
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 04 January 2023
Sec. Plant Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1042981
This article is part of the Research Topic Molecular Basis of Seed Germination and Dormancy View all 8 articles
Introduction: Light is a major environmental factor in regulating germination and post-germination development of shallow photo-dormant seeds in Nicotiana tabacum L. (tobacco). However, its molecular mechanism remains largely unclear.
Methods and results: In this study, we compared the phenotypes of the seeds germinated under light and dark, and systematically investigated their regulatory networks by integrating transcriptomic and proteomic data. Under light, the germination increased ~25%, the length of the hypocotyl shortened ~3 cm, and the apical hook disappeared. 9, 161, 342 differentially expressed genes (DEGs) and 128, 185, 81 differentially expressed proteins (DEPs) were regulated by light in the development stage of seed imbibition, radicle protrusion and cotyledon expansion respectively. 0, 19 and 1 co-up-regulated and 1, 30 and 64 co-down-regulated DEGs (DEP) were observed in the three stages, respectively. Of them, 2S albumin large chain, was down-regulated by light in imbibed seed. Oleosin 18.5 kDa (OLEO1) and Glyceraldehyde-3-phosphate dehydrogenase (GAPA1), Oxygen-evolving enhancer protein 1-1 and anchloroplastic (PSBO1), hub genes (proteins) in protein-protein interaction network (PPI), were downregulated and up-regulated in germinated seeds by light, respectively. OLEO1, a hub gene (proteins), was down-regulated by light in post-germination seedling.
Conclusion: These results systematically revealed the molecular networks regulated by light during germination and post-germination development of shallow photo-dormant tobacco seeds.
Light is one of the major environmental factors in regulating seed germination. Seeds could be divided into three categories according their response to white light. Photophilic seeds, germinate after a short time exposure to light, while photophobic seeds, only germinate under dark. Non-photoblastic seeds, germinate well in both light and darkness. Hormones ABSCISIC ACID (ABA) and GIBBERELLINS (GA) are thought to be involved in the regulation of germination in all three categories seed (Sawada et al., 2008; Guo et al., 2018; Mérai et al., 2019). In Arabidopsis, the interaction between light, ABA and GA has been elaborately illuminated during seed germination, the biosynthesis of GA is stimulated by light, and in turn high levels of GA suppresses the synthesis of ABA (Vanstraelen & Benková, 2012; de Wit et al., 2016). PHYTOCHROME B (PHYB) and (or) PHYA is activated by red (R) and (or) far red (FR) light and mediates the degradation of PHYTOCHROME INTERACTING FACTOR1 (PIF1), and that relieves the suppression of PIF1 to GA anabolism and signal, which results in GA level increasing and suppresses ABA anabolism (Oh et al., 2006; Oh et al., 2007), promoting germination. In the dark, PHYB and (or) PHYA is inactivated and allows PIF1 accumulation, leading to suppression of GA signaling and stimulation of ABA anabolism (Kim et al., 2008; Oh et al., 2009), which results in ABA levels increasing and GA levels decreasing, inhibiting germination.
Following germination, seeds alternatively develop into etiolated or green seedling when they were exposed to dark or light. Light inhibits hypocotyls elongation and promotes apical hook disappearance and cotyledon unfolding. pif mutants exhibit a constitutive photomorphogenic phenotype even in darkness, as characterized by a shorter hypocotyl, open cotyledons, and apical hook defects (Shin et al., 2009; Leivar and Quail, 2011). In the dark, PHYs are inactivated that leads to the accumulation of PIFs (Gu et al., 2017; Qiu et al., 2019) and CONSTITUTIVE PHOTOMORPHOGENIC1/SUPPRESSOR OF PHYA-105 complexes (COP1/SPAs) (Huang, 2014; Hoecker, 2017), which promotes skomorphogenesis. Upon light, PHYs are activated to regulate the degradation of PIFs and destabilization of COP1/SPA complexes (Balcerowicz et al., 2011; Pacín et al., 2013; Sheerin et al., 2015), resulting in the accumulation of HY5, which promotes photomorphogenesis (Osterlund et al., 2000). In addition, pif mutants exhibit a apical hook defects phenotype even in darkness (Shin et al., 2009; Leivar and Quail, 2011). PIFs transcription factors promote hook curvature by activating HLS1 transcription (Zhang et al., 2018).
Seed germination begins imbibition and completes at the protruding radicle, and subsequently enters post-germination stage. Their molecular mechanisms of each development stage have been extensively studied in Arabidopsis, which recognized as a model plant to study biology mechanism of plant growth. However, the molecular mechanism of germination is quite different among species, especially between photophilic, non-photoblastic and photophobic seeds. In tobacco, photodormant seeds cannot germinate in the dark, which was taken as an model plant for studying the mechanism of seed photodormancy (Leubner-Metzger, 2002; Kucera et al., 2005). In our prevent study, we noticed fresh seeds of minority varieties in tobacco (shallow-photodormant type, dark-germination is more than 50%) that matured in shading environments could germinate independent of light (Dong et al., 2022). In this study, seeds of shallow-photodormant type were selected to study the molecular mechanisms of photo/sko-regulated germination and post-germination development by integrating transcriptomic and proteomic data. In particular, the PPI network was used to reveal the role of light at seed imbibition, radicle protrusion and cotyledon expansion stages.
Seeds of tobacco variety Y85 were provided by the Tobacco Research Institute of Guizhou. All the seeds were harvested 40 days after pollination, and mechanically dried at 40 ℃ for 36 hours in a hot air dryer (Jiang su, China). After threshing, the seeds were stored at room temperature, and all the experiments were conducted within 1 month. Seeds were incubated in two environments (light expose and continue dark) and developed in three phases (DAI2, 4, 6) (Table 1), containing three replicates of 18 samples were used for transcriptome and proteome sequencing, respectively.
The seeds were first disinfected by soaking in 0.5% copper sulfate solution for 15 min. Subsequently, the seeds were rinsed three times with distilled water. Finally, the disinfected seeds were dried with absorbent paper. 0.8% agar solutions were evenly poured into petri dishes to make agar germination beds. 100 seeds were evenly placed on the surface of the agar bed using a 10*10 grid, with three replicates for each treatment. The petri dishes were subsequently placed in an artificial climate chamber (Ningbo, China) and cultivated at 25°C, 80% humidity, 12 h photoperiod or constant darkness.
After 9 days' cultivation in greenhouse, the effects of different light environment treatments on tobacco seed germination related traits (germination rate, germination potential) were investigated. The formula of germination rate is:
Germination potential = The number of germinated seeds within 7 days / The number of tested seeds × 100%;
Germination rate = The number of seeds germinated within 14 days / The number of tested seeds× 100%.
Seed or seedling phenotype was identified as described by Li et al., (2019). In brief, three replicates of 100 seeds per treat were positioned in an imaging chamber, and the images were obtained daily through the Scanalyzer HTS (German, LemnaTec Company). Images were processed using Lemna Grid, which is an image analysis component of the LemnaTec HTS system. The total perimeter and area of whole seeds (seedlings) were measured by using the images from DAI1 to DAI 9. The perimeter and (or) area of radicle, hypocotyl and cotyledons were measured by using the images from DAI6 to DAI 9 images. In addition, the caliper length of the cotyledons (reflect the curvature of hooks) was also measured during this time.
Transcriptome sequencing and data analysis refer to our previously published method (Dong et al., 2022). Briefly, total RNA of each sample (see materials section) was extracted through RNAprep pure Plant Kit (Tiangen, Beijing, China). The quality of the RNA was determined by using a Nanophotometer Spectrophotometer (IMPLEN, California, USA) and an Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA). The libraries were constructed through a TruSeq Stranded mRNA LT Sample Prep Kit (Illumina, CA, USA). The transcriptome sequencing was performed by OE Biotech Co., Ltd. (Shanghai, China). The libraries were sequenced by using an Illumina HiSeq X Ten platform, and 150 bp paired-end reads were generated. Raw reads were processed through Trimomatic (Bolger et al., 2014). Clean reads were mapped to the tobacco genome using HISAT2 (Kim et al., 2015). Fragments per kilobase million (Roberts et al., 2011) value for each gene was calculated using Cufflinks (Trapnell et al., 2010), and their read counts were obtained using HTSeq-Count (Anders et al., 2015). DEGs analysis was performed using the DESeq (2012) R package (Love et al., 2018). p-values < 0.05, fold change (FC) < 0.5 or > 2 and FPKM≥2 were used as thresholds for significantly DEGs. A principal component analysis (PCA) was performed on the expression profiles using the R(PCA) function.
Proteome sequencing and data analysis refers to Wang (2016). In Brief, ~1g sample (see materials section) was weighed, powdered in liquid nitrogen and transferred into 700 μL of extraction buffer. Then, 7×protease inhibitor solution (Roche Applied Science, Indianapolis, IN) were added, mixed completely, and shaken for 30 min at 4°C. Subsequently, the mixture was centrifuged at 12,000g for 10 min at 4°C, and supernatant was transferred to a new tube and the protein concentration was determined by the Brad-Centration method. Trypsin digestion and iTRAQ analysis were performed by OE Biotech Co., Ltd. (Shanghai, China). For the identification of DEPs and FC were calculated automatically by Mascot. For statistical analysis, a two-tailed t-test was used and a p-value was calculated for each protein, which was conducted by Mascot ,too. At last, only proteins with FC > 2 or < 0.5 and p-values < 0.05 were considered significantly different. A principal component analysis (PCA) was performed on the expression profiles using the R(PCA) function.
