
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 10 October 2022
Sec. Plant Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1022369
This article is part of the Research Topic Fruit Ripening: From Present Knowledge to Future Development, Volume II View all 9 articles
Fruit ripening is a highly regulated and complex process involving a series of physiological and biochemical changes aiming to maximize fruit organoleptic traits to attract herbivores, maximizing therefore seed dispersal. Furthermore, this process is of key importance for fruit quality and therefore consumer acceptance. In fleshy fruits, ripening involves an alteration in color, in the content of sugars, organic acids and secondary metabolites, such as volatile compounds, which influence flavor and aroma, and the remodeling of cell walls, resulting in the softening of the fruit. The mechanisms underlying these processes rely on the action of phytohormones, transcription factors and epigenetic modifications. Strawberry fruit is considered a model of non-climacteric species, as its ripening is mainly controlled by abscisic acid. Besides the role of phytohormones in the regulation of strawberry fruit ripening, a number of transcription factors have been identified as important regulators of these processes to date. In this review, we present a comprehensive overview of the current knowledge on the role of transcription factors in the regulation of strawberry fruit ripening, as well as in compiling candidate regulators that might play an important role but that have not been functionally studied to date.
Fleshy fruit ripening is an extremely complex process that involves biochemical, physiological and structural changes resulting in fruits more appealing for seed dispersal. Among these changes, fruit ripening involves an alteration in color, in the content of sugars, organic acids and secondary metabolites such as volatile compounds, which influence flavor and aroma, and the remodeling of cell walls, resulting in the softening of the fruit. The regulatory mechanisms underlying fruit ripening rely on the coordinated roles of phytohormones, transcription factors (TFs) and epigenetic modifications, which are in turn regulated by external and internal stimuli (Li et al., 2022b). Those regulatory mechanisms have been mainly studied for climacteric fruit ripening, in which tomato (Solanum lycopersicum) is the model organism. Climacteric ripening is characterized by the requirement of the phytohormone ethylene and a burst in cellular respiration, and many regulators involved in its regulation have been described so far (Klee and Giovannoni, 2011). In contrast, non-climacteric fruit ripening, for which the woodland and cultivated strawberry species (Fragaria vesca and Fragaria × ananassa respectively) have become the model, is not dependent on ethylene or a respiration burst but is mainly regulated by abscisic acid (ABA) (Jia et al., 2011; Bai et al., 2021).
Strawberry is a popular fruit crop thanks to its flavor, aroma and nutritional value, with a huge impact on the agricultural economy of many countries. Furthermore, its extracts are known to produce cytotoxic effects on several human cancer lines (Lucioli et al., 2019) and against ageing progression (Giampieri et al., 2017), therefore benefitting human health. Besides their importance both economically and as a health-promoting fruit, the development of strawberry fruits is also very interesting botanically. Thus, strawberries are achenetum-type fruits whose fleshy part develops from the flower receptacle, while the achenes, the real fruits, are derived from the fertilized carpels and dot the surface of the receptacle (Liu et al., 2020).
Besides the function of different phytohormones in the regulation of strawberry fruit ripening (Symons et al., 2012; Gu et al., 2019), the role of many TFs in strawberry ripening has been studied to date, although a comprehensive compilation of this knowledge is lacking. In this review, we will focus on how strawberry TFs regulate fruit development- and ripening-related processes like hormonal balance, flavonoids biosynthesis, carbohydrates metabolism, volatile production and cell wall modifications, which are summarized in Table 1 and Figure 1.
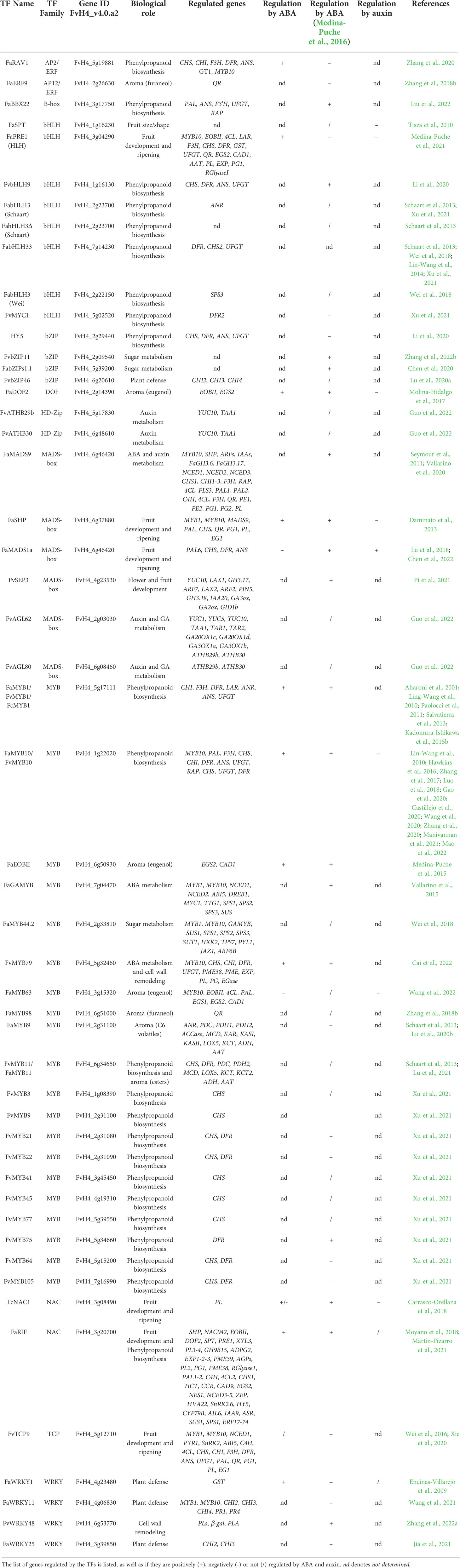
Table 1 List of strawberry fruit development- and ripening-related transcription factors and their biological role.
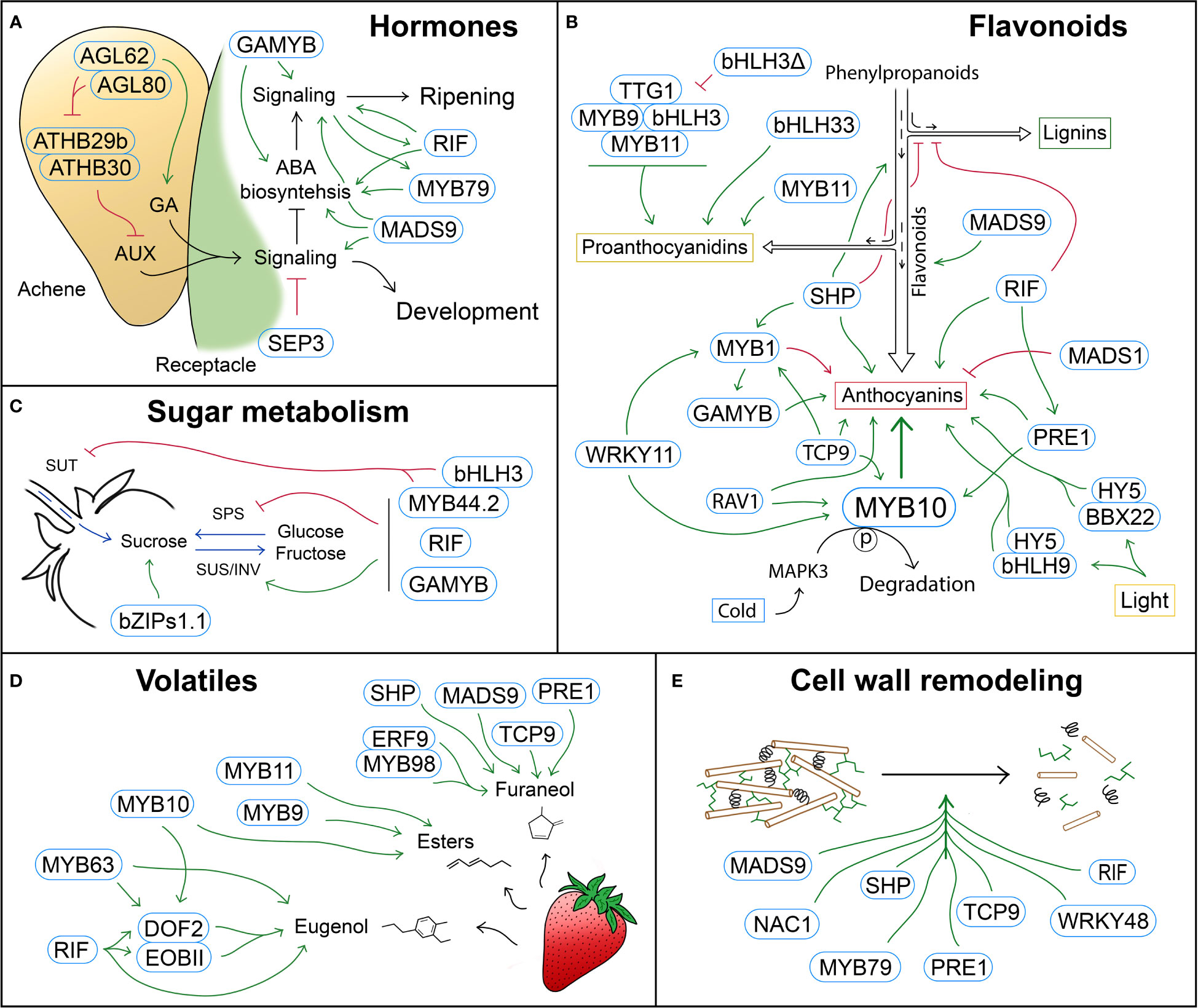
Figure 1 Schematic representation of TFs with a role in different strawberry ripening-related processes. (A) Hormone biosynthesis and signaling are regulated by complex GNRs that govern the switch from the development stages, in which auxin (AUX) and gibberellic acid (GA) promote cell fruit division and expansion, to the ripening stages, which are mainly regulated by abscisic acid (ABA). (B) Regulation of the phenylpropanoid pathway by TFs that control metabolic fluxes and modulate different branches of the pathway, such as the lignin, proanthocyanidin or anthocyanin pathways. (C) TFs involved in the regulation of sugar metabolism (SPS, SUS and INV) and transport (SUT). (D) TFs controlling the biosynthesis of volatile compounds, mainly esters, as well as furaneol and eugenol. (E) TFs regulating cell wall modification, and therefore fruit softening during ripening. Blue rounded rectangles indicate TFs. Green arrows and red block symbols denote positive and negative regulation respectively.
Once ovaries have been fertilized after pollination, auxin and gibberellic acid (GA) biosynthesis is initiated in achenes and transported to the receptacle. It has been recently and elegantly reported the role of the complex of type I MADS-box genes FvAGL62/FvAGL80 promoting auxin biosynthesis in the endosperm of fertilized seeds (Guo et al., 2022). Guo and collaborators also showed that the FvAGL62/FvAGL80-mediated auxin biosynthesis is not direct via activation of auxin biosynthesis genes such as FvYUC10 or FvTAR1 but through the repression of FvATHBs TFs, which negatively regulate those biosynthesis genes at pre-fertilization stages. Furthermore, GA biosynthesis genes were downregulated in fvagl62 mutants, supporting the role of FvAGL62 in the biosynthesis of this hormone (Figure 1A; Guo et al., 2022). The synthesis of both auxin and GAs is maintained during their first developmental stages, where the fruit increases in width and length due to an active cell division and cell expansion (Zhou et al., 2021). In addition, in these initial stages, GA promotes ABA catabolism in the receptacle by activating the expression of the cytochrome P450 monooxygenase FvCYP707A4a, which catalyses ABA’s 8’-hydroxylation, thus preventing the onset of ripening before fruit development is completed (Liao et al., 2018). Later during development, auxin and GA levels decline, as well as FvCYP707A4a expression, allowing ABA accumulation in the receptacle and the transition between fruit growth and ripening stages (Gu et al., 2019). ABA is of great importance since it is a dominant positive regulator of strawberry fruit ripening, as it has an essential role in the synchronization of the central regulatory network controlling all ripening-related processes like alterations of texture, color, sweetness, flavor and aroma (Li et al., 2022a).
