- 1Department of Biotechnology, Guru Nanak Dev University, Amritsar, India
- 2Department of Bioinformatics, Hans Raj Mahila Maha Vidyalaya, Jalandhar, India
- 3Biotechnology Division, Institute of Himalayan Bioresource Technology, Council of Scientific and Industrial Research, Palampur, HP, India
Peptidyl-prolyl cis-trans isomerases (PPIases) are ubiquitous proteins which are essential for cis-trans isomerisation of peptide bonds preceding the proline residue. PPIases are categorized into four sub-families viz., cyclophilins, FK506-binding proteins (FKBPs), parvulins and protein phosphatase 2A phosphatase activators (PTPAs). Apart from catalysing the cis-trans isomerization, these proteins have also been implicated in diverse cellular functions. Though PPIases have been identified in several important crop plants, information on these proteins, except cyclophilins, is scanty in wheat. In order to understand the role of these genes in wheat, we carried out genome-wide identification using computational approaches. The present study resulted in identification of 71 FKBP (TaFKBP) 12 parvulin (TaPar) and 3 PTPA (TaPTPA) genes in hexaploid wheat genome, which are distributed on different chromosomes with uneven gene densities. The TaFKBP and TaPar proteins, besides PPIase domain, also contain additional domains, indicating functional diversification. In silico prediction also revealed that TaFKBPs are localized to ER, nucleus, chloroplast and cytoplasm, while the TaPars are confined to cytoplasm and nucleus. The TaPTPAs, on the contrary, appear to be present only in the cytoplasm. Evolutionary studies predicted that most of the TaFKBP, TaPar and TaPTPA genes in hexaploid wheat have been derived from their progenitor species, with some events of loss or gain. Syntenic analysis revealed the presence of many collinear blocks of TaFKBP genes in wheat and its sub-genome donors. qRT-PCR analysis demonstrated that expression of TaFKBP and TaPar genes is regulated differentially by heat stress, suggesting their likely involvement in thermotolerance. The findings of this study will provide basis for further functional characterization of these genes and their likely applications in crop improvement.
Introduction
The plants cope up with different biotic and abiotic stresses through adjustments at physiological, biochemical and molecular levels, that include the production of antioxidative enzymes, accumulation of compatible solutes, signaling molecules, chaperones, etc. (Wang et al., 2003). The peptidyl-prolyl cis-trans isomerases (PPIases) are yet another class of proteins that have also been reported to play an important role in stress adaptation of plants (Sharma and Singh, 2003; Wang et al., 2003; Singh et al., 2019; Singh H et al., 2020; Thirumalaikumar et al., 2021). The PPIases are the only enzymes known that can catalyze the cis-trans transition of peptidyl-prolyl bonds. Though the peptide bonds in proteins occur in trans conformation, about 6.5% of the peptidyl-prolyl bonds occur in cis state due to the five-membered ring structure of prolyl residue (Galat and Rivière, 1998). The cis-trans transition of the peptidyl-prolyl bond is a rate-limiting step in correct folding of proteins and, hence, requires intervention of PPIases (Fischer et al., 1989) which are typical enzymes and follow the Michaelis-Menton kinetics. Depending upon their sensitivity towards immunosuppressive drugs, the PPIases are categorised into two sub-families, immunophilins and non-immunophilins. While the immunophilins comprise of cyclosporin A (CsA)-binding proteins, cyclophilins, and FK506-binding proteins, FKBPs, the non-immunophilin PPIases include structurally distinct parvulins and phospho-tyrosyl phosphatase activators (PTPAs), that are not sensitive to any of these immunosuppressive agents (Rahfeld et al., 1994). The cyclophilins and FKBPs, characterized by the presence of cyclophilin-like domain (CLD) and FK506-binding domain (FKBD), respectively, are ubiquitous proteins and have been reported in almost all organisms ranging from microbes to plants and animals (Göthel and Marahiel, 1999; Singh H et al., 2020; Yadav et al., 2022; Zhou et al., 2022). These proteins are localized in different sub-cellular organelles, viz., mitochondria, endoplasmic reticulum, cytosol and nucleus (Breiman et al., 1992; Luan et al., 1996; Xu et al., 1998; Carol et al., 2001). The cyclophilins and FKBPs are encoded by large gene families in plants. Genome-wide computational analysis revealed the presence of 21, 23, 24, 29, 30 and 38 different FKBPs in Prunus persica, Arabidopsis thaliana, Solanum lycopersicum, Oryza sativa, Zea mays, and Malus domestica, respectively (He et al., 2004; Gollan and Bhave, 2010; Wang W et al., 2012; Zhang Y et al., 2014; Dong et al., 2018; Waseem et al., 2018). Members of FKBPs have also been characterized in different fungal species such as Aspergillus spp. and Penicillium spp. (Joseph et al., 1999; Singh et al., 2021). Likewise, 28, 29, 30, 33, 38-78, 62, 83 and 91 cyclophilins have been reported in O. sativa, A. thaliana, M. domestica, Medicago truncatula, Gossypium spp., soybean, Triticum aestivum and Brassica napus, respectively (Gasser et al., 1990; Romano et al., 2004; Trivedi et al., 2012; Mainali et al., 2014; Hanhart et al., 2017; Chen et al., 2019; Singh et al., 2019; Ge et al., 2020; Wang et al., 2022). Contrary to the cyclophilins and FKBPs, the PTPAs and parvulins are less abundant and only one or two members have been reported in human, yeast and Penicillium spp. (Rahfeld et al., 1994; Lu et al., 1996; Jordens et al., 2006; Magnusdottir et al., 2006; Mueller et al., 2006; Singh et al., 2021).
In mammals, the cyclophilins and FKBPs are involved in immunosuppression, as interaction of these proteins with CsA and FK506, respectively, prevent transcription of genes encoding interleukin-2 (Liu et al., 1991; O'Keefe et al., 1992), leading to suppression of immune response. The immunophilins also regulate important cellular processes in plants, with the roles of cyclophilins discussed extensively in a recent review (Singh H et al., 2020 and references therein). Recently, an Arabidopsis cyclophilin, AtCYP18-1, was reported to play an important role in splicing of introns that are retained in response to heat stress during germination (Jo et al., 2022). Cyclophilin ROC3 was implicated in regulation of ABA-induced stomatal closure and drought stress response in Arabidopsis (Liu et al., 2021). Similarly, ectopic expression of a pigeonpea cyclophilin (CcCYP) was shown to confer tolerance against multiple abiotic stresses in transgenic rice (Juturu et al., 2021). The FKBPs too have been shown to perform diverse cellular functions viz., signal transduction (Gopalan et al., 2004), transcription (Kang et al., 2008), assembly of multiprotein complexes (Geisler and Bailly, 2007; Nigam et al., 2008; Boudko et al., 2014). protein trafficking (Luan et al., 1994), apoptosis (Meiri et al., 2010), fertility (Yu et al., 2012), biosynthesis of long chain fatty acids (Harrar et al., 2003), regulation of gene expression (Li and Luan, 2010), photosynthetic membrane assembly in plants (Lima et al., 2006) and as histone chaperones (Singh A et al., 2020). Several FKBPs also exhibit chaperonic activity that is independent of PPIase activity. For example, one of the wheat FKBPs, FKBP73, exihbited chaperonic function despite abrogation of its PPIase activity (Kurek et al., 2002). Expression of FKBP genes, FKBP62 and FKBP65, was enhanced in response to wounding, NaCl and malondialdehyde treatment in Arabidopsis (Vucich and Gasser, 1996; Weber et al., 2004; Aviezer-Hagai et al., 2007; Meiri and Breiman, 2009). Yu et al. (2017) demonstrated that constitutive expression of a Z. mays FKBP gene, ZmFKBP20-1, resulted in higher tolerance to drought and salt stress in the transgenic Arabidopsis plants. The role of a rice FKBP, OsFKBP20-1b, in stress adaptation was attributed to regulation of RNA processing through its interaction with a splicing factor OsSR45 (Park et al., 2020).
It is, thus, evident that different PPIases play important roles in growth and development processes of plants as well as in adaptation to different abiotic stresses. Earlier studies by our group demonstrated that the cyclophilin family in wheat consists of 83 genes, several of which are modulated under heat stress (Singh et al., 2019). However, except for cyclophilins (Singh et al., 2019), information on PPIases belonging to FKBPs, parvulins and PTPAs is scanty in this important crop plant. Therefore, the present study was undertaken to carry out in silico identification and characterization of FKBPs, parvulins and PTPAs in wheat and to study their regulation by thermal stress. The results of this study revealed that the wheat genome encodes for 71 FKBPs, 12 parvulins, and 3 PTPAs, and expression of several of these genes is modulated differentially by heat stress. These findings will provide impetus for further functional characterization of these gene families, and their likely applications in crop improvement through biotechnological and breeding strategies.
Materials and methods
Identification and retrieval of FKBP, parvulin and PTPA gene sequences
The different FKBPs, parvulins, and PTPAs encoded by the wheat genome were identified by using the coding (CDS) and protein sequences of different organisms for homology-based search. For FKBPs, sequences from O. sativa, A. thaliana, P. persica and M. domestica; for PTPAs, sequences from Drosophila melanogaster, Homo sapiens, Saccharomyces cerevisiae and Saccharomyces pombe; and for parvulins, sequences of A. thaliana, H. sapiens, S. cerevisiae and Escherichia coli were employed for this study. The CDS and protein sequences for different organisms were obtained from their respective databases (UniProt; Universal Protein knowledgebase; https://www.uniprot.org/, NCBI; National Centre for Biotechnology Information; https://www.ncbi.nlm.nih.gov/, RGAP; Rice Genome Annotation Project; http://rice.plantbiology.msu.edu/analyses_search_locus.shtml, TAIR; The Arabidopsis Information Resource; https://www.arabidopsis.org/, GDR; Genome Database for Rosaceae; https://www.rosaceae.org/retrieve/sequences) and from other publicly available sources (Supplementary Table S1). BLASTN and BLASTP (Ensembl Plants; https://plants.ensembl.org) programs were used for homology search with an e-value cut off of 10 and 1, respectively. The redundant sequences having the same transcript IDs were removed from the dataset. The matches based on score, coverage and percent identity were retrieved and selected for further analysis. For FKBPs, the retrieved protein sequences were used to build a Hidden Markov model (HMM; HMMER software package; Eddy, 2011). This model was used as a query for running HMM search to obtain more distantly homologous FKBPs from the wheat proteome (IWGSC RefSeq v2.0). Ensembl plants (https://plants.ensembl.org/Triticum_aestivum/Info/Index) was used to download the genomic sequences, CDS, untranslated regions (UTRs), introns, exons, cDNA as well as protein sequences of the corresponding matches.
Conserved domain search and subcellular localization prediction
Domain analysis was performed using PFAM (http://pfam.xfam.org; Finn et al., 2016), Prosite (https://prosite.expasy.org/; Sigrist et al., 2012) and conserved domain database of NCBI (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Only those members which displayed the presence of typical functional domain of their respective PPIase family were subjected to additional characterization and investigation. Graphical representation of the domains and motifs was performed using IBS1.0 stand alone program (Liu W et al., 2015). The subcellular localization was predicted by using LocTree3 protein subcellular localization prediction system (https://rostlab.org/services/loctree3/; Goldberg et al., 2014).
Sequence analysis
The deduced amino acid sequences were aligned with Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). The molecular weights and pIs of the putative PPIases were predicted by biosynthesis’s peptide property calculator web server Version 3.1 (https://www.biosyn.com/peptidepropertycalculator/peptidepropertycalculator.aspx). Amino acid sequences of the respective domains were extracted using EMBOSS: extractseq web interface (bioinformatics.nl/cgi-bin/emboss/extractseq; Olson, 2002). Active site residues of the aligned domain sequences were analysed by employing ESPript 3.0 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi; Robert and Gouet, 2014).
Gene structure, motif analysis and phylogenetic analysis of FKBPs, parvulins, and PTPAs
Gene structures were analyzed using GSDS 2.0 server (http://gsds.cbi.pku.edu.cn/; Hu et al., 2015). Conserved motifs of the protein sequences were identified with MEME Suite (https://meme-suite.org/meme/; Bailey et al., 2015), and edited using TBtools (https://github.com/CJ-Chen/TBtools; Chen et al., 2020). Mega X was employed to construct phylogenetic trees by Neighbor Joining (NJ) method using 1000 bootstrap values. Respective protein sequences of FKBPs, parvulins and PTPAs from T. aestivum, A. thaliana, O. sativa, Aegilops tauschii, Triticum dicoccoides and Triticum urartu were used for the construction of phylogenetic trees. Constructed phylogenetic trees were subsequently edited using Interactive Tree of Life web server (https://itol.embl.de/upload.cgi).
Chromosome mapping and syntenic analysis
Predicted wheat FKBP, parvulin and PTPA genes were mapped to the corresponding chromosomes using MapChart2.32 (Voorrips, 2002). The map obtained was further processed with InKscape (https://inkscape.org/) to highlight various features. Gene duplication events between T. aestivum, its progenitors (T. urartu, T. dicoccoides and A. tauschii), O. sativa and A. thaliana were analyzed with MCScanX (Multiple Collinearity Scan toolkit, https://github.com/wyp1125/MCScanX; Wang Y et al., 2012) keeping threshold values of <1e-4 and 5 for e-value and match size, respectively. The tandem gene duplications and collinear blocks identified with MCScanX were plotted using shinyCircos online application (https://venyao.shinyapps.io/shinyCircos/; Yu et al., 2017). Dual synteny plot was generated by TBtools software (https://github.com/CJ-Chen/TBtools; Chen et al., 2020).
Phylogenetic analysis
Phylogenetic tree was generated to understand the evolutionary relationship among different FKBP, parvulin and PTPA proteins. Alignment of the amino acid sequences of all the proteins was carried out by Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/; Sievers et al., 2011). The phylogenetic tree was generated with MEGA X (Molecular Evolutionary Genetics Analysis) across computing platforms (Kumar et al., 2018) using Neighbour-Joining method with 1000 bootstrap replicates. The tree was exported to the Interactive Tree of Life (https://itol.embl.de/; Letunic and Bork, 2016) for subsequent processing and annotation.
Identification of cis-regulatory elements
The upstream 2 kb promoter regions of the gene sequences were retrieved from Ensembl plants database and used as inputs to search cis-regulatory elements in the database (HSEAT, Heat Shock Element Analysis Tool; https://sourceforge.net/projects/heast/).
Effect of heat stress on expression of FKBP, parvulin and PTPA genes
Seeds of wheat (T. aestivum L.) cultivar HD2967 were surface sterilized using Tween 20 and subsequently sown in soil, and grown in the culture room at 25 ± 2 °C. For acclimation to heat stress, the seven days old seedlings were exposed to 37°C for 2 h. Heat stress was imposed by incubating the seedlings at lethal temperature of 50°C for 4.5 h with and without acclimation at 37°C for 2 h. Seedlings kept at 25 °C served as experimental control. Healthy seedlings after the treatments were collected at different time intervals and snap frozen in liquid nitrogen until further analysis.
Total RNA from the leaves was isolated using TRIZOL solution (Invitrogen, USA), and reverse transcription was performed with random hexamer primers using superscript III First-strand synthesis system kit following the manufacturer’s instructions (Invitrogen, USA). For expression analysis, primers binding to common regions of different homoeologs were designed with Primer-BLAST tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome). The quantitative real-time PCR (qRT-PCR) was performed on Ariamx real-time PCR system (Agilent Technologies, USA) with Brillant III ultra-Fast SYBR green QPCR master mix (Agilent Technologies, USA). Expression analysis was carried out in three independent biological replicates. Actin was used as an internal control for normalization and the fold change was determined using 2−ΔΔCt method (Livak and Schmittgen, 2001). The data obtained were subjected to one-way analysis of variance (ANOVA; graphpad prism).
For digital expression analysis (DGE), expression pattern of TaFKBP and TaPar genes was determined using WheatExp containing homoelogue-dependent gene expression profiles for wheat (https://wheat.pw.usda.gov/WheatExp/; Pearce et al., 2015). Protein sequences were searched against WheatExp database using tblastn and the matched hits were retrieved. RNAseq expression data from a previous work (Liu Z et al., 2015) was used as a reference for DGE analysis of the selected PPIases. The heat map of normalized (log10) expression values was generated using heatmap function from the NMF CRAN library (https://cran.r-project.org/web/packages/NMF/index.html) in RStudio version 4.0.2 (http://www.rstudio.com/; Myers et al., 2020). Hierarchical clustering of rows was performed with Euclidean distance and complete linkage.
Protein-protein interaction network
The protein sequences of TaFKBPs, TaPars and TaPTPAs were submitted to STRING (v11.5) to predict the relationships between themselves and with other proteins (Szklarczyk et al., 2019) based on orthologs from A. thaliana. The parameter “max number of interactors to show” was set to “no more than 10 interactors” while the interacting partners were subjected to k-means clustering to obtain 3 clusters. The query protein (T. aestivum) names were used to label the nodes.