In this study, the protein and mRNA were from the same reference genome (Kim et al., 2015), and the combined analysis was performed between the protein and its corresponding mRNA according to the gene ID. Cor-Up-regulated and Cor-Down-regulated represent DEGs (DEPs) that are simultaneously up-regulated and down-regulated, respectively. Gene Ontology (GO) enrichment of Cor-Up-regulated and Cor-Down-regulated DEGs (DEPs) were analyzed through the R package based on hypergeometric distribution. GeneMANIA online software (https://genemania.org/) was used to construct PPI networks and the networks can reveal the relationships between genes, including co-expression, physical interactions, co-localization and inter-gene interactions. Cor-Up-regulated and Cor-Down-regulated DEGs (DEPs) of each COM were used to construct 6 PPI networks.
Total RNA of seeds and seedlings was extracted using a TIANGEN RNA prep pure plant plus kit (Tiangen, Beijing, China). The purity and concentration of the RNA was assessed using a NanoDrop-2000 spectrophotometer (Thermo Science, MA, USA) and its integrity was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA). Purified RNA (1 µg per sample) was reverse transcribed to first-strand cDNA using a cDNA Reverse Transcription Kit (PrimeScriptTM RT Master Mix, Takara). RT-PCR was performed using TaKaRa TB Green® Premix Ex Taq™ II (Tli RnaseH Plus). Fluorescence Quantification Kit 20 µl reaction system as follows: 10 µL TB Green Premix Ex Taq (2×) (Tli RnaseH Plus), 0.4µL ROX reference dye (50×), 2µl of cDNA diluted to 40 ng/µL, 0.8 µl of upstream and downstream primers (Table S1), respectively, and made up to 20µl using double distilled water. RT-PCR reaction conditions: pre-denaturation for 0.5 min at 95°C, followed by at 95℃ for 5 s, 58℃ for 30 s and cycling 40 times. Relative expression levels of each gene were determined using a Step One Plus real-time PCR instrument with three replicates per treatment. Relative expression levels were calculated according to the published method (Livak and Schmittgen, 2001).
In this study, we compared the developmental dynamics of tobacco seeds germinated under 12h light/dark cycles and continuous dark. According to phenotype, the development process could be divided into two stages (Figure 1), the germination stage (DAI1- 1 days after imbibition to DAI4- 4 days after imbibition /DAI5- 5 days after imbibition), and the post-germination development stage (DAI6- 6 days after imbibition to DAI9- 9 days after imbibition). Seed germination finished on DAI4 in light, while it taken 5 days in dark (Figure 1).
Under light, the germination percentage was ~90%, which was significantly higher than the dark germination of ~65% (Figure 2). Before DAI2, there were no significant differences of phenotype between light and dark treatments (Figure 1). From DAI6, the areas and diameters of the seedlings grown in continuous dark were significantly higher to that of 12h light/dark treatment, but the difference gradually decreased from DAI8 onwards (Figures S1, S2).
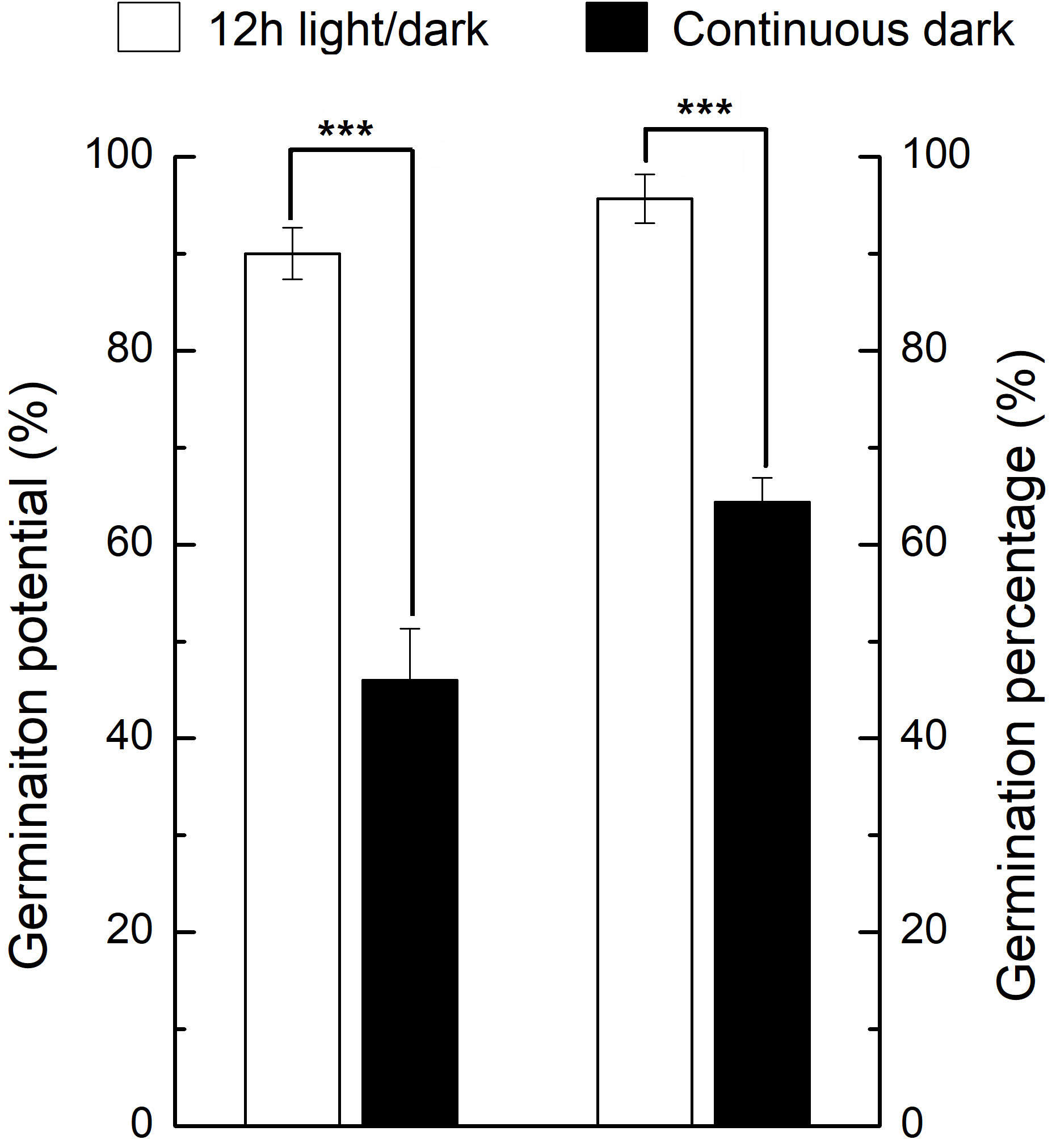
Figure 2 Germination percentage and germination potential of seeds under light and dark conditions. * indicates the significance of the test. * means significant at level 0.05; ** means significant at 0.01 level; *** means significant at 0.001 level. Same as below.
We further compared the phenotypic differences of radicles, hypocotyls and cotyledons developed under light and dark from DAI6 (radicle protrusion) to DAI9 (cotyledon expansion). Hypocotyls lengths were several times longer (Figure 3), and the cotyledon areas were significantly larger under dark (Figures 4A, B). However, cotyledon curvatures were larger under light (Figure 4C).
Subsequently, we compared transcriptomic and proteomic data of seeds germinated under light and dark, a total of 7 combinations (COMs) were accomplished (Table 1). In COM1, 9 DEGs were identified, 0 and 9 of which were up- and down-regulated (Figure S3A). 128 DEPs were identified, 60 and 68 of which were up- and down-regulated (Figure S3B). 0 and 1 DEGs (DEPs) was co-up- and co-down-regulated (Figure 5), and 0 pathways were enriched. In COM2, there were 161 DEGs, 64 and 97 of which were up- and down-regulated (Figure S3A). 185 DEPs were identified, 90 and 95 of which were up- and down-regulated (Figure S3B). 19 and 30 DEGs (DEPs) were co-up- and co-down-regulated (Figure 5), and 21 pathways were enriched (Figure 6). In COM3, 342 DEGs were identified, 32 and 310 of which were up- and down-regulated (Figure S3A). 81 DEPs were identified, 19 and 64 of which were up- and down-regulated (Figure S3B). 1 and 30 DEGs (DEPs) were co-up- and co-down-regulated, and 16 pathways were enriched (Figure S4).
