
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 09 November 2022
Sec. Plant Abiotic Stress
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1054064
This article is part of the Research Topic Abiotic and Biotic Stress in Horticultural Crops: Insight into Recent Advances in the Underlying Tolerance Mechanism View all 46 articles
Lilium pumilum is an important ornamental, culinary and medicinal bulbous plants with salt tolerance. However, salt tolerance of lily, particularly the bulb, has been studied relatively little, which brings challenges to the cultivation of lily varieties with high salt tolerance. Here, we performed transcriptome sequencing on the bulb organs of L. pumilum under salt stress treatment, analyzed differential gene expressed levels and then identified several key genes associated with salt stress tolerance at genome-wide scale. For the first time, we revealed the obvious response against salt stress for L. pumilum bulb organs, while distinct from those for root organs. Several key genes obtained through transcriptome analysis and DEG screening include NF-YB3 transcription factor, metallothionein type 2 protein, vicilin like seed storage protein and bidirectional sugar transporter SWEET14. Rather than typical ROS scavengers like superoxide dismutase, peroxidase, and glutathione transferase, non-typical ROS scavengers such as the metallothionein type 2 protein, and vicilin like seed storage protein were upregulated in our work. The bidirectional sugar transporter SWEET14 protein and the hormone signaling proteins such as E3-ubiquitin protein ligases, PYL4 and protein phosphatase 2C were also upregulated, suggesting the role of sugars and hormones in the bulb organ responses to salt stress. Co-expression analysis of the DEGs further confirmed that NF-YB3 transcription factor acted as a hub gene, suggesting that salt stress can promote flowering of L. pumilum. Taken together, we identified important candidate genes associated with salt tolerance of the L. pumilum bulb organs, which may provide the excellent basis for further in-depth salt tolerance mechanisms of the lily bulbs.
Lilium, a genus of herbaceous flowering plant growing from bulbs, is widely cultivated horticultural crop. The flowers of these plants are popular in ornamental gardens or indoor potted plants. Lilies are also used in the culinary industry and folkloric medicine (Zhou et al., 2021). Lilium pumilum, commonly known as coral lily, is native to grassy mountainous areas of northeast China which are heavily salinized region of the country. Therefore, L. pumilum is regarded as a salt tolerant plant, but the molecular mechanism of salt tolerance in L. pumilum has not been extensively studied, which limits its widespread application.
Salt stress is a major abiotic stress which adversely impacts plant growth and development (Shrivastava and Kumar, 2015). Worldwide distribution of saline-alkali land and soil salinization of non-saline land have been significantly reducing the cultivable land areas, challenging the agricultural, horticultural and forestry production. Salt accumulation in arable soils is caused by seawater and irrigation water that contains trace amounts of sodium chloride (NaCl) (Flowers and Yeo, 1995; Tester and Davenport, 2003). Damages caused by salt stress are classified into two categories of osmotic stress and ionic toxicity and several salt tolerant plants including haplotypes have evolved salt tolerance responses as follows: In early salt stress period, osmotic homeostasis was biased by accumulated salts in soil, decreasing the water absorbing ability of plants and accumulation of osmolytes such as sucrose is one of the natural responses of plants to recover the osmotic homeostasis, ultimately lowering water loss and maximizing water uptake. Later, salt ions accumulate within plant cells and cause ion toxicity, affecting intracellular metabolic processes and Na+ exclusion and vacuole compartmentalization are another typical plant responses to minimize the ion toxicity (Blumwald, 2000; Sahi et al., 2006; Munns and Tester, 2008; Flowers and Colmer, 2015). In previous studies, multiple factors have been found to play crucial roles in the above salt stress responses in plants (Zhao et al., 2021); which include histidine kinase receptor protein HK1, Ca2+ dependent signaling factors like calcium-dependent protein kinases (CDPKs), calcineurin B-like proteins (CBLs) with CBL-interacting protein kinases (CIPKs) and calmodulin-binding transcription activators (CAMTAs), transcription factors such as basic leucine zipper (bZIP), WRKY, APETALA2/ETHYLENE RESPONSE FACTOR (AP2/ERF), MYB, basic helix–loop–helix (bHLH) and NAC families, Na+ and K+ transporters like CYCLIC NUCLEOTIDE-GATED CHANNEL (CNGC) and the GLUTAMATE-LIKE RECEPTOR (GLR) families, tonoplast-localized Na+/H+ exchanger 1 (NHX1), plasma membrane-localized SALT OVERLY SENSITIVE 1 (SOS1) Na+/H+ antiporters, high-affinity K+ transporter HKT, ROS scavenging enzymes like superoxide dismutase, peroxidase, and glutathione transferase, organic osmolytes like proline, glycine betaine, sugar alcohols, polyamines, and proteins from the late embryogenesis abundant (LEA) superfamily (Deinlein et al., 2014). Phytohormones such as abscisic acid (ABA), auxin, cytokinin (CK), gibberellic acid (GA), brassinosteroids (BRs), ethylene, jasmonic acid (JA), salicyclic acid (SA) and strigolactones (SLs) are also important factors that regulate plant responses to salt stress (Zhao et al., 2021). These hormones are classified into two groups as stress hormones such as ABA, SA, JA, ethylene and growth hormones including auxin, CKs, and act in a sophisticated crosstalk (Yu et al., 2020). In addition, the mitogen activated protein kinase (MAPK) cascade has been ubiquitously found in salt stress responses of many plant species, and it is believed to function as point of convergence in plant development and signaling associated with hormones and stresses (Sinha et al., 2014).
The advent of ‘Omics’ technologies such as the transcriptome analysis have been rapidly developed and widely applied to plants, enabling the genome-wide identification of key regulatory factors in plant responses to various stresses (Imadi et al., 2015; Zhu et al., 2015; Ju et al., 2021). Transcriptome analysis via RNA-sequencing (RNA-seq) can provide dynamic nature of transcriptome at higher coverage and greater resolution than previously employed methods such as Sanger sequencing- and microarray (Kukurba and Montgomery, 2015). RNA-seq has already been successfully applied to L. pumilum and revealed key genes associated with dormancy release at cold storage condition (Wang et al., 2018) and tepal trichome development (Xin et al., 2021). The response of L. pumilum bulbs to salt stress has not yet been studied using RNA-seq. In recent years, it has been reported that transcription factors from L. pumilum can improve plant salt tolerance (Yong et al., 2019; Wang et al., 2020; Wang et al., 2022; Yan et al., 2022). But they have not yet been studied at genome-wide scale using RNA-seq. Root is the very organ that first senses and responds to salt stress and leaf is the organ that first shows apparent morphological changes upon salt stress. Therefore, most of studies on salt stress have focused on the root and leaf. Bulbous plants such as L. pumilum have unique organs called the bulb which is known to function as nutrient storage or flowering organs. It is believed that L. pumilum is salt-tolerant ornamental plant which may be related to its unique bulb organ function. But it requires evidences showing that the bulb organs help the plants to overcome stressful environments such as salt stress. Therefore, RNA-seq analysis of lily bulb organs can provide novel insights into the salt stress tolerance of L. pumilum.
In this study, we analyzed the transcriptome data of the L. pumilum bulb organs treated with salt stress in order to identify key genes associated with salt stress responses of bulb organs at genome-wide scale. The transcriptome data of L. pumilum bulb organs were generated by Illumina sequencing and Trinity de novo assembly. The genes showing differential expression (DEGs) in the L. pumilum bulb organs upon salt stress treatment were then identified by comparing the gene transcript profiles in the bulb organ samples between salt-treated and control plants at different time points of salt stress treatment, mainly focusing on identifying the candidate genes associated with transcription regulation, ROS scavenging, ion detoxification, osmolyte accumulation and hormone signaling. Putative hub genes were predicted through building co-expression network using STRING database and Cytoscape software. The expression levels of several key genes were validated by qRT-PCR. The preliminary results of this study provided the basic information on molecular mechanism underlying regulatory network for salt stress responses in the L. pumilum bulb organs that may serve as foundation for further in-depth functional studies.
L. pumilum plants were grown in laboratory at College of Landscape Architecture of Northeast Forestry University (Harbin, Heilongjiang, China). Healthy seedlings were cultured on hydroponic tank supplemented with Hoagland nutrient solution at 70% relative humidity, and 22-25 °C ambient temperature. 3 wk-old plants were treated with 150 mM NaCl for 12 h. The plants at 0 h of NaCl treatment were used as control. Bulb organ samples were collected at 2 h and 12 h, respectively.
Total RNA was extracted from the bulb organ samples of different sampling times using Trizole reagent (TG-DP441, China). The quantity and purity of the extracted RNA samples were evaluated by Bioanalyzer 2100 and RNA 100 Nano LabChip Kit (Agilent, CA, USA) with RIN number >7.0. PolyA RNA was obtained by purification of total RNA with oligo-dT magnetic beads. Then, mRNA was treated with divalent cations under temperature gradient for fragmentation into smaller size. Next, cDNA library was constructed by reverse-transcription reactions from those cleaved RNA fragments using RNA-seq sample preparation kit (Illumina, San Diego, USA). Finally, the cDNA library was sequenced on Illumina Novaseq™ 6000 platform (LC Sciences, USA). All raw reads were deposited in the Sequence Read Archive (SRA) database in NCBI with accession number PRJNA851552. Clean reads were obtained by removing adaptor contamination, low quality bases as well as undetermined bases using Cutadapt (Martin, 2011) and in-house Perl scripts and their quality was then assessed by FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Trinity 2.4.0 (Grabherr et al., 2011) was utilized to assemble the clean data. The de novo assembled transcripts were clustered and then the overlapping reads were counted to obtain unigenes for further analysis. All unigenes obtained by de novo assembly were blasted to NCBI non-redundant protein database (NR), Gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), Pfam, swiss-Prot, and eggNOG database using DIAMOND (Buchfink et al., 2015) with threshold of e-value <0.00001.
Expression levels of the unigenes were determined by calculating TPM (Transcripts Per Kilobase of exon model per Million mapped reads) (Mortazavi et al., 2008). DEGs were screened out according to the criteria of log2 (fold change) >1 or <-1 with p value <0.05 (statistically significant) using R package-edgeR (Robinson et al., 2009). The DEGs were further annotated to GO and KEGG databases for their functional enrichment analysis. In GO enrichment analysis, all the DEGs were mapped to corresponding GO terms, the gene number in each term was calculated and then the GO terms significantly enriched in DEGs were found through the hypergeometric test. KEGG enrichment analysis for the DEGs was also conducted to find enriched pathways for the DEGs. GO and KEGG analyses data were converted into scatter plots using ggplot2. DEGs at different periods (0-2 h and 2-12 h) of salt stress were compared to find out the key genes which were predicted as essential regulators for salt stress responses of the bulb organs in L.pumilum.
