- Shandong Provincial Key Laboratory of Plant Stress Research, College of Life Sciences, Shandong Normal University, Shandong, China
Homeodomain leucine zipper (HD-ZIP) proteins are plant-specific transcription factors that contain a homeodomain (HD) and a leucine zipper (LZ) domain. The highly conserved HD binds specifically to DNA and the LZ mediates homodimer or heterodimer formation. HD-ZIP transcription factors control plant growth, development, and responses to abiotic stress by regulating downstream target genes and hormone regulatory pathways. HD-ZIP proteins are divided into four subclasses (I–IV) according to their sequence conservation and function. The genome-wide identification and expression profile analysis of HD-ZIP proteins in model plants such as Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) have improved our understanding of the functions of the different subclasses. In this review, we mainly summarize and discuss the roles of HD-ZIP proteins in plant response to abiotic stresses such as drought, salinity, low temperature, and harmful metals. HD-ZIP proteins mainly mediate plant stress tolerance by regulating the expression of downstream stress-related genes through abscisic acid (ABA) mediated signaling pathways, and also by regulating plant growth and development. This review provides a basis for understanding the roles of HD-ZIP proteins and potential targets for breeding abiotic stress tolerance in plants.
Introduction
Common abiotic stresses include drought, high and low temperatures, oxidative stress, salinity, and toxic metals, and both abiotic and biotic stresses reduce crop yields, resulting in significant economic losses (Patel et al., 2017). Globally, approximately 20% and 22% of arable land is affected by drought and salinization, reducing crop yields in affected areas by more than 50% (Vinocur and Altman, 2005; Agarwal et al., 2017). Harmful metals exist as free ions or with functional groups (such as glutathione sulfhydryl groups), and their reduction produces reactive oxygen species (ROS) that ultimately lead to oxidative damage to plant cells (Kaur et al., 2021). Plants have evolved different mechanisms to deal with abiotic stresses (Sytar et al., 2021). These adaptive mechanisms involve changes in plant physiology and biochemistry, molecular and cellular levels, and are regulated by complex networks. Given the impacts of climate change and severe weather, as well as soil contamination with salt and metals, on crop production, it is important to understand the ability of plants to respond to single and combined stresses to improve crop resilience (Baillo et al., 2019).
Transcription factors (TFs) are involved in regulatory networks that regulate plant growth and development and respond to stress (Haak et al., 2017). TFs activate or repress the transcription of these target genes by binding to specific cis-acting elements within the promoters of these target genes, and participate in signal transduction pathways. In plants, about 10% of genes encode transcription factors that perform their specific functions during different growth and developmental stages of plants. Transcription factors are divided into different families according to their DNA-binding domains. Arabidopsis (Arabidopsis thaliana) has an estimated 30 transcription factor families, including 1922 transcription factors (Aglawe et al., 2012). Several types of TFs involved in abiotic stress have been cloned, including members of the Homeodomain-leucine zipper (HD-ZIP) and APETALA2, WRKY (it is characterized by a highly conserved WRKYGQK core motif at the N-terminus), NAM, ATAF and CUC (NAC), Ethylene-responsive element binding proteins (AP2/EREBP), v-myb avian myeloblastosis viral oncogene homolog (MYB), heat stress transcription factor (HSF), zinc finger proteins (ZFPs), and basic helix-loop-helix (bHLH) transcription factor families (Wang et al., 2016; Li et al., 2019e). Since transcription factors are major regulators of stress-related genes, they can be used for genetic engineering to improve stress tolerance in plants. Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) gene editing technology is an important tool used to study TFs to improve plant stress resistance. Researchers comprehensively reviewed the use of CRISPR/Cas9 to evaluate the roles of genes in environmental stress tolerance, transcriptional and translational regulation, and crop improvement (Bao et al., 2019). Many TFs respond to stress and regulate a large number of downstream genes; therefore, regulating the expression of transcription factor genes is a research hotspot (Hoang et al., 2017). In this review, we focus on plant-specific HD-ZIP TFs, which play an essential role in plant responses to abiotic stress.
The structure and classification of HD-ZIP proteins
HD-ZIP transcription factors are members of the homeobox (HB) protein family. These TFs have a highly conserved homeodomain (HD) consisting of 61 amino acids, and a leucine zipper (LZ) element tightly linked to the carboxy terminus of HD. HD can specifically bind to DNA, and LZ mediates protein dimerization, which is important for DNA recognition (Xie et al., 2021). HD-ZIP proteins are divided into four subfamilies (I–IV) according to the specificity of DNA binding, sequence similarity, other conserved motifs, and differences in physiological functions (Sharif et al., 2021). The conserved domains and different motifs of four subfamilies of HD-ZIP proteins were shown in Figure 1 (Sessa et al., 2018; Sharif et al., 2021). The HD-ZIP I protein contains a relatively poorly conserved LZ domain and a highly conserved HD domain. And its HD and LZ domains are located in the middle of the protein, which has the dual symmetric DNA sequence of CAAT(A/T)ATTG specific binding properties (Elhiti and Stasolla, 2009; Gong et al., 2019). Apart from the LZ and HD domains, members of the HD-ZIP I family differ from other family members in that they do not share domains and/or motifs. Many HD-ZIP I genes are involved in regulating signal transduction, regulating plant growth and development, and regulating plant tolerance to environmental stresses such as drought, salinity, cold, and heavy metal stress (Gong et al., 2019). Compared with HD-ZIP I, HD-ZIP II proteins have an N-terminal consensus sequence and a conserved CPSCE motif. The motif is named after five conserved amino acids (Cys, Pro, Ser, Cys and Glu). Moreover, HD-ZIP II protein has high homology with HD-ZIP I, and has specific binding properties to the double symmetrical DNA sequence of CAAT(C/G)ATTG (Elhiti and Stasolla, 2009; Gong et al., 2019). The HD domains of HD-ZIP I and II proteins have high sequence homology and both contain a highly conserved tryptophan-phenylalanine-glutamine-asparagine-arginine-arginine (WFQNRR) motif, the target DNA sequences recognized by the two differ by only one nucleotide in the middle. The specificity of this binding to a particular DNA element is primarily determined by the third helix, but the other two helices also contribute to this binding. By replacing Arg55 in helix 3 with Ala55, the protein cannot bind to DNA. Glu46 and Thr56 of helix 3 of HD-ZIP II protein are essential for this class of proteins to recognize C/G bases in DNA elements, while in HD-ZIP I protein, these two amino acid residues are replaced by Ala46 and Trp56, respectively (Ariel et al., 2007; Elhiti and Stasolla, 2009; Gong et al., 2019). The expression of HD-ZIP II protein is mainly regulated by photochemical conditions, and its conserved domain CPSCE motif is involved in regulating plant responses to light, shade and abiotic stresses (Park et al., 2013; Tognacca et al., 2021). Structurally, HD-ZIP III is very different from HD-ZIP I and II proteins, and has essentially the same domain as HD-ZIP IV protein. The HD-ZIP III and HD-ZIP IV subfamilies contain a steroidogenic acute regulatory protein-related lipid transfer (START) domain and a START-associated domain (SAD). The START domain responds to abscisic acid (ABA). However, the function of the SAD is unclear. HD-ZIP III and HD-ZIP IV proteins differ in that the C-terminus of HD-ZIP III protein contains the MEKHLA domain associated with various chemical and physical stimuli, whereas HD-ZIP IV protein does not contain this structure (Zhu et al., 2018). The HD-ZIP III protein can recognize the GTAAT(G/C)ATTAC sequence, while the HD-ZIP IV protein can interact with the TAAATG(C/T)A sequence (Elhiti and Stasolla, 2009; Gong et al., 2019).
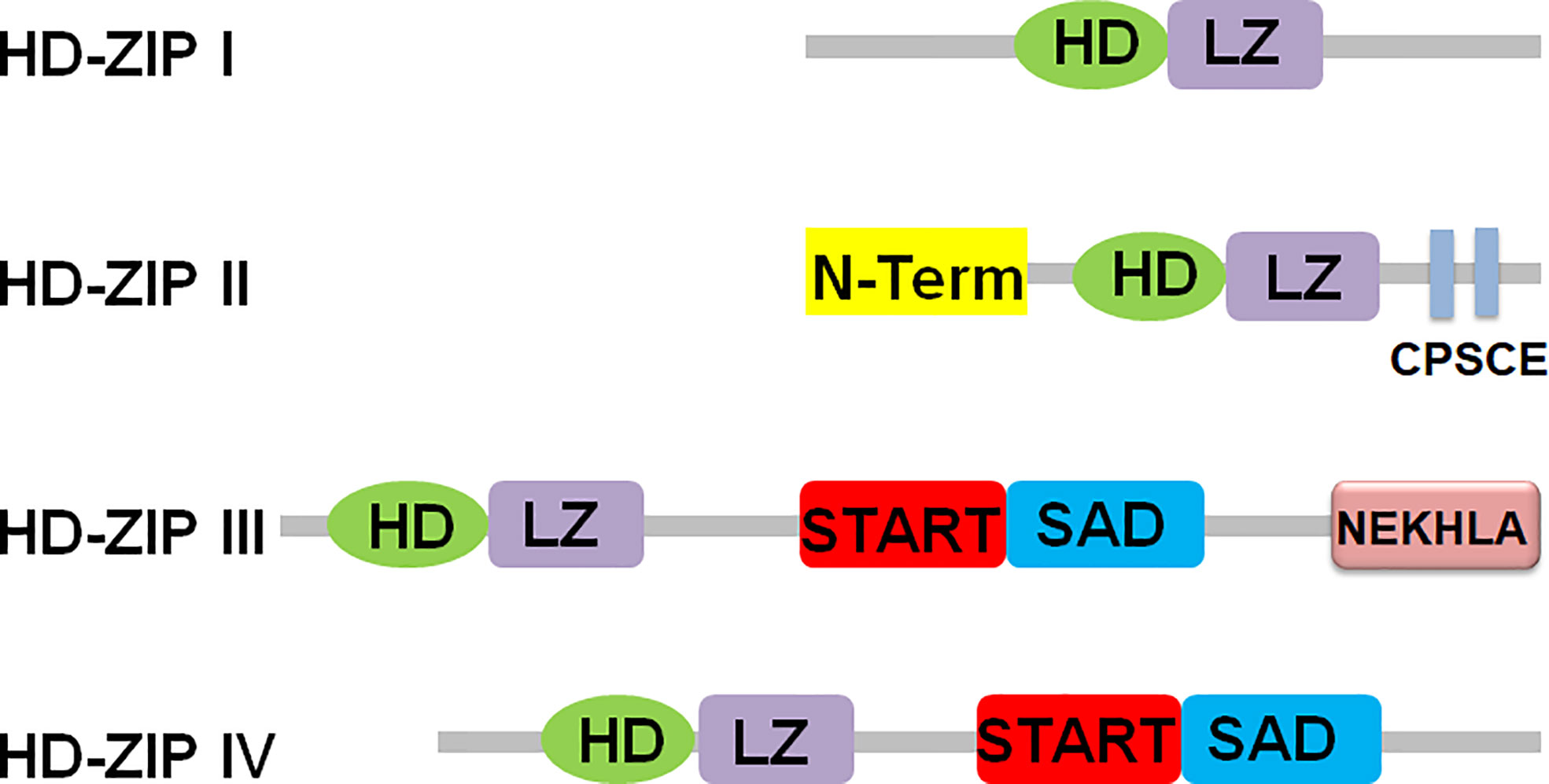
Figure 1 Schematic diagram of HD-ZIP family protein structure. HD, homeodomain; LZ, leucine-zipper; N-Term, N-term consensus sequence; START, steroidogenic acute regulatory proteinlipid transfer domain lipid transfer domain; MEKHLA, Met-Glu-Lys- His-Leu-Ala domain; SAD, START associated domain.
Genes in the same subfamily usually tend to be conserved in function because their structures are relatively similar. Different subfamilies have different gene structures and encode diverse proteins, so their roles in plant growth and development are also different. The expression of HD-ZIP genes is subject to different regulatory mechanisms. For example, HD-ZIP I family genes play critical roles in regulating plant development, regulation of signaling networks, and signaling networks triggered by endogenous and external stimuli in response to various abiotic stresses (Gong et al., 2019). HD-ZIP II family genes are involved in plant organ development, photoprotection and hormone response, and their expression is regulated by light signals (Qiu et al., 2022). Genes of the HD-ZIP III subfamily are post-transcriptionally regulated by the microRNAs (miRNAs) miR165/166 (Emery et al., 2003; Carlsbecker et al., 2010; Fan et al., 2021). At least six distinct plant hormones interact to dynamically regulate the spatiotemporal expression of miR165/166 and HD-ZIP III subfamily proteins during Arabidopsis development (Sessa et al., 2018). It has been shown that the HD-ZIP III protein directly and positively regulated the expression of HD-ZIP II genes (Turchi et al., 2015). HD-ZIP IV subfamily genes play essential roles in epidermal cell differentiation and root development (Qiu et al., 2022).
Subcellular localization of HD-ZIP proteins
A protein’s subcellular localization indicates its function. The majority identified HD-ZIP family members localize in the nucleus and directly regulate genes expression by binding to cis-elements in their target promoters (Zou et al., 2011). For example, the HD-ZIP I subfamily members AtHB12 in Arabidopsis (Shin et al., 2004), ZmHDZ1 in maize (Zea mays) (Wang et al., 2017), PtHB13 in trifoliate orange (Citrus trifoliata) (Ma et al., 2020), OsHOX12 and OsHOX14 in rice (Oryza sativa) (Shao et al., 2018), JcHDZ07 (Tang et al., 2019a) and JcHDZ16 (Tang et al., 2019b) in physic nut (Jatropha curcas), NtHD-ZIP I in tobacco (Nicotiana tabacum) (Li et al., 2019e), and CaATHB-12 in pepper (Capsicum annuum) (Zhang et al., 2020) are all nuclear-localized proteins. HD-ZIP II subfamily members HAT22/ABIG1 in Arabidopsis (Liu et al., 2016), CaHB1 in pepper (Oh et al., 2013), MaHDZII.4 and MaHDZII.7 in Musa accuminata (Yang et al., 2020), and the OCL1 in the HD-ZIP IV subfamily in maize (Depège-Fargeix et al., 2011) and ROC4 (Wei et al., 2016) in rice are also nuclear-localized proteins. Yeast one-hybrid screen identifies the HD-ZIP I transcription factor PtHB13 in citrus as an upstream regulator of FLOWERING LOCUS C (PtFLC). Citrus flowering is regulated by PtHB13 through binding to the PtFLC promoter (Ma et al., 2020). OsHOX12 and OsHOX14 belong to the HD-ZIP I family of genes that bind a specific DNA sequence AH1 CAAT (A/T) ATTG to activate its downstream reporter gene expression (Shao et al., 2018). PtHB13, OsHOX12 and OsHOX14 all function in the nucleus and directly regulate the expression of their downstream genes by binding to cis-acting elements on downstream target promoters. Under salt treatment, JcHDZ07 negatively regulates transgenes by regulating the expression of several stress-responsive genes, such as AtDREB1A, AtDREB2A, AtHKT1;1, Δ1-Pyrroline-5-carboxylate synthetase 1 (P5CS1), AtSOS1, AtSOS3, AtNHX1 and AtAPX1 (Tang et al., 2019a). This indicates that JcHDZ07 protein makes plants more sensitive to salt stress by inhibiting the expression of salt-responsive genes. HAT22/ABIG1 represses HRE2 transcription by acting on a negative cis-regulatory element in the HRE2 promoter in response to plant hypoxia and/or salt stress (Seok et al., 2022). These results indicates that HD-ZIPs localized in the nucleus mainly act as a transcription factor to regulate downstream target genes. In addition, although some individual HD-ZIP genes are localized and expressed in the nucleus, they interact with proteins in a certain way to exert their transcription factor functions. Transcriptional activation or repression by transcription factors is accompanied by changes in chromatin structure that enable transcription factors to gain access to their target promoters, a step that often requires multi-protein complexes capable of manipulating nucleosome structure. OUTER CELL LAYER1 (OCL1) is a nuclear-localized transcription factor with transcriptional activation activity that interacts with SWI3C1 resulting in chromatin opening and active transcription. SWI3C1 is a component of the SWI/SNF chromatin remodeling complex and is a C subfamily protein of plant SWI3 proteins (Depège-Fargeix et al., 2011).
