- College of Life and Environmental Sciences, Hangzhou Normal University, Hangzhou, China
Ethylene Responsive Factor (ERF) subfamily comprise the largest number of proteins in the plant AP2/ERF superfamily, and have been most extensively studied on the biological functions. Members of this subfamily have been proven to regulate plant resistances to various abiotic stresses, such as drought, salinity, chilling and some other adversities. Under these stresses, ERFs are usually activated by mitogen-activated protein kinase induced phosphorylation or escape from ubiquitin-ligase enzymes, and then form complex with nucleic proteins before binding to cis-element in promoter regions of stress responsive genes. In this review, we will discuss the phylogenetic relationships among the ERF subfamily proteins, summarize molecular mechanism how the transcriptional activity of ERFs been regulated and how ERFs of different subgroup regulate the transcription of stress responsive genes, such as high-affinity K+ transporter gene PalHKT1;2, reactive oxygen species related genes LcLTP, LcPrx, and LcRP, flavonoids synthesis related genes FtF3H and LhMYBSPLATTER, etc. Though increasing researches demonstrate that ERFs are involved in various abiotic stresses, very few interact proteins and target genes of them have been comprehensively annotated. Hence, future research prospects are described on the mechanisms of how stress signals been transited to ERFs and how ERFs regulate the transcriptional expression of stress responsive genes.
Introduction
Transcription factors (TFs) are able to interact with promoters of target genes, for activating or repressing their transcriptional expressions. As one of the largest transcription factor families in plants, the AP2/ERF (APETALA2/Ethylene Responsive Factor) plays indispensable roles in plant growth, development, hormone regulation, and especially in responses to various stresses (Tiwari et al., 2012; Lee et al., 2016; Xu et al., 2020; Yu et al., 2021). Commonly, members of the AP2/ERF family contain at least one copy of a DNA binding domain called the AP2 domain consisting of 60~70 amino acids (Liu et al., 2001; Magnani et al., 2004; Xie et al., 2019a; Zhang et al., 2020). The AP2 domain could be divided into two conservation acting elements, denominated as YRG and RAYD (Okamuro et al., 1997). The YRG element, consisting of about 20 amino acids, is a basic hydrophilic region for DNA binding. While the RAYD element, located at the C-terminus, is an amphiphilic region with the presence of an α-helix that can interact with other proteins or DNA (Owji et al., 2017; Ghorbani et al., 2020). Although the AP2 domain has been considered as plant specific, it was also found in the DNA binding domain of viral and bacterial HNH (His-Asn-His) endonuclease (A class of homing endonucleases) (Magnani et al., 2004). It is supposed that the gain of AP2 domain in plants was as a result of horizontal gene transfer via transposition and homing processes (Magnani et al., 2004).
Considering the variations in full-length of protein and in residue of AP2/ERF domain, members in this family have been categorized into series of subfamilies. Based on the sequence similarity of the Arabidopsis (Arabidopsis thaliana) annotated genome, Riechmann et al. (2000) firstly classified 144 Arabidopsis AP2/ERF members into three subfamilies, such as AP2, ERF, and RAV (Related to ABI3/VP1). Since not all ERF family members respond to ethylene induction, Sakuma et al. (2002) divided ERF proteins into two subfamilies, including the ethylene-related ERF subfamily and the ethylene-free DREB (Dehydration responsive element binding protein) subfamily. The DREB subfamily was further subdivided into six subgroups, called A-1 to A-6, while the six ERF subgroups were named as B-1 to B-6. Besides, a Soloist (few unclassified factors) subfamily was added into the AP2/ERF family (Sakuma et al., 2002). In this classification system, the AP2 subfamily contains two highly similar AP2 domains, while the ERF and DREB subfamilies each contains a single AP2 domain. Proteins of the DREB subfamily contain conserved amino acid residues at 14 (Val) and 19 (Glu) of the AP2/ERF domains, while those of ERF subfamily are Ala and Asp at position-14 and position-19 (Chen K. et al., 2022). In addition, the RAV subfamily contains an AP2 structure and a B3 structure, the Soloist subfamily contains an AP2 structural domain that differs significantly from those of other ERF transcription factors (Feng et al., 2020). Nakano et al. (2006) outlined the phylogenetic history of ERF transcription factors and groups functionally similar proteins together. They identified 147 AP2/ERF members in Arabidopsis and categorized them into four subfamilies, such as AP2, ERF, RAV, and Soloist. In which, the ERF and DREB subfamilies were combined as a novel ‘ERF subfamily’. In this study, all members of the abovementioned DREB and ERF subfamilies were included in the ERF subfamily. Based on the common amino acid sequence motifs outside of the AP2/ERF domain, the ERF subfamily was further divided into 12 groups, namely, groups I to X, VI-L (VI like) and Xb-L (Xb like) (Nakano et al., 2006; Zhu et al., 2021). To date, the classification scheme proposed by Nakano et al. (2006) is most extensively employed in literature.
The first ERF was identified as an ethylene response element binding protein from tobacco (Nicotiana tabacum L.) (Ohme-Takagi and Shinshi, 1995). With more extensive genome sequences, the identification and characterization of ERFs have been conducted in various plants. 122 ERFs have been found in Arabidopsis thaliana (Zhang et al., 2012), while there are 131, 104, 341, 166 and 323 homologues in Oryza sativa (Shao et al., 2020), Triticum aestivum (Zhuang et al., 2011), Nicotiana tabacum (Gao et al., 2020), Zea mays (Zhang J. et al., 2022) and Glycine max (Jiang et al., 2020), respectively. In addition, ERFs from different species were proven to regulate plant growth and development, immunity, and responses to various stresses (Muller and Munne-Bosch, 2015; An et al., 2020; Hong et al., 2022). Moreover, recent studies also showed that ERFs involved in the regulation of plant flavonoid synthesis (An et al., 2020; Zhao C. N. et al., 2021), which are essential to the homeostasis of ROS under abiotic stresses. However, it is still unclear how the variation of AP2/ERF domain in ERF subfamily affect their biological functions. In this study, phylogenetic analysis of the ERF subfamily in Arabidopsis thaliana, Oryza sativa, Triticum aestivum, Nicotiana tabacum, Glycine max and Zea mays will be conducted based on the sequence of the AP2/ERF domain, and the biological function of each subgroup will be specially discussed.
Phylogenetic analysis
Whole-genome protein sequences of Nicotiana tabacum, Arabidopsis thaliana, Glycine max, Oryza sativa, Triticum aestivum, and Zea may were obtained from NCBI (https://www.ncbi.nlm.nih.gov/). The Hidden Markov model (HMM) profile of the AP2 domain (PF00847) was downloaded from the PFAM database (http://pfam.xfam.org/) (Zhang et al., 2021a), and was used to search candidate ERF proteins against the abovementioned whole-genome protein sequences of six species using the software HMMER3.0. After removal of redundant and incomplete sequences in CD-HIT, the putative ERFs sequences were analyzed with SMART (http://smart.embl.de/) and NCBI CD-search (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) to confirm the presence and number of AP2 domain (Wang H. T. et al., 2022). Proteins that contain one AP2 domain but lack of B3 domain were retained as putative ERF proteins for phylogenetic analysis. A phylogenetic tree was subsequently constructed using the NJ method of MEGA 7.0, with bootstrap (1,000 replicates), and was visualized by the Interactive Tree of Life (iTOL). Finally, TBtools (Chen C. J. et al., 2020) was used to visualize the phylogenetic tree, conserved motifs and domain of AP2/ERF genes.
The phylogenetic distribution revealed that the 1117 ERFs were distributed into 8 groups which showed eight different clades labeled with Arabic numerals 1-8 and different line colors, where groups 1, 2, 3, 6, 7 and 8 could be further subdivided (Figure 1). Among the 23 subgroups, subgroup 3a has the largest number of members, consisting of 138 members. Whereas groups 3b, 5 and 8a contained only 2 members. The phylogenetic groups defined by Nakano et al. (2006) were designated as Roman numerals (I to X, VI-L, Xb-L) and were designated by 12 different colored circles outside. The white color means that the two categories do not correspond to each other. Comparing the two phylogenetic groups, we found that there are 12 ERF proteins with different classification compared to Nakano’s system, which is possibly due to conserved residues which might dominate biological functions of ERFs. For example, NP_196680.1 in subgroup IX according to Nakano was reassigned in subgroup 1b in the present phylogenetic tree. Proteins in the subgroup 1b, including the NP_196680.1, demonstrated with several conserved elements at specific positions, such as WLG, AYD, YRG and LNFP. Among them, WLG and YRG are supposed to play essential roles on biological functions of ERFs in 1b subgroup (Gao et al., 2020). However, these residues are not conserved in members of subgroup IX. Based on similar principle, NP_197901.1(II), NP_178173.1(VIII), NP_197480.1(VIII), NP_197346.1(VIII), NP_177301.1(III), NP_174636.1(III), NP_680184.1(V), NP_196720.1(V), NP_196895.1(X), NP_197357.2(IV), NP_196837.1(III) are reassigned to 1a, 2b, 2b, 3a, 3c, 3c, 3c, 3e, 7c, 7c and 8c, respectively.
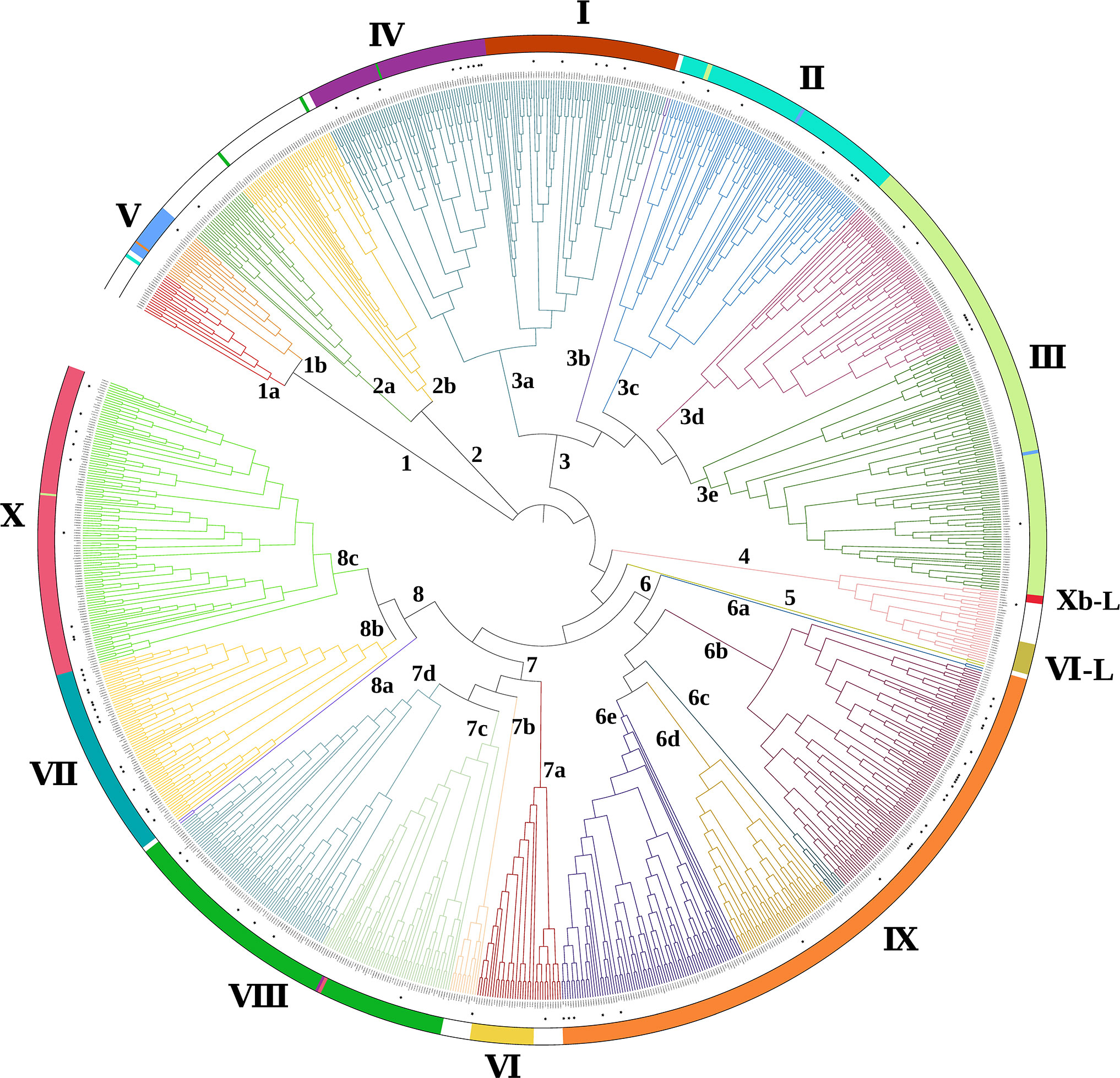
Figure 1 Phylogenetic relationships of ERF family members. The phylogenetic analysis of the ERF subfamily of Nicotiana tabacum, Arabidopsis thaliana, Glycine max, Oryza sativa, Triticum aestivum, and Zea mays by MEGA7.0 (Kumar et al., 2016), constructed the tree using the neighbor joining method and 1000 bootstrap replicates and used iTOL (https://itol.embl.de/) to visualize the tree (He et al., 2021; Magar et al., 2022). The category of functional annotated ERFs mentioned in this review which are listed in Supplementary Table S2 are labeled with black star markers. ERF protein sequences were obtained from NCBI (https://www.ncbi.nlm.nih.gov) database. The protein ID and sequence of all these ERFs are listed in Supplementary Tables S1, S3. The ERF contained in each subgroup is present in Supplemental Table S4.
ERFs of different subgroup selectively bind to particular cis-acting elements of target genes
Commonly, there are some core residues conserved in DNA binding domain of transcription factors that reflect structural and functional specificities in a certain family. A sequence alignment was used to find the conserved residues in the ERF/AP2 DNA binding domain (Nakano et al., 2006; Xie et al., 2019b) of 1117 ERF proteins (Figure 2 and Supplementary Figure S1). It was found that residues Tyr-2 (Y), Arg-3(R), Gly-4 (G), Val-5 (V), Arg-6 (R), Arg-8 (R), Arg-9 (R), Trp-10 (W), Gly-11 (G), Lys-12 (K), Trp-13 (W), Ala-14 (A), Ala-15 (A), Glu-16 (E), Ile-17 (I), Arg-18 (R), Asp-19 (D), Pro-20 (P), Arg-25 (R), Trp-27 (W), Leu-28 (L), Gly-29 (G), Glu-35 (E), Ala-37 (A), Ala-38 (A), Ala-40 (A), Asp-42 (D), Ala-44 (A), Ala-45 (A) and Gly-50 (G) are conserved among most (> 50%) ERF proteins. In addition, the residues Val-14 (V), Trp-28 (W), Leu-29 (L), Thr-33 (T), Ala-34 (A), Tyr-41 (Y) and Asn-57 (N) could also be found in about 40% ERF proteins. These conserved amino acids might determine their binding activity to different cis elements (Yamaguchi-Shinozaki and Shinozaki, 1994; Fujimoto et al., 2000). For example, the Val-14 (V) was previously proven as a key residue in the β-sheet of the ERF/AP2 domain for binding with DRE element (Zhuang et al., 2016), while Ala-37 (A) was predicted as a crucial residue of the α-helix in the DNA binding domain or as an important element for the stability of the ERF/AP2 domain (Liu et al., 2006).
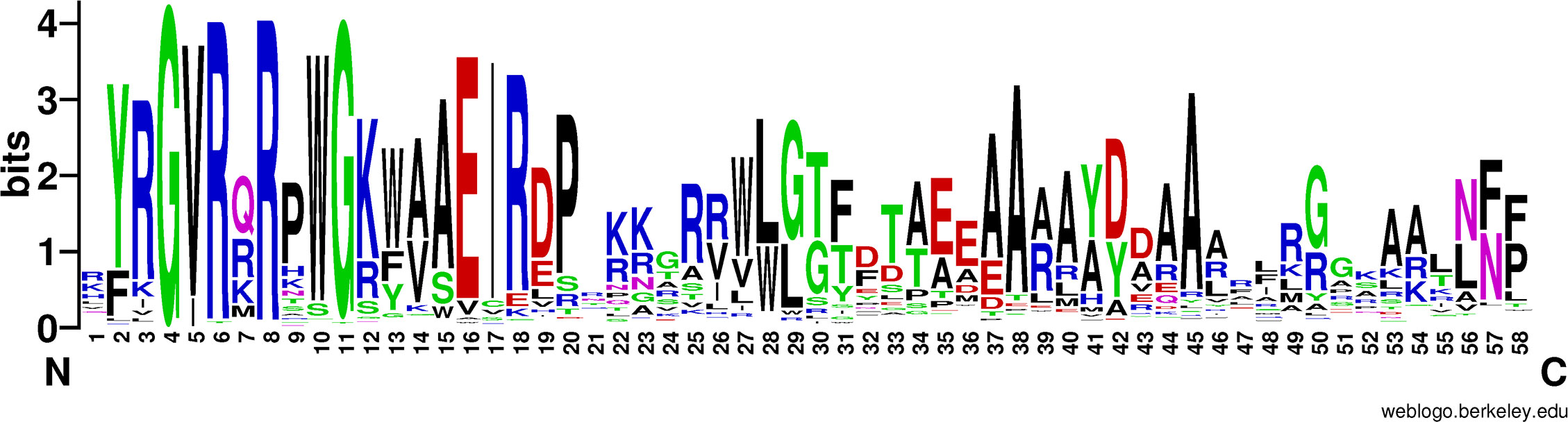
Figure 2 The motif-logo of DNA binding domains in ERF proteins. The motif-logo was plotted with multiple comparisons of 1117 AP2 domains using the software Weblogo (http://weblogo.berkeley.edu/logo.cgi). The overall height of the stack indicates the conservativeness of the sequence at that position, while the height of the letters within the stack indicates the relative frequency of each amino group at that position.
