- 1Department of Horticulture, Zhejiang University, Hangzhou, China
- 2Zhejiang Provincial Key Laboratory of Integrative Biology of Horticultural Plants, Zhejiang University, Hangzhou, China
- 3The Key Laboratory of Horticultural Plant Growth, Development and Quality Improvement, Ministry of Agriculture of China, Hangzhou, China
- 4Agricultural Experiment Station, Zhejiang University, Hangzhou, China
BBX (B-box) proteins play a vital role in light-induced anthocyanin biosynthesis. PpBBX18 was an indispensable regulator for the induction of anthocyanin biosynthesis in the peel of red pear fruit (Pyrus pyrifolia Nakai.). However, the upstream regulation of BBX genes has not been well characterized. In this study, PpZAT5, a cysteine2/histidine2-type transcription factor, was discovered as the upstream negative regulator of PpBBX18. The results showed that PpZAT5 functions as a transcriptional repressor and directly binds to the CAAT motif of PpBBX18 and inhibits its expression. PpZAT5 expression was inhibited by light, which is converse to the expression pattern of anthocyanin-related structural genes. In addition, less anthocyanin accumulated in the PpZAT5-overexpressing pear calli than in the wild-type pear calli; on the contrary, more anthocyanin accumulated in PpZAT5-RNAi pear calli. Moreover, the crucial genes involved in light-induced anthocyanin biosynthesis were markedly down-regulated in the transcriptome of PpZAT5 overexpression pear calli compared to wild-type. In conclusion, our study indicates that PpBBX18 is negatively regulated by a C2H2-type transcriptional repressor, PpZAT5, which reduces anthocyanin content in pear. The present results demonstrate an upstream molecular mechanism of PpBBX18 and provide insights into light-induced anthocyanin biosynthesis.
Introduction
Although the pear cultivars with the peel color of green, yellow, and brown are still dominant in the Chinese pear market, red Asian pears are gradually gaining market acceptance and consumer preference (Zhang et al., 2013; Sun et al., 2014). The red coloration of pear is attributed to anthocyanin accumulation, and anthocyanin has been identified as the main pigment in many fruits. Anthocyanin is synthesized by the phenylpropanoid pathway, in which PAL (phenylalanine ammonia-lyase), CHS (chalcone synthase), CHI (chalcone isomerase), F3H (flavanone 3-hydroxylase), F3’H (flavonoid 3’-hydroxylase), DFR (dihydroflavonol 4-reductase), ANS (anthocyanin synthase), and UFGT (UDP-glucose: flavonoid 3-glucosyltransferase) are the critical enzymes encoded by structure genes (Winkel-Shirley, 2001; Koes et al., 2005). R2R3-MYB, basic helix-loop-helix (bHLH), and WD40-repeat protein (WDR) regulate the expression of structural genes in the process of anthocyanin biosynthesis, and they frequently exert efficacy as the MYB-bHLH-WDR (MBW) complex (Koes et al., 2005; Jaakola, 2013; Xu et al., 2015). A series of MYB proteins related to anthocyanin biosynthesis have been identified, such as AtMYB75 (PAP1), and AtMYB90 (PAP2) in Arabidopsis (Borevitz et al., 2000; Allan and Espley, 2018), MdMYB1/A/10 in apple (Takos et al., 2006; Ban et al., 2007; Espley et al., 2007) and PpMYB10, PpMYB114 in pear (Feng et al., 2010; Yao et al., 2017).
Among the three components of the MBW complex, the expression of MYB is the most significantly regulated by various internal and external factors at both the transcriptional and translational levels, among which light is considered as the indispensable environmental factor in many plant species (Allan and Espley, 2018). In the light-signal transduction cascade, ELONGATED HYPOCOTYL 5 (HY5) is the hub transcription factor integrating various upstream light signals and regulating the expression of downstream genes, including the anthocyanin biosynthetic genes (Shin et al., 2013; An et al., 2017; Bai et al., 2019b). However, due to the lack of transcription activation motifs, HY5 proteins usually form a complex with other transcription factors. Recent studies showed that a series of BBX proteins serve as the partners of HY5 proteins, forming complex and regulating the downstream genes. In pear, PpHY5 interacts with PpBBX18 (the ortholog of Arabidopsis AtBBX21) and PpBBX16 (the ortholog of Arabidopsis AtBBX22) and activates the expression of PpMYB10. PpBBX21 (the ortholog of Arabidopsis AtBBX24/25) could interfere with forming the active transcription activator complex PpHY5-PpBBX18 and then suppress anthocyanin biosynthesis (Bai et al., 2019b).
The expressions of BBX proteins are usually highly activated by light. In Arabidopsis, the expression of LZF1/AtBBX22 is upregulated by light and largely depends on HY5 (Chang et al., 2008). Light dynamically promotes the expression of AtBBX21 at both the transcription and translation levels (Xu et al., 2016). Similarly, most BBXs related to anthocyanin biosynthesis are activated by light. Our previous research showed that PpBBX18 and PpBBX21 are upregulated under light conditions and then regulate anthocyanin biosynthesis (Bai et al., 2019b). However, the influence of light on anthocyanin biosynthesis is a fine-tuned process (Jaakola, 2013), so we speculate that the release of transcriptional suppression in light conditions might also be involved in the expression of BBX genes and further regulate anthocyanin biosynthesis.
This study applied yeast one-hybrid library screening with PpBBX18 promoter as the bait to elucidate the transcriptional regulation of PpBBX18. As a result, the PpZAT5 was obtained, its expression was inhibited by light. ZINC FINGER of ARABIDOPSIS THALIANA (ZAT) proteins belong to cysteine2/histidine2-type (C2H2-type) zinc finger protein family, harboring a motif with paired cysteine and histidine residues which together chelate a zinc ion to stabilize the domain (Shimeld, 2008). In general, the C2H2 zinc finger motif ranges in length from 25 to 30 residuals with the pattern of amino acids X-X-C-X(1–5)-C-X(12)-H-X(3–6)-H (X may be any amino acid and the numbers in brackets show the number of residues) (Shimeld, 2008). Besides zinc finger domains, a majority of C2H2 zinc finger proteins have nuclear localization signal (NLS, also called B-box), an EAR motif [L/FDLNL/F(x)P] locating at the C terminus with repression activity and L-box related to protein-protein interaction (Ciftci-Yilmaz and Mittler, 2008). In addition, most C2H2 zinc finger proteins contain a highly conserved plant-specific motif, QALGGH, which plays a critical role in DNA binding activity (Kubo et al., 1998; Takatsuji, 1998).
In Arabidopsis thaliana, 176 C2H2 zinc finger members have been identified and classified into three different sets (A, B, and C) according to the positions of the zinc finger motif and their sequences (Englbrecht et al., 2004). C2H2 zinc finger proteins play an essential role in the regulation of metabolic pathways of plants, particularly in stress response and defense activation (Englbrecht et al., 2004; Ciftci-Yilmaz and Mittler, 2008). AtZAT18 positively regulated the response to drought stress in Arabidopsis (Chan et al., 2017), whereas SLZF3 (the ortholog of Arabidopsis ZAT12) increased the accumulation of AsA and promoted the salt tolerance in tomato and Arabidopsis (Li et al., 2018). In addition, some specific C2H2-type zinc finger proteins could promote anthocyanin biosynthesis in plants. SlZF2 (the ortholog of Arabidopsis ZAT10) positively responded to salt stress, drought, and potassium chloride treatments and facilitated the accumulation of anthocyanin and malondialdehyde in tomatoes (Solanum lycopersicum) (Hichri et al., 2014). MdZAT5 was induced by various abiotic stress treatments and promoted anthocyanin biosynthesis by regulating the expressions of genes in the anthocyanin biosynthesis pathway (Wang et al., 2022). AtZAT6 participated in oxidative stress-induced anthocyanin biosynthesis in Arabidopsis by binding to the promoters of several genes associated with anthocyanin (Shi et al., 2018). Although most C2H2 zinc finger proteins regulate metabolic pathways by binding to downstream genes, the recognition site of C2H2 zinc finger protein is not entirely understood. C2H2 zinc finger protein ZPT2-2 of petunia can specifically recognize two tandemly repeated AG(C)T core sequences separated by 13 bp (Takatsuji, 1998), while Arabidopsis Di19 (one C2H2 zinc-finger protein) and AtZAT6 bind to the TACA(A/G)T element of downstream genes promoters (Liu et al., 2013; Shi et al., 2014). In pears, the binding site of C2H2 zinc finger proteins had not been reported.
In this study, we found that PpZAT5 encodes a transcriptional repressor with the EAR motif at the C terminus and its expression was suppressed by light. PpZAT5 could directly bind to CAAT motif of PpBBX18 promoter and inhibit the anthocyanin biosynthesis by repressing PpBBX18 expression. Our work revealed the upstream regulatory mechanism of PpBBX18, which would be helpful for elucidating the mechanism of pear coloration.
Materials and methods
Plant materials and treatment
Fruits were treated as previously described (Ni et al., 2020). ‘Hongzaosu’ pears (Pyrus pyrifolia × Pyrus communis) were from the orchard of the Institute of Horticulture, Henan Academy of Agricultural Sciences, Henan, China. Fruits were covered with lightproof double-layered paper bags during the growing season and bagged fruits were collected at maturity (155 days after full bloom), then immediately transported to laboratory. Bag-removed fruits were incubated for 10 days under continuous white light (60 μmol m-2 s-1) that was provided by overhead LEDs in a growth chamber set at 17°C and 80% relative humidity. After initiating the irradiation, the fruit peel was sampled at 0, 12, 24, 48, 72, 144, and 240 h. The peels of two fruits randomly sampled were collected as one biological replicate, and analyses were completed with three biological replicates (i.e., six fruits in total). The collected peels were immediately frozen in liquid nitrogen and stored at -80°C.
The pear calli induced from young fruit flesh cells of Pyrus communis ‘Clapp’s Favorite’ were used to transformation test as previously described (Bai et al., 2019a). In short, we constructed an expression vector with pCAMBIA1301-PpZAT5, pCAMBIA1301-PpZAT5-RNAi and pCAMBIA1300-PpZAT5 plasmid at first and then transformed them into Agrobacterium tumefaciens strain EHA105. Pear calli were incubated with A. tumefaciens cells harboring vectors (OD600 of 0.4-0.6) for 12-14 minutes and co-cultured for 2-3 days in darkness at 24°C. Pear calli were cultivated on solid Murashige and Skoog medium with proper hygromycin in the next 2 to 3 months and were sub-cultured every 2 weeks. For the treatment, both freshly sub-cultured transgenic and wild-type pear calli were exposed to continuous light until they turned red. Make sure every treatment was completed with at least three biological duplicates. The primers used for vector construction are listed in Supplementary Table 2.
