- 1Plant Systems Engineering Research Center, Korea Research Institute of Bioscience & Biotechnology (KRIBB), Daejeon, South Korea
- 2Department of Plant Medicine, Chungbuk National University, Cheongju, South Korea
- 3Biological Resource Center, Korea Research Institute of Bioscience & Biotechnology (KRIBB), Jeongeup, South Korea
Programmed cell death (PCD), a characteristic feature of hypersensitive response (HR) in plants, is an important cellular process often associated with the defense response against pathogens. Here, the involvement of LytB, a gene encoding 4-hydroxy-3-methylbut-2-enyl diphosphate reductase that participates in the final step of the plastid methylerythritol phosphate (MEP) pathway, in plant HR cell death was studied. In Nicotiana benthmiana plants, silencing of the NbLytB gene using virus-induced gene silencing (VIGS) caused plant growth retardation and albino leaves with severely malformed chloroplasts. In NbLytB-silenced plants, HR-related cell death mediated by the expression of either the human proapoptotic protein gene Bax or an R gene with its cognate Avr effector gene was inhibited, whereas that induced by the nonhost pathogen Pseudomonas syringae pv. syringae 61 was enhanced. To dissect the isoprenoid pathway and avoid the pleiotropic effects of VIGS, chemical inhibitors that specifically inhibit isoprenoid biosynthesis in plants were employed. Treatment of N. benthamiana plants with fosmidomycin, a specific inhibitor of the plastid MEP pathway, effectively inhibited HR-related PCD, whereas treatment with mevinolin (a cytoplasmic mevalonate pathway inhibitor) and fluridone (a carotenoid biosynthesis inhibitor) did not. Together, these results suggest that the MEP pathway as well as reactive oxygen species (ROS) generation in the chloroplast play an important role in HR-related PCD, which is not displaced by the cytosolic isoprenoid biosynthesis pathway.
1 Introduction
Hypersensitive response (HR), characterized by programmed cell death (PCD) in plants, is often observed during the response to pathogen attack. Reactive oxygen species (ROS) generation, ion flux alterations, protein modifications, and gene activation have been implicated in the regulation of pathogen-induced PCD (Dangl et al., 1996; Greenberg and Yao, 2004; Van Breusegem and Dat, 2006; Mur et al., 2008; Huysmans et al., 2017). One of the most rapid plant defense responses to pathogen attack is ROS burst, which involves the production of superoxide anion (), hydroxyl radical (•OH), and hydrogen peroxide (H2O2) at the site of attempted invasion (Byczkowski and Gessner, 1988; Apostol et al., 1989; Levine et al., 1994; Ausubel, 2005; Ye et al., 2021). Elevated ROS levels directly lead to pathogen death, thus strengthening the cell wall and acting as a signal to activate further defenses (Jabs, 1999; Torres et al., 2006). ROS are primarily generated by nicotinamide adenine dinucleotide phosphate (NADPH) oxidase in the plasma membrane (Doke et al., 1996). Other sources of ROS in plants include photosynthesis, photorespiration, and oxidative mitochondrial respiration (Foyer and Noctor, 2009). In plant cells, the chloroplast is a major source of ROS and therefore is an important player in cell death responses (Zapata et al., 2005). For example, in Tobacco mosaic virus (TMV)-infected leaves of tobacco (Nicotiana tabacum) plants, light induces H2O2 generation and cell death upon mitogen-activated protein kinase (MAPK) activation (Liu et al., 2007). Similarly, Arabidopsis thaliana plants grown under low light exhibit compromised local and systemic defense responses against infection by bacterial pathogens (Genoud et al., 2002). Light is required for the HRT (Hypersensitive Response to Turnip crinkle virus (TCV)) gene-mediated HR and resistance to TCV (Chandra-Shekara et al., 2006). Accelerated cell death 2 (ACD2), a chloroplast protein, modulates cell death and resistance to pathogen infection in Arabidopsis (Yao and Greenberg, 2006). Together, these findings suggest a link between chloroplast-derived signals and pathogen-induced PCD, although the nature of the link remains unknown.
Owing to extensive investigation, the signaling pathways of apoptosis are relatively well known in animals (Willis et al., 2003). The B cell lymphoma-2 (Bcl-2) family consists of both cell death-inducing proapoptotic proteins (e.g., Bax, Bak, Bid, Bik, Bcl-XS, Bok, and Hrk) and cell survival-promoting antiapoptotic proteins (e.g., Bcl-SL, Bcl-2, Bcl-W, Bcl-XL, Bfl-1, Mcl-1, Brag-1, and A1) (Hockenbery et al., 1993; Green and Reed, 1998; Gross et al., 1999). Proapoptotic proteins induce the loss of mitochondrial membrane potential, which results in the release of several apoptotic regulatory proteins, including cytochrome c and procaspases, as well as ROS generation. For example, translocation of Bax to the outer mitochondrial membrane is accompanied by the release of apoptogenic factors, such as cytochrome c, into the cytosol, leading to the activation of caspases, a family of cysteine proteases that control and mediate the apoptotic response (Liu et al., 1996; Jurgensmeier et al., 1998). Genomic data analysis suggests that the animal Bcl-2 gene family does not exist in plants, which implies that plants possess different PCD pathways than animals (Ameisen, 2002). However, overexpression of the animal apoptosis protein gene Bax in plants can trigger HR-like apoptosis (Kawai-Yamada et al., 2001). Moreover, in tobacco (Nicotiana benthamiana), Bax-induced PCD was accompanied by an increase in ROS production and Pathogenesis-Related (PR) gene expression, similar to that observed during TMV-induced HR (Lacomme and Santa Cruz, 1999; Kawai-Yamada et al., 2001). In addition, Arabidopsis possesses a functionally conserved homolog of the mammalian Bax Inhibitor-1 (BI-1) protein, which inhibits PCD induced by Bax in yeast and plant (Kawai et al., 1999; Kawai-Yamada et al., 2001). BI-1 is an evolutionarily conserved transmembrane protein present in the endoplasmic reticulum (ER) that, when overexpressed in plants, inhibits various PCDs induced by pathogens, heat stress, and phytotoxins, in addition to Bax (Watanabe and Lam, 2008). These results suggest that some parts of the animal and plant PCD pathways are evolutionarily conserved at the molecular level. In the past few years, BI-1 has been identified as a novel apoptosis regulatory factor belonging to the TMBIM (Transmembrane BAX Inhibitor-1 Motif-containing) protein family (Rojas-Rivera and Hetz, 2015). In mammals, proteins in this group have at least six highly conserved members, and in plants, several homologs of these proteins, in addition to BI-1, have been reported (Rojas-Rivera and Hetz, 2015). Although how BI-1 functions in cytoprotection is not yet clear, it is known to regulate calcium secretion from the ER, inositol-requiring enzyme 1 (IRE1) activity, and autophagy during ER stress in both plant and animals (Lebeaupin et al., 2020).
In the late 1990s, a new group of conserved eukaryotic apoptosis regulatory proteins, Bcl-2 associated athanogenes (BAGs), was discovered (Takayama et al., 1995; Takayama and Reed, 2001). BAGs have a common conserved region that interacts with the ATPase domain of heat shock protein 70 (Hsc70/Hsp70), called the BAG domain (BD), which is known to be involved in PCD and autophagy in various stressful environments (Takayama and Reed, 2001). Advanced bioinformatics analysis, based on protein structure rather than on primary sequence, has revealed that Arabidopsis possesses seven BAG homologous genes containing the BD (Doukhanina et al., 2006). BAG homologs are also present in other plant species and, similar to those in animals, are cytoprotective against a variety of agents that induce stress-induced PCDs, including pathogens (Williams et al., 2014). Plant BAGs, unlike animal BAGs, are localized to various subcellular organelles, and some BAGs have a calmodulin binding domain not found in animal BAGs. These results, together with the observed presence or absence in plants of other major animal apoptosis regulators, has led to speculation that the mechanisms of cell death regulation in plants are different from those in animals (Thanthrige et al., 2020).
Isoprenoids are found in all living organisms but are especially abundant and diverse in plants (Peñuelas and Munné-Bosch, 2005; Kirby and Keasling, 2009; Pulido et al., 2012). These compounds are produced from a basic five-carbon unit, isopentenyl diphosphate (IPP), and its isomer, dimethylallyl diphosphate (DMAPP). IPP and DMAPP are produced through one of two pathways: the cytosolic mevalonate (MVA) pathway and the chloroplast 1-deoxy-D-xylulose 5-phosphate (DXP)/2-C-methyl-D-erythritol 4-phosphate (MEP) pathway (Peñuelas and Munné-Bosch, 2005; Kirby and Keasling, 2009; Pulido et al., 2012). The MVA pathway has been extensively studied in various organisms, including yeast and animals; however, the MEP pathway was uncovered in plants only in recent decades. The MEP pathway, which comprises seven enzymatic steps, has been identified in eubacteria and photosynthetic organisms, including plants, algae, cyanobacteria, and diatoms (Banerjee and Sharkey, 2014). Genetic analyses revealed that the MEP pathway plays critical roles in multiple aspects of plant cell physiology (Phillips et al., 2008). However, many questions remain about the reciprocal network of isoprenoid biosynthetic precursors (IPP and DMAPP), which are synthesized independently in two intracellular compartments, the cytoplasm and the chloroplast, and the effect of these precursors on plant development. Nonetheless, no studies have yet been conducted to investigate the effect of the MEP pathway on pathogen-induced PCD in plants.
Previously, using the virus-induced gene silencing (VIGS) approach, we studied the HR involvement of pepper (Capsicum annuum) genes upregulated in response to biological stresses in N. benthamiana plants (Lee et al., 2013). Here, we report on another gene identified in the VIGS screen, which encodes 4-hydroxy-3-methylbut-2-enyl diphosphate reductase (LytB), an enzyme that catalyzes the last step of the plastid MEP pathway. Using a combination of genetic knockdown and chemical inhibition assays, we show that the MEP pathway is essential for Bax-mediated PCD as well as for HR-mediated R–Avr interaction. These findings provide important evidence showing that, in addition to ROS, the MEP pathway is involved in regulating plant HR-related PCD.
2 Materials and methods
2.1 Cloning of NbLytB
To isolate the NbLytB gene, a N. benthamiana DNA library was amplified by PCR using sequence-specific primers designed on the basis of an Expressed Sequence Tag (EST; GenBank accession no. AY497304). The PCR amplicon was purified using the Gel&PCR purification system (BioFACT, Daejeon, South Korea), and the purified product was cloned into the pGEM T-Easy vector (Promega, Madison, WI, USA). The cloned fragment was sequenced and then used to design primers for 5´-rapid amplification of cDNA ends (RACE) PCR. Then, total RNA was extracted from N. benthamiana leaves and amplified by 5´-RACE PCR, using the designed primers and Ambion’s FirstChoice® RLM RACE Kit (Ambion, Austin, TX, USA), according to the manufacturer’s instructions.