Results
Identification of gene families encoding wheat FKBPs, Parvulins and PTPAs
Bioinformatics analysis of wheat genome using Ensembl plants resulted in identification of 71 TaFKBP, 12 TaPar and 3 TaPTPA genes (Supplementary Tables S1, S2). The nomenclature adopted in this study is as used earlier for wheat cyclophilins (Singh et al., 2019). The wheat FKBP (TaFKBP), parvulin (TaPar) and PTPA (TaPTPA) proteins were suffixed with their predicted molecular weight and chromosomal localization. Localization analysis revealed that the TaFKBP genes are present on all the 21 wheat chromosomes, with the maximum number (8) on chromosome 2B and the lowest, one each, on chromosomes 3B, 4A, 4B and 4D (Figure 1). In contrast, only eight wheat chromosomes showed the presence of TaPar genes, with the chromosome 2D harbouring the maximum three members. The chromosomes 3B, 4A, 5A, 5B and 5D were predicted to contain only a single TaPar gene (Figure 1). Only a single homeologue triplet of PTPA genes was observed in wheat, which were localized on chromosome 7A, 7B and 7D, respectively. In silico analysis of one of the FKBPs, TaFKBP23-6-U, indicated the localization of this gene in an uncharacterized part of wheat genome, denoted by ‘U’ (Figure 1). Though majority of the identified genes were present as triplets due to the three progenitor genomes AA, BB and DD, contributed by the diploid species T. urartu, Aegilops speltoides and A. tauschii, respectively (Feldman and Levy, 2005), absence or existence of additional copies was also observed for some TaFKBP and TaPar genes. For example, TaFKBP29-1-2A, TaFKBP24-2-2B and TaFKBP29-2-2D, a homeologue triplet localized on chromosome 2A, 2B and 2D, depicted the presence of two additional copies on chromosome 2B (TaFKBP7-1-2B; TaFKBP16-7-2B) and one on 2D (TaFKBP20-6-2D). The gene encoding TaFKBP7-1-2B appears to be a paralogue containing partial FKBD sequence (missing N-terminal region) having more than 97℅ identities with TaFKBP29-1-2A, TaFKBP24-2-2B and TaFKBP29-2-2D proteins. Similarly, the TaPar genes, TaPar13-2-2A, TaPar13-3-2B and TaPar13-1-2D also exhibited additional copies on chromosomes 2D (TaPar12-1-2D), 3B (TaPar11-1-3B) and 4A (TaPar11-2-4A).
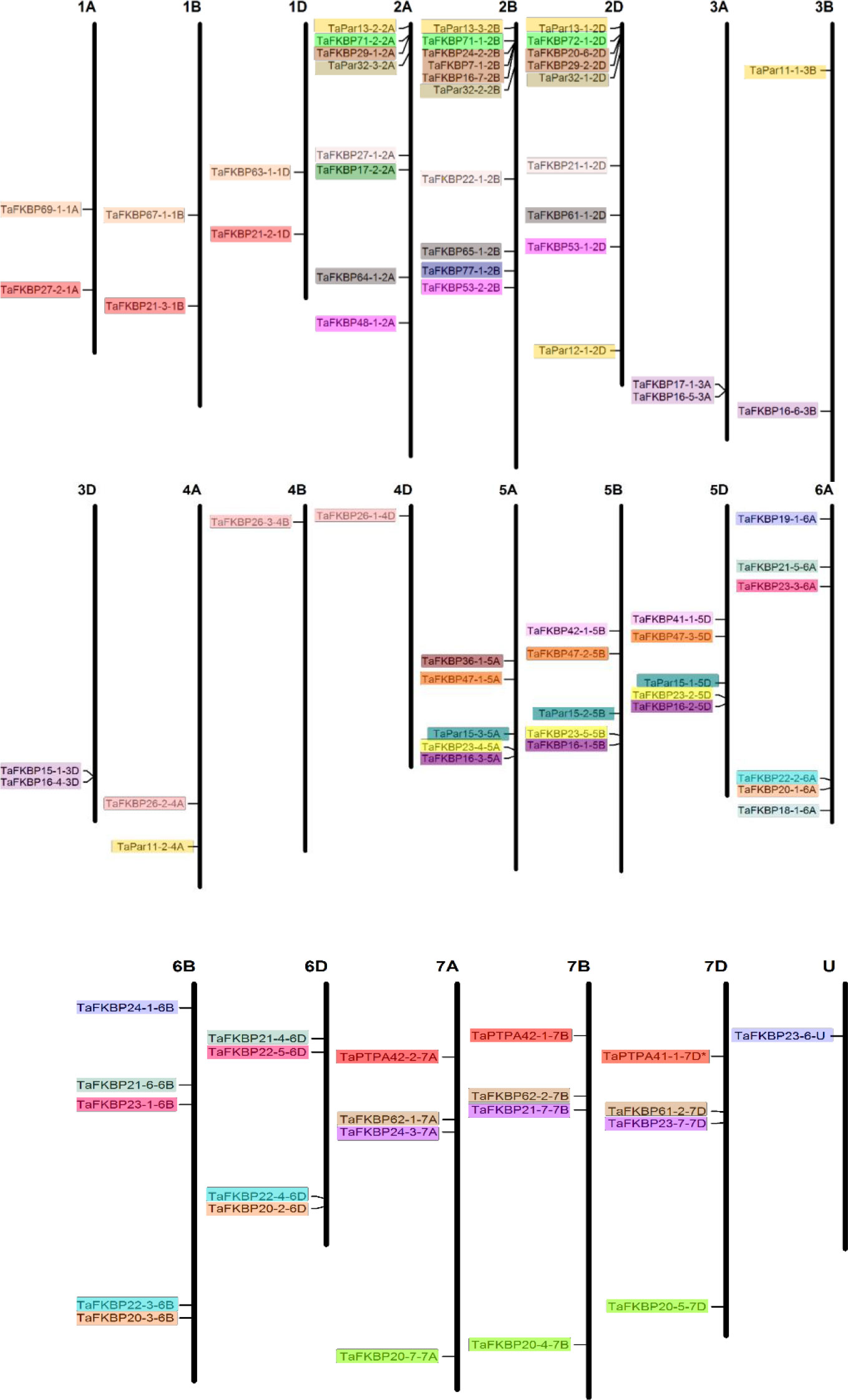
Figure 1 Chromosomal distribution of the genes encoding FKBPs, parvulins and PTPAs in wheat. The chromosome size was determined from the IWGSC1.0 assembly and is drawn to scale. Homeologous groups of genes are represented in same colors (U represents uncharacterized part of genome).
The number of amino acid residues in TaFKBPs and TaPars varies from 68 to 649, and 99 to 301, respectively, whereas the molecular weights of these sub-families range between 7.59 to 77.10 kDa, and 11.06 to 32.70 kDa, respectively (Supplementary Table S1). Genome-wide analysis revealed that only three PTPAs are encoded by the wheat genome, with TaPTPA41-1-7D* and TaPTPA42-1-7B consisting of 393 amino acid residues each, compared to 394 in TaPTPA42-2-7A. Since the CDS and amino acid sequences of TaPTPA41-1-7D* contain some unannotated residues, its exact molecular weight and pI could not be predicted. The molecular weights of TaPTPA42-1-7B and TaPTPA42-2-7A were computed as 42.46 and 42.57 kDa, respectively. Isoelectric points (pIs) of different TaFKBPs, TaPars and TaPTPAs range between 4.32 to 10.68, 6.15 to 9.92, and 6.9 to 7.1, respectively (Supplementary Table S1). While the TaFKBPs were predicted to localize to different subcellular compartments viz., ER (1), nucleus (6), chloroplast (32) and cytoplasm (32), the TaPars are likely to localize only in cytoplasm (4) and nucleus (8). On the contrary, all the three TaPTPA proteins were predicted to be present only in the cytoplasm.
Chromosome localization and synteny analysis of wheat TaFKBP, TaPar and TaPTPA genes
Gene duplication events are vital for the amplification of gene families and their subsequent evolution in wheat. A tandem duplication event occurs when two or more similar genes are found located within a 200 kb of chromosomal region (Holub, 2001). Duplication events that gave rise to TaFKBP, TaPar and TaPTPA genes were studied in O. sativa, A. thaliana, T. aestivum (AABBDD) and its progenitors, T. urartu (AA), T. dicoccoides (AABB) and A. tauschii (DD) using MCScanX (Supplementary Table S3) (Wang Y et al., 2012). Due to absence of genome sequence of the B sub-genome donor A. speltoides, wild emmer wheat (T. dicoccoides) was used as the source of this sub-genome. Neither T. aestivum nor any of its progenitors showed syntenic relationship for parvulin and the PTPA genes. Syntenic relationships were also not observed for FKBP genes between wheat, O. sativa, A. thaliana and T. urartu. Several tandem duplicated pairs of FKBP genes were identified in T. urartu (2), T. dicoccoides (2), A. tauschii (3) and T. aestivum (29) (Figure 2A; Supplementary Table S4). Microcollinearity analyses of the genome segments across different progenitor species revealed the presence of several orthologous segments sharing collinear blocks of FKBP genes (Figures 2A, B; Supplementary Table S5). T. aestivum contains four blocks of FKBP genes collinear with each of its two progenitor species, A. tauschii and T. dicoccoides. In addition, six segmentally duplicated blocks of FKBP genes were also identified in T. aestivum, compared to a single block in T. dicoccoides. These results indicate that tandem as well as segmental gene duplication events might have played an important role in the evolution of FKBP gene family in wheat. Furthermore, our results also suggest that the wheat FKBP genes are entirely derived from its progenitors since most of the wheat chromosomes, except 1B, 2A, 2B, 2D and 3D, harbor the same number of genes as are present on the corresponding chromosomes of the sub-genome donors (Supplementary Table S6). However, the chromosomes 1B, 2A, 2B, 2D and 3D depicted the presence of higher number of FKBP genes relative to the sub-genome donors, indicating enrichment due to duplication events. Another reason for this difference could be the loss of these genes in T. dicoccoides and A. tauschii during evolution. Relative to T. dicoccoides and T. aestivum, the difference in FKBP genes in T. urartu may also be ascribed to similar events. In contrast, the wheat chromosomes 1D, 4B and 5D harbor fewer number of FKBP genes than its sub-genome donors, suggesting loss during the course of evolution. Similar trend was also observed for TaPar genes on chromosomes 2A, 2B and 2D (Supplementary Table S7). The TaFKBP genes (71) showed homology with 14, 40, 20, 16, 23, 16 and 18 counterparts in T. urartu, T. dicoccoides, A. tauschii, Arabidopsis, O. sativa, P. persica and M. domestica, respectively. Similarly, different homologues of TaPars (12) were also observed in A. thaliana (3), E. coli (1), human (2), S. cerevisiae (1), T. urartu (3), T. dicoccoides (9) and A. tauschii (5). The A. thaliana, O. sativa, T. urartu and A. tauschii also depicted the presence of a single homologue of TaPTPA gene. Absence of PTPA genes in one of the wheat progenitors, T. dicoccoides suggests, that the gene encoding TaPTPA42-1-7B might have been derived from the wheat B sub-genome donor A. speltoides. Alternately, it is also likely that these genes might have been lost during evolution, as observed for 14-3-3 gene family in wheat (Shao et al., 2021). This study provides further evidence that the genes encoding FKBPs, parvulins and PTPAs are conserved across major plant species and their abundance in the wheat genome may be attributed to various duplication events during the course of evolution.
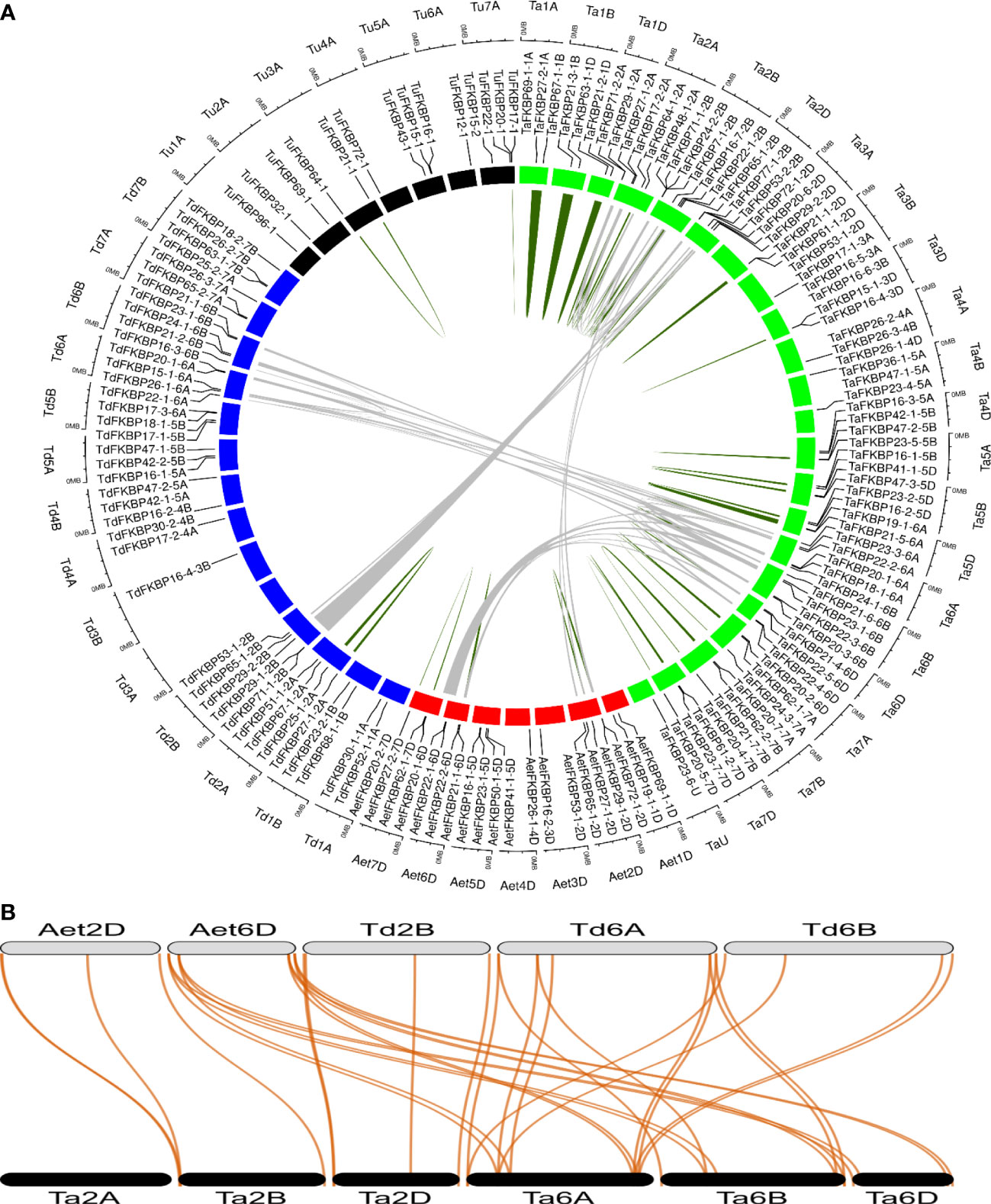
Figure 2 Collinearity analysis of TaFKBP genes using genome sequences of T. aestivum (Ta, AABBDD, green box) and progenitor species A. tauschii (AeT, DD, red box), T. urartu (Tu, AA, black box) and T. dicoccoides (Td, AABB, blue box). (A) A circular layout of TaFKBP orthologues with collinear blocks joined across the corresponding chromosomes in T. aestivum and progenitor species by grey lines. The tandemly duplicated genes are marked with dark green lines. The scale on the circle is in Megabases. (B) The specific collinear blocks between Ta (black color), Aet (grey color) and Td (grey color) are shown as a dual synteny plot. The collinear genes are connected by red lines.
Gene structure organization
Analysis of gene organization revealed significant variability in the exon-intron structure of different TaFKBP, TaPar and TaPTPA genes (Figure 3). Genes of all the three PPIase sub-families showed the presence of introns in their open reading frames (ORFs), with the number of introns varying significantly within each of the sub-families. For instance, the number of introns in the ORF of these genes ranges between 1-19 in TaFKBPs, 1-7 in TaPars and 1-2 in TaPTPAs. The genes encoding PTPAs and FKBPs exhibited homoeolog-dependent variability in the number of introns, as also observed earlier for the wheat cyclophilin genes (Singh et al., 2019). For example, TaFKBP16-7-2B and TaFKBP20-6-2D consist of six introns each in their ORFs compared to 2-10 in their other homeologues TaFKBP7-1-2B, TaFKBP24-2-2B, TaFKBP29-1-2A and TaFKBP29-2-2D. Similarly, TaPTPA42-1-7B and TaPTPA42-2-7A depicted a single intron in their ORFs, relative to two in the third homeolog, TaPTPA41-1-7D*. On the contrary, the TaPar genes showed conservation in the abundance of introns within members of the same homeologous groups. The length of introns also differs among different PPIase genes, with both the smallest (53 bp) and the largest (7738 bp) being observed in TaFKBPs, TaFKBP15-1-3D and TaFKBP67-1-1B, respectively. Within the TaPTPA gene family, the smallest (272 bp) and the largest (3302 bp) introns were observed in TaPTPA41-1-7D*. The homeologous group of TaPar genes, comprising of TaPar32-1-2D, TaPar32-2-2B and TaPar32-3-2A, depicted the smallest length (80 bp) for intron 4, while the largest (2833 bp) was observed in TaPar15-2-5B. While majority of the TaFKBPs (50) contain untranslated regions (UTRs) in both the 5’ and 3’ regions, the same could not be observed for 12 of these genes. Nine TaFKBPs exhibited introns only in their 3’ UTR and two in only 5’UTR. Absence of 5’ UTR or 3’ UTR was also observed among TaFKBPs. While 5’ UTR is lacking in six TaFKBPs (TaFKBP16-7-2B, TaFKBP20-3-6B, TaFKBP20-6-2D, TaFKBP22-5-6D, TaFKBP27-2-1A and TaFKBP67-1-1B), three TaFKBPs (TaFKBP19-1-6A, TaFKBP21-7-7B, TaFKBP77-1-2B) are devoid of 3’ UTR. Though UTR of several TaFKBPs depicted the presence of introns, none of the TaPar and TaPTPA genes exhibited this feature. The genes TaPar11-1-3B, TaPar11-2-4A, TaPTPA41-1-7D* and TaPTPA42-2-7A showed absence of both 5’ and 3’UTRs. Unlike the cyclophilin and FKBP gene families in lower organisms viz., Leptosphaeria maculans (Singh et al., 2014), that contain shorter introns and compact genomes, the length of introns is larger than exons in most of the TaFKBP, TaPar and TaPTPA genes.
