
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 06 December 2022
Sec. Plant Development and EvoDevo
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1023628
This article is part of the Research TopicContinuous Flowering: A much needed Ornamental Revolution for Floricultural CropsView all 8 articles
 Kai Zhao1*
Kai Zhao1* Yuzhen Zhou2
Yuzhen Zhou2 Yan Zheng2
Yan Zheng2 Rui-yue Zheng2
Rui-yue Zheng2 Meijuan Hu2
Meijuan Hu2 Yan Tong2
Yan Tong2 Xianmei Luo1
Xianmei Luo1 Yangting Zhang2
Yangting Zhang2 Ming-li Shen1
Ming-li Shen1Prunus mume (Rosaceae, Prunoideae) serves as an excellent ornamental woody plant with a large-temperature-range cultivation scope. Its flower buds require a certain low temperature to achieve flowering circulation. Thus, it is important to delve into the processes of flower bud differentiation and dormancy, which affected its continuous flowering. These processes are generally considered as regulation by the MADS-box homologs, SHORT VEGETATIVE PHASE (SVP), and DORMANCY-ASSOCIATED MADS-BOX (DAM). However, a precise model on their interdependence and specific function, when acting as a complex in the flower development of P. mume, is needed. Therefore, this study highlighted the integral roles of PmDAMs and PmSVPs in flower organ development and dormancy cycle. The segregation of PmDAMs and PmSVPs in a different cluster suggested distinct functions and neofunctionalization. The expression pattern and yeast two-hybrid assays jointly revealed that eight genes were involved in the floral organ development stages, with PmDAM1 and PmDAM5 specifically related to prolificated flower formation. PmSVP1–2 mingled in the protein complex in bud dormancy stages with PmDAMs. Finally, we proposed the hypothesis that PmSVP1 and PmSVP2 could combine with PmDAM1 to have an effect on flower organogenesis and interact with PmDAM5 and PmDAM6 to regulate flower bud dormancy. These findings could help expand the current molecular mechanism based on MADS-box genes during flower bud development and dormancy.
Functional plants always change our routine life in a particular way, people are willing to unfold the mechanism, and a better usage can be applied to them. One significant part is that the modern floriculture industry calls for continuous flowering. Prunus mume (Mei flower) has been grown as woody flowers with over 3,000 years of cultivation history in China. After long-term natural and artificial selection in current breeding, we can enjoy cultivars with full aroma, different petal colors, and different branch types (Zhang et al., 2018). However, as representative woody plants in Rosaceae, Mei flowers experienced seasonal flowering (Zhang et al., 2012); the continuous flowering composed by floral ontogeny, organ differentiation, and bud expansion was stopped during cold-weather days. A defense mechanism is set up to protect the plant from cold damage, called dormancy. In previous research, we found the key roles of DAM genes and raised the complex model (Zhao et al., 2018a). As per our further observation, flower bud differentiation associated with flower shapes was completed in autumn; then, all flower buds stop growing at a certain low temperature. Naturally, it could get a quick release from dormancy and bloom early in spring. These processes which impact the ornamental value of P. mume are supposed to be regulated by the MADS-box gene family. Beyond this, a crosstalk of PmCBFs and PmDAMs on phytohormones was recorded, which highlight the inverse function of gibberellic acid (GA) and abscisic acid in this process (Zhao et al., 2018b). Some studies have shown that two PmSVPs and six PmDAMs belong to the MADS-box gene family, and more members were identified in one to two clades and characterized in P. mume and species in Rosaceae (Falavigna et al., 2018). Fine transcriptomes of P. mume analysis confirm the core co-expression relations to dormancy (Li et al., 2021). Despite the fact that these genes play vital roles in the floral organ development and dormancy cycle (Sasaki et al., 2011), little is known on the functional and structural associations between PmSVPs and PmDAMs.
SVP genes, which belongs to the STMADS11 subfamily of MADS-box gene family, are a crucial flowering repressor in Arabidopsis thaliana (Blazquez and Weigel, 2000). A homologous gene of AtSVP with similar functions has been identified in different herbaceous plants, such as Brassica pekinensis (Lee et al., 2000) and Narcissus tazetta (Li et al., 2015). Moreover, SVP genes were regarded as significantly expanding in Rosaceae, as there are about four times the number of members than those in Brassicaceae (Liu et al., 2020). It implied a more complicated and sensitive control in dormancy. SVP genes are implicated in floral transition and may affect the identity of the floral meristem (Liu et al., 2018). In perennial species, several reports evaluated the function of DAM and SVP-like genes in heterologous systems. In Eucalyptus grandis, the overexpression of SVP-like gene (EgrSVP) resulted in delaying the flowering time and increasing the inflorescence (Brill and Watson, 2004). The ectopic expression of Poncirus trifoliata SVP in wild-type A. thaliana induced some changes such as delayed flowering, additional trichomes, and floral defects (Li et al., 2010). In addition, SVP-like genes have been proposed to regulate dormancy. In P. mume, the overexpression of PmSVP1 and PmSVP2 in A. thaliana resulted in changes of the floral organs, additional trichomes, leaf-like sepals, and increased rosette branches (Li et al., 2017). Six tandemly duplicated PmDAMs share a sequence homology to SVP genes, which appeared to inherit functions from the current PmSVPs. Processing a growth inhibitory effect, the overexpression genes from sweet cherries and Prunus mume displayed a relevant delay of floral organofaction (Li et al., 2017; Wang et al., 2021). Beyond these discoveries, the current omics data of transcriptome and small RNA sequencing displayed the joint roles of SVPs and DAMs in Prunus mume. All members functioned by a network control in floral bud break and dormancy cycling (Zhang et al., 2022).
The DAM genes belong to the MIKCC type of MADS-box gene. The MIKCC type of genes could influence pollen development in A. thaliana (Liu et al., 2013). PpeMADS20 and PpeMADS36 are highly expressed in pollens and play a role in floral organ formation (Wells et al., 2015). The overexpression of EeDAM1 from Euphorbia esula in Arabidopsis slightly delayed the flowering (Horvath et al., 2010). These studies could contribute to explore the role of DAM genes in regulating flower development. Furthermore, DAM genes were initially found in Ever-growing mutant of P. persica and well known as regulators associated with growth and dormancy cycle (Li et al., 2009). The expression of DAM genes of P. avium reached the highest spot during endodormancy, which is compatible with the role as repressors of endodormancy release or bud break (Rothkegel et al., 2017). By comparing the promoter regions of the DAM genes of P. trichocarpa and E. esula, it was found that some expression patterns are critical to the feedback of the stimulus factors that induce dormancy. Current inventions provided evidence that in pear alternative splicing of PpDAM1, especially PpDAM1.2 showed a predominantly higher expression than PpDAM1.1 and PpDAM1.3 in two cultivars and thus could play a crucial role in the pear flower bud dormancy process (Li et al., 2021).
MADS-box proteins can widely form multimeric complexes with other proteins to regulate plant development. In Arabidopsis, SVP proteins form complexes with some MADS-box proteins to regulate flowering time and flower development. SVP and FLC proteins form a complex which delays flowering by inhibiting the expression of the floral integrator genes FT and SOC1 as well as GA-related genes (Xie et al., 2021). PmDAM6, which interacts with PmSOC1, may participate in floral organogenesis, dormancy transition, and flowering time regulation in P. mume (Yong et al., 2021). Nevertheless, PmDAM1, PmDAM5, and PmDAM6 could form homo- and heteromeric complexes and act during the different stages of the dormancy cycle. These studies illustrate the composition of SVP-like and DAM complexes that have particular functions in flower development (Zhao et al., 2018a). Therefore, clarifying the nature of DAM and SVP-like protein complexes may help to better understand the integral function and potential regulation in flower bud differentiation and dormancy.
As a summary of the discussion above, it is interesting to find out the specific relationship between SVPs and DAMs and the neofunctionalization in this subclade. In this study, two PmSVPs and six PmDAMs originated from phylogenetic relationships were further analyzed. The expression patterns of eight genes in flower bud differentiation and dormancy were investigated by qRT-PCR. Furthermore, yeast two-hybrid assays were performed to examine the protein–protein interactions of PmSVPs and PmDAMs. Based on these results, we discussed the individual and combined function of PmSVPs and PmDAMs during flower bud development. A new model for flower organogenesis and dormancy was generated. This study may provide a theoretical basis for the improvement of flower phenotype and the regulation of flowering time in P. mume.
Mei cultivars with different flower types—’Jiangmei’, ‘Sanlun Yudie’, and ‘Subai Taige’—from Jiufeng International Plum Blossom Garden, Beijing, China (40°07′ N, 116°11′ E), were used in this study from July 2015 to February 2016. To analyze the expression pattern of PmSVPs and PmDAMs during flower bud differentiation and dormancy in three cultivars, a sample of each stage was collected every 30 days from July to February. Two samples at eight stages of early floral development with consistent appearance were collected every 57 days: one was immediately frozen in liquid nitrogen for RNA extraction, and the other was preserved in formaldehyde/acetic acid for the performance of paraffin sectioning (displayed in Figures 1, 2 and Supplementary Figure S1). Flower samples of ‘Sanlun Yudie’ planted in the Fujian Normal University were collected to clone PmSVPs and PmDAMs.
