- Beijing Key Laboratory of Growth and Development Regulation for Protected Vegetable Crops, College of Horticulture, China Agriculture University, Beijing, China
qRT-PCR is a common and key technical means to study gene expression in biological research. However, reliability and accuracy of quantification by qRT-PCR is entirely dependent on the identification of appropriate reference genes. Cucumber as an economical vegetable is widely cultivated worldwide and is subject to serious nematode infection, especially from M. incognita. Plant could employ beneficial soil bacteria in the rhizosphere to enhance plant adaptability to various stresses. In this study, the optimal reference genes in cucumber under M. incognita stress and Pseudomonas treatment were calculated and confirmed. A total of thirteen candidate reference genes were identified across three different treatments. Of these, geNorm, NormFinder and BestKeeper programs combined RefFinder software identified EF1 and UBI are the most suitable reference gene in the root knot and whole root of cucumber infected M. incognita, respectively, and CACS is the most suitable reference gene in the whole root of cucumber treated by Pseudomonas. The work first validated the most suitable reference genes for the normalization gene expression in cucumber by nematode infected or Pseudomonas inoculated, and these results would facilitate the further research on M. incognita or Pseudomonas soil rhizosphere microbe interaction with cucumber.
Introduction
Cucumber (Cucumis sativus L.) is one of the most important vegetable crops in protected cultivation, which is widely planted in the world. Cucumber has been developed as a new model species in plant biology due to its many desirable traits, including (i) small number of genes, (ii) rich diversity of sex expression, (iii) suitability for vascular biology studies short life cycle (three months from seed to seed), (iv) mixed phloem loading mechanism (Ma et al., 2019), and (v) Agrobacterium tumefaciens-mediated transformation (Li et al., 2017). The physiological, biochemical and breeding of cucumber have been studied for several decades, and it became more realistic to further study the molecular biology of cucumber as available of genomic sequence of cucumber (http://cucurbitgenomics.org/organism/2) (Huang et al., 2009) and accumulating resources in genetics and genomics.
In recent years, it has been found that root-knot nematode has caused serious damage to protected cultivation vegetable and leads to the continuous production reduction of cucumber in most areas due to growth obstacles (Jaiteh et al., 2012). So far, four kinds of root knot nematodes, including Meloidogyne javanica (M. javanica), Meloidogyne arenaria (M. arenaria), Meloidogyne incognita (M. incognita), and Meloidogyne hapla (M. hapla) are considered as serious threats to crop productions, among them, M. incognita has the widest distribution of host (Moens and Perry, 2009; Shahid et al., 2022). As its obligate biotrophic nature, M. incognita mainly relies on the host plant for nutrition and maintains the relationship with the host in a few weeks (Jones et al., 2011; Yook et al., 2011). Besides, M. incognita affects the root activity and the root absorption and transportation of water and inorganic salt ions, and hormones content like JA, SA and ABA, resulting in short aboveground plants, abnormal yellowing of leaf color, defoliation, growth weakness, wilting, and reduced yield (Mbaluto et al., 2020). There has been a lot of research on plant-nematode interaction, including how nematodes find their host, how the host perceive nematode precontact, and which host defense responses are elicited upon perception. Thus, many regulated genes have been reported in response to nematode infection, such as transporter genes, hormone related genes and cell wall embolism gens. AtACA8 (P-type ATPase gene), AtAUX1 (auxin influx transporter gene) and AtSUC1 (sucrose transporter gene) were induced significantly in Arabidopsis after infected by M. incognita though microarray analysis (Hammes et al., 2005). In rice, OsBAK1 (brassinolide co-receptor gene) was induced by M. incognita infection based on the RNA-Seq analysis (Zhou et al., 2020). Although these genes expression have also been identified by qRT-PCR, there has been no selection research of housekeeping genes (HKGs) in plant infected by M. incognita yet.
Plant growth-promoting rhizobacteria (PGPR) is a kind of important beneficial bacteria that promote plant growth and development, absorb nutrients, improve plant stress and inhibit pests (Vejan et al., 2016). Plants inoculation with PGPRs stains, such as B. subtilis GB03 and Pseudomonas spp., could enhance the tolerance for osmotic stress in plants by up-regulate the glycine betaine biosynthesis (Sandhya et al., 2010; Zhang et al., 2010). PGPR indirectly boosts plant growth rate, has also been widely reported in the study of microbial plant interaction. Pseudomonas is an important PGPR that promotes seed germination, root growth, accumulation of mineral nutrients, water use and prevention of plant diseases. Under salinity conditions, Pseudomonas was enriched in the rhizosphere and endosphere to enhance plant adaptability to salt stress through inducing the production of stress alleviating metabolites, like indole acetic acid, exopolysaccharides, and gibberellins. In this context, many functions of Pseudomonas on plants have been examined in physiology view (Kumar et al., 2019; Li et al., 2021). However, there is little research involving the molecular mechanism about the interaction between Pseudomonas and plants. Thus, the identification of appropriate reference genes will be valuable for advancing the plant-microorganism interaction.
Gene expression analysis is the basis for elucidating the molecular mechanisms of various biological processes, and quantitative real-time polymerase chain reaction (qRT-PCR) has become a common method to study gene expression due to its advantages of good repeatability, high sensitivity, high specific and high throughput (Nolan et al., 2006). However, this approach requires one or more HKGs that are stably expressed as criteria for normalizing gene expression. The ideal HKGs should be systematically evaluated in different tissues under different experimental conditions, and its expression should be stable and then used as a control for qRT-PCR analysis (Nolan et al., 2006). HKGs are a class of stably expressed genes, which can maintain the basic functions of cell division, growth and development, cell apoptosis and the whole physiological process of plant metabolism. Many HKGs, including Actin (ACT), a-Tubulin (TUA), F-box protein (F-BOX), YSL8 (mitosis protein), Ubiquitin-conjugating enzyme E2 (UBC), 60S ribosomal protein L36a/L44 (PRL36Aa), Protein phosphatase (PP2A) and Clathrin adaptor complexes medium submit family protein (CACS) had been used in cucumber (Migocka and Papierniak, 2011; Warzybok and Migocka, 2013; Joseph et al., 2018; Liang et al., 2018; Miao et al., 2019). After metal treatment, CACS expression was the most stable reference gene, and EF1 expression was the most reliable reference gene under salt, osmotic and oxidative stresses in cucumber (Migocka and Papierniak, 2011). Under different nitrogen conditions, TIP41, F-box and EF1 were the most suitable genes to normalize CsNRTs expression (Warzybok and Migocka, 2013). For Cucumber (Cucumis sativus L.), pumpkin (Cucurbita moschata Duch.) and cucumber-pumpkin grafting experiment, CACS and 40SRPS8 were the most stable reference genes in all samples (Miao et al., 2019). A mass of experiments demonstrated that HKGs cannot be utilized universally across different experimental conditions, plant species, developmental stages or even different organs within a single species. Therefore, many researchers have tried to find reliable reference genes expressed under specific experimental conditions and multi-HKGs should be used in one experiment.
Here, in order to promote the current quantification of gene expression in cucumber roots infected M. incognita and treated by Pseudomonas, thirteen candidate HKGs across seven timepoints in cucumber roots, which were infected by M. incognita and inoculated by three Pseudomonas, were assessed to identify suitable cohort of stably expressed HKGs for accurate and reliable normalization of cucumber qRT-PCR expression data, respectively. According to four procedures and some statistical methods, including geNorm (Vandesompele et al., 2002), NormFinder (Andersen et al., 2004), BestKeeper (Pfaffl et al., 2004), and RefFinder (Duan et al., 2017), the most suitable candidate HKGs were finally screened. The selected suitable reference genes will be helpful for understanding the cucumber-nematode interaction and investigating how Pseudomonas to improve plant growth on molecular level.
Materials and methods
Plant material and treatments
Cucumber (Xintaimici) was used in this study. The cucumber seeds were germinated in 25°C of darkness and sown in black pots with soil and sand (v:v, 1:1). Seedlings were grown a 75%-90% relative humidity and 16 h photoperiod (200 mmol m-2 s-1) at 28°C/day and 18°C/night for 7 days with standard Hoagland nutrient solution. Two treatments were applied in this study, which were M. incognita infection and Pseudomonas inoculation, respectively. Plant samples were divided into three categories, root knots produced by M. incognita (Treatment 1), whole root infected by M. incognita (Treatment 2) and inoculated by Pseudomonas (Treatment 3).
Nematode egg masses were collected from infested swamp cabbage roots and kept in sterile water for hatching at 28°C. Three-week-old cucumber seedlings were inoculated with freshly hatched second-stage juvenile nematode and 500 nematodes per plant. Cucumber roots infected by M. incognita (root knots and whole roots) were harvested at 0 d, 7 d, 14 d, 21 d, 28 d, and 35 d. For the Pseudomonas treatment, RH58, RH61, and RH62 strains were used, the purified beneficial rhizobacteria strains were shaken to OD600 = 1.0. The roots were collected at inoculated 72 h by Pseudomonas.
Total RNA isolation and cDNA synthesis
Cucumber root tissues were disrupted under liquid nitrogen frozen conditions. Approximately 100 mg of the ground root tissues were placed into a 1.5 mL centrifuge tube and total RNA extracted using RNeasy plant kit (Huayueyang, Beijing, China). RNA concentration and purity were examined by nucleic acid spectrophotometer (NanoDrop™ 1000, ThermoFisher Scientific, USA) and OD260/280 showed values between 1.8 and 2.0. The quality of RNA was detected by agarose gel electrophoresis. 1 ug of total RNA was used for cDNA synthesized using Fastking cDNA Dispelling RT Super Mix Kit (TIANGEN, Being, China) according to the manufacturer’s instructions. cDNA was diluted 1:5 in RNase-free water and stored at -20°C until use in qRT-PCR experiment.
