- Zhejiang Provincial Key Laboratory of Germplasm Innovation and Utilization for Garden Plants, School of Landscape and Architecture, Zhejiang A&F University, Hangzhou, Zhejiang, China
Continuous flowering is a key horticultural trait in ornamental plants, whereas the specific molecular regulation mechanism remains largely unknown. In sweet osmanthus (Osmanthus fragrans Lour.), plants based on their flowering characteristics are divided into once-flowering (OF) habit and continuous flowering (CF) habit. Here, we first described the flowering phenology shifts of OF and CF habits in sweet osmanthus through paraffin section and microscope assay. Phenotypic characterization showed that CF plants had constant new shoot growth, floral transition, and blooming for 1 year, which might lead to a continuous flowering trait. We performed the transcriptome sequencing of OF and CF sweet osmanthus and analyzed the transcriptional activity of flowering-related genes. Among the genes, three floral integrators, OfFT, OfTFL1, and OfBFT, had a differential expression during the floral transition process in OF and CF habits. The expression patterns of the three genes in 1 year were revealed. The results suggested that their accumulations corresponded to the new shoots occurring and the floral transition process. Function studies suggested that OfFT acted as a flowering activator, whereas OfBFT was a flowering inhibitor. Yeast one-hybrid assay indicated that OfSPL8 was a common upstream transcription factor of OfFT and OfBFT, suggesting the vital role of OfSPL8 in continuous flowering regulation. These results provide a novel insight into the molecular mechanism of continuous flowering.
Introduction
Flowering is a vital developmental event and mediated by complex genetic mechanisms (Chen D. et al., 2018; Chen Z. et al., 2018). Among flowering plants, most species have a seasonal flowering period, blooming once a year in a certain season (once-flowering habit, OF), whereas some plants are able to flower continuously under a favorable season (continuous flowering habit, CF), such as rose, strawberry, and orchid (Ahmad et al., 2021). Obtaining plants with an extended flowering period is a goal of many gardeners, but the molecular regulators of continuous flowering are still not very clear (Iwata et al., 2012).
In OF plants, especially in model plant Arabidopsis, six major pathways are known to control flowering time: ambient temperature, photoperiod, gibberellin, vernalization, age, and autonomous pathways (Taylor et al., 2018). Signals from the six pathways converge into several important floral integrator genes, FLOWERING LOCUS T (FT), TERMINAL FLOWER 1 (TFL1), SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1), APETALA1 (AP1), and LEAFY (LFY) (Shannon and Meeks-Wagner, 1991; Han et al., 2018; Jung et al., 2021). TFL1 and FT belong to the phosphatidylethanolamine-binding protein (PEBP) family and are key integrators of the floral transition pathways (Shannon et al., 1991). They have highly homologous sequences but act in an antagonistic manner in many species (Carmona et al., 2007; Jeon et al., 2011). TFL1, a flowering repressor, binds to FD to determine the floral transition by inhibiting the expression of LFY and AP1, which are downstream of the FT gene (Wang et al., 2021). As a downstream of FT, SOC1 is a flowering activator and can upregulate floral meristem identity genes AP1 and LFY to regulate flowering (Zhang et al., 2019).
Compared with OF plants, numerous studies on the molecular mechanism of continuous flowering have been conducted and several significant findings have been published. In rose, the CF habit is controlled by a monogenic recessive locus, and TFL1 homologues are good candidate genes (Semeniuk, 1971; Hurst, 1941). By studying seven CF roses with distinct origins, a 9-kbp-long retrotransposon was discovered in the second intron of RoKSN, which might result in continuous flowering (Kumar and Bennetzen, 1999; Cairns, 2000). Epigenetic modifications and retrotransposon insertions are also thought to be concerned with the CF behavior of rose (Bai et al., 2021; Soufflet-Freslon et al., 2021). In strawberries, the continuous flowering habit occurs in both diploid strawberries (woodland strawberry) and octoploid strawberry (garden strawberry). According to Darrow (1966), a 2-bp deletion in FvTFL1 might be a putative causal mutation for the perpetual flowering trait in woodland strawberry (Darrow, 1966; Koskela et al., 2012). In garden strawberries, a single dominant gene termed Perpetual Flowering and Runnering (PFRU) might cause continuous flowering, together with reducing the number of runners (Ahmadi et al., 1990; Gaston et al., 2013). However, this locus is not an ortholog of that containing the FvTFL1 gene in woodland strawberries, indicating the distinct genes in woodland and garden strawberries contributing to the CF phenotype. However, the specific genes of PFRU still remain unknown (Lewers et al., 2019; Hytönen and Kurokura, 2020). Recent studies have found that Arundina graminifolia, an orchid specie, gives continuous flowering throughout the year (Ahmad et al., 2021). Many essential flowering-related genes have been discovered recently, and a model of regulating continuous flowering has been developed (Ahmad et al., 2021; Li et al., 2021; Ahmad et al., 2022). The model summarizes multiple regulatory pathways, including vernalization, circadian clock, photoperiod, autonomous, and GA pathways. The above studies on continuous flowering can be adopted as a basis for future research on other plants, but the molecular mechanism of continuous flowering remains largely unclear.
Sweet osmanthus (Osmanthus fragrans Lour.), a member of the Oleaceae family, is cultivated as a famous ornamental plant in China (Wang et al., 2022). According to flowering characteristics, sweet osmanthus is divided into OF habit, such as ‘Yanhonggui’, and CF habit, like ‘Sijigui’ (Fu et al., 2020). In contrast to OF habit, which only flowers in autumn, CF habit can bloom continuously under favorable environmental conditions (Fu et al., 2020). The obvious flowering differences between OF and CF sweet osmanthus offer excellent materials for studying the regulatory mechanism of continuous flowering. According to Yang (2021), OfAP1 and OfTFL1 are the continuous flowering candidate genes in sweet osmanthus, whereas the genetic and molecular basis remains a mystery (Yang et al., 2021). Therefore, the current study analyzes transcriptome data to identify the important genes involved in the continuous flowering of sweet osmanthus. Specific results in this work will provide a guidance for future investigations into the genetic control of continuous flowering in plants.
Materials and methods
Plant materials and growth conditions
O. fragrans ‘Sijigui’ and ‘Yanhonggui’ used in this study were cultivated in a Pingshan tree nursery (Hangzhou, China, N30°15′14′′N, 119°43′39′′E). The trees used for experiment flowered normally for several years and showed similar growth under natural conditions. For ‘Sijigui’, undifferentiation buds (S0) and differentiation buds (mix of inflorescence primordium differentiation stage and small floret primordium differentiation stage, S1) in winter (flowering in winter) and early summer (blooming in autumn) were collected, and for ‘Yanhonggui’, buds with S0 and S1 stages in early summer (blooming in autumn) were collected, respectively. The collected buds from ‘Sijigui’ and ‘Yanhonggui’ were used for transcriptome sequencing and expression analysis. All materials were saved in liquid nitrogen and immediately refrigerated at -80°C prior. The methods to culture the callus from ‘Sijigui’ were described in a previous research (Zhu et al., 2022). Seeds of Arabidopsis (Col-0) were maintained in our laboratory and sown on Murashige and Skoog medium to screen transgenic plants. The seedlings were transferred to a light incubator under a 16-h light/8-h dark photoperiod at 22°C.
Phenotypic observation
To understand the flowering time and phenotypic changes of sweet osmanthus, the developmental stages of buds in 1 year were observed by stereo microscope and paraffin sections (from 15 April 2020 to 1 April 2021). A total of 10 buds from the three plants were collected every 15 days and stored in FAA fixative (3:1 ratio of 10% ethanol: glacial acetic acid) for paraffin section observation. The method of paraffin section referred to Fan et al. 2018. Samples were sliced to a 10-µm thickness, and an Axio Imager A2 positive fluorescence microscope (Carl Zeiss, Oberkochen, Germany) was used to observe and photograph the development stages of buds. At the same time, the buds were also observed through a microscope.
Illumina sequencing
For ‘Sijigui’, buds at S0 and S1 stages in winter were collected on November 15 and December 1, respectively (referring to the third flowering process in Figure 2), and the S0 and S1 buds in early summer were collected on June 1 and June 15, respectively (referring to the first flowering process in Figure 2). For ‘Yanhonggui’, buds at S0 and S1 stages in early summer were also collected on June 1 and June 15, respectively (referring to the flowering process in Figure 1A). RNA from the six tissues was extracted. Prokaryotic mRNA was enriched by eliminating rRNA using the Ribo-Zero™ Magnetic Kit, whereas eukaryotic mRNA was enriched with Oligo(dT) beads (Epicentre). With the use of random primers and fragmentation buffer, the enriched mRNA was reverse transcribed into cDNA. DNA polymerase I, RNase H, dNTP, and buffer were used to create second-strand cDNA. The cDNA fragments were then end repaired and poly(A) added. Illumina sequencing adapters were ligated after they had been purified using a QIAquick PCR extraction kit. Gene Denovo Biotechnology Co. used agarose gel electrophoresis to size-select the ligation products before PCR amplification and Illumina HiSeq™ 4000 sequencing (Guangzhou, China).
