- Faculty of Science and Technology, Free University of Bozen/Bolzano, Bolzano, Italy
The application of copper (Cu)-based fungicides for crop protection plans has led to a high accumulation of Cu in soils, especially in vineyards. Copper is indeed an essential micronutrient for plants, but relatively high concentrations in soil or other growth substrates may cause toxicity phenomena, such as alteration of the plant’s growth and disturbance in the acquisition of mineral nutrients. This last aspect might be particularly relevant in the case of nitrate , whose acquisition in plants is finely regulated through the transcriptional regulation of transporters and plasma membrane H+-ATPase in response to the available concentration of the nutrient. In this study, cucumber plants were grown hydroponically and exposed to increasing concentrations of Cu (i.e., 0.2, 5, 20, 30, and 50 µM) to investigate their ability to respond to and acquire . To this end, the kinetics of substrate uptake and the transcriptional modulation of the molecular entities involved in the process have been assessed. Results showed that the inducibility of the high-affinity transport system was significantly affected by increasing Cu concentrations; at Cu levels higher than 20 µM, plants demonstrated either strongly reduced or abolished uptake activity. Nevertheless, the transcriptional modulation of both the nitrate transporter CsNRT2.1 and the accessory protein CsNRT3.1 was not coherent with the hindered uptake activity. On the contrary, CsHA2 was downregulated, thus suggesting that a possible impairment in the generation of the proton gradient across the root PM could be the cause of the abolishment of uptake.
Introduction
Nitrogen (N) is a fundamental element for crops, and its limitations can severely affect plant development and productivity (Marschner, 2012). This nutrient can be acquired by roots from different sources, namely inorganic (e.g., nitrate— and ammonia— ) and organic (e.g., urea and amino acids) (Nacry et al., 2013; Zanin et al., 2015). However, among the inorganic N ones, represents the main source for the majority of crops cultivated in aerobic agricultural soils (Crawford and Glass, 1998). The uptake kinetics of this form, when analyzed as a function of its external concentration, show a biphasic trend (Crawford and Glass, 1998; Forde and Clarkson, 1999), ascribable to at least two transport systems, the high and low affinity transport systems (HATS and LATS, respectively) (Touraine and Glass, 1997; Filleur et al., 2001). The HATS is generally more involved in the uptake when the external concentration is lower than 0.5–1 mM, and it shows a saturable behavior, described by a Michaelis–Menten kinetic (Filleur et al., 2001). On the other hand, LATS is active when the concentration in the growth medium is higher than 0.5–1 mM, and it displays non-saturable kinetics in which the uptake rate linearly increases along with the substrate concentration (Touraine and Glass, 1997). Moreover, uptake at the level of the root plasma membrane (PM) is mediated by at least two gene families, NRT1 and NRT2 (Plett et al., 2010; Nacry et al., 2013). The NRT1 gene encodes for low-affinity transporters, whereas the NRT2 gene family encodes for high-affinity transporters (Plett et al., 2010). Furthermore, members of the NRT2 gene family are often expressed together with accessory proteins belonging to the NRT3 family, which are required for the functionality and regulation of HATS transporters (Zhou et al., 2000; Tong et al., 2005; Okamoto et al., 2006; Pii et al., 2016).
In field conditions, the concentration in soil is variable in time and space. Thus, plants have adapted in order to modulate their uptake capacity according to nutrient availability (Marschner, 2012). In fact, following the exposure of roots to (like after nitrate fertilization of crops), an upregulation of genes encoding transporters belonging to the inducible HATS (iHATS) takes place (Siddiqi et al., 1989). This phenomenon is termed induction (Jackson et al., 1973), and it results in a higher uptake rate by roots. The induction is a transient phenomenon, since the uptake rate generally reaches its maximum in a few hours or days, in herbaceous species or tree plants, respectively (Crawford and Glass, 1998; Min et al., 1998; Pii et al., 2014), and it is afterwards downregulated, due to a negative feedback inhibition (Glass et al., 2001).
Nitrate uptake by roots is secondary active transport, therefore it requires metabolic energy to be sustained (Siddiqi et al., 1990; Glass et al., 1992). Previous evidence has demonstrated that is taken up through a co-transporting mechanism along with H+, whereby the proton gradient across the PM is generated and maintained by the activity of the PM H+-ATPase (McClure et al., 1990a; McClure et al., 1990b; Glass et al., 1992; Santi et al., 1995). Several studies in the last years have shown that both the biochemical activity and the molecular regulation of PM H+-ATPase mirror the profile observed for the induction of uptake rate (Santi et al., 2003; Nikolic et al., 2012; Pii et al., 2014; Pii et al., 2016), further disclosing the essential role of these molecular entities for HATS functionality. It is interesting to point out that PM H+-ATPase involved in the mineral acquisition processes in plants represents only a subgroup, namely P3 (Pedersen et al., 2012), of a wider gene family encompassing H+-ATPase enzymes featuring different physiological functions and substrate specificity (Axelsen and Palmgren, 1998; Palmgren and Nissen, 2011). Yet, despite the redundancy of PM H+-ATPase, only specific isoforms are involved in uptake at the root level (Santi et al., 2003; Nikolic et al., 2012; Pii et al., 2014; Pii et al., 2016).
Copper (Cu) is an essential element for plants, being involved in a plethora of physiological functions such as, for instance, photosynthesis, respiration, C and N metabolism, and protection against oxidative stress (Marschner, 2012). Despite its crucial role in plants, it is required in very low amounts (Yruela, 2009). On the other hand, its excess might induce stressing conditions like alterations in photosynthesis capacity, stunted growth (Brunetto et al., 2016), root length reduction, and root tip darkening and thickening (Vinit-Dunand et al., 2002; Lequeux et al., 2010; Meier et al., 2012; Feigl et al., 2013). Moreover, when exposed to toxic Cu concentrations, plants can present nutritional disorders, displayed by an altered accumulation of macro- and micronutrients at both shoot and root levels (Baldi et al., 2018; Marastoni et al., 2019a; Marastoni et al., 2019b; Feil et al., 2020; Cesco et al., 2021). It has been recently demonstrated that a Cu excess can impair the ability of cucumber plants to take up phosphorus (P) from the growth medium by negatively affecting the expression of high-affinity transporters in roots (Feil et al., 2020). Similarly, Marastoni et al. (2019b) have recently shown that a Cu excess could differentially affect the expression of transporters involved in the uptake of micronutrients, whose extent was dependent on the tolerance level to the metal exhibited by the different grapevine rootstocks. Furthermore, previous pieces of research have indicated that Cu stress can alter N nutrition in plants, affecting both the ability of plants to take up and and the biochemical activities devoted to N assimilation (Xiong et al., 2006; Zhang et al., 2014; Hippler et al., 2018; Gong et al., 2019; Huo et al., 2020). In this context, it has been observed that prolonged Cu toxicity in Arabidopsis thaliana can downregulate the expression of NRT1 genes encoding low-affinity nitrate transporters (NRT1 family) as well as those encoding specific isoforms of the plasma membrane proton pump (i.e., AHA2) (Hippler et al., 2018). On the contrary, medium Cu excess (i.e., 5 and 10 µM) could induce the upregulation of the NRT2 gene family in 24–76 h, albeit not at sufficient levels to restore N uptake at the root level. Despite these pieces of evidence, a detailed investigation concerning the Cu effects on acquisition mechanisms has never been carried out so far.
Indeed, the increase in Cu concentration in agricultural soil is currently becoming a severe environmental problem, particularly in vineyards, mainly due to the application of cupric fungicides for crop defense plans (Cesco et al., 2021). In a context of greater sustainability in agriculture and the need to guarantee food for a growing population, it is therefore mandatory to deepen our knowledge about plant responses to Cu toxicity, particularly focusing on the dynamics of N acquisition. Despite N being a fundamental macronutrient for plants, research efforts to understand the relationships between Cu and N in this field are still very limited, and, to the best of our knowledge, the influence of Cu toxicity on the inducibility of HATS genes has not been investigated yet. For these reasons, the main aim of our research was to investigate the ability of a crop model plant, i.e., Cucumis sativus L., to respond to root application of if exposed to increasing Cu concentrations. The kinetics of substrate uptake and the transcriptional modulation of the molecular entities involved in the process have been assessed.
