
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 01 December 2022
Sec. Plant Cell Biology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1060228
This article is part of the Research TopicCooperation of Gene Regulatory Networks And Phytohormones in Cell Development and MorphogenesisView all 6 articles
 Kun Chen1†
Kun Chen1† Chang Qu1†
Chang Qu1† Xiao-yue Zhang1
Xiao-yue Zhang1 Wei Wang1
Wei Wang1 Chen-rui Gu1
Chen-rui Gu1 Gui-feng Liu1
Gui-feng Liu1 Qi-bin Yu2
Qi-bin Yu2 Chuan-ping Yang1*
Chuan-ping Yang1* Jing Jiang1*
Jing Jiang1*Leaves are one of the vegetative organs of plants that are essential for plant growth and development. PIN-FORMED (PINs) gene is an indoleacetic acid (IAA) transporter that plays a critical role in leaf development. To determine the function of BpPIN3 in leaf polarity formation in Betula pendula, the transgenic lines with BpPIN3 overexpression (OE) and BpPIN3-reduced expression (RE) were analyzed using the Agrobacterium-mediated method. The RE lines displayed the characteristics of leaf margin adaxial upward curling, with lower expression of BpPIN3 resulting in greater rolling. Tissue localization of IAA in the auxin GUS reporter system proved that auxin in the RE was mainly distributed in the secondary veins, palisade tissues, and epidermal cells in the leaf margin area. The auxin content in the leaf margin area was significantly greater than that in the main vein tissue. The cell density of the palisade tissue and the ratio of palisade tissue to spongy tissue in the curled leaf margin of the RE lines were found to be significantly decreased. RNA-seq analysis revealed that the RE hormone-signaling pathway genes were significantly enriched compared with those of the OE and WT lines; in particular, the auxin response-related genes SAURs (i.e., SAUR23, SAUR24, SAUR28, and SAUR50) and GH3.10 were found to be significantly upregulated. qRT-PCR analysis indicated that BpPIN3 expression at the leaf margin was significantly lower than that near the main vein in the RE lines. In contrast, the expression levels of SAURs and GH3.10 were significantly higher than those near the midrib. In conclusion, BpPIN3 regulates the expression of auxin response-related genes and the polar transport of auxin to change the polar form of the proximal and distal axes of birch leaves.
Leaves are important vegetative organs in plants. Leaf morphogenesis is of great significance to plant growth and development (Ichihashi and Tsukaya, 2015; Chitwood and Sinha, 2016). In plant developmental biology, polarity establishment is the core issue related to organ morphogenesis. To study the mechanism of leaf polarity formation, past studies have attempted to define the polarity of the leaf in a three-dimensional spatial axis according to the relative position relationship between leaf primordium and shoot apical meristem (SAM) (Bowman, 2000). This axis comprises the proximal-distal axis, medial-lateral axis, and adaxial-abaxial axis (Bowman, 2000; Bowman et al., 2002; Hasson et al., 2010). Of these, leaf adaxial-abaxial polarity formation is an important leaf development process that is closely related to important physiological processes such as leaf photosynthesis. Several studies have demonstrated that plant leaf adaxial-abaxial polarity formation involves complex gene regulatory networks, which mainly include adaxial-abaxial polarity genes, adaxial-abaxial boundary-related genes, small RNAs, and plant hormones. These factors regulate the leaf adaxial-abaxial polarity formation through synergistic or antagonistic effects (Eshed et al., 2004; Bonaccorso et al., 2012; Iwasaki et al., 2013; Machida et al., 2015). For example, the HD-ZIP II and HD-ZIP III subfamily members of the homeodomain-leucine zipper (HD-ZIP) transcription factor gene family are involved in the regulation of leaf adaxial surface polarity formation. Among them, REVOLUTA (REV) gene deletion mutant rev of the HD-ZIP III gene family in Arabidopsis thaliana caused abnormal leaf cell division and leaf edge rolling (McConnell et al., 2001; Otsuga et al., 2001). The double-deletion mutants or triple-deletion mutants of REV subfamily genes exhibited obvious defective phenotypes. The rev/phv double-deletion mutant causes the abaxialization of the plant and the formation of a trumpet-like structure (Prigge et al., 2005). The phb/phv/rev triple mutant caused a defective SAM, which produced only abaxial cotyledons (Emery et al., 2003). Studies have revealed that microRNAs were involved in plant proximal-abaxial polarity formation (Juarez et al., 2004; Chitwood et al., 2009). On the adaxial surface of leaves, miRNAs formed a complex with ARGONAUTE1 (AGO1), which further inhibited the translation of its potential target homeodomain/leucine zipper genes PHABULOSA (PHB) and PHAVOLUTA (PHV), which were distributed and expressed in a polar manner in the leaf primordium, thereby regulating the adaxial cell development (Kidner and Martienssen, 2004). In addition, ARP, KANADI (KAN), AUXIN RESPONSIVE FACTOR (ARF), and other family genes also play a key role in the formation of adaxial-abaxial leaf polarity. In cotton (Gossypium spp.), leaf development was regulated by the auxin response factor ARF16-1 through transcriptional regulation of KNOX2-1. The heterologous expression of this gene in Arabidopsis suggested that its overexpression led to the cracking of leaves and leaf abaxial rolling (He et al., 2021). These observations prove that plant leaf development is a complex process involving the transcription factor family, small RNAs, and several hormone-related genes that affect leaf development by regulating the leaf hormone levels.
Auxin is one of the essential hormones in plants. Auxin regulates plant growth and development by creating concentration differences in different tissues through synthetic regulation or transport regulation (Ludwig-Müller, 2011; Adamowski and Friml, 2015; Kasahara, 2016). Similarly, IAA is essential for leaf polarity formation. Auxin not only regulates cell division and promotes cell elongation but also affects the morphogenesis of individual plants, leaves, and other organs due to its concentration gradient (Ding and Friml, 2010; Ljung, 2013; Robert et al., 2015). The PIN-FORMED (PINs) family is a gene family that encodes auxin polar transport carrier elements. It plays an important role in the auxin flow between cells and mediating auxin regulation (Wisniewska et al., 2006). Different members of the family genes have different functions. In the Arabidopsis unhinged-1 (unh-1) mutant, the expression of PIN1 at the leaf margin was inhibited, which modified the auxin content in the leaves, resulting in the narrowing of the leaves and obvious serration at the leaf margin (Pahari et al., 2014). In addition, the protein PIN3 is sensitive to red light (R) and far-red light (FR). Its location within cells can determine the direction of polar auxin transport, which, in turn, affects the distribution pattern of auxin. This event impacts hypocotyl development (Keuskamp et al., 2010). The APETALA2 transcription factor DORNROSCHEN (DRN) interferes with auxin response and PIN1 expression, resulting in asymmetric auxin distribution in leaves, which further affects the establishment of adaxial-abaxial polarity in leaves (Chandler et al., 2007). The formation of local auxin concentration maximum and minimum in plants through polar auxin transport (PAT) and the establishment of auxin concentration gradient affect leaf development, with PINs being the key participants in this process. Although a sufficient number of studies have detailed how PINs affect the auxin polarity distribution in plants, there is a paucity of such studies on trees.
Birch (Betula platyphylla × B. pendula) is an important broad-leaved, fast-growing tree species in China (Wang et al., 2018). Leaves are essential organs for plant growth and development, as they provide energy through photosynthesis, respiration, and storage of nutrients. The development of leaves is a complex process that involves physiological and biochemical changes (Tsukaya, 2005). In our previous study, we discovered that the transcriptomes of leaves of birch and Betula pendula ‘Dalecarlica’ differed mainly in their auxin signal transduction genes, such as BpPINs, Aux/IAA, ARFs, and GH3s (Bian et al., 2019). Analysis of the expression characteristics of BpPINs family genes in birch indicated that 6 BpPINs had high expression in the leaves and that the expression of BpPINs was significantly positively correlated with the auxin content. The BpPINs family genes were believed to be vital for the birch leaf margin’s morphogenesis (Qu et al., 2020). The expression of BpPIN3 was the highest in the developing first leaf compared with other leaves, tissues, and organs (Kun et al., 2022). This finding suggested that BpPIN3 plays an important role in the morphogenesis and development of birch leaves. However, the mechanism by which BpPIN3 affects the leaf margin development remains unclear.
This study aimed to clarify the function of BpPIN3 by elucidating the molecular mechanism underlying leaf rolling in BpPIN3-reduced expression lines. The leaf palisade tissue (PT) cell density, leaf endogenous IAA content, and tissue localization were determined through leaf anatomical observation. The relationship between BpPIN3 and IAA was accordingly clarified. Based on the differentially expressed gene (DEG) mining through RNA-seq analysis, the key genes regulating leaf adaxial rolling were identified, and the role of BpPIN3 in leaf polarity formation was determined. These findings offer guidance for the genetic engineering of birch trees for desired traits.
Bpev01.c1162.g0001.m0001 was identified as the AtPIN3 homolog with 75.38% similarity in the birch genome. The open reading frame (ORF) of the sequence was 1953 bp, encoding a protein of 650 amino acids (Figure 1A). The primers were designed according to the full-length sequence of the ORF and are listed in Supplementary Table S1. The cDNA of birch leaves was used as a template for PCR amplification. Gel electrophoresis revealed that the amplified band was visible at 2000 bp (Figure S1A). After purification, ligation, and transformation of E. coli, single clones were sequenced. The amplified sequence was 1953 bp, which is consistent with the information in the NCBI database.