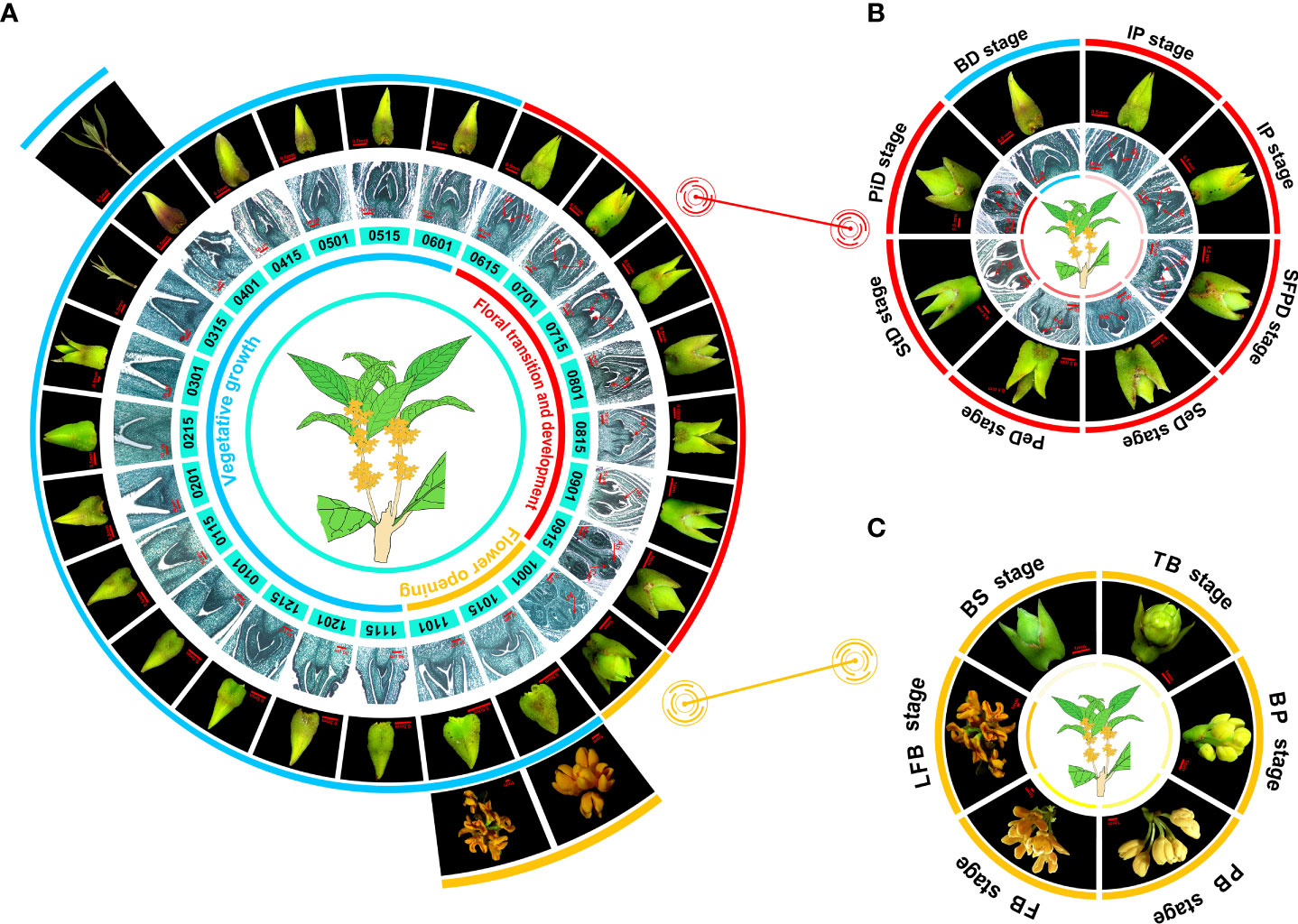
Figure 1 The flowering phenology shifts in O. fragrans ‘Yanhonggui’ for 1 year. (A) The phenotypic changes of ‘Yanhonggui’ in 1 year. Different colored lines represent different stages of development: blue represents vegetative growth; red means floral transition and development; orange represents flower opening. The numbers below the picture represent dates. For example, 0115 is January 15. (B) The floral transition and development process. BD: bud pre-differentiation stage; IP: inflorescence primordium differentiation stage; SFPD: small floret primordium differentiation stage; SeD: sepal differentiation stage; PeD: petal differentiation stage; StD: stamen differentiation stage; PiD: pistil differentiation stage. Due to the obvious change in phenotype, the IP stage was repeated. (C) The flower opening process. BS, ball-shaped stage; TB, top bracts stage; BP, bud pedicel stage; PB, primary blooming stage; FB, full blooming stage; LFB, late full blooming stage. Br, Bract; Ip, inflorescence; FP, floret primordium; Se, sepa primordium; Pe, petal primordium; St, stamen primordium; An, anther; Car, carpel.
Analysis of differentially expressed genes
To identify differentially expressed genes across samples or groups, the edgeR package (http://www.r-project.org/) was used. We identified genes with a fold change ≥2 and a false discovery rate (FDR) of <0.05 in a comparison as significant differentially expressed genes (DEGs). DEGs were then subjected to enrichment analysis of GO functions and KEGG pathways.
GO enrichment analysis
Gene Ontology enrichment analysis provided all GO terms that were significantly enriched in DEGs compared with the genome background and filtered the DEGs that correspond to biological functions. Firstly, all DEGs were mapped to GO terms in the Gene Ontology database (http://www.geneontology.org/). Then, gene numbers were calculated for every term, and significantly enriched GO terms in DEGs compared with the genome background were defined by the hypergeometric test. The estimated p-value underwent FDR correction with a threshold of ≤0.05. GO terms that fit these criteria were deemed to be considerably enriched in DEGs.
Pathway enrichment analysis
KEGG pathway enrichment analysis, comparing DEGs with the backdrop of the entire genome, revealed highly enhanced metabolic or signal transduction pathways. The calculation method is similar to that used in GO analysis above. The estimated p-value underwent FDR correction with a threshold of FDR ≤0.05. Significantly enriched pathways in DEGs were identified as those that met this requirement.
Quantitative real-time PCR analysis
Specific genes were chosen for expression analysis through qRT-PCR. The buds from the middle and upper regions of the plants were collected on the 1st and 15th of every month in 1 year (total 24 different samples). Since we paid more attention to the new shoot growth and floral transition process, buds from April 15 to June 15 were collected in the first flowering process (Figure 2). From July 1 to November 1, buds were collected in the second flowering process. From November 15 to January 1, buds were collected in the third flowering process, and buds from January 15 to April 1 were collected in the fourth flowering process. Total RNA from 24 samples during a year was extracted by the RNAprep Pure Plant Kit (Vazyme, Nanjing, China). cDNA was obtained using HiScript® III Reverse Transcriptase (Vazyme, Nanjing, China). qRT-PCR was performed by the LightCycler 480 qRT-PCR System II (Roche, Basel, Switzerland): SYBR Premix Ex Taq™ 10 µl, up- and downstream primer (10 μmol/l) 0.8 μl, cDNA 2 μl, ddH2O 6.4 μl, total 20 µl. Three biological replicates were performed. OfACT and AtACTIN2 (At3g18780) were the reference genes for sweet osmanthus and Arabidopsis, respectively.

Figure 2 The flowering phenology shifts in O. fragrans ‘Sijigui’ for 1 year. First to fourth flowerings respectively represent different flowering processes in 1 year. The numbers below the picture represent dates. For example, 0115 is January 15. Br, bract; Ip, inflorescence; FP: floret primordium; Se, sepa primordium; Pe, petal primordium; St, stamen primordium; An, anther; Car, carpel.
Transformation in ‘Sijigui’ callus and Arabidopsis
OfFT and OfBFT were inserted into the pORER4-35S-GFP vector with restriction enzyme cutting sites of NheI and XhoI (Supplementary Table S1). The fusion vector was transformed into GV3101 through a heat shock method. Then, the infection solution was configured to OD600 = 0.4–0.6 with the buffer solutions (10 mM MES, 10 mM MgCl2, and 0.1 mM AS) and kept standing at room temperature for 3 h. The callus of O. fragrans was added to the infection solution and cultured at 28°C for 0.5 h under dark conditions. The callus was washed for four times with ddH2O and cultured in symbiotic mediums with 3 days under dark conditions. After that, the callus was saved in a -80°C refrigerator for further analysis. The constructed vectors were also introduced into Arabidopsis Col-0. The MS medium containing 50 mg/l kanamycin was used to screen the transgenic plants. We obtained 18 and 15 independent transgenic lines of OfFT and OfBFT, respectively, and homozygous transgenic lines from T3 were used for further research. For expression analysis, the entire shoots of T3 homozygous transgenic lines of OfFT and OfBFT were collected at 14 and 20 days, respectively, after planting and subjection to qRT-PCR analysis.
Yeast one-hybrid assay
Fragments of the OfFT and OfBFT promoter were cloned into the pHis2 vector, and pHis2-proOfFT/OfBFT were transformed into Y187 to screen the suitable resistance concentrations of 3AT. The suitable concentrations were used to screen ‘Sijigui’ cDNA library and interaction validation. A yeast one-hybrid system was performed using the Matchmaker™ Gold Yeast Two-Hybrid System from Clontech as recommended by the manufacturer.
Dual-Glo® luciferase assay
The interaction between OfSPL8 protein and the promoter of OfFT and OfBFT was verified by the Dual-Glo® Luciferase Assay. OfSPL8 was cloned into the pORER4-35S-GFP vector, and the promoters of OfFT and OfBFT were cloned into pGreen0800II-LUC. All vectors were transformed into GV3101 (pSoup-p19). Then, 35S::OfSPL8 and proOfFT-LUC were coinjected into the leaves of tobacco, together with 35S::OfSPL8 and proOfBFT-LUC. The activities of fluorescein enzymes were measured after 3 days. The activities of firefly luciferase (LUC) and renilla luciferase (REN) were measured using the GloMax® multifunctional instrument (Promega, USA).
