- 1State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, South China Agricultural University, Guangzhou, China
- 2Root Biology Center & College of Natural Resources and Environment, South China Agricultural University, Guangzhou, China
- 3Guangdong Laboratory of Lingnan Modern Agricultural Science and Technology, Guangzhou, China
NUCLEAR FACTOR Y subunit alpha (NF-YA), together with NF-YB and NF-YC, regulates plant growth and development, as well as plant responses to biotic and abiotic stresses. Although extensive studies have examined the functions of NF-YAs in Arabidopsis thaliana, the roles of NF- YAs in Glycinme max are poorly understood. In this study, we identified a phosphorus (P) starvation-responsive NF-YA8 in soybean. The expression of GmNF-YA8 is induced by low P or low nitrogen in leaves, but not by potassium or iron starvation, respectively. GmNF-YA8 is localized in the nucleus and plasma membrane. Ectopic expression of GmNF-YA8 inhibits plant growth and delayed flowering in Arabidopsis. Exogenous application of gibberellic acid (GA) rescues the delayed flowering phenotype in Arabidopsis overexpressing GmNF-YA8 lines GmNF-YA8OE-05 and GmNF-YA8OE-20. Moreover, quantitative real time PCR (qRT-PCR) verified that overexpression of GmNF-YA8 downregulates GA20ox2 and GA3ox2 expression, but upregulates GA2ox2 and GA2ox3 that encode enzymes, which inactive bioactive GAs. Consistent with the late flowering phenotype of Arabidopsis trangenic lines that overexpress GmNF-YA8, the transcript levels of flowering-promoting genes AP1, CO, LFY, and SOC1 are reduced. In addition, overexpression of GmNF-YA8 promotes the emergence of lateral root (LR) primordium from epidermis rather than the initiation of LR in low P, and increases the LR density in low nitrogen. Our results provide insights into the roles of GmNF-YA8.
Introduction
The NUCLEAR FACTOR Y (NF-Y) heterotrimeric complexes, which consist of NUCLEAR FACTOR Y alpha (NF-YA), NUCLEAR FACTOR Y beta (NF-YB), and NUCLEAR FACTOR Y gamma (NF-YC), have been reported to bind the the CCAAT box, a cis-element presents in about 25% of eukaryotic gene promoters (Mantovani, 1998; Petroni et al., 2012). In mammals and yeast, each NF-Y subunit is encoded by a single gene, but plant genomes contains multiple genes for each subunit. For example, the Arabidopsis thaliana genome contains 10 NF-YA, 13 NF-YB, and 13 NF-YC genes. Similarly, dozens of NF-YA, NF-YB, and NF-YC genes have been identified in rice (Oryza sativa), wheat (Triticum aestivum), Brachypodium, and Glycine max (Stephenson et al., 2007; Thirumurugan et al., 2008; Cao et al., 2011). The soybean genome contains 21 GmNF-YA, 32 GmNF-YB, and 15 GmNF-YC genes (Quach et al., 2015), suggesting that these genes have more diverse roles in soybean than in Arabidopsis. However, the functions of the soybean NF-YA remain elusive.
Past studies in Arabidopsis have revealed that NF-YA genes play crucial roles in plant growth and development (Nelson et al., 2007; Li et al., 2008; Petroni et al., 2012). For example, AtNF-YAs regulate seed germination. Overexpression of AtNA-YA1 or AtNF-YA9 promotes seed germination in the presence of exogenous abscisic acid (ABA) (Siriwardana et al., 2014), but overexpression of AtNF-YA2, AtNF-YA5, AtNF-YA8, or AtNF-YA10 delays seed germination (Mu et al., 2013; Siriwardana et al., 2014). The AtNF-YA5 mutant nf-ya5 does not respond to blue light, and AtNF-YA5 appears to activate Lhcb1.3 in response to blue light (Warpeha et al., 2007). AtNF-YA2 is also documented to be involved in photomorphogenesis, and in the dark, transcription of AtNF-YA2 is repressed and AtNF-YA2 protein is degraded (Myers et al., 2016). AtNF-YA2 and AtNF-YA10 are both targets of miR169; overexpressing a miR169-resistant allele of AtNF-YA2 results in higher density of lateral root (LR), but does not affect primary root (PR) growth (Sorin et al., 2014). Overexpression of AtNF-YA2, AtNF-YA7, or AtNF-YA10 results in dwarfing of both seedlings and adult plants, and in smaller siliques and seeds (Leyva-González et al., 2012). Previous studies revealed that some AtNF-YAs are negative regulators of flowering (Wenkel et al., 2006). Overexpression of AtNF-YA1, AtNF-YA4, AtNF-YA7, or AtNF-YA9 leads to late flowering (Wenkel et al., 2006; Siriwardana et al., 2016). By contrast, AtNF-YA2 seem to be a positive regulator of flowering, and only the heterodimer of AtNF-YA2 and Atnf-YA6, but not AtNF-YA2 or AtNF-YA6, binds the CCAAT-box in promoter of FT, thus promoting FT expression and early flowering (Siriwardana et al., 2016). Whereas another study reported that AtNF-YA2, AtNF-YB2, and AtNF-YC9 bind to the promoter of SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) (Hou et al., 2014; Gnesutta et al., 2017), but REPRESSOR OF ga1-3 (RGA), a typical DELLA protein, interacts with AtNF-YA2 and prevents its binding to the promoter of flowering gene SOC1 (Hou et al., 2014).
A growing body evidence suggests that NF-YAs regulate plants’ responses to abiotic stress. In Arabidopsis, overexpressing AtNF-YA1 lines are hypersensitive to salt stress and ABA during the early post-germination growth stages (Li et al., 2013). AtNF-YA5 has been reported to be induced by drought, accordingly loss of function of AtNF-YA5 results in increased water loss in drought conditions (Li et al., 2008). AtNF-YA10 is downregulated by salinity and ABA, and overexpression of AtNF-YA10 results in increased sensitivity to salinity and decreased sensitivity to ABA (Ma et al., 2015). In rice, the abundance of OsNF-YA7 mRNA increases in response to drought, and overexpression of OsNF-YA7 increases drought tolerance in an ABA-independent manner (Lee et al., 2015). Similarly, transcription of OsNF-YA8 is induced by drought and its overexpression in rice plants improves their drought tolerance via ABA-independent approach. In contrast, OsNF- YA4 expression is not increased by drought stress, but highly induced by ABA (Lee et al., 2015). The transcript level of GmNF-YA8 is stimulated by water deficiency (Quach et al., 2015), whereas the role of which in drought tolerance need to be deciphered. In addition, AtNF-YA5, OsNF-YA8, GmNF-YA10, and GmNF-YA3 are also targeted by miR169 (Li et al., 2008; Zhao et al., 2009; Xu et al., 2013; Ni et al., 2013). These results indicate that transcription of NF-YA is tightly regulated in plants.
NF-YAs are involved in plant nutrition. In Arabidopsis, transcription of AtNF-YA2, AtNF-YA3, AtNF-YA5, or AtNF-YA8 is induced by low nitrogen (N) stress in both roots and shoots, and in line with this, the the expression of miR169 is reduced in N-starved roots and shoots (Zhao et al., 2011). Overexpressing MIR169a in Arabidopsis increases sensitivity to low N stress and decreases the total N contents of roots and leaves (Zhao et al., 2011). Microarray analysis showed that the expression of AtNF-YA2, AtNF-YA3, AtNF-YA5, AtNF-YA7, or AtNF-YA10 is stimulated by low phosphate (Pi) stress (Misson et al., 2005), and further qRT-PCR verified that AtNF-YA2, AtNF-YA7, And AtNF-YA10 are induced by Pi deficiency (Leyva-González et al., 2012). Whereas the responses of soybean NF-YA family to Pi deficiency remain elusive.
Several studies have been carried out to investigate the functions of Glycine max NF-YAs (GmNF-YAs). The 21 GmNF-YA genes can be divided into three subgroups, and seven of the genes are induced by drought (Quach et al., 2015). GmNF-YA3 is induced by ABA, NaCl, and cold, and ectopic expression of this gene (renamed as GmNF-YA12 in 2015 (Quach et al., 2015) in Arabidopsis reduces water loss by modulating the expression of AtABA1, AtABA2, AtABI1, and AtABI2, thereby increasing tolerance to drought (Ni et al., 2013). More recently, GmNF-YA5 was reported to be positive regulator during drought stress. Expression of GmNF-YA5 is induced by drought in an ABA-dependent manner, and the transgenic expression of GmNF-YA5 enhances the drought tolerance of Arabidopsis and soybean (Ma et al., 2020). GmNF-YA1a and 1b have been observed to positively regulate arbuscular mycorrhization (Schaarschmidt et al., 2013). Phosphorus (P) is one of essential macroelements for plant growth and development. Low P stress delays flowering and decreases yield of crops in agriculture (Jiang et al., 2007; Nord and Lynch, 2008). Plants cope with Pi starvation by remodeling of root system architecture with increased number of LRs and LR length, shorter PR, and denser root hair to boost the uptake of Pi from growth medium (López-Bucio et al., 2002; Pérez-Torres et al., 2008). Some classic transcription factors involved in P nutrition such as PHR1 and WRKY6 have been identified in Arabidopsis.
In 2013, We identified low Pi-responsive microRNAs and targets of them in soybean at genomic level, and found that soybean miR69 family members target NF-YAs (Xu et al., 2013). To further deepen our understanding to soybean’s responses to low P stress and screen low Pi- responsive transcription factors, we initially used RNA-seq experiment to screen long-term (14 days) Pi deficiency-responsive genes in soybean leaves at genomic level. Hundreds of genes induced or repressed by low P in soybean leaves have been identified, including dozens of transcription factor genes. Among them, one of GmNF-YAs, namely GmNF-YA8, is induced by low P. Given that low P stress delays flowering (Jiang et al., 2007) and overexpression of NF-YAs in Arabidopsis results in dwarfing and late flowering (Leyva-González et al., 2012), we are curious about the function of GmNF-YA8 in P nutrition and flowering. In this study, we confirmed that GmNF-YA8 is induced by low P stress by quantitative real time PCR (qRT-PCR) and established that the gene is also induced by low N stress. GmNF-YA8 is localized in the nucleus and plasma membrane. Ectopic expression of GmNY-YA8 in Arabidopsis delays flowering, and application of exogenous GA rescues the late flowering phenotype. Moreover, overexpression of GmNF-YA8 decreases AtGA20ox2 and AtGA3ox2 expression, and increases AtGA2ox2 and AtGA2ox3 expression. Arabidopsis plants overexpressing GmNF-YA8 shows increased LR emergence in low P, and an increased number and density of LRs in low N.
Materials and methods
Soybean and arabidopsis growth
Seeds of Glycine max genotype YC03-3 were sterilized with 10% NaClO, germinated in sands and transferred when the cotyledon was open and the apical bud had developed. To explore the responses of GmNF-YA8 to nutrient deficiencies, soybean seedlings were first cultured in complete Hoagland solution (pH 5.9) for 7 days as described (Xu et al., 2013). Then soybean seedlings with the first developed trifoliate leaf were transferred into Pi-replete (HP, 500 µM KH2PO4), HN (3 mM nitrogen), HK (2 mM potassium), HF (50 µM Fe-EDTA), Pi-deplete (LP, 25 µM KH2PO4), LN (300 µM nitrogen), LK (250 µM potassium), and LF (0 µM Fe-EDTA) media for 14 days, respectively. Soybean plants were grown in growth chambers with 16h light cycles. Nutrient solutions were aerated 15 min every 3h, and replaced with fresh nutrient solutions every 2 days. As described (Huo et al., 2018), Arabidopsis seeds were sterilized with 75% ethanol for 2 minutes followed by 100% ethanol for 2 minutes. Then seeds were sown on half strength MS media containing 1% sucrose and 0.8% agar (pH 5.8). Square petri dishes were maintained in a growth chamber with a 16h light (100 µE m-2 s-1)/8h dark cycle and with a temperature cycle of 23°C light/21°C darkness. Additionally, stratified Arabidopsis seeds were sown in soil and cultured in a growth chamber with a 16h light (100 µE m-2 s-1)/8h dark cycle and with a temperature cycle of 23° C light/21° C darkness. For the low P and low N treatment, sterilized Arabidopsis seeds were sown in half strength MS medium, and stratified in −4° C for 2 days. At 3 days after germination, the seedlings were transferred into different media that contains 500 µM KH2PO4 (HP), or 0 µM KH2PO4 (LP), or 10 mM KNO3 and 1 mM NH4NO3 (HN) or 100 µM KNO3 and 10 µM NH4NO3 (LN), respectively. After 7 days, the treated seedlings were scanned to measure the PR length, the average length of first-order LR, and the number of lateral roots and lateral root primordium.
Quantification of fresh weight, concentrations of soluble phosphate (Pi) and total phosphorus (P) of soybean seedlings.
The sampled soybean plants that were subjected to low P stress for 14 days as described were weighted to quantify the fresh weight, including root and leaves fresh weight. Accordingly, the ratio of root to shoot was determined.