In order to build co-expression network. The DEGs at 0-12 h with p <0.05 were selected and their interaction was predicted using STRING database (https://cn.string-db.org/cgi/input?sessionId=bOrBIG9AvMjL&input_page_show_search=on ). The Arabidopsis thaliana was used as reference organism in the prediction. Then, the network was built using Cytoscape. Network analysis and hub gene identification were conducted using network analyzer and cytohubba plugins, respectively, in Cytoscape.
Several key DEGs were randomly selected for qRT-PCR to verify their expression levels. RNA was extracted and then reversely transcribed into cDNA using Rever Tra Ace qPCR RT Kit (TOYOBO, Osaka, Japan) by a reaction flow of 37 °C, 15 min and 98 °C, 5 min. Primers were designed using the RealTimePCR design tool of the IDT website (http://sg.idtdna.com/scitools/Applications/RealTimePCR ) and were synthesized by Comate Bioscience (Jilin, China). Primer information was shown in Additional file. qRT-PCR was performed using triazole reagent UltraSYBR Mixture kit (CWBIO, Beijing, China). The 20 μl reaction system was as follows (Table 1):
qRT-PCR reactions were performed in Roche Light Cycler 96 (Roche, Basel, CH) according to following program: preincubation, 95 °C for 10 min; three steps amplification, 95 °C for 10 s, 60 °C for 30 s, and 72 °C for 32 s (total 40 cycles); melting, 95 °C for 15 s, 60 °C for 1 min, 95 °C for 15 s; cooling, 37 °C for 30 s. Lily Actin was used as an internal reference gene. All experiments were performed in three biological replicates. Relative expression level was calculated by the 2-ΔΔCt method.
Total RNA of the bulb samples of L. pumilum were sequenced by Illumina double-terminal sequencing technology. Three biological replicates, a total of 9 samples, were generated for each time point. The sequencing data of all samples exceeded 6 G. In total, 492572310 raw sequencing reads were obtained from the control and treated samples and were then subjected to cleaning overrepresented sequences such as Illumina adaptor sequence, generating 455124918 clean reads that were used for subsequent analyses (Table 2).
Q20, Q30, and GC content were over 98, 95, and 49%, respectively, indicating high quality of sequencing results. The sequencing data were assembled by Trinity. 137087 transcripts and 51566 unigenes were obtained, with an average length of 481 bp and 436 bp, respectively (Table 3). Length distribution and GC content of the transcripts and unigenes represented high quality of assembly works (Figure 1). The N50 value for transcript and unigene was 1132 and 1331, respectively, suggesting that the assembled data were competent for subsequent analyses.
De novo assembled sequencing data were annotated to six databases namely NR, GO, KEGG, Pfam, swiss-Prot, and eggNOG using the DIAMOND program. The annotated number of unigenes for each database is shown in Table 4.
All unigenes were annotated at least by one database. Most unigenes (26510; 51.41%) were annotated to NR. According to the annotation result to NR database, the unigenes of L. pumilum bulb showed high homology with Elaeis guineensis (23.04%) and Phoenix dactylifera (19.58%) (Figure 2).
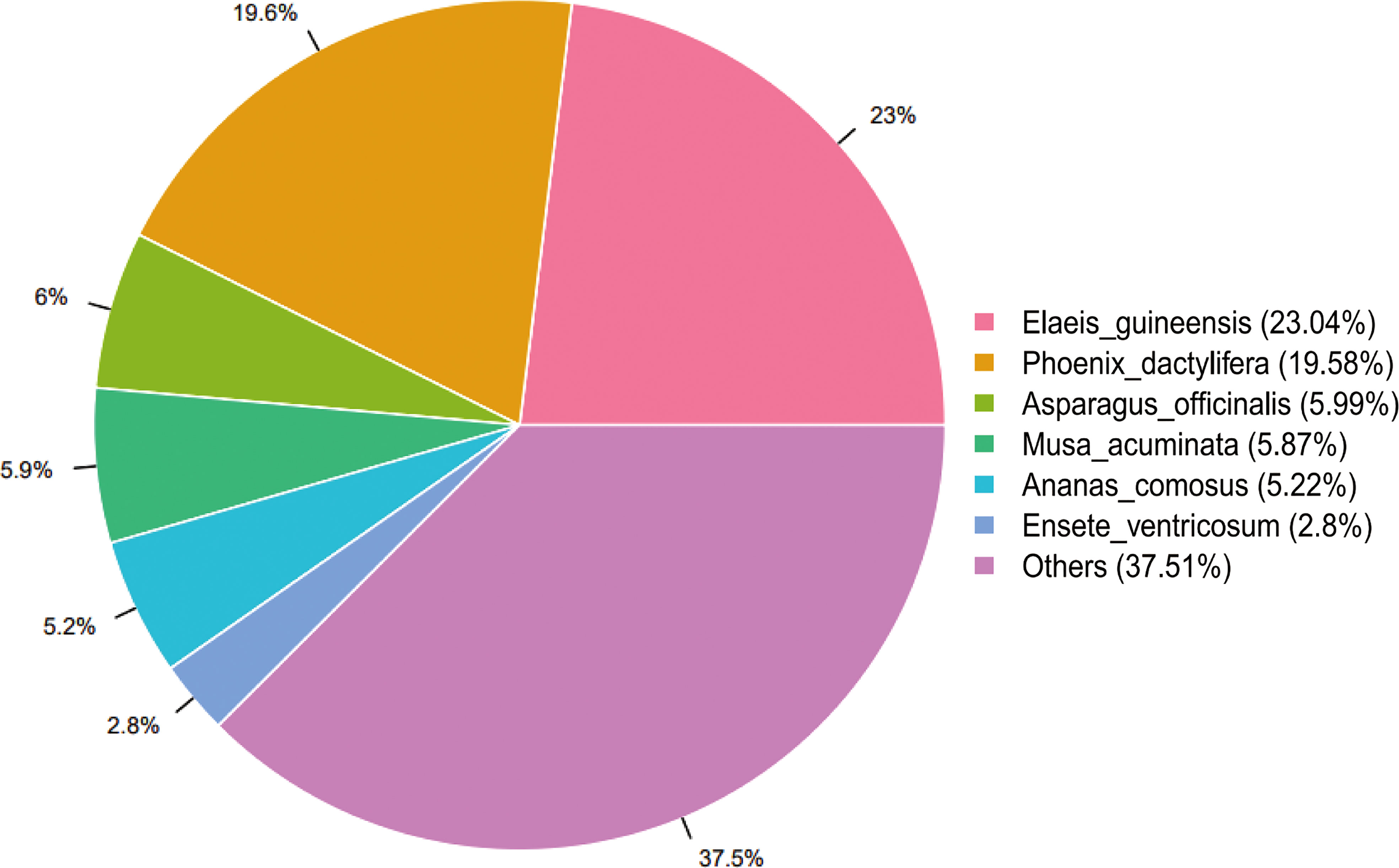
Figure 2 Species distribution for the assembled unigenes annotated to NCBI non-redundant protein database (NR). The percentages represent genetic homology degree of assembled unigenes with respective species.
All annotated unigenes were functionally classified by GO, KEGG, and eggNOG databases. In the GO ontology annotation, unigenes were grouped into 3 categories of “biological process”, “cellular component”, and “molecular function” (Figure 3). In the category of “biological process”, several activities including gene transcription and translation, protein modification and degradation, secondary metabolism, and responses to salt, cold, heat, water and oxidative stresses as well as hormones like auxin and abscisic acid were found in salt stress response of L. pumilum. In the categories of “cellular component” and “molecular function”, various cellular components and molecular functions such as nucleus, cytoplasm, membranes, chloroplast, and mitochondria and protein binding, DNA binding, metal ion binding, etc were enriched in salt stress response of L. pumilum. In the KEGG annotation, gene transcription and translation also showed the highest activities and environmental adaptation and secondary metabolilsm were the most active metabolism except for the housekeeping metabolisms such as carbohydrate, lipid, amino acid metabolisms (Figure 4). Annotation by the eggNOG database also revealed similar patterns in classification of the unigenes (Figure 5). Enrichment analysis of DEGs among the unigenes annotated by GO and KEGG will be analyzed in the next section.
Among the samples (0, 2 h, and 12 h of salt treatment), total 4096 DEGs (log2Fold change>1 or <-1) were identified by pairwise comparison of TPM values. 1772 (878: upregulated and 894: downregulated) DEGs were identified at 0-2 h period of salt stress, and 2324 (1031 upregulated and 1293 downregulated) DEGs at 2-12 h period of salt stress (Figure 6A).
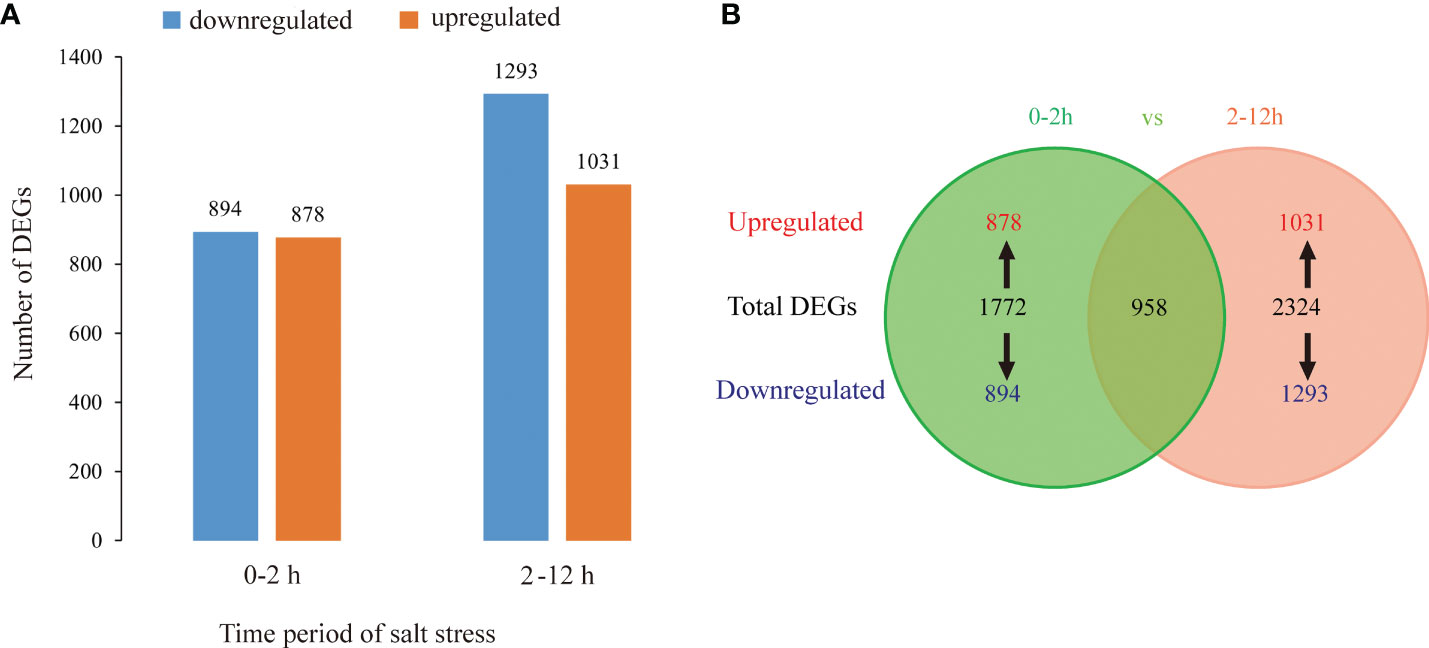
Figure 6 DEGs between L. pumilum bulb samples at different time points (0, 2, and 12 h) during salt stress. (A) Number of upregulated or downregulated DEGs in pairwise comparisons, (B) Venn diagram showing number of DEGs in different comparisons.
