- 1Key Laboratory of Biology and Genetic Improvement of Oil Crops Research Institute, Oil Crops Research Institute, Chinese Academy of Agricultural Sciences (CAAS), Wuhan, China
- 2School of Biology and Food Engineering, Guangdong University of Petrochemical Technology, Maoming, China
Brassica napus L. (B. napus) is a vital oilseed crop cultivated worldwide; low temperature (LT) is one of the major stress factors that limit its growth, development, distribution, and production. Even though processes have been developed to characterize LT-responsive genes, only limited studies have exploited the molecular response mechanisms in B. napus. Here the transcriptome data of an elite B. napus variety with LT adaptability was acquired and applied to investigate the gene expression profiles of B. napus in response to LT stress. The bioinformatics study revealed a total of 79,061 unigenes, of which 3,703 genes were differentially expressed genes (DEGs), with 2,129 upregulated and 1,574 downregulated. The Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analysis pinpointed that the DEGs were enriched in LT-stress-responsive biological functions and metabolic pathways, which included sugar metabolism, antioxidant defense system, plant hormone signal transduction, and photosynthesis. Moreover, a group of LT-stress-responsive transcription factors with divergent expression patterns under LT was summarized. A combined protein interaction suggested that a complex interconnected regulatory network existed in all detected pathways. RNA-seq data was verified using real-time quantitative polymerase chain reaction analysis. Based on these findings, we presented a hypothesis model illustrating valuable information for understanding the LT response mechanisms in B. napus.
Introduction
Brassica napus is extensively grown and distributed in the Yellow River basin in China (Ke et al., 2020). B. napus is the third most important oil crop in China (Li et al., 2021). To date, B. napus has become a research hotspot due to its commercial and ecological benefits (Liu et al., 2019). It has long been known that environmental concerns such as low temperature (LT) stress, including chilling (0–10°C) and freezing (<4°C), affect the germination, growth, development, production, and spatial distribution of crops. In recent years, winter- and semi-winter rapeseeds cultivated in the Yangtze River basin were sowed from the end of September to mid-October, which leads to threatened growth and production of rapeseed under LT stress conditions (Cong et al., 2019; Huang et al., 2020). Improvement in cold tolerance has been a major goal for the agricultural research of B. napus over the recent years; there is an urgent need to develop and cultivate early-maturing rapeseed varieties with cold resistance (Raza et al., 2021).
Over the years, available literature has reported LT-responsive genes and regulatory mechanisms in Arabidopsis (Du et al., 2017; Yu et al., 2020). The researchers focused on elucidating the LT response molecular regulatory mechanisms in different crop species such as tomato (Niu et al., 2022), apple (An et al., 2018), banana (Liu et al., 2018a), rice (Lv et al., 2017; Hang et al., 2018), rapeseed (Luo et al., 2019; Pu et al., 2019; Huang et al., 2020), and maize (Li et al., 2016b). These efforts revealed numerous genes, molecular regulators, and biological pathways to ameliorate plant LT stress effects (Shi et al., 2018; Ding et al., 2019). To cope with unfavorable environmental factors, increasing attention has been given to understanding the cold response mechanisms in oilseed crops, especially rapeseed—for example, next-generation sequencing (NGS) technology is widely applied to reveal the transcriptomic changes in rapeseed under different stresses (Xiong et al., 2022). Recently, the transcriptomic analysis identified various cold-responsive (COR) pathways that regulate photosynthesis, abscisic acid (ABA) homeostasis and transport, plant hormone signal transduction, ribosome biogenesis, MAPK signaling pathway, calcium signal transduction, and antioxidant defense systems in rapeseed (Ma et al., 2019). Different studies reported that the accumulation of different metabolites, including proline, soluble sugar, and protein contents, and antioxidant enzyme activity were rapidly increased under freezing stress (-2°C) in rapeseed (Yan et al., 2019). Dehydrins such as late embryogenesis abundant (LEA) proteins are accumulated in response to cold stress by a higher expression of LEA10 and LEA18, LEA90, and LEA104 genes in B. napus (Maryan et al., 2019). LT stress-inducible proteins are antifreeze proteins, cold-shock domain proteins, and heat shock proteins (HSPs). Under LT stress, various enzymes, such as oxidases and desaturases, are activated to scavenge the reactive oxygen species (Heidarvand and Maali, 2010; Su et al., 2021). Among different antioxidant enzymes, superoxide dismutase (SOD), peroxidase (POD), ascorbate peroxidase (APX), and catalase (CAT) act as the first line of reactive oxygen species (ROS) scavenging to protect stress-induced oxidative damage in plants (Sharma et al., 2019; Su et al., 2021).
The phenotypic and RNA-seq profiling of transgenic B. napus ectopic overexpressing Arabidopsis C-repeat/DRE-binding factor (CBF) has suggested that the CBF cold response of Arabidopsis has been maintained in rapeseed (Jaglo et al., 2001). In the grape plant, CBL-interacting protein kinases 18 act as a positive regulator of the CBF cold signaling pathway by modulating ROS homeostasis (Yu et al., 2022b). Plant-hormone-mediated pathways are also critical in plant LT stress tolerance—for example, sly-miR156e-3p mediated the posttranscriptional regulation of transcription factor S1MYB15, which positively regulates ABA-mediated cold tolerance in the tomato (Zhang et al., 2022b). In the last decade, different efforts have been made to investigate the physiological and molecular perspectives of LT response of B. napus (Du et al., 2016; Huang et al., 2018; Xin et al., 2019; Ke et al., 2020). Despite these efforts, the complex molecular mechanisms of LT stress responses in B. napus need to be further explored. The NGS profoundly helped to dissect genome-wide novel and diverse molecular response mechanisms in plants. These NGS technologies provide simple, low-cost, sensitive, accurate, and fast tools for elucidating the genome-wide regulation of desired traits (Tyczewska et al., 2016; Sharma et al., 2018). The RNA-seq technology has many successful stories in profiling the genetic architecture of LT stress tolerance, including B. rapa (Ma et al., 2019), B. oleracea (Zhang et al., 2019c), B. napus (Du et al., 2016; Ke et al., 2020), A. thaliana (Kaplan et al., 2007), Z. mays (Li et al., 2016b; Li et al., 2019), O. sativa (Guan et al., 2019), and E. japonica Lindl (Zhang et al., 2022a).
Based on the abovementioned details, the molecular picture associated with cold response has become clearer. B. napus was capable of growth at LT conditions; however, the cold responses of the early-maturing variety in a short time remain to be characterized. In this study, we assessed the dynamic changes that occur at the molecular level to elucidate the LT stress responses through biochemical and transcriptome analysis in B. napus. Biochemical analysis helps to identify the activation of different kinds of phytohormones under LT stress. The transcriptome analysis helps explore various COR gene pathways and their regulatory mechanisms operating under LT conditions in B. napus. The data generated in this study would enhance the understanding of LT response and regulatory mechanism and identify substantial genetic resources that are useful for modifications and enhancing LT tolerance in B. napus.
Materials and methods
Plant materials, growth conditions, and treatments
Rapeseed Cultivar-18, an excellent inbred winter cultivar bred by Oil Crops Research Institute, Wuhan, China, that can be successfully cultivated under LT conditions, was used in this study. The seeds were germinated on moist filter paper in petri plates under soft tube white lights at a temperature range of 25 ± 1°C, humidity of 60%, and 16-h light/8 h dark photoperiod in a growth chamber. One-week-old seedlings were transferred to plastic pots filled with a 1:1:1 mixture of peat vermiculite/moss/perlite in a growth room (16 h light/8 h dark photoperiod, with a constant temperature of 25 ±1°C for approximately 21 days. At a 4-leaves stage, healthy seedlings were selected and transferred to LT stress (4°C) conditions. Before LT treatment (0 d) and after LT conditions (1 d), leaves were sampled, immediately frozen in liquid nitrogen, and preserved at -80°C until further use. Seedlings were recovered under normal growth conditions for two days after LT stress treatment. Total RNA was extracted from a pooled sample of seedling leaves from each group.
RNA preparation and deep sequencing
RNA preparation and transcriptome analysis were performed as previously described (Raza et al., 2021). Briefly, total RNA was extracted from the leaves of LT stressed and non-stressed samples using the TRIzol kit (Invitrogen, USA). RNA concentration and quality were quantified using NanoDrop 2000 (Thermo). The RNA sample was prepared from 1 μg RNA per sample for sequencing. cDNA libraries were produced using NEBNext UltraTM RNA Library Prep Kit for Illumina (NEB, USA), following the manufacturer’s instructions at the Biomarker company (Beijing, China), and then sequenced on an Illumina platform employing paired-end technology. Clean data was further subjected to Q20, Q30, GC-content and subsequent analysis. Hisat2 tools software was used to map clean reads with the reference genome sequence of B. napus (https://www.genoscope.cns.fr/brassicanapus) (Chalhoub et al., 2014). The sequence data is publicly accessible on the National Center for Biotechnology Information database (PRJNA596550).
Differential expression analysis
Differential expression analysis of data under two conditions, LT stressed and non-stressed, was performed using the DEseq, which provided gene expression data based on the negative binomial distribution. Gene expression level was measured by the number of fragments per kilobase of exon in per million fragments mapped reads (Trapnell, 2010). Benjamini and Hochberg’s method was used to control the false discovery rate (Benjamini and Hochberg, 1995). Genes with an adjusted P-value <0.001 were considered differentially expressed genes (DEGs). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed using a hypergeometric test with a false discovery rate adjusted p-value <0.05.
Gene functional annotation
The functional annotations of all these obtained DEGs were explored using the BLASTx program to find out the NCBI non-redundant protein (NR) and Arabidopsis protein (TAIR10) databases. The DEGs were subjected to GO enrichment analysis using the GOseq R packages based on Wallenius non-central hyper-geometric distribution (Young et al., 2010). The Blast2GO program was used to categorize the GO terms into three major categories of molecular function, cellular component, and biological process (Conesa and Götz, 2008). The KEGG pathway annotation was performed using KOBAS software to elaborate the corresponding metabolic pathways (Mao et al., 2005).
Physiological and biochemical analysis
To explore the physiological and biochemical changes under LT stress conditions, the contents of hydrogen peroxide (H2O2), malondialdehyde (MDA), soluble sugar, and proline (Pro) were quantified using commercial kits purchased from SolarBio (https://solarbio.com) following the manufacturer’s protocol. The antioxidant enzyme activities, including superoxide dismutase (EC 1.15.1.1), peroxidase (EC 1.11.1.7), catalase (EC 1.11.1.6), and ascorbate peroxidase (EC 1.11.1.11), were quantified following the SolarBio kits (https://solarbio.com) guidelines. The photosynthesis parameter Fv/Fm was measured using a portable chlorophyll-fluorometer OS-30p+ (OPTI-SCIENCES, China). All physiological and biochemical parameters were measured using a spectrophotometer microplate reader (Epoch, BioTek, Instruments, USA) in triplicate biological replicates.
qRT-PCR validation
The RNA-seq data was validated through qRT-PCR analysis. The first-strand cDNA was reverse-transcribed from 1 μg DNA-free RNA using EasyScript®One-Step cDNA Synthesis SuperMix (Trans) according to the manufacturer’s instructions. Amplification was performed using a StepOnePlusReal-Time PCR System (Applied Biosystems) with a Power SYBR®Green PCR Master Mix according to the manufacturer’s instructions. The reaction protocol was as follows: 95°C for 10 m, 42 cycles at 95°C for 15 s, and 60°C for 15 s, with 10 ul as the final volume following three technical replicates. Relative expression data were analyzed using the 2–ΔΔCt algorithm using B. napus ACTIN as an internal control (Wang et al., 2014). The primers used for qRT-PCR are listed in Supplementary Table S1.
Results
Physiological and biochemical response to LT stress
LT stress has damaging effects, which impair the growth and productivity of B. napus. After a short-term 4°C stress treatment, B. napus seedlings showed >99% survival rate after 2 days of recovery (Figures 1A, B). However, 4°C-LT-stress-treated seedlings exhibited significant differences in physiological and biochemical indices compared with CK conditions. Therefore, the maximum quantum efficiency of photosystem II (PSII) Fv/Fm is reduced by 29.22% under LT stress treatment in rapeseed (Figure 1C). However, LT stress leads to the induction of H2O2 and MDA contents in plants. H2O2 plays a dual role in response to stress conditions and acts as a ROS and signaling molecule to activate the stress response mechanism in plants. However, excessive H2O2 accumulation is damaging to cells. LT stress treatment significantly increased the H2O2 and MDA levels—specifically, H2O2 increased by 16.52% and MDA increased by 29.77% (Figures 1D, E). Similarly, osmoprotectants, such as soluble sugars and proline, increased by 19.14% and 42.97% compared with CK, respectively (Figures 1F, G). These results indicate that the accumulation of osmoprotectants has a protective role in LT-stressed B. napus seedlings. In response to higher ROS, the plant defense system is activated to ameliorate the effects of LT stress compared with CK. The built-in plant defense system is comprised of various kinds of antioxidant enzymes. These antioxidant enzymes, including SOD, POD, CAT, and APX, equilibrate ROS production and are involved in the conversion to the least deleterious biochemicals within plant cells under LT stress. We observed that the LT stress significantly increased the SOD enzyme activity by 19.61%, POD enzyme activity by 15.82%, and APX enzyme activity by 19.69% compared with CK seedlings (Figures 1H–J). In contrast, CAT enzyme activity was reduced by 15.3% compared with CK (Figure 1K). These results indicate that, in B. napus seedlings, complex physiological and biochemical responses are simultaneously activated to protect them from the injurious effects of LT stress (Figure 1).
