
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 31 October 2022
Sec. Plant Symbiotic Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1044070
This article is part of the Research Topic Rhizosphere Interactions: Root Exudates and the Rhizosphere Microbiome View all 23 articles
 Chuyun Yang1,2
Chuyun Yang1,2 Zhuomi Xie1,2
Zhuomi Xie1,2 Sheng Qian1,2
Sheng Qian1,2 Junyi Zhang3
Junyi Zhang3 Zhijian Yu1,2
Zhijian Yu1,2 Mingjie Li1,2
Mingjie Li1,2 Li Gu1,2
Li Gu1,2 Shuangshuang Qin4
Shuangshuang Qin4 Zhongyi Zhang1,2*
Zhongyi Zhang1,2*Previous studies have indicated that some Rehmannia glutinosa Leucine-rich repeat receptor-like protein kinases (LRR-RLKs) are involved in the formation of replant disease. However, it remains unclear how the interaction of LRR-RLKs with a key factor, the interaction between root exudates and Fusarium oxysporum, results in formation of replant disease. In this study, the influences of root exudates, F. oxysporum and the interaction of these two factors on expression of nine R. glutinosa LRR-RLKs (RgLRRs) were analyzed. The resulting eight RgLRRs of them were highly expressed at the early stage, and rapidly declined at later stages under mixed treatment of root exudates and F. oxysporum. The functions of nine RgLRRs under root exudates, F. oxysporum and mixed treatment of root exudates and F. oxysporum were preliminarily analyzed using transient overexpression and RNAi experiments. The results showed that high expression of RgLRR19, RgLRR21, RgLRR23 and RgLRR29 could decrease the damage to root cells from the mixed treatment of root exudates and F. oxysporum, but the interference of these genes enhanced the damage levels of root cells. Based on this, stable transgenic R. glutinosa seedlings were acquired. Overexpression of RgLRR29 conferred resistance of R. glutinosa seedlings to root exudates, F. oxysporum and mixed treatment. These results indicated that the continuous proliferation of F. oxysporum supported by root exudates altered the expression patterns of RgLRRs in R. glutinosa, then disordered the growth and development of R. glutinosa, finally leading to the formation of replant disease.
Rehmannia glutinosa, a plant in the Scrophulariaceae family, is a perennial herb that is widely cultivated in China (Li and Meng, 2015). R. glutinosa contains numerous pharmacologically active compounds, including catalpol, Rehmannia glycosides, Leonurus glycosides, Rehmannia polysaccharides, amino acids and stigmasterol. R. glutinosa is an important raw material in many traditional Chinese medicines. However, during production of R. glutinosa, its yield and quality are seriously affected by replant disease. Replant disease has continuously caused damage to R. glutinosa for at least 8-10 years (Zhang et al., 2013). Replanted R. glutinosa grown slowly, and the formation of tuberous roots may be arrested by replant disease (Li Q, et al., 2017). Thus, it is necessary and urgent to solve replant disease in production of R. glutinosa. However, effective methods to control replant disease have not been found until recently.
The majority of studies have demonstrated that an imbalance of the rhizosphere micro-ecological environment mediated by plant root exudates was the primary cause of replant disease (Grotewold and Vivanco, 2003; Xuan et al., 2005; Zhang and Lin, 2009). During the growth process of replanted plants, root exudates in the rhizosphere soils are the main source of allelotoxic substances (Bertin et al., 2003; Narula et al., 2008; Li Q et al., 2014). In previous studies on R. glutinosa, some secondary metabolites including ferulic acid, syringic acid and some flavonoids in root exudates, have been identified as important allelotoxic substances (Li et al., 2012; Zhang et al., 2015; Zhang et al., 2016). In addition, recent advances suggest that iridoid and phenylethanoid glycosides are also potential allelotoxic substances (Zhang et al., 2019). During the formation of replant disease, root exudates are the determining factor regulating the microbial biodiversity of the rhizosphere (Bais et al., 2006; Tu and Wu, 2010; Haichar et al., 2014; Chen et al., 2016). The abundance of Fusarium oxysporum spores was increased significantly in replanted R. glutinosa rhizosphere soils, and thus this fungus has been identified as the crucial pathogen in the formation of replant disease in R. glutinosa (Li et al., 2013; Li et al., 2016; Chen et al., 2019). Furthermore, root exudates of R. glutinosa in replanted conditions could induce proliferation of pathogenic pathogen F. oxysporum, resulting in aggravation of R. glutinosa disease (Li et al., 2016). In addition, the proliferation of F. oxysporum inhibited salicylic acid signal transduction and promoted the formation of replant disease (Chen et al., 2019). Therefore, current opinion holds that the interaction between root exudates and F. oxysporum is closely related to the formation of replant disease of R. glutinosa (Li et al., 2013; Wu et al., 2018). However, the interaction mechanism between root exudates and F. oxysporum in the formation of replant disease remains unclear.
Previous studies found that the genes related to plant immune systems were significantly upregulated in replanted R. glutinosa and that the interaction of these genes were closely related to the abnormal growth and death of replanted R. glutinosa (Chen et al., 2019; Li et al., 2017; Yang YH, et al., 2014, Yang et al., 2015). It is worth noting that in core processes of replant disease, immune system-related proteins such as Leucine-rich repeat receptor-like protein kinases (LRR-RLKs) and pathogenesis-related protein 10 (PR10) were specifically regulated in replanted R. glutinosa (Li MJ et al., 2017; Wu et al., 2015). The immune system of plants is primarily composed of two- layers of defense, effector-triggered immunity (ETI) and pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI). The first layer, PTI, can effectively recognize PAMPs located in the cell walls using conserved regions of the genes. LRR-RLKs are a large protein family in the PTI system that play important roles in plant growth, development, and defense response (Jones and Dangl, 2006; de Lorenzo et al., 2009; Li et al., 2018). Recent research had identified 40 RgLRRs gene members by screening the full-length RgLRRs genes in the full-length transcript of R.glutinosa, and a total of 27 RgLRRs genes were found that were up-regulated in the early stages of formation of R. glutinosa replant disease (Xie et al., 2019). These results suggested that the immune response system, especially RgLRRs, may play an important role in the interaction between rhizosphere harmful microbes and allelotoxic substances. However, these RgLRRs genes were screened under complicated field cultivation conditions, and the expression of R. glutinosa genes was affected by other factors, including biotic and abiotic stressors in the soil. The key mechanism of core immunity proteins that respond to replant disease is still unknown.
In this study, the root exudates and key pathogenic microbe F. oxysporum were used as leading factors in simulated stresses of R. glutinosa under replanted conditions. The dynamic changes of pathogenic microbes in the rhizosphere soils were investigated in detail. We chose nine RgLRRs that were highly expressed in replanted R. glutinosa, and the expression and physiological index levels of the nine RgLRRs were examined in the roots of R. glutinosa. As a representative RgLRR, the function of RgLRR29 under the treatments of root exudates, F. oxysporum, and the comprehensive stress of of root exudates and F. oxysporum was studied by reverse genetic methods. This study provides an effective method to examine the physiological changes and further study the expression patterns of key RgLRRs under replanted stress. These results will provide important clues for revealing insights into the formation mechanism of replant disease in R. glutinosa.
Tissue culture seedlings of R. glutinosa “Wen85-5” were grown in a tissue culture room at the Institute of GAP for Chinese Medicinal Materials, Fujian Agriculture and Forestry University. Root exudates solution was acquired using a root exudates collection device constructed in previous study (Feng et al., 2022). An F. oxysporum strain specific to replanted R. glutinosa (coded No. CCS043) was prepared from five-day-old cultures in Potato Dextrose Broth (PDB) medium. R. glutinosa seedlings were transferred into pots (10 cm ×10 cm) filled with complex medium consisting of peat and vermiculite (v: v = 2:1) after acclimatization (Chen et al., 2019), and grown in a greenhouse at 28 ± 2°C with a photoperiod of 14 h: 10 h light: dark. According to previous studies, root exudates, conidial suspensions of F. oxysporum and a mixture of the two (root exudates and F. oxysporum, v: v= 1:1) were selected as key stress factors to treat R. glutinosa seedlings (Li et al., 2016; Chen et al., 2019; Feng et al., 2022). The concentration levels of root exudates were adjusted to 1.60 mg·mL−1 with ddH2O, while conidial suspensions of F. oxysporum were adjusted to 1×108 conidia·mL−1. Three different solutions were used to irrigate R. glutinosa in pots with same volume of solution (10 mL) every two days. Each treatment was replicated three times. Root samples treated by different factors were collected at 0, 5, 10, and 15 days (DAP 0, 5, 10, and 15). Appearance of R. glutinosa under different treatments was carefully observed every five days. Root tips of plants from different treatments were sampled and stained by Trypan blue and DAB methods. All root samples were then cleaned and stored at −80°C after being frozen in liquid nitrogen for biochemical index and qRT-PCR analyses.
Previous studies have identified 40 R. glutinosa LRR-RLKs from the screening of the early transcriptomic data (Xie et al., 2019). Of these, nine RgLRRs (RgLRR19, RgLRR21, RgLRR23, RgLRR24, RgLRR25, RgLRR26, RgLRR27, RgLRR29, and RgLRR33) were found to be able to specifically respond to replant disease in field (Xie et al., 2019). To construct overexpression vectors, the full-length open reading frames (ORFs) of RgLRRs were cloned into the entry vector pBI121-EGFP digested by Kpn I and Xho I. To build vectors for RNAi, specific fragments in the range of 100–300 bp in the ORFs of RgLRRs were amplified. The specific fragments were cloned into the entry vector pRNAiGG (Yan et al., 2012).
For subcellular localization of the nine RgLRRs, the RgLRRs were fused with EGFP. The EGFP fusion constructs were driven by the double 35S promoter. All primers used in vector construction are shown in Tables S1a–c. Agrobacterium tumefaciens strain GV3101 containing 35S::RgLRRs-EGFP and 35S::GFP (used as a control) were grown overnight in LB solution containing 50 μg·mL−1 Kan and 100 μg·mL−1 Rif, and then adjusted to OD600 = 0.8. The bacterial solution was resuspended with the injection buffer (10 mM MgCl2, 100 mM 2-morpholinoethanesulfonic acid, and 200 μM acetosyringone (AS), pH = 5.8) and injected into leaves of Nicotiana benthamiana using a needle syringe. At 48 h after injection, the EGFP signals were observed with a laser scanning confocal microscope (LEICA TCS SP8).
The rhizosphere microbial DNA was extracted using a reference extraction kit (Beijing Tianmo, TD601, China). The quantity of F. oxysporum was detected using absolute quantification PCR (Chen et al., 2019). The specific primers of F. oxysporum (ITS1-F: 5’-CTTGGTCATTTAGAGGAAGTAA-3’, ITS4-R: 5’-TCCTCCGCTTATTGATATGC-3’) were used to amplify the DNA fragments using touchdown PCR. The bright electrophoretic strips of target genes were extracted using a gel pure DNA Kit (Magen, Guangzhou, China), and were inserted into a pMD19-T vector. Then, the vector DNA solution was transformed into E. coli DH 5α. One or two single white colonies were selected to identify whether the target genes were correctly cloned into the vector. The solutions containing the appropriate size of DNA fragments were chosen to extract plasmids using a Hipure Plasmid Micro Kit (Magen, Guangzhou, China) following the manufacturer’s instructions.