2.2 VIGS assay
Wild-type (WT) tobacco (N. benthamiana) plants were grown in soil in an environmentally controlled growth room at 23°C under 16 h light/8 h dark photoperiod. VIGS was used to silence the target N. benthamiana genes as described previously (Liu et al., 2002; Moon et al., 2016). Briefly, recombinant pTRV2 plasmids and the pTRV1 vector containing RNA1, which is required for virus replication, were separately transformed into Agrobacterium tumefaciens strain GV2260. A 5 ml culture of A. tumefaciens containing each pTRV2 construct and a 20 ml culture of A. tumefaciens harboring pTRV1 were grown at 28°C for 16 h in Luria-Bertani (LB) medium (Thermo Fisher Scientific, Waltham, MA, USA) containing 100 µg/ml rifampicin (Sigma-Aldrich, St. Louis, MO, USA) and 50 µg/ml kanamycin (Sigma-Aldrich). To perform VIGS assays, A. tumefaciens cells were resuspended in infiltration buffer (10 mM MgCl2, 10 mM MES (pH 5.6), 200 µM acetosyringone (Sigma-Aldrich)) to a final optical density (OD600) of 0.5 (pTRV2) or 0.7 (pTRV1), and each pTRV2 cell suspension was mixed with the pTRV1 suspension at a 1:1 ratio. After a 4 h incubation at room temperature, the mixed Agrobacterium cultures were infiltrated into the abaxial surface of N. benthamiana leaves using a needleless syringe. A morphological phenotype of gene silencing was observed in systemic leaves at 10 days after infiltration. Agrobacterium-mediated transient expression experiments were performed on leaves with a relatively high probability of target gene silencing, based on their albino phenotype caused by silencing of the NbPDS or NbLytB gene.
2.3 Semiquantitative reverse transcriptase PCR
Total RNA was extracted from the agroinfiltrated N. benthamiana leaves using TRI Reagent (Molecular Research Center, inc., Cincinnati, OH, USA), according to the manufacturer’s instructions, and treated with DNase I (NEB, Ipswich, MA, USA). Then, cDNA was synthesized from 2 μg of total RNA using the Moloney murine leukemia virus (M-MLV) reverse transcriptase (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions. Semiquantitative RT-PCR was performed on DNA Engine® Peltier Thermal Cycler (Bio-Rad, Hercules, CA, USA) using AccuPower® PCR PreMix (Bioneer, Daejeon, Korea) and gene-specific primers (Supplementary Table 1). The PCR program consisted of 30 cycles (for phytoene desaturase (NbPDS) and NbLytB genes) and 22 cycles (for the NbActin gene; internal reference) of the following three steps: 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min. PCR products were separated on 1.0% agarose gels and visualized by ethidium bromide staining (Burton et al., 2000; Liu et al., 2002).
2.4 HR-PCD assays
A. tumefaciens cells carrying different expression constructs (pBtex:empty vector, Bax, HRT, TCV CP, Rx, PVX CP) were grown overnight in LB medium containing 100 µg/ml rifampicin and 50 µg/ml kanamycin. Subsequently, the Agrobacterium cells were precipitated, washed, and resuspended in infiltration buffer to obtain a final OD600 of 0.5. Then, the Agrobacterium cell suspensions were incubated at room temperature for 4 h before infiltration into N. benthamiana leaves. Pseudomonas syringae pv. syringae 61 (Pss61) and the hrp- mutant were grown overnight at 28°C in King’s medium (Duchefa Biochemie, Haarlem, Netherlands), and resuspended in 10 mM MgCl2 (104 cfu/ml).
2.5 Pathogen inoculation
Viral inoculum was prepared from N. benthamiana leaves infected by GFP-tagged TMV (TMV : GFP). Two weeks after VIGS, N. benthamiana plants expressing the TMV resistance N gene were rubbed with the sap of leaves inoculated with TMV : GFP and containing carborundum (Sigma-Aldrich) (Collum and Culver, 2017). All inoculated plants were maintained in the growth room at 23°C under 16 h light/8 h dark photoperiod. TMV : GFP fluorescence in the inoculated plants was observed daily using a UVP BioImaging system (UVP LLC, Upland, CA, USA), and photographs were taken using a Nikon Coolpix E8800 digital camera (Nikon, Tokyo, Japan). Two independent biological replicates were performed, with each replicate containing at least three plants.
Pseudomonas syringae pv. tabaci (Pst) was grown overnight at 28°C in King’s B medium, resuspended in 10 mM MgCl2 (104 cfu/ml), and infiltrated into the leaves of gene-silenced or inhibitor-treated plants using a 1 ml plastic needleless syringe. To measure the bacterial inoculum, the leaves of three independently silenced plants were harvested at 0, 1, 2, and 4 days post inoculation (dpi). Two 1.0 cm2 leaf disks were excised from each leaf, and leaf disks was ground in 1 ml of 10 mM MgCl2. Then, 100 µl of 10-fold serial dilutions of each leaf-disk extract was plated on King’s B-agar medium containing 50 µg/ml spectinomycin (Sigma-Aldrich), and the colonies formed were counted.
2.6 Trypan blue staining
The TB staining procedure was performed using modifications of a previously described method (Guo et al., 2019). Leaf samples were covered with lactophenol–Trypan blue (Sigma-Aldrich) mixture (10 ml of glycerol, 9.3 ml of unsaturated liquid phenol, 10 ml of lactic acid, and 10 mg of TB) and then subjected to vacuum infiltration for 5 min in a desiccator. The plate harboring leaf samples was placed in boiling water for 5 min and then incubated at room temperature for 12–16 h. After removing the staining solution, the leaves were covered with 2.5 g/ml chloral hydrate (TCI, Kasei Kogyo, Tokyo, Japan) for destaining. The destained leaves were equilibrated with 60% (w/v) glycerol, mounted on glass slides, and observed under a microscope (Zeiss, Munich, Germany).
2.7 Measurement of H2O2 levels
Production of H2O2 was detected, as described previously (Thordal-Christensen et al., 1997), using 3, 3’-diaminobenzidine (DAB) (Sigma-Aldrich), which forms a reddish-brown precipitate upon reaction with H2O2. Briefly, Bax-treated full leaves were detached at 0, 12, and 24 h postinfiltration; placed in 1 mg/ml DAB (pH 5.6); and subjected to vacuum infiltration for 10 min. Subsequently, the leaves were incubated at 23°C for 8 h under light. To remove chlorophyll and excess DAB stain, the leaves were boiled in alcohol: lactophenol (2:1) solution for 5 min. The destained leaves were rinsed three times with 50% ethanol and then stored in 50% ethanol. The DAB staining pattern and intensity were assessed visually (Samuel and Ellis, 2002).
2.8 Transmission electron microscopy analysis
Sections of leaves were obtained from 2-week-old plants subjected to VIGS and fixed for 2 h in 2.5% paraformaldehyde–glutaraldehyde mixture buffered with 0.1 M phosphate (pH 7.2). The fixed leaf sections were then postfixed for 1 h in 1% osmium tetroxide (OsO4) prepared in 0.1 M phosphate buffer (pH 7.2), and dehydrated in graded ethanol and propylene oxide. The dehydrated samples were embedded in Epon-812, and ultrathin sections were prepared using the UltraCut E ultramicrotome (Leica, Vienna, Austria). The prepared sections were stained with uranyl acetate and lead citrate (UA/Pb), and examined on a CM 20 electron microscope (Philips, Amsterdam, Netherlands).
2.9 RNA blot analysis
To perform RNA blot analysis, 10 µg of total RNA was separated electrophoretically on a 1% formaldehyde-containing agarose gel and then transferred onto a Hybond-N+ nylon membrane (GE Healthcare, Buckinghamshire, UK) by overnight capillary transfer with 20× SSC (3 M NaCl, 0.3 M sodium citrate (pH 7.0)). To determine TMV : GFP transcript levels, the RNA blot was hybridized with a 412 bp cDNA fragment of the movement protein (MP) gene of TMV, which was amplified using the TMV MP_F and TMV MP_R primers (Supplementary Table 1) and subsequently labeled with [32P]-dCTP using the Rediprime II DNA Labeling System (GE Healthcare) hybridization of the radiolabeled probe was performed as described previously (Church and Gilbert, 1984; Choi et al., 2002). Then, the membrane was subjected to two 5 min washes each with prewarmed 2× SSC and 0.1% SDS. The hybridization signal was visualized on a BAS-1800II PhosphorImager (Fuji Photo Film, Tokyo, Japan).
2.10 Chemical inhibitor treatments
Three inhibitors were used to perform the chemical inhibition assays: mevinolin (MEV; AG Scientific, San Diego, CA, USA), an inactive lactone, which inhibits 3-hydroxy-3-methylglutaryl coenzyme A reductase and thus blocks the cytosolic MVA pathway; fluridone (FLU; Duchefa), which inhibits PDS activity, thus blocking the carotenoid biosynthesis pathway; and fosmidomycin (FOS; 3-(N-hydroxyamino) propyl phosphate) (Invitrogen), which specifically inhibits (DXR) in the chloroplast. Ten millimolar stock solutions of MEV and FLU were prepared in ethanol and stored at 4°C, while that of FOS was prepared in water and stored at -20°C. To perform the chemical inhibitor treatments, working solutions of each inhibitor (100 µM MEV, 100 µM FLU, 1 mM FOS) were prepared from the corresponding stock solutions and infiltrated into entire abaxial surface of the fully expanded leaves of 4-week-old N. benthamiana plants using a 1 ml needleless syringe. Sixteen hours after the inhibitor/mock treatment, the inhibitor-treated leaves were infiltrated with elicitors (Bax, HRT/TCV CP, Rx/PVX CP) and nonhost pathogen (Pss61) for PCD and gene expression analyses.
2.11 Measurement of ion leakage
Four leaf disks (1 cm in diameter) were floated on 6 ml of distilled water for 12 h at room temperature, with shaking. After the 12 h incubation, the conductivity of the water was measured (value A) with a NeoMet conductivity meter (EC-470L; iSTEK, Seoul, Korea). Finally, the water containing leaf discs were autoclaved at 121°C for 5 min, cooled to room temperature, and then the conductivity of the water was measured (value B). Ion leakage (%) in each sample was calculated as follows: (value A/value B) × 100. Means and standard deviations of three independent biological replicates were calculated (Mittler et al., 1999).
2.12 Immunoblot analysis
To extract total proteins, leaf samples were ground in liquid nitrogen and resuspended in urea lysis buffer (8 M urea, 100 mM NaH2PO4, 10 mM Tris-HCl (pH 8.0)). The samples were then centrifuged at 13,000 rpm for 15 min at 4°C, and the supernatants were mixed with 5× SDS sample buffer. The extracted proteins were separated on 12% SDS-polyacrylamide gels and transferred to polyvinylidene difluoride (PVDF) membranes (Roche, Basel, Switzerland). To determine Bax abundance, the membranes were incubated first with anti-Bax antibody (Santa Cruz Biotechnology, Dallas TX, USA) and then with horseradish peroxidase-conjugated anti-rabbit IgG antibody (Santa Cruz Biotechnology). Finally, the Bax protein was detected using the Enhanced Chemiluminescence (ECL) Kit (Thermo Fisher Scientific).