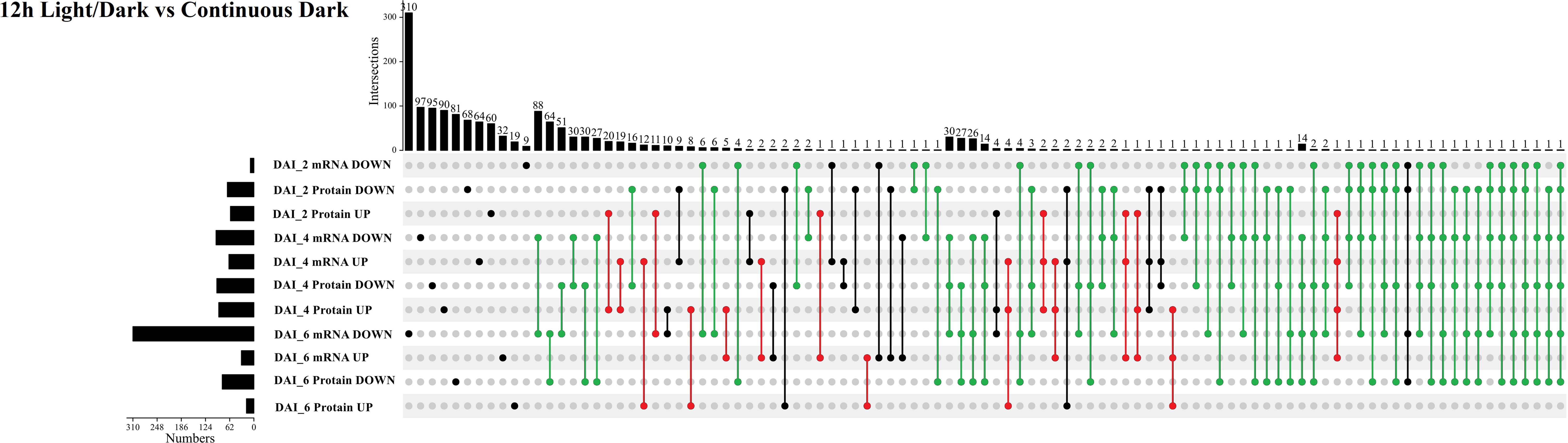
Figure 5 Multiple comparisons of light-regulated DEGs (DEPs) in seed germination and post-germination developmental stages. The histogram on the left represents the number of co-up and co-down regulated DEGs (DEPs) within each comparison. The histogram on the top represents the number of co-up or -down regulated DEGs (DEPs) from multiple comparisons. Multiple comparisons were displayed by lines and linked dots.
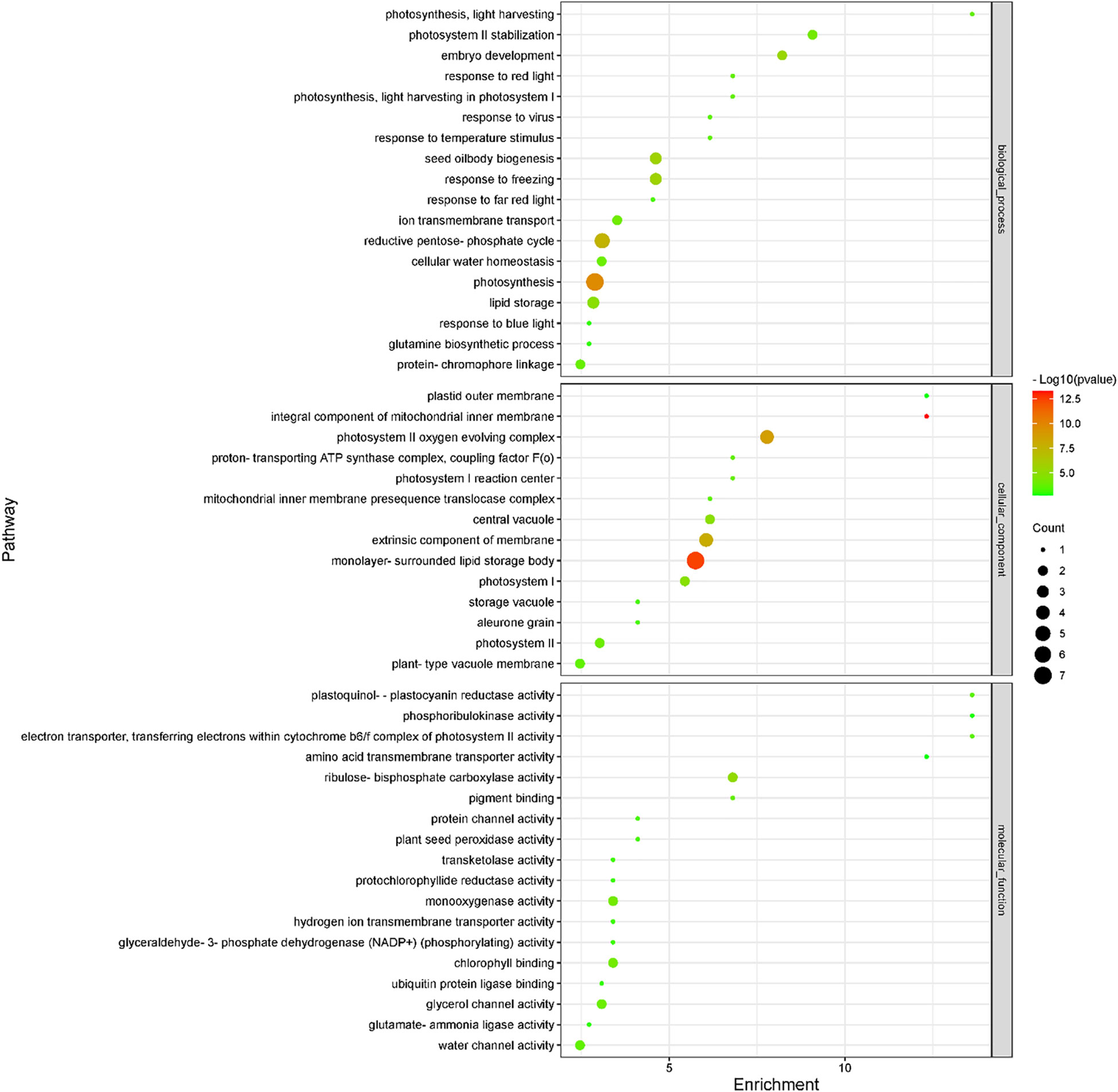
Figure 6 Enriched GO terms of light-regulated pathways at radicle protrusion stage. The size of the circle represented the number of genes enriched in the pathway, and the color of the cluster represents the confidence interval, and p-value was calculated from hypergeometric test.
We also compared transcriptomic and proteomic data in three develop stage. 445 (50) up-regulated and 224 (49) down-regulated genes (proteins) were detected in COM4 (Figure S5A). Of them, 15 co-up-regulated and 24 co-down-regulated regulated genes (proteins) were identified (Figure S5A), and 9 pathways were enriched (Figure S6). 556 (50) up-regulated and 394 (49) down-regulated genes (proteins) were detected in COM5 (Figure S5B). Of them, 12 co-upregulated and 27 co-down-regulated genes (proteins) were identified (Figure S5B), and 6 pathways were enriched (Figure S7). 214 (246) up-regulated and 702 (180) down-regulated genes (proteins) were detected in COM6 (Figure S5A). Of them, 92 co-up-regulated and 37 co-down-regulated genes (proteins) were identified (Figure S5A), and 50 pathways were enriched (Figure S8). 203 (184) up-regulated and 949 (231) down-regulated genes (proteins) were detected in COM7 (Figure S5B). Of them, 63 co-up-regulated and 111 co-down-regulated genes (proteins) were identified (Figure S5B), and 57 pathways were enriched (Figure S9).
Principal component analysis (PCA) was used to analyze the diversity and similarity of samples from both light treatment and developmental perspectives. According to light treatment, the samples of LY85_2 and DY85_2 were gathered together by DEGs and DEPs, while LY85_6 and DY85_6 were completely separated by DEGs and DEPs (Figures 7A, B). LY85_4 and DY85_4 were gathered together by DEGs (Figure 7A), but not by DEPs (Figure 7B). According to develop stage, the samples of DY85_2, DY85_4 and DY85_6 were completely separated by DEGs and DEPs (Figures 7A, B), while the samples of LY85_2 and LY85_4 were completely separated by DEPs (Figure 7B), but not by DEGs (Figure 7A). Samples of LY85_4 and LY85_6 were completely separated by DEGs and DEPs (Figures 7A, B).
