
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 17 November 2022
Sec. Plant Symbiotic Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1034230
This article is part of the Research Topic Metabolic Adjustments and Gene Expression Reprogramming for Symbiotic Nitrogen Fixation in Legume Nodules, Volume II View all 6 articles
 Ru-Jie Li1,2
Ru-Jie Li1,2 Chun-Xiao Zhang1,2
Chun-Xiao Zhang1,2 Sheng-Yao Fan1,2
Sheng-Yao Fan1,2 Yi-Han Wang1,2
Yi-Han Wang1,2 Jiangqi Wen3
Jiangqi Wen3 Kirankumar S. Mysore3
Kirankumar S. Mysore3 Zhi-Ping Xie1,2*
Zhi-Ping Xie1,2* Christian Staehelin1,2*
Christian Staehelin1,2*Nod factors secreted by nitrogen-fixing rhizobia are lipo-chitooligosaccharidic signals required for establishment of the nodule symbiosis with legumes. In Medicago truncatula, the Nod factor hydrolase 1 (MtNFH1) was found to cleave Nod factors of Sinorhizobium meliloti. Here, we report that the class V chitinase MtCHIT5b of M. truncatula expressed in Escherichia coli can release lipodisaccharides from Nod factors. Analysis of M. truncatula mutant plants indicated that MtCHIT5b, together with MtNFH1, degrades S. meliloti Nod factors in the rhizosphere. MtCHIT5b expression was induced by treatment of roots with purified Nod factors or inoculation with rhizobia. MtCHIT5b with a fluorescent tag was detected in the infection pocket of root hairs. Nodulation of a MtCHIT5b knockout mutant was not significantly altered whereas overexpression of MtCHIT5b resulted in fewer nodules. Reduced nodulation was observed when MtCHIT5b and MtNFH1 were simultaneously silenced in RNA interference experiments. Overall, this study shows that nodule formation of M. truncatula is regulated by a second Nod factor cleaving hydrolase in addition to MtNFH1.
Legumes (Fabaceae) can form a mutualistic relationship with nitrogen-fixing bacteria, called rhizobia. In formed nodules, carbon and nutrients are provided by host plants, while rhizobia differentiated into bacteroids deliver fixed nitrogen to the plant. Formation of nodules on legume roots depends on a molecular signal exchange between both symbiotic partners (Perret et al., 2000; Mergaert et al., 2020). Specific flavonoids or isoflavonoids are secreted by legume roots and interact with rhizobial NodD proteins to induce expression of bacterial nodulation (nod) genes (Liu and Murray, 2016). The nod genes are responsible for biosynthesis and secretion of Nod factors. These rhizobial signal molecules are lipo-chitooligosaccharides (modified chitin oligosaccharides, LCOs) containing β-1,4-linked N-acetylglucosamine residues with three to five sugars and a fatty acid chain at the non-reducing end. Furthermore, Nod factors with additional modifications such as a sulfate group at the reducing end are produced by many strains (Perret et al., 2000). Sinorhizobium meliloti strains overexpressing certain nod genes can secrete high amounts of 6-O-sulfated Nod factors with a C16:2 fatty acid (Lerouge et al., 1990; Schultze et al., 1992). Nod factor production is a common feature among almost all rhizobia except certain Bradyrhizobium strains (Giraud et al., 2007).
Nod factor signaling in host legumes triggers expression of symbiotic genes required for bacterial infection and nodule development. Nod factor receptor genes have been first identified in the model legumes Lotus japonicus and Medicago truncatula (Roy et al., 2020; Yang et al., 2022). When Nod factor receptor genes of L. japonicus were ectopically expressed in M. truncatula, roots could be nodulated by Mesorhizobium loti, which is a natural symbiont of L. japonicus (Radutoiu et al., 2007). Such experiments indicated that Nod factor receptors are determinants of host specific nodulation. In response to Nod factor perception, a calcium oscillation response (calcium spiking) is induced in nuclei of host cells. Calcium- and calmodulin-dependent protein kinases are the sensors of nuclear calcium spiking (Yang et al., 2022). These protein kinases can phosphorylate transcription factors such as CYCLOPS in L. japonicus (Singh et al., 2014) and GmSTF3 (TGACG-Motif Binding Factor 3) in soybean (Wang et al., 2021). Ultimately, additional transcription factors such as NIN (Nodule Inception) trigger expression of symbiotic genes required for bacterial infection and formation of nodule primordia (Roy et al., 2020; Yang et al., 2022).
Rhizobia often enter host plants via root hairs. Root hair curling results in formation of an infection pocket in which the bacteria form a microcolony (Fournier et al., 2015). Following endocytosis, infection threads are initiated which later elongate in the root hairs until they reach the cortical cells where they branch and colonize the formed nodule primordium. The formation of infection pockets is accompanied by secretion of proteins into the symbiotic interface. In M. truncatula for example, MtENOD11 (Early Nodulin 11), a proline-rich cell wall protein, has been reported to be accumulated within the infection pocket (Charron et al., 2004; Fournier et al., 2015). MtENOD11 is a suitable marker gene for studying activation of Nod factor signaling in M. truncatula roots (Charron et al., 2004).
Chitinases are enzymes that hydrolyze β-1,4 linkages in chitin (poly N-acetylglucosamine). Plant chitinases belong to the glycoside hydrolase families 18 (class III and V chitinases) and 19 (class I, II and IV chitinases) (Drula et al., 2022). Since Nod factors are also composed of β-1,4-linked N-acetylglucosamine, certain plant chitinases can cleave Nod factors thereby forming a hydrophobic cleavage product with an acyl chain and a hydrophilic carbohydrate moiety. Different modifications in Nod factors can render certain Nod factors partially or completely resistant to hydrolysis by plant chitinases (Staehelin et al., 1994a; Staehelin et al., 1994b; Goormachtig et al., 1998; Schultze et al., 1998; Ovtsyna et al., 2000; Tian et al., 2013; Zhang et al., 2016). Purified Nod factors applied to the rhizosphere of legumes were rapidly degraded and inactivated by plant hydrolases within few hours (Staehelin et al., 1994b; Heidstra et al., 1994; Staehelin et al., 1995; Ovtsyna et al., 2000; Ovtsyna et al., 2005; Tian et al., 2013; Cai et al., 2018).
Nod factor signaling induces expression of genes encoding Nod factor cleaving enzymes. In Sesbania rostrata, expression of a Nod factor cleaving class III chitinase gene was found to be induced by applied Nod factors and when roots were inoculated with Azorhizobium caulinodans (Goormachtig et al., 1998). In M. truncatula, expression of a class V chitinase gene was found to be up-regulated by inoculation with S. meliloti bacteria and Nod factor treatments (Salzer et al., 2004). Enzyme tests with recombinant protein expressed in E. coli showed that the enzyme lacks chitin-cleaving activity but can efficiently hydrolyze Nod factors. The protein was therefore named MtNFH1 (Nod factor hydrolase 1; Tian et al., 2013). MtNFH1 cleaves tetrasaccharidic and pentasaccharidic Nod factors of S. meliloti, thereby forming lipodisaccharides. Based on the known structures of the class V chitinases of tobacco (Nicotiana tabacum) and Arabidopsis thaliana, a homology modeling and substrate docking simulation was performed for MtNFH1. The results suggested a binding cleft for the Nod factor substrate that specifically accommodates the fatty acid chain (Tian et al., 2013).
Recently, a MtNFH1 knockout mutant was found to show delayed root hair infection, suggesting that too high Nod factor concentrations impair the formation of an infection thread. Furthermore, MtNFH1 mutant plants exhibited abnormal nodule branching at 20 days post inoculation (dpi). These results suggested that spatio-temporal fine-tuning of Nod factor levels affect the nodule symbiosis at early and late symbiotic stages (Cai et al., 2018). In L. japonicus, a class V chitinase gene, LjCHIT5, has been found to be implicated in the symbiosis with M. loti. LjCHIT5 knockout mutant plants formed fewer pink (nitrogen-fixing) nodules and had a defective nodule infection phenotype. Furthermore, the mutants showed reduced infection thread elongation inside nodule primordia while root hair infection was not affected (Malolepszy et al., 2018).
The differences in MtNFH1 and LjCHIT5 mutant phenotypes suggest that additional Nod factor-cleaving enzymes of M. truncatula could play a symbiotic role. In M. truncatula, the class V chitinase genes MtCHIT5b and MtCHIT5a and are most related to MtNFH1 (amino acid sequence identity of 75% and 46%, respectively). MtCHIT5b and MtNFH1 are located in tandem on chromosome 4 of the M. truncatula genome, suggesting gene duplication during evolution. Recombinant MtCHIT5a and MtCHIT5b proteins expressed in Escherichia coli can hydrolyze chitooligosaccharides but Nod factor cleaving activity has not yet been demonstrated for these proteins (Tian et al., 2013; Zhang et al., 2016). However, MtCHIT5b with a serine-to-proline substitution at the predicted substrate binding cleft was found to possess Nod factor hydrolase activity similar to MtNFH1. Conversely, a corresponding proline-to-serine substitution in MtNFH1 led to a variant that did not release detectable amounts of cleavage products from S. meliloti Nod factors (Zhang et al., 2016).
In this study, we found that recombinant MtCHIT5b expressed in E. coli could hydrolyze S. meliloti Nod factors under newly established purification and test conditions. MtCHIT5b expression in young root hairs was rapidly induced by applied Nod factors or rhizobial inoculation. Expression of MtCHIT5b with fluorescent tags in M. truncatula showed that the enzyme is mainly localized in the infection pocket of a curled root hair. Analysis of MtCHIT5b mutant plants indicated reduced Nod factor hydrolysis in the rhizosphere but no obvious symbiotic phenotype was observed. In contrast, MtCHIT5b overexpression negatively affected nodule formation. RNA interference experiments indicated that simultaneous silencing of MtCHIT5b and MtNFH1 impairs nodule formation.
Escherichia coli DH5α and BL21 (DE3) (Novagen, Madison, USA) were grown in Luria-Bertani (LB) medium (5 g L-1 yeast extract, 10 g L-1 tryptone, 10 g L-1 NaCl) at 37 °C. Sinorhizobium meliloti Rm41 and fluorescent derivatives were cultured in TY medium (3 g L-1 yeast extract, 5 g L-1 tryptone, 0.4 g L-1 CaCl2) at 27 °C. For plant inoculation, bacteria were centrifuged (4 000 rpm for 10 min) and the pellets were resuspended in 10 mM sterilized MgSO4 (OD600 ≈ 0.2). S. meliloti 1021 (pEK327) was grown in defined medium and used for Nod factor purification (Schultze et al., 1992; Tian et al., 2013). Agrobacterium rhizogenes ARqua-1 (Quandt et al., 1993) was cultured in TY medium at 27 °C. DNA constructs used for this study were cloned into the plasmids pET-28b (Novagen, Darmstadt, Germany), pBBR1MCS-2 (Kovach et al., 1995), pUB-GFP (Maekawa et al., 2008), pCAMBIA1302 and pCAMBIA1305 (http://www.cambia.org/).
To obtain red fluorescent S. meliloti bacteria (strain Rm41-mCherry), DNA containing the spectinomycin resistance gene promoter of pHP45 (Quandt and Hynes, 1993) and the mCherry gene (Shaner et al., 2004) was cloned into pBBR1MCS-2 (Kovach et al., 1995) using XhoI/XbaI. The plasmid was then mobilized into S. meliloti Rm41 by tri-parental mating (Figurski and Helinski, 1979). A similar approach was used for construction of Rm41-mTFP1 expressing mTFP1 (Ai et al., 2006).
For MtCHIT5b promoter-β-glucuronidase gene (GUS) fusion analysis, a 3 022-bp promoter sequence flanking the coding region of MtCHIT5b was amplified from genomic DNA of M. truncatula R108 and cloned into pCAMBIA1305 with BamHI/NcoI, generating a promoter-GUS fusion (MtCHIT5bp-GUS construct). Then, a red fluorescent protein (RFP) expression cassette (Chen et al., 2009) was cloned into the constructed binary vector using KpnI.
