- Department of Agriculture, Forestry, and Bioresources, Research Institute of Agriculture and Life Science, Plant Genomics and Breeding Institute, College of Agriculture and Life Science, Seoul National University, Seoul, South Korea
Several genes regulating capsaicinoid biosynthesis including Pun1 (also known as CS), Pun3, pAMT, and CaKR1 have been studied. However, the gene encoded by Pun2 in the non-pungent Capsicum chacoense is unknown. This study aimed to identify the Pun2 gene by genetic mapping using interspecific (C. chacoense × Capsicum annuum) and intraspecific (C. chacoense × C. chacoense) populations. QTL mapping using the interspecific F2 population revealed two major QTLs on chromosomes 3 and 9. Two bin markers within the QTL regions on two chromosomes were highly correlated with the capsaicinoid content in the interspecific population. The major QTL, Pun2_PJ_Gibbs_3.11 on chromosome 3, contained the pAMT gene, indicating that the non-pungency of C. chacoense may be attributed to a mutation in the pAMT gene. Sequence analysis revealed a 7 bp nucleotide insertion in the 8th exon of pAMT of the non-pungent C. chacoense. This mutation resulted in the generation of an early stop codon, resulting in a truncated mutant lacking the PLP binding site, which is critical for pAMT enzymatic activity. This insertion co-segregated with the pungency phenotype in the intraspecific F2 population. We named this novel pAMT allele pamt11. Taken together, these data indicate that the non-pungency of C. chacoense is due to the non-functional pAMT allele, and Pun2 encodes the pAMT gene.
Introduction
Pepper (Capsicum spp.) is the only natural resource producing the pungency principle capsaicinoid. Capsaicinoids are secondary metabolites in pepper that function as protection against insects and microorganisms (Tewksbury and Nabhan, 2001; Levey et al., 2006). Capsaicinoids have been used as food additives, medicine, and personal defense tools due to their pungency and their pharmacological and chemical effects (Meghvansi et al., 2010; Khan et al., 2014; Srinivasan, 2016). Some pepper cultivars produce capsinoids, which have similar chemical structure and properties to capsaicinoids, but these are almost non-pungent (Tanaka et al., 2009). Capsinoids as well as capsaicinoids have been proven to exhibit anti-cancer, anti-obesity, anti-diabetic, and antioxidant activities (Uarrota et al., 2021).
Capsaicinoids are conjugated compounds of precursors derived from the phenylpropanoid pathway and the branched-chain fatty acid pathway. In the phenylpropanoid pathway, phenylamine is converted to precursors by phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H), 4-coumaroyl-CoA ligase (4CL), p-coumaroyl shikimate/quinate 3-hydroxylase (C3H), caffeoyl-CoA 3-O-methyltransferase (COMT), and putative aminotransferase (pAMT), resulting in vanillylamine (Bennett and Kirby, 1968; Leete and Louden, 1968; Sukrasno and Yeoman, 1993; Curry et al., 1999). In the branched-chain fatty acid pathway, the final products are produced from either valine or leucine by a series of enzymes including acetolactate synthase (ALS), branched-chain amino acid aminotransferase (BCAT), ketoacyl-ACT synthase (KAS), acyl carrier protein (ACL), ketoacyl-ACP reductase (KR1), and acyl-ACP thioesterase (FAT). The products of the two pathways, vanillylamine and the branched-chain fatty acids, are condensed by capsaicin synthase (CS), an acyltransferase 3 (AT3) also known as Pun1, resulting in different capsaicinoids depending on the type of branched-chain fatty acids (Suzuki et al.,, 1981; Harwood, 1996; Markai et al., 2002; Koeda et al., 2019).
Although capsaicinoid biosynthesis has long been studied, the structural genes involved in the pathway have not been fully elucidated (Aza-González et al., 2011). The Pun1 gene was considered the only gene that affects the loss of pungency in pepper until two other loci, Pun2 and Pun3, were identified (Stellari et al., 2010; Han et al., 2019). Pun1 encodes a putative acyltransferase, which is the last enzyme in capsaicinoid biosynthesis. The encoded protein is called capsaicin synthase (CS) because vanillylamine from the phenylpropanoid pathway and the branched-chain fatty acid are condensed at the ends of two pathways. Stellari et al. (2010) discovered Pun2, a second locus affecting pungency in pepper, which was not allelic to the Pun1 locus. However, the gene encoded by the Pun2 locus is not known. The Pun3 gene encodes the transcription factor CaMYB31, which plays a role as a master regulator of capsaicinoid biosynthetic genes in pepper including Pun1 (Arce-Rodriguez and Ochoa-Alejo, 2017; Han et al., 2019; Zhu et al., 2019). The putative aminotransferase (pAMT) encodes a protein that catalyzes the formation of vanillylamine from vanillin in the phenylpropanoid pathway (Lang et al., 2009). Recently, a putative ketoacyl-ACP reductase (CaKR1) in the branched-chain fatty acid pathway was found to be related to pungency in Capsicum chinense (Koeda et al., 2019).
Capsaicinoid content is a quantitative trait in pepper that is regulated by multiple genes (Sanatombi and Sharma, 2008), and many studies have used QTL mapping to identify the genetic factors that regulate capsaicinoid content (Ben‐Chaim et al., 2006; Blum et al., 2003; Yarnes et al., 2013). The genes underlying the QTL often correspond to known structural genes (Han et al., 2018).
Recessive alleles of these genes explain the loss of pungency in Capsicum species. The first reported mutant allele of Pun1 was identified with a large 2.5 kb deletion spanning 1.8 kb of the putative promoter and 0.7 kb of the truncated first exon (Stewart et al., 2005). This deletion was widespread throughout Capsicum annuum cultivars, especially in bell peppers. The second Pun1 allele has a 4 bp deletion in the first exon region that creates an early stop codon in C. chinense (Stewart et al., 2007). In Pun13 reported in Capsicum frutescens, a premature stop codon leads to truncation in the second exon (Stellari et al., 2010). Pun14 was identified to have a single nucleotide insertion in the second exon, which generates a frameshift mutation (Kirii, et al., 2017). In non-pungent Capsicum chacoense, a recessive pun2 allele is thought to be the reason for the non-pungency (Stellari et al., 2010). The mutant allele of Pun3 was found in the non-pungent C. annuum accession with a nonsense mutation in the first exon, which leads to a premature stop codon (Han et al., 2019). More than 10 nonfunctional alleles of pAMT have been reported and were found to cause loss of pungency in Capsicum species from the results of Lang et al. (2009) that reported a nonsense mutation at pAMT in C. annuum ‘CH-19 Sweet.’ A mutation in pAMT alters capsaicinoids into capsinoids (Sutoh et al., 2006; Lang et al., 2009). Capsinoid biosynthesis is also regulated by Pun1 (Han et al., 2013).
The Pun2 locus has not been studied since it was first reported (Stellari et al., 2010). The Pun2 locus was suggested as a novel locus of pungency based on the complementation tests of Capsicum chacoense PI260433-np with other known Pun1 alleles, including Pun11, Pun12, Pun13, and Pun14 (Stewart et al., 2005; Stewart et al., 2007; Stellari et al., 2010; Kirii et al., 2018). By bulk segregant analysis, the marker ‘Hpms1-172’ was identified and was evidence that the Pun2 locus was located on the upper arm of chromosome 7 (Stellari et al., 2010).
In this study, we attempted to identify the gene encoded by the Pun2 locus using two F2 populations. First, we performed QTL mapping in an interspecific F2 population derived from a cross between C. annuum and C. chacoense PI260433-np, which contains the recessive pun2 allele. As a result, one major and one minor QTL that affected non-pungency capsaicinoid content were identified. A candidate gene in the major QTL was nominated and mapped in an intraspecific F2 population derived from a cross between the pungent and the non-pungent C. chacoense accessions. The candidate gene mapping revealed that the Pun2 locus encodes pAMT.