The role of other hormones during the regulation of strawberry ripening has also been studied, although their implications are not totally well understood. For example, ethylene is known to regulate the expression of genes involved in cell wall degradation and ripening-related metabolic pathways (Castillejo et al., 2004; Merchante et al., 2013), being likely involved at later stages of the ripening process due to its late accumulation pattern and the expression of genes related to its metabolism and signaling (Gu et al., 2019). However, its role does not seem critical since exogenous treatments with the ethylene inhibitor 1-methylcyclopropane (1-MCP) do not affect the common ripeness traits such as red color, sugar accumulation or acid loss (Reis et al., 2020). Cytokinins (CKs) dramatically increase their levels at the ripe stage due to changes in the expression of genes involved in their biosynthesis and catabolism, possibly implying a role during the last stages of ripening as well (Gu et al., 2019). Salicylic acid (SA), whose roles are typically associated with plant defense, also increases through ripening, although it is not clear its function in the regulation of this process (Kim et al., 2019). Methyl jasmonate (MeJA) accumulates during fruit development and drops through ripening, however, it has been reported an acceleration of the ripening upon its external application (Han et al., 2019). Finally, even though brassinosteroids (BRs) have been implicated in the regulation of ripening in other non-climacteric fruits such as grapes (Symons et al., 2006), their level decreases during strawberry fruit development, suggesting that they might play a role during early stages in strawberries instead (Symons et al., 2012).
The ABA biosynthetic pathway can be regulated at different levels. One of the most important enzymes in the pathway is the 9-cis-epoxycarotenoid dioxygenases (NCEDs), which use 9-cis-violaxanthin and 9′-cis-neoxanthin as substrates to form xanthoxin in a limiting reaction. Other regulatory steps include one of the first reactions in the pathway catalyzed by zeaxanthin epoxidase (ZEP) and the ABA catabolism mediated by CYP707A (Liao et al., 2018). Recently, a NAC TF named Ripening Inducing Factor (FaRIF) has been reported to play a key role in the regulation of strawberry fruit ripening (Martín-Pizarro et al., 2021). In this sense, stable RNAi silencing lines produce fruits with a significant reduction in the ABA content, which is consistent with lower expression levels of FaNCED3, FaNCED5 and FaZEP, as well as other ABA signaling genes, like the ABA-induced genes FaHVA22, FaSnRK2.6 and the ELONGATED HYPOCOTYL5 (FaHY5), a basic leucine zipper (bZIP) TF. Furthermore, the delayed ripening progress of FaRIF-silenced fruits was complemented when treated with exogenous ABA, supporting a positive feedback loop regulatory mechanism between ABA and FaRIF in the control of the ripening process. Besides ABA, the downregulation of FaRIF resulted in alterations in the expressions of genes involved in the metabolism of other hormones, including auxin and ethylene biosynthetic and signaling genes (Martín-Pizarro et al., 2021).
A similar positive regulatory feedback loop has also been reported for ABA and a MYB transcription factor, FvMYB79, which is positively regulated by ABA. Thus, FvMYB79 silencing decreases the ABA content and delays fruit ripening, a phenotype that could not be reverted even after ABA treatment (Cai et al., 2022). Another MYB transcription factor FaGAMYB has been shown to be an important regulator of strawberry fruit ripening, as it induces ABA biosynthesis by regulating NCED1 and NCED2, as well as ABI5 and DREB1, two TFs involved in ABA signaling. Interestingly, the regulation of many ripening-related genes by FaGAMYB is produced in both ABA-dependent and -independent manners, as ABA treatment in transiently FaGAMYB-silenced fruits is able to recover only part of the differentially expressed genes (Vallarino et al., 2015). Other TF with a role in ABA regulation include the TCP gene FvTCP9, whose transient overexpression and silencing result in the accumulation and reduction of the ABA content respectively due to the regulation of the expression of NCED1 and the signaling genes PYR1, SnRK2 and ABI5 (Xie et al., 2020).
Even though studies usually focus on the possible regulatory role of a TF upon ABA, other genes have been reported to regulate other hormones. FvSEP3 is a MADS-box TF involved in flower identity and organogenesis, as it is a member of the ABCE model. Its role during flower and fruit development in strawberry has been studied by the generation of stable EMS mutants and CRISPR/Cas9 lines, which presented sepaloid floral organs instead of petals and stamens, as well as shorter styles compared to those of the WT (Pi et al., 2021). These aberrant flowers could develop into parthenocarpic fruit, supporting a role of SEP genes in repressing fruit growth in strawberry. In this context, a transcriptome analysis showed that FvSEP3 may have a role in the inhibition of the very first steps of fruit development since genes involved in auxin metabolism (FvYUC10, FvGH3.17, and FvGH3.18), transport (FvLAX1 and FvPIN5) and signaling (FvARF7 and FvIAA20) pathways were upregulated in fvsep3 fruits. Furthermore, fvsep3 mutation also resulted in a misregulation of GA pathway genes, such as the biosynthetic FveGA3ox and the catabolic FvGA2ox genes, as well as the receptor FvGID1b (Pi et al., 2021). Besides its role at early stages of fruit development, FvSEP3 expression increases during ripening, suggesting a role in this process. Consistently, fruit ripening is delayed in fvsep3 mutants, although how FvSEP3 regulates this process or its putative interaction with ABA is still unknown.
Another SEPALLATA MADS-box TF, FaMADS9, has also been shown to regulate hormone metabolism and signaling in two independent studies, contributing to strawberry fruit ripening. In the first study, a significant delay in the ripening process was achieved by the silencing of the gene (Seymour et al., 2011), while in the second work, no visible ripening phenotype was observed in their RNAi lines (Vallarino et al., 2020). These differences might be due to the specificity of the silencing. Thus, in the case of Seymour and collaborators, some related MADS-box genes were downregulated in the FaMADS9-RNAi fruits, such as SEP3-, SHATTERPROOF2- and AGL6-like genes. However, in the study of Vallarino and collaborators, only FaMADS9 was downregulated in the FaMADS9-silenced lines out of the 34 MADS-box genes expressed in F. × ananassa fruit, while FaSHP was upregulated. Interestingly, the overexpression and silencing of FaSHP promote an up- and downregulation of FaMADS9, respectively (Daminato et al., 2013), suggesting that these two genes share a feedback regulatory loop. Transcriptome analysis of the stable FaMADS9-silenced fruits at two ripening stages revealed alterations in auxin metabolism and signaling. Specifically, GH3 genes, nine members out of the seventeen AUXIN RESPONSE FACTORS (ARFs) genes expressed in strawberry fruits and seven AUX/IAA, were downregulated in both or any of the ripening stages studied. Interestingly, and despite the normal fruit appearance, FaMADS9-silenced fruits showed a reduction in the ABA content, which correlates with the downregulation of FaNCED1, FaNCED2 and FaNCED3 biosynthetic genes (Vallarino et al., 2020).
Besides the knowledge of how all these different TFs regulate hormone biosynthesis and signaling (Figure 1A), several studies have also reported how some TFs are in turn regulated by different hormones (Table 1) (Encinas-Villarejo et al., 2009; Tisza et al., 2010; Daminato et al., 2013; Medina-Puche et al., 2015; Medina-Puche et al., 2016; Molina-Hidalgo et al., 2017; Carrasco-Orellana et al., 2018; Lu et al., 2018; Moyano et al., 2018; Medina-Puche et al., 2019; Xie et al., 2020; Zhang et al., 2020; Martín-Pizarro et al., 2021; Cai et al., 2022; Wang et al., 2022), highlighting the complex regulatory network between hormones and transcriptional regulators to control strawberry fruit development and ripening.
Many of the benefits that strawberry consumption contributes to human health rely on polyphenols, whose metabolic pathway has been well studied in strawberry (Fait et al., 2008; Muñoz et al., 2011; Urrutia et al., 2015). Polyphenols constitute a structurally and functionally diverse group of compounds produced through the shikimate-phenylpropanoid pathways (Bontpart et al., 2016). The end product of the shikimate pathway, the aromatic amino acid phenylalanine, is the main precursor for the biosynthesis of most polyphenols. The phenylpropanoid pathway uses phenylalanine as the initial substrate which is transformed into 4-coumaroyl-CoA by the sequential activity of the enzymes phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H) and 4-coumarate-CoA ligase (4CL). Then, the 4-coumaroyl-CoA is the branching point of the pathway, being the substrate of the flavonoid or monolignol pathway (Pott et al., 2020). Among all polyphenols, strawberry is especially rich in flavonoids. The first committed step in the flavonoid biosynthetic pathway is catalyzed by the chalcone synthase (CHS), which is followed by the chalcone isomerase (CHI) to produce the flavanone naringenin. This compound is subsequently converted into the different groups of flavonoid compounds, which differ in the degree of oxidation of the three-carbon bridge, and include anthocyanins, the main responsible for the red color of strawberry fruits, and proanthocyanidins (PAs or condensed tannins), their most abundant flavonoid (Buendía et al., 2010; Petrussa et al., 2013). In particular, anthocyanins are synthesized by a series of reactions catalyzed by flavanone 3-hydroxylase (F3H), flavonoid 3’-hydroxylase (F3’H), flavonoid 3’-5’-hydroxylase (F3’5’H), dihydroflavonol 4-reductase (DFR), anthocyanidin synthase/leucoanthocyanidin dioxygenase (ANS/LDOX) and UDP-glucose flavonoid 3-O-glucosyltransferase (UFGT). They are synthesized at the endoplasmic reticulum and later transported into the vacuole for storage mainly by glutathione S-transferases (GSTs) (Luo et al., 2018; Castillejo et al., 2020). Among the anthocyanins, pelargonidin-3-glucoside is the major pigment in strawberry receptacles, while cyanidin-3-glucoside is present in a minor content (Almeida et al., 2007; Zhao et al., 2021). Finally, for the biosynthesis of PAs, two main enzymes are involved, i.e., leucocyanidin reductase (LAR) and anthocyanidin reductase (ANR). Many TFs have been reported to be involved in the regulation of this metabolic pathway (Karlova et al., 2014). Next, we will provide an overview of their role in the biosynthesis of these polyphenolic compounds (Figure 1B).
The regulation of the phenylpropanoid pathway is largely regulated by MBW ternary complexes, which are constituted by the TFs R2R3MYB and bHLH, and a WD-repeat protein. The first TF characterized in strawberry was FaMYB1 and its regulation upon anthocyanin biosynthesis (Aharoni et al., 2001). It was firstly characterized by stable overexpressing lines in Nicotiana benthamiana (Aharoni et al., 2001) and Lotus corniculatus (Paolocci et al., 2011), where it produced a misregulation of the flavonoid biosynthetic pathway. Later, this role was also found in Fragaria chiloensis, a strawberry species that develops white/pinkish receptacles and red achenes at the fully ripe stage of the fruits. Salvatierra and collaborators showed that the white skin phenotype of F. chiloensis receptacles was reverted to red pigmented fruits by the silencing of FcMYB1, which resulted in the repression of flavonoid biosynthetic genes such as CHI, F3H, DFR, LAR and ANR, and the upregulation of ANS and UFGT. Therefore, FcMYB1 silencing is redirecting the precursors of the flavonoid pathway from the PAs branch (LAR and ANR) towards the biosynthesis of anthocyanins (ANS and UFGT), resulting in the accumulation of pelargonidin-3-glucoside (Salvatierra et al., 2013). Similar results supporting the negative regulation of anthocyanins by MYB1 were found in F. × ananassa, since the overexpression of FaMYB1 led to a downregulation of LAR and UFGT and the subsequent reduction in the total anthocyanins content (Kadomura-Ishikawa et al., 2015b).
