
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 21 September 2022
Sec. Plant Biotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1021161
This article is part of the Research TopicMulti-omics Strategies to Analyze Complex Agronomic Traits in PlantsView all 11 articles
Bamboo shoot is one of nutritious vegetables in China. However, the edible quality of fresh bamboo shoots deteriorates easily after harvest. Here, morphological, physiological, transcriptomic and microRNA sequencing analyses were conducted to investigate the postharvest characteristics of moso bamboo (Phyllostachys edulis) shoots. Rapid decreases of soluble sugars, structural polysaccharides and hydrolyzed tannins, and increases of lignin and condensed tannins were observed in the postharvest bamboo shoots. Differentially expressed genes (DEGs) and miRNAs with opposite trends were mainly enriched in structural polysaccharide metabolism, starch and sucrose metabolism and glycolysis pathways, which were consistent with the changes of carbohydrates. A co-expression network of carbohydrate metabolism was constructed, which was verified by qPCR and yeast one-hybrid (Y1H) assay. Furthermore, the function of one hub glycosyltransferase gene was validated in Arabidopsis, which confirmed that it was involved in xylan biosynthesis. These results are of great significance for revealing the carbohydrate metabolism mechanisms of postharvest bamboo shoots and provide a potential candidate gene for molecular breeding related to xylan in the future.
Bamboo shoots, a kind of immature and expanding culms generated from the nodes of bamboo pseudo-rhizomes, are usually considered as a highly desirable vegetable because of their sweet taste, crisp and crunchy texture and excellent nutrition (Singhal et al., 2013). Carbohydrates in bamboo shoots are abundant and available in multiple forms, with the largest proportion of polysaccharides including starch as an energy source, and cellulose, and hemicellulose as dietary fibers. The dietary fiber of bamboo shoots accounts for 50% of carbohydrates (Wang et al., 2020), playing a critical role in hypolipidemic and anti-obesity activities as well as the taste (Tanabe et al., 2017). However, due to the perishability of bamboo shoots, the shelf life and market value of fresh bamboo shoots have been restricted. The soluble sugar content of bamboo shoots decreased significantly at the beginning of storage, e.g., the starch content of Fargesia yunnanensis shoots decreased significantly after 3 days of storage (Yu et al., 2021). Meanwhile, the changes in phenolic content of bamboo shoots led to browning and increased lignin content, along with the phenylalanine ammonia-lyase, and cinnamyl alcohol dehydrogenase activities increased rapidly (Li X. et al., 2019).
Therefore, several studies have focused on storage and food processing technology by both physical and chemical methods. In terms of physical methods, such as cooling, UV-C, gamma radiation and hypobaric atmosphere are often applied to extend the shelf life of bamboo shoots, which prolong the edible time of bamboo shoots by reducing the accumulation of lignin and chlorophyll (Wang et al., 2019; Wen et al., 2019). As for chemical methods, hormone treatment is commonly used to keep vegetable fresh. Exogenous melatonin and diphenyliodonium iodide treatment effectively delay the lignification of bamboo shoot and reduce the rate of yellowing during cold storage (Li C. et al., 2019; Li D. et al., 2019), while brassinolide treatment alleviates chilling injury of bamboo shoots by enhanced enzyme activities related to energy and proline metabolism (Liu et al., 2016). However, even low-temperature (LT) storage cannot stop the lignification process. Moreover, many techniques cannot be applied for ordinary households, especially bamboo shoot farmers living in remote mountainous areas who have difficulty using refrigerators. In view of the mechanisms underlying these physical and chemical methods, it suggests that the regulation of carbohydrate metabolism is important for affecting bamboo shoot edible quality and shelf life. Although similar biological processes occur in other harvested vegetables and fruit, and the effect of carbohydrate metabolism on quality is relatively clear, those in bamboo shoots are still unclear.
Structural polysaccharide metabolism, starch, and sucrose metabolism and glycolysis are the main carbohydrate metabolisms. Structural polysaccharides affected the juiciness and adhesion of cells, and promoted fruit soften or granulation. Propectin and cellulose showed significant increase in postharvest granulated orange, and the genes encoding enzymes responsible for pectin and cellulose, e.g., galacturonosyltransferase (GAUT) and cellulose synthase complex were up-regulated, while that of the gene encoding polygalacturonase responsible for pectin degradation showed significant decrease (Yao et al., 2020). In postharvest kiwifruits and persimmon fruit, the protopectin and cellulose content decreased while water-soluble pectin content increased progressively, accompanied by the increased activities of UDP-xylose 4-epimerase, UDP-Glc dehydrogenase, pectinesterase and cellulase (Tian et al., 2021). Genes encoding sucrose phosphate synthase (SPS), invertase (INV), and α-amylase (AMY) showed significantly differential expression in Powell (Citrus sinensis), fresh lotus seeds and kiwifruits during storage (Sun et al., 2021; Tian et al., 2021). Those genes encoding pyruvate kinase (PK) and phosphofructokinase related to glycolysis are highly variable during storage in many kinds of fruit and vegetables, such as sugarbeet roots, lotus seeds, and kiwifruits (Tian et al., 2021). Within sucrose and starch cycle, starch synthesis and degradation are simultaneously, during the preharvest period, taking place with net breakdown thereafter (Schouten et al., 2016). In addition, transcription factors (TFs) like bHLH, MYB, ERF, and LBD play important roles in regulating postharvest physiological changes (Wu et al., 2021).
Transcriptome sequencing is a powerful tool to rapidly obtain information on the expressed fraction of a genome, including the gene expression patterns, and the small ribonucleic acid (RNAs) of the targeted genes. The molecular mechanism of some physiological changes in postharvest bamboo shoots has been preliminarily analyzed, and the pathways of secondary cell walls and hormone biosynthesis are identified (Zhang et al., 2018). It was noticed that genes involved in lignin and hemicellulose biosynthesis were up-regulated and down-regulated, respectively (Li et al., 2018; Zhang et al., 2020). In addition, NAC, bHLH, bZIP, MYB, and WRKY were identified as critical TFs for shoot postharvest senescence, in which PheNAP2 and PheNAP3 were found to promote leaf senescence in Arabidopsis (Li X. et al., 2019). The expression of jasmonic acid (JA) biosynthesis related genes was highly consistent with that of lignin precursor biosynthesis genes under low temperature (LT), indicating a LT-lignification or LT-JA-lignification regulatory pathway existed in Lei bamboo shoots (Hou et al., 2022). However, the molecular mechanism affecting edible quality associated with carbohydrates in postharvest shoots is still unclear. In this study, the characteristics related to the main carbohydrate metabolism of postharvest moso bamboo shoots were revealed by integrated analyses of anatomical observation, chemical component and enzyme activity determination, transcriptomic and microRNA sequencing, which provided new insight into the modulation affecting the quality of postharvest moso bamboo shoots.