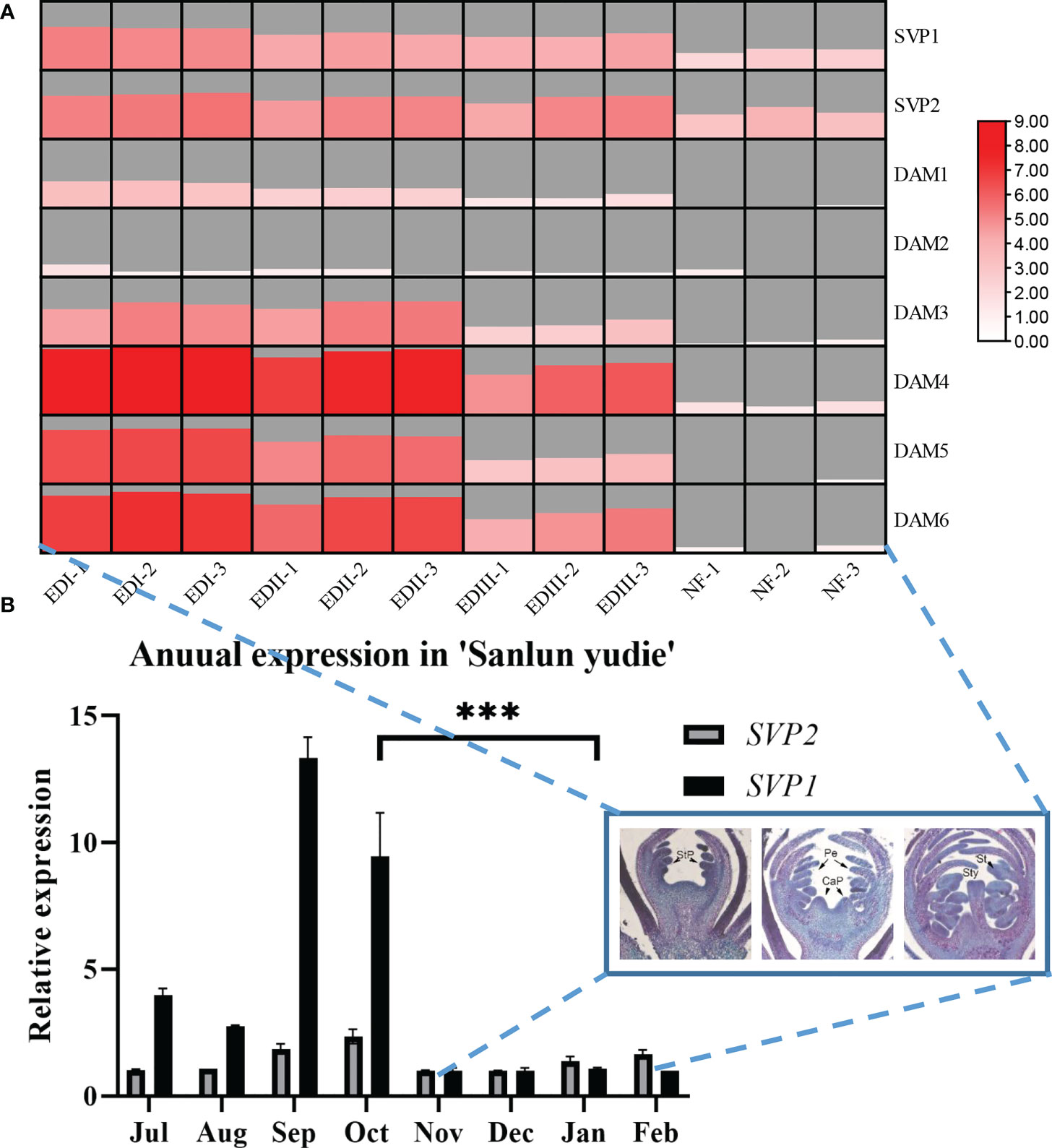
Figure 1 (A) Transcriptome data of PmSVPs and PmDAMs during four dormancy stages from 6 November 2015 to 8 March 2016. In the heat map, the samples considered were listed as follows: November 22 as endodormancy I (EDI), December 14 as endodormancy II (EDII), January 6 as endodormancy III (EDIII), and February 18 as natural flowering (NF). Data were scaled by log2, and the gray area displayed the scale size for each sample. (B) Annual expression patterns of PmSVPs in ‘Sanlun Yudie’ throughout floral growth and dormancy. The asterisks in the figure mean significance between samples.
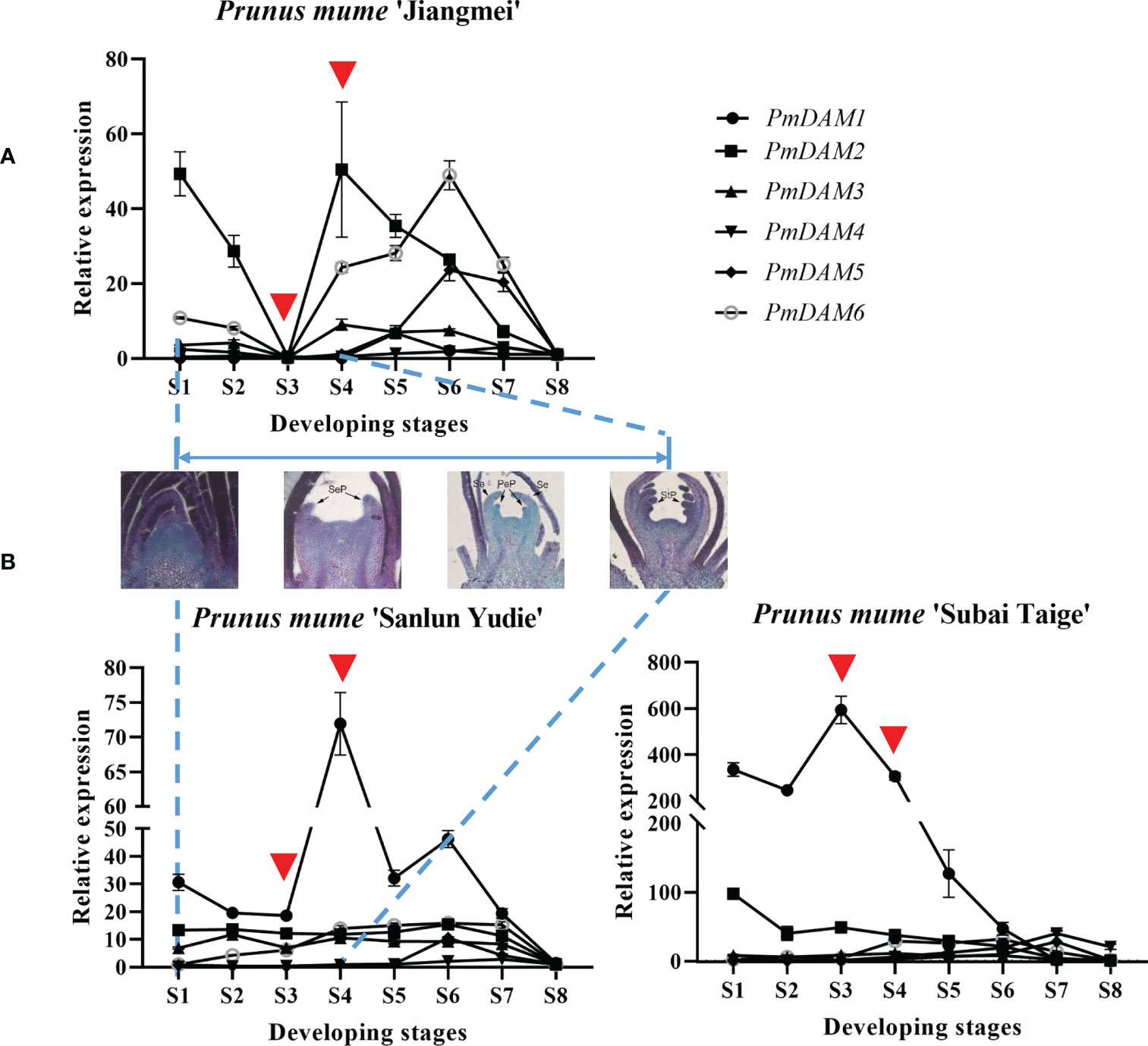
Figure 2 (A, B) Expression patterns of PmDAMs during the flower development of P. mume in the three cultivars. The middle figures show the section of S1–S4 stages. The red triangles located the core expression difference in the first four stages.