Afterwards, the most studied TF in strawberry, the key positive regulator of the anthocyanin biosynthesis MYB10, was characterized. The role of MYB10 regulating anthocyanin biosynthesis in strawberry was first reported by stable overexpression in F. × ananassa, which resulted in an increase of the anthocyanins levels in fruits, flowers, leaves and roots (Lin-Wang et al., 2010). After that, a deeper characterization was made by transient silencing FaMYB10 in F. × ananassa (Medina-Puche et al., 2014) and by stable overexpression in F. vesca (Lin-Wang et al., 2014), supporting its role promoting anthocyanin biosynthesis. Thus, MYB10 downregulation produces fruits with white flesh and skin, while overexpression leads to a greater accumulation of pelargonidin-3-glucoside and cyanidin-3-glucoside. These changes are the consequence of an altered expression of many genes related to flavonoid biosynthesis, transport and regulation, like PAL, CHS, CHI, F3H, DFR, ANS, UFGT and RAP/GST1 (GST transporter) (Lin-Wang et al., 2014; Medina-Puche et al., 2014), being the promoters of CHS, UFGT, DFR and MYB10 itself directly regulated by MYB10 (Lin-Wang et al., 2014; Li et al., 2020). However, the relationship between MYB10 and MYB1 needs further analyses. Thus, despite dual-luciferase assays have shown that they could regulate each other (Lin-Wang et al., 2014), no significant differences in gene expression for MYB10 have been observed after MYB1 overexpression/silencing and vice versa (Medina-Puche et al., 2014). Remarkably, despite the importance of MYB10 in the regulation of anthocyanin biosynthesis, no further information about the direct target genes of this TF has been clarified yet, besides those previously mentioned genes.
Different genome-scale DNA analyses have identified allelic variation in MYB10 as the main cause producing skin and flesh color variations in different species of Fragaria genus in nature (Hawkins et al., 2016; Castillejo et al., 2020; Manivannan et al., 2021). In particular, four different polymorphisms in MYB10 have been described explaining the lack of anthocyanin in fruits from different accessions of F. vesca so far: (1) a G35C (W12S) SNP (Hawkins et al., 2016; Zhang et al., 2017), (2) an insertion at the third exon (LTR retrotransposon), (3) a single nucleotide insertion at position 329, and (4) a large deletion in chromosome 1 that removes FvMYB10 (Castillejo et al., 2020). In F. × ananassa cv Camarosa, its main dominant homoelog is FaMYB10-2, which is also the allele with the highest genetic variation. In this case, the genotypes that present red-fleshed fruits have a large transposon insertion (~23kb) in the FaMYB10-2 promoter that induces a higher expression of MYB10, probably contributing with some putative regulatory cis-elements to the promoter that were identified, such as ABA-, MeJA- and sugar-responsive elements, as well as MYB binding motifs and enhancers (Castillejo et al., 2020). In the same study, a new allele of MYB10 was identified in F. chiloensis that also produces a premature stop codon due to an 8-bp insertion, generating a truncated version of the protein that lacks 54 amino acids at the C-terminal domain and that correlated with all white-fruited genotypes analyzed. The same polymorphism has been also described in the white-fruited F. × ananassa cv Snow Princess, which impairs the interaction of this truncated version of MYB10 with the WD40-repeat protein FaTTG1 (Wang et al., 2020). Interestingly, all white-fruited genotypes found in nature that have been characterized so far presented allelic variations in MYB10, supporting a convergent/parallel evolutionary mechanism to control anthocyanin biosynthesis since mutations in other regulators that lead to a general lack of flavonoids/anthocyanins in the plant could be detrimental due to their biological role against different stresses (Luo et al., 2018; Castillejo et al., 2020).
Besides MYB1 and MYB10, other MYB TFs involved in the regulation of the phenylpropanoid pathway have been characterized or proposed as candidate regulators. Among them, FvMYB79 transient overexpression and silencing up- and downregulate the expression of MYB10, CHS, CHI, DFR and UFGT (Cai et al., 2022). Another MYB TF, FaMYB63, which is related to the biosynthesis of phenylpropanoid-derived volatiles as we will discuss later, is also able to directly regulate MYB10 expression in dual-luciferase assays (Wang et al., 2022). Moreover, several bHLH TFs have been involved in this pathway to date. Among them, the stable silencing of bHLH33 produced fruits with no phenotypic effect regarding anthocyanin accumulation in F. vesca, probably due to redundancy with other bHLH TFs. However, it has been shown that FvbHLH33 strongly induces FvMYB10 regulatory activity upon DFR and UFGT promoters in transient transactivation assays, and that this activation could be reduced when FvMYB1 was co-transformed (Lin-Wang et al., 2014). Its homeolog in F. × ananassa (FabHLH33), described as a candidate for the regulation of PAs biosynthesis (Schaart et al., 2013), is able to physically interact with many members of the MYB family considered as either PA or anthocyanin biosynthesis regulators (Xu et al., 2021), as well as with FaTTG1 (Schaart et al., 2013). Another bHLH-like gene involved in this pathway is PACLOBUTRAZOL RESISTANCE 1 (FaPRE1), an atypical HLH transcription regulator characterized by the absence of the basic domain with DNA-binding activity (Medina-Puche et al., 2019). Thus, FaPRE1 transient silencing leads to a downregulation of FaMYB10 and several genes of the phenylpropanoid pathway, i.e. CHS, F3H, DFR, a putative anthocyanidin 3’-O-beta-glucosyltransferase (3´GT) and RAP/GST1, probably through the regulation of FaMYB10 (Medina-Puche et al., 2019). Consistent with these changes in gene expression, stable overexpression of FaPRE1 increased the content of anthocyanins in leaves and petioles (Medina-Puche et al., 2021).
Other MBW-forming proteins reported to play a role in the phenylpropanoid pathway include the complex constituted by the paralogs FaMYB9 and FaMYB11, and FabHLH3 (ortholog to F. vesca FvH4_2g23700) and FaTTG1, which orthologs in Arabidopsis, i.e. AtTT2, AtTT8 and AtTTG1, respectively, physically interact and regulates PA biosynthesis (Walker et al., 1999; Nesi et al., 2000; Nesi et al., 2001; Schaart et al., 2013; Xu et al., 2021). In strawberry, the expression of FaMYB9 and FaMYB11 highly correlates with those of F3’H, ANS, ANR and LAR, as well as with the total PAs content in unripe fruits (Schaart et al., 2013). This study also suggests other TFs, i.e FaMYB5, the truncated version of FabHLH3, FabHLH3Δ and FaMYC1 as regulators of PA biosynthesis. Thus, interaction assays showed that FaMYB5, whose ortholog in Arabidopsis has an inhibitory role in the regulation of PAs biosynthesis, is also able to interact with FabHLH3. Similarly, the truncated FabHLH3Δ protein might have a negative regulatory role competing with other functional bHLHs in its interaction with FaMYB1, FaMYB5, FaMYB9 and FaMYB11. Finally, FaMYC1 might be also involved in PAs biosynthesis as its gene expression correlates with PAs content (Xu et al., 2021) and it interacts with FaTTG1 and FabHLH33 (Schaart et al., 2013). Some of these phenylpropanoid/PAs-related genes are also regulated by FaGAMYB. Thus, FaMYB10, FaMYB1 expression is downregulated in FaGAMYB-RNAi fruits, while FaMYC1 and FaTTG1 are upregulated, which is also accompanied by a secondary metabolic profile characteristic of early stages of fruit ripening, i.e., lower anthocyanins (pelargonidin-3-glucoside and cyanidin-3-glucoside) and hydroxycinnamic acid derivatives, and higher PAs contents (Vallarino et al., 2015).
More components of the regulatory MBW complexes have been recently identified in the woodland strawberry. Among them, FvMYB3, FvMYB21, FvMYB22, FvMYB45, FvMYB64, FvMYB77 and FvMYB105 have been proposed as putative regulators of PAs biosynthesis, while FvMYB41 was associated with anthocyanin accumulation based on their phylogenetical relationship with MYB TFs involved in flavonoid biosynthesis in other species and the correlation of their expression with the accumulation of these compounds during fruit development and ripening (Xu et al., 2021). Protein interaction and transactivation assays demonstrated that several of the identified MYB TFs interact with bHLH proteins and promote the expression of CHS2 and DFR2 as well as PAs biosynthesis. In particular, FvMYB3, FvMYB9, FvMYB11 FvMYB21, FvMYB22, FvMYB41, FvMYB64, FvMYB75 and FvMYB105 were able to form complexes with FvbHLH3 (FvH4_2g23700), FvbHLH33, and FvMYC1. Furthermore, all those MYBs, as well as FvMYB45 and FvMYB77, but with the exception of FvMYB75, could bind to the CHS2 promoter. In the case of DFR2 promoter, FvMYB11 and FvMYB21 could bind it by themselves; FvMYB10 and FvMYB75 could bind it although only in the presence of FvMYC1; and FvMYB22, FvMYB64 and FvMYB105 also required the presence of coregulators, in particular FvbHLH3, FvbHLH33, or FvMYC1. Moreover, when these three MYBs, FvMYB22, FvMYB64 or FvMYB105, were transiently co-expressed with FvbHLH33, they also induced CHS2 and DFR2 gene expression and PAs accumulation in strawberry fruits (Xu et al., 2021). All this data supports an extraordinarily complex regulation of the phenylpropanoid pathway, in which a high number of TFs may form heterocomplexes to fine-tune the biosynthesis of polyphenolic compounds.
As previously mentioned, FaRIF constitutes a major regulator of strawberry fruit ripening, playing a key role in phenylpropanoid biosynthesis. Thus, FaRIF regulates many structural genes like PAL1, PAL2, C4H, 4CL2, CHS1, CHI2, F3H, ANS, UFGT1 and UFGT2, as well as other anthocyanin-related regulators such as PRE1. Remarkably, transcriptomic data in FaRIF-silenced lines showed that FaRIF downregulation produces a drift in the metabolic flux from the flavonoid branch to the monolignol pathway, resulting in a reduction in the anthocyanin content, but an increase in the precursors of lignin biosynthesis, such as coumaric acid and the hexose derivatives of the coumaric, caffeic and ferulic acids. Consequently, these changes produce a higher accumulation of lignin content in the fruit receptacle (Martín-Pizarro et al., 2021).
MADS-box TFs are also involved in the synthesis of polyphenolic compounds. Hence, FaMADS9 silencing results in an alteration in their content. In particular, green fruits showed a lower content of ellagitannins, galloyl, quercetin and kaempferol derivatives, and an increase levels of PAs compared to the control. At the red stages, the pelargonidin derivative compounds were increased, while the cyanidin derivatives were reduced, probably due to the downregulation of F3’H in white receptacles (Vallarino et al., 2020). Other phenylpropanoid-related genes differentially expressed in FaMADS9-RNAi lines included CHS1, CHI1, CHI3, F3H, MYB10, RAP and other three GST transporters, which were upregulated at the white stage. Furthermore, PAL1, PAL2, C4H, 4CL, F3H (Vallarino et al., 2020) and CHS (Seymour et al., 2011) were downregulated at the ripe stage. Other MADS-box, FaSHP, positively regulates the anthocyanin biosynthesis since transient FaSHP-silenced fruits present a downregulation in the expression of PAL, CHS, MYB1 MYB10 and MADS9, resulting in a decrease in the anthocyanin content, and higher levels of unripe-characteristics compounds such as caffeic acid derivatives and ellagitannins (Daminato et al., 2013). Besides these MADS-box regulators that play a positive role in the regulation of the flavonoid pathway, a negative regulator has also been identified. In particular, FaMADS1a expression decreases during ripening and its overexpression results in a delayed ripening and a reduction in the content of anthocyanin due to the downregulation of structural genes such as FaPAL6, FaCHS, FaDFR and FaANS (Lu et al., 2018). It has been recently reported the role of miR5290 in the regulation of FaMADS1a. Thus, miR5290 expression, which is induced during ripening by ABA, represses FaMADS1a, thus releasing its negative regulatory effect on anthocyanin biosynthesis (Chen et al., 2022).