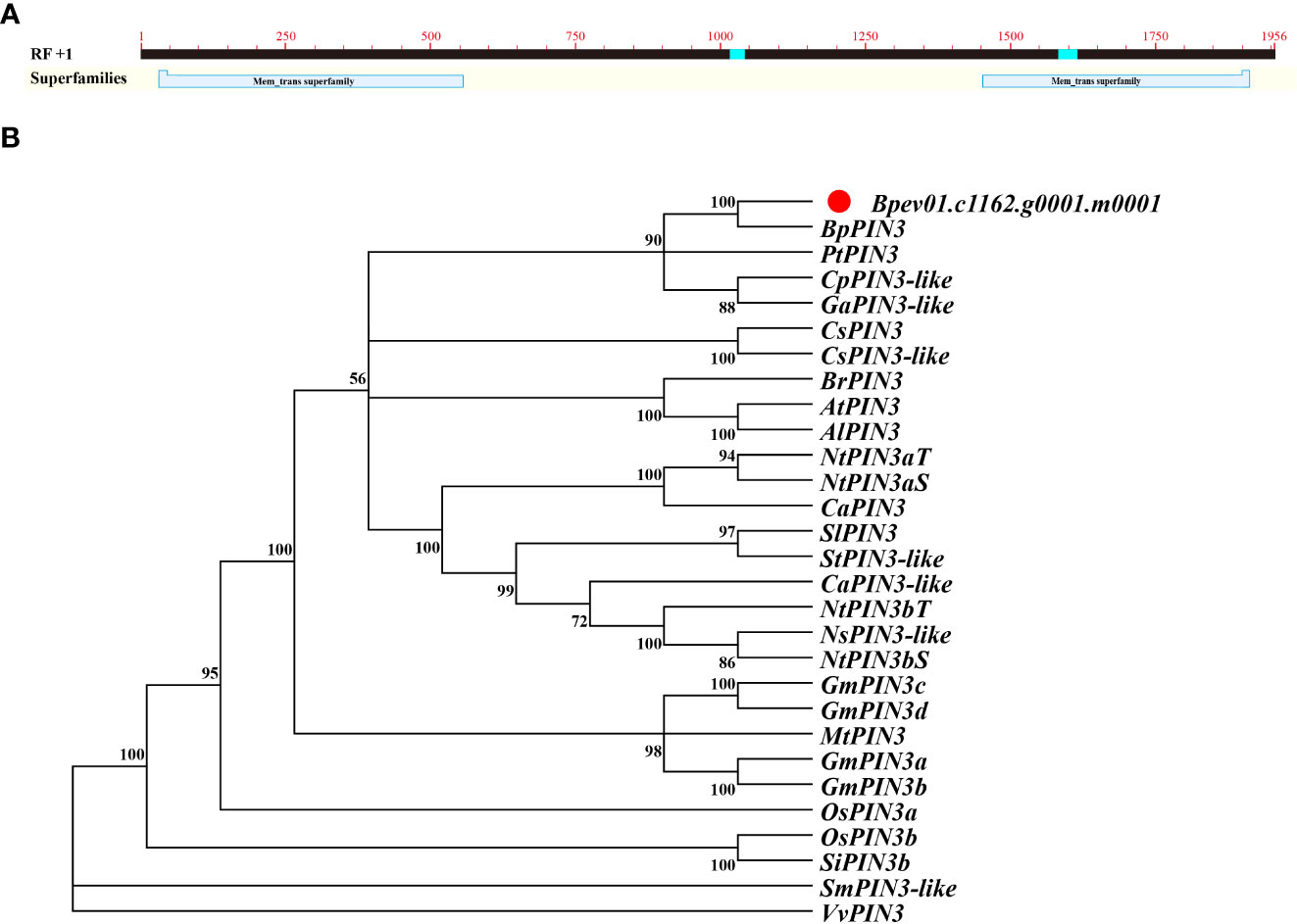
Figure 1 Sequence and phylogenetic analysis of BpPIN3 (A) BpPIN3 sequence was analyzed by BlastX; (B) Cluster analysis was performed on the orthologous sequence of BpPIN3 and PIN3 amino acid in different species.
Protein sequence alignment and phylogenetic tree analysis revealed that the Bpev01.c1162.g0001.m0001 gene shared high similarity with PIN3 from other species and 100% similarity with BpPIN3 from Betula pendula subsp.pendula. Therefore, this gene was considered the same as BpPIN3 of birch (Figures 1B; S1 B).
BpPIN3 overexpression lines (OE1–OE3) and the suppressed expression lines (RE1–RE3) were obtained through Agrobacterium-mediated zygotic embryo transformation (Figure S2A). Total DNA from the leaves of the BpPIN3 OE lines was used as the template. The 35S:: PIN3 plasmid was used as the positive control, whereas the WT birch DNA was used as the negative control. PCR amplification was performed using two ends of the BpPIN3 ORF as primers. The results revealed that the positive plasmid and the 3 transgenic lines were amplified in a single band near 2000 bp, which is consistent with the base length of BpPIN3 sequence (1953 bp). The WT strain showed no amplified band (Figure S2B).
The positive target sequence and the complementary sequence were amplified through PCR. The inserted target gene sequence and the reverse complementary sequence in OE lines showed amplified bands, which were consistent with the expected 400-bp length (Figures S2C, D). The results indicated that the target gene was integrated into the birch genome.
The gene expression of BpPIN3 in three OE lines was significantly higher than that in WT (P< 0.05), as determined through qRT-PCR. The expression of BpPIN3 in OE3 was 17.74 times higher than that in WT. The expression of BpPIN3 in the 3 RE lines was significantly lower than that in WT (P< 0.05). The expression of BpPIN3 in RE2 was the lowest and 3.85 times lower than that in WT (Figure 2A). At the same time, qRT-PCR detection of the remaining 5 BpPINs in the RE lines revealed that the expression of only BpPIN3 in the 3 RE lines was significantly lower than that in the WT lines, while those of the other 5 BpPINs were significantly upregulated or showed no significant difference (Figure S3).
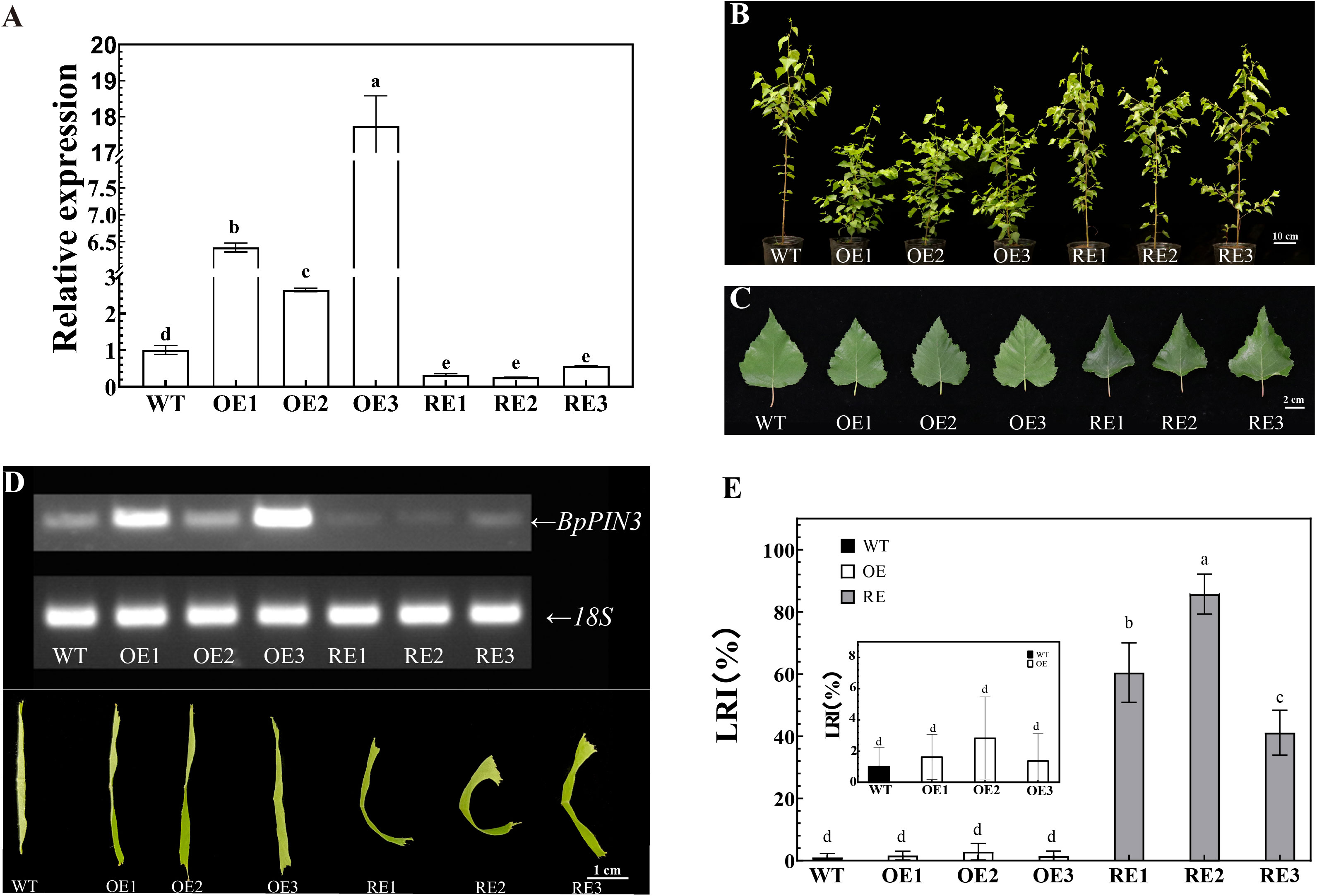
Figure 2 Leaf phenotype of RE, OE, and WT lines (A) BpPIN3 expression in the tested lines. Error bars indicate SD (mean ± SD), n = 3; (B) Phenotypic image of 13-month-old test lines, Scale bar = 10 cm; (C) Comparison of the third leaf of the tested lines, Scale bar = 2 cm; (D) Semi-quantitative RT-PCR electrophoretogram of the tested lines and the curly leaf phenotypes in different BpPIN3 transgenic lines; (E) LRI comparison of the fourth leaf of the tested lines. Error bars indicate SD (mean ± SD), n = 30. Different letters denote statistically significant differences after one-way ANOVA.
Leaves and seedling height of BpPIN3 transgenic birch were observed and measured. The leaf length, leaf width, and leaf area of the RE and OE lines were smaller than those of the WT lines, although the differences were nonsignificant (Supplementary Table S2). Although OE1-2 was significantly lower than that in the other lines, there was no significant difference in the plant height among OE3, RE, and WT lines (Supplementary Table S3). The leaves of RE lines displayed a phenomenon of adaxial upward curling in which the leaf margin curled upward (Figures 2B, C). The relationship between the expression level of BpPIN3 and the degree of leaf rolling was found to be negative (Figure 2D). The leaf rolling index (LRI) is an important index that reflects the degree of leaf rolling. Therefore, the LRI statistics indicated no significant difference between the OE lines and WT lines, and the mean values of the 3 lines were all<4%. The LRI of 3 RE lines was significantly higher than that of WT (P< 0.0001), and the LRI of RE2 lines was 85.7% (Figure 2E). The decrease in BpPIN3 expression caused adaxial leaf rolling, whereas the increase in BpPIN3 expression showed no significant effect on the leaf phenotype.
After transparentizing the mature leaves of the tested lines, palisade cells density showed no statistically significant difference between the OE lines and WT lines (Figures 3A–D). However, the palisade cells density of the RE lines was significantly lower than that of the WT and OE lines (P< 0.0001). The palisade cells density in the main vein of the RE lines was 27.42 (cells/µm2), which was 6.16% and 4.96% lower than that of the WT and OE lines, respectively (Figure 3C). The density of palisade cells in the leaf margin area was only 22.44 (cells/µm2), which was 25.47% lower than that of WT lines and 25.18% lower than that of the OE lines (Figure 3D). This finding indicated that compared with WT and OE lines, the palisade tissue cells of RE lines are loosely arranged. The closer to the leaf margin, the more obvious the degree of looseness.
