
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 10 November 2022
Sec. Plant Breeding
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1050171
This article is part of the Research Topic Root Development - Proceedings from the joint Rooting 2021 and 11th Symposium of the International Society for Root Research View all 6 articles
B3 is a class of plant-specific transcription factors with important roles in plant development and growth. Here, we identified 69 B3 transcription factors in Brachypodium distachyon that were unevenly distributed across all five chromosomes. The ARF, REM, LAV, and RAV subfamilies were grouped based on sequence characteristics and phylogenetic relationships. The phylogenetically related members in the B3 family shared conserved domains and gene structures. Expression profiles showed that B3 genes were widely expressed in different tissues and varied in response to different abiotic stresses. BdB3-54 protein from the REM subfamily was located in the nucleus by subcellular localization and processed transcriptional activation activity. Overexpression of BdB3-54 in Arabidopsis increased primary root length. Our study provides a basis for further research on the functions of BdB3 genes.
B3 transcription factors (TFs) are plant-specific and contain at least one B3 domain. The B3 domain is named according to its position in the third basic region of the maize (Zea mays L.) VIVIPAROUS-1 protein (Suzuki et al., 1997). This B3 domain of approximately 110 amino acids forms two short α-helices and seven β-barrels (Swaminathan et al., 2008). B3 TFs are classified into RAV (related to ABI3/VP1), LAV (LEAFY COTYLEDON2-ABI3-VAL), REM (Reproductive meristem), and ARF (Auxin response factor) subfamilies based on domain characteristics and phylogenetic relationships (Swaminathan et al., 2008; Yamasaki et al., 2013). ARF and LAV members contain a single B3 domain whereas REM members possess up to six B3 domains. RAV possesses one B3 domain and an AP2/ERF domain (Swaminathan et al., 2008). The recognition sequence motif 5’-TGTCTC-3’ is an ARF member (Ulmasov et al., 1997) and motifs 5’-CATGCA-3’ and 5’-CACCTG-3’ are LAV members (Ulmasov et al., 1997) and RAV members (Kagaya et al., 1999), respectively. However, the DNA-binding abilities of REM members are still not fully understood and need further investigation (Levy et al., 2002).
B3 genes are widely involved in plant growth and development. In the LAV group, Arabidopsis thaliana FUSCA3 regulates seed maturation (Luerssen et al., 1998); maize ZmABI19 is essential for the initiation of grain filling (Yang et al., 2021); and overexpression of citrus FUSCA3 promotes somatic embryogenesis (Liu et al., 2018). In the ARF group, overexpression of AtARF8 affects the development of fruit, hypocotyl and roots (Tian et al., 2004); AtARF4 regulates the regeneration of shoot meristems (Zhang et al., 2021); and OsARF8 regulates hypocotyl elongation (Yang et al., 2006). In the RAV group, rice (Oryza sativa L.) RAV members regulate flowering time (Osnato et al., 2020), whereas overexpression of strawberry (Fragaria × ananassa) FaRAV1 increases anthocyanin production (Zhang et al., 2020). In the REM group, overexpression of Arabidopsis REM16 accelerates flowering (Yu et al., 2020), and silencing of both REM34 and REM35 in Arabidopsis affects the development of reproductive organs (Caselli et al., 2019).
The B3 genes are also involved in stress and hormone responses. Arabidopsis RAV1 functions in abscisic acid (ABA) signaling by regulating the expression of ABI3, ABI4, and ABI5 in ABA signaling (Feng et al., 2014); whereas overexpression of cotton (Gossypium hirsutum L.) RAV1 in Arabidopsis [Arabidopsis thaliana (L.) Heynh.] causes sensitivity to ABA, salt, and drought stresses (Li et al., 2015). Moreover, AtARF7 is involved in hypocotyl response to auxin (Harper et al., 2000).
B3 TFs have been identified in the genomes of many plant species, including 118 in Arabidopsis, 91 in rice (Swaminathan et al., 2008), 72 in Citrus sinensis L. (Liu et al., 2020), 57 in pineapple (Ananas comosus L.) (Ruan et al., 2021), 187 in Brassica rapa L (Peng and Weselake, 2013), 81 in soybean [Glycine max (L.) Merr.] (Peng and Weselake, 2013), and 61 in castor bean (Ricinus communis L.) (Wang et al., 2022). However, little is known about Brachypodium [Brachypodium distachyon (L.) Beauv.], the model monocot. In this study, we investigated the number, structure, and classification of B. distachyon B3 TFs. We cloned the gene BdB3-54 to study its function in root development through overexpression in Arabidopsis. Our study provides a basis for further research on plant B3 genes.
The genome sequences of Brachypodium, Arabidopsis, rice, maize, sorghum (Sorghum bicolor L.), barley (Hordeum vulgare L.), wheat (Triticum aestivum L.), and foxtail millet [Setaria italica (L.) Beauv.] were obtained from Ensembl Plants (Bolser et al., 2017). Identification of B3 TFs was carried out in four steps. First, a BLAST search was performed on the obtained genome protein sequences using Arabidopsis and rice B3 protein sequences as queries (threshold: E<e-5). Second, results from the first step were used to search the B3 structural domain signature model (PF02362) from Pfam (threshold: E<e-5) (El-Gebali et al., 2019). Third, alternative splicing events and redundancies were manually removed and the NCBI-CDD interface (Marchler-Bauer et al., 2015) was used to confirm putative B3 TFs, removing those without a B3 structural domain.
The physical and chemical properties of B3 TFs were predicted using the ExPASy web server (Artimo et al., 2012), and subcellular localization of B3 proteins was predicted using CELLO (Yu et al., 2004).
MEGA7 software was used to construct the Neighbor-Joining (NJ) trees (Kumar et al., 2016) with 1,000 replications based on the full-length sequence alignment. Segmentally and tandemly duplicated events, and collinearity relationships between BdB3 and other plants were analyzed using MCScanX (Wang et al., 2012). TBtools was utilized to map positions, duplications, and collinearity relationships of the candidate genes (Chen et al., 2020).
Gene structures were predicted by GSDS 2.0 (Hu et al., 2015). Conserved protein regions were predicted using NCBI-CDD (Marchler-Bauer et al., 2015). Gene compositions were drawn using TBtools (Chen et al., 2020).
Brachypodium distachyon ecotype Bd21 was grown in an artificial climate chamber under a 16 h light (26°C; 08:00–00:00)/8 h darkness (24°C; 00:00–08:00) cycle. Roots, stems, leaves, young inflorescences, and seeds were sampled 10 d after pollination to determine different tissue expressions. Ten-day-old seedlings were subjected to simulated drought (20% PEG6000), salt (200 mM), heat (42°C), 10 μM 3-indoleacetic acid (IAA), 10 μM salicylic acid (SA), 10 μM ABA, and 10 μM jasmonic acid (JA) treatments for 2 h in hydroponic culture and sampled for different stresses. After sampling, tissues and whole seedlings were collected and stored at -80°C for RNA isolation. Total RNA was extracted using an RNA extraction kit (TIANGEN, Beijing). Next, RT-qPCR was performed in triplicate as previously described (Guo et al., 2021). Relative expression levels were calculated using the 2–ΔΔCt method and normalized to the expression of BdGAPDH (Hong et al., 2008) or AtActin 8 (Reichel et al., 2016).
The coding sequence (CDS) of BdB3-54 was amplified by PCR and cloned into pCambia-1302 and pCAMV35S-GFP with a NOS terminator, respectively. pCambia-1302-BdB3-54 and pCAMV35S-BdB3-54-GFP were transferred into the Agrobacterium tumefaciens strain GV3101 through electroporation. Homozygous transformants of Arabidopsis were obtained using the floral dip method (Clough and Bent, 1998). The transgenic lines were screened using hygromycin B solution (40 mg/L) and confirmed by PCR analysis. Third-generation seeds of transgenic lines were used for further analysis. Finally, pCAMV35S-BdB3-54-GFP was transformed into tobacco (Nicotiana tabacum L.) leaves using the GV3101 strain for subcellular localization with an Olympus IX83-FV1200 confocal microscope (Olympus, Tokyo).