Soybean seedlings were dried at 105°C for 30 min, and then oven-dried at 75°C prior to weighting. The methods described (Zhou et al., 2008) were strictly followed to measure the concentration of soluble Pi and total P.
Subcellular localization of GmNF-YA8
To explore the subcellular localization of GmNF-YA8, we constructed the fusion protein of GFP with GmNF-YA8. The open reading fragment of GmNF-YA8 was amplified and then recombined into the pMDC43 vector based on the method as described (Peng et al., 2016). Agrobacterium tumefaciens mediated transient expression in Nicotiana benthamiana leaves was conducted as described (Peng et al., 2016). The Agrobacterium GV3101 strain harboring the constructs of GFP-GmNF-YA8 was inoculated in YEP medium and incubated for 24 h at 28°C. After two days, the green fluorescence in the transformed tobacco epidermal cells was detected at 488 nm by confocal laser scanning microscopy (LSM780, Zeiss, Jena, Germany).
Extraction of genomic DNA, RNA and reserve transcription of mRNA
RNA from Arabidopsis and soybean seedlings was extracted with TRIzol method. RNA was transcribed through MLV-transcriptase. DNA was extracted with CTAB method. Other molecular experiments were based on the standard methods as described (Huo et al., 2018).
Vector of construction and transformation of Arabidopsis
The plasmid for ectopic expressing GmNF-YA8 was constructed based on standard molecular biology protocols and gateway-based system (Curtis and Grossniklaus, 2003). The open reading frame of GmNF-YA8 was amplified and cloned into the pMDC32 plasmid. Arabidopsis was transformed through floral dipping as described (Clough and Bent, 1998). Single copy T-DNA insertion transgenic lines were screened in growth media containing 50 µg/ml hygromycin. The single copy T-DNA insertion homeozygous T3 generation was used in this study. The RNA was extracted from transgenic lines and the expression levels of GmNF-YA8 was further assessed by qRT-PCR.
qRT-PCR analysis
To quantify transcript level of GmNF-YA8 under different nutrition conditions, qRT-PCR was used. The roots and leaves of soybean cultivar YC03-3 seedlings that were subjected to nutrient stresses were quickly sampled in liquid N, and stored at −80°C. Total RNA extraction and cDNA synthesis were based on standard method as described (Huo et al., 2018). Glycine max EF1a (GmEF1a) was used to normalize qRT-PCR data (Xu et al., 2013).
The expression levels of flowering- and GA-related genes, as well as four LR emergence-related genes (AtLAX3, AtPIP2;1, AtPLA1 and AtPME1) were assessed. The rosette leaves of twelve- day-old Col-0 plants and overexpression lines GmNF-YA8OE-05 and GmNF-YA8OE-20 in Col-0 background grown in soils were sampled and total RNA was extracted with Trizol method. cDNAs were obtained with reverse transcriptase-mediated reactions. The transcript abundance of the above-mentioned genes was determined via qRT-PCR, and AtEF1a was used as a reference gene to normalize qRT-PCR results (Huo et al., 2018). All specific qRT-PCR and clone primer pairs are listed in Table S1.
Quantification of bolting, flowering time, number of rosette leaves, and root-related parameters
The height of the bolt and of Arabidopsis plants was measured with a ruler. The flowering time was determined based on the number of days from sowing to the first flower opening. The number of rosette leaves was counted manually. The Arabidopsis seedlings in media were scanned, and then the PR length was determined with ImageJ. The Arabidopsis LRs were counted and LR primodium (LRP) were measured using a steromicroscopy.
Data analysis
All data were analyzed with R. Student’s t-test was employed to compare differences. R 3.0.1 package (R Core Team, 2018), ggplot2 (Wickham, 2016) and ggpubr (Kassambara, 2019) were used to draw figures, respectively.
Results
Low P inhibits soybean seedling growth, and decreases the concentration of solubale and total phosphorus in soybean roots and leaves
As reported, Pi deficiency inhibit plant growth and development. Accordingly we measure the biomass of soybean seedlings that subjected to low P for 14 days. As shown in Figure 1A, in contrast to high P, low P inhibits soybean seedling growth, the total fresh weight of soybean plant in low P is 63.7 percent of that in high P. As expected, low P increases the ratio of root to shoot (Figure 1B). Moreover we measured the contents of soluble and total P in soybean plants. Low P treatment significantly decreases the concentration of soluble Pi and total P in soybean roots and leaves, respectively. The concentration of soluble Pi in leaves and roots in high P conditions is 5.8 and 8.8 folds of that in low P, respectively (P < 0.01, Figure 1C). The concentration of total P in leaves and roots in high P is 5.8 and 1.7 times to that in low P, respectively (P < 0.01, Figure 1D).
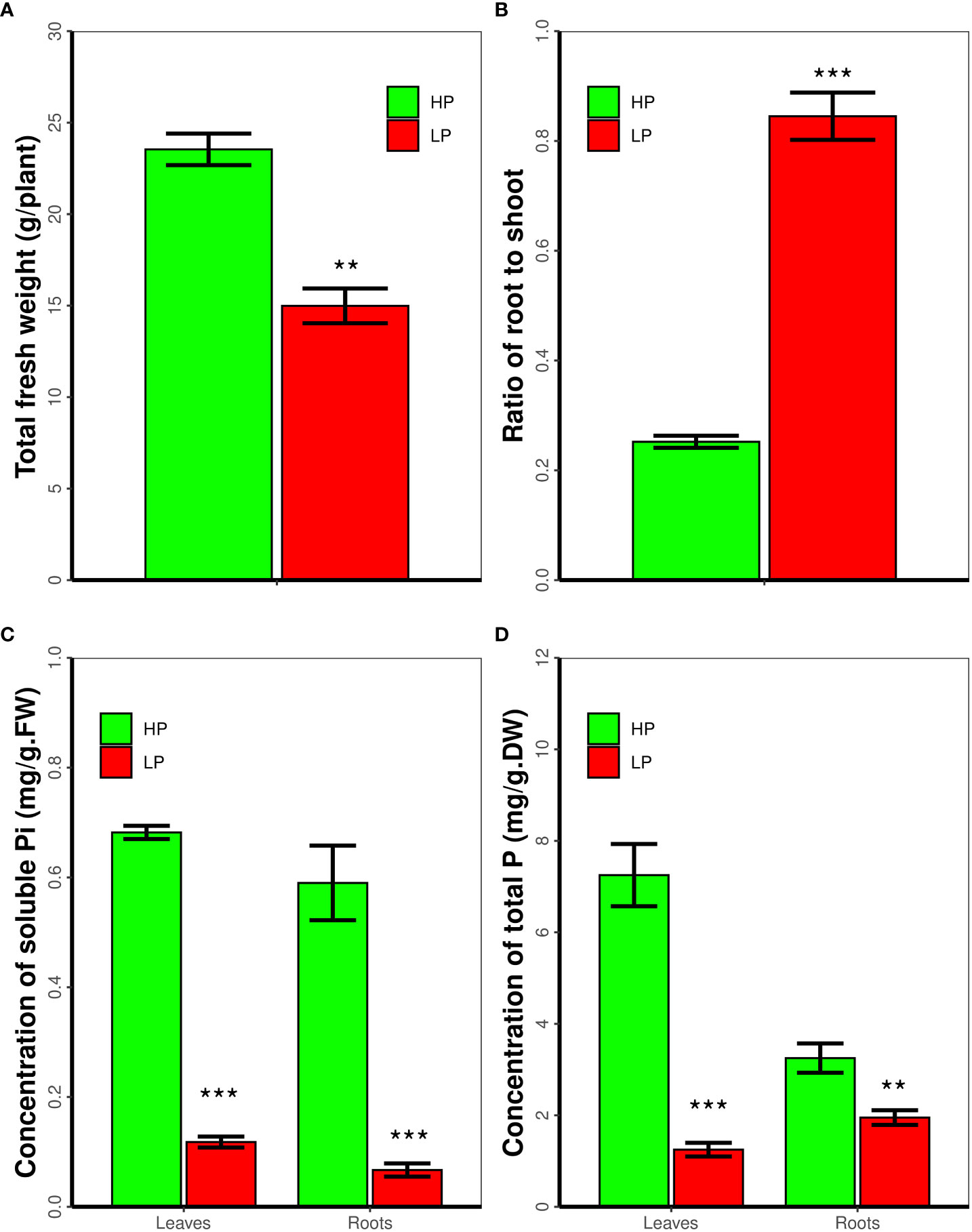
Figure 1 Low P stress inhibits soybean seedling growth and decreases the concentration of soluble Pi and total P. Seven-day-old soybean seedling were subjected to low P stress for 14 day. The Fresh weight of roots and shoots, the ratio of root to shoot, and the concentration of soluble phosphorus and total phosphorus was measured. Results are means ± SE from three independent experiments. Students’t-test was used to compare difference. **P < 0.01; ***P < 0.001.
The expression of GmNF-YA8 is induced by low P and low N
As previously reported, NF-YA genes are responsive to low P or low N stress in Arabidopsis (Zhao et al., 2011; Leyva-González et al., 2012). Although GmNF-YA8 (Glyma.09g023800) is induced by drought (Quach et al., 2015), its responses to nutrient deficiencies were not known. Based on our soybean leaf RNA-seq results (deposited in NCBI, accession number: PRJNA489734), we established that GmNF-YA8 was induced by long-term Pi starvation. Given the responsiveness of GmNF-YA8 to Pi deficiency, we want to know whether the gene responds to other nutrient deficiencies, specifically N, potassium (K), and iron (Fe). We used qRT-PCR to measure GmNF- YA8 transcript levels in leaves and roots of soybean plants that had been subjected to long-term (14 days) nutrient deficiencies. Pi starvation (LP) caused a 2.4-fold increase in GmNF-YA8 transcript levels Relative to the Pi-replete (HP) control in leaves (P < 0.05) but not in roots (Figure 2A). Under low N (LN), the transcript level of GmNF-YA8 in leaves were induced 1.19 folds relative to high N (HN) controls (P < 0.05, Figure 2B). By contrast, low K (LK) and low Fe (LF) did not induce GmNF-YA8 expression in either leaves or roots (Figure 2C, D).
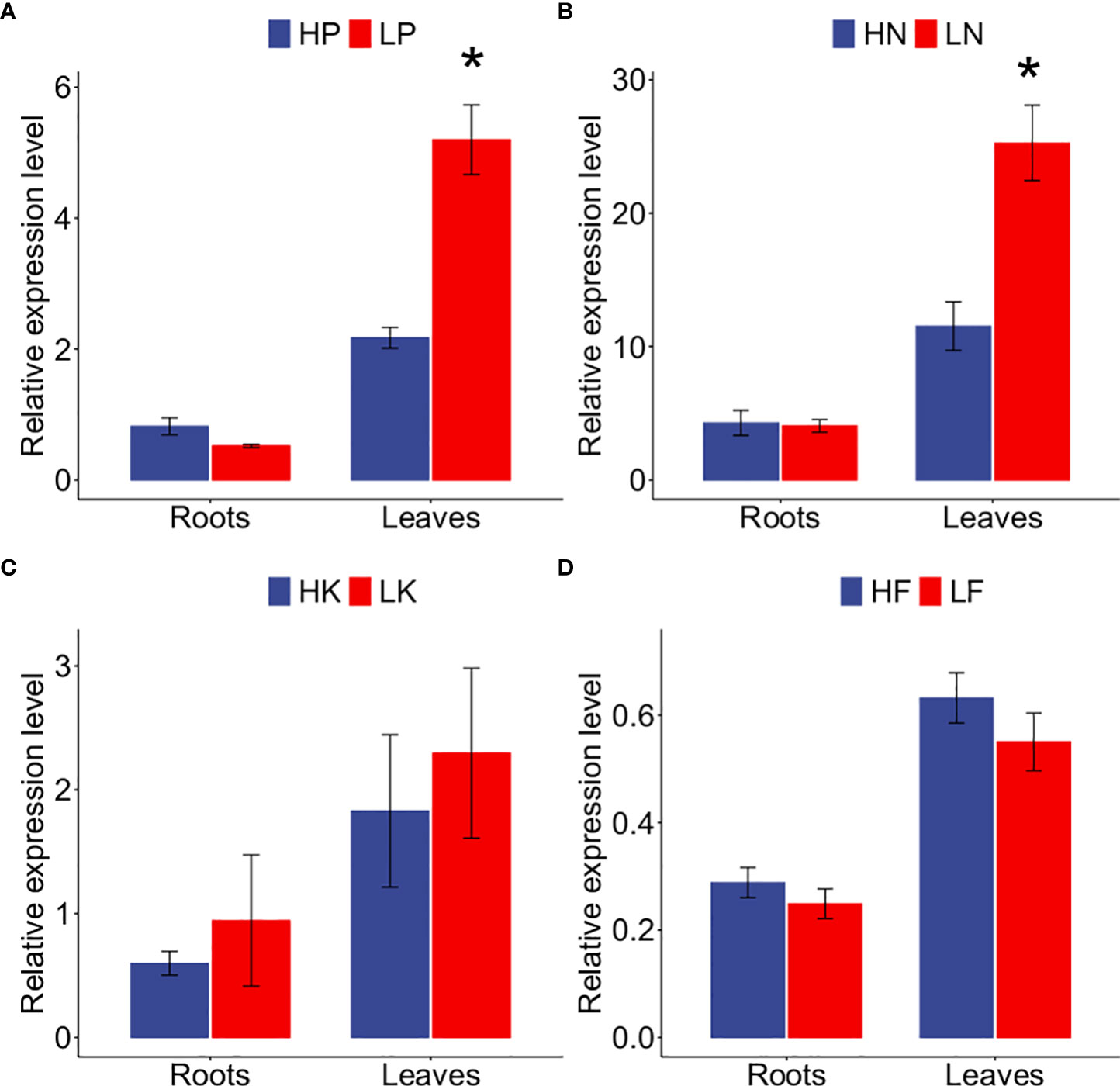
Figure 2 Responses of GmNF-YA8 to long-term nutrient starvation in soybean roots and leaves. (A) phosphorus (P) starvation; (B) nitrogen (N) starvation; (C) Potassium (K) starvation; (D) iron (Fe) starvation. HP, 500 µM KH 2 PO 4; LP, 25 µM KH2 PO4; HN, 3mM nitrogen; LN, 300 µM nitrogen, HK, 2 mM potassium; LK, 250 µM potassium; HF, 50 µM Fe-EDTA; LF, 0 µM Fe-EDTA. Results are means ± SE from 3 independent experiments. Student’s t-test were used to explore the differences between control and nutrient deficiency conditions (*P < 0.05).
