- State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A&F University, Yangling, China
Iron (Fe) plays an important role in cellular respiration and catalytic reactions of metalloproteins in plants and animals. Plants maintain iron homeostasis through absorption, translocation, storage, and compartmentalization of iron via a cooperative regulative network. Here, we showed different physiological characteristics in the leaves and roots of Malus baccata under Fe sufficiency and Fe deficiency conditions and propose that MbHY5 (elongated hypocotyl 5), an important transcription factor for its function in photomorphogenesis, participated in Fe deficiency response in both the leaves and roots of M. baccata. The gene co-expression network showed that MbHY5 was involved in the regulation of chlorophyll synthesis and Fe transport pathway under Fe-limiting conditions. Specifically, we found that Fe deficiency induced the expression of MbYSL7 in root, which was positively regulated by MbHY5. Overexpressing or silencing MbYSL7 influenced the expression of MbHY5 in M. baccata.
Introduction
Although iron content is very abundant in the earth, its main existing form is ferric iron (Fe3+), which is insoluble and difficult for plants to uptake (Jeong and Guerinot, 2009). Iron (Fe) is one of the most essential micronutrients in plants and plays an important role in whole-life processes, including chlorophyll synthesis, electron transfer, and respiration (Kobayashi and Nishizawa, 2012). Also, iron can affect physiological processes such as nitrogen metabolism, carbohydrate, and organic acid metabolism in plants (Curie and Briat, 2003; Hell and Stephan, 2003; Kobayashi and Nishizawa, 2012).
Fe deficiency can cause a series of problems in fruit production (Tagliavini et al., 1995; Alvarez-Fernandez et al., 2003; Hao et al., 2022). Therefore, revealing the sophisticated mechanism of Fe2+ uptake, transport, and homeostasis in fruit plants is important for fruit yield and quality. Fe deficiency affects a variety of physiological and biochemical reactions in the leaves and roots of fruit plants. One of the most prominent symptoms in plant is interveinal chlorosis, or veins yellowing, which leads to a reduced photosynthetic performance of fruit trees (Curie and Briat, 2003; Hao et al., 2022). About 80% of the total iron was stored in chloroplasts; although iron is not a component of chlorophyll, it is an indispensable catalyst for chlorophyll synthesis (Yang et al., 2022). Previous studies have shown that the number of thylakoid membranes decreased in the lamellar structure of the chloroplast under iron deficiency (TerBush et al., 2013). Roots under iron deficiency can form root tip swellings or increase lateral roots and/or root hairs (Morrissey and Guerinot, 2009).
Iron content in plants mainly depends on the uptake and transport of exogenous iron by roots. In plants, there are two distinct strategies for root iron uptaking (Ivanov et al., 2012). Plant species belonging to the dicot and non-graminaceous monocot lineages use Strategy I, which consists of three steps: first, proton efflux from plant cells was mediated by the P-type ATPase to decrease the pH of the rhizosphere soil, which leads to soil acidification and an increase of iron solubility. Meanwhile, Fe(III) is also chelated and mobilized by coumarin-family phenolics exported by an ABC transporter PDR9 from the cortex to the rhizosphere (Tsai and Schmidt, 2017). Next, Fe(III) is reduced to Fe(II) by ferric reduction oxidase 2 (FRO2) localized on the plasma membrane. Third, the divalent iron Fe(II) was taken up into epidermal cells by metal transporter IRT1 (Eide et al., 1996; Santi and Schmidt, 2009). Subsequently, nicotianamine synthase (NAS), yellow stripe-like (YSL), and other transporters helped Fe(II) transport to vacuoles, chloroplasts, and other organs and organelles for further utilization (Walker and Connolly, 2008). Strategy II plants (grasses) synthesize and secrete phytosiderophores (PS) which form chelates with Fe(III) in roots, and this complex was then transported into cells by YSL transporters (Curie et al., 2009). In either way, YSLs play key roles in iron transportation and acquisition. Multiple copies of YSL genes were found in the genomes of angiosperm and gymnosperm species (Chowdhury et al., 2022). AtYSL1, AtYSL3, AtYSL4, and AtYSL6 have been demonstrated to be involved in the transportation of Fe and Zn from leaves to seeds through the phloem (Murata et al., 2006; Ishimaru et al., 2010; Kumar et al., 2019). The expression of AtYSL2 was downregulated in response to iron deficiency (Zang et al., 2020). In addition, YSLs have been proposed as transporters of iron from xylem to phloem and then to young tissues (Le Jean et al., 2005; Morrissey and Guerinot, 2009). YSL2 and YSL7 have been found to be associated with the movement of Fe/Zn-NA complexes to maintain Fe homeostasis in Arabidopsis (Khan et al., 2018).
HY5 (elongated hypocotyl 5) is a member of the basic leucine zipper (bZIP) transcription factors, which is known for its key roles in light reception and transmission (Gangappa and Botto, 2016; Li et al., 2020). Moreover, HY5 has been shown to be a positive regulator in nitrate absorption, phosphate response, and copper signaling pathways (Zhang et al., 2014; Huang et al., 2015; Chen et al., 2016; Gao et al., 2021). Arabidopsis HY5 mutants contain less chlorophyll content (Oyama et al., 1997; Holm et al., 2002; Xiao et al., 2022). A recent study has shown that HY5 can bind the promoter of the FER gene in roots, which is required for the induction of iron mobilization genes, thus providing us a new perspective in understanding the regulatory mechanism of iron uptake in plants (Guo et al., 2021). However, few studies have reported the correlation of HY5 and chlorophyll synthesis genes under Fe-deficient conditions. Moreover, no report has yet been published on the regulative role of HY5 to YSL iron transporters in response to iron stress in Malus.
Malus baccata has been widely used as a cold-resistant apple rootstock, especially in Northeast China. However, M. baccata is sensitive to iron deficiency. In this study, we compared the physiological characteristics and the transcriptive features of M. baccata under Fe-sufficient/deficiency conditions in the leaves and the roots and explored the regulative role of MbHY5 to chlorophyll metabolic genes and iron transporters (MbYSL). Our results provide insight into the molecular mechanism of iron deficiency response in M. baccata.
Materials and methods
Plant material and growth conditions
M. baccata in vitro shoots were cultured on MS medium (0.5 mg/l 6-BA and 0.5 mg/l IBA) for 30 days (Hao et al., 2022). Next, seedlings (with a height ~5 cm) were transported to the rooting medium (0.5 mg/l IBA) and cultured for 30 days. Rooted seedlings were transplanted into an improved-Hoagland nutrient solution and cultured for 3 weeks. Seedlings were cultivated at 25 ± 2°C day/21 ± 1°C night with a 16-h day/8-h night photoperiod.
Measurement of chlorophyll contents and rhizosphere pH
Seedling leaves grown on Fe-sufficient (+Fe, 40 μM) and Fe-deficient (-Fe, 0 μM) for 0, 24, 72, and 144 h were sampled, respectively. Leaves were cut into pieces after cleaning and removal of the veins. Next, 0.2 g tissues was mixed with quartz sand, calcium carbonate, and 95% ethanol. The absorbance of the filtrate was measured using a spectrophotometer (Shimadzu, Kyoto, Japan) at 663 and 645 nm. The rhizosphere pH was measured using a pH meter.
FCR activity
FCR activity was determined by the Ferrozine assay. The roots were first cultivated under +Fe and -Fe conditions for 0, 72, and 144 h and were then submerged into a chromogenic medium (0.5 mM ferrozine, 0.5 mM FeNa-EDTA, 0.5 mM CaSO4, and 0.7% (w/v) agar (Schmidt et al., 2000)) and incubated in the dark for 1 h. All measurements were performed at room temperature with a Shimadzu spectrophotometer (Kyoto, Japan).
Perls staining
Fresh root, stem, and leaf tissues were collected and placed in a small box (2 cm*2 cm*2 cm), which contains an appropriate amount of OCT, with tissues submerged by an embedding agent. Next, the bottom of the box was exposed to liquid nitrogen for quick freezing. Finally, the embedded blocks were placed on a freezing microtome for slicing, with continuous slicing of 10~20 μm. Perls staining was conducted using a Prussian Blue Iron Stain Kit (Solarbio, 60533ES20). Micro-tissues were transferred into Perls solution and stained for 0.5~1 h, then they were washed with deionized water and incubated in the methanol solution (Sun et al., 2020). Imaging was performed with a volume microscope (BA210, Motic) (Jia et al., 2018).
Fe content
The roots and leaves of the M. baccata seedlings treated under +Fe and -Fe conditions (see above) at different times were sampled 1 g for each sample. The samples were first dried at 105°C for 30 min then were placed at 80°C for 72 h till the samples were completely dry. Inductively coupled plasma–optical emission spectrometry was used to determine the active iron contents (Zheng et al., 2021).