There are also some HD-ZIP transcription factors that are distributed in both the cytoplasm and the nucleus, and its distribution is determined by its unique N-terminus. For example, the HD ZIP I protein MdHB1 in apple (Malus × domestica) (Jiang et al., 2017), HD ZIP II protein ATHB17 in Arabidopsis (Zhao et al., 2017) and HD ZIP IV protein CsGL2-LIKE in cucumber (Cucumis sativus) (Cai et al., 2020). MdHB1 can interact with WD40, MYB and bHLH in the cytoplasm to function as a homodimer and indirectly regulate gene expression. MdHB1 confines MdTTG1, MdMYB10 and MdbHLH3 to the cytoplasm and indirectly represses the transcription of MdDFR and MdUFGT. Once MdHB1 is silenced, these transcription factors are released, which in turn activate MdDFR and MdUFGT expression, and promotes anthocyanin biosynthesis (Jiang et al., 2017). However, some HD-ZIP transcription factors expressed in the cytoplasm and nucleus, whose functions in the cytoplasm were not expressed, still function as transcription factors in the nucleus. For example, ATHB17 is a transcriptional repressor containing an EAR (ERF-associated repression of amphipathic)-like motif. ATHB17 positively regulates plant tolerance to oxidative, drought and salt stress. ATHB17 is a repressor of photosynthesis associated nuclear genes (PhANGs) and it has been reported to suppress transcription of 26 PhANGs under normal or salt stress conditions, among which PSBO1, LHB1B1, LHB1B2, LHCA2, and FDA6 are directly regulated by ATHB17 (Zhao et al., 2017). ATHB17 directly or indirectly represses the transcription of many PhANGs through binding to its target promoters. It was also found that ATHB17 can directly bind to the promoter of ATSIG5 to activate its transcription, and then its protein translocates to the chloroplast as an important regulator of many chloroplast genes, such as psbA, psbB, psbC, psbD, and psBT (Zhao et al., 2017). ATSIG5 is a transcription factor whose expression can be induced by various stresses including high light, salinity, osmotic stress and low temperature (Nagashima et al., 2004). Studies have shown that CsGL2-LIKE interacts with the jasmonic acid (JA) ZIM domain protein CsJAZ1 to form the HD-ZIP IV-CsJAZ1 complex involved in cucumber male flower development (Cai et al., 2020). These HD-ZIP genes function in both the cytoplasm and nucleus. This can illustrate that, in the cytoplasm, HD-ZIP proteins form heterodimers or homodimers with other proteins to promote their transcriptional specificity.
HD-ZIP proteins in plant growth and development
The HD-ZIP proteins of Arabidopsis are the most thoroughly understood (Turchi et al., 2013; Carabelli et al., 2018). Genome-wide identification and expression profiling have helped to identify and functionally characterize many other HD-ZIP proteins in different plants, such as potato (Solanum tuberosum L.) (Li et al., 2019d), cucumber (Cucumis sativus L.) (Sharif et al., 2020), tobacco (Li et al., 2019e), sesame (Sesamum indicum L.) (Wei et al., 2019), tea plant (Camellia sinensis L.) (Shen et al., 2019), Japanese apricot (Prunus mume L.) (Li et al., 2019c), physic nut, and foxtail millet (Setaria italica L.) (Chai et al., 2018). HD-ZIP proteins participate in regulating plant growth and development, such as fruit development, and maturity, anthocyanin accumulation, flowering, vascular development, epidermal cell development.
HD-ZIP proteins regulate fruit ripening by regulating cell wall degradation and ethylene biosynthesis. The HD-ZIP I genes MaHDZI.19 and MaHDZI.26, and the HD-ZIP II genes MaHDZII.4 and MaHDZII.7 in banana (Musa acuminata) are significantly up-regulated during fruit ripening. These four MaHDZs specifically localize in the nucleus and activate several maturation-related genes, including MaACO5, which is related to ethylene biosynthesis, and MaEXP2, MaEXPA10, MaPG4, and MaPL4, which are related to cell wall degradation (Yang et al., 2020). LcHB2, a member of the litchi (Litchi chinensis) HD-ZIP I subfamily, regulates fruit drop by directly activating the cell wall degradation-related genes LcCEL2 and 8, while the HD-ZIP I gene LcHB3 regulates fruit drop by promoting ethylene biosynthesis (Li et al., 2019a; Li et al., 2019b). The HD-ZIP II gene PpHB.G7 interacts with the promoters of the ethylene biosynthesis genes PpACS1 and PpACO1 to promote peach (Prunus persica) maturation by promoting ethylene biosynthesis (Gu et al., 2019). Silencing of MdHB1 leads to the accumulation of anthocyanins in apple pulp. MdHB1 normally confines MdTTG1, MdMYB10 and MdbHLH3 proteins to the cytoplasm. But when MdHB1 was silenced, these transcription factors were released, which in turn activated the expression of MdDFR and MdUFGT, and promoted anthocyanin biosynthesis, resulting in red-fleshed apple fruit (Jiang et al., 2017).
HD-ZIP proteins participate in induction of flowering and regulate flowering time. HaHB10 in the sunflower (Helianthus annuus L.) is an HD-ZIP II family gene that responds to dark and light conditions and is involved in plant flowering induction. It mediates the vegetative-reproductive transition by activating specific flowering-related genes in response to salicylic acid (Rueda et al., 2005; Dezar et al., 2011). The HD-ZIP IV subfamily member ROC4 promotes flowering in rice (Wei et al., 2016), while heterologous expression of the maize (Zea mays) HD-ZIP IV subfamily gene OCL1 in rice delayed flowering time (Depège-Fargeix et al., 2011). RICE FLOWERING LOCUS T 1 (RFT1), Heading date 3a (Hd3a), and Early heading date 1 (Ehd1) promote flowering under long-day conditions in rice, and ROC4 indirectly inhibits the expression of these genes (Wei et al., 2016). HD-ZIP proteins are also involved in regulating the fertility of germ cells. The HD-ZIP IV subfamily member CsGL2-LIKE in cucumber is expressed in male flower buds and anthers (Cai et al., 2020). CsGL2-LIKE silencing represses the expression of the flower-forming gene, the flowering locus T (FT), resulting in partial male sterility, delayed flowering, and reduced pollen and seed vigour. CsGL2-LIKE also interacts with CsJAZ1 to regulate male flower development (Cai et al., 2020).
HD-ZIP proteins participate in vascular development. Five members of the Arabidopsis HD-ZIP III subfamily ATHB8, PHAVOLUTA/AtHB9 (PHV), PHABULOSA/AtHB14 (PHB), AtHB15/CORONA (CAN), and REVOL UTA/IFL1 (REV) are associated with vascular development (Baima et al., 1995; Prigge et al., 2005). Vascular defects are pronounced in phb phv cna triple mutant stems, and the loss-of-function mutation of REV leads to defective vascular development (Prigge et al., 2005). ATHB8 overexpression lines promotes vascular cell differentiation in Arabidopsis (Baima et al., 2001).
Most HD-ZIP IV genes are specifically expressed in epidermal or sub-epidermal cells play important roles in the development and maintenance of the outer cell layer (Chew et al., 2013). Arabidopsis has 16 HD-ZIP IV subfamily members, including PROTODERMAL FACTOR2 (PDF2), ENHANCED DROUGHT TOLERANCE1/HOMEODOMAIN GLABROUS11 (AtEDT1/HDG11), HDG12, ARABIDOPSIS THALIANA MERISTEM LAYER1 (ATML1), and GLABRA2 (GL2). For example, GL2, a member of the Arabidopsis HD-ZIP IV subfamily, plays a critical role in the development of trichomes and root hairs. It acts as a negative regulator in the fate determination and initiation stage of root hairs (Song et al., 2011; Khosla et al., 2014). Kale lines overexpressing the HD-ZIP IV family gene AtEDT1/HDG11 produced more root hairs and larger root structures, increasing their tolerance to drought and osmotic stress compared to wild type (Zhu et al., 2016).
HD-ZIP genes also regulate plant organ development. The overexpression and mutant studies of ATHB13 and ATHB23 genes showed that they are involved in the regulation of inflorescence stem, silique, ovule and seed development (Ribone et al., 2015). Arabidopsis HomeoBox 1 (AtHB1) is mainly expressed in hypocotyls and roots and is up-regulated in short-day-grown seedlings (Capella et al., 2015). Compared with wild-type Arabidopsis, AtHB1 knockdown mutants and overexpression lines exhibited shorter and longer hypocotyls, respectively. And AtHB1 acts downstream of PHYTOCHROME-INTERACTING FACTOR 1 (PIF1) to promote hypocotyl elongation (Capella et al., 2015).
HD-ZIP proteins in plant abiotic stress
HD-ZIP family TFs are involved in the regulation of plant abiotic stress. In the model legume Medicago truncatula L., salt stress induces the expression of the HD-ZIP I transcription factor MtHB1 (Ariel et al., 2010). The researchers demonstrated through bioinformatics analysis that the CsHDZ gene in tea (Camellia sinensis) responds to ABA, high temperature, low temperature, high salinity and drought. The expression patterns of CsHDZ genes are related to the abiotic stress treatments they are subjected to. Expression of most selected CsHDZ genes was significantly increased under various stress treatments (Shen et al., 2019). The researchers identified 45 HD-ZIP genes in sesame (Sesamum indicum L.) seeds, named as SiHDZ01-SiHDZ45. Analysis of SiHDZ gene expression showed that these genes have different expression patterns in different tissues. More than half of SiHDZ genes were found to be differentially expressed under salt and drought stress, while HD-ZIP I and HD-ZIP II family genes play important roles in sesame response to stress (Wei et al., 2019). The HD-ZIP gene of potato (Solanum tuberosum L.) is involved in all stages of its growth and development. The study showed that 13 HD-ZIP genes of potato were expressed differently in different tissues at high temperature (37°C) and low temperature (4°C). Under 37°C high temperature treatment, the expression levels of StHOX11 and StHOX28 in leaves, StHOX25 in stems, and StHOX20, StHOX23 and StHOX43 in roots were significantly up-regulated. Under low temperature conditions, the expression of StHOX39 in stems and StHOX1 and StHOX20 in roots and leaves were significantly increased (Li et al., 2019d). Studies have shown that in Craterostigma plantagineum, the expression of all CPHB gene families is regulated by leaf and root dehydration. HD-ZIP I family members CPHB-6 and CPHB-7 accumulated in leaves at the initial stage of dehydration, but their expression decreased after prolonged dehydration. HD-ZIP I family members CPHB-4/5 and HD-ZIP II family CPHB-3 were down-regulated by dehydration in both leaves and roots. This suggests that HD-ZIP plays a role in regulating gene expression of desiccation tolerance in Craterostigma plantagineum (Deng et al., 2002).
HD-ZIP proteins in drought tolerance
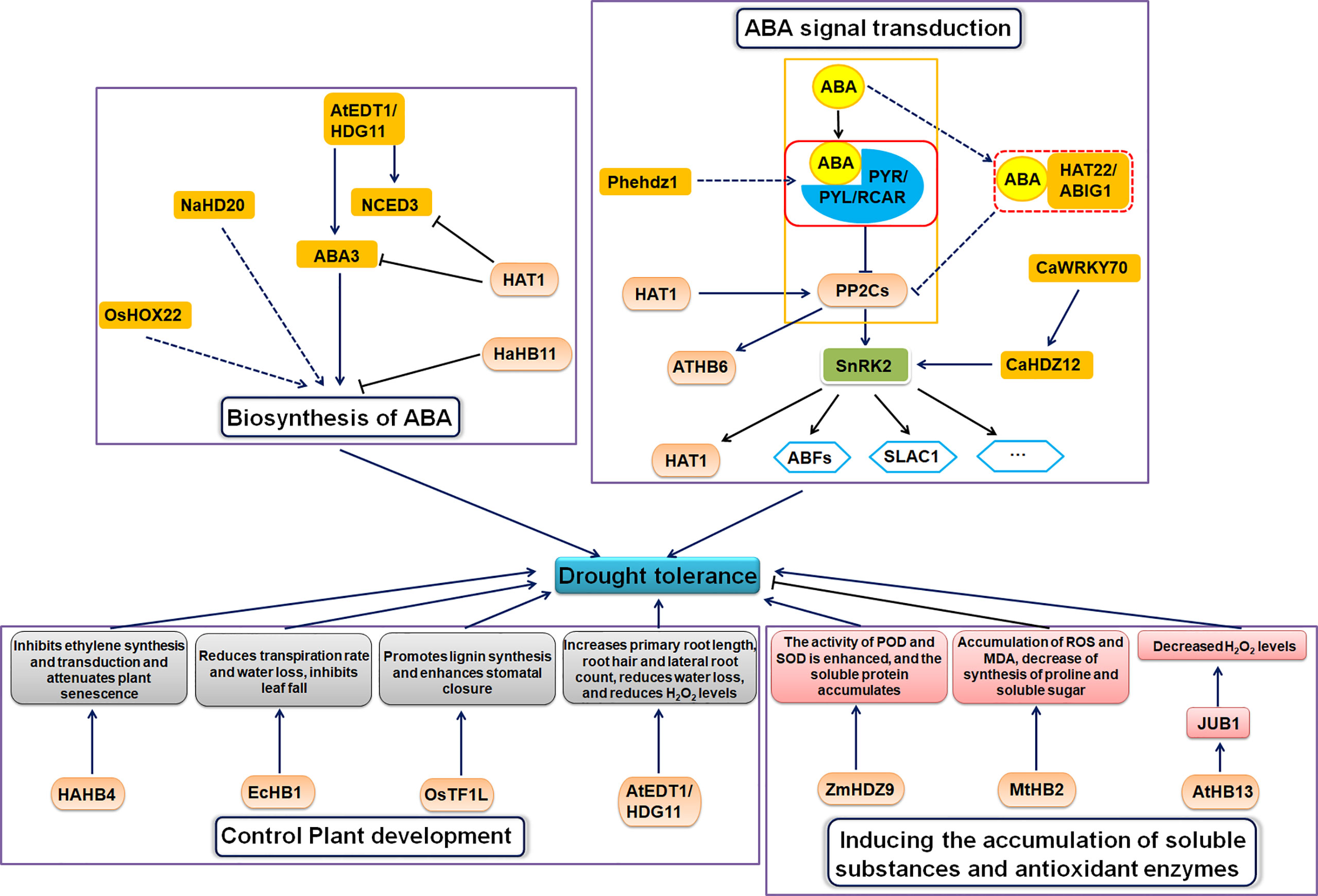
Due to global climate change, drought stress has received increasing attention as one of the most harmful abiotic stresses to plants (Yao et al., 2020). Drought stress affects plant physiology, biochemistry, and metabolism, damaging cells, stunting growth, and reducing crop yield and quality (Gupta et al., 2020). HD-ZIP proteins facilitate plant adaptation to drought stress in multiple ways (Figure 2).
HD-ZIP proteins regulate drought stress through the ABA signaling pathway
Some HD-ZIP transcription factors specifically bind ABA signaling or cis-elements in stress-responsive gene promoters to regulate drought response upon induction by exogenous ABA and drought stress (Mehmood et al., 2022).
Roles in biosynthesis of ABA
ABA is a 15-carbon sesquiterpene generated by the cleavage of carotenoids in the 2-C-Methyl-D-erythritol 4-phosphate (MEP) pathway. Short-chain acyl-CoA dehydrogenase (SCAD), zeaxanthin cyclooxygenase (ZEP), and 9-cis-epoxycarotenoid dioxygenase (NCED) play key roles in ABA biosynthesis (Marin et al., 1996; Seo and Koshiba, 2002; Marusig and Tombesi, 2020).
