- 1College of Agro-Grassland Science, Nanjing Agricultural University, Nanjing, China
- 2Institute of Botany, Jiangsu Province and Chinese Academy of Sciences, Nanjing, China
- 3Department of Horticulture, Oregon State University, Corvallis, OR, United States
Zoysia matrella is a salt-tolerant turfgrass grown in areas with high soil salinity irrigated with effluent water. Previous studies focused on explaining the regulatory mechanism of Z. matrella salt-tolerance at phenotypic and physiological levels. However, the molecular mechanism associated with salt tolerance of Z. matrella remained unclear. In this study, a high-efficient method named FOX (full-length cDNA overexpression) hunting system was used to search for salt-tolerant genes in Z. matrella. Eleven candidate genes, including several known or novel salt-tolerant genes involved in different metabolism pathways, were identified. These genes exhibited inducible expression under salt stress condition. Furthermore, a novel salt-inducible candidate gene ZmGnTL was transformed into Arabidopsis for functional analysis. ZmGnTL improved salt-tolerance through regulating ion homeostasis, reactive oxygen species scavenging, and osmotic adjustment. In summary, we demonstrated that FOX is a reliable system for discovering novel genes relevant to salt tolerance and several candidate genes were identified from Z. matrella that can assist molecular breeding for plant salt-tolerance improvement.
Introduction
Soil salinization has been an adverse environmental factor restricting plant growth and development, as well as limiting plant production and quality (Katerji et al., 2003; Zhang et al., 2013; Zhang and Shi, 2013; Zhang et al., 2022). Therefore, improving plant salt-tolerance will be crucial for crop production in large saline regions. Understanding the physiological and molecular mechanisms are beneficial for plant adaption to salt stress (Van Zelm et al., 2020). Over the past two decades, scientists have described several regulatory pathways related to plant salt tolerance, including ion balance (Ji et al., 2013; Benito et al., 2014; Almeida et al., 2017), osmotic adjustment (Flowers et al., 2015; Slama et al., 2015), and reactive oxygen species (ROS) degradation (Yang and Guo, 2018a; Yang and Guo, 2018b). These pathways have been further verified through gene function analysis, such as Salt Overly Sensitive 1-3 (SOS1-3) involved in Na+ exclusion and ion homeostasis control in many plant species (Zhu, 2000; Shi et al., 2003; Han et al., 2022), antioxidant enzymes GhSOD1 and GhCAT1 in cotton (Luo et al., 2013), and P5CS related to proline metabolism conferring salt-inducible osmotic adjustment in rice (Igarashi et al., 1997), etc. As described above, most of these salt-tolerant genes were identified from model plants or crop species, very few salt-tolerant genes have been explored from halophytes that adapt to higher salinity.
Halophytes, such as non-salt secreting type Thellungiella halophile, Salicornia brachiate, Suaeda corniculate (Mishra and Tanna, 2017), Suaeda maritima (Zhang et al., 2013) and Puccinellia tenuiflora (Han et al., 2022), and salt secreting type Limonium bicolor (Gao et al., 2021), Avicennia officinalis (Jyothi-Prakash et al., 2014) and Zoysia matrella (Chen et al., 2015), can survive from high salinity conditions with some of them even directly irrigated with saline water. Systematic screening for salt-tolerant genes from halophytes will provide valuable information for explaining the underlying molecular mechanism of their salt tolerance. Several methods such as DNA seq, RNA seq, proteomics, and metabonomics have been successfully applied for gene mining in plants (Chen et al., 2015; Yuan et al., 2015). Genes involved in the special ultrastructure of salt glands were discovered in L. bicolor through RNA seq method (Yuan et al., 2015), which has been a primary approach in most non-model plants. However, functional genes can change in post-transcriptional level which could not be detected by RNA seq. FOX system as a gain-of-function method using heterologous overexpression of full-length cDNA libraries in model plant Arabidopsis has been successfully applied for gene mining in Arabidopsis and rice (Ichikawa et al., 2006; Higuchi et al., 2011; Higuchi-Takeuchi and Matsui, 2014). For example, TsHsfA1d from Thellungiella salsuginea identified via FOX hunting system functioned as a positive regulator of heat stress response in Arabidopsis (Higashi et al., 2013). Overexpressing rice OsREX1-S screened through FOX was confirmed to enhance tolerance of host plants to cadmium (Kunihiro et al., 2014). OsCPK21 cloned from full-length cDNA overexpressed rice was involved in the positive regulation in response to abscisic acid (ABA) and salt stress (Asano et al., 2011).
Halophyte Zoysia matrella is an excellent warm season turfgrass that can growth in saline soils. Our previous research mainly focused on the salt tolerance evaluation and physiological responses of Z. matrella to salinity. Whereas the molecular mechanism of its salt tolerance is still unclear. Moreover, we identified several potential salt-genes from Z. matrella through yeast-based FOX system (Chen et al., 2015). In current study, we aimed to construct the Arabidopsis-based FOX system for further screening candidate salt-tolerant genes for future molecular breeding. In addition, we selected a novel salt-inducible candidate gene ZmGnTL (β-1,6-N-acetylglucosaminyltransferase like enzyme) for functional analysis.
Materials and methods
Full-length cDNA expression library construction of Z. matrella
The cDNA entry library plasmid was produced in our previous work (Chen et al., 2015). The library plasmid was inserted into plant expression vector pEarleyGate103 with recombination reaction system (Invitrogen, USA). The reaction products were transformed into ElectroMAX™ DH10B™ T1 competent cells by electroporation, and 50 μL of 100-fold diluted cells were plated on a LB solid medium containing 50 mg L-1 kanamycin. Kanamycin resistant bacterial colonies were counted after 12 h and transferred to 1 mL 50 mg L-1 kanamycin LB liquid medium for propagation. The recombination fragment size was estimated by polymerase chain reaction (PCR) with universal primer pair (TATCCTTCGCAAGACCCTTCCTCTA/GGTAAGTTTTCCGTATGTTGCATCA) of pEarleyGate103 vector.
Transformation of Arabidopsis, screening of salt-tolerant plants, and functional analysis of ZmGnTL
The expression library plasmid was transformed into Agrobacterium tumefaciens EHA105 competent cells by electroporation, and the transformed cells were grown in LB solid medium containing 50 mg L-1 kanamycin for three days. All kanamycin resistant colonies were collected and suspended in 5% sucrose solution plus 0.5% Silwet-L77, and then introduced into Arabidopsis thaliana accession Columbia (Col-0) via floral dip method. The T1 generation seeds were obtained and screened in Murashige and Skoog (MS) solid medium containing 25 mg L-1 ampicillin, 20 mg L-1 glufosinate ammonium and 150 mM NaCl. Salt-tolerant transgenic plants were selected and transplanted into the soil. Leaves of those transgenic plants were used for extracting DNA, and PCR was performed using universal primer pair (F1/R1: TATCCTTCGCAAGACCCTTCCTCTA/GGTAAGTTTTCCGTATGTTGCATCA) of pEarleyGate103 vector. Each PCR product was sequenced and BLASTX (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was performed using these DNA sequences to identify putative salt tolerant genes.
ZmGnTL gene was reamplified from the cDNA template of Z. matrella using ORF primer pair (ccggtcgacATGACGTCACCGGCGCCGGCGTACA/agtgaattcgtGTCACGTAGGATGACCGAGTCCGC) and then inserted into expression vector pEarleyGate103 with recombination reaction system. For further gene functional analysis, ZmGnTL was retransformed into Arabidopsis and the transgenic plants overexpressing ZmGnTL were screened following the methods of MS solid medium containing 20 mg L-1 glufosinate. gDNA-PCR and RT-PCR was detected using primer pair (F1/R1, shown in above). The seeds of WT and T3 generation homozygous lines were sterilized and planted in MS solid medium containing 0 or 120 mM NaCl, and the growth and biomass of seedlings were measured for salt tolerance analysis. In addition, the 18d old plants of WT and T3 generation transgenic Arabidopsis were treated with 150 mM and 200 mM NaCl in pots containing nutrient soil for 15d, and the phenotype was observed and physiological indexes were analyzed following the methods below.
Expression analysis of candidate salt-tolerant genes
Z. matrella were hydroponic planted in Hoagland nutrient solution and treated with 300 mM NaCl concentration. The leaves were collected at 0, 1, 3, 6, 24, 48 h and RNA were extracted using Trizol RNA Kit (Invitrogen, USA). For the reverse transcription reaction, 0.5 μg RNA was used in the reaction with PrimeScript RT reagent Kit and treated with gDNA Eraser (TaKaRa, USA). Primer pairs of candidate salt-tolerant genes and ZmACT (GenBank Number: GU290545) as a reference gene are displayed in Table 1. LightCycler 480 SYBR I master (Roche, Switzerland) was used for each 15 μL reaction, which contained 5 μL of diluted cDNA (60 ng/μl), 7.5 μL 2×SYBR I master, 0.4 μL each primer (10 μM), and 1.7 μL ddH2O. The qRT-PCR reactions were performed using a LightCycler 480 II (Roche, Switzerland) with the following cycling conditions: an initial denaturation at 95°C for 10 min, followed by 40 cycles of 95°C/15 s, 58°C/15 s and 72°C/30 s, and thereafter melting curves were produced at 60-95°C. Gene relative expression levels were measured by 2-ΔΔCt method. Each qRT-PCR analysis was performed in triplicate.
Measurement of electrolyte leakage (EL) and relative water content (RWC)
Electrolyte leakage (EL) of leaves was measured according to the method of Blum and Ebercon (1981). Briefly, about 0.2 g fresh leaves were weighed and placed into a 50 mL centrifuge tube containing 30 mL ultrapure water. The centrifuge tubes were agitated on a shaker for 24 h at room temperature, and the initial conductivity (C0) was measured with conductivity meter (Thermo, New York, USA). The tubes containing the same leaf tissue samples were then autoclaved at 121°C for 15 mins and agitated for another 24 h to measure the final conductivity (Cl). The EL was calculated as C0/Cl × l00%. About 0.2 g fresh leaves were collected to measure leaf fresh weight (FW), submerged in water for 12 h to measure leaf turgid weight (TW), then dried at 80°C for 72 h to measure leaf dry weight (DW). The RWC was calculated as (FW–DW)/(TW–DW) × 100%.
Ion content analysis
Ten days after salt stress treatment, about 0.1 g fresh leaf or root tissue of Arabidopsis was oven-dried at 80°C to a constant weight. The oven-dried tissue was then decomposed for 45 min at 160°C by a microwave (ETHOS ONE, Milestone, Italy) in a digestion solution of 3 mL 65% nitric acid. After that, the liquid was diluted to 30 mL with ultrapure water. The contents of K+ and Na+ were determined by an ICP-OES (Optima 8000, Perkin Elmer, USA).
Quantification of superoxide O2- and hydrogen peroxide (H2O2)
The production rate was measured according to Zhang et al. (2016). Briefly, leaf tissue (0.1 g) was frozen using liquid nitrogen and ground to powder, homogenized in 3 mL 65 mM phosphate buffer (PBS, pH 7.8), and centrifuged at 10000 × g for 15 min at 4°C. In the meantime, a 5:1 ratio of PBS (pH 7.8) and 10 mM hydroxylamine hydrochloride mixture was incubated at 25°C for 10 min. Then, 0.5 mL of the supernatant obtained from centrifugation was transferred to 0.5 mL mixture and incubated at 25°C for 20 min. After incubation, 1 mL 58 mM sulfonamides and 1 mL 7 mM naphthylamine were added to the mixture, and incubated at 25°C for another 20 min. Finally, 3 mL of chloroform was added to the reaction mixture, vortexed, then centrifuged at 10000 × g for 3 min. The absorbance was measured at 530 nm using a spectrophotometer (Spectronic Instruments, NY, USA). The production rate of was calculated according to the formula described by Elstner and Heupel (1976).