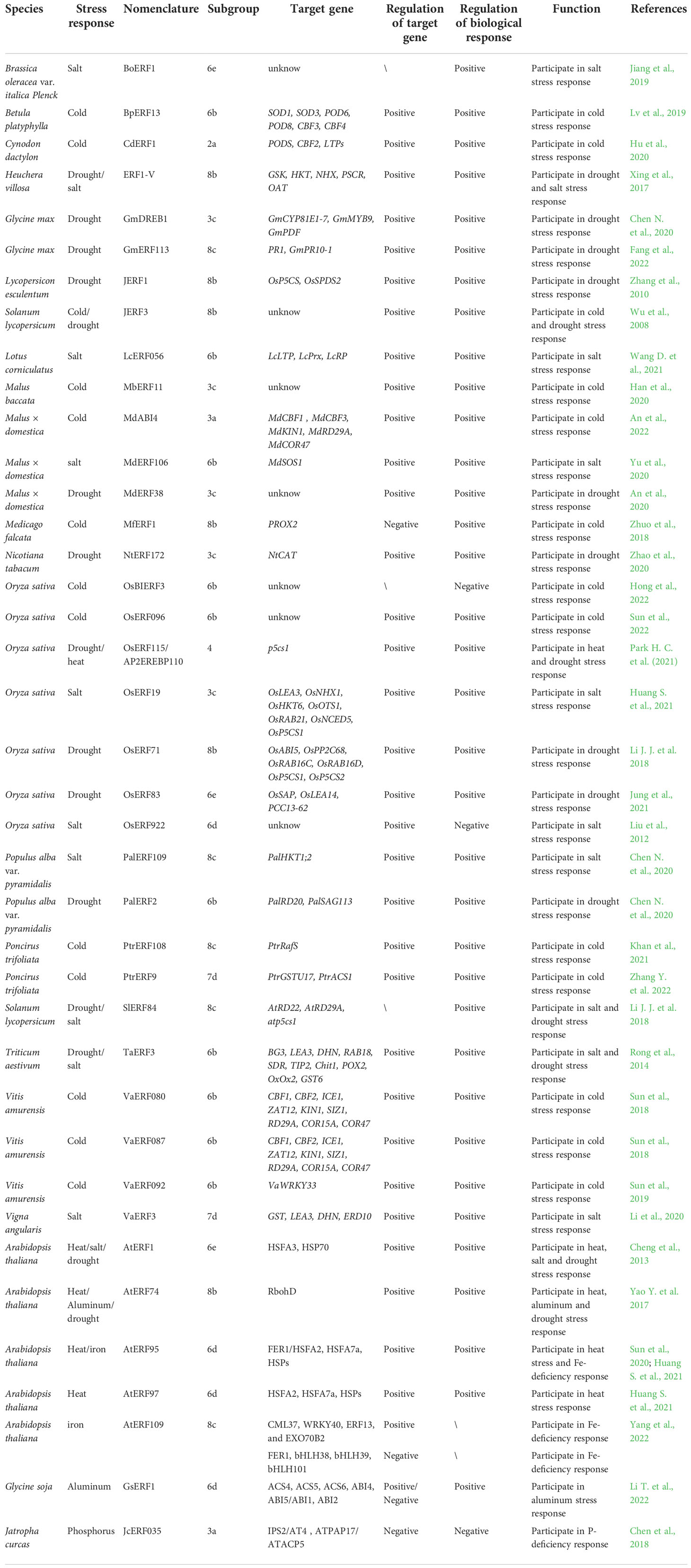
Usually, members of the ERFs subfamily prefer to recognized the Ethylene-Response Element (ERE) with GCC-box sequence (AGCCGCC) which presents in the promoters of ethylene inducible pathogenesis-related (PR) genes as well as some genes associating abiotic stresses, while the DREBs subfamily recognizes Dehydration-Responsive or C-Repeat Element (DRE/CRT) with a core motif of [(A/G)CCGAC] in promoters of target genes to confer resistance to abiotic stresses, especially drought and cold (Yamaguchi-Shinozaki and Shinozaki, 1994; Ohme-Takagi and Shinshi, 1995; Stockinger et al., 1997; Zarei et al., 2011; Yang et al., 2018; Debbarma et al., 2019; Xie et al., 2019b; Zhang J. et al., 2022). These two primary cis-elements have been identified in several plant species, such as Arabidopsis (Arabidopsis thaliana) (Yang et al., 2009), rice (Oryza sativa) (Mohanty, 2021), wheat (Triticum turgidumssp) (Xing et al., 2017), soybean (Glycine max) (Jiang et al., 2020), tomato (Lycopersicon esculentum) (Yang H. H. et al., 2021), maize (Zea mays) (Hao et al., 2020) and tobacco (Nicotiana tabacum) (Gao et al., 2020).
The GCC-box is one of the most common targets in ERF subgroups 3a, 6b, 6e, 7a, 7d, 8b and 8c. Using heteronuclear multidimensional NMR techniques, Allen et al. (1998) reported the first three-dimensional solution structure of AP2/ERF domain in AtERF1 from Arabidopsis, and the secondary structure resembled the zinc finger which contains α-helices. It was found that the α-helix and β-sheet of AP2/ERF domain can recognize the core cis-acting elements GCC-box of the ethylene-responsive promoter region in target genes (Hao et al., 1998). Actually, the β-sheet in ERFs was more important for formation of the domain-GCC box complex (Sakuma et al., 2002). Rong et al. (2014) showed that the TaERF3 protein bound to the GCC-box cis-element in the promoters of seven stress-related genes, which positively regulated the responses to drought and salinity in wheat. Han et al. (2016) demonstrated that MaERF11 bound to the GCC-box motif of three ripening-related Expansin genes (MaEXP2, MaEXP7 and MaEXP8), as well as an ethylene biosynthetic gene (MaACO1) in banana (Musa acuminata). Zhao et al. (2017) found that soybean GmERF113 bound to the GCC-box in pathogenesis-related (PR) genes, PR1 and PR10-1, and positively regulated their expressions to increase resistance to Phytophthora sojae infection. Fang et al. (2022) further showed that the GmERF113 also positively regulated the drought response in soybean by activating GmPR10-1 gene. In addition, Park H. C. et al. (2021) showed that AtERF72 was confirmed to recognize the GCC box in the promoters of several PR genes and activate their transcription, which could be enhanced by AtMPK6-induced phosphorylation. Zheng H. et al. (2021) confirmed that the interaction between SmERF73 and the GCC-box promoter elements of four tanshinone-associated genes regulated tanshinone biosynthesis in response to stress elicitors in Salvia miltiorrhiza. Besides, Zang et al. (2021) showed that the maize ZmERF061 may directly activate the expression of downstream defense-related genes by interacting with the GCC-box element in their promoter regions. In Lilium longiflorum, the heat-inducible LlERF110 may hinder the establishment of thermotolerance via being recruited to GCC-elements (Li T. et al., 2022). Zhu et al. (2022) suggested that VaERF16 from Chinese wild grape (Vitis. amurensis ‘Shuang You’) increased the transcript levels of VaPDF1.2 by binding directly to the GCC box in its promoter, enhanced resistance of grapevine to Botrytis cinerea infection.
The DRE/CRT element could be recognized by ERFs which are mainly from subgroups 3a, 3d, 3e, 6b, 6e, 7a and 8b. The DRE element was identified from the promoter region of the RD29A gene which was involved in drought resistance in Arabidopsis (Yamaguchi-Shinozaki and Shinozaki, 1994; Zhao et al., 2013). And the CRT element was a similar motif with DRE element in cold-inducible genes (Baker et al., 1994; Zhao et al., 2013). Azzeme et al. (2017) showed that the elevated accumulation of Elaeis guineensis DREB1 in transgenic seedlings regulated the expressions of eight DRE-containing genes by interacting with DRE motif in their promoters under both oxidative and cold stress. Additionally, ZmDREB1A induces ABA-independent genes like COR15A, KIN1, and KIN2 through binding their DRE/CRT sequences to further affect the expressions of dehydration and cold-responsive genes (Qin et al., 2004; Kimotho et al., 2019). Feng et al. (2019) found that zoysiagrass (Zoysia japonica) ZjDREB1.4 protein, which enhanced Arabidopsis tolerance to temperature stresses, was capable of binding specifically but weakly to the DRE/CRT element. Besides, Lee et al. (2021) demonstrated that the Arabidopsis CBF1 can bind to DRE/CRT motifs in the promoter of COR15a to confer freezing tolerance. Similarly, CBF2 and CBF3 of Arabidopsis, bind to the DRE, being also related to low-temperatures (Wu et al., 2017; Kidokoro et al., 2020). Mao et al. (2020) confirmed that SlDREBA4 specifically bound to the DRE elements of the downstream Hsp genes and contributed to heat tolerance in tomatoes (Solanum lycopersicum). However, recent study indicated that some DREB was also involved in certain processes during the plant life cycle via binding to DRE/CRT motifs. In transgenic cotton, Lu et al. (2022) suggested that not only did AmCBF1 from Ammopiptanthus mongolicus enhance cotton drought and cold stress tolerance, but it was capable of binding to the CRT/DRE elements in the upstream promoter of GhPP2C1 or GhPP2C2 and repressing their expression, which led to cotton dwarfing.
Despite the generalization shown previously that the ERFs bind to GCC-box and the DREBs bind to DRE/CRT element, an increasing number of studies have been reported that some ERFs are also capable of binding to DRE/CRT elements and vice versa. (Sun et al., 2008), implying their potential roles in abiotic stress (Eini et al., 2013; Thirugnanasambantham et al., 2015; Behera et al., 2022). For instance, Tsi1, an ERF protein from tobacco, binds specifically to the GCC and the DRE/CRT sequences, resulting in improved tolerance to salt and pathogens (Park et al., 2001). Zhang G. Y. et al. (2009) suggested that GmERF3 specifically bound to both the GCC box and DRE/CRT element to enhance the soybean’s tolerances to salinity, drought as well as pathogen infection. CaPF1, a pepper (Capsicum annuum) ERF, was proved to confer pathogen and freezing tolerance in transgenic Arabidopsis through binding to GCC box and DRE/CRT element (Yi et al., 2004). TSRF1, an ERF protein from tomato, binds to both GCC-box and DRE sequences to promote drought and osmotic tolerance in some transgenic plants (Zhang et al., 2004; Quan et al., 2010). In addition, LcERF056 from Lotus corniculatus bound to cis-element GCC-box or DRE of reactive oxygen species (ROS)-related genes to enhance plant salt tolerance (Wang D. et al., 2021). By binding to the GCC and DRE cis-elements, OsERF096 activated the expression of unknown targets to regulate cold tolerance of rice via JA-mediated signaling (Sun et al., 2022). Chen N. N. et al. (2022) indicated that PalERF2 regulated drought response in poplar (Populus alba var. pyramidalis) by binding to DRE motifs on the promoters of drought-responsive genes PalRD20 and PalSAG113. In walnut (Juglans regia), Yang G. Y. et al. (2021) revealed that JrERF2-2 effectively improved plant drought tolerance through interacting with JrWRKY7 to control the expression of GSTs by binding to GCC-box or DRE motif. Finally, IbRAP2.4 from Ipomoea batatas bound to both DRE and GCC-box elements, which promoted lateral root formation and enhanced the drought tolerance of transgenic Arabidopsis, while it inhibited storage root formation in transgenic sweet potato by comprehensively up-regulating lignin biosynthesis pathway genes (Bian et al., 2022).
In particular, some ERF and DREB members bound to both abovementioned elements in promoter of genes involved in hormone pathway, like ethylene, jasmonic acid and auxin. For instance, the tomato (Solanum lycopersicum [f. sp. Lycopersicon esculentum]) LeERF2 and DRE could form as a transcriptional complex on the promoter and activate the expression of LeACO3 for ethylene biosynthesis (Zhang Z. J. et al., 2009). In addition, Li et al. (2016) found that Apple (Malus domestica) MdERF2 interacted with the DRE motif of MdACS1 gene and suppressed its transcription, thereby inhibiting ethylene biosynthesis in ripening fruit. In Chinese flowering cabbage (Brassica rapa var. parachinensis), Tan et al. (2018) revealed that BrERF72 directly activated expressions of JA biosynthetic genes BrLOX4, BrAOC3, and BrOPR3 through binding to GCC or DRE/CRT cis-element during JA-promoted leaf senescence. Besides, Liu et al. (2018) demonstrated that tomato Sl-ERF.B3 could regulate the expression of Sl-IAA27 probably through directly binding to the typical DRE/CRT element presenting in its promoter region, while Sl-ERF.B3 was associated with ethylene and auxin signaling. The reason for their different DNA-binding specificity depends on the divergent residues in β-sheet of AP2/ERF domain (Thirugnanasambantham et al., 2015). In which, some residues dominate the binding ability of ERFs and DREBs to DRE/CRT and/or GCC-box. For example, the Val-14 and Glu-19, especially Val-14, were proven to be essential for specific binding to DRE (Sakuma et al., 2002). In addition, Liu et al. (2006) indicated that Ala-37 was reported to play a key role in binding to both GCC-box and DRE, while Sun et al. (2008) shown that the Ser-15 in the AP2/ERF domain was demonstrated to be essential for its specific binding to GCC-box. Recent studies showed that some other amino acids, such as Pro-9, His-9 and Ser-9, also have important functions (Liu et al., 2020; Zhang L. et al., 2022).
Additionally, ERFs can also recognize other cis-elements that diverge significantly from the abovementioned two motifs (Welsch et al., 2007; Shaikhali et al., 2008; Xie et al., 2019b). For example, ERF75/RAP2.2 regulated carotenoid biosynthesis pathway in Arabidopsis via directly binding to 5′-ATCTA-3′ sequences of genes PSY and PDS (Welsch et al., 2007). Besides, RAP2.2 also bound to hypoxia-responsive promoter elements (HRPE) (5′-AAACCA(G/C)(G/C)(G/C)GC-3′) to regulate Hypoxia-Responsive gene expression in Arabidopsis (Gasch et al., 2016). In addition, RRTF1/ERF109 participated in ROS homeostasis under dehydration stress through binding to GCC box-like motif (AGACGCC) of genes ZAT12 (Wang et al., 2020).
In short, ERFs recognize and bind to various cis-elements which in the promoters of target genes to participate in different regulatory processes by regulating their expression (Table 1). We found that recent researches on cis-elements of ERF mainly focused on 4 species, like Arabidopsis thaliana, Glycine max, Solanum lycopersicum and Zea mays. Moreover, most of ERFs positively regulated the expression of target genes through binding to their cis-elements, only one tenth of ERFs play negative regulatory roles. Finally, ERFs of different subgroups were inclined to recognize different cis-elements. It is suggested that the ERF members in subgroups 3a, 6b, 6e, 7a and 8b are capable of recognizing cis-elements in GCC-box and/or DRE/CRT element. The subgroups 7d and 8c are more likely to bind to GCC-box, while ERFs in subgroup 3d and 3e have more possibilities to recognize DRE/CRT element.
Regulatory mechanism of ERFs involved in their transcriptional activations
ERFs recognize specific motifs and function as activator or repressor of a particular gene. In general, activation domains identified in plant. ERFs do not have distinct sequence motifs but tend to be rich in acidic amino acids, like glutamic acid, aspartic acid. Tiwari et al. (2012) named a motif ‘EDLL’ based on the conserved glutamic acid (E), aspartic acid (D) and leucine (L) residues. The EDLL motif has the ability to activate the transcription process. For example, ORA59, an ERF from Arabidopsis, contains EDLL motif and the specific Leu residue at position 228 of the ORA59 EDLL motif mainly contributed to its transcriptional activity on AtACT gene expression (Pre et al., 2008). On the other hand, ERFs containing the ERF-associated amphiphilic repression (EAR) motif (LxLxLx or DLNxxP) are usually involved in repression mechanism (Ohta et al., 2001; Hiratsu et al., 2003). For instance, Liu J. X. et al. (2021) demonstrated that AgERF8, an EAR-type ERF from celery (Apium graveolens), negatively affected the resistance of transgenic Arabidopsis to ABA and salt stress through inhibiting downstream expression of genes.
The transcriptional activity of ERFs might also be affected by post-translational modified histone, such as acetylated- or methylated-histone (Figure 3). These modifications activate or repress transcription by generating more ‘open’ or ‘closed’ chromatin configurations, respectively (Pfluger and Wagner, 2007). A study in peanut (Achnids hypogaea) showed that inhibition of histone deacetylase (HDACs) and polyethylene glycol (PEG) treatment induced acetylation around the promoter region of AhDREB1, which promoted the transcription of AhDREB1 and improved the drought resistance in plant (Zhang B. H. et al., 2018). Anh Tuan et al. (2016) suggested that methylated histone modification induced the expression of PpEBB and regulated bud break in Japanese pear (Pyrus pyrifolia) by activating cell cycle regulatory genes.
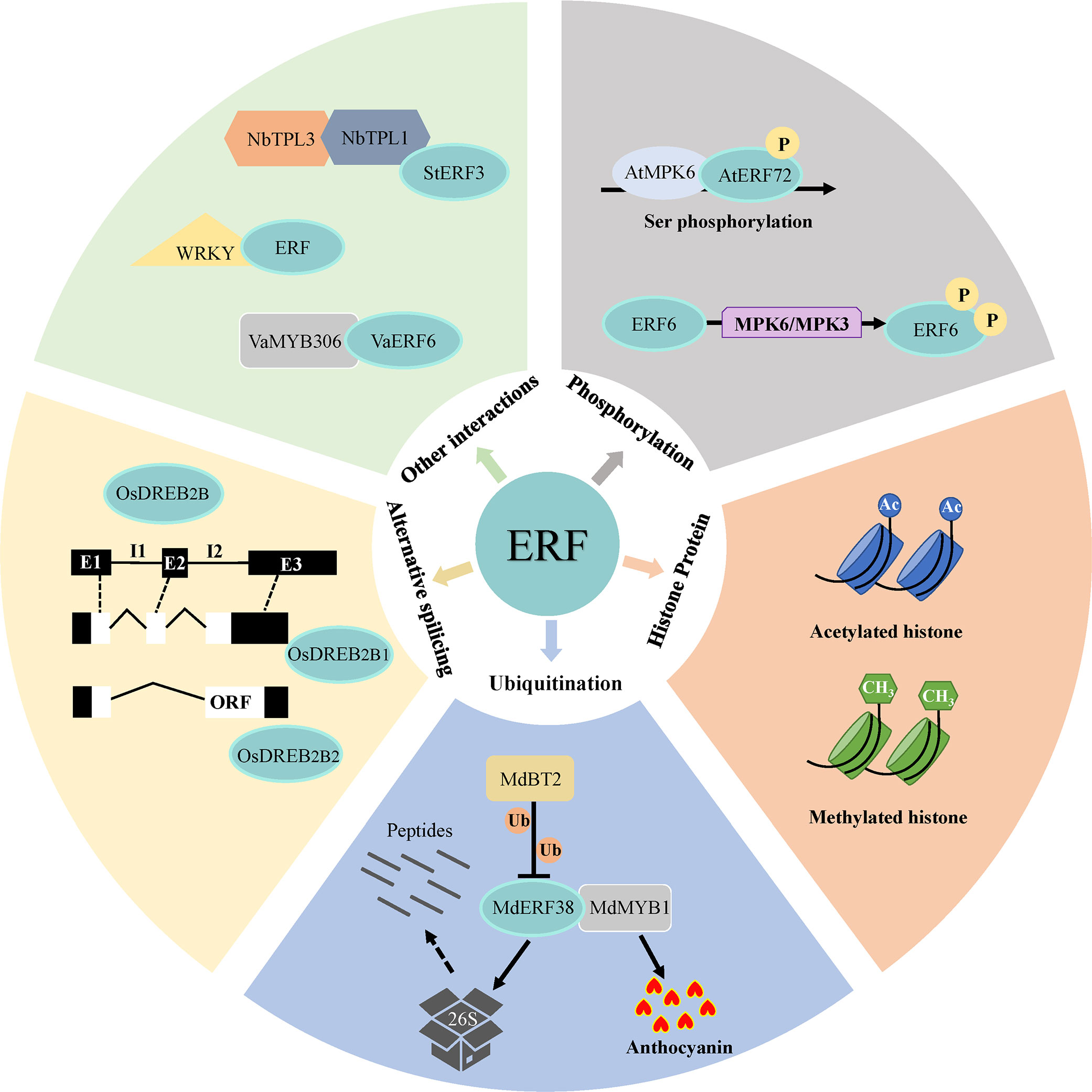
Figure 3 Different regulatory patterns of ERF transcription activation. Regulatory mechanisms of ERFs involved in their transcriptional activations include alternative splicing of their coding mRNA, activation or degradation by post-translational modifications (phosphorylation or ubiquitination), induction by chromatin configurations (such as acetylated- or methylated-histone), and regulation through combination with other nucleic proteins.
