
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 14 December 2022
Sec. Plant Abiotic Stress
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1035573
The regulation of reactive oxygen species (ROS) levels in plants is ensured by mechanisms preventing their over accumulation, and by diverse antioxidants, including enzymes and nonenzymatic compounds. These are affected by redox conditions, posttranslational modifications, transcriptional and posttranscriptional modifications, Ca2+, nitric oxide (NO) and mitogen-activated protein kinase signaling pathways. Recent knowledge about protein-protein interactions (PPIs) of antioxidant enzymes advanced during last decade. The best-known examples are interactions mediated by redox buffering proteins such as thioredoxins and glutaredoxins. This review summarizes interactions of major antioxidant enzymes with regulatory and signaling proteins and their diverse functions. Such interactions are important for stability, degradation and activation of interacting partners. Moreover, PPIs of antioxidant enzymes may connect diverse metabolic processes with ROS scavenging. Proteins like receptor for activated C kinase 1 may ensure coordination of antioxidant enzymes to ensure efficient ROS regulation. Nevertheless, PPIs in antioxidant defense are understudied, and intensive research is required to define their role in complex regulation of ROS scavenging.
Reactive oxygen species (ROS) are integral regulatory components of multiple metabolic and developmental pathways under normal and stress conditions in plants. Due to the ability to cause harmful irreversible oxidative modifications of biomolecules, ROS cellular levels must be controlled by precisely orchestrated antioxidative systems. These include effective nonenzymatic antioxidants and array of antioxidant enzymes compartmentalized into the main subcellular sites of ROS production (Mittler et al., 2022). Mechanisms regulating antioxidant enzymes are multifaceted and involve retrograde signaling, redox modifications, transcriptional, posttranscriptional and posttranslational regulations (Dvořák et al., 2021b). Another level of regulation is represented by the interaction of antioxidant enzymes with diverse proteins. Among the most important interactions in redox biology are those occurring between redox buffering proteins such as thioredoxins and glutaredoxins and their targets to reverse ROS- and NO-mediated redox modifications of target proteins (Mittler et al., 2022). Nevertheless, compelling evidence exists about protein-protein interactions (PPIs) of antioxidant enzymes with regulatory proteins such as activators, chaperones, stabilizers, but also signaling and scaffold proteins. Employing these interactions, antioxidant enzymes take part in diverse metabolic, signaling and developmental processes and transfer signals from redox homeostasis to metabolism.
Here we summarize the current knowledge about PPIs of most important antioxidant enzymes. In addition, we provide brief overview of methods for PPI identification and describe structural details of PPIs, mainly those determined by protein redox modifications.
The ability to interact with other proteins is one of the basic attributes of proteins. Many cellular processes, including metabolism and signaling, are regulated by proteins operating in protein complexes (Nussinov et al., 2013; Sudha et al., 2014). Proteins undergo either obligatory or non-obligatory PPIs. While obligatory interactions are typical for proteins that are not functional in their protomer form, non-obligatory interactions are quite common for proteins that are active also in spatially separated form (Nooren and Thornton, 2003; Seychell and Beck, 2021). In permanent interactions, protein-protein interfaces are often hydrophobic, formed by aromatic amino acid groups and hydrophobic non-polar groups that intersperse Van der Waals bonds. In non-obligatory and transient complexes, interfaces are formed by polar and charged amino acid groups and they contain a larger number of hydrogen bonds between interaction partners (Moreira et al., 2007; Perkins et al., 2010). In addition, for transient interactions, it was found that up to 75% of amino acids in the interface participate in inter- and intra-protein interactions, which reduces the frequency of structural changes of proteins. These interactions are mediated not only by polar amino acids (Asp, Glu, His, Arg) but also by hydrophobic ones (Leu, Phe, Try, Met; Jayashree et al., 2019). Interaction interfaces in transiently interacting proteins are less conserved and exhibit higher structural plasticity as those in permanently interacting proteins (Mintseris and Weng, 2005; Levy, 2010). It is estimated that 15-40% of PPIs are mediated by short linear peptide sequences (SLiMs) with a length of 3-10 amino acids. The ability to mediate interactions can be affected by reversible post-translational modifications (PTMs) of amino acid residues, which usually spatially interfere with the protein-binding domain (Blikstad and Ivarsson, 2015; Duan and Walther, 2015). Phosphorylation, acetylation, or glycosylation are often responsible for the establishment or disruption of PPIs (Ngounou Wetie et al., 2013) by changing the electrostatic or structural properties of the interaction interface (Duan and Walther, 2015). For example, phosphorylation of Tyr32 and Tyr64 of K-Ras4B GTPase alters the binding free energy of residues in the interaction interface, thus leading to generation of fewer inter-molecular hydrogen bonds and salt bridges between Ras and its interaction partner named guanine nucleotide exchange factor son of sevenless (SOS; Wang et al., 2021). Another example shows that phosphorylation of Thr37 and Thr46 residues in intrinsically disordered protein 4E-BP2 promotes formation of four-β-stranded domains by folding in Pro18-Arg62 region, and this is weakening the interaction with eukaryotic translation initiation factor 4E (eIF4E; Bah et al., 2015). In some cases, the residue subjected to PTM may be a part of a specific motif that is recognized by conserved binding modules (Yang, 2005). One of the first explored examples of a binding module is Src homology 2 (SH2) domain, which specifically recognizes phosphorylated Tyr (pTyr) residues that also form a docking site for interaction (Hubbard and Till, 2000). In addition, there are six different classes of SH2, containing proteins that recognize pTyr but also three to five C-terminal residues neighboring pTyr, collectively contributing to SH2 module specificity (Stein et al., 2009). It has also been shown that PPIs are affected by the oxidation state of a cysteine located in the proximity of the binding site (Shi et al., 2021; van Dam et al., 2021). Cysteine oxidation may lead to disruption of interaction as shown for PPI between B-cell lymphoma (Bcl-2) and extracellular signal-regulated kinases 1/2 (ERK1/2), where oxidations of Cys158 and Cys229 on Bcl-2 by hydrogen peroxide (H2O2) cause dissociation of ERK and promotion of apoptosis in human lung epithelial cells (Luanpitpong et al., 2013). Complexes constituted by intermolecular disulfide bonds might depend on the cellular redox homeostasis. Under control conditions, salicylic acid (SA) receptor nonexpresser of PR genes 1 (NPR1) is in the form of an oligomer with individual subunits bound by intermolecular disulfide bonds through Cys82 and Cys216 (Mou et al., 2003). After pathogen attack, these bonds are broken by thioredoxin (TRX3 and TRX5)-mediated reduction, NPR1 monomer is released and triggers systemic acquired resistance (SAR) in the nucleus by interacting with transcription factor GACG sequence specific binding protein 1 (TGA1), which activates expression of defense-related genes (Mou et al., 2003; Lindermayr et al., 2010). Additionally, S-nitrosylation of Cys156 supports formation of oligomers, and thus maintains long-term NPR1 homeostasis (Tada et al., 2008). Finally, TGA1-NPR1 interaction is sensitive also to oxidative modifications of TGA1. Intramolecular disulfide bond between Cys260 and Cys266 in TGA1 prevents interaction with NPR1. SA accumulation leads to their reduction, allowing interaction with the monomeric form of NPR1 (Després et al., 2003). These findings clearly point to the role of Cys redox state and its PTM in the regulation of PPIs.
The identification of PPIs relies on multiple in vivo or in vitro techniques differing in technical complexity and throughput. The most widely used low throughput methods include yeast two-hybrid assay (Y2H), bimolecular fluorescence complementation assay (BiFC) and Förster resonance energy transfer (FRET).
Yeast two-hybrid assay (Y2H) is relatively cheap, simple and fast method suitable for studying weak and transient interactions (Vinayagam et al., 2010). It is used not only for studying the interaction of a selected pair of proteins, but also for high-throughput screening of a cDNA library to reveal new interaction partners of the protein of interest (POI; Chepelev et al., 2008). However, it suffers from a high occurrence of false positive and false negative interactions (Sprinzak et al., 2003) and the inability to study membrane proteins. In addition, the heterologous system of yeast limits the studies of the PTM-dependent interactions in plants (Ngounou Wetie et al., 2019) and cannot reveal the indirect interaction of proteins in a multiprotein complex. It has to be noted that the drawbacks of Y2H have been improved by various modifications (Xing et al., 2016).
BiFC detects the interaction of two proteins, each of them being fused to one of the complementary terminal fragments of fluorescent protein (FP). If the proteins interact, complementary reconstitution of the reporter FP occurs and may be detected by fluorescence microscopy (Hu et al., 2002; Walter et al., 2004). This method enables the study of transient PPIs and provides information on the subcellular localization of the interaction. Nevertheless, the high affinity of complementary FP fragments may result in FP reconstitution without interaction of analyzed proteins, especially in cases of their overabundance. The high stability of reconstituted FP prevents studying dynamic interactions. On the other hand, the irreversibility of this complex allows identification of weak and transient interactions (Kerppola, 2006; Cui et al., 2019). BiFC has also been applied for high-throughput analyses (Miller et al., 2015).
Another widely applied in vivo method of studying PPIs is based on FRET (Förster, 1948). The principle of this low-throughput method is based on the fusion of two POIs to suitable donor or acceptor fluorophores or FPs, also called FRET pairs, which can undergo a nonradiative (dipole-dipole) transfer of energy if they are in distance less than 10 nm from each other, and detection of emission shift from donor to acceptor by confocal microscopy. The main advantage of FRET is that it enables spatio-temporal interaction studies in living cells with high precision and resolution (Cui et al., 2019; Struk et al., 2019). However, classical FRET analysis suffers from the presence of high background caused by spectral bleed-through and autofluorescence (Xing et al., 2016). Therefore, several FRET modifications suitable for in vivo studies of PPIs have been developed, such as FRET-FLIM (fluorescence lifetime imaging microscopy), FRET-APB (acceptor photobleaching), Triple-FRET, Homo-FRET and others (Xing et al., 2016; Cui et al., 2019; Strotmann and Stahl, 2022).
Proximity labelling (PL) is an in vivo high throughput approach that allows to study weak, transient or hydrophobic PPIs in living cells using enzyme-dependent labelling of proteins that are in the close proximity to POI and their subsequent MS analysis (Qin et al., 2021; Yang et al., 2021). A catalytic enzyme is fused to the POI and converts a suitable substrate into a reactive product that binds to proteins in the POI proximity. The substrate often contains biotin moieties which allows fast isolation and enrichment of labelled proteins from the mixture. Recently, a new method for PPI in cell lines based on PL was proposed (McCutcheon et al., 2020). POI is first modified by a photoproximity label in living cells. After light-triggered photocleavage, covalent labelling of proximal proteins occurs by reactive carbene of the released label. The advantage of this method is that it allows studying PPIs of redox-sensitive proteins, which are highly dependent on the redox state of the cell. This method was designed and used in the study of kelch-like ECH-associated protein 1 (KEAP1) in the HEK293T cell line so far and confirmed the presence of known but also new KEAP1 interactors (McCutcheon et al., 2020).
Co-fractionation coupled with mass spectrometry (CF-MS) is a high-throughput method allowing the detection of interacting proteins in native non-denaturing conditions, without the need for antibodies or epitope tagging of studied POI. Native protein extract is separated chromatographically and proteins in each biochemical fraction are identified by MS. PPI is confirmed when co-elution of interacting proteins occurs over multiple distinct separations (Wan et al., 2015; McWhite et al., 2020).
Affinity purification (AP) of protein complexes is suitable for PPI detection in vitro, both on low and high throughput levels. It reduces the complexity of the sample and allows the purification of protein complexes prior to MS analysis. It is based on co-purification of native or epitope-tagged POI and its interaction partners and subsequent MS/MS analysis of the eluted complex (Gingras et al., 2007). Tandem AP-MS (TAP-MS) is an approach which combines two different tags fused to POI in tandem divided by cleavage site, allowing two-step purification and reducing false positive interactions (Rigaut et al., 1999; Leene et al., 2008). The most high-throughput methods are prone to false-positive results, and it is assumed that more than half of all high-throughput data are not correct (von Mering et al., 2002; Sprinzak et al., 2003). AP-MS often leads to a detection of larger amounts of proteins that form the negative background and thus impair the reliability of the analysis. This problem is partially solved by TAP-MS, the disadvantage of which, however, is the loss of weak and transient interactions (Gavin et al., 2011). Noteworthy, even a well performed AP/TAP-MS-based PPI analysis needs to be verified. Co-immunoprecipitation (Co-IP) together with Western blotting (WB) are most frequently used methods to validate AP/TAP-MS data (Ngounou Wetie et al., 2013). Another way to confirm interaction is by reciprocal isolation, in which the role of bait and target proteins are switched and reciprocally analyzed (Miteva et al., 2013). The interaction probability might be increased by additional bioinformatic analyses, such as prediction of localization, PPI probability and co-expression analysis.
Currently, a reliable identification of PPI requires the application of at least two (but preferably three) independent techniques (Xing et al., 2016; Vadovič et al., 2019; Samakovli et al., 2020). Each method suffers from shortcomings that lead to false-positive and false-negative results, and it is therefore necessary to use proper positive and negative controls.
It is rather challenging task to isolate and identify interactors of redox-sensitive proteins or interactions conditioned by their oxidative state. Such studies are mostly based on above-mentioned methods with various modifications. An effective approach is the substitution of the putative regulatory/binding redox modified residues followed by correlation of the interaction between native and mutated form of the protein under Cys-modifying conditions. Using this principle, the interactions of Arabidopsis SKP1-like1 (ASK1) with cullin 1 (CUL1), transport inhibitor resistant 1 (TIR1) coronatine insensitive 1 (COI1) and auxin signaling F-Box (AFB) of the E3 ubiquitin ligase complex SKP1–cullin–F-boxTIR1/AFBs (SCFTIR1/AFBs; Iglesias et al., 2018) have been identified and characterized. It was found that ASK1 Cys118 and Cys37 can undergo S-nitrosylation or S-glutathionylation. Employing Y2H (using sodium nitroprusside as the NO donor) and pull-down analyses (using NO-Cys as the nitrosylating agent) with mutated versions of ASK1 (C118A and C37A) it was revealed that the interaction with the SCFTIR1/AFB or the SCFCOI1 complex proteins depends on S-nitrosylation of these Cys residues and leads to disruption of auxin and jasmonic acid (JA) signaling in Arabidopsis (Iglesias et al., 2018; Terrile et al., 2022).
An efficient approach was used during the study of the redox-dependent interactome of 2-cysteine peroxiredoxins (2-CysPRXs) in Arabidopsis. The redox conformation-specific 2-CysPRX interactome was investigated using affinity pull-down (Liebthal et al., 2020). Authors have used site-targeted mutated pseudoreduced C54S and pseudohyperoxidized C54D variants to reveal interactomes in oxidized and reduced leaf cells lysates (König et al., 2013; Liebthal et al., 2020).
Redox sensitive interactome may be also studied using H2O2-oxidized and DTT-reduced forms of tagged POIs by pull-down assay. Interactome of different redox forms of phosphatase and tensin homolog (PTEN) acting as a tumor suppressor protein during cancer development was studied by this approach (Verrastro et al., 2016; Chen et al., 2018). Thus, oxidized or reduced forms of GST-tagged PTEN were used as a bait and the HCT116 cell lysate was pulled-down to bind PTEN redox-sensitive interactome. Label-free LC-MS analysis was performed to identify interactors and results were validated by WB (Verrastro et al., 2016).
SODs catalyze the dismutation of superoxide anion to less toxic H2O2, an essential component of plant abiotic and biotic stress signaling (Castro et al., 2021; Mittler et al., 2022). Plant SOD isoforms are subcellularly compartmentalized into chloroplasts, mitochondria, cytosol, peroxisomes and apoplast, the main sites of production. In the genome of Arabidopsis, three genes encoding FeSODs (FSD1, FSD2, and FSD3), two genes coding for MnSODs (MSD1; MSD2), and three genes encoding Cu/ZnSODs (CSD1, CSD2, and CSD3) have been described (Kliebenstein et al., 1998; Pilon et al., 2011; Dvořák et al., 2021a; Chen et al., 2022).
SODs respond to changes in external conditions via transcriptional, posttranscriptional and posttranslational regulation (Dvořák et al., 2021b). One of the major factors affecting expression of FSD1 and CSD genes is the availability of Cu2+ (Yamasaki et al., 2009; Yan et al., 2017; Melicher et al., 2022). Furthermore, expression of SOD genes also depends on diverse transcription factors under changing external conditions (Dvořák et al., 2021b). SODs are also modulated by various PTMs such as phosphorylation, nitration, glutathionylation, and glycation (reviewed in Yamakura and Kawasaki, 2010; Banks and Andersen, 2019; Dvořák et al., 2021b). Moreover, this complex regulatory network is complemented by vital PPIs.
As experimentally confirmed by in-gel SOD activity assay, yeast Lys-independent aerobic growth assay and Y2H, Arabidopsis CSD isoforms are activated by interaction with copper chaperone for superoxide dismutase (CCS; Table 1), which delivers Cu2+ to their active site (Abdel-Ghany et al., 2005; Chu et al., 2005; Cohu et al., 2009; Huang et al., 2012). The activation of Cu/ZnSOD by CCS is an evolutionarily conserved process and it was confirmed for SOD1 in S. cerevisiae (Rae et al., 1999; Schmidt et al., 2000), mice and humans (Wong et al., 2000; Carroll et al., 2004). All three protein domains of CCS (N-terminal antioxidant protein 1 (ATX1)-like domain containing the copper-binding site, central domain responsible for physical interaction with Cu/ZnSOD, and C-terminal domain responsible for Cu2+ transfer to Cu/ZnSOD) show a high degree of conservation (Dreyer and Schippers, 2019), contribute to the binding activity and are essential for Cu/ZnSOD activation (Schmidt et al., 1999; Lamb et al., 2001; Chu et al., 2005). Arabidopsis CCS occurs in three splicing variants showing different subcellular localization. While the N-terminal plastid transit peptide-containing CCS320 provides specific activation of CSD2, cytosolic CSD1 and peroxisomal CSD3 are possibly activated by CCS184 or CCS229, which both contain C-terminal peroxisomal targeting peptide (Chu et al., 2005; Dreyer and Schippers, 2019). However, an existence of CCS-independent activation pathway was suggested for CSD1 and CSD3 (Huang et al., 2012). This is also supported by the absence of CCS gene in some organisms containing Cu/ZnSOD, such as Caenorhabditis elegans or Drosophila melanogaster (Jensen and Culotta, 2005; Dreyer and Schippers, 2019). The activation of CSD1 might be mediated in Arabidopsis by an interaction with DJ-1 homolog A (AtDJ-1A; Table 1) protein, as examined by isothermal titration calorimetry and BiFC (Xu et al., 2010). Genetic studies indicated the involvement of AtDJ-1A in protection against oxidative stress and high light by CSD1 activation, however AtDJ-1A may exhibit intrinsic antioxidant activity as well (Xu et al., 2010). Its human orthologue was identified as a SOD1 activating Cu2+ chaperone, potentially delivering Cu2+ to its catalytic center (Girotto et al., 2014). AtDJ-1 protein shows several conserved residues across plants, humans and bacteria. Unlike other organisms, Arabidopsis exhibit duplication of AtDJ-1, as AtDJ-1A, B and C isoforms contain two full-length DJ-1/PARK7 polypeptides (Wei et al., 2007; Xu et al., 2010). It remains to be elucidated, whether an analogous activation mechanism also occurs for CSD2 in chloroplasts by AtDJ-1B and AtDJ-1C isoforms, which are essential for plant viability and chloroplast development (Lin et al., 2011) and have chaperone activity (Lewandowska et al., 2019). It has to be noted, that Arabidopsis ccs mutant entirely lacks CSD2 activity (Cohu et al., 2009; Huang et al., 2012).
