- 1Zhejiang Provincial Key Laboratory of Plant Evolutionary Ecology and Conservation, Taizhou University, Taizhou, China
- 2Section of Maritime Space and Island Management, Yuhuan Municipal Bureau of Natural Resources and Planning, Yuhuan, China
- 3Department of Security Production Management, Taizhou Circular Economy Development Co., Ltd., Taizhou, China
Background: Kandelia obovata is an important mangrove species extensively distributed in Eastern Asia that is susceptible to low-temperature stress. NAC (NAM, ATAF1/2 and CUC2) domain proteins are transcription factors (TFs) that play various roles in plant growth and development and in the plant response to environmental stresses. Nevertheless, genome-wide analyses of K. obovata NAC genes (KoNACs) and their responses to chilling stress have rarely been studied.
Methods: The KoNAC gene family was identified and characterized using bioinformatic analysis, the subcellular location of some NAC proteins was confirmed using confocal microscopy analysis, and the KoNACs that responded to chilling stress were screened using RNA-seq and qRT-PCR analysis.
Results: A total of 79 KoNACs were identified, and they were unequally distributed across all 18 chromosomes of K. obovata. The KoNAC proteins could be divided into 16 subgroups according to the phylogenetic tree based on NAC family members of Arabidopsis thaliana. The KoNACs exhibited greater synteny with A. thaliana sequences than with Oryza sativa sequences, indicating that KoNACs underwent extensive evolution after the divergence of dicotyledons and monocotyledons. Segmental duplication was the main driving force of the expansions of KoNAC genes. Confocal microscopy analysis verified that the four randomly selected KoNACs localized to the nucleus, indicating the accuracy of the bioinformatic predictions. Tissue expression pattern analysis demonstrated that some KoNAC genes showed tissue-specific expression, suggesting that these KoNACs might be important for plant development and growth. Additionally, the expression levels of 19 KoNACs were significantly (15 positively and 4 negatively) induced by cold treatment, demonstrating that these KoNACs might play important roles during cold stress responses and might be candidate genes for the genetic engineering of K. obovata with enhanced chilling stress tolerance. Coexpression network analysis revealed that 381 coexpressed pairs (between 13 KoNACs and 284 other genes) were significantly correlated.
Conclusions: Seventy-nine KoNACs were identified in K. obovata, nineteen of which displayed chilling-induced expression patterns. These genes may serve as candidates for functional analyses of KoNACs engaged in chilling stress. Our results lay the foundation for evolutionary analyses of KoNACs and their molecular mechanisms in response to environmental stress.
Introduction
Mangrove forests are composed of woody plants that grow in subtropical and tropical coastal intertidal zones, where they play an important role in preventing wind, attenuating waves, purifying seawater, maintaining biodiversity, supplying seafood, and sequestering carbon (Trégarot et al., 2021). Mangroves grow in harsh conditions involving anaerobic soils, extreme tides, high salinity, strong winds, sea-level rise, and extreme climate events (Guo et al., 2017). Cold stress is one of the most crucial factors limiting the development, growth, and geographical distribution of mangrove plants among all of these abiotic stresses (Shen et al., 2021). Cold stress (low temperature) can be categorized into chilling stress (0−15°C) or freezing stress (< 0°C) according to its intensity (Jiang et al., 2021). Due to their inability to adapt to low temperatures, tropical and subtropical plants typically exhibit sensitivity to cold stress and are susceptible to chilling stress (Wu et al., 2019). On the Pacific coast of North America, the 50% mortality temperature threshold for black mangroves is −0.92°C (Bardou et al., 2020). In early 2008, a chilling event occurred in southern China, resulting in the withering and even death of many mangrove plants (Chen et al., 2017). Recently, a study of mangrove responses to chilling stress reported that the leaves of Bruguiera gymnorhiza showed obvious yellowing, curling, and wilting following exposure to a temperature of 5°C for 13 days (Wang et al., 2022).
Kandelia obovata is a species of the Rhizophoraceae family that was initially named K. candel in the regions of China and Japan and has now been reclassified as a new species (Sheue et al., 2003). As the dominant mangrove species in Eastern Asia, K. obovata provides significant ecological functions and valuable socioeconomic services to humans, such as providing habitat, preserving species, safeguarding shorelines, purifying water, and sequestrating carbon (Wu et al., 2020; Wei et al., 2022). K. obovata is the most cold-resistant species among mangrove plants, and the northernmost point of its natural distribution is Fuding (27° 20’ N) in Fujian Province, China (Chen et al., 2017). Recently, certain mangrove species have even expanded to higher latitudes due to climate change (Osland et al., 2013). K. obovata was introduced to Yueqing Bay (28° 20’ N), Zhejiang Province, where the propagules of adult trees can settle naturally and grow into seedlings (Lu et al., 2022). However, extreme cold climatic events exert a destructive influence on mangroves. For example, a severe chilling injury in the winter of 2015 led to different levels of defoliation and death of K. obovata in Yueqing Bay (Lu et al., 2022).
K. obovata has a remarkable capability to cope with environmental cold stress. In recent years, an increasing number of research studies have uncovered the physiological and molecular mechanisms by which K. obovata adapts to low temperature (Fei et al., 2015; Peng et al., 2015; Su et al., 2019; Fei et al., 2021a; Fei et al., 2021b). An AP2/EREBP transcription factor (TF), KcCBF3, was cloned in our previous study, and it was shown to be involved in the adaptation of K. obovata to 4°C chilling stress (Du and Li, 2019), which is in accordance with the results of Peng et al. (2020). We also found that 9 WRKY transcription factors (TFs) exhibited 4°C-induced expression patterns, implying that they may play important roles in the response of K. obovata to chilling (Du et al., 2022). TFs are proteins that can either activate or suppress the expression of target genes by binding to specific cis-elements in promoter regions or interacting with other regulatory factors (Manna et al., 2021). In the past few decades, a large number of research efforts have focused on identifying and characterizing various TFs that participate in plant stress responses in either the ABA-independent pathway or the ABA-dependent pathway, such as WRKY, AP2/EREBP, MYB, bHLH, bZIP, and NAC TFs (Wang et al., 2016).
The acronym NAC originated from the names of three genes that were initially discovered to contain a particular NAC domain: NAM (No Apical Meristem), ATAF1/ATAF2 (Arabidopsis Transcription Activation Factor 1/2), and CUC2 (Cup-shaped Cotyledon 2) (Nakashima et al., 2012). A classic NAC protein consists of a highly conserved DNA-binding NAC domain within the N-terminal region and a variable transcriptional regulatory region at the C-terminal (Shao et al., 2015). The NAC domain is a highly conserved DNA-binding domain containing approximately 150–160 amino acid residues and can be classified into five subdomains (A to E) (Ooka et al., 2003). The function of the NAC domain is relevant to DNA binding, nuclear localization, and the constitution of homodimers as well as heterodimers with other NAC domain-containing TFs (Olsen et al., 2005). In contrast, the C-terminal region of NAC proteins is highly divergent and functions as a transcriptional activator or repressor to regulate the transcription of target genes (Nakashima et al., 2012).
NAC proteins can be found in a variety of plant species and are one of the largest families of TFs in plants. The completion of high-quality plant genome sequences has provided the possibility for the genome-wide analysis of all members belonging to specific gene families (Nuruzzaman et al., 2010). Whole-genome-based studies have revealed 117 NAC genes in Arabidopsis thaliana, 151 in Oryza sativa, 74 in Vitis vinifera, 110 in Solanum tuberosum, 204 in Brassica rapa, and 152 in Zea mays, Glycine max and Nicotiana tabacum thus far (Shao et al., 2015). NACs are plant-specific proteins that play various roles in plant development, morphogenesis, and senescence (Singh et al., 2021). In addition, NAC proteins have been demonstrated to have multiple functions in the responses to biotic stresses, such as bacterial and virus infection (Nakashima et al., 2007), and abiotic stresses, such as drought, salinity, heat, cold, and waterlogging (Nuruzzaman et al., 2013). It was shown that NAC TFs regulate plant chilling resistance through CBF/DREB-dependent or CBF/DREB-independent pathways (Diao et al., 2020). In CBF/DREB-dependent pathway, NACs improve plant cold resistance by inducing the expression of CBF/DREB transcription factors and thus up-regulating the expression of COR (cold responsive) genes (Jin et al., 2017; Yong et al., 2019). However, a few NACs increase the plant resistance to cold stress by enhancing the hypersensitivity to ABA in CBF/DREB-independent pathways (Yong et al., 2019); recent study has shown that NAC can also activate the transcription of ethylene synthesis-related enzyme genes by binding to their promoters to induce ethylene synthesis and thus improve plant cold tolerance (Dong et al., 2022).
K. obovata widely distributes along the southeastern coast of China and can endure harsh conditions such as high salinity, submergence, hypoxia, or even extremely low temperatures (Fei et al., 2021b; Nizam et al., 2022). Although the important functions of NACs under abiotic stress have been widely illustrated in many plant species, information on these proteins in K. obovata is still unavailable. The genome of K. obovata was recently published, providing a valuable genomic resource for molecular research on this species (Hu et al., 2020). In the present study, we identified and characterized NAC genes (KoNACs) in the whole genome of K. obovata, surveyed their expression patterns in various tissues, and revealed putative candidate KoNAC genes under chilling stress. This study might lay a foundation for further research on the molecular mechanism of KoNAC genes in the adaptation of K. obovata to chilling stress.
Materials and methods
Identification of KoNAC genes in K obovata
The complete genome data of K. obovata were retrieved from the Genome Sequence Archive database (https://bigd.big.ac.cn/gsa/browse/CRA002395). The NAC gene sequences of A. thaliana and rice were searched and downloaded from the Arabidopsis Information Resource (TAIR) website (http://www.Arabidopsis.org) and the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/), respectively. A hidden Markov model (HMM) profile of the Pfam NAC domain (PF01849) and NAM domain (PF02365) downloaded from the Pfam database (http://pfam.sanger.ac.uk/) was employed to identify KoNAC genes by using HMMER 3.2.1 with the default parameters, and the cutoff value was set to 0.01. The putative KoNACs were further validated by screening for NAC and NAM domains in the Pfam database (http://pfam.xfam.org/) and were finally confirmed as the KoNAC proteins after removing redundant sequences with CD-HIT program (http://cd-hit.org/) and performing BLASTP searches. The physicochemical properties of the KoNAC proteins, including their molecular weight (MW) and isoelectric point (pI), were analyzed with the online software ExPASy (https://www.expasy.org). The subcellular localization of the KoNAC proteins was determined with the PSORT program (https://psort.hgc.jp/).
Phylogenetic analysis of KoNAC proteins
The NAC protein sequences of A. thaliana were obtained from the TAIR database (http://www.Arabidopsis.org). An unrooted phylogenetic tree consisting of KoNACs and NACs from A. thaliana was constructed with software MEGA6.0 using the neighbor-joining (NJ) method with 1,000 bootstrap iterations. All KoNACs were categorized into different subgroups based on the classification of NAC proteins from A. thaliana (Ooka et al., 2003).
Conserved motif and gene structure analysis
The conserved motif in the KoNAC protein sequences was searched using the Multiple Expectation Maximization for Motif Elicitation (MEME) program (http://meme-suite.org/tools/meme) with the parameters according to Song et al. (2020). The exon/intron structure pattern of the KoNAC genes was analyzed using the Gene Structure Display Server (GSDS) program (http://gsds.cbi.pku.edu.cn/) by comparing their predicted coding sequences with the corresponding full-length genomic DNA sequences. The conserved motifs and exon/intron structure were visualized using TBtools (Chen et al., 2020).
Chromosomal location, gene duplication and synteny analysis of KoNAC genes
The chromosomal localizations of all the KoNAC genes were determined from the genome annotation file and mapped using MapChart (Voorrips, 2002). The genome sequence and annotation information of A. thaliana and rice were downloaded from TAIR and Ensembl (http://plants.ensembl.org/index.html), respectively. The identification of tandem duplication and segmental duplication was detected by the MCScanX program (Wang et al., 2012) with the default parameters, in short, BLASTP alignment significance: E-value threshold = 10-5, MatchScore = 50, GapPenalty = -1, maximum number of intervening genes ≤ 25. The synonymous (Ks) and nonsynonymous (Ka) substitution rates of KoNAC-homologous genes were calculated using the online tool available at http://services.cbu.uib.no/tools/kaks, and the selection pressure endured by gene pairs was calculated according to the Ka/Ks ratio.
