- 1Center for Informational Biology, School of Life Science and Technology, University of Electronic Science and Technology of China, Chengdu, China
- 2School of Life and Environmental Sciences, Plant Breeding Institute, The University of Sydney, Cobbitty, NSW, Australia
- 3Characteristic Crops Research Institute, Chongqing Academy of Agricultural Sciences, Yongchuan, China
- 4Crop Research Institute, Sichuan Academy of Agricultural Sciences, Chengdu, China
- 5School of Agriculture, Food and Wine, The University of Adelaide, Waite, Glen Osmond, SA, Australia
Rye 6R-derived stripe rust resistance gene Yr83 in wheat background was physically mapped to fraction length (FL) 0.87-1.00 on the long arm by non-denaturing-fluorescence in situ hybridization (ND-FISH), Oligo-FISH painting and 6R-specific PCR markers.Stripe rust resistance gene Yr83 derived from chromosome 6R of rye (Secale cereale) “Merced” has displayed high resistance to both Australian and Chinese wheat stripe rust isolates. With the aim to physically map Yr83 to a more precise region, new wheat- 6R deletion and translocation lines were produced from derived progenies of the 6R(6D) substitution line. The non-denaturing fluorescence in situ hybridization (ND-FISH) patterns of 6R were established to precisely characterize the variations of 6R in different wheat backgrounds. Comparative ND-FISH analysis localized the breakpoints of 6RL chromosomes relative to Oligo-pSc200 and Oligo-pSc119.2 rich sites in deletion lines. Molecular marker and resistance analyses confirmed that Yr83 is physically located at the fraction length (FL) 0.87-1.00 of 6RL and covers the corresponding region of 806-881 Mb in the reference genome of Lo7. Oligo-FISH painting demonstrated that the region carrying Yr83 is syntenic to the distal end of long arm of homoeologous group 7 of the Triticeae genome. The developed wheat-6R lines carrying the Yr83 gene will be useful for breeding for rust resistance.
Introduction
Stripe rust (yellow rust), caused by Puccinia striiformis f. sp. tritici (Pst), is an important disease that occurs in most global wheat-growing regions (Morgounov et al., 2012; Wang and Chen, 2017). Cultivation of resistant cultivars is economical and environmentally friendly compared to chemical control. Eighty-four stripe rust resistance genes have been identified and many are being used in wheat breeding (McIntosh et al., 2021; Klymiuk et al., 2022). However, rapid emergence of virulent Pst races has rendered many resistance genes in released cultivars ineffective, which has prompted the continuing search for more durable resistance (Kou and Wang, 2010; Schwessinger, 2017). Breeding durable disease resistant cultivars relies largely on continually discovering and introducing new resistance genes into adapted varieties, especially the genes from wild species (Ellis et al., 2014; Zhang et al., 2018).
Cultivated rye (Secale cereale L.) has been a valuable source of potentially useful genes for wheat improvement (Rabinovich, 1997; Tang et al., 2011). However, reports on stripe rust resistance genes derived from Secale species are far behind the other foliar disease resistance genes identified (Spetsov and Daskalova, 2022). The chromosome arm 1RS from ‘Petkus’ rye was introduced into many wheat cultivars grown worldwide and carries stripe rust resistance gene Yr9 (Mago et al., 2005), which is no longer effective in most locations. Several rye chromosomes were found to have novel stripe rust resistance and have been incorporated into wheat backgrounds (Wang et al., 2009; Li et al., 2016; Schneider et al., 2016; An et al., 2019). Among them, chromosome 6R has displayed a number of polymorphic variations, and the 6R derived from different origins has been found to possess multiple disease-resistance genes. Several wheat-6R introgression lines with novel disease resistance gene(s) have been developed (Friebe et al., 1996; Dundas et al., 2001; An et al., 2015; Hao et al., 2018; Li et al., 2020b; Duan et al., 2022; Ashraf et al., 2022).
Asiedu et al. (1990) developed a wheat-chromosome 6R substitution line Sub6R(6D) from triticale T-701, derived from rye “Merced”, and later Dundas et al. (2001) localized the nematode resistance gene CreR to an interstitial region in the long arm of 6R. Li et al. (2020b) found that the Sub6R(6D) were highly resistant to both stripe rust and powdery mildew pathogens, and physically located the new stripe rust resistance gene Yr83 on FL 0.73-1.00 of 6RL by FISH on wheat-rye 6R translocation and deletion lines. In this study, the physical location of Yr83 on 6RL was further narrowed down by molecular cytogenetic approaches with the information from the available whole genomic sequences of Secale genomes (Bauer et al., 2017; Li et al., 2021; Rabanus-Wallace et al., 2021).
The objectives of this study were to identify new wheat - 6R deletion and translocation lines and further physically localize Yr83 gene on the specific region by FISH and molecular markers, and assess the developed wheat-6R introgression lines for wheat breeding.
Materials and methods
Plant materials
The triticale line T-701 was originally developed by CIMMYT (Li et al., 2020b), while the wheat-S. cereale 6R(6D) substitution line (Sub6R(6D)) and deletion line T6RL22 were developed at University of Adelaide, Australia (Asiedu et al., 1990; Dundas et al., 2001; Li et al., 2020a). CS-Imperial rye 6R addition line was obtained from Dr. Bernd Friebe, Wheat Genetics Resource Center, Department of Plant Pathology, Kansas State University, USA. S. cereale cv. Kustro and Weining were kindly provided by Prof. Zongxiang Tang and Huaiyu Zhang at Sichuan Agricultural University, China; Wheat cv. Mianyang 11 (MY11) and Chuanmai 42 (CM42) are maintained in our laboratory at the School of Life Science and Technology, University of Electronic Science and Technology of China.
Fluorescence in situ hybridization (FISH)
Root tips from germinated seeds were collected and treated with nitrous oxide followed by enzyme digestion, using the procedure of Han et al. (2006). Synthesized oligo-nucleotide probes Oligo-pSc200, Oligo-pSc119.2, and Oligo-pTa535 were used for identifying wheat chromosomes following the descriptions of Tang et al. (2014) and Fu et al. (2015). A new probe Oligo-248 was developed from tandem repeats database on B2DSC website (Lang et al., 2019). The tandem repeat-based oligo-nucleotide probes Oligo-k288 (Wang et al., 2019) and Oligo-D (Tang et al., 2018) for ND-FISH are listed in Table 1. After FISH using the above oligos as probes, the sequential FISH painting with bulked oligos was conducted following the description by Li and Yang (2022). Photomicrographs of FISH chromosomes were taken with an Olympus BX-53 microscope equipped with a DP-70 CCD camera. Images were processed using Photoshop 3.0 (Adobe Systems Incorporated, CA, USA).
Molecular marker analysis
DNA was extracted from young leaves using a sodium dodecyl sulfate (SDS) protocol (Li et al., 2015). The PCR markers in rye (Qiu et al., 2016), 6R-specific markers (Li et al., 2016; Du et al., 2018), PCR-based Landmark Unique Gene (PLUG) primers (Ishikawa et al., 2009; Li et al., 2013), CINAU markers (Zhang et al., 2017), and rye Lo7 genomic region-specific markers were based on searching the website of the Triticeae Multi-omics Center (http://202.194.139.32/). All primers were synthesized by Shanghai Invitrogen Biotechnology Co. Ltd. Amplified PCR products were electrophoresed on a 1.0% agarose gel as described by Li et al. (2015). The physical locations of the molecular markers on chromosomes were based on the reference genomes of Chinese Spring wheat (IWGSC 2018) and Lo7 rye (Rabanus-Wallace et al., 2021).
Stripe rust reactions
Stripe rust reactions were observed in the field at the Sichuan Academy of Agricultural Sciences Experimental Station. Ten plants were grown per 1-m row with a 25-cm spacing between rows. Bread wheat cv. MY11 planted on both sides of each experimental row served as an inoculum spreader and susceptible control after inoculation with a mixture of races CYR32, 33 and 34. Reactions evaluated at the heading and grain-filling stages were recorded on a 0–4 infection type (IT) scale according to Bariana and McIntosh (1993).