Each 0.5 g leaf tissue sample was frozen using liquid nitrogen and ground to powder, then 5 mL cold 0.1% trichloroacetic acid (TCA) was added and homogenized. The homogenate was centrifuged at 12000 × g for 15 min and 0.5 mL of the supernatant was mixed with 0.5 mL 10 mM PBS buffer (pH 7.0) and 1 mL 1 M KI. The mixture was incubated at 28°C for 15 min in dark. The absorbance was measured at 390 nm. The content of H2O2 was calculated based on a standard curve generated with known H2O2 concentrations.
The analysis of enzymatic antioxidant activity
The enzymatic antioxidant activities of superoxide dismutase (SOD), peroxidase (POD), catalase (CAT), and ascorbate peroxidase (APX) were quantified using the methods described by Zhang et al. (2016). About 0.3 g leaf tissue was frozen using liquid nitrogen, ground to powder, then homogenized in 3 mL precooled 50 mM PBS (pH 7.8) containing 1% polyvinylpyrrolidone (PVP) and 0.2 mM EDTA. Homogenates were centrifuged at 15000 × g at 4°C for 20 min, and the supernatant was saved for the following enzyme activity analyses. SOD activity was measured at 560 nm absorbance. It is defined as the amount of enzyme required to cause 50% inhibition of nitroblue tetrazolium chloride reduction (Meloni et al., 2003). POD activity was measured by determining guaiacol oxidation by H2O2 at 470 nm (Lacan and Baccou, 1998). CAT activity was measured by monitoring the disappearance of H2O2 at 240 nm (Maehly and Chance, 1954). APX activity was measured by the decrease in absorbance at 290 nm for 1 min (Nakano and Asada, 1981).
Quantification of proline, glycine betaine and soluble sugar
The proline content was measured according to the description of Abrahám et al. (2010). About 0.1 g leaf or root tissue was ground to powder using liquid nitrogen and then homogenized in 0.5 mL 3% sulfosalicylic acid. Homogenates were centrifuged at 15000 × g at room temperature for 5 min. The plant extract supernatant (100 μL) was transferred to a 1.5 mL centrifuge tube, mixed with 500 μL reaction mixture (3% sulfosalicylic acid: glacial acetic acid: Acidic ninhydrin=1: 2: 2), and incubated at 96°C for 60 min. After incubation, 1 mL toluene was added to the reaction mixture and vortexed for 20 s; the organic layer was transferred into a fresh tube after allowing the separation of the organic and water phases without disturbance for a minimum of 5 mins. The absorbance of those samples was measured at 520 nm, and proline concentration was determined by referencing to a standard curve.
The glycine betaine content was measured according to the description of Wu et al. (2018). Freeze-dried plant leaf or root (1.0 g) was ground and suspended in 25 mL 60% methanol. The extraction process was facilitated by treating the samples in an ultrasonic cleaner (Bilon, Shanghai, China) for 30 mins. Then samples were centrifuged at 15,000 × g for 8 min, and the supernatant were transferred to a new 25 mL volumetric flask and brought to volume with 60% methanol. Ten μL of each glycine betaine standard and sample was used for liquid chromatograph (Thermo fisher, Shanghai, USA) quantification at 192 nm wavelength. The glycine betaine content was determined by referencing to the standard curve.
Freeze-dried plant materials (50 mg leaf or root) were homogenized in 5 mL 80% alcohol, incubated at 30°C for 30 mins, and then centrifuged at 4500 × g at 20°C for 10 mins. The supernatant was transferred to a 50 mL centrifuge tube, mixed with 2.5 mL 80% alcohol, incubated at 30°C for 30 mins, and then centrifuged at 4500 × g at 20°C for 10 mins. This extraction step was repeated. The supernatant was transferred to a 25 mL volumetric flask and brought to volume by purified water. Then, 1 mL from the 25 mL volumetric flask was transferred to a fresh glass tube and mixed with 1 mL 23% phenol, subsequently, 5 mL 98% sulfuric acid was added and homogenized. After 15 mins, the reaction solution was cooled down to room temperature and then incubated at 30°C for 30 mins. Finally, the absorbance was measured at 490 nm, and each soluble sugar concentration was determined by referencing to its standard curve (glucose, fructose, or sucrose); and soluble sugar concentration was the sum of the three sugar contents.
Results
cDNA expression library
To evaluate the cDNA library quality, bacteria containing the library plasmid were cultured overnight on solid medium (LB + 50 mg L-1 kanamycin) resulting in 520 colonies on a plate (Figure 1A). Subsequently, 24 colonies were randomly selected for PCR to determine the inserted fragment size, and the result showed that the recombination rate was 100% and the average fragment size was 1.64 kb (Figure 1B).
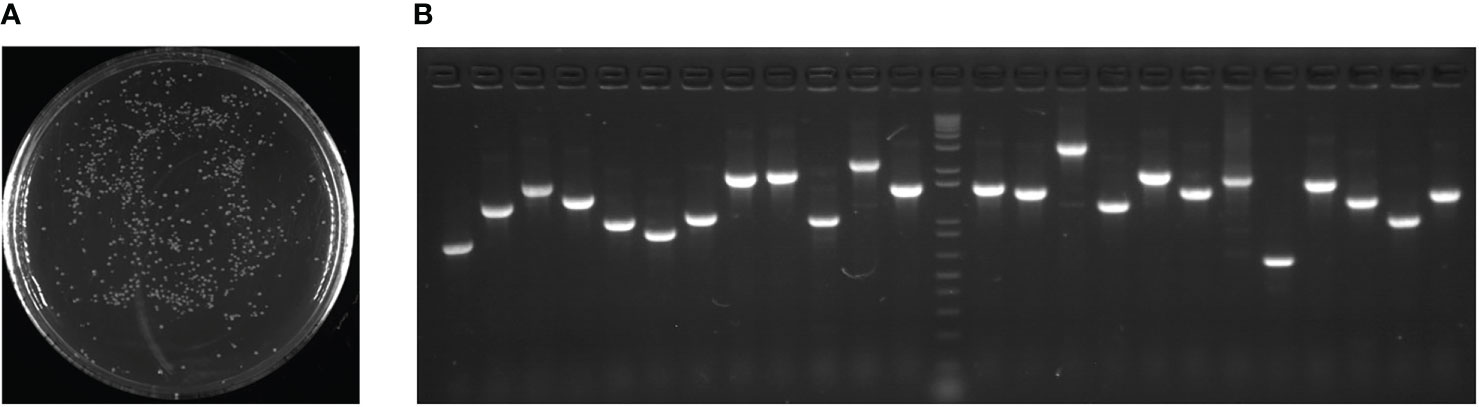
Figure 1 cDNA expression library quality assays. (A) 100-fold diluted bacteria with transformed cDNA library plasmid were cultured overnight on solid medium (LB + 50 mg L-1 kanamycin). (B) Twenty-four colonies were randomly selected for PCR to determine the size of the inserted fragment.
Salt-tolerant screening and gene mining of Arabidopsis
The constructed expression library plasmid was transformed into Arabidopsis by floral dip method, and around 5000 T1 transgenic seedlings were obtained. The harvested seeds were used to screen salt tolerant seedlings in NaCl solid medium (MS + 20 mg L-1 Basta +150 mM NaCl). Finally, 25 T2 salt-tolerant plants were obtained (Figure 2). We extracted the DNA from these T2 transgenic lines for PCR and identified 11 candidate salt-tolerance genes by sequencing (Table 2).
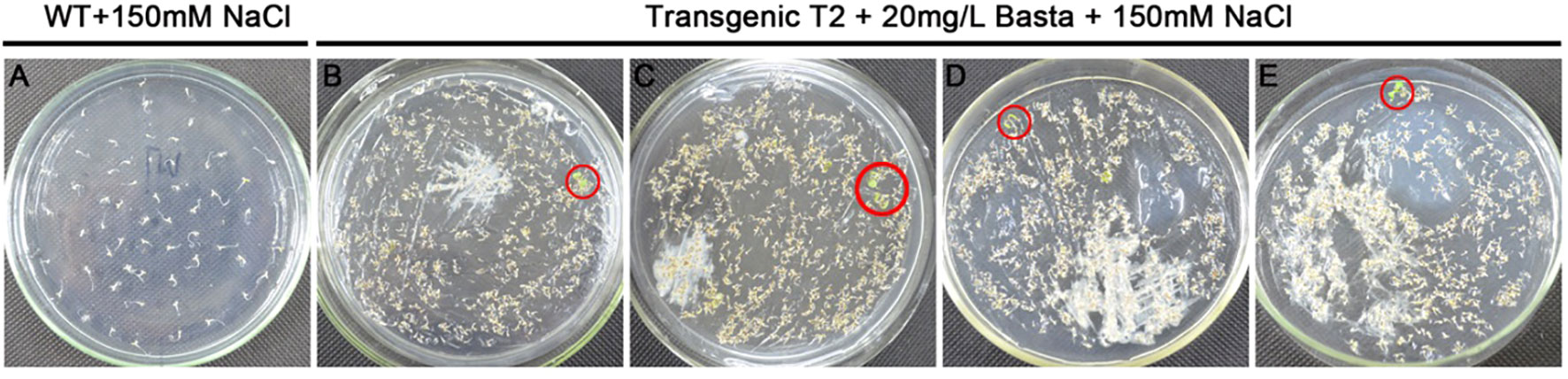
Figure 2 Screening of salt-tolerant Arabidopsis on MS plates. (A) Germination was completely inhibited with wildtype (WT) at 150 mM NaCl. (B–E) Screening transgenic lines on 20 mg L-1 Basta +150 mM NaCl MS plates, and salt-tolerant seedlings were circled in red.
Analysis of the expression of salt-tolerant genes with qRT-PCR
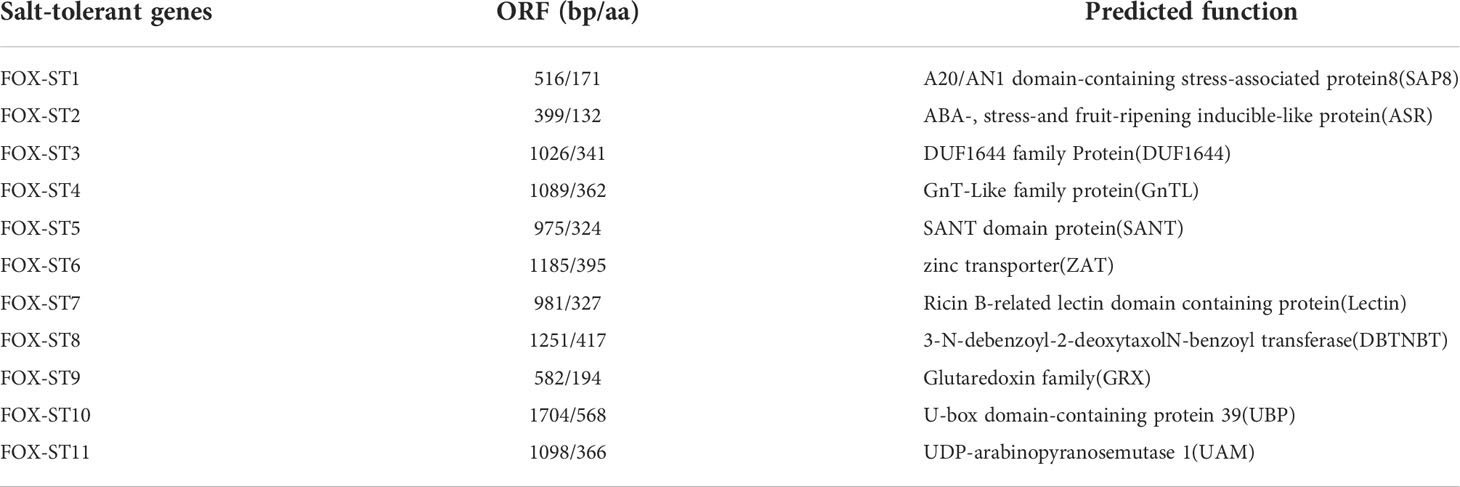
We analyzed the expression pattern of 11 candidate salt-tolerance genes response to salt stress in Z. matrella by qRT-PCR. The result showed that salt stress induced the gene expression of ZmSAP8, ZmASR, ZmDUF1644, ZmGnTL, ZmSANT, ZmZAT, ZmLectin, ZmGRX, and ZmUBP, whereas the expression of ZmDBTNBT and ZmUAM remained relatively stable (Figure 3). The expression level of ZmSAP8, ZmASR, ZmDUF1644, ZmGnTL, ZmGRX, and ZmUBP peaked at 6 h after salt stress, while the expression of ZmSANT and ZmZAT peaked at 24 h (Figure 3). ZmLectin significantly increased in expression after 1 h of salt stress, and highest expression level was observed at 48 h (Figure 3). Interestingly, the relative expression of ZmGnTL declined at 24 h after reaching the highest expression at 6 h and increased again at 48 h (Figure 3).