The moso bamboo (Phyllostachys edulis) shoots of 15 cm ∼ 20 cm in length and 5 cm ∼ 8 cm in basal diameter were harvested from a wild bamboo forest (28°45′58″N, 115°45′39″E, 399.0 m above sea level) in Nanchang, Jiangxi province, China, in December 2019. The shoots were stored under room temperature conditions (22 ± 2°C, RH = 75%) wrapped in paper bags. Middle part of the shoots in Figure 1A were collected at different storage time (S1, S2, S3, and S4 represented bamboo shoots stored for 0, 3, 6, and 12 d), one part of samples was immediately frozen in liquid nitrogen and then stored at −80°C, another part of samples was fixed with FAA and stored at 4°C until use, respectively. Three biological replicates were set for each time point. The total RNA and small RNA from 12 samples was isolated using the TRIzol and miRcute plant miRNA extraction and isolation reagent (item numbers DP504, TIANGEN Biotech Co., Ltd., Beijing), respectively.
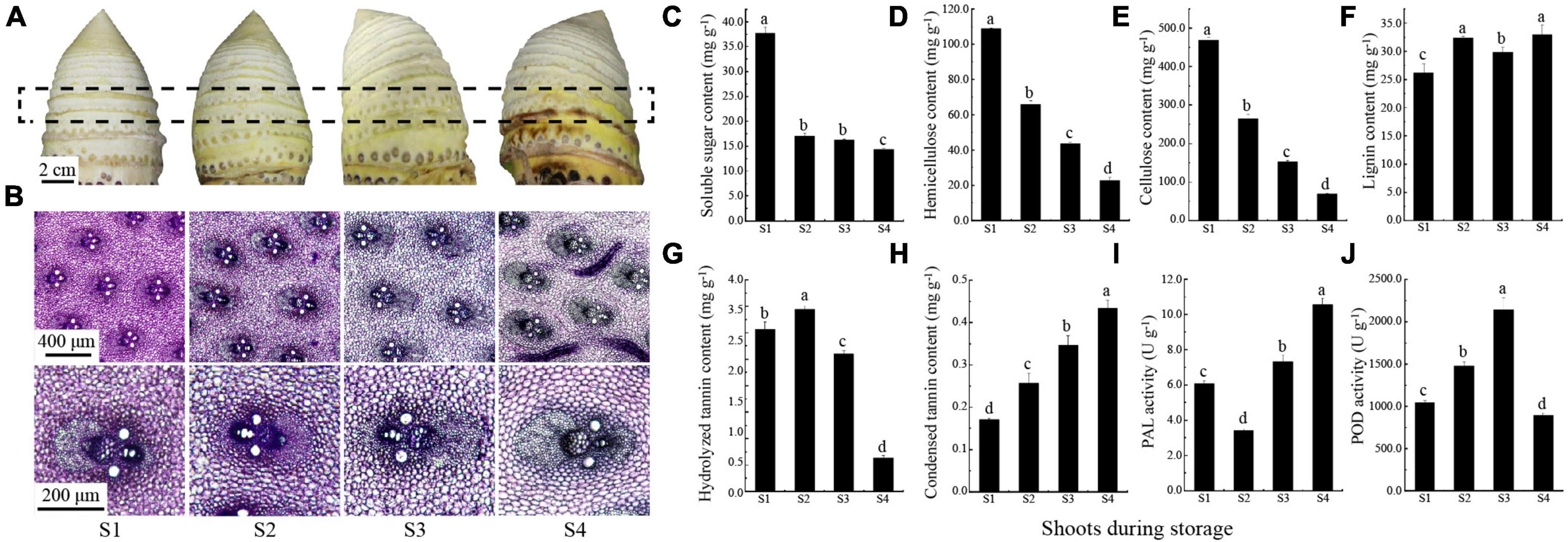
Figure 1. Changes of postharvest moso bamboo shoots. (A) Morphological changes of shoots after postharvest 0, 3, 6, and 12 d (S1: 0 d; S2: 3 d; S3: 6 d; S4: 12 d). (B) Staining intensity of vascular bundle transverse section of the boxed area in (A). (C–H) The content of soluble sugar (C), hemicellulose (D), cellulose (E), lignin (F), hydrolyzed tannins (G) and condensed tannins (H), the activities of PAL (I) and POD (J). Lowercase letters a, b, c, and d indicate significant differences (P < 0.05).
The shoot samples were stained with 5% (v/v) of toluidine blue (TBO), as described in the reference (Yang et al., 2021). The content of soluble sugar, cellulose, hemicellulose, lignin, hydrolyzed tannins, and condensed tannins, and the enzyme activity of peroxidase (POD) and L-phenylalanine ammonia lyase (PAL) were determined with detection kits (item numbers G0501, G0519, G0527, G0708, G0119, G0120, G0114, and G0107) of Grace Biotechnology Co., Ltd., JiangSu. The content of xylan was determined by immunofluorescence with detection kits (item numbers S0169O) of KETE Biotechnology Co., Ltd., Jiangsu. Three replicates were performed for each determination.
A total 12 transcriptome and 12 small RNA libraries were generated using NEBNext UltraTM RNA Library Prep Kit for Illumina (NEB, USA) and NEBNext Multiplex Small RNA Library Prep Set for Illumina (NEB, USA) following manufacturer’s recommendations, respectively. Illumina platform was used to sequence 12 high quality RNA libraries at Biomarker Technologies Corporation (Beijing, China). Low quality reads, reads with adaptors and reads with unknown bases were removed from the data sets to obtain the clean reads, which were then mapped to the reference genome of moso bamboo, followed by unigene expression level calculation with fragments per kilobase of transcript per million fragments mapped (FPKM) in each sample.
