- 1Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing, China
- 2Institute of Crop Sciences/National Key Facility for Crop Gene Resources and Genetic Improvement, Chinese Academy of Agricultural Sciences, Beijing, China
Maize (Zea mays) is one of the most important food crops in the world with the greatest global production, and contributes to satiating the demands for human food, animal feed, and biofuels. With population growth and deteriorating environment, efficient and innovative breeding strategies to develop maize varieties with high yield and stress resistance are urgently needed to augment global food security and sustainable agriculture. CRISPR-Cas-mediated genome-editing technology (clustered regularly interspaced short palindromic repeats (CRISPR)-Cas (CRISPR-associated)) has emerged as an effective and powerful tool for plant science and crop improvement, and is likely to accelerate crop breeding in ways dissimilar to crossbreeding and transgenic technologies. In this review, we summarize the current applications and prospects of CRISPR-Cas technology in maize gene-function studies and the generation of new germplasm for increased yield, specialty corns, plant architecture, stress response, haploid induction, and male sterility. Optimization of gene editing and genetic transformation systems for maize is also briefly reviewed. Lastly, the challenges and new opportunities that arise with the use of the CRISPR-Cas technology for maize genetic improvement are discussed.
Introduction
Increasing population, climate change, and environmental stresses are crucial issues threatening global food security. It is estimated that, at present, the increase in yields of the four key global crops of maize, rice, wheat, and soybean is far below the rate of 2.4% per year, which will be needed continuously up until 2050 in order to feed the ever-growing population (Ray et al., 2013).
Maize (Zea mays), an important crop with the greatest global production, can contribute to satiating the demands for human food, animal feed, and biofuels (Chavez-Arias et al., 2021). However, maize grain yield is a complex quantitative trait determined by multiple genetic and environmental factors (Lu et al., 2020; Zhang et al., 2020; Chavez-Arias et al., 2021; Malenica et al., 2021; Prasanna et al., 2021). It is estimated that the yield lost via drought stress is currently over 20% of the maize area per year (Boyer et al., 2013; Malenica et al., 2021), and via high temperatures, an average of 7.4% is lost for every 1°C increase (Lobell et al., 2013; Zhao et al., 2017). Moreover, lepidopteran pests and fungal diseases can also cause over 20% or 30% of yield loss, respectively (Liu et al., 2016). Hence, the breeding of new elite crop varieties for high yield, disease resistance and abiotic stress tolerance is essential to meet the demands for maize.
Conventional plant breeding methods based on crossbreeding have been restricted in their ability to develop elite yield varieties due to the decline in natural genetic diversity, rare germplasm resources, time-consuming, intensive labor and slow breeding process (Xiao et al., 2017; Hua et al., 2019). Genetic modification (GM) technologies, such as transgenic technologies, genome editing (GE), and molecular-assisted breeding, can efficiently overcome some problems with conventional breeding, and have shown great potential for breeding elite crops with high yields under environmental stress. GM crops have been widely planted worldwide and brought multiple benefits by increasing global grain yield and quality (Abdelrahman et al., 2018; Paul et al., 2018; Montagu, 2019; Fernie and Sonnewald, 2021). In 2015, 53.6 Mha of GM maize was cultivated, representing about 1/3 of planted maize globally. In 2020, 79% of maize in the USA was GM (Pellegrino et al., 2018; Montagu, 2019; Malenica et al., 2021). However, the transgenic technologies are constrained by biological processes such as the rate of recombination and the gene-centric nature of transgenic traits.
Recently, GE technologies including ZFN, TALEN, and CRISPR-Cas, have brought unparalleled opportunities for crop breeding by precisely manipulating genetic information on a genomic scale (Hamdan et al., 2022). In particular, the CRISPR-Cas9 system and its derivative systems such as CRISPR/Cpf1, base editing (BE) and prime editing (PE) provide powerful tools to edit the plant genome by incorporating random mutations, editing multiple loci, and inducting heritable inversions and translocations (Khan et al., 2019; Capdeville et al., 2021; Fiaz et al., 2021). Moreover, developed CRISPR-dCas9 system showed an increasingly important role in the process of gene activation and repression, epigenome editing, modulation of chromatin topology, live-cell chromatin imaging and DNA-free genetic modification. Particularly, this system enables the simultaneous activation of multiple key genes that positively control different agronomic traits, which is conducive to the rapid realization of crop genetic improvement by multigene pyramiding (Zhang et al., 2019). Owing to its high efficiency, simple operation and low cost, the CRISPR-Cas technology has rapidly shown promising potential in plant functional genomics studies and the genetic improvement of crop such as rice, wheat, soybean, maize, and potato (Rao et al., 2022). This review focuses on the application and prospects of CRISPR-Cas technology in the basic science and the creation of new germplasms in maize.
Functional annotation of maize genes using CRISPR-Cas technology
Gene-editing technology has paved an efficient and predictable way to decipher the functions of key genes and develop new germplasms in maize (Feng et al., 2016; Doll et al., 2019). Recently, the potential functions of many genes involved in maize development programs and stress responses have been well dissected using CRISPR-Cas technology (Table 1 and Figure 1). For the regulation of growth and development, ZmSMC3 has been found to be essential for sister chromatid cohesion and meiotic centromere pairing, and its knockout causes slow growth and dwarfed plants (Zhang et al., 2020). MMS21 participates in root and vegetative growth, pollen germination, and seed development by maintaining maize genome activity and stability (Zhang et al., 2021). ZmNRPC2 controls RNA polymerase III activity and the expression of multiple genes involved in kernel development, and its knockout results in significantly reduced kernel size (Zhao et al., 2020). ZmThx20 participates in the regulation of kernel size and storage protein filling in seed, and its mutation resulted in shrunken kernel (Li et al., 2021). KNR6 and AGAP are required for vesicle trafficking, and their mutations lead to severe defects in inflorescences and roots, short ears with fewer kernels, and dwarfed plants (Li et al., 2021). CC-type glutaredoxins such as MSCA1, ZmGRX2 and ZmGRX5 mediate redox status of target proteins, and the triple knockout mutants show severely suppressed ear and tassel growth and dwarfed plant (Yang et al., 2021). YIGE1 regulates inflorescence meristem size and ear length by tuning sugar and auxin signal pathways (Luo et al., 2021). ZmMIC1 participates in the growth of seedlings by affecting 5’-methylthioadenosine salvage and nicotianamine biosynthesis (Sun et al., 2022). Moreover, ZmPT7 and ZmPAT7 are essential for inorganic phosphate acquisition and tassel branch number, respectively (Wang et al., 2020).