Plasmid solutions containing DNA fragments of correct size were amplified by qRT-PCR using the plasmid primer RV-M/M13-47. The concentration of target gene DNA was measured by a Nanodrop2000 spectrophotometer (Thermo Scientific, USA) and diluted to 0,1, 2, 3, and 4 ng·μL–1. The standard curve was drawn according to the DNA concentration of the target gene and Ct value. Finally, the soil DNA extracts were detected by qRT-PCR using F. oxysporum specific primers. The copy number of F. oxysporum spores were calculated from the standard curve. Each gene was analyzed with three replicates.
The root samples were used to detect the presence of in situ accumulation of superoxide and H2O2 by staining with 3,3-diaminobenzidine (DAB) and Trypan blue, respectively. In brief, root tips 2.0–2.5 cm in length were immediately immersed in an aqueous solution of 1 mg·mL–1 DAB in 50 mmol·L–1 potassium phosphate buffer (pH=6.4) and vacuum infiltrated and incubated for 12 h in the dark according to a previous report (Zhang et al., 2017). The root tips were placed in an ethanol: lactic acid: glycerol (3:1:1) mixture to boil for 5 min, and stored in 60% glycerol. The root tips were washed three times with water before photographing 10–15 individuals randomly sampled from each group in each experiment. According to the method of Liu (Liu et al., 2016), the root tips were immersed in 10% KOH, at 90°C for 1 h; the KOH solution was discarded, and the root tips were washed with ddH2O. Then, 1 mL of 2% HCl solution was added in the centrifuge tubes allowed to stand for 90 min. The HCl solution was discarded, and 1 mL Trypan blue staining solution was added (the final concentration of Trypan blue was 10 mg·mL-1) and stained for 30 min. The root tips were removed and added to 1 mL ethanol for overnight decolorization before photography using fluorescence microscope (Leica DM5000 B, Leica Microsystems Ltd., Heerbrugg, Switzerland).
The root samples of R. glutinosa seedlings under root exudates, F. oxysporum and the mixed solution of root exudates and F. oxysporum were used in these assays. Approximately 2 g of root samples were homogenized in 3 mL of 50 mM potassium phosphate buffer (pH=7.0). The supernatant was collected by centrifugation at 10,000×g for 10 min at 4°C, and then used for the activity determination of peroxidase (POD), catalase (CAT), superoxide dismutase (SOD), and malondialdehyde (MDA). POD activity was determined as guaiacol oxidation by H2O2. SOD activity was analyzed based on the inhibition rates of the reduction of nitro blue tetrazolium (NBT), CAT activity was determined as the H2O2 consumption (Aebi, 1983). MDA content was determined by the thiobarbituric acid reaction method according to our previous report (Peng et al., 2019). A chlorophyll meter (SPAD-502, Minolta Camera Co. Japan) was used to determine the total chlorophyll content according to our previous study (Wang et al., 2021).
The content of salicylic acid (SA) was detected using a one-step double-antibody sandwich enzyme-linked immunosorbent assay (ELISA). The content of SA was measured at 450 nm using a microplate reader (BIO-Tek ELX800, USA). Calculations of the ELISA data were performed as described in Wang et al. (2012).
The overexpression and RNAi vectors of RgLRRs were constructed in this study. The resulting constructs were transformed with the Agrobacterium tumefaciens GV3101 strain using the freeze-thaw method and verified by sequencing (Sangon, Shanghai, China). The A. tumefaciens strains GV3101 harboring RgLRRs-overexpression (RgLRRs-OX) or RgLRRs-RNAi (RgLRRs-Ri) transformation constructs were grown overnight in LB culture solution. The bacteria solutions were adjusted to OD600 = 0.8 and set to 5000/rpm for 5 min, and the bacteria were resuspended with the infection solution (containing 100 μM AS). To transiently transform RgLRRs genes in isolated tuber roots of R. glutinosa, 1 cm diameter segments of root tubers of R. glutinosa were selected and washed using flowing water for 1 h, and then soaked in 75% alcohol for 60 s. The root segments were then cut into 2 mm segments with the same thickness and equal size with a sterile scalpel, and treated in 0.3 MPa for 3 h with different infection solutions using a vacuum pump, respectively. The infected root segments were dried and connected to Murashige and Skoog (MS) solid medium containing 50 mg·L–1 AS. After dark culture for two days, these root segments were transferred into 1% agarose plate that were pre-cultivated in Petri dish at 26°C. Then, 20 μL root exudates solution, 20 μL conidial suspension of F. oxysporum, or 20 μL mixed solution of root exudates and F. oxysporum were added to the root segments every 12 h. Simultaneously, the area of mycelial expansion was used to test the resistance of root segments, the antioxidant enzyme activities, MDA content, and gene expression levels were determined for different treatments of root segments.
The overexpression vector of RgLRR29 was constructed in this study. The full-length ORF of RgLRR29 was cloned into pBI121-EGFP digested by Kpn I and Xho I sites. The expression of RgLRR29 was driven by double 35S promoters. For R. glutinosa seedling regeneration, approximately 20-day-old leaves were detached from the R. glutinosa seedlings, and cut along straight lines. The 100 leaf explants were dipped into a bacterial suspension (OD600 = 0.8) of A. tumefaciens GV3101, harboring the RgLRR29 transformation construct. After 30 min, the leaf explants were blotted dry with autoclaved filter paper, placed in MS basal medium containing 100 μM AS, and cultured at 25°C in the dark for two days. Then the leaf explants were transferred into differentiation medium containing 0.5 mg·L-1 NAA and 2 mg·L-1 6-BA under a 12-h light/12-h dark photoperiod for seedling regeneration, with the medium replaced with fresh medium every 15 days until seedling regeneration.
To evaluate whether the overexpressed RgLRR29 had been integrated into the transgenic R. glutinosa genome, total genomic DNA was isolated from the leaves of the transgenic lines using the cetyltrimethylammonium bromide (CTAB) method. DNA of transgenic plants was detected through the Kan gene with specific primers (Kan-F: CGTTCCATAAATTCCCCTCG; Kan-R: ATCTCGTGATGGCAGGTTGG). The expression level of RgLRR29 was determined by qRT-PCR.
The RgLRR29-OX lines were used to assess the expression levels by qRT-PCR. Then, the RgLRR29-OX and wild-type (WT) seedlings were transplanted into pots (20 cm×18 cm) containing organic matrix nutrition soils and grown in a greenhouse under 25°C with a photoperiod of 14 h: 10 h light: dark until the roots of the R. glutinosa expanded to the harvest period. The tuberous roots of RgLRR29-OX and WT R. glutinosa plants were used to culture the next generation of plants. When the fibrous roots were developed and elongated, then the tuberous roots were removed from the plants for avoiding influence of other factors. For studying the change of phenotype and physiological response in replanted R. glutinosa, RgLRR29-OX and WT seedlings were planted in plastic pots (10 cm ×10 cm), root exudates solution, conidial suspension of F. oxysporum and the mixed solution of root exudates and F. oxysporum spores were added, and the seedlings were grown in the above-mentioned greenhouse under normal field management for fifteen days. The roots of RgLRR29-OX and WT seedlings were sampled, immediately frozen in liquid nitrogen, and stored at –80°C for further analyses regarding gene expressions and physiological indexes.
Total RNA was isolated from 100 mg of R. glutinosa roots (fresh-weight) using a plant RNA extraction kit (Nanjing Vazyme Biotech Co., Ltd.). The cDNA synthesis was performed with the Evo M-MLV Mix Kit with gDNA Clean for qRT-PCR AG11728 (Accurate biotechnology (Hunan) Co., Ltd.). Each reaction contained 10 µL of 2 × SYBR Green Pro Taq HS Premix AG11701 (Accurate Biotechnology (Hunan) Co., Ltd.), 2 µL of template cDNA and 0.4 µL of each forward and reverse primers (10 µM). The data were normalized on the basis of the 18S rRNA (DQ469606) threshold cycle (Ct) value. All primers used in this experiment are shown in Table S1d. The qRT-PCR reaction procedure was performed as follows, incubation at 95°C for 2 min followed by 40 cycles at 95°C for 5 s and 60°C for 30 s. Each gene was tested in triplicates with three technical repeats. The expression level for each sample was expressed by the 2−△△CT method (Livak and Schmittgen, 2001). The data were exhibited as the mean ± SD of three independent experiments.
Raw data were compiled and regression analyses and graphs were prepared using Microsoft Excel. Multiple comparisons (LSD) were used to evaluate the significant differences between the compared values. Each value with three replicates represented as the mean ± SD. p < 0.05 was considered as significant between any two groups.
In previous study, a specific culture device was designed to isolate high-purity of root exudates of R. glutinosa (Feng et al., 2022). In this study, root exudates of R. glutinosa were collected based on this device. The root regions of R. glutinosa seedlings were irrigated with root exudates solution, conidial suspension of F. oxysporum and the mixed solution of root exudates and F. oxysporum spores to further investigate the interaction between root exudates and F. oxysporum. At 10 days after treatments, the copy number of F. oxysporum in the comprehensive treatment of root exudates and F. oxysporum was 1.82, 2.27 and 1.36 times of those in the control (CK), single root exudates and F. oxysporum treatments, respectively. At 15 days after treatments, the copy number of F. oxysporum in the comprehensive treatment of root exudates and F. oxysporum was 3.62 times of that in the F. oxysporum treatment group (Figure 1B). Appearance and key physiological indicators of R. glutinosa seedlings were analyzed to explore the effects of different treatments on the growth of R. glutinosa. Leaves of R. glutinosa treated by the comprehensive stress of root exudates and F. oxysporum displayed deeper yellowing and serious wilting, and some leaves presented symptoms of purple leaves in comparison to the controls (Figure 1A). At 15 days after treatment, the chlorophyll contents of leaves in the comprehensive treatment of root exudates and F. oxysporum were significantly lower than in the root exudates and F. oxysporum groups. The chlorophyll content for R. glutinosa leaves under both root exudates and F. oxysporum treatment decreased gradually compared with controls, but the chlorophyll content of the comprehensive treatment of root exudates and F. oxysporum was the lowest (Figure 1C). In the process of three treatments, the content of SA in the roots of plants under the treatment of F. oxysporum and the comprehensive treatment of root exudates and F. oxysporum gradually decreased, and the content of SA under root exudates treatment was firstly increased but then decreased, and the content of SA under the comprehensive treatment of root exudates and F. oxysporum was the lowest (Figure 1D). The results were consistent with previous studies in R. glutinosa treated by single or a combination of representative allelotoxic substances (Li et al., 2016; Feng et al., 2022), again indicating that root exudates was the key factor promoting the proliferation of F. oxysporum in the R. glutinosa rhizosphere.