Differential expression analysis was performed using DESeq2 (Love et al., 2014), and genes with Fold Change (FC) ≥ 2 and False Discovery Rate (FDR) < 0.01 were assigned as differentially expressed (Tarazona et al., 2011). The putative functions of the differentially expressed genes (DEGs) and target genes were determined according to Non-redundant protein (NR) database. The cluster classification of DEGs based on expression of their transcriptome profiles were performed using OmicShare Tools1 with default parameters. The Gene Ontology (GO) enrichment analysis was carried out using R-based GOseq package to identify the over-represented functional categories, and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was implemented by KOBAS 3.0 (Xie et al., 2011). For co-expression network analysis, the weighted gene co-expression network analysis (WGCNA) package was used. Correlation analysis of gene expression was performed using programs in the BMKCloud.2 Based on the results of WGCNA, the genes in the module with the highest carbohydrate correlation were selected to construct the co-expression network. DEGs related to structural polysaccharide metabolism, starch, and sucrose metabolism and glycolysis were screened from this module by gene function annotation. The upstream sequences of these DEGs were further analyzed to identify potential regulatory TFs in turquoise module combining with biological information in PlantTFDB databases.3 The small RNA sequencing data from this study and psRNATarget database4 were used to identify potential regulatory miRNAs targeting these DEGs.
For Small RNA data analysis, clean reads of small RNA were obtained by removing reads containing adapter and ploy-N besides low-quality reads using Trimmomatic v0.35. After screening against reads shorter than 18 nt and longer than 35 nt, rRNA, tRNA, snoRNA, and repeat sequences, the redundant reads were processed into unique sequences with associated read counts for miRNA prediction by comparing with genome and known miRNAs from miRBase using Bowtie alignment tool v1.1.2 with two mismatches. Randfold tools software was used for novel miRNA secondary structure prediction. Subsequently, the mRNA targets of the identified miRNAs were predicted using psRNATarget server with default parameters. The annotation of these target genes refers to the method of DEG annotation in 2.3. The miRNAs expression determination and differentially expressed miRNAs (DEMs) analysis refers to the method of transcriptome analysis.
Primer Premier 5.0 software was used to design the specific primers based on the candidate genes and miRNA sequences (Supplementary Table 1A). The qPCR reactions were performed with the SYBR Green Master Mix in a qTOWER 2.2 system (Analytik Jena, Germany) with four technical replicates, using PeTIP41 (Fan et al., 2013) and U6 snRNA (Yang et al., 2021) as the endogenous controls for the genes and miRNAs, respectively. The qPCR conditions were as follows: 95°C for 6 min, followed by 40 cycles at 95°C for 15 s and 60°C for 15 s. The expression levels were calculated using the 2–ΔΔCT method (Livak and Schmittgen, 2001).
The 5′ UTR and promoter sequences of PH02Gene04386 (a glycosyltransferase 43 gene) were segmented into four sequences with different lengths, including 5′ UTR sequence of 600 and 1,119 bp upstream of cDNA namely 5′ UTR-600 and 5′ UTR-1119, promoter sequence of 600 and 1,200 bp upstream of mRNA namely pro-600 and pro-1200, respectively. The coding sequence (CDS) of two PeMYBs (PH02Gene12213 and PH02Gene31429) were recombined into the pGADT7-Rec2 vector. Six SMRE element (SMRE2 ∼ SMRE7) regions and four 5′ UTR/promoter sequences of PH02Gene04386 were inserted into the pHIS2 vector, respectively. Two recombinant vectors were co-transformed into the Y187 strain. The transformants were screened on Leu-, Trp-, and His-deficient medium supplemented with 20 mM of 3-amino-1,2,4-triazole for 3 d, using cotransformants containing p53:His2 with pGADT7:53 and p53:His2 with pGADT7:PeMYBs as the positive control and negative control, respectively. The primers used in yeast one-hybrid (Y1H) assay were listed in Supplementary Table 1B.
The CDS of PH02Gene04386 was isolated and cloned into the expression vector of pSuper1300. The transgenic plants overexpressing (OE) PH02Gene04386 were obtained through ectopic expression in Arabidopsis (Col-0) plants using the floral dip method, which were further verified by PCR and qPCR. Root length of transgenic and wild-type (WT) Arabidopsis seedlings was counted after 7 days growing on Murashige and Skoog medium with 50 mM xylose as the carbon source. The main inflorescence stem (1 ∼ 2 cm aboveground) of both transgenic and WT Arabidopsis plants of 6 weeks were used for histological staining with 5% (v/v) of TBO, xylan immunofluorescence localization with LM10 (Keppler and Showalter, 2010) and xylan content analysis, respectively. The fluorescence signals of FITC were recorded with a Zeiss LSM 980 confocal microscope. Images were taken with an Olympus CX31 light microscope (Tokyo, Japan).
A standard t-test is used to determine statistical significance with a 95% confidence interval. In all figures, data are represented as the mean ± standard error (SE).
During storage at room temperature, the surface of bamboo shoots changed significantly, and gradually browned with time (Figure 1A). Meanwhile, the degree of staining intensity of lignin was deepening and the polysaccharides was lightening in vascular bundle (Figure 1B), which indicated a potential change in carbohydrate and phenols content. Since the carbohydrates and phenols are considered to be important factors affecting postharvest edible quality, the content of these substances such as soluble sugar, hemicellulose, cellulose, lignin, and procyanidins in postharvest moso bamboo shoots are measured. The results showed that the continuous decreases were observed in soluble sugar from 37.75 mg g–1 (S1) to 14.36 mg g–1 (S4), hemicellulose from 108.92 mg g–1 (S1) to 22.54 mg g–1 (S4) and cellulose from 468.68 mg g–1 (S1) to 69.99 mg g–1 (S4) (Figures 1C–E). In contrast, the lignin content showed an increase from 26.23 mg g–1 (S1) to 32.91 mg g–1 (S4) (Figure 1F). The content of hydrolyzed tannin increased slightly from 3.07 mg g–1 (S1) to 3.45 mg g–1 (S2) and then significantly decreased to 0.63 mg g–1 (S4) (Figure 1G). Meanwhile, the content of condensed tannin (procyanidin) increased continuously, and doubled that of S1 (0.17 mg g–1) at the end (S4, 0.43 mg g–1) (Figure 1H). The activity of PAL was first decreased from 6.08 U g–1 (S1) to 3.41 U g–1 (S2), and then increased significantly to 10.55 U g–1 (S4) (Figure 1I). The activity of POD peaked at 2140.90 U g–1 (S3), but decreased significantly to 897.27 U g–1 at the end (S4) (Figure 1J). These results were in agreement with our current understanding of taste and flavor decline in postharvest moso bamboo shoots.