Material and methods
Plant material and growing conditions
Cucumber (C. sativus L. cv. Chinese Long) seeds were germinated for 5 days in darkness at 22°C on filter paper moistened with 0.5 mM CaSO4 and placed vertically (Feil et al., 2020). Five-day-old seedlings were transferred into 2 L plastic pots filled with 1.5 L of a full-strength nutrient solution (NS) containing 2 mM Ca(NO3)2, 0.5 mM MgSO4, 0.7 mM K2SO4, 0.1 mM KCl, 10 µM H3BO3, 0.5 µM MnSO4, 0.2 µM CuSO4, 0.5 µM ZnSO4, 0.01 µM (NH4)6Mo7O24, 80 µM Fe-EDTA, buffered at pH 6 with 0.1 mM MES KOH, continuously aerated, and renewed twice a week (Marastoni et al., 2019a).
Experiment 1: Effect of Cu toxicity on NO3− LATS and HATS
To investigate the effect of Cu toxicity on both the nitrate low affinity transport system (LATS) and the high affinity transport system (HATS), cucumber seedlings after germination were grown for 7 days in full-strength NS (see above). At the end of this cultivation period, plants were split into four groups and subjected to increasing concentrations of Cu (i.e., 0.2, 5, 25, and 50 µM) for 7 days. At the end of the growing period, in the presence of different Cu concentrations, the uptake rate was measured. Plants were removed from the NS, the root systems were washed in 0.1 mM CaSO4 solution for 5 min and then exposed for 7 min to an aerated uptake solution containing 1 mM MES-BTP, pH 6.0, and either 1 mM or 200 µM 15 to test the low affinity or high affinity range, respectively. Three independent growth experiments were performed (three biological replicates). Each sample of each biological replicate consisted of roots pooled from two plants. The experiments were run in a climate chamber under controlled conditions (14/10 h light/dark, 24/19°C, 250 µmol m−2 s−1 light intensity, and 70% relative humidity).
Experiment 2: Effect of Cu toxicity on the induction of NO3− HATS
For the HATS induction experiment, after germination, cucumber seedlings were transferred and grown for 7 days in full-strength NS (see above). At the end of this cultivation period, plants were then split into five groups and subjected to different Cu concentrations (i.e., 0.2, 5, 20, 30, and 50 µM) for 7 days; the last 4 days of this second cultivation period, cucumber plants were also deprived of N (Supplementary Figure 1). The induction of HATS was carried out by treating N-starved plants with 250 µM Ca(NO3)2 (induced plants), whilst control plants were treated with 250 µM CaSO4 (not induced plants) (Pii et al., 2019), to balance the Ca2+ supplementation. To determine uptake rate, plants were removed from the NS, the root systems were washed in 0.1 mM CaSO4 solution for 5 min and then exposed for 7 min to an aerated uptake solution containing 200 µM 15 in 1 mM MES-BTP, pH 6.0 (see below). Nitrate uptake rate was measured 0, 4, 8, 12, and 24 h after the induction treatments (HAT). At each time point, plant roots were sampled and either dried to a constant weight for isotopic analyses or frozen in liquid N2 for gene expression analyses. Three independent growth experiments were performed (three biological replicates). Each sample of each biological replicate consisted of roots pooled from two plants. The experiments were run in a climate chamber under controlled conditions (14/10 h light/dark, 24/19°C, 250 µmol m−2 s−1 light intensity, and 70% relative humidity).
Determination of nitrate uptake rate
Nitrate uptake assays were carried out using 15N-labeled calcium nitrate [Ca(15NO3)2, 60 atom% 15N, Sigma-Aldrich], as previously described (Pii et al., 2016; Pii et al., 2019). Briefly, the roots of each seedling were washed in 0.1 mM CaSO4 for 5 min and then exposed for 7 min to an aerated uptake solution composed of 100 μM Ca(15NO3)2 in 1 mM MES-BTP, pH 6.0. The roots were then rinsed for 2 min in 0.1 mM CaSO4 (Pii et al., 2016; Pii et al., 2019). Roots and shoots were then separated and dried at 70°C for 48 h until a constant weight was reached. The 15N content was determined using isotope ratio mass spectrometry analysis (Delta V isotope mass spectrometer, ThermoFisher Scientific), as previously described (Pii et al., 2019).
Bioinformatics
The identification of NRT2 and NRT3 genes in the cucumber genome (using the Cucumber (Gy14) v2 genome, http://cucurbitgenomics.org/organism/16) was based on amino acid sequence similarity with NRT2 and NRT3 proteins of other organisms, such as Vitis vinifera, A. thaliana, Populus trichocarpa, Glycine max, Solanum lycopersicum, Oryza sativa, Sorghum bicolor, and Zea mays (Pii et al., 2014). Similarly, putative PM H+-ATPase genes were identified based on amino acid sequence similarity with PM H+-ATPase of Nicotiana plumbaginifolia Viv., O. sativa L., A. thaliana (L.) Heynh. (Arango et al., 2003), V. vinifera (Pii et al., 2014), and Z. mays (Pii et al., 2016). The amino acid sequences were obtained from public databases (http://www.ncbi.nlm.nih.gov/, http://www.uniprot.org/uniprot/). The predicted sequences for NRT2, NRT3, and PM H+-ATPase in cucumber were identified through a BLASTP search (Altschul et al., 1997). By using the selected putative proteins for NRT2, NRT3, and PM H+-ATPase, a phylogenetic analysis was performed; the amino acid sequences were aligned by the ClustalW algorithm (https://www.genome.jp/tools-bin/clustalw). Phylogenetic trees were built using the Phylogenetic Interference Package program (PHYLIP; University of Washington, http://evolution.genetics.washington.edu/phylip.html), and they were visualized through FigTree software version 1.4.4.
Real-time quantitative (RT-qPCR) analysis
Total RNA was extracted from the stored frozen roots using the Spectrum Plant Total RNA Kit (Sigma-Aldrich) according to the manufacturer’s instructions. Afterwards, 1 μg of total RNA was treated with 10 U of DNAse RQ1 and used for cDNA synthesis by the ImProm-II Reverse Transcription System (Promega) and oligo(dT)15 primer as per the manufacturer’s recommendations. The quantitative real-time PCR reaction (qRT-PCR) was performed using the SsoFast EvaGreen Supermix (Bio-Rad). Gene-specific primers were designed for the target genes (Supplementary Table 1). Experiments were carried out in triplicate with the following conditions: 5 min at 95°C, followed by 40 cycles at 95°C for 30 s and 55°C for 30 s. The amplification efficiency was calculated from raw data using LinRegPCR software (Ramakers et al., 2003). For each transcript, the mean normalized expression value was calculated using the housekeeping transcript, and the relative expression ratio values were calculated by 2−ΔΔCt method according to Livak and Schmittgen (2001).
Statistical analyses
Data are presented as means ± SE, whereby n is represented by three independent biological replicates; each biological replicate consists of roots pooled from two plants. Depending on the dataset, the significance of differences among means was calculated by either Student’s t-test, one-way ANOVA with post-hoc Tukey HSD, or two-way ANOVA, as specified in figure legends. The statistical analyses and data visualization were carried out using R software v.3.6.1 and the packages listed in Supplementary Information 1.
Results
Effect of Cu toxicity on nitrate uptake (Experiment 1)
The effects of Cu toxicity on both nitrate HATS and LATS transporters were investigated by exposing plants to increasing Cu concentrations (i.e., 0.2, 5, 25, and 50 µM). As shown in Figure 1, the heavy metal did not negatively affect the uptake rate in the low affinity range; when plants were exposed to 50 µM Cu, the activity of LATS did not show a significant difference compared to control plants (i.e., plants treated with 0.2 µM Cu). On the other hand, cucumber plants treated with 5 and 25 µM Cu displayed a uptake rate that was significantly higher as compared to control samples (Figure 1). As far as HATS is concerned, the highest Cu concentration (i.e., 50 µM) completely abolished the ability of cucumber plants to acquire (Figure 1). The other Cu levels tested did not significantly affect the uptake rate compared to control samples, although starting at 5 µM Cu, a decreasing trend was observed (Figure 1).
