
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 22 November 2022
Sec. Plant Biotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1034215
This article is part of the Research Topic CRISPR-based genome editing for seed oil improvements in Brassica napus L. View all 6 articles
Seed oleic acid is an important quality trait sought in rapeseed breeding programs. Many methods exist to increase seed oleic acid content, such as the CRISPR/Cas9-mediated genome editing system, yet there is no report on seed oleic acid content improvement via this system’s precise editing of the double loci of BnFAD2. Here, a precise CRISPR/Cas9-mediated genome editing of the encoded double loci (A5 and C5) of BnFAD2 was established. The results demonstrated high efficiency of regeneration and transformation, with the rapeseed genotype screened in ratios of 20.18% and 85.46%, respectively. The total editing efficiency was 64.35%, whereas the single locus- and double locus-edited ratios were 21.58% and 78.42%, respectively. The relative proportion of oleic acid with other fatty acids in seed oil of mutants was significantly higher for those that underwent the editing on A5 copy than that on C5 copy, but it was still less than 80%. For double locus-edited mutants, their relative proportion of oleic acid was more than 85% in the T1 and T4 generations. A comparison of the sequences between the double locus-edited mutants and reference showed that no transgenic border sequences were detected from the transformed vector. Analysis of the BnFAD2 sequence on A5 and C5 at the mutated locus of double loci mutants uncovered evidence for base deletion and insertion, and combination. Further, no editing issue of FAD2 on the copy of A1 was detected on the three targeted editing regions. Seed yield, yield component, oil content, and relative proportion of oleic acid between one selected double loci-edited mutant and wild type were also compared. These results showed that although the number of siliques per plant of the wild type was significantly higher than those of the mutant, the differences in seed yield and oil content were not significant between them, albeit with the mutant having a markedly higher relative proportion of oleic acid. Altogether, our results confirmed that the established CRISPR/Cas9-mediated genome editing of double loci (A5 and C5) of the BnFAD2 can precisely edit the targeted genes, thereby enhancing the seed oleic acid content to a far greater extent than can a single locus-editing system.
Rapeseed (Brassica napus L.) is the next most important oil crop after soybean (Glycine max) in terms of both the land area under cultivation and seed yield worldwide. Rapeseed oil that is low in both erucic acid and glucosinolate is considered high-quality edible oil due to its high percentage (>90%) of C18 unsaturated fatty acids and other healthy secondary metabolites, such as vitamin E, sterol, and polyphenols (Barth, 2009; Chew, 2020; Tan et al., 2022). Among these unsaturated fatty acids, oleic acid, linoleic acid, and linolenic acid are the major constituents of seed oil. Oleic acid is recognized as a type of healthy fatty acid; for example, it can reduce blood pressure by regulating the membrane lipid structure to control G protein-mediated signaling (Terés et al., 2007; Terés et al., 2008). Furthermore, a higher oleic acid content of rapeseed oil can prolong its shelf time because of a lower ratio of oxidation in comparison with conventional oil having a low oleic acid and high linolenic acid content (Roszkowska et al., 2015; Cao et al., 2020; López et al., 2022). For example, Merrill et al. (2008) found that the oil stability index of high-oleic canola, high-oleic sunflower, and very high-oleic canola was higher than that of sunflower, soybean, canola, corn, partially hydrogenated soybean, and oleic safflower. Normally, oleic acid contributes about 55% to 65% of total fatty acids in most modern rapeseed varieties used for cultivation (Hu et al., 2006). Rapeseed has a much lower oleic acid percentage than oil crops such as soybean, sunflower, and peanut (Dhakal et al., 2014; Alberio et al., 2016; Konuskan et al., 2019). Compared with other oil crops, the advantages of rapeseed oil with high oleic acid percentage are as follows: firstly, its larger cultivation area and higher oil yield can offset the shortage due to the low oil yield of other oil crops; secondly, its higher percentage of the high level of oleic acid that is a monounsaturated fatty acid relative to polyunsaturated fatty acid, linoleic acid, and linolenic acid would make it healthier. Therefore, increasing the amount of seed oleic acid in rapeseed is an imperative breeding objective from the nutrition perspective.
Understanding the genetic control of oleic acid content is crucial to the breeding program for high oleic acid improvement in rapeseed. Previous investigations suggested that rapeseed had high heritabilities (h2 = 0.94) for the seed oleic acid content within both high and low oleic acid types (Schierholt and Becker, 2001). This suggests that the seed oleic acid of rapeseed might be controlled by a handful of genes or some major effective loci (Velasco et al., 2003), which also suggests the strong possibility to breed high oleic acid in rapeseed via biotechnological manipulation of just a few genes or loci. One attempted way to breed a high oleic acid variety relied on ethyl methanesulfonate (EMS) mutation (Spasibionek, 2006; Lee et al., 2018). The seed oleic acid content can reportedly reach at least 85% in the F4 generation after EMS mutagenesis; however, low amounts of seed were also observed despite abundant pollen, suggesting that fertility might be impaired by this method (Auld et al., 1992).
Because the EMS mutagenesis method depends on a large population for targeted trait screening, leading to laborious work for researchers, other methods are sought to be explored. Genetic engineering achieved via RNA interference (RNAi) knockdown and genome editing of critical genes controlling seed oleic acid content, to augment the seed oleic acid content, could offer an effective way forward. The FATTY ACID ELONGASE (FAE) gene is responsible for erucic acid content in rapeseed (Kanrar et al., 2006; Wu et al., 2015). Previous investigations demonstrated that fatty acid elongation mutants in rapeseed and Arabidopsis affected elongation from C18:1 to C20:1 and C20:1 to C22:1; (Kunst et al., 1992; Roscoe et al., 2001). Thus, it is possible for the genetic manipulation of BnFAE to increase C18 lipid species. Shi et al. (2015) performed RNAi knockdown of BnFAE, which resulted in the relative proportion of oleic acid with other fatty acids in seed oil increasing from 21.3% to 60.0% while decreasing the relative proportion of erucic acid from 42.5% to 2.1%. In order to further increase the relative proportion of seed oleic acid, researchers created both BnFAE1-Ri and BnFAD2-Ri lines. UnlikeBnFAE1-Ri, the relative proportion of seed oleic acid of BnFAD2-Ri increased modestly, from 21.3% to 31.9%, whereas the relative proportion of erucic acid content slightly rose from 42.3% to 45.6%. Interestingly, following hybridization between the two RNAi knockdown mutants, a higher relative proportion of oleic acid was obtained that reached 80.3% in the BnFAD2-Ri/BnFAE1-Ri lines (Peng et al., 2010; Shi et al., 2017). In addition to FAE1, Zhang et al. (2018) identified a CCCH-type transcription factor (BnZFP1) from a rapeseed subtractive hybridization library that correlated well with seed oleic acid content. Overexpressing BnZFP1 in rapeseed led to its proportion of seed oleic acid increasing to 65.8% to 75.7%, higher than the wild type’s (63.7%). However, the seed oleic acid content was significantly decreased by 4.8% when the BnZFP1 expression was interrupted in transgenic plants.
Apart from BnFAE1 and BnZFP1, the BnFAD2 gene has been studied extensively in oil crops during fatty acid biosynthesis and is directly associated with oleic acid content (Fulvio et al., 2022; Razeghi-Jahromi et al., 2022). We know that disruption of FAD2 results in a greater oleic acid content of different tissues, such as roots, leaves, and seed in Arabidopsis (Okuley et al., 1994). In rapeseed, a major locus responsible for seed oleic acid content on chromosome A5 was found and mapped, it accounting for ca. 83% of the total variation. Furthermore, BnFAD2 had four copies: BnaFAD2.A1, BnaFAD2.A5, BnaFAD2.C1, and BnaFAD2.C5 (Yang et al., 2012). Other two loci on chromosomes 3 and 9 were also detected (Zhao et al., 2008; Zhao et al., 2019). Till now, it remains unclear how many minor-effect loci controlling seed oleic acid content exist in rapeseed, but this is not important because of major-effect loci found according to a previous investigation (Yang et al., 2012). Therefore, the selection of a targeted locus is essential when pursuing the high oleic acid content improvement of rapeseed. Sivaraman et al. (2004) developed a high-oleic acid content line in a zero-erucic acid line of mustard (Brassica juncea L.) by antisense suppression of the B. rapa FAD2 gene. The relative proportion of seed oleic acid increased from 39.5%~53.4% to 69.0%~74.8%.
Recently, however, genomic editing technology has gained popularity (Bahariah et al., 2021; Vu et al., 2021). Okuzaki et al. (2018) performed CRISPR-Cas9-mediated genome editing of target gene BnFAD2_Aa, which resulted in a higher relative proportion of oleic acid, rising from 74.6% in the wild type to 80.0% in the best-performing line. Later, Huang et al. (2020) developed genome editing with the CRISPR-Cas9 system to simultaneously modify BnFAD2 multiple copies in order to further increase the seed oleic acid content; it generally reached 75.0%–80.0%, with only two lines having a seed oleic acid content greater than 80.0%. The reason why this multiple modification yielded only two lines with an oleic acid composition of more than 80% is still unknown. In this study, we hypothesized that, compared with editing of a single locus, double editing of the A5 and C5 copies of BnFAD2 via the CRISPR/Cas 9 system would significantly increase the seed oleic acid content of rapeseed.
Here, we employed the CRIPR/Cas9 system to mutate A5 and C5 copies of BnaFAD2 in rapeseed. Successful mutagenesis resulted in higher seed oleic acid content of targeted mutants at BnaFAD2 vis-à-vis the wild type. Further, the mutated trait can be stably inherited with no significant differences in seed yield and oil content. Our results suggest that precise modification of the double loci of BnFAD2 can be applied in the CRISPR/Cas9 system to create new germplasms for the purpose of breeding plants with high seed oleic acid content.