Results
Karyotyping of 6R in Sub6R(6D) and derived lines in Sichuan wheat background
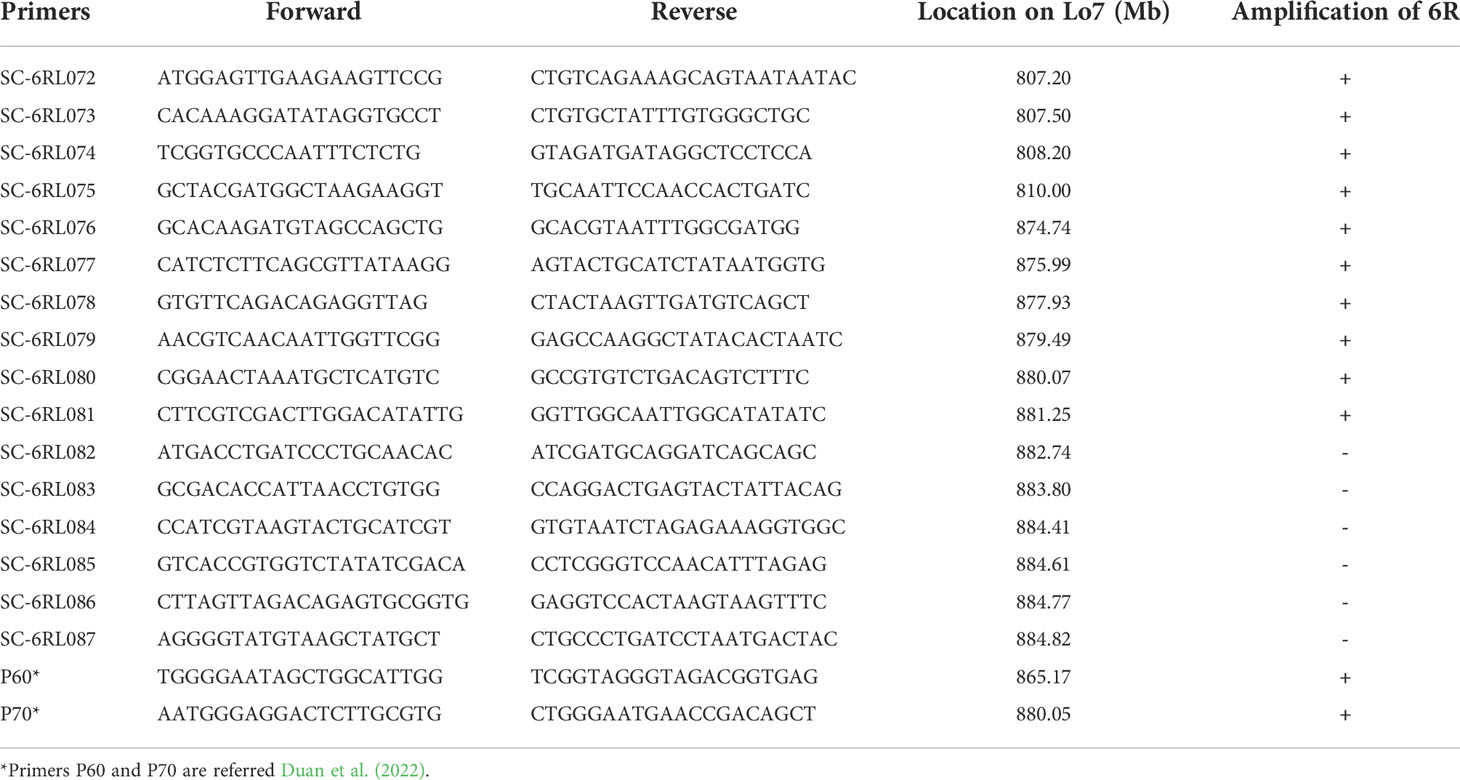
Li et al. (2020a) characterized chromosome 6R from Triticale T-701 and Sub6R(6D) by sequential multicolor FISH (mc-FISH) and genomic in situ hybridization (GISH). In the present study, ND-FISH using probe Oligo-Ku was used in place of GISH to identify the rye chromosome 6R in Sub6R(6D) (Figure 1A). ND-FISH with probes Oligo-pSc119.2 and Oligo-pSc200 was used to generate the standard karyotype of 6R. The short arm of 6R carried a strong and a weak pSc119.2 sites at telomeric and subtelomeric regions, respectively, and a strong Oligo-pSc200 site in the distal region. The 6RL arm had four Oligo-pSc119.2 and two Oligo-pSc200 signal sites, and of these the Oligo-pSc200 hybridization sites were located between the interstitial and sub-telomeric Oligo-pSc119.2 sites (Figure 1B). Li et al. (2020a) observed two strong hybridization bands on 6RL shown by GISH using rye genomic DNA as probe. Based on the comparison of the GISH banding and FISH patterns (Zhou et al., 2010), the two GISH bands on 6RL were identical to the physical positions of Oligo-pSc200 hybridization sites. A line R23 containing 6DS.6RL translocation was selected from the BC1F4 generation from Sub6R(6D) to MY11. Moreover, we developed a new ND-FISH probe Oligo-248, which gives rise to a specific hybridization site at the telomeric region of 6RL in T-701, Sub6R(6D) and line R23 (Figure 1; Figure S1). The updated ND-FISH karyotype of 6R (Figure 1G) will be helpful to characterize structural variations for chromosome 6R, especially the distal end of 6RL, in the progenies or mutant lines.
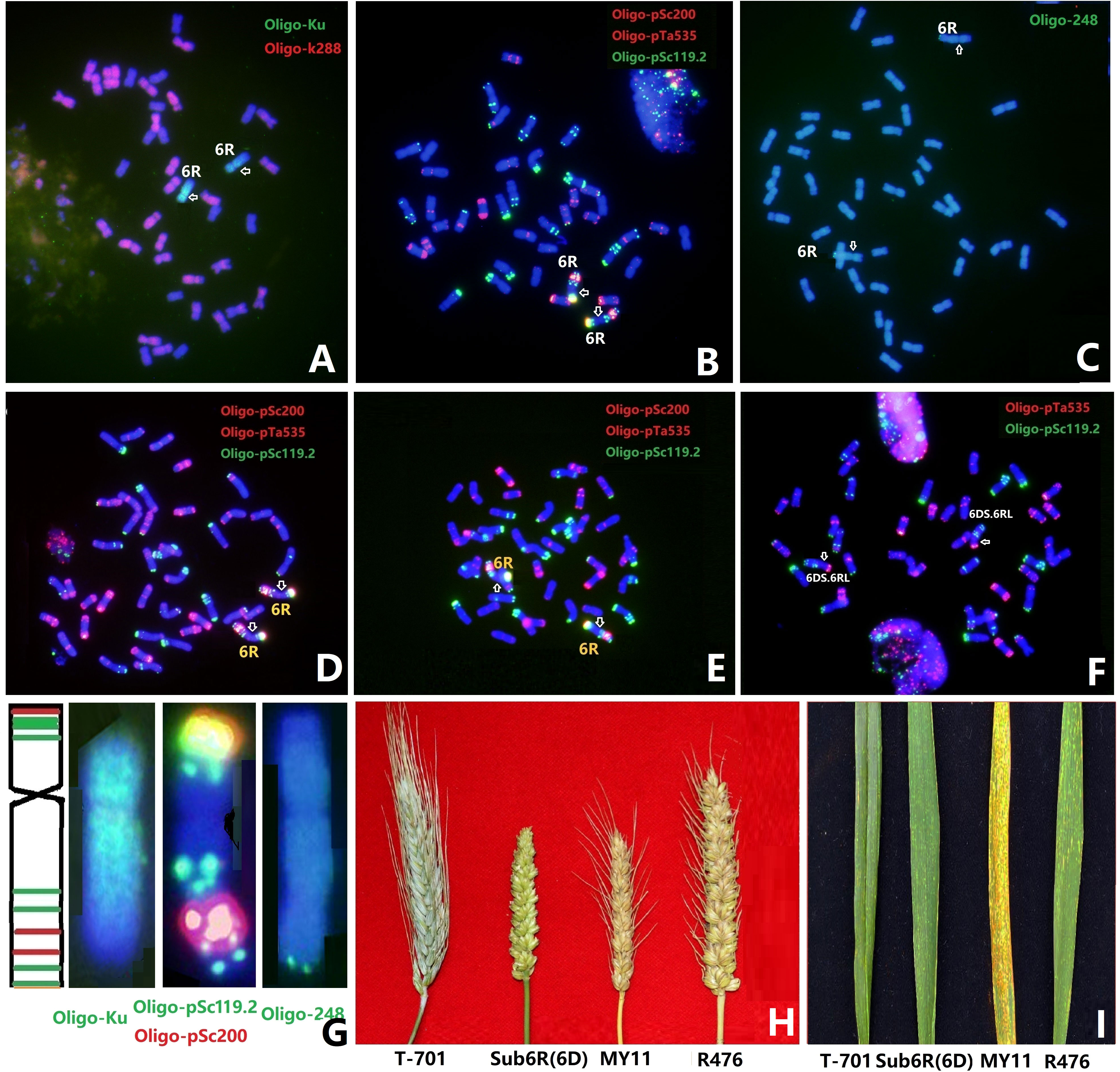
Figure 1 ND-FISH karyotyping of chromosome 6R in different wheat backgrounds. FISH of Sub6R(6D) line was performed using rye-specific oligo probe Oligo-Ku (A), Oligo-pSc119.2+ Oligo-pSc535 + Olio-pSc200 (B), and Oligo-238 (C). Lines R323 in CM42 (D), R476 in MY11 (E), and R27 with T6DS.6RL in MY11 (F) were revealed by ND-FISH using probes Oligo-pSc119.2, Oligo-pTa535, and Olio-pSc200. The karyotype of 6R is shown using multiple probes (G), the spike morphology (H) and stripe rust reaction (I) of the above wheat-6R lines and the parents are presented. Arrows pointed the positions of centromeres of 6R.
