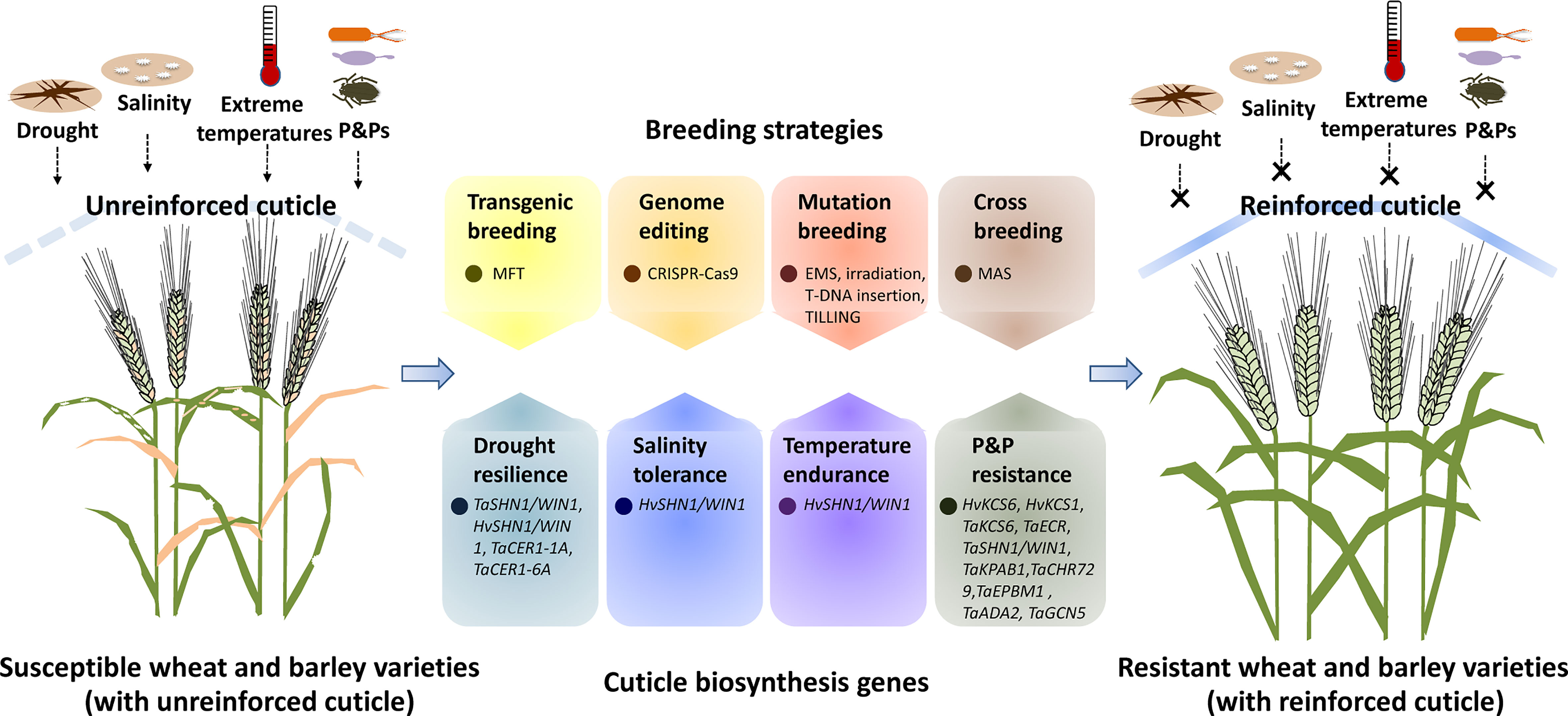
Figure 1 A simplified model for manipulating cuticle biosynthesis to improve abiotic and biotic stress tolerance in wheat and barley. Cuticle biosynthesis genes in wheat and barley get involved in the regulation of plant response to abiotic and biotic stresses such as drought, salinity, extreme temperatures, and attacks of pathogens and pests (P&Ps). Genetic manipulation of these cuticle biosynthesis genes by transgenic breeding, genome editing, mutagenesis breeding and cross breeding could reinforce the cuticle, resulting in improved abiotic and biotic stress resistance in wheat and barley.
As a protective shield covering aerial organs, the cuticle also protects plant tissues from other environmental stresses such as salinity, heat, cold, and UV radiation, which has been supported by a broad range of research on model and crop plants (Zhang et al., 2020; Abdullah et al., 2021; Busta et al., 2021; Benítez et al., 2022; González Moreno et al., 2022; Kan et al., 2022; Liu et al., 2022). Expression of eight wheat FAR genes (TaFAR1, TaFAR2, TaFAR3, TaFAR4, TaFAR5, TaFAR6, TaFAR7, and TaFAR8) are up-regulated by cold stress, and three FAR genes (TaFAR2, TaFAR3, and TaFAR4) are induced by salinity stress, suggesting that these cuticle biosynthesis FAR genes might get involved in wheat response to temperature and salt stress (Wang et al., 2015; Wang et al., 2016; Chai et al., 2018). Interestingly, heterologous expression of barley HvSHN1/WIN1 gene in tobacco led to enhanced plant tolerance to heat, salt, and drought stress, further supporting the contribution of cuticle biosynthesis to abiotic stress resilience in wheat and barley (Table 1, Figure 1) (Djemal and Khoudi, 2021).