The pGBKT7-BdB3-54 vector was constructed for yeast autoactivation assays. Then, pGBKT7-BdB3-54, negative vector pGBKT7, and positive vector pGBKT7-p53 were transformed into yeast strain Y2H. The surviving clones were grown on SD/-Trp medium, and the transformed yeast cells were diluted and dotted on SD/-Trp and SD/-Trp/-Ade/-His media. Cells were incubated at 30°C for 3 d. Primers used in this study (Supplementary Table S1) were designed using the Oligo 7 software (Rychlik, 2007).
Arabidopsis seedlings were grown in a growth chamber under a 16 h light (22°C; 08:00–00:00)/8 h darkness (20°C; 00:00–08:00) regime. Root length was measured on the tenth day and counted using the ImageJ software (Rueden et al., 2017). Photos of the root apical meristem cell on the 4-day-old plants were taken after staining with propidium iodide (PI, 0.01 mg/ml) for 1-2 min using confocal microscopy (Olympus IX83-FV1200, Japan) with a 561-nm laser for PI. Data were analyzed and plotted using the IBM SPSS Statistics software (USA). Values are shown as means ± SD, and significant differences are indicated by different letters or e-values (P <0.05, one-way ANOVA).
A comprehensive search of the six monocot plant species identified 69, 589, 99, 92, 130, and 91 B3 genes in B. distachyon, wheat, maize, foxtail millet, barley, and sorghum, respectively (Table 1). Based on the characteristics of the conserved domains and the number of B3 domains, these genes were classified into four subfamilies: 250 in ARF, 76 in RAV, 890 in REM, and 63 in LAV (Supplementary Tables 2, 3).
The 69 putative B3 TFs were unevenly distributed on five B. distachyon chromosomes with 21, 18, 12, 10, and 8 on chromosomes 1, 2, 3, 4, and 5, respectively (Figure 1). They were named BdB3-1 to BdB3-69 and validated with expressed sequence tags (ESTs) from the NCBI database. The predicted length of the BdB3 proteins ranged from 166 (BdB3-16) to 1,227 (BdB3-55) amino acids with molecular weights ranging from 18.10 (BdB3-16) to 139.50 (BdB3-55) kDa, and the genomic sequence lengths ranged from 1,837 bp (BdB3-22) to 14,692 bp (BdB3-55) (Supplementary Table 3). Protein subcellular localization prediction showed that 62 BdB3 proteins were localized in the nucleus, three in the cytoplasmic, three in the chloroplast, and one (BdB3-68) in the extracellular matrix.
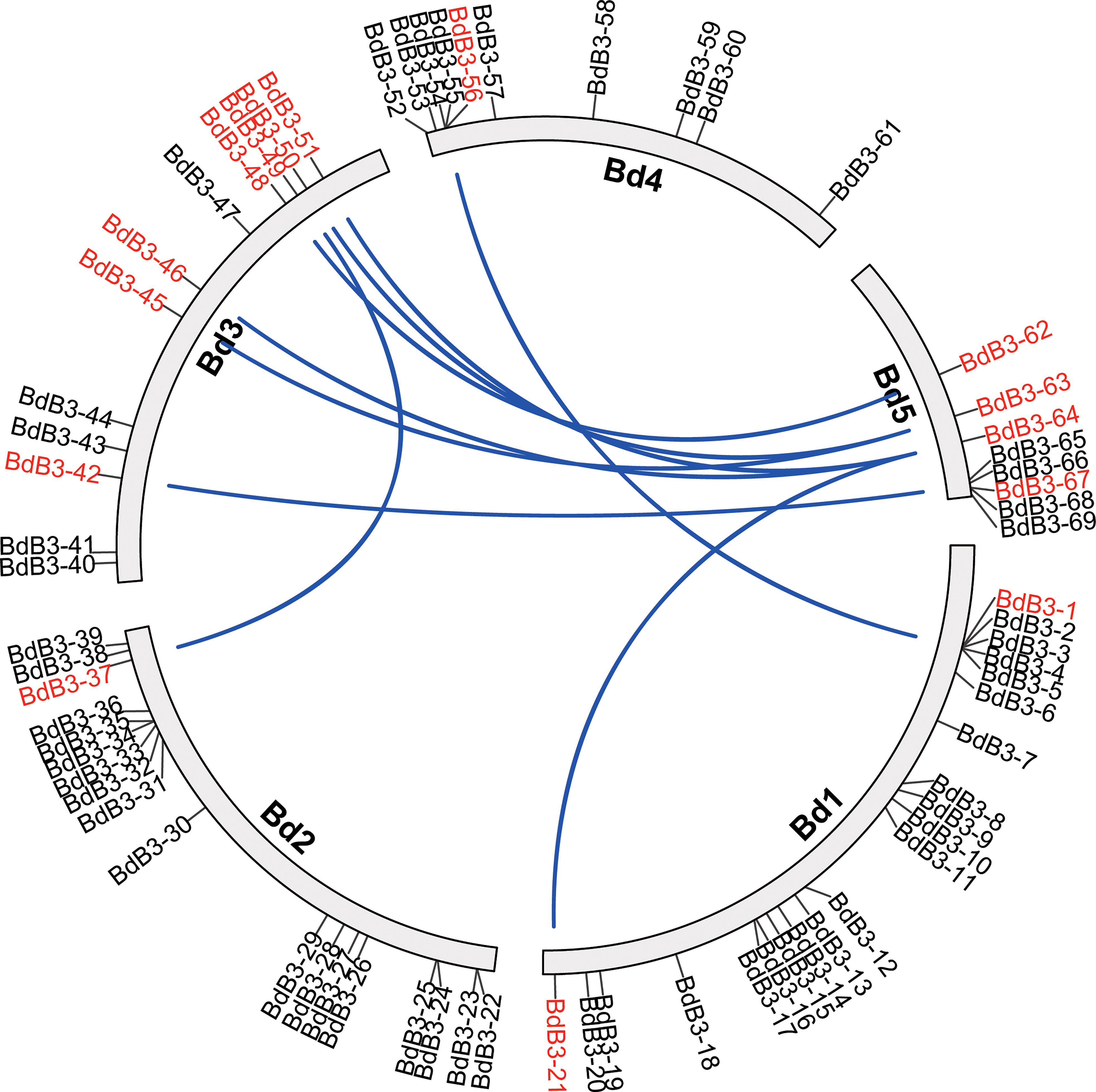
Figure 1 Chromosome location and segmental duplication of the BdB3 genes. Blue lines connect duplicated genes that are shown in red.
Gene duplication analysis detected 15 tandemly duplicated genes. They formed nine gene pairs. Among them, BdB3-64 was pared with three genes (BdB3-21, BdB3-46, and BdB3-51) and BdB3-63 was pared with two genes (BdB3-45, and BdB3-50) (Figure 1, Supplementary Table 4). Genome synteny between B. distachyon and the other plant species showed 3, 51, 52, 41, 52, and 47 BdB3 gene homologs in Arabidopsis, rice, wheat, barley, sorghum, and maize, respectively (Figure 2, Supplementary Table 5). These results suggest that BdB3 genes share similar structures and functions with orthologs in other monocot plants.