The semi-winter Brassica napus line B57-1 was used for Agrobacterium-mediated transformation. This line has proven itself highly reliable and effective in the production of transgenic plants. It has high seed oil content (51.23%) and low relative proportion of seed oleic acid (67.25%).
The transformed plants (T0) and subsequent plants with different generations (T1 and T4) for seed fatty acid analysis were grown in a greenhouse. Because there was only one plant of each mutated line in the T0 generation, there were no replications in this generation. From T1 to T4, the mutated plants and wild type (B57-1) were planted in a completely randomized block design with three replications. A mutant line of #289 was selected to compare with the wild type on the yield, yield component, oil content, and relative proportion of oleic acid with other fatty acids in seed oil. Both lines were in a completely randomized block design with three replications as well in greenhouse conditions. The area of the block was 6 m2 with a population density of 15,000 plant ha-1.
All plants were planted from October 5th to 10th in each generation. Soil was only watered before seeding, and no irrigation was employed after seeding. Plants were thinned into one plant when they were at the three-true leaf stage. Low release compound fertilizer (N-P2O5-K2O: 20-7-8, Hubei Yishizhuang Agricultural Technology Company Ltd., Yichang, China) was applied in a dose of 1,500 kg ha-1 as basal fertilizer before seeding. No top dressing was performed. The mean temperature (outside of the greenhouse because we did not record the temperature in the greenhouse) is illustrated in Figure S1 from 2019 to 2022 during the rapeseed growth season.
Three sgRNAs T1, T2, and T3, targeting both BnaA05.FAD2 and BnaC05.FAD2, were designed online (http://crispr.hzau.edu.cn/CRISPR2). The binary pYLCRIPSR/Cas9 multiplex genome targeting vector system, kindly provided by Prof. Yaoguang Liu (South China Agricultural University), included both pYLCRISPR/Cas9Pubi-H and pYLCRISPR/Cas9P35S-H and three plasmids with sgRNA cassettes driven by the promoters of AtU3d, AtU3b, and AtU6-1; this system was used for the construct assembly following a previously described methodology (Ma et al., 2015). The oligos used to construct the sgRNA vectors are listed in Table S1. The resulting construct contained a Cas9 expression cassette and sgRNA expression cassettes with target sequences and a hygromycin resistance cassette (Figure 1).

Figure 1 The binary construct P35S:Cas9-BnFAD2 with three sgRNAs driven by the U3d, U3b, and U6-1 promoters from Arabidopsis.
After verifying the fused constructs by sequencing, the resulting constructs were transformed into B. napus via the Agrobacterium tumefaciens-mediated hypocotyl method (Zhou et al., 2002). Regenerated seedlings (T0 lines) were selected according to their resistance to hygromycin and confirmed by PCR, the latter using the flanking primers PB-L and PB-R (Table S1) (Ma et al., 2015).
PCR was performed to amplify the genomic region surrounding the CRISPR target sites, using specific primers (Table S1), and the PCR fragments were directly sequenced to identify the mutations. The ensuing sequences were read through sequencing chromatograms and compared with wild-type (WT) sequences to detect the presence of any indels.
The T1 mutant lines were grown in a greenhouse. Leaves from each individual were collected to extract their DNA for PCR amplifications using the flanking primers PB-L and PB-R of vector (Table S1). Those without amplified bands of individuals were selected for further genotyping; the fragments including the indels were amplified and sequenced using specific primers (Table S1).
Total genomic DNAs were extracted from leaf material with a plant genomic DNA kit (Tiangen Biotech, Beijing, China) by following its instructions. The DNA concentration (>50 ng μl-1) was measured by a NanoDrop spectrophotometer, and fragmentation was achieved by applying sonication. Then, the fragmented DNA was purified and end-repaired and its sizes determined by gel electrophoresis. Paired-end libraries with insert sizes of 350 bp were prepared following Illumina’s standard genomic DNA library preparation procedure. Next, a control library quality for sequencing was assembled. We sequenced (based on sequencing by synthesis [SBS] technology) the whole genome of rapeseed using the Illumina NovaSeq 6000 platform (Illumina, USA).
Paired-end Illumina raw reads were cleaned by removing adaptors and barcodes and then quality-filtered using the Trimmomatic tool. Reads were trimmed from both ends, and individual bases with a Phred quality score <20 were discarded, as were any reads having more than three consecutive uncalled bases. Entire reads with a median quality score lower than 21 or less than 40 bp in length after trimming were also discarded. After completing the quality filtering process, the reads were mapped to the reference sequence (Table S2), using Bowtie2 v.2.2.6. Then, all putative FAD2 reads mapped to the reference sequence as mentioned above were used for the de novo assembly to reconstruct the FAD2 gene sequence using SPAdes 3.6.1 with iterative K-mer sizes of 55, 87, and 121. De novo assembled contigs were concatenated into larger contigs by Sequencher 5.3.2 software (Gene Codes Inc., Ann Arbor, MI, USA), which based on at least a 20-bp overlap and 98% similarity. A “genome walking” technique, using the Unix “grep” function, was implemented to find all remaining reads that could fill any gaps between contigs that did not assemble in the initial set of analyses.
To carry out the multiple-sequence alignment, DNAMAN sequence analysis software was used. The consensus sequence among all sequences is presented in this study.
Selected double-locus-edited mutants and wild types were sequenced for their BnFAD2 on each A1 and C1copy. These sequences were matched to the BnFAD2 gene with three edited regions in both A1 and C1 copies.
Seeds from five double-locus-edited mutants were collected. The samples were ground into powder with liquid nitrogen. Total RNA was extracted using the RNeasy Plant Mini Kit (QIAGEN, Hilden, Germany) including treatment with DNase I (Takara, Dalian, China). The RNA was used for cDNA synthesis using a two-step RT-PCR kit (Takara, Dalian, China) according to the manufacturer’s instructions. The resultant cDNA was subjected to reverse transcription-polymerase chain reaction (qRT-PCR) analysis (LightCycler® 480 System, Roche, Basel, Switzerland). The primers of BnFAD2on A1, C1, A5, and C5 copies and BnACTIN (housekeeping gene) are listed in Table S1. The transcript amount was calculated according to Livak and Schmittgen’s method (2001).
The WT and different-type homozygous T0, T1, and T4 mutant lines were grown in a greenhouse, each with three biological replicates except for the T0 generation. Seeds were harvested to analyze the composition of fatty acids through gas chromatography, as described by Lian et al. (2017).
Five plants were randomly selected excluded from border plants for yield component analysis in each block. Yield components including the number of branches and siliques and seed number of per silique were recorded. A 1,000-seed weight was weighed in a balance. Seed yield was recorded in a core area of each block. Seed oil content was analyzed by the near-infrared method.
Statistical analysis of the data was implemented in SPSS software (v17.0, Chicago, IL, USA). Analyses of variance (ANOVAs) of randomized complete block design were performed on the relative proportion of the seed fatty acid profile among edited mutants and wild type in T1 and T4 generation, respectively; the transcript level relative to BnACTIN of four copies of the BnFAD2 gene on A1, C1, A5, and C5 chromosome; and seed yield, yield components, oil content, and proportion of oleic acid between mutant (#289) and wild types. Mean values of the above experiment were distinguished for significant differences using Duncan’s test at an alpha probability of 0.05.
In this study, we firstly compared the regeneration efficiency of rapeseed with 23 germplasms. These results showed that the regeneration efficiency varied from 0% up to 20.18% (Table 1). Eight germplasms had zero regeneration, and three germplasms had a regeneration ratio higher than 10.00%. Therefore, we focused on B57-1, whose hypocotyl served as explant tissue for genetic transformation because of its high regeneration (Table 1, Figure 2A; Figure S2). We further analyzed the number of positive mutants using the CRISPR-Cas9-mediated genome editing of BnFAD2. In all, 282 transformants were harvested, consisting of 41 (14.54%) negative mutants and 241 (85.56%) positive mutants (Figure 2B). The latter result indicated that the CRIPSR-Cas9-mediated genome editing of BnFAD2 was successful and highly efficient.