With the aim to transfer segments of rye 6R into Sichuan wheat background, a total of 908 plants of F3 progenies between Sub6R(6D) and Sichuan wheat CM42 and MY11 were screened by ND-FISH using probes Oligo-pSc119.2 and Oligo-pSc200. Lines R323 and R476 with a pair of chromosomes 6R in CM42 and MY11 backgrounds were produced in the BC1F3 generation from Sub6R(6D) to CM42 and MY11, respectively. The ND-FISH karyotypes of the chromosomes of R323 and R476 are shown in Figures 1D, F, respectively. The telosomic chromosomes 6RS, 6RL, isochromosomes iso6RS and iso6RL were identified in the F4 progenies (Figure 2A). In addition, 25 plants (2.7%) had different wheat-6R translocations including T1DS.6RL, T2DS.6RL, T6DS.6RL, T6BS.6RL, and T6RS.6DL, indicating high frequency of breakage and re-fusion of 6R with wheat chromosomes. A homozygous T6DS.6RL translocation line R23 was developed (Figure 2). ND-FISH with the grass common centromeric repeat Oligo-CCS1 and rye-specific centromeric repeat Oligo-pAWRC1.1 probes (Figures 2B–D) suggested that line R23 contained a recombined centromere from wheat 6D and rye 6R based on different intensities of signals using the two probes. The line R476 displayed an earlier flowering time of 7-10 days and reduced plant height of 80-90 cm, with significant longer spike length compared to the parent MY11 (Figure 1H), in addition to retaining the stripe rust resistance from 6RL (Figure 1I), suggesting that “Merced” derived 6R has positive effects on agronomic traits and rust resistance in wheat background.
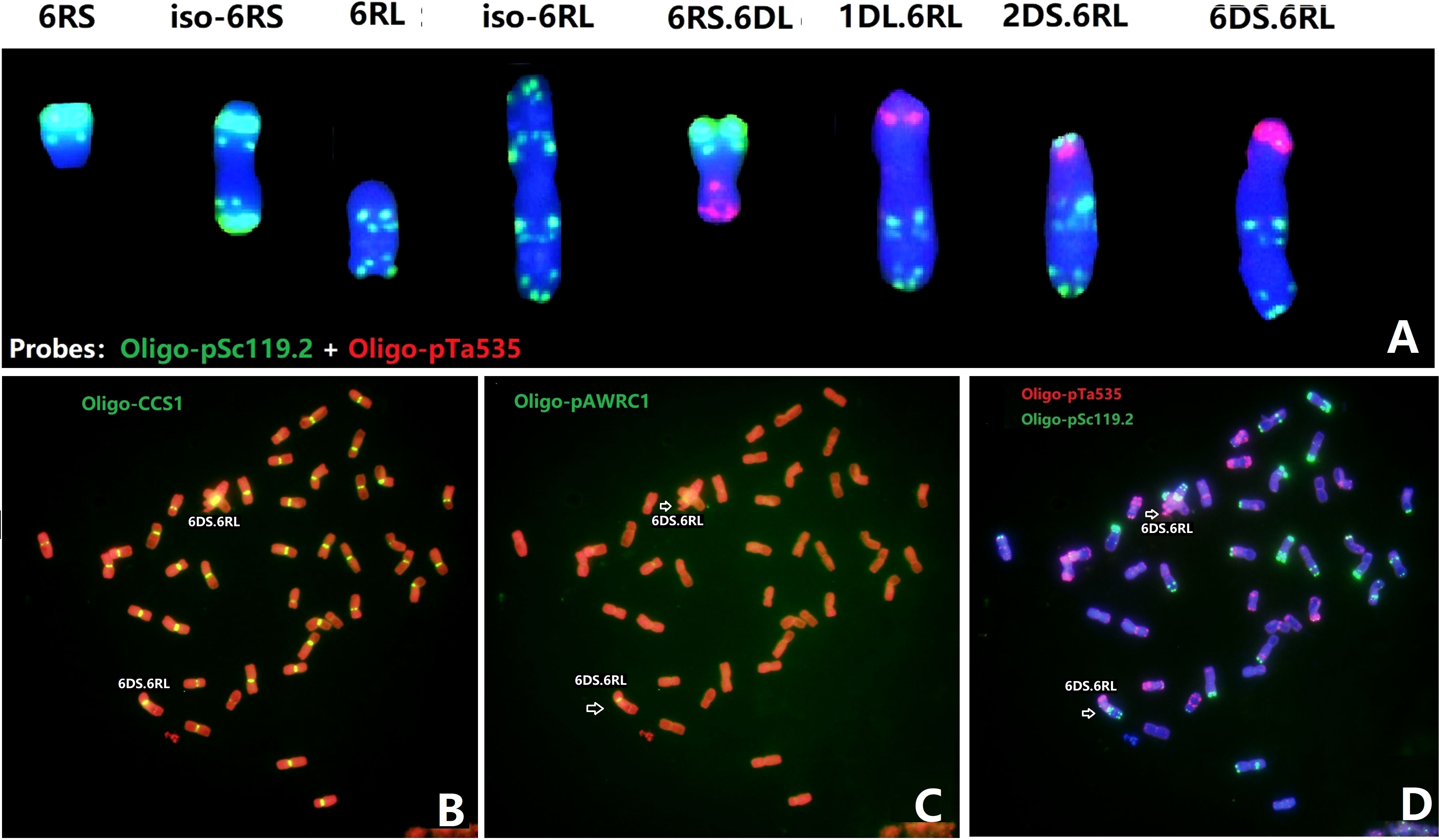
Figure 2 Sequential ND-FISH of the chromosome variations of 6R (A) and line R23 with T6DS.6RL in a MY11 background (B-D). The different types of 6R variations and wheat-6R translocations are shown by ND-FISH with probes Oligo-pSc119.2 and Oligo-pTa535 (A). The centromeric probes Oligo-CCS1 (B) and Oligo-pAWRC1.1 (C), and Oligo-pSc119.2 + Oligo-pTa535 (D), were used to show that the centromere in T6DS.6RL contains both wheat and rye centromeres.
Identification of new 6R deletion and translocation lines
With the aim to further localize the Yr83 gene, a total of 1,662 M3 plants from 195 M2:3 families of the gamma-irradiated Sub6R(6D) line were screened by ND-FISH using Oligo-pSc119.2, Oligo-pTa535, and Oligo-pSc200. Approximately, 4.0% of plants contained a modified 6R, including deletion, telosomic or isotelosomic 6R, and wheat-6R translocation chromosomes. Based on the standard karyotype of 6R (Figure 1), three types of deletions in 6RL were detected based on FISH with Oligo-pSc119.2 and Oligo-pSc200. Type 6R-1 had a deletion showing the loss of the most distal Oligo-pSc119.2 site at the terminal end of 6RL with an estimated breakpoint at FL 0.87 on 6RL (Figures 3A, E). Type 6R-2 was a larger deletion of the distal end of 6RL with the breakpoint after the second Oligo-pSc200 site (Figures 3B, E), which was estimated at the breakpoint of FL 0.82, and the deleted segment was FL 0.82-1.00. Type 6R-3 was an even bigger deletion than Type 6R-2 with the breakpoint between the two Oligo-pSc200 sites (Figures 3C, E). A line T6AL.6RLdel developed from progenies of the cross Sub6R(6D)/CS ph1b mutant Schomburgk/6R deletion (Dundas et al., 2001) was also characterized by ND-FISH with Oligo-pSc200, Oligo-pTa535, and Oligo-pSc119.2 (Figure 3D). The breakpoint was located between the two Oligo-pSc200 sites on 6RL, which is similar to that in Type 6R-3 (Figure 3C) with FL 0.73 (Figure 3E).