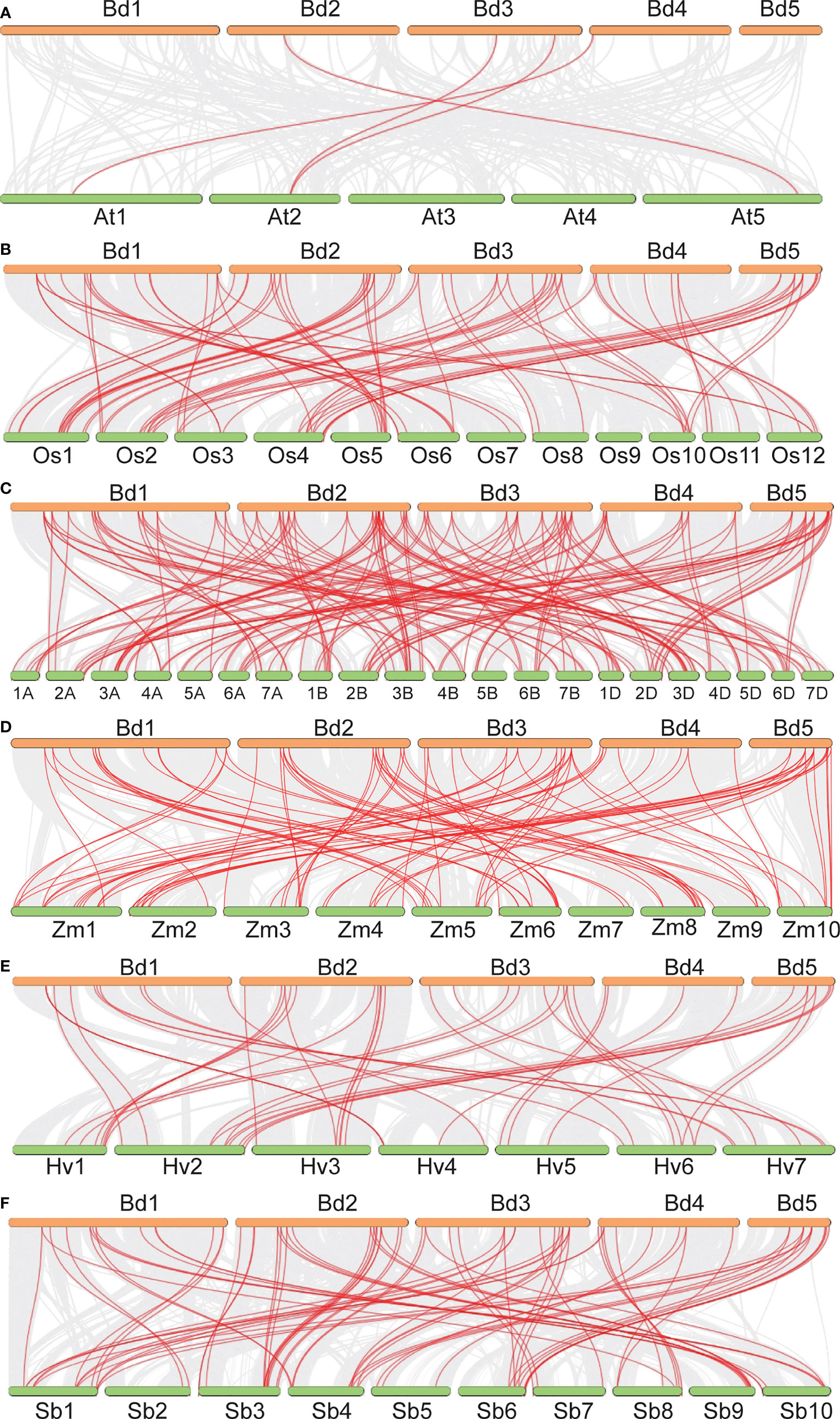
Figure 2 Synteny analysis of the B3 genes between B distachyon and A thaliana (A), O. sativa (B), T. aestivum (C), Z. mays (D), H vulgare (E), and S. bicolor (F). Gray lines in the background indicate the collinear blocks within B distachyon and other plant species, and red lines highlight syntenic B3 gene pairs.
NJ trees for the four subfamilies were constructed according to sequence characteristics to explore the phylogenetic relationships of BdB3 TFs (Figure 3A). As shown in Figure 3B, each B3 member contained at least one B3 domain; REM members contained 1 to 6 B3 domains; and ARF members contained a single B3 domain at the N-terminus and one or two Aux/IAA domains (carboxyl-terminal interaction domains). LAV members had one B3 domain at the C-terminus and two members (BdB3-6 and BdB3-7) had a CW-type zinc finger. Each RAV member contained one AP2 domain at the N-terminus and one B3 domain at the C-terminus.
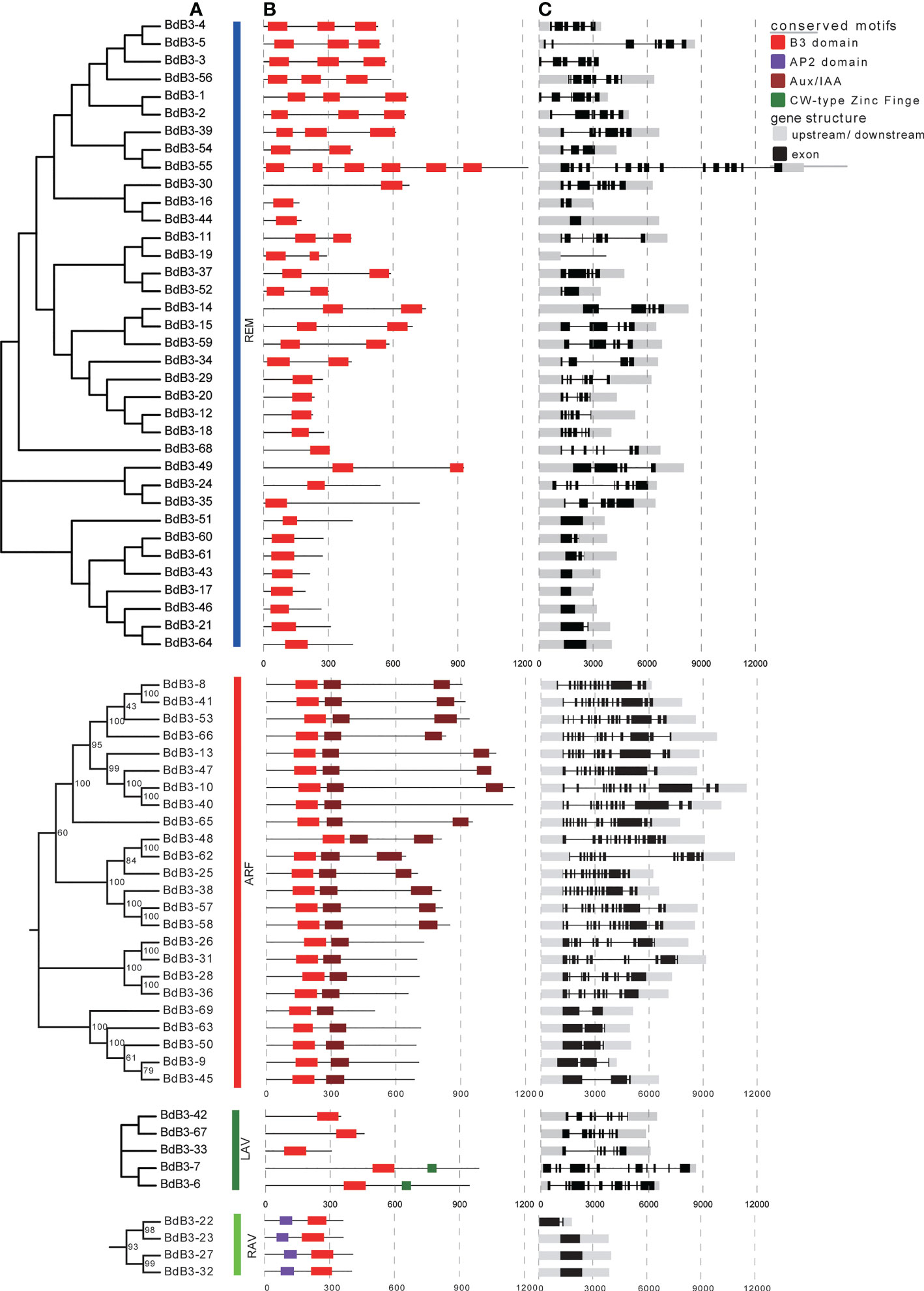
Figure 3 Phylogenetic relationships, conserved protein domains, and gene structures of B distachyon REM, RAV, ARF, and LAV members. (A) Neighbor-joining trees constructed for B3 genes from the four subfamilies. (B) The four conserved protein domains are shown in different colors. (C) Structures of BdB3 genes. Gray boxes indicate up- or down-stream structures, black boxes indicate exons, and black lines indicate introns.