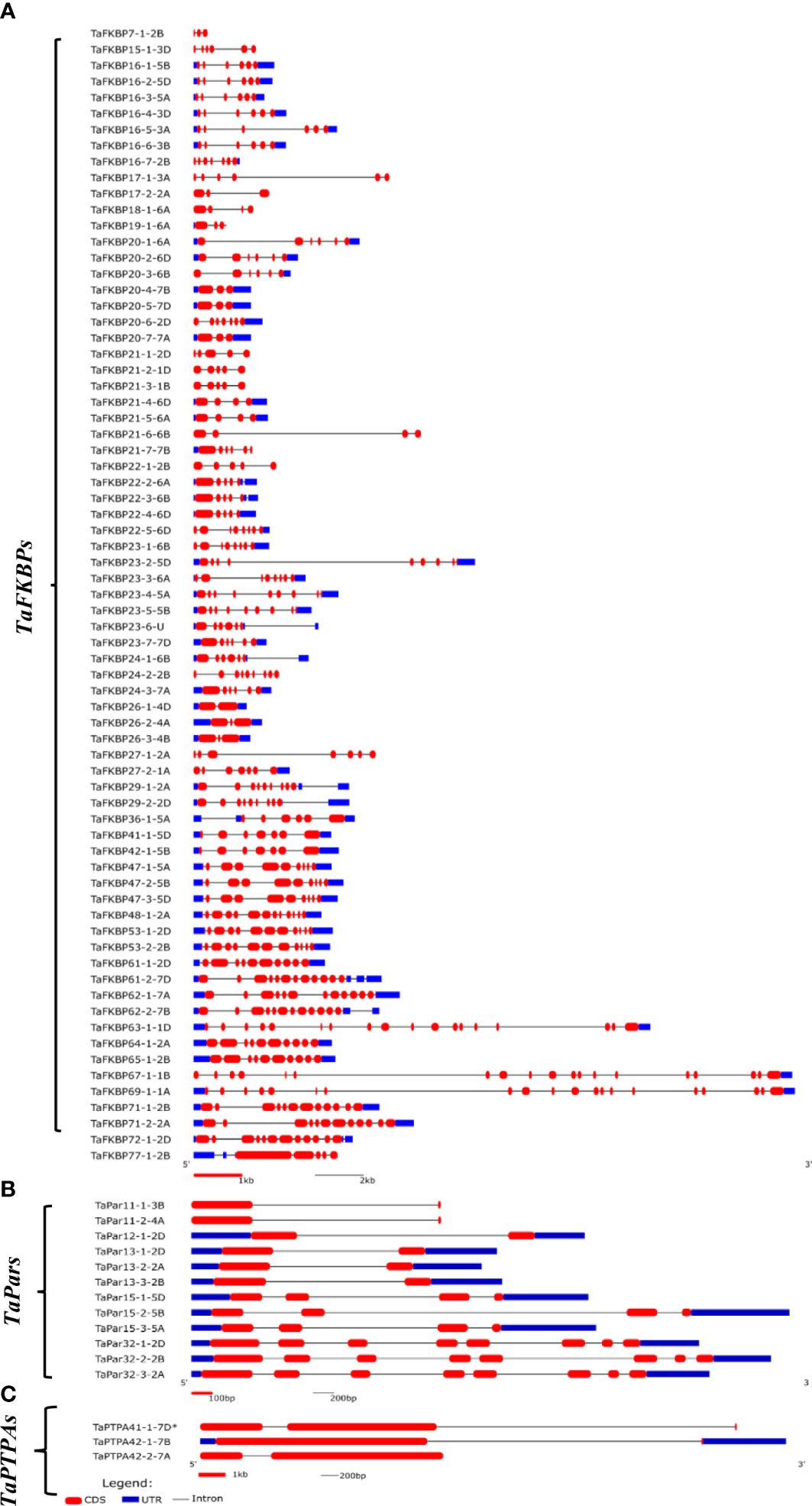
Figure 3 Structure of genes encoding FKBPs (A), Parvulins (B) and PTPAs (C) in T. aestivum. * represents exact molecular weight and pI not determined.
Domain architecture and active site residue analysis
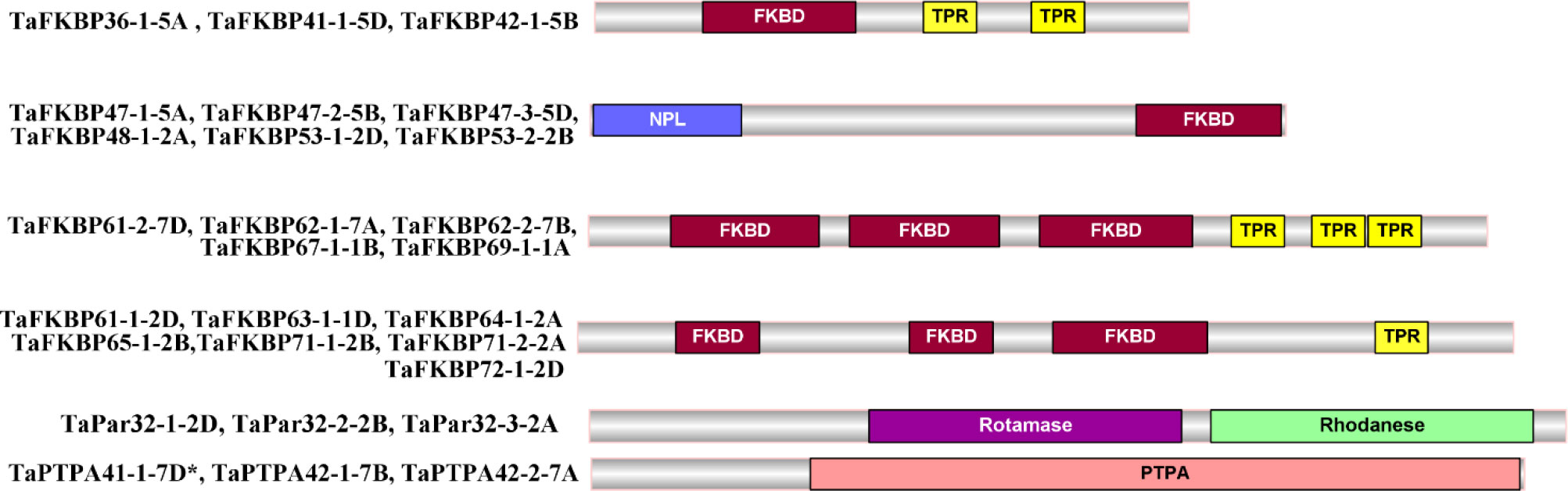
The FKBP, parvulin and PTPA proteins in hexaploid wheat contain the characteristic signature domains, with several members of former two families also containing additional domains (Figure 4). Unlike the TaFKBPs and TaPars, several of which are multi-domain proteins, the TaPTPAs consist of only the PTPA domain. Besides FKBD, 21 of the 71 TaFKBPs also demonstrated the presence of other domains, such as nucleoplasmin-like (NPL) and tetratricopeptide repeat (TPR) domain, implying diversification in their roles (Supplementary Table S1). Except for NPL domain-containing FKBPs, such as TaFKBP47-1-5A, TaFKBP47-2-5B, TaFKBP47-3-5D, TaFKBP48-1-2A, TaFKBP53-1-2D and TaFKBP53-2-2B, that exhibit FKBD at C-terminus, this domain in other multi-domain proteins is localized at the N-terminus. One of the single domain proteins, TaFKBP19-1-6A, contains a partial N-terminus FKBD. Twelve of the multi-domain FKBPs also contain TPR domain in addition to three FKBDs, indicating their likely role in protein-protein interactions. Since the NPL domain-containing FKBPs have been demonstrated to play an important role in chromatin remodelling and regulation of transcription in A. thaliana (Li and Luan, 2010), the presence of this domain in the wheat FKBPs (TaFKBP47-1-5A, TaFKBP47-2-5B, TaFKBP47-3-5D and TaFKBP48-1-2A, TaFKBP53-1-2D, TaFKBP53-2-2B) implies the role of these proteins in regulation of gene expression.
Comparison of active site residues with the human FKBP, hFKBP12, revealed the absence of His (H-87) residue in all members of FKBP family in wheat (Supplementary Table S8; Figure S1A). Similarly, compared to their human counterpart hPar14, all wheat parvulins also lack the critical residue Asp (D-74; Supplementary Table S9 and Figure S1B). Variability in other active site residues was also observed in TaFKBPs and TaPars. The parvulins TaPar11-1-3B (11.06 kDa) and TaPar11-2-4A (11.16 kDa) possess only two of the conserved active site residues i.e., His (H-42) and Ser (S-72), indicating that these proteins might be enzymatically inactive. This speculation, however, needs experimental validation. The higher molecular weight TaFKBPs, such as TaFKBP61-1-2D (61.35 kDa), TaFKBP61-2-7D (61.97 kDa), TaFKBP62-1-7A (62.01 kDa), TaFKBP62-2-7B (62.02kDa), TaFKBP63-1-1D (63.97 kDa), TaFKBP64-1-2A (64.89 kDa), TaFKBP65-1-2B (65.60 kDa), TaFKBP67-1-1B (67.96 kDa), TaFKBP69-1-1A (69.90 kDa), TaFKBP71-1-2B (71.30 kDa), TaFKBP71-2-2A (71.80 kDa) and TaFKBP72-1-2D (72.07 kDa), which contain multiple FKBDs, also depicted difference in the occurrence of active site residues in different FKBDs. This observation suggests that the FKBDs with the maximum number of conserved active site residues might be enzymatically active while others may not be functional. This speculation, though warrants experimental confirmation, is also supported by a previous study which showed that of the three FKBDs in the wheat FKBP, wFKBP73, only one of the domains was enzymatically active (Unger et al., 2010). Our analysis also revealed that the different FKBDs in TaFKBPs are conserved among different homeologues. High identity of more than 94.31% was also observed in PTPA domains of TaPTPA41-1-7D*, TaPTPA42-1-7B and TaPTPA42-2-7A (Figure 5), signifying conservation of these proteins.
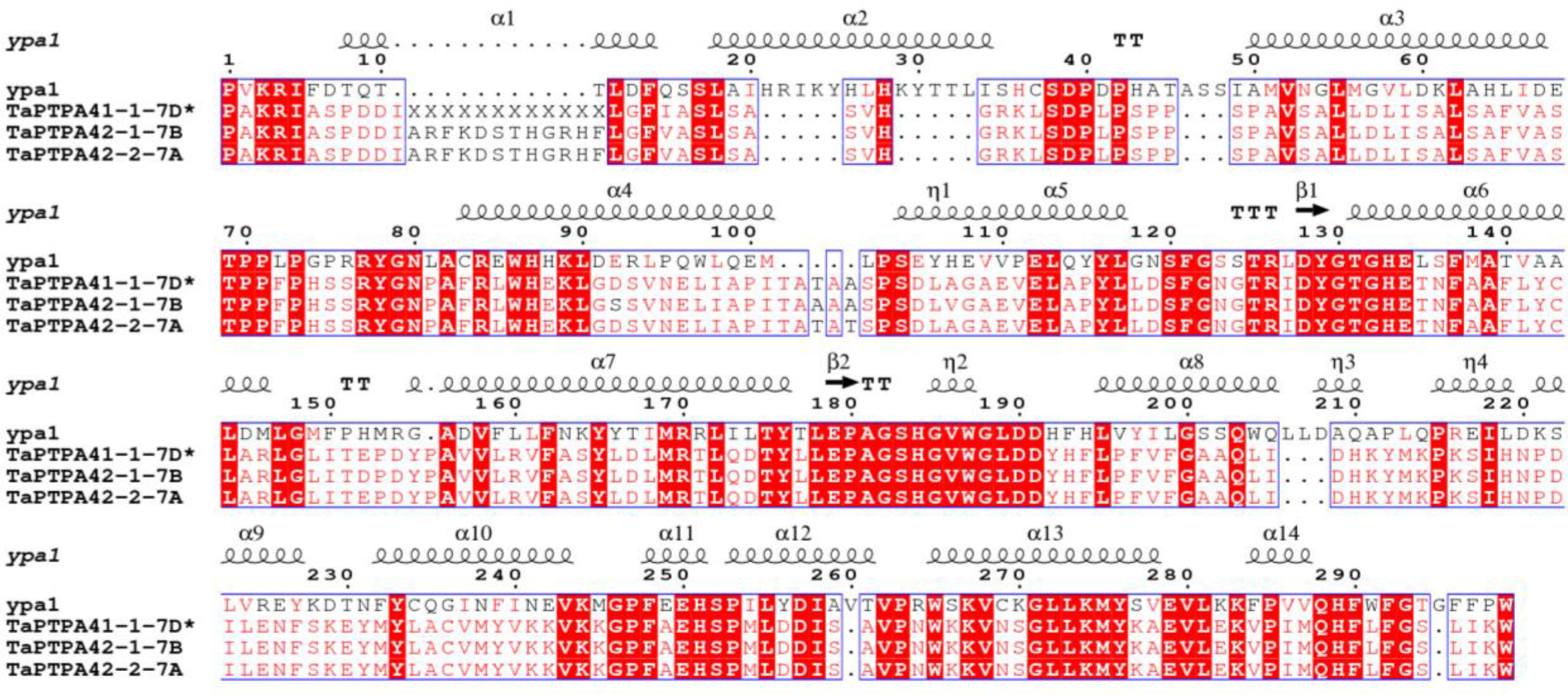
Figure 5 Multiple sequence alignment of PTPA domains of wheat PTPA proteins and yeast PTPA ypa1. * represents exact molecular weight and pI not determined.
Motif analysis
Protein sequences of TaFKBPs, TaPars and TaPTPAs were also analysed for the presence of conserved motifs. A total of 15 motifs, varying in length from 15 to 150 amino acids, were identified in TaFKBPs (Table 1; Figure 6A). The FKBD comprises of motifs 1, 3, 4, 5, 6 and 7, with the motifs 1, 3, 4 and 5 being observed in maximum number of proteins (67, 65, 56 and 59, respectively). The motifs 1, 3 and 4 consist of 21 amino acid residues each, compared to 15 residues in motif 5, with all constituting a component of FKBD. The absence of motifs 6 and 7 in TaFKBPs might be due to deletion of these sequences in their C-termini. Presence of only two motifs (1 and 7) in the highest molecular weight protein TaFKBP77-1-2B (77.10 kDa), might be due to the presence of partial FKBD in this protein. Apart from the conserved FKBDs, in silico analysis also predicted the presence of motifs in TPR (motifs 2 and 15) and NPL domains (motifs 11 and 12) of the multi-domain FKBPs. The motifs 1, 3, 4, 5 (FKBP_C) and 8 (unknown function) were observed most commonly, with 36 TaFKBPs exhibiting the presence of this combination. However, exclusive presence of motif 13 (ASP_Protease) and motif 14 (Domain of unknown function) in two different homeologue triplets i.e., TaFKBP48-1-2A, TaFKBP53-1-2D and TaFKBP53-2-2B; and TaFKBP47-1-5A, TaFKBP47-2-5B and TaFKBP47-3-5D, respectively, might be associated with some discrete functions of these proteins, that are yet to be elucidated. In another homeologue triplet, TaFKBP24-1-6B, TaFKBP23-6-U and TaFKBP19-1-6A, the former two proteins harbour motifs 1, 3, 4 (FKBP_C) and 8 (unidentified motif), while TaFKBP19-1-6A consists of only motifs 4 and 8. The presence of other motifs such as LBR tudor, cytidylate kin 2, ASP_Protease, domain of unknown function and CDC45-like protein in few of the wheat FKBPs suggests novel cellular roles that are yet to be studied. Akin to TaFKBPs, homeolog-specific variations in motifs were also observed in TaPars (Table 1; Figure 6B). On the contrary, all the three PTPAs, TaPTPA41-1-7D*, TaPTPA42-1-7B and TaPTPA42-2-7A, exhibited the same motif architecture (Table 1; Figure 6C).