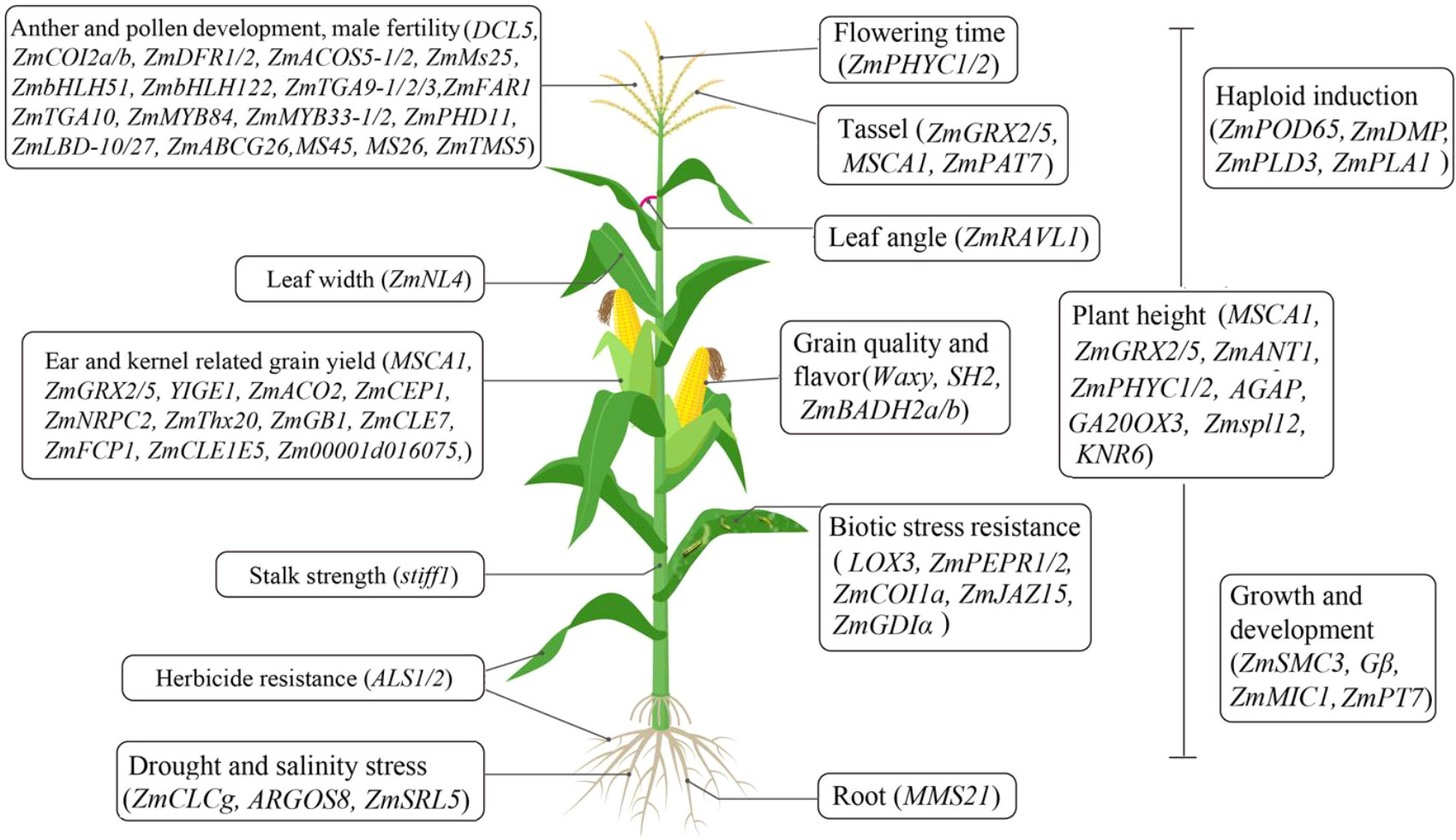
Figure 1 A schematic of the CRISPR-Cas technology used for the functional genomics study and generation of new germplasms in maize.
For stress response, ZmCLCg positively regulates chloride transport and sodium chloride stress in maize (Luo et al., 2021). ZmSRL5 is essential for maintaining cuticular wax structure and drought tolerance in maize (Pan et al., 2020). Moreover, ZmPEPR1 restricts S. exigua larval growth through regulating ZmPep3-activated foliar anti-herbivore defenses (Poretsky et al., 2020). Maize G protein β subunit (Gβ) regulates the trade-off between growth and defense response by tuning meristem size and autoimmunity (Wu et al., 2020). Additionally, other key genes have been characterized and utilized in the development of new germplasms as follows.
Generation of new maize germplasm resources using CRISPR-Cas technology
The CRISPR-Cas technology can overcome the limitations of conventional breeding due to the lack of available genetic resources and negative genetic linkage drag, and enables researchers to quickly and precisely modify target genes related to various traits in specific varieties, which has shown unique advantages in accelerating breeding process by generating new germplasms with more flexibility (Dong et al., 2019). Recently, CRISPR-Cas technology has been widely used to improve a variety of agronomic traits in different crops including rice, maize, wheat, barley and tomato (Liu et al., 2022). In maize, a series of new germplasms have been generated using the CRISPR-Cas technology. We with increased yield, improved quality and enhanced stress resistance, as well as male sterile lines, haploid inducers, and specialty corns We summarized them one by one in terms of improved yield, quality and special corns, haploid inducers, male sterile lines, and stress resistance.
Yield-related traits
Maize yield has been found to be closely associated with multiple yield-related traits including ear diameter, ear length, ear row number, ear weight, 100-kernel weight, and kernel number per row (Lu et al., 2020; Zhang et al., 2020). Thus, the editing of key genes related to these traits can be conducive to improving maize grain yield. For example, ZmCEP1 is involved in nitrate and sugar transport into the kernel, and its knockout can effectively enhance plant height, ear length, kernel size and 100-kernel weight (Xu et al., 2021). The editing of ZmACO2 can significantly increase ear length, kernel number per row, ear weight, and grain yield in hybrids (Ning et al., 2021). For key genes positively controlling agronomic traits, CRISPR-Cas genome editing of their promoters is a potential way to generate new quantitative variations for breeding (Rodriguez-Leal et al., 2017). In maize, genome editing of cis-regulatory regions within the promoters of ZmCLE7, ZmFCP1 and ZmCLE1E5 can effectively increase meristem size and multiple grain-yield-related traits (Liu et al., 2021; Chen and Tian, 2021).
It has also been shown that ideal plant architecture for high-density planting can contribute to an increase in maize grain yield (Wei et al., 2018; Cao et al., 2022).Gene editing of ZmRAVL1, a positive regulator of leaf angle, can generate an upright leaf architecture and enhance high-density maize yields (Tian et al., 2019). The knockout of ZmNL4 results in narrow leaves, which can be conducive to optimizing plant architecture for high-density planting (Gao et al., 2021). The double knockout mutant of zmphyC1 and zmphyC2 shows a moderate early-flowering phenotype under long-day conditions, providing valuable target genes to develop maize cultivars for adapting to different local environments (Li et al., 2020). Moreover, knockout of Zm00001d016075, a negative regulator of kernel row number, can increase the number of kernels per ear and grain yield (An et al., 2022). These studies have provided feasible strategies to improve maize yield via CRISPR-Cas genome editing.
Specialty corns
At present, the market demand for specialty corns, such as sweet, waxy or baby corns is increasing, and CRISPR-Cas technology provides effective ways to create these specialty germplasms (Dong et al., 2019). For example, simultaneous editing of ZmBADH2a and ZmBADH2b can generate an aromatic corn by increasing the accumulation of 2-acetyl-1-pyrroline (Wang et al., 2021). Moreover, a supersweet and waxy maize has been created by CRISPR–Cas9 editing of SH2 and WX (Dong et al., 2019). Recently, elite CRISPR–wx corn hybrids with higher yield have been developed by editing of a waxy allele in 12 inbred lines. Importantly, these CRISPR–wx corns are agronomically superior to introgressed hybrids, and are out of the scope of regulatory oversight over genetically modified organisms in the United States, Argentina, Brazil and Chile (Gao et al., 2020), and have initiated a new age in the global commercial production of gene-edited maize. These studies have indicated that CRISPR-Cas technology is a powerful tool to precisely develop various specialty corns with more flexibility in genetic background selection.
