- Department of Plant Science and Technology, Chung-Ang University, Anseong, South Korea
Acidovorax citrulli (Ac) is a gram-negative bacterium that causes bacterial fruit blotch (BFB) disease in cucurbit crops including watermelon. However, despite the great economic losses caused by this disease worldwide, Ac-resistant watermelon cultivars have not been developed. Therefore, characterizing the virulence factors/mechanisms of Ac would enable the development of effective control strategies against BFB disease. The 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase (BdpM) is known to participate in the glycolysis and gluconeogenesis pathways. However, the roles of the protein have not been characterized in Ac. To elucidate the functions of BdpmAc (Bdpm in Ac), comparative proteomic analysis and diverse phenotypic assays were conducted using a bdpmAc knockout mutant (bdpmAc:Tn) and a wild-type strain. The virulence of the mutant to watermelon was remarkably reduced in both germinated seed inoculation and leaf infiltration assays. Moreover, the mutant could not grow with fructose or pyruvate as a sole carbon source. However, the growth of the mutant was restored to levels similar to those of the wild-type strain in the presence of both fructose and pyruvate. Comparative proteomic analyses revealed that diverse proteins involved in motility and wall/membrane/envelop biogenesis were differentially abundant. Furthermore, the mutant exhibited decreased biofilm formation and twitching halo size. Interestingly, the mutant exhibited a higher tolerance against osmotic stress. Overall, our findings suggest that BdpmAc affects the virulence, glycolysis/gluconeogenesis, biofilm formation, twitching halo size, and osmotic tolerance of Ac, suggesting that this protein has pleiotropic properties. Collectively, our findings provide fundamental insights into the functions of a previously uncharacterized phosphoglycerate mutase in Ac.
Introduction
Acidovorax citrulli (Ac) is a gram-negative, rod-shaped, and seed-borne bacterium belonging to the beta subdivision of the Proteobacteria (Schaad et al., 1978; Silva et al., 2016). Ac is the causative agent of bacterial fruit blotch (BFB) disease, which affects cucurbit crops worldwide. This disease can occur at any stage of watermelon growth and the symptoms can be detected in various parts of the plant (Rahimi-Midani and Choi, 2020). When a watermelon seedling is infected by Ac, the cotyledon and leaf develop a water-soaked lesion, and the seedling wilts and dies after a few days (Burdman et al., 2005). In infected fruit, the surface of the fruit develops a water-soaked area and later becomes blackened, which is accompanied by the decay of the inner flesh of the fruit. Ac strains can be divided into two groups: Group I is mainly isolated from non-watermelon cucurbits such as melon, and Group II is mainly isolated from watermelon (Zivanovic and Walcott, 2017). BFB greatly impacts fruit production and poses a major threat to the cucurbit industry worldwide (Shrestha et al., 2013; Islam et al., 2020). Nevertheless, there are currently no effective methods to control this disease because resistant cultivars/lines have not been identified. Therefore, characterizing the virulence factors and mechanisms of Ac infection is crucial for the development of effective disease control strategies.
Diverse virulence factors of Ac have already been reported in previous studies. Many gram-negative phytopathogens use the type III secretion system (T3SS) to directly secrete effector proteins into the host cell to promote their growth or suppress host defenses (Alfano and Collmer, 2004). In Ac, hrpX and hrpG, which play key roles in the regulation of the T3SS, are essential virulence factors (Zhang et al., 2018). The type II secretion system, which acts as an enzyme or toxin secretory pathway into the extremal environment (Douzi et al., 2012), is also crucial for virulence in Ac (Zivanovic and Walcott, 2017). Gram-negative bacteria also possess type IV pili, which are flexible filaments that enable bacterial adhesion to various surfaces (Craig et al., 2004). Ac cells with mutations in type IV pili-related genes exhibited reductions in twitching motility and virulence (Rosenberg et al., 2018). Flagella are also known to contribute to the virulence of Group I strains of Ac (Bahar et al., 2011). Additionally, quorum sensing, a cell-to-cell communication system in bacteria that can regulate gene expression, is also a well-known virulence mechanism in gram-negative pathogenic bacteria including Ac (De Kievit and Iglewski, 2000; Wang et al., 2016). Previous studies have also reported that a ferric uptake regulator, yggS family pyridoxal 5’-phosphate binding protein, a bifunctional chorismate mutase/prephenate dehydratase, and a pyridoxal phosphate-dependent aminotransferase contribute to virulence in Ac (Liu et al., 2019; Kim et al., 2020; Lee et al., 2021; Wang et al., 2022). Although several studies have characterized Ac virulence, additional efforts are still needed, as many virulence-related genes in Ac have not been studied yet.
The glycolysis pathway is a catabolic process in which 6-carbon glucose molecules are converted to 2-carbon pyruvate molecules, resulting in the release of ATP (Ganapathy-Kanniappan and Geschwind, 2013). The generated pyruvate can then enter the tricarboxylic acid (TCA) cycle to generate more ATP (Rui, 2014). In contrast, the gluconeogenesis pathway is an anabolic process in which 2-carbon pyruvate molecules are converted to 6-carbon glucose molecules (Mapes and Harris, 1976). Phosphoglycerate mutases (PGMs) are critical enzymes in both glycolysis and gluconeogenesis. This enzyme catalyzes the interconversion of 3-phosphoglycerate and 2-phosphoglycerate in both the glycolysis and gluconeogenesis pathways (Davies et al., 2011). Therefore, PGMs are responsible for synthesizing both glucose and pyruvate in the glycolysis and gluconeogenesis pathways. The PGMs can be categorized into two groups without sequence similarities: cofactor-dependent phosphoglycerate mutases (dPGMs) and cofactor-independent phosphoglycerate mutases (iPGMs) (Zhang et al., 2004). dPGMs are found in all vertebrates, some invertebrates, fungi, and some gram-negative bacteria. iPGMs are found in all plants, algae, some invertebrates, fungi, and some gram-positive bacteria (Jedrzejas, 2000). However, dPGMs are needed for the activation of the 2,3-bisphosphoglycerate, whereas iPGMs are not (Fraser et al., 1999). Mice infected with a mutant strain of Staphylococcus aureus lacking iPGM exhibited milder disease symptoms (Radin et al., 2019). Moreover, the dPGM of Burkholderia phymatum plays a crucial role in the formation of root nodules in Mimosa pudica (Chen et al., 2012). However, the functions of PGMs in Ac are yet to be elucidated.
The present study characterized the functions of a putative 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase in Ac (BdpmAc) belonging to the dPGM group in the KACC17005 strain, which has an annotated genome (Park et al., 2017). To examine the functions of BdpmAc in Ac, pathogenicity and growth assays in M9 minimal media with a sole carbon source were conducted. Additionally, label-free shotgun comparative proteomic analysis was conducted to explore the potential cellular mechanisms related to BdpmAc. Based on our proteomic analysis data, biofilm formation, twitching halo size, and osmotic stress assays were investigated. Through these phenotype assays, our findings demonstrated that BdpmAc is involved in virulence and other key phenotypes in Ac.
Materials and methods
Bacterial strains and growth conditions
Ac strain KACC17005, which belongs to Group II, was used as the wild-type strain in this study. Ac was grown in Tryptic Soy Broth (30 g/L) (TSB) or TSB (30 g/L) supplemented with 1.5% Agar (TSA) at 28°C. The E. coli strains EC100D and DH5 were used to identify the Tn5 transposon insertion site and for cloning, respectively. E. coli strains were grown in Luria-Bertani broth (LB) (Tryptone 10 g/L, NaCl 10 g/L, Yeast extract 5 g/L) or LB supplemented with 1.5% agar at 37°C. In this study, M9 media was used as a minimal medium. For the selection of bacteria harboring genes of interest, appropriate antibiotics were added to each medium at the following final concentrations: rifampicin, 50 μg/mL; ampicillin, 100 μg/mL; kanamycin, 50 μg/mL; gentamicin, 10 μg/mL.
Selection of bdpmAc:Tn via Tn mutant library screening and generation of BdpmAc complemented strain
The bacterial strains and plasmids used in this study are summarized in Supplementary Table 1. An Ac mutant with decreased virulence was selected by screening the EZ-Tn™ insertional library as previously reported (Kim et al., 2020). After selecting the mutant, the Tn insertion site was identified by following the manufacturer’s instructions (Lucigen, Middleton, WI, USA). A putative 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase (Accession No. ATG93870) was disrupted by Tn and the identified mutant was named bdpmAc:Tn. To generate a complemented strain, the full-length bdpmAc gene from the wild-type strain was amplified with bdpmAc-specific primers (5’-ggtaccatgcacaaactcgtcctgat-3’ and 5’-cgtcccagggcaaggcccaccaccaccaccaccactgaagctt-3’) and cloned into the pGem-T easy vector (Promega, Madison, Wisconsin, USA) to obtain pGem-bdpmAc. The cloned gene was confirmed by Sanger sequencing. Subsequently, the confirmed gene was cloned again into the pBBR1-MCS5 plasmid (Kovach et al., 1995), which contains the LacZ promoter for expression of cloned genes, thus obtaining pMCS5-bdpmAc. Finally, pMCS5-bdpmAc was introduced into bdpmAc:Tn. This insertion was confirmed by polymerase chain reaction (PCR) with bdpmAc-specific primers. The successful insertion of pMCS5-bdpmAc resulted in the bdpmAc:Tn(BdpmAc) complemented strain. To avoid the side effects from the vector, pBBR1-MCS5 was introduced into Ac and bdpmAc:Tn to generate Ac(EV) and bdpmAc:Tn (EV), respectively. The generated strains were confirmed by PCR with pBBR1-MCS5 specific primers.