Regulation of plant-P&P interactions by cuticle biosynthesis in wheat and barley
As the first contact interphase between aerial plant organs and P&Ps, the cuticle regulates multiple processes such as pathogen (pre)penetration, plant immunity, and pest behaviors in plant-P&P interactions, which has been summarized by previous reviews (Wang et al., 2020; Arya et al., 2021; Li et al., 2022). Accumulating evidence revealed that plant cuticle governs the interactions of P&Ps with wheat and barley. Firstly, cuticular wax signals are essential for triggering the (pre)penetration development of pathogenic powdery mildew fungi in wheat and barley. Conidial germination of the barley powdery mildew fungus Blumeria graminis f. sp. hordei (Bgh) is attenuated on the barley wax biosynthetic mutant emr1 and cer-zh (Table 1, Figure 1) (Weidenbach et al., 2014; Li et al., 2018). Likewise, silencing of wheat wax biosynthesis genes TaKCS6, TaECR, TaSHN1/WIN1, TaCDK8, TaKPAB1, TaEPBM1, TaADA2, and TaGCN5 all led to the attenuated wax accumulation and compromised germination of the wheat powdery mildew fungus Blumeria graminis f. sp. tritici (Bgt) (Table 1, Figure 1) (Kong and Chang, 2018; Wang et al., 2019; Kong et al., 2020b). Notably, in vitro application of wax constituents, VLC aldehydes, stimulate the Bgh germination in a dose-dependent manner and could fully restore the Bgt germination penalty on the wheat wax biosynthetic mutant, suggesting that VLC aldehydes are the plant wax signals essential for stimulating (pre)penetration development of Blumeria graminis in wheat and barley (Hansjakob et al., 2010; Kong and Chang, 2018; Wang et al., 2019; Kong et al., 2020b). Secondly, cuticle biosynthesis contributes to the immunity of wheat and barley against some pathogens. For instance, barley resistance to Fusarium head blight (FHB) is correlated with the expressions of cuticle biosynthesis genes HvKAS2, HvCYP86A2, HvCYP89A2, HvLACS2 and HvSHN1/WIN1 in resistant cultivar (Table 1, Figure 1) (Kumar et al., 2016). Knockdown of HvSHN1/WIN1 expression by VIGS in this resistant barley cultivar resulted in reduced cuticle lipid accumulation and attenuated FHB resistance, further supporting the contribution of cuticle lipid biosynthesis to barley FHB resistance (Kumar et al., 2016). Thirdly, cuticle biosynthesis gets involved in wheat response to pest infestation. Kosma et al. reported that infestation of the pest Hessian fly leads to the up-regulation of cuticle biosynthesis genes such as TaCER3, TaCER1, TaCER4, TaKCS1, TaKCS6, TaCER5, together with the alteration in wheat wax and cutin compositions (Kosma et al., 2010). Notably, these transcriptomic and metabolic responses displayed the difference in resistant and susceptible wheat cultivars, implying that cuticle biosynthesis might play a key role in the regulation of wheat resistance against Hessian fly infestation (Kosma et al., 2010).
Strategies, limitations, and perspectives on exploiting cuticle biosynthesis for wheat and barley improvement
As a hydrophobic shield covering aerial plant organs, the cuticle contributes to plant adaptation to environmental stresses such as drought, salinity, extreme temperatures, and P&P attacks (Arya et al., 2021; Liu et al., 2022). Through analyzing leaf wax alkane and grain yield traits in five wheat cultivars released during the past six decades, Liu et al. observed a tendency to increase and a strong correlation in leaf wax alkane concentration and grain yield across the historical wheat varieties, suggesting that increased leaf wax alkane concentration has been selected in breeding efforts for wheat production improvement (Liu et al., 2019). Exploiting cuticle biosynthesis by advanced breeding strategies such as transgenic breeding, genome editing, mutation breeding, and cross breeding could provide more avenues for wheat and barley improvement (Liu et al., 2019).
Genetic engineering of cuticle biosynthesis genes could confer plant stress resistance. For instance, the over-expression of TaSHN1/WIN1 and TaCER1-6A led to wax over-accumulation and increased drought resilience in bread wheat (Table 1, Figure 1) (Bi et al., 2018; He et al., 2022). Notably, ectopic expression of Arabidopsis AtMYB96 and AtWSD1 could enhance drought tolerance of oil crop Camelina sativa, implying that cuticle biosynthesis genes identified from model plants could also be employed for the transgenic improvement of crop plants like wheat and barley (Lee et al., 2014; Abdullah et al., 2021). Due to biosafety concerns, selectable marker genes (SMGs) used for the selection of transformants should be eliminated from GM crops, which was facilitated by marker-free transgenic (MFT) strategies (Ling et al., 2016; Wang G. P. et al., 2016; Ahmad and Mukhtar, 2017; Wang et al., 2017). By employing the double right border (DRB) T-DNA vector, Cao et al. successfully constructed the marker-free and transgene insertion site-defined (MFTID) transgenic wheat plants for silencing lipoxygenase (LOX) gene (Cao et al., 2020). These MFTID transgenic wheat plants exhibited attenuated LOX gene expression, and improved grain storability, and fatty acid content, thereby paving a path for creating MFTID plants with altered cuticle traits in wheat and barley (Figure 1) (Cao et al., 2020). As recalcitrant crops, wheat and barley have low rates of transformation and regeneration (Mrízová et al., 2014; Hensel, 2020; Mirzaee et al., 2022; Wijerathna-Yapa et al., 2022). Over-expression of the WUSCHEL family gene TaWOX5 and the chimeric gene harboring wheat TaGRF4 and TaGIF1 were reported to improve wheat efficiency of transformation and regeneration respectively (Debernardi et al., 2020; Wang et al., 2022). These breakthroughs in crop transformation and regeneration would certainly facilitate the genetic engineering of the cuticle biosynthesis genes in wheat and barley.
As an advanced genome editing (GE) technique, the CRISPR-Cas9 (clustered regularly interspaced short palindromic repeats-CRISPR associated 9) system has been extensively employed for functional genomics and trait improvement in model and crop plants (Chen et al., 2019; Manghwar et al., 2019; Zhu et al., 2020; Gao, 2021). Knockout of MYS1 and MYS2, transcription repressors of DECREASE WAX BIOSYNTHESIS (DEWAX), by CRISPR-Cas9 system, resulted in the reduced wax loads and attenuated plant drought tolerance, suggesting that genome editing of cuticle biosynthesis genes could effectively alter plant stress tolerance (Liu et al., 2022c). However, the conventional application of the CRISPR-Cas9 system necessitates plant transformation and regeneration, which hinders its use in wheat and barley breeding. Interestingly, Li et al. established a tissue culture-free genome editing approach in Cas9-overexpressed (Cas9-OE) wheat plants by employing an engineered BSMV-based sgRNA (BSMV-sg) delivery vector (Li et al., 2021). By adding RNA mobility sequence in the virus vector, Chen et al. and Ellison et al. successfully enhanced the editing rate of this convenient virus-mediated gene editing system, which paved a new path for genetic manipulation of cuticle biosynthesis genes in wheat and barley (Figure 1) (Ellison et al., 2020; Chen H. et al., 2022).