Post-translational modifications (PTMs) influence ERF transcriptional activity through a series of ways including phosphorylation and ubiquitination (Figure 3). Protein phosphorylation is one of the reversible PTM of ERFs, which are controlled by kinases to phosphorylate and phosphatases to dephosphorylate substrates. MAPKs or MPKs (Mitogen-activated protein kinases), a kind of kinases, are involved in phosphorylating protein substrates to regulate cellular processes (Lee et al., 2015). For instance, Park H. C. et al. (2021) suggested that the phosphorylation of AtERF72 by AtMPKs plays roles at increased DNA binding activity and many stress signaling pathways, including heat and oxidative stress in Arabidopsis. Similarly, phosphorylation of Arabidopsis ERF6 by MPK3/MPK6 in either the gain-of-function transgenic plants or in response to Botrytis cinereal infection increases ERF6 protein stability (Meng et al., 2013). Ubiquitination is another essential PTM that affects the structure or stability of substrate proteins. In rice, the E3-ubiquitin ligase OsHOS1 targets OsEREBP1 and OsEREBP2 for degradation and modulates the expression of OsRMC, a gene involved in root mechanosensing, through the interaction with two ERFs (Lourenco et al., 2015). Besides, An et al. (2021) revealed that MdBT2 negatively modulated MdERF38-promoted anthocyanin biosynthesis by accelerating the ubiquitination-mediated degradation of the apple (Malus pumila) MdERF38 protein in response to drought stress.
Moreover, alternative splicing has been reported to impact the transcriptional activity of ERFs (Figure 3). Various ERF functional isoforms produced by alternative splicing were found, such as, rice OsDREB2A/2B (Matsukura et al., 2010), maize (Zea mays) ZmDREB2A (Qin et al., 2007), wheat (Triticum aestivum) WDREB2 (Egawa et al., 2006), and barley (Hordeum vulgare) HvDRF1 (Xue and Loveridge, 2004). It was recently revealed that plants could produce an inactive ERF form containing stop codons before the DNA binding domain during normal conditions, while under stress conditions, the exon with a premature stop codons is excluded to generate a functional transcription factor (Xie et al., 2019b).
ERFs are also capable of forming transcriptional complex (Figure 3). Interaction between these TFs are integral to transcriptional regulation. Several ERFs cooperatively recruit transcriptional co-repressors such as topless (TPL) and topless-related (TPR) (Causier et al., 2012), thereby inhibiting the expression of downstream target genes. In transgenic Nicotiana benthamiana, StERF3 from potato (Solanum tuberosum) interacted with the co-repressors NbTPL1 and NbTPL3 via the EAR motif, which facilitated the cell death (Qi et al., 2022). On the other hand, it was found that protein-protein interactions between ERF and WRKY and synergistic regulatory effects in Arabidopsis and Persimmons (Diospyros kaki Thunb) (Zhu et al., 2019). In addition, Zhu et al. (2022) demonstrated that VaERF16 from Vitis amurensis ‘Shuang You’ interacted with the MYB family transcription factor VaMYB306, and the VaERF16-VaMYB306 transcriptional complex resulted in higher transcript levels of VaPDF1.2 to enhance resistance of grapevine to Botrytis cinerea infection.
Transcriptional regulation of ERFs in response to abiotic stresses
Previous study demonstrated by high-throughput that plenty of stresses, including drought, salinity, cold have a significant induction/repression effect on the transcriptional expression of ERF genes, which proved ERF subfamily genes are extensively involved in a variety of adversity responses to stress. We collected the transcriptomic data in six plant species to verify our hypothesis on the relevance that ERF members of specific subgroup involved in particular abiotic stresses, such as salinity, cold and drought (Figure 4). The heatmap was generated by the software TBtools (Chen C. J. et al., 2020) with the 113 ERF genes transcriptome values of log2FoldChange (Log2FC) in six species under drought, cold, and salinity stress, respectively (Shankar et al., 2016; Jin et al., 2017; Li et al., 2017; Sharma et al., 2018; Guan et al., 2019; Sharmin et al., 2020; Liu X. et al., 2021; Yu et al., 2022) (Supplemental Table S5). Expression profiles of expressed genes are presented with gradient blue and red boxes, blue represents low expression and red represents high expression. Specially, grey boxes indicate no available values (NA) been found.
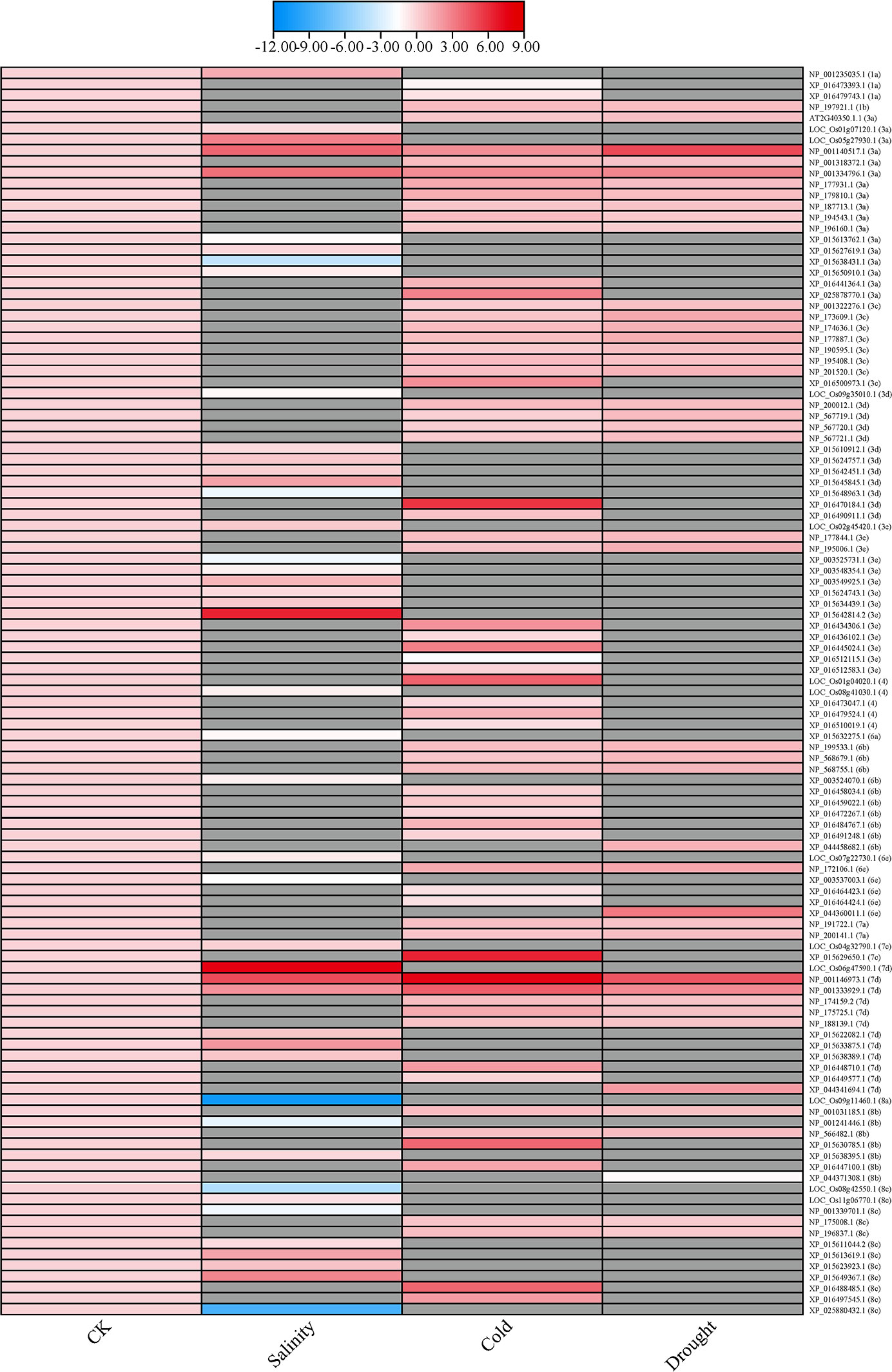
Figure 4 Heatmap of differentially expressed ERF genes in response to abiotic stresses. The heatmap was constructed by TBtools (Chen C. J. et al., 2020) with the 113 ERF genes transcriptome values of log2FoldChange (Log2FC) in six species (Arabidopsis thaliana L., Glycine max L., Nicotiana tabacum L., Oryza sativa L., Triticum aestivum L., and Zea mays L.) under drought, cold, and salinity stress, respectively (Shankar et al., 2016; Jin et al., 2017; Li et al., 2017; Sharma et al., 2018; Guan et al., 2019; Sharmin et al., 2020; Liu X. et al., 2021; Yu et al., 2022) (Supplemental Table S5).
Results showed that 113 ERF genes, from subgroups 1, 3, 4, 6, 7 and 8, were found to be induced or inhibited by these three abiotic stresses. Under salt stress, 45 ERF genes in subgroups 1a, 3a, 3d, 3e, 4, 6a, 6b, 6e, 7c, 7d, 8a, 8b and 8c exhibited with differentially expression compared to those under control condition (CK). Among them, the transcriptional level of members in subgroups 1a and 7c were elevated, while those of members in subgroups 4, 6a, 6b and 8a were declined. Under cold stress, only 8 out of 68 ERF genes expression reduced, which were distributed in 1a, 3e, 4, 6e and 7d subgroups. There were 3 ERF genes from subgroups 3e and 6b without differently expressed. Under drought stress, 42 ERF genes distributed in 1b, 3a, 3c, 3d, 3e, 6b, 6e, 7a, 7d, 8b and 8c subgroups, only a single ERF of those gene from subgroup 8b decreased in expression. On the other hand, ERF in subgroups 3c, 6b, 6d and 7 participate in multiple stresses, while ERF in subgroups 6a and 8a focus on only one stress. However, ERFs in subgroups 2b, 3b, 5, 6c and 7b have not been found to regulate these abiotic stresses response.
Interestingly, there are only 7 ERFs, out of the abovementioned 113 ERF genes, have been annotated with biological function involved in abiotic stresses. For example, CBF1 was found with increased transcription level, consistent with its positive role in regulating plant tolerance to cold stress (Lee et al., 2021). Similarly, CBF2, CBF3 and ERF74 also established mutually confirmed relationships between molecular mechanisms and transcriptome. Meanwhile, it also indicated that there are a large number of ERFs (106 out of abovementioned 113) related to abiotic stress, but no specific molecular mechanism researches of them have been carried out.
ERFs regulate plant responses to various abiotic stresses
To withstand environmental stresses, plants have evolved interconnected regulatory pathways that enable them to respond and adapt to their environments in a timely manner (Zhang H. et al., 2022) (Figure 5) (Agafonov et al., 2016; Nath and Tuteja, 2016; Yuan et al., 2016; Lievens et al., 2017; Zargara et al., 2017; Abhinandan et al., 2018; Dutta et al., 2018; Tiwari and Lata, 2018; Priya et al., 2019; Gong et al., 2020; Mahmood et al., 2020; Angulo et al., 2021; Devireddy et al., 2021; Jha et al., 2022). Plant response strategies to abiotic stresses involve changes at the molecular, cellular, biochemical, and physiological levels (Baillo et al., 2019). The response strategies could be classified to non-adaptive responses and adaptive responses, former including the detrimental changes in membrane fluidity and protein structure as well as the disruptions in enzyme kinetics and molecular interactions, latter including the repair of stress-induced damage, the re-balancing of cellular homeostasis and the adjustment of growth to levels suitable for the particular stress condition (Zhu, 2016; Zhang et al., 2020).
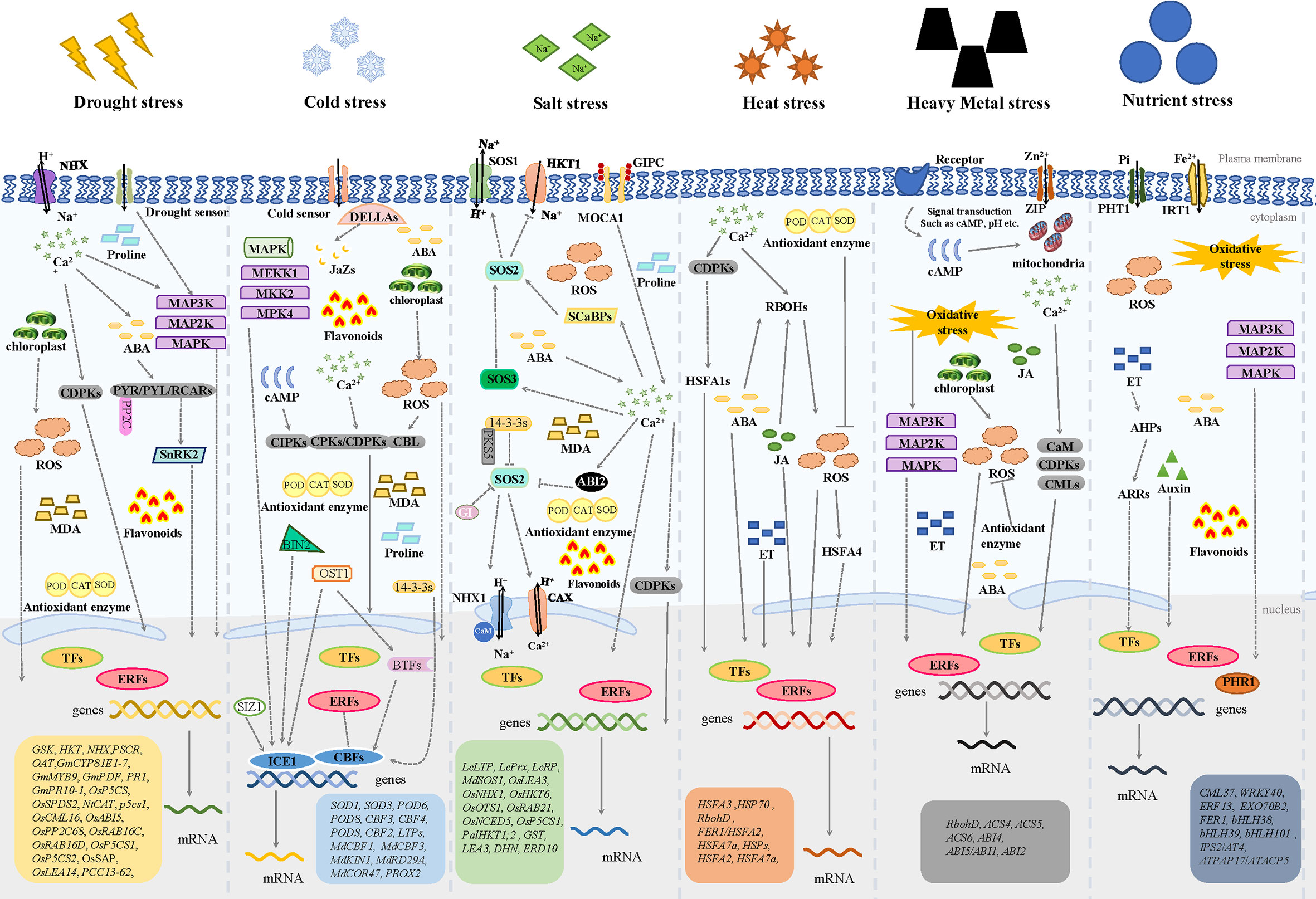
Figure 5 The mechanisms of ERF subfamily transcription factors on regulating plant responses to abiotic stresses. NHX: Na+/H+ antiporter; MAP3K: Mitogen-activated protein kinase kinase kinases; MAP2K: Mitogen-activated protein kinase kinases; MAPK: mitogen-activated protein kinase; ABA: abscisic acid; CDPK: Ca2+-dependent protein kinase; PYR/PYL/RCARs: pyrabactin resistance/pyr1-like/regulatory components of ABA receptors; SnRK2: sucrose non-fermenting-1-related protein kinase 2; PP2C: Type 2C protein phosphatase; ROS: reactive oxygen species; MDA: Malondialdehyde; POD: Peroxidase; CAT: catalase; SOD: Superoxide Dismutase; DELLAs: D-aspartic acid, E-glutamic acid, L-leucine, A-alanin; JaZs: Jasmonate ZIM-domain; MEKK1: Mitogen-activated protein kinase kinase kinases 1; MKK2: Mitogen-activated protein kinase kinases 2; MPK4: mitogen-activated protein kinase 4; cAMP: Cyclic adenosine monophosphate; CIPKs: CBL-interacting protein kinases; CPKs: Ca2+-dependent protein kinases; CBL: calcineurin B-like protein; BIN2: brassinosteroid-insensitive 2; OST1: stomatal opening factor 1; 14-3-3s: general regulatory factor, GRF; BTFs: basic transcription factors; SIZ1: SUMO E3 ligase 1; ICE1: Inducer of CBF Expression 1; CBFs: C-repeat binding factors; SOS1, SOS2, SOS3: salt overly sensitive 1, 2, 3; HKT1: high-affinity potassium transporter 1; MOCA1: monocation induced Ca2+ increases 1; GIPC: glycosyl inositol phosphorylceramide; SCaBPs: SOS3-like calcium binding proteins; ABI2: ABA insensitive 2; PKS5: SOS2-like Protein Kinase 5; GI: Gigantea; CAX: Cation/H+ exchanger antiporter; CaM: calmodulin; CMLs: calmodulin-like proteins; RBOHs: respiratory burst oxidase homologs; HSF: HEAT SHOCK FACTOR (e.g., HSFA1s, HSFA4); JA: Jasmonic acid; ET: Ethylene; PHT1: Phosphate Transporter 1; IRT1: Iron-regulated transporter 1; ZIP: ZRT/IRT-Related Protein; AHPs: Arabidopsis Histidine-containing Phosphotransmitters; ARRs: Arabidopsis Response Regulators; PHR1: phosphate starvation response 1.
