
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 12 October 2022
Sec. Functional and Applied Plant Genomics
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1021669
This article is part of the Research TopicNew Germplasm Resources, Biotechnologies, and Their Applications in Vegetable ResearchView all 5 articles
Brassica oleracea is an economically important species, including seven cultivated variants. Agrobacterium-mediated transformation of B. oleracea crops, mainly via hypocotyl and cotyledon, has been achieved in the past. However, previously established transformation methods showed low efficiency, severe genotype limitation and a prolonged period for transformants acquisition, greatly restricting its application in functional genomic studies and crop improvement. In this study, we have compared the shoot regeneration and genetic transformation efficiency of hypocotyl, cotyledon petiole and curd peduncle explants from twelve genotypes of cauliflower and broccoli. Finally, an Agrobacterium-mediated transformation method using curd peduncle as explant was established, which is rapid, efficient, and amenable to high-throughput transformation and genome editing. The average genetic transformation efficiency of this method is stable up to 11.87% and was successfully implemented in twelve different genotypes of cauliflower and broccoli and other B. oleracea crops with low genotype dependence. Peduncle explants were found to contain abundant cambial cells with a strong cell division and shoot regeneration ability, which might be why this method achieved stable and high genetic transformation efficiency with almost no genotype dependence.
Brassica oleracea is an economically important and outcrossing species domesticated from wild cabbage as early as 2000 BCE and has been classified into many unique morphotypes, including cabbage (var. capitata), kale (var. acephala), brussels sprout (var. gemmifera), Chinese kale (var. albiflora), kohlrabi (var. caulorapa), broccoli (var. italica) and cauliflower (var. botrytis) (Smyth, 1995). The crops of B. oleracea are widely grown in China, India, Italy, and other countries located mainly in Asia and Europe (FAO, 2018), which are rich in nutrients including antioxidants, vitamin A, vitamin K, β-carotene, and sulforaphane glucosinolate with anti-cancer properties (Ahmed and Ali, 2013; Chaudhary et al., 2014).
The early domestication of B. oleracea crops is generally assumed to take place in the Mediterranean and, eventually spread to different countries of Asia, Europe and America through trade (Snogerup, 1980; Branca et al., 2012). Previous studies have shown that B. oleracea species have a low genetic diversity indicating a narrow genetic base (Branca et al., 2012; Rakshita et al., 2021). Environmental stresses, diseases and insects cause substantial yield and quality losses to B. oleracea crops. Genetic engineering, including targeted genome editing technologies, could overcome sexual incompatibility barriers and improve the efficiency of conventional breeding, so it is becoming one of the most important approaches to improve their resistance to biotic and abiotic stresses. Moreover, de novo genome sequencing and chromosome assembly of B. oleracea crops were successfully completed recently, which would facilitate the cloning of target genes controlling beneficial traits (Belser et al., 2018; Sun et al., 2019; Guo et al., 2021). However, the precondition for delivering genes and genome editing is the accessibility of efficient transformation systems.
Agrobacterium-mediated transformation method is popular for transferring genes within B. oleracea crops because of the high proportion of single copy and stable transformation efficiency (Ravanfar et al., 2017). The shoot regeneration studies in B. oleracea crops have been widely reported, mostly using hypocotyls, cotyledon petioles and true leaf blocks as explants (Bhalla and Smith, 1998; Ravanfar et al., 2009; Farzinebrahimi et al., 2012; Kumar et al., 2015a; Kumar et al., 2015b; Kumar and Srivastava, 2016). The Agrobacterium-mediated transformation has been achieved firstly with several reporter or marker genes (npt-II, gus and gfp) (Christey et al., 1997; Cogan et al., 2001; Bhalla and Singh, 2008; Dhiman et al., 2014; Ravanfar and Aziz, 2014), followed by some functional genes including flowering control by SLG, FCA, CONSTANS, insect resistance by cryIA(B), cry1A(C), and delayed post-harvest yellowing or prolonged shelf-life by SAD, ACC oxidase 1, BoCLH1 (Toriyama et al., 1991; Metz et al., 1995; Cao et al., 1999; Gapper et al., 2002; Viswakarma et al., 2004; Chen et al., 2008). However, in most of the published articles, the transformation has been met with limited success. Several drawbacks remain to be solved, such as low transformation efficiency, high dependence on genotype and a prolonged period of calli induction delaying resistant shoots regeneration, which hinders further development of this technology and also prevents its application in gene editing (Kumar and Srivastava, 2016).
The purpose of this study was to establish an efficient and universal Agrobacterium tumefaciens-mediated transformation system in B. oleracea crops, especially in cauliflower and broccoli. We evaluated the shoot regeneration efficiency in different explants, including hypocotyl, cotyledon petiole and curd peduncle from twelve different genotypic lines. Subsequently, we investigated the factors affecting Agrobacterium-mediated transformation efficiency, such as genotypes, explant types, antibiotics concentration and infection duration. The optimized peduncle-based system produced a stable transformation efficiency with an average of 11.87% with almost no genotype dependence. The regenerated transgenic plants still retained the memory of passing vernalization and could flower without an additional process of vernalization, which greatly accelerated the acquisition of homozygous transgenic plants of the T1 generation. Moreover, this system was also successfully applied to other B. oleracea crops, including cabbage and Chinese kale. Overall, the system developed in the present study can be applied in functional genomics research and traits improvement of cauliflower and broccoli, which can also be extended to other B. oleracea crops, such as Chinese kale and cabbage.
Twelve pure lines of cauliflower and broccoli, including DH lines (developed through microspore culture) and inbred lines (F6-8) (Table S1) were used in the present study. The seeds of these pure lines were collected from the Institute of Vegetables of Zhejiang Academy of Agricultural Sciences, Hangzhou, Zhejiang Province, which has been engaged in the breeding of cauliflower and broccoli for more than fifteen years. For in vitro germination, seeds were surface-sterilized in 70% ethanol for 40 s and then immersed in 3% sodium hypochlorite for 10 min, followed by rinsing 3-4 times with sterile ddH2O. The seeds were then placed on the MS medium added with 20 g sucrose and 9 g agarose in one liter of solution (pH 5.8-6.0) and incubated at 22/20°C (day/night), a light cycle of 14/10 h (day/night), and light intensity of approximately 100 μmol m–2 s–1. After six days of cultivation, the explants of hypocotyls and cotyledons with petioles were cut off and used for Agrobacterium-mediated transformation. For harvesting the peduncles explants, seeds were directly sown in a multicellular tray filled with substrate and then transplanted into the soil condition in a greenhouse at the seedling stage with 3-4 true leaves. When the flower head matured, peduncles were harvested and surface-sterilized, as mentioned above. The tested cabbage and Chinese kale materials are bought from the market, named Zhonggan 21 and J1401, respectively.
Agrobacterium tumefaciens strain GV3101 harboring plasmid of pCambia1301 was used in this study. The BoTFL1 gene highly expressed in the curd development stage of cauliflower was selected to construct the target vector (Sheng et al., 2020). The coding region of BoTFL1 was amplified using cDNA generated from the curd of cauliflower (with primers of KpnI-F and KpnI-R) and was then sub-cloned into the binary plasmid vector of pCambia1301 carrying hygromycin phosphotransferase (HPT), neomycin phosphotransferase II (nptII) and GUS gene (Table S2).