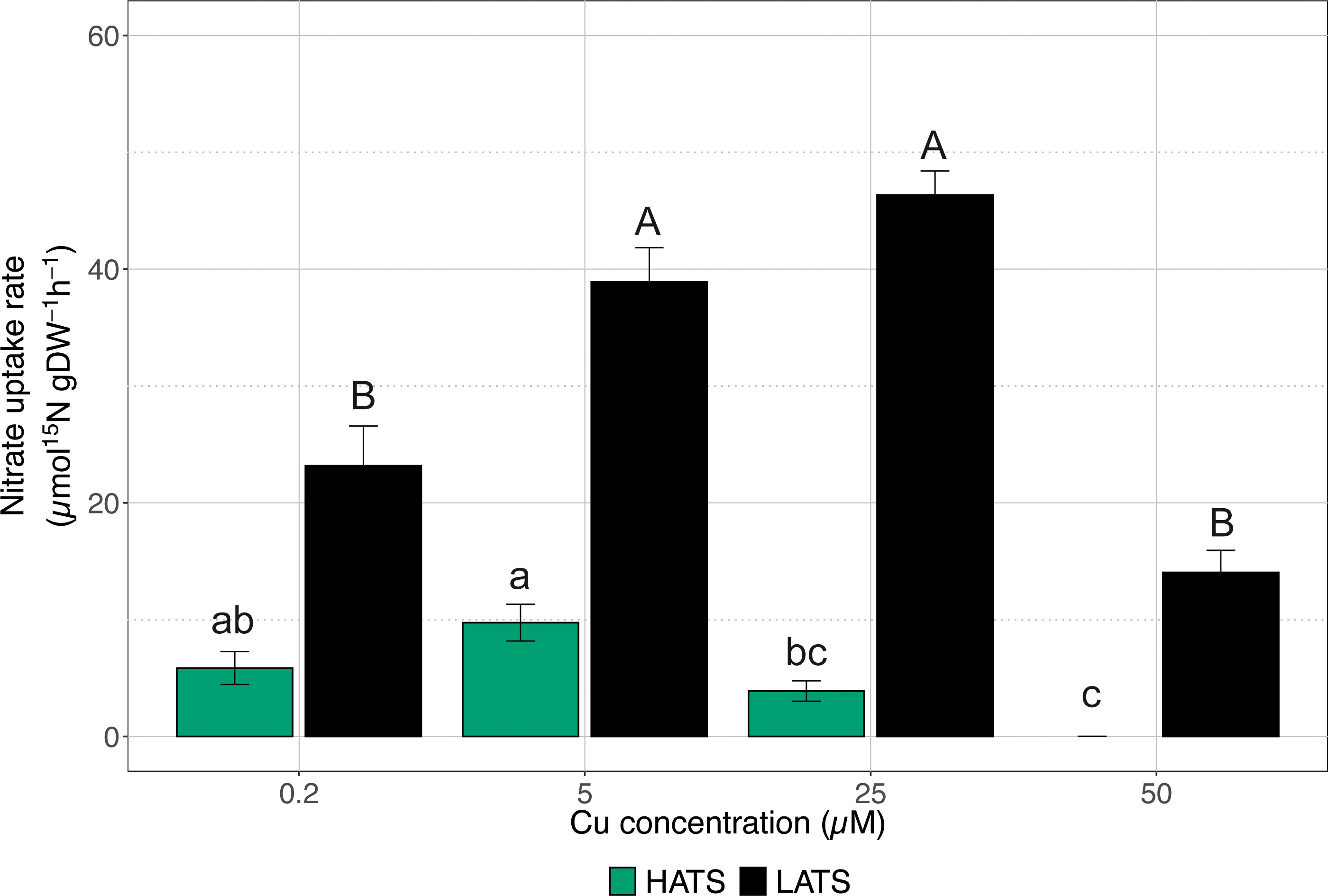
Figure 1 High-affinity (HATS) and low-affinity (LATS) uptake rates in the roots of cucumber plants. Cucumber plants have been exposed to increasing concentrations of Cu (0.2, 5, 25, 50 µM). Uptake rates were determined by placing the seedlings in either 200 μM or 1 mM 15 solution for 7 min in HATS and LATS, respectively. Data are the means ± SE of three independent biological replicates; each biological replicate was obtained by pooling two independent plants. Different letters indicate significantly different values as determined using a one-way ANOVA with Tukey post hoc tests (P<0.001).
Nitrate induction is prevented by high Cu availability (Experiment 2)
To deepen the understanding of Cu effects on the high-affinity uptake system, acquisition rates were measured in cucumber seedlings, either not-induced or induced with 500 µM , at different times (0, 4, 8, 12, and 24 HAT) that were previously exposed to increasing Cu concentrations (i.e., 0.2, 5, 20, 30, and 50 µM) (Supplementary Figure 1). In control plants, the uptake rate showed the expected behavior following treatment with . Induced plants displayed a significant increase in the nutrient uptake velocity already at 4 HAT and reached its maximum at 8 HAT, as compared to not induced seedlings. Afterwards, the uptake rate declined, and at 12 and 24 HAT, it was comparable to the values obtained in not-induced plants (Figure 2). In plants exposed to 5 and 20 µM Cu, the induction with caused an increase in the nutrient uptake rate, which reaches its highest value at 4 HAT and declines soon after to rate values comparable to those of not-induced plants (Figure 2). Interestingly, the maximum levels of uptake rates obtained in induced plants exposed to 5 and 20 µM Cu are comparable to those observed in control samples. On the other hand, when the Cu concentration was increased to 30 µM, the enhancement in uptake rate following induction was completely abolished. Nevertheless, the constitutive uptake activity of not-induced plants was maintained to the same extent with respect to induced plants exposed to either 0.2, 5, or 20 µM Cu (Figure 2). Consistently with the results obtained in the previous experiment (Figure 1), 50 µM Cu in the nutrient solution not only prevented the upregulation of uptake rate following induction, but it also abolished the ability of cucumber plants to take up from the growth substrate (Figure 2).
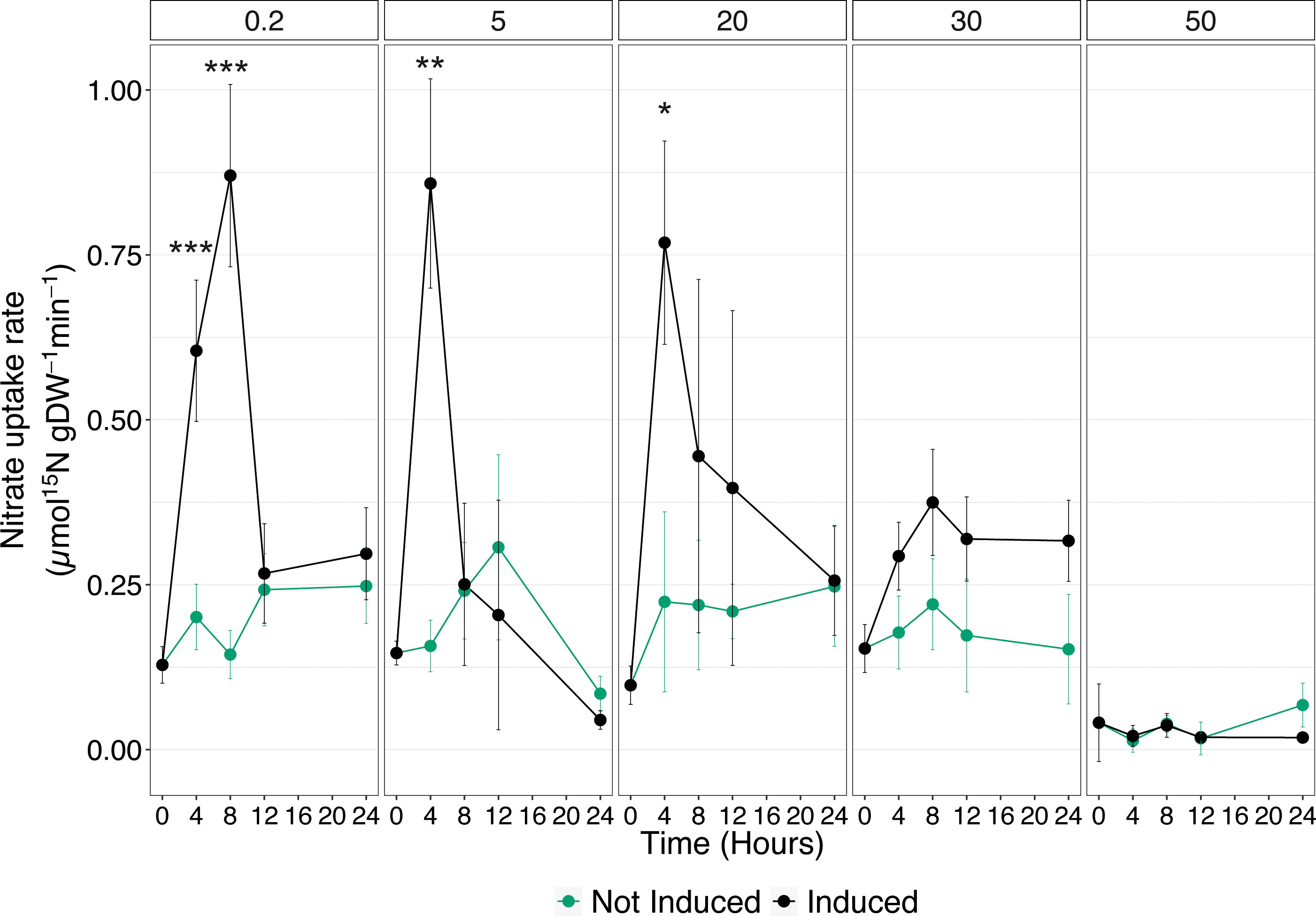
Figure 2 High-affinity uptake rates in the roots of cucumber plants. Plants assessed have been either induced or not induced, and exposed to increasing concentrations of Cu (0.2, 5, 20, 30, and 50 µM). Uptake rates were determined by placing the seedlings in 200 μM 15 solution for 7 min. Data are the means ± SE of three independent biological replicates; each biological replicate was obtained by pooling two independent plants. The statistical significance at each time point was determined using Student’s t-test: *, P<0.05; **, P<0.01; ***, P<0.001.