We examined the clustering patterns of DEGs for different periods (0-2 h and 2-12 h) of salt stress by clustering the DEGs according to the similarity in their functions, metabolic processes or cellular pathways. In our study, TPM of a differential gene was displayed as Z-value, and the cluster analysis was performed using 100 DEGs with the smallest p value. The clustering analysis (Figure 7) showed that 3 biological replicates for all samples have good repeatability.
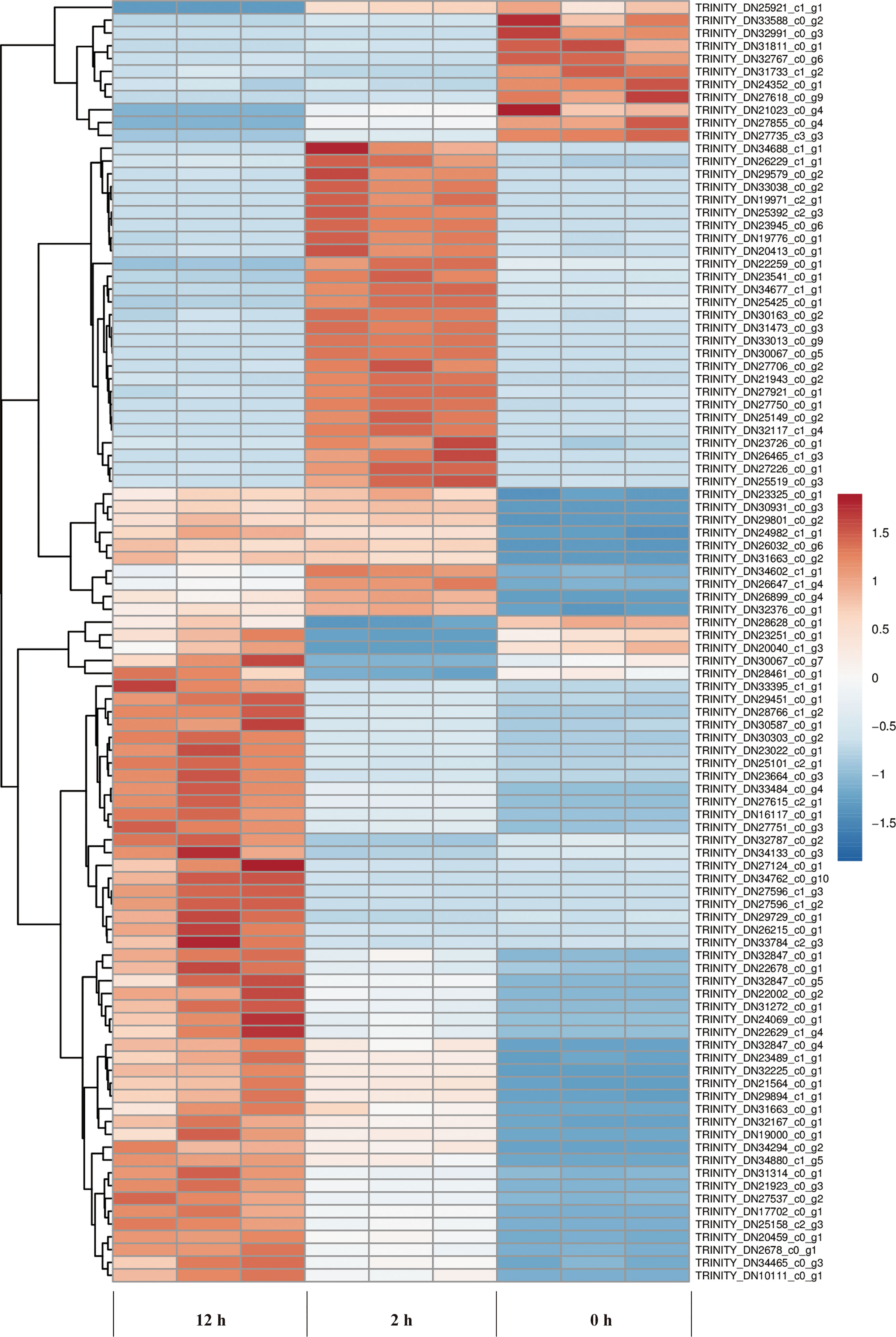
Figure 7 Cluster analysis of DEGs at different periods (0, 2, and 12 h) of salt stress. The abscissa represents the three independent biological replicates for the control (0 h), 2 h and 12 h duration of salt stress treatment, respectively. The ordinate represents the ID of a hundred genes with smallest p value. Color gradient from blue to red represents gene expression level from low to high.
The DEGs at the initial stage (0-2 h period) and later stage (2-12 h period) during salt stress were subjected to GO enrichment analysis (Figure 8).
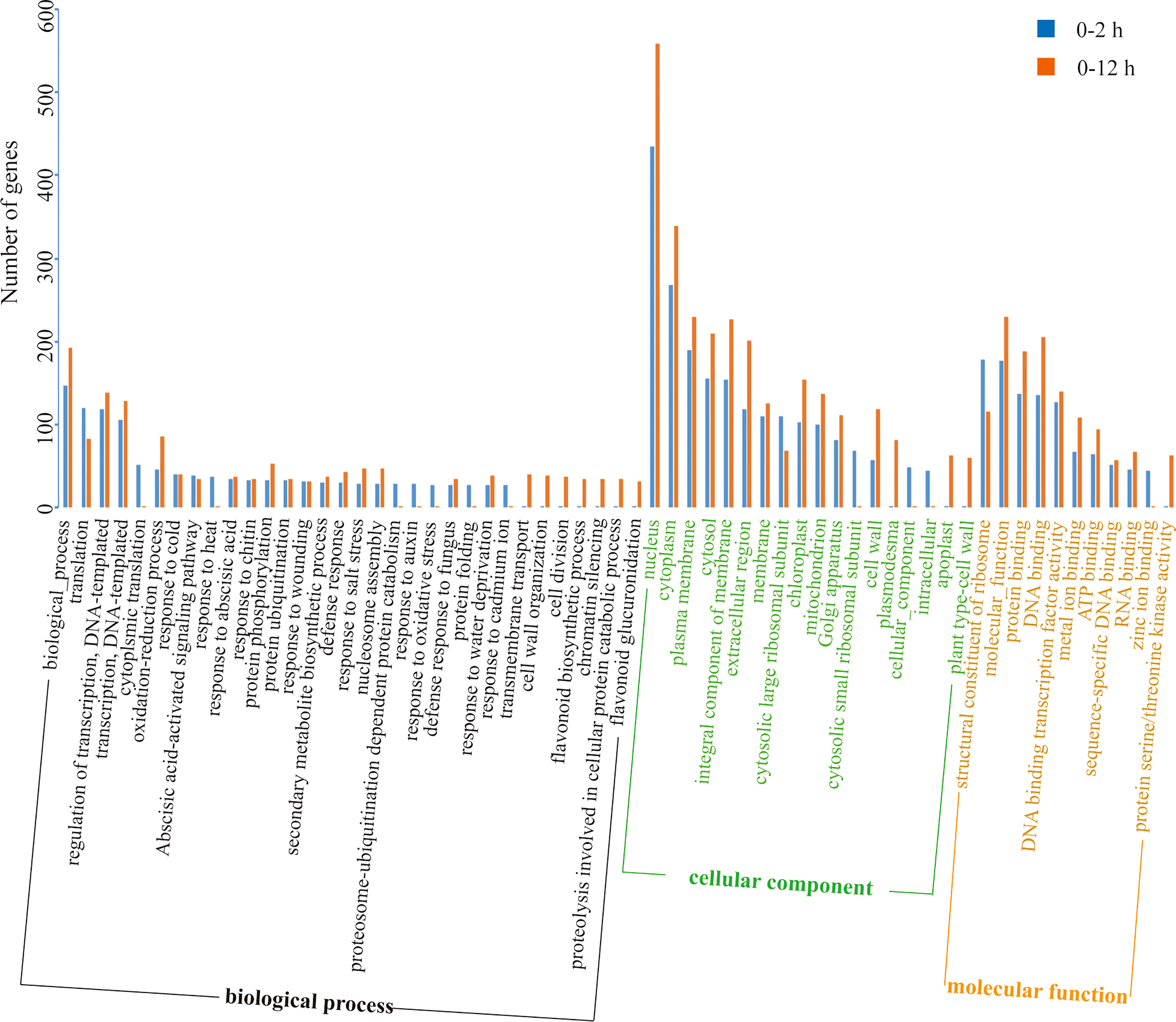
Figure 8 GO enrichment analysis of DEGs of two different periods (0-2 h and 2-12 h) during salt stress.
DEGs were highly enriched in the GO functions such as “biological process”, “translation”, “DNA-templated transcription and its regulation”, “oxidation-reduction processes”, “nucleus”, “cytoplasm”, “membrane”, “protein binding”, “DNA binding”, etc. The different enrichment results at the initial stage (0-2 h) and the later stage (2-12 h) indicated that different genetic programs controlled the response of lily bulb to salt stress in this study. As the salt stress prolongs, the genes that belong to the GO functions including “cytoplasmic translation”, “response to heat”, “proteosome-ubiquitination dependent protein catabolism”, “proteolysis involved in cellular protein catabolic process”, “response to auxin”, “response to oxidative stress”, “protein folding”, “response to cadmium ion”, “transmembrane transport”, “cell wall organization”, “cell division”, “chromatin silencing”, “flavonoid biosynthetic process”, “flavonoid glucuronidation”, “cytosolic small ribosomal subunit”, “plasmodesma”, “cellular component”, “intracellular”, “apoplast”, “plant type-cell wall”, “zinc ion binding”, and “protein serine/threonine kinase activity” showed dramatically differential expression. Therefore, it is obvious that the complicated responses elicited by salt stress includes a wide-range of cellular activities and the genetic program of the plant (lily bulb) has a temporal differential pattern in which the aforementioned DEGs are regulated differently depending on time.