Cis-Acting Regulatory Element (CARE) analysis
The 2-kb upstream sequences of the transcription start site of the KoNAC genes were extracted and submitted to the PlantCARE online server (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to predict the putative CAREs engaged in plant development, phytohormone, and abiotic stress responses. The frequency of each CARE in KoNAC genes was counted, and the 20 most common CAREs in KoNAC genes were visualized using TBtools (Chen et al., 2020).
Experimental determination of the subcellular localization of several KoNACs
To experimentally analyze the subcellular localization of KoNACs, the cDNAs of four randomly selected KoNACs (KoNAC15, KoNAC27, KoNAC54, and KoNAC58) were cloned into the expression vector CAM-turbo-GFP-FLAG and fused in frame with a GFP or mCherry gene. The primers used for subcellular localization are listed in Supplementary Table S1. The recombinant vector and a mock vector were introduced into A. thaliana leaf protoplasts, which were then imaged with a laser scanning confocal microscope (Olympus FV1000 viewer, Tokyo, Japan). Arabidopsis leaf protoplast isolation and PEG-mediated transformation were performed as previously described (Yoo et al., 2007).
Expression profiles of KoNAC genes in different tissues and in response to cold stress based on transcriptome data
Transcriptome data of K. obovata leaves in response to cold stress were downloaded from the National Center for Biotechnology Information (NCBI) database (Accession number: PRJNA678025). Transcriptome data of different tissues of K. obovata were also downloaded from the NCBI database (Accession number: PRJNA416402). In addition to the root, stem, leaf, and fruit tissues, the expression levels of the KoNAC genes were investigated in flower, pistil, stamen, sepal, and fruit. The relative gene expression values were expressed as transcripts per million (TPM), and all of the transcriptome data were converted into log2 (TPM + 1) values (Zeng et al., 2020). For expression analysis of genes in response to cold stress, the differentially expressed KoNACs were chosen based on the criteria of | log2 (fold-change) ≥1 | and P < 0.05. The expression levels of the KoNAC genes were visualized in a heatmap using TBtools software (Chen et al., 2020).
Plant material and treatments
Mature and pest-free propagules of K. obovata were picked from Maoyan Island (28°13′N, 121°10′E), Yuhuan, Zhejiang Province, China. They were washed and grown in river sand in a growth chamber at a temperature of 25°C, 75% humidity, and a photoperiod of 14 h light/10 h dark. The seedlings were watered once a week with Hoagland solution, and they were exposed to 4°C chilling stress for 0, 1, 3, or 12 hours at the six-leaf stage. Then, the leaves were collected and immediately frozen in liquid nitrogen and stored at -80°C in an ultralow temperature freezer for qRT-PCR. All treatments were designed with three independent biological replications.
Validation of KoNAC gene expression levels via qRT-PCR
Total RNA was extracted from frozen leaves using an RNASimple Total RNA Kit (Tiangen, Beijing, China) and then used for cDNA synthesis with a TIANScript cDNA Kit (Tiangen, Beijing, China). The concentration of cDNA was determined with a NanoDrop 1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Then, qRT-PCR was carried out on a CFX96 Touch real-time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA) with SuperReal PreMix Plus (SYBR Green) (Tiangen, Beijing, China). The primers used for qRT-PCR are shown in Supplementary Table S2. The relative gene expression levels of selected KoNACs were determined with the 2-ΔΔCT method (Livak and Schmittgen, 2001) using 18S rRNA of K. obovata as the internal reference gene. The data are presented as the mean ± standard error; statistical significance was assessed by one-way ANOVA followed by Duncan’s multiple range tests at P < 0.05.
Prediction of coexpression networks among KoNAC genes and potential regulated genes
To further explore the potential activators or repressors associated with KoNACs, the Pearson correlation coefficients (PCCs) between KoNACs and non-KoNAC genes were calculated based on their expression levels using the R package. The absolute value of the PCC ≥ 0.95 and P value < 0.001 was set as the screening cutoff, and genes that met this criterion were regarded as potential correlated regulators. Cytoscape software (version 3.6.1) was used to visualize the network.
Results
Identification and characterization of KoNAC genes
Members of the KoNAC family were identified in the K. obovata genome via HMM searches with profiles PF01849 and PF02365, which represent the NAC and NAM domains, respectively. A total of 79 candidate genes were further identified by local BLASTP searches and were designated KoNAC1 to KoNAC79 based on their order on the chromosomes (Supplementary Table S3). Basic information of the 79 KoNAC genes, including the length of their CDSs and the lengths, MWs, pIs and subcellular localizations of their encoded proteins, was analyzed in the present study. The KoNAC CDS lengths ranged from 408 (KoNAC29) to 1,941 bp (KoNAC18), with an average length of 1,021 bp. The MWs ranged from 15.53 kDa (KoNAC29) to 73.58 kDa (KoNAC18), with an average of 44.58 kDa. The theoretical pIs varied from 4.46 (KoNAC53) to 9.63 (KoNAC11). The subcellular localizations of the 79 KoNAC proteins were predicted to be the nucleus (64), cytoplasm (7), chloroplast (4), mitochondrion (2), vacuole (1) and extracellular space (1).
Phylogenetic analysis and classification of KoNAC proteins
To explore the evolutionary relationships of the KoNAC members, an unrooted phylogenetic tree was constructed by using the protein sequences of KoNACs and NACs from A. thaliana. The results showed that the 79 KoNAC proteins could be divided into 16 subgroups, including ATAF, ANAC3, NAP, ONAC022, NAM, NAC1, OsNAC7, ANAC001, TIP, ANAC011, NAC2, ONAC003, ANAC063, SENU5, and two unclassified KoNACs according to their homology with NAC proteins in A. thaliana; however, no KoNAC member from the TERN or OSNAC8 subfamily was identified (Figure 1). Among the 79 KoNAC proteins, the members belonging to the NAC and NAM subfamilies were the most abundant, with 10 members, whereas the ANAC3 subfamily contained the fewest KoNAC proteins, with only one member. The phylogenetic tree data indicated that KoNAC proteins showed some diversity, which was consistent with reports from A. thaliana (Ooka et al., 2003).
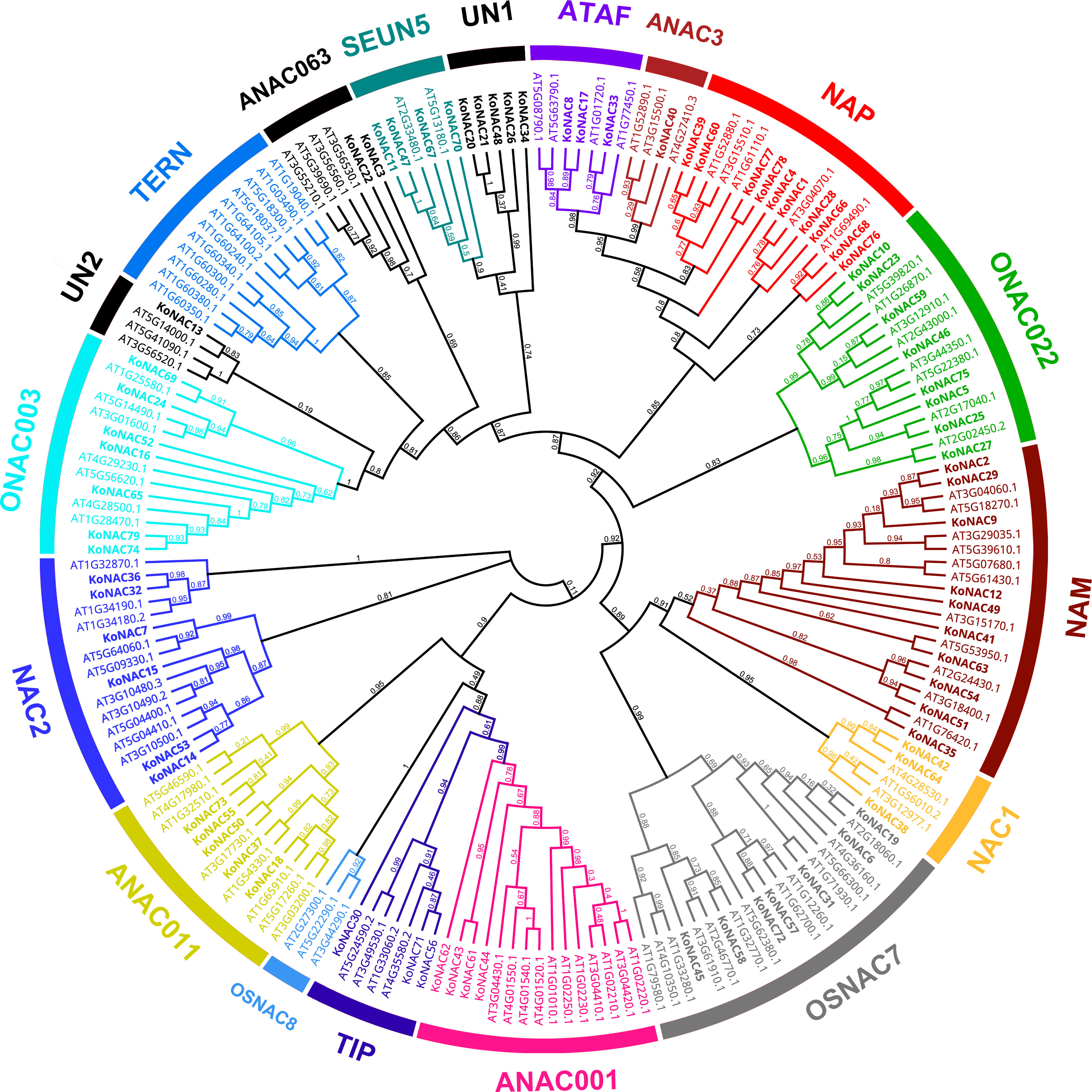
Figure 1 Phylogenetic relationships among NAC proteins identified in Kandelia obovata and Arabidopsis thaliana. The neighbor-joining (NJ) unrooted phylogenetic tree was constructed according to the NAC protein sequences of K. obovata and A. thaliana using the program MEGA 6.0. The subfamilies are labeled and indicated with different colors, and the unclassified KoNACs are represented by the abbreviation “UN”.
Conserved motifs of protein KoNACs and gene structure of KoNACs
To obtain more insights into the functional regions of KoNAC proteins, the conserved motifs were identified with the MEME program. A total of 20 conserved motifs were revealed, designated motifs 1–20 (Supplementary Table S4). These conserved motifs ranged in length from 8 to 50 amino acid residues. Furthermore, motifs 5, 3 and 1 were the conserved regions with the highest occurrence probability, and the frequencies of occurrence were 78, 66 and 62, respectively (Figure 2A). KoNAC26 contained only one type of motif, whereas KoNAC77 and KoNAC78 contained the greatest number of motifs (10 types). Generally, KoNAC proteins that were clustered in the same subgroups shared similar motif compositions, indicating that the same subgroups of genes showed similar functions.
