- Chongqing Key Laboratory of Molecular Biology of Plant Environmental Adaptations, College of Life Sciences, Chongqing Normal University, Chongqing, China
Phosphatidylethanolamine-binding proteins (PEBP) family plays important roles in regulating plant flowering time and morphogenesis. However, geneme-wide identification and functional analysis of PEBP genes in the rigorous short-day plant Perilla frutescens (PfPEBP) have not been studied. In this study, 10 PfPEBP were identified and divided into three subfamilies based on their phylogenetic relationships: FT-like, TFL1-like and MFT-like. Gene structure analysis showed that all PfPEBP genes contain 4 exons and 3 introns. Motifs DPDxP and GIHR essential for anion-binding activity are highly conserved in PfPEBP. A large number of light-responsive elements were detected in promoter regions of PfPEBP. Gene expression of PfFT1 exhibited a diurnal rhythm. It was highly expressed in leaves under the short-day photoperiod, but higher in flowers and seeds under the long-day photoperiod. Overexpression of PfFT1 in Arabidopsis thaliana not only promoted early flowering of Col-0 or Ler, but also rescued the late flowering phenotype of ft-1 mutant. We concluded that PfFT1 promotes early flowering by regulating the expression of flowering-related genes AtAP1, AtLFY, AtFUL and AtSOC1. In conclusion, our results provided valuable information for elucidating the functions of PfPEBP in P. frutescens and shed light on the promoting effect of PfFT1 on flowering.
Introduction
Phosphatidylethanolamine-binding proteins (PEBP) are widely-distributed in animals, plants and microorganisms. They contain a conserved PEBP domain and play important roles in regulating plant flowering, seed development and germination (Yu et al., 2019; Cheng et al., 2021). Currently, the PEBP gene family is divided into three subfamilies: MOTHER OF FT AND TFL1-like (MFT-like), FLOWERING LOCUS T-like (FT-like), and TERMINAL FLOWERING 1-like (TFL1-like). MFT-like was thought to be the common ancestor of FT-like and TFL1-like, which emerged during the evolution of seed plants (Hedman et al., 2009). MFT genes were highly expressed in seeds and involved in seed development and germination by regulating abscisic acid (ABA) and gibberellin (GA) signaling pathways (Xi et al., 2010). FT-like and TFL1-like subfamily members were involved in the regulation of flowering time and morphogenesis. The TFL1-like subfamily includes TFL1, CENTRORADIALIS (CEN) and BROTHER OF FT AND TFL1 (BFT). TFL1 could inhibit the formation of flower primordia, thus delaying flowering. TFL1 inhibited flowering by binding to the bZIP type transcription factor FLOWERING LOCUS D (FD) (Kaneko-Suzuki et al., 2018; Freytes et al., 2021; Zuo et al., 2021). Introduction of apple (Malus × domestica) MdTFL1 or MdTFL2 genes into Arabidopsis could significantly delay flowering and increase the number of rosette leaves and plant height (Zuo et al., 2021). Overexpression of PgTFL1 and PgCENa of pomegranate (Punica granatum) were able to suppress the flowering defect of Arabidopsis tfl1-14 mutant (Patil et al., 2018). In rice (Oryza sativa), OsCEN and Hd3a (Heading date 3a, an ortholog of FT protein) competitively bind to OsFD and regulate flowering (Kaneko-Suzuki et al., 2018).
The FT-like subfamily includes two members, FT and TWIN SISTER OF FT (TSF). The FT-FD complex can activate the expression of flowering-related genes, such as FRUITFULL (FUL), LEAFY (LFY), APETALA1 (AP1) and SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1) (Abe et al., 2005; Wigge et al., 2005; Lee and Lee, 2010). In the LD plant Arabidopsis, overexpression of AtFT could significantly rescue the delayed flowering of ft-1 mutant (Ruiz-García et al., 1997; Kardailsky et al., 1999). In the short-day (SD) plant rice, Hd3a-OsFD1 complex could activate the expression of OsMADS15 (a homologue of AP1), and promoted the flowering (Taoka et al., 2011). At present, homologous genes of FT have been identified in several higher plant species, including pear (Pyrus communis) (Zhang M. et al., 2021), cotton (Gossypium hirsutum) (Wang et al., 2019), sugarcane (Saccharum spp.) (Venail et al., 2022), rice (Kaneko-Suzuki et al., 2018), and Arabidopsis (Wigge et al., 2005). Growing evidences showed that FT genes play crucial roles in plant flowering. In tobacco (Nicotiana tabacum), NtFT1, NtFT2 and NtFT3 inhibited flowering, while NtFT4 could promote flowering (Harig et al., 2012), which was caused the mutations of three key amino acids in the PEBP domain. Functional analysis revealed that the sugarcane ScFT3 gene could rescue the late flowering phenotype of the Arabidopsis ft-10 mutant, but ScFT5 could not (Venail et al., 2022). In addition, FT genes can also regulate developmental processes other than the floral transition. In onion (Allium cepa), AcFT1, AcFT5 and AcFT6 regulate bulb formation under long-day (LD) conditions (Rashid et al., 2019). Two FT-like paralogues (StSP3D and StSP6A) could respond to different environmental cues and regulate flowering and tubers formation in potato (Navarro et al., 2011).
Perilla (Perilla frutescens) seed oil is rich in unsaturated fatty acids. The content of α- linolenic acid is up to 60%, which has important application in medicine and food industry (Liao et al., 2018). Additionally, Perilla is a rigorous SD plant (King and Zeevaart, 1973), making it an ideal species for exploring the effect of photoperiod on flowering. The PEBP gene family of P. frutescens (PfPEBP) has not been identified and their functions are still unknown. The recently released P. frutescens genome provides important data for solving above questions (Zhang Y. et al., 2021). In this study, the members of PfPEBP genes were identified and systematically analyzed at the genome-wide level, including phylogenetic relationship, gene structure and chromosomal distribution. The effect of photoperiod on PfFT1 gene expression, as well as its diurnal rhythm and tissue specificity, was analyzed. Its role in promoting flowering was further explored by transforming PfFT1 into Arabidopsis. In conclusion, our findings will help to understand the molecular mechanism of the regulatory roles of PfFT1 in flowering control of Perilla.
Materials and methods
Identification of PEBP gene family members in P. frutescens
The annotated genome data of P. frutescens was obtained from the GenBank (https://www.ncbi.nlm.nih.gov/genbank/). Amino acid sequences of Arabidopsis and rice PEBPs were retrieved from the TAIR (http://www.arabidopsis.org/) and RGAP (http://rice.uga.edu/) databases, respectively. To identify PfPEBP, the Hidden Markov Model (HMM) profile of the PBP domain (PF01161) was obtained from the Pfam database and used as the query. In addition, amino acid sequences encoded by the genome of P. frutescens were also searched using HMMER v3.3.2 (E-value ≤ 1.0 × e−5). BLASP was performed against P. frutescens genome using Arabidopsis PEBPs as queries (E-value ≤ 1.0 × e−5). Finally, PfPEBP candidates were submitted to CD-Search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), SMART (http://smart.embl-heidelberg.de/) and Pfam (http://pfam.xfam.org/search) for conserved domain analysis.
Sequence characterization
The basic physicochemical properties of PfPEBP were analyzed using the ProtParam tool in Expasy (https://www.expasy.org/), including putative molecular weight (MW), isoelectric point (pI), and grand average of hydropathicity (GRAVY). Subcellular localizations were predicted using the Plant-mPLoc prediction tool (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/). The chromosomal mapping was performed based on genome annotation information and visualized with the TBtools (Chen et al., 2020). Gene structure was analyzed using the GSDS 2.0 server (http://gsds.gao-lab.org/). Finally, the conserved motifs were predicted using the MEME Suite (https://meme-suite.org/meme/tools/meme) with the following parameters: optimum width 10–50 amino acids, any number of repetitions of a motif, and maximum number of motifs set at 5.
Multiple sequence alignments and phylogenetic analysis
Amino acid sequences of PfPEBP were aligned using the Clustal Omega and visualized using the Jalview. Phylogenetic tree of 25 PEBP proteins from Perilla, Arabidopsis and rice were constructed using the Neighbor-Joining (N-J) method (1000 bootstrap replicates) in MEGA X based on JTT+G substitution matrix. The phylogenetic tree was visualized with the iTOL tool (https://itol.embl.de).
Cis-acting elements analysis
To understand the cis-acting elements in the promoter region of PfPEBP, a 2000 bp upstream fragment of the initiation codon (ATG) was retrieved and predicted by the PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). In addition, the cis-acting elements of rice OsFTL1, OsFTL2, OsFTL3, and Arabidopsis AtFT were also predicted and visualized using the TBtools.
Prediction of protein-protein interaction (PPI) network
The homologous proteins of PfFTs and PfTFL1s in Arabidopsis were determined by BLASTP. Subsequently, prediction of PPI networks with PfFTs and PfTFL1s as hub proteins were performed using STRING v11.5 (https://string-db.org/) (Szklarczyk et al., 2019).
