- 1Department of Molecular Genetics, Dong-A University, Busan, South Korea
- 2School of Life Sciences, Anhui Agricultural University, Hefei, China
- 3Graduate School of Applied Bioscience, Dong-A University, Busan, South Korea
- 4Institut für Biologie, Fachgebiet Mikrobiologie, Universität Kassel, Kassel, Germany
The Elongator complex in eukaryotes has conserved tRNA modification functions and contributes to various physiological processes such as transcriptional control, DNA replication and repair, and chromatin accessibility. ARABIDOPSIS ELONGATOR PROTEIN 4 (AtELP4) is one of the six subunits (AtELP1–AtELP6) in Arabidopsis Elongator. In addition, there is an Elongator-associated protein, DEFORMED ROOTS AND LEAVES 1 (DRL1), whose homolog in yeast (Kti12) binds tRNAs. In this study, we explored the functions of AtELP4 in plant-specific aspects such as leaf morphogenesis and evolutionarily conserved ones between yeast and Arabidopsis. ELP4 comparison between yeast and Arabidopsis revealed that plant ELP4 possesses not only a highly conserved P-loop ATPase domain but also unknown plant-specific motifs. ELP4 function is partially conserved between Arabidopsis and yeast in the growth sensitivity toward caffeine and elevated cultivation temperature. Either single Atelp4 or drl1-102 mutants and double Atelp4 drl1-102 mutants exhibited a reduction in cell proliferation and changed the adaxial–abaxial polarity of leaves. In addition, the single Atelp4 and double Atelp4 drl1-102 mutants showed remarkable downward curling at the whole part of leaf blades in contrast to wild-type leaf blades. Furthermore, our genetic study revealed that AtELP4 might epistatically act on DRL1 in the regulation of cell proliferation and dorsoventral polarity in leaves. Taken together, we suggest that AtELP4 as part of the plant Elongator complex may act upstream of a regulatory pathway for adaxial–abaxial polarity and cell proliferation during leaf development.
Introduction
Leaf formation in plants requires intricate and multiple mechanisms to determine various factors including leaf size and shape, phyllotaxis, heteroblasty, and structural asymmetry. These mechanisms are managed through genetic programs adapted by leaf function and environmental cues during evolutionary progression. To accomplish leaf development, plants undergo dynamic sequences of regulatory processes: from loss of meristematic stem cell identity, leaf initiation, establishment of leaf polarity (adaxial–abaxial, medial–lateral, and distal–proximal axis), cytoplasmic growth, cell proliferation and expansion, endoreduplication, cell type-specific differentiation such as guard cells, vascular tissue, and trichomes, to mature leaves (Kalve et al., 2014). During adaxial/abaxialization among these processes, leaves construct the adaxial side, which is adjacent to the shoot apical meristem (SAM), the upper sun-exposed side in mature leaves, and the abaxial side, which is farther from SAM, the lower shaded side in mature leaves. Leaf adaxial/abaxialization specializes leaf external and internal structure according to their function; the adaxial portion is built up into compactly packed and chloroplast-rich cells and more trichomes for optimal light capture and defense, whereas the abaxial portion is built up into loosely packed cells and more guard cells and stoma for gas exchange. The vascular bundle has also discrimination between adaxial and abaxial sides, showing different positioning of xylem and phloem cells. Therefore, the acquisition of adaxial/abaxial identity in leaf structure is necessary for the performance of leaf function. Numerous studies have shown that the expression of class III homeodomain-leucine zipper (HD-ZIP III) genes, PAHBULOSA (PHB), PHAVOLUTA (PHV), and REVOLUTA (REV) determines the identity of the adaxial domain (McConnell et al., 2001), while the expression of YABBY (YAB) and KANADI (KAN) gene families determines the identity of the abaxial domain (Eshed et al., 2004). In addition, the expression of adaxial- or abaxial-determinant genes is regulated not only by each other through negative feedback (Tsukaya, 2013) but also by small RNAs and auxin (Pekker et al., 2005; Alvarez et al., 2006).
The growth imbalance between the adaxial side and the abaxial side causes defects in leaf flatness, and leaf epinasty or hyponasty (upward or downward curling, respectively) (Sandalio et al., 2016). These morphological changes are triggered by alteration in auxin accumulation or gradient (Enders and Strader, 2015). However, it is not well known how auxin controls the leaf flatness or the growth difference between the adaxial/abaxial sides.
Apart from the differential transcription of protein-encoding genes, an emerging mechanism to regulate protein levels is translational modulation. The latter involves the modification of tunable mRNA transcripts (MoTT) with their decoding coupled to tRNA modifications, in particular anticodon wobble uridine (U34) bases. The Elongator complex was originally identified as a transcription factor in yeast (S. cerevisiae) (Otero et al., 1999; Wittschieben et al., 1999). Meanwhile, however, it is widely accepted that the primary and conserved role of Elongator from yeast to plants and humans lies with tRNA modification and mRNA translation (Otero et al., 1999; Hawkes et al., 2002; Huang et al., 2005; Mehlgarten et al., 2010; Nelissen et al., 2010; Johansson et al., 2018; Nakai et al., 2019; Abbassi et al., 2020). Accordingly, the Elongator complex (Elp1–Elp6) is key to a pathway that adds 5-carboxy-methyl (cm5) groups to several tRNA anticodons (Selvadurai et al., 2014; Glatt et al., 2016; Lin et al., 2019; Dauden et al., 2019). Among others, these can be further modified to 5-methoxy-carbonyl-methyl (mcm5) groups or 2-thio derivatives (mcm5s2) thereof in concert with U34 methylase and thiolase activities (Schaffrath and Leidel, 2017; Johansson et al., 2018; Sokołowski et al., 2018).
The Elongator complex consists of two subcomplexes, each comprising three subunits: Elp123 and Elp456 (Dauden et al., 2019). In yeast, there is an additional protein (Kti12/Tot4), which associates with Elongator (Frohloff et al., 2001; Mehlgarten et al., 2017). It binds tRNA and is essential for the tRNA modification activity, which resides in the Elongator catalytic subunit Elp3 (Svejstrup, 2007; Glatt et al., 2016; Krutyhołowa et al., 2019; Abbassi et al., 2020). Elongator components in eukaryotes are structurally conserved (Dauden et al., 2017a; Dauden et al., 2017b), and previous studies had reported that in yeast, two copies each of Elp4, Elp5, and Elp6 form a RecA-ATPase-like ring structure with roles in substrate recognition during Elongator-mediated tRNA modification (Glatt et al., 2012; Glatt et al., 2016; Dauden et al., 2017a; Dauden et al., 2017b). Despite their similarity in structure and tRNA modification activity, Elongator complexes from yeast, mammals, and plants may functionally differ in various physiological processes and contexts (Nelissen et al., 2005; Close et al., 2006; Chen et al., 2009; Nelissen et al., 2010; Nakai et al., 2019).
Previously, we isolated and characterized Arabidopsis DEFORMED ROOTS AND LEAVES 1 (DRL1), a gene homologous to yeast Kluyveromyces lactis TOXIN INSENSITIVE 12/TOXIN TARGET 4 (KTI12/TOT4), which encodes an Elongator-associated tRNA binding protein (Mehlgarten et al., 2017; Krutyhołowa et al., 2019). DRL1 shows a partially conserved function to yeast Kti12 and is involved in the regulation of leaf polarity and cell proliferation of leaves of Arabidopsis thaliana. In this study, we characterized AtELP4 functions. Although we could not detect binding between AtELP4 and DRL1 or other individual plant Elongator subunits and DRL1, we found that an Atelp4 mutant showed narrow and abaxialized leaves very similar to those of a drl1-102 mutant. We suggest that AtELP4 as part of the Elongator complex in Arabidopsis plays critical roles in the establishment of leaf polarity, especially adaxialization through a collaboration with DRL1 and regulation of leaf cell proliferation.
Materials and methods
Phylogeny relationship, domain, and motif analysis
All ELP4 full-length protein sequences from various eukaryotes identified and verified conserved domain through Pfam (https://pfam.xfam.org/) (Finn et al., 2006), NCBI Conserved Domain Database (CDD) database (https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml), and SMART tool (http://smart.emblheidelberg.de) (Letunic et al., 2012). In addition, identified ELP4 genes were aligned using ClustalX software with default parameters (Thompson et al., 1997). Finally, the phylogeny was constructed by using the neighbor-joining method (NJM). The amino acid sequences of ELP4 proteins used in the alignment were retrieved from the NCBI GenBank database (https://www.ncbi.nlm.nih.gov/). The GenBank accession numbers of the amino acid sequences are A. thaliana (Q9C778.1), Ostreococcus tauri (XP_003082942.1), Marchantia polymorpha subsp. ruderalis (BBN04784.1), Physcomitrella patens (XP_024385693.1), Selaginella moellendorffii (XP_024534079.1), Amborella trichopoda (XP_020520905.1), Nymphaea colorata (XP_031482638.1), Zea mays (NP_001131483.1), Sorghum bicolor (XP_021304923.1), O. sativa Japonica Group (XP_015643899.1), Brachypodium distachyon (XP_003563291.1), Phalaenopsis equestris (XP_020577487.1), Macadamia integrifolia (XP_042486942.1), Rosa chinensis (XP_040364853.1), Glycine max (KAH1257688.1), Medicago truncatula (XP_013443887.1), Gossypium hirsutum (XP_016734795.1), Hibiscus syriacus (KAE8729835.1), Vitis vinifera (RVW57326.1), Helianthus annuus (XP_035832566.1), Manihot esculenta (XP_021624621.1), Cucumis sativus (XP_031743242.1), Ipomoea nil (XP_019185341.1), Nicotiana tabacum (XP_016448924.1), Brassica napus (XP_022558390.1), Salvia hispanica (XP_047969651.1), Solanum lycopersicum (XP_025883858.1), S. cerevisiae (NP_015224), Schizosaccharomyces pombe (Q9USP1), K. lactis (XP_455048), Dictyostelium discoideum (Q54XS0.1), Hydra vulgaris (XP_047135159.1), Caenorhabditis elegans (NP_001370473.1), Strongylocentrotus purpuratus (XP_030844920.1), Aplysia californica (XP_005105912.1), Drosophila melanogaster (Q9VMQ7), Xenopus laevis (NP_001088363.1), Danio rerio (NP_001017638.1), Mus musculus (NP_076365.2), and Homo sapiens (KAI2559206.1). Subsequently, conserved motifs of ELP4 in plants, yeasts, and animals were identified by using the MEME online tool (http://meme-suite.org/tools/meme) with the other default parameters and five maximum numbers of motifs (Bailey et al., 2015).
Syntenic relationship analysis
Sequence similarity of ELP4 genes between the plant and yeast was carried through circoletto software (http://tools.bat.infspire.org/circoletto/) (Darzentas, 2010). ELP4 gene sequences were used as queries against plants and yeast. The E-value was maintained constant at 1 × 10−10 and displays the sequences that produced hits based on percentage (%) identity (blue color 70%, green color 80%, orange color 90%, and red color 100%) with other default parameters.
Cis-acting elements and gene ontology annotation analysis
The promoter sequences (ATG start codon with 1,500 bp) of ELP4 genes were estimated through the online PlantCARE database (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002). CELLO2GO was used to identify the gene ontology (GO) annotations of the whole ELP4 genes. The CELLO platform (http://cello.life.nctu.edu.tw/) uses hierarchical vocabularies to connect genes and GO terms. Functional enrichment analysis of ELP4 genes was carried out by using DAVID 6.8 (http://david.ncifcrf.gov/) online tools. The gene ontology enrichment was categorized into three main groups: biological process (BP), cellular component (CC), and molecular function (MF).
Yeast strains, media, and general methods
Yeast strains were from S. cerevisiae wild-type BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and ELP4/KTI9/TOT7 deletion mutant Y02150 (As BY4741, but elp4Δ) (Brachmann et al., 1998). At 30°C, yeast strains were consistently cultivated on standard rich (YPD) and minimal (SC minimal) growth media. For the test of thermosensitivity and caffeine sensitivity, yeast stains grew on YPD media with or without caffeine at 30°C or 38°C.
Yeast complementation test
Arabidopsis and yeast ELP4 coding regions were amplified using the EcoRI site at the 5′ end and BamHI site at the 3′ end to validate AtELP4 in yeast elp4Δ deletion mutant by using the following primers EcoRI-AtELP4‐for (5′‐GGA ATT CCA TGG CTG CAC CAA ACG TTC GTA G‐3′) and BamHI-AtELP4-rev (5′‐CGG GGA TCC CGT CAA AAA TCT AGT GCT CCG G‐3′), and DNA fragment was cloned into pBluescript SK(-) vector. The AtELP4 fragments eluted from EcoRI and BamHI digestion were subcloned into the EcoRI–BglII site of the pTU1 vector, which were overexpressed under the control of the TDH3 promoter and CMK1 terminator in the pTU1 vector. Yeast elp4Δ cells were transformed with pTU1 empty vector and AtELP4-harboring pTU1 using electroporation. Transformants were selected on a minimal medium lacking uracil (SD-U) (Jun et al., 2015).