Quantitative real-time PCR and public RNA-seq data analysis
Total RNA was extracted from the roots of M. baccata seedlings and was purified using the RNAprep Pure Plant Kit (TIANGEN, Beijing, China) according to the manufacturer’s instructions. cDNA was prepared from total RNA using the HiScript II 1st Strand cDNA Synthesis Kit (+gDNA wiper) (Vazyme, Nanjing, Chain). The LightCycler® 480 II system (Roche) was used for the qPCR assay, and the primers are listed in Supplementary Table 5. The relative expression of each gene was calculated based on the 2-△△Ct method.
A total of 30 groups of RNA-seq data from a project (PRJNA598053) was used to analyze the expression pattern of chlorophyll synthesis and iron transporter genes under Fe sufficiency and Fe deficiency conditions (0, 24, and 72 h) (Sun et al., 2020) (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA598053/ ). Data for the project were downloaded from the NCBI database, including roots and leaves. The expression abundance of the leaves and roots genes was calculated using the FPKM value, and the relative expression level is shown as log2 (fold change) values.
Plasmid construction and GUS histochemical staining
The full length of the MbHY5-coding sequence was inserted into the PRI101 (AN) vector. The promoters (upstream ~2 kb) of MbYSL7 or MbYSL2 were cloned respectively into the pCAMBIA1391 vector with the GUS reporter (Li et al., 2021b). Histochemical GUS staining of Nicotiana benthamiana leaves was conducted as previously described (Liu et al., 2002; Sun et al., 2020). The samples were incubated for 24 h at 37°C. Chlorophyll was removed by washing the samples with 70% (v/v) ethanol for 2 days. Imaging was performed with a volume microscope (MZ10F, Leica).
Transient expression
The full length of the MbYSL7-coding sequence was amplified without the stop codon using the specific primer pairs (Supplementary Table 5) and was inserted into the PRI101 (AN) vector with the 35S promoter. In order to repress the expression of MbYSL7, the pTRV-MbYSL7 vector was constructed as previously described (Sun et al., 2020; Hao et al., 2022). The MbYSL7-overexpression and VIGS vectors were transformed into Agrobacterium tumefaciens cells (GV3101). Infected apple seedlings were placed in a dark place for 2 days and then were transferred to normal light conditions for 1 day. Seedlings grown on Fe-sufficient and Fe-deficient conditions for 0, 24, 72, and 144 h were sampled and then stored at -80°C for RNA extraction.
Yeast one-hybrid assay
The full-length MbHY5 CDS sequence was inserted into pB42AD (AD vector), while the MbYSL7 or MbYSL2 protein-binding sites (CACGTG) were inserted into pLacZi (BD vector). The fusion vectors were transformed into the yeast EYG48 strain (Li et al., 2021b; Wu et al., 2021).
Phylogenetic tree
Homologous YSL gene sequences of M. domestica, M. baccata, and Arabidopsis thaliana were aligned using ClustalX version 2.0 (Jeanmougin et al., 1998). The phylogenetic tree was constructed in MEGA (version 11) (Tamura et al., 2021) with the Neighbor-Joining method (bootstrap replicates = 100).
Co-expression gene network analysis
In order to identify key genes involved in Fe deficiency in M. baccata, chlorophyll synthesis-related genes and iron homeostasis-related genes were selected, and their expression patterns under Fe deficiency were investigated based on the transcriptome data. Subsequently, their co-expressed genes were predicted using the AppleMDO database (network analysis) (http://bioinformatics.cau.edu.cn/AppleMDO/) (Da et al., 2019). Finally, these genes (503 genes in the leave samples and 693 genes in root samples) were selected to construct the co-expression network using Cytoscape 3.8.0 (Shannon et al., 2003; Zhao et al., 2017).
Statistical analysis and diagram drawing
Statistical analyses were executed using GraphPad Prism. The correlation of MbHY5 and chlorophyll synthesis- and roots iron homeostasis-related genes was calculated using the Pearson correlation (Lv et al., 2021). All statistical analyses were performed by one-way ANOVA test, with p ≤ 0.05 considered as significantly different among different samples. Diagrams illustrating the mechanism of chlorophyll synthesis and Fe acquisition were created using BioRender (https://biorender.com/) (Therby-Vale et al., 2022).
Results
M. baccata leaves and roots are sensitive to Fe deficiency
The chlorophyll content of M. baccata leaves showed a continual decrease from 0 to 144 h (Figure 1A) under -Fe treatments. After 144 h, the rhizosphere pH of -Fe treatment was lower than that of +Fe treatment, but with no statistically significant differences (Figure 1B). The results indicated that iron deficiency caused lower chlorophyll content in the leaves and a decrease in rhizosphere pH. Meanwhile, as for the content of active Fe in the leaves, it decreased from 104 to 42 mg/kg·DW after 144-h Fe deficiency stress. Similarly, its content in the roots also decreased from 923 to 284 mg/kg·DW (Figures 1C, D).
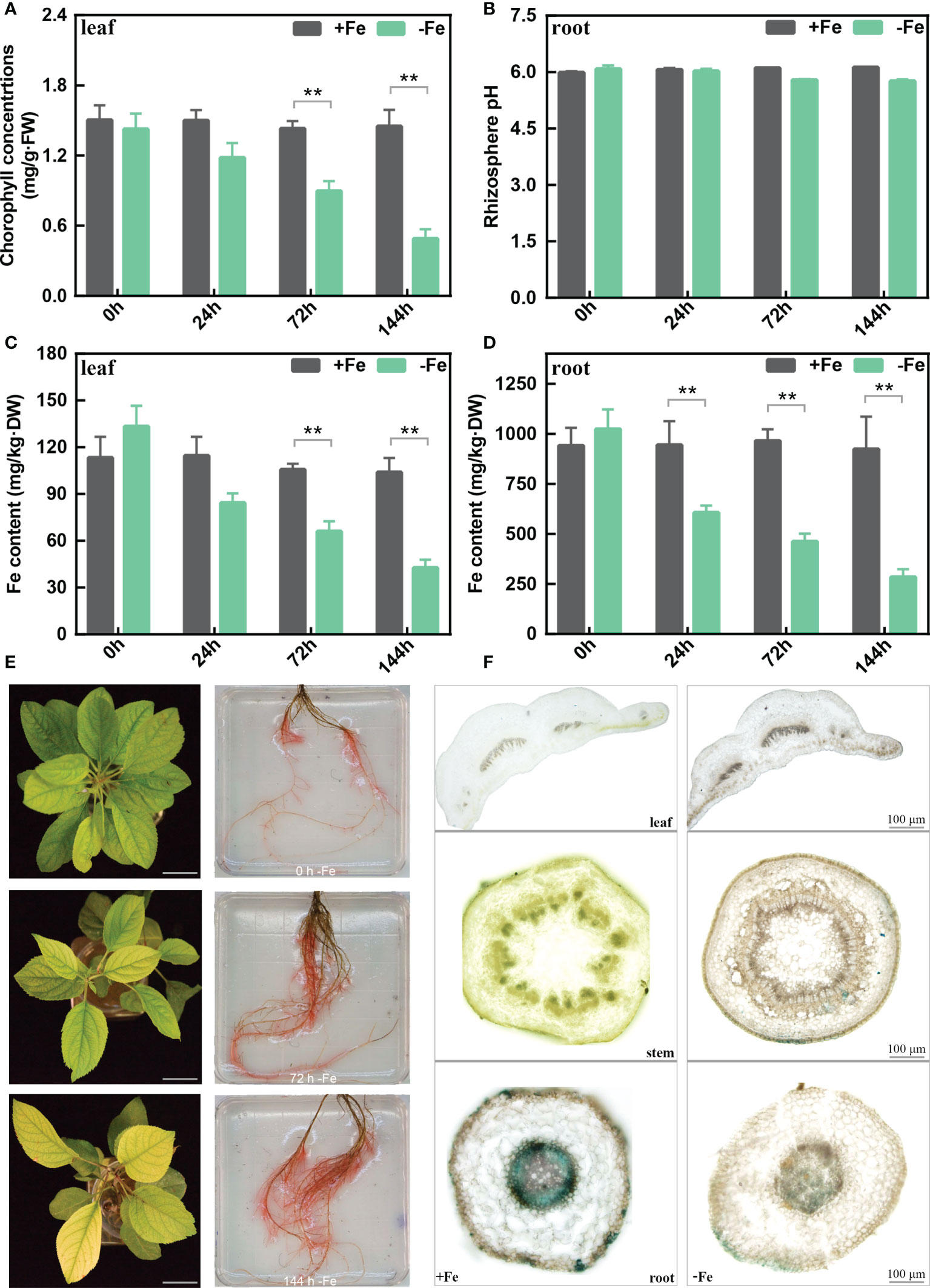
Figure 1 The physiological changes in the leaves and roots of M. baccata seedlings under Fe-deficient and Fe-sufficient conditions. (A) Chlorophyll concentration in leaves. (B) Rhizosphere pH in roots. (C) Fe content in leaves. (D) Fe content in roots. (E) Chlorosis extent in leaves and corresponding FCR activities in roots under 0, 72, and 144 h (scale bar: 0.5 cm). (F) Perls staining of different tissues, including leaf, stem, and root (scale bar: 100 μm). Asterisks indicate statistically significant differences (**p < 0.01). Error bars denote ± SD (biological replicates = 3).