Virulence assay
Citrullus lanatus var. vulgaris line SBA seeds, which were kindly provided by Partner Seed Company (Gimje, Korea), were used for the virulence assay. Two inoculation assays were performed in this study: germinated-seed inoculation and leaf infiltration. The germinated seed inoculation assay was conducted as previously reported (Kim et al., 2020). Briefly, the watermelon seeds were germinated on water-soaked filter paper for two days. The Ac strains were cultured on TSA at 28°C, harvested using cotton swabs, and suspended in 10 mM MgCl2. The bacterial suspension was adjusted to an OD600nm of 0.3 (approximately 108 colony forming units (CFUs)/mL), and serially diluted (10-2) with the same buffer to reach a 106 CFU/mL concentration. The germinated watermelon seeds were soaked with the prepared bacterial suspension and incubated with gentle agitation for 1 hour at 22°C. The inoculated seeds were sown in 50 pot trays with autoclaved soil and grown for 7 days. The disease severity of inoculated watermelon seedlings was measured every day for 7 days. The disease severity was evaluated based on a 0–2 scale, where 0 indicates no symptoms, 1 indicates the occurrence of spots, and 2 indicates wilting. The disease index was calculated as follows: [(number of plants with no symptoms)×0 + (number of plants with spots)×1 + (number of plants presenting wilting)×2]/Total number of plants. Five independent experiments with ten biological replicates were conducted per strain. The leaf infiltration experiments were conducted as previously reported (Lee et al., 2021). Briefly, the watermelon seeds were grown in a growth chamber until they reached the four true leaves stage. A bacterial suspension was then prepared with 105 CFU/mL (final concentration) in 10 mM MgCl2. The prepared inoculant was directly infiltrated into the first and second true leaves of the watermelon seedlings using a needleless syringe. To monitor the bacterial growth, the infiltrated leaves were punched using 0.4 cm diameter cork borers, after which two leaf disks were ground in 200 µL of 10 mM MgCl2 using tissue grinders to extract the bacterial cells. The extracted bacterial cells were serially diluted, dotted onto TSA containing the proper antibiotics, and incubated at 28°C for two days. Five independent experiments were conducted with three biological replicates per strain.
Growth assay
The growth of Ac strains was measured in TSB and M9 containing diverse carbon sources, and appropriate antibiotics were added to the media. For TSB, the Ac strains were grown on TSA (1.5% agar). The grown bacterial cells were harvested using cotton swabs and washed twice with distilled water. After washing, each strain was resuspended and adjusted to an OD600nm of 0.3 (approximately 108 CFU/mL). The adjusted bacterial cells were diluted (10-3) with TSB to achieve a 105 CFU/mL concentration and incubated in a shaking incubator at 28°C. Bacterial growth was monitored every 12 hours for five days using a spectrophotometer. For minimal media, Ac(EV), bdpmAc:Tn(EV), and bdpmAc:Tn(BdpmAc) were grown in six different conditions with appropriate antibiotics (M9 with glucose, M9 with fructose, M9 with pyruvate, M9 with fructose and pyruvate, M9 with mannitol, and M9 with sucrose). The final concentration of each supplemented carbon source was 0.4% in the medium. In M9 with fructose and pyruvate, each carbon source was added at a 0.2% concentration to adjust the entire carbon source to 0.4%. The bacterial cells were washed twice and adjusted to an OD600nm of 0.05 in the minimal media and cell growth was monitored for ten days at 24 h intervals. For this assay, three biological replicates were examined for each strain, and at least four independent experiments were conducted.
Comparative proteomics
Proteomic analysis was performed at the BT Research Facility Center, Chung-Ang University. The Ac and bdpmAc:Tn strains were used for label-free shotgun comparative proteomic analysis. Sample preparation, protein extraction, and peptide generation/quantification were performed as reported previously (Kim et al., 2020). Concretely, the three biological replicates from each sample (a total of six samples) were used for the analysis. Bacterial cells were grown in TSB, collected at an OD600nm of 0.6 (approximately 2×108 CFU/mL) using the centrifuge (7,300 g, 15 minutes, 28 °C), and washed twice with 50 mM Tris-HCl (pH 7.8). The washed cells were suspended in lysis buffer (6 M guanidine HCl, 10 mM dithiothreitol, 50 mM Tris-HCl pH 7.8) and sonicated with an ultrasonic processor (Colo Parmer, Vernon Hills, IL, USA). After sonication, the total soluble proteins were precipitated using trichloroacetic acid and treated with trypsin to obtain tryptic digested peptides. The peptides were cleaned using a Sep-Pak Vac 1cc tC18 cartridge (Waters, Milford, MA, USA) and quantified using the BCA assay kit (Thermo Fisher Scientific, Rockford, IL, USA). Next, 1 μg of the samples was analyzed using a split-free nano LC system (EASY-nLC II; Thermo Fisher Scientific, Bremen, Germany) linked to an LTQ Velos Pro instrument (Thermo Fisher Scientific) under nanospray ion mode. To separate the tryptic digested peptides, the samples were loaded onto a 7.5 cm column packed with MAGIC C18AQ 200A (Michrom BioResources, Auburn, CA, USA) and eluted for 420 minutes (300 nL/min flow rate) under the following water/acetonitrile gradient conditions: buffer A, 100% water with 0.1% formic acid; buffer B, 100% acetonitrile with 0.1% formic acid; 7% of buffer B for 5 minutes, 35% of buffer B for 380 minutes, 80% of buffer B for 10 min, and 7% of buffer B for 25 minutes. To obtain the full mass spectra, six data-dependent ms/ms scans were employed with an m/z 300–2000 mass range. The ion charge state selection was allowed for 2+ and 3+. Dynamic exclusion was permitted under the following conditions: repeat count, 1; repeat duration, 0.5 minutes; elimination duration, 3 minutes. The maximum value of the six most intense ions from each full mass scan was sequentially selected for fragmentation. The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez- Riverol et al., 2022) partner repository with the dataset identifier PXD035156.
Proteins were identified and quantified according to a previously established protocol (Kim et al., 2020). The Thermo Proteome Discoverer software (ver. 1.3.0.399) coupled with the SEQUEST search algorithm was used to identify proteins based on Ac strain KACC17005 genome data (Accession No. CP023687) obtained from the NCBI database. The target-decoy strategy was used in this analysis to improve reliability (Elias and Gygi, 2007). Two missed cleavages were allowed and the results were deemed significant at a 0.01 false discovery rate. For mass accuracy, the precursor was 100 ppm and the probability score was more than 20. After identifying the proteins, the Scaffold 4 software (Proteome Software, Portland, OR, USA) was used to conduct comparative analyses. The peptide spectrum matches (PSMs) were used for comparison (Choi et al., 2008). PSMs from each protein were normalized to the total PSMs from each biological replicate. The average value of the PSMs from the three biological replicates was used to identify differentially abundant (more than 2-fold change) proteins between Ac and bdpmAc:Tn. Pairwise comparisons were conducted via Student’s t-test (P<0.05). Clusters of orthologous groups (COG) analysis was used for classifying the differentially abundant proteins (Tatusov et al., 2000).
Osmotic stress assay
For the osmotic stress assay, the Ac strains were exposed to 2.5% NaCl and 40% sorbitol. Both assays were conducted as previously described (Kim et al., 2020; Park et al., 2021). In the NaCl treatment, the bacterial suspension was adjusted to an OD600nm of 0.1 (approximately 3 × 107 CFU/mL) and exposed to 2.5% NaCl for 20 minutes and 40% sorbitol for 40 minutes in TSB. Then, the exposed cells were serially diluted, dotted on TSA (1.5% agar) with appropriate antibiotics, and incubated at 28°C for 2 days, after which the bacterial colonies were counted. TSB alone was used as a negative control. To calculate survivability, we obtained the ratio of the viable cells from the treatments to those from the control. Three biological replicates were analyzed for each strain and seven and five independent experiments were conducted for NaCl and sorbitol, respectively.
Biofilm formation
Biofilm formation capacity was measured as described in a previous study (Kim et al., 2020) Ac strains were suspended in TSB, adjusted to an OD600nm of 0.3 (108 CFU/mL), and diluted (10-3) (105 CFU/mL). The prepared suspension was cultured in a 96-well polyvinyl chloride plate for one and three days at 28°C without agitation. After incubation, the cell culture supernatants were removed and washed one time using autoclaved water. Crystal violet (0.1%) was used to stain the remaining cells for 30 minutes. After washing the cells two times with autoclaved water, the stained cells were resuspended in 95% ethanol with gentle shaking. After 20 minutes, the resuspended cells were measured using a Spectramax 190 microplate reader (Molecular Devices, Sunnyvale, CA, USA) at 590 nm. A total of 20 biological replicates of each strain were used in this assay and eight independent experiments were conducted.
Twitching halo
The twitching halo production assay was conducted as described in a previous study (Kim et al., 2020). Ac strains were cultured in TSA, harvested, and adjusted to an OD600nm of 0.3 (approximately 108 CFU/mL). Next, 5 μL of bacterial suspension was dotted on TSA (0.5% agar) and the samples were incubated at 28°C. After two days, the diameter of the colonies and the diameter of the twitching halo were measured under a LEICA M205 C microscope (LEICA, Wetzlar, Germany). Three biological replicates of each strain were employed and eight independent experiments were conducted.
Statistical analysis
Pairwise comparisons were conducted via Student’s t-test or one-way analysis of variance (ANOVA) with Tukey’s HSD (honestly significant difference) test using the SPSS 12.0K software (SPSS, Inc., Chicago, IL, USA). A p-value < 0.05 was considered statistically significant.