Figure 3 ND-FISH of wheat-rye 6R lines from irradiated progenies of Sub6R(6D) lines. The wheat-6R lines contains different types of 6RL deletions (A–D). The diagrams show the FISH patterns of the 6RL deletions by ND-FISH of Oligo-pSc119.2 (green) + Oligo-pTa535 (red) + Oligo-pSc200 (red) as probes (E).
Sequential ND-FISH using probes Oligo-k288 or Oligo-D and Oligo-Ku (Table 1), as well as Oligo-pSc119.2 and Oligo-pTa535, was used to detect the breakpoints between wheat and 6R. Probe Oligo-k288 hybridizes specifically to the A and B-genome chromosomes, while Oligo-D hybridizes specifically to D-genome chromosomes. Two reciprocal Robertsonian translocations involving 7A-6R and 7D-6R were observed in 0.06% plants, while 12 types of non-Robertsonian wheat-6R translocations were detected in 2.13% plants (Figure S2). The breakpoints at FL 0.50 of 6RS, and FL 0.50 and FL 0.73 on 6RL occurred at the highest frequencies for the non-Robertsonian translocations with wheat chromosomes.
Confirmation of the breakpoints on 6RL deletions by PCR markers
In total, 190 PLUG markers (Ishikawa et al., 2009), 321 CINAU markers (Zhang et al., 2017), and 190 markers specific for 6RL in Kustro rye (Qiu et al., 2016) were used to amplify DNA from the wheat lines MY11, Sub6R(6D), T6DS.6RL, R266, R76 and R367. The additional 16 6RL specific primers (Table 2) were designed with reference to the location of genome sequence of Lo7 6RL (Rabanus-Wallace et al., 2021). As shown in Figure 4, the breakpoints in R266 (and T6AL.6RL22) were physically located in the region between 720.56-723.16 Mb by markers Ku-6RL142 and Ku-6RL112. The breakpoint of R76 was physically located in the region between 784.09-786.82 Mb by markers Ku-6RL416 and Ku-6RL912. The breakpoint of R367 was physically located 806.17-807.21 Mb by markers Ku-6RL17 and SC-6RL072 (Figure 4). In the distal region of 6RL, markers SC-6RL082 to SC-6RL087 (Table 2) did not amplify in Sub6R(6D), but amplified in CS-Imperial 6R addition. It is likely that the present 6R may have lost the region of 882-885 Mb at terminal end of 6RL compared to Lo7 rye genome or possibly the two rye cultivars were highly divergent in the region. The association between the unique hybridization of probe Oligo-248 in ND-FISH (Figure S1) and the apparent loss of the 882-885 Mb region of 6RL needs to be further studied. The deletion in R367 was thus about 75 Mb of 6RL (806–881 Mb), corresponding to the reference genome of Lo7.
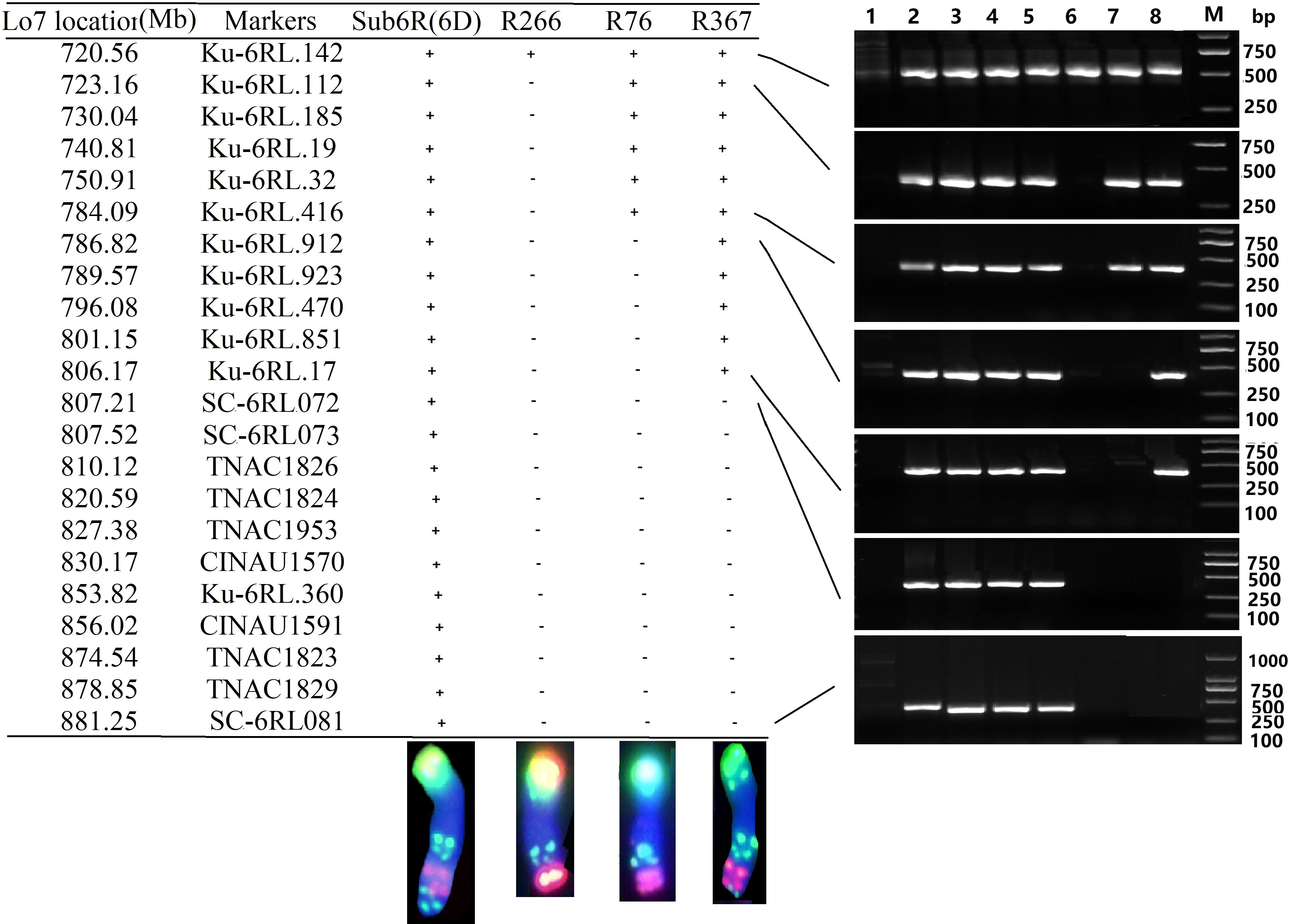
Figure 4 Physical locations of chromosome-specific markers on Sub6R(6D), 6RL deletion lines R266, R76, and R376. The marker location (left) was determined in reference to the genome of Lo7 (Mb). “+” represents amplification, while “-” represents no amplification of 6RL specific bands. The plant materials 1-8 used (right) were CS, Weining rye, T-701, Sub6R(6D), R23, R266, R76, and R367. The chromosomes were subjected to ND-FISH using Oligo-pS119.2 (green) and Oligo-pSc200 (red).
Line Sub6R(6D) and T6DS.6RL translocation line R23 were analyzed with ND-FISH using Oligo-pSc119.2 and Oligo-pTa535 as probes, and then FISH painting with the bulk oligo probes Synt6 and Synt7 (Figure 5). FISH painting with Synt6 showed that the entire 6RS and a proximal region of 6RL had distinct signals, suggesting homoeology to the wheat group 6 chromosomes. Comparing the karyotype of 6RL by ND-FISH with probes Oligo-pSc119.2 and Oligo-pTa535, it seems that the tandem repetitive sequence localizations of the two Oligo-pSc119.2 sites were likely to be in the Synt7 regions in 6RL of Sub6R(6D) and R23. The FISH painting with probe Synt7 hybridized to the distal region of 6RL at about FL 0.82-1.00, indicating that this region is homoeologous to wheat group 7 chromosomes.