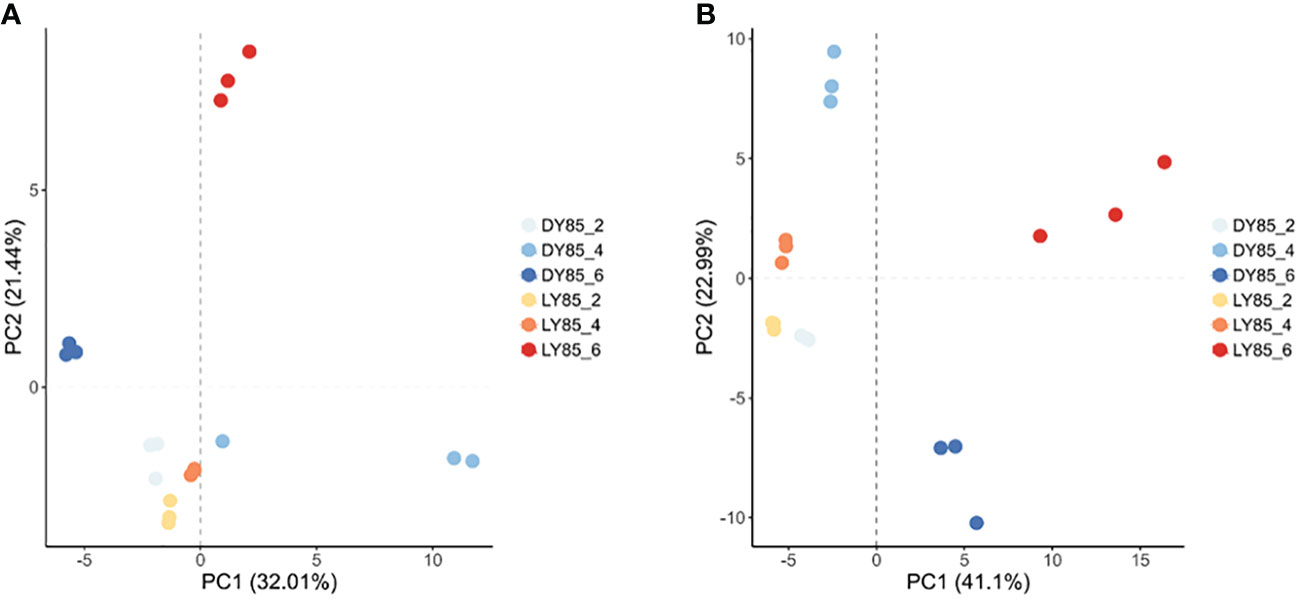
Figure 7 PCA plots of light-regulated germination and post-germination development. They were plotted with DEG (A) and DEP (B), respectively. The consistency and diversity of the samples were observed through PCA. Each point in the figure represents a repetition, and different colors represent the difference of light treatment or development stage.
The expression patterns of co-up- and down-regulated DEGs (DEPs) were analyzed by using UpSetR, and a total of 64 patterns were identified among COM1, COM2 and COM3 (Figure 5). 47 patterns were identified between COM4 and COM6 (Figure S5A) and 53 patterns between COM5 and COM7 (Figure S5B). GeneMANIA software was used to analyze associations in terms of co-expression, physical interactions and predicted pathway, and 6 PPI networks were constructed (Figure 8 and Figure S10). GO analysis was used to identify of the cellular components of each COM (Table 1), and 6 component diagrams were constructed (Figure 9 and Figure S11).
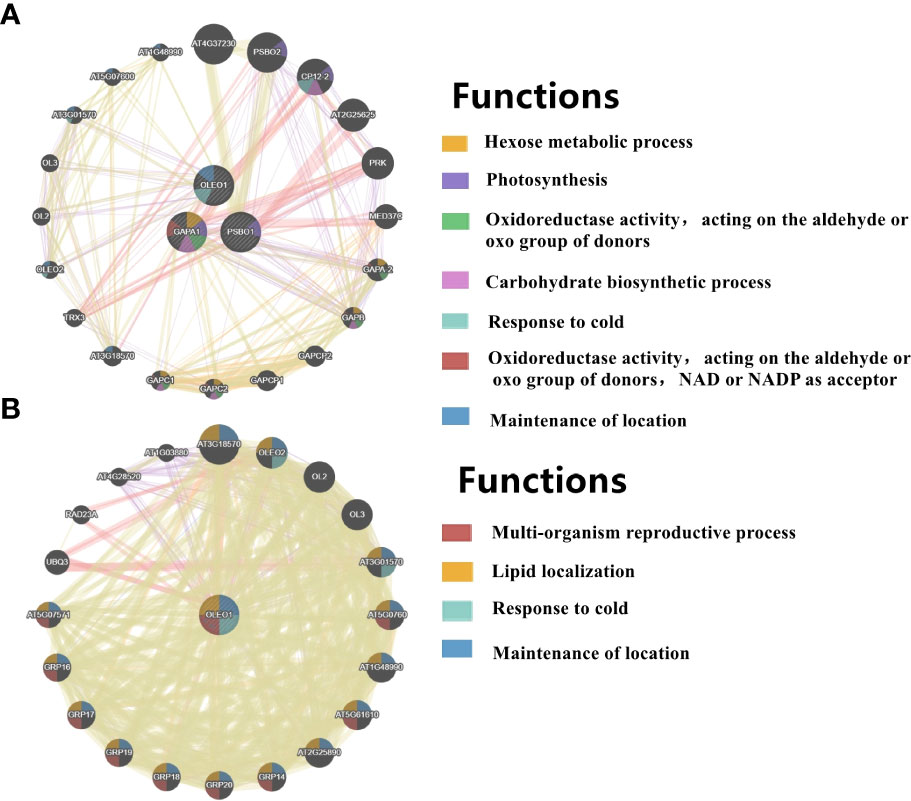
Figure 8 PPI networks of light-regulated germination and post-germination development, (A) radicle protrusion and (B) cotyledon expansion stage. The protein-protein interaction network for DEGs (DEPs) was analyzed using the GeneMANIA database during seed germination and post-germination development. At least 20 most frequently changed neighboring genes were shown. Each network node represented a gene. The node color represented the possible functions of these respective genes.
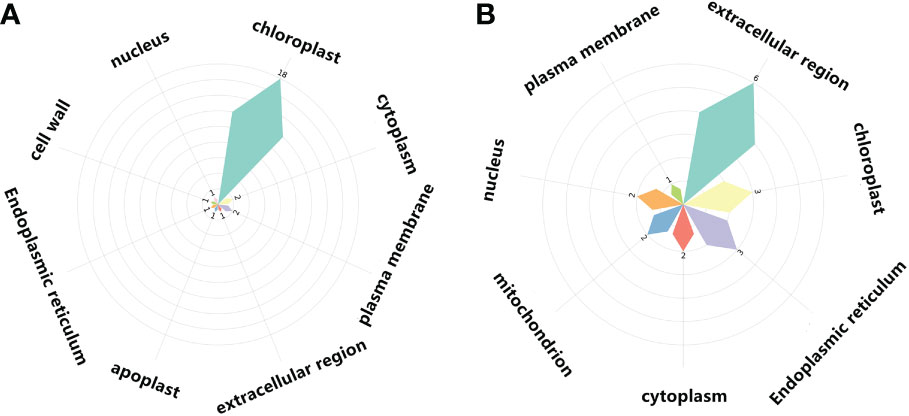
Figure 9 Cellular components of light-regulated germination and post-germination development, (A) Radicle protrusion and (B) cotyledon expansion stage. Petal size showed the number of cellular components enriched, and the number of signaling pathways enriched in cellular components was also listed.
In COM1, we only identified one co-expressed genes (proteins) of the 2S albumin large chain, which was down-regulated by light in imbibed seed. 2S albumin is clearly as a seed storage protein and abundantly expressed in endosperm, whose biological functions include food reserve for germination and as an inhibitor of α-amylase to inhibit pre-germination during seed development. Because insufficient number of co-expressed genes (proteins) was identified, PPI network and cellular components diagrams cannot be established in COM1.
In COM2, GAPA1, PSBO1, OLEO1 were the hubs of these 23 genes (proteins) (Figure 8A). Of them, GAPA1 and PSBO1 were up-regulated, while OLEO1 was down-regulated by light (Table 2). Their biological functions mainly included regulation of hexose metabolism process, photosynthesis, oxidoreductase activity, and so on (Figure 8A). For instance, GAPA1, GAPA2, GAPAB, GAPC1 and GAPC2 were consider to be involved in regulating of hexose metabolic process, and they participated in regulating photosynthesis together with PSBO2 and CP12-2. Notably, 18 of these 23 genes (proteins) were localized in the chloroplast (Figure 9A).