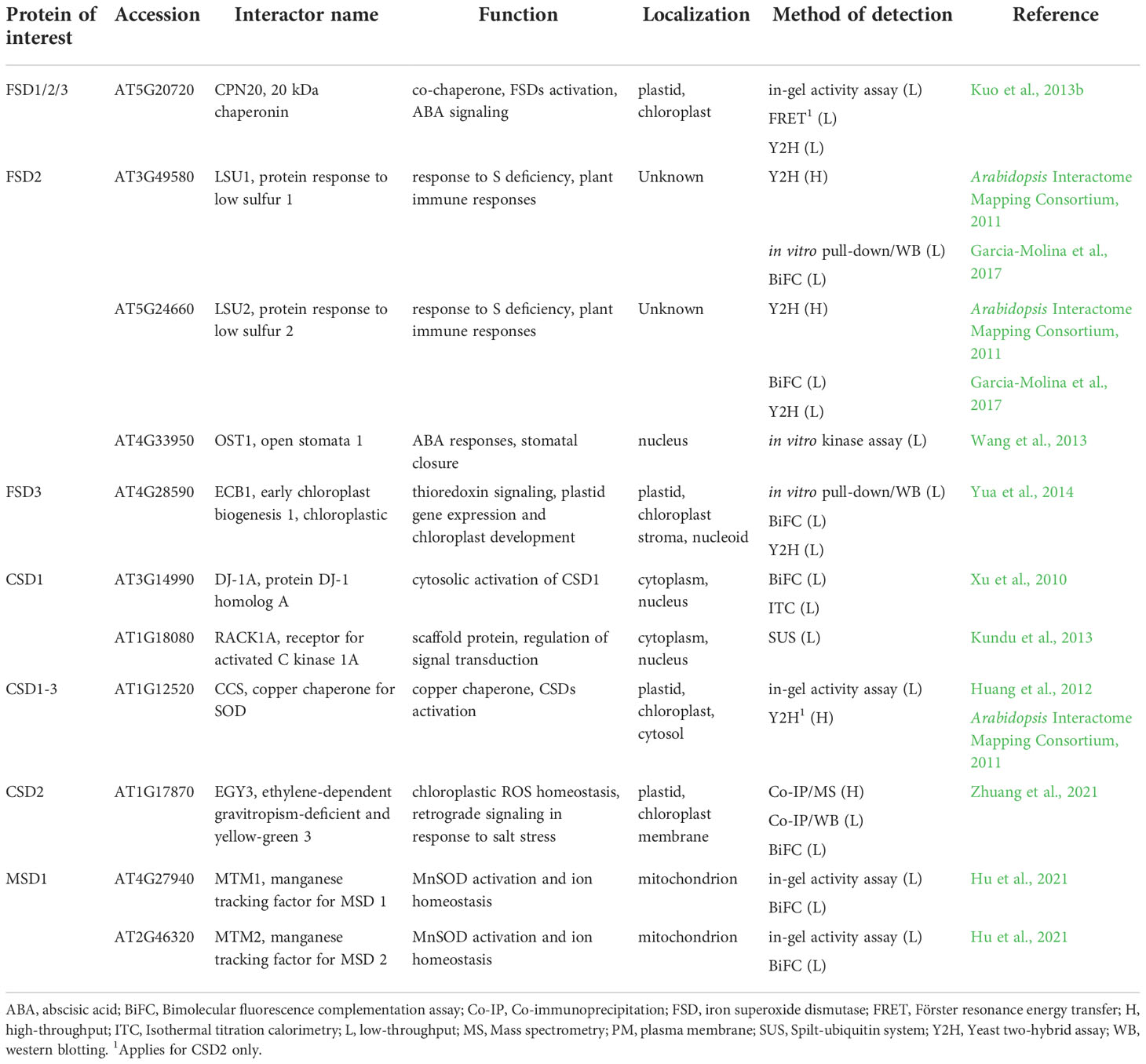
Table 1 List of Arabidopsis superoxide dismutases interaction partners found by low-throughput methods with their respective function and localization.
Recently, Co-IP coupled to either MS (Co-IP/MS) or WB (Co-IP/WB) and BiFC assay indicated the interaction of CSD2 with ethylene-dependent gravitropism-deficient and yellow-green 3 (EGY3), a chloroplast membrane-associated ATP-independent metalloprotease (Zhuang et al., 2021). EGY3 is a pseudoprotease showing a high homology with the family of site-2-proteases (S2P), which are involved in proteolytic cleavage of membrane-anchored transcription factors at the plasma membrane. These proteases occur in prokaryotes, mammals and plants, however, EGY3 was identified only in plants with high conservation in the plant kingdom (Adamiec et al., 2017; Adamiec et al., 2020). EGY3 expression is strongly induced by salt, oxidative stress (Zhuang et al., 2021), high light, and high temperature stress (Adamiec et al., 2022). The stress-induced interaction of CSD2 with M50-like domain of EGY3 stabilizes CSD2 as indicated by decreased abundance of CSD2 under salt stress and high light conditions in egy3 mutants. The CSD2-EGY3 interaction is important for H2O2 pool maintenance to ensure chloroplast-nucleus retrograde signaling (Zhuang et al., 2021; Adamiec et al., 2022). The Cu2+-dependence of this regulation remains to be revealed.
Several other interactions of CSDs were identified by large-scale OMICs-based analyses (Table 1; Table S1) including heavy metal-associated isoprenylated plant protein 20 (HIPP20; Arabidopsis Interactome Mapping Consortium, 2011), receptor for activated C kinase 1 (RACK1A; Kundu et al., 2013), both for CSD1 only, chitin elicitor receptor kinase 1 (CERK1; Le et al., 2014) for both CSD1 and CSD2, and polyubiquitin 3 (UBQ3; Kim et al., 2013) for CSD1 and CSD3.
Chitin elicitor receptor kinase 1 (CERK1), a pattern recognition receptor responsive to chitin and salt stress (Espinoza et al., 2017), triggers immune responses accompanied with massive ROS production (Erwig et al., 2017). In addition to CSD1 and CSD2, CERK1 putatively interacts with ascorbate peroxidase 1 (APX1), thioredoxin H4 and putative glutaredoxin C4, as detected by a Y2H screen (Table S1; Le et al., 2014). Interactions likely take place at the cytoplasmic kinase domain of CERK1, which may possibly synchronize the ROS production with antioxidant defense during plant biotic interactions and salt stress response. The reliability of these findings has to be further verified in planta.
Heavy metal-associated isoprenylated plant proteins (HIPPs) are metallochaperones localized in cytosol, endoplasmic reticulum (ER), and nucleus. They are involved in heavy metal homeostasis and detoxification mechanisms, protein folding, removal of non-native proteins by ER-associated degradation, plant-pathogen interactions, and transcriptional responses to cold and drought (de Abreu-Neto et al., 2013; Cowan et al., 2018; Guo et al., 2021). HIPPs typically contain one or two metal binding domains (de Abreu-Neto et al., 2013) present also in CCS structure (N-terminal ATX1-like domain). Based on their structural similarities with SOD-activating CCS proteins (de Abreu-Neto et al., 2013), HIPPs might be considered as putative CSDs activators.
The precise mechanism of Fe2+ delivery to the active site of FeSOD was not identified yet. Nevertheless, chloroplastic FeSOD activation is known to be mediated by interaction with chloroplast-localized chaperonin 20 (CPN20), as confirmed by in-gel SOD activity assay, FRET and Y2H (Table 1; Kuo et al., 2013b). In vitro experiments proved the CPN20 ability to bind Fe2+ and incorporate Fe2+ to the FSD1-holoenzyme (Kuo et al., 2013a). CPN20 is involved in the regulation of abscisic acid (ABA) signaling (Zhang et al., 2013b; Guo et al., 2015; Liang et al., 2021) and its overexpression confers increased FSD1 activity in Arabidopsis. Additionally, CPN20 likely activates FSD2 and FSD3 as well, since activities of all FSD isoenzymes increased in the presence of CPN20 in vitro (Kuo et al., 2013b). The function of this interaction has not been revealed so far. It is quite likely, that it is specific for the regulation of chloroplastic FeSODs with the potential impact on ROS signaling. The mechanism of FSD1 activation in cytosol is still not uncovered and needs further investigation.
FSD2 and FSD3 (exclusively localized in chloroplasts) are components of a plastid-encoded RNA polymerase (PEP) complex involved in chloroplast development and maintenance (Pfannschmidt et al., 2015). Both isoforms possess ROS scavenging activity and might protect nucleoids from oxidative damage (Myouga et al., 2008). Additionally, in vitro pull-down assay, BiFC and Y2H showed that FSD3 interacts with early chloroplast biogenesis 1 (ECB1; Table 1). ECB1 has, thioredoxin activity and regulates chloroplastic gene expression by affecting the redox status of the PEP complex (Yua et al., 2014). The presence of both FSD2 and FSD3 in the PEP complex and an interaction with ECB1 indicate the possible role of FSD2 and FSD3 in the regulation of chloroplast gene expression. This is supported by the pale green phenotypes of fsd2 and fsd3 single mutants and by the arrested chloroplast development in fsd2/fsd3 double mutant (Myouga et al., 2008). FSD3 is expressed in two splicing variants (FSD3 and FSD3S) with equal N-termini, but different C-termini, showing different localizations in chloroplasts. FSD3 is localized only to chloroplast nucleoids, while FSD3S was found in entire organelle. Of note, only the FSD3 splicing variant rescued the fsd3 phenotype (Lee et al., 2019). Based on these results, phenotypes of both fsd2 and fsd3 mutants may suggest an involvement of FSD2 and FSD3 in the regulation of PEP complex, perhaps leading to impaired expression of genes involved in photosynthesis. The question, whether this function is dependent on FSD2 and FSD3 enzymatic activity, still remains unanswered.
FSD2 activity is linked to plant stress responses through the interaction with low sulphur upregulated 1 (LSU1), which was revealed by in vitro pull-down assay, Y2H and BiFC (Table 1; Garcia-Molina et al., 2017). LSUs represent a plant-specific gene family, and recent phylogenetic analysis demonstrated that they belong to a Spermatophyta-specific gene family (Uribe et al., 2022). LSUs are expressed under various abiotic and biotic stress conditions and coordinate plant immune responses via PPIs during simultaneous exposure of plants to abiotic stresses and pathogen attack (Garcia-Molina et al., 2017; Niemiro et al., 2020). LSU1 is localized predominantly in guard cells, and its interaction increased enzymatic activity of FSD2. The LSU1-FSD2 interaction might be implicated in stomatal closure by regulating H2O2 levels or remodeling gene expression via activation of ROS-responsive genes (Garcia-Molina et al., 2017). Interestingly, the guard cell chloroplast relocation of LSU1 upon treatment with Pseudomonas syringae virulence effectors hampers FSD2-LSU1 interaction. LSU1 downregulation is connected with increased disease susceptibility in plants exposed to abiotic stresses while overexpression leads to improved disease resistance (Garcia-Molina et al., 2017). Therefore, the FSD2-LSU1 interaction and following ROS regulation are crucial in combinatorial plant responses to biotic and abiotic stress (Garcia-Molina et al., 2017).
In addition to the aforementioned interacting partners of FSDs, several large-scale, OMICs-based PPI analyses suggested interaction with other proteins having diverse functions (Table S1). These included chloroplast import (translocase of chloroplast 132 (TOC132); Dutta et al., 2014) and protein degradation (UBQ3; Igawa et al., 2009) for FSD1. This SOD isoform might be also a component of a G-protein complex, since it was shown to interact with guanine nucleotide-binding protein subunit beta (AGB1), guanine nucleotide-binding protein alpha-1 subunit (GPA1), and an effector of its beta subunit called acireductone dioxygenase 2 (ARD2; Friedman et al., 2011; Klopffleisch et al., 2011). FSD2 has been suggested to interact with cysteine/histidine-rich c1 domain family protein, btb/poz domain protein, ubiquitin-conjugating enzyme 27 (UBC27; Arabidopsis Interactome Mapping Consortium, 2011), btb/poz and math domain-containing protein 3 (BPM3; Altmann et al., 2020), open stomata 1 (OST1; Wang et al., 2013), and UBQ3 (Kim et al., 2013), which are localized in cytosol. The suggested interacting proteins are not localized or functionally connected to chloroplast. Therefore, it is unlikely that these proteins interact with the mature FSD2 protein. Nevertheless, the interaction of above-mentioned proteins with the nascent FSD2 protein cannot be ruled out. Teosinte branched 1/cycloidea/pcf (TCPs) are highly conserved plant-specific transcription factors with preferential localization in cytosol and nucleus (Martín-Trillo and Cubas, 2010). It was suggested by Y2H approach, that FSD2 interacts with TCP13 and TCP15, both possessing chloroplast-targeting sequence (Table S1; Baba et al., 2001; Wagner and Pfannschmidt, 2006; Martín-Trillo and Cubas, 2010). TCP13 is involved in the regulation of gene expression in chloroplasts (Baba et al., 2001) and plays important roles in leaf and root growth during abiotic stress (Urano et al., 2022). Similar roles have been suggested for TCP15 (Wagner and Pfannschmidt, 2006; Martín-Trillo and Cubas, 2010). FSD2-TCP interaction implies additional possible mechanism of FSD2-mediated regulation of chloroplastic genes expression.
The first mechanism of mitochondrial MnSOD activation was described for yeast SOD2, which is activated by interaction with manganese trafficking factor for mitochondrial SOD2 (MTM1; Luk et al., 2003). Recently, the functional homologs of yeast MTM1 (yMTM1) were identified in Arabidopsis. AtMTM1 and AtMTM2 are mitochondrial carriers with conserved transmembrane sequences interacting with MSD1, as found by both in-gel SOD activity assay and BiFC (Table 1). AtMTM1 and AtMTM2 can activate yeast SOD2 in yMTM1-mutant cells, which points to their high homology with yMTM1 (Hu et al., 2021). Additionally, both MTM isoforms are involved in Mn2+ and Fe2+ homeostasis, regulation of flowering time and root elongation (Hu et al., 2021; Hu and Jinn, 2022), the latter function being also associated to MSD1 (Morgan et al., 2008).
High-throughput CF-MS analysis identified several important metabolic proteins as potential MSD1 interactors (Table S1; McWhite et al., 2020). Notably, interactions with enzymes of tricarboxylic acid cycle, such as mitochondrial malate dehydrogenases (mMDHs) and succinate–CoA ligase subunits (SCS-alpha-1/2 and SCS-beta), may be related to the repression of tricarboxylic acid cycle flux in isolated mitochondria in MSD1 RNAi lines (Morgan et al., 2008).
Taken together, SODs may regulate gene expression via PPIs, and are essential constituents of the ROS signaling pathways, at least partially by affecting subcellular pools of H2O2.
Catalases (CATs) are iron-containing homo-tetrameric metalloenzymes that catalyze the decomposition of H2O2 to water and oxygen and play a central role in the regulation of H2O2-signaling during plant responses to abiotic and biotic stresses (Yuan et al., 2017; Su et al., 2018; Yang et al., 2019a). Three main CAT families evolved during the evolution, including heme-typical CATs, catalase-peroxidases, and minor manganese CATs. Typical CATs represent a most abundant group occurring in most living organisms, including eubacteria, archaebacteria, protista, fungi, animals and plants (Zamocky et al., 2008; Pan et al., 2022). Although plant CATs are recognized as peroxisomal proteins, the presence of CATs has been also detected in nucleus (Al-Hajaya et al., 2022), cytoplasm, mitochondria and chloroplasts (Mullen et al., 1997). Three CAT isoforms (CAT1, CAT2, and CAT3) documented in Arabidopsis have diverse developmental and stress-related functions (Palma et al., 2020). Expression pattern of CATs is tightly regulated by numerous transcription factors (Wang and Chu, 2020; Chu et al., 2021; Dvořák et al., 2021b; Song et al., 2021). PTMs, such as phosphorylation, glycation, Tyr-nitration, acetylation, S-nitrosation regulate CATs functions (Zou et al., 2015; Palma et al., 2020; Zhang et al., 2020). Notably, CATs are frequently bound and inhibited by pathogen effectors leading to pathogen spread and mitigation of host immunity (Murota et al., 2017; Sun et al., 2017; Jiao et al., 2021; Yuan et al., 2021).
CATs interact with numerous proteins of diverse functions. CATs peroxisomal targeting is mediated by their interaction with peroxisomal targeting signal 1 receptor (PEX5; Table 2; Oshima et al., 2008; Freitas et al., 2011). CATs are subsequently released by PEX14, allowing CAT tetramerisation (Freitas et al., 2011). A recent study revealed the importance of a C-terminal heme binding domain of CAT for its peroxisomal targeting (Fujikawa et al., 2019).
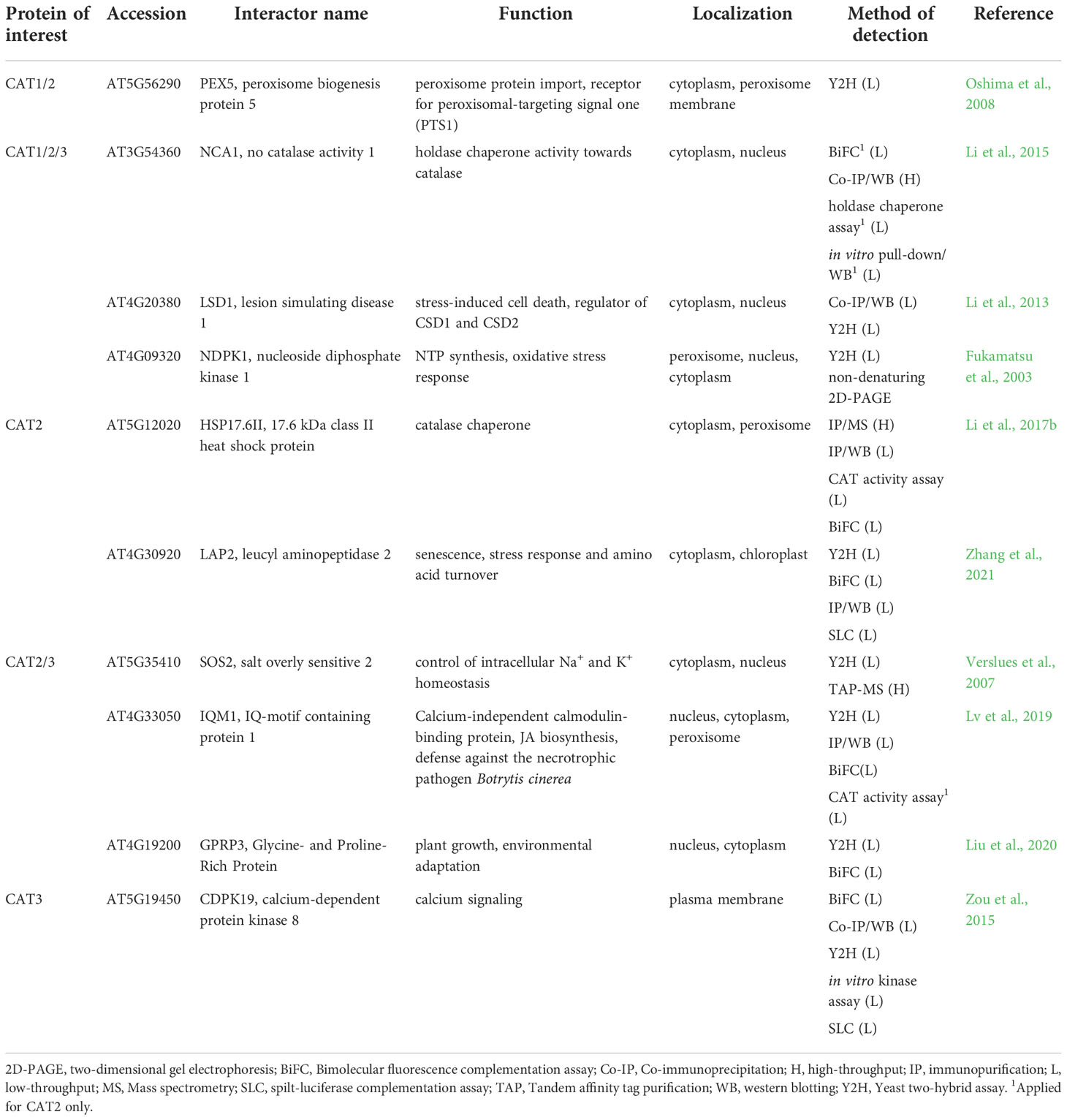
Table 2 Interaction partners of Arabidopsis catalases found by low-throughput methods with their respective function and localization.