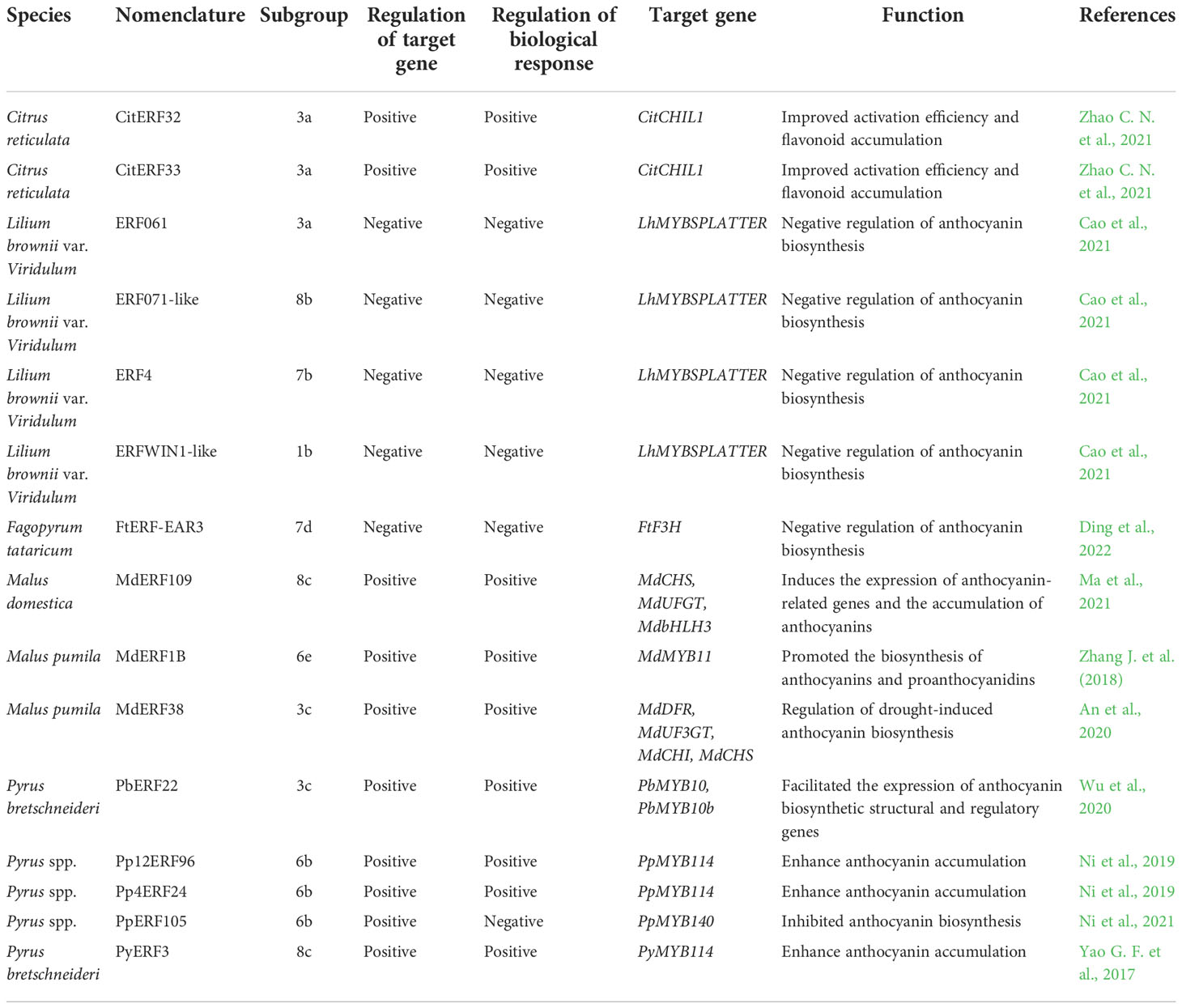
Increasing studies support the involvement of ERFs in abiotic stress, including drought, salinity, cold, high temperature, heavy metal toxicity and nutrition stresses which responses by controlling the activation of stress-response genes (Klay et al., 2018; Debbarma et al., 2019). To uncover the potential roles of ERFs from different subgroups in different abiotic stresses, most of the functionally annotated ERF TFs were categorized into the above-mentioned 8 subfamilies (Figure 1 and Table 2). More than half of the ERFs subfamilies are involved in abiotic stress responses. Among them, the six subfamilies 2, 3, 4, 6, 7 and 8 are involved in drought, salt, cold and other stress responses, and their detailed subgroups are 2a, 3a, 3c, 3d, 3e, 4, 6b, 6d, 6e, 7a, 7d, 8b and 8c. Most of the drought responsive members were from 6b and 8b. The great majority of ERFs related salinity were distributed in 6b subgroup. The ERFs subgroup mainly involved in clod stress response were 3d and 6b. In addition, five subgroups were found to regulate the heat tolerance of plants, including 3e, 4, 6d, 6e and 8b. ERFs of subgroups 6d and 8b are involved in the response to heavy metal toxicity. There are three subgroups of ERFs participating in nutrition stress, including subgroups 3a, 6d, and 8c. In short, ERFs of subgroups 3, 6 and 8 seem to participate in most kinds of abiotic stresses, such as drought, salinity, cold, heat and nutrition stresses.
ERFs involved in drought stress
Drought is one of the most serious abiotic stresses that could adversely hinder plant growth, development and productivity (Ali et al., 2021). Drought stress usually causes water deficit in plant, which is embodied in height decreased, leaf wilting, number and area of leaves changed (Yang X. Y. et al., 2021). At the physiological and biochemical level, the balance of ROS homeostasis in plant was broken down due to the excessive accumulation (Zhao J. Q. et al., 2021). Furthermore, the increasing reactive oxygen free radicals make plant cells suffer oxidative stress (Guo et al., 2018; Yang X. Y. et al., 2021).
It seems that ERFs could modulate the transcriptional expression of drought-responsive gene for production of osmolyte or regulate the plant ability on ROS scavenging (Table 2). On the one hand, ERFs modulate the expression of osmolyte synthesis genes to confer drought tolerance (Wang et al., 2018). Zhang et al. (2010) found that JERF1, a tomato ERF protein, significantly enhanced drought tolerance of transgenic rice through increasing the synthesis of the osmolyte proline. Besides, An et al. (2020) found that MdERF38, an apple ERF protein, promoted anthocyanin biosynthesis in response to drought stress. In rice, OsERF115/AP2EREBP110 enhance drought tolerance by elevating the expression level of a proline biosynthesis P5CS1 gene (Park S. I. et al., 2021). In wheat, Rong et al. (2014) showed that TaERF3 positively regulated stress-related genes which increasing the accumulation of proline and chlorophyll thereby enhancing the drought tolerance. ERF1-V in wheat (Heuchera villosa) improved drought tolerance via modulating P5CR and OAT involved in the proline synthesis (Xing et al., 2017). GmDREB1 could confer drought tolerance of soybean by increasing the photosynthetic efficiency, the accumulation of osmoregulation substances, and the synthesis of melatonin (Chen K. et al., 2022). On the other hand, ERFs could also enhance the ROS scavenging ability of plants under drought condition. Drought induced NtERF172 was proven to positively promote the catalase (CAT)-mediated hydrogen peroxide scavenging in tobacco (Zhao et al., 2020). SlERF84-overexpressed tomato could elevate both superoxide dismutase (SOD) and peroxidase (POD) activities under drought stress (Li Z. et al., 2018).
Recent studies showed that ERFs regulate the plant drought-tolerant responses mainly through the abscisic acid (ABA) signaling pathways (Table 2). ABA is responsible for drought stress tolerance via its capacity to enhance stomatal closure and regulate the expression of drought stress-responsive genes (Takahashi et al., 2020). For instance, the expression of the ERF-type transcription factor OsERF83 was induced by ABA, and that rice overexpressing OsERF83 showed a stronger drought tolerance (Jung et al., 2021). PalERF2 from poplar is recruited to up-regulate the transcription of PalRD20 (a stress-inducible caleosin and positively regulates stomatal closure) and down-regulate the expression of PalSAG113, a repressor of the ABA pathway, resulting in enhanced tolerance to drought (Chen N. N. et al., 2022). Li J. J. et al. (2018) demonstrated that OsERF71 played a positively affect drought tolerance of rice by enhancing the expression of genes (such as OsABI5, OsPP2C68, OsRAB16C and OsRAB16D) associated with ABA signaling and proline biosynthesis. Fang et al. (2022) recently showed that the GmERF113 improves the drought tolerance of soybean by downregulating the abscisic acid 8’-hydroxylase 3 (GmABA8’-OH 3), associated with upregulating of SOD and POD activities.
These researches mentioned above indicate that a large number of ERFs regulate plant responses to drought in ABA-dependent manner. Through the ABA pathway, plants could then synthesize series of osmolyte and accumulate various ROS-scavenging enzymes.
ERFs involved in salinity stress
Salinity is a widespread abiotic stress that constrains plant growth (Wang D. et al., 2021). Under salt stress, plants suffer osmotic stress and ionic toxicity (Wang H. et al., 2022). Osmotic and ionic stresses further exert detrimental effects on plants, such as oxidative stress (Yang and Guo, 2018). Recently, many ERFs have been found to improve plant tolerance to salinity (Table 2).
Many ERFs could regulate plants’ tolerance to salinity by modulating Na+/K+ homeostasis. In rice, OsERF922 might negatively regulated plant tolerance to salt stress by destroying Na+/K+ homeostasis and mediating ABA-signaling pathway (Liu et al., 2012), while OsERF19 could regulate the expression of the salt-responsive Na+/H+ antiporter OsNHX1 and the high-affinity K+ transporter OsHKT6 and OsOTS1 (Huang S. et al., 2021). Besides, PalERF109 enhanced poplar salt tolerance through upregulating a high-affinity K+ transporter (HKT) gene PalHKT1;2 (Chen N. et al., 2020). In addition, MdERF106 associated with MdMYB63, promoted the expression of downstream MdSOS1 and further improved the Na+ expulsion under salt stress in apple (Malus × domestica) (Yu et al., 2020).
ERFs could also regulate the expression of antioxidant enzyme for scavenging salinity induced ROS. The positive regulator LcERF056 is found to enhance salt tolerance in Lotus corniculatus by directly upregulating ROS-related genes LcLTP, LcPrx, and LcRP (Wang D. et al., 2021). In broccoli (Brassica oleracea var. italica Plenck), BoERF1 significantly reduced the content of H2O2 and increased the activities of CAT, POD and SOD, thereby improving salt resistance of plant (Jiang et al., 2019). In transgenic Arabidopsis, overexpression of VaERF3 from Vigna angularis resulted in higher levels of proline accumulation and lower malondialdehyde (MDA) and ROS contents under salinity stress conditions (Li et al., 2020).
Thus, ERFs are involved in modulating the salt tolerance of plant by modulating Na+/K+ homeostasis and regulating the expression of antioxidant enzyme. But the stress-related molecular regulatory network is known to complex and mostly unexplored, the various roles of ERFs in maintaining ion homeostasis or metabolic balance in plants under salt stress may need further concern.
ERFs involved in cold stress
Cold stress, one of the most major abiotic stresses, can be generally categorized into chilling and freezing (Li W. Y. et al., 2022). Both of them usually reduce the fluidity of cell membrane, affect the stability of proteins, break the intracellular ion homeostasis in plant (Ding et al., 2019). Furthermore, the burst of ROS caused by cold produced the osmotic stress and oxidative stress, which result in cell damage and even death (Hu et al., 2020). Actually, plants have evolved sophisticated mechanisms (Table 2) to withstand cold stress (Zheng S. et al., 2021).
Increasing studies have demonstrated that ERFs regulate the activities of antioxidant enzyme and ROS-scavenging to change the tolerance of cold in plant. Wu et al. (2008) demonstrated that JERF3 reduced the accumulation of ROS, which enhanced adaptation to freezing in tobacco. Sun et al. (2018) showed that both VaERF080 and VaERF087 increased antioxidant enzyme activities and regulated the expression of cold-related genes CBF1, CBF2, ICE1, ZAT12, KIN1, SIZ1, RD29A, COR15A, and COR47, which improved the cold tolerance in transgenic Arabidopsis. Lv et al. (2019) found that overexpression of BpERF13 up-regulated SOD1, SOD3, POD6, POD8, CBF3 and CBF4 genes and down-regulated the accumulation of ROS to resist oxidative stress, thus enhancing the cold tolerance of birch (Betula platyphylla). Hu et al. (2020) showed that overexpression of CdERF1 in bermudagrass (Cynodon dactylon) positively regulated cold response by activating cold stress-related genes PODs, CBF2 and LTPs. In transgenic Arabidopsis, heterologous expression Malus baccata MbERF11 contributed to cold stress response probably by promoting the ability to scavenge ROS (Han et al., 2020). Zhang Y. et al. (2022) revealed that (Poncirus trifoliata) PtrERF9 acted downstream of ethylene signaling and functioned positively in cold tolerance via modulation of ROS homeostasis by regulating PtrGSTU17 gene. Moreover, varieties of physiological and biochemical reactions could be regulated by ERFs to influence the adaptation of cold in plant. Zhuo et al. (2018) suggested that MfERF1 from Medicago falcata conferred cold tolerance through polyamine turnover, antioxidant protection and proline accumulation. Khan et al. (2021) revealed that PtrERF108, a positive regulator of cold tolerance, is attributed to its role in the modulation of raffinose content by transcriptionally regulating the PtrRafS gene. In rice, OsBIERF3 significantly decreased the contents of proline to suppress the cold stress response (Hong et al., 2022).
Besides, ERFs neutralize the damage of cold stress to plant through hormone signaling pathways. Sun et al. (2019) revealed that VaERF092 regulated the transcriptional expression of VaWRKY33 and further enhanced cold stress tolerance of grape by regulation of hormone metabolism. Sun et al. (2022) suggested that a new module, the miR1320-OsERF096, regulates cold tolerance of rice by repressing the JA-mediated cold signaling pathway. An et al. (2022) found that MdABI4 integrated jasmonic acid and abscisic acid signals to precisely modulate cold tolerance in apple through the JAZ-ABI4-ICE1-CBF regulatory cascade.
These researches mentioned above attest the principal roles of ERFs in plant suffering cold stress, inducing the expression of genes involved in hormone signaling pathways, elevating the activities of antioxidant enzyme and ROS-scavenging. However, it is worth mentioning that the regulatory roles of plant ERFs in cold tolerance are far from clear as only a very few of them have been explicitly characterized, relative to a large number of genes in this superfamily (Khan et al., 2021).
ERFs involved in other abiotic stresses
Many other abiotic stresses, such as high temperature, heavy metal toxicity and nutrition deficiency, also cause plant growth inhibition, damage, and in the most severe cases, cell death, resulting in major crop yield losses worldwide (Gechev and Petrov, 2020). In order to adapt to these stresses, plants must sense the changes of the temperature, concentrations of heavy metal and mineral nutrient both externally and internally, and generate physiological and morphological responses via a series of metabolic processes including scavenging of ROS and biosynthesis of hormones (e.g., ethylene, jasmonic acid).
Among them, high temperature usually decreases the biosynthesis of auxin and cytokinin, and further impede growth and development of plants. Besides, heat stress could induce phase transition of cell membrane and elevate the accumulation of excess ROS, which leads to oxidative stress (Hasanuzzaman et al., 2013a; Hasanuzzaman et al., 2013b; Li and Howell, 2021). The Arabidopsis AtERF1, an upstream component in both jasmonic acid and ethylene signaling, was showed to activate HSFA3 and HSP70 expression and enhanced the thermotolerance (Cheng et al., 2013). Yao Y. et al. (2017) demonstrated that AtERF74 directly binds to the promoter of RbohD and activates its expression for ROS elimination under heat stresses in Arabidopsis. Under heat stress, AtERF95 can physically interact with AtERF97 for regulating a common set of target genes, including known heat-responsive genes and directly bind to the promoter of HSFA2 (Sun et al., 2020; Huang J. Y. et al., 2021).
Heavy metals, such as aluminum (Al) and zinc (Zn), at elevated concentrations produce severe toxicity symptoms in plants, directly interacting with sulfhydryl group of functional proteins, which disrupts their structure and function, and thus, renders them inactive (Janicka-Russak et al., 2008; Sharma and Dietz, 2009; DalCorso et al., 2013). ERF-VII transcription factors are usually key regulators of the molecular response to hypoxia (van Dongen and Licausi, 2015). Carbonare et al. (2019) found that poplar (Populus spp.) ERF-VII Pop_ERFB2-1 could regulate the expression of hypoxia-responsive genes under high intracellular Zn concentrations. Li L. et al. (2022) suggested that overexpression of GsERF1 may enhance aluminum tolerance of Arabidopsis through an ethylene-mediated pathway and/or ABA signaling pathway. Yao Y. et al. (2017) demonstrated that Arabidopsis AtERF74 enhance plant tolerance to aluminum toxicity dependent on the ERF74-RbohD-ROS signal pathway.
14 essential mineral nutrient elements are required for the optimal growth and development of plants, such as phosphorus (Pi) and iron (Fe), etc (White and Brown, 2010). Plants suffered from Phosphorus (P) deficiency will experience a strong reduction of primary root growth and an arrest of cell division as well as the loss of the quiescent center identity (Sanchez-Calderon et al., 2005; Sanchez-Calderon et al., 2006; Svistoonoff et al., 2007). Chen et al. (2018) indicated that down-regulation of the JcERF035 gene might contribute to the regulation of root system architecture and both biosynthesis and accumulation of anthocyanins in aerial tissues of Arabidopsis under low Pi conditions. Yang et al. (2022) suggested that Arabidopsis AtERF109 is a negative regulator of the leaf response to Fe deficiency. Sun et al. (2020) found that Arabidopsis AtERF95, formed as complex with EIN3, could specifically binds to promoter GCC-box and transactivates of FER1 expression, and consequently regulate sensitivity to Fe deficiency during seedling establishment.
