
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 20 October 2022
Sec. Plant Bioinformatics
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1047090
This article is part of the Research Topic Cucurbitaceae: Multi-omics, Functional Analysis, and Molecular Breeding View all 15 articles
 Jinqiang Yan1,2
Jinqiang Yan1,2 Bin Liu3
Bin Liu3 Zhenqiang Cao1
Zhenqiang Cao1 Lin Chen1,2
Lin Chen1,2 Zhaojun Liang1
Zhaojun Liang1 Min Wang1,2
Min Wang1,2 Wenrui Liu1,2
Wenrui Liu1,2 Yu'e Lin1*
Yu'e Lin1* Biao Jiang1,2*
Biao Jiang1,2*Photosynthesis, a fundamental process for plant growth and development, is dependent on chloroplast formation and chlorophyll synthesis. Severe disruption of chloroplast structure results in albinism of higher plants. In the present study, we report a cucumber albino alc mutant that presented white cotyledons under normal light conditions and was unable to produce first true leaf. Meanwhile, alc mutant could grow creamy green cotyledons under dim light conditions but died after exposure to normal light irradiation. No chlorophyll and carotenoid were detected in the alc mutant grown under normal light conditions. Using transmission electron microscopy, impaired chloroplasts were observed in this mutant. The genetic analysis indicated that the albino phenotype was recessively controlled by a single locus. Comparative transcriptomic analysis between the alc mutant and wild type revealed that genes involved in chlorophyll metabolism and the methylerythritol 4-phosphate pathway were affected in the alc mutant. In addition, three genes involved in chloroplast development, including two FtsH genes and one PPR gene, were found to have negligible expression in this mutant. The quality of RNA sequencing results was further confirmed by real-time quantitative PCR analysis. We also examined 12 homologous genes from alc mutant in other plant species, but no genetic variation in the coding sequences of these genes was found between alc mutant and wild type. Taken together, we characterized a cucumber albino mutant with albinism phenotype caused by chloroplast development deficiency and this mutant can pave way for future studies on plastid development.
Chloroplasts, which are DNA-containing organelles play crucial roles in attuning plant development and plant interaction with environmental cues. At the time of illumination, the chloroplast develops from proplastids via the process of photomorphogenesis (Pogson et al., 2015; Chan et al., 2016). The chloroplast is the site of photosynthesis and production of hormones (e.g., abscisic acid, jasmonic acid, and salicylic acid, and other major metabolites (Unlu et al., 2014). Its abnormal development or accumulation of pigments inside itself could affect photosynthesis and further disrupt plant growth and biomass yield (Wang et al., 2016; Shi et al., 2017; Xiong et al., 2017; Gotoh et al., 2018).
Leaf color mutation has been widely reported in many plant species. In most of the chlorophyll-less mutants and chlorophyll-deficient mutants, a sudden increase in the production of reactive oxygen species (ROS) was detected after exposure to light conditions (Sakowska et al., 2018; Li et al., 2019). The ROS accumulation induced by excessive light could effectuate oxidative damage in plants, resulting in leaf bleaching or leading to plant death. Natural or induced albino mutants were frequently identified and characterized among different kinds of leaf color mutations, especially in Arabidopsis and rice. For example, some formation of albino mutants was affected by environmental factors and conditionally green-revertible. Disruption of OsABCI8 resulted in the development of albino leaves in rice under continuous rainy days; nevertheless, the leaves gradually turned green following rainy days (Zeng et al., 2017). Mutations in the gene OsTCD5 encoding a monooxygenase, or OsTCD11 encoding the ribosomal small subunit protein S6 in chloroplasts (RPS6) resulted in a temperature-sensitive albino mutant; the leaves displayed albinism at low temperatures but turned green at high temperatures (Wang et al., 2016). Mutation in gene FLN2 encoding fructokinase-like protein2 shows opposite phenotype; the fln2 mutant is albino at high temperatures (Qiu et al., 2018). There are other mutants that only exhibit albino phenotype at certain development stages. Disruption of a pentatricopeptide repeat protein causes an albino phenotype during the seedling stage but the plants are able to turn green during plant growth and development (Su et al., 2012). A mutation in SEEDLING PLASTID DEVELOPMENT1 resulted in albino cotyledons, but these plants appeared similar to the wild type plants once the initial true leaves developed and the seedlings were transferred to the soil (Ruppel et al., 2011).
Somatic albino mutants were also described with expression of genes involved in chlorophyll biosynthesis and chloroplast development affected in mutated leaves (Ma et al., 2018; Lu et al., 2019). Some lethal mutations are caused by deficiency in chloroplast development. Loss of function of DXS1 leads to an albino phenotype in tomato with premature lethality performance (Garcia-Alcazar et al., 2017). A single-nucleotide mutation in the plastid ribosomal protein produces abnormal chloroplasts and causes seedling lethality in rice (Zhao et al., 2016).
Cucumber (Cucumis sativus L.), belonging to the Cucurbitacea family is an important vegetable crop worldwide. Cucumber albino mutation was only reported by Iida and Amano in 1991 (Iida and Amano, 1991), which was induced by irradiation. However, no further studies have been carried out since then. In the present study, we reported a spontaneous mutation of an albino mutant from the cucumber inbred line “g32”, which exhibited white cotyledons and hypocotyl, and died before developing any true leaves. We aim to decipher the potential mechanism of albinism formation in this cucumber albino mutant via cytologic, genetic and transcriptomica characterization.
Cucumber inbred line g32 was used in this study. The inbred line was provided by Vegetable Research Institute, Guangdong Academy of Agricultural Sciences Guangzhou, China. Seeds from a self-pollinated g32 cucumber fruit were soaked in water for 4 h and then kept in the incubator with moderate humidity at 28 °C for germination. Thereafter, germinated seeds were planted in a plug tray either under artificial light irradiation (LED light model, defined as normal light) or dim light in the greenhouse of Vegetable Research Institute, Guangdong Academy of Agricultural Sciences. Dim light treatment was performed in a black plastic bag covered homemade growth chamber. Light intensity of normal light and of dim light was 8000 LUX and 35 LUX, respectively. Seven-day-old seedlings of wild type and alc mutant were used for phenotypic evaluation, fluorescence microscopy, transmission electron microscopy analysis, and high-throughput RNA sequencing.
To determine the chlorophyll a (Chla), chlorophyll b (Chlb), total chlorophyll (Chl), and carotenoid (Car) contents, wild type and alc mutant seedlings grown under normal and dim light conditions were compared. A total of 0.1 g cotyledon from each sample was cut into small pieces and transferred into 10 mL 80% (v/v) acetone and kept in dark until the tissue turned white. Each sample was analyzed in three biological replicates. For each sample, the absorbance was measured at 663, 645, and 470 nm thrice, respectively. Concentrations of Chla, Chlb, Chl, and Car were calculated as described previously (Lichtenthaler, 1987).
For fluorescence microscopy, the abaxial epidermis of cotyledons was used. Chloroplast autofluorescence (red) was captured under Zeiss LSM710 (Germany) confocal microscope with the following settings: excitation at 633 nm, emission at 647–721 nm. Data were analyzed using software ZEN (2010).
Cotyledons of wild type and alc mutant seedlings grown under dim light conditions and normal light irradiation were analyzed by TEM. All the cotyledon samples were cut into 1–2 mm2 sections and fixed in 2.5% glutaraldehyde and 4% paraformaldehyde in phosphate buffer (pH 6.8–7.2) under vacuum for 3 h. After washing with phosphate buffer the samples were fixed in 1% osmium tetroxide (OsO4) for 3 h and again washed with phosphate buffer. The samples were dehydrated through a series of ethanol concentration. The samples were infiltrated with an increasing ratio of Spurr’s resin dilutions [25%, 50%, 75%, and 100% (v/v)] to substitute ethanol, and finally embedded in Spurr’s resin. After cutting, the sections were viewed under a HitachiH-7700 (Hitachi) transmission electron microscope.
Total RNA was extracted from cotyledons of 7-day-old wild type and alc mutant seedlings using Trizol Kit (Promega, USA) according to the manufacturer’s instructions. Extracted RNA was treated with RNase-free DNase I (TaKaRa, Japan) to remove residual DNA. RNA degradation and contamination were monitored on 1% agarose gels. RNA purity was checked using the NanoPhotometer® spectrophotometer (IMPLEN, CA, USA) and RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA).
Cotyledons of wild type and alc mutant seedlings grown under normal light conditions were collected for high-throughput sequencing. Each sample was analyzed in three biological replicates. A total of 1 µg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, USA) following manufacturer’s recommendations. Library preparations were sequenced on an Illumina Novaseq platform and 150 bp paired-end reads were generated. Thereafter, reads with adaptors, reads with unknown bases, as well as low quality reads were removed to generate clean reads. The remaining high-quality clean reads were mapped to Cucumber (Chinese Long) Reference Genome v2 (http://www.cucurbitgenomics.org/organism/2).
The mapped reads of each sample were assembled using StringTie (v1.3.3b) 17 in a reference-based approach, and featureCounts v1.5.0-p3 18 was used to count the reads numbers mapped to each gene. Fragments Per Kilobase of transcript sequence per Millions base pairs (FPKM) of each gene was calculated based on the gene length and read count mapped to this gene. Differential expression analysis of two groups was performed using the DESeq2 R package (1.16.1) 19. The resulting P-values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted P-value (padj) <0.05 and |log2(FoldChange)| > 2 were assigned as differentially expressed genes (DEGs). To functionally annotate the DEGs, Gene Ontology (GO, http://www.geneontology.org/) and Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg/) annotation of the unigenes were analyzed using clusterProfiler R package.