Exon numbers in B3 genes ranged from one to 16 (Figure 3C). Each subgroup had a different number of exons; RAV members contained 1-2 exons, whereas the REM members had 1-15. Additionally, 22 members contained more than 5 exons, and all LAV members had more than 7 exons. The exon number in ARF members varied greatly with five having 2 or 3 exons, and the other members having more than 10 exons. This structural diversity implies diverse functions for BdB3 genes.
Twenty BdB3 genes, including 3 RAV, 6 ARF, 4 LAV, and 7 REM members, were analyzed for expressions levels in different tissues (roots, stems, leaves, young inflorescences, and seeds sampled 10 days after pollination) using RT-qPCR. Expression of these BdB3 genes was detected in all tissues (Figure 4, Figure S1A). For example, the LAV genes BdB3-7 and BdB3-67 were highly expressed in seeds, whereas BdB3-33 and BdB3-42 were highly expressed in roots and inflorescences, respectively. Further, the REM genes BdB3-12, -30, -39, and -49 had high expression levels in inflorescences, whereas BdB3-37, -49, and -54 showed high expression levels in roots.
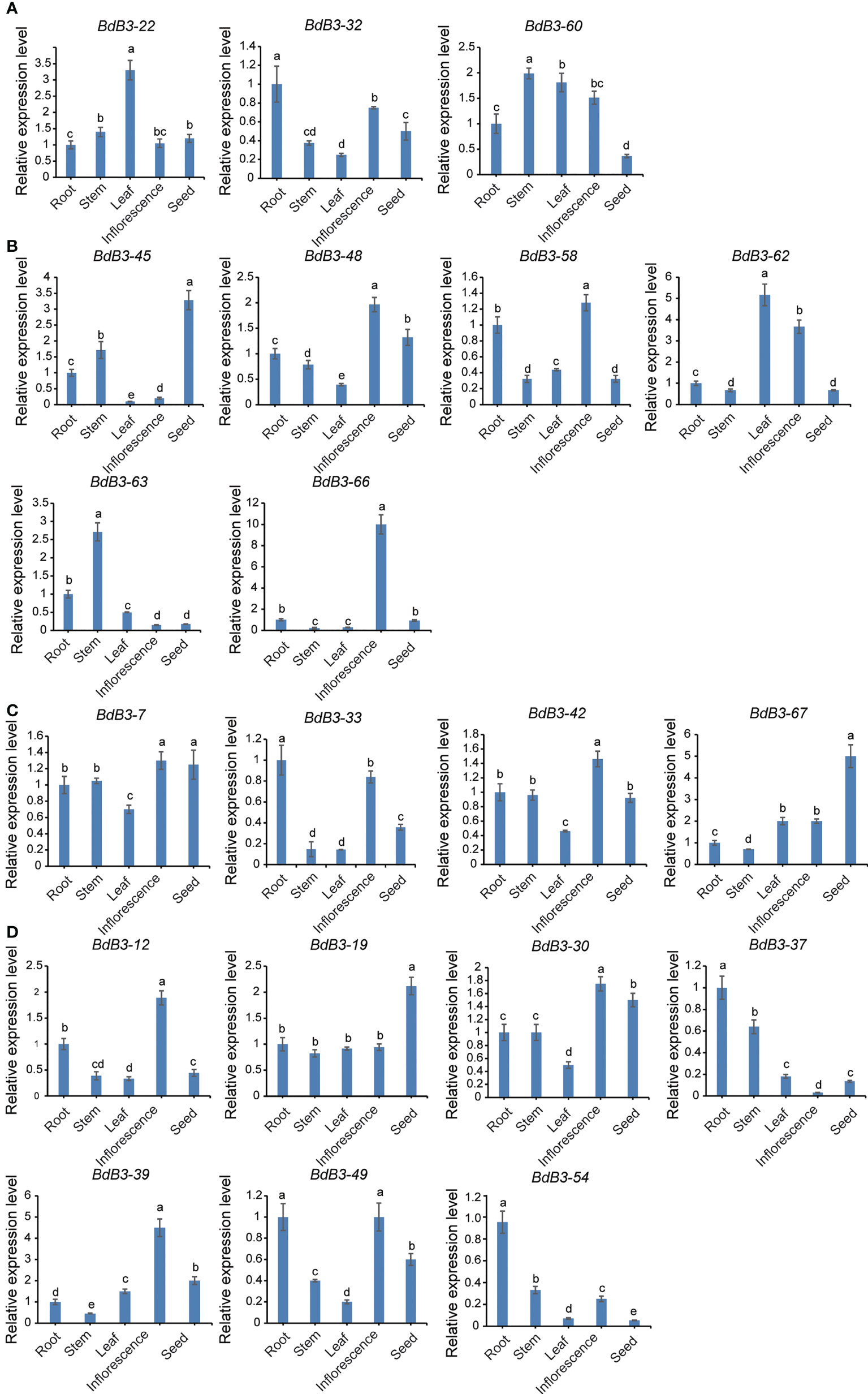
Figure 4 Expression patterns of BdB3 genes in different tissues. (A–D) represent the expression patterns of RAV, ARF, LAV, and REM genes. Abscissas represent different tissues, including roots, stems, leaves, early inflorescences, and seeds at 10 days after pollination. Ordinates represent relative expression levels. Transcript levels of BdB3 genes were normalized to those of BdGAPDH, and expression levels of root tissues were set to 1. Data are shown as means ± SE (n = 3) Letters above the bars indicate significant differences (P <0.05, one-way ANOVA)..
The expression of selected BdB3 genes under abiotic and hormonal stresses varied considerably compared to the control (no treatment) (Figure 5, Figures S1B, C). For the RAV family, genes BdB3-22 and BdB3-32 were significantly down-regulated by IAA and SA, and BdB3-22 and BdB3-32 were significantly down-regulated by salinity and heat. JA had the greatest impact on the expression of BdB3-32, whereas BdB3-60 was up-regulated by IAA, SA, and ABA. Three (BdB3-45, -58, and -63) and five (BdB3-45, -48, -58, -63, and -66) ARF genes were highly expressed under IAA and SA treatments, respectively. Five genes (BdB3-45, -48, -58, -63, -66), four genes (BdB3-45, -48, -58, and -66), and three genes (BdB3-48, -58, and -63) were up-regulated under drought, heat, and salinity stresses. BdB3-62 was down-regulated by all stress conditions. All LAV genes were down-regulated by IAA. BdB3-7 was up-regulated under all different treatments apart from IAA. Among REM genes, heat stress had the greatest impact on their expressions with BdB3-12, -30, -37, -39, and -49 being up-regulated and genes BdB3-19 and -54 being down-regulated. Hormones including IAA, SA, ABA, and JA significantly regulated the expressions of BdB3-12, -30, -37, -39, and -49.