Results
Flowering phenology shifts of OF habit in sweet osmanthus
To disclose the OF habit flowering characteristics in sweet osmanthus, O. fragrans ‘Yanhonggui’ was selected and buds at the upper half of a branch were continuously taken for follow-up observation. The microscope and paraffin sections were used, and the observation continued throughout the year. We found that the annual growth cycle of ‘Yanhonggui’ could be divided into three periods: vegetative growth, floral transition and development, and flower opening (Figure 1A). After the flowers had faded in the autumn, vegetative growth commenced and lasted until the buds entered the floral transition in the next early June (early summer). Additionally, it was discovered that during vegetative development, buds grew slowly with no significant changes in phenotype during the whole winter but quickly generated new shoots in the spring. The buds in new shoots would continue to grow in spring and enter floral transition in early summer and bloom in autumn (Figure 1A). According to paraffin sections, the floral transition and development period occurred from June to September. The whole period was further divided into seven stages: bud pre-differentiation stage (BD), inflorescence primordium differentiation stage (IP), small floret primordium differentiation stage (SFPD), sepal differentiation stage (SeD), petal differentiation stage (PeD), stamen differentiation stage (StD), and pistil differentiation stage (PiD). Due to the obvious change in phenotype, the IP stage was repeated (Figure 1B). Based on the phenotypic of the buds, it suggested that during the floral development process, the apical bracts of the buds were dehisced gradually, together with the expansion of the whole buds. Interestingly, although the buds had finished the floral development process (having well-developed pollen grains), they could not bloom immediately until when the ambient temperature was cooler in autumn. The blooming time of ‘Yanhonggui’ is not fixed each year because of the variable cooling time in the fall. Our previous research revealed that ambient temperature had significant effects on flower opening in ‘Yanhonggui’ (Fu et al., 2020). Therefore, we believed that the actual blooming time in ‘Yanhonggui’ depends more on the flower opening than the floral transition. Similar to the floral transition and development period, the flower opening period, based on phenotypic differences, was also made up of different stages. Here, we presented the six stages with the most significant phenotypic changes to introduce the changes of phenotype: ball-shaped stage (BS), top bracts stage (TB), bud pedicel stage (BP), primary blooming stage (PB), full blooming stage (FB), and late full blooming stage (LFB) (Figure 1C).
Flowering phenology shifts of CF habit in sweet osmanthus
Compared with ‘Yanhonggui’, although ‘Sijigui’ had a similar floral transition and development process in early summer and flower opening process in autumn, it could bloom constantly from October to the following June. According to the observations using microscope and paraffin sections, new shoot growth and floral development occurred concurrently and mainly had four times in 1 year, with the most vigorous flowering in autumn (Figure 2). Paraffin sections revealed that the main floral transition events lasted about 2.5 months (first flowering), 1.5 months (second flowering), 1 month (third flowering), and 1.5 months (fourth flowering), respectively. The floral transition processes began in early June, mid-September, early December, and early February and ended in late August, early November, early January, and mid-March, respectively. In addition to the differences in the time required for flower bud differentiation, ‘Sijigui’ had discrepancies in the structure of inflorescences in different seasons. It produced cymose inflorescence in autumn and racemose inflorescence in winter and spring, which had a pedicure at the base of flowers (Figure 2). The results suggested that compared with autumn, the molecular mechanism of flowering regulation might have a difference in winter and spring in ‘Sijigui’.
We also discovered that numerous buds generated in early July were unable to undergo floral transition throughout the whole summer until the cooler autumn (Figure 2). Ambient temperature had a vital role in flowering regulation in sweet osmanthus (Zhu et al., 2019). We hypothesized that ambient temperature might be involved in continuous flowering regulation of CF habit, and high temperatures in the summer impeded floral transition and led to few or no blooms of “Sijigui” in the summer.
Illumina sequencing and analysis of DEGs
To further study the possible regulatory mechanism of continuous flowering in sweet osmanthus, we collected three biological replicate samples of S0 and S1 from ‘Sijigui’ in winter and early summer and S0 and S1 from ‘Yanhonggui’ in early summer for transcriptome sequencing (Figure 3A). It generated about 966.8 million reads from six development stages. Each sample produced about 6.2–9.6 Gb clean data with an average of 26.2 million clean reads (Supplementary Table S2). We filtered 94,529 differentially expressed unigenes (DEGs), which were annotated using NR, SWISSPORT, KOG, and KEGG databases. The covering annotation data were 48.6%, 31.86%, 27.30%, and 21.23%, respectively (Supplementary Table S3).
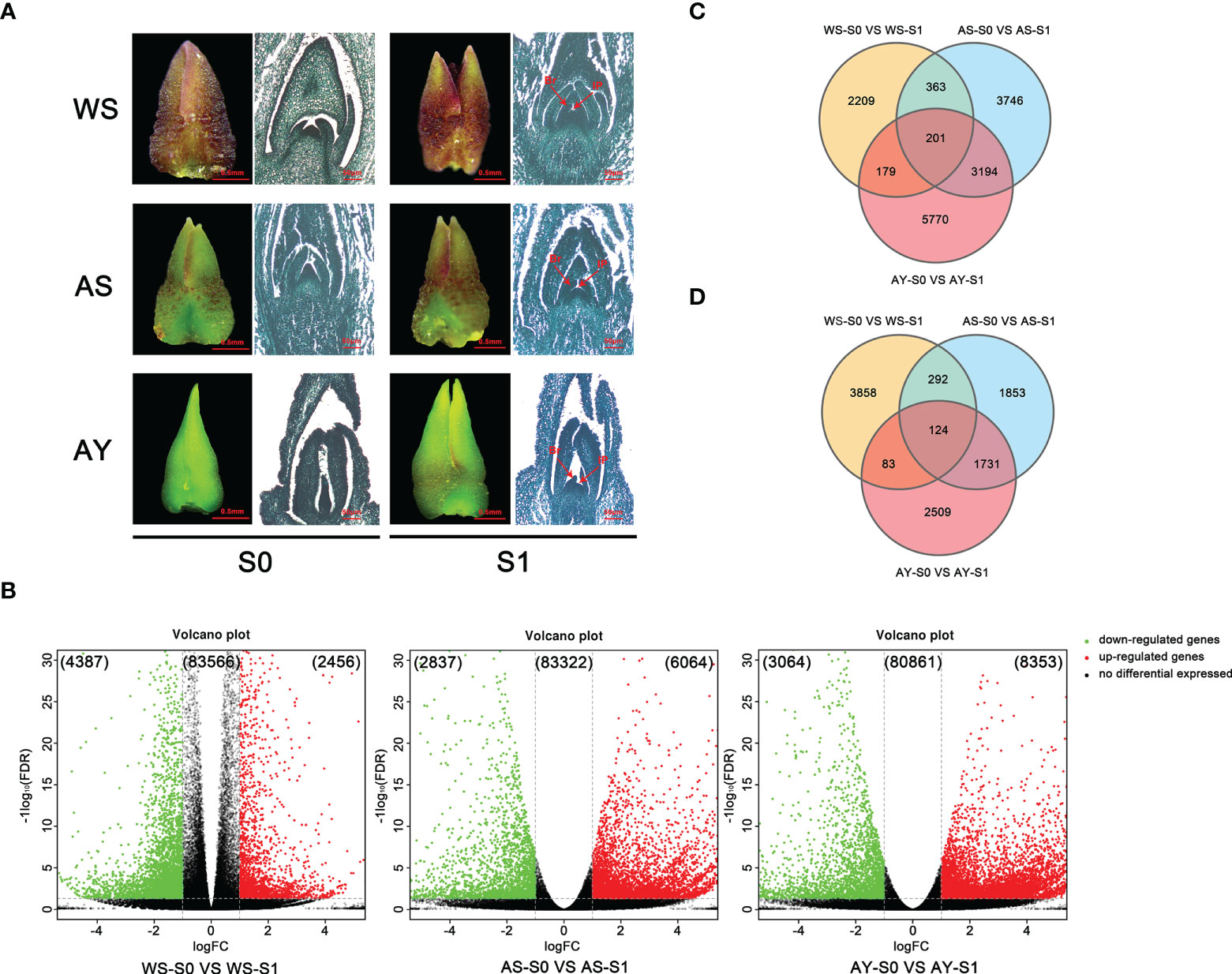
Figure 3 Comparative analysis of DEGs during the floral transition process in OF and CF habits. (A) Samples for transcriptome sequencing analysis. WS: buds blooming in winter from ‘Sijigui’; AS: buds blooming in autumn from ‘Sijigui’; AY: buds blooming in autumn from ‘Yanhonggui’. S0: undifferentiation buds; S1: differentiation buds (mix of inflorescence primordium differentiation stage and small floret primordium differentiation stage). Br: bract; Ip: inflorescence; FP: floret primordium. (B) The expression trends of DEGs in the three comparisons. Green circles: downregulated genes; red circles: upregulated genes; black circles: non-differentially expressed genes. (C) Venn diagram of the number of upregulated genes among the three comparisons. (D) Venn diagram of the number of downregulated genes among the three comparisons.
Detailed comparative analysis of the DEGs was performed in three comparisons, namely, WS-S0 vs. WS-S1, AS-S0 vs. AS-S1, and AY-S0 vs. AY-S1 (P value ≤0.01 and |log2Ratio| ≥ 1). We found that 4,387 and 2,456 unigenes were down- or upregulated, respectively, in the WS-S0 vs. WS-S1 comparison, whereas the unigenes down- or upregulated in the AS-S0 vs. AS-S1 combination were 2,837 and 6,064, respectively. Meanwhile, 3,064 and 8,353 unigenes in the AY-S0 vs. AY-S1 comparison were down- or upregulated, respectively (Figure 3B). In summary, upregulated unigenes were more than downregulated unigenes during the three comparisons. In addition, only 201 DEGs were all upregulated in three comparisons, and 2,209, 3,746, and 5,570 DEGs were upregulated in the WS-S0 vs. WS-S1, AS-S0 vs. AS-S1, and AY-S0 vs. AY-S1 comparisons, respectively (Figure 3C). Simultaneously, 124 DEGs were downregulated in three comparisons, and 3,858, 1,853, and 2,509 DEGs in the WS-S0 vs. WS-S1, AS-S0 vs. AS-S1, and AY-S0 vs. AY-S1 comparisons were downregulated, respectively (Figure 3D).
GO enrichment and KEGG pathway analysis of DEGs
In all three comparisons, ‘metabolic process’, ‘cellular process’, and ‘catalytic activity’ were the most significantly enriched GO terms in the biological process. In the cellular component, ‘cell’, ‘cell part’, and ‘membrane’ were highly enriched in the WS-S0 vs. WS-S1 comparison, whereas ‘cell’, ‘cell part’, and ‘organelle’ were significantly enriched in the other two comparisons. In the molecular function, ‘Catalytic activity’ and ‘Binding’ were obviously enriched among the three comparisons (Supplementary Figure S1). There were 20,065 unigenes mapped into 132 KEGG pathways, with 121, 128, and 131 KEGG metabolic pathways in the WS-S0 vs. WS-S1, AS-S0 vs. AS-S1, and AY-S0 vs. AY-S1 comparisons, respectively (Supplementary Table S4).