To further understand the molecular mechanisms of above changes, 12 transcriptome libraries were constructed, with 325,040,670 clean reads generated. The clean data were mapped to the reference genome of moso bamboo with the mapping ratio varying from 96.16 to 96.66% (Supplementary Table 2A). Meanwhile, 7,458 unigenes were identified, among which 5,061 were functionally annotated (Supplementary Table 2B). A total 12,553 DEGs were identified by comparing the samples (Supplementary Figures 1A,C). Annotation analysis showed that there were 1,046 DEGs encoding TFs belonging to 44 families, in which MYB (112), bHLH (91), AP2/ERF (80), WRKY (79), and NAC (70) were the most over-represented TF families (Supplementary Figure 1B).
We performed expression analysis of the DEGs, and grouped genes according to the similar expression of their transcriptome profiles. A total of six clusters were identified (Figures 2A–C), and most of the DEGs belonged to two kinds of clusters: down-regulated (cluster I, 3,596 DEGs) and up-regulated (cluster II, 2,723 DEGs). In cluster I, GO and KEGG analyses showed that the DEGs were enriched in carbohydrate metabolism (Figures 2D,E). Different from cluster I, DEGs in cluster II were related to secondary metabolism (Figures 2F,G). The results of GO and KEGG analyses in cluster III showed that DEGs were enriched in a variety of pathways, including photosynthesis, carbohydrate and secondary metabolism (Supplementary Figures 2A,B). Considering the functional terms in combination with morphological characteristics, we proposed that the decrease of soluble sugar and structural polysaccharides along with the increase of lignin and procyanidins may be due to the expression changes of DEGs involved in carbohydrate metabolism and secondary metabolism. Among them, carbohydrate-related DEGs were the most abundant, which was consistent with the significant changes in carbohydrate content.
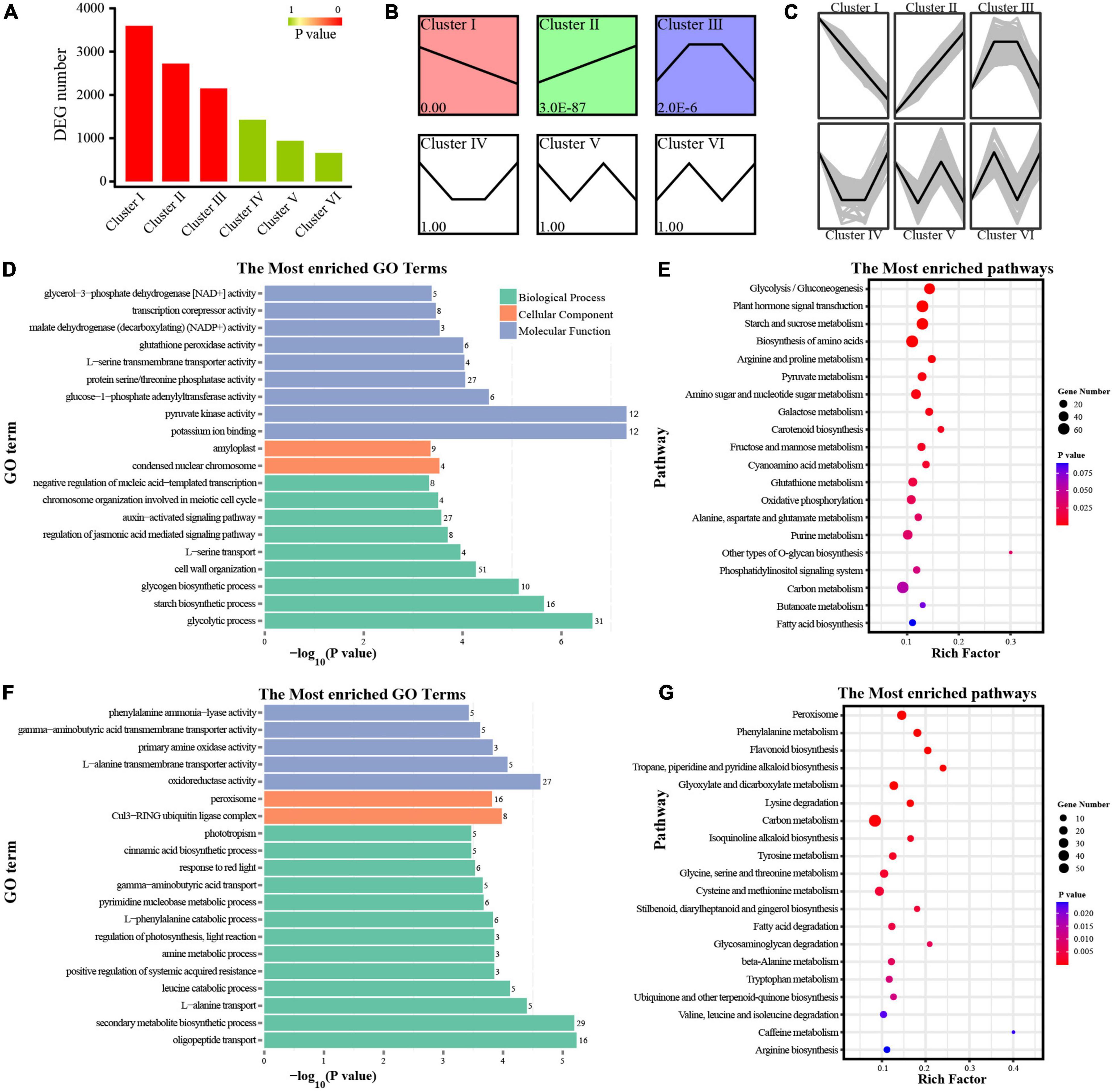
Figure 2. Function enrichment analysis of DEGs. (A) DEG number in different clusters. (B) Different clusters based on P-values. (C) DEGs with similar expression trends in a specific cluster. (D–G) GO and KEGG analyses of DEGs in cluster I (D,E), cluster II (F,G). Rich Factor represents the value of enrichment factor which is the quotient of foreground value (the number of DEGs), and the larger the value, the more significant enrichment; Coloring indicates P-value with higher in red and lower in blue, and the lower P-value, the more significantly enriched. Point size indicates the number of DEGs.