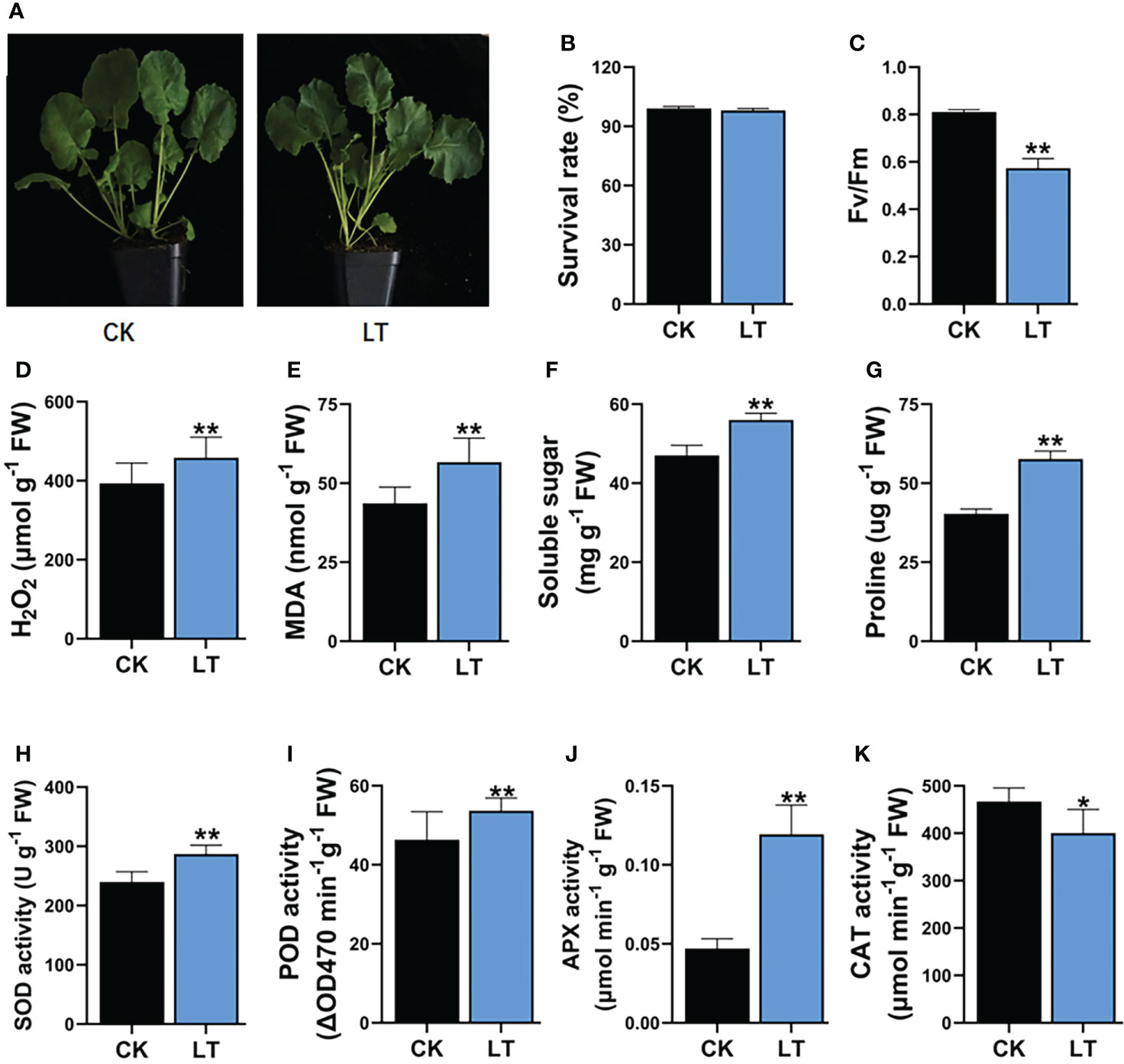
Figure 1 Impact of LT stress treatment on physiology and biochemical indexes in rapeseed. (A) Plant phenotype, (B) survival rate, (C) maximum quantum efficiency of photosystem II (PSII) Fv/Fm, (D) hydrogen peroxide (H2O2), (E) malondialdehyde (MDA), (F) soluble sugar, (G) proline contents, (H) superoxide dismutase (SOD) 1124 activity, (I) peroxidase (POD) activity, (J) ascorbate peroxidase (APX) activity, (K) 1125 Catalase (CAT) activity. Asterisks show significant levels at **P ≤ 0.01, *P ≤ 0.05.
RNA-seq and data quality control analysis
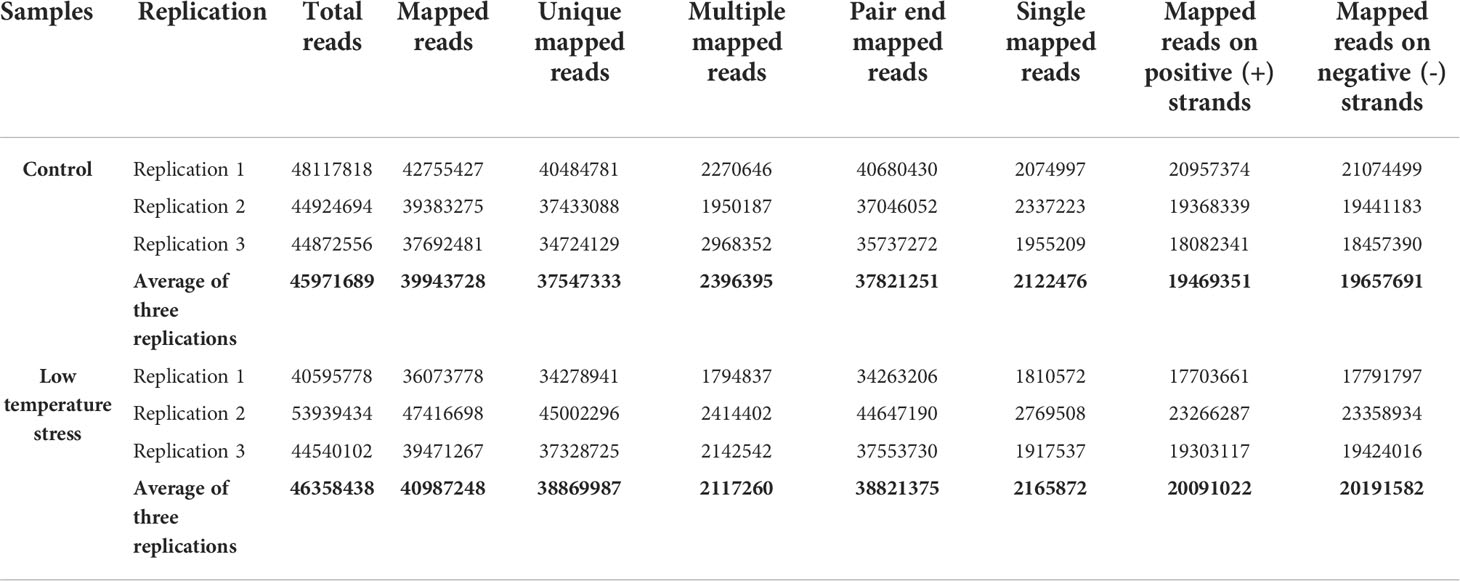
The current study used B. napus plants as experimental material to reveal transcriptome changes under LT stress. At the four-leaves stage, healthy plants were subjected to 4°C for 1 day under 16/8-h photo/dark period for LT treatment. The morphological changes in the leaves of B. napus plants were recorded. The phenotypic changes indicated that B. napus plants were challenged by LT stress (Figure 1A). Subsequently, the leaves were harvested, and two kinds of cDNA libraries were constructed from LT-stressed and CK samples. The cDNA libraries were sequenced using Illumina technology with 10× depth. After trimming the low-quality reads, averages of 45,971,689 and 46,358,438 clean reads were obtained from the CK and LT samples, respectively. The Q30 for all sequenced libraries was greater than 95%, and the GC content of each treatment was ≥47%. Averages of about 39,943,728 (86.84%) and 40,987,248 (88.46%) clean reads were successfully mapped to the reference genome from the CK and LT samples, respectively (Table 1). A total of 7,9061 unigenes were procured.
Annotation of the B. napus transcriptome
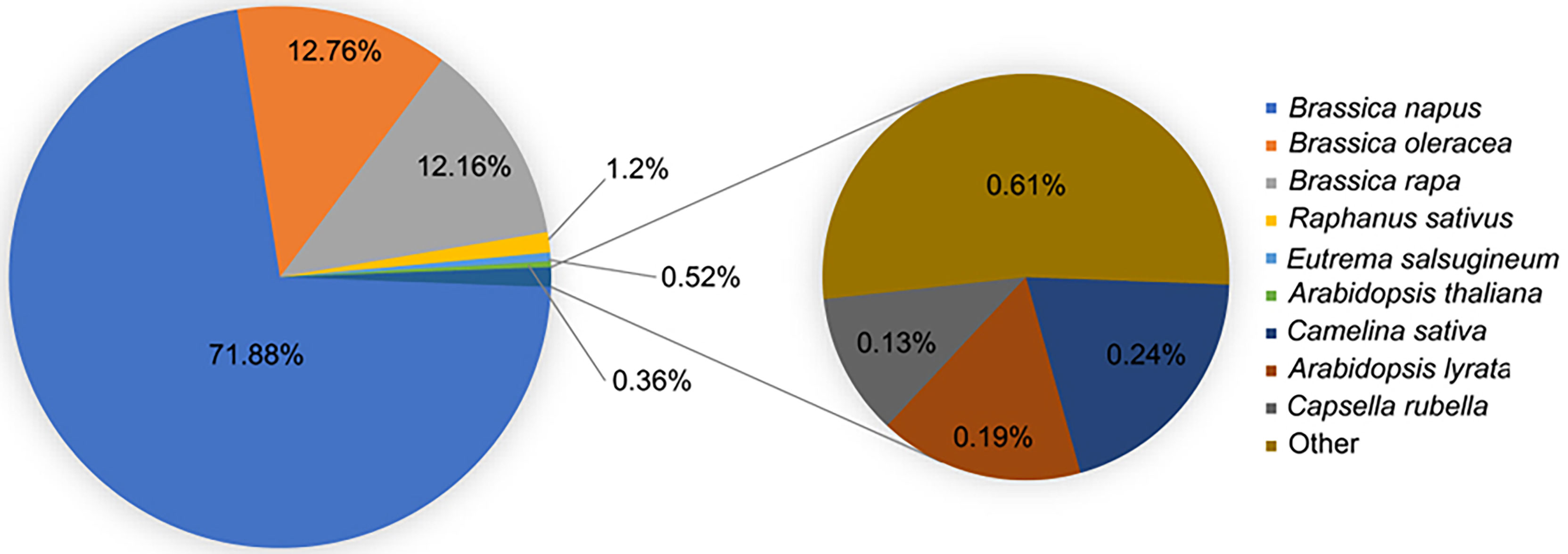
B. napu unigenes were annotated using the BLAST function against the NR and TAIR freely available databases. B. napus best matched with B. napus in the NR database compared with B. oleracea, B. rapa, Raphanus sativus, and other related species, which demonstrated that it was immensely homologous with B. napus (Figure 2). GO analysis was performed to categorize the function of each unigene. In total, 79,061 unigenes were successfully annotated to 55 GO terms. These terms were categorized into 55 GO terms, including 20 groups in biological processes, 14 in molecular functions, and 12 in cellular components. The biological process terms were enriched in cellular processes, metabolic processes, single organism process, responses to stimulus, and biological regulation. The enriched terms in molecular functions were binding, catalytic activity, nucleic acid binding transcription factor activity, transporter activity, and structural molecule activity. Among the cellular component terms, the five most significant classifications were cell, cell part, organelle, membrane, and organelle part (Supplementary Figure S1; Supplementary Table S2). In addition to GO analysis, the KEGG pathway analysis revealed several essential pathways involved in LT stress response. In total, 18,993 unigenes were successfully assigned to 119 pathways using KOBAS software. The major KEGG pathways were enriched in ribosome biogenesis, plant hormone signal transduction, carbon metabolism, biosynthesis of amino acids, starch and sucrose metabolism, protein processing in the endoplasmic reticulum, spliceosome, and endocytosis (Supplementary Table S3).
Identification and analysis of DEGs
In order to evaluate the response to LT stress in C18, differential expression analysis was performed between samples before cytokinin (CK) and after LT treatment (LT stress). In the comparison between two groups, a total of 3,703 were identified as significant DEGs in the threshold of log2(fold change) ≥1 or ≤-1 (P-value ≤ 0.05) (Supplementary Table S4). To characterize the biological functions of the DEGs, we performed GO biological process (GO-BP) enrichment analysis on all upregulated and downregulated DEGs (Supplementary Table S5). The results showed that upregulated DEGs were mostly involved in water deprivation response, pyrimidine ribonucleotide biosynthetic process, hyperosmotic salinity response, cold response, RNA methylation, heat acclimation, chitin response, defense response, abscisic acid response, protein import into the nucleus, and response to jasmonic acid. While the downregulated DEGs mainly participated in chitin response, glucosinolate biosynthetic process, carboxylic acid catabolic process, light stimulus, and respiratory burst involved in defense response were the most enriched terms. As shown above, diverse changes and adaptations in biological reactions may occur in B. napus in response to LT stress.
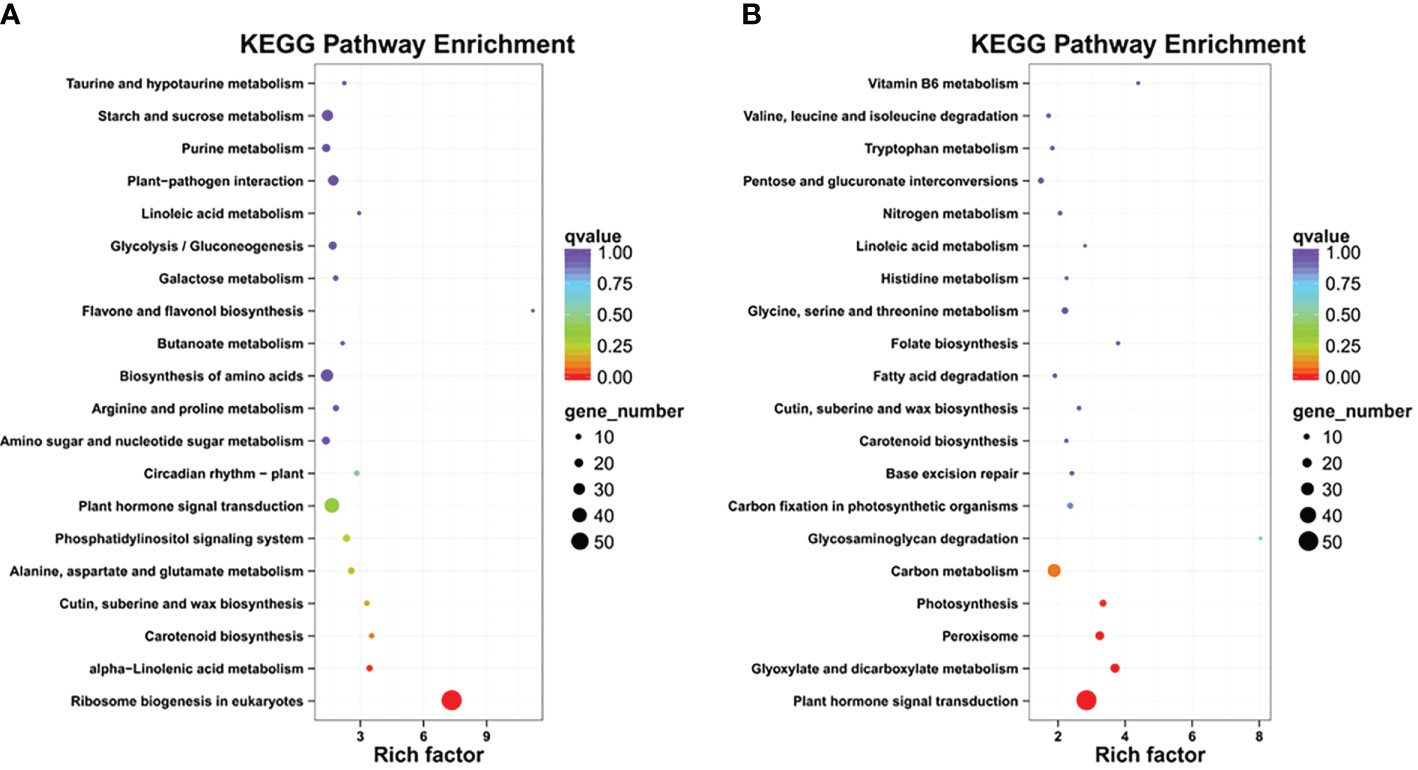
To further categorize the important LT-stress-related pathways, the KEGG pathway enrichment analysis was performed for upregulated and downregulated DEGs (Supplementary Table S6). The major KEGG pathways for upregulated DEGs were ribosome biogenesis and alpha-linolenic acid metabolism, while downregulated DEGs were enriched in plant hormone signal transduction, glyoxylate and dicarboxylate metabolism, peroxisome, photosynthesis, and carbon metabolism, which suggested that LT stress has harmful effects on various biological processes, and B. napus simultaneously initiates its defense though complex biosynthesis and metabolic pathways (Figures 3A, B).