To uncover the potential roles of ERFs from different subgroups in response to various abiotic stresses. Most of the functionally annotated ERFs were categorized into the abovementioned 23 subgroups. We found that nearly half of the ERF subgroups have not been reported to be involved in any abiotic stress, such as subgroup 1a, 1b, 2b, 3b, 5, 6a, 6c, 7b, 7c and 8a. The remaining subgroups were revealed to participate in response to at least one kind of abiotic stresses. Interestingly, members in subgroup 2a are thought to exclusively regulate cold stress, while members in subgroup 3a, 3c, 3d, 6b, 8b and 8c were found to be able to take over the regulations of drought and cold responses, respectively. Similarly, members in subgroup 3e are recognized to exclusively regulate heat stress and members in subgroup 7a are thought to exclusively modulate salinity, while members in subgroup 6d, 6e and 8b are involved in heat and salt stresses. Meanwhile, Members of subgroup 3c, 6b, 6e and 8b were found to regulate the drought and salt tolerance of plants and members of subgroup 3c, 6b, 7d and 8b are involved in cold and salinity responses. ERF members of 4 subgroups (3c, 6b, 8b, 8c) regulate the molecular mechanism of plant responded to three kinds of stresses, including drought, salt and cold. In addition, ERFs of subgroup 8b seem to participate in most kinds of abiotic stresses, such as drought, salinity, chilling, Heat and heavy metal stress. Moreover, subgroup 6d of ERFs participated in salt, heat, heavy metal and nutrition stress. So, it’s true that the variation of AP2/ERF domain in ERF subfamily affect their biological functions related to abiotic stresses.
ERFs regulate plant tolerances to abiotic stress mainly through modulation the syntheses of antioxidative metabolites
Adverse conditions such as drought, salinity, cold and other abiotic stresses usually induce the accumulation of ROS that are detrimental to plant growth and development (Miller et al., 2010; Dreyer and Dietz, 2018). Excessive ROS would lead to increased levels of cell death, thus inhibiting plant growth and reducing crop productivity. There are two main ROS scavenging systems that evolved in plants, including enzymatic and non-enzymatic scavenging system. The enzymatic scavenging system is commonly constituted with SOD, ascorbate peroxidase (APX), and glutathione reductase (GR) (Wang C. L. et al., 2021). Besides, plants depend on non-enzymatic pathways to scavenge several highly toxic ROS, such as 1O2 and •OH, that cannot be scavenged by enzymatic antioxidant systems (Das and Roychoudhury, 2014; Morales and Munne-Bosch, 2019). Generally, the non-enzymatic scavenging system contains several antioxidative metabolites, including MDA and proline, etc. Recently, increasing studies found that flavonoids, a large group of natural metabolites with variable phenolic structures, play crucial roles to scavenge free radical activity to reduce oxidative stress in plants (Pi et al., 2016; Pi et al., 2018; Pi et al., 2019).
Flavonoids represent a wide array of plant secondary metabolites which present C6-C3-C6 structure (Dias et al., 2021). According to their multifarious structures, flavonoids can be further divided into flavanones, flavones, flavonols, isoflavonoids, anthocyanidins and proanthocyanidins (PAs) (Zhao C. N. et al., 2021). In recent years, more and more attention has been paid to the functions of flavonoids. Ferreyra et al. (2012) indicated that flavonoids are involved in plant growth and development processes such as aroma, coloration and signaling (Zhang J. et al., 2009; Jeon et al., 2022; Mahon et al., 2022), while Dong et al. (2020) emphasized that flavonoids also exhibit specific stress resistance function in response to abiotic stresses, such as drought, salinity, cold, heavy metals and other abiotic stresses (Ding et al., 2019; Ghori et al., 2019; Chourasia et al., 2021; Razi and Muneer, 2021). It is worth noting that flavonoids often generated as scavengers to free radicals which always increasingly accumulate in plant suffering abiotic stresses (Zheng et al., 2022). Excessive free radicals are known to severely cause plant death for its strong oxidant effects (Nauser and Gebicki, 2019). Acting as effective antioxidants, flavonoids contribute to eliminating the oxidative free radicals, which is benefiting from the hydroxyl groups in flavonoids (Speisky et al., 2022). Flavonoids biosynthesis is controlled by diverse enzymes, such as CHS (chalcone synthase), CHI (chalcone isomerase), DFR (dihydroflavonol 4-reductase), F3H (flavanone 3-hydroxylase), F3’H (flavonoid 3’-hydroxylase), F3’5’H (flavonoid 3’,5’-hydroxylase), UFGT (UDP-glucose:flavonoid 3-glucosyltransferase), and ANS (anthocyanin synthase) (Fu et al., 2021). It is reported that numerous transcription factors including MYB, ERF, WRKY and bHLH have been found to affect the synthesis of flavonoids by regulating the expressions of these target genes (Ding et al., 2022). In order to reveal the relationship between ERFs and flavonoid biosynthesis, annotated functional ERFs are present in Table 3. It was found that ERFs in subgroups 1b, 3a, 3c, 6b, 6e, 7b, 7d, 8b and 8c play crucial roles in the modulation of flavonoids metabolism.
ERFs are involved in modulation of flavonoids biosynthesis through the co-regulation of transcription factors, especially by the interaction with MBW (MYB-bHLH-WDR) complex (Kirschner, 2022). The Chinese pear (Pyrus bretschneideri) PyERF3 was found to interact with PyMYB114 and its partner PybHLH3 to form a new complex (ERF3-MYB114-bHLH3), hence, to co-regulate anthocyanin biosynthesis (Yao G. F. et al., 2017). Besides, Ni et al. (2019) demonstrated that Pp4ERF24 and Pp12ERF96 promoted anthocyanin biosynthesis in ‘Red Zaosu’ pear (Pyrus spp.) via enhancing the interaction between PpbHLH3 and PpMYB114 as well as the expression of PpMYB114-induced PpUFGT gene. Additionally, the ‘Zaosu’ pear PbERF22 might regulate anthocyanin biosynthesis by enhancing the activation effects of PbMYB10 and PbMYB10b on the PbUFGT promoter (Wu et al., 2020). In apple, Zhang J. et al. (2018) revealed that not only did MdERF1B interact with MdMYB9/11 proteins, but also bound to their promoters to activate the expression of MdLAR, MdANR, and MdANS, which induced PAs and anthocyanin production. In Citrus reticulata, CitERF33 formed a transcription complex with CitRAV1 to strongly enhance the flavonoid accumulation efficiency (Zhao C. N. et al., 2021). Ni et al. (2021) indicated that PpERF105 inhibited anthocyanin biosynthesis in pear through activating PpMYB140 capable of interacting with bHLH3 and bHLH33 to form the repressive PpMYB140/bHLH3 or bHLH33/WD-repeat [M(140)BW] complex. Furthermore, recent studies have revealed that some ERFs alone could bind directly to the promoter of flavonoids biosynthesis genes. Ding et al. (2022) illustrated that FtERF-EAR3 inhibited the expression of FtF3H through binding to the GCC-box in FtF3H promoter, which decreased flavonoids accumulation in Fagopyrum tataricum. Zhao C. N. et al. (2021) demonstrated that CitERF32 and CitERF33 activated the transcription of CitCHIL1 in Citrus and Arabidopsis for significantly enhancing the accumulation of flavanones and flavones. Ma et al. (2021) found that MdERF109 promoted light-induced anthocyanin biosynthesis by directly binding to promoters of anthocyanin-related genes MdCHS, MdUFGT, and MdbHLH3 in apple. Besides, An et al. (2020) showed that apple MdERF38 was able to promote the expression of anthocyanin biosynthetic genes MdDFR, MdUF3GT, MdCHI and MdCHS under drought stress. Cao et al. (2021) suggested that four lily (Lilium brownii var. Viridulum) transcription factors, ERF4, ERF WIN1-like, ERF061 and ERF071-like, might negatively regulate anthocyanin accumulation by directly modulating LhMYBSPLATTER gene. In short, a series of ERFs were confirmed to bind directly to promoters of genes involved in flavonoids synthesis and regulate their transcription under abiotic stresses. Current reports focus mainly on anthocyanidins synthesis in these processes, it is still not clear whether and how ERFs regulate the accumulation of flavonoids in other subclasses.
Comprehensively correlating subgroups and their biological functions, we conclude that the ERFs from 3a, 3d, 3e, 6b, 6e, 7a, 7d, 8b and 8c subgroup engage in drought, salinity, cold, heat, heavy metal and nutrition stress via binding to the GCC-box or/and DRE/CRT element of stress responsive genes. Notably, some of these subgroups could also induce flavonoid biosynthesis. We conjecture that ERF transcription factors in 8c subgroup are capable of binding to GCC-box or DRE/CRT element in drought, salinity, and cold and nutrition responsive genes to further modulate the synthesis of flavonoids, which regulates the tolerance of plant suffering corresponding stress. In like manner, ERF subgroup 6b are able to interact with GCC-box or DRE/CRT element in promoter of stress responsive genes to participate in flavonoid biosynthesis under drought, cold and salinity stress, while the 3a subgroup relates to drought and cold stress. Similarly, ERFs members of subgroup 6e and 7d tends to recognize GCC-box or DRE/CRT element of salinity-responsive genes to change the biosynthesis of flavonoids, when 6e and 7d are associated with drought and cold stress, respectively. Meanwhile, it also makes sense that ERFs in subgroup 3c regulate the synthesis of flavonoids in response to drought, salinity and cold stress. Nevertheless, our conjecture still needs further confirmation to reveal the molecular mechanism during the process.
Conclusion
In this review, a comprehensive analysis of the ERF subfamily regarding the phylogenetic relationships, conserved motifs, cis-acting elements, stress response and regulation mechanism of ERF transcriptional activity was performed. ERFs are plant specific transcription factors, which play an important role in abiotic stresses, such as drought, salinity, chilling and some other adversities. Under these stresses, A series of post-translational modifications such as phosphorylation and ubiquitination affect the transcriptional activity of ERFs. ERFs are activated by mitogen-activated protein kinase induced phosphorylation, forming stable complexes with other transcriptional regulators and structural protein, then binding to cis-element in promoter regions of stress responsive genes. Generally, most ERFs were reported to bind specifically to the GCC cis-element, while the DREBs recognizes DRE/CRT cis-element to confer resistance to abiotic stresses. Beyond that, ERFs also modulate the synthesis of diverse metabolites, including proline, malondialdehyde and flavonoids etc. Act as an antioxidative agent, flavonoids are capable of scavenging ROS generated in plants during abiotic stresses.
ERF is a critical downstream component of the ethylene signaling pathway. Though previous transcriptome results suggested that large numbers of ERF genes of different subgroups play varied roles in response to abiotic stresses, very few interact proteins and target genes of them have been comprehensively annotated, and the molecular mechanism how stress signals been transited to ERFs and how ERFs regulate the transcriptional expression of stress responsive genes remains poorly understood and need further exploration.
Author contributions
YW, XL, JZ, HZ, ST, WX, JP, and FY analyzed the phylogenetic relationships of ERF family members. EP conceived the original idea for the review. All authors wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Science Foundation of China (grants no. 31970286 and 31301053 to EP), the Natural Science Foundation of Zhejiang Province (grants no. LY22C020001 and LY17C020004 to EP) and the Hangzhou Science and Technology Bureau (grant no. 20170432B01 to EP).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1042084/full#supplementary-material
References
Abhinandan, K., Skori, L., Stanic, M., Hickerson, N. M. N., Jamshed, M., Samuel, M. A. (2018). Abiotic stress signaling in wheat – an inclusive overview of hormonal interactions during abiotic stress responses in wheat. Front. Plant Sci. 9, 734. doi: 10.3389/fpls.2018.00734
Agafonov, O., Selsto, C. H., Thorsen, K., Xu, X. M., Drengstig, T., Ruoff, P. (2016). The organization of controller motifs leading to robust plant iron homeostasis. PloS One 11, e0147120. doi: 10.1371/journal.pone.0147120
Ali, E. F., El-Shehawi, A. M., Ibrahimn, O. H. M., Abdul-Hafeez, E. Y., Moussa, M. M., Hassan, F. A. S. (2021). A vital role of chitosan nanoparticles in improvisation the drought stress tolerance in catharanthus roseus (L.) through biochemical and gene expression modulation. Plant Physiol. Biochem. 161, 166–175. doi: 10.1016/j.plaphy.2021.02.008
Allen, M. D., Yamasaki, K., Ohme-Takagi, M., Tateno, M., Suzuki, M. (1998). A novel mode of DNA recognition by a β-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 17, 5484–5496. doi: 10.1093/emboj/17.18.5484
Angulo, M., Garcia, M. J., Alcantara, E., Perez-Vicente, R., Romera, F. J. (2021). Comparative study of several fe deficiency responses in the arabidopsis thaliana ethylene insensitive mutants ein2-1 and ein2-5. Plants-Basel 10, 262. doi: 10.3390/plants10020262
Anh Tuan, P., Bai, S., Saito, T., Imai, T., Ito, A., Moriguchi, T. (2016). Involvement of EARLY BUD-BREAK, an AP2/ERF transcription factor gene, in bud break in Japanese pear (Pyrus pyrifolia nakai) lateral flower buds: Expression, histone modifications and possible target genes. Plant Cell Physiol. 57, 1038–1047. doi: 10.1093/pcp/pcw041
An, J. P., Wang, X. F., Zhang, X. W., You, C. X., Hao, Y. J. (2021). Apple BT2 protein negatively regulates jasmonic acid-triggered leaf senescence by modulating the stability of MYC2 and JAZ2. Plant Cell Environ. 44, 216–233. doi: 10.1111/pce.13913
An, J. P., Xu, R. R., Liu, X., Su, L., Yang, K., Wang, X. F., et al. (2022). Abscisic acid insensitive 4 interacts with ICE1 and JAZ proteins to regulate ABA signaling-mediated cold tolerance in apple. J. Exp. Bot. 73, 980–997. doi: 10.1093/jxb/erab433
An, J. P., Zhang, X. W., Bi, S. Q., You, C. X., Wang, X. F., Hao, Y. J. (2020). The ERF transcription factor MdERF38 promotes drought stress-induced anthocyanin biosynthesis in apple. Plant J. 101, 573–589. doi: 10.1111/tpj.14555
Azzeme, A. M., Abdullah, S. N. A., Aziz, M. A., Wahab, P. E. M. (2017). Oil palm drought inducible DREB1 induced expression of DRE/CRT- and non-DRE/CRT-containing genes in lowland transgenic tomato under cold and PEG treatments. Plant Physiol. Biochem. 112, 129–151. doi: 10.1016/j.plaphy.2016.12.025
Baillo, E. H., Kimotho, R. N., Zhang, Z., Xu, P. (2019). Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes 10, 771. doi: 10.3390/genes10100771
Baker, S. S., Wilhelm, K. S., Thomashow, M. F. (1994). The 5′-region of arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 24, 701–713. doi: 10.1007/BF00029852
Behera, T. K., Krishna, R., Ansari, W. A., Aamir, M., Kumar, P., Kashyap, S. P., et al. (2022). Approaches involved in the vegetable crops salt stress tolerance improvement: Present status and way ahead. Front. Plant Sci. 12, 787292. doi: 10.3389/fpls.2021.787292
Bian, X. F., Kim, H. S., Kwak, S. S., Zhang, Q., Liu, S., Ma, P. Y., et al. (2022). Different functions of IbRAP2.4, a drought-responsive AP2/ERF transcription factor, in regulating root development between arabidopsis and sweetpotato. Front. Plant Sci. 13, 820450. doi: 10.3389/fpls.2022.820450
Cao, Y. W., Bi, M. M., Yang, P. P., Song, M., He, G. R., Wang, J., et al. (2021). Construction of yeast one-hybrid library and screening of transcription factors regulating LhMYBSPLATTER expression in Asiatic hybrid lilies (Lilium spp.). BMC Plant Biol. 21, 563. doi: 10.1186/s12870-021-03347-1
Carbonare, L. D., White, M. D., Shukla, V., Francini, A., Perata, P., Flashman, E., et al. (2019). Zinc excess induces a hypoxia-like response by inhibiting cysteine oxidases in poplar roots. Plant Physiol. 180, 1614–1628. doi: 10.1104/pp.18.01458
Causier, B., Ashworth, M., Guo, W., Davies, B. (2012). The TOPLESS interactome: a framework for gene repression in arabidopsis. Plant Physiol. 158, 423–438. doi: 10.1104/pp.111.186999