In traditional mutation breeding, genetic mutations were induced by chemical, physical and biological agents such as ethyl methanesulfonate (EMS), X-rays, gamma rays, fast neutrons, and T-DNAs. Compared with genetic engineering and genome editing, this traditional mutation breeding based on random mutagenesis is labor-intensive, time-consuming, and less effective (Holme et al., 2019). As an advanced strategy in targeted mutation breeding, targeting induced local lesions (TILLING) deploys saturated mutagenesis and high-throughput screening approaches to efficiently introduce single-nucleotide mutation to any genomic regions like cuticle biosynthesis genes (McCallum et al., 2000; Kurowska et al., 2011; Chen et al., 2014). A drought-insensitive TILLING line 1 (ditl1) mutant was recently identified from the rice TILLING mutant population and was revealed to harbor mutation in the cuticle biosynthesis-related WSD1-like gene (Choi et al., 2022). These induced mutations with desired cuticle traits could be introduced into elite cultivars of wheat and barley via cross breeding facilitated by advanced marker-assisted selection (MAS) approaches (Figure 1) (Hickey et al., 2019; Thudi et al, 2021).
Although the genetic manipulation of some cuticle biosynthesis genes could enhance abiotic and biotic stress tolerance in wheat and barley, many challenges need to be overcome regarding the exploitation of cuticle biosynthesis for wheat and barley improvement. For instance, present cuticle phenotyping techniques such as GC-MS (gas chromatography-mass spectrometry) and MALDI (matrix assisted laser desorption/ionization) imaging are low throughput and time-consuming, and high-throughput and high-precision methods needed to be developed for identifying wheat and barley mutants with cuticle traits (Petit et al., 2017). Furthermore, over-expression of cuticle biosynthesis genes usually enhances plant stress resilience with yield failure due to altered metabolic and energy flux. It is, therefore, vital for breeders to identify new cuticle biosynthesis genes conferring plant stress resilience without yield penalty. Moreover, strict policy regulations have been imposed on GMOs (genetically modified organisms) in some countries, and these regulations needed to be modified for placing wheat and barley varieties developed by genetic engineering and/or genome editing of cuticle biosynthesis genes into markets (Wolt and Wolf, 2018; Eriksson et al., 2020; Redden, 2021; Hundleby et al., 2022).
Concluding remarks
Herein, we provide an overview of recent progress in the understanding of cuticle biosynthesis in wheat and barley, and highlight the contribution of cuticle biosynthesis in the adaptation of wheat and barley to environmental challenges. Current strategies and limitations on exploiting cuticle biosynthesis for wheat and barley improvement are discussed. As depicted in Figure 1, genetic manipulation of cuticle biosynthesis genes by transgenic breeding, genome editing, mutation breeding, and cross breeding could result in cuticle reinforcement and lead to improved performance of wheat and barley under stressful environments. Although the past decades have seen unprecedented proceedings in the exploration and exploitation of cuticle biosynthesis, we still have a long way to go toward fully understanding cuticle biosynthesis in wheat and barley. For instance, most of the characterized cuticle biosynthesis genes come from model plants, while wheat and barley genes involved in the biosynthesis of cuticular lipids, especially cutin monomers, are poorly understood. Furthermore, cuticle compositions of wheat and barley vary along with plant developmental stages and environmental conditions, but the response mechanism of cuticle biosynthesis to developmental and environmental cues remains to be uncovered in wheat and barley. Moreover, the cuticle plays a vital role in plant tolerance to abiotic stresses, but the functions and mechanisms of cuticle biosynthesis in the adaptation of wheat and barley to salinity, temperature, and UV stresses remain to be disclosed. In addition, wax signals from wheat and barley cuticles are revealed to facilitate conidial germination of powdery mildew, but whether and how cuticle biosynthesis regulates interactions of wheat and barley with other P&Ps such as bacterial pathogens and pests is still unknown. With advances in the understanding of cuticle biosynthetic machinery in wheat and barley, manipulating cuticle biosynthesis would certainly promote crop improvement for stress resilience and disease resistance.
Author contributions
CC and XW wrote this manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Natural Science Foundation of Shandong Province (ZR2022MC008, ZR2017BC109), National Natural Science Foundation of China (31701412), the Qingdao Science and Technology Bureau Fund (17-1-1-50-jch) and Qingdao University Fund (DC1900005385).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdullah, H. M., Rodriguez, J., Salacup, J. M., Castañeda, I. S., Schnell, D. J., Pareek, A., et al. (2021). Increased cuticle waxes by overexpression of WSD1 improves osmotic stress tolerance in Arabidopsis thaliana and camelina sativa. Int. J. Mol. Sci. 22, 5173. doi: 10.3390/ijms22105173
Agrawal, R., Jiří, F., Thakur, J. K. (2021). The kinase module of the mediator complex: an important signaling processor for the development and survival of plants. J. Exp. Bot. 72, 224–240. doi: 10.1093/jxb/eraa439
Aharoni, A., Dixit, S., Jetter, R., Thoenes, E., van Arkel, G., Pereira, A. (2004). The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16, 2463–2480. doi: 10.1105/tpc.104.022897