Identification of flowering-related genes
Our data identified 1,990 transcription factors (TFs) from 58 TF families and found some flowering-related families, including the ERF/AP2, C2H2, BHLH, NAC, MYB, WRKY, bZIP, COL, MADS and PEBP families (Supplementary Table S5). The expression patterns of the TFs from the above families were analyzed (Figure 4). In the ERF/AP2 family, three of them (ERF25, ERF110, ERF61) were significantly upregulated in comparison AY-S0 vs. AY-S1(C3), and ERF20 was downregulated in the C3 comparison, whereas the expression of two genes (AP2-1, AP2-7) was decreased in the AS-S0 vs. AS-S1(C2) and C3 comparisons, accompanied by induction of two genes (AIL1, ANT). Six genes from the C2H2 family were obviously expressed. Three genes (SteA, RPN4, C2H2-7) were increased in C2 and C3 comparisons, and one gene (ZAT5) was decreased. Two genes (C2H2-32, C2H2-45) were just upregulated in the C3 comparison. In the BHLH family, the expressions of BHLH35, BHLH13, and BHLH6 were promoted in the WS-S0 vs. WS-S1(C1) comparison and inhibited in the C2 and C3 comparisons. The accumulation of BHLH90 was increased in the C2 and C3 comparisons. Only two genes in the NAC family had a significant expression, in which NAC56 was inhibited and NAC83 was promoted respectively in the C2 and C3 comparisons, whereas in the MYB family, two genes (MYB113 and MYB12) were all downregulated in the C2 and C3 comparisons. It was found that in the WRKY family, WRKY28 was upregulated in all three comparisons and WRKY6 was upregulated in the C2 and C3 comparisons. WRKY14 and WRKY72 were accumulated in the C1 comparison, and WRKY72 showed a significant reduction in the C2 and C3 comparisons. In the B-ZIP family, ATHB12 and ATHB52 exhibited opposite expression patterns in both C2 and C3 comparisons. ATHB12 was increased in the C2 and C3 comparisons and ATHB52 was decreased. Eleven genes in the MADS family had an obvious expression, and seven of them (AP1, DEFA, SEP3, GLO, AG-1, MADS6, SEP1) were upregulated in three comparisons. AG-2 and MADS1 expressions were increased in the C2 and C3 comparisons, whereas AGL8 was decreased in the same comparisons. AGL12 was only upregulated in the C3 comparison. In the COL family, the expression of COL14 was promoted in three comparisons. COL5 was increased in the C2 comparison and decreased in the C1 comparison. Four genes with a significant expression in the PEBP family were found, and two genes (FT and MFT) were upregulated in all three comparisons, especially in the C2 and C3 comparisons. BFT has a specific high expression in the C1 comparison. TFL1 was downregulated in three comparisons. In addition to the above families, we identified an LFY homologous gene, which was highly expressed in C2 and C3 comparisons, without obvious change in the C1 comparison.

Figure 4 Comparative analysis of DEGs during the floral transition process in OF and CF habits. Red means the upregulation, and green means the downregulation. C1, C2, and C3 represent WS-S0 vs. WS-S1, AS-S0 vs. AS-S1, and AY-S0 vs. AY-S1 comparisons.
Expression patterns of PEBP family genes correlated with CF habit in ‘Sijigui’
Previous studies indicated that several genes from the PEBP family had important roles in CF habit, especially the homologous genes of TFL1 and FT (Koskela et al., 2012; Ahmad et al., 2022). Here, we focused on the homologous genes of FT (Unigene0030305, OfFT), TFL1 (Unigene0041633, OfTFL1), and BFT (Unigene24810, OfBFT), which were significantly expressed in three comparisons, whereas there was no difference in the DNA sequence of the three genes in OF and CF habits (Supplementary Figure S2A). We further analyzed their expression patterns during 1 year in ‘Sijigui’ (Figure 5A). It showed that the three genes had distinct expression characteristics. They all had four different expression peaks during a year, and the expression patterns of OfTFL1 and OfBFT were similar but opposite to OfFT. Moreover, referring to the flowering phenology shifts of ‘Sijigui’, each new shoot growth was accompanied by a high expression of OfTFL1 and OfBFT and a low expression of OfFT, whereas OfFT was upregulated during the floral transition processes, with downregulation of OfTFL1 and OfBFT. The results suggested that in ‘Sijigui’, both OfTFL1 and OfBFT might play important roles in new shoot growth, and OfFT might be involved in the floral transition process. Amino acid sequence alignment showed that OfTFL1 contained two conservative residues (His84 and Asp139), which could distinguish TFL1 homologous genes from FT homologous (Figure 5B). Moreover, OfBFT had the same conservative residues as with OfTFL1. Phylogenetic analysis suggested that OfTFL1, OfBFT, and OfFT were closely related to the TFL1 subfamily, BFT subfamily, and FT subfamily, respectively (Supplementary Figure S2B).
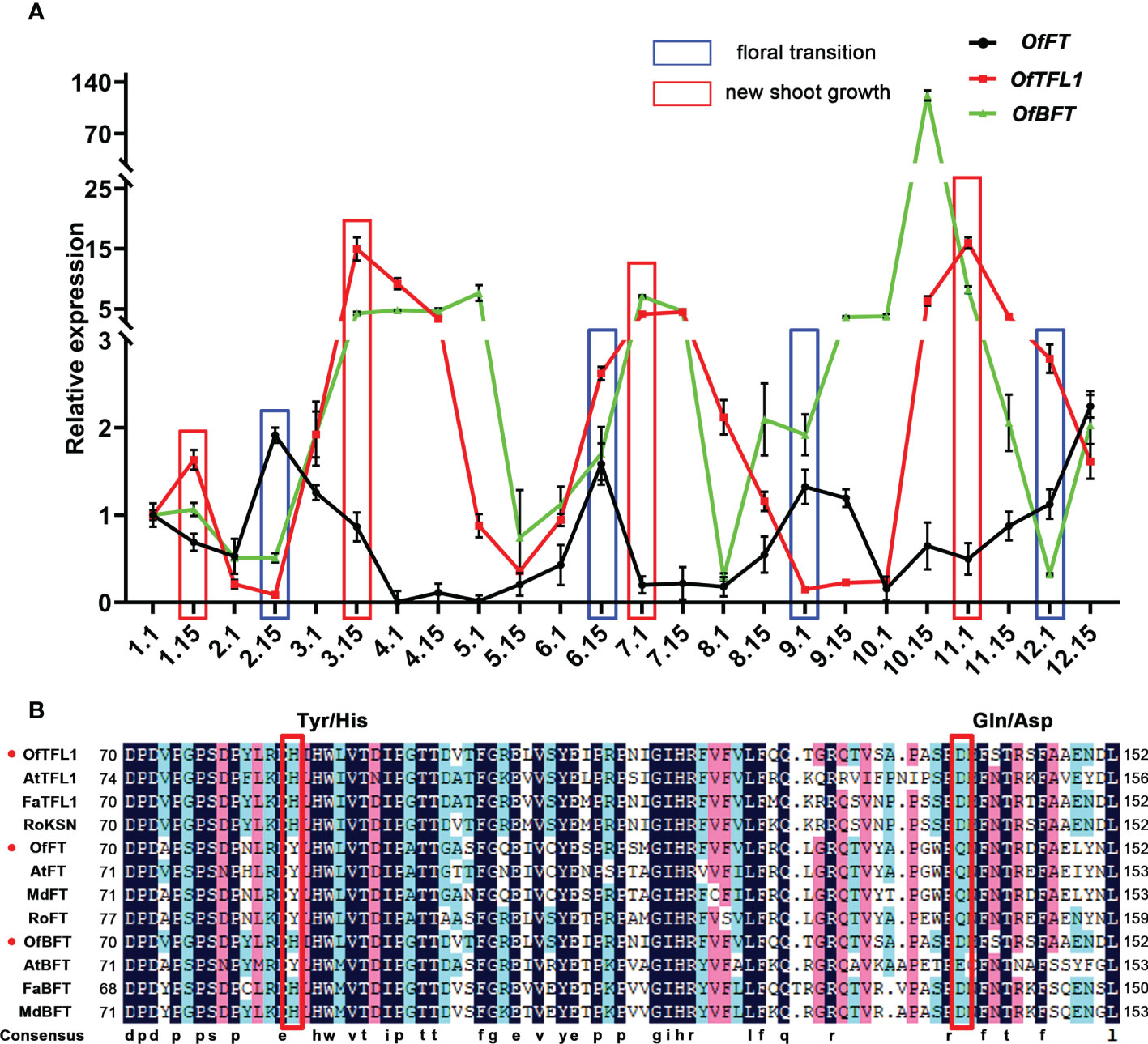
Figure 5 Isolation and expression analysis of OfFT, OfBFT, and OfTFL1 in OF and CF habits. (A) Expression patterns of OfFT, OfBFT, and OfTFL1 in 1 year. Red boxes represent the time new shoots occur. Blue boxes represent the time of floral transition. Means ±SD, n = 3. (B) Multiple amino acid sequence alignment of TFL1 and FT proteins in different species. Red box indicates Tyr/His and Gln/Asp that differed between TFL1 and FT proteins.