Anthocyanin measurement
Total anthocyanin contents of red pear peel and pear calli were measured as previously described by Ni (Ni et al., 2021). Briefly, 0.1 g powder of frozen pear calli or peels was soaked in 1mL of methanol: acetic acid (99:1, v/v) solution and incubated overnight at 4°C in darkness. And then, the absorbance of the extracting solution was measured by a DU800 spectrophotometer (Beckman Coulter, Brea, CA, USA) at 530, 620, and 650 nm. The anthocyanin content of pear calli was calculated as the following formula: ([A530-A650]-0.2×[A650-A620])/0.1.
RNA extraction and quantitative reverse-transcription PCR
RNA of pear peel and pear calli was extracted following a modified cetyl-trimethyl-ammonium bromide method (Zhang et al., 2013). According to the instruction of PrimeScript™ RT Reagent Kit with gDNA Eraser (TaKaRa, https://www.takarabio.com/), first-strand cDNA was synthesized with 1μg RNA served as the template, and the qRT-PCR assay was conducted by a CFX Connect™ real-time PCR system (Bio-Rad, https://www.bio-rad.com/) and then adjusted cDNA concentrations to make sure that amount of template for the qPCR analysis was equal. The relative expression level of genes was calculated using the 2-△△Ct method with the pear Actin gene (PpActin; GenBank accession No. JN684184) as the reference controls, and the analyses were carried out with three biological replicates. The primers used were designed by the Primer-BLAST online tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) and are listed in Supplementary Table 2.
Subcellular localization
A vector containing pCAMBIA1300-PpZAT5 plasmid and the empty vector (i.e., GFP alone) as control was constructed and then transformed into Agrobacterium tumefaciens strain GV3101. A. tumefaciens cells harboring pCAMBIA1300-PpZAT5 and empty vector were injected into N. benthamiana (mCherry) leaves, respectively. Fluorescence signals were detected by an A1 confocal laser scanning microscope (Nikon, Tokyo, Japan) at 36-72 h after the injection. The primers used are listed in Supplementary Table 2.
Trans-acting activity assay
The full length of PpZAT5 CDS was cloned to the pGBKT7 (BD) vector containing the VP16 fragment. And PpERF105-BD and PpERF105-VP16BD recombinant plasmid as well as positive (VP16-BD) and negative (BD) controls were transformed into yeast (Saccharomyces cerevisiae) strain AH109 cells independently with the Yeastmaker™ Yeast Transformation System 2 (TaKaRa). The yeast transformants were selected in plates containing a selective synthetic dextrose medium lacking tryptophan (SD/-Trp). The positive yeast transformants were further screened on plates containing SD/-Trp/-His medium supplemented with X-α-gal (SD/-Trp/-His/X-α-gal). The trans-activity of PpZAT5 was assessed according to the blue color of yeast strain AH109 growing on SD/-Trp/-His medium. The quantitative analysis of trans-activity based on β-galactosidase activity was measured as previously described by Bai (Bai et al., 2019b) with at least three biological replicates for each measurement. The primers used are listed in Supplementary Table 2.
Yeast one-hybrid assay
Yeast one-hybrid assays were performed following the manufacturer’s protocol of Matchmaker Gold Yeast One-Hybrid System Kit (TaKaRa, Otsu, Japan). Promoter fragments were ligated to the pAbAi vector and linearized, while PpZAT5 CDSs were cloned to the pGADT7 vector (AD). pAbAi vectors were transformed into the yeast strain Y1HGold and selected on plates containing a selective synthetic dextrose medium lacking uracil (SD/-Ura). And then prey vectors were transformed into Y1HGold caring pAbAi vector and detected on SD/-Leu/AbA plates. The primers used are listed in Supplementary Table 2.
Dual-luciferase assay
Dual-luciferase assays were carried out as previously described (Bai et al., 2019a). The full-length CDS of PpZAT5 was cloned to the pGreenII 0029 62-SK vector, while PpBBX18 promoter sequence was inserted into the pGreenII 0800-LUC vector. Both vectors were transformed into A. tumefaciens strain GV3101, and then A. tumefaciens harboring pGreenII 0029 62-SK vector and pGreenII 0800-LUC vector were injected with N. benthamiana leaves in 10:1 ratio. The Dual-Luciferase Reporter Assay System (E710, Promega, Madison, WI, USA) was used to analyze the firefly luciferase and Renilla luciferase activities with Modulus Luminometers (GloMax96, Promega). The analysis was carried out in three independent experiments with at least six biological replicates per assay. The primers used are listed in Supplementary Table 2.
Electrophoretic mobility shift assay
EMSAs were conducted as previously described (Bai et al., 2019b). Briefly, oligonucleotide probes labeled with biotin at the 3’ end were synthesized (Huagen, http://www.huagene.cn) and then annealed to complementary oligonucleotides (95°C for 5 min, 70°C for 20 min, and cooled it down to room temperature and set aside). Following the instructions of LightShift Chemiluminescent EMSA Kit (Thermo Scientific), both binding buffer [2.5% glycerol, 5 mM MgCl2, 50 ng poly (deoxyinosinic-deoxycytidylic) acid, 0.05% NP-40] with and without proteins were prepared for incubation of 30 fmol of the labeled probe at room temperature for 10 min. The non-labeled probe was added to reactions in different volumes for the competition assays. After SDS-PAGE electrophoresis, the DNA was transferred to a nylon membrane (Millipore, http://www.merckmillipore.com/), and the anti-biotin antibody provided in the kit was used for biotin-labeled probes detection.
RNA-seq data analysis
We used the wild-type and pCambia1301-PpZAT5 pear calli for RNA-Seq analysis. The calli were incubated under dark or light for two days for RNA-Seq. The calli of three different lines were simultaneously treated and used as biological replicates. The total RNAs were extracted as described above. Ten micrograms of total RNA each were used for next-generation sequencing. Novogene (Beijing, China) performed the library construction and sequencing using the HiSeq X (Illumina, San Diego, CA) platform with a 150-bp pair-end strategy. The clean reads were mapped to the European pear genome sequence (Chagne et al., 2014) http://www.rosaceae.org) using HISAT2 (Pertea et al., 2016) with default parameters. The reads were then assembled into transcripts and compared with reference gene models using StringTie (Pertea et al., 2016). The differentially expressed gene analysis was performed using DESeq2 (Love et al., 2014).
Results
Identification and characterization of PpZAT5
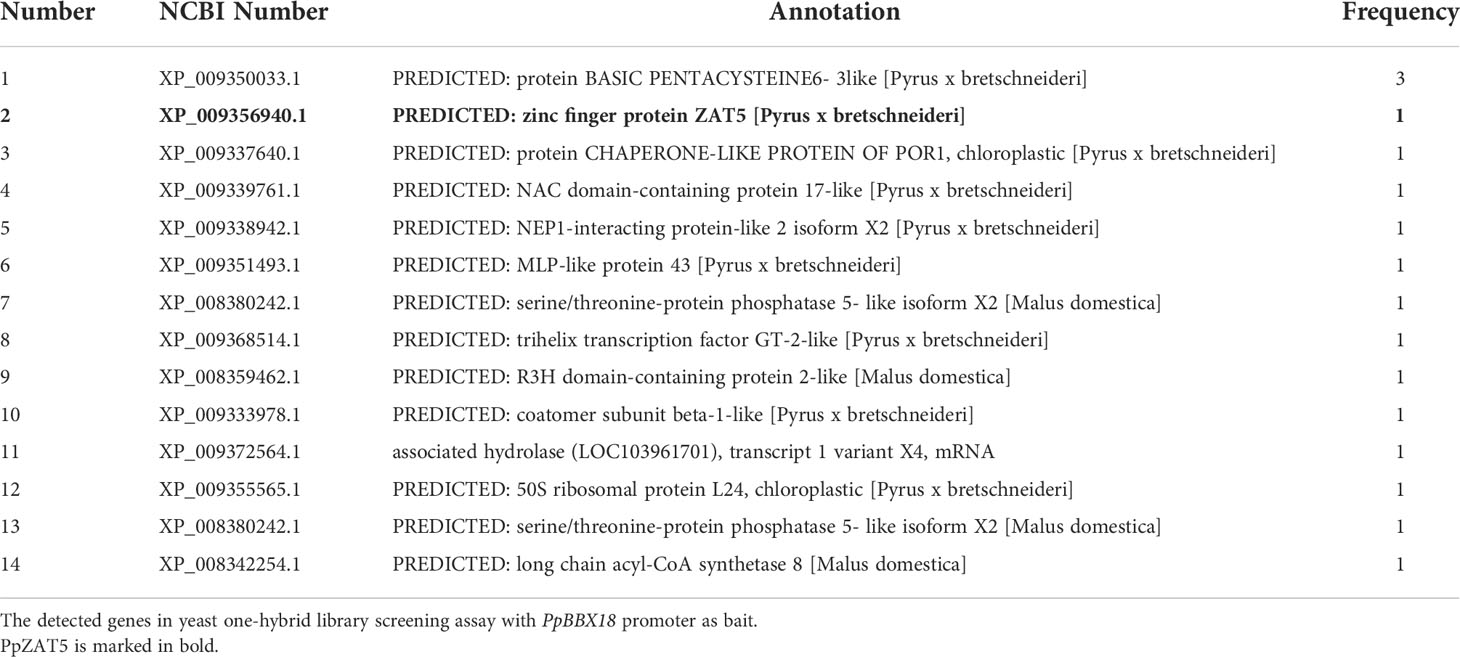
The functions of BBX proteins in light-induced anthocyanin biosynthesis have been well studied, but their upstream regulatory mechanism is still unclear. To figure out the upstream gene regulatory network of PpBBX18, we established a yeast one-hybrid system with PpBBX18 promoter as bait based on the cDNA library of red pear peel (Pyrus pyrifolia Nakai). Following bioinformatics sequencing and analysis, 14 cDNA sequences were identified (Table 1), among which PpZAT5 was selected for further analysis since C2H2 zinc finger proteins play vital roles in regulating metabolic pathways of plants, including anthocyanin biosynthesis.