To confirm RNA-seq results, 12 DEGs were selected for qRT-PCR validation. First strand cDNA synthesis was performed using TransScript All-in-One First-Strand cDNA Synthesis SuperMix for qPCR (Transgen, China) with 1 μg of RNA used for high-throughput sequencing. Quantitative RT-PCR was carried out using 0.2 μg cDNA using PerfectStart Green qPCR Supermix (Transgen, China) according to the manufacturer’s instructions. Reactions were performed and analyzed on CFX96 Real-Time PCR Detection System. Three biological replicates and three technical replicates per sample were performed for each gene. Gene expression was normalized against α‐TUBULIN (TUA) gene (Liu et al., 2016). Primers used are listed in Supplementary Table 1.
We observed a few naturally occurring albino seedlings during the reproduction of cucumber inbred line “g32”, which is a southern China type cucumber. The cotyledons of these seedlings were small and entirely white, with short and white hypocotyl and short primary root (Figure 1). The mutant was named as alc (albino cotyledon) thereafter. The mutants dried out and died in a few days after emerging from the substrate without growing any true leaves.

Figure 1 Growth phenotype of alc and wild type from the progenies of a cucumber inbred line g32. (A) Different cotyledons of alc and wild type seedlings. (B) Morphological difference between alc and wild type seedlings.
It was noteworthy that occasionally cotyledons of a few alc mutants that were still inside the shell showed subtle greenish; this led us to propose that the occurrence of greenish cotyledons in alc mutants may be affected by light. Therefore, we performed three sets of experiments to validate this hypothesis (Figure 2A). In the first experiment, all the seeds were grown under dim light conditions at all times. After emerging from the substrate, alc mutants presented creamy green cotyledons with complete white hypocotyl, while wild type seedlings showed yellowish-green cotyledons and hypocotyl (Figure 2B). In the continually dim light environment, both alc mutant and wild-type plants spindled and died without growing first true leaf. In the second experiment, after the seeds emerged from the substrate, they were first allowed to grow under normal light conditions until we were able to distinguish between alc mutant and wild-type plants. We then transferred them to dim light conditions and observed that the cotyledons of alc mutants remained whitish, without turning to cream green (Figures 2C1, C2). In the last experiment, the seeds were grown under dim light conditions until seedlings emerged from substrate, and then they were exposed to normal light condition (Figure 2A3). After 5 h of light exposure, the green color in the cotyledon of alc mutant started to degrade (Figure 2D1). The cotyledon shrunk and dried out after exposure to light for 30 h (Figure 2D2). Therefore, we conclude that alc is a light sensitive albino mutant. Normal light irradiation is a lethal factor for alc mutant and, the damage caused by light is irreversible.
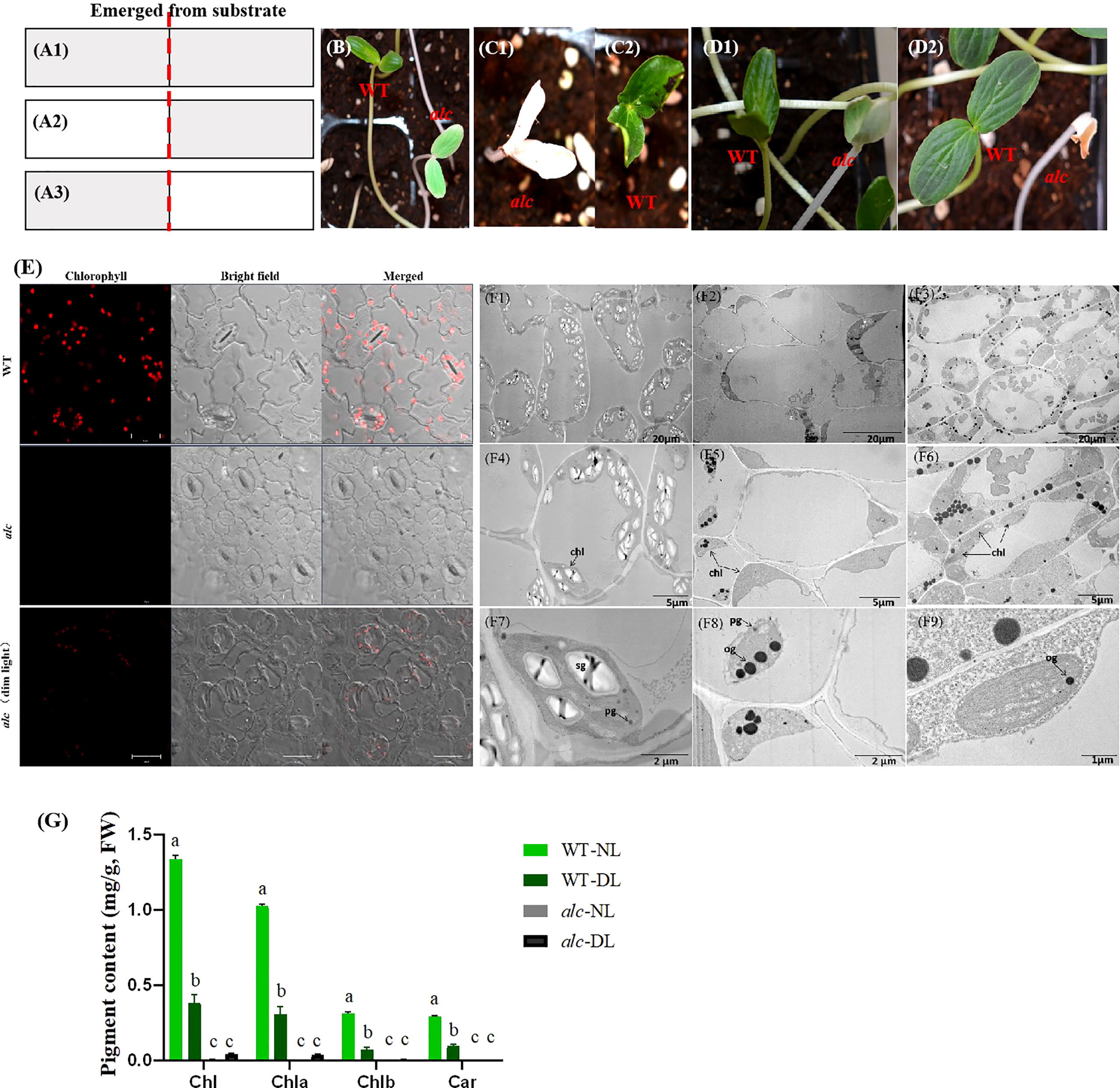
Figure 2 Short-lived chloroplast recovery in alc mutant under dim light condition. (A1–A3) Schematic illustration of light treatments of WT and alc seedlings. A1 Plants were treated with continuous dim light before and after they emerged from substrate. A2 First, plants were treated with normal light irradiation, then after emerging from substrate, they were moved to dim light condition. A3 First, plants were treated under dim light, then after emerging from substrate, they were moved to normal light condition. (B) Phenotype of WT and alc seedlings under indicated condition that described in a1. (C) Phenotype of WT (C1) and alc (C2) seedlings under indicated conditions that described in A2. (D) Phenotype of WT and alc seedlings under indicated conditions that described in A3. D1 Phenotype of WT and alc seedlings exposed to normal light after 5 hours. D2 Phenotype of WT and alc seedlings exposed to normal light after 30 hours. (E) Fluorescence microscopy images of cotyledon abaxial epidermis of wild type, alc mutant cucumber seedlings grown under normal light condition and alc mutant seedlings grown under dim light condition. Bar: 20μm. (F1–F9) Transmission electron microscopy of cotyledons from alc mutant and wild type seedlings. F1, F4 An overview of cotyledon cells of wild type grown under normal light condition. (F2–F5) An overview of cotyledon cells of alc mutant grown under normal light condition. F3, F6 An overview of cotyledon cells of alc mutant grown under dim light condition. (F7–F9) Enlarged views of chloroplast ultrastructure of wild type, alc mutant grown under normal light condition and alc mutant grown under dim light condition, respectively. chl, chloroplast sg, starch granules pg, plastoglobuli og, osmiophilic plastoglobuli. (G) Chlorophyll content and carotenoid content of wild type and alc mutant grown under normal light and dim light conditions. WT-NL, wild type grown under normal light condition. WT-DL, wild type grown under dim light condition alc-NL, alc mutant grown under normal light condition alc-DL, alc mutant grown under dim light condition. Chla, chlorophyll a; Chlb, chlorophyll b; Chl, total chlorophyll.
Most of the albino phenotypes in other plants lack chlorophyll and carotenoids. Thus, we examined the chlorophyll and carotenoid content of cotyledons from both wild type and alc mutant seedlings that grew under normal light and dim light conditions. Fluorescence microscopy revealed that chlorophyll fluorescence in wildtype was more intense than that observed in alc mutants grown under dim light conditions (Figure 2E; presented in red). As expected, no chlorophyll fluorescence was detected in alc mutants that grew under normal light conditions. Chlorophyll content was measured based on the fluorescence intensity (Figure 2G1). No carotenoid was detected in alc mutants grown under normal light conditions; however, a small amount, 0.03 mg/g (fresh weight), was detected in alc mutants grown under dim light conditions (Figure 2G2).
Since most of the chlorophyll content in plants are in chloroplast, we further investigated the chloroplast ultrastructure in the cotyledons of wild type and alc mutant seedlings that grew under both light and dim light conditions using TEM. In the cotyledons of wild type seedlings, we observed numerous well-developed, crescent-shaped chloroplasts with stroma thylakoids, grana thylakoids, starch granules, and plastoglobuli within the membranes Figures 2F1, F4, F7). In contrast, the chloroplasts in the alc mutant decreased dramatically in number and showed abnormal shapes that lacked stroma and grana thylakoids, but contained osmiophilic plastoglobulis in the inner membrane system (Figures 2F2, F5, F8). The alc mutants that grew under dim light conditions comprised stroma thylakoid as well as osmiophilic plastoglobulis (Figures 2F3, F6, F9).