Haploid inducers
Doubled haploid technology can create perfectly homozygous individuals by rapidly fixing the recombinant haploid genomes on homogenous progeny, overcoming various constraints in genetic improvement and enabling rapid evaluation of phenotypic traits in maize. If a doubled haploid technology is combined with CRISPR-Cas technology, the breeders can perform faster and more precise crop breeding (Jacquier et al., 2020; Jacquier et al., 2021). In maize, the editing of ZmPOD65, ZmPLD3, ZmPLA1 and ZmDMP using CRISPR-Cas technology has successfully generated haploids, providing an approach to unravel the molecular mechanisms of haploid induction and the creation of various haploid inducers (Liu et al., 2017; Zhong et al., 2019; Li et al., 2021; Jiang et al., 2022). Furthermore, using a CRISPR-Cas9 cassette, haploid-inducer mediated genome editing was established to generate genome-edited haploids in the elite lines, which can greatly accelerate maize breeding by rapidly creating pure lines with desirable traits (Wang et al., 2019).. These studies provide potential ways to clarify the molecular mechanisms of haploid induction and breeding of haploid inducers.
Male sterility lines
Male sterility plays an important role in hybrid seed production. Using CRISPR-Cas technology, new male sterile lines have been created utilizing male fertility genes including ZmTMS5, ZmMs7, MS8, Ms26 and Ms45 (Svitashev et al., 2015; Li et al., 2017; Chen et al., 2018). Recently, a simple next-generation hybrid seed production system was created to generate a nuclear-genetic-male-sterility (GMS) line by knocking out ZmMS26 and its MGM maintainer line simultaneously, enabling effective production of sortable hybrid seeds labeled by a red fluorescent protein (DsRED) (Qi et al., 2020). Moreover, ZmCOI2a/b, ZmDFR1, ZmDFR2, ZmACOS5-1/2, DCL5, ZmABCG26, ZmFAR1, ZmMs25, ZmbHLH51, ZmbHLH122, ZmTGA9-1/2/3, ZmTGA10, ZmMYB33-1/2, ZmMYB84, ZmPHD11, and ZmLBD10/27 are required for the anther and pollen development, and they have been confirmed, and will be utilized, as potential target genes for the development of novel male-sterile lines using CRISPR-Cas technology (Teng et al., 2020; Zhang et al., 2021; Jiang et al., 2021; Jiang et al., 2021; Qi et al., 2022; Liu et al., 2022).
Stress resistance
Biotic and abiotic stresses cause devastating crop yield loss worldwide. Maize resistance is a complex quantitative trait determined by multiple genes and various environmental factors, and only few identified genes and rare elite germplasm resources are available for maize resistance breeding (Yang et al., 2017). Recently, an effective way for the creation of novel germplasm resources and breeding resistant varieties has been successfully applied via gene editing technology. For example, the knockout of lox3 could enhance maize durable resistance to Ustilago maydis (DC.) Corda by triggering reactive oxygen species bursts (Pathi et al., 2020). CRISPR-Cas9 targeted editing of ZmGDIα can strongly increase maize resistance against maize rough dwarf virus without agronomic penalty (Liu et al., 2022). ZmCOIa, by interacting with ZmJAZ15, antagonistically modulates maize immunity to Gibberella stalk rot (Fusarium graminearum Schwabe (teleomorph Gibberella zeae); GSR), and the knockout of ZmCOIa can enhance maize resistance to GSR (Ma et al., 2021).These studies provide a good indication of the potential of gene editing technology in the breeding of maize cultivars with increased resistance to fungal pathogens or insects.
Multiple abiotic stresses including drought, extreme temperature, flooding, and soil conditions seriously affect maize production. Compared with traditional breeding methods, genome editing technologies have shown considerable potential in the breeding of stress-tolerant variants (Chavez-Arias et al., 2021; Malenica et al., 2021; Prasanna et al., 2021; Chennakesavulu et al., 2021). Stalk lodging caused by various environmental stresses is a major threat to maize yield and quality, and developing maize lines with high stalk strength has become of major interest to breeders to ensure high and stable yield (Armarego-Marriott, 2020). stiff1 is a negative regulator of maize stalk strength, its edited allele with 2 bp deletion, caused a frameshift and an early-stop translation, conferred CRISPR-edited plants with a stronger stalk, which contributes to high density planting and preventing stalk lodging (Zhang et al., 2020). Moreover, semidwarf maize plants have been generated by editing ZmGA20ox3 using CRISPR-Cas9 technology, which might be useful in the creation of new germplasm with stronger lodging resistance and suitable for high-density planting (Zhang et al., 2020). For drought tolerance, precisely editing the promoter sequence of ARGOS8 leads to its upregulated expression and enhances maize grain yield under drought stress (Shi et al., 2017). Thus, CRISPR-Cas technology paves the way for the generation of novel germplasm sources for breeding stress-tolerant maize.
Challenges and prospects
Future and industrialization prospect of CRISPR-Cas edited maize
Breeding elite maize lines with multiple traits is a time-consuming and low efficiency process. Genome editing technology has revolutionized cereal crop improvement, and will shape future agricultural practices by integrating with traditional breeding (Hisano et al., 2021; Matres et al., 2021). In particular, CRISPR-Cas technology has shown the most exciting potential for the rapid and precise development of desirable varieties with multiple agronomic traits and stress tolerance (Figure 1). Now, the generation of single- and multiple-gene mutagenesis by CRISPR-Cas has become a powerful tool for functional genomics studies, to generate new germplasm resources, and to accelerate high-precision breeding processes in maize (Svitashev et al., 2015; Feng et al., 2016; Zhu et al., 2016; Agarwal et al., 2018; Doll et al., 2019; Liu et al., 2020). Notably, the combination of CRISPR-Cas technology with haploid induction or male sterility will help rapidly and predictably integrate desirable agronomic traits into new elite varieties.
It should be noted that many CRISPR-Cas-manipulated genes have been reported to improve maize traits, but very few lines have been validated by field trials and only CRISPR-waxy maize hybrids have had a limited commercial launch, in the Midwestern United States (Gao et al., 2020). Many transformed plants have only been tested for their potential breeding value in model systems or glasshouses, which may result in poor availability to develop elite maize lines suitable for field planting, and they might encounter negative effects in the field (Simmons et al., 2021). Thus, field testing is essential to evaluate whether the reported gene effects can be finally utilized in elite germplasm and maize production. To date, only 1671 genes, representing about 4.4% of the maize genome, have been tested to evaluate their field performance, and only 22 gene leads have been validated (Xiao et al., 2017; Simmons et al., 2021; Shikha et al., 2021). Thus, it is urgent to accelerate field testing of existing CRISPR-Cas-edited maize materials.
In agriculture, CRISPR-Cas technology has accelerated the process of breeding new crop varieties with improved traits, and increasing CRISPR-edited crops have been on the market, but their future depends on a scientific global regulatory framework (Ahmad et al., 2021). Among numerous CRISPR-edited maize germplasms, only limited precommercial launch of CRISPR-waxy hybrids was conducted in the Midwestern USA (Gao et al., 2020). Thus, it is more important to reframe existing regulatory policies to develop more scientific and technical criteria for the commercial production of CRISPR-Cas edited crops worldwide (Zhang et al., 2020; Ahmad et al., 2021). In addition, although CRISPR has potential to bring disruptive innovations for maize breeding, gene edited maize should consider the need of practical production and market demand to breed locally adapted and preferred crop varieties.
Improved CRISPR-Cas technologies and transformation systems for maize
Three major obstacles to the use of CRISPR technology in maize is the low gene editing frequency, low genetic transformation efficiency and few maize materials amenable for transformation. Recently, many progresses have been made in the improvement of CRISPR-Cas technology and transformation methods for maize (Table 2).