Candidate reference genes selection and qRT-PCR assay
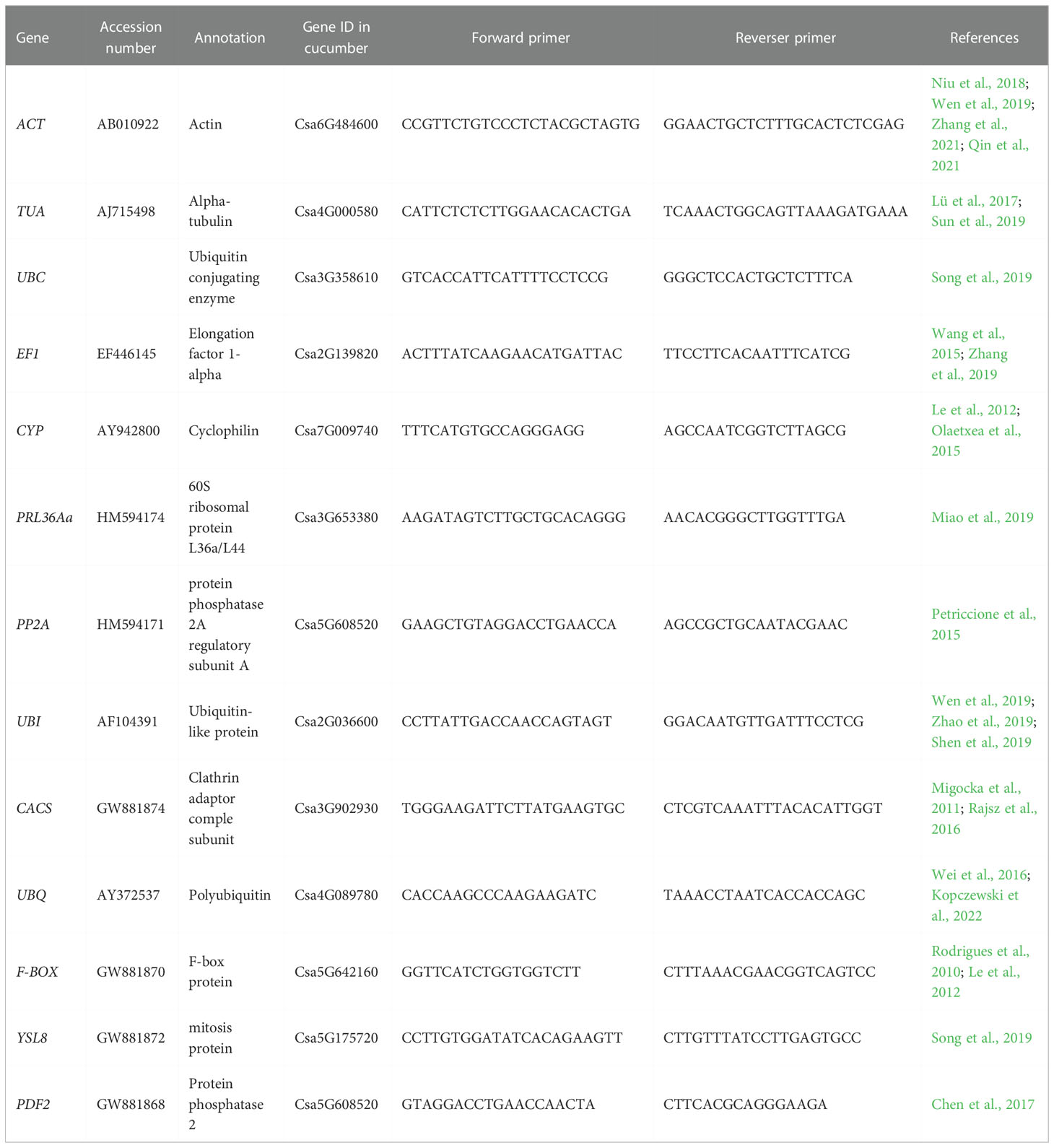
A total of thirteen potential reference genes were selected in this study. According to previous studies, the expression of these candidate genes is relatively stable in plant growth and development processes, or under biotic or abiotic stresses. The sequences of candidate reference genes were acquired from the GenBank and Cucumber genome database. The primer pairs (Table 1) were completely consistent with previous study (Migocka and Papierniak, 2011; Miao et al., 2019). The primers specificity of thirteen candidate reference genes were detected by PCR and 1% agarose gel electrophoresis (Figure 1). qRT-PCR experiment was performed on an ABI 7500 Real Time PCR system (Applied Biosystems, USA) using SYBR® Green I (ChamQ SYBR qPCR Master Mix) (Vazyme, Beijing, China). Each 10 μl reaction mixture contained 2 μl of cDNA template, 5 μl of SYBR® Green I, 0.2 μl of each primer, and 2.6 μl of ddH2O. The qRT-PCR reaction system was as follow: 94°C for 30 s, 40 cycles of 94°C for 5 s and 60°C for 34 s.
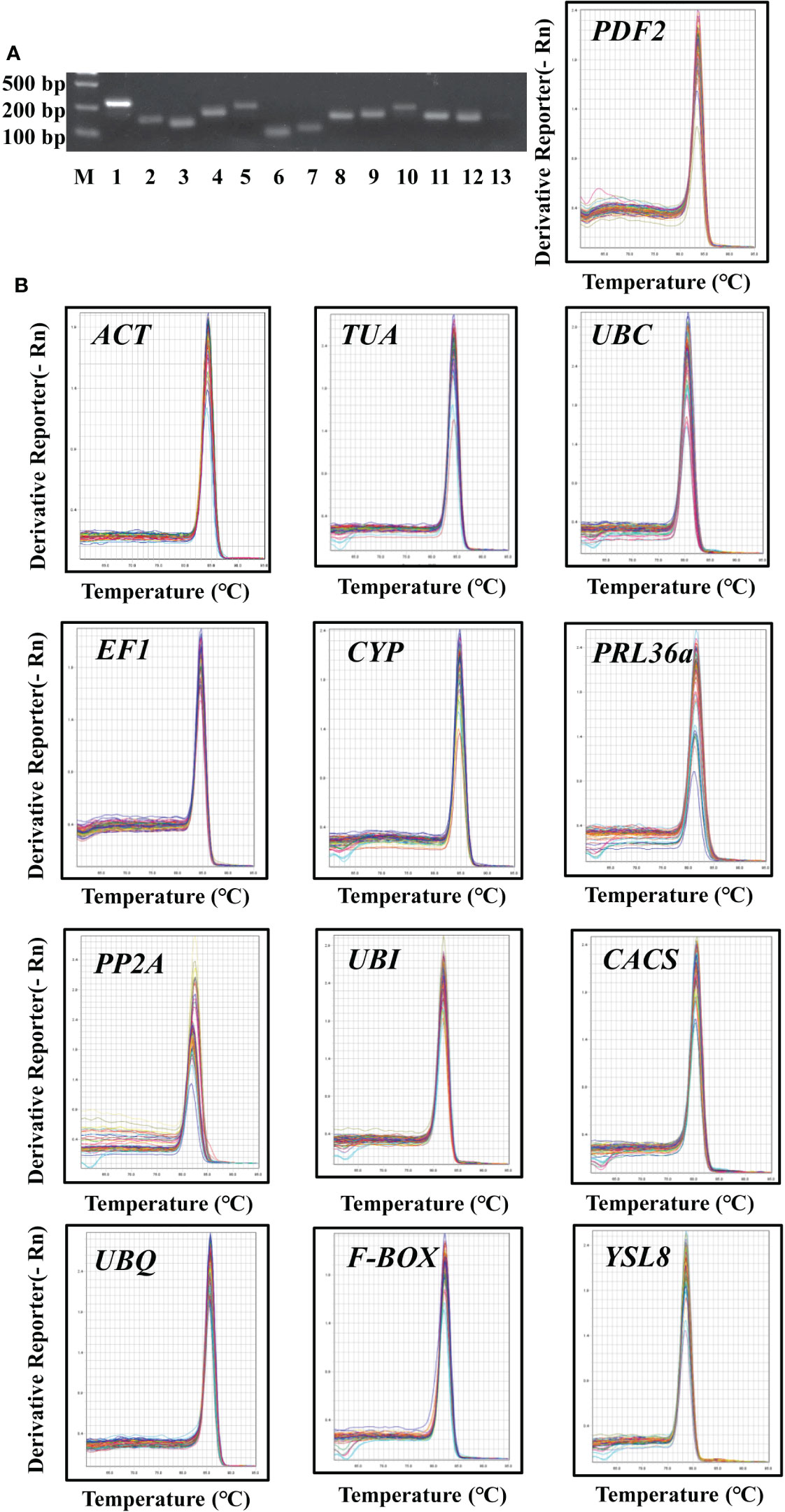
Figure 1 Primer specificity of candidate reference genes. (A), Agarose gel analysis of RT-PCR generated amplicons for each of the thirteen assessed candidate reference genes. M, marker; 1-13, ACT, TUA, UBC, EF1, CYP, PRL36Aa, PP2A, UBI, CACS, UBQ, F-BOX, YSL8, PDF2. (B), Melt curve analysis of the thirteen assessed candidate reference genes across different samples in cucumber roots showed a single peak for each primer pair at a specific annealing temperature.
Gene expression stability analysis
A boxplot of cycle threshold (CT) values to evaluate the expression levels of thirteen candidate reference genes. To evaluate the expression stability of thirteen candidate reference genes under different experiment treatments, four statistical algorithms, including geNorm (Vandesompele et al., 2002), NormFinder (Andersen et al., 2004), BestKeeper (Pfaffl et al., 2004), and RefFinder, were used. In the geNorm program, M value represents the expression stability of calculated genes, and V value represents the pair average variation of measured genes. The lower the M value of the reference gene, the higher expression stability, and the M value ≥1.5 indicates that the expression of candidate genes is unstable (Vandesompele et al., 2002). The NormFinder program firstly obtains the expression stability value of each gene, then selects the most appropriate reference gene according to the stability value of candidate genes (Andersen et al., 2004). BestKeeper can be used to compare the expression levels of 10 candidate reference genes in 100 samples at most. BestKeeper can calculate the standard deviation (SD) and coefficient of variation (CV) of each gene and determine the expression stability of candidate genes by comparing SD and CV (Pfaffl et al., 2004). Finally, a comprehensive ranking of candidate genes was produced by RefFinder (Duan et al., 2017). RefFinder is a web-based tool that integrates geNorm, Normalfinder, BestKeeper and ΔCT method program, which provides the overall ranking of candidate genes through geometric average of attributed weights of each algorithm.