Three types of explants including, hypocotyl, cotyledon petiole and peduncle, were pre-cultured for 5 d in MS1-1and MS1-2 medium, respectively, at the temperature of 22/20°C (day/night), a light cycle of 14/10 h (day/night), and light intensity of approximately 100 μmol m–2 s–1. Then, all the explants were infected by the plasmid-harboring Agrobacterium with a bacterial concentration of OD600 = 0.8-1.0 for different time duration. After 2 days of co-culture in the dark on MS1-1and 2 medium, all the explants were transferred to MS2-1 and 2 medium for 7 d of recovery culture, respectively. After that, all the explants were transferred to the selection medium of MS3-1 and 2 medium, respectively, and the mediums were renewed every fifteen days until the resistant shoots emerged. Then, the hygromycin-resistant plants were cut from the explants and transferred to an MS4 medium to induce root growth. The rooted plantlets were transplanted into a small nutrient bowl filled with substrate and cultured in an artificial climate chamber for two weeks with a temperature of 22/20°C (day/night), a light cycle of 14/10 h (day/night), and light intensity of approximately 100 μmol m–2 s–1. Then the plantlets were transplanted in the soil condition in a specially isolated greenhouse and continued to grow until the flowering and seed setting stage. The detailed components of all the above culture mediums are shown in Table S3.
Putative transformants were pre-screened by PCR. A common cetyltrimethylammonium bromide (CTAB) method was used to extract the genomic DNA of each hygromycin-resistant plant (Doyle, 1987). The constructed plasmid vector of pCambia1301 and untransformed wild-type plants were used as positive and negative controls, respectively. A specific primer set, forward 5’- CGATTGCGTCGCATCGACC -3’ and reverse 5’ - TTCTACAACCGGTCGCGGAG -3’, was selected to amplify a 0.58 kb fragment specific to the HPT gene. PCR was carried out in a total volume of 20 μL reaction mixture comprising 12.5 μL of the PCR mix (Takara, ExTaq), 1 µL of genomic DNA (50-100 ng/μL), 0.75 μL of each primer and 5 μL of double distilled sterile water. The PCR cycle conditions were as follows: 5 min, 94°C; 20 s, 94°C, 20 s, 58°C, and 30 s, 72°C for 30 cycles; and 10 min, 72°C. The amplified products of the HPT gene were electrophoresed in 1% agarose gels and viewed under a UV transilluminator.
The PCR-positive plants were selected for GUS staining assay for further confirmation of the insertion of the target T-DNA into the recipient genome. The untransformed plants were selected as the negative control. At the stage of in vitro culture, one leaf from each plantlet was sampled and immersed in a 2-mM X-Gluc solution (Leagene, DP0013) overnight at 30°C. At the curd maturity stage and blooming, the small branches, buds and flowers were cut and immersed in the same X-Gluc solution, which was vacuumed for 30 minutes (0.01 MPa), and then kept overnight at 30°C. After staining, the tissues were washed in 75% ethanol three to five times until no chlorophyll content was visible. The explants with at least one blue spot were determined to possess the transient GUS expression.
To further ascertain the microstructure of the explant transverse incision sites clearly, a cryo-SEM was applied (HITACHI S-3000N&Quorum PP3000T, Japan and England) according to the procedures presented by Kotula et al. (2019). After 30 minutes of Agrobacterium infection and one day of co-cultivation, the interface of explants was cross-sectioned and cryo-fixed in a carbon film copper grid with liquid nitrogen. The carrier grid with samples was then quickly delivered to a cryo-preparation chamber under vacuum. After that, the frozen samples were sublimated for 6 min at − 80°C and then delivered to an SEM chamber and conducted for microstructure observations.
The fresh explants were cut into approximately 1 cm long segments and then fixed in Farmer’s Fixative (3:1 ethanol: acetic acid) for 24 h at 4°C. The subsequent procedures of dehydration, embedment and section were conducted as described by Oi et al. (2022). Images were taken under a D72 light microscope (Olympus). Three biological replicates were performed for each explant.
Factors of the genotypes and explants were evaluated in this study. At least three biological replicates for each factor were analyzed, and each biological replicate consisted of 50-60 explants for shoot regeneration and 80-90 explants for Agrobacterium-mediated transformation. The SPSS software of version 16.0 was selected for statistical analysis of all the experimental data in this study, and the significant differences among treatments were set at P < 0.05.
Two accessions (C90IL-9 of cauliflower and B80IL-2 of broccoli) were selected to evaluate the shoot regeneration efficiency of three types of explant, including hypocotyl, cotyledon petiole and curd peduncle. Hypocotyl and cotyledon petiole were excised from seedlings grown on a sterile medium for 6 d after seed sterilization (Figure 1A). The curd peduncle was taken from the mature curd of cauliflower and broccoli. After sterilization, the curd peduncles were cut into small segments of about 0.5 cm long and placed on the shoot induction medium. According to the diameter of the peduncles cross-section, they were divided into four grades; 0.1-0.2 cm (grade 1), 0.3-0.5 cm (grade 2), 0.6-0.8 cm (grade 3) and 0.9-1.3 cm (grade 4) (Figures 1B, C). The hypocotyls and cotyledon petioles were cultivated on callus induction medium for 21 d and then cultured in regeneration medium for another 30-40 d until successful shoot regeneration (Figure 1A). For the accession of C90IL-9, the shoot regeneration efficiencies of the hypocotyl and cotyledon petiole explants were 85.13% and 80.69%, and they developed 3.52 and 5.35 shoots per explant, respectively. The shoot regeneration efficiencies of both explants from the accession of B80IL-2 were 90.16% and 80.52%, respectively, producing 3.82 and 6.17 shoots per explant (Table 1). The curd peduncle explants of both accessions cultured on a bud induction medium for 25-30 days can directly regenerate buds without callus induction (Figures 1B, C). The shoot regeneration efficiency of curd peduncles at grade 2 and grade 3 were higher, which could achieve more than 94% and produce 7.79 to 11.21 shoots per explant.