ZAT proteins in pear, apple, and strawberry genomes were identified using the blast algorithm using Arabidopsis ZAT1 to ZAT7 as queries and then established the phylogenetic tree. The phylogenetic analysis revealed that PpZAT5 was a homologous protein of AtZAT5 (Figure 1A). Moreover, multiple sequence alignment of some C2H2-type zinc finger proteins conserved domains showed that PpZAT5 contained two zinc finger conserved domains in 369 bp to 447 bp and 633 bp to 705 bp, as well as an EAR motif located at the C terminus as shown in Figure 1B. EAR motif that is located at the C terminus of PpZAT5 is generally related to repression activity, which suggests PpZAT5 is a candidate transcriptional repressor.
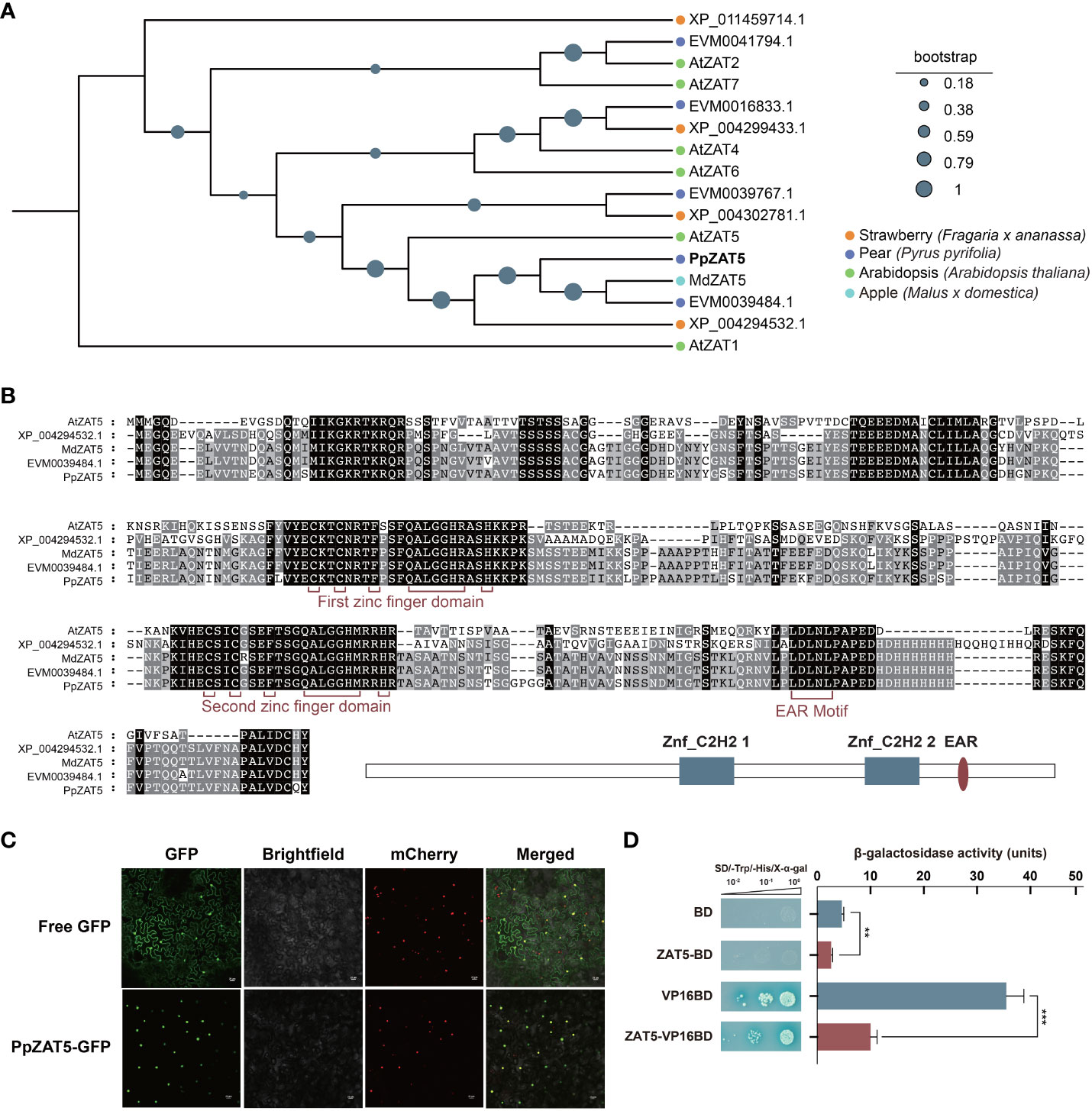
Figure 1 Bioinformatics analysis and trans-acting activity of PpZAT5. (A) Phylogenetic analysis of the C2H2-type zinc finger proteins from pear and other plant species obtained through blast algorithm using Arabidopsis ZAT1 to ZAT7 as queries. Pear (Pyrus pyrifolia); Apple (Malus x domestica): MdZAT5(MD03G1128800); Arabidopsis (Arabidopsis thaliana): AtZAT1(AT2G46800.1), AtZAT2(AT2G17180.1), AtZAT4(AT2G45120.1), AtZAT5(AT2G28200.1), AtZAT6(AT5G04340.1), AtZAT7(AT3G46090.1); Strawberry (Fragaria x ananassa). (B) Alignment of C2H2-type zinc finger proteins from pear, strawberry and apple. (C) Subcellular localization of PpZAT5 expressed in tobacco leaf cells. Scale bars = 25 μm. (D) Trans-acting activity of PpZAT5 transformed into yeast cells. The β-galactosidase activities reflected the trans-acting activities. Error bars for trans-acting activity represent the standard deviation of three independent experiments each. Asterisks indicate significant differences (two-tailed Student’s t-test, **P < 0.01, ***P < 0.001).
A subcellular localization assay of PpZAT5 showed that yellow fluorescent signals appeared only in the nuclei after transiently expressing PpZAT5-GFP fusion protein in the leaves of tobacco (Nicotiana benthamiana) plants with mCherry in the nucleus. In contrast, the fluorescence signal of GFP alone was observed throughout the cells following the expression of empty GFP protein (Figure 1C). We further analyzed the trans-acting activity of PpZAT5 in the yeast system. The AH109 yeast cells containing PpZAT5 fused to the DNA-binding domain (BD) failed to grow on the selective medium (SD/-Trp/-His) supplemented with X-α-gal, while the positive control with VP16-BD grew well and turned to blue. The fusion of PpZAT5 with the DNA-binding domain showed less β-galactosidase activity than that of the construct harboring only BD, and the positive control VP16BD showed higher β-galactosidase activity than the fusion of PpZAT5 and VP16BD (Figure 1D). Hence, PpZAT5 appears to be a nuclear-localized transcriptional repressor.
The expression pattern of PpZAT5 is negatively related to that of anthocyanin biosynthetic genes
Our previous transcriptome data of wild-type pear calli treated under continuously light conditions (Premathilake et al., 2020) revealed the low expressions of PpZAT5 after 12 and 48 h in contrast to the increased expressions of anthocyanin-related structural genes after 48 h (Supplementary Figure 1). To further explore the expression pattern of PpZAT5 under light conditions, the PpZAT5 expression was assessed in ‘Hongzaosu’ pear fruit under continuous light conditions for 240 h. The anthocyanin accumulation started after 72 h and was significantly enhanced until the end of the continuous light treatment (Figures 2A, B). Meanwhile, the expression levels of anthocyanin biosynthesis-related genes PpUFGT, PpANS, and PpBBX18 peaked at 12 h after starting the light exposure and then down-regulated soon after 24 h (Figure 2D), which was opposite to the expression of PpZAT5. PpZAT5 expression was down-regulated after 12 h from starting to expose continuous light and upregulated gradually after 24 h (Figure 2C). These results indicate that PpZAT5 responses to the light and might be related to light-induced anthocyanin accumulation.
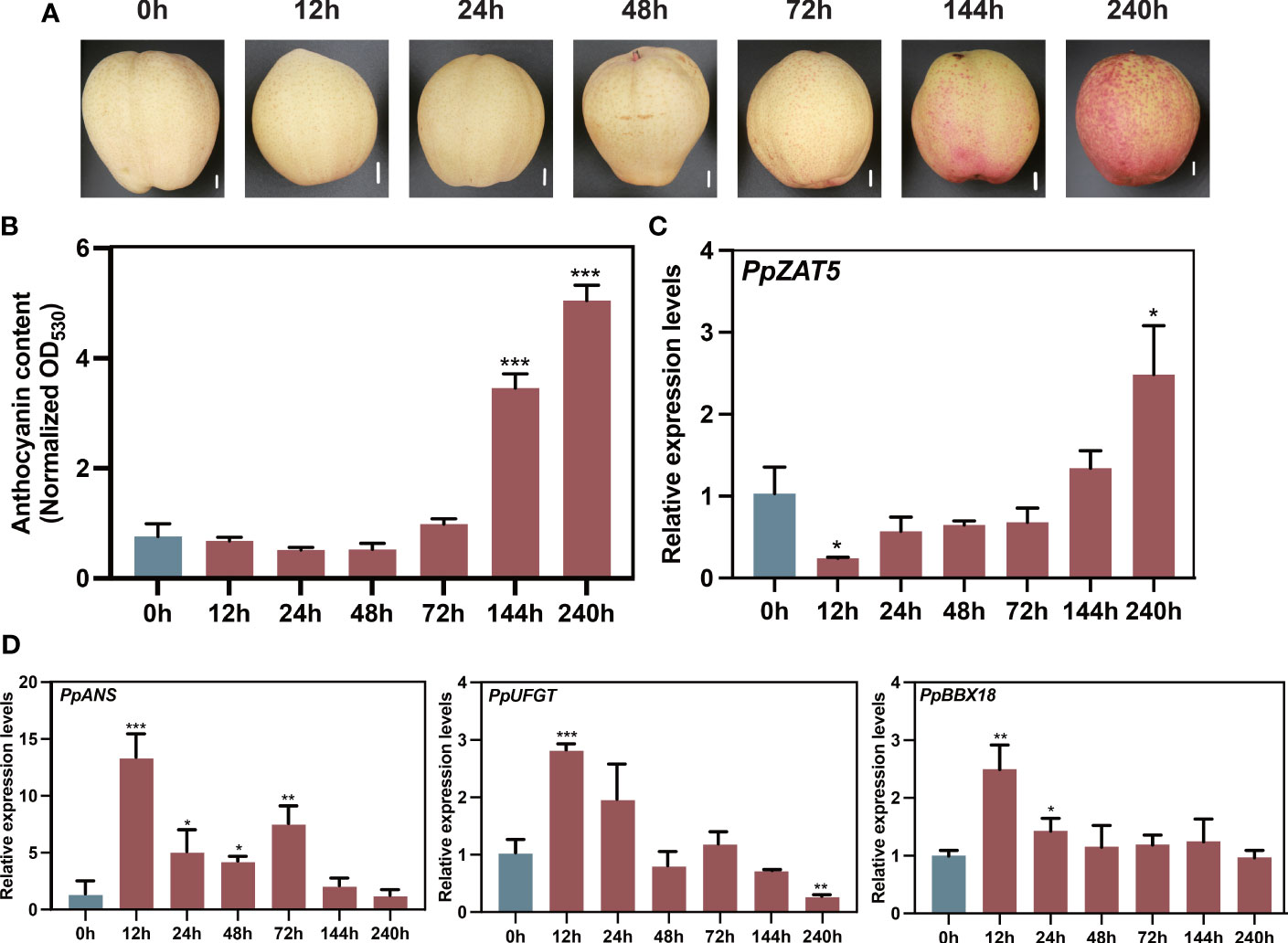
Figure 2 Light-responsive PpZAT5 expression. (A) Anthocyanin accumulation in the pear, ‘Hongzaosu’ fruit peel after the light treatment. Scale bars = 1 cm. (B) Changes in the fruit peel anthocyanin contents during the light treatment. (C) PpZAT5 expression pattern during light treatment. (D) PpANS, PpUFGT expression patterns during light treatment. Error bars for anthocyanin contents and expression levels represent the standard deviation of three independent experiments. Asterisks indicate significant differences (two-tailed Student’s t-test, *P < 0.05, **P < 0.01, ***P < 0.001).
