- 1Laboratory of Microbiological Monitoring and Bioremediation of Soils, All-Russian Research Institute for Agricultural Microbiology, Pushkin, Russia
- 2Laboratory of Soil Biology and Biochemistry, V.V. Dokuchaev Soil Science Institute, Moscow, Russia
- 3Department of Applied Ecology, St. Petersburg State University, Saint-Petersburg, Russia
- 4Gregor Mendel Institute, Austrian Academy of Sciences, Vienna BioCenter, Vienna, Austria
Nodule bacteria (rhizobia), N2-fixing symbionts of leguminous plants, represent an excellent model to study the fundamental issues of evolutionary biology, including the tradeoff between microevolution, speciation, and macroevolution, which remains poorly understood for free-living organisms. Taxonomically, rhizobia are extremely diverse: they are represented by nearly a dozen families of α-proteobacteria (Rhizobiales) and by some β-proteobacteria. Their genomes are composed of core parts, including house-keeping genes (hkg), and of accessory parts, including symbiotically specialized (sym) genes. In multipartite genomes of evolutionary advanced fast-growing species (Rhizobiaceae), sym genes are clustered on extra-chromosomal replicons (megaplasmids, chromids), facilitating gene transfer in plant-associated microbial communities. In this review, we demonstrate that in rhizobia, microevolution and speciation involve different genomic and ecological mechanisms: the first one is based on the diversification of sym genes occurring under the impacts of host-induced natural selection (including its disruptive, frequency-dependent and group forms); the second one—on the diversification of hkgs under the impacts of unknown factors. By contrast, macroevolution represents the polyphyletic origin of super-species taxa, which are dependent on the transfer of sym genes from rhizobia to various soil-borne bacteria. Since the expression of newly acquired sym genes on foreign genomic backgrounds is usually restricted, conversion of resulted recombinants into the novel rhizobia species involves post-transfer genetic changes. They are presumably supported by host-induced selective processes resulting in the sequential derepression of nod genes responsible for nodulation and of nif/fix genes responsible for symbiotic N2 fixation.
1 Introduction
The trade-off between the diversification processes occurring at different phylogenetic levels—intra-species (microevolution), species, and super-species (macroevolution)—represents a puzzling issue in evolutionary biology. According to the Synthetic Theory of Evolution (STE) initially developed for the sexually reproducing organisms, speciation and macroevolution represent an extension of microevolution: local populations, biotypes, and subspecies formed under the impacts of natural, mostly individual (Darwinian) selection are presumably transformed into novel species, genera, and higher ranked taxa (Dobzhansky, 1951; Timofeeff-Ressovsky et al., 1977). However, Philiptschenko (1927) who coined the terms “microevolution” and “macroevolution,” as well as Koonin (2011) supposed that sufficiently different genetic mechanisms were responsible for these processes.
For eukaryotic organisms, tradeoff between micro- and macro-evolution remains obscure since adaptive impacts of macroevolutionary events, for example of macromutations in the master genes (e.g., in homeotic genes controlling the developmental regulation) are difficult to quantify in free-living plants or animals (Theißen, 2006). Symbiotic models are useful to address the adaptive impacts of developmental innovations resulting from the integration of hosts with microbial partners, which is best studied using the examples of root nodules in legumes (Bhattacharjee et al., 2015) and of light organs in squids (Peyer et al., 2014).
Broad opportunities to address the trade-off between different levels of evolution are provided by root nodule bacteria (rhizobia), N2-fixing symbionts of legumes (Fabaceae) from the Rosid I clade of dicot plants. These bacteria include a range of polyphyletically originated families and over a hundred species, mostly from α-proteobacteria (Rhizobiales) and some β-proteobacteria (e.g., Paraburkholderia) (Young, 1996; Zakhia and de Lajudie, 2001; Berrada and Fikri-Benbrahim, 2014; Wang et al., 2019). Several categories of symbiotically specialized (sym) genes of different origins were revealed in rhizobia including nod (synthesis of lipo-chito-oligosaccharidic Nod factors, NFs eliciting the root nodule development) (Gottfert, 1993; Debellé et al., 2001; Shamseldin, 2013), nif (synthesis of nitrogenase enzyme catalyzing N2 reduction to ammonium) and fix (energy supply of nitrogenase, nif gene regulation) (Shamseldin, 2013).
The extant rhizobia species may be classified into two categories emerged at different evolutionary stages (Figure 1): (1) primary (ancestral) species, emerged from free-living N2 fixers by genome rearrangements, resulting in formation of sym (nod + nif/fix) gene networks; (2) secondary (derived) species, originated via transfer of sym genes into various soil and plant-associated bacteria (Provorov and Andronov, 2016). The primary rhizobia represented by slow-growing Bradyrhizobium species close to Rhodopseudomonas presumably acquired the ability for in planta N2 fixation by allocating some photosynthesis-regulating genes into the nitrogenase-regulating fix network (Section 3.1).
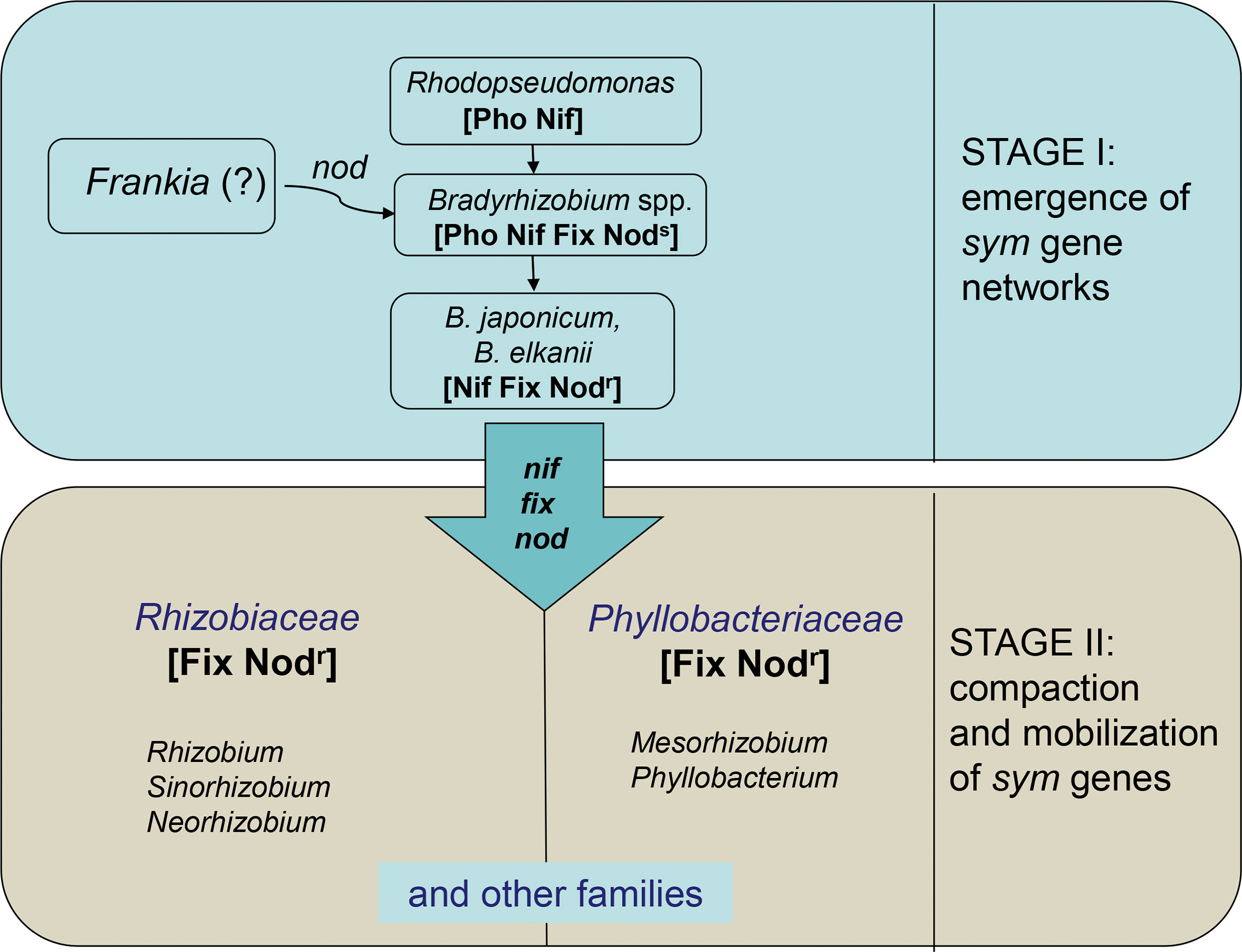
Figure 1 Rhizobia natural history: a two-stage scenario. Stage I: conversion of free-living diazotrophs (Rhodopseudomonas) into the primary rhizobia (Bradyrhizobium) via establishing the symbiotically specialized (sym) gene networks including genes for nodulation (nod) or N2 fixation (nif/fix). Stage II: emergence of secondary rhizobia via dissemination of sym genes among various families (e.g., Rhizobiaceae, Phyllobacteriaceae) of soil and plant-associated bacteria. Microbial phenotypes include: Pho, photosynthesis; Nif, free-living N2 fixation; Fix, symbiotic N2 fixation; Nods, stem nodulation (crack entry); Nodr, root nodulation (root hair entry).
The best studied secondary rhizobia are represented by fast-growing Neorhizobium, Rhizobium, and Sinorhizobium species (Rhizobiaceae) close to Agrobacterium, which is often addressed as the same genus as Rhizobium (Berrada and Fikri-Benbrahim, 2014; Ormeño-Orrillo et al., 2015; Wang et al., 2019). These symbiotically specialized bacteria are devoid of the ability to fix N2 ex planta, suggesting their origin via sym gene transfer from the preexisting rhizobia species. Transfer of large plasmids harboring sym genes as well as of chromosomally located sym islands may be responsible for the emergence of novel rhizobia genotypes and the distribution of sym genes in the microbial communities populating natural and agricultural ecosystems (Ding and Hynes, 2009). This transfer may stimulate recombination between IS copies, which are multiple in the symbiotically specialized genome regions providing the evolutionary important variation in rhizobia populations (Arashida et al., 2022).
An intriguing example of secondary rhizobia is represented by Azorhizobium caulinodans close to the free-living diazotroph Xanthobacter autotrophicus (Lee et al., 2008). This symbiont of tropical legume Sesbania rostrata combines the ability to fix N2 not only in planta, but also ex planta supporting the bacteria growth on N-free media (Boogerd et al., 1994). The genomic location of the nod cluster on a chromosomal island suggests the origin of A. caulinodans from a free-living N2 fixer via horizontal transfer of the nod gene cluster from a rhizobia species (Lee et al., 2008).
Rhizobia evolution involves two genomic strategies: “gain-and-loss of sym genes” and “compaction of sym gene clusters.” The gain of new sym genes from non-symbiotic networks occurred via horizontal gene transfer (HGT) and duplication–divergence (DD) mechanisms. In addition to fix genes, some nod genes have been recruited into symbiotic networks via the DD mechanism. For example, the nodD gene controlling expression of the nod regulon was obviously derived from the lysM-araC family of transcriptional regulators by acquiring the ability to percept plant-released flavonoids (Hassan and Mathesius, 2012). However, “common” nod genes (nodABC) which encode for the oligochitin part of the NF molecule were probably transferred from Frankia, the ancient N2 fixing symbiont of Rosid I plants, to some (presumably primary) rhizobia (Persson et al., 2015), which perhaps transmitted these genes to the other legume symbionts (HGT-based emergency of rhizobia).