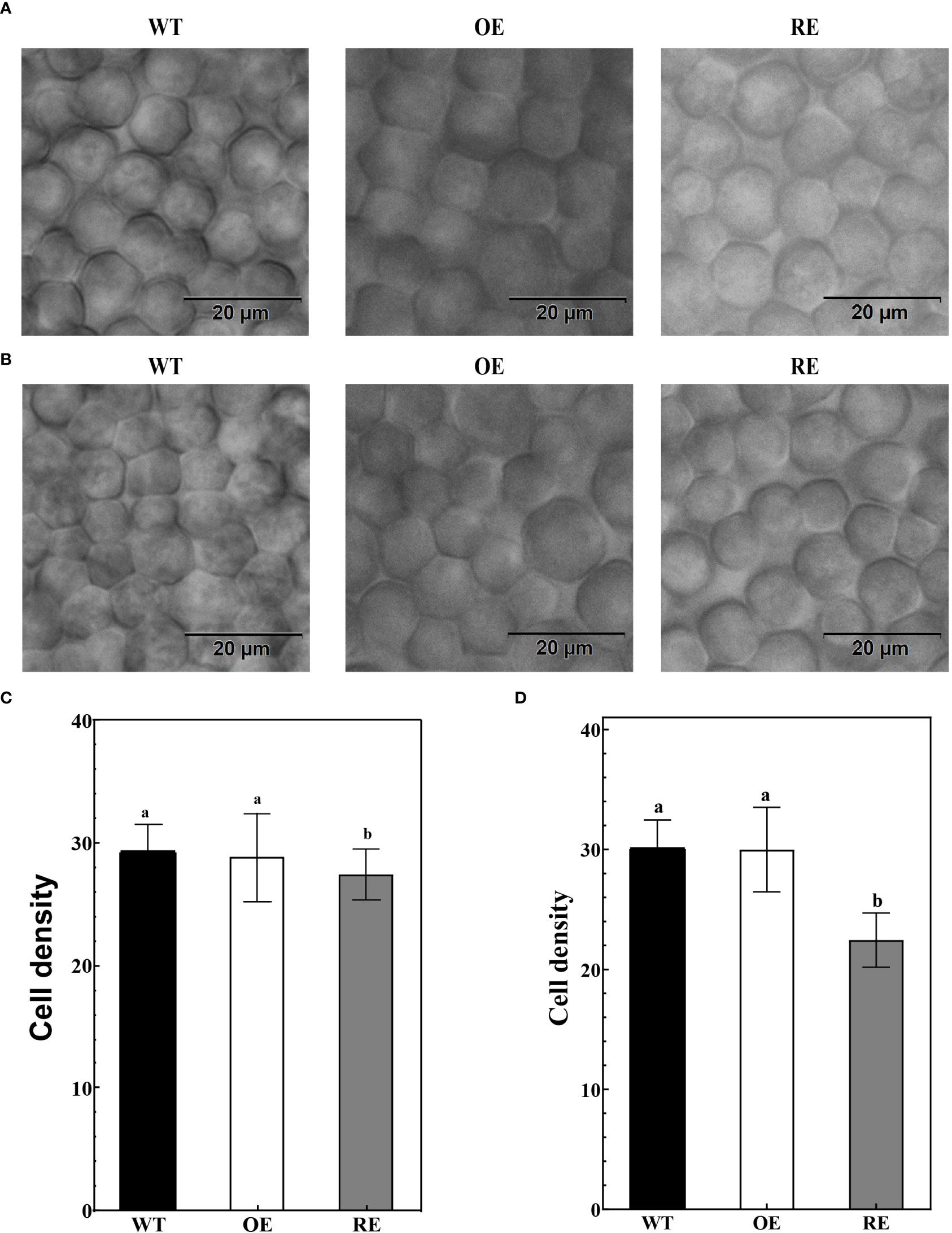
Figure 3 Leaf palisade tissues of the RE, OE, and WT lines (A) Observation of the palisade tissue cells in the main vein area; (B) Leaf margin palisade tissue cell observation; (C) The number of cells per unit area of the palisade tissues in the main vein area; Error bars indicate SD, n = 70; (D) Number of cells per unit area of the leaf margin palisade tissues. Error bars indicate SD, n = 70. Different letters denote statistically significant differences after one-way ANOVA.
The anatomical observation indicated no significant difference between the OE lines and WT lines in terms of the palisade/spongy ratio and PT density at the leaf margin curling site. However, the palisade/spongy ratio and PT density of the RE lines were 33.33% and 30.1% lower, respectively, than those of the WT lines (Figures 4A–C).
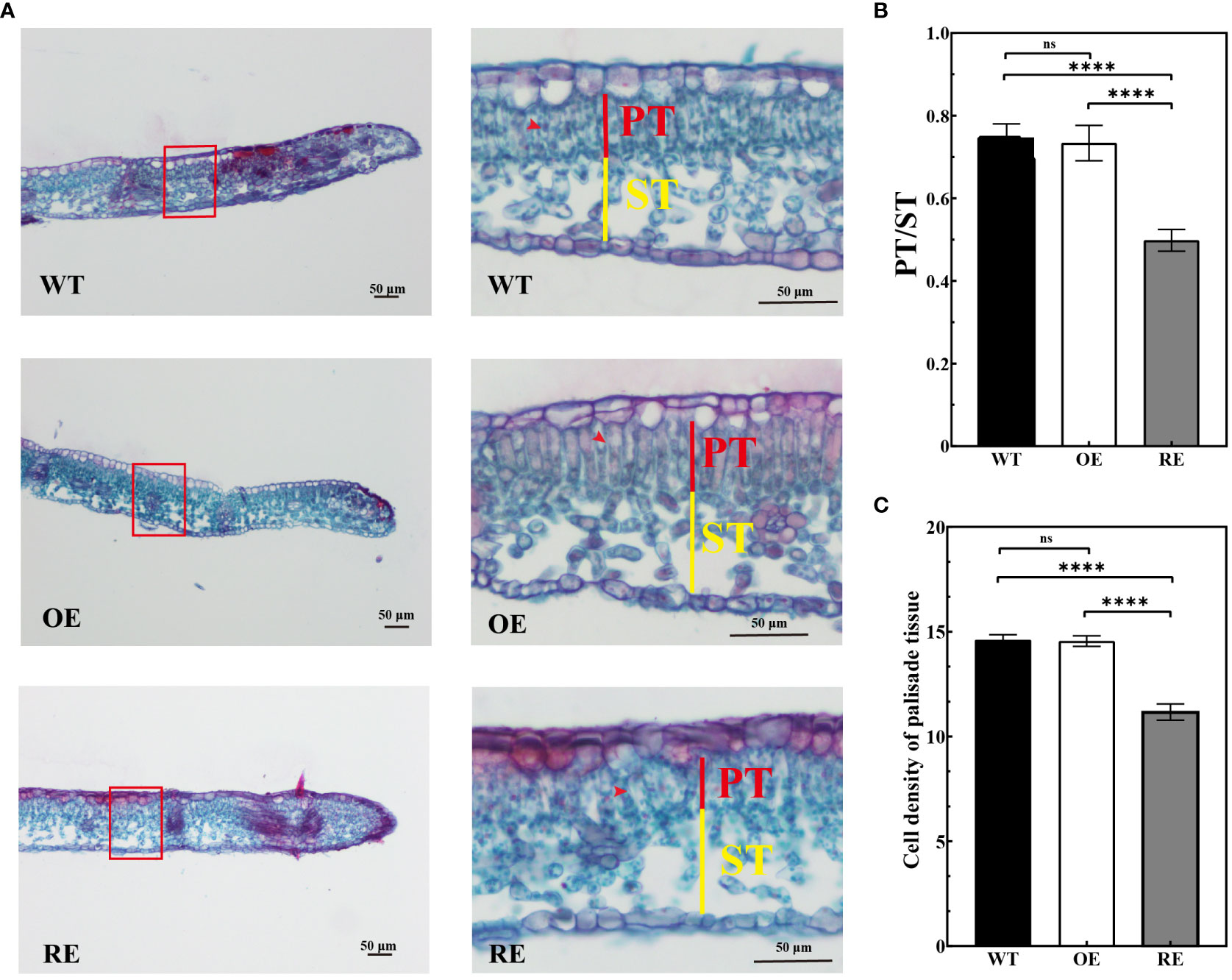
Figure 4 Leaf margin section observation of the RE, OE, and WT lines (A) Leaf margin sections of RE, WT, and OE lines, Scale bar = 50 μm; (B) Marginal zone ratio of the test lines; (C) Cell density of palisade tissues under the transverse view of the leaf margin area. Error bars indicate SD, n = 30. Data were analyzed by one-way ANOVA. ****p< 0.0001 and ns, no significance.
To determine whether the morphology of the leaves of the RE lines was changed after flattening, 13 landmarks (Figure S4A) were marked on the leaves of the tested lines, and the original coordinate matrix was transformed into a covariance matrix (Figure S4B) by using Procrustes of MorphoJ. The results of principal component analysis (PCA) revealed that the principal components PC1 and PC2 accounted for only 47.33% of all leaf type differences, whereas PC1 (28.78%) could not significantly distinguish each strain (Figures S4C, D). This finding indicated that the leaf type of RE, OE, and WT in the flat state did not change significantly. The adaxial rolling of the leaves of RE lines did not induce changes in their leaf type.
To identify the DEGs in BpPIN3 transgenic birch, RNA-seq analysis was performed on mature leaves of the RE, OE, and WT line, and the differentially expressed genes (DEGs) were analyzed based on the following criteria: p< 0.05 and|Log2Foldchange| ≥ 1. PCA of the transcriptome data clearly displayed differences among all transgenic lines. Each point represents the transcriptome of a different sample (Figure S5). As the leaf shape of OE and WT lines is the same, the analysis only focuses on the related DEGs in WT-RE and OE-RE. The results revealed that there were 686 DEGs in the intersection, of which 381 DEGs were upregulated and 305 DEGs were downregulated in the RE lines when compared with WT lines (Figure 5A). GO enrichment of the DEGs in the intersection displayed significant enrichment pathways mainly in the cell surface receptor signaling pathway, plant organ development, regulation of the hormone levels, and response to jasmonic acid (Figures 5B; S6). Through KEGG enrichment analysis of the intersection, 686 DEGs were found to be enriched in 5 pathways including unsaturated fatty acid biosynthesis, tryptophan metabolism, hormone signal transduction, lysine degradation, and zeatin biosynthesis (Figure 5C). Further analysis of the 5 enriched KEGG pathways revealed that the hormone signal transduction pathway was mainly related to the auxin signal pathway, and the 5 genes including GH3.10 and SAURs in this pathway displayed an upregulated expression trend in the RE lines (Figure 5D). This finding indicated that auxin response-related genes were significantly highly expressed in the RE lines. The expression of these 5 DEGs was verified by quantitative real-time reverse transcription PCR (qRT-PCR) analysis, which was consistent with the DEG data (Figure S7).