Other families of TFs have also been shown to regulate strawberry fruit color. FaRAV1 is an AP2/ERF TF, which transient overexpression promotes anthocyanin accumulation due to the promotion of CHS, CHI, F3H, DFR, ANS and GT1 expression both directly and indirectly, as it is also a direct positive regulator of MYB10 (Zhang et al., 2020). Another TF, FvTCP9, not only regulates ABA biosynthesis, as previously discussed, but also promotes anthocyanin biosynthesis, supported by transient overexpression and silencing assays that resulted in the upregulation and downregulation respectively of structural genes such as C4H, 4CL, CHS, CHI, F3H, DFR, ANS, UFGT and the TFs MYB1 and MYB10 (Xie et al., 2020). Furthermore, FvTCP9 interacts with FvMYC1, so it has been proposed that FvTCP9 might regulate this metabolic pathway in a direct way as a partner of FvMYC1 and indirectly promoting ABA biosynthesis (Xie et al., 2020).
Environmental conditions such as light and temperature are known to modulate flavonoid biosynthesis in fruits (Takos et al., 2006; Kadomura-Ishikawa et al., 2015a; Xu et al., 2018; Zhang et al., 2018a), and several regulators of these processes have been described recently. Among them, it has been reported the role of the bZIP TF FvHY5, and FvbHLH9 in the regulation of anthocyanin biosynthesis in response to light (Li et al., 2020). Thus, both TFs are induced by light in strawberries (Xu et al., 2018) and their transient overexpression promote anthocyanin biosynthesis, interacting to form an heterodimeric complex that directly binds to the promoters of CHS and DFR (Li et al., 2020). In a similar way, the B-Box TF FaBBX22 is able to form heterodimers with FaHY5 and regulate PAL, ANS, F3’H, UFGT and RAP gene expression in a light-dependent manner, resulting in a higher accumulation of anthocyanins (Liu et al., 2022).
Low temperatures result in a reduction in the anthocyanin content in strawberry. It has been recently reported the role of MITOGEN-ACTIVATED PROTEIN KINASE3 (FvMAPK3), which mediates the response to low temperature phosphorylating MYB10 and CHS proteins. This modification results in a reduction of MYB10 transcriptional activity and the proteasome-mediated degradation of CHS1, therefore negatively regulating the anthocyanin biosynthesis (Mao et al., 2022).
The role of flavonoids has been well described regarding plant defense (Shah and Smith, 2020), so usually biosynthetic genes from this pathway are analyzed when studying plant response against pathogens. In this context, FaWRKY11, which is a positive regulator of fruit resistance against Botrytis cinerea, can also promote the expression of MYB1 and MYB10, although fruits in which this TF were transiently overexpressed or silenced did not display any evident color phenotype (Wang et al., 2021). In a similar way, FaWRKY1, another positive regulator of plant resistance, is able to induce GST in A. thaliana (Encinas-Villarejo et al., 2009), while FaWRKY25 is a negative regulator of CHI2 and CHI3 (Jia et al., 2021). Besides the role of these WRKY TF, a bZIP-like protein, FvbZIP46 has also been shown to positively induce fruit resistance against B. cinerea and positively regulate CHI2, CHI3 and CHI4 gene expression (Lu et al., 2020a). Although these genes are known to contribute to plant defense, the effect of misregulating FvbZIP46 on anthocyanin production has not been reported yet.
Sugar content in strawberry fruits is one of the most important traits as it is the main determinant of consumer preferences (Yan et al., 2018). It does not only affect the sweetness perception, but the ratios between sugars and organic acids play an important role in the final flavor (Fait et al., 2008; Fan et al., 2021). Glucose, fructose and sucrose are the main soluble sugars in strawberry, although sucrose is the sugar with the highest increase during ripening (Fait et al., 2008). Sucrose biosynthesis starts in the cytosol with the combination of fructose 6-phosphate and UDP-glucose to form sucrose 6-phosphate by the sucrose phosphate synthase (SPS). In the next step sucrose 6-phosphate is dephosphorylated by sucrose phosphate phosphatase (SPP) to form sucrose, the principal sugar transported from photosynthetic to sink tissues. Inside sink cells, sucrose can be hydrolyzed either to glucose and fructose by invertases in an irreversible reaction, or to fructose and UDP-glucose by sucrose synthase (SUS), which catalyses a reversible reaction instead (Stein and Granot, 2019). Moreover, sugars are known to be important regulators of many processes as they present signaling properties, including strawberry fruit ripening (Jia et al., 2013; Jia et al., 2016).
Among the TFs described to play a role in the regulation of sugar metabolism in strawberry fruits (Figure 1C), FaGAMYB has been shown to promote sugar biosynthesis. Thus, FaGAMYB-silenced fruits displayed a decreased sucrose content compared with the control, consistent with the downregulation of FaSPS1, FaSPS2 and FaSPS3, and the upregulation of FaSUS (Vallarino et al., 2015). Another MYB TF, FaMYB44.2 has been shown to play an important negative role. Its transient overexpression leads to a decrease in the content of glucose, fructose and sucrose, which is explained by the altered expression of genes related to sucrose biosynthesis, degradation and transport, such as FaSUS1, FaSPS1/2/3 and FaSUT1. Besides, other genes that can alter sucrose metabolism indirectly, like hexokinase 2 (FaHXK2) and trehalose-6-phosphate synthase 7 (FaTPS7) were also differentially expressed. Furthermore, FaMYB44.2 directly interacts with FaSPS3, FaSUS1 and FaHXK2 promoters and forms protein complexes with other members of its family, i.e., FaMYB44.1, FaMYB44.3, as well as with phenylpropanoid-related TFs such as FaMYB1, FabHLH3 (FvH4_2g22150, which is different to the previously named FabHLH3), FabHLH33 and FaTTG1. In the current proposed model, FaMYB44.2 and FabHLH3 form a complex that negatively regulates FaSPS3 expression and therefore sucrose biosynthesis. However, Wei and collaborators also showed that this repression is impaired by FaMYB10, which competes for FabHLH3 binding during ripening (Wei et al., 2018). Despite the role in the regulation of sugar metabolism, the misregulation of FaMYB44.2 also results in an alteration of different organic acids and volatile compounds as well as the expression of genes involved in other ripening-related processes, such as anthocyanin biosynthesis (FaMYB1 and FaMYB10), hormone signaling (FaPYL1, FaJAZ1 or FaARF6B), and general ripening regulation (FaGAMYB).
FaRIF was also reported to be involved in sugar metabolism (Martín-Pizarro et al., 2021). Thus FaRIF-downregulated fruits were also affected in their sugar metabolism, producing a higher accumulation of glucose and fructose, and a reduction in the sucrose content, which is supported by the downregulation of FaSPS1 and the upregulation of FaSUS1. Furthermore, genes encoding glycolytic and fermentation enzymes are downregulated by FaRIF, supporting an important role in the regulation of the aerobic/anaerobic balance that changes during strawberry fruit ripening (Wang et al., 2017; Martín-Pizarro et al., 2021). Finally, FaMADS9 also positively promotes sugar accumulation during ripening since RNAi fruits for this TF showed a reduction of Brix content as the consequence of a reduced content of sucrose, glucose and fructose (Vallarino et al., 2020). Furthermore, this study showed that FaMADS9 regulates starch degradation, which is important during strawberry fruit development (Souleyre et al., 2004). Thus, the levels of maltose and isomaltose, disaccharides produced by the hydrolysis of starch, and some amylases-encoding enzymes were altered in FaMADS9-RNAi fruits, being therefore disrupted in these fruits the degradation of starch to fuel the fruit growth and ripening (Vallarino et al., 2020).
Finally, two members of the bZIP TF family have been reported to positively regulate sugar accumulation. Hence, the heterologous overexpression of FvbZIP11 in tomato fruits produced a higher content of total soluble solids and sugars (Zhang et al., 2022b) while transiently overexpressing FabZIPs1.1 in strawberry fruits induced a greater accumulation of sucrose (Chen et al., 2020).
Volatile organic compounds (VOCs) influence strawberry flavor and aroma, essential traits for fruit quality. More than 360 VOCs have been identified in strawberry fruit, but it is generally considered that only a part of them are able to influence the organoleptic properties. These compounds constitute a diverse group, including esters, aldehydes, alcohols, ketones, terpenes, furanones and alkanes. During strawberry ripening most acids, esters, furan, ketones, lactones and terpenes increase their levels, while most alcohols, aldehydes, alkanes, and furanones exhibit a decrease (Lu et al., 2020b). Among all groups, esters are the most represented compounds in ripe fruits (Song and Forney, 2008), ranging from 25% to 90% of total volatiles, and contributing as the main source of fruity and floral odors (Yan et al., 2018) and with a sweetness-enhancing ability (Fan et al., 2021). In contrast, C6 aldehydes have been identified as the major compounds in immature fruits (Song and Forney, 2008).
Many volatile compounds, including esters, are derived from fatty acids (FAs) through the LOX pathway, which starts with the transformation of linoleic (18:2) and linolenic (18:3) acids into their hydroperoxide derivatives by β-oxidation carried out by lipoxygenases (LOXs) (Lu et al., 2022). Hydroperoxide isomers can be further metabolized to aldehydes by hydroperoxide lyase (HPL), which in turn are reduced by alcohol dehydrogenase (ADH) to form alcohols, that can finally be the substrate of alcohol acyl transferase (AAT) to produce esters (Lu et al., 2021).
Besides the role regulating PAs content, the paralogs FaMYB9 and FaMYB11 also regulate the content of volatile C6 and esters compounds respectively (Lu et al., 2020; Lu et al., 2021). Thus, FaMYB9, whose expression decreases during the ripening process, is a positive regulator of C6 volatiles since its silencing leads to a general reduction of these compounds (Lu et al., 2020). Specifically, FaMYB9 regulates the content in hexanal and (E)-2-hexenal, which can contribute to the grassy flavor of unripe fruits (Larsen and Poll, 1990; Du et al., 2011), and in methyl isovalerate, which contributes to fruity notes (Alstrup et al., 2020). Lu and collaborators also found that FaMYB9 protein physically interacts with FaLOX5, regulating as well its expression and that of other LOX pathway genes. Similarly, FaMYB11 also induce FaLOX5 expression by directly binding to its promoter. Furthermore, FaMYB11 also promotes the expression of FaADH and FaAAT among other genes of the FA biosynthetic pathway. Therefore, FaMYB11 transient up- and downregulation modify the volatile composition, mainly in the content of aldehydes and esters (Lu et al., 2021). FvMYB10 is also able to regulate esters production. Hence, stable FvMYB10 overexpression lines have been shown to produce fruits with a higher content of the esters butyl and hexyl acetate, which level contributes to consumer preferences (Klee and Tieman, 2018). MYB10 overexpression also leads to increased levels of ethyl butanoate, but a reduction in the content of octyl acetate. In contrast, FvMYB10-silenced fruits only showed a significant increase of 2-heptanone, which also contributes to flavor and consumer preferences (Lin-Wang et al., 2014; Klee and Tieman, 2018).
In strawberry, furanone-derived volatiles are mainly represented by 4-hydroxy-2,5-dimethyl-3(2H)-furanone (HDMF, furaneol) and 2,5-dimethyl-4-methoxy-3(2)H-furanone (DMMF, mesifurane), which are considered to generate a caramel-like aroma (Larsen and Poll, 1990) and contributes to the sweetness of the fruit (Fan et al., 2021). Although the biosynthetic pathway has not yet been totally elucidated, 4-hydroxy-5-methyl-2-methylene-3(2H)-furanone (HMMF) has been identified as the immediate precursor of HDMF, catabolized by the enzyme quinone oxidoreductase (QR). In this context, a MYB TF, FaMYB98, has been identified in a yeast-one-hybrid screening as a direct regulator of FaQR transcription. Moreover, it has been found that FaQR expression synergistically increases when FaMYB98 forms a complex with the Ethylene-Response Factor protein FaERF9, which depends on the former to promote FaQR transcription and the biosynthesis of furaneol (Zhang et al., 2018b). Furthermore, other ripening-related TFs have been found to regulate FaQR expression. Thus, FaQR is downregulated when FaMADS9, FaSHP, FaPRE1 and FvTCP9 are silenced, supporting a positive role of these regulators on the biosynthesis of furaneol (Seymour et al., 2011; Daminato et al., 2013; Medina-Puche et al., 2019; Xie et al., 2020).