The impact of gene loss on rhizobia evolution may be illustrated by nifV, which was revealed in phototrophic Bradyrhizobium genotypes. This gene encodes for the synthesis of homocitrate, a precursor for MoFe-cofactor of nitrogenase, which is usually supplied by plants, e.g., under the control of FEN1 in Lotus japonicus. Inactivation of FEN1 results in the loss of N2 fixation, which is restored after the introduction of nifV into Lotus-nodulating Mesorhizobium loti (Terpolilli et al., 2012). The nif gene losses occurred in many symbionts, e.g., primary rhizobia (Bradyrhizobium) retained 15–17 nif genes typical for free-living N2-fixers, while only 7–8 nif genes were found in the secondary rhizobia (Rhizobium, Sinorhizobium), which are not capable of ex planta N2 fixation (Pini et al., 2011). In later groups, a range of negative symbiosis regulators were revealed which inactivation by Tn5 insertions results in increased N2-fixing activity or nodulation competitiveness (Provorov et al., 2014; Onishchuk et al., 2017).
Compaction of sym gene arrangement was indicated in both primary and secondary rhizobia. The majority of Bradyrhizobium strains harbor sym genes in several chromosomal loci, while in some strains these genes are clustered on Sym plasmids (Okazaki et al., 2015). A compact arrangement was found in Mesorhizobium species in which nod and nif genes are clustered in chromosomal islands transferred in bacterial populations as conjugative transposons (Sullivan et al., 2002). In Rhizobiaceae species, an important factor for the high mobility of sym genes is represented by their clustering on the large Sym plasmids (pSyms) typical for Rhizobium and Sinorhizobium spp. (Poole et al., 2018). In R. leguminosarum bv. viciae, ancestral (A) strains isolated from nodules of Vavilovia formosa, a relict legume close to the common ancestor of the Fabeae tribe, possess a more scattered plasmid sym cluster (>90 kb) than the derived (D) strains isolated from Pisum and Vicia species (<60 kb) (Chirak et al., 2019). The A → D transitions involved a range of “gain-and-loss” events that presumably resulted in improved fitness in R. leguminosarum strains (Table 1).
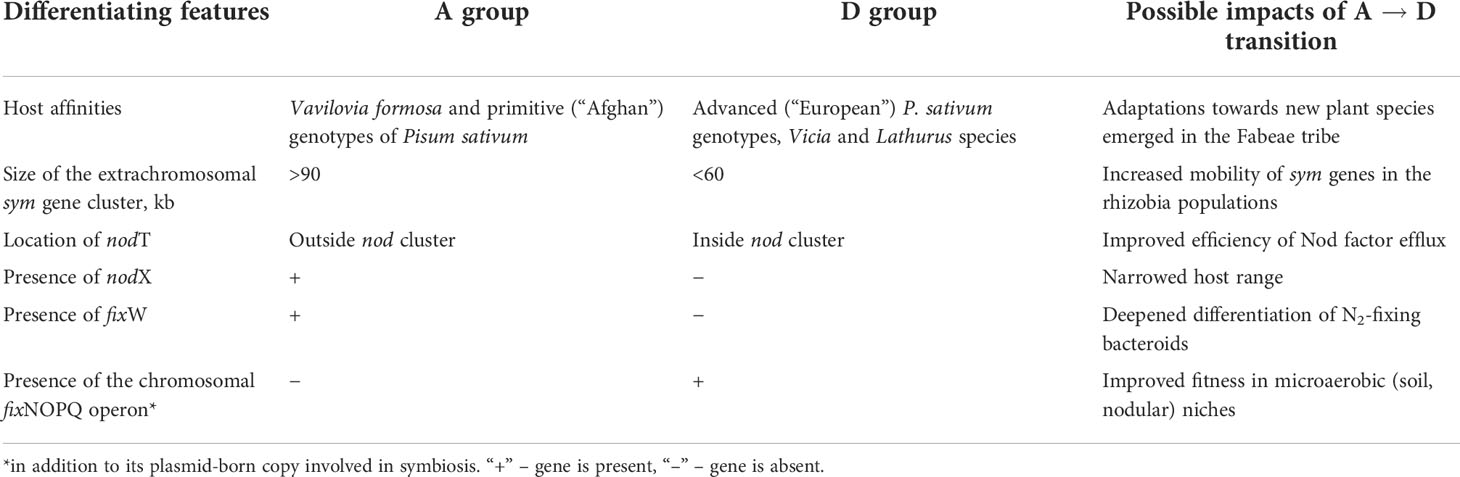
Table 1 Differentiation of ancestral (A) and derived (D) groups of Rhizobium leguminosarum bv. viciae genotypes [from Chirak et al. (2019)].
2 Microevolution and speciation: Divergence of rhizobia genomes
According to STE initially developed for sexually reproducing eukaryotic organisms, microevolution and speciation represent a continuum of biodivergence processes which are elicited by natural selection based on the differential ability of competing genotypes to produce fertile progeny (Dobzhansky, 1951; Timofeeff-Ressovsky et al., 1977). In contrast to microevolution, speciation results in a reproductive barrier: eukaryotic species are usually addressed as genetically closed systems in which recombination is restricted by intra-species hybridization, while the import of new genes via HGT is negligible (Katz, 2015).
For prokaryotes, these approaches are not valid since their reproduction is uncoupled from recombination, which occurs via parasexual processes resulting in HGT. Prokaryotic speciation does not mean a genetic barrier: gene exchange can be implemented between distant organisms, e.g., between bacteria and archaea (Fuchsman et al., 2017). The HGT-based gene flow provides the genomic cohesion of prokaryotic populations since the intensity of this flow is correlated to the relatedness of genotypes involved and may be used to suggest a biological species concept for prokaryotes which is at least partly analogous to the concept used for eukaryotes (Bobay, 2020). Importantly, prokaryotic species possess open genetic systems with pangenomes differentiated into the stable core parts which are transmitted vertically and the variable accessory parts for which HGT is intensive. The pangenome analysis may provide a new definition of prokaryotic species based on the identification of lineage-specific gene sets. While being similar to the classical biological definition based on allele flow, this definition does not rely on DNA similarity levels and does not require analysis of homologous recombination (Moldovan and Gelfand, 2018).
2.1 Divergence of core and accessory genomes
Suitable models to address the trade-off between intra-species diversification and speciation are represented by fast-growing rhizobia from the Rhizobiaceae family characterized by narrow host ranges towards the Galegoid legumes (Figure 2). We demonstrated (Kimeklis et al., 2018; Kimeklis et al., 2019) that in Rhizobium leguminosarum, the symbiotically contrasting biovars viciae (associated with the legume tribe Fabeae) and trifolii (associated with clovers from the Trifolieae tribe) are either not diverged for hkgs (p-distance analysis) or their divergence is sufficiently lower than for sym genes (group separation analysis).
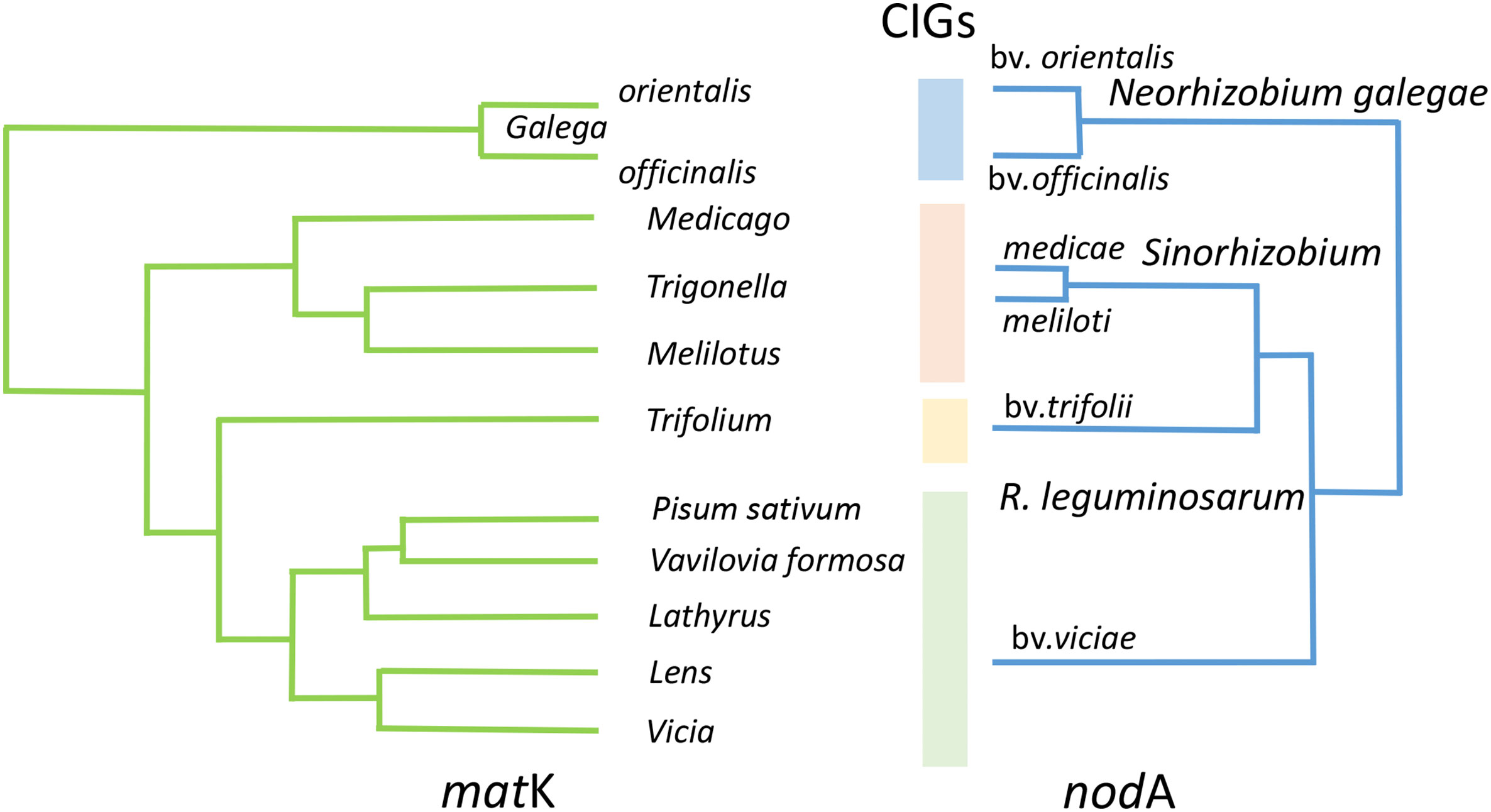
Figure 2 Phylogenetic congruence revealed in fast-growing rhizobia for nodA gene towards Galegoid legumes for matK gene. Within the cross-inoculation groups (CIGs), a completely developed symbioses are formed which are usually characterized by active N2 fixation; between CIGs, the under-developed non-N2-fixing nodules may be formed rarely. Stylized phylogenetic trees were adapted using a graphic redactor from Azani et al. (2017) (matK) and Haukka et al. (1998) (nodA) with the reduced number of nodes and the preserved initial topologies.