As for optimizing CRISPR-CAS technology to improve gene editing efficiency, a type I-E CRISPR-Cascade tool for gene activation, a ViMeBox (VIsual Maize Editing toolBox) high-frequency gene targeting method, and a seed fluorescence reporter (SFR)-assisted CRISPR-Cas9 system have been established in maize, providing effective and rapid methods to generate genome-edited plants (Young et al., 2019; Barone et al., 2020; Xu et al., 2021; Yan et al., 2021). Further, a highly efficient prime-editing tool, unrestricted by the strict NG protospacer adjacent motif (PAM) sequences, has been created by optimizing pegRNA expression in maize (Jiang et al., 2020). A selectable marker-free site-specific gene insertion can efficiently increase the frequency of targeted gene insertion (Peterson et al., 2021). Recently, CRISPR-Cas technologies have successfully manipulated targeted large fragment insertion/inversion and chromosomal crossovers in maize, providing promising methods to develop new maize varieties with multiple-traits stacking by breaking linkage drag, generating genetic diversity and facilitating trait introgression (Lee and Wang, 2020; Schwartz et al., 2020; Gao et al., 2020; Kouranov et al., 2022).
Traditional transgenic technology by overexpressing specific genes is an important technology to verify gene function and developing new crop materials with the improved traits, but its wide application is limited due to its difficulty in realizing the multi-gene polymerization and the strict regulation of genetically modified crops. Recently, the developed CRISPR-based gene activation technology has offered powerful tool for the simultaneous activation of multiple genes in a way superior to traditional transgenic technology, which provides new strategy for the manipulation of crop genetic improvement using the key genes positively controlling specific agronomic traits (Ding et al., 2022). For example, a CRISPR/dCas9-based toolkit was established to precisely and effectively activate expression of specific target genes in maize (Qi et al., 2022).
Moreover, off-target effects are frequently reported and widely existed in genome editing plants. A variety of approaches including improved CRISPR-Cas systems and new methods have been developed to eliminate the potential for off-target editing (Young et al., 2019; Modrzejewski et al., 2020; Sturme et al., 2022). It has been reported that the off-target editing in CRISPR-Cas edited maize plants could be mitigated using specifically designing guide RNAs differed from other genomic locations (Young et al., 2019). Moreover, a PAM-less/free high-efficiency adenine base editor toolbox (PhieABE toolbox) can effectively prevent off-target editing in rice, which shows wide applications in plant functional genomics and crop improvement (Tan et al., 2022).
Currently, Agrobacterium-mediated transformation is widely used to generate gene-edited plants, but this has major constraints due to few maize inbred lines amenable for transformation, low transformation efficiency and lower editing frequency (Ayar et al., 2013; Peterson et al., 2021; Hunter, 2021). Recently, several Agrobacterium-mediated transformation methods have been improved for maize genome-editing. For example, an agrobacterium-mediated CRISPR-Cas9 platform for high-efficiency genome editing has been used in a maize Hi II line (Char et al., 2017; Lee et al., 2019). Additionally, the Fast-Flowering Mini-Maize (FFMM) line A, previously unsuitable for genetic transformation, has now been bred for transformable lines by hybridizing it with a transformable genotype high Type-II callus parent A (Hi-II A) with line A of FFMM (McCaw et al., 2020). Moreover, a ternary vector system-based platform, employing WUS and Bbm for Agrobacterium-mediated transformation, can effectively increase the efficiency of transformation and gene editing, and widen the range of maize lines available for transformation (Zhang et al., 2019; Julkowska, 2019). In addition, the gene editing rate of CRISPR-Cas12a using particle bombardment was increased to over 60% in an elite maize variety (Dong et al., 2021). Thus, continuous optimization of transformation methods and gene editing systems is a prerequisite for the promotion of the application of gene editing technology in maize.
Author contributions
YW, QT, LP, HZ, and XL were contributed to the writing of this review and approved it for publication.
Funding
This work was supported by Nanfan special project, CAAS (Grant No: YBXM15) and the Central Public interest Scientific Institution Basal Research Fund (No.1610392022012).
Acknowledgments
This work was funded by the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences, the major projects of national science and technology (2019ZX08013010-015), and the Central Public-interest Scientific Institution Basal Research Fund (No. 1610392022012). The authors would like to express their gratitude to EditSprings (https://www.editsprings.cn) for the expert linguistic services provided.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdelrahman, M., Al-Sadi, A. M., Pour-Aboughadareh, A., Burritt, D. J., Tran, L. P. (2018). Genome editing using CRISPR/Cas9-targeted mutagenesis: An opportunity for yield improvements of crop plants grown under environmental stresses. Plant Physiol. Biochem. 131, 31–36. doi: 10.1016/j.plaphy.2018.03.012
Agarwal, A., Yadava, P., Kumar, K., Singh, I., Kaul, T., Pattanayak, A., et al. (2018). Insights into maize genome editing via CRISPR/Cas9. Physiol. Mol. Biol. Pla 24 (2), 175–183. doi: 10.1007/s12298-017-0502-3
Ahmad, A., Munawar, N., Khan, Z., Qusmani, A. T., Khan, S. H., Jamil, A., et al. (2021). An outlook on global regulatory landscape for genome-edited crops. Int. J. Mol. Sci. 22 (21), 11753. doi: 10.3390/ijms222111753
An, Y., Chen, L., Li, Y. X., Li, C., Shi, Y., Zhang, D., et al. (2022). Fine mapping qKRN5.04 provides a functional gene negatively regulating maize kernel row number. TAG Theor. Appl. Genet 135 (6), 1997–2007. doi: 10.1007/s00122-022-04089-w
Armarego-Marriott, T. (2020). Stiffening stems: Identification of the stiff1 gene involved in maize stalk strength. Plant Cell 32 (1), 12. doi: 10.1105/tpc.19.00852
Ayar, A., Wehrkamp-Richter, S., Laffaire, J. B., Le Goff, S., Levy, J., Chaignon, S., et al. (2013). Gene targeting in maize by somatic ectopic recombination. Plant Biotechnol. J. 11 (3), 305–314. doi: 10.1111/pbi.12014