2.13 Chlorophyll fluorescence measurements
Two weeks after VIGS, fully expanded leaves were detached from three N. benthamiana plants silenced for the indicated genes. The detached leaves were dark-adapted for 30 min, and then the maximum photochemical efficiency of photosystem II (PSII; Fv/Fm), the level of photoprotective quenching of fluorescence (1–qP, where qP is the level of photochemical quenching of PSII, also referred to as Fq′/Fv′), recovery of nonphotochemical quenching (NPQ), and light response curves of the quantum yield of PSII (ΦPSII) were measured as described previously (Choi et al., 2002). The experiments were performed at room temperature.
3 Results
3.1 Silencing of NbLytB suppresses Bax-mediated PCD in Nicotiana benthamiana
In previous study, silencing of the pepper LytB gene resulted in an albino phenotype, similar to the phenotype induced by silencing of the phytoene desaturase (PDS) gene (Liu et al., 2002), which is widely used as positive control in VIGS experiments; however, the silencing of these two genes had opposite effects on Bax-induced PCD (data not shown). This prompted us to further investigate why these two genotypes exhibiting similar defects in chloroplast development respond differently to Bax-induced cell death. First, the LytB gene homolog was isolated from the N. benthamiana cDNA library, and the cDNA fragment (737–1,147 nt) was cloned into the pTRV2 vector (Supplementary Figures 1 and 2). The resultant construct as well as pTRV2:GFP (negative control) and pTRV2:NbPDS (positive control) (Moon et al., 2016) were used to perform VIGS experiments. Consistent with the results of previous VIGS studies using MEP pathway genes (Page et al., 2004), the NbLytB-silenced N. benthamiana plants exhibited growth arrest and their newly emerged leaves showed an albino phenotype and wavy edges. The albino leaf phenotype of NbLytB-silenced plants was very similar to that of NbPDS-silenced plants (Supplementary Figure 3A). Furthermore, semiquantitative RT-PCR showed that the endogenous NbLytB and NbPDS mRNA levels were greatly reduced in silenced plants (Supplementary Figure 3B), thus confirming the silencing of target genes.
To determine the role of NbLytB in HR cell death, the human Bax gene was transiently overexpressed in NbLytB-silenced N. benthamiana plants through Agrobacterium-mediated transformation (Lee et al., 2013). In GFP- and NbPDS-silenced plants generated using the corresponding VIGS vectors, yellowing or necrosis was observed in all leaves at the site of Bax infiltration at 3 days post inoculation (dpi), whereas in the mock treatment site expressing the empty vector (pBtex), symptoms were mild or completely absent (Figure 1A). On the other hand, little or weak cell death was observed in the leaves of NbLytB-silenced plants (Figure 1A). In addition, cell death in silenced leaves was detected by staining each leaf with Trypan blue (TB) at 36 h after Bax infiltration (Figure 1B). Trypan blue enters cells with damaged membranes and binds to intracellular proteins, turning the cells blue (Koch and Slusarenko, 1990). Therefore, live (unstained) cells can be distinguished from dead (blue) cells under a microscope. Compared with the GFP-silenced (control) leaves, the number of dead (blue) cells was dramatically lower in NbLytB-silenced leaves and higher in NbPDS-silenced leaves (Figure 1B); this is consistent with the results shown in Figure 1A. Since the leaves of both NbPDS- and NbLytB-silenced plants showed the albino phenotype, we performed DAB staining to compare the ROS levels between the two genotypes (Figure 1C). ROS functions as a secondary messenger that mediates PCD in plants, and its accumulation is a hallmark of HR (Lamb and Dixon, 1997; Coll et al., 2011). The production of H2O2 in plant tissues can be visualized by staining with DAB, which is oxidized by H2O2, resulting in the formation of a brown precipitate (Thordal-Christensen, 1997). In GFP-silenced plants, the intensity of brown staining in the leaf area infiltrated with Bax was the strongest at 24 hours post inoculation (hpi) compared with the other time points (Figure 1C). On the other hand, the leaves of NbLytB-silenced plants showed the strongest brown staining immediately after Bax infiltration, and the level of H2O2 in these leaves was slightly lower than that in GFP-silenced leaves at 24 hpi. The NbPDS-silenced plants showed increased production of H2O2 at 0 and 24 hpi, and the level of H2O2 accumulation in NbPDS-silenced plants was considerably higher than that observed in NbLytB- and GFP-silenced plants. The strong brown color observed in NbPDS- and NbLytB-silenced plants immediately after agroinfiltration (0 hpi) was presumably caused by the production of H2O2 in response to wounding stress induced by agroinfiltration (Suzuki and Mittler, 2012) rather than by Bax expression. In addition, we suspect that the stronger DAB staining of both VIGSed plants relative to that of GFP-silenced control plants was because they contained malfunctioning chloroplasts that weakened their ROS scavenging function. Overall, the increase or decrease in cell death observed in NbPDS- and NbLytB-silenced plants, respectively, appears to be related to the generation of excess H2O2.
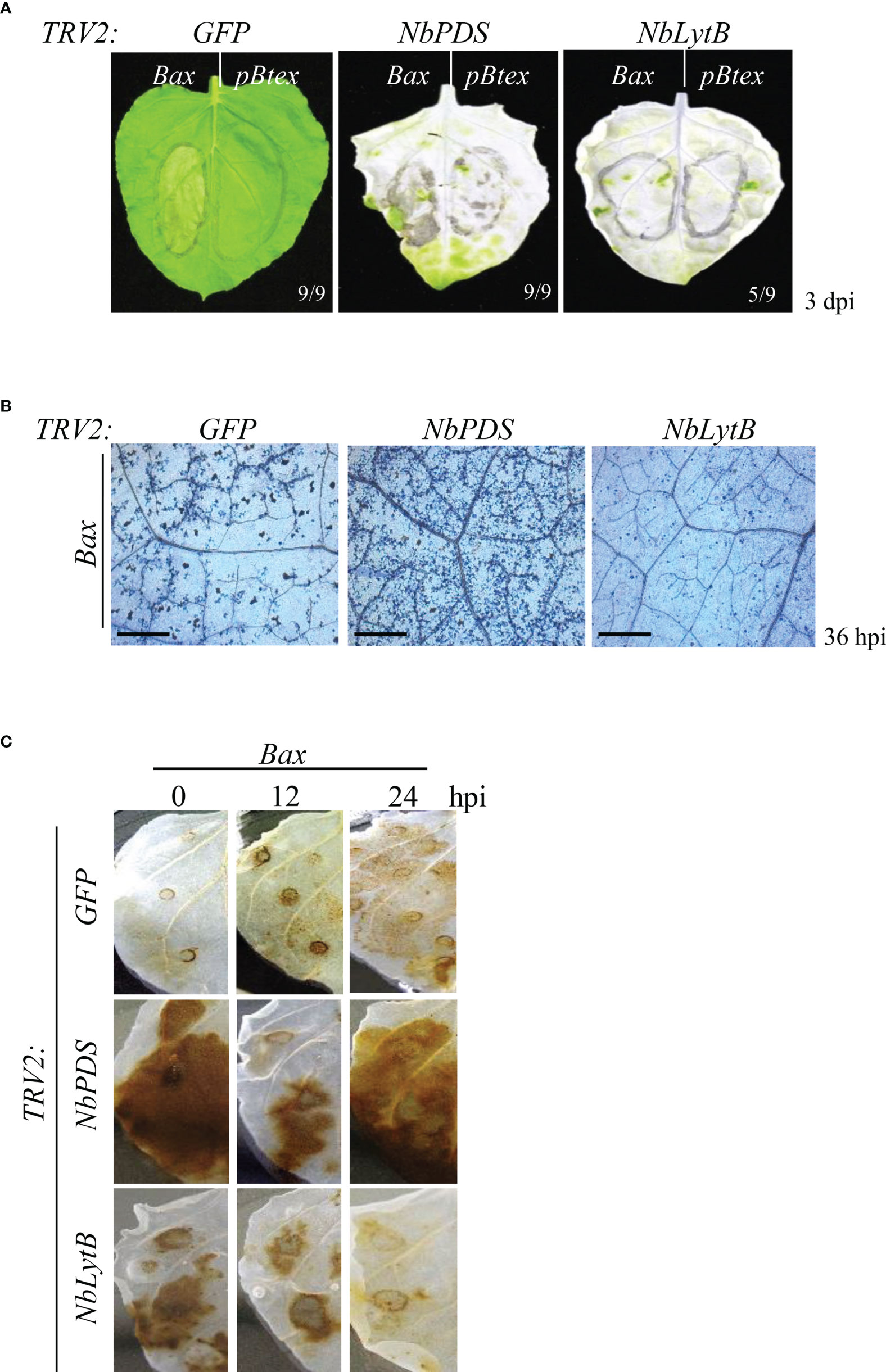
Figure 1 NbLytB is required for Bax-mediated PCD in N. benthamiana (A) Bax-mediated PCD in TRV2:GFP (control) plants, and NbPDS- and NbLytB-silenced plants. Areas infiltrated with Agrobacterium containing the empty pBtex vector or the Bax expression vector are outlined with a circle on each leaf. Pictures were taken 3 dpi. (B) Visualization of cell death in the leaves of GFP-, NbPDS-, and NbLytB-silenced plants by TB staining at 36 h after Bax infiltration. Scale bar = 20 µm (C) Impact of NbLytB on H2O2 accumulation by Bax-mediated PCD. H2O2 accumulation in GFP-, NbPDS-, and NbLytB-silenced leaves was visualized by DAB staining at the indicated time points after infiltration with Agrobacterium expressing Bax.
To determine whether NbLytB is also involved in plant pathogen-related PCD, cell death responses induced by nonhost pathogen infection or interaction between the R gene and its cognate effector were monitored in the silenced plants (Supplementary Figure 3C). In this experiment, the bean bacterial pathogen Pss61, which carries the hrp gene cluster and induces HR in N. benthamiana leaves (Alfano et al., 1997), was used as the nonhost pathogen, while its hrp-deficient mutant (Pss61hrp-) was used as a non-HR induction control. Additionally, the R genes HRT and Rx (Resistance to Potato virus X (PVX)) were used in combination with their cognate effectors TCV coat protein (T-CP) and PVX CP (P-CP), respectively (Cooley et al., 2000; Bendahmane et al., 2002), while a CP mutant that did not induce R gene-mediated HR was used as a non-HR induction control (Moon et al., 2016). GFP-silenced plants (control) showed apparent cell death at 36 hpi in all PCD induction treatments (Supplementary Figure 3C). NbLytB-silenced plants suppressed Rx- and HRT-mediated HR, similar to the inhibitory response of Bax-induced cell death. On the other hand, in response to Pss61, the NbLytB- and NbPDS-silenced plants displayed a similar cell death phenotype to that of GFP-silenced control plants (Supplementary Figure 3C). These results indicate that the inhibition of PCD caused by NbLytB silencing is not common to all PCDs, and suggest that the role of chloroplasts in PCD may differ depending on the PCD elicitor.