Eugenol is another important volatile compound contributing to strawberry fruit aroma, and it derives from the phenylpropanoid pathway (Medina-Puche et al., 2015). Eugenol biosynthesis starts with the transformation of feruloyl-CoA into coniferyl aldehyde by the cinnamoyl-CoA reductase (CCR). Coniferyl aldehyde is then reduced by the cinnamyl alcohol dehydrogenase (CAD) to generate coniferyl alcohol, which is converted to coniferyl acetate by a coniferyl alcohol acetyltransferase (CAAT), and finally transformed to eugenol by the Eugenol Synthase (EGS) (Rastogi et al., 2013). Several TFs have been identified as regulators of eugenol biosynthesis. Among them, the MYB TF FaEOBII positively regulates eugenol production, as its silencing produces a reduction of its content due to the downregulation of the structural genes FaCAD1 and FaEGS2. A transactivation assay also showed that FaEOBII directly binds to FvCAD1 promoter. Interestingly, FaEOBII is in turn positively regulated by FaMYB10 (Lin-Wang et al., 2014; Medina-Puche et al., 2015). Thus, FaMYB10 also contributes to the regulation of the branch of the phenylpropanoid pathway responsible for the biosynthesis of these volatile compounds since, besides FaEOBII, it regulates the expression of CCR and CAD genes (Medina-Puche et al., 2014). Another MYB-like TF involved in eugenol biosynthesis is FaMYB63, which is able not only to directly regulate structural genes of the pathway, i.e., FaCAD1, FaEGS1, and FaEGS2, but also indirectly positively regulating FaMYB10 and FaEOBII expression (Wang et al., 2022).
Another eugenol-related TF is FaDOF2, a protein belonging to the plant-specific DOF (DNA binding one zinc finger) family. FaDOF2 positively regulates FaEOBII and FaEGS2 gene expression, probably by direct binding to their promoters, as they present several binding sites that can be recognized by FaDOF2 (Molina-Hidalgo et al., 2017). Interestingly, FaDOF2 and FaEOBII constitute a positive feedback loop, since FaDOF2 expression is also positively modulated by FaEOBII. Furthermore, FaDOF2 interacts with FaEOBII generating a complex that fine-tunes the expression of genes involved in eugenol production. FaPRE1 has also been described to positively regulate the expression of the regulator FaEOBII as well as that of the structural genes FaCAD1 and FaEGS2 and two alcohol acyl transferases involved in esters biosynthesis (FaAAT1-2) (Medina-Puche et al., 2019). Finally, FaRIF is also involved in this pathway, since it promotes the expression of eugenol-related genes, both in a direct way by inducing the FaEGS2 expression, and indirectly through the activation of FaEOBII and FaDOF2 (Martín-Pizarro et al., 2021). Furthermore, FaRIF has also been suggested as a regulator for the production of the terpenic volatile compounds linalool and nerolidol, since the responsible gene for their biosynthesis, NEROLIDOL SYNTHASE1 (FaNES1), was downregulated in FaRIF silenced fruits (Martín-Pizarro et al., 2021).
In summary, the biosynthesis of different aroma-related compounds is regulated by a complex gene regulatory network (GNR) involving different types of TFs (Figure 1D).
Fruit softening is a complex process that includes cell wall disassembly and degradation of the middle lamella, leading to a lower cell-to-cell adhesion. The primary cell wall is mainly formed by a cellulose microfibril network, a glycan matrix, and a pectin network, held together and cross-linked with other wall components (Posé et al., 2011). Pectin is the most abundant class of macromolecule within the primary cell wall and the middle lamella matrixes, and its solubilization is known to be the most consistent feature during strawberry fruit softening (Posé et al., 2011). The role of many enzymes during cell wall disassembly has been characterized specifically regarding fruit ripening processes. The activity of enzymes like pectate lyases (PL), polygalacturonases (PG), pectin esterase (PE), pectin methylesterase (PME), β-1,4-glucanases (EGase), expansins (Exp), β-xylosidases (Xyl), β-galactosidase (β-Gal), and α-arabinofuranosidases (Ara) modify different polysaccharides, especially matrix glycans and pectins (Posé et al., 2011).
A number of TFs have been deeply characterized in their role regulating cell wall composition (Figure 1E). Among them, a WRKY-type protein, FvWRKY48, whose expression increases during strawberry fruit ripening, regulates the chemical properties of the cell wall during this process (Zhang et al., 2022a). Thus, this TF regulates the reduction of homogalacturonan (HG) pectin polymer, since stable overexpressing and silencing lines produce fruits that present lower and higher content respectively in the middle lamella and tricellular junction zone. These changes are reflected in altered fruit firmness and ripening progress of those transgenic lines and are explained by the positive regulation of three PL genes and one β-Gal by FvWRK48. Furthermore, yeast-one-hybrid, EMSA, and ChIP-qPCR assays identified a direct interaction of FvWRKY48 in the promoter region of one of the PL genes, FvPLA, whose overexpression and silencing is able to mimic the HG content phenotype in the FvWRKY48 misexpression.
FaRIF has also been found to be a central regulator of cell wall composition. Stable FaRIF-silenced and overexpression lines develop firmer and softer receptacles respectively, supporting a role of this TF in promoting fruit softening during ripening. The general role of FaRIF in the regulation of cell wall composition is confirmed by a transcriptome analysis in fruits from those transgenic lines that showed a misregulation of cell wall degradation-related genes such as FaXYL3, FaPL2, FaPL3, FaPL4, FaGHB15 (EGase), and the PGs FaPG1 and FaADPG2. Moreover, the expression of enzymes responsible for modifying the cell wall coding genes was also altered, including FaEXP1, FaEXP2, FaEXP3, FaPME38, FaPME39 and different FaAGPs, which encodes for arabino galactan-proteins (Martín-Pizarro et al., 2021). Besides FaRIF, another NAC protein, FcNAC1 has been shown to directly induce PL expression, although its function in cell wall remodeling has not been studied yet (Carrasco-Orellana et al., 2018). Another direct regulation has been described for FvMYB79, which besides regulating the expression of phenylpropanoid-related genes, binds to the promoter of FaPME38 activating its expression (Cai et al., 2022). Furthermore, the expression of PME, EXP, PL, PG and EGase genes was significantly downregulated in FvMYB79-RNAi fruits, and upregulated when this TF was overexpressed (Cai et al., 2022). Similar transient experiments allowed to identified cell wall-related genes differentially expressed in FvTCP9 silenced and overexpression lines, including β-Gal1/2/3 and EXP1/2/5 (Xie et al., 2020).
Among the MADS-box proteins involved in strawberry fruit softening, FaMADS9 might promote this process during ripening, since its downregulation increased fruit firmness in the work of Seymour and collaborators (Seymour et al., 2011), but not in Vallarino’s study (Vallarino et al., 2020). Again, and as previously mentioned, this difference might be the consequence of the possible off-target effects over other related MADS-box genes in Seymour’s work. Nevertheless, both studies identified differences in the expression of genes related to cell wall modifications in FaMADS9-RNAi fruits, which showed an upregulation of PE2 (Seymour et al., 2011) and PG2 (Vallarino et al., 2020), and a downregulation of cellulase, PL1 and PE1 in red fruits (Seymour et al., 2011), and an upregulation of PG1 and PG2 in white fruits (Vallarino et al., 2020), supporting a role of this TF in the modulation of the cell wall composition. Besides the role of the MADS-like protein FaMADS9, transient silencing and overexpression of FaSHP altered the expression of PG1, PL and EG1 (Daminato et al., 2013), although its role in the regulation of fruit firmness or cell wall composition has not been clarified yet.
Finally, FaPRE1 has also been proposed to control cell wall metabolism. Thus, FaPRE1 downregulation negatively regulates FaPG1 and FaRGlyaseI and produces an upregulation of a number of cell wall genes whose transcription is usually higher in immature fruits (Medina-Puche et al., 2019). Although FaPRE1 expression is receptacle-specific, its ectopic overexpression leads to an elongation of vegetative organs, probably due to the regulation of many genes that encode cell wall-modifying enzymes (Medina-Puche et al., 2021). However, the role of FaPRE1 in the regulation of fruit softening during strawberry fruit ripening requires further investigation.
A number of transcriptome studies have been performed in order to identify whole strawberry TF families. These studies are usually accompanied by a brief characterization of the family and their putative role in the regulation of different processes, like their regulatory role in hormone metabolism, flavonoid biosynthesis or response against pathogens. In these studies, different members of these families have been proposed as candidate regulators of specific traits related to fruit ripening. For example, the NAC TF family was studied in F. × ananassa regarding its putative role in fruit development and ripening (Moyano et al., 2018). Thus, the expression of FaNAC006, FaNAC021, FaNAC022, FaNAC035 (FaRIF) and FaNAC042 is induced during fruit development and ripening. Furthermore, their expression was downregulated after fruits were treated with 1-Nordihydroguaiaretic acid (NDGA), an inhibitor of ABA biosynthesis, supporting their potential role as regulators of strawberry fruit ripening, although only FaRIF has been functionally validated so far among these candidate genes (Martín-Pizarro et al., 2021). Furthermore, FaNAC022 and FaNAC042 were suggested as regulators of vascular tissue and secondary cell wall development, and FaNAC006 and FaNAC092 as fruit senescence regulators (Moyano et al., 2018).
Other members of MBW complexes have been proposed as putative regulators of anthocyanin biosynthesis based on their ripening-related pattern expression and function of their putative orthologs, such as FabHLH17, FabHLH25, FabHLH27, FabHLH29, FabHLH40, FabHLH80 and FabHLH98 from the bHLH family (Zhao et al., 2018), and FaMYB28, FaMYB54 (MYB1) and FaMYB576 from the MYB family (Liu et al., 2021), as well as FvMYB33 as a possible PAs biosynthesis regulator (Shulaev et al., 2011). In a similar way, the GRAS TFs were proposed as regulators of strawberry fruit ripening. In particular, FvGRAS27 was also proposed as a regulator of anthocyanin accumulation and FvGRAS54 as a general regulator of ripening (Chen et al., 2019). Regarding the TCP family, FvTCP12 and FvTCP17 could also be involved in regulating ripening-related processes as their expression shows an increasing pattern during ripening and are highly induced after ABA treatment (Wei et al., 2016). Finally, FaERF3, FaERF6 and FaERF71a were also proposed as ripening regulators due to their expression pattern (Sánchez-Sevilla et al., 2017). In conclusion, there are a large number of putative regulators of strawberry fruit ripening which role has not been validated yet, so their study will shed light on their contribution to this process.
In this work, we have summarized the role of a large number of TFs that have been studied in relation to the control of different processes related to strawberry fruit ripening to date. However, besides all this knowledge acquired so far, the precise regulation of each specific process involved during strawberry fruit development and ripening is still poorly understood. One explanation is the complexity itself of strawberry as a model plant, which hinders, and even prevents in the case of the octoploid species, to perform genetic studies to understand the relationship between those regulators. Therefore, most studies are based on the phenotypical and molecular characterization of either RNAi or overexpression stable transgenic lines, or on transient assays, where resulting transcriptome changes can be followed. However, no study has been performed so far to identify the direct targets and consensus DNA-binding sequences of any TF genome-wide with assays like DAP (DNA Affinity Purification) or ChIP (Chromatin Immunoprecipitation) sequencing, with the exception of a targeted analysis by ChIP-qPCR to identify the direct regulation of PLA by FvWRKY48 (Zhang et al., 2022a) and an in vitro DNA binding assay that identified the consensus sequence of FaDOF2 (Molina-Hidalgo et al., 2017). This is the consequence of the complexity of the ChIP methodology in general and in strawberry in particular, considering that stable tagged-TF overexpression lines in order to use efficient commercial antibodies are not easy to obtain, nor the ChIP protocol to optimize. On top of that, the polyploidy of F. × ananassa is another important challenge that complicates genomic studies. However, we believe that the in vitro approach DAP-seq will become a very useful and informative alternative that will be widely used in the identification of direct target genes of strawberry TFs in the near future. Finally, it has been recently and successfully applied the CRISPR/Cas9 genome-editing tool in strawberry plants (Zhou et al., 2018; Gao et al., 2020; Pi et al., 2021; Mao et al., 2022), including the octoploid species (Martín-Pizarro et al., 2019; Wilson et al., 2019). This tool brings new opportunities to deeply characterize TFs, avoiding misleading conclusions about their role that might arise in either knockdown assays or with spontaneous mutations. For example, the TFs RIPENING INHIBITOR (RIN), NON-RIPENING (NOR), and COLORLESS NON-RIPENING (CNR) have traditionally been considered master regulators of tomato fruit ripening (Seymour et al., 2013). However, CRISPR/Cas9 knockout lines for these TFs resulted in mutants where the developing fruits displayed a more subtle ripening phenotype than the original spontaneous mutants, which have been shown to be gain-of-function (rin) or dominant-negative (nor and cnr) mutations (Ito et al., 2017; Gao et al., 2019; Wang et al., 2019). These results question the role of just a few TFs as the upstream master regulators of fruit ripening, and suggest a more complex network of TFs underlying the control of this process in tomato, and probably in other species such as strawberry. This highlights the promise of CRISPR/Cas9 mediated dissection of molecular processes in crop species like strawberry and, together with the integration of transcriptome studies and ChIP-seq/DAP-seq analyses, will open a new horizon in the discovery and characterization of the regulatory networks that control strawberry fruit ripening.