Bioinformatics
With the aim of gaining a deeper insight into the influence of increasing concentrations of Cu on the molecular mechanisms involved in uptake in cucumber plants, the transcriptional modulation of the molecular entities involved in the response to induction, namely NRT2, NRT3, and PM H+-ATPase genes, has been investigated.
Genes encoding for putative NRT2 and NRT3 proteins in the C. sativus genome were identified on the basis of protein sequence homology with members of the NRT2 family of V. vinifera, A. thaliana, P. trichocarpa, G. max, S. lycopersicum, O. sativa, S. bicolor, and Z. mays (Pii et al., 2014). This approach allowed the identification of two proteins (Cucsa.268720.1 and Cucsa.286270.1) encoding for putative high affinity transporters; the phylogenetic analysis showed that both clustered with previously characterized high affinity transporters belonging to other dicot species (Supplementary Figure 2). In particular, Cucsa.268720.1, hereafter referred to as CsNRT2.1, clustered in a branch of the phylogenetic tree formed by different transporters isolated from P. trichocarpa, V. vinifera, and G. max genomes, yet displaying a higher degree of homology with GmNRT2.1, whereas Cucsa.286270.1, hereafter referred to as CsNRT2.5, showed the highest homology with NRT2.5 transporters of both dicots and monocots (Supplementary Figure 2). The same approach allowed the isolation of a single protein, Cucsa.098830.1, encoding a putative NRT3 orthologous gene in the cucumber genome. The phylogenetic analysis showed that Cucsa.098830.1 clustered together with the other proteins belonging to dicots and featuring a higher degree of homology with LjNAR2.1 (Supplementary Figure 3).
The identification of putative PM H+-ATPase has been achieved as previously described (Pii et al., 2016). The Blastp analysis carried out on C. sativus genome allowed the retrieval of 10 sequences putatively encoding PM H+-ATPase, whereas the phylogenetic analysis showed that three sequences (Cucsa,158480.1, Cucsa.197480.1, and Cucsa.097990.1) belong to subfamily I, four sequences (Cucsa.311000.1, Cucsa.161790.1, Cucsa.081200.1, and Cucsa.089200.1) to subfamily II and three sequences (Cucsa.132660.1, Cucsa.350780.1, and Cucsa.096210.1) to subfamily IV (Supplementary Figure 4).
Gene expression analyses
The quantification of gene expression has been carried out at key time points (i.e., 0, 8, and 24 HAT) during the induction experiment, in accordance with the results obtained in the assessment of uptake rate. Considering the high-affinity transporters, the gene expression analysis confirmed that CsNRT2.1 in control plants (i.e., 0.2 µM Cu) was modulated as expected (Figure 3A). In fact, CsNRT2.1 did not show any alteration in the mRNA abundance in not induced plants throughout the time-course experiment, whilst in induced plants it presented a significant induction at 8 HAT and a reduction in the expression levels at 24 HAT, indeed mirroring uptake rate data (Figure 3A). Similar responses have also been observed in plants exposed to 20 and 30 µM Cu (Figure 3A). On the other hand, in plants treated with 5 µM Cu, induction significantly upregulated CsNRT2.1 expression at 8 HAT, and it was maintained at the same levels until 24 HAT (Figure 3A). When plants were exposed to 50 µM Cu, CsNRT2.1 was downregulated at 8 HAT and upregulated at 24 HAT with respect to not-induced cucumber plants (Figure 3A). Interestingly, the gene CsNRT3.1 encoding the accessory protein displayed very similar transcriptional regulation as compared to CsNRT2.1 (Figure 3B), confirming the coordinated activity of these two molecular entities in the induction of uptake in plants (Pii et al., 2016).
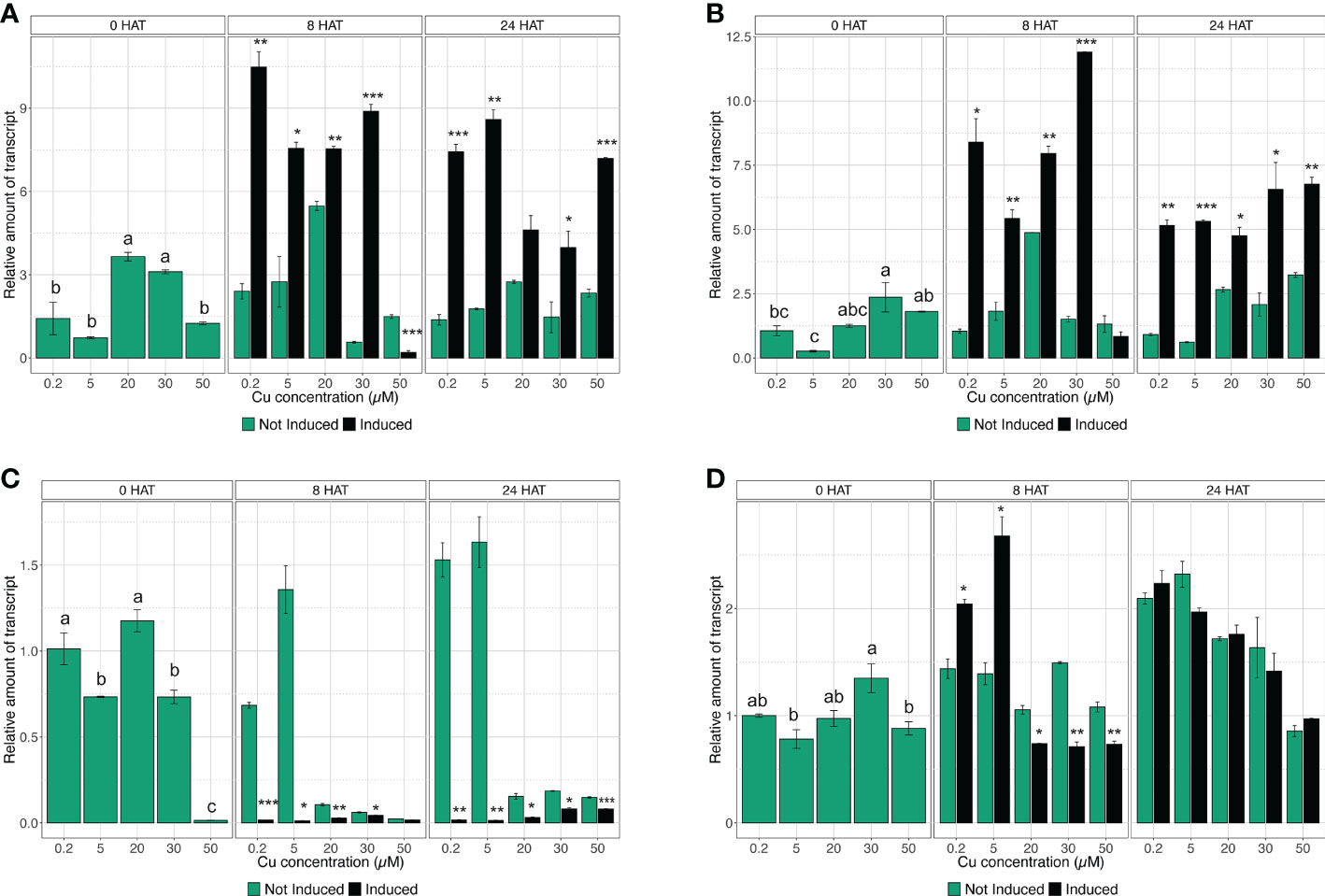
Figure 3 Time-course expression analysis of CsNRT2.1, CsNRT3.1, CsNRT2.3, and CsHA2. The expression levels of CsNRT2.1 (A), CsNRT3.1 (B), CsNRT2.5 (C), and CsHA2 (D) were assessed by qRT-PCR in the roots of cucumber plants, either not-induced or induced with and subjected to increasing concentration of Cu (0.2, 5, 20, 30, and 50 µM), at 0, 8, and 24 HAT. The data are normalized to the translation elongation factor isoform 1-alpha. The relative expression ratios were calculated using not induced control roots sampled before the treatments (not induced, 0.2 µM Cu at 0 HAT). Data are means (±SE), n = 3. Different letters within a time-point indicate significantly different values as determined using one-way ANOVA with Tukey post hoc tests (P<0.001). The statistical significance between not-induced and induced samples was tested by Student’s t-test: *, P<0.05; **, P<0.01; ***, P<0.001).