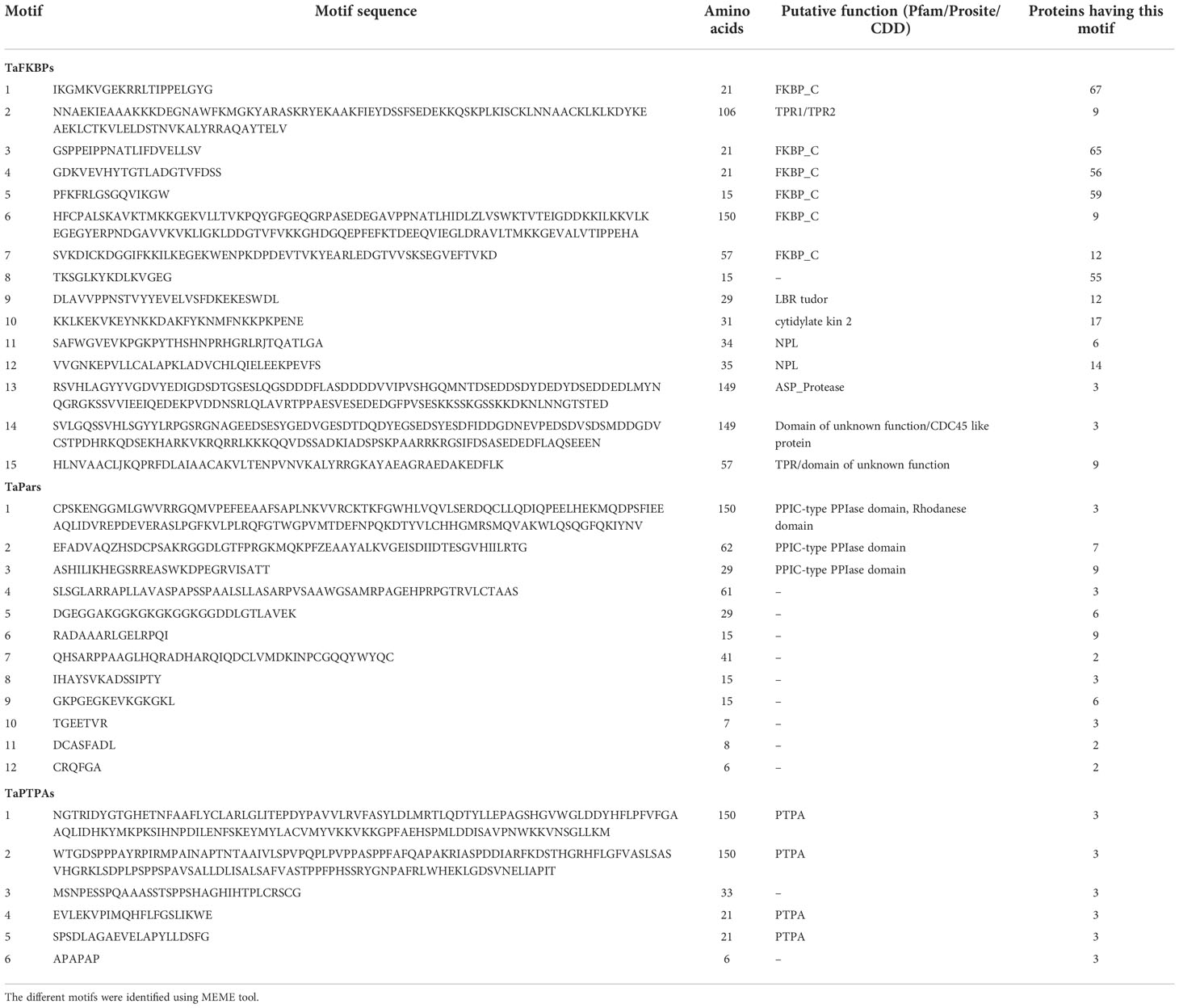
Table 1 Comparative analysis of different conserved motifs in FKBP, parvulin and PTPA proteins in T. aestivum.
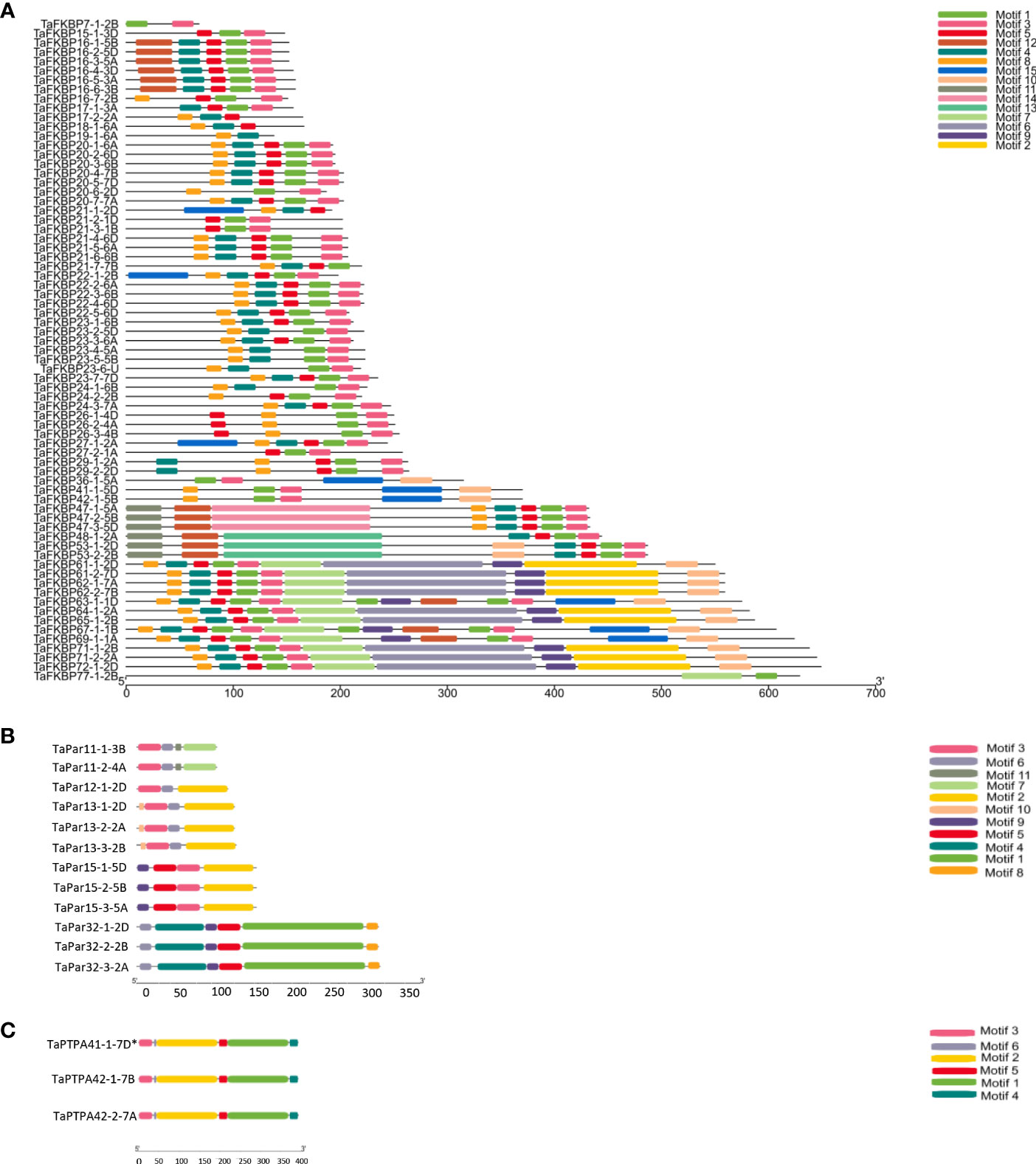
Figure 6 Organization of different conserved motifs in FKBP (A), parvulin (B) and PTPA (C) proteins of T. aestivum.
Phylogenetic analysis of wheat FKBP, parvulin and PTPA proteins
To understand evolutionary relationships among FKBPs, parvulins and PTPAs of different plant species, we constructed separate phylogenetic tree for each of these protein families using alignments of amino acid sequences from wheat, its progenitors, and other plant species. For FKBPs, the phylogenetic tree constructed using NJ method in MEGA X was based on alignment of 196 FKBD-containing proteins from T. aestivum (70), T. urartu (14), T. dicoccoides (40), A. tauschii (20), A. thaliana (23) and O. sativa (29) (Figure 7A). Similarly, for parvulins also, the phylogenetic tree included 40 different parvulin protein sequences from T. aestivum (12), T. urartu (3), T. dicoccoides (9), A. tauschii (4), A. thaliana (3), O. sativa (4), H. sapiens (2), S. cerevesiae (1) and E. coli (2) (Figure 7B). For PTPAs, a total of seven proteins from T. aestivum (3), T. urartu (1), A. tauschii (1), A. thaliana (1) and O. sativa (1) were included for constructing the phylogenetic tree (Figure 7C).
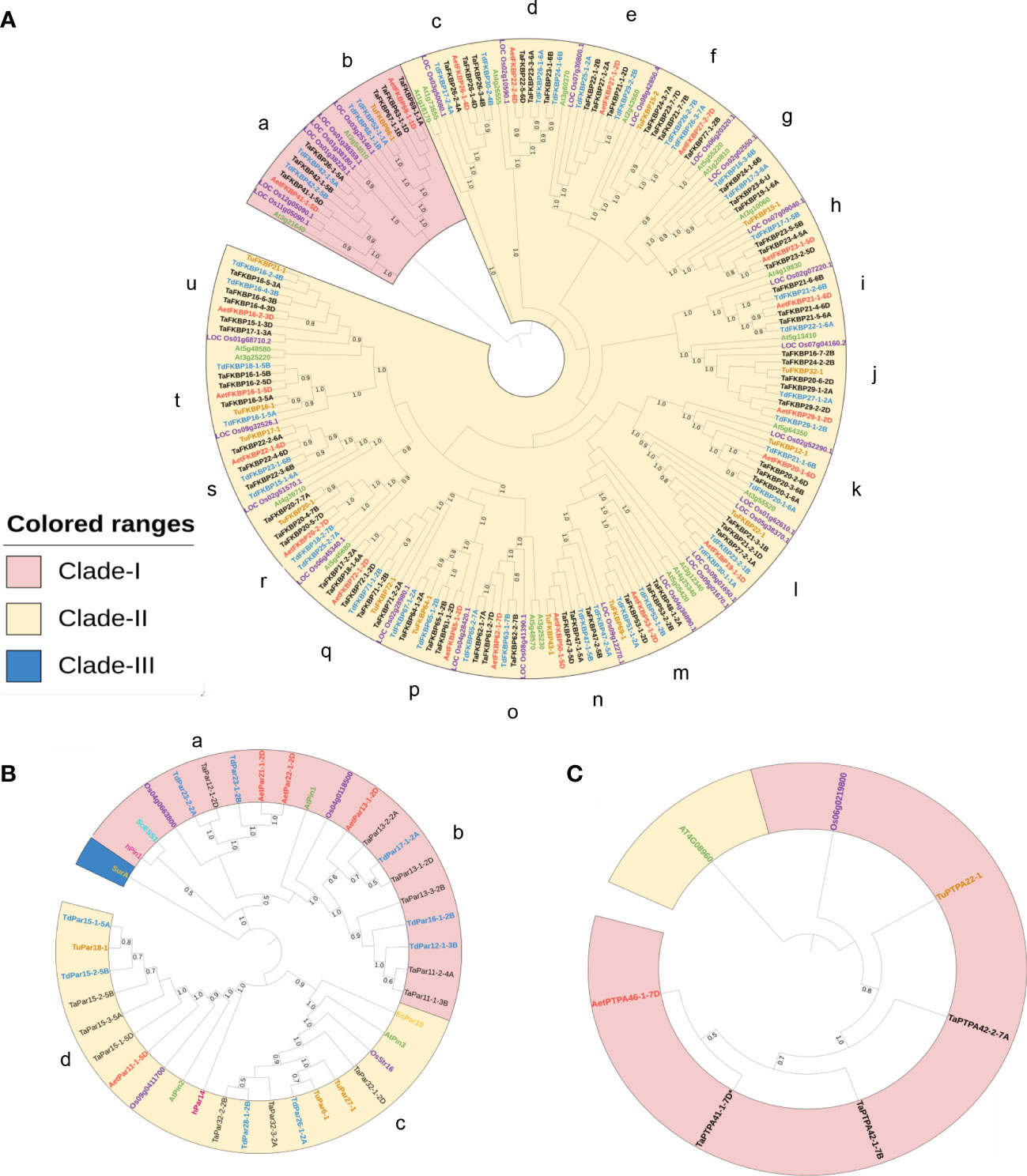
Figure 7 Phylogenetic analysis of FKBP (A), Parvulin (B) and PTPA (C) proteins of T. aestivum (black), T. urartu (orange), T. dicoccoides (blue), A. tauschii (red), A. thaliana (green) and O. sativa (purple), H. sapiens (pink), S. cerevisiae (blue) and E. coli (yellow). Clustal Omega was used to perform the multiple sequence alignment. The phylogenetic tree was constructed by Neighbor-Joining (NJ) method of MEGA X standalone application with 1000 bootstrap replicates. The generated tree was annotated using iTOL online server. Different clades are highlighted with different colors.
Phylogenetic analysis classified the TaFKBPs into two different clades (Clades-I and II), which were further divided into various sub-groups consisting of homologous FKBPs from different plant species. Clade-I consisted of 21 members compared to 175 in Clade-II. The ultimate sub-groups (labeled a to u) comprised all the TaFKBP homeologue triads that were clustered together with their progenitor species and homologues from other plant species. The close clustering of homeologue triad was also observed for cyclophilins and 14-3-3 gene families in common wheat (Singh et al., 2019; Shao et al., 2021). While the sub-groups a, c, d, e, g, i and o lacked FKBP homologues from T. urartu (A genome), the sub-groups h, q and u showed the absence of FKBP homologues from T. dicoccoides (A genome). The clustering pattern depicted in the phylogenetic tree prompts us to hypothesise that the FKBP genes on chromosomes 1A, 2A, 4A, 5A, 6A and 7A of the common wheat either evolved from T. dicoccoides, or the corresponding genes present in T. urartu were later lost during the course of evolution. Conservation of most of the homeologue triplets implies that members of the TaFKBP gene family were not subjected to significant changes during the course of evolution and these genes in hexaploid wheat have been acquired from its progenitors due to polyploidy. The presence of FKBP homologues from O. sativa (monocot) and A. thaliana (dicot) in each of the sub-groups implies existence of these genes even before divergence of monocots and dicots. Furthermore, the grouping of FKBPs appears to be domain specific and also influenced by the localization pattern, and the arrangement of introns and exons in the corresponding genes. For example, Clade-I consisted of multi-domain FKBPs possessing TPR and FKBDs. One of its two sub-clades contained proteins with three FKBDs and one TPR domain, with their localization predicted to be in the nucleus. However, all other members with similar domain architecture, but localized to cytosol, formed a separate sub-clade in Clade-II. Members containing multiple domains formed different sub-groups in Clade-II (for example sub-groups m, n, o, p, q). On the other hand, most of the FKBPs consisting of only a single FKBD were clustered together based on their localization pattern. This implies that the localization signal (Supplementary Table S1) and conservation of the underlying structure (Figure 3A) might have played an important role in the functional divergence of FKBPs in common wheat.
The phylogenetic tree of TaPars depicted three major clades viz., Clades-I, II and III. The Clades I and II comprised of 19 and 20 members, respectively, while the third clade contained only a single parvulin, surA, from E. coli, and was differentiated from other two clades that consisted of various eukaryotic parvulins. Both Clade-I and Clade-II were further divided into sub-clades (labeled a-d), with all the sub-groups, except sub-group a that consisted of only a single member TaPar12-1-2D from D genome of T. aestivum, represented by triplet homeologues. The sub-group b consisted of two additional proteins from T. aestivum (TaPar11-1-3B and TaPar11-2-4A), encoded by a gene each on chromosomes 3B and 4A. Except for sub-group d, none of the other sub-groups depicted the presence of members representing each of the three wheat sub-genomes. For example, sub-groups a and b lacked representatives from T. urartu, while sub-group c was devoid of corresponding homologue from A. tauschii. Computational analysis categorised the PTPA proteins into two groups, Clades-I and II, with the former comprising of closely clustered PTPA proteins from different species (T. aestivum, T. urartu, A. tauschii and O. sativa), while Clade-II included PTPA from dicot A. thaliana. These observations suggest that in contrast to FKBP genes, the PTPA genes might have diverged after differentiation of monocots from dicots.
Expression analysis of TaFKBP, TaPar and TaPTPA genes in response to heat stress
The effect of heat stress on expression of TaFKBP, TaPar and TaPTPA genes was investigated in response to various heat stress treatments using qRT-PCR (Figures 8–10). The survivability of the wheat seedlings was followed, as described earlier, after exposure to lethal temperature (50 °C) without and with acclimation of seedlings at 37 °C for 2 h (Singh et al., 2019). The seedlings that were acclimated at 37°C for 2 h before heat stress at 50°C for 4.5 h showed higher survival (80%) as compared to the seedlings that were exposed to lethal heat stress at 50°C without acclimation (19%), indicating acquisition of thermotolerance at sub-lethal temperature (Singh et al., 2019). For expression analysis, common primers for 28 different homeolog TaFKBP groups (71 genes), five for TaPars (12 genes) and a single primer pair for the TaPTPAs (3 genes) were designed and validated by conventional PCR. Of these, 14 and nine primer pairs, representing 43 and nine different TaFKBPs and TaPars, respectively, from different homeolog groups depicted amplification and were used further for qRT-PCR analysis (Supplementary Table S10). The TaPTPA genes, however, did not show amplification.