For subcellular localization, DNA encoding MtCHIT5b fused to mCherry (MtCHIT5b:mCherry) was amplified by overlap extension PCR using genomic DNA of M. truncatula R108 and mCherry DNA as a template. The amplicon was then inserted with XbaI/KpnI into pUB-GFP containing a Ubiquitin gene promoter from L. japonicus, which possesses strong promoter activity in M. truncatula (Lei et al., 2015; Kovács et al., 2022). Furthermore, a construct encoding MtCHIT5b fused to GFP with an upstream MtCHIT5b promoter sequence was used for subcellular localization. The 3 022-bp MtCHIT5b promoter sequence (amplified from genomic DNA of M. truncatula R108) was cloned into pCAMBIA1302 with BamHI/NcoI. Next, the coding region of MtCHIT5b was amplified from genomic DNA of M. truncatula R108 and the overhangs were removed with NcoI. The Hieff Clone™ Plus Multi One Step Cloning Kit (Yeasen, Shanghai, China) was then used to insert the MtCHIT5b coding sequence between the promoter sequence and the GFP tag. For an additional plasmid, an RFP expression cassette (Chen et al., 2009) was inserted into the binary vector with the help of KpnI.
Binary vectors with RNAi constructs were used to silence MtCHIT5b/MtNFH1 expression in M. truncatula roots. A DNA fragment containing a 613-bp sequence of MtCHIT5b fragment was amplified from genomic DNA of M. truncatula R108 and inserted with EcoRI/SmaI into a pBluescript vector containing a DNA spacer region. The reverse complement fragment was also amplified and inserted into the vector with BamHI/XbaI. The whole dsRNA1 cassette was then excised with HindIII/SacI and inserted into pCAMBIA1305. Finally, an RFP expression cassette (Chen et al., 2009) was cloned into the HindIII site of the constructed vector. In addition, a similar binary vector was constructed which contains the 667-bp dsRNA2 sequence derived from MtNFH1 (Tian et al., 2013).
To constitutively overexpress MtCHIT5b in roots of M. truncatula, the coding sequence of MtCHIT5b was amplified from genomic DNA of M. truncatula R108 and inserted into pUB-GFP with XbaI/KpnI. The promoter-MtCHIT5b construct was then PCR amplified and inserted into pCAMBIA1305 carrying an RFP expression cassette with the help of the cloning kit mentioned above.
Construction of plasmids used for expression of MtCHIT5b and MtNFH1 proteins in E. coli has been reported previously (Tian et al., 2013; Zhang et al., 2016). Further information on strains and plasmids used in this study can be found in Supplementary Table 1. Primers are listed in Supplementary Table 2.
Medicago truncatula ecotype R108 and Jemalong A17 were used in this study. The Tnt1 insertion mutant lines NF16543 (chit5b) and NF11260 (nfh1-3b) were obtained from the M. truncatula Tnt1 mutant collection currently hosted at the Institute of Agricultural Biosciences, Oklahoma State University, Ardmore, USA (Tadege et al., 2008; Pislariu et al., 2012). In contrast to the previously described nfh1-3 mutant (Cai et al., 2018), nfh1-3b inoculated with S. meliloti did not form branched nodules at 3 weeks post inoculation (wpi) under the experimental conditions used in this study. The nodulation signaling mutant dmi3-1 of ecotype Jemalong A17 (TRV25; Sagan et al., 1998) was kindly provided by Dr. Clare Gough (INRAE-CNRS, Castanet-Tolosan, France).
M. truncatula seeds were scarified with sandpaper, sterilized in 10% commercial bleach for 5 min and finally rinsed in sterile water for several times. The seeds were then placed on inverted 1.2% (v/v) water agar plates in the dark at 4 °C for 48 h and later at 23 ± 1 °C for 18 h. Unless otherwise specified, seedlings were grown on 1% (v/v) agar plates containing BNM (buffered nodulation medium) (Ehrhardt et al., 1992). The lower part of the plate was covered with aluminum foil. The plates were then placed in a growth room at an angle of ~60°. Special growth conditions were established for root transformation and nodulation experiments. All plants were kept in an air-conditioned growth room at 23 ± 1 °C under 16/8-h light/dark conditions as described previously (Cai et al., 2018).
Binary vectors were transformed into competent A. rhizogenes ARqua-1 by electroporation (1 800 V for 5 ms). Cells were sprayed on TY plates containing 50 mg L-1 kanamycin and 50 mg L-1 streptomycin and incubated at 27 °C. Formed colonies were confirmed by PCR. Hairy root transformation was performed according to the protocol described by Boisson-Dernier et al. (2001) with minor modifications. At the stage of emerging hairy roots, seedlings were transferred to solid HRE plates (Díaz et al., 2005) supplemented with 300 μg/mL timentin and kept under growth conditions as reported (Cai et al., 2018). Ten days later, seedlings were analyzed by fluorescence microscopy and non-transgenic roots lacking RFP fluorescence were removed. A similar transformation efficiency was observed for the different binary vectors used in this study.
Genomic DNA was isolated from young leaves of M. truncatula (R108 wild-type plants and mutant lines) using cetyltrimethylammonium bromide. To confirm the exact Tnt1 insertion sites in mutant plants, the isolated DNA was used as template for PCRs using primers designed from the flanking regions of the Tnt1 insertion sites. Information on used primers is provided in Supplementary Table 2.
Purification of Nod factors from S. meliloti 1021 (pEK327) was performed as described previously (Staehelin et al., 1994b; Tian et al., 2013) with the exception that Amberlite XAD-2 resin (20-60 mesh; Supelco, Merck, Kenilworth, USA) instead of n-butanol was used for the initial purification step with bacterial culture supernatants. For purification of NodSm-IV(C16:2, S) and NodSm-V(C16:2, S), the culture supernatant was adjusted to pH 9.5 and shaken at 27 °C for 24 h. Amberlite XAD-2 resin was extensively washed with acetone, then with methanol and final equilibrated with water. Subsequently, 40 g of the conditioned resin was added to 1 L of the prepared bacterial culture supernatant. After incubation overnight (27 °C; 150 rpm), the resin material was collected with a metal mesh and extensively washed with water. Using a vacuum filtration system, the Nod factors were eluted from the resin with 40 mL methanol and then with 30 mL acetone. The filtrates were collected and dried in a rotavapor system. Nod factors were then fractionated by reverse-phase HPLC using a Nova Pak C18 column (3.9 x 150 mm, particle size 4 µm; Waters, Milford, USA) and 30% or 35% (v/v) acetonitrile/water containing 40 mM ammonium acetate. Fractions containing Nod factors were collected and dried using a speed-vac concentrator. C18 Sep-Pak cartridges (Waters, Milford, USA) were used for desalting purified Nod factors. Briefly, purified Nod factors were dissolved in 5 mL water containing 0.5% (v/v) dimethyl sulfoxide (DMSO) and then loaded onto the cartridges which were washed with methanol and conditioned with water. After washing with 10 mL water, the Nod factors were eluted with 5 mL methanol. The desalted Nod factors were dried in a speed-vac concentrator and re-dissolved in 1 mL water containing 0.5% (v/v) DMSO.
E. coli strain BL21 (DE3) carrying pET-28b containing MtCHIT5b without predicted signal peptide (Zhang et al., 2016) was used to obtain enzymatically active MtCHIT5b with an N-terminal His-tag. A single colony was cultured in 3 mL of liquid LB supplemented with 50 μg mL-1 kanamycin on a shaker (200 rpm at 37 °C). After incubation (12 h), 2 mL of the obtained preculture were used for inoculation of 400 mL LB (containing 50 μg mL-1 kanamycin) and the bacteria were cultured under the same growth conditions. When the optical density (OD600) reached 0.5, isopropyl-β-D-thiogalactopyranoside was added to obtain a final concentration of 0.4 mM. The cultures were then kept on the shaker at 18 °C for 20 h. Finally, the cultures were centrifuged (4 000 rpm at 4 °C for 15 min) and the bacterial pellets were collected. MtCHIT5b was purified under native conditions. Briefly, each bacterial pellet was resuspended in 8 mL lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 7.5) supplemented with 1 mg/mL lysozyme. Next, the mixture was sonicated on ice with an ultrasonic processor (Ningbo Scientz Biotechnology Co., Ningbo, China; 7 s burst followed by 53 s cooling step for 18 min, 60 W). After sonication, the suspension was gently shaken at 100 rpm for 30 min and then subjected to centrifugation (12 000 rpm at 4 °C for 20 min). The clear supernatant was collected and mixed with 0.2 mL Ni-NTA resin beads (TransGen Biotech, Beijing, China), which were conditioned by washing with lysis buffer (4 °C). After incubation (100 rpm at 4 °C for 45 min), the beads were placed into a column and washed with 16 mL wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 7.5). Finally, MtCHIT5b was eluted from the beads with 0.3 mL elution buffer (50 mM NaH2PO4, 300 mM NaCl, 600 mM imidazole, adjusted to pH 7.5). The elution fraction was immediately mixed with an equal volume of glycerol and a 0.47-fold volume of 500 mM sodium acetate buffer (pH 4.0) to obtain a final pH of 7.0.
Proteins were analyzed by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Protein samples were supplemented with loading buffer (TransGen Biotech, Beijing, China), followed by boiling at 100 °C for 10 min. After electrophoresis, gels were stained with Commassie Brilliant Blue R-250. Photos were taken with a digital camera (Nikon, Nikon, Japan). For protein quantification, different concentrations of bovine serum albumin (BSA) were used as standards. A standard curve was drawn using corresponding signal intensities of protein bands as quantified by ImageJ software (NIH, Bethesda, USA). Western blot analysis was performed with nitrocellulose membranes temporarily stained with Ponceau S dye to visualize transferred proteins. The membranes were incubated with an anti-His antibody (1:10 000 dilution; TransGen Biotech, Beijing, China). A goat-anti-mouse IgG antibody coupled to horseradish peroxidase (1:10 000 dilution; TransGen Biotech, Beijing, China) served as second antibody. The membranes were developed with an enhanced chemiluminescence kit (FDbio Science, Hangzhou, China).
If not otherwise specified, enzyme assays with Nod factors and test proteins were performed in 25 mM potassium phosphate buffer (pH 7.0) containing 0.5% DMSO and 10 μg mL-1 BSA at 37 °C. Typically, 3 nmol of a purified Nod factor substrate (NodSm-IV(C16:2, S), NodSm-V(C16:2, S) or NodSm-IV(C16:2, Ac, S)) was incubated with a 0.25 μg mL-1 MtCHIT5b sample (pH 7.0) in a given reaction volume. After incubation for different times (37 °C), samples were extracted by an equal volume of n-butanol and the collected butanol phase was dried using a speed-vac concentrator. The dried samples were resuspended in 1.2 μL DMSO and finally analyzed by HPLC using a Nova Pak C18 column and 35% (v/v) acetonitrile/water containing 40 mM ammonium acetate as mobile phase (Staehelin et al., 1994b). The chemical structures of obtained acylated cleavage products, the lipodisaccharide II(C16:2) and the O-acetylated lipodisaccharide II(C16:2, Ac), were confirmed by MALDI-TOF mass spectrometry. The instrument (Bruker, Bremen, Germany) was operating in the positive-ion mode and 2,5-dihydroxybenzoic acid was used as matrix. The Km and Vmax values of MtCHIT5b for the Nod factor substrates were determined using GraphPad Software (Dotmatics, Boston, USA). The velocity values were calculated from reactions with no more than 25% cleavage product formation.
Five-day old M. truncatula seedlings or plants with transformed roots were placed into glass plates (6 cm in diameter) filled with 7 mL of liquid BNM containing purified Nod factors and 0.5% (v/v) DMSO. Control plants were treated with BNM containing 0.5% DMSO. After incubation for 6 or 24 h in the dark (23 ± 1 °C), plants were used for real-time quantitative PCR (RT-qPCR) analysis or subjected to GUS staining.
Total RNA of plant material was extracted using the Plant Total RNA Purification Kit (GeneMark, Taiwan, China) followed by cDNA synthesis with HiScript II Q RT SuperMix (Vazyme, Nanjing, China). RT-qPCR was performed with a LightCycler 480 instrument using the LightCycler 480 SYBR Green I Master Reaction Mix (Roche, Shanghai, China). The reaction conditions were as follow: (1) 95 °C for 5 min; (2) 45 cycles: 95 °C for 20 s, 55 °C for 20 s, 72 °C for 40 s; (3) 95 °C for 5 s, 60 °C for 60 s and 95 °C for 10 s. RNAs were extracted in triplicate (three biological replicates) and each corresponding cDNA was amplified three times (three technical replicates). Medicago elongation factor was used as a reference gene. The Ct values were obtained with the help of the LightCycler 480 software. Primers used for RT-qPCR are shown in Supplementary Table 2.