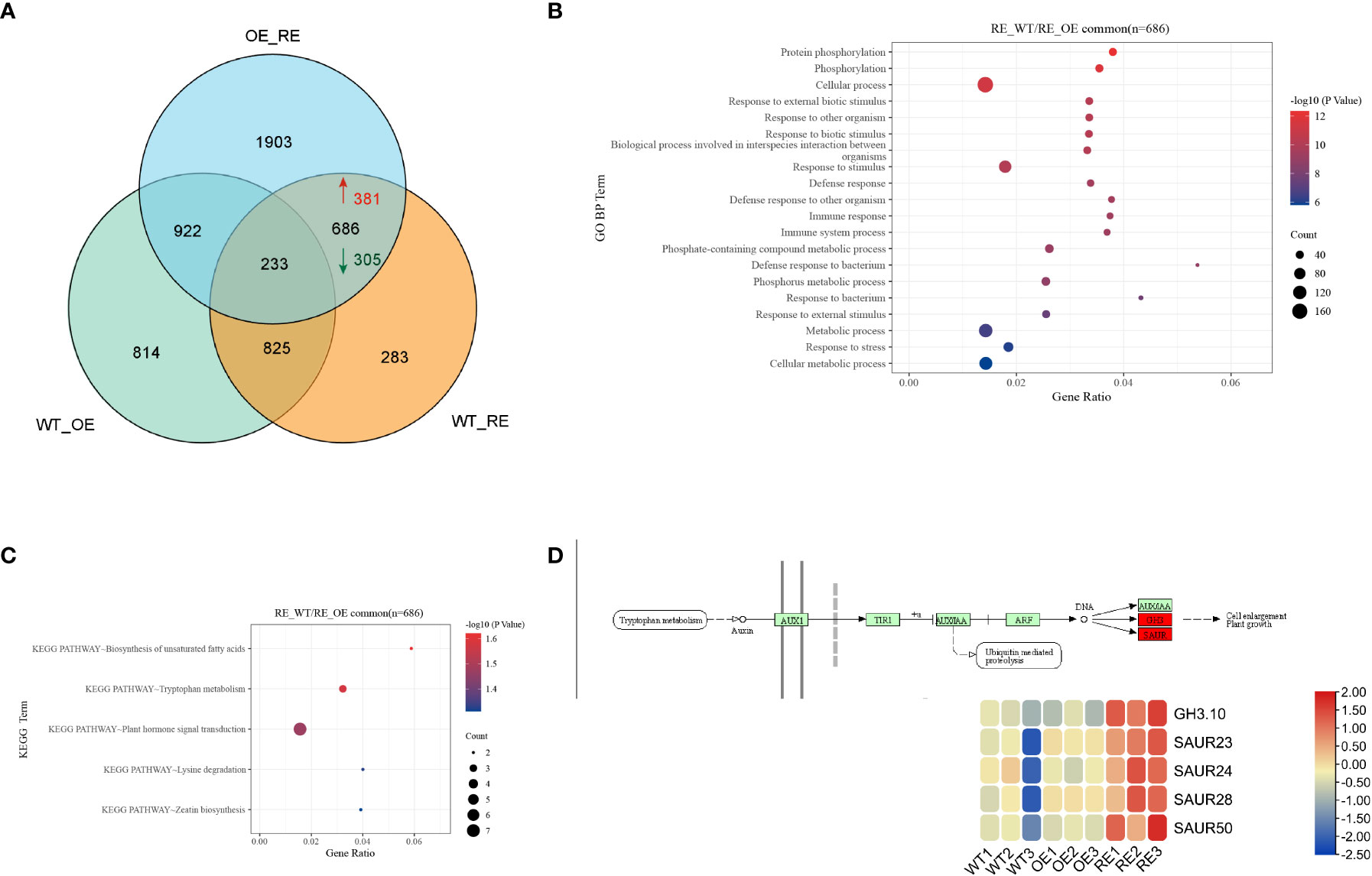
Figure 5 Transcriptome sequencing analysis of the RE, OE, and WT lines (A) Veen diagram of DEGs among RE, OE, and WT lines; (B) Target interval GO enrichment analysis; (C) Target interval KEGG enrichment analysis; (D) Heat map of gene expressions in the auxin signaling pathway.
RNA-seq analysis revealed that the expression of IAA response-related genes in the leaves of RE lines increased significantly. To determine whether the increased expression of these genes affected the auxin content in the RE lines, the IAA contents in the top buds and the first leaves of RE, OE, and WT lines were measured through high-performance liquid chromatography-mass spectrometry (ESI-HPLC-MS/MS) (Figure 6A). The results indicated that the IAA content of the RE lines was significantly higher than that of the WT lines, whereas that of the OE lines was significantly lower than that of the WT lines (P< 0.0001). The expression of BpPIN3 in the top buds and the first leaves of the RE lines was significantly lower than that of the WT lines, while it was significantly higher in the OE lines than in the WT lines (Figure 6B). The correlation analysis between the expression of BpPIN3 and IAA content demonstrated that the expression of BpPIN3 in the RE, OE, and WT lines was significantly negatively correlated with the IAA content (P = 0.0004) (Figure 6C).
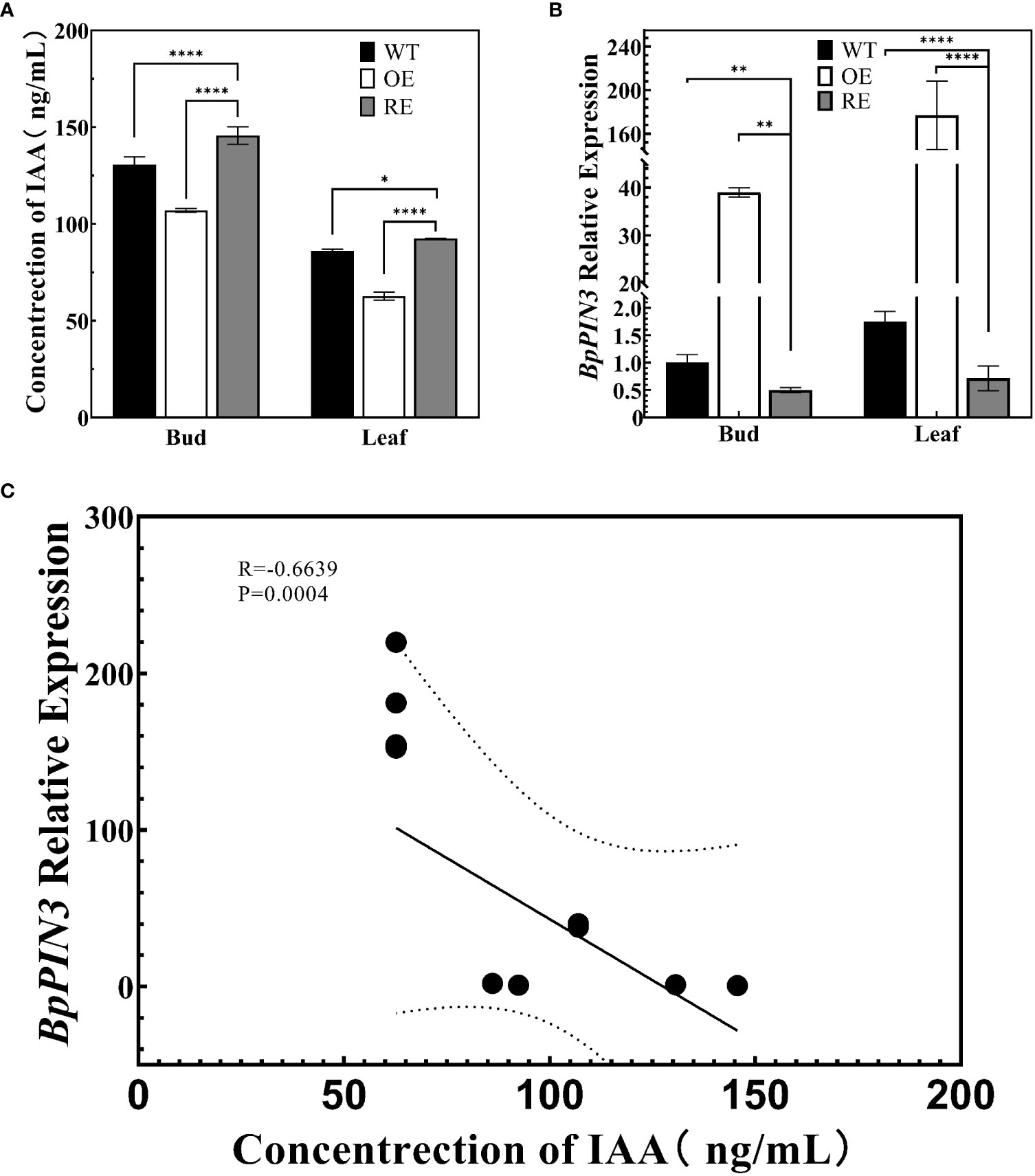
Figure 6 Auxin content and BpPIN3 expression in apical buds and leaves of the tested lines (A) Auxin content in the shoot apex and leaves of the RE, OE, and WT lines. (B) Relative expression of BpPIN3 in the shoot apex and leaves of the RE, OE, and WT lines. (C) Pearson’s correlation analysis of auxin and BpPIN3 expression. Data were analyzed by one-way ANOVA. Error bars indicate SD, n = 3. *p< 0.05; **p< 0.01; ****p< 0.0001.
To clarify whether the change in the auxin content causes leaf rolling in the RE lines, the IAA reporter vector proDR5:: GUS was introduced into the genome of RE2 lines as the receptor, resulting in the RE (proDR5:: GUS) lines. The WT (proDR5:: GUS) line was set as the control. The auxin signal was visualized by GUS staining of transgenic lines to understand the distribution of IAA in the leaves of the RE lines (Figure 7A). The observation of the stained leaves revealed that GUS staining was almost negligible in the leaf tips of the WT lines. GUS staining of the leaf tip of the RE strain was more obvious than that of the WT (Figure 7B). The tissue section observation revealed that the GUS staining sites of the WT strain were mainly concentrated in the main vein and the secondary vein of the leaf margin, and the color was uniform. The GUS staining of the RE lines was lighter in the main vein area, whereas, in the leaf margin area, it was deeper than that in the main vein area. GUS staining was also clearly visible in the epidermal cells and PTs at the leaf tip. Since GUS staining represents the distribution of IAA, the IAA content in the secondary veins of the leaf margin of the RE lines was significantly higher than that in the main veins, epidermal cells, and PTs containing IAA. No significant difference was noted in the IAA content between the main vein and the secondary vein of the WT lines, and there was no IAA distribution in the epidermal cells of the leaf tip.
