- 1State Key Laboratory of Rice Biology and Key Lab of the Ministry of Agriculture for Nuclear Agricultural Sciences, Institute of Nuclear Agricultural Sciences, Zhejiang University, Hangzhou, China
- 2Hainan Institute, Zhejiang University, Yazhou Bay Science and Technology City, Sanya, China
High temperature (HT) during grain filling had adverse influences on starch synthesis. In this study, the influences of HT on resistant starch (RS) formation in rice were investigated. Most genes in ssIIIa mutants especially in RS4 were upregulated under Normal Temperature (NT) while downregulated under HT when compared with those of wild parent R7954. ssIIIa mutants had higher RS content, more lipid accumulation, higher proportion of short chains of DP 9–15, and less long chains of DP ≥37. ssIIIa mutation exacerbated the influences of HT on starch metabolite and caused larger declines in the expression of BEI, BEIIa, BEIIb, and SSIVb when exposed to HT. HT reduced the contents of total starch and apparent amylose significantly in wild type but not in mutants. Meanwhile, lipids were enriched in all varieties, but the amounts of starch–lipid complexes and the RS content were only heightened in mutants under HT. HT led to greatest declines in the amount of DP 9–15 and increases in the proportion of fb3 (DP ≥37); the declines and increases were all larger in mutants, which resulted in varied starch crystallinity. The increased long-chain amylopectin and lipids may be the major contributor for the elevated RS content in mutants under HT through forming more starch–lipid complexes (RSV).
Introduction
Rice (Oryza sativa) is an extremely important cereal and is the main food for more than 50% of the population worldwide. As the major components in rice grains, starch mainly comprises amylose and amylopectin molecules. The ratio of amylose to amylopectin and the structure of starch granules are key factors influencing rice quality. Meanwhile, amylose content and the distributions of branch lengths in both amylose and amylopectin influence starch digestibility (Syahariza et al., 2014; Raigond et al., 2015). The rate and extent of digestibility of starch in the small intestine determine the eventual glucose level in blood, which is of great interest in the context of worldwide health concerns.
Although starch is a homopolysaccharide of glucose, a proportion of starch, namely, resistant starch (RS), is not digested in the small intestine and passed into the large intestine to produce short-chain fatty acids (Englyst and Hudson, 1996; Hu et al., 2016). Due to its digestion resistance, RS is beneficial for inflammatory bowel disease, glycemic index, cholesterol levels, coronary heart disease, and other conditions (Shen et al., 2022). Based on different origins and characteristics, RS can be classified into RSI, RSII, RSIII, RSIV, and RSV five types. RSII consists of native starch granules with a compact structure, majorly exists in raw potatoes, peas, and green bananas (Sajilata et al., 2006), and can become digestible after traditional cooking. RSIII consists of retrograded starches that are principally recrystallized amylose produced after heating starchy food, such as in rice. RSV is a complex of amylose and lipids that improve antidigestion capabilities through forming double helical structures (Hasjim et al., 2010).
Food high in RS is associated with improved cardiovascular health, gut health, and glycemic response, while RS contents in rice and other cereal crops are generally below 3% (Xia et al., 2018; Kumar et al., 2018) and affected by amylose content, starch granule structure, fine structure of amylopectin, and other metabolites such as lipids and sugars (Shen et al., 2022). Apparent amylose content (AAC) and amylopectin with degree of polymerization (DP) 8–12 have been found to be positively related to RS content (Shu et al., 2006; Shu et al., 2007; Shen et al., 2022). RS is decisive in determining the glycemic index of rice (Kumar et al., 2018). Nowadays, several genes involved in starch biosynthesis have been found to be responsible for RS formation in rice grains. The Wx gene may regulate RS formation by tuning the amylose content (Itoh et al., 2003; You et al., 2022). BEIIb may play an important role in RS formation through modulating the distribution of long and intermediate debranched amylopectin chains (Butardo et al., 2011; Guo et al., 2020; Miura et al., 2021) and tuning other metabolites such as sugar, lipid, and protein contents (Baysal et al., 2020). be1be2b double mutant shows higher RS content than the be2b mutant (Miura et al., 2021). SSIIIa has been verified to be a key gene responsible for the RS content; the loss-of-function of SSIIIa gives rise to a high RS content in rice grain, but the effect of SSIIIa on RS content depends partly on the functional Wx gene (Wxa) (Zhou et al., 2016). Double repression of SSIIa and SSIIIa resulted in higher AAC and RS content (Zhang et al., 2011).
In a preliminary study, we developed a series of rice mutants high in RS (Shu et al., 2007; Shu et al., 2009; Ding et al., 2019). These mutants have significantly lower digestibility (Shu et al., 2009; Zhou et al., 2016) and show great potentialities in lowering the serum glucose level of Type II diabetes patients. However, the absolute content of RS in these mutants showed fluctuations among different growing years or locations (data not published). Bao et al. (2020) also found that the RS content of three chalky mutants derived from R9311 differed significantly between two environments (Hangzhou and Hainan). Among environmental factors, temperature is the most important for rice kernel development. It is proposed that temperature may influence the RS formation and the functional properties of high-RS rice.
With global warming, high temperature (HT) during grain filling has become a great threat for rice production. HT during grain filling can lead to reduced yield, increased chalkiness, and varied amylose content (AC) and starch structure (Zhang et al., 2016). AC has been found either increased or decreased when exposed to HT depending on rice variety (Cheng et al., 2005; Lin et al., 2010; Liu et al., 2021). Zhao et al. (2020) found that ssIIIa-RNAi affected the susceptibility of grain chalky occurrence to HT exposure due to the relatively sensitive response of ADP-glucose pyrophosphorylase (AGPase) and SSI to HT exposure and induced significant decreases in AAC and a larger decline in the proportion of DP <12 under HT relative to wild type (WT). Although the formation of RS in rice grain has been investigated in detail from the intrinsic properties of starch, storage and processing conditions, and external factors such as non-starch components (Shen et al., 2022), few concerns have been paid on the growth environment such as temperature. In view of extreme climate change, understanding the influences of temperature on RS formation will provide valuable clues for breeding high-RS rice with improved quality. In the present study, genetic SSIIIa mutations in high-RS rice mutants RS111 and RS4 were detected. The main reserve substances in rice under two growth conditions with different day/night temperatures are measured. Furthermore, the expression levels of genes involved in starch synthesis, amylopectin chain length (CL) distribution, X-ray diffraction (XRD) analysis and Fourier transform infrared (FTIR) spectroscopy, and thermal and pasting properties of starch were investigated. In addition, a possible mechanism of temperature affecting RS formation was proposed. The results in this study give some evidence for the impacts of temperature on RS formation in rice grain and provide alternative thoughts on improving the quality of rice high in RS under global climate warming.
Materials and methods
Plant materials and growth conditions
Three indica rice varieties with different RS contents were used in this study, including R7954 (WT) and RS111 and RS4 (mutants). RS111 was derived from R7954 through 300 Gy 60-Cobalt γ-ray mutagenesis, and RS4 was obtained from RS111 by irradiation (Shu et al., 2006; Shu et al., 2009); the two mutants have been planted for more than 15 generations. All varieties were grown in artificial climatic chambers with a relative humidity of 80% under a photon flux density of 630–720 μmol·m-1·s-1. The growth cabinets were maintained at an average air temperature of 25°C (NT, ranging from 20°C to 30°C, 12-h day/12-h night cycles) until flowering. Then, half of the plants were transferred to another growth cabinet with an average air temperature of 30°C (HT, ranging from 25°C to 35°C, 12-h day/12-h night cycles). Individual panicles were labeled on the day of flowering. Grains at 8 days after flowering (DAF) were collected and immediately frozen in liquid nitrogen and stored at -80°C until use. Mature grains were harvested and dried under 37°C, then the glume and aleurone were detached using Satake rice machine (Satake Corp., Japan). After polishing, the white rice was ground into flour, passed through a 150-μm sieve and stored in a drier until use.
DNA extraction and sequencing
The total genomic DNA of rice leaf was extracted using Tris physiological saline (TPS) (1 M Tris-HCl pH 8, 0.5 M EDTA pH 8, 5 M KCl) according to the procedure modified by Xin et al. (2003). The PCR products amplified with SSIIIa CAPS primer were digested using restriction enzyme TSP509i as previously reported (Zhou et al., 2016) and separated using 2% agarose gel. Genomic SSIIIa was amplified using gSSIIIa primer, and the PCR products were sequenced using Genetic Analyzer (3730XL-96, ABI, USA). All primers used are listed in Table S1.
RNA extraction and quantitative real-time PCR
The total RNA from dehulled seeds at 8 DAF was extracted using Plant RNA Extraction Kit (9769, Takara, Japan), and cDNA was synthesized using FastKing RT Kit (KR116, TIANGEN Biotech, China) following the manufacturer’s protocol. Quantitative real-time PCR (qRT-PCR) was performed with SYBR Green Premix Pro Taq HS qPCR Kit (AG11701, Accurate Biotech, China) on the CFX96 Real-time system (Bio-Rad, USA). The rice Actin gene (LOC_Os0350885) was used as internal reference. qPCR primers of 27 starch-related genes were referred to Ohdan et al. (2005) and listed in Table S1.
Sample preparation and starch extraction
Starch was isolated using NaOH as follows. The flour and NaOH solution (0.3%, w/v) were mixed (1:20, w/v) thoroughly, shaking at 180 rpm and 4°C for 16 h. Then, the mixture was filtered with a 150-μm sieve, and the filtered solution was centrifuged at 5,000 g for 10 min. The yellow layer in the precipitation was scraped carefully, then the precipitation was suspended with ddH2O, and the pH of the slurry was adjusted to neutral. Repeated centrifugation and rinsing with ddH2O were done several times until the precipitation was clearly white. Then, the obtained starch was dried at 37°C, ground, and filtered with a 150-μm sieve. The isolated starch was used for XRD, FTIR, differential scanning calorimetry (DSC), and amylopectin chain length (CL) profile analyses.