Some HD-ZIP TFs alter plant tolerance to drought stress by altering ABA biosynthesis. Virus-induced gene silencing (VIGS) experiments show that the HD-ZIP I gene NaHD20 promotes the expression of dehydration response genes NaNCED1, NaOSM1 and NaLTP1 and the accumulation of ABA in tobacco (Nicotiana attenuata) leaves under water stress (Ré et al., 2011). OsHOX22 encodes a rice HD-ZIP I transcription factor that binds to the CAAT(G/C)ATTG element and acts as a transcriptional activator, requiring the HD and ZIP domains (Zhang et al., 2012). OsHOX22-overexpressing transgenic rice lines have increased sensitivity to ABA. Down-regulation of OsHOX22 reduces ABA content and enhances drought tolerance in plants, whereas OsHOX22 overexpression leads to increased ABA synthesis and accumulation and reduces drought tolerance (Zhang et al., 2012). The experimental results suggest that OsHOX22 negatively regulates drought tolerance by increasing ABA biosynthesis. The HD-ZIP II gene homeodomain-leucine zipper protein 1 (HAT1) is involved in the regulation of drought tolerance in Arabidopsis (Tan et al., 2018). The hat1 and hat3 mutants have enhanced drought tolerance (Tan et al., 2018). HAT1 directly binds to the promoters of the ABA biosynthesis genes ABA3 and NCED3 to negatively regulate their expression, resulting in reduced ABA accumulation (Tan et al., 2018). The expression of the sunflower HD-ZIP I gene HaHB11 was up-regulated under ABA and drought treatments, and heterologous overexpression of HaHB11 enhanced drought tolerance in Arabidopsis and alfalfa (Medicago sativa) (Cabello et al., 2017). HaHB11 differentially regulates genes involved in ABA biosynthesis (ABA1 and ABA2), ABA-independent signaling pathways (COR15A and COR47), and cytoprotection (EM6 and RD29B) (Ding et al., 2009; Msanne et al., 2011; Cabello et al., 2017). Heterologously overexpressing the Arabidopsis HD-ZIP IV gene AtEDT1/HDG11 enhances drought tolerance in Chinese kale (Brassica oleracea var. alboglabra) (Zhu et al., 2016). NCED3 and LOS5/ABA3, which encode key ABA biosynthesis enzymes, were up-regulated in the transgenic kale line under non-stress and drought stress. Drought stress also up-regulates the stress marker gene RD29A, the proline synthesis gene P5CS, and the stress tolerance-related gene LEA in this transgenic line. Furthermore, the ABA/stress-related genes NAC3, NAC5, SNAC1, DREB2, ABI3, and ABI5 were also significantly up-regulated in the AtEDT1/HDG11-overexpression lines (Zhu et al., 2016).
Roles in ABA signal transduction
The ABA signaling pathway consists of RCAR/PYR/PYL receptor proteins, PP2C protein phosphatases, SnRK2 kinases (such as SnRK2.2, SnRK2.3, and SnRK2.6), and their target substrates (Umezawa et al., 2010; Guo et al., 2011; Yao et al., 2020). When ABA binds to and activates its receptor, the receptor forms a trimer complex with PP2Cs, thus inhibiting PP2C activity and abolishing its repression of SnRK2s (Yao et al., 2020). Active SnRK2s further activate the transcription factor ABFs/AREB1 [The basic leucine zippers (bZIPs) transcription factor], thereby initiating the expression of downstream genes.
Some other HD-ZIP transcription factors regulate drought tolerance in plants through the ABA signaling pathway. ATHB6 is an HD-ZIP I family gene that negatively regulates ABA signaling and is a target of the protein phosphatase ABA Insensitive 1 (ABI1), acting downstream of ABI1. ABI1, a protein phosphatase 2C, is a key component in ABA signaling (Himmelbach et al., 2002). HAT1 promotes the expression of PP2Cs and negatively regulates the ABA-mediated drought response and ABA signaling (Tan et al., 2018). HAT1 is a also substrate of the SnRK2.3 kinase, and phosphorylation by SnRK2.3 promoted HAT1 degradation (Tan et al., 2018). Drought stress induces ABA production, which inhibits PP2C activity, releasing SnRK2s to further activate downstream HAT1 by phosphorylation (Tan et al., 2018). CaHDZ12 is an HD-ZIP I transcription factor from chickpea (Cicer arietinum), and CaHDZ12 is expressed in roots and stems under drought stress (Sen et al., 2017). CaHDZ12 causes histone acetylation at chromatin to promote decondensation and subsequent gene transcription. CaHDZ12 functions in ABA-dependent signaling and promotes osmolyte production, reduces intracellular ROS levels and induces expression of SnRK2 kinase and other stress-related genes in response to abiotic stress tolerance in chickpea (Sen et al., 2017). Further research found that, the WRKY transcription factor CaWRKY70 acts upstream of CaHDZ12 and negatively regulates its transcription by binding to the W-box(es) in the promoter region of CaHDZ12 (Sen et al., 2017). The wheat (Triticum aestivum) HD-ZIP I gene TaHDZ5-6A positively regulates the drought stress response (Li et al., 2020). Under drought stress, transgenic Arabidopsis heterologously overexpressing TaHDZ5-6A has a lower water loss rate than wild-type, and stomatal closure and proline levels increased faster in transgenic plants than in wild type (Li et al., 2020). DREB2A, RD29A, RD29B, RD26, and PP2CA were up-regulated in the transgenic Arabidopsis compared with wild type Arabidopsis in the absence of stress, and were further up-regulated in the transgenic Arabidopsis under drought stress. This suggests that TaHDZ5-6A may regulate the transcription of these ABA signaling genes under drought conditions, thereby inducing rapid stomatal closure and reducing water loss (Li et al., 2020). HD-ZIP proteins regulate drought stress through ABA signal transduction and MAPKs signaling pathway. Overexpressing a moso bamboo (Phyllostachys edulis) HD-ZIP I gene, Phehdz1, in rice caused a significant increase in proline as well as a significant decrease in malondialdehyde (MDA) content relative to wild-type plants after drought treatment, and Phehdz1-overexpressing rice plants had enhanced drought tolerance (Gao et al., 2021). The Phehdz1 promoter region contains four ABA-response elements (ABREs), and Phehdz1 overexpression increased rice sensitivity to exogenous ABA; therefore, Phehdz1 may enhance drought tolerance through ABA signaling (Gao et al., 2021). The expression of MAPK signaling pathway genes, such as MPK2, MPK4, MPK6, WRKY33, CIPK25, and PR1, changed significantly in Phehdz1-overexpressing rice under drought stress (Gao et al., 2021). These results suggest that Phehdz1 may affect secondary metabolism via integrated ABA and MAPK signaling pathways to increase drought tolerance (Gao et al., 2021). After drought treatment, expression of the stress- and ABA-responsive genes AP37, DREB1, ERF3, OsMYB3, OsNAC10, and OsWRKY47 was higher in transgenic lines compared to wild-type plants (Gao et al., 2021). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis revealed that many differentially expressed genes (DEGs) were associated with metabolic pathways, such as phenylpropane biosynthesis and diterpene biosynthesis, suggesting that Phehdz1 overexpression negatively impacts the response to drought stress, especially phenylpropane biosynthesis (Dong et al., 2020).
The expression of HD-ZIP II protein HAT22/ABA-INSENSITIVE GROWTH 1 (ABIG1) increased with ABA and drought treatment, and it shared a similar regulatory pattern with two other known ABA signaling genes, PYL6 and CIPK12, suggesting that HAT22/ABIG1 may be part of the ABA signaling pathway. Additionally, HAT22/ABIG1 is required for ABA-mediated growth inhibition and leaf yellowing, suggesting that HAT22/ABIG1 is involved in plant responses to drought stress (Liu et al., 2016).
HD-ZIP proteins regulate drought tolerance by controlling plant development
Multiple aspects of plant morphology and growth process affect drought tolerance, including leaf area, stomatal density, vascular structure, root development and plant senescence.
The sunflower HD-ZIP I subfamily HAHB4, most similar to the Arabidopsis genes ATHB7 and ATHB12, is positively regulated by drought, ethylene and ABA (Manavella et al., 2008). HAHB4 is involved in a conserved mechanism associated with ethylene-mediated senescence that increases plant tolerance to drought stress. Because overexpressing HAHB4 transgenic Arabidopsis exhibited strong tolerance to water stress and lower sensitivity to external ethylene, the overexpression lines entered the senescence pathway later. Studies have shown that the expression of HAHB4 has inhibitory effects on ethylene synthesis-related genes (such as ACO and SAM) and ethylene signaling-related genes (such as ERF2 and ERF5) (Manavella et al., 2006; Manavella et al., 2008).
The HD-ZIP II gene EcHB1 positively regulates drought tolerance in Eucalyptus, which may be related to whole-tree transpiration changes. EcHB1 overexpression inhibits defoliation in Eucalyptus under drought stress, thus promoting growth (Sasaki et al., 2019). In the EcHB1-2-overexpressing lines, stem diameter was reduced and the number of leaves per stem area increased, but there was minimal effect on stomatal conductance (gs) (Sasaki et al., 2019). Stomatal density and size influence gs (Kiyomizu et al., 2019), so the small change in gs probably occurred because stomatal density and size were not changed (Sasaki et al., 2019). Since the leaf area was significantly reduced (0.5-fold), the leaf number was slightly increased (1.3-fold), and the tree-base transpiration rate was decreased (0.6-fold), this reduced the overall transpiration rate and moisture loss, which may prevent defoliation (Sasaki et al., 2019). Reduced defoliation during drought stress may aid rapid recovery after rehydration. In addition, the vascular structure of the stem of the EcHB1 overexpression line changed, the stem radial diameter decreased, and the cell wall thickness increased (Sasaki et al., 2019). These changes increase xylem cell stiffness, but decrease the water conductivity of stems and therefore also reduce water loss during drought stress (Lachenbruch and McCulloh, 2014; Santini et al., 2016). In conclusion, EcHB1 overexpression enhanced drought tolerance by reducing leaf area (Sasaki et al., 2019).
HD-ZIP proteins improve drought tolerance by increasing lignin biosynthesis. Rice OsTF1L belongs to the HD-ZIP IV subfamily (Bang et al., 2019). Lignin biosynthesis genes, such as PMEI, were up-regulated in OsTF1L-overexpressing lines and down-regulated in OsTF1L-silenced lines. Additionally, genes involved in the drought response and stomatal movement, such as ONAC022, was also up-regulated in OsTF1L-overexpression plants (Bang et al., 2019). PMEI encodes an enzyme that inhibits pectin demethyl esterification, and when overexpressed, can improve drought tolerance (An et al., 2008). ONAC022 is a rice NAC transcription factor that regulates stomatal closure to improve plant drought tolerance. OsTF1L overexpression enhances stomatal closure and improves plant drought tolerance (Hong et al., 2016; Bang et al., 2019). Researchers have identified five direct target genes of OsTF1L. Among them, Nodulin protein, DHHC4, poxN/PRX38, and CASPL5B1 are related to lignin synthesis, and the AAA-type ATPase is involved in drought tolerance. These five genes were up-regulated when OsTF1L was transiently expressed in rice protoplasts (Ranocha et al., 2010; Roppolo et al., 2011; Qi et al., 2013; Roppolo et al., 2014; Bang et al., 2019). In conclusion, OsTF1L enhances drought tolerance by regulating the expression of drought-responsive genes, stomatal movement and lignin biosynthesis (Bang et al., 2019).
Sunflower HaHB11 encodes the HD-ZIP I transcription factor. Heterologous overexpression of HaHB11 resulted in a leaf-rolled phenotype and longer roots in Arabidopsis compared to wild type. These plants experience increased stem lignification, faster stomatal closure and less water loss under drought stress (Cabello et al., 2017). HD-ZIP IV gene AtEDT1/HDG11 overexpression in kale increased primary root length, increased the number of root hairs and lateral roots, decreased water loss, and reduced H2O2 levels to enhance drought tolerance (Zhu et al., 2016). Arabidopsis AtEDT1/HDG11 is a HD-ZIP IV family gene. In addition, studies have shown that overexpressing AtHDG11 in wild-type Arabidopsis and tobacco have altered stomatal density and root development, as well as increased ABA content and improved photosynthesis, resulting in increased drought tolerance and biomass (Yu et al., 2008). Other studies have also shown that AtHDG11 improves drought tolerance in sweetpotato, tall fescue, rice, cotton and poplar due to a series of beneficial changes at the morphological and physiological level of transgenic plants, such as developed root systems and reduced stomatal density (Cao et al., 2009; Ruan et al., 2012; Yu et al., 2013; Yu et al., 2016).
HD-ZIP proteins regulate drought tolerance by inducing the accumulation of soluble substances and antioxidant enzymes
In addition to decreasing water loss by changes in plant morphology, plants counter drought stress by accumulating soluble substances such as proline; these soluble substances improve water retention and protect cell membranes (Jiang et al., 2020; Zhang et al., 2021a). Plants also increase cellular antioxidant capacity to limit damage due to reactive oxygen species production.
Heterologous overexpression of the maize HD-ZIP I family gene ZmHDZ9 in Arabidopsis leads to higher peroxidase (POD) and superoxide dismutase (SOD) activities and more soluble protein accumulation under drought stress (Qiu et al., 2022). Overexpression of the Arabidopsis HD-ZIP I gene AtHB13 and heterologous overexpression of the sunflower homolog HaHB1 in Arabidopsis enhanced drought tolerance (Hanson et al., 2001; Ebrahimian-Motlagh et al., 2017). AtHB13 may improve drought tolerance by directly regulating the expression of JUNGBRUNNEN1 (JUB1), which encodes a NAC transcription factor that regulates H2O2 levels (Ebrahimian-Motlagh et al., 2017). MtHB2 is an alfalfa HD-ZIP I family gene, and Arabidopsis lines heterologously overexpressing MtHB2 abolished the synthesis of proline and soluble sugars, increased the accumulation of ROS and MDA, resulting in reduced osmotic regulation and greater oxidative damage under stress than in the wild type (Song et al., 2012). Therefore, HD-ZIP proteins function in positively and negatively regulating plant responses to drought stress; understanding the downstream regulatory networks underlying these positive and negative effects has important implications for improving drought tolerance in crops without incurring yield penalties.
HD-ZIP proteins in salinity stress responses
High salinity inhibits plant growth and reduces crop productivity by limiting plants’ ability to take up water and causing toxic ionic imbalances and ROS accumulation. HD-ZIP transcription factors regulate salt stress responses through multiple pathways (Wang et al., 2017; Tang et al., 2019a).
HD-ZIP proteins participate in the salt stress response by regulating ion homeostasis, compatible solute concentrations, and ROS scavenging
Plant salt stress responses include activating ion channels to regulate ion homeostasis, accumulating solutes to mitigate osmotic effects, and (as with drought stress responses), activating antioxidant defenses to limit oxidative damage.
The apple HD-ZIP I gene MdHB7-like positively regulates the salt tolerance of apple. Under salt stress, the relative electrolyte leakage (REL) and MDA content of leaves of MdHB7-like overexpressing lines were reduced compared with wild type (Zhao et al., 2021). Furthermore, the transgenic apple had lower H2O2 and concentrations and higher SOD and POD activities; SOS1, SOS2, SOS3, and NHX1 expression was higher; the proline and soluble sugar content was significantly increased; and the Na+/K+ ratio in roots, stems, and leaves was significantly lower in the MdHB7-like overexpression lines than in the wild type (Zhao et al., 2021). Under salt and drought stress, cotton lines heterologously overexpressing AtHDG11 significantly reduced MDA levels and elevated SOD and CAT activities, thus protecting transgenic plants from oxidative damage by enhancing their ROS scavenging ability (Yu et al., 2016). Proline and soluble sugars are two important and effective compatible osmo-regulatory substances in higher plants and play an important role in the oxidative stress response of plants (Yu et al., 2016). Analysis of wild type and JcHDZ07-overexpressing lines under salt stress revealed that transgenic Arabidopsis overexpressing JcHDZ07 exhibited lower proline content, lower CAT and SOD activities, and lower survival rates than wild-type. This indicated that overexpression of JcHDZ07 enhanced the sensitivity of transgenic Arabidopsis to salinity stress (Tang et al., 2019a). JcHDZ16 is a HD-ZIP I gene in physic nut. Compared with wild-type, rice lines heterologous overexpressing JcHDZ16 had decreased proline content and activities of antioxidant enzymes such as catalase (CAT) and SOD and increased REL and MDA content, thus, the sensitivity of transgenic rice to salt stress was improved. In addition, the salt stress-responsive genes OsHKT1;1, OsAPX2, OsDREB2A and OsP5CS were expressed at lower levels in rice overexpressing JcHDZ16 compared with wild type, suggesting that JcHDZ16 negatively regulates ion balance and ROS scavenging under salt stress (Tang et al., 2019b). Overexpression of PsnHDZ63, the HD-ZIP I gene of Populus simonii × P. nigra, improves the salt tolerance of poplar (Guo et al., 2021). Under salt stress, PsnHDZ63 positively regulates SOD and POD activities and reduces MDA content (Guo et al., 2021). Tomato HD-ZIP I subfamily gene SlHB2, whose expression is induced by ABA, 1-aminocyclopropane-1-carboxylic acid (ACC), methyl-jasmonic acid (MeJA), drought, high salt, low temperature and damage. SlHB2 negatively regulates plant drought and high salinity tolerance. Compared with the wild type, under drought and salt stress, the chlorophyll and water contents in the leaves of the tomato seedlings of the SlHB2-RNAi line were higher, and the water loss rate and MDA content were lower, indicating that the SlHB2-RNAi line enhanced its stress resistance (Hu et al., 2017). The HD-ZIP II gene CaHB1 of pepper enhances salt tolerance when heterologously expressed in transgenic tomato plants (Oh et al., 2013). The photosynthetic capacity was higher, and dehydrin transcripts were up-regulated 3- to 5-fold in the CaHB1-overexpressing lines compared to wild type plants (Oh et al., 2013).