Result
Primer specificity of the candidate reference genes
In order to detect the specificity of primers, all primers were tested using RT-PCR approach and the construction of melt curves. For each prime pair, a single amplicon was observed by agarose gel analysis (Figure 1A). The melting curve of DNA refers to the curve of the degradation degree of DNA double helix structure with the increase of temperature. The melting curve was generated by monitoring the fluorescence signal. Different DNA sequence has different melt temperature (Tm) value, so it represents the specificity of amplification products. In this study, the amplification Tm value of each candidate gene was analyzed by qRT-PCR, thirteen candidate genes had specific characteristic peak at melting curve, respectively (Figure 1B). These results indicated that each primer pair of thirteen candidate genes was specific to targeted region.
Expression analysis of candidate reference genes
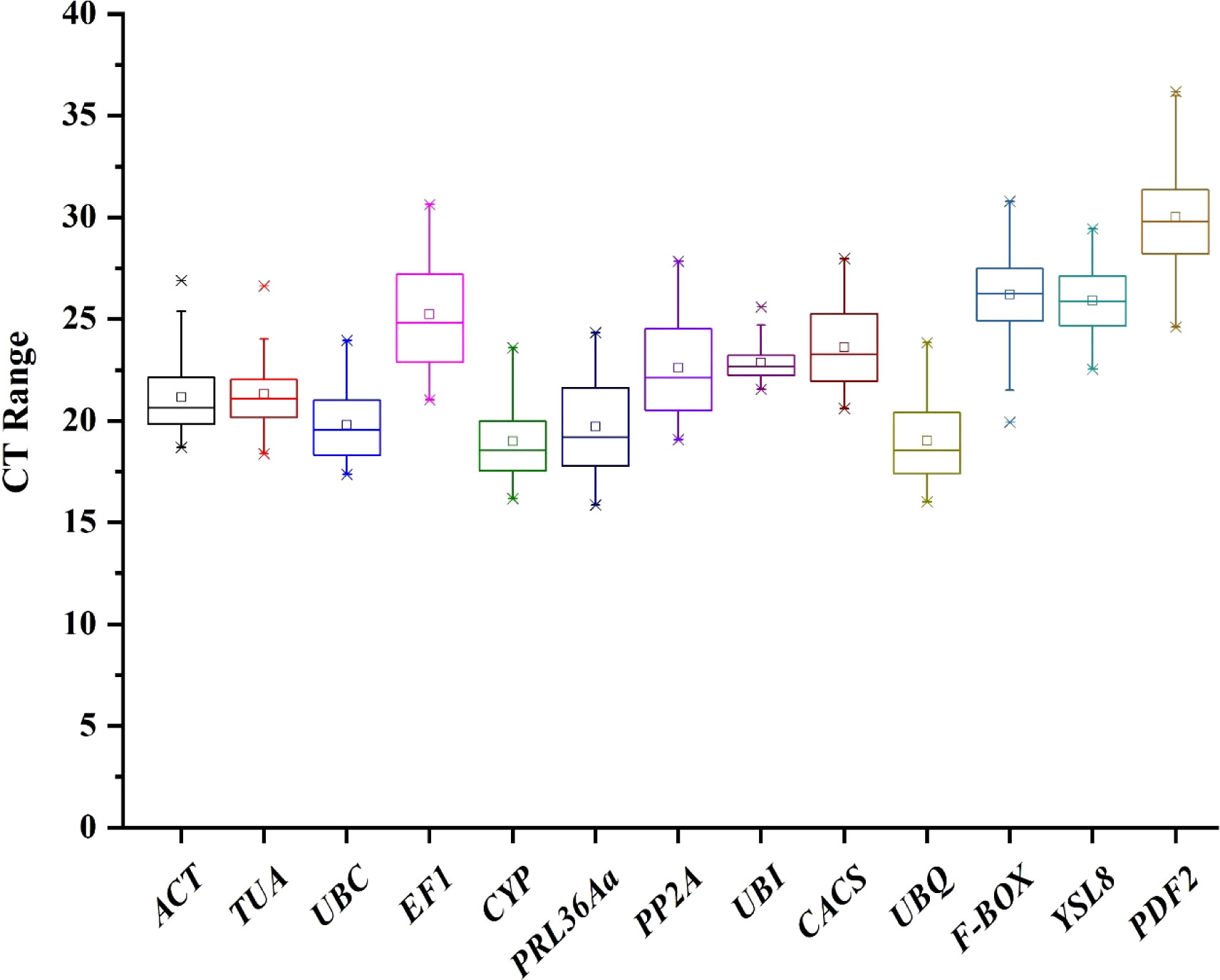
In order to evaluate the expression stability of thirteen candidate genes, expression change of candidate genes under three experimental conditions were detected: Treatment 1, cucumber root knots were sampled after infected by M. incognita; Treatment 2, cucumber whole roots were sampled after infected by M. incognita; Treatment 3, cucumber whole roots were sampled after inoculated by Pseudomonas. The quantification cycle (Ct) values of thirteen candidate genes were used for data analysis in Figure 2. Ct value indicates the transcriptional quantity and with a lower Ct value indicating a more abundant target transcript. The Ct values of thirteen candidate genes in three treatments were calculated, respectively (Supplement Table 1). In treatment 1, CYP with the lowest mean Ct value (17.54), 28.11 for PDF2 with the highest mean Ct value, and other eleven gens were primary distributed between 17 and 25. In treatment 2, similar trend was found that CYP and PDF2 with the lowest (20.38) and highest (30.98) average Ct value, respectively. However, in treatment 3, the smallest mean Ct value was 17.76 for UBQ and the highest mean Ct value 32.37 for PDF2. The box plot in Figure 2 provides an expression level overview of thirteen candidate genes combining three treatments, the results showed that the mean Ct value varied from 18.99 for CYP, to 30.01 for PDF2.
The expression stability of candidate reference genes
To identify the most suitable reference genes with stable expression across all assessed samples in experiment, three frequently used statistical algorithms, containing geNorm, NormFinder and BestKeeper, were applied. geNorm evaluates reference gene average suitability value (M) based on the relative quantitative data (2-ΔCt, ΔCt = each corresponding Ct value - the minimum Ct value) of each candidate gene (Vandesompele et al., 2002). The lower value of M reflected more stable gene expression. In treatment 1, based on the geNorm program, the stability ranking of 13 tested candidate genes was: TUA/UBC > ACT > UBI > EF1 > UBQ > CACS > PP2A > PRL36Aa > CYP > PDF2 > YSL8 > F-BOX (Figure 3A). Among the thirteen candidate genes, TUA and UBC genes with low M values were the most stable candidate genes, while of F-BOX gene with high M values was the most unstable candidate gene compared with the other twelve genes. For whole roots infected by nematode (Treatment 2), the order of gene expression stability was: UBC/RPL36Aa > UBI > ACT > TUA > CACS > PP2A > PDF2 > EF1 > UBQ > F-BOX > CYP > YSL8 (Figure 3B). PL36Aa and UBC genes were the most stable genes, but the expression of YSL8 was unstable as the M value ≥ 1.5 (Vandesompele et al., 2002), thus YSL8 was not suitable as a reference gene in this treatment. For Pseudomonas treatment (Treatment 3), the order of gene expression stability was: UBC/PP2A > CACS >TUA > F-BOX > CYP > UBI > ACT > RPL36Aa > PDF2 > UBQ > YSL8 > EF1 (Figure 3C). Based on the M values, UBC and PP2A were the most stable genes, while EF1 was the worst one compared with the other twelve candidate genes. The pairwise variation value (Vn/Vn+1) could be calculated by geNorm, which determined the optimal number of reference genes. According to Vandesompele et al. (2002), Vn/Vn+1<0.15 indicates the optimal number of reference genes is achieved. In treatment 1, except for V2/3 and V3/4, the other value of Vn/Vn+1<0.15, V2/3 was already below the threshold in treatment 2 and treatment 3 (Figure 4).