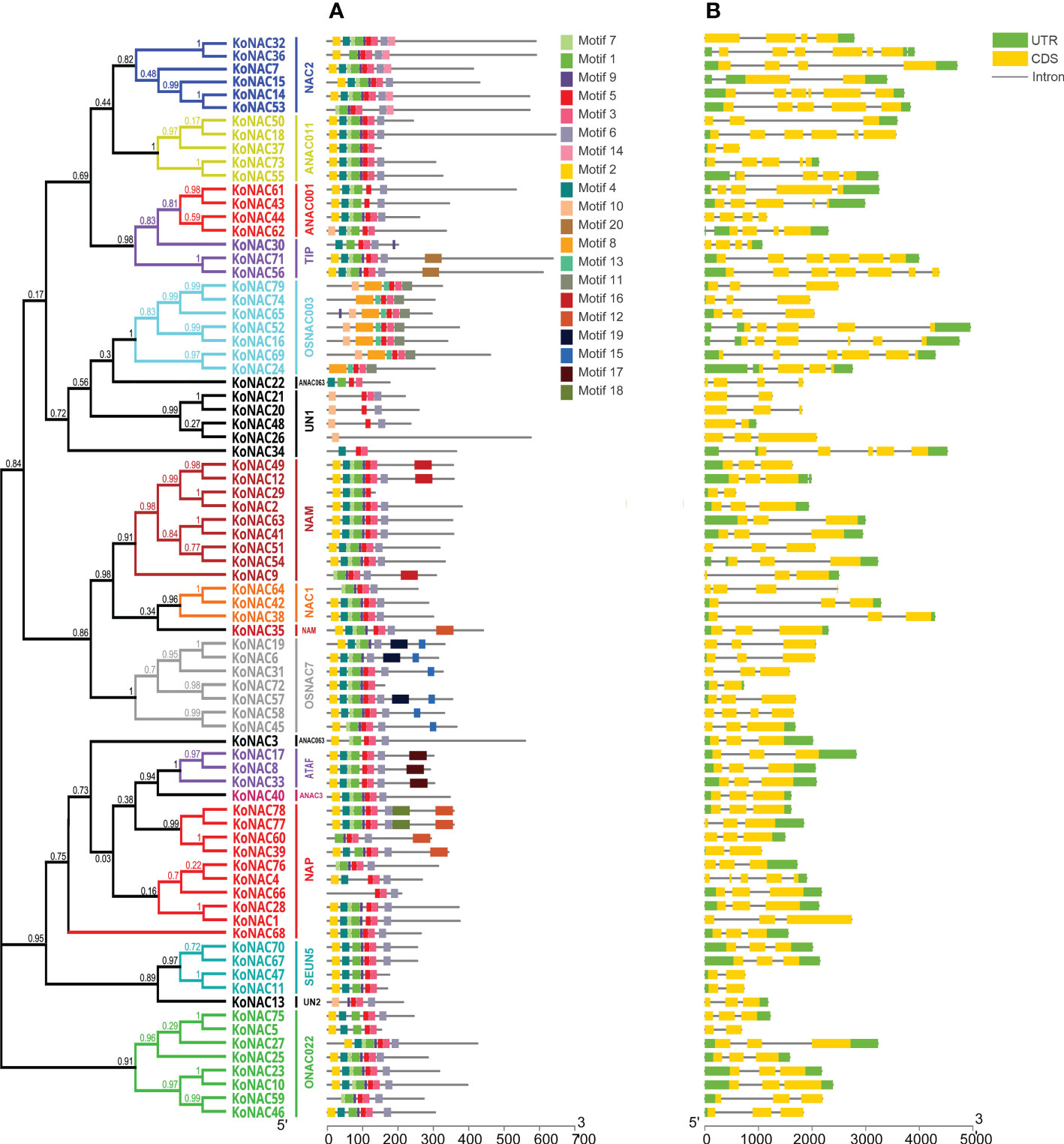
Figure 2 Conserved motifs of KoNACs and gene structures of KoNACs according to their phylogenetic relationships. The gene structures of KoNACs (A) were predicted with GSDS software. A green box represents the untranslated region (UTR), a yellow box represents an exon, and a black line represents an intron. The conserved motifs (B) were identified by the MEME web server. Each motif is indicated by a colored box, and the location of each motif can be estimated using the scale at the bottom.
To reveal the structural characteristics of the KoNAC genes, the intron–exon distribution pattern was analyzed using the program GSDS. The genomic structure of the KoNACs showed great variation, with the number of introns varying from 1 to 7 (Figure 2B). Among the 79 KoNAC genes, 9 (11.39%) had one exon, more than half (45, 56.96%) had two exons, and only 3 genes had more than 6 exons (KoNAC16, KoNAC56 and KoNAC36, with 6, 6 and 7 exons, respectively).
Chromosomal location, gene duplication, and synteny analysis of KoNAC genes
To clarify the distribution of KoNAC genes on the chromosomes of K. obovata, the program MapChart was used to map the chromosomal locations of all the identified KoNAC genes (Figure 3). The distribution of the 79 KoNACs throughout the 18 chromosomes was uneven, and there was no correlation between the number of genes on each chromosome and their length. Chromosome 8 contained the greatest number of KoNAC genes (10, 12.66%), and chromosomes 7, 14 and 16 harbored only one KoNAC gene each, whereas there were no KoNAC genes on chromosome 18.
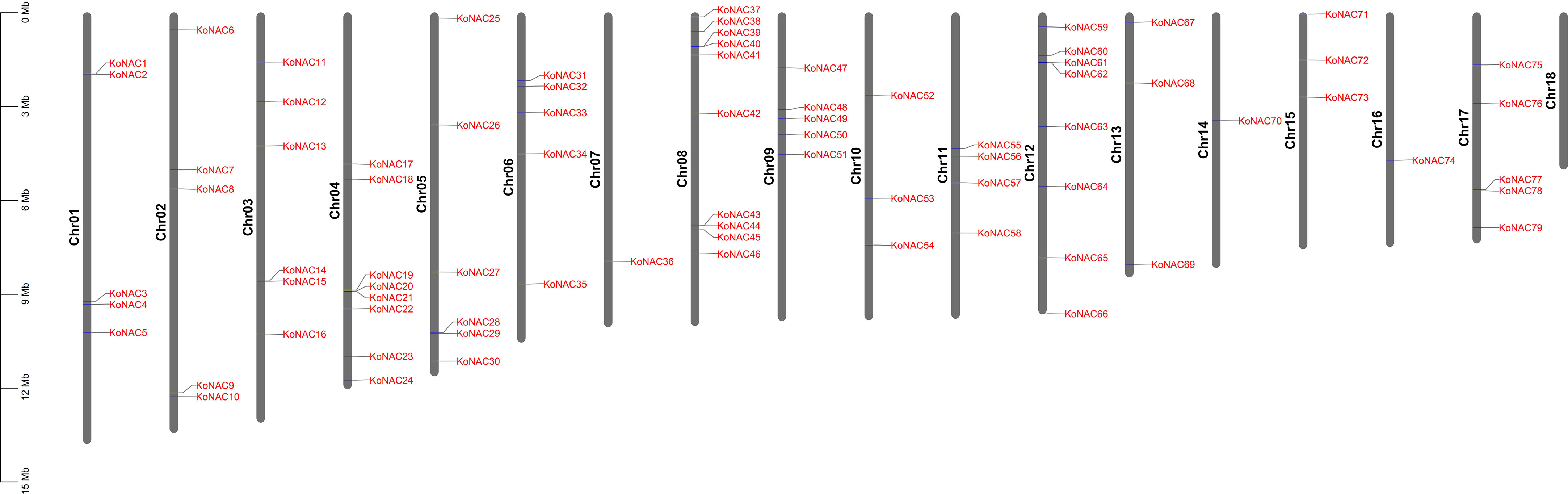
Figure 3 Distribution of 79 KoNAC genes on 18 chromosomes. Vertical bars represent the chromosomes of Kandelia obovata. The chromosome number is on the left of each chromosome. The scale on the left represents the length of the chromosome.
To determine duplication events in the KoNAC genes, a synteny analysis was carried out using software MCScanX. Forty-nine pairs of segmentally duplicated genes were found among the 79 KoNAC members. Segmentally duplicated genes were the most common on chromosome 12, followed by chromosome 8, which contained 17 and 15 pairs, respectively (Figure 4, Supplementary Table S5). Four pairs of tandemly duplicated genes were identified in the KoNAC gene family, including KoNAC20/21, KoNAC43/44, KoNAC61/62, and KoNAC77/78, and the tandemly duplicated gene pairs were present on chromosomes 4, 8, 12, and 17, respectively (Figure 4, Supplementary Table S5). The results suggested that segmental duplication was mostly responsible for the expansion of the KoNAC genes.
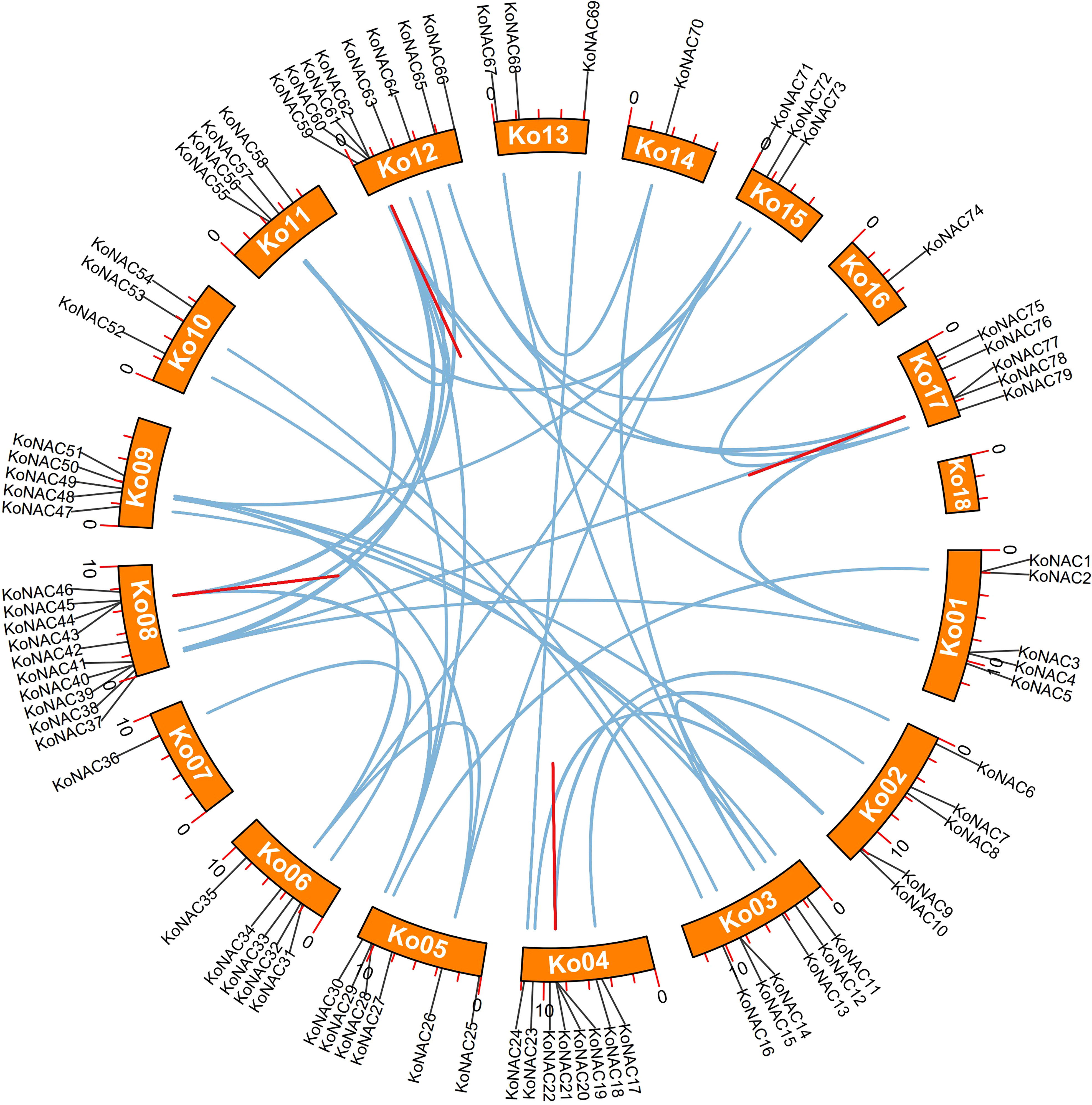
Figure 4 Schematic representations of the interchromosomal relationships of KoNAC genes. The blue lines indicate segmentally duplicated KoNAC gene pairs, and the red lines indicate tandemly duplicated KoNAC gene pairs.
To better understand the selective pressure on the KoNAC genes, the Ka/Ks ratios of the duplicated gene pairs were determined. All the segmentally and tandemly duplicated KoNAC gene pairs showed a Ka/Ks value < 1 (Supplementary Table S6), suggesting that strong purifying selection may have had a role in the evolution of the KoNAC gene family.
To further explore the evolutionary relationships of the KoNAC genes, comparative syntenic maps of the A. thaliana and Oryza sativa genes were constructed. The results indicated that 24 KoNAC genes showed collinearity relationships with 24 NAC genes in A. thaliana and 10 NAC genes in O. sativa. The number of orthologous gene pairs between K. obovata and A. thaliana O. sativa was 30, and that between K. obovata and O. sativa was 18 (Figure 5, Supplementary Table S7), suggesting that NAC genes underwent significant evolution and duplication after the divergence of dicotyledonous and monocotyledonous plants.