Plant materials and treatments
Seeds of Perilla and Arabidopsis [Col-0 (Columbia ecotype), Ler (Landsberg erecta ecotype) and ft-1 mutant (Ler ecotype)] were kept in the Key Laboratory of Oil Peony Germplasm Innovation and Utilization of Chongqing Normal University. Perilla seeds were sown in plastic pots with nutrient soil and coarse soil (1:2, v:v) and grown in a incubator: 24°C/20°C (Light/Dark), 16 h/8 h (LD: Light/Dark) or 8 h/16 h (SD: Light/Dark). The cultivation and growth conditions of Arabidopsis (Col-0, Ler and ft-1) were performed as described by Xu et al. (Xu et al., 2022).
Photoperiod effects on expression of PfFT1 gene in different tissues
When grown to 6 true leaves, leaves were collected every 4 h for 48 h and used for diurnal rhythmicity analysis of PfFT1 expression. To analyze the tissue specific expression patterns of PfFT1 under different photoperiods, the roots, stems, leaves, flowers at flowering stage were collected at 56/141 days after sown (DAS), respectively. Mature seeds were collected at 86 DAS (SD condition), 171 DAS (LD condition). All samples were snap-frozen in liquid nitrogen and stored at −80°C until use.
To investigate the expression profiles of PfFT1 in different tissues and photoperiodic conditions, quantitative real-time polymerase chain reaction (RT-qPCR) was performed. Total RNA was extracted using the RNAprep pure Plant Kit (Tiangen, Beijing) and reverse transcribed into cDNA using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Japan). The RT-qPCR reaction system (20µL) contains 10 ng cDNA, 10 µL SYBR Green Supermix (BioRad, USA), 1 µL primer pairs (10 µM/each), and ddH2O. The reaction conditions were 95 °C for 2 min, followed by 40 cycles at 95 °C for 10 s, 60 °C for 20 s and 72 °C for 20 s. Perilla PfActin7 was used as the internal reference gene. Three biological and three technical replicates (n=3×3) were performed for each sample. Primers used in this study were listed in Supplementary Table S1. The relative expression level of PfFT1 was calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001).
Gene cloning, plant transformation, and transgenic lines screening
The full-length coding sequence of PfFT1 was amplified using the leaf cDNA as the template. The PCR reaction system contains 1 µL cDNA, 10 µL Premix Taq enzyme (TAKARA, Japan), 1 µL primer pairs (10 µM/each), and 8 µL ddH2O. The reaction conditions were 94 °C for 5 min, followed by 30 cycles at 94°C for 30 s, 57°C for 30 s and 72°C for 60 s, and a final extension at 72 °C for 10 min. All PCR products were detected by 1% agarose gel electrophoresis, and purified using the SanPrep Column DNA Gel Extraction Kit (Sangon, Shanghai). The recombinant vector pCAMBIA1303-PfFT1 was constructed using the In-Fusion HD Cloning Kit (Takara, Japan) following the manufacturer’s instructions, and then transformed into Agrobacterium tumefdciens GV3101 (WEIDI, China) by the freeze-thaw method. Arabidopsis Col-0, Ler and ft-1 were transformed using the floral dip method (Clough and Bent, 1998). The harvested plant seeds were planted in 1/2 MS medium containing 20 mg/L hygromycin B. Plants that can grow 4 or more true leaves were considered as positive seedlings and transplanted to soil. Leaf genomic DNA was extracted and amplified against the T-DNA region of the pCAMBIA1303 vector to detect the target gene PfFT1. The transgenic Arabidopsis was then stained using the GUS stain Kit (Coolaber, China). Plants validated by the above two steps were regarded as the positive T0 generation. Following the same protocol, the T3 generation without trait segregation was considered as a homozygous transgenic line and used for further studies.
Measurement of flowering time and expression level of flowering-related genes
To analyze the effect of PfFT1 on flowering, wild-type Col-0, transgenic Col-0 (TC); ft-1 mutant, complemented mutant (CM); Ler, and transgenic Ler (TL) plants were grown in an artificial climate chamber under the LD condition (22°C/20°C, Light/Dark). At least 10 plants from each line were selected to record the bolting and flowering time, number of rosette leaves at flowering.
Total RNA was extracted from leaves of 20-day-old Arabidopsis (transgenic or non-transgenic line) grown under the LD conditions, and reverse transcribed into cDNA and used for gene expression analysis of PfFT1, AtFT, AtAP1, AtFUL, AtSOC1 and AtLFY. The RT-qPCR reaction system (20µL) consisted 10 ng cDNA, 10 µL SYBR Green Supermix, 1 µL primer pairs (10 µM/each), and ddH2O. The reaction condition was 95 °C for 1 min, followed by 40 cycles at 95 °C for 10 s, 60 °C for 30 s and 72 °C for 20 s. Arabidopsis AtActin8 was used as the internal reference gene. Three biological and three technical replicates (n=3×3) were performed for each sample. Primers were listed in Supplementary Table S1. The relative gene expression levels were calculated using the 2−ΔΔCT method.
Statistical analysis
All data were analyzed by Student’s t-test using IBM SPSS v26.0. Significant difference was considered when P < 0.05. Graphs were plotted using GraphPad Prism v8.0.
Results
Identification and characterization of PEBP family genes in P. frutescens
A total of 10 PfPEBP genes were identified and named according to the similarity with the homologous genes in Arabidopsis, including 2 PfFT, 4 PfTFL1, 2 PfCEN and 2 PfMFT (Supplementary Table S2). The corresponding protein length ranges from 149 (PfCEN2) to 176 (PfMFT1) amino acids, the molecular weight of PfPEBP ranges from 17.0 to 19.9 kDa, and the pI ranges from 7.74 to 9.51. All GRAVY values were less than 0, indicating that all PfPEBP are hydrophobic. Subcellular localization prediction showed that PfFTs, PfTFL1s and PfCENs, and PfMFTs were located in the nucleus, cytoplasm, cytoplasm-nucleus, respectively (Supplementary Table S2).
To understand gene structural diversities of PfPEBP genes, chromosomal location, number of exons and introns, and distribution of conserved domains were investigated. Chromosome mapping showed that 10 PfPEBP genes were distributed on Chr02, Chr05, Chr11, Chr14, Chr15, Chr17 and Chr18 (Figure S1). The phylogenetic relationships of the PfPEBP family was shown in Figure 1A. Each PfPEBP gene was composed of 4 exons and 3 introns (Figure 1B). Motifs 2-5 were conserved in all PfPEBP (Figure 1C). Only PfCEN2 lacks motif 1. Each motif contained 11-50 amino acids (Figure 1D). Members in the same subfamily shared a similar motif pattern, suggesting that they might have similar functions. Comparison analysis revealed that the conserved PEBP domain contained complete motifs 2-5, indicating that they are crucial for their functions, and also explaining the conserved domain patterns of PfPEBP members.
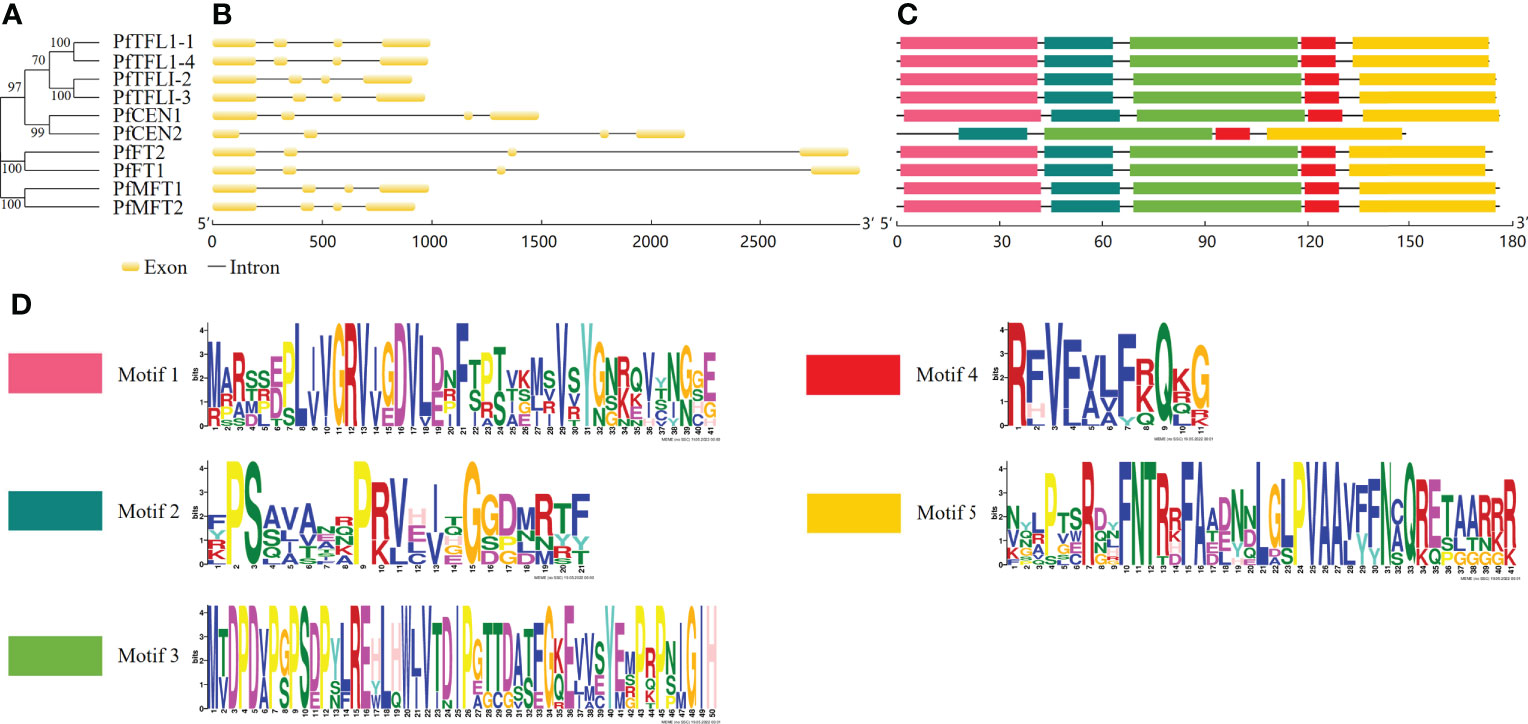
Figure 1 Gene structures and motifs of PEBP genes in P. frutescens. (A) The phylogenetic tree was constructed based on amino acid sequences of 10 PfPEBP. (B) Gene structures of PfPEBP genes was constructed using the GSDS 2.0 tools. Yellow boxes indicate exons and black lines represent introns. (C) Motif composition of PfPEBP. The conserved motif numbers in the MEME prediction was set to 5. (D) Sequence logos of five conserved motifs. The overall height of each stack indicates the sequence conservation at that position.