Materials and methods
Plant materials and mapping population constructions
A non-pungent Capsicum species indigenous to South America, C. chacoense ‘PI260433-np,’ and a pungent Korean landrace C. annuum ‘Jeju’ were used as parental lines to construct an interspecific F2 population (PJ) for QTL analysis. PI260433-np is known to be non-pungent despite a functional allele of Pun1 (Stellari et al., 2010). A total of 178 F2 plants were grown in the field of Seoul National University farm (Suwon, Republic of Korea) in 2019. The non-pungent PI260433-np was crossed with the pungent ‘PI260433-p’ to construct an intraspecific F2 population (PP). C. chacoense PI260433 is a wild pepper that shows a polymorphism in fruit pungency (Tewksbury et al., 2006; Tewksbury et al., 2008). A total of 113 F2 plants from a cross between PI260433-p and PI260433-np were grown in the greenhouse at Seoul National University (Suwon, Korea) from 2021 to 2022. An allelism test was carried out by crossing C. chacoense ‘PI260433-np’ with non-pungent cultivars C. annuum ‘YCM334’ and C. annuum ‘ECW30R.’ If two recessive genes are allelic, they will fail to complement each other in the F1 hybrids.
Genomic DNA (gDNA) extraction
gDNA was extracted from young leaves of plant materials using a modified cetyltrimethylammonium bromide (CTAB) method (Han et al., 2018). Young leaf tissues were frozen in liquid nitrogen and finely ground using a 5 mm steel bead with a vortex mixer (DAIHAN Scientific, Wonju, Korea). The concentration and quality of extracted gDNA were measured with a Nanodrop spectrophotometer (BioTek, Winooski, VT, USA) and diluted to a final concentration of 20 ng/μL in triple distilled water (TDW).
Phenotyping with Gibb’s screening and HPLC analysis
Mature green fruits harvested from each plant were sampled. Placenta tissues were used for Gibb’s analysis (Jeong et al., 2012). Placenta tissues were placed on filter paper and the same volume of 2,6-dichloroquinon-4-chloroimide (Gibb’s reagent; Sigma-Aldrich, Saint Louis, MI, USA) was sprayed. The filter paper with sprayed spots was steamed for 30 s with ammonia gas. If the color changed to blue, the samples were determined to be pungent. For HPLC analysis, at least three pepper fruits at the mature green (MG) stage were harvested, and the placental tissue was separated from each fruit. For the ‘PP’ F2 population, harvest was done in the same manner, but the whole fruits including seeds and the pericarp were for HPLC analysis. Pre-processed samples were analyzed by HPLC at the National Instrumentation Center for Environmental Management (NICEM; Seoul, Korea) to measure the capsaicinoid and capsinoid contents. Total capsaicinoids were calculated as the sum of capsaicin and dihydrocapsaicin content, and total capsinoids were calculated as the sum of capsiate and dihydrocapsiate contents.
GBS library preparation and sequencing
Genomic DNA of ‘PJ’ F2 individuals and three replications of each parent were used to construct the Illumina sequencing library for GBS followed by Truong et al. (2012). Genomic DNAs were digested with EcoRI and MseI, and then MseI adapters and EcoRI adapters with different barcodes were used to ligate DNA fragments. Each sample with the same quantity of adapter-ligated DNA fragments was pooled for sequencing. Single-end sequencing was performed on one line of an Illumina Hiseq 2000 (Illumina, San Diego, CA, USA) at Macrogen Inc. (Seoul, Republic of Korea).
SNP analysis
The adapter and barcodes were removed using CLC genomic workbench software version 8.0 (CLC Bio, Aarhus, Denmark). The reference genome of C. annuum ‘Dempsey’ (Lee et al., 2022 was used to align trimmed reads using Burrows-Wheeler Aligner version 0.7.12 (Li, 2013). To convert the alignment files into BAM files, Sequence Alignment/Map (SAM) tools version 1.1 was used. Picard Tools version 1.119 was then used to manipulate the SAM files and perform duplicate marking and sorting. The Genome Analysis Toolkit (GATK) UnifiedGenotyper version 3.3, with the criteria of a QUAL value larger than 30 and a minimum depth of 3, was used to further sort and filter SNPs.
Bin map construction
The construction of a bin map was done using the modified sliding-window approach to reduce variant calling errors and the calculation of recombination breakpoints (Han et al., 2016). SNPs with non-polymorphic and missing data were removed. The ratio of SNPs with both parental genotypes was calculated for each window, defined as 25 linked SNPs, and the overall genotype of each window was decided. SNPs with ratios over 0.7 were defined as paternal and maternal genotypes; an SNP ratio between 0.3 and 0.7 was defined as heterozygous genotype. Construction of linkage maps was conducted with Carthagene software (De Givry et al., 2005). The criteria for the construction of a linkage map were a LOD score threshold of 4.0 and a maximum distance of 50 cM. The calculation of distance between bin markers was done with the Kosambi mapping function. The final linkage map was drawn using MapChart2.3 software (Voorips, 2002). The C. annuum ‘Dempsey’ reference genome was used to construct the bin map. The comparison with the physical location of bin markers was carried out using MapChart2.3 software.
Genetic mapping of Pun2
A high-density genetic map from GBS data and phenotype data for capsaicin and dihydrocapsaicin contents in placenta tissue were used for QTL analysis. Composite interval mapping (CIM) was performed using Windows QTL cartographer v2.5 (Wang et al., 2012). The LOD threshold was determined using 1,000 permutations with a 5% probability for each chromosome and trait. The phenotypic variation proportion explained by each QTL was explained and estimated using the R2 (%) value.
Sequence analysis and marker tests
PCR was performed using EzPCR™ XO 5x Master Mix (Elpis, Korea) with 100 ng of gDNA template and 0.5 μL of 10 pmol gene-specific primers for Pun1, Pun3, CaKR1, and pAMT in a total volume of 25 μL. The PCR conditions were as follows: one cycle of pre-denaturation at 95°C for 3 min and 25–28 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 1 min. The resulting amplicons were separated on a 1% agarose gel. Sanger sequencing was performed at Macrogen (Seoul, Korea), and the nucleotide sequences were analyzed by Lasergene’s SeqMan program (DNASTAR, Madison, WI, USA). For genotyping of a novel pamt allele, a KASP marker set based on an indel in the 8th exon of pAMT was developed: 5’-(FAM)-GCTTCCTCCAATGAATCAAGCATCA-3’ (FAM-tailed forward primer), 5’-(HEX)-GCTTCCTCCAATGCATCAAAAATTGA-3’, and 5’- CCTGGCAAGTGATAGGCCCAATAA-3’ (common reverse primer). Thermocycling and endpoint genotyping for the KASP assays were performed in a Roche LC480 (Roche Applied Science, Indianapolis, IN, USA). The composition of the KASP mixture was as follows: 100 ng of plant DNA was mixed with 5 µL KASP V4.0 2X Mastermix (LGC Genomics, UK), 0.14 µL of the gene-specific KASP primer mix, and 0.06 µL MgCl2. The thermal cycling condition was 94°C hot-start for 15 min, followed by 10 cycles of 94°C for 20 s and 59°C for 60 s; the annealing temperature was decreased at a rate of 0.2°C/cycle, followed by 26 cycles at 94°C for 20 s and 57°C for 60 s and 3 cycles of 94°C for 20 s and 59°C for 60 s. The cooling cycle was at 37°C for 2 min, and the final fluorescent value was read at the same temperature as for endpoint genotyping.
Results
Characterization of parental lines
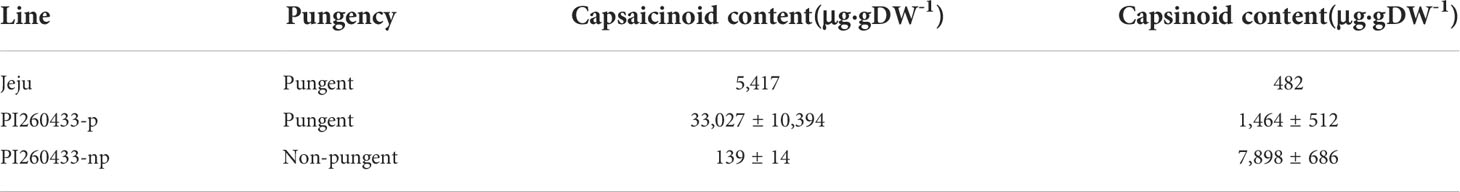
In HPLC analysis, the capsaicinoid content of Jeju was 5,417.5 2 μg·gDW-1 in placenta tissue and capsinoid content was 482.2 μg·gDW-1. The pungent PI260433-p contained 33,027 μg·gDW-1 of capsaicinoids, whereas the non-pungent line PI260433-np contained 139 μg·gDW-1 of capsaicinoids (Table 1). In contrast, the total capsinoid content in PI260433-np was higher than the content in PI260433-p: the total capsinoid content in PI260433-p and PI260433-np was 1,464 ± 512 μg·gDW-1 and 7,898 ± 686 μg·gDW-1, respectively. If Pun1 or Pun3 was not functioning in PI260433-np, which might result in no capsaincinoid accumulation, there would be no capsinoid biosynthesis either. These data indicate that the low pungency in PI260433-np might be related to a mutation of pAMT.