Results
BdpmAc is an essential virulence factor of Ac
A Tn5-insertional mutant library generated with the strain KACC17005 was screened to identify genes associated with the virulence of Ac. From this library, we selected one mutant strain that did not cause BFB disease symptoms in watermelon and linked this phenotype to a gene encoding a putative 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase (BdpmAc) (Accession No. ATG93870). To confirm the involvement of BdpmAc in the virulence of Ac, both germinated seed inoculation and leaf infiltration experiments were conducted. Furthermore, Ac(EV), bdpmAc:Tn(EV), and bdpmAc:Tn(BdpmAc) were constructed to conduct pathogenicity tests. Ac(EV) and bdpmAc:Tn(EV) were the wild-type strains and bdpmAc:Tn was the strain carrying the empty pBBR1-MCS5 vector. bdpmAc:Tn(BdpmAc) was the bdpmAc:Tn strain that carried the original bdpmAc on the pBBR1-MCS5 vector. As illustrated in Figures 1A, B, the bdpmAc:Tn(EV) strain induced little to no disease symptoms. Conversely, the Ac(EV) strain induced severe wilting symptoms, and the bdpmAc:Tn(BdpmAc) strain induced some spots and wilting symptoms. Seven days after inoculation, the disease severity index of the Ac(EV) strain was 2, whereas that of bdpmAc:Tn(EV) was 0.1. The complemented strain recovered to some extent, and the disease severity was 1.5. Similar results were observed in the leaf infiltration experiments compared to the germinated seed inoculation experiments (Figures 1C, D). Leaves infiltrated with Ac(EV) became dark and exhibited wilting symptoms. However, the leaves infiltrated with bdpmAc:Tn(EV) exhibited very mild wilting symptoms. The complemented strain induced similar symptoms in the inoculated leaves to those of the Ac(EV) strain. Furthermore, the bdpmAc:Tn(EV) strain in the inoculated watermelon leaf produced fewer CFUs than Ac(EV) and bdpmAc:Tn(BdpmAc) at 0, 2, 4, 6, and 8 days after inoculation. Therefore, our findings indicated that BdpmAc was essential for virulence.
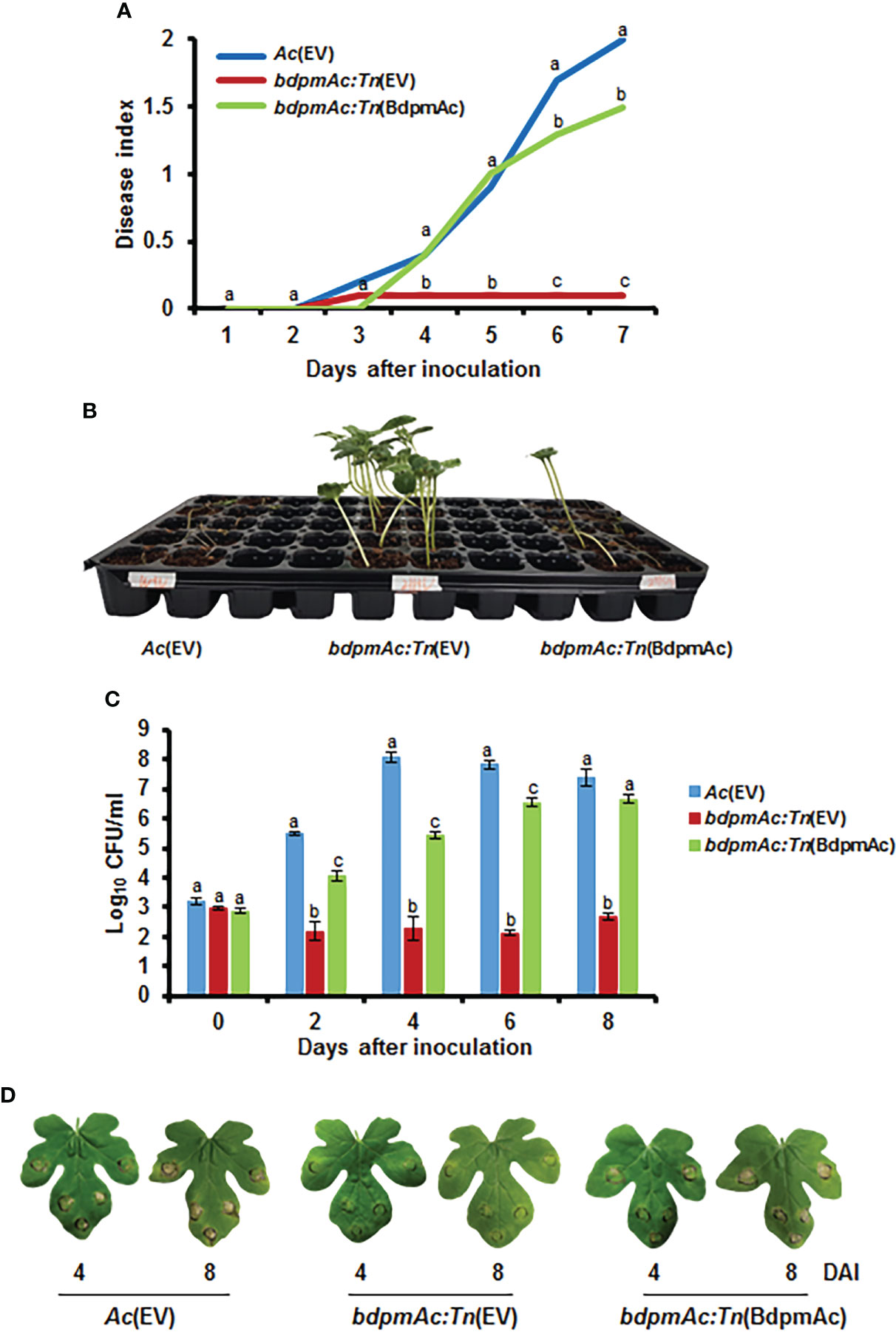
Figure 1 Virulence assay for Ac(EV), bdpmAc:Tn(EV), and bdpmAc:Tn(BdpmAc). (A) Disease index of the inoculated germinated seeds at 7 days after inoculation. Disease index: [(number of plants with no symptoms)×0 + (number of plants with spots)×1 + (number of plants presenting wilting)×2]/Total number of plants. (B) Photograph of the germinated seed inoculation. The image was taken 7 days after inoculation. Ten biological replicates were used for each strain. (C) Variations in cell numbers in the leaf infiltration experiments after 8 days using the colony counting method. Three biological replicates with three technical replicates were evaluated for each strain. (D) Photograph of the leaf infiltration experiment. The images were taken 4 and 8 days after inoculation. Different letters above the error bars indicate significant differences according to ANOVA (p<0.05) with Tukey’s HSD test and the error bars indicate the standard errors of means. Five independent experiments were conducted, all of which showed similar patterns.
BdpmAc is required for glycolysis and gluconeogenesis
Sequencing analyses confirmed that the above-described phenotypes were attributable to changes in the bdpmAc gene that encodes a putative 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase, which is a key enzyme in the glycolysis and gluconeogenesis pathways. Therefore, it can be assumed that BdpmAc may be involved in carbon source metabolism or anabolism, and thus its growth in minimal media with single carbon sources was investigated. To confirm the utilization of carbon sources, the growth of Ac strains in M9 minimal media with each glycolysis precursor (glucose, fructose, mannitol, sucrose) and gluconeogenic precursor (pyruvate) was monitored. Before conducting the experiments in the M9 medium, growth curves were measured in rich media (TSB). Ac(EV) reached the exponential growth phase in TSB after 12 hours of incubation and later reached the stationary stage (around OD600nm 2) at 72 hours. The complemented strain showed a similar growth curve. The growth of bdpmAc:Tn(EV) was slightly decreased compared to Ac(EV) and bdpmAc:Tn(BdpmAc). At 12 hours, there were no visible differences from other strains. However, its growth rate began to decrease after 12 hours at the stationary stage and the OD600nm was less than 2 (Supplementary Figure 1A). Next, the Ac(EV), bdpmAc:Tn(EV), and bdpmAc:Tn(BdpmAc) strains were examined in the presence of glycolysis precursors (glucose and fructose). In M9 with glucose, the Ac(EV) strain grew consistently well. In contrast, the growth of the bdpmAc:Tn(EV) strain was markedly reduced compared to Ac(EV) and the complemented strain. The growth of the complemented strain recovered to similar levels to those of the Ac(EV) strain (Figure 2A). In the M9 with fructose, Ac(EV) and bdpmAc:Tn(BdpmAc) grew well but bdpmAc:Tn(EV) did not (Figure 2B). In M9 supplemented with pyruvate, the Ac(EV) strain grew consistently well. However, the bdpmAc:Tn(EV) strain exhibited poor growth rates and often showed a tendency to decline. The growth of the complemented strain recovered to levels similar to those of the Ac(EV) strain (Figure 2C). As illustrated in Figures 2B, C, the bdpmAc:Tn(EV) strain did not grow well. Therefore, growth assays were conducted in M9 media containing both fructose and pyruvate. Interestingly, the growth of bdpmAc:Tn(EV) recovered to levels similar to those of Ac(EV) and the complemented strain (Figure 2D). Lastly, we examined the growth of Ac strains with mannitol produced via hydrogenation of fructose and sucrose, which is composed of glucose and fructose (Supplementary Figures 1B, D). None of the examined strains grew well in this medium. These results demonstrate that BdpmAc is essential for both glycolysis and gluconeogenesis in Ac.