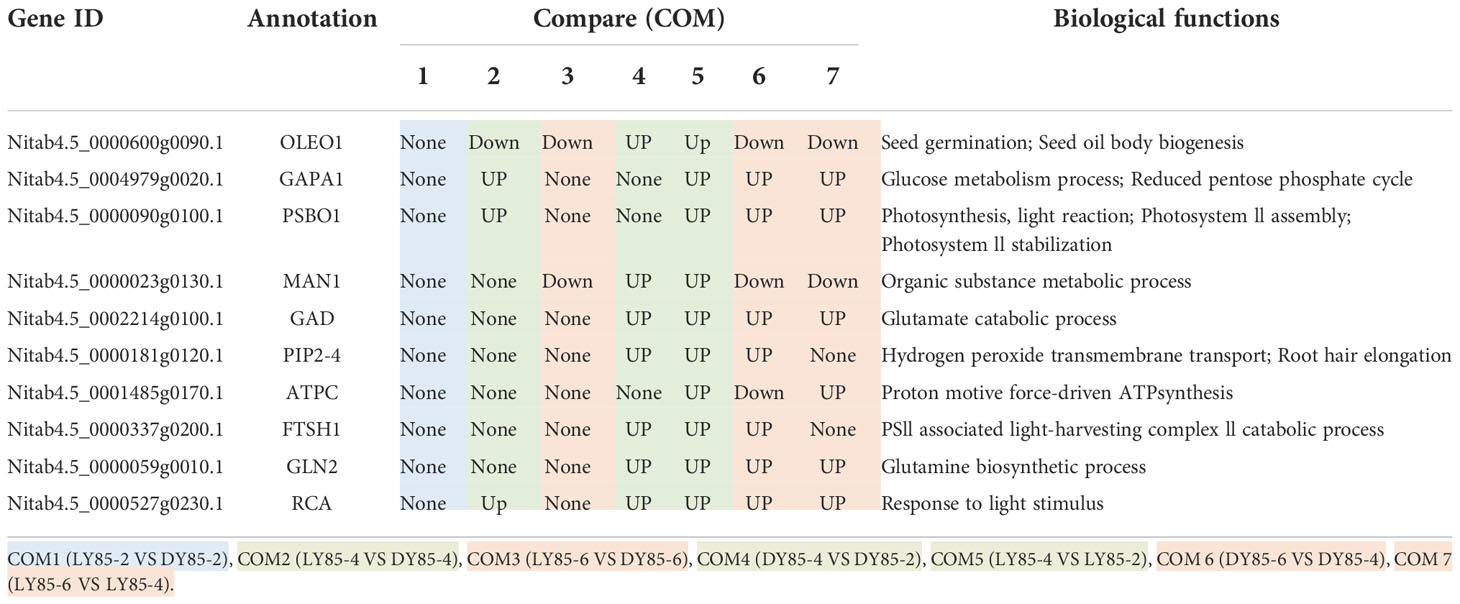
Table 2 Expression patterns of hub genes (proteins) during germination and post-germination development in light and dark.
In COM4 and COM5, the identical PPI network was constructed. Mannan endo-1,4-beta-mannosidase 1 (MAN1) was the hub of these 21 genes (proteins) respectively (Figures S10A, C), which was up-regulated at radicle protrusion stage in both light and dark (Table 2). MAN1, together with MAN3, MAN4, MAN5, MAN6 and MAN7, were predicted to regulate mannosidase activity and hydrolase activity of hydrolyzing O-glycosyl compounds and glycosyl bonds (Figures S10A, B). 6, 6, 5, 5 and 4 genes (proteins) were localized in cytoplasm, extracellular region, nucleus, chloroplast and endoplasmic reticulum, while 7, 7, 5, 5 and 4 genes (proteins) were localized in cytoplasm, nucleus, chloroplast, endoplasmic reticulum, extracellular region (Figures S11A, B).
In COM3, OLEO1 was the hub of these 21 genes (proteins), which was down-regulated by light (Table 2). OLEO1, together with other genes were predicted to regulate maintenance of location, response to cold, multi-organism reproductive process, lipid localization (Figure 8B). 6, 3 and 3 genes (proteins) were localized in extracellular region, chloroplast and endoplasmic reticulum (Figure 9B).
In COM6, PSBO1, GAPB, CPBP1, RCA, GLN2, GAPA1, FTSH1, ATPC, RRF, PIP2-4, MAN1, RPL2, GAD1, and PSBS were the hubs of these 34 genes (proteins) (Figure S10C), and their expression patterns were shown in Table 2. The functions of 34 genes (proteins) are mainly involved in regulating glucose metabolic process, photosynthesis, generation of precursor metabolites and energy, and so on (Figure S10C). 66, 13, and 11 genes (proteins) were localized in chloroplast, ribosome, cytoplasm (Figure S11C).
In COM7, PSBO1, GAPB, CPBP1, PSBQ1, RCA, GLN2, ATPC, GAPA1, MAN1, OLEO1, MAN5, RPL2 and PSBS were the hubs of these 33 genes (proteins) (Figure S10D), and their expression patterns were shown in Table 2. The functions of 33 (gene) proteins are mainly involved in regulating photosynthesis, generation of precursor metabolites and energy, light harvesting (photosynthesis), and so on (Figure S10D). 49, 15 and 14 genes (proteins) were localized in chloroplast, extracellular region, cytoplasm (Figure S11D).
Overall, the PPI networks differed more in developmental stages than in light treatment during seed germination and post-germination development. 7 of the 10 hub genes (proteins) were up-regulated from seed imbibition to radicle protrusion under either light or dark conditions. 7 of the 10 hub genes (proteins) were up-or down-regulated from radicle protrusion to cotyledon expansion under either light or dark conditions. However, 0, 4 and 2 hub genes (proteins) up-or down-regulated by light at seed imbibition, radicle protrusion and cotyledon expansion, respectively. The effect of light on seed imbibition was significantly less than on radicle protrusion and cotyledon unfolding from the PPI networks.
To validate the accuracy of the sequencing data, the expression levels of 14 genes were analyzed by qRT-PCR analysis, including 9 up-regulated and 5 down-regulated genes. 7 of the 9 up-regulated genes and 3 of the 5 down-regulated genes were shown consistent expression trends (Figure S12). Pearson correlation coefficient shows that there is correlation between transcriptome data and QRT-PCR validation results (Figure 10). These results reinforce the reliability of the sequencing data.
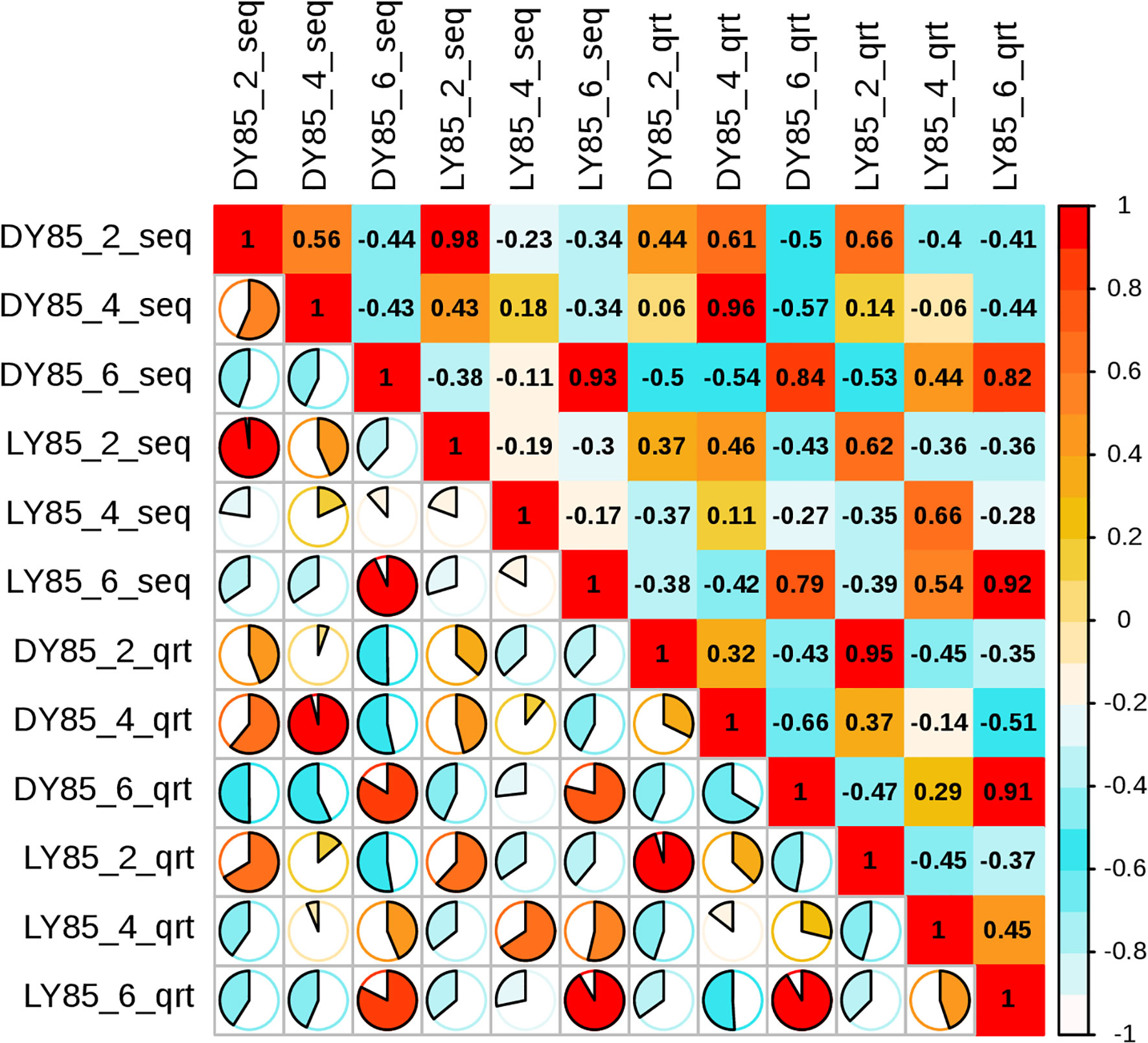
Figure 10 Pearson correlation analysis between relative transcript levels by qRT-PCR and transcriptome expression levels by FPKM value.