Phenotyping and genotyping of an interspecific population
For evaluation of the pungency in the ‘PJ’ F2 population, Gibb’s analysis was first conducted at least two times using mature green stage fruits. If the color of samples turned blue, it was determined to be pungent, while samples with no color changes were determined to be non-pungent. Of 171 F2 samples, 116 were pungent, and 55 samples were non-pungent. The segregation ratio of non-pungent vs. pungent peppers in the F2 population was 2.11:1 (Table S1). For more accurate phenotype analysis, HPLC analysis of 120 F2 plants was conducted to confirm the Gibb’s analysis results. The mean capsaicinoid content of the placenta tissue of the F2 population was 1,561 ± 193 μg·gDW-1and the mean capsinoid content was 534 ± 64 μg·gDW-1 (Table 2). Plants with a capsaicinoid content more than 100 μg·gDW- were considered pungent, whereas plants with a content less than 100 μg·gDW- were considered non-pungent. The segregation ratio in phenotype by HPLC analysis was 1.72:1 (Table S1). The distribution of both capsaicinoid and capsinoid content exhibited a positive skew; the positive skew implies the mean is greater than the median (Figure 1).
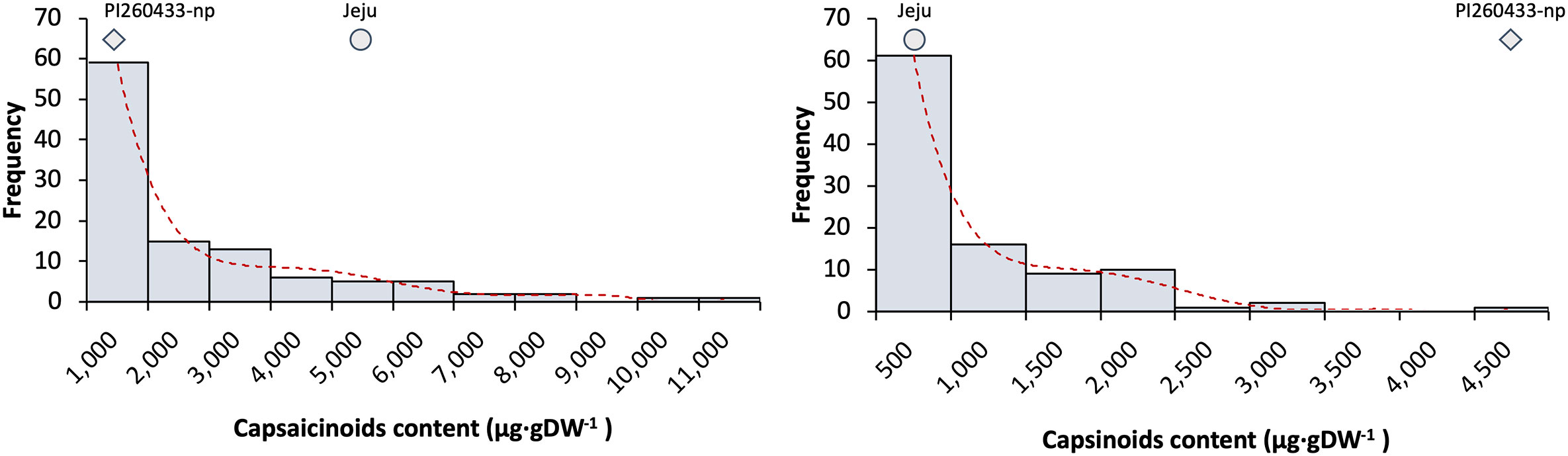
Figure 1 Frequency distribution of capsaicinoid and capsinoid contents in the ‘PJ’ F2 population. The x-axis indicates the number of plants (frequency), and the y-axis indicates the range of capsaicinoid (left) or capsinoid (right) amounts.
Genotyping of the ‘PJ’ F2 population was carried out with GBS. EcoR1/Mse1-digested DNA was used for the construction of GBS libraries. The average number of reads per sample was 2,803,611, and the reads were aligned to the C. annuum ‘Dempsey’ reference genome (Lee et al., 2022). A total of 10,136 SNPs was obtained from GBS analysis (Table S2). The density of SNPs showed differences among the chromosomes (Figure S1).
To construct a linkage map, the modified sliding window approach was used to correct missing data and genotyping errors (Huang et al., 2009; Chen et al., 2014; Han et al., 2016). To determine recombination breakpoints, 25 consecutive SNPs were considered as one sliding window. CarthaGene software was used to construct a genetic linkage map. The map consisted of 1,422 bins with an average genetic distance of 3.86 cM. Among the 12 linkage groups, the genetic distance of chromosome 4 was the longest at 752 cM, and the distance of chromosome 2 was the shortest at 234.1 cM (Table S3). The bin map developed using 1,422 bin markers was aligned to the C. annuum ‘Dempsey’ reference genome to compare the physical position of the bins (Figure S2).
QTLs associated with capsaicinoid content
QTLs controlling the total capsaicinoid content were detected in the ‘PJ’ F2 population. Two phenotype data sets and a high-density bin map were used to identify genetic factors. The QTLs associated with the pungency level by Gibbs’s analysis were mainly located on chromosomes 3 and 9, as shown in Figure 2A. A total of 13 and 14 QTLs for the capsaicinoid contents were detected on chromosomes 3 and 9, respectively (Table 3). Of 27 QTLs, 18 showed negative additive effects. Among all detected QTLs, Pun2_PJ_Gibbs_3.11 on chromosome 3 showed the highest LOD and R2 value, explaining 69% of the total phenotypic variations in the ‘PJ’ F2 population. To compare the physical location of QTLs, the C. annuum ‘Dempsey’ reference genome (Lee et al., 2022) was used. By comparing the physical positions of the detected QTLs on chromosomes 3 and 9 with the copies of the pAMT gene, we found that the major QTL Pun2_PJ_Gibbs_3.11 was located adjacent to the position of the copy of the pAMT gene on chromosome 3 (Figure S3). To validate the detected QTLs, phenotype data from HPLC analysis were used. Although several relatively high peaks were observed on three chromosomes including 3 and 9, no significant QTLs were detected, as was detected by Gibbs’s analysis (Figure 2B).