The qRT-PCR analysis carried out on CsNRT2.3 showed that, in control plants (i.e., 0.2 µM Cu) and in plants treated with 5 µM Cu, the gene did not respond to induction with , yet showed a downregulation at both 8 and 24 HAT in comparison to not-induced plants (Figure 3C). As far as 20 and 30 µM Cu-treated plants are concerned, CsNRT2.3 was downregulated in not-induced and induced plants at both 8 and 24 HAT with respect to plants at 0 HAT (Figure 3C). At a concentration of 50 µM Cu, CsNRT2.3 always resulted in downregulation to control plants at 0 HAT (Figure 3C).
After a preliminary analysis, only four of the 10 genes putatively encoding PM H+-ATPase, namely Cucsa.089200.1 (already known as CsHA2), Cucsa.081200.1 (already known as CsHA3), Cucsa.161790.1, and Cucsa.311000.1, were shown to be expressed in the root tissue of cucumber plants. Nonetheless, the qRT-PCR analyses highlighted that only CsHA2 was modulated during uptake induction (Figure 3D and data not shown). The assessment of gene expression levels allowed demonstrating that, before induction (i.e., 0 HAT), the different Cu concentrations applied to cucumber plants did not affect the modulation of CsHA2, as it was not altered as compared to control plants (Figure 3D). The treatment with caused in both 0.2 and 5 µM Cu-supplemented plants a significant upregulation in CsHA2 at 8 HAT, which is consistent with the induction observed in both CsNRT2.1, CsNRT3.1, and the uptake rate (Figures 2, 3). On the contrary, at higher Cu concentrations, namely 20, 30, and 50 µM, the expression levels of CsHA2 in induced plants were lower than those of not induced plants at 8 HAT and not significantly modulated at 24 HAT (Figure 3D).
Discussion
In the last few years, the relationship between soil contamination with heavy metals and the responses of agricultural crops exposed to such stressing conditions has been investigated from different points of view. The accumulation of Cu in soils often represents a consequence of anthropogenic activities, among which is the intensive use of cupric fungicides for the defense of fruit crops (Rooney et al., 2006; Brunetto et al., 2016; Hippler et al., 2016; Cesco et al., 2021). It is estimated that the average Cu concentration of the Earth’s crust ranges between 6 and 80 mg kg−1 (Brunetto et al., 2016); nevertheless, the distribution of Cu between the solid phase and the soil solution is dependent on the physical and chemical properties of soils, which regulate precipitation/dissolution, adsorption/desorption, and redox reactions (Brunetto et al., 2016). On these bases, the concentration of Cu in the soil solution of arable soils lies between 15 nM and 5 µM (Cesco et al., 2021). However, several surveys indicate that Cu concentrations in the superficial soil horizon can often reach 200 mg kg−1, depending on anthropogenic activities, and sometimes exceed 1,000–3,000 mg kg−1 in contaminated soil (Flores-Vélez et al., 1996; Mirlean et al., 2007), suggesting that the bioavailable Cu concentrations may be significantly higher than 5 µM. Despite being an essential plant micronutrient (Yruela, 2009), relatively high concentrations of Cu in the growth substrate can induce toxicity phenomena in plants, which are generally shown through reduced vegetative growth, the alteration of biochemical processes (e.g., photosynthesis, lipid peroxidation), and an imbalanced acquisition of mineral nutrients (Baldi et al., 2018; Marastoni et al., 2019a; Marastoni et al., 2019b; Feil et al., 2020). Previous pieces of evidence have highlighted that different plant species grown on substrates featuring medium-high (e.g., 0.2 to 20 µM) Cu concentrations were impaired in their ability to acquire N, either or , and to assimilate it (Xiong et al., 2006; Zhang et al., 2014; Hippler et al., 2018; Gong et al., 2019; Huo et al., 2020). In particular, Cu toxicity was shown, over the long term, to affect the transcriptional modulation of genes encoding both high affinity and low affinity transporters and to hinder the activity of nitrated reductase (NR) and nitrite reductase (NiR), responsible for the first steps of nitrate assimilation (Xiong et al., 2006; Zhang et al., 2014; Hippler et al., 2018; Gong et al., 2019; Huo et al., 2020). In any case, it should be highlighted that a certain degree of variation in the responses has been observed in the different plant species investigated.
In cucumber plants, exposure over a long period to increasing Cu concentrations (at least up to 25 µM) apparently has a promoting effect on the activity of the LATS transport system (Figure 1), even though the highest concentration tested (i.e., 50 µM) had a detrimental effect on the nitrate uptake rate (Figure 1). These observations are in agreement with the findings of Hippler et al. (2018), according to which the exposure of A. thaliana plants to medium-high concentrations of Cu could induce significant downregulation of genes belonging to the NRT1 family. On the other hand, the same authors showed that Cu toxicity could increase the expression of genes belonging to the NRT2 family, although uptake was prevented over the period of time considered (Hippler et al., 2018). Indeed, these observations support our results, according to which uptake in the high affinity range was depressed by Cu concentrations higher than 25 µM (Figure 1). However, in natural conditions, the availability of in the growth substrate is subject to fluctuations, so plants had to adapt by evolving the induction phenomenon to maximize their ability to acquire the nutrient when it was available (Jackson et al., 1973). However, regardless of the importance of this aspect for plant nutrition, to the best of our knowledge, investigations about the influence of Cu toxicity on the inducibility of the HATS transport system in crop plants were lacking.