Ahmad, N., Mukhtar, Z. (2017). Genetic manipulations in crops: Challenges and opportunities. Genomics 109, 494–505. doi: 10.1016/j.ygeno.2017.07.007
Arya, G. C., Sarkar, S., Manasherova, E., Aharoni, A., Cohen, H. (2021). The plant cuticle: an ancient guardian barrier set against long-standing rivals. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.663165
Benítez, J. J., González Moreno, A., Guzmán-Puyol, S., Heredia-Guerrero, J. A., Heredia, A., Domínguez, E. (2022). The response of tomato fruit cuticle membranes against heat and light. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.807723
Berhin, A., de Bellis, D., Franke, R. B., Buono, R. A., Nowack, M. K., Nawrath, C. (2019). The root cap cuticle: A cell wall structure for seedling establishment and lateral root formation. Cell 176, 1367–1378.e8. doi: 10.1016/j.cell.2019.01.005
Berhin, A., Nawrath, C., Hachez, C. (2022). Subtle interplay between trichome development and cuticle formation in plants. New Phytol. 233, 2036–2046. doi: 10.1111/nph.17827
Bhanot, V., Fadanavis, S. V., Panwar, J. (2021). Revisiting the architecture, biosynthesis and functional aspects of the plant cuticle: there is more scope. Environ. Exp. Bot. 183, 104364. doi: 10.1016/j.envexpbot.2020.104364
Bi, H., Luang, S., Li, Y., Bazanova, N., Borisjuk, N., Hrmova, M., et al. (2017). Wheat drought-responsive WXPL transcription factors regulate cuticle biosynthesis genes. Plant Mol. Biol. 94, 15–32. doi: 10.1007/s11103-017-0585-9
Bi, H., Luang, S., Li, Y., Bazanova, N., Morran, S., Song, Z., et al. (2016). Identification and characterization of wheat drought-responsive MYB transcription factors involved in the regulation of cuticle biosynthesis. J. Exp. Bot. 67, 5363–5380. doi: 10.1093/jxb/erw298
Bi, H., Shi, J., Kovalchuk, N., Luang, S., Bazanova, N., Chirkova, L., et al. (2018). Overexpression of the TaSHN1 transcription factor in bread wheat leads to leaf surface modifications, improved drought tolerance, and no yield penalty under controlled growth conditions. Plant Cell Environ. 41, 2549–2566. doi: 10.1111/pce.13339
Broun, P., Poindexter, P., Osborne, E., Jiang, C. Z., Riechmann, J. L. (2004). WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 101, 4706–4711. doi: 10.1073/pnas.0305574101
Busta, L., Schmitz, E., Kosma, D. K., Schnable, J. C., Cahoon, E. B. (2021). A co-opted steroid synthesis gene, maintained in sorghum but not maize, is associated with a divergence in leaf wax chemistry. Proc. Natl. Acad. Sci. U. S. A. 118, e2022982118. doi: 10.1073/pnas.2022982118
Cao, X., Dong, Z., Tian, D., Dong, L., Qian, W., Liu, J., et al. (2020). Development and characterization of marker-free and transgene insertion sitedefined transgenic wheat with improved grain storability and fatty acid content. Plant Biotechnol. J. 18, 129–140. doi: 10.1111/pbi.13178
Chai, G., Li, C., Xu, F., Li, Y., Shi, X., Wang, Y., et al. (2018). Three endoplasmic reticulum-associated fatty acyl-coenzyme a reductases were involved in the production of primary alcohols in hexaploid wheat (Triticum aestivum l.). BMC Plant Biol. 18, 41. doi: 10.1186/s12870-018-1256-y
Chang, Y. N., Zhu, C., Jiang, J., Zhang, H., Zhu, J. K., Duan, C. G. (2020). Epigenetic regulation in plant abiotic stress responses. J. Integr. Plant Biol. 62, 563–580. doi: 10.1111/jipb.12901
Chen, L., Hao, L., Parry, M. A., Phillips, A. L., Hu, Y. G. (2014). Progress in TILLING as a tool for functional genomics and improvement of crops. J. Integr. Plant Biol. 56, 425–443. doi: 10.1111/jipb.12192
Chen, G., Komatsuda, T., Ma, J. F., Nawrath, C., Pourkheirandish, M., Tagiri, A., et al. (2011). An ATP-binding cassette subfamily G full transporter is essential for the retention of leaf water in both wild barley and rice. Proc. Natl. Acad. Sci. U. S. A. 108, 12354–12359. doi: 10.1073/pnas.1108444108
Chen, G., Sagi, M., Weining, S., Krugman, T., Fahima, T., Korol, A. B., et al. (2004). Wild barley eibi1 mutation identifies a gene essential for leaf water conservation. Planta 219, 684–693. doi: 10.1007/s00425-004-1277-7
Chen, H., Su, Z., Tian, B., Liu, Y., Pang, Y., Kavetskyi, V., et al. (2022). Development and optimization of a barley stripe mosaic virus-mediated gene editing system to improve Fusarium head blight resistance in wheat. Plant Biotechnol. J. 20, 1018–1020. doi: 10.1111/pbi.13819
Chen, K., Wang, Y., Zhang, R., Zhang, H., Gao, C. (2019). CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 667–697. doi: 10.1146/annurev-arplant-050718-100049
Chen, J., Yang, S., Fan, B., Zhu, C., Chen, Z. (2022). The mediator complex: a central coordinator of plant adaptive responses to environmental stresses. Int. J. Mol. Sci. 23, 6170. doi: 10.3390/ijms23116170
Choi, S. Y., Lee, Y. J., Seo, H. U., Kim, J. H., Jang, C. S. (2022). Physio-biochemical and molecular characterization of a rice drought-insensitive TILLING line 1 (ditl1) mutant. Physiol. Plant 174, e13718. doi: 10.1111/ppl.13718
Daryanto, S., Wang, L., Jacinthe, P. A. (2016). Global synthesis of drought effects on maize and wheat production. PloS One 11, e0156362. doi: 10.1371/journal.pone.0156362
Debernardi, J. M., Tricoli, D. M., Ercoli, M. F., Hayta, S., Ronald, P., Palatnik, J. F., et al. (2020). A GRF-GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat. Biotechnol. 38, 1274–1279. doi: 10.1038/s41587-020-0703-0
Debono, A., Yeats, T. H., Rose, J. K., Bird, D., Jetter, R., Kunst, L., et al. (2009). Arabidopsis LTPG is a glycosylphosphatidylinositol-anchored lipid transfer protein required for export of lipids to the plant surface. Plant Cell 21, 1230–1238. doi: 10.1105/tpc.108.064451