Transgenic roots transformed with the MtCHIT5bp-GUS construct (or nodules obtained from inoculation with S. meliloti Rm41) were immersed in a solution (pH 7.0) containing 100 mM NaH2PO4, 10 mM EDTA disodium salt, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, 0.1% Triton-X100 and 0.5 mg mL-1 5-bromo-4-chloro-3-indoxyl-β-D-glucuronic acid cyclohexylammonium salt (X-Gluc). The samples were exposed to a vacuum for 25 min and then incubated at 37 °C for different periods. Finally, the stained tissue was analyzed with a Nikon Eclipse Ni-E microscope. For sectioning of nodules, GUS-stained samples were fixed in 1.25% glutaraldehyde phosphate buffer (pH 5.7) under a vacuum for 1 h. Samples were embedded in 4% (w/v) agrose and 70-μm sections were obtained using a LeicaVT1200S vibrating blade microtome.
For subcellular localization of MtCHIT5b, the protein with a fluorescent tag was expressed in M. truncatula roots with the help of A. rhizogenes. The transformed roots were inoculated with Rm41, Rm41-mCherry or Rm41-mTFP1 (OD600 ≈ 0.2). Fluorescence in infected root hairs was observed using a Zeiss ImagerZ1 fluorescence microscope with filters as recommended by the supplier (Carl Zeiss AG, Oberkochen, Germany). Furthermore, subcellular localization of MtCHIT5b was predicted by using the ProtComp 9.0 server (http://www.softberry.com/berry.phtml).
Five-day-old M. truncatula seedlings grown on BNM agar plates were transferred to 1-mL plastic syringes (Li et al., 2019). The syringes were filled with 300 μL BNM containing 0.5% (v/v) DMSO and 0.5 μM NodSm-IV(C16:2, S) to activate Nod factor signaling. After incubation in the dark (23 ± 1 °C for 18 h), the seedlings were transferred to new 1-mL syringes filled with 900 μL BNM containing 0.5% (v/v) DMSO and 5.4 μM NodSm-IV(C16:2, S). After incubation for 2 h, the seedlings were removed and the Nod factor substrate and formed II(C16:2) were extracted with an equal volume of n-butanol. Dried materials from three seedlings were combined to obtain a sample for HPLC analysis. In total, three HPLC samples per treatment were analyzed.
Germinated M. truncatula seedlings and plants with transformed roots were transferred to sterile 300-mL plastic jar units. The upper jar contained vermiculite/expanded clay at a 3:1 (v/v) ratio and the lower jar contained sterilized B&D nutrient solution (Broughton and Dilworth, 1971) supplemented with 1 mM KNO3. Each jar unit contained a single plant. After 7 days, plants were inoculated with 2 mL of a rhizobial suspension (OD600 ≈ 0.2) or mock-inoculated with 2 mL of 10 mM sterilized MgSO4. The substrate of the upper jar units was then covered with sterilized quartz stones. At the time of harvest, the nodules were counted and the nodule biomass was determined for each plant.
The M. truncatula genes MtCHIT5b and MtNFH1 show 75% amino acid sequence identity (without predicted signal peptide) (Zhang et al., 2016). In vitro activity tests with recombinant proteins expressed in E. coli showed that MtNFH1 can hydrolyze S. meliloti Nod factors into lipodisaccharides (Tian et al., 2013), while no detectable activity was found for MtCHIT5b in a previous study (Zhang et al., 2016). However, further enzyme tests in our laboratory indicated that His-tagged MtCHIT5b expressed in E. coli does not completely lack Nod factor cleaving activity. In these experiments, the previously constructed plasmid pET-MtCHIT5b and the empty vector pET-28b were freshly transformed into E. coli BL21 (DE3). The protein extracts were subjected to nickel affinity chromatography under native conditions using a lysis buffer of pH 7.5 to keep the pH significantly lower than the isoelectric point of His-tagged MtCHIT5b (pI 9.12). Proteins eluted from nickel beads with imidazole were supplemented with glycerol and adjusted to pH 7.0 with an acetate buffer to obtain a pH identical to the reaction buffer. SDS-PAGE gel analysis showed that the recombinant MtCHIT5b could be successfully eluted from the nickel column. The presence of the protein in the elution fraction was further confirmed by immunoblot analysis with an anti-His antibody (Figure 1A).
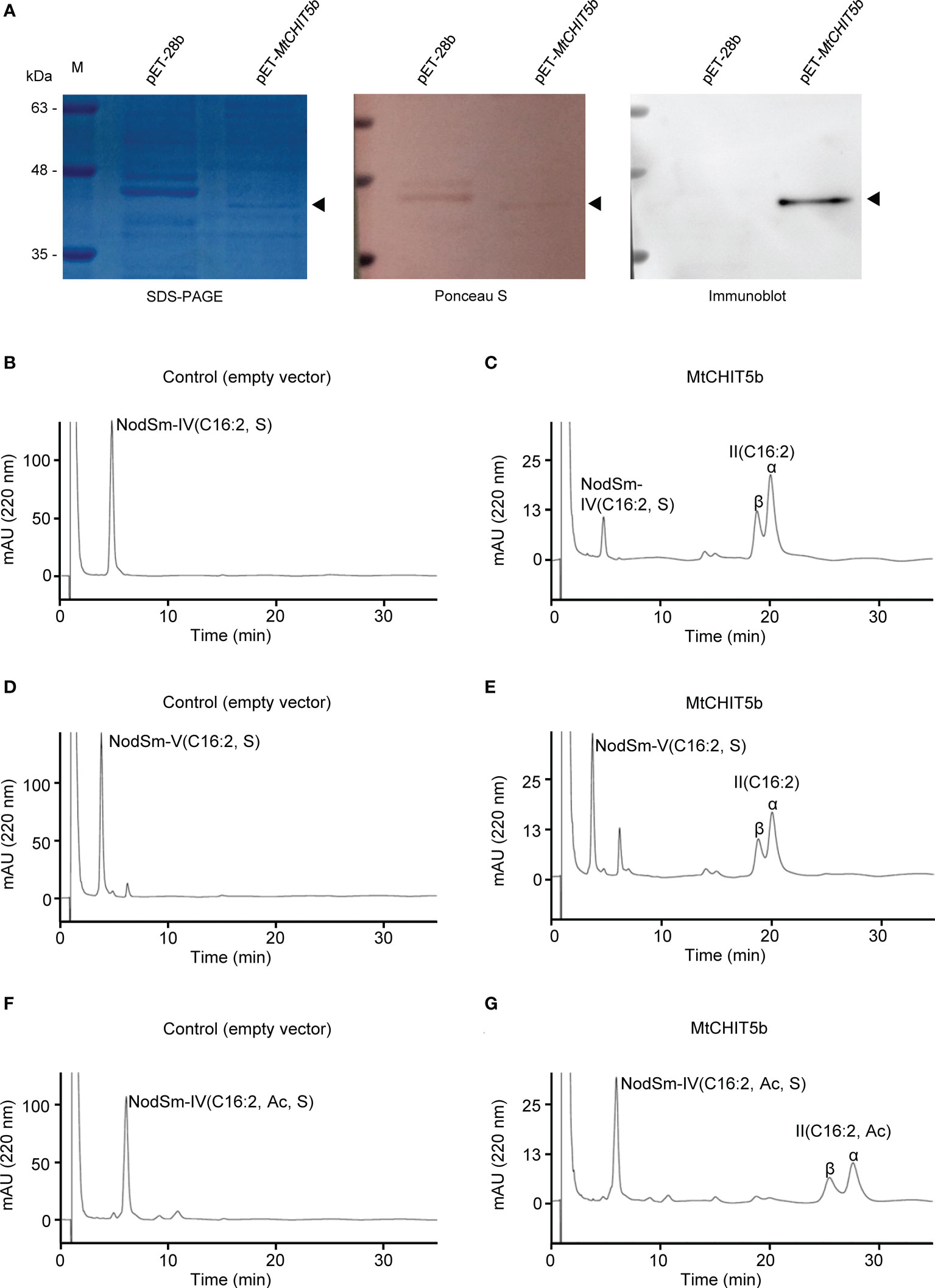
Figure 1 Preparation of recombinant MtCHIT5b protein and activity tests with Nod factor substrates. (A) Proteins eluted from nickel beads were separated on SDS-PAGE and stained with Coomassie Brilliant Blue R-250. Ponceau staining of the membrane served as a loading control. Immunoblot analysis was performed with an anti-His antibody (B-G) Activity tests with obtained MtCHIT5b (0.25 μg mL-1) and Nod factors (60 μM) in a 100-μL test system. After incubation (37 °C) for 12 h, the Nod factors and acylated cleavage products were extracted with n-butanol and analyzed by reverse-phase HPLC. (B) Incubation of control proteins from E coli carrying the empty vector pET-28b with NodSm-IV(C16:2, S). (C) Incubation of MtCHIT5b with NodSm-IV(C16:2, S) resulting in II(C16:2) formation. (D) Incubation of empty vector control proteins with NodSm-V(C16:2, S). (E) Incubation of MtCHIT5b with NodSm-V(C16:2, S) resulting in II(C16:2) formation. (F) Incubation of empty vector control proteins with NodSm-IV(C16:2, Ac, S). (G) Incubation of MtCHIT5b with NodSm-IV(C16:2, Ac, S) resulting in II(C16:2, Ac) formation.
For Nod factor hydrolysis tests, recombinant MtCHIT5b (freshly prepared from E. coli) was incubated with tetrameric NodSm-IV(C16:2, S), pentameric NodSm-V(C16:2, S), or O-acetylated tetrameric NodSm-IV(C16:2, Ac, S). The Nod factor substrates and acylated cleavage products were then subjected to HPLC analysis using a C18 column. The obtained chromatograms showed that MtCHIT5b could release II(C16:2) from either NodSm-IV(C16:2, S) or NodSm-V(C16:2, S) (Figures 1B-E). Hydrolysis of NodSm-IV(C16:2, Ac, S) by MtCHIT5b resulted in formation of the II(C16:2, Ac) cleavage product (Figures 1F, G). The fractions containing II(C16:2) or II(C16:2, Ac) were collected and their molecular weights were confirmed by mass spectrometry (Supplementary Figure 1).
To obtain kinetic data, freshly prepared MtCHIT5b was incubated with a given Nod factor substrate at various concentrations. The protein amount of MtCHIT5b for these tests was determined by ImageJ software using a SDS-PAGE gel with different amounts of BSA as standards. Substrate-velocity curves are shown in Supplementary Figure 2 and GraphPad Software was used to deduce corresponding Michaelis-Menten constants (Km values). MtCHIT5b displayed a lower Km value for NodSm-V(C16:2, S) than for NodSm-IV(C16:2, S) and NodSm-IV(C16:2, Ac, S), indicating a higher affinity of the enzyme to NodSm-V(C16:2, S). Furthermore, the activity of MtCHIT5b was calculated under substrate saturation conditions to obtain maximum velocity (Vmax) and catalytic rate constant (kcat) values. Highest kcat and kcat/Km values were obtained for NodSm-IV(C16:2, Ac, S), which is the most abundant S. meliloti Nod factor (Table 1). Collectively, these results show that MtCHIT5b is an enzyme that can hydrolyze NodSm-IV(C16:2, S), NodSm-V(C16:2, S) and NodSm-IV(C16:2, Ac, S) under the experimental conditions used in this study.