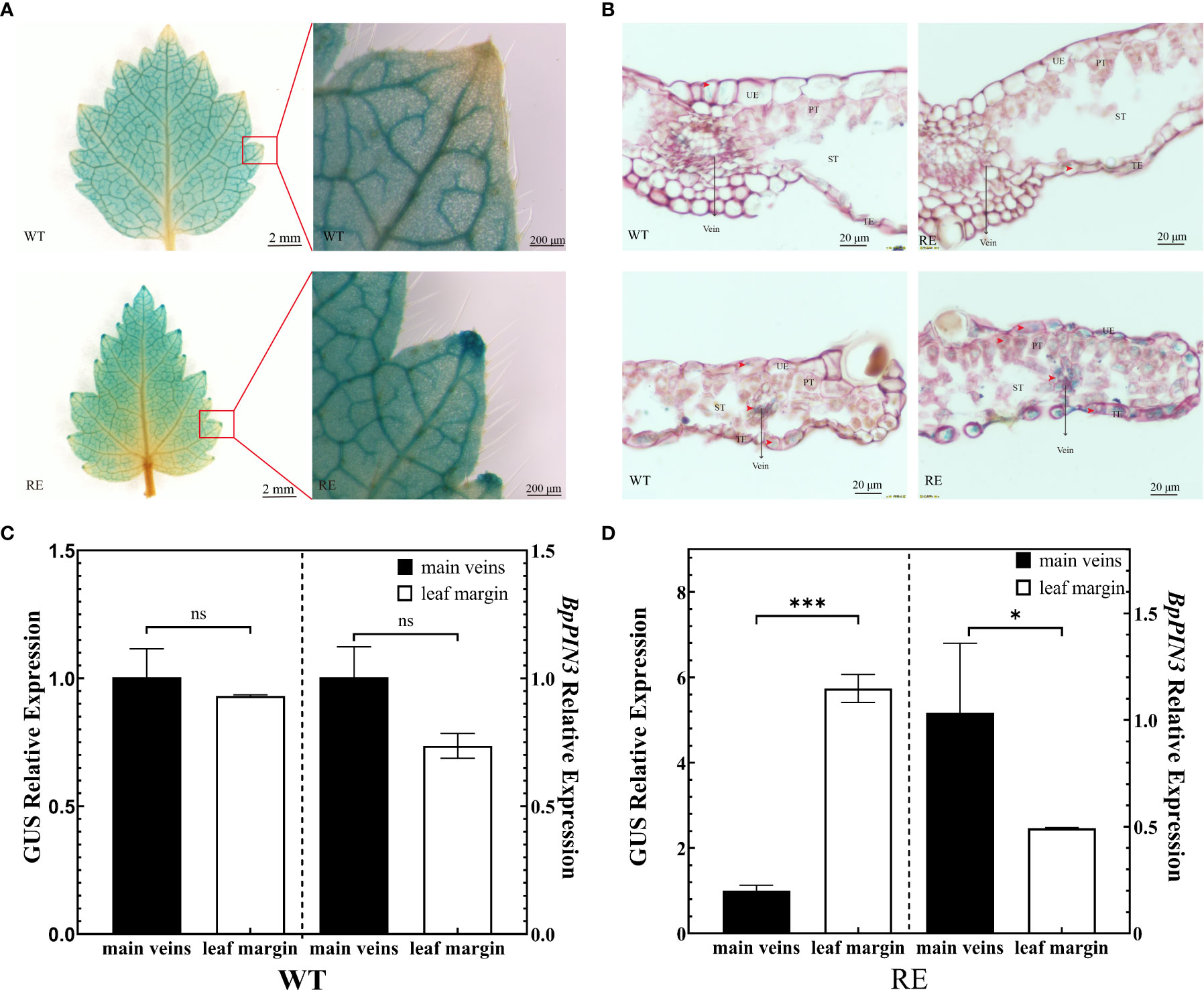
Figure 7 Asymmetric distribution of auxin in the leaves from the RE lines (A) GUS staining of the leaves from the RE and WT lines, Scale bar = 2 mm and enlarged at the leaf margin, Scale bar = 200 μm; (B) Slice observation after GUS staining in the main vein region and leaf margin region, Scale bar = 20 μm; (C) GUS and BpPIN3 expressions in the WT lines; (D) GUS and BpPIN3 expressions in the RE lines. Error bars indicate SD, n = 3. Data were analyzed by Tukey’s tests. *p< 0.05; ***p< 0.001. ns, not significantly different.
The asymmetric distribution of auxin in plants is caused by the polar transport of auxin by PINs. To understand the relationship between the asymmetric distribution of IAA content and the expression of BpPIN3 in leaves, the leaves in the main vein region and leaf margin region of the RE (proDR5:: GUS) and WT (proDR5:: GUS) lines were used as materials to determine the GUS enzyme activity. The relative expression levels of GUS and BpPIN3 were analyzed by qRT-PCR. The results revealed no significant change in the GUS activity in the leaf margin and main vein region of the WT lines; however, GUS activity in the leaf margin of the RE lines was significantly higher than that in the main vein region (Figure S8). GUS and BpPIN3 expressions in the main vein and leaf margin of the RE lines were significantly different (P< 0.05) (Figures 7C, D). However, no significant difference was noted between the WT lines (Figures 7C, D). The correlation analysis revealed a significant negative correlation between BpPIN3 and GUS expression (P< 0.0001) (Figure S9), implying that the decrease in BpPIN3 expression in the leaf margin of the RE lines may be the reason for the increase in IAA content.
RNA-seq revealed the expression levels of auxin response genes, GH3.10 and SAURs, in the auxin signaling pathway that was enriched by the significantly upregulated DEGs between the RE lines and OE and WT lines. These genes are known to be directly related to the auxin content, and an increase in their expression can increase the auxin content to a certain extent. To further explore whether these genes are related to the change in auxin content in the leaf margin of the RE lines, the expression levels of these genes in different parts of the RE lines were determined. The expression levels of each gene in the leaf margin of the RE lines were found to be higher than those in the main vein area (Figure S10), which possibly led to an increase in the auxin content in the leaf margin, consistent with the results of GUS staining.
The morphogenesis of normal leaves requires the establishment and coordination of the polarity of the adaxial-abaxial, adaxial-lateral, and basal-apical axes (Li et al., 2005; Liu et al., 2011). Abnormalities in the development of adaxial-abaxial polarity can cause an upward or downward rolling phenotype of the leaves (Moon and Hake, 2011). Some key genes that impact leaf rolling by controlling the development of leaf adaxial-abaxial polarity have been discovered. SHALLOT-LIKE1(sll1) mutation in rice (Oryza sativa) causes leaves to roll up along the adaxial side by reducing the number of vesicular cells in that specific area. Conversely, the overexpression of SLL1 leads to a reduction in the vesicular and sclerenchyma cells, resulting in leaf rolling (Zhang et al., 2009; Xiang et al., 2012). AS1 deficiency inhibits the formation of initial cells in the lateral organs, causing leaf rolling (Byrne et al., 2000). ASYMMETRIC LEAVES2 (AS2) inhibits the transcription of the abaxial-determinant genes ETTIN/ARF3, KANADI2, and YABBY5 and affects the establishment of adaxial-abaxial polarity, resulting in leaf rolling (Kojima et al., 2011). The leaf-rolling phenotype is regulated by one or more genes, often requiring synergy with the auxin transport carrier-related genes (Hibara et al., 2009; Zhang et al., 2009). Among them, PINs are involved in auxin polar transport and play a crucial role in leaf development (Scarpella et al., 2006; Chandler et al., 2007; Pahari et al., 2014). In Arabidopsis, leaf flattening requires the polar auxin transport. PIN inhibition led to flattening defects (Legris et al., 2021). Nevertheless, PIN3, which is a member of the PIN family, has rarely been reported to be involved in the establishment of leaf adaxial-abaxial polarity. The leaves of the mtpin1/mtpin3 double mutant displayed an abnormal abaxial surface only in Medicago truncatula (Zhang et al., 2020). In our study, BpPIN3 suppressed expression transgenic lines (RE lines) showed leaf rolling; however, this phenotype was distinct from that of the mtpin1/mtpin3 double mutant in that the adaxial showed an upward rolling (Figure 2C). This observation may be attributed to the different leaf structure of M. truncatula and birch, and the different effects of PIN3. Moreover, we found that the expression of the remaining 5 BpPINs in the RE lines did not decrease significantly (Figure S3). The expression of BpPIN3 exhibited a negative relationship with the LRI values in the leaves of the RE lines (Figure 2C). We, accordingly, speculated that the phenotype of leaf adaxial upward curling was caused by the inhibition of BpPIN3 in the RE lines. Further study is thus warranted to determine the effect of BpPIN3 on the polarity development in birch leaves.
Auxin is an important plant hormone, and its precise distribution is crucial for the regulation of local auxin concentration and the formation of an auxin concentration gradient in plants. This auxin distribution pattern is mainly regulated by PINs (Adamowski and Friml, 2015). The auxin concentration in different parts of the leaves is controlled by plants through PAT, which control the leaf morphology (Reinhardt et al., 2003). Asymmetric polarity of auxin can influence the development of leaf polarity and may lead to the development of a leaf-rolling phenotype in plants (Bowman et al., 2002; Liu et al., 2011). In this study, the auxin GUS reporter system was used to analyze the auxin distribution in the RE lines. It was found that auxin was mainly distributed in the epidermal cells and PTs of the leaf margin (Figures 7C, D; S8). In Arabidopsis, auxin was synthesized and aggregated at the leaf margin during leaf development and further transported by the auxin transport carrier PINs from the convergence point to the secondary veins, mesophyll, and other parts of the leaf, eventually triggering cell proliferation and vascular bundle formation for the complete leaf development (Scarpella et al., 2006). During this transport process, the change in auxin concentration affected the development of PTs and spongy tissues of the leaves and then regulated the polarity formation of the adaxial-abaxial axis. The icu6/AXR3 mutant of Arabidopsis demonstrated an abnormal accumulation of auxin in the PTs at the edge of leaves, which decreased the cell size and thus the curling of the adaxial surface of leaves (Pérez-Pérez et al., 2010). Our study revealed that the number of cells per unit area of PTs and the ratio of PTs to spongy tissues in the RE lines decreased significantly in the curled portion of the leaf margin, whereas no significant changes were observed in the OE lines (Figures 3D; 4B, C). Based on the loose arrangement of PTs in the leaf margin of the RE lines, the normal arrangement of PTs seems extremely important to maintain the polarity of leaves. We, accordingly, speculated that the normal expression of BpPIN3 is beneficial to maintain the normal arrangement of PTs of birch.
The establishment of the adaxial-abaxial polarity of plant leaves is a complex process regulated by interactions among genes, small RNAs, and plant hormones (Moon and Hake, 2011). In this study, 5 DEGs were significantly enriched in the hormone signal transduction pathway by RNA-seq analysis, all of which were significantly upregulated. These enriched genes included GH3.10 and 4 SAURs genes (i.e., SAUR23, SAUR 24, SAUR 28, and SAUR 50). The expression of these genes in the leaf margin tissues of the RE lines was significantly higher than that in the middle vein tissue (Figure S10), whereas the expression of BpPIN3 showed the opposite trend (Figure 7C). As an important auxin transport carrier, PINs respond to external stimuli to form a dynamic and strictly controlled auxin gradient in plants. The synergy of feedback mechanisms requires the rapid removal of auxin from cells when the auxin gradient is established (Friml et al., 2002; Woodward and Bartel, 2005). This process is mainly affected by GH3s (Vieten et al., 2005; Hayashi et al., 2021). SAUR proteins influence plant growth and development by altering auxin transport to adjust the auxin levels (Kant and Rothstein, 2009; Chae et al., 2012). In Arabidopsis, the overexpression of AtSAUR19 increased auxin transport in hypocotyls, whereas the knockout of AtSAUR19 decreased auxin transport (Spartz et al., 2012). A similar phenomenon was observed in AtSAUR41/63 (Chae et al., 2012; Spartz et al., 2012; Kong et al., 2013). Increasing IAA content in the leaves promoted the expression of GH3.5 (WES), resulting in the rolling of the abaxial surface of leaves in Arabidopsis (Park et al., 2007). Increasing free auxin in Arabidopsis dominant mutant yucca leads to increasing IAA inducible gene expressions, such as IAA/AUX, SAURs, and GH3s (Zhao et al., 2001). The high concentration of IAA promotes the rapid expression of GH3 and SAURs. In this study, the auxin content in the leaf margins of the RE lines was higher than that in the main veins, and SAURs were significantly upregulated at the leaf margin. The increase in auxin content may be attributed to the increase in SAURs expression in the leaf margin, and the increase of auxin content leads to the rapid induction of GH3.10, which leads to the amino acid reaction of IAA. As the expression of BpPIN3 in the leaf margin of the RE lines was significantly reduced, the normal IAA transport could not be completed, eventually leading to the formation of an auxin gradient between the leaf margin and the main veins of the RE lines.