Arabidopsis CATs are activated by no catalase activity 1 (NCA1), a cytosolic chaperone protein. Interactions between CATs and NCA1 were uncovered by Co-IP/WB and for CAT2 further validated by BiFC, in vitro pull-down assay coupled to WB and holdase chaperone assay (Table 2; Li et al., 2015). CAT2 interacts explicitly with the C-terminal tetratricopeptide repeat-like helical domain, while the N-terminal RING-finger domain of NCA1, containing zinc ion, is required for its activation. CAT2 activity increased 10-fold after interaction with NCA1 in vitro. In addition, nca1 mutants exhibit increased sensitivity to salt, cold and oxidative stress (Li et al., 2015), suggesting that NCA1-CAT interaction might be important for ROS homeostasis. Moreover, reverse genetic studies indicated that NCA1-CAT interaction may regulate immunity-triggered autophagy (Hackenberg et al., 2013). Based on the known chaperone function of NCA1, it is likely that NCA1-CAT pair may mediate optimal folding of nascent CAT in the cytosol. The NCA1 gene is present solely in plants and in some species such as Oryza sativa or Nicotiana tabacum it exists in two copies (Liu et al., 2019). Interestingly, both gene products retained the ability to bind CAT (Liu et al., 2019). These results point to the evolutionary conservation of CAT-NCA1 interaction.
Another CAT activation mechanism is based on CAT2 interaction with small heat shock protein Hsp17.6CII, a peroxisome-localized catalase chaperone, as examined by BiFC, Co-IP/MS and Co-IP/WB (Table 2; Li et al., 2017b). Small heat shock proteins (sHSPs) are ubiquitous chaperones involved in stress responses of prokaryotic and eukaryotic cells (reviewed in Waters, 2013; Haslbeck and Vierling, 2015). Overexpression of Hsp17.6CII in nca1 mutant did not lead to increased stress resistance and CAT activity, indicating that Hsp17.6CII activates CAT2 in an NCA1-dependent manner. Moreover, overexpression of Hsp17.6CII in wild type led to increased CAT activity and transgenic lines exhibited increased tolerance to abiotic stresses (Li et al., 2017b). Finally, cassava chaperone MeHSP90.9 has been identified by Y2H, BiFC and pull-down assay combined with WB as a positive regulator of MeCAT1 activity which plays an important regulatory role during drought stress resistance (Wei et al., 2020). These results highlight HSPs as another important part of CATs regulation, mainly in stress conditions.
Multiple low and high-throughput examinations proved that all three CATs interact with zinc finger protein lesion simulating disease 1 (LSD1; Table 2) which is localized in the nucleus and cytoplasm. LSDs are present exclusively in Viridiplantae with highly conserved structural domains (Li et al., 2013; Cabreira et al., 2015). LSD1 is a scaffold protein and transcriptional regulator during biotic and abiotic stress responses of plants. Moreover, it plays an essential role as a negative regulator of programmed cell death (PCD) and its numerous PPIs are redox-dependent (Li et al., 2013; Czarnocka et al., 2017). The interaction between the LSD1 and CATs required the presence of all three LSD1-like zinc finger motifs and is involved in light-dependent runaway cell death and hypersensitive response cell death regulation in Arabidopsis (Li et al., 2013). Based on the high conservation of LSD1 in plants it is likely that this regulation mechanism also occurs in other plant species (Cabreira et al., 2015), however, no other experimental evidence has been reported to this date. Nevertheless, a recent study reported that interaction of cassava MeLSD3 with MeAPX2 promoted its activity and increased antioxidant efficiency (Zeng et al., 2022). Therefore, LSDs may ensure the coordination of antioxidant defense during cell death.
CATs involvement in antioxidant defense might be mediated by the interaction with nucleoside diphosphate kinase (NDPK), a housekeeping enzyme which catalyzes interconversion of nucleoside diphosphates and triphosphates (Dorion and Rivoal, 2018). This interaction, important for development, stress and light responses, was identified in Arabidopsis by Y2H and non-denaturing two-dimensional gel electrophoresis (Table 2; Fukamatsu et al., 2003). It was confirmed in fungus Neurospora crassa (Yoshida et al., 2006; Wang et al., 2007; Lee et al., 2009) and Pisum sativum (Haque et al., 2010), showing high degree of conservation. NDPK2 and CAT2/3 form a complex with salt overly sensitive 2 (SOS2), known as a class 3 sucrose-nonfermenting 1-related kinase as revealed by Y2H and TAP-MS analyses (Table 2; Verslues et al., 2007). This complex is involved in crosstalk between salt stress response and ROS signaling (Verslues et al., 2007). NDPK is known to mediate the expression of antioxidant enzymes such as SODs, peroxidases, CAT, APX, thioredoxin reductase and peroxiredoxin in plants (Yang et al., 2003; Tang et al., 2008; Zhang et al., 2017). Thus, it might be hypothesized that NDPKs govern antioxidants also by other mechanisms, for example, by interaction with mitogen-activated protein kinases MPK3 and MPK6 (Moon et al., 2003), which may initiate expression of antioxidant enzymes (Dvořák et al., 2021b).
CAT1 might also bind to two proteins, which interact with target of rapamycin (TOR) kinase suggested by large scale TAP-MS (Table S2; Van Leene et al., 2019). Both regulatory-associated protein of mTOR 1B (RAPTOR 1B) and lethal with Sec thirteen 8/G protein β subunit-like (LST8/GβL), bear a WD-40 repeat motif, indicating possible affinity of CAT to this domain conferring PPIs or protein-DNA interactions. TOR kinase integrates nutrient-, energy- and stress-related cues with growth and metabolic outputs while above mentioned interactors are supposed to control target specificity and the stability of TOR complexes (Rexin et al., 2015).
CAT2 and CAT3 interact with the calmodulin-binding protein IQ-motif containing protein 1 (IQM1) as confirmed by Y2H, Co-IP/WB, BiFC and CAT activity assays (Table 2). IQM1 modulates CAT2 activity which indirectly stimulates the JA biosynthetic enzymes such as acyl-CoA oxidases in response to Botrytis cinerea infection (Lv et al., 2019). On the other hand, IQM1 negatively regulates plant ABA responses and stomatal movement by affecting ROS levels (Zhou et al., 2012). Proteins homologous to IQM1 were found in rice and most OsIQM members are responsive to ABA, polyethylene glycol and salt, implying their involvement in stress responses (Fan et al., 2021).
CAT2 is stabilized during salt and drought stresses by an interaction with leucine aminopeptidase 2 (LAP2), which was shown by several independent methods such as Y2H, BiFC, IP/WB, and spilt-luciferase complementation assay (Table 2). It was found that this interaction is important also for stability and increased activity of LAP2 in order to increase gamma aminobutyric acid (GABA) content as a response to salt and osmotic stress (Zhang et al., 2021). AtLAP2 is homologous to tomato LAP-A and LAP-N, which act as molecular chaperones and facilitate the stability of proteins during stresses (Scranton et al., 2012), implying that tomato LAPs share the CAT stabilization ability with LAPs of Arabidopsis.
AP-based interaction studies identified two members of 14-3-3 protein family (Table S2), 14-3-3-like protein GF14 omega (GRF2; Chang et al., 2009) and 14-3-3-like protein GF14 psi (GRF3; Shin et al., 2011), as another putative interaction partners of CAT2 and CAT3, respectively. 14-3-3 proteins bind multiple metabolic proteins and proteins involved in signaling in order to regulate plant development and biotic and abiotic stress responses (Roberts et al., 2002). GRF2 is involved in responses to salt stress by regulating H+-ATPase activity that provides proton gradient for Na+/H+ antiporter SOS1, a transport protein regulated by SOS2, another interaction partner of CATs (Verslues et al., 2007; Yang et al., 2019b). This points to the close crosstalk of CATs with the SOS pathway.
High-throughput assays indicated several nuclear CAT interactors (Table S2), which may shed light on the recently discovered nuclear targeting of CAT (Al-Hajaya et al., 2022). CAT isoforms might be associated with the nuclear pore complex (NPC) as indicated by the possible interaction of all three CATs with nuclear pore complex protein 43 (NUP43; also known as nucleoporin). It belongs to WD-40 repeat protein family and plays a role in nucleocytoplasmic transport (Tamura et al., 2010). Interestingly, CAT3 binds to Nup93/Nic96 nucleoporin interacting component-containing protein, a component of the NPC (Li and Gu, 2020) and with RNA export factor 1 (RAE1), a protein binding to NUP98, involved in mRNA export and cell division (Meier and Brkljacic, 2009). These data point to possible role of CAT in nucleocytoplasmic transport.
Other nuclear CAT interactors involve histone H3 acetyltransferase (increased DNA methylation 1 (IDM1); applied for CAT3) and defective in meristem silencing 3 (DMS3), both involved in epigenetic modifications (Kanno et al., 2008).
APX is a central enzyme of the ascorbate-glutathione cycle, a major pathway of chloroplastic H2O2 elimination and a redox balancing mechanism in adverse environmental conditions. Within this cycle, APX decomposes H2O2 using ascorbate as an electron donor. Ascorbate is recycled by either monodehydroascorbate reductase (MDAR) or dehydroascorbate reductase (DHAR; Bartoli et al., 2017).
APXs are responsive to external (abiotic and biotic stresses) and internal (phytohormones) stimuli. This enzyme encoded by several compartmentalized isoforms is regulated on transcriptional and posttranslational level (Maruta et al., 2016). The expression of cytosolic APX strongly depends on the redox state of plastoquinone pool, acting as a sensor of light intensity (Karpinski et al., 1997). APXs are targeted by redox buffering proteins such as TRXs (Gelhaye et al., 2006; Pérez-Pérez et al., 2017) or nucleoredoxin (Kneeshaw et al., 2017). In addition, APX functions are fine-tuned by PPIs. APXs are bound by chloroplast-targeted APX-related (APX-R) proteins, which are structurally related to APXs (Lazzarotto et al., 2011) and operate in ascorbate-independent manner (Lazzarotto et al., 2021). The functional relevance of this interaction remains to be elucidated.
An intricate interaction was detected between cytosolic NtcAPX and cell division cycle 48 (NtCDC48) in tobacco by high-throughput Co-IP/MS (Rosnoblet et al., 2017), FRET-FLIM and pull-down assays (Bègue et al., 2019). A chaperone-like CDC48, a protein evolutionary highly conserved in eukaryotes and archaea, regulates the proteasome-mediated degradation and autophagy during development as well as abiotic and biotic stress responses (Park et al., 2008; Stolz et al., 2011; Bègue et al., 2017; Rosnoblet et al., 2021). It undergoes oxidative modifications and can segregate and remodel its interacting partners from complexes or cellular structures (Rosnoblet et al., 2021). Some interactions of CDC48 described in archaea and eukaryotes are also evolutionarily conserved (Barthelme and Sauer, 2013). NtCDC48 negatively affects NtcAPX, since its overexpression suppresses NtcAPX expression, abundance, and activity during elicitation with cryptogein and heat shock. Thus, NtCDC48-NtcAPX interaction contributes to cAPX protein turnover (Bègue et al., 2019). Furthermore, according to high-throughput Co-IP/MS interaction studies NtCDC48 is supposed to interact with NtMnSOD and NtCAT1 (Rosnoblet et al., 2017), pointing to its broader impact on antioxidant defense. Interestingly, AtCDC48A controls the degradation of intrachloroplastic proteins during oxidative stress and is vital for methyl viologen-induced oxidative stress tolerance (Li et al., 2022). Indeed, abundances of cytosolic, stromal and thylakoid APX are hypersensitive to highly oxidative conditions induced by prolonged exposure to methyl viologen (Melicher et al., 2022). Thus, CDC48 may govern APX, SOD and CAT degradation to modulate plant antioxidant defense and may play a key role as a regulator of the cellular redox status.
Using Co-IP/MS, APX1 was found to interact with plasma membrane-localized aquaporins PIP2A, PIP1B and ammonium transporter 1.3 (AMT1;3; Bellati et al., 2016; Table S3). In addition, high throughput Y2H study indicated an interaction with CERK1, which also resides in plasma membrane (Table S3; Le et al., 2014). Although the false positivity of these identifications cannot be ruled out, they may interact with diverse cytosolic proteins via their cytosolic loops, as it was described for PIPs (Roche and Törnroth-Horsefield, 2017). APX1 co-purified with nucleolar A2-type cyclin CYCA2.3 in a TAP-MS experiment (Table S3; Boudolf et al., 2009). However, due to different localizations, the above-mentioned interaction remains to be validated by additional experiments.
Low-throughput Co-IP and Y2H assays revealed, that two peroxisomal membrane-bound APX isoforms APX3 and APX5 interact with cytosol- and nucleus-localized ankyrin repeat-containing protein 2A (AKR2A; Table 3; Shen et al., 2010). AKR2A interacts via N-terminal PEST (named after Pro-Glu-Ser-Thr) domain with the C-terminal transmembrane domain of nascent APXs to maintain their stability and to prevent protein aggregation (Shen et al., 2010). Indeed, AKR2A may bind and stabilize nascent proteins targeted to diverse organelles, such as chloroplast outer envelope membrane (OEM) proteins, contributing to chloroplast biogenesis and protein trafficking (Bae et al., 2008; Kim et al., 2015). AKR2A overexpression leads to upregulation of enzymes involved in H2O2 signaling, including APX1 and secretory peroxidase isoforms (Hu et al., 2020), indicating that AKR2A may have a broader impact on antioxidant defense. Of note, AKR2A undergoes oxidative modifications on Cys259 (S-nitrosylation and S-sulfenylation) and on Cys266 (S-sulfenylation; Hu et al., 2015; Huang et al., 2019), enabling possible feedback regulation of APXs and rapid responsivity to fluctuating cellular redox homeostasis. Plant AKR2A comprises four conserved amino acid regions, including the above-mentioned PEST sequence, two central domains, and a C-terminal ankyrin repeat domain (ARD). Interestingly, the PEST sequence exists only in land plants, while the C-terminal ARD domain has the highest conservation across prokaryotes, eukaryotes, green algae and land plants (Kim et al., 2014). This suggests that APX3-AKR2A interaction occurs solely in land plants.
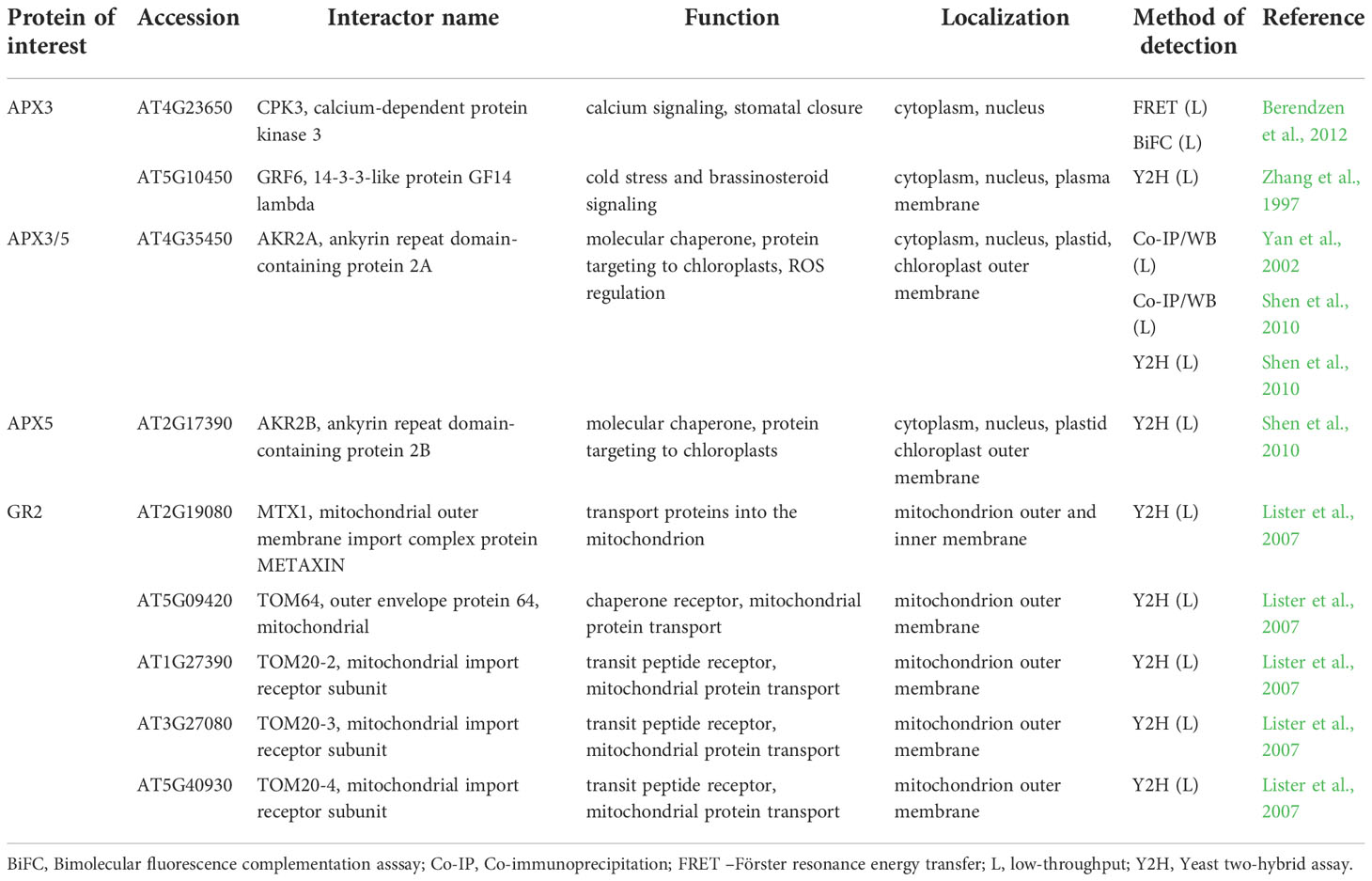
Table 3 Interaction partners of Arabidopsis ascorbate-glutathione cycle enzymes found by low-throughput methods with their respective function and localization.
According to low-throughput Y2H and Co-IP/WB analyses, both AKR2A and APX3 interact with a 14-3-3-like protein GF14 lambda (GRF6; Table 3; Zhang et al., 1997; Yan et al., 2002). Although GRF6 is not required for ANKR2A-APX3 interaction, it is proposed that this complex modulates the APX3-mediated antioxidant defense during stress responses.
Considering that interaction of ANKR2A-APX3 occurs in the cytosol, the positive effect of ANKR2A on APX3 stability might allow APX3 to interact with other cytosolic proteins, such as Ca2+-dependent protein kinase 3 (CPK3; Berendzen et al., 2012), thus linking APX3 with Ca2+ signaling. The function of this interaction, examined by FRET-FLIM and BiFC assay combined with flow cytometry (Berendzen et al., 2012), was not described in detail. CPKs were identified in plants and protozoans, but not in animal kingdom or yeast (Harmon et al., 2000), and are involved in various signaling pathways during biotic and abiotic stress responses. In Arabidopsis, CPK3 modulates salt stress response by phosphorylation of vacuolar two-pore K+ channel 1 (AtTPK1; Latz et al., 2013). Notably, AtTPK1 interacts with GRF6 via its pSer42 residue (Latz et al., 2007), which is phosphorylated by CPK3. As noted above, CPK3 and GRF6 are verified APX3 interaction partners (Table 3), therefore APX3, GRF6 and CPK3 may form a complex involved in the regulation of salt stress response.
The interactions of stromal sAPX and thylakoid tAPX have not been resolved so far.
MDARs, enzymes using NADH and NADPH as electron donors compartmentalize into cytosol, chloroplasts, mitochondria, and peroxisomes (Tanaka et al., 2021) and are crucial for plant abiotic and biotic stress responses (Li et al., 2010; Yeh et al., 2019). They are regulated on transcriptional and posttranscriptional levels (Feng et al., 2014). Nevertheless, the mechanism of their activation is only scarcely known. MDAR1 and MDAR2 are subjected to phosphorylation and nitration in response to diverse stimuli (Begara-Morales et al., 2015; Dvořák et al., 2021b) and they are modulated also by thioredoxins (Vanacker et al., 2018).
As shown by a large-scale study applying CF-MS (Table S3; McWhite et al., 2020), MDAR1/2/3 and 6 belong to a multiprotein complex comprised mainly of metabolic proteins. These include two isoforms of mitochondrial malate dehydrogenase, essential for central metabolism, photorespiration and redox homeostasis between organelles (Sun et al., 2019). In addition, mitochondrial succinyl-CoA synthetase activity is present as well. Transaldolase 2 (TRA2), regulating the balance of metabolites in the pentose-phosphate pathway, and proteins involved in carbohydrate partitioning, sucrose synthesis, and cell wall biogenesis are also components of this complex, namely, two isoforms of phosphoglucomutase 2 and 3 (PGM2 and PGM3), cytosolic and plastidial isoform of triosephosphate isomerase (TPI and TIM, respectively), and UTP-glucose-1-phosphate uridylyltransferase 1 and 2 (UGP1 and UGP2). Thus, MDARs might stand for unique enzymes adapting the primary metabolism to redox homeostasis perceived by ascorbate-glutathione pathway.