CS-G, DP, and CM-P conceived and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the European Research Council (ERC-2014-Stg 638134 to DP), the Spanish Ministry of Science and Innovation (RTI2018-097309-A-I00 and PID2021-123677OB-I00 to DP) and the Junta de Andalucía (UMA20-FEDERJA-093 and POSTDOC21_00893 to CM-P). We thank Plan Propio from the University of Málaga for financial support.
We thank Danelle K. Seymour for her suggestions to improve the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aharoni, A., De Vos, C. H. R., Wein, M., Sun, Z., Greco, R., Kroon, A., et al. (2001). The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 28, 319–332. doi: 10.1046/j.1365-313X.2001.01154.x
Almeida, J. R. M., D’Amico, E., Preuss, A., Carbone, F., de Vos, C. H. R., Deiml, B., et al. (2007). Characterization of major enzymes and genes involved in flavonoid and proanthocyanidin biosynthesis during fruit development in strawberry (Fragaria × ananassa). Arch. Biochem. Biophys. 465, 61–71. doi: 10.1016/j.abb.2007.04.040
Alstrup, J., Petersen, M. A., Larsen, F. H., Münchow, M. (2020). The effect of roast development time modulations on the sensory profile and chemical composition of the coffee brew as measured by nmr and dhs-gc–ms. Beverages 6, 1–14. doi: 10.3390/beverages6040070
Bai, Q., Huang, Y., Shen, Y. (2021). The physiological and molecular mechanism of abscisic acid in regulation of fleshy fruit ripening. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.619953
Bontpart, T., Marlin, T., Vialet, S., Guiraud, J. L., Pinasseau, L., Meudec, E., et al. (2016). Two shikimate dehydrogenases, VvSDH3 and VvSDH4, are involved in gallic acid biosynthesis in grapevine. J. Exp. Bot. 67, 3537–3550. doi: 10.1093/jxb/erw184
Buendía, B., Gil, M. I., Tudela, J. A., Gady, A. L., Medina, J. J., Soria, C., et al. (2010). HPLC-MS analysis of proanthocyanidin oligomers and other phenolics in 15 strawberry cultivars. J. Agric. Food Chem. 58, 3916–3926. doi: 10.1021/jf9030597
Cai, J., Mo, X., Wen, C., Gao, Z., Chen, X., Xue, C. (2022). FvMYB79 positively regulates strawberry fruit softening via transcriptional activation of FvPME38. Int. J. Mol. Sci. 23, 101. doi: 10.3390/ijms23010101
Carrasco-Orellana, C., Stappung, Y., Mendez-Yañez, A., Allan, A. C., Espley, R. V., Plunkett, B. J., et al. (2018). Characterization of a ripening-related transcription factor FcNAC1 from fragaria chiloensis fruit. Sci. Rep. 8, 1–12. doi: 10.1038/s41598-018-28226-y
Castillejo, C., de la Fuente, J. I., Iannetta, P., Botella, M.Á., Valpuesta, V. (2004). Pectin esterase gene family in strawberry fruit: Study of FaPE1, a ripening-specific isoform. J. Exp. Bot. 55, 909–918. doi: 10.1093/jxb/erh102
Castillejo, C., Waurich, V., Wagner, H., Ramos, R., Oiza, N., Muñoz, P., et al. (2020). Allelic variation of MYB10 is the major force controlling natural variation in skin and flesh color in strawberry (Fragaria spp.) fruit. Plant Cell 32, 3723–3749. doi: 10.1105/tpc.20.00474
Chen, H., Li, H., Lu, X., Chen, L., Liu, J., Wu, H. (2019). Identification and expression analysis of GRAS transcription factors to elucidate candidate genes related to stolons, fruit ripening and abiotic stresses in woodland strawberry (Fragaria vesca). Int. J. Mol. Sci. 20, 1–24. doi: 10.3390/ijms20184593
Chen, Q., Tang, Y. M., Wang, Y., Sun, B., Chen, T., Lei, D. Y., et al. (2020). Enhance sucrose accumulation in strawberry fruits by eliminating the translational repression of FabZIPs1.1. Sci. Hortic. (Amsterdam). 259, 108850. doi: 10.1016/j.scienta.2019.108850
Chen, R., Mao, L., Guan, W., Wei, X., Huang, Z., Wu, Y. (2022). ABA-mediated miR5290 promotes anthocyanin biosynthesis by inhibiting the expression of FaMADS1 in postharvest strawberry fruit. Postharvest Biology and Technology. 189, 111934. doi: 10.1016/j.postharvbio.2022.111934.
Daminato, M., Guzzo, F., Casadoro, G. (2013). A SHATTERPROOF-like gene controls ripening in non-climacteric strawberries, and auxin and abscisic acid antagonistically affect its expression. J. Exp. Bot. 64, 3775–3786. doi: 10.1093/jxb/ert214
Du, X., Plotto, A., Baldwin, E., Rouseff, R. (2011). Evaluation of volatiles from two subtropical strawberry cultivars using GC-olfactometry, GC-MS odor activity values, and sensory analysis. J. Agric. Food Chem. 59, 12569–12577. doi: 10.1021/jf2030924
Encinas-Villarejo, S., Maldonado, A. M., Amil-Ruiz, F., De Los Santos, B., Romero, F., Pliego-Alfaro, F., et al. (2009). Evidence for a positive regulatory role of strawberry (Fragaria× ananassa) FaWRKY1 and arabidopsis at WRKY75 proteins in resistance. J. Exp. Bot. 60, 3043–3065. doi: 10.1093/jxb/erp152
Fait, A., Hanhineva, K., Beleggia, R., Dai, N., Rogachev, I., Nikiforova, V. J., et al. (2008). Reconfiguration of the achene and receptacle metabolic networks during strawberry fruit development. Plant Physiol. 148, 730–750. doi: 10.1104/pp.108.120691
Fan, Z., Plotto, A., Bai, J., Whitaker, V. M. (2021). Volatiles influencing sensory attributes and Bayesian modeling of the soluble solids–sweetness relationship in strawberry. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.640704
Gao, Q., Luo, H., Li, Y., Liu, Z., Kang, C. (2020). Genetic modulation of RAP alters fruit coloration in both wild and cultivated strawberry. Plant Biotechnol. J. 18, 1550–1561. doi: 10.1111/pbi.13317
Gao, Y., Zhu, N., Zhu, X., Wu, M., Cai-Zhong, J., Grierson, D., et al. (2019). Diversity and redundancy of the ripening regulatory networks revealed by the fruitENCODE and the new CRISPR/Cas9 CNR and NOR mutants. Hortic. Res. 6, 39. doi: 10.1038/s41438-019-0122-x
Giampieri, F., Alvarez-Suarez, J. M., Cordero, M. D., Gasparrini, M., Forbes-Hernandez, T. Y., Afrin, S., et al. (2017). Strawberry consumption improves aging-associated impairments, mitochondrial biogenesis and functionality through the AMP-activated protein kinase signaling cascade. Food Chem. 234, 464–471. doi: 10.1016/j.foodchem.2017.05.017
Gu, T., Jia, S., Huang, X., Wang, L., Fu, W., Huo, G., et al. (2019). Transcriptome and hormone analyses provide insights into hormonal regulation in strawberry ripening. Planta 250, 145–162. doi: 10.1007/s00425-019-03155-w
Guo, L., Luo, X., Li, M., Joldersma, D., Plunkert, M., Liu, Z. (2022). Mechanism of fertilization-induced auxin synthesis in the endosperm for seed and fruit development. Nat. Commun. 13, 3985. doi: 10.1038/s41467-022-31656-y
Han, Y., Chen, C., Yan, Z., Li, J., Wang, Y. (2019). The methyl jasmonate accelerates the strawberry fruits ripening process. Sci. Hortic. (Amsterdam). 249, 250–256. doi: 10.1016/j.scienta.2019.01.061
Hawkins, C., Caruana, J., Schiksnis, E., Liu, Z. (2016). Genome-scale DNA variant analysis and functional validation of a SNP underlying yellow fruit color in wild strawberry. Sci. Rep. 6, 1–11. doi: 10.1038/srep29017
Ito, Y., Nishizawa-Yokoi, A., Endo, M., Mikami, M., Shima, Y., Nakamura, N., et al. (2017). Re-evaluation of the rin mutation and the role of RIN in the induction of tomato ripening. Nat. Plants 3, 866–874. doi: 10.1038/s41477-017-0041-5
Jia, H. F., Chai, Y. M., Li, C. L., Lu, D., Luo, J. J., Qin, L., et al. (2011). Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol. 157, 188–199. doi: 10.1104/pp.111.177311
Jia, H., Jiu, S., Zhang, C., Wang, C., Tariq, P., Liu, Z., et al. (2016). Abscisic acid and sucrose regulate tomato and strawberry fruit ripening through the abscisic acid-stress-ripening transcription factor. Plant Biotechnol. J. 14, 2045–2065. doi: 10.1111/pbi.12563
Jia, H., Wang, Y., Sun, M., Li, B., Han, Y., Zhao, Y., et al. (2013). Sucrose functions as a signal involved in the regulation of strawberry fruit development and ripening. New Phytol. 198, 453–465. doi: 10.1111/nph.12176
Jia, S., Wang, Y., Zhang, G., Yan, Z., Cai, Q. (2021). Strawberry FaWRKY25 transcription factor negatively regulated the resistance of strawberry fruits to botrytis cinerea. Genes (Basel). 12, 1–17. doi: 10.3390/genes12010056
Kadomura-Ishikawa, Y., Miyawaki, K., Takahashi, A., Masuda, T., Noji, S. (2015a). Light and abscisic acid independently regulated FaMYB10 in fragaria × ananassa fruit. Planta 241, 953–965. doi: 10.1007/s00425-014-2228-6
Kadomura-Ishikawa, Y., Miyawaki, K., Takahashi, A., Noji, S. (2015b). RNAi-mediated silencing and overexpression of the FaMYB1 gene and its effect on anthocyanin accumulation in strawberry fruit. Biol. Plant 59, 677–685. doi: 10.1007/s10535-015-0548-4
Karlova, R., Chapman, N., David, K., Angenent, G. C., Seymour, G. B., De Maagd, R. A. (2014). Transcriptional control of fleshy fruit development and ripening. J. Exp. Bot. 65, 4527–4541. doi: 10.1093/jxb/eru316
Kim, J., Lee, J. G., Hong, Y., Lee, E. J. (2019). Analysis of eight phytohormone concentrations, expression levels of ABA biosynthesis genes, and ripening-related transcription factors during fruit development in strawberry. J. Plant Physiol. 239, 52–60. doi: 10.1016/j.jplph.2019.05.013
Klee, H. J., Giovannoni, J. J. (2011). Genetics and control of tomato fruit ripening and quality attributes. Annu. Rev. Genet. 45, 41–59. doi: 10.1146/annurev-genet-110410-132507