In conclusion, BpPIN3 regulates the auxin response-related gene expression and auxin polar transport in birch leaves, thereby regulating the polarity formation of the proximal and distal axes of the leaves (Figure 8). BpPIN3 expression was significantly downregulated, and the expression of BpSAURs was significantly upregulated in the leaf margin tissue cells of the BpPIN3 RE lines. This event promoted the increase in IAA content and subsequently induced the expression of BpGH3.10. Although BpGH3.10 is involved in the synthesis of free IAA amino acid. However, only a part of IAA formed IAA-Glu and IAA-Asp amino acids, and most of IAA was retained in the upper and lower epidermal cells and secondary veins (Figures 7C; S10). Because the expression level of BpPIN3 in the leaf margin tissue cells of the RE lines was significantly lower than that in the middle vein, the accumulated IAA in this area could not be transported to the other parts of the leaf in a timely manner. Consequently, the asymmetric distribution of the IAA content in the leaf margin area was found to be significantly higher than that in the main vein area. Meanwhile, the density of PT cells was reduced and the arrangement was loose in the leaf margin area. The adaxial surface of the leaf was found to be curled.
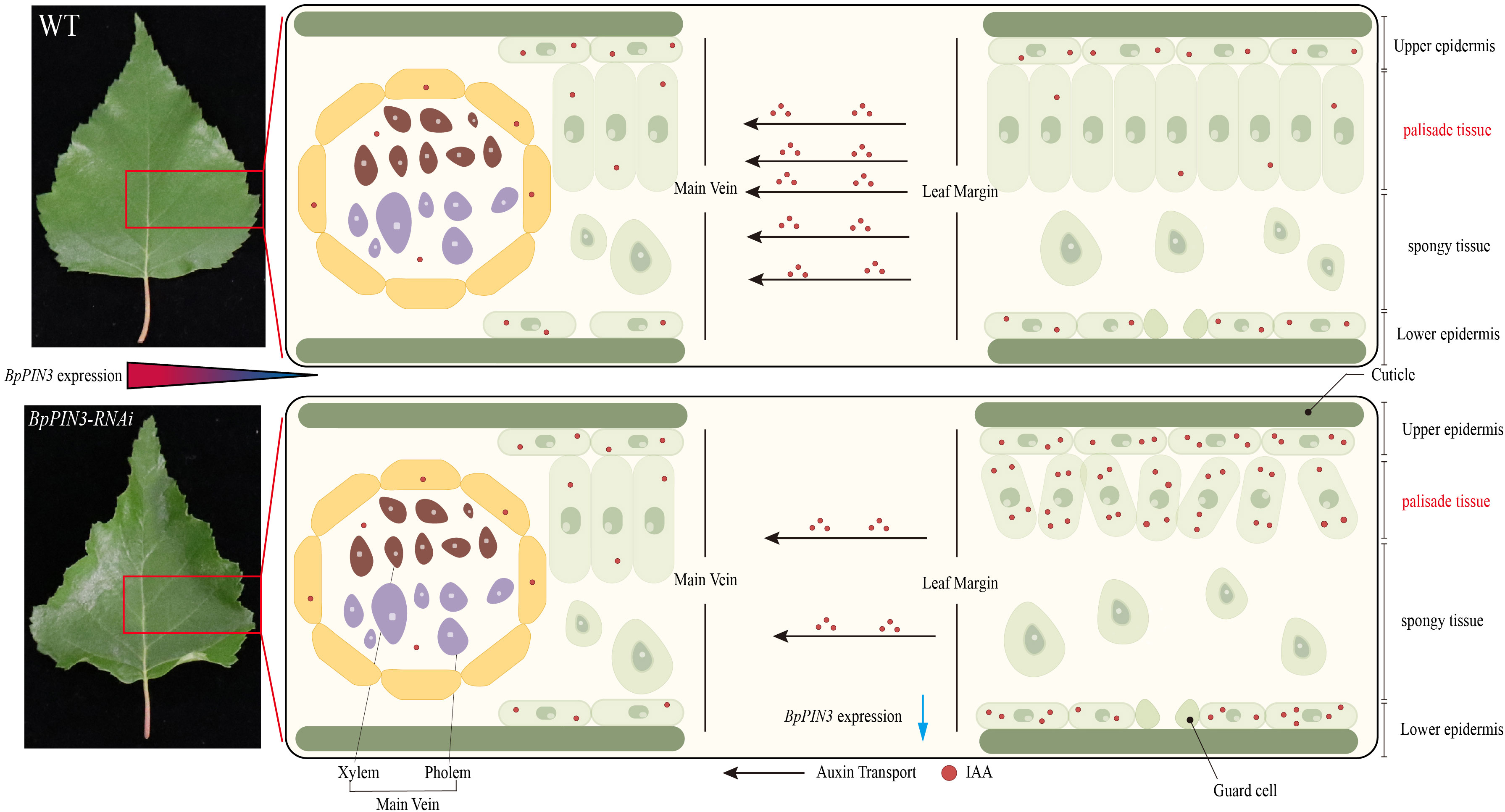
Figure 8 The schematic diagram of BpPIN3 regulating a leaf adaxial upward curl. Blue arrows indicate the decreased content or function.
BpPIN3 gene was cloned from birch, and the mature seeds of birch were used as transgenic plant material. The seeds were collected from a self-pollinated plus tree at Birch Suk improved variety base at the Northeast Forestry University (126°64’E, 45°72’N). After the bracts and periodicals were removed and sealed, the dried, mature inflorescences of birch were stored in a refrigerator at −20°C.
The main vectors included pPROKII and pFGC5941, and the strains were EHA105 and EHA105 (proDR5:: GUS). The strains were preserved in our laboratory.
The mRNA sequence of PIN3 of birch was obtained by inputting the GenBank login MG198788.1 of BpPIN3 in the NCBI (https://www.ncbi.nlm.nih.gov/). The upstream and downstream primers were designed according to the mRNA sequence. cDNA of birch leaves was used as the template for PCR amplification. The PCR product was purified and ligated to the pCloneEZ vector to create a recombinant plasmid. After transformation, the monoclonal was picked and sequenced by Beijing Qingke Biotechnology Co., Ltd. The ORF and conserved domain of BpPIN3 were determined using the ORF search and the conserved domain search service tools of NCBI, respectively. Based on the BlastX alignment, NCBI was searched for protein sequences similar to BpPIN3.
The orthologous genes of BpPIN3 were searched on NCBI, and a Neighbor-Joining phylogenetic tree was constructed using ClustalX 1.83 and MEGA7.0.
The overexpression vector was constructed according to the ORF sequence of BpPIN3. Upstream and downstream primers were designed to contain BamHI and KpnI restriction sites, respectively (Supplementary Table S1). The cDNA isolated from the birch leaves was used as a template for PCR amplification. The PCR product and pPROKII plant expression vector were digested with BamHI and KpnI, respectively. The PCR product and pPROKII plant expression vector were ligated with T4 ligase overnight at 16°C. The positive recombinant plasmid (35S:: PIN3) was obtained by screening on a 50 mg/L Kan resistance plate. The desired sequence was obtained after sequencing, and the positive recombinant plasmid was transformed into Agrobacterium EHA105 through electroporation (Huang et al., 2014).
Suppression expression vector was constructed using the NCBI Conserved Domains tool, which predicted 200-bp specific sequences of BpPIN3. These sequences were used as the target sequence in the RNAi vector to design forward and reverse primers (Supplementary Table S4). The forward and reverse fragments were assembled into pFGC5941 with restriction enzymes and through ligation (Li et al., 2021). Electroporation of Agrobacterium EHA105 yielded bacteria engineered for EHA105 (PIN3-RNAi). The transgenic birch plants were obtained via Agrobacterium- mediated zygotic embryo transformation (Huang et al., 2014).
The culture medium formula is depicted in Supplementary Table S1. The temperature in a tissue culture room was maintained at 25 ± 2°C, with a 16/8-h photoperiod and a light intensity of 1000–1500 lx.
Transgenic birch and wild-type birch (WT) lines were used to extract DNA using the DNAquick Plant System (TIANGEN). The PIN3-RNAi and 35S:: PIN3 plasmid were used as the positive control, whereas the WT line DNA was used as the negative control. PCR detection of transgenic lines was performed with specific primers listed in Supplementary Table S5 (Huang et al., 2014).
The transplantation of transgenic and WT birch tissue culture root seedlings was performed in a 25-hole seedling tray measuring 40 × 40 cm2. This procedure was performed in late May after the seedlings were cultured in a soil matrix medium. The plants included BpPIN3-reduced expression lines (RE1-RE3), overexpression lines (OE1-OE3), and the WT birch. In total, there were more than 100 transplanted plants. In the beginning of May of the second year, 30 seedlings showing the same growth vigor from each line were selected and transplanted into a 28 × 28 cm2 seedling pot. A mixture of peat, black soil, and vermiculite in a ratio of 3:2:1 was used as the substrate. All flowerpots were filled with the same amount of substrate and placed in a plastic greenhouse under the same management practice in the Birch Seed Base of the Northeast Forestry University.
Total RNA was extracted from the test lines by using the RNA extraction kit (RNeasy Plant Mini Kit, Biotake) and then reverse transcribed into cDNA by using the ReverTreAce® qPCR RT Kit (Toyobo, Osaka, Japan). SYBR Green PCR master mix (Toyobo) was used to perform real-time quantitative PCR (qRT-PCR) on the ABI 7500 Real-Time PCR system. Bp18S rRNA was used as an appropriate internal reference gene, and gene-specific primers were designed using Oligo7 software (Supplementary Table S6). The qPCR results were analyzed using the 2-ΔΔCT method, and each reaction was repeated thrice (Huang et al., 2014).
The healthy leaves of plants from the RE, OE, and WT lines were selected, and 20–30 plants were selected from each line. The highly mature leaves (fourth leaves) of each plant were selected. The leaf images were captured using a scanner and analyzed using ImageJ software. This allowed for the measurement of leaf length (Ll), leaf width (Lw), natural leaf margin distance (Ln), leaf perimeter (LP), and leaf area (La). The ratios Ll/petiole length (Pl) and Ll/Lw, as well as the LRI, were also calculated.
where Lw is the widest width of the leaf at full extension and distance, and Ln is the distance between the leaf margins under natural conditions. Cell density determination of leaf PTs
The mature leaves (the fourth leaf) of the RE, OE, and WT lines were vacuum-fixed in the FAA solution for 48 h and then placed in a chloral hydrate solution (chloral hydrate 200 g; glycerin 20 g; water, 50 mL). Palisade cells near the main vein and the leaf margin were observed under an optical microscope (OLYMPUS BX43F). Cell density was measured using image analysis software (Image J) (Schneider et al., 2012). Each measurement was repeated thrice, and the leaves of three individuals of each line were used. The number of cells in the four fields of view was measured for each part. The cell density was the number of cells in a unit area (50 μm2).