Two PmSVPs and six PmDAMs have been identified in previous studies (Li et al., 2017; Zhao et al., 2018a). The total RNA was extracted using EASYspin plus plant RNA rapid extraction kit (RN38 Aidlab, Beijing), following the manufacturer’s directions. Potentially contaminating gDNA was removed using RNase-free DNase I (M6106, Aidlab, Promega). The cDNA was synthesized and purified using Fast Quant RT Kit (KR106, Tiangen, Beijing). Plasmids were extracted by normal methods. The primers and annealing temperature values of PCRs are presented in Supplementary Table S1.
The DNA sequences were extracted from Prunus mume Genome Project (http://prunusmumegenome.bjfu.edu.cn/). The SVP1 (Pm002166) and SVP2 (Pm022002) promoters were obtained from the position of 2,000 bp before the promoter initiation codon. Cis-acting elements were predicted by Promoter 2.0 Prediction Server (https://services.healthtech.dtu.dk/service.php?Promoter-2.0). The details are listed in Supplementary Data 4.
Two PmSVP proteins and six PmDAM proteins were aligned with 28 homologous protein sequences from other plants (eight in P. persica, five in P. pseudocerasus, seven in Pyrus pyrifolia, four in M. domestica, and four in Camellia sinensis) through DNAMAN 7.0 software with default parameters. The GenBank accession numbers of all sequences are shown in Supplementary Data 2. A phylogenetic tree based on the abovementioned proteins and the other 19 type II MADS-box proteins in P. mume (Supplementary Data 3) was constructed by using the maximum-likelihood method of MEGA11 program. The parameters of this tree were set to default, except for the bootstrap values which were set to 1,000.
The expression patterns of PmSVPs and PmDAMs in different flower bud development stages were determined, all by quantitative real-time PCR following previous methods. Primers were designed for cross-introns and ensured gene specificity. The primer amplification efficiency (standard curve) was calculated and guaranteed to be within 95%–105%. The intrinsic PmPP2A gene was used as the internal control, and the relative expression levels were computed using the 2-ΔΔCt method (Ding et al., 2020). All qRT-PCR experiments were performed with three biological duplications, and each duplication was repeated in triplicate. The primers of qRT-PCR are shown in Supplementary Table S2. A comparison between samples and the drawings of relative expressions was conducted by GraphPad Prism; ANOVA was chosen as the statistical approach. The transcriptome data used for the expression were obtained from the dormancy research of P. mume (Zhang et al., 2018). The heat map was drawn by the software of TBtools (Chen et al., 2020).
The cDNA of PmDAMs and PmSVPs was amplified by PCR with gene-specific primers (Supplementary Table S3). These sequences were cloned into the pGBKT7 (bait) vectors and pGADT7 (prey) vectors (Clonetech, United States) at the EcoRI and BamHI sites, respectively, using InFusion HD Cloning Kit System. All baits were tested for autoactivation and toxicity. The yeast two-hybrid assays were performed according to the previous method (Zhao et al., 2018a). The screening for protein–protein interaction was applied in triplicate.
We obtained two SVP (PmSVP1 and PmSVP2) and six DAM genes (PmDAM1, PmDAM2, PmDAM3, PmDAM4, PmDAM5, and PmDAM6) in the P. mume genome to ascertain their phylogenetic positions and sequence differences. PmSVP1–2 had 687- and 672-bp open reading frames (ORFs), respectively, whereas PmDAM1-6 contained 708-, 723-, 708-, 669-, 705-, and 726-bp ORFs. The coding sequences of eight genes were nearly similar, encoding between 222 and 241 amino acids (Supplementary Data 1). Multiple sequence alignment of SVPs and DAMs in different species was accomplished by DNAMAN program. The result showed that two PmSVPs and six PmDAMs proteins presented four major domains (Figure 3). MADS domain was highly conserved at N terminal, whereas K domain was moderately conserved and I domain was less conserved. In addition, eight protein sequences have conservative ERF-associated amphiphilic repression motifs at C terminal. Upon comparison with other homologous proteins, PmSVPs and PmDAMs have a higher homology with the SVPs and DAMs protein sequence of P. percica and P. pseudocerasus in the Rosaceae.
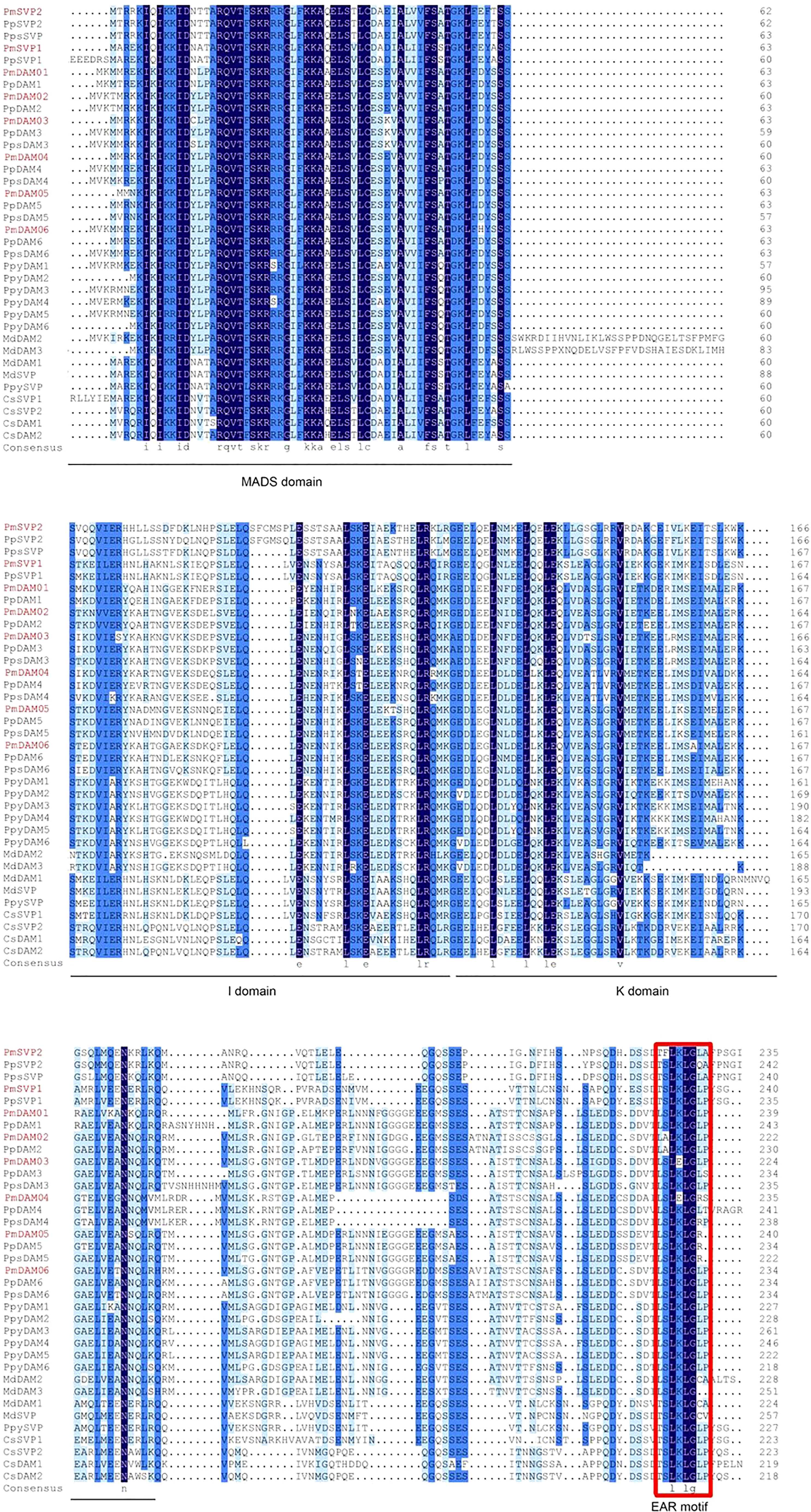
Figure 3 Multiple sequences alignment of DAM and SVP genes from P. mume and other species. The MADS domain, I domain, and K domain are shown by lines at the bottom of the alignment. The ethylene-responsive element-binding factor-associated amphiphilic repression motif is denoted by the red rectangle. The GenBank accession numbers of the genes used in the alignment are shown in Supplementary Data S2.
The phylogenetic tree exposed that two PmSVPs and six PmDAMs belong to the SVP subclade of MADs-box gene family, and they were subdivided into two groups (Figure 4). On one hand, DAM group located in the proximal end of the evolutionary tree. On the other side, in the SVP group, PmSVP1 and PmSVP2 belong to two different branches, clustered with SVP and DAM from other species, respectively. In the far-side group, DAM proteins also gathered and subdivided into two groups. PmDAM4, PmDAM5, and PmDAM6 formed a cluster, while PmDAM1, PmDAM2, and PmDAM3 were congregated in the other, which has been discussed in the previous research (Zhao et al., 2018a). Consistently with the results of multiple sequence alignment, PmSVPs and PmDAMs were clustered with others from P. persica and P. pseudocerasus.