Barone, P., Wu, E., Lenderts, B., Anand, A., Gordon-Kamm, W., Svitashev, S., et al. (2020). Efficient gene targeting in maize using inducible CRISPR-Cas9 and marker-free donor template. Mol. Plant 13 (8), 1219–1227. doi: 10.1016/j.molp.2020.06.008
Boyer, J. S., Byrne, P., Cassman, K. G., Cooper, M., Delmer, D., Greene, T., et al. (2013). The US drought of 2012 in perspective: A call to action. Glob Food Secur-Agr 2 (3), 139–143. doi: 10.1016/j.gfs.2013.08.002
Cao, Y., Zhong, Z., Wang, H., Shen, R. (2022). Leaf angle: a target of genetic improvement in cereal crops tailored for high-density planting. Plant Biotechnol. J. 20 (3), 426–436. doi: 10.1111/pbi.13780
Capdeville, N., Merker, L., Schindele, P., Puchta, H. (2021). Sophisticated CRISPR/Cas tools for fine-tuning plant performance. J. Plant Physiol. 257, 153332. doi: 10.1016/j.jplph.2020.153332
Char, S. N., Neelakandan, A. K., Nahampun, H., Frame, B., Main, M., Spalding, M. H., et al. (2017). An agrobacterium-delivered CRISPR/Cas9 system for high-frequency targeted mutagenesis in maize. Plant Biotechnol. J. 15 (2), 257–268. doi: 10.1111/pbi.12611
Chavez-Arias, C. C., Ligarreto-Moreno, G. A., Ramirez-Godoy, A., Restrepo-Diaz, H. (2021). Maize responses challenged by drought, elevated daytime temperature and arthropod herbivory stresses: A physiological, biochemical and molecular view. Front. In Plant Sci. 12. doi: 10.3389/fpls.2021.702841
Chennakesavulu, K., Singh, H., Trivedi, P. K., Jain, M., Yadav, S. R. (2021). State-of-the-Art in CRISPR technology and engineering drought, salinity, and thermo-tolerant crop plants. Plant Cell Rep 41 (3), 815–831. doi: 10.1007/s00299-021-02681-w
Chen, Q., Tian, F. (2021). Towards knowledge-driven breeding. Nat. Plants 7 (3), 242–243. doi: 10.1038/s41477-021-00864-7
Chen, R. R., Xu, Q. L., Liu, Y., Zhang, J. J., Ren, D. T., Wang, G. Y., et al. (2018). Generation of transgene-free maize Male sterile lines using the CRISPR/Cas9 system. Front. In Plant Sci. 9. doi: 10.3389/fpls.2018.01180
Ding, X., Yu, L., Chen, L., Li, Y., Zhang, J., Sheng, H., et al. (2022). Recent progress and future prospect of CRISPR/Cas-derived transcription activation (CRISPRa) system in plants. Cells 11 (19), 3045. doi: 10.3390/cells11193045
Doll, N. M., Gilles, L. M., Gerentes, M. F., Richard, C., Just, J., Fierlej, Y., et al. (2019). Single and multiple gene knockouts by CRISPR-Cas9 in maize. Plant Cell Rep. 38 (4), 487–501. doi: 10.1007/s00299-019-02378-1
Dong, S., Qin, Y. L., Vakulskas, C. A., Collingwood, M. A., Marand, M., Rigoulot, S., et al. (2021). Efficient targeted mutagenesis mediated by CRISPR-Cas12a ribonucleoprotein complexes in maize. Front. Genome Ed 3, 670529. doi: 10.3389/fgeed.2021.670529
Dong, L., Qi, X. T., Zhu, J. J., Liu, C. L., Zhang, X., Cheng, B. J., et al. (2019). Supersweet and waxy: meeting the diverse demands for specialty maize by genome editing. Plant Biotechnol. J. 17 (10), 1853–1855. doi: 10.1111/pbi.13144
Feng, C., Yuan, J., Wang, R., Liu, Y., Birchler, J. A., Han, F. (2016). Efficient targeted genome modification in maize using CRISPR/Cas9 system. J. Genet. Genomics 43 (1), 37–43. doi: 10.1016/j.jgg.2015.10.002
Fernie, A. R., Sonnewald, U. (2021). Plant biotechnology for sustainable agriculture and food safety. J. Plant Physiol. 261, 153416. doi: 10.1016/j.jplph.2021.153416
Fiaz, S., Ahmar, S., Saeed, S., Riaz, A., Mora-Poblete, F., Jung, K. H. (2021). Evolution and application of genome editing techniques for achieving food and nutritional security. Int. J. Mol. Sci. 22 (11), 5585. doi: 10.3390/ijms22115585
Gao, H., Gadlage, M. J., Lafitte, H. R., Lenderts, B., Yang, M., Schroder, M., et al. (2020). Superior field performance of waxy corn engineered using CRISPR-Cas9. Nat. Biotechnol. 38 (5), 579–581. doi: 10.1038/s41587-020-0444-0
Gao, H., Mutti, J., Young, J. K., Yang, M., Schroder, M., Lenderts, B., et al. (2020). Complex trait loci in maize enabled by CRISPR-Cas9 mediated gene insertion. Front. Plant Sci. 11, 535. doi: 10.3389/fpls.2020.00535
Gao, L., Yang, G., Li, Y., Sun, Y., Xu, R., Chen, Y., et al. (2021). A kelch-repeat superfamily gene, ZmNL4, controls leaf width in maize (Zea mays l.). Plant J. 107 (3), 817–830. doi: 10.1111/tpj.15348
Guan, H. H., Chen, X. J., Wang, K. L., Liu, X. Y., Zhang, D. F., Li, Y. X., et al. (2022). Genetic variation in ZmPAT7 contributes to tassel branch number in maize. Int. J. Mol. Sci. 23 (5), 2586. doi: 10.3390/ijms23052586
Hamdan, M. F., Mohd Noor, S. N., Abd-Aziz, N., Pua, T. L., Tan, B. C. (2022). Green revolution to gene revolution: Technological advances in agriculture to feed the world. Plants (Basel) 11 (10), 1297. doi: 10.3390/plants11101297
Hisano, H., Abe, F., Hoffie, E. R., Kumlehn, J. (2021). Targeted genome modifications in cereal crops. Breed. Sci. 71 (4), 405–416. doi: 10.1270/jsbbs.21019
Hua, K., Zhang, J., Botella, J. R., Ma, C., Kong, F., Liu, B., et al. (2019). Perspectives on the application of genome-editing technologies in crop breeding. Mol. Plant 12 (8), 1047–1059. doi: 10.1016/j.molp.2019.06.009
Hunter, C. T. (2021). CRISPR/Cas9 targeted mutagenesis for functional genetics in maize. Plants (Basel) 10 (4), 723. doi: 10.3390/plants10040723
Jacquier, N. M. A., Gilles, L. M., Martinant, J. P., Rogowsky, P. M., Widiez, T. (2021). Maize in planta haploid inducer lines: A cornerstone for doubled haploid technology. Methods Mol. Biol. 2288, 25–48. doi: 10.1007/978-1-0716-1335-1_2
Jacquier, N. M. A., Gilles, L. M., Pyott, D. E., Martinant, J. P., Rogowsky, P. M., Widiez, T. (2020). Puzzling out plant reproduction by haploid induction for innovations in plant breeding. Nat. Plants 6 (6), 610–619. doi: 10.1038/s41477-020-0664-9
Jiang, Y., An, X., Li, Z., Yan, T., Zhu, T., Xie, K., et al. (2021). CRISPR/Cas9-based discovery of maize transcription factors regulating male sterility and their functional conservation in plants. Plant Biotechnol. J. 19 (9), 1769–1784. doi: 10.1111/pbi.13590
Jiang, Y. Y., Chai, Y. P., Lu, M. H., Han, X. L., Lin, Q., Zhang, Y., et al. (2020). Prime editing efficiently generates W542L and S621I double mutations in two ALS genes in maize. Genome Biol. 21 (1), 257. doi: 10.1186/s13059-020-02170-5