HD-ZIP transcription factors regulate salt tolerance through the ABA signaling pathway
ABA regulates plant responses to many abiotic stresses and its effects on drought and salt stress tolerance target similar types of damage induced by the two stresses, such as water balance and oxidate stress.
GhHB1 is a cotton (Gossypium hirsutum) HD-ZIP I gene. Studies have shown that the expression of cotton GhHB1 is up-regulated under ABA and NaCl treatments, suggesting that it may be involved in plant responses to salt stress and ABA signaling (Ni et al., 2008). Reduced salt tolerance in rice overexpressing the maize HD-ZIP I gene ZmHDZ1 (Wang et al., 2017). In ZmHDZ1-overexpressing seedlings, exogenous ABA up-regulated ZmHDZ1 expression and the transgenic seedlings had increased ABA sensitivity compared to WT (Wang et al., 2017). These results demonstrate that ZmHDZ1 negatively regulates salt stress tolerance via an ABA-dependent signal transduction pathway (Wang et al., 2017). Salt stress and ABA treatment induced the expression of maize HD-ZIP I gene ZmHDZ10. Overexpression of ZmHDZ10 in rice enhanced its resistance to drought and salt stress and increased its sensitivity to ABA. In addition, transgenic Arabidopsis overexpressing ZmHDZ10 exhibited increased drought and salt tolerance and altered expression of stress/ABA-responsive genes, such as RD29B, P5CS1, Responsive to Dehydration 22 (RD22) and ABI1. These results suggest that the transcriptional regulator ZmHDZ10 can enhance salt and drought tolerance in plants through an ABA-dependent signaling pathway (Zhao et al., 2014). OsHOX22 and OsABI5 negatively regulate plant salt tolerance, while OsLEA3 and OsRAB16A are downstream abiotic stress response genes that are often used as markers (Duan and Cai, 2012; Zhang et al., 2012; Li et al., 2016). Compared with wild-type, ZmHDZ1-overexpressing rice showed higher expression of OsHOX22 and OsABI5, but lower expression of OsRAB16A and OsLEA3 (Wang et al., 2017). OsABI5 regulates ABA signaling, and its overexpression in rice increased sensitivity to salt stress (Zou et al., 2008). The proline content of alfalfa overexpressing the HD-ZIP I gene MsHB7 was lower than that of wild-type alfalfa, which means that overexpression of MsHB7 increases the sensitivity to salt stress (Li et al., 2022). The expression levels of the ABA-responsive genes MsSnRK2.6 and MsABI1 were lower in MsHB7-overexpressing lines after salt treatment compared to wild type (Li et al., 2022). MsHB7 is induced via an ABA-dependent signaling pathway under salt stress. In addition to down-regulation of MsSnRK2.6 and MsABI1, MsHB7 inhibits the antioxidant enzyme-encoding genes MsCatalase4 and MsPxdC (Li et al., 2022). P5CS is the rate-limiting enzyme in proline biosynthesis, and P5CS expression is also dependent on ABA signaling under salt stress (Verslues and Bray, 2006). Transcript levels of MsP5CS2 in MsHB7-overexpressing lines were lower compared to those in wild type, suggesting that MsHB7 down-regulates MsP5CS2 through an ABA-dependent signaling pathway (Li et al., 2022). The pepper HD-ZIP I genes CaHDZ03 and CaHDZ10 are salt-inducible expression genes (Zhang et al., 2021b). CAHDZ03 contains two closely connected ABA action elements, indicating its potential role in the ABA signaling pathway (Zhang et al., 2021b).
Heterologously overexpressing the pepper HD-ZIP II gene CaHB1 in tomato improved salt tolerance. Compared with wild-type, CaHB1-overexpressing tomato lines had higher photosynthetic capacity and also up-regulated the transcript level of dehydrin (Oh et al., 2013). CaHB1 was not induced by ABA, implying that it may function in an ABA-independent manner.
Taken together, HD ZIP transcription factors play an essential role in plant salt resistance. It participates in salt stress response by enhancing ion homeostasis, osmotic stress and antioxidant capacity. HD-ZIP TFs can also be involved in the regulation of salt stress responses through ABA-mediated signaling. In addition, HD ZIP transcription factors were also found to be involved in plant responses to salt stress through a pathway independent of ABA signaling, such as regulating photosynthesis.
HD-ZIP proteins in low-temperature stress
Cold stress limits plant development (Aslam et al., 2022) and leads to ROS and MDA production in cells. Low temperature stress affects cell membrane fluidity, consequently hindering photosynthesis, metabolism, and material transport (Janská et al., 2010; Ye et al., 2017). Expression profiling under abiotic stresses revealed that all 22 SlHZI genes in tomato respond to cold stress (Zhang et al., 2014b). Five HD-ZIP I genes were isolated from wheat, from TaHDZipI-1 to TaHDZipI-5 (Kovalchuk et al., 2016). Among them, when TaHDZipI-2 is overexpressed in barley (Hordeum vulgare), it affects cold tolerance by regulating cold-regulated expression of gene (COR gene) HvTMC-AP3, a putative chloroplastic amino acid selective channel protein (Kovalchuk et al., 2016). TaHDZipI-5 is induced by cold and expressed at higher levels in flowers and early developing grains, indicating that TaHDZipI-5 participates in cold tolerance during flowering (Yang et al., 2018). Using computer genome search, the researchers identified 37 HD-ZIP genes in Dendrobium officinale (DoHDZs), 34 DoHDZs were expressed under cold treatment, and 14 of them were significantly differentially expressed compared to control conditions, among them, 9 down-regulated genes and 5 up-regulated genes. The expression level of HD-ZIP I gene DHDZ5 was significantly down-regulated under cold treatment, while HD-ZIP I gene DoHDZ9 and HD-ZIP II gene DoHDZ12 were significantly up-regulated, suggesting that they may be key candidates for cold stress response (Yang et al., 2022).
HD-ZIP proteins participate in the low-temperature response by regulating cell membrane stability
The sunflower HD-ZIP I gene HaHB1 and its Arabidopsis homolog AtHB13 are associated with cold tolerance (Gong et al., 2019). Under freezing conditions, some antifreeze proteins (AFPs), such as PR2/PR4, accumulate in the apoplast of HaHB1-overexpressing Arabidopsis to improve low-temperature tolerance (Gong et al., 2019). In addition, transient transfection of sunflower and soybean (Glycine max) with HaHB1 and AtHB13 up-regulated their homologous target genes in these plants, indicating conservation of the cold-response pathway. This suggests that HaHB1 and AtHB13 are involved in cell membrane stability (Cabello et al., 2012; Cabello et al., 2017).
HD-ZIP proteins regulate low-temperature tolerance through ROS scavenging
Pepper CaATHB-12 encodes an HD-ZIP I transcription factor (Zhang et al., 2020). CaATHB-12-silenced fruit had high expression and activity of antioxidant enzymes, such as SOD and POD, suggesting its negative role in ROS scavenging. In addition, CaATHB-12 regulates carotenoid biosynthesis; CaATHB-12-silenced pepper fruit had significantly lower carotenoid contents than wild-type fruit. Carotenoids participate in ROS scavenging (Lado et al., 2015). The expression levels of the stress-responsive genes AtDREB2A, AtMYB44, AtRD29A and AtGPX3 were all lower in plants overexpressing CaATHB-12 after cold stress than in wild type (Zhang et al., 2020). Transgenic Arabidopsis increased carotenoids, flavonoids, and phenolic compounds under cold stress by upregulating carotenoid biosynthesis genes (CaPSY and CaLCYB) (Zhang et al., 2020). Heterologous overexpression of CaPSY results in increased carotenoid accumulation in transgenic Arabidopsis (Rodriguez-Uribe et al., 2012; Zhang et al., 2020). CaATHB-12 overexpression increases the carotenoid content under normal conditions; however, CaATHB-12 overexpression impairs ROS scavenging under cold stress (Zhang et al., 2020).
HD-ZIP proteins in regulating harmful metal stress
Aluminum (Al) stress due to excessive Al3+ limits plant growth and crop production in acidic soils. The earliest and most striking feature of Al poisoning is rapidly inhibited root elongation, which leads to reduced root size, restricted uptake of mineral nutrients and water, and ultimately reduced yield (Liu et al., 2014). The root transition zone (TZ) is located between the apical meristem of the root tip and the basal elongation zone, and this TZ senses Al toxicity (Rangel et al., 2007; Yang et al., 2014; Liu et al., 2020). Al mainly accumulates in the cell wall and changes cell wall properties, reduces its extensibility, and also prevents cell expansion in the root elongation zone (Horst et al., 2010). Cell wall pectin and hemicellulose bind Al (Blamey et al., 1990; Chang et al., 1999; Yang et al., 2011). Cell wall composition and structural modifications, especially those of pectin and xyloglucan oligosaccharides, affect the Al binding capacity and are important for Al stress tolerance (Lan et al., 2021).
Only a few TFs have been reported to be involved in Al tolerance. Two HD-ZIP I proteins, AtHB7 and AtHB12, were associated with Al tolerance via phenotypic analysis of overexpression lines and loss-of-function mutants (Söderman et al., 1996; Liu et al., 2020). AtHB7 and AtHB12 are closely related but their functions in Al resistance are diametrically opposed; AtHB7 has a positive role and AtHB12 has a negative role (Liu et al., 2020). AtHB7 and AtHB12 have opposite regulatory effects on Al stress tolerance, which is mediated by affecting Al accumulation in root cell walls. ATHB12 increased the expression of the cell wall-related genes DWARF4 (DWF4) and EXPANSIN A10 (EXPA10) (Hur et al., 2015; Liu et al., 2020). Al accumulated in athb7 roots, but did not accumulate in athb12 roots. AtHB7 and ATHB12 form heterodimers and appear to inhibit each other in response to Al stress. The athb7 athb12 double mutants are similar to the wild type in Al tolerance. Therefore, AtHB7 and AtHB12 may compete to regulate downstream Al-response genes (Liu et al., 2020). In addition, manganese (Mn) toxicity in acidic soils is also an important factor limiting crop yield. The researchers identified 63 and 87 Mn toxicity-response genes in Citrus sinensis and Citrus grandis, respectively. Citrus grandis is not resistant to Mn toxicity, while Citrus sinensis is resistant to Mn toxicity. Among them, manganese toxicity-induced root HD-ZIP I proteins (TDF #170-1 and 170-1k) showed a trend of up-regulation in both species (Zhou et al., 2016).
The heavy metal cadmium (Cd) is toxic to most organisms, including plants and humans (Ding et al., 2018). Cd is easily absorbed by plant roots, causing severe structural and functional changes, inhibiting seed germination and root growth, and Cd stress also increases ROS production (Farid et al., 2013; Ali et al., 2014; Rehman et al., 2022). HD-ZIP genes and their upstream microRNAs (miRNAs) are involved in Cd accumulation and tolerance. Under Cd stress, rice miR166 was down-regulated, while its target, the HD-ZIP III gene OsHB4, was up-regulated. Overexpressing miR166 in rice under Cd stress improved growth compared to the wild type, which was related to decreased Cd-induced oxidative damage and a significant increase in rice chlorophyll content. Overexpressing miR166 reduced Cd transport between roots and shoots (Ding et al., 2018). Accumulation of Cd in shoots and grains of rice mainly depends on the transport of xylem from roots to shoots (Uraguchi et al., 2009). This indicates that OsHB4 is a target of miR166, and OsHB4 regulates Cd translocation and tolerance in rice. Other genes (such as OsHMA2 and OsHMA3) may also be involved in Cd absorption, storage, transport, and detoxification in rice.
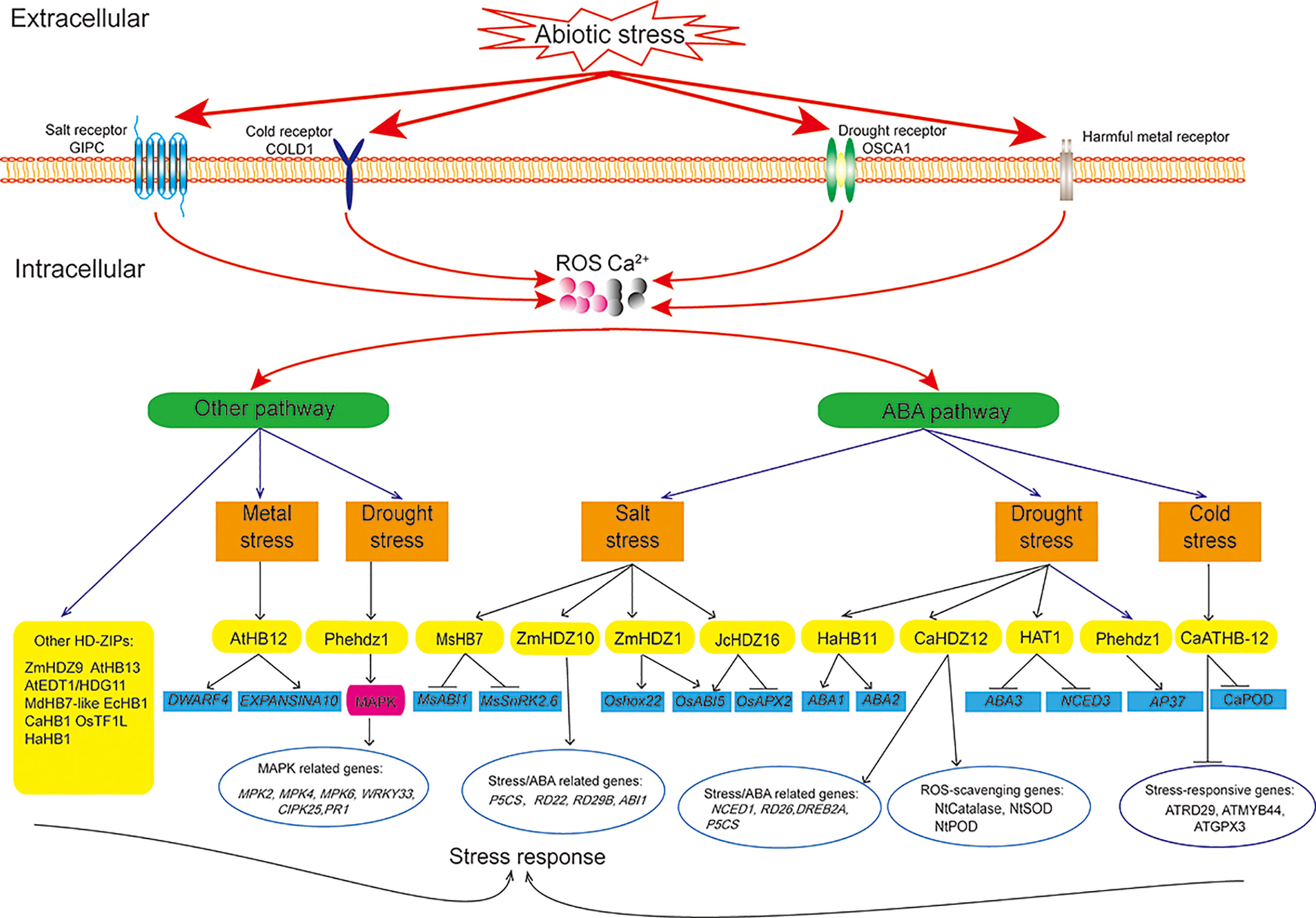
Mechanism of HD-ZIP proteins in regulating abiotic stress tolerance
Plants rapidly respond to adverse conditions via interconnected networks controlled by signaling cascades. Stress response involves signal perception and transduction and expression of stress-responsive genes (Baillo et al., 2019). Upon sensing a signal, two major signaling cascades in plants, the mitogen-activated protein kinase (MAPK) and calcium-dependent protein kinase (CDPK) pathways, are activated (Hernandez-Garcia and Finer, 2014; Erpen et al., 2018). ROS participate in signal transduction through MAPKs, and Ca2+ participates in signal transduction through the CDPK pathway, which then induce multiple pathways through the kinase cascade (Gilroy et al., 2016; Qi et al., 2018). These activated pathways regulate the expression of HD-ZIP genes, which then bind to cis-elements on target gene promoters to regulate downstream gene expression and subsequent biosynthesis of secondary metabolites (Baillo et al., 2019). Therefore, HD-ZIP transcription factors play a key role in responding to stress by directly regulating downstream target genes or interacting with other transcription factors (Kurusu et al., 2015; Gong et al., 2019; Chen and Yang, 2020; Qiu et al., 2022).