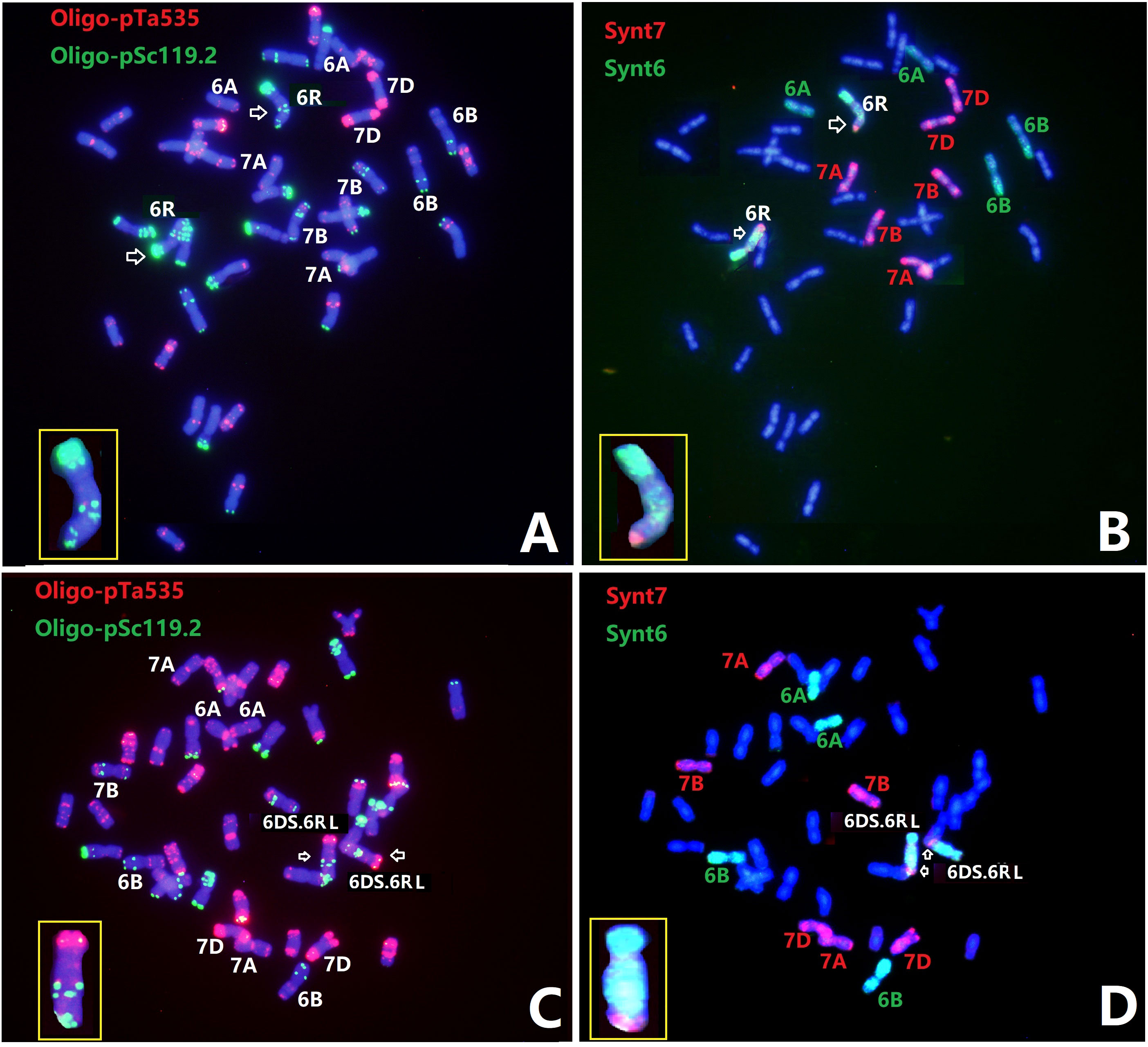
Figure 5 ND-FISH and Oligo-FISH painting of lines Sub6R(6D) (A, B) and R23 (C, D). Probes Oligo-pSc119.2 + Oligo-pTa535 (A, C) and Synt6 + Synt7 (B, D) were used. The Synt7 hybridization sites corresponded to the distal two Oligo-pSc119.2 sites in 6RL.
Stripe rust resistance and physical location of Yr83
When inoculated with a mixture of Pst races (CYR32, CYR33, and CYR34) in the field, the wheat parents CM42 and MY11 were susceptible, while line Sub6R(6D) and T6DS.6RL line R23 were resistant. The homozygous 6RL deletion lines R266, R76 and R367 were susceptible to Pst at adult plant stage (Figure S3). The results indicated that the stripe rust resistance Yr83 in 6RL was located in FL 0.87-1.00, which corresponded to 806.26-881.00 Mb region in the rye genome of Lo7.
Discussion
Over 21,000 rye accessions are maintained in genebanks worldwide, and of these approximately 35% are landraces and wild species (FAO, 2010). Hexaploid triticale contains wheat and rye genomes, and shows relatively superior vigor and reproductive stability (Lukaszewski and Gustafson, 1987; Cheng and Murata, 2002). Studies on genome-wide diversity have revealed diversified genetic structures in rye and Triticale germplasm (Bolibok-Bragoszewska et al., 2014; Sun et al., 2022; Cao et al., 2022). Among the rye chromosomes, the S. cereale chromosome 6R has displayed rapid structural change and high levels of heterogeneity of heterochromatin blocks, which has been revealed by FISH using tandem repeat probes on different rye cultivars (Guo et al., 2019). The FISH and GISH patterns using probes pSc119.2 and pSc200 showed three different karyotypes of 6R (Figure S4). The FISH patterns of 6R in Weining rye (Li et al., 2021) and Kustro rye (Li et al., 2016) had four pSc119.2 and two pSc200 sites, which is identical to the 6R in Sub6R(6D) from Merced rye (Type 6R-I, Figure S4). Two hybridization sites of pSc119.2 and three sites of pSc200 sites were observed on the 6RL of YT2 (Han et al., 2022), which is similar to the FISH patterns of CS-Imperial 6R (Type 6R-II, Figure S4). Three pSc119.2 and two pSc200 sites on 6RL were observed in Jinzhouheimai (Wu et al., 2018), AR106BONE (Duan et al., 2022), and rye cv. Qinling (Hao et al., 2018), which is typical of 6R-III (Figure S4). In addition, our previous study on S. africanum 6Rafr displayed a completely different karyotype compared to cultivated rye 6R (Li et al., 2020a), which has three pSc119.2 sites and a lack of pSc200 hybridization (Figure S4). Probe Oligo-248 was found to specifically hybridize to the 6RL terminal region of T-701, Sub6R(6D) and the T6DS.6RL line R23 (Figure 1 and Figure S1). The same pattern was also observed on 6RL in rye AR106BONE, but not in Imperial, Kustro, or Weining rye. Therefore, both molecular and cytogenetic evidence suggests that different sources of 6RL may provide diversity for future transfer of novel genetic variation to wheat.
The 6R chromosomes from different cultivated rye genotypes contain disease resistance genes with potential for wheat improvement. The 6RL chromosomes from rye cv. Prolific (Heun and Friebe, 1990; Friebe et al., 1994), JZHM (Wang et al., 2010), German White (An et al., 2015; Han et al., 2022), and Kustro (Li et al., 2016; Du et al., 2018) have powdery mildew resistance genes. Wu et al. (2018) localized PmJZHM6RL with 12 molecular markers to FL 0.51-1.00 of 6RL. Hao et al. (2018) reported the powdery mildew gene Pm56 on 6RS from rye cv. Qinling. Han et al. (2022) located the PmYT2 gene in the sub-telomeric region of 6RL, the physical region being at the 67 Mb interval corresponding to 890.09–967.51 Mb in Weining rye and 784-836 Mb in Lo7. The breakpoint at 784 Mb of Lo7 is similar to that in R76 in this study, about FL 0.82 of 6RL (Figure 4). The allelism between the Pm gene in “Merced” rye and Pm20 (Friebe et al., 1994), PmYT2 (Han et al., 2022) and PmJZHM (Wang et al., 2010) need to be tested. With respect to the stripe rust resistance, the Imperial rye 6R added in Chinese Spring and the Kustro rye 6R derived addition line 18T142 were both susceptible to stripe rust races CYR32-34 (ZX Tang, personnel communication). Li et al. (2020a) localized Yr83 on 6RL FL 0.73-1.00 using Sub6R(6D) derived deletion lines that conferred stripe rust resistance to multiple Chinese and Australian pathotypes. The present study further localized the Yr83 gene on the FL 0.87-1.00 of 6RL, which corresponds to 806-881 Mb in 6R of Lo7. Recently, Duan et al. (2022) physically located a fragment of about 37 Mb (corresponding to 848-885 Mb of Lo7) in the telomeric region of 6RL derived from rye line AR106BONE carrying Yr6RArL with high resistance to stripe rust CYR34. The allelism between Yr83 and Yr6RArL needs to be tested. Ashraf et al. (2022) identified a new stripe rust gene YrSLU on a small translocation of T6DS.6DL.6RL.6DL from 6RL of rye SLU126, and mapped it in the terminal region of 6RL. Our previous identified S. africanum 6Rafr derived stripe rust resistance gene was located on FL 0.95-1.00 of 6RafrS (Li et al., 2020a). These 6R chromosomes from cultivated and wild rye carrying stripe rust resistances are worthwhile for further wheat-rye introgression and germplasm enhancement.
