
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 24 November 2022
Sec. Functional and Applied Plant Genomics
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1060276
This article is part of the Research TopicDiscovery and Exploration of Genes Related to Important Agronomic Traits in Fruit TreesView all 17 articles
 Yanwei Ouyang1†
Yanwei Ouyang1† Xiumei Zhang2†
Xiumei Zhang2† Yongzan Wei3
Yongzan Wei3 Yukun He1
Yukun He1 Xiaohan Zhang1
Xiaohan Zhang1 Ziqiong Li1
Ziqiong Li1 Can Wang1
Can Wang1 Hongna Zhang1,2*
Hongna Zhang1,2*Flowering is an important factor to ensure the success of plant reproduction, and reasonable flowering time is crucial to the crop yield. BBX transcription factors can regulate several growth and development processes. However, there is little research on whether BBX is involved in flower formation and floral organ development of pineapple. In this study, AcBBX5, a BBX family gene with two conserved B-box domains, was identified from pineapple. Subcellular localization analysis showed that AcBBX5 was located in the nucleus. Transactivation analysis indicated that AcBBX5 had no significant toxic effects on the yeast system and presented transcriptional activation activity in yeast. Overexpression of AcBBX5 delayed flowering time and enlarged flower morphology in Arabidopsis. Meanwhile, the expression levels of AtFT, AtSOC1, AtFUL and AtSEP3 were decreased, and the transcription levels of AtFLC and AtSVP were increased in AcBBX5-overexpressing Arabidopsis, which might lead to delayed flowering of transgenic plants. Furthermore, transcriptome data and QRT-PCR results showed that AcBBX5 was expressed in all floral organs, with the high expression levels in stamens, ovaries and petals. Yeast one-hybrid and dual luciferase assay results showed that AcBBX5 bound to AcFT promoter and inhibited AcFT gene expression. In conclusion, AcBBX5 was involved in flower bud differentiation and floral organ development, which provides an important reference for studying the functions of BBX and the molecular regulation of flower.
Flowering, the symbol of vegetative development to reproductive development is an important factor to ensure the success of plant reproduction (Khan et al., 2014). The transition from vegetative stage to reproductive stage is precisely regulated by external environmental signals and internal developmental states (Albani and Coupland, 2010; Wang et al., 2021). Appropriate flowering time is crucial for the crop yield and quality (Roux et al., 2006). Flowering studies are the most extensive in Arabidopsis, and mainly involved photoperiod, vernalization, gibberellin, autonomic, ambient temperature and age-related pathways (Albani and Coupland, 2010; He, 2012; Cheng et al., 2017). Flowering is a very complex physiological process, which is regulated and coordinated by multiple genes (Liljegren et al., 1999).
BBX transcription factors have a wide range of functions and play important roles in flower initiation, light morphogenesis, anthocyanin synthesis and abiotic stress tolerance (Almada et al., 2009; An et al., 2020; Li et al., 2021). For example, CmBBX24 regulates flowering time and tolerance to freezing and drought stress in chrysanthemum by regulating GA biosynthesis (Yang et al., 2014). AtBBX20 interacts with HY5 to activate the gene expression and promote light morphogenesis (Wei et al., 2016). MdBBX22 and MdBBX33 in apple are involved in MDHY5-mediated signal transduction and regulate anthocyanin accumulation (An et al., 2019; Plunkett et al., 2019). CONSTANS (CO) in Arabidopsis is the first BBX protein to be identified in plants. Flowering locus T (FT) genes receive signals from the photoperiodic regulatory center CO under long day conditions, which drives the transition from vegetative to reproductive growth in Arabidopsis (Putterill et al., 1995). However, CO could also inhibit FT-induced flowering by affecting TERMINAL FLOWER1 (TFL1) expression under short-day conditions (Luccioni et al., 2019). Interestingly, in rice, Hd1 is a CO homolog that promotes flowering under short-day conditions but inhibits flowering under long-day conditions (Yano et al., 2000; Kojima et al., 2002; Hayama et al., 2003). In flower formation pathway, BBX protein is strongly conserved among different plants. In addition to CO, other BBX proteins are also involved in the regulation of flower formation in plants. In Arabidopsis, both AtBBX6 and AtBBX24 are positive regulators of flower formation (Hassidim et al., 2009; Li et al., 2014), while AtBBX4, AtBBX7 and AtBBX32 delay flower formation (Cheng and Wang, 2005; Tripathi et al., 2017). OsCOL4, OsBBX14 and OsCOL9 in rice delayed the heading through repressing the Ehd1 pathway under SD and LD conditions (Lee et al., 2010; Bai et al., 2016; Liu et al., 2016). BvCOL1 in sugar beet also causes early flowering under LD conditions (Chia et al., 2008).
Pineapple is one of the world’s famous tropical fruits. Spraying ethephon to induce flower formation is currently the most widely used method in pineapple production (Min and Bartholomew, 1997). At present, there are some excellent pineapple varieties that cannot be widely promoted in the market due to the difficulty in regulating the maturation time (Turnbull et al., 1999). Therefore, studying the molecular mechanism of flower formation induced by ethylene in pineapple can provide theoretical support for perinatal regulation and new variety cultivation. In recent years, several genes regulating flowering such as AcERS and AcETR (Li et al., 2016), AcERF (Zhang et al., 2021a), AcTrihelix (Wang et al., 2022), AcBBX (Ouyang et al., 2022) have been isolated from pineapple, but however, the research on its functional mechanism is still not in-depth.
BBX genes are involved in the determination of flowering time through photoperiod, and there are few studies on flower formation in response to the hormone regulation (Putterill et al., 1995; Yano et al., 2000). Previous studies have found that AcBBX5 was found in the pineapple BBX family study to have an expression peak at 12 h and 7 w after ethylene induced flowering of pineapple. AcBBX5 may be involved in ethylene induced flower formation and flower morphogenesis in pineapple (Ouyang et al., 2022). Here, AcBBX5 was identified from the BBX family analysis of pineapple, and found it may regulate flower formation of pineapple. The sequence characteristics and expression characteristics of AcBBX5 were analyzed by bioinformatics, subcellular localization and transcriptional activation assay. AcBBX5 overexpression, expression characteristics and regulatory mechanism were further analyzed. All these data lay a foundation for the further study of floral formation regulation network in pineapple.
Ananas comosus L. cv. Comte de Paris was used as experimental material, and grown in pineapple resource nursery, Zhanjiang, China. The uniform pineapple plants (15-month-old) were treated with 30 mL 200 mg/L ethephon to induce flowering, and the control group with the same amount of water instead. The flower organs including petals, ovary, stamens, sepals and styles were collected separately in pineapple. All samples were performed with three biological replications and immediately frozen in liquid nitrogen and then stored at −80 °C until further use.
Total RNA was extracted with Polysaccharide Polyphenol Plant RNA Extraction Kit (Huayueyang, China) according to the manufacturer’s instructions. After detecting the concentration and quality of RNA by NanoDrop™ One/OneC Spectrophotometer (Thermo Fisher Scientific, USA), reverse transcription was carried out through the Revert Aid First-Strand cDNA Synthesis Kit (Thermo Fisher Scientific, USA). RT-qPCR was performed with ChamQ Universal SYBR qPCR Master Mix (Vazyme, China) on LightCycler 480 II (Roche, Switzerland). AcActin was used as an internal reference gene of pineapple. The reaction conditions for RT-qPCR consisted of predegeneration (95°C for 2 min), circular reaction (95 °C for 10 s, 58 °C for 30 s, 40 cycles) and Dissociation curve (95 °C for 15 s, 60 °C for 60 s, 95 °C for 15 s). All experiments incorporated three biological samples and three technical replicates. The relative expression levels of each gene were evaluated by using the 2−ΔΔCt method (Rao et al., 2013).
The AcBBX5 protein sequence of pineapple was obtained from pineapple Genome Database (http://pineapple.zhangjisenlab.cn/pineapple/html/index.html) (Xu et al., 2018). The protein sequences of other species were all derived from NCBI (National Center for Biotechnology Information). MEME online site was used to analyze the conserved motifs (Bailey et al., 2009). The phylogenetic tree was constructed by neighbor-joining (NJ) method in MEGA 6.0 software, and the bootstrap value was set at 1000 replications (Hall, 2013).
The coding sequence of AcBBX5 was introduced into the pCAMBIA2300-GFP vector digested with Kpn I and Xba I restriction enzymes to generate the construct 35S::AcBBX5-GFP. After sequencing correctly, it was transformed into Agrobacterium tumefaciens strain GV3101. The epidermal cell transformation of tobacco leaves was injected with the A. tumefaciens carrying out with the recombinant vector. After incubation in the dark for 12 h at 25 °C, the tobaccos were transferred to normal growth for 24–36 h. The fluorescence signal was observed by a confocal scanning microscope Ax-io-Imager_LSM-800 (Zeiss, Germany) under excitation of 488 nm.