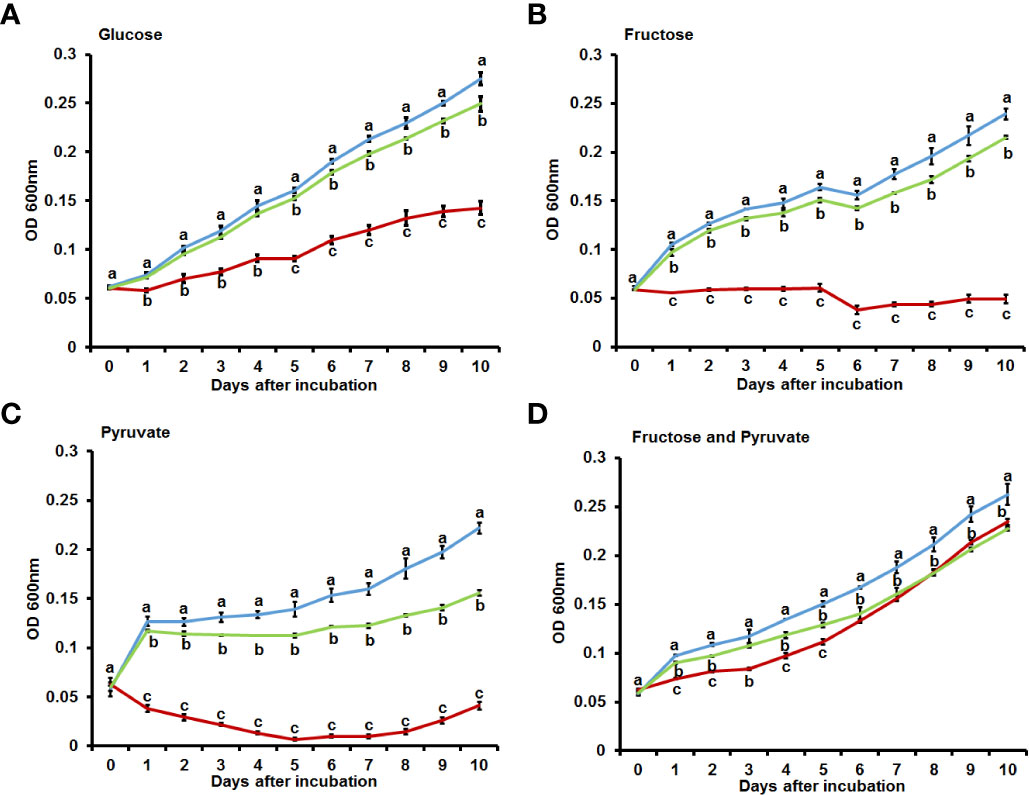
Figure 2 Growth assay in M9 minimal media with diverse carbon sources. Bacterial growth in M9 minimal media with (A) glucose (0.4%), (B) fructose (0.4%), (C) pyruvate (0.4%), and (D) fructose (0.2%) and pyruvate (0.2%) as carbon sources. The values at OD600nm were measured for 10 days at 24 h intervals. The error bars from three biological replicates indicate the standard deviation and the different characters above the error bars indicate significant differences according to ANOVA (p<0.05) with Tukey’s HSD test. At least four independent experiments were performed and all experiments displayed similar patterns.
Proteomic analysis
To explore the potential cellular mechanisms affected by BdpmAc, a label-free shotgun comparative proteomic analysis followed by COG categorization was conducted using Ac and bdpmAc:Tn. The numbers of the detected PSMs in the three biological replicates from Ac were similar to those of bdpmAc:Tn (65,333–65,715) (Supplementary Table 2). However, more proteins were identified in bdpmAc:Tn than in Ac (Supplementary Table 2). A total of 929 and 1,045 proteins were detected in all three biological replicates from Ac and bdpmAc:Tn, respectively (Supplementary Table 2). Next, these proteins were subjected to comparative proteomic analysis, which showed that 39 and 137 proteins were only detected in Ac and bdpmAc:Tn, and 11 and 52 proteins were more abundant (over 2-fold) in these strains, respectively (Figure 3A). These differentially abundant proteins were categorized using COG analysis (Figure 3B; Supplementary Table 3 and 4). BdpmAc was only detected in the wild-type but not in the bdpmAc:Tn mutant (Supplementary Tables 3, 4), indicating that the comparative proteomic analysis was correctly conducted. The numbers of classified proteins in most categories in bdpmAc:Tn were higher than those in Ac. Proteins belonging to two categories, J (translation) and L (replication and repair), were more abundant in Ac. Interestingly, diverse proteins related to carbohydrate metabolism as well as cell wall/membrane/envelop biogenesis were identified in the comparative proteomic analysis. Additionally, proteins associated with cell movement including pili motility proteins were abundantly detected. The results of our comparative proteomic analysis suggest that BdpmAc is involved in diverse phenotypes in Ac including carbohydrate metabolism, biofilm formation, and twitching motility.
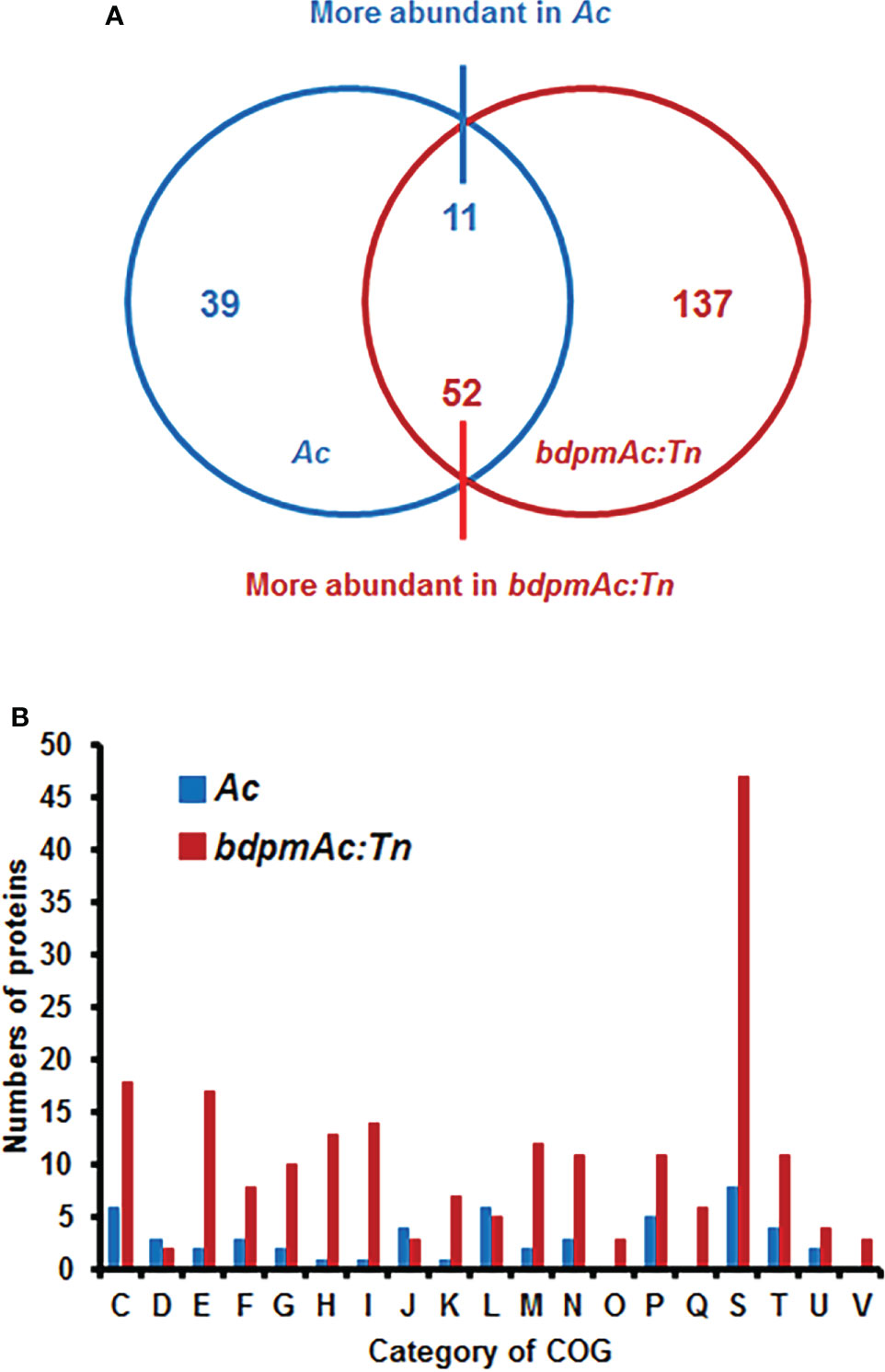
Figure 3 Comparative proteomic analysis between Ac and bdpmAc:Tn. (A) Venn diagram of the differentially (over two-fold change) abundant proteins identified in the comparative proteomic analysis. A total of 39 and 137 proteins were only found in Ac and bdpmAc:Tn, and 11 and 52 proteins were more abundant in these strains, respectively. (B) Categorization of the differentially abundant proteins using the clusters of orthologous groups (COGs). Abbreviations:C, Energy production and conversion; D, Cell cycle control and mitosis; E, Amino acid metabolism and transport; F, Nucleotide metabolism and transport; G, Carbohydrate metabolism and transport; H, Coenzyme metabolism; I, Lipid metabolism; J, Translation; K, Transcription; L, Replication and repair; M, Cell wall/membrane/envelop biogenesis; N, Cell motility; O, Post-translational modification, protein turnover, chaperone functions; P, Inorganic ion transport and metabolism; Q, Secondary structure; S, Function unknown; T, Signal transduction; U, Intracellular trafficking and secretion; V, Defense mechanisms.