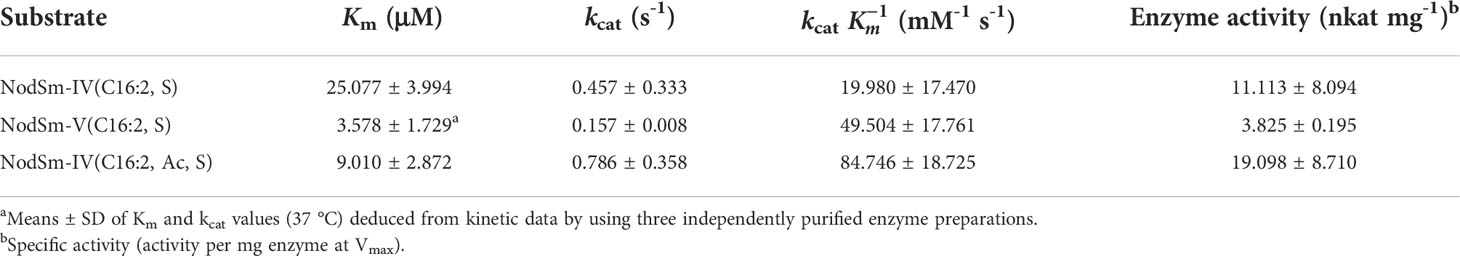
Table 1 Michaelis-Menten constants (Km), catalytic rate constants (kcat) and specific activity of MtCHIT5b for different Nod factor substrates.
MtNFH1 gene expression in M. truncatula roots was found to be stimulated by root treatments with purified Nod factors (Cai et al., 2018). We therefore performed an RT-qPCR experiment to examine whether Nod factors show a similar effect on MtCHIT5b expression. In agreement with previously performed RNA sequencing data (Roux et al., 2014), MtCHIT5b expression was considerably up-regulated when roots were treated with different concentrations of NodSm-IV(C16:2, Ac, S) for 6 h (Figure 2A). A similar increase of transcripts was also measured for MtNFH1 as reported earlier (Cai et al., 2018). In contrast, expression of MtCHIT5a, another class V chitinase gene of M. truncatula, was not affected by the Nod factor treatment (Supplementary Figure 3). Next, a 3 022-bp promoter sequence of MtCHIT5b was fused with the GUS gene to study tissue specific gene expression (MtCHIT5bp-GUS construct). In addition, the vector contained an RFP expression cassette for detection of the transgenic roots. Roots transformed with A. rhizogenes carrying the constructed binary vector were treated with Nod factors or inoculated with S. meliloti Rm41. GUS staining of untreated roots did not result in blue coloration reflecting MtCHIT5b promoter activity. However, strong blue coloration in young roots was observed when NodSm-IV(C16:2, Ac, S) was applied at a concentration range from 10-6 M to 10-10 M (Figure 2B). Similarly, inoculation with Rm41 caused induction of GUS activity in root hairs and the apex of nodule primordia (Figure 2C). Blue coloration was occasionally also observed in the central cylinder of roots treated with Nod factors or inoculated with Rm41. Nod factor treatments of dmi3-1, a non-nodulating M. truncatula Jemalong A17 mutant deficient in Nod factor signaling (Sagan et al., 1998), showed no effects on MtCHIT5b expression as determined by RT-qPCR analysis. Expression of the early nodulin gene MtENOD11 was also analyzed in this experiment (Figure 2D). Overall, induction of MtCHIT5b expression correlated with rhizobial infection during early symbiotic stages and was dependent on Nod factor signaling.
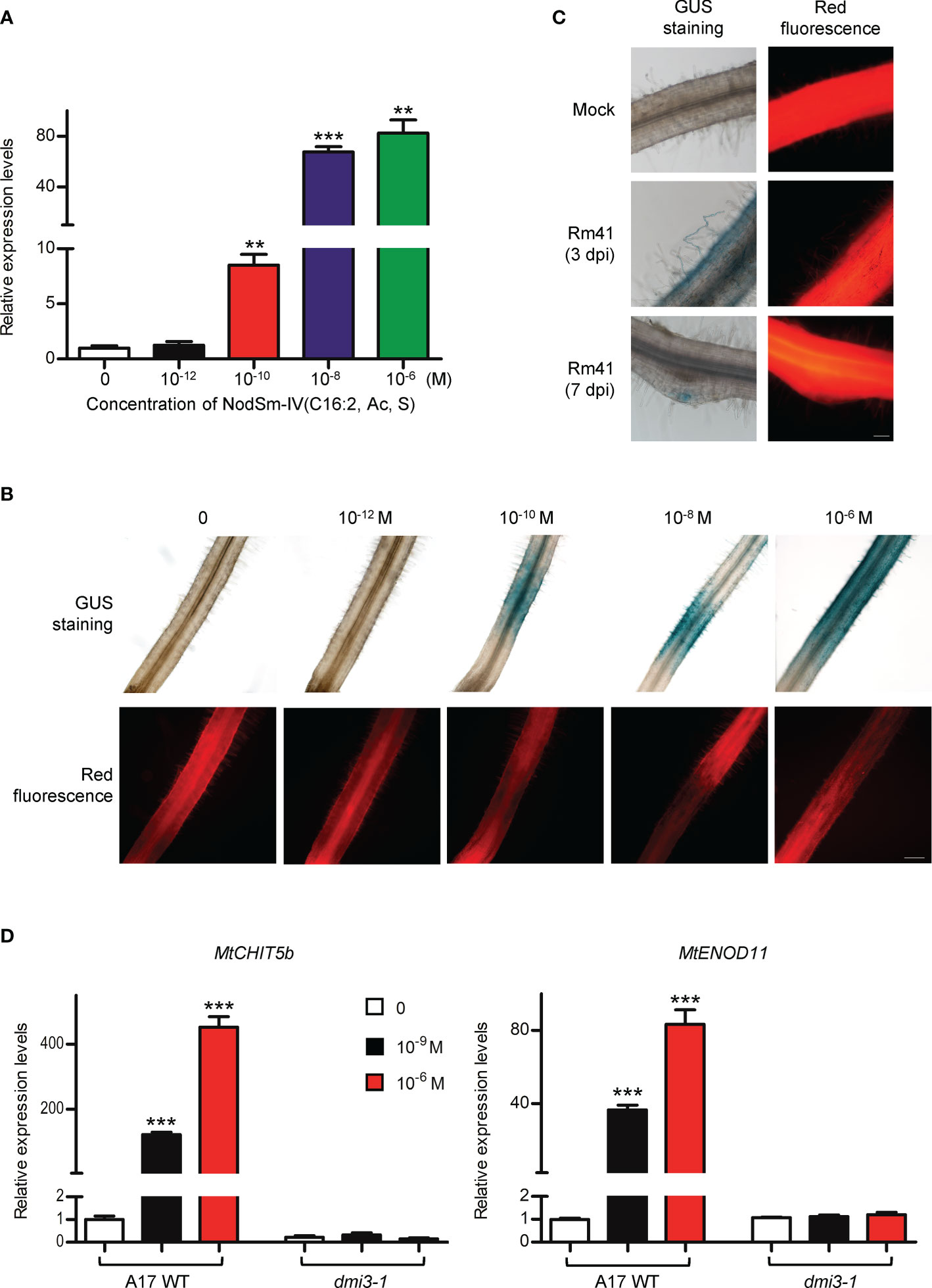
Figure 2 MtCHIT5b expression in roots of M. truncatula is induced by applied Nod factors and in response to S. meliloti inoculation. (A) R108 seedlings were treated with different concentrations of NodSm-IV(C16:2, Ac, S) dissolved in liquid BNM containing 0.5% (v/v) DMSO. After incubation (6 h in the dark), MtCHIT5b expression in roots was determined by RT-qPCR (7 seedlings per RNA extraction; n = 3). Data indicate means ± SE of normalized expression values (mean value of control set to one). The asterisks indicate significantly increased MtCHIT5b expression compared to control roots without Nod factor treatment (Student’s t-test; **, P< 0.01; ***, P< 0.001). (B, C) Promoter activity of MtCHIT5b in roots transformed with a MtCHIT5bp-GUS construct. Co-expressed RFP was used to identify transgenic roots. Plants were treated with different concentrations of NodSm-IV(C16:2, Ac, S) (B) or used for an inoculation experiment with S. meliloti Rm41 (mock-inoculation with 10 mM MgSO4; plants harvested at 3 dpi or 7 dpi) (C). Transgenic roots showing red fluorescence were subjected to GUS staining. Bar = 200 μm in (B) and 100 μm in (C). (D) Seedlings of Jemalong A17 wild-type plants and the nodulation signaling mutant dmi3-1 were treated with indicated concentrations of NodSm-IV(C16:2, Ac, S) dissolved in liquid BNM containing 0.5% (v/v) DMSO. After incubation (6 h in the dark), MtCHIT5b and MtENOD11 expression in roots was determined by RT-qPCR (7 seedlings per RNA extraction; n = 3). Data indicate means ± SE of normalized expression values (mean value of control set to one). The asterisks indicate significantly increased expression compared to control roots without Nod factor treatment (Student’s t-test; ***, P< 0.001).
To examine MtCHIT5b expression in nodules, plants were inoculated with Rm41 and nodules were harvested at 3 wpi. Isolated RNA was then used for RT-qPCR analysis. For comparison, MtCHIT5b expression in roots and leaves of these plants was also analyzed. Highest accumulation of MtCHIT5b transcripts was found for the nodules (Figure 3A). To further characterize MtCHIT5b expression in nodules, roots were transformed with the MtCHIT5bp-GUS construct and then inoculated with Rm41. GUS staining of nodules harvested at 3 wpi showed blue coloration in the apical region containing the infection zone. Nodules harvested at 4 wpi exhibited strong blue coloration in vascular bundles (Figures 3B-D). These results show that MtCHIT5b is expressed in mature nodules in accordance with previously reported RNA sequencing data from different nodule zones (Roux et al., 2014; Supplementary Figure 4).
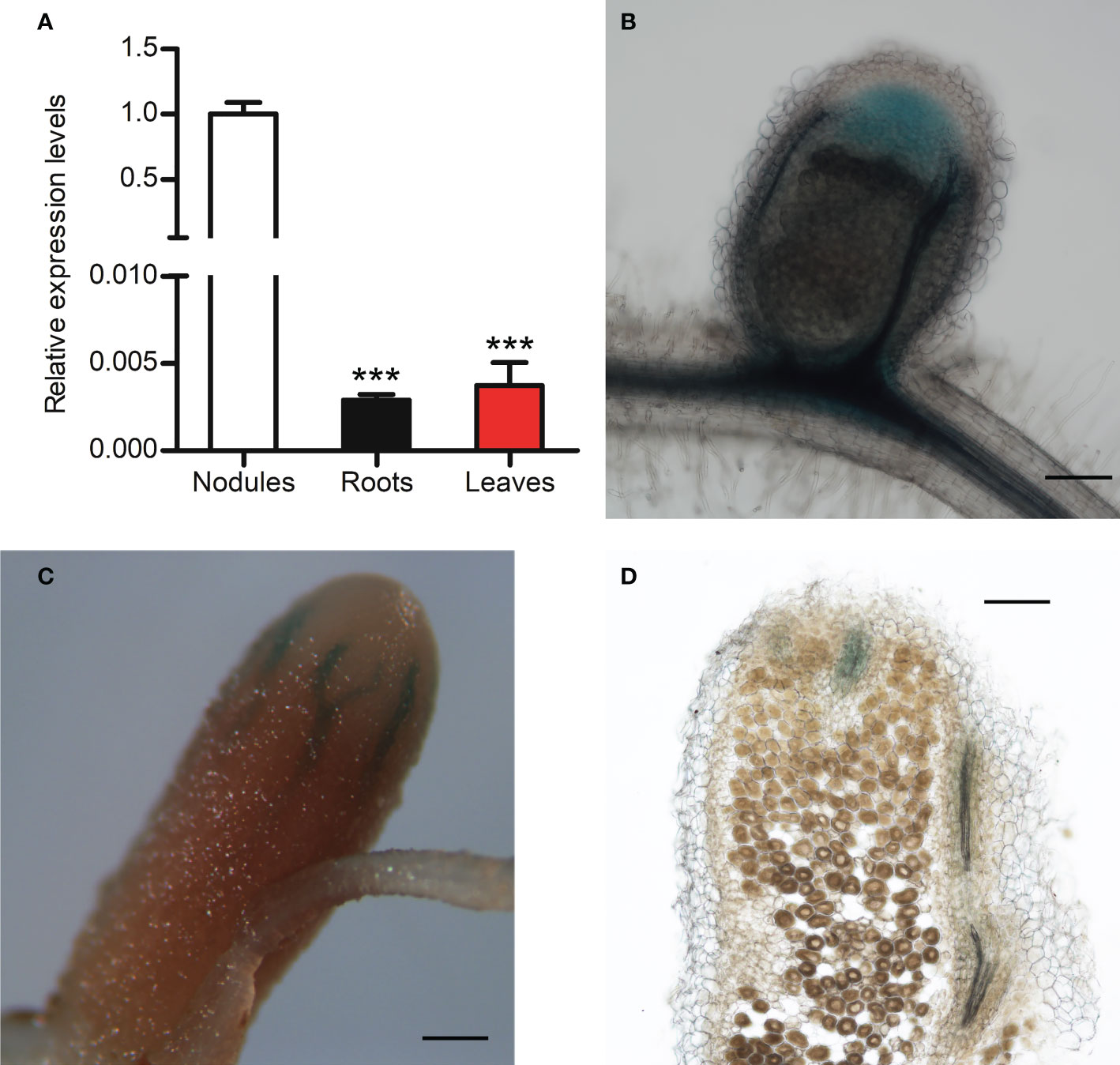
Figure 3 Analysis of MtCHIT5b expression in M. truncatula nodules. (A) RT-qPCR analysis of MtCHIT5b transcript levels in nodules of M. truncatula R108 inoculated with S. meliloti Rm41 and harvested at 3 wpi. For comparison, MtCHIT5b expression was also analyzed in roots and leaves of these plants. Data indicate means ± SE of normalized values (mean value of nodules set to one) from three independent RNA extractions (n = 3) (Student’s t-test; ***, P< 0.001). (B-D) GUS staining analysis of R108 nodules expressing the MtCHIT5bp-GUS construct at 3 wpi (B) and 4 wpi (C, D). The picture in (D) shows a sectioned nodule. Bars = 200 μm in (B, D); 500 μm in (C).