Interactions between the ABA pathway and other pathways (Yao et al., 2020), and interactions between ABA-dependent and ABA-independent pathways (Zhang et al., 2012; Cabello et al., 2017; Gong et al., 2019; Tang et al., 2019b) are involved in the stress response. ABA is rapidly synthesized under different stresses (Marcolino-Gomes et al., 2013). HD-ZIP proteins inhibit or promote ABA synthesis. HAT1 negatively regulates drought tolerance in Arabidopsis by directly binding to the promoters of the ABA biosynthesis genes ABA3 and NCED (Tan et al., 2018). HaHB11 positively regulates drought tolerance by regulating the ABA biosynthesis genes ABA1 and ABA2 (Ding et al., 2009; Msanne et al., 2011; Cabello et al., 2017). CaHDZ12 positively regulates drought tolerance by decreasing CaPP2C expression (Sen et al., 2017). MsHB7 negatively regulates salt tolerance by inhibiting the expression of ABA-response genes MsSnRK2.6 and MsABI1 (Li et al., 2022). ZmHDZ1 negatively regulates salt tolerance in rice by increasing Oshox22 and OsABI5 expression (Wang et al., 2017). In addition to the ABA signaling pathway, HD-ZIP transcription factors such as Phehdz1 may also affect secondary metabolism through an integration of ABA and MAPK signaling pathways to regulate drought tolerance (Gao et al., 2021).
Abiotic stresses stimulate ROS production in plant cells. HD-ZIP proteins also improve stress tolerance through promoting ROS scavenging. ZmHDZ9-overexpression in Arabidopsis resulted in higher SOD and POD activities under drought stress (Qiu et al., 2022). AtHB13 directly regulates the expression of downstream JUB1 to reduce cellular H2O2 levels under drought stress (Ebrahimian-Motlagh et al., 2017). JcHDZ16 negatively regulates salt tolerance by decreasing OsAPX2 expression and increasing OsABI5 expression (ZHANG et al., 2014a; Tang et al., 2019b). HD-ZIP proteins protect macromolecules and subcellular structures by promoting the accumulation of osmotic substances, such as proline (Szabados and Savouré, 2010). For example, MdHB7-like increased proline content in plants under salt stress, while AtEDT1/HDG11 increased proline content in plants under drought conditions (Zhu et al., 2016; Zhao et al., 2021).
HD-ZIP transcription factors also promote stress tolerance by regulating plant growth and development. EcHB1 altered the anatomical structure of the stem vasculature, reduced the stem radial diameter, and increased cell wall thickness to improve drought stress (Sasaki et al., 2019). OsTF1L regulates lignin biosynthesis and enhances pore closure to improve drought (Bang et al., 2019). CaHB1-overexpression reduced chlorophyll loss under salt stress (Oh et al., 2013). HaHB1 enhanced cold tolerance by regulating cell membrane stability (Gong et al., 2019). AtHB12 negatively regulates tolerance to Al by increasing the expression of cell wall-related genes (EXPANDINA10 and DWARF4) (Liu et al., 2020). Pectin and hemicellulose in the cell wall bind Al to ameliorate Al toxicity (Blamey et al., 1990; Chang et al., 1999; Yang et al., 2011; Hur et al., 2015; Liu et al., 2020).
Different stresses may provoke similar responses. One HD-ZIP transcription factor may regulate multiple downstream genes, and one stress-response gene may be regulated by multiple HD-ZIP transcription factors. For example, PheHDZ1 regulated the expression of numerous stress- and ABA-response genes, such as AP37, DREB1, ERF3, OsMYB3, OsNAC10, and OsWRKY47, to improve drought tolerance (Gao et al., 2021). CaHDZ12 regulated the expression of tobacco ABA- and stress-response genes (NtNCED1, NtRD26, NtDREB2A, and NtP5CS) and antioxidant enzyme genes (NtCatalase, NtSOD, and NtPOD) to improve drought tolerance in transgenic tobacco (Sen et al., 2017). The expression of stress-response genes (AtDREB2A, AtMYB44, AtRD29A, and ATGPX3) were lower in CaATHB-12-overexpression transgenic Arabidopsis under cold stress (Zhang et al., 2020).
Based on the above discussion on different subfamily proteins in stress tolerance, the conservative and different role of different subfamilies HD-ZIP proteins in resistance to abiotic stress in different species such as Arabidopsis, maize and rice was intuitively shown in a phylogenetic tree (Figure 3). Through phylogenetic tree analysis, it was found that HD-ZIP I proteins played an extremely important role in plant response to abiotic stress. Most HD-ZIP I subfamily proteins in plants such as model plant Arabidopsis and model crop maize respond to abiotic stresses such as salinity, drought, and osmosis through ABA-dependent or ABA-independent pathways (Hanson et al., 2001; Himmelbach et al., 2002; Wang et al., 2017). Followed by less HD-ZIP II subfamily proteins also play a role in plant responses to various abiotic stresses (Liu et al., 2016; Li et al., 2019d). Although HD-ZIP III and HD-ZIP IV subfamily genes are also characterized under stress, such as HD-ZIP III gene StHOX39 is highly expressed under low temperature conditions (Li et al., 2019f). Overexpression of HD-ZIP IV gene OsTF1L and AtEDT1/HDG11 both improved the drought tolerance of transgenic plants (Zhu et al., 2016; Bang et al., 2019). The HD-ZIP III and HD-ZIP IV genes are mainly involved in the regulation of plant development (Baima et al., 1995; Di Cristina et al., 1996; Masucci et al., 1996; Prigge et al., 2005; Khosla et al., 2014; Zhu et al., 2016).
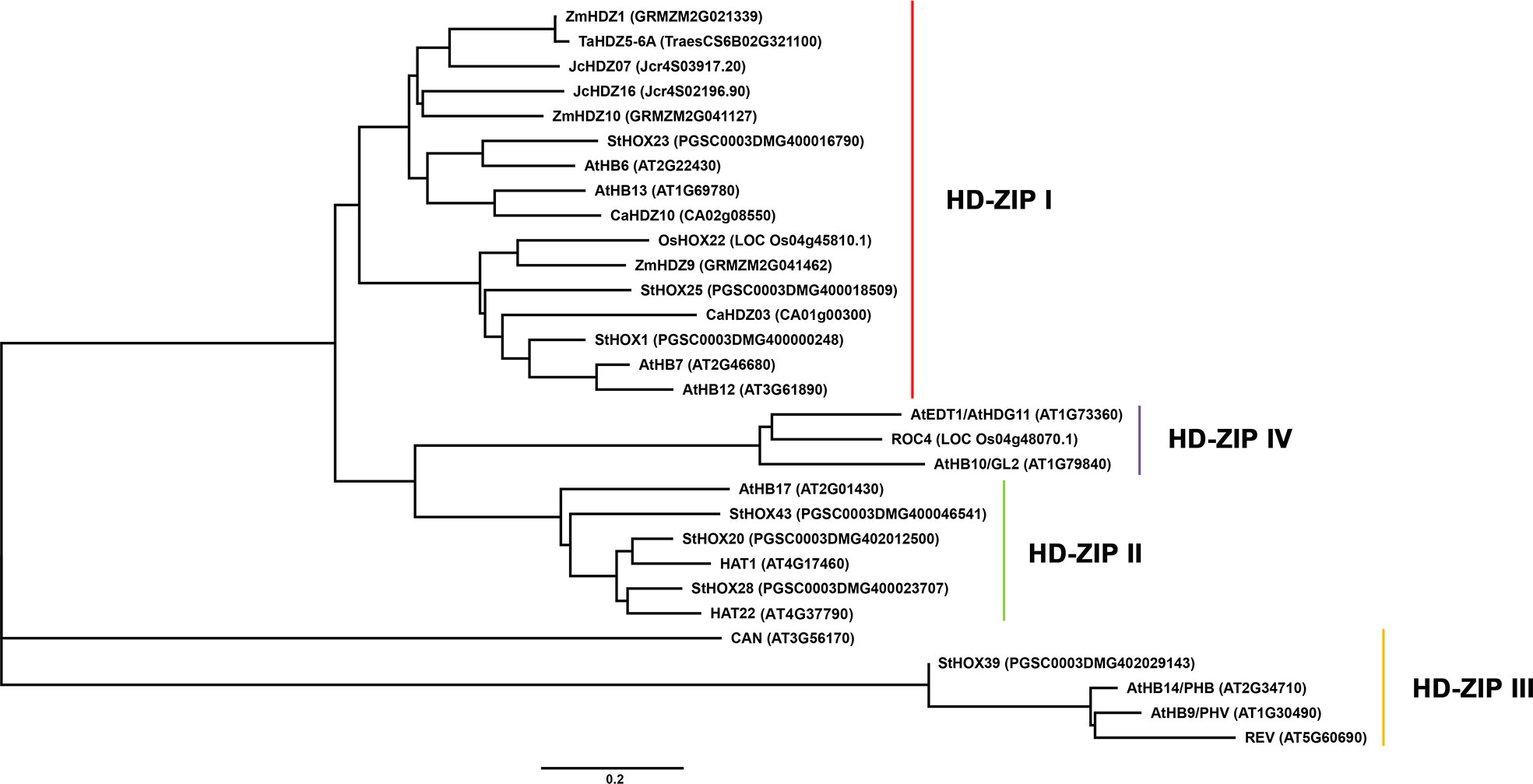
Figure 3 Phylogenetic tree analysis of HD-ZIP proteins in different species. The amino acid sequences of HD-ZIP proteins in different species were obtained from NCBI (https://www.ncbi.nlm.nih.gov/). MEGA 5.1 was used to construct a Neighbor-Joining phylogenetic tree with 1,000 bootstrap replicates.
Through the above sorting and summary, the functions and regulation mechanisms of different types of HD-ZIP proteins involved in abiotic stress were detailed in Supplementary Table 1. And the regulatory network of HD-ZIP proteins in abiotic stress response was shown in Figure 4.
Conclusions and future prospects
Considering that the world’s population is expected to reach 9 billion by 2050, improving crop tolerance to abiotic stresses is essential to meet the world’s projected food and energy needs. TFs have important roles in stress tolerance. Significant progress has been made in the role of transcription factor families in abiotic and biotic stress responses. In particular, HD-ZIP proteins play important roles in plant development and tolerance to abiotic stresses.
In recent years, great progress has been made in the study of plant HD-ZIP proteins. Since KNOTTED1 was first isolated from maize, researchers have now identified many HD-ZIP transcription factors from various plants. Although HD-ZIP family members have been extensively investigated in model plants and model crops, the molecular mechanisms in regulating abiotic stress are still insufficient. The upstream and downstream regulatory mechanisms of many HD-ZIP transcription factors remain unclear. Although genetic analysis shows that their stress response depends on the activation or inhibition of downstream target genes, they may also be regulated by their upstream genes, or work together through interacting proteins. However, scientific questions such as what is the upstream regulatory signal, how to form interacting proteins, how to regulate the expression of downstream genes, and what the regulatory network is all need to be solved. In addition, the response of HD-ZIP proteins to stress stimuli is extremely complex. Overexpressing a single HD-ZIP transcription factor may promote or inhibit the expression of multiple downstream genes. Different transcription factors may regulate the same gene, and a single transcription factor may respond to different abiotic stresses (Baillo et al., 2019). Identifying individual transcription factor genes at the molecular level is challenging. Multiple transcription factors can be studied by combining research methods and stresses to clarify the interactions between different transcription factors, rather than studying a single transcription factor and stress. Therefore, the development of biotechnologies such as CRISPR-Cas9, genome-wide association analysis, genome editing and single-cell sequencing will be very helpful to study the mechanism of HD-ZIP proteins in different species to improve plant abiotic stress tolerance. Chromatin immunoprecipitation followed by sequencing (ChIP-seq) and other biochemical technologies can be used to study the accurate binding sites of HD-ZIP transcription factors, and find the downstream functional genes regulated by them and the signal transduction pathways involved in plant stress tolerance.
Furthermore, it is important to study the mechanisms of stress adaptation in xerophytes and halophytes, especially HD-ZIP TFs in these plants. Due to the long-term growth and development of plants under adversity, stress tolerant plants have developed adaptability in their morphological anatomy, physiology and biochemistry, and can complete their life history under extreme adversity conditions. It is possible to study the stress tolerance function and mechanism of HD-ZIP transcription factors in xerophytes and halophytes. Isolating these key stress tolerance genes and transforming crops is of great significance for improving the stress tolerance of crops. Therefore, elucidating the molecular network regulation mechanism of HD-ZIP transcription factors in stress tolerant plants is also the focus of future scientific research, which can lay a foundation for the subsequent analysis of the biological functions of important plant transcription factors, and at the same time help to provide theoretical support and technical guidance for efficient screening of new crop varieties with resistance to abiotic stress.
Many plant developmental traits are also closely related to stress tolerance. HD-ZIP proteins are involved in the differentiation of plant trichomes and root hairs, and trichomes and root hairs participate in stress tolerance (Kang et al., 2014; Yang et al., 2015). Many HD-ZIP proteins participate in anthocyanins biosynthesis, and anthocyanins are involved in numerous abiotic stresses (Mbarki et al., 2018; Cirillo et al., 2021). In particular, the proteins of the HD-ZIP III and HD-ZIP IV subfamilies play a more important role in plant development. In the follow-up studies, the developmental phenotype and plant stress tolerance can be combined to better study the mechanism of these different HD-ZIP proteins in stress tolerance.
HD-ZIP proteins with better stress tolerance effect have been identified and characterized in more and more model plants and model crops. However, most of these studies were conducted in the laboratory. Under field conditions, crops are subject to a more complex and harsh environments. Therefore, there is an urgent need to test candidate these HD-ZIP proteins under field conditions to improve abiotic stress tolerance. Although analyzing plants overexpressing transcription factor genes under specific stresses is very informative, we need to assess the impact of these approaches on crop yield and productivity. Therefore, future research should not only explore whether stress-related transcription factor genes improve stress tolerance and growth, but also their effects on crop yield under natural field conditions.
Authors contributions
YL and ZY wrote this manuscript. YZ, JG, LL and CW participated in the writing and modification of this manuscript. GH and BW conceptualized the idea. All authors read and approved the final manuscript.
Funding
This work was supported by National Natural Science Research Foundation of China (project No. 32000209 and 32170301), Natural Science Research Foundation of Shandong Province (project No. ZR2020QC031) and China Postdoctoral Science Foundation (project No. 2020M672114).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1027071/full#supplementary-material
Abbreviations
TF, transcription factor; HD-ZIP, homeodomain leucine zipper protein; HD, homologous domain; LZ, leucine zipper; DNA, Deoxyribonucleic acid; mRNA, Messenger RNA; ROS, reactive oxygen species; HB, homeobox; N-Term, N-term consensus sequence; START, steroidogenic acute regulatory proteinlipid transfer domain lipid transfer domain; MEKHLA, Met-Glu-Lys- His-Leu-Ala domain; SAD, START associated domain; PCR, polymerase chain reaction; ABA, Abscisic acid; MDA, malondialdehyde; ABRE, ABA response element; qRT-PCR, Quantitative Real-time PCR; WT, wild-type; DEGs, differentially expressed genes; KEGG, Kyoto Encyclopedia of Genes and Genomes; ChIP, Chromatin Immunoprecipitation; RNA, Ribonucleic acid; RNA-seq, RNA-sequencing; ChIP-seq, ChIP-sequencing; H2O2, hydrogen peroxide; SOD, superoxide dismutase; POD, peroxidase; REL, relative electrolyte leakage; CAT, catalase; AFPs, antifreeze proteins; Al, Aluminum; TZ, transition zone; EZ, elongation zone; Cd Cadmium; miRNAs, microRNAs; Ca2+, calcium ions; MAPKs, mitogen-activated protein kinases; CDPK, calcium-dependent protein kinase; MEP, 2-C-Methyl-D-erythritol 4-phosphate; ZEP, Zeaxanthin cyclooxygenase; NCED, 9-cis-epoxycarotenoid dioxygenase; SCAD, short-chain acyl-CoA dehydrogenase.