To summarize, the above results indicated that the chloroplast development was impaired in alc mutants grown under normal light conditions during seedling development. Moreover, normal light may be lethal to the alc mutant because it interrupts thylakoid biogenesis, as observed by the presence of thylakoid in the alc mutant grown in dim light conditions but not in those grown under normal light conditions.
As the alc mutant died in the seedling stage, we could not obtain homozygous seeds. Therefore, we considered the mutant parental line “g32” as F1 and used its self-pollinated seeds to investigate the inheritance pattern of the albino phenotype. We planted a total of 123 seeds from the self-pollinated “g32” cucumber and put them under normal light conditions, of which 116 seeds germinated successfully (germination rate 94.3%). Thirty-two and eighty-four germinated seedlings showed albino and wild type phenotype, respectively. Chi-square analysis (χ2 = 0.414, p = 0.520) indicated that the segregation ratio between albino and wild type was 1:3. Therefore, the albino trait is likely controlled by a single recessive nuclear gene.
The transcriptomes of cotyledons from the alc mutant and wild type seedlings were examined by RNA-seq, each with three biological replicates. Overall, 97,869,948 to 123,516,412 clean reads were obtained after filtering low quality reads. After mapping to the cucumber reference genome 9930 v2 (Huang et al., 2009; Li et al., 2011), 21,664 transcripts were identified. High correlation coefficients among the replicates demonstrated the consistency of the transcriptional changes within each sample (Figure 3A). In total, 1,256 genes were upregulated and 1,584 were downregulated in the alc mutant compared to the wild type cotyledons (|log2FC| ≥ 2) (Figure 3B; Supplementary Table 2). Based on the annotation, DEGs were annotated using GO and KEGG pathway to identify the significantly enriched biological processes and pathways between the alc mutant and wild type. In total, 2,175 DEGs were classified into 814 GO terms belonging to three categories: biological process, cellular component, and molecular function (Supplementary Table 3). Cellular carbohydrate biosynthetic process (GO:0034637, p=0.00010) and cellular carbohydrate metabolic process (GO:0044262, p=0.00017) were the most significantly enriched biological processes (Supplementary Figure 1). As shown in Figure 4, 171 and 196 DEGs were identified as transmembrane transport (GO:0055085) and transporter activity (GO:0005215), respectively, which were among the most enriched GO terms. (Supplementary Table 3). A total of, 957 DEGs were assigned to 110 KEGG pathways (Supplementary Table 4), among which, top 20 enriched KEGG pathways are illustrated in Figure 4. Carbon metabolism (KEGG: csv01200), phenylpropanoid biosynthesis (KEGG: csv00940), and starch and sucrose metabolism (KEGG: csv00500) pathways contained 69, 50, and 44 DEGs, respectively (Supplementary Table 4).
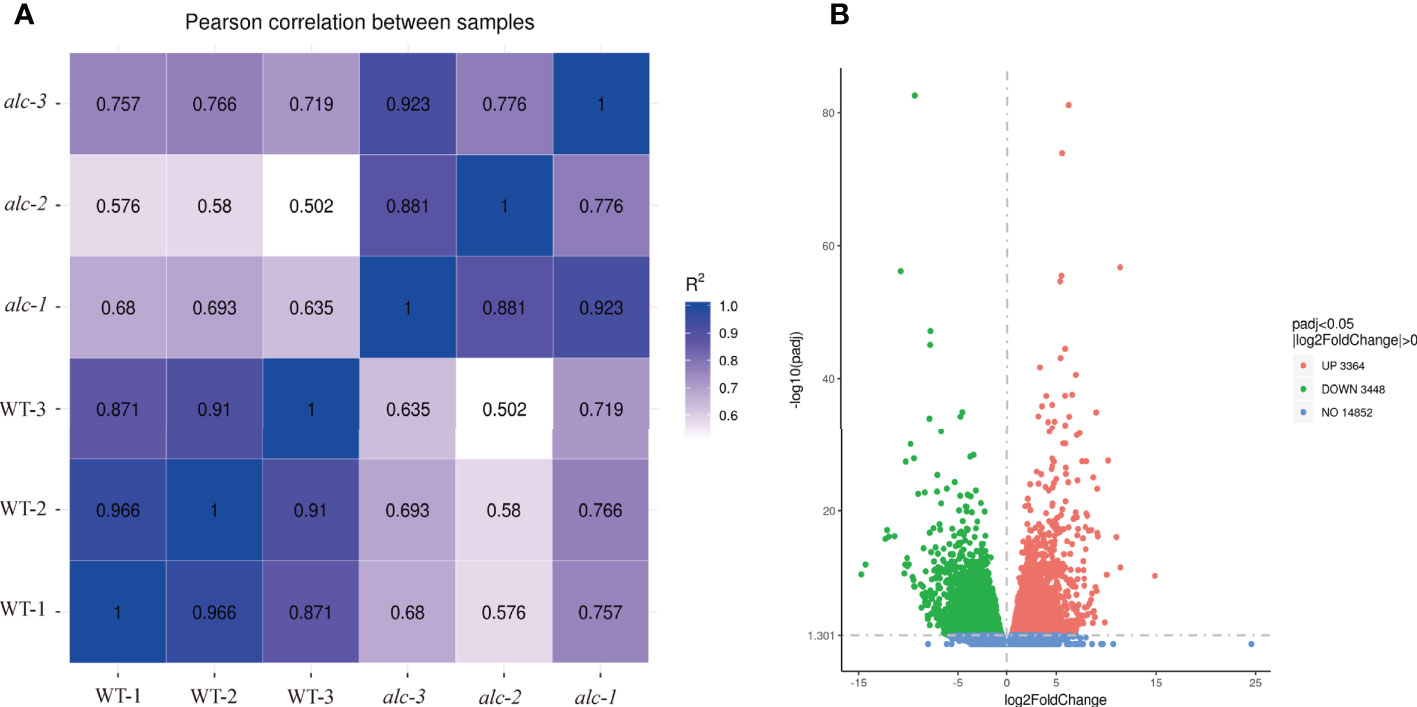
Figure 3 Diagrams illustrating correlations/distances among transcriptomes and the number of differentially expressed genes in alc mutant and wild type seedlings. (A) Correlation matrix and cluster dendrogram of the whole dataset of the mapped reads. The analysis was performed by comparing the values of the entire transcriptome of all two samples with three biological replicates. Correlation analysis (coefficients R2) and hierarchical cluster analysis were performed using R software. Dark blue color indicated a stronger correlation and light blue weaker (R2). (B) Volcano plot showing DEGs between alc mutant and wild type. X-axis represented log2(Fold Change) and y-axis represents -log10 (padj). Red, green and blue dots represented up-regulated, down-regulated and not DEGs, respectively.
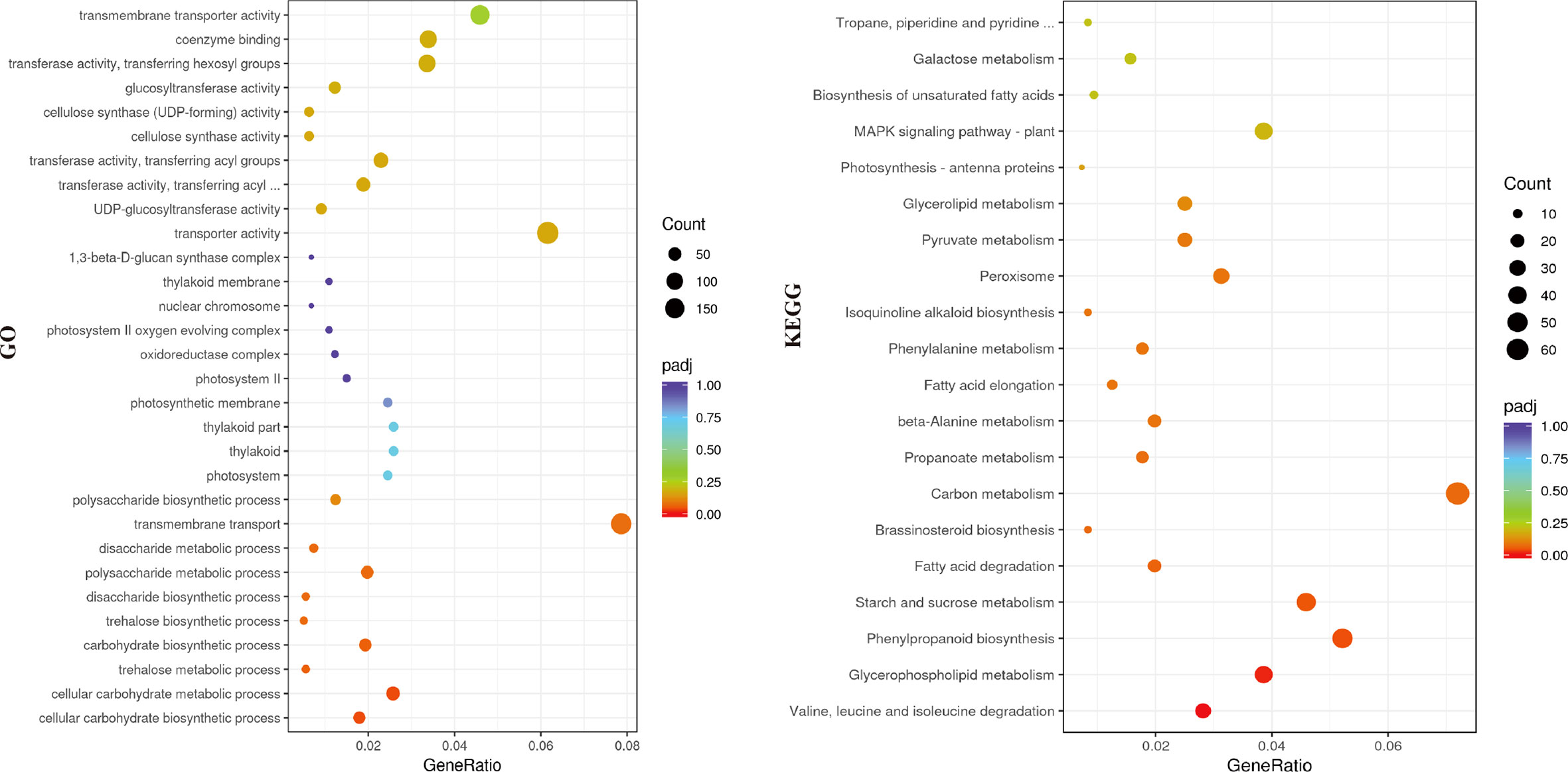
Figure 4 Genes enriched in different categories in the GO (left) and KEGG (right) analysis. X-axis represented the gene ratio of enriched genes among the background genes in different categories and y-axis represented the GO or KEGG terms. A high padj-value was represented by red, and a low value represented by purple. The size of the bubble represented number of genes annotated to each term.