Jiang, Y., Li, Z., Liu, X., Zhu, T., Xie, K., Hou, Q., et al. (2021). ZmFAR1 and ZmABCG26 regulated by microRNA are essential for lipid metabolism in maize anther. Int. J. Mol. Sci. 22 (15), 7916. doi: 10.3390/ijms22157916
Jiang, C., Sun, J., Li, R., Yan, S., Chen, W., Guo, L., et al. (2022). A reactive oxygen species burst causes haploid induction in maize. Mol. Plant 15 (6), 943–955. doi: 10.1016/j.molp.2022.04.001
Julkowska, M. M. (2019). Restriction release: Improved maize transformation efficiency. Plant Physiol. 181 (4), 1397–1397. doi: 10.1104/pp.19.01311
Khan, M. Z., Zaidi, S. S., Amin, I., Mansoor, S. (2019). A CRISPR way for fast-forward crop domestication. Trends Plant Sci. 24 (4), 293–296. doi: 10.1016/j.tplants.2019.01.011
Kouranov, A., Armstrong, C., Shrawat, A., Sidorov, V., Huesgen, S., Lemke, B., et al. (2022). Demonstration of targeted crossovers in hybrid maize using CRISPR technology. Commun. Biol. 5 (1), 53. doi: 10.1038/s42003-022-03004-9
Lee, K., Wang, K. (2020). Level up to chromosome restructuring. Nat. Plants 6 (6), 600–601. doi: 10.1038/s41477-020-0669-4
Lee, K., Zhu, H., Yang, B., Wang, K. (2019). An agrobacterium-mediated CRISPR/Cas9 platform for genome editing in maize. Methods Mol. Biol. 1917, 121–143. doi: 10.1007/978-1-4939-8991-1_10
Li, Y., Lin, Z., Yue, Y., Zhao, H. M., Fei, X. H., Lizhu, E., et al. (2021). Loss-of-function alleles of ZmPLD3 cause haploid induction in maize. Nat. Plants 7 (12), 1579–1588. doi: 10.1038/s41477-021-01037-2
Li, P., Li, Z., Xie, G., Zhang, J. (2021). Trihelix transcription factor ZmThx20 is required for kernel development in maize. Int. J. Mol. Sci. 22 (22), 12137. doi: 10.3390/ijms222212137
Liu, H., Chen, W., Li, Y., Sun, L., Chai, Y., Chen, H., et al. (2022). CRISPR/Cas9 technology and its utility for crop improvement. Int. J. Mol. Sci. 23 (18), 10442. doi: 10.3390/ijms231810442
Liu, L., Gallagher, J., Arevalo, E. D., Chen, R., Skopelitis, T., Wu, Q., et al. (2021). Enhancing grain-yield-related traits by CRISPR-Cas9 promoter editing of maize CLE genes. Nat. Plants 7 (3), 287–294. doi: 10.1038/s41477-021-00858-5
Liu, Q. S., Hallerman, E., Peng, Y. F., Li, Y. H. (2016). Development of bt rice and bt maize in China and their efficacy in target pest control. Int. J. of Mol. Sci. 17 (10), 1561. doi: 10.3390/ijms17101561
Liu, H. J., Jian, L., Xu, J., Zhang, Q., Zhang, M., Jin, M., et al. (2020). High-throughput CRISPR/Cas9 mutagenesis streamlines trait gene identification in maize. Plant Cell 32 (5), 1397–1413. doi: 10.1105/tpc.19.00934
Liu, C. L., Kong, M., Yang, F., Zhu, J. J., Qi, X. T., Weng, J. F., et al. (2022). Targeted generation of null mutants in ZmGDI alpha confers resistance against maize rough dwarf disease without agronomic penalty. Plant Biotechnol. J. 20 (5), 803–805. doi: 10.1111/pbi.13793
Liu, C. X., Li, X., Meng, D. X., Zhong, Y., Chen, C., Dong, X., et al. (2017). A 4-bp insertion at ZmPLA1 encoding a putative phospholipase a generates haploid induction in maize. Mol. Plant 10 (3), 520–522. doi: 10.1016/j.molp.2017.01.011
Liu, W. Y., Lin, H. H., Yu, C. P., Chang, C. K., Chen, H. J., Lin, J. J., et al. (2020). Maize ANT1 modulates vascular development, chloroplast development, photosynthesis, and plant growth. Proc. Natl. Acad. Sci. U.S.A. 117 (35), 21747–21756. doi: 10.1073/pnas.2012245117
Liu, X. Z., Zhang, S. W., Jiang, Y. L., Yan, T. W., Fang, C. W., Hou, Q. C., et al. (2022). Use of CRISPR/Cas9-based gene editing to simultaneously mutate multiple homologous genes required for pollen development and Male fertility in maize. Cells 11 (3), 439. doi: 10.3390/cells11030439
Li, Q., Wu, G., Zhao, Y., Wang, B., Zhao, B., Kong, D., et al. (2020). CRISPR/Cas9-mediated knockout and overexpression studies reveal a role of maize phytochrome c in regulating flowering time and plant height. Plant Biotechnol. J. 18 (12), 2520–2532. doi: 10.1111/pbi.13429
Li, J., Zhang, H. W., Si, X. M., Tian, Y. H., Chen, K. L., Liu, J. X., et al. (2017). Generation of thermosensitive male-sterile maize by targeted knockout of the ZmTMS5 gene. J. Genet. and Genomics 44 (9), 465–468. doi: 10.1016/j.jgg.2017.02.002
Li, M., Zhao, R., Du, Y., Shen, X., Ning, Q., Li, Y., et al. (2021). The coordinated KNR6-AGAP-ARF1 complex modulates vegetative and reproductive traits by participating in vesicle trafficking in maize. Cells 10 (10). doi: 10.3390/cells10102601
Lobell, D. B., Hammer, G. L., McLean, G., Messina, C., Roberts, M. J., Schlenker, W. (2013). The critical role of extreme heat for maize production in the united states. Nat. Clim Change 3 (5), 497–501. doi: 10.1038/nclimate1832
Luo, Y., Zhang, M., Liu, Y., Liu, J., Li, W., Chen, G., et al. (2022). Genetic variation in YIGE1 contributes to ear length and grain yield in maize. New Phytol 234 (2), 513–526. doi: 10.1111/nph.17882
Luo, M. J., Zhang, Y. X., Li, J. N., Zhang, P. P., Chen, K., Song, W., et al. (2021). Molecular dissection of maize seedling salt tolerance using a genome-wide association analysis method. Plant Biotechnol. J. 19 (10), 1937–1951. doi: 10.1111/pbi.13607
Lu, X., Zhou, Z. Q., Yuan, Z. H., Zhang, C. S., Hao, Z. F., Wang, Z. H., et al. (2020). Genetic dissection of the general combining ability of yield-related traits in maize. Front. In Plant Sci. 11. doi: 10.3389/fpls.2020.00788
Malenica, N., Dunic, J. A., Vukadinovic, L., Cesar, V., Simic, D. (2021). Genetic approaches to enhance multiple stress tolerance in maize. Genes (Basel) 12 (11),1760. doi: 10.3390/genes12111760
Ma, L., Sun, Y., Ruan, X., Huang, P. C., Wang, S., Li, S., et al. (2021). Genome-wide characterization of jasmonates signaling components reveals the essential role of ZmCOI1a-ZmJAZ15 action module in regulating maize immunity to gibberella stalk rot. Int. J. Mol. Sci. 22 (2), 870. doi: 10.3390/ijms22020870
Matres, J. M., Hilscher, J., Datta, A., Armario-Najera, V., Baysal, C., He, W., et al. (2021). Genome editing in cereal crops: an overview. Transgenic Res. 30 (4), 461–498. doi: 10.1007/s11248-021-00259-6
McCaw, M. E., Lee, K., Kang, M., Zobrist, J. D., Azanu, M. K., Birchler, J. A., et al. (2020). Development of a transformable fast-flowering mini-maize as a tool for maize gene editing. Front. Genome Ed 2, 622227. doi: 10.3389/fgeed.2020.622227