BdpmAc is related to biofilm formation
Our comparative proteomic analyses identified several differentially abundant proteins associated with cell wall/membrane/envelop biogenesis. The bacterial cell wall/membrane is known to be closely related to biofilm formation (Bucher et al., 2015). Additionally, a previous study reported that a phosphoglycerate mutase knockout mutant of Stenotrophomonas maltophilia exhibited a decreased biofilm formation capacity (Ramos-Hegazy et al., 2020). Therefore, we next sought to compare the biofilm formation capacity of Ac(EV), bdpmAc:Tn(EV), and bdpmAc:Tn(BdpmAc) using a 96-well PVC microplate assay. The biofilm formation capacity of the Ac strains was evaluated and measured at 1 and 3 days after incubation (DAI). At 1 DAI, biofilm formation significantly differed between Ac(EV) and bdpmAc:Tn(EV) (Figure 4A). Specifically, the biofilm formation ability of bdpmAc:Tn(EV) decreased markedly (over 5.9-fold) compared to Ac(EV). In contrast, the biofilm formation capacity of bdpmAc:Tn(BdpmAc) was similar to that of Ac(EV). The results observed at 3 DAI were similar to those observed at 1 DAI (Figure 4B). The biofilm formation capacity of bdpmAc:Tn(EV) was lower (over 3.2-fold) than that of Ac(EV) and bdpmAc:Tn(BdpmAc) on day 3. Therefore, our findings demonstrated that BdpmAc is involved in biofilm formation.
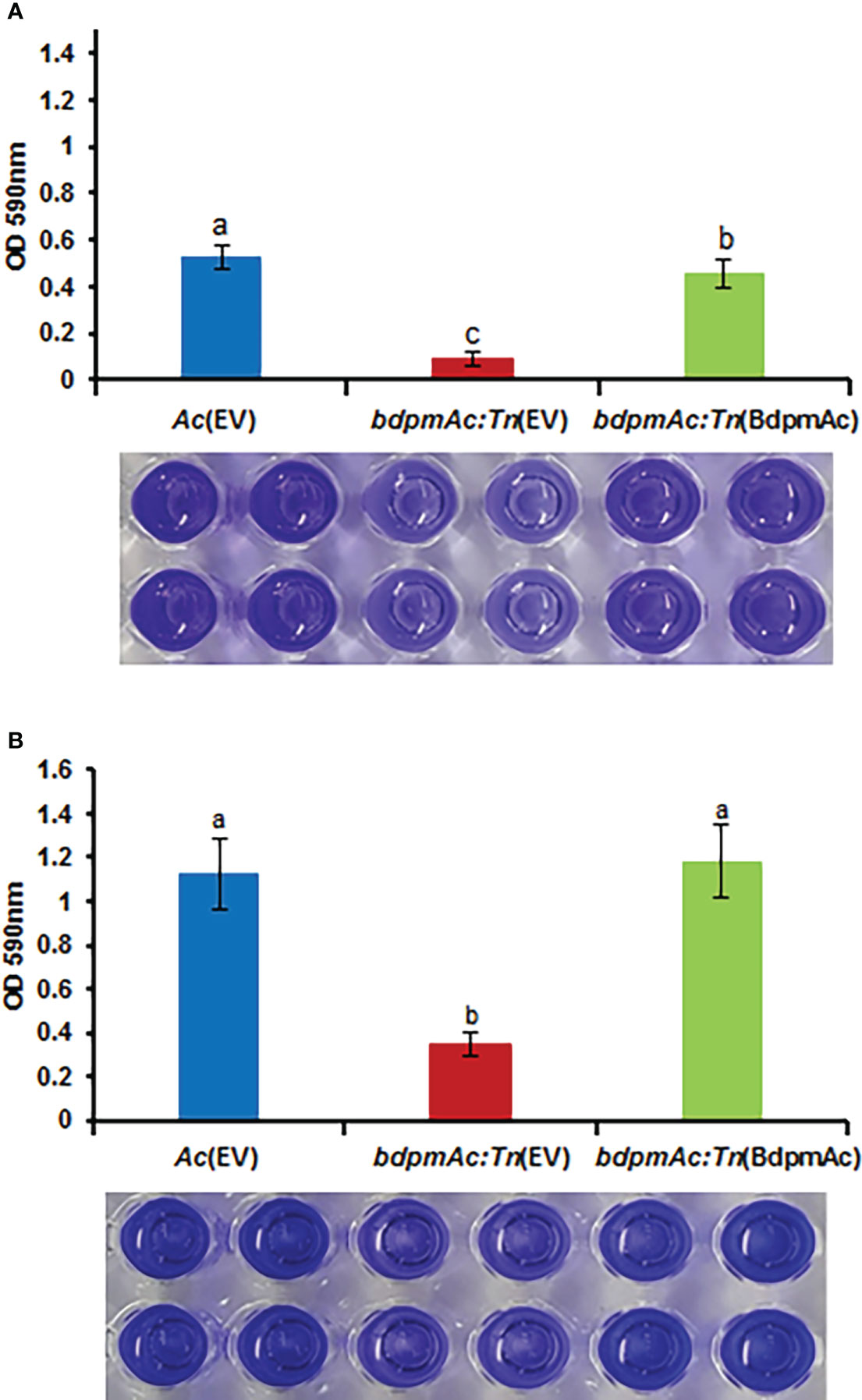
Figure 4 Biofilm formation in Ac(EV), bdpmAc:Tn(EV), and bdpmAc:Tn(BdpmAc). Ac strains were incubated in polyvinyl chloride 96-well plates for (A) one and (B) three days after incubation. For quantification, the formed biofilm was stained using 0.1% crystal violet. The stained biofilm was dissolved in 95% ethanol and measured using a UV spectrophotometer at OD590nm. The error bars indicate the standard deviations of twenty biological replicates and different characters above the error bars indicate significant differences according to ANOVA (p<0.05) with Tukey’s HSD test. Eight independent experiments were conducted, all of which exhibited similar patterns.
BdpmAc:Tn(EV) showed reduced twitching halos
Many proteins involved in cell motility were detected in the COG categorization. Similar to other gram-negative plant pathogenic bacteria, Ac possesses two types of appendages (pili and flagella) as motility organs. However, the Group II strain of Ac rarely showed flagella-dependent motility and mostly used pili-dependent motility (Bahar et al., 2010). Therefore, to verify if BdpmAc is related to cell motility, the twitching motility (i.e., pili-dependent motility) assay was conducted. This assay was conducted in TSB with 0.5% agar and the sizes of the bacterial colonies and twitching halos were evaluated. The colony sizes of Ac(EV), bdpmAc:Tn(EV), and bdpmAc:Tn(BdpmAc) were 0.73 cm, 0.7 cm, and 0.7 cm, respectively (Figure 5), all of which were deemed to be statistically equal. However, there was a significant difference in the twitching halos between Ac(EV) and bdpmAc:Tn(EV). The twitching halos of bdpmAc:Tn(EV) were smaller (1.26 cm) compared to those of Ac(EV) (1.85 cm). In contrast, the twitching halo size of the complemented strain was restored (1.83 cm) and was similar to that of the Ac(EV) strain. These results demonstrated that BdpmAc affects the twitching halo size of Ac.
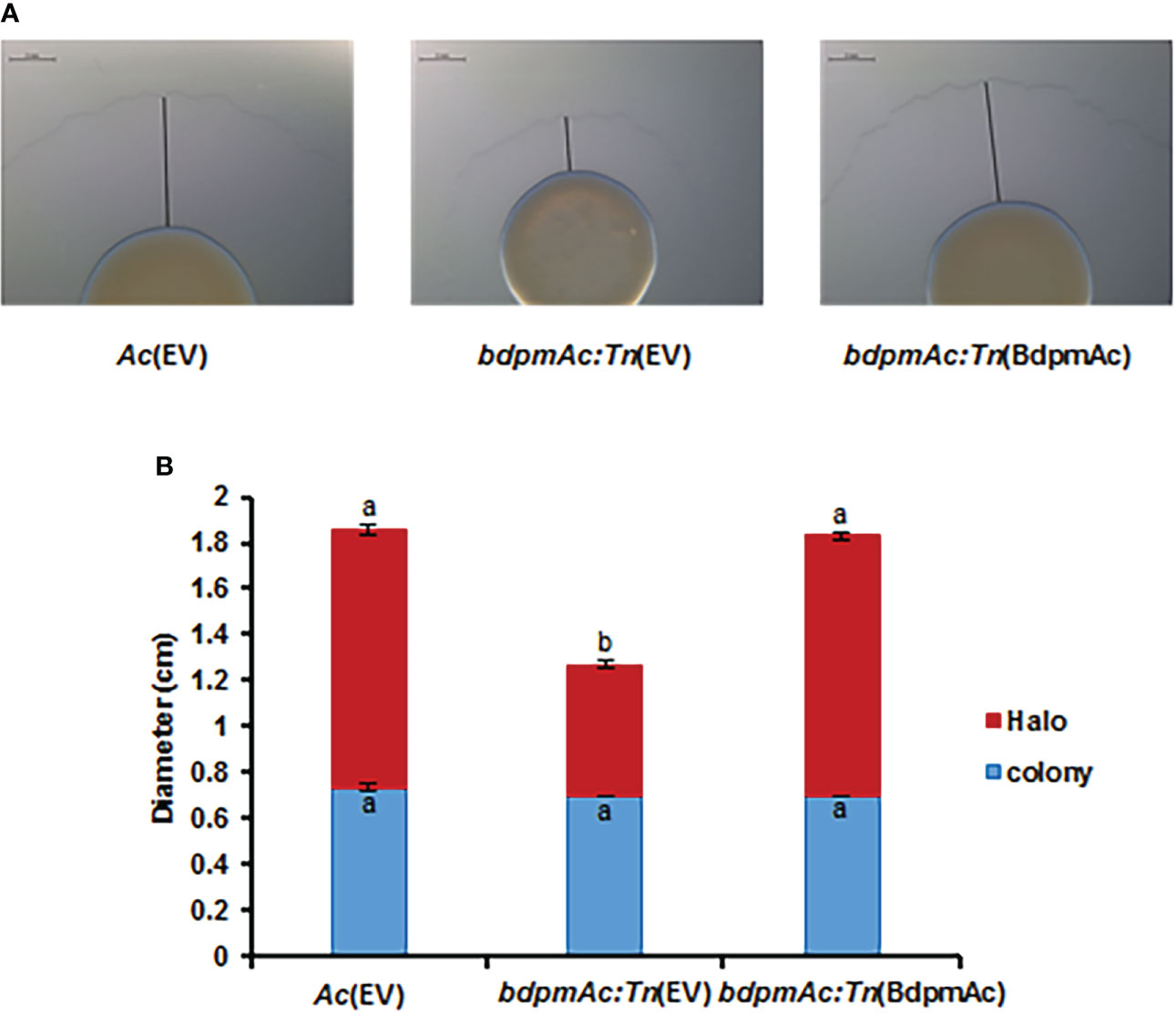
Figure 5 Twitching halo size in Ac(EV), bdpmAc:Tn(EV), and bdpmAc:Tn(BdpmAc). (A) Observation of twitching halo size under a stereoscopic microscope. The black lines indicate the length of the twitching halo. Scale bar: 2 mm. (B) Measurement of the colony and twitching halo sizes. The blue and red bars indicate the diameter of the colony and twitching halo diameter, respectively. The error bars indicate the standard deviations of three biological replicates. Different characters indicate significant differences according to ANOVA (p<0.05) with Tukey’s HSD test. Eight independent experiments were carried out, all of which exhibited similar patterns.