A yeast assay system was used to examine the transcriptional activity of AcBBX5. Nde I and Sal I were selected as restriction sites, and the coding sequences of AcBBX5 were inserted into the bait vector pGBKT7 by homologous recombination. Following the manufacturer’s protocol, the recombinant vectors were transferred into AH109 strain yeast and cultured on SD/-Trp medium at 30 °C for 2–3 days. Single colonies were selected for amplification culture and then transferred to SD/-Trp and SD/-Trp/-His/-Ade solid media for further culture. Three days later, X-α -Gal was added to observe whether the colony was blue.
Arabidopsis thaliana (ecotype Colombia) grown under conditions of 21 ± 1°C (day/night, 16/8 h) and used for heterologous transformation in this study. pCAMBIA2300-GFP-AcBBX5 vectors (the recombinant vectors were used for subcellular localization) were used to transform into Arabidopsis via the floral dip method (Clough and Bent, 1998). Empty vectors were transformed as control. The seeds of the T0 generation were harvested and screened on plates containing MS medium with 50 mg/L kanamycin sulfate and then further verified the transgenic plants by PCR amplification. The T3 generation transgenic Arabidopsis lines were used for subsequent phenotype observation and functional analysis. To detect the expression levels of the AcBBX5 and some flowering-related genes in transgenic and control plants, 28-day-old seedlings of both transgenic and control Arabidopsis plants were collected for qRT-PCR analysis.
To investigate the function of the AcBBX5 protein, we first identified and cloned AcBBX5 from the ‘Paris’ pineapple by RT-PCR. The coding sequence of AcBBX5 was 603 bp in length, encoding 197 amino acid residues. In order to better understand the properties of AcBBX5, the amino acid sequence of AcBBX5 was analyzed. The results showed that AcBBX5 and its homologous proteins in other species all contained two B-box conserved domains, and contained motif 1, motif 2 and motif 4 in conserved motif analysis (Figure 1A). However, compared with homologous protein sequences of other species, AcBBX5 sequence was significantly shorter and lacks motif 3, which was common in other sequences. Interestingly, the positions of motifs 1 and 2 in the sequence almost overlapped with those of the two B-box conserved domains, and both motifs 1 and 2 might represent B-box conserved domains. Furthermore, multiple sequence alignments with homologous amino acid sequences in Arabidopsis, rice, pear, cucumber and tomato revealed that the amino acid sequence similarity between AcBBX5 and homologous proteins in other species were 24.8% (AtBBX22), 24.53% (CsaBBX14), 24.27% (OsBBX16), 22.93% (PbBBX18), and 25.60% (SlBBX22), respectively (Figure S1). The low sequence similarity is probably due to the absence of a fragment at the C-terminal of AcBBX5 relative to homologous sequences of other species.
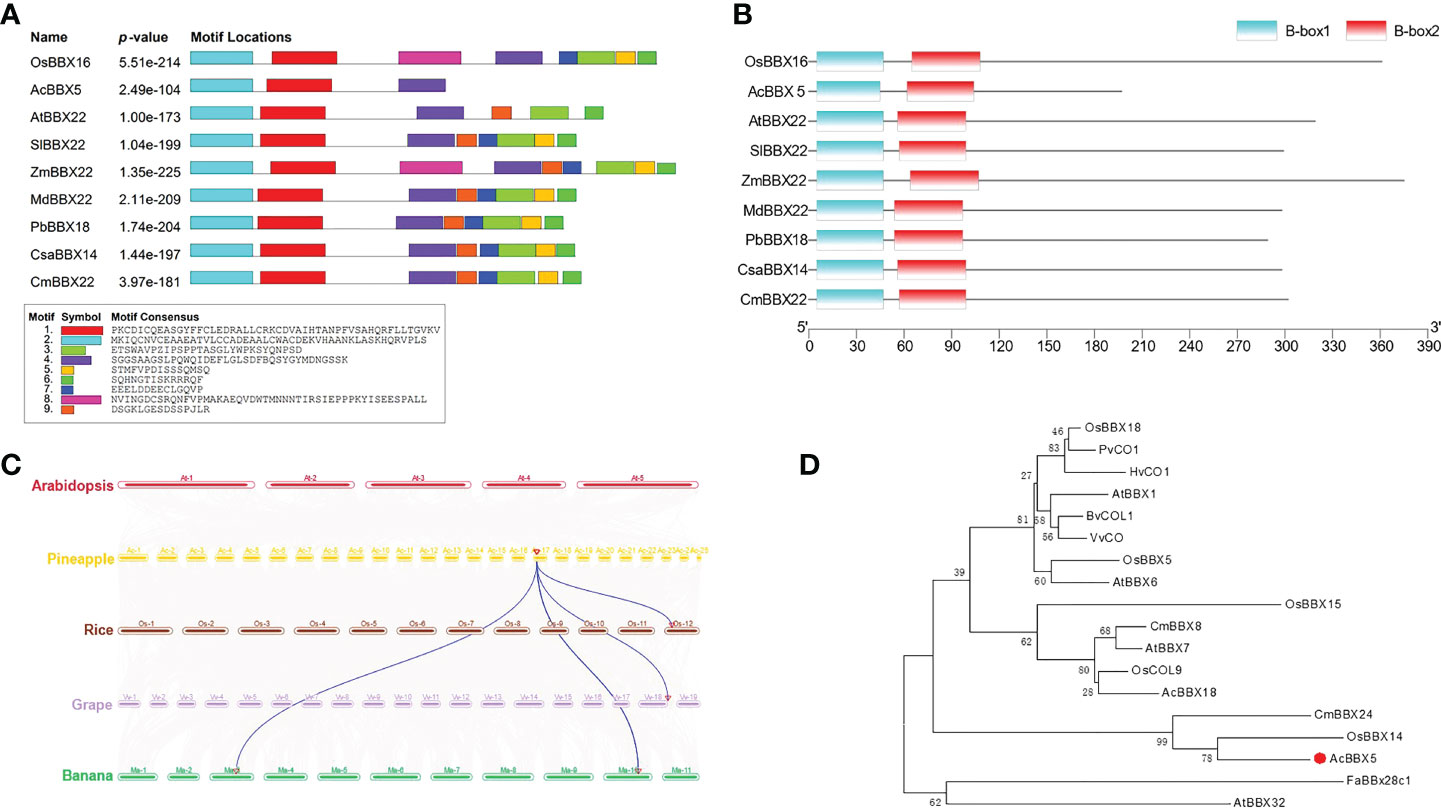
Figure 1 Characterization of AcBBX5 sequence in pineapple. (A) Motif analysis of AcBBX5 proteins. Different colored boxes represent the different types of motifs. (B) Domain analysis of AcBBX5 proteins. (C) Syntenic relationships of AcBBX5 in pineapple with rice, grape and banana. (D) Phylogenetic analyses of AcBBX5. Phylogenetic tree based on BBX proteins involved in flower formation regulation.
The function of BBX protein has been studied in many plants. Collinearity analysis of pineapple with rice, grape, and banana revealed that pineapple AcBBX5 has one homologous gene pair in rice and banana, but two in grape (Figure 1). In order to investigate the role of AcBBX5 protein in flower formation, 18 BBX proteins that have been confirmed to be involved in flower formation regulation were selected to construct phylogenetic tree (Figure 1 and Table S1). Among them, there were six BBX proteins promoting flowering (BvCOL1, AcBBX18, HvCO1, VvCO, AtBBX6 and CmBBX8) and 9 delayed flowering proteins (AtBBX32, AtBBX7, BvCOL1, CmBBX24, FaBBx28c1, OsBBX14, OsBBX15, OsCOL9, OsBBX27 and PvCO1). AtBBX1 and OsBBX18 are genes that can both promote and inhibit flower formation. Phylogenetic tree analysis showed that AcBBX5 and delayed flowering proteins (CmBBX24 and OsBBX14) clustered in the same branch, suggesting that their functions may be similar. AcBBX5 may have the function of delaying flower formation as CmBBX24 and OsBBX14.
Subcellular localization information is of great significance to our understanding of protein function. To investigate the subcellular localization of the AcBBX5 protein, 35S::AcBBX5-GFP protein was transiently expressed in Nicotiana benthamiana leaves (Figure 2). Under confocal microscope, the GFP fluorescence of the control vector was distributed in the nucleus and the cell membrane, while the fluorescence signals of 35S::AcBBX5-GFP were detected in only the nucleus. The results indicate that AcBBX5 localizes in the nucleus, and may act as a transcription factor in the nucleus to participate in the transcriptional regulation of other gene expression.