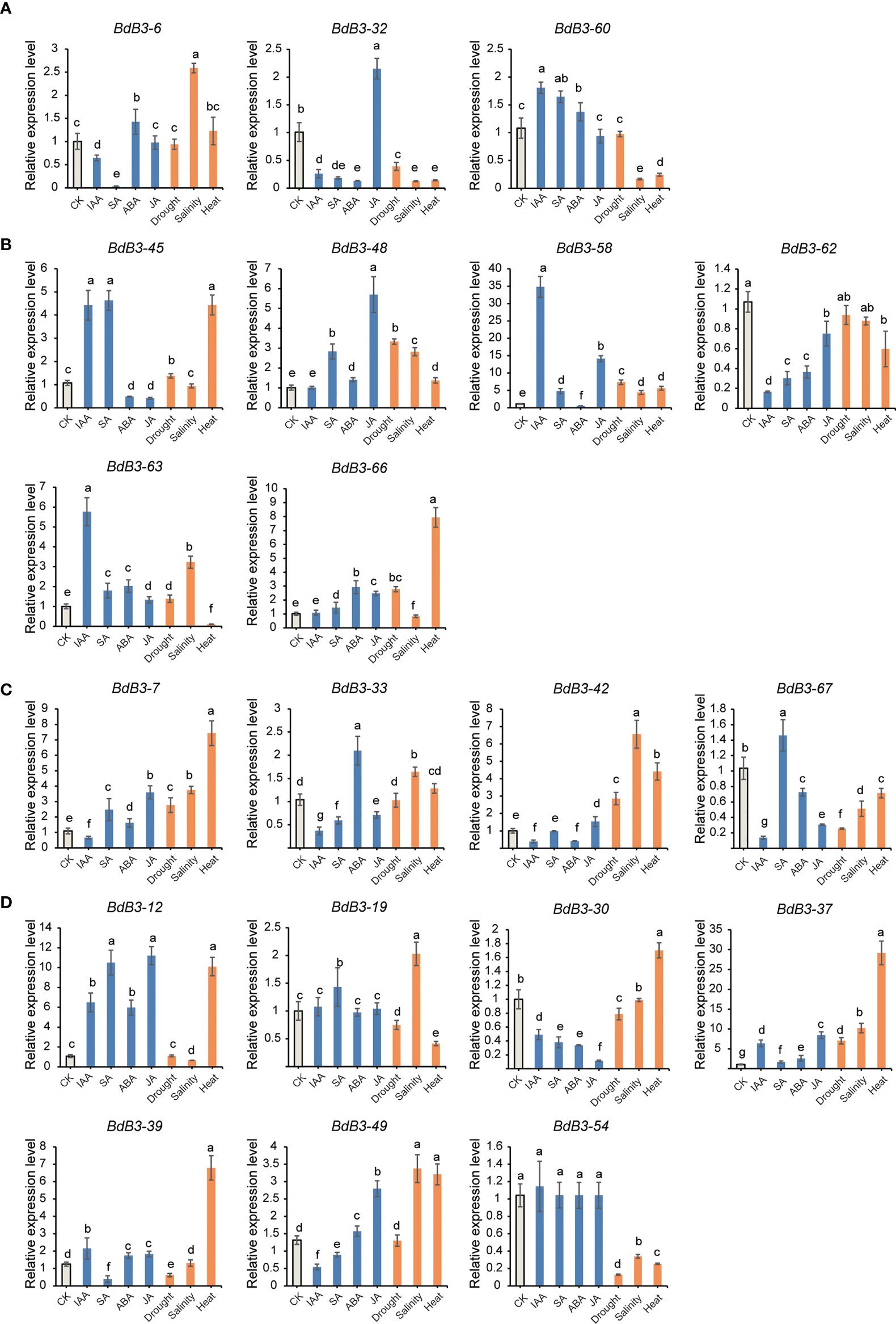
Figure 5 Expression patterns of BdB3 genes under different hormonal and abiotic stress conditions. (A–D) represent the expression patterns of RAV, ARF, LAV, and REM members. Abscissas represents different stress of hormone treatments. CK, non-treated. Ordinates represent the relative expression levels. Data are means ± SE (n = 3). Letters above the bars indicate significant differences (P <0.05, one-way ANOVA).
BdB3-54 belongs to the REM subfamily and its functions have rarely been investigated. Subcellular localization analysis predicted that BdB3-54 protein was localized in the nucleus. When transiently expressed in tobacco leaves, BdB3-54 fusion protein signals overlapped the DAPI signal confirming that BdB3-54 protein was located in the nucleus (Figure 6A).
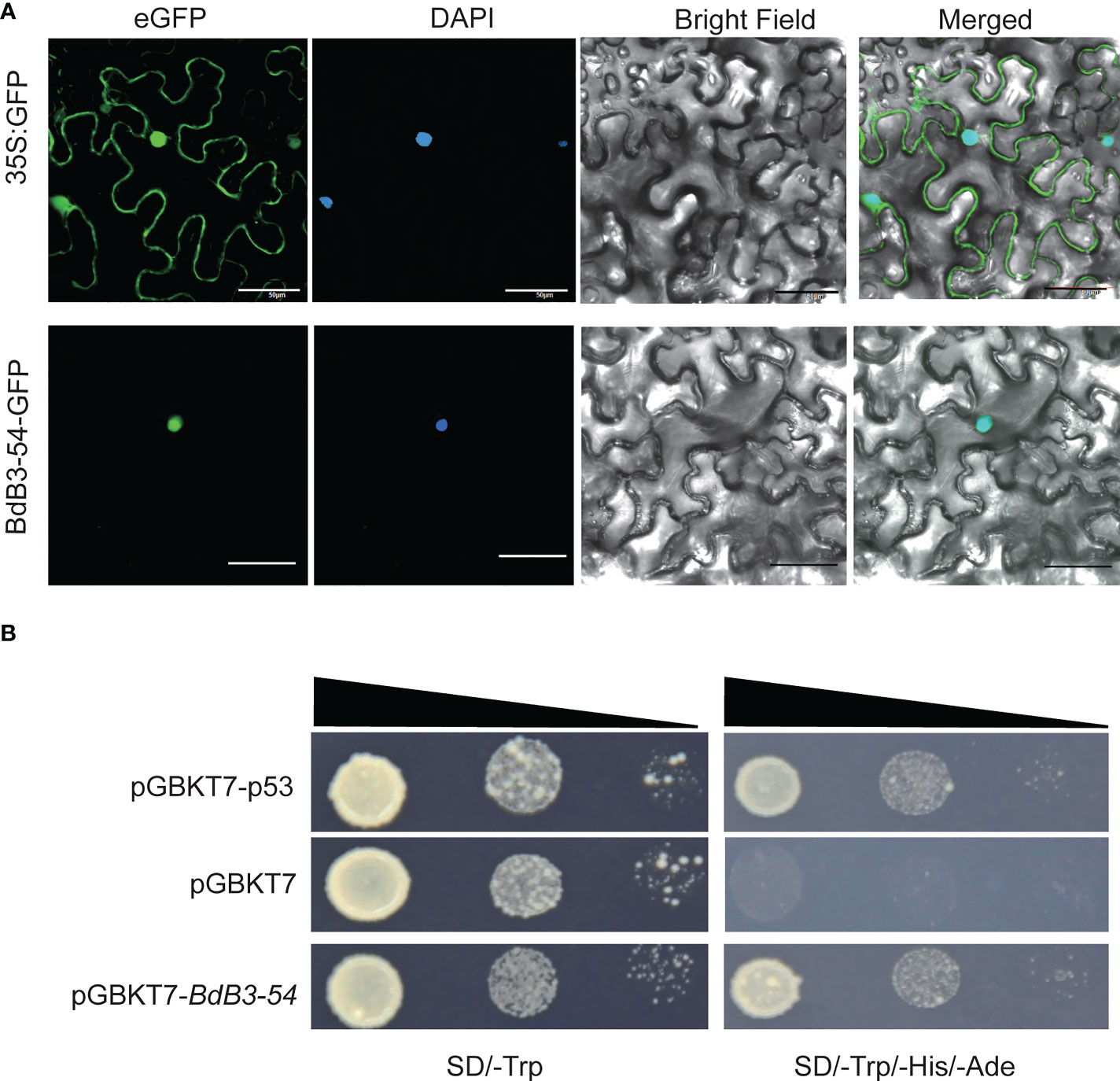
Figure 6 Subcellular localization and transactivation assay of BdB3-54. (A). BdB3-54 protein was transiently expressed in tobacco (Nicotiana benthamiana L.) leaves to determine its subcellular localization; bar, 50 μm. (B). BdB3-54 transactivation assay. BdB3-54 was ligated to the pGBKT7 vector, and transformed yeast cells were screened on SD/-Trp and SD/-Trp-His-Ade media.