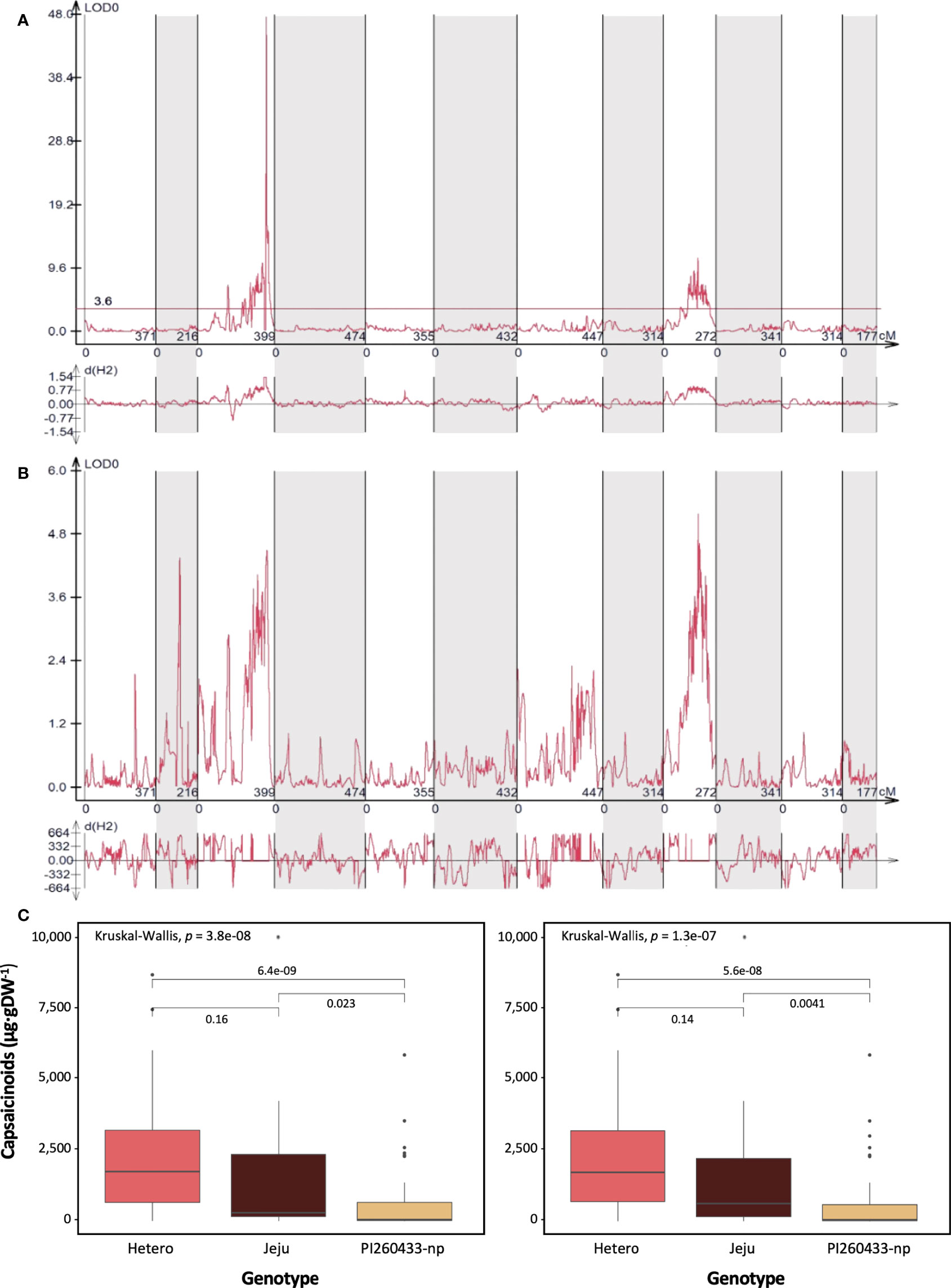
Figure 2 QTL mapping of capsaicinoid content in the ‘PJ’ F2 population. (A) The detected QTLs were associated with the phenotypic data from Gibb’s analysis. The most dominant QTL was located on chromosome 3, and the other QTL was detected on chromosome 9. (B) The detected QTLs were associated with the phenotypic data from HPLC analysis. No significant QTLs were detected. (C) Comparison of QTLs and SNPs associated with capsaicinoid content and box plots of capsaicinoid content regulated by two markers representing the two QTLs. Bin markers used for box plot were Bin7_25_1M_249000000 (left) and Bin10_25_1M_204000000 (right) in the ‘PJ’ F2 population.
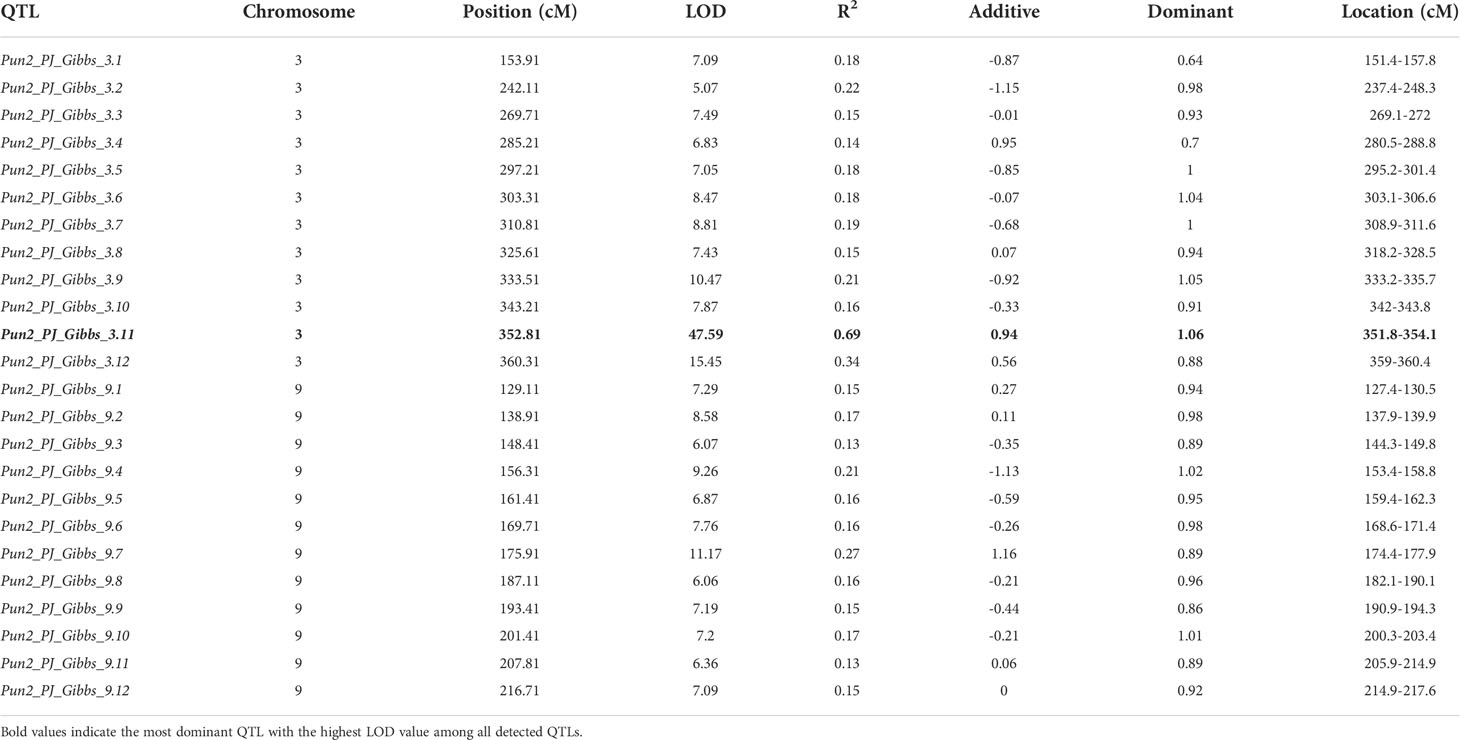
Table 3 Quantitative trait loci (QTL) associated with the pungency level measured by Gibbs’s analysis of the placental tissues in the ‘PJ’ F2 population.
To validate the effect of the QTLs, individual plants in the ‘PJ’ F2 population were sorted into three groups according to genotype (‘PI260433-np’, ‘Jeju,’ or heterozygote) at bin markers located within the QTLs. The box plot was drawn with Bin7_25_1M_249000000 for the QTL detected on chromosome 3 and Bin10_25_1M_204000000 for the QTL detected on chromosome 9. All markers on both chromosomes were highly associated with the capsaicinoid content (Figure 2C). This result indicated that the QTLs of the pungency level measured by Gibb’s analysis correlated with the actual capsaicinoid content.
Identification of nucleotide sequence variations in Pun1, Pun3, CaKR1, and pAMT
To confirm whether pAMT, which is located at the major QTL position on chromosome 3, contributes to the difference in pungency of C. chacoense, we investigated the genetic variations of the pAMT gene between the two C. chacoense accessions, PI260433-p and PI260433-np. The full-length sequences of pAMT were obtained by Sanger sequencing and compared, and a 7 bp insertion (5’-AATCAAG-3’) in the 8th exon of pAMT was found in PI260433-np but not in PI260433-p (Figure 3A). This insertion caused an early stop codon, resulting in a truncated translation product lacking the pyridoxal 5-phosphate (PLP) binding domain vital for pAMT enzymatic activity (Figure 3B). This mutation has never been reported in Capsicum spp. and was named pamt11. There were no variations in the gDNA sequence between PI260433-p and PI260433-np in other structural genes including Pun1, Pun3, and CaKR1 except for several SNPs and InDels that altered some amino acid sequences (Figures S4–S6). Therefore, the difference in fruit pungency between PI260433-np and PI260433-p may due to the mutation of pAMT.