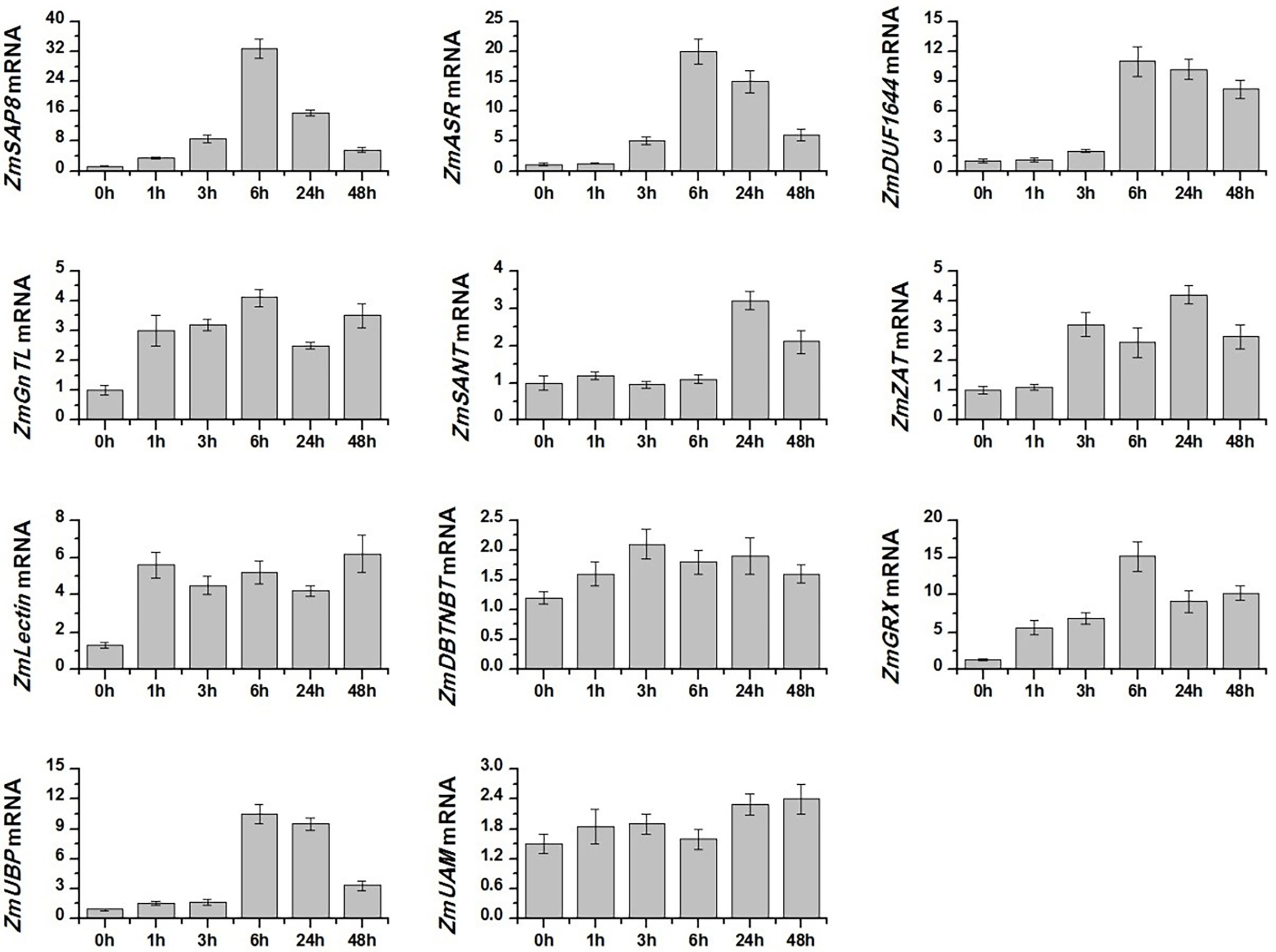
Figure 3 Relative expression levels of candidate salt-tolerant genes of Zoysia matrella treated with 300 mM NaCl. Data are shown as means ± SE of five biological replicates.
Sequence analysis of ZmGnTL
To elucidate the potential role of ZmGnTL in the abiotic stress response of Arabidopsis, we cloned the sequence of ZmGnTL. The sequence length of ZmGnTL is 1089 bp, encoding 362 amino acids. Clustering analysis with related genes from rice and Arabidopsis suggested that those gene sequences can be divided into three groups (Group I, II, and III). In Group III, ZmGnTL was shown to have high sequence homology with Os03g44580 and AtGnTL (AT3G52060) (Figure 4A). A multiple sequence alignment revealed that ZmGnTL contained an amino-terminal signal peptide and a catalytic domain (GnT), which is important for glycosylation. In addition, Pfam database (http://pfam.xfam.org) analysis revealed that a conserved acid Glu279 site in the GnT domain may be critical for its activity (Figures 4B, C).
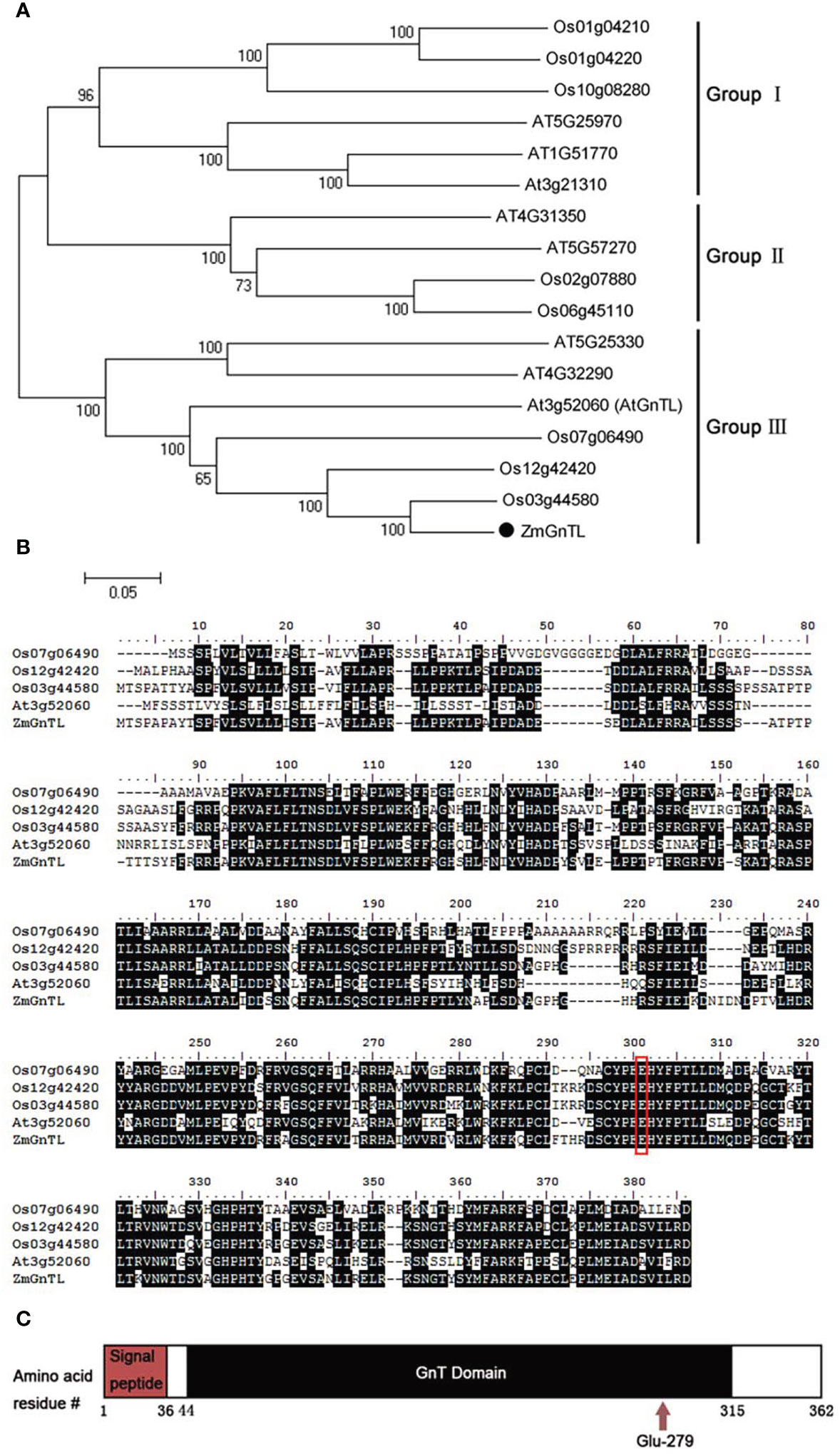
Figure 4 Sequence analysis of ZmGnTL. (A) Clustering analysis with related genes from rice and Arabidopsis. (B) Multiple sequence alignment analysis. (C) Pfam database predicted conserved domain.
Overexpression of ZmGnTL enhances salt-tolerance of Arabidopsis
The expression of ZmGnTL increased substantially in Arabidopsis subjected to salt stress (Figure 3). It is plausible that ZmGnTL plays an important role in salt adaptation. To further confirm this hypothesis, we re-transformed ZmGnTL into Arabidopsis, and further evaluated the salt tolerance of transgenic Arabidopsis lines. The gDNA-PCR and RT-PCR analysis indicated that ZmGnTL successfully transformed into Arabidopsis (Figure 5A). The overexpressed lines (OX) under salt stress exhibited healthier phenotypes than wildtype (WT) in both MS medium and soil matrix. Whereas ZmGnTL-OX and WT were similar in growth under normal condition (Figures 5B, D). The biomass of OX lines was higher than WT under salt stress (Figure 5C), and the RWC content was also significantly higher in ZmGnTL-OX plants (Figure 5F). Additionally, both OX3 and OX4 exhibited lower EL levels compared with WT under salt stress (Figure 5E).