Djemal, R., Khoudi, H. (2021). The barley SHN1-type transcription factor HvSHN1 imparts heat, drought and salt tolerances in transgenic tobacco. Plant Physiol. Biochem. 164, 44–53. doi: 10.1016/j.plaphy.2021.04.018
Domínguez, E., Heredia-Guerrero, J. A., Heredia, A. (2017). The plant cuticle: old challenges, new perspectives. J. Exp. Bot. 68, 5251–5255. doi: 10.1093/jxb/erx389
Elejalde-Palmett, C., Martinez San Segundo, I., Garroum, I., Charrier, L., De Bellis, D., Mucciolo, A., et al. (2021). ABCG transporters export cutin precursors for the formation of the plant cuticle. Curr. Biol. 31, 2111–2123.e9. doi: 10.1016/j.cub.2021.02.056
Ellison, E. E., Nagalakshmi, U., Gamo, M. E., Huang, P. J., Dinesh-Kumar, S., Voytas, D. F. (2020). Multiplexed heritable gene editing using RNA viruses and mobile single guide RNAs. Nat. Plants 6, 620–624. doi: 10.1038/s41477-020-0670-y
Eriksson, D., Custers, R., Edvardsson Björnberg, K., Hansson, S. O., Purnhagen, K., Qaim, M., et al. (2020). Options to reform the European union legislation on GMOs: risk governance. Trends Biotechnol. 38, 349–351. doi: 10.1016/j.tibtech.2019.12.016
Fich, E. A., Segerson, N. A., Rose, J. K. (2016). The plant polyester cutin: biosynthesis, structure, and biological roles. Annu. Rev. Plant Biol. 67, 207–233. doi: 10.1146/annurev-arplant-043015-111929
Gao, C. (2021). Genome engineering for crop improvement and future agriculture. Cell 184, 1621–1635. doi: 10.1016/j.cell.2021.01.005
González Moreno, A., de Cózar, A., Prieto, P., Domínguez, E., Heredia, A. (2022). Radiationless mechanism of UV deactivation by cuticle phenolics in plants. Nat. Commun. 13, 1786. doi: 10.1038/s41467-022-29460-9
Greer, S., Wen, M., Bird, D., Wu, X., Samuels, L., Kunst, L., et al. (2007). The cytochrome P450 enzyme CYP96A15 is the midchain alkane hydroxylase responsible for formation of secondary alcohols and ketones in stem cuticular wax of Arabidopsis. Plant Physiol. 145, 653–667. doi: 10.1104/pp.107.107300
Haas, M., Schreiber, M., Mascher, M. (2019). Domestication and crop evolution of wheat and barley: Genes, genomics, and future directions. J. Integr. Plant Biol. 61, 204–225. doi: 10.1111/jipb.12737
Hansjakob, A., Bischof, S., Bringmann, G., Riederer, M., Hildebrandt, U. (2010). Very-long-chain aldehydes promote in vitro prepenetration processes of blumeria graminis in a dose- and chain length-dependent manner. New Phytol. 188, 1039–1054. doi: 10.1111/j.1469-8137.2010.03419.x
Haslam, T. M., Haslam, R., Thoraval, D., Pascal, S., Delude, C., Domergue, F., et al. (2015). ECERIFERUM2-LIKE proteins have unique biochemical and physiological functions in very-long-chain fatty acid elongation. Plant Physiol. 167, 682–692. doi: 10.1104/pp.114.253195
Haslam, T. M., Kunst, L. (2021). Arabidopsis ECERIFERUM2-LIKEs are mediators of condensing enzyme function. Plant Cell Physiol. 61, 2126–2138. doi: 10.1093/pcp/pcaa133
He, J., Li, C., Hu, N., Zhu, Y., He, Z., Sun, Y., et al. (2022). ECERIFERUM1-6A is required for the synthesis of cuticular wax alkanes and promotes drought tolerance in wheat. Plant Physiol. doi: 10.1093/plphys/kiac394
Hen-Avivi, S., Savin, O., Racovita, R. C., Lee, W. S., Adamski, N. M., Malitsky, S., et al. (2016). A metabolic gene cluster in the wheat W1 and the barley cer-cqu loci determines β-diketone biosynthesis and glaucousness. Plant Cell 28, 1440–1460. doi: 10.1105/tpc.16.00197
Hensel, G. (2020). Genetic transformation of triticeae cereals - summary of almost three- decade's development. Biotechnol. Adv. 40, 107484. doi: 10.1016/j.biotechadv.2019.107484
Hickey, L. T., Hafeez, A, N., Robinson, H., Jackson, S. A., Leal-Bertioli, S., Tester, M., et al. (2019). Breeding crops to feed 10 billion. Nat. Biotechnol. 37, 744–754. doi: 10.1038/s41587-019-0152-9
Holme, I. B., Gregersen, P. L., Brinch-Pedersen, H. (2019). Induced genetic variation in crop plants by random or targeted mutagenesis: convergence and differences. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01468
Hong, L., Brown, J., Segerson, N. A., Rose, J. K., Roeder, A. H. (2017). CUTIN SYNTHASE 2 maintains progressively developing cuticular ridges in Arabidopsis sepals. Mol. Plant 10, 560–574. doi: 10.1016/j.molp.2017.01.002
Hundleby, P., D'Aoust, M. A., Finkle, C., Atkins, J., Twyman, R. M. (2022). Regulation of molecular farming products. Methods Mol. Biol. 2480, 313–333. doi: 10.1007/978-1-0716-2241-4_17
Hura, T. (2020). Wheat and barley: acclimatization to abiotic and biotic stress. Int. J. Mol. Sci. 21, 7423. doi: 10.3390/ijms21197423
Ichino, T., Yazaki, K. (2022). Modes of secretion of plant lipophilic metabolites via ABCG transporter-dependent transport and vesicle-mediated trafficking. Curr. Opin. Plant Biol. 66, 102184. doi: 10.1016/j.pbi.2022.102184
Ingram, G., Nawrath, C. (2017). The roles of the cuticle in plant development: organ adhesions and beyond. J. Exp. Bot. 68, 5307–5321. doi: 10.1093/jxb/erx313
Jacott, C. N., Boden, S. A. (2020). Feeling the heat: developmental and molecular responses of wheat and barley to high ambient temperatures. J. Exp. Bot. 1, 5740–5751. doi: 10.1093/jxb/eraa326
Kan, Y., Mu, X. R., Zhang, H., Gao, J., Shan, J. X., Ye, W. W., et al. (2022). TT2 controls rice thermotolerance through SCT1-dependent alteration of wax biosynthesis. Nat. Plants 8, 53–67. doi: 10.1038/s41477-021-01039-0
Kannangara, R., Branigan, C., Liu, Y., Penfield, T., Rao, V., Mouille, G., et al. (2007). The transcription factor WIN1/SHN1 regulates cutin biosynthesis in Arabidopsis thaliana. Plant Cell 19, 1278–1294. doi: 10.1105/tpc.106.047076