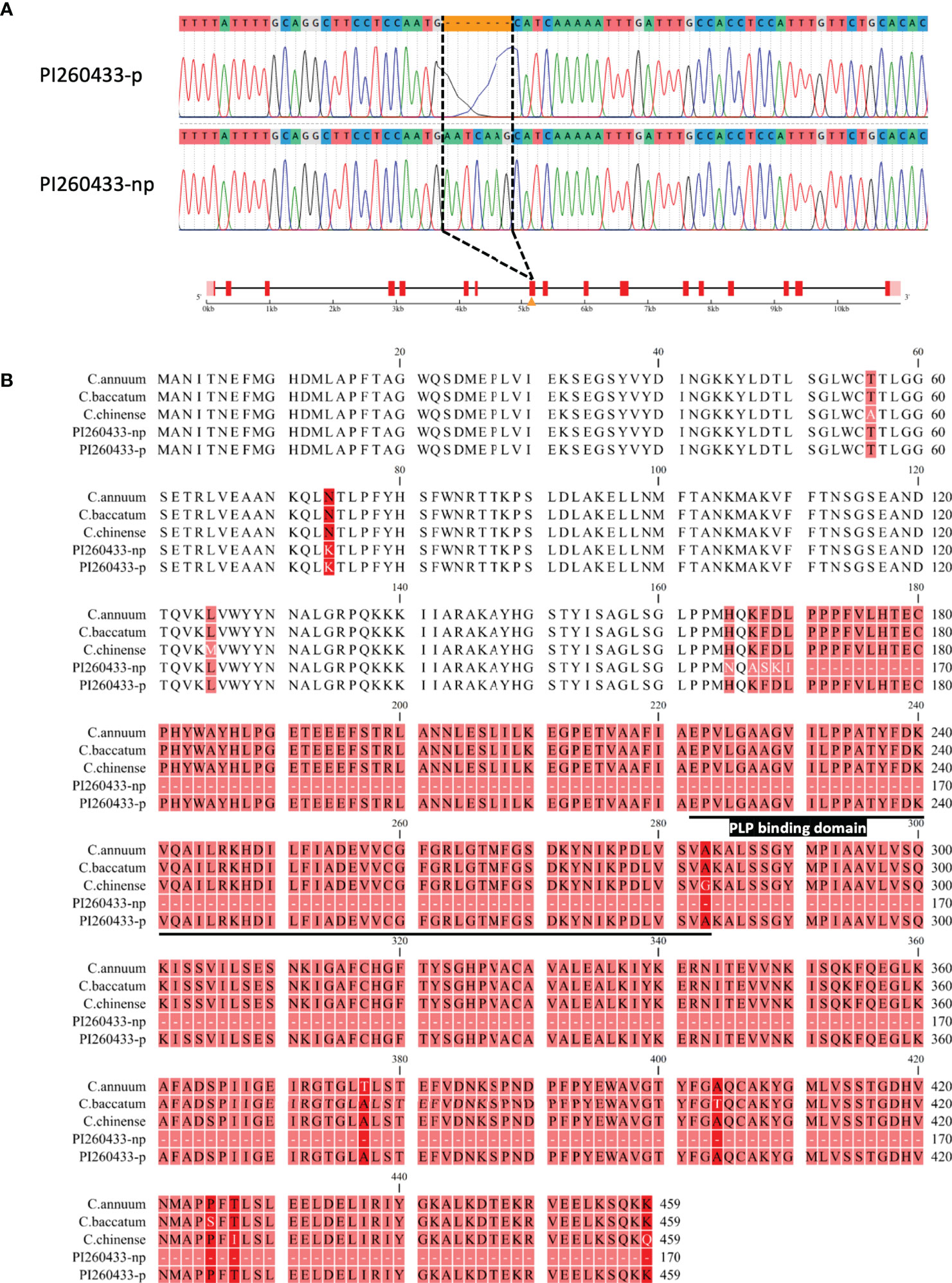
Figure 3 Schematic structure of pamt11. (A) Gene structure of pAMT. A sequence of 7 bp (5’-AATCAAG-3’) is inserted in the 8th exon of pAMT in PI260433-np. Boxes and lines indicate exons and introns, respectively. (B) Amino acid sequences of translated proteins of pAMT. The pyridoxal 5-phosphate (PLP) binding domain is underlined.
Co-segregation of a novel pamt allele with capsinoid contents in an F2 population
A KASP marker based on the novel 7 bp indel of pAMT was designed to test the association of this mutation and pungency in an F2 mapping population obtained from a cross between PI260433-p and PI260433-np. A total of 113 F2 plants was genotyped by KASP assay. The F2 population was segregated into three genotypes, two homozygous genotypes (pAMTP/pAMTP, pAMTNP/pAMTNP), and one heterozygous genotype (pAMTP/pAMTNP). The segregation ratio of three genotypes, pAMTP/pAMTP, pAMTP/pAMTNP, and pAMTNP/pAMTNP, in the F2 population was 43:41:29 (or 1.48:1.41:1). The HPLC analysis of the ‘PP’ F2 population showed that the segregation ratio of phenotype was 2.88:1 (pungent: non-pungent) with a p-value of 0.86 (>0.05) (Table 4). Comparison between thepungency phenotype and the genotype of pAMT confirmed that the capsaicinoid and capsinoid contents co-segregated with pamt11 (Figure 4). Taken together, these results suggest that the non-pungency of C. chacoense ‘PI260433-np’ is due to pamt11, and Pun2 encodes the pAMT gene.
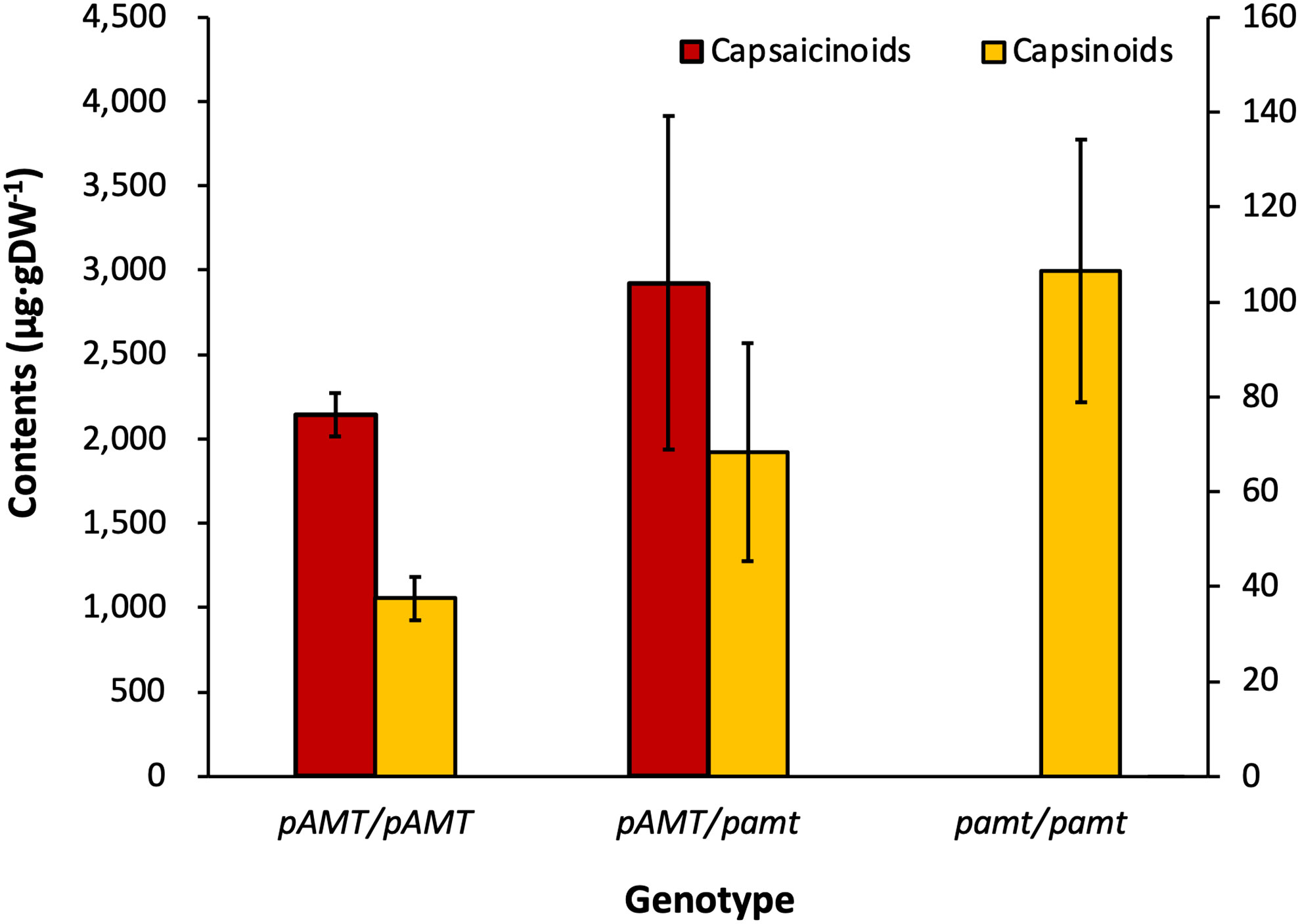
Figure 4 Capsaicinoid and capsinoid contents by genotypes of pAMT in the ‘PP’ F2 population. The capsaicinoid content was not able to be determined in the group of homozygous pamt11, whereas the capsinoid content was the largest among all genotype groups.