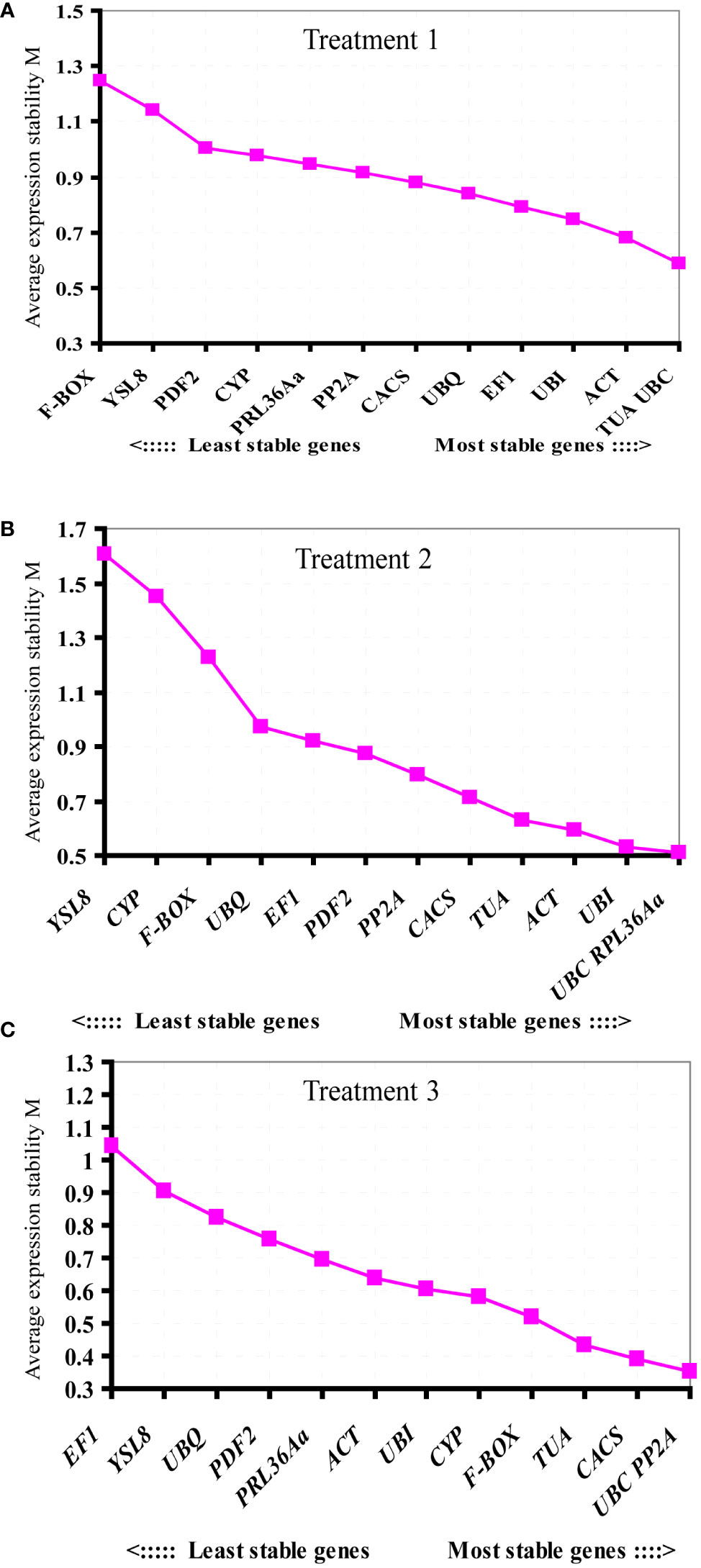
Figure 3 Ranking of the candidate reference genes according to geNorm analysis. The M value showing gene expression stability was calculated in three different treatments. (A) root knot treated by M. incognita (Treatment 1), (B) whole root treated by M. incognita (Treatment 2), and (C) whole root treated by Pseudomonas (Treatment 3).
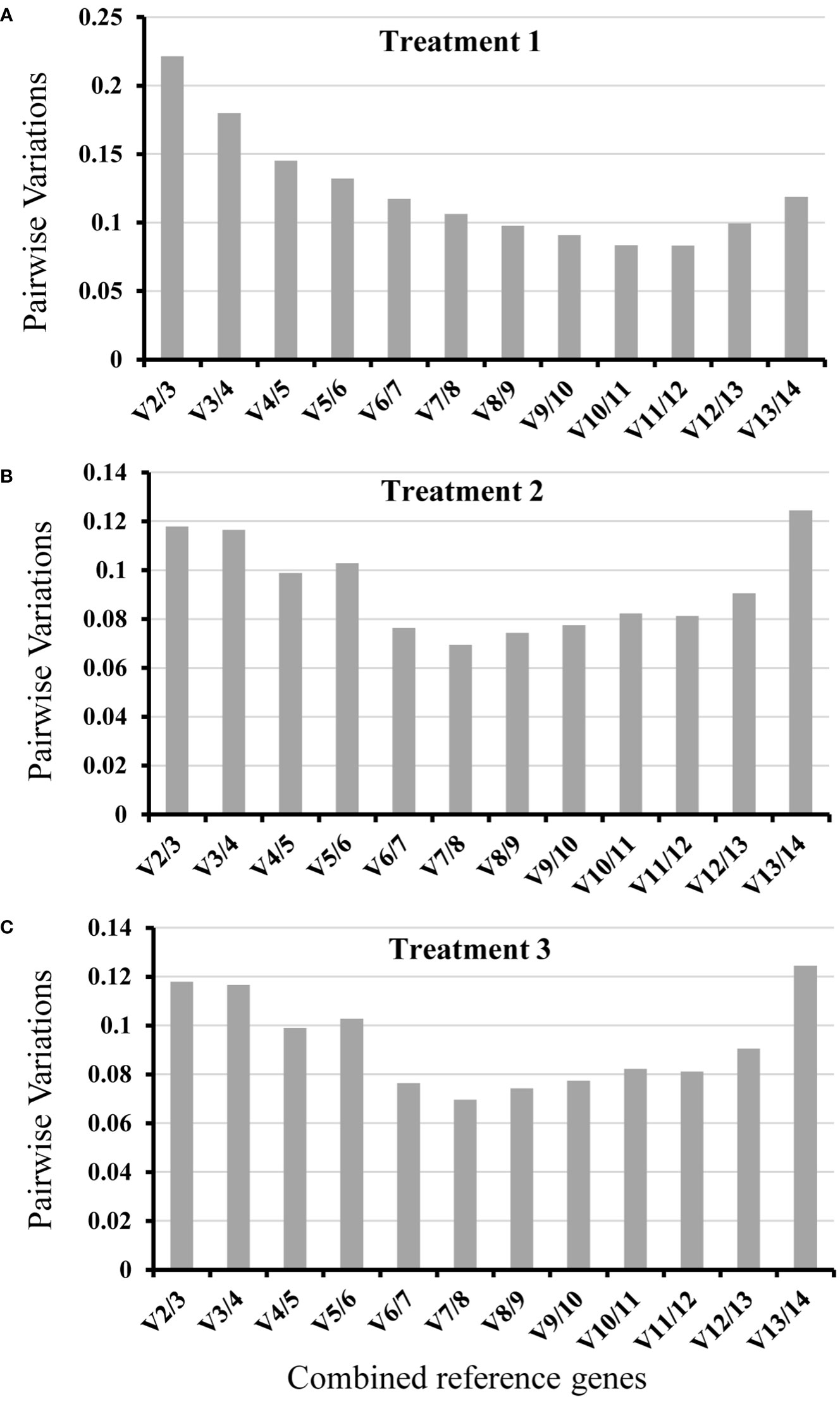
Figure 4 Pairwise variation (V) of candidate reference genes calculated by geNorm. The Vn/Vn+1 value calculated reflecting gene expression stability of thirteen candidate reference genes in three different treatments. (A), Treatment 1: root knot samples caused by M. incognita; (B), Treatment 2: whole root infected by M. incognita; (C), Treatment 3: whole root treated by Pseudomonas.
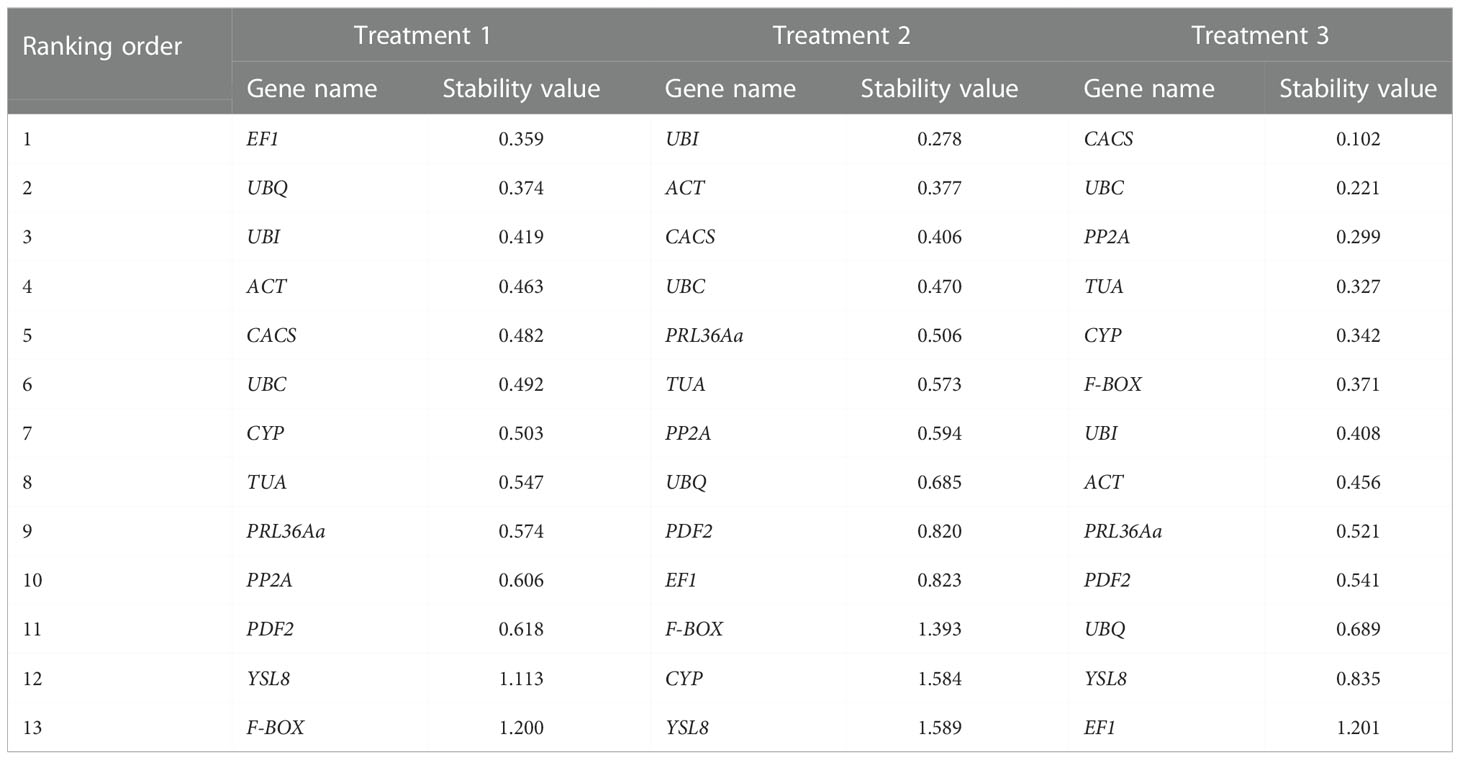
Another program for analyzing the stability of reference genes, NormFinder uses a mathematical model to describe the expression values measured by RT-PCR, analyzes the sample subgroups separately, estimates the expression changes within and between groups, and then synthesizes into a stable value, that is the stable value of candidate gene. The judgment standard is that the most appropriate candidate gene showed minimum value calculated by NormFinder. In treatment 1, based on the NormFinder program analysis, the stability sequence of the thirteen tested candidate genes was: EF1 > UBQ > UBI > ACT > CACS > UBC > CYP > TUA > PRL36Aa > PP2A > PDF2 > YSL8 > F-BOX (Table 2). Among the thirteen genes, EF1 was the most stable gene with the lowest value 0.357, and F-BOX was the most unreliable one. In treatment 2, the expression stability of candidate gene was: UBI > ACT > CACS > UBC > PRL36Aa > TUA > PP2A > UBQ > PDF2 > EF1 > F-BOX > CYP > YSL8 (Table 2). The results showed that UBI was the most reliable gene with the lowest value of 0.278, and the maximum value of YSL8 was 1.589, which was the most unreliable gene compared with the other candidate genes. In treatment 3, the sequence of candidate gene expression stability was: CACS > UBC > PP2A > TUA > CYP > F-BOX > UBI > ACT > PRL36Aa > PDF2 > UBQ > YSL8 > EF1 (Table 2). Among the thirteen genes, CACS with the minimum value 0.102 suggested it was the most reliable gene, while the value of EF1 was the highest (1.201).