We further measured the FCR activity of the roots to better understand the iron acquisition processes. Fe-deficient roots showed higher FCR activity in contrast with Fe-sufficient roots at different treatment times (Figure 1E). Moreover, Perls staining results showed that tissues (leave, stem, and root) from Fe-sufficient conditions showed stronger Fe3+ staining than Fe-deficient ones (Figure 1F). Interestingly, it also showed that Fe deficiency induces a sharp decrease of Fe3+ in xylem and phloem (Figure 1F). In conclusion, these results revealed that iron deficiency induced morphological and biochemical changes in M. baccata, including decreases in chlorophyll content, rhizosphere pH, and active iron content in the leaves and roots.
Iron deficiency induced the expression of chlorophyll synthesis genes in leaves
We hypothesized that the well-known light-responsive gene HY5 or PIF genes may have participated in the regulation of the chlorophyll synthesis process (Figure 2A). Indeed, we detected a series of chlorophyll metabolic genes from RNA-seq analysis under Fe deficiency, including Glu-tRNA reductase (HEMA), Glu 1-semialdehyde (GSA), uroporphyrinogen III synthase (UROS), chlorophyll synthase (CHLG), GUN, Chla oxygenase (CAO), protochlorophyllide oxidoreductase (PPO), and divinyl reductase (DVR). The results showed that HEMA1-1, HEMA1-2, CHLG1-1, CHLI, PPO5, CAO1-2, and CRD1 were highly expressed in all treatment times (Figure 2B). In contrast, the gene expressions of DVR, CHLG1-2, CLH1-1, UROS, and CLH were significantly lower in leaves (Figure 2B). Specially, the expression levels of PPO3, PPO9, CHLM1-1, CLH1, PPO8, GUN, GSA1-2, CHLM1-2, HEMA1-3, CAO1-2, CHLG1-1, and HEMA1-1 were significantly changed under Fe deficiency. We constructed a gene co-expression network to investigate the correlation of HY5 or PIF genes and the chlorophyll biosynthesis-related genes. The results showed that HY5, PIF1, PIF3, HMEA, GSA, and GUN form a complicated co-expression network in regulating chlorophyll biosynthesis (Figure 2C; Supplementary Table 1). Moreover, the expression levels of HY5 were positively correlated with those of most chlorophyll biosynthesis-related genes, such as HMEA, GSA1-2, CAO, CHLI, PPO, and GUN. The Pearson correlation coefficients between HY5 and these genes ranged from 0.54 to 0.78. In contrast, UROS, CHLG1-2, DVR, and CLH1-1 were only slightly correlated or did not correlate with HY5 (Figure 2D).
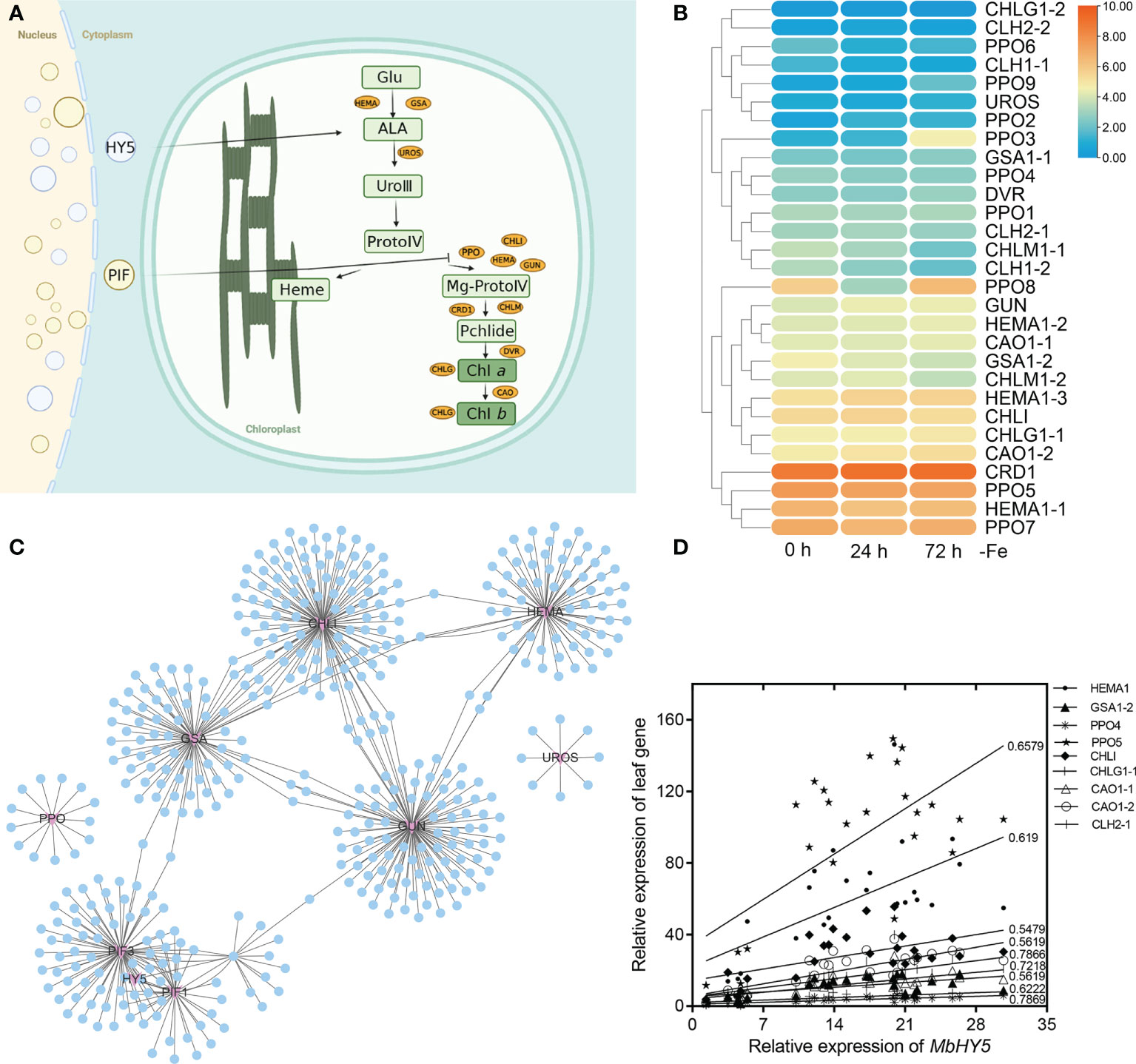
Figure 2 MbHY5 was associated with the chlorophyll synthesis genes in the leaves under Fe deficiency. (A) The assumption model of the MbHY5 gene participating the regulation of chlorophyll synthesis. Ovals represent chlorophyll biosynthesis-related genes. HEMA: glutamyl-tRNA reductase, GSA: glutamate-1-semialdehyde 2,1-aminotransferase, CHLH, Mg-chelatase, CHLM, Mg-protoporphyrin IX methyltransferase, CRD, Mg-protoporphyrin IX monomethylester cyclase; DVR, divinyl chlorophyllide a 8-vinyl-reductase; CAO, Chla oxygenase; CHLG, chlorophyll synthase; PPO, protochlorophyllide oxidoreductase. The model was drawn by BioRender (https://biorender.com/ ). (B) The heatmap showing the expression of chlorophyll biosynthesis-related genes in Fe-deficient conditions. (C) The co-expression network of HY5/PIF and chlorophyll biosynthesis-related genes. (D) Pearson correlation coefficients between MbHY5 and chlorophyll biosynthesis-related genes.
Analysis of the expression profiles of Strategy I-related genes under iron deficiency
Under iron deficiency conditions, Malus baccata, similar to other dicots, use Strategy I to acquire Fe in roots. We summarized the key genes reported in transferring and regulating Fe2+ transportation from the rhizosphere into root cells, including AHA2, FRO2, PDR9, IRT1, bHLH100/101, OPT3, and FIT (Ito and Gray, 2006; Satbhai et al., 2017; Khan et al., 2018; Lv et al., 2021; Pei et al., 2022) (Figure 3A). Under Fe deficiency, most of these genes were highly induced in roots, especially for PDR1, HY5, YSL7, FDR2, and FER genes (Figure 3B).