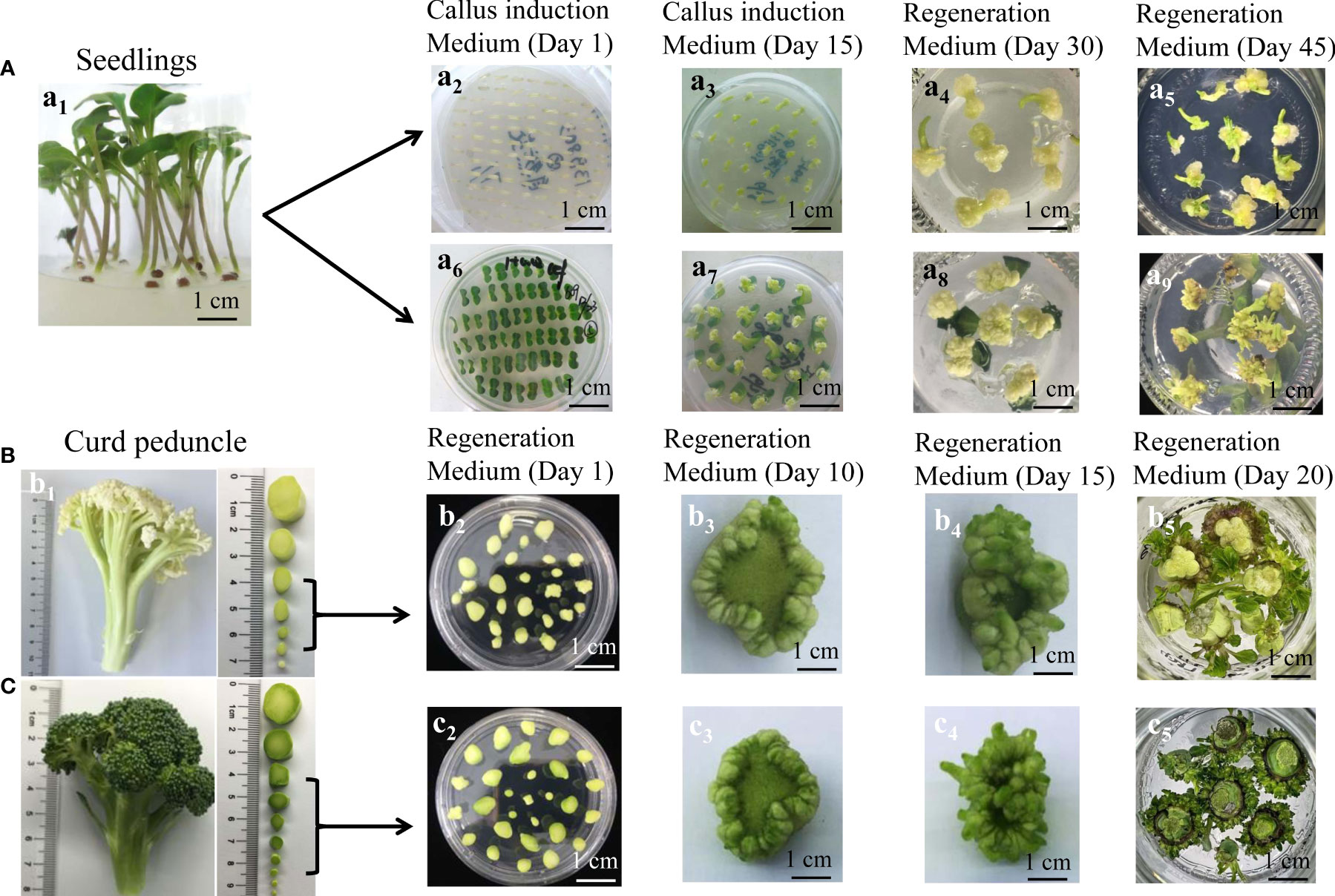
Figure 1 The shoot regeneration from different explants of C90IL-9 and B80IL-2. (A) shoot regeneration from hypocotyl and cotyledon petiole explants of C90IL-9. (A1), seedlings grown on sterile medium for 6 d after seed sterilization, Calli induction and shoot regeneration from hypocotyl (A2-5) and cotyledon petiole (A6-9) from day 1 to day 45. (B) and (C) shoots directly regenerated from curd peduncle explants. A branch from a mature curd of C90IL-9 (B1) and B80IL-2 (C1), respectively, and the peduncles with cross-sectional diameter of 0.3-0.8 cm were selected for shoot induction culture from day 1 to day 20 [(B2-5) of C90IL-9 and (C2-5) of B80IL-2].
Genotype dependence is the main limiting factor for successful shoot regeneration and Agrobacterium-mediated transformation in Brassica crops. In order to improve the universality of this protocol, in the present investigation, we have carried out the regeneration assays for twelve core breeding lines of broccoli and cauliflower with different ecotypes and genotypes. For the explants of hypocotyl and cotyledon petiole, significant differences were observed between the accessions. B80IL-2 attained the highest shoot regeneration efficiency of 90.09%, and C60DH-3 showed the lowest of 52.43%. Peduncle explants exhibited overall high regeneration efficiency ranging from 93.26% to 98.34%, with an average of 96.29%, with no significant differences among the accessions (Figure 2, Table S4). In general, achieving more than 50% regeneration efficiency is regarded as efficient in genetic transformation experiments. Therefore, the next step of Agrobacterium-mediated genetic transformation was carried out for all the genotypes and different explants so as to select the optimal combination of explant and genotype.
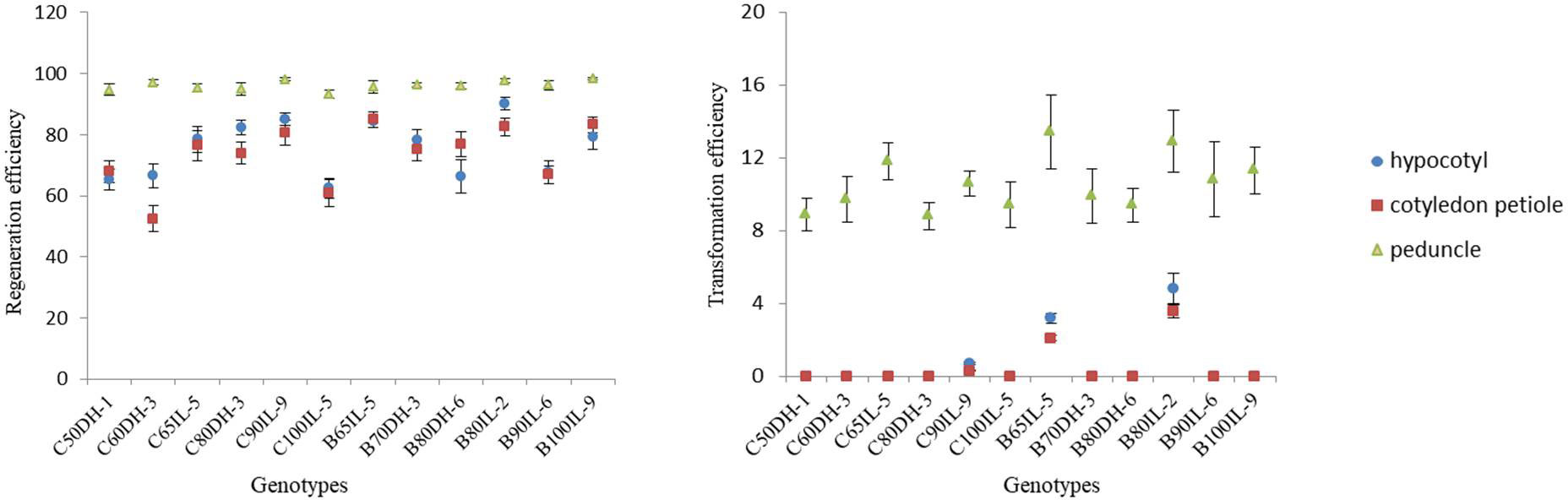
Figure 2 Effect of genotypes on the shoot regeneration and transformation efficiency of different explants. The regeneration and transformation rate were calculated as the percentage of the explants regenerated shoots and positive regenerated plants/total explants, respectively, for each genotype.
For Agrobacterium-mediated transformation, we selected the over-expression vector pCMABIA-1301 with the homologous gene of TERMINAL FLOWER 1 in cauliflower and broccoli (BoTFL1) and marker gene GUS, which are under the control of CaMV35S promoter. Three types of explants, including hypocotyl, cotyledon petiole and curd peduncle from the above twelve genotypes of cauliflower and broccoli, were used for Agrobacterium-mediated transformation. The tolerance of peduncle explants to Agrobacterium tumefaciens was significantly higher than that of hypocotyl and cotyledon. After 10 minutes of Agrobacterium infection, the browning rate of hypocotyl and cotyledon increased significantly and exceeded 46% after 20 minutes of immersion. The peduncle explants could withstand the infection time of 30 minutes, and then the browning rate increased with the extension of infection time. After 60 minutes of infection, the browning rate of the curd peduncle reached about 40% (Table S5). Therefore, the infection time of 10 minutes for hypocotyls and cotyledons and 30 minutes for peduncle were considered to be most suitable for all the genotypes. The regenerated buds from various explants of cauliflower and broccoli were sensitive to hygromycin, which at a concentration of 10 mg/L could inhibit more than 80% of the regeneration efficiency. The results of three independent genetic transformation experiments showed that over 60% of the survived plantlets were positive for both PCR and GUS activity under the selection of 10 mg/l hygromycin.