Importantly, within the host-specific biovars, variation for hkgs is much more pronounced than for sym genes suggesting that hkg-dependent speciation is not correlated to symbiotic diversification (Kimeklis et al., 2019). The complementary data were obtained using ANI technique suggesting that hkgs are diverged dramatically within a local R. leguminosarum population resulting in several genomic (cryptic) species which include bv. viciae and bv. trifolii strains (Kumar et al., 2015). Therefore, cryptic speciation represents a sympatric process which does not represent an extension of symbiotic diversification; they constitute two parallel pathways of divergent evolution (Figure 3).
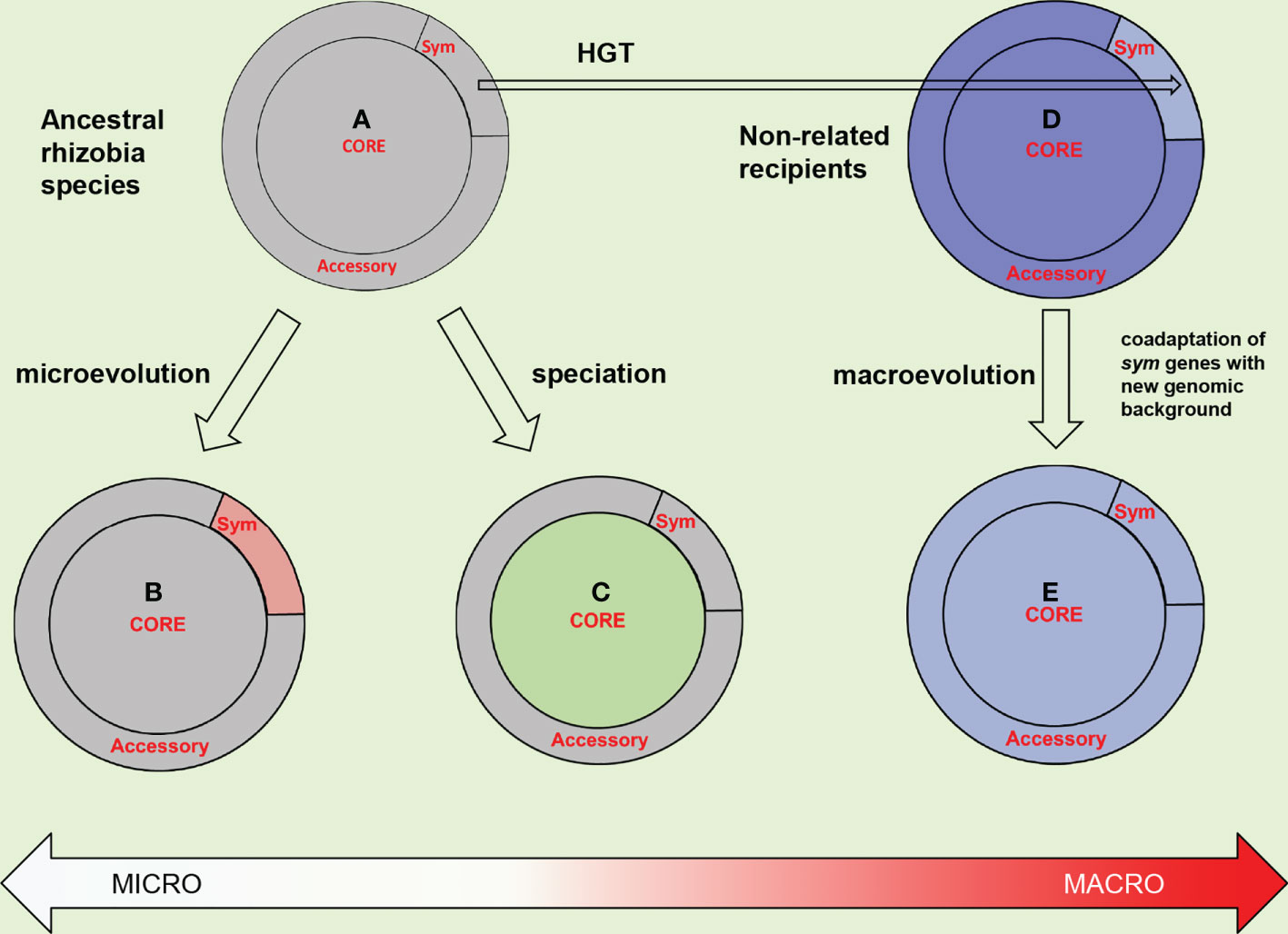
Figure 3 Trade-off between microevolution, speciation, and macroevolution in rhizobia. The proposed evolutionary paths are presented for an initial rhizobia species, which genome is differentiated into core and accessory parts, the later includes symbiotically specialized (sym) genes (A) Microevolution involves the divergence of sym genes (changed from gray for rosy) resulted in a host-specific (sym)biovar (B). Speciation involves the divergence of core genes (changed for green) resulted in a cryptic (genomic) species (C). Macroevolution is elicited by horizontal sym gene transfer (HGT) from rhizobia to genome of a non-related recipient (dark blue) (D). Coadaptation of the acquired sym genes with the foreign genomic background (changed for light blue) (E) involve the host-induced selective pressures presented in Figure 5.
The symbiosis-independent sympatric diversification of hkgs may be responsible for the emergence of novel species in different rhizobia groups. For example, a range of Bradyrhizobium species emerged in the Chinese center of soybean origin and retained similar host ranges. B. japonicum and B. elkanii strains produce N2-fixing nodules with Glycine spp. and other legumes from the symbiotically promiscuous Phaseoleae tribe, including Cajanus, Phaseolus, and Vigna (Lee et al., 2008).
Similarly, divergence of two sister Sinorhizobium (Ensifer) species, S. meliloti and S. medicae for the core genome markers occurred in spite of overlapping the geographic distribution and host ranges: these species differ for the ability to nodulate only some diploid medics, e.g. Medicago polymorpha (Rome et al., 1996; Bailly et al., 2006). Divergence for hkgs is also much stronger than for sym genes within Neorhizobium galegae host-specific biovars orientalis and officinalis, in which symbiotic N2 fixation is restricted to Galega orientalis and G. officinalis, respectively (Karasev et al., 2019).
Similar trends were revealed for plant pathogenic bacteria, in which the evolutionary dynamics of virulence genes differed greatly from the dynamics of hkgs. For example, in Pseudomonas syringae, differentiation into several dozens of host-specific pathovars determined by vir genes from accessory genomes differs greatly from phylogenetic groups determined by the core genome (Sarkar and Guttman, 2004; Słomnicka et al., 2015).
2.2 Ecological factors of rhizobia evolution
These factors may be classified into selective and stochastic ones operating in plant-associated rhizobia populations at the individual level (bacterial cells infecting the root hairs) or at the group level (cell groups maintained in individual plants or in different nodules formed on the same plant). The selective pressures may be positive or negative with respect to shifts in particular genotype frequencies resulting in their increase or decrease. With respect to gene structure, selective pressures may be differentiated into the purifying (stabilizing) and driving ones using the ratio of non-synonymous (dN) to synonymous (dS) substitutions: at dN/dS<1, selection is purifying, at dN/dS >1 it is driving while at dN/dS ≈ 1, the neutral, selection-independent evolution occurs (Kryazhimskiy and Plotkin, 2008).
2.2.1 Host-specific diversification: Individual selection
Since in the majority of sym genes, expression is inducible under symbiotic conditions, the evolution of these genes is dependent on hosts. The resultant co-evolution of the partners is based on cross-regulation of genes encoding for host-symbiont recognition (e.g., bacterial nod genes for NF synthesis and the plant NFR genes for NF perception) (Broghammer et al., 2012) or for their metabolic integration (e.g., bacterial nif/fix genes for nitrogenase synthesis and plant GS/GOGAT/AAT genes for ammonium assimilation) (Betti et al., 2012). The ability of rhizobia genotypes for in-planta multiplication is based greatly on the production of NFs, eliciting the development of nodular niches for the propagation of bacteria. This selection may be highly efficient since the numbers of rhizobia cells released from the decayed nodules may exceed manifold their numbers in the soil (Yan et al., 2014).
Evolution of rhizobia populations under impacts of hosts may be presented as an interplay of Darwinian Selection (DaS) dependent on multiplication rates of co-inoculated genotypes, and of Frequency-dependent Selection (FdS) dependent also on the genotypic ratios in the inoculum (root-associated population) (Provorov and Vorobyov, 2006). Since soil rhizobia populations are highly polymorphic, severe competition occurs between virulent strains for occupation of nodular niches which are rich in C nutrients. Experimentally, a non-linear dynamics of rhizobia genotypic ratios was demonstrated for two-strain competition (Amarger and Lobreau, 1982):
N1: N2= c(I1:I2 )a , where:
I1 and I2 are cell numbers of competing strains in the inoculum, N1 and N2—numbers in nodules formed by these strains; c is constant which may be either more or less than 1, demonstrating the strain competitiveness; a is constant which is uniformly less than 1 (usually, 0, 2< a< 0, 8).
From this dependence, it is evident that competition for nodulation results in negative FdS in favor of rare genotypes since N1:N2 > I1:I2 at I1< I2 (Provorov and Vorobyov, 2000; Onishchuk et al., 2017). The impact of FdS on mutualism evolution may be represented by an increased population diversity stimulated by the bacterial circulation in plant–soil systems. Deep amplicone sequencing of nodA gene libraries in R. leguminosarum populations composed of bv. viciae and bv. trifolii strains allowed us (Andronov et al., 2015) to address the host-induced selective pressures at two levels—genotypic (operational taxonomic units, OTUs identified at 97%–98% similarity) and haplotypic (individual sequences within the same OTU). We demonstrated that the number of OTUs identified for the nodA gene in nodules is lower than in soil since the avirulent and non-competitive genotypes are excluded from in planta multiplication. However, within symbiotically active OTUs, the diversity of haplotypes is usually increased, suggesting that the rare virulent strains are supported by FdS.
Additional selective pressures were induced during interaction of rhizobia populations with several hosts due to the preferential choice of some bacterial genotypes by legume plants (Andronov et al., 2015; Batstone et al., 2020). It was demonstrated that in R. leguminosarum the host preference pertains nod region located on sym plasmid, not the chromosomally located 16S–23S rRNA region suggesting the impacts of host plants on the microevolution of the symbionts, not on their speciation (Jorrin and Imperial, 2015). Due to the choice of the partners, co-vegetating legume hosts induce Disruptive Selection (DiS), resulting in a narrowed host range, which may be correlated with an improved N2 fixation rate (Provorov and Vorobyov, 2010).
Importantly, host preference does not pertain to the nif genes, suggesting that hosts do not select symbionts directly for improved N2-fixing activity (Westhoek et al., 2017). However, host preference may result in an increased nodulation competitiveness of rhizobia strains which provide the indirect selective pressures for N2-fixing genotypes (Simonsen and Stinchcombe, 2014). Direct selection in favor of these genotypes may be due to sanctions against non-N2-fixing clones which occupy some nodules (Westhoek et al., 2017) or to stimulating propagation of active N2-fixers which occupy other nodules on the same root, collectively resulting in group-level (inter-deme, kin) selection for improved mutualism efficiency (Section 2.2.2). Analysis of the combined operation of FdS and DiS in the R. leguminosarum population composed of viciae and trifolii biovars may be done via comparison of subpopulations residing in rhizospheric and nodular niches of alternative hosts. It appeared that the p-distance between rhizosperic subpopulations of Vicia/Lathyrus and Trifolium is close to the sum of two p-distances between rhizospheric and nodular subpopulations calculated for each host (Figure 4), suggesting the nearly equal inputs of FdS and DiS in the symbiotic diversification of R. leguminosarum genotypes.