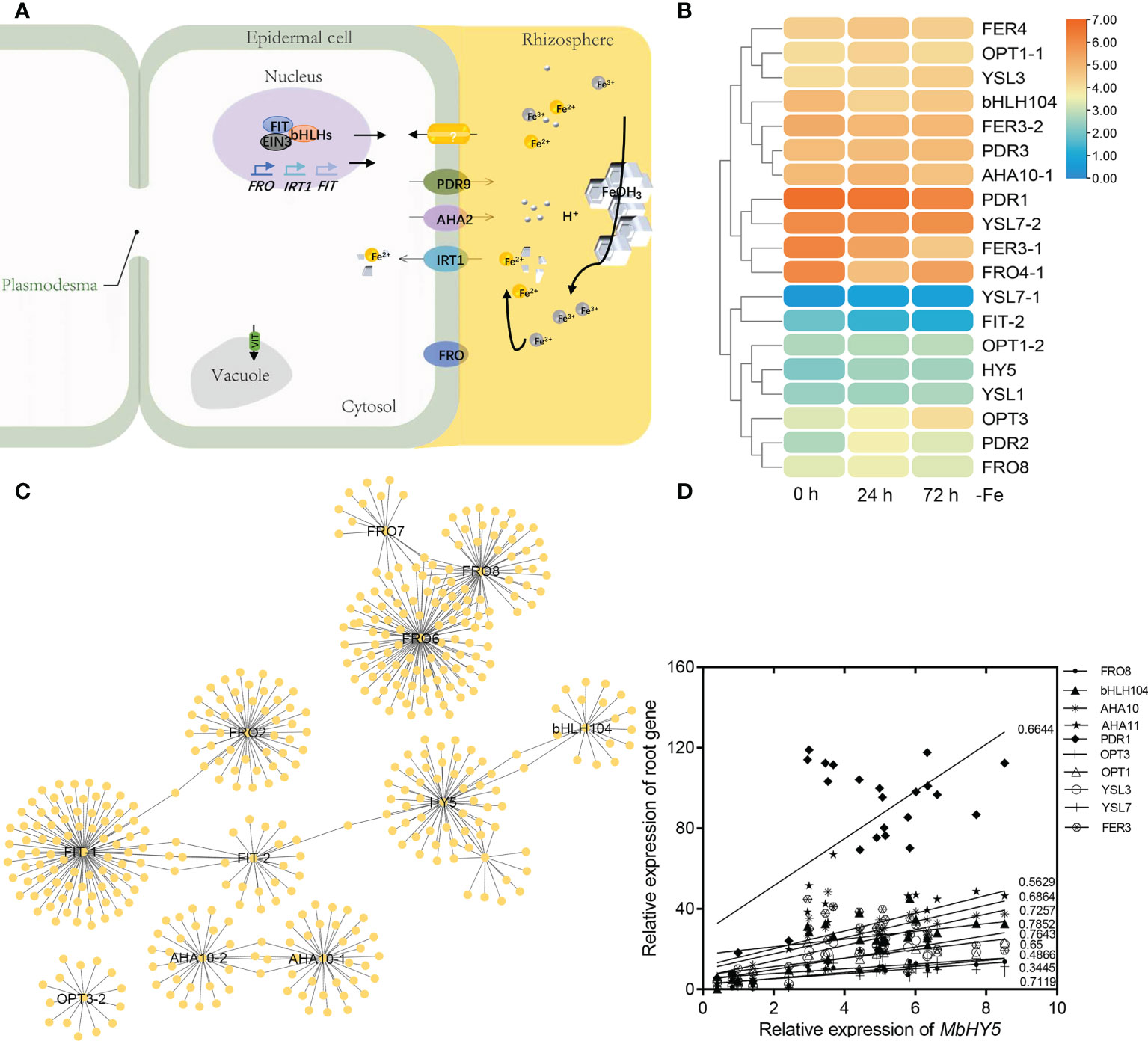
Figure 3 MbHY5 is related with Fe acquisition and transportation-related genes in roots in Fe-deficient conditions. (A) A model showing the acquisition and transportation of Fe in the roots of dicot plants under Fe deficiency. Ovals represent Fe homeostasis-related genes. (B) The heatmap showing the expression levels of the Fe homeostasis-related genes. (C) The co-expression network of HY5 and iron responsive genes in roots. (D) Pearson correlation coefficients between MbHY5 and Fe homeostasis-related genes.
In order to analyze the regulatory network of iron homeostasis genes in roots, a total of 693 iron homeostasis-related genes in roots were selected to construct the co-expression network, and the results showed that MbHY5–bHLH04–FIT–FRO2 constructed the biggest module, indicating that MbHY5 plays an essential role in regulation iron homeostasis in roots (Figure 3C; Supplementary Table 2). Pearson correlation analysis further showed that iron homeostasis-related genes differentially expressed in root under Fe deficiency were significantly positively related with MbHY5, including OPT3, PDR1, bHLH104, YSL, and AHA10 (Figure 3D). The correlation coefficients ranged from 0.45 to 0.78 (Figure 3D).
MbHY5 directly promotes the expression of MbYSL7 in response to Fe deficiency
We found that the expressions of YSL2 and YSL7 were highly related to HY5 (r = 0.7693 and 0.7119, respectively, Pearson correlation) (Figure 4A). The phylogenetic tree showed that each of the apple YSL genes clustered with its closely related homologous genes in Arabidopsis (Figure 4B). Previous studies have shown that HY5 can bind to the promoters of SlFER and AtBTS and induce the expression of a series of iron-uptaken genes under iron-deficient conditions (Guo et al., 2021; Mankotia et al., 2022).
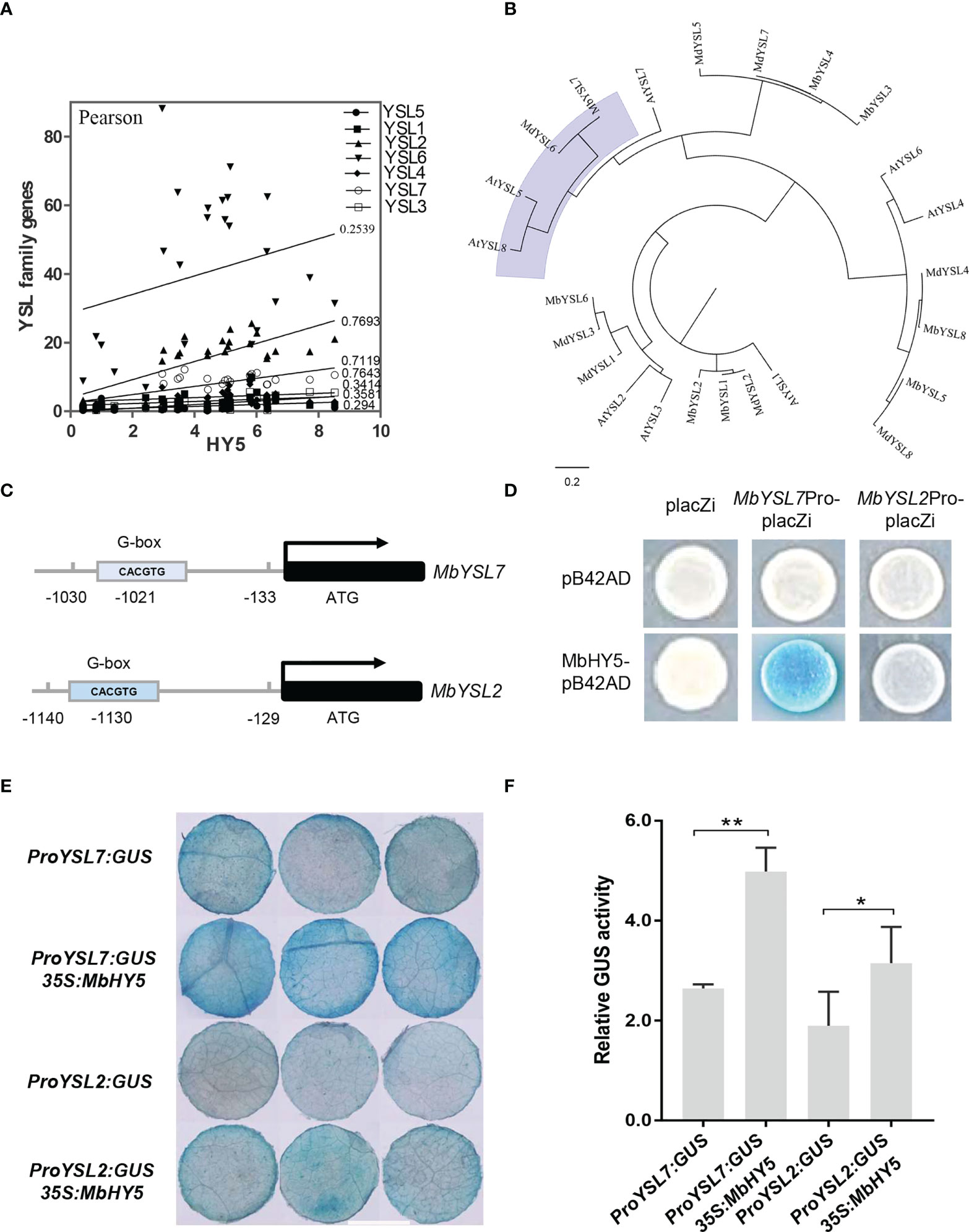
Figure 4 MbHY5 regulates MbYSL7 under Fe deficiency. (A) Pearson correlation coefficients of MbHY5 and YSL family genes. (B) The phylogenetic tree of the YSL proteins in three species, including Arabidopsis, M. baccata, and M. domestica. (C) Putative HY5-binding site (G-box) was found in the promoter region of MbYSL7 and MbYSL2, respectively. (D) Y1H assay. Coding sequence of MbHY5 was inserted into pB42AD while the promoter region of MbYSL7 or MbYSL2 was inserted into pLacZi, respectively. (E, F) GUS staining and GUS enzyme activity of transient transformations of tobacco leaves (scale bar = 1 cm); gene constructs used for the transformations were labeled. Error bars indicate SDs (biological replicates = 3), asterisks indicate statistically significant differences (*p < 0.05, **p < 0.01).