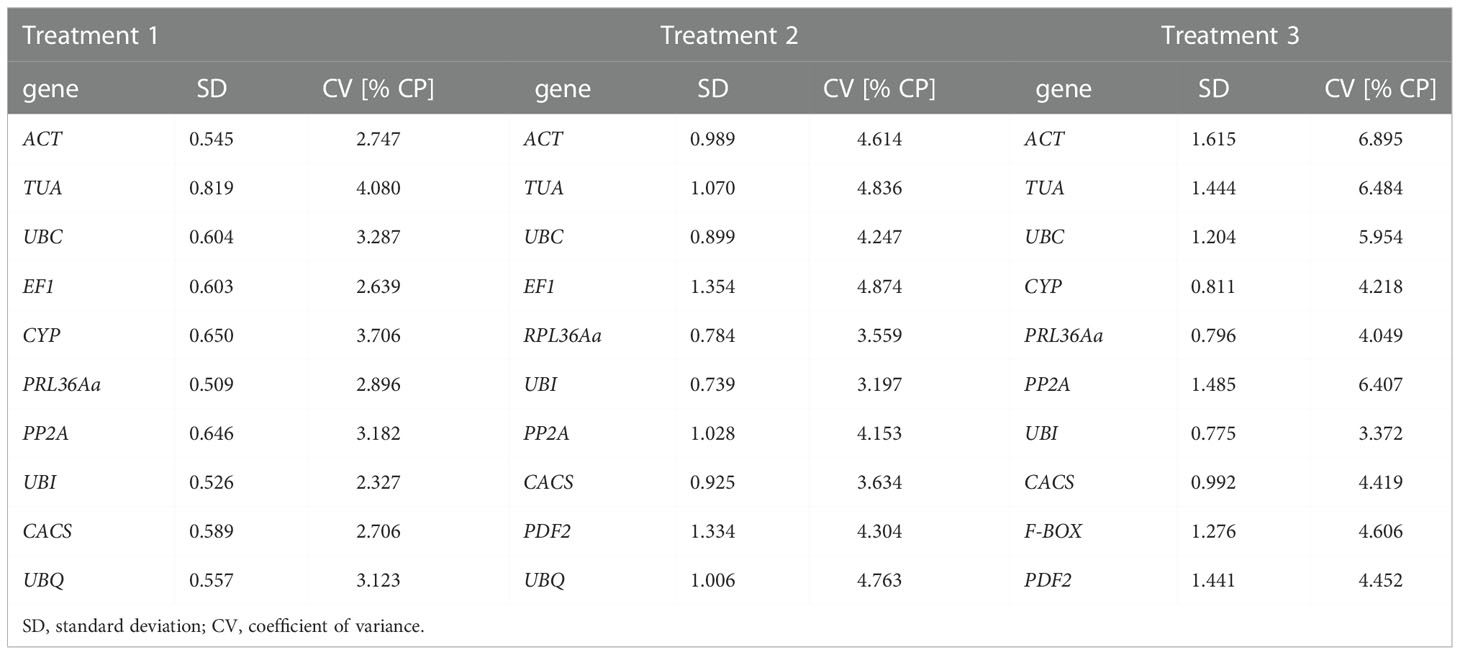
BestKeeper program can only compare the expression levels of up to 10 candidate genes in 100 samples, generally, the analysis of candidate genes by BestKeeper program is based on geNorm program and NormFinder. The criterion of BestKeeper program for determining the stability of candidate genes was that the smaller SD (Standard Deviation) and CV (Coefficient of variation), the better stability. When SD > 1, the expression of candidate gene was unstable. Based on geNorm Program and NormFinder analysis, the top-ranked ten candidate genes were selected for further analyzing by BestKeeper. In treatment 1, top ten candidate genes, including UBI, PRL36Aa, ACT, UBQ, CACS, EF1, UBC, PP2A, CYP and TUA, were selected for further analysis. In treatment 2, ten genes, including ACT, TUA, UBC, EF1, RPL36Aa, UBI, PP2A, CACS, PDF2, UBQ, were selected for BestKeeper analysis. In treatment 3, ten candidate genes including ACT, TUA, UBC, CYP, PRL36Aa, PP2A, UBI, CACS, F-BOX, PDF2 for further study (Table 2, Table 3).
Based on BestKeeper program, in treatment 1, the stability sequence of the ten tested candidate reference genes was PRL36Aa > UBI > ACT > UBQ > CACS > EF1 > UBC > PP2A > CYP > TUA, among them, PRL36Aa gene was the most stable reference gene with the minimum value of SD compared with other genes (Table 3, Table 4). In treatment 2, the stability order of the top ten candidate genes was: UBI > PRL36Aa > CACS > UBC > ACT > UBQ > PP2A > TUA > PDF2 > EF1 (Table 3, Table 4). According to the BestKeeper program analysis, the expression of UBI, RPL36Aa, UBC, CACS and ACT were stable with SD < 1, and UBI was the most stable one. In treatment 3, these genes expression stability order from high to low was: UBI > PRL36Aa > CYP > CACS > UBC > F-BOX > PDF2 > TUA > PP2A > ACT (Table 3, Table 4). Among them, the expression of UBI, PRL36Aa and CYP were reliable and UBI was the most reliable one. On the contrary, the expression of F-BOX, UBC, CACS, TUA, PP2A, ACT and PDF2 were unstable in treatment 3.
Comprehensive evaluation of the expression stability of candidate reference genes
Three specialized analysis programs, NormFinder, GeNorm, and BestKeeper, gave the similar but slightly different ranks for thirteen tested candidate genes. To normalize the gradate, a comprehensive analysis that regraded the expression stability of thirteen tested genes was performed. RefFinder was selected to make the comprehensive analysis. According to RefFinder analysis, in treatment 1, the order of candidate reference gene stability was: EF1 > UBI > ACT > TUA > CACS > UBC > PRL36Aa > PP2A > CYP > UBQ > PDF2 > YSL8 > F-BOX (Table 4). Among thirteen genes, EF1 was the most stable reference gene and F-BOX was the most unstable one compare with the others candidate genes. In treatment 2, the order of candidate reference gene was: UBI > PRL36Aa >UBC > ACT > CACS > PP2A > UBQ > TUA > EF1 > PDF2 > F-BOX > CYP > YLS8 (Table 4). Among them, UBI was the most reliable reference gene, while YLS8 was the most unreliable gene. In treatment 3, the reliability sequence was: CACS > CYP > UBI > UBC > PRL36Aa > PP2A > TUA > F-BOX > ACT > PDF2 > UBQ > YSL8 > EF1 (Table 4), CACS was the most stable one, while EF1 was the worst one compared with other candidate genes. These results indicated that UBI and CACS were relatively stable genes compared with other candidate genes and they could be extensive used in three different treatments.
Validation of selected candidate reference genes
To validate the suitability of the suitable reference gens, the top two and the bottom two ranked genes following RefFinder analysis were selected to generate an expression profile for two genes in each one treatment. CsJAZ3 and CsPIN2 expression were quantified across all root knots and whole roots samples infected by M. incognita using selected candidate reference genes (the top two: EF1 and UBI in treatment 1, UBI and PRL36Aa in treatment 2; the bottom two: YSL8 and F-BOX in treatment 1, CYP and YLS8 in treatment 2); CsTIP1 and CsSOS1 expression were quantified in whole root samples inoculated by Pseudomonas through the top two (CACS and CYP) and the bottom two (YSL8 and EF1). When applying the two best candidate reference genes for expression normalization, qRT-PCR revealed that CsJAZ3 and CsPIN2 transcript abundance increased and decreased, respectively, in root knot and whole root samples at 14-day after M. incognita infection (Figure 5A, C); CsTIP1 and CsSOS1 expressions were decreased and increased, respectively, in whole roots inoculated by Pseudomonas. When the two best candidate reference genes combination was used to normalize CsJAZ3/CsPIN2 and CsTIP1/CsSOS1 expression in cucumber roots treated by M. incognita and Pseudomonas, respectively, similar expression profiles were observed (Figure 5A, C).