According to the genetic transformation process shown in Figure S1, the transgene-positive plants were obtained only from the hypocotyl and cotyledon petiole explants of the three genotypes, and the transformation efficiency was as low as 0.3-4.8%. Conversely, transgenics were achieved from peduncle explants of all the genotypes, and the average transformation efficiency ranged from 8.8 to 13.4%. Additionally, Agrobacterium-mediated transformation of hypocotyl and cotyledon petiole explants took a longer time, about 70-80 days, to obtain transgene-positive plants because of the requirement of a long recovery time after Agrobacterium infection and an extended period of callus induction and growth. In contrast, the peduncle explants could successfully regenerate transgene-positive plants within 35-40 days because of their strong tolerance to Agrobacterium tumefaciens, rapid recovery of post-infection, and the ability to regenerate buds directly with almost no callus induction.
A total of 38 and 56 lines of broccoli and cauliflower, respectively, with positive PCR and GUS activity, were transplanted in the soil condition (Figure 3A). During blooming, the second round of PCR and GUS activity detection tests were carried out to verify the authenticity of the transgenic plants. Interestingly, eight cauliflower plants lost the target band in PCR detection, whose leaves and buds also showed no GUS activity, indicating that they lost the target T-DNA during growth. The other 48 cauliflower plants preserved the target T-DNA insertion based on PCR and GUS staining results (Figures 3B-b1, C). However, all broccoli plants amplified the target band in the second round of PCR detection, and the sepals, petals and anthers showed strong GUS activity, confirming stable T-DNA insertion within the broccoli genome (Figures 3B-b2, D, E).

Figure 3 PCR analysis and histochemical GUS staining of transgenic plants at the stages of aseptic seedlings (A) and flowering after transplanted into soil condition in greenhouse (B–E). (A1) hygromycin resistant shoots, (A2) hygromycin resistant plants regenerated adventitious roots, a3, PCR analysis of transgenic plants using a pair of specific primers for HPT gene and a target band of 580 bp was amplified from the positive plants. Histochemical GUS staining for the leaves of transgenic seedlings and those with blue spots were positive plants (A4, A5). (B1) and (B2), the positive plants of cauliflower (B1) and broccoli (B2) identified at seedling stage were transplanted into the soil for the second round of PCR analysis. The second round of histochemical GUS staining analysis of cauliflower curd (C1, negative control, (C2-5) transgenic plants) and broccoli flowers (D1 and C1, negative control, D2-5 and E2-5, transgenic plants). w, water. NC, negative control. PC, positive control. RP, regenerated hygromycin resistant plants.
Peduncle explants are similar to young stems and have a nearly cylindrical shape. The section of the peduncle explant is nearly circular, and the circumferential layer near the epidermis contains a large number of meristem cells, which possess extremely strong meristematic ability (Figures 4A, B). Under hormonal stimulation, shoot emergence from the meristem cells can be seen at the incision site with the naked eye after 10 days of culture, and the whole plant can be regenerated after 25-30 days. After Agrobacterium tumefaciens infection, cambial cells were observed to have a strong recovery ability. Shoot regeneration was restored approx. 10 days after infection, and the resistant plants were regenerated 35-40 days after infection. Conversely, the microtubule tissue of hypocotyl and cotyledon petiole was underdeveloped. From the cross-sectional pictures, a large area of loosely arranged epidermal cells with large vacuoles and few vascular bundles in the center was observed (Figure 4C). Therefore, the direct regeneration ability of hypocotyl and cotyledon petiole was found to be poor, which required a process of callus induction to regenerate shoots.
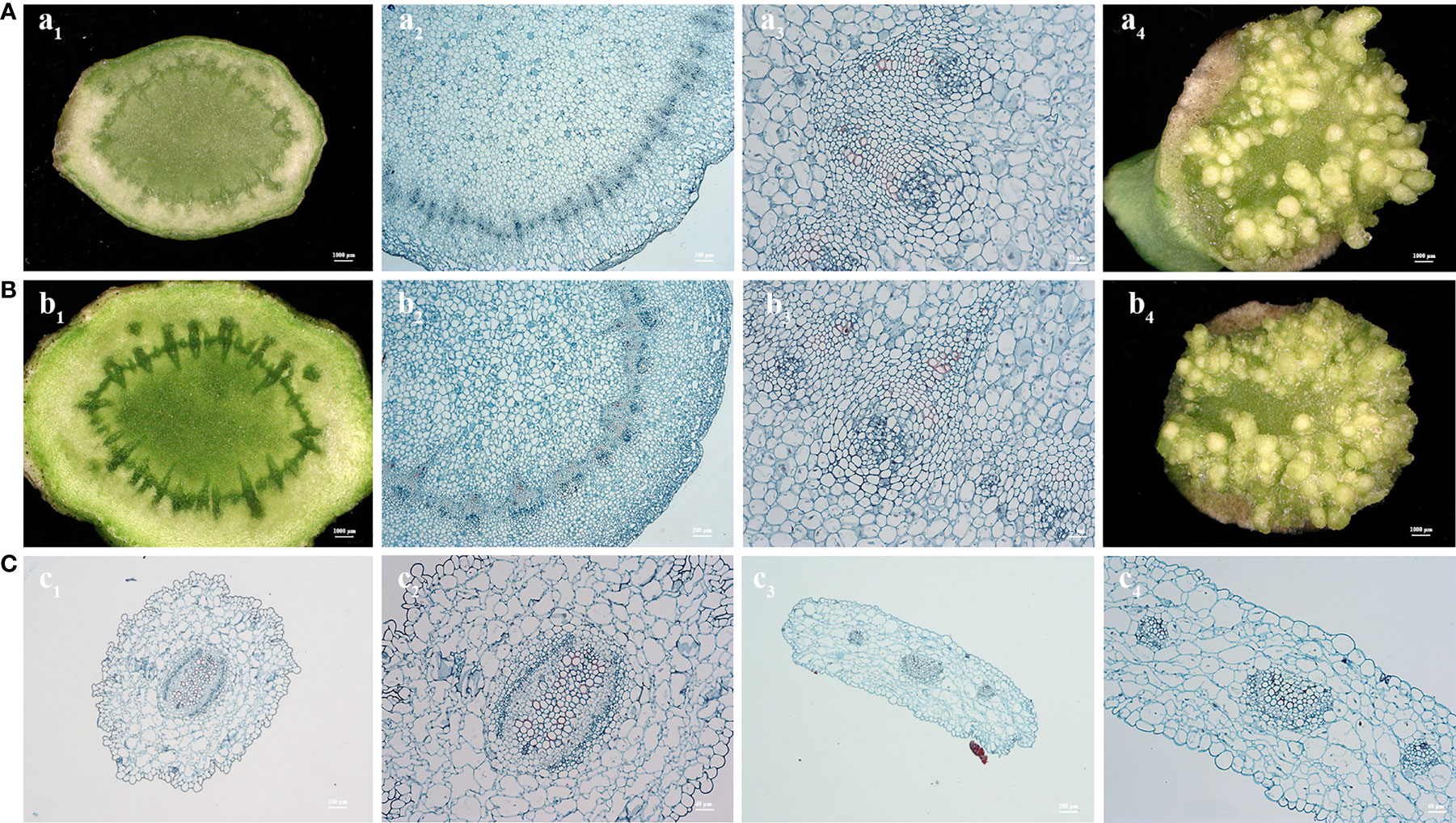
Figure 4 Cross section observation of paraffin embedded tissues from explants of curd peduncle (A, B), hypocotyl and cotyledon petiole (C). Cross section of fresh peduncle explants of C90IL-9 (A1-3) and B80IL-2 (B1-3). Shoot induction of peduncle of C90IL-9 (A4) and B80IL-2 (B4) after 10 days of culture in shoot induction medium. Cross section of fresh hypocotyl (C1-2) and cotyledon petiole (C3-4) explants of C90IL-9.