Plant materials and growth conditions
Individual Arabidopsis homozygous T-DNA insertion mutants (Columbia-0 (Col-0) background) in AtELP4 (At3g11220, SALK 079193/Atelp4) and DRL1 (At1g13870, SALK 056915/drl1-102) were obtained from the Arabidopsis Biological Resource Center (ABRC; https://abrc.osu.edu). The homozygote genotype of insertion mutants was confirmed by amplification of T-DNA in T-DNA inserted genomic DNA region using T-DNA and gene-specific primers. The double mutant was germinated by crossing single mutants, isolated in the F2 progeny, and confirmed by the abovementioned PCR-based method.
Plants were grown in a Murashige–Skoog (MS) media containing 0.43% MS medium salt mixture, 2% sucrose, and 0.2% gellan gum (pH 6.3) and transferred to a mixture of soil, vermiculite, and pearlite (2:1:1) under long-day conditions (16-h light/8-h dark, 50–100 μE/m2/s) at 22°C. Seeds were surface-sterilized, vernalized for 3 days at 4°C, and then germinated on MS media (Jun et al., 2015).
Anatomical analyses
The mature third rosette leaves were collected from plants on 21 DAS and fixed with fixation solution (5% (v/v) acetic acid, 5% (v/v) formaldehyde, and 45% (v/v) ethanol in water) under a vacuum. Then, they were dehydrated and cleared as described (Jun et al., 2019). For transverse sectional analysis of leaves, the widest region of dehydrated leaves was embedded in Technovit 7100 resin (Kulzer & Co. GmbH, Wehrheim, Germany) and sliced transversely by using a microtome (Jun et al., 2019). Slices were stained with toluidine blue [1% toluidine blue in phosphate-buffered saline (PBS)], dried, and then observed.
Leaf length and width were measured using from tip to base and the widest part of the leaf blade from 10 mature third rosette leaves of individual plants on 21 DAS. Palisade cells in cleared leaves and transverse sectional slices were observed by light microscopy (Axioskop2, Carl Zeiss, Germany). Cell numbers were counted in the region from mid-vein to leaf margin of the widest region in leaves, and cell size and intercellular airspace were measured from palisade cells of the central region between mid-vein and leaf margin at the widest region in leaves from six mature third rosette leaves of the individual plant on 21 DAS.
The NIH IMAGE software ImageJ was used to analyze the data, and the Statistical Package for the Social Sciences program (SPSS 13.0, SPSS Inc. Chicago) was used for statistical analysis.
Measurement of chlorophyll content and photosynthetic efficiency
To estimate total chlorophyll, fresh weight 0.1 g of leaf blade was taken from 28-day-old plants and soaked in 2 ml of 90% aqueous acetone at 25°C in the dark for 12 h until being cleared by centrifugation for 1 min at 5,000 rpm. The absorbance of the supernatant was measured at wavelengths 645, 663, and 750 nm using a spectrophotometer. Chlorophyll a and b contents were estimated according to the formula of Ritchie (Ritchie, 2006).
Photosynthesis efficiency was calculated from 10 plants by measuring chlorophyll fluorescence imaging with the Handy FluorCam fluorescence imaging equipment (Photon Systems Instruments, Brno, Czech Republic). Subsequently, Fv/Fm value was estimated from individual plants using FluorCam7 software according to the manufacturer’s instructions (Arena et al., 2020).
RNA isolation and qRT-PCR analysis of gene expression
Total RNA was isolated from rosette leaf blades of 21-day-old plants and yeast using the RNeasy mini kit (Qiagen, Hilden, Germany), and cDNA was synthesized by reverse transcription using the Reverse TraAce-a-First strand cDNA synthesis kit (TOYOBO, Tokyo, Japan). The quantitative RT-PCR (qRT-PCR) was performed using SYBR® Premix Ex TaqTM II (TAKARA, Otsu, Japan) and Bio-Rad CFX96TM Real-Time System (Bio-Rad, Hercules, CA, USA) with three technical replicates. Primers were designed by Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi) for qRT-PCR and are listed in Supplementary Table 5. PCR thermocycles were run using the following conditions: denaturation at 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s. The normalized expression level of each gene was calculated using the ΔΔCq method and β-tubulin (TUB4) gene for Arabidopsis, and S. cerevisiae ACTIN 1 (ScACT1) gene for yeast was used as a control.
DNA ploidy analysis
The mature third rosette leaf of plants on 21 DAS was harvested, and nuclei of leaves were extracted in Otto buffer (Otto, 1992; Doležel and Göhde, 1995) by chopping. Isolated nuclei were stained with propidium iodide (PI). Finally, DNA ploidy was analyzed with the flow cytometer, Cytomics FC500 flow cytometer (Beckman Coulter, Brea, CA, USA).
Results
Phylogeny relationship, evolutionary, conserved motif, and shared plant-specific domain analysis
As mentioned above, the Elongator complex was originally identified as a transcription factor in yeast (S. cerevisiae) (Otero et al., 1999; Wittschieben et al., 1999). Elongator components in eukaryotes are structurally conserved (Dauden et al., 2017a,b; Otero et al., 1999). Petrakis et al. (2005) suggested a weak, salt-labile interaction between yeast Kti12 and Elp4, although they did not prove the presence of Elp4 in the highly purified Kti12. Based on these findings, we attempted to examine the functions of AtELP4 and its genetic interaction with DRL1, a gene homologous to yeast KTI12. At first initially, we performed an amino acid alignment and phylogenetic analysis of ELP4 from various eukaryotes including yeast, human, and plants using MEGA7 software. ClustalX pairwise alignment of AtELP4 amino acid sequences with ELP4s from several species indicated that AtELP4 proteins displayed approximately 60% identity with plant ELP4 proteins, but only about 20% identity was observed with yeast or animal Elp4 proteins. In detail, AtELP4 was approximately 65% identical with other dicot plant ELP4 proteins (V. vinifera, C. sativus, G. max, and S. lycopersicum) and about 57% identical with monocot plant ELP4 proteins (O. sativa and Z. mays). Moreover, AtELP4 was just 20% identical to yeast or human Elp4 proteins (Supplementary Table 1).
However, we also performed the structural analysis of ELP4 using NCBI (Conserved Domain Database) (https://www.ncbi.nlm.nih.gov/cdd) to find conserved motifs of the ELP4 from various eukaryotes. ELP4 from eukaryotes shared PAXNEB domain, which is found in the histone acetyltransferase complex of RNA polymerase II holoenzyme. Since human and mouse PAXNEB genes were identified as yeast ELP4 homolog due to their significant homology (Winkler et al., 2001); PAXNEB domain (accession no. PF05625, https://www.ebi.ac.uk/interpro/entry/pfam/PF05625/) became a common/representative domain of an RNA polymerase II Elongator protein subunit. PAXNEB domain is known as the P-loop ATPase motif of RecA or RAD families, which catalyze recombination reactions using ATP-dependent DNA binding and DNA-dependent ATP hydrolase activities (Ponting, 2002; Nelissen et al., 2005; Glatt et al., 2012). Especially, PAXNEB domain in plant ELP4 homologs was conserved with KaiC-like domain (accession no. CD01124, https://www.ncbi.nlm.nih.gov/Structure/cdd/cd01124), which was found in KaiC, a circadian clock protein in cyanobacteria and possesses autophosphorylation activity (E-value < 0.1).
MEME online tool was used to uncover unknown motifs among ELP4 from plants, yeast, and animals (http://meme-suite.org/). A minimum of five motifs were used in the motif analysis along with other default parameters. A phylogenetic tree was built in order to better comprehend the makeup of the 27 plants, 3 yeast, and 10 animal species. The phylogeny was classified into three groups (plants, animals, and yeast) (Figure 1). We investigated the conserved motif distribution on ELP4. ELP4 proteins in the three groups share the same conserved motifs, which supports the phylogenetic tree conclusions (Figure 1). However, ELP4 proteins contain a variety of conserved motifs in each group. Motif 1 was identified as conserved motifs and could be traced in all groups (plant, animal, and yeast). Motifs 2 and 3 were found in plants and animals, not in yeast, showing specific features in multicellular organisms. Interestingly, motif 4 and motif 5 were present only in ELP4 of plants, indicating that these families have some unique evolutionary processes and functionalities (Figure 1 and Supplementary Table 2). These results of phylogenic analysis and conserved motif search revealed that ELP4 might play not only representative roles but also species-specific roles according to organisms, although ELP4 homologs are evolutionarily conserved.
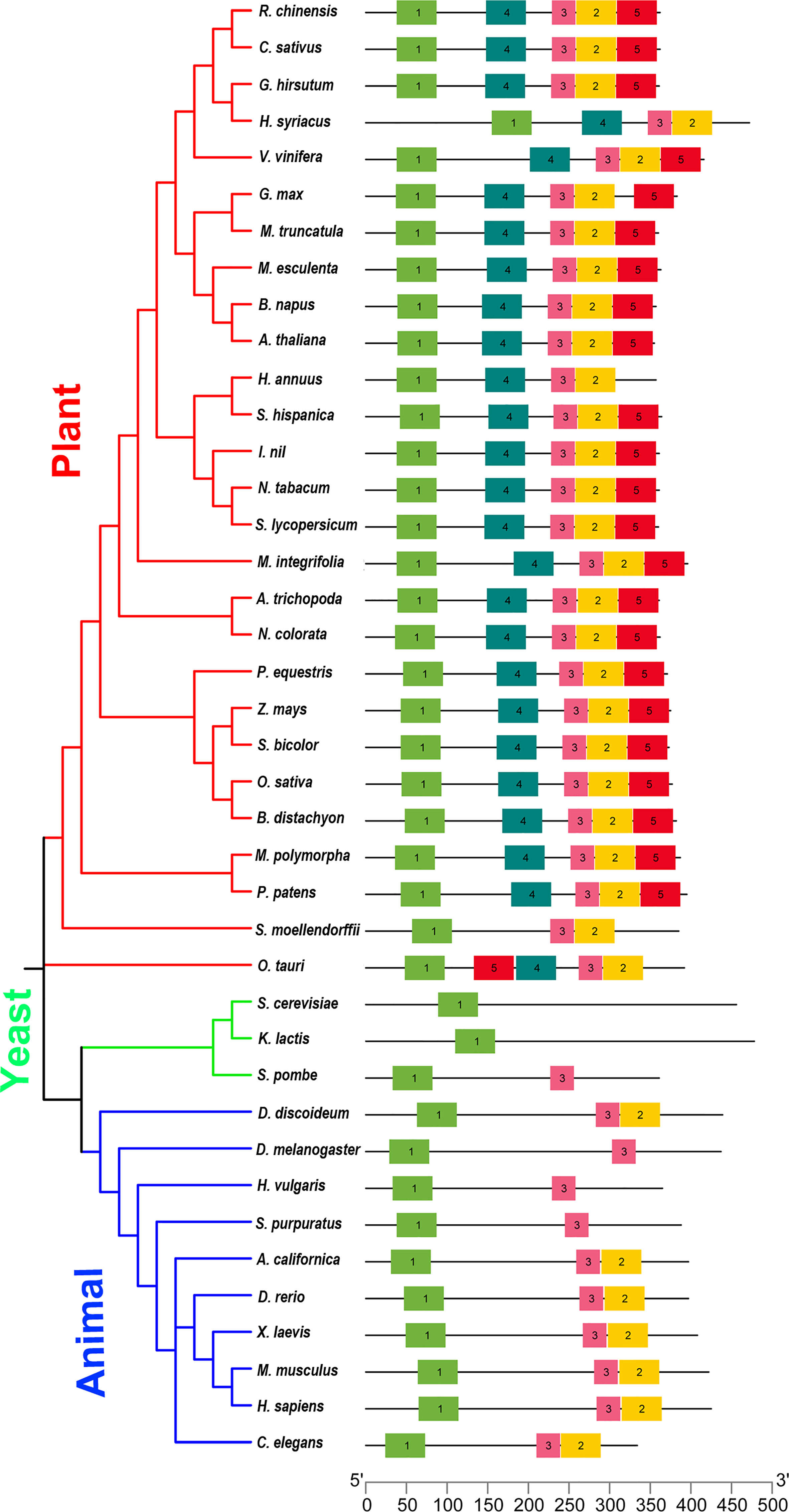
Figure 1 The phylogeny and conserved motif analysis and classified into three groups: plant, yeast, and animal. ELP4 protein motif composition is represented by distinct colored boxes with appropriate motif numbers and figure legends at the top.