Transcription factors’ response to LT stress
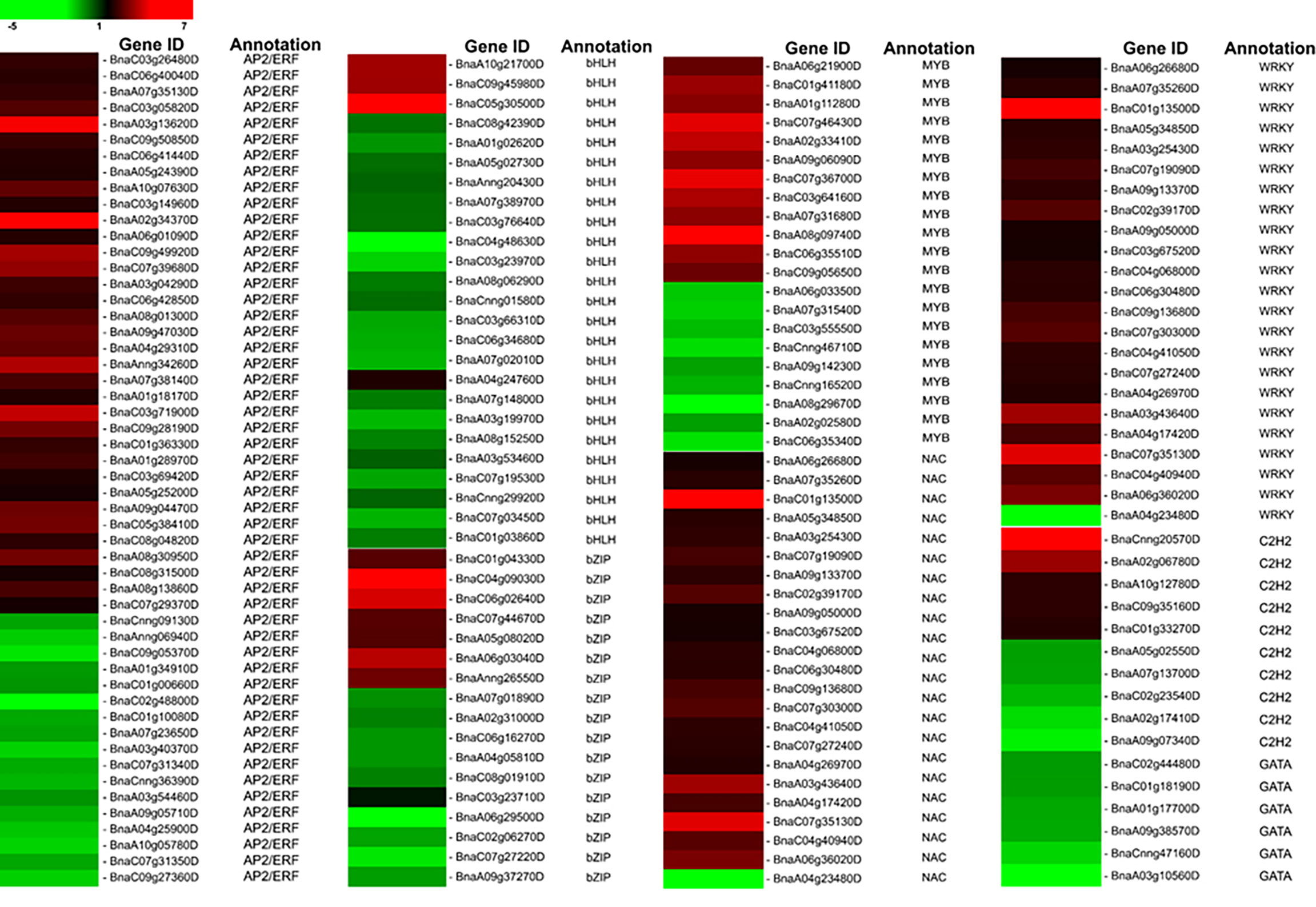
Transcription factors (TFs) recognize and bind to cis DNA elements—small regulatory sequences typically composed of non-coding DNA sequences (Wittkopp and Kalay, 2012)—in the promoter region to regulate the expression of downstream genes that have crucial functions in various plant stress responses. In the current study, 176 DEGs belonging to eight TF families that respond to biotic or abiotic stresses were determined (Figure 4). Specifically, 52 AP2/ERF members were identified, including 35 upregulated genes (BnaC03g26480D, BnaC06g40040D, BnaA07g35130D, BnaA03g13620D, etc.) and 17 downregulated genes (BnaA01g34910D, BnaA07g23650D, BnaA10g05780D, etc.). In our data, the basic helix–loop–helix (bHLH) family was the second largest that consist of 25 members, including three upregulated genes (BnaA10g21700D, BnaC09g45980D, and BnaC05g30500D) and 22 downregulated genes. We detected 21 MYB family members, including 12 upregulated and nine downregulated genes that respond to LT stress in B. napus. In the current study, it is also evident that WRKY TFs play significant regulatory roles in B. napus under LT stress (Figure 4). We found that 23 WRKY TFs respond to LT stress; among them, 22 members have induced expression, and only one member, BnaA04g23480D, was repressed under LT stress. Similarly, a total of 22 NAC TFs were found in this study, including 19 upregulated members and two downregulated members (BnaCnng30500D and BnaA04g24740D). In addition, among all identified basic leucine zipper transcription factor (bZIP TF) family members, seven were upregulated, while 11 were downregulated. Thus, it is suggested that the strong activation or repression of transcription factors by LT stress may promote the defense response in B. napus.
Analysis of DEGs related to potential pathways
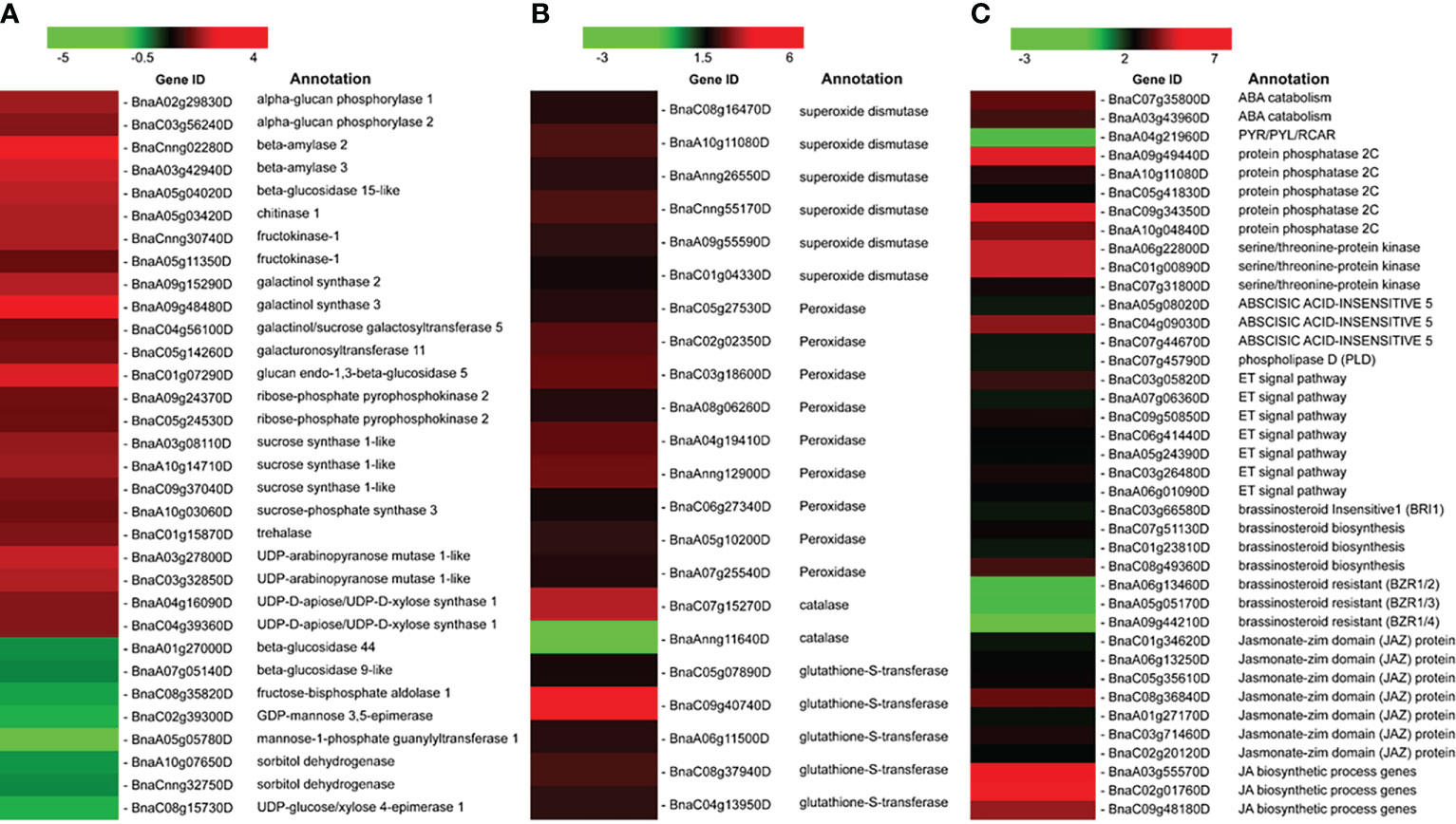
When plants were exposed to cold stress, dynamic changes in starch content occurred. Many genes involving starch and sucrose metabolism have been previously proven to respond to LT stress, such as ADP-glucose pyrophosphorylase, β-amylase, and sucrose synthases, and were detected (Sicher, 2011; Yu et al., 2022a). As summarized in the heat map (Figure 5), different combinations of starch-degrading enzymes operate under LT stresses, and most of them were upregulated—for instance, we found the activation of three genes encoding sucrose synthase 1-like (BnaA03g08110D, BnaC09g37040D, and BnaA10g14710D) that promotes the catabolization of sucrose to uridine di-phosphoglucose and fructose. Various genes involved in galactose metabolism were also upregulated, such as BnaA09g15290D, BnaA09g48480D, and BnaC04g56100D, which encode galactinol synthase 2, galactinol synthase 3, and galactinol/sucrose galactosyltransferase 5. These results are consistent with previous reports (Ciereszko et al., 2001; Baud et al., 2004). ROS such as hydrogen peroxide (H2O2), superoxide (O2-), singlet oxygen (1O2), and hydroxyl radical (OH-) are quickly produced from multiple organelles under LT stress (Qi et al., 2018), known as toxic agents, and also perceived as the second messengers by plant cells and trigger rapid responses. Here almost all DEGs belonging to the ROS-scavenging enzyme system were upregulated, including six SOD members, one CAT member, nine POD members, and five glutathione S-transferase (GST) members (Figure 5), except only one CAT gene (BnaAnng11640D) that was downregulated, suggesting that these enzymes were actively expressed to form a complex antioxidant defense under short-time LT stress. Phytohormones include auxins, ABA, gibberellins (GA), CK, ethylene (ET), salicylic acid, jasmonates (JA), brassinosteroids (BR), and strigolactones, and they play critical roles in helping the plants to adapt to temperature stresses (Verma et al., 2016). As shown in Figure 5, 14 DEGs involved in the ABA signal pathway have an induced expression in response to LT stress, followed by one gene that showed a repressed expression. In addition, seven ET signal pathway-related and 10 JA signal pathway-related DEGs were upregulated. Three DEGs belonging to the BR signal pathway were significantly downregulated; however, three BR biosynthesis pathway genes were upregulated. Therefore, the abovementioned results indicated that the ABA, ET, BR, and JA signaling pathways were activated in the LT stress response of B. napus.
Interaction network between B. napus LT-stress-related genes
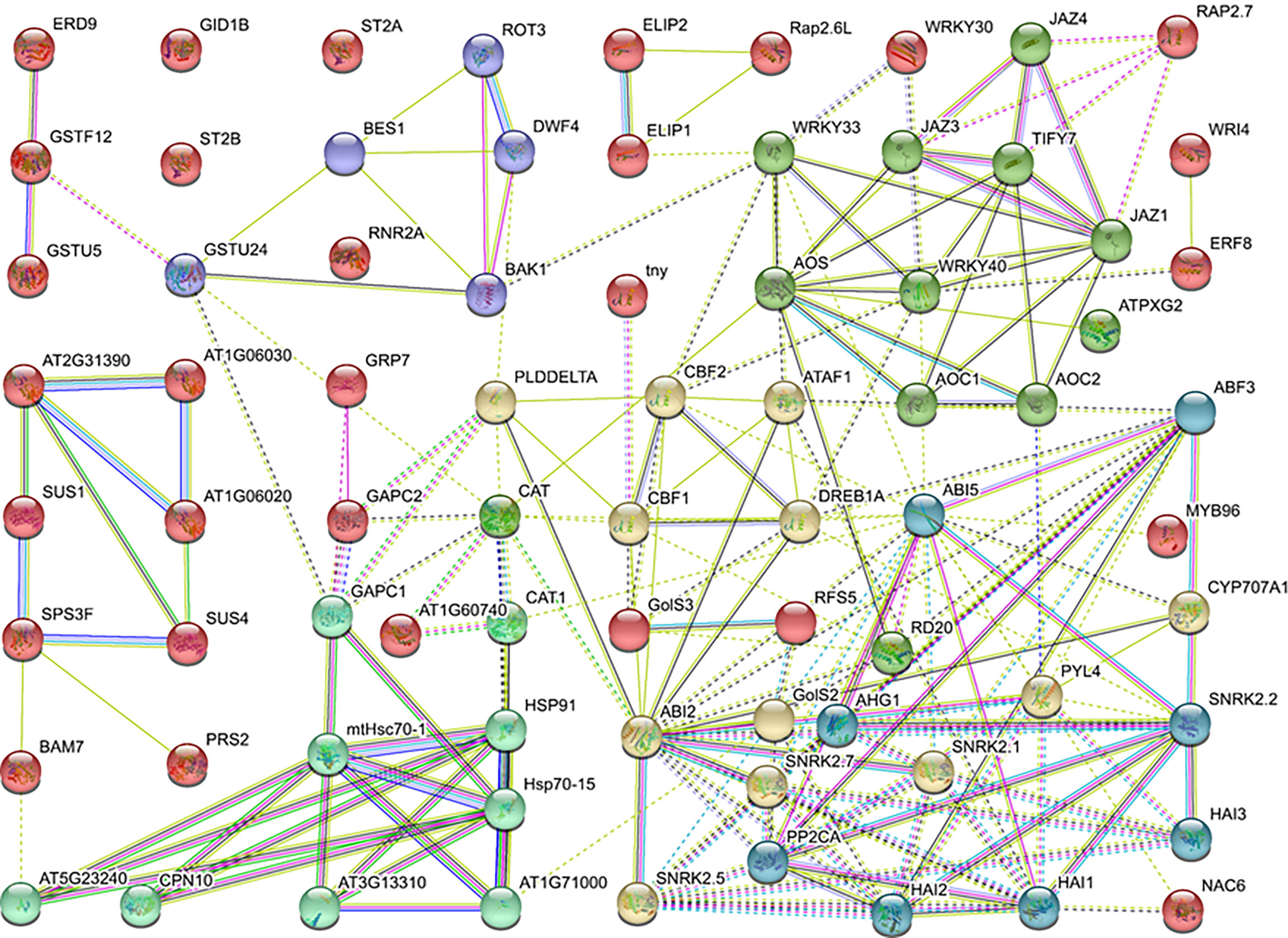
To elaborate the genes’ role in regulating LT response at the protein level, the protein–protein interactions network was constructed using DEGs enriched in GO or KEGG terms, including sugar metabolism, hormone, antioxidant activity, transcription factors, and photosynthesis. The homologs of potential DEGs were referred to the Arabidopsis database and subjected to STRING database analysis. As the results demonstrated, the homologs of DEGs were divided into six expression patterns, and the interaction lines exhibited different types of association among genes. Specifically, PYR/PYL/RCAR-PP2C-SnRK2-ABF, known as the ABA-dependent pathway, was detected. Meanwhile, nearly half of the genes were associated with each other and regulated the CBFs and corresponding stress-related genes. Besides the abovementioned data, JAZ/TIFYs pathway genes were detected, which indicates that the jasmonic acid signal pathway may participate in the quick cold response of B. napus (Figure 6).