A transactivation assay was performed to test the transcriptional activation activity of BdB3-54 using Y2H assays. Yeast cells carrying the pGBKT7-BdB3-54 plasmid grew well on the defective SD/-Trp-His-Ade medium, which was similar to pGABKT7-p53, a positive control plasmid. In contrast, yeast cells carrying the negative control pGBKT-7 showed much less growth (Figure 6B). These results indicated that BdB3-54 had transcriptional self-activation activity.
Expression pattern analysis showed that BdB3-54 was highly expressed in root tissues (Figure 4D). To determine its role in root development, two transgenic Arabidopsis lines overexpressing BdB3-54 driven by the CaMV35S promoter were generated (Figure 7A). The primary root lengths of transgenic plants overexpressing BdB3-54 were significantly longer than the wild-type (WT) Col-0 (Figures 7B, C). To explore potential factors leading to the longer primary root lengths of transgenic plants, we examined the root apical meristem cell size on the 4-day-old plants. With the same number of cells, the transgenic lines occupied a larger area (Figure S2). We also investigated the gene expression of four root development-related genes in the WT and the transgenic lines AtWOX5, AtARF7, AtARF19, and AtEXPA4. As shown in Figure 7D, the expression levels of these genes were significantly higher in transgenic plants than in the WT (P <0.05). These results indicated that ectopic expression of BdB3-54 regulated primary root growth in Arabidopsis.
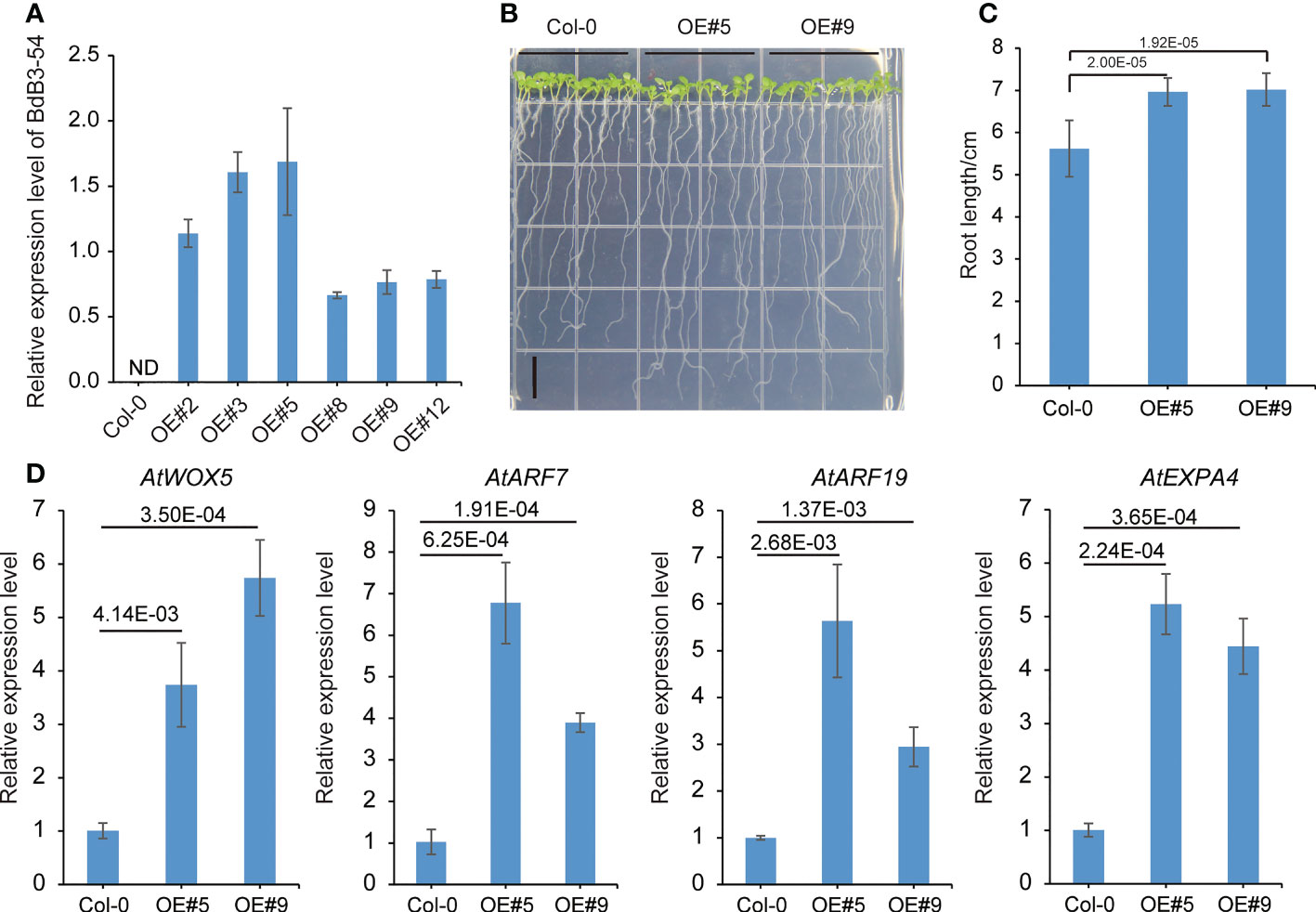
Figure 7 Root phenotypes of Arabidopsis lines overexpressing BdB3-54. (A) Relative expression levels of BdB3-54 in six transgenic T3 lines. ND, expression not detected. (B) Phenotypes of Arabidopsis lines overexpressing BdB3-54 (10-day-old plants). Scale bar, 1 cm. (C) Statistical results of root length. (D) Relative expression levels of four root-development genes in Arabidopsis. Data are means ± SE (n = 3). Numbers above the horizontal line indicate the significance of differences (one-way ANOVA).
We identified 69 B3 genes in the B. distachyon genome. The proportion of B3 genes in the B. distachyon genome was approximately 0.20%, which was less than that in rice (0.24%), wheat (0.38%), Arabidopsis (0.43%), and other monocot species (Table 1) (Swaminathan et al., 2008). The gene number of three subfamilies, excluding ARF members, was lower than that in other plant species, suggesting that gene loss had occurred during evolution.
The B3 genes in B. distachyon were grouped into REM, ARF, LAV, and RAV subfamilies according to protein characteristics and the number of B3 domains. Each subfamily member shares a similar domain composition and gene structure. Gene structures for different subfamilies showed significant variation in intron number and length, indicating that these B3 genes might have undergone intron loss or gain during evolution. Phylogenetic analysis of different subfamilies in previous studies indicated that the same clade members also shared similar gene components, including gene structure and conserved domains, suggesting conserved functions and common origins (Bhattacharjee et al., 2015; Verma and Bhatia, 2019).
Genes perform functions according to their expression in different tissues, and gene expression patterns reflect the gene function. For example, many Arabidopsis B3 genes, such as ARF3, ARF5, ARF6, ARF8, and ARF9, have diverse functions in the development of carpels, floral parts, and fruit, as well as lateral roots (Li et al., 2016; Zhang et al., 2018). AtVAL-1, -2, and -3 proteins are required for seed germination (Suzuki et al., 2007; Jia et al., 2013; Schneider et al., 2016). Tissue-specific expression patterns in the present study indicated that BdB3 genes were expressed in all tissues examined, and members in different subfamilies showed different expression patterns, further indicating their functional diversity in plant growth and development. Genome-wide expression pattern analyses showed that wheat, citrus, and cotton B3 genes also had different expression patterns in different tissues (Liu and Zhang, 2017; Liu et al., 2020; Luo et al., 2022).
BdB3 genes displayed significant differential expression under different abiotic stress and hormone conditions, suggesting crucial roles in response to stress. Hormones such as IAA, ABA, SA, and JA are known to regulate stress-related pathways as plants grow and develop (Verma et al., 2016). Most BdB3 genes responded differentially under different hormones, suggesting that hormones specifically regulated the expression of BdB3 genes under certain conditions. Similarly, hormones also regulate the expression of plant B3 genes in other species, such as Arabidopsis, chickpea, and citrus, indicating that the functions of B3 genes are diverse but conserved across plant species (Verma and Bhatia, 2019; Liu et al., 2020).