Major reserves and RS measurement
AAC and total starch content were measured according to the methods described by Shu et al. (2014). The total protein was measured using Digestor (DT208, FOSS, Denmark) and automatic Kjeldahl nitrogen analyzer (Foss8400, FOSS, Denmark) based on the principle of Kjeldahl method. Lipid content was measured using a fat analyzer (SOX406, Hanon Group, China) based on Soxhlet extraction method.
RS content was measured according to Shu et al. (2014), but the released glucose content was measured using Glucose Assay Kit (Ningbo Saike Biological Technology, China). The glucose content was converted to starch content by a factor of 0.9.
Amylopectin chain length distribution
Amylopectin was extracted from starch using a fractionation method according to the procedure described by Kong et al. (2008). Purified amylopectin was debranched with pullulanase (E-PULKP, Megazyme, USA) and isoamylase (E-ISAMYHP, Megazyme, USA) using the method described by Kong et al. (2016). The CL distributions of debranched amylopectin were determined using high-performance anion-exchange chromatography (HPAEC) (DionexICS-5000+, Sunnyvale, CA, USA) with BioLC gradient pump and a pulsed amperometric detector (PAD) (Kong et al., 2016).
X-ray diffraction
The X-ray powder diffractometer (Bruker D8 Advance, Brucker, Germany) was used for XRD analysis. Starch samples were packed into a round glass sample holder and scanned from 4° to 70° 2θ with a step of 0.02°/2 s. The relative crystallinity (RC; %) was calculated using MDI Jade 6.5.
Fourier transform infrared spectroscopy
Starch granule short-range ordered structure was detected with the FTIR spectrometer (NICOLET iS50FT-IR, Thermo Scientific, USA). The resolution of the instrument is 4 cm-1, and the scanning range is from 4,000 cm-1 to 400 cm-1. The relative absorbances at 1,045, 1,022 and 995 cm-1 were extracted from the deconvoluted spectra and were measured from the baseline to the peak height.
Differential scanning calorimetry
The thermal properties of starch were determined using DSC (Q20, TA Instruments, Newcastle, DE, USA) according to the method described by Shu et al. (2009). The enthalpy change (ΔH), onset temperature (To), peak temperature (Tp), and conclusion temperature (Tc) were calculated using Universal Analysis 2000 program (Version 4.4A).
Rapid viscosity analysis
The pasting properties of rice flours were measured using a rapid visco analyzer (Model 4500, Perten Instruments, Sweden) according to the procedure described by Bao et al. (2004). Peak viscosity (PV), hot paste viscosity (HPV), cold paste viscosity (CPV), breakdown value (BD = PV-HPV), and setback value (SB = CPV-PV) were recorded.
Statistical analysis
All experiments have biological triplicates. One-way analysis of variance followed by the least significant difference (LSD) test (P < 0.05 or P < 0.01) was performed using SPSS 22 (IBM, USA).
Results
Genotypes of RS111 and RS4
SSIIIa has been confirmed to be a key gene for RS formation in rice, as RS111 and RS4 are both derived from R7954 as b10, whether they are allelic ssIIIa mutations was detected first. Compared with R7954, RS4 and RS111 carried a G/A mutation in the fifth intron of SSIIIa, which caused varied splicing and led to a 4-bp deletion in the coding sequence (Figures 1A, B). With the SSIIIa CAPS marker developed based on this Single Nucleotide Polymorphism (SNP), the PCR products were digested into two shorter products by TSP509i (Figure 1C). This proved that RS4 and RS111 carried the same mutation in SSIIIa gene as b10 (Zhou et al., 2016) and are two allelic ssIIIa mutants.
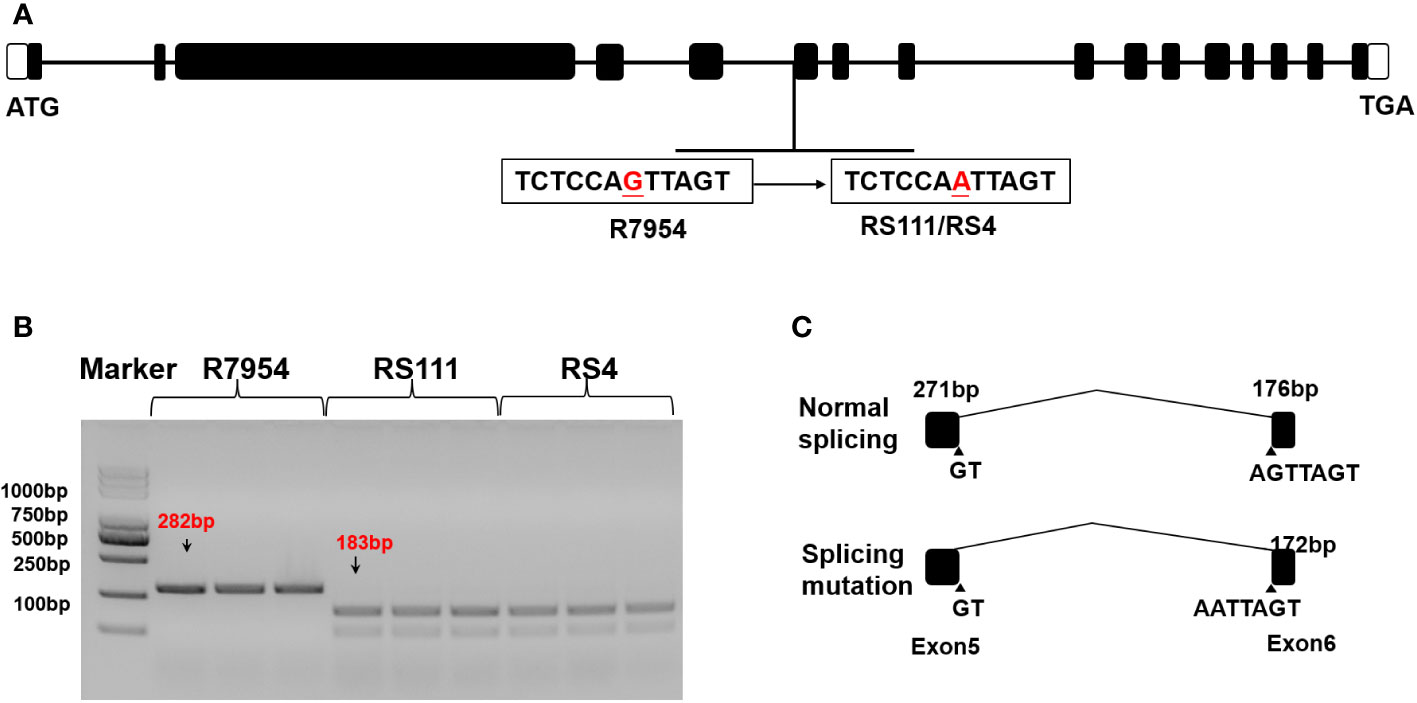
Figure 1 SSIIIa sequence variations in wild type (WT) (R7954) and mutants (RS111 and RS4). (A) Genomic sequencing of WT and mutants. (B) Genotyping of SSIIIa alleles using the SSIIIa CAPS marker. (C) Varied splicing of transcripts in WT and mutants.
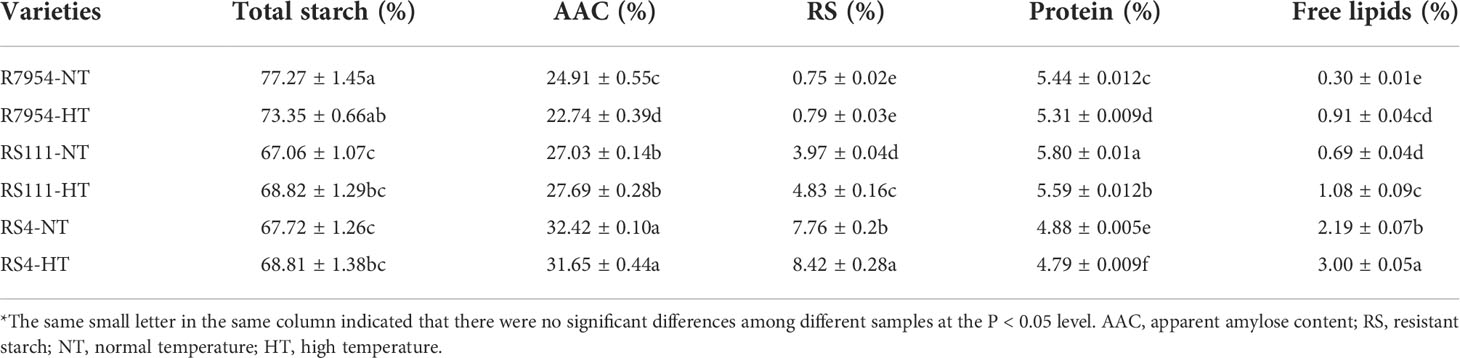
Main reserve substances in the grains under different temperatures
The main reserves in grains of mutants RS111 and RS4 differed significantly from those in R7954, such as decreased starch content, elevated AAC, RS and lipid contents (Table 1). While RS4 and RS111 also showed differences in the contents of the main components, except total starch (Table 1). The RS content in RS4 under NT reached up to 7.76% and the lipid content was high up to 2.19%, which was almost two times and three times as high as those in RS111, respectively. In addition, the protein in RS4 was the least but that in RS111 was the highest. Meanwhile, RS4 showed almost transparent grain under NT, although both mutants showed serious chalk under HT (Figure S1). It is noteworthy that although both RS4 and RS111 had the same ssIIIa mutations, other genes related to starch or lipid metabolites might be mutated in RS4.