Validation of transcriptome data by qRT-PCR analysis
To validate the transcriptome data, 32 candidate genes were randomly selected and subjected to qRT-PCR analysis. These target genes belong to various functional categories of the KEGG pathways, including photosynthesis, sugar metabolism, plant hormone signal transduction, anti-oxidant defense systems, and transcription factors. Finally, we compared the RNA-seq expression level with the qRT-PCR results. The qRT-PCR results were consistent with the RNA-seq, confirming the reliability of the transcriptome results (Figure 7).
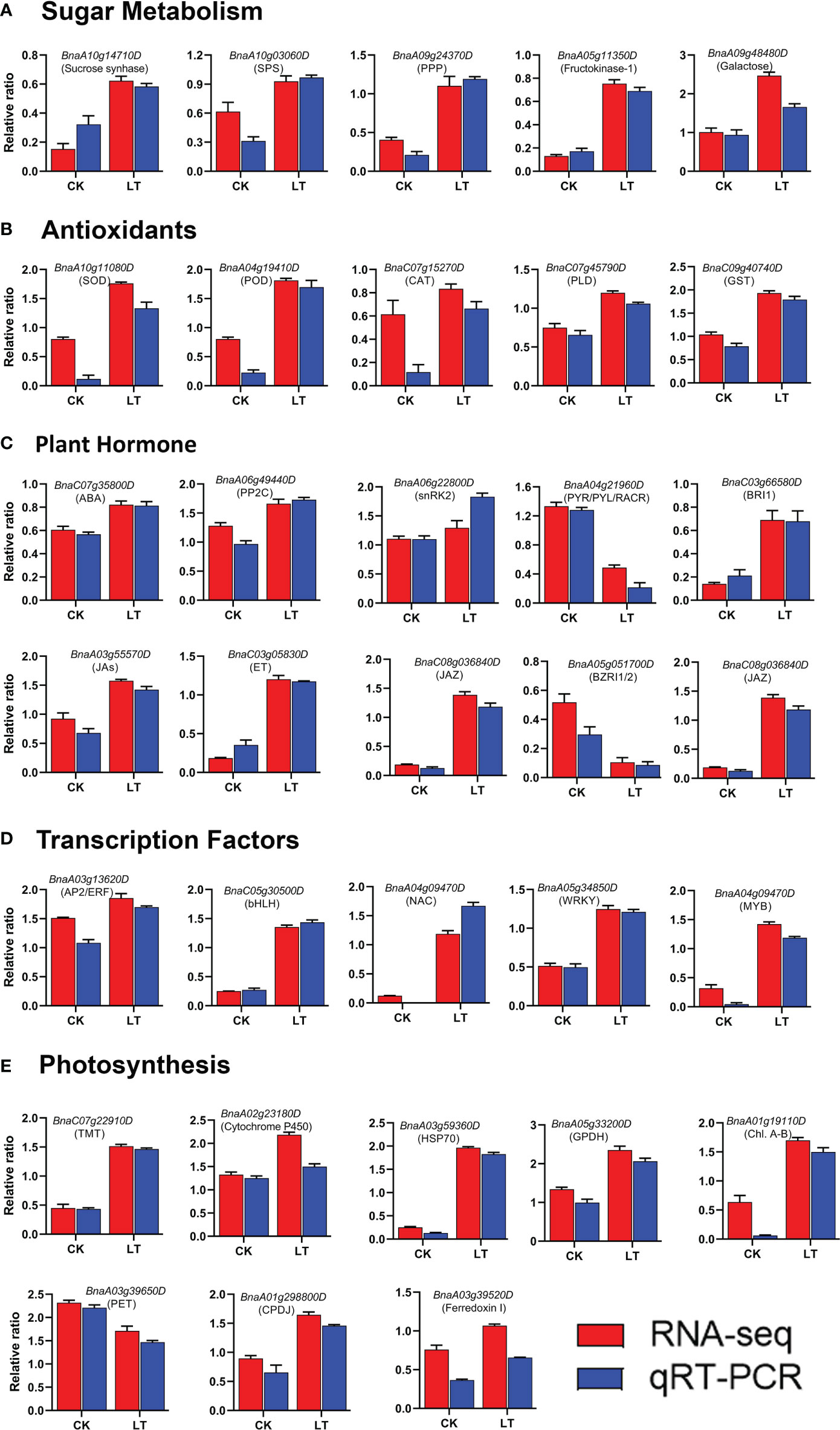
Figure 7 qRT-PCR validation of RNA-seq data: (A) Sugar metabolism, (B) Antioxidants, (C) Plant Hormone, (D) Transcription factors, (E) Photosynthesis.
Discussion
To dissect the LT response mechanisms in B. napus, various LT-related candidate DEGs were explored from transcriptomic data, and their mRNA abundance was investigated in LT stress response processes. In this study, our primary focus was on those DEGs involved in sugar metabolism (starch, sucrose, and carbohydrate), photosynthesis, plant antioxidant defense system, plant hormone signaling networks, and TFs. The LT stress had adverse effects on photosynthesis. Under LT stress, the cellular respiration process of mitochondria and the photosynthesis process in chloroplast generate oxidative stress and lead to ROS stockpiling (Oelze et al., 2008; Janmohammadi et al., 2015). Oxidative stress due to higher levels of ROS accumulation causes damage to DNA, lipids, and proteins (Schieber and Chandel, 2014). In B. napus, photosynthesis-related genes were mostly downregulated and took part in the light reaction and the Calvin cycle. Decreased photosynthesis activity is directly linked with reduced plant productivity under LT stress conditions. In addition to downregulated genes, few genes were induced and might have played a significant role in protecting the photosynthesis system under LT stress in B. napus. Genes involved in phytohormone signaling networks, sugar metabolism, and antioxidants had variable expression levels, which showed the complexity of the molecular regulation network in response to LT stress (Bari and Jones, 2009; Li et al., 2016a; Ding et al., 2019).
Sugar metabolism, source of energy, and keeping osmotic homeostasis
Previous studies demonstrated that soluble sugars accumulate in response to stress; however, they may play multiple roles under stress. Sugar molecules act as biochemical components for cold acclimation and protect plant cells from damage (Gilmour et al., 2000). Sugars also function as stress signal molecules and trigger a series of signal transduction and defense reactions (Eveland and Jackson, 2012). Sucrose synthase and sucrose phosphate synthase are essential enzymes responsible for metabolic energy in the sugar metabolism pathway (Fugate et al., 2019; Stein and Granot, 2019). During LT stress, Pi accumulation decreased in the cytoplasm (Strand et al., 1999), and altered Pi level signals led to the activation of enzymes in the sucrose synthesis pathway (Hurry et al., 2000). Thus, we have focused on the starch and sucrose metabolism pathway in B. napus and several genes related to sugar metabolism surveyed under LT stress conditions (Figure 5) and verified their expression pattern through qRT-PCR (Figure 7). Different kinds of sugars accumulated in plants, such as sucrose, glucose, raffinose, and fructose, which are involved in LT tolerance (Gu et al., 2018). Sucrose synthase involved in sucrose catabolism and sucrose phosphate synthase involved in biosynthesis are important enzymes primarily responsible for metabolic energy in the sugar metabolism pathway. The sucrose catabolization process acts as a source of ATP or cell wall biosynthesis (Fugate et al., 2019). Therefore, sucrose synthase enzymes are mainly localized in cell walls, mitochondria, and vacuoles (Stein and Granot, 2019). Similarly, upregulated sucrose synthase genes were reported under LT stress in Arabidopsis (Ciereszko et al., 2001; Baud et al., 2004). Furthermore, one sucrose phosphate synthase gene (BnaA10g03060D) was upregulated, which describes the possible role of sucrose biosynthesis under LT stress in B. napus (Figure 5A). Similarly, our biochemical analysis revealed the higher accumulation of soluble sugar and proline under LT stress (Figures 1F, G). A previous study has reported that up to 50% of the activity of sucrose phosphate synthase gene was induced in wheatgrass under LT stress (Jaikumar et al., 2016). In rice, the interaction of calcium-dependent protein kinase OsCPK17 with sucrose phosphate synthase and plasma membrane intrinsic protein is compulsory for LT stress response (Almadanim et al., 2017). Fructokinase plays an essential role in sugar metabolism, signaling, and growth and also has osmosis-protective effects in LT stress (Li et al., 2017b). A high expression of fructokinase genes has been proven to enhance LT tolerance in Arabidopsis (Su et al., 2018). In this study, two genes encoding fructokinase-1 (BnaCnng30740D and BnaA05g11350D) were upregulated in B. napus under LT stress (Figure 5A). Activation of these fructokinase genes suggested a critical role in LT stress tolerance in B. napus.
The overexpression of galactinol synthase (GolS) genes enhanced LT stress tolerance in Ammopiptanthus nanus (Liu et al., 2016). GolS catalyzes the raffinose family oligosaccharide biosynthetic pathway (Salvi et al., 2016). In the current study, various genes involved in galactose metabolism were also upregulated (Figure 5A). Similarly, pentose phosphate pathway genes were induced, such as BnaA09g24370D and BnaC05g24530D which encode ribose-phosphate pyrophosphokinase 2, which was chloroplastic under LT stress (Figure 5A). It has been reported that pentose phosphate pathway genes are hub genes involved in rapid rapeseed seed germination under cold stress (Luo et al., 2019). We also observed multiple up- and downregulated genes, such as beta-D-xylosidase 4, involved in amino sugar and nucleotide sugar metabolism. Hence, when exposed to LT stress, numerous genes involved in sucrose–starch metabolism were induced or repressed, which may lead to the accumulation of osmoprotectants such as sucrose, glucose, and fructose, subsequently maintaining osmotic protection or providing necessary energy under LT stress conditions.
Antioxidant defense system: A cellular cushion against LT stress
A higher accumulation of ROS after LT stress always causes oxidative damage to cellular organs, such as nucleic acid, lipids, and proteins (Suzuki and Mittler, 2006; Schieber and Chandel, 2014). Accordingly, we observed higher H2O2 levels in LT-stressed B. napus seedlings (Figure 1D). In addition to oxidative damage, ROS-mediated activated enzymatic and non-enzymatic ROS scavenging systems detoxify and relieve the cellular oxidative stress level (Miller et al., 2010; Sies et al., 2017). In B. napus during LT stress, the ROS-scavenging enzymatic antioxidant system consists of POD, SOD, CAT, APX, GST, and glutathione peroxidase (GPX) (Yan et al., 2019). The non-enzymatic antioxidant systems included the reduced form of glutathione system, vitamin E\vitamin C system, and secondary metabolites or antioxidants, such as carotenoids, steroids, flavonoids, and anthocyanins. In B. napus, ROS homeostasis—by utilizing these antioxidant systems—protects cellular oxidative stress injury. SOD is known as the first line of defense for ROS-scavenging systems. SOD converts damaging free radicals to less harmful products (H2O2 and O2) in cells through the dismutation process (Zhang et al., 2019b). The induced expression of SOD genes upon cold exposure was reported in Medicago truncatula (Song et al., 2018) and Setaria italica (Wang et al., 2018b). In plants, SODs are mainly divided into three classes based on metal co-factors: copper- and zinc-containing superoxide dismutase (Cu/Zn-SODs), iron superoxide dismutase (Fe-SOD), and manganese superoxide dismutase (Mn-SOD). Different classes of SODs are distributed at different positions or in organs in the cell. Cu/Zn-SODs are generally considered eukaryotic enzymes mainly distributed in the cytosol and chloroplast (Kroll et al., 1995; Wang et al., 2018a). Our findings indicate that various SOD genes (BnaC08g16470D, BnaA10g11080D, BnaAnng26550D, BnaCnng55170D, BnaA09g55590D, and BnaC01g04330D) were induced under LT stress conditions, indicating that it may have LT response regulation in B. napus (Figure 5B). Similarly, we observed an induced SOD activity in LT-stressed seedlings through biochemical analysis, which indicates that SOD activity is genetically controlled (Figure 1H). POD and CAT catalyzed H2O2 into simple H2O in cells. Ascorbate peroxidase enzymes detoxify H2O2 in plant cells. APX is involved in the ascorbate–glutathione cycle to catalyze the H2O2 into H2O using ascorbate as a specific electron donor. APX enzymes are mainly located in subcellular compartments, such as chloroplasts, cytosol, mitochondria, and peroxisome (Caverzan et al., 2012). In our results, the higher activation of APX enzyme activity depicted a protective role from the accumulated H2O2 and MDA by converting them into less harmful components or exclusion from the cellular environment (Figures 1D, E). In this study, POD genes (BnaC05g27530D, BnaC02g02350D, BnaC03g18600D, BnaA08g06260D, BnaA04g19410D, BnaAnng12900D, BnaC06g27340D, BnaA05g10200D, and BnaA07g25540D) were significantly upregulated upon LT stress exposure in B. napus. Accordingly, our biochemical analysis revealed the induction of POD activity in LT-stressed B. napus seedlings, which indicates a close relationship between the expression of these POD genes and POD enzyme activity (Figures 1, 5B). One of the CAT genes’ expressions (BnaC07g15270D) was upregulated, while another (BnaAnng11640D) was repressed. POD and CAT genes have variable responses to the LT stress, which implicated that these genes may play divergent roles during LT stress response in B. napus (Figure 5B). We found that CAT enzyme activity was suppressed in LT stress seedlings (Figure 1K). These results also indicate that rapeseed A genome genes might play a critical role in the CAT enzyme activity compared with the B genome. Similarly, GST and GPX respond to environmental stress and act as scavengers of ROS and oxidizing radicals (Milla et al., 2003; Xu et al., 2015). In this study, the expression of GST genes (BnaC05g07890D, BnaC09g40740D, BnaA06g11500D, BnaC08g37940D, and BnaC04g13950D) was significantly enhanced, depicting their role in LT stress (Figure 5B). In one sentence, these findings elaborated various ROS signaling pathways and provided insight into antioxidant genes, which formed a complex antioxidant defense system under LT stress conditions in B. napus.