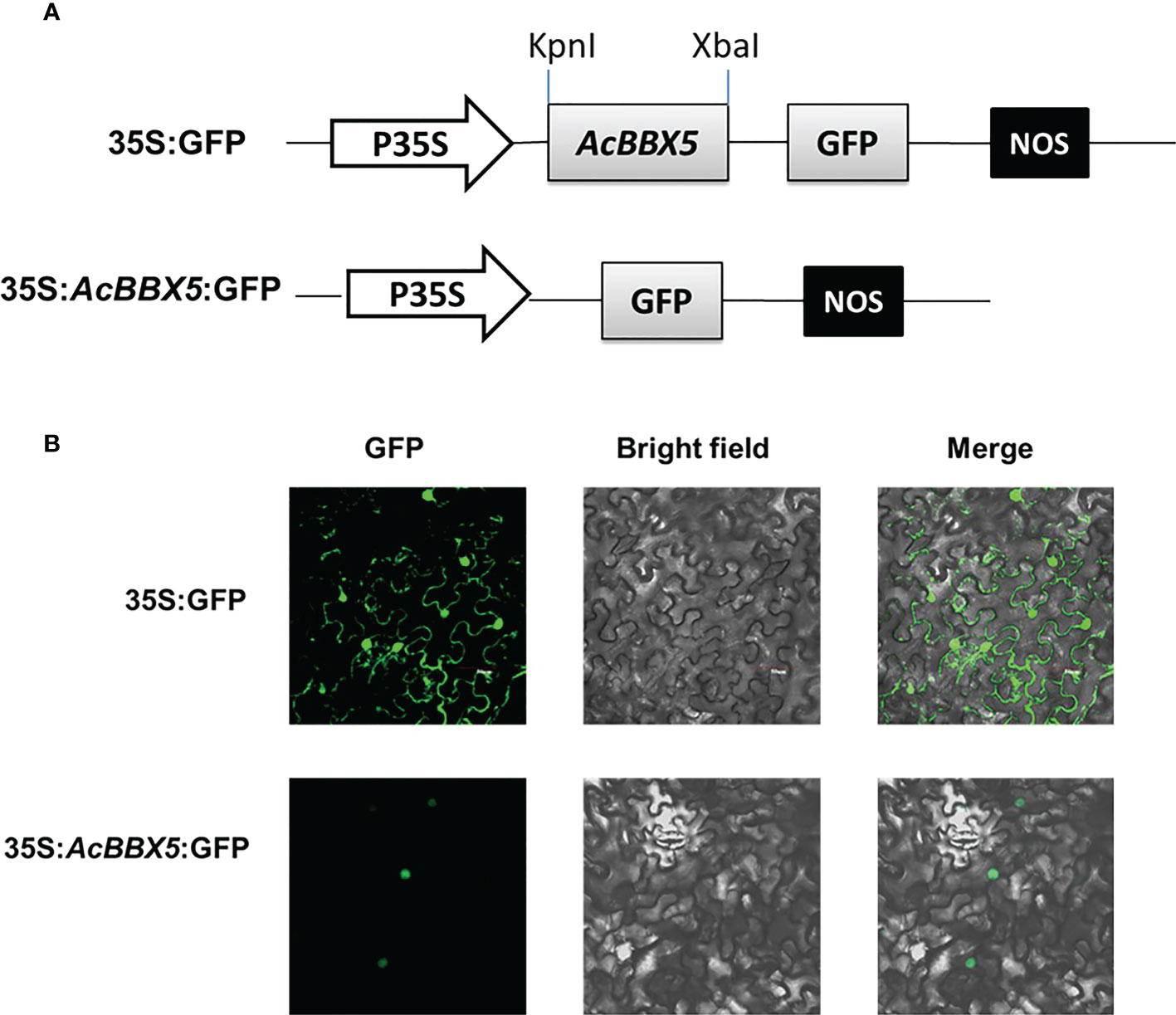
Figure 2 Subcellular localization of AcBBX5 in tobacco leaf cells. (A) Vectors for AcBBX5 subcellular localization analysis. (B) AcBBX5 was localized in tobacco leaf epidermal cells. 2300-GFP and 2300-AcBBX5-GFP plasmids were transformed into tobacco. The dark field, bright field and merge field were shown in the left, middle and right panels, respectively.
The detection of transcriptional activation and yeast toxicity provides a foundation for further exploring the mechanism of AcBBX5. The complete coding region of AcBBX5 was fused to the GAL4-binding domain in the pGBKT7 vector, and expressed in the yeast strain Y2H. The pGADT7-T and pGBKT7-53 vectors served as the positive control, while the empty pGBKT7 vector was negative control. As shown in Figure 3, the positive control strains and the yeast cells with the pGBKT7-AcBBX5 vectors grew extremely well on both SD/-Trp and SD/-Trp/-His/-Ade medium, and could turn blue on medium coated with X-α-gal, whereas those negative control strains were unable to grow on SD/-Trp/-His/-Ade medium, which confirms that AcBBX5 has no significant toxic effects on the yeast system and exhibited transcriptional activation activity in yeast.
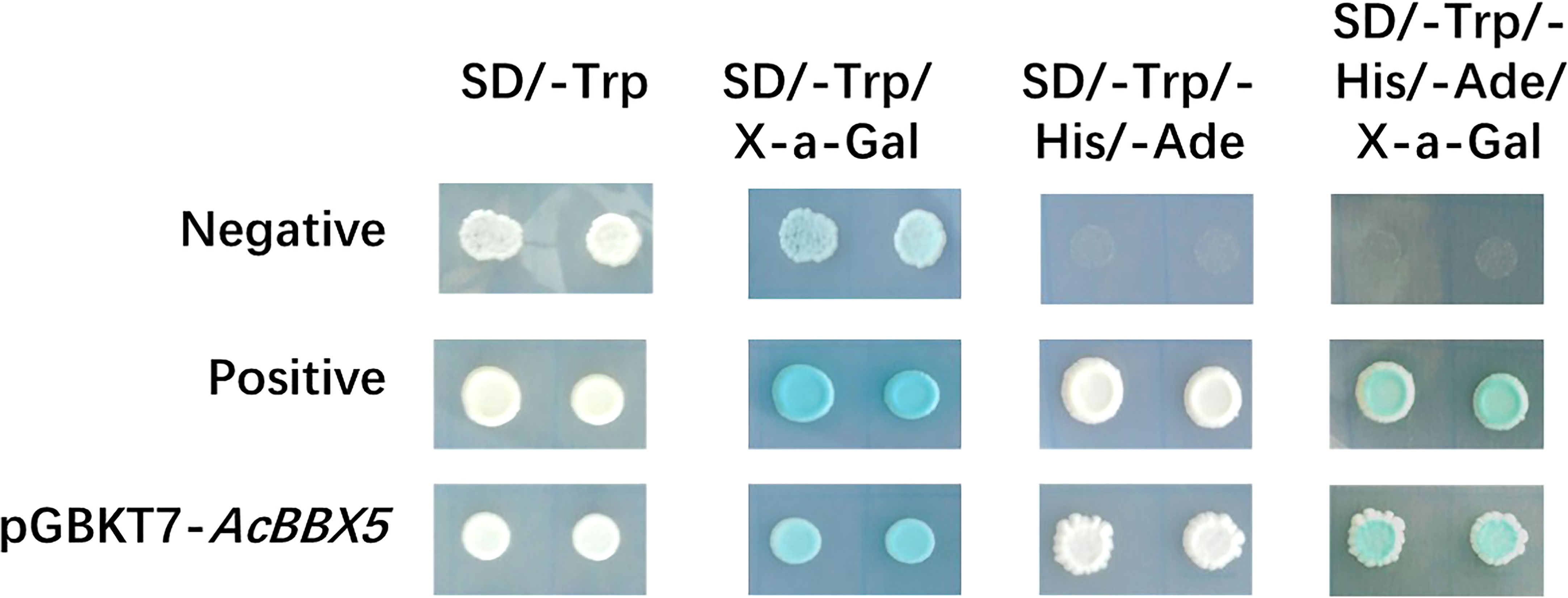
Figure 3 Transcriptional activation activity analysis of AcBBX5 in yeast. The empty vector pGBKT7 was transformed into yeast as the negative control; pGADT7-T and pGBKT7-53 were co-transformed into yeast was used as a positive control.