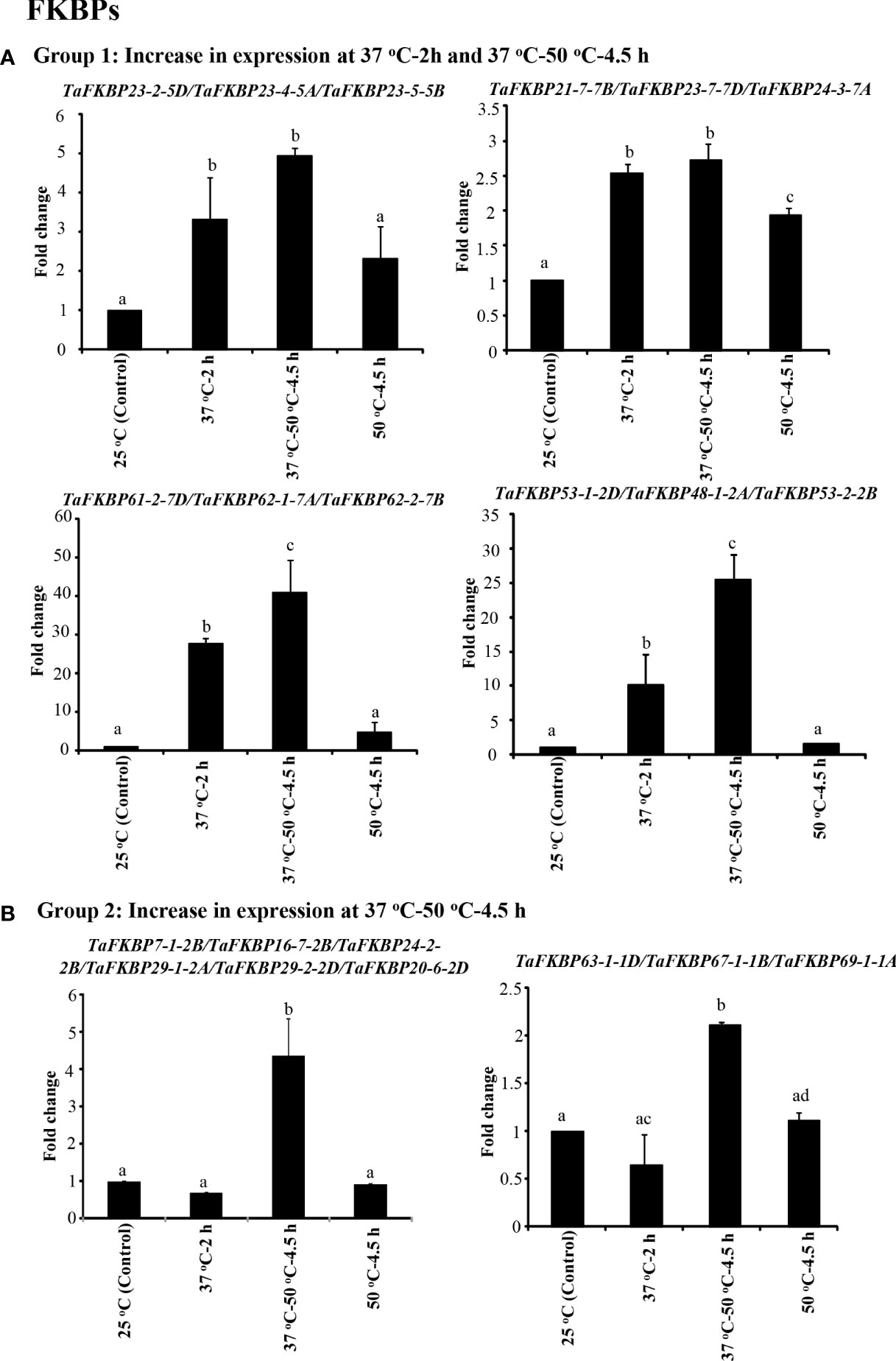
Figure 8 qRT-PCR analysis of FKBP genes in the seven days old wheat seedlings. (A) Genes whose expression was induced during acclimation phase (37°C-2 h) and was maintained for further 4.5 h on subsequent exposure to 50 °C. (B) Genes that registered an increase in expression only at 37 °C-50 °C-4.5 h. Actin was used as reference. The values depict the mean of three biological replicates ± standard error and the different lowercase letters denote significant difference between the treatments at P ≤ 0.001 (Tukey-HSD test; α=0.05) (/represents homeologs).
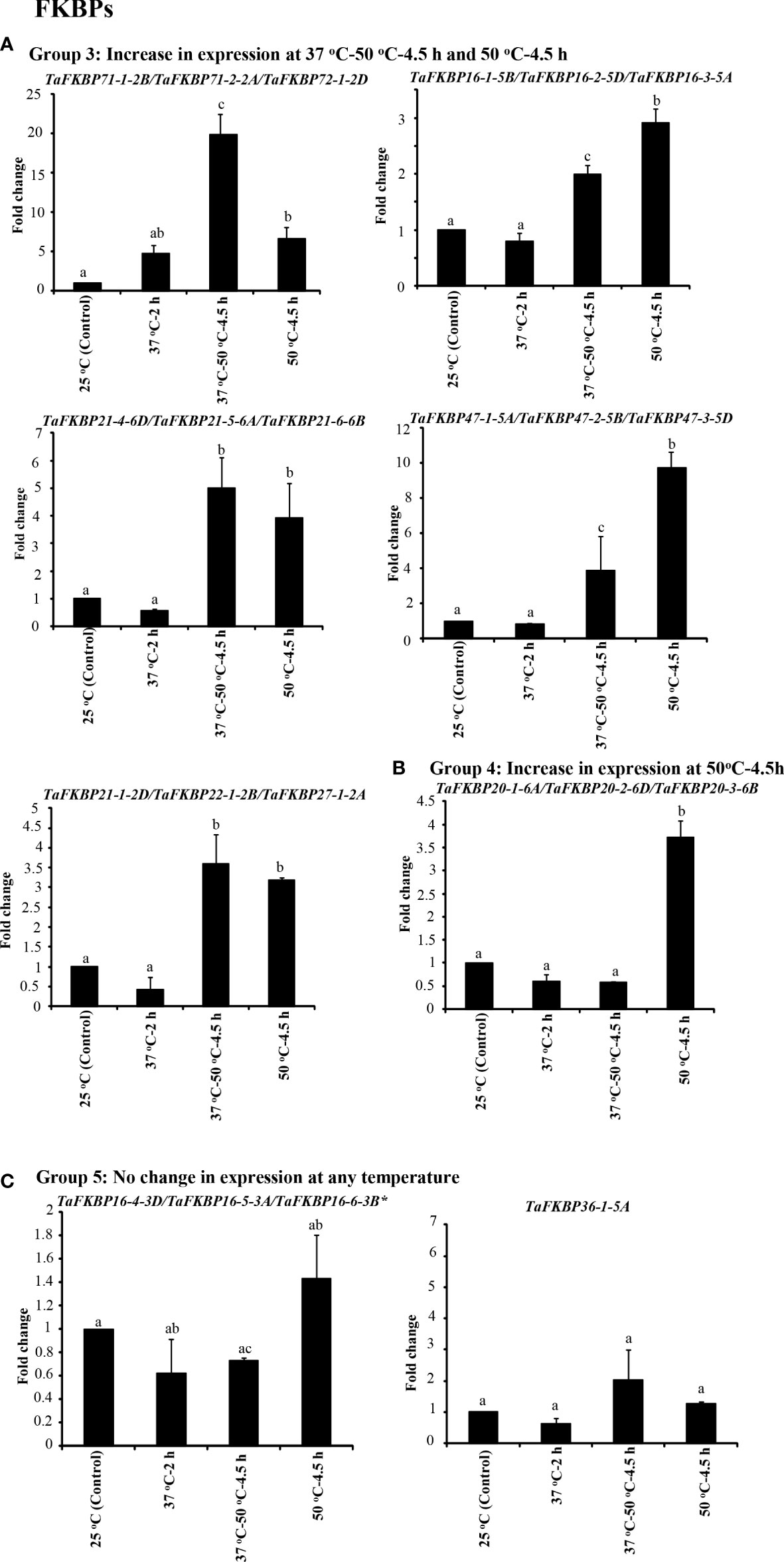
Figure 9 qRT-PCR analysis for studying the effect of heat stress on expression of TaFKBP genes in the seven days old wheat seedlings. (A) Genes showing upregulation in response to both direct heat stress (50 °C) and after acclimation (37 °C-50 °C-4.5 h), (B) only direct heat stress (50 °C) and (C) unaffected by heat stress. Actin was used as reference. The values depict the mean of three biological replicates ± standard error and the different lowercase letters denote significant difference between the treatments at P ≤ 0.001 and P ≤ 0.05* (Tukey-HSD test; α=0.05) (/represents homeologs).
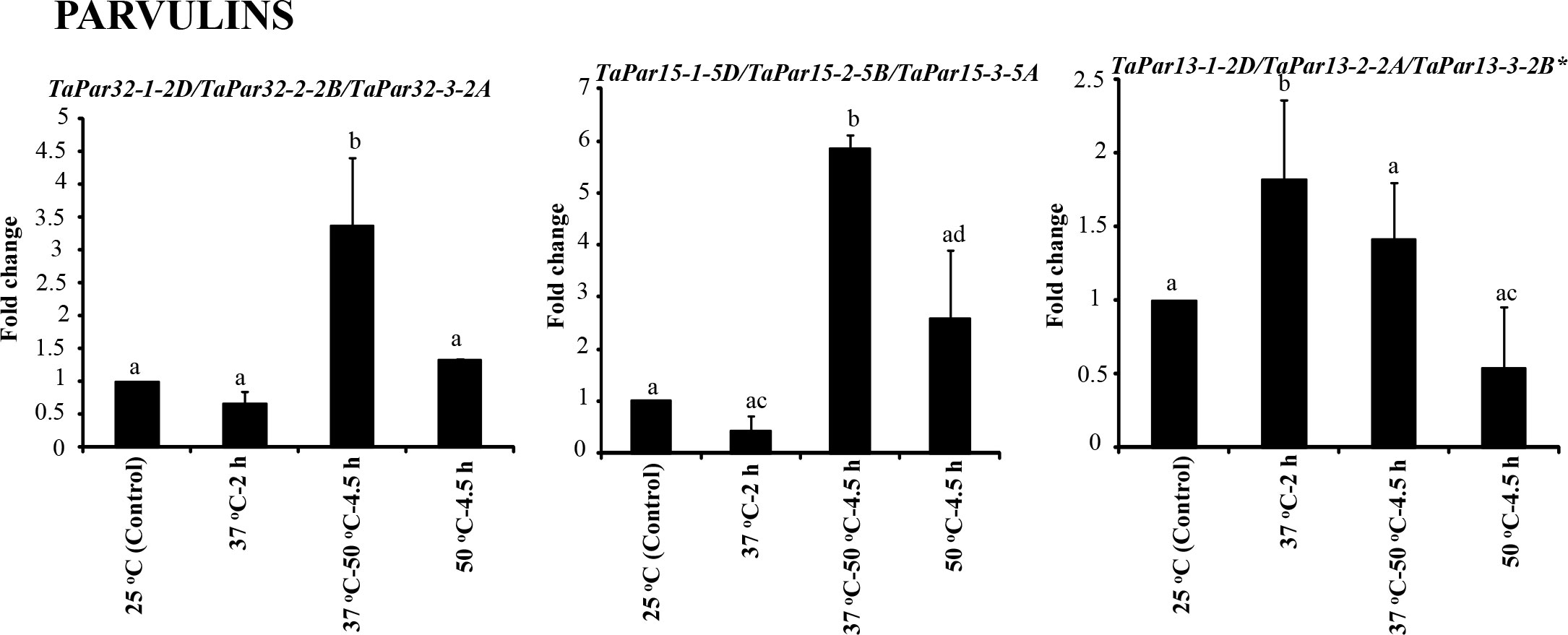
Figure 10 Effect of heat stress on expression of genes encoding parvulins in seven days old wheat seedlings using Actin as reference. The values depict the mean of three biological replicates ± standard error and the different lowercase letters denote significant difference between the treatments at P ≤ 0.001 and P ≤ 0.05* (Tukey-HSD test; α=0.05) (/represents homeologs).
On the basis of expression profile, the TaFKBP genes were classified into five different groups (Figures 8, 9). The expression of Group 1 genes, except TaFKBP53-1-2D/TaFKBP48-1-2A/TaFKBP53-2-2B and TaFKBP61-2-7D/TaFKBP62-1-7A/TaFKBP62-2-7B, after increasing during acclimation (37 °C for 2 h) was also maintained under subsequent lethal heat stress (37 °C-50 °C-4.5 h), indicating the likely role of these genes in thermotolerance (Figure 8A). Among the Group 1 genes, highest expression after acclimation (27.7-fold) and in response to subsequent lethal heat stress (40.8-fold) was observed for TaFKBP61-2-7D/TaFKBP62-1-7A and TaFKBP62-2-7B, respectively. On the contrary, TaFKBP21-7-7B/TaFKBP23-7-7D and TaFKBP24-3-7A registered the lowest increase in transcript level after acclimation (2.5-fold) and at 37 °C-50 °C-4.5 h (2.7-fold). However, these genes were the only members of Group 1 which depicted significantly higher expression compared to control 25 °C when exposed to direct heat stress at 50 °C for 4.5 h (Figure 8A).
The expression of Group 2 genes, compared to control, was enhanced significantly only when the thermal stress (50 °C-4.5 h) was imposed after acclimation (Figure 8B). Exposure to acclimation temperature (37 °C-2 h) and direct heat stress had no appreciable effect on the expression of these genes (Figure 8B). The genes that depicted an increase in expression in response to both direct heat stress and after acclimation were categorised in Group 3 (Figure 9A). Among the Group 3 genes, the increase in transcript abundance by heat stress at 50 °C ranged from 2.9-fold (TaFKBP16-1-5B/TaFKBP16-2-5D/TaFKBP16-3-5A) to 9.7-fold (TaFKBP47-1-5A/TaFKBP47-2-5B/TaFKBP47-3-5D) without acclimation, and from 1.9-fold (TaFKBP16-1-5B/TaFKBP16-2-5D/TaFKBP16-3-5A) to 19.9-fold (TaFKBP71-1-2B/TaFKBP71-2-2A/TaFKBP72-1-2D) after acclimation (37 °C-50 °C-4.5 h). The expression of these genes was not affected by sub-lethal temperature (37 °C for 2 h).
Contrary to Group 1, 2 and 3, the Group 4 comprised a single TaFKBP triplet, TaFKBP20-1-6A/TaFKBP20-2-6D/TaFKBP20-3-6B, that showed increase in transcript level (3.7-fold) only in response to direct exposure to 50 °C but was unaffected by any other temperature regime (Figure 9B). The genes TaFKBP16-4-3D/TaFKBP16-5-3A/TaFKBP16-6-3B and TaFKBP36-1-5A, whose expression was not responsive to any of the temperature treatments, were grouped into Group 5 (Figure 9C). Expression analysis of TaPar genes revealed that compared to 25 °C control, the transcript levels of TaPar32-1-2D/TaPar32-2-2B/TaPar32-3-2A and TaPar15-1-5D/TaPar15-2-5B/TaPar15-3-5A were upregulated by heat stress after acclimation (37 °C-50 °C-4.5 h), but were not responsive to direct heat stress (50 °C for 4.5 h) (Figure 10). On the contrary, the expression of TaPar13-1-2D/TaPar13-2-2A/TaPar13-3-2B was enhanced only during acclimation (37 °C for 2 h), but was not affected by direct thermal stress.
Expression analysis of TaFKBPs and TaPars was also performed digitally under heat (40 °C for 1 h and 6 h) and drought stress conditions using RNAseq expression data (Figure 11; Liu Z et al., 2015). Heatmap analysis revealed differential modulation of several members of the TaFKBP gene family under heat stress. Of the 71 different TaFKBP genes, TaFKBP64-1-2A and TaFKBP65-1-2B showed substantial upregulation at 40 °C over control (Figure 11A). The significant increase in expression of TaFKBP62-1-7A/TaFKBP62-2-7B during acclimation phase (37 °C-50 °C-4.5 h) and their subsequent downregulation at higher temperature (50 °C-4.5 h), as observed in our study, is also supported by heatmap analysis which showed upregulation of these genes only when exposed to 40 °C for only 1 h, with prolonged stress for 6 h resulting in a decrease (Figure 11A). Induction of Group 3 genes TaFKBP71-2-2A/TaFKBP72-1-2D (Figure 9A) during acclimation is also supported by digital expression analysis, which showed higher expression after heat stress. Similarly, decrease in expression of TaPar13-2-2A and TaPar13-3-2B genes in response to direct heat stress (50 °C) is also consistent with the results of digital analysis that revealed downregulation of these genes at 40 °C (Figure 11B). Difference in the expression profile of several other genes, such as TaFKBP20-1-6A/TaFKBP20-2-6D/TaFKBP20-3-6B, observed between the present study and digital analysis, may be ascribed to difference in the temperature regime and the genotype used.