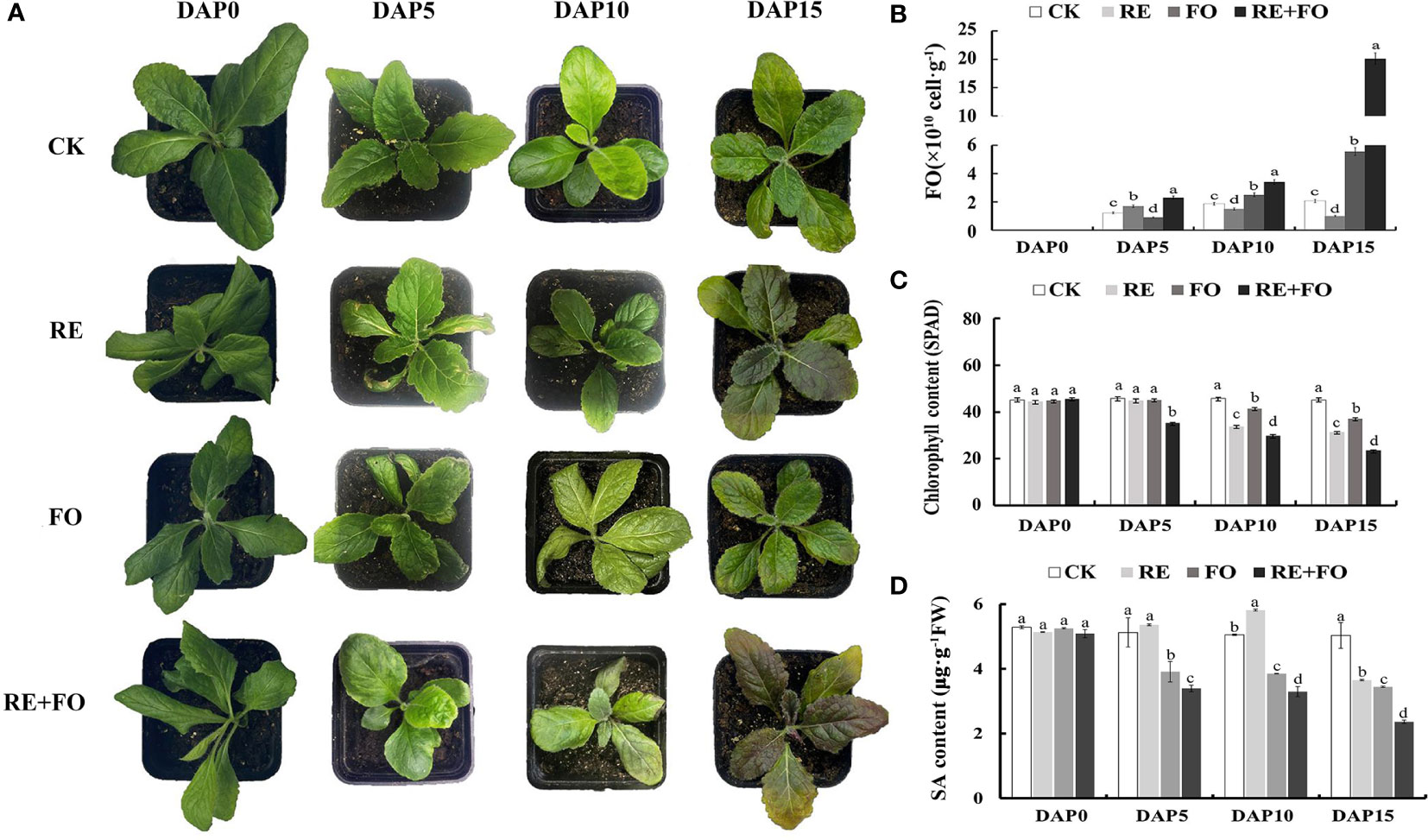
Figure 1 Morphological differences, Fusarium oxysporum proliferation, chlorophyll content and Salicylic acid (SA) content analysis of Rehmannia glutinosa seedlings under root exudates, F. oxysporum and comprehensive treatment of root exudates and F. oxysporum. (A) Phenotypic changes of R. glutinosa seedlings under root exudates, F. oxysporum and comprehensive treatment of root exudates and F. oxysporum. There were three biological replicates for each treatment; (B) Analysis of F.oxysporum proliferation in R. glutinosa rhizosphere soils under root exudates, F. oxysporum and comprehensive treatment of root exudates and F. oxysporum; (C) Chlorophyll content analysis of R. glutinosa under root exudates, F. oxysporum and comprehensive treatment of root exudates and F. oxysporum. (D) SA content analysis of R. glutinosa under root exudates, F. oxysporum and comprehensive treatment of root exudates and F. oxysporum. DAP, Days after planting; CK, Normalplanting “Wen85-5” R; glutinosa; RE, the treatment of root exudates; FO, the treatment of F. oxysporum; RE+FO; the comprehensive treatment of root exudates and F. oxysporum. Different letters represent a significant difference at P < 0.05.
The antioxidant enzyme activities of R. glutinosa seedlings roots were determined to study the damage level of R. glutinosa seedlings in treatments of root exudates, F. oxysporum and comprehensive stress of root exudates and F. oxysporum. The trends of POD and CAT activities were similar to SOD activities showing an upward trend in the early treated stages (days 1–5) and a downward trend at days 5–15 in comparison to controls (Figure 2A). Simultaneously, the MDA content of R. glutinosa seedlings under different treatments increased gradually with increasing treatment time under each treatment. Overall, the comprehensive treatment of root exudates and F. oxysporum significantly promoted the accumulation of reactive oxygen species (ROS) in R. glutinosa seedlings compared to treatment with single root exudates or F. oxysporum.
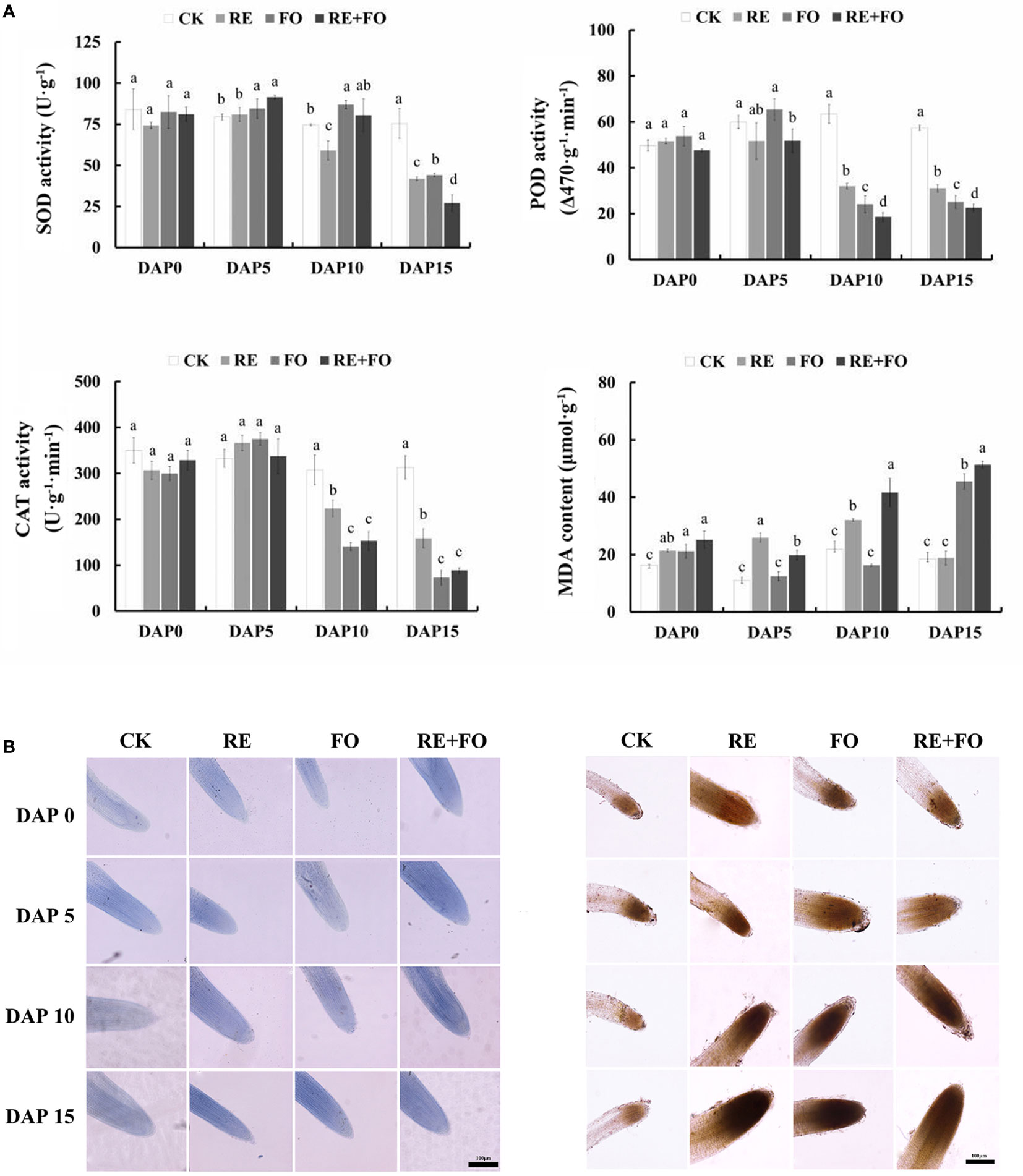
Figure 2 Changes in physiological indexes (A) and reactive oxygen species (ROS) levels (B) of Rehmannia glutinosa seedlings under root exudates, F. oxysporum and comprehensive stress of root exudates and F. oxysporum. (A): Measurement of SOD, POD, CAT and detection of MDA in R. glutinosa roots under root exudates, F. oxysporum and comprehensive stress of root exudates and F. oxysporum; (B): ROS levels of Rehmannia glutinosa root tip stained with Trypan blue (left) and DAB (right) mediated by the treatments of root exudates, F. oxysporum and comprehensive stress of root exudates and F. oxysporum. Bar = 100 μm. DAP, Days after planting; RE, the treatment of root exudates; FO, the treatment of F.oxysporum; RE+FO, the comprehensive treatment of root exudates and F. oxysporum. Different letters represent a significant difference at p < 0.05.
To further determine the oxidative damage of R. glutinosa seedlings under root exudates, F. oxysporum and comprehensive stress of the two factors visually, the oxidative damages levels of plant cells in R. glutinosa roots were detected by Trypan blue and DAB staining. The results suggested that the levels of damage of the root cells in R. glutinosa treated with root exudates, F. oxysporum and comprehensive stress of the two factors gradually deepened with increasing of treatment time compared to controls (Figure 2B). The reactive oxygen species content could be detected by the color depth in the root tips of R. glutinosa. It is worth noting that the root tips of plants treated with comprehensive stress of root exudates and F. oxysporum were the most severely damaged compared with the controls, root exudates or F. oxysporum treatments. These results indicated that the comprehensive treatment of root exudates and F. oxysporum increased the damage levels of F. oxysporum to R. glutinosa.
To reveal the molecular functions of these RgLRR genes, the nine RgLRRs was further cloned from R. glutinosa roots. To determine the localization of RgLRRs in cells, the complete coding regions of the nine RgLRRs were fused to the N-terminus of EGFP. The 35S::GFP-RgLRRs constructs were transiently expressed in leaves of N. benthamiana, and the green fluorescence from fusion proteins of 35S::GFP-RgLRRs was detected by fluorescence microscopy. The resulting green fluorescence were clearly observed in the cell membranes of N. benthamiana leaves (Figure 3). The results indicated that these RgLRRs of R. glutinosa were all located in the cellular membranes, similar to the cellular position of most plant LRR-RLK proteins.
