- 1College of Plant Science and Technology, Beijing University of Agriculture, Beijing, China
- 2Bei Jing Bei Nong Enterprise Management Co., Ltd, Beijing, China
A strawberry RIPK1, a leu-rich repeat receptor-like protein kinase, is previously demonstrated to be involved in fruit ripening as a positive regulator; however, its role in vegetable growth remains unknown. Here, based on our first establishment of Agrobacterium-mediated transformation of germinating seeds in diploid strawberry by FvCHLH/FvABAR, a reporter gene that functioned in chlorophyll biosynthesis, we got FvRIPK1-RNAi mutants. Downregulation of FvRIPK1 inhibited plant morphogenesis, showing curled leaves; also, this silencing significantly reduced FvABAR and FvABI1 transcripts and promoted FvABI4, FvSnRK2.2, and FvSnRK2.6 transcripts. Interestingly, the downregulation of the FvCHLH/ABAR expression could not affect FvRIPK1 transcripts but remarkably reduced FvABI1 transcripts and promoted FvABI4, FvSnRK2.2, and FvSnRK2.6 transcripts in the contrast of the non-transgenic plants to the FvCHLH/FvABAR-RNAi plants, in which chlorophyll contents were not affected but had abscisic acid (ABA) response in stomata movement and drought stress. The distinct expression level of FvABI1 and FvABI4, together with the similar expression level of FvSnRK2.2 and FvSnRK2.6 in the FvRIPK1- and FvABAR/CHLH-RNAi plants, suggested that FvRIPK1 regulated plant morphogenesis probably by ABA signaling. In addition, FvRIPK1 interacted with FvSnRK2.6 and phosphorylated each other, thus forming the FvRIPK1–FvSnRK2.6 complex. In conclusion, our results provide new insights into the molecular mechanism of FvRIPK1 in plant growth.
Highlights
Based on our first establishment of Agrobacterium-mediated transformation of germinating seeds in diploid strawberry (Fragaria vesca), we find that the strawberry ripk1 mutant is stunted, demonstrating that FvRIPK1 is essential to plant morphogenesis by multiple abscisic acid (ABA) signaling. FvRIPK1 interacts with FvSnRK2.6, and they phosphorylate each other. Our findings provide new insights into a molecular mechanism of FvRIPK1 in plant growth and environment response.
Introduction
Strawberry is a member of the genus Fragaria (Rosaceae), which is not only an important fruit crop worldwide but is also regarded as a model plant for studying non-climacteric fruit ripening. In the past years, much progress has been made in identification of gene function via traditional stable and transient transgenic systems in strawberry; however, more or less limits of both have been reported (Guidarelli and Baraldi, 2015; Haddadi et al., 2015; Zhao et al., 2019). Thus, development of a rapid, efficient, and stable transformation system is urgent in strawberry for both research and agricultural improvement.
Although strawberry leaf tissue has been widely studied to develop efficient transformation and regeneration methods through somatic tissue or callus formation (Nehra et al., 1990; El Mansouri et al., 1996; Haymes and Davis, 1997; Balokhina et al., 2000; Jimenez-Bermudez et al., 2002; Agius et al., 2003; Oosumi et al., 2006; Qin et al., 2008; Zhang et al., 2014; Song et al., 2019), the regeneration of transgenic strawberry plants takes a long time from transformation to identification (Barcelo et al., 1998; Folta and Dhingra, 2006; Haddadi et al., 2015). For example, it takes at least 15 months from transformation to gaining transgenic fruits for gene function analysis in ripening (Hoffmann et al., 2006; Miyawaki et al., 2012). In addition, although the transient transformation technology in strawberry fruits provides a rapid tool to study gene function in ripening, remarkable limits are existent in fruit transient transgenic system (Guidarelli and Baraldi, 2015; Zhao et al., 2019).
In addition to the widely used leaf disk transformation (Horsch et al., 1985; Cappelletti et al., 2015; Song et al., 2019), other tissues are also used for gene transformation (Potrykus, 1990; Hiei et al., 1994; Ishida et al., 1996; Clough and Bent, 1998; Bent, 2006; Zhang et al., 2006; Harwood, 2014; Ishida et al., 2014; Ratanasut et al., 2017; Van Eck, 2018; Song et al., 2019). The mature and immature embryo-generated calli are used to develop transgenic plants in rice, maize, wheat, and barley (Potrykus, 1990; Hiei et al., 1994; Ishida et al., 1996; Ishida et al., 2014). Notably, a non-tissue culture-based method, floral dip transformation, is widely and efficiently used in Arabidopsis as a model system (Clough and Bent, 1998; Bent, 2006; Zhang et al., 2006). The Agrobacterium-mediated transformation of germinating seeds (AMTGS) of Arabidopsis, another non-tissue culture approach, is also early reported (Feldmann and Marks, 1987). In recent years, the AMTGS of kenaf (Hibiscus cannabinus) seeds have been developed within 3 months with 6% transformation efficiency by the efficient, fast, reliable stable, non-tissue culture transgenic method (Hanana et al., 2018). Therefore, to rapidly identify gene function in strawberry, an AMTGS of diploid strawberry “Hawaii-4” (Fragaria vesca) is necessary to be established.
It is known that selection of a good reporter gene contributes to early identification of transgenic lines. In the past years, the H subunit of magnesium chelatase magnesium chelatase (CHLH), an enzyme involved in chlorophyll biosynthesis, is chosen as a reporter gene, in that its silencing phenotype was clearly observed through chimeric phenotypes with yellow/white leaves (Hiriart et al., 2002; Hiriart et al., 2003; Pignatta et al., 2007; Park et al., 2016). In addition, the plant CHLH has multiple functions, such as plastid-signaling trigger (Mochizuki et al., 2001), anti-sigma factor in sugar catabolism (Osanai et al., 2009), molecular circadian clock switch (Legnaioli et al., 2009), and putative abscisic acid (ABA) receptor (ABAR; Shen et al., 2006; Wu et al., 2009; Shang et al., 2010; Du et al., 2012; Zhang et al., 2013; Jiang et al., 2014; Liang et al., 2015; Bi et al., 2019). Because CHLH and ABAR are the same gene in this article, we will call it as ABAR in the later section. Notably, using FaABAR as a bait in a yeast two-hybrid (Y2H) assay, FaRIPK1 was recently screened, and it is a nucleus-localized and ABA-binding leu-rich repeat receptor-like protein kinase (LRR-RLKs); furthermore, the two proteins interact in the nucleus and serve as the receptor complex to regulate strawberry fruit ripening (Hou et al., 2018). However, its role in plant vegetable growth is yet unknown.
In this study, on the basis of ABAR function in chlorophyll biosynthesis, an AMTGS in diploid strawberry was first established, by which the function of RIPK1 in vegetative growth is subsequently explored for the first time. In addition, the RIPK1 expression was investigated in the abar lines, in which the chlorophyll level is not significantly downregulated; meanwhile, the ABAR expression was also detected in the ripk1 lines. Moreover, at the protein level, FvRIPK1 interacted with the ABA signaling component FvSnRK2.6 and phosphorylates each other. Finally, the action mechanism of the RIPK1 and ABAR was discussed.
Materials and methods
Plant materials and germination
The diploid strawberry Hawaii-4 (F. vesca) plants are grown in a green house at 25°C–28°C. The seeds were taken from ripe fruits, and the residual receptacle was removed; then the seeds were put at room temperature to dry for 1–2 days. The dried seeds are placed into a 2-ml tube and stored at 4°C for use. Seeds were sterilized by 1% sodium hypochlorite for 8 min and washed with sterile water three times. The sterilized seeds were transferred onto an Murashige and Skoog (MS) germination medium [4.43 g/L MS (PhytoTech Labs M519), 2% sucrose (Sangon, China), 0.8% agar (Sangon, China), pH 5.8] and left at 4°C for 24 h to break dormancy. The dormancy-broken seeds were treated with three manners: light culture in a light incubator (PRX-350C, Sdfu, China), dark culture wrapped with tin foil, and light culture after 2 days of dark culture. The seed culture condition in a light incubator is as a 16-h photoperiod with a light intensity of 30,000 LX (250 μmol·m−2·s−1) at 24°C. Each treatment was inoculated with 20 seeds (three repeats) for seed germination.
Vector construction and Agrobacterium tumefaciens transformation
Two vectors were used to optimize transformation parameters, namely, a GV3101 strain carrying a pCAMBIA 1300 vector with a green fluorescent protein (GFP) reporter gene or carrying a pBI121 vector with a β-glucuronidase (GUS) reporter gene and a kanamycin (Kana) resistance gene.
The RNAi recombinant plasmid was constructed using the Gateway system, and target genes were silenced in strawberry fruit by intron splicing RNA (ihpRNA). The 465-bp FvABAR and 595-bp FvRIPK1 were amplified and cloned into the pDONR™ 221 vector (Invitrogen, Beijing, China) and then into the pK7GWIWG2 (II) RR vector using the Gateway LR Clonase II enzyme (Invitrogen 11791-020, USA) combination technology. The pK7GWIWG2 (II) RR vector contains the red fluorescence protein (DsRed) reporter gene and the kanamycin resistance gene. The recombinant plasmid was transformed into A. tumefaciens GV3101, and a single colony was picked and placed in a Luria broth (LB) medium containing antibiotics 20 μg/ml rifampicin (Sangon, A600812, China), 50 μg/ml gentamicin (Sangon, A100304, China), and 100 μg/ml spectinomycin (Sangon, B541016, China).
The single colony of GV3101 strains (GV3101 strain of pCAMBIA 1300 and GV3101 strain of pBI121) was put into an LB medium containing antibiotics 20 μg/ml rifampicin, 50 μg/ml gentamicin, 100 μg/ml kanamycin (Sangon, A506636, China) at 28°C and 180 rpm overnight. The bacteria were collected by centrifugation at 6,000 rpm for 5 min and suspended using MS infection [4.43 g/L MS, 2% sucrose, 200 μmol/L Acetosyringone (As, Sigma-Aldrich, D134406, USA)], pH 5.8, to prepare an Agrobacterium infection solution. The primers used are listed in Supplemental Table S1.
Optimization of parameters related to infection efficiency
The preimbibed seeds were placed in an Agrobacterium infection solution and cocultured at 120 rpm, 28°C for 24 h, and then the seeds were placed in an MS germination medium and wrapped in tin foil at 25°C for 3 days. Subsequently, the seeds were further placed in an MS selection medium [4.43 g/L MS, 2% sucrose, 75 mg/L kanamycin, 200 mg/L Timentin Termetin (PhytoTech Labs T869), 0.8% agar, pH 5.8]. After 3 days, GFP/DsRed fluorescent tag and GUS reporter gene were used for early screening and statistics. After 2 weeks, the seedlings with fluorescence were transplanted into quartz sand, covered with cling film for 3 days to prevent water loss, and watered with the kanamycin-added nutrient solution to allow the seedlings to grow under appropriate selection pressure at condition with a greenhouse at 24°C, light/dark 16/8 h.
To determine factors related to infection efficiency, we performed a series of experiments, namely, optical density (OD) values (OD at 600 nm, OD600 = 0.5, 1, 1.5, and 2), infection time (12, 24, and 36 h), and seed germination status (initial germination without white root tip, germination with white root tip, and germination with cotyledons). The optimal infection parameters were determined by investigation of GFP using stereo fluorescence microscope (ZEISS-Axiocam 506 color) and a GUS histochemical stain kit (Solarbio, G3060). Infection efficiency was calculated from the proportion of plants with GFP fluorescent protein expression or the proportion of plants with GUS expression. During the infection process, 20 seeds were placed in each Petri dish, and three replicates were performed.
Determination of appropriate Kanamycin concentration for screening
Natural sterile seeds are cultured on the MS selection medium containing different concentrations of kanamycin with 0, 25, 50, 75 and 100 mg/L in order to determine the most suitable antibiotic screening concentration. Twenty seeds were placed in each Petri dish, and the experiment was done with three repetitions.