We are also interested in the transcript level of GmNF-YA8 in soybean organs. Based on the public data (www.soybase.org), the transcripts of GmNF-YA8 in flower is the highest, followed by that in one-cm pod, young leaf, and root. the expression of GmNA-YA8 is also detected in seeds and nodule (Figure 3). This suggests that GmNF-YA8 is constitutively expressed in soybean organ and possibly play important role in growth and development. In addition, based on the public RNA-seq data (http://ipf.sustech.edu.cn/pub/soybean/), we found that GmNA-YA8 is induced by low P, and respond to dehydration, and day length (Supplementary Sheet 1). These results imply that GmNF-YA8 plays versatile roles in response to environmental factors.
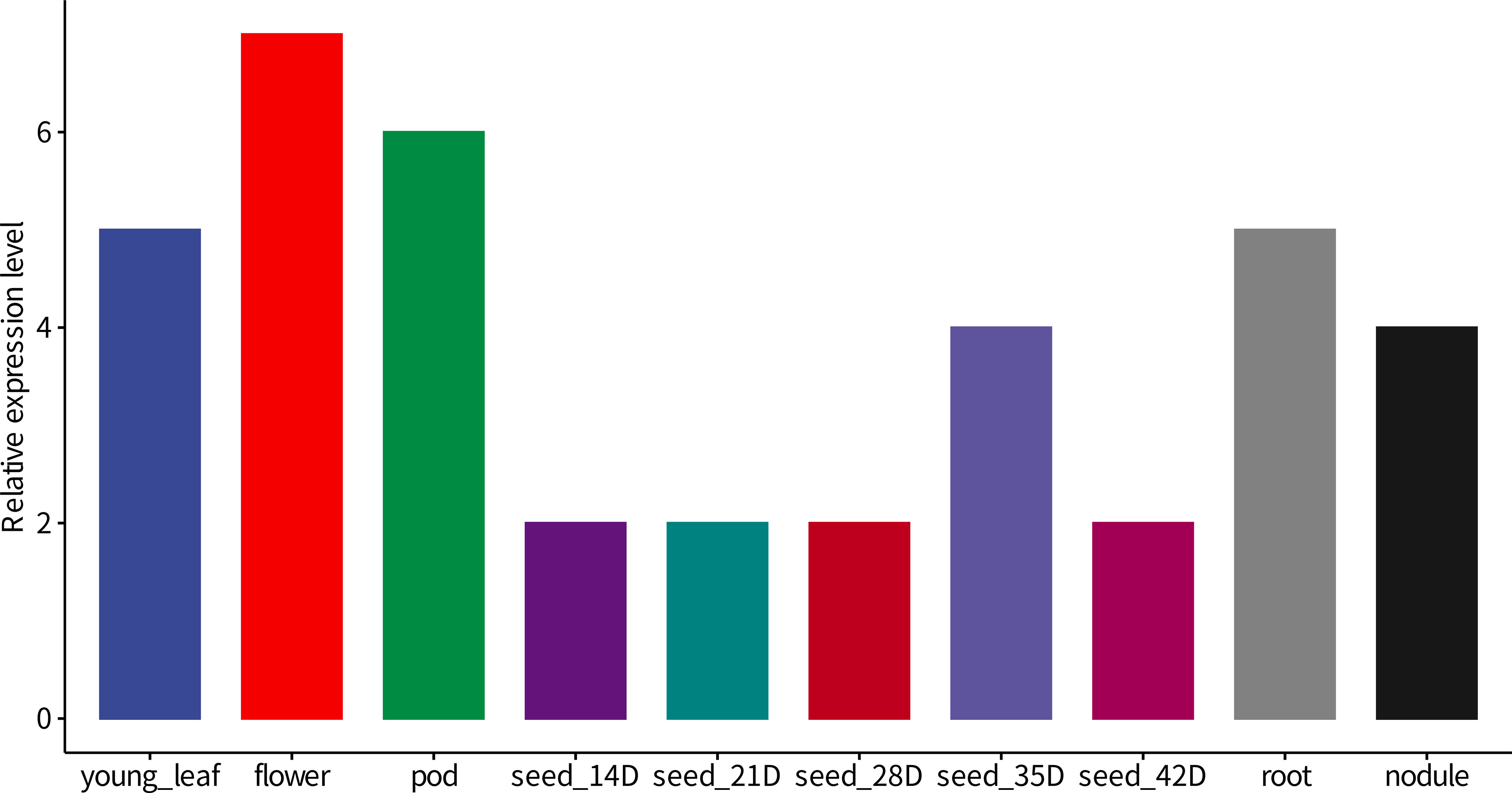
Figure 3 The relative transcript level of GmNF-YA8 in different soybean organs Base on public RNA-seq data (www.soybase.org), the relative transcript level Reads Per Kilobase Per Million mapped reads (RPKM) of GmNF-YA8 was shown. POD, one-cm long pod; D, Day after fertilization.
GmNF-YA8 is localized to the nucleus and cell membrane
Bioinformatics analysis indicates that GmNF-YA8 encodes a protein with 219 amino acids (www.phytozome.org), which is predicated by PSORT (www.psort.org) to be localized in the nucleus. Given that NF-YAs act as transcription factor in plant cell (Li et al., 2008), we are curious about the subcellular localization of GmNF-YA8. To this end, we constructed the fusion protein of GFP-GmNF-YA8 using the gateway vector pMDC43. We then transformed Nicotiana benthamiana leaf epidermal cells with Agrobacterium tumefaciens GV3101 that contains the construct encoding GFP-GmNF-YA8. After two days, we detected the green fluorescence signals in the nucleus and plasma membrane by laser confocal microscopy (Figure 4). This suggests that GmNF-YA8 is localized in the nucleus and plasma membrane.
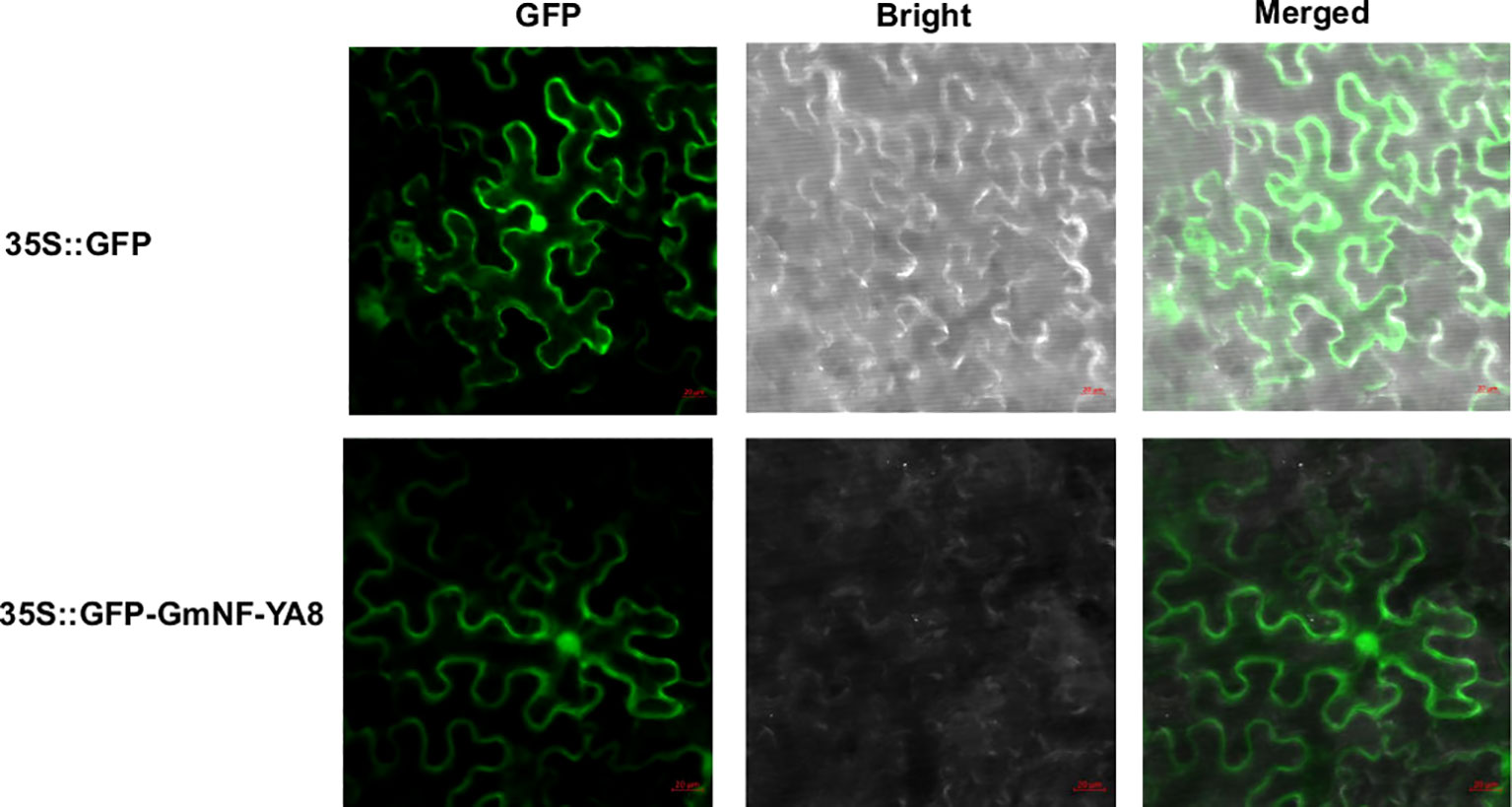
Figure 4 GmNF-YA8 is localized in the nucleus and plasma membrane. GFP signal was detected in transformed Nicotiana benthamiana leaf epidermis cells at 488 nm by laser confocal miscroscopy. GFP: GFP channel; Bright: light channel; Merged: the overlap of GFP channel and light channel. 35S::GFP: pMDC43 empty vector containing GFP; 35S::GFP-GmNF-YA8: pMDC43- GmNF-YA8 vector containing the fusion of GFP with GmNF-YA8.
Ectopic expression of GmNF-YA8 in Arabidopsis inhibits stem growth and delays bolting and flowering
To determine the function of GmNF-YA8, we overexpressed GmNF-YA8, driven by the constitutive 35S promoter in Arabidopsis Col-0 wild-type plants. By screening for hygromycin resistance, we identified dozens of T1 transgenic lines. To identify single T-DNA insertion transgenic Arabidopsis lines, the T2 seeds from individual T1 plants were sown in half strength MS medium with 25 µg/mL hygromycin and screened after 10 days based on the segregation ratio (3:1) of hygromycin resistance gene. Accordingly, homozygous transgenic lines were also determined in the T3 generation based on hygromycin resistance phenotype. We thus selected two single-copy T- DNA insertion homozygous lines, namely GmNF-YA8-05 and GmNF-YA8-20, and used qRT-PCR to verify that they were transgenic lines (Figure S1). Thus these two lines were used for subsequent experiments.
Under long-day (16h light) conditions, bolting was delayed in the transgenic expressing lines relative to non-transformed Col-0 plants (Figure 5A). When the stems of Col-0 plants were one-cm high, no obvious bolts were seen in GmNF-YA8OE-05 and GmNF-YA8OE-20 plants. Moreover, Col-0 plants had fewer rosette leaves at the time of bolting than did GmNF-YA8OE-05 and GmNF-YA8OE-20 plants when they bolted (Figure 5B).