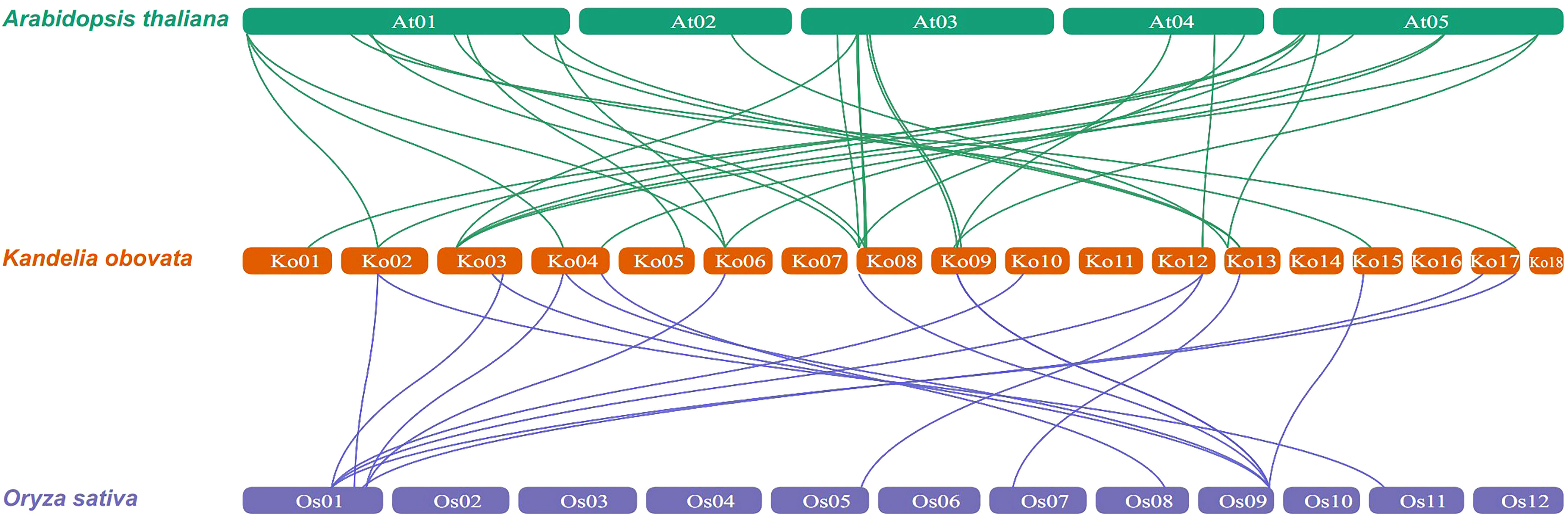
Figure 5 Synteny analysis of NAC genes between Kandelia obovata and two representative plant species (Arabidopsis thaliana and Oryza sativa). Green and blue lines represent syntenic NAC gene pairs of K. obovata and A. thaliana and O. sativa, respectively.
Cis-elements in the promoter regions of KoNAC genes
To analyze the likely cis-elements of the KoNAC genes, the 2-kb upstream promoter region sequences of the KoNACs were obtained and used to search the PlantCARE database. A number of cis-elements related to developmental processes, phytohormone responses, and abiotic stress responses were identified (Figure 6, Supplementary Figure S1). The cis-elements related to plant development mainly included light-responsive elements and meristem-specific elements. The cis-elements involved in hormone responses included abscisic acid (ABA), gibberellin (GA), methyl jasmonate (MeJA), and ethylene response elements. Among the cis-elements related to stress responses, low-temperature response, anaerobic induction, and wound response elements were the most abundant motifs. The results suggested that KoNACs might play an important role under the control of hormones during the development and stress responses of K. obovata.
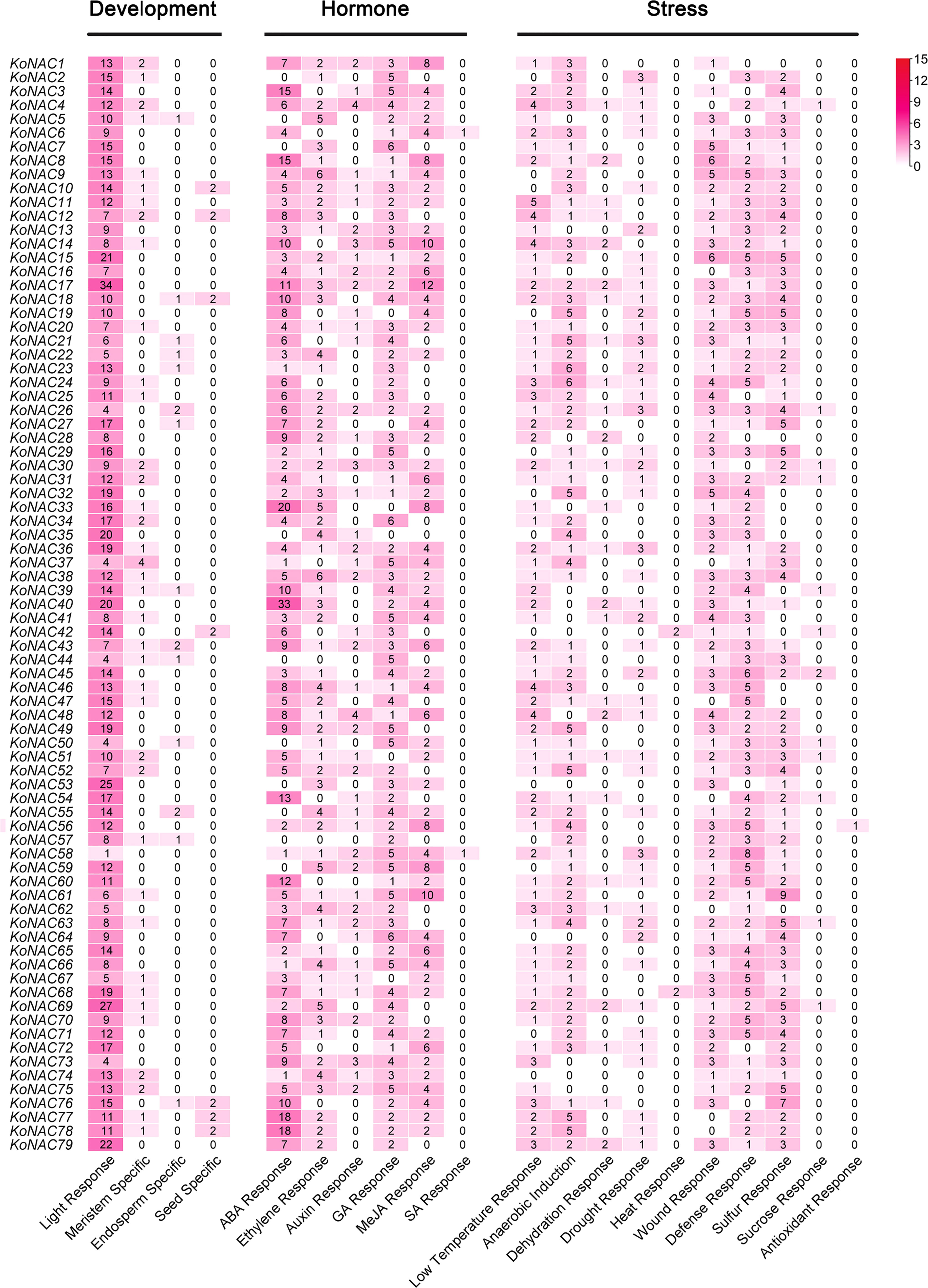
Figure 6 The number of each type of cis-acting element in the promoter region of each KoNAC gene. Annotation of the cis-elements: ABA, abscisic acid; GA, gibberellin; MeJA, methyl jasmonate; SA, salicylic acid.
Confirmation of the subcellular localization of several KoNAC proteins
To experimentally determine the subcellular localization of KoNACs, four members, KoNAC15, KoNAC27, KoNAC54, and KoNAC58, were randomly selected from different clades (NAC2, ONAC022, NAM, and OsNAC7, respectively) according to the previous phylogenetic tree, and their CDS were cloned into an expression vector containing the GFP tag driven by the CaMV35S promoter. Confocal microscopy analysis showed that the fluorescent signal of GFP was distributed in the plasma membrane, cytoplasm and nucleus of the protoplasts, while GFP-fused KoNAC15, KoNAC27, KoNAC54, and KoNAC58 localized only to the nucleus (Figure 7). Our results were consistent with the in silico subcellular localization prediction of KoNACs (Supplementary Table S3).
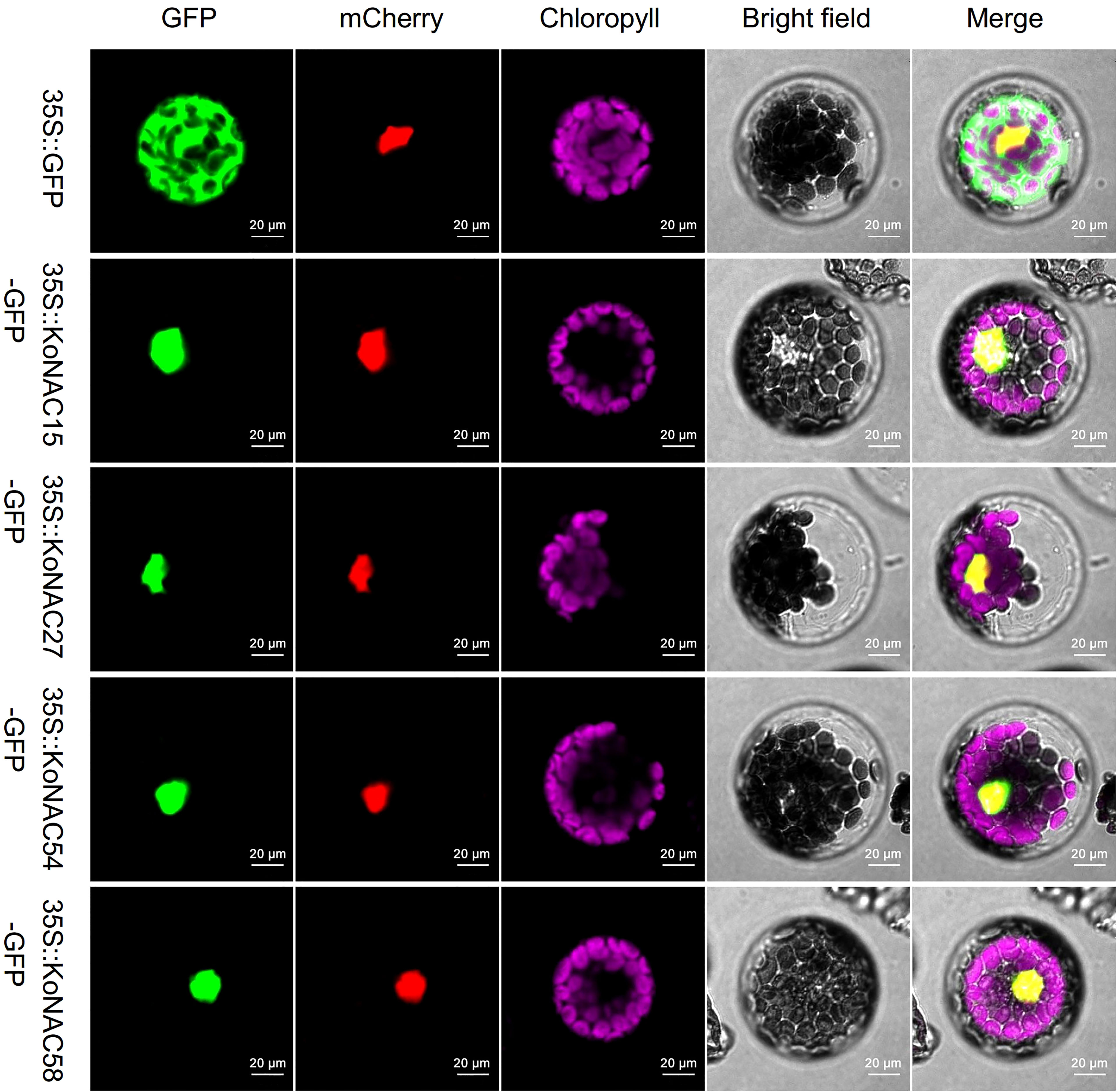
Figure 7 KoNAC-GFP fusion vectors were used to determine the subcellular localization of several KoNACs in Arabidopsis thaliana protoplast cells. The 35S::EGFP (green) construct used as a negative control was extensively observed in both the nucleus and the cytoplasm of A. thaliana protoplasts. A nuclear-localized 35S::mCherry protein (red) was applied to mark the nucleus. Chlorophyll autofluorescence (purple) demonstrates the location of chloroplasts. Scale bar = 0.02 mm.