A gene construct encoding MtCHIT5b fused to the red fluorescent protein mCherry (driven by a Ubiquitin promoter from L. japonicus) was constitutively expressed in roots of M. truncatula to examine subcellular localization of MtCHIT5b. The transformed roots were then inoculated with S. meliloti Rm41 expressing the fluorescent protein mTFP1 (monomeric Teal Fluorescent Protein 1). Microscopy analysis of the inoculated roots showed that fluorescence signals emitted by the constitutively expressed MtCHIT5b:mCherry fusion protein are associated with curled root hairs, indicating that the protein was secreted into the rhizosphere and that the protein levels were increased in the infection pocket (Figures 4A-D). To substantiate these findings, roots were transformed with a construct encoding MtCHIT5b fused to green fluorescent protein (GFP), which was driven by a native promoter sequence of MtCHIT5b (3 033-bp sequence upstream of the ATG start codon). When roots were inoculated with Rm41, infected root hairs showed green fluorescence at their tips. Inoculation tests with Rm41 expressing mCherry indicated that fluorescence signals emitted by MtCHIT5b: GFP and mCherry partially co-localize in the region of the infection pocket, resulting in yellow signals in the overlay picture (Figures 4E-J). Neighboring root hairs without rhizobial infection did not show green fluorescence (Figures 4K-N). Control roots expressing GFP driven by the CaMV 35S promoter showed only faint background fluorescence in infection pockets when infected by rhizobia (Figures 4O-R). Overall, these findings indicate that MtCHIT5b is a secreted protein which comes into direct contact with Nod factor producing bacteria in the infection pocket. MtCHIT5b trafficking along the secretory pathway is also supported by bioinformatic prediction using SignalP-6.0 and the ProtComp 9.0 server (Supplementary Figure 5).
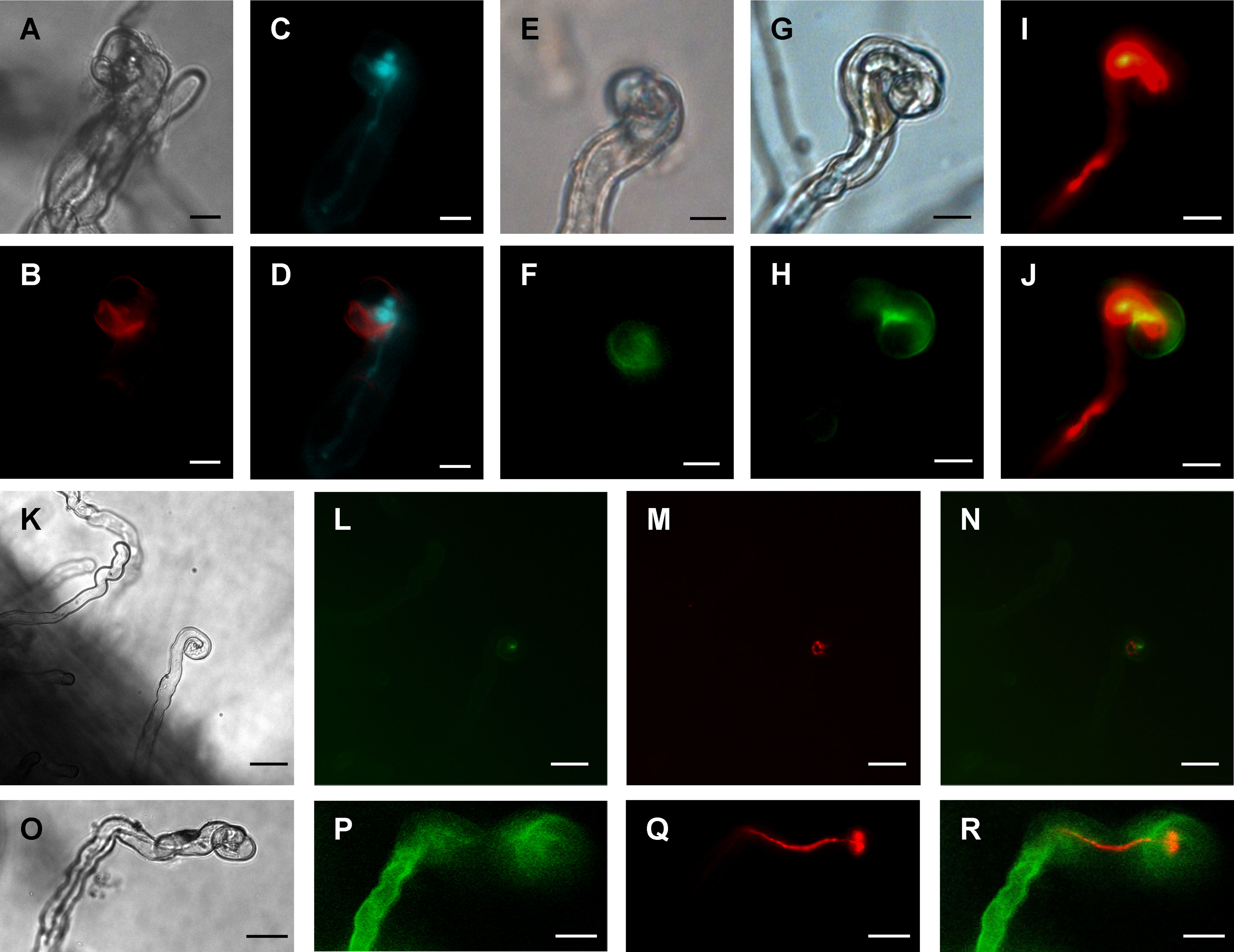
Figure 4 Subcellular localization of fluorescent MtCHIT5b in root hairs of M. truncatula. (A-D) Transgenic roots of R108 plants expressing MtCHIT5b:mCherry driven by a Ubiquitin gene promoter from L. japonicus were inoculated with S. meliloti Rm41-mTFP1. Root hairs were microscopically analyzed under bright field conditions (A), for red fluorescence emission to detect the MtCHIT5b:mCherry fusion protein (B), and for emission of teal fluorescent signals to visualize Rm41-mTFP1 bacteria (C). The overlay image (D) was obtained from (B, C) and indicates partial co-localization of MtCHIT5b:mCherry with teal fluorescent rhizobia in the infection pocket. (E-N) Transgenic roots expressing MtCHIT5b:GFP driven by the MtCHIT5b native promoter were inoculated with S. meliloti Rm41 (E, F) or S. meliloti Rm41-mCherry (G-N). Root hairs were analyzed under bright field conditions (E, G, K), for green fluorescent signals emitted by MtCHIT5b:GFP (F, H, L), and for red fluorescent signals emitted by Rm41-mCherry (I, M). The overlay images (J, N) indicate co-localization of MtCHIT5b:GFP with red fluorescent rhizobia in the infection pocket (J derived from H and I; N from L and M). (O-R) Similar analysis was performed for transgenic control roots expressing GFP driven by the CaMV 35S promoter. Roots were inoculated with S. meliloti Rm41-mCherry and root hairs were analyzed under bright field conditions (O) and for emission of green (P) or red (Q) fluorescent signals. The overlay image (R) was obtained from (P, Q). Bars = 10 μm in (A-J) and 25 μm in (K-R).
To explore a possible MtCHIT5b function in symbiosis, R108 wild-type plants and a MtCHIT5b mutant were compared. The Tnt1 retrotransposon mutant line NF16543 was obtained from the mutant collection at Oklahoma State University and homozygous mutant plants were identified by PCR analysis. The Tnt1 insertion of these chit5b mutant plants is located 401-bp downstream of the ATG start codon of MtCHIT5b. In parallel, nfh1-3b mutant plants were PCR-selected from the NF11260 line, which was previously used to obtain nfh1-3 (Cai et al., 2018). Compared to wild-type plants, chit5b plants showed significantly reduced MtCHIT5b transcript levels in roots treated with NodSm-IV(C16:2, S). Likewise, MtNFH1 expression was reduced in nfh1-3b roots (Supplementary Figure 6).
The mutant plants were then characterized for their ability to cleave NodSm-IV(C16:2, S) in the rhizosphere. Roots of 5-day-old seedlings of wild-type, chit5b and nfh1-3b plants were pretreated with NodSm-IV(C16:2, S) for 18 h to induce hydrolase activity. The roots were then examined for rhizospheric Nod factor cleavage activity using the same Nod factor as substrate. After incubation, the seedlings were removed, and the incubation medium was extracted with n-butanol. The conversion of NodSm-IV(C16:2, S) into II(C16:2) was then detected by reverse-phase HPLC. Under the used test conditions, chit5 and nfh1-3b plants showed both reduced Nod factor degradation activity as compared to wild-type plants. Quantitative analysis of HPLC peaks indicated that the sum of the chit5 and nfh1-3b activities was comparable to the activity of wild-type plants. Thus, the secreted MtCHIT5b and MtNFH1 proteins appear to contribute additively to the hydrolysis of Nod factors in the rhizosphere (Figure 5).