In addition, KEGG pathway annotation of the DEGs in two different periods (0-2 h vs 2-12 h) during salt stress showed that the DEGs were associated with more than 30 pathways and were highly enriched in the pathways associated with “Ribosomes”, “Plant hormone signal transduction”, “Phenylpropanoid biosynthesis”, “MAPK signaling pathway-plant”, and “Starch and sucrose metabolism” (Figure 9). DEG enrichment results at two different time periods (0-2 h and 2-12 h) also revealed the differences in pathway types and gene numbers, which might be related to temporally different responses to salt stress at 0-2 h and 2-12 h periods.
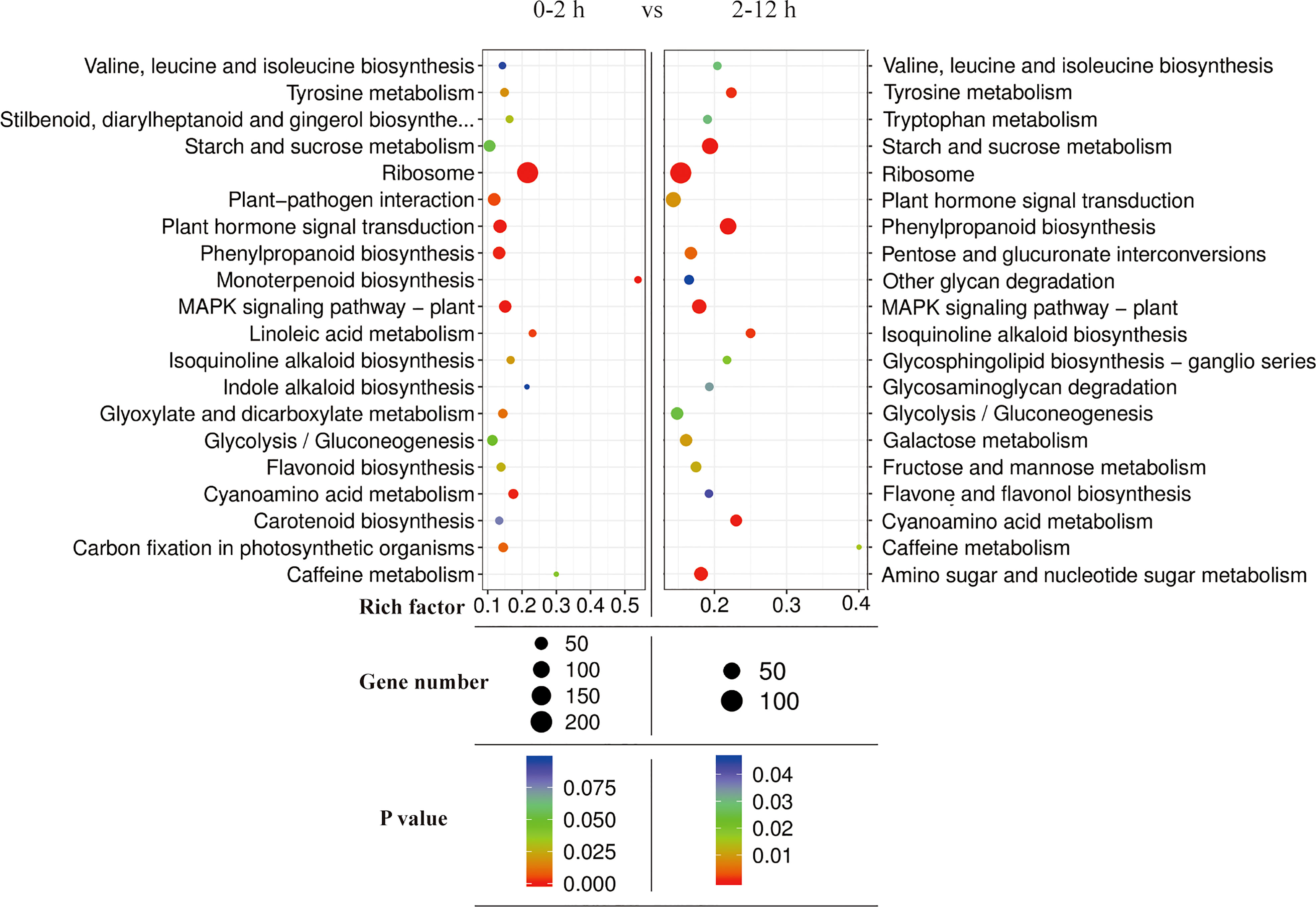
Figure 9 KEGG enrichment analysis of DEGs of two different periods (0-2 h and 2-12 h) during salt stress.
Then, we focused on the 958 DEGs which were co-induced throughout the entire period of salt stress (0-12 h) (Figure 6B). Among them, we identified various transcription factors belonging to AP2/ERF, bHLH, bZIP, HD2/ZIP2, HSF, MYB, NAC, NF-YB, and WRKY families (Figure 10), among which AP2/ERF, bHLH, WRKY transcription factors take the highest proportion. A few unknown proteins with DNA-binding transcription factor activity also showed remarkably significant differential expression patterns during salt stress. Of all, NF-YB3 transcription factor showed high and constant upregulation during the throughout entire salt stress period (0-12 h). Besides, bHLH35*, bHLH13, ERF053, ERF1A, ERF1B, and WRKY28 showed remarkable upregulation albeit not constant for entire period. bHLH35*, bHLH13, ERF053, ERF1A, and ERF1B were highly upregulated at the initial period (0-2 h) during salt stress treatment and then declined, indicating their roles in Lily bulb’s primary responses to salt stress. In contrast, WRKY28, WRKY50*, and bHLH41 were downregulated during the initial period, and then upregulated later, suggesting that they might have distinguishing roles other than bHLH35*, bHLH13, ERF053, ERF1A, and ERF1B.
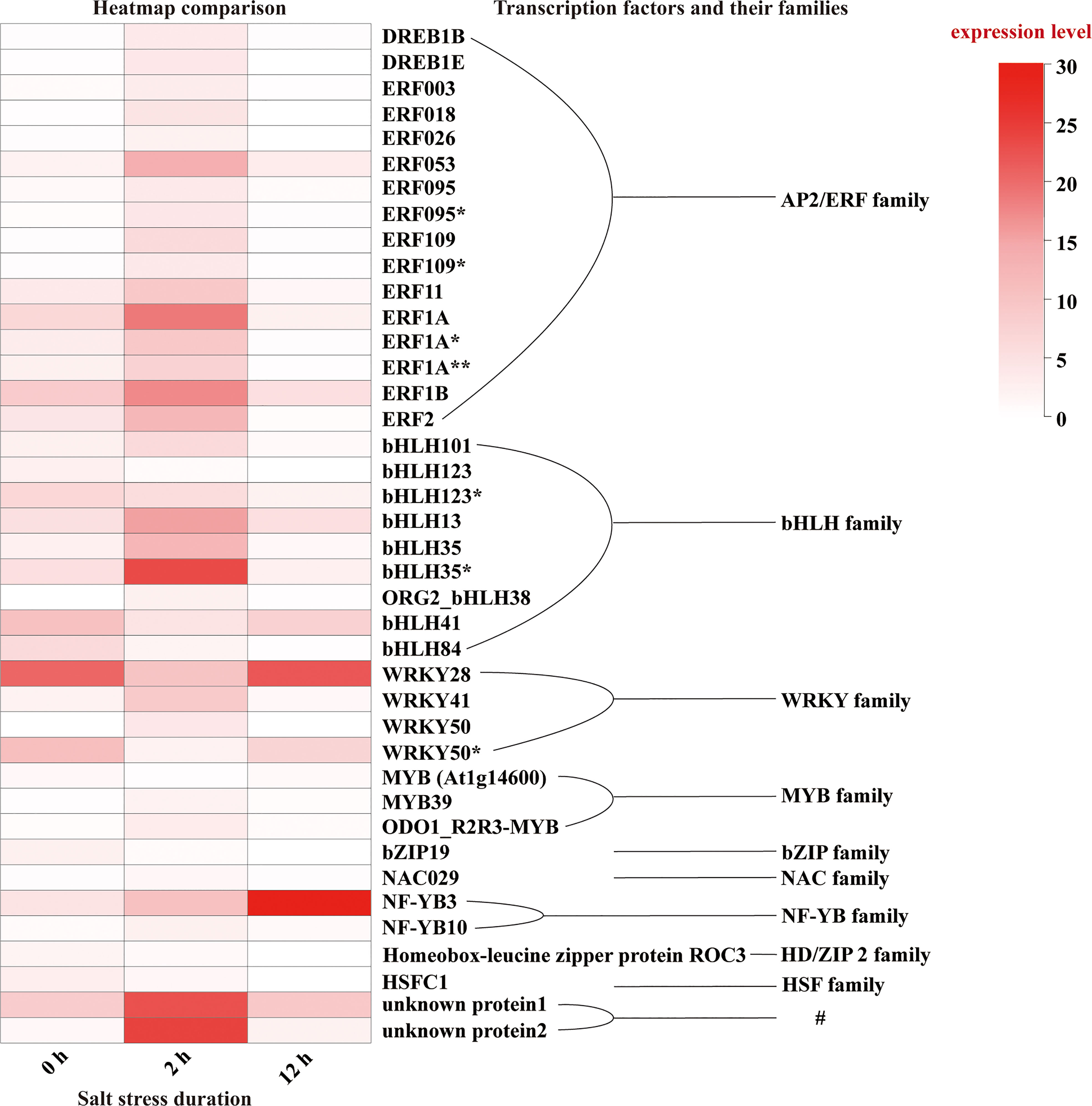
Figure 10 Expression pattern of the transcription factors identified in DEGs. “*” indicates the identical gene ID but different species and “#” means that they have DNA-binding transcription factor activity although they were unknown proteins.
In addition, 48 of the 958 DEGs showed the continuous upregulation or downregulation during entire period (0-12 h), and only 25 genes with annotated information showed temporal differential expression (Figure 11).