As widely assessed in previous studies (Kronzucker et al., 1995; Pii et al., 2014; Pii et al., 2016; Pii et al., 2019), the induction phenomenon caused a transient increase in the uptake rate in roots of control plants (i.e., 0.2 µM Cu), reaching the highest values at 8 HAT and showing afterwards a decrease back towards values close to those of not induced plants (Figure 2). The observed behavior in the uptake dynamics is determined by the transcriptional modulation of high-affinity transporter genes (NRT2 gene family), the accessory protein (NRT3), and plasma membrane (PM) H+-ATPases (McClure et al., 1990a; McClure et al., 1990b; Glass et al., 1992; Santi et al., 1995; Pii et al., 2014; Pii et al., 2016; Pii et al., 2019). As expected, the qRT-PCR analyses confirmed that in control plants the increase in the uptake rate was sustained by an upregulation at 8 HAT of the genes involved in the process, particularly CsNRT2.1, CsNRT3.1 and CsHA2, encoding for a nitrate transporter, the accessory protein, and a PM H+-ATPase, respectively, while a negative feedback regulation was detected at 24 HAT (Figure 3). On the other hand, qRT-PCR studies also highlighted that CsNRT2.3 was not involved in the response to induction, at least in our experimental conditions (Figure 3).
With increasing Cu concentrations (i.e., 5 and 20 µM) in the growth medium, the peak of uptake rate due to induction was anticipated at 4 HAT, followed by a steep downregulation of the phenomenon (Figure 2). Nevertheless, the detected values of uptake rate were comparable to those obtained in induced plants exposed to physiological levels of Cu. On the contrary, supplementation with higher concentrations of Cu (i.e., 30 and 50 µM) completely abolished the plant response to induction, as if they were insensitive to the presence of in the external medium (Figure 2). Interestingly, a similar effect has also been observed by Rizzardo and co-workers (2012), who demonstrated an impairment in induction in maize plants treated with 10 µM Cadmium (Cd). However, the impairment in induction observed in 30 and 50 µM Cu-treated plants was not mirrored by the transcriptional modulation of genes. In fact, in plants grown at 30 µM Cu, CsNRT2.1 and CsNRT3.1 were strongly upregulated in induced plants with respect to not-induced ones at both 8 and 24 HAT (Figure 3), thus suggesting that the ability of plants to sense in the external medium was not compromised in these conditions. On the other hand, when plants were exposed to the highest Cu concentration (i.e., 50 µM), CsNRT2.1 and CsNRT3.1 showed a delayed upregulation in induced plants, being in fact significantly more expressed only at 24 HAT (Figure 3); however, such transcriptional regulation did not correspond to a significant increase in the uptake rate in induced plants (Figure 2).
The plasma membrane (PM) of epidermal root cells represents the first cellular structure that is the target for heavy metal toxicity in soil, which is well known to cause membrane lipid damage and alter the ionic homeostasis capacity of cells (Fodor et al., 1995). Considering that the activity of PM H+-ATPases is at the base of the electrochemical gradient across the membrane energizing the nutrient acquisition from the soil solution, it is clear that the regulation and functionality of these enzymes might be particularly crucial for plants exposed to heavy metal toxicity (Pedersen et al., 2012). In our experimental model, when plants were treated with 5 µM Cu, CsHA2 was upregulated at 8 HAT in response to induction, as in induced samples not treated with excessive amounts of Cu (Figure 3). Notwithstanding, CsHA2 induction levels in 5 µM Cu-treated plants were higher compared to reference samples (i.e., induced plants treated with 0.2 µM Cu), as though the slightly higher heavy metal concentration in the growth medium might require a further contribution of PM H+-ATPase to maintain the root cell ion homeostasis. Indeed, previous studies have demonstrated that cucumber roots exposed to 10 µM Cu or Cd for a short period of time (i.e., 6 days) showed a higher expression of different CsHA isoforms and an increased proton pumping activity, further corroborating the role of these enzymes in the response to enhanced concentrations of heavy metals in the growth medium (Janicka-Russak et al., 2008; Janicka-Russak et al., 2012). The exposure to higher concentrations of Cu (20, 30, and 50 µM) compromised the responsiveness of CsHA2 (Figure 3). At 8 HAT, the gene was in fact downregulated in treated plants as compared to not-induced ones, while the expression levels were not significantly modulated in plants analyzed at 24 HAT (Figure 3). It is worth noting that loss in the responsiveness of PM H+-ATPase transcriptional regulation, which is normally observed during the induction (Santi et al., 2003; Nikolic et al., 2012; Pii et al., 2016), is coherent with the impaired uptake in plants treated with higher Cu concentrations (Figure 2). However, information concerning the transcriptional regulation of CsHA following the exposure to toxic concentration of Cu is very little. Janicka-Russak et al. (2008), by applying a semi-quantitative RT-PCR approach, showed that CsHA3 transcript abundance was not significantly affected by exposing plants to 100 µM Cu for 2 h. Later, similar results were obtained via quantitative RT-PCR analyses carried out on different isoforms of CsHA isolated from cucumber plants exposed to 10 µM Cu for 6 days, also demonstrating no influence of Cu toxicity on PM H+-ATPase enzyme abundance (Janicka-Russak et al., 2012). Despite this, it has been shown that the transcriptional regulation of PM H+-ATPase can be negatively affected in maize plants treated with 10 µM Cd, further suggesting a possible interference of heavy metals in these mechanisms and also considering the demonstrated impact on HATS induction (Rizzardo et al., 2012). On the other hand, an inhibitory effect due to short-term exposure to Cu on PM H+-ATPase functionality was demonstrated in the roots of different plant species (Kennedy and Gonsalves, 1989; Fodor et al., 1995). More recently, Astolfi and co-workers (2003; 2005) reported inhibition of activity of PM H+-ATPase in oat and maize roots after long-term treatment (7 and 21 days) with Cd.
Conclusions
This research study was aimed at understanding the effects of excessive Cu availability in the rhizosphere of C. sativus L. plants on the biochemical and molecular mechanisms devoted to the acquisition of , particularly focusing on the induction of the high affinity transport system (HATS). When the concentration in the substrate was higher than 1 mM, Cu did not show any negative influence on nutrient uptake but rather promoted it at lower Cu concentrations. On the contrary, the inducibility of HATS was significantly affected by increasing Cu concentrations, which indeed showed a detrimental impact by preventing cucumber plants from the uptake of . Especially in the cases of Cu concentrations higher than 20 µM, cucumber plants demonstrated either a strongly reduced or abolished uptake activity, albeit the transcriptional modulation of both the nitrate transporter CsNRT2.1 and the accessory protein CsNRT3.1 was coherent with the expected induction. Nevertheless, in these conditions, genes encoding PM H+-ATPase (i.e., CsHA2) were downregulated instead of induced, thus suggesting that a possible impairment in the generation of the proton gradient across the root PM could be the cause of the abolishment of uptake.
These findings are particularly relevant considering that, as mentioned above, the bioavailable Cu concentration in contaminated soils may exceed 5 µM level that has been pointed out as a threshold for toxicity, above which also a severe impairment in the uptake of N and P (Feil et al., 2020) has been demonstrated. Despite the need for additional investigation to better understand the molecular bases underpinning the interaction between Cu and the transcriptional regulation of PM H+-ATPase, the data presented further highlight that increasing concentrations of this heavy metal in agricultural soils can represent an issue to be addressed in the near future, not only for its documented toxic effects on crops but also for its ability to inhibit or impair the uptake of macro and micronutrients by other plants, consequently impacting on the efficiency and sustainability of the agricultural practice of crop fertilization with nitrate.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
Experimental Design: SC and YP. Experiments execution and data collection: SF and MA. Data analyses and visualization: MA and YP. Data interpretation: MA, SC, and YP. Manuscript writing and critical revision: SF, MA, SC, and YP. Financial support: YP. All authors contributed to the article and approved the submitted version.