References
Agarwal, P. K., Gupta, K., Lopato, S., Agarwal, P. (2017). Dehydration responsive element binding transcription factors and their applications for the engineering of stress tolerance. J. Exp. Bot. 68, 2135–2148. doi: 10.1093/jxb/erx118
Aglawe, S. B., Fakrudin, B., Patole, C. B., Bhairappanavar, S. B., Koti, R. V., Krishnaraj, P. U. (2012). Quantitative RT-PCR analysis of 20 transcription factor genes of MADS, ARF, HAP2, MBF and HB families in moisture stressed shoot and root tissues of sorghum. Physiol. Mol. Biol. Plants 18, 287–300. doi: 10.1007/s12298-012-0135-5
Ali, B., Qian, P., Jin, R., Ali, S., Khan, M., Aziz, R., et al. (2014). Physiological and ultra-structural changes in Brassica napus seedlings induced by cadmium stress. Biol. plantarum 58, 131–138. doi: 10.1007/s10535-013-0358-5
An, S. H., Sohn, K. H., Choi, H. W., Hwang, I. S., Lee, S. C., Hwang, B. K. (2008). Pepper pectin methylesterase inhibitor protein CaPMEI1 is required for antifungal activity, basal disease resistance and abiotic stress tolerance. Planta 228, 61–78. doi: 10.1007/s00425-008-0719-z
Ariel, F., Diet, A., Verdenaud, M., Gruber, V., Frugier, F., Chan, R., et al. (2010). Environmental regulation of lateral root emergence in Medicago truncatula requires the HD-zip I transcription factor HB1. Plant Cell 22, 2171–2183. doi: 10.1105/tpc.110.074823
Ariel, F. D., Manavella, P. A., Dezar, C. A., Chan, R. L. (2007). The true story of the HD-zip family. Trends Plant Sci. 12, 419–426. doi: 10.1016/j.tplants.2007.08.003
Aslam, M., Fakher, B., Ashraf, M. A., Cheng, Y., Wang, B., Qin, Y. (2022). Plant low-temperature stress: Signaling and response. Agronomy 12, 702. doi: 10.3390/agronomy12030702
Baillo, E. H., Kimotho, R. N., Zhang, Z., Xu, P. (2019). Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes 10, 771. doi: 10.3390/genes10100771
Baima, S., Nobili, F., Sessa, G., Lucchetti, S., Ruberti, I., Morelli, G. (1995). The expression of the athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121, 4171–4182. doi: 10.1242/dev.121.12.4171
Baima, S., Possenti, M., Matteucci, A., Wisman, E., Altamura, M. M., Ruberti, I., et al. (2001). The Arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol. 126, 643–655. doi: 10.1104/pp.126.2.643
Bang, S. W., Lee, D. K., Jung, H., Chung, P. J., Kim, Y. S., Choi, Y. D., et al. (2019). Overexpression of OsTF1L, a rice HD-zip transcription factor, promotes lignin biosynthesis and stomatal closure that improves drought tolerance. Plant Biotechnol. J. 17, 118–131. doi: 10.1111/pbi.12951
Bao, A., Burritt, D. J., Chen, H., Zhou, X., Cao, D., Tran, L. P. (2019). The CRISPR/Cas9 system and its applications in crop genome editing. Crit. Rev. Biotechnol. 39, 321–336. doi: 10.1080/07388551.2018.1554621
Blamey, F., Edmeades, D., Wheeler, D. (1990). Role of root cation–exchange capacity in differential aluminum tolerance of lotus species. J. Plant Nutr. 13, 729–744. doi: 10.1080/01904169009364112
Cabello, J. V., Arce, A. L., Chan, R. L. (2012). The homologous HD-zip I transcription factors HaHB1 and AtHB13 confer cold tolerance via the induction of pathogenesis-related and glucanase proteins. Plant J. 69, 141–153. doi: 10.1111/j.1365-313X.2011.04778.x
Cabello, J. V., Giacomelli, J. I., Gómez, M. C., Chan, R. L. (2017). The sunflower transcription factor HaHB11 confers tolerance to water deficit and salinity to transgenic Arabidopsis and alfalfa plants. J. Biotechnol. 257, 35–46. doi: 10.1016/j.jbiotec.2016.11.017
Cai, Y., Bartholomew, E. S., Dong, M., Zhai, X., Yin, S., Zhang, Y., et al. (2020). The HD-ZIP IV transcription factor GL2-LIKE regulates male flowering time and fertility in cucumber. J. Exp. Bot. 71, 5425–5437. doi: 10.1093/jxb/eraa251
Cao, Y. J., Wei, Q., Liao, Y., Song, H. L., Li, X., Xiang, C. B., et al. (2009). Ectopic overexpression of AtHDG11 in tall fescue resulted in enhanced tolerance to drought and salt stress. Plant Cell Rep. 28, 579–588. doi: 10.1007/s00299-008-0659-x
Capella, M., Ribone, P. A., Arce, A. L., Chan, R. L. (2015). Arabidopsis thaliana HomeoBox 1 (AtHB1), a homedomain-leucine zipper I (HD-zip I) transcription factor, is regulated by PHYTOCHROME-INTERACTING FACTOR 1 to promote hypocotyl elongation. New Phytol. 207, 669–682. doi: 10.1111/nph.13401
Carabelli, M., Possenti, M., Sessa, G., Ruzza, V., Morelli, G., Ruberti, I. (2018). Arabidopsis HD-zip II proteins regulate the exit from proliferation during leaf development in canopy shade. J. Exp. Bot. 69, 5419–5431. doi: 10.1093/jxb/ery331
Carlsbecker, A., Lee, J. Y., Roberts, C. J., Dettmer, J., Lehesranta, S., Zhou, J., et al. (2010). Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465, 316–321. doi: 10.1038/nature08977
Chai, W., Si, W., Ji, W., Qin, Q., Zhao, M., Jiang, H. (2018). Genome-wide investigation and expression profiling of HD-zip transcription factors in foxtail millet (Setaria italica L.). BioMed. Res. Int. 2018, 8457614. doi: 10.1155/2018/8457614
Chang, Y. C., Yamamoto, Y., Matsumoto, H. (1999). Accumulation of aluminium in the cell wall pectin in cultured tobacco (Nicotiana tabacum L.) cells treated with a combination of aluminium and iron. Plant Cell Environ. 22, 1009–1017. doi: 10.1046/j.1365-3040.1999.00467.x
Chen, Q., Yang, G. (2020). Signal function studies of ROS, especially RBOH-dependent ROS, in plant growth, development and environmental stress. J. Plant Growth Regul. 39, 157–171. doi: 10.1007/s00344-019-09971-4
Chew, W., Hrmova, M., Lopato, S. (2013). Role of homeodomain leucine zipper (HD-zip) IV transcription factors in plant development and plant protection from deleterious environmental factors. Int. J. Mol. Sci. 14, 8122–8147. doi: 10.3390/ijms14048122
Cirillo, V., D’amelia, V., Esposito, M., Amitrano, C., Carillo, P., Carputo, D., et al. (2021). Anthocyanins are key regulators of drought stress tolerance in tobacco. Biology 10, 139. doi: 10.3390/biology10020139
Deng, X., Phillips, J., Meijer, A. H., Salamini, F., Bartels, D. (2002). Characterization of five novel dehydration-responsive homeodomain leucine zipper genes from the resurrection plant Craterostigma plantagineum. Plant Mol. Biol. 49, 601–610. doi: 10.1023/A:1015501205303
Depège-Fargeix, N., Javelle, M., Chambrier, P., Frangne, N., Gerentes, D., Perez, P., et al. (2011). Functional characterization of the HD-ZIP IV transcription factor OCL1 from maize. J. Exp. Bot. 62, 293–305. doi: 10.1093/jxb/erq267
Dezar, C. A., Giacomelli, J. I., Manavella, P. A., Ré, D. A., Alves-Ferreira, M., Baldwin, I. T., et al. (2011). HAHB10, a sunflower HD-zip II transcription factor, participates in the induction of flowering and in the control of phytohormone-mediated responses to biotic stress. J. Exp. Bot. 62, 1061–1076. doi: 10.1093/jxb/erq339
Di Cristina, M., Sessa, G., Dolan, L., Linstead, P., Baima, S., Ruberti, I., et al. (1996). The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. The Plant journal : for cell and molecular biology 10 (3), 393–402. doi: 10.1046/j.1365-313x.1996.10030393.x
Ding, Y., Gong, S., Wang, Y., Wang, F., Bao, H., Sun, J., et al. (2018). MicroRNA166 modulates cadmium tolerance and accumulation in rice. Plant Physiol. 177, 1691–1703. doi: 10.1104/pp.18.00485
Ding, Z., Li, S., An, X., Liu, X., Qin, H., Wang, D. (2009). Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J. Genet. Genomics 36, 17–29. doi: 10.1016/S1673-8527(09)60003-5
Dong, N. Q., Sun, Y., Guo, T., Shi, C. L., Zhang, Y. M., Kan, Y., et al. (2020). UDP-Glucosyltransferase regulates grain size and abiotic stress tolerance associated with metabolic flux redirection in rice. Nat. Commun. 11, 2629. doi: 10.1038/s41467-020-16403-5
Duan, J., Cai, W. (2012). OsLEA3-2, an abiotic stress induced gene of rice plays a key role in salt and drought tolerance. PloS One 7, e45117. doi: 10.1371/journal.pone.0045117
Ebrahimian-Motlagh, S., Ribone, P. A., Thirumalaikumar, V. P., Allu, A. D., Chan, R. L., Mueller-Roeber, B., et al. (20172118). JUNGBRUNNEN1 confers drought tolerance downstream of the HD-zip I transcription factor AtHB13. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.02118
Elhiti, M., Stasolla, C. (2009). Structure and function of homodomain-leucine zipper (HD-zip) proteins. Plant Signal Behav. 4, 86–88. doi: 10.4161/psb.4.2.7692
Emery, J. F., Floyd, S. K., Alvarez, J., Eshed, Y., Hawker, N. P., Izhaki, A., et al. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13, 1768–1774. doi: 10.1016/j.cub.2003.09.035
Erpen, L., Devi, H. S., Grosser, J. W., Dutt, M. (2018). Potential use of the DREB/ERF, MYB, NAC and WRKY transcription factors to improve abiotic and biotic stress in transgenic plants. Plant Cell Tissue Organ Culture (PCTOC) 132, 1–25. doi: 10.1007/s11240-017-1320-6
Fan, Y., Li, W., Li, Z., Dang, S., Han, S., Zhang, L., et al. (2021). Examination of the transcriptional response to LaMIR166a overexpression in Larix kaempferi (Lamb.) Carr. Biol. (Basel) 10, 576. doi: 10.3390/biology10070576
Farid, M., Shakoor, M. B., Ehsan, S., Ali, S., Zubair, M., Hanif, M. (2013). Morphological, physiological and biochemical responses of different plant species to Cd stress. Int. J. Chem. Biochem. Sci. 3, 1–44.