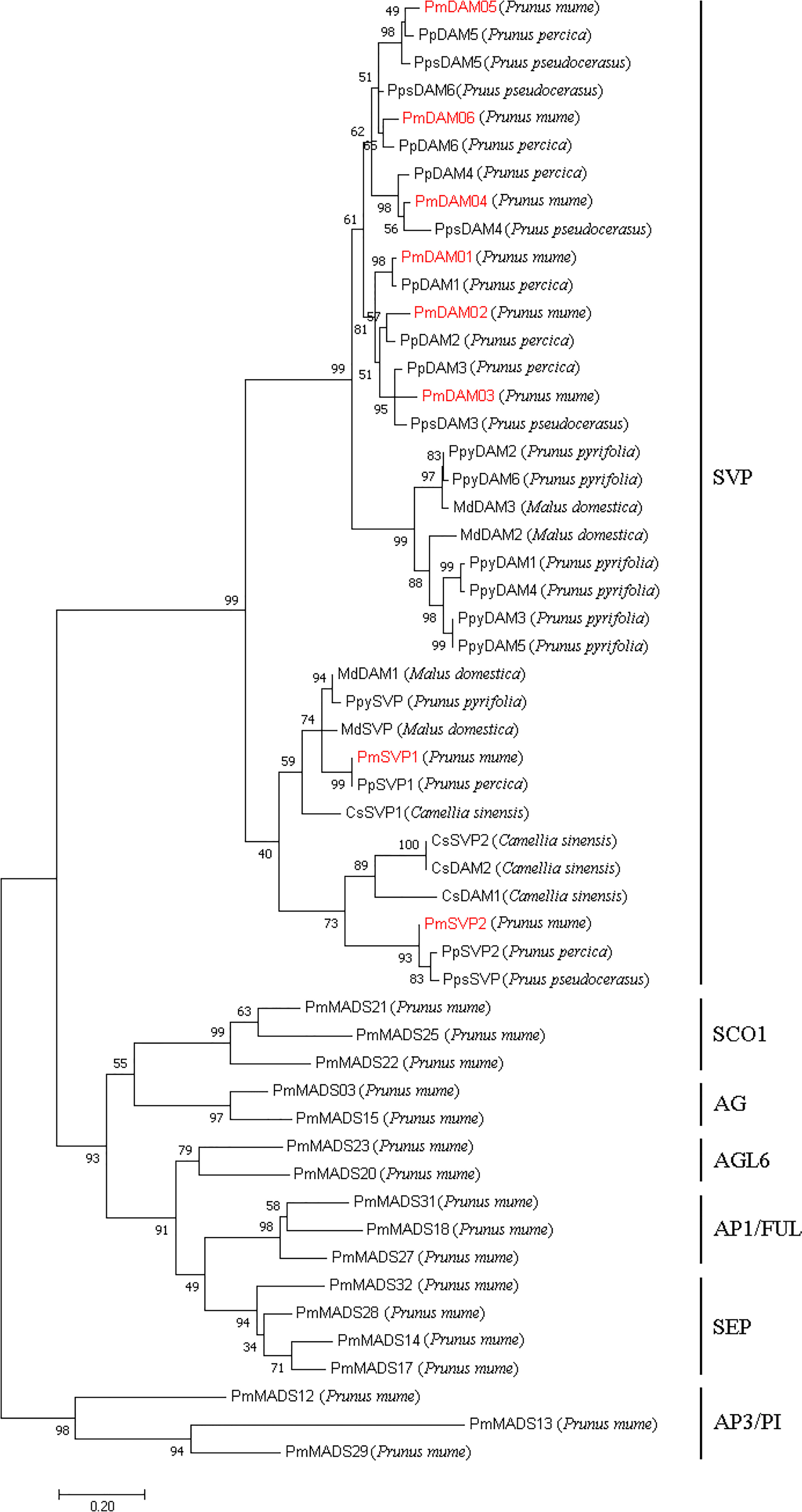
Figure 4 Phylogenetic tree of DAM and SVP proteins and other 17 type II MADS-box proteins in P. mume. The sequences of these proteins are shown in Supplementary Data S3. The types of P. mume MADS-box proteins in different clusters were named according to the phylogenetic analysis of the MADS-box gene family (Xu et al., 2014). The numbers above the branches represent the bootstrap value. DAMs and SVPs in P. mume are colored red.
According to the dormancy cycle of P. mume, dormancy occurs from September to October, and then the plant keeps dormant from November to January, finally breaking its dormancy in February. To explore the role of two PmSVPs and six PmDAMs in flower organ development and dormancy, the expression patterns of the eight genes were investigated by quantitative RT-PCR. From the annual relative expression pattern (Figure 1B), we found that PmSVP1 displayed a rather high rhythm in September and October, and PmSVP2 was expressed higher in the months of July to October. These two genes were active in warm months, and from November to February the expressions were quite low. Combined with the transcriptome data (Figure 1A), in the dormancy stage (EDI–EDIII), DAM gene, especially PmDAM4-6, reflected higher expressions than PmSVP1–2. This suggested that the main working members in the dormancy stage were DAM gene, and we also found that, in this period, the floral bud did little differentiation process (Figure 1B), the section pictures included basal tissues, and most processes finished in the months of July to October.
With the expectation into the development stages, paraffin sections were done to exhibit general flower bud differentiation (Supplementary Figure S1), we obtained eight typical development stages, in which the process of flower bud differentiation lasted from July to November. These included undifferentiated stage (S1), flower primordium differentiation stage (S2), sepal differentiation stage (S3), petal differentiation stage (S4), stamen differentiation stage (S5), pistil differentiation stage (S6), ovary development (S7), and pollen formation (S8). To show the function differences, we quantified eight SVP homologous genes in three cultivars, namely, ‘Jiangmei’, ‘Sanlun Yudie’, and ‘Subai Taige’. These three cultivars own similar characteristics (Figure 5A), except for the petal numbers. Indeed ‘Jiangmei’ has the least petals and ‘Subai Taige’ the most (Figure 5B). This last characteristic was the result of prime flower bud differentiation. PmSVP1–2 were relatively highly expressed in the first seven stages of ‘Sanlun Yudie’ and ‘Subai Taige’ (Figure 5C), but as for ‘Jiangmei’, especially in S3 stage, the expression of PmSVP1–2 was quite low. The said results showed accordance with the petal numbers of the three cultivars.
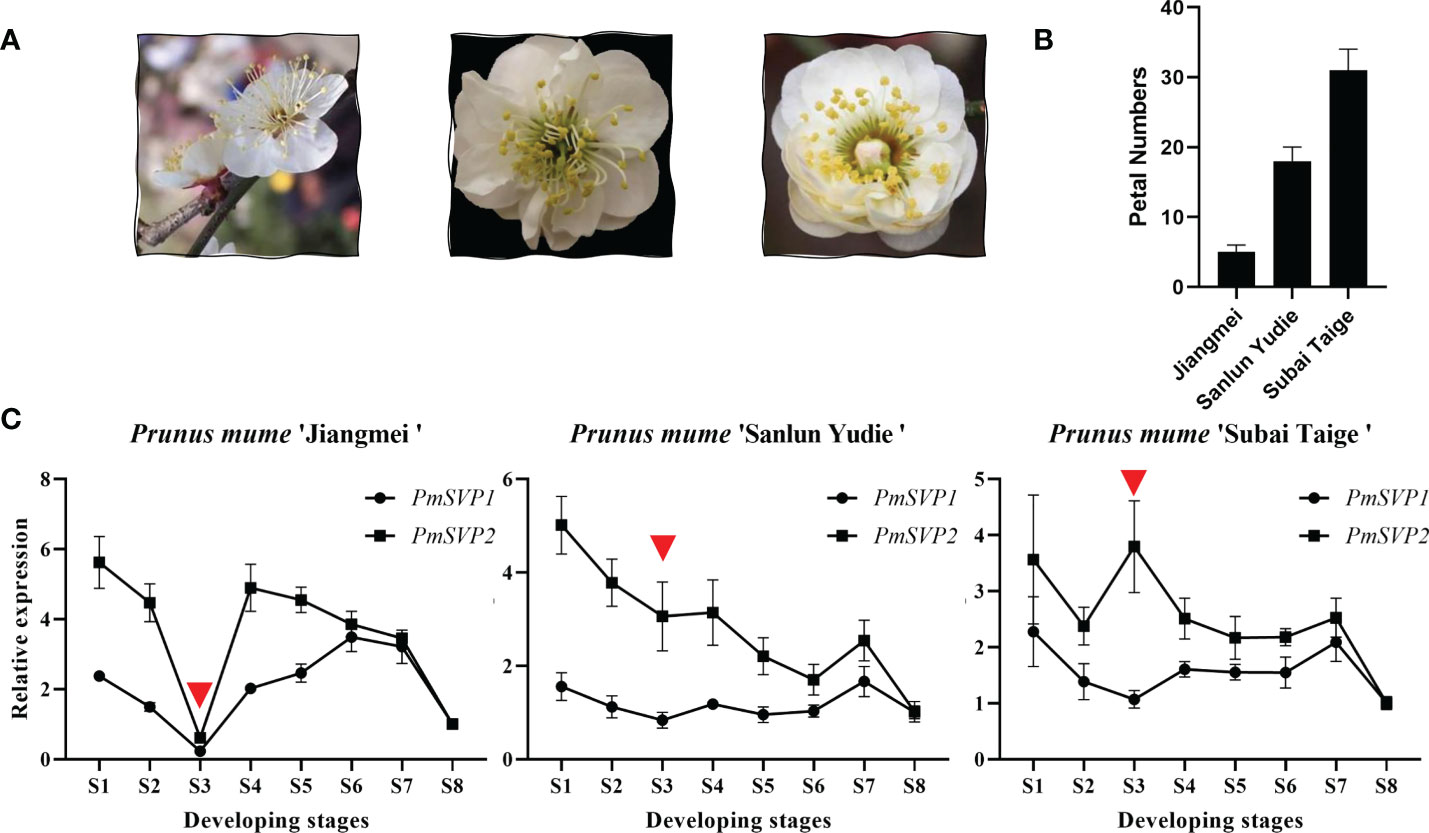
Figure 5 (A) Final shape of the three cultivars. (B) Petal numbers of the three cultivars. (C) Expression patterns of PmSVPs during the flower development of P. mume. Eight development stages of P. mume flower buds: undifferentiation (S1), flower primordium formation (S2), sepal initiation (S3), petal initiation (S4), stamen initiation (S5), pistil initiation (S6), ovule development (S7), and anther development (S8).