Porphyrin and chlorophyll metabolism (KEGG: csv00860) was a significantly enriched pathway in KEGG analysis. Many key enzymes involved in this pathway showed distinct expression profile between the alc mutant and the wild type. The expression of most genes, including HEMB (Csa2G401270), HEME (Csa4G082410, Csa5G218840), HEMF (Csa4G056670), HEMG (Csa6G007980), CHLG (Csa4G311220), and CAO (Csa6G385090), was slightly higher in the alc mutant than in the wild type (Figure 5A; Supplementary Table 5). However, POR (Csa4G638340) was downregulated in the alc mutant, presenting an opposite expression pattern to that of other genes (Figure 5A; Supplementary Table 5).
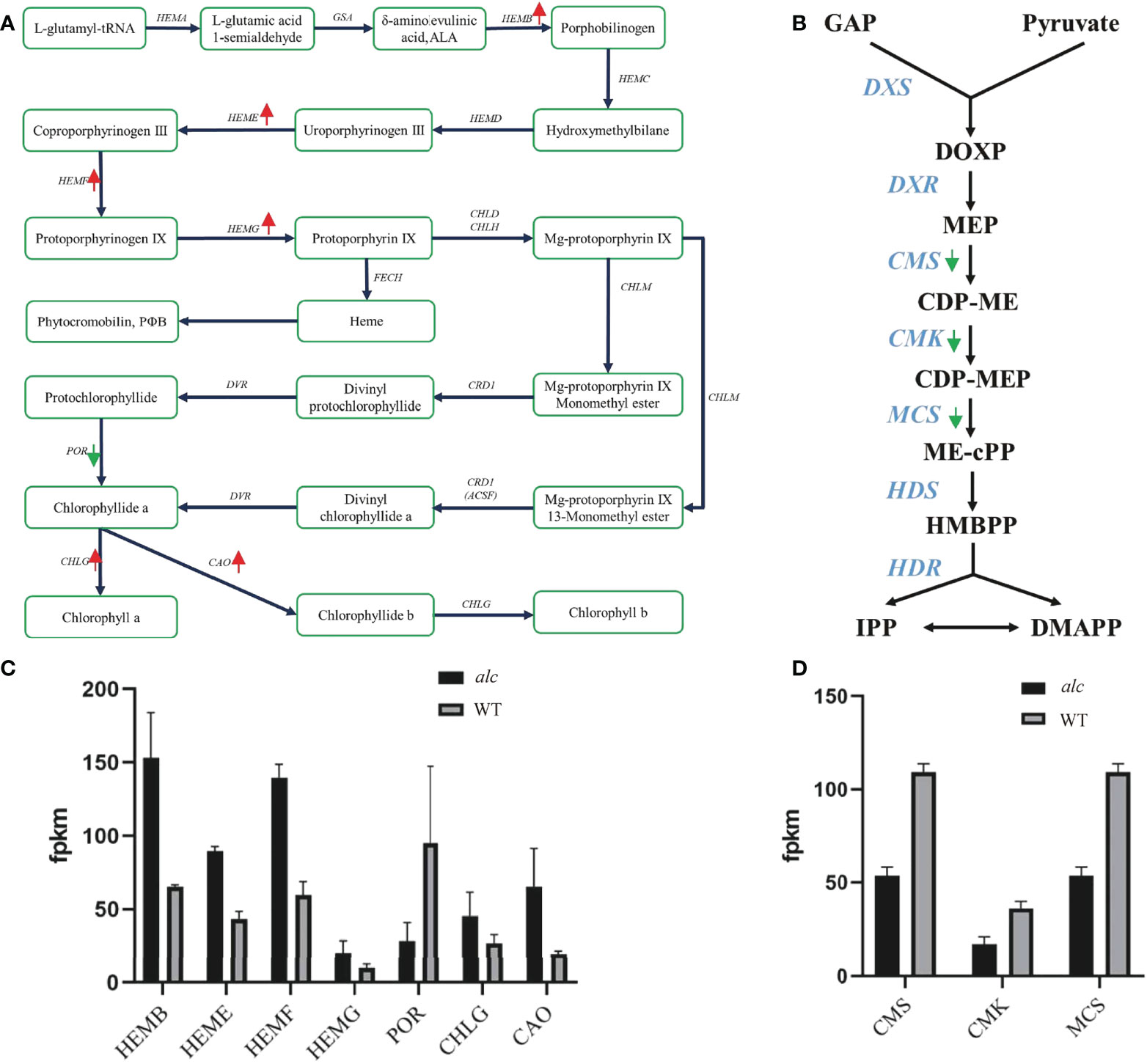
Figure 5 Differential expression of genes involved in chlorophyll metabolism and Methylerythritol 4-phosphate (MEP) pathway. (A, B) Diagram showing major genes in chlorophyll metabolism and MEP pathway, respectively. Red arrows indicated up-regulated genes and green arrows indicated down-regulated genes in alc mutant. (C, D) Expression profile of DEGs in chlorophyll metabolism and MEP pathway achieved by RNA-seq. The mean FPKM (fragments per kilo base of transcript per million mapped reads) values for the DEGs were calculated from three biological replicates for each genotype. Error bars indicated standard deviations. GAP, glyceraldehyde-3-phosphate; DOXP, 1-deoxy-D-xylulose-5-phosphate; MEP, 2-C-methyl-D-erythritol-4-phosphate; CDP-ME, 4-diphosphocytidyl-2-C-methyl-D-erythritol; CDP-MEP, 4-diphosphocytidyl-2-C-methyl-D-erythritol-2-phosphate; ME-cPP, 2-C-methyl-D-erythritol-2,4-C-cyclodiphosphate; HMBPP, 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate; IPP, isopentenyl diphosphate DMAPP, dimethylallyl diphosphate.
The methylerythritol 4-phosphate (MEP) pathway is mainly involved in the production of isoprenoid precursors; isopentenyl diphosphate (IPP) and dimethylallyl diphosphate in photosynthetic eukaryotes (Cordoba et al., 2009). As shown in Figure 5B, there were at least seven key enzymes involved in the MEP pathway. Four candidate genes encoding CMS (Csa3G113320 and Csa4G049620), CMK (Csa1G600780), and MCS (Csa4G049620) were downregulated in the alc mutant (Supplementary Table 5).
Chloroplast-localized FtsH proteins are crucial for the biogenesis of thylakoid membranes (Wagner et al., 2012). However, we observed negligible expression of the two FtsH genes (Csa6G504470 and Csa6G504480) in the alc mutant (Supplementary Table 2). Some pentatricopeptide repeat (PPR) proteins are involved in plastid gene expression and can also affect chloroplast development (Myouga et al., 2010). The expression of PPR gene (Csa5G189930) in the alc mutant was more than 3.5-fold higher than that in the wildtype (Supplementary Table 2).
Nineteen DEGs involved in thylakoid related functional activities, including thylakoid (GO:0009579), thylakoid part (GO:0044436), and thylakoid membrane (GO:0042651), were highlighted (Figure 6; Supplementary Table 3). Three PsbPs genes (Csa2G030040, Csa1G088470, and Csa1G181310) were upregulated in the alc mutant, while 16 other genes, namely, PsbR (Csa4G064020), PsaG/PsaK (Csa3G060980, Csa6G525340), PsbO (Csa6G488340), PsaD (Csa3G147780), PsaH (Csa3G483830), PsbP (Csa4G063440), PsaE (Csa2G079660), PsbY (Csa5G592810), PsbW (Csa7G378440), PsaN (Csa6G483300), PsbQ (Csa1G066480, Csa3G414060), PsbX (Csa1G595840), PsaF (Csa1G714680), and PetM (Csa7G075020) were downregulated in the alc mutant (Supplementary Table 2).