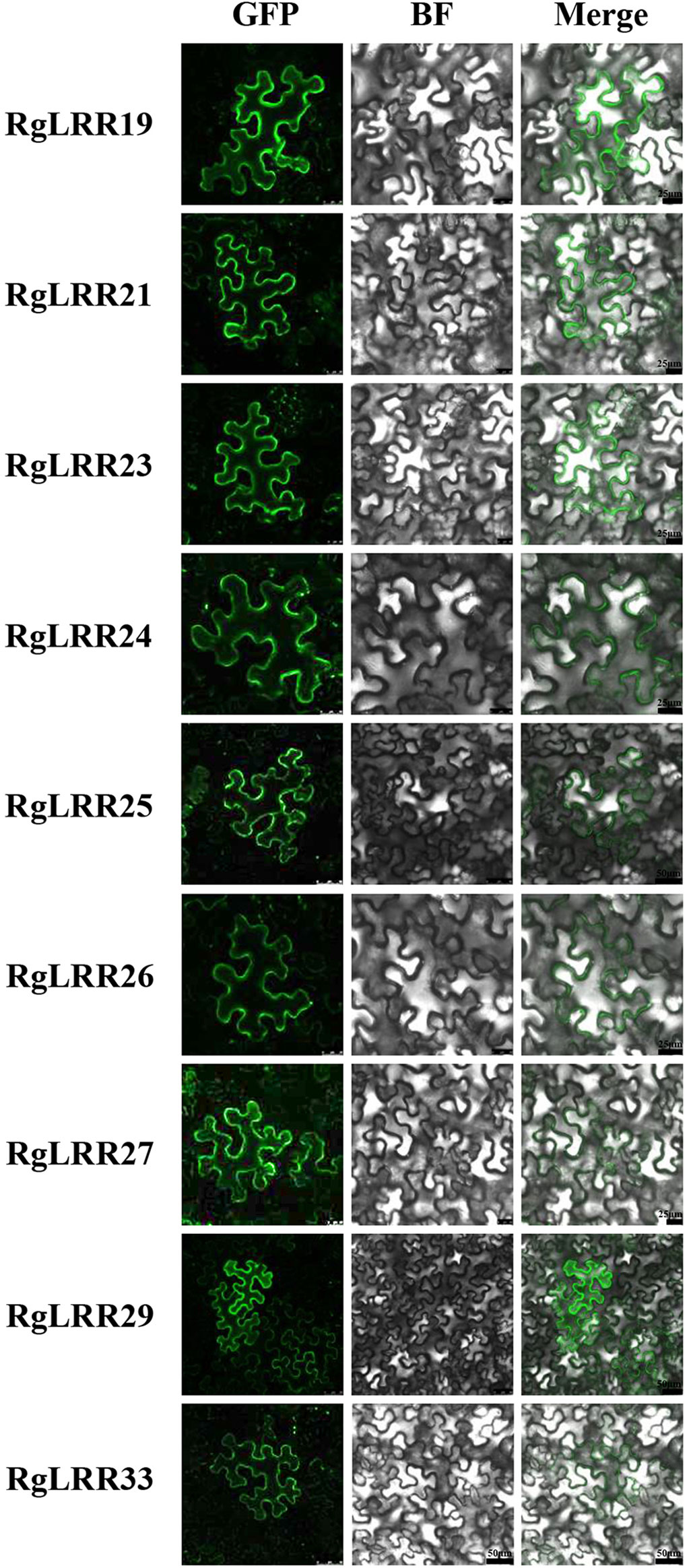
Figure 3 Subcellular localization of RgLRRs in Nicotiana benthamiana. GFP, Green fluorescence protein; BF, Bright field. There were five replicates for each experiment.
To examine the effects of root exudates, F. oxysporum, and their interaction in R. glutinosa, the expression patterns of RgLRRs in R. glutinosa under the treatments of root exudates, F. oxysporum and comprehensive stress of them were analyzed by qRT-PCR. The results showed that the expression patterns of the nine RgLRRs could be divided into three groups according to their expression patterns (Figure 4). The first group included RgLRR33 was weakly significantly expressed during the treatments. The second group consisted of RgLRR19, RgLRR21, RgLRR25, RgLRR26, RgLRR27, and RgLRR29, that were highly expressed at early stages and finally down-regulated at 15 days after three treatments. In the third group, the expression levels of RgLRR23 and RgLRR24 grew steadily. The expression trends of RgLRRs were consistent in that they were up-regulated at first but decreased gradually with increasing of F. oxysporum treatment time. However, RgLRR21, RgLRR23, RgLRR26, RgLRR29, and RgLRR33 were down-regulated at 10 days after the treatment of F. oxysporum, and other RgLRRs were down-regulated at 15 days after the treatment of F. oxysporum. From the expression profiling of all three groups, except for RgLRR33, the other eight RgLRRs all responded significantly to the treatment of comprehensive stress of root exudates and F. oxysporum.
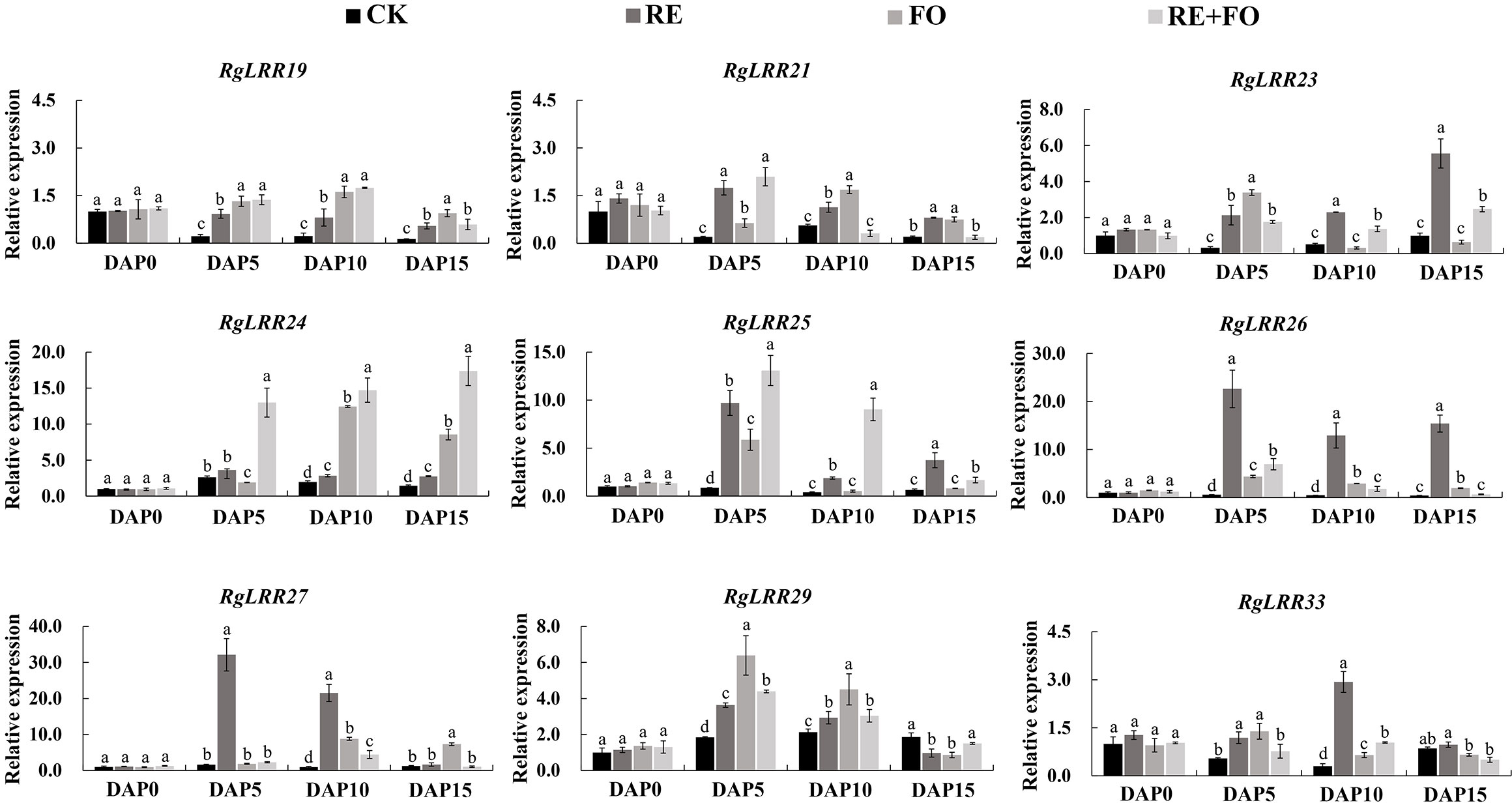
Figure 4 Expression patterns of crucial RgLRRs in Rehmannia glutinosa under the treatments of root exudates, F. oxysporum and comprehensive stress of root exudates and F. oxysporum. DAP, Days after planting; RE, the treatment of root exudates; FO, the treatment of F. oxysporum; RE+FO,the comprehensive treatment of root exudates and F. oxysporum. Different letters represent a significant difference at p < 0.05.
To preliminarily identify the functions of RgLRRs in different key replant disease factor stresses, nine RgLRRs (RgLRR19, RgLRR21, RgLRR23, RgLRR24, RgLRR25, RgLRR26, RgLRR27, RgLRR29 and RgLRR33) were transiently transformed into root segments and further treated by root exudates, F. oxysporum and comprehensive stress of them. The phenotypic changes from the transient RgLRRs overexpression and RNAi in R. glutinosa root segments under the treatments of root exudates, F. oxysporum and comprehensive stress of them were further observed (Figure S1). The results showed that the transient overexpression of RgLRRs genes in root segments of R. glutinosa except RgLRR33 showed higher resistance to these three treatments, but transient RNAi of RgLRRs genes resulted in the root segments (Figure 5A), which suffering increased damage, indicating that the transient overexpression of RgLRRs genes had an effect on the resistance to these stresses. In addition, after transient overexpression of different RgLRRs genes, the root segments showed different levels of resistance to the treatments of root exudates, F. oxysporum and comprehensive stress of them. The transient overexpression of RgLRR26 and RgLRR33 was associated with higher resistance to the treatment of root exudates. However, RgLRR19, RgLRR25, RgLRR26 and RgLRR29 may also play positive roles in the resistance to the effects to the treatment of F. oxysporum. In addition, transient overexpression of RgLRR19, RgLRR21, RgLRR23 and RgLRR29 resulted in the lowest degree of damage under the comprehensive treatment of root exudates and F. oxysporum (Figure 5B).