To investigate the changes in carbohydrate metabolism, the expression patterns of DEGs in three key metabolic pathways were further analyzed. The result showed that DEGs were remarkably enriched in the pathway related to nucleotide sugar metabolism. Most of the DEGs were down-regulated, especially those involved in pectin and xylan biosynthesis, while the DEGs involved in arabinose biosynthesis were up-regulated during storage (Figure 3). Three DEGs encoding UDP-apiose/xylose synthase (AXS) and four DEGs encoding UDP-glucose 6-dehydrogenase (UGDH, EC:1.1.1.22) enriched in nucleotide sugar metabolism pathway were identified, with all three AXS DEGs and three of four UGDH DEGs down-regulated significantly. In addition, one DEG encoding UDP-arabinose 4-epimerase (UXE, EC:5.1.3.5) involved in arabinose biosynthesis was identified and its expression increased with storage time.
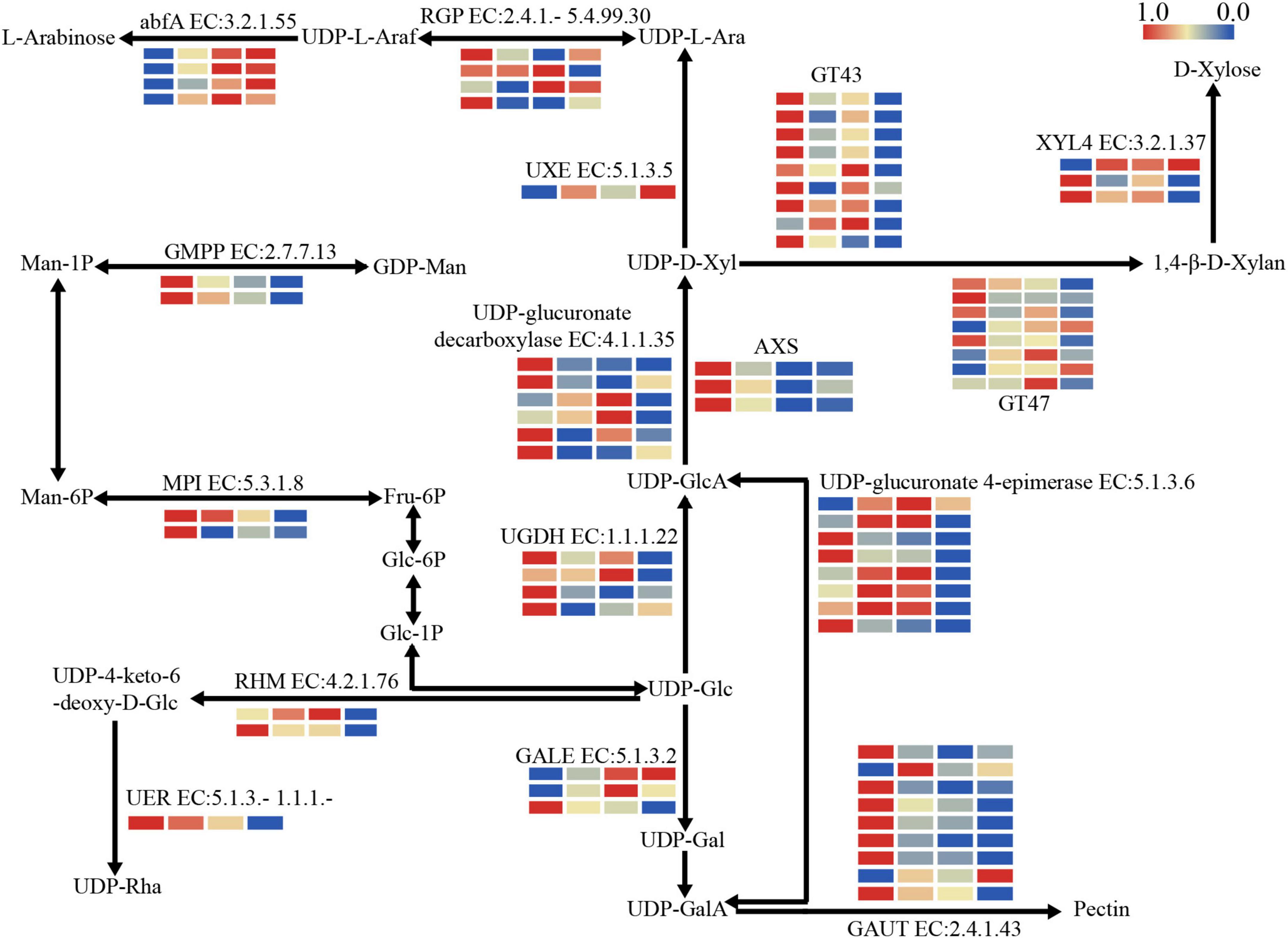
Figure 3. The expression patterns of DEGs involved in structural polysaccharide metabolism. abfA, alpha-L-arabinofuranosidase; AXS, UDP-apiose/xylose synthase; GALE, UDP-glucose 4-epimerase; GAUT, α-1,4-galacturonosyltransferase; GT43, glycosyltransferase 43; GT47, glycosyltransferase 47; RGP, eversibly glycosylated polypeptide; RHM, UDP-glucose 4,6-dehydratase; UER, 3,5-epimerase/4-reductase; UGDH, UDP-glucose 6-dehydrogenase; UXE, UDP-arabinose 4-epimerase; XYL4, xylan 1,4-beta-xylosidase. The color bar indicates log2-based fragments per kilobase per million (FPKM) with higher in red and lower in blue.
Further, the DEGs related to nucleotide sugar polymerization were identified, such as those encoding glycosyltransferase family 43 (GT43, EC:2.4.2.-) and glycosyltransferase family 47 (GT47, EC:2.4.2.-) associated with xylan biosynthesis. Most of nine GT43 DEGs were down-regulated significantly during storage, while the expression trends were significantly different among GT47 DEGs, in which the expression of PH02Gene05811 was significantly down-regulated and PH02Gene12063 was first up-regulated and then down-regulated. In addition, the DEGs involved in pectin synthesis were also identified. For example, DEGs encoding GAUT (EC:2.4.1.43) changed significantly during storage, with most of them showing a significant decrease at S2 immediately.