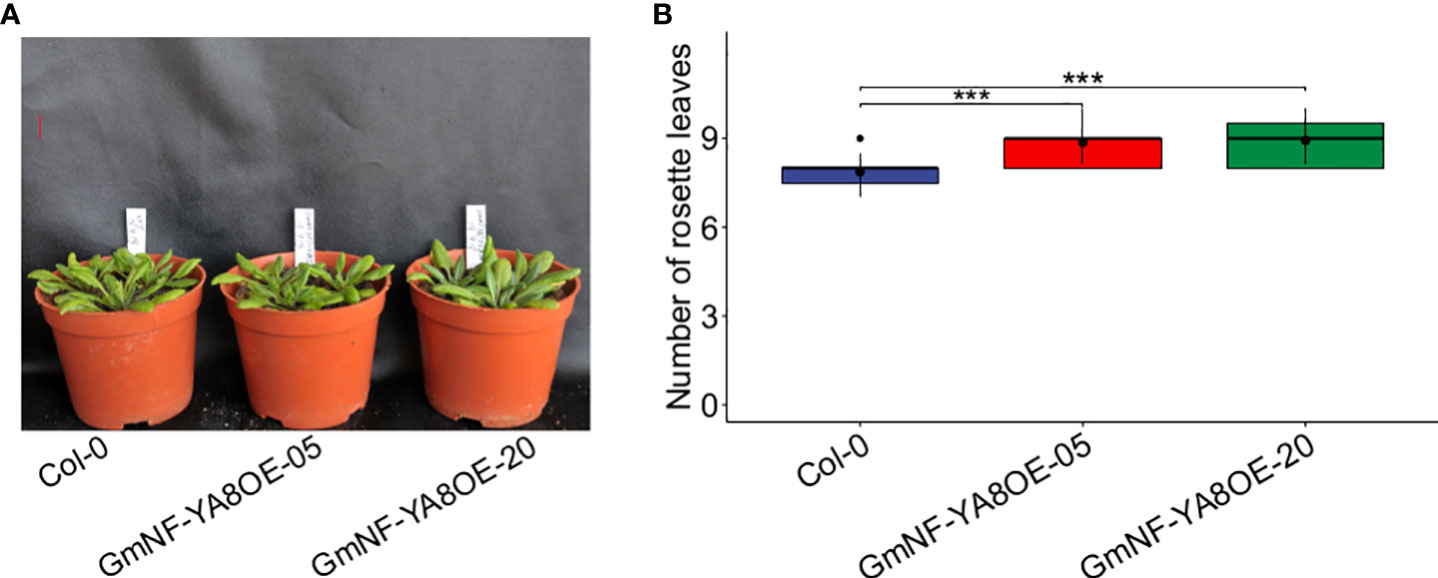
Figure 5 Ectopic expression of GmNF-YA8 delays bolting in Arabidopsis (A) Photos were taken when the bolt in 20-day-old Col-0 plants was one-cm high, the transgenic lines had not yet bolted. GmNF-YA8OE-05 and GmNF-YA8OE-20 indicates the transgenic lines that overexpress GmNF- YA8, respectively. Scale bar, 1 cm. (B) The number of rosettes was counted in each 12 Col-0, GmNF-YA8OE- (B) 05, and GmNF-YA8OE-20 plants respectively, when the bolt was one-cm high. Student’s t-test was used to compare the difference between Col-0, GmNF-YA8OE-05 and GmNF-YA8OE-20 plants (* P< 0.05; ***P < 0.001).
As the bolting of overexpressing GmNF-YA8 lines was delayed (Figure 5A), we deduced that flowering time of overexpressing GmNF-YA8 lines is also delayed. To verify this, we further explored the effect of ectopic expression of GmNF-YA8 on flowering time, defined as the number of days when the first flower is opening as described by previous studies (Wenkel et al., 2006; Siriwardana et al., 2016). In control conditions (no GA treatment), GmNF-YA8OE-05 and GmNF-YA8OE-20 plants flowered later than Col-0 plants (P < 0.05; Figure 6A, B). Application of 100 µM GA3 promoted flowering in Col-0, GmNF-YA8OE-05, and GmNF-YA8OE-20 plants, and abolished the difference between the control and transgenic plants (Figure 6B). The plant height was also measured when the first flower of the plants had open. In contrast to Col-0 seedlings, the plant height of GmNF-YA8OE-05 and GmNF-YA8OE-20 seedlings was shorter in control conditions (no GA treatment), application of GA obviously increased plant height of GmNF-YA8OE-05 and GmNF-YA8OE-20 plants and masked the difference between Col-0, GmNF- YA8OE-05 and GmNF-YA8OE-20 plants (Figure 6C). Collectively, these results indicate that ectopic expression of GmNF-YA8 delays flowering by decreasing endogenous GA levels.
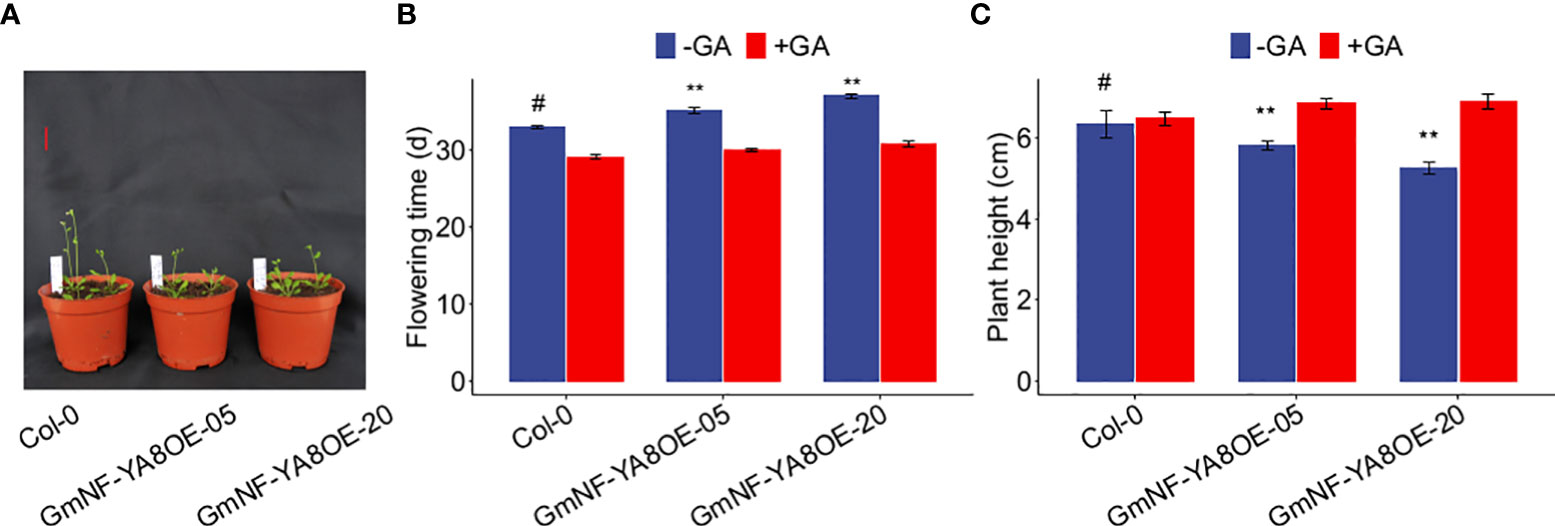
Figure 6 Ectopic expression GmNF-YA8 delays flowering in Arabidopsis (A) Photographs were taken when the first flower had opened in Col-0, but not in GmNF-YA8OE-05 and GmNF-YA8OE-20 plants. Scale bar, 2 cm. (B) The flowering time was measured when the first flower was opened in Col-0, GmNF-YA8OE-05 and GmNF-YA8OE-20, respectively. (C) Plant height was measured when the first flower was opened. Results are means ± SE from each 12 Col-0, GmNF-YA8OE-05 and GmNF-YA8OE-20 plants, respectively. Difference between Col-0 and the transgenic lines GmNF-YA8OE-05 and GmNF-YA8OE-20 were compared using Student’s t-test (**P < 0.01; #, the control).
We also measured plant height at 25 and 35 days after germination (DAG). Under normal conditions (no GA treatment), GmNF-YA8OE-05 and GmNF-YA8OE-20 plants were shorter than Col-0 plants at 25 DAG (Figures 7A, B). The height of GmNF-YA8OE-05 and GmNF-YA8OE-20 plants was 88.49% and 86.21% s of Col-0 plants, respectively (Figure 7B). Interestingly, there was no significant difference between the height of Col-0, GmNF-YA8OE-05, and GmNF-YA8OE-20 plants at 35 DAG. Application of exogenous 100 µM GA3 increased the stem growth of Col-0, GmNF-YA8OE-05, and GmNF-YA8OE-20 plants, more obviously in GmNF-YA8OE-20 plants (Figures 7C, D). These results indicate that ectopic expression of GmNF-YA8 in Arabidopsis inhibits stem growth in young plants but does not affect GA responsiveness.
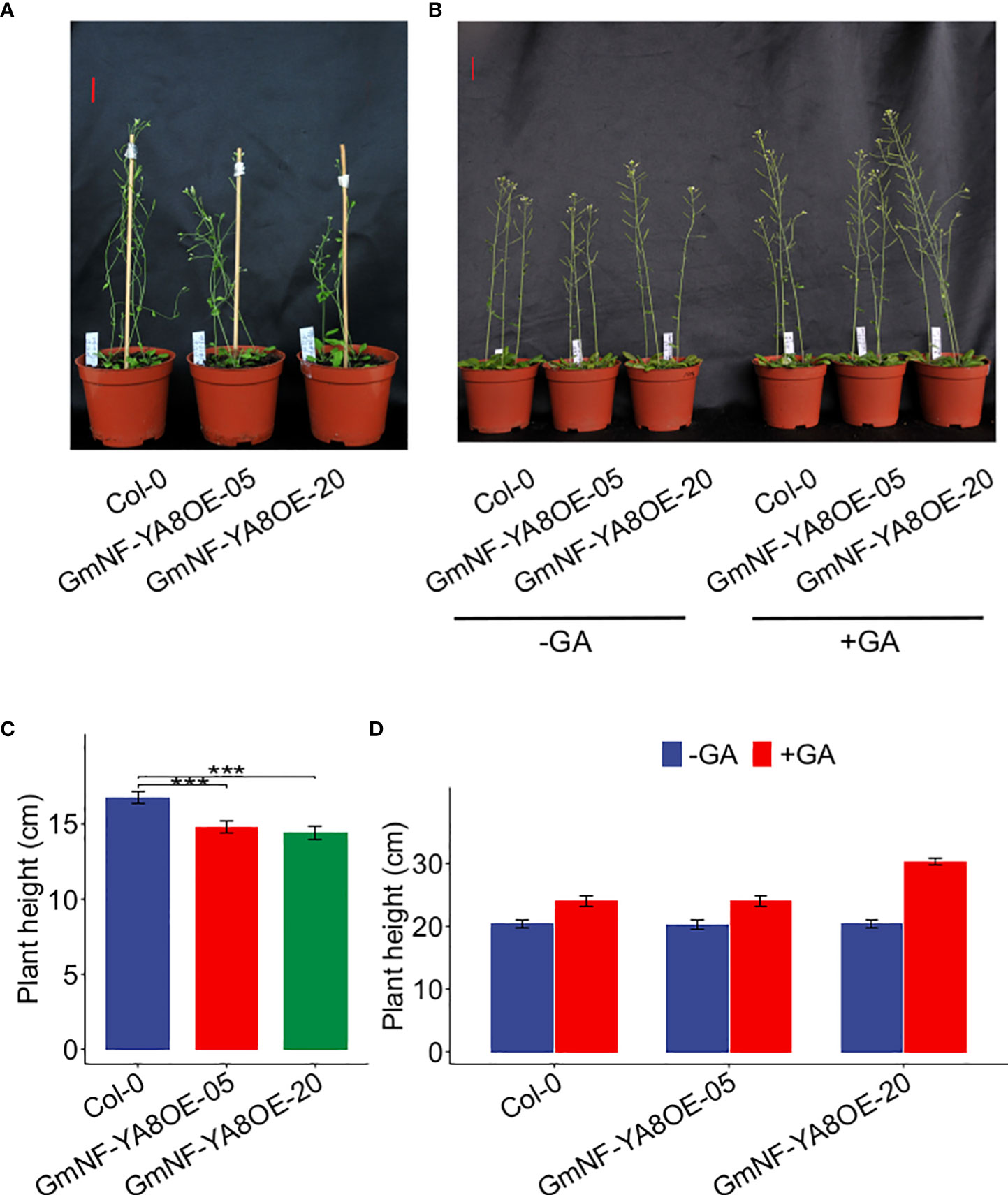
Figure 7 Ectopic expression of GmNF-YA8 decreases Arabidopsis plant height The plant height of the non-transformed Col-0 control and transgenic lines GmNF-YA8OE-05 and GmNF- YA8OE-20 was measured on the 25th day after germination (DAG) (A, B) and 35 DAG (C, D), respectively. Results are means ± SE from each 12 Col-0, GmNF-YA8OE-05, and GmNF-YA8OE-20 plants, respectively. Student’s t-test was used to compare the differences between Col-0 and the transgenic lines GmNF-YA8OE-05 and GmNF-YA8OE-20 (***P < 0.001). Scale bar, 2 cm.