Under HT, the protein content was decreased in all three varieties and the lipid contents were all increased. The lipid content in R7954 increased up to 0.91%, close to that in RS111, and the lipid content in RS4 reached up to 3.0%. However, the total starch content and AAC in R7954 were reduced by 3.92% and 2.17%, respectively, while those in RS111 and RS4 are similar as those at NT. HT elevated the RS content only in RS111 and RS4 but not in R7954; the RS content increased from 3.97% to 4.83% and from 7.76% to 8.42% in RS111 and RS4, respectively (Table 1). This indicated that HT played different influences on the storage accumulations among R7954, RS111, and RS4, although they have the same genetic background.
Expression pattern of genes involved in starch synthesis
Among the detected 27 starch synthesis-related genes, the expression of SSIIIa was significantly downregulated in both RS111 and RS4, indicating that the mutation of SSIIIa diminished its transcript. Compared with R7954, most genes in mutants especially in RS4 were upregulated under NT, while these were downregulated under HT (Figure 2). However, the expressions of SSIIIb, AGPL4, and SSIIa were lower in RS4 than those in RS111 and R7954; the expression level of SSIIa in RS111 was the highest under both NT and HT (Figure 2A).
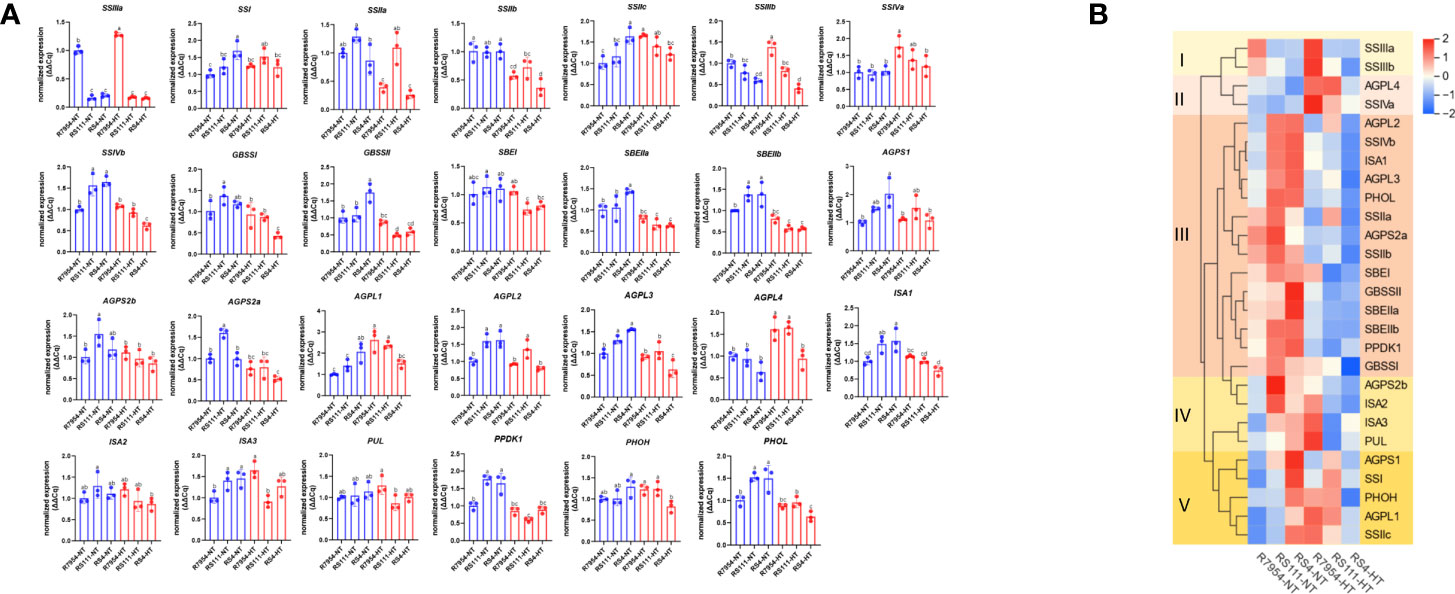
Figure 2 Expression levels of different genes in the three varieties under normal temperature (NT) and high temperature (HT) (A) and the clustering according to their expression levels (B). The same small letter indicated that there were no significant differences among different samples at the P < 0.05 level.
Upon HT, the expressions of starch-related genes showed different responses among R7954, RS111, and RS4. According to their expressions under NT and HT, all genes tested could be categorized into five groups (Figure 2B). SSIIIa and SSIIIb exhibited higher expressions in R7954 at HT than those at NT but showed similar or even lower expressions in RS111 and RS4 at HT. AGPL4 and SSIVa were upregulated under HT in all three varieties. Group III contains most of the genes, including SSIIa, BEI, and SBEIIb; the expressions of these genes in the three varieties all declined to a certain extent under HT, although the attenuated expression of some genes in R7954 was slight. ssIIIa mutation increased the susceptibility of genes, such as GBSSI, SSIVb, ISA1, BEIIb, and BEIIa, to HT exposure, bringing up larger declines in the expression of most genes detected.
Amylopectin chain length distributions
Based on the DP, the side chains of amylopectin can be classified into four fractions, namely, fa with DP 6–12, fb1 with DP 13–24, fb2 with DP 25–36, and fb3 with DP ≥37, which correspond to A, B1, B2, and B3 together with longer chains, respectively (Nakamura et al., 1997). The proportions of fa, fb1, fb2, and fb3 from the three varieties under NT and HT were presented in Table 2. Among the three varieties, RS4 had the highest proportion of fa and the lowest amount of fb3, followed by RS111 and R7954. High-RS mutants RS111 and RS4 had reduced average chain length of amylopectin (Table 2). The varietal differences in amylopectin CL among R7954, RS111, and RS4 were consistent at either NT or HT; mutants had more chains of DP 9–15 and DP 20–30, with the greatest increases in DP 9–12 and less chains of DP ≥37 (Figures 3A, B). These results were basically similar to previous results (Fujita et al., 2007; Shu et al., 2007; Zhao et al., 2020). Under HT, the amounts of fa and fb1 were decreased and fb2 and fb3 were increased for all three varieties (Table 2), with the greatest decreases in DP 9–19 and increases in DP ≥37 (Figure 3C). Obvious decreases in fa under HT were also found in ssIIIa-RNAi rice (Zhao et al., 2020). It is noteworthy that the increase or decrease of the proportion of each chain at HT did not correspond to the borders of the fractions. HT resulted in an increase in the proportion of DP 20–30 in R7954 but not in RS111 and RS4 (Figure 3C). Several studies also found that HT decreased the proportion of fa and increased the proportions of fb2 and fb3 (Umemoto et al., 1999; Jiang et al., 2003; Zhang et al., 2016). The similar trends of decline in the proportion of chains with DP 9–19 in all three varieties indicated that the mechanism causing the temperature-dependent variation of CL is different from the one specifically regulating the proportions of fa and fb1 caused by the mutation of ssIIIa.
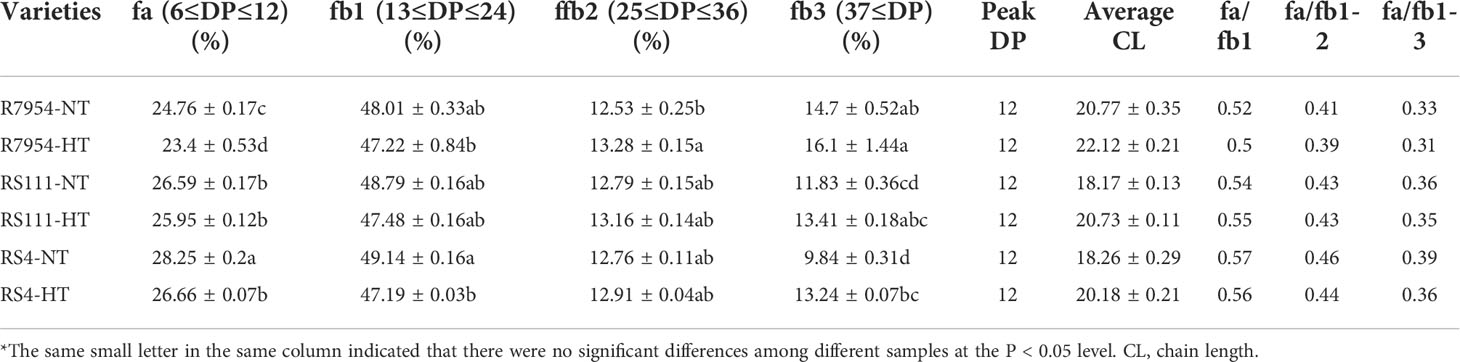
Table 2 Chain length distribution of amylopectin in all rice materials, as determined by HPAEC-PAD analysis*.