Kim, J., Kim, R. J., Lee, S. B., Chung Suh, M. (2022). Protein-protein interactions in fatty acid elongase complexes are important for very-long-chain fatty acid synthesis. J. Exp. Bot. 73, 3004–3017. doi: 10.1093/jxb/erab543
Kim, R. J., Lee, S. B., Pandey, G., Suh, M. C. (2022). Functional conservation of an AP2/ERF transcription factor in cuticle formation suggests an important role in the terrestrialization of early land plants. J. Exp. Bot. doi: 10.1093/jxb/erac360
Kong, L., Chang, C. (2018). Suppression of wheat TaCDK8/TaWIN1 interaction negatively affects germination of Blumeria graminis f.sp. tritici by interfering with very-long-chain aldehyde biosynthesis. Plant Mol. Biol. 96, 165–178. doi: 10.1007/s11103-017-0687-4
Kong, L., Liu, Y., Zhi, P., Wang, X., Xu, B., Gong, Z., et al. (2020a). Origins and evolution of cuticle biosynthetic machinery in land plants. Plant Physiol. 184, 1998–2010. doi: 10.1104/pp.20.00913
Kong, L., Zhi, P., Liu, J., Li, H., Zhang, X., Xu, J., et al. (2020b). Epigenetic activation of Enoyl-CoA reductase by an acetyltransferase complex triggers wheat wax biosynthesis. Plant Physiol. 183, 1250–1267. doi: 10.1104/pp.20.00603
Kosma, D. K., Nemacheck, J. A., Jenks, M. A., Williams, C. E. (2010). Changes in properties of wheat leaf cuticle during interactions with Hessian fly. Plant J. 63, 31–43. doi: 10.1111/j.1365-313X.2010.04229.x
Kosma, D. K., Rowland, O. (2016). Answering a four decade-old question on epicuticular wax biosynthesis. J. Exp. Bot. 67, 2538–2540. doi: 10.1093/jxb/erw144
Kumar, V., Thakur, J. K., Prasad, M. (2021). Histone acetylation dynamics regulating plant development and stress responses. Cell Mol. Life Sci. 78, 4467–4486. doi: 10.1007/s00018-021-03794-x
Kumar, A., Yogendra, K. N., Karre, S., Kushalappa, A. C., Dion, Y., Choo, T. M. (2016). WAX INDUCER1 (HvWIN1) transcription factor regulates free fatty acid biosynthetic genes to reinforce cuticle to resist Fusarium head blight in barley spikelets. J. Exp. Bot. 67, 4127–4139. doi: 10.1093/jxb/erw187
Kurdyukov, S., Faust, A., Nawrath, C., Bär, S., Voisin, D., Efremova, N., et al. (2006). The epidermis-specific extracellular BODYGUARD controls cuticle development and morphogenesis in Arabidopsis. Plant Cell 18, 321–339. doi: 10.1105/tpc.105.036079
Kurowska, M., Daszkowska-Golec, A., Gruszka, D., Marzec, M., Szurman, M., Szarejko, I., et al. (2011). TILLING: a shortcut in functional genomics. J. Appl. Genet. 52, 371–390. doi: 10.1007/s13353-011-0061-1
Lee, R. (2011). The outlook for population growth. Science 333, 569–573. doi: 10.1126/science.1208859
Lee, S., Fu, F., Xu, S., Lee, S. Y., Yun, D. J., Mengiste, T. (2016). Global regulation of plant immunity by histone lysine methyl transferases. Plant Cell 28, 1640–1661. doi: 10.1105/tpc.16.00012
Lee, S. B., Kim, H., Kim, R. J., Suh, M. C. (2014). Overexpression of arabidopsis MYB96 confers drought resistance in Camelina sativa via cuticular wax accumulation. Plant Cell Rep. 33, 1535–1546. doi: 10.1007/s00299-014-1636-1
Lee, S. B., Suh, M. C. (2015). Advances in the understanding of cuticular waxes in Arabidopsis thaliana and crop species. Plant Cell Rep. 34, 557–572. doi: 10.1007/s00299-015-1772-2
Lee, S. B., Suh, M. C. (2022). Regulatory mechanisms underlying cuticular wax biosynthesis. J. Exp. Bot. 73, 2799–2816. doi: 10.1093/jxb/erab509
Lee, S. B., Yang, S. U., Pandey, G., Kim, M. S., Hyoung, S., Choi, D., et al. (2020). Occurrence of land-plant-specific glycerol-3-phosphate acyltransferases is essential for cuticle formation and gametophore development in physcomitrella patens. New Phytol. 225, 2468–2483. doi: 10.1111/nph.16311
Levy, A. A., Feldman, M. (2022). Evolution and origin of bread wheat. Plant Cell 4, 2549–2567. doi: 10.1093/plcell/koac130
Lewandowska, M., Keyl, A., Feussner, I. (2020). Wax biosynthesis in response to danger: its regulation upon abiotic and biotic stress. New Phytol. 227, 698–713. doi: 10.1111/nph.16571
Li, M., Yang, Z., Chang, C. (2022). Susceptibility is new resistance: wheat susceptibility genes and exploitation in resistance breeding. Agriculture 12, 1419. doi: 10.3390/agriculture12091419
Li, H., Chang, C. (2021). Evolutionary insight of plant cuticle biosynthesis in bryophytes. Plant Signal. Behav. 16, 1943921. doi: 10.1080/15592324.2021.1943921
Li, C., Chen, G., Mishina, K., Yamaji, N., Ma, J. F., Yukuhiro, F., et al. (2017). A GDSL-motif esterase/acyltransferase/lipase is responsible for leaf water retention in barley. Plant Direct. 1, e00025. doi: 10.1002/pld3.25
Li, C., Haslam, T. M., Krüger, A., Schneider, L. M., Mishina, K., Samuels, L., et al. (2018). The β-Ketoacyl-CoA synthase HvKCS1, encoded by cer-zh, plays a key role in synthesis of barley leaf wax and germination of barley powdery mildew. Plant Cell Physiol. 59, 806–822. doi: 10.1093/pcp/pcy020
Li, T., Hu, J., Sun, Y., Li, B., Zhang, D., Li, W., et al. (2021). Highly efficient heritable genome editing in wheat using an RNA virus and bypassing tissue culture. Mol. Plant 14, 1787–1798. doi: 10.1016/j.molp.2021.07.010
Li, T., Sun, Y., Liu, T., Wu, H., An, P., Shui, Z., et al. (2019). TaCER1-1A is involved in cuticular wax alkane biosynthesis in hexaploid wheat and responds to plant abiotic stresses. Plant Cell Environ. 42, 3077–3091. doi: 10.1111/pce.13614
Li, S., Wang, X., He, S., Li, J., Huang, Q., Imaizumi, T., et al. (2016). CFLAP1 and CFLAP2 are two bHLH transcription factors participating in synergistic regulation of AtCFL1-mediated cuticle development in Arabidopsis. PloS Genet. 12, e1005744. doi: 10.1371/journal.pgen.1005744