3.1.1 NbLytB is involved in both basal defense and R gene-mediated defense responses
Next, we asked whether HR inhibition caused by the silencing of NbLytB was associated with the disease resistance response of plants. First, to observe the R gene-mediated disease resistance response, N. benthamiana plants carrying the TMV resistance gene N were subjected to VIGS to silence NbPDS, NbLytB, and NbSGT1, and then inoculated with GFP-tagged TMV (TMV : GFP) (Erickson et al., 1999; Liu et al., 2002). Silencing of NbSGT1 was performed as a positive control to loss of TMV resistance by the N gene (Peart et al., 2002). At 7 days after TMV : GFP inoculation, fluorescent green spots were clearly observed in the inoculated leaves of NbPDS-, NbLytB-, and NbSGT1-silenced plants, and the size and number of these spots increased in the order of NbSGT1-, NbLytB-, and NbPDS-silenced plants. However, no GFP foci were observed in the inoculated leaves of GFP-silenced control plants (Figure 2A). TMV : GFP fluorescence was not observed in the upper leaves of all gene-silenced plants, except NbSGT1-silenced plants, at 14 dpi (data not shown). The N gene-mediated HR was observed in the inoculated leaves at 3 dpi via TB staining. Compared with the GFP-silenced control leaves, the size of TB-stained cells was increased in NbPDS- and NbSGT1-silenced leaves but slightly reduced in NbLytB-silenced leaves (Figure 2A). In addition, RNA gel blot analysis revealed virus accumulation in NbLytB- and NbSGT1-silenced plants (Figure 2B). These results indicate that NbLytB is potentially involved in HR and resistance responses mediated by the N gene.
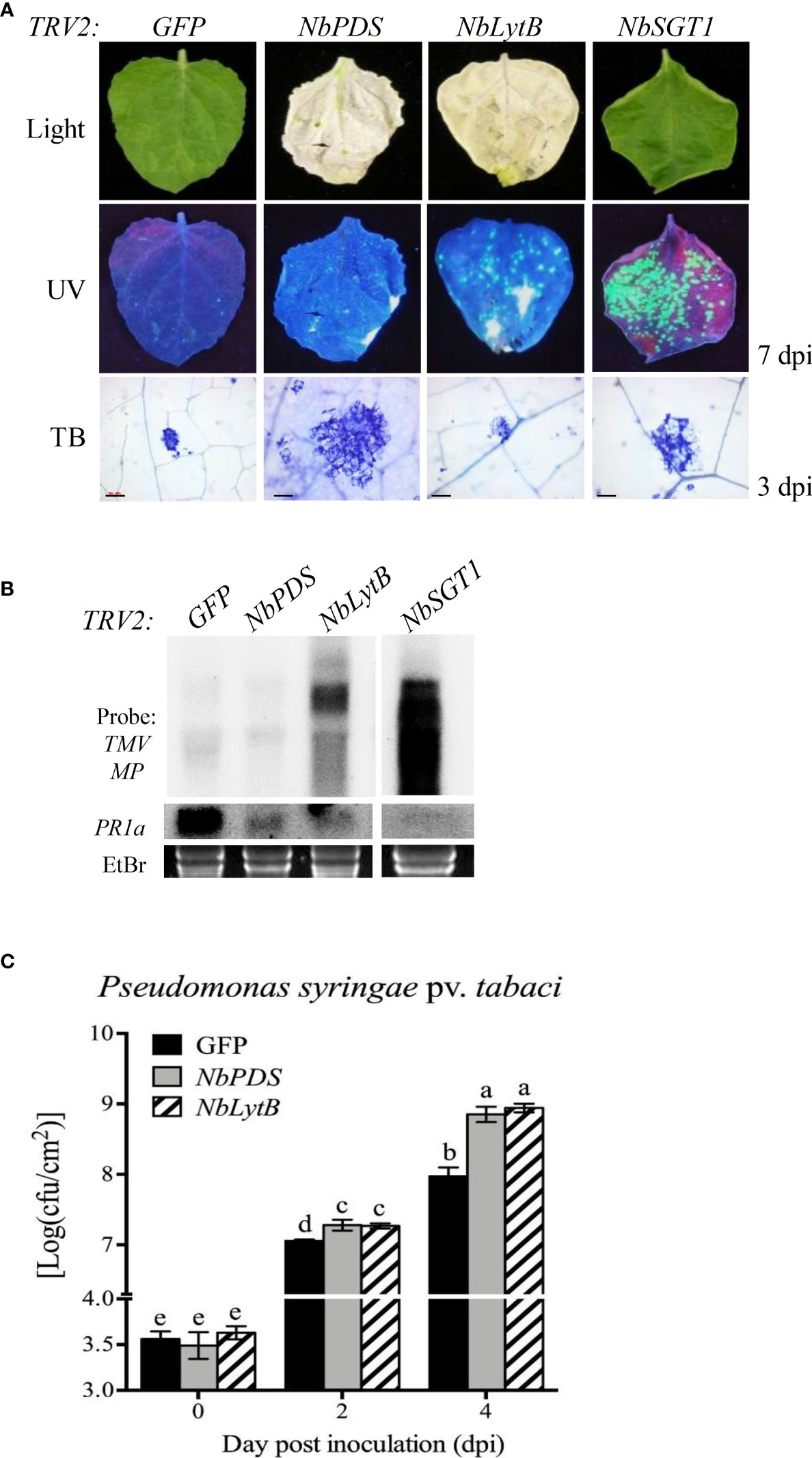
Figure 2 NbLytB silencing suppresses N gene-mediated resistance to TMV and increases the susceptibility of N. benthamiana plants to the bacterial pathogen Pst (A) Effect of NbLytB on the TMV N gene-mediated HR-PCD and on the resistance to TMV : GFP infection. Images show HR-PCD (top panel) and TMV : GFP fluorescence (middle panel) at 7 dpi. The leaves of GFP-, NbPDS-, NbLytB-, and NbSGT1-silenced transgenic N. benthamiana plants expressing the N gene were inoculated with TMV : GFP at 2 weeks after VIGS. HR-PCD was observed by TB staining at 3 dpi (bottom panel). Scale bars = 100 µm. NbSGT1-silenced plants were used as a positive control for TMV : GFP infection. WL, white light; UV, ultraviolet light. (B) RNA gel blot showing the accumulation of the MP of TMV : GFP and the expression of PR1 in inoculated leaves collected at 6 dpi. The rRNA bands stained with ethidium bromide served as a loading control. Similar results were obtained in two independent experiments. (C) Growth of Pst in GFP-, NbPDS-, and NbLytB-silenced N. benthamiana plants. The leaves of gene-silenced plants were inoculated with Pst, and bacterial growth was evaluated at 0, 2, and 4 dpi. Data represent mean ± standard deviation (SD) of at least three independent biological replicates, with each replicate containing six leaf disks. Different letters indicate significant differences (P < 0.05; Student’s t-test). Three independent experiments were performed with similar results.
To further evaluate the role of NbLytB in plant basal defense responses, the silenced leaves were inoculated with Pseudomonas syringae pv. tabaci (Pst), a disease-causing bacterial pathogen of N. benthamiana (Ramegowda et al., 2013). The NbPDS- and NbLytB-silenced plants showed 7–8-fold higher bacterial population than the GFP-silenced control plants (Figure 2C). However, no difference was observed in the degree of disease susceptibility between NbPDS- and NbLytB-silenced plants, suggesting that reduction in the basal resistance of these plants might be related to the lack of chloroplast function.
3.1.2 Structural defects in chloroplasts and reduced photosynthetic efficiency in NbLytb-silenced plants
To study whether the differences in HR between NbPDS- and NbLytB-silenced plants were caused by the differences in their chloroplast morphology, we examined the morphology of intracellular organelles in NbPDS-, NbLytB-, and GFP-silenced leaves by TEM (Figure 3A). The chloroplasts of GFP-silenced control plants showed a well-developed thylakoid membrane with large starch granules. However, in the albino sectors of NbPDS- and NbLytB-silenced leaves, chloroplasts were small in size, irregular in shape, and contained poorly developed thylakoid membranes, consistent with a previous report (Page et al., 2004). The chloroplasts of NbLytB-silenced leaves contained more vesicles than stroma and thylakoids (Figure 3A). In addition, the maximum efficiency of PSII photochemistry, i.e., the ratio of variable to maximal fluorescence (Fv/Fm), was drastically decreased in NbPDS- and NbLytB-silenced plants compared with the GFP-silenced control plants (Figure 3B). Taken together, these results indicate that HR inhibition by NbLytB silencing is not appreciably associated with chloroplast differentiation or a reduction in photosynthetic function.
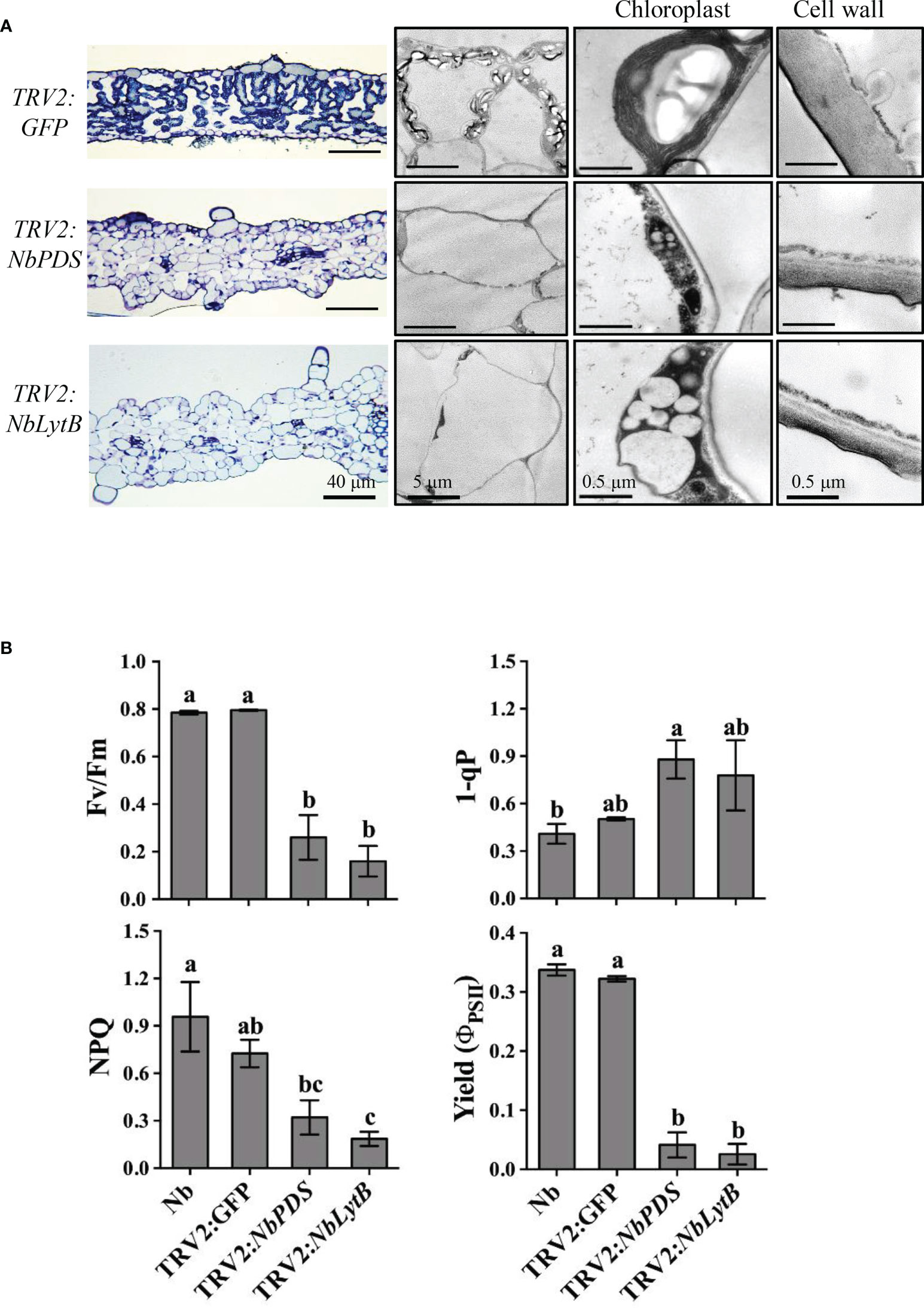
Figure 3 Downregulation of NbLytB disrupts chloroplast morphology and reduces photosynthetic efficiency. (A) Transmission electron microscope (TEM) images showing the ultrastructure of chloroplasts in the leaves of TRV2:GFP plants (control), and NbPDS- and NbLytB-silenced plants. Enlarged images show details of the chloroplasts and the cell wall. (B) Silencing of NbLytB reduces photosynthetic efficiency. The photosystem II (PSII) photochemistry of the detached leaves of unsilenced N. benthamiana (Nb) and GFP-, NbPDS-, and NbLytB-silenced N. benthamiana plants was examined by measuring the photochemical efficiency of PSII (Fv/Fm), reduced state of PSII (1-qP), nonphotochemical quenching of absorbed light (NPQ), and effective quantum efficiency of PSII (Yield). Data represent mean ± standard deviation (SD) of three replicates. Three independent experiments were performed with similar results.