Identification of transgenic plants by PCR
In order to confirm transformation of T-DNA in diploid strawberry genome, leaves of 1-month-old transgenic strawberry plants with red light were collected, and genomic DNA was extracted by Plant Genomic DNA Extraction Kit (Aidlab Biotech, DN15, China). The leaves of wild-type plants were used as control. The fragments of anti-Kana and DsRed genes were amplified by PCR. The primers used are listed in Supplemental Table S2.
Stomata are sensitive to exogenous abscisic acid treatment
Six leaflets were selected from each seedling, and six sampled parts from each leaflet were used to determine SPAD values using a chlorophyll meter (Konica-Minolta Holdings, Inc., SPAD-502Plus). To explore the function of FvABAR in ABA response in stomata movement rather than chlorophyll synthesis, three transgenic plants with no significant difference in chlorophyll content from the control were selected to observe whether the stomata were sensitive to exogenous ABA stimulation in the buffer in an MES-KCl buffer solution [KCl 50 mmol/L, 2-(N-morpholino)ethanesulfonic acid (MES) 10 mmol/L, CaC12 0.1 mmol/L, pH 5.8].
ABA promotes stomata closure test: The stomata are opened by a light suspension culture in a light incubator at 24°C for 6 h, and then some leaves are incubated in an MES-KCl buffer solution for 3 h, then transferred to an MES-KCl buffer solution containing 50 μM ABA for 3 h. ABA inhibition stomatal opening test: In a 24°C light incubator, wrap the Petri dish in a dark suspension culture for 6 h to close the stomata. After that, the foil was peeled off, a part of the leaves was incubated in an MES-KCl buffer solution for 3 h, and the other part was transferred to an MES-KCl buffer solution containing 50 μM ABA for 3 h.
Observing pores with a transparent nail polish: polishing transparent nail evenly on the back of leaf, air drying for 5–8 min, stripping the lower epidermis with nail polish by hand, placing it in the center of glass slides, and then covering with coverslips. The prepared slides were observed under an optical microscope and photographed to measure the stomatal conductance. Six fields of view were selected for each treatment in the test, and five stomata apertures were measured in each field of view. Each treatment was repeated three times.
Drought test of FvABAR-RNAi plants
Three transgenic plants and wild-type plants selected above were subjected to a drought test. Before wither, the plants were immediately transferred to a tray for watering. The experiment was repeated three times
Real-time PCR
Total RNA from leaves of about 40-day transgenic and wild-type plants were extracted using an Omega RNA extraction kit (Omega Bio-tek, USA) according to the manufacturer’s protocols. To generate first-strand cDNA, 400 ng of total RNA was reverse-transcribed using the Trans kit (Transgen, China) according to the manufacturer’s protocols. The first-strand cDNA was used as a template for PCR amplification for real-time PCR on a LightCycler 96 real-time PCR system (Roche Diagnostics GmbH, Mannheim City, Germany) using TransStart Top Green qPCR SuperMix (Transgen, China). The reactions of 10 µl contained 5 µl qPCR SuperMix, 0.25 µl forward specific primer (10 µM; Sangon, China), 0.25 µl reverse specific primer (10 µM; Sangon, China), 2 µl cDNA template, and 2.5µl ddH2O. Actin was used as a reference gene. Relative gene expression was analyzed by LightCycler® 96 SW 1.1.The primers used for real-time PCR are shown in Supplemental Table S3. The experiment was repeated three times.
Kanamycin resistance test of T0 generation seeds
Twenty seeds were obtained from a single inbred T0 generation, sprinkled on quartz sand, and irrigated with kanamycin (75 mg/L). After 10 days of culture, the sensitivity of the plants to kanamycin resistance was observed, and DNA molecules were analyzed for the resistant seedlings. The primers used are listed in Supplemental Table S2.
Yeast two‐hybrid assay
The Coding Sequence (CDS) of FvSnRK2.6 was cloned into a pGBKT7 vector, and the CDS of FvRIPK1, FvRIPK1-N, and FvRIPK1-KD was cloned into a pGADT7 vector. The plasmids were cotransfected into the yeast strain Gold. The transfected yeast cells were placed on an SD/‐Leu‐Trp medium and incubated at 30°C for 3 days. The grown yeast was transferred to an SD/‐Leu‐Trp (‐LW) medium and an SD/‐Ade‐His‐Leu‐Trp (LWHA) medium and incubated for another 3 days.
Firefly luciferase complementation imaging assay
The CDS of FvSnRK2.6 was cloned into pCAMBIA1300‐NLUC and that of FvRIPK1, FvRIPK1-N, and FvRIPK1-KD was cloned into pCAMBIA1300‐CCLUC. The constructs were transformed into Agrobacterium GV3101 and coinjected into Nicotiana benthamiana leaves. After 48–72 h, the luciferase (LUC) signal was captured by a cold charge-coupled device camera.
Bimolecular fluorescence complementation assay
Bimolecular fluorescence complementation (BiFC) assays were performed as described previously (Waadt et al., 2008). FvSnRK2.6 or FvABI1 was cloned into pSPYCE, and FvRIPK1 was cloned into pSPYNE173. The constructions were coinjected into N. benthamiana leaves. After 48–72 h, the GFP signal was observed under ZEISS LSM 710.
In vitro phosphorylation assay
Recombinant proteins GST-FvRIPK1, GST-FvRIPK1K843R, MBP-FvSnRK2.6, and MBP-FvSnRK2.6G35R were purified from BL21 Escherichia coli and subjected to an in vitro phosphorylation assay. About 1 μg GST-FvRIPK1 was mixed with 2 μg MBP-FvSnRK2.6G35R, and the mixed proteins were incubated in a kinase buffer (2 mmol/L Tris‐HCl, pH 7.5, 1 μmol/L CaCl2, 0.2 mmol/L dithiothreitol, 0.5 mmol/L MgCl2, 0.1 mmol/L MnCl2), with 50 μmol/L cold ATP and 1 μCi γ‐32P‐ATP at 30°C for 30 min. The reactions were stopped by adding 4 μl 5× SDS loading buffer, and proteins were separated by 8% SDS‐PAGE. The protein gel was stained, decolorized, wrapped tightly with plastic wrap, and then put into the phosphor screen. Finally, radioactivity was detected by a Typhoon 9410 imager.
Results
Optimization of infection parameters on the basis of germination seeds
To establish rapid and stable transgenic system in strawberry, the AMTGS in diploid strawberry was first studied based on both various infection parameters and previous reports (Feldmann and Marks, 1987; Hanana et al., 2018). The seed germination rates were not significantly different under three light treatments, namely, light culture, dark culture, and light culture after dark culture for 2 days. When the seeds had imbibed for 4, 5, and 6 days, a few, half, and 90% of seeds could germinate, respectively (Figure 1A). Thus, light culture was used in seed germination. To confirm optimal kanamycin concentration for screening, sensitivity of the natural seeds to kanamycin is determined by adding different concentrations of kanamycin to the medium. With an increase in kanamycin concentrations at 0, 25, 50, 75, and 100 mg/L during a 15-day cultivation in an MS selection medium, the natural seeds germinated correspondingly by 95% germination seedlings with normal green seedling, 91.7% germination seedlings with light green cotyledons, 41.7% germination seedlings with no full extension cotyledons, 20% germination seedlings with yellow or white cotyledons, and 5% germination seedlings with browned or died cotyledons, respectively (Figures 1B, C). Thereby, 75 mg/L kanamycin in an MS selection medium was used as an antibiotic selection.
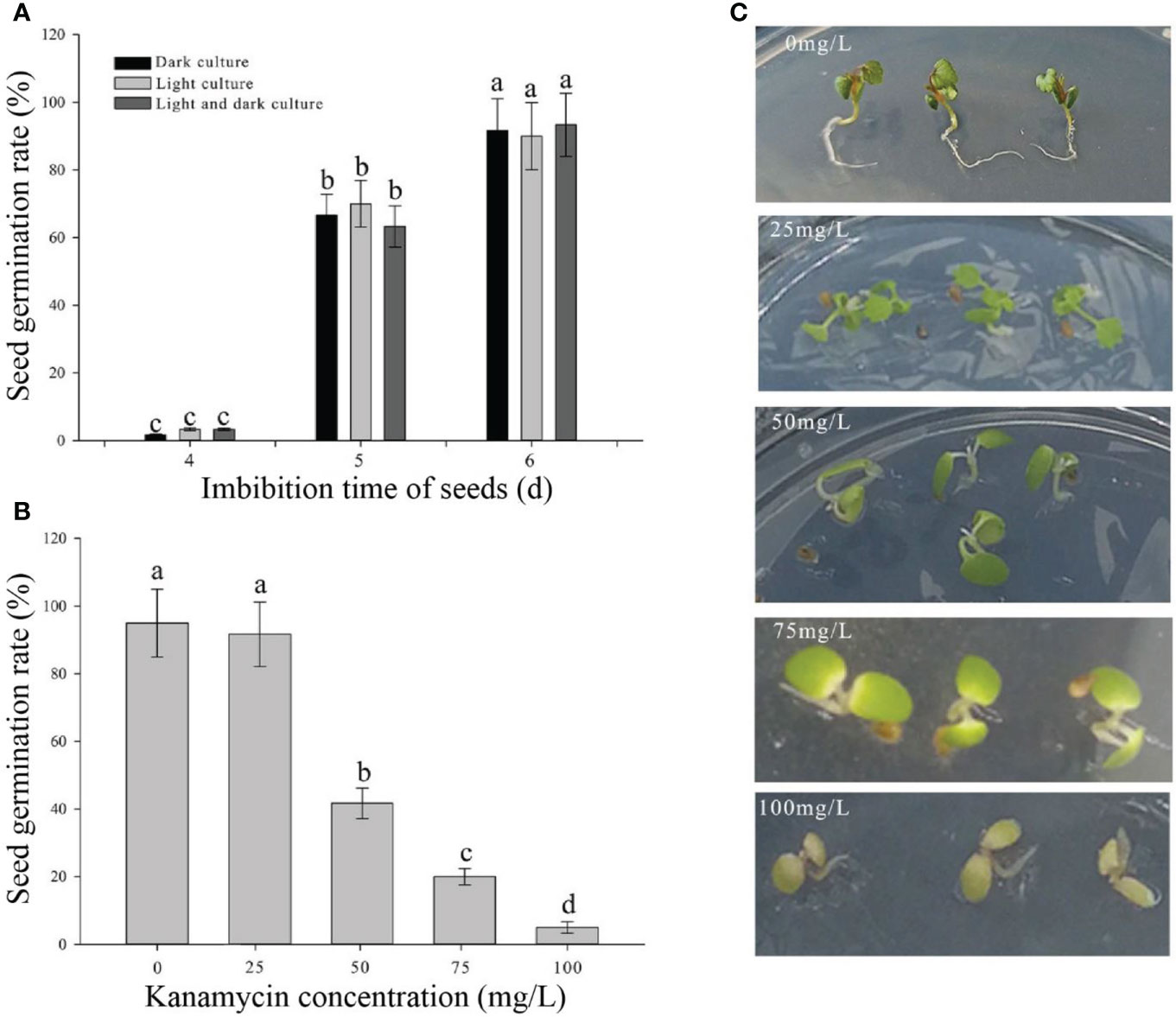
Figure 1 Determination of culture parameters in germination seeds and the optimal kanamycin screening concentration. (A) Seed germination rates in different light culture conditions and imbibing time. (B) Seed germination rates in different kanamycin concentrations after a 15-day cultivation. (C) Phenotypes in different kanamycin concentrations after a 15-day cultivation. Each treatment was treated with 20 seeds. The error bars represent the standard error (n = 3). Different letters indicate statistically significant differences at p < 0.05 as determined by Duncan’s test.