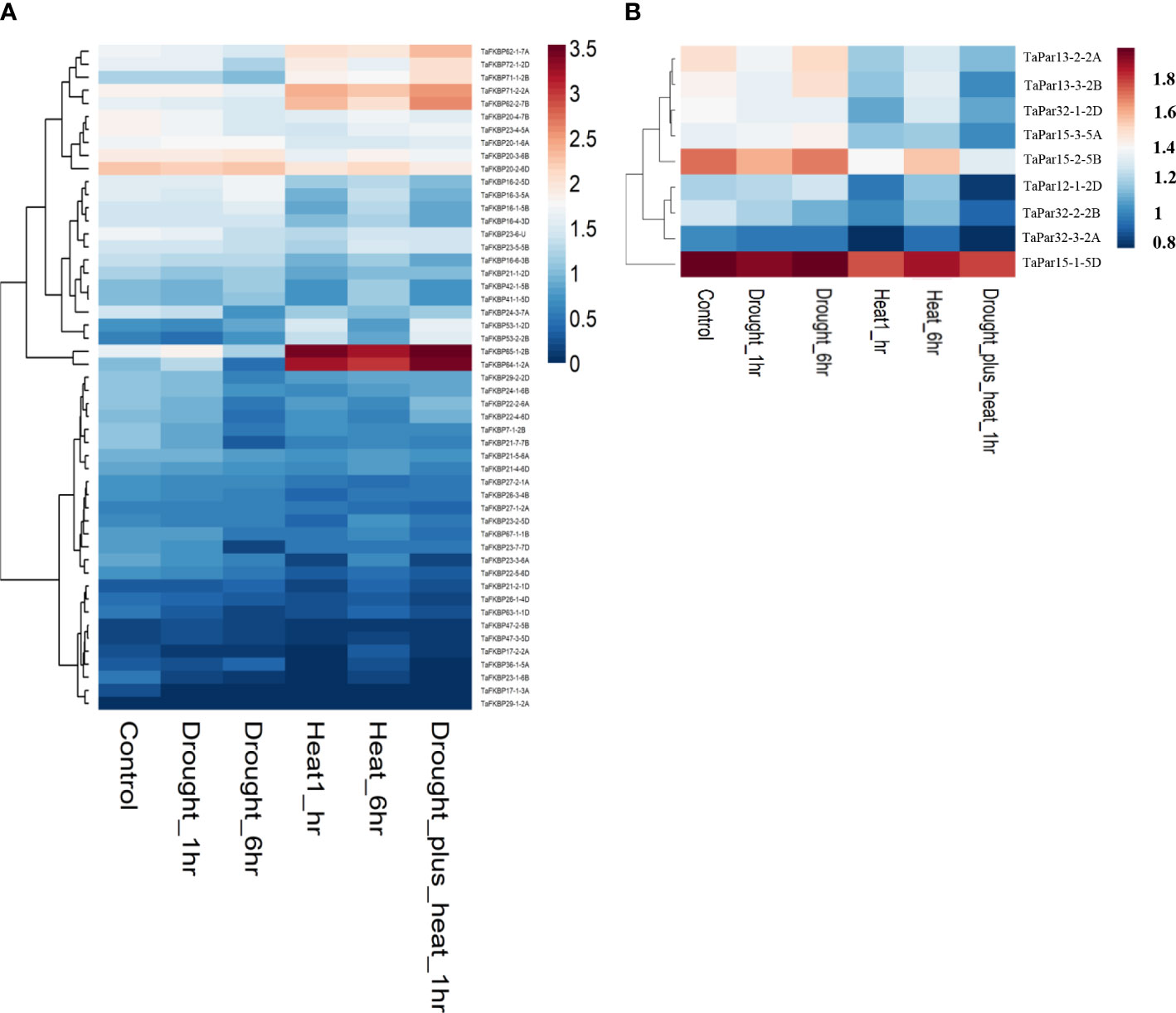
Figure 11 Heatmap showing expression of wheat FKBP (A) and parvulin (B) encoding genes under heat and drought stress conditions. Expression values were derived from the data available at WheatExp database (https://wheat.pw.usda.gov/WheatExp/) pertaining to treatment of one week old seedlings with heat stress at 40°C, drought or both for 1 h and 6 h duration (Liu Z et al., 2015). The scaled color gradient varies from blue (downregulation) to red (upregulation).
Heat shock elements have been implicated in regulation of genes by thermal stress (Schὂffl et al., 1998). Therefore, to understand the role of cis-regulatory elements in the stress-responsiveness of different genes, 2.0 kb upstream regions of these genes were examined for the identification of heat stress responsive elements (Supplementary Table S11). These analyses resulted in identification of different heat shock elements (HSEs), such as stress response element (STRE), TTC rich type 1, TTC rich type 2, TTC rich type 3, TTC rich type 4, Gap type 1, Gap type 2, Gap type 3 and Perfect HSE (Qazi et al., 2020), which with the exception of few members, were observed in varying number (0-23) in the upstream regions of all genes. The perfect HSEs (Shakeel et al., 2011) were identified in the upstream regions of only seven TaFKBP genes (TaFKBP20-1-6A, TaFKBP22-1-2B, TaFKBP23-5-5B, TaFKBP61-1-2D, TaFKBP77-1-2B, TaPTPA41-1-7D* and TaPTPA42-2-7A), out of which significant heat-induced increase in expression was observed only for TaFKBP20-1-6A (Group 4) on exposure to direct heat stress at 50 °C, and for TaFKBP22-1-2B (Group 3) at both 37 °C-50 °C-4.5 h and 50 °C-4.5 h (Figure 8). Similarly, the increase in expression observed at 37 °C-50 °C-4.5 h for the TaPar homeolog triplet TaPar15-1-5D/TaPar15-2-5B and TaPar15-3-5A could be due to the presence of HSEs present as STREs. Though the Group 5 genes did not show any change in transcript levels under any of the heat stress conditions, the upstream region of these genes also showed the presence of stress responsive cis elements, suggesting their likely role under different stress conditions.
Protein-protein interaction network
The PPI networks of TaFKPBs, TaPars and TaPTPAs (Figure 12) were constructed using the STRING database (Szklarczyk et al., 2019). In total, TaFKBPs matched to 19 distinct A. thaliana orthologs, with identities ranging from 34.9 to 91.1% (Supplementary Table S12). As observed, the PPI network of FKBPs consisted of 29 nodes and 170 edges (Figure 12A), with the former showing varied degree of interaction ranging from 2 to 23 (Supplementary Table S13). Among TaFKBPs, the highest degree of interaction (19) was exhibited by TaFKBP20-3-6B and TaFKBP20-7-7A, while the lowest (6) was shown by TaFKBP47-3-5D and TaFKBP53-2-2B, suggesting that the FKBP proteins were highly linked with other proteins and are likely to play important roles in different biological processes. The k-means clustering grouped all the interacting partners into three clusters of size 13, 12 and 4 (Supplementary Table S14). The 12 TaPars matched to 3 distinct A. thaliana orthologs, with identities of 47.5 to 90.6% (Supplementary Table S12). The PPI network of TaPars (Figure 12B) consisted of 13 nodes and 24 edges, and the degree of interaction of nodes varied from 1 to 6 (Supplementary Table S13). The highest degree of interaction (7) among these proteins was exhibited by TaPar15-3-5A, and the lowest (4) by TaPar32-3-2A. The k-means clustering grouped all the interacting partners into three clusters of size 13, 12 and 4 (Supplementary Table S14). On the other hand, the three TaPTPA proteins matched a single A. thaliana, homologue with identity ranging from 57.8 to 59.1% (Supplementary Table S12). The PPI network of PTPAs (Figure 12C) consisted of 11 nodes and 47 edges with varied degree of interaction (Supplementary Table S13). TaPTPAs were clustered into three groups containing 3 to 4 members. As observed for TaFKBPs and TaPars, the PPI network of TaPTPAs also formed clusters with interacting partners, indicating their probable functions in the biological systems (Supplementary Table S14).
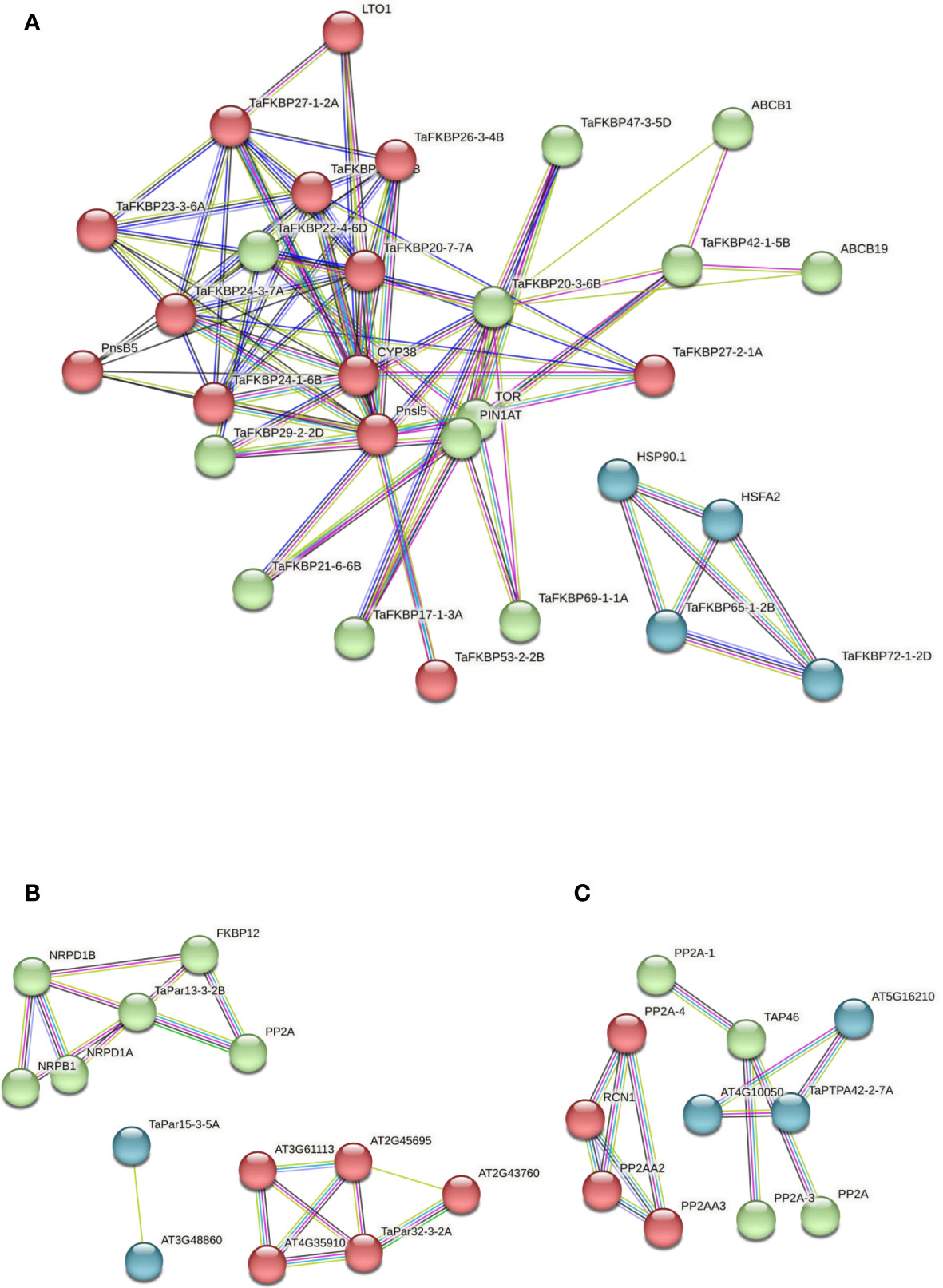
Figure 12 Protein-protein interaction (PPI) networks of wheat FKPBs (A), parvulins (B), and PTPAs (C), constructed using the STRING database version 11.5 (https://string-db.org/; Szklarczyk et al., 2019). The clusters obtained using k-means clustering in each of the protein family are colored in red, green and blue. The network nodes represent proteins and the edges represent associations. The color of the edge line indicates the type of evidence i.e., violet: fusion, green: neighborhood, blue: cooccurrence, purple: experimental, yellow: text mining, light blue: database, black: coexpression.
Discussion
The FKBPs, parvulins and PTPAs are imperative for cis-trans isomerisation of the peptidyl-prolyl bonds, and several studies have revealed their essential role in diverse cellular processes (Hanes et al., 1989; Luan et al., 1994; Lu et al., 1996; Janssens and Goris, 2001; Van Hoof and Goris, 2003; Gopalan et al., 2004; Meiri et al., 2010; Yu et al., 2012; Singh et al., 2021). The availability of whole genome sequences has enabled detailed in silico analysis and characterization of these gene families in different plant species. Though previous studies characterized FKBPs in several plants (He et al., 2004; Gollan and Bhave, 2010; Wang W et al., 2012; Zhang X et al., 2014; Dong et al., 2018; Waseem et al., 2018), information on this important family of genes, until this study, was lacking in wheat. The present analysis revealed that the hexaploid wheat genome encodes for 71 different TaFKBP genes, which are unevenly distributed on the 21 different chromosomes. On the contrary, the TaPTPA and TaPar genes are confined to a single or only few chromosomes, respectively. The expansion of FKBP gene family in wheat appears to be the result of different duplication events, such as whole genome, segmental and tandem duplications. Synteny analysis indicated the presence of many collinear blocks of TaFKBP genes in wheat and its sub-genome donors. In addition, the presence of triplet homeologues for most of the TaFKBPs suggest that whole genome duplication due to polyploidization is likely the major force behind evolution of FKBP gene family in wheat, which was also reported for cyclophilin, 14-3-3 and invertase genes (Singh et al., 2019; Shao et al., 2021; Wang et al., 2021). However, compared to their corresponding sub-genome donors, the difference in prevalance of FKBP genes on some hexaploid wheat chromosomes indicates either a gain due to duplication or loss during the course of evolution (Shao et al., 2021).
Phylogenetic analysis showed strong clustering of homoeologue triplets of TaFKBP, TaPar and TaPTPA proteins, with overall grouping influenced by localization patterns and arrangement of sequence motifs. The monocot and dicot homologues of FKBPs and parvulins were clustered together in each of the sub-groups, indicating the existence of these genes in plants even before divergence of the two (Figures 7A, B). Similar observation was also reported for the invertase gene family in common wheat (Wang et al., 2021). On the contrary, the monocot and dicot PTPAs were clustered separately, indicating the acquisition of these genes by wheat genome after differentiation of dicots and monocots. Structural analysis of the genes revealed that the length of exons, which is variable in TaFKBP genes, could be related to their orthologs. This is evident, since the length of exon 3 (34 bp), 4 (62 bp) and 5 (53 bp) in homeologues TaFKBP20-1-6A, TaFKBP20-2-6D, and TaFKBP20-3-6B is similar to the length of exon 2, 3 and 4 in their A. thaliana ortholog AtFKBP12 (Dong et al., 2018). Further, the length of these exons is also consistent with their other orthologs in B. rapa, O. sativa, P. persica, S. lycopersicum and M. domestica (Dong et al., 2018). The uniform pattern of exons was also observed for all the TaPar genes, and it corresponds with their A. thaliana orthologs AtPIN1, AtPIN2 and AtPIN3 (He et al., 2004). As reported for FKBP genes of S. lycopersicum (Waseem et al., 2018), considerable diversity was also noticed in the 5’ and 3’ UTR regions of TaFKBPs.
As observed earlier for cyclophilin proteins in wheat, the FKBPs also show variability in their domain architecture and localization, implying functional diversification (Singh et al., 2019). The presence of TPR domains in majority of the multi-domain FKBPs suggests their likely role in protein-protein interactions (Kurek et al., 2002). Two of the homeologous groups of FKBPs, TaFKBP47-1-5A/TaFKBP47-2-5B/TaFKBP47-3-5D, and TaFKBP48-1-2A/TaFKBP53-1-2D/TaFKBP53-2-2B also contain an NPL-domain along with the FKBD. It is speculated that as reported for AtFKBP53, which contains an NPL-domain (Li and Luan, 2010), these wheat FKBPs might also be acting as histone chaperones, which however, requires experimental validation. Different types of structural motifs observed in TaFKBPs indicate towards their functional specificities. For instance, the presence of motifs 2 and 15 suggests their likely participation in protein-protein interactions, since other multi-domain FKBPs viz., wFKBP73 and wFKBP77 in wheat (Reddy et al., 1998), AtFKBP62 in A. thaliana (Meiri and Breiman, 2009) and human FKBP52 (Pirkl and Buchner, 2001), which contain these sequences, were implicated in interaction with the molecular chaperone HSP90 via their TPR domain. Similarly, the existence of the motifs 11 and 12 in the N-terminus NPL-domain of two groups of homeologues (TaFKBP47-1-5A/TaFKBP47-2-5B/TaFKBP47-3-5D, and TaFKBP48-1-2A/TaFKBP53-1-2D/TaFKBP53-2-2B) suggests that these proteins may be acting as histone chaperons, as observed for their orthologous proteins AtFKBP53 in A. thaliana (Li and Luan, 2010), and FPR3 and FPR4 in S. cerevisiae (Benton et al., 1994; Kuzuhara and Horikoshi, 2004). The motif 10 (cytidylate kinase 2) was observed in several (17) FKBPs implying their likely role in pyrimidine biosynthesis, as reported for the cytidylate kinase protein Cmk (Wojtkiewicz et al., 2020). The Aspartic_Protease (ASP_Protease) motif predicted in a homeolog triplet TaFKBP48-1-2A/TaFKBP53-1-2D/TaFKBP53-2-2B indicate the role of these proteins in biotic and abiotic stress responses in plants, since these proteases are known to regulate the pathogenesis-related proteins (Rodrigo et al., 1989; Rodrigo et al., 1991), and nitrogen remobilization under water stress conditions (de Carvalho et al., 2001). The homeologues TaFKBP47-1-5A/TaFKBP47-2-5B/TaFKBP47-3-5D, predicted to harbour motif 14 (Domain of unknown function/CDC45 like protein), may be involved in cell cycle regulation, as demonstrated for the AtCDC45 in A. thaliana (Stevens et al., 2004).
All wheat parvulins showed the presence of the characteristic rotamase domain, but the multi-domain parvulins TaPar32-1-2D/TaPar32-2-2B/TaPar32-3-2A also contain an additional rhodanese-like domain towards C-terminus, akin to their A. thaliana ortholog AtPin3 (He et al., 2004). The different TaPars exhibited homeolog-dependent differences with respect to the different motifs. For instance, the occurrence of motif 12 was observed only in TaPar32-1-2D and TaPar32-3-2A, but not in their third homeolog TaPar32-2-2B. Similarly, motifs 3 (PPIC-type PPIase domain) and 6 (function not known) were observed most commonly (TaPar11-1-3B, TaPar11-2-4A, TaPar12-1-2D, TaPar13-1-2D, TaPar13-2-2A, TaPar13-3-2B), while Motifs 7 and 11 (functions not known) were noted only in few of these proteins (TaPar11-1-3B, TaPar11-2-4A). It is likely that the differential presence of the motifs might signify functional divergence in the roles of different parvulin homeologues.