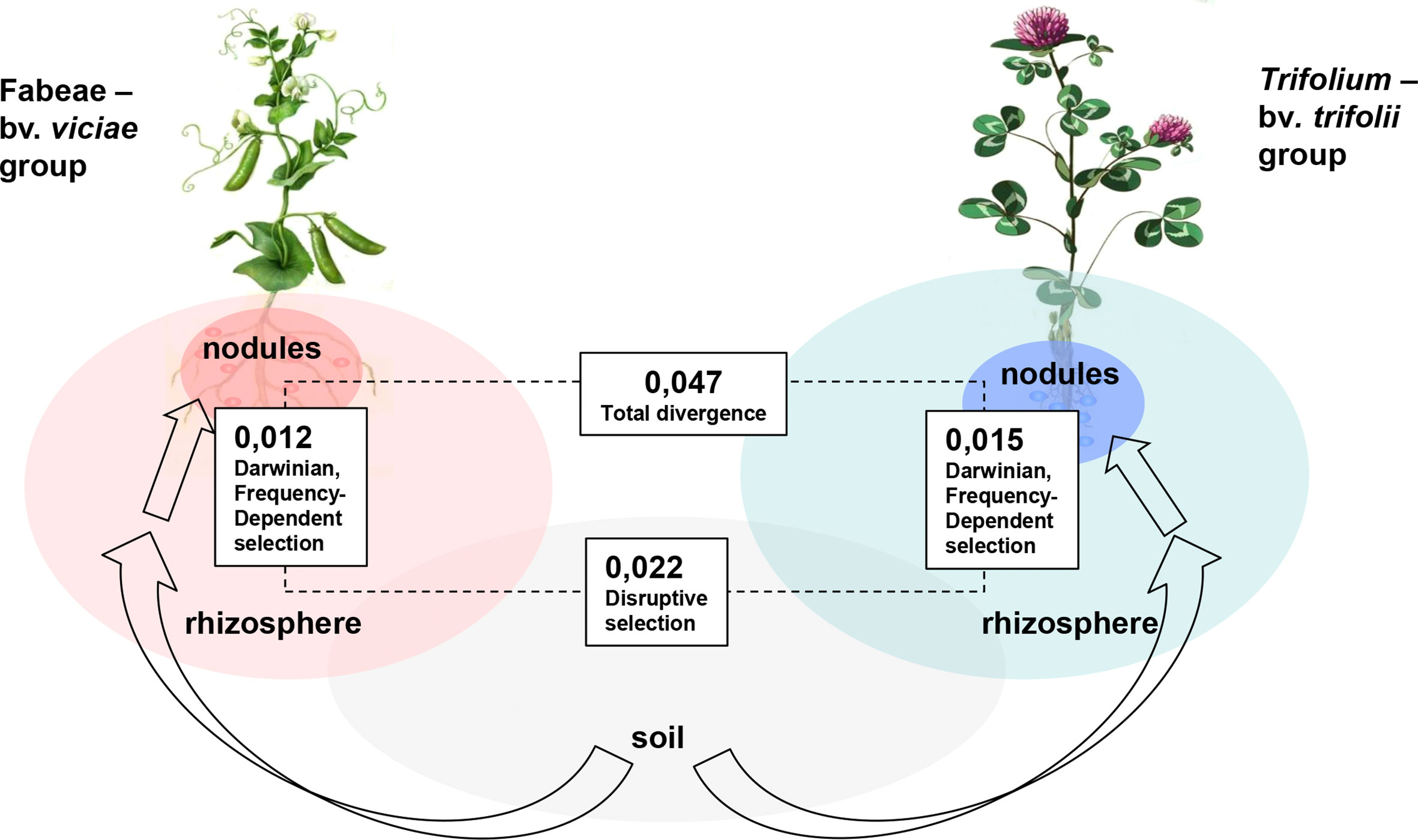
Figure 4 Diversification of Rhizobium leguminosarum population elicited by the natural selection pressures in symbiotic systems [adapted from Andronov et al. (2015)]. Arrows represent the bacteria migration into symbiotic niches, figures – p-distances between nodA in subpopulations formed in different niches under the impacts of host-induced selection: (i) Disruptive selection is elicited by rhizobia migration from soil to rhizospheres of hosts representing different cross-inoculation groups formed by the Fabeae tribe with bv. viciae and by Trifolium spp. with bv. trifolii; and (ii) Darwinian and Frequency-dependent selection are elicited by migration of bacteria from rhizospheres to nodules in each cross-inoculation group.
Host-induced selection in rhizobia populations results in the coevolution of the partners represented by shifts in population structure in a symbiotic organism under the impacts of population changes in its host (Janzen, 1980). These shifts result in the “evolutionary molding” (Igolkina et al., 2019), expressed as the congruent nod and NFR phylogenies and matching the diversity of NFs in rhizobial populations to the diversity of plant receptors. A pronounced congruence was also revealed when the phylogenies of galegoid legumes and their fast-growing symbionts (the Rhizobiaceae) were compared (Figure 2), but was not revealed for the higher ranked taxa of the partners (Provorov, 1998), probably due to multiple changes in host ranges induced by HGT in rhizobia populations (Section 3.1).
2.2.2 Symbiotic N2 fixation: Group selection
According to the concept of symbiosis, coined by de Bary (1879), it represents a continuum of mutualistic and antagonistic interactions, which are similar in their mechanisms and are evolutionary interconnected. Being the founder of phytopathology, de Bary assigned a leading role in this continuum to parasitism, which can be sometimes reorganized into mutualism under the impacts of selective pressures favoring reciprocally beneficial cooperation (Dobzhansky, 1951). Lewis (1974) suggested that these reorganizations were related to nutritional strategies in symbiotic microbes that evolved from necrotrophic to biotrophic and symbiotrophic ones.
Simulation of population dynamics in symbiotic organisms suggests that individual selection explains readily the evolution of antagonism but not of mutualism (Wyatt et al., 2013), which is sometimes addressed as a side-effect of individual adaptations (Smith, 1989). Specifically, DaS models fail to explain the rhizobia evolution for irreversible differentiation into N2-fixing bacteroids, which may be addressed as temporary organelles of plant cells (Coba de la Peña et al., 2017), since the bacteroid operation is considered “altruistic” towards the legume hosts (Provorov, 2021).
In order to reconcile the theory of natural selection with the evolution of beneficial cooperation, one can suggest that the selective pressures favoring mutualism are implemented within the endosymbiotic microbial populations under host impacts. In nodular symbiosis, these pressures should be related to the positive feedbacks of partners: the intra-nodular rhizobia groups (which are often represented by clonal progenies of individual cells) if fixing N2 actively, obtain a preferential C supply from hosts which support not only the nitrogenase activity but also the in planta propagation of N2-fixing bacterial genotypes (Udvardi and Kahn, 1992). The population structures of micro-symbionts favorable for this selection result from inoculation mechanisms evolved from the crack entry that leads to the mixed rhizobial infection, towards the root hair entry by individual cells or micro-colonies which favors the “clonal endophytes” (Brewin, 1998; Sprent, 2001; Brewin, 2004).
Experimental evidence for preferential C supply of Fix+ soybean nodules, which may be either provided with or devoid of N2 (Denison and Kiers, 2004), suggests a positive in planta selection in favor of the clonally propagated Fix+ genotypes. This propagation may be also supported by negative selection against Nod+Fix− genotypes due to host-induced “sanctions” based on nutrient restrictions or on defense reactions (Denison, 2000). Collectively, these mechanisms may promote group (inter-deme) selection in favor of N2-fixing genotypes within the nodular rhizobia populations.
3 Macroevolution: Divergence based on gene transfer
For eukaryotic organisms, the emergence of superspecies taxa (macroevolution) is usually considered as an extension of microevolution and speciation as adaptive processes dependent on natural selection. Rhizobia provide the opportunity to address the validity of this approach for prokaryotes by dissecting the genomic and ecological mechanisms of their macroevolution. It involves the emergence of symbiotic N2-fixers from free-living bacteria acquiring the sym gene systems via two processes: (i) genomic rearrangements in the ancestral N2 fixers (Divergence of core and accessory genomes); and (ii) transfer of sym genes from rhizobia to diverse soil-borne bacteria (Figure 1).
3.1 HGT-based emergency of rhizobia
As we indicated previously, emergence of primary rhizobia (Bradyrhizobium) from free-living phototrophic N2-fixers (Rhodopseudomonas) involved: (i) allocation of some photosynthesis-controlling genes into nitrogenase-controlling network in free-living bacteria which resulted in the root-associated genotypes to be used for rice crop fertilization (Maeda, 2022) and may be further evolved into phototrophic stem-nodulating rhizobia (see below); (ii) acquisition of nod genes via HGT resulted in root-nodulating genotypes (Ding and Hynes, 2009; Arashida et al., 2022). The hypothesis of the direct filiation of Rhodopsedomonas into primary rhizobia (Bradyrhizobium) is supported by the transitional forms represented by phototrophic bradyrhizobial strains devoid of nod genes (Mornico et al., 2012). A possibility to consider the legume-nodulating β-proteobacteria (e.g., Paraburkholderia) as the primary rhizobia may be discussed since some representatives of this bacterial group were identified as endosymbionts of Glomerymycotan fungi (Pawlowska et al., 2018), the ancient symbionts of land plants forming arbuscular mycorrhiza which perhaps donated some of their bacterial symbionts to the plant hosts (Provorov and Shtark, 2014).
The first process involves the conversion of some photosynthesis-controlling genes into nitrogenase-controlling ones (e.g., cc3NOPQ into fixNOPQ) (Rey and Harwood, 2010). This reorganization resulted in photosynthetically active Bradyrhizobium genotypes nodulating the stems in some tropical legumes (e.g., Aeschynomene) via the crack entry without using NFs typical for majority of rhizobia (Sprent, 2001). The NF synthesis has been acquired by bradyrhizobial species (e.g., B. japonicum, B. elkanii) nodulating the legumes via root hair infection. In these bacteria, phototrophy was functionally substituted by the ability to use plant photosynthesis products. The resulted bradyrhizobia often retain the ex planta nif gene expression, but they are usually not capable of diazotrophic growth due to low free-living nitrogenase activity (Wongdee et al., 2018). The emergence and evolution of primary rhizobia involved the enlargement of individual genomes and of pangenomes, which was based on the extension of their accessory parts (Table 2).