Plant hormone signal network to encounter LT stress
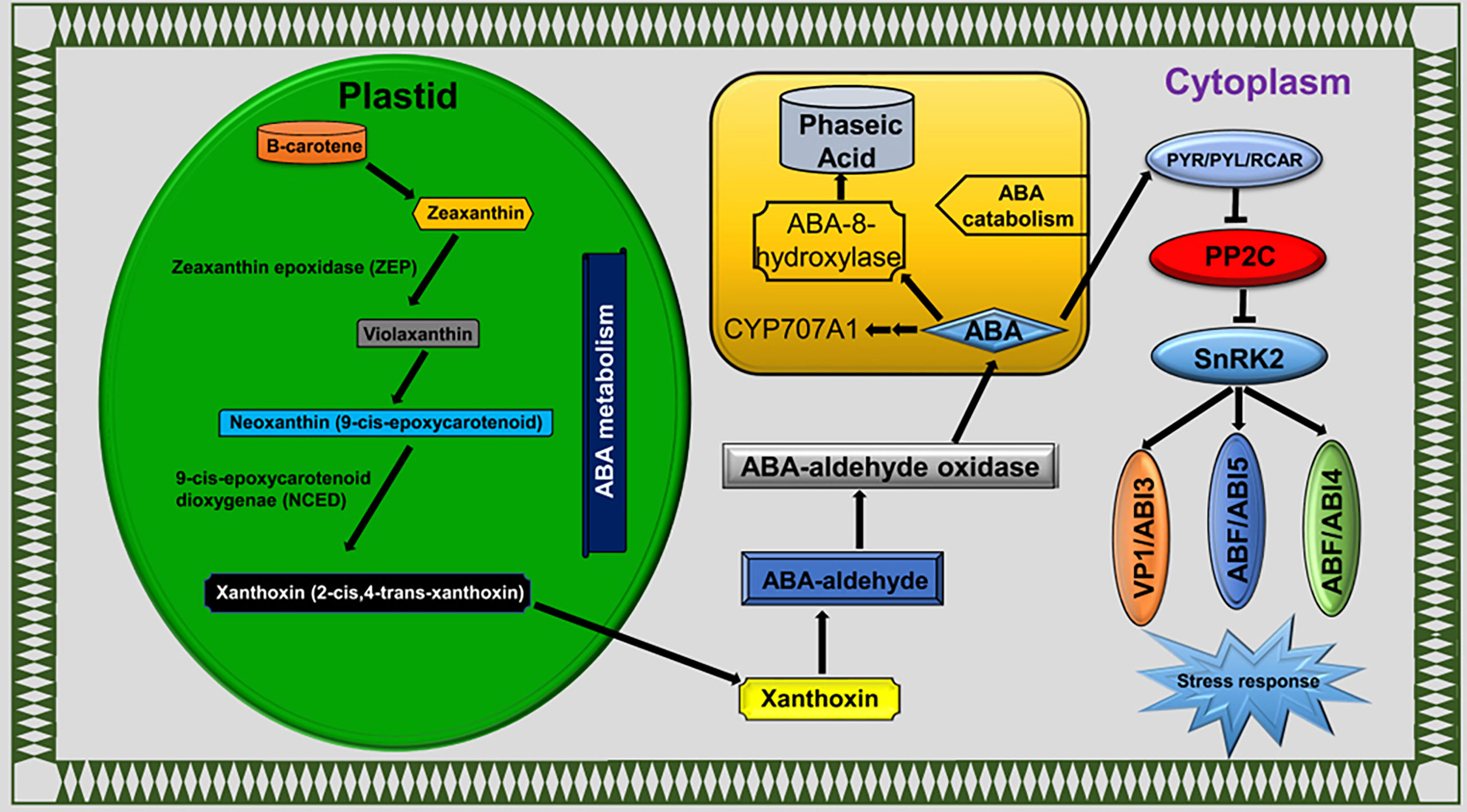
Plant hormones were extensively studied under various environmental cues, including LT stress conditions in plants. Phytohormones have key functions in stress responses, such as to initiate a series of signal events and activate the expression of stress-responsive genes (Ku et al., 2018). Calcium sensors in cellular membranes respond to external stimuli to activate downstream signaling events and are influenced by ABA (Edel and Kudla, 2016). The plant responds to LT stress via ABA-dependent and ABA-independent pathways (Gusta et al., 2005; Ishitani et al., 1997). ABA accumulation mitigates the LT stress effects in rice (Zhao et al., 2015) and bermudagrass (Huang et al., 2017). Interestingly, exogenous ABA treatment reduced the H2O2, electrolyte leakage and MDA contents in bermudagrass compared with non-ABA-treated plants under LT stress conditions (Huang et al., 2017). In this study, we detected the ABA biosynthesis, catabolism, and signaling pathway genes as DEGs (Figure 8). Our findings indicated that genes involved in ABA catabolism, such as CYP707A1/2 (BnaC07g35800D and BnaA03g43960D), were activated in response to LT stress in B. napus (Figure 5C). CRY707A encodes cytochrome P450 monooxygenase that catalyzes the ABA into 8′-hydroxy ABA (Seiler et al., 2011). During the biosynthesis process, ABA repressed the expression of CYP707A genes but increased the expression of NCED. Variation in CYP707A expression level changed the cellular ABA levels and accordingly repressed or induced the NCED expression level. ABA catabolism and biosynthesis work antagonistically, which shows a strong feedback and feedforward loop mechanism to limit or enhance the ABA contents in cells. In addition to ABA accumulation, CYP707A works as a hub to coordinate with auxin, GA, and ABA signals (Liao et al., 2018). The variable expression level of CYP707A genes suggested that ABA catabolism and biosynthesis work side by side in response to LT stress in B. napus (Figure 4). The ABA signaling pathway is a double-negative regulatory system consisting of proteins SNF1-related protein kinase 2 (SnRK2), type 2C protein phosphatase (PP2C), and ABA receptors pyrabactin resistance 1 (PYR1)/PYR1‐like (PYLs)/regulatory components of ABA receptors (RCAR) family (Hauser et al., 2011). In the absence of ABA, PP2C-mediated dephosphorylation repressed SnRK2 kinases to block signal transduction. In response to external stimuli, ABA-receptor PYR/PYL/RCAR protein inactivates PP2C, resulting in SnRK2 activation. Activated SnRK2 causes the upregulation of bZIP group TFs to increase the mRNA level of downstream ABA-responsive genes (Sirichandra et al., 2009; Li et al., 2011) (Figure 8). ABA-insensitive 5 (ABI5) is a bZIP TF that regulates ABA-dependent seed germination, growth, and development under environmental constraints. ABI regulates the expression of downstream ABA pathway genes that consist of the ABSCISIC ACID RESPONSE ELEMENT motif in the promoter region (Skubacz et al., 2016). The Arabidopsis mutant for ABI5 genes exhibited a reduced ABA level and seed dormancy, while complementation of ABI5 rescues the expression of ABA-responsive genes and dormancy. Similarly, the exogenous application of ABA reduces the cold sensitivity of the ABI mutant of Arabidopsis (Wu et al., 2015). Phospholipase D (PLD) involved in ABA signaling is a key regular of plant growth, development, and abiotic stress in B. napus (Lu et al., 2019). In this study, PYR/PYL/RCAR (BnaA04g21960D) was significantly downregulated. Meanwhile, PP2C (BnaA09g49440D, BnaA10g04840D, BnaA10g11080D, BnaC05g41830D, and BnaC09g34350D), SnRK2 (BnaA06g22800D, BnaC01g00890D, and BnaC07g31800D), ABI5 (BnaA05g08020D, BnaC04g09030D, and BnaC07g44670D), and PLD gene (BnaC07g45790D) were found to be upregulated under LT stress (Figure 5C). These findings suggested that the ABA signaling pathway actively responds to LT stress in B. napus.
The phytohormone ethylene (ET), C2H4, is an important regulator of the cold stress response. It is debated whether the effect of ET response to LT stress is negative or positive in plants (Hu et al., 2016). ET negatively affects freezing tolerance as the repression of ET biosynthesis enhances freezing tolerance in Medicago truncatula seedlings (Zhao et al., 2014). On the other side, ET accumulation positively regulates cold tolerance in grapevine (Sun et al., 2016). These results indicate that ET accumulation imparts LT stress tolerance in species-specific manners. The mechanism of the ethylene signaling pathway from ethylene recognition at the cellular membrane to transcriptional regulation in the nucleus and the post-transcriptional level was well reported (Yang et al., 2015; Ku et al., 2018). CTR1 modulates the ethylene signaling pathway through a direct interaction with ET receptors. Active CTR1 can repress EIN2, but EIN2 repression is rescued in the presence of ET. The EIN2 translocates from the cell membrane and activates EIN3/EIL1 in the nucleus. EIN3 and EIL1 are key TFs that modulate ET-responsive gene expression (Ju et al., 2012). The EIN3 targets the ETHYLENE RESPONSE FACTOR1 in Arabidopsis. The ubiquitin/proteasomal degradation system regulated the EIN3 and EIL1 levels in Arabidopsis. EIN2 is compulsory for mediating ethylene-induced EIN3/EIL1 expression and EIN3-binding F-box protein (EBF1/2) degradation (An et al., 2010). We observed several ET signal pathway-related genes—BnaC03g05820D, BnaA07g06360D, BnaC09g50850D, BnaC06g41440D, BnaA05g24390D, BnaC03g26480D, and BnaA06g01090D—which were differentially expressed and may participate in B. napus LT stress response (Figure 5C).
Plant steroids such as brassinosteroids (BR) bind to protein receptors in membranes, regulate membrane structure and growth, and enhance abiotic stress tolerance (Filek et al., 2017). Brassinosteroids enhanced cold stress tolerance in Medicago truncatula (Arfan et al., 2019). In this study, genes related to BR signal pathways were deduced, such as brassinosteroid insensitive 1 (BRI1) precursor (BnaC03g66580D), brassinosteroid biosynthesis (BnaC07g51130D, BnaC01g23810D, and BnaC08g49360D), and brassinosteroid resistant (BZR1/2) (BnaA06g13460D, BnaA05g05170D, and BnaA09g44210D), which were differentially expressed in LT-stressed B. napus (Figure 5C). BRI1 is a transmembrane receptor kinase. BRI1 is required for BR perception at the cell membrane surface and signal transduction via phosphorylation of brassinosteroid signaling kinase. BIN2 phosphorylates BZR1 and BZR2 to downregulate the BR-responsive genes (Zhang et al., 2014). In Arabidopsis, overexpression of BZR1 and knockdown of BIN2 genes enhanced the freezing tolerance via CBF-dependent and CBF-independent pathways (Li et al., 2017a).
Similarly, the phytohormone JA signaling pathway played a key role in signaling and activating the downstream CBF/DREB1 pathway during cold stress in Arabidopsis (Zheng et al., 2018). Exogenous JA effectively increased the freezing tolerance of Artemisia annua (Liu et al., 2017b). Various genes encoding the Jasmonate-zim domain (JAZ) protein (BnaC01g34620D, BnaA06g13250D, BnaC05g35610D, BnaC08g36840D, BnaA01g27170D, BnaC03g71460D, and BnaC02g20120D) were induced under LT stress in this study (Figure 5C). The JAZ proteins bind to the promoter’s region of TFs, such as MYC, to repress jasmonate signaling (Figure 6) (Katsir et al., 2008). CORONATINE INSENSITIVE 1 (COI1) works as a JA receptor and is an important part of the SCF–CoI1 ubiquitin E3 ligase complex. COI1 used the ubiquitin/proteasome pathway to degrade the JAZ protein. JA biosynthetic process genes (BnaA03g55570D, BnaC02g01760D, and BnaC09g48180D) are activated under LT stress (Figure 5C). These results implied that jasmonate signal pathways might function in B. napus LT response processes. In a nutshell, the LT stress response of B. napus variety C18 was a complicated network involving the ABA, JA, BR, and ET signal pathways.
Transcription factor: efficient regulators of genomes in response to LT stress
Different TF families played an important role in enhancing LT stress tolerance, such as APETALA2/ethylene response factor (AP2/ERF), bZIP, WRKY, and MYB in plants. The AP2/ERF TF family gene members are important regulators in stress response (Mizoi et al., 2012). In a previous study, 132 AP2/ERF TFs were found to differentially express and respond to LT stress in rapeseed (Du et al., 2016). The bHLH TF family is the second largest protein family in plants that respond to multiple stresses including drought, salt, and especially cold stress (Sun et al., 2018; Li et al., 2020). MYB TFs exhibited diverse roles in growth, development, anthocyanin accumulation, hormone responses, and cold stress tolerance (An et al., 2020). WRKY TFs played an active role in plant defense against biotic and abiotic challenges. The regulation of WRKY TF enhanced cold tolerance in grapevine, and the heterologous expression of WRKY enhanced Arabidopsis’ cold tolerance (Sun et al., 2019). A significant induction of WRKY TFs demonstrates the prominent roles of this family in B. napus LT stress response (Figure 4). Similarly, NAC TFs engaged in multiple biological processes, including signal transduction, development, and biotic and abiotic stress responses. The plant cold stress response of the NAC TF family was evident in Prunus mume (Zhuo et al., 2018), pepper (Hou et al., 2020), and rice (Pradhan et al., 2019). In previous studies, the role of bZIP TFs in cold stress tolerance was described, such as in B. oleracea (Hwang et al., 2016), Magnolia wufengensis (Deng et al., 2019), and rice (Pradhan et al., 2019). In addition, other TFs such as C2H2, G2-like, GATA, HSF, Trihelix, and ZF-HD, either up- or downregulated, were found in response to LT stress in B. napus. The relative expression of LT-stress-responsive TFs was verified by qRT-PCR (Figure 7). The differential expression of numerous TFs suggested that relatively complex networks activated in response to cold stress in B. napus are regulated at the transcriptional level to fine-tune various biological pathways to enhance LT stress tolerance.