After a 3-day culture in the selection medium, the GFP could be observed in the successfully transgenic plants (Figures 2A, B). Similarly, the not successfully infected seedlings had no GUS expression (Figures 2C, E), whereas the successfully infected seedlings showed blue in the roots and true leaves with GUS expression (Figures 2D, F). Base on the observation of both GFP and GUS expressions in transgenic seedlings, a 1.5-OD600 value of the Agrobacterium infection solution was optimum infection concentration (Figure 2G). The germination seeds with white root tips had the highest infection efficiency (Figure 2H) during the 24-h infection (Figure 2I).
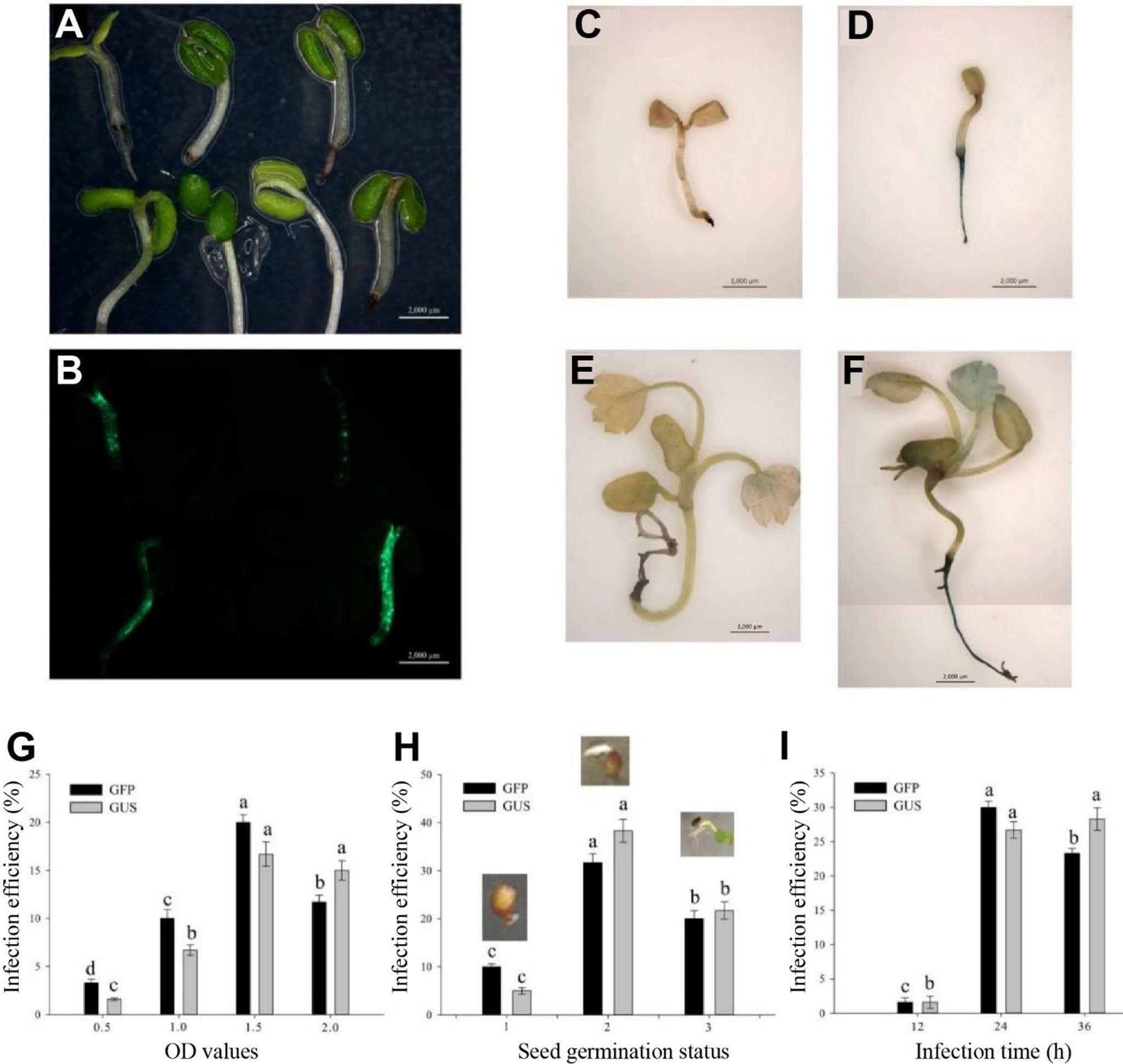
Figure 2 Determination of influence factors in Agrobacterium-mediated transformation of germinating seeds. (A) Infection seedlings after a 3-day culture. (B) GFP expression in the 3-day culture infection seedlings. (C, E) No GUS blue in the uninfection seedlings. (D, F) GUS blue in successfully infected seedlings. (C, D) Infection seedlings after a 3-day culture. (E, F) Infection seedlings after a 2-week culture. (G) Effect of optical density (OD) values on infection efficiency. (H) Effect of seed germination status on infection efficiency including initial germination, white root tip and green cotyledon stages. (I) Effect of infection time on infection efficiency. Each treatment was treated with 20 seeds. The error bars represent the standard error (n = 3). Different letters indicate statistically significant differences at p < 0.05 as determined by Duncan’s test. Bars = 0.2 cm.
Taken together, the optimum parameters of AMTGS in diploid strawberry are as follows: the germination seeds with white tip, 75 mg/L kanamycin in an MS selection medium, 1.5-OD600 value of the Agrobacterium infection solution, and 24-h infection in a light culture.
Establishment of a rapid and stable transgenic system by the Agrobacterium-mediated transformation of germinating seeds by a reporter gene of FvABAR in strawberry
In addition to using the GFP and GUS as early transgenic positive selection, the ABAR encoding the H subunit of magnesium chelatase involved in chlorophyll biosynthesis is chosen as a reporter gene owing to its silencing phenotypes with yellow/white leaves easy to be observed (Hiriart et al., 2002; Hiriart et al., 2003; Pignatta et al., 2007; Park et al., 2016).
Based on the optimal transformation conditions obtained above, we have transformed the germination seeds with white radicles of diploid strawberry “Hawaii-4” through the GV3101 strain carrying the pK7GWIWG2 (II) RR + FvABAR vector. After a 3-day infection under dark culture, the seeds were transferred to an MS selection medium. After 3 days, the seeds were screened by DsRed fluorescence. After 2 weeks, the fluorescent seedlings were transferred to quartz sand and watered using a nutrient solution containing 75 mg/L kanamycin. The whole processes for transformation of germination strawberry seeds were shown in Figure 3. One month after transplanting, in comparison to the control, the FvABAR-RNAi plants showed yellow spots (Figure 4A, line 1, phenotype 1) or half green and half white in a leaf (Figure 4A, line 2, phenotype 2). The PCR was performed to detect both kanamycin and DsRed gene fragments in the transgenic plants. The expected 400-bp Kana and 964-bp DsRed products were detected in five FvABAR-RNAi plants but not in the wild-type plants (Figures 4B, C). The RT-PCR result showed that ABAR was knocked down in FvABAR-RNAi Line1 (Figures 4D). And the infection efficiency of phenotype 1 and 2 was about 20% or 12% (Figures 4E). These results showed that on basis of the report genes including GFP, GUS, DsRed, and ABAR, a rapid and stable transgenic system by the AMTGS is success.
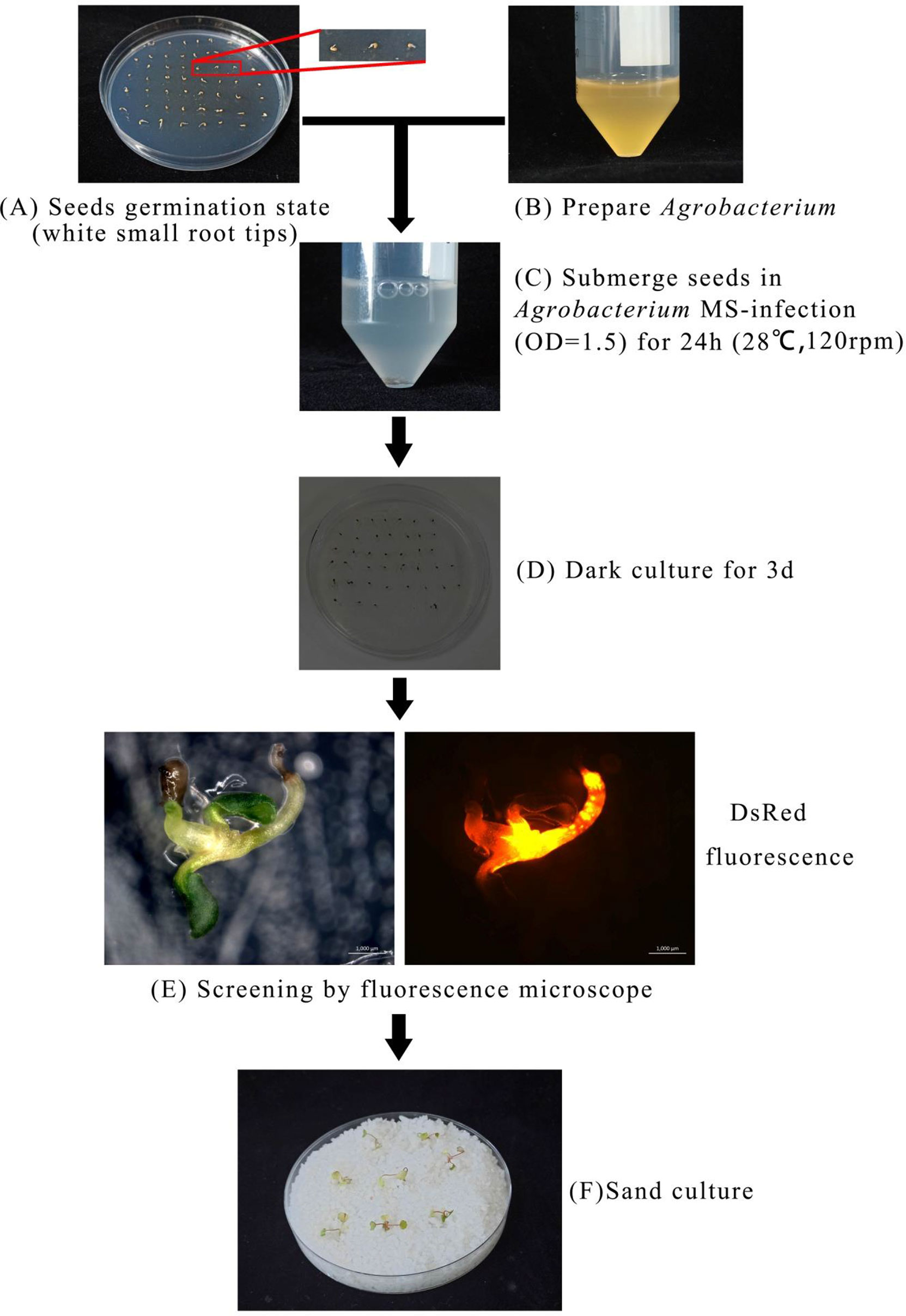
Figure 3 Establishment of a rapid and stable transgenic system by the Agrobacterium-mediated transformation of germinating seeds. The preimbibed seeds were placed in an Agrobacterium infection solution and cocultured at 120 rpm, 28°C for 24 h, (A–C) and then the seeds were placed in an MS germination medium and wrapped in tin foil at 25°C for 3 days (D). Subsequently, the seeds were further placed in an MS selection medium [4.43 g/L MS, 2% sucrose, 75 mg/L kanamycin, 200 mg/L Timentin (PhytoTech Labs T869), 0.8% agar, pH 5.8]. After 3 days, GFP/DsRed fluorescent tag and GUS reporter gene were used for early screening and statistics (E). After 2 weeks, the seedlings with fluorescence were transplanted into quartz sand, covered with cling film for 3 days to prevent water loss, and watered with the kanamycin-added nutrient solution to allow the seedlings to grow under appropriate selection pressure at a condition with a greenhouse at 24°C, light/dark 16/8 h (F).