PpZAT5 directly bound to the promoter of PpBBX18 and repressed the expression
Given that PpZAT5 was discovered by yeast one-hybrid library screening assay with PpBBX18 promoter as bait, we speculated that it might bind to the promoter of PpBBX18. To prove this speculation, four promoter fragments of PpBBX18 (ranging from 1174 bp to 455 bp) were constructed for activity analysis. The results showed that PpZAT5 could bind to promoter fragments of PpBBX18 from - 455 bp to 0 bp (Figure 3A). Then, we divided the 455 bp fragment into four parts, and the yeast one-hybrid assay result revealed that PpZAT5 could bind to the 58 bp DNA fragment from -312 bp to -254 bp (Figure 3B). Based on the 58 bp promoter fragment analysis, we found two CAAT-box motifs.
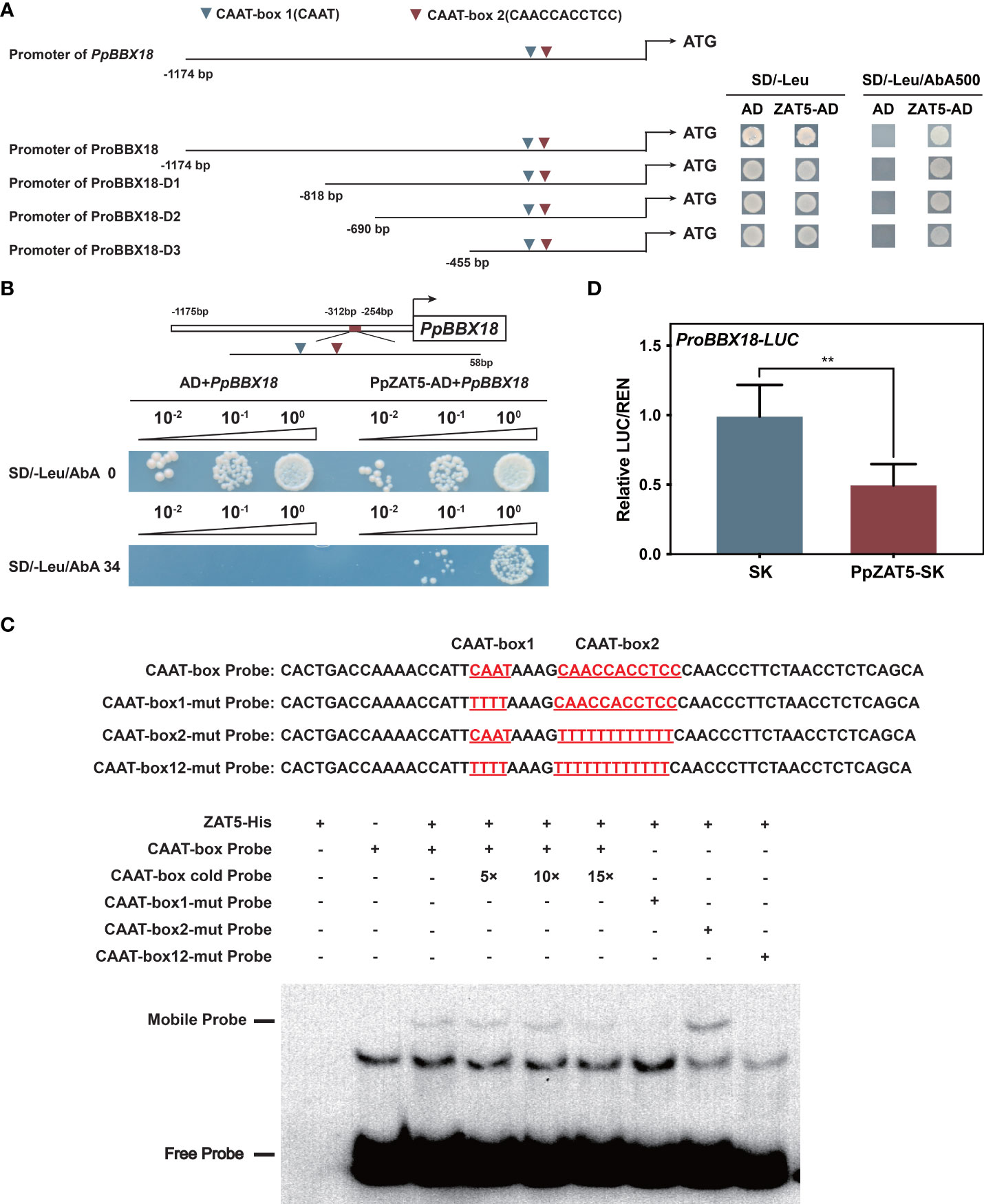
Figure 3 PpZAT5 was directly bound to the promoter of PpBBX18 and repressed its expression. (A) Yeast one-hybrid assays of PpZAT5 on a series of fragments of the PpBBX18 promoter. Red triangles indicate CAAT-box 1 (CAAT); Green triangles indicate CAAT-box 2 (CAACCACCTCC) (B) Yeast one-hybrid assays of PpZAT5 on a 58bp fragment of the PpBBX18 promoter, which contains two CAAT-box. (C) Direct binding of PpZAT5 to CAAT-box1 of the PpBBX18 promoter in vitro. (D) A dual-luciferase assay verified that PpZAT5 reduce PpBBX18 promoter activity. Error bars for dual-luciferase assays represent the standard deviation of three independent experiments each with six technical replicates. Asterisks indicate significant differences (two-tailed Student’s t-test, **P < 0.01).
We further verified the binding activity in electrophoretic mobility shift assay (EMSA) with the 58 bp fragment as a probe. We found that the combination band of PpZAT5 protein and the biotin-labeled probe is getting fainter with adding the cold probes, which confirmed that PpZAT5 could bind directly to the PpBBX18 promoter (Figure 3C). In addition, the PpZAT5 protein failed to bind to the PpBBX18 promoter with a mutation to CAAT-box 1 or both CAAT-box 1 and CAAT-box 2, but mutation to CAAT-box 2 could not influence the binding between them (Figure 3C), which verified that PpZAT5 protein could bind to CAAT-box 1 of PpBBX18 promoter. Furthermore, dual-luciferase assays showed that PpZAT5 significantly diminished transcriptional activation activity of PpBBX18 in contrast to the control, which demonstrated that PpZAT5 could repress PpBBX18 expression (Figure 3D). To sum up, PpZAT5 could bind to CAAT-box 1 of the PpBBX18 promoter directly and then inhibit the transcriptional activation activity of PpBBX18.
Involvement of PpZAT5 in the regulation of anthocyanin biosynthesis
To explore the function of PpZAT5 in anthocyanin biosynthesis, we generated a construct harboring the full-length coding sequence (CDS) of PpZAT5 under the control of the cauliflower mosaic virus 35S promoter (35S: PpZAT5). The construct was transformed into pear calli, while the wild-type pear calli were used as a control. Quantitative RT-PCR analysis showed that the expression levels of PpZAT5 were significantly high in transgenic calli lines, PpZAT5-OE #3, #6 (Figure 4A). Both fresh wild-type pear calli and PpZAT5-expressing transgenic calli lines were treated in continuous light conditions until they turned red after 5 days. We found that lower-level of anthocyanin accumulated in PpZAT5-expressing transgenic calli lines compared to wild-type pear calli after 5 days of light treatment and under dark, no anthocyanin was accumulated in both wild-type and overexpressed lines (Figures 4B, C). Meanwhile, we found that compared to wild-type pear calli, the expressions of anthocyanin biosynthesis structural genes PpF3H, PpANS, and PpUFGT and light-responsive PpBBX18 were endogenously down-regulated in PpZAT5-OE #3, #6 calli lines in contrast to wild-type calli (Figure 4D).