The cross-sectional diameter of the peduncle explant ranged from 0.3 to 0.8 cm, the area of which is much larger than that of the hypocotyl and cotyledon petiole (Figure 1). The results of freeze electron microscopy showed that the peduncle incision was normal after pre-culture, which was effective for full contact with Agrobacterium tumefaciens. After Agrobacterium infection and co-cultured for 1 day, a large number of Agrobacterium tumefaciens were found in the vascular bundle near the cambium cells of peduncle explants (Figure 5). This might have happened due to the release of chemotactic substances such as acetosyringone by peduncle explants inducing the accumulation of Agrobacterium tumefaciens, thereby increasing the transformation rate of incision cells. In contrast, the incisions of hypocotyl and cotyledon petiole were greatly shrunken after pre-culture, which largely reduced the contact area with Agrobacterium. In addition, no accumulation of Agrobacterium was found in the wounds of hypocotyl and cotyledon petiole explants (Figure 5).
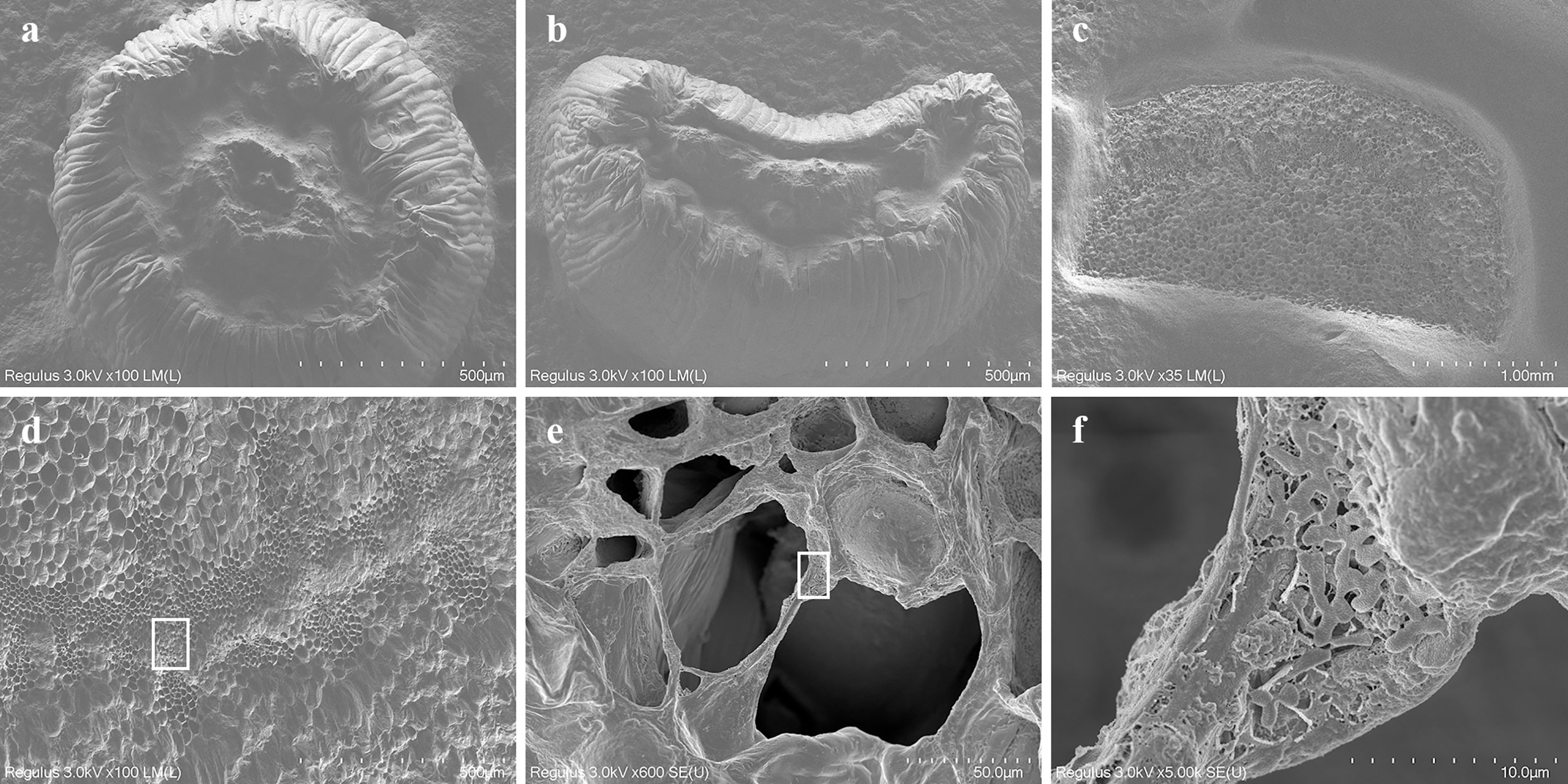
Figure 5 Cryo-electron microscopic observation of incisional tissues of three different explants. The incision morphology of hypocotyl (A), cotyledon petiole (B) and curd peduncle (C) after infected with Agrobacterium tumefaciens and co-cultured for one day. (D–F) incision morphology of peduncle explants under different magnification and many Agrobacterium tumefaciens were found in the vascular bundle near the cambium cells (F).
B. oleracea crops go through a typical type of vernalization process, in which the plants need to grow to the mature vegetative stage and then undergo a period of low-temperature treatment to promote flowering. Peduncle explants derived from the curds in the reproductive growth phase could remember their prior vernalization and then pass it on through mitosis. Therefore, the transgenic plants regenerated from the explants of the curd peduncle do not need to undergo the process of vernalization and can bloom as long as there are proper illumination and temperature. g acceFor the late maturinssions, the plants regenerated from peduncle explants started showing curds (cauliflower) and buds (broccoli) at the 9-10 leaf stage, and the flowering time was significantly earlier in these plants. Moreover, the regenerated plants of some early and medium maturing genotypes bloom directly in sterile culture bottles (Figure 6).

Figure 6 Transgenic plants regenerated from peduncle explants can directly grow curds and flower without going through low-temperature of vernalization. (A) resistant shoot screening of the peduncle explants from C60DH-3. (B, C) the regenerated plants of B65IL-5 (B) and C60DH-3 (C) budded in the tissue culture bottles. (D) transgenic plants of C90IL-9 were transplanted into soil, and the curd began to appear after 8-9 true leaves were grown. (E) transgenic plants of B100IL-9 were transplanted into soil and budded after growing 9-10 true leaves.
The Agrobacterium-mediated genetic transformation technique based on peduncle explants established in this study was further tested in cabbage and Chinese kale to determine the universality of this method in other B. oleracea vegetable crops. The young budding lateral branches were selected and excised into different segments with a diameter of 0.3-0.8 cm. The sterilization and transformation methods followed the procedures presented in Figure S1. Resistant buds were regenerated from the tender stem explants of cabbage approx. 30 days after Agrobacterium infection. After the PCR amplification using primers specific to HPT gene and GUS histochemical staining of the regenerated plants, fifteen transgene-positive plants were obtained from 147 tender stem explants, and the average genetic transformation efficiency was 10.21%. The degree of lignification of the tender stem of Chinese kale was higher, and the browning rate after Agrobacterium infection was a little more severe compared with those of cabbage. However, the regeneration of transgenic-positive plants was also successfully obtained by this method, and the genetic transformation efficiency was about 6.82% (Figure 7).