Ectopic expression GmNF-YA8 affects the transcription of GA biosynthesis- and deactivation-related genes
As well documented, GAs play important roles in stem growth and flowering and delaying leaf senescence (Hedden and Thomas, 2012; Colebrook et al., 2014). Here we established that ectopic expression of GmNF-YA8 delayed flowering of Arabidopsis (Figures 6A, B), and that application of GA rescued flowering (Figure 6B). We thus deduced that ectopic expression of GmNF-YA8 affects GA biosynthesis and/or deactivation. To reveal the underlying molecular mechanisms, we measured the expression levels of GA biosynthesis- and deactivation-related genes in the rosette leaves of 12-day-old Arabidopsis seedlings. As reported, GA20ox and GA3ox are involved in the biosynthesis of active GAs, whereas GA2ox catalyzes the conversion of bioactive GA1 and GA4 to inactive GAs such as GA8 and GA34 (Ogawa et al., 2003; Hedden and Thomas, 2012). We found that transcript levels of GA20ox2, but not GA20ox1 and GA20ox3 were significantly lower in GmNF- YA8OE-5 and GmNF-YA8OE-20 plants than that in Col-0 plants (Figure 8A). In addition, the transcript abundance of GA3ox2, but not GA3ox1, was much lower in GmNF-YA8OE-5 and GmNF- YA8OE-20 plants than that in Col-0 plants (Figure 8B). As for GA2ox expression, ectopic expression of GmNF-YA8 increased the transcript levels of GA2ox2 and GA2ox3, but not GA2ox1 (Figure 8C). These data suggest that ectopic expression of GmNF-YA8 decreases the production of active GAs and promotes the conversion of active GAs to inactive GAs.
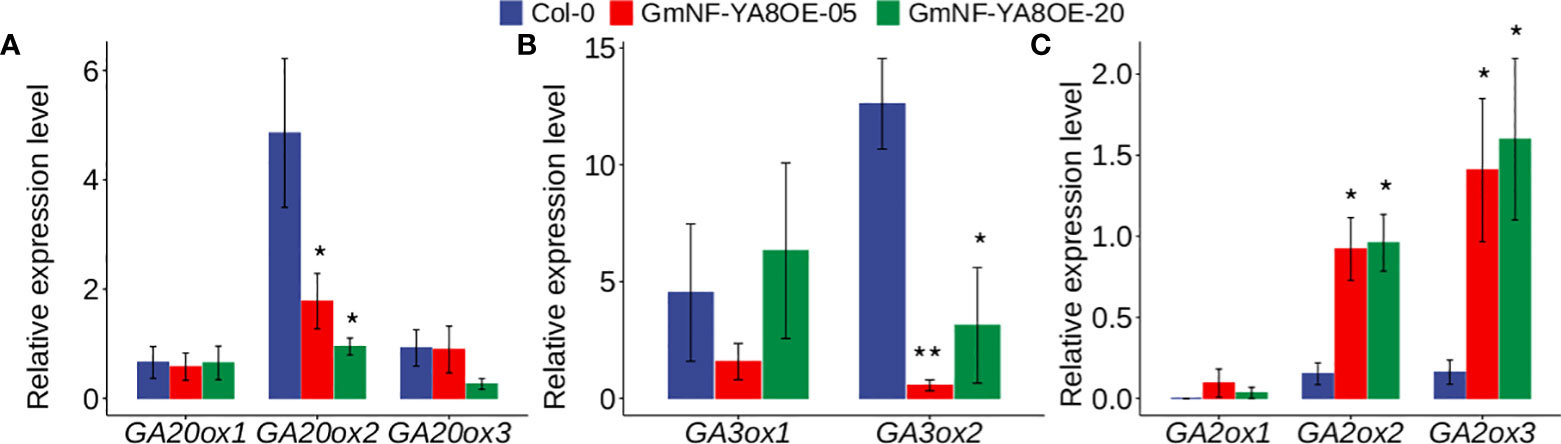
Figure 8 Effects of overexpressing GmNF-YA8 in Arabidopsis on the transcript levels of GA biosynthesis- and deactivation-related genes. (A): GA20ox gene family; (B): GA3ox gene family; (C):GA2ox gene family. Results are means ± SE from three independent experiments. Student’s t-test was used to compare the difference between Col-0, and transgenic lines GmNF-YA8OE-05 and GmNF-YA8OE-20 (*P < 0.05; **P < 0.01).
Ectopic expression of GmNF-YA8 affects the transcription of flowering-related genes
Previous studies have shown that NF-YAs positively or negatively regulate flowering in Arabidopsis (Siriwardana et al., 2016). As revealed, APETALA1 (AP1), CONSTANS (CO), FLOWERING LOCUS T (FT), LEAFY (LFY), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1), and VRN (Chandler et al., 1996; Blazquez et al., 1998; Ng and Yanofsky, 2001; Levy et al., 2002; Moon et al., 2003) promote flowering, whereas FLOWERING LOCUS C (FLC) inhibits flowering in Arabidopsis (Michaels and Amasino, 1999; Bastow et al., 2004). Given that ectopic expression of soybean NF-YA8 delayed flowering in Arabidopsis (Figure 4), we wondered whether ectopic expression of GmNF-YA8 represses flowering by modulating the expression of flowering-related genes. As shown, the transcripts of AP1, CO, LFY, and SOC1 were significantly downregulated in GmNF-YA8OE-05 and GmNF-YA8OE-20 relative to Col-0, whereas FLC was significantly upregulated in GmNF-YA8OE-20 line relative to Col-0 (Figure 9). These results indicate that GmNF-YA8 delays flowering probably by decreasing the transcription of AP1, CO, LFY, and SOC1.
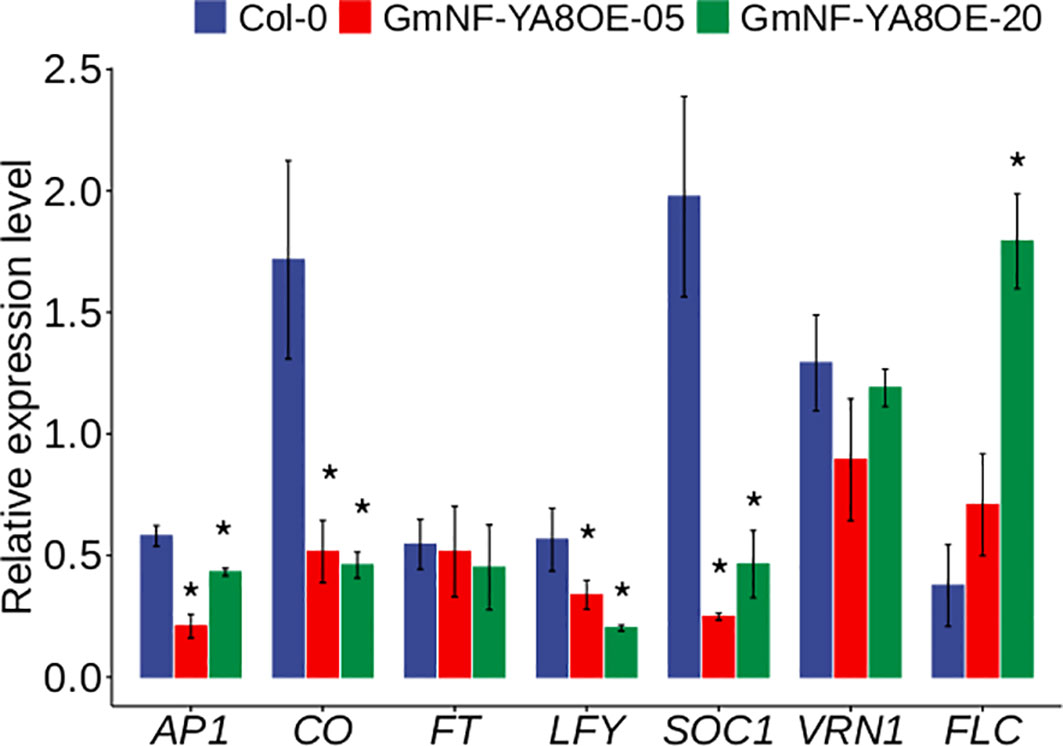
Figure 9 Effects of ectopic expression of GmNF-YA8 on the expression levels of flowering-related genes in Arabidopsis. Results are means ± SE from three independent experiments. Student’s t- tests was used to compare the differences between Col-0 and the transgenic lines GmNF-YA8OE- 05 and GmNF-YA8OE-20 (*P < 0.05).
Ectopic expression of GmNF-YA8 promotes lateral root emergence under low P
In soybean, GmNF-YA8 expression was induced by low P and low N (Figure 2A, B). We thus investigated whether ectopic expression of GmNF-YA8 affects low P responses in Arabidopsis, in particular the remodeling of root system architecture (López-Bucio et al., 2002). To reveal the potential roles of GmNF-YA8 in root growth and development under low P conditions, we grew the overexpressing GmNF-YA8 lines in high P and low P and measured the PR length, the number of LRs and LR density. Consistent with previous studies (Jiang et al., 2007), low P clearly inhibited PR growth in Col-0, GmNF-YA8OE-05, and GmNF-YA8OE-20 plants, respectively (Figure 10A). In low P, the number of LR was significantly increased in the transgenic lines by 13.42% in GmNF- YA8OE-05 and 14.44% in GmNF-YA8OE-20 relative to Col-0 (P < 0.05, Figure 10B), but in high P there was no difference between Col-0 and transgenic lines. Additionally, the LR density in GmNF- YA8OE-05 and GmNF-YA8OE-20 was higher than in Col-0 (P < 0.05, Figure 10C). There are three possibilities to explain overexpressing GmNF-YA8 results in more emerged LRs: by increasing initiation of LR primordium (LRP), by stimulating the penetration of LRP from epidermis, or by both mechanisms. To distinguish these possibilities, we counted the total number of LRP and the number of emerged LR. No difference was observed in the total number of LRP between Col-0 and the transgenic lines, but the percentage of emerged LRs was 85% in GmNF-YA8OE-05 (P < 0.01) and 89% in GmNF-YA8OE-20 (P < 0.001), in contrast to 73% in Col-0 (Figure 10D). These results suggest that GmNF-YA8 does not promote initiation of LRP but does promote the emergence of LRs from the epidermis in low P conditions.
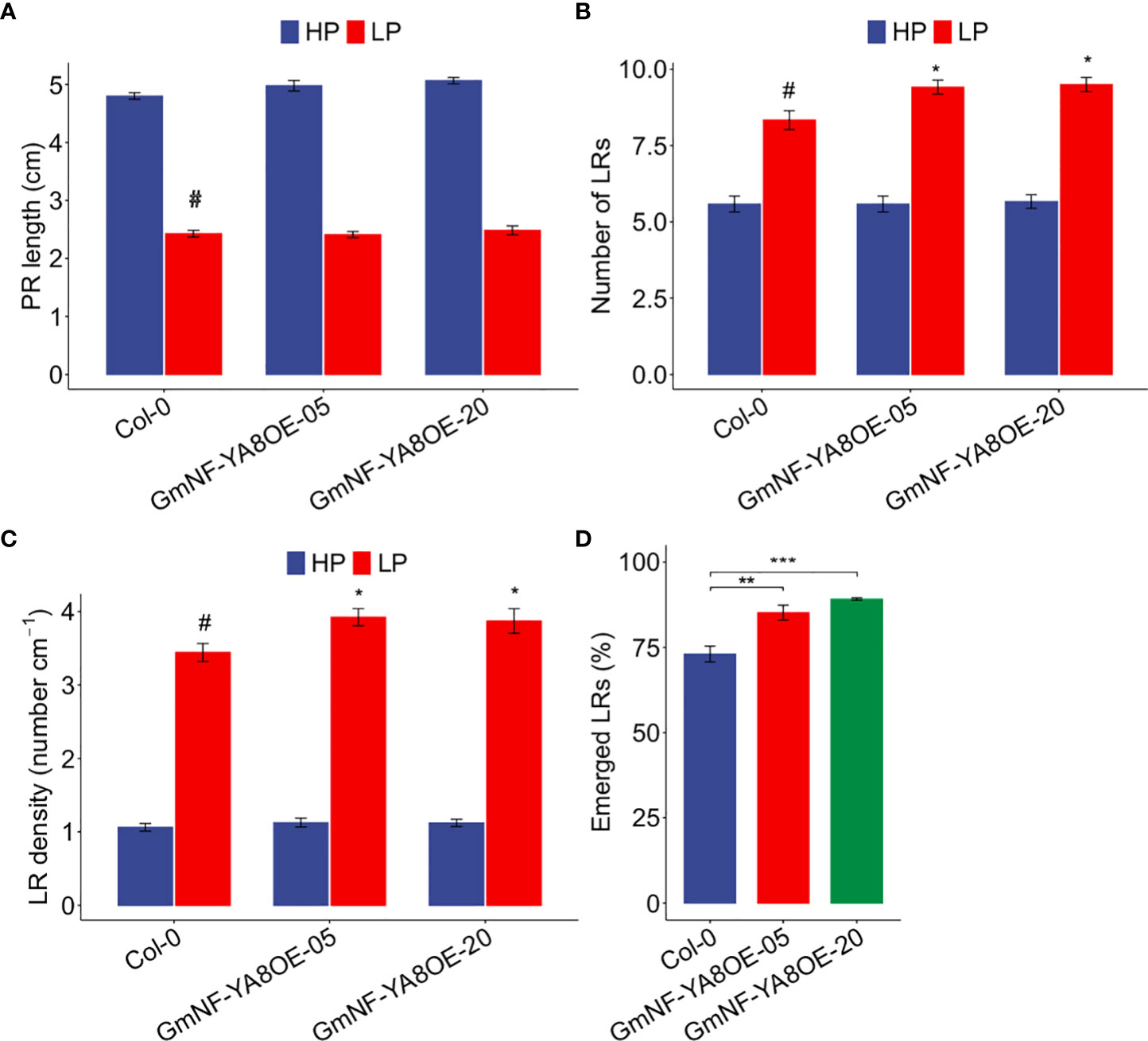
Figure 10 Ectopic expression of GmNF-YA8 increases number of lateral roots (LRs) under low P (LP) conditions, and boosted the percentage of emerged LRs under LP. (A)Primary root (PR) length; (B) Number of LRs; (C) LR density; (D) Emerged LRs (%). Results are means ± SE of three independent experiments. Student’s t-test was used to compare the differences between Col-0 and the transgenic lines GmNF-YA8OE-05 and GmNF-YA8OE-20 (*P < 0.05; **P < 0.01; ***P < 0.001; #, the control).