Various studies have reported that B3 genes are involved in plant root growth and development. For examples, the LAV member FUSCA3 interacts with LEC2 to control the formation of lateral roots in Arabidopsis (Tang et al., 2017); RAV member GmRAV1 is an important positive regulator involved in promoting root regeneration in Arabidopsis and soybean (Zhang et al., 2019); Ectopic expression of TaARF4-A in Arabidopsis leads to shortened primary root length (Wang et al., 2019); AtARF7 and AtARF19 regulate the formation of lateral roots through the activation of LBD/ASL genes (Okushima et al., 2007). Our study showed that BdB3-54 contained two B3 domains and acted as a TF in B. distachyon, which is supported by the fact that a BdB3-54-GFP fusion protein was localized in the nucleus and had transcriptional activity in yeast cells.
BdB3-54 was highly expressed in root tissue, and overexpression of the gene increased root length when compared to the WT. These observations suggest that the B3 genes are involved in root development and growth, and the longer root length in transgenic plants is mainly due to enlarged cells. Root development-related genes, such as expansins (EXP), WUS-related homeobox genes (WOXs), and ARFs, were also detected during our investigation of BdB3-54 function. Among these root development-related genes, AtWOX5 is expressed in the quiescent center and affects root development (Kong et al., 2015); AtARF7 and AtARF19 regulate the formation of lateral root formation via direct activation of the downstream genes (Okushima et al., 2007); and AtEXPA4 is involved in root elongation (Liu et al., 2021). Expression of AtWOX5, AtARF7, AtARF19, and AtEXPA4 was up-regulated in a transgenic Arabidopsis line carrying BdB3-54 suggesting that BdB3-54 regulates the expression of other root development-related genes. B. distachyon, wheat, and rice all belong to pooideae. The function of BdB3-54 in root development indicated that it can be used for molecular breeding in cereal crops
In conclusion, 69 B3 genes were identified in the B. distachyon genome. These genes were expressed in different plant tissues and showed different responses to various stresses. Further study on one of the highly expressed genes, BdB3-54, indicated that this gene functions as a TF and has an important role in root development.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
JG analyzed the data and wrote the manuscript. HL and KD helped to carry out the experiments. PG and XY contributed to writing the manuscript. MZ and WS contributed to experimental design, provided advice for data analysis, and assisted in writing the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the National Key R&D Program of China (2021YFD1901101), the Key Science and Technology Program of Shanxi Province, China (20210140601026), the National Natural Science Foundation of China (31901541) and the Project of Biological Breeding of Shanxi Agricultural University, China (YZGC102). The funding body did not exert influence on the design of the study, collection, analysis, and interpretation of data, or in writing the manuscript.
We gratefully acknowledge help from Professor Hongjie Li (Chinese Academy of Agricultural Sciences) and Robert A. McIntosh (University of Sydney), with English editing.
The authors declare that the research was conducted in the absence of any commercial or financial commitments that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1050171/full#supplementary-material
Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G., de Castro, E., et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucleic. Acids Res. 40, W597–W603. doi: 10.1093/nar/gks400
Bhattacharjee, A., Ghangal, R., Garg, R., Jain, M. (2015). Genome-wide analysis of homeobox gene family in legumes: identification, gene duplication and expression profiling. PloS One 10, e0119198. doi: 10.1371/journal.pone.0119198
Bolser, D. M., Staines, D. M., Perry, E., Kersey., P. J. (2017). Ensembl plants: Integrating tools for visualizing, mining, and analyzing plant genomic data. Methods Mol. Biol. 1533, 1–31. doi: 10.1007/978-1-4939-6658-5_1
Caselli, F., Beretta, V. M., Mantegazza, O., Petrella, R., Leo, G., Guazzotti, A., et al. (2019). REM34 and REM35 control female and male gametophyte development in Arabidopsis thaliana. Front. Plant Sci. 10, 1351. doi: 10.3389/fpls.2019.01351
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The pfam protein families database in 2019. Nucleic. Acids Res. 47, D427–Dd32. doi: 10.1093/nar/gky995
Feng, C. Z., Chen, Y., Wang, C., Kong, Y. H., Wu, W. H., Chen., Y. F. (2014). Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J. 80, 654–668. doi: 10.1111/tpj.12670
Guo, J., Bai, X., Dai, K., Yuan, X., Guo, P., Zhou, M., et al. (2021). Identification of GATA transcription factors in Brachypodium distachyon and functional characterization of BdGATA13 in drought tolerance and response to gibberellins. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.763665
Harper, R. M., Stowe-Evans, E. L., Luesse, D. R., Muto, H., Tatematsu, K., Watahiki, M. K., et al. (2000). The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell 12, 757–770. doi: 10.1105/tpc.12.5.757
Hong, S. Y., Seo, P. J., Yang, M. S., Xiang, F., Park., C. M. (2008). Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol. 8, 112. doi: 10.1186/1471-2229-8-112
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., Gao., G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Jia, H., McCarty, D. R., Suzuki, M. (2013). Distinct roles of LAFL network genes in promoting the embryonic seedling fate in the absence of VAL repression. Plant Physiol. 163 (3), 1293–1305. doi: 10.1104/pp.113.220988
Kagaya, Y., Ohmiya, K., Hattori., T. (1999). RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic. Acids Res. 27, 470–478. doi: 10.1093/nar/27.2.470
Kong, X., Lu, S., Tian, H., Ding, Z. (2015). WOX5 is shining in the root stem cell niche. Trends Plant Sci. 20, 601–603. doi: 10.1016/j.tplants.2015.08.009
Kumar, S., Stecher, G., Tamura., K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Levy, Y. Y., Mesnage, S., Mylne, J. S., Gendall, A. R., Dean., C. (2002). Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science 297, 243–246. doi: 10.1126/science.1072147
Li, X. J., Li, M., Zhou, Y., Hu, S., Hu, R., Chen, Y., et al. (2015). Overexpression of cotton RAV1 gene in Arabidopsis confers transgenic plants high salinity and drought sensitivity. PloS One 10, e0118056. doi: 10.1371/journal.pone.0118056
Liu, Z., Ge, X. X., Qiu, W. M., Long, J. M., Jia, H. H., Yang, W., et al. (2018). Overexpression of the CsFUS3 gene encoding a B3 transcription factor promotes somatic embryogenesis in Citrus. Plant Sci. 277, 121–131. doi: 10.1016/j.plantsci.2018.10.015
Liu, Z., Ge, X. X., Wu, X. M., Xu, Q., Atkinson, R. G., Guo., W. W. (2020). Genome-wide analysis of the citrus B3 superfamily and their association with somatic embryogenesis. BMC Genomics 21, 305. doi: 10.1186/s12864-020-6715-9
Liu, W., Xu, L., Lin, H., Cao, J. (2021). Two expansin genes, AtEXPA4 and AtEXPB5, are redundantly required for pollen tube growth and atexpa4 is involved in primary root elongation in Arabidopsis thaliana. Genes (Basel) 12, 249. doi: 10.3390/genes12020249
Liu, C., Zhang, T. (2017). Expansion and stress responses of the AP2/EREBP superfamily in cotton. BMC Genomics 18, 118. doi: 10.1186/s12864-017-3517-9