3.1.3 Suppression of HR by fosmidomycin
LytB catalyzes the last step of the MEP pathway in chloroplasts. To investigate the role of the MEP pathway in HR and rule out the pleiotropic effects of VIGS, we used chemical inhibitors that specifically inhibit the isoprenoid biosynthesis pathway in plants. Three chemical inhibitors were selected (FOS, FLU, MEV) (Figure 4A). FOS specifically inhibits DXR, an enzyme required to convert DXP to MEP in the chloroplast (Zeidler et al., 1998), which induces an overall inhibitory effect on chloroplast isoprenoid synthesis, as observed in NbLytB-silenced plants. MEV inhibits 3-hydroxy-3-methylglutaryl coenzyme A reductase, which blocks the cytosolic MVA isoprenoid biosynthesis pathway (Hemmerlin et al., 2003). Finally, FLU inhibits PDS activity and blocks the carotenoid biosynthetic pathway, inducing a PDS-silencing-like phenotype (Chae et al., 2004).
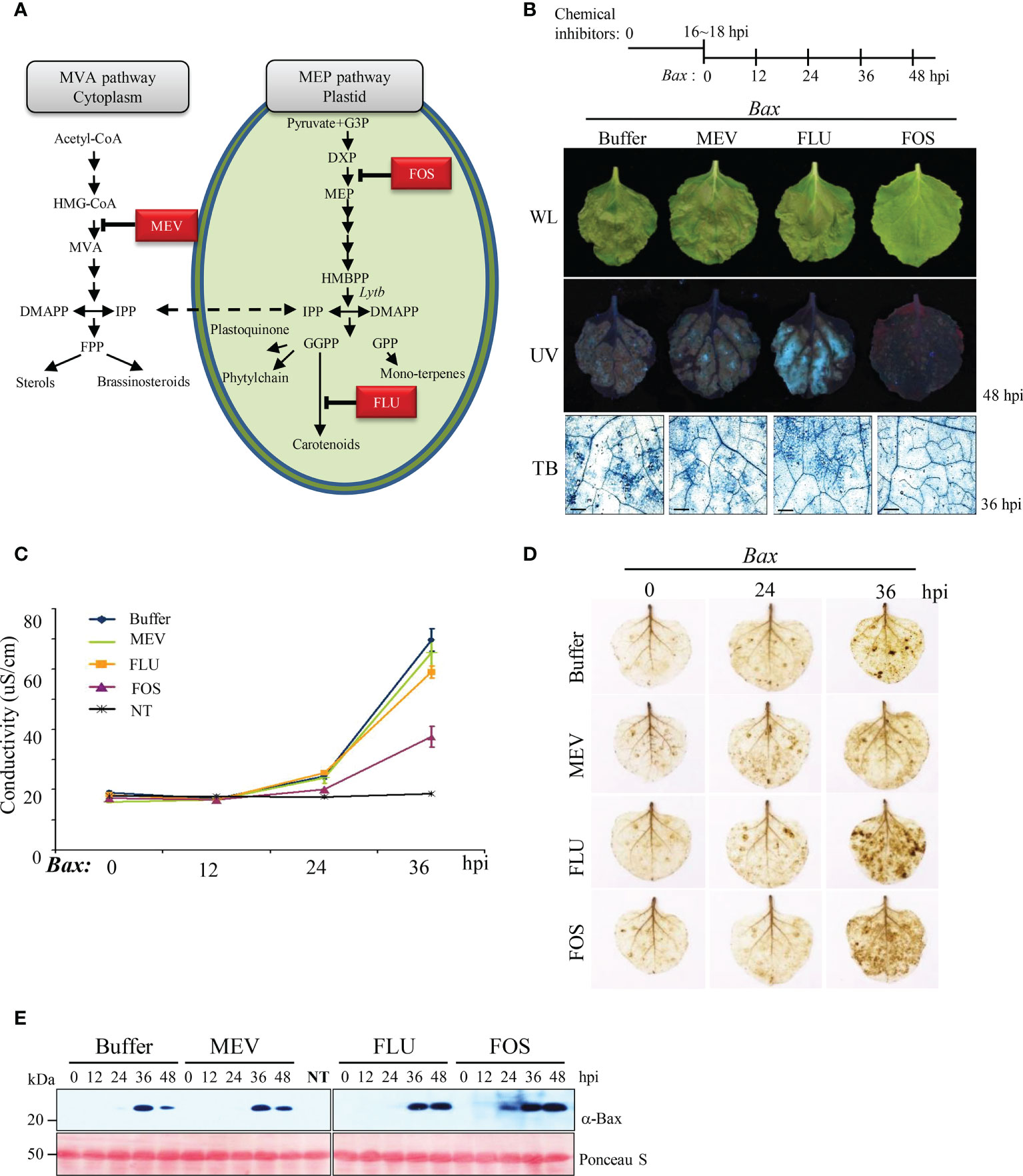
Figure 4 MEP pathway is involved in Bax-mediated PCD (A) Isoprenoid biosynthesis in plant cell. The first intermediate specific to each pathway is boxed. HMG-CoA, 3-Hydroxy -3-methylglutaryl CoA; MVA, mevalonic acid; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; FPP, farnesyl diphosphate; GA-3-P, glyceraldehyde 3-phosphate; DOXP, 1-deoxy-D-xylulose-5-phosphate; MEP, 2-C-methyl-D- erythritol 4-phosphate; HMBPP, 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate; GGPP, geranylgeranyl diphosphate; GPP, geranyl diphosphate. The steps inhibited by MEV, FOS, and FLU are indicated. (B) Effect of selected inhibitors on Bax-mediated PCD. A schematic diagram of the experimental procedure used to study Bax-mediated PCD in plants pretreated with inhibitors of the MEP pathway is shown on the top. Pictures were taken under white light (WL; top panel) and ultraviolet light (UV; middle panel) at 48 hpi. Bax-mediated PCD was observed by TB staining at 36 hpi (bottom panel). Scale bar = 20 µm. (C) Ion leakage assay of leaves treated with chemical inhibitors and then infiltrated with Bax. Data represent means ± standard error (SE) of six leaf disks per genotype. NT indicates buffer-treated N. benthamiana without Bax infiltration. (D) DAB staining of inhibitor-treated N. benthamiana leaves at the indicated time points after infiltration with Agrobacterium expressing Bax. (E) Western blotting analysis of Bax expression in buffer- (control) and inhibitor-treated N. benthamiana leaves collected at 0, 12, 24, 36, and 48 hpi using anti-Bax antibody. Positive staining of inhibitor-treated leaves with anti-Bax antibody confirmed agroinfiltration with Bax. Rubisco (loading control) was detected by Ponceau S.
The treatment concentrations of FOS (1 mM) and FLU (100 µM) were adjusted to avoid plant cell damage and ensure that the yellowing of upper leaves caused by these treatments was similar (Supplementary Figure 4A). First, we monitored Bax-mediated HR cell death induced by each chemical inhibitor in N. benthamiana leaves. HR was clearly inhibited in FOS-treated leaves compared with the mock-treated (control) and other inhibitor-treated leaves (Figure 4B; Supplementary Figure 4B). These results were also confirmed through TB staining and quantitative analysis of ion leakage (Figure 4B, C). On the other hand, H2O2 production was not reduced in FOS-treated leaves, unlike VIGS, and the level of H2O2 in FOS-treated leaves was similar to that in FLU-treated leaves (Figure 4D). The accumulation of Bax protein in the chemical inhibitor-treated leaves was assessed by western blot analysis, which confirmed that Bax protein levels were not altered by inhibitor treatments (Figure 4E).
Next, we examined whether the MEP pathway also modulates R gene-mediated HR-PCD and/or nonhost pathogen-induced PCD. Coexpression of HRT/T-CPWT and Rx/P-CPWT induced HR-PCD in the buffer-, MEV-, and FLU-treated plants but not in FOS-treated plants (Figure 5A). PCD induced by Pss61 was similar to the buffer-treated control in all inhibitor-treated leaves (Figure 5A). The R gene-induced PCD was further supported by the microscopic observation of the cell death response in TB-stained leaves (Figure 5B).