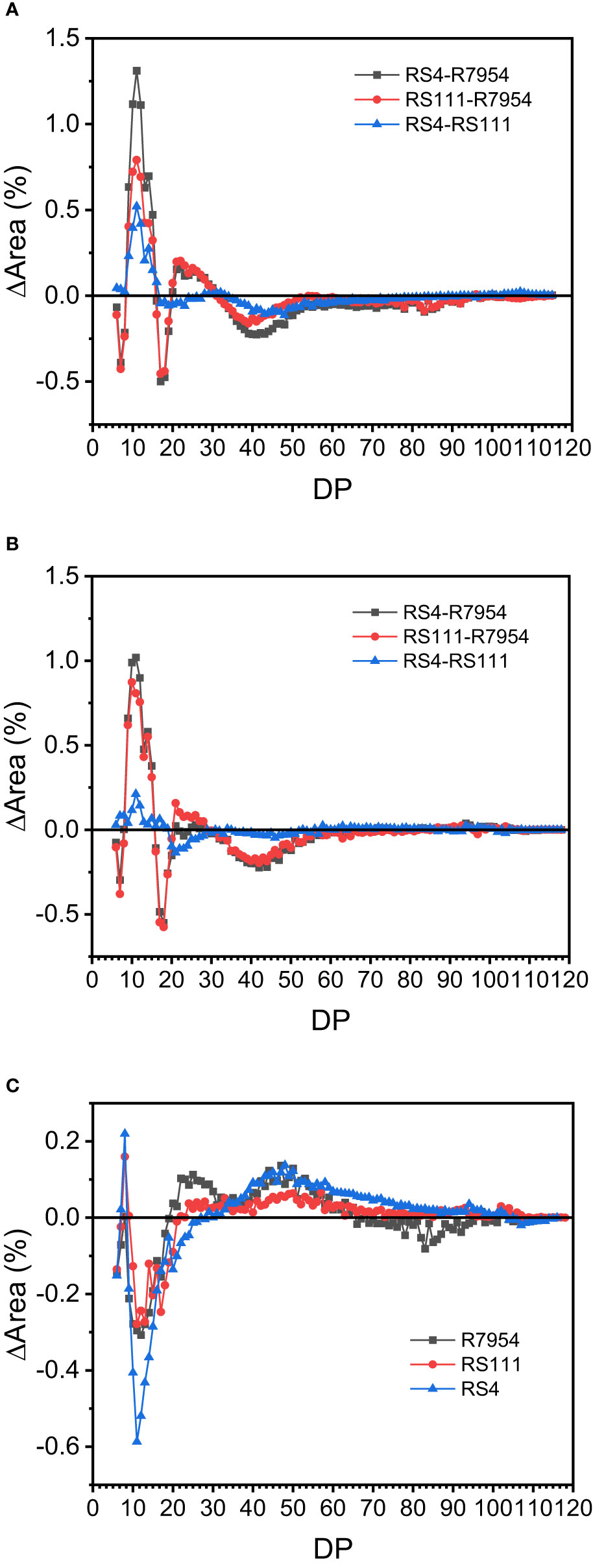
Figure 3 Comparison of the chain length distribution patterns of amylopectin among rice at normal temperature (NT) (A), high temperature (HT) (B), and between HT and NT (C).
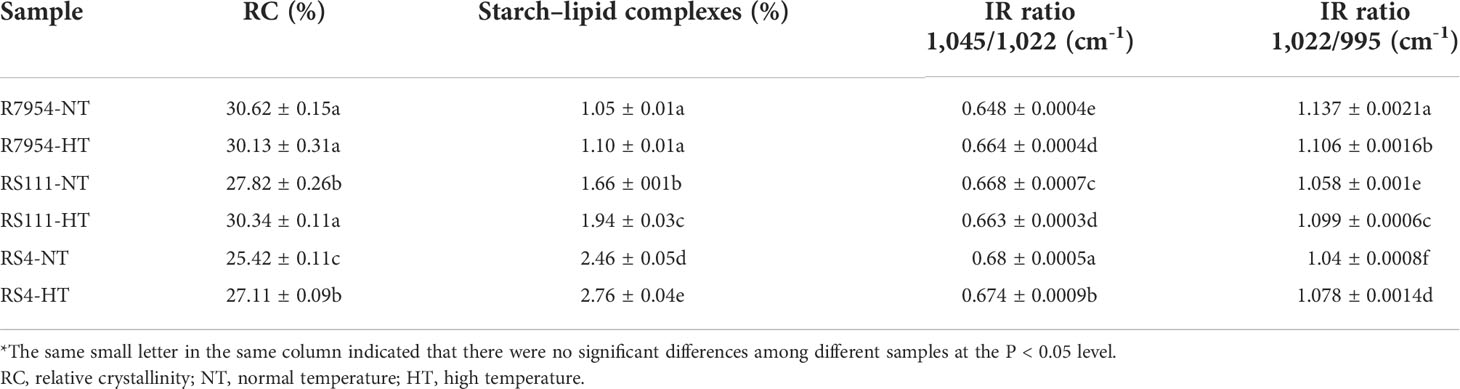
Long-range and short-range ordered structure
XRD can be used to measure the helices that are packed in regular arrays (in the long-range distance) forming crystallinity (Man et al., 2012). The XRD pattern of all starches showed diffraction peaks at ca. 15°, 23° and an unresolved doublet at ca. 17° and 18° (Figure 4A), which are the typical A-type crystalline pattern (Pérez and Bertoft, 2010). In addition, all XRD patterns showed a diffraction peak around 2θ 20°, which indicated the existence of the amylose–lipid complex (Putseys et al., 2010). RS4 had the lowest RC, but the most starch–lipid complexes, especially under HT (Table 3). This might be due to the higher lipid content and AAC in RS4 and RS111 (Table 1). Under HT, RC and the starch–lipid complexes of RS111 and RS4 were elevated but not for R7954 (Table 3).
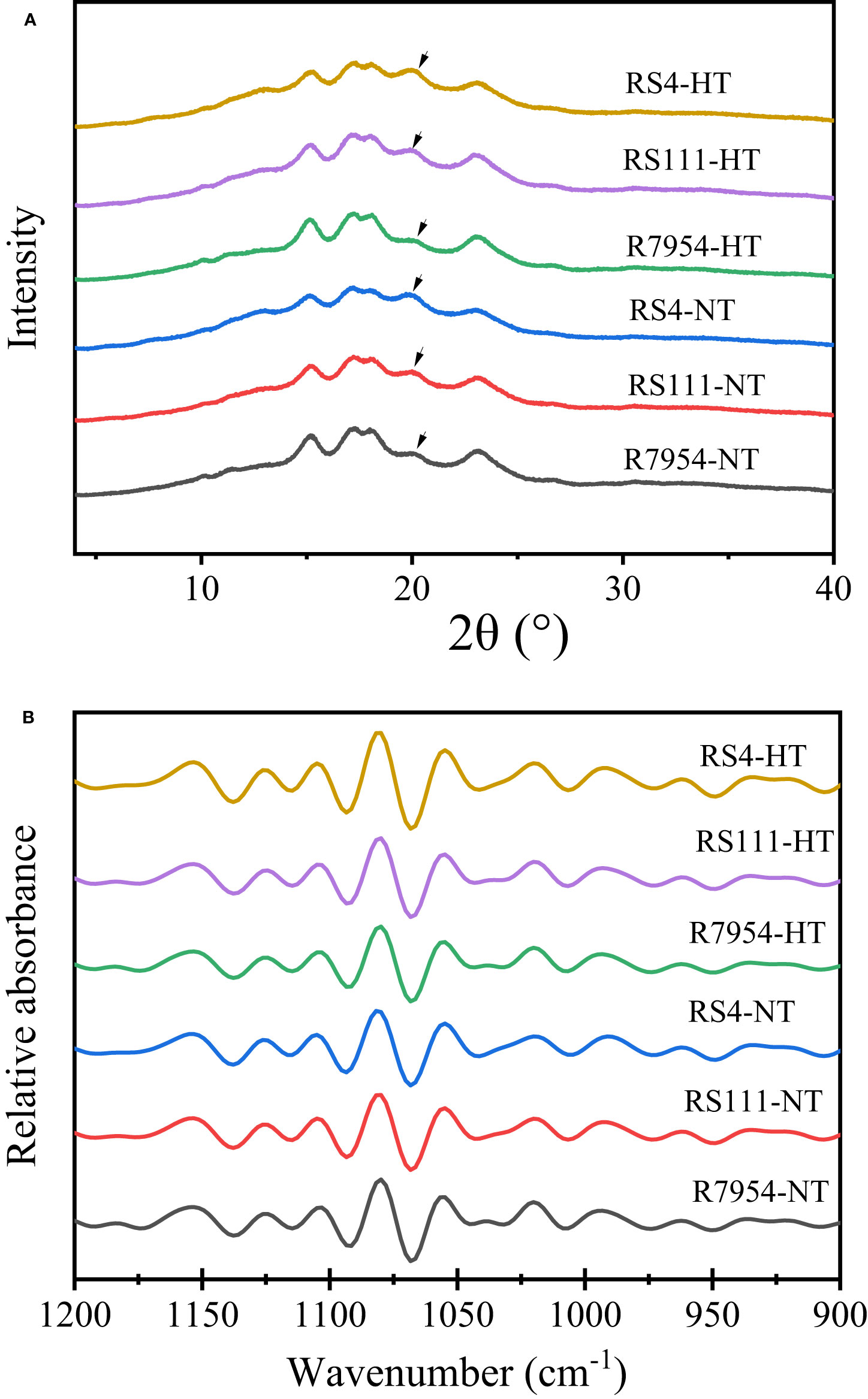
Figure 4 X-ray diffraction (XRD) spectra (A) and deconvoluted Fourier transform infrared (FTIR) spectroscopy spectra (B) of starches from wild type (WT) and mutants at normal temperature (NT) and high temperature (HT). The arrow in panel (A) indicated the peaks of starch–lipid complexes.