In starch and sucrose metabolic pathways, biosynthesis-related DEGs were down-regulated significantly at S2, e.g., three DEGs encoding starch synthase (glgA, EC:2.4.1.21) and four DEGs encoding SPS (EC:2.4.1.14), while hydrolysis-related DEGs were relatively up-regulated at S2 and S3 (Supplementary Figure 3), e.g., eight DEGs encoding amylase (AMY, EC:3.2.1.1, BMY, EC:3.2.1.2) and six DEGs encoding fructosidase (INV, EC:3.2.1.26). As for Glycolysis, the DEGs encoding enzymes involved in irreversible reactions were down-regulated rapidly at S2, e.g., hexokinase (HK, EC:2.7.1.1) and PK (EC:2.7.1.40) DEGs. However, there were some exceptions, e.g., three DEGs encoding fructose-bisphosphate aldolase (ALDO, EC:4.1.2.13), which were first up-regulated and reached a peak at S3, and then down-regulated (Supplementary Figure 4).
A total 363.6 million of 418.9 million reads were generated after quality control (Supplementary Table 3A). The length distribution of the unique small RNA reads indicated that 24 nt were the most abundant (36.6%) (Supplementary Table 3B). Further, 47 unique conserved miRNAs and 415 novel miRNAs were identified, with lengths varied from 19 to 25 nt (Supplementary Figures 5A,B), which agreed with many other RNA-Seq experiments (Chen et al., 2020; Li et al., 2021). Of these miRNAs, 451 pre-miRNAs corresponding to 462 mature miRNAs were identified, which were grouped into 93 families (Supplementary Figure 5C and Supplementary Table 3C). The majority of mature miRNAs were characterized by a 5′-uridine residue and the proportion of A and U bases exceed 50%, which was consistent with the characteristics of plant miRNAs attributed to DCL1 cleavage and AGO1 association (Rajagopalan et al., 2006).
The majority of the identified miRNAs were expressed in more than one sample. In total, 182 miRNAs showed differential expression patterns across different combinations, including 26 known miRNAs and 156 novel miRNAs. There were 34 miRNAs with significant expression changes in moso bamboo shoots at S2, S3, and S4 compared with those at S1 (Figures 4A,B). We merged the DEMs with similar expression profiles to perform cluster analysis. A total of 84 DEMs were clustered into four major clusters with P-value less than 0.05 (Figure 4C). The DEMs in cluster I were up-regulated and those in cluster III were down-regulated, while those in cluster II were up-regulated firstly and then down-regulated during storage (Figure 4D). The opposite expression trends between DEMs and DEGs suggested that they might have a potential negative regulatory relationship.
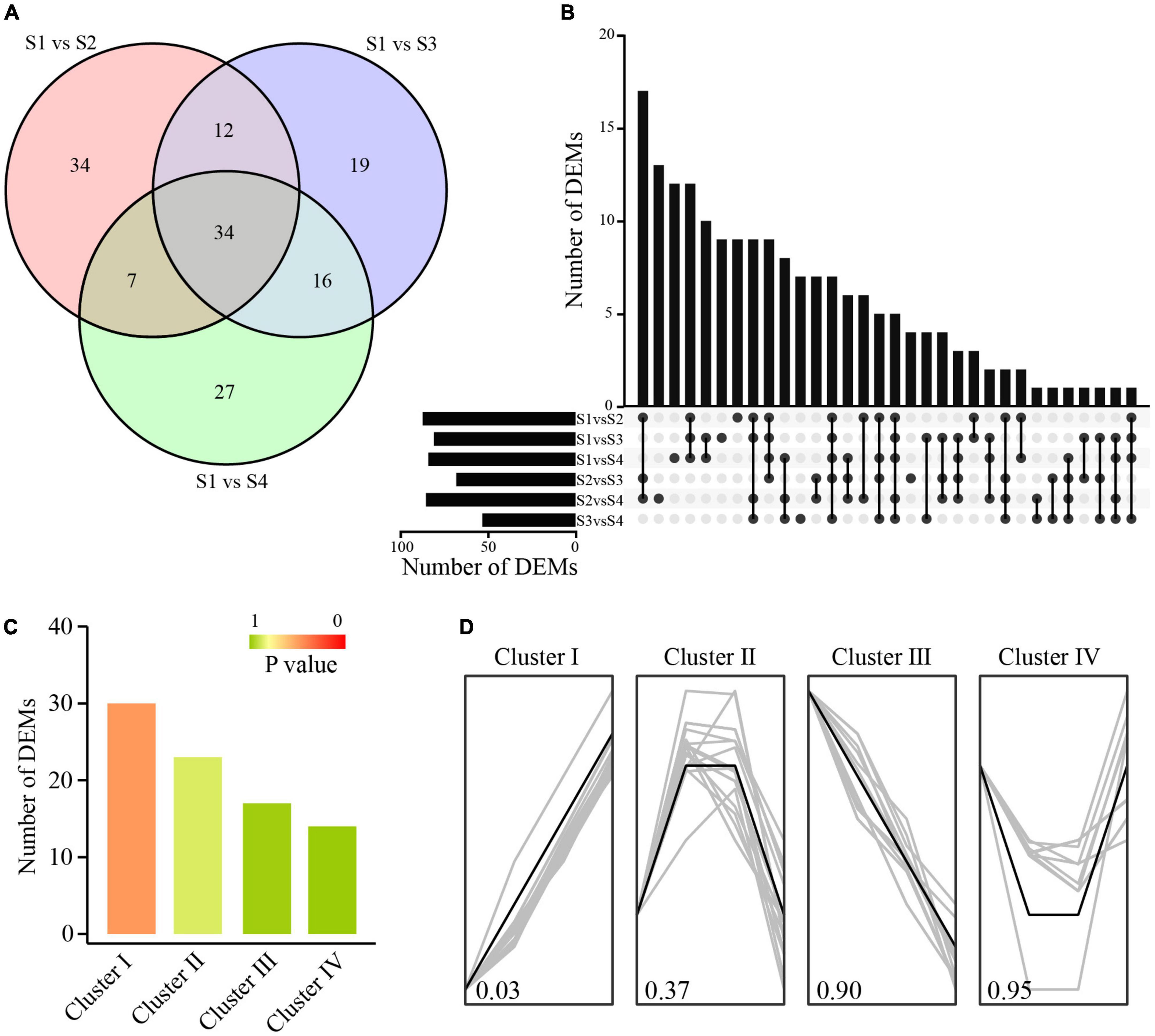
Figure 4. Identification and cluster analysis of DEMs. (A) Venn diagram of DEMs. (B) Number of DEMs between different comparison groups. (C) DEM number in different clusters. (D) DEMs with similar expression trends in a specific cluster.