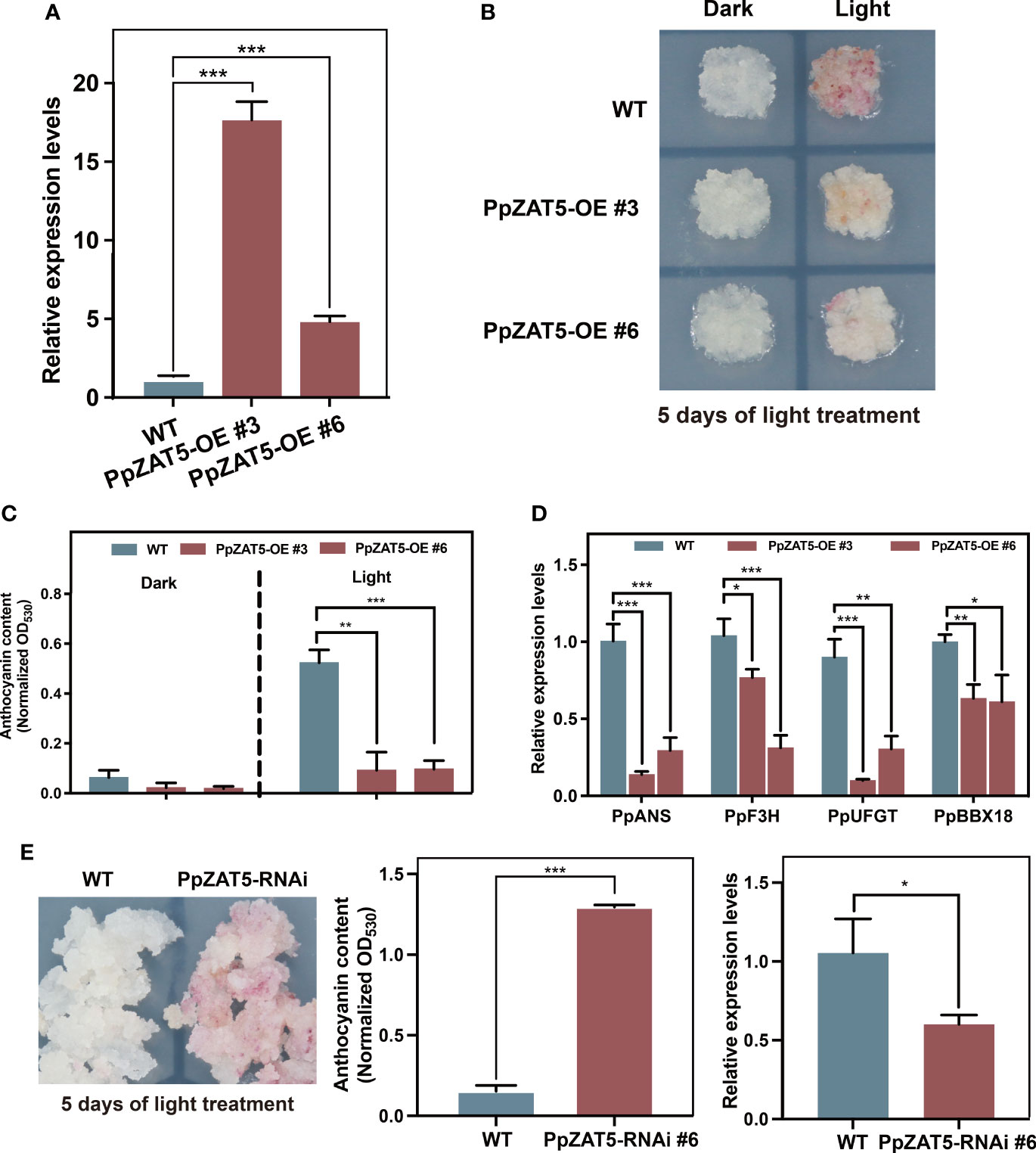
Figure 4 Overexpression and RNA interference (RNAi) of PpZAT5 in pear calli. (A) The expression level of PpZAT5 in overexpression pear calli. (B) Less anthocyanin accumulate in PpZAT5-OE pear calli. The pear calli were incubated under strong light at 17°C for 5 days. (C) Anthocyanin contents in PpZAT5-OE pear calli under dark and light treatment. (D) Expression levels of anthocyanin-related genes in pear calli under light treatment. (E) RNA interference (RNAi) of PpZAT5 promotes anthocyanin accumulation in pear calli. Error bars for anthocyanin contents and expression levels represent the standard deviation of three independent experiments. Asterisks indicate significant differences (two-tailed Student’s t-test, *P < 0.05, **P < 0.01, ***P < 0.001).
To validate, we also generated PpZAT5-RNA interference (RNAi) pear calli. In response to 5 days of continuous light treatment, more anthocyanin accumulated in the PpZAT5-RNAi than in the wild-type pear calli (Figure 4E). Moreover, to further prove the function of PpZAT5 in anthocyanin biosynthesis, we generated PpZAT5-overexpressing pear calli with the different expression vectors. The results showed that PpZAT5-overexpressing pear calli with expression vector pCAMBIA1301or pCAMBIA1300 both accumulated less anthocyanin than wild-type after 5 days of continuous light treatment (Supplementary Figure 2). Taken together, the results indicate that PpZAT5 regulates anthocyanin biosynthesis by inhibiting PpBBX18 transcription.
Transcriptome analysis of PpZAT5 overexpression pear calli
To further analyze the function of PpZAT5, transgenic calli (35S: PpZAT5) and wild-type calli were treated under dark and continuous light conditions for 5 days. Then the wild-type and PpZAT5-overexpression calli were subjected to RNA-Seq analysis. The results showed that light treatment induced the transcription changes of more than 8000 genes, among which 4005 genes overlapped with PpZAT5-overexpression calli samples. On the other hand, 4895 genes were identified as light-induced differentially expressed genes in PpZAT5-overexpression calli specifically, which might be induced by light depending on PpZAT5 (Figure 5A). The KEGG enrichment analysis was carried out in the differential expression genes of wild-type and PpZAT5-overexpression calli under light treatment. The results showed that related biological-related pathways of anthocyanin biosynthesis had relatively high P-values, such as flavonoid biosynthesis pathway, phenylpropanoid biosynthesis pathway, transcription factors, and others (Figure 5B). In addition, the crucial genes involved in light-induced anthocyanin biosynthesis, including transcription factors such as PpBBX18, PpBBX16, PpbHLH3, PpbHLH33, PpMYB10, and structure genes such as PpCHS, PpCHI, PpF3H, PpDFR, PpANS were markedly down-regulated in the transcriptome of PpZAT5 overexpression pear calli comparing to wild-type (Figure 5C; Supplementary Table 1). In addition, transcriptome analysis showed differentially-expressed genes (DEGs) involved in the phytohormone pathway, including auxin, salicylic acid, and abscisic acid. Most of the DEGs were upregulated in PpZAT5 overexpressed pear calli under dark conditions, but no differences were found between wild-type and PpZAT5 overexpressed pear calli under light conditions (Figure 5D). These results further confirmed that PpZAT5 was involved in the anthocyanin biosynthesis pathways and might also regulate the phytohormone pathway/metabolomics.
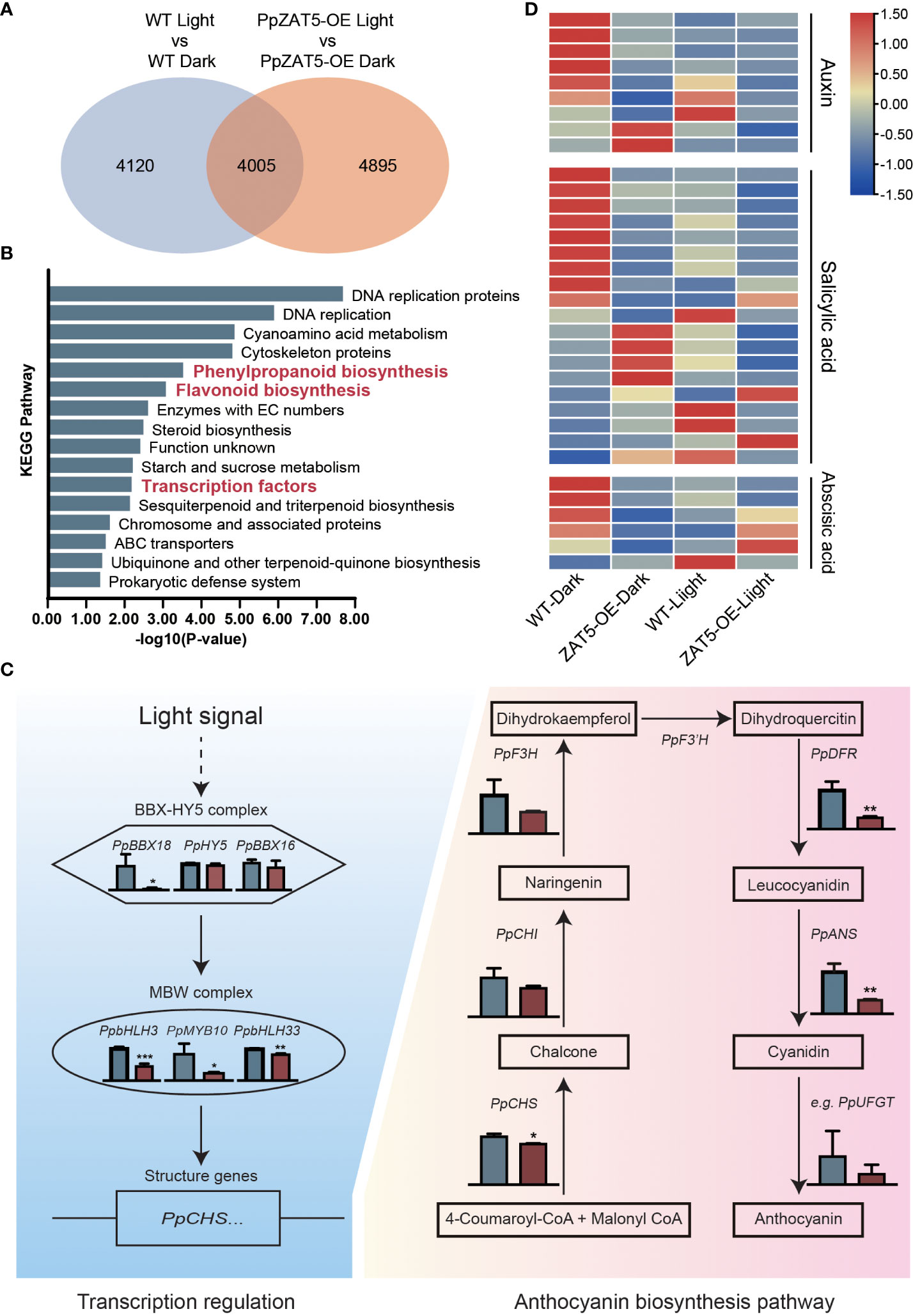
Figure 5 RNA-Seq analysis of PpZAT5 overexpression calli under light conditions. (A) The Venn graph of the numbers of differential expression genes between wild-type and PpZAT5 overexpression calli under dark/light conditions. (B) The enrichment analysis of KEGG pathways in the differential expression genes between wild-type and PpZAT5 overexpression calli under light condition. (C) FPKM value of genes involved in light-induced anthocyanin biosynthesis in wild-type and PpZAT5-OE pear calli under light treatment. Blue color represents wild-type pear calli, red color represents PpZAT5 overexpression pear calli. (D) The heatmap of differential expression genes involved in the phytohormone pathway between wild-type and PpZAT5 overexpression calli under dark and light conditions. Error bars for the FPKM value represent the standard deviation of three independent experiments. Asterisks indicate significant differences (two-tailed Student’s t-test, *P < 0.05, **P < 0.01, ***P < 0.001).