Discussion
The pungency of pepper fruit is controlled by single genes and QTLs. The pAMT gene was proposed to encode a protein that catalyzes vanillin to vanillylamine in the capsaicinoid biosynthesis pathway (Curry et al., 1999; Blum et al., 2003; Abraham-Juárez et al., 2008). A mutation of pAMT leads to a loss of pungency with the production of capsinoids instead of capsaicinoids (Sutoh et al., 2006; Lang et al., 2009). The loss of the pungency gene pun2 was previously reported in C. chacoense ‘PI260433-np’ (Stellari et al., 2010). In this study, we found that PI260433-np contained approximately 200 times lower levels of capsaicinoids and approximately 5 times higher levels of capsinoids than PI260433-p. PI260433-np did not result in a complete loss of pungency, as a small amount of capsaicinoids was detected, demonstrating that Pun1 in PI260433-np encodes a functional protein. pAMT mutants produce a trace amount of capsaicinoids, whereas Pun1 mutants completely lose pungency (Han et al., 2013; Tanaka et al., 2015). However, PI260433-np produced a notable amount of capsaicinoids, which indicated a malfunctioning pAMT gene.
To locate the Pun2 locus, we first performed QTL mapping using an interspecific population derived from a cross between the non-pungent C. chacoense ‘PI260433-np’ and pungent C. annuum ‘Jeju,’ and two major QTLs were detected on chromosomes 3 and 9, where homologs of pAMT are located. One major QTL on chromosome 3 was located at the exact position of one of the pAMT copies. On chromosome 9, there was another copy of pAMT, but the position did not exactly match the QTL region. We postulated that the pAMT homolog on chromosome 3 might be the Pun2 gene. To test this hypothesis, a candidate analysis using pAMT was performed, and the results identified a 7 bp insertion at the 8th exon in pAMT of ‘PI260433-np.’ This insertion causes a frameshift mutation leading to early termination of pAMT translation. This pAMT allele, named pamt11, co-segregated with the non-pungency in the F2 population derived from a cross between PI260433-p and PI260433-np. In this population, the non-pungency phenotype was segregated as a single gene rather than a quantitative trait, as was observed in the interspecific population. Interestingly, the capsaicinoid content was higher in heterozygous genotype of pAMT/pamt than in homozygous pAMT/pAMT. This may be due to the heterotic effect in intraspecific population (Naves et al., 2022). To date, several copies of pAMT have been reported: two on chromosome 3, one on chromosome 7, and one on chromosome 9 (Blum et al., 2003; Mazourek et al., 2009; Scossa et al., 2019). The positions of pAMT copies are frequently associated QTLs for capsaicinoid content. Ben-Chaim et al., 2006) reported two QTLs associated with the capsaicinoid content on chromosome 7 in an interspecific population derived from C. annuum and C. frutescens (Ben-Chaim et al., 2006). The position of one of two QTLs, cap7.2, was near pAMT on chromosome 7. Han et al. (2018) also reported that one of the QTLs for capsaicinoid content corresponded to the pAMT gene on chromosome 3. These results demonstrate that the pAMT genes on different chromosomes affect capsaicinoid biosynthesis.
We noted that the Pun2 position reported in our study is different from previous reports. Stellari et al. (2010) reported that Pun2 was located on the upper arm of chromosome 7; however, they were unable to show the exact position of the gene because the lack of polymorphic markers around the gene hindered mapping.
No nonfunctional pAMT gene in C. chacoense has been reported. Three and one pAMT mutant alleles were reported in C. annuum and C. frutescens, respectively (Lang et al., 2009; Tanaka et al., 2010a; Park et al., 2015; Tsurumaki and Sasanuma, 2019). Six pAMT mutant alleles were found in C. chinense (Tanaka et al., 2010b; Koeda et al., 2014; Tanaka et al., 2015; Tanaka et al., 2018). As C. chacoense is a wild Capsicum species, this species may have untapped traits that might be lost during the domestication of other Capsicum species (Mongkolporn and Taylor, 2011). The nonfunctional pAMT allele may be a valuable genetic resource for the breeding of high capsinoids cultivars.
In summary, we discovered a novel non-functional pAMT allele and identified the Pun2 locus, which may encode the pAMT gene. This novel allele may be used as a breeding material to produce capsinoids in chili peppers.
Data availability statement
The data presented in the study are deposited in the NABIC repository (https://nabic.rda.go.kr/nolog/NV-0748-000001/snpVcfView.do), accession number NV-0748.
Author contributions
SY and D-GL contributed equally as the first authors of this work. SY wrote the manuscript and carried out the phenotyping and genotyping in the intraspecific population, the development of the molecular marker set, and data analysis. At the same time, D-GL conducted QTL mapping in the interspecific population. SB contributed to phenotyping, data analysis, and the development of figure sets. J-PH and SJ contributed to data processing and analysis. B-CK supervised the overall processes and revised the manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2021R1A2C2007472). This work was supported by a grant from the ‘New Breeding Technologies Development Program (Project No. PJ0165432022)’, Rural Development Administration, Republic of Korea.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1039393/full#supplementary-material
Supplementary Figure 1 | SNP densities of the ‘PJ’ F2 population. The scale bar on the right indicates the number of SNPs within a 1 Mb window size.
Supplementary Figure 2 | Comparison of the genetic map of the ‘PJ’ F2 population with the physical map. The scale bar on the left shows the genetic map position (cM).
Supplementary Figure 3 | The comparison between the detected QTLs and the pAMT genes on chromosomes 3 and 9. The yellow box and the red arrow indicate the location of the QTL and the pAMT gene, respectively.
Supplementary Figure 4 | Amino acid sequence of the translated protein of Pun1. Amino acid sequences were obtained from the reference genome of C. annuum ‘Takanotsume,’ C. baccatum ‘PBC81,’ and C. chinense ‘Habanero.’ The different residues are marked in white text in black boxes.
Supplementary Figure 5 | Amino acid sequence of the translated protein of Pun3. Amino acid sequences were obtained from the reference genomes of C. annuum ‘Takanotsume’ and C. baccatum ‘PBC81.’ The different residues are marked in white text in black boxes.
Supplementary Figure 6 | Amino acid sequences of the translated protein of CaKR1. Amino acid sequences were obtained from the reference genomes of C. annuum ‘Takanotsume,’ C. baccatum ‘PBC81,’ and C. chinense ‘Habanero.’ The different residues are marked in white text in black boxes.
References
Abraham-Juárez, M., Rocha-Granados, M., López, M. G., Rivera-Bustamante, R. F., Ochoa-Alejo, N., et al (2008). Virus-induced silencing of Comt, pAmt and Kas genes results in a reduction of capsaicinoid accumulation in chili pepper fruits. Planta 227 (3), 681–695. doi: 10.1007/s00425-007-0651-7
Arce-Rodríguez, M. L., Ochoa-Alejo, N. (2017). An R2R3-MYB transcription factor regulates capsaicinoid biosynthesis. Plant Physiol. 174 (3), 1359–1370. doi: 10.1104/pp.17.00506
Aza-González, C., Núñez-Palenius, H. G., Ochoa-Alejo, N. (2011). Molecular biology of capsaicinoid biosynthesis in chili pepper (Capsicum spp.). Plant Cell Rep. 30 (5), 695–706. doi: 10.1007/s00299-010-0968-8
Bennett, D. J., Kirby, G. W. (1968). Constitution and biosynthesis of capsaicin. J. Chem. Soc. C.: Organ. 4, 442–446. doi: 10.1039/j39680000442
Ben-Chaim, A., Borovsky, Y., Falise, M., Mazourek, M., Kang, B. C., Paran, I., et al. (2006). QTL analysis for capsaicinoid content in Capsicum. Theor. Appl. Genet. 113 (8), 1481–1490.