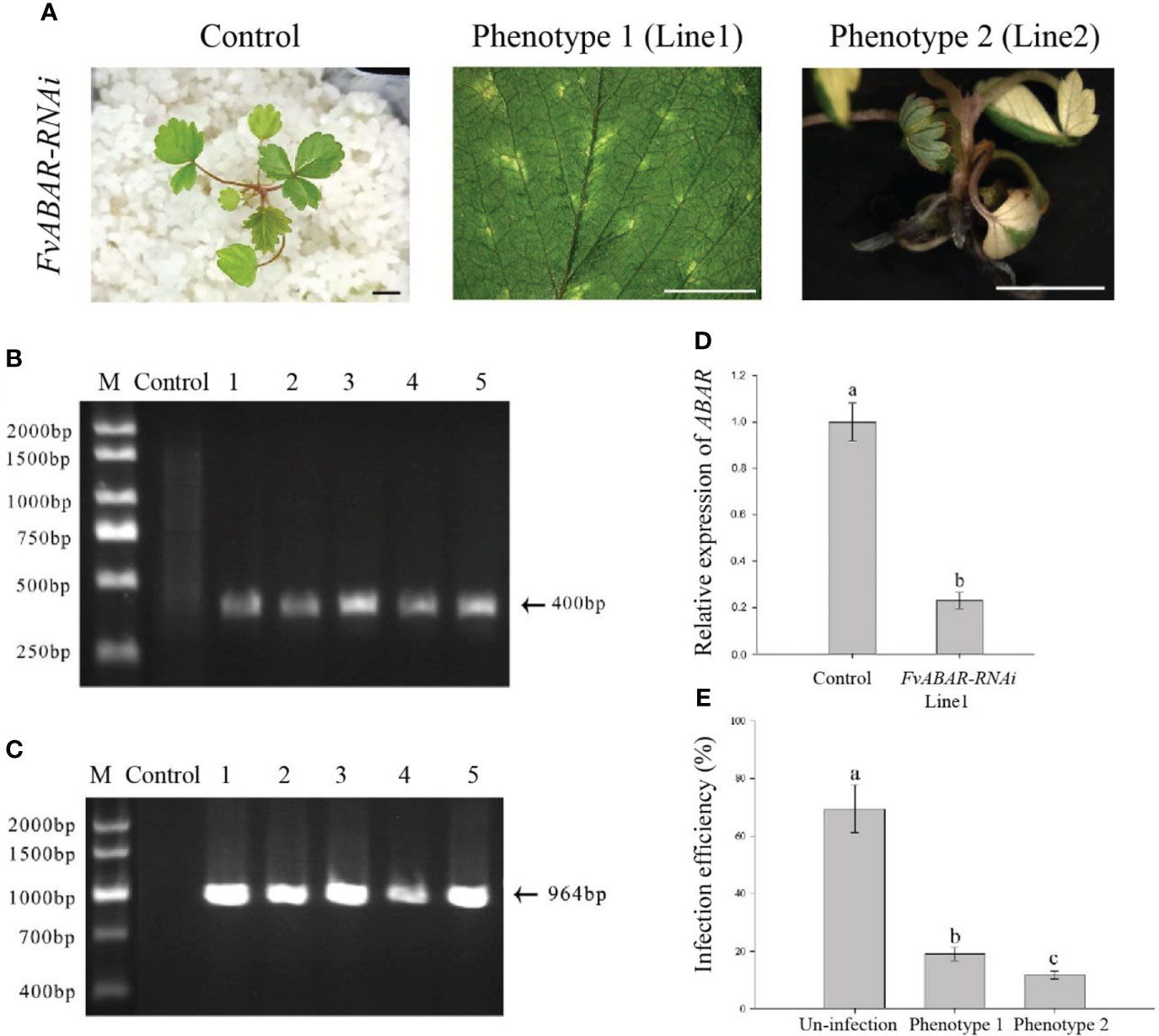
Figure 4 Phenotype of the FvABAR-RNAi plants in strawberry. (A) Loss of green color in the FvABAR-RNAi plants. (B) The 400-bp Kana products were detected in five FvABAR-RNAi plants (lines 1–5) but not in the wild-type plants (control) by qPCR. (C) The 964-bp DsRed products were detected in five FvABAR-RNAi plants (lines 1–5) but not in the wild-type plants (line 1) by PCR. The expected 400-bp Kana and 964-bp DsRed products were detected in five FvABAR-RNAi plants but not in the wild-type plants. Bars = 0.5 cm. (D) FvABAR gene expression of transgenic plants and wild-type plants. (E) Proportion of infected seedlings showing phenotype 1 and 2 plants to total infected plants.
Phenotypes of FvRIPK1-silencing plants by the Agrobacterium-mediated transformation of germinating strawberry seeds
Based on the processes of FvABAR-silencing transgenic plants by the Agrobacterium-mediated transformation of germinating strawberry seeds (Figure 3), the GV3101 strain carrying the pK7GWIWG2 (II) RR + FvRIPK1 vector were used for transformation of the germination seeds. One month after transplanting, in comparison to the control, FvRIPK1-RNAi plants were stunted, showing curled leaves (Figure 5A). The phenotype of line 1 differed from that of line 2 maybe because their infected site in seeds was different. But the transgenic plants finally died and survived no more than 7 weeks after transplanting for some unknown reasons, so we cannot get the next generation to perform the assays. The PCR was performed to detect both kanamycin and DsRed gene fragments in the transgenic plants. The expected 400-bp Kana and 964-bp DsRed products were detected in five FvRIPK1-RNAi plants but not in the wild-type plants (Figures 5B, C). These results showed that FvRIPK1 is essential to strawberry plant growth.
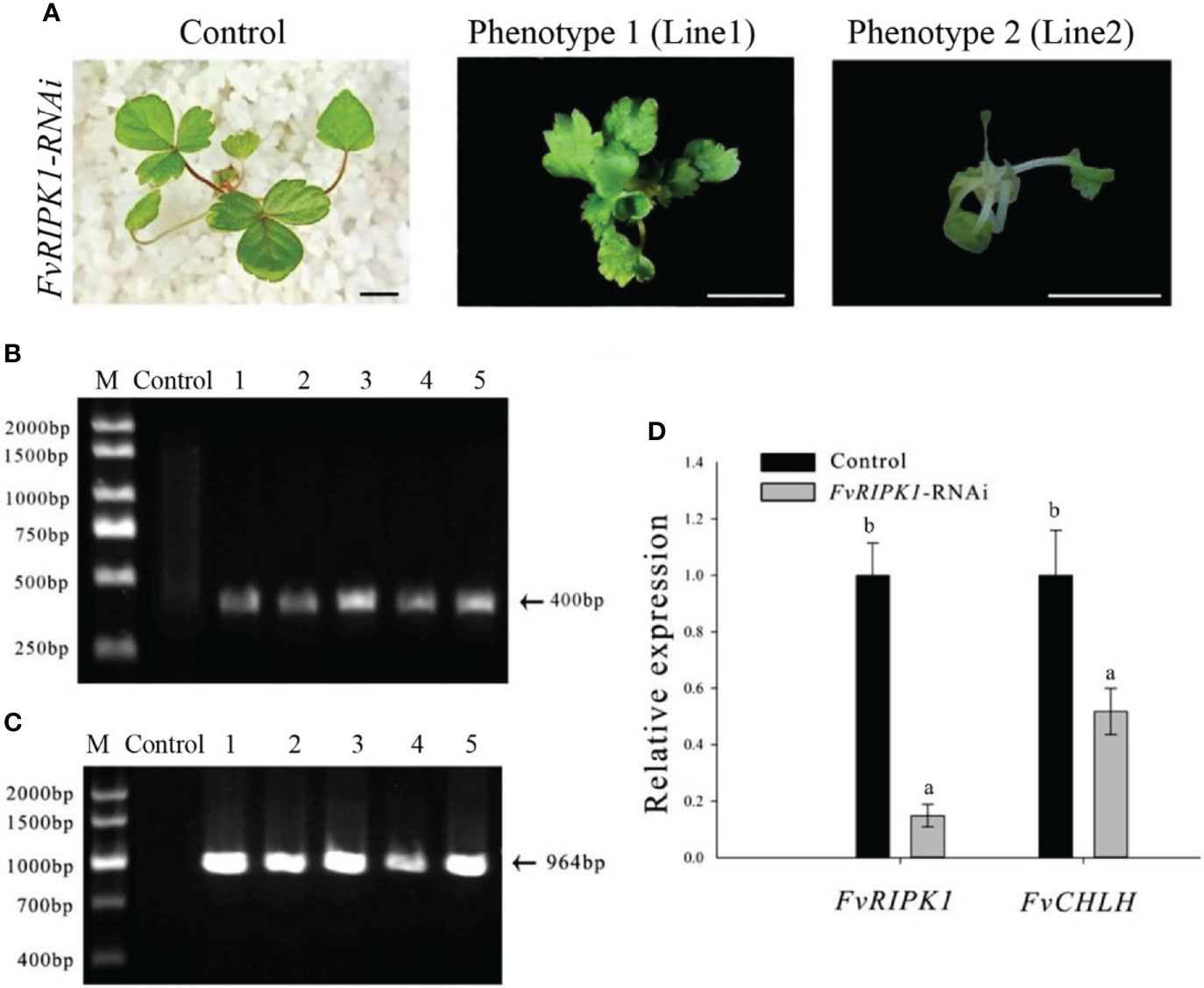
Figure 5 Phenotype of the FvRIPK1-RNAi plants in strawberry. (A) Deformed leaves with normal green color in the FvRIPK1-RNAi plants. (B) The 400-bp Kana products were detected in five FvRIPK1-RNAi plants (lines 2–6) but not detected in the wild-type plants (line 1) by qPCR. (C) The 964-bp DsRed products were detected in five FvRIPK1-RNAi plants (lines 2–6) but not detected in the wild-type plants (line 1) by qPCR. (D) The FvABAR expression was significantly downregulated in the FvRIPK1-RNAi plants. The error bars represent the standard error (n = 3). Different letters indicate statistically significant differences at p < 0.05 as determined by Duncan’s test. Bars = 0.5 cm.
Given the interaction between strawberry ABAR and RIPK1 (Hou et al., 2018), FvABAR expression was investigated in the FvRIPK1-RNAi plants. The results showed that the FvABAR expression levels were significantly downregulated in the FvRIPK1-RNAi line 1 plant, in which the FvRIPK1 transcripts remarkably downregulated in comparison to the control (Figure 5D). The result indicates that FvRIPK1 is related to FvABAR. Because of the FvRIPK1-RNAi plants with normal green leaves (Figure 5A) that the decrease of the FvABAR expression in the FvRIPK1-RNAi plants is mainly related to plant morphogenesis rather than chlorophyll synthesis (Figure 5), it may also be related to ABA signaling.
Downregulation of FvRIPK1 transcripts inhibits FvABAR expression while not vice versa, but both are involved in ABA signaling
To explore the relationship of FvRIPK1 with FvABAR at expression levels and whether both are involved in ABA signaling in strawberry, the FvABAR-transgenic plants had no significant decrease in chlorophyll contents (Figure 6A) but had ABA response in stomata movement (Figure 6B) and drought stress (Figure 6C) in comparison to the wild plants. Thus, the FvABAR transgenic plants were used to assess a role of FvABAR in ABA signaling including aba insensitive 1 (ABI1), aba insensitive 4 (ABI4), SNF1-related protein kinase 2.2 (SnRK2.2), SNF1-related protein kinase 2.6 (SnRK2.6), NAC domain-containing protein 72-like (NAC72), and dehydration-responsive protein RD22 (RD22) (Shen et al., 2006; Gu et al., 2017). The results showed that the downregulation of the FvABAR expression could not affect FvRIPK1 transcripts but significantly promoted FvABI4, FvSnRK2.2, and FvSnRK2.6 transcripts and significantly reduced FvABI1, FvNAC72, and FvRD22 transcripts (Figure 7A). The downregulation of the FvRIPK1 expression could reduce FvABAR transcripts; significantly promoted FvABI1, FvSnRK2.2, FvSnRK2.6, and FvNAC72 transcripts; and significantly inhibited FvABI1 and FvRD22 transcripts (Figure 7B). The results that the downregulation of the FvABAR expression could not affect FvRIPK1 transcripts, suggesting the role of FvABAR in ABA signaling was probably partially independent of FvRIPK1, whereas the downregulation of FvRIPK1 expression reduced FvABAR transcripts, suggesting FvRIPK1 in ABA signaling somehow is dependent of FvABAR.