Chromosome region-specific markers and the recently complete genomic sequences of rye are valuable resources for targeting introgressed chromatin from different cultivated and wild rye accessions to wheat (Tang et al., 2011; Qiu et al., 2016; Li et al., 2016; Li et al., 2021; Rabanus-Wallace et al., 2021). The evidence supports that the present-day cultivated rye 6RL contains the translocated segments from 3R and 7R, and these chromosome re-arrangements were clearly defined by comparative molecular markers (Devos et al., 1993; Li et al., 2013; Rabanus-Wallace et al., 2021). The centromeric regions of 6R were predicted at 290-300 Mb region of Lo7 genome by the physical location of pAWRC1.1 repeats using B2DSC web site (Lang et al., 2019). Li et al. (2020a) located Yr83 on FL 0.73-1.00 of 6RL, which is the distal 27% of the 6RL arm. The present study demonstrated this breakpoint to be at 720.56-723.16 Mb. The markers revealed that the FL 0.73-1.00 of 6RL included the homoeologous groups 3 and 7 re-arranged regions (Li et al., 2020a). Our study produced a deletion line R376 with a breakpoint at about 805 Mb of Lo7 genome, which was at FL 0.87-1.00 of 6RL, and Oligo-FISH painting confirmed that the fragment of the last two Oligo-pSc119.2 sites of 6RL belonged to linkage group 7 (Figure 5). Comprehensive FISH studies on cytogenetic stocks of 6RL deletions and genomic regions of the Lo7 genome have confirmed that the fine physical map of Yr83 within FL 0.87-1.00 (806-881 Mb) was syntenic to linkage group 7. Therefore, the putative candidate genes for Yr83, and also for Yr6RArL, YtSLU, and PmYT2, might be syntenic with the functional conserved genes from the linkage group 7 of the Triticeae genomes.
Rye chromosome 6R contained the highest number of predicted NBS-LRR genes than any other rye chromosomes (Rabanus-Wallace et al., 2021). The 6RL distal region with homology to linkage group 7 region has accumulated NLR genes (Qian et al., 2021). Several genes for rust and powdery mildew resistance genes were characterized in the NLR rich gene clusters in the telomeric regions of wheat group 7 chromosomes, such as Pm1 (Hewitt et al., 2021). Moreover, the Pm60 gene (TraesCS7A02G553800) from wild emmer wheat (Zou et al., 2018; Wu et al., 2021) was also homologous to SECCE6Rv1G0450040, a NBS gene located at 865,626,675 to 865,630,526 bp of 6RL terminal region. Further investigation will be conducted on the expression of rye specific NLR genes in Triticale, wheat-rye addition and introgression lines under stripe rust challenge (Khalil et al., 2015; Zhao et al., 2022). The development of EMS mutants for the Sub6R(6D) and T6DS.6RL lines, and the subsequent target-sequence enrichment and sequencing (TEnSeq) pipeline for Yr83 are undergoing (Zhang et al., 2020).
Substitution line Sub6R(6D) carrying Yr83 conferred stripe rust resistance in China and Australia, and also displayed high levels of resistance to cereal cyst nematode and powdery mildew at adult plant stage (Dundas et al., 2001; Li et al., 2020a). We have shown that the 6R from Sub6R(6D) in a Sichuan wheat background has improved the agronomic traits, including the increase of spike length and reduced plant height (Figure 1). The translocation lines T6DS.6RL and T6AS.6RL as well as T7A-6R and T7D-6R (Figure S2) will be used as the starting lines for small translocation segment induction. The two Robertsonian translocations T6DS.6RL and T6AS.6RL might be able to be used directly in wheat breeding if no obvious linkage drag could be observed to introgress multiple disease resistances from rye 6R for wheat improvement.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
ZY, PZ, and ID conceived the project and designed the experiments. GL, QM, YZ, and JL performed the experiments, GL, YZ, and EY analyzed the data, ZY and GL wrote the paper. All authors read and approved the manuscript.
Funding
The research was funded by the International Cooperation Project (2022YFH0012) of the Science and Technology Department of Sichuan, China, National Natural Science Foundation of China (31971886), and Guide Foundation for Research Institutions Performance Incentive in Chongqing (No. cstc2021jxjl80021).
Acknowledgments
We are thankful to Jie Zhang (Biotechnology and Nuclear Technology Research Institute, Sichuan Academy of Agricultural Sciences) for helping us irradiate the seeds used in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1035784/full#supplementary-material
Supplementary Figure 1 | Sequential ND-FISH of T-701 (A, D) Sub6R(6D) (B, E) and T6DS.6RL line R23 (C, F) with Oligo-pSc119.2 + Oligo-pTa535 (A, B, C) and Oligo-248 (D, E, F) respectively. The arrows point to the FISH signal of Oligo-248 in the terminal region of 6RL.
Supplementary Figure 2 | Different types of wheat-6R chromosome translocations were identified in the M3 progenies using ND-FISH. Oligo-Ku and Oligo-D (A) and Oligo-pSc119.2 + Oligo-pTa535 (B) were used to show chromosome T5DS.5DL-6RL (A, B). Different types of translocations were showed in (C), with breakpoints indicated by arrowheads (C).
Supplementary Figure 3 | The stripe rust responses of deletion lines R266, R367, and R76, translocation line R23, and substitution line Sub6R(6D). All three deletion lines were susceptible, whereas the translocation and substitution lines were highly resistant.
Supplementary Figure 4 | FISH patterns of chromosome 6R by ND-FISH with Oligo-pSc119.2 (green) + Oligo-pSc200 (red) as probes. Three different karyotypes were observed for cultivated rye, which were distinctively different from S. africanum.