Chen, C. J., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y. H., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cheng, M. C., Liao, P. M., Kuo, W. W., Lin, T. P. (2013). The arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 162, 1566–1582. doi: 10.1104/pp.113.221911
Chen, N. N., Qin, J. J., Tong, S. F., Wang, W. W., Jiang, Y. Z. (2022). One AP2/ERF transcription factor positively regulates pi uptake and drought tolerance in poplar. Int. J. Mol. Sci. 23, 5241. doi: 10.3390/ijms23095241
Chen, K., Tang, W. S., Zhou, Y. B., Chen, J., Xu, Z. S., Ma, R., et al. (2022). AP2/ERF transcription factor GmDREB1 confers drought tolerance in transgenic soybean by interacting with GmERFs. Plant Physiol. Biochem. 170, 287–295. doi: 10.1016/j.plaphy.2021.12.014
Chen, N., Tong, S., Tang, H., Zhang, Z., Liu, B., Lou, S., et al. (2020). The PalERF109 transcription factor positively regulates salt tolerance via PalHKT1;2 in populus alba var. pyramidalis. Tree Physiol. 40, 717–730. doi: 10.1093/treephys/tpaa018
Chen, Y. B., Wu, P. Z., Zhao, Q. Q., Tang, Y. H., Chen, Y. P., Li, M. R., et al. (2018). Overexpression of a phosphate starvation response AP2/ERF gene from physic nut in arabidopsis alters root morphological traits and phosphate starvation-induced anthocyanin accumulation. Front. Plant Sci. 9, 1186. doi: 10.3389/fpls.2018.01186
Chourasia, K. N., Lal, M. K., Tiwari, R. K., Dev, D., Kardile, H. B., Patil, V. U., et al. (2021). Salinity stress in potato: Understanding physiological, biochemical and molecular responses. Life-Basel 11, 545. doi: 10.3390/life11060545
DalCorso, G., Manara, A., Furini, A. (2013). An overview of heavy metal challenge in plants: from roots to shoots. Metallomics 5, 1117–1132. doi: 10.1039/c3mt00038a
Das, K., Roychoudhury, A. (2014). Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2, 53. doi: 10.3389/fenvs.2014.00053
Debbarma, J., Sarki, Y. N., Saikia, B., Boruah, H. P. D., Singha, D. L., Chikkaputtaiah, C. (2019). Ethylene response factor (ERF) family proteins in abiotic stresses and CRISPR–Cas9 genome editing of ERFs for multiple abiotic stress tolerance in crop plants: A review. Mol. Biotechnol. 61, 153–172. doi: 10.1007/s12033-018-0144-x
Devireddy, A. R., Tschaplinski, T. J., Tuskan, G. A., Muchero, W., Chen, J. G. (2021). Role of reactive oxygen species and hormones in plant responses to temperature changes. Int. J. Mol. Sci. 22, 8843. doi: 10.3390/ijms22168843
Dias, M. C., Pinto, D., Silva, A. M. S. (2021). Plant flavonoids: Chemical characteristics and biological activity. Molecules 26, 5377. doi: 10.3390/molecules26175377
Ding, M. Q., He, Y. Q., Zhang, K. X., Li, J. B., Shi, Y. L., Zhao, M. Y., et al. (2022). JA-induced FtBPM3 accumulation promotes FtERF-EAR3 degradation and rutin biosynthesis in tartary buckwheat. Plant J. 111, 323–334. doi: 10.1111/tpj.15800
Ding, Y. L., Shi, Y. T., Yang, S. H. (2019). Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 222, 1690–1704. doi: 10.1111/nph.15696
Dong, N. Q., Sun, Y. W., Guo, T., Shi, C. L., Zhang, Y. M., Kan, Y., et al. (2020). UDP-Glucosyltransferase regulates grain size and abiotic stress tolerance associated with metabolic flux redirection in rice. Nat. Commun. 11, 2629. doi: 10.1038/s41467-020-16403-5
Dreyer, A., Dietz, K. J. (2018). Reactive oxygen species and the redox-regulatory network in cold stress acclimation. Antioxidants 7, 169. doi: 10.3390/antiox7110169
Dutta, S., Mitra, M., Agarwal, P., Mahapatra, K., De, S., Sett, U., et al. (2018). Oxidative and genotoxic damages in plants in response to heavy metal stress and maintenance of genome stability. Plant Signaling Behav. 13, e1460048. doi: 10.1080/15592324.2018.1460048
Egawa, C., Kobayashi, F., Ishibashi, M., Nakamura, T., Nakamura, C., Takumi, S. (2006). Differential regulation of transcript accumulation and alternative splicing of a DREB2 homolog under abiotic stress conditions in common wheat. Genes Genet. Syst. 81, 77–91. doi: 10.1266/ggs.81.77
Eini, O., Yang, N., Pyvovarenko, T., Pillman, K., Bazanova, N., Tikhomirov, N., et al. (2013). Complex regulation by Apetala2 domain-containing transcription factors revealed through analysis of the stress-responsive TdCor410b promoter from durum wheat. PloS One 8, e58713. doi: 10.1371/journal.pone.0058713
Fang, X., Ma, J., Guo, F. C., Qi, D. Y., Zhao, M., Zhang, C. Z., et al. (2022). The AP2/ERF GmERF113 positively regulates the drought response by activating GmPR10-1 in soybean. Int. J. Mol. Sci. 23, 8159. doi: 10.3390/ijms23158159
Feng, K., Hou, X. L., Xing, G. M., Liu, J. X., Duan, A. Q., Xu, Z. S., et al. (2020). Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 40, 750–776. doi: 10.1080/07388551.2020.1768509
Feng, W. Q., Li, J., Long, S. X., Wei, S. J. (2019). A DREB1 gene from zoysiagrass enhances arabidopsis tolerance to temperature stresses without growth inhibition. Plant Sci. 278, 20–31. doi: 10.1016/j.plantsci.2018.10.009
Ferreyra, M. L. F., Rius, S. P., Casati, P. (2012). Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 3, 222. doi: 10.3389/fpls.2012.00222
Fujimoto, S. Y., Ohta, M., Usui, A., Shinshi, H., Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box–mediated gene expression. Plant Cell. 12, 393–404. doi: 10.1105/tpc.12.3.393
Fu, M. Y., Yang, X., Zheng, J. R., Wang, L., Yang, X. Y., Tu, Y., et al. (2021). Unraveling the regulatory mechanism of color diversity in camellia japonica petals by integrative transcriptome and metabolome analysis. Front. Plant Sci. 12, 685136. doi: 10.3389/fpls.2021.685136
Gao, Y., Han, D., Jia, W., Ma, X. H., Yang, Y. X., Xu, Z. C. (2020). Molecular characterization and systematic analysis of NtAP2/ERF in tobacco and functional determination of NtRAV-4 under drought stress. Plant Physiol. Biochem. 156, 420–435. doi: 10.1016/j.plaphy.2020.09.027
Gasch, P., Fundinger, M., Muller, J. T., Lee, T., Bailey-Serres, J., Mustroph, A. (2016). Redundant ERF-VII transcription factors bind to an evolutionarily conserved cis-motif to regulate hypoxia-responsive gene expression in arabidopsis. Plant Cell. 28, 160–180. doi: 10.1105/tpc.15.00866
Gechev, T., Petrov, V. (2020). Reactive oxygen species and abiotic stress in plants. Int. J. Mol. Sci. 21, 7433. doi: 10.3390/ijms21207433
Ghorbani, R., Zakipour, Z., Alemzadeh, A., Razi, H. (2020). Genome-wide analysis of AP2/ERF transcription factors family in brassica napus. Physiol. Mol. Biol. Plants. 26, 1463–1476. doi: 10.1007/s12298-020-00832-z
Ghori, N. H., Ghori, T., Hayat, M. Q., Imadi, S. R., Gul, A., Altay, V., et al. (2019). Heavy metal stress and responses in plants. Int. J. Environ. Sci. Technol. 16, 1807–1828. doi: 10.1007/s13762-019-02215-8
Gong, Z. Z., Xiong, L. M., Shi, H. Z., Yang, S. H., Herrera-Estrella, L. R., Xu, G. H., et al. (2020). Plant abiotic stress response and nutrient use efficiency. Sci. China-Life. Sci. 63, 635–674. doi: 10.1007/s11427-020-1683-x
Guan, S. X., Xu, Q., Ma, D. R., Zhang, W. Z., Xu, Z. J., Zhao, M. H., et al. (2019). Transcriptomics profiling in response to cold stress in cultivated rice and weedy rice. Gene 685, 96–105. doi: 10.1016/j.gene.2018.10.066
Guo, R., Shi, L. X., Jiao, Y., Li, M. X., Zhong, X. L., Gu, F. X., et al. (2018). Metabolic responses to drought stress in the tissues of drought-tolerant and drought-sensitive wheat genotype seedlings. AoB. Plants 10, ply016. doi: 10.1093/aobpla/ply016
Han, D. G., Han, J. X., Yang, G. H., Wang, S., Xu, T. L., Li, W. H. (2020). An ERF transcription factor gene from malus baccata (L.) borkh, MbERF11, affects cold and salt stress tolerance in arabidopsis. Forests 11, 514. doi: 10.3390/f11050514
Han, Y. C., Kuang, J. F., Chen, J. Y., Liu, X. C., Xiao, Y. Y., Fu, C. C., et al. (2016). Banana transcription factor MaERF11 recruits histone deacetylase MaHDA1 and represses the expression of MaACO1 and expansins during fruit ripening. Plant Physiol. 171, 1070–1084. doi: 10.1104/pp.16.00301
Hao, D. Y., Ohme-Takagi, M., Sarai, A. (1998). Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plant. J. Biol. Chem. 273, 26857–26861. doi: 10.1074/jbc.273.41.26857
Hao, L. D., Shi, S. B., Guo, H. B., Li, M., Hu, P., Wei, Y. D., et al. (2020). Genome-wide identification and expression profiles of ERF subfamily transcription factors in zea mays. PeerJ 8, e9551. doi: 10.7717/peerj.9551
Hasanuzzaman, M., Nahar, K., Alam, M. M., Roychowdhury, R., Fujita, M. (2013a). Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 14, 9643–9684. doi: 10.3390/ijms14059643
Hasanuzzaman, M., Nahar, K., Fujita, M. (2013b). Extreme temperatures, oxidative stress and antioxidant defense in plants. Abiotic. Stress—Plant. Responses. Appl. Agricult. pp, 169–205. doi: 10.5772/54833
He, S., Hao, X., He, S., Hao, X., Zhang, P., Chen, X. (2021). Genome-wide identification, phylogeny and expression analysis of AP2/ERF transcription factors family in sweet potato. BMC Genomics 22, 748. doi: 10.1186/s12864-021-08043-w
Hiratsu, K., Matsui, K., Koyama, T., Ohme-Takagi, M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in arabidopsis. Plant J. 34, 733–739. doi: 10.1046/j.1365-313x.2003.01759.x
Hong, Y. B., Wang, H., Gao, Y. Z., Bi, Y., Xiong, X. H., Yan, Y. Q., et al. (2022). ERF transcription factor OsBIERF3 positively contributes to immunity against fungal and bacterial diseases but negatively regulates cold tolerance in rice. Int. J. Mol. Sci. 23, 606. doi: 10.3390/ijms23020606
Huang, S., Ma, Z., Hu, L., Huang, K., Zhang, M., Zhang, S., et al. (2021). Involvement of rice transcription factor OsERF19 in response to ABA and salt stress responses. Plant Physiol. Biochem. 167, 22–30. doi: 10.1016/j.plaphy.2021.07.027
Huang, J. Y., Zhao, X. B., Burger, M., Wang, Y. R., Chory, J. (2021). Two interacting ethylene response factors regulate heat stress response. Plant Cell. 33, 338–357. doi: 10.1093/plcell/koaa026
Hu, Z. R., Huang, X. B., Amombo, E., Liu, A., Fan, J. B., Bi, A. Y., et al. (2020). The ethylene responsive factor CdERF1 from bermudagrass (Cynodon dactylon) positively regulates cold tolerance. Plant Sci. 294, 110432. doi: 10.1016/j.plantsci.2020.110432
Janicka-Russak, M., Kabala, K., Burzynski, M., Klobus, G. (2008). Response of plasma membrane h+-ATPase to heavy metal stress in cucumis sativus roots. J. Exp. Bot. 59, 3721–3728. doi: 10.1093/jxb/ern219
Jeon, J. S., Rybka, D., Carreno-Quintero, N., De Vos, R., Raaijmakers, J. M., Etalo, D. W. (2022). Metabolic signatures of rhizobacteria-induced plant growth promotion. Plant Cell Environ. 45, 3086–3099. doi: 10.1111/pce.14385
Jha, U. C., Nayyar, H., Siddique, K. H. M. (2022). Role of phytohormones in regulating heat stress acclimation in agricultural crops. J. Plant Growth Regul. 41, 1041–1064. doi: 10.1007/s00344-021-10362-x
Jiang, M., Ye, Z. H., Zhang, H. J., Miao, L. X. (2019). Broccoli plants over-expressing an ERF transcription factor gene BoERF1 facilitates both salt stress and sclerotinia stem rot resistance. J. Plant Growth Regul. 38, 1–13. doi: 10.1007/s00344-018-9799-6
Jiang, W. B., Zhang, X. J., Song, X. W., Yang, J. F., Pang, Y. Z. (2020). Genome-wide identification and characterization of APETALA2/Ethylene-responsive element binding factor superfamily genes in soybean seed development. Front. Plant Sci. 11, 566647. doi: 10.3389/fpls.2020.566647
Jin, J. J., Zhang, H., Zhang, J. F., Liu, P. P., Chen, X., Li, Z. F., et al. (2017). Integrated transcriptomics and metabolomics analysis to characterize cold stress responses in nicotiana tabacum. BMC Genomics 18, 496. doi: 10.1186/s12864-017-3871-7
Jung, S. E., Bang, S. W., Kim, S. H., Seo, J. S., Yoon, H. B., Kim, Y. S., et al. (2021). Overexpression of OsERF83, a vascular tissue-specific transcription factor gene, confers drought tolerance in rice. Int. J. Mol. Sci. 22, 7656. doi: 10.3390/ijms22147656
Khan, M., Hu, J. B., Dahro, B., Ming, R. H., Zhang, Y., Wang, Y., et al. (2021). ERF108 from poncirus trifoliata (L.) raf. functions in cold tolerance by modulating raffinose synthesis through transcriptional regulation of PtrRafS. Plant J. 108, 705–724. doi: 10.1111/tpj.15465
Kidokoro, S., Kim, J.-S., Ishikawa, T., Suzuki, T., Shinozaki, K., Yamaguchi-Shinozaki, K. (2020). DREB1A/CBF3 is repressed by transgene-induced DNA methylation in the arabidopsis ice1-1 mutant. Plant Cell. 32, 1035–1048. doi: 10.1105/tpc.19.00532
Kimotho, R. N., Baillo, E. H., Zhang, Z. (2019). Transcription factors involved in abiotic stress responses in maize (Zea mays l.) and their roles in enhanced productivity in the post genomics era. PeerJ 7, e7211. doi: 10.7717/peerj.7211
Kirschner, G. K. (2022). Flavonoids make buckwheat a superfood – new insights into their biosynthesis. Plant J. 111, 321–322. doi: 10.1111/tpj.15800
Klay, I., Gouia, S., Liu, M., Mila, I., Khoudi, H., Bernadac, A., et al. (2018). Ethylene response factors (ERF) are differentially regulated by different abiotic stress types in tomato plants. Plant Sci. 274, 137–145. doi: 10.1016/j.plantsci.2018.05.023
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lee, J., Eschen-Lippold, L., Lassowskat, I., Bottcher, C., Scheel, D. (2015). Cellular reprogramming through mitogen-activated protein kinases. Front. Plant Sci. 6, 940. doi: 10.3389/fpls.2015.00940
Lee, D. K., Jung, H., Jang, G., Jeong, J. S., Kim, Y. S., Ha, S. H., et al. (2016). Overexpression of the OsERF71 transcription factor alters rice root structure and drought resistance. Plant Physiol. 172, 575–588. doi: 10.1104/pp.16.00379
Lee, E. S., Park, J. H., Wi, S. D., Kang, C. H., Chi, Y. H., Chae, H. B., et al. (2021). Redox-dependent structural switch and CBF activation confer freezing tolerance in plants. Nat. Plants. 7, 914–922. doi: 10.1038/s41477-021-00944-8
Li, P. C., Cao, W., Fang, H. M., Xu, S. H., Yin, S. Y., Zhang, Y. Y., et al. (2017). Transcriptomic profiling of the maize (Zea mays l.) leaf response to abiotic stresses at the seedling stage. Front. Plant Sci. 8, 290. doi: 10.3389/fpls.2017.00290
Licausi, F., Kosmacz, M., Weits, D., Giuntoli, B., Giorgi, F., Voesenek, L., et al. (2011). Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479, 419–22. doi: 10.1038/nature10536
Lievens, L., Pollier, J., Goossens, A., Beyaert, R., Staal, J. (2017). Abscisic acid as pathogen effector and immune regulator. Front. Plant Sci. 8, 587. doi: 10.3389/fpls.2017.00587
Li, W. L., Fu, Y. R., Lv, W. Q., Zhao, S. C., Feng, H., Shao, L. Y., et al. (2022). Characterization of the early gene expression profile in populus ussuriensis under cold stress using PacBio SMRT sequencing integrated with RNA-seq reads. Tree Physiol. 42, 646–663. doi: 10.1093/treephys/tpab130
Li, J. J., Guo, X., Zhang, M. H., Wang, X., Zhao, Y., Yin, Z. G., et al. (2018). OsERF71 confers drought tolerance via modulating ABA signaling and proline biosynthesis. Plant Sci. 270, 131–139. doi: 10.1016/j.plantsci.2018.01.017
Li, Z. X., Howell, S. H. (2021). Heat stress responses and thermotolerance in maize. Int. J. Mol. Sci. 22, 948. doi: 10.3390/ijms22020948
Li, T., Jiang, Z. Y., Zhang, L. C., Tan, D. M., Wei, Y., Yuan, H., et al. (2016). Apple (Malus domestica) MdERF2 negatively affects ethylene biosynthesis during fruit ripening by suppressing MdACS1 transcription. Plant J. 88, 735–748. doi: 10.1111/tpj.13289
Li, L., Li, X. G., Yang, C., Cheng, Y. B., Cai, Z. D., Nian, H., et al. (2022). GsERF1 enhances arabidopsis thaliana aluminum tolerance through an ethylene-mediated pathway. BMC Plant Biol. 22, 258. doi: 10.1186/s12870-022-03625-6
Li, Z., Tian, Y., Xu, J., Fu, X., Gao, J., Wang, B., et al. (2018). A tomato ERF transcription factor, SlERF84, confers enhanced tolerance to drought and salt stress but negatively regulates immunity against pseudomonas syringae pv. tomato DC3000. Plant Physiol. Biochem. 132, 683–695. doi: 10.1016/j.plaphy.2018.08.022
Liu, M. C., Chen, Y., Chen, Y., Shin, J. H., Mila, I., Audran, C., et al. (2018). The tomato ethylene response factor sl-ERF.B3 integrates ethylene and auxin signaling via direct regulation of sl-Aux/IAA27. New Phytol. 219, 631–640. doi: 10.1111/nph.15165
Liu, D., Chen, X., Liu, J., Ye, J., Guo, Z. (2012). The rice ERF transcription factor OsERF922 negatively regulates resistance to magnaporthe oryzae and salt tolerance. J. Exp. Bot. 63, 3899–3911. doi: 10.1093/jxb/ers079