OfFT functioned as a flowering promoting gene in ‘Sijigui’ and Arabidopsis
To elucidate the biological function of OfFT, we overexpressed it in the callus of ‘Sijigui’, driven with the cauliflower mosaic virus (CaMV) 35S promoter, and measured the expression levels of OfFT and some flowering-related genes. It showed that compared with WT, OfFT was highly expressed in the callus (Figure 6A). According to previous studies, FT could promote flowering through upregulating the expression of AP1 and LFY (Carmona et al., 2007; Jeon et al., 2011). Here, the accumulation of OfAP1 and OfLFY in OfFT-overexpressed callus was also detected, and it showed that both the genes were induced (Figure 6A). We also verified the role of OfFT in flowering regulation in wild-type Arabidopsis (Col-0). Homozygous lines of the T3 generation were chosen for the phenotypic observation. The transgenic plants displayed early flowering phenotypes with less rosette leaves than wild-type plants under long-day conditions (Figure 6B). The entire shoots of T3 homozygous transgenic lines of OfFT were collected at 14 days after planting, and the expression levels of AtFT, AtAP1 and AtLFY in transgenic plants were investigated. Compared with Col-0, it indicated that the expressions of AtAP1 and AtLFY were increased in transgenic plants, whereas the expression levels of AtFT had no significant change (Figure 6C, Supplementary Figure S3A). All results showed that OfFT functioned as a positive regulator of flowering by inducing the expression of flowering-related genes.
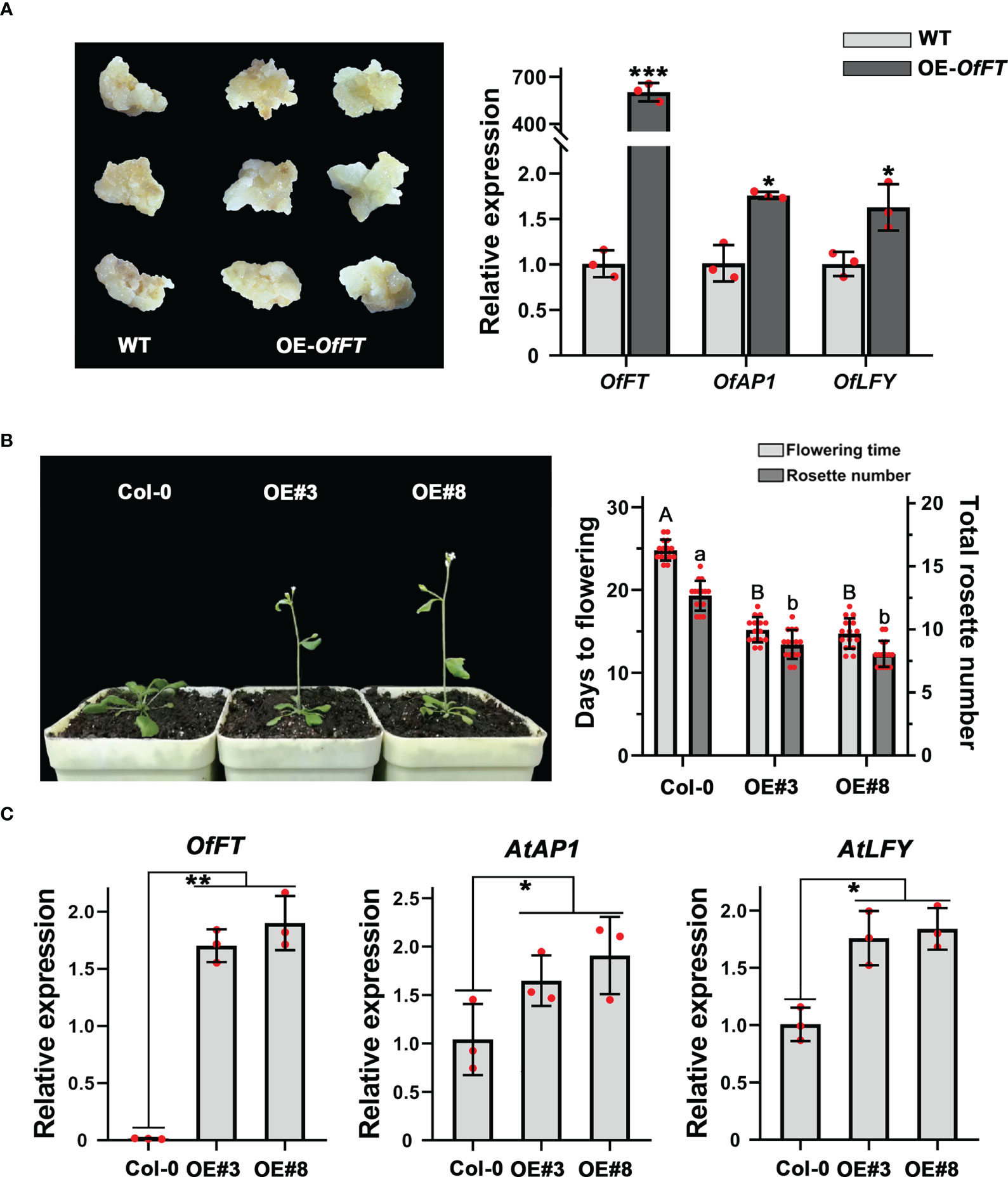
Figure 6 OfFT functioned as a flowering activator in both O. fragrans and Arabidopsis. (A) The expression levels of flowering-related genes in overexpression ‘Sijigui’ callus. Data were the means ±SD, n = 3, Student’s t test, *p < 0.05 and ***p < 0.001. (B) Phenotypic analyses of transgenic Arabidopsis plants overexpressing OfFT. Uppercase letters represent significance of flowering time compared with mock-treated control. Lowercase letters represent significance of rosette number compared with mock-treated control. Means ± SD, n = 3, *p < 0.05, **p < 0.01. For Figure 6: (C) The expression levels of flowering-related genes in transgenic plants. Means ±SD, n = 3. *p < 0.05, **p < 0.01.
OfBFT functioned as a flowering suppressor in ‘Sijigui’ and Arabidopsis
To reveal the role of OfBFT in flowering regulation, we also overexpressed it in the callus of ‘Sijigui’ and Arabidopsis and performed qRT-PCR to confirm the transformation efficiency. It showed that OfBFT was effectively accumulated in the callus. In flowering time control of Arabidopsis, BFT is functionally redundant with TFL1, which determines the floral transition by inhibiting the expression of LFY and AP1 (Ryu et al., 2014). In this study, overexpression of OfBFT resulted in the downregulated expression of OfAP1 and OfLFY (Figure 7A). In Arabidopsis, the ectopic expression of OfBFT affected the reproductive development process with a late-flowering phenotype and more rosette leaves under long-day conditions (Figure 7B). We then also studied the expression levels of the vital flowering-related genes AtBFT, AtAP1, and AtLFY. The entire shoots of T3 transgenic lines were collected at 20 days after planting, and the mRNA levels were analyzed. Compared with Col-0, it demonstrated that the expression of AtBFT was not obviously changed, whereas AtAP1 and AtLFY were significantly decreased in transgenic plants (Figure 7C, Supplementary Figure S3B). All results showed that OfBFT was a flowering suppressor by reducing the accumulation of flowering activating genes.
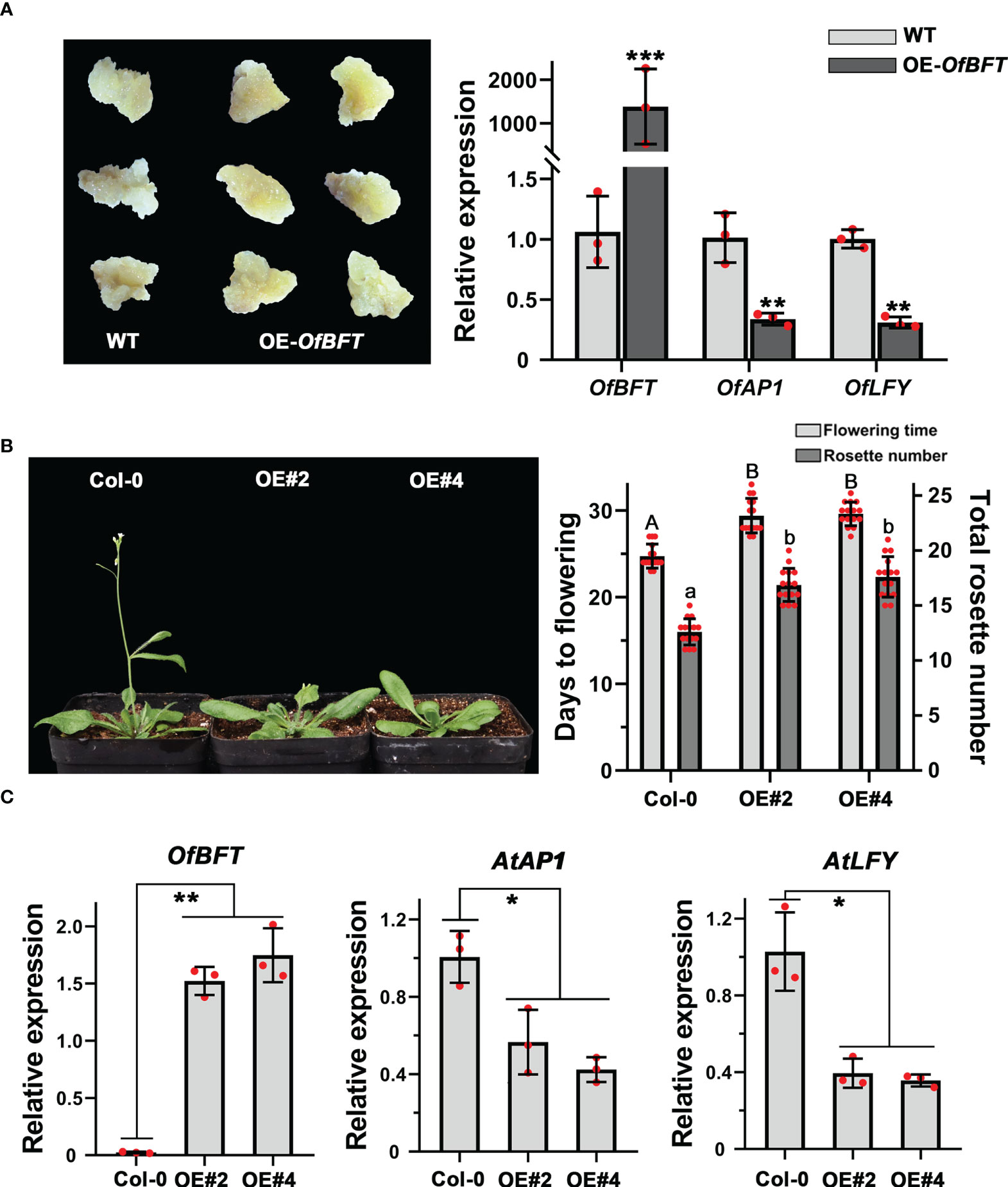
Figure 7 OfBFT functioned as a flowering suppressor in both O. fragrans and Arabidopsis. (A) The expression levels of flowering-related genes in overexpression ‘Sijigui’ callus. Data were the means ±SD, n = 3, Student’s t test, **p < 0.01 and ***p < 0.001. (B) Phenotypic analyses of transgenic Arabidopsis plants overexpressing OfBFT. Uppercase letters represent significance of flowering time compared with mock-treated control. Lowercase letters represent significance of rosette number compared with mock-treated control. Means ± SD, n = 3, *p < 0.05, **p < 0.01. (C) The expression levels of flowering-related genes in transgenicplants. Means ±SD, n = 3. *p < 0.05, **p < 0.01.