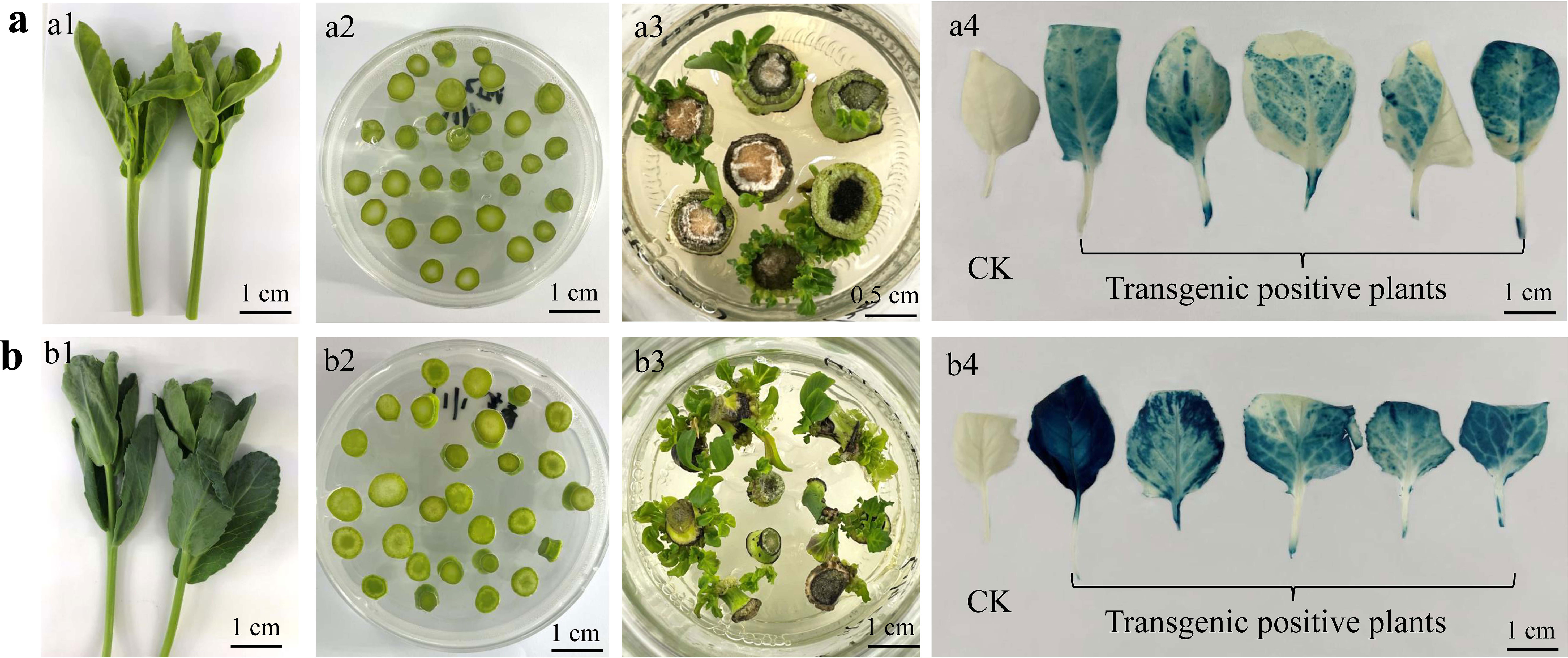
Figure 7 Transgenic plants regenerated from tender stems of lateral branches of Chinese kale (A) and cabbage (B). The tender stems of lateral branches (A1, B1), which were cut into segments with a diameter of 0.3-0.8 cm (A2, B2). Resistant buds were regenerated from the tender stem explants (A3, B3) and the histochemical GUS staining of the leaves from transgenic positive plants (A4, B4).
The tissue culture-based transformation technique has been widely used in gene editing and transgenesis to promote plant genomics studies. Although Brassica vegetables respond well to tissue culture and several published reports have demonstrated the successful regeneration of transformed plants in broccoli and cauliflower, the effectiveness of transformation is significantly correlated with the genotypes, and only a few genotypes have been reported to be successfully transformed (Chakrabarty et al., 2002; Chen et al., 2008; Ravanfar and Aziz, 2014). Moreover, many factors such as explant type, Agrobacterium concentration and immersion time also greatly influence the regeneration and transformation efficiency (Sharma and Srivastava, 2014; Kumar and Srivastava, 2016). Therefore, the establishment of an efficient, stable and general transformation system independent of genotype has always been one of the key points of genetic research in Brassica crops.
Hypocotyl and cotyledon petiole derived from sterile seedlings were the two most commonly used explants in genetic transformation research of B. oleracea crops, and some successful transformation events using the two types of explant have also been presented in several reports (Chakrabarty et al., 2002; Chen et al., 2008). However, these two types of explants-based genetic transformation systems showed strong genotype dependence, and the reproducibility of the experiments was not high (Chen et al., 2008). In general, genotypes with lower genetic transformation efficiency exhibited poorer experimental repeatability. The results of this study were in agreement with such an observation. Among the twelve genotypes, only three accessions regenerated transgene-positive plants with the transformation efficiency ranging from 0.3 to 4.8%. Additionally, hypocotyl and cotyledon petiole explants also require a process of dedifferentiation to form calli, which are then redifferentiated and eventually regenerate shoots (Wagoner et al., 1992; Suri et al., 2005). Dedifferentiation and redifferentiation phages of plant tissue culture is a long process, which greatly prolongs the time to regenerate positive plants and increases the frequency of potential somatic variants (Metz et al., 1995; Chen et al., 2001).
The peduncle explant used in the present study is a fleshy stem located below the inflorescence meristem and a small flower bud in the mature curd of cauliflower and broccoli. Timothy et al. (1995) and Chen et al. (2001) reported the successful regeneration of transformants using peduncle explant in broccoli, which exhibited higher genetic transformation efficiency than the hypocotyl explant. The findings of our study also confirmed the above observation. The peduncle explants from twelve genotypes of cauliflower and broccoli obtained up to 95% shoot regeneration efficiency, and all the accessions successfully regenerated transgenic plants with transformation efficiency ranging from 8.8 to 13.4%. Moreover, the peduncle-based genetic transformation technique established in this study is stable and can be applied to different genotypes of B. oleracea crops.
Peduncle explants, similar to young stems, contain typical tissues of the xylem, phloem and middle cambium. The cambium cells possess vigorous cell division and shoot regeneration ability (Liu et al., 2014; Alaguero-Cordovilla et al., 2021). In this study, we observed that many shoots have regenerated from the cambium cells of peduncle explants by direct organogenesis with no or few calli induction during the processes of Agrobacterium elimination and hygromycin selection. Due to the elimination of the callus induction process, the time required for shoot regeneration from peduncle explants was greatly shortened, and the vitrification rate and potential genomic variation rate of the regenerated shoots were lower than those of hypocotyl and cotyledon petiole explants (Chen et al., 2001). Moreover, peduncle explants express strong tolerance to Agrobacterium tumefaciens, which might be due to the support of developed vascular tissue, and the extension of Agrobacterium-infection time could improve the genetic transformation efficiency to a certain extent. Therefore, the developed cambium cells and microtubule tissue might be the reason for achieving high genetic transformation efficiency. These tissue-specific organ characteristics are less affected by genotype. Therefore, the peduncle explant-based transformation system established in this study could be applied to different genotypes of B. oleracea crops, including cauliflower and broccoli.
The species of B. oleracea includes seven cultivated variants. Since 2014, the whole genome sequences of cabbage (var. capitata) (Liu et al., 2014), kale (var. acephala) (Parkin et al., 2014), broccoli (var. italica) (Belser et al., 2018) and cauliflower (var. botrytis) (Sun et al., 2019; Guo et al., 2021), etc, have been published in succession, providing important references for the genetic localization and cloning of the target genes controlling important agronomic traits. Since then, the research on important gene mining and their regulatory mechanism of B. oleracea crops has entered a period of rapid development. A series of genetic loci or genes controlling plant growth, resistance to biotic and abiotic stresses, quality composition and fertility, etc., have been excavated (Li et al., 2021; Shaw et al., 2021; Zhang et al., 2021; Han et al., 2021; Ren et al., 2022; Alemán et al., 2022). Therefore, the effective transformation method established in this study will greatly accelerate the functional validation and regulatory mechanism study of the target genes controlling important agronomic traits of B. oleracea crops and promote rapid gene pyramiding to develop excellent target materials.