Acknowledgments
We acknowledge the Open Access Publishing Fund of the Free University of Bozen-Bolzano.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1034425/full#supplementary-material
References
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Arango, M., Gévaudant, F., Oufattole, M., Boutry, M. (2003). The plasma membrane proton pump ATPase: the significance of gene subfamilies. Planta 216, 355–365. doi: 10.1007/s00425-002-0856-8
Astolfi, S., Zuchi, S., Chiani, A., Passera, C. (2003). In vivo and in vitro effects of cadmium on H+ATPase activity of plasma membrane vesicles from oat (Avena sativa l.) roots. J. Plant Physiol. 160, 387–393. doi: 10.1078/0176-1617-00832
Astolfi, S., Zuchi, S., Passera, C. (2005). Effect of cadmium on H+ATPase activity of plasma membrane vesicles isolated from roots of different s-supplied maize (Zea mays l.) plants. Plant Sci. 169, 361–368. doi: 10.1016/j.plantsci.2005.03.025
Axelsen, K. B., Palmgren, M. G. (1998). Evolution of substrate specificities in the p-type ATPase superfamily. J. Mol. Evol. 46, 84–101. doi: 10.1007/PL00006286
Baldi, E., Miotto, A., Ceretta, C. A., Quartieri, M., Sorrenti, G., Brunetto, G., et al. (2018). Soil-applied phosphorous is an effective tool to mitigate the toxicity of copper excess on grapevine grown in rhizobox. Sci. Hortic. (Amsterdam). 227, 102–111. doi: 10.1016/j.scienta.2017.09.010
Brunetto, G., Bastos de Melo, G. W., Terzano, R., Del Buono, D., Astolfi, S., Tomasi, N., et al. (2016). Copper accumulation in vineyard soils: Rhizosphere processes and agronomic practices to limit its toxicity. Chemosphere 162, 293–307. doi: 10.1016/j.chemosphere.2016.07.104
Cesco, S., Pii, Y., Borruso, L., Orzes, G., Lugli, P., Mazzetto, F., et al. (2021). A smart and sustainable future for viticulture is rooted in soil: How to face cu toxicity. Appl. Sci. 11, 1–21. doi: 10.3390/app11030907
Crawford, N. M., Glass, A. D. M. (1998). Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 3, 389–395. doi: 10.1016/S1360-1385(98)01311-9
Feigl, G., Kumar, D., Lehotai, N., Tugyi, N., Molnár, Á., Ördög, A., et al. (2013). Physiological and morphological responses of the root system of Indian mustard (Brassica juncea l. czern.) and rapeseed (Brassica napus l.) to copper stress. Ecotoxicol. Environ. Saf. 94, 179–189. doi: 10.1016/j.ecoenv.2013.04.029
Feil, S. B., Pii, Y., Valentinuzzi, F., Tiziani, R., Mimmo, T., Cesco, S. (2020). Copper toxicity affects phosphorus uptake mechanisms at molecular and physiological levels in Cucumis sativus plants. Plant Physiol. Biochem. 157, 138–147. doi: 10.1016/j.plaphy.2020.10.023
Filleur, S., Dorbe, M.-F., Cerezo, M., Orsel, M., Granier, F., Gojon, A., et al. (2001). An Arabidopsis T-DNA mutant affected in Nrt2 genes is impaired in nitrate uptake. FEBS Lett. 489, 220–224. doi: 10.1016/S0014-5793(01)02096-8
Flores-Vélez, L. M., Ducaroir, J., Jaunet, A. M., Robert, M. (1996). Study of the distribution of copper in an acid sandy vineyard soil by three different methods. Eur. J. Soil Sci. 47, 523–532. doi: 10.1111/j.1365-2389.1996.tb01852.x
Fodor, E., Szabó-Nagy, A., Erdei, L. (1995). The effects of cadmium on the fluidity and h+-ATPase activity of plasma membrane from sunflower and wheat roots. J. Plant Physiol. 147, 87–92. doi: 10.1016/S0176-1617(11)81418-5
Forde, B. G., Clarkson, D. T. (1999). “Nitrate and ammonium nutrition of plants: physiological and molecular perspectives,” in Advances in Botanical Research. Ed. Callow, J. A. (Academic Press), 1–90. doi: 10.1016/S0065-2296(08)60226-8
Glass, A. D. M., Brito, D. T., Kaiser, B. N., Kronzucker, H. J., Kumar, A., Okamoto, M., et al. (2001). Nitrogen transport in plants, with an emphasis on the regulation of fluxes to match plant demand. J. Plant Nutr. Soil Sci. 164, 199–207. doi: 10.1002/1522-2624(200104)164:2<199::AID-JPLN199>3.0.CO;2-K
Glass, A. D. M., Shaff, J. E., Kochian, L. V. (1992). Studies of the uptake of nitrate in barley: IV. electrophysiology. Plant Physiol. 99, 456–463. doi: 10.1104/pp.99.2.456
Gong, Q., Wang, L., Dai, T., Zhou, J., Kang, Q., Chen, H., et al. (2019). Effects of copper on the growth, antioxidant enzymes and photosynthesis of spinach seedlings. Ecotoxicol. Environ. Saf. 171, 771–780. doi: 10.1016/j.ecoenv.2019.01.016
Hippler, F. W. R., Cipriano, D. O., Boaretto, R. M., Quaggio, J. A., Gaziola, S. A., Azevedo, R. A., et al. (2016). Citrus rootstocks regulate the nutritional status and antioxidant system of trees under copper stress. Environ. Exp. Bot. 130, 42–52. doi: 10.1016/J.ENVEXPBOT.2016.05.007
Hippler, F. W. R., Mattos-Jr, D., Boaretto, R. M., Williams, L. E. (2018). Copper excess reduces nitrate uptake by arabidopsis roots with specific effects on gene expression. J. Plant Physiol. 228, 158–165. doi: 10.1016/j.jplph.2018.06.005
Huo, K., Shangguan, X., Xia, Y., Shen, Z., Chen, C. (2020). Excess copper inhibits the growth of rice seedlings by decreasing uptake of nitrate. Ecotoxicol. Environ. Saf. 190, 110105. doi: 10.1016/J.ECOENV.2019.110105
Jackson, W. A., Flesher, D., Hageman, R. H. (1973). Nitrate uptake by dark-grown corn seedlings: Some characteristics of apparent induction. Plant Physiol. 51, 120–127. doi: 10.1104/pp.51.1.120
Janicka-Russak, M., Kabała, K., Burzyński, M. (2012). Different effect of cadmium and copper on h+-ATPase activity in plasma membrane vesicles from Cucumis sativus roots. J. Exp. Bot. 63, 4133–4142. doi: 10.1093/jxb/ers097
Janicka-Russak, M., Kabała, K., Burzyński, M., Kłobus, G. (2008). Response of plasma membrane h+-ATPase to heavy metal stress in Cucumis sativus roots. J. Exp. Bot. 59, 3721–3728. doi: 10.1093/jxb/ern219
Kennedy, C. D., Gonsalves, F. A. N. (1989). The action of divalent zn, cd, Hg, Cu and Pb ions on the ATPase activity of a plasma membrane fraction isolated from roots of Zea mays. Plant Soil 117, 167–175. doi: 10.1007/BF02220709
Kronzucker, H. J., Siddiqi, M. Y., Glass, A. D. M. (1995). Kinetics of influx in spruce. Plant Physiol. 109, 319–326. doi: 10.1104/pp.109.1.319
Lequeux, H., Hermans, C., Lutts, S., Verbruggen, N. (2010). Response to copper excess in Arabidopsis thaliana: Impact on the root system architecture, hormone distribution, lignin accumulation and mineral profile. Plant Physiol. Biochem. 48, 673–682. doi: 10.1016/j.plaphy.2010.05.005
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Marastoni, L., Sandri, M., Pii, Y., Valentinuzzi, F., Brunetto, G., Cesco, S., et al. (2019a). Synergism and antagonisms between nutrients induced by copper toxicity in grapevine rootstocks: Monocropping vs. intercropping. Chemosphere 214, 563–578. doi: 10.1016/j.chemosphere.2018.09.127
Marastoni, L., Sandri, M., Pii, Y., Valentinuzzi, F., Cesco, S., Mimmo, T. (2019b). Morphological root responses and molecular regulation of cation transporters are differently affected by copper toxicity and cropping system depending on the grapevine rootstock genotype. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00946