A G-box (CACGTG) element was found in each of the promoters of MbYSL2 and MbYSL7, which allows HY5 binding (Figure 4C). Y1H analysis showed that MbHY5 can directly bind to the promoter of MbYSL7, but not that of MbYSL2 (Figure 4D). Transient transformation of tobacco leaves with proMbYSL7:GUS showed lower GUS activity than co-transformation with 35S:MbHY5 (Figures 4E, F). Similarly, co-transformation of 35S:MbHY5 and proMbYSL2:GUS showed slightly higher GUS activity than the transformation of proMbYSL2:GUS only (Figures 4E, F). In conclusion, these data suggested that MbHY5 functions as a positive and direct regulator of MbYSL7.
Expression of MbYSL7 in transient transgenic apple seedlings
To further investigate whether MbYSL7 was involved in regulating Fe deficiency responses in apple, we made transient transformed lines of apple seedlings with overexpression vector and VIGS vector, respectively. As we can see, compared with the control line, the expression levels of MbYSL7 were highly induced in the transient transformed apple seedling lines of 35S:MbYSL7-1, -2, and -3 (Figure 5A). Under –Fe treatment, the expressions of MbYSL7 and MbHY5 were highly increased in MbYSL7 overexpression lines, compared with the control lines (Figure 5B). The expression of MbYSL7 was greatly reduced in pTRV : MbYSL7-1 (Figure 5C). Specifically, the expression of MbYSL7 slightly increased at the 144-h -Fe treatment, compared with that of the 72-h treatment. In comparison, the expression level of MbHY5 was lowest at the initial -Fe treatment but greatly induced from 24 h onward (Figure 5D). Similar to MbHY5, we found that MbYSL7 was positively related with chlorophyll synthesis-related genes as well, including PPO5, GSA1-2, and HEMA (Supplementary Table 3). In addition, we observed that MbYSL7 positively correlated with most of Fe homeostasis genes in root either, such as AHA10, bHLH104, and PDR2; the correlation coefficients ranged from 0.40 to 0.92 (Supplementary Table 4).
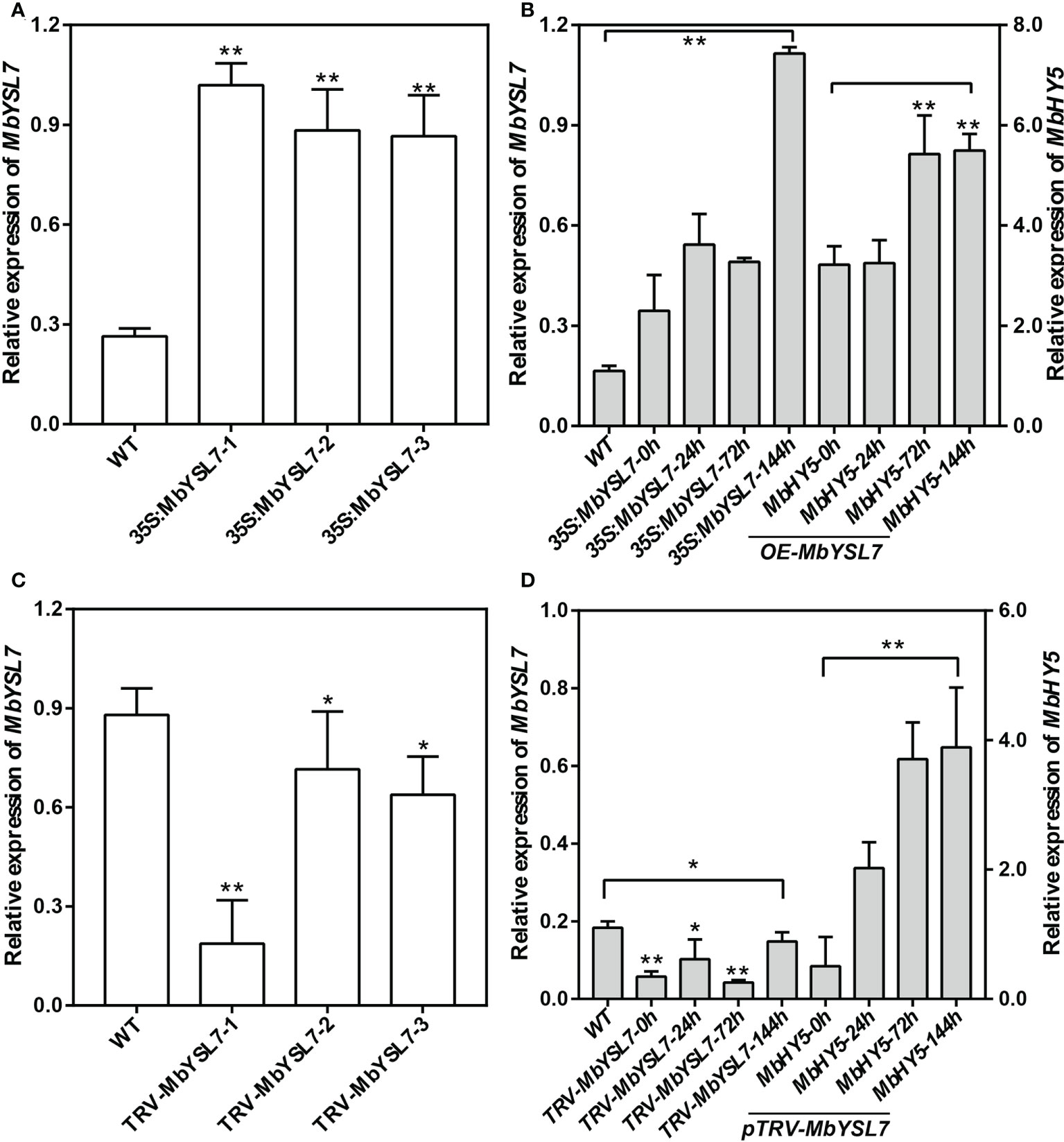
Figure 5 Expression of MbHY5 in transient transgenic M. baccata seedlings overexpressing or silencing MbYSL7. (A) Relative expression levels of MbYSL7 in transgenic lines overexpressing MbYSL7. (B) Relative expression levels of MbYSL7 and MbHY5 in the roots of the overexpression lines under Fe deficiency. (C) Relative expression levels of MbYSL7 in transgenic lines silencing MbYSL7. (D) Relative expression levels of MbYSL7 and MbHY5 in the roots of the overexpression lines under Fe deficiency. Error bars indicate SDs (biological replicates = 3), asterisks indicate statistically significant differences (*p < 0.05, **p < 0.01).
Discussion
In plants, iron deficiency leads to chlorosis caused by a reduced chlorophyll biosynthesis (Li et al., 2021a). Chlorophyll content decreased dramatically in chlorosis leaves under Fe deficiency (Figure 1), which is in agreement with the findings in citrus and grapes (Chen et al., 2004; Jin et al., 2017). Iron deficiency increased ferric chelate reduction (FCR) activity and decreased the rhizosphere pH of the apple roots (Figure 1). Also, we observed a reduction of active Fe content in the leaves and roots under iron deficiency. Perls staining is a reliable chemical method to stain the iron trivalent in tissues; ferric iron reacts with potassium ferrocyanide and generates blue insoluble compounds (Lv et al., 2021; Hao et al., 2022). Under Fe deficiency, a lower ferric iron content was observed compared to that of the Fe-sufficient treatment (Figure 1).