In conclusion, we have developed a stable, effective and genotype-independent Agrobacterium-mediated transformation system with the advantages of resistant shoots acquirement within a short period of 35-40 days, which could be successfully applied to different genotypes of broccoli and cauliflower, and even other species of B. oleracea. The established method has laid the foundation for further gene function studies through genetic transformation and mutant creation.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Conceptualization, HG and XS; methodology, XS and HY; software, YS; writing—original draft preparation, XS; writing—review and editing, XS, JW, and HY; supervision, HG; project administration, HG and JW. All authors have read and agreed to the published version of the manuscript.
This work was funded by the Science and Technology Department of Zhejiang Province for Key Agriculture Development (2021C02065-4-1, 2021C02065-4-4), the Key R&D Program in Zhejiang Province (2021C02042, 2022C02051), Natural Science Foundation of Zhejiang Province (LD22C150002) and Project of Zhejiang Academy of Agricultural Sciences.
We appreciate Dr. Sun Bo (College of Horticulture, Sichuan Agricultural University, China) for the guidance and help in the process of experiment.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1021669/full#supplementary-material
Ahmed, F. A., Ali, R. F. M. (2013). Bioactive compounds and antioxidant activity of fresh and processed white cauliflower. BioMed. Res. Int. 2013, 367819. doi: 10.1155/2013/367819
Alaguero-Cordovilla, A., Sánchez-García, A. B., Ibáñez, S., Albacete, A., Cano, A., Acosta, M., et al. (2021). An auxin-mediated regulatory framework for wound-induced adventitious root formation in tomato shoot explants. Plant Cell Environ. 44 (5), 1642–1662. doi: 10.1111/pce.14001
Alemán, B. J., Qin, J., Cai, C. C., Zou, C. M., Bucher, J., Paulo, M. J., et al. (2022). Genetic dissection of morphological variation in rosette leaves and leafy heads in cabbage (Brassica oleracea var. capitata). Theor. Appl. Genet. doi: 10.1007/s00122-022-04205-w
Belser , C., Istace, B., Denis, E., Dubarry, M., Baurens, F. C., Falentin, C., et al. (2018). Chromosome-scale assemblies of plant genomes using nanopore long reads and optical maps. Nat. Plants. 4, 879–887. doi: 10.1038/s41477-018-0289-4
Bhalla, P. L., Singh, M. B. (2008). Agrobacterium-mediated transformation of Brassica napus and Brassica oleracea. Nat. Protoc. 3, 181–189. doi: 10.1038/nprot.2007.527
Bhalla, P. L., Smith, N. A. (1998). Comparison of shoot regeneration potential from seedling explants of Australian cauliflower (Brassica oleracea var. botrytis) varieties. Aust. J. Agric. Res. 49, 1261–1266. doi: 10.1071/A98067
Branca, F., Argento, S., Tribulato, A. (2012). Assessing genetic reserves in Sicily (Italy): the Brassica wild relatives case study. Agrobiodiversity Conserv., 52–58. doi: 10.1079/9781845938512.0052
Cao, J., Tang, J. D., Strizhov, N., Shelton, A. M., Earle, E. D. (1999). Transgenic broccoli with high levels of bacillus thuringiensis Cry1C protein control diamondback moth larvae resistant to Cry1A or Cry1C. Mol. Breed. 5 (2), 131–141. doi: 10.1023/A:1009619924620
Chakrabarty, R., Viswakarma, N., Bhat, S. R., Kirti, P. B., Singh, B. D., Chopra, V. L. (2002). Agrobacterium-mediated transformation of cauliflower: optimization of protocol and development of bt-transgenic cauliflower. J. Biosci. 27, 495–502. doi: 10.1007/BF02705046
Chaudhary, A., Sharma, U., Vig, A. P., Singh, B., Arora, S. (2014). Free radical scavenging, antiproliferative activities and profiling of variations in the level of phytochemicals in different parts of broccoli (Brassica oleracea var. italica). Food Chem. 148, 373–380. doi: 10.1016/j.foodchem.2013.10.042
Chen, L. O., Chin, H. L., Kelkarc, S. M., Chang, Y. M., Shawa, J. F. (2008). Transgenic broccoli (Brassica oleracea l. var. italica) with antisense chlorophyllase (BOCLH1) delays post harvest yellowing. Plant Sci. 174, 25–31. doi: 10.1016/j.plantsci.2007.09.006
Chen, L. F., Hwang, J. Y., Charng, Y. Y., Sun, C. W., Yang, S. F. (2001). Transformation of broccoli (Brassica oleracea var. italica) with gene via agrobacterium tumefaciens for post-harvest yellowing retardation. Mol. Breed. 7, 243–257. doi: 10.1023/A:1011357320259
Christey, M. C., Sinclair, B. K., Braun, R. H., Wyke, L. (1997). Regeneration of transgenic vegetable brassicas (B. oleracea and b. campestris) via ri-mediated transformation. Plant Cell Rep. 16, 587–593. doi: 10.1007/BF01275497
Cogan, N., Harvey, E., Robinson, H., Lynn, J., Pin, D., Newbury, H. J., et al. (2001). The effects of anther culture and plant genetic background on Agrobacterium rhizogenes–mediated transformation of commercial cultivars and derived double haploid Brassica oleracea. Plant Cell Rep. 20, 755–762. doi: 10.1007/s00299-001-0395-y
Dhiman, K., Verma, S., Srivastava, D. K. (2014). Plant regeneration, genetic transformation and expression of gus gene in broccoli. Veg Sci. 41, 129–134.
Doyle, J. J. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bulletin. 19, 11–15.
FAO (2018) (Rome: Food and Agriculture Organization of the United Nations). Available at: http://faostat.fao.org (Accessed 15 May 2022).
Farzinebrahimi, R., Taha, R. M., Fadainasab, M., Mokhtar, S. (2012). In vitro plant regeneration, antioxidant and antibacterial studies on broccoli, Brassica oleracea var. italica. Pak J. Bot. 44, 2117–2122. doi: 10.1111/tpj.12008
Gapper, N. E., McKenzie, M. J., Christey, M. C., Braun, R. H., Coupe, S. A., Lill, R. E., et al. (2002). Agrobacterium tumefaciens-mediated transformation to alter ethylene and cytokinin biosynthesis in broccoli. Plant Cell Tissue Organ Cult. 70, 41–50. doi: 10.1023/A:1016057208267
Guo, N., Wang, S. Y., Gao, L., Liu, Y. M., Wang, X., Lai, E. H., et al. (2021). Genome sequencing sheds light on the contribution of structural variants to Brassica oleracea diversification. BMC Biol. 19, 93. doi: 10.1186/s12915-021-01031-2
Han, F. Q., Huang, J. J., Xie, Q., Liu, Y. M., Fang, Z. Y., Yang, L. M., et al. (2021). Genetic mapping and candidate gene identification of BoGL5, a gene essential for cuticular wax biosynthesis in broccoli. BMC Genomics 22 (1), 811. doi: 10.1186/s12864-021-08143-7
Kotula, L., Clode, P. L., de la Cruz Jimenez, J., Colmer, T. D. (2019). Salinity tolerance in chickpea is associated with the ability to ‘exclude’ Na from leaf mesophyll cells. J. Exp. Bot. 70, 4991–5002. doi: 10.1093/jxb/erz241
Kumar, P., Gambhir, G., Gaur, A., Srivastava, D. K. (2015b). Molecular analysis of genetic stability in tissue culture raised plants of broccoli (Brassica oleracea l. var. italica). Curr. Sci. 109, 1470–1475.