Liu, J. X., Wu, B., Feng, K., Li, M. Y., Duan, A. Q., Shen, D., et al. (2021). A celery transcriptional repressor AgERF8 negatively modulates abscisic acid and salt tolerance. Mol. Genet. Genomics 296, 179–192. doi: 10.1007/s00438-020-01738-x
Liu, X., Yang, X. X., Zhang, B. (2021). Transcriptome analysis and functional identification of GmMYB46 in soybean seedlings under salt stress. PeerJ 9, e12492. doi: 10.7717/peerj.12492
Liu, Q., Zhang, G., Chen, S. (2001). Structure and regulatory function of plant transcription factors. Chin. Sci. Bull. 46, 271–278. doi: 10.1007/BF03187184
Liu, Y. D., Zhang, L., Chen, L. J., Pang, S. Q., Zheng, Q., Quan, S. W., et al. (2020). Genome-wide identification, expression and potential function analysis of the ERF and DREB subfamily members in tomato. Res. Square. 580, 1303–1308. doi: 10.21203/rs.3.rs-126159/v1
Liu, Y., Zhao, T. J., Liu, J. M., Liu, W. Q., Liu, Q., Yan, Y. B., et al. (2006). The conserved Ala37 in the ERF/AP2 domain is essential for binding with the DRE element and the GCC box. FEBS Lett. 580, 1303–1308. doi: 10.1016/j.febslet.2006.01.048
Li, W. Y., Wang, C., Shi, H. H., Wang, B., Wang, J. X., Liu, Y. S., et al. (2020). Genome-wide analysis of ethylene-response factor family in adzuki bean and functional determination of VaERF3 under saline-alkaline stress. Plant Physiol. Biochem. 147, 215–222. doi: 10.1016/j.plaphy.2019.12.019
Li, T., Wu, Z., Xiang, J., Zhang, D. H., Teng, N. J. (2022). Overexpression of a novel heat-inducible ethylene-responsive factor gene LlERF110 from lilium longiflorum decreases thermotolerance. Plant Sci. 319, 111246. doi: 10.1016/j.plantsci.2022.111246
Lourenco, T. F., Serra, T. S., Cordeiro, A. M., Swanson, S. J., Gilroy, S., Saibo, N. J., et al. (2015). The rice E3-ubiquitin ligase HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 modulates the expression of ROOT MEANDER CURLING, a gene involved in root mechanosensing, through the interaction with two ETHYLENE-RESPONSE FACTOR transcription factors. Plant Physiol. 169, 2275–2287. doi: 10.1104/pp.15.01131
Lu, J. C., Wang, L. H., Zhang, Q. Q., Ma, C. X., Su, X. F., Cheng, H. M., et al. (2022). AmCBF1 transcription factor regulates plant architecture by repressing GhPP2C1 or GhPP2C2 in gossypium hirsutum. Front. Plant Sci. 13, 914206. doi: 10.3389/fpls.2022.914206
Lv, K. W., Li, J., Zhao, K., Chen, S., Nie, J., Zhang, W. L., et al. (2019). Overexpression of an AP2/ERF family gene, BpERF13, in birch enhances cold tolerance through upregulating CBF genes and mitigating reactive oxygen species. Plant Sci. 292, 110375. doi: 10.1016/j.plantsci.2019.110375
Magar, M. M., Liu, H., Yan, G. (2022). Genome-wide analysis of AP2/ERF superfamily genes in contrasting wheat genotypes reveals heat stress-related candidate genes. Front. Plant Sci. 13, 853086. doi: 10.3389/fpls.2022.853086
Magnani, E., Sjolander, K., Hake, S. (2004). From endonucleases to transcription factors: evolution of the AP2 DNA binding domain in plants. Plant Cell. 16, 2265–2277. doi: 10.1105/tpc.104.023135
Mahmood, T., Khalid, S., Abdullah, M., Ahmed, Z., Shah, M. K. N., Ghafoor, A., et al. (2020). Insights into drought stress signaling in plants and the molecular genetic basis of cotton drought tolerance. Cells 9, 105. doi: 10.3390/cells9010105
Mahon, E. L., de Vries, L., Jang, S. K., Middar, S., Kim, H., Unda, F., et al. (2022). Exogenous chalcone synthase expression in developing poplar xylem incorporates naringenin into lignins. Plant Physiol. 188, 984–996. doi: 10.1093/plphys/kiab499
Mao, L. Z., Deng, M. H., Jiang, S. R., Zhu, H. S., Yang, Z. G., Yue, Y. L., et al. (2020). Characterization of the DREBA4-type transcription factor (SlDREBA4), which contributes to heat tolerance in tomatoes. Front. Plant Sci. 11, 554520. doi: 10.3389/fpls.2020.554520
Matsukura, S., Mizoi, J., Yoshida, T., Todaka, D., Ito, Y., Maruyama, K., et al. (2010). Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol. Genet. Genomics 283, 185–196. doi: 10.1007/s00438-009-0506-y
Ma, H. Y., Yang, T., Li, Y., Zhang, J., Wu, T., Song, T. T., et al. (2021). The long noncoding RNA MdLNC499 bridges MdWRKY1 and MdERF109 function to regulate early-stage light-induced anthocyanin accumulation in apple fruit. Plant Cell. 33, 3309–3330. doi: 10.1093/plcell/koab188
Meng, X., Xu, J., He, Y., Yang, K. Y., Mordorski, B., Liu, Y., et al. (2013). Phosphorylation of an ERF transcription factor by arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell. 25, 1126–1142. doi: 10.1105/tpc.112.109074
Miller, G., Suzuki, N., Ciftci-Yilmaz, S., Mittler, R. (2010). Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 33, 453–467. doi: 10.1111/j.1365-3040.2009.02041.x
Mohanty, B. (2021). Promoter architecture and transcriptional regulation of genes upregulated in germination and coleoptile elongation of diverse rice genotypes tolerant to submergence. Front. Genet. 12, 639654. doi: 10.3389/fgene.2021.639654
Morales, M., Munne-Bosch, S. (2019). Malondialdehyde: Facts and artifacts. Plant Physiol. 180, 1246–1250. doi: 10.1104/pp.19.00405
Muller, M., Munne-Bosch, S. (2015). Ethylene response factors: A key regulatory hub in hormone and stress signaling. Plant Physiol. 169, 32–41. doi: 10.1104/pp.15.00677
Nakano, T., Suzuki, K., Fujimura, T., Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in arabidopsis and rice. Plant Physiol. 140, 411–432. doi: 10.1104/pp.105.073783
Nath, M., Tuteja, N. (2016). NPKS uptake, sensing, and signaling and miRNAs in plant nutrient stress. Protoplasma 253, 767–786. doi: 10.1007/s00709-015-0845-y
Nauser, T., Gebicki, J. M. (2019). Fast reaction of carbon free radicals with flavonoids and other aromatic compounds. Arch. Biochem. Biophys. 674, 108107. doi: 10.1016/j.abb.2019.108107
Ni, J. B., Bai, S. L., Zhao, Y., Qian, M. J., Tao, R. Y., Yin, L., et al. (2019). Ethylene response factors Pp4ERF24 and Pp12ERF96 regulate blue light-induced anthocyanin biosynthesis in ‘Red zaosu’ pear fruits by interacting with MYB114. Plant Mol. Biol. 99, 67–78. doi: 10.1007/s11103-018-0802-1
Ni, J. B., Premathilake, A. T., Gao, Y. H., Yu, W. J., Tao, R. Y., Teng, Y. W., et al. (2021). Ethylene-activated PpERF105 induces the expression of the repressor-type R2R3-MYB gene PpMYB140 to inhibit anthocyanin biosynthesis in red pear fruit. Plant J. 105, 167–181. doi: 10.1111/tpj.15049
Ohme-Takagi, M., Shinshi, H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell. 7, 173–182. doi: 10.1105/tpc.7.2.173
Ohta, M., Matsui, K., Hiratsu, K., Shinshi, H., Ohme-Takagi, M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell. 13, 1959–1968. doi: 10.1105/tpc.010127
Okamuro, J. K., Caster, B., Villarroel, R., Montagu, M. V., Jofuku, K. D. (1997). The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. United. States America. 94, 7076–7081. doi: 10.1073/pnas.94.13.7076
Owji, H., Hajiebrahimi, A., Seradj, H., Hemmati, S. (2017). Identification and functional prediction of stress responsive AP2/ERF transcription factors in brassica napus by genome-wide analysis. Comput. Biol. Chem. 71, 32–56. doi: 10.1016/j.compbiolchem.2017.09.004
Park, S. I., Kwon, H. J., Cho, M. H., Song, J. S., Kim, B. G., Baek, J., et al. (2021). The OsERF115/AP2EREBP110 transcription factor is involved in the multiple stress tolerance to heat and drought in rice plants. Int. J. Mol. Sci. 22, 7181. doi: 10.3390/ijms22137181
Park, H. C., Park, B. O., Kim, H. S., Kim, S. H., Lee, S. W., Chung, W. S. (2021). AtMPK6-induced phosphorylation of AtERF72 enhances its DNA binding activity and interaction with TGA4/OBF4 in arabidopsis. Plant Biol. (Stuttg). 23, 11–20. doi: 10.1111/plb.13196
Park, J. M., Park, C. J., Lee, S. B., Ham, B. K., Shin, R., Paek, K. H. (2001). Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell. 13, 1035–1046. doi: 10.1105/tpc.13.5.1035
Pfluger, J., Wagner, D. (2007). Histone modifications and dynamic regulation of genome accessibility in plants. Curr. Opin. Plant Biol. 10, 645–652. doi: 10.1016/j.pbi.2007.07.013
Pi, E. X., Qu, L. Q., Hu, J. W., Huang, Y. Y., Qiu, L. J., Lu, H., et al. (2016). Mechanisms of soybean roots’ tolerances to salinity revealed by proteomic and phosphoproteomic comparisons between two cultivars. Mol. Cell. Proteomics. 15, 266–288. doi: 10.1074/mcp.M115.051961
Pi, E. X., Xu, J., Li, H. H., Fan, W., Zhu, C. M., Zhang, T. Y., et al. (2019). Enhanced salt tolerance of rhizobia-inoculated soybean correlates with decreased phosphorylation of the transcription factor GmMYB183 and altered flavonoid biosynthesis. Mol. Cell. Proteomics. 18, 2225–2243. doi: 10.1074/mcp.RA119.001704
Pi, E. X., Zhu, C. M., Fan, W., Huang, Y. Y., Qu, L. Q., Li, Y. Y., et al. (2018). Quantitative phosphoproteomic and metabolomic analyses reveal GmMYB173 optimizes flavonoid metabolism in soybean under salt stress. Mol. Cell. Proteomics. 17, 1209–1224. doi: 10.1074/mcp.RA117.000417
Pre, M., Atallah, M., Champion, A., De Vos, M., Pieterse, C. M., Memelink, J. (2008). The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 147, 1347–1357. doi: 10.1104/pp.108.117523
Priya, M., Dhanker, O. P., Siddique, K. H. M., HanumanthaRao, B., Nair, R. M., Pandey, S., et al. (2019). Drought and heat stress-related proteins: an update about their functional relevance in imparting stress tolerance in agricultural crops. Theor. Appl. Genet. 132, 1607–1638. doi: 10.1007/s00122-019-03331-2
Qin, F., Kakimoto, M., Sakuma, Y., Maruyama, K., Osakabe, Y., Tran, L. S. P., et al. (2007). Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in zea mays l. Plant J. 50, 54–69. doi: 10.1111/j.1365-313X.2007.03034.x
Qin, F., Sakuma, Y., Li, J., Liu, Q., Li, Y. Q., Shinozaki, K., et al. (2004). Cloning and functional analysis of a novel DREB1/CBF transcription factor involved in cold-responsive gene expression in zea mays l. Plant Cell Physiol. 45, 1042–1052. doi: 10.1093/pcp/pch118
Qi, Y., Yang, Z., Sun, X., He, H., Guo, L., Zhou, J., et al. (2022). Heterologous overexpression of StERF3 triggers cell death in nicotiana benthamiana. Plant Sci. 315, 111149. doi: 10.1016/j.plantsci.2021.111149
Quan, R. D., Hu, S. J., Zhang, Z. L., Zhang, H. W., Zhang, Z. J., Huang, R. F. (2010). Overexpression of an ERF transcription factor TSRF1 improves rice drought tolerance. Plant Biotechnol. J. 8, 476–488. doi: 10.1111/j.1467-7652.2009.00492.x
Razi, K., Muneer, S. (2021). Drought stress-induced physiological mechanisms, signaling pathways and molecular response of chloroplasts in common vegetable crops. Crit. Rev. Biotechnol. 41, 669–691. doi: 10.1080/07388551.2021.1874280
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C., Keddie, J., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. doi: 10.1126/science.290.5499.2105
Rong, W., Qi, L., Wang, A. Y., Ye, X. G., Du, L. P., Liang, H. X., et al. (2014). The ERF transcription factor TaERF3 promotes tolerance to salt and drought stresses in wheat. Plant Biotechnol. J. 12, 468–479. doi: 10.1111/pbi.12153
Sakuma, Y., Liu, Q., Dubouzet, J. G., Abe, H., Shinozaki, K., Yamaguchi-Shinozaki, K. (2002). DNA-Binding specificity of the ERF/AP2 domain of arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009. doi: 10.1006/bbrc.2001.6299
Sanchez-Calderon, L., Lopez-Bucio, J., Chacon-Lopez, A., Cruz-Ramirez, A., Nieto-Jacobo, F., Dubrovsky, J. G., et al. (2005). Phosphate starvation induces a determinate developmental program in the roots of arabidopsis thaliana. Plant Cell Physiol. 46, 174–184. doi: 10.1093/pcp/pci011
Sanchez-Calderon, L., Lopez-Bucio, J., Chacon-Lopez, A., Gutierrez-Ortega, A., Hernandez-Abreu, E., Herrera-Estrella, L. (2006). Characterization of low phosphorus insensitive mutants reveals a crosstalk between low phosphorus-induced determinate root development and the activation of genes involved in the adaptation of arabidopsis to phosphorus deficiency. Plant Physiol. 140, 879–889. doi: 10.1104/pp.105.073825
Shaikhali, J., Heiber, I., Seidel, T., Stroher, E., Hiltscher, H., Birkmann, S., et al. (2008). The redox-sensitive transcription factor Rap2.4a controls nuclear expression of 2-cys peroxiredoxin a and other chloroplast antioxidant enzymes. BMC Plant Biol. 8, 48. doi: 10.1186/1471-2229-8-48
Shankar, R., Bhattacharjee, A., Jain, M. (2016). Transcriptome analysis in different rice cultivars provides novel insights into desiccation and salinity stress responses. Sci. Rep. 6, 23719. doi: 10.1038/srep23719
Shao, W. J., Ao, T., Lang, M. L. (2020). Research advances on the mechanism of AP2/ERF transcriptional factors in response to abiotic stresses in plants. Mol. Plant Breeding. 18, 4981–4988. doi: 10.13271/j.mpb.018.004981
Sharma, S. S., Dietz, K. J. (2009). The relationship between metal toxicity and cellular redox imbalance. Trends Plant Sci. 14, 43–50. doi: 10.1016/j.tplants.2008.10.007
Sharma, R., Singh, G., Bhattacharya, S., Singh, A. (2018). Comparative transcriptome meta-analysis of arabidopsis thaliana under drought and cold stress. PloS One 13, e0203266. doi: 10.1371/journal.pone.0203266
Sharmin, R. A., Bhuiyan, M. R., Lv, W. H., Yu, Z. P., Chang, F. G., Kong, J. J., et al. (2020). RNA-Seq based transcriptomic analysis revealed genes associated with seed-flooding tolerance in wild soybean (Glycine soja sieb. & zucc.). Environ. Exp. Bot. 171, 103906. doi: 10.1016/j.envexpbot.2019.103906
Speisky, H., Shahidi, F., de Camargo, A. C., Fuentes, J. (2022). Revisiting the oxidation of flavonoids: Loss, conservation or enhancement of their antioxidant properties. Antioxidants 11, 133. doi: 10.3390/antiox11010133
Stockinger, E. J., Gilmour, S. J., Thomashow, M. F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the c-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. U.S.A. 94, 1035–1040. doi: 10.1073/pnas.94.3.1035
Sun, Y., Li, J. Q., Yan, J. Y., Yuan, J. J., Li, G. X., Wu, Y. R., et al. (2020). Ethylene promotes seed iron storage during arabidopsis seed maturation via ERF95 transcription factor. J. Integr. Plant Biol. 62, 1193–1212. doi: 10.1111/jipb.12986
Sun, M. Z., Shen, Y., Chen, Y., Wang, Y., Cai, X. X., Yang, J. K., et al. (2022). Osa-miR1320 targets the ERF transcription factor OsERF096 to regulate cold tolerance via JA-mediated signaling. Plant Physiol. 189, 2500–2516. doi: 10.1093/plphys/kiac208
Sun, S., Yu, J. P., Chen, F., Zhao, T. J., Fang, X. H., Li, Y. Q., et al. (2008). TINY, a dehydration-responsive element (DRE)-binding protein-like transcription factor connecting the DRE- and ethylene-responsive element-mediated signaling pathways in arabidopsis. J. Biol. Chem. 283, 6261–6271. doi: 10.1074/jbc.M706800200
Sun, X. M., Zhang, L. L., Wong, D. C. J., Wang, Y., Zhu, Z. F., Xu, G. Z., et al. (2019). The ethylene response factor VaERF092 from amur grape regulates the transcription factor VaWRKY33, improving cold tolerance. Plant J. 99, 988–1002. doi: 10.1111/tpj.14378
Sun, X. M., Zhu, Z. F., Zhang, L. L., Fang, L. C., Zhang, J. S., Wang, Q. F., et al. (2018). Overexpression of ethylene response factors VaERF080 and VaERF087 from vitis amurensis enhances cold tolerance in arabidopsis. Sci. Horticult. 243, 320–326. doi: 10.1016/j.scienta.2018.08.055
Svistoonoff, S., Creff, A., Reymond, M., Sigoillot-Claude, C., Ricaud, L., Blanchet, A., et al. (2007). Root tip contact with low-phosphate media reprograms plant root architecture. Nat. Genet. 39, 792–796. doi: 10.1038/ng2041
Takahashi, F., Kuromori, T., Urano, K., Kazuko, Y.-S., Kazuo, S. (2020). Drought stress responses and resistance in plants: From cellular responses to long-distance intercellular communication. Front. Plant Sci. 11, 556972. doi: 10.3389/fpls.2020.556972
Tan, X. L., Fan, Z. Q., Shan, W., Yin, X. R., Kuang, J. F., Lu, W. J., et al. (2018). Association of BrERF72 with methyl jasmonate-induced leaf senescence of Chinese flowering cabbage through activating JA biosynthesis-related genes. Horticult. Res. 5, 22. doi: 10.1038/s41438-018-0028-z
Thirugnanasambantham, K., Durairaj, S., Saravanan, S., Karikalan, K., Muralidaran, S., Islam, V. I. H. (2015). Role of ethylene response transcription factor (ERF) and its regulation in response to stress encountered by plants. Plant Mol. Biol. Reporter. 33, 347–357. doi: 10.1007/s11105-014-0799-9
Tiwari, S. B., Belachew, A., Ma, S. F., Young, M., Ade, J., Shen, Y., et al. (2012). The EDLL motif: a potent plant transcriptional activation domain from AP2/ERF transcription factors. Plant J. 70, 855–865. doi: 10.1111/j.1365-313X.2012.04935.x