In order to estimate the biological function of these miRNAs, their target genes were identified. A total of 13,748 miRNA-target pairs were identified by TargetFinder and psRNATarget, consisting of 424 miRNAs and 6,024 genes. There were 998 target genes annotated to 125 KEGG pathways. Carbohydrate metabolism and terpenoid and polyketide metabolism were dominant pathways (Supplementary Figure 6), which indicated that transcriptional regulation of carbohydrate metabolism might have important effects on the physiological and biochemical changes in postharvest moso bamboo shoots.
The co-expression network was comprehensively investigated using a WGCNA to predict the role of genes involved in carbohydrate metabolism. A total 4,048 genes with eigengene connectivity (kEM) value more than 0.7 were clustered into four major distinct modules (Supplementary Figure 7A). The turquoise module had a high correlation with sugar (0.99), cellulose (0.96), hemicellulose (0.96), and xylan (0.82), indicating that the genes in this module may be involved in carbohydrate metabolism of postharvest moso bamboo shoots. The correlation between the module and the compounds and expression trends of DEGs were shown in Supplementary Figures 7B,C. The expression of genes and their miRNA pairs in the turquoise module was further analyzed (Figure 5A). Combining the analysis of biological process, motif information, WGCNA, and coherent miRNA-target pairs, a network containing 15 TFs, 78 genes and 30 miRNAs was established, which supported that these TFs and miRNAs were significantly coupled with multiple key enzyme genes related to structural polysaccharides metabolism, starch and sucrose metabolism and glycolysis (Figure 6B). The gene families involved in xylan metabolism were the most numerous, including GT43, GT47, AXS, XYL, and UXS families.
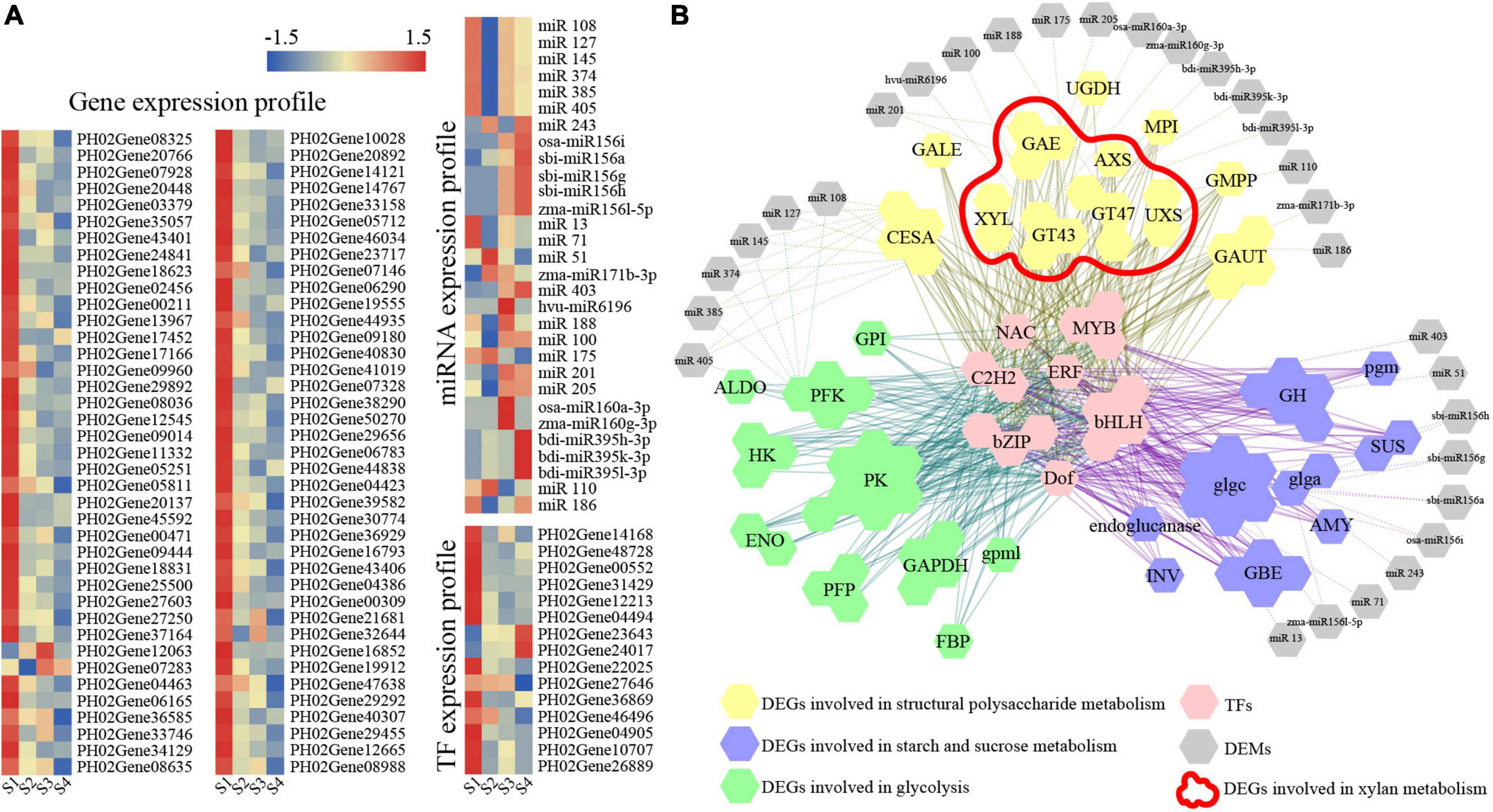
Figure 5. Analysis of DEGs and DEMs involved in carbohydrate metabolism of postharvest moso bamboo shoots. (A) Expression patterns of DEGs and DEMs. (B) Visualization of co-expression relationships between DEGs and DEMs. The color bar indicates log2-based fragments per kilobase per million (FPKM) with higher in red and lower in blue.