Expression analysis of TaFKBP, TaPar and TaPTPA genes
The expression of TaFKBP, TaPar and TaPTPA genes was analysed in response to heat stress to understand their likely role in thermotolerance of wheat seedlings. These results revealed that the expression of different TaFKBP and TaPar genes in wheat is modulated differentially by heat stress, which is in accordance with the earlier studies in O. sativa, Z. mays, M. domestica, and Penicillium (Nigam et al., 2008; Yu et al., 2012; Dong et al., 2018; Waseem et al., 2018; Singh et al., 2021). For instance, the heat stress-induced increase in TaFKBP61-2-7D/TaFKBP62-1-7A/TaFKBP62-2-7B (Group 1) was also observed for their maize ortholog, ZmFKBP62a (Yu et al., 2012). Similarly, the increase in expression of TaFKBP63-1-1D/TaFKBP67-1-1B/TaFKBP69-1-1A (Group 2) after exposure to lethal heat stress following acclimation (37 °C-50 °C-4.5 h) is consistent with the temperature-induced increase in expression of ZmFKBP72 in maize (Yu et al., 2012). Apart from the heat-upregulated members, the TaFKBP and TaPar families in wheat also possess genes that are downregulated under thermal stress, indicating functional divergence in their roles, which might be contributing to maintenance of homeostasis under stress conditions. The identification of commonly found heat shock elements in the promoter regions of heat-stress inducible genes, such as TaFKBP20-1-6A (Group 4) and TaFKBP21-1-2D/TaFKBP27-1-2A (Group 3), suggests their putative role in stress response, that, however, needs to be explored further by overexpression and knockdown approaches. Of the different FKBP genes analyzed, TaFKBP61-2-7D/TaFKBP62-1-7A/TaFKBP62-2-7B (Group 1) showed highest increase in transcript levels during acclimation (27.7-fold) and lethal heat stress following acclimation (40.8-fold). These proteins are approximately 98.03% identical with the wFKBP73, that in addition to its PPIase activity was also shown to have chaperonic function (Supplementary Figure S2A; Kurek et al., 2002). Therefore, it is likely that as observed for the wheat cyclophilin TaCYPA-1 (Kaur et al., 2016), the enhanced levels of TaFKBP61-2-7D/TaFKBP62-1-7A/TaFKBP62-2-7B under heat stress might be providing protection against heat-induced damage by preventing protein aggregation and mediating protein folding through their chaperone/PPIase activity. As observed for TaFKBPs, expression of TaPars was also affected differentially by heat stress, prompting us to speculate that besides developmental regulation, these proteins might have an important role in stress response of plants. Though expression of none of the three TaPTPA homeologues was detected in the present study, role of these genes can’t be ruled out and, hence, requires further analysis.
Protein-Protein interaction network
The PPI networks of TaFKPBs, TaPars and TaPTPAs demonstrated that these proteins interact among themselves as well as with other proteins, suggesting their involvement in diverse biological functions (Figure 12). The PPI network of FKBPs showed that the TaFKBP65-1-2B and TaFKBP72-1-2D might be playing essential roles in response to abiotic stress (Figure 12A), since both of them share the network cluster with heat shock protein 90-1 (HSP90.1) and heat shock transcription factor A-2 (HSFA2), indicating their likely role in regulation of thermotolerance in plants, as reported for ROF1 in A. thaliana (AtFKBP62) (Meiri and Breiman, 2009). The role of TaFKBP65-1-2B and TaFKBP72-1-2D in thermotolerance is also substantiated by their enhanced expression in heat stress, as revealed by heatmap analysis (Figure 11A). Similarly, the putative interaction of TaFKBP42-1-5B with ABC transporter B family members ABCB1 and ABCB19 is consistent with the previous study reporting interaction of Arabidopsis ortholog AtFKBP42 (TWD1) with vacuolar ABC transporters, which is important for auxin transport (Geisler et al., 2003; Geisler et al., 2004).
Computational analysis also predicted the interaction between TaPar13-3-2B and serine/threonine- protein phosphatase 2α (PP2A), implying its role in regulation of cell cycle in plants (Yeh and Means, 2007; Figure 12B). The putative interaction between TaPTPA42-2-7A and HEAT (huntingtin-elongation-A subunit-TOR) repeat-containing protein (AT5G16210) indicates its association with the PP2A scaffolding subunit that comprises of 15 tandem repeats of HEAT motif (Walter et al., 1989; Hemmings et al., 1990; Figure 12C). Though the PPI network analyses indicate the role of TaFKBPs, TaPars and TaPTPA in different cellular functions, understanding the mechanisms underlying these processes requires detailed analysis of in vivo interactions by BiFC/FRET approaches.
Conclusion
To conclude, the genes encoding different FKBP, parvulin and PTPA proteins were identified for the first time in wheat genome. Further, characterization of gene structure, domain architecture, motif analysis, cis-regulatory elements and evolutionary analysis was also carried out. The results of this study suggest functional divergence of these proteins, that needs to be elucidated by experimental approaches. The expression profiling of these genes at the seedling stage revealed that the TaFKBP and TaPar genes in wheat are differentially regulated in response to heat stress, implying their role in stress response. The findings of this study will provide the basis for further functional characterization of these gene families in wheat, that may lead to their application in improvement of crop plants through breeding and biotechnological approaches.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
PS: conceived idea, designed the experiments, reviewing and editing of the manuscript; HS: designed the bioinformatic study, participated in writing of manuscript, data analysis; AS: conducted bioinformatic analysis and performed qRT-PCR analysis of genes, writing of manuscript, data curation and analysis; KK: helped in qRT-PCR analysis of genes, data analysis; AK: contributed in writing of manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
We gratefully acknowledge Department of Biotechnology, Government of India, New Delhi for supporting the MSc. Biotechnology Teaching Program. AS is thankful to Department of Science and Technology (DST), Govt. of India for providing the DST-INSPIRE research fellowship (Registration number. IF190202; Sanction order: DST/INSPIRE Fellowship/2019/IF190202, Dated: 30-11-2020). KK acknowledges University Grant Commission, New Delhi, Government of India for the Rajiv Gandhi National Fellowship (Sanction number: F1-17.1/2017-18/RGNF-2017-18-SC-PUN-44307/)Sa-III/website, Dated: 28-07-2017). We also acknowledge the support provided by the Department of Science and Technology, Government of India in the form of the “Fund for Improvement of Science & Technology Infrastructure (FIST)” grant (D. O. No. SR/FST/PG College/2009) to develop computational resources at the Department of Bioinformatics, Hans Raj Mahila MahaVidyalaya, Jalandhar. The authors acknowledge Dr. Sanjay Kumar, Director CSIR- Institute of Himalayan Bioresource Technology, Palampur for the institutional support. Thanks are also due to Prof. Wen Yao, National Key Laboratory of Crop Genetic Improvement, National Center of Plant Gene Research (Wuhan), Huazhong Agricultural University, Wuhan, China, College of Life Sciences, Henan Agricultural University, Zhengzhou, Chin for his help in fine tuning Circos plots developed through the shinyCircos application.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1053524/full#supplementary-material
References
Aviezer-Hagai, K., Skovorodnikova, J., Galigniana, M., Farchi-Pisanty, O., Maayan, E., Bocovza, S., et al. (2007). Arabidopsis immunophilins ROF1 (AtFKBP62) and ROF2 (AtFKBP65) exhibit tissue specificity, are heat-stress induced, and bind HSP90. Plant Mol. Biol. 63, 237–255. doi: 10.1007/s11103-006-9085-z
Bailey, T. L., Johnson, J., Grant, C. E., Noble, W. S. (2015). The MEME suite. Nucl. Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Benton, A. L., Sivan, A. B., Hamsher, K. D., Varney, N. R., Spreen, O. (1994). Contributions to neuropsychological assessment: A clinical manual (USA: Oxford University Press).
Boudko, S. P., Ishikawa, Y., Nix, J., Chapman, M. S., Bächinger, H. P. (2014). Structure of human peptidyl-prolyl cis–trans isomerase FKBP22 containing two EF-hand motifs. Protein Sci. 23, 67–75. doi: 10.1002/pro.2391
Breiman, A., Fawcett, T. W., Ghirardi, M. L., Mattoo, A. K. (1992). Plant organelles contain distinct peptidylprolyl cis-trans isomerases. J. Biol. Chem. 267, 21293–21296. doi: 10.1002/pro.2391
Carol, R. J., Breiman, A., Erel, N., Vittorioso, P., Bellini, C. (2001). PASTICCINO1 (AtFKBP70) is a nuclear-localised immunophilin required during Arabidopsis thaliana embryogenesis. Plant Sci. 161, 527–535. doi: 10.1016/S0168-9452(01)00437-X
Carvalho, M. H. C., d’Arcy-Lameta, A., Roy-Macauley, H., Gareil, M., El Maarouf, H., Pham-Thi, A. T., et al. (2001). Aspartic protease in leaves of common bean (Phaseolus vulgaris l.) and cowpea (Vigna unguiculata l. Walp): enzymatic activity, gene expression and relation to drought susceptibility. FEBS Lett. 492, 242–246. doi: 10.1016/S0014-5793(01)02259-1
Chen, Q., Chen, Q. J., Sun, G. Q., Zheng, K., Yao, Z. P., Han, Y. H., et al. (2019). Genome-wide identification of cyclophilin gene family in cotton and expression analysis of the fibre development in Gossypium barbadense. Int. J. Mol. Sci. 20, 349. doi: 10.3390/ijms20020349
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Dong, Q., Mao, K., Duan, D., Zhao, S., Wang, Y., Wang, Q., et al. (2018). Genome-wide analyses of genes encoding FK506-binding proteins reveal their involvement in abiotic stress responses in apple. BMC Genomics 19, 1–16. doi: 10.1186/s12864-018-5097-8
Eddy, S. R. (2011). Accelerated profile HMM searches. PloS Comp. Biol. 7, e1002195. doi: 10.1371/journal.pcbi.1002195
Feldman, M., Levy, A. A. (2005). Allopolyploidy–a shaping force in the evolution of wheat genomes. Cytogenet. Genome Res. 109, 250–258. doi: 10.1159/000082407
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2016). The pfam protein families database: towards a more sustainable future. Nucl. Acids Res. 44, 279–285. doi: 10.1093/nar/gkv1344
Fischer, G., Wittmann-Liebold, B., Lang, K., Kiefhaber, T., Schmid, F. X. (1989). Cyclophilin and peptidyl-prolyl cis-trans isomerase are probably identical proteins. Nature 337, 476–478. doi: 10.1038/337476a0
Galat, A., Rivière, S. (1998). Peptidyl-prolyl cis/trans isomerases: Protein profile (Oxford University Press Oxford).
Gasser, C. S., Gunning, D. A., Budelier, K. A., Brown, S. M. (1990). Structure and expression of cytosolic cyclophilin/peptidyl-prolyl cis-trans isomerase of higher plants and production of active tomato cyclophilin in. Escherichia coli. Proc. Natl. Acad. Sci. 87, 9519–9523. doi: 10.1073/pnas.87.24.9519
Geisler, M., Bailly, A. (2007). Tete-a-tete: the function of FKBPs in plant development. Trends Plant Sci. 12, 465–473. doi: 10.1016/j.tplants.2007.08.015
Geisler, M., Girin, M., Brandt, S., Vincenzetti, V., Plaza, S., Paris, N., et al. (2004). Arabidopsis immunophilin-like TWD1 functionally interacts with vacuolar ABC transporters. Mol. Bio. Cell. 15, 3393–3405. doi: 10.1091/mbc.E03–11–0831
Geisler, M., Kolukisaoglu, H. U., Bouchard, R., Billion, K., Berger, J., Saal, B., et al. (2003). TWISTED DWARF1, a unique plasma membrane-anchored immunophilin-like protein, interacts with arabidopsis multidrug resistance-like transporters AtPGP1 and AtPGP19. Mol. Bio. Cell. 14, 4238–4249. doi: 10.1091/mbc.e02-10-0698
Ge, L., Zhang, K., Cao, X., Weng, Y., Liu, B., Mao, P., et al. (2020). Sequence characteristics of Medicago truncatula cyclophilin family members and function analysis of MsCYP20-3B involved in axillary shoot development. Mol. Biol. Rep. 47, 907–919. doi: 10.1007/s11033-019-05183-x
Goldberg, T., Hecht, M., Hamp, T., Karl, T., Yachdav, G., Ahmed, N., et al. (2014). LocTree3 prediction of localization. Nucl. Acids Res. 42, 350–355. doi: 10.1093/nar/gku396
Gollan, P. J., Bhave, M. (2010). Genome-wide analysis of genes encoding FK506-binding proteins in rice. Plant Mol. Biol. 72, 1–16. doi: 10.1007/s11103-009-9547-1
Gopalan, G., He, Z., Balmer, Y., Romano, P., Gupta, R., Héroux, A., et al. (2004). Structural analysis uncovers a role for redox in regulating FKBP13, an immunophilin of the chloroplast thylakoid lumen. Proc. Natl. Acad. Sci. U.S.A. 101, 13945–13950. doi: 10.1073/pnas.0405240101
Göthel, S., Marahiel, M. (1999). Peptidyl-prolyl cis-trans isomerases, a superfamily of ubiquitous folding catalysts. CMLS Cell. Mol. Life Sci. 55, 423–436. doi: 10.1007/s000180050299
Hanes, S. D., Shank, P. R., Bostian, K. A. (1989). Sequence and mutational analysis of ESS1, a gene essential for growth in Saccharomyces cerevisiae. Yeast 5, 55–72. doi: 10.1002/yea.320050108
Hanhart, P., Thieß, M., Amari, K., Bajdzienko, K., Giavalisco, P., Heinlein, M., et al. (2017). Bioinformatic and expression analysis of the Brassica napus l. cyclophilins. Sci. Rep. 7, 1–17. doi: 10.1038/s41598-017-01596-5
Harrar, Y., Bellec, Y., Bellini, C., Faure, J. D. (2003). Hormonal control of cell proliferation requires PASTICCINO genes. Plant Physiol. 132, 1217–1227. doi: 10.1002/yea.320050108
He, Z., Li, L., Luan, S. (2004). Immunophilins and parvulins. superfamily of peptidyl prolyl isomerases in Arabidopsis. Plant Physiol. 134, 1248–1267. doi: 10.1104/pp.103.031005
Hemmings, B. A., Adams-Pearson, C., Maurer, F., Mueller, P., Goris, J., Merlevede, W., et al. (1990). Alpha.-and. beta.-forms of the 65-kDa subunit of protein phosphatase 2A have a similar 39 amino acid repeating structure. Biochemistry 29, 3166–3173. doi: 10.1021/bi00465a002
Holub, E. B. (2001). The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2, 516–527. doi: 10.1038/35080508
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Janssens, V., Goris, J. (2001). Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. 353, 417–439. doi: 10.1042/0264-6021:3530417
Jo, S. H., Park, H. J., Lee, A., Jung, H., Park, J. M., Kwon, S. Y., et al. (2022). The arabidopsis cyclophilin CYP18-1 facilitates PRP18 dephosphorylation and the splicing of introns retained under heat stress. Plant Cell 34, 2383–2403. doi: 10.1093/plcell/koac131
Jordens, J., Janssens, V., Longin, S., Stevens, I., Martens, E., Bultynck, G., et al. (2006). The protein phosphatase 2A phosphatase activator is a novel peptidyl-prolyl cis-trans-isomerase. J. Biol. Chem. 281, 6349–6357. doi: 10.1074/jbc.M507760200
Joseph, J. D., Heitman, J., Means, A. R. (1999). Molecular cloning and characterization of Aspergillus nidulans cyclophilin b. Fungal Genet. Biol. 27, 55–66. doi: 10.1006/fgbi.1999.1131
Juturu, V. N., Ramesh, P., Gopalakrishna, M., Mallikarjuna, G., Talakayala, A., Puli, C. O. R., et al. (2021). Pigeonpea cyclophilin (CcCYP) enhances abiotic stress tolerance in rice. Rice Sci. 28, 3.