Syntenic relationship between plant and yeast
We evaluate the collinearity relationship among plant and yeast species. The 27 plants (A. trichopoda, A. thaliana, B. distachyon, B. napus, C. sativus, G. max, G. hirsutum, H. annuus, H. syriacus, M. integrifolia, M. esculenta, M. polymorpha subsp ruderalis, M. truncatula, N. tabacum, N. colorata, O. sativa, P. equestris, P. patens, R. chinensis, S. hispanica, S. moellendorffii, S. lycopersicum, S. bicolor, V. vinifera, Z. mays, O. tauri, and I. nil) and three yeast (S. pombe, S. cerevisiae, and K. lactis) were subjected to microsynteny analysis to better understand the evolutionary and origin mechanism of ELP4 genes. Synteny analysis in plants and yeast revealed a substantial association between duplication, expression, gene evolution, triplication, and function. A. thaliana gene sequence demonstrated synteny with the B. napus gene sequence, with the red color indicating that these genes have 100% similarities (Figure 2). Furthermore, yeast (S. pombe, S. cerevisiae, and K. lactis) had synteny with each other and among plants, which was identical in orange (90%) and blue (70%) colors (Figure 2). ELP4 genes belonging to plants are mostly connected via red lines, showing more than 100% identity among plants. As yeast is considered the progenitor of the plants, the same thing was supported by our results where some plant sequences were 70% identical to yeast genes and are shown in blue color in Figure 2. Our result suggests that there are collinearity relationships between the different plant and yeast gene sequences, suggesting a potential evolutionary mechanism.

Figure 2 The circos tool was used to determine the synteny relationship of ELP4 genes between the plant and yeast. The colors blue, green, orange, and red represent ≤50%, ≤70%, ≤90%, and >90% similarity, respectively.
Cis-acting elements analysis
The potential cis-acting factors were identified by using PlantCARE to better comprehend the expression control mechanism of ELP4 genes in plants and yeasts (Supplementary Table 3). These cis-acting elements were categorized mainly into three biological processes: biotic/abiotic stress responsiveness, phytohormones, and plant growth/development. The majority of ELP4 genes included the following cis-acting elements: ABRE, AAGAA-motif, TCA-element, ARE, TCA, MYB, LTR, I-box, 02-site, MYB-binding site, CAT-Box, TATA-box, G-box, and Sp1 (Figure 3). Stress-related cis-elements MYB, LTRs, and ARE, which are associated with drought, low temperature, and anaerobic induction, respectively, were stress-related cis-elements that were investigated. I-box, Sp1, Myb-binding site, and G-Box were distributed across the promoter regions (responsible for light).
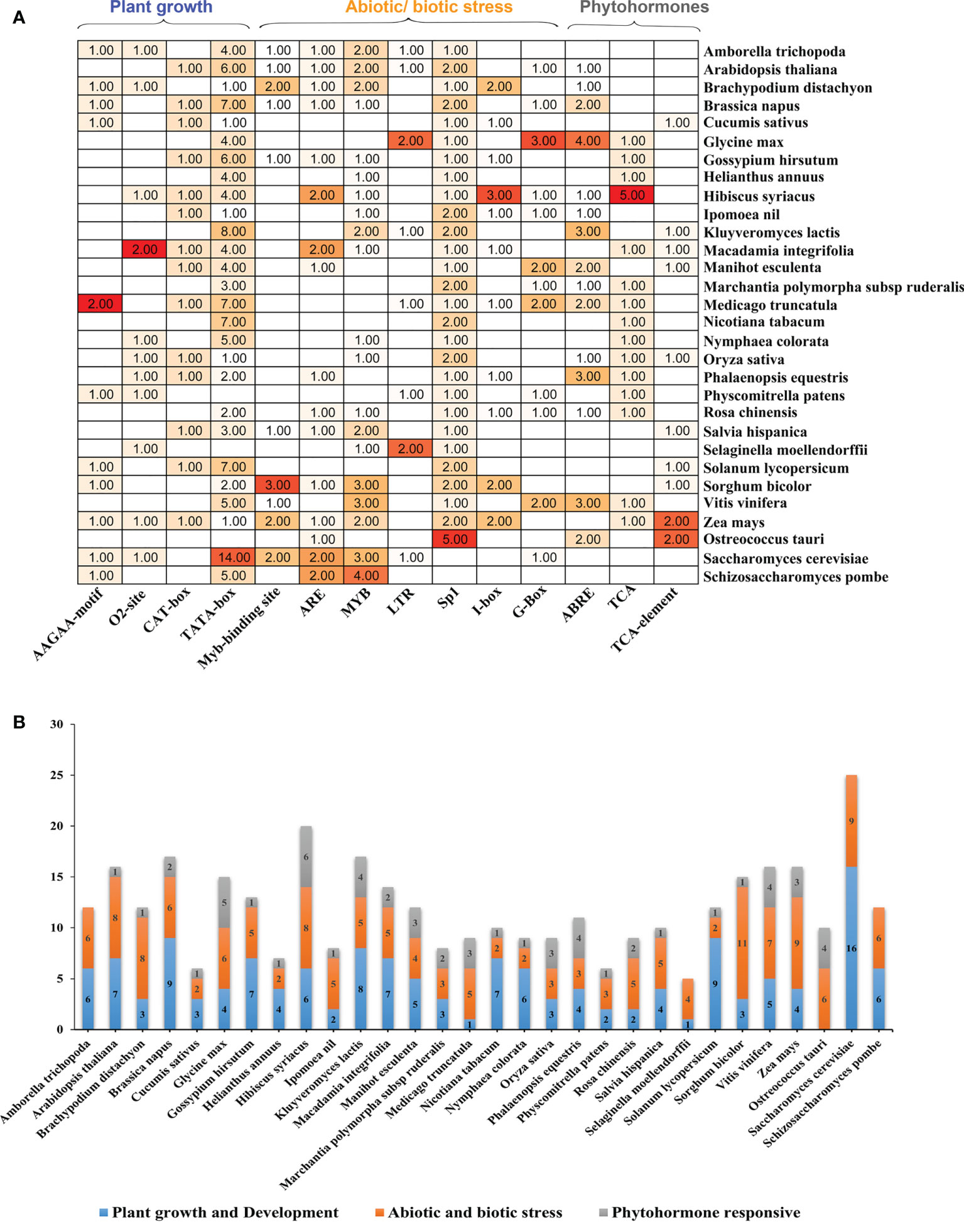
Figure 3 Cis-acting elements in ELP4 promoter of various plants and yeast. (A) Cis-acting elements with functional similarity on the upstream region of ELP4 genes are demonstrated by multiple colors. (B) In each category (plant growth development, abiotic/biotic stress, and phytohormone responsive), present cis-acting elements represent with different colors.
Phytohormone-related cis-elements, such as the TCA-element, TCA, and ABRE, were also found, which are associated with abscisic acid and salicylic acid (Figure 3 and Supplementary Table 3). O2-site (related in zinc metabolic control), AAGAA-motif (development-related), CAT-box (associated to meristem expression), and TATA-box (transcription start core promoter) are in the growth/development category. Moreover, Sp1 and TATA-box covered the highest portion, while in phytohormonal response, ABRE contributes the maximum portion. These results indicate that ELP4 genes had versatile functionality in plants and yeast.
Functional enrichment analysis
The putative functions of the ELP4 protein are anticipated by utilizing GO annotation analysis. Depending on amino acid similarities, ELP4 proteins were categorized into 23 distinct classes and split into three major ontologies: cellular component, molecular function, and biological process (Supplementary Table 4). In molecular function annotation, ELP4 protein was predicted to be most functional in transferase activity (51.67%), followed by enzyme regulator activity (48.33%). In cellular component annotation, ELP4 protein was annotated with protein complex, cytoplasm, organelle, intracellular, nucleus, and cell, with 13.67% followed by nucleoplasm (12.56%) and cytosol (6.68%). Moreover, in biological process annotation, predicted ELP4 protein was annotated with cellular nitrogen compound metabolic process and biosynthetic process along with the same percentage (12.95%), while signal transduction and stress responsiveness contributed equally (11.70%). However, cellular protein modification and chromosomal organization annotated 11.28%, while embryo development, reproduction, and anatomical structure development contributed equally (5.89%). Catabolic, small molecule, and nucleobase-containing compound catabolic processes contribute minimally (1.84%), while the tRNA metabolic process alone contributed 4.98% (Figure 4).
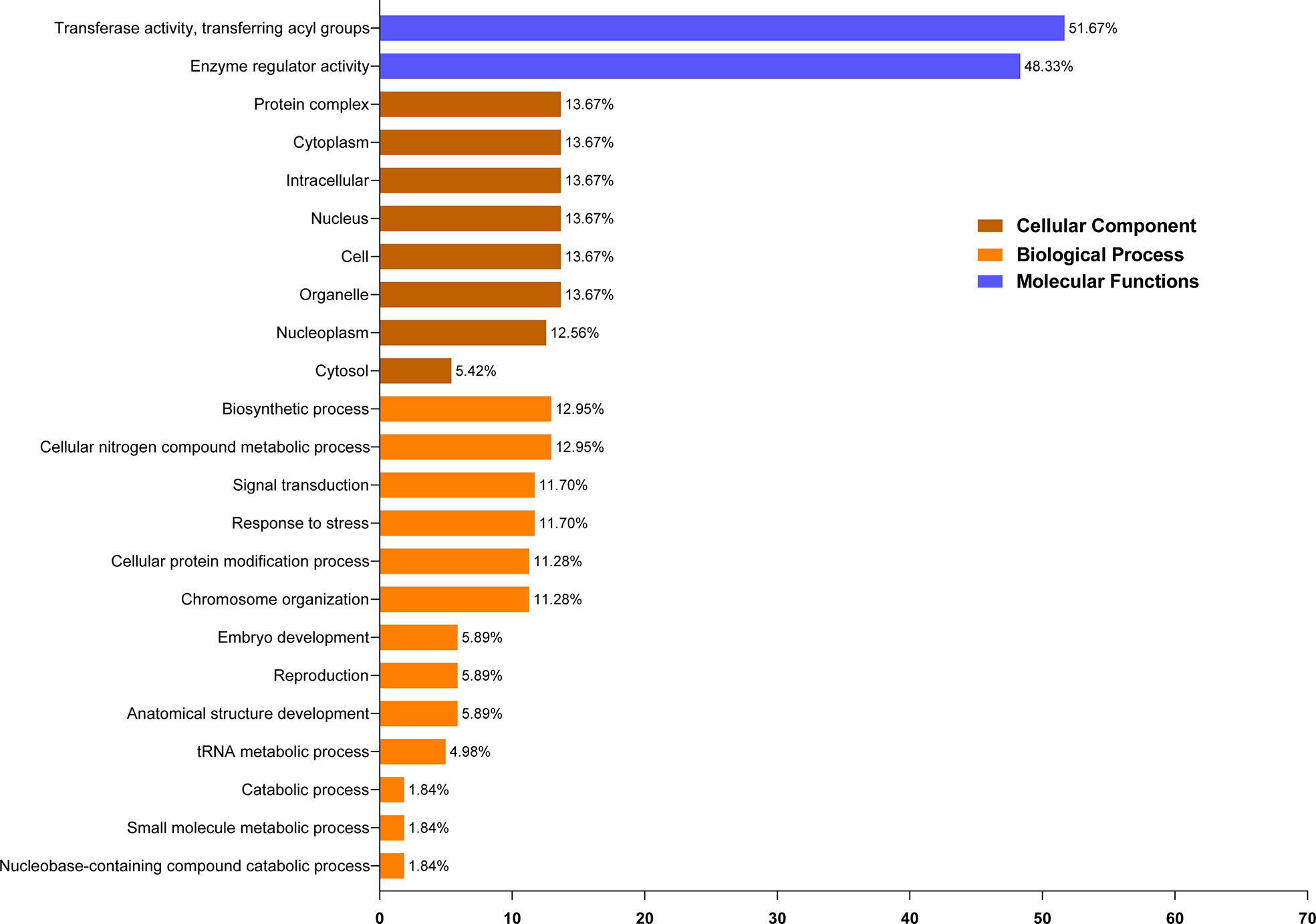
Figure 4 GO enrichment analysis of ELP4 genes. Based on functions, these are classified into three groups (molecular functions, biological processes, and cellular components) and represented through different colors. GO, gene ontology.