Lateral root emergence from epidermis is influenced by LIKE AUXIN-UPTAKE CARRIERS3 (LAX3), PECTIN METHYL ESTERASE1 (PME1, PECTIN LYASES1 (PLA1, and PLASMA MEMBERANE INTRINSIC PROTEIN2;1 (PIP2;1) (Laskowski et al., 2006; Swarup et al., 2008; Péret et al., 2012; Vilches-Barro and Maizel, 2015) in Arabidopsis. As revealed, AtPME1 catalyzes the degradation of pectin (Lavenus et al., 2013). PLA1 transcript levels are very high when LRP penetrate from epidermis (Laskowski et al., 2006). As an auxin influx carrier, LAX3 is responsible for the accumulation of auxin in cortex and epidermal cells and upregulates the expression of XTR6 that is involved in LR growth and development (Swarup et al., 2008; Péret et al., 2012). By contrast, transcription of AtPIP2;1 is repressed when LRP emerged from epidermis (Péret et al., 2012; Lavenus et al., 2013). To gain a better understanding to how GmNF-YA8 influences LR emergence, we examined the expression of these four genes in PR of wild type Col-0 and overexpressing GmNF-YA8 plants that were cultured in high P and low P for 7 days. Because no LRP were observed in the first 0.4 cm of the primary root tip, we discarded this region of the PR and collected the remainder for extraction of total RNA. There was no difference in the transcript abundance of AtPME1, AtPLA1, AtLAX3, and AtPIP2;1 between the transgenic lines GmNF- YA8OE-05, GmNF-YA8OE-20 and Col-0 (Figure S2). Considering that these four genes are tightly regulated at the temporal and spatial levels, future experiments will need to measure their transcript levels in epidermis, cortex, and endodermis.
Ectopic expression of GmNF-YA8 increases lateral root number, length, and density under low N
NF-YAs and miR169 have been reported to be involved in low N responses (Zhao et al., 2011). Because GmNF-YA8 was induced by low N (Figure 1B), we thus explored the role of this gene in low N responses. We grew Col-0, GmNF-YA8OE-05, and GmNF-YA8OE-20 plants in high N and low N, and then measured their root-related parameters. In high N, we observed no difference between Col-0 and the ectopic expression lines in PR length (Figure 11A), number of LRs (Figure 11B), average length of the first-order (primary) LRs in each seedling (Figure 11C), and LR density (Figure 11D). In low N, by contrast, the number of LRs was increased by 58.02% in GmNF-YA8OE05 (P < 0.05) and by 107.40% in GmNF-YA8OE-20 plants (P < 0.01, Figure 11B) relative to Col-0; the average length of first-order LRs in each seedling was boosted by 20.35% in GmNF-YA8OE-05 (P < 0.05) and by 42.04% in GmNF-YA8OE-20 plants (P < 0.01, Figure 11C); and LR density was increased by 56.77% in GmNF-YA8OE-05 (P < 0.01) and by 70.15% in GmNF-YA8OE-20 seedlings (P < 0.001, Figure 11D). These results suggest that GmNF-YA8 play active roles in remodeling of root system in response to N deficiency.
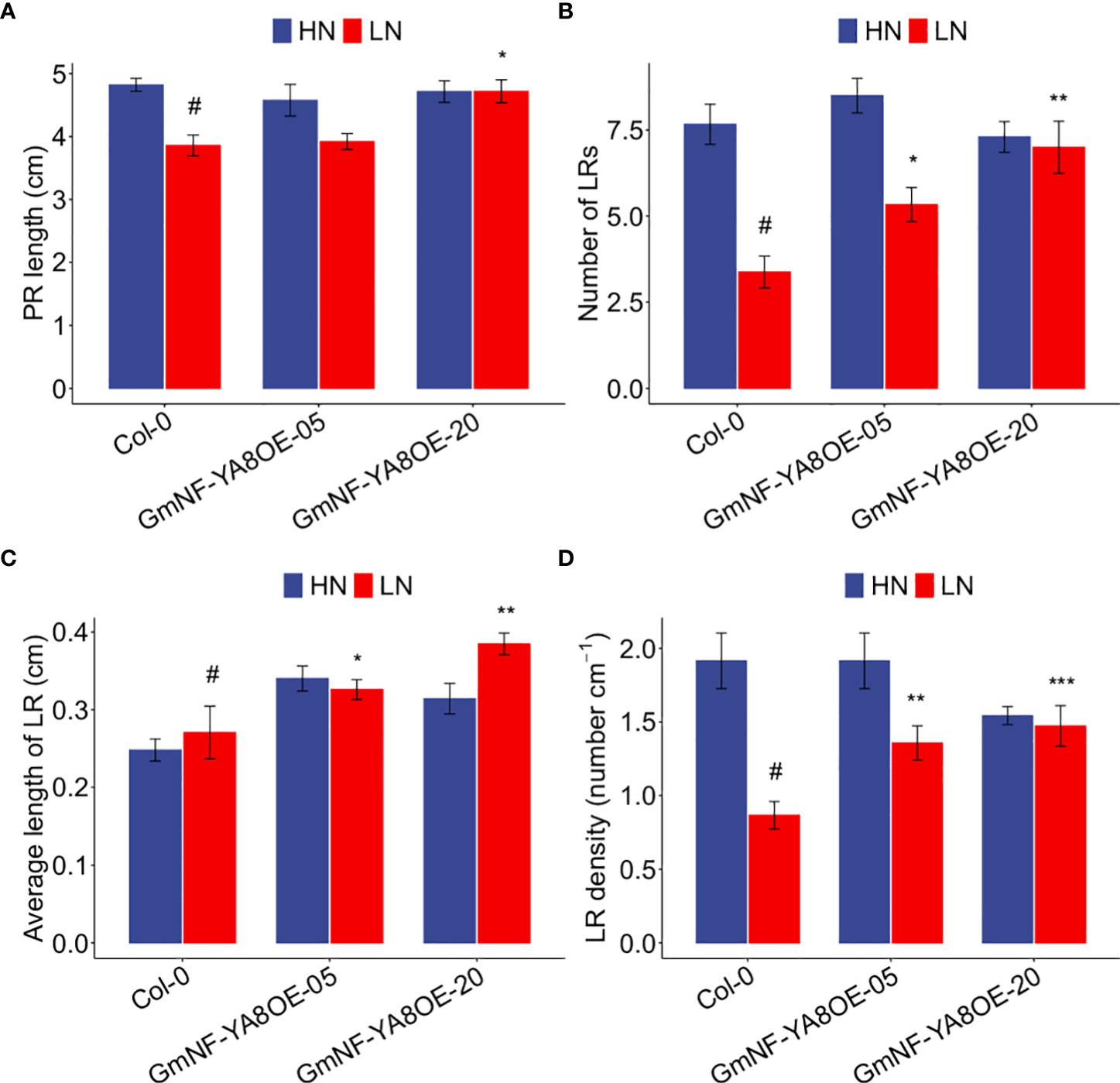
Figure 11 Ectopic expression of GmNF-YA8 increases number of lateral roots (LRs), average length of first-order LRs, and LR density in low N. (A) PR length; (B) LR number; (C) average length of first-order LRs; (D) LR density. Results are means ± SE from three independent experiments. Student’s t-test was used to assess the differences between Col-0 and the transgenic lines GmNF-YA8OE-05 and GmNF-YA8OE-20 (*P < 0.05; **P < 0.01; ***P < 0.001; #, the control).
Discussion
It is well documented that NF-YA interacts with NF-YB and NF-YC to form heterotrimeric transcription factors that regulate the expression of genes involved in plant growth, development, and stress tolerance (Siriwardana et al., 2016). The Glycine max genome encodes 21 NF-YAs, 32 NF-YBs, and 15 NF-YCs (Quach et al., 2015), with the potential to form a huge number of different heterotrimeric combinations. Past studies demonstrated that expression of GmNF-YA3 is induced by drought in soybean and that ectopic expression of GmNF-YA3 in Arabidopsis increases drought tolerance (Ni et al., 2013). In addition, overexpressing GmNF-YA5 enhances tolerance of Arabidopsis and soybean to drought, respectively (Ma et al., 2020). In this study, we revealed that low P stress increased the ratio of root to shoot (Figure 1B), and decreased the concentration of soluble Pi and total P in soybean roots or leaves (Figure 1C, D). GmNF-YA8 is induced by low P and low N stress in soybean leaves (Figure 2), and GmNF-YA8 is localized in the nucleus and plasma membrane (Figure 4). We further showed that overexpression of GmNF-YA8 in Arabidopsis delays bolting (Figure 5), flowering (Figure 6), and stem growth (Figure 7) via modulating the expression of GA biosynthesis- and flowering-related genes (Figure 8-9). Ectopic expression of GmNF-YA8 in Arabidopsis promoted LR emergence in low P (Figure 10),as well as boosting LR growth and density under low N conditions (Figure 11). To our knowledge,this is the first report on the role of soybean NF-YA in flowering, and root growth and development in low P.
NF-YAs are key players in plant nutrition. Microarray analysis indicates that AtNF-YA2, AtNF-YA3, AtNF-YA5, AtNF-YA7, and AtNF-YA10 are induced by low P availability. An increase in AtNF-YA2, AtNF-YA8, and AtNF-YA10 transcript levels was detected by qRT-PCR 4 days after germination in low P, low N, and high sucrose conditions. AtNF-YA3 and AtNF-YA5 showed a similar response to high sucrose but no change in response to low N and low P conditions (Leyva-González et al., 2012). Our initial RNA-seq results revealed that GmNF-YA8 was induced in soybean leaves by P starvation (deposited with NCBI, accession number: PRJNA489734). In addition, our RNA-seq analysis showed that Glyma.15G129900, another NF-YA gene, is also induced by low P in soybean leaves. In the present study, qRT-PCR confirmed that GmNF-YA8 was induced in leaves but not roots in low P (Figure 2A). These results suggest that GmNF-YA8 modulates systemic responses of soybean to Pi deficiency. Of course, the identification of low Pi- responsive NF-YAs in soybean roots is of importance to understand the role of GmNF-YA in local P signaling. In Arabidopsis, the expression of AtNF-YA2, AtNF-YA3, AtNF-YA5, and AtNF-YA8 is induced by N starvation (Wang et al., 2003; Zhao et al., 2011). Consistently, we revealed that GmNF-YA8 is induced by lone-term low N stress (Figure 1B). Of note, the 2-kb-long promoter regions of AtNRT1.1 and AtNRT2.1 contain CCAAT cis-elements (Zhao et al., 2011), suggesting that these genes are regulated by NF-YAs in Arabidopsis. Up to now, it is not clear whether GmNF-YA8 regulates the transcription of nitrate transporter in soybean. Our findings imply that the involvement of NF-YAs in N and P signaling is conserved across plant species. Considering the responses of GmNF-YA8 to P and N starvation, identifying genes that are regulated by GmNF-YA8 at the transcription level is crucial to understand the roles of GmNF-YA8 in P and N nutrition, especially those GA biosynthesis- and flowering-related genes in soybean. As reported, plant transcription factor NF-YAs bind CCAAT motif to regulate gene transcription. Give that overexpressing GmNF-YA8 alters the expression level of GA- and flowering-related genes in Arabidopsis, we thus analyzed the CCAAT cis-element in the promoter region of related Arabidopsis genes. As expected, one to nine CCAAT (or ATTGG) motifs were found in tested genes (Table S2). For instance, one CCAAT motif is found in promoter of AtGA2ox1, nine CCAAT motifs in promoter of AtGA3ox1 (Table S2). Thus we deduced that overexressing GmNF-YA8 delays flowering via modulating the activities of GA-and flowering-related genes at transcription level. Moreove it is also necessary to screen transcription factors that directly bind the cis-element in the promoter of GmNF-YA8.