HY5 has been found to be involved in the metabolism of nitrogen (N), phosphorus (P), copper (Cu), sulfur (S), etc. (Zhang et al., 2014; Gangappa and Botto, 2016; Yang et al., 2020; Gao et al., 2021). In Arabidopsis, HY5 regulates the expression of key nitrogen signaling genes including NIA1, NIR1, NRT1.1, NRT2.1, and AMT1;2 (Jonassen et al., 2008; Jonassen et al., 2009; Yanagisawa, 2014; Chen et al., 2016; Xiao et al., 2022). In apple, NIA2 and NRT1.1 were positively regulated by HY5 in promoting nitrate assimilation (An et al., 2017). Nevertheless, few studies have reported its function in Fe uptake and homeostasis. In Arabidopsis, HY5 regulates BTS in response to Fe deficiency. Similar results were also found in tomato, in which the HY5-FER pathway could be involved in Fe metabolism (Guo et al., 2021; Mankotia et al., 2022). In the present study, we firstly found that MbHY5 was significantly changed in M. baccata under Fe deficiency. HY5 plays essential roles in photosynthetic pigment synthesis in light responses (Liu et al., 2017; Liu et al., 2020). It regulates the expression of chlorophyll-related genes in leaves, including HEMA1, GUN4, CAO, PORC, and CHLH (Toledo-Ortiz et al., 2014; Job and Datta, 2021). In addition, HY5 can regulate the genes involved in maintaining iron homeostasis, such as FRO2, FIT, IRTI, and PYE in roots (Mankotia et al., 2022). Further analysis found that MbHY5 participated in the regulation of chlorophyll synthesis in the leaves and iron acquisition in the roots under iron deficiency (Figures 2 and 3). Our results enriched the regulatory mechanism of HY5 in plants in response to Fe deficiency.
YSL genes have been found to participate in plant metal uptake, such as Cu and Fe (Ishimaru et al., 2010; Zheng et al., 2012; Dai et al., 2018). In Arabidopsis, AtYSL1-3 and AtYSL6-8 were responsive under Fe deficiency conditions; among them, some were characterized as long-distance signaling media or Fe(II)-NA transporters (Waters et al., 2006; Castro-Rodriguez et al., 2021). Previously, MtYSL7, AtYSL7, and GmYSL7 were identified and characterized as peptide transporters without further functional annotation (Castro-Rodriguez et al., 2021; Gavrin et al., 2021). Our results suggested that MbYSL7 plays an important role under Fe deficiency. Interestingly as evidenced by our Y1H and the transient co-transformation assays, MbYSL7 was positively regulated by MbHY5. Overall, we propose that MbHY5-YSL7 was involved in regulating the genes involved in chlorophyll synthesis and iron transportation, in both the leaves and the roots, to alleviate iron deficiency-caused chlorosis and to promote Fe transportation (Figure 6).
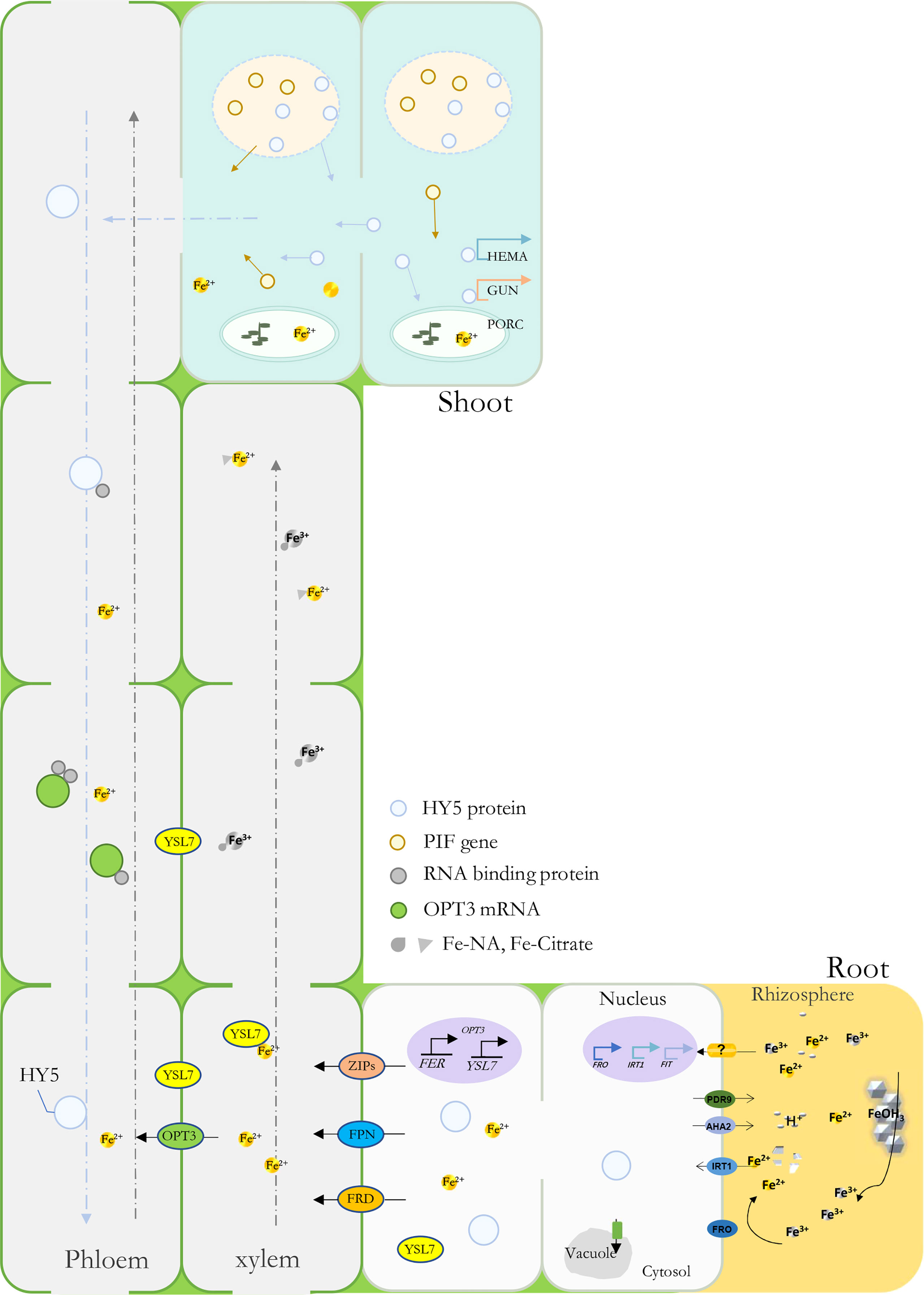
Figure 6 A model depicting MbHY5 as an important regulative transcription factor by regulating chlorophyll synthesis-related genes in the leaves, and Fe acquisition and transportation-related genes in the roots under Fe deficiency.
Conclusion
Contrasting differences of chlorophyll content and the concentration of active iron were observed under +Fe and -Fe conditions in M. baccata. We propose that MbHY5 functions as a vital transcription factor in regulating chlorophyll synthesis and Fe transportation. Lastly, MbHY5 directly regulates the expression of MbYSL7 in roots under Fe deficiency.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
YS designed the experiment, analyzed the data, and drafted the manuscript. YS, JWL, and PF prepared the materials and performed the bioinformatics analysis. YL, JKL, and FY helped with the qRT-PCR analysis. YZ, FM, and TZ edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the National Natural Science Foundation of China (32102311 and 32102338), the China Postdoctoral Science Foundation (2021M690129), the Chinese Universities Scientific Fund (2452020265 and 2452021133), and the Xinjiang Production and Construction Corps Key Laboratory of Protection and Utilization of Biological Resources in Tarim Basin (BRZD2105).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1035233/full#supplementary-material
References
Alvarez-Fernandez, A., Paniagua, P., Abadia, J., Abadia, A. (2003). Effects of fe deficiency chlorosis on yield and fruit quality in peach (Prunus persica l. batsch). J. Agr. Food Chem. 51 (19), 5738–5744. doi: 10.1021/jf034402c
An, J. P., Qu, F. J., Yao, J. F., Wang, X. N., You, C. X., Wang, X. F., et al. (2017). The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res-England. 4, 17056. doi: 10.1038/hortres.2017.23
Castro-Rodriguez, R., Escudero, V., Reguera, M., Gil-Diez, P., Quintana, J., Prieto, R. I., et al. (2021). Medicago truncatula yellow stripe-Like7 encodes a peptide transporter participating in symbiotic nitrogen fixation. Plant Cell Environ. 44 (6), 1908–1920. doi: 10.1111/pce.14059
Chen, L. S., Smith, B. R., Cheng, L. L. (2004). CO2 assimilation, photosynthetic enzymes, and carbohydrates of 'Concord' grape leaves in response to iron supply. J. Am. Soc Hortic. Sci. 129 (5), 738–744. doi: 10.21273/JASHS.129.5.0738
Chen, X. B., Yao, Q. F., Gao, X. H., Jiang, C. F., Harberd, N. P., Fu, X. D. (2016). Shoot-to-root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr. Biol. 26 (5), 640–646. doi: 10.1016/j.cub.2015.12.066
Chowdhury, R., Nallusamy, S., Shanmugam, V., Loganathan, A., Muthurajan, R., Sivathapandian, S. K., et al. (2022). Genome-wide understanding of evolutionary and functional relationships of rice yellow stripe-like (YSL) transporter family in comparison with other plant species. Biologia 77 (1), 39–53. doi: 10.1007/s11756-021-00924-5
Curie, C., Briat, J. F. (2003). Iron transport and signaling in plants. Annu. Rev. Plant Biol. 54, 183–206. doi: 10.1146/annurev.arplant.54.031902.135018
Curie, C., Cassin, G., Couch, D., Divol, F., Higuchi, K., Jean, M., et al. (2009). Metal movement within the plant: contribution of nicotianamine and yellow stripe 1-like transporters. Ann. Bot-London. 103 (1), 1–11. doi: 10.1093/aob/mcn207
Dai, J., Wang, N., Xiong, H., Qiu, W., Nakanishi, H., Kobayashi, T., et al. (2018). The yellow stripe-like (YSL) gene gunctions in internal copper transport in peanut. Genes 9, 635. doi: 10.3390/genes9120635
Da, L. L., Liu, Y., Yang, J. T., Tian, T., She, J. J., Ma, X. L., et al. (2019). AppleMDO: a multi-dimensional omics database for apple co-expression networks and chromatin states. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01333
Eide, D., Broderius, M., Fett, J., Guerinot, M. L. (1996). A novel iron-regulated metal transporter from plants identified by functional expression in yeast. P. Natl. Acad. Sci. U.S.A. 93 (11), 5624–5628. doi: 10.1073/pnas.93.11.5624