Tiwari, S., Lata, C. (2018). Heavy metal stress, signaling, and tolerance due to plant-associated microbes: An overview. Front. Plant Sci. 9, 452. doi: 10.3389/fpls.2018.00452
van Dongen, J. T., Licausi, F. (2015). Oxygen sensing and signaling. Annu. Rev. Plant Biol. 66, 345–367. doi: 10.1146/annurev-arplant-043014-114813
Wang, C. L., Deng, Y. Z., Liu, Z. S., Liao, W. B. (2021). Hydrogen sulfide in plants: Crosstalk with other signal molecules in response to abiotic stresses. Int. J. Mol. Sci. 22, 12068. doi: 10.3390/ijms222112068
Wang, J., Nan, N., Shi, L., Li, N., Huang, S., Zhang, A., et al. (2020). Arabidopsis BRCA1 represses RRTF1-mediated ROS production and ROS-responsive gene expression under dehydration stress. New Phytol. 228, 1591–1610. doi: 10.1111/nph.16786
Wang, H. T., Ni, D. Q., Shen, J. C., Deng, S. S., Xuan, H. D., Wang, C. C., et al. (2022). Genome-wide identification of the AP2/ERF gene family and functional analysis of GmAP2/ERF144 for drought tolerance in soybean. Front. Plant Sci. 13, 848766. doi: 10.3389/fpls.2022.848766
Wang, D., Sun, Z. M., Hu, X. X., Xiong, J. B., Hu, L. Z., Xu, Y. D., et al. (2021). The key regulator LcERF056 enhances salt tolerance by modulating reactive oxygen species-related genes in lotus corniculatus. BMC Plant Biol. 21, 605. doi: 10.1186/s12870-021-03336-4
Wang, S. J., Zhou, B. R., Yao, W. J., Jiang, T. B. (2018). PsnERF75 transcription factor from populus simonii × p. nigra confers salt tolerance in transgenic arabidopsis. J. Plant Biol. 61, 61–71. doi: 10.1007/s12374-017-0450-z
Wei, C., Li, M., Cao, X., Jin, Z., Zhang, C., Xu, M., et al. (2022). Linalool synthesis related PpTPS1 and PpTPS3 are activated by transcription factor PpERF61 whose expression is associated with DNA methylation during peach fruit ripening. Plant Sci 317, 111–200. doi: 10.1016/j.plantsci
Welsch, R., Maass, D., Voegel, T., Dellapenna, D., Beyer, P. (2007). Transcription factor RAP2.2 and its interacting partner SINAT2: stable elements in the carotenogenesis of arabidopsis leaves. Plant Physiol. 145, 1073–1085. doi: 10.1104/pp.107.104828
White, P. J., Brown, P. H. (2010). Plant nutrition for sustainable development and global health. Ann. Bot. 105, 1073–1080. doi: 10.1093/aob/mcq085
Wu, J., Folta, K. M., Xie, Y. F., Jiang, W. M., Lu, J., Zhang, Y. L. (2017). Overexpression of muscadinia rotundifolia CBF2 gene enhances biotic and abiotic stress tolerance in arabidopsis. Protoplasma 254, 239–251. doi: 10.1007/s00709-015-0939-6
Wu, T., Liu, H. T., Zhao, G. P., Song, J. X., Wang, X. L., Yang, C. Q., et al. (2020). Jasmonate and ethylene-regulated ethylene response factor 22 promotes lanolin-induced anthocyanin biosynthesis in ‘Zaosu’ pear (Pyrus bretschneideri rehd.) fruit. Biomolecules 10, 278. doi: 10.3390/biom10020278
Wu, L. J., Zhang, Z. J., Zhang, H. W., Wang, X., Huang, R. F. (2008). Transcriptional modulation of ethylene response factor protein JERF3 in the oxidative stress response enhances tolerance of tobacco seedlings to salt, drought, and freezing. Plant Physiol. 148, 1953–1963. doi: 10.1104/pp.108.126813
Xie, Z., Nolan, T., Jiang, H., Tang, B., Zhang, M., Li, Z., et al. (2019a). The AP2/ERF transcription factor TINY modulates brassinosteroid-regulated plant growth and drought responses in arabidopsis. Plant Cell. 31, 1788–1806. doi: 10.1105/tpc.18.00918
Xie, Z., Nolan, T. M., Jiang, H., Yin, Y. H. (2019b). AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in arabidopsis. Front. Plant Sci. 10, 228. doi: 10.3389/fpls.2019.00228
Xing, L. P., Di, Z. C., Yang, W. W., Liu, J. Q., Li, M. N., Wang, X. J., et al. (2017). Overexpression of ERF1-V from haynaldia villosa can enhance the resistance of wheat to powdery mildew and increase the tolerance to salt and drought stresses. Front. Plant Sci. 8, 1948. doi: 10.3389/fpls.2017.01948
Xue, G. P., Loveridge, C. W. (2004). HvDRF1 is involved in abscisic acid-mediated gene regulation in barley and produces two forms of AP2 transcriptional activators, interacting preferably with a CT-rich element. Plant J. 37, 326–339. doi: 10.1046/j.1365-313x.2003.01963.x
Xu, S. Q., Yao, S. C., Huang, R. S., Tan, Y., Huang, D. (2020). Transcriptome-wide analysis of the AP2/ERF transcription factor gene family involved in the regulation of gypenoside biosynthesis in gynostemma pentaphyllum. Plant Physiol. Biochem. 154, 238–247. doi: 10.1016/j.plaphy.2020.05.040
Yamaguchi-Shinozaki, K., Shinozaki, K. (1994). A novel cis-acting element in an arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. 6, 251–264. doi: 10.1105/tpc.6.2.251
Yang, Y. Q., Guo, Y. (2018). Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 217, 523–539. doi: 10.1111/nph.14920
Yang, C. L., Huang, Y. T., Schmidt, W., Klein, P., Chan, M. T., Pan, I. C. (2022). Ethylene response Factor109 attunes immunity, photosynthesis, and iron homeostasis in arabidopsis leaves. Front. Plant Sci. 13, 841366. doi: 10.3389/fpls.2022.841366
Yang, R. C., Liu, J., Lin, Z., Sun, W., Zhang, Y. Q. (2018). ERF transcription factors involved in salt response in tomato. Plant Growth Regul. 84, 1–10. doi: 10.1007/s10725-017-0362-4
Yang, X. Y., Lu, M. Q., Wang, Y. F., Wang, Y. R., Liu, Z. J., Chen, S. (2021). Response mechanism of plants to drought stress. Horticulturae 7, 50. doi: 10.3390/horticulturae7030050
Yang, G. Y., Peng, S. B., Wang, T. Y., Gao, X. Q., Li, D. P., Li, M. G., et al. (2021). Walnut ethylene response factor JrERF2-2 interact with JrWRKY7 to regulate the GSTs in plant drought tolerance. Ecotoxicol. Environ. Saf. 228, 112945. doi: 10.1016/j.ecoenv.2021.112945
Yang, H. H., Sun, Y. G., Wang, H. X., Zhao, T. T., Xu, X. Y., Jiang, J. B., et al. (2021). Genome-wide identification and functional analysis of the ERF2 gene family in response to disease resistance against stemphylium lycopersici in tomato. BMC Plant Biol. 21, 72. doi: 10.1186/s12870-021-02848-3
Yang, S., Wang, S., Liu, X., Yu, Y., Yue, L., Wang, X., et al. (2009). Four divergent arabidopsis ethylene-responsive element-binding factor domains bind to a target DNA motif with a universal CG step core recognition and different flanking bases preference. FEBS J. 276, 7177–7186. doi: 10.1111/j.1742-4658.2009.07428.x
Yao, Y., He, R. J., Xie, Q. L., Zhao, X. H., Deng, X. M., He, J. B., et al. (2017). ETHYLENE RESPONSE FACTOR 74 (ERF74) plays an essential role in controlling a respiratory burst oxidase homolog d (RbohD)-dependent mechanism in response to different stresses in arabidopsis. New Phytol. 213, 1667–1681. doi: 10.1111/nph.14278
Yao, G. F., Ming, M. L., Allan, A. C., Gu, C., Li, L. T., Wu, X., et al. (2017). Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J. 92, 437–451. doi: 10.1111/tpj.13666
Yi, S. Y., Kim, J. H., Joung, Y. H., Lee, S., Kim, W. T., Yu, S. H., et al. (2004). The pepper transcription factor CaPF1 confers pathogen and freezing tolerance in arabidopsis. Plant Physiol. 136, 2862–2874. doi: 10.1104/pp.104.042903
Yuan, L. L., Zhang, M., Yan, X., Bian, Y. W., Zhen, S. M., Yan, Y. M. (2016). Dynamic phosphoproteome analysis of seedling leaves in brachypodium distachyon l. reveals central phosphorylated proteins involved in the drought stress response. Sci. Rep. 6, 35280. doi: 10.1038/srep35280
Yu, L., Liu, W., Guo, Z., Li, Z., Jiang, H., Zou, Q., et al. (2020). Interaction between MdMYB63 and MdERF106 enhances salt tolerance in apple by mediating Na(+)/H(+) transport. Plant Physiol. Biochem. 155, 464–471. doi: 10.1016/j.plaphy.2020.08.017
Yu, W. C., Yu, Y., Wang, C., Zhang, Z. J., Xue, Z. H. (2021). Mechanism by which salt stress induces physiological responses and regulates tanshinone synthesis. Plant Physiol. Biochem. 164, 10–20. doi: 10.1016/j.plaphy.2021.04.011
Yu, Y., Yu, M., Zhang, S. X., Song, T. Q., Zhang, M. F., Zhou, H. W., et al. (2022). Transcriptomic identification of wheat AP2/ERF transcription factors and functional characterization of TaERF-6-3A in response to drought and salinity stresses. Int. J. Mol. Sci. 23, 3272. doi: 10.3390/ijms23063272
Zang, Z. Y., Wang, Z., Zhao, F. X., Yang, W., Ci, J. B., Ren, X. J., et al. (2021). Maize ethylene response factor ZmERF061 is required for resistance to exserohilum turcicum. Front. Plant Sci. 12, 630413. doi: 10.3389/fpls.2021.630413
Zarei, A., Korbes, A. P., Younessi, P., Montiel, G., Champion, A., Memelink, J. (2011). Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in arabidopsis. Plant Mol. Biol. 75, 321–331. doi: 10.1007/s11103-010-9728-y
Zargara, S. M., Gupta, N., Nazir, M., Mahajan, R., Malik, F. A., Sofi, N. R., et al. (2017). Impact of drought on photosynthesis: Molecular perspective. Plant Gene. 11, 154–159. doi: 10.1016/j.plgene.2017.04.003
Zhang, G. Y., Chen, M., Li, L. C., Xu, Z. S., Chen, X. P., Guo, J. M., et al. (2009). Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J. Exp. Bot. 60, 3781–3796. doi: 10.1093/jxb/erp214
Zhang, L., Chen, L. J., Pang, S. Q., Zheng, Q., Quan, S. W., Liu, Y. F., et al. (2022). Function analysis of the ERF and DREB subfamilies in tomato fruit development and ripening. Front. Plant Sci. 13, 849048. doi: 10.3389/fpls.2022.849048
Zhang, J., Liao, J. Y., Ling, Q. Q., Xi, Y., Qian, Y. X. (2022). Genome-wide identification and expression profiling analysis of maize AP2/ERF superfamily genes reveal essential roles in abiotic stress tolerance. BMC Genomics 23, 125. doi: 10.1186/s12864-022-08345-7
Zhang, Z., Li, F., Li, D., Zhang, H., Huang, R. (2010). Expression of ethylene response factor JERF1 in rice improves tolerance to drought. Planta 232, 765–774. doi: 10.1007/s00425-010-1208-8
Zhang, Y., Ming, R. H., Khan, M., Wang, Y., Dahro, B., Xiao, W., et al. (2022). ERF9 of poncirus trifoliata (L.) raf. undergoes feedback regulation by ethylene and modulates cold tolerance via regulating a glutathione s-transferase U17 gene. Plant Biotechnol. J. 20, 183–200. doi: 10.1111/pbi.13705
Zhang, J., Shi, S. Z., Jiang, Y. N., Zhong, F., Liu, G. Y., Yu, C. M., et al. (2021a). Genome-wide investigation of the AP2/ERF superfamily and their expression under salt stress in Chinese willow (Salix matsudana). Plant Biol. 9, e11076. doi: 10.7717/peerj.11076
Zhang, J., Subramanian, S., Stacey, G., Yu, O. (2009). Flavones and flavonols play distinct critical roles during nodulation of medicago truncatula by sinorhizobium meliloti. Plant J. 57, 171–183. doi: 10.1111/j.1365-313X.2008.03676.x
Zhang, B. H., Su, L. C., Hu, B., Li, L. (2018). Expression of AhDREB1, an AP2/ERF transcription factor gene from peanut, is affected by histone acetylation and increases abscisic acid sensitivity and tolerance to osmotic stress in arabidopsis. Int. J. Mol. Sci. 19, 1441. doi: 10.3390/ijms19051441
Zhang, J. Y., Wang, Q. J., Guo, Z. R. (2012). [Progresses on plant AP2/ERF transcription factors]. Yi. Chuan. 34, 835–847. doi: 10.3724/sp.j.1005.2012.00835
Zhang, J., Xu, H. F., Wang, N., Jiang, S. H., Fang, H. C., Zhang, Z. Y., et al. (2018). The ethylene response factor MdERF1B regulates anthocyanin and proanthocyanidin biosynthesis in apple. Plant Mol. Biol. 98, 205–218. doi: 10.1007/s11103-018-0770-5
Zhang, H. B., Zhang, D. B., Chen, J., Yang, Y. H., Huang, Z. J., Huang, D. F., et al. (2004). Tomato stress-responsive factor TSRF1 interacts with ethylene responsive element GCC box and regulates pathogen resistance to ralstonia solanacearum. Plant Mol. Biol. 55, 825–834. doi: 10.1007/s11103-004-2140-8
Zhang, Z. J., Zhang, H. W., Quan, R. D., Wang, X. C., Huang, R. F. (2009). Transcriptional regulation of the ethylene response factor LeERF2 in the expression of ethylene biosynthesis genes controls ethylene production in tomato and tobacco. Plant Physiol. 150, 365–377. doi: 10.1104/pp.109.135830
Zhang, H., Zhao, Y., Zhu, J. K. (2020). Thriving under stress: How plants balance growth and the stress response. Dev. Cell. 55, 529–543. doi: 10.1016/j.devcel.2020.10.012
Zhang, H., Zhu, J., Gong, Z., Zhu, J. K. (2022). Abiotic stress responses in plants. Nat. Rev. Genet. 23, 104–119. doi: 10.1038/s41576-021-00413-0
Zhao, Y. L., Chang, X., Qi, D. Y., Dong, L. D., Wang, G. J., Fan, S. J., et al. (2017). A novel soybean ERF transcription factor, GmERF113, increases resistance to phytophthora sojae infection in soybean. Front. Plant Sci. 8, 299. doi: 10.3389/fpls.2017.00299
Zhao, Q., Hu, R. S., Liu, D., Liu, X., Wang, J., Xiang, X. H., et al. (2020). The AP2 transcription factor NtERF172 confers drought resistance by modifying NtCAT. Plant Biotechnol. J. 18, 2444–2455. doi: 10.1111/pbi.13419
Zhao, C. N., Liu, X. J., Gong, Q., Cao, J. P., Shen, W. X., Yin, X. R., et al. (2021). Three AP2/ERF family members modulate flavonoid synthesis by regulating type IV chalcone isomerase in citrus. Plant Biotechnol. J. 19, 671–688. doi: 10.1111/pbi.13494
Zhao, K., Shen, X. J., Yuan, H. Z., Liu, Y., Liao, X., Wang, Q., et al. (2013). Isolation and characterization of dehydration-responsive element-binding factor 2C (MsDREB2C) from malus sieversii roem. Plant Cell Physiol. 54, 1415–1430. doi: 10.1093/pcp/pct087
Zhao, J. Q., Wang, X. F., Pan, X. B., Jiang, Q. Q., Xi, Z. M. (2021). Exogenous putrescine alleviates drought stress by altering reactive oxygen species scavenging and biosynthesis of polyamines in the seedlings of Cabernet sauvignon. Front. Plant Sci. 12, 767992. doi: 10.3389/fpls.2021.767992
Zheng, Y. Z., Deng, G., Zhang, Y. C. (2022). Multiple free radical scavenging reactions of flavonoids. Dyes. Pigments. 198, 109877. doi: 10.1016/j.dyepig.2021.109877
Zheng, H., Jing, L., Jiang, X. H., Pu, C. J., Zhao, S. S., Yang, J., et al. (2021). The ERF-VII transcription factor SmERF73 coordinately regulates tanshinone biosynthesis in response to stress elicitors in salvia miltiorrhiza. New Phytol. 231, 1940–1955. doi: 10.1111/nph.17463
Zheng, S., Su, M., Wang, L., Zhang, T., Wang, J., Xie, H., et al. (2021). Small signaling molecules in plant response to cold stress. J. Plant Physiol. 266, 153534. doi: 10.1016/j.jplph.2021.153534
Zhu, J. K. (2016). Abiotic stress signaling and responses in plants. Cell 167, 313–324. doi: 10.1016/j.cell.2016.08.029
Zhuang, J., Chen, J. M., Yao, Q. H., Xiong, F., Sun, C. C., Zhou, X. R., et al. (2011). Discovery and expression profile analysis of AP2/ERF family genes from triticum aestivum. Mol. Biol. Rep. 38, 745–753. doi: 10.1007/s11033-010-0162-7
Zhuang, J., Li, M. Y., Wu, B., Liu, Y. J., Xiong, A. S.. (2016). Arg156 in the AP2-domain exhibits the highest binding activity among the 20 individuals to the GCC box in BnaERF-B3-hy15, a mutant ERF transcription factor from Brassica napus. Front Plant Sci 7, 1603. doi: 10.3389/fpls.2016.01603
Zhu, P. H., Chen, Y., Zhang, J. F., Wu, F., Wang, X. F., Pan, T., et al. (2021). Identification, classification, and characterization of AP2/ERF superfamily genes in masson pine (Pinus massoniana lamb.). Sci. Rep. 11, 5441. doi: 10.1038/s41598-021-84855-w
Zhu, Q. G., Gong, Z. Y., Huang, J., Grierson, D., Chen, K. S., Yin, X. R. (2019). High-CO2/Hypoxia-Responsive transcription factors DkERF24 and DkWRKY1 interact and activate DkPDC2 promoter. Plant Physiol. 180, 621–633. doi: 10.1104/pp.18.01552
Zhuo, C. L., Liang, L., Zhao, Y. Q., Guo, Z. F., Lu, S. Y. (2018). A cold responsive ethylene responsive factor from medicago falcata confers cold tolerance by up-regulation of polyamine turnover, antioxidant protection, and proline accumulation. Plant Cell Environ. 41, 2021–2032. doi: 10.1111/pce.13114
Keywords: ERF subfamily, AP2/ERF superfamily, flavonoids, abiotic stresses, cis-elements
Citation: Wu Y, Li X, Zhang J, Zhao H, Tan S, Xu W, Pan J, Yang F and Pi E (2022) ERF subfamily transcription factors and their function in plant responses to abiotic stresses. Front. Plant Sci. 13:1042084. doi: 10.3389/fpls.2022.1042084
Received: 12 September 2022; Accepted: 09 November 2022;
Published: 30 November 2022.
Edited by:
Kamrun Nahar, Sher-e-Bangla Agricultural University, BangladeshReviewed by:
Huapeng Zhou, Sichuan University, ChinaVivek Verma, Central University of Rajasthan, India
Copyright © 2022 Wu, Li, Zhang, Zhao, Tan, Xu, Pan, Yang and Pi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Erxu Pi, erxupi@hznu.edu.cn
†These authors have contributed equally to this work
 Ying Wu†
Ying Wu†