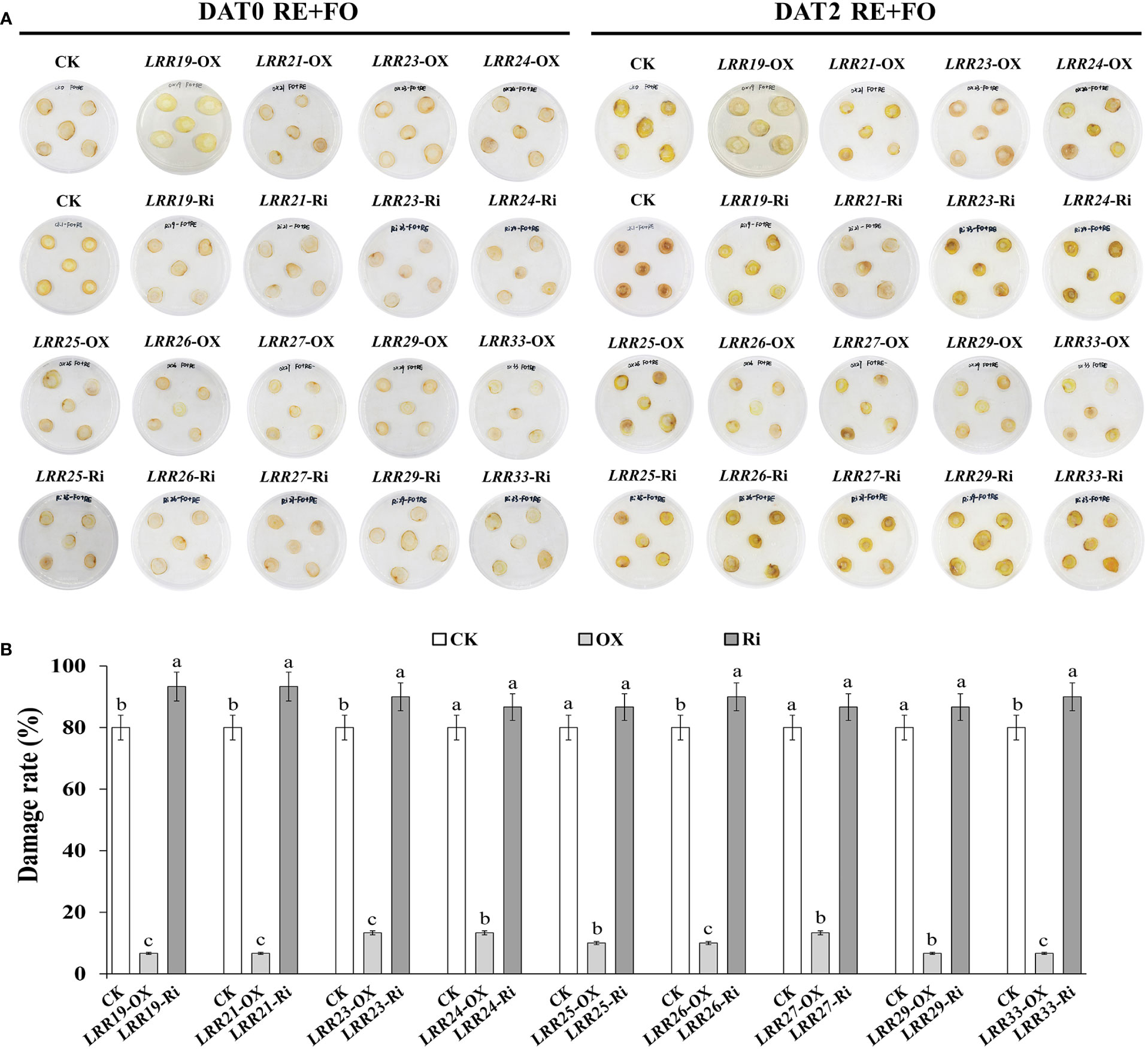
Figure 5 Phenotypic changes (A) and damage rate (B) of root segments with transient overexpression and interference of different RgLRRs under the treatment of comprehensive stress of root exudates and F. oxysporum. OX, Overexpression; Ri, RNA interference; CK, Empty vector as control; DAT, Days after treatment; RE+FO, the comprehensive treatment of root exudates and F. oxysporum. Different letters above bars indicated significant differences at P = 0.05 using Student-Newman-Keuls multiple comparisons.
To further explore the effects of these treatments on R. glutinosa root segments with transiently overexpressed and interfered RgLRRs, the antioxidant enzyme activities and MDA content were analyzed. As a result, the activities of SOD, POD and CAT in root segments with transient overexpressed RgLRRs were higher than those treated with empty vectors in R. glutinosa root segments, with lower content of MDA in root segments with transient overexpression of RgLRRs. It can be seen that RgLRR19-OX, RgLRR21-OX, RgLRR23-OX and RgLRR29-OX had the lowest damage degree of hydrogen peroxide through trypan blue staining results of root slices (Figure 6A). The opposite results occurred after RNAi of RgLRRs (Figures S1, S2). It was worth noting that under the comprehensive treatment of root exudates and F. oxysporum, the antioxidant enzyme activities of root segments with transient overexpressed RgLRR19, RgLRR21, RgLRR23 and RgLRR29 were higher than those of controls, and the MDA contents were lower (Figure 6B). These results indicated that the expression levels of RgLRRs may be related to resistance of the root cells to root exudates and F. oxysporum.
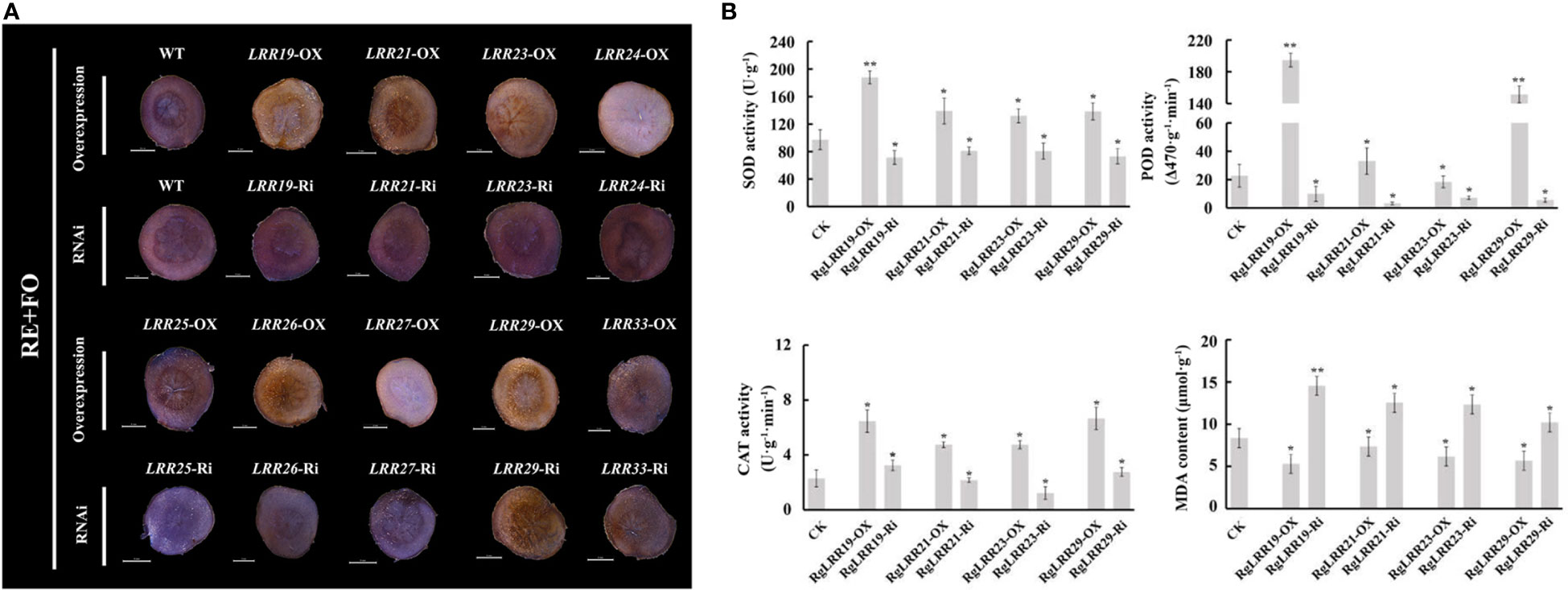
Figure 6 Changes of root segments with transient overexpression and RNAi of nine RgLRRs under comprehensive treatment of root exudates and F. oxysporum (A), Trypan blue staining of root segments with transient overexpression and RNAi of nine RgLRRs under comprehensive treatment of root exudates and F. oxysporum; (B), The physiological indexes analysis of root segments with transient overexpression and RNAi of crucial RgLRRs under comprehensive treatment of root exudates and F. oxysporum. OX, Overexpression; Ri, RNA interference; CK, Empty vector as control; RE+FO, the comprehensive treatment of root exudates and F. oxysporum. Asterisks indicate significant difference compared with the corresponding controls (*, P < 0.05; **, P < 0.01).
As a representative RgLRR, in this experiment, RgLRR29 was overexpressed in tissue-cultured R. glutinosa seedlings via Agrobacterium-mediated transformation using the leaf disc method. Leaf explant of R. glutinosa infected by Agrobacterium were regenerated on plates with resistance. The resulting 30-day shoots infected by Agrobacterium with RgLRR29 with resistance emerged from leaf explants. These shoots were grown to form complete culture seedling of R. glutinosa with over expressed RgLRR29 (RgLRR29-OX) for 90 days (Figure 7A). Then after three weeks, the root morphogenesis of WT and transgenic R. glutinosa shoots cultured in rooting medium was established (Figure 7B). After these candidate transgenic RgLRR29-OX lines were fully acclimatized, they were transferred into pots with peat and vermiculite matrix. Simultaneously, PCR was used to validate the positive RgLRR29-OX lines in these new R. glutinosa seedling that emerged from the tuberous roots. The results showed that 952 bp of the kanamycin gene were specifically amplified from the DNA of 4 transgenic plants (RgLRR29-OX-S1, -S2, -S3 and -S4) while the WT (wild-type) seedlings showed no amplification (Figure 7C). Simultaneously, qRT-PCR was used to analyze the expression levels of RgLRR29 in RgLRR29-OX lines and wild lines. Compared with the WT, the expression levels of the RgLRR29 in OX-S1#, OX-S2#, OX-S3#, and OX-S4# increased by 75, 18, 14, and 64-fold, respectively. Resulting the RgLRR29-OX-S1 and RgLRR29-OX-S4 lines presented higher expression levels of RgLRR29 compared with WT seedlings (Figure 7D). ROS accumulation was significantly induced in RgLRR29-OX leaves compared with WT plant leaves of the same age (Figure 7E). In this study, we found that plant height, the number of adventitious roots and the rooting rate of RgLRR29-overexpression plants were significantly lower than those of WT plants when they were cultured in MS medium (Figure 7F). These results indicated that transgenic R. glutinosa seedlings of overexpression RgLRR29 were successfully acquired, thereby providing materials for verifying its function in the treatments of root exudates, F. oxysporum and comprehensive stress of root exudates and F. oxysporum.