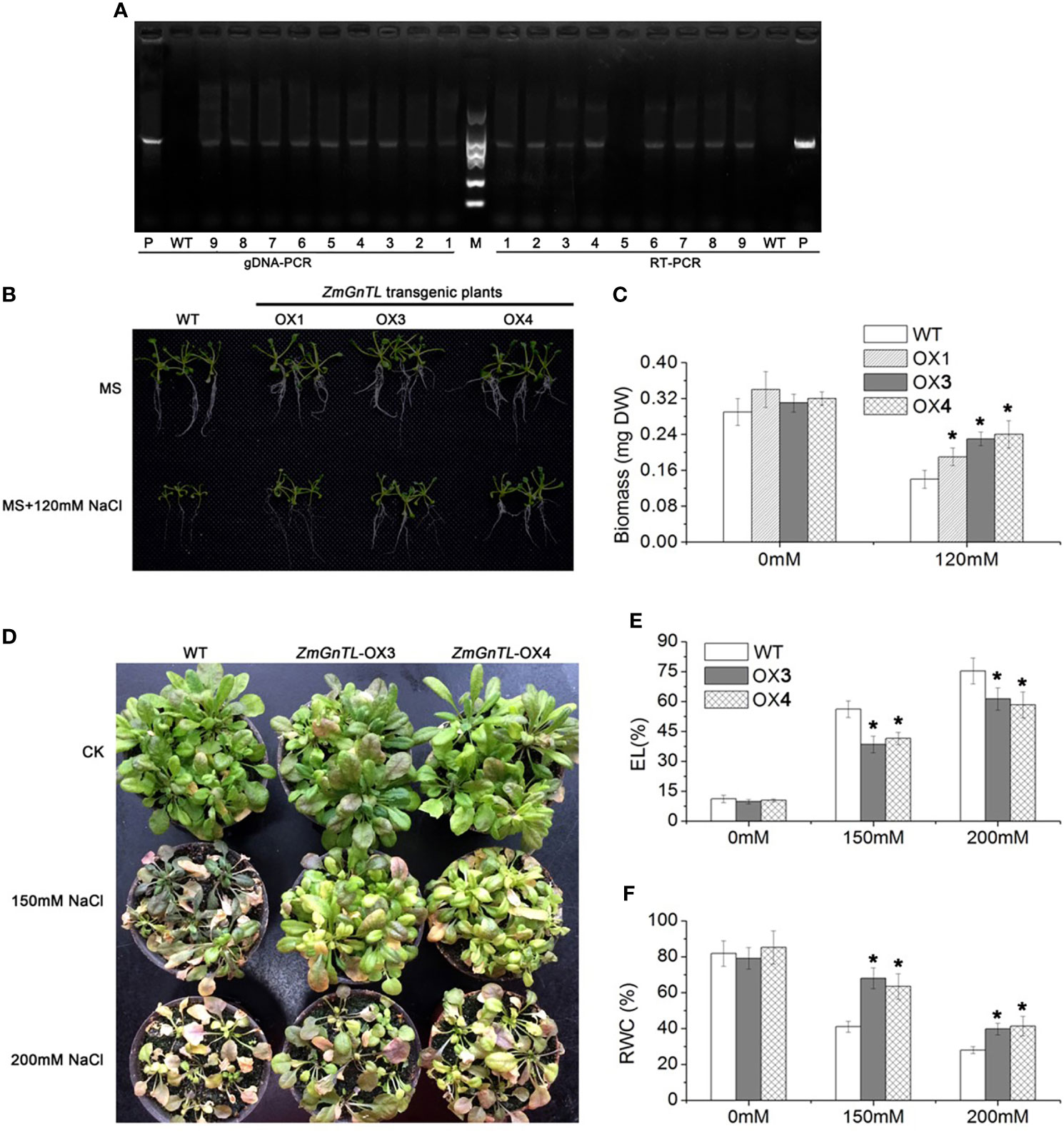
Figure 5 Overexpression of ZmGnTL enhanced the salt-tolerance in Arabidopsis. (A) gDNA -PCR and RT-PCR analysis of transgenetic lines, P (ZmGnTL plasmid), WT (wild type), 1-9 (transgenetic lines). (B) A comparison of salt-tolerant phenotypes and wildtype (WT) grown on MS medium. (C) Biomass of Arabidopsis seedlings grown on MS medium. (D) A comparison of salt-tolerant phenotypes and WT in soil matrix. (E, F) Electrolytic leakage (EL) and relative water content (RWC) of Arabidopsis plants. Data are shown as means ± SE of three to five biological replicates, and * indicated statistical significance at P< 0.05.
The physiological changes of ZmGnTL transgenic Arabidopsis under salt stress
To clarify the physiological changes of ZmGnTL-OX plants under salt stress, we measured the change of Na+ and K+ of ZmGnTL-OX and WT plants under different salinity treatments (0, 150, and 200 mM). Under control condition, the K+ and Na+ contents were not significantly different between ZmGnTL-OX lines and WT Arabidopsis plants. Whereas the K+ content was increased observably in ZmGnTL-OX plants, while there is no significant difference in Na+ content between ZmGnTL-OX plants and WT. (Figure 6). Therefore, the K+/Na+ ratio was higher in ZmGnTL-OX lines than that in WT under salt stress (Figure 6).
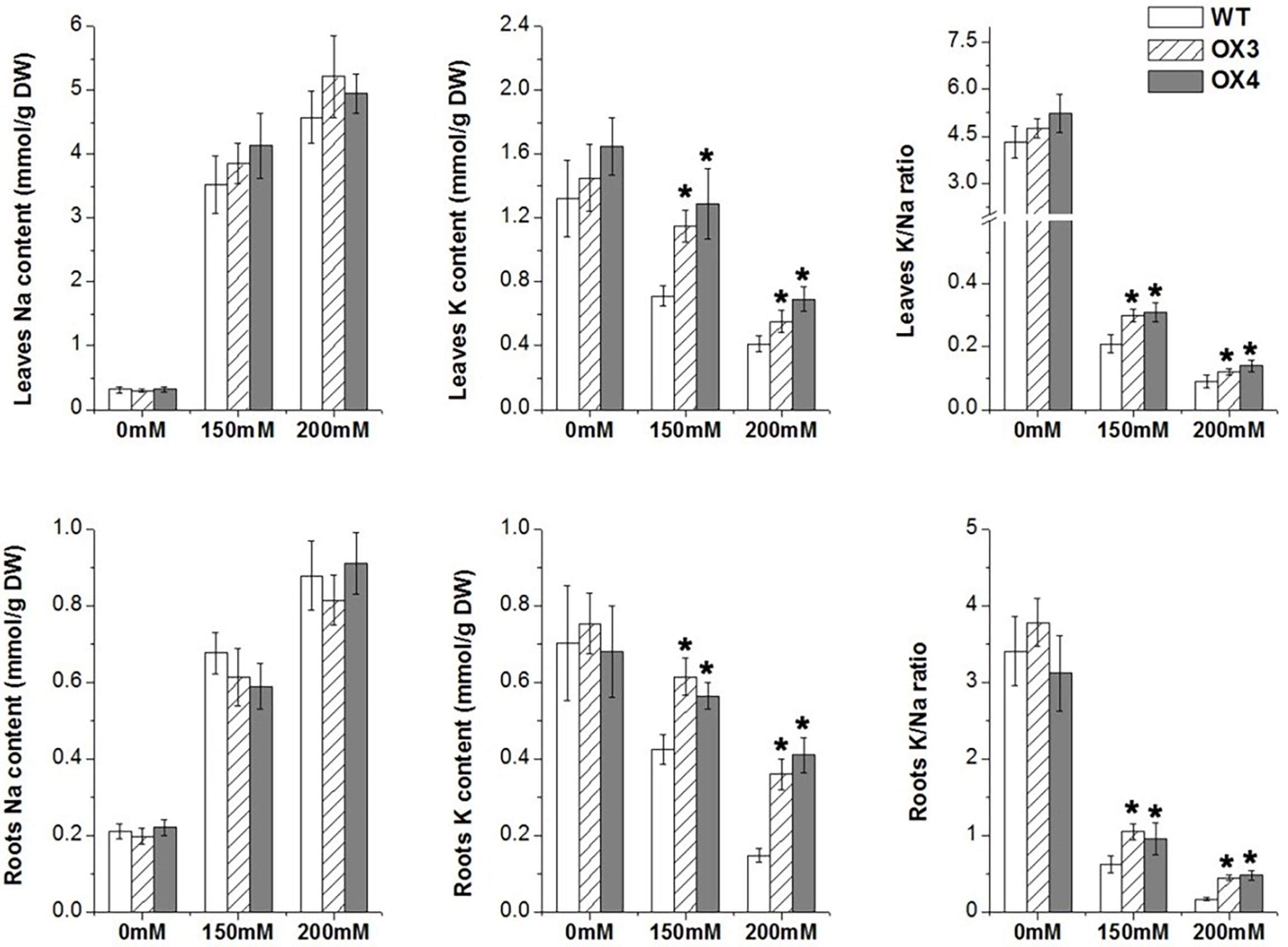
Figure 6 Contents of Na+ and K+ in leaves and roots of ZmGnTL transgenic Arabidopsis. ZmGnTL-OX plants and WT were treated with 0 mM, 150 mM, and 200 mM NaCl for 10 days. Data are shown as means ± SE of three to five biological replicates, and * indicated statistical significance at *P< 0.05.
We also analyzed the change of antioxidation system and osmolytes of ZmGnTL-OX and WT plants under salt stress. The result showed that the overexpression of ZmGnTL significantly decreased the contents of and H2O2 under salt stress (Figure 7). The enzyme activities of SOD, POD, CAT and APX were not different between ZmGnTL-OX lines and WT under normal growth condition (Figure 7). However, the activity levels of antioxidant enzymes SOD and APX were substantially higher in ZmGnTL-OX lines than those in WT Arabidopsis under salt stress, although the activities of POD and CAT were not affected by the overexpression (Figure 7). Proline and glycine betaine contents were also not significantly different in roots and leaves between ZmGnTL-OX lines and WT under normal condition (Figure 8). On the contrary, the accumulation of proline and glycine betaine was increased in leaves of ZmGnTL-OX plants under both 150 and 200 mM NaCl compared with WT. The contents of proline and glycine in root were significantly different under 150 mM NaCl but not under 200 mM NaCl (Figure 8). There was not significantly difference in soluble sugar contents in both roots and leaves between ZmGnTL-OX lines and WT under both normal and salt stress conditions (Figure 8).
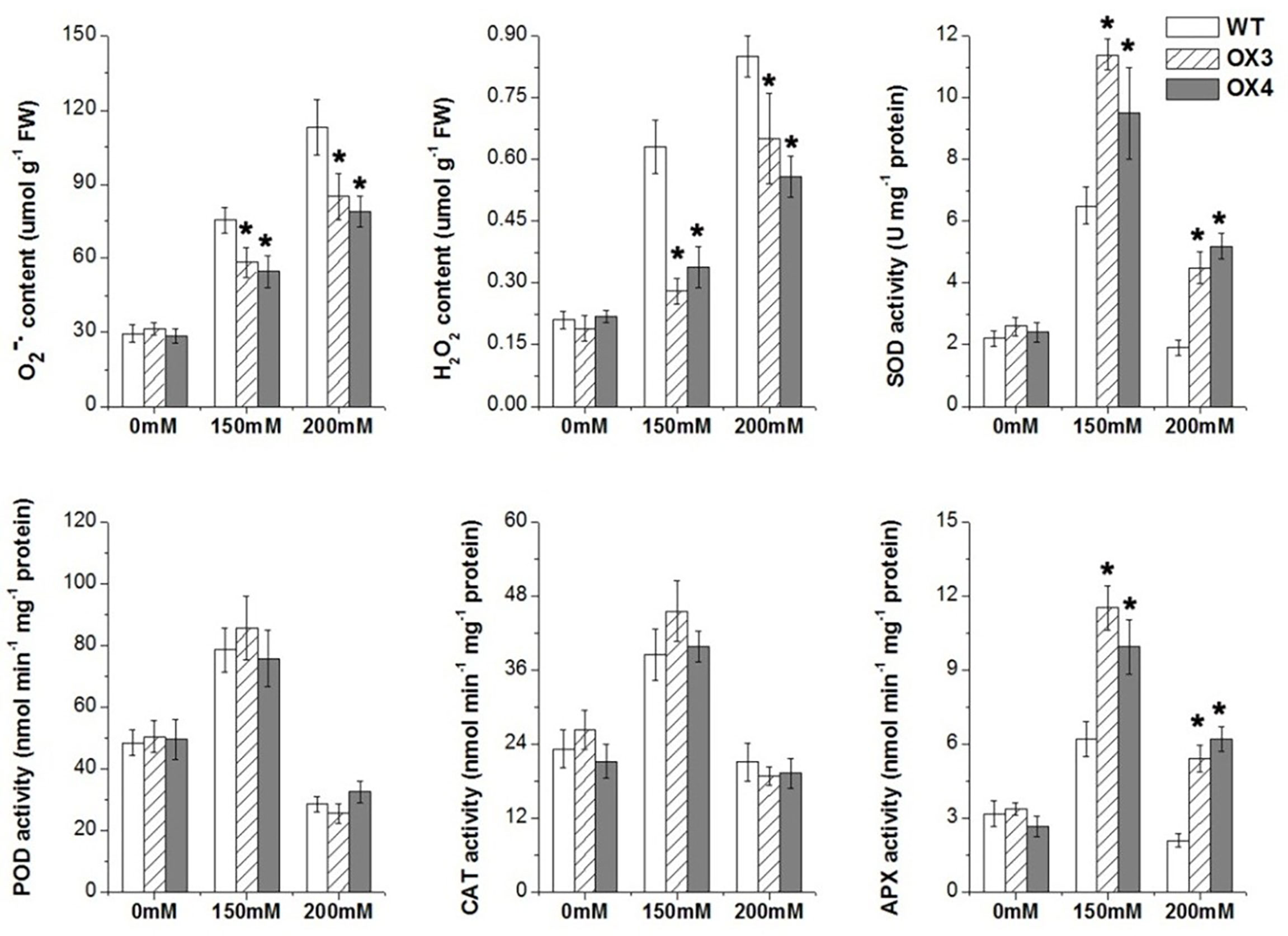
Figure 7 Reactive oxygen species contents and antioxidant enzyme activities of ZmGnTL transgenic Arabidopsis. ZmGnTL-OX plants and WT were treated with 0 mM, 150 mM, and 200 mM NaCl for 10 days. Data are shown as means ± SE of three to five biological replicates, and * indicated statistical significance at *P< 0.05.