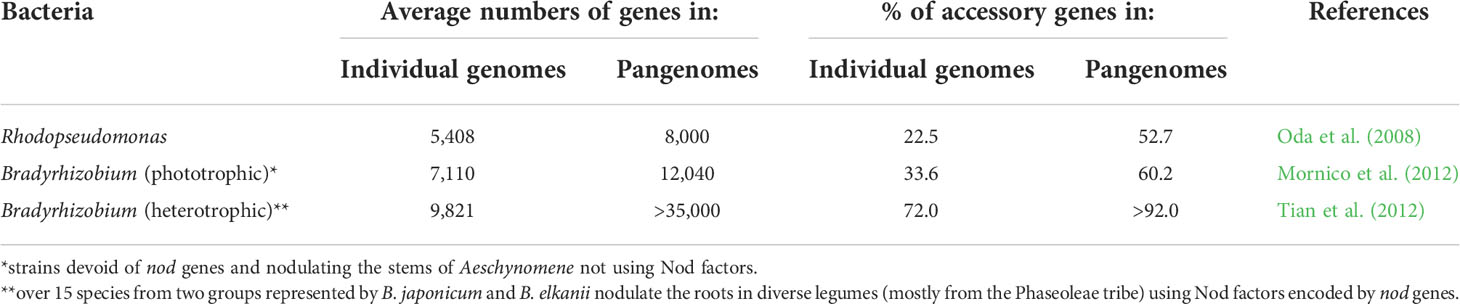
Table 2 Genomic features of free-living (Rhodopseudomonas) and symbiotic (Bradyrhizobium) members of the Bradyrhizobiaceae.
The nod genes encoding for the NF synthesis may be acquired by rhizobia from actinobacteria Frankia, which are ancient N2-fixing symbionts of Rosid I dicots. Specifically, some Frankia strains possess common nodABC genes which are activated during host (Datisca glomerata) nodulation and are functionally interchangeable with the rhizobial nod genes (Persson et al., 2015). Importantly, NodA-like acyl transferases are found in diverse actinobacteria, while in α-proteobacteria these enzymes are restricted to rhizobia. When acquired nodABC, ancestral rhizobia possibly substituted Frankia in the endosymbiotic niches due to a more rapid multiplication of unicellular α-proteobacteria as compared to multicellular actinobacteria (Provorov and Vorobyov, 2010).
Subsequent evolution of the Rhizobiales was presumably due to HGT-based polyphyletic emergence of multiple rhizobia taxa induced by co-migration of symbionts with their hosts into the novel areas wherein sym genes were donated by introduced bacteria to the local ones (Table 3). This evolution is well documented for symbionts of polebean (Phaseolus vulgaris), which in the Central- and Southern-American centers of origin is associated with R. tropici and R. etli (Eardly et al., 1995; Bernal and Graham, 2001; Aguilar et al., 2004). In Europe where the polebean was introduced in Columbian times, a broad spectrum of new symbionts emerged, including R. gallicum, R. giardinii, and R. leguminosarum bv. phaseoli (Amarger et al., 1994; Laguerre et al., 2001; Martínez-Romero, 2003). These rhizobia harbor a range of nod markers common to R. tropici and R. etli, suggesting the similarities of NFs synthesized by ancestral and derived polebean symbionts (Laguerre et al., 2001).
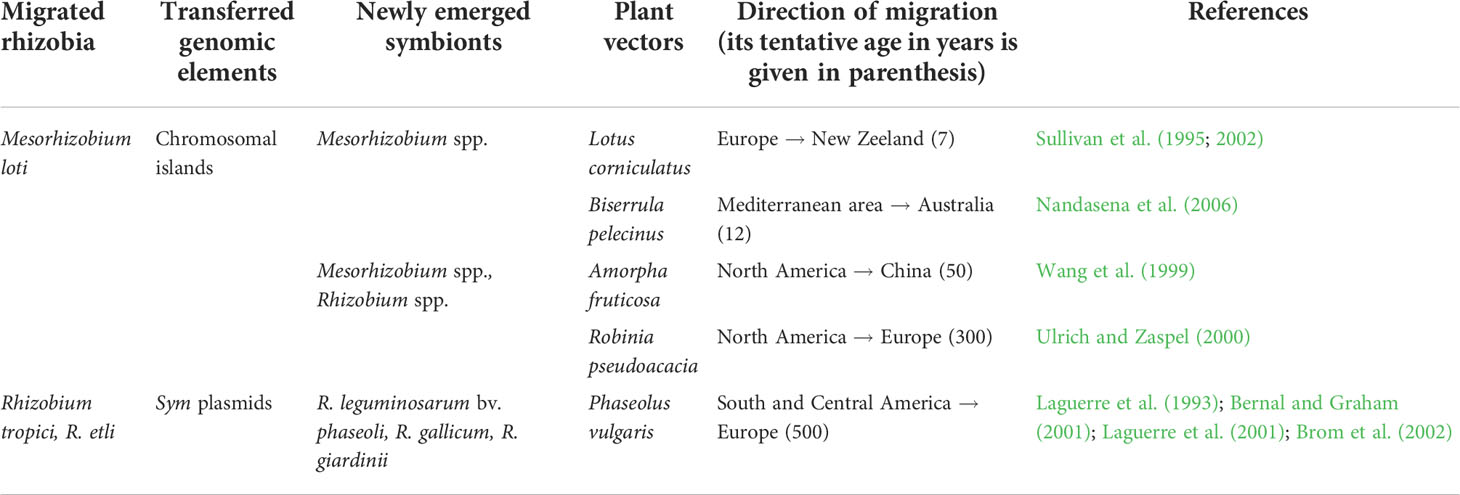
Table 3 Emergence of novel symbionts of legumes by transfer of sym genes to local bacteria from rhizobia migrated to the novel ecological areas with the help of plant vectors.
A rapid generation of novel symbionts via recombination of introduced rhizobia with local bacteria was demonstrated for Mesorhizobium spp. harboring sym genes in chromosomal islands, which may be transmitted to local bacteria as the conjugative transposons via type 4 secretion systems (Sullivan et al., 2002). This transmission was demonstrated for the trefoil (Lotus corniculatus) rhizobia introduced from Europe to New Zealand, wherein the novel symbiont populations were established in a few years (Sullivan et al., 1995; Sullivan et al., 2002). Similar processes accompanied the co-introduction of Mesorhizobium spp. with Robinia pseudoacacia from North America to Europe, Amorpha fruticosa from North America to China, and Biserrula pelecinus from the Mediterranean area to Australia (Table 3).
3.2 sym gene activation
Obviously, the HGT-based emergence of novel rhizobia species may be restricted by the poor expression of transferred sym genes in the foreign genomic background. For example, after pSym transfer from Rhizobium and Sinorhizobium species to closely related agrobacteria, low virulent, non-N2-fixing recombinants usually emerge. Few reports are available on the emergency of N2-fixing recombinants via transfer of pSyms into agrobacteria from the broad-host-range rhizobia, e.g., from R. tropici (Rogel et al., 2001). The recombinant genotypes resulting from sym gene transfer from rhizobia to distant bacteria are mostly non-virulent (reviewed in Provorov and Vorobyov (2010)). Therefore, additional genomic changes accompanied by host-induced selective pressures were required to support the symbiotically active strains in which the newly acquired sym genes were functional (Figure 5).
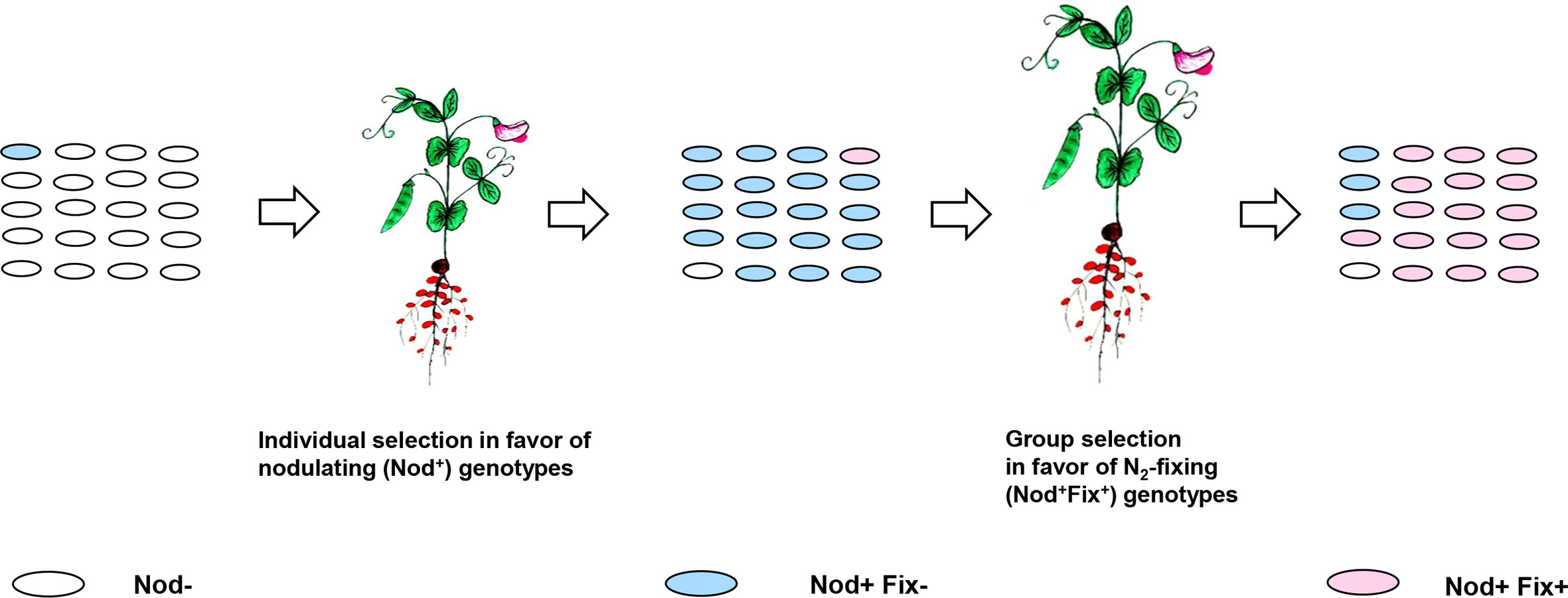
Figure 5 Derepression of the newly acquired sym genes in recombinant genotypes supported by host-induced selective pressures. Individual selection (Section 2.2.1) in a population of non-nodulating (Nod−) recombinants generated by transfer of sym genes results in an increased frequency of Nod+Fix− genotypes generated via nod gene derepression. Group selection (introduced in Section 2.2.2) occurs in favor of N2-fixing (Nod+Fix+) genotypes (with the derepressed nif/fix genes) propagated actively in N2-fixing nodules due to their preferential carbon supply.
Analysis of host-induced selective processes involved in rhizobia evolution (Section 2.2) suggests that conversion of non-virulent recombinants into active nodulators may be dependent on individual selection in favor of virulent genotypes with derepressed nod genes, providing the ability of bacteria to actively propagate in planta. Positive selection for nod gene derepression (Nod+ phenotype) may be highly effective if the rare virulent rhizobia cells are picked by plants from mixtures with numerous non-virulent bacteria (Clúa et al., 2018).
However, Nod+ strains supported by individual selection may be represented mostly by non-N2-fixing cheaters, which require additional genetic changes to be transformed into N2-fixing (Fix+) mutualists. The relevant selective pressures possibly operate in planta at the level of intra-nodular clones: Fix+ genotypes may be supported by group (inter-deme) selection (Provorov, 2021). It favors strains with nif/fix gene derepression propagated due to the active supply of bacteria with the plant photosynthesis products based on their exchange for N2 fixation products (Figure 5).