This phenomenon also happened to the expression of DAM genes. The expression of PmDAM4–6 in different flower types was relatively low compared with PmDAM1. PmDAM4, PmDAM5, and PmDAM6 showed similar expression profiles from stage 4 to stage 8 (S4–S8). The transcription of PmDAM4 and PmDAM5 was highly expressed in S5-S7, and PmDAM6 had a high expression during S4–S7 (Figure 2B). PmDAM1 performed noticeably well in the stages of S1–S8, and a clinal diversity was observed in S3 and S4 (Figures 2A, B), which is the core stage of petal formation (Figure 2B; S1–S4 paraffin sections). However, in ‘Jiangmei’, PmDAM1 was less expressed with other DAMs in S3 stages. In the garret-like flower type ‘Subai Taige’, PmDAM1–3 were relatively highly expressed in S1–S6, and PmDAM1 reached a peak in S3. It is obvious that the expression levels of PmDAM1 were significantly higher than those of the other genes and had similar patterns with ‘Sanlun Yudie’.
The expression difference between SVPs may be the result of promoter elements. We obtained 2,000 nucleotide sequences and analyzed the binding elements (Figure 6). Some sites like MYB, MYC, and ABRE were found, and a total of 89 elements were displayed as short lines. In the elements, most were TAT-box, six were MYB binding sites, and two were MYC elements. One ABRE recognizing site was found in the promoter of PmSVP1 at the location of 1,296–1,301. This may be the reason that PmSVP1 was expressed higher than PmSVP2 in this research.
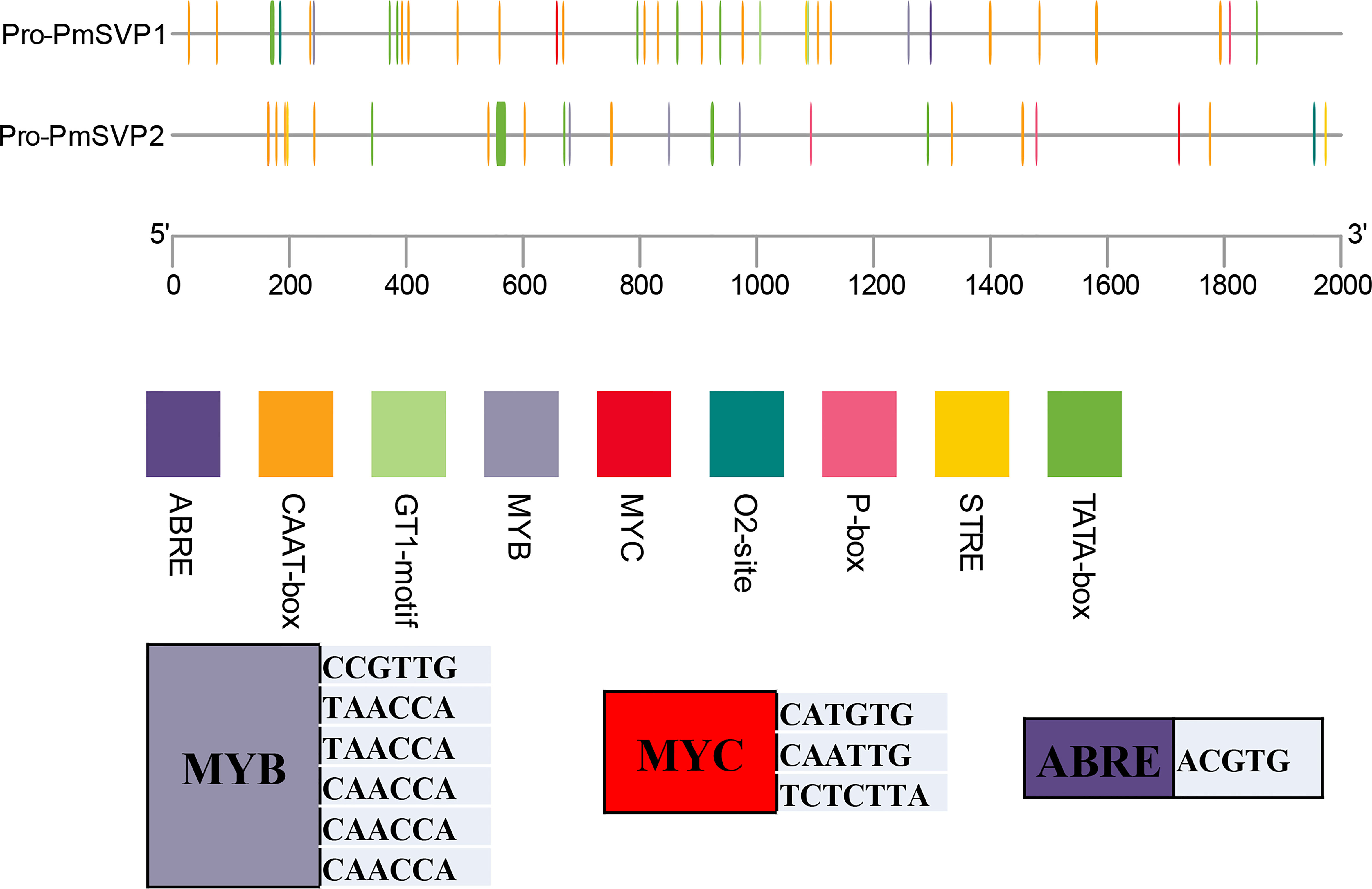
Figure 6 The 2k promoter elements of PmSVPs. Stubs listed from the 5′ end to the 3′ end show the location of each motif as recognized by the Plant Care online program. Colored blocks show the cis elements of different genes. At the bottom is the list of main recognition sequences.
Yeast two-hybrid assays were performed to investigate the interaction relationships between proteins among PmSVPs and PmDAMs. All of the eight bait proteins had no autoactivation activity and toxicity from the results of growth in two-type auxotrophic medium, and the transformed yeast formed a bacterial colony with all four levels of initial concentration. PmSVP1, PmSVP2, PmDAM1, and PmDAM5 could form heterodimers with each other, including strong interactions and weak results (Figure 7). These heterodimers showed unequal interactive capability. PmDAM1 and PmDAM5 could strongly dimerize with PmSVP1. PmSVP1 interacted with PmDAM1 moderately, while the interaction between PmSVP1 and PmDAM5 was weak. The abilities of PmSVP2 to interact with PmDAM1 or PmDAM5 were moderate. The Y2H confirmed the dimerization between each two molecules.