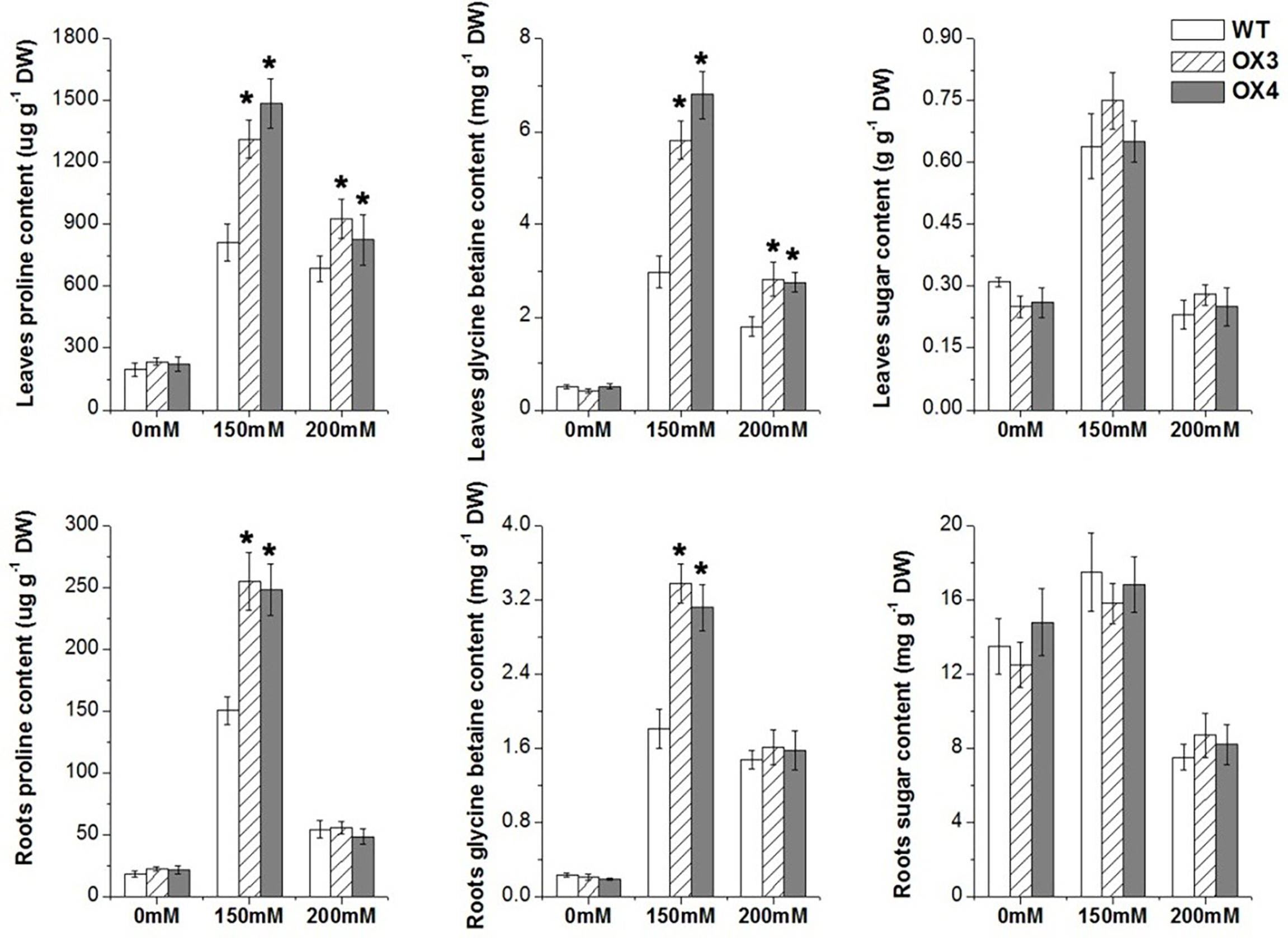
Figure 8 Osmolyte contents in leaves and roots of ZmGnTL transgenic Arabidopsis. ZmGnTL-OX plants and WT were treated with 0 mM, 150 mM, and 200 mM NaCl for 10 days. Data are show as means ± SE of three to five biological replicates, and * indicated statistical significance at *P< 0.05.
ZmGnTL influenced the expression of salt tolerant genes
To explore the molecular mechanism of ZmGnTL in regulating salt stress, WT and ZmGnTL-OX Arabidopsis plants were subjected to 24 h of salt stress to analyze the expression of genes associated with ion transport (AKT1, NHX1, VP1, and KUP7) (Hirsch et al., 1998; Apse et al., 2003; Li et al., 2005; Han et al., 2016), antioxidation (APX1 and Mn-SOD) (Li et al., 2019; Chen et al., 2022), and osmotic adjustment (PDH, P5CS, CMO, and BADH) (Peng et al., 1996; Strizhov et al., 1997; Fitzgerald et al., 2009; Luo et al., 2012). The expression of AtNHX1 was significantly higher in ZmGnTL-OX lines than that in WT roots and leaves under salt stress, while the expression was similar under normal condition (Figure 9). In the roots, the expression of AtAKT1 was significantly higher in ZmGnTL-OX lines compared with that in WT under salt stress (Figure 9). Overexpression of ZmGnTL increased the expression of AtAPX1 under 150 mM NaCl and AtMn-SOD under both 150 and 200 mM NaCl conditions, but their expression levels were lower and similar between ZmGnTL-OX lines and WT under normal condition (Figure 10). Under salt stress, genes related to osmotic stress regulation, such as AtP5CS and AtBADH, were also up-regulated in ZmGnTL-OX transgenic lines, the expression of AtPDH was similar between ZmGnTL-OX lines and WT under both 0 and 200 mM NaCl conditions but significantly lower in ZmGnTL-OX lines under 100 mM NaCl (Figure 10).
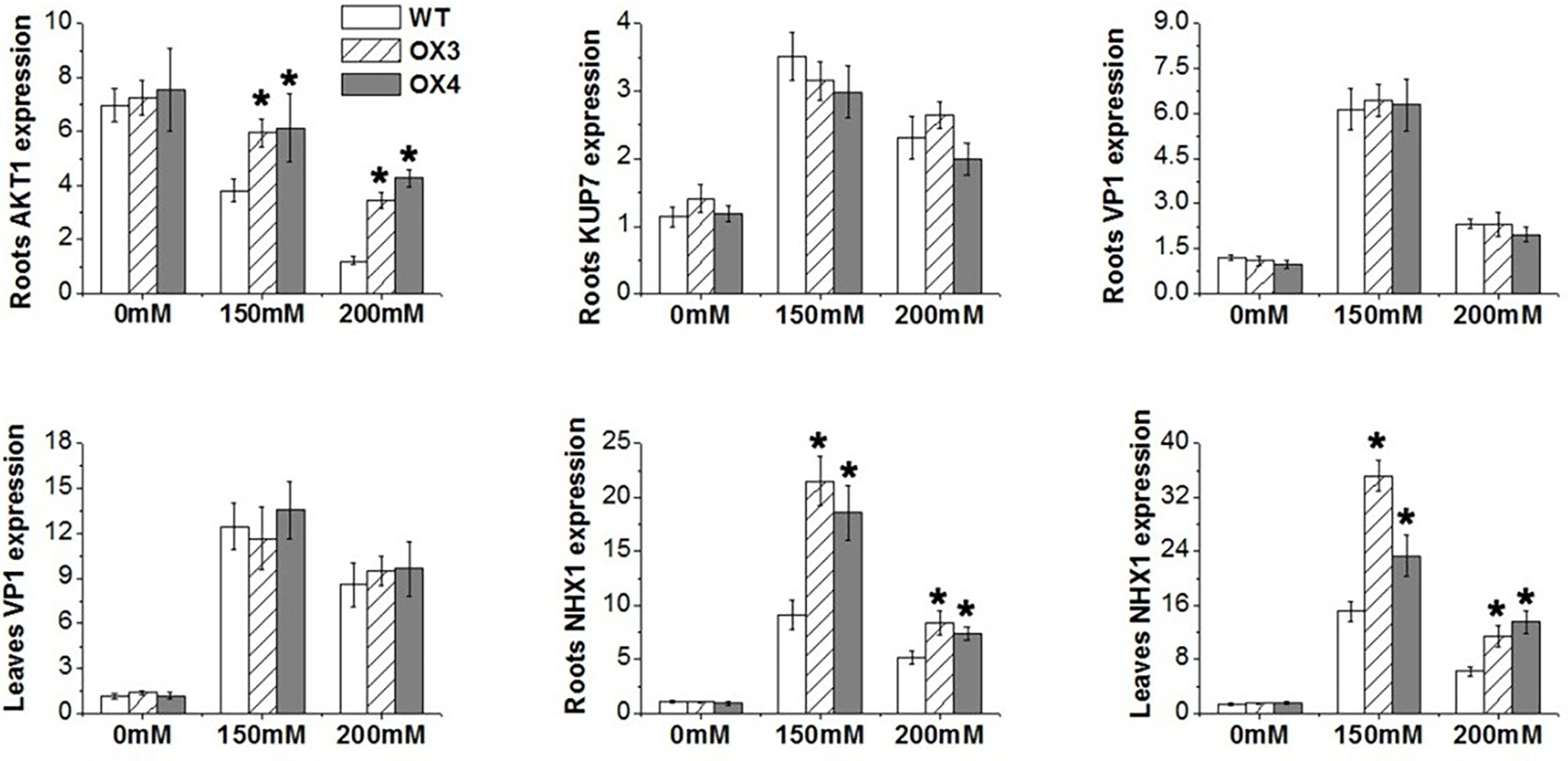
Figure 9 The relative expression of genes regulating ions balance in Arabidopsis under salt stress. ZmGnTL-OX plants and WT were treated with 0 mM, 150 mM, and 200 mM NaCl for 48 h. Data are shown as means ± SE of five biological replicates, and * indicated statistical significance at *P< 0.05.