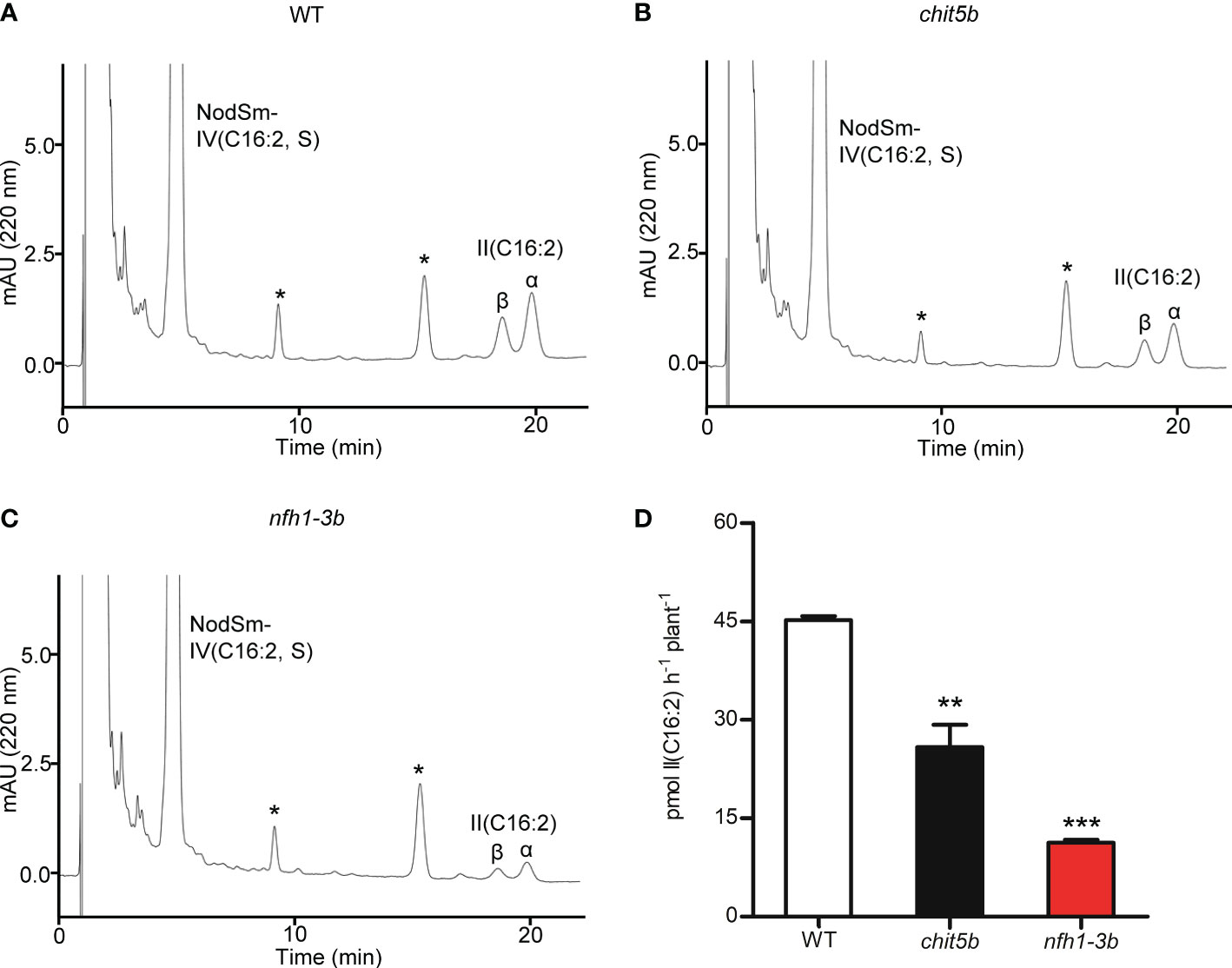
Figure 5 Nod factor degradation in the rhizosphere of M. truncatula wild-type, chit5b and nfh1-3b mutant plants. NodSm-IV (C16:2, S) hydrolysis was analyzed in the rhizosphere of R108 wild-type, chit5b and nfh1-3b seedlings. Roots were pretreated with 0.5 μM NodSm-IV (C16:2, S) for 18 h and then assayed for 2 h with 5.4 μM NodSm-IV (C16:2, S). Formation of II (C16:2) was analyzed by reverse-phase HPLC. (A-C) Representative chromatograms showing NodSm-IV(C16:2, S) and II (C16:2) from wild-type (A), chit5b (B) and nfh1-3b (C) seedlings. Asterisks indicate nonidentified root compounds. (D) Hydrolytic activities deduced from HPLC chromatograms. Data indicate means ± SE (3 seedlings per sample; n = 3). The asterisks indicate significantly reduced cleavage activity in chit5b and nfh1-3b compared to wild-type plants (Student’s t test; **, P< 0.01; ***, P< 0.001).
The wild-type and mutant plants were then inoculated with S. meliloti Rm41 to determine their nodulation phenotype. Compared to wild-type plants, chit5 and nfh1-3b showed a similar nodule number and nodule biomass at 3 wpi, indicating that these mutants were not impaired in nodule formation under the used test conditions (Supplementary Figure 7).
An RNA interference (RNAi) approach was used to silence MtCHIT5b and MtNFH1 in roots of M. truncatula plants. The coding sequences of MtCHIT5b and MtNFH1 show 80% nucleotide identity at the mRNA level. RNAi-silenced roots were generated by A. rhizogenes-mediated transformation using a binary vector containing an RNAi construct derived from the MtCHIT5b sequence in the sense and antisense orientations (named dsRNA1). In an additional experiment, an RNAi construct derived from the MtNFH1 sequence (named dsRNA2) was used (Tian et al., 2013). For detection of transformed roots, the binary vectors also contained an RFP expression cassette. RT-qPCR analysis of red fluorescent roots showed that expression of MtCHIT5b and MtNFH1 in wild-type plants was reduced in roots transformed with the dsRNA1 or dsRNA2 constructs. Due to a possible compensation effect, a slightly higher expression level in these roots was determined for the chitinase gene MtCHIT5a (Zhang et al., 2016), which is most related to MtCHIT5b/MtNFH1 (Figures 6A, B). Moreover, when treated with Nod factors, dsRNA2-transformed roots showed reduced transcript levels not only for MtCHIT5b but also for MtENOD11, suggesting that Nod factor signaling was partially impaired (Supplementary Figure 8). Wild-type and nfh1-3b roots, both transformed with the dsRNA1 construct, were then used for nodulation tests with S. meliloti Rm41. Non-transgenic roots lacking co-expressed RFP were identified by fluorescence microscopy and removed prior to inoculation. Compared to the empty vector control, the nodule frequency (number of nodules per root biomass) was 5.4-fold lower for dsRNA1-transformed wild-type roots and 3.4-fold lower for dsRNA1-transformed nfh1-3b roots (Figure 6C). Similarly, a 2.2-fold lower nodule frequency was observed for dsRNA2-transformed wild-type roots and a 5.9-fold lower one for dsRNA2-transformed chit5b roots (Figure 6D). When calculated on a whole-plant basis, nodule numbers and nodule biomass values were significantly reduced in dsRNA1- or dsRNA2-transformed roots of all examined genotypes (Supplementary Figure 9). Overall, the performed RNAi experiments indicate that simultaneous silencing of MtCHIT5b and MtNFH1 impairs nodule formation in M. truncatula plants.
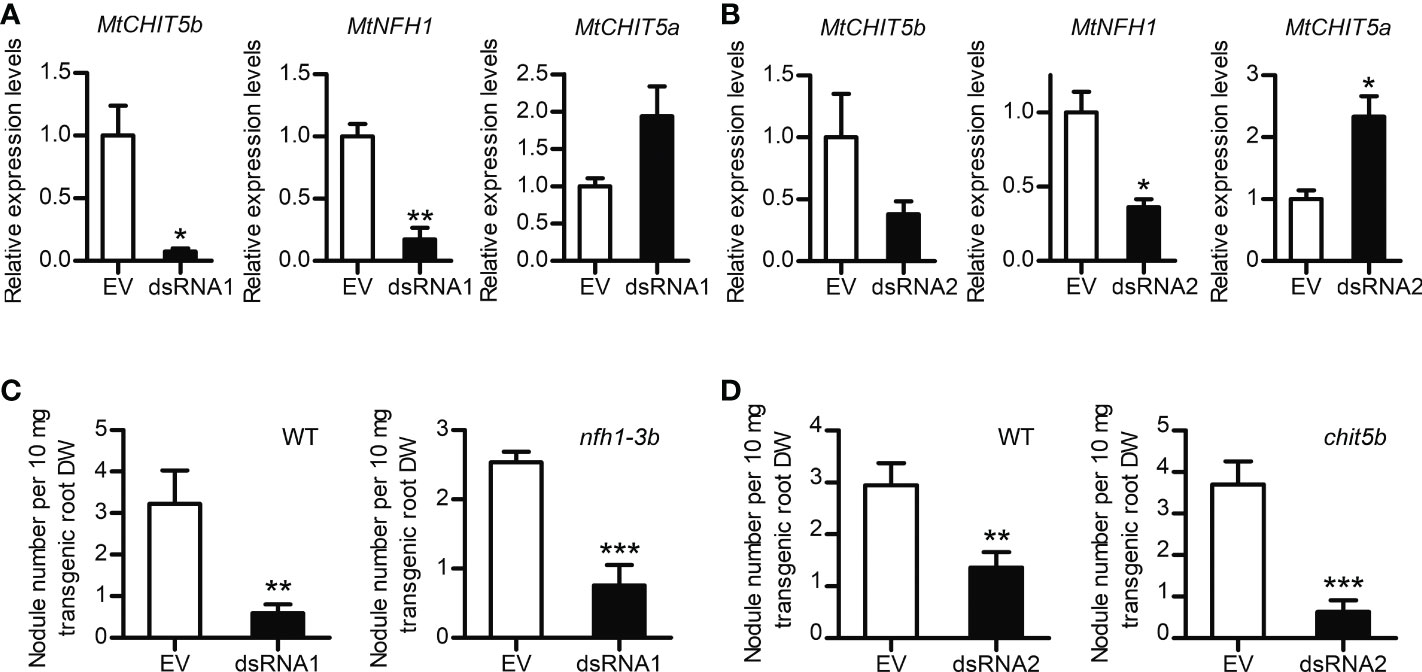
Figure 6 Nodulation phenotype of M. truncatula roots silenced in MtCHIT5b/MtNFH1 expression by the dsRNA1 and dsRNA2 constructs. (A, B) RT-qPCR analysis of MtCHIT5b, MtNFH1 and MtCHIT5a expression in R108 wild-type roots transformed with the dsRNA1 construct (A). or the dsRNA2 construct (B). Roots transformed with the empty vector (EV) containing RFP alone served as a control. Plants were harvested at 17 days post A. rhizogenes-mediated transformation and co-expression of RFP allowed identification of transgenic roots. Data indicate means ± SE of normalized values (n = 3; mean expression value of the empty vector control set to one) (Student’s t-test; *, P< 0.05; **, P< 0.01). (C, D) Nodule formation on roots of R108 wild-type (WT), nfh1-3b or chit5b mutant plants transformed with the dsRNA1 construct (C) or the dsRNA2 construct (D). Roots transformed with the EV served as a control. Plants with red fluorescent roots were inoculated with S. meliloti Rm41. At the time of harvest (4 wpi), the nodule frequency per root dry weight (DW) was determined for each plant. Data indicate means ± SE (Student’s t test; **, P< 0.01; ***, P< 0.001). Data shown in panel (C) were obtained from 16 transformed WT roots (EV, n = 7; dsRNA1, n = 9) and from 20 transformed nfh1-3b mutant roots (EV, n = 9; dsRNA1, n = 11). Data shown in panel D were obtained from 17 transformed WT roots (EV, n = 9; dsRNA2, n = 8) and 22 transformed chit5b mutant roots (EV, n = 10; dsRNA2, n = 12).
To study the effects of MtCHIT5b-overexpression, a binary vector containing the MtCHIT5b coding sequence driven by a Ubiquitin promoter from L. japonicus was constructed. A. rhizogenes strains carrying this vector or the empty vector (containing RFP) were then used to transform roots of M. truncatula R108 plants. RT-qPCR analysis indicated constitutive overexpression of MtCHIT5b in red fluorescent roots while expression of MtNFH1 and MtCHIT5a was not altered (Supplementary Figure 10A-C). When treated with Nod factors, the transformed roots also exhibited strong overexpression of MtCHIT5b, whereas transcript levels of MtENOD11 were lower than in control plants (Supplementary Figure 10D-E). These findings suggest that the applied Nod factors were rapidly degraded by the MtCHIT5b-overexpressing roots. For nodulation tests with S. meliloti Rm41, non-transgenic roots (lacking co-expressed RFP) were removed before inoculation. Compared to the empty vector control, the nodule frequency (number of nodules per root biomass) was reduced in plants with MtCHIT5b-overexpressing roots. When calculated on a whole-plant basis, plants with roots overexpressing MtCHIT5b formed fewer nodules and the nodule biomass was also lower (Figure 7). Thus, overexpression of MtCHIT5b and MtCHIT5b/MtNFH1 silencing in M. truncatula roots resulted in a similar symbiotic phenotype.