Modrzejewski, D., Hartung, F., Lehnert, H., Sprink, T., Kohl, C., Keilwagen, J., et al. (2020). Which factors affect the occurrence of off-target effects caused by the use of CRISPR/Cas: A systematic review in plants. Front. Plant Sci. 11, 574959. doi: 10.3389/fpls.2020.574959
Montagu, M. V. (2019). The future of plant biotechnology in a globalized and environmentally endangered world. Genet. Mol. Biol. 43 (1 suppl 2), e20190040. doi: 10.1590/1678-4685-GMB-2019-0040
Ning, Q., Jian, Y., Du, Y., Li, Y., Shen, X., Jia, H., et al. (2021). An ethylene biosynthesis enzyme controls quantitative variation in maize ear length and kernel yield. Nat. Commun. 12 (1), 5832. doi: 10.1038/s41467-021-26123-z
Pan, Z., Liu, M., Zhao, H., Tan, Z., Liang, K., Sun, Q., et al. (2020). ZmSRL5 is involved in drought tolerance by maintaining cuticular wax structure in maize. J. Integr. Plant Biol. 62 (12), 1895–1909. doi: 10.1111/jipb.12982
Pathi, K. M., Rink, P., Budhagatapalli, N., Betz, R., Saado, I., Hiekel, S., et al. (2020). Engineering smut resistance in maize by site-directed mutagenesis of LIPOXYGENASE 3. Front. Plant Sci. 11, 543895. doi: 10.3389/fpls.2020.543895
Paul, M. J., Nuccio, M. L., Basu, S. S. (2018). Are GM crops for yield and resilience possible? Trends Plant Sci. 23 (1), 10–16. doi: 10.1016/j.tplants.2017.09.007
Pellegrino, E., Bedini, S., Nuti, M., Ercoli, L. (2018). Impact of genetically engineered maize on agronomic, environmental and toxicological traits: a meta-analysis of 21 years of field data. Sci. Rep. 8 (1), 3113. doi: 10.1038/s41598-018-21284-2
Peterson, D., Barone, P., Lenderts, B., Schwartz, C., Feigenbutz, L., St Clair, G., et al. (2021). Advances in agrobacterium transformation and vector design result in high-frequency targeted gene insertion in maize. Plant Biotechnol. J. 19 (10), 2000–2010. doi: 10.1111/pbi.13613
Poretsky, E., Dressano, K., Weckwerth, P., Ruiz, M., Char, S. N., Shi, D., et al. (2020). Differential activities of maize plant elicitor peptides as mediators of immune signaling and herbivore resistance. Plant J. 104 (6), 1582–1602. doi: 10.1111/tpj.15022
Prasanna, B. M., Cairns, J. E., Zaidi, P. H., Beyene, Y., Makumbi, D., Gowda, M., et al. (2021). Beat the stress: breeding for climate resilience in maize for the tropical rainfed environments. Theor. and Appl. Genet. 134 (6), 1729–1752. doi: 10.1007/s00122-021-03773-7
Qi, X., Gao, H., Lv, R., Mao, W., Zhu, J., Liu, C., et al. (2022)CRISPR/dCas-mediated gene activation toolkit development and its application for parthenogenesis induction in maize. Plant Commun., 100449. doi: 10.1016/j.xplc.2022.100449
Qi, X. L., Guo, S. W., Wang, D., Zhong, Y., Chen, M., Chen, C., et al. (2022). ZmCOI2a and ZmCOI2b redundantly regulate anther dehiscence and gametophytic male fertility in maize. Plant J. 110 (3), 849–862. doi: 10.1111/tpj.15708
Qi, X., Zhang, C., Zhu, J., Liu, C., Huang, C., Li, X., et al. (2020). Genome editing enables next-generation hybrid seed production technology. Mol. Plant 13 (9), 1262–1269. doi: 10.1016/j.molp.2020.06.003
Rao, Y., Yang, X., Pan, C., Wang, C., Wang, K. (2022). Advance of clustered regularly interspaced short palindromic repeats-Cas9 system and its application in crop improvement. Front. Plant Sci. 13, 839001. doi: 10.3389/fpls.2022.839001
Ray, D. K., Mueller, N. D., West, P. C., Foley, J. A. (2013). Yield trends are insufficient to double global crop production by 2050. PloS One 8 (6), e66428. doi: 10.1371/journal.pone.0066428
Rodriguez-Leal, D., Lemmon, Z. H., Man, J., Bartlett, M. E., Lippman, Z. B. (2017). Engineering quantitative trait variation for crop improvement by genome editing. Cell 171 (2), 470–480 e478. doi: 10.1016/j.cell.2017.08.030
Schwartz, C., Lenderts, B., Feigenbutz, L., Barone, P., Llaca, V., Fengler, K., et al. (2020). CRISPR-Cas9-mediated 75.5-Mb inversion in maize. Nat. Plants 6 (12), 1427–1431. doi: 10.1038/s41477-020-00817-6
Shi, J. R., Gao, H. R., Wang, H. Y., Lafitte, H. R., Archibald, R. L., Yang, M. Z., et al. (2017). ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 15 (2), 207–216. doi: 10.1111/pbi.12603
Shikha, K., Shahi, J. P., Vinayan, M. T., Zaidi, P. H., Singh, A. K., Sinha, B. (2021). Genome-wide association mapping in maize: status and prospects. 3 Biotech. 11 (5), 244. doi: 10.1007/s13205-021-02799-4
Simmons, C. R., Lafitte, H. R., Reimann, K. S., Brugiere, N., Roesler, K., Albertsen, M. C., et al. (2021). Successes and insights of an industry biotech program to enhance maize agronomic traits. Plant Sci. 307, 110899. doi: 10.1016/j.plantsci.2021.110899
Sturme, M. H. J., van der Berg, J. P., Bouwman, L. M. S., De Schrijver, A., de Maagd, R. A., Kleter, G. A., et al. (2022). Occurrence and nature of off-target modifications by CRISPR-cas genome editing in plants. ACS Agric. Sci. Technol. 2 (2), 192–201. doi: 10.1021/acsagscitech.1c00270
Sun, W. T., Zhou, X. J., Chen, C., Zhang, X., Tian, X. L., Xiao, K., et al. (2022). Maize interveinal chlorosis 1 links the yang cycle and fe homeostasis through nicotianamine biosynthesis. Plant Physiol. 188 (4), 2131–2145. doi: 10.1093/plphys/kiac009
Svitashev, S., Young, J. K., Schwartz, C., Gao, H., Falco, S. C., Cigan, A. M. (2015). Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 169 (2), 931–945. doi: 10.1104/pp.15.00793
Tan, J., Zeng, D., Zhao, Y., Wang, Y., Liu, T., Li, S., et al. (2022). PhieABEs: a PAM-less/free high-efficiency adenine base editor toolbox with wide target scope in plants. Plant Biotechnol. J. 20 (5), 934–943. doi: 10.1111/pbi.13774
Teng, C., Zhang, H., Hammond, R., Huang, K., Meyers, B. C., Walbot, V. (2020). Dicer-like 5 deficiency confers temperature-sensitive male sterility in maize. Nat. Commun. 11 (1), 2912. doi: 10.1038/s41467-020-16634-6
Tian, J. G., Wang, C. L., Xia, J. L., Wu, L. S., Xu, G. H., Wu, W. H., et al. (2019). Teosinte ligule allele narrows plant architecture and enhances high-density maize yields. Science 365 (6454), 658. doi: 10.1126/science.aax5482
Wang, F., Cui, P. J., Tian, Y., Huang, Y., Wang, H. F., Liu, F., et al. (2020). Maize ZmPT7 regulates pi uptake and redistribution which is modulated by phosphorylation. Plant Biotechnol. J. 18 (12), 2406–2419. doi: 10.1111/pbi.13414
Wang, Y., Liu, X., Zheng, X., Wang, W., Yin, X., Liu, H., et al. (2021). Creation of aromatic maize by CRISPR/Cas. J. Integr. Plant Biol. 63 (9), 1664–1670. doi: 10.1111/jipb.13105