4 Towards a multilevel classification of evolutionary processes (conclusion)
In this paper, we use nodule bacteria as a model to study the interplay of microevolutionary, speciation, and macroevolutionary processes. For eukaryotic organisms, their trade-off is conventionally considered as hierarchic and reductionist: speciation and macroevolution are addressed as an extension of microevolution (Dobzhansky, 1951; Koonin, 2009). According to STE, these processes are driven by individual selection, which supports genotypes with high fitness measured as the production of fertile progeny (Timofeeff-Ressovsky et al., 1977; Koonin, 2011). However, different genetic mechanisms were proposed for micro- and macro-evolution by J. Philiptschenko (1927), who suggested that in eukaryotic organisms these processes are associated with reorganizations of nuclear and cytoplasmic genes, respectively.
We suggest that diverse evolutionary strategies are implemented in rhizobia at different phylogenetic levels and result in the gene, genomic, and phylogenomic reorganizations responsible for the rhizobia microevolution, speciation, and macroevolution (Table 4). Divergent evolution based on genomic reorganizations and driven by disruptive selection occurs mostly at the species and subspecies levels, while at the superspecies level, HGT-based (reticular) evolution is implemented. These reorganizations are elicited by host-induced and environmentally-dependent selective factors, which include symbiosis-specific forms of natural selection operating at the individual and group levels. The impact of these factors is evident also for the macroevolutionary processes responsible for conversion of HGT-born recombinants into novel rhizobia species (Figure 5). Importantly, in rhizobia, the symbiotically specific natural selection pressures induce modifications of sym gene clusters, which represent the most active evolutionary part of the dispensable (accessory) gene pool driving evolution of the whole bacterial genomes.
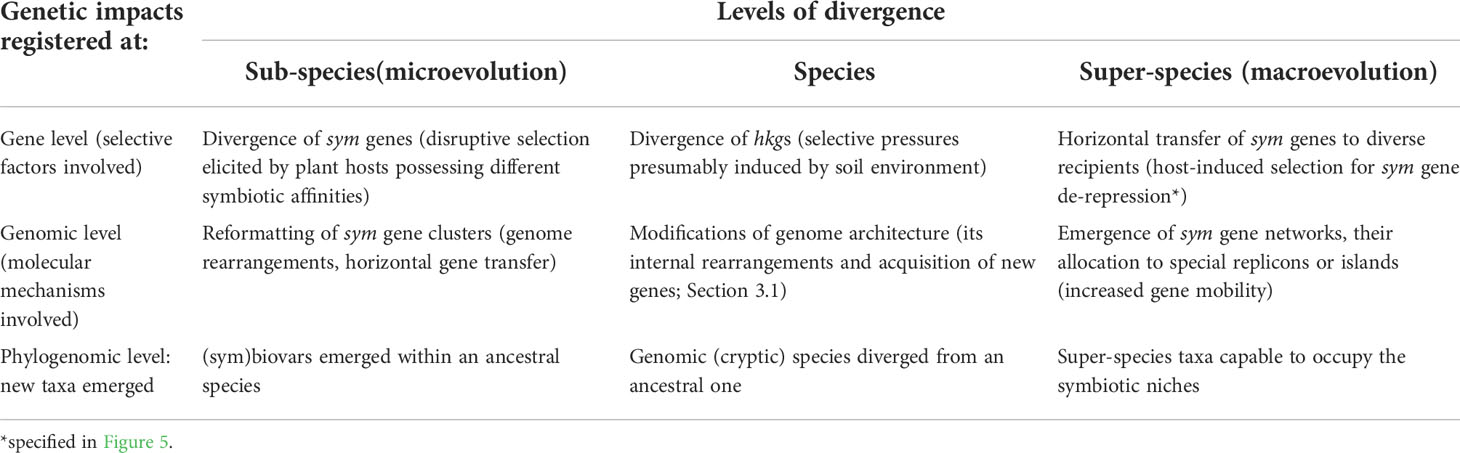
Table 4 Multilevel classification of evolutionary processes in rhizobia based on reorganizations of symbiotically specialized (sym) and of housekeeping genes (hkg).
Rhizobia provide broad opportunities to study the coevolution of partners in mutualistic symbioses. According to the definition coined by Janzen (1980), coevolution involves the inter-dependent changes in population structures of tightly interacting species. Up to now, co-evolutionary processes have been studied mostly in antagonistic symbioses wherein “gene-for-gene” interactions between parasites and hosts are implemented (Jones and Dangl, 2006). These interactions are controlled by individual selection operating in the Darwinian and frequency-dependent forms; it usually results in the coordinated oscillations in frequencies of virulence and resistance genes encoding the specificity of interactions between the partners (Luijckx et al., 2013).
A range of similar molecular and ecological mechanisms involved in the coevolution of the partners were revealed in mutualistic symbioses. In the legume–rhizobia system, co-evolution may be represented as “evolutionary molding,” matching the rhizobia population diversity for nod genes encoding for NF synthesis to the host diversity for NF-specific receptors (Igolkina et al., 2019). This co-evolution results in the narrowing specificity of the interactions of the partners, which may be correlated to an increased benefit of their co-operation (Provorov and Vorobyov, 2010).
For future research, it would be interesting to address the legume-rhizobia coevolution for the components of cooperative metabolic pathways, linking the bacterial nif/fix genes for nitrogenase synthesis with the plant GS/GOGAT/AAT genes for assimilation of fixed nitrogen or the bacterial genes encoding for catabolism of C compounds with the plant genes responsible for providing these compounds for bacteroids (Udvardi and Poole, 2013). From a perspective, application of the rhizobia–legume model will allow us to represent the coevolution of the partners for signaling and metabolic interactions as the natural history of an integral holobiont possessing a hologenome encoding for the cooperative adaptations of tightly integrated organisms to adverse environments (Zilber-Rosenberg and Rosenberg, 2008). Specifically, legume genes for hosting rhizobia may be addressed as homeotic (master) genes since they allow plants to switch on the novel developmental program providing the adaptively valuable symbiotrophic N nutrition (Bhattacharjee et al., 2015).
Speaking generally, symbiotic models provide clear examples of punctuated evolution (Gould and Eldredge, 1993), since the hosting of symbiotic microbes represents the rapid evolutionary bursts in contrast to gradual evolution suggested by the conventional models of natural selection (Gould, 1989). In symbiotic systems, hypothetical “hopeful monsters” may be replaced by actual “successful cooperators” in which the increased fitness is due to functional and structural innovations resulting from the integration of hosts with symbiotic microbes into the holobiont/hologenome units (Theis et al., 2016). In these units, the non-friendly, competitive, and antagonistic interactions of partners may be reorganized into beneficial symbioses, providing the broad prospects for adaptive and progressive coevolution of partners (Douglas, 2014).
Author contributions
NP—project conceptualization, manuscript preparation, and funding acquisition. EA—project conceptualization and manuscript editing. AK—manuscript preparation. OO—collection of literature and manuscript proofreading. AI—project conceptualization and data processing. EK—project conceptualization, data processing. All authors contributed to the article and approved the submitted version.
Funding
This study is supported by the Russian Science Foundation, grant 19-16-00081P.
Acknowledgments
Authors are grateful for the contribution of the Russian Collection of Agricultural Microorganisms (RCAM, WDCM 966 supervised by Dr. V. Safronova) and the Centre for Genomic Technologies, Proteomics and Cell Biology in ARRIAM.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aguilar, O. M., Riva, O., Peltzer, E. (2004). Analysis of Rhizobium etli and its symbiosis with Phaseolus vulgaris supports coevolution in centers of host diversification. Proc. Natl. Acad. Sci. U.S.A. 101 (37), 13548–13553. doi: 10.1073/pnas.0405321101
Amarger, N., Bours, M., Revoy, F., Allard, M. R., Laguerre, G. (1994). Rhizobium tropici nodulates field-growing Phaseolus vulgaris in France. Plant Soil. 161, 147–156. doi: 10.1007/BF00046386
Amarger, N., Lobreau, J. P. (1982). Quantitative study of nodulation competitiveness in Rhizobium strains. Appl. Environ. Microbiol. 44 (3), 583–588. doi: 10.1128/aem.44.3.583-588.1982
Andronov, E. E., Igolkina, A. A., Kimeklis, A. K., Vorobyov, N. I., Provorov, N. A. (2015). Characteristics of natural selection in populations of nodule bacteria (Rhizobium leguminosarum) interacting with different host plants. Russ. J. Genet. 51, 949–956. doi: 10.1134/S1022795415100026
Arashida, H., Odake, H., Sugawara, M., Noda, R., Kakizaki, K., Ohkubo, S., et al. (2022). Evolution of rhizobial symbiosis islands through insertion sequence-mediated deletion and duplication. ISME J. 16, 112–121. doi: 10.1038/s41396-021-01035-4
Azani, N., Babineau, M., Bailey, C. D., Banks, H., Barbosa, A. R., Pinto, R. B., et al. (2017). A new subfamily classification of the leguminosae based on a taxonomically comprehensive phylogeny: The legume phylogeny working group (LPWG). Taxon. 66, 44–77. doi: 10.12705/661.3
Bailly, X., Olivieri, I., de Mita, S., Cleyet-Marel, J.-C., Bena, G. (2006). Recombination and selection shape the molecular diversity pattern of nitrogen-fixing Sinorhizobium sp. associated to Medicago. Molec. Ecol. 15, 2719–2734. doi: 10.1111/j.1365-294X.2006.02969.x
Batstone, R. T., Peters, M. A. E., Simonsen, A. K., Stinchcombe, J. R., Frederickson, M. E. (2020). Environmental variation impacts trait expression and selection in the legume–rhizobium symbiosis. Amer. J. Bot. 107, 195–208. doi: 10.1002/ajb2.1432
Bernal, G., Graham, P. H. (2001). Diversity on the rhizobia associated with Phaseolus vulraris l. @ in Ecuador, the comparison with Mexican bean rhizobia. Canad. J. Microbiol. 47 (6), 526–534. doi: 10.1139/w01-037
Berrada, H., Fikri-Benbrahim, K. (2014). Taxonomy of the rhizobia: current perspectives. Br. Microbiol. Res. J. 4 (6), 16–639. doi: 10.9734/BMRJ/2014/5635
Betti, M., García-Calderón, M., Pérez-Delgado, C. M., Credali, A., Estivill, G., Galván, F., et al. (2012). Glutamine synthetase in legumes: recent advances in enzyme structure and functional genomics. Int. J. Mol. Sci. 13, 7994–8024. doi: 10.3390/ijms13077994
Bhattacharjee, A., Ghangal, R., Garg, R., Jain, M. (2015). Genome-wide analysis of homeobox gene family in legumes: Identification, gene duplication and expression profiling. PloS One 10 (3), e0119198. doi: 10.1371/journal.pone.0119198
Bobay, L.-M. (2020). “The prokaryotic species concept and challenges,” in The pangenome: Diversity, dynamics and evolution of genomes. Eds. Tettelin, H., Medini, D. (Cham, CH: Springer).
Boogerd, F. C., Ferdinandy-van Vlerken, M. M., Mawadza, C., Pronk, A. F., Stouthamer, A. H., van Verseveld, H. W. (1994). Nitrogen fixation and hydrogen metabolism in relation to the dissolved oxygen tension in chemostat cultures of the wild type and a hydrogenase-negative mutant of Azorhizobium caulinodans. Appl. Environ. Microbiol. 60 (6), 1859–1866. doi: 10.1128/aem.60.6.1859-1866.1994
Brewin, N. J. (1998). “Tissue and cell invasion by rhizobium: The structure and development of infection threads and symbiosomes,” in The rhizobiaceae: Molecular biology of model plant-associated bacteria. Eds. Spaink, H. P., Kondorosi, A., Hooykaas, P. J. J. (Dordrecht, the Netherlands: Springer), 441–452.