Light is one of the important environmental factors in the life cycle of higher plants. Photothermal receptor phytochromes integrate light and temperature signals (Jung et al., 2016; Legris et al., 2016), thereby regulating plant growth and development to adapt to the environment. For instance, light‐activated phytochrome to regulate the degradation of its direct targets PIFs, resulting in increasing of GA level and decreasing of ABA level (Oh et al., 2006; Oh et al., 2007), promoting germination. Light‐activated phytochrome regulate the degradation of PIFs and the destabilize of COP1/SPA complexes (Al-Sady et al., 2006; Shen et al., 2007; Huang, 2014; Hoecker, 2017)), resulting in the accumulation of HY5 (Osterlund et al., 2000), which modulates photomorphogenesis. Modulate of light, PHYs and PIFs plays important roles in both germination and post-germination development of Arabidopsis seed. However, the molecular mechanisms of germination do differ among seed categories. Photophilic seeds germinate well under light exposure, while photophobic seeds favor germinate in the dark. In this study, we systematically analyzed the molecular mechanisms of light regulated germination and post-germination development in shallow photodormant tobacco seeds.
Germination of photophilic seeds promoted by light that had been reported in many species, such as Arabidopsis (Borthwick et al., 1952; Shinomura et al., 1996; Hennig et al., 2001), Lactuca sativa Linn (lettuce) (Toyomasu et al., 1998; Sawada et al., 2008), Nicotiana tabacum L (tobacco) (Wang et al., 2021; Dong et al., 2022), and so on. In this study, we noticed shallow photodormant seeds have higher germination percentage and faster speed under light. It is well known that GA and ABA antagonistically regulate seed germination (Bentsink and Koornneef, 2008). The interaction among light, ABA and GA has been elaborately illuminated during seed germination in Arabidopsis, the biosynthesis of GA is stimulated by light, and in turn high levels of GA suppresses the synthesis of ABA (Vanstraelen and Benková, 2012; de Wit et al., 2016). Unexpectedly, no genes (proteins) related to this pathway were found to be differentially expressed by combined transcriptome and proteome analysis in this study.
In Arabidopsis, light promotes the expression of GA3ox1 (GA biosynthesis) and CYP707A2 (ABA catabolism) and inhibits the transcription of the GA2ox2 (GA catabolism) and NCED6, 9 (ABA biosynthesis), resulting in a high GA/ABA ratio and consequently promoting seed germination (Kim et al., 2008; Gabriele et al., 2009; Née et al., 2017). In this study, the expression of NtGA3ox2 and NtCYP707A1 is promoted, and NtNCED6 is suppressed by light (Fig S13). In addition, the expressions of GIBBERELLIC ACID-IN-SENSITIVE (GAI, a negative regulator of GA signal) (Oh et al., 2007) and ABSCISIC ACID-INSENSITIVE 3, 5 (ABI3, 5, positive regulators of ABA signal) (Oh et al., 2009; Kim et al., 2016) are inhibited by light to promote seed germination in Arabidopsis. In this study, the expression of NtGAI and NtABI3, 5 is suppressed by light (Figure S13). Similar results were noticed in other photophilic-seeded species, such as lettuce (Toyomasu et al., 1998; Sawada et al., 2008), and tobacco (Wang et al., 2021; Dong et al., 2022).
Therefore, we believed that the light signaling pathway also plays a key regulatory role in the germination of shwallow photodormant seeds. In our previous study, a large number of DEGs in this signaling pathway were detected during germination of shwallow photodormant seeds in tobacco by signal transcriptomic sequencing (Dong et al., 2022). Why these DEGs (DEPs) could not be detected in this study? Possibly because less number of DEPs was detected by proteomics sequencing and some DEGs or (DEPs) were filtered by the combined analysis of transcriptome and proteome.
It has been hypothesized that light regulates germination of tobacco seed in two aspects (Leubner-Metzger, 2003). On the one hand, light activates phytochromes to release photodormancy and promote germination. On the other hand, light promotes endosperm rupture in non-photodormant seeds. Several specific target enzymes, such as EXP (EXPANSIN), XTH (XYLOGLUCANENDO-TRANSGLYCOSYLASE/HYDROLASE), and βGLUI (CLASS β-1, 3-GLUCANASE), have been proposed to promote germination by inducing endosperm weakening in Arabidopsis and tobacco (Steinbrecher and Leubner-Metzger, 2016). The expressions of genes required for cell wall loosening, such as EXP and XTH, are repressed to inhibit seed germination under dark in Arabidopsis (Oh et al., 2009). The expression of NtXTH was upregulated by light to promote the germination of fresh seeds in tobacco (Wang et al., 2021). The above studies suggest that light can promote the weakening of endosperm during seed germination by inducing the expression of genes involved in regulating cell wall hydrolysis.
In this study, 6 MANs were up-regulated during radicle protrusion, whose functions were predicted to regulate mannosidase activity and hydrolase activity of hydrolyzing O-glycosyl compounds and glycosyl bonds. βGLUI is induced prior to the rupture of the micropylar endosperm, which is required for germination of tobacco seeds (Leubner-Metzger and Meins, 2000; Petruzzelli et al.,1999), and light is hypothesized to be involved in the induction of βGLUI (Wu et al., 2001). However, expression levels of MANs were not significantly improved by light. So, we speculated that cell wall hydrolysis is required for seed germination, and there may be two types of hydrolases, one is regulated by light, and the other not be affected by light. This also helps explain that nearly half of the shallow photodormant seeds can germinate in dark, while light can improve the germination percentage and accelerate germination speed.
Seed germination begins with imbibition and completes at radicle protrusion, and then enters the stage of seedling development, including hypocotyl elongation, cotyledon expansion and de-yellowing, etc. In soil, seeds germinate in dark and develop following a dark-adapted biology program named skotomorphogenesis, which is characterized by long hypocotyls and etiolated cotyledons that contain etioplasts. When the seedlings emerge from the soil, the developmental program changes into photomorphogenesis, hypocotyl elongation is inhibited, and the cotyledons turn green and develop functional chloroplasts (Pogson et al., 2015). In the present study, similar phenotypes were noticed when the seeds were incubated in light and dark.
From quiescent seeds to imbibed seeds (DAI2), the shoot apical meristem and epidermis are completely devoid of chlorophyll, regardless of their light environment (Tejos et al., 2010; Yadav et al., 2019). In this study, the phenotypic difference was not significant on DAI2, and photosynthesis-related genes were not differentially expressed by light. When the radicle protrudes, photosynthesis-related genes (proteins) were up-regulated or down-regulated by light, followed by a green hypocotyl phenotype. In direct photomorphogenesis, cell-specificity of chloroplast biogenesis of mostly cotyledons starts after the emergence of radicle from the seed coat (Yadav, 2019). In this study, multiple regulators involved in photosynthesis, such as GAPA1, 2, GAPAB, GAPC1, 2, PSBO2 and CP12-2, were noticed to be regulated by light at radicle protrusion, but not at cotyledon expansion stage. Notably, 18 of 23 hut genes (proteins) are located in the chloroplast at radicle protrusion stage.
Therefore, we speculated that chloroplast development genes were highly expressed after radicle protrusion, which may lay the foundation for photosynthesis in subsequent seedling development.
Light is an important signal factor that involved in regulating of chloroplast biogenesis during post-germination development. It requires the coordinated expression of chloroplast biogenesis genes encoded in nuclear and plastid genomes (Liebers et al., 2022). PHYB are activated by light to promote the degradation of PIF3, which represses the nuclear-encoded components of the plastid transcriptional machinery required for transcription of the plastid-encoded photosynthesis genes (Dubreuil et al., 2017). The light-PHYs-PIFs module not only plays an important role in seed germination and post-germination development, but also participates in the regulation of chloroplast development. It has been widely concerned the relationship between seedling morphogenesis and chloroplast biogenesis. For instance, SCO1 is critical for plastid development during embryogenesis, and its mutants sco1-2 and sco1-3 display a seedling lethal phenotype. Seedlings of sco1 mutants typically die shortly after germination due to improper chloroplast development (Ruppel and Hangarter, 2007). However, little is known about whether abnormalities in precursor, corpus luteum or chloroplast development affect seed germination. In addition, it remains unknown whether chloroplast or its precursors are involved in the regulation of seed germination.
Light is an important signaling factor, which is widely involved in the regulation of germination and early seedling development in shallow photodormant tobacco seeds, including increased germination rate, inhibition of hypocotyl elongation, promotion of cotyledon unfolding, and so on. 9, 161, 342 differentially expressed genes (DEGs) and 128, 185, 81 differentially expressed proteins (DEPs) were identified at the stage of seed imbibition, radicle protrusion and cotyledon expansion, respectively. PCA results of these DEGs (DEPs) showed that samples of imbibed seeds and germinated seeds were gathered for a short distance, while the samples of post-germination seedling were completely separated from them. 0, 19 and 1 co-up-regulated, while 1, 30 and 64 co-down-regulated DEGs (DEPs) were identified at the stage of seed imbibition, radicle protrusion and cotyledon expansion, respectively, and 0, 16 and 21 pathways were enriched by using these co-up or co-down-regulated DEGs (DEPs). Oleosin 18.5 kDa (OLEO1), Glyceraldehyde-3-phosphate dehydrogenase (GAPA1), Oxygen-evolving enhancer protein 1-1 and anchloroplastic (PSBO1) were the hub genes (proteins) of these co-up- and co-down-regulated DEGs (DEPs) at radicle protrusion stage, 18 of these 23 PPI (genes) proteins were localized in the chloroplast. OLEO1 was the hub genes (proteins) of these co-up- and co-down-regulated DEGs (DEPs) at cotyledon expansion stage, and the cellular components of the more gene (protein) localization were extracellular region, nucleus and endoplasmic reticulum. The above results systematically analyzed the regulatory mechanism of light during germination and post-germination development in shallow photodormant tobacco seeds.