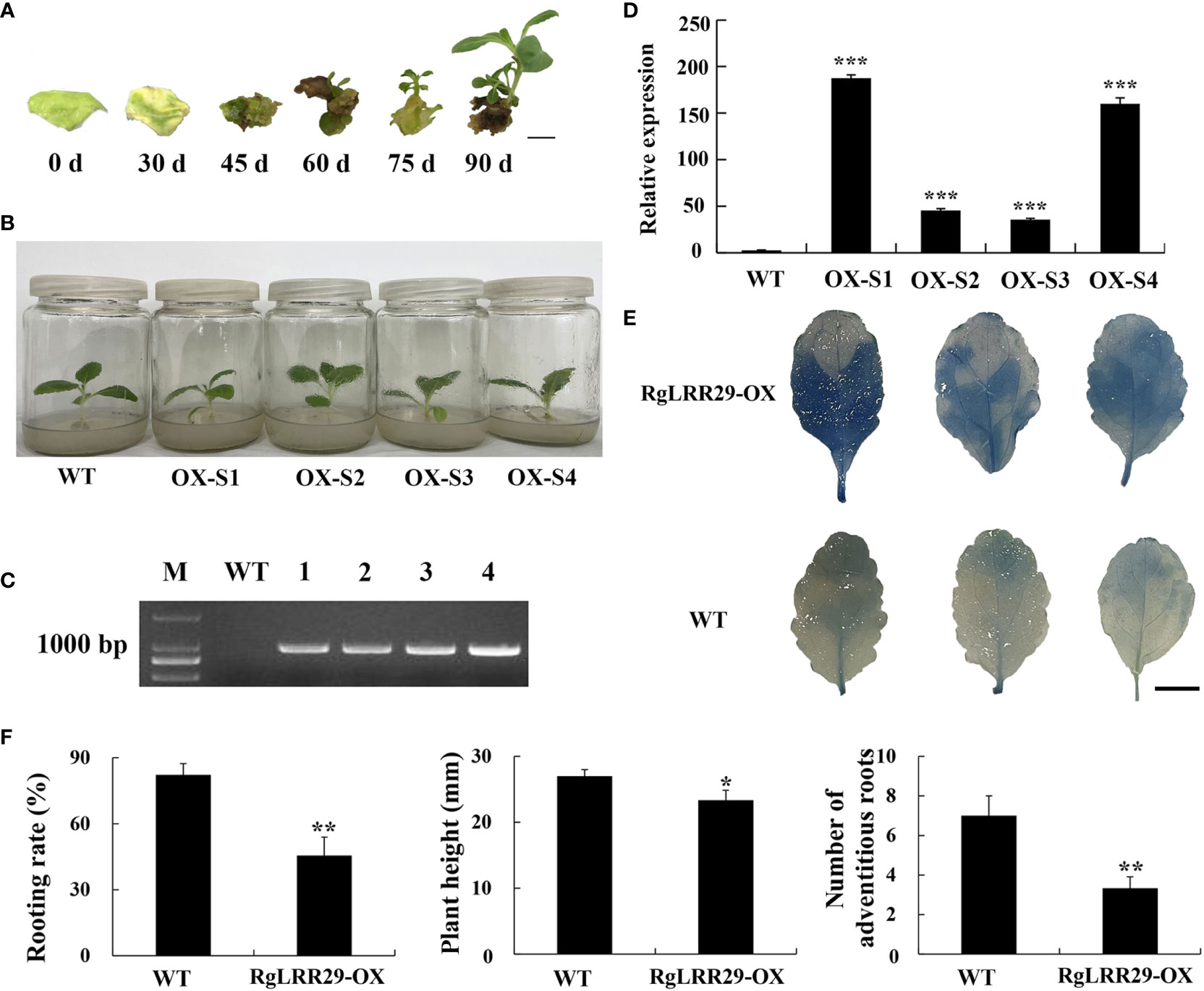
Figure 7 Confirmation of positive RgLRR29-overpression Rehmannia glutinosa. (A) Regeneration process of RgLRR29-overexpression R. glutinosa plants. Bar = 1cm; (B) Morphology of RgLRR29-overexpression R. glutinosa plants cultured in MS medium for three weeks. OX-S1, OX-S2, OX-S3, and OX-S4 represent different lines of RgLRR29-overexpression R. glutinosa plants. Wild-type (WT) plants of the same age are shown as the control; (C) PCR products for the positive screening of transgenic lines; Lanes M and WT represented the DL2000 size marker and WT plants, respectively; Lanes 1–4 represented RgLRR29-overexpression lines 1 to 4, respectively; (D) Expression patterns of different RgLRR29-overexpression lines (OX-S1, OX-S2, OX-S3, OX-S4) in roots. The error bars represent the standard error (n = 3) (***: p < 0.001); (E) ROS accumulation in RgLRR29-overexpression and WT R. glutinosa plants. Trypan blue staining was used to detect ROS accumulation in the leaves of 2-week-old RgLRR29-overexpression and WT R. glutinosa plants; (F) The rooting rate, Plant height and the number of adventitious roots of WT and RgLRR29-overexpression R. glutinosa plants. The rooting rate was calculated from three independent experiments. More than 15 plants were subjected to root induction each time. Other data are shown for one representative result of three independent experiments; the results are shown as the average ± SE (n = 5). *P < 0.05 and **P < 0.01 indicate significant differences based on the t-test.
After 120 days, RgLRR29-OX-S1, RgLRR29-OX-S4, and WT R. glutinosa seedlings generated tuberous roots (Figure 8A). The diameter of roots of RgLRR29-overexpressing plants was less than WT plants (Figure 8B). However, the root length of RgLRR29-overexpressing plants was longer than WT plants (Figure 8C). To further confirmed the biological function of RgLRR29-OX in R. glutinosa under the treatments of root exudates, F. oxysporum and comprehensive stress of root exudates and F. oxysporum, RgLRR29-OX and WT seedlings were cultivated in pots and irrigated with root exudates solution, conidial suspensions of F. oxysporum, and a mixture of the two in the root regions. After 15 days of treatment, the transgenic plants displayed significantly less wilting and lower degree of root browning and rot symptoms than the WT seedlings. Under the treatments of root exudates, F. oxysporum and comprehensive stress of root exudates and F. oxysporum, the antioxidant system including SOD, POD, and CAT in RgLRR29-OX also showed higher activities compared with WT seedlings. In contrast, the content of MDA was lower than WT under the treatments of root exudates, F. oxysporum and comprehensive stress of root exudates and F. oxysporum (Figure 8D), and it may positively regulate the immune defense of R. glutinosa against replant disease. The results indicated that RgLRR29-OX showed higher resistance to root exudates, F. oxysporum, and the double stress compared with WT seedling.
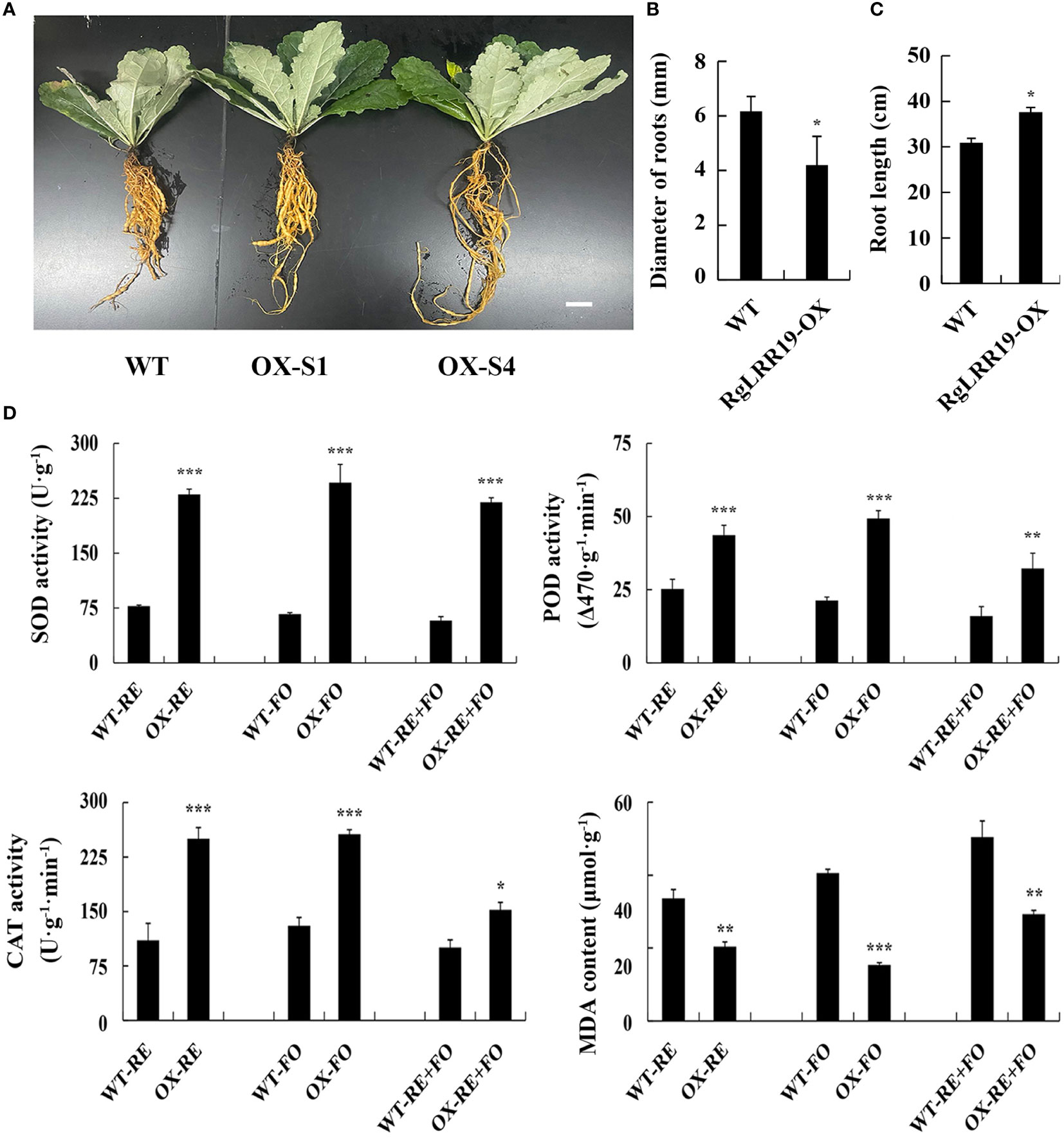
Figure 8 RgLRR29-overexpression improves plant resistance against root exudates, F. oxysporum and comprehensive stress of root exudates and F. oxysporum. (A–C), Phenotypic analysis of the WT and transgenic lines. Bar = 2 cm. (D) Changes of physiological indexes in WT and RgLRR29-overexpression Rehmannia glutinosa plants under the treatments of root exudates, F. oxysporum and comprehensive stress of root exudates and F. oxysporum. RE, the treatment of root exudates; FO, the treatment of F. oxysporum; RE+FO, the comprehensive treatment of root exudates and F. oxysporum. The error bars represent the standard error (n = 3) (*: p < 0.05; **: p < 0.01; ***: p < 0.001).
Increasing evidence has suggested that replant disease is the results of the comprehensive effects of various factors in the “plant-soil-microbes” system. Among these, the main causes that contribute to the occurrence of replant disease have been found to be allelotoxic substances, microbes, and the imbalance of rhizosphere microecology induced by their interaction (Bertin et al., 2003; Li et al., 2014; Yang RX et al., 2014; Ji et al., 2021; Feng et al., 2022). In other words, allelotoxic substances promote continuously the proliferation of harmful microbes in rhizosphere soils, which then infect the replanted plants and disturb their normal growth (Zhang and Lin, 2009; Dong et al., 2016; Zhang et al., 2016; Westerveld and Shi, 2021). Root exudates and litter mixtures are main sources of allelotoxic substances, which play important roles in regulating microbial community (Jin et al., 2022). Root exudates were be focused on numerous studies of replant disease (Zhang et al., 2013). In this study, we also indicated that root exudates of R. glutinosa in replanted condition could induce chemotaxis proliferation of its pathogenic pathogen F. oxysporum. It was verified once again that the imbalance of microecological environment in the rhizosphere soils caused by root exudates possibly is an important factor causing R. glutinosa replanted disease.