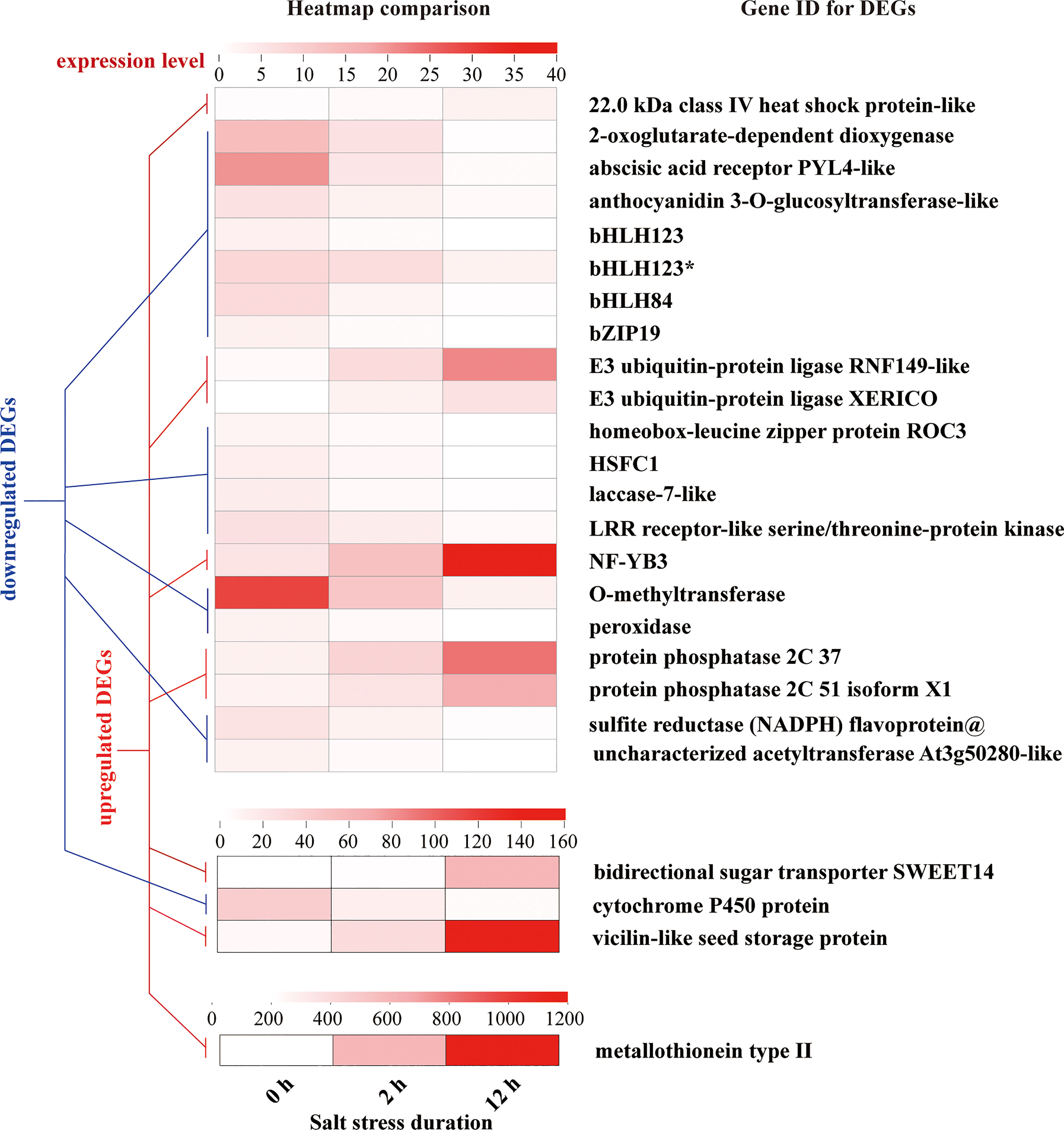
Figure 11 DEGs with the tendency of constant upregulation or downregulation during entire period (0-12 h) of salt stress. Each Gene ID of DEGs represents putative or predicted name based on one discovered in other eukaryotic organisms. “*” indicates the identical gene ID but different species.
We classified these genes into 6 categories: stress responses (heat shock protein, 2-oxoglutarate-dependent dioxygenase, laccase, LRR receptor-like serine/threonine protein kinase, O-methyltransferase, peroxidase, cytochrome P450 protein, metallothionein type 2), hormone signaling (abscisic acid receptor PYL, E3 ubiquitin-protein ligases, protein phosphatases), primary metabolism for energy conservation (sulfite reductase flavoprotein), secondary metabolite production (anthocyanidin 3-O-glucosyltransferase, O-methyltransferase, acetyltransferase, bidirectional sugar transporter SWEET14), seed development (vicilin like seed storage protein), and transcription factors (bHLH123, bHLH84, bZIP19, homeobox-leucine zipper protein ROC3, HSFC1, NF-YB3). In the whole process of salt stress, the expression of bHLH84, bHLH123, bHLH123*, bZIP19, homeobox-leucine zipper protein ROC3, HSFC1 was downregulated, and the expression of NF-YB3 was upregulated 3-6 times. Among other DEGs, metallothionein type 2 protein showed unusually high expression levels and strong upregulation in response to salt stress treatment.
Total 1039 DEGs at 0- 12 h (p< 0.05) were chosen as input data for prediction of their interaction in STRING database. Using Cytoscape, we input the output file of STRING search and obtained a network with 432 DEGs and their 1126 interacting events. We further ranked them and identified putative hub genes using cytohubba plugin (Figure 12).
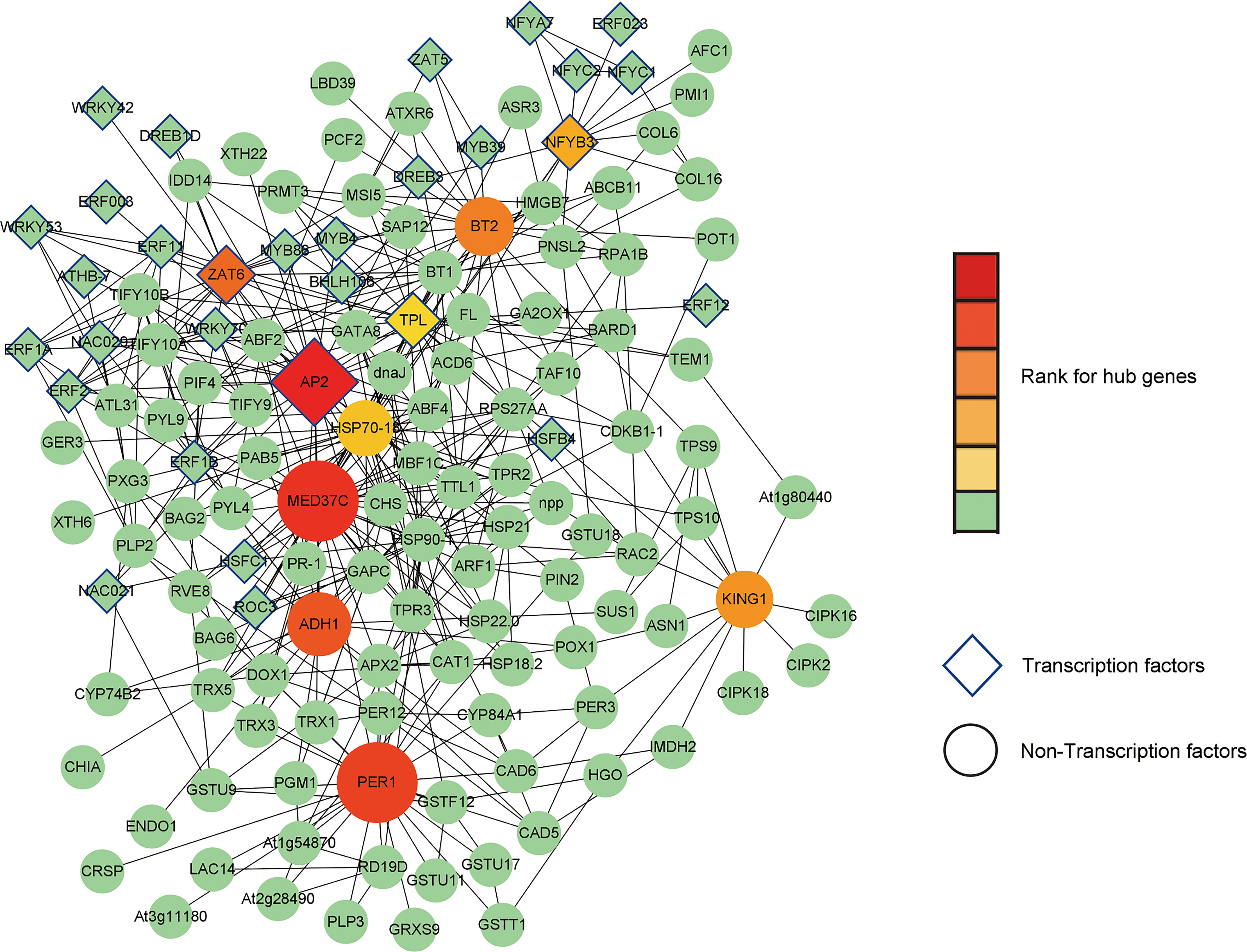
Figure 12 Ten putative hub genes and their interacting proteins. Transcription factors and non-Transcription factors are shaped as diamond and ellipse, respectively. Larger size and darker red color represent higher rank, while green color annotates lower rank.
From co-expression network, we identified ten putative hub genes including AP2 (TRINITY_DN33762_c0_g4), MED37C (TRINITY_DN32748_c0_g1), PER1 (TRINITY_N30585_c1_g4), ADH1 (TRINITY_DN34595_c0_g1), ZAT6 (TRINITY_DN29763_c0_g1), BT2 (TRINITY_DN28127_c0_g3), KING1 (TRINITY_DN32088_c0_g2), NF-YB3 (TRINITY_DN19275_c0_g1), HSP70-18 (TRINITY_DN21721_c1_g4), TPL (TRINITY_DN30465_c0_g4). Among them, TPL and NF-YB3 showed relatively higher log2FC values (7.76 and 2.98, respectively) in our RNA-seq data, suggesting that they play inductive and key roles in the response of lily bulbs to salt stress.
To verify accuracy and reliability of the RNA-seq data, we randomly selected twelve genes and further quantified their expression levels by qRT-PCR. The results showed that the expression patterns of the DEGs determined through qRT-PCR were highly consistent with the RNA-seq data (Figure 13).