Role of photosynthesis system in LT stress response
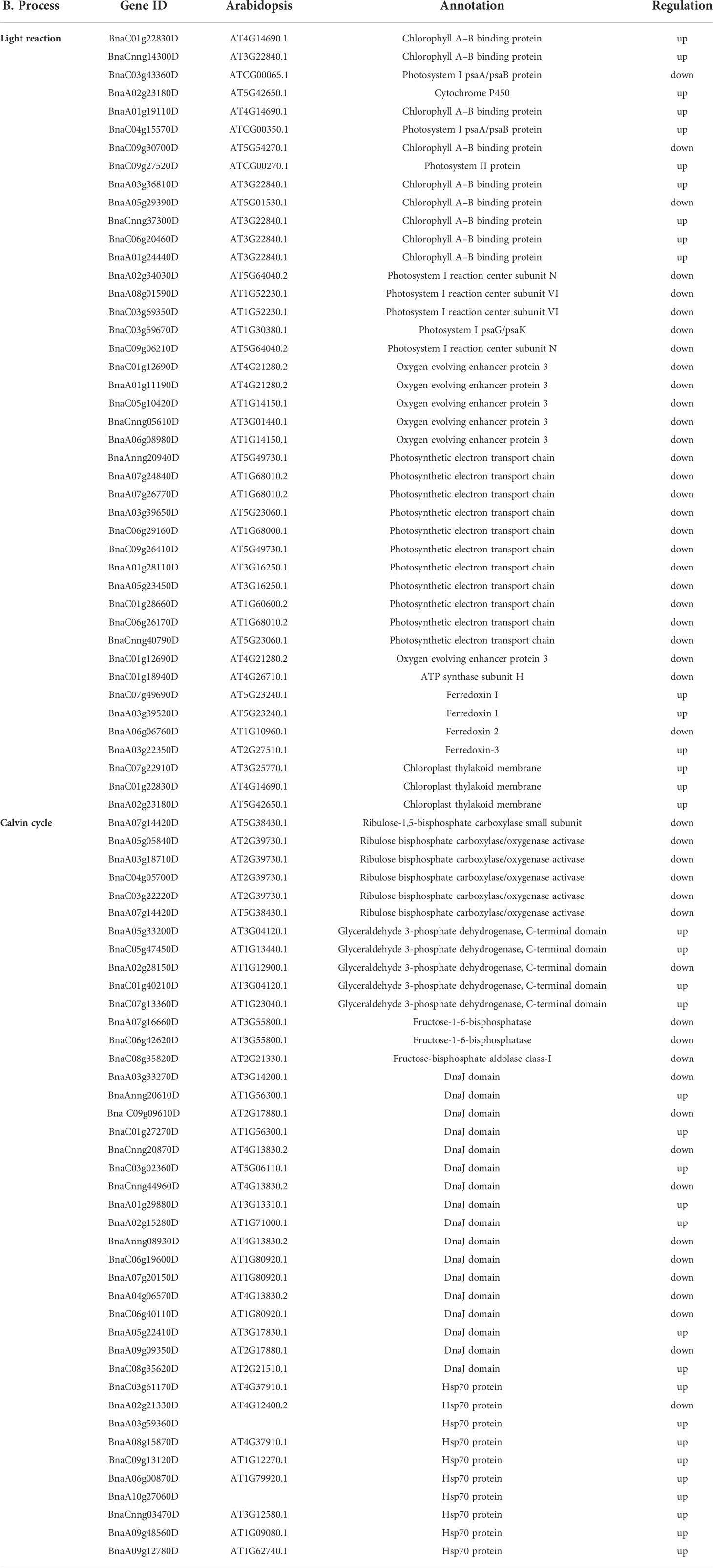
The chloroplast, a photosynthesis organ, is acutely sensitive to LT stress. Thus, LT stress halts the photosynthesis process in plants (Ma et al., 2018). Photosystem II (PSII) is a crucial component of a photosynthetic machinery that is actively inactivated under stress, and the accumulation of ROS halts the repairing mechanism of PSII in plants (Nishiyama and Murata, 2014). LT stress reduces the photosynthetic capacity by affecting the electron transfer in PSII and the efficiency of CO2 fixation to photosynthates (Paredes and Quiles, 2015). The repair mechanism of PSII involves the replacement of damaged D1 protein by newly synthesized D1 protein at the expense of ATP in the degradation and synthesis of D1 (Murata and Nishiyama, 2018). In this study, primarily genes related to the photosynthesis pathway were repressed, which indicated that LT stress halts the photosynthesis system in B. napus (Table 2). Accordingly, we also found that the maximum quantum efficiency of PSII Fv/Fm was reduced in LT-stressed seedlings, indicating a direct relationship with photosynthesis pathway genes (Figure 1C). Most of these genes are essential components of the light reaction and the Calvin cycle process of photosynthesis. During light reaction, sunlight was utilized by chlorophyll pigments to synthesize the high-energy compounds ATP and NADPH (Armbruster et al., 2017). The Calvin cycle is a light-independent redox reaction to fix carbon dioxide into the sugar glucose molecules and gaseous detoxification during photosynthesis (Sun et al., 2015; Nowicka and Kruk, 2018). Repression of genes involved in photosynthetic components were reported in tea (Shi et al., 2019), hibiscus (Paredes and Quiles, 2015), and rice (Liu et al., 2018b) under LT stress. Nonetheless, we also observed that numerous genes in the photosynthetic apparatus were induced during LT stress in B. napus. Among upregulated genes, genes were related to chlorophyll A–B-binding protein (BnaC01g22830D, BnaA01g19110D, and BnaA01g24440D), cytochrome P450, thylakoid membrane, photosynthetic reaction center protein (BnaC09g27520D), and ferredoxin I (BnaC07g49690D and BnaA03g39520D), while genes related to photosynthetic electron transport (BnaA03g39650D) were downregulated. Besides the abovementioned details, two gene families like chloroplastic chaperone protein dnaJ (BnaA01g29880D and BnaA02g15280D) and heat shock protein 70 (HSP70) members (BnaC03g61170D, BnaA03g59360D, and BnaA06g00870D) were induced. Under stress, DnaJ proteins function for protein homeostasis and protein complex stabilization. Transgenic tomato plants overexpressing DnaJ proteins exhibited maximum efficiency and stability of PSII and D1 protein complexes under cold stress conditions. During cold stress, HSP70 was discovered as the partner of DnaJ proteins (Kong et al., 2014). Ferredoxin molecules are sensitive to stress. Engineering of tobacco chloroplasts by an isofunctional protein of ferredoxin (a cyanobacterial flavodoxin) enhanced the tobacco plant tolerance to multiple stresses, including chilling (Zurbriggen et al., 2008). In our study, some genes related to glyceraldehyde-3-phosphate dehydrogenase (GAPDH)—such as BnaA05g33200D, BnaC05g47450D, and BnaC01g40210D—were induced (Table 2). GAPDH is involved in cellular metabolism and generates energy. GAPDH was induced in potato tubers under cold stress (Liu et al., 2017a). The ubiquitous enzyme adenylate kinase interacts with the chloroplast GAPDH, forming a stable complex that regulates the ATP/NADPH ratio inside the chloroplast to optimize the Calvin–Benson cycle in response to environmental stress (Zhang et al., 2018). Then, we performed qRT-PCR to verify the expression pattern of photosynthesis pathway-related DEGs, which further confirmed the reliability of the RNA-seq data (Figure 7). Thus, differential expression of photosynthetic system genes indicates that they may play vital roles in response to LT stress in B. napus. The exact mechanism of these factors remains unresolved and provides new directions for future LT stress studies.
Protein–protein interaction networks in LT stress response
Protein interaction networks regulate various cellular functions, such as signal transduction, metabolic pathways, organ formation, cell cycle regulation, and plant defense (Bray, 2006; Zhang et al., 2010). The protein–protein interactions of the sugar metabolism, hormone, antioxidant activity, transcription factors, and photosynthesis-related genes revealed that the protein domains physically interact through a complex network. Specifically, the SnRK2 family contains key regulators of cellular osmotic stress and ABA responses in plants. Here the SNF1-related protein kinase 2 (SnRK2), ABI, AOS, JAZ, and HSP families shared a maximum number of interaction lines (Figure 6). CYP707A1 encodes abscisic acid 8’-hydroxylases, which is indispensable for seed dormancy and germination in Arabidopsis (Okamoto et al., 2006). Similarly, ABI5 plays a significant role in the regulation of seed germination, seedling growth, development, and abiotic stresses (Zinsmeister et al., 2016). In the ABA signal pathway, the ABI5 protein directly interacts with CYP707A1, ABI2, PP2C phosphatases, PYR/PYL/RCAR receptors, and SnRK2 kinases. The interaction of these proteins suggested that the expression of these genes is tightly associated with LT stress response. Based on these findings, ABI5 is a hub protein under LT response in B. napus which directly or indirectly interacts with antioxidants, TFs, ABA signaling, and regulation of LT-stress-related genes along with various other proteins. In Arabidopsis, alginate oligosaccharide (AOS) induced resistance to the pathogen by a salicylic-acid-mediated signaling pathway (Zhang et al., 2019a). Jasmonate and related signaling compounds have a significant role in plant immunity and development. WRKY1 negatively regulates the ABA signal pathway through JAZ1 and ABI1 in drought response (Luo et al., 2020). Moreover, AOS functions in fine-tuning JA formation and improving LT stress tolerance (Pi et al., 2009; Liu et al., 2017b). In this study, the JAZ protein directly interacts with WRKY, AOS, and other essential genes. These results showed that a complex interactive network at the protein level was activated in response to LT stress response in B. napus. Within the network, some proteins act as a hub to activate or repress the function of other related proteins, which helped to cope with the LT stress in B. napus.
Conclusion
B. napus is the major oil crop with the largest planting area in China. However, its productivity and cultivation area are severely affected by LT stress. In the current study, the transcriptomic study helped to identify various LT-stress-responsive pathways and molecular events. A set of potential DEGs that respond to LT stress was discovered. Different expressions of the key systems indicated that they might play vital roles in response to LT stress in B. napus. It is noticeable that the B. napus response to LT stress is multiplex, not only regulating the genetic architecture but also activating the antioxidant and osmoprotectant system to improve the LT stress tolerance (Figure 9). A deep understanding of these molecular events will provide new avenues for LT stress tolerance improvement in B. napus.
Data availability statement
The data presented in the study are deposited in the National Center for Biotechnology 136 Information (NCBI) repository, accession number PRJNA596550.
Author contributions
MH and YL conceived the idea and wrote the manuscript. MH, DL, and ZL performed the experiments. DL and XD helped in the literature searches and data analysis. YC, XZ, GL, and YL supervised the work, reviewed and edited the manuscript, and managed funding resources. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Key Research Program & Technology Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS-ZDRW202109 and CAAS-ZDRW202105), the National Nature Science Foundation of China (32072131), the Key Research and Development projects in Hubei Province (2022BBA0038), and the Science and Technology Innovation Project of the Chinese Academy of Agricultural Sciences (CAAS), China, and Fundamental Research Funds for the Central Nonprofit Scientific Institution (OCRI, CAAS, 1610172018010).
Acknowledgments
MH is thankful to CSC for providing a Ph.D. scholarship and to OCRI-CAAS for research environment. MH is thankful to GYL for article language improvement and providing APC. We would also like to thank all the members of the Key Lab of Biology and Genetic Improvement of Oil Crops, OCRI-CAAS, Wuhan, China, for their support throughout the study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1050995/full#supplementary-material
References
Almadanim, M. C., Alexandre, B. M., Rosa, M. T. G., Sapeta, H., Leitao, A. E., Ramalho, J. C., et al. (2017). Rice calcium-dependent protein kinase OsCPK17 targets plasma membrane intrinsic protein and sucrose-phosphate synthase and is required for a proper cold stress response. Plant Cell Environ. 40, 1197–1213. doi: 10.1111/pce.12916
An, J. P., Li, R., Qu, F. J., You, C. X., Wang, X. F., Hao, Y. J. (2018). An apple NAC transcription factor negatively regulates cold tolerance via CBF-dependent pathway. J. Plant Physiol. 221, 74–80. doi: 10.1016/j.jplph.2017.12.009
An, J. P., Wang, X. F., Zhang, X. W., Xu, H. F., Bi, S. Q., You, C. X., et al. (2020). An apple MYB transcription factor regulates cold tolerance and anthocyanin accumulation and undergoes MIEL1-mediated degradation. Plant Biotech. J. 18, 337–353. doi: 10.1111/pbi.13201
An, F., Zhao, Q., Ji, Y., Li, W., Jiang, Z., Yu, X., et al. (2010). Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding f-box 1 and 2 that requires EIN2 in arabidopsis. Plant Cell. 22, 2384–2401. doi: 10.1105/tpc.110.076588
Arfan, M., Zhang, D. W., Zou, L. J., Luo, S. S., Tan, W. R., Zhu, T., et al. (2019). Hydrogen peroxide and nitric oxide crosstalk mediates brassinosteroids induced cold stress tolerance in medicago truncatula. Int. J. Mol. Sci. 20, 1–15. doi: 10.3390/ijms20010144
Armbruster, U., Correa Galvis, V., Kunz, H. H., Strand, D. D. (2017). The regulation of the chloroplast proton motive force plays a key role for photosynthesis in fluctuating light. Curr. Opin. Plant Biol. 37, 56–62. doi: 10.1016/j.pbi.2017.03.012
Bari, R., Jones, J. D. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69, 473–488. doi: 10.1007/s11103-008-9435-0
Baud, S., Vaultier, M. N., Rochat, C. (2004). Structure and expression profile of the sucrose synthase multigene family in arabidopsis. J. Exp. Bot. 55, 397–409. doi: 10.1093/jxb/erh047
Benjamini, Y., Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc Ser. 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Bray, S. J. (2006). Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7, 678–689. doi: 10.1038/nrm2009
Caverzan, A., Passaia, G., Rosa, S. B., Ribeiro, C. W., Lazzarotto, F., Margis-Pinheiro, M. (2012). Plant responses to stresses: Role of ascorbate peroxidase in the antioxidant protection. Genet. Mol. Biol. 35, 1011–1020. doi: 10.1590/S1415-47572012000600016
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wang, X., et al. (2014). Plant genetics. early allopolyploid evolution in the post-neolithic brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Ciereszko, I., Johansson, H., Kleczkowski, L. A. (2001). Sucrose and light regulation of a cold-inducible UDP-glucose pyrophosphorylase gene via a hexokinase-independent and abscisic acid-insensitive pathway in arabidopsis. Biochem. J. 354, 67–72. doi: 10.1042/bj3540067
Conesa, A., Götz, S. (2008). Blast2GO: A comprehensive suite for functional analysis in plant genomics. Inter J. Plant Genomics 2008, 619832. doi: 10.1155/2008/619832
Cong, R. H., Zhang, Z., Lu, J. W. (2019). Climate impacts on yield of winter oilseed rape in different growth regions of the Yangtze river basin. Chin. J. Oil Crop Science. 41, 894–903. doi: 10.19802/j.issn.1007-9084.2019046
Deng, S., Ma, J., Zhang, L., Chen, F., Sang, Z., Jia, Z., et al. (2019). De novo transcriptome sequencing and gene expression profiling of magnolia wufengensis in response to cold stress. BMC Plant Biol. 19, 321. doi: 10.1186/s12870-019-1933-5
Ding, Y., Shi, Y., Yang, S. (2019). Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phyto. 222, 1690–1704. doi: 10.1111/nph.15696
Du, C., Hu, K., Xian, S., Liu, C., Fan, J., Tu, J., et al. (2016). Dynamic transcriptome analysis reveals AP2/ERF transcription factors responsible for cold stress in rapeseed (Brassica napus l.). Mol. Genet. Genomics 291, 1053–1067. doi: 10.1007/s00438-015-1161-0
Du, X. Z., Jin, Z. P., Liu, D. M., Yang, G. D., Pei, Y. X. (2017). Hydrogen sulfide alleviates the cold stress through MPK4 in arabidopsis thaliana. Plant Physiol. Bioch. 120, 112–119. doi: 10.1016/j.plaphy.2017.09.028
Edel, K. H., Kudla, J. (2016). Integration of calcium and ABA signaling. Curr. Opin. Plant Biol. 33, 83–91. doi: 10.1016/j.pbi.2016.06.010
Eveland, A. L., Jackson, D. P. (2012). Sugars, signalling, and plant development. J. Exp. Bot. 63, 3367–3377. doi: 10.1093/jxb/err379
Filek, M., Rudolphi-Skorska, E., Sieprawska, A., Kvasnica, M., Janeczko, A. (2017). Regulation of the membrane structure by brassinosteroids and progesterone in winter wheat seedlings exposed to low temperature. Steroids 128, 37–45. doi: 10.1016/j.steroids.2017.10.002
Fugate, K. K., Eide, J. D., Martins, D. N., Grusak, M. A., Deckard, E. L., Finger, F. L. (2019). Colocalization of sucrose synthase expression and sucrose storage in the sugarbeet taproot indicates a potential role for sucrose catabolism in sucrose accumulation. J. Plant Physiol. 240, 153016. doi: 10.1016/j.jplph.2019.153016
Gilmour, S. J., Sebolt, A. M., Salazar, M. P., Everard, J. D., Thomashow, M. F. (2000). Overexpression of the arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 124, 1854–1865. doi: 10.1104/pp.124.4.1854
Guan, S., Xu, Q., Ma, D., Zhang, W., Xu, Z., Zhao, M., et al. (2019). Transcriptomics profiling in response to cold stress in cultivated rice and weedy rice. Gene 685, 96–105. doi: 10.1016/j.gene.2018.10.066
Gu, H., Lu, M., Zhang, Z., Xu, J., Cao, W., Miao, M. (2018). Metabolic process of raffinose family oligosaccharides during cold stress and recovery in cucumber leaves. J. Plant Physiol. 224-225, 112–120. doi: 10.1016/j.jplph.2018.03.012
Gusta, L. V., Trischuk, R., Weiser, C. J. (2005). Plant Cold Acclimation: The Role of Abscisic Acid. Journal of Plant Growth Regulation. 24:308–318.