With FTIR, short-range ordered structure of starch granule can be observed, the ratio of absorption at 1,045/1,022 cm-1 and 1,022/995 cm-1 can reflect the degree of order structure and the proportion of amorphous to ordered carbohydrate structure in starch granule external region, respectively (Sevenou et al., 2002). All three varieties showed similar FTIR profiles under both NT and HT; no new peaks appeared (Figure 4B). RS111 and RS4 showed higher 1,045/1,022 cm-1 ratio but lower 1,022/995 cm-1 ratio under NT (Table 3), indicating that there were more short-range order helical structure in the external region and less proportion of amorphous to ordered structure in the mutants. Under HT, the 1,045/1,022 cm-1 ratio of R7954 was elevated but that of RS111 and RS4 lessened (Table 3), indicating that the short-range ordered structure especially in the external region of the starch granule in RS111 and RS4 was slightly attenuated. HT played more apparent influences on starch ordered structure in mutants than in R7954, especially on long-range ordered structure.
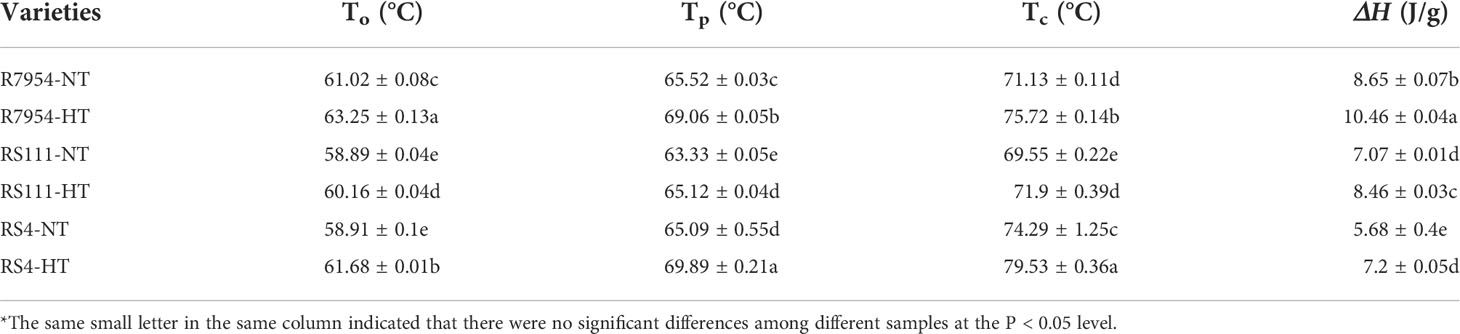
Thermal properties and pasting properties
At NT, RS111 and RS4 had lower To, Tp, and ΔH than R7954. Compared with the values of To (58.91°C–61.02°C), Tp (63.33°C–65.52°C), Tc (69.55°C–74.29°C), and ΔH (5.68–8.65 J/g) at NT, it is obvious that HT significantly elevated the gelatinization temperature (GT) and ΔH (Table 4).
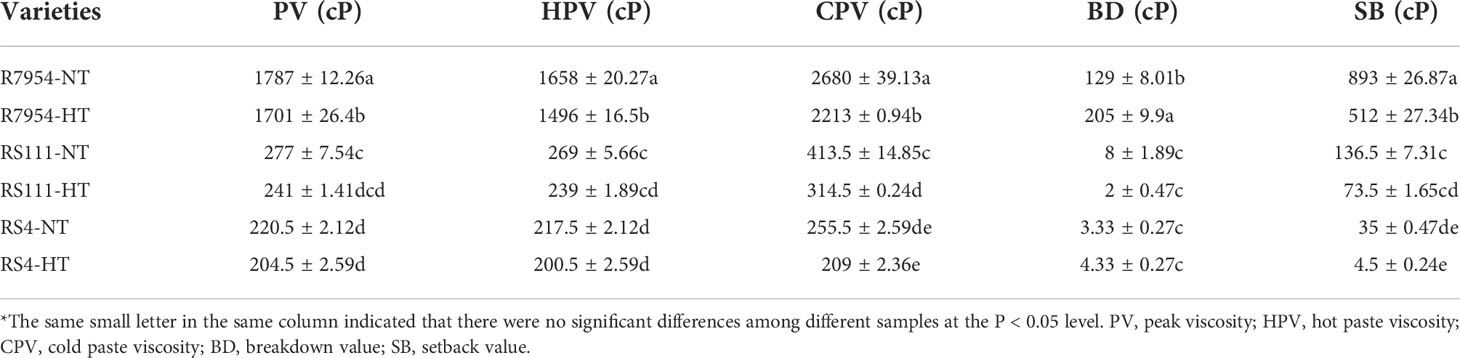
As for the paste viscosity, RS111 and RS4 showed significantly lower paste viscosity than R7954 regardless of whether at NT or HT (Table 5). The PV of R7954, RS111, and RS4 at NT was 1,787 cP, 277 cP, and 200.5 cP, respectively. BD and SB of RS111 and RS4 were also lower than those of R7954. Other ssIIIa mutants also showed very low viscosity (Fujita et al., 2007; Zhang et al., 2011). Under HT, the PV, HPV, CPV, and SB values were decreased in all three varieties, indicating that HT contributed greatly to pasting properties of rice starches, and the cooking quality of rice grain decreased under HT. Decreased paste viscosity of rice under HT had also been observed by Zhang et al. (2020).
Discussion
The influences of ssIIIa mutation and high temperature on RS and other accumulations in rice grains
The SSIIIa gene has been identified as the key gene for RS formation in rice grains (Zhou et al., 2016). RS111 and RS4 are two ssIIIa allelic mutants, and the increased AAC and RS content in RS111 and RS4 (Table 1) might be majorly due to the mutation of SSIIIa (Figure 1). However, RS4 had higher RS and lipid contents (Table 1) that contribute to its greatest starch–lipid complexes (Table 3), as AAC can complex with lipids to form RSV (Hasjim et al., 2010; Zhou et al., 2016). This indicated that, besides SSIIIa, other genes involved in starch synthesis or lipid metabolites might also contribute to RS formation in rice grains.
During grain development, an increment in temperature of 1.6°C–3.1°C can disturb the accumulation of storage materials (Liu et al., 2021). Previous studies showed that HT can increase protein content (Lin et al., 2010) and was unfavorable for lipid accumulation during rice grain ripening. However, HT showed negative influences on protein content and stimulated lipid accumulation in this study (Table 1), although the decreased protein content is small in absolute values. This might be due to different varieties studied. Variations in AAC under HT may also be dependent on rice varieties. Cheng et al. (2005) found that HT increased the AC in high-AC varieties, while Liu et al. (2021) found that AC decreased under elevated temperatures. The different responses of AAC in R7954 and mutants RS111 and RS4 to HT (Table 1) might be because of the increased sensitivity of amyloplast enzymes in RS111 and RS4 exposed to HT. AAC has been thought to be a key determinant for RS formation; higher AAC always related to higher RS content (Raigond et al., 2015). The inconsistent decreases or increases in AAC and RS in the three varieties under HT indicated that, besides amylose, other factors such as lipids also play important roles in RS formation under HT.
Impacts of ssIIIa mutation and high temperature on the expressions of genes involved in starch synthesis and amylopectin fine structure
SSIIIa is a key enzyme responsible for the elongation of long branched amylopectin; it might regulate the whole cascade of starch biosynthesis through the formation of large enzyme complexes with other enzymes (Crofts et al., 2012; Crofts et al., 2015). Loss-of-function mutation of SSIIIa results in abnormal starch metabolites in RS111 and RS4, resulting in a lower total starch content, higher AAC and lipid content, and more proportion of DP 9–15 and DP 20–30 when compared with those of R7954 (Figure 3). The enhanced expression of SSIIa in RS111 might partly compensate for the loss-of-function of SSIIIa, as they play partially overlapping roles during starch synthesis (Zhang et al., 2011), which might partially explain the differences in CL distribution of amylopectin between RS111 and RS4 (Figure 2).
Generally, HT leads to a declined expression of many starch biosynthesis genes such as GBSSI, BEs, cyPPDK (Jiang et al., 2003; Yamakawa et al., 2007; Zhang et al., 2021) and SSs like SSIIIa, SSII, and SSIV (Liu et al., 2021). SSI, SSIIa, SSIIIa, BEI, and BEIIb function specifically in elongating or synthesizing A chains, B1 chains, B2 chains, or longer chains (Nakamura et al., 2005; Fujita et al., 2006; Fujita et al., 2007; Jeon et al., 2010). The decreased transcripts of starch synthase and branching enzyme genes (Figure 2) in all varieties resulted in decreases in the amount of fa and fb1 under HT (Table 2, Figure 3) (Jiang et al., 2003). Meanwhile, lower expression levels of SSI, SSIIc, and some other genes in RS4 under HT (Figure 2) might be responsible for its noticeable decreases in amylopectin short chains. The more pronounced decreases in the expression of several genes such as BEIIb, BEI and BEIIa in RS111 and RS4 indicated that ssIIIa mutation exacerbated the influences of HT on starch metabolites.
Influences of high temperature on starch crystalline
Crystallinity was influenced majorly by fine amylopectin structure and AC (Man et al., 2014; Bertoft, 2017). More AAC and shorter amylopectin usually lead to a defective crystallite (Luo et al., 2021). The lower RC in RS111 and RS4 might be due to their higher AAC (Table 1) and more short chains of amylopectin (Table 2). The increased RC in RS4 and RS111 at HT was consistent with the pronouncedly decreased amount of fa and fb1 and increased proportion of fb2 and fb3 at HT (Table 2). Although amylopectin long chains are of benefit for the formation of double helices, complexing with non-starchy compounds increased short-range ordered structure (Chi et al., 2021). The increased starch–lipid complexes in RS111 and RS4 might contribute to the increased 1,045/1,022 cm-1 ratio (Table 3).