The data presented in the study are deposited in the NCBI repository, accession number PRJNA914116, https://www.ncbi.nlm.nih.gov/sra/PRJNA914116.
ZL supervised the research. QL, MZ, SD and PY prepared the samples and performed the experiments. ZL, QL, and JZ analyzed the data. ZL, EL and QL wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the National Natural Science Foundation of China (31860420, 32060512), the Major project of Guizhou Tobacco Company [201904], the Science and Technology Plan Project of Guizhou Province ([2018]5781). The authors declare that this study received funding from Guichou Tobacco Company. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article, or the decision to submit it for publication.
We sincerely thank MedSCI for linguistic editing and proofreading during the preparation of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1042981/full#supplementary-material
Supplementary Figure 1 | Perimeter dynamic curve of seed germinated under light and dark.
Supplementary Figure 2 | Area dynamic of seed germinated under light and dark.
Supplementary Figure 3 | Number of DEGs (DEPs) of seed germinated under light and dark.
Supplementary Figure 4 | Enriched GO terms of light-regulated pathways at cotyledon expansion stage. The size of the circle represents the number of genes enriched in the pathway, and the color of the cluster represents the confidence interval, and p-value was calculated from hypergeometric test.
Supplementary Figure 5 | Multiple comparisons of light-regulated DEGs (DEPs) during seed germination and post-germination development. The histogram on the left represents the number of co-up and co-down regulated DEGs (DEPs) within each comparison. The histogram on the top represents the number of co-up or -down regulated DEGs (DEPs) from multiple comparisons. Multiple comparisons are displayed by lines and linked dots.
Supplementary Figure 6 | Enriched GO terms of DEGs (DEPs) from sko-germinated seeds/sko-imbibed seeds. The size of the circle represents the number of genes enriched in the pathway, and the color of the cluster represents the confidence interval, and p-value was calculated from hypergeometric test.
Supplementary Figure 7 | Enriched GO terms of DEGs (DEPs) from sko-developed seedling/sko-germinated seeds. The size of the circle represents the number of genes enriched in the pathway, and the color of the cluster represents the confidence interval, and p-value was calculated from hypergeometric test.
Supplementary Figure 8 | Enriched GO terms of DEGs (DEPs) from photo-germinated seeds/photo-imbibed seeds. The size of the circle represents the number of genes enriched in the pathway, and the color of the cluster represents the confidence interval, and p-value was calculated from hypergeometric test.
Supplementary Figure 9 | Enriched GO terms of DEGs (DEPs) from photo-developed seedling/photo-germinated seeds. The size of the circle represents the number of genes enriched in the pathway, and the color of the cluster represents the confidence interval, and p-value was calculated from hypergeometric test.
Supplementary Figure 10 | PPI network of light-regulated seeds and seedlings. (A) sko-germinated seeds/sko-imbibed seeds (B) photo-germinated seeds/photo-imbibed seeds (C) sko-developed seedling/sko-germinated seeds. (D) photo-developed seedling/photo-germinated seeds. The protein-protein interaction network for DEGs (DEPs) was analyzed using the GeneMANIA data base during seed germination and post-germination development. At least 20 most frequently changed neighboring genes are shown. Each network node represents a gene. The node color represents the possible functions of these respective genes.
Supplementary Figure 11 | Cellular components from different development stages seeds. (A) sko-germinated seeds/sko-imbibed seeds (B) photo-germinated seeds/photo-imbibed seeds (C) sko-developed seedling/sko-germinated seeds. (D) photo-developed seedling/photo-germinated seeds. Petal size shows the number of cellular components enriched, and the number of signaling pathways enriched in cellular components is also listed.
Supplementary Figure 12 | Expression levels of genes involved in photosynthesis during germination and post-germination development.
Supplementary Figure 13 | Expression levels of genes involved in ABA or GA metabolism or signal during germination.
Al-Sady, B., Ni, W., Kircher, S., Schäfer, E., Quail, P. H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23, 439–446. doi: 10.1016/j.molcel.2006.06.011
Anders, S., Pyl, P. T., Huber, W. (2015). HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Balcerowicz, M., Fittinghoff, K., Wirthmueller, L., Maier, A., Fackendahl, P., Fiene, G., et al. (2011). Light exposure of arabidopsis seedlings causes rapid de-stabilization as well as selective post-translational inactivation of the repressor of photomorphogenesis SPA2: Functional divergence between SPA1 and SPA2. Plant J. 65, 712–723. doi: 10.1111/j.1365-313X.2010.04456.x
Bentsink, L., Koornneef, M. (2008). Seed dormancy and germination. Arabidopsis Book 6, e0119. doi: 10.1199/tab.0119
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Borthwick, H. A., Hendricks, S. B., Parker, M. W., Toole, E. H., Toole, V. K. (1952). A reversible photoreaction controlling seed germination. Proc. Natl. Acad. Sci. U.S.A. 38, 662–666. doi: 10.1073/pnas.38.8.662
de Wit, M., Galvão, V. C., Fankhauser, C. (2016). Light-mediated hormonal regulation of plant growth and development. Annu. Rev. Plant Biol. 67, 513–537. doi: 10.1146/annurev-arplant-043015-112252
Dong, S., ling Liu, Y., Zhang, M., Zhang, J., Wang, J. H., Li, Z. H. (2022). Maternal light environment interacts with genotype in regulating seed photodormancy in tobacco. Environ. Exp. Bot. 194, 104745. doi: 10.1016/j.envexpbot.2021.104745
Dubreuil, C., Ji, Y., Strand, Å., Grönlund, A. (2017). A quantitative model of the phytochrome-PIF light signalling initiating chloroplast development. Sci. Rep. 7, 13884. doi: 10.1038/s41598-017-13473-2
Gabriele, S., Rizza, A., Martone, J., Circelli, P., Costantino, P., Vittorioso, P. (2009). The dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1: DAG1 represses seed germination via PIL5 signalling. Plant J. 61, 312–323. doi: 10.1111/j.1365-313X.2009.04055.x
Gu, D., Chen, C.-Y., Zhao, M., Zhao, L., Duan, X., Duan, J., et al. (2017). Identification of HDA15-PIF1 as a key repression module directing the transcriptional network of seed germination in the dark. Nucleic Acids Res. 45, 7137–7150. doi: 10.1093/nar/gkx283
Guo, G., Liu, X., Sun, F., Cao, J., Huo, N., Wuda, B., et al. (2018). Wheat miR9678 affects seed germination by generating phased siRNAs and modulating abscisic Acid/Gibberellin signaling. Plant Cell 30, 796–814. doi: 10.1105/tpc.17.00842
Hennig, L., Poppe, C., Sweere, U., Martin, A., Schäfer, E. (2001). Negative interference of endogenous phytochrome b with phytochrome a function in Arabidopsis. Plant Physiol. 125, 1036–1044. doi: 10.1104/pp.125.2.1036
Hoecker, U. (2017). The activities of the E3 ubiquitin ligase COP1/SPA, a key repressor in light signaling. Curr. Opin. Plant Biol. 7, 63–69. doi: 10.1016/j.pbi.2017.03.015
Huang, X. (2014). Beyond repression of photomorphogenesis: role switching of COP/DET/FUS in light signaling. Curr. Opin. Plant Biol. 8, 96–103. doi: 10.1016/j.pbi.2014.07.003
Jung, J.-H., Domijan, M., Klose, C., Biswas, S., Ezer, D., Gao, M., et al. (2016). Phytochromes function as thermosensors in. Arabidopsis Sci. 354, 886–889. doi: 10.1126/science.aaf6005
Kim, J., Kang, H., Park, J., Kim, W., Yoo, J., Lee, N., et al. (2016). PIF1-interacting transcription factors and their binding sequence elements determine the in vivo targeting sites of PIF1. Plant Cell 28, 1388–1405. doi: 10.1105/tpc.16.00125
Kim, D., Langmead, B., Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kim, D. H., Yamaguchi, S., Lim, S., Oh, E., Park, J., Hanada, A., et al. (2008). SOMNUS, a CCCH-type zinc finger protein in Arabidopsis , negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell 20, 1260–1277. doi: 10.1105/tpc.108.058859