Discussion
PpZAT5 suppresses anthocyanin biosynthesis
Generally, C2H2 zinc finger proteins act as positive regulators in anthocyanin biosynthesis and respond to abiotic stress (Englbrecht et al., 2004; Ciftci-Yilmaz and Mittler, 2008). In Arabidopsis, SlZF2 (the ortholog of Arabidopsis ZAT10) was rapidly induced by abiotic stresses and overexpression of SlZF2 promoted anthocyanin accumulation (Hichri et al., 2014). AtZAT6 responding to hydrogen peroxide (H2O2), positively regulated oxidative stress-induced anthocyanin and total flavonoid biosynthesis (Shi et al., 2018). A recent study showed that MdZAT5 responded to various abiotic stresses and promoted anthocyanin biosynthesis by increasing the expressions of anthocyanin-related genes (Wang et al., 2022). Like AtZAT5, the genes mentioned above and PpZAT5 all belong to subgroup Cl-2i of C2H2 zinc finger proteins. However, our study showed that PpZAT5 functions as a transcriptional repressor to inhibit anthocyanin accumulation in pear (Figure 6). The phylogenetic analysis and multiple sequence alignment showed that MdZAT5 (MD03G1128800) is the homolog of another pear gene (EVM0039484.1), not PpZAT5 (EVM0021417.1) (Figures 1A, B). This gene is located on chromosome 3, while PpZAT5 is on chromosome 11 of pear (Pyrus pyrifolia). Functional verification tests revealed the inhibitory effects of PpZAT5 on anthocyanin biosynthesis in PpZAT5-overexpression and RNAi pear calli (Figure 4). Transcriptome analysis of the wild-type and PpZAT5-OE pear calli indicated that PpZAT5 modulates the expression of TF genes related to light-induced anthocyanin biosynthesis and structural genes of anthocyanin biosynthesis passway (Figure 5C). The inhibitory effect of PpZAT5 on anthocyanin accumulation might contribute to the counter-balancing mechanism of fine-tuning anthocyanin biosynthesis by cooperating with other positive regulators, which enriches the network of light-regulated anthocyanin biosynthesis.
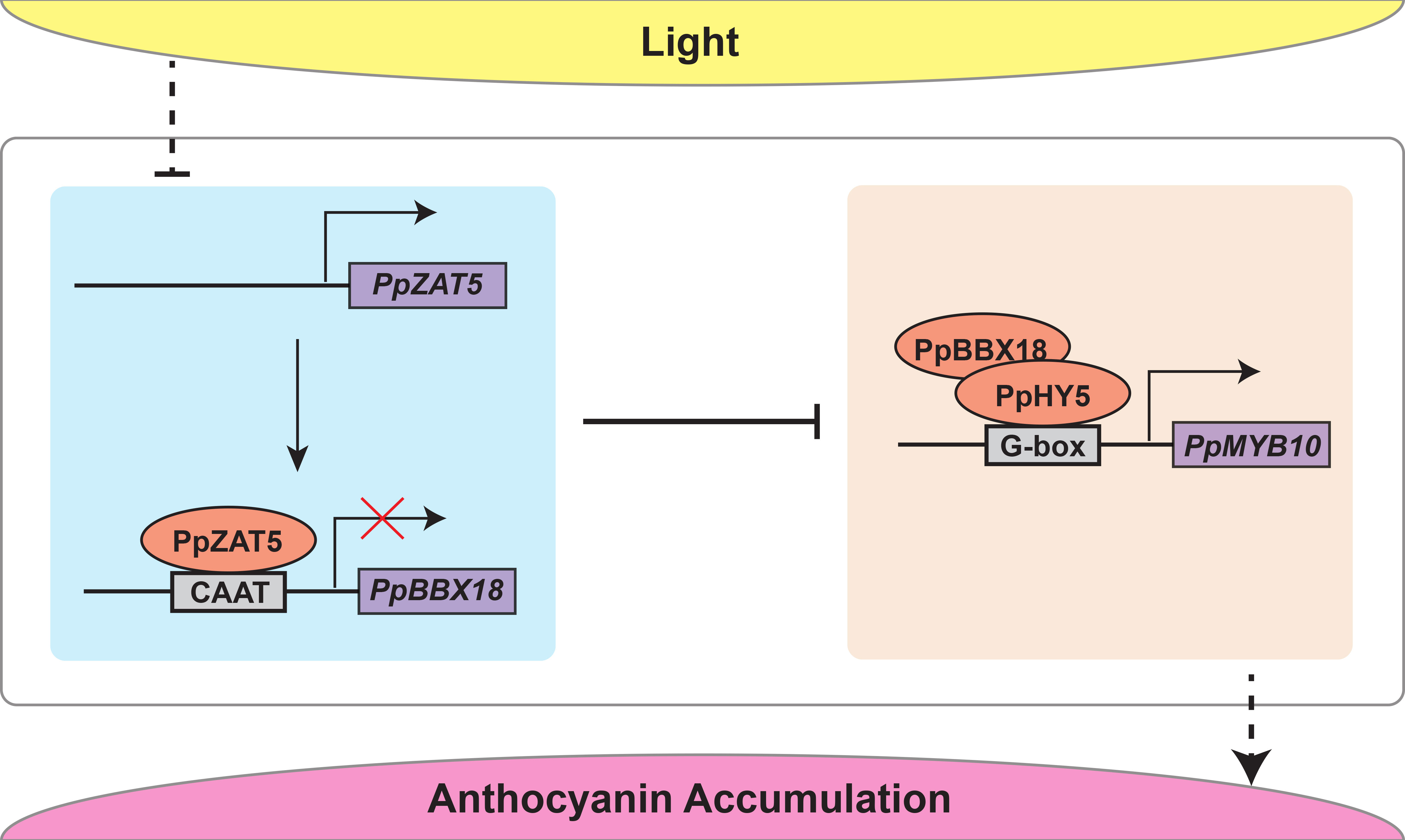
Figure 6 The proposed model indicates that under continuous light treatment, PpZAT5 downregulates the expression of PpBBX18 by directly bound to its promoter and ultimately represses the signaling of light-induced anthocyanin accumulation in pear.
Light promotes anthocyanin biosynthesis by inhibiting the expression of PpZAT5, a transcription repressor
The process of anthocyanin biosynthesis is fine-tuned. At present, the repressor MYB proteins and some BBX proteins have been studied a lot, they antagonize with positive regulators and fine-tune anthocyanin accumulation. In our study, the PpZAT5 expression pattern is converse to anthocyanin-related genes. When transferred to light conditions from dark, PpZAT5 expression was repressed at first and recovered gradually (Figure 2C), while anthocyanin-related genes were strongly activated early and gradually decreased (Figure 2D). Taking account of the interaction between PpZAT5 and PpBBX18 as well as their function, we speculated that the inhibitory effect of PpZAT5 on anthocyanin biosynthesis might be in the late period of light treatment and PpZAT5 is a braking gene in the process of anthocyanin biosynthesis.
PpZAT5 expression was repressed by light (Figure 2C), which disinhibited the effect on PpBBX18 and promoted anthocyanin biosynthesis. Interestingly, light inhibition on transcription repressors has not been reported, but inhibition of light on transcription activators is universal. In Arabidopsis, HDA19 and SNLs are the crucial negative regulators of the light signaling pathway, and their expressions are repressed by light (Jing et al., 2021). BR is one of the important inhibitory factors of photomorphogenesis, and light suppresses the expression of key BR biosynthesis genes. Their expressions were significantly lower in light-grown seedlings than in seedlings grown in the dark (Zhang et al., 2021). Light signal inhibits the DNA binding ability of ARF6 and ARF8 by regulating direct interaction between photoreceptor CRY1and phyB with ARF6 and ARF8. In addition, light signal inhibits the transcriptional activation of ARF by promoting the accumulation of Aux/IAA protein and the interaction between Aux/IAA and ARF to inhibit auxin signal, thus promoting photomorphogenesis (Mao et al., 2020). In our study, light repressed the expression of PpZAT5, and transcriptome analysis of PpZAT5 overexpression pear calli showed that it might participate in the phytohormone pathway, including auxin, salicylic acid, and abscisic acid, speculated that PpZAT5 might be repressed by light similarly.