Amino acid alignment and conserved domain analysis
Amino acid sequence alignment showed that the identities between PfFTs and PfTFL1s were more than 55% (Figure 2). All PfPEBP have conserved motif DPDxP and GxHR, which are critical for anion-binding. Particularly, motif GxHR has a preference for Ile residues. The conserved motif LYN in PfFTs was localized in segment C, which was essential for the enzymatic activity. Fragment D of PfFTs contains the motif SGTGGR, while that of PfTFL1s were replaced by the motif TAARRR. Further analysis showed that all PfFTs had the key amino acid residues Tyr84, Gln139, but they were replaced by His85, Asp142 or Asn142 in PfTFL1s.
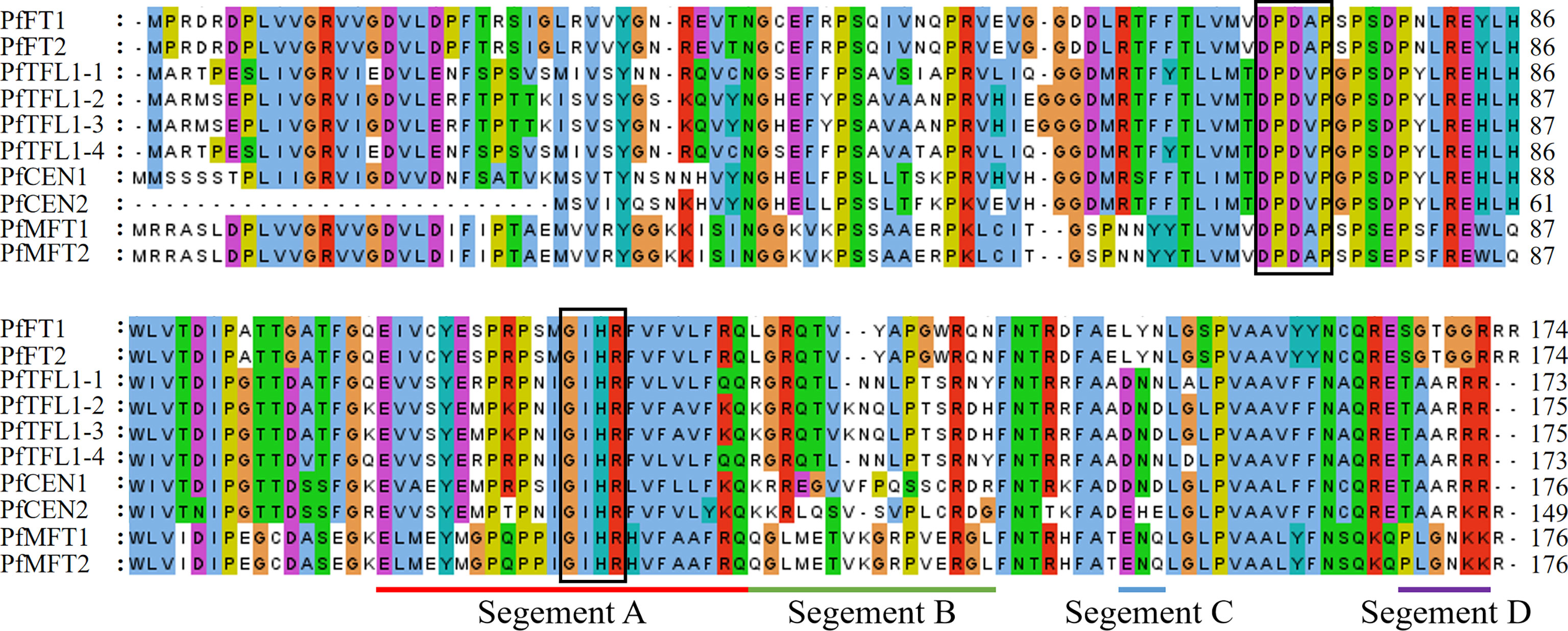
Figure 2 Multiple sequence alignment of PEBP in P. frutescens. Conserved motif DPDxP and GIHR was marked with black frame, respectively. Segment A–D was underlined with different colors.
Phylogenetic analysis
To explore the evolutionary relationship of the PfPEBP family, a phylogenetic tree of PEBP from P. frutescens, A. thaliana, and O. sativa was constructed. The results showed that 35 PEBP proteins were clustered into three subfamilies: FT-like, TFL1-like and MFT-like (Figure 3). The FT-like subfamily has 18 members, including AtFT, OsFTLs, and PfFTs. The TFL1-like subfamily has 12 members, including AtCEN, AtTFL1, AtBFT, OsCENs, PfCENs and PfTFL1s. The MFT-like subfamily has five members, including AtMFT, OsMFTs, and PfMFTs.
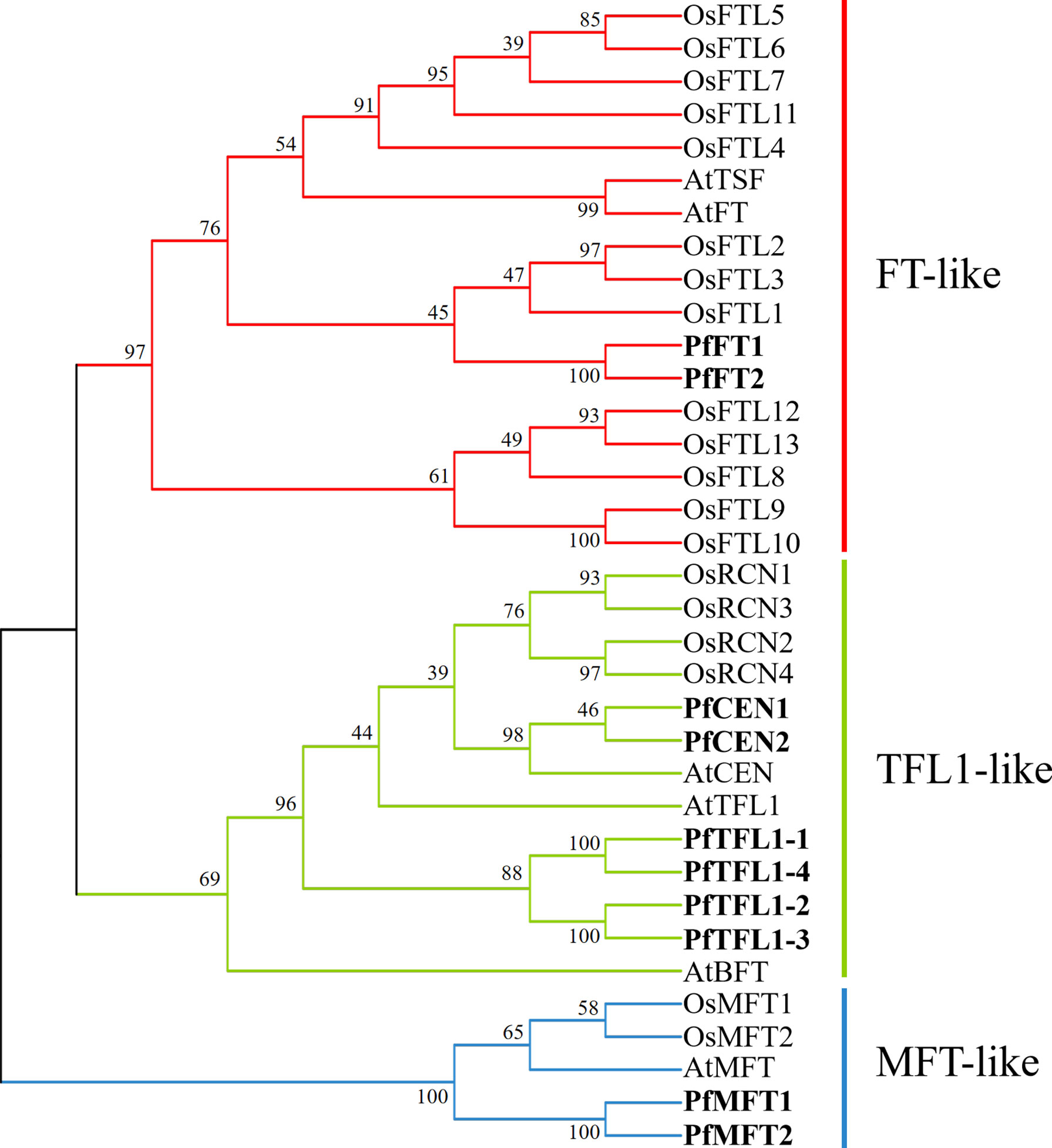
Figure 3 Phylogenetic relationship of PEBP in P. frutescens, A. thaliana, and O. sativa. The phylogenetic tree was constructed using the Neighbor-Joining method in MEGAX with 1000 bootstrap replicates. FT-like, TFL1-like, and MFT-like subfamilies were marked by red, green, and blue, respectively. PfPEBP were shown in bold font. GenBank accession number of each sequence was listed in Supplementary Table S2 and S3.