OfSPL8 could bind to the promoter of OfFT and OfBFT to regulate their expression
A yeast one-hybrid assay was performed to screen the potential upstream transcription factors through the promoter of OfFT and OfBFT, respectively. The ‘Sijigui’ cDNA library was built previously in our lab. Interestingly, we found that OfSPL8 was a common upstream transcription factor of OfFT and OfBFT. Because of the existence of false positives, the binding among OfSPL8, proOfFT, and proOfBFT was investigated through yeast one-hybrid and dual-luciferase (dual-LUC) assay. It showed that both the cells harboring OfSPL8 in the pGADT7 recombinant vector and proFT or proBFT in the pHis2 recombinant vector were grown normally on TDO (20 mM 3-AT) solid medium (Figure 8A). The results of the dual-LUC assay suggested that OfSPL8 could significantly induce the transcriptional activity of the OfFT promoter (Figure 8B) and obviously inhibit the activity of the OfBFT promoter (Figure 8C). These results revealed that OfSPL8 could bind to the promoter of OfFT and OfBFT to regulate their expression in ‘Sijigui’.
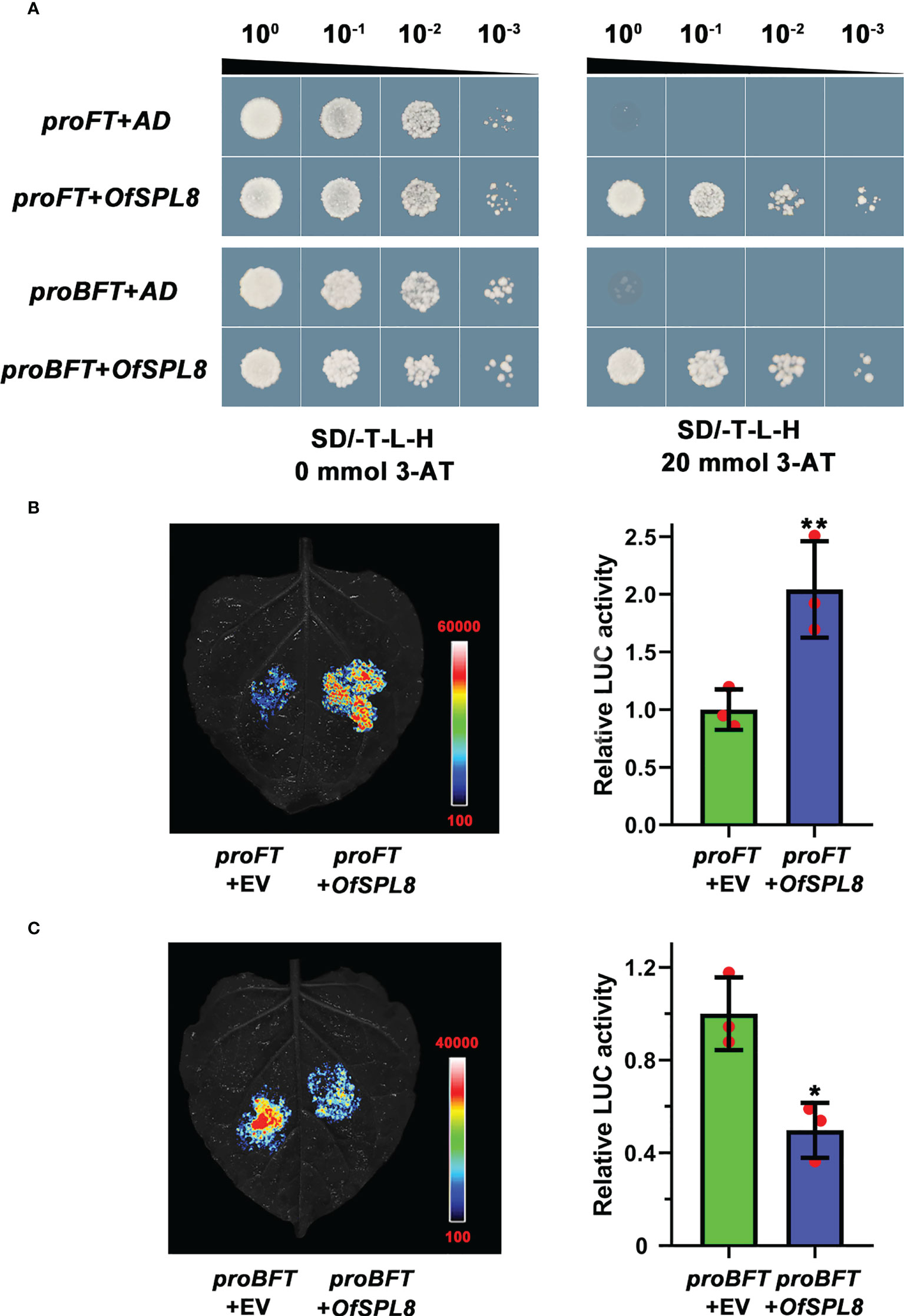
Figure 8 The binding between OfSPL8, proOfFT, and proOfTFL1. (A) Yeast one-hybrid analysis of proOfFT, proOfBFT, and OfSPL8. (B) Dual-LUC analysis of proOfFT and OfSPL8. Student’s t test means ± SD, n = 3, **p < 0.01. (C) Dual-LUC analysis of proOfBFT and OfSPL8. Student’s t test means ±SD, n = 3, *p < 0.05.
Discussion
Flowering, transitioning from vegetative growth to reproductive growth, is an important life activity for plants (Chen D. et al., 2018; Chen Z. et al., 2018). Most plants flower once a year, and only a few plants bloom in multiple seasons, such as Osmanthus fragrans var. ‘Sijigui’, Jasminum sambac, Fuchsia hybrida, Rosa chinensis, and Citrus limon (Yang et al., 2021). Continuous flowering (CF) is one of the most important horticultural traits for ornamental plants, although there is only limited scientific understanding about this phenomenon. Based on current studies, not only flowering habit but also some important development processes have differences between OF and CF habits, including the length of juvenile stage and inflorescence architecture (Iwata et al., 2012).
In rose, the primary shoots of OF plants remained to have indeterminate growth and only flowered on axillary secondary shoots (Vries, 1976), whereas in CF roses, plants had a short juvenile stage and bloomed rapidly after seed germination. Moreover, all shoots had determinate growth with a terminal inflorescence in CF roses (Vries, 1976). In OF woodland strawberry, the terminal meristem differentiated into an inflorescence, and the axillary meristems maintained vegetative development, which might develop into runners (Koskela et al., 2018), whereas in CF habit, runner growth was inhibited and the axillary meristems could differentiate into inflorescences under favorable conditions (Koskela et al., 2018). A CF mutant, super long blooming1 (slb1), was found in Liriodendron chinense (Sheng et al., 2021). The slb1 mutant had a prolonged flowering phase and performed floral transition and flowering at the same season (Sheng et al., 2021). In OF habit of sweet osmanthus, flower buds differentiated in summer and bloomed in autumn. After flowering, some vegetative buds grew slowly and then formed new shoots in the next spring (Figure 1). Although CF habit had the same flowering process in autumn as OF habit, it had continuity of vegetative growth and formed favorable buds for the following floral transition and flowering (Figure 2). Additionally, like CF roses, CF sweet osmanthus also had a terminal inflorescence on new shoots. According to the difference between the OF and CF sweet osmanthus, the ability to induce new shoot growth might be closely related to continuous flowering habit.
Many TF families are involved in floral transition and flowering time regulation (Chen D. et al., 2018). Here, we identified some TFs from several flowering-related families, including the ERF/AP2, C2H2, BHLH, NAC, MYB, WRKY, bZIP, COL, MADS, and PEBP families. The MADS, ERF/AP2, and C2H2 families were the top three families with the most differential expression TFs in our study (Figure 4). In flowering plants, MADS-box genes are master regulators of the floral transition process (Kennedy et al., 2018). We identified that 11 genes in the MADS family had an obvious expression among the three comparisons. AP1, DEFA, SEP3, GLO, AG-1, MADS6, and SEP1 were upregulated in all three comparisons, and AG-2 and MADS1 expressions were increased in the C2 and C3 comparisons, whereas AGL12 was only upregulated in the C3 comparison. In Arabidopsis, AtAP1 was a flowering activator and functioned as a downstream of the AtFT gene (Wang et al., 2021). AtSEPs and AtAG were members of the ABCDE genetic model, which participated in floral organ identity (Irish, 2017). In sweet cherries, PavSEP and PavAP1 proteins also interacted with PavSVP to regulate floral transition (Wang et al., 2021). The ERF/AP2 family is one of the largest TF families in the plant kingdom, which is widely involved in the plant development process in Arabidopsis, rice, and barley (Sharman, 1994; Mehrnia et al., 2013; Ren et al., 2013). In our study, we found that eight genes from the ERF/AP2 family (four in the ERF subfamily and four in the AP2 subfamily) had a significant expression (Figure 4). In rice, several genes from the C2H2 family participate in controlling flowering time (Wu et al., 2019), whereas six genes having different expression patterns were found to have obvious expression among three comparisons in sweet osmanthus. It indicated the different roles of genes in the flowering regulation of OF and CF habits. The BHLH family is the second largest transcription factor family in plants, and in Arabidopsis, BHLH48 and BHLH60 regulated flowering under long-day conditions through the GA pathway (Li et al., 2017). The expressions of BHLH35, BHLH13, and BHLH6 were just promoted in the C1 comparison and inhibited in the C2 and C3 comparisons, suggesting that the three genes might be involved in the floral transition process in winter of CF habit (Figure 4). In sweet cherry, woodland strawberry, and Arabidopsis, the NAC, MYB, and WRKY families played roles during the floral transition process (Villar et al., 2020; Lei et al., 2020). In our study, only two genes in both the NAC and MYB families had a significant expression, together with four genes in the WRKY family. In the COL family, the expression of COL14 was promoted in three comparisons, whereas COL5 was decreased in the C1 comparison. In CF habit rose, RcCOL4 and RcCOL5 were in response to different photoperiods to regulate continuous flowering (Lu et al., 2020). Moreover, a LFY homologous gene was highly expressed in C2 and C3 comparisons. Similar to AtAP1, AtLFY was also involved in flowering regulation and floral organ identity (Wang et al., 2021).