Previous studies have found that AcBBX5 had a high expression peak at 7 w after ethylene induction, which was the stage of floret development (Ouyang et al., 2022). To evaluate whether AcBBX5 is involved in flower formation, overexpressed Arabidopsis transgenic lines of AcBBX5 were constructed. Transgenic plants carrying pCAMBIA2300-GFP empty vector were used as negative control. Three highly expressed lines were selected from T3 homozygous positive transgenic lines (OE-3, OE-9 and OE-13) by PCR amplification and QRT-PCR to study their characteristics (Figure 4A). All three AcBBX5 transgenic lines flowered significantly later than control Arabidopsis, and rosette numbers were significantly higher than those of control. In the long day (16 h light/8 h dark) condition, the control plants with about 18 rosette leaves began to blossom about 18 days after transplanting. The overexpressed OE-3, OE-9 and OE-13 bloomed at about 58, 50 and 29 days after transplanting, and the number of rosette leaves was about 37, 25 and 23, respectively (Figure 4B). We also detected the transcript levels of AcBBX5 in T3 lines of OE-3, OE-9 and OE-13 Arabidopsis respectively, and found that expression levels of AcBBX5 in different transgenic lines were significantly increased. Interestingly, the flowering time was negatively associated with the expression of AcBBX5 in transgenic Arabidopsis plants (Figure 5). These results indicated that overexpression of AcBBX5 in Arabidopsis leaded to a serious delay in flowering time.
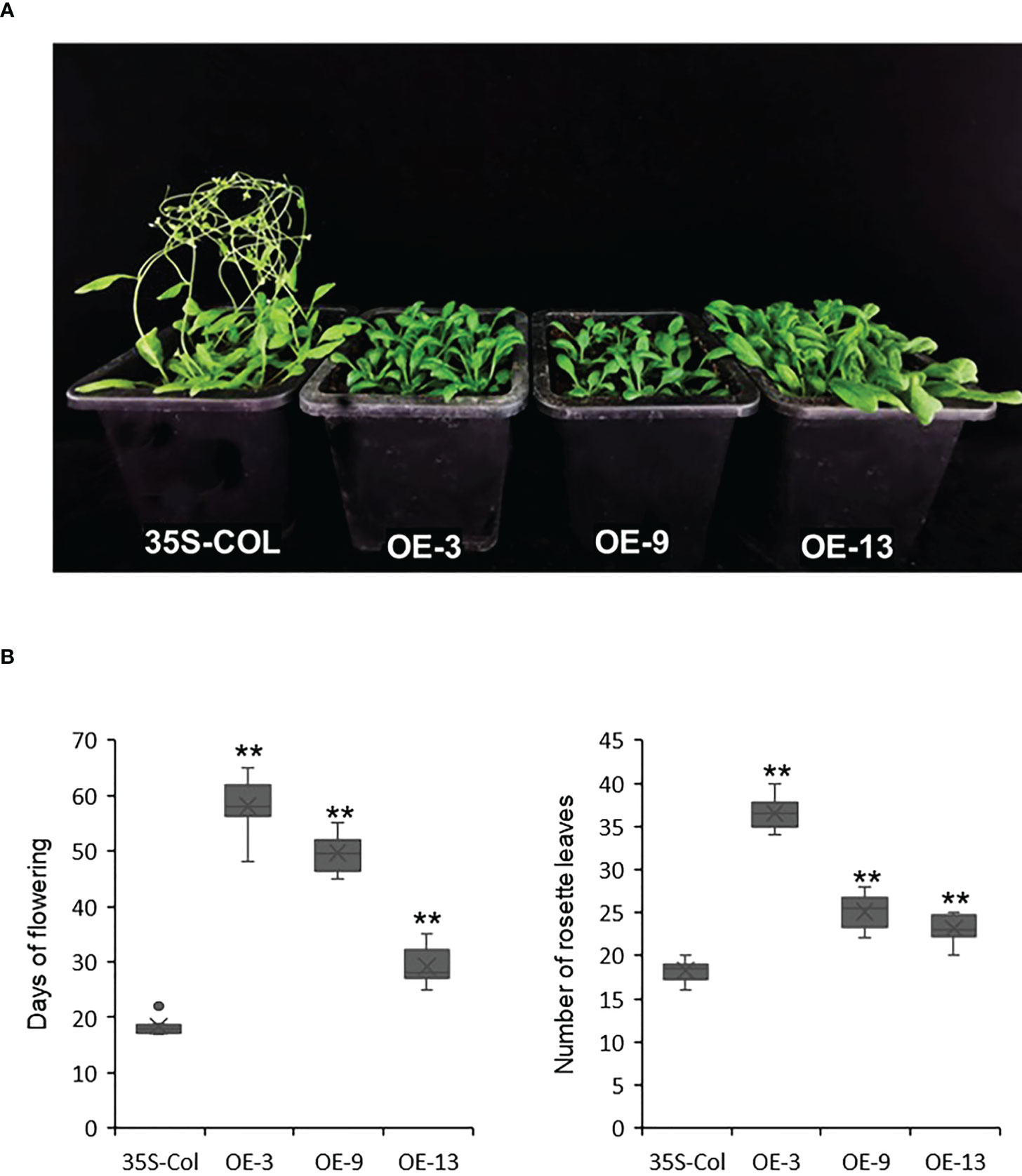
Figure 4 Ectopic expression of AcBBX5 in Arabidopsis. (A) Phenotypes analysis of T3 transgenic Arabidopsis with AcBBX5. (B) Days to flowering and number of rosette leaves at flowering in transgenic plants (n=8). Asterisks indicate significant differences. (*P < 0.1, **P < 0.05, based on Student’s t-test).
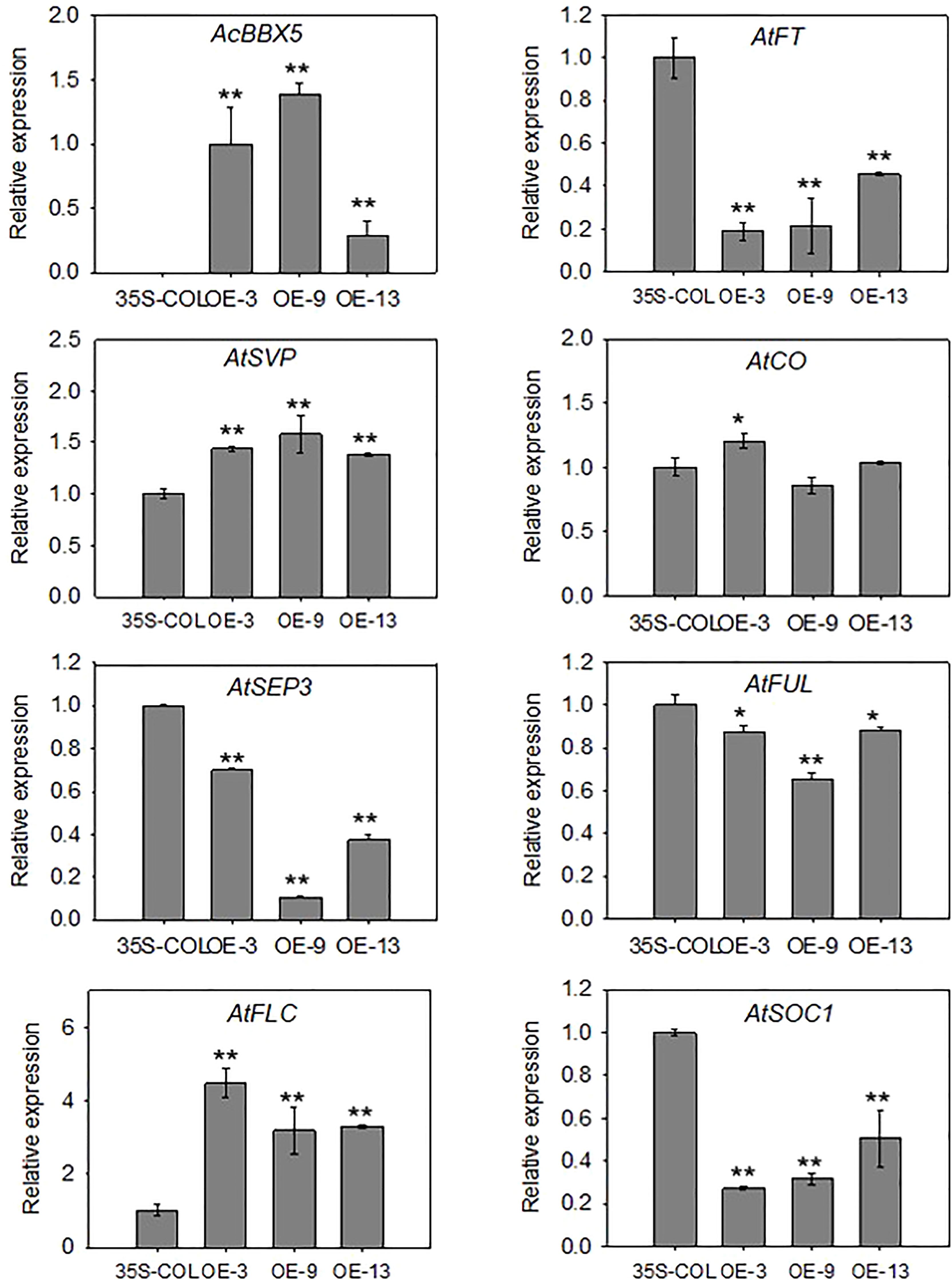
Figure 5 Expression analysis in transgenic Arabidopsis plants. RT-qPCR analysis of AcBBX5 and genes involved in flowering, including AtSEP3, AtSVP, AtCO, AtFLC, AtSOC1, AtFUL and AtFT, in transgenic Arabidopsis plants (n=3). Asterisks indicate significant differences. (*P < 0.1, **P < 0.05, based on Student’s t-test).