Some studies on the mechanism of replant disease have indicated that the interaction affects each other among allelotoxic substances, harmful microbes and immune-related proteins are closely related to the death of replanted R. glutinosa (Wu et al., 2013; Zhang et al., 2013). To reveal the roles of immune-related proteins during the formation of replant disease, this study analyzed in detail the effects of allelotoxic substances using root exudates collected using specific device (Feng et al., 2022) and harmful microbes on R. glutinosa seedlings from tissue culture under controlled conditions. Using tissue culture seedlings and controlled conditions could avoid the interference of other biotic or abiotic stress factors in the field. In previous studies of replant disease, single or several representative kinds of allelotoxic substances and rhizosphere microbes were used to simulate the interactions between two factors in rhizosphere soils (Zhang et al., 2015; Li et al., 2016). Root exudates have significant allelopathic activity and are the main source of allelotoxic substances (Liu et al., 2010; Zhang et al., 2015). These results more effectively reflected the interactions between allelotoxic substances and harmful microbes compared to replacing several specific allelotoxic substances by root exudates. Among nine RgLRRs in R. glutinosa, there were at least eight RgLRRs that responded to the comprehensive stress of root exudates and F. oxysporum. A total of six RgLRRs (RgLRR19, RgLRR21, RgLRR25, RgLRR26, RgLRR27, and RgLRR29) were highly expressed in the early stages and decreased in the later stages under the stresses of root exudates and F. oxysporum. It is worth noting that their expression patterns showed similar trends with the expression patterns of RgLRRs at different developmental stages of replanted R. glutinosa in natural fields (Xie et al., 2019). This result suggested that early infection of F. oxysporum in R. glutinosa activated the expression of RgLRRs, but with the continuous infection triggered by root exudates, the plant immune system was gradually inhibited. In conclusion, these results preliminarily confirmed that the interaction between root exudates and rhizosphere microbes affected the expression of key immune-related proteins in R. glutinosa during the formation of replant disease.
Previous studies have demonstrated that the function of plant immune proteins will be gradually diminished during the process of F. oxysporum infection of R. glutinosa (Chen et al., 2019). In the field, the continuous accumulation of allelochemicals in rhizosphere soils of replanted plants has led to the proliferation of harmful microbes in the rhizosphere (Zhang et al., 2013; Jiao et al., 2015; Dong et al., 2016; Zhou and Wu, 2018; Westerveld and Shi, 2021). During this, the functions of plant immune proteins will be seriously diminished in comparison to responses against single pathogenic microbes, making replanted plants more susceptible to disease infection. LRR-RLKs have been identified in many plant species and have been implicated in regulating the processes of plant growth, development, and responses to biotic and/or abiotic stresses (Jones and Dangl, 2006; de Lorenzo et al., 2009; Halter et al., 2014). Most of the RLKs identified as being involved in plant defense are of the LRR-RLK class including the rice Xa21 protein and the Arabidopsis Flagellin Sensitive 2 (FLS2) and bacterial translation elongation factor EF-Tu receptor (EFR) (Song et al., 1995; Chinchilla et al., 2007; Schoonbeek et al., 2015). During the interaction of plants and microbes, overexpression of LRR-RLK genes can increase plant resistance to pathogens. For example, overexpression of OsSERK1 in two rice cultivars led to an increase in host resistance to a blast fungus (Hu et al., 2005). GbSOBIR1 played a critical role in Gossypium barbadense resistance to Verticillium dahliae (Zhou et al., 2018). Overexpression of MdBAK1 in Malus domestica inhibited colonization of F. oxysporum in host plants (Liu et al., 2022). Therefore, the enhancement of the activity of immune-related proteins might increase the resistance of replanted plants to harmful microbes. In this study, root segments displaying transient overexpression of RgLRR19, RgLRR21, RgLRR23, or RgLRR29 showed higher resistance levels to a mixed stress of RE and FO. Furthermore, stable overexpression of RgLRR29 effectively improved resistance of R. glutinosa under the treatments of root exudates, F. oxysporum and comprehensive stress of root exudates and F. oxysporum. There existed a trade-off between the immune responses and plant growth and development, so overexpression of RgLRR29 affected the root expansion. However, it has been shown that RgLRR29-OX could decrease the mortality levels of R.glutinosa seedlings compared to WT seedlings. These results suggested that RgLRR proteins may be key proteins in the interactions between root exudates and plant, and that the activity degree of RgLRRs determined the damage degree of replanted R. glutinosa. In addition, this study preliminarily indicated that activity of RgLRRs weakened with the proliferation of F. oxysporum induced by root exudates in replanted R.glutinosa.
In conclusion, this study confirmed that root exudates could induce F. oxysporum to colonize in the rhizosphere soils of R. glutinosa and promote the formation of replant disease. RgLRRs played important roles in the process of replant disease. Through the overexpression of key RgLRR, we found that the overexpressing R. glutinosa seedlings had enhanced resistance to the combined stresses of root exudates and F. oxysporum. Hence, this study preliminarily confirmed the interaction among plant immune key proteins, allelopathic substances, and F. oxysporum, thereby providing a key breakthrough for further revealing the mechanism of R. glutinosa replant disease. In addition, the acquisition and identification of an RgLRR protein has provided a key clue for preventing or reducing the harmful effects of replant disease.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
CY and ZX conceived the study. ZZ supervised this research. CY, SQ, ZY, and ZX performed the experiments. ML, LG, JZ and SSQ provided technical help. CY and ZX analyzed the data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This research was financially supported by the National Natural Science Foundation of China (Grant No. 81573538), the Natural Science Foundation of Fujian Province (2020J01531), the Distinguished Youth Fund of Fujian Agriculture and Forestry University (Kxjq20010), and the Special Fund for Science and Technology Innovation of Fujian Agriculture and Forestry University (CXZX2020010A).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1044070/full#supplementary-material
Supplementary Figure 1 | Phenotypic changes of root segments with transient overexpression and interference of different RgLRRs under the treatments of root exudates, F. oxysporum and comprehensive stress of root exudates and F. oxysporum. OX, Overexpression; Ri, RNA interference; CK, Empty vector as control; DAT, Days after treatment; RE, the treatment of root exudates; FO, the treatment of F. oxysporum; RE+FO, the comprehensive treatment of root exudates and F. oxysporum; There were three replicates for each treatment.
Supplementary Figure 2 | Trypan blue staining analysis of root segments with transient overexpression and RNAi of nine RgLRRs under the treatments of root exudates, F. oxysporum and comprehensive stress of root exudates and F. oxysporum. CK, Empty vector as control; RE, the treatment of root exudates; FO, the treatment of F. oxysporum; RE+FO, the comprehensive treatment of root exudates and F. oxysporum; Bar=5 mm, There were three replicates for each treatment.
Supplementary Figure 3 | Antioxygen enzyme activities and MDA content analysis of root segments with transient overexpression and RNAi of nine RgLRRs under the treatments of root exudates, F. oxysporum and comprehensive stress of root exudates and F. oxysporum. OX, Overexpression; Ri, RNA interference; CK, Empty vector as control.
Aebi, H. E. (1983). Catalase in: Methods of enzymatic analysis. (Bergmeyer, H.U. (Ed.)). Verlag Chemie Weinhem. 3, 273–286. doi: 10.1016/B978-0-12-091302-2.50032-3
Bais, H. P., Weir, T. L., Perry, L. G., Gilroy, S., Vivanco, J. M. (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57 (01), 233–266. doi: 10.1146/annurev.arplant.57.032905.105159
Bertin, C., Yang, X., Weston, L. A. (2003). The role of root exudates and allelochemicals in the rhizosphere. Plant Soil. 256 (01), 67–83. doi: 10.1023/A:1026290508166
Chen, A. G., Gu, L., Xu, N., Feng, F., Zhang, Z. (2019). NB-LRRs not responding consecutively to Fusarium oxysporum proliferation caused replant disease formation of Rehmannia glutinosa. Int. J. Mol. Sci. 20 (13), e3203. doi: 10.3390/ijms20133203
Chen, A. G., Li, M. J., Zhang, B., Wang, F. J., Gu, L., Feng, F. J., et al. (2016). Review on catastrophe mechanism of medicinal plant and its rhizosphere microecosystem mediated by consecutive monoculture. Mod. Chin. Med. 18, 239–245. doi: 10.13313/j.issn.1673-4890.2016.2.026
Chinchilla, D., Zipfel, C., Robatzek, S., Kemmerling, B., Nürnberger, T., Jones, J. D. G., et al. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 448, 497–500. doi: 10.1038/nature05999
de Lorenzo, L., Merchan, F., Laporte, P., Thompson, R., Clarke, J., Sousa, C., et al. (2009). A novel plant leucine-rich repeat receptor kinase regulates the response of Medicago truncatula roots to salt stress. Plant Cell. 21 (2), 668–680. doi: 10.1105/tpc.108.059576
Dong, L. L., Xu, J., Feng, G. Q., Li, X. W., Chen, S. L. (2016). Soil bacterial and fungal community dynamics in relation to Panax notoginseng death rate in a continuous cropping system. Sci. Rep. 6, 31802. doi: 10.1038/srep31802
Feng, F. J., Yang, C. Y., Li, M. J., Zhan, S. Y., Liu, H. Y., Chen, A. G., et al. (2022). Key molecular events involved in root exudates-mediated replanted disease of Rehmannia glutinosa. Plant Physiol. Biochem. 172, 135–150. doi: 10.1016/j.plaphy.2022.01.014
Grotewold, E., Vivanco, J. M. (2003). Root exudation and rhizosphere biology. Plant Physiol. 132, 44–51. doi: 10.1104/pp.102.019661
Haichar, F. Z., Santaella, C., Heulin, T., Achouak, W. (2014). Root exudates mediated interactions belowground. Soil Biol. Biochem. 77, 69–80. doi: 10.1016/j.soilbio.2014.06.017
Halter, T., Imkampe, J., Mazzotta, S., Wierzba, M., Postel, S., Bücherl, C., et al. (2014). The leucine-rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr. Biol. 24, 134–143. doi: 10.1016/j.cub.2013.11.047