The PEBP family is involved in regulating flowering time and plant architecture (Karlgren et al., 2011). In plants, the PEBP family can be divided into three main clades: FT-like, TFL1-like, and MFT-like subfamilies (Wickland et al., 2015). FT and TWIN SISTER OF FT (TSF) are two members of the FT-like subfamily, which acts as a vital signaling molecule to promote flowering (Cao et al., 2016). FT-like genes, identified in many species, were mainly accumulated in leaves and then delivered to the buds to control the switch from vegetative growth to reproductive growth (Corbesier et al., 2007). Additionally, in the CF mutant of Liriodendron chinense, a unique FT splicing variant with intron retention specific to slb1 mutants represents the vital role of the FT gene in CF habit (Sheng et al., 2021). Unlike FT-like genes, TFL1 and BFT are classified into the TFL1-like clade and are usually known as flowering repressors (Wickland and Hanzawa, 2015). It has also been revealed that TFL1 in various species function in regulating seed development and inflorescence development and affecting flowering habit (Koskela and Hytönen, 2018; Zhang et al., 2020). This study identified the homologous genes of FT, TFL1, and BFT in both OF and CF habit, whereas the DNA sequence had no difference (Figure 4, Supplementary Figure S2A). Further studies showed that the expression patterns of the three genes were related to the continuous flowering habit of sweet osmanthus (Figure 4). In both rose and woodland strawberries, TFL1 homologues are good candidate genes for CF habit (Hurst, 1941; Koskela et al., 2012). In kiwifruit, Acbft lines displayed an ever-growing phenotype and increased branching, suggesting the role of BFT in new branch growth (Herath et al., 2022). Here, the functions of OfFT and OfBFT were analyzed in ‘Sijigui’ and Arabidopsis. The results showed that OfFT was a flowering promoting gene in both ‘Sijigui’ and Arabidopsis by increasing the expression of AP1 and LFY genes (Figure 6), whereas OfBFT functioned as a flowering suppressor through downregulating the expressions of AP1 and LFY (Figure 7). Interestingly, OfSPL8 was also found as a common upstream transcriptional regulator of OfFT and OfBFT and was involved in regulating their expression (Figure 8). It indicated the important role of OfSPL8 in CF habit of sweet osmanthus. SPLs have a beneficial function in plant floral transition and inflorescence development through regulating the flower meristem identity gene (Unte, 2003; Zhang et al., 2007). In Arabidopsis, SPLs are key components of the aging pathway in regulating the transition from plant vegetative growth to reproductive development (Wang, 2014). SPLs also act as activators of flowering by regulating flower meristem identity genes in Arabis alpina (Akhter et al., 2022).
In conclusion, these findings first revealed the flowering phenology shifts in OF and CF habits of sweet osmanthus. The role of several floral integration genes in CF habit were also analyzed. Our work might provide a new light on the regulatory pathway of continuous flowering.
Data availability statement
The data presented in the study are deposited in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SAMN31364653.
Author contributions
HZ, BD, and QF designed the research. QW, GG, XC, XL, YW, and SZ performed the experiments. QW, GG XC, and XL analyzed the data. QW and GG wrote the manuscript. HZ revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Natural Science Foundation of China (Grant Nos. 32072615 and 31902057), the major scientific and technological project of Zhejiang Province (2021C02071), and the molecular mechanism of FT/TFL1-mediated floral transition in response to gibberellin in Osmanthus ‘Sijigui’ (32201617).
Acknowledgments
We would like to thank Wei Zhou (Zhejiang Chinese Medical University) for providing the pHis2 and pGreen0800-LUC plasmids.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1049479/full#supplementary-material
References
Ahmadi, H., Bringhurst, R. S., Voth, V. (1990). Modes of inheritance of photoperiodism in fragaria. J. Amer. Soc Hortic. Sci. 115, 146–152. doi: 10.1016/0304-4238(90)90115-U
Ahmad, S., Lu, C., Gao, J., Ren, R., Wei, Y., Wu, J., et al. (2021). Genetic insights into the regulatory pathways for continuous flowering in a unique orchid arundina graminifolia. BMC Plant Biol. 21, 587–599. doi: 10.1186/s12870-021-03350-6
Ahmad, S., Peng, D., Zhou, Y., Zhao, K. (2022). The genetic and hormonal inducers of continuous flowering in orchids: An emerging view. Cells 11, 657–673. doi: 10.3390/cells11040657
Akhter, S., Westrin, K. J., Zivi, N., Nordal, V., Kretzschmar, W., Delhomme, N., et al. (2022). Cone-setting in spruce is regulated by conserved elements of theage-dependent flowering pathway. New Phytol. doi: 10.1111/nph.18449
Bai, M., Liu, J., Fan, C., Chen, Y., Chen, H., Lu, J., et al. (2021). KSN heterozygosity is associated with continuous flowering of Rosa rugosa purple branch. Hortic. Res. 8, 26–36. doi: 10.1038/s41438-021-00464-8
Cairns, T. (2000). “Modern roses XI,” in The world encyclopedia of roses (San Diego: Academic press).
Cao, K., Cui, L., Zhou, X., Ye, L., Zou, Z., Deng, S., et al. (2016). Four tomato FLOWERING LOCUS T-like proteins act antagonistically to regulate floral initiation. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.01213
Carmona, M. J., Calonje, M., Martínez-Zapater, J. M. (2007). The FT/TFL1 gene family in grapevine. Plant Mol. Biol. 63, 637–650. doi: 10.1007/s11103-006-9113-z
Chen, Z., Han, Y., Ning, K., Ding, Y., Zhao, W., Yan, S., et al. (2018). Inflorescence development and the role of LsFT in regulating bolting in lettuce (Lactuca sativa l.). Front. Plant Sci. 8. doi: 10.3389/fpls.2017.02248
Chen, D., Yan, W., Fu, L. Y., Kaufmann, K. (2018). Architecture of gene regulatory networks controlling flower development in arabidopsis thaliana. Nat. Commun. 31, 4534–4546. doi: 10.1038/s41467-018-06772-3
Corbesier, L., Vincent, C., Jang, S., Fornara, F., Fan, Q., Searle, I., et al. (2007). FT protein movement contributes to long-distance signaling in floral induction of arabidopsis. Science 316, 1030–1033. doi: 10.1126/science.1141752
Darrow, G. M. (1966). The strawberry history, breeding and physiology (New York, NY, USA: Holt, Reinhart and Winston).