Gao, Y., Liu, H., Zhang, K., Li, F., Wu, M., Xiang, Y. (2021). A moso bamboo transcription factor, Phehdz1, positively regulates the drought stress response of transgenic rice. Plant Cell Rep. 40, 187–204. doi: 10.1007/s00299-020-02625-w
Gilroy, S., Białasek, M., Suzuki, N., Górecka, M., Devireddy, A. R., Karpiński, S., et al. (2016). ROS, calcium, and electric signals: key mediators of rapid systemic signaling in plants. Plant Physiol. 171, 1606–1615. doi: 10.1104/pp.16.00434
Gong, S., Ding, Y., Hu, S., Ding, L., Chen, Z., Zhu, C. (2019). The role of HD-zip class I transcription factors in plant response to abiotic stresses. Physiol. Plant 167, 516–525. doi: 10.1111/ppl.12965
Gu, C., Guo, Z.-H., Cheng, H.-Y., Zhou, Y.-H., Qi, K.-J., Wang, G.-M., et al. (2019). A HD-ZIP II HOMEBOX transcription factor, PpHB. G7, mediates ethylene biosynthesis during fruit ripening in peach. Plant Sci. 278, 12–19. doi: 10.1016/j.plantsci.2018.10.008
Guo, Q., Jiang, J., Yao, W., Li, L., Zhao, K., Cheng, Z., et al. (2021). Genome-wide analysis of poplar HD-zip family and over-expression of PsnHDZ63 confers salt tolerance in transgenic Populus simonii ×P. nigra. Plant Sci. 311, 111021. doi: 10.1016/j.plantsci.2021.111021
Guo, J., Yang, X., Weston, D. J., Chen, J. G. (2011). Abscisic acid receptors: Past, present and future †. J. Integr. Plant Biol. 53, 469–479. doi: 10.1111/j.1744-7909.2011.01044.x
Gupta, A., Rico-Medina, A., Caño-Delgado, A. I. (2020). The physiology of plant responses to drought. Science 368, 266–269. doi: 10.1126/science.aaz7614
Haak, D. C., Fukao, T., Grene, R., Hua, Z., Ivanov, R., Perrella, G., et al. (2017). Multilevel regulation of abiotic stress responses in plants. Front. Plant Sci. 8, 1564. doi: 10.3389/fpls.2017.01564
Hanson, J., Johannesson, H., Engström, P. (2001). Sugar-dependent alterations in cotyledon and leaf development in transgenic plants expressing the HDZhdip gene ATHB13. Plant Mol. Biol. 45, 247–262. doi: 10.1023/A:1006464907710
Hernandez-Garcia, C. M., Finer, J. J. (2014). Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 217, 109–119. doi: 10.1016/j.plantsci.2013.12.007
Himmelbach, A., Hoffmann, T., Leube, M., Höhener, B., Grill, E. (2002). Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J. 21, 3029–3038. doi: 10.1093/emboj/cdf316
Hoang, X. L. T., Nhi, D. N. H., Thu, N. B. A., Thao, N. P., Tran, L. P. (2017). Transcription factors and their roles in signal transduction in plants under abiotic stresses. Curr. Genomics 18, 483–497. doi: 10.2174/1389202918666170227150057
Hong, Y., Zhang, H., Huang, L., Li, D., Song, F. (2016). Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Front. Plant Sci. 7, 4. doi: 10.3389/fpls.2016.00004
Horst, W. J., Wang, Y., Eticha, D. (2010). The role of the root apoplast in aluminium-induced inhibition of root elongation and in aluminium resistance of plants: a review. Ann. Bot. 106, 185–197. doi: 10.1093/aob/mcq053
Hu, J. T., Chen, G. P., Yin, W. C., Cui, B. L., Yu, X. H., Lu, Y., et al. (2017). Silencing of SlHB2 improves drought, salt stress tolerance, and induces stress-related gene expression in tomato. J. Plant Growth Regul. 36, 578–589. doi: 10.1007/s00344-017-9664-z
Hur, Y. S., Um, J. H., Kim, S., Kim, K., Park, H. J., Lim, J. S., et al. (2015). Arabidopsis thaliana homeobox 12 (ATHB 12), a homeodomain-leucine zipper protein, regulates leaf growth by promoting cell expansion and endoreduplication. New Phytol. 205, 316–328. doi: 10.1111/nph.12998
Janská, A., Maršík, P., Zelenková, S., Ovesná, J. (2010). Cold stress and acclimation–what is important for metabolic adjustment? Plant Biol. 12, 395–405. doi: 10.1111/j.1438-8677.2009.00299.x
Jiang, Y., Liu, C., Yan, D., Wen, X., Liu, Y., Wang, H., et al. (2017). MdHB1 down-regulation activates anthocyanin biosynthesis in the white-fleshed apple cultivar 'Granny smith'. J. Exp. Bot. 68, 1055–1069. doi: 10.1093/jxb/erx029
Jiang, D., Lu, B., Liu, L., Duan, W., Chen, L., Li, J., et al. (2020). Exogenous melatonin improves salt stress adaptation of cotton seedlings by regulating active oxygen metabolism. PeerJ 8, e10486. doi: 10.7717/peerj.10486
Kang, J. H., Mcroberts, J., Shi, F., Moreno, J. E., Jones, A. D., Howe, G. A. (2014). The flavonoid biosynthetic enzyme chalcone isomerase modulates terpenoid production in glandular trichomes of tomato. Plant Physiol. 164, 1161–1174. doi: 10.1104/pp.113.233395
Kaur, R., Das, S., Bansal, S., Singh, G., Sardar, S., Dhar, H., et al. (2021). Heavy metal stress in rice: Uptake, transport, signaling, and tolerance mechanisms. Physiol. Plant 173, 430–448. doi: 10.1111/ppl.13491
Khosla, A., Paper, J. M., Boehler, A. P., Bradley, A. M., Neumann, T. R., Schrick, K. (2014). HD-Zip proteins GL2 and HDG11 have redundant functions in Arabidopsis trichomes, and GL2 activates a positive feedback loop via MYB23. Plant Cell 26, 2184–2200. doi: 10.1105/tpc.113.120360
Kiyomizu, T., Yamagishi, S., Kume, A., Hanba, Y. T. (2019). Contrasting photosynthetic responses to ambient air pollution between the urban shrub Rhododendron × pulchrum and urban tall tree Ginkgo biloba in Kyoto city: stomatal and leaf mesophyll morpho-anatomies are key traits. Trees 33, 63–77. doi: 10.1007/s00468-018-1759-z
Kovalchuk, N., Chew, W., Sornaraj, P., Borisjuk, N., Yang, N., Singh, R., et al. (2016). The homeodomain transcription factor TaHDZipI-2 from wheat regulates frost tolerance, flowering time and spike development in transgenic barley. New Phytol. 211, 671–687. doi: 10.1111/nph.13919
Kurusu, T., Kuchitsu, K., Tada, Y. (2015). Plant signaling networks involving Ca2+ and Rboh/Nox-mediated ROS production under salinity stress. Front. Plant Sci. 6, 427. doi: 10.3389/fpls.2015.00427
Lachenbruch, B., Mcculloh, K. A. (2014). Traits, properties, and performance: how woody plants combine hydraulic and mechanical functions in a cell, tissue, or whole plant. New Phytol. 204, 747–764. doi: 10.1111/nph.13035
Lado, J., Rodrigo, M. J., Cronje, P., Zacarías, L. (2015). Involvement of lycopene in the induction of tolerance to chilling injury in grapefruit. Postharvest Biol. Technol. 100, 176–186. doi: 10.1016/j.postharvbio.2014.10.002
Lan, Y., Chai, Y., Xing, C., Wu, K., Wang, L., Cai, M. (2021). Nitric oxide reduces the aluminum-binding capacity in rice root tips by regulating the cell wall composition and enhancing antioxidant enzymes. Ecotoxicol Environ. Saf. 208, 111499. doi: 10.1016/j.ecoenv.2020.111499
Li, C., Ma, X., Huang, X., Wang, H., Wu, H., Zhao, M., et al. (2019a). Involvement of HD-ZIP I transcription factors LcHB2 and LcHB3 in fruitlet abscission by promoting transcription of genes related to the biosynthesis of ethylene and ABA in litchi. Tree Physiol. 39, 1600–1613. doi: 10.1093/treephys/tpz071
Li, C., Zhao, M., Ma, X., Wen, Z., Ying, P., Peng, M., et al. (2019b). The HD-zip transcription factor LcHB2 regulates litchi fruit abscission through the activation of two cellulase genes. J. Exp. Bot. 70, 5189–5203. doi: 10.1093/jxb/erz276
Li, L., Zheng, T., Zhuo, X., Li, S., Qiu, L., Wang, J., et al. (2019c). Genome-wide identification, characterization and expression analysis of the HD-zip gene family in the stem development of the woody plant Prunus mume. PeerJ 7, e7499. doi: 10.7717/peerj.7499
Li, S., Yue, W., Wang, M., Qiu, W., Zhou, L., Shou, H. (2016). Mutation of OsGIGANTEA leads to enhanced tolerance to polyethylene glycol-generated osmotic stress in rice. Front. Plant Sci. 7, 465. doi: 10.3389/fpls.2016.00465
Li, S., Chen, N., Li, F., Mei, F., Wang, Z., Cheng, X., et al. (2020). Characterization of wheat homeodomain-leucine zipper family genes and functional analysis of TaHDZ5-6A in drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 20, 1–23. doi: 10.1186/s12870-020-2252-6
Li, W., Dong, J., Cao, M., Gao, X., Wang, D., Liu, B., et al. (2019d). Genome-wide identification and characterization of HD-ZIP genes in potato. Gene 697, 103–117. doi: 10.1016/j.gene.2019.02.024
Li, W., Dong, J., Cao, M., Gao, X., Wang, D., Liu, B, et al. (2019f). Genome-wide identification and characterization of HD-ZIPgenes in potato. Gene 697, 103–117.
Li, X., Hou, Y., Li, M., Zhang, F., Yi, F., Kang, J., et al. (2022). Overexpression of an ABA-inducible homeodomain-leucine zipper I gene MsHB7 confers salt stress sensitivity to alfalfa. Ind. Crops Products 177, 114463. doi: 10.1016/j.indcrop.2021.114463
Li, Y., Bai, B., Wen, F., Zhao, M., Xia, Q., Yang, D. H., et al. (2019e). Genome-wide identification and expression analysis of HD-ZIP I gene subfamily in Nicotiana tabacum. Genes (Basel) 10, 575. doi: 10.3390/genes10080575
Liu, J., Piñeros, M. A., Kochian, L. V. (2014). The role of aluminum sensing and signaling in plant aluminum resistance. J. Integr. Plant Biol. 56, 221–230. doi: 10.1111/jipb.12162
Liu, T., Longhurst, A. D., Talavera-Rauh, F., Hokin, S. A., Barton, M. K. (2016). The Arabidopsis transcription factor ABIG1 relays ABA signaled growth inhibition and drought induced senescence. Elife 5, e13768. doi: 10.7554/eLife.13768.019
Liu, Y., Xu, J., Guo, S., Yuan, X., Zhao, S., Tian, H., et al. (2020). AtHB7/12 regulate root growth in response to aluminum stress. Int. J. Mol. Sci. 21, 4080. doi: 10.3390/ijms21114080
Ma, Y.-J., Li, P.-T., Sun, L.-M., Zhou, H., Zeng, R.-F., Ai, X.-Y., et al. (2020). HD-ZIP I transcription factor (PtHB13) negatively regulates citrus flowering through binding to FLOWERING LOCUS c promoter. Plants-Basel 9 (1), 114. doi: 10.3390/plants9010114
Manavella, P. A., Arce, A. L., Dezar, C. A., Bitton, F., Renou, J. P., Crespi, M., et al. (2006). Cross-talk between ethylene and drought signalling pathways is mediated by the sunflower hahb-4 transcription factor. Plant J. 48, 125–137. doi: 10.1111/j.1365-313X.2006.02865.x
Manavella, P. A., Dezar, C. A., Ariel, F. D., Drincovich, M. F., Chan, R. L. (2008). The sunflower HD-zip transcription factor HAHB4 is up-regulated in darkness, reducing the transcription of photosynthesis-related genes. J. Exp. Bot. 59, 3143–3155. doi: 10.1093/jxb/ern170
Marcolino-Gomes, J., Rodrigues, F. A., Oliveira, M. C. N., Farias, J. R. B., Neumaier, N., Abdelnoor, R. V., et al. (2013). Expression patterns of GmAP2/EREB-like transcription factors involved in soybean responses to water deficit. PloS One 8, e62294. doi: 10.1371/journal.pone.0062294
Marin, E., Nussaume, L., Quesada, A., Gonneau, M., Sotta, B., Hugueney, P., et al. (1996). Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J. 15, 2331–2342. doi: 10.1002/j.1460-2075.1996.tb00589.x
Marusig, D., Tombesi, S. (2020). Abscisic acid mediates drought and salt stress responses in Vitis vinifera—a review. Int. J. Mol. Sci. 21, 8648. doi: 10.3390/ijms21228648
Masucci, J. D., Rerie, W. G., Foreman, D. R., Zhang, M., Galway, M. E., Marks, M. D., et al (20208648). The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development (Cambridge, England), 122 (4), 1253–1260. doi: 10.1242/dev.122.4.1253
Mbarki, S., Sytar, O., Zivcak, M., Abdelly, C., Cerda, A., Brestic, M. (20181518). Anthocyanins of coloured wheat genotypes in specific response to SalStress. Molecules 23, 1518. doi: 10.3390/molecules23071518
Mehmood, M., Khan, M. J., Khan, M. J., Akhtar, N., Mughal, F., Shah, S. T. A., et al. (2022). Systematic analysis of HD-ZIP transcription factors in sesame genome and gene expression profiling of SiHD-ZIP class I entailing drought stress responses at early seedling stage. Mol. Biol. Rep. 49, 2059–2071. doi: 10.1007/s11033-021-07024-2
Msanne, J., Lin, J., Stone, J. M., Awada, T. (2011). Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta 234, 97–107. doi: 10.1007/s00425-011-1387-y
Nagashima, A., Hanaoka, M., Shikanai, T., Fujiwara, M., Kanamaru, K., Takahashi, H., et al. (2004). The multiple-stress responsive plastid sigma factor, SIG5, directs activation of the psbD blue light-responsive promoter (BLRP) in Arabidopsis thaliana. Plant Cell Physiol. 45, 357–368. doi: 10.1093/pcp/pch050
Ni, Y., Wang, X., Li, D., Wu, Y., Xu, W., Li, X. (2008). Novel cotton homeobox gene and its expression profiling in root development and in response to stresses and phytohormones. Acta Biochim. Biophys. Sin. (Shanghai) 40, 78–84. doi: 10.1111/j.1745-7270.2008.00371.x
Oh, S.-K., Yoon, J., Choi, G. J., Jang, H. A., Kwon, S.-Y., Choi, D. (2013). Capsicum annuum homeobox 1 (CaHB1) is a nuclear factor that has roles in plant development, salt tolerance, and pathogen defense. Biochem. Biophys. Res. Commun. 442, 116–121. doi: 10.1016/j.bbrc.2013.11.019
Park, M. Y., Kim, S. A., Lee, S. J., Kim, S. Y. (2013). ATHB17 is a positive regulator of abscisic acid response during early seedling growth. Mol. Cells 35, 125–133. doi: 10.1007/s10059-013-2245-5
Patel, K. G., Thankappan, R., Mishra, G. P., Mandaliya, V. B., Kumar, A., Dobaria, J. R. (20171881). Transgenic peanut (Arachis hypogaea L.) overexpressing mtlD gene showed improved photosynthetic, physio-biochemical, and yield-parameters under soil-moisture deficit stress in lysimeter system. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01881
Prigge, M. J., Otsuga, D., Alonso, J. M., Ecker, J. R., Drews, G. N., Clark, S. E. (2005). Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17, 61–76. doi: 10.1105/tpc.104.026161
Qi, B., Doughty, J., Hooley, R. (2013). A golgi and tonoplast localized s-acyl transferase is involved in cell expansion, cell division, vascular patterning and fertility in Arabidopsis. New Phytol. 200, 444–456. doi: 10.1111/nph.12385
Qi, J., Song, C. P., Wang, B., Zhou, J., Kangasjärvi, J., Zhu, J. K., et al. (2018). Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J. Integr. Plant Biol. 60, 805–826. doi: 10.1111/jipb.12654
Qiu, X., Wang, G., Abou-Elwafa, S. F., Fu, J., Liu, Z., Zhang, P., et al. (2022). Genome-wide identification of HD-ZIP transcription factors in maize and their regulatory roles in promoting drought tolerance. Physiol. Mol. Biol. Plants 28, 425–437. doi: 10.1007/s12298-022-01147-x
Rangel, A. F., Rao, I. M., Horst, W. J. (2007). Spatial aluminium sensitivity of root apices of two common bean (Phaseolus vulgaris L.) genotypes with contrasting aluminium resistance. J. Exp. Bot. 58, 3895–3904. doi: 10.1093/jxb/erm241
Ranocha, P., Denancé, N., Vanholme, R., Freydier, A., Martinez, Y., Hoffmann, L., et al. (2010). Walls are thin 1 (WAT1), an Arabidopsis homolog of Medicago truncatula NODULIN21, is a tonoplast-localized protein required for secondary wall formation in fibers. Plant J. 63, 469–483. doi: 10.1111/j.1365-313X.2010.04256.x
Ré, D. A., Dezar, C. A., Chan, R. L., Baldwin, I. T., Bonaventure, G. (2011). Nicotiana attenuata NaHD20 plays a role in leaf ABA accumulation during water stress, benzylacetone emission from flowers, and the timing of bolting and flower transitions. J. Exp. Bot. 62, 155–166. doi: 10.1093/jxb/erq252
Rehman, S., Chattha, M. U., Khan, I., Mahmood, A., Hassan, M. U., Al-Huqail, A. A., et al. (2022). Exogenously applied trehalose augments cadmium stress tolerance and yield of mung bean (Vigna radiata L.) grown in soil and hydroponic systems through reducing Cd uptake and enhancing photosynthetic efficiency and antioxidant defense systems. Plants 11, 822. doi: 10.3390/plants11060822
Ribone, P. A., Capella, M., Chan, R. L. (2015). Functional characterization of the homeodomain leucine zipper I transcription factor AtHB13 reveals a crucial role in Arabidopsis development. J. Exp. Bot. 66, 5929–5943. doi: 10.1093/jxb/erv302