In order to better understand the molecular mechanism of AcBBX5 involved in flowering time regulation, the cDNAs of AcBBX5 transgenic Arabidopsis lines as template were used to analyze the genes involved in flower regulation in Arabidopsis by QRT-PCR (Figure 5). Seven genes involved in the determination of flowering time and flower development were searched from the Arabidopsis flowering database (http://www.phytosystems.ulg.ac.be/florid/). Compared with the control plants, the expression levels of FT, SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1), FRUITFULL (FUL) and SEPALLATA3 (SEP3) genes promoting flower formation in Arabidopsis were inhibited in overexpressed lines, while the expression levels of FLOWERING LOCUS C (FLC) and SHORT VEGETATIVE PHASE (SVP) genes negatively regulating flower formation were significantly increased in transgenic Arabidopsis plants. These results further confirmed that AcBBX5 repressed flower formation in Arabidopsis by regulating the expression of other floral genes and this might suggest the possibility that AcBBX5 represses flowering in Pineapple.
In addition to delay flowering, florets were also larger in AcBBX5 transgenic lines compared to control lines at full flowering (Figure 6). To test whether AcBBX5 is involved in floral organ development, transcriptome data and QRT-PCR were used to analyze floral organs of pineapple, including sepal, ovary, stamen, petal and pistil. The results demonstrated that AcBBX5 was specifically expressed in the floral organ with higher expression levels in stamen and petals. The results showed that AcBBX5 was specifically expressed in floral organs, with higher expression levels in stamens and petals. The phenotypic characteristics of AcBBX5 were evaluated by comparing the sepal and petal sizes of AcBBX5 and control (35S-COL) flowers. Compared with the control plants, the width of petals and sepals in the transgenic plants was larger, suggesting that AcBBX5 was also involved in floral organ development.
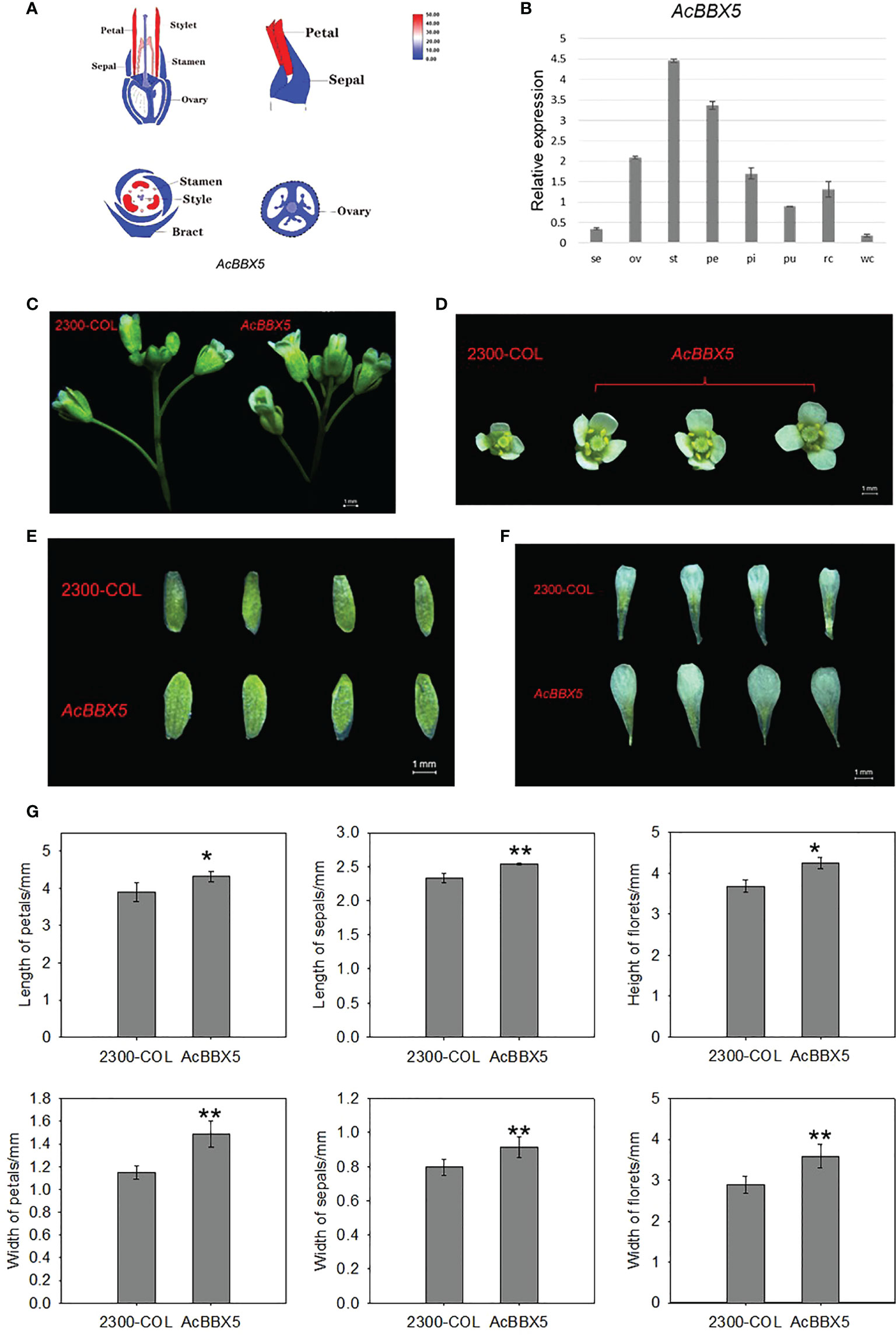
Figure 6 Phenotypes and expression characteristics of floral organs in transgenic Arabidopsis. (A) Transcriptome data. (B) RT-QPCR. Se, Sepal; Pe, Petal; St, Stamen; Ov, ovary; St, Style; Pu, Fruit; Rc, Root; and Wc, Leaf. (C–F) Phenotypes characteristics of floral organs. (G) Statistical analysis of floret height, floret width, petal length, petal width, sepal length and sepal width of AcBBX5 transgenic lines and control lines (n=3). Asterisks indicate significant differences. (*P < 0.1, **P < 0.05, based on Student’s t-test).
To further identify the genes directly regulated by AcBBX5 in pineapple flower formation, yeast one-hybrid assay was performed to determine whether AcBBX5 could bind to the promoter of AcFT. PB42AD-AcBBX5 and Placzi-proAcFT vectors were combined to transform yeast strain EGY48. PB42AD/Placzi- proAcFT and Placzi/PB42AD-AcBBX5 were used as negative control. The transformed yeast showed blue color in SD/Trp-Ura- medium containing X-Gal, but the negative control did not. This result indicated that AcBBX5 could bind to the promoter of AcFT (Figure 7A). In addition, the reporter (pGreenII0800-LUC-proAcFT) and effector plasmid (35S:AcBBX5) were constructed and a dual luciferase (LUC) assay was performed in N. benthamiana leaf cells to examine whether AcBBX5 could activate the AcFT promoter. As expected, in the LUC reporter assays, the fluorescence signal was significantly attenuated in sites infected by AcBBX5 and AcFT promoters compared with the negative control, and the activity of the promoter expressed by the LUC/REN ratio was significantly reduced relative to the control (Figure 7B). In conclusion, AcBBX5 can bind to the AcFT promoter and inhibit its expression, thereby inhibiting the flower formation of pineapple.