Hang, R., Wang, Z., Deng, X., Liu, C., Yan, B., Yang, C., et al. (2018). Ribosomal RNA biogenesis and its response to chilling stress in oryza sativa. Plant Physiol. 177, 381–397. doi: 10.1104/pp.17.01714
Hauser, F., Waadt, R., Schroeder, J. I. (2011). Evolution of abscisic acid synthesis and signaling mechanisms. Curr. Biol. 21, R346–R355. doi: 10.1016/j.cub.2011.03.015
Heidarvand, L., Maali, A. R. (2010). What happens in plant molecular responses to cold stress? Acta Physiol. Plant 32, 419–431. doi: 10.1007/s11738-009-0451-8
Hou, X. M., Zhang, H. F., Liu, S. Y., Wang, X. K., Zhang, Y. M., Meng, Y. C., et al. (2020). The NAC transcription factor CaNAC064 is a regulator of cold stress tolerance in peppers. Plant Sci. 291, 110346. doi: 10.1016/j.plantsci.2019.110346
Huang, Y., Hussain, M. A., Luo, D., Xu, H., Zeng, C., Havlickova, L., et al. (2020). A brassica napus reductase gene dissected by associative transcriptomics enhances plant adaption to freezing stress. Front. Plant Sci. 11, 971. doi: 10.3389/fpls.2020.00971
Huang, X., Shi, H., Hu, Z., Liu, A., Amombo, E., Chen, L., et al. (2017). ABA is involved in regulation of cold stress response in bermudagrass. Front. Plant Sci. 8, 1613. doi: 10.3389/fpls.2017.01613
Huang, Z., Zhao, N., Qin, M., Xu, A. (2018). Mapping of quantitative trait loci related to cold resistance in brassica napus l. J. Plant Physiol. 231, 147–154. doi: 10.1016/j.jplph.2018.09.012
Hu, Z., Fan, J., Chen, K., Amombo, E., Chen, L., Fu, J. (2016). Effects of ethylene on photosystem II and antioxidant enzyme activity in Bermuda grass under low temperature. Photosynth Res. 128, 59–72. doi: 10.1007/s11120-015-0199-5
Hurry, V., Strand, A., Furbank, R., Stitt, M. (2000). The role of inorganic phosphate in the development of freezing tolerance and the acclimatization of photosynthesis to low temperature is revealed by the pho mutants of arabidopsis thaliana. Plant J. 24, 383–396. doi: 10.1046/j.1365-313x.2000.00888.x
Hwang, I., Manoharan, R. K., Kang, J. G., Chung, M. Y., Kim, Y. W., Nou, I. S. (2016). Genome-wide identification and characterization of bZIP transcription factors in brassica oleracea under cold stress. BioMed. Res. Int. 2016, 4376598. doi: 10.1155/2016/4376598
Ishitani, M., Xiong, L., Stevenson, B., Zhu, J. K. (1997). Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell. 9, 1935–1949.
Jaglo, K. R., Kleff, S., Amundsen, K. L., Zhang, X., Haake, V., Zhang, J. Z., et al. (2001). Components of the arabidopsis c-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in brassica napus and other plant species. Plant Physiol. 127, 910–917. doi: 10.1104/pp.010548
Jaikumar, N. S., Snapp, S. S., Sharkey, T. D. (2016). Older thinopyrum intermedium (Poaceae) plants exhibit superior photosynthetic tolerance to cold stress and greater increases in two photosynthetic enzymes under freezing stress compared with young plants. J. Exp. Bot. 67, 4743–4753. doi: 10.1093/jxb/erw253
Janmohammadi, M., Zolla, L., Rinalducci, S. (2015). Low temperature tolerance in plants: changes at the protein level. Phytochemistry 117, 76–89. doi: 10.1016/j.phytochem.2015.06.003
Ju, C., Yoon, G. M., Shemansky, J. M., Lin, D. Y., Ying, Z. I., Chang, J., et al. (2012). CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in arabidopsis. Proc. Natl. Acad. Sci. U S A. 109, 19486–19491. doi: 10.1073/pnas.1214848109
Kaplan, F., Kopka, J., Sung, D. Y., Zhao, W., Popp, M., Porat, R., et al. (2007). Transcript and metabolite profiling during cold acclimation of arabidopsis reveals an intricate relationship of cold-regulated gene expression with modifications in metabolite content. Plant J. 50, 967–981. doi: 10.1111/j.1365-313X.2007.03100.x
Katsir, L., Schilmiller, A. L., Staswick, P. E., He, S. Y., Howe, G. A. (2008). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci U S A. 105, 7100–7105.
Ke, L., Lei, W., Yang, W., Wang, J., Gao, J., Cheng, J., et al. (2020). Genome-wide identification of cold responsive transcription factors in Brassica napus l. BMC Plant Biol. 20, 62. doi: 10.1186/s12870-020-2253-5
Kong, F., Deng, Y., Zhou, B., Wang, G., Wang, Y., Meng, Q. (2014). A chloroplast-targeted DnaJ protein contributes to maintenance of photosystem II under chilling stress. J. Exp. Bot. 65, 143–158. doi: 10.1093/jxb/ert357
Kroll, J. S., Langford, P. R., Wilks, K. E., Keil, A. D. (1995). Bacterial [Cu,Zn]-superoxide dismutase: phylogenetically distinct from the eukaryotic enzyme, and not so rare after all! Microbiology 141, 2271–2279. doi: 10.1099/13500872-141-9-2271
Ku, Y. S., Sintaha, M., Cheung, M. Y., Lam, H. M. (2018). Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int. J. Mol. Sci. 19, 1–35. doi: 10.3390/ijms19103206
Liao, X., Li, M., Liu, B., Yan, M., Yu, X., Zi, H., et al. (2018). Interlinked regulatory loops of ABA catabolism and biosynthesis coordinate fruit growth and ripening in woodland strawberry. Proc. Natl. Acad. Sci. U S A. 115, E11542–E11550. doi: 10.1073/pnas.1812575115
Li, H., Dong, Y., Chang, J., He, J., Chen, H., Liu, Q., et al. (2016a). High-throughput MicroRNA and mRNA sequencing reveals that MicroRNAs may be involved in melatonin-mediated cold tolerance in citrullus lanatus l. Front. Plant Sci. 7, 1231. doi: 10.3389/fpls.2016.01231
Li, W., Huai, X., Li, P., Raza, A., Mubarik, M. S., Habib, M., et al. (2021). Genome-wide characterization of glutathione peroxidase (GPX) gene family in rapeseed (Brassica napus l.) revealed their role in multiple abiotic stress response and hormone signaling. Antioxidants (Basel). 10. doi: 10.3390/antiox10091481
Li, Z., Hu, G., Liu, X., Zhou, Y., Li, Y., Zhang, X., et al. (2016b). Transcriptome sequencing identified genes and gene ontologies associated with early freezing tolerance in maize. Front. Plant Sci. 7, 1477. doi: 10.3389/fpls.2016.01477
Li, C., Jia, H., Chai, Y., Shen, Y. (2011). Abscisic acid perception and signaling transduction in strawberry: a model for non-climacteric fruit ripening. Plant Signal. Behav. 6, 1950–1953. doi: 10.4161/psb.6.12.18024
Li, Y., Li, L., Ding, W., Li, H., Shi, T., Yang, X., et al. (2020). Genome-wide identification of osmanthus fragrans bHLH transcription factors and their expression analysis in response to abiotic stress. Environ. Exp. Botany. 172, 103990. doi: 10.1016/j.envexpbot.2020.103990
Li, N. N., Qian, W. J., Wang, L., Cao, H. L., Hao, X. Y., Yang, Y. J., et al. (2017b). Isolation and expression features of hexose kinase genes under various abiotic stresses in the tea plant (Camellia sinensis). J. Plant Physiol. 209, 95–104. doi: 10.1016/j.jplph.2016.11.007
Liu, W. H., Cheng, C. Z., Chen, F. L., Ni, S. S., Lin, Y. L., Lai, Z. X. (2018a). High-throughput sequencing of small RNAs revealed the diversified cold-responsive pathways during cold stress in the wild banana (Musa itinerans). BMC Plant Biol. 18, 1–26. doi: 10.1186/s12870-018-1483-2
Liu, T., Fang, H., Liu, J., Reid, S., Hou, J., Zhou, T., et al. (2017a). Cytosolic glyceraldehyde-3-phosphate dehydrogenases play crucial roles in controlling cold-induced sweetening and apical dominance of potato (Solanum tuberosum l.) tubers. Plant Cell Environ. 40, 3043–3054. doi: 10.1111/pce.13073
Liu, C., Feng, Z. C., Xiao, T. H., Ma, X. M., Zhou, G. S., Huang, F. H., et al. (2019). Development, potential and adaption of Chinese rapeseed industry. Chin. J. Oil Crop Science. 41, 485–489. doi: 10.7505/j.issn.1007-9084.2019.04.001
Liu, X., Lan, J., Huang, Y., Cao, P., Zhou, C., Ren, Y., et al. (2018b). WSL5, a pentatricopeptide repeat protein, is essential for chloroplast biogenesis in rice under cold stress. J. Exp. Bot. 69, 3949–3961. doi: 10.1093/jxb/ery214
Liu, W., Wang, H., Chen, Y., Zhu, S., Chen, M., Lan, X., et al. (2017b). Cold stress improves the production of artemisinin depending on the increase in endogenous jasmonate. Biotechnol. Appl. Biochem. 64, 305–314. doi: 10.1002/bab.1493
Liu, Y., Zhang, L., Chen, L., Ma, H., Ruan, Y., Xu, T., et al. (2016). Molecular cloning and expression of an encoding galactinol synthase gene (AnGolS1) in seedling of ammopiptanthus nanus. Sci. Rep. 6, 36113. doi: 10.1038/srep36113
Li, H., Ye, K., Shi, Y., Cheng, J., Zhang, X., Yang, S. (2017a). BZR1 positively regulates freezing tolerance via CBF-dependent and CBF-independent pathways in arabidopsis. Mol. Plant 10, 545–559. doi: 10.1016/j.molp.2017.01.004
Li, P., Zheng, T., Li, L., Zhuo, X., Jiang, L., Wang, J., et al. (2019). Identification and comparative analysis of the CIPK gene family and characterization of the cold stress response in the woody plant prunus mume. PeerJ. 7, e6847. doi: 10.7717/peerj.6847
Lu, S., Fadlalla, T., Tang, S., Li, L., Ali, U., Li, Q., et al. (2019). Genome-wide analysis of phospholipase d gene family and profiling of phospholipids under abiotic stresses in brassica napus. Plant Cell Physiol. 60, 1556–1566. doi: 10.1093/pcp/pcz071
Luo, X., Li, C., He, X., Zhang, X., Zhu, L. (2020). ABA signaling is negatively regulated by GbWRKY1 through JAZ1 and ABI1 to affect salt and drought tolerance. Plant Cell Rep. 39, 181–194. doi: 10.1007/s00299-019-02480-4
Luo, T., Xian, M., Zhang, C., Zhang, C., Hu, L., Xu, Z. (2019). Associating transcriptional regulation for rapid germination of rapeseed (Brassica napus l.) under low temperature stress through weighted gene co-expression network analysis. Sci. Rep. 9, 55. doi: 10.1038/s41598-018-37099-0
Lv, Y., Yang, M., Hu, D., Yang, Z., Ma, S., Li, X., et al. (2017). The OsMYB30 transcription factor suppresses cold tolerance by interacting with a JAZ protein and suppressing β-amylase expression. Plant Physiol. 173, 1475–1491. doi: 10.1104/pp.16.01725
Ma, X., Chen, C., Yang, M., Dong, X., Lv, W., Meng, Q. (2018). Cold-regulated protein (SlCOR413IM1) confers chilling stress tolerance in tomato plants. Plant Physiol. Biochem. 124, 29–39. doi: 10.1016/j.plaphy.2018.01.003
Ma, L., Coulter, J. A., Liu, L., Zhao, Y., Chang, Y., Pu, Y., et al. (2019). Transcriptome analysis reveals key cold-Stress-Responsive genes in winter rapeseed (Brassica rapa L.). Int. J. Mol. Sci. 20, 1–19. doi: 10.3390/ijms20051071
Mao, X. Z., Cai, T., Olyarchuk, J. G., Wei, L. P. (2005). Automated genome annotation and pathway identification using the KEGG orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793. doi: 10.1093/bioinformatics/bti430
Maryan, K. E., Lahiji, H. S., Farrokhi, N., Komeleh, H. H. (2019). Analysis of brassica napus dehydrins and their Co-expression regulatory networks in relation to cold stress. Gene Expr Patterns. 31, 7–17. doi: 10.1016/j.gep.2018.10.002
Miller, G., Suzuki, N., Ciftci-Yilmaz, S., Mittler, R. (2010). Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 33, 453–467. doi: 10.1111/j.1365-3040.2009.02041.x
Mizoi, J., Shinozaki, K., Yamaguchi-Shinozaki, K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 86–96. doi: 10.1016/j.bbagrm.2011.08.004
Murata, N., Nishiyama, Y. (2018). ATP is a driving force in the repair of photosystem II during photoinhibition. Plant Cell Environ. 41, 285–299. doi: 10.1111/pce.13108
Nishiyama, Y., Murata, N. (2014). Revised scheme for the mechanism of photoinhibition and its application to enhance the abiotic stress tolerance of the photosynthetic machinery. Appl. Microbiol. Biotechnol. 98, 8777–8796. doi: 10.1007/s00253-014-6020-0
Niu, X., Lu, H., Fan, Y., Wang, W., Yuan, Y., Hawkins, M., et al. (2022). Manipulation of the transcription factor SlNAC1 for improved tolerance to abiotic stress in tomato. Plant Cell Environ. 1–14. doi: 10.1111/pce.14437
Nowicka, B., Kruk, J. (2018). [Genetic engineering as a method for the improvement of photosynthesis]. Postepy Biochem. 64, 13–20. doi: 10.18388/pb.2018_100
Oelze, M. L., Kandlbinder, A., D., K. J. (2008). Redox regulation and over reduction control in the photosynthesizing cell: complexity in redox regulatory networks. BBA Gen. Subj. 1780, 1261–1272. doi: 10.1016/j.bbagen.2008.03.015
Okamoto, M., Kuwahara, A., Seo, M., Kushiro, T., Asami, T., Hirai, N., et al. (2006). CYP707A1 and CYP707A2, which encode abscisic acid 8'-hydroxylases, are indispensable for proper control of seed dormancy and germination in arabidopsis. Plant Physiol. 141, 97–107. doi: 10.1104/pp.106.079475
Paredes, M., Quiles, M. J. (2015). The effects of cold stress on photosynthesis in hibiscus plants. PloS One 10, e0137472. doi: 10.1371/journal.pone.0137472
Pi, Y., Jiang, K., Cao, Y., Wang, Q., Huang, Z., Li, L., et al. (2009). Allene oxide cyclase from camptotheca acuminata improves tolerance against low temperature and salt stress in tobacco and bacteria. Mol. Biotechnol. 41, 115–122. doi: 10.1007/s12033-008-9106-z
Pradhan, S. K., Pandit, E., Nayak, D. K., Behera, L., Mohapatra, T. (2019). Genes, pathways and transcription factors involved in seedling stage chilling stress tolerance in indica rice through RNA-seq analysis. BMC Plant Biol. 19, 352. doi: 10.1186/s12870-019-1922-8
Pu, Y., Liu, L., Wu, J., Zhao, Y., Bai, J., Ma, L., et al. (2019). Transcriptome profile analysis of winter wapeseed (Brassica napus L.) in response to freezing stress, reveal potentially connected events to freezing stress. Int. J. Mol. Sci. 20, 1–24. doi: 10.3390/ijms20112771