Previous studies indicated that slower digestibility of processed starch is usually related to the short-range ordered structures; however, the reassembled ordered molecular aggregation architecture showed more pronounced starch digestibility. Compared with long-range ordered structures, short-range ordered structures exhibited a weaker effect on starch digestibility (Chi et al., 2021). The increased RC in RS4 and RS111 under HT might partly contribute to their enhanced RS content, as an increase in the degree of crystallinity can result in a reduction in starch digestibility in vitro and in vivo (Rombo et al., 2001; García-Rosas et al., 2009), and the amorphous matrix might be more susceptible to enzyme attack (Chung et al., 2006).
Influences of high temperature on starch thermal and paste viscosity
GT has been found to be related to crystallinity associated with molecular order (Table 3). The amount of short-chain amylopectin (DP ≤12) was negatively correlated with GT; starch with relatively high levels of long branch chains (13 ≤ DP ≤ 24) requires higher temperatures for complete dissociation (Zhang et al., 2020). The higher AAC (Table 1) and the more proportion of fa in mutants (Table 2, Figure 3) indicated that they have shorter double helices formed between adjacent chains in the crystalline lamellae (Jeon et al., 2010), leading to a lower starch GT and ΔH (Table 4). The enhanced GT under HT may be due to the increased crystallites and double helices (Table 3). Under HT, the increased proportion of amylopectin long chains that span at least two crystalline lamellae and more ordered structures can form more stable double helices; hence, a higher GT would be required to disorder the ordered structure (Witt et al., 2012). A previous study also found that HT exposure led to a significantly higher GT (Pang et al., 2021). Furthermore, the variations of GT between NT and HT were larger than those among varieties (Table 4), indicating that the influences of temperature on GT were more obvious than that of ssIIIa mutation.
The very low viscosity of RS111 and RS4 might be due to their high AAC and lipids (Table 1), as lipids and amylose complexes formed during cooking can render entanglements with amylopectin molecules and restrict the swelling of granules, causing incomplete gelatinization during cooking (Sun et al., 2018), which results in a higher pasting temperature and a lower PV (Ai and Jane, 2015). Increased lipids under HT (Table 1) might be responsible partially for the decreased viscosity. Previous studies have found that Wx and SSIIa were the main genes involved in many grain quality properties such as gelatinization and paste viscosity (Kharabian-Masouleh et al., 2012); significantly different expressions of GBSSI and SSIIa among R7954, RS4, and RS111 at both NT and HT might explain the different viscosity among them.
Proposed regulations of high temperature on RS formation in rice
Rice high in RS had a relatively higher proportion of DP 8–12 and less DP ≥37 (Shu et al., 2007). Among the three varieties, the higher the RS content, the most the proportion of DP 9–15 and the least the proportion of DP ≥37. While under HT, the amylopectin chains with DP 9–15 in RS4 and RS111 were greatly decreased and chains with DP ≥37 were increased, but the RS contents in RS111 and RS4 under HT were significantly increased (Table 1). The incongruent correlation between RS and DP 9–15 and DP ≥37 under HT indicated that the mechanism causing enhanced RS under HT should be different from that caused by loss of function of SSIIIa, which might be due to the differences in stability or temperature preference of starch synthase and starch branching enzyme (Umemoto et al., 1999).
The synthesis of amylose and amylopectin might compete for substrates and enzymes (Han et al., 2019) and maintain equilibrium under normal conditions. While inactivation of SSIIIa results in varied transcripts of genes related to starch synthesis (Figure 2), leading to more substrates synthetized by GBSSI into amylose (Figure 5). Furthermore, inactive SSIIIa facilitated the synthesis of DP 10–15 chains from short DP 6–9 chains of A chains and DP 20–30 chains from DP 16–19 chains of B1 chains of amylopectin by enhancing the endogenous SSI activity and reduced the formation of DP >35 chains (Fujita et al., 2007) (Table 2, Figure 5B). Enhanced transcripts of SSIVb or other enzymes might partially compensate for the elongation of B2-B4 in the ssIIIa mutant (Figure 5). Concurrently, the increased expression levels of BEs such as BEI and BEIIb (Figure 2) might increase the branching frequency and enhance the proportion of short chains further (Table 2, Figure 5). Less proportion of short-chain DP 10–15 is an important determinant for high RS (Table 1) (Shu et al., 2007; Shu et al., 2009). Additionally, pyruvate phosphate dikinase (PPDK) presented in the complex with AGPase may enable G-1-P channel to lipid, while the starch biosynthetic complexes are proposed to inhibit the activity of PPDK and AGPase, the absence of SSIIIa enhanced the expression of PPDK (Figure 2) and promoted lipid accumulation (Figure 5). The high levels of amylose and lipids in mutants especially in RS4 can form more amylose–lipid complexes (RSV) (Table 3).
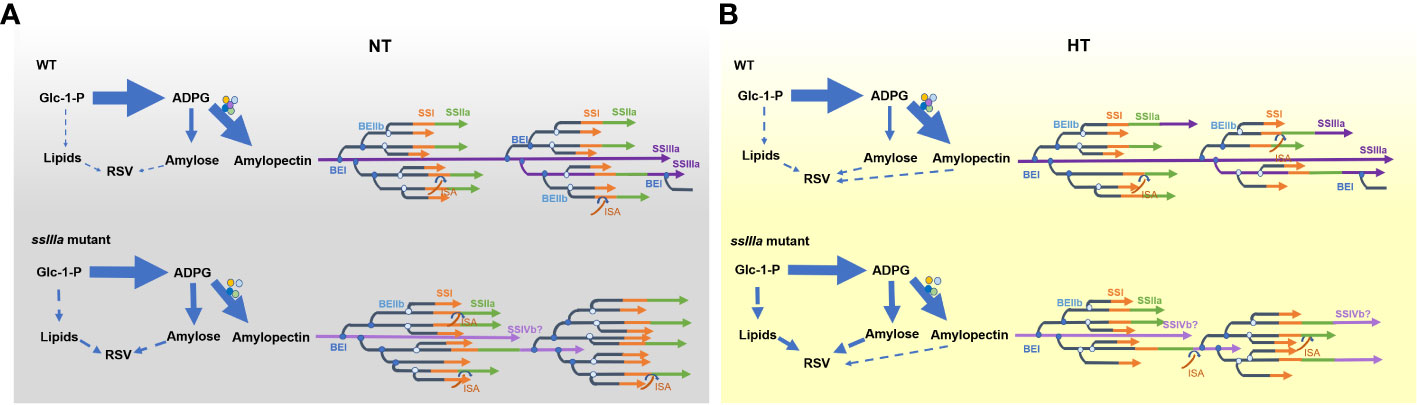
Figure 5 Proposed processes of RS formation in rice. (A) The loss-of-function mutation of SSIIIa causes repartition of carbohydrate, breaks the equibrium synthesis of amylose and amylopectin, and leads to higher AAC and lipid content that promote the formation of starch–lipid complexes. (B) Loss-of-function mutation of SSIIIa leads to more short chains of DP 9–15 both under NT and high temperature (HT). HT decreases the branching, resulting in less proportion of short chains and more long chains. The increased long chains and enhanced lipid content under HT stimulate more RS accumulation.
Under HT, the altered transcripts of several genes in R7954 led to aberrant starch synthesis (Table 1). Fan et al. (2019) found that the ratio of SBE activity to that of GBSS was increased under HT. Downregulation of the Wx gene due to transcript inhibition or posttranscriptional regulation (Zhang et al., 2021) caused less synthesis of amylose, while for ssIIIa mutants, although the expression levels of most genes were also reduced, amylopectin and amylose biosynthesis maintained a relative balance, but the starch enzyme complexes might be disrupted further due to the lower activity of most amyloplast enzymes and more carbon were partitioned into lipid metabolites, leading to the synthesis of more lipids (Table 1, Figure 5). Furthermore, decreases in the transcripts of BEI and BEIIb (Figure 2) resulted in lower branching frequency (Figure 5) and produced fewer short chains of DP ≤19, with the greatest decreases in chains of DP 9–19 (Figure 3). The increased transcripts of SSIIIa in R7954 and SSIVb in mutants under HT (Figure 2) might also partially contribute to the increased proportion of long chains (DP >37) (Figure 3, Table 2), reflected as intensified crystallinity in mutants (Table 3). Longer amylopectin side chains might also be involved in RS formation, although the short chains of DP 9–15 are the major determinants for RS properties, as retrograded amylopectin may also reduce enzyme susceptibility (Eerlingen and Delcour, 1995). Moreover, aside from amylose, long chains of amylopectin may also complex with the increased free lipids and lead to increases in the contents of RSV at HT (Table 3). The molecular mechanisms of RS produced in high-RS rice mutants under HT might not be uniform with that under NT. Trimming the fine structure of amylopectin and regulating lipid metabolites through biological means might be alternative ways to obtain rice rich in RS with acceptable palatability under global climate change.