NF-YAs have been report to be nuclear protein. For instance, AtNF-YA5, GmNF-YA3 and ZmNF-YA3 are localized in nucleus (Li et al., 2008; Ni et al., 2013; Su et al., 2018). Consistently, here we found that GmNF-YA8 is localized in nucleus by tobacco leaf transient transformation experiment (Figure 4), indicating GmNF-YA8 seem to act as transcription factor. On the other hand, the GFP-GmNF-YA8 fusion protein can also be detected in the cell membrane (Figure 4). We noticed that some transcription factors are localized to plasma membrane of plant cells (Liu et al., 2014; Yang et al., 2020).
As reported, overexpression of AtNF-YA2, AtNF-YA8, or AtNF-YA10 in Arabidopsis results in a dwarf phenotype (Leyva-González et al., 2012). In line with this, we observed that ectopic expression of GmNF-YA8 in Arabidopsis delayed bolting (Figure 5) and decreased seedling height, especially during the early stages of growth (Figure 6, Figure 7). The application of GA3 rescues the slow stem growth (Figure 6, Figure 7). Moreover, we detected a decrease in the transcript levels of GA biosynthesis-related genes such as GA20ox2 and GA3ox2, and an increase in those of GA2ox2 and GA2ox3 that are involved in GA inactivation (Figure 8). Thus, Arabidopsis plants overexpressing GmNF-YA8 might have reduced GA biosynthesis and increased GA metabolism. Moreover, genes encoding xyloglucan endotransglucosylase-hydrolases (XTHs) and expansins (EXPs), which are involved in cell elongation, have been observed to be strongly downregulated in overexpressing AtNF-YA2, AtNF-YA7, or AtNF-YA10 Arabidopsis lines (Leyva-González et al., 2012). Hence, it remains to be determined whether the levels of XTH and EXP transcripts are also reduced in Arabidopsis plants overexpressing GmNF-YA8.
Previous studies revealed that NF-YA positively or negatively regulate flowering (Siriwardana et al., 2016). Here we revealed that ectopic expression of GmNF-YA8 in Arabidopsis delays flowering (Figure 6B). Consistently, overexpressing AtNF-YA7 or NF-YA9 also delays flowering in Arabidopsis (Siriwardana et al., 2016). However, overexpressing AtNF-YA2 or NF-YA6 results in early flowering (Siriwardana et al., 2016). Interestingly, ZmNF-YA3 forms complex with CO-like and FPF1, and thus binds the promoter of FT like 12 to promote flowering in maize (Su et al., 2018). AtNF-YA2 and AtNF-YA6 heterotrimerize with NF-YB2 and NF-YC3 in vitro to bind the -5.3 kb CCAAT box of FLOWERING LOCUS T, thus promoting flowering (Siriwardana et al., 2016). Interestingly, the transcription of flowering-related genes such as AP1, CO, LFY, and SOC1 was reduced by the overexpression of GmNF-YA8 (Figure 9). Hence, it is possible that GmNF-YA8 together with NF-YB and NF-YC, even other NF-YA, regulates flowering via its binding to the cis- element of GA biosynthesis- and flowering-related genes in Arabidopsis. Further ChIP-seq experiments will be used to verify this.
As reported, low P stress results in the reduction of bioactive GA (GA4) via upregulating GA20ox1 and GA2ox2, and application of exogenous GA promotes Arabidopsis flowering under low P conditions (Jiang et al., 2007). Moreover, suboptimal P availability delays bolting, flowering and maturation in different Arabidopsis genotypes grown in soils (Nord and Lynch, 2008). Here, we revealed that long-term low P stress induced GmNF-YA8 expression in soybean leaves but not roots (Figure 1), and overexpressing GmNF-YA8 delays flowering in Arabidopsis (Figure 6). Hence we hypothesize that GmNF-YA8 is involved in the flowering process of soybean when subjected to low P stress. Low P stress promotes LR emergence and LR density (López-Bucio et al., 2002; Pérez-Torres et al., 2008). We observed that overexpressing GmNF-YA8 increased the density of LRs when compared to Col-0 plants in low P (Figure 10). However, no difference was detected in the total number of LRP between Col-0 and transgenic lines (Figure 10). Ectopic expression of GmNF-YA8 only stimulated the emergence of LRP but not the initiation of LRP (Figure 10). We also explored the expression of AtPME1, AtPLA1, AtLAX3, and AtPIP2 in low P, which are involved in LR emergence. To our surprise, the transcript abundance of these genes were not significantly affected by overexpressing GmNF-YA8 (Figure S2). AtPME1, AtPLA1, AtPLA2, and AtEXP1 are induced by auxin when LRPs are emerging or have emerged from the cortex and epidermis cells (Laskowski et al., 2006; Swarup et al., 2008), whereas AtPIP2;1 is downregulated by auxin in cortex cells during LR emergence stage (Péret et al., 2012). Thus, LR emergence-related genes are tightly regulated in a spatial-and temporal-dependent manner. Here, we just detected the expression levels of AtPLA1, AtPME1, AtLAX3, and AtPIP2;1 in the rooting zone (Figure S2). Hence, we can not rule out the possibility that these genes are expressed at different levels in the epidermis, cortex, and endodermis cells between Col-0 and GmNF-YA8OE-05 and GmNF-YA8OE-20 line. Of note, no difference was detected between Col-0 and the overexpressing GmNF-YA8 lines (GmNF-YA8OE-05 and GmNF-YA8OE-20) in high N (10 mM KNO3 plus 1 mm NH4NO3) with respect to PR length, average length of first-order (or primary) LRs, number of LRs, and LR density. Whereas the number of LRs, average length of first-order LRs, LR density were boosted in transgenic lines under low N conditions (0.1 mM KNO3 plus 0.01 mm NH4NO3) (Figure 11). This suggests that GmNF-YA8 might improve the tolerance of transgenic Arabidopsis to low N stress via regulating LR emergence and growth. Different results have been reported based on different experimental systems; for instance, LR elongation and PR growth are decreased by high N (1 mM nitrate), but LR density does not appear to be affected by nitrate availability (0.01-1 mM nitrate) (Linkohr et al., 2002). High N (10 mM KNO3 plus 5 mM glutamine) decreases the number of LRs, LR length and density in Arabidopsis Col-0, but low N (0.5 mM KNO3 plus 5 mM glutamine) lightly increases the number and density of LRP and LRs (Tranbarger et al., 2003).
In Arabidopsis, AtNF-YA2 and AtNF-YA10 modulates LR development. Both AtNF-YA2 and AtNF-YA10 are highly expressed in the pericycle, followed in vasculature of elongation zone of PR. Overexpression of miR169-resistant AtNF-YA2 leads to the increase in emerged LRs and total LRPs (Sorin et al., 2014). In Lotus, LjNF-YA1 and LjNF-YB1 have been observed to regulate LR development. Overexpression of LjNF-YA1 results in abnormal LR tips, and extra cell division is taken place in the pericycle that is the origin of LRP (Siefers et al., 2009). Although we did not detect the expression domains of GmNF-YA8 in PR and LR at tissue level, overexpression of GmNF-YA8 has no role in PR and LR growth and development in normal nutrient conditions (Figure 10-11). Given that Glycine max genome has 21 NF-YAs, it is necessary to screen other NF-YAs that regulate soybean root growth and development via detailed transcriptome analysis together with gene editing.
Taken together, we have characterized the functions of GmNF-YA8 gene via ectopic expression. Ectopic overexpressing GmNF-YA8 in Arabidopsis delayed bolting, flowering, and repressing stem growth via affecting the expression of GA-biosynthesis-related genes (GA20ox2, GA20ox3, GA3ox2, GA20ox2, GA20ox3) and flowering-related genes (AP1, CO, LFY, and SOC1), and promoted LR emergence in low P and low N. Hence, GmNF-YA8 appears to have potential applications in breeding soybean with improved N and P effiiciency in agriculture.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
SO, ZX participated in the study design, carried out the experiments and data analysis. CM, BL carried out the experiments and analyzed data. JW conceived, designed experiments, analyzed data, and authored the manuscript. All authors have contributed to the article and approved the submitted manuscript.
Funding
This study was partially supported by National Key Research and Development Program of China (No.2021YFF1000500) and Natural Science Foundation of Guangdong Province, China (No. 2017A030313102).
Acknowledgments
We thank Jennifer Mach for her comments and help in English writing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1033938/full#supplementary-material
Supplementary Figure 1 | Identification of over-expressing GmNG-YA8 in Arabidopsis wild type Col-0 background. The #5 (GmNF-YA8OE-05) and #20 (GmNF-YA8OE-20) indicate the two single copy T-DNA insertion lines. Total RNA was extracted from Col-0 and transgenic Arabidopsis lines, respectively. The quantitative real-time PCR (qRT-PCR) was employed to explore the transcript level of GmNF-YA8 in Arabidopsis. Arabidopsis EF1a was used as house-keeping gene to normalize PCR results.
Supplementary Figure 2 | Transcript levels of AtLAX3, AtPME1, AtPIP2, and AtPLA1 in rooting zone of PR.
Supplementary Table 1 | List of primer pairs used in this study.
Supplementary Table 2 | List of CCAAT motifs in the promoter region of GA- and flowering-related genes.
Supplementary Sheet 1 | Fold change of GmNF-YA8 to different treatments. RNA-seq data is from public soybean RNA-seq database (http://ipf.sustech.edu.cn/pub/soybean/), which includes 4,085 Glycine max RNA-Seq libraries.