Li, S. B., Xie, Z. Z., Hu, C. G., Zhang, J. Z. (2016). A review of auxin response factors (ARFs) in plants. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00047
Luerssen, H., Kirik, V., Herrmann, P., Miséra., S. (1998). FUSCA3 encodes a protein with a conserved VP1/AB13-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J. 15, 755–764. doi: 10.1046/j.1365-313x.1998.00259.x
Luo, Y. X., Chen, S. K., Wang, P. D., Peng, D., Zhang, X., Li, H. F., et al. (2022). Genome-wide analysis of the RAV gene family in wheat and functional identification of TaRAV1 in salt stress. Int. J. Mol. Sci. 23, 8834. doi: 10.3390/ijms23168834
Marchler-Bauer, A., Derbyshire, M. K., Gonzales, N. R., Lu, S., Chitsaz, F., Geer, L. Y., et al. (2015). CDD: NCBI’s conserved domain database. Nucleic. Acids Res. 43, D222–D226. doi: 10.1093/nar/gku1221
Okushima, Y., Fukaki, H., Onoda, M., Theologis, A., Tasaka., M. (2007). ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19, 118–130. doi: 10.1105/tpc.106.047761
Osnato, M., Matias-Hernandez, L., Aguilar-Jaramillo, A. E., Kater, M. M., Pelaz, S. (2020). Genes of the RAV family control heading date and carpel development in rice. Plant Physiol. 183, 1663–1680. doi: 10.1104/pp.20.00562
Peng, F. Y., Weselake, R. J. (2013). Genome-wide identification and analysis of the B3 superfamily of transcription factors in brassicaceae and major crop plants. Theor. Appl. Genet. 126, 1305–1319. doi: 10.1007/s00122-013-2054-4
Reichel, M., Liao, Y., Rettel, M., Ragan, C., Evers, M., Alleaume, A. M., et al. (2016). In planta determination of the mRNA-binding proteome of Arabidopsis etiolated seedling. Plant Cell 28, 2435–2452. doi: 10.1105/tpc.16.00562
Ruan, C. C., Chen, Z., Hu, F. C., Fan, W., Wang, X. H., Guo, L. J., et al. (2021). Genome-wide characterization and expression profiling of B3 superfamily during ethylene-induced flowering in pineapple (Ananas comosus l.). BMC Genomics 22, 561. doi: 10.1186/s12864-021-07854-1
Rueden, C. T., Schindelin, J., Hiner, M. C., DeZonia, B. E., Walter, A. E., Arena, E. T., et al. (2017). ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinf. 18, 529. doi: 10.1186/s12859-017-1934-z
Rychlik, W. (2007). OLIGO 7 primer analysis software. Methods Mol. Biol. 402, 35–60. doi: 10.1007/978-1-59745-528-2_2
Schneider, A., Aghamirzaie, D., Elmarakeby, H., Poudel, A. N., Koo, A. J., Heath, L. S., et al. (2016). Potential targets of VIVIPAROUS1/ABI3-LIKE1 (VAL1) repression in developing Arabidopsis thaliana embryos. Plant J. 85, 305–319. doi: 10.1111/tpj.13106
Suzuki, M., Kao, C. Y., McCarty., D. R. (1997). The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell 9, 799–807. doi: 10.1105/tpc.9.5.799
Suzuki, M., Wang, H. H., McCarty, D. R. (2007). Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol. 143, 902–911. doi: 10.1104/pp.106.092320
Swaminathan, K., Peterson, K., Jack., T. (2008). The plant B3 superfamily. Trends Plant Sci. 13, 647–655. doi: 10.1016/j.tplants.2008.09.006
Tang, L. P., Zhou, C., Wang, S. S., Yuan, J., Zhang, X. S., Su., Y. H. (2017). FUSCA3 interacting with LEAFY COTYLEDON2 controls lateral root formation through regulating YUCCA4 gene expression in Arabidopsis thaliana. New Phytol. 213, 1740–1754. doi: 10.1111/nph.14313
Tian, C. E., Muto, H., Higuchi, K., Matamura, T., Tatematsu, K., Koshiba, T., et al. (2004). Disruption and overexpression of auxin response factor 8 gene of Arabidopsis affect hypocotyl elongation and root growth habit, indicating its possible involvement in auxin homeostasis in light conditions. Plant J. 40, 333–343. doi: 10.1111/j.1365-313X.2004.02220.x
Ulmasov, T., Hagen, G., Guilfoyle., T. J. (1997). ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865–1868. doi: 10.1126/science.276.5320.1865
Verma, S., Bhatia, S. (2019). A comprehensive analysis of the B3 superfamily identifies tissue-specific and stress-responsive genes in chickpea (Cicer arietinum l.). 3 Biotec. 9, 346. doi: 10.1007/s13205-019-1875-5
Verma, V., Ravindran, P., Kumar, P. P. (2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16, 86. doi: 10.1186/s12870-016-0771-y
Wang, W. B., Ao, T., Zhang, Y. Y., Wu, D., Xu, W., Han, B., et al. (2022). Genome-wide analysis of the B3 transcription factors reveals that RcABI3/VP1 subfamily plays important roles in seed development and oil storage in castor bean (Ricinus communis). Plant Divers. 44, 201–212. doi: 10.1016/j.pld.2021.06.008
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic. Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Wang, J., Wang, R., Mao, X., Li, L., Chang, X., Zhang, X., et al. (2019). TaARF4 genes are linked to root growth and plant height in wheat. Ann. Bot. 124, 903–915. doi: 10.1093/aob/mcy218
Yamasaki, K., Kigawa, T., Seki, M., Shinozaki, K., Yokoyama., S. (2013). DNA-Binding domains of plant-specific transcription factors: Structure, function, and evolution. Trends Plant Sci. 18, 267–276. doi: 10.1016/j.tplants.2012.09.001
Yang, T., Guo, L., Ji, C., Wang, H., Wang, J., Zheng, X., et al. (2021). The B3 domain-containing transcription factor ZmABI19 coordinates expression of key factors required for maize seed development and grain filling. Plant Cell 33, 104–128. doi: 10.1093/plcell/koaa008
Yang, J. H., Han, S. J., Yoon, E. K., Lee., W. S. (2006). Evidence of an auxin signal pathway, microRNA167-ARF8-GH3, and its response to exogenous auxin in cultured rice cells. Nucleic. Acids Res. 34, 1892–1899. doi: 10.1093/nar/gkl118
Yu, C. S., Lin, C. J., Hwang., J. K. (2004). Predicting subcellular localization of proteins for gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 13, 1402–1406. doi: 10.1110/ps.03479604
Yu, Y., Qiao, L., Chen, J., Rong, Y., Zhao, Y., Cui, X., et al. (2020). Arabidopsis REM16 acts as a B3 domain transcription factor to promote flowering time via directly binding to the promoters of SOC1 and FT. Plant J. 103, 1386–1398. doi: 10.1111/tpj.14807
Zhang, Z., Shi, Y., Ma, Y., Yang, X., Yin, X., Zhang, Y., et al. (2020). The strawberry transcription factor FaRAV1 positively regulates anthocyanin accumulation by activation of FaMYB10 and anthocyanin pathway genes. Plant Biotechnol. J. 18, 2267–2279. doi: 10.1111/pbi.13382
Zhang, K., Wang, R., Zi, H., Li, Y., Cao, X., Li, D., et al. (2018). AUXIN RESPONSE FACTOR3 rulates floral meristem determinacy by repressing cytokinin biosynthesis and signaling. Plant Cell 30, 324–346. doi: 10.1105/tpc.17.00705
Zhang, M. M., Zhang, H. K., Zhai, J. F., Zhang, X. S., Sang, Y. L., Cheng., Z. J. (2021). ARF4 regulates shoot regeneration through coordination with ARF5 and IAA12. Plant Cell Rep. 40, 315–325. doi: 10.1007/s00299-020-02633-w
Keywords: B3 transcription factor, Brachypodium distachyon, genome-wide, root development, transgenesis
Citation: Guo J, Liu H, Dai K, Yuan X, Guo P, Shi W and Zhou M (2022) Identification of Brachypodium distachyon B3 genes reveals that BdB3-54 regulates primary root growth. Front. Plant Sci. 13:1050171. doi: 10.3389/fpls.2022.1050171
Received: 21 September 2022; Accepted: 27 October 2022;
Published: 10 November 2022.
Edited by:
Tianzhen Zhang, Zhejiang University, ChinaReviewed by:
Youping Wang, Yangzhou University, ChinaCopyright © 2022 Guo, Liu, Dai, Yuan, Guo, Shi and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weiping Shi, c2hpd2VpOTY4QHN4YXUuZWR1LmNu; Meixue Zhou, bWVpeHVlLnpob3VAdXRhcy5lZHUuYXU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.