Gangappa, S. N., Botto, J. F. (2016). ). the multifaceted roles of HY5 in plant growth and development. Mol. Plant 9 (10), 1353–1365. doi: 10.1016/j.molp.2016.07.002
Gao, Y. Q., Bu, L. H., Han, M. L., Wang, Y. L., Li, Z. Y., Liu, H. T., et al. (2021). Long-distance blue light signalling regulates phosphate deficiency-induced primary root growth inhibition. Mol. Plant 14 (9), 1539–1553. doi: 10.1016/j.molp.2021.06.002
Gavrin, A., Loughlin, P. C., Brear, E., Griffith, O. W., Bedon, F., Grotemeyer, M. S., et al. (2021). Soybean yellow stripe-like 7 is a symbiosome membrane peptide transporter important for nitrogen fixation. Plant Physiol. 186 (1), 581–598. doi: 10.1093/plphys/kiab044
Guo, Z. X., Xu, J., Wang, Y., Hu, C. Y., Shi, K., Zhou, J., et al. (2021). The phyBdependent induction of HY5 promotes iron uptake by systemically activating FER expression. EMBO Rep. 22 (7), e51944. doi: 10.15252/embr.202051944
Hao, P. B., Lv, X. M., Fu, M. M., Xu, Z., Tian, J., Wang, Y., et al. (2022). Long-distance mobile mRNA CAX3 modulates iron uptake and zinc compartmentalization. EMBO Rep. 23 (5), e53698. doi: 10.15252/embr.202153698
Hell, R., Stephan, U. W. (2003). Iron uptake, trafficking and homeostasis in plants. Planta 216 (4), 541–551. doi: 10.1007/s00425-002-0920-4
Holm, M., Ma, L. G., Qu, L. J., Deng, X. W. (2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Gene Dev. 16 (10), 1247–1259. doi: 10.1101/gad.969702
Huang, L. F., Zhang, H. C., Zhang, H. Y., Deng, X. W., Wei, N. (2015). HY5 regulates nitrite reductase 1 (NIR1) and ammonium transporter1;2 (AMT1;2) in Arabidopsis seedlings. Plant Sci. 238, 330–339. doi: 10.1016/j.plantsci.2015.05.004
Ishimaru, Y., Masuda, H., Bashir, K., Inoue, H., Tsukamoto, T., Takahashi, M., et al. (2010). Rice metal-nicotianamine transporter, OsYSL2, is required for the long-distance transport of iron and manganese. Plant J. 62 (3), 379–390. doi: 10.1111/j.1365-313X.2010.04158.x
Ito, H., Gray, W. M. (2006). A gain-of-function mutation in the Arabidopsis pleiotropic drug resistance transporter PDR9 confers resistance to auxinic herbicides. Plant Physiol. 142 (1), 63–74. doi: 10.1104/pp.106.084533
Ivanov, R., Brumbarova, T., Bauer, P. (2012). Fitting into the harsh reality: regulation of iron-deficiency responses in dicotyledonous plants. Mol. Plant 5 (1), 27–42. doi: 10.1093/mp/ssr065
Jeanmougin, F., Thompson, J. D., Gouy, M., Higgins, D. G., Gibson, T. J. (1998). Multiple sequence alignment with clustal x. Trends Biochem. Sci. 23 (10), 403–405. doi: 10.1016/s0968-0004(98)01285-7
Jeong, J., Guerinot, M. L. (2009). Homing in on iron homeostasis in plants. Trends Plant Sci. 14 (5), 280–285. doi: 10.1016/j.tplants.2009.02.006
Jia, D. J., Shen, F., Wang, Y., Wu, T., Xu, X. F., Zhang, X. Z., et al. (2018). Apple fruit acidity is genetically diversified by natural variations in three hierarchical epistatic genes: MdSAUR37, MdPP2CH and MdALMTII. Plant J. 95 (3), 427–443. doi: 10.1111/tpj.13957
Jin, L. F., Liu, Y. Z., Du, W., Fu, L. N., Hussain, S. B., Peng, S. A. (2017). Physiological and transcriptional analysis reveals pathways involved in iron deficiency chlorosis in fragrant citrus. Tree Genet. Genomes. 13 (3), 1104. doi: 10.1007/s11295-017-1136-x
Job, N., Datta, S. (2021). PIF3/HY5 module regulates BBX11 to suppress protochlorophyllide levels in dark and promote photomorphogenesis in light. New Phytol. 230 (1), 190–204. doi: 10.1111/nph.17149
Jonassen, E. M., Lea, U. S., Lillo, C. (2008). HY5 and HYH are positive regulators of nitrate reductase in seedlings and rosette stage plants. Planta 227 (3), 559–564. doi: 10.1007/s00425-007-0638-4
Jonassen, E. M., Sevin, D. C., Lillo, C. (2009). The bZIP transcription factors HY5 and HYH are positive regulators of the main nitrate reductase gene in Arabidopsis leaves, NIA2, but negative regulators of the nitrate uptake gene NRT1.1. J. Plant Physiol. 166 (18), 2071–2076. doi: 10.1016/j.jplph.2009.05.010
Khan, M. A., Castro-Guerrero, N. A., McInturf, S. A., Nguyen, N. T., Dame, A. N., Wang, J. J., et al. (2018). Changes in iron availability in Arabidopsis are rapidly sensed in the leaf vasculature and impaired sensing leads to opposite transcriptional programs in leaves and roots. Plant Cell Environ. 41 (10), 2263–2276. doi: 10.1111/pce.13192
Kobayashi, T., Nishizawa, N. K. (2012). Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 63, 131–152. doi: 10.1146/annurev-arplant-042811-105522
Kumar, A., Kaur, G., Goel, P., Bhati, K. K., Kaur, M., Shukla, V., et al. (2019). Genome-wide analysis of oligopeptide transporters and detailed characterization of yellow stripe transporter genes in hexaploid wheat. Funct. Integr. Genomic. 19 (1), 75–90. doi: 10.1007/s10142-018-0629-5
Le Jean, M., Schikora, A., Mari, S., Briat, J. F., Curie, C. (2005). A loss-of-function mutation in AtYSL1 reveals its role in iron and nicotianamine seed loading. Plant J. 44 (5), 769–782. doi: 10.1111/j.1365-313X.2005.02569.x
Li, J., Cao, X. M., Jia, X. C., Liu, L. Y., Cao, H. W., Qin, W. Q., et al. (2021a). Iron deficiency leads to chlorosis through impacting chlorophyll synthesis and nitrogen metabolism in Areca catechu l. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.710093
Li, X., Shen, F., Xu, X. Z., Zheng, Q. B., Wang, Y., Wu, T., et al. (2021b). An HD-ZIP transcription factor, MxHB13, integrates auxin-regulated and juvenility-determined control of adventitious rooting in Malus xiaojinensis. Plant J. 107 (6), 1663–1680. doi: 10.1111/tpj.15406
Li, J., Terzaghi, W., Gong, Y. Y., Li, C. R., Ling, J. J., Fan, Y. Y., et al. (2020). Modulation of BIN2 kinase activity by HY5 controls hypocotyl elongation in the light. Nat. Commun. 11 (1), 1592. doi: 10.1038/s41467-020-15394-7
Liu, L. L., Lin, N., Liu, X. Y., Yang, S., Wang, W., Wan, X. C. (2020). From chloroplast biogenesis to chlorophyll accumulation: the interplay of light and hormones on gene expression in Camellia sinensis cv. shuchazao leaves. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00256
Liu, X. Q., Li, Y., Zhong, S. W. (2017). Interplay between light and plant hormones in the control of Arabidopsis seedling chlorophyll biosynthesis. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01433
Liu, Y. L., Schiff, M., Marathe, R., Dinesh-Kumar, S. P. (2002). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30 (4), 415–429. doi: 10.1046/j.1365-313x.2002.01297.x
Lv, X. M., Sun, Y. Q., Hao, P. B., Zhang, C. K., Tian, J., Fu, M. M., et al. (2021). RBP differentiation contributes to selective transmissibility of OPT3 mRNAs. Plant Physiol. 187 (3), 1587–1604. doi: 10.1093/plphys/kiab366
Mankotia, S., Singh, D., Monika, K., Meena, H., Meena, V., Yadav, R. K., et al. (2022). Elongated hypocotyl 5 (HY5) regulates BRUTUS (BTS) to maintain iron homeostasis in Arabidopsis thaliana. bioRxiv 2022.04.26.489524. doi: 10.1101/2022.04.26.489524