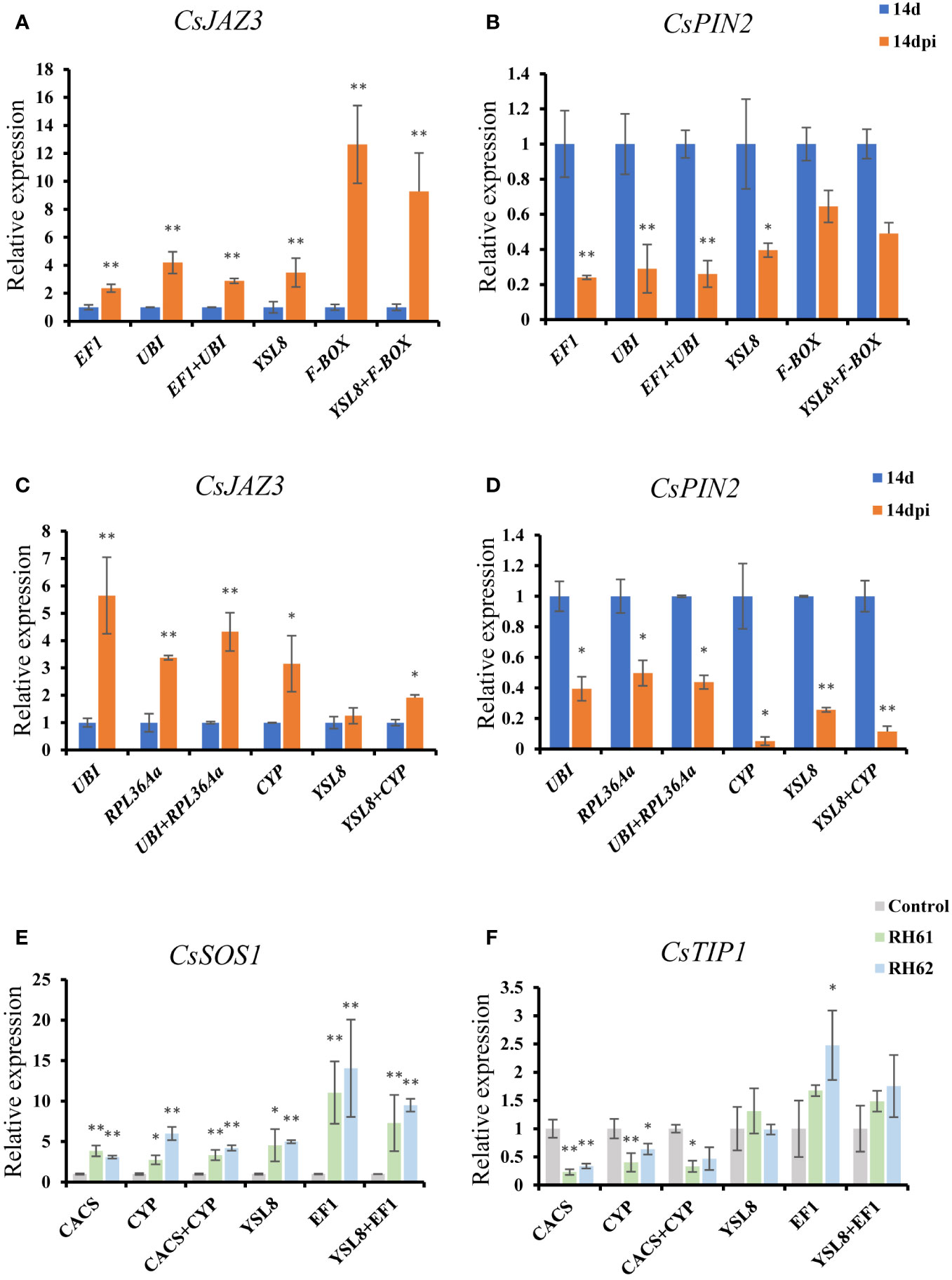
Figure 5 Profiling of CsJAZ3, CsPIN2, CsTIP1 and CsSOS1 expression in cucumber roots of treatment 1, 2 and 3 using the most suitable and least suitable sets of candidate reference genes. (A, B), normalized CsJAZ3 and CsPIN2 expression using the most suitable candidate reference genes (EF1 and UBI) and the least suitable candidate genes (YSL8 and F-BOX) in treatment 1; (C, D), normalized CsJAZ3 and CsPIN2 expression using the most suitable candidate reference genes (UBI and RPL36Aα) and the least suitable candidate genes (YSL8 and CYP) in treatment 2; (E, F), normalized CsTIP1 and CsSOS1 expression using the most suitable candidate reference genes (CACS and CYP) and the least suitable candidate genes (YSL8 and EF1) in treatment 3. Abbreviation: 14 d, 14 day; 14 dpi, 14 days post infection by M. incognita. RH61 and RH62 are two kinds of Pseudomonas. "*"and"**" indicate a significant difference from the those based on t-test, with a P-value of 0.05 and 0.01 respectively.
Besides, the least stably candidate genes ranked by RefFinder also be used for qRT-PCR normalization analysis. Interestingly, when YSL8 as reference gene to normalize the expression of CsJAZ3 and CsPIN2, the results were similar with the top two candidate reference genes used in treatment 1 and 2; in treatment 1, when F-BOX as reference gene, CsJAZ3 expression increased sharply and CsPIN2 expression showed no change in root knot samples (Figure 5B); in treatment 2, CsJAZ3 and CsPIN2 expression profile showed similar result with the top two candidate reference genes when used CYP individual and CYP/YSL8 combination as reference genes for normalization (Figure 5D); in treatment 3, when YSL8 and EF1 as candidate reference genes, CsTIP1 expression had no change with or without Pseudomonas treatment, while CsSOS1 transcript level was increased like the results of CACS/CYP as reference genes (Figure 5E, F), and similar results were also showed when using YSL8 and EF1 combination as reference genes for normalization (Figure 5E, F).
Discussion
Cucumber as an economical vegetable is widely cultivated worldwide and is subject to serious nematode infection, especially from M. incognita. Plant could employ beneficial soil bacteria in the rhizosphere to enhance plant adaptability to various stresses, and Pseudomonas had been confirmed that play a role in increasing plant tolerance to abiotic stress (Li et al., 2021). It is important to add the knowledge regarding the response mechanism of plant to infection by M. incognita and the regulation mechanism by beneficial soil bacteria Pseudomonas. qRT-PCR is an important and convenient tool for detecting gene expression and gene function. However, selecting appropriate reference genes is necessary to ensure the stability and accuracy of qRT-PCR. In this study, the optimal reference genes in cucumber under M. incognita stress and Pseudomonas treatment were first calculated and confirmed.
Thirteen candidate reference genes ACT (Actin), TUA (alpha-tubulin), UBC (Ubiquitin conjugating enzyme), EF1 (Elongation factor 1-alpha), CYP (Cyclophilin), PRL36Aa (60S ribosomal protein L36a/L44), PP2A (protein phosphatase 2A regulatory subunit A), UBI (Ubiquitin-like protein), CACS (Clathrin adaptor comple subunit), UBQ (Polyubiquitin), F-BOX (F-box protein), YSL8 (mitosis protein), PDF2 (Protein phosphatase 2) were selected in this study (Warzybok and Migocka, 2013; Li et al., 2013; Gan et al., 2017). The stability orders of candidate reference genes were showed according to four
The stability of candidate reference genes was analyzed according to four algorithms and a comprehensive order regraded the expression stability of thirteen tested genes was showed. In treatment 1, EF1 and UBI were the top two suitable candidate reference genes, and YSL8 and F-BOX were the least suitable genes according to RefFinder analysis. Thus EF1, UBI, YSL8 and F-BOX were selected as reference genes to generate expression profiling for CsJAZ3 and CsPIN2. The results showed that the expression trend of CsJAZ3 and CsPIN2 were consistent when using EF1, UBI, YSL8 as reference genes, while their expression is different when F-BOX as reference gene (Figure 5). In addition, the result that CsJAZ3 and CsPIN2 expression were increased and decreased, respectively, is consistent with their orthologous gene expression in tomato and Arabidopsis after M. incognita infection (Hammes et al., 2005; Shukla et al., 2018). YSL8 was the bottom two in the order of thirteen candidate genes in treatment 1, however, the M value of YSL8 evaluated by geNorm was less than 1.5 which is a stability evaluation value. It is reasonable to speculate that, except F-BOX, other twelve candidate genes might be used as HKG in treatment 1 experiment. In treatment 2, CYP and YSL8 were the bottom two candidate genes according to comprehensive analysis, the result was different when CYP as reference gene for normalization (Figure 5). Although the expression trend of CsJAZ3 and CsPIN2 was correspond when the top two candidate genes (UBI and RPL36Aa) as reference gene and when YSL8 as reference gene to normalize, YSL8 still was not a good choice because YSL8 even ranked latter than CYP and its M value is bigger than 1.5 which represents YSL8 is an unreliable reference gene. In addition, ACT are suitable reference genes in root knot and whole root samples after M. incognita infection, but they were not suitable for a reference gene under Pseudomonas treatment (Table 4, Figure 3). In previous research, ACT has been employed to normalize gene expression in several species infected by nematode, including Arabidopsis and tomato (Hewezi et al., 2010; Uehara et al., 2010; Vijayapalani et al., 2018; Xu et al., 2019; Huang et al., 2022), although none of the research involves HKGs screening in nematode infection experiment. It might be reasonable to speculate that ACT can be used as a universal HKG to normalize gene expression level in different species under nematode infection treatment.
Pseudomonas had been certified that could enhance plant adaptability to salt stress through inducing the production of stress alleviating metabolites, thus two salt-response related genes, CsTIP1 (aquaporin protein) and CsSOS1 (salt overly sensitive gene), were selected to identify the stability of candidate reference genes. In treatment 3, M values of thirteen candidate genes were less than 1.5, however, the bottom two genes (YSL8 and EF1) as reference genes for normalization showed different expression of CsTIP1and CsSOS1 compared with the top two candidate genes (CACS and CYP). Different from above result, EF1 could as a suitable gene in Arabidopsis thaliana, Nicotiana benthamiana and Solanum tuberosum under abiotic stress including heat, cold, hormones, dehydration and biotic stress including Pseudoperonospora cubensis (Nicot et al., 2005; Remans et al., 2008; Catinot et al., 2008). EF1 in cucumber is also the most reliable expression of reference gene in leaves, stems and roots under Cucumber green mottle mosaic virus (CGMMV) treatment (Liang et al., 2018). When CACS and CYP as reference genes to normalize the expression of CsTIP1 and CsSOS1, the results were consistent with that TIP1 and SOS1 were reduced and induced, respectively, by salt stress (Zhu et al., 2009; Zhang et al., 2020), and the result also provided molecular evidence for Pseudomonas inoculation could improve plant tolerance for salt stress. These results suggested that HKG cannot be utilized universally across different plant species, experimental conditions, or even different environmental conditions with a single species.