Expression profiles of KoNACs in different tissues
The expression profiles of all 79 KoNACs in different tissues, such as the roots, stems, leaves, flowers, sepals, pistils, stamens, and fruit, were investigated based on K. obovata transcriptomic data that were publicly available. Generally, the average expression level of KoNAC genes was highest in roots, followed by stamens, and lowest in pistils (Figure 8A). Some KoNAC genes, such as KoNAC25, KoNAC33, KoNAC36, KoNAC67, KoNAC70, KoNAC71, and KoNAC75, showed high expression in roots; however, KoNAC8, KoNAC14, KoNAC17, KoNAC33, KoNAC34, KoNAC40, KoNAC60, and KoNAC61 presented higher expression levels in stamens. Additionally, KoNAC3, KoNAC4, KoNAC21, KoNAC26, KoNAC37, KoNAC41, KoNAC42, KoNAC44, and KoNAC76 were negligibly expressed in most of the tissues evaluated (Figure 8B). The variable expression patterns of KoNACs in different tissues may indicate that KoNAC genes play different roles during the tissue development process.
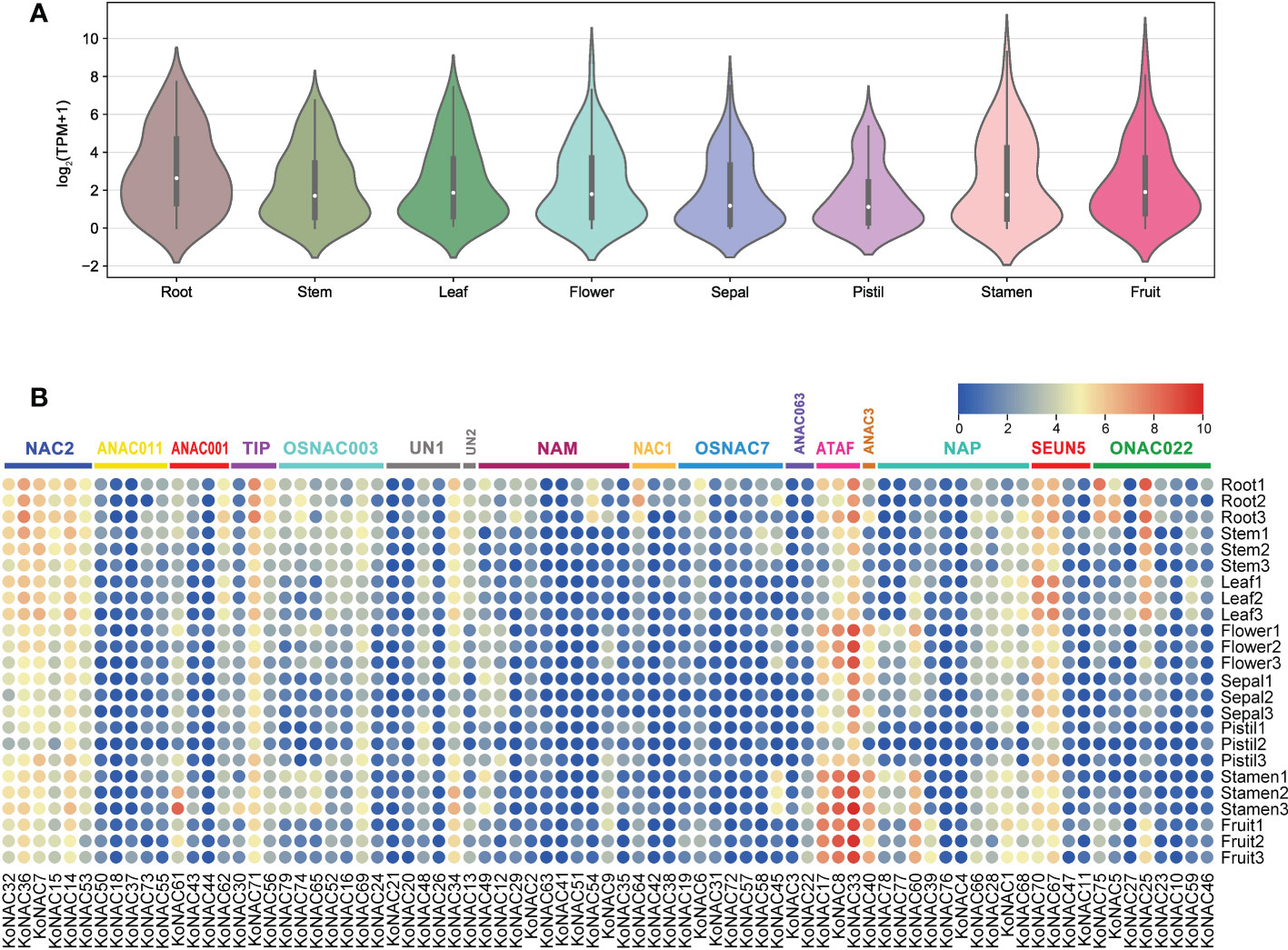
Figure 8 Expression of KoNAC genes in Kandelia obovata. (A) Bean plot of the expression levels of all KoNACs in different tissues. (B) The transcript levels of the KoNAC genes in 8 tissues of K. obovata were investigated based on publicly available transcriptomic data. The color scale shows increasing expression levels from blue to red.
Expression pattern of KoNACs in response to chilling stress
To evaluate the potential roles of KoNACs in response to cold stress, the expression pattern of KoNACs in the leaves of K. obovata was examined after being subjected to 5°C low temperature stress based on transcriptome data from NCBI database. In general, the average expression level of KoNACs did not change significantly compared with the control when subjected to the first time cold treatment; however, the second time and fourth time cold treatments significantly increased the expression of KoNACs (Figure 9A). The expression levels of most KoNAC genes, particularly KoNAC5, KoNAC8, KoNAC13, KoNAC17, KoNAC25, KoNAC30, KoNAC33, KoNAC40, KoNAC43, KoNAC46, KoNAC49, KoNAC59, KoNAC68, KoNAC75, and KoNAC79, were significantly upregulated more than twofold after chilling treatment. However, the expression levels of KoNAC16, KoNAC52, KoNAC61, and KoNAC62 decreased with increasing chilling stress treatment time, particularly in the fourth time treatment, and the expression levels were significantly lower than those in the control (Figure 9B).
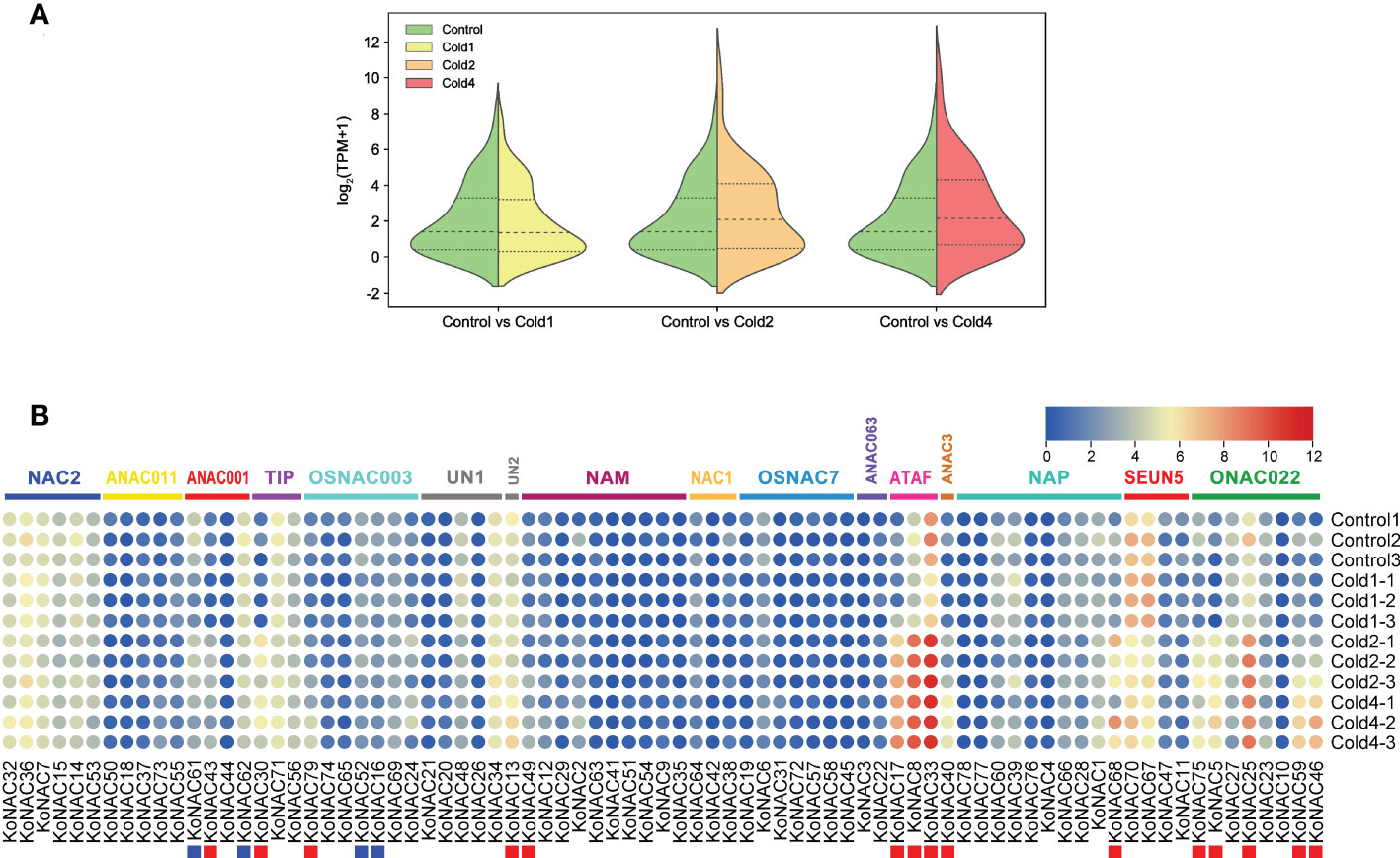
Figure 9 Expression of KoNAC genes in Kandelia obovata. (A) Bean plot of all KoNACs before and after 4°C chilling stress. (B) The transcript levels of KoNAC genes in response to cold stress were investigated based on publicly available transcriptomic data. Ten-month-old seedlings of K. obovata were treated, and the leaves were collected for transcriptome analysis. Cold1, the first time cold treatment, 2/5°C (light/day) for 4 d, then recovery at 22/27°C (light/day) for 15 d; Cold2, the second time cold treatment, continue 2/5°C (light/day) for 4 d, then recovery at 22/27°C (light/day) for 15 d; Cold3, the third time treatment, continue 2/5°C (light/day) for 4 d, then recovery at 22/27°C (light/day) for 15 d; Cold4, the fourth time treatment, continue 2/5°C (light/day) for 4 d. The red and blue squares indicate significantly enhanced and repressed expression under chilling stress, respectively (p < 0.05).
To identify the potential KoNACs that are important to low-termpearture tolerance, nine upregulated KoNACs were chosen, and their relative expression levels were quantified by qRT-PCR. The expression of all the selected KoNAC genes was upregulated when subjected to 4°C stress for 3 h or 12 h compared to the control (Figure 10). Under low-temperature stress, the expression of KoNAC5, KoNAC17, KoNAC25 and KoNAC68 increased initially but then decreased, peaking at 3 h after 4°C treatment, and their expression levels were significantly higher than those in the control. However, the expression levels of KoNAC8, KoNAC30, KoNAC40, KoNAC46 and KoNAC59 continued to increase after 4°C treatment and peaked at 12 h.