Blum, E., Mazourek, M., O’connell, M., Curry, J., Thorup, T., Liu, K., et al. (2003). Molecular mapping of capsaicinoid biosynthesis genes and quantitative trait loci analysis for capsaicinoid content in Capsicum. Theor. Appl. Genet. 108 (1), 79–86. doi: 10.1007/s00122-003-1405-y
Chen, Z., Wang, B., Dong, X., Liu, H., Ren, L., Chen, J., et al. (2014). An ultra-high density bin-map for rapid QTL mapping for tassel and ear architecture in a large F2 maize population. BMC Genomics 15 (1), 1–10.
Curry, J., Aluru, M., Mendoza, M., Nevarez, J., Melendrez, M., O’Connell, M. A. (1999). Transcripts for possible capsaicinoid biosynthetic genes are differentially accumulated in pungent and non-pungent capsicum spp. Plant Sci. 148 (1), 47–57. doi: 10.1016/S0168-9452(99)00118-1
De Givry, S., Bouchez, M., Chabrier, P., Milan, D., Schiex, T. (2005). CARHTA GENE: multipopulation integrated genetic and radiation hybrid mapping. Bioinformatics 21 (8), 1703–1704. doi: 10.1093/bioinformatics/bti222
Han, K., Jang, S., Lee, J. H., Lee, D. G., Kwon, J. K., Kang, B. C. (2019). A MYB transcription factor is a candidate to control pungency in Capsicum annuum. Theor. Appl. Genet. 132 (4), 1235–1246. doi: 10.1007/s00122-018-03275-z
Han, K., Jeong, H. J., Sung, J., Keum, Y. S., Cho, M. C., Kim, J. H., et al. (2013). Biosynthesis of capsinoid is controlled by the Pun1 locus in pepper. Mol. Breed. 31 (3), 537–548. doi: 10.1007/s11032-012-9811-y
Han, K., Jeong, H. J., Yang, H. B., Kang, S. M., Kwon, J. K., Kim, S., et al. (2016). An ultra-high-density bin map facilitates high-throughput QTL mapping of horticultural traits in pepper (Capsicum annuum). DNA Res. 23 (2), 81–91.
Han, K., Lee, H. Y., Ro, N. Y., Hur, O. S., Lee, J. H., Kwon, J. K., et al. (2018). QTL mapping and GWAS reveal candidate genes controlling capsaicinoid content in Capsicum. Plant Biotechnol. J. 16 (9), 1546–1558. doi: 10.1111/pbi.12894
Harwood, J. L. (1996). Recent advances in the biosynthesis of plant fatty acids. Biochim. Biophys. Acta (BBA)-Lipids. Lipid Metab. 1301 (1-2), 7–56. doi: 10.1016/0005-2760(95)00242-1
Huang, X., Feng, Q., Qian, Q., Zhao, Q., Wang, L., Wang, A., et al. (2009). High-throughput genotyping by whole-genome resequencing. Genome Res. 19 (6), 1068–1076.
Jeong, H. J., Hwang, D. Y., Ahn, J. T., Chun, J. Y., Han, K. E., Lee, W. M., et al. (2012). Development of a simple method for detecting capsaicinoids using gibb’s reagent in pepper. Hortic. Sci. Technol. 30 (3), 294–300. doi: 10.7235/hort.2012.12011
Khan, F. A., Mahmood, T., Ali, M., Saeed, A., Maalik, A. (2014). Pharmacological importance of an ethnobotanical plant: Capsicum annuum l. Natural Product. Res. 28 (16), 1267–1274. doi: 10.1080/14786419.2014.895723
Kirii, E., Goto, T., Yoshida, Y., Yasuba, K. I., Tanaka, Y. (2017). Non-pungency in a Japanese chili pepper landrace (Capsicum annuum) is caused by a novel loss-of-function Pun1 allele. Hortic. J. 86 (1), 61–69.
Kirii, E., Goto, T., Yoshida, Y., Yasuba, K. I., Tanaka, Y. (2018). Nonpungency in a Japanese chili pepper landrace (Capsicum annuum) is caused by a novel loss-of-function Pun1 allele. Hortic. J. MI–148. doi: 10.2503/hortj.MI-148
Koeda, S., Sato, K., Saito, H., Nagano, A. J., Yasugi, M., Kudoh, H., et al. (2019). Mutation in the putative ketoacyl-ACP reductase CaKR1 induces loss of pungency in Capsicum. Theor. Appl. Genet. 132 (1), 65–80. doi: 10.1007/s00122-018-3195-2
Koeda, S., Sato, K., Tomi, K., Tanaka, Y., Takisawa, R., Hosokawa, M., et al. (2014). Analysis of non-pungency, aroma, and origin of a Capsicum chinense cultivar from a Caribbean island. J. Japanese. Soc. Hortic. Sci. 83, 244–251. doi: 10.2503/jjshs1.CH-105
Lang, Y., Kisaka, H., Sugiyama, R., Nomura, K., Morita, A., Watanabe, T., et al. (2009). Functional loss of pAMT results in biosynthesis of capsinoids, capsaicinoid analogs, in capsicum annuum cv. CH-19 sweet. Plant J. 59 (6), 953–961. doi: 10.1111/j.1365-313X.2009.03921.x
Lee, J. H., Venkatesh, J., Jo, J., Jang, S., Kim, G. W., Kim, J. M., et al. (2022). High-quality Chromosome-scale Genomes Facilitate Effective Identification of Large Structural Variations in Hot and Sweet Peppers. Hortic. Res. doi: 10.1093/hr/uhac210
Leete, E., Louden, M. C. (1968). Biosynthesis of capsaicin and dihydrocapsaicin in Capsicum frutescens. J. Am. Chem. Soc. 90 (24), 6837–6684. doi: 10.1021/ja01026a049
Levey, D. J., Tewksbury, J. J., Cipollini, M. L., Carlo, T. A. (2006). A field test of the directed deterrence hypothesis in two species of wild chili. Oecologia 150 (1), 61–68. doi: 10.1007/s00442-006-0496-y
Markai, S., Marchand, P. A., Mabon, F., Baguet, E., Billault, I., Robins, R. J. (2002). Natural deuterium distribution in branched-chain medium-length fatty acids is nonstatistical: A site-specific study by quantitative 2H NMR spectroscopy of the fatty acids of capsaicinoids. ChemBioChem 3 (2-3), 212–218. doi: 10.1002/1439-7633(20020301)3:2/3<212::AID-CBIC212>3.0.CO;2-R
Mazourek, M., Pujar, A., Borovsky, Y., Paran, I., Mueller, L., Jahn, M. M., et al (2009). A dynamic interface for capsaicinoid systems biology. Plant Physiol. 150 (4), 1806–1821. doi: 10.1104/pp.109.136549
Meghvansi, M. K., Siddiqui, S., Khan, M. H., Gupta, V. K., Vairale, M. G., Gogoi, H. K., et al. (2010). Naga chilli: a potential source of capsaicinoids with broad-spectrum ethnopharmacological applications. J. Ethnopharmacol. 132 (1), 1–14. doi: 10.1016/j.jep.2010.08.034
Mongkolporn, O., Taylor, P. W. (2011). “Capsicum,” in Wild crop relatives: genomic and breeding resources (Berlin, Heidelberg: Springer), 43–57.
Naves, E. R., Scossa, F., Araújo, W. L., Nunes-Nesi, A., Fernie, A. R., Zsögön, A. (2022). Heterosis for capsacinoids accumulation in chili pepper hybrids is dependent on parent-of-origin effect. Sci. Rep. 12 (1), 1–9. doi: 10.1038/s41598-022-18711-w
Park, Y. J., Nishikawa, T., Minami, M., Nemoto, K., Iwasaki, T., Matsushima, K. (2015). A low-pungency S3212 genotype of Capsicum frutescens caused by a mutation in the putative aminotransferase (p-AMT) gene. Mol. Genet. Genomics 290 (6), 2217–2224. doi: 10.1007/s00438-015-1071-1
Sanatombi, K., Sharma, G. J. (2008). Capsaicin content and pungency of different Capsicum spp. cultivars. Notulae Botanicae Horti Agrobotanici Cluj-Napoca 36 (2), 89–90.