Qi, J., Song, C. P., Wang, B., Zhou, J., Kangasjärvi, J., Zhu, J. K., et al. (2018). Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J. Integr. Plant Biol. 60, 805–826. doi: 10.1111/jipb.12654
Raza, A., Su, W., Hussain, M. A., Mehmood, S. S., Zhang, X., Cheng, Y., et al. (2021). Integrated analysis of metabolome and transcriptome reveals insights for cold tolerance in rapeseed (Brassica napus l.). Front. Plant Sci. 12, 721681. doi: 10.3389/fpls.2021.721681
Salvi, P., Saxena, S. C., Petla, B. P., Kamble, N. U., Kaur, H., Verma, P., et al. (2016). Differentially expressed galactinol synthase(s) in chickpea are implicated in seed vigor and longevity by limiting the age induced ROS accumulation. Sci. Rep. 6, 35088. doi: 10.1038/srep35088
Schieber, M., Chandel, N. S. (2014). ROS function in redox signaling and oxidative stress. Curr. Biol. 24, R453–R462. doi: 10.1016/j.cub.2014.03.034
Seiler, C., Harshavardhan, V. T., Rajesh, K., Reddy, P. S., Strickert, M., Rolletschek, H., et al. (2011). ABA biosynthesis and degradation contributing to ABA homeostasis during barley seed development under control and terminal drought-stress conditions. J. Exp. Bot. 62, 2615–2632. doi: 10.1093/jxb/erq446
Sharma, T. R., Devanna, B. N., Kiran, K., Singh, P. K., Arora, K., Jain, P., et al. (2018). Status and prospects of next generation sequencing technologies in crop plants. Curr. Issues Mol. Biol. 27, 1–36. doi: 10.21775/cimb.027.001
Sharma, A., Shahzad, B., Kumar, V., Kohli, S. K., Sidhu, G. P. S., Bali, A. S., et al. (2019). Phytohormones regulate accumulation of osmolytes under abiotic stress. Biomolecules 9, 1–36. doi: 10.3390/biom9070285
Shi, Y., Cai, Z., Li, D., Lu, J., Ye, J., Liang, Y., et al. (2019). Effect of freezing on photosystem II and assessment of freezing tolerance of tea cultivar. Plants 8, 1–14. doi: 10.3390/plants8100434
Shi, Y., Ding, Y., Yang, S. (2018). Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 23, 623–637. doi: 10.1016/j.tplants.2018.04.002
Sicher, R. (2011). Carbon partitioning and the impact of starch deficiency on the initial response of arabidopsis to chilling temperatures. Plant Sci. 181, 167–176. doi: 10.1016/j.plantsci.2011.05.005
Sies, H., Carsten, B., Dean, P. J. (2017). Oxidative stress. Annu. Rev. Biochem. 86, 715–748. doi: 10.1146/annurev-biochem-061516-045037
Sirichandra, C., Gu, D., Hu, H. C., Davanture, M., Lee, S., Djaoui, M., et al. (2009). Phosphorylation of the arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett. 583, 2982–2986. doi: 10.1016/j.febslet.2009.08.033
Skubacz, A., Daszkowska-Golec, A., Szarejko, I. (2016). The role and regulation of ABI5 (ABA-insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front. Plant Sci. 7, 1884. doi: 10.3389/fpls.2016.01884
Song, J., Zeng, L., Chen, R., Wang, Y., Zhou, Y. (2018). In silico identification and expression analysis of superoxide dismutase (SOD) gene family in medicago truncatula. 3 Biotech. 8, 348. doi: 10.1007/s13205-018-1373-1
Stein, O., Granot, D. (2019). An overview of sucrose synthases in plants. Front. Plant Sci. 10, 95. doi: 10.3389/fpls.2019.00095
Strand, A., Hurry, V., Henkes, S., Huner, N., Gustafsson, P., Gardeström, P., et al. (1999). Acclimation of arabidopsis leaves developing at low temperatures. increasing cytoplasmic volume accompanies increased activities of enzymes in the Calvin cycle and in the sucrose-biosynthesis pathway. Plant Physiol. 119, 1387–1398. doi: 10.1104/pp.119.4.1387
Su, C., Chen, K., Ding, Q., Mou, Y., Yang, R., Zhao, M., et al. (2018). Proteomic analysis of the function of a novel cold-regulated multispanning transmembrane protein COR413-PM1 in arabidopsis. Int. J. Mol. Sci. 19, 1–22. doi: 10.3390/ijms19092572
Sun, X., Wang, Y., Sui, N. (2018). Transcriptional regulation of bHLH during plant response to stress. Biochem. Biophys. Res. Commun. 503, 397–401. doi: 10.1016/j.bbrc.2018.07.123
Sun, H., Zhang, W., Tang, L., Han, S., Wang, X., Zhou, S., et al. (2015). Investigation of the role of the calvin cycle and C1 metabolism during HCHO metabolism in gaseous HCHO-treated petunia under light and dark conditions using 13C-NMR. Phytochem. Anal. 26, 226–235. doi: 10.1002/pca.2556
Sun, X., Zhang, L., Wong, D. C. J., Wang, Y., Zhu, Z., Xu, G., et al. (2019). The ethylene response factor VaERF092 from amur grape regulates the transcription factor VaWRKY33, improving cold tolerance. Plant J. 99, 988–1002. doi: 10.1111/tpj.14378
Sun, X., Zhao, T., Gan, S., Ren, X., Fang, L., Karungo, S. K., et al. (2016). Ethylene positively regulates cold tolerance in grapevine by modulating the expression of ETHYLENE RESPONSE FACTOR 057. Sci. Rep. 6, 24066. doi: 10.1038/srep24066
Su, W., Raza, A., Gao, A., Jia, Z., Zhang, Y., Hussain, M. A., et al. (2021). Genome-wide analysis and expression profile of superoxide dismutase (SOD) gene family in rapeseed (Brassica napus l.) under different hormones and abiotic stress conditions. Antioxidants (Basel). 10. doi: 10.3390/antiox10081182
Suzuki, N., Mittler, R. (2006). Reactive oxygen species and temperature stresses: A delicate balance between signaling and destruction. Physiologia Plantarum. 126, 45–51. doi: 10.1111/j.0031-9317.2005.00582.x
Trapnell, C. (2010). Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. . 28, 511–515. doi: 10.1038/nbt.1621
Tyczewska, A., Gracz, J., Kuczynski, J., Twardowski, T. (2016). Deciphering the soybean molecular stress response via high-throughput approaches. Acta Biochim. Pol. 63, 631–643. doi: 10.18388/abp.2016_1340
Verma, V., Ravindran, P., Kumar, P. P. (2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16, 86. doi: 10.1186/s12870-016-0771-y
Wang, Z., Chen, Y., Fang, H. D., Shi, H. F., Chen, K. P., Zhang, Z. Y., et al. (2014). Selection of reference genes for quantitative reverse-transcription polymerase chain reaction normalization in brassica napus under various stress conditions. Mol. Genet. Genomics 289, 1023–1035. doi: 10.1007/s00438-014-0853-1
Wang, L. B., Wang, L., Zhang, Z. E., Ma, M., Wang, R. Z., Qian, M., et al. (2018a). Genome-wide identification and comparative analysis of the superoxide dismutase gene family in pear and their functions during fruit ripening. Postharvest Biol. Technol. 143, 68–77. doi: 10.1016/j.postharvbio.2018.04.012
Wang, T., Song, H., Zhang, B., Lu, Q., Liu, Z., Zhang, S., et al. (2018b). Genome-wide identification, characterization, and expression analysis of superoxide dismutase (SOD) genes in foxtail millet (Setaria italica l.). Biotech. 8, 486. doi: 10.1007/s13205-018-1502-x
Wittkopp, P. J., Kalay, G. (2012). Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 13, 59–69. doi: 10.1038/nrg3095
Wu, J., Seng, S., Sui, J., Vonapartis, E., Luo, X., Gong, B., et al. (2015). Gladiolus hybridus ABSCISIC ACID INSENSITIVE 5 (GhABI5) is an important transcription factor in ABA signaling that can enhance gladiolus corm dormancy and arabidopsis seed dormancy. Front. Plant Sci. 6, 960. doi: 10.3389/fpls.2015.00960
Xin, H., Xianchao, N., Pan, X., Wei, L., Min, Y., Yu, K., et al. (2019). Comparative transcriptome analyses revealed conserved and novel responses to cold and freezing stress in Brassica napus l. G3 (Bethesda). 9, 2723–2737. doi: 10.1534/g3.119.400229
Xiong, H., Wang, R., Jia, X., Sun, H., Duan, R. (2022). Transcriptomic analysis of rapeseed (Brassica napus. l.) seed development in xiangride, qinghai plateau, reveals how its special eco-environment results in high yield in high-altitude areas. Front. Plant Sci. 13, 927418. doi: 10.3389/fpls.2022.927418
Yang, C., Lu, X., Ma, B., Chen, S. Y., Zhang, J. S. (2015). Ethylene signaling in rice and arabidopsis: conserved and diverged aspects. Mol. Plant 8, 495–505. doi: 10.1016/j.molp.2015.01.003
Yan, L., Shah, T., Cheng, Y., Lu, Y., Zhang, X. K., Zou, X. L. (2019). Physiological and molecular responses to cold stress in rapeseed (Brassica napus l.). J. Integr. Agr. 18, 2742–2752. doi: 10.1016/S2095-3119(18)62147-1
Young, M. D., Wakefield, M. J., Smyth, G. K. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. Evol. 11, 1–12. doi: 10.1186/gb-2010-11-2-r14
Yu, H., Kong, X., Huang, H., Wu, W., Park, J., Yun, D. J., et al. (2020). STCH4/REIL2 confers cold stress tolerance in arabidopsis by promoting rRNA processing and CBF protein translation. Cell Rep. 30, 229–242. doi: 10.1016/j.celrep.2019.12.012
Yu, J., Xu, S., Liu, X., Li, T., Zhang, D., Teng, N., et al. (2022a). Starch degradation and sucrose accumulation of lily bulbs after cold storage. Int. J. Mol. Sci. 23, 1–17. doi: 10.3390/ijms23084366
Yu, Q., Zheng, Q., Shen, W., Li, J., Yao, W., Xu, W. (2022b). Grape CIPK18 acts as a positive regulator of CBF cold signaling pathway by modulating ROS homeostasis. Environ. Exper Bot. 203, 105063. doi: 10.1016/j.envexpbot.2022.105063
Zhang, J., An, H., Zhang, X., Xu, F., Zhou, B. (2022a). Transcriptomic analysis reveals potential gene regulatory networks under cold stress of loquat (Eriobotrya japonica lindl.). Front. Plant Sci. 13, 944269. doi: 10.3389/fpls.2022.944269
Zhang, Y., Gao, P., Yuan, J. S. (2010). Plant protein-protein interaction network and interactome. Curr. Genomics 11, 40–46. doi: 10.2174/138920210790218016
Zhang, C., Howlader, P., Liu, T., Sun, X., Jia, X., Zhao, X., et al. (2019a). Alginate oligosaccharide (AOS) induced resistance to pst DC3000 via salicylic acid-mediated signaling pathway in arabidopsis thaliana. Carbohydr Polym. 225, 115221. doi: 10.1016/j.carbpol.2019.115221
Zhang, Y., Launay, H., Liu, F., Lebrun, R., Gontero, B. (2018). Interaction between adenylate kinase 3 and glyceraldehyde-3-phosphate dehydrogenase from chlamydomonas reinhardtii. FEBS J. 285, 2495–2503. doi: 10.1111/febs.14494
Zhang, J., Li, X. M., Lin, H. X., Chong, K. (2019b). Crop improvement through temperature resilience. Annu. Rev. Plant Biol. 70, 753–780. doi: 10.1146/annurev-arplant-050718-100016
Zhang, L., Song, J., Lin, R., Tang, M., Shao, S., Yu, J., et al. (2022b). The SlMYB15 transcription factor targeted by sly-miR156e-3p positively regulates ABA-mediated cold tolerance in tomato. J. Exp. Bot. erac370. doi: 10.1093/jxb/erac370
Zhang, W., Wang, S., Yu, F., Tang, J., Shan, X., Bao, K., et al. (2019c). Genome-wide characterization and expression profiling of SWEET genes in cabbage (Brassica oleracea var. capitata l.) reveal their roles in chilling and clubroot disease responses. BMC Genomics 20, 93. doi: 10.1186/s12864-019-5454-2
Zhang, D., Ye, H., Guo, H., Johnson, A., Zhang, M., Lin, H., et al. (2014). Transcription factor HAT1 is phosphorylated by BIN2 kinase and mediates brassinosteroid repressed gene expression in arabidopsis. Plant J. 77, 59–70. doi: 10.1111/tpj.12368
Zhao, M., Liu, W., Xia, X., Wang, T., Zhang, W. H. (2014). Cold acclimation-induced freezing tolerance of medicago truncatula seedlings is negatively regulated by ethylene. Physiol. Plant 152, 115–129. doi: 10.1111/ppl.12161
Zhao, J., Zhang, S., Yang, T., Zeng, Z., Huang, Z., Liu, Q., et al. (2015). Global transcriptional profiling of a cold-tolerant rice variety under moderate cold stress reveals different cold stress response mechanisms. Physiol. Plant 154, 381–394. doi: 10.1111/ppl.12291
Zheng, Y., Luo, L., Wei, J., Chen, Q., Yang, Y., Hu, X., et al. (2018). The glutamate receptors AtGLR1.2 and AtGLR1.3 increase cold tolerance by regulating jasmonate signaling in arabidopsis thaliana. Biochem. Biophys. Res. Commun. 506, 895–900. doi: 10.1016/j.bbrc.2018.10.153
Zhuo, X., Zheng, T., Zhang, Z., Zhang, Y., Jiang, L., Ahmad, S., et al. (2018). Genome-wide analysis of the NAC transcription factor gene family reveals differential expression patterns and cold-stress responses in the woody plant prunus mume. Genes 9, 1–22. doi: 10.3390/genes9100494
Zinsmeister, J., Lalanne, D., Terrasson, E., Chatelain, E., Vandecasteele, C., Vu, B. L., et al. (2016). ABI5 is a regulator of seed maturation and longevity in legumes. Plant Cell. 28, 2735–2754. doi: 10.1105/tpc.16.00470
Keywords: abiotic stress, low temperature stress, RNA sequencing, DEGs, transcription factors, photosynthesis, antioxidants
Citation: Hussain MA, Luo D, Zeng L, Ding X, Cheng Y, Zou X, Lv Y and Lu G (2022) Genome-wide transcriptome profiling revealed biological macromolecules respond to low temperature stress in Brassica napus L. Front. Plant Sci. 13:1050995. doi: 10.3389/fpls.2022.1050995
Received: 22 September 2022; Accepted: 14 October 2022;
Published: 14 November 2022.
Edited by:
Muhammad Ahsan Altaf, Hainan University, ChinaReviewed by:
Quaid Hussain, Zhejiang Agriculture & Forestry University, ChinaIftikhar Ali, Zhengzhou University, China
Muhammad Furqan Ashraf, UiT The Arctic University of Norway, Norway
Taimoor Hassan Farooq, Bangor College China, China
Copyright © 2022 Hussain, Luo, Zeng, Ding, Cheng, Zou, Lv and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Lv, bHZ5YW4zMjJAMTYzLmNvbQ==; Guangyuan Lu, bHV3aXpAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Muhammad Azhar Hussain
Muhammad Azhar Hussain Dan Luo1†
Dan Luo1† Liu Zeng
Liu Zeng Xiaoyu Ding
Xiaoyu Ding Yong Cheng
Yong Cheng Xiling Zou
Xiling Zou Yan Lv
Yan Lv