PpZAT5 with EAR motif may act as a transcription repressor
Phylogenetic analysis showed that PpZAT5 is the homolog of Arabidopsis AtZAT5; meanwhile, multiple sequence alignment showed that PpZAT5 possesses two dispersed C2H2-type zinc fingers and an EAR motif related to repression activity (Figures 1A, B). EAR (ERF-associated amphiphilic repression) motif is the first active repressing motif found in plants and is in the C-terminal region of genes. It is the most representative transcriptional repression motif in plants (Ohta et al., 2001; Kagale et al., 2010). The repression capacity of NtERF3, an ERF protein from tobacco, is completely abolished when deleted C-terminal EAR motif region. Likewise, the deletion of C-terminal EAR motif regions eliminates the inhibitory effect of ZAT10 and ZAT11 on downstream genes in Arabidopsis (Ohta et al., 2001). A subset of zinc finger proteins (ZFPs) with EAR motifs have been identified as active repressors in various plant species (Kagale et al., 2010). Transcription repressors of C2H2-type zinc finger proteins containing EAR-motif at the C terminus act as a crucial negative regulator in the growth and development of plants (Singh et al., 2019). In Arabidopsis, AtZAT10 is induced rapidly and transiently by cold and then represses a classical stress-response gene RD29A transcription by binding to its promoter (Lee et al., 2002). AtZAT12 downregulated the expression of the CBF genes that play significant roles in the cold response pathway, indicating it is involved in a negative regulatory circuit to regulate cold tolerance of Arabidopsis (Vogel et al., 2004). Moreover, AtZAT12 loss-of-function lines are more resistant to heat stress, demonstrating that AtZAT12 functions as a repressor to influence the heat resistance of Arabidopsis (Davletova et al., 2005). In our study, PpZAT5 was localized at nuclear and showed strong trans-repression activity in yeast compared to the positive control VP16 (Figures 1C, D), which confirmed it functions as a transcription repressor in plant physiological metabolism. The transcriptional repressing activity of PpZAT5 might be due to the EAR motif at the C-terminal region.
PpZAT5 represses the expression of PpBBX18 by binding to the CAAT motif
Both internal and external factors influence anthocyanin biosynthesis in plants, and light was thought to be a vital external factor for anthocyanin biosynthesis (Sun et al., 2014; Bulgakov et al., 2017). A series of BBX proteins have been confirmed to regulate photomorphogenesis and light-induced anthocyanin biosynthesis pathways (Holtan et al., 2011; Xu et al., 2016; Zhang et al., 2017; Job et al., 2018). According to the number of B-box and CCT domains, BBX proteins can be divided into five types, from I to V in Arabidopsis (Khanna et al., 2009). Most BBX proteins of type IV are involved in HY5-dependent photomorphogenesis. HY5 has been shown to functionally interact with B-box-containing proteins such as BBX21-BBX22 and BBX24-BBX25 in Arabidopsis. BBX21 and BBX22 promote HY5 function in photomorphogenesis by binding to its promoter directly, while BBX24 and BBX25 suppress photomorphogenesis through direct physical interaction (Gangappa and Botto, 2016). Our previous study has proved that PpBBX18 could provide the trans-acting activity for PpHY5 as a result, positively regulated anthocyanin biosynthesis in pear (Bai et al., 2019b). In our study, dual-luciferase assays and yeast one-hybrid deletion assay showed that PpZAT5 repressed the expression of PpBBX18 by binding to a 58bp DNA fragment of PpBBX18 promoter (Figures 3A, B, D). Therefore, overexpression of PpZAT5 in pear calli inhibited anthocyanin accumulation while PpZAT5-RNAi pear calli accumulated more anthocyanin (Figure 4). These results indicate that PpZAT5 with the EAR motif suppresses anthocyanin biosynthesis by repressing the expression of PpBBX18.
The recognition mechanism of C2H2 zinc finger proteins to target DNA is not thoroughly studied. The function of C2H2 zinc finger proteins in regulating downstream genes expression began with the ZPT2-2 identified in petunia, and ZPT2-type proteins recognized two tandemly repeated AGT sequences separated by about 10 bp (Takatsuji et al., 1992; Takatsuji, 1998). AtZAT10 recognizes AG(C)T sequences separated by 3 bp or 4 bp (Sakamoto et al., 2004). In Arabidopsis, some C2H2-type zinc finger proteins bind to TACA(A/G)T element that is different from ZPT2-type proteins. Di19 (one C2H2 zinc-finger protein) could promote the expression of PR1, PR2, and PR5 by binding to the TACA(A/G)T element of their promoters, resulting in increased drought tolerance of Arabidopsis (Liu et al., 2013). AtZAT6 also binds to the TACAAT element of downstream gene promoters such as EDS1, PAD4, PR1, PR2, and PR5 and then increases their expression to modulate the biotic and abiotic stress responses (Shi et al., 2014). In our study, the electrophoretic mobility shift assay (EMSA) verified PpZAT5 binding to the CAAT-box 1 (CAAT) motif of the PpBBX18 promoter (Figure 3C). Compared to TACA(A/G)T, CAAT-box 1 (CAAT) has two fewer bases, which verified that the CAAT motif might be the core site of PpZAT5 binding to the PpBBX18 promoter. Our study gives possibilities for C2H2-type zinc finger proteins to regulate more potential downstream genes.
In a word, our previous studies have shown that PpBBX18 interacts with PpHY5 via two B-box domains and then forms a heterodimer, in which PpHY5 binds to the G-box motif of PpMYB10 and PpBBX18 provides the trans-acting activity, thus inducing transcription of PpMYB10 as well as anthocyanin biosynthesis (Bai et al., 2019b). In this study, we found that PpZAT5 inhibits anthocyanin biosynthesis by binding to CAAT motif of PpBBX18 promotor and then represses its expression (Figure 6).
Data availability statement
The datasets presented in this study are publicly available. This data can be found with the accession No. PRJNA871954 in SRA database of NCBI.
Author contributions
SB and YT conceived and supervised the research work. LZ performed most experiments and wrote the manuscript. RT started the experiments. SW helped to revise the manuscript. YG helped to analyze the RNA-Seq data. LW, SY, XZ, WY, XW, KL provided technical assistance. JN helped to write the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by Zhejiang Provincial Natural Science Foundation of China under Grant No. LR22C150001, the National Natural Science Foundation of China (32072545 to YT), and the China Agriculture Research System of MOF and MARA.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1022034/full#supplementary-material
References
Allan, A. C., Espley, R. V. (2018). MYBs drive novel consumer traits in fruits and vegetables. Trends Plant Science. 23, 693–705. doi: 10.1016/j.tplants.2018.06.001
An, J. P., Qu, F. J., Yao, J. F., Wang, X. N., You, C. X., Wang, X. F., et al. (2017). The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Horticulture. Res. 4, 17023. doi: 10.1038/hortres.2017.23
Bai, S. L., Tao, R. Y., Tang, Y. X., Yin, L., Ma, Y. J., Ni, J. B., et al. (2019a). BBX16, a b-box protein, positively regulates light-induced anthocyanin accumulation by activating MYB10 in red pear. Plant Biotechnol. J. 17, 1985–1997. doi: 10.1111/pbi.13114
Bai, S. L., Tao, R. Y., Yin, L., Ni, J. B., Yang, Q. S., Yan, X. H., et al. (2019b). Two b-box proteins, PpBBX18 and PpBBX21, antagonistically regulate anthocyanin biosynthesis via competitive association with Pyrus pyrifolia ELONGATED HYPOCOTYL 5 in the peel of pear fruit. Plant J. 100, 1208–1223. doi: 10.1111/tpj.14510
Ban, Y., Honda, C., Hatsuyama, Y., Igarashi, M., Bessho, H., Moriguchi, T. (2007). Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol. 48, 958–970. doi: 10.1093/pcp/pcm066
Borevitz, J. O., Xia, Y., Blount, J., Dixon, R. A., Lamb, C. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 12, 2383–2393. doi: 10.1105/tpc.12.12.2383
Bulgakov, V. P., Avramenko, T. V., Tsitsiashvili, G. S. (2017). Critical analysis of protein signaling networks involved in the regulation of plant secondary metabolism: focus on anthocyanins. Crit. Rev. Biotechnol. 37, 685–700. doi: 10.3109/07388551.2016.1141391
Chagne, D., Crowhurst, R. N., Pindo, M., Thrimawithana, A., Deng, C., Ireland, H., et al. (2014). The draft genome sequence of European pear (Pyrus communis l. 'Bartlett'). PLoS One 9, e92644. doi: 10.1371/journal.pone.0092644
Chang, C. S., Li, Y. H., Chen, L. T., Chen, W. C., Hsieh, W. P., Shin, J., et al. (2008). LZF1, a HY5-regulated transcriptional factor, functions in arabidopsis de-etiolation. Plant J. 54, 205–219. doi: 10.1111/j.1365-313X.2008.03401.x
Chan, Z. L., Wang, Q. F., Yang, L., Quan, W. L., Li, J. Z., Zhang, L. H., et al. (2017). The arabidopsis Cys2/His2 zinc finger transcription factor ZAT18 is a positive regulator of plant tolerance to drought stress. J. Exp. Botany. 68, 2991–3005. doi: 10.1093/jxb/erx157
Ciftci-Yilmaz, S., Mittler, R. (2008). The zinc finger network of plants. Cell. Mol. Life Sci. 65, 1150–1160. doi: 10.1007/s00018-007-7473-4
Davletova, S., Schlauch, K., Coutu, J., Mittler, R. (2005). The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in arabidopsis. Plant Physiol. 139, 847–856. doi: 10.1104/pp.105.068254
Englbrecht, C. C., Schoof, H., Bohm, S. (2004). Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics 5, 39. doi: 10.1186/1471-2164-5-39
Espley, R. V., Hellens, R. P., Putterill, J., Stevenson, D. E., Kutty-Amma, S., Allan, A. C. (2007). Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 49, 414–427. doi: 10.1111/j.1365-313X.2006.02964.x