Analysis of cis-acting regulatory elements
To further understand the regulatory patterns of PfPEBP genes, the cis-regulatory elements within a fragment 2000 bp upstream of the start codon (ATG) of each gene were analyzed. Based on their functions, they were divided into three categories: growth and development, hormone responses, and biotic/abiotic stress responses (Figure 4A and Supplementary Table S4). Light-responsive elements, such as Box4 and G-Box were present in the promoter of all PfPEBP genes, suggesting that they are light signal reponse regulators. Circadian-responsive element (circadian) only occurred the promoter of PfMFT genes, whereas the anaerobic-inducible element (GC-motif) was only present in the promoter of PfTFL1 and PfMFT genes. Further analysis found that the distribution of hormone response elements was quite different. The auxin responsive element (AuxRR-core) was only found in PfFT and PfMFT genes. These findings revealed that the PfPEBP genes could respond light, hormones and stresses.
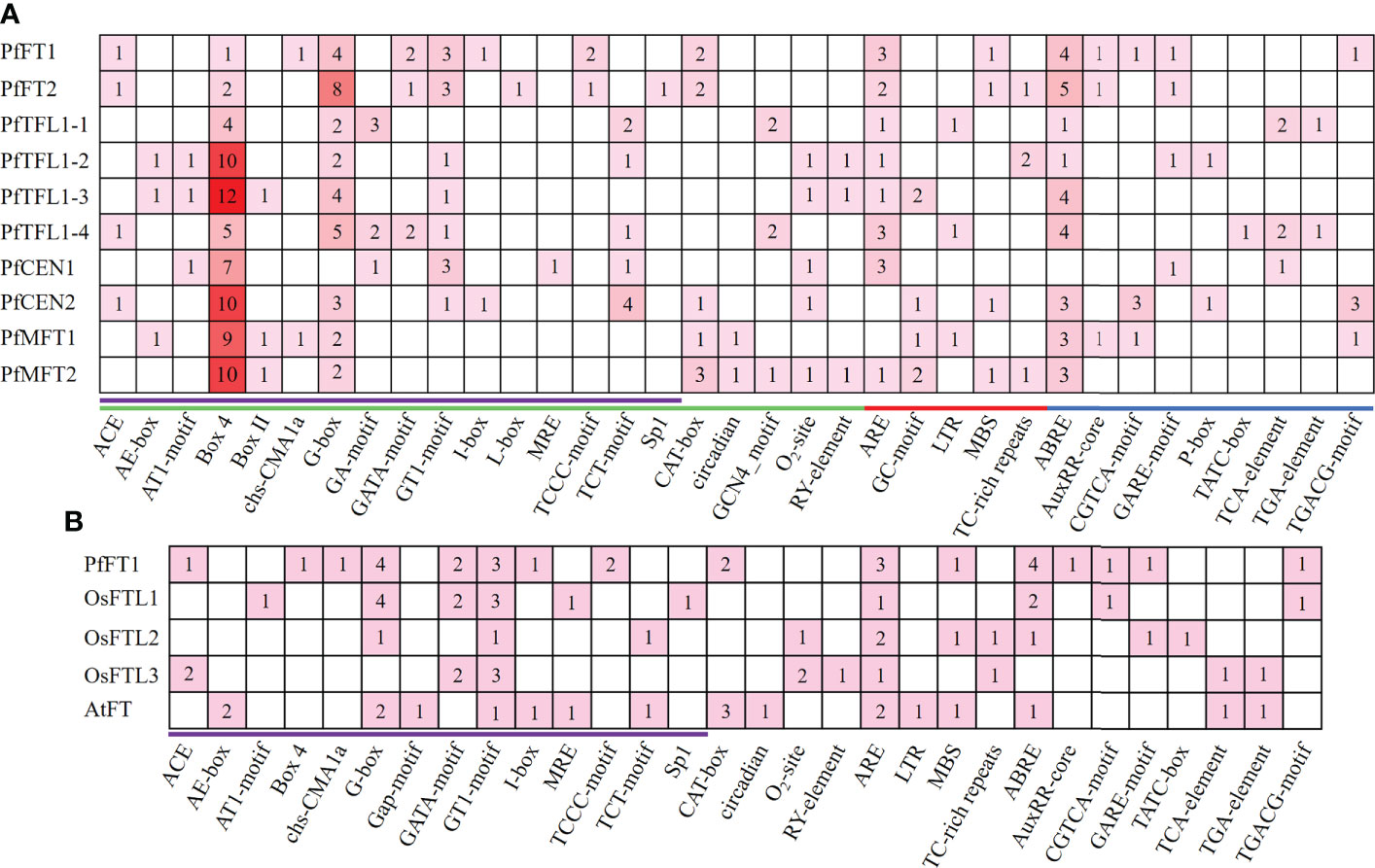
Figure 4 Cis-regulatory elements analysis. (A) Cis-regulatory elements analysis was performed in a 2000 bp promoter region of PfPEBP genes. Green, red, and blue lines indicate growth and development, biotic/abiotic stress responses, and hormone responsive element, respectively. Light-response elements were marked in purple. The numbers and colors represent the number of specific cis-acting elements in a gene. The larger the number, the darker the color, and the smaller the number, the lighter the color. (B) Cis-regulatory elements in the promoter region of PfFT1, OsFTL1–3, AtFT. Light-response elements were marked in purple.
Comparion analysis revealed that Arabidopsis (LD plant) has specific light-response elements, such as AE-Box and Gap-motif, while Perilla and rice (SD plant) have specific light-response elements, such as ACE, AT1-motif, Box-4, Chs-CMA1a, GATA-motif, TCCC-motif, and sp1 (Figure 4B and Supplementary Table S5). Differences in cis-acting elements in promoter regions of PEBP genes in LD and SD plants might lead to their different responses to photoperiod.
PPI network
Comparison of amino acid sequences is helpful to understand functional similarities, amino acid sequence identities between PfFTs and AtFT was 78.16%, and that of PfTFL1s and AtTFL1 was 68.60-72.83% (Figure S2 and Supplementary Table S6). High sequence identity and conservation of key motifs provided an important basis for PPI network prediction, which provides a more intuitive understanding of their functions. STRING analysis showed that both PfFTs and PfTFL1s potentially interact with FD, AP1, LFY, CO, AGL20, DECOY, RPL23/L15e, and RPL29. In addition, PfFTs also interacted with FLC, GI, and SVP (Figure 5). This result suggested that PfFTs and PfTFL1s may have different functions in regulating flowering through similar pathways.