Kumar, P., Gaur, A., Srivastava, D. K. (2015a). Morphogenic response of leaf and petiole explants of broccoli using thidiazuron. J. Crop Improv. 29, 432–446. doi: 10.1080/15427528.2015.1046626
Kumar, P., Srivastava, D. K. (2016). Biotechnological advancement in genetic improvement of broccoli (Brassica oleracea l. var. italica), an important vegetable crop. Biotechnol. Lett. 38 (7), 1049–1063. doi: 10.1007/s10529-016-2080-9
Li, Z. S., Liu, Y. M., Yuan, S. X., Han, F. Q., Fang, Z. Y., Yang, L. M., et al. (2021). Fine mapping of the major QTLs for biochemical variation of sulforaphane in broccoli florets using a DH population. Sci. Rep. 11 (9004), 1–11. doi: 10.1038/s41598-021-88652-3
Liu, S., Liu, Y., Yang, X., Tong, C., Edwards, D., Parkin, I. A., et al. (2014). The brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 5 (1), 3930. doi: 10.1038/ncomms4930
Liu, B., Wang, L., Zhang, J., Li, J., Zheng, H. Q., Chen, J., et al. (2014). WUSCHEL-related homeobox genes in Populus tomentosa: diversified expression patterns and a functional similarity in adventitious root formation. BMC Genomics 15, 296. doi: 10.1186/1471-2164-15-2960
Metz, T. D., Dixit, R., Earle, E. D. (1995). Agrobacterium tumefaciens-mediated transformation of broccoli (Brassica oleracea var. italica) and cabbage (B. oleracea var. capitata). Plant Cell Rep. 15, 287–292. doi: 10.1007/BF00193738
Oi, T., Clode, P. L., Taniguchi, M., Colmer, T. D., Kotula, L. (2022). Salt tolerance in relation to elemental concentrations in leaf cell vacuoles and chloroplasts of a C4 monocotyledonous halophyte. Plant Cell Environ. 45 (5), 1490–1506. doi: 10.1111/pce.14279
Parkin, I. A., Koh, C., Tang, H. B., Robinson, S. J., Kagale, S., Clarke, W. E., et al. (2014). Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid brassica oleracea. Genome Biol. 15 (6), R77. doi: 10.1186/gb-2014-15-6-r77
Rakshita, K. N., Singh, S., Verma, V. K., Sharma, B. B., Saini, N., Iquebal, M. A., et al. (2021). Agro-morphological and molecular diversity in different maturity groups of Indian cauliflower (Brassica oleracea var. botrytis ). PloS One 16 (12), e0260246. doi: 10.1371/journal.pone.0260246
Ravanfar, S. A., Aziz, M. A. (2014). Shoot tip regeneration and optimization of Agrobacterium tumefaciens-mediated transformation of broccoli (Brassica oleracea var. italica). cv. green marvel. Plant Biotechnol. 9, 27–36. doi: 10.1007/s11816-014-0340-5
Ravanfar, S. A., Aziz, M. A., Kadir, M. A., Rashid, A. A., Sirchi, M. H. T. (2009). Plant regeneration of Brassica oleracea var. Italic (broccoli) cv. green marvel was affected by plant growth regulators. Afr J. Biotechnol. 8, 2523–2528. doi: 10.4314/ajb.v8i11.60750
Ravanfar, S. A., Orbovic, V., Moradpour, M., Abdul, A. M., Karan, R., Wallace, S., et al. (2017). Improvement of tissue culture, genetic transformation, and applications of biotechnology to Brassica. Biotechnol. Genet. Eng. Rev. 33, 1–25. doi: 10.1080/02648725.2017.1309821
Ren, W. J., Si, J. C., Chen, L., Fang, Z. Y., Zhuang, M., Lv, H. H., et al. (2022). Mechanism and utilization of ogura cytoplasmic male sterility in cruciferae crops. Int. J. Mol. Sci. 23 (16). doi: 10.3390/ijms23169099
Sharma, P., Srivastava, D. K. (2014). Effect of hygromycin on tomato cultures and its application in Agrobacterium–mediated tomato transformation. Crop Improv. 41, 43–49.
Shaw, R. K., Shen, Y. S., Zhao, Z. Q., Sheng, X. G., Wang, J. S., Yu, H. F., et al. (2021). Brassica oleracea molecular breeding strategy and challenges towards improvement of downy mildew resistance in cauliflower. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.667757
Sheng, X. G., Zhao, Z. Q., Wang, J. S., Yu, H. F., Shen, Y. S., Gu, H. H. (2020). Identification of Brassica oleracea orthologs of the PEBP family and their expression patterns in curd development and flowering in cauliflower. Biotechnol. Biotec Eq. 34 (1), 605–613. doi: 10.1080/13102818.2020.1790418
Smyth, D. R. (1995). Flower development: Origin of the cauliflower. Curr. Biol. 5 (4), 361–363. doi: 10.1016/S0960-9822(95)00072-8
Snogerup, S. (1980). The wild forms of the Brassica oleracea group (2n = 18) and their possible relations to the cultivated ones. Brassica Crops Wild allies. 1, 121–132.
Sun, D. L., Wang, C. G., Zhang, X. L., Zhang, W. L., Jiang, H. M., Yao, X. W., et al. (2019). Draft genome sequence of cauliflower (Brassica oleracea l. var. botrytis) provides new insights into the c genome in Brassica species. Hortic. Res. 6, 82. doi: 10.1038/s41438-019-0164-0
Suri, S. S., Saini, A. R. K., Ramawat, K. G. (2005). High frequency regeneration and Agrobacterium tumefaciens mediated transformation of broccoli (Brassica oleracea var. italica). Eur. J. Hort Sci. 70, 71–78.
Timothy, D. M., Richard, T. R., Juliet, D. T., Anthony, M. S., Elizabeth, D. E. (1995). Transgenic broccoli expressing a bacillus thuringiensis insecticidal crystal protein: implications for pest resistance management strategies. Mol. Breed. 4, 309–317. doi: 10.1007/BF01248408
Toriyama, K., Stein, J. C., Nasrallah, M. E., Nasrallah, J. B. (1991). Transformation of Brassica oleracea with an S-locus gene from B. campestris changes the self-incompatibility phenotype. Theor. Appl. Genet. 81, 769–776. doi: 10.1007/BF00224988
Viswakarma, N., Bhattacharya, R. C., Chakrabarty, R., Dargan, S., Bhat, S. R., Kirti, P. B., et al. (2004). Insect resistance of transgenic plants i.e. broccoli (‘Pusa broccoli KTS-1’) expressing a synthetic cryIA(b) gene. J. Hortic. Sci. Biotechnol. 79, 182–188. doi: 10.1080/14620316.2004.11511746
Wagoner, W. J., Kellogg, J. A., Bestwick, R. K., Stamp, J. A. (1992). Superior regeneration and Agrobacterium infectability of broccoli and cauliflower tissues and the identification of a procedure for the generation of transgenic plants. HortSci. 27, 620–621. doi: 10.21273/HORTSCI.27.6.620f
Keywords: Brassica oleracea, cauliflower, broccoli, curd peduncle explants, direct shoot regeneration, effective transformation
Citation: Sheng X, Yu H, Wang J, Shen Y and Gu H (2022) Establishment of a stable, effective and universal genetic transformation technique in the diverse species of Brassica oleracea. Front. Plant Sci. 13:1021669. doi: 10.3389/fpls.2022.1021669
Received: 17 August 2022; Accepted: 15 September 2022;
Published: 12 October 2022.
Edited by:
Mengyao Li, Sichuan Agricultural University, ChinaReviewed by:
Changwei Zhang, Nanjing Agricultural University, ChinaCopyright © 2022 Sheng, Yu, Wang, Shen and Gu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Honghui Gu, Z3VoaEB6YWFzLmFjLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.