ELP4 functions between Arabidopsis and yeast are partially conserved
To identify shared functions between yeast and Arabidopsis, we performed a complementation investigation utilizing AtELP4 protein in a yeast elp4Δ mutant. Yeast elp4Δ mutant exhibited resistance toward zymocin and γ-toxin, slow cell growth, thermosensitivity above 38°C, and hypersensitivity toward caffeine (Jablonowski et al., 2001). To confirm the functional complementation of AtELP4 in yeast elp4Δ mutant, we constructed the yeast expression vector AtELP4 harboring pTU1 and transformed it into yeast BY4741-derived elp4Δ mutant, Y02150. Finally, RT-PCR and qRT-PCR were used to validate AtELP4 expression in yeast elp4Δ mutant (Supplementary Figure 1). Each transgenic elp4Δ mutant containing an empty vector and overexpressing AtELP4 (Y02150:pTU-empty #1 and Y02150:pTU-AtELP4 #2, respectively) was chosen for further investigation. Compared to the retardation of cell growth of elp4Δ mutant Y02150 at normal conditions (30°C), AtELP4 expression in Y02150 did not rescue to wild type, BY4741 (Figure 5A). For a complementation assay for thermosensitivity and caffeine sensitivity, yeast lines grew at 38°C or in caffeine-containing media. Even though it did not fully restore the growth rate of the wild type, Y02150 that expressed AtELP4 grew faster at 38°C than Y02150 (Figure 5B). This result indicates that AtELP4 expression in Y02150 partially rescued the growth defects at 38°C (Figure 5B). However, AtELP4 expression in Y02150 did not absolutely rescue the sensitivity toward caffeine (Figure 5C). These results exhibited that the function of AtELP4 gene is partially conserved in that of yeast ELP4 gene.
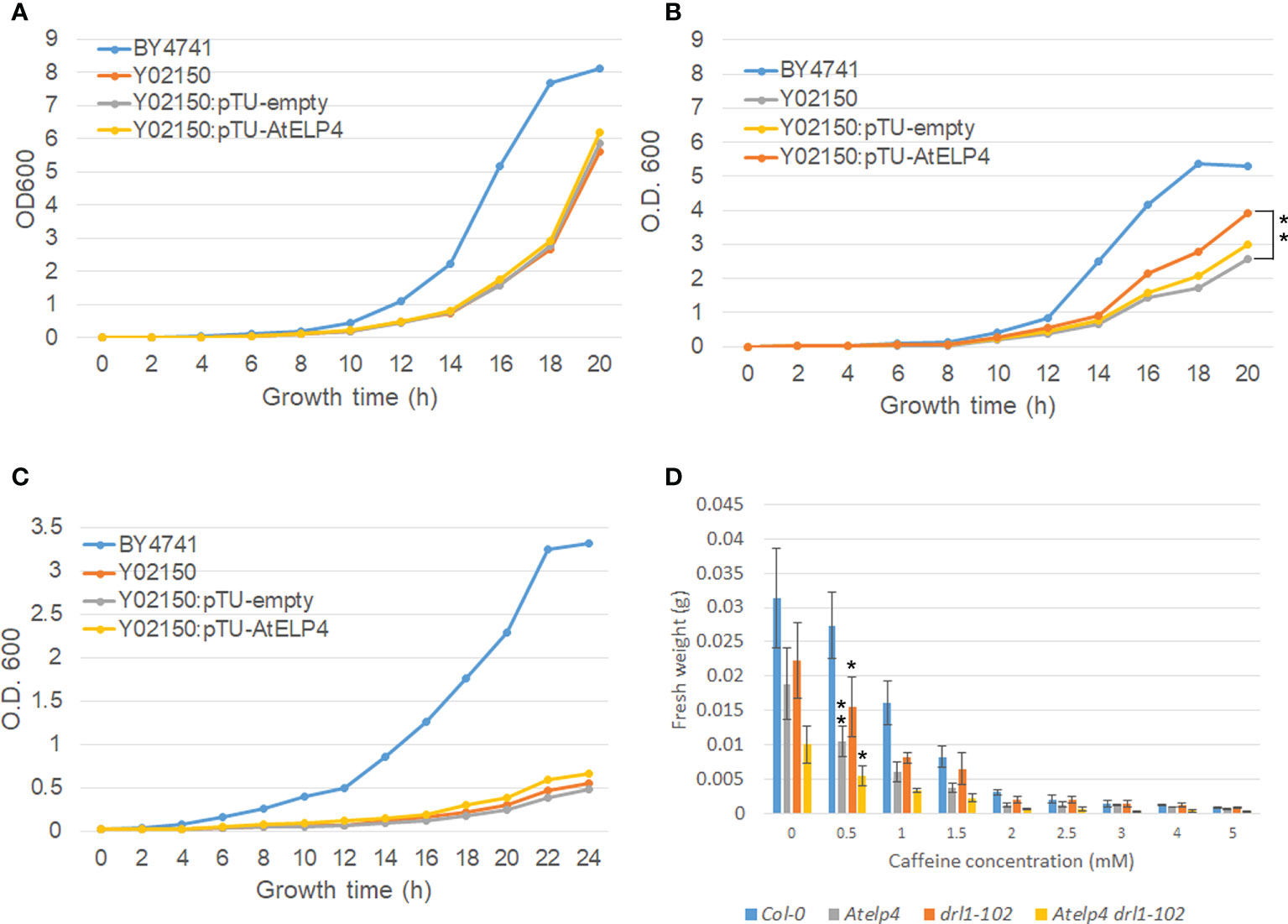
Figure 5 Complementation test of yeast elp4Δ mutant and caffeine sensitivity of Arabidopsis Atelp4 and drl1-102 single and double mutants. (A–C) Complementation of yeast elp4Δ mutant, Y02150 by Arabidopsis ELP4 expression in thermosensitivity and caffeine sensitivity. Wild-type BY4741, elp4Δ mutant Y02150, and the empty vector pTU1 or ELP4-driven pTU1 introduced Y02150 transgenic lines were grown in YPD media. For test of thermosensitivity, yeast lines were grown at (A) 30°C and (B) 38°C. Asterisks indicate significant differences compared to Y02150 at 20 h using a Student’s t-test (**, p-value <0.01). (C) For test of caffeine sensitivity, yeast lines were grown in YPD media containing 7.5 mM of caffeine. Yeast growth was determined by measuring optical densities at 2-h intervals over a period of 20 h using a spectrophotometer at 600 nm. (D) Caffeine sensitivity of Arabidopsis wild type, and Atelp4, drl1-102, and Atelp4 drl1-102 mutants. Plants were grown in MS media containing caffeine concentration in the range of 0 to 5 mM for 21 DAS, and then fresh weight of plants was measured. Ten individuals in each plant were collected, and data are shown as the means with SEs. Asterisks in panels B and D indicate significant differences compared to no treatment of each plant group using a Student’s t-test (*p-value <0.05; **p-value <0.01).
Caffeine sensitivity of Arabidopsis DRL1 and AtELP4 mutations
As mentioned above, ELP4 and KTI12 mutation in yeast triggered the growth defect by caffeine (Jun et al., 2015; Mehlgarten et al., 2017), and caffeine is well known as a cytokinesis inhibitor that disrupts cell plate in dividing cells of plants. To confirm the functional similarity of DRL1 and ELP4 between Arabidopsis and yeast, we observed the alteration of caffeine sensitivity by single and double ELP4 and DRL1 mutants, Atelp4 and drl1-102 in Arabidopsis. We measured the fresh weight of the plant shoot part that grew on the media containing each caffeine concentration series on 14 DAS. All plants of wild type and single and double mutants showed a decrease in fresh weight depending on caffeine concentration (Figure 5D). However, there was a variation in the extent of weight reduction between wild type and mutants. Compared to the normal condition, the fresh weight of each single and Atelp4 drl1-102 double mutants on the 0.5 mM of caffeine decreased to 70%, 55%, and 55%, whereas that of wild type decreased up to 87% (Figure 5D). The hypersensitivity toward caffeine by DRL1 and AtELP4 mutation in Arabidopsis indicates that DRL1 and ELP4 between Arabidopsis and yeast were conserved functionally, in the viewpoint of sensitivity of caffeine.
Atelp4 mutant had traits similar to drl1-102 mutant but showed severe phenotype during leaf development
We characterized Atelp4 single mutant and Atelp4 drl1-102 double mutant of Arabidopsis to study the Elongator complex’s role in leaf development. In the previous report, we described that the drl1-101 mutant formed trumpet-like and filamentous leaves, which had defects in adaxial–abaxial polarity and cell division (Cho et al., 2007). Compared with the wild type (Col-0), the drl1-102 mutant showed narrow and downward-curling (epinastic) leaf blades and an imprecise boundary between the leaf blade and petiole. Atelp4 mutant also showed narrow leaf blades with severely decreased size and unclear marginal boundary between blade and petiole; Atelp4 drl1-102 double mutant had a similar phenotype in leaves compared to Atelp4 single mutant, exhibiting a severe reduction in leaf width and serration in leaf margin (Figures 6A, B). The total area of the mature third leaf of Atelp4 and drl1-102 reduced up to 54.2% and 39.8% of that of the wild type (Figure 6C). Similar results were observed for the Atelp4 drl1-102 double mutant (39.2% of that of the wild type) (Figure 6C). The reduction in leaf width (70.8% to 59.3%) was more severe than the decrease in leaf length (82.8% to 75.1% in the mutants), relative to the wild type (Figures 6D, E). Due to more reduced leaf width than leaf length, the leaf index of Atelp4 and drl1-102 single and double mutants was higher (1.7, 1.9, and 1.8, respectively) than that of wild type (1.4) (Figure 6F), indicating narrower leaf blades in mutants. However, there was no significant difference between the three mutants. Our genetic approaches indicate that AtELP4 is epistatic to DRL1.
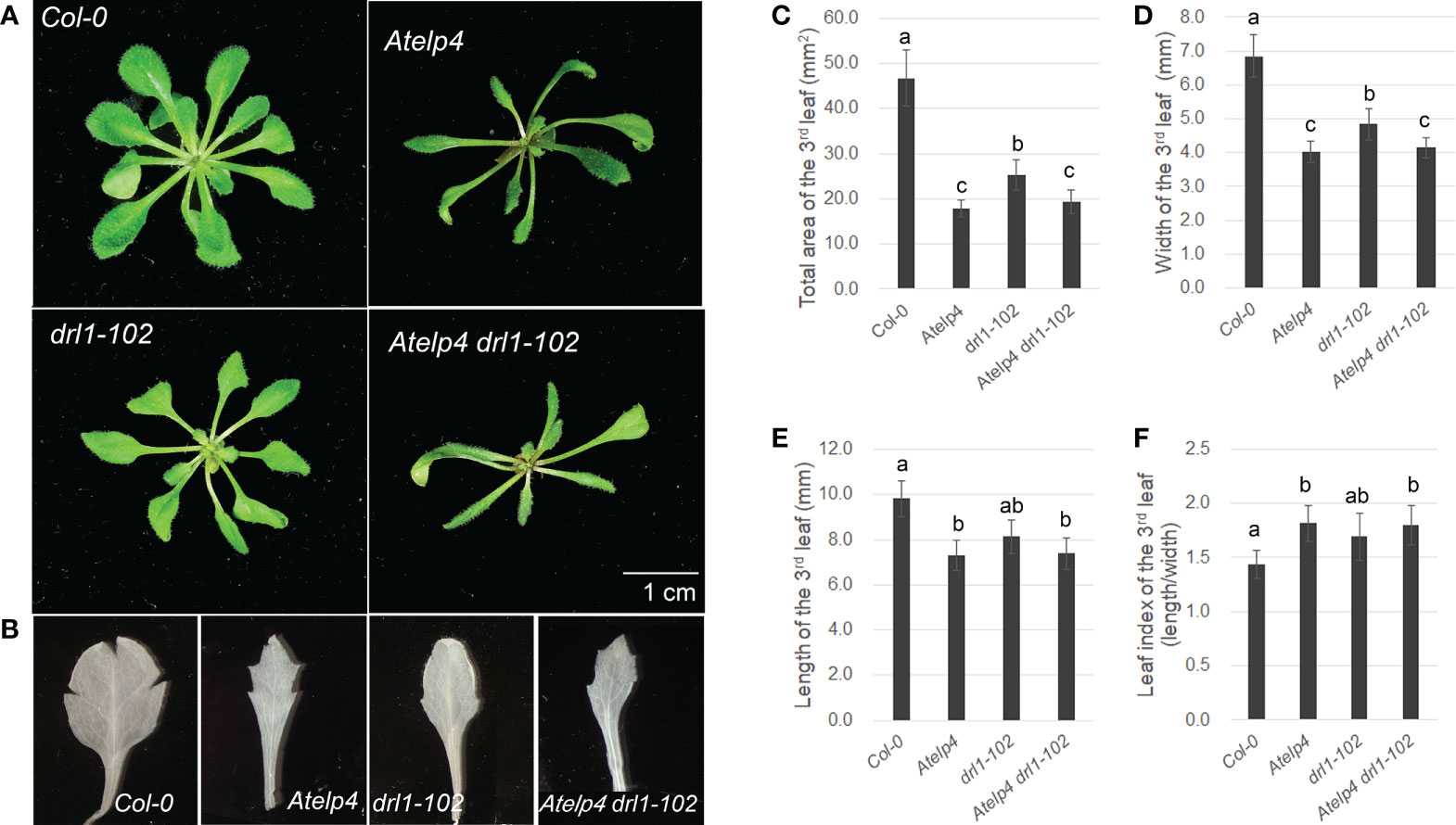
Figure 6 Plant morphology of Atelp4 and drl1-102 single and double mutants. Morphology of (A) plants on 35 DAS and (B) the mature third rosette leaves on 21 DAS of wild type, and Atelp4, drl1-102, and Atelp4 drl1-102 mutants. Scale bar indicates 1 cm. (C) Total area, (D) width, (E) length, and (F) leaf index (the ratio of length to width of leaf blade) on the mature third rosette leaves in plants on 21 DAS. Leaves were independently collected from 10 plants, and data are shown as the means with SEs. Different lowercase letters in panels C–F indicate significant differences between plants, according to ANOVA and Tukey’s multiple range test (p-value <0.01).