In conclusion, RS4 and RS111 are two ssIIIa allelic mutants as b10 (Zhou et al., 2016). Meanwhile, other mutations responsible for RS formation might exist in RS4, as the RS in RS4 was significantly higher than that in RS111. Loss-of-function mutation of SSIIIa boosted the expressions of most genes involved in starch biosynthesis under NT, such as Phol, SSI, BEIIb, and SSIVb. The varied expressions of genes involved in starch biosynthesis in ssIIIa mutants caused the increases in AAC and lipid content, more proportion of DP 9–15, and less proportion of DP >37, which finally led to enhanced RS. However, most of the genes upregulated under NT in ssIIIa mutants especially in RS4 were downregulated under HT, such as GBSSI, SSI, BEI, BEIIb, and ISA. The decreased activities of SSs and BEs under HT caused greatest decreases in chains of DP 9–19 and produced more proportion of long chains (DP >37) in all materials, but only reduced AAC and total starch content in R7954. Moreover, the varied transcripts of genes involved in starch biosynthesis under HT channeled more carbon flux to lipid synthesis, promoting lipid accumulation. The varied starch structure and lipid content under HT resulted in increased crystallinity, GT, ΔH and reduced viscosity. The high AAC and enhanced long amylopectin chains and lipid content in RS111 and RS4 under HT might boost the starch–lipid complexes further and contribute to the elevated RS.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
YZ: Investigation, Data curation, Formal analysis, Writing-original draft. ZC: Investigation, Data curation, Formal analysis, Writing-original draft. SJ: Investigation. JC: Investigation. DW: Funding acquisition, material bred. XS: Conceptualization, Formal analysis, Writing-review and editing, Supervision and Funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This project was financially supported by grants from the National Key Research and Development Program of China (2021YFF100020), Functional Rice Breeding and germplasm enhancement (2022C02011, 2021C02063), National Natural Science Foundation of China (32072038) and Administration Bureau of Yazhou Bay Science Technology City (SKJC-2020-02-010).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1059749/full#supplementary-material
References
Ai, Y., Jane, J. L. (2015). Gelatinization and rheological properties of starch. Starch/Staerke 67, 213–224. doi: 10.1002/star.201400201
Bao, J., Kong, X., Xie, J., Xu, L. (2004). Analysis of genotypic and environmental on rice starch. 1. apparent amylose content, pasting viscosity, and gel texture. J. Agric. Food Chem. 52, 6010–6016. doi: 10.1021/jf049234i
Bao, J., Ying, Y., Zhou, X., Xu, Y., Wu, P., Xu, F., et al. (2020). Relationships among starch biosynthesizing protein content, fine structure and functionality in rice. Carbohydr. Polym. 237, 116118. doi: 10.1016/j.carbpol.2020.116118
Baysal, C., He, W., Drapal, M., Villorbina, G., Medina, V., Capell, T., et al. (2020). Inactivation of rice starch branching enzyme IIb triggers broad and unexpected changes in metabolism by transcriptional reprogramming. Proc. Natl. Acad. Sci. U. S. A. 117, 26503–26512. doi: 10.1073/pnas.2014860117
Bertoft, E. (2017). Understanding starch structure: Recent progress. Agronomy 7, 56. doi: 10.3390/agronomy7030056
Butardo, V. M., Fitzgerald, M. A., Bird, A. R., Gidley, M. J., Flanagan, B. M., Larroque, O., et al. (2011). Impact of down-regulation of starch branching enzyme IIb in rice by artificial microRNA-and hairpin RNA-mediated RNA silencing. J. Exp. Bot. 62, 4927–4941. doi: 10.1093/jxb/err188
Cheng, F., Zhong, L., Zhao, N., Liu, Y., Zhang, G. (2005). Temperature induced changes in the starch components and biosynthetic enzymes of two rice varieties. Plant Growth Regul. 46, 87–95. doi: 10.1007/s10725-005-7361-6
Chi, C., Li, X., Huang, S., Chen, L., Zhang, Y., Li, L., et al. (2021). Basic principles in starch multi-scale structuration to mitigate digestiblity: A review. Trends Food Sci. Technol. 109, 154–168. doi: 10.1016/j.tifs.2021.01.024
Chung, H. J., Lim, H. S., Lim, S. T. (2006). Effect of partial gelatinization and retrogradation on the enzymatic digestion of waxy rice starch. J. Cereal Sci. 43, 353–359. doi: 10.1016/j.jcs.2005.12.001
Crofts, N., Abe, K., Aihara, S., Itoh, R., Nakamura, Y., Itoh, K., et al. (2012). Lack of starch synthase IIIa and high expression of granule-bound starch synthase I synergistically increase the apparent amylose content in rice endosperm. Plant Sci., 193–194, 62–69. doi: 10.1016/j.plantsci.2012.05.006
Crofts, N., Abe, N., Oitome, N. F., Matsushima, R., Hayashi, M., Tetlow, I. J., et al. (2015). Amylopectin biosynthetic enzymes from developing rice seed form enzymatically active protein complexes. J. Exp. Bot. 66, 4469–4482. doi: 10.1093/jxb/erv212
Ding, Y., Huang, J., Zhang, N., Rasmussen, S. K., Wu, D., Shu, X. (2019). Physiochemical properties of rice with contrasting resistant starch content. J. Cereal Sci. 89, 102815. doi: 10.1016/j.jcs.2019.102815
Eerlingen, R., Delcour, J. (1995). Formation, analysis, structure and properties of type III enzyme resistant starch. J. Cereal Sci. 22, 129–138. doi: 10.1016/0733-5210(95)90042-X
Englyst, H. N., Hudson, G. J. (1996). The classification and measurement of dietary carbohydrates. Food Chem. 57, 15–21. doi: 10.1016/0308-8146(96)00056-8
Fan, X., Li, Y., Lu, Y., Zhang, C., Li, E., Li, Q., et al. (2019). The interaction between amylose and amylopectin synthesis in rice endosperm grown at high temperature. Food Chem. 301, 125258. doi: 10.1016/j.foodchem.2019.125258
Fujita, N., Yoshida, M., Asakura, N., Ohdan, T., Miyao, A., Hirochika, H., et al. (2006). Function and characterization of starch synthase I using mutants in rice. Plant Physiol. 140, 1070–1084. doi: 10.1104/pp.105.071845
Fujita, N., Yoshida, M., Kondo, T., Saito, K., Utsumi, Y., Tokunaga, T., et al. (2007). Characterization of SSIIIa-deficient mutants of rice: the function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. 144, 2009–2023.doi: 10.1104/pp.107.102533
García-Rosas, M., Bello-Pérez, A., Yee-Madeira, H., Ramos, G., Flores-Morales, A., Mora-Escobedo, R. (2009). Resistant starch content and structural changes in maize (Zea mays) tortillas during storage. Starch/Staerke 61, 414–421. doi: 10.1002/star.200800147
Guo, D., Ling, X., Zhou, X., Li, X., Wang, J., Qiu, S., et al. (2020). Evaluation of the quality of a high-resistant starch and low-glutelin rice (Oryza sativa l.) generated through CRISPR/Cas9-mediated targeted mutagenesis. J. Agric. Food Chem. 68, 9733–9742. doi: 10.1021/acs.jafc.0c02995
Han, H., Yang, C., Zhu, J., Zhang, L., Bai, Y., Li, E., et al. (2019). Competition between granule bound starch synthase and starch branching enzyme in starch biosynthesis. Rice 12, 96–105. doi: 10.1186/s12284-019-0353-3
Hasjim, J., Lee, S. O., Hendrich, S., Setiawan, S., Ai, Y., Jane, J. L. (2010). Characterization of a novel resistant-starch and its effects on postprandial plasma-glucose and insulin responses. Cereal Chem. 87, 257–262. doi: 10.1094/CCHEM-87-4-0257
Hu, Y., Leu, R. K. L., Christophersen, C. T., Somashekar, R., Conlon, M. A., Meng, X. Q., et al. (2016). Manipulation of the gut microbiota using resistant starch is associated with protection against colitis-associated colorectal cancer in rats. Carcinogenesis 37, 366–375. doi: 10.1093/carcin/bgw019
Itoh, K., Ozaki, H., Okada, K., Hori, H., Takeda, Y., Mitsui, T. (2003). Introduction of wx transgene into rice wx mutants leads to both high- and low-amylose rice. Plant Cell Physiol. 44, 473–480. doi: 10.1093/pcp/pcg068
Jeon, J. S., Ryoo, N., Hahn, T. R., Walia, H., Nakamura, Y. (2010). Starch biosynthesis in cereal endosperm. Plant Physiol. Biochem. 48, 383–392. doi: 10.1016/j.plaphy.2010.03.006
Jiang, H., Dian, W., Wu, P. (2003). Effect of high temperature on fine structure of amylopectin in rice endosperm by reducing the activity of the starch branching enzyme. Phytochemistry 63, 53–59. doi: 10.1016/S0031-9422(03)00005-0
Kharabian-Masouleh, A., Waters, D. L. E., Reinke, R. F., Ward, R., Henry, R. J. (2012). SNP in starch biosynthesis genes associated with nutritional and functional properties of rice. Sci. Rep. 2, 1–9. doi: 10.1038/srep00557
Kong, X., Bertoft, E., Bao, J., Corke, H. (2008). Molecular structure of amylopectin from amaranth starch and its effect on physicochemical properties. Int. J. Biol. Macromol. 43, 377–382. doi: 10.1016/j.ijbiomac.2008.07.018
Kong, X., Kasapis, S., Zhu, P., Sui, Z., Bao, J., Corke, H. (2016). Physicochemical and structural characteristics of starches from Chinese hull-less barley cultivars. Int. J. Food Sci. Technol. 51, 509–518. doi: 10.1111/ijfs.12984
Kumar, A., Sahoo, U., Baisakha, B., Okpani, O. A., Ngangkham, U., Parameswaran, C., et al. (2018). Resistant starch could be decisive in determining the glycemic index of rice cultivars. J. Cereal Sci. 79, 348–353. doi: 10.1016/j.jcs.2017.11.013