References
An, D., Ma, P., Zheng, Q., Fu, S., Li, L., Han, F., et al. (2019). Development and molecular cytogenetic identification of a new wheat-rye 4R chromosome disomic addition line with resistances to powdery mildew, stripe rust and sharp eyespot. Theor. Appl. Genet. 132, 257–272. doi: 10.1007/s00122-018-3214-3
An, D., Zheng, Q., Luo, Q., Ma, P., Zhang, H., Li, L., et al. (2015). Molecular cytogenetic identification of a new wheat-rye 6R chromosome disomic addition line with powdery mildew resistance. PloS One 10, e0134534. doi: 10.1371/journal.pone.0134534
Ashraf, R., Johansson, E., Vallenback, P., Steffenson, B. J., Bajgain, P., Rahmatov, M. (2022). Identification of a small translocation from 6R possessing stripe rust resistance to wheat. Plant Dis. doi: 10.1094/PDIS-07-22-1666-RE
Asiedu, R., Fisher, J. M., Driscoll, C. J. (1990). Resistance to Heterodera avenae in the rye genome of triticale. Theor. Appl. Genet. 79, 331–336. doi: 10.1007/bf01186075
Bariana, H. S., McIntosh, R. A. (1993). Cytogenetic studies in wheat. XV. location of rust resistance genes in VPM1 and their genetic linkage with other disease resistance genes in chromosome 2A. Genome 36, 476–482. doi: 10.1139/g93-065
Bauer, E., Schmutzer, T., Barilar, I., Mascher, M., Gundlach, H., Martis, M. M., et al. (2017). Towards a whole-genome sequence for rye (Secale cereale l.). Plant J. 89, 853–869. doi: 10.1111/tpj.13436
Bolibok-Bragoszewska, H., Targonska, M., Bolibok, L., Kilian, A., Rakoczy-Trojanowska, M. (2014). Genome-wide characterization of genetic diversity and population structure in Secale. BMC Plant Biol. 14, 184. doi: 10.1186/1471-2229-14-184
Cao, D., Wang, D., Li, S., Li, Y., Hao, M., Liu, B. (2022). Genotyping-by-sequencing and genome-wide association study reveal genetic diversity and loci controlling agronomic traits in tritical. Theor. Appl. Genet. 135, 1705–1715. doi: 10.1007/s00122-022-04064-5
Cheng, Z., Murata, M. (2002). Loss of chromosomes 2R and 5RS in octoploid triticale selected for agronomic traits. Genes Genet. Syst. 77, 23–29. doi: 10.1266/ggs.77.23
Devos, K. M., Atkinson, M. D., Chinoy, C. N., Francis, H. A., Harcourt, R. L., Koebner, R. M. D., et al. (1993). Chromosomal rearrangements in the rye genome relative to that of wheat. Theor. Appl. Genet. 85, 673–680. doi: 10.1007/BF00225004
Duan, Y., Luo, J., Yang, Z., Li, G., Tang, Z., Fu, S. (2022). The physical location of stripe rust resistance genes on chromosome 6 of rye (Secale cereale l.) AR106BONE. Front. Plant Sci. 13, 928014. doi: 10.3389/fpls.2022.928014
Dundas, I. S., Frappell, D. E., Crack, D. M., Fisher, J. M. (2001). Deletion mapping of a nematode resistance gene on rye chromosome 6R in wheat. Crop Sci. 41, 1771–1778. doi: 10.2135/cropsci2001.1771
Du, H., Tang, Z., Duan, Q., Tang, S., Fu, S. (2018). Using the 6RLKu minichromosome of rye (Secale cereale l.) to create wheat-rye 6D/6RLKu small segment translocation lines with powdery mildew resistance. Int. J. Mol. Sci. 19, 3933. doi: 10.3390/ijms19123933
Ellis, J. G., Lagudah, E. S., Spielmeyer, W., Dodds, P. N. (2014). The past, present and future of breeding rust resistant wheat. Front. Plant Sci. 24, 641. doi: 10.3389/fpls.2014.00641
FAO (2010). The second report on the state of the world’s plant genetic (Rome: Resources for Food and Agriculture).
Friebe, B., Heun, M., Tuleen, N., Zeller, F. J., Gill, B. S. (1994). Cytogenetically monitored transfer of powdery mildew resistance from rye into wheat. Crop Sci. 34, 621–625. doi: 10.2135/cropsci1994.0011183x003400030003x
Friebe, B., Jiang, J., Raupp, W. J., McIntosh, R. A., Gill, B. S. (1996). Characterization of wheat-alien translocations conferring resistance to diseases and pests: current status. Euphytica 91, 59–87. doi: 10.1007/BF00035277
Fu, S., Chen, L., Wang, Y., Li, M., Yang, Z., Qiu, L., et al. (2015). Oligonucleotide probes for ND-FISH analysis to identify rye and wheat chromosomes. Sci. Rep. 5, 10552. doi: 10.1038/srep10552
Guo, J., Lei, Y., Zhang, H., Song, D., Liu, X., Cao, Z., et al. (2019). Frequent variations in tandem repeats pSc200 and pSc119.2 cause rapid chromosome evolution of open-pollinated rye. Mol. Breeding. 39, 133. doi: 10.1007/s11032-019-1033-0
Han, F., Lamb, J. C., Birchler, A. (2006). High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc. Natl. Acad. Sci. U.S.A. 103, 3238–3243. doi: 10.1073/pnas.0509650103
Han, G., Yan, H., Wang, J., Cao, L., Liu, S., Li, X., et al. (2022). Molecular cytogenetic identification of a new wheat-rye 6R addition line and physical localization of its powdery mildew resistance gene. Front. Plant Sci. 13, 889494. doi: 10.3389/fpls.2022.889494
Hao, M., Liu, M., Luo, J., Fan, C., Yi, Y., Zhang, L., et al. (2018). Introgression of powdery mildew resistance gene Pm56 on rye chromosome arm 6RS into wheat. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01040
Heun, M., Friebe, B. (1990). Introgression of powdery mildew resistance from rye into wheat. Phytopathology 80, 242–245. doi: 10.1094/Phyto-80-242
Hewitt, T., Müller, M. C., Molnár, I., Mascher, M., Holušová, K., Šimková, H., et al. (2021). A highly differentiated region of wheat chromosome 7AL encodes a Pm1a immune receptor that recognizes its corresponding AvrPm1a effector from. Blumeria graminis. New Phytol. 229, 2812–2826. doi: 10.1111/nph.17075
Ishikawa, G., Nakamura, T., Ashida, T., Saito, M., Nasuda, S., Endo, T., et al. (2009). Localization of anchor loci representing five hundred annotated rice genes to wheat chromosomes using PLUG markers. Theor. Appl. Genet. 118, 499–514. doi: 10.1007/s00122-008-0916-y
Khalil, H. B., Ehdaeivand, M. R., Xu, Y., Laroche, A., Gulick, P. J. (2015). Identification and characterization of rye genes not expressed in allohexaploid triticale. BMC Genomics 16, 281. doi: 10.1186/s12864-015-1480-x
Klymiuk, V., Chawla, H. S., Wiebe, K., Ens, J., Fatiukha, A., Govta, L., et al. (2022). Discovery of stripe rust resistance with incomplete dominance in wild emmer wheat using bulked segregant analysis sequencing. Commun. Biol. 5, 826. doi: 10.1038/s42003-022-03773-3
Kou, Y., Wang, S. (2010). Broad-spectrum and durability: understanding of quantitative disease resistance. Curr. Opin. Plant Biol. 13, 181–185. doi: 10.1016/j.pbi.2009.12.010
Lang, T., Li, G., Wang, H., Yu, Z., Chen, Q., Yang, E., et al. (2019). Physical location of tandem repeats in the wheat genome and application for chromosome identification. Planta 249, 663–675. doi: 10.1007/s00425-018-3033-4
Li, J., Dundas, I., Dong, C., Li, G., Trethowan, R., Yang, Z., et al. (2020b). Identification and characterization of a new stripe rust resistance gene Yr83 on rye chromosome 6R in wheat. Theor. Appl. Genet. 133, 1095–1107. doi: 10.1007/s00122-020-03534-y
Li, J., Endo, T. R., Saito, M., Ishikawa, G., Nakamura, T., Nasuda, S. (2013). Homoeologous relationship of rye chromosome arms as detected with wheat PLUG markers. Chromosoma 122, 555–564. doi: 10.1007/s00412-013-0428-7
Li, M., Tang, Z., Qiu, L., Wang, Y., Tang, S., Fu, S. (2016). Identification and physical mapping of new PCR-based markers specific for the long arm of rye (Secale cereale l.) chromosome 6. J. Genet. Genomics 43, 199–206. doi: 10.1016/j.jgg.2015.11.005
Li, G., Tang, L., Yin, Y., Zhang, A., Yu, Z., Yang, E., et al. (2020a). Molecular dissection of Secale africanum chromosome 6Rafr in wheat enabled localization of genes for resistance to powdery mildew and stripe rust. BMC Plant Biol. 20, 134. doi: 10.1186/s12870-020-02351-1
Li, G., Yang, Z. (2022). Oligo-FISH paints in triticeae. Curr. Protoc. 2, e364. doi: 10.1002/cpz1.364
Li, G., Zhang, T., Yu, Z., Wang, H., Yang, E., Yang, Z. (2021). An efficient oligo-FISH painting system for revealing chromosome rearrangements and polyploidization in triticeae. Plant J. 105, 978–993. doi: 10.1111/tpj.15081
Li, G., Zhang, H., Zhou, L., Gao, D., Lei, M., Zhang, J., et al. (2015). Molecular characterization of Sec2 loci in wheat-Secale africanum derivatives demonstrates genomic divergence of Secale species. Int. J. Mol. Sci. 16, 8324–8336. doi: 10.3390/ijms16048324
Lukaszewski, A. J., Gustafson, J. P. (1987). Plant Breeding Reviews. In: Janick J (ed) Cytogenetics of triticale, vol 5. AVI Publishing, New York. pp 41–93. doi: 10.1002/9781118061022.ch3
Mago, R., Miah, H., Lawrence, G. J., Wellings, C. R., Spielmeyer, W., Bariana, H. S., et al. (2005). High-resolution mapping and mutation analysis separate the rust resistance genes Sr31, Lr26 and Yr9 on the short arm of rye chromosome 1. Theor. Appl. Genet. 112, 41–50. doi: 10.1007/s00122-005-0098-9
McIntosh, R. A., Dubcovsky, J., Rogers, W. J., Xia, X., Raupp, W. J. (2021). Catalogue of gene symbols for wheat: 2021 supplement. Annu. Wheat Newsletter. 67, 104–113.