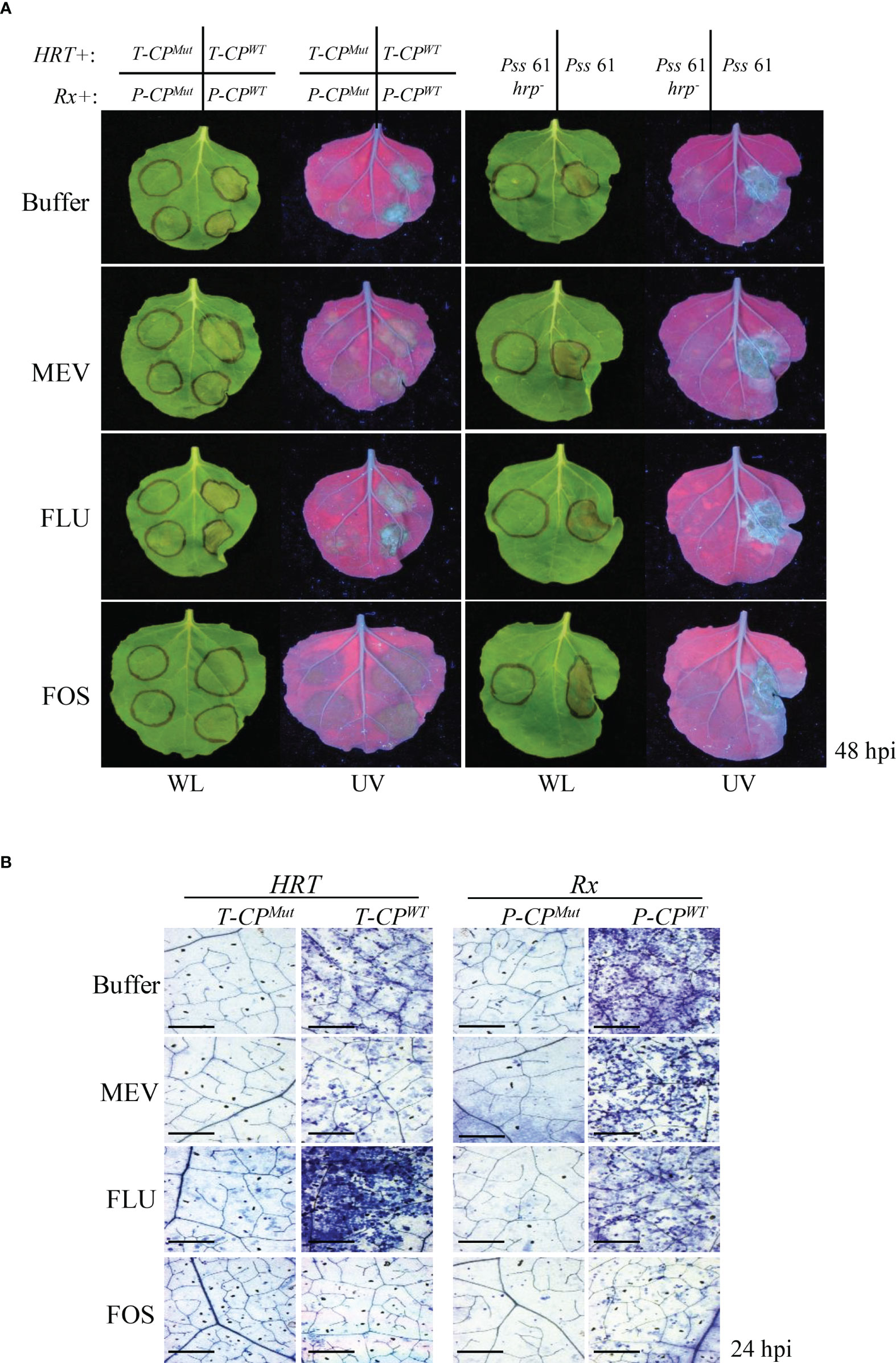
Figure 5 Inhibition of the MEP pathway suppresses R/Avr coexpression-mediated HR- related PCD (A and B) R gene-mediated and nonhost-induced plant PCD assays. (A) Left panel: Agrobacterium strains carrying the R gene were co-infiltrated with those carrying the corresponding Avr gene into N. benthamiana leaves 16 h after the buffer (control) or inhibitor treatment. T-CPWT,; T-CPMut, mutant CP of TCV (negative control); P-CPWT; P-CPMut, mutant CP of PVX (negative control). Right panel: bean (nonhost) bacterial pathogen Pss61 and its hrp-deficient mutant (Pss61hrp-; negative control) were inoculated into N. benthamiana leaves 16 h after the buffer (control) or inhibitor treatment. The PCD responses at the infiltration sites were evaluated at 48 hpi, and pictures were taken. WL, white light; UV, ultraviolet light. (B) Evaluation of cell death in the leaves of inhibitor-treated plants at 24 h after HRT/T-CPWT or Mut or Rx/P-CPWT or Mut infiltration by TB staining. The leaves of buffer-, MEV-, and FLU-treated plants showed numerous dead cells, whereas those of FOS-treated plants showed few dead cells. Scale bar = 20 µm.
To examine how the chemical inhibitor treatment affects the basal plant defense response, the inhibitor-treated plants were inoculated with Pst and monitored for bacterial growth (Supplementary Figure 4C). Plants treated with FOS and FLU showed increased susceptibility to Pst compared to the mock- or MEV-treated plants, suggesting that chloroplast is essential for an effective basal resistance response to pathogens.
4 Discussion
In this dissection of the novel mechanisms of HR-related PCD in plants, the plant LytB gene was identified as being involved in Bax- and R–Avr interaction-induced HR cell death. Silencing of NbLytB in N. benthamiana plants efficiently suppressed Bax-mediated PCD and HR as well as defense responses against pathogens such as Pst and TMV : GFP. Consistent with the results of silencing NbLytB, the FOS-mediated inhibition of IPP biosynthesis in plastids interfered with Bax- and R–Avr interaction-induced PCD and defense responses following pathogen infection. These results reveal that the IPP metabolite in plastids plays critical roles in HR PCD associated with plant immunity.
VIGS utilizes the short interfering RNA (siRNA)-induced antiviral defense mechanism of plants and is considered a very suitable technique for studying the functional genomics of polyploid plants, such as N. benthamiana, whose entire genome sequence information is not well known. Carotenoids are essential for protecting the photosynthetic system against photooxidative damage (Bartley and Scolnik, 1995; Niyogi, 1999). Null mutation of the LytB gene homolog IspH in Arabidopsis resulted in an albino phenotype and severely impaired chloroplast development (Ahn and Pai, 2008; Jeon et al., 2010). Since ROS production is highly correlated with HR, the inhibition of HR PCD in NbLytB-silenced plants was suggested to be due to a decrease in chloroplast ROS production. However, the abnormalities in chloroplast morphology and the decrease in photosynthetic efficiency observed in NbLytB-silenced plants were very similar to those observed in NbPDS-silenced plants in which suppression of HR PCD was not observed (Supplementary Figure 3). In addition, MEP inhibitor treatment suppressed HR PCD but did not reduce H2O2 production relative to that in the buffer-treated control plants (Figures 4 and 5). Taken together, these results suggest that HR PCD inhibition in NbLytB-silenced plants is more strongly associated with lower MEP pathway metabolite levels than with the amount of chloroplast-generated ROS.
IPP, used by prenyltransferases as a building block for the sequential elongation of hydrocarbon skeletons, is synthesized either via the cytosolic MVA pathway or the plastidic MEP pathway (Peñuelas and Munné-Bosch, 2005; Pulido et al., 2012). The importance of the cytoplasmic MVA pathway for the immune system of animals has long been emphasized because isoprenoids produced from the MVA pathway are converted into various secondary lipids and used for the post-translational modifications of proteins (Buhaescu and Izzedine, 2007; Akula et al., 2016). However, in plant immune responses, it is not yet known whether each of the two independent IPP production pathways have distinct immune response mechanisms. According to the results of the current study, Bax overexpression- and R–Avr interaction-induced HR-related PCD was dependent on plastid MEP pathway metabolites, and did not appear to be complemented by cytosolic MVA pathway metabolites. Unlike mammals, most known plant pattern recognition receptors (PRRs) are localized to the plasma membrane (PM). Because protein post-translational modifications are important for PM localization, it will be interesting to observe the role of MEP-derived IPP in the subcellular localization of these key PRRs (Macho and Zipfel, 2014). Among plant BAGs, BAG6 is localized to vacuoles and induces cell death, unlike other BAGs (Kang et al., 2006). Future studies directed at determining whether the reduction of plastid-derived IPP affects intracellular organelle targeting of specific defense-related proteins and PCD regulatory proteins would help elucidate the role of plastid-derived IPPs in plant immunity.
Overexpression of BI-1 in Arabidopsis thaliana suppresses Bax-induced cell death without inhibiting ROS production (Kawai-Yamada et al., 2004). Further studies showed that BI-1 partially inhibits Bax-induced cell death by regulating Ca2+ homeostasis of the ER at a stage following ROS generation (Ihara-Ohori et al., 2007). These results are very similar to those obtained using FOS-treated plants, in which Bax-induced HR is suppressed even in the presence of ROS generation (Figure 4). The ER stress response is closely related to R gene-mediated HR (Williams et al., 2014). Silencing of ER chaperones involved in ER-quality control, such as calreticulins in N. benthamiana, compromises receptor-like protein Cf-mediated immune responses (Liebrand et al., 2012), and binding immunoglobulin protein (BiP)-silenced tobacco plants show reduced HRT-mediated HR (Moon et al., 2016). Together, these results raise the possibility that MEP-derived IPPs are linked with the ER stress responses to modulate HR-PCD. Future studies will be needed to determine whether the reduction of MEP-derived IPP induces a stress response in the ER.
Polyisoprenoids are secondary isoprenoid metabolites, and although their biological functions have not yet been clearly elucidated, it is assumed that they serve as lipid carriers in the process of protein glycosylation in plants (Surmacz and Swiezewska, 2011). Arabidopsis mutants with reduced biosynthesis of the polyisoprenoid dolicol show reduced protein glycosylation, increased sensitivity to darkness, and an early senescence phenotype (Zhang et al., 2008). The 3-hydroxy-3-methylglutaryl CoA reductase gene, an important regulator of the MVA pathway, is strongly expressed in tobacco leaves during the TMV resistance response (Kang et al., 1998), and the expression of two key enzymes in the MEP pathway, 1-deoxyxylulose 5-phosphate synthase and DXR, is increased in rice following treatment with a chitin elicitor (Okada et al., 2007). These results suggest that polyisoprenoids involved in plant defense are likely ‘mosaic compounds’ originating from both the MEP and/or MVA pathways (Skorupinska-Tudek et al., 2008). Further studies are needed to determine whether the reduction of MEP pathway-generating IPP caused by NbLytb silencing affects the production of specific polyisoprenoids and whether this results in an ER stress response associated with the accumulation of immature proteins in the ER.
In conclusion, these results demonstrate that the plastid MEP pathway is involved in plant defense responses, supporting the importance of the chloroplast-produced IPP in HR-PCD in plants. Further experiments are required to understand how this pathway is integrated with upstream and downstream pathways, and to obtain further insights into the complex regulatory mechanisms underlying PCD and disease resistance in plants.
Data availability statement
All of the data supporting the results of this study can be found in either the article/Supplementary Materials together with the accession numbers of the nucleotide sequences deposited in GenBank (NCBI).
Author contributions
SL and JP conceived and designed the study; SL performed VIGS and chemical inhibitor treatment experiments; SJ, CH, and JL performed cloning, RT-PCR, and TB staining. SL, SJ, CH, JL, and JP wrote the manuscript; SL, SJ, BC, and JP revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the KRIBB Initiative Program (KGM9942213 and KGM5372221) and the Basic Research Program of National Research Foundation of Korea grant (NRF-2017R1A2B4012820 and NRF-2020R1I1A2075335 to JP) of the Korean government (MSIP: Ministry of Science, ICT, and Future Planning).