Recently, new reference gene CsARF (ADP ribosylation factor 1) in cucumber had been identified, its expression was stable and could be a suitable housekeeping gene in cucumber stems infected by Pectobacterium Brasiliense (Yuan et al., 2022). In addition to those identified reference genes, continuing exploration and validation of new housekeeping genes being pushed forward for different organs, various developmental stages, under different environmental stresses of plants, thus data acquired by qRT-PCR would be more accuracy and reliable.
In conclusion, EF1 is the most suitable reference gene in the root knot samples of cucumber infected by root knot nematode, although EF1 has never been considered in previous plant-nematode studies. UBI is the most suitable reference gene in the whole root samples of cucumber under root-knot nematode stress, and CACS is the most suitable reference gene in the whole root samples of cucumber treated by Pseudomonas. Through BestKeeper, NormFinder, and geNorm analysis, UBI, ACT and CACS were used simultaneously as reference genes no matter in whole root or root knot samples of cucumber infected by M. incognita. CACS, CYP and UBI were suitable reference genes in Pseudomonas treated cucumber whole root samples. Among them, UBI and CACS are relatively stable and could be utilized in three experiments conditions.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
YT and LG conceived and designed the experiments. TJ, SM, XW and ML performed the experiments and collected the data. TJ and SM executed the data analyses. All authors contributed to the interpretation of the results. TJ, SM, YT and LG wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the National Natural Science Foundation of China (No. 31972478), National Key Research and Development Program of China (No. 2019YFD1001900), the China Agriculture Research System (NO. CARS-23) and Beijing Innovation Consortium of Agriculture Research System (NO. BAIC01-2022).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1061921/full#supplementary-material
References
Andersen, C. L., Jensen, J. L., Orntoft, T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. doi: 10.1158/0008-5472.CAN-04-0496
Catinot, J., Buchala, A., Abou-mansour, E., Métraux, J. P. (2008). Salicylic acid production in response to biotic and abiotic stress depends on isochorismate in Nicotiana benthamiana. FEBS Lett. 582, 473–478. doi: 10.1016/j.febslet.2007.12.039
Chen, W., Shao, J., Ye, M., Yu, K., Bednarek, S., Duan, X., et al. (2017). Blueberry VcLON 2, a peroxisomal LON protease, is involved in abiotic stress tolerance. Environ. Exp. Bot. 134, 1–11. doi: 10.1016/j.envexpbot.2016.10.008
Duan, M., Wang, J., Zhang, X., Yang, H., Wang, H., Qiu, Y., et al. (2017). Identification of optimal reference genes for expression analysis in radish (Raphanus sativus l.) and its relatives based on expression stability. Front. Plant Sci. 8, 1605. doi: 10.3389/fpls.2017.01605
Gan, D., Zhan, M., Yang, F., Zhang, Q., Hu, K., Xu, W., et al. (2017). Expression analysis of argonaute, dicer-like, and RNA-dependent RNA polymerase genes in cucumber (Cucumis sativus l.) in response to abiotic stress. J. Genet. 96 (2), 235–249. doi: 10.1007/s12041-017-0758-y
Hammes, U. Z., Schachtman, D. P., Berg, R. H., Nielsen, E., Koch, W., McIntyre, L. M., et al. (2005). Nematode-induced changes of transporter gene expression in arabidopsis roots. Mol. Plant-Microbe Interact. 18 (12), 1247–1257. doi: 10.1094/MPMI-18-1247
Hewezi, T., Howe, P. J., Maier, T. R., Hussey, R. S., Mitchum, M. G., Davis, E. L., et al. (2010). Arabidopsis spermidine synthase is targeted by an effector protein of the cyst nematode Heterodera schachtii. Plant Physiol. 152 (2), 968–984. doi: 10.1104/pp.109.150557
Huang, S., Li, R., Zhang, Z., Li, L., Gu, X., Fan, W., et al. (2009). The genome of the cucumber, Cucumis sativus l. Nat. Genet. 41, 1275–1281. doi: 10.1038/ng.475
Huang, H., Zhao, W., Qiao, H., Li, C., Sun, L., Yang, R., et al. (2022). SlWRKY45 interacts with jasmonate-ZIM domain proteins to negatively regulate defense against the root-knot nematode Meloidogyne incognita in tomato. Horticult. Res. 9, uhac197. doi: 10.1093/hr/uhac197
Jaiteh, F., Kwoseh, C., Akromah, R. (2012). Evaluation of tomato genotypes for resistance to root-knot nematodes. Afr. Crop Sci. J. 20, 41–49. doi: 10.4314/ACSJ.V20I1
Jones, L. M., De Giorgi, C., Urwin, P. E. (2011). “C. elegans as a resource for studies on plant-parasitic nematodes,” in Genomics and molecular genetics of plant–nematode interactions. Eds. Gheysen, G., Fenoll, C., Jones, J. T. (UK: Springer Science and Business Media, B.V), 175–220. doi: 10.1007/978-94-007-0434-3_10
Joseph, J. T., Poolakkalody, N. J., Shah, J. M. (2018). Plant reference genes for development and stress response studies. J. Biosci. 43 (1), 173–187. doi: 10.1007/s12038-017-9728-z
Kopczewski, T., Kuźniak, E., Kornaś, A., Rut, G., Nosek, M., Ciereszko, I., et al. (2022). Local and systemic changes in photosynthetic parameters and antioxidant activity in cucumber challenged with Pseudomonas syringae pv lachrymans. Int. J. Mol. Sci. 21 (17), 6378. doi: 10.3390/ijms21176378
Kumar, M., Etesami, H., Kumar, V. (2019). Saline soil-based agriculture by halotolerant microorganisms (Singapore: Springer Nature Singapore Pte Ltd).
Le, D. T., Aldrich, D. L., Valliyodan, B., Watanabe, Y., Ha, C.V., Nishiyama., R, et al. (2012). Evaluation of candidate reference genes for normalization of quantitative RT-PCR in soybean tissues under various abiotic stress conditions. PloS One 7, e46487. doi: 10.1371/annotation/6a5108f5-50f8-418e-854d-8f3eb94e6fc0
Liang, C., Hao, J., Meng, Y., Luo, L., Li, J. (2018). Identifying optimal reference genes for the normalization of microRNA expression in cucumber under viral stress. PloS One 13 (3), e0194436. doi: 10.1371/journal.pone.0194436
Li, P., Chen, L., Zhou, Y., Xia, X., Shi, K., Chen, Z., et al. (2013). Brassinosteroids-induced systemic stress tolerance was associated with increased transcripts of several defence-related genes in the phloem in Cucumis sativus. PloS One 8, 1–8. doi: 10.1371/journal.pone.0066582
Li, H., La, S., Zhang, X., Gao, L, Tian, Y. (2021). Salt-induced recruitment of specific root-associated bacterial consortium capable of enhancing plant adaptability to salt stress. Multidiscip. J. Microbial. Ecol. J. 15 (10), 2865–2882. doi: 10.1038/s41396-021-00974-2
Li, X., Ma, S., Shan, N., Zhang, X. L., Sui, X. L., Zhang, Z. X.. (2017). A protocol for agrobacterium-mediated transformation of cucumber (Cucumis sativus l.) from cotyledon explants. Protocol. exch. 6159, 1–19. doi: 10.1038/protex.2017.107
Lü, J., Sui, X., Ma, S., Li, X., Liu, H., Zhang, Z.. (2017). Suppression of cucumber stachyose synthase gene (CsSTS) inhibits phloem loading and reduces low temperature stress tolerance. Plant Mol. Biol. 95 (1-2), 1–15. doi: 10.1007/s11103-017-0621-9
Ma, S., Sun, L. L., Sui, X. L., Li, Y., Chang, Y., Fan, J., et al. (2019). Phloem loading in cucumber: combined symplastic andapoplastic strategies. Plant J. 98, 391–404. doi: 10.1111/tpj.14224
Mbaluto, C. M., Ahmad, E. M., Fu, M., Martínez-Medina, A., van Dam, N. M. (2020). The impact of spodoptera exigua herbivory on Meloidogyne incognita induced root responses depends on the nematodeslife cycle stages. AoB Plants 12, plaa029. doi: 10.1093/aobpla/plaa029
Miao, L., Qin, X., Gao, L. H., Li, Q., Li, S., He, C., et al. (2019). Selection of reference genes for quantitative real-time PCR analysis in cucumber (Cucumis sativus l.), pumpkin (Cucurbita moschata duch.) and cucumber-pumpkin grafted plants. Peer J. 7, e6536. doi: 10.7717/peerj.6536
Migocka, M., Papierniak, A. (2011). Identification of suitable reference genes for studying gene expression in cucumber plants subjected to abiotic stress and growth regulators. Mol. Breed. 28 (3), 343–357. doi: 10.1007/s11032-010-9487-0
Moens, M., Perry, R. N. (2009). Migratory plant endoparasitic nematodes: a group rich in contrasts and divergence. Annu. Rev. Phytopathol. 47, 313–332. doi: 10.1146/annurev-phyto-080508-081846
Nicot, N., Hausman, J. F., Hoffmann, L., Evers, D. (2005). Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 56, 2907–2291. doi: 10.1093/jxb/eri285
Niu, H., Liu, X., Tong, C., Wang, H., Li, S., Lu, L., et al. (2018). The WUSCHEL-related homeobox1 gene of cucumber regulates reproductive organ development. J. Exp. Bot. 69 (22), 5373–5387. doi: 10.1093/jxb/ery329