Li, C., Liu, C., Ma, X., Wang, A., Duan, R., Nawrath, C., et al. (2015). Characterization and genetic mapping of eceriferum-ym (cer-ym), a cutin deficient barley mutant with impaired leaf water retention capacity. Breed. Sci. 65, 327–332. doi: 10.1270/jsbbs.65.327
Li, F., Wu, X., Lam, P., Bird, D., Zheng, H., Samuels, L., et al. (2008). Identification of the wax ester synthase/acyl-coenzyme a: diacylglycerol acyltransferase WSD1 required for stem wax ester biosynthesis in. Arabidopsis. Plant Physiol. 148, 97–107. doi: 10.1104/pp.108.123471
Ling, F., Zhou, F., Chen, H., Lin, Y. (2016). Development of marker-free insect-resistant indica rice by Agrobacterium tumefaciens-mediated co-transformation. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01608
Liu, X., Feakins, S. J., Ma, X. F., Anderson, J. D., Vidal, E., Blancaflor, E. B. (2019). Crop breeding has increased the productivity and leaf wax n-alkane concentration in a series of five winter wheat cultivars developed over the last 60 years. J. Plant Physiol. 243, 153056. doi: 10.1016/j.jplph.2019.153056
Liu, Q., Huang, H., Chen, Y., Yue, Z., Wang, Z., Qu, T., et al. (2022c). Two arabidopsis MYB-SHAQKYF transcription repressors regulate leaf wax biosynthesis via transcriptional suppression on DEWAX. New Phytol. doi: 10.1111/nph.18498
Liu, L., Jose, S. B., Campoli, C., Bayer, M. M., Sánchez-Diaz, M. A., McAllister, T., et al. (2022a). Conserved signalling components coordinate epidermal patterning and cuticle deposition in barley. Nat. Commun. 13, 6050. doi: 10.1038/s41467-022-33300-1
Liu, L., Wang, X., Chang, C. (2022b). Toward a smart skin: Harnessing cuticle biosynthesis for crop adaptation to drought, salinity, temperature, and ultraviolet stress. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.961829
Manghwar, H., Lindsey, K., Zhang, X., Jin, S. (2019). CRISPR/Cas system: recent advances and future prospects for genome editing. Trends Plant Sci. 24, 1102–1125. doi: 10.1016/j.tplants
Mao, X., Weake, V. M., Chapple, C. (2019). Mediator function in plant metabolism revealed by large-scale biology. J. Exp. Bot. 70, 5995–6003. doi: 10.1093/jxb/erz372
McCallum, C. M., Comai, L., Greene, E. A., Henikoff, S. (2000). Targeting induced local lesions IN genomes (TILLING) for plant functional genomics. Plant Physiol. 123, 439–442. doi: 10.1104/pp.123.2.439
Ménard, R., Verdier, G., Ors, M., Erhardt, M., Beisson, F., Shen, W. H. (2014). Histone H2B monoubiquitination is involved in the regulation of cutin and wax composition in Arabidopsis thaliana. Plant Cell Physiol. 55, 455–466. doi: 10.1093/pcp/pct182
Mirzaee, M., Osmani, Z., Frébortová, J., Frébort, I. (2022). Recent advances in molecular farming using monocot plants. Biotechnol. Adv. 58, 107913. doi: 10.1016/j.biotechadv.2022.107913
Mrízová, K., Holasková, E., Öz, M. T., Jiskrová, E., Frébort, I., Galuszka, P. (2014). Transgenic barley: a prospective tool for biotechnology and agriculture. Biotechnol. Adv. 32, 137–157. doi: 10.1016/j.biotechadv.2013.09.011
Pascal, S., Bernard, A., Deslous, P., Gronnier, J., Fournier-Goss, A., Domergue, F., et al. (2019). Arabidopsis CER1-LIKE1 functions in a cuticular very-long-chain alkane-forming complex. Plant Physiol. 179, 415–432. doi: 10.1104/pp.18.01075
Patwari, P., Salewski, V., Gutbrod, K., Kreszies, T., Dresen-Scholz, B., Peisker, H., et al. (2019). Surface wax esters contribute to drought tolerance in Arabidopsis. Plant J. 98, 727–744. doi: 10.1111/tpj.14269
Petit, J., Bres, C., Mauxion, J. P., Bakan, B., Rothan, C. (2017). Breeding for cuticle- associated traits in crop species: traits, targets, and strategies. J. Exp. Bot. 68, 5369–5387. doi: 10.1093/jxb/erx341
Philippe, G., De Bellis, D., Rose, J., Nawrath, C. (2022). Trafficking processes and secretion pathways underlying the formation of plant cuticles. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.786874
Philippe, G., Sørensen, I., Jiao, C., Sun, X., Fei, Z., Domozych, D. S., et al. (2020). Cutin and suberin: assembly and origins of specialized lipidic cell wall scaffolds. Curr. Opin. Plant Biol. 55, 11–20. doi: 10.1016/j.pbi.2020.01.008
Pighin, J. A., Zheng, H., Balakshin, L. J., Goodman, I. P., Western, T. L., Jetter, R., et al. (2004). Plant cuticular lipid export requires an ABC transporter. Science 306, 702–704. doi: 10.1126/science.1102331
Redden, R. (2021). Genetic modification for agriculture-proposed revision of GMO regulation in Australia. Plants 10, 747. doi: 10.3390/plants10040747
Reynoud, N., Petit, J., Bres, C., Lahaye, M., Rothan, C., Marion, D., et al. (2021). The complex architecture of plant cuticles and its relation to multiple biological functions. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.782773
Rowland, O., Zheng, H., Hepworth, S. R., Lam, P., Jetter, R., Kunst, L. (2006). CER4 encodes an alcohol-forming fatty acyl-coenzyme a reductase involved in cuticular wax production in Arabidopsis. Plant Physiol. 142, 866–877. doi: 10.1104/pp.106.086785
Samarah, H. N. (2005). Effects of drought stress on growth and yield of barley. Agron. Sustain. Dev. 25, 145–149. doi: 10.1051/agro:2004064
Savary, S., Willocquet, L., Pethybridge, S. J., Esker, P., McRoberts, N., Nelson, A. (2019). The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439. doi: 10.1038/s41559-018-0793-y
Schneider, L. M., Adamski, N. M., Christensen, C. E., Stuart, D. B., Vautrin, S., Hansson, M., et al. (2016). The Cer-cqu gene cluster determines three key players in a β-diketone synthase polyketide pathway synthesizing aliphatics in epicuticular waxes. J. Exp. Bot. 67, 2715–2730. doi: 10.1016/j.jplph.2020.153351