Arabidopsis has two cytosolic (DHAR1 and DHAR2) and one chloroplastic (DHAR3) DHAR isoforms. They are highly responsive to a wide range of stimuli and their overexpression confers plant resistance to abiotic stresses (Chen and Gallie, 2006; Noshi et al., 2017). Their expression and activity are linked to MAPK signaling, Ca2+ levels, and expression of transcription factors (Dvořák et al., 2021b). However, direct regulation of DHARs by TFs was not experimentally proved so far. In addition, DHARs undergo S-glutathionylation (Dixon et al., 2005) and have been identified as thioredoxin targets (Hägglund et al., 2008).
DHAR1 interactors have been found mainly by large-scale study exploiting a mating-based split-ubiquitin system accommodated for membrane protein interactions with other membrane or cytosolic proteins (Table S3; Jones et al., 2014). This approach identified 46 potential DHAR1 membrane-localized interacting partners, mainly involving receptors and transporters. DHAR1 may interact with 5 members of multidrug and toxic compound extrusion (MATE) protein family involved in plant resistance to multiple cytotoxic compounds, including antibiotics and heavy metals. They are also capable of transporting flavonoids or anthocyanins and have developmental functions (Remy and Duque, 2014; Suzuki et al., 2015; Miyauchi et al., 2017). Two of these interactions (DTX20 and DTX14) were verified by in planta by GFP split assay (Jones et al., 2014), increasing the reliability of the interaction identification.
DHAR1 may also interact with nitrate transporter 1.12 (NTR1.12), glucose transporter 1 (GLT1; both interactions proved in planta), proteins transporting phosphoenolpyruvate (pep)/phosphate (phosphoenolpyruvate (pep)/phosphate translocator 2; (PPT2)), H+ or Na+ (cation/H+ antiporter 27; (CHX27)), GABA (putative GABA transporter 2 (GAT2)), L-cystine (cystinosin homolog), sucrose (bidirectional sugar transporter SWEET9) and UDP-xylose (UDP-xylose transporter 1 (UXT1); Table S3; Jones et al., 2014). This implies possible novel functions of DHAR1, linking redox homeostasis to membrane integrity ensuring transmembrane transport of different compounds. Cytosolic DHAR1, DHAR2 and DHAR3 interactions have not been studied so far.
Glutathione reductases (GRs) are catalyzing the NADPH dependent recovery of reduced glutathione consumed by DHAR activity. Thus, they substantially contribute to glutathione recycling. Arabidopsis GR isoforms are localized in the cytosol, peroxisomes (GR1), chloroplasts and mitochondria (GR2; Gill et al., 2013). They are important for ROS regulation under diverse stimuli (Müller-Schüssele et al., 2020). According to a large-scale study exploiting CF-MS (Table S3; McWhite et al., 2020), GR1 and GR2 are members of a complex containing two isoforms of protein disulfide isomerase-like proteins PDIL1-1 and PDIL1-2, proteins involved in oxidative protein folding (Fan et al., 2022) and two chloroplastic transketolase isoforms TKL1 and TKL2 involved in carbon metabolism (Rocha et al., 2014). The cytosolic and membrane anchored aminopeptidase M1 (APM1), a potential interactor of GR1 and GR2, is involved in auxin transport as knockout mutant in APM1 shows irregular cell divisions during embryogenesis (Peer et al., 2009). Other proteins represent enzymes involved in primary (Dihydroxy-acid dehydratase (DHAD), 2-hydroxyacyl-CoA lyase (HACL), pyridoxine/pyridoxamine 5’-phosphate oxidase 1 (PPOX1), sulfoquinovosyldiacylglycerol 1 (SQD1)) or secondary metabolism (chorismate synthase (EMB1144)). Additionally, GR2 interacts with components of mitochondrial protein transport (Table 3) including TOM20 mitochondrial import receptor subunits TOM20-2/3 and 4, mitochondrial outer envelope protein 64 (TOM64) and mitochondrial outer membrane import complex protein METAXIN (Table 3; Lister et al., 2007), which is in accordance to GR2 mitochondrial localization.
RACK1 comprises seven repetitive WD-40 motifs with β-propeller structure that confers PPI capabilities (Adams et al., 2011; Figure 1A). It is conserved in all eukaryotic organisms with high DNA sequence identity and exhibits high sequence and structural conservation among diverse plant species (Iwasaki et al., 1995; McKhann et al., 1997; Chen et al., 2006a; Figure 1B). It is involved in plant developmental processes as well as responses to biotic and abiotic stresses (Zhang et al., 2013a; Islas-Flores et al., 2015). In Arabidopsis, three RACK1 isoforms are encoded by RACK1A, RACK1B and RACK1C genes (Chen et al., 2006a), while there are two isoforms in rice (OsRACK1A and OsRACK1B; Nakashima et al., 2008) and one in alfalfa (Msgb1; McKhann et al., 1997). It is localized to cytosol and nucleus where it may interact with various proteins (Speth et al., 2013). Currently, BioGRID, a database of protein, genetic and chemical interactions (Oughtred et al., 2021) reports 295 protein interactors for Arabidopsis RACK1A and at least 17 of them were validated by more than one PPI method so far.
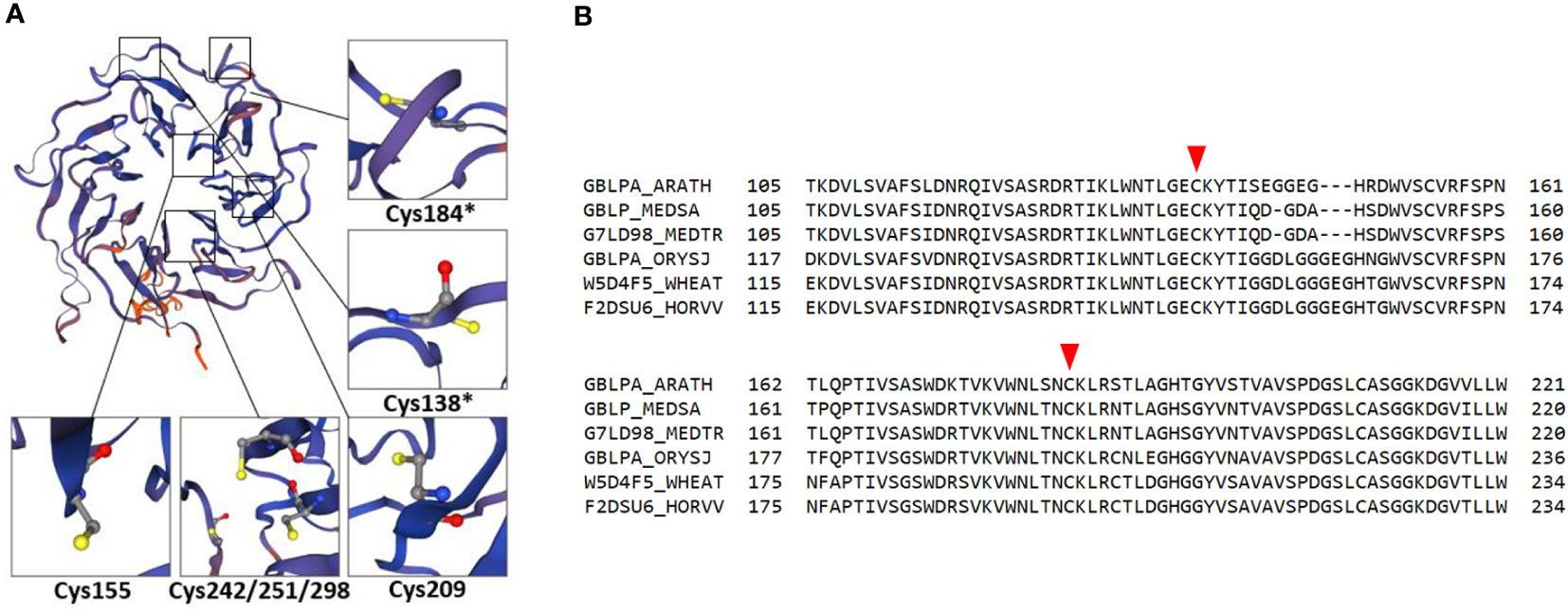
Figure 1 Redox-active cysteines in RACK1A amino acid sequence. (A) RACK1A tertiary structure model with detail view of cysteine residues. Asterisks indicate redox-active cysteine residues with high surface accessibility. The model is based on maltose-binding protein fused with RACK1A from Arabidopsis and was built using Expasy SWISS-MODEL software (Bienert et al., 2017; Waterhouse et al., 2018). (B) Alignment of RACK1A amino acid sequence ranging from 105 to 221 amino acids from Arabidopsis with orthologous proteins in different plant species. Orthologous proteins from Oryza sativa (GBLPA_ORYSJ), Hordeum vulgare (F2DSU6_HORVV), Triticum aestivum (W5D4F5_WHEAT), Medicago sativa (GBLP_MEDSA), and Medicago truncatula (G7LD98_MEDTR) are aligned to Arabidopsis RACK1A (GBLPA_ARATH) protein sequence. Red arrowheads indicate redox-active conserved cysteines.
In plants, RACK1 is a crucial regulator of ABA responses during seed germination, root growth and cotyledon greening (Guo et al., 2019). RACK1 affects ABA signaling through the interaction with eukaryotic translation initiation factors (Guo et al., 2011) or proteins involved in miRNA biogenesis (Speth et al., 2013). Genetic modification of RACK1A expression is associated with alteration of genes involved in ABA biosynthesis (Zhang et al., 2018). In addition, RACK1 serves as a scaffold for MAPK cascade in response to bacterial protease IV and ArgC in Arabidopsis (Cheng et al., 2015).
A growing evidence shows that RACK1 significantly contributes to ROS homeostasis by regulating either ROS generation or scavenging. RACK1 interacts with NADPH oxidase, a plasma-membrane localized superoxide anion-generating enzyme implicated in plant immune responses, abiotic stress tolerance and development (Sagi and Fluhr, 2006). OsRACK1A activates NADPH oxidase encoded by RbohB gene by recruiting its activator Rac1 to Rac1-immune complex as found by low-throughput Y2H, split-ubiquitin and Co-IP assays, and leads to increased ROS production (Nakashima et al., 2008). ROS generation mediated by RACK1-RBOH interaction is important for ABA responses as well (Nakashima et al., 2008; Zhang et al., 2014; Li et al., 2017a).
In addition to NADPH oxidases, genetic modification of RACK1 often causes alterations of enzymes involved in ROS decomposition. For example, downregulation of OsRACK1A enhanced SOD activity, decreased lipid peroxidation and increased polyethylene glycol-mediated drought stress tolerance in rice (Li et al., 2009). The impact of RACK1 on antioxidants might be broader, because soybean GmRACK1-RNAi lines displayed higher CAT, SOD, APX and GST transcript levels and SOD, POD and CAT activities during drought stress. On the other hand, GmRACK1 overexpressing lines exhibited reduced SOD, POD and CAT activities, which correlated with increased ROS levels (Li et al., 2018).
As revealed by high throughput studies, one possible mechanism how RACK1 may modulate antioxidant enzymes is through PPIs (Table 4). Antioxidant enzymes, including those involved in detoxification of superoxide and H2O2 were identified as putative RACK1-interacting partners (Table 4). Cytoplasmic and nuclear localization of RACK1A suggests that this scaffold protein should be effective and functionally important mainly in these subcellular compartments. It might be expected, that interactions of RACK1s with antioxidant enzymes will modulate their activity or enable/prevent interaction with other regulatory factors.
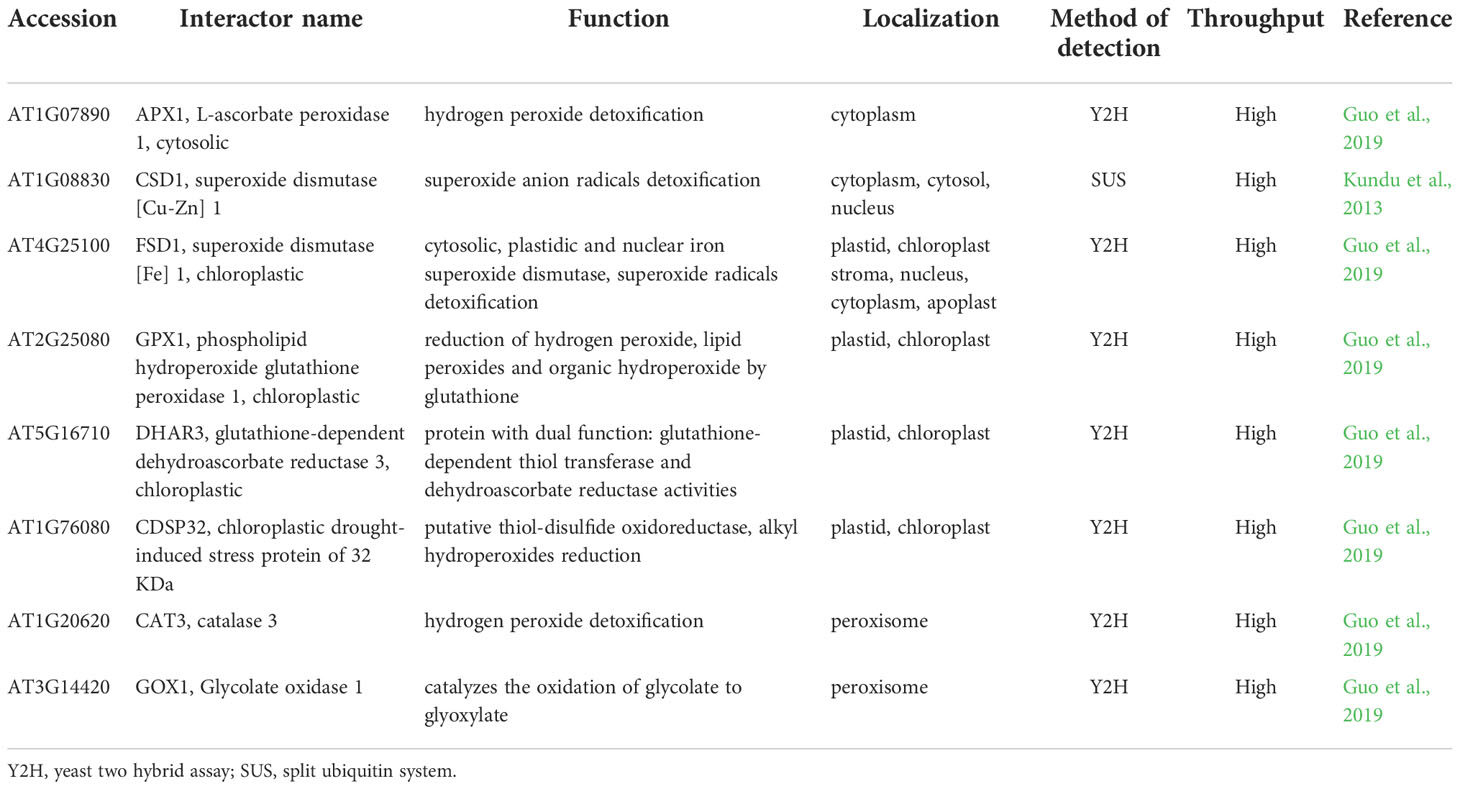
Table 4 Known protein-protein interactions of receptor for activated C kinase 1 (RACK1) with antioxidant enzymes.
According to high-throughput Y2H PPI study, RACK1A may interact with cytosolic APX1 (Guo et al., 2019; Table 4). Noteworthy, both RACK1A and APX1 are involved in plant responses to ABA. APX1 promoter possesses an ABA response element (ABRE; Saxena et al., 2020) and APX1 is thought to be regulated by ABA. During simultaneous exposure to drought and heat stress, APX1 is likely responsible for regulation of H2O2 level important for stomata closure (Zandalinas et al., 2016). Additionally, APX1 knock-out mutant exerts suppressed levels of RACK1A transcript in Arabidopsis, indicating the link between APX1 and RACK1A in H2O2 signaling pathways (Pnueli et al., 2003). These data indicate possible involvement of RACK1-APX1 interaction in plant ABA signaling.
RACK1 is also linked to the Cu2+-dependent regulation of SODs. CSDs and FSD1 expressions are regulated in Cu2+-dependent manner via SPL7 and miR398 (Pilon et al., 2011). Interestingly, RACK1 isoforms are involved in the biogenesis of miR398, leading to negative regulation of CSD1 and CSD2 (Speth et al., 2013). This possibly occurs during salt stress conditions or ABA response, judging from the fact, that miRNA398 levels are decreased under salt stress conditions, resulting in enhanced CSD1 and CSD2 expression (Jagadeeswaran et al., 2009). The possible mechanistic link between RACK1A and FSD1 is supposed by the similar germination response of fsd1 and rack1a mutants to salt stress (Guo et al., 2009; Dvořák et al., 2021a). However, RACK1 interacts with FSD1 and CSD1 also directly as revealed by high-throughput Y2H (FSD1) and low-throughput split-ubiquitin assay (CSD1; Table 4; Table S1; Kundu et al., 2013; Guo et al., 2019).
Previous high- and low-throughput Y2H, BiFC and pull-down interaction studies indicate that both RACK1A and FSD1 interact with components of G-protein complex. While RACK1A interacts with regulator of G-protein signaling 1(AtRGS1), AGB1 and guanine nucleotide-binding protein subunit gamma 1 and 2 (AGG1/2), FSD1 interacts with AtRGS1, GPA1 and AGB1 (Klopffleisch et al., 2011; Olejnik et al., 2011; Cheng et al., 2015). AtRGS1 accelerates the intrinsic GTP hydrolysis rate of GPA1, thus acting as a repressor of the active state of GPA1 (Chen et al., 2003). AtRGS1 was shown to be involved in glucose- and ABA-mediated signaling during seed germination and is responsible for the induction of ABA-responsive and ABA biosynthetic genes (Chen et al., 2006b).
The proposed interactions of RACK1A with other redox-related plastidic enzymes (glutathione peroxidase 1 (GPX1), DHAR3, chloroplastic drought-induced stress protein of 32 KDa (CDSP32) and peroxisomal enzymes (CAT3, glycolate oxidase 1 (GOX1); Guo et al., 2019) studied by high-throughput Y2H assay have not been validated and their function is not known (Table 4). RACK1A may not serve only as a scaffold mediating the interaction of other proteins during the redox responses, but may also stabilize the interaction partner and assist during its transport to the specific target organelle. In mouse melanocytes, interaction of VPS9-ankyrin repeat protein (VARP) with RACK1 leads to VARP stabilization (Marubashi et al., 2016). In human pulmonary artery endothelial cells, RACK1 interacts with TGF-β inhibited membrane-associated protein (TIMAP) and farnesyl transferase and ensures TIMAP prenylation and localization to the plasma membrane (Boratkó et al., 2013). It must be noted, that the reliability of Arabidopsis RACK1A PPIs with plastidic and peroxisomal proteins is questionable and they must be subjected to thorough validation.
A significant body of evidence shows that RACK1 is sensitive to oxidative/abiotic stress on mRNA and protein levels. Methyl viologen, a well-known oxidative stress inducer, enhanced the activity of the RACK1A promoter in Arabidopsis and overexpression of RACK1 in rice led to increased sensitivity to this stress (Fennell et al., 2021). Soybean GmRACK1 gene is downregulated by H2O2, suggesting its involvement in ROS signaling (Li et al., 2018). In addition, RACK1A homo-dimerizes under UV-B-induced oxidative stress as found by heterologous expression of AtRACK1A in yeast (Sabila et al., 2016). Notably, RACK1 protein may undergo redox PTMs. A redox proteomic study identified a reduction of conserved redox-sensitive cysteine residues in wheat RACK1 in response to ABA (Bykova et al., 2011). Specific redox-sensitive cysteine residues in the Arabidopsis RACK1A amino acid sequence can undergo S-nitrosylation (Cys138, Cys209; Hu et al., 2015), S-sulfenylation (Cys138, Cys 184; Huang et al., 2019) or S-sulfhydration (Cys138; Aroca et al., 2017), as proposed by proteomic studies. Bioinformatic modeling and predictions suggested that two of these cysteine residues (Cys138; Cys184) are redox-sensitive, have high surface accessibility (Figure 1A), and are conserved across plant kingdom (Figure 1B). These modifications may represent a possible regulatory mechanism by which RACK1 links external conditions with ROS homeostasis. Nevertheless, their physiological relevance remains unknown in plants. It may be expected that cysteine redox modifications modulate RACK1A function and have specific consequences in the plant stress tolerance. Oxidation of RACK1A may affect its localization, role in ABA signaling and MAPK scaffolding. In addition, it may also change the overall antioxidant capacity in the cell.