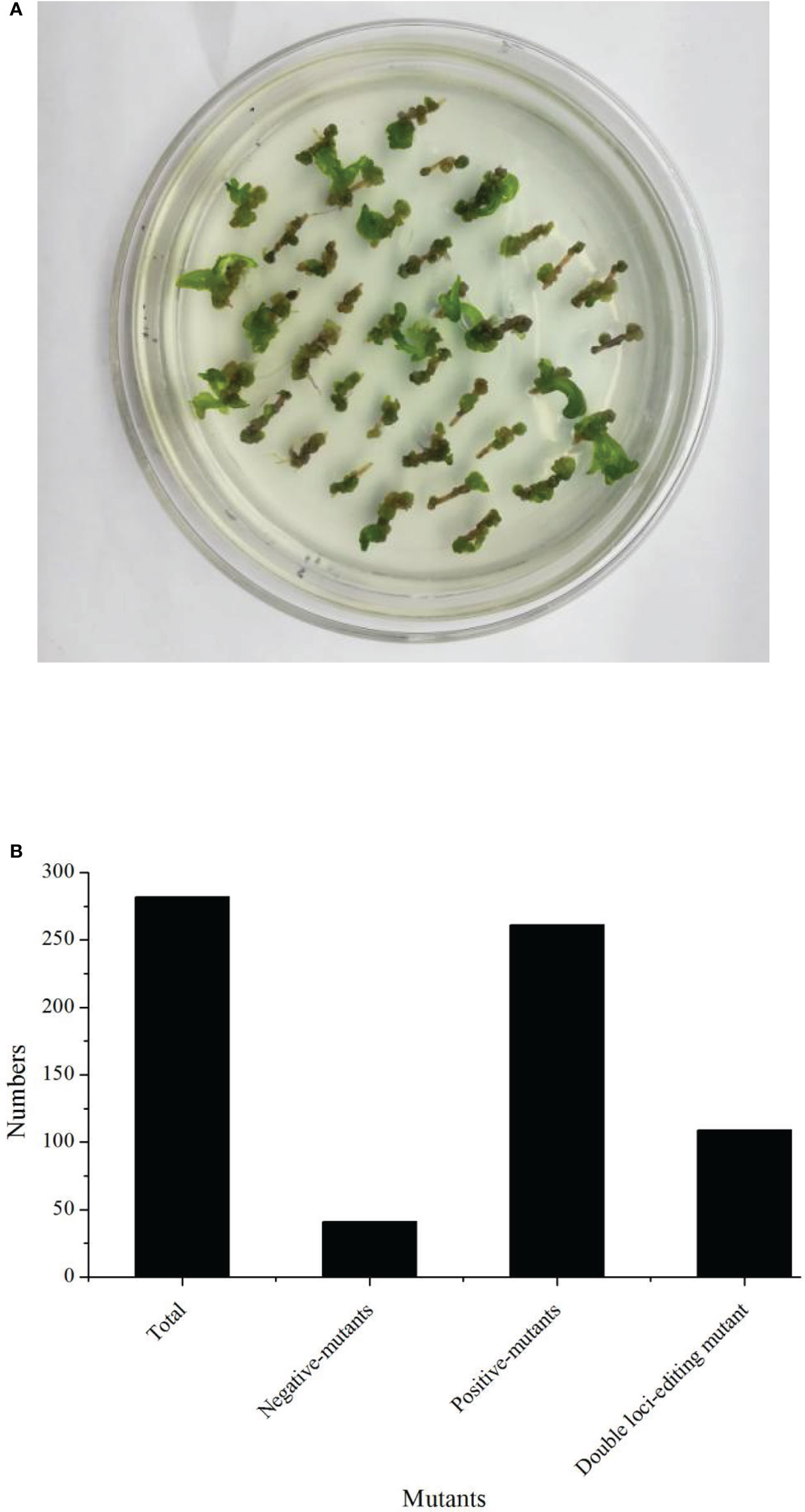
Figure 2 Regeneration status and transformation efficiency of the genotype B57-1. (A) Callus from the hypocotyl of B57-1. (B) Numbers of plants that underwent total transformation, negative- and positive-mutants.
Four editing types could be identified in the current CRIPSR-Cas9-mediated genome editing of the BnFAD2 system. The first type was the wild type, in that BnFAD2 went unedited by the CRISPR-Cas9-mediated genome editing system (Figure 3A). In this type, all peaks of different kinds of base were sharp and clear. The second type was that cells partially edited (Figure 3B). In this type, all the sharp and clear peaks the same as the wild type were observed, which was dominant. Further, some unequal small peaks were observed. As a callus develops into plant, the phenotype is similar to the wild type but some mutated tissues exist. Thus, the plant is called chimera. The third type was homozygous, in that the copy was simultaneously edited in a homologous chromosome (Figure 3C). In this type, mutated bases can be found in two homologous chromosomes as compared with the wild type (Figure 3C). The fourth was heterozygous, whereby a homologous chromosome was mutated but another one was not mutated (Figure 3D). In this type, mutated bases can be found in one of the homologous chromosomes. The sequence of another homologous chromosome was the same as the wild type (Figure 3D).
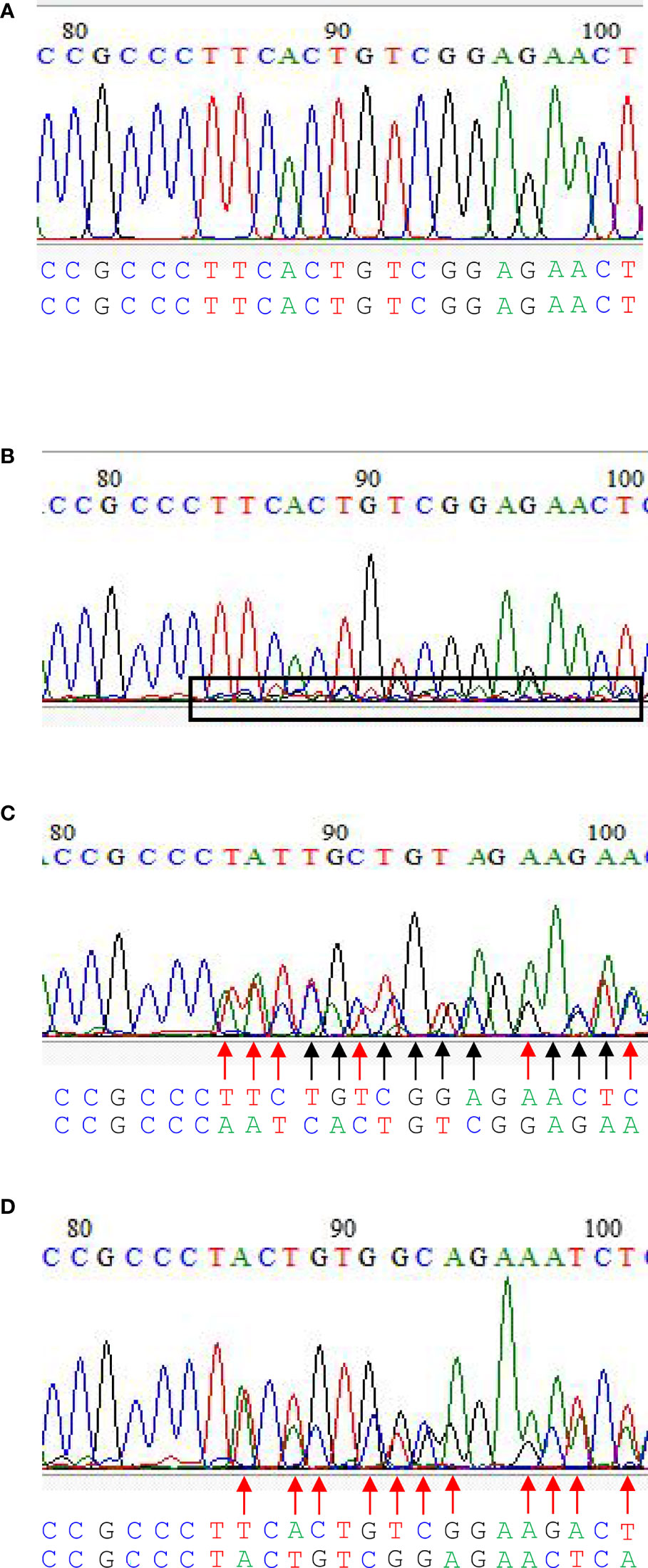
Figure 3 The editing types in CRIPSR/Cas9-mediated genome editing of the double loci of BnFAD2. (A) Wild type, (B) cells partially edited, (C) homozygous edited, and (D) heterozygous edited. In (B), all the sharp and clear peaks similar to the wild type were observed, which were dominant. Further, some unequal small peaks were observed within the box with a black border. As a callus develops into plant, the phenotype is similar to the wild type, but some mutated tissues exist. Sequences below the figure with the black arrow indicate the base mutated in both homologous chromosomes, while those with the red arrow indicate the base mutated only in one of the homologous chromosomes. Homozygous edited type showed base mutations on both homologous chromosomes compared with the wild type (C). Heterozygous edited type showed base mutations only on one of the homologous chromosome.
Overall, 216 plants were sequenced to determine the editing efficiency of the CRIPSR-Cas9-mediated genome editing of the BnFAD2 system. Results showed that 77 lines were not edited by this system (Figure 4A). The number of single locus- and double-locus-edited lines was 30 and 109, respectively. Therefore, the total editing efficiency of the CRIPSR-Cas9-mediated genome editing of the BnFAD2 system was 64.35%, for which the single- and double-locus editing efficiencies were 21.58% and 78.42%, respectively (Figure 4A). The PCR results for the edited lines are depicted in Figure 4B. Evidently, the edited lines were detected in the targeted band after the electrophoresis.