Atelp4 and drl1-102 mutants are defective in cell proliferation and the adaxial–abaxial polarity patterning during leaf development
We used paradermal images and transverse sections to investigate the cellular phenotype at the widest part of the third leaf on the 21 (DAS). This was performed to mitigate the cellular changes in the leaves (Jun et al., 2019). Leaves of drl1-102 mutant showed reductions in cell number and size, 62.0% and 75.4%, respectively, compared to those of the wild type (Figures 7C, D). Atelp4 mutant showed reduced cell number and size, 61.7% and 51.5% in Atelp4, respectively. Atelp4 drl1-102 mutant also showed a reduction of cell number and size up to 58.2% and 52.0%, respectively, compared to the wild type (Figures 7A, C, D). These data indicated that DRL1 and AtELP4 positively regulated cell numbers in a similar way. From the viewpoint of cell proliferation and expansion, AtELP4 was epistatic to DRL1.
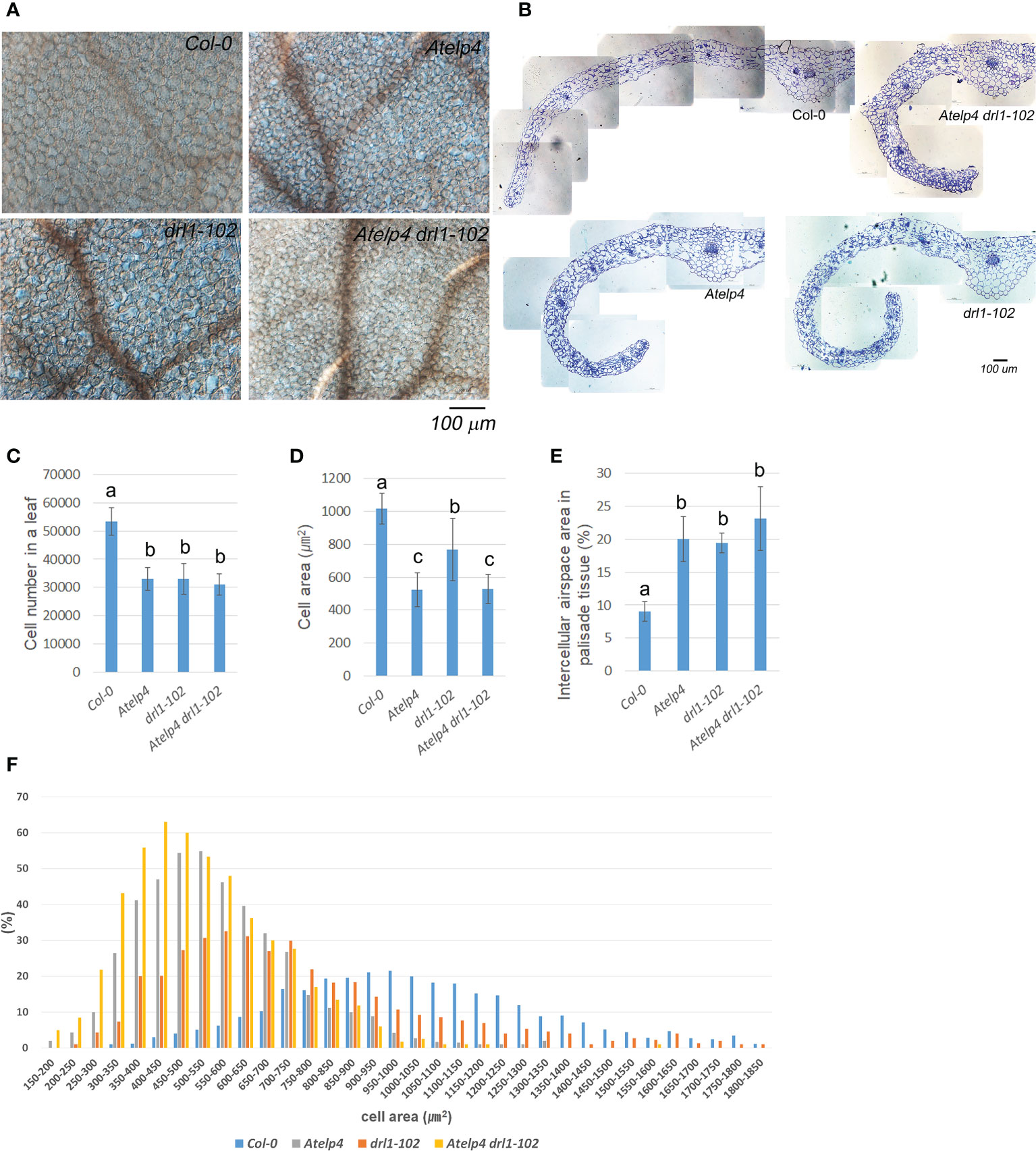
Figure 7 Internal morphology in leaves and statistical analysis of palisade cells from Atelp4 and drl1-102 single and double mutants. (A) Paradermal images of palisade cells in the mature third rosette leaves in wild type, Atelp4, drl1-102, and Atelp4 drl1-102 on 21 DAS. (B) Transverse section of the middle region in the mature third leaves. Scale bar = 100 μm. (C) Cell number, (D) cell size, and (E) proportion of intercellular airspace in the mature third rosette leaves on 21 DAS. (F) Area distribution of palisade cells in the central region between leaf mid-vein and margin in the widest part of leaf blade. Leaves were independently collected from six plants, and data are shown as the means with SEs. Different lowercase letters in panels (C–E) indicate significant differences between plants, according to ANOVA and Tukey’s multiple range test (p-value <0.01).
In the distribution of palisade cell size in leaves, the drl1-102 mutant showed wider bell-shaped distribution than the wild type (Figure 7F). These results indicated that the cell size of drl1-102 leaves was more irregular than that of the wild type. However, the distribution of cell size in leaves from Atelp4 and Atelp4 drl1-102 showed sharper bell-shaped distribution, indicating some suppression processes in cell expansion. The measured cell area of wild type and Atelp4 and drl1-102 single and double mutants approximately follows a lognormal distribution with an average of 1,018.0, 767.6, 524.0, and 529.6 μm2 with a standard deviation of 92.6, 189.0, 104.3, and 89.0 μm2, respectively (Figure 7F).
In addition, leaves of each single and double mutant had increased intracellular airspace between palisade mesophyll cells of the adaxial subepidermal layer (Figures 7B, E). The proportion of intercellular airspace of Col-0, Atelp4, drl1-102, and Atelp4 drl1-102 was 11.1%, 17.6%, 18.5%, and 23.4% to the area of the central region between leaf mid-vein and margin in the widest part of the leaf blade, respectively, and showed an increment of intercellular airspace in the adaxial subepidermal layer in each single and double mutants (Figure 7E). In detail, intercellular airspace in drl1-102, Atelp4, and Atelp4 drl1-102 increased by 215.4%, 221.6%, and 256.1%, respectively, compared to that of the wild type (Figure 7E). These findings suggested that in mutants, the palisade’s adaxial side was transformed into an abaxial-like structure.
In the transverse section, mutants exhibited downward curling at the whole leaf blade, an increase in intercellular space/distance on the adaxial side, and a larger portion of phloem tissue in vascular bundles, compared to those of the wild type (Figure 7B). Especially, at Atelp4 and drl1-102, each single and double mutant showed remarkable downward curling at the whole part of the leaf blades, whereas the wild type showed downward curling at the marginal part of the leaf blades (Figure 7B).
Decreased chlorophyll content in Atelp4 and drl1-102 mutants
To explore further evidence of the abaxialization of leaves, we measured chlorophyll concentration and photosynthetic efficiency. Total chlorophyll in 1 mg of leaf in drl1-102, Atelp4, and Atelp4 drl1-102 mutants was reduced to 53.3%, 48.7%, and 59.5%, respectively, compared to wild type (Figure 8A). However, reduction in chlorophyll content had no effect on phenotype and photosynthesis efficiency in mutant leaves, with no variation in Fv/Fm value when compared to wild type (Figures 8B, C, D). Presumably, the residual amount of chlorophyll in mutants was sufficient to support a normal rate of photosynthesis containing light capturing.
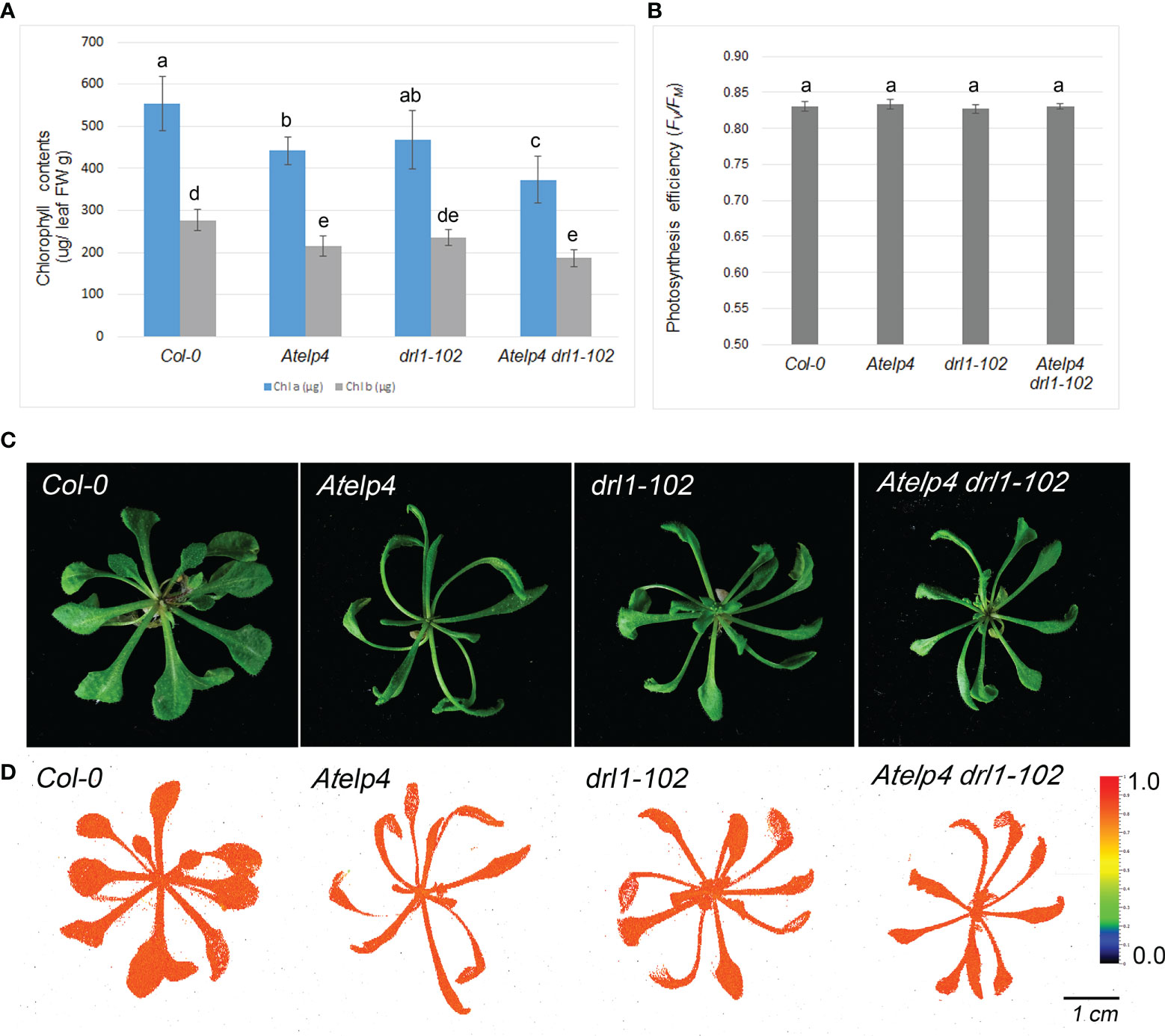
Figure 8 Photosynthesis efficiency and chlorophyll contents of Atelp4 and drl1-102 single and double mutants. (A) Chlorophyll content. (B) Photosynthesis efficiency (Fv/Fm ratio) (n = 10) measured by FluorCam, (C) phenotypes of plants, and (D) false color image of photosynthesis efficiency (Fv/Fm ratio) captured by FluorCam. The color box in panel D means photosynthesis efficiency from 0 (minimum value, blue) to 1.0 (maximum value, red). Data are shown as means of eight plants with SEs. Different lowercase letters in panels A and B indicate significant differences between plants, according to ANOVA and Tukey’s multiple range test (p-value <0.01).