Klee, H. J., Tieman, D. M. (2018). The genetics of fruit flavour preferences. Nat. Rev. Genet. 19, 347–356. doi: 10.1038/s41576-018-0002-5
Larsen, M., Poll, L. (1990). Odour thresholds of some important aroma compounds in strawberries. Z Leb. Unters. Forsch. 191, 129–131. doi: 10.1007/BF01201770
Liao, X., Li, M., Liu, B., Yan, M., Yu, X., Zi, H., et al. (2018). Interlinked regulatory loops of ABA catabolism and biosynthesis coordinate fruit growth and ripening in woodland strawberry. Proc. Natl. Acad. Sci. 115, E11542–E11550. doi: 10.1073/pnas.1812575115
Li, B.-J., Grierson, D., Shi, Y., Chen, K.-S. (2022a). Roles of abscisic acid in regulating ripening and quality of strawberry, a model non-climacteric fruit. Hortic. Res. 9, uhac089. doi: 10.1093/hr/uhac089
Li, X., Wang, X., Zhang, Y., Zhang, A., You, C.-X. (2022b). Regulation of fleshy fruit ripening: From transcription factors to epigenetic modifications. Hortic. Res. 9, uhac013. doi: 10.1093/hr/uhac013
Lin-Wang, K., Bolitho, K., Grafton, K., Kortstee, A., Karunairetnam, S., McGhie, T. K., et al. (2010). An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in rosaceae. BMC Plant Biol. 10, 50. doi: 10.1186/1471-2229-10-50
Lin-Wang, K., McGhie, T. K., Wang, M., Liu, Y., Warren, B., Storey, R., et al. (2014). Engineering the anthocyanin regulatory complex of strawberry (Fragaria vesca). Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00651
Liu, Z., Ma, H., Jung, S., Main, D., Guo, L. (2020). Developmental mechanisms of fleshy fruit diversity in rosaceae. Annu. Rev. Plant Biol. 71, 547–573. doi: 10.1146/annurev-arplant-111119-021700
Liu, J., Wang, J., Wang, M., Zhao, J., Zheng, Y., Zhang, T., et al. (2021). Genome-wide analysis of the R2R3-MYB gene family in fragaria × ananassa and its function identification during anthocyanins biosynthesis in pink-flowered strawberry. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.702160
Liu, Y., Ye, Y., Wang, Y., Jiang, L., Yue, M., Tang, L., et al. (2022). B-box transcription factor FaBBX22 promotes light-induced anthocyanin accumulation in strawberry ( fragaria × ananassa ). Int. J. Mol. Sci. 23, 7757. doi: 10.3390/ijms23147757
Li, Y., Xu, P., Chen, G., Wu, J., Liu, Z., Lian, H. (2020). FvbHLH9 functions as a positive regulator of anthocyanin biosynthesis by forming a HY5-bHLH9 transcription complex in strawberry fruits. Plant Cell Physiol. 61, 826–837. doi: 10.1093/pcp/pcaa010
Lucioli, S., Pastorino, F., Nota, P., Ballan, G., Frattarelli, A., Fabbri, A., et al. (2019). Extracts from cell suspension cultures of strawberry (Fragaria x ananassa duch): Cytotoxic effects on human cancer cells. molecules 24, 1738. doi: 10.3390/molecules24091738
Lu, W., Chen, J., Ren, X., Yuan, J., Han, X., Mao, L., et al. (2018). One novel strawberry MADS-box transcription factor FaMADS1a acts as a negative regulator in fruit ripening. Sci. Hortic. (Amsterdam). 227, 124–131. doi: 10.1016/j.scienta.2017.09.042
Lu, H., Li, L., Xu, Y., Li, D., Li, G., Yan, Y., et al. (2022). FaLEC2 repressing FaLOX2 promoter involved in the metabolism of LOX-derived volatiles during strawberry ripening. Sci. Hortic. 303, 111188. doi: 10.1016/j.scienta.2022.111188
Lu, H., Luo, Z., Li, D., Jiang, Y., Li, L. (2021). FaMYB11 promotes the accumulation of volatile esters by regulating FaLOX5 during strawberry (Fragaria × ananassa) ripening. Postharvest. Biol. Technol. 178, 111560. doi: 10.1016/j.postharvbio.2021.111560
Lu, H., Luo, Z., Wang, L., Liu, W., Li, D., Belwal, T., et al. (2020b). FaMYB9 is involved in the regulation of C6 volatile biosynthesis in strawberry. Plant Sci. 293, 110422. doi: 10.1016/j.plantsci.2020.110422
Luo, H., Dai, C., Li, Y., Feng, J., Liu, Z., Kang, C. (2018). Reduced anthocyanins in petioles codes for a GST anthocyanin transporter that is essential for the foliage and fruit coloration in strawberry. J. Exp. Bot. 69, 2595–2608. doi: 10.1093/jxb/ery096
Lu, B., Wang, Y., Zhang, G., Feng, Y., Yan, Z., Wu, J., et al. (2020a). Genome-wide identification and expression analysis of the strawberry FvbZIP gene family and the role of key gene FabZIP46 in fruit resistance to Gray mold. Plants (Basel) 9, 1199. doi: 10.3390/plants9091199
Manivannan, A., Han, K., Lee, S. Y., Lee, H., Hong, J. P., Kim, J., et al. (2021). Genome-wide analysis of MYB10 transcription factor in fragaria and identification of QTLs associated with fruit color in octoploid strawberry. Int. J. Mol. Sci. 22, 12587. doi: 10.3390/ijms222212587
Mao, W., Han, Y., Chen, Y., Sun, M., Feng, Q., Li, L., et al. (2022). Low temperature inhibits anthocyanin accumulation in strawberry fruit by activating FvMAPK3-induced phosphorylation of FvMYB10 and degradation of chalcone synthase 1. Plant Cell 34, 1226–1249. doi: 10.1093/plcell/koac006
Martín-Pizarro, C., Triviño, J. C., Posé, D. (2019). Functional analysis of the TM6 MADS-box gene in the octoploid strawberry by CRISPR/Cas9-directed mutagenesis. J. Exp. Bot. 70, 949–961. doi: 10.1093/jxb/ery400
Martín-Pizarro, C., Vallarino, J. G., Osorio, S., Meco, V., Urrutia, M., Pillet, J., et al. (2021). The NAC transcription factor FaRIF controls fruit ripening in strawberry. Plant Cell 33, 1574–1593. doi: 10.1093/plcell/koab070
Medina-Puche, L., Blanco-Portales, R., Molina-Hidalgo, F. J., Cumplido-Laso, G., García-Caparrós, N., Moyano-Cañete, E., et al. (2016). Extensive transcriptomic studies on the roles played by abscisic acid and auxins in the development and ripening of strawberry fruits. Funct. Integr. Genomics 16, 671–692. doi: 10.1007/s10142-016-0510-3
Medina-Puche, L., Cumplido-Laso, G., Amil-Ruiz, F., Hoffmann, T., Ring, L., Rodríguez-Franco, A., et al. (2014). MYB10 plays a major role in the regulation of flavonoid/phenylpropanoid metabolism during ripening of fragaria × ananassa fruits. J. Exp. Bot. 65, 401–417. doi: 10.1093/jxb/ert377
Medina-Puche, L., Martínez-Rivas, F. J., Molina-Hidalgo, F. J., García-Gago, J. A., Mercado, J. A., Caballero, J. L., et al. (2021). Ectopic expression of the atypical HLH FaPRE1 gene determines changes in cell size and morphology. Plant Sci. 305, 110830. doi: 10.1016/j.plantsci.2021.110830
Medina-Puche, L., Martínez-Rivas, F. J., Molina-Hidalgo, F. J., Mercado, J. A., Moyano, E., Rodríguez-Franco, A., et al. (2019). An atypical HLH transcriptional regulator plays a novel and important role in strawberry ripened receptacle. BMC Plant Biol. 19, 1–16. doi: 10.1186/s12870-019-2092-4
Medina-Puche, L., Molina-Hidalgo, F. J., Boersma, M., Schuurink, R. C., López-Vidriero, I., Solano, R., et al. (2015). An R2R3-MYB transcription factor regulates eugenol production in ripe strawberry fruit receptacles. Plant Physiol. 168, 598–614. doi: 10.1104/pp.114.252908
Merchante, C., Vallarino, J. G., Osorio, S., Aragüez, I., Villarreal, N., Ariza, M. T., et al. (2013). Ethylene is involved in strawberry fruit ripening in an organ-specific manner. J. Exp. Bot. 64, 4421–4439. doi: 10.1093/jxb/ert257
Molina-Hidalgo, F. J., Medina-Puche, L., Cañete-Gómez, C., Franco-Zorrilla, J. M., López-Vidriero, I., Solano, R., et al. (2017). The fruit-specific transcription factor FaDOF2 regulates the production of eugenol in ripe fruit receptacles. J. Exp. Bot. 68, 4529–4543. doi: 10.1093/jxb/erx257
Moyano, E., Martínez-Rivas, F. J., Blanco-Portales, R., Molina-Hidalgo, F. J., Ric-Varas, P., Matas-Arroyo, A. J., et al. (2018). Genome-wide analysis of the NAC transcription factor family and their expression during the development and ripening of the fragaria × ananassa fruits. Plos One 13, 1–23. doi: 10.1371/journal.pone.0196953
Muñoz, C., Sánchez-Sevilla, J. F., Botella, M. A., Hoffmann, T., Schwab, W., Valpuesta, V. (2011). Polyphenol composition in the ripe fruits of fragaria species and transcriptional analyses of key genes in the pathway. J. Agric. Food Chem. 59, 12598–12604. doi: 10.1021/jf203965j
Nesi, N., Debeaujon, I., Jond, C., Pelletier, G., Caboche, M., Lepiniec, L. (2000). The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in arabidopsis siliques. Plant Cell 12, 1863–1878. doi: 10.1105/tpc.12.10.1863
Nesi, N., Jond, C., Debeaujon, I., Caboche, M., Lepiniec, L. (2001). The arabidopsis TT2 gene encodes and R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13, 2099. doi: 10.2307/3871430
Paolocci, F., Robbins, M. P., Passeri, V., Hauck, B., Morris, P., Rubini, A., et al. (2011). The strawberry transcription factor FaMYB1 inhibits the biosynthesis of proanthocyanidins in lotus corniculatus leaves. J. Exp. Bot. 62, 1189–1200. doi: 10.1093/jxb/erq344
Petrussa, E., Braidot, E., Zancani, M., Peresson, C., Bertolini, A., Patui, S., et al. (2013). Plant flavonoids-biosynthesis, transport and involvement in stress responses. Int. J. Mol. Sci. 14, 14950–14973. doi: 10.3390/ijms140714950
Pi, M., Hu, S., Cheng, L., Zhong, R., Cai, Z., Liu, Z., et al. (2021). The MADS-box gene FveSEP3 plays essential roles in flower organogenesis and fruit development in woodland strawberry. Hortic. Res. 8, 1–15. doi: 10.1038/s41438-021-00673-1
Posé, S., García-Gago, J. A., Santiago-Doménech, N., Pliego-Alfaro, F., Quesada, M. A., Mercado, J. A. (2011). Strawberry fruit softening: Role of cell wall disassembly and its manipulation in transgenic plants. Genes Genomes Genomics 5, 40–48.