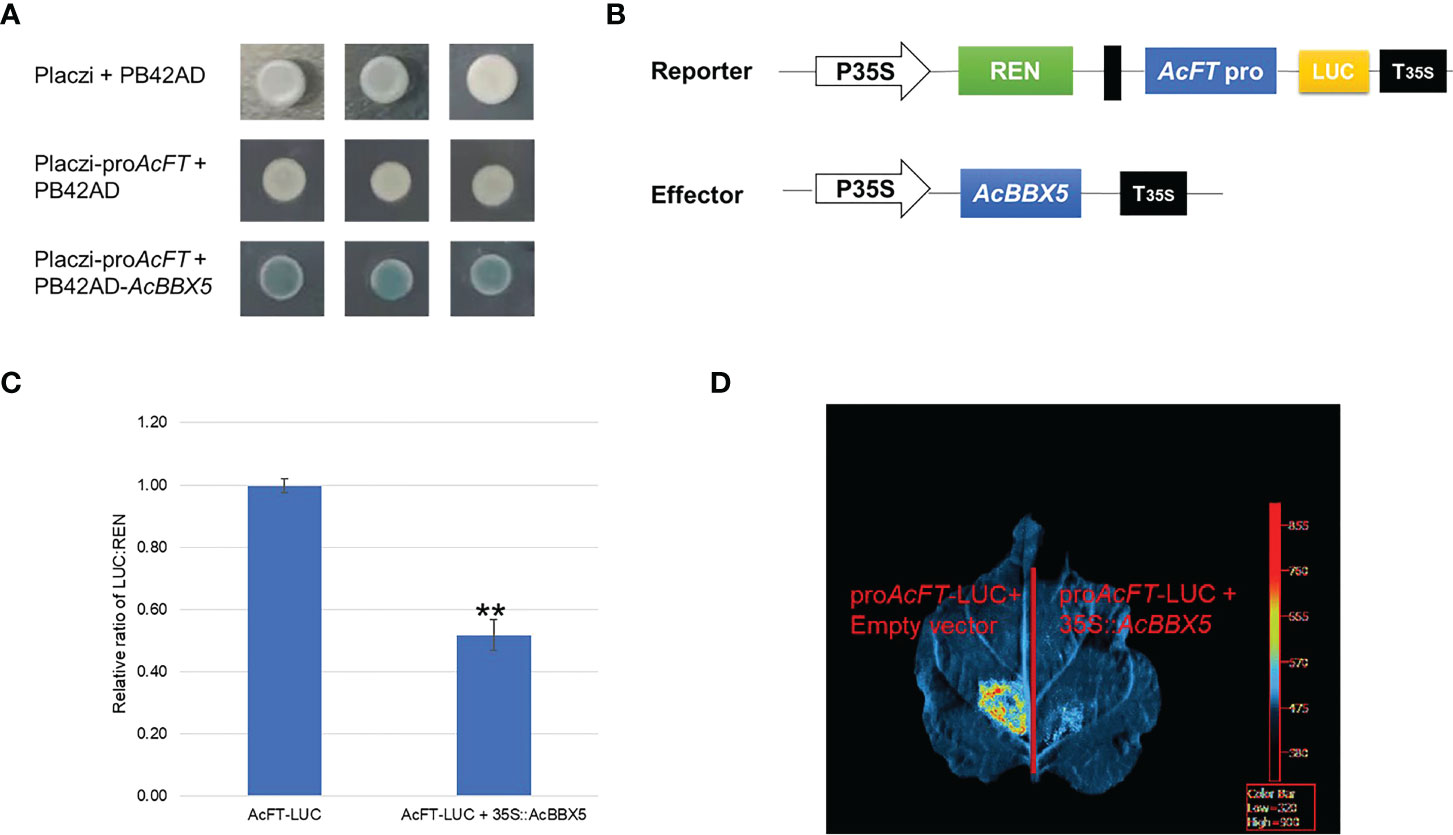
Figure 7 AcBBX5 binds to AcFT promoter and negatively regulates its expression. (A) Yeast one-hybrid assay to test whether AcBBX5 could directly bind to the promoters of AcFT (B) Schematic diagram of effector and reporter structures used for dual luciferase assay. LUC, firefly luciferase. REN, Renilla luciferase. P35S and T35S, the promoter and terminator of CaMV35S, respectively. (C) The comparison of luciferase activity (n=3). Asterisks indicate significant differences. (*P < 0.1, **P < 0.05, based on Student’s t-test). (D) Representative bioluminescence image of AcBBX5 activation on the AcFT promoter in tobacco leaves.
The BBX gene family is widely involved in plant growth and development and response to the environment. AcBBX5 is a member of the pineapple AcBBX family (Ouyang et al., 2022). Here, conserved domain analysis confirmed that AcBBX5 and other orthologous proteins contain two conserved B-box domains at the N-terminus (Figure 1A). Motif analysis identified two conserved B-box domains containing a different conserved motif (Figure 1B). Based on the difference in amino acid sequence identity of the b-box motif and the specificity of zinc binding amino acid residues, it is divided into two types: B-box 1 and B-box 2 (Crocco and Botto, 2013). Plant B-box domains can form heterodimers within the BBX protein family or with other proteins, and play an important role in mediating protein interactions and regulating the gene expression (Gangappa et al., 2013). For example, PpBBX18 in pear forms a heterodimer with PpHY5 via two b-box domains, thereby inducing PpMYB10 transcription and regulating anthocyanin biosynthesis (Bai et al., 2019). Subcellular localization analysis showed that AcBBX5 protein was located in the nucleus of epidermal cells of Nicotiana benthamiana leaves (Figure 2) and had transcriptional activation activity in yeast (Figure 3), suggesting that AcBBX5 gene has general characteristics of transcription factor and may be involved in the transcription level of other genes. However, Figure S1 shows that the sequence length of AcBBX5 was significantly shorter than the others. In addition, except for AcBBX5, which has only 3 conserved motifs, other homologous proteins have at least 6 conserved motifs (Figure 1A). These imply that the function of AcBBX5 may be different from that of homologous genes in other species.
BBX genes are involved in the determination of flowering time (Putterill et al., 1995; Yano et al., 2000). AtBBX24, CmBBX8 and HvCO1 are the BBX genes that have been shown to promote flower formation. AtBBX24 overexpression not only reduces the expression level of FLC, but also activates FT and SOC1 expression, which leads to early flowering in Arabidopsis under long- and short-day conditions (Li et al., 2014). In summer chrysanthemum, overexpression of CmBBX8 accelerated flowering under long- and short-day conditions. CmFTL1 can act as floral inducer in long day conditions, and CmBBX8 promotes flowering through binding with CORE element (CCACA) of the CmFTL1 promoter (Wang et al., 2020). In barley, HvCO1 overexpression up-regulated HvFT1, which may promote flowering by activating HvFT1. But it has no promoting effect on Arabidopsis thaliana (Armstead et al., 2005). In this study, overexpression of AcBBX5 gene in Arabidopsis delayed flowering time (Figure 4). BBX has also been reported to inhibit floral formation in other species. In rice, OsBBX14 delays heading date via different ways under long and short-day conditions. OsBBX14 delays heading date by promoting the expression of Hd1 under long day conditions. However, under short day conditions, it acts as a repressor of Ehd1 to delay heading date (Bai et al., 2016). CmBBX24 may inhibit flowering in Chrysanthemum Morifolium by negatively regulating the expression of GA biosynthetic genes (GA20ox and GA3ox) and photoperiodic flowering pathway genes (GI, PRR5, CO, FT and SOC1) (Yang et al., 2014). OsBBX14 and CmBBX24, which inhibit flower-forming genes, cluster on the same branch of the evolutionary tree with AcBBX5 (Figure 1D), suggesting that genes of the same classification or more closely related genes may have similar functions.
Molecular studies on flowering have focused on FT, FLC, CO and SOC1 genes. In addition to AtBBX24 (Li et al., 2014), which has been shown to regulate FLC, FT and SOC1, AtBBX7/AtCOL9 has been reported to inhibit CO and FT expression and delay floral transition in Arabidopsis (Cheng and Wang, 2005). The heterodimerization between BBX28 and CO affects the activation of FT transcription by CO, which negatively regulates Arabidopsis flower formation (Liu et al., 2020). In this study, the relative expression levels of positive regulators of flower formation (FT, SEP3, SOC1 and FUL) were inhibited in Arabidopsis transgenic lines, while negative regulators of flower formation (FLC and SVP) were higher than control lines (Figure 5). This further confirmed that AcBBX5 overexpression inhibited flowering in Arabidopsis. However, the relative expression of CO in the Arabidopsis transgenic lines was almost no different from that in the control, indicating that AcBBX5 does not negatively regulate flower formation by inhibiting CO expression.