The statistical analysis revealed that modifications in Atelp4 mutant leaves and cells were more severe than those in drl1-102 single mutant and similar with those in Atelp4 drl1-102 double mutant. These results indicate that DRL1 might act upstream of AtELP4 in two processes: regulation of cell proliferation and adaxial–abaxial polarity establishment.
AtELP4 and DRL1 control the expression of genes that regulate leaf polarity and cell proliferation of leaves
We previously described that drl1-101 mutant (No-0 background) showed suppression of cell proliferation and alteration in adaxial–abaxial polarity establishment through RT-PCR.
To verify that Atelp4 leaves underwent abaxilization in the adaxial side and inhibition of cell proliferation, we next examined the expression level of leaf polarity- or cell proliferation-relative genes by using qRT-PCR.
Atelp4, drl1-102, and Atelp4 drl1-102 mutants showed higher expression levels of the YAB family and abaxialization marker, KAN genes, and lower expression levels in adaxialization marker genes, HD-ZIP III genes, PHB, PHV, and REV (Figure 9). In addition, leaf polarity specifying gene, ASYMMETRIC LEAVES 2 (AS2), expression was decreased in AtELP4 mutation-dependent manner, while leaf abaxial identity specifying genes, AUXIN RESPONSE FACTOR 4 (ARF4), expression was increased in single and double mutants, compared to wild type (Supplementary Figure 2).
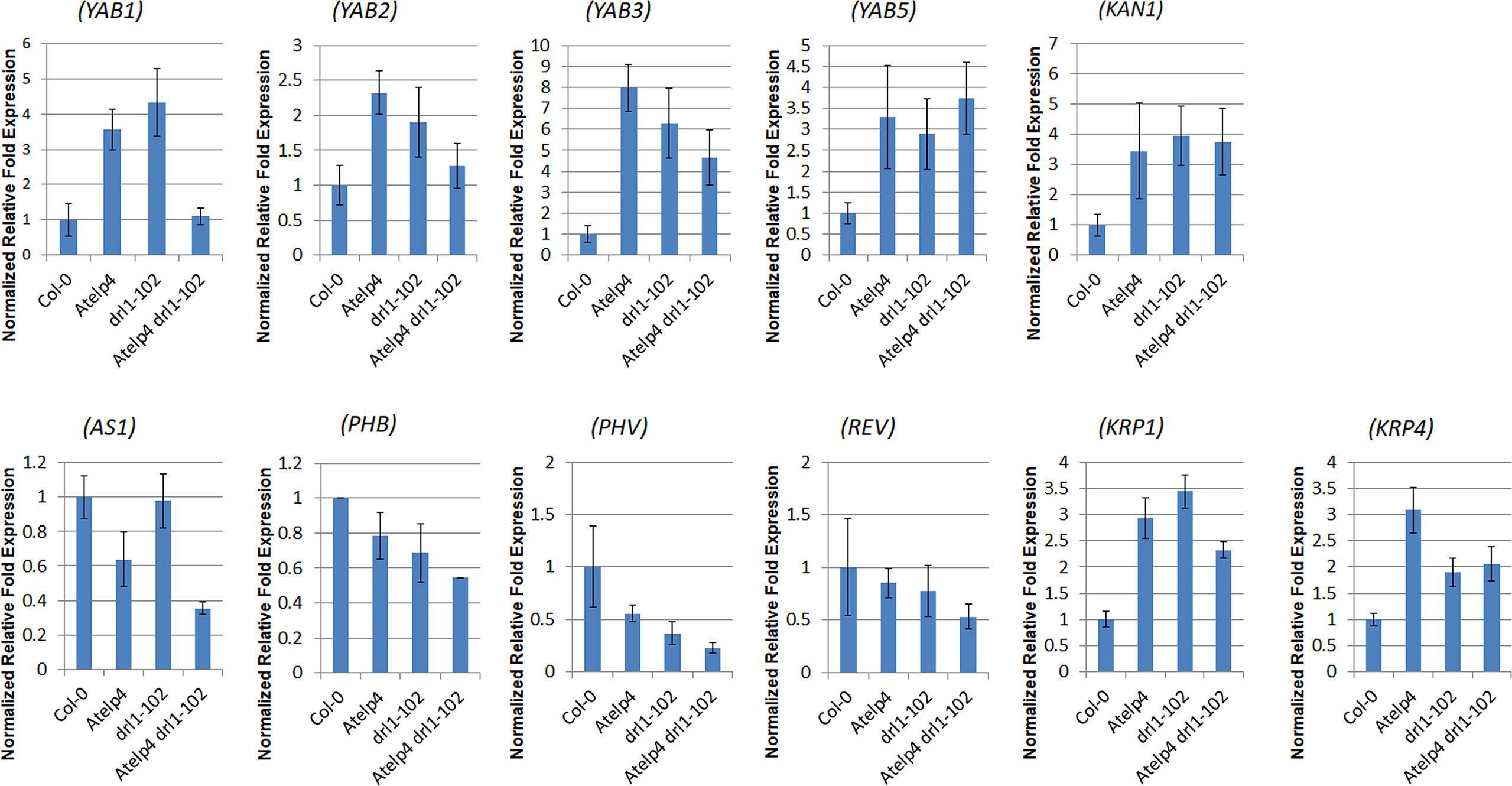
Figure 9 Expression level of leaf polarity- and cell proliferation-relative genes in Atelp4 and drl1-102 single and double mutants. Identical amounts of total RNA isolated from 21-day-old rosette leaves of wild type (Col-0), Atelp4, drl1-102, and Atelp4 drl1-102 were subjected to qRT-PCR experiments to analyze the expression level of leaf polarity- and cell proliferation-relative genes. The expression level of analyzed genes was normalized by that of TUB4. Primer sequences of used genes are described in Supplementary Table 5.
We also analyzed the expression level of regulating genes of cell proliferation. Atelp4 and drl1-102 showed a weak increment in CYCB1;1 and CYCD3;1 expression, which is well known as positive cell cycle markers promoting cell division, compared to wild type (Supplementary Figure 2). Because these expressions were opposite to expected results in cell proliferation in mutants, we examined the expression level of KIP-RELATED PROTEINS (KRPs), which act as a cell cycle inhibitor. Single and double mutants displayed increase in KRP1 and KRP4 transcripts, compared to the wild type (Figure 9). According to the gene expression map of Arabidopsis eFP Brower (www.bar.utoronto.ca, Winter et al., 2007), KRP1 and KRP4 were strongly expressed in leaves, whereas other KRPs expressed much less than KRP1 and KRP4 in leaves. Therefore, we speculated that DRL1 and AtELP4 might take part in the control of the cell cycle through transcript regulation of cell cycle regulators.
AtELP4 and DRL1 regulate endoreduplication processes during leaf development
During the development of Arabidopsis leaves, the cell cycle for cell division and proliferation is known to transit to the endoreduplication cycle repeating DNA replication without entering mitosis (Jun et al., 2013). To understand the function of the Elongator complex on the development of mesophyll cells, we performed a DNA ploidy analysis to examine the nuclear ploidy in the third leaves of Atelp4 and drl1-102.
In 21-day-old wild-type plants (Col-0), DNA ploidy distribution in leaves reached an equal portion between the amount of 2C and 4C and the amount of more than 8C. In contrast, Atelp4 and drl1-102 showed more portion of the amount of 2C and 4C than that of 8C. 2C and 4C represent the G1 and G2 phases of dividing cells, respectively, while the appearance and accumulation of 8C and more DNA ploidy represent the departure from the cell cycle and activation of endoreduplication (Supplementary Figure 3). Hence, this result indicates that endoreduplication is delayed in Atelp4 and drl1-102.
Discussion
Conserved functions of ELP4 and DRL1 between Arabidopsis and yeast
Elongator in eukaryotes is a highly conserved complex with tRNA modification output that contributes to multiple cellular functions (Nelissen et al., 2005; Nakai et al., 2019). Among the six subunits of the Elongator complex, ELP4 forms a RecA-ATPase-like fold that assembles with ELP5 and ELP6 into a heterohexameric ring (Glatt et al., 2012). Functionally, the yeast (Elp456)2 subcomplex specifically recognizes and interacts with tRNA (Glatt et al., 2012). Mutations of yeast Elongator subunits and its associated protein Kti12 caused growth retardation at 38°C and sensitivity to caffeine treatment (Krogan and Greenblatt 2001; Jun et al., 2015; Mehlgarten et al., 2017). Our results showed that AtELP4 expression partially rescued growth retardation at 38°C of yeast elp4Δ mutant (Figure 5B). In our previous study, we demonstrated that plant DRL1 expression also partially rescued growth retardation at 38°C of the yeast kti12Δ mutant (Jun et al., 2015), indicating that the gene functions are partially conserved between plant DRL1 and yeast KTI12. Collectively, these cross-complementation analyses suggested that AtELP4 and DRL1 of Arabidopsis were closely related in function to their yeast homologs.
In contrast, we found one unknown motif specifically in plant ELP4, although further studies are necessary to prove this motif as a plant-specific binding or catalytic site. Previous research suggested that Kti12 motifs preserved in plant ortholog DRL1 may be involved in cofactor binding (Jun et al., 2015). We revealed that DRL1 is unable to fully replace the function of its yeast counterpart, Kti12, whereas hybrid domain fusion proteins between the two can be based on genetic cross-complementation analysis (Mehlgarten et al., 2017). Non-conserved plant-specific regions in DRL1 have been proposed to facilitate a selective physical or functional interaction with particular Elongator subunits from Arabidopsis.
Recently, it was demonstrated that ELP4 mutation impairs tRNA binding and diminishes Elongator activity, causing neurological defects in human cells (Gaik et al., 2022). Structural studies of mammalian Elongator provided evidence that in higher eukaryotes, Elp123 and Elp456 subcomplexes differ in a cell-type specific function (Gaik et al., 2022). This may be in contrast to unicellular yeast models suggesting species-specific sequence variation in Elongator subunits as a cause of evolutionary differences between lower and higher eukaryotes.
Elongator complex might control the expression of adaxial–abaxial determinant genes
Our findings show that the distribution pattern of palisade cell size in drl1-102 mutant leaves is more bell-shaped than in wild type (Figure 7F), indicating that the cell size of drl1-102 leaves was more irregular than that of wild type. In addition, leaves of Atelp4 and drl1-102 single and double mutants had increased intracellular airspace between palisade mesophyll cells of the adaxial subepidermal layer (Figure 7E). Furthermore, our results showed that loss-of-function mutation in AtELP4 or DRL1 induced not only increased expression levels of abaxial determinants, YAB and KAN families, but also decreased, expressing levels of adaxial determinants, AS2 and HD-ZIP III transcription factors, indicating that Elongator complex might regulate both the repression of the transcription of YAB and KAN families and the activation of the transcription of AS1 and HD-ZIP III genes. Nelissen et al. (2005) reported that Atelp3 mutant was abaxialized in the adaxial side of leaves, showing the narrow leaves with less, larger, and more irregularly shaped palisade cells with more intercellular spaces. Kojima et al. (2011) also reported that AtELP3 represses the expression of abaxial determinant genes, although it was unclear how AtELP3 regulates the transcription of abaxial determinant genes. These results supported our suggestion of the regulation of the dorsoventrality function by the Elongator complex. Recently, Nakai et al. (2019) reported that the tRNA modification function of Elongator is not required for the control of leaf abaxial–adaxial polarity.
Activation of YAB1/FIL transcription is shown to require the interaction of a specific mediator, MONOPTEROS (MP/ARF5), and SWI/SNF complex and recruitment on YAB loci at flower primordium initiation (Wu et al., 2015). (Nelissen et al. 2010) suggested based on ChIP analysis that the Holo-Elongator complex did not operate as a general transcription regulator but rather was involved in the acetylation of target genes. Exactly, they reported histone H3 lysine 14 acetylation in the coding regions of auxin repressor Short Hypocotyl 2 (SHY2; called also IAA3) and auxin influx carrier LAX2 genes to be under the control of Holo-Elongator (Nelissen et al., 2010). These findings showed that Elongator-dependent chromatin remodeling may regulate leaf polarity by activating or suppressing the expression of adaxial–abaxial determining genes via the auxin signaling cascade. Our results showed a reduction in AS1 expression in Atelp4 single and Atelp4 drl1-102 double mutants, suggesting the Elongator complex affects the expression of AS1. Taken together, we suggest that the Elongator complex might act upon a regulatory pathway for the expression of adaxial or abaxial determinant genes. However, the precise mode of Elongator action within the regulation of leaf patterning remains to be shown.