Thudi, M., Palakurthi, R., Schnable, J. C., Chitikineni, A., Dreisigacker, S., Mace, E., et al. (2021). Genomic resources in plant breeding for sustainable agriculture. J Plant Physiol. 257, 153351. doi: 10.1016/j.jplph.2020.153351
Todd, J., Post-Beittenmiller, D., Jaworski, J. G. (1999). KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J. 17, 119–130. doi: 10.1046/j.1365-313x.1999.00352.x
von Wettstein-Knowles, P. (1972). Genetic control of β-diketone and hydroxy-β-diketone synthesis in epicuticular waxes of barley. Planta 106, 113–130. doi: 10.1007/BF00383991
von Wettstein-Knowles, P. (2017). The polyketide components of waxes and the cer-cqu gene cluster encoding a novel polyketide synthase, the β-diketone synthase, DKS. Plants (Basel). 6. doi: 10.3389/fpls.2017.00621
Wang, X., Kong, L., Zhi, P., Chang, C. (2020). Update on cuticular wax biosynthesis and its roles in plant disease resistance. Int. J. Mol. Sci. 21, 5514. doi: 10.3390/ijms21155514
Wang, K., Liu, H., Du, L., Ye, X. (2017). Generation of marker-free transgenic hexaploid wheat via an Agrobacterium-mediated co-transformation strategy in commercial Chinese wheat varieties. Plant Biotechnol. J. 15, 614–623. doi: 10.1111/pbi.12660
Wang, K., Shi, L., Liang, X., Zhao, P., Wang, W., Liu, J., et al. (2022). The gene TaWOX5 overcomes genotype dependency in wheat genetic transformation. Nat. Plants 8, 110–117. doi: 10.1038/s41477-021-01085-8
Wang, Y., Wang, M., Sun, Y., Wang, Y., Li, T., Chai, G., et al. (2015). FAR5, a fatty acyl-coenzyme a reductase, is involved in primary alcohol biosynthesis of the leaf blade cuticular wax in wheat (Triticum aestivum l.). J. Exp. Bot. 66, 1165–1178. doi: 10.1093/jxb/eru457
Wang, M., Wang, Y., Wu, H., Xu, J., Li, T., Hegebarth, D., et al. (2016). Three TaFAR genes function in the biosynthesis of primary alcohols and the response to abiotic stresses in Triticum aestivum. Sci. Rep. 6, 25008. doi: 10.1038/srep25008
Wang, T., Xing, J., Liu, X., Yao, Y., Hu, Z., Peng, H., et al. (2018). GCN5 contributes to stem cuticular wax biosynthesis by histone acetylation of CER3 in Arabidopsis. J. Exp. Bot. 69, 2911–2922. doi: 10.1093/jxb/ery077
Wang, G. P., Yu, X. D., Sun, Y. W., Jones, H. D., Xia, L. Q. (2016). Generation of marker- and/or backbone-free transgenic wheat plants via agrobacterium-mediated transformation. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01324
Wang, X., Zhi, P., Fan, Q., Zhang, M., Chang, C. (2019). Wheat CHD3 protein TaCHR729 regulates the cuticular wax biosynthesis required for stimulating germination of Blumeria graminis f.sp. tritici. J. Exp. Bot. 70, 701–713. doi: 10.1093/jxb/ery377
Weidenbach, D., Jansen, M., Franke, R. B., Hensel, G., Weissgerber, W., Ulferts, S., et al. (2014). Evolutionary conserved function of barley and arabidopsis 3-KETOACYL-CoA SYNTHASES in providing wax signals for germination of powdery mildew fungi. Plant Physiol. 166, 1621–1633. doi: 10.1104/pp.114.246348
Wijerathna-Yapa, A., Ramtekey, V., Ranawaka, B., Basnet, B. R. (2022). Applications of in vitro tissue culture technologies in breeding and genetic improvement of wheat. Plants 11, 2273. doi: 10.3390/plants11172273
Wolt, J. D., Wolf, C. (2018). Policy and governance perspectives for regulation of genome edited crops in the united states. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01606
Xue, D., Zhang, X., Lu, X., Chen, G., and Chen, Z. H (2017). Molecular and evolutionary mechanisms of cuticular wax for plant drought tolerance. Front. Plant Sci. 8, 621. doi: 10.3389/fpls.2017.00621
Yang, X., Zhao, H., Kosma, D. K., Tomasi, P., Dyer, J. M., Li, R., et al. (2017). The acyl desaturase CER17 is involved in producing wax unsaturated primary alcohols and cutin monomers. Plant Physiol. 173, 1109–1124. doi: 10.1104/pp.16.01956
Yeats, T. H., Rose, J. K. (2013). The formation and function of plant cuticles. Plant Physiol. 163, 5–20. doi: 10.1104/pp.113.222737
Zhang, P., Wang, R., Yang, X., Ju, Q., Li, W., Lü, S., et al. (2020). The R2R3-MYB transcription factor AtMYB49 modulates salt tolerance in Arabidopsis by modulating the cuticle formation and antioxidant defence. Plant Cell Environ. 43, 1925–1943. doi: 10.1111/pce.13784
Zhi, P., Chang, C. (2021). Exploiting epigenetic variations for crop disease resistance improvement. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.692328
Zhu, H., Li, C., Gao, C. (2020). Applications of CRISPR-cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 21, 661–677. doi: 10.1038/s41580-020-00288-9
Zhu, Y., Schluttenhoffer, C. M., Wang, P., Fu, F., Thimmapuram, J., Zhu, J. K., et al. (2014). CYCLIN-DEPENDENT KINASE8 differentially regulates plant immunity to fungal pathogens through kinase-dependent and -independent functions in Arabidopsis. Plant Cell 26, 4149–4170. doi: 10.1105/tpc.114.128611
Keywords: wheat, barley, breeding, cutin, wax, cuticle, abiotic stress, biotic stress
Citation: Wang X and Chang C (2022) Exploring and exploiting cuticle biosynthesis for abiotic and biotic stress tolerance in wheat and barley. Front. Plant Sci. 13:1064390. doi: 10.3389/fpls.2022.1064390
Received: 08 October 2022; Accepted: 24 October 2022;
Published: 10 November 2022.
Edited by:
Laura Siracusa, National Research Council (CNR), ItalyReviewed by:
Shiyou Lu, Hubei University, ChinaCopyright © 2022 Wang and Chang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cheng Chang, Y2NAcWR1LmVkdS5jbg==
 Xiaoyu Wang
Xiaoyu Wang Cheng Chang
Cheng Chang