Yeast one-hybrid and dual luciferase assay results found that AcBBX5 is transcriptional activator (Figure 3), but it can bind to the promoter of FT gene and inhibit its expression (Figure 7). Similarly, previous studies on transcription factors in other species have also confirmed that transcription factors with transcriptional activation activity can also act as repressors under certain conditions. In grape hyacinth, MaBBX51 interfered with the binding of MaHY5 to the promoters of MaMybA and MaDFR, thereby inhibiting anthocyanin biosynthesis (Zhang et al., 2022). VvMYB30 in grapevine competes with activator VvMYB14 to bind to common binding sites in VvSTS15/21 promoter, controls stilbene biosynthesis in grapevine (Mu et al., 2022). In grape, VvMYB30 competed with the activator VvMYB14 to bind to characteristic binding sites in the VvSTS15/21 promoter to effect stilbene biosynthesis (Mu et al., 2022). In Arabidopsis, AtBBX24 inhibited anthocyanin accumulation by interfering with the binding of HY5 to the promoter of related genes in anthocyanin biosynthesis (Job et al., 2018). AtBBX19 negatively regulated flowering time by interfering with CO binding to FT through physical interaction with CO proteins (Wang et al., 2014). In view of this, BBX5 may act as a weak activator, which binds to the FT promoter of the target gene and occupies a limited binding site to block the binding of some strong activators, thus inhibiting the expression of FT gene. This speculation still needs further systematic experimental verification.
A trait is regulated by multiple genes, and a gene may be involved in the regulation of multiple traits. Here, AcBBX5 may not only regulate the flower formation of pineapple (Figure 4), but also promote the growth and development of floral organs (Figure 6). BBX gene is one of the many genes involved in flower organ growth and development. However, there are few studies on the BBX gene in flower organs. In rose, RhBBX28 is a key player in regulating petal senescence, and overexpression of RhBBX28 produces smaller flowers than WT (Zhang et al., 2021b). In the present study, AcBBX5 overexpression also showed differences in flower size in Arabidopsis. But in contrast to RhBBX28, overexpression of AcBBX5 produced larger flowers than the control (Figure 6). Transcriptome and quantitative data showed that AcBBX5 was highly expressed in petals, suggesting that AcBBX5 may play a role in petal development. Further studies and sufficient experimental evidence are needed to prove the specific regulatory mechanism.
In this study, AcBBX5, a member of the BBX family, is identified as a negative regulator of floral formation in Arabidopsis and may be involved in floral organ development. Subcellular localization and transcriptional activation analysis showed that AcBBX5 was located in the nucleus and had transcriptional activation potential. The relative expressions of FT, SOC1, FUL and SEP3 were decreased, and FLC and SVP were increased in AcBBX5-overexpressing Arabidopsis. In addition, AcBBX5 was expressed in different floral organs of pineapple, but highly expressed in stamens, ovary and petals. Yeast one-hybrid and a dual luciferase assay results confirmed that AcBBX5 bound to AcFT promoter in pineapple and inhibited the expression of AcFT gene. These data will enrich the known regulatory network of BBX in different plants and provide information on the regulation of flowering and floral organ development in pineapple.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
YO and HZ wrote the manuscript and completed the experiments. XZ and HZ designed the experiment and contributed to data. YH took charge the experimental materials treatment and collection. YW investigated the data analyze of the study. YO and CW carried out RNA extraction and QRTPCR verification. XZ and ZL constructed the vectors and completed the transgenic experiments. HZ and XZ conceived the study. All authors contributed to the article and approved the submitted version.
The project was funded by the National Natural Science Fund of China (31872079 and 32160687), the National Key R&D Program of China (2019YFD1001105 and 2018YFD1000504), the Natural Science Foundation of Hainan Province (321RC467 and 322MS013), the major science and technology project of Hainan Province (ZDKJ2021014), and the Scientific Research Start-up Fund Project of Hainan University (KYQD-ZR-20090).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1060276/full#supplementary-material
Supplementary Figure 1 | Protein alignment of AcBBX5 and its homologs from other species.
Albani, M. C., Coupland, G. (2010). Comparative analysis of flowering in annual and perennial plants. Curr. Top. Dev. Biol. 91, 323–348. doi: 10.1016/S0070-2153(10)91011-9
Almada, R., Cabrera, N., Casaretto, J. A., Ruiz-Lara, S., González Villanueva, E. (2009). VvCO and VvCOL1, two CONSTANS homologous genes, are regulated during flower induction and dormancy in grapevine buds. Plant Cell Rep. 28, 1193–1203. doi: 10.1007/s00299-009-0720-4
An, J., Wang, X., Zhang, X., Bi, S., You, C., Hao, Y. (2019). MdBBX 22 regulates UVB-induced anthocyanin biosynthesis through regulating the function of MdHY 5 and is targeted by MdBT2 for 26S proteasome-mediated degradation. Plant Biotechnol. J. 17, 2231–2233. doi: 10.1111/pbi.13196
An, J., Wang, X., Zhang, X., You, C., Hao, Y. (2020). Apple b-box protein BBX37 regulates jasmonic acid-mediated cold tolerance through the JAZ-BBX37-ICE1-CBF pathway and undergoes MIEL1-mediated ubiquitination and degradation. New Phytol. 229, 2707–2729. doi: 10.1111/nph.17050
Armstead, I. P., Skøt, L., Turner, L. B., Skøt, K., Donnison, I. S., Humphreys, M. O., et al. (2005). Identification of perennial ryegrass (Lolium perenne (L.) and meadow fescue (Festuca pratensis (Huds.)) candidate orthologous sequences to the rice Hd1(Se1) and barley HvCO1 CONSTANS-like genes through comparative mapping and microsynteny. New Phytol. 167, 239–247. doi: 10.1111/j.1469-8137.2005.01392.x
Bailey, T., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bai, S., Tao, R., Yin, L., Ni, J., Yang, Q., Yan, X., et al. (2019). Two b-box proteins, PpBBX18 and PpBBX21, antagonistically regulate anthocyanin biosynthesis via competitive association with pyrus pyrifolia ELONGATED HYPOCOTYL 5 in the peel of pear fruit. Plant J. 100, 1208–1223. doi: 10.1111/tpj.14510
Bai, B., Zhao, J., Li, Y., Zhang, F., Zhou, J., Chen, F., et al. (2016). OsBBX14 delays heading date by repressing florigen gene expression under long and short-day conditions in rice. Plant Sci. 247, 25–34. doi: 10.1016/j.plantsci.2016.02.017
Cheng, X., Wang, Z. (2005). Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in arabidopsis thaliana. Plant J. 43, 758–768. doi: 10.1111/j.1365-313X.2005.02491.x
Cheng, J., Zhou, Y., Lv, T., Xie, C., Tian, C. (2017). Research progress on the autonomous flowering time pathway in arabidopsis. Physiol. Mol. Biol. Plants. 23, 477–485. doi: 10.1007/s12298-017-0458-3
Chia, T. Y., Müller, A., Jung, C., Mutasa-Göttgens, E. S. (2008). Sugar beet contains a large CONSTANS-LIKE gene family including a CO homologue that is independent of the early-bolting (B) gene locus. J. Exp. Bot. 59, 2735–2748. doi: 10.1093/jxb/ern129
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x
Crocco, C. D., Botto, J. F. (2013). BBX proteins in green plants: insights into their evolution, structure, feature and functional diversification. Gene 531, 44–52. doi: 10.1016/j.gene.2013.08.037
Gangappa, S. N., Crocco, C. D., Johansson, H., Datta, S., Hettiarachchi, C., Holm, M., et al. (2013). The arabidopsis b-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell 25, 1243–1257. doi: 10.1105/tpc.113.109751
Hall, B. G. (2013). Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 30, 1229–1235. doi: 10.1093/molbev/mst012