Elongator may be required for the determination of leaf flatness
Our results proclaimed that drl1-102, Atelp4 single, and Atelp4 drl1-102 double mutants had defects in the flatness of leaf blades, showing narrow and epinastic leaves (downward curling/curvature of leaf blade margin to abaxial side; downward curling along to the longitudinal axes) (Figure 7B). Leaf epinasty/hyponasty (downward curling or upward curling, respectively) was caused by the coordinated anisotropy of growth including cell division and expansion in epidermal, mesophyll parenchyma, and vascular tissues in leaves (Sandalio et al., 2016). The curling of leaves is known to be caused by differences or imbalances of cell proliferation or expansion rate between epidermis or mesophyll cell layers of adaxial and abaxial sides in leaves. In our results, abaxialized in the adaxial side of leaves from Atelp4, drl1-102, and Atelp4 drl1-102 mutants caused not only a reduction in cell number and size of palisade mesophyll cells but also an increase in intercellular airspace in the adaxial subepidermal layer, suggesting an alteration in flexibility/strength of the adaxial side (Figure 7E).
Consistent with the epinastic phenotype of leaves from Atelp4 and drl1-102 mutants, overexpression of YAB1 or YAB3 resulted in narrow and epinastic leaves (Siegfried et al., 1999). Loss of function in AtELP3 also showed narrow and epinastic leaves (Kojima et al., 2011). Additionally, the loss of function of AS1, AS2, or HD-ZIP III and a REV feedback regulator, ZPR overexpression, triggered downward curling with abaxialized leaves (Wenkel et al., 2007). These studies supported our suggestion that the Elongator complex may regulate leaf curling.
In contrast, auxin accumulation and gradients are factors controlling leaf epinasty. Exogenous auxin application or genetic alteration of auxin biosynthesis or signaling genes have been reported to present epinastic or hyponastic leaves by the disruption of auxin homeostasis (Keller and Van Volkenburgh, 1997; Keller and Van Volkenburgh, 1998; Kawano et al., 2003). Auxin overproduction caused by a reduction in free IAA or a decrease in auxin responsiveness causes leaf epinasty, according to genetic studies of rooty (King et al., 1995), superroot2 (Delarue et al., 1998), yucca (Zhao et al., 2001), and iaaM (Romano et al., 1995), whereas elevated auxin level or increased auxin response causes leaf hyponasty from the genetic studies of icu6 (Pérez-Pérez et al., 2010) and iamt1-D (Qin et al., 2005). iamt1-D, gain-of-function indole-3-acetic acid (IAA) carboxyl methyltransferase1 (IAMT1), converts free IAA to methyl-IAA in vitro, showing that reduction of free IAA has narrow and hyponastic leaves, whereas RNAi transgenic plants had epinastic leaves (Qin et al., 2005).
Although we do not yet have direct evidence of auxin level changes or response to auxin in leaf morphogenesis in Atelp4 mutant, our results presented that Elongator might be a linker between auxin response and the establishment of leaf polarity. These complicated multiple functions enable the Elongator to play an important role in auxin signaling (Nelissen et al., 2010, Leitner et al., 2015).
Elongator function in tRNA modification may be required for proper leaf development
Modification at the anticodon wobble position (U34) of tRNALys, tRNAGlu, and tRNAGln is in charge of fine-tuning mRNA translation efficiency and rate (Nakai et al., 2019). It is reported that the plant Elongator cooperates with two UBIQUITIN-RELATED MODIFIER 1 (URM1)-like proteins involved in sulfur modification of tRNA at U34 (Nakai et al., 2019). Lack of URM1-like proteins or AtELP3 exhibited an increase in intercellular airspace, a decrease in chlorophyll contents, and a delay of endoreduplication, suggesting the importance of U34 modification in leaf morphogenesis (Nakai et al., 2019). Similarly, our results showed that Atelp4 and drl1-102 single and double mutants had increased intercellular airspace and decreased cell number. Atelp4 and drl1-102 mutants also showed reduced chlorophyll contents and a delay of endoreduplication in leaves (Figure 8A and Supplementary Figure 3).
In summary, we suggest that AtELP4 as part of the Elongator complex in Arabidopsis plays critical roles in cell proliferation, leaf flatness, and establishment of leaf polarity through a collaboration with DRL1 during leaf morphogenesis. While the tRNA modification function clearly is important for proper leaf morphogenesis, the Elongator complex may also be required to contribute to leaf development in plants. Further studies will have to await the molecular mechanism and mode of action of individual Elongator subunits to elucidate Elongator’s diverse and versatile functions in plants.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
K-HC, RS and G-TK conceived and designed the study. SEJ, MAM, RS, and G-TK wrote the manuscript. SEJ, MAM, TYH, and YSK performed the investigation and SEJ, MAM, and G-TK analyzed data. All authors reviewed the results and approved the final version of the manuscript.
Funding
This research was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No. 2021R1A2C1006572), by the Basic Science Research Program through the NRF funded by the Ministry of Education (No. 2020R1A6A1A03047729 and No. 2020R1I1A1A01054294), and by the Green Fusion Technology Program funded by Ministry of Environment.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1033358/full#supplementary-material
Supplementary Figure 1 | Expression level of AtELP4 and ScELP4 in yeast wild type (BY4741), elp4Δ mutant (Y02150), three lines of pTU1-empty vector harboring elp4Δ mutant (#1, #5, and #8), and two lines of pTU1-AtELP4 harboring elp4Δ mutant (#2 and #29) by (A) semi qRT-PCR and (B) qRT-PCR. To confirm ELP4 silence of Y02150, the expression level of endogenous Saccharomyces cerevisiae ELP4 (ScELP4) was estimated. The expression level was normalized to that of ScACT1.
Supplementary Figure 2 | Expression level of AtELP4, DRL1, and other genes by qRT-PCR in Atelp4 and drl1-102 single and double mutants. Identical amounts of total RNA isolated from 21-day-old seedlings of wild type (Col-0), Atelp4, drl1-102, and Atelp4 drl1-102 were subjected to qRT-PCR experiments to analyze the expression level of leaf polarity- and cell proliferation-relative genes. The expression level of analyzed genes was normalized by that of TUB4. Primer sequences of used genes are described in SUPPLEMENTARY TABLE 5.
Supplementary Figure 3 | Distribution of DNA ploidy on the mature 3rd rosette leaves of plants on 21 DAS.
References
Abbassi, N. E., Biela, A., Glatt, S., Lin, T.-Y. (2020). How elongator acetylates tRNA bases. Int. J. Mol. Sci. 21, 8209. doi: 10.3390/ijms21218209
Alvarez, J. P., Pekker, I., Goldshmidt, A., Blum, E., Amsellem, Z., Eshed, Y. (2006). Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 18 (5), 1134–1151. doi: 10.1105/tpc.105.040725
Arena, C., Conti, S., Francesca, S., Melchionna, G., Hájek, J., Barták, M., et al. (2020). Eco-physiological screening of different tomato genotypes in response to high temperatures: A combined field-to-Laboratory approach. Plants 9 (4), 508. doi: 10.3390/plants9040508
Bailey, T. L., Johnson, J., Grant, C. E., Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43 (W1), W39–W49. doi: 10.1093/nar/gkv416
Brachmann, C. B., Davies, A., Cost, G. J., Caputo, E., Li, J., Hieter, P., et al. (1998). Designer deletion strains derived from saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14 (2), 115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2
Chen, Y. T., Hims, M. M., Shetty, R. S., Mull, J., Liu, L., Leyne, M., et al. (2009). Loss of mouse ikbkap, a subunit of elongator, leads to transcriptional deficits and embryonic lethality that can be rescued by human IKBKAP. Mol. Cell. Biol. 29 (3), 736–744. doi: 10.1128/MCB.01313-08
Cho, K.-H., Choi, H., Seki, M., Jun, S. E., Yi, Y. B., Shinozake, K., et al. (2007). DRL1 regulates adaxial leaf patterning and shoot apical meristem activity in Arabidopsis. J. Plant Biol. 50, 467–474. doi: 10.14348/molcells.2015.2297
Close, P., Hawkes, N., Cornez, I., Creppe, C., Lambert, C. A., Rogister, B., et al. (2006). Transcription impairment and cell migration defects in elongator-depleted cells: implication for familial dysautonomia. Mol. Cell 22 (4), 521–531. doi: 10.1016/j.molcel.2006.04.017
Darzentas, N. (2010). Circoletto: visualizing sequence similarity with circos. Bioinf. (Oxford England) 26 (20), 2620–2621. doi: 10.1093/bioinformatics/btq484
Dauden, M. I., Jaciuk, M., Muller, C. W., Glatt, S. (2017b). Structural asymmetry in the eukaryotic elongator complex. FEBS Lett. 592, 502–515. doi: 10.1002/1873-3468.12865
Dauden, M. I., Jaciuk, M., Weis, F., Lin, T.-Y., Kleindienst, C., Abbassi, N. E. H., et al. (2019). Molecular basis of tRNA recognition by the elongator complex. Sci. Adv. 5, eaaw2326. doi: 10.1126/sciadv.aaw2326
Dauden, M. I., Kosinski, J., Kolaj-Robin, O., Desfosses, A., Ori, A., Faux, C., et al. (2017a). Architecture of the yeast elongator complex. EMBO Rep. 18, 264–279. doi: 10.15252/embr.201643353
Delarue, M., Prinsen, E., Onckelen, H. V., Caboche, M., Bellini, C. (1998). Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J. 14 (5), 603–611. doi: 10.1046/j.1365-313x.1998.00163.x
Doležel, J., Göhde, W. (1995). Sex determination in dioecious plants melandrium album and m. rubrum using high-resolution flow cytometry. Cytometry 19, 103–106. doi: 10.1002/cyto.990190203
Enders, T. A., Strader, L. C. (2015). Auxin activity: Past, present, and future. Am. J. Bot. 102 (2), 180–196. doi: 10.3732/ajb.1400285
Eshed, Y., Izhaki, A., Baum, S. F., Floyd, S. K., Bowman, J. L. (2004). Asymmetric leaf development and blade expansion in Arabidopsisare mediated by KANADI and YABBY activities. Development 131, 12, 2997–3006. doi: 10.1242/dev.01186
Finn, R. D., Mistry, J., Schuster-Böckler, B., Griffiths-Jones, S., Hollich, V., Lassmann, T., et al. (2006). Pfam: clans, web tools and services. Nucleic Acids Res. 34, D247–D251. doi: 10.1093/nar/gkj149
Frohloff, F., Fichtner, L., Jablonowski, D., Breunig, K. D., Schaffrath, R.. (2001). Saccharomyces cerevisiae Elongator mutations confer resistance to the Kluyveromyces lactis zymocin. EMBO J. 20 (8), 1993–2003. doi: 10.1093/emboj/20.8.1993
Gaik, M., Kojic, M., Stegeman, M. R., Öncü-Öner, T., Kościelniak, A., Jones, A., et al. (2022). Functional divergence of the two elongator subcomplexes during neurodevelopment. EMBO Mol. Med. 14 (7), e15608. doi: 10.15252/emmm.202115608
Glatt, S., Létoquart, J., Faux, C., Taylor, N. M., Séraphin, B., Müller, C. W. (2012). The elongator subcomplex Elp456 is a hexameric RecA-like ATPase. Nat. Struct. Mol. Biol. 19 (3), 314–320. doi: 10.1038/nsmb.2234
Glatt, S., Zabel, R., Kolaj-Robin, O., Onuma, O. F., Baudin, F., Graziadei, A., et al. (2016). Structural basis for tRNA modification by Elp3 from Dehalococcoides mccartyi. Nat. Struct. Mol. Biol. 23, 794–802. doi: 10.1038/nsmb.3265