Fan, L., Chen, M., Dong, B., Wang, N., Yu, Q., Wang, X., et al. (2018). Transcriptomic analysis of flower bud differentiation in magnolia sinostellata. Genes 9, 212–226. doi: 10.3390/genes9040212
Fu, J., Zhang, C., Liu, Y., Pang, T., Dong, B., Gao, X., et al. (2020). Transcriptomic analysis of flower opening response to relatively low temperatures in osmanthus fragrans. BMC Plant Biol. 20, 337–352. doi: 10.1186/s12870-020-02549-3
Gaston, A., Perrotte, J., Lerceteau-Köhler, E., Rousseau-Gueutin, M., Petit, A., Hernould, M., et al. (2013). PFRU, a single dominant locus regulates the balance between sexual and asexual plant reproduction in cultivated strawberry. J. Exp. Bot. 64, 1837–1848. doi: 10.1093/jxb/ert047
Han, R., Truco, M. J., Lavelle, D. O., Michelmore, R. W. (2021). A composite analysis of flowering time regulation in lettuce. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.632
Herath, D., Voogd, C., Mayo-Smith, M., Yang, B., Allan, A., Putterill, J., et al. (2022). CRISPR-Cas9-mediated mutagenesis of kiwifruit BFT genes results in an evergrowing but not early flowering phenotype. Plant Biotechnol. J., 1–13. doi: 10.1111/pbi.13888
Hytönen, T., Kurokura, T. (2020). Control of flowering and runnering in strawberry. Hortic. J. 89, 96–107. doi: 10.2503/hortj.UTD-R011
Irish, V. (2017). The ABC model of floral development. Curr. Biol. 27, R887–R890. doi: 10.1016/j.cub.2017.03.045
Iwata, H., Gaston, A., Remay, A., Thouroude, T., Jeauffre, J., Kawamura, K., et al. (2012). The TFL1 homologue KSN is a regulator of continuous flowering in rose and strawberry. Plant J. 69, 116–125. doi: 10.1111/j.1365-313X.2011.04776.x
Jung, H., Lee, A., Jo, S. H., Park, H. J., Jung, W., Kim, H., et al. (2021). Nitrogen signaling genes and SOC1 determine the flowering time in a reciprocal negative feedback loop in Chinese cabbage (Brassica rapa l.) based on CRISPR/Cas9-mediated mutagenesis of multiple BrSOC1 homologs. Int. J. Mol. Sci. 22, 4631–4650. doi: 10.3390/ijms22094631
Karlgren, A., Gyllenstrand, N., Kallman, T., Sundstrom, J. F., Moore, D., Lascoux, M, et al. (2011). Evolution of the PEBP gene family in plants: functional diversification in seed plant evolution. Plant Physiol. 156, 1967–1977. doi: 10.1104/pp.111.176206
Kennedy, A., Pan, S., Schilling, S., Melzer, R. (2018). MADS-box genes and crop domestication: the jack of all traits. J. Exp. Bot. 69, 1447–1469. doi: 10.1093/jxb/erx479
Koskela, E. A., Hytönen, T. (2018). “Control of flowering in strawberries,” in The genomes of rosaceous berries and their wild relatives. compendium of plant genomes. Eds. Hytönen, T., Graham, J., Harrison, R. (Cham: Springer), 35–48. doi: 10.1007/978-3-319-76020-9_4
Koskela, E. A., Mouhu, K., Albani, M. C., Kurokura, T., Rantanen, M., Sargent, D., et al. (2012). Mutation in TERMINAL FLOWER1 reverses the photoperiodic requirement for flowering in the wild strawberry, fragaria vesca. Plant Physiol. 159, 1043–1054. doi: 10.1104/pp.112.196659
Kumar, A., Bennetzen, J. L. (1999). Plant retrotransposons. Annu. Rev. Genet. 33, 479–532. doi: 10.1146/annurev.genet.33.1.479
Lei, Y., Sun, Y., Wang, B., Yu, S., Dai, H., Li, H., et al. (2020). Woodland strawberry WRKY71 acts as a promoter of flowering via a transcriptional regulatory cascade. Hortic. Res. 7, 137–150. doi: 10.1038/s41438-020-00355-4
Lewers, K. S., Castro, P., Hancock, J. F., Weebadde, C. K., Die, J. V., Rowland, L., et al. (2019). Evidence of epistatic suppression of repeat fruiting in cultivated strawberry. BMC Plant Biol. 19, 386–403. doi: 10.1186/s12870-019-1984-7
Li, Y., Wang, H., Li, X., Liang, G., Yu, D. (2017). Two DELLA-interacting proteins bHLH48 and bHLH60 regulate flowering under long-day conditions in arabidopsis thaliana. J. Exp. Bot. 68, 2757–2767. doi: 10.1093/jxb/erx143
Li, Y., Zhang, B., Wang, Y., Gong, X., Yu, H. (2021). DOTFL1 affects the floral transition in orchid dendrobium chao praya smile. Plant Physiol. 186, 2021–2036. doi: 10.1093/plphys/kiab200
Lu, J., Sun, J., Jiang, A., Bai, M., Fan, C., Liu, J., et al. (2020). Alternate expression of CONSTANS-LIKE 4 in short days and CONSTANS in long days facilitates day-neutral response in Rosa chinensis. J. Exp. Bot. 71, 4057–4068. doi: 10.1093/jxb/eraa161
Mehrnia, M., Balazadeh, S., Zanor, M. I., Mueller-Roeber, B. (2013). EBE, an AP2/ERF transcription factor highly expressed in proliferating cells, affects shoot architecture in arabidopsis. Plant Physiol. 162, 842–857. doi: 10.1104/pp.113.214049
Ren, D. Y., Li, Y. F., Zhao, F. M., Sang, X. C., Shi, J. Q., Wang, N., et al. (2013). MULTI-FLORET SPIKELET1, which encodes an AP2/ERF protein determines SPIKELET meristem fate and sterile lemma identity in rice. Plant Physiol. 162, 872–884. doi: 10.1104/pp.113.216044
Ryu, J. Y., Lee, H. J., Seo, P. J., Jung, J. H., Ahn, J. H., Park, C., et al. (2014). The arabidopsis floral repressor BFT delays flowering by competing with FT for FD binding under high salinity. Mol. Plant 7, 377–387. doi: 10.1093/mp/sst114
Semeniuk, P. (1971). Inheritance of recurrent blooming in Rosa wichuraiana. J. Hered. 62, 203–204. doi: 10.1093/jhered/62.3.203-a
Shannon, S., Meeks-Wagner, D. R. (1991). A mutation in the arabidopsis TFL1 gene affects inflorescence meristem development. Plant Cell. 3, 877–892. doi: 10.2307/3869152
Sharman, B. C. (1994). Branched heads in wheat and wheat hybrids. Nature 153, 497–508. doi: 10.1038/153497a0
Sheng, Y., Zhaodong Hao, Z., Peng, Y., Liu, S., Hu, L., Shen, Y., et al. (2021). Morphological, phenological, and transcriptional analyses provide insight into the diverse flowering traits of a mutant of the relic woody plant liriodendron chinense. Hortic. Res. 8, 174–189. doi: 10.1038/s41438-021-00610-2
Soufflet-Freslon, V., Araou, E., Jeauffre, J., Thouroude, T., Chastellier, A., Michel, G., et al. (2021). Diversity and selection of the continuous-flowering gene, RoKSN, in rose. Hortic. Res. 8, 76–86. doi: 10.1038/s41438-021-00512-3
Taylor, M., Tornqvist, C. E., Zhao, X., Grabowski, P., Doerge, R., Ma, J., et al. (2018). Genome-wide association study in pseudo-F2 populations of switchgrass identifies genetic loci affecting heading and anthesis dates. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01250
Unte, U. S. (2003). SPL8, an SBP-box gene that affects pollen sac development in arabidopsis. Plant Cell 15, 1009–1019. doi: 10.1105/tpc.010678
Villar, L., Lienqueo, I., Llanes, A., Rojas, P., Jorge Perez, J., Correa, F., et al. (2020). Comparative transcripto-mic analysis reveals novel roles of transcription factors and hormones during the flowering induction and floral bud differentiation in sweet cherry trees (Prunus avium l. Cv Bing) PloS One 15, e0230110–e0230137. doi: 10.1371/journal.pone.0230110
Wang, J. W. (2014). Regulation of flowering time by the miR156-mediated age pathway. J. Exp. Bot. 65, 4723–4730. doi: 10.1093/jxb/eru246
Wang, Y. H., He, X. H., Yu, H. X., Mo, X., Fan, Y., Fan, Z., et al. (2021). Overexpression of four MiTFL1 genes from mango delays the flowering time in transgenic arabidopsis. BMC Plant Biol. 21, 407–422. doi: 10.1186/s12870-021-03199-9
Wang, Y. G., Zhang, C., Xu, B., Fu, J. X., Du, Y. X., Fan, Q., et al. (2022). Temperature regulation of carotenoid accumulation in the petals of sweet osmanthus via modulating expression of carotenoid biosynthesis and degradation genes. BMC Genomics 23, 418–429. doi: 10.1186/s12864-022-08643-0
Wickland, D. P., Hanzawa, Y. (2015). The FLOWERING LOCUS T/TERMINAL FLOWER 1 gene family: functional evolution and molecular mechanisms. Mol. Plant 8, 983–997. doi: 10.1016/j.molp.2015.01.007
Wu, Q., Bai, X., Zhao, W., Shi, X., Xiang, D., Wan, Y., et al. (2019). Investigation into the underlying regulatory mechanisms shaping inflorescence architecture in chenopodium quinoa. BMC Genomics 20, 658–677. doi: 10.1186/s12864-019-6027-0
Yang, J., Wang, S., Xia, H., Guo, P., Wang, Y., Shang, F., et al. (2021). Comparative analysis of candidate genes for multi-season flowering in two varieties of sweet osmanthus. Sci. Horti. 285, 110175–110180. doi: 10.1016/j.scienta.2021.110175
Zhang, S., Gottschalk, C., Nocker, S. (2019). Genetic mechanisms in the repression of flowering by gibberellins in apple (Malus x domestica borkh.). BMC Genomics 20, 747. doi: 10.1186/s12864-019-6090-6
Zhang, B., Li, C., Li, Y., Yu, H. (2020). Mobile TERMINAL FLOWER1 determines seed size in arabidopsis. Nat. Plants 6, 1146–1157. doi: 10.1038/s41477-020-0749-5
Zhang, Y., Schwarz, S., Saedler, H., Huijser, P. (2007). SPL8, a local regulator in a subset of gibberellin-mediated developmental processes in arabidopsis. Plant Mol. Biol. 63, 429–439. doi: 10.1007/s11103-006-9099-6
Zhu, S., Fang, Q., Wang, Y., Zhong, S., Dong, B., Zhao, H., et al. (2022). OfSPL11 gene from osmanthus fragrans promotes plant growth and oxidative damage reduction to enhance salt tolerance in arabidopsis. Horticulturae 8, 412. doi: 10.3390/horticulturae8050412
Keywords: flowering regulation, continuous flowering, transcription factors, transcriptome, Osmanthus fragrans
Citation: Wang Q, Gao G, Chen X, Liu X, Dong B, Wang Y, Zhong S, Deng J, Fang Q and Zhao H (2022) Genetic studies on continuous flowering in woody plant Osmanthus fragrans. Front. Plant Sci. 13:1049479. doi: 10.3389/fpls.2022.1049479
Received: 20 September 2022; Accepted: 10 October 2022;
Published: 03 November 2022.
Edited by:
Tangchun Zheng, Beijing Forestry University, ChinaReviewed by:
Xiang Gao, Northeast Normal University, ChinaSagheer Ahmad, Guangdong Academy of Agricultural Sciences, China
Yuanzheng Yue, Nanjing Forestry University, China
Guilin Chen, Inner Mongolia University, Hohhot, China
Copyright © 2022 Wang, Gao, Chen, Liu, Dong, Wang, Zhong, Deng, Fang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongbo Zhao, emhhb2hiQHphZnUuZWR1LmNu; Qiu Fang, MTU5OTYzMDU4OTJAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Qianqian Wang†
Qianqian Wang† Yiguang Wang
Yiguang Wang Hongbo Zhao
Hongbo Zhao