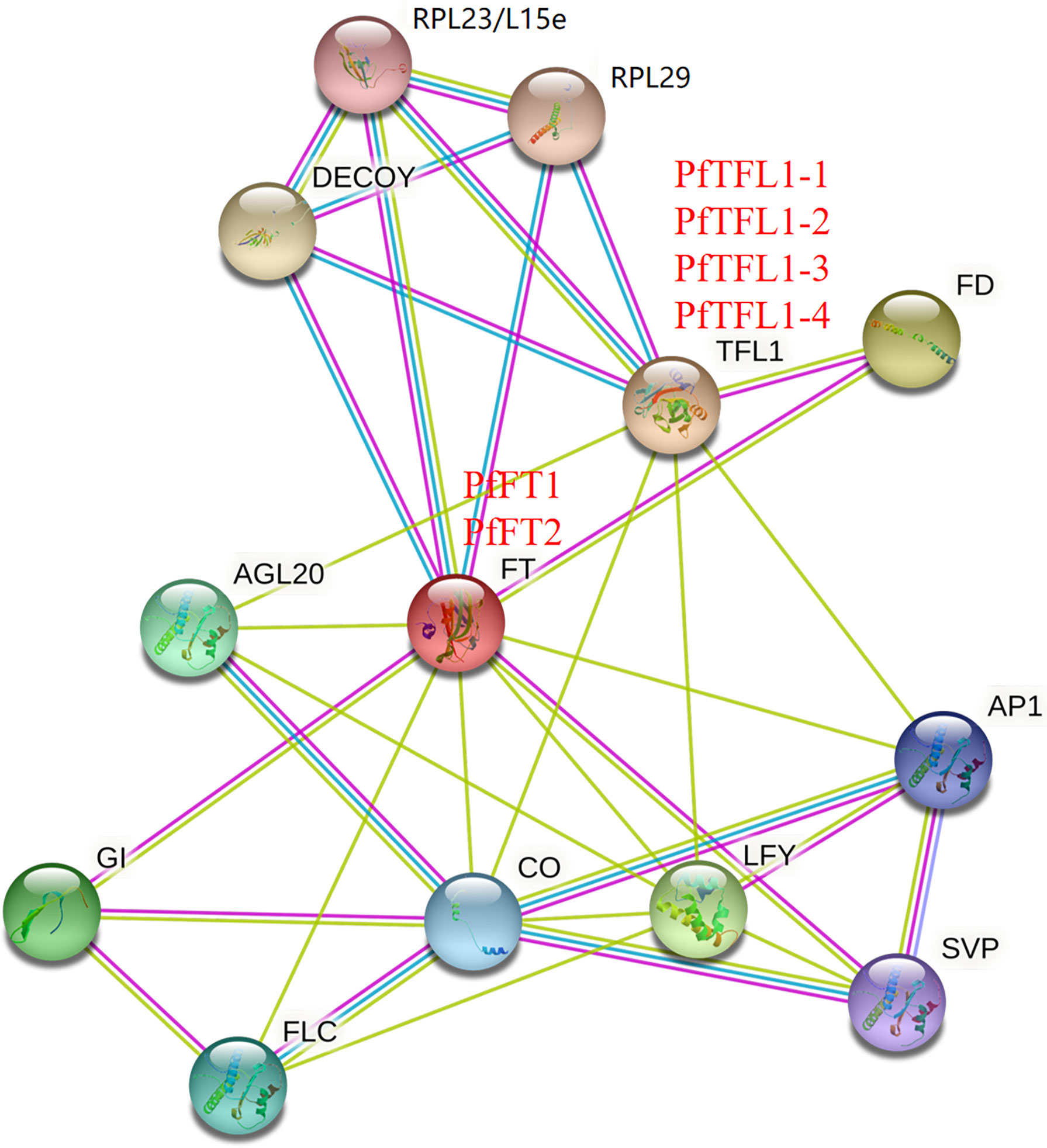
Figure 5 Protein-protein interaction network. The PPI network was predicted based on the homologous proteins in Arabidopsis. RPL23/L15e, Ribosomal protein L23/L15e family protein; RPL29, Ribosomal protein L29 family protein; DECOY, DECOY; FD, FLOWERING LOCUS D; TFL1, TERMINAL FLOWER 1; FT, FLOWERING LOCUS T; AGL20, AGAMOUS-LIKE 20; AP1, APETALA1; GI, GIGANTEA; CO, CONSTANS; LFY, LEAFY; SVP, SHORT VEGETATIVE PHASE; FLC, FLOWERING LOCUS C.
Photoperiod effects on expression patterns of PfFT1 genes in different tissues
To understand effects of photoperiod on PfFT1 expression, its expression within 48 h under LD or SD conditions was analyzed by RT-qPCR (Figure 6A). The result revealed that PfFT1 have specific rhythmic expression patterns under different photoperiod conditions. Under the SD conditions, its expression level was increased with the increase of light duration within 0-8 h, then gradually decreased under the dark condition (8-24 h). Within 24-48h, its expression level peaked after 8 h of light (32 h), but it was higher than that within 0-24 h. Under the LD condition, the expression level reached a peak after 4 h light exposure (significantly lower than the peak value under the SD condition), and then gradually decreased to a very low level. In conclusion, the expression level of PfFT1 under the LD condition was extremely low, which strongly supported that the Perilla is a SD plant.
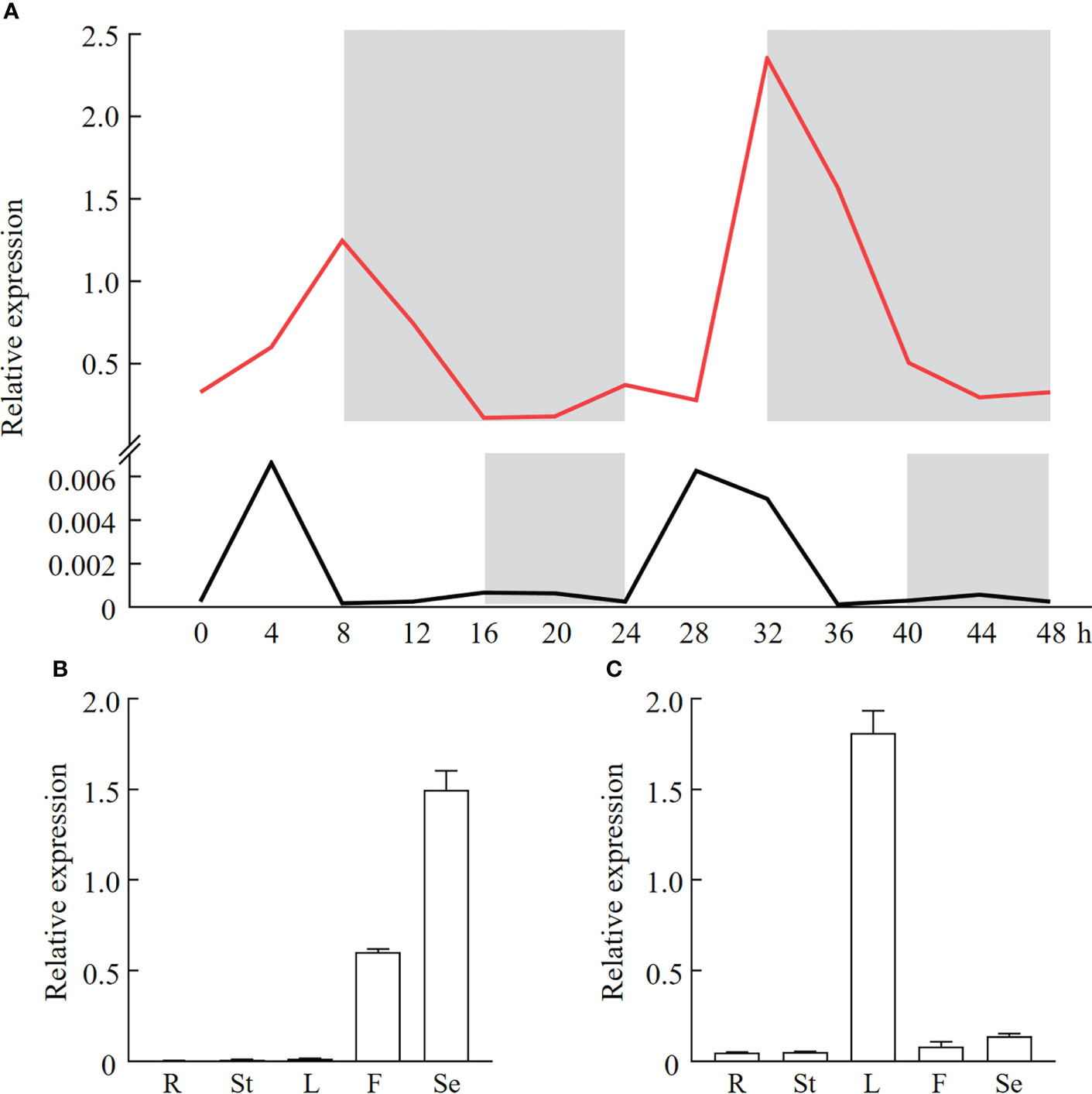
Figure 6 Diurnal expression pattern and tissue specificity analysis of PfFT1 in P. frutescens. (A) Diurnal expression pattern of PfFT1 in leaves under the SD (red line) or LD (black line) photoperiod. The dark duration was shaded with gray. Relative expression level of PfFT1 in different tissues under the SD (B) or LD (C) photoperiod. R, roots; St, stems; L, leaves; F, flowers; Se, seeds. The roots, stems, leaves, flowers at flowering stage (56/141 DAS) were collected, and mature seeds were collected at 86/171 DAS under SD and LD condition, respectively.
To further explore the relationship between the PfFT1 expression and flowering, its tissue expression pattern under different photoperiods was analyzed (Figures 6B, C). Under the LD condition, the highest expression was detected in seeds, then followed by flowers, while it was extremely low in roots, stems and leaves. Under the SD condition, highest expression was found in leaves, followed by flowers and seeds. But the expression was very low in roots and stems. These results suggested that PfFT1 could promote flowering under SD conditions, whereas it might also regulate seed development under LD conditions.