Lin, C. J., Li, C. Y., Lin, S. K., Yang, F. H., Huang, J. J., Liu, Y. H., et al. (2010). Influence of high temperature during grain filling on the accumulation of storage proteins and grain quality in rice (Oryza sativa l.). J. Agric. Food Chem. 58, 10545–10552. doi: 10.1021/jf101575j
Liu, W., Yin, T., Zhao, Y., Wang, X., Wang, K., Shen, Y., et al. (2021). Effects of high temperature on rice grain development and quality formation based on proteomics comparative analysis under field warming. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.746180
Luo, X., Cheng, B., Zhang, W., Shu, Z., Wang, P., Zeng, X. (2021). Structural and functional characteristics of japonica rice starches with different amylose contents. CYTA - J. Food 19, 532–540. doi: 10.1080/19476337.2021.1927194
Man, J., Lin, G., Wang, Z., Wang, Y., Liu, Q., Wei, C. (2014). Different structures of heterogeneous starch granules from high-amylose rice. J. Agric. Food Chem. 62, 11254–11263. doi: 10.1021/jf503999r
Man, J., Yang, Y., Zhang, C., Zhou, X., Dong, Y., Zhang, F., et al. (2012). Structural changes of high-amylose rice starch residues following in vitro and in vivo digestion. J. Agric. Food Chem. 60, 9332–9341. doi: 10.1021/jf302966f
Miura, S., Koyama, N., Crofts, N., Hosaka, Y., Abe, M., Fujita, N. (2021). Generation and starch characterization of non-transgenic BEI and BEIIb double mutant rice (Oryza sativa) with ultra-high level of resistant starch. Rice 14, 3. doi: 10.1186/s12284-020-00441-0
Nakamura, Y., Francisco, P. B., Jr., Hosaka, Y., Sato, A., Sawada, T., Kubo, A., et al. (2005). Essential amino acids of starch synthase IIa differentiate amylopectin structure and starch quality between japonica and indica rice varieties. Plant Mol. Biol. 58, 213–227. doi: 10.1007/sl
Nakamura, Y., Kubo, A., Shimamune, T., Matsuda, T., Harada, K., Satoh, H. (1997). Correlation between activities of starch debranching enzyme and α-polyglucan structure in endosperms of sugary-1 mutants of rice. Plant J. 12, 143–153. doi: 10.1046/j.1365-313X.1997.12010143.x
Ohdan, T., Francisco, P. B., Sawada, T., Hirose, T., Terao, T., Satoh, H., et al. (2005). Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J. Exp. Bot. 56, 3229–3244. doi: 10.1093/jxb/eri292
Pang, Y., Hu, Y., Bao, J. (2021). Comparative phosphoproteomic analysis reveals the response of starch metabolism to high-temperature stress in rice endosperm. Int. J. Mol. Sci. 22. doi: 10.3390/ijms221910546
Pérez, S., Bertoft, E. (2010). The molecular structures of starch components and their contribution to the architecture of starch granules: A comprehensive review. Starch/Staerke 62, 389–420. doi: 10.1002/star.201000013
Putseys, J. A., Lamberts, L., Delcour, J. A. (2010). Amylose-inclusion complexes: Formation, identity and physico-chemical properties. J. Cereal Sci. 51, 238–247. doi: 10.1016/j.jcs.2010.01.011
Raigond, P., Ezekiel, R., Raigond, B. (2015). Resistant starch in food: A review. J. Sci. Food Agric. 95, 1968–1978. doi: 10.1002/jsfa.6966
Rombo, G. O., Taylor, J. R. N., Minnaar, A. (2001). Effect of irradiation, with and without cooking of maize and kidney bean flours, on porridge viscosity and in vitro starch digestibility. J. Sci. Food Agric. 81, 497–502. doi: 10.1002/jsfa.838
Sajilata, M. G., Singhal, R. S., Pushpa, R. K. (2006). Resistant starch — a review. Compr. Rev. Food Sci. Food Saf. 5, 1–17. doi: 10.1111/j.1541-4337.2006.tb00076.x
Sevenou, O., Hill, S. E., Farhat, I. A., Mitchell, J. R. (2002). Organisation of the external region of the starch granule as determined by infrared spectroscopy. Int. J. Biol. Macromol. 31, 79–85. doi: 10.1016/S0141-8130(02)00067-3
Shen, L., Li, J., Li, Y. (2022). Resistant starch formation in rice: Genetic regulation and beyond. Plant Commun. 3, 100329. doi: 10.1016/j.xplc.2022.100329
Shu, X., Jia, L., Gao, J., Song, Y., Zhao, H., Nakamura, Y., et al. (2007). The influences of chain length of amylopectin on resistant starch in rice (Oryza sativa l.). Starch - Stärke 59, 504–509. doi: 10.1002/star.200700640
Shu, X., Jiao, G., Fitzgerald, M. A., Yang, C., Shu, Q., Wu, D. (2006). Starch structure and digestibility of rice high in resistant starch. Starch/Staerke 58, 411–417. doi: 10.1002/star.200600501
Shu, X., Jia, L., Ye, H., Li, C., Wu, D. (2009). Slow digestion properties of rice different in resistant starch. J. Agric. Food Chem. 57, 7552–7559. doi: 10.1021/jf900988h
Shu, X., Sun, J., Wu, D. (2014). Effects of grain development on formation of resistant starch in rice. Food Chem. 164, 89–97. doi: 10.1016/j.foodchem.2014.05.014
Sun, J., Wang, Y., Zhang, X., Rasmussen, S. K., Jiang, X., Song, W., et al. (2018). Dependence of physiochemical, functional and textural properties of high-resistant starch rice on endogenous nonstarch polysaccharides. Int. J. Food Sci. Technol. 53, 1079–1086. doi: 10.1111/ijfs.13686
Syahariza, Z. A., Sar, S., Warren, F. J., Zou, W., Hasjim, J., Tizzotti, M. J., et al. (2014). Erratum: The importance of amylose and amylopectin fine structures for starch digestibility in cooked rice grains. Food Chem. 136, 617–618. doi: 10.1016/j.foodchem.2012.08.053
Umemoto, T., Nakamura, Y., Satoh, H., Terashima, K. (1999). Differences in amylopectin structure between two rice varieties in relation to the effects of temperature during grain-filling. Starch/Staerke 51, 58–62. doi: 10.1002/(SICI)1521-379X(199903)51:2<58::AID-STAR58>3.0.CO;2-J
Witt, T., Doutch, J., Gilbert, E. P., Gilbert, R. G. (2012). Relations between molecular, crystalline, and lamellar structures of amylopectin. Biomacromolecules 13, 4273–4282. doi: 10.1021/bm301586x
Xia, J., Zhu, D., Wang, R., Cui, Y., Yan, Y. (2018). Crop resistant starch and genetic improvement : a review of recent advances. Theor. Appl. Genet. 131, 2495–2511. doi: 10.1007/s00122-018-3221-4
Xin, Z., Velten, J. P., Oliver, M. J., Burke, J. J. (2003). High-throughput DNA extraction method suitable for PCR. Biotechniques 34, 820–826. doi: 10.2144/03344rr04
Yamakawa, H., Hirose, T., Kuroda, M., Yamaguchi, T. (2007). Comprehensive expression profiling of rice grain filling-related genes under high temperature using DNA microarray. Plant Physiol. 144, 258–277. doi: 10.1104/pp.107.098665
You, H., Liang, C., Zhang, O., Xu, H., Xu, L., Chen, Y., et al. (2022). Variation of resistant starch content in different processing types and their starch granules properties in rice. Carbohydr. Polym. 276, 118742. doi: 10.1016/j.carbpol.2021.118742
Zhang, G., Cheng, Z., Zhang, X., Guo, X., Su, N., Jiang, L., et al. (2011). Double repression of soluble starch synthase genes SSIIa and SSIIIa in rice ( Oryza sativa l.) uncovers interactive effects on the physicochemical properties of starch. Genome 54, 448–459. doi: 10.1139/g11-010
Zhang, H., Xu, H., Jiang, Y., Zhang, H., Wang, S., Wang, F., et al. (2021). Genetic control and high temperature effects on starch biosynthesis and grain quality in rice. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.757997
Zhang, C., Yang, Y., Chen, Z., Chen, F., Pan, L., Lu, Y., et al. (2020). Characteristics of grain physicochemical properties and the starch structure in rice carrying a mutated ALK/SSIIa gene. J. Agric. Food Chem. 68, 13950–13959. doi: 10.1021/acs.jafc.0c01471
Zhang, C., Zhou, L., Zhu, Z., Lu, H., Zhou, X., Qian, Y., et al. (2016). Characterization of grain quality and starch fine structure of two japonica rice (Oryza sativa) cultivars with good sensory properties at different temperatures during the filling stage. J. Agric. Food Chem. 64, 4048–4057. doi: 10.1021/acs.jafc.6b00083
Zhao, Q., Ye, Y., Han, Z., Zhou, L., Guan, X., Pan, G., et al. (2020). SSIIIa-RNAi suppression associated changes in rice grain quality and starch biosynthesis metabolism in response to high temperature. Plant Sci. 294. doi: 10.1016/j.plantsci.2020.110443
Keywords: resistant starch, high temperature, amylopectin, chain length distribution, rice
Citation: Zhou Y, Cheng Z, Jiang S, Cen J, Wu D and Shu X (2022) High temperature boosts resistant starch content by altering starch structure and lipid content in rice ssIIIa mutants. Front. Plant Sci. 13:1059749. doi: 10.3389/fpls.2022.1059749
Received: 02 October 2022; Accepted: 21 October 2022;
Published: 18 November 2022.
Edited by:
Xinyang Wu, China Jiliang University, ChinaReviewed by:
Changquan Zhang, Yangzhou University, ChinaYihua Wang, Nanjing Agricultural University, China
Xiangjin Wei, State Key Laboratory of Rice Biology and Chinese National Center for Rice Improvement (CAAS), China
Copyright © 2022 Zhou, Cheng, Jiang, Cen, Wu and Shu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoli Shu, c2h1eGxAemp1LmVkdS5jbg==
†These authors have contributed equally to this work
 Yufeng Zhou
Yufeng Zhou Zhenfeng Cheng1,2†
Zhenfeng Cheng1,2† Shuo Jiang
Shuo Jiang Xiaoli Shu
Xiaoli Shu