Brewin, N. J. (2004). Plant cell wall remodeling in the Rhizobium-legume symbiosis. Crit. Rev. Plant Sci. 23 (4), 293–316. doi: 10.1080/07352680490480734
Broghammer, A., Krusell, L., Blaise, M., Sauer, J., Sullivan, J. T., Maolanon, N., et al. (2012). Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc. Natl. Acad. Sci. U.S.A. 109, 13859–13864. doi: 10.1073/pnas.1205171109
Brom, S., Girard, L., Garcia-de los Santos, A., Sanjuan-Pinilla, J. M., Olivares, J., Sanjuan, J. (2002). Conservation of plasmid-encoded traits among bean-nodulating Rhizobium species. Appl. Environ. Microbiol. 68 (5), 2555–2561. doi: 10.1128/AEM.68.5.2555-2561.2002
Chirak, E. R., Kimeklis, A. K., Karasev, E. S., Kopat, V. V., Safronova, V. I., Belimov, A. A., et al. (2019). Search for ancestral features in genomes of Rhizobium leguminosarum bv. viciae strains isolated from the relict legume Vavilovia formosa. Genes. 10, 990. doi: 10.3390/genes10120990
Clúa, J., Roda, C., Zanetti, M. E., Blanco, F. A. (2018). Compatibility between legumes and rhizobia for establishment of a successful nitrogen-fixing symbiosis. Genes. 9, 125. doi: 10.3390/genes9030125
Coba de la Peña, T., Fedorova, E., Pueyo, J. J., Lucas, M. M. (2017). The symbiosome: legume and rhizobia co-evolution toward a nitrogen-fixing organelle? Front. Plant Sci. 8. doi: 10.3389/fpls.2017.02229
Debellé, F., Moulin, L., Mangin, B., Dénarié, J., Boivin, C. (2001). Nod genes and nod signals and the evolution of the Rhizobium-legume symbiosis. Acta Biochim. Pol. 48 (2), 359–365. doi: 10.18388/abp.2001_3921
Denison, R. F. (2000). Legume sanctions and the evolution of symbiotic cooperation by rhizobia. Am. Nat. 156 (6), 567–576. doi: 10.1086/316994
Denison, R. F., Kiers, E. T. (2004). Lifestyle alternatives for rhizobia: mutualism, parasitism and foregoing symbiosis. FEMS Microbiol. Lett. 237 (2), 187–193. doi: 10.1111/j.1574-6968.2004.tb09695.x
Ding, H., Hynes, M. F. (2009). Plasmid transfer systems in the rhizobia. Can. J. Microbiol. 55 (8), 917–927. doi: 10.1139/w09-056
Douglas, A. E. (2014). Symbiosis as a general principle in eukaryotic evolution. Cold Spring Harb. Perspect. Biol. 6 (2), a016113. doi: 10.1101/cshperspect.a016113
Eardly, B. D., Wang, F.-S., Whittam, T. S., Selander, R. K. (1995). Species limits in Rhizobium populations that nodulate the common bean (Phaseolus vulgaris). Appl. Environ. Microbiol. 61 (2), 507–512. doi: 10.1128/aem.61.2.507-512.1995
Fuchsman, C. A., Collins, R. E., Rocap, G., Brazelton, W. J. (2017). Effect of the environment on horizontal gene transfer between bacteria and archaea. PeerJ. 5, e3865. doi: 10.7717/peerj.3865
Gottfert, M. (1993). Regulation and function of rhizobial nodulation genes. FEMS Microbiol. Rev. 104, 39–64. doi: 10.1111/j.1574-6968.1993.tb05863.x
Gould, S. J. (1989). Wonderful life: The burgess shale and the nature of history (New York: W.W. Norton & Company).
Gould, S. J., Eldredge, N. (1993). Punctuated equilibrium comes of age. Nature. 366 (6452), 223–227. doi: 10.1038/366223a0
Hassan, S., Mathesius, U. (2012). The role of flavonoids in root-rhizosphere signalling: Opportunities and challenges for improving plant-microbe interactions. J. Exp. Bot. 63, 3429–3444. doi: 10.1093/jxb/err430
Haukka, K., Lindström, K., Young, J. P. (1998). Three phylogenetic groups of nodA and nifH genes in Sinorhizobium and Mesorhizobium isolates from leguminous trees growing in Africa and Latin America. Appl. Environ. Microbiol. 64 (2), 419–426. doi: 10.1128/AEM.64.2.419-426.1998
Igolkina, A. A., Bazykin, G. A., Chizhevskaya, E. P., Provorov, N. A., Andronov, E. E. (2019). Matching population diversity of rhizobial nodA and legume NFR5 genes in plant-microbe symbiosis. Ecol. Evol. 9, 10377–10386. doi: 10.1002/ece3.5556
Janzen, D. H. (1980). When is it coevolution? Evolution 34, 409–616. doi: 10.1111/j.1558-5646.1980.tb04849.x
Jones, J. D. G., Dangl, J. L. (2006). The plant immune system. Nature. 444, 323–329. doi: 10.1038/nature05286
Jorrin, B., Imperial, J. (2015). Population genomics analysis of legume host preference for specific rhizobial genotypes in the Rhizobium leguminosarum bv. viciae symbioses. Mol. Plant Microbe Interact. 28 (3), 310–318. doi: 10.1094/MPMI-09-14-0296-FI
Karasev, E. S., Andronov, E. E., Aksenova, T. S., Tupikin, A. E., Provorov, N. A., et al. (2019). Evolution of goat's rue rhizobia (Neorhizobium galegae): an analysis of the polymorphism of the nitrogen fixation genes and the genes of nodule formation. Russ. J. Genet. 55, 234–238. doi: 10.1134/S1022795419020078
Katz, L. A. (2015). Recent events dominate interdomain lateral gene transfers between prokaryotes and eukaryotes and, with the exception of endosymbiotic gene transfers, few ancient transfer events persist. Philos. Trans. R Soc Lond. B Biol. Sci. 370 (1678), 20140324. doi: 10.1098/rstb.2014.0324
Kimeklis, A. K., Chirak, E. R., Kuznetsova, I. G., Sazanova, A. L., Safronova, V. I., Belimov, A. A., et al. (2019). Rhizobia isolated from the relict legume Vavilovia formosa represent a genetically specific group within Rhizobium leguminosarum biovar viciae. Genes. 10, 991. doi: 10.3390/genes10120991
Kimeklis, A. K., Kuznetsova, I. G., Sazanova, A. L., Safronova, V. I., Belimov, A. A., Onishchuk, O. P., et al. (2018). Divergent evolution of symbiotic bacteria: rhizobia of the relic legume Vavilovia formosa form an isolated group within Rhizobium leguminosarum bv. viciae. Russ. J. Genet. 54, 866–870. doi: 10.1134/S1022795418070062
Koonin, E. V. (2009). The origin at 150: is a new evolutionary synthesis in sight? Trends Genet. 25, 473–475. doi: 10.1016/j.tig.2009.09.007
Koonin, E. V. (2011). Logic of chance, the nature and origin of biological evolution (Upper Saddle River, NJ: FT Press).
Kryazhimskiy, S., Plotkin, J. B. (2008). The population genetics of dN/dS. PloS Genet. 4, e1000304. doi: 10.1371/journal.pgen.1000304
Kumar, N., Lad, G., Giuntini, E., Kaye, M. E., Udomwong, P., Shamsani, N. J., et al. (2015). Bacterial genospecies that are not ecologically coherent: population genomics of Rhizobium leguminosarum. Open Biol. 5, 140133. doi: 10.1098/rsob.140133
Laguerre, G., Geniaux, E., Mazurier, S. I., Rodrigues, C. R., Amarger, N. (1993). Conformity and diversity among field isolates of Rhizobium leguminosarum bv. viciae, bv. trifolii and bv. phaseoli revealed by DNA hybridization using chromosome and plasmid probes. Canad. J. Microbiol. 39, 412–419. doi: 10.1139/m93-060
Laguerre, G., Nour, S. M., Macheret, V., Sanjuan, J., Drouin, P., Amarger, N. (2001). Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiology. 147 (Pt4), 981–993. doi: 10.1099/00221287-147-4-981
Lee, K. B., De Backer, P., Aono, T., Liu, C. T., Suzuki, S., Suzuki, T., et al. (2008). The genome of the versa-tile nitrogen fixer Azorhizobium caulinodans ORS571. BMC Genomics 9, 271. doi: 10.1186/1471-2164-9-271
Lewis, D. H. (1974). “Microorganisms and plants: the evolution of parasitism and mutualism,” in Evolution of microbial world. proc. 24th symp. Soc. general microbiol (Cambridge: Univ. Press), 367–392.
Luijckx, P., Fienberg, H., Duneau, D., Ebert, D. (2013). A matching-allele model explains host resistance to parasites. Curr. Biol. 23 (12), 1085–1088. doi: 10.1016/j.cub.2013.04.064
Maeda, I. (2022). Potential of phototrophic purple nonsulfur bacteria to fix nitrogen in rice fields. Microorganisms. 10 (1), 28. doi: 10.3390/microorganisms10010028
Martínez-Romero, E. (2003). Diversity of Rhizobium-phaseolus vulgaris symbiosis: overview and perspectives. Plant Soil. 252, 11–23. doi: 10.1023/A:1024199013926
Moldovan, M. A., Gelfand, M. S. (2018). Pangenomic definition of prokaryotic species and the phylogenetic structure of prochlorococcus spp. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00428
Mornico, D., Miché, L., Béna, G., Nouwen, N., Verméglio, A., Vallenet, D., et al. (2012). Comparative genomics of Aeschynomene symbionts: insights into the ecological lifestyle of nod-independent photosynthetic bradyrhizobia. Genes. 3 (1), 35–61. doi: 10.3390/genes3010035
Nandasena, K. G., O’Hara, G. W., Tiwari, R. P., Howieson, J. G. (2006). Rapid in situ evolution of nodulating strains for Biserrula pelecinus l. through lateral transfer of a symbiosis island from the original mesorhizobial inoculant. Appl. Environ. Microbiol. 72, 7365–7367. doi: 10.1128/AEM.00889-06
Oda, Y., Larimer, F. W., Chain, P. S., Malfatti, S., Shin, M. V., Vergez, L. M., et al. (2008). Multiple genome sequences reveal adaptations of a phototrophic bacterium to sediment microenvironments. Proc. Natl. Acad. Sci. U.S.A. 105 (47), 18543–18548. doi: 10.1073/pnas.0809160105
Okazaki, S., Noisangiam, R., Okubo, T., Kaneko, T., Oshima, K., Hattori, M. (2015). Genome analysis of a novel Bradyrhizobium sp. DOA9 carrying a symbiotic plasmid. PloS One 10, e0117392. doi: 10.1371/journal.pone.0117392
Onishchuk, O. P., Vorobyov, N. I., Provorov, N. A. (2017). Nodulation competitiveness of nodule bacteria: genetic control and adaptive significance: Review. Appl. Biochem. Microbiol. 53, 131–139. doi: 10.1134/S0003683817020132
Ormeño-Orrillo, E., Servín-Garcidueñas, L. E., Rogel, M. A., González, V., Peralta, H., Mora, J., et al. (2015). Taxonomy of rhizobia and agrobacteria from the rhizobiaceae family in light of genomics. System. Appl. Microbiol. 38 (4), 287–291. doi: 10.1016/j.syapm.2014.12.002
Pawlowska, T. E., Gaspar, M. L., Lastovetsky, O. A., Mondo, S. J., Real-Ramirez, I., Shakya, E., et al. (2018). Biology of fungi and their bacterial endosymbionts. Annu. Rev. Phytopathol. 56, 289–309. doi: 10.1146/annurev-phyto-080417-045914
Persson, T., Battenberg, K., Demina, I. V., VigilStenman, T., Vanden Heuvel, B., Pujic, P., et al. (2015). Candidatus frankia datiscae Dg1, the actinobacterial microsymbiont of Datisca glomerata, expresses the canonical nod genes nodABC in symbiosis with its host plant. PloS One 10, e0127630. doi: 10.1371/journal.pone.0127630
Peyer, S. M., Pankey, M. S., Oakley, T. H., McFall-Ngai, M. J. (2014). Eye-specification genes in the bacterial light organ of the bobtail squid Euprymna scolopes, and their expression in response to symbiont cues. Mech. Dev. 131, 111–126. doi: 10.1016/j.mod.2013.09.004
Pini, F., Galardini, M., Bazzicalupo, M., Mengoni, A. (2011). Plant-bacteria association and symbiosis: Are there common genomic traits in alphaproteobacteria? Genes. 2 (4), 1017–1032. doi: 10.3390/genes2041017
Poole, P., Ramachandran, V., Terpolilli, J. (2018). Rhizobia: From saprophytes to endosymbionts. Nat. Rev. Microbiol. 16, 291–303. doi: 10.1038/nrmicro.2017.171
Provorov, N. A. (1998). Coevolution of rhizobia with legumes: facts and hypotheses. Symbiosis. 24 (3), 337–367.