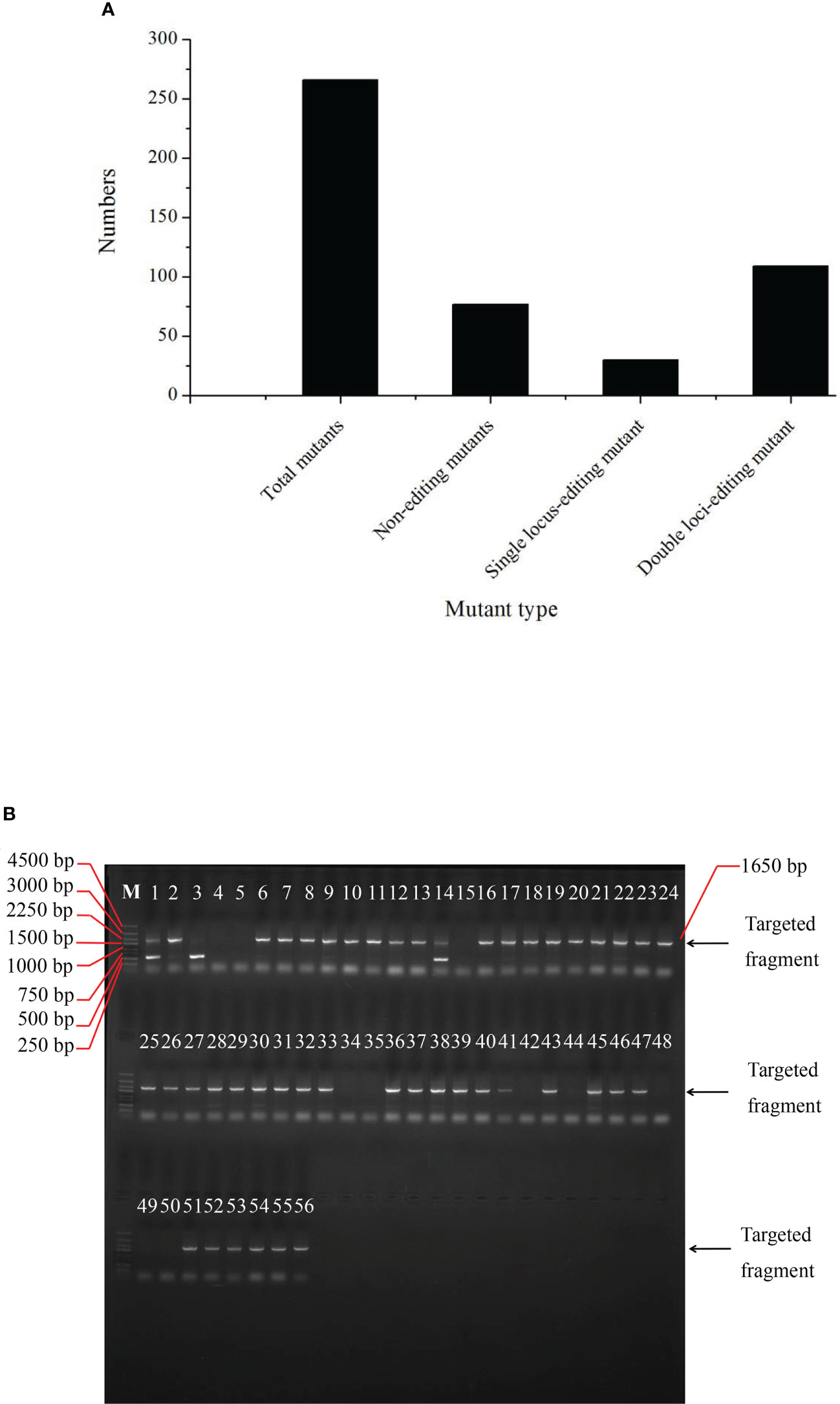
Figure 4 Edited mutant types in the T0 generation. (A) Numbers of total mutants, non-edited mutant, single locus-edited mutant, and double loci-edited mutant. (B) PCR amplification results of T0 generation indicating whether the plants numbered from 1 through 56 were edited.
We then analyzed the seed fatty acid profile in T0 seeds of the wild-type and genome-edited mutants (Table 2). The relative proportion of oleic acid with other fatty acids in seed oil was significantly increased in the genome-edited lines whether the line was single locus- or double loci-edited. However, the effect of double-locus-edited mutants on the relative proportion of seed oleic acid was stronger than that of single-locus-edited lines. The relative proportion of seed oleic acid in three single-locus-edited lines, #122, #216, and #294, were below 80%, but it respectively increased by 8.94%, 13.90%, and 15.32%, when compared with the wild type. Furthermore, the two lines #216 and #294 edited on the A5 copy had a similar relative proportion of seed oleic acid that surpassed that of line #122, whose C5 copy had been edited. This suggested that the copy of BnFAD2 on A5 exerted a greater effect upon seed oleic acid than did the C5 copy. Regarding the double-locus-edited lines, their relative proportion of seed oleic acid spanned 86.99% to 89.61%. The average relative proportion of seed oleic acid of 13 lines was increased by 28.96% vis-à-vis the wild type. As for other fatty acid compounds, the relative proportion of palmitic acid, stearic acid, arachidic acid, and arachidonic acid was each slightly affected by CRIPSR-Cas9-mediated genome editing of the BnFAD2 system. Furthermore, the erucic acid contents in all mutant lines and wild types were undetectable. The relative proportion of linoleic acid and linolenic acid decreased drastically in the mutants especially for double-locus-edited ones. Averagely, the relative proportions of linoleic acid and linolenic acid were decreased by 81.93% and 63.29%, respectively. Clearly, the bolstered seed oleic acid content arose from the reduction in linolenic acid and linoleic acid contents, albeit differently between single locus- and double loci-edited lines due to the disruption of BnFAD2. For the effect of mutation type on seed fatty acid profile in edited mutants, no difference in the proportion of seed oleic acid, linolenic acid, and linoleic acid was observed between a mutant (#202) with a heterozygous type in C5 copy and a wild type. Further, heterozygous type in both-locus (A5/C5)-edited mutants (#106, #112, and #129) showed a decreased proportion of oleic acid, which ranged from 75.97% to 81.86%. The proportion of oleic acid was higher than the wild type but substantially reduced as compared to the homozygous ones (Table 2). The result indicated that homozygous mutants contributed more to the percent increase in seed oleic acid compared to the wild type.
To further confirm whether the mutants can be stably inherited from the generations of T0, the relative proportion of seed oleic acid was measured in the T1 and T4 generations (Figures 5 and 6; Table S3 and S4). According to these results, the relative proportion of seed oleic acid of both the single-locus- and double-locus-edited lines showed that this trait could be stably inherited. Yet, the relative proportion of seed oleic acid of single locus-edited lines decreased more in the T1 generation, by 5.27%, 4.29%, and 2.44% in lines #122, #215, and #294, respectively, but only slightly from the T1 to T4 generation in those lines. In stark contrast to single-locus-edited lines, double-locus-edited lines exhibited a very stable level of the relative proportion of seed oleic acid with evidence of little variation. The results suggested that the double-locus mutation was more advantageous to the genetic stability of seed oleic acid content than was single locus editing in this study.
To further understand the types of editing that give rise to the mutants, five double-edited mutants were selected and their sequences of BnaFAD2 copies on the A5 and C5 chromosomes were assayed (Table 3). Three editing types of the BnaFAD2 copy on chromosome A5 were distinguishable. The first one entailed a deletion of bases with different base numbers, such as for lines #206 and #342. The second type was insertion of bases such as line #238, at which a base, T, was inserted into one of the homologous chromosomes. The third type was a hybrid, featuring both insertion and deletion of bases such as in lines #230 and #240. The #230 line had the same editing mode whereas different editing modes were discernible in the #240 line, consisting of deletion and insertion/deletion.
Four editing types of the BnaFAD2 copy on chromosome C5 were found. The first was deletion of bases with different base numbers, such as for lines #206 and #238. The second type was the hybrid (both insertion and deletion of bases) such as the #230 line. The third was large-fragment deletion, such as in the case of line #240 for which a fragment having 348 bases was deleted. The fourth type was large-fragment insertion, for example, as happened in the #342 line, with the insertion of a fragment comprising 400 bases. These results suggested that not solely one type but rather multiple editing modes can generate a seed high oleic acid content.
To confirm whether the mutants from T1 generation contained the fragment of the transformed vector, five mutants were selected (#407, #397, #486, #510, and #382) for re-sequencing analysis. Results showed that although sequencing reads can be matched to the sequence of the targeted gene, most could only match to ca. the 1,287th position therein. Furthermore, at this position, only 30 bp can be reliably matched. Hence, it may be concluded that no reads can be matched to the vector sequence (Table 4). The finding suggested that the segregation of mutants happened without the fragment from the transformed vector.
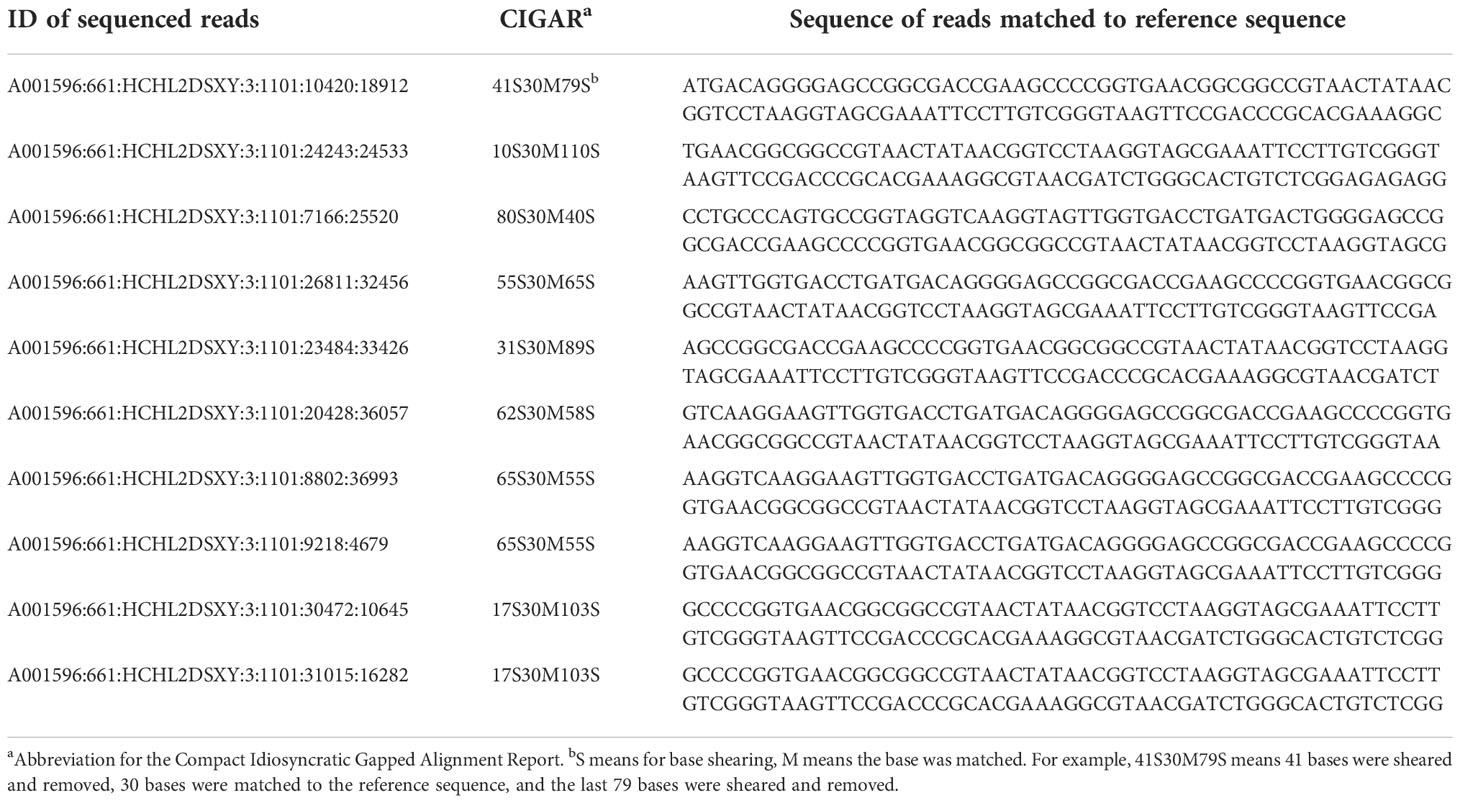
Table 4 Transgenic analysis on the mutants via matching between sequenced reads and the reference sequence at the 1,287th position.