Acknowledgments
We thank Dr. Dinesh Kumar for providing the pTRV1 and pTRV2 constructs, Sir David Baulcombe for providing Rx, PVX CP, and PVX CP mutant clones, and Dr. Youn-il Park, who helped measure the photosynthetic efficiency of VIGSed plants.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1032682/full#supplementary-material
References
Ahn, C. S., Pai, H. S. (2008). Physiological function of IspE, a plastid MEP pathway gene for isoprenoid biosynthesis, in organelle biogenesis and cell morphogenesis in nicotiana benthamiana. Plant Mol. Biol. 66, 503–517. doi: 10.1007/s11103-007-9286-0
Akula, M. K., Shi, M., Jiang, Z., Foster, C. E., Miao, D., Li, A. S., et al. (2016). Control of the innate immune response by the mevalonate pathway. Nat. Immunol. 17, 922–929. doi: 10.1038/ni.3487
Alfano, J. R., Klm, H. S., Delaney, T. P., Collmer, A. (1997). Evidence that the pseudomonas syringae pv. syringae hrp-linked hrmA gene encodes an avr-like protein that acts in an hrp-dependent manner within tobacco cells. Mol. Plant Microbe Interact. 10, 580–588. doi: 10.1094/MPMI.1997.10.5.580
Ameisen, J. C. (2002). On the origin, evolution, and nature of programmed cell death: a timeline of four billion years. Cell Death Differentiation 9, 367–393. doi: 10.1038/sj.cdd.4400950
Apostol, I., Heinstein, P. F., Low, P. S. (1989). Rapid stimulation of an oxidative burst during elicitation of cultured plant cells : Role in defense and signal transduction. Plant Physiol. 90, 109–116. doi: 10.1104/pp.90.1.109
Ausubel, F. M. (2005). Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol. 6, 973–979. doi: 10.1038/ni1253
Banerjee, A., Sharkey, T. D. (2014). Methylerythritol 4-phosphate (MEP) pathway metabolic regulation. Nat. Prod Rep. 31, 1043–1055. doi: 10.1039/C3NP70124G
Bartley, G. E., Scolnik, P. A. (1995). Plant carotenoids: pigments for photoprotection, visual attraction, and human health. Plant Cell 7, 1027–1038. doi: 10.1105/tpc.7.7.1027
Bendahmane, A., Farnham, G., Moffett, P., Baulcombe, D. C. (2002). Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the rx locus of potato. Plant J. 32, 195–204. doi: 10.1046/j.1365-313X.2002.01413.x
Buhaescu, I., Izzedine, H. (2007). Mevalonate pathway: a review of clinical and therapeutical implications. Clin. Biochem. 40, 575–584. doi: 10.1016/j.clinbiochem.2007.03.016
Burton, R. A., Gibeaut, D. M., Bacic, A., Findlay, K., Roberts, K., Hamilton, A., et al. (2000). Virus-induced silencing of a plant cellulose synthase gene. Plant Cell 12, 691–706. doi: 10.1105/tpc.12.5.691
Byczkowski, J. Z., Gessner, T. (1988). Biological role of superoxide ion-radical. Int. J. Biochem. 20, 569–580. doi: 10.1016/0020-711X(88)90095-X
Chae, S. H., Yoneyama, K., Takeuchi, Y., Joel, D. M. (2004). Fluridone and norflurazon, carotenoid-biosynthesis inhibitors, promote seed conditioning and germination of the holoparasite orobanche minor. Physiol. Plant 120, 328–337. doi: 10.1111/j.0031-9317.2004.0243.x
Chandra-Shekara, A. C., Gupte, M., Navarre, D., Raina, S., Raina, R., Klessig, D., et al. (2006). Light-dependent hypersensitive response and resistance signaling against turnip crinkle virus in arabidopsis. Plant J. 45, 320–334. doi: 10.1111/j.1365-313X.2005.02618.x
Choi, S. M., Jeong, S. W., Jeong, W. J., Kwon, S. Y., Chow, W. S., Park, Y. I. (2002). Chloroplast Cu/Zn-superoxide dismutase is a highly sensitive site in cucumber leaves chilled in the light. Planta 216, 315–324. doi: 10.1007/s00425-002-0852-z
Church, G. M., Gilbert, W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. U.S.A. 81, 1991–1995. doi: 10.1073/pnas.81.7.1991
Coll, N. S., Epple, P., Dangl, J. L. (2011). Programmed cell death in the plant immune system. Cell Death Differentiation 18, 1247–1256. doi: 10.1038/cdd.2011.37
Collum, T. D., Culver, J. N. (2017). Tobacco mosaic virus infection disproportionately impacts phloem associated translatomes in arabidopsis thaliana and nicotiana benthamiana. Virology 510, 76–89. doi: 10.1016/j.virol.2017.07.002
Cooley, M. B., Pathirana, S., Wu, H. J., Kachroo, P., Klessig, D. F. (2000). Members of the arabidopsis HRT/RPP8 family of resistance genes confer resistance to both viral and oomycete pathogens. Plant Cell 12, 663–676. doi: 10.1105/tpc.12.5.663
Dangl, J. L., Dietrich, R. A., Richberg, M. H. (1996). Death don't have no mercy: Cell death programs in plant-microbe interactions. Plant Cell 8, 1793–1807. doi: 10.2307/3870230
Doke, N., Miura, Y., Sanchez, L. M., Park, H. J., Noritake, T., Yoshioka, H., et al. (1996). The oxidative burst protects plants against pathogen attack: mechanism and role as an emergency signal for plant bio-defence–a review. Gene 179, 45–51. doi: 10.1016/S0378-1119(96)00423-4
Doukhanina, E. V., Chen, S., van der Zalm, E., Godzik, A., Reed, J., Dickman, M. B. (2006). Identification and functional characterization of the BAG protein family in arabidopsis thaliana*. J. Biol. Chem. 281, 18793–18801. doi: 10.1074/jbc.M511794200
Erickson, F. L., Holzberg, S., Calderon-Urrea, A., Handley, V., Axtell, M., Corr, C., et al. (1999). The helicase domain of the TMV replicase proteins induces the n-mediated defence response in tobacco. Plant J. 18, 67–75. doi: 10.1046/j.1365-313X.1999.00426.x
Foyer, C. H., Noctor, G. (2009). Redox regulation in photosynthetic organisms: signaling, acclimation, and practical implications. Antioxid Redox Signal 11, 861–905. doi: 10.1089/ars.2008.2177
Genoud, T., Buchala, A. J., Chua, N. H., Metraux, J. P. (2002). Phytochrome signalling modulates the SA-perceptive pathway in arabidopsis. Plant J. 31, 87–95. doi: 10.1046/j.1365-313X.2002.01338.x
Greenberg, J. T., Yao, N. (2004). The role and regulation of programmed cell death in plant-pathogen interactions. Cell Microbiol. 6, 201–211. doi: 10.1111/j.1462-5822.2004.00361.x
Green, D. R., Reed, J. C. (1998). Mitochondria and apoptosis. Science 281, 1309–1312. doi: 10.1126/science.281.5381.1309
Gross, A., Mcdonnell, J. M., Korsmeyer, S. J. (1999). BCL-2 family members and the mitochondria in apoptosis. Genes Dev. 13, 1899–1911. doi: 10.1101/gad.13.15.1899
Guo, W. L., Chen, B. H., Guo, Y. Y., Yang, H. L., Mu, J. Y., Wang, Y. L., et al. (2019). Improved powdery mildew resistance of transgenic nicotiana benthamiana overexpressing the cucurbita moschata CmSGT1 gene. Front. Plant Sci. 10, 955. doi: 10.3389/fpls.2019.00955
Hemmerlin, A., Hoeffler, J. F., Meyer, O., Tritsch, D., Kagan, I. A., Grosdemange-Billiard, C., et al. (2003). Cross-talk between the cytosolic mevalonate and the plastidial methylerythritol phosphate pathways in tobacco bright yellow-2 cells. J Biol Chem 278 (29), 26666–76. doi: 10.1074/jbc.M302526200
Hockenbery, D. M., Oltvai, Z. N., Yin, X. M., Milliman, C. L., Korsmeyer, S. J. (1993). Bcl-2 functions in an antioxidant pathway to prevent apoptosis. Cell 75, 241–251. doi: 10.1016/0092-8674(93)80066-N
Huysmans, M., Lema A, S., Coll, N. S., Nowack, M. K. (2017). Dying two deaths — programmed cell death regulation in development and disease. Curr. Opin. Plant Biol. 35, 37–44. doi: 10.1016/j.pbi.2016.11.005
Jabs, T. (1999). Reactive oxygen intermediates as mediators of programmed cell death in plants and animals. Biochem. Pharmacol. 57, 231–245. doi: 10.1016/S0006-2952(98)00227-5
Ihara-Ohori, Y., Nagano, M., Muto, S., Uchimiya, H., Kawai-Yamada, M. (2007). Cell death suppressor Arabidopsis bax inhibitor-1 is associated with calmodulin binding and ion homeostasis. Plant Physiol. 143(2), 650–60. doi: 10.1104/pp.106.090878
Jeon, Y., Hwang, A. R., Hwang, I., Pai, H. S. (2010). Silencing of NbCEP1 encoding a chloroplast envelope protein containing 15 leucine-rich-repeats disrupts chloroplast biogenesis in nicotiana benthamiana. Mol. Cells 29, 175–183. doi: 10.1007/s10059-010-0011-5
Jurgensmeier, J. M., Xie, Z., Deveraux, Q., Ellerby, L., Bredesen, D., Reed, J. C. (1998). Bax directly induces release of cytochrome c from isolated mitochondria. Proc. Natl. Acad. Sci. U.S.A. 95, 4997–5002. doi: 10.1073/pnas.95.9.4997
Kang, C. H., Jung, W. Y., Kang, Y. H., Kim, J. Y., Kim, D. G., Jeong, J. C., et al. (2006). AtBAG6, a novel calmodulin-binding protein, induces programmed cell death in yeast and plants. Cell Death Differentiation 13, 84–95. doi: 10.1038/sj.cdd.4401712
Kang, M. K., Park, K. S., Choi, D. (1998). Coordinated expression of defense-related genes by TMV infection or salicylic acid treatment in tobacco. Mol. Cells 8, 388–392.