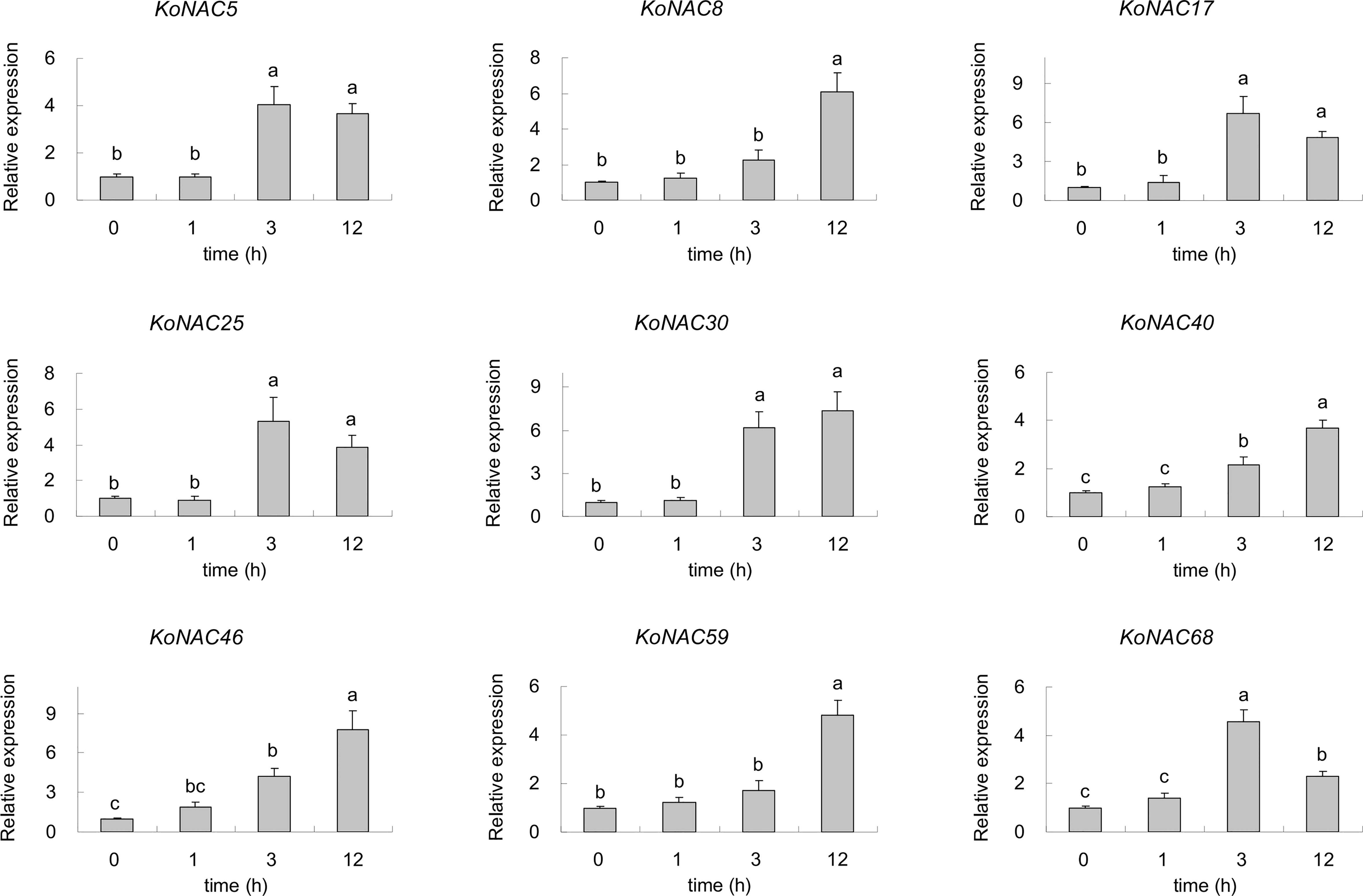
Figure 10 The expression profiles of nine potential KoNACs in response to chilling stress. The relative expression levels of nine KoNACs were measured in plants subjected to a temperature of 4°C for 0, 1, 3, and 12 h. The transcript levels of the selected genes were assessed via qRT-PCR and normalized to 18S rRNA levels. The error bars represent the standard errors. The values with the same letter are not significantly different according to Duncan’s multiple range test (P < 0.05; n =3).
Significantly coexpressed gene pairs between KoNACs and mRNAs
Genome-wide gene expression profiling of KoNACs and mRNAs from the leaves of K. obovata treated with low temperature was conducted to find coexpressed gene pairs. The possible target mRNAs were determined using the Pearson correlation test for all abovementioned KoNAC genes whose expression significantly increased after chilling treatment. There was a correlation between 284 significantly expressed mRNAs and 13 KoNACs (|PCC| > 0.95; P < 0.001); all 381 coexpressed pairs were positively correlated except for KoNAC8-GWHTACBH001948 and KoNAC33-GWHTACBH018122, which were negatively correlated (Supplementary Figure S2; Supplementary Table S8). Overall, our results implied that the KoNACs might regulate the responses of these possible target genes to low-temperature stress mainly in a positive manner.
Discussion
Identification and evolutionary analysis of KoNAC genes
Many studies have reported that low temperatures are vital to the survival and distribution of mangroves (Krauss et al., 2008; Sippo et al., 2018; Fei et al., 2021a). NAC proteins are a class of TFs particular to plants that are vital for multiple plants during growth and development processes as well as in various biotic or abiotic defense responses (Nuruzzaman et al., 2013). An increasing number of NAC genes have been identified with the ongoing completion of genome sequencing in different species, such as A. thaliana, rice, tomato, barley, wheat, cotton, and potato (Yuan et al., 2019). Nevertheless, few NAC TFs engaged in the chilling stress response have been researched in K. obovata thus far. In the present study, the characteristics of KoNAC genes at the genome level were investigated, and their tissue expression profiles and responses to chilling stress were investigated. The goal of our study was to screen candidate KoNAC genes potentially engaged in the low-temperature response and provide a basis for the further elucidation of the functions and molecular mechanisms of KoNAC genes in response to chilling stress.
In the current study, seventy-nine KoNAC genes were identified in total in the whole genome of K. obovata, whose genome size was 180 Mb. It has been reported that the genomes of A. thaliana, O. sativa and Fagopyrum tataricum, with sizes of 125 Mb, 466 Mb and 489 Mb, contain 117, 151 and 80 NAC genes, respectively (Liu et al., 2019). The numbers of NAC genes are nearly the same in K. obovata and F. tataricum; however, the genome size of F. tataricum is more than 2.7 times that of K. obovata, so there is no correlation between the number of NAC genes in various plants and the size of their genome. Cannon et al. (2004) proposed that gene duplication can be considered the primary driving force of gene family expansion and evolution, where the main duplication patterns are segmental duplication and tandem duplication. It has been reported that tandem duplications play important roles in the expansion of the NAC family in O. sativa (Nuruzzaman et al., 2010), Solanum tuberosum (Singh et al., 2013) and Eucalyptus grandis (Hussey et al., 2015). Segmental duplications appear to be more dominant in the expansion of the NAC family, as observed in Panicum miliaceum (Shan et al., 2020), Raphanus sativus (Huang et al., 2022), and Vigna radiate (Tariq et al., 2022).
Our results indicated that 49 segmental duplications and 4 tandem duplications were detected in 79 KoNAC genes, which implied that segmental duplication was the main driving force for the expansion of the KoNAC gene family, similar to findings in V. radiate (Tariq et al., 2022). It was reported that K. obovata has undergone two polyploidization events: an ancient polyploidization event shared with most eudicots (γ-event) and a recent whole-genome duplication (WGD)/segmental duplication shared with other Rhizophoreae plants (Hu et al., 2020). However, larger segmental duplications occupy much of the A. thaliana genome, in which at least four large-scale duplication events occurred during the formative period in the diversification of angiosperms (100 to 200 million years ago) (Vision et al., 2000). This may be the reason why the Arabidopsis genome is smaller but has more NAC members than that of K. obovata.
The unrooted NJ tree based on NAC protein sequences of K. obovata and A. thaliana was constructed to explain the phylogenetic relationships, and all 79 KoNAC genes were divided into 16 subgroups according to their sequence homology with A. thaliana (Ooka et al., 2003). However, our results were not consistent with findings in other plant species, such as Dactylis glomerata (14 subgroups) (Yang et al., 2021), Fagopyrum tataricum (15 subgroups) (Liu et al., 2019), Chenopodium quinoa (14 subgroups) (Li et al., 2019), and Theobroma cacao (12 subgroups) (Shen et al., 2020), suggesting that NAC proteins exhibit diversity in different species. These differences may be associated with the fact that the above four species are all terrestrial plants, while K. obovata is a mangrove plant. Thus, more NAC transcription factors may have evolved over a long period of time to allow the mangrove species to adapt to harsher habitats characterized by high salinity, submergence, and hypoxia.
The number of introns in the KoNAC genes varied from 1 to 7, similar to numbers reported in many plant species (Liu et al., 2019; Shan et al., 2020; Song et al., 2022). In general, the deletion or insertion of introns can result in a loss of gene function. Interestingly, the intron numbers of KoNAC genes were significantly negatively correlated with the percent changes in their expression levels after the first cold treatment (Supplementary Figure S3). Jeffares et al. (2008) showed that genes with rapidly changing expression levels in response to stress present significantly lower intron densities in A. thaliana; perhaps the splicing of two or more introns requires more time than transcription and becomes rate limiting. Our research is consistent with the results of Jeffares, indicating that more attention should be given to the KoNACs with fewer introns if the research goal is to focus on genes that are immediately responsive to environmental stress.
Expression profiles and functional prediction of KoNAC genes
As one of the largest families of TFs, NACs act as positive or negative regulators that regulate the responses of plant species such as A. thaliana and rice to biotic and abiotic stresses (Ooka et al., 2003; Nuruzzaman et al., 2013). In the present study, expression analyses revealed that KoNAC7, KoNAC14, KoNAC33, KoNAC67, and KoNAC71 showed constitutively high expression levels in root, stem, leaf, pistil, stamen, flower, sepal, and fruit, implying that they may play important roles in different tissues of the plant; this result is similar to the finding that FtNAC14 from F. tataricum was most abundantly expressed in both stem and leaf, FtNAC43, FtNAC46 and FtNAC58 were highly expressed in the root; however, FtNAC70 showed the highest expression level in the flower (Liu et al., 2019). Moreover, KoNAC45 and KoNAC54 specifically displayed high expression in reproductive or vegetative organs, showing high expression in stamens and roots, respectively; a similar result was found in Picea wilsonii, in which PwNAC30 exhibits tissue-specific expression, mainly in roots, leaves, pistils, and pods but not in stamens (Liang et al., 2020). Taken together, our results suggested the differential and specific roles of KoNAC family members during K. obovata growth and development.
Many studies have been conducted to clarify the function of NAC TFs in reactions to various environmental stresses, such as salinity, drought, heat and flooding (Fang et al., 2015; Yuan et al., 2020; Song et al., 2022). In the present study, the average expression level of the KoNACs was highest in roots, and a large number of KoNAC genes showed high levels of expression, such as KoNAC25, KoNAC33, KoNAC36, KoNAC64, KoNAC67, KoNAC70, KoNAC71, and KoNAC75 (Figure 8B). The plant root is an organ that directly contacts the soil and is the first line of defense for maintaining plant productivity under soil abiotic stress (Suralta et al., 2018). Liu et al. (2016) reported that the overexpression of A. thaliana ATAF1 (AT1g01720) in rice increased its root length, shoot height and tolerance to salt. KoNAC33 is an ortholog of ATAF1 (Figure 1), and K. obovata survives in harsh environments characterized by conditions such as submergence, hypoxia, and salinity (Fei et al., 2021b; Nizam et al., 2022); thus, the high expression levels of KoNACs may alleviate the effects of environmental stresses. Further studies on KoNACs are needed to determine their functions and regulatory mechanisms.