Promotion effect of PfFT1 gene on flowering in Arabidopsis
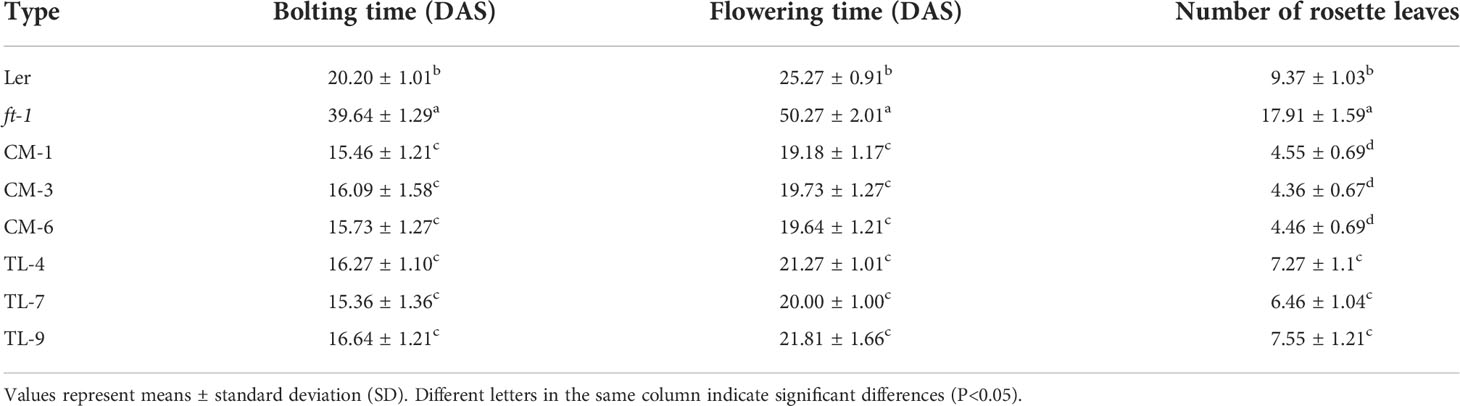
To study the regulatory role of PfFT1 on flowering, PfFT1 gene driven by CaMV 35S promoter was transferred into Arabidopsis Col-0, Ler, and ft-1, respectively (Figures S3A, B). After hygromycin screening, GUS staining and PCR identification, a total of 18 TC, 10 CM and 15 TL transgenic lines were obtained (Figures S3C–F). Subsequently, three T3 plants of each transgenic line were selected based on the expression level of PfFT1 for further analysis. Compared with the controls, all transgenic plants exhibited varying degrees of early flowering (Figures 7A–C). Compared with the wild-type Col-0, the bolting and flowering time of TC lines were 5-8 day earlier with 2-3 less rosette leaves at flowering (Table 1). Overexpression of PfFT1 not only promoted early flowering of Col-0, but also rescued the flowering time defect of ft-1 mutant. All CM lines started bolting at day 15-16, the flowering time was 30 d earlier than that of ft-1 line, and the number of rosette leaves at flowering was only 4-5. However, the ft-1 mutant bloomed around day 50 with 18 rosette leaves (Table 2). These results indicated that the overexpression of PfFT1 could complemented the delayed flowering in ft-1 mutant. In addition, it was found that the bolting and flowering time of TL lines were 3-5 d earlier than that of Ler type (Table 2).

Figure 7 Flowering traits of transgenic Arabidopsis lines. (A) From left to right are Col-0, TC-2, TC-7, and TC-8 (29th DAS). (B) From left to right are Ler, ft-1, CM-1, CM-3, and CM-6 (20th DAS). (C) From left to right are Ler, TL-4, TL-7, and TL-9 (22th DAS).
Expression analysis of flowering-related endogenous genes in Arabidopsis
RT-qPCR analysis showed that PfTF1 was expressed in all TC, CM and TL lines (Figures S4A–C), but was not detected in wild-type Col-0, and Ler type. Expression of AtFT and PfFT1 were not detected in ft-1 mutant (Figures S4B, E).
To verify whether the introduction of PfFT1 could regulate the expression of endogenous flowering-related genes in Arabidopsis, the expression level of AtAP1, AtFUL, AtSOC1 and AtLFY were analyzed (Figures 8A–L). In TC lines, high expression of AtSOC1 and AtAP1 was observed (Figures 8A, G). Compared with the ft-1 mutant, AtAP1, AtFUL, AtSOC1 and AtLFY were significantly up-regulated in CM lines (Figures 8B, E, H, K). Compared with the non-transgenic ft-1 line, the flowering time of transgenic lines was significantly earlier. It was also earlier than the average flowering time of Ler (25.27 ± 0.91) (Table 2). Compared with the non-transgenic Ler, AtAP1 was highly expressed in OL lines, but AtSOC1 was less expressed (Figures 8C, I).
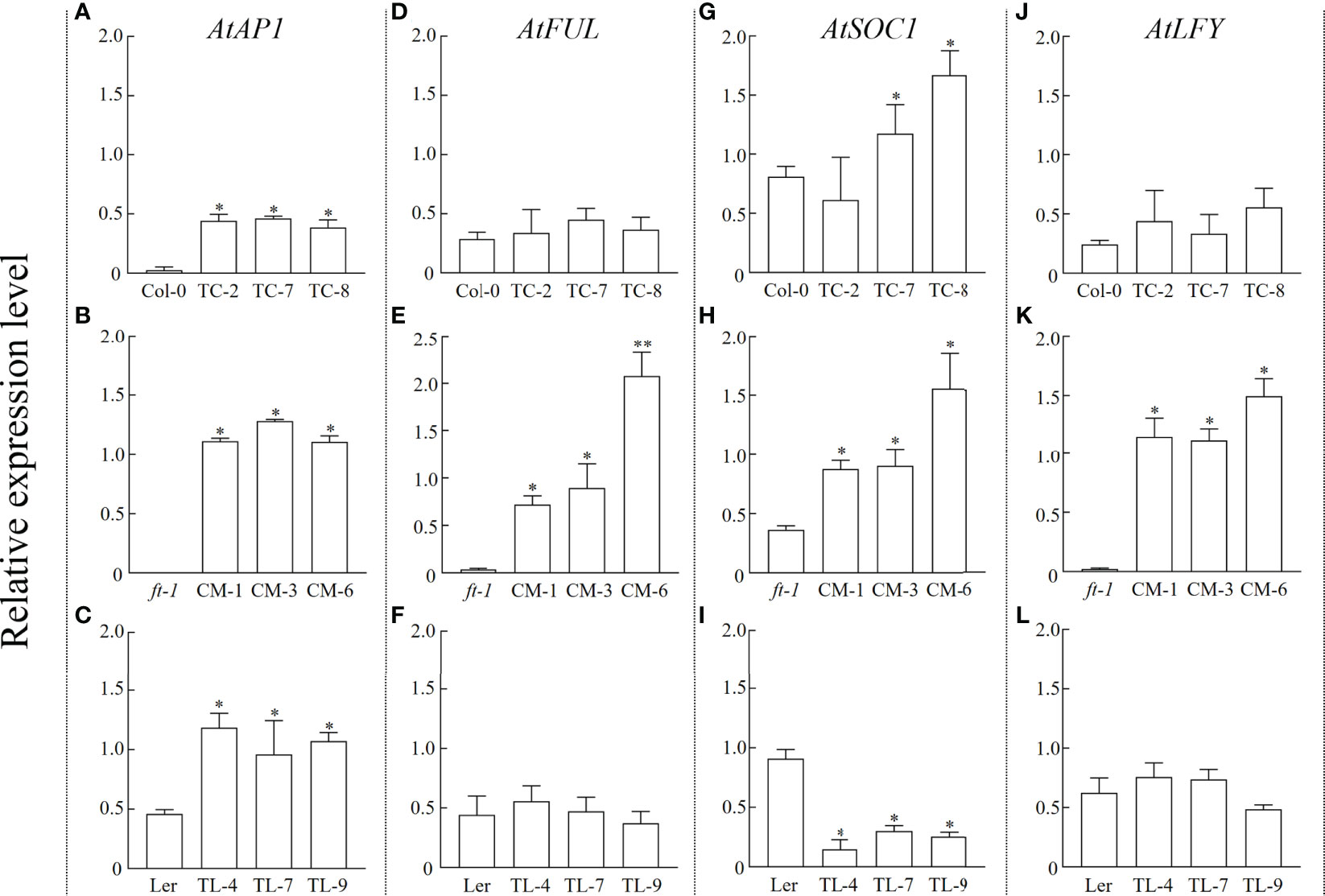
Figure 8 Relative expression level of endogenous flowering related genes in Arabidopsis. Expression levels of AtAP1, AtFUL, AtSOC1, and AtLFY in Col-0 and TC lines (A, D, G, J), ft-1 and CM lines (B, E, H, K), Ler and TL lines (C, F, I, L), respectively * means p < 0.05.