Kucera, B., Cohn, M. A., Leubner-Metzger, G. (2005). Plant hormone interactions during seed dormancy release and germination. Seed Sci. Res. 15, 281–307. doi: 10.1079/SSR2005218
Legris, M., Klose, C., Burgie, E. S., Rojas, C. C. R., Neme, M., Hiltbrunner, A., et al. (2016). Phytochrome b integrates light and temperature signals in Arabidopsis. Science 354, 897–900. doi: 10.1126/science.aaf5656
Leivar, P., Quail, P. H. (2011). PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16, 19–28. doi: 10.1016/j.tplants.2010.08.003
Leubner-Metzger, G. (2002). Seed after-ripening and over-expression of class I beta-1,3-glucanase confer maternal effects on tobacco testa rupture and dormancy release. Planta 215, 959–968. doi: 10.1007/s00425-002-0837-y
Leubner-Metzger, G. (2003). Functions and regulation of β-1,3-glucanases during seed germination, dormancy release and after-ripening. Seed Sci. Res. 13, 17–34. doi: 10.1079/SSR2002121
Leubner-Metzger, G., Meins, F. (2000). Sense transformation reveals a novel role for class I β-1,3-glucanase in tobacco seed germination: Role of β-1,3-glucanase in seed germination. Plant J. 23, 215–221. doi: 10.1046/j.1365-313x.2000.00773.x
Liebers, M., Cozzi, C., Uecker, F., Chambon, L., Blanvillain, R., Pfannschmidt, T. (2022). Biogenic signals from plastids and their role in chloroplast development. J. Exp. Bot., erac344. doi: 10.1093/jxb/erac344
Li, Z., Liu, Y., Liu, R. (2019). Integrating multiple-capsule traits quantitative evaluation of seed maturity by 3D phenotypic platform in nicotiana tabacum. horts 54, 993–997. doi: 10.21273/HORTSCI13915-19
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Love, M. I., Soneson, C., Patro, R. (2018). Swimming downstream: statistical analysis of differential transcript usage following salmon quantification. F1000Res 7, 952. doi: 10.12688/f1000research.15398.3
Mérai, Z., Graeber, K., Wilhelmsson, P., Ullrich, K. K., Arshad, W., Grosche, C., et al. (2019). Aethionema arabicum: a novel model plant to study the light control of seed germination. J. Exp. Bot. 70, 3313–3328. doi: 10.1093/jxb/erz146
Née, G., Xiang, Y., Soppe, W. J. (2017). The release of dormancy, a wake-up call for seeds to germinate. Curr. Opin. Plant Biol. 35, 8–14. doi: 10.1016/j.pbi.2016.09.002
Oh, E., Kang, H., Yamaguchi, S., Park, J., Lee, D., Kamiya, Y., et al. (2009). Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21, 403–419. doi: 10.1105/tpc.108.064691
Oh, E., Yamaguchi, S., Hu, J., Yusuke, J., Jung, B., Paik, I., et al. (2007). PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19, 1192–1208. doi: 10.1105/tpc.107.050153
Oh, E., Yamaguchi, S., Kamiya, Y., Bae, G., Chung, W.-I., Choi, G. (2006). Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 47, 124–139. doi: 10.1111/j.1365-313X.2006.02773.x
Osterlund, M. T., Hardtke, C. S., Wei, N., Deng, X. W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466. doi: 10.1038/35013076
Pacín, M., Legris, M., Casal, J. J. (2013). COP1 re-accumulates in the nucleus under shade. Plant J. 75, 631–641. doi: 10.1111/tpj.12226
Petruzzelli, L., Kunz, C., Waldvogel, R., Meins, Jr. F., Leubner-Metzger, G. (1999). Distinct ethylene- and tissue-specific regulation of 1,3-glucanases and chitinases during pea seed germination. Planta 209, 195–201. doi: 10.1007/s004250050622
Pogson, B. J., Ganguly, D., Albrecht-Borth, V. (2015). Insights into chloroplast biogenesis and development. Biochim. Biophys. Acta (BBA) - Bioenergetics 1847, 1017–1024. doi: 10.1016/j.bbabio.2015.02.003
Qiu, Y., Li, M., Kim, R. J.-A., Moore, C. M., Chen, M. (2019). Daytime temperature is sensed by phytochrome b in Arabidopsis through a transcriptional activator HEMERA. Nat. Commun. 10, 140. doi: 10.1038/s41467-018-08059-z
Roberts, A., Trapnell, C., Donaghey, J., Rinn, J. L., Pachter, L. (2011). Improving RNA-seq expression estimates by correcting for fragment bias. Genome Biol. 12, R22. doi: 10.1186/gb-2011-12-3-r22
Ruppel, N. J., Hangarter, R. P. (2007). Mutations in a plastid-localized elongation factor G alter early stages of plastid development in Arabidopsis thaliana. BMC Plant Biol. 10, 37. doi: 10.1186/1471-2229-7-37
Sawada, Y., Katsumata, T., Kitamura, J., Kawaide, H., Nakajima, M., Asami, T., et al. (2008). Germination of photoblastic lettuce seeds is regulated via the control of endogenous physiologically active gibberellin content, rather than of gibberellin responsiveness. J. Exp. Bot. 59, 3383–3393. doi: 10.1093/jxb/ern192
Sheerin, D. J., Menon, C., zur Oven-Krockhaus, S., Enderle, B., Zhu, L., Johnen, P., et al. (2015). Light-activated phytochrome a and b interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27, 189–201. doi: 10.1105/tpc.114.134775
Shen, Y., Khanna, R., Carle, C. M., Quail, P. H. (2007). Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 145, 1043–1051. doi: 10.1104/pp.107.105601
Shin, J., Kim, K., Kang, H., Zulfugarov, I. S., Bae, G., Lee, C.-H., et al. (2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. U.S.A. 106, 7660–7665. doi: 10.1073/pnas.0812219106
Shinomura, T., Nagatani, A., Hanzawa, H., Kubota, M., Watanabe, M., Furuya, M. (1996). Action spectra for phytochrome a- and b-specific photoinduction of seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 93, 8129–8133. doi: 10.1073/pnas.93.15.8129
Steinbrecher, T., Leubner-Metzger, G. (2016). The biomechanics of seed germination. EXBOTJ, erw428. doi: 10.1093/jxb/erw428
Tejos, R. I., Mercado, A. V., Meisel, L. A. (2010). Analysis of chlorophyll fluorescence reveals stage specific patterns of chloroplast-containing cells during Arabidopsis embryogenesis. Biol. Res. 43, 99–111. doi: 10.4067/S0716-97602010000100012
Toyomasu, T., Kawaide, H., Mitsuhashi, W., Inoue, Y., Kamiya, Y. (1998). Phytochrome regulates gibberellin biosynthesis during germination of photoblastic lettuce seeds. Plant Physiol. 118, 1517–1523. doi: 10.1104/pp.118.4.1517
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Vanstraelen, M., Benková, E. (2012). Hormonal interactions in the regulation of plant development. Annu. Rev. Cell Dev. Biol. 28, 463–487. doi: 10.1146/annurev-cellbio-101011-155741
Wang, J. (2016). iTRAQ protein profile analysis provides integrated insight into mechanisms of tolerance to TMV in tobacco (Nicotiana tabacum). J. Proteomics 10, 21–30. doi: 10.1016/j.jprot.2015.11.009
Wang, Y., Zhang, M., Dong, S., Liu, Y.-L., Li, Z.-H. (2021). The opposite roles of white light in regulating germination of fresh and aged seed in tobacco. Plants 10, 2457. doi: 10.3390/plants10112457
Wu, C.-T., Leubner-Metzger, G., Meins, F., Bradford, K. J. (2001). Class I β-1,3-Glucanase and chitinase are expressed in the micropylar endosperm of tomato seeds prior to radicle emergence. Plant Physiol. 126, 1299–1313. doi: 10.1104/pp.126.3.1299
Yadav, D., Zemach, H., Belausov, E., Charuvi, D. (2019). Initial proplastid-to-chloroplast differentiation in the developing vegetative shoot apical meristem of Arabidopsis. Biochem. Biophys. Res. Commun. 519, 391–395. doi: 10.1016/j.bbrc.2019.09.019
Yadav, A., Bakshi, S., Yadukrishnan, P., Lingwan, M., Dolde, U., Wenkel, S., et al. (2019). The B-Box-containing microProtein miP1a/BBX31 regulates photomorphogenesis and UV-B protection. Plant Physiol. 179, 1876–1892. doi: 10.1104/pp.18.01258
Keywords: seed germination, imbibition, radicle protrusion, cotyledon expansion, transcriptome, proteome
Citation: Liu Q, Li Z, Zhang M, Dong S, Yang P, Zhang J and Loades E (2023) Systematic analysis of photo/sko-regulated germination and post-germination development of shallow photodormant seeds in Nicotiana tabacum L.. Front. Plant Sci. 13:1042981. doi: 10.3389/fpls.2022.1042981
Received: 13 September 2022; Accepted: 22 November 2022;
Published: 04 January 2023.
Edited by:
Yifeng Wang, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Guangwu Zhao, Zhejiang Agriculture and Forestry University, ChinaCopyright © 2023 Liu, Li, Zhang, Dong, Yang, Zhang and Loades. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenhua Li, emhsaTNAZ3p1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.