Kang, C. B., Hong, Y., Dhe-Paganon, S., Yoon, H. S. (2008). FKBP family proteins: immunophilins with versatile biological functions. Neurosignals 16, 318–325. doi: 10.1159/000123041
Kaur, G., Singh, S., Dutta, T., Kaur, H., Singh, B., Pareek, A., et al. (2016). The peptidyl-prolyl cis-trans isomerase activity of the wheat cyclophilin, TaCypA-1, is essential for inducing thermotolerance in Escherichia coli. Biochimie Open 2, 9–15. doi: 10.1016/j.biopen.2015.11.003
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evo. 35, 1547. doi: 10.1093/molbev/msy096
Kurek, I., Pirkl, F., Fischer, E., Buchner, J., Breiman, A. (2002). Wheat FKBP73 functions in vitro as a molecular chaperone independently of its peptidyl prolyl cis-trans isomerase activity. Planta 215, 119–126. doi: 10.1007/s00425-001-0722-0
Kuzuhara, T., Horikoshi, M. (2004). A nuclear FK506-binding protein is a histone chaperone regulating rDNA silencing. Nat. Struct. Mol. Biol. 11, 275–283. doi: 10.1038/nsmb733
Letunic, I., Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucl. Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290
Li, H., Luan, S. (2010). AtFKBP53 is a histone chaperone required for repression of ribosomal RNA gene expression in Arabidopsis. Cell Res. 20, 357–366. doi: 10.1038/cr.2010.22
Lima, A., Lima, S., Wong, J. H., Phillips, R. S., Buchanan, B. B., Luan, S. (2006). A redox-active FKBP-type immunophilin functions in accumulation of the photosystem II supercomplex in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 103, 12631–12636. doi: 10.1073/pnas.0605452103
Liu, J., Farmer, Jr. J.D., Lane, W. S., Friedman, J., Weissman, I., Schreiber, S. L. (1991). Calcineurin is a common target of cyclophilin-cyclosporin a and FKBP-FK506 complexes. Cell 66, 807–815. doi: 10.1016/0092-8674(91)90124-h
Liu, H., Shen, J., Yuan, C., Lu, D., Acharya, B. R., Wang, M., et al. (2021). The cyclophilin ROC3 regulates ABA-induced stomatal closure and the drought stress response of Arabidopsis thaliana. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.668792
Liu, W., Xie, Y., Ma, J., Luo, X., Nie, P., Zuo, Z., et al. (2015). IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics 31, 3359–3361. doi: 10.1093/bioinformatics/btv362
Liu, Z., Xin, M., Qin, J., Peng, H., Ni, Z., Yao, Y., et al. (2015). Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and sdrought acclimation in wheat (Triticum aestivum l.). BMC Plant Biol. 5, 152. doi: 10.1186/s12870-015-0511-8
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luan, S., Albers, M. W., Schreiber, S. L. (1994). Light-regulated, tissue-specific immunophilins in a higher plant. Proc. Natl. Acad. Sci. U.S.A. 91, 984–988. doi: 10.1073/pnas.91.3.984
Luan, S., Kudla, J., Gruissem, W., Schreiber, S. L. (1996). Molecular characterization of a FKBP-type immunophilin from higher plants. Proc. Natl. Acad. Sci. U.S.A. 93, 6964–6969. doi: 10.1073/pnas.93.14.6964
Lu, Y. F., Tomizawa, K., Moriwaki, A., Hayashi, Y., Tokuda, M., Itano, T., et al. (1996). Calcineurin inhibitors, FK506 and cyclosporin a, suppress the NMDA receptor-mediated potentials and LTP, but not depotentiation in the rat hippocampus. Brain Res. 729, 142–146. doi: 10.1016/0006-8993(96)00568-9
Magnusdottir, A., Stenmark, P., Flodin, S., Nyman, T., Hammarstrom, M., Ehn, M., et al. (2006). The crystal structure of a human PP2A phosphatase activator reveals a novel fold and highly conserved cleft implicated in protein-protein interactions. J. Biol. Chem. 281, 22434–22438. doi: 10.1074/jbc.C600100200
Mainali, H. R., Chapman, P., Dhaubhadel, S. (2014). Genome-wide analysis of cyclophilin gene family in soybean (Glycine max). BMC Plant Biol. 14, 282. doi: 10.1186/s12870-014-0282-7
Meiri, D., Breiman, A. (2009). Arabidopsis ROF1 (FKBP62) modulates thermotolerance by interacting with HSP90.1 and affecting the accumulation of HsfA2-regulated sHSPs. Plant J. 59, 387–399. doi: 10.1111/j.1365-313X.2009.03878.x
Meiri, D., Tazat, K., Cohen-Peer, R., Farchi-Pisanty, O., Aviezer-Hagai, K., Avni, A., et al. (2010). Involvement of Arabidopsis ROF2 (FKBP65) in thermotolerance. Plant Mol. Biol. 72, 191–203. doi: 10.1007/s11103-009-9561-3
Mueller, J. W., Kessler, D., Neumann, D., Stratmann, T., Papatheodorou, P., Hartmann-Fatu, C., et al. (2006). Characterization of novel elongated parvulin isoforms that are ubiquitously expressed in human tissues and originate from alternative transcription initiation. BMC Mol. Biol. 7, 1–9. doi: 10.1186/1471-2199-7-9
Myers, T. A., Chanock, S. J., Machiela, M. J. (2020). LDlinkR: an R package for rapidly calculating linkage disequilibrium statistics in diverse populations. Front. Genet 11, 157. doi: 10.3389/fgene.2020.00157
Nigam, N., Singh, A., Sahi, C., Chandramouli, A., Grover, A. (2008). SUMO-conjugating enzyme (Sce) and FK506-binding protein (FKBP) encoding rice (Oryza sativa l.) genes: genome-wide analysis, expression studies and evidence for their involvement in abiotic stress response. Mol. Genet. Genom. 279, 371–383. doi: 10.1007/s00438-008-0318-5
O'Keefe, S. J., Tamura, J. I., Kincaid, R. L., Tocci, M. J., O'Neill, E. A. (1992). FK-506-and CsA-sensitive activation of the interleukin-2 promoter by calcineurin. Nature 357, 692–694. doi: 10.1038/357692a0
Olson, S. A. (2002). Emboss opens up sequence analysis. Brief. Bioinform. 3, 87–91. doi: 10.1093/bib/3.1.87
Park, H. J., You, Y. N., Lee, A., Jung, H., Jo, S. H., Oh, N., et al. (2020). OsFKBP20-1b interacts with the splicing factor OsSR45 and participates in the environmental stress response at the post-transcriptional level in rice. Plant J. 102, 992–1007. doi: 10.1111/tpj.14682
Pearce, S., Vazquez-Gross, H., Herin, S. Y., Hane, D., Wang, Y., Gu, Y. Q., et al. (2015). WheatExp: an RNA-seq expression database for polyploid wheat. BMC Plant Biol. 15, 299. doi: 10.1186/s12870-015-0692-1
Pirkl, F., Buchner, J. (2001). Functional analysis of the Hsp90-associated human peptidyl prolyl cis-trans isomerases FKBP51, FKBP52 and Cyp40. J. Mol. Biol. 308, 795–806. doi: 10.1006/jmbi.2001.4595
Qazi, S. R., Ahmad, S., Shakeel, S. N. (2020). HSEAT: a tool for plant heat shock element analysis, motif identification and analysis. Curr. Bioinform. 15, 196–203. doi: 10.2174/1574893614666190102151956
Rahfeld, J. U., Rücknagel, K. P., Schelbert, B., Ludwig, B., Hacker, J., Mann, K., et al. (1994). Confirmation of the existence of a third family among peptidyl-prolyl cis-trans isomerases amino acid sequence and recombinant production of parvulin. FEBS Lett. 352, 180–184. doi: 10.1016/0014-5793(94)00932-5
Reddy, R. K., Kurek, I., Silverstein, A. M., Chinkers, M., Breiman, A., Krishna, P. (1998). High-molecular-weight FK506-binding proteins are components of heat-shock protein 90 heterocomplexes in wheat germ lysate. Plant Physiol. 118, 1395–1401. doi: 10.1104/pp.118.4.1395
Robert, X., Gouet, P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucl. Acids Res. 42, W320–W324. doi: 10.1093/nar/gku316
Rodrigo, I., Vera, P., Conejero, V. (1989). Degradation of tomato pathogenesis-related proteins by an endogenous aspartyl endoproteinase. Eur. J. Biochem. 184, 663–669. doi: 10.1111/j.1432-1033.1989.tb15064.x
Rodrigo, I., Vera, P., Van Loon, L. C., Conejero, V. (1991). Degradation of tobacco pathogenesis-related proteins: evidence for conserved mechanisms of degradation of pathogenesis-related proteins in plants. Plant Physiol. 95, 616–622. doi: 10.1104/pp.95.2.616
Romano, P. G. N., Horton, P., Gray, J. E. (2004). The arabidopsis cyclophilin gene family. Plant Physiol. 134, 1268–1282. doi: 10.1104/pp.103.022160
Schὂffl, F., Prandl, R., Reindl, A. (1998). Regulation of the heat-shock response. Plant Physiol. 117, 1135–1141. doi: 10.1104/pp.117.4.1135
Shakeel, S., Haq, N. U., Heckathorn, S. A., Hamilton, E. W., Luthe, D. S. (2011). Ecotypic variation in chloroplast small heat-shock proteins and related thermotolerance in Chenopodium album. Plant Physiol. Biochem. 49, 898–908. doi: 10.1016/j.plaphy.2011.05.002
Shao, W., Chen, W., Zhu, X., Zhou, X., Jin, Y., Zhan, C., et al. (2021). Genome-wide identification and characterization of wheat 14-3-3 genes unravels the role of TaGRF6-a in salt stress tolerance by binding MYB transcription factor. Int. J. Mol. Sci. 22, 1904. doi: 10.3390/ijms22041904
Sharma, A. D., Singh, P. (2003). Effect of water stress on expression of a 20 kD cyclophilin-like protein in drought susceptible and tolerant cultivars of sorghum. J. Plant Biochem. Biotechnol. 12, 77–80. doi: 10.1007/BF03263165
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol. Syst. Biol. 7, 539. doi: 10.1038/msb.2011.75
Sigrist, C. J., De Castro, E., Cerutti, L., Cuche, B. A., Hulo, N., Bridge, A., et al. (2012). New and continuing developments at PROSITE. Nucl. Acids Res. 41, 344–347. doi: 10.1093/nar/gks1067
Singh, A. K., Datta, A., Jobichen, C., Luan, S., Vasudevan, D. (2020). AtFKBP53: a chimeric histone chaperone with functional nucleoplasmin and PPIase domains. Nucl. Acids Res. 48, 1531–1550. doi: 10.1093/nar/gkz1153
Singh, M., Kaur, K., Sharma, A., Kaur, R., Joshi, D., Chatterjee, M., et al. (2021). Genome-wide characterization of peptidyl-prolyl cis-trans isomerases in Penicillium and their regulation by salt stress in a halotolerant P. oxalicum. Sci. Rep. 11, 1–19. doi: 10.1038/s41598-021-91602-8
Singh, H., Kaur, K., Singh, S., Kaur, P., Singh, P. (2019). Genome-wide analysis of cyclophilin gene family in wheat and identification of heat stress responsive members. Plant Gene 19, 100197. doi: 10.1016/J.PLGENE.2019.100197
Singh, H., Kaur, K., Singh, M., Kaur, G., Singh, P. (2020). Plant cyclophilins: Multifaceted proteins with versatile roles. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.585212
Singh, K., Zouhar, M., Mazakova, J., Rysanek, P. (2014). Genome wide identification of the immunophilin gene family in Leptosphaeria maculans: a causal agent of blackleg disease in oilseed rape (Brassica napus). Omics J. Integr. Biol. 18, 645–657. doi: 10.1089/omi.2014.0081
Stevens, R., Grelon, M., Vezon, D., Oh, J., Meyer, P., Perennes, C., et al. (2004). A CDC45 homolog in Arabidopsis is essential for meiosis, as shown by RNA interference-induced gene silencing. Plant Cell 16, 99–113. doi: 10.1105/tpc.016865
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucl. Acids Res. 47, D607–D613. doi: 10.1093/nar/gky1131
Thirumalaikumar, V. P., Gorka, M., Schulz, K., Masclaux-Daubresse, C., Sampathkumar, A., Skirycz, A., et al. (2021). Selective autophagy regulates heat stress memory in arabidopsis by NBR1-mediated targeting of HSP90. 1 and ROF1. Autophagy 17, 2184–2199. doi: 10.1080/15548627.2020.1820778
Trivedi, D. K., Yadav, S., Vaid, N., Tuteja, N. (2012). Genome wide analysis of cyclophilin gene family from rice and arabidopsis and its comparison with yeast. Plant Signal. Behav. 7, 1653–1666. doi: 10.4161/psb.22306
Unger, T., Dym, O., Albeck, S., Jacobovitch, Y., Bernehim, R., Marom, D., et al. (2010). Crystal structure of the three FK506 binding protein domains of wheat FKBP73: evidence for a unique wFK73_2 domain. J. Struct. Funct. Genomics 11, 113–123. doi: 10.1007/s10969-010-9085-8
Van Hoof, C., Goris, J. (2003). Phosphatases in apoptosis: to be or not to be, PP2A is in the heart of the question. Biochim. Biophys. Acta 1640, 97–104. doi: 10.1016/S0167-4889(03)00029-6
Voorrips, R. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. Heredity 93, 77–78. doi: 10.1093/jhered/93.1.77
Vucich, V. A., Gasser, C. S. (1996). Novel structure of a high molecular weight FK506 binding protein from Arabidopsis thaliana. Mol. Gen. Genet. 252, 510–517. doi: 10.1007/BF02172397
Walter, G., Ferre, F., Espiritu, O., Carbone-Wiley, A. (1989). Molecular cloning and sequence of cDNA encoding polyoma medium tumor antigen-associated 61-kDa protein. Proc. Natl. Acad. Sci. 86, 8669–8672. doi: 10.1073/pnas.86.22.8669
Wang, W. W., Ma, Q., Xiang, Y., Zhu, S. W., Cheng, B. J. (2012). Genome-wide analysis of immunophilin FKBP genes and expression patterns in Zea mays. Genet. Mol. Res. 11, 1690–1700. doi: 10.4238/2012.June.25.2
Wang, X. F., Qiao, Z. W., Wang, D. R., Wang, X., You, C. X. (2022). Genome-wide identification and stress response analysis of cyclophilin gene family in apple (Malus domestica).
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucl. Acids Res. 40, 49–49. doi: 10.1093/nar/gkr1293
Wang, W., Vinocur, B., Altman, A. (2003). Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218, 1–14. doi: 10.1007/s00425-003-1105-5
Wang, C., Wang, G., Qu, X., Zhang, X., Ji, W., Zhang, H. (2021). Genome-wide identification and expression analysis of the invertase gene family in common wheat. bioRxiv. doi: 10.1101/2021.12.29.474404
Waseem, M., Ahmad, F., Habib, S., Gao, Y., Li, Z. (2018). Genome-wide identification of FK506-binding domain protein gene family, its characterization, and expression analysis in tomato (Solanum lycopersicum l.). Gene 678, 143–154. doi: 10.1016/j.gene.2018.08.021
Weber, H., Chételat, A., Reymond, P., Farmer, E. E. (2004). Selective and powerful stress gene expression in Arabidopsis in response to malondialdehyde. Plant J. 37, 877–888. doi: 10.1111/j.1365-313x.2003.02013.x
Wojtkiewicz, P., Biernacka, D., Gorzelak, P., Stupak, A., Klein, G., Raina, S. (2020). Multicopy suppressor analysis of strains lacking cytoplasmic peptidyl-prolyl cis-trans isomerases identifies three new PPIase activities in Escherichia coli that includes the DksA transcription factor. Int. J. Mol. Sci. 21, 5843. doi: 10.3390/ijms21165843
Xu, Q., Liang, S., Kudla, J., Luan, S. (1998). Molecular characterization of a plant FKBP12 that does not mediate action of FK506 and rapamycin. Plant J. 15, 511–519. doi: 10.1046/j.1365-313x.1998.00232.x
Yadav, S., Centola, M., Glaesmann, M., Pogoryelov, D., Ladig, R., Heilemann, M., et al. (2022). Cyclophilin anaCyp40 regulates photosystem assembly and phycobilisome association in a cyanobacterium. Nat. Commun. 13, 1–17. doi: 10.1038/s41467-022-29211-w
Yeh, E., Means, A. (2007). PIN1, the cell cycle and cancer. Nat. Rev. Cancer 7, 381–388. doi: 10.1038/nrc2107
Yu, Y., Li, Y., Jia, F., Zhao, M., Li, W., Sun, Q., et al. (2017). ZmFKBP20-1 improves the drought and salt tolerance of transformed. Arabidopsis. J. Plant Biol. 60, 558–570. doi: 10.1007/s12374-017-0262-1
Yu, Y., Zhang, H., Li, W., Mu, C., Zhang, F., Wang, L., et al. (2012). Genome-wide analysis and environmental response profiling of the FK506-binding protein gene family in maize (Zea mays l.). Gene 498, 212–222. doi: 10.1016/j.gene.2012.01.094
Zhang, X., Deng, Y., Chang, H., Ji, C., Zhang, M., Peng, J., et al. (2014). The specific and rapid labeling of cell surface proteins with recombinant FKBP-fused fluorescent proteins. Protein Cell. 5, 800–803. doi: 10.1007/s13238-014-0090-8
Zhang, Y., Han, J., Liu, D., Wen, X., Li, Y., Tao, R., et al. (2014). Genome-wide identification and analysis of FK506-binding protein gene family in peach (Prunus persica). Gene 536, 416–424. doi: 10.1016/j.gene.2013.10.059
Zhou, Q. Q., Wang, Y., Hu, J. J., Zhang, L., Li, J. B., Xu, Y. J., et al. (2022). Identification and characterization of a cyclophilin a gene from Chinese shrimp fenneropenaeus chinensis: sequence features and expression profiles. Invertebr. Surviv. J., 19, 105–114. doi: 10.25431/1824-307X/isj.v19i1.105-114
Keywords: FK506-binding proteins, parvulins, phospho-tyrosyl phosphatase activators, heat stress, wheat
Citation: Suri A, Singh H, Kaur K, Kaachra A and Singh P (2022) Genome-wide characterization of FK506-binding proteins, parvulins and phospho-tyrosyl phosphatase activators in wheat and their regulation by heat stress. Front. Plant Sci. 13:1053524. doi: 10.3389/fpls.2022.1053524
Received: 25 September 2022; Accepted: 23 November 2022;
Published: 15 December 2022.
Edited by:
Bhanu Prakash Petla, International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), IndiaReviewed by:
Venura Herath, University of Peradeniya, Sri LankaHarmeet Kaur, National Institute for Plant Biotechnology, Indian Council of Agricultural Research, India
Bharati Pandey, Panjab University, India
Copyright © 2022 Suri, Singh, Kaur, Kaachra and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Prabhjeet Singh, c2luZ2hwcmFiaGplZXQ2MkBnbWFpbC5jb20=
†These authors have contributed equally to this work
 Anantika Suri
Anantika Suri Harpreet Singh
Harpreet Singh Kirandeep Kaur1†
Kirandeep Kaur1† Prabhjeet Singh
Prabhjeet Singh