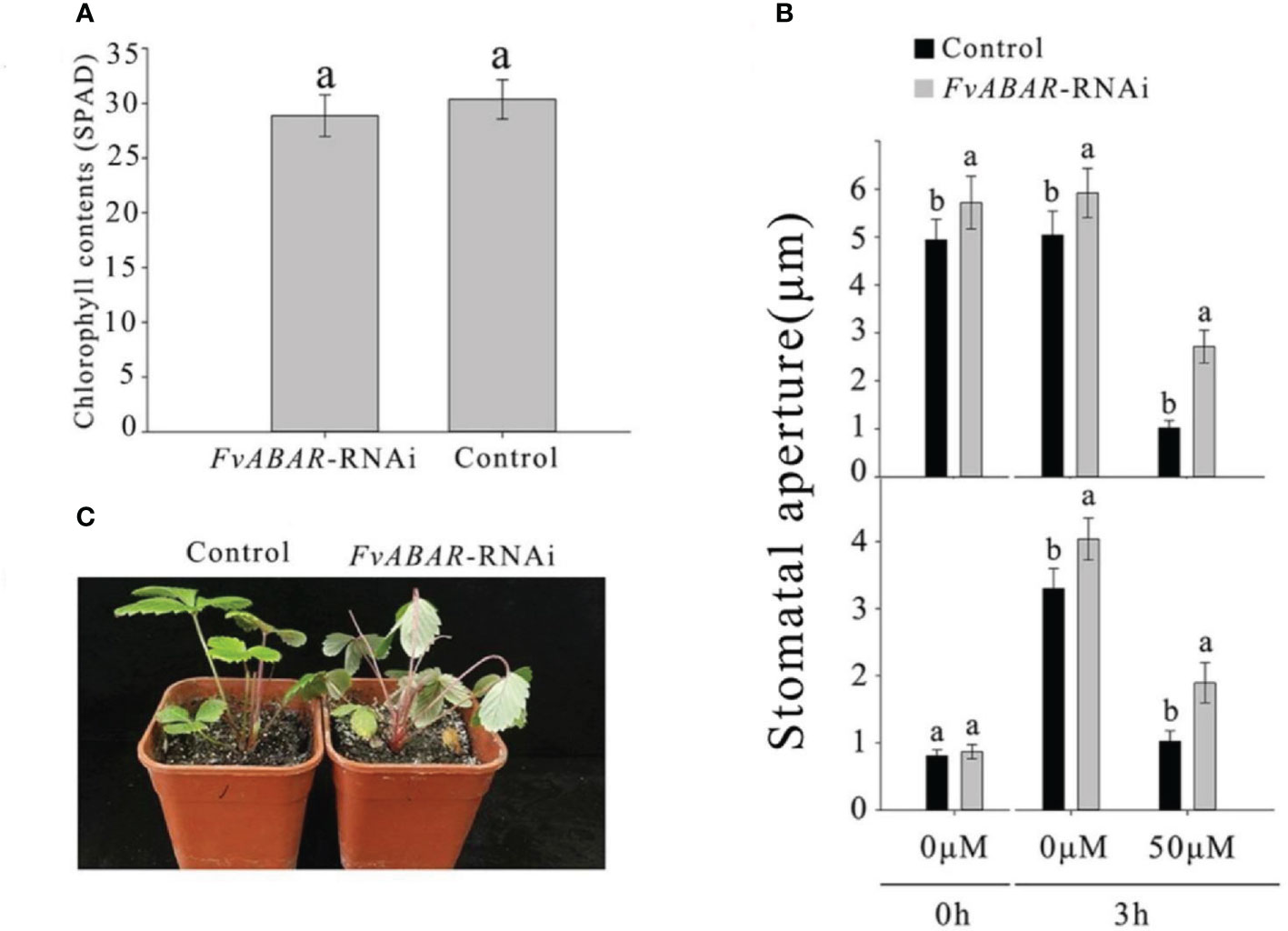
Figure 6 FvABAR plays a role in drought stress and stomatal movement in response to ABA. (A) The FvABAR-RNAi plants with no significant difference in chlorophyll contents from the wild plants. (B) Stomatal movement of the FvABAR-RNAi plants responsive to ABA. In the ABA-promoted stomatal closure test (top plot), the stomata are completely opened after 6 h of light culture. Then, adding 50 μM of ABA for 3 h of light culture, the stomata completely closed, and the RNAi plant showed insensitivity to ABA in stomata closure compared with the control. In the ABA inhibition stomatal opening, the stomata are completely closed after 6 h of dark culture. Then, adding 50 μM of ABA for 3 h of light culture, the stomata completely opened, and the RNAi plant showed insensitivity to ABA in stomata opening compared with the control. (C) Plant phenotypes of the wild-type control plants and FvABAR transgenic plants under 2 weeks of free water. The error bars represent the standard error (n = 3). Different letters indicate statistically significant differences at p < 0.05 as determined by Duncan’s test.
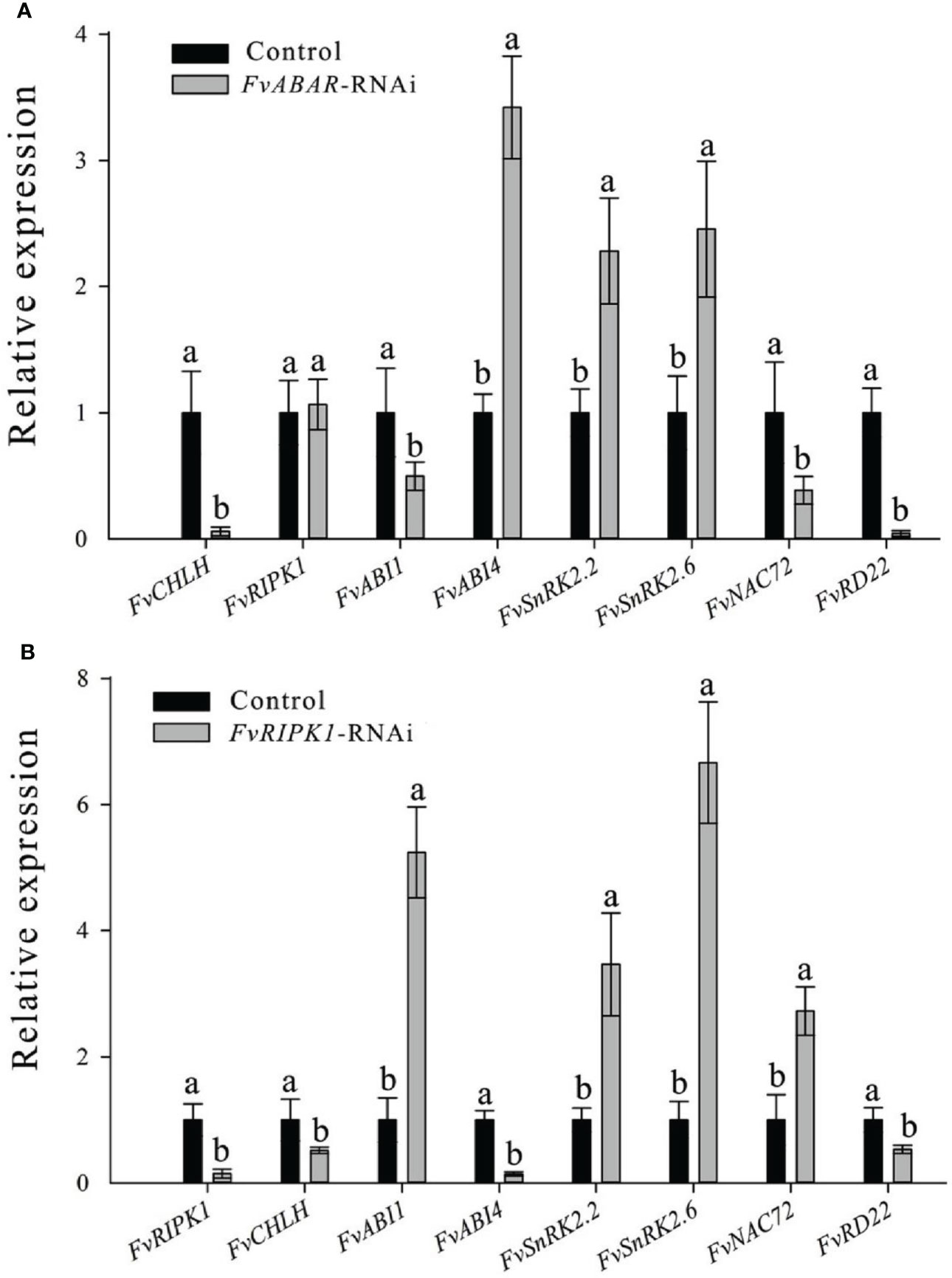
Figure 7 Alteration of the FvABAR or FvRIPK1 expression affects transcripts of several ABA-responsive and drought-related genes in the RNAi plants. RTPCR results showd the FvCHLH, FvRIPK1, FvABI1, FvABI4, FvSnRK2.2, FvSnRK2.6, FvNAC72 and FvRD22 gene expression in FvABAR-RNAi strawberry (A) and in FvRIPK1-RNAi strawberry (B). Actin mRNA was used as an internal control. The error bars represent the standard error (n = 3). Different letters indicate statistically significantdifferences at p < 0.05 as determined by Duncan’s test.
T1 seedlings of FvABAR-RNAi plants retains kanamycin resistance
The seeds of wild-type plants and T1 generations of FvABAR transgenic plants were spread on the quartz sand soaked in the nutrient solution containing 75 mg/L kanamycin. In the presence of kanamycin, the seeds of wild-type plants cannot germinate or the cotyledons turn yellow 10 days after germination, and the true leaves could not develop. T1-resistant seedlings could germinate on quartz sand with kanamycin and continued to grow normally (Figure 8A). The kanamycin-resistant bands and DsRed bands were detected in T1 generations (Figure 8B). These results demonstrate that the transgenic plants can be obtained by seed transformation and the relative genetic characteristics may be transferred to T1 progeny.