These data show that RACK1 may link cellular redox homeostasis under salt stress conditions or during ABA response to modulate antioxidant defense through PPIs (Figure 2). However, further functional studies are essential to support this hypothesis.
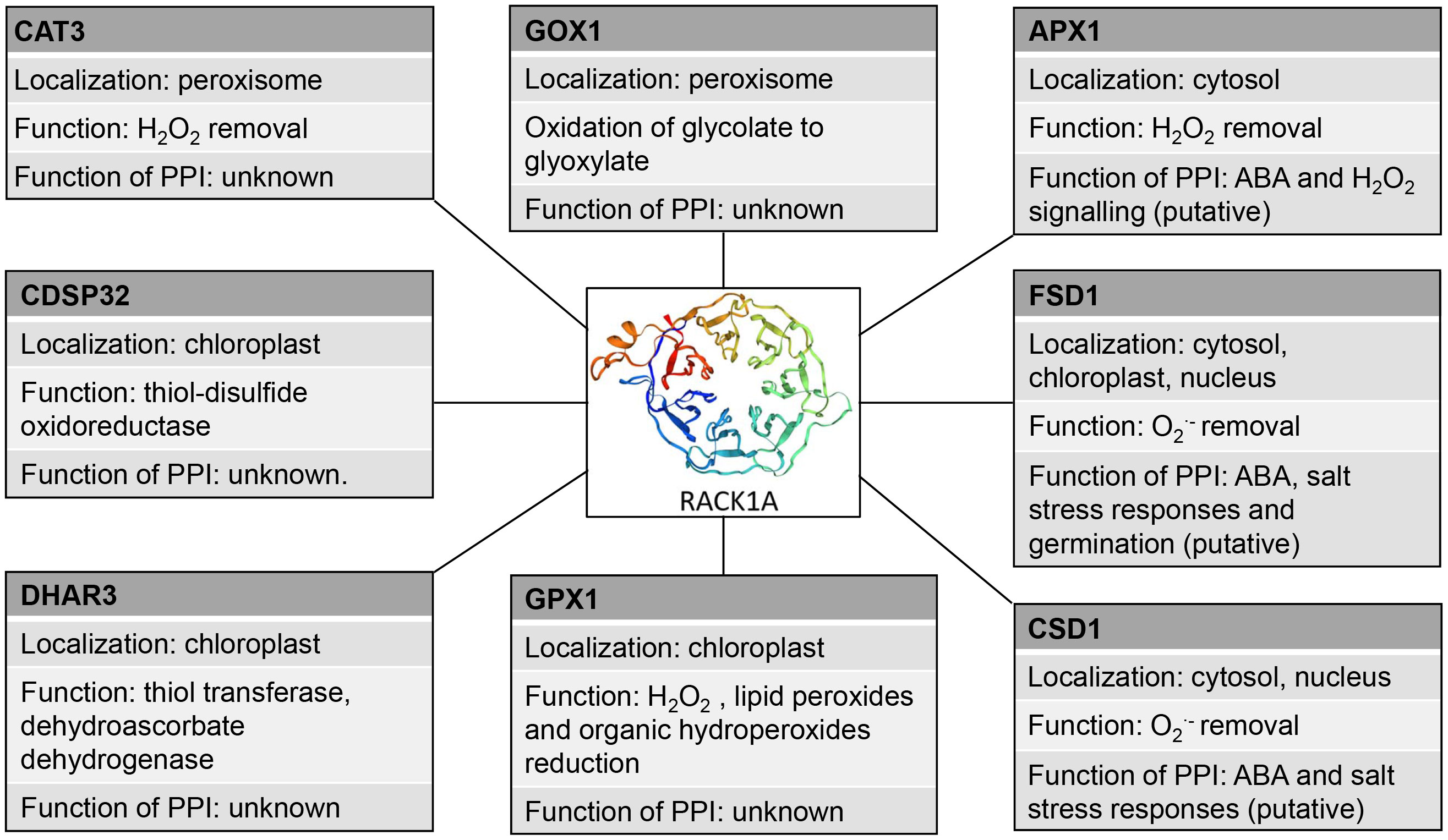
Figure 2 Schematic depiction of interaction partners of RACK1A with their antioxidant molecular functions and proposed functions of the interactions. PPI: protein-protein interaction, CAT3, catalase 3, GOX1 - Glycolate oxidase 1, APX1, ascorbate peroxidase 1, CDSP32 - chloroplastic drought-induced stress protein of 32 KDa, FSD1, iron superoxide dismutase, CSD1 Cu/Zn superoxide dismutase, GPX1, glutathuione peroxidase 1, DHAR3, dehydroascorbate reductase 3.
Our review shows that PPIs of antioxidant enzymes represent an important part of the machinery of ROS regulation. Through PPIs, these enzymes are regulated by folding, stabilization, degradation and activation, which have crucial consequences in ROS accumulation as well as plant stress tolerance. On the other hand, PPIs may link ROS scavenging with diverse metabolic and physiological processes. Importantly, proteins such as CDC48, LSDs and RACK1 may integrate regulation of multiple antioxidant enzymes, thus ensuring their synchronization and homeostasis. Nevertheless, PPIs in plant antioxidant defense are understudied and require further intensive research. It is of eminent importance to employ modern techniques of PPI identification combined with genetic and functional genetic studies. This should reveal the spatial and temporal orchestration of posttranslational, transcriptional and translational regulation of antioxidant enzymes employing PPIs.
PM, PD and TT drafted the manuscript which was revised and edited by TT and JŠ. All authors approved the final version of the manuscript. All authors contributed to the article and approved the submitted version.
This research was funded by Grant No. 19-00598S from the Czech Science Foundation GAČR and by the ERDF project “Plants as a tool for sustainable global development” (No.CZ.02.1.01/0.0/0.0/16_019/0000827).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1035573/full#supplementary-material
Abdel-Ghany, S. E., Burkhead, J. L., Gogolin, K. A., Andrés-Colás, N., Bodecker, J. R., Puig, S., et al. (2005). AtCCS is a functional homolog of the yeast copper chaperone Ccs1/Lys7. FEBS Lett. 579, 2307–2312. doi: 10.1016/j.febslet.2005.03.025
Adamiec, M., Ciesielska, M., Zalaś, P., Luciński, R. (2017). Arabidopsis thaliana intramembrane proteases. Acta Physiol. Plant 39, 146. doi: 10.1007/s11738-017-2445-2
Adamiec, M., Dobrogojski, J., Wojtyla, Ł., Luciński, R. (2022). Stress-related expression of the chloroplast EGY3 pseudoprotease and its possible impact on chloroplasts’ proteome composition. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.965143
Adamiec, M., Misztal, L., Kasprowicz-Maluśki, A., Luciński, R. (2020). EGY3: homologue of S2P protease located in chloroplasts. Plant Biol. (Stuttg) 22, 735–743. doi: 10.1111/plb.13087
Adams, D. R., Ron, D., Kiely, P. A. (2011). RACK1, a multifaceted scaffolding protein: Structure and function. Cell Commun. Signal 9, 22. doi: 10.1186/1478-811X-9-22
Al-Hajaya, Y., Karpinska, B., Foyer, C. H., Baker, A. (2022). Nuclear and peroxisomal targeting of catalase. Plant Cell Environ. 45, 1096–1108. doi: 10.1111/pce.14262
Altmann, M., Altmann, S., Rodriguez, P. A., Weller, B., Elorduy Vergara, L., Palme, J., et al. (2020). Extensive signal integration by the phytohormone protein network. Nature 583, 271–276. doi: 10.1038/s41586-020-2460-0
Arabidopsis Interactome Mapping Consortium (2011). Evidence for network evolution in an arabidopsis interactome map. Science 333, 601–607. doi: 10.1126/science.1203877
Aroca, A., Benito, J. M., Gotor, C., Romero, L. C. (2017). Persulfidation proteome reveals the regulation of protein function by hydrogen sulfide in diverse biological processes in arabidopsis. J. Exp. Bot. 68, 4915–4927. doi: 10.1093/jxb/erx294
Baba, K., Nakano, T., Yamagishi, K., Yoshida, S. (2001). Involvement of a nuclear-encoded basic helix-loop-helix protein in transcription of the light-responsive promoter of psbD. Plant Physiol. 125, 595–603. doi: 10.1104/pp.125.2.595
Bae, W., Lee, Y. J., Kim, D. H., Lee, J., Kim, S., Sohn, E. J., et al. (2008). AKR2A-mediated import of chloroplast outer membrane proteins is essential for chloroplast biogenesis. Nat. Cell. Biol. 10, 220–227. doi: 10.1038/ncb1683
Bah, A., Vernon, R. M., Siddiqui, Z., Krzeminski, M., Muhandiram, R., Zhao, C., et al. (2015). Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature 519, 106–109. doi: 10.1038/nature13999
Banks, C. J., Andersen, J. L. (2019). Mechanisms of SOD1 regulation by post-translational modifications. Redox Biol. 26, 101270. doi: 10.1016/j.redox.2019.101270
Barthelme, D., Sauer, R. T. (2013). Bipartite determinants mediate an evolutionarily conserved interaction between Cdc48 and the 20S peptidase. Proc. Natl. Acad. Sci. U. S. A. 110, 3327–3332. doi: 10.1073/pnas.1300408110
Bartoli, C. G., Buet, A., Gergoff Grozeff, G., Galatro, A., Simontacchi, M. (2017). ““Ascorbate-glutathione cycle and abiotic stress tolerance in plants,”,” in Ascorbic acid in plant growth, development and stress tolerance. Eds. Hossain, M. A., Munné-Bosch, S., Burritt, D. J., Diaz-Vivancos, P., Fujita, M., Lorence, A. (Cham: Springer International Publishing), 177–200. doi: 10.1007/978-3-319-74057-7_7
Begara-Morales, J. C., Sánchez-Calvo, B., Chaki, M., Mata-Pérez, C., Valderrama, R., Padilla, M. N., et al. (2015). Differential molecular response of monodehydroascorbate reductase and glutathione reductase by nitration and S -nitrosylation. J. Exp. Bot. 66, 5983–5996. doi: 10.1093/jxb/erv306
Bègue, H., Besson-Bard, A., Blanchard, C., Winckler, P., Bourque, S., Nicolas, V., et al. (2019). The chaperone-like protein CDC48 regulates ascorbate peroxidase in tobacco. J. Exp. Bot. 70, 2665–2681. doi: 10.1093/jxb/erz097
Bègue, H., Jeandroz, S., Blanchard, C., Wendehenne, D., Rosnoblet, C. (2017). Structure and functions of the chaperone-like p97/CDC48 in plants. Biochim. Biophys. Acta Gen. Subj. 1861, 3053–3060. doi: 10.1016/j.bbagen.2016.10.001
Bellati, J., Champeyroux, C., Hem, S., Rofidal, V., Krouk, G., Maurel, C., et al. (2016). Novel aquaporin regulatory mechanisms revealed by interactomics. Mol. Cell Prot. 15, 3473–3487. doi: 10.1074/mcp.M116.060087
Berendzen, K. W., Böhmer, M., Wallmeroth, N., Peter, S., Vesić, M., Zhou, Y., et al. (2012). Screening for in planta protein-protein interactions combining bimolecular fluorescence complementation with flow cytometry. Plant Methods 8, 25. doi: 10.1186/1746-4811-8-25
Bienert, S., Waterhouse, A., de Beer, T. A. P., Tauriello, G., Studer, G., Bordoli, L., et al (2017). The SWISS-MODEL Repository - new features and functionality. Nucleic Acids Res. 45, D313–D319. doi: 10.1093/nar/gkw1132
Blikstad, C., Ivarsson, Y. (2015). High-throughput methods for identification of protein-protein interactions involving short linear motifs. Cell Commun. Signal 13, 38. doi: 10.1186/s12964-015-0116-8
Boratkó, A., Gergely, P., Csortos, C. (2013). RACK1 is involved in endothelial barrier regulation via its two novel interacting partners. Cell Commun. Signal 11, 2. doi: 10.1186/1478-811X-11-2
Boudolf, V., Lammens, T., Boruc, J., Van Leene, J., Van Den Daele, H., Maes, S., et al. (2009). CDKB1;1 forms a functional complex with CYCA2;3 to suppress endocycle onset. Plant Physiol. 150, 1482–1493. doi: 10.1104/pp.109.140269
Bykova, N. V., Hoehn, B., Rampitsch, C., Banks, T., Stebbing, J.-A., Fan, T., et al. (2011). Redox-sensitive proteome and antioxidant strategies in wheat seed dormancy control. Proteomics 11, 865–882. doi: 10.1002/pmic.200900810
Cabreira, C., Cagliari, A., Bücker-Neto, L., Margis-Pinheiro, M., de Freitas, L. B., Bodanese-Zanettini, M. H. (2015). The phylogeny and evolutionary history of the lesion simulating disease (LSD) gene family in viridiplantae. Mol. Genet. Genomics 290, 2107–2119. doi: 10.1007/s00438-015-1060-4
Carroll, M. C., Girouard, J. B., Ulloa, J. L., Subramaniam, J. R., Wong, P. C., Valentine, J. S., et al. (2004). Mechanisms for activating Cu- and zn-containing superoxide dismutase in the absence of the CCS Cu chaperone. Proc. Natl. Acad. Sci. U. S. A. 101, 5964–5969. doi: 10.1073/pnas.0308298101
Castro, B., Citterico, M., Kimura, S., Stevens, D. M., Wrzaczek, M., Coaker, G. (2021). Stress-induced reactive oxygen species compartmentalization, perception and signalling. Nat. Plants 7, 403–412. doi: 10.1038/s41477-021-00887-0
Chang, I.-F., Curran, A., Woolsey, R., Quilici, D., Cushman, J., Mittler, R., et al. (2009). Proteomic profiling of tandem affinity purified 14-3-3 protein complexes in arabidopsis thaliana. Proteomics 9, 2967–2985. doi: 10.1002/pmic.200800445
Chen, C.-Y., Chen, J., He, L., Stiles, B. L. (2018). PTEN: Tumor suppressor and metabolic regulator. Front. Endocrinol. 9. doi: 10.3389/fendo.2018.00338
Chen, Z., Gallie, D. R. (2006). Dehydroascorbate reductase affects leaf growth, development, and function. Plant Physiol. 142, 775–787. doi: 10.1104/pp.106.085506
Cheng, Z., Li, J.-F., Niu, Y., Zhang, X.-C., Woody, O. Z., Xiong, Y., et al. (2015). Pathogen-secreted proteases activate a novel plant immune pathway. Nature 521, 213–216. doi: 10.1038/nature14243
Chen, Y., Ji, F., Xie, H., Liang, J., Zhang, J. (2006b). The regulator of G-protein signaling proteins involved in sugar and abscisic acid signaling in arabidopsis seed germination. Plant Physiol. 140, 302–310. doi: 10.1104/pp.105.069872
Chen, H., Lee, J., Lee, J.-M., Han, M., Emonet, A., Lee, J., et al. (2022). MSD2, an apoplastic Mn-SOD, contributes to root skotomorphogenic growth by modulating ROS distribution in arabidopsis. Plant Sci. 317, 111192. doi: 10.1016/j.plantsci.2022.111192
Chen, J.-G., Ullah, H., Temple, B., Liang, J., Guo, J., Alonso, J. M., et al. (2006a). RACK1 mediates multiple hormone responsiveness and developmental processes in arabidopsis. J. Exp. Bot. 57, 2697–2708. doi: 10.1093/jxb/erl035
Chen, J.-G., Willard, F. S., Huang, J., Liang, J., Chasse, S. A., Jones, A. M., et al. (2003). A seven-transmembrane RGS protein that modulates plant cell proliferation. Science 301, 1728–1731. doi: 10.1126/science.1087790
Chepelev, N., Chepelev, L., Alamgir, M. D., Golshani, A. (2008). Large-Scale protein-protein interaction detection approaches: Past, present and future. Biotechnol. Equip. 22, 513–529. doi: 10.1080/13102818.2008.10817505
Chu, C.-C., Lee, W.-C., Guo, W.-Y., Pan, S.-M., Chen, L.-J., Li, H.-M., et al. (2005). A copper chaperone for superoxide dismutase that confers three types of copper/zinc superoxide dismutase activity in arabidopsis. Plant Physiol. 139, 425–436. doi: 10.1104/pp.105.065284
Chu, X., Wang, J.-G., Li, M., Zhang, S., Gao, Y., Fan, M., et al. (2021). HBI transcription factor-mediated ROS homeostasis regulates nitrate signal transduction. Plant Cell 33, 3004–3021. doi: 10.1093/plcell/koab165
Cohu, C. M., Abdel-Ghany, S. E., Gogolin Reynolds, K. A., Onofrio, A. M., Bodecker, J. R., Kimbrel, J. A., et al. (2009). Copper delivery by the copper chaperone for chloroplast and cytosolic copper/zinc-superoxide dismutases: regulation and unexpected phenotypes in an arabidopsis mutant. Mol. Plant 2, 1336–1350. doi: 10.1093/mp/ssp084
Cowan, G. H., Roberts, A. G., Jones, S., Kumar, P., Kalyandurg, P. B., Gil, J. F., et al. (2018). Potato mop-top virus co-opts the stress sensor HIPP26 for long-distance movement. Plant Physiol. 176, 2052–2070. doi: 10.1104/pp.17.01698
Cui, Y., Zhang, X., Yu, M., Zhu, Y., Xing, J., Lin, J. (2019). Techniques for detecting protein-protein interactions in living cells: principles, limitations, and recent progress. Sci. China Life Sci. 62, 619–632. doi: 10.1007/s11427-018-9500-7
Czarnocka, W., van der Kelen, K., Willems, P., Szechyńska-Hebda, M., Shahnejat-Bushehri, S., Balazadeh, S., et al. (2017). The dual role of LESION SIMULATING DISEASE 1 as a condition-dependent scaffold protein and transcription regulator: Insight into the LSD1 molecular function. Plant Cell Environ. 40, 2644–2662. doi: 10.1111/pce.12994
de Abreu-Neto, J. B., Turchetto-Zolet, A. C., de Oliveira, L. F. V., Zanettini, M. H. B., Margis-Pinheiro, M. (2013). Heavy metal-associated isoprenylated plant protein (HIPP): characterization of a family of proteins exclusive to plants. FEBS J. 280, 1604–1616. doi: 10.1111/febs.12159
Després, C., Chubak, C., Rochon, A., Clark, R., Bethune, T., Desveaux, D., et al. (2003). The arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15, 2181–2191. doi: 10.1105/tpc.012849
Dixon, D. P., Skipsey, M., Grundy, N. M., Edwards, R. (2005). Stress-induced protein s-glutathionylation in arabidopsis. Plant Physiol. 138, 2233–2244. doi: 10.1104/pp.104.058917
Dorion, S., Rivoal, J. (2018). Plant nucleoside diphosphate kinase 1: A housekeeping enzyme with moonlighting activity. Plant Signal. Behav. 13, e1475804. doi: 10.1080/15592324.2018.1475804
Dreyer, B. H., Schippers, J. H. M. (2019). “Copper-zinc superoxide dismutases in plants: Evolution, enzymatic properties, and beyond,” in Annual plant reviews online, ed. Roberts, J. A. (Atlanta, GA: American Cancer Society), 933–968. doi: 10.1002/9781119312994.apr0705
Duan, G., Walther, D. (2015). The roles of post-translational modifications in the context of protein interaction networks. PloS Comput. Biol. 11, e1004049. doi: 10.1371/journal.pcbi.1004049
Dutta, S., Teresinski, H. J., Smith, M. D. (2014). A split-ubiquitin yeast two-hybrid screen to examine the substrate specificity of atToc159 and atToc132, two arabidopsis chloroplast preprotein import receptors. PloS One 9, e95026. doi: 10.1371/journal.pone.0095026
Dvořák, P., Krasylenko, Y., Ovečka, M., Basheer, J., Zapletalová, V., Šamaj, J., et al. (2021a). In vivo light-sheet microscopy resolves localisation patterns of FSD1, a superoxide dismutase with function in root development and osmoprotection. Plant Cell Environ. 44, 68–87. doi: 10.1111/pce.13894
Dvořák, P., Krasylenko, Y., Zeiner, A., Šamaj, J., Takáč, T. (2021b). Signaling toward reactive oxygen species-scavenging enzymes in plants. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.618835
Erwig, J., Ghareeb, H., Kopischke, M., Hacke, R., Matei, A., Petutschnig, E., et al. (2017). Chitin-induced and CHITIN ELICITOR RECEPTOR KINASE1 (CERK1) phosphorylation-dependent endocytosis of arabidopsis thaliana LYSIN MOTIF-CONTAINING RECEPTOR-LIKE KINASE5 (LYK5). New Phytol. 215, 382–396. doi: 10.1111/nph.14592
Espinoza, C., Liang, Y., Stacey, G. (2017). Chitin receptor CERK1 links salt stress and chitin-triggered innate immunity in arabidopsis. Plant J. 89, 984–995. doi: 10.1111/tpj.13437
Fan, T., Lv, T., Xie, C., Zhou, Y., Tian, C. (2021). Genome-wide analysis of the IQM gene family in rice (Oryza sativa l.). Plants (Basel) 10, 1949. doi: 10.3390/plants10091949
Fan, F., Zhang, Q., Zhang, Y., Huang, G., Liang, X., Wang, C., et al. (2022). Two protein disulfide isomerase subgroups work synergistically in catalyzing oxidative protein folding. Plant Physiol. 188, 241–254. doi: 10.1093/plphys/kiab457
Feng, H., Wang, X., Zhang, Q., Fu, Y., Feng, C., Wang, B., et al. (2014). Monodehydroascorbate reductase gene, regulated by the wheat PN-2013 miRNA, contributes to adult wheat plant resistance to stripe rust through ROS metabolism. Biochim. Biophys. Acta Gene Regul. Mech. 1839, 1–12. doi: 10.1016/j.bbagrm.2013.11.001
Fennell, H. W. W., Ullah, H., van Wijnen, A. J., Lewallen, E. A. (2021). Arabidopsis thaliana and oryza sativa receptor for activated c kinase 1 (RACK1) mediated signaling pathway shows hypersensitivity to oxidative stress. Plant Gene 27, 100299. doi: 10.1016/j.plgene.2021.100299
Förster, T. H. (1948). Zwischenmolekulare energiewanderung und fluoreszenz. Ann. Phys. 437, 55–75. doi: 10.1002/andp.19484370105
Freitas, M. O., Francisco, T., Rodrigues, T. A., Alencastre, I. S., Pinto, M. P., Grou, C. P., et al. (2011). PEX5 protein binds monomeric catalase blocking its tetramerization and releases it upon binding the n-terminal domain of PEX14. J. Biol. Chem. 286, 40509–40519. doi: 10.1074/jbc.M111.287201