McClure, P. R., Kochian, L. V., Spanswick, R. M., Shaff, J. E. (1990a). Evidence for cotransport of nitrate and protons in maize roots: I. effects of nitrate on the membrane potential. Plant Physiol. 93, 281–289. doi: 10.1104/pp.93.1.281
McClure, P. R., Kochian, L. V., Spanswick, R. M., Shaff, J. E. (1990b). Evidence for cotransport of nitrate and protons in maize roots: II. measurement of and h+ fluxes with ion-selective microelectrodes. Plant Physiol. 93, 290–294. doi: 10.1104/pp.93.1.290
Meier, S., Alvear, M., Borie, F., Aguilera, P., Ginocchio, R., Cornejo, P. (2012). Influence of copper on root exudate patterns in some metallophytes and agricultural plants. Ecotoxicol. Environ. Saf. 75, 8–15. doi: 10.1016/j.ecoenv.2011.08.029
Min, X., Siddiqi, M. Y., Guy, R. D., Glass, A. D. M., Kronzucker, H. J. (1998). Induction of nitrate uptake and nitrate reductase activity in trembling aspen and lodgepole pine. Plant Cell Environ. 21, 1039–1046. doi: 10.1046/j.1365-3040.1998.00340.x
Mirlean, N., Roisenberg, A., Chies, J. O. (2007). Metal contamination of vineyard soils in wet subtropics (southern Brazil). Environ. Pollut. 149, 10–17. doi: 10.1016/j.envpol.2006.12.024
Nacry, P., Bouguyon, E., Gojon, A. (2013). Nitrogen acquisition by roots: physiological and developmental mechanisms ensuring plant adaptation to a fluctuating resource. Plant Soil 270, 1–29. doi: 10.1007/s11104-013-1645-9
Nikolic, M., Cesco, S., Monte, R., Tomasi, N., Gottardi, S., Zamboni, A., et al. (2012). Nitrate transport in cucumber leaves is an inducible process involving an increase in plasma membrane h+-ATPase activity and abundance. BMC Plant Biol. 12, 66. doi: 10.1186/1471-2229-12-66
Okamoto, M., Kumar, A., Li, W., Wang, Y., Siddiqi, M. Y., Crawford, N. M., et al. (2006). High-affinity nitrate transport in roots of Arabidopsis depends on expression of the NAR2-like gene AtNRT3.1. Plant Physiol. 140, 1036–1046. doi: 10.1104/pp.105.074385
Palmgren, M. G., Nissen, P. (2011). P-type ATPases. Annu. Rev. Biophys. 40, 243–266. doi: 10.1146/annurev.biophys.093008.131331
Pedersen, C. N. S., Axelsen, K. B., Harper, J. F., Palmgren, M. G. (2012). Evolution of plant p-type ATPases. Front. Plant Sci. 3. doi: 10.3389/fpls.2012.00031
Pii, Y., Aldrighetti, A., Valentinuzzi, F., Mimmo, T., Cesco, S. (2019). Azospirillum brasilense inoculation counteracts the induction of nitrate uptake in maize plants. J. Exp. Bot. 70, 1313–1324. doi: 10.1093/jxb/ery433
Pii, Y., Alessandrini, M., Dall’Osto, L., Guardini, K., Prinsi, B., Espen, L., et al. (2016). Time-resolved investigation of molecular components involved in the induction of high affinity transport system in maize roots. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01657
Pii, Y., Alessandrini, M., Guardini, K., Zamboni, A., Varanini, Z. (2014). Induction of high-affinity uptake in grapevine roots is an active process correlated to the expression of specific members of the NRT2 and plasma membrane h+-ATPase gene families. Funct. Plant Biol. 41, 353–365. doi: 10.1071/FP13227
Plett, D., Toubia, J., Garnett, T., Tester, M., Kaiser, B. N., Baumann, U. (2010). Dichotomy in the NRT gene families of dicots and grass species. PloS One 5, e15289. doi: 10.1371/journal.pone.0015289
Ramakers, C., Ruijter, J. M., Deprez, R. H. L., Moorman, A. F. M. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339, 62–66. doi: 10.1016/S0304-3940(02)01423-4
Rizzardo, C., Tomasi, N., Monte, R., Varanini, Z., Nocito, F. F., Cesco, S., et al. (2012). Cadmium inhibits the induction of high-affinity nitrate uptake in maize (Zea mays l.) roots. Planta 236, 1701–1712. doi: 10.1007/s00425-012-1729-4
Rooney, C. P., Zhao, F. J., McGrath, S. P. (2006). Soil factors controlling the expression of copper toxicity to plants in a wide range of European soils. Environ. Toxicol. Chem. 25, 726–732. doi: 10.1897/04-602R.1
Santi, S., Locci, G., Monte, R., Pinton, R., Varanini, Z. (2003). Induction of nitrate uptake in maize roots: Expression of a putative high-affinity nitrate transporter and plasma membrane h+-ATPase isoforms. J. Exp. Bot. 54, 1851–1864. doi: 10.1093/jxb/erg208
Santi, S., Locci, G., Pinton, R., Cesco, S., Varanini, Z. (1995). Plasma membrane h+-ATPase in maize roots induced for uptake. Plant Physiol. 109, 1277–1283. doi: 10.1104/pp.109.4.1277
Siddiqi, M. Y., Glass, A. D. M., Ruth, T. J., Fernando, M. (1989). Studies of the regulation of nitrate influx by barley seedlings using 13. Plant Physiol. 90, 806–813. doi: 10.1104/pp.90.3.806
Siddiqi, M. Y., Glass, A. D. M., Ruth, T. J., Rufty, T. W. (1990). Studies of the uptake of nitrate in barley: I. kinetics of 13 influx. Plant Physiol. 93, 1426–1432. doi: 10.1104/pp.93.4.1426
Tong, Y., Zhou, J.-J., Li, Z., Miller, A. J. (2005). A two-component high-affinity nitrate uptake system in barley. Plant J. 41, 442–450. doi: 10.1111/j.1365-313X.2004.02310.x
Touraine, B., Glass, A. (1997). and fluxes in the chl1-5 mutant of Arabidopsis thaliana. Plant Physiol. 114, 137–144. doi: 10.1104/pp.114.1.137
Vinit-Dunand, F., Epron, D., Alaoui-Sossé, B., Badot, P. M. (2002). Effects of copper on growth and on photosynthesis of mature and expanding leaves in cucumber plants. Plant Sci. 163, 53–58. doi: 10.1016/S0168-9452(02)00060-2
Xiong, Z. T., Liu, C., Geng, B. (2006). Phytotoxic effects of copper on nitrogen metabolism and plant growth in Brassica pekinensis rupr. Ecotoxicol. Environ. Saf. 64, 273–280. doi: 10.1016/j.ecoenv.2006.02.003
Yruela, I. (2009). Copper in plants: acquisition, transport and interactions. Funct. Plant Biol. 36, 409–430. doi: 10.1071/FP08288
Zanin, L., Zamboni, A., Monte, R., Tomasi, N., Varanini, Z., Cesco, S., et al. (2015). Transcriptomic analysis highlights reciprocal interactions of urea and nitrate for nitrogen acquisition by maize roots. Plant Cell Physiol. 56, 532–548. doi: 10.1093/pcp/pcu202
Zhang, L., Pan, Y., Lv, W., Xiong, Z. (2014). Physiological responses of biomass allocation, root architecture, and invertase activity to copper stress in young seedlings from two populations of Kummerowia stipulacea (maxim.) makino. Ecotoxicol. Environ. Saf. 104, 278–284. doi: 10.1016/j.ecoenv.2014.03.013
Keywords: copper, nitrate induction, cucumber, high affinity transport system, NRT genes, PM H+-ATPase, gene expression
Citation: Feil SB, Zuluaga MYA, Cesco S and Pii Y (2023) Copper toxicity compromises root acquisition of nitrate in the high affinity range. Front. Plant Sci. 13:1034425. doi: 10.3389/fpls.2022.1034425
Received: 01 September 2022; Accepted: 29 December 2022;
Published: 20 January 2023.
Edited by:
Corina Carranca, Instituto Nacional Investigaciao Agraria e Veterinaria (INIAV), PortugalReviewed by:
Manoj Shrivastava, Indian Agricultural Research Institute (ICAR), IndiaDavid Fernández Calviño, University of Vigo, Spain
Copyright © 2023 Feil, Zuluaga, Cesco and Pii. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Youry Pii, eW91cnkucGlpQHVuaWJ6Lml0
†These authors have contributed equally to this work
 Sebastian B. Feil†
Sebastian B. Feil† Monica Yorlady Alzate Zuluaga
Monica Yorlady Alzate Zuluaga Stefano Cesco
Stefano Cesco Youry Pii
Youry Pii