Because there were four copies of BnFAD2 identified in previous research (Huang et al., 2020), their sequences in the selected lines were further compared to confirm whether the copies of BnFAD2 on A1 and C1 were indeed edited. Results showed that the three editing targeted regions lacked any sequence changes, suggesting no editing occurred on A1 (Table 5; Figure S3). Concerning the copy on the C1 locus, the two editing targeted regions from positions 236 to 258 and from positions 728 to 750 both had one base in each line that differed from the sequence of BnFAD2 (Table 6; Figure S4). However, this differing base in all edited lines was the same as that in the wild type. Therefore, we may tentatively presume no editing occurred in either region. For another region, this spanning the positions 575 to 596, a deletion/insertion occurred in two places, at the 575th and 592nd positions. At the 575th position, three mutant lines incurred a deletion of “C”, the same as for the wild type. Hence, this position was difficult to judge whether it was an editing position. However, at the 592nd position, in two lines there was an “A” insertion. This position was regarded as edited position. At the 596th position, two lines showed the base substitution of “C” to “T” and this position could be regarded as edited position.

Table 5 Sequence comparison among references of BnFAD2, wild type, and mutants in the targeted editing region of the A1 copy.

Table 6 Sequence comparison among references of BnFAD2, wild type, and mutants in the targeted editing region of the C1 copy.
To confirm whether the expression of the edited gene, BnFAD2, was affected in A1, C1, A5, and C5 copies in seeds of five selected double-edited mutants using wild type as a control, a qRT-PCR analysis was performed (Figure 7). Results showed that the difference in the transcript level of BnFAD2 in the A5 and C5 copies was not significant in the wild type. The same trend was found in the A1 and C1 copies. However, the transcript levels of BnFAD2 in both A1 and C1 copies were significantly lower than those in the A5 and C5 copies in the wild type. Interestingly, the transcript levels of BnFAD2 of four copies in all mutants were significantly reduced as compared with the wild type. The results suggested that the expression of the BnFAD2 gene was markedly inhibited after the gene was edited.
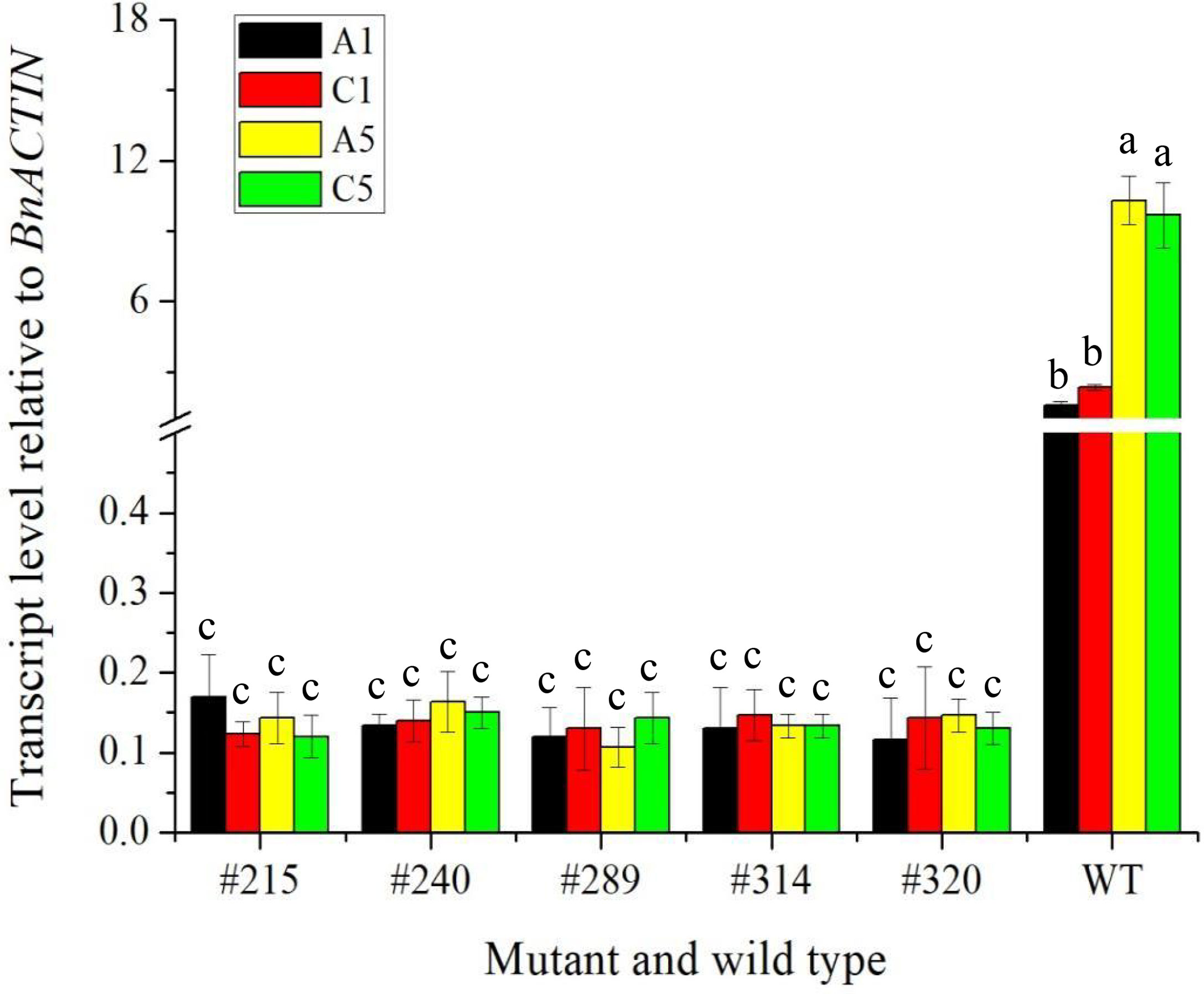
Figure 7 The transcript level relative to BnACTIN of four copies of the BnFAD2 gene on A1, C1, A5, and C5 chromosome in the five edited mutants and wild type via qPCR. The different lowercase above the histogram indicates a significant difference at the 0.05 probability level.
To further understand whether genome editing affected the seed yield, oil content, or oleic acid content, a line (#320) (Figure S5) was selected to compare those traits with the wild type (Table 7). No significant differences for the seed yield component, including plant branch numbers, seed numbers per silique, and 1000-seed weight, were found. Compared with the wild type, the number of siliques per plant was significantly lower in the edited line (reduced by 3.11%). Seed yield and seed oil content were similar between the edited line and the wild type.
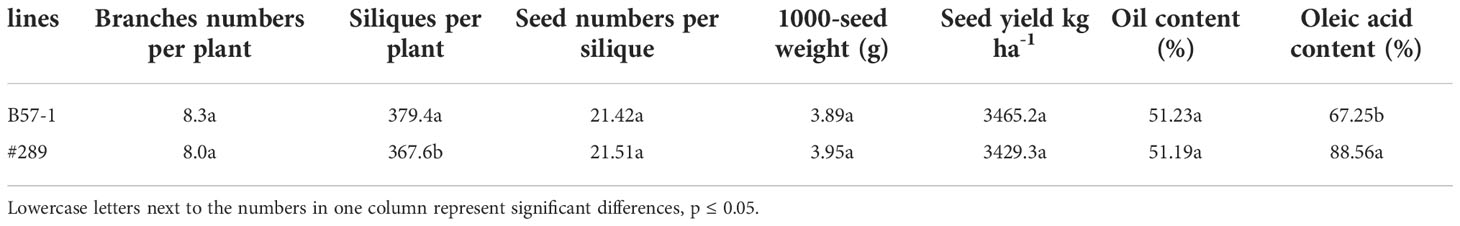
Table 7 Seed yield, yield components, seed oil content, and proportion of oleic acid between wild type (B57-1) and a selected mutant (#289).
Although there are many ways by which one or more traits were enhanced in crops to increase crop yield and quality, the efficiency among these methods can be drastically different (Saker and Singh, 2015; Khosa et al., 2016). Mutations introduced via physical, chemical, or biotechnological methods are useful tools in crop breeding (Mahadtanapuk et al., 2014; Jung and Till, 2021; Tamilzharasi et al., 2022). In order to increase seed oleic acid content, previous investigations indicate two methods that have been extensively applied, which are chemical mutation (EMS) and biological mutation (Auld et al., 1992; Sivaraman et al., 2004; Peng et al., 2010; Lee et al., 2018). Physical mutation from irradiation using 60Co and γ rays was not reported to improve the seed oleic acid content in rapeseed. This may be due to the low possibility of a desirable mutation ensuing because the mutation mechanism is a fragment disruption of the chromosome that results in a random form (Gaulden and Read, 1978). Furthermore, the disrupted chromosome fragment carrying the target gene will correspondingly discard many closely linked genes, which could affect other traits positively or negatively. Unlike physical mutation in crops, chemical mutation has been applied more frequently to rapeseed, including its seed oleic acid content. Over the decades, many studies have demonstrated that EMS mutagenesis provides an effective way to improve seed oleic acid content as well as other traits (Lee et al., 2018; Patel et al., 2022). Furthermore, many researchers were able to obtain plants whose seed oleic acid content was more than 85% (Auld et al., 1992). Nevertheless, EMS mutagenesis also occurs in a random form, and to be successful, it requires a large screening population, which is laborious work.