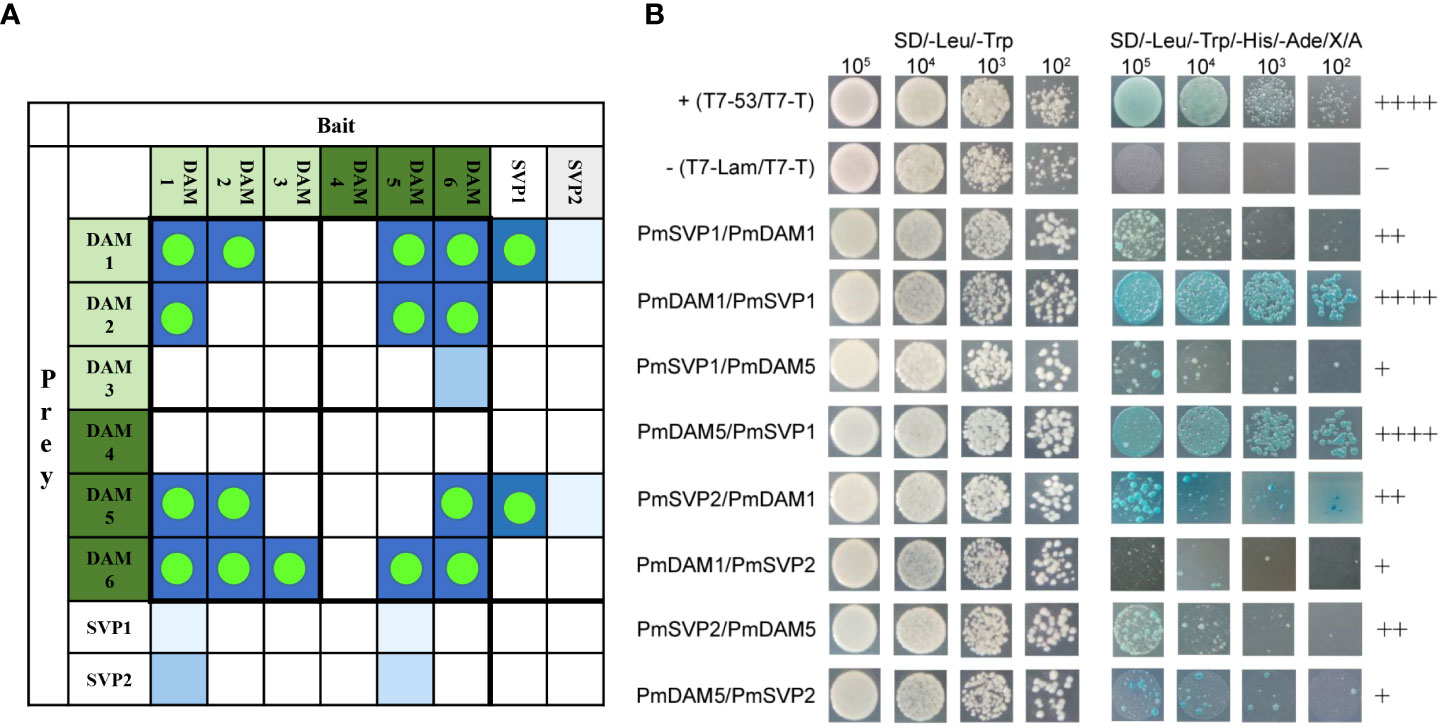
Figure 7 (A) The interaction summary among the DAM and SVP proteins, green dots highlighted the PPI results, blue blocks displayed the probability of the interactions. (B) The detailed yeast two-hybrid assays among PmSVPs and PmDAMs. To determine the interaction model of PmSVPs and PmDAMs, Yeast two-hybrid assays were performed. T7-53/T7-T was positive control, and T7-Lam/T7-T was negative control. The symbol (+) represents the capacity of the reaction. The more numbers of the symbol (+), stronger is the capacity of the reaction.
SVP-like genes, members of MADS-box gene, evolved into multiple paralogs in a wide range of species. In P. mume, there are two SVP genes (PmSVP1 and PmSVP2) and six tandemly duplicated DAM genes (PmDAM1–PmDAM6) found in the genome data. These eight genes are conserved with other SVP genes in Rosaceae, especially showing quite similar structures with SVP-like genes in P. percica. They comprised a characteristic arrangement of four domains. More differences were observed for the C-terminal region, which assign these proteins with binding capacity of different intensity (Smaczniak et al., 2012). Members from P. mume, P. percica, and P. pseudocerasus were congregated; they are reported to undergo their own duplication events in Prunus, which are different from the whole-genome duplication events shared by Malus and Pyrus (Jimenez et al., 2009). Furthermore, SVP genes and DAM genes were separated into two groups. The segregation of PmDAMs and PmSVPs into different clusters indicated that neofunctionalization and subfunctionalization may occur between these genes during the gene duplication events in Prunus. Similar results were presented in peach. The tandem duplication of PmSVPs and its ability to form a protein complex lead to functional diversity during evolution, which may have been beneficial for responding to variable environments to regulate flowering. The SVP-like gene family has been shown to perform diverse functions between different species. In Arabidopsis, SVP and AGL24 are implicated in floral transition and development. SVP genes suppress the flowering process (Yu et al., 2004). Overexpression of PmSVP1 and PmSVP2 in Arabidopsis resulted in floral abnormalities, suggesting similar roles in floral development in P. mume (Li et al., 2017). The PmSVP and PmDAM genes have also been implicated in dormancy regulation. The expression profile could help us to further infer their function during flower development and dormancy. From July to November, two Mei cultivars PmSVP1–2 were relatively highly expressed in the first seven stages, indicating that PmSVP1–2 were likely involved in the whole process of flower bud differentiation. As we have reported in the previous research (Zhao et al., 2018a), the expression levels of PmDAM1–3 were relatively high in the flower bud differentiation process, while PmDAM4–6 have a relatively high expression in the overlapping stage when the flower bud went through differentiation and dormancy process in ‘Subai Taige’. These seasonal expression profiles had a high degree of similarity to those in ‘Sanlun Yudie’ and other plants such as peach and apple. In the early flower bud differentiation of proliferation flower ‘Subai Taige’ and triple-petal flower ‘Sanlun Yudie’, PmDAM1 was significantly detected in S3 and S4, respectively. This suggested that PmDAM1 may be involved in sepal differentiation and petal initiation. As the temperature drops, PmDAM4–6 gradually increased. PmDAM5 was high in S7 in ‘Subai Taige’ and S6 in ‘Sanlun Yudie’. This gene may regulate pistil variation. PmDAM1 and PmDAM5 might be the main reasons for the formation of multiple petals and upper flower. Meanwhile, PmSVP1–2 and PmDAM4–6 were prominently expressed in late flower bud differentiation, the overlapping flower bud differentiation, and the dormancy process.
PPI were considered extremely conserved in basal eudicots, even for the most recent common ancestor of extant angiosperms (Liu et al., 2010; Li et al., 2015). While containing an interaction motif, variation in MADS-box may be a potent driver of floral developmental evolution through protein–protein interactions (Bartlett, 2017). PmSVPs and PmDAMs could form heterodimers with each other in P. mume by yeast two-hybrid assay; a similar result was also recently demonstrated in wheat. TFs could act in protein complexes, and both their activity and the sites to which they bind are likely to be strongly modulated by the presence or absence of other members of the complex (Mateos et al., 2015). Here we captured the functions of PmSVPs and PmDAMs during flower organogenesis and dormancy, especially for PmSVP1 and PmDAM1, combining this research and previous studies. In P. mume, flower bud differentiation begins in July, and PmDAM1 forms a homologous dimer with PmSVP2. During the next stage, PmDAM2 began to accumulate, PmDAM1 may be involved in the flower primordial and sepal petal differentiation by forming a homologous dimer or heterodimer with PmDAM2 and PmSVP2. Then, PmDAM3 and PmDAM6 started to participate in petal and stamen differentiation. PmDAM3 could form dimers with PmDAM6 and PmSVP2, which may mainly regulate the double petals. For pistil and ovary development, also the overlapping of flower bud differentiation and dormancy, PmDAM5 and PmDAM6 act in these stages as the temperature drops. PmDAM5, PmDAM6, and PmSVP1–2 could form a heterodimer to transform plants from flower bud differentiation to dormancy. Lastly, a homodimer of PmDAM6 was in the lead during the dormancy stage. Taken together, with the change of temperature, a dynamic interaction between PmSVPs and PmDAMs could regulate floral development and then switch into dormancy. However, a vexatious problem comes up—the aggregation morphology—whether the SVPs and DAMs functioned as dimers or multimers was still not verified in our experiment. Previous research supplied hints that SVP could form a higher complex due to the flexible C-terminal residues (Wang et al., 2015). This may seriously affect the detailed regulation manners, and in the further research, interactome or Y3H may help deepen our insights into this complex action. Therefore, we raised a hypothesis based on our foundation to describe the roles that PmSVPs and PmDAMs lead to promote tissue differentiation and plant dormancy (Figure 8). After the floral bud appearance, SVP1 gradually accumulated and mingled in the complex by DAM1 and 2 by grabbing DAM1. This led the tissue differentiation to proceed. Between September and October, with the temperature falling, DAM genes began to induce. The presence of SVP1 made it possible to maintain a higher complex, including SVP1, DAM1, DAM5, and DAM6. This helped not to perform the full function of DAM6 and DAM5, but we believed that the protein commixture also slowed down the cell activities. When the temperature finally came to a certain degree, the expressions of SVP were stopped. The disappearance of SVP released the complex. Thus, the domination of DAM6 facilitated the termination of physical activity and the inner situation switch, including dehydration and glucose accumulation, which help plant cells survive in a freezing environment.