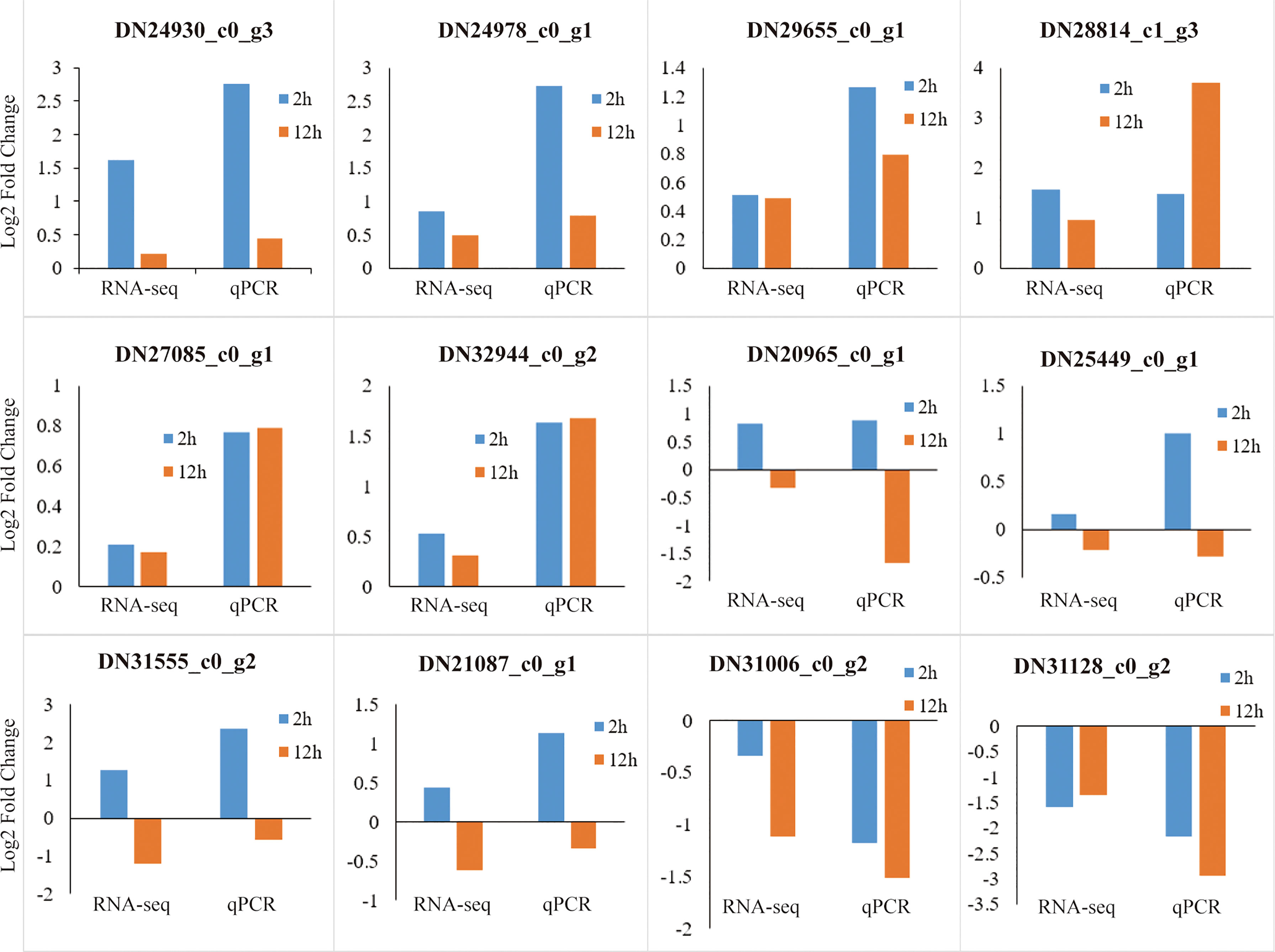
Figure 13 Verification of RNA-seq data through qRT-PCR. The abscissa represents comparison of the DEG expression level at 2 h (blue) and 12 h (red brown) to that at 0 h determined by RNA-seq and qRT-PCR, respectively. The ordinate represents the Log2 Fold Change values of the DEGs.
Salt stress affects plant physiology mainly through ion toxicity, osmotic pressure and oxidative stress. Plants respond to these threats by regulating physiological and biochemical activities to adapt to or resist salt stress (Liang et al., 2018; Zhao et al., 2021). Among them, typical activities are accumulation of osmolyte compounds, ion-selective transmembrane transport and intracellular compartmentalization, ROS scavenging, and upregulation of salt tolerance genes including sensor genes, signaling genes and transcription factors (Shabala et al., 2015; Kumar et al., 2017; Liang et al., 2018; Zhao et al., 2021). Since the roots are the first organs to sense the salt stress (Duan et al., 2014; Karlova et al., 2021; Li et al., 2021), the research on salt stress have focused on the response of roots to salt stress, and then extends to other organs and tissues, mainly leaves and stems. The ornamental bulbous plants such as the lily have unique organs called the bulb that functions as nutrient or flowering organs. L. pumilum is considered to be a salt-tolerant ornamental plant, which may be related to the unique bulb organs function of L. pumilum. But it still lacks evidences showing that the bulb organs help plants overcome stressful environments such as salt stress. Therefore, we analyzed the L. pumilum bulb transcriptome data and identified the key genes regulating the bulb’s unique responses against salt stress at genome-wide scale.
The L. pumilum bulb transcriptome data at different time points (0, 2, or 12 h after salt stress treatment) were obtained by Illumina sequencing of total RNA samples, and analyzed. A total of 51566 unigenes were found and 1772 and 2324 DEGs were identified at initial (0-2 h) and later (2-12 h) periods, respectively. GO and KEGG enrichment analysis for these DEGs indicated that expression patterns of DEGs and KEGG annotated pathways showed slight difference between 0-2 h and 2-12 h. Among the DEGs, 958 DEGs were co-expressed during entire period (0-12 h) of salt stress, and their expression patterns were analyzed. It was found that many unigenes annotated as transcription factors were involved in the responses to salt stress. They belong to AP2/ERF, bHLH, bZIP, HD2/ZIP2, HSF, MYB, NAC, NF-YB and WRKY families. Among them, NF-YB3 transcription factor showed the most intense upregulation throughout entire period of salt stress (Figure 10). Beside the transcription factors, the unigenes annotated as metallothionein type 2 protein, vicilin like seed storage protein and bidirectional sugar transporter SWEET14 showed high expression levels and intense upregulation during entire period of salt stress.
From Figures 8, 9, 11 it is obvious that stress response of the bulb organ was distinct from the typical responses such as ion-selective transmembrane transport and intracellular compartmentalization and ROS scavenging. For instance, in the transcriptome data of bulb organs, a number of genes responsible for responses to oxidative stress were identified as DEGs, 57 of which were predicted to be typical ROS scavengers, such as superoxide dismutase, peroxidase, and glutathione transferase, but most DEGs were downregulated. Some DEGs were upregulated, but their expression level was very low. In the 57 DEGs, 9 DEGs were co-expressed during entire period (both 0-2 h and 2-12 h) of salt stress and their expression patterns was shown in Figure 14. They also showed low expression levels and no intense upregulation. The effects of salt stress on bulb organs were different from those in roots, and bulb organs showed different responses, which may be related to the highly upregulated NF-YB3 transcription factor, metallothionein type 2 protein, vicilin like seed storage protein and bidirectional sugar transporter SWEET14. (Figure 11). NF-YB3 transcription factor is a subunit of the CCAAT-box binding factor family proteins (also called the Nuclear Factor Y). It is one of the large and ubiquitous transcription factors in all eukaryotic organisms including yeast, plant and animal, and plays important roles in flowering promotion, embryogenesis and seed maturation (Kumimoto et al., 2008; Laloum et al., 2013; Myers and III, 2018). NF-YB3 has also been reported to play a role in regulating plant responses to abiotic stresses such as drought, nutrient deficiency, salt, and temperature stress (Zhang et al., 2015; Yang et al., 2017; Sato et al., 2019; Dai et al., 2021; Zheng et al., 2021). In this study, we found that the expression of NF-YB3 transcription factor was upregulated in bulb organs under salt stress, suggesting that it was involved in regulation of salt stress. Co-expression network analysis also confirmed that NF-YB3 acted as a hub gene during salt stress (Figure 12). NF-YB3 is known to regulate not only the salt tolerance, but also the flowering time (Hou et al., 2014; Zhang et al., 2021). Thus, upregulation of NF-YB3 in lily bulb organs upon salt stress can promote flowering of L. pumilum, but it requires further efforts to elucidate whether salt stress eventually promotes flowering in lily bulb.
Meanwhile, metallothionein type 2 protein, vicilin like seed storage protein were intensely upregulated. Metallothionein type 2 protein is a well-known metal ion binding protein that is characterized by high cysteine residue (Coyle et al., 2002). It plays significant roles in the detoxification and homeostasis of metal ions and ROS scavenging (Joshi et al., 2015; Jin et al., 2017; Babaei-Bondarti and Shahpiri, 2020), and these roles have already been demonstrated in L. pumilum (Wang et al., 2019). Metallothionein proteins are known to be regulated by several transcription factors such as the TFIID complex comprising TATA-binding protein associated factors, metal−responsive element (MRE)-binding transcription factor (MTF)-1, NF-Y family transcription factors (Takahashi, 2015). Vicilin like seed storage protein is a seed-specific protein that acts against biotic stress factors such as fungus and microbes (Vieira Bard et al., 2014; Jimenez Lopez, 2020; Ateeq et al., 2022; Bertonceli et al., 2022). Vicilin is also known to have the superoxide dismutase activity (Shikhi et al., 2018), which is able to resist against oxidative stress by scavenging ROS. Therefore, the “cavity” in anti-oxidation due to low activity of typical ROS scavengers can be filled by abundant metallothionein proteins and vicilin like seed storage proteins. The bidirectional sugar transporter SWEET14 was also the DEG with intense upregulation (Figure 11). SWEET protein was first discovered in plants as sugar transporters that mediate transmembrane sugar transport and is known to play diverse physiological roles in flower, fruit, seed development, gibberellin transportation, and apoplasmic phloem loading (Jeena et al., 2019; Ji et al., 2022). Sugars also can act as osmolytes that resist against osmotic imbalance during salt stress (Darko et al., 2019; Jeena et al., 2019) and upregulation of SWEET14 might consequently contribute to accumulation of sugars, thereby resisting osmotic damage from salt stress in this work.