Accordingly, the application of biotechnological mutation is becoming increasingly attractive because of its high efficiency and accuracy and comparatively less labor involved. It is clear that RNAi can increase seed oleic acid content by reducing the erucic acid content via manipulation of BnFAE (Shi et al., 2015); however, the content of seed oleic acid increased to no more than ca. 65%, yet it could not go higher despite the manipulation of BnFAE. We know that after low erucic acid resource is introduced into other germplasms, all of the released rapeseed varieties should feature a low erucic acid content, and currently released rapeseed varieties do have a seed oleic acid content about 65%. Therefore, as is, there is nearly no room to improve seed oleic acid content by further manipulation of BnFAE. Another phenomenon should be highlighted: when manipulating BnFAD2 against a high erucic acid background, although the seed oleic acid content significantly increased, the seed erucic acid increased slightly in comparison with the wild type (Shi et al., 2017). The phenomenon is interesting in that when BnFAE was knocked down in plants, the contribution of increased oleic acid content was from the reduction in erucic acid content. However, when BnFAD2 was knocked down in plants with high erucic acid content, the contribution of increased oleic acid content was from the reduction in linoleic acid and linolenic acid contents (Shi et al., 2017). Thus, the mechanism of the manipulation on BnFAE and BnFAD2 should be different.
In addition to RNAi, genome editing by the CRISPR/Cas 9 system is another powerful biotechnological system for crop trait improvement. In the CRISPR/Cas9 system, selection of appropriate genotypes with high efficiency of regeneration is essential. In rapeseed, Westar is a robust genotype for transformation and it is widely used in research, such as gene function analysis (Kang et al., 2021; Hong et al., 2022). However, because Westar is not a variety from China, obtaining a desirable yield and quality is often not possible due to suboptimal growing conditions for it in China. Therefore, we screened new germplasms to find one capable for high efficiency of transformation, this being a semi-winter type of rapeseed. Apart from the screened germplasm in our study, other researchers have also used their own crop genotype instead of Westar (Shi et al., 2015; Huang et al., 2020). Because of the high efficiency of transformation, the CRISPR/Cas9 system was successfully established with double-locus editing. For editing efficiency in our study, it had a high percentage of 64.35% and the main type was double-locus-edited mutants (Figure 4A). It was reported that editing efficiency varied drastically from 5.3% to 100% per transformant with different vectors to edit three genes in rapeseed (Yang et al., 2017). For another example, the editing efficiency of pFAD2_Aa1 and pFAD2_Aa2 vectors in rapeseed was 5% and 50%, respectively (Okuzaki et al., 2018). In addition to selection of vector, the targeting efficiency of sgRNA was also affected by other factors such as target sequence and secondary structure of the targeted sgRNA (Ma et al., 2015; Scheben et al., 2017). In our result, the higher editing efficiency is possibly relevant to the high specificity of the sgRNA sequence, which can avoid off-target events greatly. Secondly, the selected editing site is suitable for nucleotide cutting with no disturbance by other proteins and Cas9 can easily approach to the site. However, the speculation required further experimental evidence. Therefore, selection of an appropriate vector, specific sgRNA sequence, and editing sites might be crucial to boost editing efficiency in rapeseed. For the edited mutation type, four types were observed in our study, which was in accordance with a previous study (Nakazato et al., 2021). It was reported that complex editing patterns were found including complete, partially, and entirely absent editing in seedlings of Arabidopsis. However, the ratio of chimeric mutation was low, and the percentage of homozygous mutants reached 86.4% in T1 plants (Nakazato et al., 2021). Thus, it is reasonable to emphasize on the homozygous mutants in the study.
Although Okuzaki et al. (2018) reported that seed oleic acid content can be increased to 80.0% by editing of BnFAD2 in the CRISPR/Cas9 system, it is unclear how many or which copies were actually edited. It is quite possible it was the A5 copy because the reported seed oleic acid content was 79.0%~80.0% which concurs with our results (Table 3, Figures 5 and 6) and previous investigation (Huang et al., 2020). In the present results, the A5 locus of BnFAD2 elicited a greater effect than did its C5 locus, which was consistent with previous results (Huang et al., 2020). Although our goal was to edit two loci on A5 and C5, the double-locus editing efficiency was high, reaching 78.4% in the mutants. Furthermore, the seed oleic acid content of all selected mutants with double loci exceeded 85%. In addition to the A5 and C5 loci, researchers have identified two other copies on A1 and C1 (Yang et al., 2012; Huang et al., 2020). Our results show that no editing issues arose for A1 in this study. As for the C1 copy, only one position showed an insertion with two mutant lines, leaving other regions and positions unedited. These results suggest it is credible to use the precise design of the double-locus editing system. Regardless, it was suggested that seed oleic acid content can be higher than that attained by single-locus editing (Huang et al., 2020); however, they only got two lines with the relative proportion of seed oleic acid >80% with both loci edited on A5 and C5. The reasons for this discrepancy are unclear because the mutation type was unknown for double edited loci in the previous study. In our study, many types of mutation were found. Multiple mutation forms were also found in other investigations (Okuzaki et al., 2018; Huang et al., 2020; Trinh et al., 2022). The results suggest that the mutation type may not be a decisive factor for determining the augmented seed oleic acid content. Although genome editing with the CRISPR/Cas9 system is an efficient method in crop trait improvement, the system has a risk of off-target resulting undesirable mutations because of the similar sequences between on-target and off-target sites (Park et al., 2022; Wu et al., 2022). Many strategies were attempted to reduce genome-wide off-target events and made great achievements (Fu et al., 2014; Kleinstiver et al., 2016; Zhang et al., 2019). As a result, considerable reports showed that off-target events were not detected in the CRISPR/Cas9 system including improvement of seed oleic acid content in rapeseed (Jia et al., 2017; Okuzaki et al., 2018; Wang et al., 2018; Zhang et al., 2019; Huang et al., 2020). Non-detection of off-target events in the previous study of genome editing of BnFAD2 might be due to a reliable CISPR/Cas9 system and small numbers of BnFAD2 copy. Therefore, we did not perform off-target analysis in the current study because of its low possibility.
Because the usage of the CRISPR/Cas9 system is designed to target one trait, it is not a desirable outcome if other important crop traits such as yield and quality are reduced. Therefore, we also compared the seed yield and yield component between the wild type and mutant with high seed oleic acid content. We found no significant differences in terms of seed yield and oil content. However, the number of siliques decreased significantly in the mutant. The yield of rapeseed is composed of the number of siliques, seed number per silique, and seed weight. Here, the markedly decreased number of siliques did not lead to the significantly decreased seed yield in the mutant. One of the possible reasons was that yield components including the number of siliques, seed number per silique, and seed weight analysis were on a single-plant-basis method. Those siliques in a plant usually contain different amounts of seed. Further, seed number per silique was not measured from all of the siliques. Therefore, the reliability of yield components may be lower in our traditional analysis. Differently from yield component analysis, seed yield analysis was based on the population level, with the seeds from all plants and all siliques in a block. Therefore, it should be more reliable for the result of seed yield. For the proportion of seed oleic acid content, the edited mutant was significantly higher than that in the wild type. According to fatty acids changed in the edited mutants, it can be easily found that the contribution of a higher oleic acid proportion was mainly due to the reduction in linolic acid content in one-locus-edited mutants. However, both reduction in linolenic acid and linolic acid content in double-locus-edited mutants led to a very high proportion of oleic acid. Fatty acid desaturase 2 is an important enzyme that catalyzes oleic acid converting into linolenic acid (Lee et al., 2013). Results of the gene expression analysis of BnFAD2 showed that the transcript level was very low when BnFAD2 was knocked out in the edited mutants. Thus, the conversion of oleic acid into linolenic acid was blocked resulting in the higher oleic acid content in single-locus-edited mutants. However, in the double-locus-edited mutants, the linoleic acid content was also decreased. It might be due to the influence on the expression of FAD3, which is responsible for the conversion from linolenic acid to linoleic acid (Wallis and Browse, 2002). However, further experiment is needed to clarify this inference. These results indicated that the derived mutant via editing can be directly used for further production in additional breeding programs because of the similar seed yield trait to the wild type.
In conclusion, we screened a genotype with high regeneration and transformation efficiency that we used for establishing the CRISPR/Cas9-mediating genome editing of double loci of BnFAD2. The stronger effect of the A5 locus than C5 locus upon seed oleic acid content was demonstrated, and further improvement of seed oleic acid content was successful by editing both loci. The mutants underwent precise editing of their A5 and C5 copies. Importantly, whereas the resulting mutant had higher seed oleic acid content, its seed yield and oil content were not affected by genome editing.
The original contributions presented in the study are publicly available. This data can be found here: NCBI, PRJNA879079.
SH designed the experiment and revised the experiment. HL performed the experiment (all) and wrote the manuscript. BL, PH, LH, and BX performed the transformation experiment and PCR detection of mutants. YR performed the fatty acid analysis and yield and oil content comparison between wild types and mutants. LJ and YZ assisted on the CRISPR/Cas9 system establishment. All authors approved the final version of the article.
This work was supported by the National Natural Science Foundation of China (32130076), the earmarked fund for the China Agriculture Research System (CARS-12), and the Zhejiang Key Laboratory of Digital Dry Land Crops (2022E10012).