Rodriguez-Uribe, L., Guzman, I., Rajapakse, W., Richins, R. D., O’connell, M. A. (2012). Carotenoid accumulation in orange-pigmented Capsicum annuum fruit, regulated at multiple levels. J. Exp. Bot. 63, 517–526. doi: 10.1093/jxb/err302
Roppolo, D., Boeckmann, B., Pfister, A., Boutet, E., Rubio, M. C., Dénervaud-Tendon, V., et al. (2014). Functional and evolutionary analysis of the CASPARIAN STRIP MEMBRANE DOMAIN PROTEIN family. Plant Physiol. 165, 1709–1722. doi: 10.1104/pp.114.239137
Roppolo, D., De Rybel, B., Dénervaud Tendon, V., Pfister, A., Alassimone, J., Vermeer, J. E., et al. (2011). A novel protein family mediates casparian strip formation in the endodermis. Nature 473, 380–383. doi: 10.1038/nature10070
Ruan, L., Chen, L. J., Chen, Y. H., He, J. L., Zhang, W., Gao, Z. L., et al. (2012). Expression of Arabidopsis HOMEODOMAIN GLABROUS 11 enhances tolerance to drought stress in transgenic sweet potato plants. J. Plant Biol. 55, 151–158. doi: 10.1007/s12374-011-9198-z
Rueda, E. C., Dezar, C. A., Gonzalez, D. H., Chan, R. L. (2005). Hahb-10, a sunflower homeobox-leucine zipper gene, is regulated by light quality and quantity, and promotes early flowering when expressed in Arabidopsis. Plant Cell Physiol. 46, 1954–1963. doi: 10.1093/pcp/pci210
Santini, N. S., Cleverly, J., Faux, R., Lestrange, C., Rumman, R., Eamus, D. (2016). Xylem traits and water-use efficiency of woody species co-occurring in the Ti tree basin arid zone. Trees 30, 295–303. doi: 10.1007/s00468-015-1301-5
Sasaki, K., Ida, Y., Kitajima, S., Kawazu, T., Hibino, T., Hanba, Y. T. (2019). Overexpressing the HD-zip class II transcription factor EcHB1 from Eucalyptus camaldulensis increased the leaf photosynthesis and drought tolerance of Eucalyptus. Sci. Rep. 9, 14121. doi: 10.1038/s41598-019-50610-5
Sen, S., Chakraborty, J., Ghosh, P., Basu, D., Das, S. (2017). Chickpea WRKY70 regulates the expression of a homeodomain-leucine zipper (HD-zip) I transcription factor CaHDZ12, which confers abiotic stress tolerance in transgenic tobacco and chickpea. Plant Cell Physiol. 58, 1934–1952. doi: 10.1093/pcp/pcx126
Seo, M., Koshiba, T. (2002). Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 7, 41–48. doi: 10.1016/S1360-1385(01)02187-2
Seok, H. Y., Tran, H. T., Lee, S. Y., Moon, Y. H. (2022). AtERF71/HRE2, an Arabidopsis AP2/ERF transcription factor gene, contains both positive and negative cis-regulatory elements in its promoter region involved in hypoxia and salt stress responses. Int. J. Mol. Sci. 23, 5310. doi: 10.3390/ijms23105310
Sessa, G., Carabelli, M., Possenti, M., Morelli, G., Ruberti, I. (2018). Multiple links between HD-zip proteins and hormone networks. Int. J. Mol. Sci. 19, 4047. doi: 10.3390/ijms19124047
Shao, J., Haider, I., Xiong, L., Zhu, X., Hussain, R. M. F., Övernäs, E., et al. (2018). Functional analysis of the HD-zip transcription factor genes Oshox12 and Oshox14 in rice. PloS One 13, e0199248. doi: 10.1371/journal.pone.0199248
Sharif, R., Raza, A., Chen, P., Li, Y., El-Ballat, E. M., Rauf, A., et al. (2021). HD-ZIP gene family: Potential roles in improving plant growth and regulating stress-responsive mechanisms in plants. Genes (Basel) 12, 1256. doi: 10.3390/genes12081256
Sharif, R., Xie, C., Wang, J., Cao, Z., Zhang, H., Chen, P., et al. (2020). Genome wide identification, characterization and expression analysis of HD-ZIP gene family in Cucumis sativus L. under biotic and various abiotic stresses. Int. J. Biol. Macromol (20) 158, 502–520. doi: 10.1016/j.ijbiomac.2020.04.124
Shen, W., Li, H., Teng, R., Wang, Y., Wang, W., Zhuang, J. (2019). Genomic and transcriptomic analyses of HD-zip family transcription factors and their responses to abiotic stress in tea plant (Camellia sinensis). Genomics 111, 1142–1151. doi: 10.1016/j.ygeno.2018.07.009
Shin, D., Koo, Y. D., Lee, J., Lee, H. J., Baek, D., Lee, S., et al. (2004). Athb-12, a homeobox-leucine zipper domain protein from Arabidopsis thaliana, increases salt tolerance in yeast by regulating sodium exclusion. Biochem. Biophys. Res. Commun. 323, 534–540. doi: 10.1016/j.bbrc.2004.08.127
Söderman, E., Mattsson, J., Engström, P. (1996). The Arabidopsis homeobox gene ATHB-7 is induced by water deficit and by abscisic acid. Plant J. 10, 375–381. doi: 10.1046/j.1365-313X.1996.10020375.x
Song, S., Chen, Y., Zhao, M., Zhang, W.-H. (2012). A novel Medicago truncatula HD-zip gene, MtHB2, is involved in abiotic stress responses. Environ. Exp. Bot. 80, 1–9. doi: 10.1016/j.envexpbot.2012.02.001
Song, S. K., Ryu, K. H., Kang, Y. H., Song, J. H., Cho, Y. H., Yoo, S. D., et al. (2011). Cell fate in the Arabidopsis root epidermis is determined by competition between WEREWOLF and CAPRICE. Plant Physiol. 157, 1196–1208. doi: 10.1104/pp.111.185785
Sytar, O., Ghosh, S., Malinska, H., Zivcak, M., Brestic, M. (2021). Physiological and molecular mechanisms of metal accumulation in hyperaccumulator plants. Physiol. Plant 173, 148–166. doi: 10.3390/genes12081256
Szabados, L., Savouré, A. (2010). Proline: a multifunctional amino acid. Trends Plant Sci. 15, 89–97. doi: 10.1016/j.tplants.2009.11.009
Tang, Y., Bao, X., Wang, S., Liu, Y., Tan, J., Yang, M., et al. (2019a). A physic nut stress-responsive HD-zip transcription factor, JcHDZ07, confers enhanced sensitivity to salinity stress in transgenic Arabidopsis. Front. Plant Sci. 10,942. doi: 10.3389/fpls.2019.00942
Tang, Y., Wang, J., Bao, X., Liang, M., Lou, H., Zhao, J., et al. (2019b). Genome-wide identification and expression profile of HD-ZIP genes in physic nut and functional analysis of the JcHDZ16 gene in transgenic rice. BMC Plant Biol. 19, 298. doi: 10.1186/s12870-019-1920-x
Tan, W., Zhang, D., Zhou, H., Zheng, T., Yin, Y., Lin, H. (2018). Transcription factor HAT1 is a substrate of SnRK2. 3 kinase and negatively regulates ABA synthesis and signaling in Arabidopsis responding to drought. PloS Genet. 14, e1007336. doi: 10.1371/journal.pgen.1007336
Tognacca, R. S., Carabelli, M., Morelli, G., Ruberti, I., Botto, J. F. (2021). ATHB2 is a negative regulator of germination in Arabidopsis thaliana seeds. Sci. Rep. 11, 9688. doi: 10.1038/s41598-021-88874-5
Turchi, L., Baima, S., Morelli, G., Ruberti, I. (2015). Interplay of HD-zip II and III transcription factors in auxin-regulated plant development. J. Exp. Bot. 66, 5043–5053. doi: 10.1093/jxb/erv174
Turchi, L., Carabelli, M., Ruzza, V., Possenti, M., Sassi, M., Peñalosa, A., et al. (2013). Arabidopsis HD-zip II transcription factors control apical embryo development and meristem function. Development 140, 2118–2129. doi: 10.1242/dev.092833
Umezawa, T., Nakashima, K., Miyakawa, T., Kuromori, T., Tanokura, M., Shinozaki, K., et al. (2010). Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol. 51, 1821–1839. doi: 10.1093/pcp/pcq156
Uraguchi, S., Mori, S., Kuramata, M., Kawasaki, A., Arao, T., Ishikawa, S. (2009). Root-to-shoot Cd translocation via the xylem is the major process determining shoot and grain cadmium accumulation in rice. J. Exp. Bot. 60, 2677–2688. doi: 10.1093/jxb/erp119
Verslues, P. E., Bray, E. A. (2006). Role of abscisic acid (ABA) and Arabidopsis thaliana ABA-insensitive loci in low water potential-induced ABA and proline accumulation. J. Exp. Bot. 57, 201–212. doi: 10.1093/jxb/erj026
Vinocur, B., Altman, A. (2005). Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr. Opin. Biotechnol. 16, 123–132. doi: 10.1016/j.copbio.2005.02.001
Wang, H., Wang, H., Shao, H., Tang, X. (2016). Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Front. Plant Sci. 7, 67. doi: 10.3389/fpls.2016.00067
Wang, Q., Zha, K., Chai, W., Wang, Y., Liu, B., Jiang, H., et al. (2017). Functional analysis of the HD-zip I gene ZmHDZ1 in ABA-mediated salt tolerance in rice. J. Plant Biol. 60, 207–214. doi: 10.1007/s12374-016-0413-9
Wei, J., Choi, H., Jin, P., Wu, Y., Yoon, J., Lee, Y.-S., et al. (2016). GL2-type homeobox gene Roc4 in rice promotes flowering time preferentially under long days by repressing Ghd7. Plant Sci. 252, 133–143. doi: 10.1016/j.plantsci.2016.07.012
Wei, M., Liu, A., Zhang, Y., Zhou, Y., Li, D., Dossa, K., et al. (2019). Genome-wide characterization and expression analysis of the HD-zip gene family in response to drought and salinity stresses in sesame. BMC Genomics 20, 748. doi: 10.1186/s12864-019-6091-5
Xie, L., Yan, T., Li, L., Chen, M., Hassani, D., Li, Y., et al. (2021). An HD-ZIP-MYB complex regulates glandular secretory trichome initiation in Artemisia annua. New Phytol. 231, 2050–2064. doi: 10.1111/nph.17514
Yang, C., Gao, Y., Gao, S., Yu, G., Xiong, C., Chang, J., et al. (2015). Transcriptome profile analysis of cell proliferation molecular processes during multicellular trichome formation induced by tomato Wov gene in tobacco. BMC Genomics 16, 868. doi: 10.1186/s12864-015-2099-7
Yang, Z.-B., Geng, X., He, C., Zhang, F., Wang, R., Horst, W. J., et al. (2014). TAA1-regulated local auxin biosynthesis in the root-apex transition zone mediates the aluminum-induced inhibition of root growth in Arabidopsis. Plant Cell 26, 2889–2904. doi: 10.1105/tpc.114.127993
Yang, Y., Luang, S., Harris, J., Riboni, M., Li, Y., Bazanova, N., et al. (2018). Overexpression of the class I homeodomain transcription factor TaHDZipI-5 increases drought and frost tolerance in transgenic wheat. Plant Biotechnol. J. 16, 1227–1240. doi: 10.1111/pbi.12865
Yang, Y.-Y., Shan, W., Kuang, J.-F., Chen, J.-Y., Lu, W.-J. (2020). Four HD-ZIPs are involved in banana fruit ripening by activating the transcription of ethylene biosynthetic and cell wall-modifying genes. Plant Cell Rep. 39, 351–362. doi: 10.1007/s00299-019-02495-x
Yang, Q., Xiang, W., Li, Z., Nian, Y., Fu, X., Zhou, G., et al. (2022). Genome-wide characterization and expression analysis of HD-ZIP gene family in dendrobium officinale. Front. Genet. 13, 797014. doi: 10.3389/fgene.2022.797014
Yang, J. L., Zhu, X. F., Peng, Y. X., Zheng, C., Li, G. X., Liu, Y., et al. (2011). Cell wall hemicellulose contributes significantly to aluminum adsorption and root growth in Arabidopsis. Plant Physiol. 155, 1885–1892. doi: 10.1104/pp.111.172221
Yao, T., Zhang, J., Xie, M., Yuan, G., Tschaplinski, T. J., Muchero, W., et al. (2020). Transcriptional regulation of drought response in Arabidopsis and woody plants. Front. Plant Sci. 11, 572137. doi: 10.3389/fpls.2020.572137
Ye, Y., Ding, Y., Jiang, Q., Wang, F., Sun, J., Zhu, C. (2017). The role of receptor-like protein kinases (RLKs) in abiotic stress response in plants. Plant Cell Rep. 36, 235–242. doi: 10.1007/s00299-016-2084-x
Yu, H., Chen, X., Hong, Y. Y., Wang, Y., Xu, P., Ke, S. D., et al. (2008). Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell 20, 1134–1151. doi: 10.1105/tpc.108.058263
Yu, L., Chen, X., Wang, Z., Wang, S., Wang, Y., Zhu, Q., et al. (2013). Arabidopsis enhanced drought tolerance1/HOMEODOMAIN GLABROUS11 confers drought tolerance in transgenic rice without yield penalty. Plant Physiol. 162, 1378–1391. doi: 10.1104/pp.113.217596
Yu, L. H., Wu, S. J., Peng, Y. S., Liu, R. N., Chen, X., Zhao, P., et al. (2016). Arabidopsis EDT1/HDG11 improves drought and salt tolerance in cotton and poplar and increases cotton yield in the field. Plant Biotechnol. J. 14, 72–84. doi: 10.1111/pbi.12358
Zhang, Z., Chen, X., Guan, X., Liu, Y., Chen, H., Wang, T., et al. (2014b). A genome-wide survey of homeodomain-leucine zipper genes and analysis of cold-responsive HD-zip I members’ expression in tomato. Bioscience Biotechnology Biochem. 78, 1337–1349. doi: 10.1080/09168451.2014.923292
Zhang, Q., Cui, M., Xin, X., Ming, X., Jing, L., Wu, J.-X. (2014a). Overexpression of a cytosolic ascorbate peroxidase gene, OsAPX2, increases salt tolerance in transgenic alfalfa. J. Integr. Agric. 13, 2500–2507. doi: 10.1016/S2095-3119(13)60691-7
Zhang, S., Haider, I., Kohlen, W., Jiang, L., Bouwmeester, H., Meijer, A. H., et al. (2012). Function of the HD-zip I gene Oshox22 in ABA-mediated drought and salt tolerances in rice. Plant Mol. Biol. 80, 571–585. doi: 10.1007/s11103-012-9967-1
Zhang, Z., Tariq, A., Zeng, F., Chai, X., Graciano, C. (2021a). Involvement of soluble proteins in growth and metabolic adjustments of drought-stressed Calligonum mongolicum seedlings under nitrogen addition. Plant Biol. 23, 32–43. doi: 10.1111/plb.13190
Zhang, R.-X., Zhu, W.-C., Cheng, G.-X., Yu, Y.-N., Li, Q.-H., Ul Haq, S., et al. (2020). A novel gene, CaATHB-12, negatively regulates fruit carotenoid content under cold stress in Capsicum annuum. Food Nutr. Res. 64, 10.29219/fnr.v64.3729. doi: 10.29219/fnr.v64.3729
Zhang, Z., Zhu, R., Ji, X., Li, H. J., Lv, H., Zhang, H. Y. (2021b). Genome-wide characterization and expression analysis of the HD-ZIP gene family in response to salt stress in pepper. Int. J. Genomics 2021, 8105124. doi: 10.1155/2021/8105124
Zhao, P., Cui, R., Xu, P., Wu, J., Mao, J. L., Chen, Y., et al. (2017). ATHB17 enhances stress tolerance by coordinating photosynthesis associated nuclear gene and ATSIG5 expression in response to abiotic stress. Sci. Rep. 7, 45492. doi: 10.1038/srep45492
Zhao, Y., Ma, Q., Jin, X., Peng, X., Liu, J., Deng, L., et al. (2014). A novel maize homeodomain-leucine zipper (HD-zip) I gene, Zmhdz10, positively regulates drought and salt tolerance in both rice and Arabidopsis. Plant Cell Physiol. 55, 1142–1156. doi: 10.1093/pcp/pcu054
Zhao, S., Wang, H., Jia, X., Gao, H., Mao, K., Ma, F. (2021). The HD-zip I transcription factor MdHB7-like confers tolerance to salinity in transgenic apple (Malus domestica). Physiologia Plantarum 172, 1452–1464. doi: 10.1111/ppl.13330
Zhou, C.-P., Li, C.-P., Liang, W.-W., Guo, P., Yang, L.-T., Chen, L.-S. (2016). Identification of manganese-toxicity-responsive genes in roots of two citrus species differing in manganese tolerance using cDNA-AFLP. Trees 31, 813–831. doi: 10.1007/s00468-016-1507-1
Zhu, Y., Song, D., Xu, P., Sun, J., Li, L. (2018). Identification of manganese-toxicity-responsive genes in roots. Plant Biotechnol. J. 16, 808–817. A HD and Chen, L.-S. (2016). doi: 10.1111/pbi.12830
Zhu, Z., Sun, B., Xu, X., Chen, H., Zou, L., Chen, G., et al. (2016). Overexpression of AtEDT1/HDG11 in Chinese kale (Brassica oleracea var. alboglabra) enhances drought and osmotic stress tolerance. Front. Plant Sci. 7, 1285. doi: 10.3389/fpls.2016.01285
Zou, M., Guan, Y., Ren, H., Zhang, F., Chen, F. (2008). A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol. Biol. 66, 675–683. doi: 10.1007/s11103-008-9298-4
Keywords: HD-ZIP protein, transcription factor, abiotic stress, plant growth and development, molecular mechanism
Citation: Li Y, Yang Z, Zhang Y, Guo J, Liu L, Wang C, Wang B and Han G (2022) The roles of HD-ZIP proteins in plant abiotic stress tolerance. Front. Plant Sci. 13:1027071. doi: 10.3389/fpls.2022.1027071
Received: 24 August 2022; Accepted: 26 September 2022;
Published: 12 October 2022.
Edited by:
Weiqiang Li, RIKEN, JapanCopyright © 2022 Li, Yang, Zhang, Guo, Liu, Wang, Wang and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Baoshan Wang, YnN3YW5nQHNkbnUuZWR1LmNu; Guoliang Han, Z2xfaGFuQHNkbnUuZWR1LmNu
†These authors have contributed equally to this work
 Yuxia Li
Yuxia Li Zongran Yang†
Zongran Yang† Yuanyuan Zhang
Yuanyuan Zhang Lili Liu
Lili Liu Chengfeng Wang
Chengfeng Wang Baoshan Wang
Baoshan Wang Guoliang Han
Guoliang Han