Kawai, M., Pan, L., Reed, J. C., Uchimiya, H. (1999). Evolutionally conserved plant homologue of the bax inhibitor-1 (BI-1) gene capable of suppressing bax-induced cell death in yeast 1. FEBS Lett. 464, 143–147. doi: 10.1016/S0014-5793(99)01695-6
Kawai-Yamada, M., Jin, L., Yoshinaga, K., Hirata, A., Uchimiya, H. (2001). Mammalian bax-induced plant cell death can be down-regulated by overexpression of arabidopsis bax inhibitor-1 (AtBI-1). Proc. Natl. Acad. Sci. U.S.A. 98, 12295–12300. doi: 10.1073/pnas.211423998
Kawai-Yamada, M., Ohori, Y., Uchimiya, H. (2004). Dissection of Arabidopsis Bax inhibitor-1 suppressing Bax-, hydrogen peroxide-, and salicylic acid-induced cell death. Plant Cell. 16(1), 21–32. doi: 10.1105/tpc.014613
Kirby, J., Keasling, J. D. (2009). Biosynthesis of plant isoprenoids: Perspectives for microbial engineering. Annu. Rev. Plant Biol. 60, 335–355. doi: 10.1146/annurev.arplant.043008.091955
Koch, E., Slusarenko, A. (1990). Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2, 437–445. doi: 10.1105/tpc.2.5.437
Lacomme, C., Santa Cruz, S. (1999). Bax-induced cell death in tobacco is similar to the hypersensitive response. Proc. Natl. Acad. Sci. U.S.A. 96, 7956–7961. doi: 10.1073/pnas.96.14.7956
Lamb, C., Dixon, R. A. (1997). The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 251–275. doi: 10.1146/annurev.arplant.48.1.251
Lebeaupin, C., Blanc, M., Vallée, D., Keller, H., Bailly-Maitre, B. (2020). BAX inhibitor-1: between stress and survival. FEBS J. 287, 1722–1736. doi: 10.1111/febs.15179
Lee, J. H., Kim, Y. C., Choi, D., Park, J. M. (2013). Identification of novel pepper genes involved in bax- or INF1-mediated cell death responses by high-throughput virus-induced gene silencing. Int. J. Mol. Sci. 14 (11), 22782–95. doi: 10.3390/ijms141122782
Levine, A., Tenhaken, R., Dixon, R., Lamb, C. (1994). H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79, 583–593. doi: 10.1016/0092-8674(94)90544-4
Liebrand, T. W., Smit, P., Abd-El-Haliem, A., De Jonge, R., Cordewener, J. H., America, A. H., et al. (2012). Endoplasmic reticulum-quality control chaperones facilitate the biogenesis of cf receptor-like proteins involved in pathogen resistance of tomato. Plant Physiol. 159, 1819–1833. doi: 10.1104/pp.112.196741
Liu, X., Kim, C. N., Yang, J., Jemmerson, R., Wang, X. (1996). Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell 86, 147–157. doi: 10.1016/S0092-8674(00)80085-9
Liu, Y., Ren, D., Pike, S., Pallardy, S., Gassmann, W., Zhang, S. (2007). Chloroplast-generated reactive oxygen species are involved in hypersensitive response-like cell death mediated by a mitogen-activated protein kinase cascade. Plant J. 51, 941–954. doi: 10.1111/j.1365-313X.2007.03191.x
Liu, Y., Schiff, M., Marathe, R., Dinesh-Kumar, S. P. (2002). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for n-mediated resistance to tobacco mosaic virus. Plant J. 30, 415–429. doi: 10.1046/j.1365-313X.2002.01297.x
Macho, A. p., Zipfel, C. (2014). Plant PRRs and the activation of innate immune signaling. Mol. Cell 54, 263–272. doi: 10.1016/j.molcel.2014.03.028
Mittler, R., Herr, E. H., Orvar, B. L., Van Camp, W., Willekens, H., Inze, D., et al. (1999). Transgenic tobacco plants with reduced capability to detoxify reactive oxygen intermediates are hyperresponsive to pathogen infection. Proc. Natl. Acad. Sci. U.S.A. 96, 14165–14170. doi: 10.1073/pnas.96.24.14165
Moon, J. Y., Lee, J. H., Oh, C.-S., Kang, H.-G., Park, J. M. (2016). Endoplasmic reticulum stress responses function in the HRT-mediated hypersensitive response in nicotiana benthamiana. Mol Plant Pathol. 17, 1382–1397. doi: 10.1111/mpp.12369
Mur, L. A., Kenton, P., Lloyd, A. J., Ougham, H., Prats, E. (2008). The hypersensitive response; the centenary is upon us but how much do we know? J. Exp. Bot. 59, 501–520. doi: 10.1093/jxb/erm239
Niyogi, K. K. (1999). Photoprotection revisited: Genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 333–359. doi: 10.1146/annurev.arplant.50.1.333
Okada, A., Shimizu, T., Okada, K., Kuzuyama, T., Koga, J., Shibuya, N., et al. (2007). Elicitor induced activation of the methylerythritol phosphate pathway toward phytoalexins biosynthesis in rice. Plant Mol. Biol. 65, 177–187. doi: 10.1007/s11103-007-9207-2
Page, J. E., Hause, G., Raschke, M., Gao, W., Schmidt, J., Zenk, M. H., et al. (2004). Functional analysis of the final steps of the 1-deoxy-D-xylulose 5-phosphate (DXP) pathway to isoprenoids in plants using virus-induced gene silencing. Plant Physiol. 134, 1401–1413. doi: 10.1104/pp.103.038133
Peart, J. R., Lu, R., Sadanandom, A., Malcuit, I., Moffett, P., Brice, D. C., et al. (2002). Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl. Acad. Sci. U.S.A. 99, 10865–10869. doi: 10.1073/pnas.152330599
Peñuelas, J., Munné-Bosch, S. (2005). Isoprenoids: an evolutionary pool for photoprotection. Trends Plant Sci. 10, 166–169. doi: 10.1016/j.tplants.2005.02.005
Phillips, M. A., León, P., Boronat, A., Rodríguez-Concepción, M. (2008). The plastidial MEP pathway: unified nomenclature and resources. Trends Plant Sci. 13, 619–623. doi: 10.1016/j.tplants.2008.09.003
Pulido, P., Perello, C., Rodriguez-Concepcion, M. (2012). New insights into plant isoprenoid metabolism. Mol. Plant 5, 964–967. doi: 10.1093/mp/sss088
Ramegowda, V., Senthil-Kumar, M., Ishiga, Y., Kaundal, A., Udayakumar, M., Mysore, K. S. (2013). Drought stress acclimation imparts tolerance to sclerotinia sclerotiorum and pseudomonas syringae in nicotiana benthamiana. Int. J. Mol. Sci. 14, 9497–9513. doi: 10.3390/ijms14059497
Rojas-Rivera, D., Hetz, C. (2015). TMBIM protein family: ancestral regulators of cell death. Oncogene 34, 269–280. doi: 10.1038/onc.2014.6
Samuel, M. A., Ellis, B. E. (2002). Double jeopardy: both overexpression and suppression of a redox-activated plant mitogen-activated protein kinase render tobacco plants ozone sensitive. Plant Cell 14, 2059–2069. doi: 10.1105/tpc.002337
Skorupinska-Tudek, K., Poznanski, J., Wojcik, J., Bienkowski, T., Szostkiewicz, I., Zelman-Femiak, M., et al. (2008). Contribution of the Mevalonate and Methylerythritol Phosphate Pathways to the Biosynthesis of Dolichols in Plants. Journal of Biological Chemistry 283 (30), 21024–21035
Surmacz, L., Swiezewska, E. (2011). Polyisoprenoids – secondary metabolites or physiologically important superlipids? Biochem. Biophys. Res. Commun. 407, 627–632. doi: 10.1016/j.bbrc.2011.03.059
Suzuki, N., Mittler, R. (2012). Reactive oxygen species-dependent wound responses in animals and plants. Free Radic. Biol. Med. 53, 2269–2276. doi: 10.1016/j.freeradbiomed.2012.10.538
Takayama, S., Reed, J. C. (2001). Molecular chaperone targeting and regulation by BAG family proteins. Nat. Cell Biol. 3, E237–E241. doi: 10.1038/ncb1001-e237
Takayama, S., Sato, T., Krajewski, S., Kochel, K., Irie, S., Milian, J. A., et al. (1995). Cloning and functional analysis of BAG-1: A novel bcl-2-binding protein with anti-cell death activity. Cell 80, 279–284. doi: 10.1016/0092-8674(95)90410-7
Thanthrige, N., Jain, S., Bhowmik, S. D., Ferguson, B. J., Kabbage, M., Mundree, S., et al. (2020). Centrality of BAGs in Stress responses, and host defense. Trends Plant Sci. 25, 1131–1140. doi: 10.1016/j.tplants.2020.04.012
Thordal-Christensen, H. Z., Wei, Z., Collinge, Y. (1997). Subceullar localization of H2O2 in plants, H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11, 1187–1194. doi: 10.1046/j.1365-313X.1997.11061187.x
Torres, M. A., Jones, J. D., Dangl, J. L. (2006). Reactive oxygen species signaling in response to pathogens. Plant Physiol. 141, 373–378. doi: 10.1104/pp.106.079467
Van Breusegem, F., Dat, J. F. (2006). Reactive oxygen species in plant cell death. Plant Physiol. 141, 384–390. doi: 10.1104/pp.106.078295
Watanabe, N., Lam, E. (2008). BAX inhibitor-1 modulates endoplasmic reticulum stress-mediated programmed cell death in arabidopsis*. J. Biol. Chem. 283, 3200–3210. doi: 10.1074/jbc.M706659200
Williams, B., Verchot, J., Dickman, M. B. (2014). When supply does not meet demand-ER stress and plant programmed cell death. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00211
Willis, S., Day, C. L., Hinds, M. G., Huang, D. C. (2003). The bcl-2-regulated apoptotic pathway. J. Cell Sci. 116, 4053–4056. doi: 10.1242/jcs.00754
Yao, N., Greenberg, J. T. (2006). Arabidopsis accelerated cell death2 modulates programmed cell death. Plant Cell 18, 397–411. doi: 10.1105/tpc.105.036251
Ye, C., Zheng, S., Jiang, D., Lu, J., Huang, Z., Liu, Z., et al. (2021). Initiation and execution of programmed cell death and regulation of reactive oxygen species in plants. Int. J. Mol. Sci. 22 (23), 12942. doi: 10.3390/ijms222312942
Zapata, J. M., Guera, A., Esteban-Carrasco, A., Martin, M., Sabater, B. (2005). Chloroplasts regulate leaf senescence: delayed senescence in transgenic ndhF-defective tobacco. Cell Death Differ 12, 1277–1284. doi: 10.1038/sj.cdd.4401657
Zeidler, J., Schwender, J., Muller, C., Wiesner, J., Weidemeyer, C., Beck, E., et al. (1998). Inhibition of the non-mevalonate 1-deoxy-D-xylulose-5-phosphate pathway of plant isoprenoid biosynthesis by fosmidomycin. Z Naturforsch. Teil C 53, 980–986. doi: 10.1515/znc-1998-11-1208
Keywords: bax-mediated cell death, chloroplast, hypersensitive response, LytB, methylerythritol phosphate pathway, reactive oxygen species
Citation: Lee S, Jo SH, Hong CE, Lee J, Cha B and Park JM (2022) Plastid methylerythritol phosphate pathway participates in the hypersensitive response-related cell death in Nicotiana benthamiana. Front. Plant Sci. 13:1032682. doi: 10.3389/fpls.2022.1032682
Received: 31 August 2022; Accepted: 07 October 2022;
Published: 26 October 2022.
Edited by:
Li Bo Han, Fujian Agriculture and Forestry University, ChinaReviewed by:
Xinyang Wu, China Jiliang University, ChinaAhmad A. Omar, University of Florida, United States
Copyright © 2022 Lee, Jo, Hong, Lee, Cha and Park. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jeong Mee Park, am1wYXJrQGtyaWJiLnJlLmty
†Present address: Sanghun Lee, Department of Botany and Plant Pathology, Purdue University, West Lafayette, IN, United States
Chi-Eun Hong, Department of Herbal Crop Research, National Institute of Horticultural and Herbal Science (NIHHS), Eumseong, Chungbuk, South Korea
 Sanghun Lee
Sanghun Lee Sung Hee Jo
Sung Hee Jo Chi Eun Hong
Chi Eun Hong Jiyoung Lee
Jiyoung Lee Byeongjin Cha2
Byeongjin Cha2 Jeong Mee Park
Jeong Mee Park