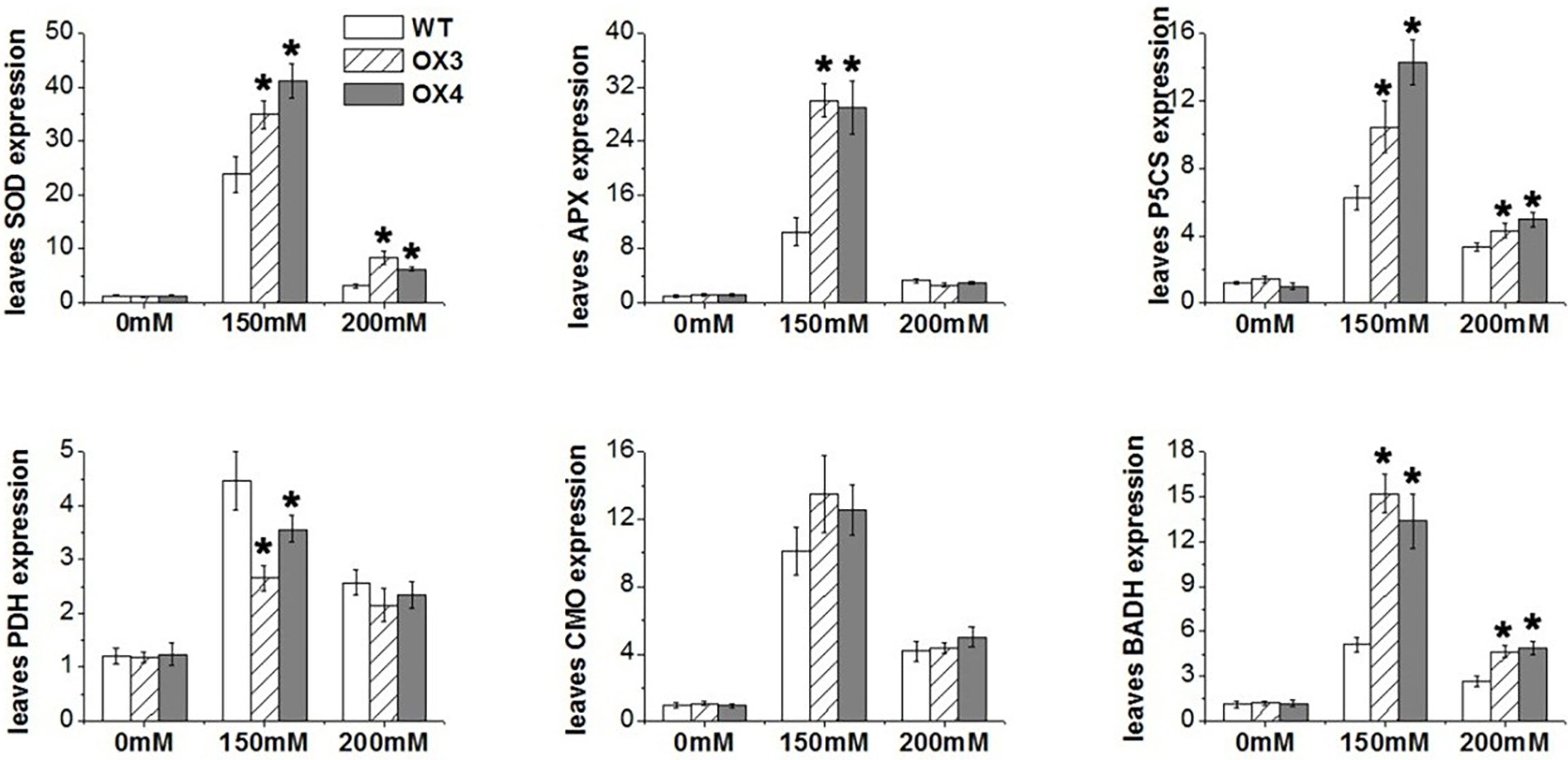
Figure 10 The relative expression of genes regulating antioxidant enzyme and osmolytes in Arabidopsis under salt stress. ZmGnTL-OX plants and WT were treated with 0 mM, 150 mM, and 200 mM NaCl for 48 h. Data are shown as means ± SE of five biological replicates, and * indicated statistical significance at *P< 0.05.
Discussion
Candidate salt-tolerant genes from Z. matrella
Salt stress leads to various physiological and molecular changes and impedes plant growth. To alleviate the damage of high concentrations of sodium, many genes are involved in regulating the salt stress under various mechanisms. In this study, 11 salt-tolerant genes from Z. matrella were identified. Many of these genes have been reported in various important biological processes.
The stress-associated protein 8 (SAP8) was reported as an osmotic stress-responsive transcription factor (Kanneganti and Gupta, 2008). Previous research showed that several SAP proteins are regulated by drought and salinity stress (Kanneganti and Gupta, 2008). Another gene ASR was associated with ABA in regulating stress and fruit ripening (Yoon et al., 2021); this gene family has been identified to responding to abiotic stresses and ABA in maize and rice but is absent in Arabidopsis (Zhang et al., 2015; Yoon et al., 2021). For example, a study has demonstrated that overexpression of OsASR1 and OsASR3 improved drought and salinity tolerance in transgenic rice (Joo et al., 2013). Glutaredoxins (GRXs) are small disulfide oxidoreductases that catalyze the reduction of disulfide bridges (Rouhier et al., 2008). Studies on the function of GRXs in plants have mainly focused on model plants, and it has been shown that GRXs are involved in the stress response and hormone signaling (Morita et al., 2015; El-Kereamy et al., 2015; Verma et al., 2016; Li et al., 2021). Zinc transporter (ZAT) is a type of zinc finger proteins (ZFPs) which are transcriptional regulators in plants (Han et al., 2020). In rice, ZAT was reported to regulate the expression of several genes that involved in ROS signaling when plants were under salt stress (Sun et al., 2010). Recently, GhZAT34 and GhZAT79 genes from Gossypium hirsutum were found to enhance salt tolerance in Arabidopsis and cotton (Rehman et al., 2021).
Lectins are a group of structurally diverse proteins which are defined as carbohydrate binding proteins and further divided into 25 subfamilies. Lectin plays an important role in response to abiotic or biotic stimuli (Vierbuchen, 1991; Naithani et al., 2021). In this study, we also identified a Glycosyltransferases (GTs) family gene GnTL in Z. matrella. GTs family protein is required for protein glycosylation (Vogt and Jones, 2000), and studies have shown that overexpressing genes (UGT85A5 and UGT87A2) from this family increased salt-tolerance in Arabidopsis and tobacco (Sun et al., 2013; Li et al., 2017). The protein encoded in Arabidopsis AtGnTL (AT3G52060) is involved in plasmodesmata interaction (Zalepa-King and Citovsky, 2013), but the function of AtGnTL related to salt tolerance has not been fully described.
Using the FOX system, we also identified an interesting gene, DUF1644, which belong to DUFs (domains of unknown functions) families (Bateman et al., 2010). The DUF1644 gene family is highly conserved in plants, but the biological function is unclear. Recently, a salt-induced gene, OsSIDP366 (stress induced DUF1644 family protein), was found in rice, and overexpressing OsSIDP366 significantly improved the salt tolerance of rice (Guo et al., 2016). Additionally, we identified ZmSANT gene. SANT domain protein was reported to be associated with chromatin remodeling, histone acetylation and deacetylation, but the biological function is unknown (Boyer et al., 2004; Marcum and Radhakrishnan, 2019). Fox system is reliable and efficient for screening up-regulated genes under salt stress or other abiotic stresses. Using this method, we successfully identified many candidate genes for further studying the mechanism of salt tolerance in halophytes.
Overexpression of ZmGnTL improved the salt tolerance of Arabidopsis
GnTL genes belong to the glycosyltransferase superfamily and are crucial in glycan synthesis (Fukuda and Hindsgaul, 1994) by adding the oligosaccharide side chains to glycoproteins (Siddiqui et al., 2005). This protein has been found in more than 19 plant species (Zalepa-King and Citovsky, 2013). However, many of the GnTL genes are not clearly characterized in their functions. In this study, we identified a gene ZmGnTL using a plant cDNA library screening method (FOX) under salt stress. Furthermore, we demonstrated the function of ZmGnTL in Arabidopsis salt tolerance. Salt stress severely inhibited the growth of Arabidopsis. The overexpression of ZmGnTL alleviated the damage of salt stress (Figure 5C), and the transgenic plants showed greater seedlings biomass and RWC content under salt treatment than WT (Figures 5B, E). Based on the data from the current study, we did not observe any advantages of the transgenic Arabidopsis lines overexpressing ZmGnTL growing under non-stress condition; those transgenic plants showed normal growth and similar genotypes as WT under non-salt stress condition, which was further supported by our results of similar physiological and gene expression measurements.
Under salt stress, higher Na+ accumulation in plant leads to the disruption of ion homeostasis. Since excessive Na+ often leads to K+ deficiency, plants need to modulate the Na+/K+ homeostasis through maintaining high K+/Na+ ratio (Park et al., 2016). The potassium transporters, such as the inward-rectifier K+ channel Arabidopsis K transporter (AKT1) plays an important role in K+ uptake and transport in the root (Nieves-Cordones et al., 2014). In this study, we found that overexpression of ZmGnTL increased K+ content and K+/Na+ ratio (Figure 6) through up-regulating the expression of AtAKT1 and AtNHX1 (a Na+/H+ antiporter, which transports Na+ away from the cytosol to vacuolar) under salt stress to maintain the ion balance (Figure 9).
Under salt stress, ion imbalance and water deficiency in the plant cell cause osmotic stress. Salt stress induces the reduction in cell turgor pressure, shrinkage of the plasma membrane, and physical alteration of the cell wall (Park et al., 2016). In order to alleviate the damage of osmotic stress, plants activate the osmolyte (such as proline, polyols, and sugars) accumulation under salt stress (Yang and Guo, 2018a). Our results demonstrated that overexpression of ZmGnTL increased proline and glycine betaine contents in response to salt stress (Figure 8). In addition, gene expression experiment also revealed that the increased expression of AtP5CS (proline biosynthesis) and AtBADH (betaine aldehyde dehydrogenase) in transgenic lines compared with WT under salt stress; however, the expression of AtPDH was decreased (Figure 10). Proline dehydrogenase (PDH) is functioned to remove free proline and prevent excessive proline accumulation after salt stress. A reciprocal regulation of P5CS and PDH was previously described to control the levels of proline during and after osmotic stress (Peng et al., 1996).
In plants, salt-stress-triggered ion stress and osmotic stress cause metabolism imbalance and toxic accumulation of ROS, which lends to oxidative damages (Yang and Guo, 2018b). Plant cells sense the accumulated ROS and respond rapidly by using regulatory mechanisms to scavenge ROS and activate a series of downstream adaptive responses (Park et al., 2016; Van Zelm et al., 2020). Several studies have shown that the activities of ROS scavenging enzymes and antioxidants are triggered by salt stress stimuli. For example, the APX and CAT are activated by salt stress to alleviate oxidative stresses (Sofo et al., 2015; Choudhury et al., 2017). In our study, the content of ROS and H2O2 were significantly lower in transgenic Arabidopsis compared to WT. These findings were also supported by the increased expression of SOD and APX genes in synthesizing ROS detoxifying proteins under salt stress (Figure 6). Therefore, our results indicated that ZmGnTL could effectively increase the content of antioxidant enzymes and help plants to alleviate the ROS toxicity caused by salt stress.