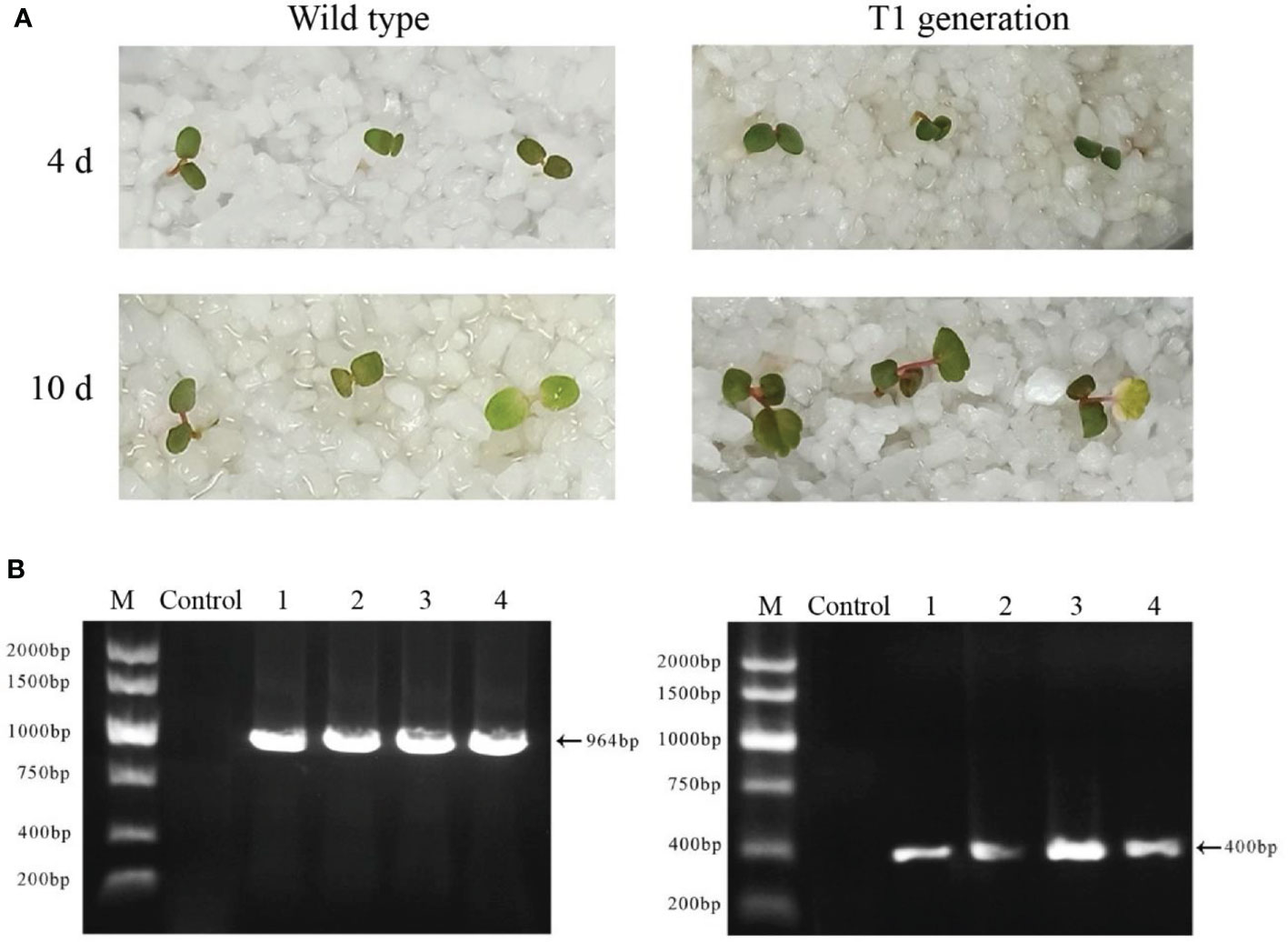
Figure 8 Isolation and identification of Ti-resistant plants. (A) The phenotype of wild-type plant seeds germinated for 4 and 10 days under the irrigation of a 75-mg/L kanamycin nutrient solution (left); the phenotype of T0 generation of FvABAR-RNAi plant seeds germinated for 4 and 10 days under the irrigation of a 75-mg/L kanamycin nutrient solution (right). (B) DsRed gene (left) and kanamycin resistance gene (right) of T1 generation–resistant seedlings of FvABAR-RNAi plants. Line 1 is wild-type plants, line 2 is positive control, and lines 3–5 are T1 resistant seedlings.
FvRIPK1 interacts with FvSnRK2.6 and phosphorylates each other
As shown in the results, the downregulation of FvRIPK1 influenced the transcription level of FvSnRK2.6 (Figure 7B). However, whether FvRIPK1 has a relationship with FvSnRK2.6 at the protein level is still unknown. Based on the study before (Hou et al., 2018), we divided FvRIPK1 into two domains, namely, FvRIPK1-N-termianl (1-763 aa, FvRIPK1-N) and FvRIPK1 kinase domain (764-1154 aa, FvRIPK1-KD). First, we performed the Y2H assay to test for the interaction between FvRIPK1 and FvSnRK2.6. We found that FvRIPK1-KD was sufficient to bind FvSnRK2.6 (Figure 9A). Next, we used firefly luciferase complementation imaging assay for protein interaction and detected a robust luciferase signal between FvSnRK2.6 and FvRIPK1 or FvRIPK1-KD (Figure 9B). Moreover, FvRIPK1 interacted with FvSnRK2.6 in the nucleus, not with FvABI1 in the BiFC assay (Figure 9C). FvRIPK1 and FvSnRK2.6 are both protein kinases, so we next check the phosphorylation relationship between them. We got the kinase-dead form GST-FvRIPK1K843R or MBP-FvSnRK2.6G35R to perform the in vitro phosphorylation assay. The results showed that FvRIPK1 can phosphorylate FvSnRK2.6 and vice versa (Figure 9D), but the phosphorylation signal was weak. In all, FvRIPK1 interacts with FvSnRK2.6, and they phosphorylate each other.
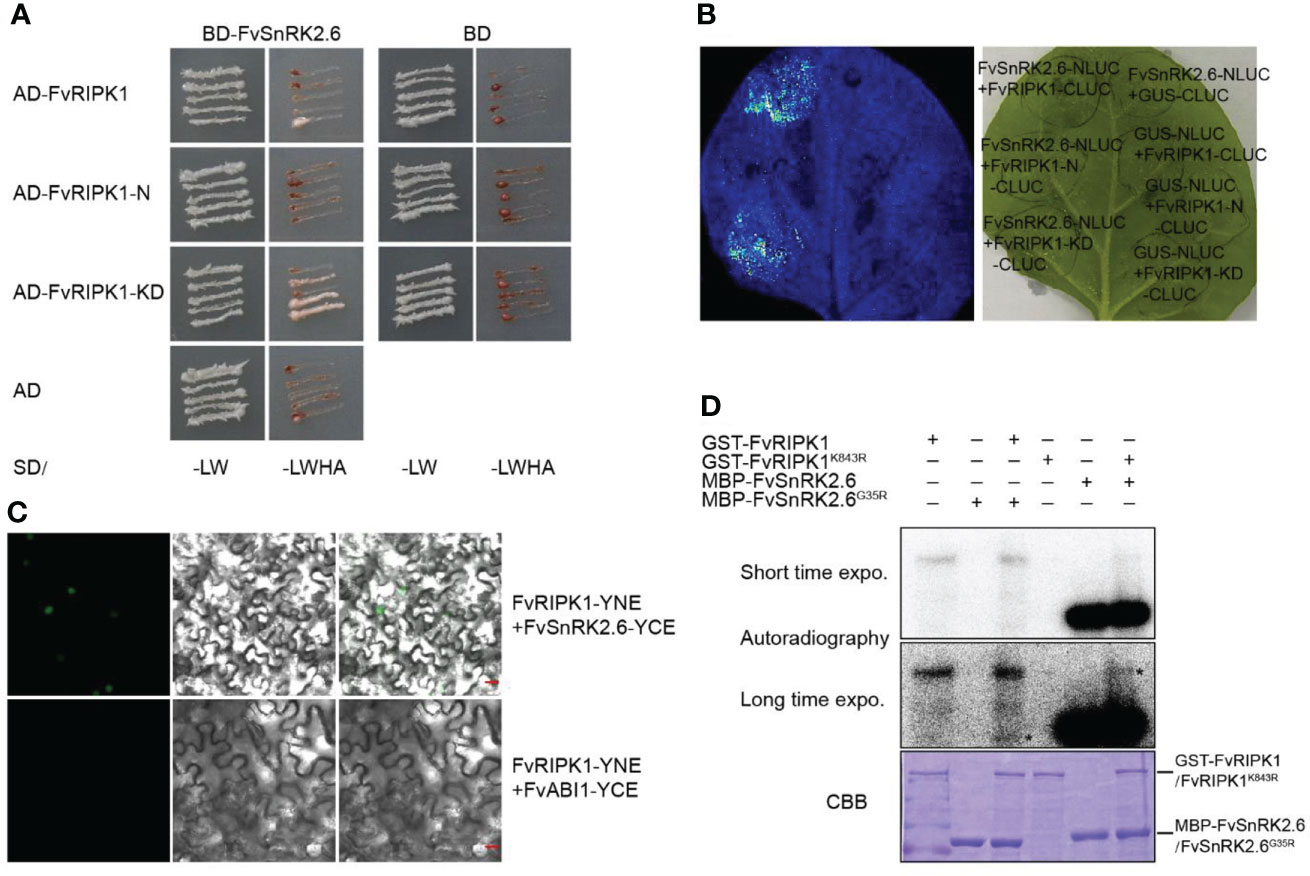
Figure 9 FvRIPK1 interacts with FvSnRK2.6 and phosphorylates each other. (A) The kinase domain (KD) domain of FvRIPK1 interacts with FvSnRK2.6 in the yeast two-hybrid (Y2H) assay. −LW, synthetic dropout medium without Leu and Trp; −LWHA, synthetic dropout medium without Leu, Trp, His, and Ade. (B) The luciferase complementation imaging (LCI) assay shows that FvRIPK1 or FvRIPK1-KD interacts with FvSnRK2.6. (C) The bimolecular fluorescence complementation (BiFC) assay indicates that FvRIPK1 interacts with FvSnRK2.6, not with FvABI1. (D) FvRIPK1 and FvSnRK2.6 phosphorylate each other in vitro phosphorylation assay.
Discussion
First establishment of rapid stable transgenic system by the Agrobacterium-mediated transformation of early germination seeds in strawberry
Strawberry is not only considered as a model plant for the Rosaceae family (Folta and Davis, 2006) but also regarded as an ideal model plant for studying non-climacteric fruit ripening (Li et al., 2011). Thus, in the past years, strawberry transient and stable genetic transformations have been extensively studied (Nehra et al., 1990; El Mansouri et al., 1996; Haymes and Davis, 1997; Balokhina et al., 2000; Jimenez-Bermudez et al., 2002; Agius et al., 2003; Oosumi et al., 2006; Qin et al., 2008; Zhang et al., 2014; Haddadi et al., 2015; Song et al., 2019). However, some limits exist in the present transformation system, and it is necessary to develop a rapid and stable transformation system in strawberry.
Notably, in addition to the widely used Arabidopsis floral dip (Clough and Bent, 1998; Bent, 2006; Zhang et al., 2006), leaf disk (Horsch et al., 1985) and the mature/immature embryos (Potrykus, 1990; Van Eck, 2018), the AMTGS of Arabidopsis and kenaf is ever reported (Feldmann and Marks, 1987; Hanana et al., 2018). In reference to the reports, we have also established a rapidly stable transgenic system in strawberry by the Agrobacterium-mediated transformation of early germination seeds using reporter genes as early (GFP and GUS) and later (DsRed and ABAR) screening as follows: (1) using seeds from ripe fruits; (2) the seeds are required to dry at room temperature; (3) the seeds are required to be fully imbibed; (4) using germination seeds with initial white root tips; (5) shaking inoculation of the initial germination seeds by the Agrobacterium MS infection solution (1.5 of OD600) for 24 h; (6) the infected seeds were darkly cultivated on the MS medium for 3 days at 24°C; (7) the dark culture seeds were transferred onto the MS selection medium to cultivate for 3 days; (8) the 3-day cultured seedlings were early screened by fluorescence; (9) after 2 weeks, the fluorescent seedlings were transferred to quartz sand.
Taken together, our first establishment of the fast (within 4 months), high-efficient (over 10% transformation efficiency), and stable transgenic system by germination seeds in strawberry is a power and useful non-tissue culture approach, which is helpful to promote the study of non-climacteric fruit ripening, strawberry genetic engineering breakthroughs, and molecular breeding development.
The distinct and coordinate role of FvABAR and FvRIPK1 in vegetable growth and ABA signaling in strawberry
In the model plant Arabidopsis, ABAR serves as multiple functions, such as the H subunit of magnesium chelatase, plastid-signaling trigger, anti-sigma factor, circadian clock switch, and putative ABA receptor (Mochizuki et al., 2001; Shen et al., 2006; Legnaioli et al., 2009; Osanai et al., 2009; Wu et al., 2009; Shang et al., 2010; Du et al., 2012; Zhang et al., 2013; Jiang et al., 2014; Liang et al., 2015; Bi et al., 2019). Notably, in recent years, much progress has been made in understanding the role of ABAR in fruit ripening (Jia et al., 2010; Jia et al., 2011; Medina-Puche et al., 2014; Kadomura-Ishikawa et al., 2015; Zhang et al., 2017; Hou et al., 2018). In peach, silencing of PaABAR expression inhibits chlorophyll biosynthesis and fruit ripening (Jia et al., 2010; Jia et al., 2011). In strawberry, silencing of FaABAR expression inhibits fruit ripening (Jia et al., 2011). More recent studies have proved that FaRIPK1 (a leucine-rich repeat receptor-like protein kinase) can interact with FaABAR to regulate strawberry fruit ripening (Hou et al., 2018). However, the distinct and coordinate role of FvABAR and FvRIPK1 in vegetable growth and ABA signaling in strawberry remains unknown.