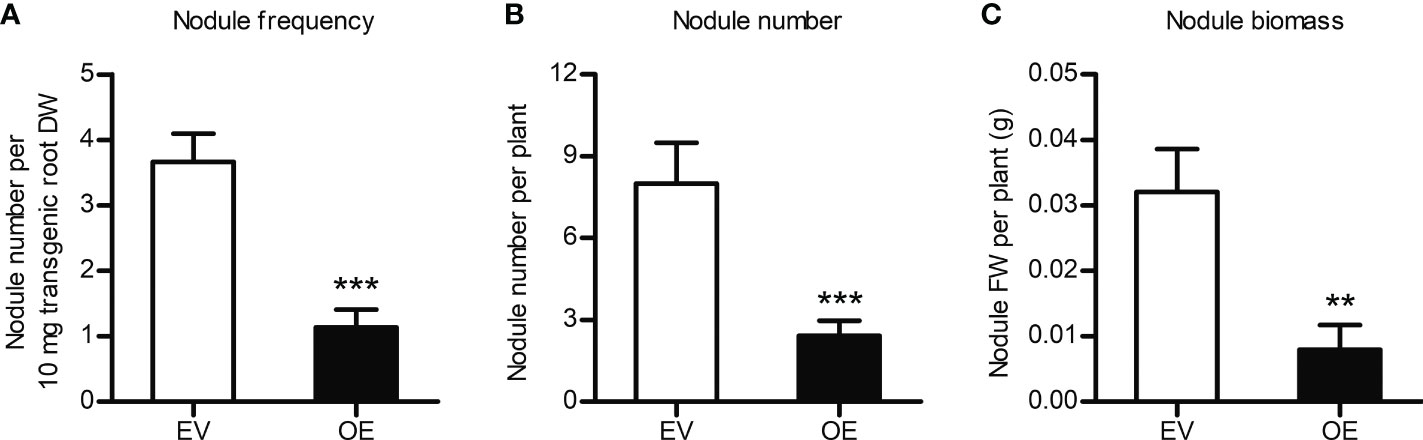
Figure 7 Constitutive overexpression of MtCHIT5b impairs nodulation of M. truncatula. R108 plants constitutively overexpressing MtCHIT5b in roots (OE) were obtained by A rhizogenes-mediated transformation. Plants with red fluorescent roots (co-expression of RFP) were inoculated with S. meliloti Rm41 and harvested at 4 wpi. Plants transformed with the empty vector containing RFP alone (EV) served as a control. Data indicate means ± SE (EV, n = 8; OE, n = 12) (Student’s t test; **, P< 0.01; ***, P< 0.001). (A) Number of formed nodules per root dry weight (DW). (B) Number of formed nodules per plant. (C) Biomass (fresh weight, FW) of formed nodules per plant.
We report in this study that MtCHIT5b expressed in E. coli possesses Nod factor-cleaving activity under newly established test conditions. Like recombinant MtNFH1 (Tian et al., 2013), MtCHIT5b was found to release lipodisaccharides from S. meliloti Nod factors in vitro. Detection of such an activity was surprising because MtCHIT5b expressed in E. coli was reported to be only active when the serine at the predicted substrate binding cleft is substituted to proline (Zhang et al., 2016). In the present study, the His-tagged MtCHIT5b protein (pI 9.12) was extracted from E. coli cells at pH 7.5, while pH 8.0 was used previously (Zhang et al., 2016). Moreover, His-tagged MtCHIT5b (eluted from the nickel column with imidazole) was supplemented with glycerol and immediately adjusted to pH 7.0 with acetate buffer. Subsequent Nod factor hydrolysis was performed in a potassium phosphate buffer (pH 7.0), whereas a sodium acetate buffer (pH 5.0) was used previously (Zhang et al., 2016). In comparison with MtNFH1 (Tian et al., 2013), MtCHIT5b showed lower Nod factor hydrolase activity but displayed higher affinity to Nod factors (lower Km values) under the test conditions used in this work.
Our tests with Nod factors added to roots of M. truncatula seedlings indicated that MtCHIT5b contributes to Nod factor hydrolysis in the rhizosphere, particularly when roots of 5-day-old seedlings are incubated in 1-mL syringes filled with BNM. Previous studies were performed with younger seedlings and used Jensen medium for such tests (Staehelin et al., 1994b; Staehelin et al., 1995; Tian et al., 2013; Cai et al., 2018). The sum of rhizospheric Nod factor hydrolase activity of the chit5b and nfh1-3b mutants could reach the activity level of wild-type plants, suggesting that MtCHIT5b and MtNFH1 are the two major Nod factor-cleaving enzymes in the rhizosphere. In this context, it is worth noting that no extracellular Nod factor cleaving activity was observed for roots of young nfh1-3 mutant seedlings treated with Nod factors (Cai et al., 2018), suggesting that MtCHIT5b expression is weak at such an early stage.
MtCHIT5b and MtNFH1 appear to have a similar gene expression pattern and subcellular localization. A Nod factor treatment of M. truncatula roots resulted in increased MtCHIT5b transcript accumulation within few hours, indicating that expression of MtCHIT5b is regulated by Nod factor signaling as reported for MtNFH1 previously (Cai et al., 2018). Induction of MtCHIT5b expression by Nod factors was stronger than that of MtNFH1 in our experiment (Figure 2A; Supplementary Figure 3), suggesting that the promoters of these genes differ in Nod factor-responsive elements. Furthermore, the results of our promoter-GUS fusion analysis indicated that seedlings inoculated with S. meliloti exhibit strongly induced MtCHIT5b promoter activity in emerging root hairs and on the tips of nodule primordia. Microscopic analysis of S. meliloti-infected seedlings expressing MtCHIT5b fused to a fluorescent protein showed strong fluorescence signals in the infection pocket of a curled root hair. A similar subcellular localization was observed for MtNFH1 (Cai et al., 2018). Thus, both enzymes appear to be co-localized with the rhizobial microcolony in the infection pocket. These observations suggest that Nod factor levels in the infection pocket are simultaneously reduced by both enzymes. In older M. truncatula plants, formed nodules showed relatively high expression levels of MtCHIT5b. These findings are largely in line with the RNA sequencing results obtained by Roux et al. (2014). The accumulation of MtCHIT5b transcripts in nodules indicates that the enzyme may participate in symbiotic processes in the infection zone of mature nodules.
RNAi-mediated silencing of MtCHIT5b/MtNFH1 led to a considerable reduction in nodule formation (Figure 6). Because of the high similarity between the MtCHIT5b/MtNFH1 nucleotide sequences, the dsRNA1 and dsRNA2 constructs caused silencing of both genes in the transformed roots. On the other hand, nodulation tests with chit5, nfh1-3b and wild-type plants showed no obvious differences, indicating that knockout of a single hydrolase gene was not sufficient to affect nodule formation under the test conditions used (Supplementary Figure 7). Overall, we conclude that MtCHIT5b and MtNFH1 show additive effects on Nod factor hydrolysis in the rhizosphere and cooperatively promote nodule formation. In other words, MtCHIT5b and MtNFH1 act redundantly with respect to nodule formation. We hypothesize that high concentrations of Nod factors impair nodule formation and that inactivation of excess amounts of Nod factors by MtCHIT5b and MtNFH1 promotes nodule formation. A permanent activation of Nod factor signaling by too high Nod factor concentrations could possibly impair establishment of symbiosis. Furthermore, it cannot be ruled out that high concentrations of Nod factors induce plant immunity and autoregulation responses which are known to negatively affect nodulation (Cao et al., 2017; Cai et al., 2018; Li et al., 2022). The experiments shown in Figure 2 indicate that MtCHIT5b expression was stimulated by inoculation with rhizobia and by Nod factor concentrations as low as 10-10 M. These findings indicate that the concentration of Nod factors produced by rhizobia in the rhizosphere was at least 10-10 M. We suggest that Nod factor levels in the infection pocket are much higher.
While MtCHIT5b and MtNFH1 must be co-silenced to negatively affect nodule formation in M. truncatula, mutation of the single chitinase gene LjCHIT5 was sufficient to impair nodule formation in L. japonicus (Malolepszy et al., 2018). LjCHIT5 can apparently hydrolyze Nod factors of M. loti (Malolepszy et al., 2018) whereas cleavage of S. meliloti Nod factors by recombinant LjCHIT5 was not observed in vitro (Zhang et al., 2016). These data indicate that Nod factor cleaving enzymes in different legumes possess different substrate specificities. It is worth noting in this context that LjCHIT5 is more related to MtCHIT5a than to the MtCHIT5b/MtNFH1 genes, which belong to a separate clade in a constructed phylogenetic tree (Supplementary Figure 11). Thus, it appears that M. truncatula and L. japonicus have recruited different class V chitinase genes for nodule symbiosis during evolution.
Roots constitutively overexpressing MtCHIT5b showed reduced nodule formation in our study. Overexpression of MtCHIT5b likely caused excessive hydrolysis of Nod factors, resulting in too low signal concentrations and thus reduced bacterial infection. Inoculation of plants with rhizobia expressing a bacterial chitinase gene and thus producing less Nod factor is another strategy to examine the symbiotic role of Nod factor levels. Using this approach, alfalfa plants inoculated with S. meliloti expressing the chiB gene of Serratia marcescens showed delayed nodulation (Krishnan et al., 1999). Taken together, these results indicate that the amount of Nod factors required for infection and subsequent nodulation must reach a minimum level.
In conclusion, the data obtained in this work show that MtCHIT5b contributes to hydrolysis of S. meliloti Nod factors. MtCHIT5b and MtNFH1 cooperate to adjust Nod factors to an optimal concentration range for nodulation.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
R-JL, C-XZ, S-YF and Y-HW performed research. JW and KM screened for Tnt1 insertion mutants and provided seeds. R-JL, C-XZ, S-YF, Y-HW, Z-PX and CS analyzed data. R-JL, Z-PX and CS conceived the work and wrote the paper. All authors contributed to the article and approved the submitted version.
This work was financially supported by the National Natural Science Foundation of China (grants 32161133026 and 31670241), the Science Foundation of the State Key Laboratory of Biocontrol and the Guangdong Provincial Key Laboratory of Plant Resources (grant 2020B1212060027).
We are grateful to Yu-Feng Wu, Qi Sun, Wen-Hui Hu, Chun-Lian Li, Christian Wagner and Yan Wang (Sun Yat-sen University) for their help with various aspects of this work. We thank Eva Kondorosi (Biological Research Center, Hungarian Academy of Sciences, Szeged, Hungary) for S. meliloti strain 1021 (pEK327) and Clare Gough (INRAE-CNRS, Castanet-Tolosan, France) for the dmi3-1 mutant.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1034230/full#supplementary-material
Ai, H. W., Henderson, J. N., Remington, S. J., Campbell, R. E. (2006). Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging. Biochem. J. 400, 531–540. doi: 10.1042/BJ20060874
Boisson-Dernier, A., Chabaud, M., Garcia, F., Bécard, G., Rosenberg, C., Barker, D. G. (2001). Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 14, 695–700. doi: 10.1094/MPMI.2001.14.6.695
Broughton, W. J., Dilworth, M. J. (1971). Control of leghaemoglobin synthesis in snake beans. Biochem. J. 125, 1075–1080. doi: 10.1042/bj1251075
Cai, J., Zhang, L. Y., Liu, W., Tian, Y., Xiong, J. S., Wang, Y. H., et al. (2018). Role of the Nod factor hydrolase MtNFH1 in regulating nod factor levels during rhizobial infection and in mature nodules of Medicago truncatula. Plant Cell 30, 397–414. doi: 10.1105/tpc.17.00420
Cao, Y., Halane, M. K., Gassmann, W., Stacey, G. (2017). The role of plant innate immunity in the legume-rhizobium symbiosis. Annu. Rev. Plant Biol. 68, 535–561. doi: 10.1146/annurev-arplant-042916-041030
Charron, D., Pingret, J. L., Chabaud, M., Journet, E. P., Barker, D. G. (2004). Pharmacological evidence that multiple phospholipid signaling pathways link Rhizobium nodulation factor perception in Medicago truncatula root hairs to intracellular responses, including Ca2+ spiking and specific ENOD gene expression. Plant Physiol. 136, 3582–3593. doi: 10.1104/pp.104.051110
Chen, S., Songkumarn, P., Liu, J., Wang, G. L. (2009). A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol. 150, 1111–1121. doi: 10.1104/pp.109.137125
Díaz, C. L., Grønlund, M., Schlaman, H. R., Spaink, H. P. (2005). “Induction of hairy roots for symbiotic gene expression studies,” in Lotus japonicus handbook. Ed. Márquez, A. J. (Dordrecht: Springer), 261–277.