For the anatomical observation, mature leaves (fourth leaves) from the RE, OE, and WT lines were selected. The leaves with the widest leaf margins of 1 × 1 cm2 were sliced using the conventional paraffin section method (Liu et al., 2013). Each line was repeated thrice, and the leaves from 3 plants were selected for each repetition. The thickness of the leaf tissue slice was 10 μm. Safranine-fast green staining was performed, and the tissue was observed under an optical microscope (OLYMPUS BX43F). The leaf PT, sponge tissue (ST) thickness, PT cell density, and PT cell density were determined using Image J (Schneider et al., 2012). Statistics were gathered from 3 slices per plant.
The calculation formula used was as follows:
PT cell density is the number of cells per unit length (100 μm).
The RE, OE, and WT lines showing consistent growth were selected, and the apical buds and one leaf were respectively picked. Twelve ramets were collected from each line. Endogenous phytohormones were extracted from the samples using a mixture of isopropanol, water, and hydrochloric acid. The content of endogenous phytohormones in plants was determined using the UPLC-MS/MS platform composed of the Agilent 1290 High-performance Liquid Chromatograph and Absie QTRAP 6500 + mass spectrometer. Cavens Detection Technology Co., Ltd. provided both auxin standards and antibodies. To maintain a high degree of precision, the samples were run thrice.
The leaves of tissue culture seedlings from the BpPIN3 RE lines were used. The leaves pre-cultured for 2–3 days were inoculated with EHA105 (proDR5::GUS) bacterial solution at OD600 of approximately 0.5 for 10–20 min. Under the screening condition of 50 mg/L hygromycin B (Hyg), the RE (proDR5:: GUS)-resistant calli were obtained. The resistant calli were inoculated into the differentiation medium containing 50 mg/L Hyg to induce the formation of adventitious buds, and multiple subculture seedlings were obtained. A rooting culture was performed on the solid medium containing WPM + 0.4 mg/L IBA. After 10 days, GUS staining was performed on the whole seedlings (Jefferson et al., 1987). After GUS staining, the leaves were cut and photographed using a stereomicroscope. The leaves of RE and WT lines after GUS staining were sliced by using the conventional paraffin method (Liu et al., 2013). Each line was repeated thrice, and the leaves of 3 individual plants were selected for each repetition. The thickness of the slice was 10 μm, and 0.02% ruthenium red was used for staining. After neutral resin sealing, the optical microscope (OLYMPUS BX43F) was used for observation and photography.
The leaf margin and the main vein region of the RE and WT lines were used as materials. Quantitative measurements of the GUS activity were conducted using the Plant GUS ELISA Kit (Cat. No. JN0; Jining Shiye, Shanghai, China).
RNA was extracted from the mature leaves of the RE, OE, and WT lines and treated with DNaseI. Three biological replicates per line were established. Each biological replicate had 6 individual leaves. RNA sequencing was performed by the BGI Group on the DNBSEQ-T7 sequencing platform. Bowtie2 was used for a comparative analysis of the birch reference genome after the removal of low-quality reads (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml). Based on the RNA-seq data, DEGs were calculated using the maximum expectation algorithm (Tusher et al., 2001). In addition, the DEGs were annotated based on the following databases: NCBI/Nr (http://www.ncbi.nlm.nih.gov), NCBINt (http://www.ncbi.nlm.nih.gov), Swiss-Prot (http://www.uniprot.org/keywords/), and KEGG pathway database (http://www.genome.jp/kegg). The expression level of each gene was calculated using FPKM (Florea et al., 2013). EdgeR was used to define the DEGs between each two-sample comparison, with the thresholds set as follows: the false discovery rate (FDR) ≤ 0.01, Log2FC ≥ 1, and Log2FPKM ≥ 1 (Edge R package; version 3.28.1). The target genes were first annotated in the GO database (http://www.geneontology.org/) and then classified into the corresponding functional categories to evaluate their biological functions. In addition, the KEGG pathway enrichment analysis was performed using the KEGG pathway database (http://www.genome.jp/kegg).
One-way or two-way analysis of variance was used for statistical analysis. Tukey’s method was used for multiple comparisons. Data are expressed as the mean ± standard deviation (SD). The difference is denoted in four levels, significant *p< 0.05, highly significant **p< 0.01, extremely significant ***p< 0.001, and highly extremely significant ****p< 0.0001.TBtools (Chen et al., 2020), SPSS22.0, and GraphPad Prism (GraphPad Prism 8; GraphPad) were used to analyze and plot the data.
BpPIN3 is involved in leaf polarity formation in birch. The decrease in BpPIN3 expression led to an increase in the auxin response-related gene expression in birch leaves, which affected the polar transport of auxin, resulting in a difference in auxin concentration in the leaves. The excessive auxin at the leaf margin modified the structure of PT cells, leading to upward curling at the leaf margin.
The original contributions presented in the study are publicly available. This data can be found here: NCBI, PRJNA885271.
KC, G-FL and JJ designed the research, KC and CQ conducted the experiments, KC conducted the data analysis and wrote the manuscript, KC, WW and C-RG performed transcriptome data analysis, X-YZ, Q-BY, CY and JJ revised the manuscript, X-YZ participated some partial index measurement. Q-BY has carried on the experiment instruction and the experimental design consummation. All authors contributed to the article and approved the submitted version.
This research was supported by the National Key R&D Program of China during the 14th Five-year Plan Period (2021YFD2200103) and Heilongjiang Touyan Innovation Team Program (Tree Genetics and Breeding Innovation Team).
Thanks for the technical platform and support provided by the State Key Laboratory of Tree Genetics and Breeding, Northeast Forestry University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1060228/full#supplementary-material
Adamowski, M., Friml, J. (2015). PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell 27 (1), 20–32. doi: 10.1105/tpc.114.134874
Bian, X., Qu, C., Zhang, M., Li, Y., Han, R., Jiang, J., et al. (2019). Transcriptome sequencing to reveal the genetic regulation of leaf margin variation at early stage in birch. Tree Genet. Genomes 15 (1), 1–17. doi: 10.1007/s11295-018-1312-7
Bonaccorso, O., Lee, J. E., Puah, L., Scutt, C. P., Golz, J. F. (2012). FILAMENTOUS FLOWER controls lateral organ development by acting as both an activator and a repressor. BMC Plant Biol. 12, 176. doi: 10.1186/1471-2229-12-176
Bowman, J. L. (2000). Axial patterning in leaves and other lateral organs. Curr. Opin. Genet. Dev. 10 (4), 399–404. doi: 10.1016/s0959-437x(00)00103-9
Bowman, J. L., Eshed, Y., Baum, S. F. (2002). Establishment of polarity in angiosperm lateral organs. Trends Genet. 18 (3), 134–141. doi: 10.1016/s0168-9525(01)02601-4
Byrne, M. E., Barley, R., Curtis, M., Arroyo, J. M., Dunham, M., Hudson, A., et al. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in arabidopsis. Nature 408 (6815), 967–971. doi: 10.1038/35050091
Chae, K., Isaacs, C. G., Reeves, P. H., Maloney, G. S., Muday, G. K., Nagpal, P., et al. (2012). Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. Plant J. 71 (4), 684–697. doi: 10.1111/j.1365-313X.2012.05024.x
Chandler, J. W., Cole, M., Flier, A., Grewe, B., Werr, W. (2007). The AP2 transcription factors DORNROSCHEN and DORNROSCHEN-LIKE redundantly control arabidopsis embryo patterning via interaction with PHAVOLUTA. Development 134 (9), 1653–1662. doi: 10.1242/dev.001016
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chitwood, D. H., Nogueira, F. T., Howell, M. D., Montgomery, T. A., Carrington, J. C., Timmermans, M. C. (2009). Pattern formation via small RNA mobility. Genes Dev. 23 (5), 549–554. doi: 10.1101/gad.1770009
Chitwood, D. H., Sinha, N. R. (2016). Evolutionary and environmental forces sculpting leaf development. Curr. Biol. 26 (7), R297–R306. doi: 10.1016/j.cub.2016.02.033
Ding, Z., Friml, J. (2010). Auxin regulates distal stem cell differentiation in arabidopsis roots. Proc. Natl. Acad. Sci. U.S.A. 107 (26), 12046–12051. doi: 10.1073/pnas.1000672107
Emery, J. F., Floyd, S. K., Alvarez, J., Eshed, Y., Hawker, N. P., Izhaki, A., et al. (2003). Radial patterning of arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13 (20), 1768–1774. doi: 10.1016/j.cub.2003.09.035
Eshed, Y., Izhaki, A., Baum, S. F., Floyd, S. K., Bowman, J. L. (2004). Asymmetric leaf development and blade expansion in arabidopsis are mediated by KANADI and YABBY activities. Development 131 (12), 2997–3006. doi: 10.1242/dev.01186
Florea, L., Song, L., Salzberg, S. L. (2013). Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Res 2, 188. doi: 10.12688/f1000research.2-188.v2
Friml, J., Wiśniewska, J., Benková, E., Mendgen, K., Palme, K. (2002). Lateral relocation of auxin efflux regulator PIN3 mediates tropism in arabidopsis. Nature 415 (6873), 806–809. doi: 10.1038/415806a
Hasson, A., Blein, T., Laufs, P. (2010). Leaving the meristem behind: the genetic and molecular control of leaf patterning and morphogenesis. C. R. Biol. 333 (4), 350–360. doi: 10.1016/j.crvi.2010.01.013
Hayashi, K. I., Arai, K., Aoi, Y., Tanaka, Y., Hira, H., Guo, R., et al. (2021). The main oxidative inactivation pathway of the plant hormone auxin. Nat. Commun. 12 (1), 6752. doi: 10.1038/s41467-021-27020-1
He, P., Zhang, Y., Li, H., Fu, X., Shang, H., Zou, C., et al. (2021). GhARF16-1 modulates leaf development by transcriptionally regulating the GhKNOX2-1 gene in cotton. Plant Biotechnol J 19 (3), 548–562. doi: 10.1111/pbi.13484