RNA-seq results indicated that the expression levels of some KoNACs in the leaves of K. obovata changed significantly under low-temperature stress. Among these KoNACs, fifteen members were found to be upregulated more than twofold, but only 4 were downregulated. Similar outcomes were discovered in Pyrus pyrifolia (Ahmad et al., 2018), P. bretschneideri (Gong et al., 2019), and Prunus mume (Zhuo et al., 2018), meaning that NACs might act as positive or negative regulators. Overexpression of LlNAC2 (from Lilium lancifolium) and CaNAC035 (from Capsicum annuum) in A. thaliana enhances tolerance to chilling stress (Yong et al., 2019; Zhang et al., 2020), and the same function was also reported in a study of the SlNAC1 gene from Suaeda liaotungensis (Li et al., 2014). Previous studies have offered convincing support for the phylogenetic analysis-based prediction of the functions of several TF gene family members (Yuan et al., 2020). Both LlNAC2 and CaNAC035 belong to the ATAF subfamily, and these genes are homologous to KoNAC8, KoNAC17, and KoNAC33; moreover, SlNAC1 is clustered into the TIP subgroup and is homologous to KoNAC30, indicating that KoNAC8, KoNAC17, KoNAC30, and KoNAC33 may play an important role in the response to chilling stress in K. obovata.
In PCC analysis, a total of 10 WRKY, 4 ERF, 1 GATA, 2 MYB-related proteins and 2 zinc finger TFs were found to be positively correlated with the expression of NACs, suggesting that a variety of TFs were involved in the adaptation process when K. obovata was subjected to low-temperature stress. Recently, Fei et al. (2022) reported that KoWRKY40 was highly induced in leaves and roots under 5°C cold stress, ectopic overexpression of the KoWRKY40 gene in A. thaliana significantly enhanced the low-temperature tolerance of transgenic plants and increased the expression of antioxidant enzyme and COR genes. KoWRKY40 is highly homologous to GWHTACBH004052 in this study, which is significantly and positively correlated with the expression of KoNAC25, KoNAC8 and KoNAC33. In addition, Sun et al. (2019) showed that the transcription factor VaERF092 regulates the expression of VaWRKY33 from Vitis amurensis by binding to the promoter GCC-box of VaWRKY33, leading to the enhancement of transgenic grape calli to cold stress. Therefore, further studies conducted to elucidate the precise regulatory mechanisms of TF genes in response to chilling stress should consider the interactions between TFs above.
Conclusion
A total of 79 KoNAC genes were systematically identified and characterized in K. obovata, and their expression patterns in eight tissues and their expression profiles under low temperature were analyzed. These TFs could be categorized into 16 subgroups based on the phylogenetic tree constructed with NAC proteins of A. thaliana. Gene chromosomal location and synteny analysis showed that segmental duplication was the main driving force of the expansion of KoNAC genes. RNA-seq data indicated tissue-specific expression of some NACs, and 15 members were upregulated significantly in response to cold stress, but only 4 were downregulated. The KoNACs with significantly changed expression levels may be candidate genes for the genetic engineering of K. obovata with enhanced stress tolerance. Our research provides systematic information about KoNAC genes and will facilitate more functional studies of prospective KoNAC members in response to abiotic stress.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: https://bigd.big.ac.cn/gsa/browse/CRA002395.
Author contributions
ZD and JL designed the research study. ZD, SY, DY, YZ, and ZC performed the experiments. ZD, YT, and WS analyzed the data. ZD and JL wrote the manuscript. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the Public Welfare Project of Science Technology Department of Zhejiang Province (LGN21C160007), the Science and Technology Project of Department of Natural Resources of Zhejiang Province (2021-43), Taizhou Science and Technology Project (22nya05), and the Project of Yuhuan Municipal Bureau of Natural Resources and Planning (BY-YHGQCG-2021-04).
Conflict of interest
Author ZC was employed by Taizhou Circular Economy Development Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1048822/full#supplementary-material
Supplementary Figure 1 | cis-acting elements in each KoNAC. The distribution of 20 cis-acting elements in the 2000 bp upstream promoter region is shown. The different types of cis-acting elements are represented by different colors, as shown on the right.
Supplementary Figure 2 | Coexpression network of KoNACs with other genes in Kandelia obovata under chilling stress. The coexpression network was established between 13 KoNACs and 284 significantly expressed mRNAs whose absolute Pearson correlation coefficient values were equal to or greater than 0.95. Within this coexpression network, 379 and 2 pairs were positively and negatively correlated, respectively. The yellow circles represent KoNAC genes, while the blue rectangles represent the coexpressed genes. The solid gray lines represent positive correlations, while the dashed green lines represent negative correlations.
Supplementary Figure 3 | The percent change in the expression levels of KoNACs under the first cold treatment is negatively correlated with the number of introns. The percent change in the expression levels of KoNACs under the first cold treatment was calculated with the formula (E1-E0)*100%/E0. E1 and E0 indicate the expression levels of KoNACs under the first cold treatment and in the control, respectively.
References
Ahmad, M., Yan, X., Li, J., Yang, Q., Jamil, W., Teng, Y., et al. (2018). Genome wide identification and predicted functional analyses of NAC transcription factors in Asian pears. BMC Plant Biol. 18, 214. doi: 10.1186/s12870-018-1427-x
Bardou, R., Parker, J. D., Feller, I. C., Cavanaugh, K. C. (2020). Variability in the fundamental versus realized niches of north American mangroves. J. Biogeography. 48, 160–175. doi: 10.1111/jbi.13990
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4, 10. doi: 10.1186/1471-2229-4-10
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, L., Wang, W., Li, Q. Q., Zhang, Y., Yang, S., Osland, M. J., et al. (2017). Mangrove species' responses to winter air temperature extremes in China. Ecosphere. 8, e01865. doi: 10.1002/ecs2.1865
Diao, P., Chen, C., Zhang, Y., Meng, Q., Lv, W., Ma, N. (2020). The role of NAC transcription factor in plant cold response. Plant Signaling Behavior. 15, 1785668. doi: 10.1080/15592324.2020.1785668
Dong, Y., Tang, M., Huang, Z., Song, J., Xu, J., Ahammed, G. J., et al. (2022). The miR164a-NAM3 module confers cold tolerance by inducing ethylene production in tomato. Plant J. 111, 440–456. doi: 10.1111/tpj.15807
Du, Z., Li, J. (2019). Expression, purification and molecular characterization of a novel transcription factor KcCBF3 from Kandelia candel. Protein Expression Purification. 153, 26–34. doi: 10.1016/j.pep.2018.08.006
Du, Z., You, S., Zhao, X., Xiong, L., Li, J. (2022). Genome-wide identification of WRKY genes and their responses to chilling stress in Kandelia obovata. Front. Genet. 13, 875316. doi: 10.3389/fgene.2022.875316
Fang, Y., Liao, K., Du, H., Xu, Y., Song, H., Li, X., et al. (2015). A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J. Exp. Botany. 66, 6803–6817. doi: 10.1093/jxb/erv386
Fei, J., Wang, Y.-S., Cheng, H., Sun, F.-L., Sun, C.-C. (2021a). Comparative physiological and proteomic analyses of mangrove plant Kandelia obovata under cold stress. Ecotoxicology. 30, 1826–1840. doi: 10.1007/s10646-021-02483-6
Fei, J., Wang, Y.-S., Cheng, H., Su, Y.-B., Zhong, Y., Zheng, L. (2021b). Cloning and characterization of KoOsmotin from mangrove plant Kandelia obovata under cold stress. BMC Plant Biol. 21, 10. doi: 10.1186/s12870-020-02746-0
Fei, J., Wang, Y.-S., Cheng, H., Su, Y.-B., Zhong, Y.-J., Zheng, L. (2022). The Kandelia obovata transcription factor KoWRKY40 enhances cold tolerance in transgenic Arabidopsis. BMC Plant Biol. 22, 274. doi: 10.1186/s12870-022-03661-2
Fei, J., Wang, Y.-S., Jiang, Z.-Y., Cheng, H., Zhang, J.-D. (2015). Identification of cold tolerance genes from leaves of mangrove plant Kandelia obovata by suppression subtractive hybridization. Ecotoxicology 24, 1686–1696. doi: 10.1007/s10646-015-1486-9
Gong, X., Zhao, L., Song, X., Lin, Z., Gu, B., Yan, J., et al. (2019). Genome-wide analyses and expression patterns under abiotic stress of NAC transcription factors in white pear (Pyrus bretschneideri). BMC Plant Biol. 19, 161. doi: 10.1186/s12870-019-1760-8
Guo, W., Wu, H., Zhang, Z., Yang, C., Hu, L., Shi, X., et al. (2017). Comparative analysis of transcriptomes in rhizophoraceae provides insights into the origin and adaptive evolution of mangrove plants in intertidal environments. Front. Plant Sci. 8, 795. doi: 10.3389/fpls.2017.00795
Huang, Y., Cui, L., Chen, W., Liu, Z., Yuan, W., Zhu, F., et al. (2022). Comprehensive analysis of NAC transcription factors and their expressions during taproot coloration in radish (Raphanus sativus L.). Scientia Horticulturae. 299, 111047. doi: 10.1016/j.scienta.2022.111047
Hussey, S. G., Saïdi, M. N., Hefer, C. A., Myburg, A. A., Grima-Pettenati, J. (2015). Structural, evolutionary and functional analysis of the NAC domain protein family in Eucalyptus. New Phytologist. 206, 1337–1350. doi: 10.1111/nph.13139
Hu, M.-J., Sun, W.-H., Tsai, W.-C., Xiang, S., Lai, X.-K., Chen, D.-Q., et al. (2020). Chromosome-scale assembly of the Kandelia obovata genome. Horticulture Res. 7, 75. doi: 10.1038/s41438-020-0300-x
Jeffares, D. C., Penkett, C. J., Bähler, J. (2008). Rapidly regulated genes are intron poor. Trends Genet. 24, 375–378. doi: 10.1016/j.tig.2008.05.006
Jiang, J., Hou, R., Yang, N., Li, L., Deng, J., Qin, G., et al. (2021). Physiological and TMT-labeled proteomic analyses reveal important roles of sugar and secondary metabolism in Citrus junos under cold stress. J. Proteomics. 237, 104145. doi: 10.1016/j.jprot.2021.104145
Jin, C., Li, K.-Q., Xu, X.-Y., Zhang, H.-P., Chen, H.-X., Chen, Y.-H., et al. (2017). A novel NAC transcription factor, PbeNAC1, of Pyrus betulifolia confers cold and drought tolerance via interacting with PbeDREBs and activating the expression of stress-responsive genes. Front. Plant Science. 8, 1049. doi: 10.3389/fpls.2017.01049
Krauss, K. W., Lovelock, C. E., McKee, K. L., López-Hoffman, L., Ewe, S. M., Sousa, W. P. (2008). Environmental drivers in mangrove establishment and early development: A review. Aquat. Botany. 89, 105–127. doi: 10.1016/j.aquabot.2007.12.014
Liang, K.-h., Wang, A.-b., Yuan, Y.-h., Miao, Y.-h., Zhang, L.-y. (2020). Picea wilsonii NAC transcription factor PwNAC30 negatively regulates abiotic stress tolerance in transgenic Arabidopsis. Plant Mol. Biol. Reporter. 38, 554–571. doi: 10.1007/s11105-020-01216-z
Li, F., Guo, X., Liu, J., Zhou, F., Liu, W., Wu, J., et al. (2019). Genome-wide identification, characterization, and expression analysis of the NAC transcription factor in Chenopodium quinoa. Genes. 10, 500. doi: 10.3390/genes10070500