Drula, E., Garron, M.-L., Dogan, S., Lombard, V., Henrissat, B., Terrapon, N. (2022). The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res. 50, D571–D577. doi: 10.1093/nar/gkab1045
Ehrhardt, D. W., Atkinson, E. M., Long, S. R. (1992). Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti Nod factors. Science 256, 998–1000. doi: 10.1126/science.10744524
Figurski, D. H., Helinski, D. R. (1979). Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76, 1648–1652. doi: 10.1073/pnas.76.4.1648
Fournier, J., Teillet, A., Chabaud, M., Ivanov, S., Genre, A., Limpens, E., et al. (2015). Remodeling of the infection chamber before infection thread formation reveals a two-step mechanism for rhizobial entry into the host legume root hair. Plant Physiol. 167, 1233–1242. doi: 10.1104/pp.114.253302
Giraud, E., Moulin, L., Vallenet, D., Barbe, V., Cytryn, E., Avarre, J. C., et al. (2007). Legumes symbioses: Absence of nod genes in photosynthetic bradyrhizobia. Science 316, 1307–1312. doi: 10.1126/science.1139548
Goormachtig, S., Lievens, S., De Velde, W. V., Van Montagu, M., Holsters, M. (1998). Srchi13, a novel early nodulin from Sesbania rostrata, is related to acidic class III chitinases. Plant Cell 10, 905–915. doi: 10.1105/tpc.10.6.905
Heidstra, R., Geurts, R., Franssen, H., Spaink, H. P., Van Kammen, A., Bisseling, T. (1994). Root hair deformation activity of nodulation factors and their fate on Vicia sativa. Plant Physiol. 105, 787–797. doi: 10.1104/pp.105.3.787
Kovach, M. E., Elzer, P. H., Hill, D. S., Robertson, G. T., Farris, M. A., Roop, R. M., et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176. doi: 10.1016/0378-1119(95)00584-1
Kovács, S., Kiss, E., Jenei, S., Fehér-Juhász, E., Kereszt, A., Endre, G. (2022). The Medicago truncatula IEF gene is crucial for the progression of bacterial infection during symbiosis. Mol. Plant Microbe Interact. 35, 401–415. doi: 10.1094/MPMI-11-21-0279-R
Krishnan, H. B., Kim, K. Y., Krishnan, A. H. (1999). Expression of a Serratia marcescens chitinase gene in Sinorhizobium fredii USDA191 and Sinorhizobium meliloti RCR2011 impedes soybean and alfalfa nodulation. Mol. Plant Microbe Interact. 12, 748–751. doi: 10.1094/MPMI.1999.12.8.748
Lei, M. J., Wang, Q., Li, X., Chen, A., Luo, L., Xie, Y., et al. (2015). The small GTPase ROP10 of Medicago truncatula is required for both tip growth of root hairs and Nod factor-induced root hair deformation. Plant Cell 27, 806–822. doi: 10.1105/tpc.114.135210
Lerouge, P., Roche, P., Faucher, C., Maillet, F., Truchet, G., Promé, J. C., et al. (1990). Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344, 781–784. doi: 10.1038/344781a0
Li, Y., Pei, Y., Shen, Y., Zhang, R., Kang, M., Ma, Y., et al. (2022). Progress in the self-regulation system in legume nodule development-AON (autoregulation of nodulation). Int. J. Mol. Sci. 23, 6676. doi: 10.3390/ijms23126676
Liu, C. W., Murray, J. D. (2016). The role of flavonoids in nodulation host-range specificity: an update. Plants 5, 33. doi: 10.3390/plants5030033
Li, R. J., Wang, Y. H., Cai, J., Liu, J., Xie, Z. P., Staehelin, C. (2019). “In vivo analysis of rhizosphere enzyme activities by the use of plastic syringes,” in Methods in rhizosphere biology research. Eds. Reinhardt, D., Sharma, A. K. (Singapore: Springer), 19–25.
Maekawa, T., Kusakabe, M., Shimoda, Y., Sato, S., Tabata, S., Murooka, Y., et al. (2008). Polyubiquitin promoter-based binary vectors for overexpression and gene silencing in Lotus japonicus. Mol. Plant Microbe Interact. 21, 375–382. doi: 10.1094/MPMI-21-4-0375
Malolepszy, A., Kelly, S., Sorensen, K. K., James, E. K., Kalisch, C., Bozsoki, Z., et al. (2018). A plant chitinase controls cortical infection thread progression and nitrogen-fixing symbiosis. Elife 7, e38874. doi: 10.7554/eLife.38874
Mergaert, P., Kereszt, A., Kondorosi, E. (2020). Gene expression in nitrogen-fixing symbiotic nodule cells in Medicago truncatula and other nodulating plants. Plant Cell 32, 42–68. doi: 10.1105/tpc.19.00494
Ovtsyna, A. O., Dolgikh, E. A., Kilanova, A. S., Tsyganov, V. E., Borisov, A. Y., Tikhonovich, I. A., et al. (2005). Nod factors induce Nod factor cleaving enzymes in pea roots. Genetic and pharmacological approaches indicate different activation mechanisms. Plant Physiol. 139, 1051–1064. doi: 10.1104/pp.105.061705
Ovtsyna, A. O., Schultze, M., Tikhonovich, I. A., Spaink, H. P., Kondorosi, E., Kondorosi, A., et al. (2000). Nod factors of Rhizobium leguminosarum bv. viciae and their fucosylated derivatives stimulate a Nod factor cleaving activity in pea roots and are hydrolyzed in vitro by plant chitinases at different rates. Mol. Plant Microbe Interact. 13, 799–807. doi: 10.1094/MPMI.2000.13.8.799
Perret, X., Staehelin, C., Broughton, W. J. (2000). Molecular basis of symbiotic promiscuity. Microbiol. Mol. Biol. Rev. 64, 180–201. doi: 10.1128/MMBR.64.1.180-201.2000
Pislariu, C. I., D. Murray, J., Wen, J., Cosson, V., Muni, R. R. D., Wang, M., et al. (2012). A Medicago truncatula tobacco retrotransposon insertion mutant collection with defects in nodule development and symbiotic nitrogen fixation. Plant Physiol. 159, 1686–1699. doi: 10.1104/pp.112.197061
Quandt, J., Hynes, M. F. (1993). Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127, 15–21. doi: 10.1016/0378-1119(93)90611-6
Quandt, H. J., Pühler, A., Broer, I. (1993). Transgenic root nodules of Vicia hirsuta: a fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol. Plant Microbe Interact. 6, 699–706. doi: 10.1094/MPMI-6-699
Radutoiu, S., Madsen, L. H., Madsen, E. B., Jurkiewicz, A., Fukai, E., Quistgaard, E. M., et al. (2007). LysM domains mediate lipochitin–oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J. 26, 3923–3935. doi: 10.1038/sj.emboj.7601826
Roux, B., Rodde, N., Jardinaud, M. F., Timmers, T., Sauviac, L., Cottret, L., et al. (2014). An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J. 77, 817–837. doi: 10.1111/tpj.12442
Roy, S., Liu, W., Nandety, R. S., Crook, A., Mysore, K. S., Pislariu, C. I., et al. (2020). Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 32, 15–41. doi: 10.1105/tpc.19.00279
Sagan, M., de Larambergue, H., Morandi, D. (1998). “Genetic analysis of symbiosis mutants in Medicago truncatula,” in Biological nitrogen fixation for the 21st century. Eds. Elmerich, C., Kondorosi, A., Newton, W. E. (Dordrecht, The Netherlands: Kluwer Academic Publishers), 317–318. doi: 10.1007/978-94-011-5159-7
Salzer, P., Feddermann, N., Wiemken, A., Boller, T., Staehelin, C. (2004). Sinorhizobium meliloti-induced chitinase gene expression in Medicago truncatula ecotype R108-1: a comparison between symbiosis-specific class V and defence-related class IV chitinases. Planta 219, 626–638. doi: 10.1007/s00425-004-1268-8
Schultze, M., Quiclet-Sire, B., Kondorosi, E., Virelizer, H., Glushka, J. N., Endre, G., et al. (1992). Rhizobium meliloti produces a family of sulfated lipooligosaccharides exhibiting different degrees of plant host specificity. Proc. Natl. Acad. Sci. U. S. A. 89, 192–196. doi: 10.1073/pnas.89.1.192
Schultze, M., Staehelin, C., Brunner, F., Genetet, I., Legrand, M., Fritig, B., et al. (1998). Plant chitinase/lysozyme isoforms show distinct substrate specificity and cleavage site preference towards lipochitooligosaccharide nod signals. Plant J. 16, 571–580. doi: 10.1046/j.1365-313x.1998.00326.x
Shaner, N. C., Campbell, R. E., Steinbach, P. A., Giepmans, B. N., Palmer, A. E., Tsien, R. Y. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22, 1567–1572. doi: 10.1038/nbt1037
Singh, S., Katzer, K., Lambert, J., Cerri, M., Parniske, M. (2014). CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 15, 139–152. doi: 10.1016/j.chom.2014.01.011
Staehelin, C., Granado, J., Müller, J., Wiemken, A., Mellor, R. B., Felix, G., et al. (1994a). Perception of Rhizobium nodulation factors by tomato cells and inactivation by root chitinases. Proc. Natl. Acad. Sci. U. S. A. 91, 2196–2200. doi: 10.1073/pnas.91.6.2196
Staehelin, C., Schultze, M., Kondorosi, E., Kondorosi, A. (1995). Lipo-chitooligosaccharide nodulation signals from Rhizobium meliloti induce their rapid degradation by the host plant alfalfa. Plant Physiol. 108, 1607–1614. doi: 10.1104/pp.108.4.1607
Staehelin, C., Schultze, M., Kondorosi, E., Mellor, R. B., Boller, T., Kondorosi, A. (1994b). Structural modifications in Rhizobium meliloti Nod factors influence their stability against hydrolysis by root chitinases. Plant J. 5, 319–330. doi: 10.1111/j.1365-313X.1994.00319.x
Tadege, M., Wen, J., He, J., Tu, H., Kwak, Y., Eschstruth, A., et al. (2008). Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 54, 335–347. doi: 10.1111/j.1365-313X.2008.03418.x
Tian, Y., Liu, W., Cai, J., Zhang, L. Y., Wong, K. B., Feddermann, N., et al. (2013). The nodulation factor hydrolase of Medicago truncatula: characterization of an enzyme specifically cleaving rhizobial nodulation signals. Plant Physiol. 163, 1179–1190. doi: 10.1104/pp.113.223966
Wang, T., Guo, J., Peng, Y., Lyu, X., Liu, B., Sun, S., et al. (2021). Light-induced mobile factors from shoots regulate rhizobium-triggered soybean root nodulation. Science 374, 65–71. doi: 10.1126/science.abh2890
Yang, J., Lan, L., Jin, Y., Yu, N., Wang, D., Wang, E. (2022). Mechanisms underlying legume–rhizobium symbioses. J. Integr. Plant Biol. 64, 244–267. doi: 10.1111/jipb.13207
Keywords: Medicago truncatula, Sinorhizobium meliloti, Nod factors, Nod factor hydrolysis, symbiosis
Citation: Li R-J, Zhang C-X, Fan S-Y, Wang Y-H, Wen J, Mysore KS, Xie Z-P and Staehelin C (2022) The Medicago truncatula hydrolase MtCHIT5b degrades Nod factors of Sinorhizobium meliloti and cooperates with MtNFH1 to regulate the nodule symbiosis. Front. Plant Sci. 13:1034230. doi: 10.3389/fpls.2022.1034230
Received: 01 September 2022; Accepted: 25 October 2022;
Published: 17 November 2022.
Edited by:
Attila Kereszt, Eötvös Lóránd Research Network, HungaryReviewed by:
Yongjia Zhong, Fujian Agriculture and Forestry University, ChinaCopyright © 2022 Li, Zhang, Fan, Wang, Wen, Mysore, Xie and Staehelin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhi-Ping Xie, eGllenBpbmdAbWFpbC5zeXN1LmVkdS5jbg==; Christian Staehelin, Y3N0QG1haWwuc3lzdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.