Conclusion
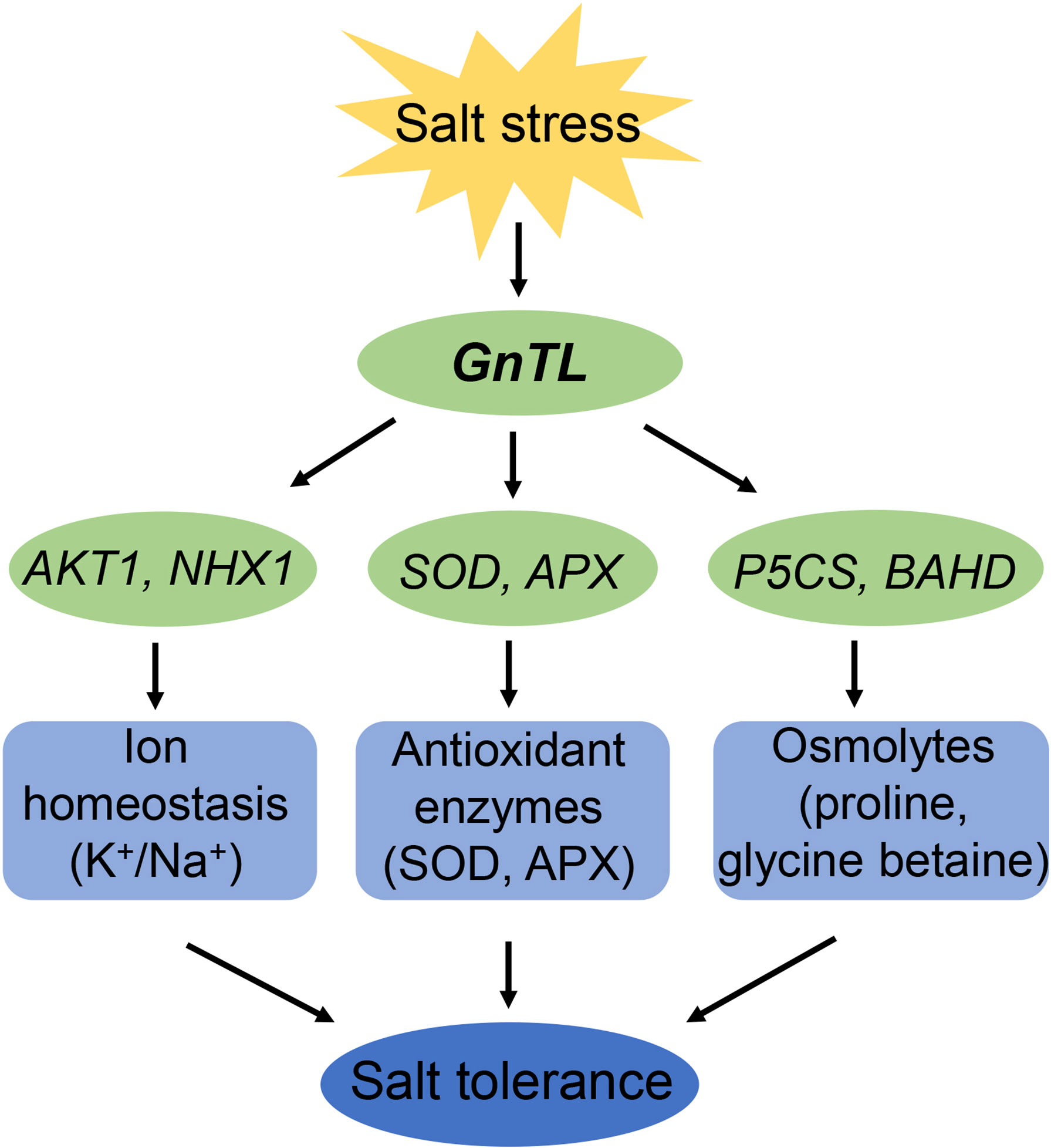
In summary, 11 new salt tolerance candidate genes from Z. matrella were identified by FOX system. Among those genes, we analyzed the function of ZmGnTL in Arabidopsis in response to salt stress. Overexpression of ZmGnTL significantly up-regulated the expression of K+ transporter gene, AKT1, tonoplast Na+/H+ antiporter gene, NHX1, SOD, APX, P5CS, and BADH, and down-regulated the expression of proline dehydrogenase gene, PDH. Our results suggested that ZmGnTL was involved in alleviating ion toxicity, and oxidative and osmotic stress under salt stress (Figure 11). ZmGnTL could be an important target gene for improving crop salt tolerance through genetic engineering.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Author contributions
YC, JiL, and JZ designed the experiments. YZ, JZ, and JuL performed the experiments. YZ, YC, and WK analyzed the data with suggestions by JC, HG, and JuL. YZ, YC, and RW wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (31301806, 31872953 and 31672193).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abrahám, E., Hourton-Cabassa, C., Erdei, L., Szabados, L. (2010). Methods for determination of proline in plants. Methods Mol. Biol. 639, 317–331. doi: 10.1007/978-1-60761-702-0_20
Almeida, D. M., Oliveira, M. M., Saibo, N. J. M. (2017). Regulation of na+ and k+ homeostasis in plants: towards improved salt stress tolerance in crop plants. Genet. Mol. Biol. 40, 326–345. doi: 10.1590/1678-4685-GMB-2016-0106
Apse, M. P., Sottosanto, J. B., Blumwald, E. (2003). Vacuolar cation/H+ exchange, ion homeostasis, and leaf development are altered in a T-DNA insertional mutant of AtNHX1, the arabidopsis vacuolar Na+/H+ antiporter. Plant J. 36, 229–239. doi: 10.1046/j.1365-313x.2003.01871.x
Asano, T., Hakata, M., Nakamura, H., Aoki, N., Komatsu, S., Ichikawa, H., et al. (2011). Functional characterisation of OsCPK21, a calcium-dependent protein kinase that confers salt tolerance in rice. Plant Mol. Biol. 75, 179–191. doi: 10.1007/s11103-010-9717-1
Bateman, A., Coggill, P., Finn, R. D. (2010). DUFs: families in search of function. Acta Crystallogr. Sect. F. Struct. Biol. Cryst. Commun. 66, 1148–1152. doi: 10.1107/S1744309110001685
Benito, B., Haro, R., Amtmann, A., Cuin, T. A., Dreyer, I. (2014). The twins k+ and na+ in plants. J. Plant Physiol. 171, 723–731. doi: 10.1016/j.jplph.2013.10.014
Blum, A., Ebercon, A. (1981). Cell membrane stability as a measure of drought and heat tolerance in wheat1. Crop Sci. 21, 43–47. doi: 10.2135/cropsci1981.0011183X002100010013x
Boyer, L. A., Latek, R. R., Peterson, C. L. (2004). The SANT domain: a unique histone-tail-binding module? Nat. Rev. Mol. Cell Biol. 5, 158–163. doi: 10.1038/nrm1314
Chen, H., Lee, J., Lee, J. M., Han, M., Emonet, A., Lee, J. (2022). MSD2, an apoplastic Mn-SOD, contributes to root skotomorphogenic growth by modulating ROS distribution in arabidopsis. Plant Sci. 317, 111192. doi: 10.1016/j.plantsci.2022.111192
Chen, Y., Zong, J., Tan, Z., Li, L., Hu, B., Chen, C. (2015). Systematic mining of salt-tolerant genes in halophyte-zoysia matrella through cDNA expression library screening. Plant Physiol. Biochem. 89, 44–52. doi: 10.1016/j.plaphy.2015.02.007
Choudhury, F. K., Rivero, R. M., Blumwald, E., Mittler, R. (2017). Reactive oxygen species, abiotic stress and stress combination. Plant J. 90, 856–867. doi: 10.1111/tpj.13299
El-Kereamy, A., Bi, Y. M., Mahmood, K., Ranathunge, K., Yaish, M. W., Nambara, E. (2015). Overexpression of the CC-type glutaredoxin, OsGRX6 affects hormone and nitrogen status in rice plants. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00934
Elstner, E. F., Heupel, A. (1976). Inhibition of nitrite formation from hydroxylammonium-chloride: a simple assay for superoxide dismutase. Anal. Biochem. 70, 616–620. doi: 10.1016/0003-2697(76)90488-7
Fitzgerald, T. L., Waters, D. L., Henry, R. J. (2009). Betaine aldehyde dehydrogenase in plants. Plant Biol. 11, 119–130. doi: 10.1111/j.1438-8677.2008.00161.x
Flowers, T. J., Munns, R., Colmer, T. D. (2015). Sodium chloride toxicity and the cellular basis of salt tolerance in halophytes. Ann. Bot. 115, 419–431. doi: 10.1093/aob/mcu217
Gao, Y. G., Zhao, B. Q., Jiao, X. M., Chen, M., Wang, B. S., Yuan, F. (2021). Coupled development of aalt glands, stomata, and pavement cells in limonium bicolor. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.745422
Guo, C., Luo, C. K., Guo, L. J., Li, M., Guo, X. L., Zhang, Y. X., et al. (2016). OsSIDP366, a DUF1644 gene, positively regulates responses to drought and salt stresses in rice. J. Integr. Plant Biol. 58, 492–502. doi: 10.1111/jipb.12376
Han, G. L., Lu, C. X., Guo, J. R., Qiao, Z. Q., Sui, N., Qiu, N. W., et al. (2020). C2H2 zinc finger proteins: master regulators of abiotic stress responses in plants. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00115
Han, Q. Q., Wang, Y. P., Li, J., Li, J., Yin, X. C., Jiang, X. Y., et al. (2022). The mechanistic basis of sodium exclusion in puccinellia tenuiflora under conditions of salinity and potassium deprivation. Plant J. 112, 322–338. doi: 10.1093/mp/sst024
Han, M., Wu, W., Wu, W. H., Wang, Y. (2016). Potassium transporter KUP7 is involved in k+ acquisition and translocation in arabidopsis root under k+-limited conditions. Mol. Plant 9, 437–446. doi: 10.1016/j.molp.2016.01.012
Higashi, Y., Ohama, A., Ishikawa, T., Katori, T., Shimura, A., Kusakabe, K., et al (2013). HsfA1d, a protein identified via FOX hunting using Thellungiella salsuginea cDNAs, improves heat tolerance by regulating heat stress responsive gene expression. Mol. Plant 6, 411–22. doi: 10.1093/mp/sst024
Higuchi, M., Kondou, Y., Ichikawa, T., Matsui, M. (2011). Full-length cDNA overexpressor gene hunting system (FOX hunting system). Methods Mol. Biol. 678, 77–89. doi: 10.1007/978-1-60761-682-5_7
Higuchi-Takeuchi, M., Matsui, M. (2014). Screening for gene function using the FOX (full-length cDNA overexpressor gene) hunting system. Methods Mol. Biol. 1056, 201–210. doi: 10.1007/978-1-62703-592-7_19
Hirsch, R. E., Lewis, B. D., Spalding, E. P., Sussman, M. R. (1998). A role for the AKT1 potassium channel in plant nutrition. Science 280, 918–921. doi: 10.1126/science.280.5365.918
Ichikawa, T., Nakazawa, M., Kawashima, M., Iizumi, H., Kuroda, H., Kondou, Y., et al. (2006). The FOX hunting system: an alternative gain-of-function gene hunting technique. Plant J. 48, 974–985. doi: 10.1111/j.1365-313X.2006.02924.x
Igarashi, Y., Yoshiba, Y., Sanada, Y., Yamaguchi-Shinozaki, K., Wada, K., Shinozaki, K. (1997). Characterization of the gene for delta1-pyrroline-5-carboxylate synthetase and correlation between the expression of the gene and salt tolerance in oryza sativa l. Plant Mol. Biol. 33, 857–865. doi: 10.1023/a:1005702408601
Ji, H. T., Pardo, J. M., Batelli, G., Van Oosten, M. J., Bressan, R. A., Li, X. (2013). The salt overly sensitive (SOS) pathway: established and emerging roles. Mol. Plant 6, 275–286. doi: 10.1093/mp/sst017
Joo, J., Lee, Y. H., Kim, Y. K., Nahm, B. H., Song, S. I. (2013). Abiotic stress responsive rice ASR1 and ASR3 exhibit different tissue-dependent sugar and hormone-sensitivities. Mol. Cells 35, 421–435. doi: 10.1007/s10059-013-0036-7
Jyothi-Prakash, P. A., Mohanty, B., Wijaya, E., Lim, T. M., Lin, Q., Loh, C. S., et al. (2014). Identification of salt gland-associated genes and characterization of a dehydrin from the salt secretor mangrove avicennia officinalis. BMC Plant Biol. 14, 291. doi: 10.1186/s12870-014-0291-6
Kanneganti, V., Gupta, A. K. (2008). Overexpression of OsiSAP8, a member of stress associated protein (SAP) gene family of rice confers tolerance to salt, drought and cold stress in transgenic tobacco and rice. Plant Mol. Biol. 66, 445–462. doi: 10.1007/s11103-007-9284-2
Katerji, N., van Hoorn, J. M., Hamdy, A., Mastrorilli, M. (2003). Salinity effect on crop development and yield, analysis of salt tolerance according to several classification methods. Agric. Water Manag. 62, 37–66. doi: 10.1016/S0378-3774(03)00005-2
Kunihiro, S., Kowata, H., Kondou, Y., Takahashi, S., Matsui, M., Berberich, T., et al. (2014). Overexpression of rice OsREX1-s, encoding a putative component of the core general transcription and DNA repair factor IIH, renders plant cells tolerant to cadmium- and UV-induced damage by enhancing DNA excision repair. Planta 239, 1101–1111. doi: 10.1007/s00425-014-2042-1
Lacan, D., Baccou, J. C. (1998). High levels of antioxidant enzymes correlate with delayed senescence in nonnetted muskmelon fruits. Planta 204, 377–382. doi: 10.1007/s004250050269
Li, Z. Q., Li, J. T., Bing, J., Zhang, G. F. (2019). The role analysis of APX gene family in the growth and developmental processes and in response to abiotic stresses in arabidopsis thaliana. Hereditas 41, 534–547. doi: 10.16288/j.yczz.19-026
Li, T. T., Li, M. Z., Jiang, Y. M., Duan, X. W. (2021). Genome-wide identification, characterization and expression profile of glutaredoxin gene family in relation to fruit ripening and response to abiotic and biotic stresses in banana (Musa acuminata). Int. J. Biol. Macromol. 170, 636–651. doi: 10.1016/j.ijbiomac.2020.12.167
Li, P., Li, Y. J., Wang, B., Yu, H. M., Li, Q., Hou, B. K. (2017). The arabidopsis UGT87A2, a stress-inducible family 1 glycosyltransferase, is involved in the plant adaptation to abiotic stresses. Physiol. Plant 159, 416–432. doi: 10.1111/ppl.12520