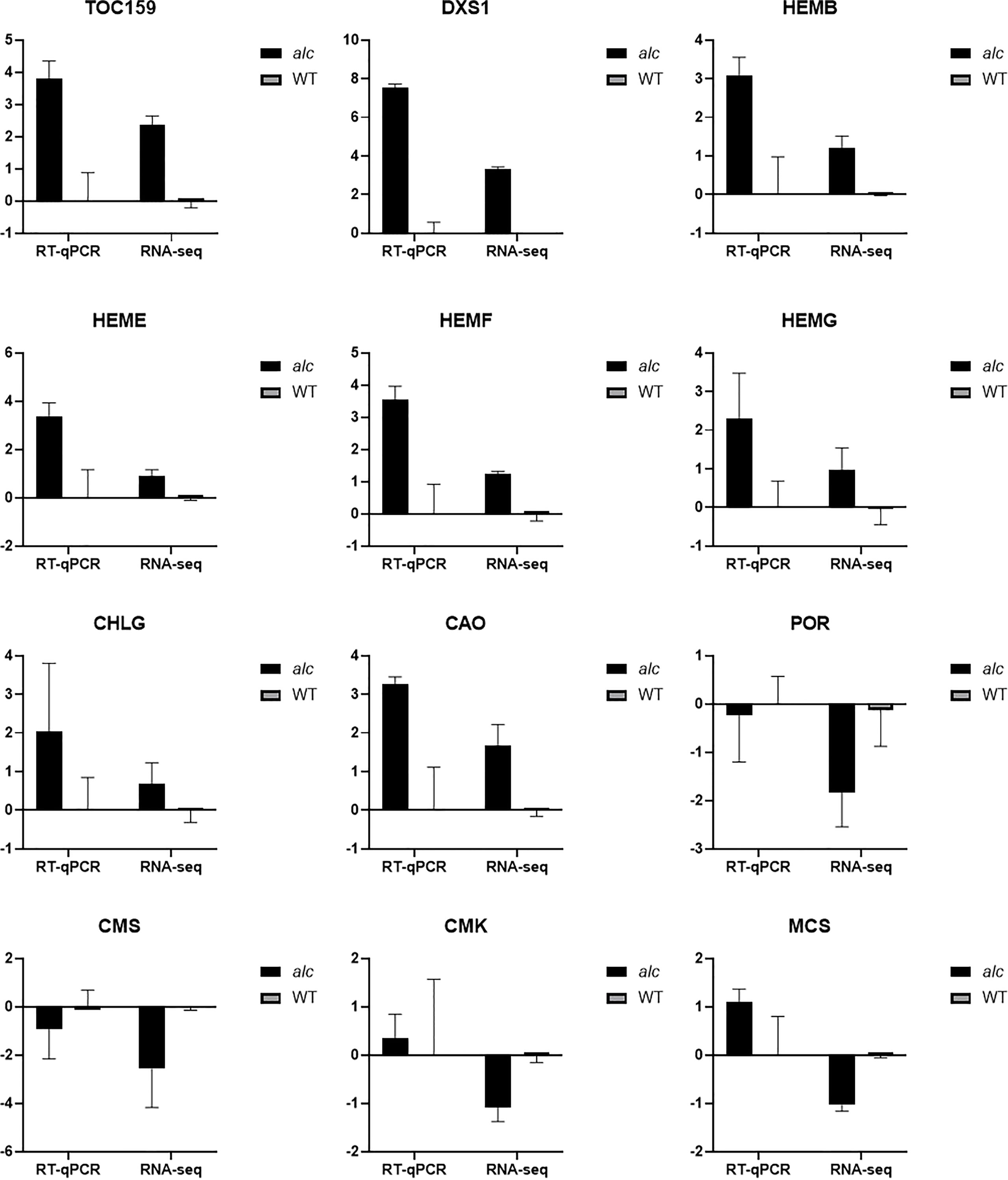
Figure 6 Expression profile of selected differentially expressed genes between alc mutant and wild type from RNA-seq result achieved by qRT-PCR. X-axis represented gene name and y-axis represented relative expression (–ΔΔCt) value of each gene. Data are shown as means (n = 3). Error bars indicated standard deviations.
Previous studies have reported that mutations of FRUCTOKINASE-LIKE PROTEIN in barley and rice (Qin et al., 2015; Lv et al., 2017; He et al., 2018), RPL21c (chloroplast 50S ribosomal protein L21) in Arabidopsis and rice (Yin et al., 2012; Lin et al., 2015), EMB (embryo-defective) in Arabidopsis (Huang et al., 2009; Liang et al., 2010; Ye et al., 2017; Chen et al., 2018), PDS3 (phytoene desaturase) in Arabidopsis (Qin et al., 2007), TOC159 (Translocase of chloroplast 159) in Arabidopsis (Kakizaki et al., 2009; Shanmugabalaji et al., 2018), DXS1 (1-deoxy-D-xylulose-5-phosphate synthase 1) in tomato (Garcia-Alcazar et al., 2017), and PurD (phosphoribosylamine–glycine ligase) in rice (Zhang et al., 2018) can cause albinism. The coding sequences (CDSs) of the above mentioned homologous genes of alc mutant and wild type were compared using our transcriptome data. However, no variant was found among these genes in thealc mutant and wildtype. Among these genes, only TOC159 (Csa4G001670) and DXS1 (Csa3G114510) showed more abundant expression in alc mutant than in the wild-type (Supplementary Table 6).
Twelve genes including seven genes involved in chlorophyll metabolism, three genes from the MEP pathway, TOC159, and DXS1 were selected for RT-qPCR verification (the information of the genes is listed in Supplementary Table 1). The relative expression (–ΔΔCt) of each gene was calculated using corresponding wild type gene expressions as control. The correlation between the relative expression value and RNA-seq result (log2FoldChange) of the alc mutant was determined. The correlation between RT-qPCR and RNA-seq data was 0.9780 (P < 0.0001, ****), indicating the reliability of our RNA-seq results.
Albinism occurs among different living organisms ranging from human beings to animals as well as higher plants (Kamaraj and Purohit, 2014; Galante Rocha de Vasconcelos et al., 2017; Le Ret et al., 2018). In the present study, some albino seedlings were observed in the progenies of a self-pollinated cucumber fruit during seed increase period of a cucumber inbred line named “g32”. The mutant was named alc, which presented white cotyledons and hypocotyl, and died before developing first true leaf under normal light conditions. In most studies, albino plants were caused by lack of chlorophyll and impaired chloroplast development (Liang et al., 2010; Zhu et al., 2015; Le Ret et al., 2018). Consistent with most albino mutants, lack of chlorophyll and defective chloroplast development was also observed in the alc mutant (Figure 2). In wild-type seedlings, ultrastructure of chloroplast was well presented with compactly arranged chloroplasts, while few or even no chloroplasts were observed in albino seedlings. Furthermore, an abnormal chloroplast ultrastructure lacking starch granules and thylakoids, but with osmiophilic plastoglobuli was observed in the alc mutant (Figure 4). Osmiophilic plastoglobuli generally appear as a result of the degradation of thylakoid membranes under stress (Wang et al., 2016) Therefore, we propose that irradiation with normal light might act as an abiotic stress cue for alc mutant, leading to the degradation of thylakoid membranes.
Interestingly, we found that the cucumber alc mutant differed from the reported fln1, rpl21c, emb, pds3, toc159, dxs1, and purd albino mutants as this mutant presented green cotyledons under dim light conditions. Additionally, we checked the homologous gene sequence of these albino related genes in cucumber, but found no variation within the coding sequences of the alc mutant and wild type, suggesting that alc might be a novel light sensitive albino mutant. Similar phenotype was reported in pap7-1 albino mutant of Arabidopsis (Grübler et al., 2017). Pap7-1 mutant had an albino cotyledon and died grown under light conditions; however, under dark conditions, pap7-1 could grow when supplied with sucrose supplemented medium (Grübler et al., 2017). However, the molecular mechanism describing the exact nature of pap7-1 is still unknown.
Since all the seeds were planted under the same culture condition, we could exclude the possible of environmental influences on the albino phenotype of the alc mutant. Genetic analysis revealed that this albino phenotype was controlled by a recessive locus. Phenotypic evaluation of the previous generation of “g32” was also performed but albino seedlings were not observed (data not shown), confirming the recessive inheritance of this albino allele. Albinism is not a desired phenomenon in plant breeding since it could affect plant growth as well as production. However, this alc mutant is of great importance to us for detection of new genes controlling plastid development.
Transcriptome analysis has been extensively applied to identify major genes and dissect regulatory networks involved in albinism by using different albino mutants (Shi et al., 2017; Li et al., 2017; Ma et al., 2018). In our study, transcriptome analysis revealed that the expression of genes involved in chloroplast development, chlorophyll metabolism, MEP pathway, and other genes, such as glutathione S-transferase, was altered in the alc mutant. This transcriptional modification suggests that these genes play important roles for the albino phenotype of the alc mutant.
Light plays a crucial role in plant development. In light-sensitive mutants, a sudden increase in ROS production would occur under excessive light and induce oxidative damage leading to leaf bleaching. Arabidopsis ch1 is a Chlb deficient mutant and is devoid of photosystem II (PSII) Chl-protein antenna complexes. Thus, oxidizing side of PSII is impaired and ch1 is sensitive to photooxidative stress (Havaux et al., 2007). Nearly all DEGs in the Photosynthesis-antenna proteins pathway (Supplementary Table 4, KEGG: csv00196) were downregulated in the alc mutant, indicating a weak functionality of core reaction center. Peroxisomes are organelles that contribute to the reduction of oxidative stress (del Río et al., 2006). Most of genes in Peroxisome pathway (24 out of 30) (Supplementary Table 4, KEGG: csv04146) were upregulated in the alc mutant. Under normal light irradiation for wild-type but excessive for alc, the alc might produce more ROS and lead to bleaching and finally died as ch1 (Ramel et al., 2013). Since pap7-1 mutant could be arrested under very dim light (Grübler et al., 2017), we tried to culture the mutant under dim light condition but failed. Further studies are needed to confirm whether the morphology and flowering of the mutant are similar to that of pap7-1 grown under sucrose supplemented medium and dim light conditions.