Besides, E3-ubiquitin protein ligases were significantly upregulated during salt stress (Figure 11). E3-ubiquitin ligase mediated protein ubiquitination is one of the main post-translation modification pathways for protein which regulates various intracellular processes such as hormone signaling and stress resistance (Duplan and Rivas, 2014; Shu and Yang, 2017; Mandal et al., 2018; Kelley, 2018). The E3-ubiquitin ligases are known to regulate salt stress by participating in SOS pathway, MAPK Cascade, ABA signaling pathway, flowering pathway and ROS homeostasis (Wang et al., 2022). In our study, MAPK pathway was significantly activated during salt stress (Figure 9) and this might be associated with upregulation of E3-ubiquitin ligases. The enzymes also serve as central regulators of perception, regulation and biosynthesis of hormones such as auxin, brassinosteroid, cytokinin, ethylene, gibberellic acid, jasmonate, salicylic acid, and strigolactone (Kelley, 2018). In addition, PYL4 and protein phosphatase 2C are the major proteins in ABA signaling pathway; ABA inhibits PP2C through PYL4 (an ABA receptor protein), thereby releases SnRKs which then regulate energy homeostasis by promoting catabolism and repressing anabolism, thus actively inhibiting plant growth (Park et al., 2009; Zhang et al., 2020). Our study reported the downregulation of PYL4 and upregulation of protein phosphatase (Figure 11) which might lead to inactivation of ABA signaling, eventually relieving growth inhibition effect of ABA.
Bulb organs are different from roots that can directly sense and respond to salt stress, so the response of the bulb to salt stress would indirect and not exactly the same as the responses of root to salt stress. Substantiating this, our work provided the evidences that revealed bulb organ-unique responses against salt stress in L. pumilum, which are different from the previous results in roots or leaves. Altogether, lily bulb organs showed obvious salt tolerance that was evidenced by upregulation of non-typical ROS scavengers (such as the metallothionein type 2 protein) and osmolyte accumulation-related proteins (vicilin like seed storage protein, bidirectional sugar transporter SWEET14). Besides, phytohormone signaling factors (E3-ubiquitin protein ligases and protein phosphatase 2C) were upregulated, suggesting that salt tolerance of the lily plants requires hormone signaling. Among the DEG transcription factors including DREB, ERF, bHLH, bZIP, HD2/ZIP2, HSF, MYB, NAC, NF-YB, and WRKY (Figure 10), NF-YB3 was the only factor that showed full period upregulation. Co-expression analysis further confirmed that NF-YB3 is a putative hub factor that regulates lily bulb response against salt stress. NF-YB3 is a well-known transcription factor that promotes flowering in plants and Lily bulb is a reproductive organ of L. pumilum. Therefore, upregulation of NF-YB3 in the bulbs under salt stress can promote flowering of lily. Further researches are still required to unveil which upstream signaling factors induce metallothionein type 2 protein, vicilin like seed storage protein, bidirectional sugar transporter SWEET14, E3-ubiquitin protein ligases and protein phosphatase 2C, as well as which downstream genes of NF-YB3 transcription factor are induced during salt stress to govern NF-YB3-mediated salt stress responses or flowering signals in L. pumilum.
As a wildflower plant with high salt-alkali tolerance, L. pumilum is regarded as an important germplasm resource for the lily stress resistance breeding in achieving efficient gardening. The bulbs are the important organs for lily propagation and other uses; flowers are developed from them and more importantly, they can be used for culinary and medicinal purposes. Besides, these unique bulb organs are believed to play significant roles in regulating high salt-alkali tolerance of L. pumilum, albeit lacking evidences.
Our work for the first time revealed that bulb organs of L. pumilum respond significantly against salt stress, which is different from root organs. Through transcriptome analysis, we identified DEGs associated with salt stress and then screened several key genes with intense upregulation degree. They include NF-YB3 transcription factor, metallothionein type 2 protein, vicilin like seed storage protein and bidirectional sugar transporter SWEET14. It has been commonly accepted that stress responses mainly lead to increase in ROS level and always include upregulation of typical ROS scavengers like superoxide dismutase, peroxidase, and glutathione transferase, however, they were not differentially expressed and rather the non-typical ROS scavengers such as the metallothionein type 2 protein, and vicilin like seed storage protein were upregulated in our work. Imbalance in osmotic pressure caused by salt stress is another type of damage and plants normally respond with the accumulation of osmolytes such as sucrose. Upregulation of the bidirectional sugar transporter SWEET14 protein reported in this study may promote the sucrose accumulation favorable for maintaining osmotic homeostasis under salt stress. We also reported that the hormone signaling proteins such as E3-ubiquitin protein ligases, PYL4 and protein phosphatase 2C were upregulated, suggesting the role of hormones in the bulb organ responses to salt stress. Besides, co-expression analysis of the DEGs confirmed that NF-YB3 transcription factor acted as a hub gene. Overall, these results lay a foundation for further research on the response mechanism of bulb to salt stress in L. pumilum under salt stress, which has important reference value for studying the molecular mechanism of regulation network of lily coping with different abiotic environmental stress, and provide key genes information for tolerance breeding of lily.
qRT-PCR Primer information was shown in Additional file.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA851552.
KS, UP, and SS conceived and designed the experiments. KS and SS conducted the experiments. KS, UP, YW, and HY elaborated and analyzed experimental data, YZ, KS, and UP wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Fundamental Research Funds for the Central Universities [grant numbers 2572019BK04]; and the Natural Science Foundation of Heilongjiang Province [grant numbers LH2019C004].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1054064/full#supplementary-material
Ateeq, M., Adeel, M. M., Kanwal, A., Tahir Ul Qamar, M., Saeed, A., Khaliq, B., et al. (2022). In silico analysis and functional characterization of antimicrobial and insecticidal vicilin from moth bean (vigna aconitifolia (jacq.) marechal) seeds. Molecules 27, 3251. doi: 10.3390/molecules27103251
Babaei-Bondarti, Z., Shahpiri, A. (2020). A metallothionein type 2 from avicennia marina binds to iron and mediates hydrogen peroxide balance by activation of enzyme catalase. Phytochemistry 176, 112396. doi: 10.1016/j.phytochem.2020.112396
Bertonceli, M. A. A., Oliveira, A. E. A., Ferreira, A. T. S., Perales, J., Fernandes, K. V. S. (2022). A vicilin-like protein extracted from clitoria fairchildiana cotyledons was toxic to callosobruchus maculatus (coleoptera: chrysomelidae). Pesticide Biochem. Physiol. 184, 105129. doi: 10.1016/j.pestbp.2022.105129
Blumwald, E. (2000). Sodium transport and salt tolerance in plants. Curr. Opin. Cell Biol. 12, 431–434. doi: 10.1016/S0955-0674(00)00112-5
Buchfink, B., Xie, C., Huson, D. H. (2015). Fast and sensitive protein alignment using diamond. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Coyle, P., Philcox, J. C., Carey, L. C., Rofe, A. M. (2002). Metallothionein: the multipurpose protein. Cell Mol. Life Sci. 59, 627–647. doi: 10.1007/s00018-002-8454-2
Dai, J., Hu, A., Zhang, J., Liao, W., Ma, H., Wu, J.-Z., et al. (2021). Nf-yb-mediated active responses of plant growth under salt and temperature stress in eucalyptus grandis. Plants 10, 1107. doi: 10.3390/plants10061107
Darko, E., Végh, B., Khalil, R., Marček, T., Szalai, G., Pál, M, et al. (2019). Metabolic responses of wheat seedlings to osmotic stress induced by various osmolytes under iso-osmotic conditions. PloS One 14, e226151. doi: 10.1371/journal.pone.0226151
Deinlein, U., Stephan, A. B., Horie, T., Luo, W., Xu, G., Schroeder, J. I., et al. (2014). Plant salt-tolerance mechanisms. Trends Plant Sci. 19, 371–379. doi: 10.1016/j.tplants.2014.02.001
Duan, L., Sebastian, J., Dinneny, J. R. (2014). “Salt-stress regulation of root system growth and architecture in arabidopsis seedlings,”. Plant cell expansion methods and protocols. Ed. Estevez, J. M. (New York, NY: Humana Press) 105–122.
Duplan, V., Rivas, S. (2014). E3 ubiquitin-ligases and their target proteins during the regulation of plant innate immunity. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00042
Flowers, T. J., Colmer, T. D. (2015). Plant salt tolerance: adaptations in halophytes. Ann. Bot. 115, 327–331. doi: 10.1093/aob/mcu267
Flowers, T. J., Yeo, A. R. (1995). Breeding for salinity resistance in crop plants: where next? Aust. J. Plant Physiol. 22, 875–884. doi: 10.1071/PP9950875
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I, et al. (2011). Full-length transcriptome assembly from rna-seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Hou, X., Zhou, J., Liu, C., Liu, L., Shen, L., Yu, H., et al. (2014). Nuclear factor y-mediated h3k27me3 demethylation of the soc1 locus orchestrates flowering responses of arabidopsis. Nat. Commun. 5, 4601. doi: 10.1038/ncomms5601
Imadi, S. R., Kazi, A. G., Ahanger, M. A., Gucel, S., Ahmad, P. (2015). Plant transcriptomics and responses to environmental stress: an overview. J. Genet. 94, 525–537. doi: 10.1007/s12041-015-0545-6
Jeena, G. S., Kumar, S., Shukla, R. K. (2019). Structure, evolution and diverse physiological roles of sweet sugar transporters in plants. Plant Mol. Biol. 100, 351–365. doi: 10.1007/s11103-019-00872-4
Jimenez Lopez, J. C. (2020). Narrow-leafed lupin (lupinus angustifolius l.) β-conglutin: a multifunctional family of proteins with roles in plant defence, human health benefits, and potential uses as functional food. Legume Sci. 2, e33. doi: 10.1002/leg3.33
Jin, S., Xu, C., Li, G., Sun, D., Li, Y., Wang, X, et al. (2017). Functional characterization of a type 2 metallothionein gene, ssmt2, from alkaline-tolerant suaeda salsa. Sci. Rep. 7, 17194. doi: 10.1038/s41598-017-18263-4
Ji, J., Yang, L., Fang, Z., Zhang, Y., Zhuang, M., Lv, H, et al. (2022). Plant sweet family of sugar transporters: structure, evolution and biological functions. Biomolecules 12, 205. doi: 10.3390/biom12020205
Joshi, R., Pareek, A., Singla-Pareek, S. L. (2015). “Plant metallothioneins: classification, distribution, function, and regulation,”. Plant metal interaction emerging remediation techniques Ed. Ahmad, P. (Elsevier Inc) 239–261.