BdpmAc is associated with tolerance to osmotic stress
As indicated by our COG analyses, proteins categorized in group M (cell wall/membrane/envelope biosynthesis) were more abundant in bdpmAc:Tn compared to Ac. The bacterial cell envelopes are essential for stabilizing the turgor pressure (Rojas et al., 2018). These findings suggest that bdpmAc:Tn is more tolerant to osmotic stress than Ac. To test this hypothesis, the three strains were exposed to NaCl and sorbitol. First, the three strains were exposed to 2.5% NaCl in TSB for 20 minutes. Ac(EV) exhibited a 9.6% survival rate, whereas bdpmAc:Tn(EV) had a 15.3% survival rate (Figure 6A). Moreover, the survival rate of the complemented strain was also lower than that of bdpmAc:Tn(EV) (3.75%). Next, Ac was exposed to 40% sorbitol in TSB for 40 minutes and the results of this assay were consistent with those of the NaCl stress experiments. The Ac(EV) strain exhibited a survival rate of 64%, whereas bdpmAc:Tn(EV) exhibited a 93.6% survival rate (Figure 6B). The survival rate of the complemented strain was also lower than that of bdpmAc:Tn(EV) (57%). These data indicate that BdpmAc is associated with in tolerance to osmotic stress.
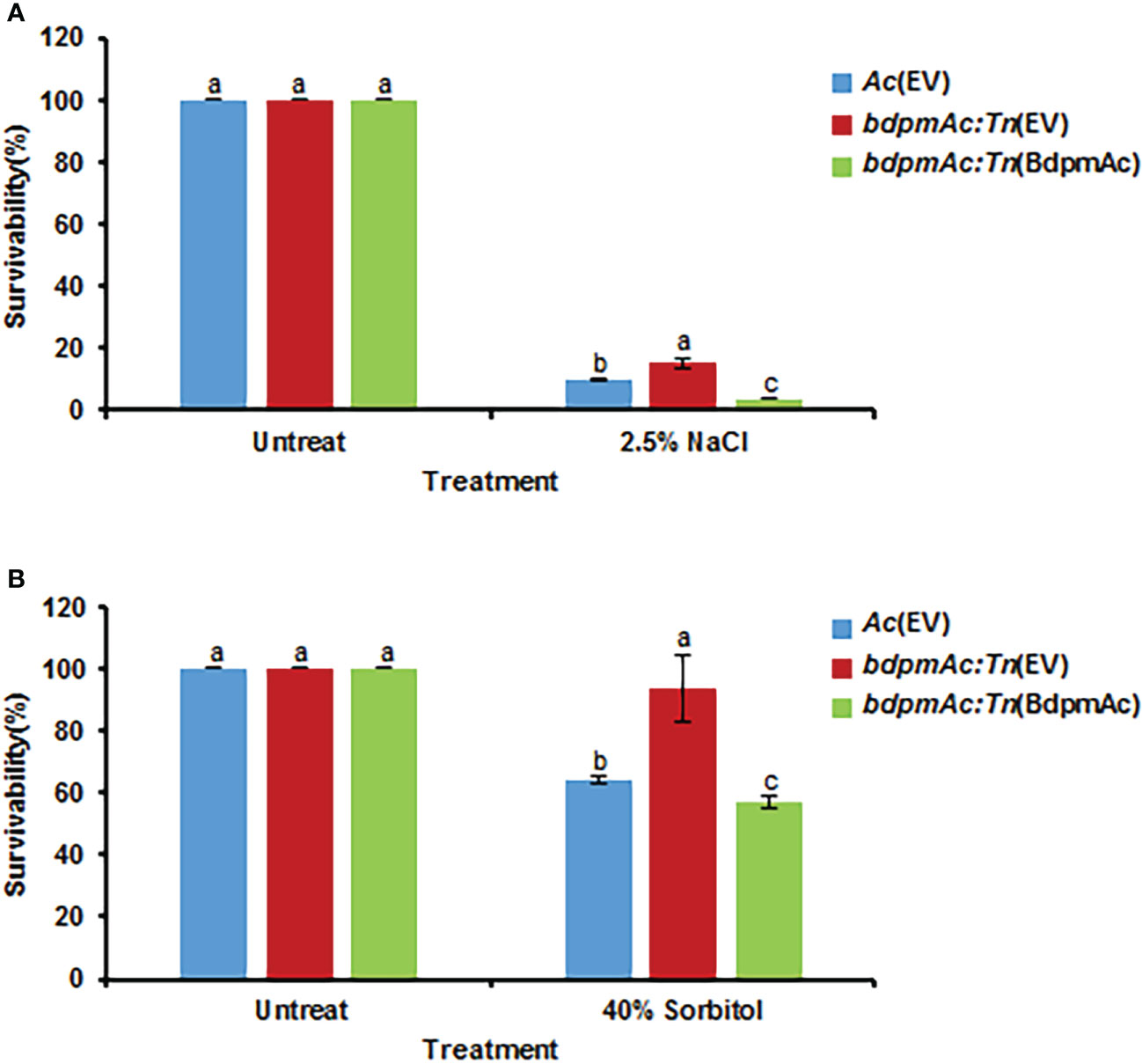
Figure 6 Osmotic stress tolerance. The tolerance of the Ac strains to osmotic stresses was evaluated with (A) 2.5% NaCl and (B) 40% sorbitol in TSB. Bacterial cell survival rates were evaluated via colony counting. The survival rate was calculated as the ratio of the viable cell numbers from the stress conditions to those of the untreated samples. The error bars indicate the standard error of the mean from three biological replicates with three technical replicates and different characters above the bars indicate significant differences according to ANOVA (p<0.05) with Tukey’s HSD Test. Seven and five independent experiments were conducted for NaCl and sorbitol, respectively. All experiments showed similar patterns.
Discussion
PGMs are key enzymes in both the glycolysis and gluconeogenesis pathways. These enzymes can be classified into two groups: iPGMs and dPGMs. Moreover, a previous study reported that iPGM was involved in the virulence of certain bacteria (Radin et al., 2019). However, the involvement of dPGMs in bacterial virulence is rarely reported. Using two different pathogenicity assays, our study demonstrated that BdpmAc, which belongs to the dPGM group, is indispensable for virulence in Ac. Compared with Ac(EV) and bdpmAc:Tn(BdpmAc), our findings also demonstrated that bdpmAc:Tn(EV) did not grow well in M9 medium with glucose or fructose, which are precursors in the glycolysis pathway. In agreement with our observations, a dPGM knockout mutant in Burkholderia phymatum also showed similar growth patterns in a previous study (Chen et al., 2012). Interestingly, bdpmAc:Tn(EV) did not grow in M9 with fructose but exhibited moderate to low growth rates in M9 supplemented with glucose. The glycolysis pathway is the first line in carbohydrate metabolism and it produces pyruvate, which is required to generate ATP via the TCA cycle (Harris and Harper, 2015). Kersters et al. reported that some bacteria were able to use the Entner-Doudoroff pathway, which is another glucose-catabolizing pathway (Kersters and De Ley, 1968). This pathway can produce pyruvate from glucose without PGMs. Interestingly, a previous studies has reported that this pathway was less efficient than the normal glycolysis pathway in terms of ATP production (Stettner and Segrè, 2013). This means that some bacteria possess an alternative and less efficient pathway to use glucose. Therefore, bdpmAc:Tn(EV) was likely able to grow slightly in the presence of glucose without BdpmAc using the aforementioned pathway instead of the normal glycolysis pathway. Phosphogluconate dehydratases and keto-deoxy-phosphogluconate aldolases are known to be required for the Entner-Doudoroff pathway (Conway, 1992). Interestingly, the wild-type Ac strain KACC17005 used in this study possesses homologs for the aforementioned enzymes (ATG92716 and ATG92717) (Park et al., 2017). Therefore, it appears that bdpmAc:Tn(EV) can use the low-efficiency Entner-Doudoroff pathway, resulting in slow growth rates in the presence of glucose. Unlike the above-described alternative glucose pathway, alternative pathways for fructose metabolism have not been reported in bacteria. Therefore, we could deduce that the Ac KACC17005 strain does not possess an alternative pathway for the utilization of fructose because bdpmAc:Tn(EV) is not able to grow in the presence of fructose as a sole carbon source.