Discussion
In plants, the PEBP family is a class of proteins with the conserved PEBP domain, and involved in inflorescence structure and seed development, and germination (Yu et al., 2019; Schmidt et al., 2020; Zhang et al., 2020). However, their functions in Perilla have not been studied. Therefore, studies on the PfPEBP family will be helpful for the genetic breeding and improvement of yield traits of P. frutescens.
In this study, a total of 10 PfPEBP genes were identified in the P. frutescens genome, and all consist of 4 exons and 3 introns. Phylogenetic analysis showed that PfPEBP were clustered into three subfamilies: FT-like, TFL1-like and MFT-like. Members in a same subfamily have the conserved motif DPDxP and GxHR, indicating that they are evolutionarily conserved. Previous studies found that the substitution of key amino acids in the PEBP domain (Tyr85→His88; Gln140→Asp144) resulted in the opposite roles of FT and TFL1 in regulating plant flowering (Hanzawa et al., 2005; Ahn et al., 2006). The amino acid residues encoded by the fourth exon of FT and TFL1 determine their functional specificity. The corresponding part was divided into four fragments. Segment B and C (LYN motif) are crucial for FT-induced flowering (Ahn et al., 2006).
Prediction of PPI networks helps to explore potential roles of PfPEBP. In the photoperiod regulation pathway, the photoreceptor receives the light signal and transmits to CONSTANS (CO) through the diurnal clock. In this pathway, CO acts as a transcriptional activator to induce plant flowering, while FLC acts as a flowering inhibitor through the vernalization pathway (Luo et al., 2019). In addition, CO and FLC also regulate the expression of downstream genes FT, SOC1 (also known as AGL20), and LFY. A previous study showed that the transcription factor SHORT VEGETATIVE PHASE (SVP) binds to the CArG motif in the promoter of FT gene and negatively regulates its expression, thereby regulating plant flowering (Lee et al., 2007). During the vegetative growth stage, the TFL-FD complex can inhibit the expression of LFY gene in the meristem through specifically binding to the G-box motif in the second exon, thereby maintaining the vegetative properties of the meristem (Yamaguchi, 2021). PfFT1 could promote flowering probably by regulating the expression of downstream flowering-related genes AP1, LFY, FUL and SOC1 (Figure 8).
In the SD plant Perilla and rice, the promoter regions of PfPEBP genes contain specific light-response elements, such as ACE, AT1-motif, Box-4. Therefore, we speculated that they might be involved in regulating flowering. Studies have shown that the flower formation signal integrated by the photoperiod can induce the flower formation by activating FT expression. In rice, the highest expression of Hd3a was detected in leaves under the SD condition (Izawa et al., 2002; Tamaki et al., 2007). In the LD plant Arabidopsis, its highest expression level was also found in leaves under the LD condition (Wigge et al., 2005). In this study, the same expression pattern of PfFT1 was detected in leaves with a diurnal rhythm under the SD condition. It was also showed that the expression level of FT genes was closely related to seasonal flowering of soybean (Glycine max), poplar (Populus deltoides) and loquat (Eriobotrya japonica) (Hsu et al., 2006; Reig et al., 2017; Lee et al., 2021). Our results revealed that the expression of PfFT1 in P. frutescens had obvious diurnal rhythm under the SD or LD condition. A previous study has shown that PfFT1, a key protein for flowering induction, is synthesized in leaves and transported to the meristem of the shoot apex to induce flower formation (Kaneko-Suzuki et al., 2018). Under the SD condition, the high expression of PfFT1 in leaves is expected as Perilla is a SD plant.
Our results indicated that the expression of PfFT1 could promote flowering. The flowering time of the transgenic TC lines were 6-8 day earlier than that of the non-transgenic plants Col-0. Its overexpression could rescue the late flowering phenotype of ft-1 mutant, and the flowering time was 5-6 day earlier than that of Ler type. Similarly, its overexpression in Ler type resulted in 2-3 day earlier flowering. Studies have shown that overexpression of FT gene could promote the development of floral organs. In cassava (Manihot esculenta), transgenic overexpression of the endogenous MeFT1 resulted in early flowering by recruiting downstream floral meristem identity genes (MeAP1, MeLFY and MeSOC1) in shoot apical tissues (Odipio et al., 2020). The expression of LsFT was involved in the bolting of different lettuce (Lactuca sativa) varieties. Knockdown of LsFT by RNA interference significantly delayed bolting in lettuce (Chen et al., 2018). In addition, overexpression of LsFT in Arabidopsis could rescue the late-flowering phenotype of ft-2 mutant (Chen et al., 2018). In Medicago truncatula, MtFTa1 and MtFDa were considered as key flowering regulators, MtFDa was essential for floral transition and secondary inflorescence development (Cheng et al., 2021). In this study, early flowering of transgenic Arabidopsis was positively correlated with the expression levels of floral meristem recognition genes AtAP1, AtLFY and AtFUL, which was consistent with the findings in M. esculenta (Odipio et al., 2020). Our findings lay a foundation for further researches on PfPEBP genes in regulating flowering of P. frutescens.
Conclusion
Ten PfPEBP genes were identified in the rigorous SD plant Perilla, and divided into three subfamilies (FT-like, TFL1-like, and MFT-like) based on their phylogenetic relationships and gene structure characteristics. Cis-regulatory element analysis revealed that the promoter region of each PfPEBP genes contains many light-response elements. The expression of PfFT1 is regulated by the photoperiod with tissue specificity and diurnal rhythm. The transgenic Arabidopsis lines overexpressing PfFT1 gene exhibit an early flowering phenotype. We speculated that the molecular mechanism of flowering promotion by PfFT1 is to activate expressions of regulatory genes AtAP1, AtFUL, AtSOC1 and AtLFY. In conclusion, results laid a foundation for elucidating the molecular mechanism of PfFT1 gene regulating flowering and genetic traits improvement of P. frutescens.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
HX, YH, and TZ designed the experiments and wrote the manuscript. HX, XG, and DL executed the experiments and prepared the Figures. HX, XG, GL, and JL analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the National Natural Science Foundation of China (Project No. 31171588), Science and Technology Research Program of Chongqing Municipal Education Commission (Project No. KJZD-K202200508). This work was also supported by the Graduate Research Innovation Project of Chongqing Normal University (Project No. YKC21033).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1026696/full#supplementary-material
Supplementary Figure 1 | Chromosome distribution of PfPEBP genes. The chromosomal locations of PfPEBP genes were mapped based on genome annotation. The scale is in mage bases (Mb).
Supplementary Figure 2 | Multiple sequence alignment for PPI network prediction. Multiple sequence alignment of PfFTs and AtFT (A), PfTFL1s and AtTFL1 (B) was performed by Clustal Omega.
Supplementary Figure 3 | Cloning of PfFT1 genes and identification of transgenic Arabidopsis. (A) Agrose gel electrophoresis of PfFT1 amplified from Perilla cDNA. M, DNA Marker (from bottom to top) were 100, 250, 500, 750, 1000, and 2000 bp, sequentially; 1, Fragment amplified with PfFT1-F and PfFT1-R primers. (B) Detailed information of vector pCAMBIA1303-PfFT1. (C) Transgenic lines screened on the medium with hygromycin B. (D) Identification of transgenic lines by PCR amplification of target PfFT1 gene. M: DNA Marker; 1-3, line OE-2,OE-7,OE-8, 4-6, line CM-1,CM-3,CM-6, 7-9, line OL-4,OL-7, OL-9, 10-13, line Col, ft-1, Ler, and control (ddH2O). (E) Identification of transgenic lines by GUS staining.
Supplementary Figure 4 | Expression analysis of PfFT1 and endogenious gene AtFT in transgenic Arabidopsis. Expression level of PfFT1, AtFT1 in Col-0 and TC line (A, D), ft-1 and CM line (B, E), Ler and TL line (C, F), respectively.