Ju, Y., Min, Z., Zhang, Y., Zhang, K., Liu, M., Fang, Y-l, et al. (2021). Transcriptome profiling provide new insights into the molecular mechanism of grapevine response to heat, drought, and combined stress. Scientia Hortic. 286, 110076. doi: 10.1016/j.scienta.2021.110076
Karlova, R., Boer, D., Hayes, S., Testerink, C. (2021). Root plasticity under abiotic stress. Plant Physiol. 187, 1057–1070. doi: 10.1093/plphys/kiab392
Kelley, D. R. (2018). E3 ubiquitin ligases: key regulators of hormone signaling in plants. Mol. Cell. Proteomics 17, 1047–1054. doi: 10.1074/mcp.MR117.000476
Kukurba, K. R., Montgomery, S. B. (2015). Rna sequencing and analysis. Cold Spring Harbor Protoc. 2015, p84970. doi: 10.1101/pdb.top084970
Kumar, J., Singh, S., Singh, M., Srivastava, P. K., Mishra, R. K., Singh, V. P., et al. (2017). Transcriptional regulation of salinity stress in plants: a short review. Plant Gene 11, 160–169. doi: 10.1016/j.plgene.2017.04.001
Kumimoto, R. W., Adam, L., Hymus, G. J., Repetti, P. P., Reuber, T. L., Marion, C. M., et al. (2008). The nuclear factor y subunits nf-yb2 and nf-yb3 play additive roles in the promotion of flowering by inductive long-day photoperiods in arabidopsis. Planta 228, 709–723. doi: 10.1007/s00425-008-0773-6
Laloum, T., De Mita, S., Gamas, P., Baudin, M., Niebel, A. (2013). Ccaat-box binding transcription factors in plants: y so many? Trends Plant Sci. 18, 157–166. doi: 10.1016/j.tplants.2012.07.004
Liang, W., Ma, X., Wan, P., Liu, L. (2018). Plant salt-tolerance mechanism: a review. Biochem. Biophys. Res. Commun. 495, 286–291. doi: 10.1016/j.bbrc.2017.11.043
Li, P., Yang, X., Wang, H., Pan, T., Wang, Y., Xu, Y., et al. (2021). Genetic control of root plasticity in response to salt stress in maize. Theor. Appl. Genet. 134, 1475–1492. doi: 10.1007/s00122-021-03784-4
Mandal, A., Sharma, N., Muthamilarasan, M., Prasad, M. (2018). Ubiquitination: a tool for plant adaptation to changing environments. Nucleus 61, 253–260. doi: 10.1007/s13237-018-0255-6
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput seqeuncing reads. Embnet J. 17, 1–3. doi: 10.14806/ej.17.1.200
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by rna-seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Munns, R., Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Myers, Z. A., III, B. F. H. (2018). Nuclear factor-y: still complex after all these years? Curr. Opin. Plant Biol. 45, 96–102. doi: 10.1016/j.clinmicnews.2015.01.008
Park, S., Fung, P., Nishimura, N., Jensen, D. R., Fujii, H., Zhao, Y., et al. (2009). Abscisic acid inhibits type 2c protein phosphatases via the pyr/pyl family of start proteins. Science 324, 1068–1071. doi: 10.1126/science.1173041
Robinson, M. D., McCarthy, D. J., Smyth, G. K. (2009). Edger: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Sahi, C., Singh, A., Blumwald, E., Grover, A. (2006). Beyond osmolytes and transporters: novel plant salt-stress tolerance-related genes from transcriptional profiling data. Physiol Plantarum 127, 1–9. doi: 10.1111/j.1399-3054.2005.00610.x
Sato, H., Suzuki, T., Takahashi, F., Shinozaki, K., Yamaguchi-Shinozaki, K. (2019). Nf-yb2 and nf-yb3 have functionally diverged and differentially induce drought and heat stress-specific genes. Plant Physiol. 180, 1677–1690. doi: 10.1104/pp.19.00391
Shabala, S., Wu, H., Bose, J. (2015). Salt stress sensing and early signalling events in plant roots: current knowledge and hypothesis. Plant Sci. 241, 109–119. doi: 10.1016/j.plantsci.2015.10.003
Shikhi, M., Nair, D. T., Salunke, D. M. (2018). Structure-guided identification of function: role of capsicum annuum vicilin during oxidative stress. Biochem. J. 475, 3057–3071. doi: 10.1042/BCJ20180520
Shrivastava, P., Kumar, R. (2015). Soil salinity: a serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 22, 123–131. doi: 10.1016/j.sjbs.2014.12.001
Shu, K., Yang, W. (2017). E3 ubiquitin ligases: ubiquitous actors in plant development and abiotic stress responses. Plant Cell Physiol. 58, 1461–1476. doi: 10.1093/pcp/pcx071
Sinha, A. K., Jaggi, M., Raghuram, B., Tuteja, N. (2014). Mitogen-activated protein kinase signaling in plants under abiotic stress. Plant Signaling Behav. 6, 196–203. doi: 10.4161/psb.6.2.14701
Takahashi, S. (2015). Positive and negative regulators of the metallothionein gene (review). Mol. Med. Rep. 12, 795–799. doi: 10.3892/mmr.2015.3459
Tester, M., Davenport, R. (2003). Na+ tolerance and na+ transport in higher plants. Ann. Bot. 91, 503–527. doi: 10.1093/aob/mcg058
Vieira Bard, G. C., Nascimento, V. V., Oliveira, A. E. A., Rodrigues, R., Da Cunha, M., Dias, G. B., et al. (2014). Vicilin-like peptides from capsicum baccatum l. seeds are α-amylase inhibitors and exhibit antifungal activity against important yeasts in medical mycology. Biopolymers 102, 335–343. doi: 10.1002/bip.22504
Wang, Y., Cao, S., Guan, C., Kong, X., Wang, Y., Cui, Y., et al. (2020). Overexpressing the nac transcription factor lpnac13 from lilium pumilum in tobacco negatively regulates the drought response and positively regulates the salt response. Plant Physiol. Biochem. 149, 96–110. doi: 10.1016/j.plaphy.2020.01.036
Wang, Y., Cui, Y., Liu, B., Wang, Y., Sun, S., Wang, J., et al. (2022). Lilium pumilum stress-responsive nac transcription factor lpnac17 enhances salt stress tolerance in tobacco. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.993841
Wang, L., Fan, L., Zhao, J., Xu, C., Jin, S. (2019). Abiotic stress resistance analysis of lilium pumilum overexpressing the lpmt2 gene. J. Agric. Sci. 11, 1–12. doi: 10.5539/jas.v11n18p76
Wang, S., Lv, X., Zhang, J., Chen, D., Chen, S., Fan, G., et al. (2022). Roles of e3 ubiquitin ligases in plant responses to abiotic stresses. Int. J. Mol. Sci. 23, 2308. doi: 10.3390/ijms23042308
Wang, W., Su, X., Tian, Z., Liu, Y., Zhou, Y., He, M., et al. (2018). Transcriptome profiling provides insights into dormancy release during cold storage of lilium pumilum. BMC Genomics 19, 196. doi: 10.1186/s12864-018-4536-x
Xin, Y., Pan, W., Chen, X., Liu, Y., Zhang, M., Chen, X., et al. (2021). Transcriptome profiling reveals key genes in regulation of the tepal trichome development in lilium pumilum d.c. Plant Cell Rep. 40, 1889–1906. doi: 10.1007/s00299-021-02753-x
Yang, M., Zhao, Y., Shi, S., Du, X., Gu, J., Xiao, K., et al. (2017). Wheat nuclear factor y (nf-y) b subfamily gene tanf-yb3;l confers critical drought tolerance through modulation of the aba-associated signaling pathway. Plant Cell Tissue Organ Culture (Pctoc) 128, 97–111. doi: 10.1007/s11240-016-1088-0
Yan, H., Liu, B., Cui, Y., Wang, Y., Sun, S., Wang, J., et al. (2022). Lpnac6 reversely regulates the alkali tolerance and drought tolerance of lilium pumilum. J. Plant Physiol. 270, 153635. doi: 10.1016/j.jplph.2022.153635
Yong, Y., Zhang, Y., Lyu, Y. (2019). A stress-responsive nac transcription factor from tiger lily (llnac2) interacts with lldreb1 and llzhfd4 and enhances various abiotic stress tolerance in arabidopsis. Int. J. Mol. Sci. 20, 3225. doi: 10.3390/ijms20133225
Yu, Z., Duan, X., Luo, L., Dai, S., Ding, Z., Xia, G., et al. (2020). How plant hormones mediate salt stress responses. Trends Plant Sci. 25, 1117–1130. doi: 10.1016/j.tplants.2020.06.008
Zhang, P., Li, X., Wang, Y., Guo, W., Chachar, S., Riaz, A., et al. (2021). Prmt6 physically associates with nuclear factor y to regulate photoperiodic flowering in arabidopsis. Abiotech 2, 403–414. doi: 10.1007/s42994-021-00065-y
Zhang, T., Zhang, D., Liu, Y., Luo, C., Zhou, Y., Zhang, L., et al. (2015). Overexpression of a nf-yb3 transcription factor from picea wilsonii confers tolerance to salinity and drought stress in transformed arabidopsis thaliana. Plant Physiol. Biochem. 94, 153–164. doi: 10.1016/j.plaphy.2015.05.001
Zhang, H., Zhao, Y., Zhu, J. K. (2020). Thriving under stress: how plants balance growth and the stress response. Dev. Cell 55, 529–543. doi: 10.1016/j.devcel.2020.10.012
Zhao, S., Zhang, Q., Liu, M., Zhou, H., Ma, C., Wang, P., et al. (2021). Regulation of plant responses to salt stress. Int. J. Mol. Sci. 22, 4609. doi: 10.3390/ijms22094609
Zheng, X., Zhang, H., Zhang, L., Xu, F., Shi, L., Wang, S., et al. (2021). Identification and comprehensive analysis of the nuclear factor-y family genes reveal their multiple roles in response to nutrient deficiencies in brassica napus. Int. J. Mol. Sci. 22, 10354. doi: 10.3390/ijms221910354
Zhou, J., An, R., Huang, X. (2021). Genus lilium: a review on traditional uses, phytochemistry and pharmacology. J. Ethnopharmacol 270, 113852. doi: 10.1016/j.jep.2021.113852
Keywords: Lilium pumilum, salt stress, bulb organ, transcriptome analysis, ornamental plant
Citation: So K, Pak U, Sun S, Wang Y, Yan H and Zhang Y (2022) Transcriptome profiling revealed salt stress-responsive genes in Lilium pumilum bulbs. Front. Plant Sci. 13:1054064. doi: 10.3389/fpls.2022.1054064
Received: 26 September 2022; Accepted: 21 October 2022;
Published: 09 November 2022.
Edited by:
Awadhesh Kumar, National Rice Research Institute (ICAR), IndiaReviewed by:
S. Prashant Jeevan Kumar, Directorate of Floricultural Research (ICAR), IndiaCopyright © 2022 So, Pak, Sun, Wang, Yan and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanni Zhang, dGRjcXRnenlAMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.