Ac(EV) and bdpmAc:Tn(BdpmAc) could grow well in the presence of pyruvate, which is a gluconeogenic and TCA cycle precursor, whereas the growth of bdpmAc:Tn(EV) was inhibited. In a previous study, the growth of a dPGM knockout mutant of B. phymatum was restored when provided with a mixture of both glycolysis and gluconeogenesis precursors in minimal media (Chen et al., 2012). In agreement with this observation, the growth rates of bdpmAc:Tn(EV) were comparable to those of Ac(EV) and bdpmAc:Tn(BdpmAc). These results indicate that BdpmAc is required for both the glycolysis and gluconeogenesis pathways. Additionally, neither of the strains grew well in the presence of mannitol and sucrose, which are high molecular weight compounds. These findings indicate that Ac cannot use high molecular weight compounds for glycolysis and therefore could not utilize either of the aforementioned compounds as a carbon source. Previous studies have also reported that Ac cannot use mannitol and sucrose as sole carbon sources (Melo et al., 2014). Taken together, our findings suggested that Ac possesses an alternative pathway for glucose metabolism, and BdpmAc is a key enzyme for the glycolysis and gluconeogenesis pathways in Ac. The growth of bdpmAc:Tn(EV) in rich media (TSB) was slightly attenuated. Furthermore, in the comparative proteomic analysis, the proteins belonging to group D (cell cycle control/cell division/chromosome partitioning) in Ac were more abundant than those in bdpmAc:Tn. These findings indicate that BdpmAc may affect bacterial multiplication and reproduction. However, the virulence of the mutant was dramatically reduced in both the germinated seed assay and the leaf infiltration assay. Therefore, it can be speculated that there are other mechanisms related to BdpmAc functions that may contribute to the virulence of Ac.
Felgner et al. demonstrated that AroA was required for amino acid synthesis and other cellular processes including virulence in Salmonella enterica serovar Typhimurium (Felgner et al., 2016). Similarly, another study reported that a putative bifunctional chorismate mutase/prephenate dehydratase, CmpAc, is involved in virulence in Ac and other mechanisms such as the biosynthesis of phenylalanine, twitching, biofilm formation, motility, and tolerance to stresses (Kim et al., 2020). These studies demonstrate that one protein is associated with several distinct cellular mechanisms, meaning that it displays pleiotropic effects. Likewise, our comparative proteomic analyses and phenotypic assays also demonstrated that BdpmAc possesses pleiotropic effects in Ac.
Our proteomic analyses revealed that the abundance of diverse proteins categorized in group N (cell motility) was altered by BdpmAc. Similar to other bacteria, Ac also exhibits flagella- and pili-dependent motility. However, Ac strains categorized in Group II exhibited only pili-dependent motility (i.e., twitching motility) but not flagella-dependent motility in laboratory conditions (Bahar et al., 2010). Because the wild-type KACC17005 strain used in this study belongs to Group II (Kim et al., 2020), we characterized twitching motility in rich medium and found that bdpmAc:Tn(EV) exhibited a reduction in twitching motility. This reduction in twitching motility might explain the low virulence of the mutant. Other studies also support this speculation. For instance, the loss of genes associated with twitching motility decreased the virulence of Ac in two previous studies (Bahar et al., 2009; Rosenberg et al., 2018). Twitch motility is known to be involved in biofilm formation, which is an important determinant of virulence in prokaryotic organisms (Davey and O’toole, 2000). A previous study demonstrated that the pgm mutant of Stenotrophomonas maltophilia displayed an impaired biofilm formation capacity (Ramos-Hegazy et al., 2020). In agreement with the aforementioned report, the biofilm formation ability of bdpmAc:Tn(EV) was dramatically reduced compared to the wild-type and the complemented strains. Taken together, our findings suggest that BdpmAc affects twitching motility and biofilm formation, which may in turn affect the virulence of Ac. Interestingly, many proteins classified in Group M (cell wall/membrane/envelope biosynthesis) were identified in the comparative proteomic analysis. Additionally, bdpmAc:Tn(EV) exhibited a higher tolerance against NaCl- and sorbitol-induced osmotic stress, thus demonstrating that BdpmAc is involved in cell wall/membrane functions. Virulence and tolerance to osmotic stress are known to be positively correlated in gram-negative bacteria (Park et al., 2019; Kim et al., 2020). However, the tolerance of bdpmAc:Tn(EV) against osmotic stress was accompanied by a decrease in virulence in our study. Although the mechanisms behind this seemingly conflicting phenomenon remain unclear, we can speculate that BdpmAc is involved in other cellular processes and mechanisms in addition to modulating virulence.
Using comparative proteomic analysis and phenotypic assays, we demonstrated that a putative 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase, BdpmAc, is indispensable for virulence, glycolysis, and gluconeogenesis in Ac. Furthermore, BdpmAc is also related to biofilm formation and twitching motility. We also showed that BdpmAc may possess pleiotropic effects in Ac. Particularly, this study suggests that BdpmAc, which is involved in carbohydrate metabolism, may also be an important virulence factor. Therefore, our findings provide novel insights into the functions of a phosphoglycerate-dependent phosphoglycerate mutase, which could be a potential target to develop anti-virulence agents to control BFB.
Data availability statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2022) partner repository with the dataset identifier PXD035156.
Author contributions
S-WH conceived the study. S-WH and JCL designed the experiments. JCL, JWL, YC, and JC conducted the experiments. JCL and S-WH analyzed the data and prepared the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No. NRF-2020R1A2C1013040), Republic of Korea. This research was also supported by the Chung-Ang University Graduate Research Scholarship in 2022 to JWL.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1039420/full#supplementary-material
Supplementary Figure 1 | Growth assay in TSB and M9 minimal media with mannitol and sucrose. Bacterial growth in (A) TSB and M9 minimal media with (B) mannitol (0.4%) and (C) sucrose (0.4%). The values at OD600nm were evaluated for five and ten days in TSB and minimal media, respectively. The error bars from three biological replicates indicate the standard deviation and different characters above the error bars indicate significant differences according to ANOVA (p<0.05) with Tukey’s HSD test. At least four independent experiments were performed, all of which exhibited similar patterns.
References
Alfano, J. R., Collmer, A. (2004). Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42, 385–414. doi: 10.1146/annurev.phyto.42.040103.110731
Bahar, O., de la Fuente, L., Burdman, S. (2010). Assessing adhesion, biofilm formation and motility of Acidovorax citrulli using microfluidic flow chambers. FEMS Microbiol. Lett. 312, 33–39. doi: 10.1111/j.1574-6968.2010.02094.x
Bahar, O., Goffer, T., Burdman, S. (2009). Type IV pili are required for virulence, twitching motility, and biofilm formation of Acidovorax avenae subsp. citrulli. Mol. Plant-Microbe Interact. 22, 909–920. doi: 10.1094/MPMI-22-8-0909
Bahar, O., Levi, N., Burdman, S. (2011). The cucurbit pathogenic bacterium Acidovorax citrulli requires a polar flagellum for full virulence before and after host-tissue penetration. Mol. Plant-Microbe Interact. 24, 1040–1050. doi: 10.1094/MPMI-02-11-0041
Bucher, T., Oppenheimer-Shaanan, Y., Savidor, A., Bloom-Ackermann, Z., Kolodkin-Gal, I. (2015). Disturbance of the bacterial cell wall specifically interferes with biofilm formation. Environ. Microbiol. Rep. 7, 990–1004. doi: 10.1111/1758-2229.12346
Burdman, S., Kots, N., Kritzman, G., Kopelowitz, J. (2005). Molecular, physiological, and host-range characterization of Acidovorax avenae subsp citrulli isolates from watermelon and melon in Israel. Plant Dis. 89, 1339–1347. doi: 10.1094/PD-89-1339
Chen, W.-M., Prell, J., James, E., Sheu, D.-S., Sheu, S.-Y. (2012). Effect of phosphoglycerate mutase and fructose 1,6-bisphosphatase deficiency on symbiotic Burkholderia phymatum. Microbiol. (Reading England) 158, 1127–1136. doi: 10.1099/mic.0.055095-0
Choi, H., Fermin, D., Nesvizhskii, A. I. (2008). Significance analysis of spectral count data in label-free shotgun proteomics. Mol. Cell. proteomics: MCP 7, 2373–2385. doi: 10.1074/mcp.M800203-MCP200
Conway, T. (1992). The entner-doudoroff pathway: history, physiology and molecular biology. FEMS Microbiol. Rev. 9, 1–27. doi: 10.1111/j.1574-6968.1992.tb05822.x
Craig, L., Pique, M. E., Tainer, J. A. (2004). Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2, 363–378. doi: 10.1038/nrmicro885
Davey, M. E., O'toole, G. A. (2000). Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. reviews: MMBR 64, 847–867. doi: 10.1128/MMBR.64.4.847-867.2000