Provorov, N. A. (2021). Genetic individuality and inter-species altruism: modelling of symbiogenesis using different types of symbiotic bacteria. Biol. Commun. 66 (1), 65–71. doi: 10.21638/spbu03.2021.108
Provorov, N. A., Andronov, E. E. (2016). Evolution of root nodule bacteria: Reconstruction of the speciation processes resulting from genomic rearrangements in a symbiotic system. Microbiology. 85, 131–139. doi: 10.1134/S0026261716020156
Provorov, N. A., Onishchuk, O. P., Yurgel, S. N., Kurchak, O. N., Chizhevskaya, E. P., Vorobyov, N. I., et al. (2014). Construction of highly-effective symbiotic bacteria: evolutionary models and genetic approaches. Russ. J. Genet. 50, 1125–1136. doi: 10.1134/S1022795414110118
Provorov, N. A., Shtark, O. (2014). Directed evolution of fungi and plants in the symbiotic systems. Mycol. Phytopathol. 48 (3), 151–160.
Provorov, N. A., Vorobyov, N. I. (2000). Population genetics of rhizobia: construction and analysis of an “infection and release” model. J. Theor. Biol. 205 (1), 105–119. doi: 10.1006/jtbi.2000.2051
Provorov, N. A., Vorobyov, N. I. (2006). Interplay of Darwinian and frequency-dependent selection in the host-associated microbial populations. Theor. Popul. Biol. 70 (3), 262–272. doi: 10.1016/j.tpb.2006.06.002
Provorov, N. A., Vorobyov, N. I. (2010). Evolutionary genetics of plant-microbe symbioses. Ed. Tikhonovich, I. A. (New York: NOVA Science Publishers).
Rey, F. E., Harwood, C. S. (2010). FixK, a global regulator of microaerobic growth, controls photosynthesis in Rhodopseudomonas palustris. Mol. Microbiol. 75 (4), 1007–1020. doi: 10.1111/j.1365-2958.2009.07037.x
Rogel, M. A., Hernandez-Lucas, I., Kuykendall, L. D., Balkwill, D. L., Martinez-Romero, E. (2001). Nitrogen-fixing nodules with Ensifer adhaerens harboring Rhizobium tropici symbiotic plasmids. Appl. Environ. Microbiol. 67 (7), 3264–3268. doi: 10.1128/AEM.67.7.3264-3268.2001
Rome, S., Fernandez, M. P., Brunel, B., Normand, P., Cleyet-Marel, J. C. (1996). Sinorhizobium medicae sp. nov., isolated from annual Medicago spp. Int. J. Syst. Bacteriol. 46, 972–980. doi: 10.1099/00207713-46-4-972
Sarkar, S. F., Guttman, D. S. (2004). Evolution of the core genome of Pseudomonas syringae, a highly clonal, endemic plant pathogen. Appl. Environ. Microbiol. 70, 1999–2012. doi: 10.1128/AEM.70.4.1999-2012.2004
Shamseldin, A. (2013). The role of different genes involved in symbiotic nitrogen fixation - review. GJBB. 8, 84–94. doi: 10.5829/idosi.gjbb.2013.8.4.82103
Simonsen, A. K., Stinchcombe, J. R. (2014). Standing genetic variation in host preference for mutualist microbial symbionts. Proc. Biol. Sci. 281 (1797), 20142036. doi: 10.1098/rspb.2014.2036
Słomnicka, R., Olczak-Woltman, H., Bartoszewski, G., Niemirowicz-Szczytt, K. (2015). Genetic and pathogenic diversity of Pseudomonas syringae strains isolated from cucurbits. Eur. J. Plant Pathol. 141, 1–14. doi: 10.1007/s10658-014-0524-4
Sullivan, J. T., Patrick, H. N., Lowther, W. L., Scot, D. B., Ronson, C. W. (1995). Nodulating strains of Rhizobium loti arise through chromosomal symbiotic gene transfer in the environment. Proc. Natl. Acad. Sci. U.S.A. 92 (19), 8985–8989. doi: 10.1073/pnas.92.19.8985
Sullivan, J. T., Trzebiatowski, J. R., Cruickshank, R. W., Gouzy, J., Brown, S. D., Elliot, R. M. (2002). Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J. Bacteriol. 184 (11), 3086–3095. doi: 10.1128/JB.184.11.3086-3095.2002
Terpolilli, J. J., Hood, G. A., Poole, P. S. (2012). What determines the efficiency of N2-fixing Rhizobium-legume symbioses? Adv. Microb. Physiol. 60, 325–389. doi: 10.1016/B978-0-12-398264-3.00005-X
Theißen, G. (2006). The proper place of hopeful monsters in evolutionary biology. Theory Biosci. 124 (3-4), 349–369. doi: 10.1016/j.thbio.2005.11.002
Theis, K. R., Dheilly, N. M., Klassen, J. L., Brucker, R. M., Baines, J. F., Bosch, T. C., et al. (2016). Getting the hologenome concept right: an eco-evolutionary framework for hosts and their microbiomes. mSystems. 1 (2), e00028–e00016. doi: 10.1128/mSystems.00028-16
Tian, C. F., Zhou, Y. L., Zhang, Y. M., Li, Q. Q., Zhang, Y. Z., Li, D. F., et al. (2012). Comparative genomics of rhizobia nodulating soybeans suggests extensive recruitment of lineage-specific genes in adaptations. Proc. Natl. Acad. Sci. U.S.A. 109 (22), 8629–8634. doi: 10.1073/pnas.1120436109
Timofeeff-Ressovsky, N. W., Jablokov, A. V., Glotov, N. V. (1977). Grundriss der populationslehre (Jena: Gustav Fisher Verlag).
Udvardi, M. K., Kahn, M. L. (1992). Evolution of the (Brady)Rhizobium-legume symbiosis: why do bacteroids fix nitrogen? Symbiosis. 14, 87–101.
Udvardi, M., Poole, P. S. (2013). Transport and metabolism in legume-rhizobia symbioses. Annu. Rev. Plant Biol. 64, 201–225. doi: 10.1146/annurev-arplant-050312-120235
Ulrich, A., Zaspel, I. (2000). Phylogenetic diversity of rhizobial strains nodulating Robinia pseudoacacia l. Microbiology. 146 (Pt11), 2997–3005. doi: 10.1099/00221287-146-11-2997
Wang, E. T., Tian, C. F., Chen, W. F., Young, J. P. W., Chen, W. X. (2019). Ecology and evolution of rhizobia (principles and applications) (Singapore: Springer). doi: 10.1007/978-981-32-9555-1
Wang, E. T., van Berkum, P., Sui, X. H., Beyene, D., Chen, W., Martinez-Romero, E. (1999). Diversity of rhizobia associated with Amorpha fruticosa isolated from Chinese soils and description of Mesorhizobium amorphae sp. nov. Int. J. Syst. Bacteriol. 49 (1), 51–65. doi: 10.1099/00207713-49-1-51
Westhoek, A., Field, E., Rehling, F., Mulley, G., Webb, I., Poole, P. S., et al. (2017). Policing the legume-rhizobium symbiosis: A critical test of partner choice. Sci. Rep. 7 (1), 1419. doi: 10.1038/s41598-017-01634-2
Wongdee, J., Boonkerd, N., Teaumroong, N., Tittabutr, P., Giraud, E. (2018). Regulation of nitrogen fixation in Bradyrhizobium sp. strain DOA9 involves two distinct NifA regulatory proteins that are functionally redundant during symbiosis but not during free-living growth. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.01644
Wyatt, G. A. K., West, S. A., Gardner, A. (2013). Can natural selection favour altruism between species? J. Evol. Biol. 26 (9), 1854–1858. doi: 10.1111/jeb.12195
Yan, J., Han, X. Z., Ji, Z. J., Li, Y., Wang, T. E., Xie, Z. H., et al. (2014). Abundance and diversity of soybean-nodulating rhizobia in black soil are impacted by land use and crop management. Appl. Environ. Microbiol. 80, 5394–5402. doi: 10.1128/AEM.01135-14
Young, J. P. W. (1996). Phylogeny and taxonomy of rhizobia. Plant Soil. 186, 45–52. doi: 10.1007/BF00035054
Zakhia, F., de Lajudie, P. (2001). Taxonomy of rhizobia. Agronomie. 21, 569–576. doi: 10.1051/agro:2001146
Keywords: rhizobia, micro- and macro-evolution, speciation, natural selection, plant–microbe symbioses, evolutionary genomics, symbiotic N2 fixation, leguminous plants
Citation: Provorov NA, Andronov EE, Kimeklis AK, Onishchuk OP, Igolkina AA and Karasev ES (2022) Microevolution, speciation and macroevolution in rhizobia: Genomic mechanisms and selective patterns. Front. Plant Sci. 13:1026943. doi: 10.3389/fpls.2022.1026943
Received: 24 August 2022; Accepted: 06 October 2022;
Published: 25 October 2022.
Edited by:
Ricardo Aroca, Experimental Station of Zaidín (CSIC), SpainReviewed by:
Alessio Mengoni, University of Florence, ItalyJosé David Flores Félix, Universidade da Beira Interior, Portugal
Copyright © 2022 Provorov, Andronov, Kimeklis, Onishchuk, Igolkina and Karasev. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nikolay A. Provorov, provorovnik@yandex.ru
 Nikolay A. Provorov
Nikolay A. Provorov