Three genes involved in chloroplast development were found to show different transcript levels between the alc mutant and wild type. Two of these genes belong to FtsH family, which is essential for chloroplast development. For example, both ftsh1 ftsf5 and ftsh2 ftsh8 double mutants developed white seedlings with disrupted chloroplast development (Zaltsman et al., 2005). Significantly fewer FstH transcripts were detected in the alc mutant as compared to those in wild-type cucumber. Entratricopeptide Repeat Protein Pigment-Defective Mutant2 (PDM2), that encodes a PPR protein, is required for chloroplast development by regulation of plastid gene expression (Du et al., 2017). We detected very low expression of a PPR gene in the alc mutant. The extremely low expression of chloroplast related genes might impact chloroplast development and result in an albino phenotype. Key genes involved in chlorophyll metabolism, the MEP pathway, and thylakoid function were also affected in the alc mutant. Many genes in the chlorophyll metabolism pathway, including HEMB, HEME, HEMF, HEMG, and CHLG were slightly up regulated in the alc mutant. The increased expression of these genes was also observed in other albino mutants. Most of the chlorophyll-biosynthesis related genes, such as HEMC, HEME and CHLG were upregulated in white leaves compared with those in green leaves of Ananas comosus var. Bracteatus (Li et al., 2017). Similar results were observed in the wheat mta albino mutant where the expression of HEME was upregulated (Shi et al., 2017). The decreased expression of most genes might be regulated by a feedback mechanism. POR, a key light-dependent enzyme, is essential to chlorophyll biosynthesis where it catalyzes protochlorophyllide to chlorophyllide (Yang and Cheng, 2004; Zhang et al., 2019). POR is crucial for plant growth and development because por mutant and POR RNAi line displayed reduced chlorophyll content and severe photoautotrophic growth defects (Kim and Apel, 2012; Paddock et al., 2012). The expression level of POR was low in the alc mutant, the mta albino wheat mutant (Shi et al., 2017), and the complete white leaves of Ananas comosus var. Bracteatus (Li et al., 2017), indicating that the downregulation of POR might have an impact on chlorophyll synthesis in the alc mutant. The MEP pathway is essential for the biosynthesis of photosynthesis-related compounds, such as carotenoids, chlorophylls, gibberellins, and abscisic acid, which are of vital importance for plant development and metabolism (Flores-Perez et al., 2008). The mutation of genes in the pathway impaired the biosynthesis of these compounds, disrupted chloroplast development, and resulted in abnormal plant morphology, especially in leaf color (Xing et al., 2010; Chandran et al., 2016). IspD (CMS), IspE (CMK), and IspF (MCS) are the third, fourth and fifth enzymes in the MEP pathway, respectively (Figure 5B). Related mutants possessed yellow or albino leaves with arrested development of chloroplasts (Hsieh et al., 2008; Chen et al., 2018; Huang et al., 2018). In this study, the expression of CMS, CMK and MCS in the alc mutant was lower than that in the wildtype. The low-level expression may contribute to the disruption of chloroplast development. Thylakoid related genes corresponded to reaction centers of PSI and PSII where photochemical reactions occur and convert light energy into chemical energy. Almost all genes related to thylakoid were downregulated in the alc mutant, demonstrating the decrease of photosynthesis viability.
Isocitrate lyase and malate synthase are two enzymes unique to the glyoxylate cycle, which is considered essential for postgerminative growth and seedling establishment (Eastmond et al., 2000). Under normal light conditions, the enzyme activities in the glyoxylate cycle decreased rapidly as seedlings become photosynthetic, whereas those under dark conditions maintained a continuously high level of enzyme activity (Trelease et al., 1971; Becker et al., 1978; He et al., 2016). In this study, the abundance of isocitrate lyase (Csa2G420990) and malate synthase (Csa1G050360) was higher in the mutant than in the wildtype (Figure 6). The wild-type exhibited autotrophic growth with a high activity of peroxidase (Csa4G285740) and chlorophyll A-B binding protein (Csa3G664560), while the alc mutant was unable to perform photosynthesis due to the lack of functional chloroplasts. Glutathione S-transferase have been reported to mainly function in response to biotic and abiotic stresses, such as oxidative stress (Horváth et al., 2015), temperature stress (Roxas et al., 1997; Liang et al., 2018) and different pathogen invasion (Sytykiewicz et al., 2014; Gong et al., 2018; Ducker et al., 2019). The high concentration of glutathione S-transferase can result in decreased chlorophyll content (Jiang et al., 2010). When the alc mutant is exposed to light, which is lethal rather than beneficial, it expressed (Csa4G304240, Csa4G303170) (Figure 6) more glutathione S-transferase than the wild type; this might also promote chlorophyll degradation.
In conclusion, a cucumber albino mutant alc was cytologically, genetically and transcriptomically characterized in this study. Our results demonstrated that the albino phenotype of the mutant was mainly due to the disability in chlorophyll synthesis and chloroplast development, which resulted in no chlorophyll content in the cotyledons and finally seedlings died as not being able to photosynthesize. In this study, we could not determine the causal mutation of alc. However, Bulk Segregant Analysis (BSA), using albino and green seedling pools along with map-based cloning would be useful to finely mapping the mutated gene in future studies.
The RNA-seq datasets presented in this study can be accessed through National Center for Biotechnology Information (NCBI) BioProject database under accession number PRJNA685868.
JY, BL, YL, and BJ: conceptualization and writing—review and editing; JY, ZC, LC, ZL, MW, and WL: formal analysis and investigation; JY: writing—original draft preparation. All authors have read and approved the final manuscript.
This study was supported by Key-Area Research and Development Program of Guangdong Province (2020B020220001), The Discipline Team Construction Project of GDAAS (202103TD), the Training Plan for Young and Middle-aged Discipline Leaders of GDAAS (R2020PY-JG003) and Talent Introduction Plan of GDAAS (R2021YJ-YB2004). The funding bodies have no role in the study design, data analysis and interpretation, and manuscript writing, but just provide the financial supports.
The authors thank associate professor Yuhui Wang from College of Horticulture at Nanjing Agricultural University for her assistance in language editing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1047090/full#supplementary-material
Supplementary Table 1 | Primers used for qRT-PCR validation.
Supplementary Table 2 | Up-regulated and down-regualted genes in albino mutant.
Supplementary Table 3 | GO enrichment analysis of differentially expressed genes between albino mutant and wild type.
Supplementary Table 4 | KEGG analysis of differentially expressed genes between albino mutant and wild type.
Supplementary Table 5 | Differentially expressed genes in chlorophyll metabolism and MEP pathway between albino mutant and wild type.
Supplementary Table 6 | Differentially expressed genes of reported albino genes between albino mutant and wild type.
Supplementary Figure 1 | Histogram of GO enrichment analysis.
Supplementary Figure 2 | A heat map showing expression patterns of genes related to chlorophyll metabolism, chloroplast formation and Methylerythritol 4-phosphate (MEP) pathway.
Becker, W. M., Leaver, C. J., Weir, E. M., Riezman, H. (1978). Regulation of glyoxysomal enzymes during germination of cucumber: I. developmental changes in cotyledonary protein, RNA, and enzyme activities during germination. Plant Physiol. 62 (4), 542–549. doi: 10.1104/pp.62.4.542
Chandran, A. K. N., Lee, G. S., Yoo, Y. H., Yoon, U. H., Ahn, B. O., Yun, D. W., et al. (2016). Functional classification of rice flanking sequence tagged genes using MapMan terms and global understanding on metabolic and regulatory pathways affected by dxr mutant having defects in light response. Rice (N Y) 9 (1), 17. doi: 10.1186/s12284-016-0089-2
Chan, K. X., Phua, S. Y., Crisp, P., McQuinn, R., Pogson, B. J. (2016). Learning the languages of the chloroplast: Retrograde signaling and beyond. Annu. Rev. Plant Biol. 67, 25–53. doi: 10.1146/annurev-arplant-043015-111854
Chen, H., Li, S., Li, L., Hu, H., Zhao, J. (2018). Arabidopsis EMB1990 encoding a plastid-targeted YlmG protein is required for chloroplast biogenesis and embryo development. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00181