Davies, D. R., Staker, B. L., Abendroth, J. A., Edwards, T. E., Hartley, R., Leonard, J., et al. (2011). An ensemble of structures of Burkholderia pseudomallei 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase. acta crystallographica. Section F Struct. Biol. crystallization Commun. 67, 1044–1050. doi: 10.1107/S1744309111030405
De Kievit, T. R., Iglewski, B. H. (2000). Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68, 4839. doi: 10.1128/IAI.68.9.4839-4849.2000
Douzi, B., Filloux, A., Voulhoux, R. (2012). On the path to uncover the bacterial type II secretion system. philosophical transactions of the royal society of London. Ser. B Biol. Sci. 367, 1059–1072. doi: 10.1098/rstb.2011.0204
Elias, J. E., Gygi, S. P. (2007). Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214. doi: 10.1038/nmeth1019
Felgner, S., Frahm, M., Kocijancic, D., Rohde, M., Eckweiler, D., Bielecka, A., et al. (2016). aroA-deficient salmonella enterica serovar typhimurium is more than a metabolically attenuated mutant. mBio 7, e01220–e01216. doi: 10.1128/mBio.01220-16
Fraser, H. I., Kvaratskhelia, M., White, M. F. (1999). The two analogous phosphoglycerate mutases of Escherichia coli. FEBS Lett. 455, 344–348. doi: 10.1016/S0014-5793(99)00910-2
Ganapathy-Kanniappan, S., Geschwind, J. F. H. (2013). Tumor glycolysis as a target for cancer therapy: Progress and prospects. Mol. Cancer 12, 152. doi: 10.1186/1476-4598-12-152
Harris, R. A., Harper, E. T. (2015). “Glycolytic pathway,” in eLS (Chichester: John Wiley & Sons, Ltd), 1–8. doi: 10.1002/9780470015902.a0000619.pub3
Islam, R., Hossain, M. R., Kim, H. T., Nath, U. K., Abuyusuf, M., Jung, H. J., et al. (2020). Molecular characterization of Acidovorax citrulli strain NIHHS15-280 causing bacterial fruit blotch disease in Korea and screening of resistance sources in melon. Horticulture Environ. Biotechnol. 61, 115–126. doi: 10.1007/s13580-019-00190-0
Jedrzejas, M. J. (2000). Structure, function, and evolution of phosphoglycerate mutases: comparison with fructose-2,6-bisphosphatase, acid phosphatase, and alkaline phosphatase. Prog. Biophysics Mol. Biol. 73, 263–287. doi: 10.1016/S0079-6107(00)00007-9
Kersters, K., De Ley, J. (1968). The occurrence of the entner-doudoroff pathway in bacteria. Antonie van Leeuwenhoek 34, 393–408. doi: 10.1007/BF02046462
Kim, M., Lee, J., Heo, L., Han, S.-W. (2020). Putative bifunctional chorismate mutase/prephenate dehydratase contributes to the virulence of Acidovorax citrulli. Front. Plant Sci. 11, 569552. doi: 10.3389/fpls.2020.569552
Kovach, M. E., Elzer, P. H., Hill, D. S., Robertson, G. T., Farris, M. A., Roop, R. M., 2nd, et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176. doi: 10.1016/0378-1119(95)00584-1
Lee, J., Heo, L., Han, S. W. (2021). Comparative proteomic analysis for a putative pyridoxal phosphate-dependent aminotransferase required for virulence in Acidovorax citrulli. Plant Pathol. J. 37, 673–680. doi: 10.5423/PPJ.NT.09.2021.0139
Liu, J., Tian, Y. L., Zhao, Y. Q., Zeng, R., Chen, B. H., Hu, B. S., et al. (2019). Ferric uptake regulator (FurA) is required for Acidovorax citrulli virulence on watermelon. Phytopathology 109, 1997–2008. doi: 10.1094/PHYTO-05-19-0172-R
Mapes, J. P., Harris, R. A. (1976). Inhibition of gluconeogenesis and lactate formation from pyruvate by N6, O2'-dibutyryl adenosine 3':5'-monophosphate. J. Biol. Chem. 251, 6189–6196. doi: 10.1016/S0021-9258(20)81843-2
Melo, L. A., Tebaldi, N. D., Mehta, A., Marques, A. S. A. (2014). Comparing Acidovorax citrulli strains from melon and watermelon: Phenotypic characteristics, pathogenicity and genetic diversity. Trop. Plant Pathol. 39, 154–162. doi: 10.1590/S1982-56762014000200006
Park, H. J., Jung, B., Lee, J., Han, S. W. (2019). Functional characterization of a putative DNA methyltransferase, EadM, in Xanthomonas axonopodis pv. glycines by proteomic and phenotypic analyses. Sci. Rep. 9, 2446. doi: 10.1038/s41598-019-38650-3
Park, H. J., Seong, H. J., Lee, J., Heo, L., Sul, W. J., Han, S. W. (2021). Two DNA methyltransferases for site-specific 6mA and 5mC DNA modification in Xanthomonas euvesicatoria. Front. Plant Sci. 12, 621466. doi: 10.3389/fpls.2021.621466
Park, H.-J., Seong, H. J., Sul, W. J., Oh, C.-S., Han, S.-W. (2017). Complete genome sequence of Acidovorax citrulli strain KACC17005, a causal agent for bacterial fruit blotch on watermelon. Korean J. Microbiol. 53, 340–341. doi: 10.7845/kjm.2017.7084
Perez-Riverol, Y., Bai, J., Bandla, C., Garcia-Seisdedos, D., Hewapathirana, S., Kamatchinathan, S., et al. (2022). The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552. doi: 10.1093/nar/gkab1038
Radin, J. N., Kelliher, J. L., Solórzano, P. K. P., Grim, K. P., Ramezanifard, R., Slauch, J. M., et al. (2019). Metal-independent variants of phosphoglycerate mutase promote resistance to nutritional immunity and retention of glycolysis during infection. PloS Pathog. 15, e1007971–e1007971. doi: 10.1371/journal.ppat.1007971
Rahimi-Midani, A., Choi, T. J. (2020). Transport of phage in melon plants and inhibition of progression of bacterial fruit blotch. Viruses-Basel 12, v12040477. doi: 10.3390/v12040477
Ramos-Hegazy, L., Chakravarty, S., Anderson, G. G. (2020). Phosphoglycerate mutase affects Stenotrophomonas maltophilia attachment to biotic and abiotic surfaces. Microbes Infection 22, 60–64. doi: 10.1016/j.micinf.2019.08.001
Rojas, E. R., Billings, G., Odermatt, P. D., Auer, G. K., Zhu, L., Miguel, A., et al. (2018). The outer membrane is an essential load-bearing element in gram-negative bacteria. Nature 559, 617–621. doi: 10.1038/s41586-018-0344-3
Rosenberg, T., Salam, B. B., Burdman, S. (2018). Association between loss of type IV pilus synthesis ability and phenotypic variation in the cucurbit pathogenic bacterium Acidovorax citrulli. Mol. Plant-Microbe Interact. 31, 548–559. doi: 10.1094/MPMI-12-17-0324-R
Rui, L. Y. (2014). Energy metabolism in the liver. Compr. Physiol. 4, 177–197. doi: 10.1002/cphy.c130024
Schaad, N. W., Sowell, G., Goth, R. W., Colwell, R. R., Webb, R. E. (1978). Pseudomonas pseudoalcaligenes subsp citrulli sub-Nov. Int. J. Systematic Bacteriology 28, 117–125. doi: 10.1099/00207713-28-1-117
Shrestha, R. K., Rosenberg, T., Makarovsky, D., Eckshtain-Levi, N., Zelinger, E., Kopelowitz, J., et al. (2013). Phenotypic variation in the plant pathogenic bacterium Acidovorax citrulli. PloS One 8, 0073189. doi: 10.1371/journal.pone.0073189
Silva, G. M., Souza, R. M., Yan, L. C., Junior, R. S., Medeiros, F. H. V., Walcott, R. R. (2016). Strains of the group I lineage of Acidovorax citrulli, the causal agent of bacterial fruit blotch of cucurbitaceous crops, are predominant in Brazil. Phytopathology 106, 1486–1494. doi: 10.1094/PHYTO-05-16-0205-R
Stettner, A. I., Segrè, D. (2013). The cost of efficiency in energy metabolism. Proc. Natl. Acad. Sci. 110, 9629. doi: 10.1073/pnas.1307485110
Tatusov, R. L., Galperin, M. Y., Natale, D. A., Koonin, E. V. (2000). The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28, 33–36. doi: 10.1093/nar/28.1.33
Wang, T., Guan, W., Huang, Q., Yang, Y., Yan, W., Sun, B., et al. (2016). Quorum-sensing contributes to virulence, twitching motility, seed attachment and biofilm formation in the wild type strain AAC-5 of Acidovorax citrulli. Microbial Pathogenesis 100, 133–140. doi: 10.1016/j.micpath.2016.08.039
Wang, Y. J., Zhao, Y. Q., Xia, L. M., Chen, L., Liao, Y. J., Chen, B. H., et al. (2022). yggS encoding pyridoxal 5 '-phosphate binding protein is required for Acidovorax citrulli virulence. Front. Microbiol. 12, 783862. doi: 10.3389/fmicb.2021.783862
Zhang, Y. H., Foster, J. M., Kumar, S., Fougere, M., Carlow, C. K. S. (2004). Cofactor-independent phosphoglycerate mutase has an essential role in Caenorhabditis elegans and is conserved in parasitic nematodes. J. Biol. Chem. 279, 37185–37190. doi: 10.1074/jbc.M405877200
Zhang, X., Zhao, M., Yan, J., Yang, L., Yang, Y., Guan, W., et al. (2018). Involvement of hrpX and hrpG in the virulence of Acidovorax citrulli strain Aac5, causal agent of bacterial fruit blotch in cucurbits. Front. Microbiol. 9, 507–507. doi: 10.3389/fmicb.2018.00507
Keywords: acidovorax citrulli, bacterial fruit blotch, proteomics, 3-bisphosphoglycerate-dependent phosphoglycerate mutase, virulence
Citation: Lee J, Lee J, Cho Y, Choi J and Han S-W (2022) A putative 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase is involved in the virulence, carbohydrate metabolism, biofilm formation, twitching halo, and osmotic tolerance in Acidovorax citrulli. Front. Plant Sci. 13:1039420. doi: 10.3389/fpls.2022.1039420
Received: 08 September 2022; Accepted: 14 October 2022;
Published: 09 November 2022.
Edited by:
Hyong Woo Choi, Andong National University, South KoreaReviewed by:
Rikky Rai, University of Florida, United StatesMee Kyung Sang, National Institute of Agricultural Science, South Korea
Copyright © 2022 Lee, Lee, Cho, Choi and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sang-Wook Han, c3doYW5AY2F1LmFjLmty
 Jongchan Lee
Jongchan Lee Jeongwook Lee
Jeongwook Lee Yongmin Cho
Yongmin Cho Junhyeok Choi
Junhyeok Choi Sang-Wook Han
Sang-Wook Han