Morrissey, J., Guerinot, M. L. (2009). Iron uptake and transport in plants: the good, the bad, and the lonome. Chem. Rev. 109 (10), 4553–4567. doi: 10.1021/cr900112r
Murata, Y., Ma, J. F., Yamaji, N., Ueno, D., Nomoto, K., Iwashita, T. (2006). A specific transporter for iron(III)-phytosiderophore in barley roots. Plant J. 46 (4), 563–572. doi: 10.1111/j.1365-313X.2006.02714.x
Oyama, T., Shimura, Y., Okada, K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Gene Dev. 11 (22), 2983–2995. doi: 10.1101/gad.11.22.2983
Pei, D., Hua, D. P., Deng, J. P., Wang, Z. F., Song, C. P., Wang, Y., et al. (2022). Phosphorylation of the plasma membrane h+-ATPase AHA2 by BAK1 is required for ABA-induced stomatal closure in Arabidopsis. Plant Cell. 34 (7), 2708–2729. doi: 10.1093/plcell/koac106
Santi, S., Schmidt, W. (2009). Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol. 183 (4), 1072–1084. doi: 10.1111/j.1469-8137.2009.02908.x
Satbhai, S. B., Setzer, C., Freynschlag, F., Slovak, R., Kerdaffrec, E., Busch, W. (2017). Natural allelic variation of FRO2 modulates Arabidopsis root growth under iron deficiency. Nat. Commun. 8, 15603. doi: 10.1038/ncomms15603
Schmidt, W., Tittel, J., Schikora, A. (2000). Role of hormones in the induction of iron deficiency responses in Arabidopsis roots. Plant Physiol. 122 (4), 1109–1118. doi: 10.1104/pp.122.4.1109
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13 (11), 2498–2504. doi: 10.1101/gr.1239303
Sun, Y. Q., Hao, P. B., Lv, X. N., Tian, J., Wang, Y., Zhang, X. Z., et al. (2020). A long non-coding apple RNA, MSTRG.85814.11, acts as a transcriptional enhancer of SAUR32 and contributes to the fe-deficiency response. Plant J. 103 (1), 53–67. doi: 10.1111/tpj.14706
Tagliavini, M., Rombola, A. D., Marangoni, B. (1995). Response to iron-deficiency stress of pear and quince genotypes. J. Plant Nutr. 18 (11), 2465–2482. doi: 10.1080/01904169509365077
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11 molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38 (7), 3022–3027. doi: 10.1093/molbev/msab120
TerBush, A. D., Yoshida, Y., Osteryoung, K. W. (2013). FtsZ in chloroplast division: structure, function and evolution. Curr. Opin. Cell Biol. 25 (4), 461–470. doi: 10.1016/j.ceb.2013.04.006
Therby-Vale, R., Lacombe, B., Rhee, S. Y., Nussaume, L., Rouached, H. (2022). Mineral nutrient signaling controls photosynthesis: focus on iron deficiency-induced chlorosis. Trends Plant Sci. 27 (5), 502–509. doi: 10.1016/j.tplants.2021.11.005
Toledo-Ortiz, G., Johansson, H., Lee, K. P., Bou-Torrent, J., Stewart, K., Steel, G., et al. (2014). The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PloS Genet. 10 (6), e1004416. doi: 10.1371/journal.pgen.1004416
Tsai, H. H., Schmidt, W. (2017). Mobilization of iron by plant-borne coumarins. Trends Plant Sci. 22 (6), 538–548. doi: 10.1016/j.tplants.2017.03.008
Walker, E. L., Connolly, E. L. (2008). Time to pump iron: iron-deficiency-signaling mechanisms of higher plants. Curr. Opin. Plant Biol. 11 (5), 530–535. doi: 10.1016/j.pbi.2008.06.013
Waters, B. M., Chu, H. H., DiDonato, R. J., Roberts, L. A., Eisley, R. B., Lahner, B., et al. (2006). Mutations in Arabidopsis yellow stripe-Like1 and yellow stripe-Like3 reveal their roles in metal ion homeostasis and loading of metal ions in seeds. Plant Physiol. 141 (4), 1446–1458. doi: 10.1104/pp.106.082586
Wu, B., Shen, F., Wang, X., Zheng, W. Y., Xiao, C., Deng, Y., et al. (2021). Role of MdERF3 and MdERF118 natural variations in apple flesh firmness/crispness retainability and development of QTL-based genomics-assisted prediction. Plant Biotechnol. J. 19 (5), 1022–1037. doi: 10.1111/pbi.13527
Xiao, Y. T., Chu, L., Zhang, Y. M., Bian, Y. T., Xiao, J. H., Xu, D. Q. (2022). HY5: a pivotal regulator of light-dependent development in higher plants. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.800989
Yanagisawa, S. (2014). Transcription factors involved in controlling the expression of nitrate reductase genes in higher plants. Plant Sci. 229, 167–171. doi: 10.1016/j.plantsci.2014.09.006
Yang, Z. H., Chen, Z. X., He, N., Yang, D., Liu, M. D. (2022). Effects of silicon and iron application on arsenic absorption and physiological characteristics of rice (Oryza sativa l.). B. Environ. Contam. Tox. 108 (6), 1046–1055. doi: 10.1007/s00128-022-03476-9
Yang, C., Shen, W. J., Yang, L. M., Sun, Y., Li, X. B., Lai, M. Y., et al. (2020). HY5-HDA9 module transcriptionally regulates plant autophagy in response to light-to-dark conversion and nitrogen starvation. Mol. Plant 13 (3), 515–531. doi: 10.1016/j.molp.2020.02.011
Zang, J., Huo, Y. Q., Liu, J., Zhang, H. R., Liu, J., Chen, H. B. (2020). Maize YSL2 is required for iron distribution and development in kernels. J. Exp. Bot. 71 (19), 5896–5910. doi: 10.1093/jxb/eraa332
Zhang, H. Y., Zhao, X., Li, J. G., Cai, H. Q., Deng, X. W., Li, L. (2014). MicroRNA408 is critical for the HY5-SPL7 gene network that mediates the coordinated response to light and copper. Plant Cell. 26 (12), 4933–4953. doi: 10.1105/tpc.114.127340
Zhao, T., Holmer, R., de Bruijn, S., Angenent, G. C., van den Burg, H. A., Schranz, M. E. (2017). Phylogenomic synteny network analysis of MADS-box transcription factor genes reveals lineage-specific transpositions, ancient tandem duplications, and deep positional conservation. Plant Cell. 29 (6), 1278–1292. doi: 10.1105/tpc.17.00312
Zheng, X. D., Chen, H. F., Su, Q. F., Wang, C. H., Sha, G. L., Ma, C. Q., et al. (2021). Resveratrol improves the iron deficiency adaptation of Malus baccata seedlings by regulating iron absorption. BMC Plant Biol. 21 (1), 433. doi: 10.1186/s12870-021-03215-y
Keywords: Malus baccata, iron deficiency, chlorophyll synthesis, Fe transporter, regulatory network, MbHY5
Citation: Sun Y, Luo J, Feng P, Yang F, Liu Y, Liang J, Wang H, Zou Y, Ma F and Zhao T (2022) MbHY5-MbYSL7 mediates chlorophyll synthesis and iron transport under iron deficiency in Malus baccata. Front. Plant Sci. 13:1035233. doi: 10.3389/fpls.2022.1035233
Received: 02 September 2022; Accepted: 27 September 2022;
Published: 19 October 2022.
Edited by:
Bowen Liang, Hebei Agricultural University, ChinaReviewed by:
Xu Xiaozhao, China Agricultural University, ChinaJia-Yu Xue, Nanjing Agricultural University, China
Copyright © 2022 Sun, Luo, Feng, Yang, Liu, Liang, Wang, Zou, Ma and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tao Zhao, dGFvLnpoYW9AbndhZnUuZWR1LmNu; Fengwang Ma, ZndtNjRAbndzdWFmLmVkdS5jbg==; Yangjun Zou, eWFuZ2p1bnpvdUAxMjYuY29t
 Yaqiang Sun
Yaqiang Sun Jiawei Luo
Jiawei Luo Fengwang Ma
Fengwang Ma Tao Zhao
Tao Zhao