Friedman, E. J., Wang, H. X., Jiang, K., Perovic, I., Deshpande, A., Pochapsky, T. C., et al. (2011). Acireductone dioxygenase 1 (ARD1) is an effector of the heterotrimeric G protein β subunit in arabidopsis. J. Biol. Chem. 286, 30107–30118. doi: 10.1074/jbc.M111.227256
Fujikawa, Y., Suekawa, M., Endo, S., Fukami, Y., Mano, S., Nishimura, M., et al. (2019). Effect of mutation of c-terminal and heme binding region of arabidopsis catalase on the import to peroxisomes. Biosci. Biotechnol. Biochem. 83, 322–325. doi: 10.1080/09168451.2018.1530094
Fukamatsu, Y., Yabe, N., Hasunuma, K. (2003). Arabidopsis NDK1 is a component of ROS signaling by interacting with three catalases. Plant Cell Physiol. 44, 982–989. doi: 10.1093/pcp/pcg140
Garcia-Molina, A., Altmann, M., Alkofer, A., Epple, P. M., Dangl, J. L., Falter-Braun, P. (2017). LSU network hubs integrate abiotic and biotic stress responses via interaction with the superoxide dismutase FSD2. J. Exp. Bot. 68, 1185–1197. doi: 10.1093/jxb/erw498
Gavin, A.-C., Maeda, K., Kühner, S. (2011). Recent advances in charting protein–protein interaction: mass spectrometry-based approaches. Curr. Opin. Biotechnol. 22, 42–49. doi: 10.1016/j.copbio.2010.09.007
Gelhaye, E., Navrot, N., Macdonald, I. K., Rouhier, N., Raven, E. L., Jacquot, J.-P. (2006). Ascorbate peroxidase-thioredoxin interaction. Photosynth. Res. 89, 193–200. doi: 10.1007/s11120-006-9100-x
Gill, S. S., Anjum, N. A., Hasanuzzaman, M., Gill, R., Trivedi, D. K., Ahmad, I., et al. (2013). Glutathione and glutathione reductase: a boon in disguise for plant abiotic stress defense operations. Plant Physiol. Biochem. 70, 204–212. doi: 10.1016/j.plaphy.2013.05.032
Gingras, A.-C., Gstaiger, M., Raught, B., Aebersold, R. (2007). Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell Biol. 8, 645–654. doi: 10.1038/nrm2208
Girotto, S., Cendron, L., Bisaglia, M., Tessari, I., Mammi, S., Zanotti, G., et al. (2014). DJ-1 is a copper chaperone acting on SOD1 activation. J. Biol. Chem. 289, 10887–10899. doi: 10.1074/jbc.M113.535112
Guo, J., Hu, Y., Zhou, Y., Zhu, Z., Sun, Y., Li, J., et al. (2019). Profiling of the receptor for activated c kinase 1a (RACK1a) interaction network in arabidopsis thaliana. Biochem. Biophys. Res. Commun. 520, 366–372. doi: 10.1016/j.bbrc.2019.09.142
Guo, P., Jiang, S., Bai, C., Zhang, W., Zhao, Q., Liu, C. (2015). Asymmetric functional interaction between chaperonin and its plastidic cofactors. FEBS J. 282, 3959–3970. doi: 10.1111/febs.13390
Guo, J., Wang, S., Valerius, O., Hall, H., Zeng, Q., Li, J.-F., et al. (2011). Involvement of arabidopsis RACK1 in protein translation and its regulation by abscisic acid. Plant Physiol. 155, 370–383. doi: 10.1104/pp.110.160663
Guo, J., Wang, J., Xi, L., Huang, W.-D., Liang, J., Chen, J.-G. (2009). RACK1 is a negative regulator of ABA responses in arabidopsis. J. Exp. Bot. 60, 3819–3833. doi: 10.1093/jxb/erp221
Guo, T., Weber, H., Niemann, M. C. E., Theisl, L., Leonte, G., Novák, O., et al. (2021). Arabidopsis HIPP proteins regulate endoplasmic reticulum-associated degradation of CKX proteins and cytokinin responses. Mol. Plant 14, 1918–1934. doi: 10.1016/j.molp.2021.07.015
Hackenberg, T., Juul, T., Auzina, A., Gwizdz, S., Malolepszy, A., van der Kelen, K., et al. (2013). Catalase and NO CATALASE ACTIVITY1 promote autophagy-dependent cell death in arabidopsis. Plant Cell 25, 4616–4626. doi: 10.1105/tpc.113.117192
Hägglund, P., Bunkenborg, J., Maeda, K., Svensson, B. (2008). Identification of thioredoxin disulfide targets using a quantitative proteomics approach based on isotope-coded affinity tags. J. Proteome Res. 7, 5270–5276. doi: 10.1021/pr800633y
Haque, M. E., Yoshida, Y., Hasunuma, K. (2010). ROS resistance in pisum sativum cv. Alaska: the involvement of nucleoside diphosphate kinase in oxidative stress responses via the regulation of antioxidants. Planta 232, 367–382. doi: 10.1007/s00425-010-1173-2
Harmon, A. C., Gribskov, M., Harper, J. F. (2000). CDPKs, a kinase for every Ca2+ signal? Trends Plant Sci. 5, 154–159. doi: 10.1016/S1360-1385(00)01577-6
Haslbeck, M., Vierling, E. (2015). A first line of stress defense: small heat shock proteins and their function in protein homeostasis. J. Mol. Biol. 427, 1537–1548. doi: 10.1016/j.jmb.2015.02.002
Huang, C.-H., Kuo, W.-Y., Weiss, C., Jinn, T.-L. (2012). Copper chaperone-dependent and -independent activation of three copper-zinc superoxide dismutase homologs localized in different cellular compartments in arabidopsis. Plant Physiol. 158, 737–746. doi: 10.1104/pp.111.190223
Huang, J., Willems, P., Wei, B., Tian, C., Ferreira, R. B., Bodra, N., et al. (2019). Mining for protein s-sulfenylation in arabidopsis uncovers redox-sensitive sites. Proc. Natl. Acad. Sci. U. S. A. 116, 21256–21261. doi: 10.1073/pnas.1906768116
Hubbard, S. R., Till, J. H. (2000). Protein tyrosine kinase structure and function. Annu. Rev. Biochem. 69, 373–398. doi: 10.1146/annurev.biochem.69.1.373
Hu, W., Chen, L., Qiu, X., Wei, J., Lu, H., Sun, G., et al. (2020). AKR2A participates in the regulation of cotton fibre development by modulating biosynthesis of very-long-chain fatty acids. Plant Biotechnol. J. 18, 526–539. doi: 10.1111/pbi.13221
Hu, C.-D., Chinenov, Y., Kerppola, T. K. (2002). Visualization of interactions among bZIP and rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9, 789–798. doi: 10.1016/S1097-2765(02)00496-3
Hu, J., Huang, X., Chen, L., Sun, X., Lu, C., Zhang, L., et al. (2015). Site-specific nitrosoproteomic identification of endogenously s-nitrosylated proteins in arabidopsis. Plant Physiol. 167, 1731–1746. doi: 10.1104/pp.15.00026
Hu, S.-H., Jinn, T.-L. (2022). Impacts of Mn, fe, and oxidative stressors on MnSOD activation by AtMTM1 and AtMTM2 in arabidopsis. Plants (Basel) 11, 619. doi: 10.3390/plants11050619
Hu, S.-H., Lin, S.-F., Huang, Y.-C., Huang, C.-H., Kuo, W.-Y., Jinn, T.-L. (2021). Significance of AtMTM1 and AtMTM2 for mitochondrial MnSOD activation in arabidopsis. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.690064
Igawa, T., Fujiwara, M., Takahashi, H., Sawasaki, T., Endo, Y., Seki, M., et al. (2009). Isolation and identification of ubiquitin-related proteins from arabidopsis seedlings. J. Exp. Bot. 60, 3067–3073. doi: 10.1093/jxb/erp134
Iglesias, M. J., Terrile, M. C., Correa-Aragunde, N., Colman, S. L., Izquierdo-Álvarez, A., Fiol, D. F., et al. (2018). Regulation of SCFTIR1/AFBs E3 ligase assembly by s-nitrosylation of arabidopsis SKP1-like1 impacts on auxin signaling. Redox Biol. 18, 200–210. doi: 10.1016/j.redox.2018.07.003
Islas-Flores, T., Rahman, A., Ullah, H., Villanueva, M. A. (2015). The receptor for activated c kinase in plant signaling: Tale of a promiscuous little molecule. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.01090
Iwasaki, Y., Komano, M., Ishikawa, A., Sasaki, T., Asahi, T. (1995). Molecular cloning and characterization of cDNA for a rice protein that contains seven repetitive segments of the trp-asp forty-amino-acid repeat (WD-40 repeat). Plant Cell Physiol. 36, 505–510. doi: 10.1093/oxfordjournals.pcp.a078786
Jagadeeswaran, G., Saini, A., Sunkar, R. (2009). Biotic and abiotic stress down-regulate miR398 expression in arabidopsis. Planta 229, 1009–1014. doi: 10.1007/s00425-009-0889-3
Jayashree, S., Murugavel, P., Sowdhamini, R., Srinivasan, N. (2019). Interface residues of transient protein-protein complexes have extensive intra-protein interactions apart from inter-protein interactions. Biol. Direct 14, 1. doi: 10.1186/s13062-019-0232-2
Jensen, L. T., Culotta, V. C. (2005). Activation of CuZn superoxide dismutases from Caenorhabditis elegans does not require the copper chaperone CCS. J. Biol. Chem. 280, 41373–41379. doi: 10.1074/jbc.M509142200
Jiao, Z., Tian, Y., Cao, Y., Wang, J., Zhan, B., Zhao, Z., et al. (2021). A novel pathogenicity determinant hijacks maize catalase 1 to enhance viral multiplication and infection. New Phytol. 230, 1126–1141. doi: 10.1111/nph.17206
Jones, A. M., Xuan, Y., Xu, M., Wang, R.-S., Ho, C.-H., Lalonde, S., et al. (2014). Border control–a membrane-linked interactome of arabidopsis. Science 344, 711–716. doi: 10.1126/science.1251358
Kanno, T., Bucher, E., Daxinger, L., Huettel, B., Böhmdorfer, G., Gregor, W., et al. (2008). A structural-maintenance-of-chromosomes hinge domain–containing protein is required for RNA-directed DNA methylation. Nat. Genet. 40, 670–675. doi: 10.1038/ng.119
Karpinski, S., Escobar, C., Karpinska, B., Creissen, G., Mullineaux, P. M. (1997). Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in arabidopsis during excess light stress. Plant Cell 9, 627–640. doi: 10.1105/tpc.9.4.627
Kerppola, T. K. (2006). Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protoc. 1, 1278–1286. doi: 10.1038/nprot.2006.201
Kim, D. H., Lee, J.-E., Xu, Z.-Y., Geem, K. R., Kwon, Y., Park, J. W., et al. (2015). Cytosolic targeting factor AKR2A captures chloroplast outer membrane-localized client proteins at the ribosome during translation. Nat. Commun. 6, 6843. doi: 10.1038/ncomms7843
Kim, D. H., Park, M.-J., Gwon, G. H., Silkov, A., Xu, Z.-Y., Yang, E. C., et al. (2014). An ankyrin repeat domain of AKR2 drives chloroplast targeting through coincident binding of two chloroplast lipids. Dev. Cell 30, 598–609. doi: 10.1016/j.devcel.2014.07.026
Kim, D.-Y., Scalf, M., Smith, L. M., Vierstra, R. D. (2013). Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in arabidopsis. Plant Cell 25, 1523–1540. doi: 10.1105/tpc.112.108613
Kliebenstein, D. J., Monde, R. A., Last, R. L. (1998). Superoxide dismutase in arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiol. 118, 637–650. doi: 10.1104/pp.118.2.637
Klopffleisch, K., Phan, N., Augustin, K., Bayne, R. S., Booker, K. S., Botella, J. R., et al. (2011). Arabidopsis G-protein interactome reveals connections to cell wall carbohydrates and morphogenesis. Mol. Syst. Biol. 7, 532. doi: 10.1038/msb.2011.66
Kneeshaw, S., Keyani, R., Delorme-Hinoux, V., Imrie, L., Loake, G. J., Le Bihan, T., et al. (2017). Nucleoredoxin guards against oxidative stress by protecting antioxidant enzymes. Proc. Natl. Acad. Sci. U.S.A. 114, 8414–8419. doi: 10.1073/pnas.1703344114
König, J., Galliardt, H., Jütte, P., Schäper, S., Dittmann, L., Dietz, K.-J. (2013). The conformational bases for the two functionalities of 2-cysteine peroxiredoxins as peroxidase and chaperone. J. Exp. Bot. 64, 3483–3497. doi: 10.1093/jxb/ert184
Kundu, N., Dozier, U., Deslandes, L., Somssich, I. E., Ullah, H. (2013). Arabidopsis scaffold protein RACK1A interacts with diverse environmental stress and photosynthesis related proteins. Plant Signal Behav. 8, e24012. doi: 10.4161/psb.24012
Kuo, W.-Y., Huang, C.-H., Jinn, T.-L. (2013a). Chaperonin 20 might be an iron chaperone for superoxide dismutase in activating iron superoxide dismutase (FeSOD). Plant Signal. Behav. 8, e23074. doi: 10.4161/psb.23074
Kuo, W. Y., Huang, C. H., Liu, A. C., Cheng, C. P., Li, S. H., Chang, W. C., et al. (2013b). CHAPERONIN 20 mediates iron superoxide dismutase (FeSOD) activity independent of its co-chaperonin role in arabidopsis chloroplasts. New Phytol. 197, 99–110. doi: 10.1111/j.1469-8137.2012.04369.x
Lamb, A. L., Torres, A. S., O’Halloran, T. V., Rosenzweig, A. C. (2001). Heterodimeric structure of superoxide dismutase in complex with its metallochaperone. Nat. Struct. Biol. 8, 751–755. doi: 10.1038/nsb0901-751
Latz, A., Becker, D., Hekman, M., Müller, T., Beyhl, D., Marten, I., et al. (2007). TPK1, a Ca(2+)-regulated arabidopsis vacuole two-pore k(+) channel is activated by 14-3-3 proteins. Plant J. 52, 449–459. doi: 10.1111/j.1365-313X.2007.03255.x
Latz, A., Mehlmer, N., Zapf, S., Mueller, T. D., Wurzinger, B., Pfister, B., et al. (2013). Salt stress triggers phosphorylation of the arabidopsis vacuolar k+ channel TPK1 by calcium-dependent protein kinases (CDPKs). Mol. Plant 6, 1274–1289. doi: 10.1093/mp/sss158
Lazzarotto, F., Teixeira, F. K., Rosa, S. B., Dunand, C., Fernandes, C. L., de Vasconcelos Fontenele, A., et al. (2011). Ascorbate peroxidase-related (APx-r) is a new heme-containing protein functionally associated with ascorbate peroxidase but evolutionarily divergent. New Phytol. 191, 234–250. doi: 10.1111/j.1469-8137.2011.03659.x
Lazzarotto, F., Wahni, K., Piovesana, M., Maraschin, F., Messens, J., Margis-Pinheiro, M. (2021). Arabidopsis APx-r is a plastidial ascorbate-independent peroxidase regulated by photomorphogenesis. Antioxidants 10, 65. doi: 10.3390/antiox10010065
Le, M. H., Cao, Y., Zhang, X.-C., Stacey, G. (2014). LIK1, a CERK1-interacting kinase, regulates plant immune responses in arabidopsis. PloS One 9, e102245. doi: 10.1371/journal.pone.0102245
Lee, S., Joung, Y. H., Kim, J.-K., Do Choi, Y., Jang, G. (2019). An isoform of the plastid RNA polymerase-associated protein FSD3 negatively regulates chloroplast development. BMC Plant Biol. 19, 524. doi: 10.1186/s12870-019-2128-9
Leene, J. V., Witters, E., Inzé, D., Jaeger, G. D. (2008). Boosting tandem affinity purification of plant protein complexes. Trends Plant Sci. 13, 517–520. doi: 10.1016/j.tplants.2008.08.002
Lee, B., Yoshida, Y., Hasunuma, K. (2009). Nucleoside diphosphate kinase-1 regulates hyphal development via the transcriptional regulation of catalase in neurospora crassa. FEBS Lett. 583, 3291–3295. doi: 10.1016/j.febslet.2009.09.026
Levy, E. D. (2010). A simple definition of structural regions in proteins and its use in analyzing interface evolution. J. Mol. Biol. 403, 660–670. doi: 10.1016/j.jmb.2010.09.028
Lewandowska, A., Vo, T. N., Nguyen, T.-D. H., Wahni, K., Vertommen, D., Van Breusegem, F., et al. (2019). Bifunctional chloroplastic DJ-1B from arabidopsis thaliana is an oxidation-robust holdase and a glyoxalase sensitive to H2O2. Antioxidants 8, 8. doi: 10.3390/antiox8010008
Liang, L., Wang, Q., Song, Z., Wu, Y., Liang, Q., Wang, Q., et al. (2021). O-Fucosylation of CPN20 by SPINDLY derepresses abscisic acid signaling during seed germination and seedling development. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.724144
Li, D.-H., Chen, F.-J., Li, H.-Y., Li, W., Guo, J.-J. (2018). The soybean GmRACK1 gene plays a role in drought tolerance at vegetative stages. Russ. J. Plant Physiol. 65, 541–552. doi: 10.1134/S1021443718040155
Li, Y., Chen, L., Mu, J., Zuo, J. (2013). LESION SIMULATING DISEASE1 interacts with catalases to regulate hypersensitive cell death in arabidopsis. Plant Physiol. 163, 1059–1070. doi: 10.1104/pp.113.225805
Liebthal, M., Schuetze, J., Dreyer, A., Mock, H.-P., Dietz, K.-J. (2020). Redox conformation-specific protein-protein interactions of the 2-cysteine peroxiredoxin in arabidopsis. Antioxidants 9, E515. doi: 10.3390/antiox9060515
Li, X., Gu, Y. (2020). Structural and functional insight into the nuclear pore complex and nuclear transport receptors in plant stress signaling. Curr. Opin. Plant Biol. 58, 60–68. doi: 10.1016/j.pbi.2020.10.006
Li, G., Li, J., Hao, R., Guo, Y. (2017b). Activation of catalase activity by a peroxisome-localized small heat shock protein Hsp17.6CII. J. Genet. Genomics 44, 395–404. doi: 10.1016/j.jgg.2017.03.009
Li, J., Liu, J., Wang, G., Cha, J.-Y., Li, G., Chen, S., et al. (2015). A chaperone function of NO CATALASE ACTIVITY1 is required to maintain catalase activity and for multiple stress responses in arabidopsis. Plant Cell 27, 908–925. doi: 10.1105/tpc.114.135095
Li, D., Liu, H., Yang, Y., Zhen, P., Liang, J. (2009). Down-regulated expression of RACK1 gene by RNA interference enhances drought tolerance in rice. Rice Sci. 16, 14–20. doi: 10.1016/S1672-6308(08)60051-7
Lindermayr, C., Sell, S., Müller, B., Leister, D., Durner, J. (2010). Redox regulation of the NPR1-TGA1 system of arabidopsis thaliana by nitric oxide. Plant Cell 22, 2894–2907. doi: 10.1105/tpc.109.066464
Lin, J., Nazarenus, T. J., Frey, J. L., Liang, X., Wilson, M. A., Stone, J. M. (2011). A plant DJ-1 homolog is essential for arabidopsis thaliana chloroplast development. PloS One 6, e23731. doi: 10.1371/journal.pone.0023731
Li, D.-H., Shen, F.-J., Li, H.-Y., Li, W. (2017a). Kale BoRACK1 is involved in the plant response to salt stress and peronospora brassicae gaumann. J. Plant Physiol. 213, 188–198. doi: 10.1016/j.jplph.2017.03.014