Wang, B. B., Zhu, L., Zhao, B. B., Zhao, Y. P., Xie, Y. R., Zheng, Z. G., et al. (2019). Development of a haploid-inducer mediated genome editing system for accelerating maize breeding. Mol. Plant 12 (4), 597–602. doi: 10.1016/j.molp.2019.03.006
Wei, H., Zhao, Y., Xie, Y., Wang, H. (2018). Exploiting SPL genes to improve maize plant architecture tailored for high-density planting. J. Exp. Bot. 69 (20), 4675–4688. doi: 10.1093/jxb/ery258
Wu, Q. Y., Xu, F., Liu, L., Char, S. N., Ding, Y. Z., Je, B. I., et al. (2020). The maize heterotrimeric G protein beta subunit controls shoot meristem development and immune responses. P Natl. Acad. Sci. U.S.A. 117 (3), 1799–1805. doi: 10.1073/pnas.1917577116
Xiao, Y. J., Liu, H. J., Wu, L. J., Warburton, M., Yan, J. B. (2017). Genome-wide association studies in maize: Praise and stargaze. Mol. Plant 10 (3), 359–374. doi: 10.1016/j.molp.2016.12.008
Xu, R., Li, Y., Sui, Z., Lan, T., Song, W., Zhang, M., et al. (2021). A c-terminal encoded peptide, ZmCEP1, is essential for kernel development in maize. J. Exp. Bot. 72 (15), 5390–5406. doi: 10.1093/jxb/erab224
Xu, J., Yin, Y., Jian, L., Han, B., Chen, Z., Yan, J., et al. (2021). Seeing is believing: a visualization toolbox to enhance selection efficiency in maize genome editing. Plant Biotechnol. J. 19 (5), 872–874. doi: 10.1111/pbi.13575
Yang, Q., Balint-Kurti, P., Xu, M. (2017). Quantitative disease resistance: Dissection and adoption in maize. Mol. Plant 10 (3), 402–413. doi: 10.1016/j.molp.2017.02.004
Yang, R., Xu, F., Wang, Y. M., Zhong, W. S., Dong, L., Shi, Y. N., et al. (2021). Glutaredoxins regulate maize inflorescence meristem development via redox control of TGA transcriptional activity. Nat. Plants 7 (12), 1589. doi: 10.1038/s41477-021-01029-2
Yan, Y., Zhu, J., Qi, X., Cheng, B., Liu, C., Xie, C. (2021). Establishment of an efficient seed fluorescence reporter-assisted CRISPR/Cas9 gene editing in maize. J. Integr. Plant Biol. 63 (9), 1671–1680. doi: 10.1111/jipb.13086
Young, J. K., Gasior, S. L., Jones, S., Wang, L., Navarro, P., Vickroy, B., et al. (2019). The repurposing of type I-e CRISPR-cascade for gene activation in plants. Commun. Biol. 2, 383. doi: 10.1038/s42003-019-0637-6
Young, J., Zastrow-Hayes, G., Deschamps, S., Svitashev, S., Zaremba, M., Acharya, A., et al. (2019). CRISPR-Cas9 editing in maize: Systematic evaluation of off-target activity and its relevance in crop improvement. Sci. Rep. 9 (1), 6729. doi: 10.1038/s41598-019-43141-6
Zhang, J., Augustine, R. C., Suzuki, M., Feng, J., Char, S. N., Yang, B., et al. (2021). The SUMO ligase MMS21 profoundly influences maize development through its impact on genome activity and stability. PloS Genet. 17 (10), e1009830. doi: 10.1371/journal.pgen.1009830
Zhang, J., Feng, C., Su, H., Liu, Y., Liu, Y., Han, F. (2020). The cohesin complex subunit ZmSMC3 participates in meiotic centromere pairing in maize. Plant Cell 32 (4), 1323–1336. doi: 10.1105/tpc.19.00834
Zhang, X. X., Guan, Z. R., Li, Z. L., Liu, P., Ma, L. L., Zhang, Y. C., et al. (2020). A combination of linkage mapping and GWAS brings new elements on the genetic basis of yield-related traits in maize across multiple environments. Theor. and Appl. Genet. 133 (10), 2881–2895. doi: 10.1007/s00122-020-03639-4
Zhang, D., Hussain, A., Manghwar, H., Xie, K., Xie, S., Zhao, S., et al. (2020). Genome editing with the CRISPR-cas system: an art, ethics and global regulatory perspective. Plant Biotechnol. J. 18 (8), 1651–1669. doi: 10.1111/pbi.13383
Zhang, S., Wu, S., Niu, C., Liu, D., Yan, T., Tian, Y., et al. (2021). ZmMs25 encoding a plastid-localized fatty acyl reductase is critical for anther and pollen development in maize. J. Exp. Bot. 72 (12), 4298–4318. doi: 10.1093/jxb/erab142
Zhang, J., Zhang, X., Chen, R., Yang, L., Fan, K., Liu, Y., et al. (2020). Generation of transgene-free semidwarf maize plants by gene editing of gibberellin-Oxidase20-3 using CRISPR/Cas9. Front. Plant Sci. 11, 1048. doi: 10.3389/fpls.2020.01048
Zhang, Z. H., Zhang, X., Lin, Z. L., Wang, J., Liu, H. Q., Zhou, L. N., et al. (2020). A Large transposon insertion in the stiff1 promoter increases stalk strength in Maize([OPEN]). Plant Cell 32 (1), 152–165. doi: 10.1105/tpc.19.00486
Zhang, Q., Zhang, Y., Lu, M. H., Chai, Y. P., Jiang, Y. Y., Zhou, Y., et al. (2019). A novel ternary vector system united with morphogenic genes enhances CRISPR/Cas delivery in Maize(1)([OPEN]). Plant Physiol. 181 (4), 1441–1448. doi: 10.1104/pp.19.00767
Zhao, C., Liu, B., Piao, S., Wang, X., Lobell, D. B., Huang, Y., et al. (2017). Temperature increase reduces global yields of major crops in four independent estimates. Proc. Natl. Acad. Sci. U.S.A. 114 (35), 9326–9331. doi: 10.1073/pnas.1701762114
Zhao, H., Qin, Y., Xiao, Z., Li, Q., Yang, N., Pan, Z., et al. (2020). Loss of function of an RNA polymerase III subunit leads to impaired maize kernel development. Plant Physiol. 184 (1), 359–373. doi: 10.1104/pp.20.00502
Zhao, B., Xu, M., Zhao, Y., Li, Y., Xu, H., Li, C., et al. (2022). Overexpression of ZmSPL12 confers enhanced lodging resistance through transcriptional regulation of D1 in maize. Plant Biotechnol. J. 20 (4), 622–624. doi: 10.1111/pbi.13787
Zhong, Y., Liu, C. X., Qi, X. L., Jiao, Y. Y., Wang, D., Wang, Y. W., et al. (2019). Mutation of ZmDMP enhances haploid induction in maize. Nat. Plants 5 (6), 575–580. doi: 10.1038/s41477-019-0443-7
Keywords: CRISPR-Cas technology, gene editing, maize, gene function, germplasms, variety improvement
Citation: Wang Y, Tang Q, Pu L, Zhang H and Li X (2022) CRISPR-Cas technology opens a new era for the creation of novel maize germplasms. Front. Plant Sci. 13:1049803. doi: 10.3389/fpls.2022.1049803
Received: 21 September 2022; Accepted: 25 November 2022;
Published: 16 December 2022.
Edited by:
Anshu Alok, University of Minnesota Twin Cities, United StatesReviewed by:
Mariana Rocha Maximiano, Universidade Católica Dom Bosco, BrazilLiliana Medina-Aparicio, Universidad Nacional Autónoma de México, Mexico
Chunsheng Xue, Shenyang Agricultural University, China
Purva Bhalothia, Birla Institute of Scientific Research, India
Copyright © 2022 Wang, Tang, Pu, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haiwen Zhang, emhhbmdoYWl3ZW5AY2Fhcy5jbg==; Xinhai Li, bGl4aW5oYWlAY2Fhcy5jbg==
 Youhua Wang
Youhua Wang Qiaoling Tang1
Qiaoling Tang1 Li Pu
Li Pu Haiwen Zhang
Haiwen Zhang