References
Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., et al. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056. doi: 10.1126/science.1115983
Ahn, J. H., Miller, D., Winter, V. J., Banfield, M. J., Lee, J. H., Yoo, S. Y., et al. (2006). A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J. 25, 605–614. doi: 10.1038/sj.emboj.7600950
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cheng, X., Li, G., Krom, N., Tang, Y., Wen, J. (2021). Genetic regulation of flowering time and inflorescence architecture by MtFDa and MtFTa1 in Medicago truncatula. Plant Physiol. 185, 161–178. doi: 10.1093/plphys/kiaa005
Chen, Z., Han, Y., Ning, K., Ding, Y., Zhao, W., Yan, S., et al. (2018). Inflorescence development and the role of LsFT in regulating bolting in lettuce (Lactuca sativa l.). Front. Plant Sci. 8. doi: 10.3389/fpls.2017.02248
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Freytes, S. N., Canelo, M., Cerdán, P. D. (2021). Regulation of flowering time: When and where? Curr. Opin. Plant Biol. 63, 102049. doi: 10.1016/j.pbi.2021.102049
Hanzawa, Y., Money, T., Bradley, D. (2005). A single amino acid converts a repressor to an activator of flowering. Proc. Natl. Acad. Sci. U. S. A. 102, 7748–7753. doi: 10.1073/pnas.0500932102
Harig, L., Beinecke, F. A., Oltmanns, J., Muth, J., Müller, O., Rüping, B., et al. (2012). Proteins from the FLOWERING LOCUS T-like subclade of the PEBP family act antagonistically to regulate floral initiation in tobacco. Plant J. 72, 908–921. doi: 10.1111/j.1365-313X.2012.05125.x
Hedman, H., Källman, T., Lagercrantz, U. (2009). Early evolution of the MFT-like gene family in plants. Plant Mol. Biol. 70, 359–369. doi: 10.1007/s11103-009-9478-x
Hsu, C. Y., Liu, Y., Luthe, D. S., Yuceer, C. (2006). Poplar FT2 shortens the juvenile phase and promotes seasonal flowering. Plant Cell 18, 1846–1861. doi: 10.1105/tpc.106.041038
Izawa, T., Oikawa, T., Sugiyama, N., Tanisaka, T., Yano, M., Shimamoto, K. (2002). Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev. 16, 2006–2020. doi: 10.1101/gad.999202
Kaneko-Suzuki, M., Kurihara-Ishikawa, R., Okushita-Terakawa, C., Kojima, C., Nagano-Fujiwara, M., Ohki, I., et al. (2018). TFL1-like proteins in rice antagonize rice FT-like protein in inflorescence development by competition for complex formation with 14-3-3 and FD. Plant Cell Physiol. 59, 458–468. doi: 10.1093/pcp/pcy021
Kardailsky, I., Shukla, V. K., Ahn, J. H., Dagenais, N., Christensen, S. K., Nguyen, J. T., et al. (1999). Activation tagging of the floral inducer FT. Science 286, 1962–1965. doi: 10.1126/science.286.5446.1962
King, R. W., Zeevaart, J. A. (1973). Floral stimulus movement in Perilla and flower inhibition caused by noninduced leaves. Plant Physiol. 51, 727–738. doi: 10.1104/pp.51.4.727
Lee, S. H., Choi, C. W., Park, K. M., Jung, W. H., Chun, H. J., Baek, D., et al. (2021). Diversification in functions and expressions of soybean FLOWERING LOCUS t genes fine-tunes seasonal flowering. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.613675
Lee, J., Lee, I. (2010). Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 61, 2247–2254. doi: 10.1093/jxb/erq098
Lee, J. H., Yoo, S. J., Park, S. H., Hwang, I., Lee, J. S., Ahn, J. H. (2007). Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 21, 397–402. doi: 10.1101/gad.1518407
Liao, B., Hao, Y., Lu, J., Bai, H., Guan, L., Zhang, T. (2018). Transcriptomic analysis of Perilla frutescens seed to insight into the biosynthesis and metabolic of unsaturated fatty acids. BMC Genomics 19, 213. doi: 10.1186/s12864-018-4595-z
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luo, X., Chen, T., Zeng, X., He, D., He, Y. (2019). Feedback regulation of FLC by FLOWERING LOCUS t (FT) and FD through a 5' FLC promoter region in Arabidopsis. Mol. Plant 12, 285–288. doi: 10.1016/j.molp.2019.01.013
Navarro, C., Abelenda, J. A., Cruz-Oró, E., Cuéllar, C. A., Tamaki, S., Silva, J., et al. (2011). Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 478, 119–122. doi: 10.1038/nature10431
Odipio, J., Getu, B., Chauhan, R. D., Alicai, T., Bart, R., Nusinow, D. A., et al. (2020). Transgenic overexpression of endogenous FLOWERING LOCUS T-like gene MeFT1 produces early flowering in cassava. PloS One 15, e0227199. doi: 10.1371/journal.pone.0227199
Patil, H. B., Chaurasia, A. K., Azeez, A., Krishna, B., Subramaniam, V. R., Sane, A. P., et al. (2018). Characterization of two TERMINAL FLOWER1 homologs PgTFL1 and PgCENa from pomegranate (Punica granatum l.). Tree Physiol. 38, 772–784. doi: 10.1093/treephys/tpx154
Rashid, M., Cheng, W., Thomas, B. (2019). Temporal and spatial expression of Arabidopsis gene homologs control daylength adaptation and bulb formation in onion (Allium cepa l.). Sci. Rep. 9, 14629. doi: 10.1038/s41598-019-51262-1
Reig, C., Gil-Muñoz, F., Vera-Sirera, F., García-Lorca, A., Martínez-Fuentes, A., Mesejo, C., et al. (2017). Bud sprouting and floral induction and expression of FT in loquat [Eriobotrya japonica (Thunb.) lindl.]. Planta 246, 915–925. doi: 10.1007/s00425-017-2740-6
Ruiz-García, L., Madueño, F., Wilkinson, M., Haughn, G., Salinas, J., Martínez-Zapater, J. M. (1997). Different roles of flowering-time genes in the activation of floral initiation genes in Arabidopsis. Plant Cell 9, 1921–1934. doi: 10.1105/tpc.9.11.1921
Schmidt, F. J., Zimmermann, M. M., Wiedmann, D. R., Lichtenauer, S., Grundmann, L., Muth, J., et al. (2020). The major floral promoter NtFT5 in tobacco (Nicotiana tabacum) is a promising target for crop improvement. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01666
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613. doi: 10.1093/nar/gky1131
Tamaki, S., Matsuo, S., Wong, H. L., Yokoi, S., Shimamoto, K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316, 1033–1036. doi: 10.1126/science.1141753
Taoka, K., Ohki, I., Tsuji, H., Furuita, K., Hayashi, K., Yanase, T., et al. (2011). 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476, 332–335. doi: 10.1038/nature10272
Venail, J., da Silva Santos, P. H., Manechini, J. R., Alves, L. C., Scarpari, M., Falcão, T., et al. (2022). Analysis of the PEBP gene family and identification of a novel FLOWERING LOCUS t orthologue in sugarcane. J. Exp. Bot. 73, 2035–2049. doi: 10.1093/jxb/erab539
Wang, M., Tan, Y., Cai, C., Zhang, B. (2019). Identification and expression analysis of phosphatidy ethanolamine-binding protein (PEBP) gene family in cotton. Genomics 111, 1373–1380. doi: 10.1016/j.ygeno.2018.09.009
Wigge, P. A., Kim, M. C., Jaeger, K. E., Busch, W., Schmid, M., Lohmann, J. U., et al. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309, 1056–1059. doi: 10.1126/science.1114358
Xi, W., Liu, C., Hou, X., Yu, H. (2010). MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 22, 1733–1748. doi: 10.1105/tpc.109.073072
Xu, H., Li, D., Hao, Y., Guo, X., Lu, J., Zhang, T. (2022). Genome-wide analysis of DGAT gene family in Perilla frutescens and functional characterization of PfDGAT2-2 and PfDGAT3-1 in Arabidopsis. Plant Sci 324, 111426. doi: 10.1016/j.plantsci.2022.111426
Yamaguchi, N. (2021). LEAFY, a pioneer transcription factor in plants: A mini-review. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.701406
Yu, X., Liu, H., Sang, N., Li, Y., Zhang, T., Sun, J., et al. (2019). Identification of cotton MOTHER OF FT AND TFL1 homologs, GhMFT1 and GhMFT2, involved in seed germination. PloS One 14, e0215771. doi: 10.1371/journal.pone.0215771
Zhang, B., Li, C., Li, Y., Yu, H. (2020). Mobile TERMINAL FLOWER1 determines seed size in Arabidopsis. Nat. Plants 6, 1146–1157. doi: 10.1038/s41477-020-0749-5
Zhang, M., Li, P., Yan, X., Wang, J., Cheng, T., Zhang, Q. (2021). Genome-wide characterization of PEBP family genes in nine Rosaceae tree species and their expression analysis in P. mume. BMC Ecol. Evol. 21, 32. doi: 10.1186/s12862-021-01762-4
Zhang, Y., Shen, Q., Leng, L., Zhang, D., Chen, S., Shi, Y., et al. (2021). Incipient diploidization of the medicinal plant Perilla within 10,000 years. Nat. Commun. 12, 5508. doi: 10.1038/s41467-021-25681-6
Keywords: Perilla frutescens, flowering locus T, PfFT1, PEBP family, flowering time
Citation: Xu H, Guo X, Hao Y, Lu G, Li D, Lu J and Zhang T (2022) Genome-wide characterization of PEBP gene family in Perilla frutescens and PfFT1 promotes flowering time in Arabidopsis thaliana. Front. Plant Sci. 13:1026696. doi: 10.3389/fpls.2022.1026696
Received: 24 August 2022; Accepted: 26 October 2022;
Published: 18 November 2022.
Edited by:
Rafael Lozano, University of Almeria, SpainReviewed by:
Marta Santalla, Spanish National Research Council (CSIC), SpainStephen Jackson, University of Warwick, United Kingdom
Copyright © 2022 Xu, Guo, Hao, Lu, Li, Lu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tao Zhang, emh0MjE4OEAxMjYuY29t
 Huaxiang Xu
Huaxiang Xu Xi Guo
Xi Guo Junxing Lu
Junxing Lu