Chen, N., Wang, P., Li, C., Wang, Q., Pan, J., Xiao, F., et al. (2018). A single nucleotide mutation of the IspE gene participating in the MEP pathway for isoprenoid biosynthesis causes a green-revertible yellow leaf phenotype in rice. Plant Cell Physiol. 59 (9), 1905–1917. doi: 10.1093/pcp/pcy108
Cordoba, E., Salmi, M., Leon, P. (2009). Unravelling the regulatory mechanisms that modulate the MEP pathway in higher plants. J. Exp. Bot. 60 (10), 2933–2943. doi: 10.1093/jxb/erp190
del Río, L. A., Corpas, F. J., Sandalio, L. M., Palma, J. M., Gómez, M., Barroso, J. B. (2002). Reactive oxygen species, antioxidant systems and nitric oxide in peroxisomes. J. Exp. Bot. 53 (372), 1255–1272. doi: 10.1093/jexbot/53.372.1255
Ducker, R., Zollner, P., Lummen, P., Ries, S., Collavo, A., Beffa, R. (2019). Glutathione transferase plays a major role in flufenacet resistance of ryegrass (Lolium spp.) field populations. Pest Manag Sci. 75 (11), 3084–3092. doi: 10.1002/ps.5425
Du, L., Zhang, J., Qu, S., Zhao, Y., Su, B., Lv, X., et al. (2017). The Pentratricopeptide repeat protein pigment-defective Mutant2 is involved in the regulation of chloroplast development and chloroplast gene expression in arabidopsis. Plant Cell Physiol. 58 (4), 747–759. doi: 10.1093/pcp/pcx004
Eastmond, P. J., Germain, V., Lange, P. R., Bryce, J. H., Smith, S. M., Graham, I. A. (2000). Postgerminative growth and lipid catabolism in oilseeds lacking the glyoxylate cycle. Proc. Natl. Acad. Sci. U.S.A. 97 (10), 5669–5674. doi: 10.1073/pnas.97.10.5669
Flores-Perez, U., Sauret-Gueto, S., Gas, E., Jarvis, P., Rodriguez-Concepcion, M. (2008). A mutant impaired in the production of plastome-encoded proteins uncovers a mechanism for the homeostasis of isoprenoid biosynthetic enzymes in arabidopsis plastids. Plant Cell 20 (5), 1303–1315. doi: 10.1105/tpc.108.058768
Galante Rocha de Vasconcelos, F. T., Hauzman, E., Dutra Henriques, L., Kilpp Goulart, P. R., de Faria Galvao, O., Sano, R. Y., et al. (2017). A novel nonsense mutation in the tyrosinase gene is related to the albinism in a capuchin monkey (Sapajus apella). BMC Genet. 18 (1), 39. doi: 10.1186/s12863-017-0504-8
Garcia-Alcazar, M., Gimenez, E., Pineda, B., Capel, C., Garcia-Sogo, B., Sanchez, S., et al. (2017). Albino T-DNA tomato mutant reveals a key function of 1-deoxy-D-xylulose-5-phosphate synthase (DXS1) in plant development and survival. Sci. Rep. 7, 45333. doi: 10.1038/srep45333
Gong, Q., Yang, Z., Chen, E., Sun, G., He, S., Butt, H. I., et al. (2018). A phi-class glutathione s-transferase gene for verticillium wilt resistance in gossypium arboreum identified in a genome-wide association study. Plant Cell Physiol. 59 (2), 275–289. doi: 10.1093/pcp/pcx180
Gotoh, E., Suetsugu, N., Yamori, W., Ishishita, K., Kiyabu, R., Fukuda, M., et al. (2018). Chloroplast accumulation response enhances leaf photosynthesis and plant biomass production. Plant Physiol. 178 (3), 1358–1369. doi: 10.1104/pp.18.00484
Grübler, B., Merendino, L., Twardziok, S. O., Mininno, M., Allorent, G., Chevalier, F., et al. (2017). Light and plastid signals regulate different sets of genes in the albino mutant Pap7-1. Plant Physiol. 175 (3), 1203–1219. doi: 10.1104/pp.17.00982
Havaux, M., Dall'osto, L., Bassi, R. (2007). Zeaxanthin has enhanced antioxidant capacity with respect to all other xanthophylls in arabidopsis leaves and functions independent of binding to PSII antennae. Plant Physiol. 145 (4), 1506–1520. doi: 10.1104/pp.107.108480
He, D., Damaris, R. N., Fu, J., Tu, J., Fu, T., Xi, C., et al. (2016). Differential molecular responses of rapeseed cotyledons to light and dark reveal metabolic adaptations toward autotrophy establishment. Front. Plant Sci. 14. doi: 10.3389/fpls.2016.00988
He, L., Zhang, S., Qiu, Z., Zhao, J., Nie, W., Lin, H., et al. (2018). FRUCTOKINASE-LIKE PROTEIN 1 interacts with TRXz to regulate chloroplast development in rice. J. Integr. Plant Biol. 60 (2), 94–111. doi: 10.1111/jipb.12631
Horváth, E., Bela, K., Papdi, C., Gallé, Á., Szabados, L., Tari, I., et al. (2015). The role of arabidopsis glutathione transferase F9 gene under oxidative stress in seedlings. Acta Biol. Hung 66 (4), 406–418. doi: 10.1556/018.66.2015.4.5
Hsieh, M. H., Chang, C. Y., Hsu, S. J., Chen, J. J. (2008). Chloroplast localization of methylerythritol 4-phosphate pathway enzymes and regulation of mitochondrial genes in ispD and ispE albino mutants in arabidopsis. Plant Mol. Biol. 66 (6), 663–673. doi: 10.1007/s11103-008-9297-5
Huang, S., Li, R., Zhang, Z., Li, L., Gu, X., Fan, W., et al. (2009). The genome of the cucumber, cucumis sativus l. Nat. Genet. 41 (12), 1275–1281. doi: 10.1038/ng.475
Huang, R., Wang, Y., Wang, P., Li, C., Xiao, F., Chen, N., et al. (2018). A single nucleotide mutation of IspF gene involved in the MEP pathway for isoprenoid biosynthesis causes yellow-green leaf phenotype in rice. Plant Mol. Biol. 96 (1-2), 5–16. doi: 10.1007/s11103-017-0668-7
Huang, X., Zhang, X., Yang, S. (2009). A novel chloroplast-localized protein EMB1303 is required for chloroplast development in arabidopsis. Cell Res. 19 (10), 1205–1216. doi: 10.1038/cr.2009.84
Iida, S., Amano, E. (1991). Mutants induced by pollen irradiation in cucumber. Cucurbit Genet. Coop. Rpt 14, 32–33.
Jiang, H. W., Liu, M. J., Chen, I. C., Huang, C. H., Chao, L. Y., Hsieh, H. L. (2010). A glutathione s-transferase regulated by light and hormones participates in the modulation of arabidopsis seedling development. Plant Physiol. 154 (4), 1646–1658. doi: 10.1104/pp.110.159152
Kakizaki, T., Matsumura, H., Nakayama, K., Che, F. S., Terauchi, R., Inaba, T. (2009). Coordination of plastid protein import and nuclear gene expression by plastid-to-nucleus retrograde signaling. Plant Physiol. 151 (3), 1339–1353. doi: 10.1104/pp.109.145987
Kamaraj, B., Purohit, R. (2014). Mutational analysis of oculocutaneous albinism: A compact review. BioMed. Res. Int. 2014, 905472. doi: 10.1155/2014/905472
Kim, C., Apel, K. (2012). Arabidopsis light-dependent NADPH: protochlorophyllide oxidoreductase a (PORA) is essential for normal plant growth and development: an addendum. Plant Mol. Biol. 80 (2), 237–240. doi: 10.1007/s11103-012-9944-8
Le Ret, M., Belcher, S., Graindorge, S., Wallet, C., Koechler, S., Erhardt, M., et al. (2018). Efficient replication of the plastid genome requires an organellar thymidine kinase. Plant Physiol. 178 (4), 1643–1656. doi: 10.1104/pp.18.00976
Liang, D., Gao, F., Ni, Z., Lin, L., Deng, Q., Tang, Y., et al. (2018). Melatonin improves heat tolerance in kiwifruit seedlings through promoting antioxidant enzymatic activity and glutathione s-transferase transcription. Molecules 23 (3), 584. doi: 10.3390/molecules23030584
Liang, Q., Lu, X., Jiang, L., Wang, C., Fan, Y., Zhang, C. (2010). EMB1211 is required for normal embryo development and influences chloroplast biogenesis in arabidopsis. Physiol. Plant 140 (4), 380–394. doi: 10.1111/j.1399-3054.2010.01407.x
Lichtenthaler, H. K. (1987). Chlorophylls and carotenoids: pigment of photosynthetic biomembranes. Method Enzymol. 148, 350–382. doi: 10.1016/0076-6879(87)48036-1
Li, X., Kanakala, S., He, Y., Zhong, X., Yu, S., Li, R., et al. (2017). Physiological characterization and comparative transcriptome analysis of white and green leaves of Ananas comosus var. bracteatus. PloS One 12 (1), e0169838. doi: 10.1371/journal.pone.0169838
Li, Z., Mo, W., Jia, L., Xu, Y. C., Tang, W., Yang, W., et al. (2019). Rice FLUORESCENT1 is involved in the regulation of chlorophyll. Plant Cell Physiol. 60 (10), 2307–2318. doi: 10.1093/pcp/pcz129
Lin, D., Jiang, Q., Zheng, K., Chen, S., Zhou, H., Gong, X., et al. (2015). Mutation of the rice ASL2 gene encoding plastid ribosomal protein L21 causes chloroplast developmental defects and seedling death. Plant Biol. (Stuttg) 17 (3), 599–607. doi: 10.1111/plb.12271
Liu, B., Liu, X., Yang, S., Chen, C., Xue, S., Cai, Y., et al. (2016). Silencing of the gibberellin receptor homolog, CsGID1a, affects locule formation in cucumber (Cucumis sativus) fruit. New Phytol. 210 (2), 551–563. doi: 10.1111/nph.13801
Li, Z., Zhang, Z., Yan, P., Huang, S., Fei, Z., Lin, K. (2011). RNA-Seq improves annotation of protein-coding genes in the cucumber genome. BMC Genomics 12, 540. doi: 10.1186/1471-2164-12-540
Lu, M., Han, J., Zhu, B., Jia, H., Yang, T., Wang, R., et al. (2019). Significantly increased amino acid accumulation in a novel albino branch of the tea plant (Camellia sinensis). Planta 249 (2), 363–376. doi: 10.1007/s00425-018-3007-6