Scossa, F., Roda, F., Tohge, T., Georgiev, M. I., Fernie, A. R. (2019). The hot and the colorful: understanding the metabolism, genetics and evolution of consumer preferred metabolic traits in pepper and related species. Crit. Rev. Plant Sci. 38 (5-6), 339–381. doi: 10.1080/07352689.2019.1682791
Srinivasan, K. (2016). Biological activities of red pepper (Capsicum annuum) and its pungent principle capsaicin: a review. Crit. Rev. Food Sci. Nutr. 56 (9), 1488–1500. doi: 10.1080/10408398.2013.772090
Stellari, G. M., Mazourek, M., Jahn, M. M. (2010). Contrasting modes for loss of pungency between cultivated and wild species of Capsicum. Heredity 104 (5), 460–471. doi: 10.1038/hdy.2009.131
Stewart, C., Jr., Kang, B. C., Liu, K., Mazourek, M., Moore, S. L., Yoo, E. Y., et al. (2005). The Pun1 gene for pungency in pepper encodes a putative acyltransferase. Plant J. 42 (5), 675–688. doi: 10.1111/j.1365-313X.2005.02410.x
Stewart, C., Jr., Mazourek, M., Stellari, G. M., O’Connell, M., Jahn, M. (2007). Genetic control of pungency in c. chinense via the Pun1 locus. J. Exp. Bot. 58 (5), 979–991. doi: 10.1093/jxb/erl243
Sukrasno, N., Yeoman, M. M. (1993). Phenylpropanoid metabolism during growth and development of Capsicum frutescens fruits. Phytochemistry 32 (4), 839–844. doi: 10.1016/0031-9422(93)85217-F
Sutoh, K., Kobata, K., Yazawa, S., Watanabe, T. (2006). Capsinoid is biosynthesized from phenylalanine and valine in a non-pungent pepper, Capsicum annuum l. cv. CH-19 sweet. Biosci. Biotechnol. Biochem. 70 (6), 1513–1516. doi: 10.1271/bbb.50665
Suzuki, T., Kawada, T., Iwai, K., et al (1981). Biosynthesis of acyl moieties of capsaicin and its analogues from valine and leucine in capsicum fruits. Plant Cell Physiol. 22, 23–32. doi: 10.1093/oxfordjournals.pcp.a076142
Tanaka, Y., Fukuta, S., Koeda, S., Goto, T., Yoshida, Y., Yasuba, K. I. (2018). Identification of a novel mutant pAMT allele responsible for low-pungency and capsinoid production in chili pepper: accession ‘No. 4034’(Capsicum chinense). Hortic. J. 87 (2), 222–228. doi: 10.2503/hortj.OKD-115
Tanaka, Y., Hosokawa, M., Miwa, T., Watanabe, T., Yazawa, S. (2010a). Newly mutated putative-aminotransferase in nonpungent pepper (Capsicum annuum) results in biosynthesis of capsinoids, capsaicinoid analogues. J. Agric. Food Chem. 58 (3), 1761–1767. doi: 10.1021/jf903282r
Tanaka, Y., Hosokawa, M., Miwa, T., Watanabe, T., Yazawa, S. (2010b). Novel loss-of-function putative aminotransferase alleles cause biosynthesis of capsinoids, nonpungent capsaicinoid analogues, in mildly pungent chili peppers (Capsicum chinense). J. Agric. Food Chem. 58 (22), 11762–11767. doi: 10.1021/jf1019642
Tanaka, Y., Hosokawa, M., Otsu, K., Watanabe, T., Yazawa, S. (2009). Assessment of capsiconinoid composition, nonpungent capsaicinoid analogues, in Ccapsicum cultivars. J. Agric. Food Chem. 57 (12), 5407–5412. doi: 10.1021/jf900634s
Tanaka, Y., Sonoyama, T., Muraga, Y., Koeda, S., Goto, T., Yoshida, Y., et al. (2015). Multiple loss-of-function putative aminotransferase alleles contribute to low pungency and capsinoid biosynthesis in Capsicum chinense. Mol. Breed. 35 (6), 1–13. doi: 10.1007/s11032-015-0339-9
Tewksbury, J. J., Manchego, C., Haak, D. C., Levey, D. J. (2006). Where did the chili get its spice? biogeography of capsaicinoid production in ancestral wild chili species. J. Chem. Ecol. 32 (3), 547–564. doi: 10.1007/s10886-005-9017-4
Tewksbury, J. J., Nabhan, G. P. (2001). Directed deterrence by capsaicin in chillies. Nature 412 (6845), 403–404. doi: 10.1038/35086653
Tewksbury, J. J., Reagan, K. M., Machnicki, N. J., Carlo, T. A., Haak, D. C., Peñaloza, A. L. C., et al. (2008). Evolutionary ecology of pungency in wild chilies. Proc. Natl. Acad. Sci. U.S.A. 105 (33), 11808–11811. doi: 10.1073/pnas.0802691105
Truong, H. T., Ramos, A. M., Yalcin, F., de Ruiter, M., van der Poel, H. J., Huvenaars, K. H., et al. (2012). Sequence-based genotyping for marker discovery and co-dominant scoring in germplasm and populations. PLos ONE 7 (5), e37565. doi: 10.1371/journal.pone.0037565
Tsurumaki, K., Sasanuma, T. (2019). Discovery of novel unfunctional pAMT allele pamt10 causing loss of pungency in sweet bell pepper (Capsicum annuum l.). Breed. Sci. 69 (1), 133–142. doi: 10.1270/jsbbs.18150
Uarrota, V. G., Maraschin, M., de Bairros, A. D. F. M., Pedreschi, R. (2021). Factors affecting the capsaicinoid profile of hot peppers and biological activity of their non-pungent analogs (Capsinoids) present in sweet peppers. Crit. Rev. Food Sci. Nutr. 61 (4), 649–665. doi: 10.1080/10408398.2020.1743642
Voorrips, R. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Heredity. 93 (1), 77–78. doi: 10.1093/jhered/93.1.77
Wang, S. C. J. B. (2005) Windows QTL cartographer 2.5. Available at: http://statgen.ncsu.edu/qtlcart/WQTLCart.htm.
Wang, S., Basten, C. J., Zeng, Z.-B. (2012). Windows QTL Cartographer 2.5 (Raleigh, NC: Department of Statistics, North Carolina State University). Available at: http://statgen.ncsu.edu/qtlcart/WQTLCart.htm.
Yarnes, S. C., Ashrafi, H., Reyes-Chin-Wo, S., Hill, T. A., Stoffel, K. M., Van Deynze, A. (2013). Identification of QTLs for capsaicinoids, fruit quality, and plant architecture-related traits in an interspecific Capsicum RIL population. Genome 56 (1), 61–74.
Keywords: Capsaicinoid biosynthesis, Pun2, pAMT, Capsicum chacoense, QTL mapping
Citation: Yi S, Lee D-G, Back S, Hong J-P, Jang S, Han K and Kang B-C (2022) Genetic mapping revealed that the Pun2 gene in Capsicum chacoense encodes a putative aminotransferase. Front. Plant Sci. 13:1039393. doi: 10.3389/fpls.2022.1039393
Received: 08 September 2022; Accepted: 05 October 2022;
Published: 01 November 2022.
Edited by:
Congli Wang, Northeast Institute of Geography and Agroecology, (CAS), ChinaReviewed by:
Zhangsheng Zhu, South China Agricultural University, ChinaNeftali Ochoa-Alejo, Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional, Mexico
Zhaoming Qi, Northeast Agricultural University, China
Copyright © 2022 Yi, Lee, Back, Hong, Jang, Han and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Byoung-Cheorl Kang, Yms1NEBzbnUuYWMua3I=
 Shieun Yi
Shieun Yi Do-Gyeong Lee
Do-Gyeong Lee Seungki Back
Seungki Back Ju-Pyo Hong
Ju-Pyo Hong Siyoung Jang
Siyoung Jang Byoung-Cheorl Kang
Byoung-Cheorl Kang