Great appreciation is given to the editor and reviewers’ critical comments on the improvement of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1034215/full#supplementary-material
Alberio, C., Izquierdo, N. G., Galella, T., Zuil, S., Reid, R., Zambelli, A., et al. (2016). A new sunflower high oleic mutation confers stable oil grain fatty acid composition across environments. Eur. J. Agron. 73, 25–33. doi: 10.1016/j.eja.2015.10.003
Auld, D. L., Heikkinen, M. K., Erickson, D. A., Sernyk, J. L., Romero, J. E. (1992). Rapeseed mutants with reduced levels of polyunsaturated fatty acids and increased levels of oleic acid. Crop Sci. 32, 657–662. doi: 10.2135/cropsci1992.0011183X003200030016x
Bahariah, B., Masani, M. Y. A., Rasid, O. A., Parveez, G. K. A. (2021). Multiplex CRISPR/Cas9-mediated genome editing of the FAD2 gene in rice: a model genome editing system for oil palm. J. Genet. Eng. Biotechn. 19, 86. doi: 10.1186/s43141-021-00185-4
Barth, C. A. (2009). Nutritional value of rapeseed oil and its high oleic/low linolenic variety-a call for differentiation. Eur. J. Lipid Sci. Technol. 111, 953–956. doi: 10.1002/ejlt.200900019
Cao, J., Jiang, X., Chen, Q. Y., Zhang, H., Sun, H. H., Zhang, W. M., et al. (2020). Oxidative stabilities of olive and camellia oils: Possible mechanism of aldehydes formation in oleic acid triglyceride at high temperature. LWT 118, 108858. doi: 10.1016/j.lwt.2019.108858
Chew, S. C. (2020). Cold-pressed rapeseed (Brassica napus) oil: chemistry and functionality. Food Res. Int. 131, 108997. doi: 10.1016/j.foodres.2020.108997
Dhakal, K. H., Jung, K., Chae, J., Shannon, J. H., Lee, J. (2014). Variation of unsaturated fatty acids in soybean sprout of high oleic acid accessions. Food Chem. 164, 70–73. doi: 10.1016/j.foodchem.2014.04.113
Fulvio, F., Martinelli, T., Bassolino, L., Pietrella, M., Paris, R. (2022). A single point mutation in a member of FAD2 mulitgene family resulted in the generation of a high oleic line of Silybum marianum (L.) gaertn. Ind. Crop Prod. 182, 114930. doi: 10.1016/j.indcrop.2022.114930
Fu, Y., Sander, J. D., Reyon, D., Cascio, V. M., Joung, J. K. (2014). Improving CRISPR-cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 32, 279–284. doi: 10.1038/nbt.2808
Gaulden, M. E., Read, C. B. (1978). Linear dose-response of acentric chromosome fragments down to 1 r of X-rays in grasshopper neuroblasts, a potential mutagen-test system. Mutat. Res/Fund. Mol. M. 49, 55–60. doi: 10.1016/0027-5107(78)90077-5
Hong, Y., Xia, H., Li, X., Fan, R. Y., Li, Q., Ouyang, Z. W., et al. (2022). Brassica napus BnaNTT1 modulates ATP homeostasis in plastids to sustain metabolism and growth. Cell Rep. 40, 111060. doi: 10.1016/j.celrep.2022.111060
Huang, H. B., Cui, T. T., Zhang, L. L., Yang, Q. Y., Yang, Y., Xie, K. B., et al. (2020). Modification of fatty acid profile through targeted mutation at BnFAD2 gene with CRISPR/Cas9-mediated gene editing in Brassica napus. theor. Appl. Genet. 133, 2401–2411. doi: 10.1007/s00122-020-03607-y
Hu, X. Y., Sullivan-Gilbert, M., Gupta, M., Thompson, S. A. (2006). Mapping of the loci controlling oleic and linolenic acid contents and development of fad2 and fad3 allele-specific markers in canola (Brassica napus l.). Theor. Appl. Genet. 113, 497–507. doi: 10.1007/s00122-006-0315-1
Jia, H. G., Zhang, Y. Z., Orbović, V., Xu, J., White, F. F., Jones, J. B., et al. (2017). Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotech. J. 15, 817–823. doi: 10.1111/pbi.12677
Jung, C., Till, B. (2021). Mutagenesis and genome editing in crop improvement: perspectives for the global regulatory landscape. Trends Plant Sci. 26, 1258–1269. doi: 10.1016/j.tplants.2021.08.002
Kang, Y., Liu, W., Guan, C. Y., Guan, M., He, X. (2021). Evolution and functional diversity of lipoxygenase (LOX) genes in allotetraploid rapeseed (Brassica napus l.). Int. J. Biol. Macromol. 188, 844–854. doi: 10.1016/j.ijbiomac.2021.08.082
Kanrar, S., Venkateswari, J., Dureja, P., Kirti, P. B., Chopra, V. L. (2006). Modification of erucic acid content in Indian mustard (Brassica juncea) by up-regulation and down-regulation of the Brassica juncea FATTY ACID ELONGATION1 (BjFAE1) gene. Plant Cell Rep. 25, 148–155. doi: 10.1007/s00299-005-0068-3
Khosa, J. S., McCallum, J., Dhatt, A. S., Macknight, R. C. (2016). Enhancing onion breeding using molecuar tools. Plant Breed. 135, 9–20. doi: 10.1111/pbr.12330
Kleinstiver, B. P., Pattanayak, V., Prew, M. S., Tsai, S. Q., Nguyen, N. T., Zheng, Z. L., et al. (2016). High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–496. doi: 10.1038/nature16526
Konuskan, D. B., Arslan, M., Oksuz, A. (2019). Physicochemical properties of cold pressed sunflower, peanut, rapeseed, mustard and olive oils grown in the Eastern Mediterranean region. Saudi J. Biol. Sci. 26, 340–344. doi: 10.1016/j.sjbs.2018.04.005
Kunst, L., Taylor, D. C., Underhill, E. W. (1992). Fatty acid elongation in developing seeds of Arabidopsis thaliana. Plant Physiol. Biochem. 30, 425–434.
Lee, Y., Park, W., Kim, K., Jang, Y., Lee, J., Cha, Y., et al. (2018). EMS-induced mutation of an endoplasmic reticulum oleate desaturase gene (FAD2-2) results in elevated oleic acid content in rapeseed (Brassica napus l.). Euphytica 214, 28. doi: 10.1007/s10681-017-2106-y
Lee, K., Sohn, S. I., Jung, J. H., Kim, S. H., Roh, K., Kim, J., et al. (2013). Functional analysis and tissue-differential expression of four FAD2 genes in amphidipolid Brassica napus derived from Brassica rapa and Brassica oleracea. Gene 531, 253–262. doi: 10.1016/j.gene.2013.08.095
Lian, J., Lu, X., Yin, N., Ma, L., Lu, J., Liu, X., et al. (2017). Silencing of BnTT1 family genes affects seed flavonoid biosynthesis and alters seed fatty acid composition in Brassica napus. Plant Sci. 254, 32–47. doi: 10.1016/j.plantsci.2016.10.012
Livak, J. K., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
López, P. L., Marchesino, M. A., Grosso, N. R., Olmedo, R. H. (2022). Comparative study of accelerated assays for dermination of equivalent days in the shelf life of roasted high oleic peanuts: Chemical and volatile oxidation indicators in accelerated and room temperature conditions. Food Chem. 373, 131479. doi: 10.1016/j.foodchem.2021.131479
Mahadtanapuk, S., Teraarusiri, W., Nanakorn, W., Yu, L. D., Thongkumkoon, P., Anuntalabhochai, S. (2014). A new ion-bean-mutation effect application in identifciation of gene involved in bacterial antagonism of fungal infection of ornamental crops. Nucl. Instrum. Meth. B 326, 209–213. doi: 10.1016/j.nimb.2013.10.036
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., et al. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284. doi: 10.1016/j.molp.2015.04.007
Merrill, L. I., Pike, O. A., Ogden, L. V., Dunn, M. L. (2008). Oxidative stability of conventional and high-oleic vegetable oils with added antioxidants. J. Am. Oil Chem. Soc 85, 771–776. doi: 10.1007/s11746-008-1256-4
Nakazato, I., Okuno, M., Yamamoto, H., Tamura, Y., Itoh, T., Shikanai, T., et al. (2021). Targeted base editing in the plastid genome of Arabidopsis thaliana. nat. Plant 7, 906–913. doi: 10.1038/s41477-021-00954-6
Okuley, J., Lightner, J., Feldmann, K., Yadav, N., Lark, E., Browse, J. (1994). Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 6, 147–158. doi: 10.1105/tpc.6.1.147
Okuzaki, A., Ogawa, T., Koizuka, C., Kaneko, K., Inaba, M., Imamura, J., et al. (2018). CRISPR/Cas9-mediated genome editing of the fatty acid desaturase 2 gene in Brassica napus. Plant Physiol. Biochem. 131, 63–69. doi: 10.1016/j.plaphy.2018.04.025
Park, S. B., Uchida, T., Tilson, S., Hu, Z. Y., Ma, C. D., Leek, M., et al. (2022). A dual conditional CRISPR-Cas9 system to activate gene editing and reduce off-target effects in human cells. Mol. Ther. Nucl. Acids 28, 656–669. doi: 10.1016/j.omtn.2022.04.013
Patel, J., Chandnani, R., Khanal, S., Adhikari, J., Brown, N., Chee, P. W., et al. (2022). Pyramiding novel EMS-generated mutant alleles to improve fiber quality components of elite upland cotton germplasm. Ind. Crop Prod. 178, 114594. doi: 10.1016/j.indcrop.2022.114594