Hibara, K., Obara, M., Hayashida, E., Abe, M., Ishimaru, T., Satoh, H., et al. (2009). The ADAXIALIZED LEAF1 gene functions in leaf and embryonic pattern formation in rice. Dev. Biol. 334 (2), 345–354. doi: 10.1016/j.ydbio.2009.07.042
Huang, H., Wang, S., Jiang, J., Liu, G., Li, H., Chen, S., et al. (2014). Overexpression of BpAP1 induces early flowering and produces dwarfism in betula platyphylla × betula pendula. Physiol. Plant 151 (4), 495–506. doi: 10.1111/ppl.12123
Ichihashi, Y., Tsukaya, H. (2015). Behavior of leaf meristems and their modification. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.01060
Iwasaki, M., Takahashi, H., Iwakawa, H., Nakagawa, A., Ishikawa, T., Tanaka, H., et al. (2013). Dual regulation of ETTIN (ARF3) gene expression by AS1-AS2, which maintains the DNA methylation level, is involved in stabilization of leaf adaxial-abaxial partitioning in arabidopsis. Development 140 (9), 1958–1969. doi: 10.1242/dev.085365
Jefferson, R. A., Kavanagh, T. A., Bevan, M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 (13), 3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x
Juarez, M. T., Kui, J. S., Thomas, J., Heller, B. A., Timmermans, M. C. (2004). microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428 (6978), 84–88. doi: 10.1038/nature02363
Kant, S., Rothstein, S. (2009). Auxin-responsive SAUR39 gene modulates auxin level in rice. Plant Signal Behav. 4 (12), 1174–1175. doi: 10.4161/psb.4.12.10043
Kasahara, H. (2016). Current aspects of auxin biosynthesis in plants. Biosci. Biotechnol. Biochem. 80 (1), 34–42. doi: 10.1080/09168451.2015.1086259
Keuskamp, D. H., Pollmann, S., Voesenek, L. A., Peeters, A. J., Pierik, R. (2010). Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proc. Natl. Acad. Sci. U.S.A. 107 (52), 22740–22744. doi: 10.1073/pnas.1013457108
Kidner, C. A., Martienssen, R. A. (2004). Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428 (6978), 81–84. doi: 10.1038/nature02366
Kojima, S., Iwasaki, M., Takahashi, H., Imai, T., Matsumura, Y., Fleury, D., et al. (2011). ASYMMETRIC LEAVES2 and elongator, a histone acetyltransferase complex, mediate the establishment of polarity in leaves of arabidopsis thaliana. Plant Cell Physiol. 52 (8), 1259–1273. doi: 10.1093/pcp/pcr083
Kong, Y., Zhu, Y., Gao, C., She, W., Lin, W., Chen, Y., et al. (2013). Tissue-specific expression of SMALL AUXIN UP RNA41 differentially regulates cell expansion and root meristem patterning in arabidopsis. Plant Cell Physiol. 54 (4), 609–621. doi: 10.1093/pcp/pct028
Kun, C., Gonggui, F., Huaizhi, M., Jing, J. (2022). Analysis of the promoter sequence and response characteristics of the BpPIN3 gene in betula platyphylla. Bull. Botanical. Res. 42 (04), 592–601. doi: 10.7525/j.issn.1673-5102.2022.04.009
Legris, M., Szarzynska-Erden, B. M., Trevisan, M., Allenbach Petrolati, L., Fankhauser, C. (2021). Phototropin-mediated perception of light direction in leaves regulates blade flattening. Plant Physiol. 187 (3), 1235–1249. doi: 10.1093/plphys/kiab410
Li, Y., Gu, C., Gang, H., Zheng, Y., Liu, G., Jiang, J. (2021). Generation of a golden leaf triploid poplar by repressing the expression of GLK genes. Forest. Res. 1 (1), 1–7. doi: 10.48130/fr-2021-0003
Liu, Z., Jia, L., Wang, H., He, Y. (2011). HYL1 regulates the balance between adaxial and abaxial identity for leaf flattening via miRNA-mediated pathways. J. Exp. Bot. 62 (12), 4367–4381. doi: 10.1093/jxb/err167
Liu, Y. J., Xiu, Z. H., Meeley, R., Tan, B. C. (2013). Empty pericarp5 encodes a pentatricopeptide repeat protein that is required for mitochondrial RNA editing and seed development in maize. Plant Cell 25 (3), 868–883. doi: 10.1105/tpc.112.106781
Li, H., Xu, L., Wang, H., Yuan, Z., Cao, X., Yang, Z., et al. (2005). The putative RNA-dependent RNA polymerase RDR6 acts synergistically with ASYMMETRIC LEAVES1 and 2 to repress BREVIPEDICELLUS and MicroRNA165/166 in arabidopsis leaf development. Plant Cell 17 (8), 2157–2171. doi: 10.1105/tpc.105.033449
Ljung, K. (2013). Auxin metabolism and homeostasis during plant development. Development 140 (5), 943–950. doi: 10.1242/dev.086363
Ludwig-Müller, J. (2011). Auxin conjugates: their role for plant development and in the evolution of land plants. J. Exp. Bot. 62 (6), 1757–1773. doi: 10.1093/jxb/erq412
Machida, C., Nakagawa, A., Kojima, S., Takahashi, H., Machida, Y. (2015). The complex of ASYMMETRIC LEAVES (AS) proteins plays a central role in antagonistic interactions of genes for leaf polarity specification in arabidopsis. Wiley. Interdiscip. Rev. Dev. Biol. 4 (6), 655–671. doi: 10.1002/wdev.196
McConnell, J. R., Emery, J., Eshed, Y., Bao, N., Bowman, J., Barton, M. K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411 (6838), 709–713. doi: 10.1038/35079635
Moon, J., Hake, S. (2011). How a leaf gets its shape. Curr. Opin. Plant Biol. 14 (1), 24–30. doi: 10.1016/j.pbi.2010.08.012
Otsuga, D., DeGuzman, B., Prigge, M. J., Drews, G. N., Clark, S. E. (2001). REVOLUTA regulates meristem initiation at lateral positions. Plant J. 25 (2), 223–236. doi: 10.1046/j.1365-313x.2001.00959.x
Pahari, S., Cormark, R. D., Blackshaw, M. T., Liu, C., Erickson, J. L., Schultz, E. A. (2014). Arabidopsis UNHINGED encodes a VPS51 homolog and reveals a role for the GARP complex in leaf shape and vein patterning. Development 141 (9), 1894–1905. doi: 10.1242/dev.099333
Park, J. E., Park, J. Y., Kim, Y. S., Staswick, P. E., Jeon, J., Yun, J., et al. (2007). GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in arabidopsis. J. Biol. Chem. 282 (13), 10036–10046. doi: 10.1074/jbc.M610524200
Pérez-Pérez, J. M., Candela, H., Robles, P., López-Torrejón, G., del Pozo, J. C., Micol, J. L. (2010). A role for AUXIN RESISTANT3 in the coordination of leaf growth. Plant Cell Physiol. 51 (10), 1661–1673. doi: 10.1093/pcp/pcq123
Prigge, M. J., Otsuga, D., Alonso, J. M., Ecker, J. R., Drews, G. N., Clark, S. E. (2005). Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in arabidopsis development. Plant Cell 17 (1), 61–76. doi: 10.1105/tpc.104.026161
Qu, C., Bian, X., Han, R., Jiang, J., Yu, Q., Liu, G. (2020). Expression of BpPIN is associated with IAA levels and the formation of lobed leaves in betula pendula 'Dalecartica'. J. Forest. Res. 31 (1), 87–97. doi: 10.1007/s11676-018-0865-5
Reinhardt, D., Pesce, E. R., Stieger, P., Mandel, T., Baltensperger, K., Bennett, M., et al. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426 (6964), 255–260. doi: 10.1038/nature02081
Robert, H. S., Crhak Khaitova, L., Mroue, S., Benková, E. (2015). The importance of localized auxin production for morphogenesis of reproductive organs and embryos in arabidopsis. J. Exp. Bot. 66 (16), 5029–5042. doi: 10.1093/jxb/erv256
Scarpella, E., Marcos, D., Friml, J., Berleth, T. (2006). Control of leaf vascular patterning by polar auxin transport. Genes Dev. 20 (8), 1015–1027. doi: 10.1101/gad.1402406
Schneider, C. A., Rasband, W. S., Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9 (7), 671–675. doi: 10.1038/nmeth.2089
Spartz, A. K., Lee, S. H., Wenger, J. P., Gonzalez, N., Itoh, H., Inzé, D., et al. (2012). The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 70 (6), 978–990. doi: 10.1111/j.1365-313X.2012.04946.x
Tsukaya, H. (2005). Leaf shape: genetic controls and environmental factors. Int. J. Dev. Biol. 49 (5-6), 547–555. doi: 10.1387/ijdb.041921ht
Tusher, V. G., Tibshirani, R., Chu, G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U.S.A. 98 (9), 5116–5121. doi: 10.1073/pnas.091062498
Vieten, A., Vanneste, S., Wisniewska, J., Benková, E., Benjamins, R., Beeckman, T., et al. (2005). Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132 (20), 4521–4531. doi: 10.1242/dev.02027
Wang, X., Zhao, D., Liu, G., Yang, C., Teskey, R. O. (2018). Additive tree biomass equations for betula platyphylla suk. plantations in northeast China. Ann. For. Sci. 75 (2), 1–9. doi: 10.1007/s13595-018-0738-2
Wisniewska, J., Xu, J., Seifertová, D., Brewer, P. B., Ruzicka, K., Blilou, I., et al. (2006). Polar PIN localization directs auxin flow in plants. Science 312 (5775), 883. doi: 10.1126/science.1121356
Woodward, A. W., Bartel, B. (2005). Auxin: regulation, action, and interaction. Ann. Bot. 95 (5), 707–735. doi: 10.1093/aob/mci083
Xiang, J. J., Zhang, G. H., Qian, Q., Xue, H. W. (2012). Semi-rolled leaf1 encodes a putative glycosylphosphatidylinositol-anchored protein and modulates rice leaf rolling by regulating the formation of bulliform cells. Plant Physiol. 159 (4), 1488–1500. doi: 10.1104/pp.112.199968
Zhang, X., Liu, L., Wang, H., Gu, Z., Liu, Y., Wang, M., et al. (2020). MtPIN1 and MtPIN3 play dual roles in regulation of shade avoidance response under different environments in medicago truncatula. Int. J. Mol. Sci. 21 (22), 8742. doi: 10.3390/ijms21228742
Zhang, G. H., Xu, Q., Zhu, X. D., Qian, Q., Xue, H. W. (2009). SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell 21 (3), 719–735. doi: 10.1105/tpc.108.061457
Keywords: Betula pendula, BpPIN3, IAA transport, Adaxial-abaxial, leaf
Citation: Chen K, Qu C, Zhang X-y, Wang W, Gu C-r, Liu G-f, Yu Q-b, Yang C-p and Jiang J (2022) Molecular mechanism of leaf adaxial upward curling caused by BpPIN3 suppression in Betula pendula. Front. Plant Sci. 13:1060228. doi: 10.3389/fpls.2022.1060228
Received: 03 October 2022; Accepted: 17 November 2022;
Published: 01 December 2022.
Edited by:
Yongwei Sun, Inner Mongolia University, ChinaReviewed by:
Wenwei Lin, Fujian Agricultural And Forest University, ChinaCopyright © 2022 Chen, Qu, Zhang, Wang, Gu, Liu, Yu, Yang and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuan-ping Yang, eWFuZ2NwQG5lZnUuZWR1LmNu; Jing Jiang, amlhbmdqaW5nQG5lZnUuZWR1LmNu
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.