Hassidim, M., Harir, Y., Yakir, E., Kron, I., Green, R. M. (2009). Over-expression of CONSTANS-LIKE 5 can induce flowering in short-day grown arabidopsis. Planta 230, 481–491. doi: 10.1007/s00425-009-0958-7
Hayama, R., Yokoi, S., Tamaki, S., Yano, M., Shimamoto, K. (2003). Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422, 719–722. doi: 10.1038/nature01549
He, Y. (2012). Chromatin regulation of flowering. Trends Plant Sci. 17, 556–562. doi: 10.1016/j.tplants.2012.05.001
Job, N., Yadukrishnan, P., Bursch, K., Datta, S., Johansson, H. (2018). Two b-box proteins regulate photomorphogenesis by oppositely modulating HY5 through their diverse c-terminal domains. Plant Physiol. 176, 2963–2976. doi: 10.1104/pp.17.00856
Khan, M. R., Ai, X. Y., Zhang, J. Z. (2014). Genetic regulation of flowering time in annual and perennial plants. Wiley Interdiscip Rev. RNA. 5, 347–359. doi: 10.1002/wrna.1215
Kojima, S., Takahashi, Y., Kobayashi, Y., Monna, L., Sasaki, T., Araki, T., et al. (2002). Hd3a, a rice ortholog of the arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43, 1096–1105. doi: 10.1093/pcp/pcf156
Lee, Y. S., Jeong, D. H., Lee, D. Y., Yi, J., Ryu, C. H., Kim, S. L., et al. (2010). OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB. Plant J. 63, 18–30. doi: 10.1111/j.1365-313X.2010.04226.x
Liljegren, S. J., Gustafson-Brown, C., Pinyopich, A., Ditta, G. S., Yanofsky, M. F. (1999). Interactions among APETALA1, LEAFY, and TERMINAL FLOWER1 specify meristem fate. Plant Cell 11, 1007–1018. doi: 10.1105/tpc.11.6.1007
Li, C., Pei, J., Yan, X., Cui, X., Tsuruta, M., Yi., L., et al. (2021). A poplar b-box protein PtrBBX23 modulates the accumulation of anthocyanins and proanthocyanidins in response to high light. Plant Cell Environ. 44, 3015–3033. doi: 10.1111/pce.14127
Li, F., Sun, J., Wang, D., Bai, S., Clarkeand, A. K., Holm, M. (2014). The b-box family gene STO (BBX24) in arabidopsis thaliana regulates flowering time in different pathways. PloS One 9, e87544. doi: 10.1371/journal.pone.0087544
Liu, H., Gu, F., Dong, S., Liu, W., Wang, H., Chen, Z., et al. (2016). CONSTANS-like 9 (COL9) delays the flowering time in oryza sativa by repressing the Ehd1 pathway. Biochem. Biophys. Res. Commun. 479, 173–178. doi: 10.1016/j.bbrc.2016.09.013
Liu, Y., Lin, G., Yin, C., Fang, Y. (2020). B-box transcription factor 28 regulates flowering by interacting with constans. Sci. Rep. 10, 17789. doi: 10.1038/s41598-020-74445-7
Li, Y., Wu, Q., Huang, X., Liu, S., Zhang, H., Zhang, Z., et al. (2016). Molecular cloning and characterization of four genes encoding ethylene receptors associated with pineapple (Ananas comosus l.) flowering. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00710
Luccioni, L., Krzymuski, M., Sánchez-Lamas, M., Karayekov, E., Cerdán, P. D., Casal, J. J. (2019). ). CONSTANS delays arabidopsis flowering under short days. Plant J. 97, 923–932. doi: 10.1111/tpj.14171
Min, X. J., Bartholomew, D. P. (1997). Temperature affects ethylene metabolism and fruit initiation and size of pineapple. Acta Hortic. 425, 329–338. doi: 10.17660/actahortic.1997.425.36
Mu, H., Li, Y., Yuan, L., Jiang, J., Wei, Y., Duan, W., et al. (2022). MYB30 and MYB14 form a repressor-activator module with WRKY8 that controls stilbene biosynthesis in grapevine. Plant Cell, koac308. doi: 10.1093/plcell/koac308
Ouyang, Y., Pan, X., Wei, Y., Wang, J., Xu, X., He, Y., et al. (2022). Genome-wide identification and characterization of the BBX gene family in pineapple reveals that candidate genes are involved in floral induction and flowering. Genomics 114, 110397. doi: 10.1016/j.ygeno.2022.110397
Plunkett, B. J., Henry-Kirk, B., Friend, A., Diack, R., Helbig, S., Mouhu, K., et al. (2019). Apple b-box factors regulate light-responsive anthocyanin biosynthesis genes. Sci. Rep. 9, 17762. doi: 10.1038/s41598-019-54166-2
Putterill, J., Robson, F., Lee, K., Simon, R., Coupland, G. (1995). The CONSTANS gene of arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847–857. doi: 10.1016/0092-8674(95)90288-0
Rao, X., Huang, X., Zhou, Z., Lin, X. (2013). An improvement of the 2– ΔΔCT method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinforma Biomath. 3, 71–85. doi: 10.1089/cmb.2012.0279
Roux, F., Touzet, P., Cuguen, J., Le Corre, V. (2006). How to be early flowering: an evolutionary perspective. Trends Plant Sci. 11, 375–381. doi: 10.1016/j.tplants.2006.06.006
Tripathi, P., Carvallo, M., Hamilton, E. E., Preuss, S., Kay, S. A. (2017). Arabidopsis b-BOX32 interacts with CONSTANS-LIKE3 to regulate flowering. Proc. Natl. Acad. Sci. U S A. 114, 172–177. doi: 10.1073/pnas.1616459114
Turnbull, C. G., Sinclair, E. R., Anderson, K. L., Nissen, R. J., Shorter, A. J., Lanham, T. E. (1999). Routes of ethephon uptake in pineapple (Ananas comosus) and reasons for failure of flower induction. J. Plant Growth Regul. 18, 145–152. doi: 10.1007/pl00007062
Wang, C. Q., Guthrie, C., Sarmast, M. K., Dehesh, K. (2014). ). BBX19 interacts with CONSTANS to repress FLOWERING LOCUS T transcription, defining a flowering time checkpoint in arabidopsis. Plant Cell 26, 3589–3602. doi: 10.1105/tpc.114.130252
Wang, Y., He, X., Yu, H., Mo, X., Fan, Y., Fan, Z., et al. (2021). Overexpression of four MiTFL1 genes from mango delays the flowering time in transgenic arabidopsis. BMC Plant Biol. 21, 407. doi: 10.1186/s12870-021-03199-9
Wang, J., Ouyang, Y., Wei, Y., Kou, J., Zhang, X., Zhang, H. (2022). Identification and characterization of trihelix transcription factors and expression changes during flower development in pineapple. Horticulturae 8, 894. doi: 10.3390/horticulturae8100894
Wang, L., Sun, J., Ren, L., Zhou, M., Han, X., Ding, L., et al. (2020). CmBBX8 accelerates flowering by targeting CmFTL1 directly in summer chrysanthemum. Plant Biotechnol. J. 18, 1562–1572. doi: 10.1111/pbi.13322
Wei, C., Chien, C., Ai, L., Zhao, J., Zhang, Z., Li, K. H., et al. (2016). The arabidopsis b-BOX protein BZS1/BBX20 interacts with HY5 and mediates strigolactone regulation of photomorphogenesis. J. Genet. Genomics 43, 555–563. doi: 10.1016/j.jgg.2016.05.007
Xu, H., Yu, Q., Shi, Y., Hua, X., Tang, H., Yang, L., et al. (2018). PGD: Pineapple genomics database. Hortic. Res. 5, 66. doi: 10.1038/s41438-018-0078-2
Yang, Y., Ma, C., Xu, Y., Wei, Q., Imtiaz, M., Lan, H., et al. (2014). A zinc finger protein regulates flowering time and abiotic stress tolerance in chrysanthemum by modulating gibberellin biosynthesis. Plant Cell 26, 2038–2054. doi: 10.1105/tpc.114.124867
Yano, M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T., et al. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the arabidopsis flowering time gene CONSTANS. Plant Cell 12, 2473–2484. doi: 10.1105/tpc.12.12.2473
Zhang, H., Pan, X., Liu, S., Lin, W., Li, Y., Zhang, X. (2021a). Genome-wide analysis of AP2/ERF transcription factors in pineapple reveals functional divergence during flowering induction mediated by ethylene and floral organ development. Genomics 113, 474–489. doi: 10.1016/j.ygeno.2020.10.040
Zhang, H., Wang, J., Tian, S., Hao, W., Du, L. (2022). Two b-box proteins, MaBBX20 and MaBBX51, coordinate light-induced anthocyanin biosynthesis in grape hyacinth. Int. J. Mol. Sci. 23, 5678. doi: 10.3390/ijms23105678
Keywords: pineapple (Ananas comosus (L.) Merr.), B-box, expression profiling, flowering, floral organ
Citation: Ouyang Y, Zhang X, Wei Y, He Y, Zhang X, Li Z, Wang C and Zhang H (2022) AcBBX5, a B-box transcription factor from pineapple, regulates flowering time and floral organ development in plants. Front. Plant Sci. 13:1060276. doi: 10.3389/fpls.2022.1060276
Received: 03 October 2022; Accepted: 03 November 2022;
Published: 24 November 2022.
Edited by:
Ting Wu, China Agricultural University, ChinaReviewed by:
Hyo-Jun Lee, Korea Research Institute of Bioscience and Biotechnology (KRIBB), South KoreaCopyright © 2022 Ouyang, Zhang, Wei, He, Zhang, Li, Wang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongna Zhang, MTM2OTI0NzY5NzlAMTM5LmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.