Nolan, T., Hands, R. E., Bustin, S. A. (2006). Quantification of mRNA using real-time RT- PCR. Nat. Protoc. 1, 1559–1582. doi: 10.1038/nprot.2006.236
Olaetxea, M., Mora, V., Bacaicoa, E., Garnica, M., Fuentes, M., Casanova, E., et al. (2015). Abscisic acid regulation of root hhydraulic conductivity and aquaporin gene expression is crucial to the plant shoot growth eenhancement caused by rhizosphere humic acids. Plant Physiol. 169 (4), 2587–2596. doi: 10.1104/pp.15.00596
Petriccione, M., Mastrobuoni, F., Zampella, L., Scortichini, M. (2015). Reference gene selection for normalization of RT-qPCR gene expression data from actinidia deliciosa leaves infected with Pseudomonas syringae pv. Sci. Rep. 5, 1696. doi: 10.1038/srep16961
Pfaffl, M. W., Tichopad, A., Prgomet, C., Neuvians, T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: best-keeper- excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515. doi: 10.1023/B:BILE.0000019559.84305.47
Qin, N., Gao, Y., Cheng, X., Yang, Y., Wu, J., Wang, J., et al. (2021). Genome-wide identification of CLE gene family and their potential roles in bolting and fruit bearing in cucumber (Cucumis sativus l.). BMC Plant Biol. 21 (1), 143. doi: 10.1186/s12870-021-02900-2
Rajsz, A., Warzybok, A., Migocka, M. (2016). Genes encoding cucumber full-size ABCG proteins show different responses to plant growth regulators and sclareolide. Plant Mol. Biol. Rep. 34, 720–736. doi: 10.1007/s11105-015-0956-9
Remans, T., Smeets, K., Opdenakker, K., Mathijsen, D., Vangronsveld, J., Cuypers, A. (2008). Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta 227, 1343–1349. doi: 10.1007/s00425-008-0706-4
Rodrigues, F., Marcelino-guimaraes, F. C., Lima, A., Abdelnoor, R. V., Margis, R. (2010). The use of microRNAs as reference genes for quantitative polymerase chain reaction in soybean. Anal. Biochem. 406, 185–192. doi: 10.1016/j.ab.2010.07.020
Sandhya, V., Ali Sk, Z., Grover, M., Reddy, G., Venkateswarlu, B. (2010). Effect of plant growth promoting pseudomonas spp. 0n compatible solutes antioxidant status and plant growth of maize under drought stress. Plant Growth Regul. 62, 21–30. doi: 10.1007/s10725-010-9479-4
Shahid, M., Gowen, S. R., Burhan, M. (2022). Studies on the possible role of plant host on the development of root-knot nematode, Meloidogyne javanica and Pasteuria penetrans as affected by different harvesting dates. Plant Prot. 6 (2), 133–141. doi: 10.33804/pp.006.02.4207
Shen, J., Zhang, Y., Ge, D., Wang, Z., Song, W., Gu, R., et al. (2019). CsBRC1 inhibits axillary bud outgrowth by directly repressing the auxin efflux carrier CsPIN3 in cucumber. Proc. Natl. Acad. Sci. U S A. 116 (34), 17105–17114. doi: 10.1073/pnas.1907968116
Shukla, N., Yadav, R., Kaur, P., Rasmussen, S., Goel, S., Agarwal, M., et al. (2018). Transcriptome analysis of root-knot nematode (Meloidogyne incognita)-infected tomato (Solanum lycopersicum) roots reveals complex gene expression profiles and metabolic networks of both host and nematode during susceptible and resistance responses. Mol. Plant Pathol. 19 (3), 615–633. doi: 10.1111/mpp.12547
Song, Y., Wang, Y., Guo, D., Jing, L. (2019). Selection of reference genes for quantitative real-time PCR normalization in the plant pathogen Puccinia rasiliens schw. BMC Plant Biol. 19 (1), 20. doi: 10.1186/s12870-019-1629-x
Sun, L., Sui, X., Lucas, W. J., Li, Y., Feng, S., Ma, S., et al. (2019). Down-regulation of the sucrose transporter CsSUT1 causes male sterility by altering carbohydrate supply. Plant Physiol. 180 (2), 986–997. doi: 10.1104/pp.19.00317
Uehara, T., Sugiyama, S., Matsuura, H., Arie, T., Masuta, C. (2010). Resistant and susceptible responses in tomato to cyst nematode are differentially regulated by salicylic acid. Plant Cell Physiol. 51 (9), 1524–1536. doi: 10.1093/pcp/pcq109
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3 (0034), 1–11. doi: 10.1186/gb-2002-3-7-research0034
Vejan, P., Abdullah, R., Khadiran, T., Ismail, S., Nasrulhaq Boyce, A. (2016). Role of plant growth promoting rhizobacteria in agricultural sustainability-a review. Molecules 21 (5), 573. doi: 10.3390/molecules21050573
Vijayapalani, P., Hewezi, T., Pontvianne, F., Baum, T. J. (2018). An effector from the Cyst nematode heterodera schachtii derepresses host rRNA genes by altering histone acetylation. Plant Cell. 30 (11), 2795–2812. doi: 10.1105/tpc.18.00570
Wang, J., Pan, C., Wang, Y., Ye, L., Wu, J., Chen, L., et al. (2015). Genome-wide identification of MAPK, MAPKK, and MAPKKK gene families and transcriptional profiling analysis during development and stress response in cucumber. BMC Genomics 16 (1), 386. doi: 10.1186/s12864-015-1621-2
Warzybok, A., Migocka, M. (2013). Reliable reference genes for normalization of gene expression in cucumber grown under different nitrogen nutrition. PloS One 8 (9), e72887. doi: 10.1371/journal.pone.0072887
Wei, G., Tian, P., Zhang, F., Qin, H., Miao, H., Chen, Q., et al. (2016). Integrative analyses of nontargeted volatile profiling and transcriptome data provide molecular insight into VOC diversity in cucumber plants (Cucumis sativus). Plant Physiol. 172 (1), 603–618. doi: 10.1104/pp.16.01051
Wen, C., Zhao, W., Liu, W., Yang, L., Wang, Y., Liu, X., et al. (2019). CsTFL1 inhibits determinate growth and terminal flower formation through interaction with CsNOT2a in cucumber. Development 146 (14), dev180166. doi: 10.1242/dev.180166
Xu, X., Fang, P., Zhang, H., Chi, C., Song, L., Xia, X., et al. (2019). Strigolactones positively regulate defense against root-knot nematodes in tomato. J. Exp. Bot. 70 (4), 1325–1337. doi: 10.1093/jxb/ery439
Yook, K., Harris, T. W., Tamberlyn, B., Cabunoc, A., Chan, J., Chen, W. J., et al. (2011). Wormbase 2012: more genomics, more data, new website. Nucleic Acids Res. 40, 735–741. doi: 10.1093/nar/gkr954
Yuan, L., Zhao, Y., Xie, H., et al. (2022). Selection and evaluation of suitable reference genes for quantitative gene expression analysis during infection of Cucumis sativus with Pectobacterium rasiliense. J. Appl. Microbiol. 132 (5), 3717–3734. doi: 10.1111/jam.15481
Zhang, H., Li, S., Yang, L., Cai, G., Chen, H., Gao, D., et al. (2021). Gain-of-function of the 1-aminocyclopropane-1-carboxylate synthase gene ACS1G induces female flower development in cucumber gynoecy. Plant Cell. 33 (2), 306–321. doi: 10.1093/plcell/koaa018
Zhang, H., Murzello, C., Sun, Y., Kim, M. S., Xie, X., Jeter, R. M., et al. (2010). Choline and osmotic-stress tolerance induced in arabidopsis by the soil microbe Bacillus subtilis (GB03). Mol. Plant Microbe Interact. 23 (8), 1097–1104. doi: 10.1094/MPMI-23-8-1097
Zhang, F., Qin, Z., Zhou, X., Xin, M., Li, S., Luan, J.. (2019). Expression and functional analysis of the propamocarb-related gene CsMAPEG in cucumber. BMC Plant Biol. 19 (1), 371. doi: 10.1186/s12870-019-1971-z
Zhang, T., Shi, Z., Zhang, X., Zheng, S., Wang, J., Mo, J., et al. (2020). Alleviating effects of exogenous melatonin on salt stress in cucumber. Sci. Hortic. 262, 109070. doi: 10.1016/j.scienta.2019.109070
Zhao, J., Jiang, L., Che, G., Pan, Y., Li, Y., Hou, Y., et al. (2019) A Functional Allele of CsFUL1 Regulates Fruit Length through Repressing CsSUP and Inhibiting Auxin Transport in Cucumber. Plant Cell. 31 (6), 1289–1307. doi: 10.1105/tpc.18.00905
Zhou, Y., Zhao, D., Shuang, L., Xiao, D., Xuan, Y., Duan, Y., et al. (2020). Transcriptome analysis of rice roots in response to root-knot nematode infection. Int. J. Mol. Sci. 21 (3), 848. doi: 10.3390/ijms21030848
Keywords: Cucumis sativus, qRT-PCR, reference gene, Meloidogyne incognita, Pseudomonas
Citation: Ji T, Ma S, Liang M, Wang X, Gao L and Tian Y (2022) Reference genes identification for qRT-PCR normalization of gene expression analysis in Cucumis sativus under Meloidogyne incognita infection and Pseudomonas treatment. Front. Plant Sci. 13:1061921. doi: 10.3389/fpls.2022.1061921
Received: 05 October 2022; Accepted: 14 November 2022;
Published: 15 December 2022.
Edited by:
Carolina Escobar, University of Castilla-La Mancha, SpainReviewed by:
Erika V.S. Albuquerque, Brazilian Agricultural Research Corporation (EMBRAPA), BrazilTariq Mukhtar, Pir Mehr Ali Shah Arid Agriculture University, Pakistan
Copyright © 2022 Ji, Ma, Liang, Wang, Gao and Tian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongqiang Tian, dGlhbnlxMTk4NEBjYXUuZWR1LmNu; Lihong Gao, Z2FvbGhAY2F1LmVkdS5jbg==
†These authors have contributed equally to this work
 Tingting Ji
Tingting Ji Si Ma
Si Ma Meiting Liang
Meiting Liang Xueyun Wang
Xueyun Wang Lihong Gao
Lihong Gao Yongqiang Tian
Yongqiang Tian