In this study, using our newly established rapid stable genetic system based on Agrobacterium-mediated transformation of germination seeds, the transgenic FvABAR lines with no significant decrease in chlorophyll contents and with ABA response in stomata movement were selected for the study (Figure 6), demonstrating that FvABAR plays a role in ABA signaling. We further have demonstrated a main role of FvRIPK1 in plant morphogenesis rather than chlorophyll synthesis (Figure 5). Interestingly, the downregulation of the FvABAR expression could not affect the FvRIPK1 expression, but significantly reduced the FvABI1 expression and promoted the FvSnRK2.2 and FvSnRK2.6 expressions (Figure 7), suggesting that FvABAR plays a role in ABA signaling independent of FvRIPK1. The downregulation of the FvRIPK1 expression significantly inhibited the FvABAR expression and also significantly promoted the FvSnRK2.2 and FvSnRK2.6 expressions (Figure 7), suggesting that the role of FvRIPK1 is somehow dependent of FaABAR. Notably, the FvSnRK2.2 and FvSnRK2.6 expressions were both significantly upregulated in the FvABAR- and FvRIPK1-RNAi plants (Figure 7), suggesting that FvSnRK2.2 and FvSnRK2.6 play an important role in coordinate-regulation of ABA signaling. Indeed, a previously report finds a link between ABAR and SnRK2.6/OST1 in guard cell signaling in response to ABA in Arabidopsis (Liang et al., 2015). It is also previously reported that FaSnRK2.6 is a negative regulator of strawberry fruit development (Han et al., 2015). Thus, the higher expression of FvSnRK2.6 in the FvABAR- and FvRIPK1-RNAi promoted a negative ABA signaling. In addition, we found that FvRIPK1 interacts with FvSnRK2.6 in the nucleus, and they phosphorylate each other (Figure 9). But the phosphorylation sites and the regulatory mechanism of the complex FvRIPK1–FvSnRK2.6 in fruit ripening are needed to be identified. Together, the higher expression of FaABI1 in the FvRIPK1-RNAi plants (Figure 7) suggests that FvABI1 plays a distinct role in FvRIPK1-mediated ABA signaling.
In the context, we propose a model for ABA multiple signaling linked by SnRK2.6 in strawberry (Figure 10). ABA is perceived by PYR/PYLs and the putative ABA receptor complex of ABAR/RIPK1. SnRK2.6 is the shared component between the classical “ABA-PYR-PP2Cs-SnRK2.6-ABI4” and the “ABA-ABAR/RIPK1-SnRK2.6/ABI4” signaling pathways. We hypothesize that the two ABA signaling pathways help plants to keep a balance between growth and adaptation to the environment. Given that RIPK1 is essential to plant morphogenesis, this model provided new insights into a molecular mechanism of ABA in plant growth and environment response.
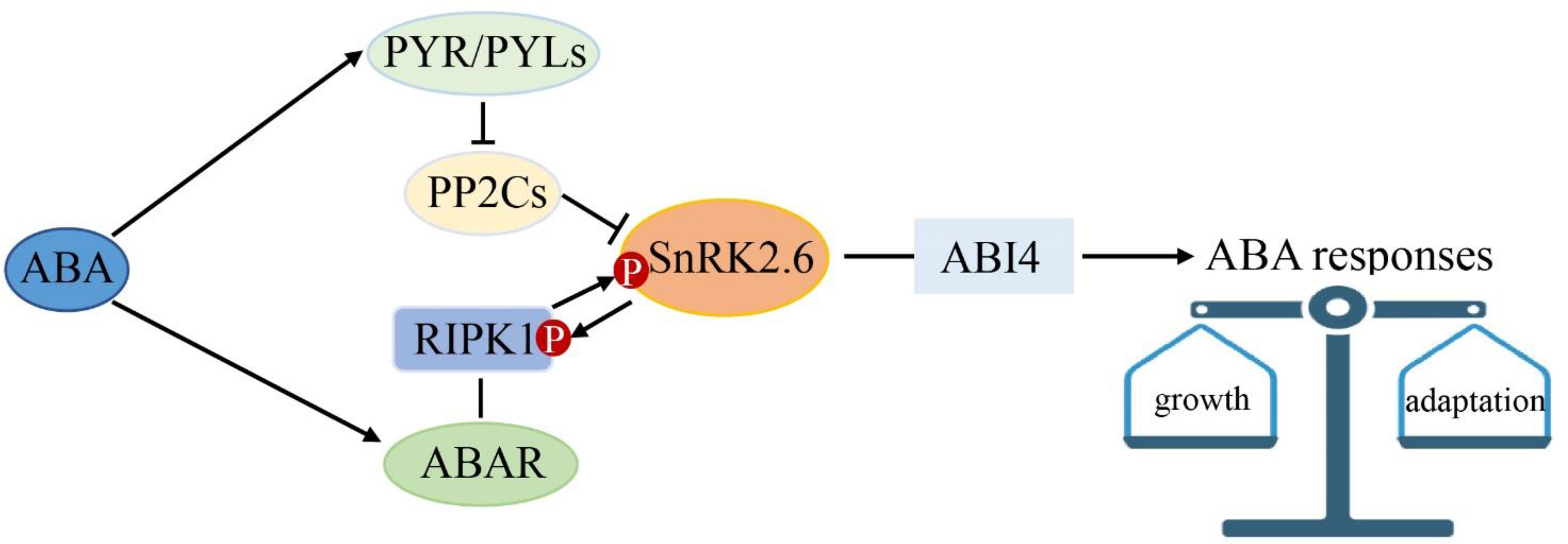
Figure 10 SnRK2.6 is the center crosstalk in multiple abscisic acid (ABA) signaling. ABA is perceived by PYR1 and the ABA receptor complex of ABAR/RIPK1. SnRK2.6 is the shared component between the classical “ABA-PYR1/PYLs-PP2Cs-SnRK2s-ABI4” and the “ABA-ABAR/RIPK1-SnRK2s-ABI4” ABA signaling pathways. This model provided new insights into a molecular mechanism of ABA in plant growth and environment response.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
X-XC, X-JG and FG do experiments, Y-YS and J-XG wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (Projects 32072516 and 32030100) and the Beijing Natural and Science Foundation Scientific Research Key Program of the Beijing Commission of Education (Grant No. KZ202010020028).
Conflict of interest
Author XC was employed by Bei Jing Bei Nong Enterprise Management Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1026571/full#supplementary-material
References
Agius, F., Gonzalez-Lamothe, R., Caballero, J. L., Munoz-Blanco, J., Botella, M. A., Valpuesta, V. (2003). Engineering increased vitamin c levels in plants by overexpression of a d-galacturonic acid reductase. Nat. Biotechnol. 21, 177–181. doi: 10.1038/nbt777
Balokhina, N. V., Kaliayeva, M. A., BuryanovYa, I. (2000). The elaboration of a shoot regeneration system for the genetic transformation of the wild strawberry ( Fragaria vesca l.). Biotekhnologiya 16, 46–51.
Barcelo, M., El-Mansouri, I., Mercado, J. A., Quesada, M. A., Pliego Alfaro, F. (1998). Regeneration and transformation via agrobacterium tumefaciens of the strawberry cultivar chandler. Plant Cell Tiss Org. Cul 54, 29–36. doi: 10.1023/A:1006031527413
Bent, A. (2006). Arabidopsis thaliana floral dip transformation method. Methods Mol. Biol. 343, 87–103. doi: 10.1385/1-59745-130-4:87
Bi, C., Ma, Y., Jiang, S. C., Mei, C., Wang, X. F., Zhang, D. P. (2019). Arabidopsis translation initiation factors eIFiso4G1/2 link repression of mRNA cap-binding complex eIFiso4F assembly with RNA-binding protein SOAR1-mediated ABA signaling. New Phyto 223, 1388–1406. doi: 10.1111/nph.15880
Cappelletti, R., Sabbadini, S., Mezzetti, B. (2015). Strawberry (Fragaria × ananassa). Methods Mol. Biol. 1224, 217–227. doi: 10.1007/978-1-4939-1658-0_18
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Du, S. Y., Zhang, X. F., Lu, Z., Xin, Q., Wu, Z., Jiang, T., et al. (2012). Roles of the different components of magnesium chelatase in abscisic acid signal transduction. Plant Mol. Biol. 80, 519–537. doi: 10.1007/s11103-012-9965-3
El Mansouri, L., Mercadi, J. A., Valpuesta, V., Lopez-Aranda, J. M., Pliegi-Alfaro, F., Quesada, M. A. (1996). Shoot regeneration and Agrobacterium -mediated transformation of Fragaria vesca l. Plant Cell Rep. 15, 642–646. doi: 10.1007/BF00232469
Feldmann, K. A., Marks, M. D. (1987). Agrobacterium-mediated transformation of germinating seeds of Arabidopsis thaliana: a non-tissue culture approach. Mol. Gen. Genet. 208, 1–9. doi: 10.1007/BF00330414
Folta, K. M., Davis, T. M. (2006). Strawberry genes and genomics. Crit. Rev. Plant Sci. 25, 399–415. doi: 10.1007/978-1-4419-0763-9_9
Folta, K. M., Dhingra, A. (2006). Invited review: Transformation of strawberry: The basis for translational genomics in Rosaceae. In vitro cell. Dev. Biol.-Plant 42, 482–490. doi: 10.1079/IVP2006807
Fujii, H., Chinnusamy, V., Rodrigues, A., Rubio, S., Antoni, R., Park, S. Y., et al. (2009). In vitro reconstitution of an abscisic acid signalling pathway. Nature 462, 660–664. doi: 10.1038/nature08599
Gu, X., Gao, Z., Wang, X., Qiao, Y., Chen, Y. (2017). Rdreb1bi, enhances drought tolerance by activating aqp-related genes in transgenic strawberry. Plant Physiol. Biochem. 119, 33–42. doi: 10.1016/j.plaphy
Guidarelli, M., Baraldi, E. (2015). Transient transformation meets gene function discovery: the strawberry fruit case. Front. Plant Sci. 12 6, 444. doi: 10.3389/fpls.2015.00444
Haddadi, F., Aziz, M. A., Abdullah, S. N., Tan, S. G., Kamaladini, H. (2015). An efficient Agrobacterium-mediated transformation of strawberry cv. Camarosa by dual plasmid system. Molecules 20, 3647–3666. doi: 10.3390/molecules20033647
Hanana, M., Ayadi, R., Mzid, R., Khouja, M. L., Hanachi, A. S., Hamrouni, L. (2018). Efficient method of seed transformation via agrobacterium tumefaciens for obtaining transgenic plants of Hibiscus cannabinus l. Ind. Crop Prod 113, 274–282. doi: 10.1016/j.indcrop.2018.01.050
Han, Y., Dang, R., Li, J., Jiang, J., Zhang, N., Jia, M., et al. (2015). SUCROSE NONFERMENTING1-RELATED PROTEIN KINASE2.6, an ortholog of OPEN STOMATA1, is a negative regulator of strawberry fruit development and ripening. Plant Physiol. 167, 915–930. doi: 10.1104/pp.114.251314
Harwood, W. A. (2014). A protocol for high-throughput Agrobacterium-mediated barley transformation. Methods Mol. Biol. 1099, 251–260. doi: 10.1007/978-1-62703-715-0_20
Haymes, K. M., Davis, T. M. (1997). Agrobacterium -mediated transformation of ‘Alpine’ Fragaria vesca and transmission of transgenes to R1 progeny. Plant Cell Rep. 17, 279–283. doi: 10.1007/s002990050392
Hiei, Y., Ohta, S., Komari, T., Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa l.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. doi: 10.1046/j.1365-313X.1994.6020271.x
Hiriart, J. B., Aro, E. M., Lehto, K. (2003). Dynamics of the VIGS-mediated chimeric silencing of the Nicotiana benthamiana ChlH gene and of the tobacco mosaic virus vector. Mol. Plant Microbe Interact. 16, 99–106. doi: 10.1094/MPMI.2003.16.2.99
Hiriart, J. B., Lehto, K., Tyystjärvi, E., Junttila, T., Aro, E. M. (2002). Suppression of a key gene involved in chlorophyll biosynthesis by means of virus-inducing gene silencing. Plant Mol. Biol. 50, 213–224. doi: 10.1023/A:1016000627231
Hoffmann, T., Kalinowski, G., Schwab, W. (2006). RNAi-induced silencing of gene expression in strawberry fruit (Fragaria x ananassa) by agroinfiltration: a rapid assay for gene function analysis. Plant J. 48, 818–826. doi: 10.1111/j.1365-313X.2006.02913.x