Liu, M., Ma, Z., Sun, W., Huang, L., Wu, Q., Tang, Z., et al. (2019). Genome-wide analysis of the NAC transcription factor family in tartary buckwheat (Fagopyrum tataricum). BMC Genomics 20, 113. doi: 10.1186/s12864-019-5500-0
Liu, Y., Sun, J., Wu, Y. (2016). Arabidopsis ATAF1 enhances the tolerance to salt stress and ABA in transgenic rice. J. Plant Res. 129, 955–962. doi: 10.1007/s10265-016-0833-0
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods. 25, 402–408. doi: 10.1006/meth.2001.1262
Li, X.-l., Yang, X., Hu, Y.-x., Yu, X.-d., Li, Q.-l. (2014). A novel NAC transcription factor from Suaeda liaotungensis k. enhanced transgenic Arabidopsis drought, salt, and cold stress tolerance. Plant Cell Rep. 33, 767–778. doi: 10.1007/s00299-014-1602-y
Lu, W.-X., Zhang, B.-H., Zhang, Y.-Y., Yang, S.-C. (2022). Differentiation of cold tolerance in an artificial population of a mangrove species, Kandelia obovata, is associated with geographic origins. Front. Plant Science. 12, 695746. doi: 10.3389/fpls.2021.695746
Manna, M., Thakur, T., Chirom, O., Mandlik, R., Deshmukh, R., Salvi, P. (2021). Transcription factors as key molecular target to strengthen the drought stress tolerance in plants. Physiologia Plantarum. 172, 847–868. doi: 10.1111/ppl.13268
Nakashima, K., Takasaki, H., Mizoi, J., Shinozaki, K., Yamaguchi-Shinozaki, K. (2012). NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 97–103. doi: 10.1016/j.bbagrm.2011.10.005
Nakashima, K., Tran, L.-S.P., Van Nguyen, D., Fujita, M., Maruyama, K., Todaka, D., et al (2007). Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 51, 617–630. doi: 10.1111/j.1365-313X.2007.03168.x
Nizam, A., Meera, S. P., Kumar, A. (2022). Genetic and molecular mechanisms underlying mangrove adaptations to intertidal environments. iScience. 25, 103547. doi: 10.1016/j.isci.2021.103547
Nuruzzaman, M., Manimekalai, R., Sharoni, A. M., Satoh, K., Kondoh, H., Ooka, H., et al. (2010). Genome-wide analysis of NAC transcription factor family in rice. Gene. 465, 30–44. doi: 10.1016/j.gene.2010.06.008
Nuruzzaman, M., Sharoni, A. M., Kikuchi, S. (2013). Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 4, 248. doi: 10.3389/fmicb.2013.00248
Olsen, A. N., Ernst, H. A., Leggio, L. L., Skriver, K. (2005). NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Science. 10, 79–87. doi: 10.1016/j.tplants.2004.12.010
Ooka, H., Satoh, K., Doi, K., Nagata, T., Otomo, Y., Murakami, K., et al. (2003). ). comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10, 239–247. doi: 10.1093/dnares/10.6.239
Osland, M. J., Enwright, N., Day, R. H., Doyle, T. W. (2013). Winter climate change and coastal wetland foundation species: Salt marshes vs. mangrove forests in the southeastern united states. Global Change Biol. 19, 1482–1494. doi: 10.1111/gcb.12126
Peng, Y.-L., Wang, Y.-S., Fei, J., Sun, C.-C. (2020). Isolation and expression analysis of two novel c-repeat binding factor (CBF) genes involved in plant growth and abiotic stress response in mangrove Kandelia obovata. Ecotoxicology 29, 718–725. doi: 10.1007/s10646-020-02219-y
Peng, Y.-L., Wang, Y.-S., Fei, J., Sun, C.-C., Cheng, H. (2015). Ecophysiological differences between three mangrove seedlings (Kandelia obovata, Aegiceras corniculatum, and Avicennia marina) exposed to chilling stress. Ecotoxicology. 24, 1722–1732. doi: 10.1007/s10646-015-1488-7
Shan, Z., Jiang, Y., Li, H., Guo, J., Dong, M., Zhang, J., et al. (2020). Genome-wide analysis of the NAC transcription factor family in broomcorn millet (Panicum miliaceum L.) and expression analysis under drought stress. BMC Genomics 21, 96. doi: 10.1186/s12864-020-6479-2
Shao, H., Wang, H., Tang, X. (2015). NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Front. Plant Sci. 6, 902. doi: 10.3389/fpls.2015.00902
Shen, Z.-J., Qin, Y.-Y., Luo, M.-R., Li, Z., Ma, D.-N., Wang, W.-H., et al. (2021). Proteome analysis reveals a systematic response of cold-acclimated seedlings of an exotic mangrove plant Sonneratia apetala to chilling stress. J. Proteomics. 248, 104349. doi: 10.1016/j.jprot.2021.104349
Shen, S., Zhang, Q., Shi, Y., Sun, Z., Zhang, Q., Hou, S., et al. (2020). Genome-wide analysis of the NAC domain transcription factor gene family in Theobroma cacao. Genes. 11, 35. doi: 10.3390/genes11010035
Sheue, C.-R., Liu, H.-Y., Yong, J. W. H. (2003). Kandelia obovata (Rhizophoraceae), a new mangrove species from Eastern Asia. Taxon. 52, 287–294. doi: 10.2307/3647398
Singh, S., Koyama, H., Bhati, K. K., Alok, A. (2021). The biotechnological importance of the plant-specific NAC transcription factor family in crop improvement. J. Plant Res. 134, 475–495. doi: 10.1007/s10265-021-01270-y
Singh, A. K., Sharma, V., Pal, A. K., Acharya, V., Ahuja, P. S. (2013). Genome-wide organization and expression profiling of the NAC transcription factor family in potato (Solanum tuberosum L.). DNA Res. 20, 403–423. doi: 10.1093/dnares/dst019
Sippo, J. Z., Lovelock, C. E., Santos, I. R., Sanders, C. J., Maher, D. T. (2018). Mangrove mortality in a changing climate: An overview. Estuarine Coast. Shelf Sci. 215, 241–249. doi: 10.1016/j.ecss.2018.10.011
Song, Y., Cui, H., Shi, Y., Xue, J., Ji, C., Zhang, C., et al. (2020). Genome-wide identification and functional characterization of the Camelina sativa WRKY gene family in response to abiotic stress. BMC Genomics 21, 786. doi: 10.1186/s12864-020-07189-3
Song, H., Liu, Y., Dong, G., Zhang, M., Wang, Y., Xin, J., et al. (2022). Genome-wide characterization and comprehensive analysis of NAC transcription factor family in Nelumbo nucifera. Front. Genet. 13, 901838. doi: 10.3389/fgene.2022.901838
Sun, X., Zhang, L., Wong, D. C. J., Wang, Y., Zhu, Z., Xu, G., et al. (2019). The ethylene response factor VaERF092 from amur grape regulates the transcription factor VaWRKY33, improving cold tolerance. Plant J 99, 988–1002. doi: 10.1111/tpj.14378
Suralta, R. R., Kano-Nakata, M., Niones, J. M., Inukai, Y., Kameoka, E., Tran, T. T., et al. (2018). Root plasticity for maintenance of productivity under abiotic stressed soil environments in rice: Progress and prospects. Field Crops Res. 220, 57–66. doi: 10.1016/j.fcr.2016.06.023
Su, W., Ye, C., Zhang, Y., Hao, S., Li, Q. Q. (2019). Identification of putative key genes for coastal environments and cold adaptation in mangrove Kandelia obovata through transcriptome analysis. Sci. Total Environment. 681, 191–201. doi: 10.1016/j.scitotenv.2019.05.127
Tariq, R., Hussain, A., Tariq, A., Khalid, M. H. B., Khan, I., Basim, H., et al. (2022). Genome-wide analyses of the mung bean NAC gene family reveals orthologs, co-expression networking and expression profiling under abiotic and biotic stresses. BMC Plant Biol. 22, 343. doi: 10.1186/s12870-022-03716-4
Trégarot, E., Caillaud, A., Cornet, C. C., Taureau, F., Catry, T., Cragg, S. M., et al. (2021). Mangrove ecological services at the forefront of coastal change in the French overseas territories. Sci. Total Environment. 763, 143004. doi: 10.1016/j.scitotenv.2020.143004
Vision, T. J., Brown, D. G., Tanksley, S. D. (2000). The origins of genomic duplications in Arabidopsis. Science. 290, 2114–2117. doi: 10.1126/science.290.5499.2114
Voorrips, R. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Heredity. 93, 77–78. doi: 10.1093/jhered/93.1.77
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Wang, H., Wang, H., Shao, H., Tang, X. (2016). Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Front. Plant Science. 7, 67. doi: 10.3389/fpls.2016.00067
Wang, S.-M., Wang, Y.-S., Su, B.-Y., Zhou, Y.-Y., Chang, L.-F., Ma, X.-Y., et al. (2022). Ecophysiological responses of five mangrove species (Bruguiera gymnorrhiza, Rhizophora stylosa, Aegiceras corniculatum, Avicennia marina, and Kandelia obovata) to chilling stress. Front. Mar. Science. 9, 846566. doi: 10.3389/fmars.2022.846566
Wei, M.-Y., Li, H., Zhong, Y.-H., Shen, Z.-J., Ma, D.-N., Gao, C.-H., et al. (2022). Transcriptomic analyses reveal the effect of nitric oxide on the lateral root development and growth of mangrove plant Kandelia obovata. Plant Soil. 472, 543–564. doi: 10.1007/s11104-021-05271-7
Wu, M., He, Z., Fung, S., Cao, Y., Guan, D., Peng, Y., et al. (2020). Species choice in mangrove reforestation may influence the quantity and quality of long-term carbon sequestration and storage. Sci. Total Environment. 714, 136742. doi: 10.1016/j.scitotenv.2020.136742
Wu, Y., Müller, M., Bai, T., Yao, S., Gailing, O., Liu, Z. (2019). Transcriptome profiling in Camellia japonica var. decumbens for the discovery of genes involved in chilling tolerance under cold stress. Ann. For. Res. 62, 51–68. doi: 10.15287/afr.2018.1311
Yang, Z., Nie, G., Feng, G., Han, J., Huang, L., Zhang, X. (2021). Genome-wide identification, characterization, and expression analysis of the NAC transcription factor family in orchardgrass (Dactylis glomerata L.). BMC Genomics 22, 178. doi: 10.1186/s12864-021-07485-6
Yong, Y., Zhang, Y., Lyu, Y. (2019). A stress-responsive NAC transcription factor from tiger lily (LlNAC2) interacts with LlDREB1 and LlZHFD4 and enhances various abiotic stress tolerance in Arabidopsis. Int. J. Mol. Sci. 20, 3225. doi: 10.3390/ijms20133225
Yoo, S.-D., Cho, Y.-H., Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Yuan, C., Li, C., Lu, X., Zhao, X., Yan, C., Wang, J., et al. (2020). Comprehensive genomic characterization of NAC transcription factor family and their response to salt and drought stress in peanut. BMC Plant Biol. 20, 454. doi: 10.1186/s12870-020-02678-9
Yuan, X., Wang, H., Cai, J., Li, D., Song, F. (2019). NAC transcription factors in plant immunity. Phytopathol. Res. 1, 3. doi: 10.1186/s42483-018-0008-0
Zeng, Z., Lu, J., Wu, D., Zuo, R., Li, Y., Huang, H., et al. (2020). Poly(ADP-ribose) glycohydrolase silencing-mediated H2B expression inhibits benzo(a)pyrene-induced carcinogenesis. Environ. Toxicology. 36, 291–297. doi: 10.1002/tox.23034
Zhang, H., Ma, F., Wang, X., Liu, S., Saeed, U. H., Hou, X., et al. (2020). Molecular and functional characterization of CaNAC035, an NAC transcription factor from pepper (Capsicum annuum L.). Front. Plant Science. 11, 14. doi: 10.3389/fpls.2020.00014
Keywords: chilling stress, expression profiles, gene duplication, Kandelia obovata, NAC gene family, phylogenetic analysis
Citation: Du Z, You S, Yang D, Tao Y, Zhu Y, Sun W, Chen Z and Li J (2022) Comprehensive analysis of the NAC transcription factor gene family in Kandelia obovata reveals potential members related to chilling tolerance. Front. Plant Sci. 13:1048822. doi: 10.3389/fpls.2022.1048822
Received: 21 September 2022; Accepted: 03 November 2022;
Published: 17 November 2022.
Edited by:
Xiyin Wang, North China University of Science and Technology, ChinaReviewed by:
Yuhui Liu, Gansu Agricultural University, ChinaJin Zhao, Agricultural University of Hebei, China
Copyright © 2022 Du, You, Yang, Tao, Zhu, Sun, Chen and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junmin Li, bGlqbXR6Y0AxMjYuY29t
 Zhaokui Du
Zhaokui Du Shixian You2
Shixian You2 Yutian Tao
Yutian Tao Yunxiao Zhu
Yunxiao Zhu Junmin Li
Junmin Li