Hu, H., Xiong, L., Yang, Y. (2005). Rice SERK1 gene positively regulates somatic embryogenesis of cultured cell and host defense response against fungal infection. Planta. 222, 107–117. doi: 10.1007/s00425-005-1534-4
Jiao, X. L., Bi, X. B., Gao, W. W. (2015). Allelopathic effect of p-coumaric acid on American ginseng and its physiological mechanism. Acta Ecol. Sin. 35, 3006–3013. doi: 10.5846/stxb201306141707
Ji, L., Nasir, F., Tian, L., Chang, J. J., Sun, Y., Zhang, J. F., et al. (2021). Outbreaks of root rot disease in different aged American ginseng plants are associated with field microbial dynamics. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.676880
Jin, X., Wang, Z. L., Wu, F. Z., Li, X. G., Zhou, X. G. (2022). Litter mixing alters microbial decomposer community to accelerate tomato root litter decomposition. Microbiology Spectrum. 10 (3), e00186–22. doi: 10.1128/spectrum.00186-22
Jones, J, D, G., Dangl, J. L. (2006). The plant immune system. Nature. 444 (7117), 323–329. doi: 10.1038/nature05286
Li, X. G., Ding, C. F., Hua, K., Zhang, T. L., Zhang, Y. N., Zhao, L., et al. (2014). Soil sickness of peanuts is attributable to modifications in soil microbes induced by peanut root exudates rather than to direct allelopathy. Soil Biol. Biochem. 78, 149–159. doi: 10.1016/j.soilbio.2014.07.019
Li, Z. F., He, C. L., Wang, Y., Li, M. J., Dai, Y. J., Wang, T., et al. (2016). Enhancement of trichothecene mycotoxins of Fusarium oxysporum by ferulic acid aggravates oxidative damage in Rehmannia glutinosa libosch. Sci. Rep. 6, 33962. doi: 10.1038/srep33962
Li, H. W., Meng, X. L. (2015). Research progress on chemical constituents and pharmacological activities of Rehmannia glutinosa. Drug Eval. Res. 38, 218–228. doi: 10.7501/j.issn.1674-6376.2015.02.022
Li, X. X., Salman, A., Guo, C., Yu, J., Cao, S. X., Gao, X. M., et al. (2018). Identification and characterization of LRR-RLK family genes in potato reveal their involvement in peptide signaling of cell fate decisions and biotic/abiotic stress responses. Cells. 7 (9), e120. doi: 10.3390/cells7090120
Liu, P., Wan, S. B., Jiang, L. H., Wang, C. B., Liu, Z. H., Zhao, H. J., et al. (2010). Autotoxic potential of root exudates of peanut (Arachis hypogaea l.). Allelopathy J. 26 (2), 197–205. doi: 10.1007/s10457-010-9321-z
Liu, F., Xu, Y. J., Jiang, H. H., Jiang, C. S., Du, Y. B., Gong, C., et al. (2016). Systematic identification, evolution and expression analysis of the Zea mays PHT1 gene family reveals several new members involved in root colonization by arbuscular mycorrhizal fungi. Int. J. Mol. Sci. 17 (6), e930. doi: 10.3390/ijms17060930
Liu, X. X., Xu, S. Z., Wang, X. P., Xin, L., Wang, L. S., Mao, Z. Q., et al. (2022). MdBAK1 overexpression in apple enhanced resistance to replant disease as well as to the causative pathogen fusarium oxysporum. Plant Physiol. Bioch. 179, 144–157. doi: 10.1016/j.plaphy.2022.03.014
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–△△Ct method. Methods. 25, 402–408. doi: 10.1006/meth.2001.1262
Li, Q., Yang, S. P., Cui, G. L., Huang, J. G., Li, L. Y., Cheng, Y. Y. (2017). Microbial biomass, enzyme activity and composition of the fungal community in rhizospheric soil cropped with Artemisia annua for several years. Acta Prataculturae Sinica. 1, 34–42. doi: 10.11686/cyxb2016091
Li, M. J., Yang, Y. H., Feng, F. J., Zhang, B., Chen, S. Q., Yang, C. Y., et al. (2017). Differential proteomic analysis of replanted Rehmannia glutinosa roots by iTRAQ reveals molecular mechanisms for formation of replant disease. BMC Plant Biol. 17, 116. doi: 10.1186/s12870-017-1060-0
Li, Z. F., Yang, Y. Q., Wu, L. K., Shu, Y., Zhao, Y. P., Huang, W. M., et al. (2013). Isolation of highly pathogenic pathogens and identification of formal specials of Rehmannia glutinosa l. Chin. J. Eco Agricult. 11, 1426–1433. doi: 10.3724/SP.J.1011.2013.01426
Li, Z. F., Yang, Y. Q., Xie, D. F., Zhu, L. F., Zhang, Z. G., Lin, W. X. (2012). Identification of autotoxic compounds in fibrous roots of Rehmannia (Rehmannia glutinosa libosch.). PLos One 7 (1), e28806. doi: 10.1371/journal.pone.0028806
Narula, N., Kothe, E., Behl, R. K. (2008). Role of root exudates in plant-microbe interactions. J. Appl. Bot. Food Qual. 82 (02), 122–130. doi: 10.1614/IPSM-08-126.1
Peng, X., Wu, H., Chen, H., Zhang, Y., Qiu, D., Zhang, Z. (2019). Transcriptome profiling reveals candidate flavonol-relatedgenes of Tetrastigma hemsleyanum under cold stress. BMC Geno. 20, 687. doi: 10.1186/s12864-0196045-y
Schoonbeek, H. J., Wang, H. H., Stefanato, F. L., Craze, M., Bowden, S., Wallington, E., et al. (2015). Arabidopsis EF-tu receptor enhances bacterial disease resistance in transgenic wheat. New Phytol. 206, 606–613. doi: 10.1111/nph.13356
Song, W. Y., Wang, G. L., Chen, L. L., Kim, H. S., Pi, L. Y., Holsten, T., et al. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene. Xa21. Sci. 270, 1804–1806. doi: 10.1126/science.270.5243.18
Tu, S. X., Wu, J. (2010). A review on research methods of root exudates. Ecol. Environ. Sci. 19 (9), 2493–2500. doi: 10.16258/j.cnki.1674-5906.2010.10.021
Wang, Y., Li, B., Du, M. W., Eneji, A. E., Wang, B. M., et al. (2012). Mechanism of phytohormone involvement in feedback regulation of cotton leaf senescence induced by potassium deficiency. J. Exp. Bot. 63(16), 5887–5901. doi: 10.1093/jxb/ers238
Wang, Y. H., Xie, Z. M., Wang, X. H., Peng, X., Zheng, J. P. (2021). Fluorescent carbon-dots enhance light harvesting and photosynthesis by overexpressing PsbP and PsiK genes. J. Nanobiotechnol. 19, 260. doi: 10.1186/S12951-021-01005-0
Westerveld, S. M., Shi, F. (2021). The history, etiology, and management of ginseng replant disease: a Canadian perspective in review. Can. J. Plant Science. 101 (06), 886–901. doi: 10.1139/CJPS-2021-0106
Wu, L. K., Huang, W. M., Wang, J. Y., Wu, H. M., Chen, J., Qin, X. J., et al. (2015). Diversity analysis of rhizosphere microflora of wild R. glutinosa grown in monocropping for different years. Acta Agronomica Sinica. 2, 308–317. doi: 10.3724/SP.J.1006.2015.00308
Wu, L. K., Li, Z. F., Li, J., Azam, Khan., M., Huang, W. M., et al. (2013). Assessment of shifts in microbial community structure and catabolic diversity in response to Rehmannia glutinosa monoculture. Appl. Soil Ecol. 67, 1–9. doi: 10.1016/j.apsoil.2013.02.008
Wu, L. K., Wang, J. Y., Wu, H. M., Chen, J., Xiao, Z. G., Qin, X. J., et al. (2018). Comparative metagenomic analysis of rhizosphere microbial community composition and functional potentials under Rehmannia glutinosa consecutive monoculture. Int. J. Mol. Sci. 19 (8), e2394 doi: 10.3390/ijms19082394
Xie, Z. M., Yang, C. Y., Chen, A. G., Li, M. J., Gu, L., Zhang, J. Y., et al. (2019). Identification and expression analysis of leucine-rich repeat receptor-like kinase family reveals the roles of resistance proteins during formation of replant disease in Rehmannia glutinosa. Int. J. Agri Biol. 22 (3), 487–496. doi: 10.17957/IJAB/15.1090
Xuan, T. D., Shinkichi, T., Khanh, T. D., Min, C. I. (2005). Biological control of weeds and plant pathogens in paddy rice by exploiting plant allelopathy: an overview. Crop Prot. 24, 197–206. doi: 10.1016/j.cropro.2004.08.004
Yang, R. X., Gao, Z. G., Liu, X., Yao, Y., Cheng, Y. (2014). Root exudates from muskmelon (Cucumis melon. l) induce autotoxicity and promote growth of fusarium oxysporum f. sp. melonis. Allelopathy J. 33 (2), 175–187. doi: 10.4025/actasciagron.v36i2.14570
Yang, Y. H., Li, M. J., Chen, X. J., Wang, P. F., Wang, F. Q., Lin, W. X., et al. (2014). De novo characterization of the Rehmannia glutinosa leaf transcriptome and analysis of gene expression associated with replanting disease. Mol. Breeding. 34, 905–915. doi: 10.1007/s11032-014-0084-5
Yang, Y. H., Li, M. J., Li, X. Y., Chen, X. J., Lin, W. X., Zhang, Z. Y. (2015). Transcriptome-wide identification of the genes responding to replanting disease in Rehmannia glutinosa l. roots. Mol. Biol. Rep. 42, 881–892. doi: 10.1007/s11033-014-3825-y
Yan, P., Shen, W. T., Gao, X. Z., Li, X. Y., Zhou, P., Jun, D. (2012). High-throughput construction of intron-containing hairpin RNA vectors for RNAi in plants. PLos One 7 (5), e3818. doi: 10.1371/journal.pone.0038186
Zhang, C., Chen, H., Cai, T. C., Deng, Y., Zhuang, R. R., Zhang, N., et al. (2017). Overexpression of a novel peanut NBS-LRR gene AhRRS5 enhances disease resistance to Ralstonia solanacearum in tobacco. Plant Biotechnol. J. 15 (1), 39–55. doi: 10.1111/pbi.12589
Zhang, Z. Y., Li, M. J., Chen, X. J., Wu, L. K., Li, J., Wang, F. Q., et al. (2013). Research advancement and control strategy of consecutive monoculture problem of Rehmannia glutinosa l. Mod. Chin. Med. 15, 38–44. doi: 10.13313/j.issn.1673-4890.2013.01.00
Zhang, B., Li, X. Z., Feng, F. J., Gu, L., Zhang, J. Y., Zhang, L. J., et al. (2015). Correlation of allelopathy of Rehmannia glutinosa root exudates and their phenolic acids contents. J. Chin. Medicinal Mater 4), 5. doi: 10.13863/j.issn1001-4454.2015.04.002
Zhang, Z. Y., Lin, W. X. (2009). Continuous cropping obstacle and allelopathic autotoxicity of medicinal plants. Chin. J. 17, 189–196. doi: 10.3724/SP.J.1011.2009.00189
Zhang, B., Li, X. Z., Wang, F. Q., Li, M. J., Zhang, J. Y., Gu, L., et al. (2016). Assaying the potential autotoxins and microbial community associated with Rehmannia glutinosa, replant problems based on its a’utotoxic circle’. Plant Soil. 407 (1-2), 307–322. doi: 10.1007/s11104-016-2885-2
Zhang, B., Weston, P. A., Gu, L., Zhang, B. Y., Li, M. J., Wang, F. Q., et al. (2019). Identification of phytotoxic metabolites released from Rehmannia glutinosa suggest their importance in the formation of its replant problem. Plant Soil. 441 (1–2), 439–454. doi: 10.1007/s11104-019-04136-4
Zhou, Y., Sun, L. Q., Wassan, G. M., He, X., Shaban, M., Zhang, L., et al. (2018). GbSOBIR1 confers verticillium wilt resistance by phosphorylating the transcriptional factor gbbhlh171 in Gossypium barbadense. Plant Biotechnol. J. 17 (1), 152–163. doi: 10.1111/pbi.12954
Keywords: replant disease, root exudates, Fusarium oxysporum, LRR-RLKs, immune proteins
Citation: Yang C, Xie Z, Qian S, Zhang J, Yu Z, Li M, Gu L, Qin S and Zhang Z (2022) Functional analysis of Rehmannia glutinosa key LRR-RLKs during interaction of root exudates with Fusarium oxysporum reveals the roles of immune proteins in formation of replant disease. Front. Plant Sci. 13:1044070. doi: 10.3389/fpls.2022.1044070
Received: 14 September 2022; Accepted: 13 October 2022;
Published: 31 October 2022.
Edited by:
Xingang Zhou, Northeast Agricultural University, ChinaReviewed by:
Niraj Agarwala, Gauhati University, IndiaCopyright © 2022 Yang, Xie, Qian, Zhang, Yu, Li, Gu, Qin and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongyi Zhang, enl6aGFuZ0BmYWZ1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.