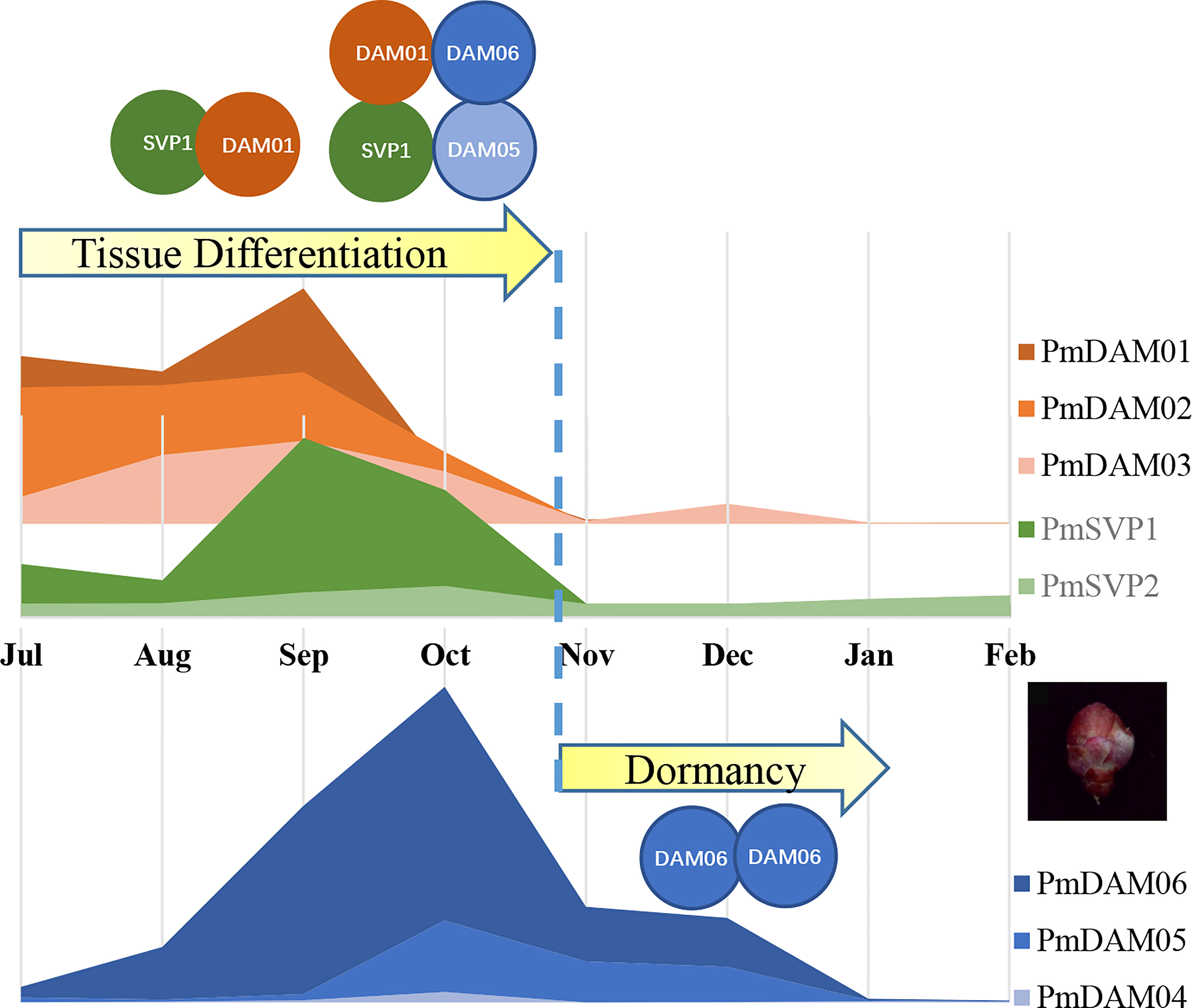
Figure 8 The assumed regulatory model for PmSVPs and PmDAMs in floral tissue differentiation and dormancy. Orange and blue lumps represent the expression changes of DAM 1–6, while green lumps stand for the expression of SVP 1–2. The curves were drawn based on the mean value of relative expression. The collision between circles displays the protein–protein interaction among SVPs and DAMs.
After the investigation of the evolutionary expression profiles and combinatorial activity of PmSVPs and PmDAMs, we rebuild the interaction model, taking a glimpse on their crucial roles in flower development control. Their activities change when they act individually or as a complex. This behavior confers flexibility to the regulatory network to prevent the premature development and growth of flowers during the unfavorable winter periods. This study could provide additional understanding on the comprehensive functions of PmSVPs and PmDAMs in the switch of flower bud development and dormancy, especially illustrating that PmSVP1 promoted floral tissue differentiation by maintaining the complex with PmDAM1. This research could provide a reference for revealing how floral morphological diversity arose and how dormancy regulation was effected.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
KZ and YuZ designed the whole experiments; KZ, YaZ, YuZ wrote the manuscript. YaZ, MH, YT, RZ, MS collected necessary samples; YuZ, XL, YZ, RZ, MS, handle the experiments and analyzed the data. KZ performed the analyses of sequencing data and was responsible for figure compiling and organization. All authors read and approved the final manuscript.
This research was supported by the National Natural Science Foundation of China (no. 31901353), the Natural Science Foundation of Fujian province (no. 2022J01639), and the scientific research innovation program “Xiyuanjiang River Scholarship” from the College of Life Sciences, Fujian Normal University.
We would like to thank the editor and the reviewers for their helpful comments on the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1023628/full#supplementary-material
Bartlett, M. E. (2017). Changing MADS-box transcription factor protein-protein interactions as a mechanism for generating floral morphological diversity. Integr. Comp. Biol. 57 (6), 1312–1321. doi: 10.1093/icb/icx067
Blazquez, M. A., Weigel, D. (2000). Integration of floral inductive signals in arabidopsis. Nature 404 (6780), 889–892. doi: 10.1038/35009125
Brill, E. M., Watson, J. M. (2004). Ectopic expression of a eucalyptus grandis SVP orthologue alters the flowering time of arabidopsis thaliana. Funct. Plant Biol. 31 (3), 217–224. doi: 10.1071/FP03180
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Ding, A., Bao, F., Zhang, T., Yang, W., Wang, J., Cheng, T., et al. (2020). Screening of optimal reference genes for qRT-PCR and preliminary exploration of cold resistance mechanisms in prunus mume and prunus sibirica varieties. Mol. Biol. Rep. 47 (9), 6635–6647. doi: 10.1007/s11033-020-05714-x
Falavigna, V. D. S., Guitton, B., Costes, E., Andres, F. (2018). I Want to (Bud) break free: The potential role of DAM and SVP-like genes in regulating dormancy cycle in temperate fruit trees. Front. Plant Sci. 9, 1990. doi: 10.3389/fpls.2018.01990
Horvath, D. P., Sung, S., Kim, D., Chao, W., Anderson, J. (2010). Characterization, expression and function of DORMANCY ASSOCIATED MADS-BOX genes from leafy spurge. Plant Mol. Biol. 73 (1-2), 169–179. doi: 10.1007/s11103-009-9596-5
Jimenez, S., Lawton-Rauh, A. L., Reighard, G. L., Abbott, A. G., Bielenberg, D. G. (2009). Phylogenetic analysis and molecular evolution of the dormancy associated MADS-box genes from peach. BMC Plant Biol. 9, 81. doi: 10.1186/1471-2229-9-81
Lee, H., Suh, S. S., Park, E., Cho, E., Ahn, J. H., Kim, S. G., et al. (2000). The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in arabidopsis. Genes Dev. 14 (18), 2366–2376. doi: 10.1101/gad.813600
Li, Z. G., Reighard, G. L., Abbott, A. G., Bielenberg, D. G. (2009). Dormancy-associated MADS genes from the EVG locus of peach [Prunus persica (L.) batsch] have distinct seasonal and photoperiodic expression patterns. J. Exp. Bot. 60 (12), 3521–3530. doi: 10.1093/jxb/erp195
Liu, J., Ren, M., Chen, H., Wu, S., Yan, H., Jalal, A., et al. (2020). Evolution of SHORT VEGETATIVE PHASE (SVP) genes in rosaceae: Implications of lineage-specific gene duplication events and function diversifications with respect to their roles in processes other than bud dormancy. Plant Genome 13 (3), e20053. doi: 10.1002/tpg2.20053
Liu, X., Sun, Z., Dong, W., Wang, Z., Zhang, L. (2018). Expansion and functional divergence of the SHORT VEGETATIVE PHASE (SVP) genes in eudicots. Genome Biol. Evol. 10 (11), 3026–3037. doi: 10.1093/gbe/evy235
Liu, C., Teo, Z. W. N., Bi, Y., Song, S. Y., Xi, W. Y., Yang, X. B., et al. (2013). A conserved genetic pathway determines inflorescence architecture in arabidopsis and rice. Dev. Cell 24 (6), 612–622. doi: 10.1016/j.devcel.2013.02.013
Liu, C. J., Zhang, J., Zhang, N., Shan, H. Y., Su, K. M., Zhang, J. S., et al. (2010). Interactions among proteins of floral MADS-box genes in basal eudicots: Implications for evolution of the regulatory network for flower development. Mol. Biol. Evol. 27 (7), 1598–1611. doi: 10.1093/molbev/msq044
Li, X. F., Wu, W. T., Zhang, X. P., Qiu, Y., Zhang, W., Li, R., et al. (2015). Narcissus tazetta SVP-like gene NSVP1 affects flower development in arabidopsis. J. Plant Physiol. 173, 89–96. doi: 10.1016/j.jplph.2014.08.017
Li, J., Yan, X., Ahmad, M., Yu, W., Song, Z., Ni, J., et al. (2021). Alternative splicing of the dormancy-associated MADS-box transcription factor gene PpDAM1 is associated with flower bud dormancy in 'Dangshansu' pear (Pyrus pyrifolia white pear group). Plant Physiol. Biochem. 166, 1096–1108. doi: 10.1016/j.plaphy.2021.07.017