Li, J. S., Yang, H. B., Peer, W. A., Richter, G., Blakeslee, J., Bandyopadhyay, A., et al. (2005). Arabidopsis h+-PPase AVP1 regulates auxin-mediated organ development. Science 310, 121–125. doi: 10.1126/science.1115711
Luo, D., Niu, X. L., Yu, J. D., Yan, J., Gou, X. J., Lu, B. R., et al. (2012). Rice choline monooxygenase (OsCMO) protein functions in enhancing glycine betaine biosynthesis in transgenic tobacco but does not accumulate in rice (Oryza sativa l. ssp. japonica). Plant Cell Rep. 31, 1625–1635. doi: 10.1007/s00299-012-1276-2
Luo, X. L., Wu, J. H., Li, Y. B., Nan, Z. R., Guo, X., Wang, Y. X., et al. (2013). Synergistic effects of GhSOD1 and GhCAT1 overexpression in cotton chloroplasts on enhancing tolerance to methyl viologen and salt stresses. PloS One 8, e54002. doi: 10.1371/journal.pone.0054002
Maehly, A. C., Chance, B. (1954). The assay of catalases and peroxidases. Methods Biochem. Anal. 1, 357–424. doi: 10.1002/9780470110171.ch14
Marcum, R. D., Radhakrishnan, I. (2019). Inositol phosphates and core subunits of the Sin3L/Rpd3L histone deacetylase (HDAC) complex up-regulate deacetylase activity. J. Biol. Chem. 294, 13928–13938. doi: 10.1074/jbc.RA119.009780
Meloni, D. A., Oliva, Marco, A., Martinez, C. A., Cambraia, J. (2003). Photosynthesis and activity of superoxide dismutase, peroxidase and glutathione reductase in cotton under salt stress. Environ. Exp. Bot. 49, 69–76. doi: 10.1016/S0098-8472(02)00058-8
Mishra, A., Tanna, B. (2017). Halophytes: Potential resources for salt stress tolerance genes and promoters. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00829
Morita, S., Yamashita, Y., Fujiki, M., Todaka, R., Nishikawa, Y., Hosoki, A., et al. (2015). Expression of a rice glutaredoxin in aleurone layers of developing and mature seeds: subcellular localization and possible functions in antioxidant defense. Planta 242, 1195–1206. doi: 10.1007/s00425-015-2354-9
Naithani, S., Komath, S. S., Nonomura, A., Govindjee, G. (2021). Plant lectins and their many roles: Carbohydrate-binding and beyond. J. Plant Physiol. 266, 153531. doi: 10.1016/j.jplph.2021.153531
Nakano, Y., Asada, K. (1981). Hydrogen peroxide is scavenged by ascorbate specific peroxidase in spinach chloroplasts. Plant Cell Physiol. 22, 867–880. doi: 10.1093/oxfordjournals.pcp.a076232
Nieves-Cordones, M., Alemán, F., Martínez, V., Rubio, F. (2014). K+ uptake in plant roots. the systems involved, their regulation and parallels in other organisms. J. Plant Physiol. 171, 688–695. doi: 10.1016/j.jplph.2013.09.021
Park, H. J., Kim, W. Y., Yun, D. J. (2016). A new insight of salt stress signaling in plant. Mol. Cells 39, 447–459. doi: 10.14348/molcells.2016.0083
Peng, Z., Lu, Q., Verma, D. P. S. (1996). Reciprocal regulation of delta 1-pyrroline-5-carboxylate synthetase and proline dehydrogenase genes controls proline levels during and after osmotic stress in plants. Mol. Gen. Genet. 253, 334–341. doi: 10.1007/pl00008600
Rehman, A., Wang, N., Peng, Z., He, S. P., Zhao, Z. B., Gao, Q., et al. (2021). Identification of C2H2 subfamily ZAT genes in gossypium species reveals GhZAT34 and GhZAT79 enhanced salt tolerance in arabidopsis and cotton. Int. J. Biol. Macromol. 184, 967–980. doi: 10.1016/j.ijbiomac.2021.06.166
Rouhier, N., Lemaire, S. D., Jacquot, J. P. (2008). The role of glutathione in photosynthetic organisms: emerging functions for glutaredoxins and glutathionylation. Annu. Rev. Plant Biol. 59, 143–166. doi: 10.1146/annurev.arplant.59.032607.092811
Shi, H., Lee, B. H., Wu, S. J., Zhu, J. K. (2003). Overexpression of a plasma membrane na+ /H+ antiporter gene improves salt tolerance in arabidopsis thaliana. Nat. Biotechnol. 21, 81–85. doi: 10.1038/nbt766
Siddiqui, S. F., Pawelek, J., Handerson, T., Lin, C. Y., Dickson, R. B., Rimm, D. L., et al. (2005). Coexpression of beta1,6-n-acetylglucosaminyltransferase V glycoprotein substrates defines aggressive breast cancers with poor outcome. Cancer Epidemiol. Biomarkers Prev. 14, 2517–2523. doi: 10.1158/1055-9965
Slama, I., Abdelly, C., Bouchereau, A., Flowers, T., Savouré, A. (2015). Diversity, distribution and roles of osmoprotective compounds accumulated in halophytes under abiotic stress. Ann. Bot. 115, 433–447. doi: 10.1093/aob/mcu239
Sofo, A., Scopa, A., Nuzzaci, M., Vitti, A. (2015). Ascorbate peroxidase and catalase activities and their genetic regulation in plants subjected to drought and salinity stresses. Int. J. Mol. Sci. 16, 13561–13578. doi: 10.3390/ijms160613561
Strizhov, N., Abrahám, E., Okrész, L., Blickling, S., Zilberstein, A., Schell, J., et al. (1997). Differential expression of two P5CS genes controlling proline accumulation during salt-stress requires ABA and is regulated by ABA1, ABI1 and AXR2 in arabidopsis. Plant J. 12, 557–569. doi: 10.1046/j.1365-313x.1997.00557.x
Sun, S. J., Guo, S. Q., Yang, X., Bao, Y. M., Tang, H. J., Sun, H., et al. (2010). Functional analysis of a novel Cys2/His2-type zinc finger protein involved in salt tolerance in rice. J. Exp. Bot. 61, 2807–2818. doi: 10.1093/jxb/erq120
Sun, Y. G., Wang, B., Jin, S. H., Qu, X. X., Li, Y. J., Hou, B. K. (2013). Ectopic expression of arabidopsis glycosyltransferase UGT85A5 enhances salt stress tolerance in tobacco. PloS One 8, e59924. doi: 10.1371/journal.pone.0059924
Van Zelm, E., Zhang, Y. X., Testerink, C. (2020). Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 71, 403–433. doi: 10.1146/annurev-arplant-050718-100005
Verma, P. K., Verma, S., Pande, V., Mallick, S., Deo Tripathi., R., Dhankher, O. P., et al. (2016). Overexpression of rice glutaredoxin OsGrx_C7 and OsGrx_C2.1 reduces intracellular arsenic accumulation and increases tolerance in arabidopsis thaliana. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00740
Vierbuchen, M. (1991). Lectin receptors. Curr. Top. Pathol. 83, 271–361. doi: 10.1007/978-3-642-75515-6_10
Vogt, T., Jones, P. (2000). Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci. 5, 380–386. doi: 10.1016/s1360-1385(00)01720-9
Wu, Y. J., Pang, L., Li, M., Li, L. X. (2018). Determination of betaine in lycii fructus by HPLC-CAD. Shandong. Chem. Industry. 47, 67–69. doi: 10.19319/j.cnki.issn.1008-021x.2018.12.026
Yang, Y. Q., Guo, Y. (2018a). Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 217, 523–539. doi: 10.1111/nph.14920
Yang, Y. Q., Guo, Y. (2018b). Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 60, 796–804. doi: 10.1111/jipb.12689
Yoon, J. S., Kim, J. Y., Kim, D. Y., Seo, Y. W. (2021). A novel wheat ASR gene, TaASR2D, enhances drought tolerance in brachypodium distachyon. Plant Physiol. Biochem. 159, 400–414. doi: 10.1016/j.plaphy.2020.11.014
Yuan, F., Lyu, M. J., Leng, B. Y., Zheng, G. Y., Feng, Z. T., Li, P. H., et al. (2015). Comparative transcriptome analysis of developmental stages of the limonium bicolor leaf generates insights into salt gland differentiation. Plant Cell Environ. 38, 1637–1657. doi: 10.1111/pce.12514
Zalepa-King, L., Citovsky, V. (2013). A plasmodesmal glycosyltransferase-like protein. PloS One 8, e58025. doi: 10.1371/journal.pone.0058025
Zhang, Y. T., Hou, K., Qian, H., Gao, Y. Y., Fang, Y., Xiao, S., et al. (2022). Characterization of soil salinization and its driving factors in a typical irrigation area of Northwest China. Sci. Total. Environ. 837, 155808. doi: 10.1016/j.scitotenv.2022.155808
Zhang, L. L., Hu, W., Wang, Y., Feng, R. J., Zhang, Y. D., Liu, J. H., et al. (2015). The MaASR gene as a crucial component in multiple drought stress response pathways in arabidopsis. Funct. Integr. Genomics 15, 2472–2460. doi: 10.1007/s10142-014-0415-y
Zhang, J., Li, H. B., Xu, B., Li, J., Huang, B. R. (2016). Exogenous melatonin suppresses dark-induced leaf senescence by activating the superoxide dismutase-catalase antioxidant pathway and down-regulating chlorophyll degradation in excised leaves of perennial ryegrass (Lolium perenne l.). Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01500
Zhang, J. L., Shi, H. Z. (2013). Physiological and molecular mechanisms of plant salt tolerance. Photosynth. Res. 115, 1–22. doi: 10.1007/s11120-013-9813-6
Zhang, J. L., Wang, S. M., Flowers, T. J. (2013). Differentiation of low-affinity na+ uptake pathways and kinetics of the effects of k+ on na+ uptake in the halophyte suaeda maritima. Plant Soil. 368, 629–640. doi: 10.1007/s11104-012-1552-5
Keywords: Zoysia matrella, FOX hunting, salt-tolerant genes, ZmGnTL, regulatory mechanism
Citation: Zheng Y, Zong J, Liu J, Wang R, Chen J, Guo H, Kong W, Liu J and Chen Y (2022) Mining for salt-tolerant genes from halophyte Zoysia matrella using FOX system and functional analysis of ZmGnTL. Front. Plant Sci. 13:1063436. doi: 10.3389/fpls.2022.1063436
Received: 07 October 2022; Accepted: 03 November 2022;
Published: 17 November 2022.
Edited by:
Jin-Lin Zhang, Lanzhou University, ChinaReviewed by:
Liang Chen, University of Chinese Academy of Sciences, ChinaXiqing Ma, China Agricultural University, China
Copyright © 2022 Zheng, Zong, Liu, Wang, Chen, Guo, Kong, Liu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu Chen, Y3l1ODAxMDI3QGFsaXl1bi5jb20=
†These authors have contributed equally to this work
 Yuying Zheng1†
Yuying Zheng1† Junqin Zong
Junqin Zong Jingbo Chen
Jingbo Chen Hailin Guo
Hailin Guo Jianxiu Liu
Jianxiu Liu Yu Chen
Yu Chen