Lister, R., Carrie, C., Duncan, O., Ho, L. H. M., Howell, K. A., Murcha, M. W., et al. (2007). Functional definition of outer membrane proteins involved in preprotein import into mitochondria. Plant Cell 19, 3739–3759. doi: 10.1105/tpc.107.050534
Liu, J., Cui, L., Xie, Z., Zhang, Z., Liu, E., Peng, X. (2019). Two NCA1 isoforms interact with catalase in a mutually exclusive manner to redundantly regulate its activity in rice. BMC Plant Biol. 19, 105. doi: 10.1186/s12870-019-1707-0
Liu, X., Wang, X., Yan, X., Li, S., Peng, H. (2020). The glycine- and proline-rich protein AtGPRP3 negatively regulates plant growth in arabidopsis. Int. J. Mol. Sci. 21, 6168. doi: 10.3390/ijms21176168
Li, F., Wu, Q.-Y., Sun, Y.-L., Wang, L.-Y., Yang, X.-H., Meng, Q.-W. (2010). Overexpression of chloroplastic monodehydroascorbate reductase enhanced tolerance to temperature and methyl viologen-mediated oxidative stresses. Physiol. Plant 139, 421–434. doi: 10.1111/j.1399-3054.2010.01369.x
Li, J., Yuan, J., Li, Y., Sun, H., Ma, T., Huai, J., et al. (2022). The CDC48 complex mediates ubiquitin-dependent degradation of intra-chloroplast proteins in plants. Cell Rep. 39, 110664. doi: 10.1016/j.celrep.2022.110664
Luanpitpong, S., Chanvorachote, P., Stehlik, C., Tse, W., Callery, P. S., Wang, L., et al. (2013). Regulation of apoptosis by bcl-2 cysteine oxidation in human lung epithelial cells. Mol. Biol. Cell 24, 858–869. doi: 10.1091/mbc.e12-10-0747
Luk, E., Carroll, M., Baker, M., Culotta, V. C. (2003). Manganese activation of superoxide dismutase 2 in saccharomyces cerevisiae requires MTM1, a member of the mitochondrial carrier family. Proc. Natl. Acad. Sci. U. S. A. 100, 10353–10357. doi: 10.1073/pnas.1632471100
Lv, T., Li, X., Fan, T., Luo, H., Xie, C., Zhou, Y., et al. (2019). The calmodulin-binding protein IQM1 interacts with CATALASE2 to affect pathogen defense. Plant Physiol. 181, 1314–1327. doi: 10.1104/pp.19.01060
Martín-Trillo, M., Cubas, P. (2010). TCP Genes: a family snapshot ten years later. Trends Plant Sci. 15, 31–39. doi: 10.1016/j.tplants.2009.11.003
Marubashi, S., Ohbayashi, N., Fukuda, M. (2016). A varp-binding protein, RACK1, regulates dendrite outgrowth through stabilization of varp protein in mouse melanocytes. J. Invest. Dermatol. 136, 1672–1680. doi: 10.1016/j.jid.2016.03.034
Maruta, T., Sawa, Y., Shigeoka, S., Ishikawa, T. (2016). Diversity and evolution of ascorbate peroxidase functions in chloroplasts: More than just a classical antioxidant enzyme? Plant Cell Physiol 57, 1377–1386. doi: 10.1093/pcp/pcv203
McCutcheon, D. C., Lee, G., Carlos, A., Montgomery, J. E., Moellering, R. E. (2020). Photoproximity profiling of protein–protein interactions in cells. J. Am. Chem. Soc 142, 146–153. doi: 10.1021/jacs.9b06528
McKhann, H. I., Frugier, F., Petrovics, G., Coba de la Peña, T., Jurkevitch, E., Brown, S., et al. (1997). Cloning of a WD-repeat-containing gene from alfalfa (Medicago sativa): a role in hormone-mediated cell division? Plant Mol. Biol. 34, 771–780. doi: 10.1023/A:1005899410389
McWhite, C. D., Papoulas, O., Drew, K., Cox, R. M., June, V., Dong, O. X., et al. (2020). A pan-plant protein complex map reveals deep conservation and novel assemblies. Cell 181, 460–474.e14. doi: 10.1016/j.cell.2020.02.049
Meier, I., Brkljacic, J. (2009). Adding pieces to the puzzling plant nuclear envelope. Curr. Opin. Plant Biol. 12, 752–759. doi: 10.1016/j.pbi.2009.09.016
Melicher, P., Dvořák, P., Krasylenko, Y., Shapiguzov, A., Kangasjärvi, J., Šamaj, J., et al. (2022). Arabidopsis iron superoxide dismutase FSD1 protects against methyl viologen-induced oxidative stress in a copper-dependent manner. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.823561
Miller, K. E., Kim, Y., Huh, W.-K., Park, H.-O. (2015). Bimolecular fluorescence complementation (BiFC) analysis: Advances and recent applications for genome-wide interaction studies. J. Mol. Biol. 427, 2039–2055. doi: 10.1016/j.jmb.2015.03.005
Mintseris, J., Weng, Z. (2005). Structure, function, and evolution of transient and obligate protein–protein interactions. Proc. Natl. Acad. Sci. U.S.A. 102, 10930–10935. doi: 10.1073/pnas.0502667102
Miteva, Y. V., Budayeva, H. G., Cristea, I. M. (2013). Proteomics-based methods for discovery, quantification, and validation of protein-protein interactions. Anal. Chem. 85, 749–768. doi: 10.1021/ac3033257
Mittler, R., Zandalinas, S. I., Fichman, Y., Van Breusegem, F. (2022). Reactive oxygen species signalling in plant stress responses. Nat. Rev. Mol. Cell Biol. 23, 663–679. doi: 10.1038/s41580-022-00499-2
Miyauchi, H., Moriyama, S., Kusakizako, T., Kumazaki, K., Nakane, T., Yamashita, K., et al. (2017). Structural basis for xenobiotic extrusion by eukaryotic MATE transporter. Nat. Commun. 8, 1633. doi: 10.1038/s41467-017-01541-0
Moon, H., Lee, B., Choi, G., Shin, D., Prasad, D. T., Lee, O., et al. (2003). NDP kinase 2 interacts with two oxidative stress-activated MAPKs to regulate cellular redox state and enhances multiple stress tolerance in transgenic plants. Proc. Natl. Acad. Sci. U.S.A. 100, 358–363. doi: 10.1073/pnas.252641899
Moreira, I. S., Fernandes, P. A., Ramos, M. J. (2007). Hot spots–a review of the protein-protein interface determinant amino-acid residues. Proteins 68, 803–812. doi: 10.1002/prot.21396
Morgan, M. J., Lehmann, M., Schwarzländer, M., Baxter, C. J., Sienkiewicz-Porzucek, A., Williams, T. C. R., et al. (2008). Decrease in manganese superoxide dismutase leads to reduced root growth and affects tricarboxylic acid cycle flux and mitochondrial redox homeostasis. Plant Physiol. 147, 101–114. doi: 10.1104/pp.107.113613
Mou, Z., Fan, W., Dong, X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113, 935–944. doi: 10.1016/S0092-8674(03)00429-X
Mullen, R. T., Lee, M. S., Trelease, R. N. (1997). Identification of the peroxisomal targeting signal for cottonseed catalase. Plant J. 12, 313–322. doi: 10.1046/j.1365-313x.1997.12020313.x
Müller-Schüssele, S. J., Wang, R., Gütle, D. D., Romer, J., Rodriguez-Franco, M., Scholz, M., et al. (2020). Chloroplasts require glutathione reductase to balance reactive oxygen species and maintain efficient photosynthesis. Plant J. 103, 1140–1154. doi: 10.1111/tpj.14791
Murota, K., Shimura, H., Takeshita, M., Masuta, C. (2017). Interaction between cucumber mosaic virus 2b protein and plant catalase induces a specific necrosis in association with proteasome activity. Plant Cell Rep. 36, 37–47. doi: 10.1007/s00299-016-2055-2
Myouga, F., Hosoda, C., Umezawa, T., Iizumi, H., Kuromori, T., Motohashi, R., et al. (2008). A heterocomplex of iron superoxide dismutases defends chloroplast nucleoids against oxidative stress and is essential for chloroplast development in arabidopsis. Plant Cell 20, 3148–3162. doi: 10.1105/tpc.108.061341
Nakashima, A., Chen, L., Thao, N. P., Fujiwara, M., Wong, H. L., Kuwano, M., et al. (2008). RACK1 functions in rice innate immunity by interacting with the Rac1 immune complex. Plant Cell 20, 2265–2279. doi: 10.1105/tpc.107.054395
Ngounou Wetie, A. G., Sokolowska, I., Channaveerappa, D., Dupree, E. J., Jayathirtha, M., Woods, A. G., et al. (2019). Proteomics and non-proteomics approaches to study stable and transient protein-protein interactions. Adv. Exp. Med. Biol. 1140, 121–142. doi: 10.1007/978-3-030-15950-4_7
Ngounou Wetie, A. G., Sokolowska, I., Woods, A. G., Roy, U., Loo, J. A., Darie, C. C. (2013). Investigation of stable and transient protein-protein interactions: Past, present, and future. Proteomics 13, 538–557. doi: 10.1002/pmic.201200328
Niemiro, A., Cysewski, D., Brzywczy, J., Wawrzyńska, A., Sieńko, M., Poznański, J., et al. (2020). Similar but not identical–binding properties of LSU (Response to low sulfur) proteins from arabidopsis thaliana. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.01246
Nooren, I. M. A., Thornton, J. M. (2003). Diversity of protein-protein interactions. EMBO J. 22, 3486–3492. doi: 10.1093/emboj/cdg359
Noshi, M., Yamada, H., Hatanaka, R., Tanabe, N., Tamoi, M., Shigeoka, S. (2017). Arabidopsis dehydroascorbate reductase 1 and 2 modulate redox states of ascorbate-glutathione cycle in the cytosol in response to photooxidative stress. Biosci. Biotechnol. Biochem. 81, 523–533. doi: 10.1080/09168451.2016.1256759
Nussinov, R., Ma, B., Tsai, C.-J. (2013). A broad view of scaffolding suggests that scaffolding proteins can actively control regulation and signaling of multienzyme complexes through allostery. Biochim. Biophys. Acta - Proteins Proteom. 1834, 820–829. doi: 10.1016/j.bbapap.2012.12.014
Olejnik, K., Bucholc, M., Anielska-Mazur, A., Lipko, A., Kujawa, M., Modzelan, M., et al. (2011). Arabidopsis thaliana nudix hydrolase AtNUDT7 forms complexes with the regulatory RACK1A protein and gamma subunits of the signal transducing heterotrimeric G protein. Acta Biochim. Pol. 58, 609–616. doi: 10.18388/abp.2011_2231
Oshima, Y., Kamigaki, A., Nakamori, C., Mano, S., Hayashi, M., Nishimura, M., et al. (2008). Plant catalase is imported into peroxisomes by Pex5p but is distinct from typical PTS1 import. Plant Cell Physiol. 49, 671–677. doi: 10.1093/pcp/pcn038
Oughtred, R., Rust, J., Chang, C., Breitkreutz, B.-J., Stark, C., Willems, A., et al. (2021). The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 30, 187–200. doi: 10.1002/pro.3978
Palma, J. M., Mateos, R. M., López-Jaramillo, J., Rodríguez-Ruiz, M., González-Gordo, S., Lechuga-Sancho, A. M., et al. (2020). Plant catalases as NO and H2S targets. Redox Biol. 34, 101525. doi: 10.1016/j.redox.2020.101525
Pan, L., Luo, Y., Wang, J., Li, X., Tang, B., Yang, H., et al. (2022). Evolution and functional diversification of catalase genes in the green lineage. BMC Genomics 23, 411. doi: 10.1186/s12864-022-08621-6
Park, S., Rancour, D. M., Bednarek, S. Y. (2008). In planta analysis of the cell cycle-dependent localization of AtCDC48A and its critical roles in cell division, expansion, and differentiation. Plant Physiol. 148, 246–258. doi: 10.1104/pp.108.121897
Peer, W. A., Hosein, F. N., Bandyopadhyay, A., Makam, S. N., Otegui, M. S., Lee, G.-J., et al. (2009). Mutation of the membrane-associated M1 protease APM1 results in distinct embryonic and seedling developmental defects in arabidopsis. Plant Cell 21, 1693–1721. doi: 10.1105/tpc.108.059634
Pérez-Pérez, M. E., Mauriès, A., Maes, A., Tourasse, N. J., Hamon, M., Lemaire, S. D., et al. (2017). The deep thioredoxome in chlamydomonas reinhardtii: New insights into redox regulation. Mol. Plant 10, 1107–1125. doi: 10.1016/j.molp.2017.07.009
Perkins, J. R., Diboun, I., Dessailly, B. H., Lees, J. G., Orengo, C. (2010). Transient protein-protein interactions: structural, functional, and network properties. Structure 18, 1233–1243. doi: 10.1016/j.str.2010.08.007
Pfannschmidt, T., Blanvillain, R., Merendino, L., Courtois, F., Chevalier, F., Liebers, M., et al. (2015). Plastid RNA polymerases: orchestration of enzymes with different evolutionary origins controls chloroplast biogenesis during the plant life cycle. J. Exp. Bot. 66, 6957–6973. doi: 10.1093/jxb/erv415
Pilon, M., Ravet, K., Tapken, W. (2011). The biogenesis and physiological function of chloroplast superoxide dismutases. Biochim. Biophys. Acta 1807, 989–998. doi: 10.1016/j.bbabio.2010.11.002
Pnueli, L., Liang, H., Rozenberg, M., Mittler, R. (2003). Growth suppression, altered stomatal responses, and augmented induction of heat shock proteins in cytosolic ascorbate peroxidase (Apx1)-deficient arabidopsis plants. Plant J. 34, 187–203. doi: 10.1046/j.1365-313X.2003.01715.x
Qin, W., Cho, K. F., Cavanagh, P. E., Ting, A. Y. (2021). Deciphering molecular interactions by proximity labeling. Nat. Methods 18, 133–143. doi: 10.1038/s41592-020-01010-5
Rae, T. D., Schmidt, P. J., Pufahl, R. A., Culotta, V. C., O’Halloran, T. V. (1999). Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science 284, 805–808. doi: 10.1126/science.284.5415.805
Remy, E., Duque, P. (2014). Beyond cellular detoxification: a plethora of physiological roles for MDR transporter homologs in plants. Front. Physiol. 5. doi: 10.3389/fphys.2014.00201
Rexin, D., Meyer, C., Robaglia, C., Veit, B. (2015). TOR signalling in plants. Biochem. 470, 1–14. doi: 10.1042/BJ20150505
Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., Séraphin, B. (1999). A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032. doi: 10.1038/13732
Roberts, M. R., Salinas, J., Collinge, D. B. (2002). 14-3-3 proteins and the response to abiotic and biotic stress. Plant Mol. Biol. 50, 1031–1039. doi: 10.1023/A:1021261614491
Rocha, A. G., Mehlmer, N., Stael, S., Mair, A., Parvin, N., Chigri, F., et al. (2014). Phosphorylation of Arabidopsis transketolase at Ser428 provides a potential paradigm for the metabolic control of chloroplast carbon metabolism. Biochem. 458, 313–322. doi: 10.1042/BJ20130631
Roche, J. V., Törnroth-Horsefield, S. (2017). Aquaporin protein-protein interactions. Int. J. Mol. Sci. 18, E2255. doi: 10.3390/ijms18112255
Rosnoblet, C., Bègue, H., Blanchard, C., Pichereaux, C., Besson-Bard, A., Aimé, S., et al. (2017). Functional characterization of the chaperon-like protein Cdc48 in cryptogein-induced immune response in tobacco. Plant Cell Environ. 40, 491–508. doi: 10.1111/pce.12686
Rosnoblet, C., Chatelain, P., Klinguer, A., Bègue, H., Winckler, P., Pichereaux, C., et al. (2021). The chaperone-like protein Cdc48 regulates ubiquitin-proteasome system in plants. Plant Cell Environ. 44, 2636–2655. doi: 10.1111/pce.14073
Sabila, M., Kundu, N., Smalls, D., Ullah, H. (2016). Tyrosine phosphorylation based homo-dimerization of arabidopsis RACK1A proteins regulates oxidative stress signaling pathways in yeast. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00176
Sagi, M., Fluhr, R. (2006). Production of reactive oxygen species by plant NADPH oxidases. Plant Physiol. 141, 336–340. doi: 10.1104/pp.106.078089
Samakovli, D., Tichá, T., Vavrdová, T., Ovečka, M., Luptovčiak, I., Zapletalová, V., et al. (2020). YODA-HSP90 module regulates phosphorylation-dependent inactivation of SPEECHLESS to control stomatal development under acute heat stress in arabidopsis. Mol. Plant 13, 612–633. doi: 10.1016/j.molp.2020.01.001
Saxena, S. C., Salvi, P., Kamble, N. U., Joshi, P. K., Majee, M., Arora, S. (2020). Ectopic overexpression of cytosolic ascorbate peroxidase gene (Apx1) improves salinity stress tolerance in brassica juncea by strengthening antioxidative defense mechanism. Acta Physiol. Plant 42, 45. doi: 10.1007/s11738-020-3032-5
Schmidt, P. J., Kunst, C., Culotta, V. C. (2000). Copper activation of superoxide dismutase 1 (SOD1) in vivo. role for protein-protein interactions with the copper chaperone for SOD1. J. Biol. Chem. 275, 33771–33776. doi: 10.1074/jbc.M006254200
Schmidt, P. J., Rae, T. D., Pufahl, R. A., Hamma, T., Strain, J., O’Halloran, T. V., et al. (1999). Multiple protein domains contribute to the action of the copper chaperone for superoxide dismutase. J. Biol. Chem. 274, 23719–23725. doi: 10.1074/jbc.274.34.23719
Scranton, M. A., Yee, A., Park, S.-Y., Walling, L. L. (2012). Plant leucine aminopeptidases moonlight as molecular chaperones to alleviate stress-induced damage. J. Biol. Chem. 287, 18408–18417. doi: 10.1074/jbc.M111.309500
Seychell, B. C., Beck, T. (2021). Molecular basis for protein–protein interactions. Beilstein J. Org. Chem. 17, 1–10. doi: 10.3762/bjoc.17.1
Shen, G., Kuppu, S., Venkataramani, S., Wang, J., Yan, J., Qiu, X., et al. (2010). ANKYRIN REPEAT-CONTAINING PROTEIN 2A is an essential molecular chaperone for peroxisomal membrane-bound ASCORBATE PEROXIDASE3 in arabidopsis. Plant Cell 22, 811–831. doi: 10.1105/tpc.109.065979
Shin, R., Jez, J. M., Basra, A., Zhang, B., Schachtman, D. P. (2011). 14-3-3 proteins fine-tune plant nutrient metabolism. FEBS Lett. 585, 143–147. doi: 10.1016/j.febslet.2010.11.025
Shi, T., Polderman, P. E., Pagès-Gallego, M., van Es, R. M., Vos, H. R., Burgering, B. M. T., et al. (2021). p53 forms redox-dependent protein-protein interactions through cysteine 277. Antioxidants 10, 1578. doi: 10.3390/antiox10101578
Song, R.-F., Li, T.-T., Liu, W.-C. (2021). Jasmonic acid impairs arabidopsis seedling salt stress tolerance through MYC2-mediated repression of CAT2 expression. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.730228