Hawkes, N. A., Otero, G., Winkler, G. S., Marshall, N., Dahmus, M. E., Krappmann, D., et al. (2002). Purification and characterization of the human elongator complex. J. Biol. Chem. 277 (4), 3047–3052. doi: 10.1074/jbc.M110445200
Huang, B., Johansson, M. J. O., Byström, A. S. (2005). An early step in wobble uridine tRNA modification requires the elongator complex. RNA 11, 424–436. doi: 10.1261/rna.7247705
Jablonowski, D., Frohloff, F., Fichtner, L., Stark, M. J., Schaffrath, R. (2001). Kluyveromyces lactis zymocin mode of action is linked to RNA polymerase II function via elongator. Mol. Microbiol. 42 (4), 1095–1105. doi: 10.1046/j.1365-2958.2001.02705.x
Johansson, M. J. O., Xu, F., Byström, A. S. (2018). Elongator-a tRNA modifying complex that promotes efficient translational decoding. biochim. biophys. Acta Gene Regul. Mech. 1861, 401–408. doi: 10.1016/j.bbagrm.2017.11.006
Jun, S. E., Cho, K.-H., Hwang, J. Y., Abdel-Fattah, W., Hammermeister, A., Schaffrath, R., et al. (2015). Comparative analysis of the conserved functions of arabidopsis DRL1 and yeast KTI12. Mol. Cells 38 (3), 243–250. doi: 10.14348/molcells.2015.2297
Jun, S. E., Kim, J. H., Hwang, J. Y., Huynh Le, T. T., Kim, G.-T. (2019). ORESARA15 acts synergistically with ANGUSTIFOLIA3 and separately from AINTEGUMENTA to promote cell proliferation during leaf growth. Int. J. Mol. Sci. 21 (1), 241. doi: 10.3390/ijms21010241
Jun, S. E., Okushima, Y., Nam, J., Umeda, M., Kim, G.-T. (2013). Kip-related protein 3 is required for control of endoreduplication in the shoot apical meristem and leaves of arabidopsis. Mole. Cells 35 (1), 47–53. doi: 10.1007/s10059-013-2270-4
Kalve, S., De Vos, D., Beemster, G. T. (2014). Leaf development: a cellular perspective. front. Plant Sci. 5. doi: 10.3389/fpls.2014.00362
Kawano, N., Kawano, T., Lapeyrie, F. (2003). Inhibition of the indole-3-acetic acid-induced epinastic curvature in tobacco leaf strips by 2,4-dichlorophenoxyacetic acid. Ann. Bot. 91 (4), 465–471. doi: 10.1093/aob/mcg043
Keller, C. P., Van Volkenburgh, E. (1997). Auxin-induced epinasty of tobacco leaf tissues (A nonethylene-mediated response). Plant Physiol. 113 (2), 603–610. doi: 10.1104/pp.113.2.603
Keller, C. P., Van Volkenburgh, E. (1998). Evidence that auxin-induced growth of tobacco leaf tissues does not involve cell wall acidification. Plant Physiol. 118 (2), 557–564. doi: 10.1104/pp.118.2.557
King, J. J., Stimart, D. P., Fisher, R. H., Bleecker, A. B. (1995). Mutation altering auxin homeostasis and plant morphology in arabidopsis. Plant Cell 7 (12), 2023–2037. doi: 10.1105/tpc.7.12.2023
Kojima, S., Iwasaki, M., Takahashi, H., Imai, T., Matsumura, Y., Fleury, D., et al. (2011). ASYMMETRIC LEAVES1 and elongator, a histone acetyltransferase complex, mediate the establishment of polarity in leaves of Arabidopsis thaliana. Plant Cell Physiol. 52, 1259–1273. doi: 10.1093/pcp/pcr083
Krogan, N. J., Greenblatt, J. F. (2001). Characterization of a six-subunit holo-elongator complex required for the regulated expression of a group of genes in Saccharomyces cerevisiae. Mol. Cell Biol. 21 (23), 8203–8212. doi: 10.1128/MCB.21.23.8203-8212.2001
Krutyhołowa, R., Hammermeister, A., Zabel, R., Abdel-Fattah, W., Reinhardt-Tews, A., Helm, M., et al. (2019). Kti12, a PSTK-like tRNA dependent ATPase essential for tRNA modification by elongator. Nucleic Acids Res. 47, 4814–4830. doi: 10.1093/nar/gkz190
Leitner, J., Retzer, K., Malenica, N., Bartkeviciute, R., Lucyshyn, D., Jäger, G., et al. (2015). Meta-regulation of Arabidopsis auxin responses depends on tRNA maturation. Cell Rep. 11 (4), 516–26. doi: 10.1016/j.celrep.2015.03.054
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30 (1), 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Doerks, T., Bork, P. (2012). SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40 (D1), D302–D305. doi: 10.1093/nar/gkr931
Lin, T.-Y., Abbassi, N. E. H., Zakrzewski, K., Chramiec-Głąbik, A., Jemioła-Rzemińska, M., Glatt, S. (2019). The elongator subunit Elp3 is a non-canonical tRNA acetyltransferase. Nat. Commun. 10, 625. doi: 10.1038/s41467-019-08579-2
McConnell, J. R., Emery, J., Eshed, Y., Bao, N., Bowman, J., Barton, M. K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411 (6838), 709–713. doi: 10.1038/35079635
Mehlgarten, C., Jablonowski, D., Wrackmeyer, U., Tschitschmann, S., Sondermann, D., Jager, G., et al. (2010). Elongator function in tRNA wobble uridine modification is conserved between yeast and plants. Mol. Microbiol. 76, 1082–1094. doi: 10.1111/j.1365-2958.2010.07163.x
Mehlgarten, C., Prochaska, H., Hammermeister, A., Abdel-Fattah, W., Wagner, M., Krutyhołowa, R., et al. (2017). Use of a yeast tRNase killer toxin to diagnose Kti12 motifs required for tRNA modification by elongator. Toxins 9 (9), 272. doi: 10.3390/toxins9090272
Nakai, Y., Horiguchi, G., Iwabuchi, K., Harada, A., Nakai, M., Hara-Nishimura, I., et al. (2019). tRNA wobble modification affects leaf cell development in Arabidopsis thaliana. Plant Cell Physiol. 60 (9), 2026–2039. doi: 10.1093/pcp/pcz064
Nelissen, H., De Groeve, S., Fleury, D., Neyt, P., Bruno, L., Bitonti, M. B., et al. (2010). Plant elongator regulates auxin-related genes during RNA polymerase II transcription elongation. Proc. Natl. Acad. Sci. U.S.A. 107 (4), 1678–1683. doi: 10.1073/pnas.0913559107
Nelissen, H., Fleury, D., Bruno, L., Robles, P., De Veylder, L., Traas, J., et al. (2005). The elongate mutants identify a functional elongator complex in plants with a role in cell proliferation during organ growth. Proc. Natl. Acad. Sci. U. S. A. 102, 7754–7759. doi: 10.1073/pnas.0502600102
Otero, G., Fellows, J., Li, Y., de Bizemont, T., Dirac, A. M. G., Gustafsson, C. M., et al. (1999). Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell 3 (1), 109–118. doi: 10.1016/S1097-2765(00)80179-3
Otto, F. (1992). “Preparation and staining of cells for high-resolution DNA analysis,” in Flow cytometry and cell sorting. Ed. Radbruch, A. (Berlin: Springer), 101–104. doi: 10.1007/978-3-662-02785-1_8
Pekker, I., Alvarez, J. P., Eshed, Y. (2005). Auxin response factors mediate arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17, 2899–2910. doi: 10.1105/tpc.105.034876
Pérez-Pérez, J. M., Candela, H., Robles, P., López-Torrejón, G., del Pozo, J. C., Micol, J. L. (2010). A role for AUXIN RESISTANT3 in the coordination of leaf growth. Plant Cell Physiol. 51 (10), 1661–1673. doi: 10.1093/pcp/pcq123
Petrakis, T. G., Søgaard, T. M. M., Erdjument-Bromage, H., Tempst, P., Svejstrup, J. Q. (2005). Physical and functional interaction between elongator and the chromatin-associated Kti12 protein. J. Biol. Chem. 280, 19454–19460. doi: 10.1074/jbc.M413373200
Ponting, C. P. (2002). Novel domains and orthologues of eukaryotic transcription elongation factors. Nucleic Acids Res. 30, 3642–3652. doi: 10.1093/nar/gkf498
Qin, G., Gu, H., Zhao, Y., Ma, Z., Shi, G., Yang, Y., et al. (2005). An indole-3-acetic acid carboxyl methyltransferase regulates arabidopsis leaf development. Plant Cell 17 (10), 2693–2704. doi: 10.1105/tpc.105.034959
Ritchie, R. (2006). Consistent sets of spectrophotometric chlorophyll equations for acetone, methanol and ethanol solvents. Photosynth. Res. 89 (1), 27–41. doi: 10.1007/s11120-006-9065-9
Romano, C. P., Robson, P. R. H., Smith, H., Estelle, M., Klee, H. (1995). Transgene-mediated auxin overproduction in arabidopsis: hypocotyl elongation phenotype and interactions with the hy6-1 hypocotyl elongation and axr1 auxin-resistant mutants. Plant Mol. Biol. 27, 1071–1083. doi: 10.1007/BF00020881
Sandalio, L. M., Rodríguez-Serrano, M., Romero-Puertas, M. C. (2016). Leaf epinasty and auxin: A biochemical and molecular overview. Plant Sci. 253, 187–193. doi: 10.1016/j.plantsci.2016.10.002
Schaffrath, R., Leidel, S. A. (2017). Wobble uridine modifications – a reason to live, a reason to die?RNA Biol. 14, 1209–1222. doi: 10.1080/15476286.2017.1295204
Selvadurai, K., Wang, P., Seimetz, J., Huang, R. H. (2014). Archaeal Elp3 catalyzes tRNA wobble uridine modification at C5 via a radical mechanism. Nat. Chem. Biol. 10, 810–812. doi: 10.1038/nchembio.1610
Siegfried, K. R., Eshed, Y., Baum, S. F., Otsuga, D., Drews, G. N., Bowman, J. L. (1999). Members of the YABBY gene family specify abaxial cell fate in arabidopsis. Development 126 (18), 4117–4128. doi: 10.1242/dev.126.18.4117
Sokołowski, M., Klassen, R., Bruch, A., Schaffrath, R., Glatt, S. (2018). Cooperativity between different tRNA modifications and their modification pathways. Biochim. Biophys. Acta Gene Regul. Mech. 1861 (4), 409–418. doi: 10.1016/j.bbagrm.2017.12.003
Svejstrup, J. Q. (2007). Contending with transcriptional arrest during RNAPII transcript elongation. Trends Biochem. Sci. 32 (4), 165–171. doi: 10.1016/j.tibs.2007.02.005
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25 (24), 4876–4882. doi: 10.1093/nar/25.24.4876
Wenkel, S., Emery, J., Hou, B. H., Evans, M. M., Barton, M. K. (2007). A feedback regulatory module formed by LITTLE ZIPPER and HD-ZIPIII genes. Plant Cell 19 (11), 3379–3390. doi: 10.1105/tpc.107.055772
Winkler, G. S., Petrakis, T. G., Ethelberg, S., Tokunaga, M., Erdjument-Bromage, H., Tempst, P., et al (2001). RNA polymerase II elongator holoenzyme is composed of two discrete subcomplexes. J. Biol. Chem. 276, 32743–32749. doi: 10.1074/jbc.M105303200
Winter, D., Vinegar, B., Nahal, H., Ammar, R., Wilson, G. V., Provart, N. J. (2007). An “Electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PloS One 2, e718. doi: 10.1371/journal.pone.0000718
Wittschieben, B. O., Otero, G., de Bizemont, T., Fellows, J., Erdjument-Bromage, H., Ohba, R., et al. (1999). A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell. 4, 123–128. doi: 10.1016/s1097-2765(00)80194-x
Wu, M. F., Yamaguchi, N., Xiao, J., Bargmann, B., Estelle, M., Sang, Y., et al. (2015). Auxin-regulated chromatin switch directs acquisition of flower primordium founder fate. eLife 4, e09269. doi: 10.7554/eLife.09269
Keywords: AtELP4, cell proliferation, DRL1, Elongator complex, leaf polarity
Citation: Jun SE, Cho K-H, Manzoor MA, Hwang TY, Kim YS, Schaffrath R and Kim G-T (2022) AtELP4 a subunit of the Elongator complex in Arabidopsis, mediates cell proliferation and dorsoventral polarity during leaf morphogenesis. Front. Plant Sci. 13:1033358. doi: 10.3389/fpls.2022.1033358
Received: 31 August 2022; Accepted: 20 September 2022;
Published: 21 October 2022.
Edited by:
Yunpeng Cao, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Quaid Hussain, Zhejiang Agriculture & Forestry University, ChinaAwais Shakoor, Universitat de Lleida, Spain
Muhammad Ahsan Altaf, Hainan University, China
Copyright © 2022 Jun, Cho, Manzoor, Hwang, Kim, Schaffrath and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gyung-Tae Kim, a2ltZ3RAZG9uZ2EuYWMua3I=
†Present address: Kiu-Hyung Cho, Gyeongbuk Institute for Bioindustry, Andong, Gyeongbuk, South Korea
‡These authors have contributed equally to this work and share first authorship
 Sang Eun Jun
Sang Eun Jun Kiu-Hyung Cho1†‡
Kiu-Hyung Cho1†‡ Muhammad Aamir Manzoor
Muhammad Aamir Manzoor Raffael Schaffrath
Raffael Schaffrath Gyung-Tae Kim
Gyung-Tae Kim