Morgounov, A., Tufan, H. A., Sharma, R., Akin, B., Bagci, A., Braun, H. J., et al. (2012). Global incidence of wheat rusts and powdery mildew during 1969-2010 and durability of resistance of winter wheat variety bezostaya 1. Eur. J. Plant Pathol. 132, 323–340. doi: 10.1007/s10658-011-9879-y
Qian, L., Wang, Y., Chen, M., Liu, J., Lu, R., Zou, X., et al. (2021). Genome-wide identification and evolutionary analysis of NBS-LRR genes from Secale cereale. Front. Genet. 12. doi: 10.3389/fgene.2021.771814
Qiu, L., Tang, Z., Li, M., Fu, S. (2016). Development of new PCR-based markers specific for chromosome arms of rye (Secale cereale l.). Genome 59, 159–165. doi: 10.1139/gen-2015-0154
Rabanus-Wallace, M. T., Hackauf, B., Mascher, M., Lux, T., Wicker, T., Gundlach, H., et al. (2021). Chromosome-scale genome assembly provides insights into rye biology, evolution, and agronomic potential. Nat. Genet. 53, 564–573. doi: 10.1038/s41588-021-00807-0
Rabinovich, S. (1997). “Importance of wheat-rye translocations for breeding modern cultivars of triticum aestivum l,” in Wheat: Prospects for global improvement, vol. 6 . Ed. Braun, H. J. (Netherlands: Springer), 401–418. doi: 10.1007/978-94-011-4896-2_55
Schneider, A., Rakszegi, M., Molnár-Láng, M., Szakács, É (2016). Production and cytomolecular identification of new wheat-perennial rye (Secale cereanum) disomic addition lines with yellow rust resistance (6R) and increased arabinoxylan and protein content (1R, 4R, 6R). Theor. Appl. Genet. 129, 1045–1059. doi: 10.1007/s00122-016-2682-6
Schwessinger, B. (2017). Fundamental wheat stripe rust research in the 21st century. New Phytol. 213, 1625–1631. doi: 10.1111/nph.14159
Spetsov, P., Daskalova, N. (2022). Resistance to pathogens in wheat-rye and triticale genetic stocks. J. Plant Pathol. 104, 99–114. doi: 10.1007/s42161-021-01019-5
Sun, Y., Shen, E., Hu, Y., Wu, D., Feng, Y., Lao, S., et al. (2022). Population genomic analysis reveals domestication of cultivated rye from weedy rye. Mol. Plant 15, 552–561. doi: 10.1016/j.molp.2021.12.015
Tang, Z., Ross, K., Ren, Z., Yang, Z., Zhang, H., Chikmawati, T., et al. (2011). “Wealth of wild species: Role in plant genome elucidation and improvement - secale,” in Wild crop relatives: Genomic and breeding resources cereals. Ed. Kole, C. (Berlin, Germany: Springer), 367–395.
Tang, S., Tang, Z., Qiu, L., Yang, Z., Li, G., Lang, T., et al. (2018). Developing new oligo probes to distinguish specific chromosomal segments and the a, b, d genomes of wheat (Triticum aestivum l.) using ND-FISH. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01104
Tang, Z., Yang, Z., Fu, S. (2014). Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J. Appl. Genet. 55, 313–318. doi: 10.1007/s13353-014-0215-z
Wang, M., Chen, X. (2017). “Stripe rust resistance,” in Stripe rust, Springers Science+Business media B.V. Dordrecht, The Netherlands Eds. Chen, X., Kang, Z., 353–558.
Wang, H., Yu, Z., Li, G., Yang, Z. (2019). Diversified chromosome rearrangements detected in a wheat- Dasypyrum breviaristatum substitution line induced by gamma-ray irradiation. Plants 8, 175. doi: 10.3390/plants8060175
Wang, C., Zheng, Q., Li, L., Niu, Y., Wang, H., Li, B., et al. (2009). Molecular cytogenetic characterization of a new T2BL.1RS wheat-rye chromosome translocation line resistant to stripe rust and powdery mildew. Plant Dis. 93, 124–129. doi: 10.1094/PDIS-93-2-0124
Wang, D., Zhuang, L., Sun, L., Feng, Y., Pei, Z., Qi, Z. (2010). Allocation of a powdery mildew resistance locus to the chromosome arm 6RL of Secale cereale l. cv. ‘Jingzhouheimai’. Euphytica 176, 157–166. doi: 10.1007/s10681-010-0199-7
Wu, N., Li, M., Sun, H., Cap, Z., Liu, P., Ding, T., et al. (2018). RNA-Seq facilitates development of chromosome-specific markers and transfer of rye chromatin to wheat. Mol. Breeding. 38, 6. doi: 10.1007/s11032-017-0762-1
Wu, Q., Zhao, F., Chen, Y., Zhang, P., Zhang, H., Guo, G., et al. (2021). Bulked segregant CGT-seq-facilitated map-based cloning of a powdery mildew resistance gene originating from wild emmer wheat (Triticum dicoccoides). Plant Biotech. J. 19, 1288–1290. doi: 10.1111/pbi.13609
Zhang, R., Fan, Y., Kong, L., Wang, Z., Wu, J., Xing, L., et al. (2018). Pm62, an adult-plant powdery mildew resistance gene introgressed from Dasypyrum villosum chromosome arm 2VL into wheat. Theor. Appl. Genet. 131, 2613–2620. doi: 10.1007/s00122-018-3176-5
Zhang, X., Wei, X., Xiao, J., Yuan, C., Wu, Y., Cao, A., et al. (2017). Whole genome development of intron targeting (IT) markers specific for Dasypyrum villosum chromosomes based on next-generation sequencing technology. Mol. Breeding. 37, 115. doi: 10.1007/s11032-017-0710-0
Zhang, J., Zhang, P., Dodds, P., Lagudah, E. (2020). How target-sequence enrichment and sequencing (TEnSeq) pipelines have catalyzed resistance gene cloning in the wheat-rust pathosystem. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00678
Zhao, F., Niu, K., Tian, X., Du, W. (2022). Triticale improvement: mining of genes related to yellow rust resistance in triticale based on transcriptome sequencing. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.883147
Zhou, J., Yang, Z., Li, G., Liu, C., Tang, Z., Zhang, Y., et al. (2010). Diversified chromosomal distribution of the tandemly repeated sequences reveal evolutionary trends in Secale (Poaceae). Plant Syst. Evol. 287, 49–56. doi: 10.1007/s00606-010-0288-z
Keywords: FISH, rye, wheat, stripe rust, Yr83
Citation: Li G, Li J, Zhang Y, Ma Q, Yang E, Zhang P, Dundas I and Yang Z (2022) Molecular and cytogenetic dissection of stripe rust resistance gene Yr83 from rye 6R and generation of resistant germplasm in wheat breeding. Front. Plant Sci. 13:1035784. doi: 10.3389/fpls.2022.1035784
Received: 03 September 2022; Accepted: 20 September 2022;
Published: 10 October 2022.
Edited by:
Hongxiang Ma, Yangzhou University, ChinaReviewed by:
Yuanfeng Hao, Institute of Crop Sciences (CAAS), ChinaHuagang He, Jiangsu University, China
Copyright © 2022 Li, Li, Zhang, Ma, Yang, Zhang, Dundas and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peng Zhang, cGVuZy56aGFuZ0BzeWRuZXkuZWR1LmF1; Ian Dundas, aWFuLmR1bmRhc0BhZGVsYWlkZS5lZHUuYXU=; Zujun Yang, eWFuZ3p1anVuQHVlc3RjLmVkdS5jbg==
 Guangrong Li
Guangrong Li Jianbo Li
Jianbo Li Yao Zhang1
Yao Zhang1 Qiang Ma
Qiang Ma Peng Zhang
Peng Zhang Ian Dundas
Ian Dundas Zujun Yang
Zujun Yang