Speth, C., Willing, E.-M., Rausch, S., Schneeberger, K., Laubinger, S. (2013). RACK1 scaffold proteins influence miRNA abundance in arabidopsis. Plant J. 76, 433–445. doi: 10.1111/tpj.12308
Sprinzak, E., Sattath, S., Margalit, H. (2003). How reliable are experimental protein–protein interaction data? J. Mol. Biol. 327, 919–923. doi: 10.1016/S0022-2836(03)00239-0
Stein, A., Pache, R. A., Bernadó, P., Pons, M., Aloy, P. (2009). Dynamic interactions of proteins in complex networks: a more structured view. FEBS J. 276, 5390–5405. doi: 10.1111/j.1742-4658.2009.07251.x
Stolz, A., Hilt, W., Buchberger, A., Wolf, D. H. (2011). Cdc48: a power machine in protein degradation. Trends Biochem. Sci. 36, 515–523. doi: 10.1016/j.tibs.2011.06.001
Strotmann, V. I., Stahl, Y. (2022). Visualization of in vivo protein–protein interactions in plants. J. Exp. Bot. 73, 3866–3880. doi: 10.1093/jxb/erac139
Struk, S., Jacobs, A., Sánchez Martín-Fontecha, E., Gevaert, K., Cubas, P., Goormachtig, S. (2019). Exploring the protein-protein interaction landscape in plants. Plant Cell Environ. 42, 387–409. doi: 10.1111/pce.13433
Sudha, G., Nussinov, R., Srinivasan, N. (2014). An overview of recent advances in structural bioinformatics of protein–protein interactions and a guide to their principles. Prog. Biophys. Mol. Biol. 116, 141–150. doi: 10.1016/j.pbiomolbio.2014.07.004
Sun, X., Han, G., Meng, Z., Lin, L., Sui, N. (2019). Roles of malic enzymes in plant development and stress responses. Plant Signal. Behav. 14, e1644596. doi: 10.1080/15592324.2019.1644596
Sun, Y., Li, P., Deng, M., Shen, D., Dai, G., Yao, N., et al. (2017). The ralstonia solanacearum effector RipAK suppresses plant hypersensitive response by inhibiting the activity of host catalases. Cell Microbiol. 19, e12736. doi: 10.1111/cmi.12736
Su, T., Wang, P., Li, H., Zhao, Y., Lu, Y., Dai, P., et al. (2018). The arabidopsis catalase triple mutant reveals important roles of catalases and peroxisome-derived signaling in plant development. J. Integr. Plant Biol. 60, 591–607. doi: 10.1111/jipb.12649
Suzuki, M., Sato, Y., Wu, S., Kang, B.-H., McCarty, D. R. (2015). Conserved functions of the MATE transporter BIG EMBRYO1 in regulation of lateral organ size and initiation rate. Plant Cell 27, 2288–2300. doi: 10.1105/tpc.15.00290
Tada, Y., Spoel, S. H., Pajerowska-Mukhtar, K., Mou, Z., Song, J., Wang, C., et al. (2008). Plant immunity requires conformational charges of NPR1 via s-nitrosylation and thioredoxins. Science 321, 952–956. doi: 10.1126/science.1156970
Tamura, K., Fukao, Y., Iwamoto, M., Haraguchi, T., Hara-Nishimura, I. (2010). Identification and characterization of nuclear pore complex components in arabidopsis thaliana. Plant Cell 22, 4084–4097. doi: 10.1105/tpc.110.079947
Tanaka, M., Takahashi, R., Hamada, A., Terai, Y., Ogawa, T., Sawa, Y., et al. (2021). Distribution and functions of monodehydroascorbate reductases in plants: comprehensive reverse genetic analysis of arabidopsis thaliana enzymes. Antioxidants 10, 1726. doi: 10.3390/antiox10111726
Tang, L., Kim, M. D., Yang, K.-S., Kwon, S.-Y., Kim, S.-H., Kim, J.-S., et al. (2008). Enhanced tolerance of transgenic potato plants overexpressing nucleoside diphosphate kinase 2 against multiple environmental stresses. Transgenic Res. 17, 705–715. doi: 10.1007/s11248-007-9155-2
Terrile, M. C., Tebez, N. M., Colman, S. L., Mateos, J. L., Morato-López, E., Sánchez-López, N., et al. (2022). S-nitrosation of E3 ubiquitin ligase complex components regulates hormonal signalings in arabidopsis. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.794582
Urano, K., Maruyama, K., Koyama, T., Gonzalez, N., Inzé, D., Yamaguchi-Shinozaki, K., et al. (2022). CIN-like TCP13 is essential for plant growth regulation under dehydration stress. Plant Mol. Biol. 108, 257–275. doi: 10.1007/s11103-021-01238-5
Uribe, F., Henríquez-Valencia, C., Arenas-M, A., Medina, J., Vidal, E. A., Canales, J. (2022). Evolutionary and gene expression analyses reveal new insights into the role of LSU gene-family in plant responses to sulfate-deficiency. Plants (Basel) 11, 1526. doi: 10.3390/plants11121526
Vadovič, P., Šamajová, O., Takáč, T., Novák, D., Zapletalová, V., Colcombet, J., et al. (2019). Biochemical and genetic interactions of phospholipase d alpha 1 and mitogen-activated protein kinase 3 affect arabidopsis stress response. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00275
Vanacker, H., Guichard, M., Bohrer, A.-S., Issakidis-Bourguet, E. (2018). Redox regulation of monodehydroascorbate reductase by thioredoxin y in plastids revealed in the context of water stress. Antioxidants 7, 183. doi: 10.3390/antiox7120183
van Dam, L., Pagès-Gallego, M., Polderman, P. E., van Es, R. M., Burgering, B. M. T., Vos, H. R., et al. (2021). The human 2-cys peroxiredoxins form widespread, cysteine-dependent- and isoform-specific protein-protein interactions. Antioxidants 10, 627. doi: 10.3390/antiox10040627
Van Leene, J., Han, C., Gadeyne, A., Eeckhout, D., Matthijs, C., Cannoot, B., et al. (2019). Capturing the phosphorylation and protein interaction landscape of the plant TOR kinase. Nat. Plants 5, 316–327. doi: 10.1038/s41477-019-0378-z
Verrastro, I., Tveen-Jensen, K., Woscholski, R., Spickett, C. M., Pitt, A. R. (2016). Reversible oxidation of phosphatase and tensin homolog (PTEN) alters its interactions with signaling and regulatory proteins. Free Radic. Biol. Med. 90, 24–34. doi: 10.1016/j.freeradbiomed.2015.11.004
Verslues, P. E., Batelli, G., Grillo, S., Agius, F., Kim, Y.-S., Zhu, J., et al. (2007). Interaction of SOS2 with nucleoside diphosphate kinase 2 and catalases reveals a point of connection between salt stress and H2O2 signaling in Arabidopsis thaliana. Mol. Cell. Biol. 27, 7771–7780. doi: 10.1128/MCB.00429-07
Vinayagam, A., Stelzl, U., Wanker, E. E. (2010). Repeated two-hybrid screening detects transient protein–protein interactions. Theor. Chem. Acc. 125, 613–619. doi: 10.1007/s00214-009-0651-8
von Mering, C., Krause, R., Snel, B., Cornell, M., Oliver, S. G., Fields, S., et al. (2002). Comparative assessment of large-scale data sets of protein–protein interactions. Nature 417, 399–403. doi: 10.1038/nature750
Wagner, R., Pfannschmidt, T. (2006). Eukaryotic transcription factors in plastids–bioinformatic assessment and implications for the evolution of gene expression machineries in plants. Gene 381, 62–70. doi: 10.1016/j.gene.2006.06.022
Walter, M., Chaban, C., Schütze, K., Batistic, O., Weckermann, K., Näke, C., et al. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438. doi: 10.1111/j.1365-313X.2004.02219.x
Wan, C., Borgeson, B., Phanse, S., Tu, F., Drew, K., Clark, G., et al. (2015). Panorama of ancient metazoan macromolecular complexes. Nature 525, 339–344. doi: 10.1038/nature14877
Wang, Y., Chu, C. (2020). S-nitrosylation control of ROS and RNS homeostasis in plants: The switching function of catalase. Mol. Plant 13, 946–948. doi: 10.1016/j.molp.2020.05.013
Wang, Y., Ji, D., Lei, C., Chen, Y., Qiu, Y., Li, X., et al. (2021). Mechanistic insights into the effect of phosphorylation on ras conformational dynamics and its interactions with cell signaling proteins. Comput. Struct. Biotechnol. J. 19, 1184–1199. doi: 10.1016/j.csbj.2021.01.044
Wang, P., Xue, L., Batelli, G., Lee, S., Hou, Y.-J., Van Oosten, M. J., et al. (2013). Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. U. S. A. 110, 11205–11210. doi: 10.1073/pnas.1308974110
Wang, N., Yoshida, Y., Hasunuma, K. (2007). Catalase-1 (CAT-1) and nucleoside diphosphate kinase-1 (NDK-1) play an important role in protecting conidial viability under light stress in neurospora crassa. Mol. Genet. Genom. 278, 235–242. doi: 10.1007/s00438-007-0244-y
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46(W1), W296–W303. doi: 10.1093/nar/gky427
Waters, E. R. (2013). The evolution, function, structure, and expression of the plant sHSPs. J. Exp. Bot. 64, 391–403. doi: 10.1093/jxb/ers355
Wei, Y., Liu, W., Hu, W., Yan, Y., Shi, H. (2020). The chaperone MeHSP90 recruits MeWRKY20 and MeCatalase1 to regulate drought stress resistance in cassava. New Phytol. 226, 476–491. doi: 10.1111/nph.16346
Wei, Y., Ringe, D., Wilson, M. A., Ondrechen, M. J. (2007). Identification of functional subclasses in the DJ-1 superfamily proteins. PloS Comput. Biol. 3, e10. doi: 10.1371/journal.pcbi.0030010
Wong, P. C., Waggoner, D., Subramaniam, J. R., Tessarollo, L., Bartnikas, T. B., Culotta, V. C., et al. (2000). Copper chaperone for superoxide dismutase is essential to activate mammalian Cu/Zn superoxide dismutase. Proc. Natl. Acad. Sci. U. S. A. 97, 2886–2891. doi: 10.1073/pnas.040461197
Xing, S., Wallmeroth, N., Berendzen, K. W., Grefen, C. (2016). Techniques for the analysis of protein-protein interactions in vivo. Plant Physiol. 171, 727–758. doi: 10.1104/pp.16.00470
Xu, X. M., Lin, H., Maple, J., Bjorkblom, B., Alves, G., Larsen, J. P., et al. (2010). The arabidopsis DJ-1a protein confers stress protection through cytosolic SOD activation. J. Cell Sci. 123, 1644–1651. doi: 10.1242/jcs.063222
Yamakura, F., Kawasaki, H. (2010). Post-translational modifications of superoxide dismutase. Biochim. Biophys. Acta 1804, 318–325. doi: 10.1016/j.bbapap.2009.10.010
Yamasaki, H., Hayashi, M., Fukazawa, M., Kobayashi, Y., Shikanai, T. (2009). SQUAMOSA promoter binding protein-like 7 is a central regulator for copper homeostasis in arabidopsis. Plant Cell 21, 347–361. doi: 10.1105/tpc.108.060137
Yan, J., Chia, J.-C., Sheng, H., Jung, H., Zavodna, T.-O., Zhang, L., et al. (2017). Arabidopsis pollen fertility requires the transcription factors CITF1 and SPL7 that regulate copper delivery to anthers and jasmonic acid synthesis. Plant Cell 29, 3012–3029. doi: 10.1105/tpc.17.00363
Yang, X.-J. (2005). Multisite protein modification and intramolecular signaling. Oncogene 24, 1653–1662. doi: 10.1038/sj.onc.1208173
Yang, Z., Mhamdi, A., Noctor, G. (2019a). Analysis of catalase mutants underscores the essential role of CATALASE2 for plant growth and day length-dependent oxidative signalling: Plant catalases. Plant Cell Environ. 42, 688–700. doi: 10.1111/pce.13453
Yang, K. A., Moon, H., Kim, G., Lim, C. J., Hong, J. C., Lim, C. O., et al. (2003). NDP kinase 2 regulates expression of antioxidant genes in arabidopsis. Proc. Jpn. Acad. Ser. B Phys. Biol. 79B, 86–91. doi: 10.2183/pjab.79B.86
Yang, Z., Wang, C., Xue, Y., Liu, X., Chen, S., Song, C., et al. (2019b). Calcium-activated 14-3-3 proteins as a molecular switch in salt stress tolerance. Nat. Commun. 10, 1199. doi: 10.1038/s41467-019-09181-2
Yang, X., Wen, Z., Zhang, D., Li, Z., Li, D., Nagalakshmi, U., et al. (2021). Proximity labeling: an emerging tool for probing in planta molecular interactions. Plant Commun. 2, 100137. doi: 10.1016/j.xplc.2020.100137
Yan, J., Wang, J., Zhang, H. (2002). An ankyrin repeat-containing protein plays a role in both disease resistance and antioxidation metabolism. Plant J. 29, 193–202. doi: 10.1046/j.0960-7412.2001.01205.x
Yeh, H.-L., Lin, T.-H., Chen, C.-C., Cheng, T.-X., Chang, H.-Y., Lee, T.-M. (2019). Monodehydroascorbate reductase plays a role in the tolerance of chlamydomonas reinhardtii to photooxidative stress. Plant Cell Physiol. 60, 2167–2179. doi: 10.1093/pcp/pcz110
Yoshida, Y., Ogura, Y., Hasunuma, K. (2006). Interaction of nucleoside diphosphate kinase and catalases for stress and light responses in Neurospora crassa. FEBS Lett. 580, 3282–3286. doi: 10.1016/j.febslet.2006.01.096
Yua, Q.-B., Ma, Q., Kong, M.-M., Zhao, T.-T., Zhang, X.-L., Zhou, Q., et al. (2014). AtECB1/MRL7, a thioredoxin-like fold protein with disulfide reductase activity, regulates chloroplast gene expression and chloroplast biogenesis in arabidopsis thaliana. Mol. Plant 7, 206–217. doi: 10.1093/mp/sst092
Yuan, H., Jin, C., Pei, H., Zhao, L., Li, X., Li, J., et al. (2021). The powdery mildew effector CSEP0027 interacts with barley catalase to regulate host immunity. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.733237
Yuan, H.-M., Liu, W.-C., Lu, Y.-T. (2017). CATALASE2 coordinates SA-mediated repression of both auxin accumulation and JA biosynthesis in plant defenses. Cell Host Microbe 21, 143–155. doi: 10.1016/j.chom.2017.01.007
Zamocky, M., Furtmüller, P. G., Obinger, C. (2008). Evolution of catalases from bacteria to humans. Antioxid. Redox Signal. 10, 1527–1548. doi: 10.1089/ars.2008.2046
Zandalinas, S. I., Balfagón, D., Arbona, V., Gómez-Cadenas, A., Inupakutika, M. A., Mittler, R. (2016). ABA is required for the accumulation of APX1 and MBF1c during a combination of water deficit and heat stress. J. Exp. Bot. 67, 5381–5390. doi: 10.1093/jxb/erw299
Zeng, H., Xu, H., Wang, H., Chen, H., Wang, G., Bai, Y., et al. (2022). LSD3 mediates the oxidative stress response through fine-tuning APX2 activity and the NF-YC15-GSTs module in cassava. Plant J. 110, 1447–1461. doi: 10.1111/tpj.15749
Zhang, D., Chen, L., Li, D., Lv, B., Chen, Y., Chen, J., et al. (2014). OsRACK1 is involved in abscisic acid- and H2O2-mediated signaling to regulate seed germination in rice (Oryza sativa, l.). PloS One 9, e97120. doi: 10.1371/journal.pone.0097120
Zhang, D., Chen, L., Lv, B., Liang, J. (2013a). The scaffolding protein RACK1: A platform for diverse functions in the plant kingdom. J. Plant Biol. Soil. Health 1, 7–13. doi: 10.13188/2331-8996.1000003
Zhang, X.-F., Jiang, T., Wu, Z., Du, S.-Y., Yu, Y.-T., Jiang, S.-C., et al. (2013b). Cochaperonin CPN20 negatively regulates abscisic acid signaling in arabidopsis. Plant Mol. Biol. 83, 205–218. doi: 10.1007/s11103-013-0082-8
Zhang, S., Li, C., Ren, H., Zhao, T., Li, Q., Wang, S., et al. (2020). BAK1 mediates light intensity to phosphorylate and activate catalases to regulate plant growth and development. Int. J. Mol. Sci. 21, 1437. doi: 10.3390/ijms21041437
Zhang, J., Movahedi, A., Sang, M., Wei, Z., Xu, J., Wang, X., et al. (2017). Functional analyses of NDPK2 in Populus trichocarpa and overexpression of PtNDPK2 enhances growth and tolerance to abiotic stresses in transgenic poplar. Plant Physiol. Biochem. 117, 61–74. doi: 10.1016/j.plaphy.2017.05.019
Zhang, Y., Wang, L.-F., Li, T.-T., Liu, W.-C. (2021). Mutual promotion of LAP2 and CAT2 synergistically regulates plant salt and osmotic stress tolerance. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.672672
Zhang, H., Wang, J., Nickel, U., Allen, R. D., Goodman, H. M. (1997). Cloning and expression of an arabidopsis gene encoding a putative peroxisomal ascorbate peroxidase. Plant Mol. Biol. 34, 967–971. doi: 10.1023/a:1005814109732
Zhang, D., Wang, Y., Shen, J., Yin, J., Li, D., Gao, Y., et al. (2018). OsRACK1A, encodes a circadian clock-regulated WD40 protein, negatively affect salt tolerance in rice. Rice (N Y) 11, 45. doi: 10.1186/s12284-018-0232-3
Zhou, Y.-P., Duan, J., Fujibe, T., Yamamoto, K. T., Tian, C.-E. (2012). AtIQM1, a novel calmodulin-binding protein, is involved in stomatal movement in arabidopsis. Plant Mol. Biol. 79, 333–346. doi: 10.1007/s11103-012-9915-0
Zhuang, Y., Wei, M., Ling, C., Liu, Y., Amin, A. K., Li, P., et al. (2021). EGY3 mediates chloroplastic ROS homeostasis and promotes retrograde signaling in response to salt stress in arabidopsis. Cell Rep. 36, 109384. doi: 10.1016/j.celrep.2021.109384
Zou, J.-J., Li, X.-D., Ratnasekera, D., Wang, C., Liu, W.-X., Song, L.-F., et al. (2015). Arabidopsis CALCIUM-DEPENDENT PROTEIN KINASE8 and CATALASE3 function in abscisic acid-mediated signaling and H2O2 homeostasis in stomatal guard cells under drought stress. Plant Cell 27, 1445–1460. doi: 10.1105/tpc.15.00144
Keywords: plants, antioxidant enzymes, protein-protein interactions, stress response, reactive oxygen species, receptor for activated C kinase 1
Citation: Melicher P, Dvořák P, Šamaj J and Takáč T (2022) Protein-protein interactions in plant antioxidant defense. Front. Plant Sci. 13:1035573. doi: 10.3389/fpls.2022.1035573
Received: 02 September 2022; Accepted: 14 November 2022;
Published: 14 December 2022.
Edited by:
Anket Sharma, Zhejiang Agriculture and Forestry University, ChinaReviewed by:
Anna Kulik, Polish Academy of Sciences, PolandCopyright © 2022 Melicher, Dvořák, Šamaj and Takáč. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tomáš Takáč, dG9tYXMudGFrYWNAdXBvbC5jeg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.