References
Bastow, R., Mylne, J. S., Lister, C., Lippman, Z., Martienssen, R. A., Dean, C. (2004). Vernalization requires epigenetic silencing of flc by histone methylation. Nature 427, 164–167. doi: 10.1038/nature02269
Blazquez, M., Green, R., Nilsson, O., Sussman, M., Weigel, D. (1998). Gibberellins promote flowering of Arabidopsis by activating the leafy promoter. Plant Cell 10, 791–800. doi: 10.2307/3870665
Cao, S., Kumimoto, R. W., Siriwardana, C. L., Risinger, J. R., Holt, B. F. (2011). Identification and characterization of NF-Y transcription factor families in the monocot model plant brachypodium distachyon. PloS One 6, e21805. doi: 10.1371/journal.pone.0021805
Chandler, J., Wilson, A., Dean, C. (1996). Arabidopsis mutants showing an altered response to vernalization. Plant journal: Cell Mol. Biol. 10, 637–644. doi: 10.1046/j.1365-313X.1996.10040637.x
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Colebrook, E. H., Thomas, S. G., Phillips, A. L., Hedden, P. (2014). The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 217, 67–75. doi: 10.1242/jeb.089938
Curtis, M. D., Grossniklaus, U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469. doi: 10.1104/pp.103.027979
Gnesutta, N., Kumimoto, R. W., Swain, S., Chiara, M., Siriwardana, C., Horner, D. S., et al. (2017). Constans imparts DNA sequence specificity to the histone fold NF-YB/NF-YC dimer. Plant Cell 29, 1516–1532. doi: 10.1105/tpc.16.00864
Hedden, P., Thomas, S. G. (2012). Gibberellin biosynthesis and its regulation. Biochem. J. 444, 11–25. doi: 10.1042/bj20120245
Hou, X., Zhou, J., Liu, C., Liu, L., Shen, L., Yu, H. (2014). Nuclear factor y-mediated h3k27me3 demethylation of the SOC1 locus orchestrates flowering responses of arabidopsis. Nat. Commun. 5, 4601. doi: 10.1038/ncomms5601
Huo, W., Li, B., Kuang, J., He, P., Xu, Z., Wang, J. (2018). Functional characterization of the steroid reductase genes GMDET2A and GMDET2B from Glycine max. Int. J. Mol. Sci. 19. doi: 10.3390/ijms19030726
Jiang, C., Gao, X., Liao, L., Harberd, N. P., Fu, X. (2007). Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-della signaling pathway in Arabidopsis. Plant Physiol. 145, 1460–1470. doi: 10.1104/pp.107.103788
Laskowski, M., Biller, S., Stanley, K., Kajstura, T., Prusty, R. (2006). Expression profiling of auxin-treated arabidopsis roots: toward a molecular analysis of lateral root emergence. Plant Cell Physiol. 47, 788–792. doi: 10.1093/pcp/pcj043
Lavenus, J., Goh, T., Roberts, I., Guyomarc’h, S., Lucas, M., De Smet, I., et al. (2013). Lateral root development in arabidopsis: fifty shades of auxin. Trends Plant Sci. 18, 450–458. doi: 10.1016/j.tplants.2013.04.006
Lee, D.-K., Kim, H. I., Jang, G., Chung, P. J., Jeong, J. S., Kim, Y. S., et al. (2015). The nf-ya transcription factor OsNF-YA7 confers drought stress tolerance of rice in an abscisic acid independent manner. Plant sci.: an Int. J. Exp. Plant Biol. 241, 199–210. doi: 10.1016/j.plantsci.2015.10.006
Levy, Y. Y., Mesnage, S., Mylne, J. S., Gendall, A. R., Dean, C. (2002). Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science 297, 243–246. doi: 10.1126/science.1072147
Leyva-González, M. A., Ibarra-Laclette, E., Cruz-Ramírez, A., Herrera-Estrella, L. (2012). Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by arabidopsis NF-YA family members. PloS One 7, e48138. doi: 10.1371/journal.pone.0048138
Li, Y.-J., Fang, Y., Fu, Y.-R., Huang, J.-G., Wu, C.-A., Zheng, C.-C. (2013). NFYA1 is involved in regulation of post germination growth arrest under salt stress in Arabidopsis. PloS One 8, e61289. doi: 10.1371/journal.pone.0061289
Linkohr, B. I., Williamson, L. C., Fitter, A. H., Leyser, H. O. (2002). Nitrate and phosphate availability and distribution have different effects on root system architecture of arabidopsis. Plant J. 29, 751–760. doi: 10.1046/j.1365-313X.2002.01251.x
Li, W.-X., Oono, Y., Zhu, J., He, X.-J., Wu, J.-M., Iida, K., et al. (2008). The arabidopsis NFYA5 transcription factor is regulated transcriptionally and post transcriptionally to promote drought resistance. Plant Cell 20, 2238–2251. doi: 10.1105/tpc.108.059444
Liu, B., Ouyang, Z., Zhang, Y., Li, X., Hong, Y., Huang, L., et al. (2014). Tomato NAC transcription factor SLSRN1 positively regulates defense response against biotic stress but negatively regulates abiotic stress response. PloS One 9, e102067. doi: 10.1371/journal.pone.0102067
López-Bucio, J., Hernández-Abreu, E., Sánchez-Calderón, L., Nieto-Jacobo, M. F., Simpson, J., Herrera-Estrella, L. (2002). Phosphate availability alters architecture and causes changes in hormone sensitivity in the arabidopsis root system. Plant Physiol. 129, 244–256. doi: 10.1104/pp.010934
Mantovani, R. (1998). A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 26, 1135–1143. doi: 10.1093/nar/26.5.1135
Ma, X.-J., Yu, T.-F., Li, X.-H., Cao, X.-Y., Ma, J., Chen, J., et al. (2020). Overexpression of GmNFYA5 confers drought tolerance to transgenic arabidopsis and soybean plants. BMC Plant Biol. 20, 123. doi: 10.1186/s12870-020-02337-z
Ma, X., Zhu, X., Li, C., Song, Y., Zhang, W., Xia, G., et al. (2015). Overexpression of wheat NF-YA10 gene regulates the salinity stress response in arabidopsis thaliana. Plant Physiol. biochem.: PPB 86, 34–43. doi: 10.1016/j.plaphy.2014.11.011
Michaels, S., Amasino, R. (1999). Flowering locus c encodes a novel mads domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956. doi: 10.2307/3870827
Misson, J., Raghothama, K. G., Jain, A., Jouhet, J., Block, M. A., Bligny, R., et al. (2005). A genome-wide transcriptional analysis using Arabidopsis thaliana affymetrix gene chips determined plant responses to phosphate deprivation. Proc. Natl. Acad. Sci. United States America 102, 11934–11939. doi: 10.1073/pnas.0505266102
Moon, J., Suh, S.-S., Lee, H., Choi, K.-R., Hong, C. B., Paek, N.-C., et al. (2003). The SOC1 mads- box gene integrates vernalization and gibberellin signals for flowering in arabidopsis. Plant journal: Cell Mol. Biol. 35, 613–623. doi: 10.1046/j.1365-313X.2003.01833.x
Mu, J., Tan, H., Hong, S., Liang, Y., Zuo, J. (2013). Arabidopsis transcription factor genes NF-YA1, 5, 6, and 9 play redundant roles in male gametogenesis, embryogenesis, and seed development. Mol. Plant 6, 188—201. doi: 10.1093/mp/sss061
Myers, Z. A., Kumimoto, R. W., Siriwardana, C. L., Gayler, K. K., Risinger, J. R., Pezzetta, D., et al. (2016). Nuclear factor y, subunit c (NF-YC) transcription factors are positive regulators of photomorphogenesis in Arabidopsis thaliana. PloS Genet. 12, e1006333. doi: 10.1371/journal.pgen.1006333
Nelson, D. E., Repetti, P. P., Adams, T. R., Creelman, R. A., Wu, J., Warner, D. C., et al. (2007). Plant nuclear factor y (NF-Y) b subunits confer drought tolerance and lead to improved corn yieldson water-limited acres. Proc. Natl. Acad. Sci. United States America 104, 16450–16455. doi: 10.1073/pnas.0707193104
Ng, M., Yanofsky, M. (2001). Activation of the arabidopsis b class homeotic genes by APETALA1. Plant Cell 13, 739–753. doi: 10.2307/3871337
Ni, Z., Hu, Z., Jiang, Q., Zhang, H. (2013). Gmnfya3, a target gene of mir169, is a positive regulator of plant tolerance to drought stress. Plant Mol. Biol. 82, 113–129. doi: 10.1007/s11103-013-0040-5
Nord, E. A., Lynch, J. P. (2008). Delayed reproduction in Arabidopsis thaliana improves fitness in soil with suboptimal phosphorus availability. Plant Cell Environ. 31, 1432–1441. doi: 10.1111/j.1365-3040.2008.01857.x
Ogawa, M., Hanada, A., Yamauchi, Y., Kuwahara, A., Kamiya, Y., Yamaguchi, S. (2003). Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15, 1591–1604. doi: 10.1105/tpc.011650
Peng, S., Tao, P., Xu, F., Wu, A., Huo, W., Wang, J. (2016). Functional characterization of soybean Glyma04g39610 as a brassinosteroid receptor gene and evolutionary analysis of soybean brassinosteroid receptors. Int. J. Mol. Sci. 17. doi: 10.3390/ijms17060897
Péret, B., Li, G., Zhao, J., Band, L. R., Voß, U., Postaire, O., et al. (2012). Auxin regulates aquaporin function to facilitate lateral root emergence. Nat. Cell Biol. 14, 991–998. doi: 10.1038/ncb2573
Pérez-Torres, C.-A., López-Bucio, J., Cruz-Ramírez, A., Ibarra-Laclette, E., Dharmasiri, S., Estelle, M., et al. (2008). Phosphate availability alters lateral root development in arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell 20, 3258–3272. doi: 10.1105/tpc.108.058719
Petroni, K., Kumimoto, R. W., Gnesutta, N., Calvenzani, V., Fornari, M., Tonelli, C., et al. (2012). The promiscuous life of plant nuclear factor y transcription factors. Plant Cell 24, 4777–4792. doi: 10.1105/tpc.112.105734
Quach, T. N., Nguyen, H. T. M., Valliyodan, B., Joshi, T., Xu, D., Nguyen, H. T. (2015). Genome-wide expression analysis of soybean NF-Y genes reveals potential function in development and drought response. molecular genetics and genomics. MGG 290, 1095—1115. doi: 10.1007/s00438-014-0978-2
R Core Team (2018). R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing).
Schaarschmidt, S., Gresshoff, P. M., Hause, B. (2013). Analyzing the soybean transcriptome during autoregulation of mycorrhization identifies the transcription factors GmNF-YA1A/B as positive regulators of arbuscular mycorrhization. Genome Biol. 14, R62. doi: 10.1186/gb-2013-14-6-r62
Siefers, N., Dang, K. K., Kumimoto, R. W., Bynum, W. E., Tayrose, G., Holt, B. F. (2009). Tissue-specific expression patterns of arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 149, 625–641. doi: 10.1104/pp.108.130591
Siriwardana, C. L., Gnesutta, N., Kumimoto, R. W., Jones, D. S., Myers, Z. A., Mantovani, R., et al. (2016). Nuclear factor Y, subunit a (NF-YA) proteins positively regulate flowering and act through flowering locus t. PloS Genet. 12, e1006496. doi: 10.1371/journal.pgen.1006496
Siriwardana, C. L., Kumimoto, R. W., Jones, D. S., Holt, B. F. (2014). Gene family analysis of the arabidopsis NF-YA transcription factors reveals opposing abscisic acid responses during seed germination. Plant Mol. Biol. Rep. 32, 971–986. doi: 10.1007/s11105-014-0704-6
Sorin, C., Declerck, M., Christ, A., Blein, T., Ma, L., Lelandais-Brière, C., et al. (2014). A mir169 isoform regulates specific NF-YA targets and root architecture in arabidopsis. New Phytol. 202, 1197–1211. doi: 10.1111/nph.12735
Stephenson, T. J., McIntyre, C. L., Collet, C., Xue, G.-P. (2007). Genome-wide identification and expression analysis of the NF-Y family of transcription factors in Triticum aestivum. Plant Mol. Biol. 65, 77–92. doi: 10.1007/s11103-007-9200-9
Su, H., Cao, Y., Ku, L., Yao, W., Cao, Y., Ren, Z., et al. (2018). Dual functions of ZmNF-YA3 in photoperiod-dependent flowering and abiotic stress responses in maize. J. Exp. Bot. 69, 5177–5189. doi: 10.1093/jxb/ery299
Swarup, K., Benková, E., Swarup, R., Casimiro, I., Péret, B., Yang, Y., et al. (2008). The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 10, 946–954. doi: 10.1038/ncb1754
Thirumurugan, T., Ito, Y., Kubo, T., Serizawa, A., Kurata, N. (2008). Identification, characterization and interaction of HAP family genes in rice. Mol. Genet. genomics: MGG 279, 279–289. doi: 10.1007/s00438-007-0312-3
Tranbarger, T. J., Al-Ghazi, Y., Muller, B., Teyssendier, D. L. S., Doumas, P., Touraine, B. (2003). Transcription factor genes with expression correlated to nitrate-related root plasticity of Arabidopsis thaliana. Plant Cell Environ. 26, 459–469. doi: 10.1046/j.1365-3040.2003.00977.x
Vilches-Barro, A., Maizel, A. (2015). Talking through walls: mechanisms of lateral root emergence in Arabidopsis thaliana. Curr. Opin. Plant Biol. 23, 31–38. doi: 10.1016/j.pbi.2014.10.005
Wang, R., Okamoto, M., Xing, X., Crawford, N. M. (2003). Microarray analysis of the nitrate response in arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol. 132, 556–567. doi: 10.1104/pp.103.021253
Warpeha, K. M., Upadhyay, S., Yeh, J., Adamiak, J., Hawkins, S. I., Lapik, Y. R., et al. (2007). The GCR1, GPA1, PRN1, NF-Y signal chain mediates both blue light and abscisic acid responses in Arabidopsis. Plant Physiol. 143, 1590–1600. doi: 10.1104/pp.106.089904
Wenkel, S., Turck, F., Singer, K., Gissot, L., Le Gourrierec, J., Samach, A., et al. (2006). Constans and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18, 2971–2984. doi: 10.1105/tpc.106.043299
Xu, F., Liu, Q., Chen, L., Kuang, J., Walk, T., Wang, J., et al. (2013). Genome-wide identification of soybean microRNAs and their targets reveals their organ-specificity and responses to phosphate starvation. BMC Genomics 14, 66. doi: 10.1186/1471-2164-14-66
Yang, J. H., Lee, K.-H., Du, Q., Yang, S., Yuan, B., Qi, L., et al. (2020). A membrane- associated NAC domain transcription factor XVP interacts with TDIF co-receptor and regulates vascular meristem activity. New Phytol. 226, 59–74. doi: 10.1111/nph.16289
Zhao, M., Ding, H., Zhu, J.-K., Zhang, F., Li, W.-X. (2011). Involvement of MiR169 in the nitrogen-starvation responses in arabidopsis. New Phytol. 190, 906–915. doi: 10.1111/j.1469-8137.2011.03647.x
Zhao, B., Ge, L., Liang, R., Li, W., Ruan, K., Lin, H., et al. (2009). Members of mir-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Mol. Biol. 10, 29. doi: 10.1186/1471-2199-10-29
Keywords: GmNF-YA8, flowering, phosphorus deficiency, lateral roots, gibberellic acid
Citation: Ou S, Xu Z, Mai C, Li B and Wang J (2022) Ectopic expression of GmNF-YA8 in Arabidopsis delays flowering via modulating the expression of gibberellic acid biosynthesis- and flowering-related genes and promotes lateral root emergence in low phosphorus conditions. Front. Plant Sci. 13:1033938. doi: 10.3389/fpls.2022.1033938
Received: 01 September 2022; Accepted: 26 September 2022;
Published: 20 October 2022.
Edited by:
Zhijian Chen, Chinese Academy of Tropical Agricultural Sciences, ChinaReviewed by:
Yiyong Zhu, Nanjing Agricultural University, ChinaKuaifei Xia, South China Botanical Garden, Chinese Academy of Sciences (CAS), China
Copyright © 2022 Ou, Xu, Mai, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinxiang Wang, amlueHdhbmdAc2NhdS5lZHUuY24=
†These authors have contributed equally to this work
 Siyan Ou1,2,3†
Siyan Ou1,2,3† Jinxiang Wang
Jinxiang Wang