Feng, S., Wang, Y., Yang, S., Xu, Y., Chen, X. (2010). Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta. 232, 245–255. doi: 10.1007/s00425-010-1170-5
Gangappa, S. N., Botto, J. F. (2016). The multifaceted roles of HY5 in plant growth and development. Mol. Plant 9, 1353–1365. doi: 10.1016/j.molp.2016.07.002
Hichri, I., Muhovski, Y., Zizkova, E., Dobrev, P. I., Franco-Zorrilla, J. M., Solano, R., et al. (2014). The Solanum lycopersicum zinc Finger2 cysteine-2/histidine-2 repressor-like transcription factor regulates development and tolerance to salinity in tomato and arabidopsis. Plant Physiol. 164, 1967–1990. doi: 10.1104/pp.113.225920
Holtan, H. E., Bandong, S., Marion, C. M., Adam, L., Tiwari, S., Shen, Y., et al. (2011). BBX32, an arabidopsis b-box protein, functions in light signaling by suppressing HY5-regulated gene expression and interacting with STH2/BBX21. Plant Physiol. 156, 2109–2123. doi: 10.1104/pp.111.177139
Jaakola, L. (2013). New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Science. 18, 477–483. doi: 10.1016/j.tplants.2013.06.003
Jing, Y., Guo, Q., Lin, R. (2021). The SNL-HDA19 histone deacetylase complex antagonizes HY5 activity to repress photomorphogenesis in arabidopsis. New Phytologist. 229, 3221–3236. doi: 10.1111/nph.17114
Job, N., Yadukrishnan, P., Bursch, K., Datta, S., Johansson, H. (2018). Two b-box proteins regulate photomorphogenesis by oppositely modulating HY5 through their diverse c-terminal domains. Plant Physiol. 176, 2963–2976. doi: 10.1104/pp.17.00856
Kagale, S., Links, M. G., Rozwadowski, K. (2010). Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in arabidopsis. Plant Physiol. 152, 1109–1134. doi: 10.1104/pp.109.151704
Khanna, R., Kronmiller, B., Maszle, D. R., Coupland, G., Holm, M., Mizuno, T., et al. (2009). The Arabidopsis b-box zinc finger family. Plant Cell. 21, 3416–3420. doi: 10.1105/tpc.109.069088
Koes, R., Verweij, W., Quattrocchio, F. (2005). Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Science. 10, 236–242. doi: 10.1016/j.tplants.2005.03.002
Kubo, K. I., Sakamoto, A., Kobayashi, A., Rybka, Z., Kanno, Y., Nakagawa, H., et al. (1998). Cys2/His2 zinc-finger protein family of petunia: Evolution and general mechanism of target-sequence recognition. Nucleic Acids Res. 26, 608–615. doi: 10.1093/nar/26.2.608
Lee, H., Guo, Y., Ohta, M., Xiong, L., Stevenson, B., Zhu, J. K. (2002). ). LOS2, a genetic locus required for cold-responsive gene transcription encodes a bi-functional enolase. EMBO J. 21, 2692–2702. doi: 10.1093/emboj/21.11.2692
Li, Y., Chu, Z., Luo, J., Zhou, Y., Cai, Y., Lu, Y., et al. (2018). The C2H2 zinc-finger protein SlZF3 regulates AsA synthesis and salt tolerance by interacting with CSN5B. Plant Biotechnol. J. 16, 1201–1213. doi: 10.1111/pbi.12863
Liu, W. X., Zhang, F. C., Zhang, W. Z., Song, L. F., Wu, W. H., Chen, Y. F. (2013). Arabidopsis Di19 functions as a transcription factor and modulates PR1, PR2, and PR5 expression in response to drought stress. Mol. Plant 6, 1487–1502. doi: 10.1093/mp/sst031
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Mao, Z., He, S., Xu, F., Wei, X., Jiang, L., Liu, Y., et al. (2020). Photoexcited CRY1 and phyB interact directly with ARF6 and ARF8 to regulate their DNA-binding activity and auxin-induced hypocotyl elongation in Arabidopsis. New Phytologist. 225, 848–865. doi: 10.1111/nph.16194
Ni, J. B., Premathilake, A. T., Gao, Y. H., Yu, W. J., Tao, R. Y., Teng, Y. W., et al. (2021). Ethylene-activated PpERF105 induces the expression of the repressor-type R2R3-MYB gene PpMYB140 to inhibit anthocyanin biosynthesis in red pear fruit. Plant J. 105, 167–181. doi: 10.1111/tpj.15049
Ni, J. B., Zhao, Y., Tao, R. Y., Yin, L., Gao, L., Strid, A., et al. (2020). Ethylene mediates the branching of the jasmonate-induced flavonoid biosynthesis pathway by suppressing anthocyanin biosynthesis in red Chinese pear fruits. Plant Biotechnol. J. 18, 1223–1240. doi: 10.1111/pbi.13287
Ohta, M., Matsui, K., Hiratsu, K., Shinshi, H., Ohme-Takagi, M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell. 13, 1959–1968. doi: 10.1105/tpc.010127
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and ballgown. Nat. Protoc. 11, 1650–1667. doi: 10.1038/nprot.2016.095
Premathilake, A. T., Ni, J. B., Shen, J. Q., Bai, S. L., Teng, Y. W. (2020). Transcriptome analysis provides new insights into the transcriptional regulation of methyl jasmonate-induced flavonoid biosynthesis in pear calli. BMC Plant Biol. 20, 388. doi: 10.1186/s12870-020-02606-x
Sakamoto, H., Maruyama, K., Sakuma, Y., Meshi, T., Iwabuchi, M., Shinozaki, K., et al. (2004). Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol. 136, 2734–2746. doi: 10.1104/pp.104.046599
Shi, H., Liu, G., Wei, Y., Chan, Z. (2018). The zinc-finger transcription factor ZAT6 is essential for hydrogen peroxide induction of anthocyanin synthesis in Arabidopsis. Plant Mol. Biol. 97, 165–176. doi: 10.1007/s11103-018-0730-0
Shimeld, S. M. (2008). C2H2 zinc finger genes of the gli, zic, KLF, SP, wilms' tumour, huckebein, snail, ovo, spalt, odd, blimp-1, fez and related gene families from Branchiostoma floridae. Dev. Genes And. Evolution. 218, 639–649. doi: 10.1007/s00427-008-0248-6
Shin, D. H., Choi, M., Kim, K., Bang, G., Cho, M., Choi, S. B., et al. (2013). HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Letters. 587, 1543–1547. doi: 10.1016/j.febslet.2013.03.037
Shi, H., Wang, X., Ye, T., Chen, F., Deng, J., Yang, P., et al. (2014). The Cysteine2/Histidine2-type transcription factor ZINC FINGER OF ARABIDOPSIS THALIANA6 modulates biotic and abiotic stress responses by activating salicylic acid-related genes and C-REPEAT-BINDING FACTOR genes in arabidopsis. Plant Physiol. 165, 1367–1379. doi: 10.1104/pp.114.242404
Singh, P., Mathew, I. E., Verma, A., Tyagi, A. K., Agarwal, P. (2019). Analysis of rice proteins with DLN repressor Motif/S. Int. J. Mol. Sci. 20, 1600–1614. doi: 10.3390/ijms20071600
Sun, Y. W., Qian, M. J., Wu, R. Y., Niu, Q. F., Teng, Y. W., Zhang, D. (2014). Postharvest pigmentation in red Chinese sand pears (Pyrus pyrifolia nakai) in response to optimum light and temperature. Postharvest. Biol. Technology. 91, 64–71. doi: 10.1016/j.postharvbio.2013.12.015
Takatsuji, H. (1998). Zinc-finger transcription factors in plants. Cell. Mol. Life Sci. 54, 582–596. doi: 10.1007/s000180050186
Takatsuji, H., Mori, M., Benfey, P. N., Ren, L., Chua, N. H. (1992). Characterization of a zinc finger DNA-binding protein expressed specifically in petunia petals and seedlings. EMBO J. 11, 241–249. doi: 10.1002/j.1460-2075.1992.tb05047.x
Takos, A. M., Jaffe, F. W., Jacob, S. R., Bogs, J., Robinson, S. P., Walker, A. R. (2006). Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 142, 1216–1232. doi: 10.1104/pp.106.088104
Vogel, J. T., Zarka, D. G., Van Buskirk, H. A., Fowler, S. G., Thomashow, M. F. (2004). Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of arabidopsis. Plant J. 41, 195–211. doi: 10.1111/j.1365-313X.2004.02288.x
Wang, D. R., Yang, K., Wang, X., Lin, X. L., Rui, L., Liu, H. F., et al. (2022). Overexpression of MdZAT5, an C2H2-type zinc finger protein, regulates anthocyanin accumulation and salt stress response in apple calli and Arabidopsis. Int. J. Mol. Sci. 23, 1897–1913. doi: 10.3390/ijms23031897
Winkel-Shirley, B. (2001). Flavonoid biosynthesis. a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126, 485–493. doi: 10.1104/pp.126.2.485
Xu, W., Dubos, C., Lepiniec, L. (2015). Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Science. 20, 176–185. doi: 10.1016/j.tplants.2014.12.001
Xu, D., Jiang, Y., Li, J., Lin, F., Holm, M., Deng, X. W. (2016). BBX21, an Arabidopsis b-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation. Proc. Natl. Acad. Sci. 113, 7655–7660. doi: 10.1073/pnas.1607687113
Yao, G., Ming, M., Allan, A. C., Gu, C., Li, L., Wu, X., et al. (2017). Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J. 92, 437–451. doi: 10.1111/tpj.13666
Zhang, X., Huai, J., Shang, F., Xu, G., Tang, W., Jing, Y., et al. (2017). A PIF1/PIF3-HY5-BBX23 transcription factor cascade affects photomorphogenesis. Plant Physiol. 174, 2487–2500. doi: 10.1104/pp.17.00418
Zhang, D., Qian, M. J., Yu, B., Teng, Y. W. (2013). Effect of fruit maturity on UV-b-induced post-harvest anthocyanin accumulation in red Chinese sand pear. Acta Physiologiae. Plantarum. 35, 2857–2866. doi: 10.1007/s11738-013-1319-5
Keywords: light, anthocyanin biosynthesis, PpBBX18, C2H2-type zinc finger proteins, PpZAT5, Pyrus pyrifolia, red pear
Citation: Zhang L, Tao R, Wang S, Gao Y, Wang L, Yang S, Zhang X, Yu W, Wu X, Li K, Ni J, Teng Y and Bai S (2022) PpZAT5 suppresses the expression of a B-box gene PpBBX18 to inhibit anthocyanin biosynthesis in the fruit peel of red pear. Front. Plant Sci. 13:1022034. doi: 10.3389/fpls.2022.1022034
Received: 18 August 2022; Accepted: 13 September 2022;
Published: 11 October 2022.
Edited by:
Serena Varotto, University of Padua, ItalyReviewed by:
Zhang Zongying, Shandong Agricultural University, ChinaYuan-Yuan Li, Shandong Agricultural University, China
Copyright © 2022 Zhang, Tao, Wang, Gao, Wang, Yang, Zhang, Yu, Wu, Li, Ni, Teng and Bai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuanwen Teng, eXd0ZW5nQHpqdS5lZHUuY24=; Songling Bai, c29uZ2xpbmdiYWlAemp1LmVkdS5jbg==
 Lu Zhang
Lu Zhang Ruiyan Tao1,2,3
Ruiyan Tao1,2,3 Shulin Yang
Shulin Yang Junbei Ni
Junbei Ni Yuanwen Teng
Yuanwen Teng Songling Bai
Songling Bai