Horsch, R. B., Fry, J. E., Hoffmann, N. L., Eichholtz, D., Rogers, S. G., Fraley, R. T. (1985). A simple and general-method for transferring genes into plants. Science 227, 1229–1231. doi: 10.1126/science.227.4691.1229
Hou, B. Z., Xu, C., Shen, Y. Y. (2018). A leu-rich repeat receptor-like protein kinase, FaRIPK1, interacts with the ABA receptor, FaABAR, to regulate fruit ripening in strawberry. J. Exp. Bot. 69, 1569–1582. doi: 10.1093/jxb/erx488
Ishida, Y., Saito, H., Ohta, S., Hiei, Y., Komari, T., Kumashiro, T. (1996). High efficiency transformation of maize (Zea mays l.) mediated by Agrobacterium tumefaciens. Nat. Biotechnol. 14, 745–750. doi: 10.1038/nbt0696-745
Ishida, Y., Tsunashima, M., Hiei, Y., Komari, T. (2014). Wheat (Triticum aestivum l.) transformation using immature embryos Agrobacterium. Methods mol biol 1223, 189–198. doi: 10.1007/978-1-4939-1695-5_15
Jia, H. F., Chai, Y. M., Li, C. L., Qin, L., Shen, Y. Y. (2011). Cloning and characterization of the h subunit of a magnesium chelatase gene (PpCHLH) in peach. J. Plant Growth Regul. 30, 445–455. doi: 10.1007/s00344-011-9207-y
Jia, H. F., Guo, J. X., Qin, L., Shen, Y. Y. (2010). Virus-induced PpCHLH gene silencing in peach leaves (Prunus persica). J. Hortic. Sci. Biotechnol. 85, 528–532. doi: 10.1080/14620316.2010.11512709
Jiang, S. C., Mei, C., Wang, X. F., Zhang, D. P. (2014). A hub for ABA signaling to the nucleus: significance of a cytosolic and nuclear dual-localized PPR protein SOAR1 acting downstream of mg-chelatase h subunit. Plant Signal Behav. 9, .11. doi: 10.4161/15592316.2014.972899
Jimenez-Bermudez, S., Redondo, N. J., Munoz-Blanco, J., Caballero, J. L., Lopez-Aranda, J. M., Valpuesta, V., et al. (2002). Manipulation of strawberry fruit softening by antisense expression of a pectate lyase gene. Plant Physiol. 128, 751–759. doi: 10.1104/pp.010671
Kadomura-Ishikawa, Y., Miyawaki, K., Takahashi, A., Masuda, T., Noji, S. (2015). Light and abscisic acid independently regulated FaMYB10 in Fragaria x ananassa fruit. Planta 241, 953–965. doi: 10.1007/s00425-014-2228-6
Legnaioli, T., Cuevas, J., Mas, P. (2009). TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J. 28, 3745–3757. doi: 10.1038/emboj.2009.297
Liang, S., Lu, K., Wu, Z., Jiang, S. C., Yu, Y. T., Bi, C., et al. (2015). A link between magnesium-chelatase h subunit and sucrose nonfermenting 1 (SNF1)-related protein kinase SnRK2.6/OST1 in arabidopsis guard cell signalling in response to abscisic acid. J. Exp. Bot. 66, 6355–6369. doi: 10.1093/jxb/erv341
Li, C. L., Jia, H. F., Chai, Y. M., Shen, Y. Y. (2011). Abscisic acid perception and signaling transduction in strawberry: A model for non-climacteric fruit ripening. Plant Signal Behav. 6, 1950–11953. doi: 10.4161/psb.6.12.18024
Medina-Puche, L., Cumplido-Laso, G., Amil-Ruiz, F., Hoffmann, T., Ring, L., Rodriguez-Franco, A., et al. (2014). MYB10 plays a major role in the regulation of flavonoid/phenylpropanoid metabolism during ripening of Fragaria x ananassa fruits. J. Exp. Bot. 65, 401–417. doi: 10.1093/jxb/ert377
Miyawaki, K., Fukuoka, S., Kadomura, Y., Hamaoka, H., Mito, T., Ohuchi, H., et al. (2012). Establishment of a novel system to elucidate the mechanisms underlying light-induced ripening of strawberry fruit with an Agrobacterium-mediated RNAi technique. Plant Biotechnol. 29, 271–277. doi: 10.5511/plantbiotechnology.12.0406a
Mochizuki, N., Brusslan, J. A., Larkin, R., Nagatani, A., Chory, J. (2001). Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of mg-chelatase h subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. 98, 2053–2058. doi: 10.1073/pnas.98.4.2053
Nehra, N. S., Chibbar, R. N., Kartha, K. K., Datla, R. S. S., Crosby, W. L., Stushnoff, C. (1990). Genetic transformation of strawberry by Agrobacterium tumefaciens using a leaf disk regeneration system. Plant Cell Rep. 9, 293–298. doi: 10.1007/BF00232854
Oosumi, T., Gruszewski, H. A., Blischak, L. A., Baxter, A. J., Wadl, P. A., Shuman, J. L., et al. (2006). High-efficiency transformation of the diploid strawberry ( Fragaria vesca ) for functional genomics. Planta 223, 1219–1230. doi: 10.1007/s00425-005-0170-3
Osanai, T., Imashimizu, M., Seki, A., Sato, S., Tabata, S., Imamura, S., et al. (2009). ChlH, the h subunit of the mg-chelatase, is an anti-sigma factor for SigE in synechocystis sp. Proc. Natl. Acad. Sci. 106, 6860–6865. doi: 10.1073/pnas.0810040106
Park, S. H., Choi, H., Kim, S., Cho, W. K., Kim, K. H. (2016). Development of virus-induced gene expression and silencing vector derived from grapevine Algerian latent virus. Plant Pathol. J. 32, 371–376. doi: 10.5423/PPJ.NT.11.2015.0237
Pignatta, D., Kumar, P., Turina, M., Dandekar, A., Falk, B. W. (2007). Quantitative analysis of efficient endogenous gene silencing in Nicotiana benthamiana plants using tomato bushy stunt virus vectors that retain the capsid protein gene. Mol. Plant Microbe Interact. 20, 609–618. doi: 10.1094/MPMI-20-6-0609
Potrykus, I. (1990). Gene transfer to cereals: an assessment. Nat. Biotechnol. 8, 535–542. doi: 10.1038/nbt0690-535
Qin, Y., Jaime, A., da Silva, T., Zhang, L., Zhang, S. (2008). Transgenic strawberry: state of art for improved traits. Biotechnol. Adv. 26, 219–232. doi: 10.1016/j.biotechadv.2007.12.004
Ratanasut, K., Rod-In, W., Sujipuli, K. (2017). In planta Agrobacterium-mediated transformation of rice. Rice Sci. 24, 181–186. doi: 10.1016/j.rsci.2016.11.001
Shang, Y., Yan, L., Liu, Z. Q., Cao, Z., Mei, C., Xin, Q., et al. (2010). The mg-chelatase h subunit of arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22, 1909–1935. doi: 10.1105/tpc.110.073874
Shen, Y. Y., Wang, X. F., Wu, F. Q., Du, S. Y., Cao, Z., Shang, Y., et al. (2006). The mg-chelatase h subunit is an abscisic acid receptor. Nature 443, 823–826. doi: 10.1038/nature05176
Song, G. Q., Prieto, H., Orbovic, V. (2019). Agrobacterium-mediated transformation of tree fruit crops: Methods, progress, and challenges. Front. Plant Sci. 10, 226. doi: 10.3389/fpls.2019.00226
Van Eck, J. (2018). The status of setaria viridis transformation: Agrobacterium-mediated to floral dip. Front. Plant Sci. 9, 652. doi: 10.3389/fpls.2018.00652
Waadt, R., Schmidt, L. K., Lohse, M., Hashimoto, K., Bock, R., Kudla, J. (2008). Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56, 505–516. doi: 10.1111/j.1365-313X.2008.03612.x
Wu, F. Q., Xin, Q., Cao, Z., Liu, Z. Q., Du, S. Y., Mei, C., et al. (2009). The magnesium-chelatase h subunit binds abscisic acid and functions in abscisic acid signaling: new evidence in Arabidopsis. . Plant Physiol 150, 1940–1954. doi: 10.1104/pp.109.140731
Zhang, Q., Folta, K. M., Davis, T. M. (2014). Somatic embryogenesis, tetraploidy, and variant leaf morphology in trasgenic diploid strawberry (Fragaria vesca subspecies vesca ‘Hawaii 4’). BMC Plant Biol. 14, 23. doi: 10.1186/1471-2229-14-23
Zhang, X., Henriques, R., Lin, S. S., Niu, Q. W., Chua, N. H. (2006). Agrobacterium-mediated transformation of arabidopsis thaliana using the floral dip method (641–646: Nat. Protoc 1). doi: 10.1038/nprot.2006.97
Zhang, S. H., Hou, B. Z., Chai, L., Yang, A. Z., Yu, X. Y., Shen, Y. Y. (2017). Sigma factor FaSigE positively regulates strawberry fruit ripening by ABA. Plant Growth Regul. 83, 417–427. doi: 10.1007/s10725-017-0308-x
Zhang, X. F., Jiang, T., Wu, Z., Du, S. Y., Yu, Y. T., Jiang, S. C., et al. (2013). Cochaperonin CPN20 negatively regulates abscisic acid signaling in Arabidopsis. Plant Mol. Biol. 83, 205–218. doi: 10.1007/s11103-013-0082-8
Keywords: strawberry, Agrobacterium-mediated transformation of germinating seeds, reporter gene, CHLH/ABAR, RIPK1, ABA, morphogenesis, environmental response
Citation: Chen X-X, Gu X-J, Gao F, Guo J-X and Shen Y-Y (2022) The protein kinase FvRIPK1 regulates plant morphogenesis by ABA signaling using seed genetic transformation in strawberry. Front. Plant Sci. 13:1026571. doi: 10.3389/fpls.2022.1026571
Received: 24 August 2022; Accepted: 11 October 2022;
Published: 31 October 2022.
Edited by:
Zhanwu Dai, Institute of Botany (CAS), ChinaReviewed by:
Bingbing Li, China Agricultural University, ChinaDa-Long Guo, Henan University of Science and Technology, China
Copyright © 2022 Chen, Gu, Gao, Guo and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuanyue Shen, c2ZtbkB0b20uY29t; Jiaxuan Guo, Z3VvamlheHVhbmdqeEAxNjMuY29t
†These authors have contributed equally to this work
 Xuexue Chen
Xuexue Chen Xiaojiao Gu
Xiaojiao Gu Fan Gao
Fan Gao Jiaxuan Guo
Jiaxuan Guo Yuanyue Shen
Yuanyue Shen