Li, L., Yu, X. X., Guo, C. C., Duan, X. S., Shan, H. Y., Zhang, R., et al. (2015). Interactions among proteins of floral MADS-box genes in nuphar pumila (Nymphaeaceae) and the most recent common ancestor of extant angiosperms help understand the underlying mechanisms of the origin of the flower. J. Systematics Evol. 53 (4), 285–296. doi: 10.1111/jse.12148
Li, Z. M., Zhang, J. Z., Mei, L., Deng, X. X., Hu, C. G., Yao, J. L. (2010). PtSVP, an SVP homolog from trifoliate orange (Poncirus trifoliata l. raf.), shows seasonal periodicity of meristem determination and affects flower development in transgenic arabidopsis and tobacco plants. Plant Mol. Biol. 74 (1-2), 129–142. doi: 10.1007/s11103-010-9660-1
Li, P., Zheng, T., Zhang, Z., Liu, W., Qiu, L., Wang, J., et al. (2021). Integrative identification of crucial genes associated with plant hormone-mediated bud dormancy in prunus mume. Front. Genet. 12, 698598. doi: 10.3389/fgene.2021.698598
Li, Y. S., Zhou, Y. Z., Yang, W. R., Cheng, T. R., Wang, J., Zhang, Q. X. (2017). Isolation and functional characterization of SVP-like genes in prunus mume. Scientia Hortic. 215, 91–101. doi: 10.1016/j.scienta.2016.12.013
Mateos, J. L., Madrigal, P., Tsuda, K., Rawat, V., Richter, R., Romera-Branchat, M., et al. (2015). Combinatorial activities of SHORT VEGETATIVE PHASE and FLOWERING LOCUS c define distinct modes of flowering regulation in arabidopsis. Genome Biol. 16, 31. doi: 10.1186/s13059-015-0597-1
Rothkegel, K., Sanchez, E., Montes, C., Greve, M., Tapia, S., Bravo, S., et al. (2017). DNA Methylation and small interference RNAs participate in the regulation of MADS-box genes involved in dormancy in sweet cherry (Prunus avium l.). Tree Physiol. 37 (12), 1739–1751. doi: 10.1093/treephys/tpx055
Sasaki, R., Yamane, H., Ooka, T., Jotatsu, H., Kitamura, Y., Akagi, T., et al. (2011). Functional and expressional analyses of PmDAM genes associated with endodormancy in Japanese apricot. Plant Physiol. 157 (1), 485–497. doi: 10.1104/pp.111.181982
Smaczniak, C., Immink, R. G. H., Angenent, G. C., Kaufmann, K. (2012). Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development 139 (17), 3081–3098. doi: 10.1242/dev.074674
Wang, J., Hou, C., Huang, J., Wang, Z., Xu, Y. (2015). SVP-like MADS-box protein from carya cathayensis forms higher-order complexes. Plant Physiol. Biochem. 88, 9–16. doi: 10.1016/j.plaphy.2015.01.002
Wang, J., Jiu, S., Xu, Y., Sabir, I. A., Wang, L., Ma, C., et al. (2021). SVP-like gene PavSVP potentially suppressing flowering with PavSEP, PavAP1, and PavJONITLESS in sweet cherries (Prunus avium l.). Plant Physiol. Biochem. 159, 277–284. doi: 10.1016/j.plaphy.2020.12.013
Wells, C. E., Vendramin, E., Tarodo, S. J., Verde, I., Bielenberg, D. G. (2015). A genome-wide analysis of MADS-box genes in peach [Prunus persica (L.) batsch]. BMC Plant Biol. 15, 41. doi: 10.1186/s12870-015-0436-2
Xie, L., Zhang, Y., Wang, K., Luo, X. M., Xu, D. A., Tian, X. L., et al. (2021). TaVrt2, an SVP-like gene, cooperates with TaVrn1 to regulate vernalization-induced flowering in wheat. New Phytol. 231 (2), 834–848. doi: 10.1111/nph.16339
Xu, Z., Zhang, Q., Sun, L., Du, D., Cheng, T., Pan, H., et al. (2014). “Genome-wide identification, characterisation and expression analysis of the MADS-box gene family in Prunus mume.” Molecular genetics and genomics: MGG 289 (5), 903–920. doi: 10.1007/s00438-014-0863-z
Yong, X., Zhou, Y. Z., Zheng, T. C., Zhao, K., Ahmad, S., Li, Y. S., et al. (2021). PmSOC1s and PmDAMs participate in flower bud dormancy of prunus mume by forming protein complexes and responding to ABA. Eur. J. Hortic. Sci. 86 (5), 480–492. doi: 10.17660/eJHS.2021/86.5.4
Yu, H., Ito, T., Wellmer, F., Meyerowitz, E. M. (2004). Repression of AGAMOUS-LIKE 24 is a crucial step in promoting flower development. Nat. Genet. 36 (2), 157–161. doi: 10.1038/ng1286
Zhang, M., Cheng, W. H., Yuan, X., Wang, J., Cheng, T. R., Zhang, Q. X. (2022). Integrated transcriptome and small RNA sequencing in revealing miRNA-mediated regulatory network of floral bud break in prunus mume. Front. Plant Sci. 13, 931454. doi: 10.3389/fpls.2022.931454
Zhang, Q., Chen, W., Sun, L., Zhao, F., Huang, B., Yang, W., et al. (2012). The genome of prunus mume. Nat. Commun. 3, 1318. doi: 10.1038/ncomms2290
Zhang, Q., Zhang, H., Sun, L., Fan, G., Ye, M., Jiang, L., et al. (2018). The genetic architecture of floral traits in the woody plant prunus mume. Nat. Commun. 9 (1), 1702. doi: 10.1038/s41467-018-04093-z
Zhang, Z., Zhuo, X., Zhao, K., Zheng, T., Han, Y., Yuan, C., et al. (2018). Transcriptome profiles reveal the crucial roles of hormone and sugar in the bud dormancy of prunus mume. Sci. Rep. 8 (1), 5090. doi: 10.1038/s41598-018-23108-9
Zhao, K., Zhou, Y., Ahmad, S., Xu, Z., Li, Y., Yang, W., et al. (2018a). Comprehensive cloning of prunus mume dormancy associated MADS-box genes and their response in flower bud development and dormancy. Front. Plant Sci. 9, 17. doi: 10.3389/fpls.2018.00017
Keywords: Prunus mume, SVP gene, DAM gene, floral organ development, dormancy, yeast two-hybrid assays
Citation: Zhao K, Zhou Yz, Zheng Y, Zheng R-y, Hu Mj, Tong Y, Luo Xm, Zhang Yt and Shen M-l (2022) The collaborative mode by PmSVPs and PmDAMs reveals neofunctionalization in the switch of the flower bud development and dormancy for Prunus mume. Front. Plant Sci. 13:1023628. doi: 10.3389/fpls.2022.1023628
Received: 20 August 2022; Accepted: 20 October 2022;
Published: 06 December 2022.
Edited by:
Fengxi Yang, Guangdong Academy of Agricultural Sciences (GDAAS), ChinaReviewed by:
Izhar Hussain, The University of Haripur, PakistanCopyright © 2022 Zhao, Zhou, Zheng, Zheng, Hu, Tong, Luo, Zhang and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kai Zhao, emhhb2thaUBmam51LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.