Lv, Y., Shao, G., Qiu, J., Jiao, G., Sheng, Z., Xie, L., et al. (2017). White leaf and panicle 2, encoding a PEP-associated protein, is required for chloroplast biogenesis under heat stress in rice. J. Exp. Bot. 68 (18), 5147–5160. doi: 10.1093/jxb/erx332
Ma, Q., Li, H., Zou, Z., Arkorful, E., Lv, Q., Zhou, Q., et al. (2018). Transcriptomic analyses identify albino-associated genes of a novel albino tea germplasm 'Huabai 1'. Hortic. Res. 5, 54. doi: 10.1038/s41438-018-0053-y
Myouga, F., Akiyama, K., Motohashi, R., Kuromori, T., Ito, T., Iizumi, H., et al. (2010). The chloroplast function database: a large-scale collection of arabidopsis Ds/Spm- or T-DNA-tagged homozygous lines for nuclear-encoded chloroplast proteins, and their systematic phenotype analysis. Plant J. 61 (3), 529–542. doi: 10.1038/s41438-018-0053-y
Paddock, T., Lima, D., Mason, M. E., Apel, K., Armstrong, G. A. (2012). Arabidopsis light-dependent protochlorophyllide oxidoreductase a (PORA) is essential for normal plant growth and development. Plant Mol. Biol. 78 (4-5), 447–460. doi: 10.1007/s11103-012-9873-6
Pogson, B. J., Ganguly, D., Albrecht-Borth, V. (2015). Insights into chloroplast biogenesis and development. Biochim. Biophys. Acta 1847 (9), 1017–1024. doi: 10.1016/j.bbabio.2015.02.003
Qin, D., Dong, J., Xu, F., Guo, G., Ge, S., Xu, Q., et al. (2015). Characterization and fine mapping of a novel barley stage green-revertible albino gene (HvSGRA) by bulked segregant analysis based on SSR assay and specific length amplified fragment sequencing. BMC Genomics 16, 838. doi: 10.1186/s12864-015-2015-1
Qin, G., Gu, H., Ma, L., Peng, Y., Deng, X. W., Chen, Z., et al. (2007). Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res. 17 (5), 471–482. doi: 10.1038/cr.2007.40
Qiu, Z., Kang, S., He, L., Zhao, J., Zhang, S., Hu, J., et al. (2018). The newly identified heat-stress sensitive albino 1 gene affects chloroplast development in rice. Plant Sci. 267, 168–179. doi: 10.1016/j.plantsci.2017
Ramel, F., Ksas, B., Akkari, E., Mialoundama, A. S., Monnet, F., Krieger-Liszkay, A., et al. (2013). Light-induced acclimation of the arabidopsis chlorina1 mutant to singlet oxygen. Plant Cell 25 (4), 1445–1462. doi: 10.1105/tpc.113.109827
Roxas, V. P., Smith, R. K., Jr., Allen, E. R., Allen, R. (1997). Overexpression of glutathione s-transferase/glutathione peroxidase enhances the growth of transgenic tobacco seedlings during stress. Nat. Biotechnol. 15 (10), 988–991. doi: 10.1038/nbt1097-988
Ruppel, N. J., Logsdon, C. A., Whippo, C. W., Inoue, K., Hangarter, R. P. (2011). A mutation in arabidopsis seedling plastid development1 affects plastid differentiation in embryo-derived tissues during seedling growth. Plant Physiol. 155 (1), 342–353. doi: 10.1104/pp.110.161414
Sakowska, K., Alberti, G., Genesio, L., Peressotti, A., Delle Vedove, G., Gianelle, D., et al. (2018). Leaf and canopy photosynthesis of a chlorophyll deficient soybean mutant. Plant Cell Environ. 41 (6), 1427–1437. doi: 10.1111/pce.13180
Shanmugabalaji, V., Chahtane, H., Accossato, S., Rahire, M., Gouzerh, G., Lopez-Molina, L., et al. (2018). Chloroplast biogenesis controlled by DELLA-TOC159 interaction in early plant development. Curr. Biol. 28 (16), 2616–2623. doi: 10.1016/j.cub.2018.06.006
Shi, K., Gu, J., Guo, H., Zhao, L., Xie, Y., Xiong, H., et al. (2017). Transcriptome and proteomic analyses reveal multiple differences associated with chloroplast development in the spaceflight-induced wheat albino mutant mta. PloS One 12 (5), e0177992. doi: 10.1371/journal.pone.0177992
Su, N., Hu, M. L., Wu, D. X., Wu, F. Q., Fei, G. L., Lan, Y., et al. (2012). Disruption of a rice pentatricopeptide repeat protein causes a seedling-specific albino phenotype and its utilization to enhance seed purity in hybrid rice production. Plant Physiol. 159 (1), 227–238. doi: 10.1104/pp.112.195081
Sytykiewicz, H., Chrzanowski, G., Czerniewicz, P., Sprawka, I., Łukasik, I., Goławska, S., et al. (2014). Expression profiling of selected glutathione transferase genes in Zea mays (L.) seedlings infested with cereal aphids. PloS One 9 (11), e111863. doi: 10.1371/journal.pone.0111863
Trelease, R. N., Becker, W. M., Gruber, P. J., Newcomb, E. H. (1971). Microbodies (Glyoxysomes and peroxisomes) in cucumber cotyledons: Correlative biochemical and ultrastructural study in light- and dark-grown seedlings. Plant Physiol. 48 (4), 461–475. doi: 10.1104/pp.48.4.461
Unlu, C., Drop, B., Croce, R., van Amerongen, H. (2014). State transitions in chlamydomonas reinhardtii strongly modulate the functional size of photosystem II but not of photosystem I. Proc. Natl. Acad. Sci. U.S.A. 111 (9), 3460–3465. doi: 10.1073/pnas.1319164111
Wagner, R., Aigner, H., Funk, C. (2012). FtsH proteases located in the plant chloroplast. Plant Physiol. 145 (1), 203–214. doi: 10.1111/j.1399-3054.2011.01548.x
Wang, Y., Wang, C., Zheng, M., Lyu, J., Xu, Y., Li, X., et al. (2016). WHITE PANICLE1, a Val-tRNA synthetase regulating chloroplast ribosome biogenesis in rice, is essential for early chloroplast development. Plant Physiol. 170 (4), 2110–2123. doi: 10.1104/pp.15.01949
Wang, C., Xu, W., Jin, H., Zhang, T., Lai, J., Zhou, X., et al. (2016). A putative chloroplast-localized Ca(2+)/H(+) antiporter CCHA1 is involved in calcium and pH homeostasis and required for PSII function in arabidopsis. Mol. Plant 9 (8), 1183–1196. doi: 10.1016/j.molp.2016.05.015
Wang, Y., Zhang, J., Shi, X., Peng, Y., Li, P., Lin, D., et al. (2016). Temperature-sensitive albino gene TCD5, encoding a monooxygenase, affects chloroplast development at low temperatures. J. Exp. Bot. 67 (17), 5187–5202. doi: 10.1093/jxb/erw287
Xing, S., Miao, J., Li, S., Qin, G., Tang, S., Li, H., et al. (2010). Disruption of the 1-deoxy-D-xylulose-5-phosphate reductoisomerase (DXR) gene results in albino, dwarf and defects in trichome initiation and stomata closure in arabidopsis. Cell Res. 20 (6), 688–700. doi: 10.1038/cr.2010.54
Xiong, D., Huang, J., Peng, S., Li, Y. (2017). A few enlarged chloroplasts are less efficient in photosynthesis than a large population of small chloroplasts in arabidopsis thaliana. Sci. Rep. 7 (1), 5782. doi: 10.1038/s41598-017-06460-0
Yang, J., Cheng, Q. (2004). Origin and evolution of the light-dependent protochlorophyllide oxidoreductase (LPOR) genes. Plant Biol. (Stuttg) 6 (5), 537–544. doi: 10.1055/s-2004-821270
Ye, L. S., Zhang, Q., Pan, H., Huang, C., Yang, Z. N., Yu, Q. B. (2017). EMB2738, which encodes a putative plastid-targeted GTP-binding protein, is essential for embryogenesis and chloroplast development in higher plants. Physiol. Plant 161 (3), 414–430. doi: 10.1111/ppl.12603
Yin, T., Pan, G., Liu, H., Wu, J., Li, Y., Zhao, Z., et al. (2012). The chloroplast ribosomal protein L21 gene is essential for plastid development and embryogenesis in arabidopsis. Planta 235 (5), 907–921. doi: 10.1007/s00425-011-1547-0
Zaltsman, A., Ori, N., Adam, Z. (2005). Two types of FtsH protease subunits are required for chloroplast biogenesis and photosystem II repair in arabidopsis. Plant Cell 17 (10), 2782–2790. doi: 10.1105/tpc.105.035071
Zeng, X., Tang, R., Guo, H., Ke, S., Teng, B., Hung, Y. H., et al. (2017). A naturally occurring conditional albino mutant in rice caused by defects in the plastid-localized OsABCI8 transporter. Plant Mol. Biol. 94 (1-2), 137–148. doi: 10.1007/s11103-017-0598-4
Zhang, T., Feng, P., Li, Y., Yu, P., Yu, G., Sang, X., et al. (2018). VIRESCENT-ALBINO LEAF 1 regulates leaf colour development and cell division in rice. J. Exp. Bot. 69 (20), 4791–4804. doi: 10.1093/jxb/ery250
Zhang, S., Heyes, D. J., Feng, L., Sun, W., Johannissen, L. O., Liu, H., et al. (2019). Structural basis for enzymatic photocatalysis in chlorophyll biosynthesis. Nature 574 (7780), 722–725. doi: 10.1038/s41586-019-1685-2
Zhao, D. S., Zhang, C. Q., Li, Q. F., Yang, Q. Q., Gu, M. H., Liu, Q. Q. (2016). A residue substitution in the plastid ribosomal protein L12/AL1 produces defective plastid ribosome and causes early seedling lethality in rice. Plant Mol. Biol. 91 (1-2), 161–177. doi: 10.1007/s11103-016-0453-z
Keywords: albino mutant, cucumber, recessive, chloroplast deficiency, transcriptome
Citation: Yan J, Liu B, Cao Z, Chen L, Liang Z, Wang M, Liu W, Lin Y and Jiang B (2022) Cytological, genetic and transcriptomic characterization of a cucumber albino mutant. Front. Plant Sci. 13:1047090. doi: 10.3389/fpls.2022.1047090
Received: 17 September 2022; Accepted: 03 October 2022;
Published: 20 October 2022.
Edited by:
Qiusheng Kong, Huazhong Agricultural University, ChinaReviewed by:
Rong Zhou, Aarhus University, DenmarkCopyright © 2022 Yan, Liu, Cao, Chen, Liang, Wang, Liu, Lin and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu'e Lin, bGlueXVlQGdkYWFzLmNu; Biao Jiang, amlhbmdiaWFvQGdkYWFzLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.