Pott, D. M., Vallarino, J. G., Cruz-Rus, E., Willmitzer, L., Sánchez-Sevilla, J. F., Amaya, I., et al. (2020). Genetic analysis of phenylpropanoids and antioxidant capacity in strawberry fruit reveals mQTL hotspots and candidate genes. Sci. Rep. 10, 1–15. doi: 10.1038/s41598-020-76946-x
Rastogi, S., Kumar, R., Chanotiya, C. S., Shanker, K., Gupta, M. M., Nagegowda, D. A., et al. (2013). 4-coumarate: CoA ligase partitions metabolites for eugenol biosynthesis. Plant Cell Physiol. 54, 1238–1252. doi: 10.1093/pcp/pct073
Reis, L., Forney, C. F., Jordan, M., Munro Pennell, K., Fillmore, S., Schemberger, M. O., et al. (2020). Metabolic profile of strawberry fruit ripened on the plant following treatment with an ethylene elicitor or inhibitor. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00995
Salvatierra, A., Pimentel, P., Moya-León, M. A., Herrera, R. (2013). Increased accumulation of anthocyanins in fragaria chiloensis fruits by transient suppression of FcMYB1 gene. Phytochemistry 90, 25–36. doi: 10.1016/j.phytochem.2013.02.016
Sánchez-Sevilla, J. F., Vallarino, J. G., Osorio, S., Bombarely, A., Posé, D., Merchante, C., et al. (2017). Gene expression atlas of fruit ripening and transcriptome assembly from RNA-seq data in octoploid strawberry (Fragaria × ananassa). Sci. Rep. 7, 1–13. doi: 10.1038/s41598-017-14239-6
Schaart, J. G., Dubos, C., Romero de la Fuente, I., van Houwelingen, A. M. M. L., de Vos, R. C. H., Jonker, H. H., et al. (2013). Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria × ananassa) fruits. New Phytol. 197, 454–467. doi: 10.1111/nph.12017
Seymour, G. B., Ostergaard, L., Chapman, N. H., Knapp, S., Martin, C. (2013). Fruit development and ripening. Annu. Rev. Plant Biol. 64, 219–241. doi: 10.1146/annurev-arplant-050312-120057
Seymour, G. B., Ryder, C. D., Cevik, V., Hammond, J. P., Popovich, A., King, G. J., et al. (2011). A SEPALLATA gene is involved in the development and ripening of strawberry (Fragaria×ananassa duch.) fruit, a non-climacteric tissue. J. Exp. Bot. 62, 1179–1188. doi: 10.1093/jxb/erq360
Shah, A., Smith, D. L. (2020). Flavonoids in agriculture: Chemistry and roles in, biotic and abiotic stress responses, and microbial associations. Agronomy 10, 1209. doi: 10.3390/agronomy10081209
Shulaev, V., Sargent, D. J., Crowhurst, R. N., Mockler, T. C., Folkerts, O., Delcher, A. L., et al. (2011). The genome of woodland strawberry (Fragaria vesca). Nat. Genet. 43, 109–116. doi: 10.1038/ng.740
Song, J., Forney, C. F. (2008). Flavour volatile production and regulation in fruit. Can. J. Plant Sci. 88, 537–550. doi: 10.4141/CJPS07170
Souleyre, E. J. F., Iannetta, P. P. M., Ross, H. A., Hancock, R. D., Shepherd, L. V. T., Viola, R., et al. (2004). Starch metabolism in developing strawberry (Fragaria x ananassa) fruits. Physiol. Plant 121, 369–376. doi: 10.1111/j.0031-9317.2004.0338.x
Stein, O., Granot, D. (2019). An overview of sucrose synthases in plants. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00095
Symons, G. M., Chua, Y.-J., Ross, J. J., Quittenden, L. J., Davies, N. W., Reid, J.B (2012). Hormonal changes during non-climacteric ripening in strawberry. J. Exp. Bot. 63, 4741–4750. doi: 10.1093/jxb/ers147
Symons, G. M., Davies, C., Shavrukov, Y., Dry, I. B., Reid, J. B., Thomas, M. R. (2006). Grapes on steroids. brassinosteroids are involved in grape berry ripening. Plant Physiol. 140, 150–158. doi: 10.1104/pp.105.070706
Takos, A. M., Jaffé, F. W., Jacob, S. R., Bogs, J., Robinson, S. P., Walker, A.R (2006). Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 142, 1216–1232. doi: 10.1104/pp.106.088104
Tisza, V., Kovács, L., Balogh, A., Heszky, L., Kiss, E. (2010). Characterization of FaSPT, a SPATULA gene encoding a bHLH transcriptional factor from the non-climacteric strawberry fruit. Plant Physiol. Biochem. 48, 822–826. doi: 10.1016/j.plaphy.2010.08.001
Urrutia, M., Schwab, W., Hoffmann, T., Monfort, A. (2015). Genetic dissection of the (poly)phenol profile of diploid strawberry (Fragaria vesca) fruits using a NIL collection. Plant Sci. 242, 151–168. doi: 10.1016/j.plantsci.2015.07.019
Vallarino, J. G., Merchante, C., Sánchez-Sevilla, J. F., de Luis Balaguer, M. A., Pott, D. M., Ariza, M. T., et al. (2020). Characterizing the involvement of FaMADS9 in the regulation of strawberry fruit receptacle development. Plant Biotechnol. J. 18, 929–943. doi: 10.1111/pbi.13257
Vallarino, J. G., Osorio, S., Bombarely, A., Casañal, A., Cruz-Rus, E., Sánchez-Sevilla, J. F., et al. (2015). Central role of FaGAMYB in the transition of the strawberry receptacle from development to ripening. New Phytol. 208, 482–496. doi: 10.1111/nph.13463
Walker, A. R., Davison, P. A., Bolognesi-Winfield, A. C., James, C. M., Srinivasan, N., Blundell, T. L., et al. (1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in arabidopsis, encodes a WD40 repeat protein. Plant Cell 11, 1337–1349. doi: 10.1105/tpc.11.7.1337
Wang, S., Shi, M., Zhang, Y., Pan, Z., Xie, X., Zhang, L., et al. (2022). The R2R3-MYB transcription factor FaMYB63 participates in regulation of eugenol production in strawberry. Plant Physiol. 188, 2146–2165. doi: 10.1093/plphys/kiac014
Wang, R., Tavano, E. C., da, R., Lammers, M., Martinelli, A. P., Angenent, G. C., et al. (2019). Re-evaluation of transcription factor function in tomato fruit development and ripening with CRISPR/Cas9-mutagenesis. Sci. Rep. 9, 1–10. doi: 10.1038/s41598-018-38170-6
Wang, H., Zhang, H., Yang, Y., Li, M., Zhang, Y., Liu, J., et al. (2020). The control of red colour by a family of MYB transcription factors in octoploid strawberry (Fragaria × ananassa) fruits. Plant Biotechnol. J. 18, 1169–1184. doi: 10.1111/pbi.13282
Wang, Y., Zhao, F., Zhang, G., Jia, S., Yan, Z. (2021). FaWRKY11 transcription factor positively regulates resistance to botrytis cinerea in strawberry fruit. Sci. Hortic. (Amsterdam). 279, 109893. doi: 10.1016/j.scienta.2020.109893
Wang, Q. H., Zhao, C., Zhang, M., Li, Y. Z., Shen, Y. Y., Guo, J. X. (2017). Transcriptome analysis around the onset of strawberry fruit ripening uncovers an important role of oxidative phosphorylation in ripening. Sci. Rep. 7, 1–11. doi: 10.1038/srep41477
Wei, W., Hu, Y., Cui, M. Y., Han, Y. T., Gao, K., Feng, J. Y. (2016). Identification and transcript analysis of the TCP transcription factors in the diploid woodland strawberry fragaria vesca. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01937
Wei, L., Mao, W., Jia, M., Xing, S., Ali, U., Zhao, Y., et al. (2018). FaMYB44.2, a transcriptional repressor, negatively regulates sucrose accumulation in strawberry receptacles through interplay with FaMYB10. J. Exp. Bot. 69, 4805–4820. doi: 10.1093/jxb/ery249
Xie, Y. G., Ma, Y. Y., Bi, P. P., Wei, W., Liu, J., Hu, Y., et al. (2020). Transcription factor FvTCP9 promotes strawberry fruit ripening by regulating the biosynthesis of abscisic acid and anthocyanins. Plant Physiol. Biochem. 146, 374–383. doi: 10.1016/j.plaphy.2019.11.004
Xu, P., Wu, L., Cao, M., Ma, C., Xiao, K., Li, Y., et al. (2021). Identification of MBW complex components implicated in the biosynthesis of flavonoids in woodland strawberry. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.774943
Xu, P., Zawora, C., Li, Y., Wu, J., Liu, L., Liu, Z., et al. (2018). Transcriptome sequencing reveals role of light in promoting anthocyanin accumulation of strawberry fruit. Plant Growth Regul. 86, 121–132. doi: 10.1007/s10725-018-0415-3
Yan, J.-W., Ban, Z.-J., Lu, H.-Y., Li, D., Poverenov, E., Luo, Z.-S., et al. (2018). The aroma volatile repertoire in strawberry fruit: a review. J. Sci. Food Agric. 98, 4395–4402. doi: 10.1002/jsfa.9039
Zhang, Y., Li, S., Chen, Y., Liu, Y., Lin, Y., Li, M., et al. (2022b). Heterologous overexpression of strawberry bZIP11 induces sugar accumulation and inhibits plant growth of tomato. Sci. Hortic. (Amsterdam). 292, 110634. doi: 10.1016/j.scienta.2021.110634
Zhang, Y., Liu, Y., Hu, W., Sun, B., Chen, Q., Tang, H. (2018a). Anthocyanin accumulation and related gene expression affected by low temperature during strawberry coloration. Acta Physiol. Plant 40, 1–8. doi: 10.1007/s11738-018-2767-8
Zhang, Z., Shi, Y., Ma, Y., Yang, X., Yin, X., Zhang, Y., et al. (2020). The strawberry transcription factor FaRAV1 positively regulates anthocyanin accumulation by activation of FaMYB10 and anthocyanin pathway genes. Plant Biotechnol. J. 18, 2267–2279. doi: 10.1111/pbi.13382
Zhang, Y., Yin, X., Xiao, Y., Zhang, Z., Li, S., Liu, X., et al. (2018b). An ETHYLENE RESPONSE FACTOR-MYB transcription complex regulates furaneol biosynthesis by activating QUINONE OXIDOREDUCTASE expression in strawberry. Plant Physiol. 178, 189–201. doi: 10.1104/pp.18.00598
Zhang, J., Zhang, Y., Dou, Y., Li, W., Wang, S., Shi, W., et al. (2017). Single nucleotide mutation in FvMYB10 may lead to the yellow fruit in fragaria vesca. Mol. Breed. 37, 1–10. doi: 10.1007/s11032-017-0625-9
Zhang, W., Zhao, S., Gu, S., Cao, X., Zhang, Y., Niu, J., et al. (2022a). FvWRKY48 binds to the pectate lyase FvPLA promoter to control fruit softening in fragaria vesca research article. Plant Physiol. 186, 1037–1049. doi: 10.1093/plphys/kiac091
Zhao, F., Li, G., Hu, P., Zhao, X., Li, L., Wei, W., et al. (2018). Identification of basic/helix-loop-helix transcription factors reveals candidate genes involved in anthocyanin biosynthesis from the strawberry white-flesh mutant. Sci. Rep. 8, 1–15. doi: 10.1038/s41598-018-21136-z
Zhao, F., Song, P., Zhang, X., Li, G., Hu, P., Aslam, A., et al. (2021). Identification of candidate genes influencing anthocyanin biosynthesis during the development and ripening of red and white strawberry fruits via comparative transcriptome analysis. PeerJ 9, e10739. doi: 10.7717/peerj.10739
Zhou, J., Sittmann, J., Guo, L., Xiao, Y., Huang, X., Pulapaka, A., et al. (2021). Gibberellin and auxin signaling genes RGA1 and ARF8 repress accessory fruit initiation in diploid strawberry. Plant Physiol. 185, 1059–1075. doi: 10.1093/PLPHYS/KIAA087
Keywords: strawberry, Fragaria vesca, Fragaria × ananassa, transcription factors, development, ripening
Citation: Sánchez-Gómez C, Posé D and Martín-Pizarro C (2022) Insights into transcription factors controlling strawberry fruit development and ripening. Front. Plant Sci. 13:1022369. doi: 10.3389/fpls.2022.1022369
Received: 18 August 2022; Accepted: 21 September 2022;
Published: 10 October 2022.
Edited by:
José Manuel Palma, Estación Experimental del Zaidín, (CSIC), SpainReviewed by:
Zhang Zongying, Shandong Agricultural University, ChinaCopyright © 2022 Sánchez-Gómez, Posé and Martín-Pizarro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David Posé, ZHBvc2VAdW1hLmVz; Carmen Martín-Pizarro, Y21tYXJ0aW5AdW1hLmVz
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.