Peng, Q., Hu, Y., Wei, R., Zhang, Y., Guan, C. Y., Ruan, Y., et al. (2010). Simultaneous silencing of FAD2 and FAE1 genes affects both oleic acid and erucic acid contents in Brassica napus seeds. Plant Cell Rep. 29, 317–325. doi: 10.1007/s00299-010-0823-y
Razeghi-Jahromi, F., Parvini, F., Zarei, A., Hosseini-Mazinani, M. (2022). Sequence characterization and temporal expression analysis of differnt SADs and FAD2-2 genes in two Iranian olive cultivars. Sci. Hortic. 305, 111415. doi: 10.1016/j.scienta.2022.111415
Roscoe, T. J., Lessire, R., Puyaubert, J., Renard, M., Delseny, M. (2001). Mutations in the fatty acid elongation 1 gene are associated with a loss of β-ketoacyl-CoA synthase activity in low erucic acid rapeseed. FEBS Lett. 492, 107–111. doi: 10.1016/S0014-5793(01)02243-8
Roszkowska, B., Tańska, M., Czaplicki, S., Konopka, I. (2015). Variation in the composition and oxidative stability of commercial rapeseed oils during their shelf life. Eur. J. Lipid Sci. Technol. 117, 673–683. doi: 10.1002/ejlt.201400271
Saker, A., Singh, M. (2015). Improving breeding efficiency through application of appropriate experimental designs and analysis models: A case of lentil (Lens culinaris medikus subsp. culinaris) yield trials. Field Crop Res. 179, 26–34. doi: 10.1016/j.fcr.2015.04.007
Scheben, A., Wolter, F., Batley, J., Puchta, H., Edwards, D. (2017). Towards CRISPR/Cas crops-bringing together genomics and genome editing. New Phytol. 216, 682–698. doi: 10.1111/nph.14702
Schierholt, A., Becker, H. C. (2001). Environmental variability and heritability of high oleic acid content in winter oilseed rape. Plant Breed. 120, 63–66. doi: 10.1046/j.1439-0523.2001.00549.x
Shi, J. H., Lang, C. X., Wang, F. L., Wu, X. L., Liu, R. H., Zheng, T., et al. (2017). Depressed expression of FAE1 and FAD2 genes modifies fatty acid profiles and storage compounds accumulation in Brassica napus seeds. Plant Sci. 263, 177–182. doi: 10.1016/j.plantsci.2017.07.014
Shi, J. H., Lang, C. X., Wu, X. L., Liu, R. H., Zheng, T., Zhang, D. Q., et al. (2015). RNAi knockdown of fatty acid enlongase1 alters fatty acid composition in Brassica napus.Biochem. Bioph. l Res. Co. 466, 518–5252. doi: 10.1016/j.bbrc.2015.09.062
Sivaraman, I., Arumugam, N., Sodhi, Y. S., Gupta, V., Mukhopadhyay, A., Pradhan, A. K., et al. (2004). Development of high oleic and low linoleic acid transgenics in a zero erucic acid Brassica juncea l. (Indian mustard) line by antisense suppression of the fad2 gene. Mol. Breed. 13, 365–375. doi: 10.1023/B:MOLB.0000034092.47934.d6
Spasibionek, S. (2006). New mutants of winter rapeseed (Brassica napus l.) with changed fatty acid composition. Plant Breed 125, 259–267. doi: 10.1111/j.1439-0523.2006.01213.x
Tamilzharasi, M., Dharmalingam, K., Venkatesan, T., Jegadeesan, S., Palaniappan, J. (2022). Mutagenic effectiveness and efficiency of gamma rays and combinations with EMS in the induction of macro mutations in blackgram (Vigna mungo (L.) hepper). Appl. Radiat. Isotopes 188, 110382. doi: 10.1016/j.apradiso.2022.110382
Tan, M., Chen, C., Fu, X., Cui, F. J., Zhang, H. B., Ye, P. P., et al. (2022). Roasting treatments affect physicochemical, aroma and nutritional quality of strong fragrant rapeseed oil. J. Food Compos. Anal. 111, 104648. doi: 10.1016/j.jfca.2022.104648
Terés, S., Barceló-Coblijn, G., Alemany, R., Benet, M., Escribá, P. V. (2007). Oleic acid is responsible for the blood pressure reduction induced by olive oil through its “membrane-lipid therapy”. action. Chem. Phys. Lipids 149, s71–s72. doi: 10.1016/j.chemphyslip.2007.06.162
Terés, S., Barceló-Coblijn, G., Benet, M., Álvarez, R., Bressani, R., Halver, J. E., et al. (2008). Oleic acid content is responsible for the reduction in blood pressure induced by olive oil. Proc. Natl. Acad. Sci. U.S.A. 105, 13811–13816. doi: 10.1073/pnas.0807500105
Trinh, D. D., Le, N. T., Bui, T. P., Le, T. N. T., Nguyen, C. X., Chu, H. H., et al. (2022). A sequential transformation method for validating soyben genome editing by CRISPR/Cas9 system. Saudi J. Biol. l Sci. 29, 103420. doi: 10.1016/j.sjbs.2022.103420
Velasco, L., Fernández-Martínez, J. M., De Haro, A. (2003). Inheritance of increased oleic acid concentration in high-erucic acid Ethiopian mustard. Crop Sci. 43, 106–109. doi: 10.2135/cropsci2003.1060
Vu, T. V., Doan, D. T. H., Kim, J., Sung, W. Y., Tran, M. T., Song, Y. J., et al. (2021). CRISPR/Cas-based precision genome editing via microhomology-mediated end joining. Plant Biotech. J. 19, 230–239. doi: 10.1111/pbi.13490
Wallis, J., Browse, J. (2002). Mutants of Arabidopsis reveal many roles for membrane lipids. Prog. Lipid Res. 41, 254–278. doi: 10.1016/S0163-7827(01)00027-3
Wang, X. H., Tu, M. X., Wang, D. J., Liu, J. W., Li, Y. J., Li, Z., et al. (2018). CRISPR/Cas9-mediated efficient targeted mutagenesis in grape in the first generation. Plant Biotechnol. J. 16, 844–855. doi: 10.1111/pbi.12832
Wu, L., Jia, Y., Wu, G., Lu, C. (2015). Molecular evidence for blocking erucic acid synthesis in rapeseed (Brassica napus l.) by a two-base-pair deletion in FAE1 (fatty acid elongase 1). J. Inter. Agr. 14, 1251–1260. doi: 10.1016/S2095-3119(14)60853-4
Wu, Y. C., Ren, Q. R., Zhong, Z. H., Liu, G. Q., Han, Y. S., Bao, Y., et al. (2022). Genome-wide analysis of PAM-relaxed Cas9 genome editors reveal substantial off-target effects by ABE8e in rice. Plant Biotechnol. J. 20, 1670–1682. doi: 10.1111/pbi.13838
Yang, Q. Y., Fan, C. C., Guo, Z. H., Qin, J., Wu, J. Z., Li, Q. Y., et al. (2012). Identification of FAD2 and FAD3 genes in Brassica napus genome and development of allele-specific markers for high oleic acid and low linolenic acid contents. Theor. Appl. Genet. 125, 715–729. doi: 10.1007/s00122-012-1863-1
Yang, H., Wu, J. J., Tang, T., Liu, K. D., Dai, C. (2017). CRISPR/Cas9-mediated genome editing efficiency creates specific mutations at multiple loci using one sgRNA in Brassica napus. Sci. Rep. 7, 7489. doi: 10.1038/s41598-017-07871-9
Zhang, J., Chen, L., Zhang, J., Wang, Y. (2019). Drug inducible CRISPR/Cas systems. Comput. Struc. Biotechnol. J. 17, 1171–1177. doi: 10.1016/j.csbj.2019.07.015
Zhang, Z. Z., Hua, L., Gupta, A., Tricoli, D., Edwards, K. J., Yang, B., et al. (2019). Development of an agrobacterium-delivered CRISPR/Cas9 system for wheat genome editing. Plant Biotech. J. 17, 1623–1635. doi: 10.1111/pbi.13088
Zhang, H. Q., Zhang, Z. Q., Xiong, T., Xiong, X. H., Wu, X. M., Guan, C. Y., et al. (2018). The CCCH-type transcription factor BnZFP1 is a positive regulator to control oleic acid levels through the expression of diacylglycerol O-acyltransferase1 gene in Brassica napus. Plant Physiol. Biochem. 132, 633–640. doi: 10.1016/j.plaphy.2018.10.011
Zhao, J. Y., Dimov, Z., Becker, H. C., Ecke, W., Möllers, C. (2008). Mapping QTL controlling fatty acid composition in a double haploid rapeseed population segregating for oil content. Mol. Breed. 21, 115–125. doi: 10.1007/s11032-007-9113-y
Zhao, Q., Wu, J., Cai, G. Q., Yang, Q. Y., Shahid, M., Fan, C. C., et al. (2019). A novel quantitative trait locus on chromosome A9 controlling oleic acid content in Brassica napus. Plant Biotechnol. J. 17, 2313–2324. doi: 10.1111/pbi.13142
Keywords: CRISPR/Cas9, genome editing, mutation, oleic acid, rapeseed, sequencing, yield
Citation: Liu H, Lin B, Ren Y, Hao P, Huang L, Xue B, Jiang L, Zhu Y and Hua S (2022) CRISPR/Cas9-mediated editing of double loci of BnFAD2 increased the seed oleic acid content of rapeseed (Brassica napus L.). Front. Plant Sci. 13:1034215. doi: 10.3389/fpls.2022.1034215
Received: 01 September 2022; Accepted: 17 October 2022;
Published: 22 November 2022.
Edited by:
Nazim Hussain, Independent researcher, Sharjah, United Arab EmiratesReviewed by:
Joaquín J. Salas, Institute for Fats (CSIC), SpainCopyright © 2022 Liu, Lin, Ren, Hao, Huang, Xue, Jiang, Zhu and Hua. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuijin Hua, c2podWExQDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.