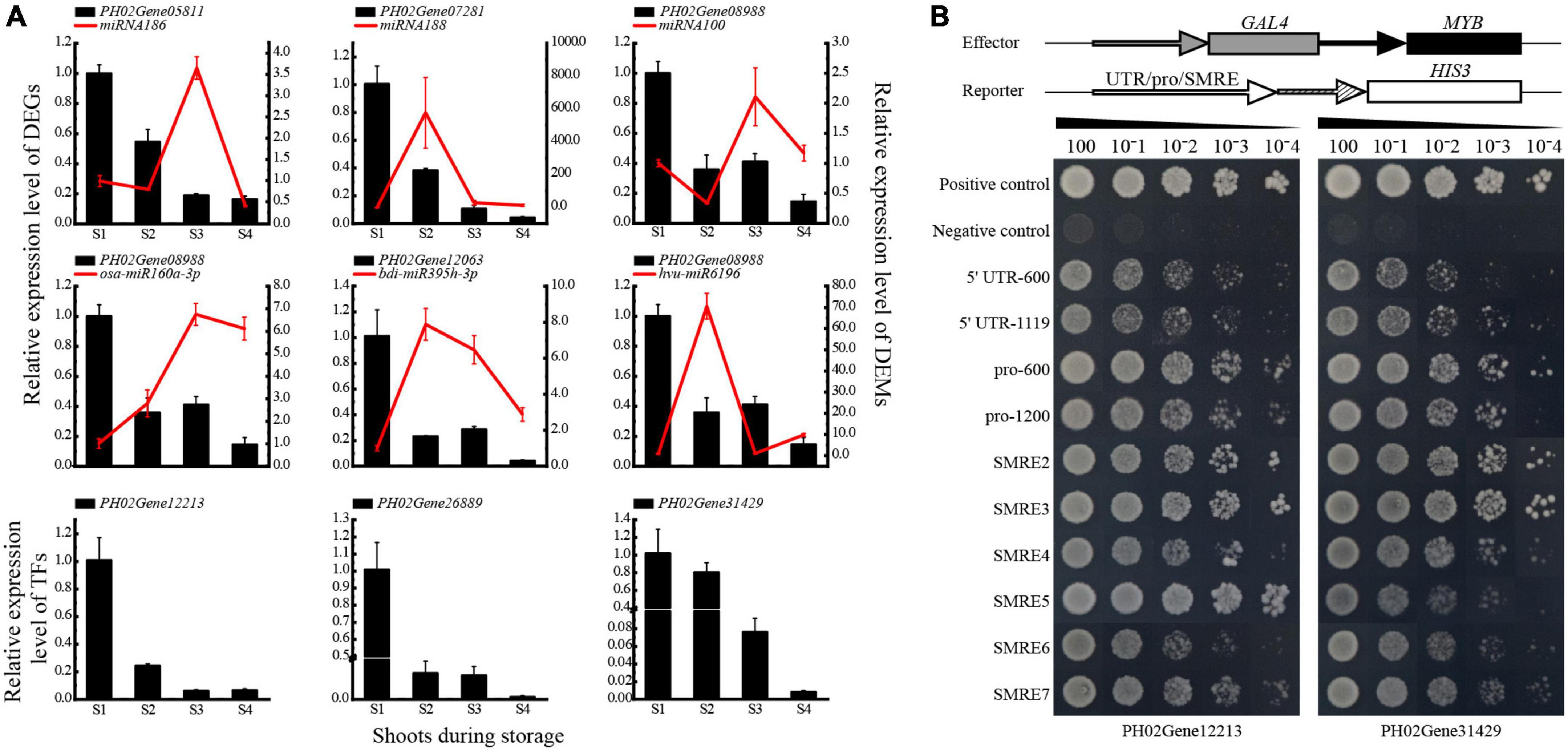
Figure 6. Validation of the genetic elements in the network of carbohydrate metabolism. (A) qPCR analysis of DEGs and DEMs. (B) Regulatory relationship analysis between MYBs (PH02Gene12213 and PH02Gene31429) and PH02Gene04386 by yeast one-hybrid assay. The red solid lines represent the miRNAs, the black blocks represent the structure genes targeted by miRNAs and MYBs. S1, S2, S3, and S4 represent bamboo shoots stored for 0, 3, 6, and 12 d.
To validate the regulatory network, qPCR and Y1H assays were conducted. Opposite expression trends of miRNA and mRNA in each pair were observed, which was similar to the results of the high-throughput sequencing. Most miRNAs were up-regulated significantly, while their target genes were down-regulated significantly at S2 or S3 (Figure 6A). Additionally, the qPCR results of key TFs, such as two MYBs of PH02Gene12213 and PH02Gene31429, showed similar expression patterns to their target genes, with trends decreasing continuously along with storage time. These results indicated that the enzyme genes might be regulated positively by the TFs and negatively by the miRNAs.
To further validate the regulatory relationship between TFs and genes, the enriched xylan biosynthesis-related gene PH02Gene04386, which was predicted to be a target of MYBs (PH02Gene12213 and PH02Gene31429), was selected for validation of Y1H assay (Figure 6B). The results showed that the MYBs could bind to the 5′ UTR, promoter sequences and SMRE elements of PH02Gene04386. The yeast transformants harboring pro-600 grew best, followed by the transformants harboring pro-1200 and triple SMRE elements. In contrast, the negative control could not grow on the nutritional screening medium. These results indicated that PH02Gene12213 and PH02Gene31429 could bind to the promoter of PH02Gene04386 and might further regulate its expression.
PH02Gene04386 was a hub gene in the regulatory network and homologous gene of IRX9, which contributed to the xylan biosynthesis in Arabidopsis. The function of PH02Gene04386 was investigated in Arabidopsis, and three OE lines (L1, L3, and L9) were used for further analysis (Supplementary Figure 8). The results showed that the primary root length of OE plants was significantly greater than that of WT on 50 mM xylose medium (P < 0.05) (Figure 7A). The staining intensity was higher and the vascular bundle cell wall was more thickness in OE flower stems than those of WT (Figures 7B,C). To verify the effect of PH02Gene04386 on the xylan content and distribution in stems of Arabidopsis inflorescence, immunofluorescence localization and xylan content analysis were performed using OE and WT plants. The results showed that the fluorescence intensity was higher and the distribution range of xylan antibody (LM10) was broader in OE plants than in WT (Figure 7D). The chemical assay also supported the results that the xylan content was higher in OE plants than in WT (Figure 7E), indicating that overexpression of PH02Gene04386 could promote xylan biosynthesis in Arabidopsis. These results supported that PH02Gene04386 was the hub gene in the regulatory network involved in xylan biosynthesis.
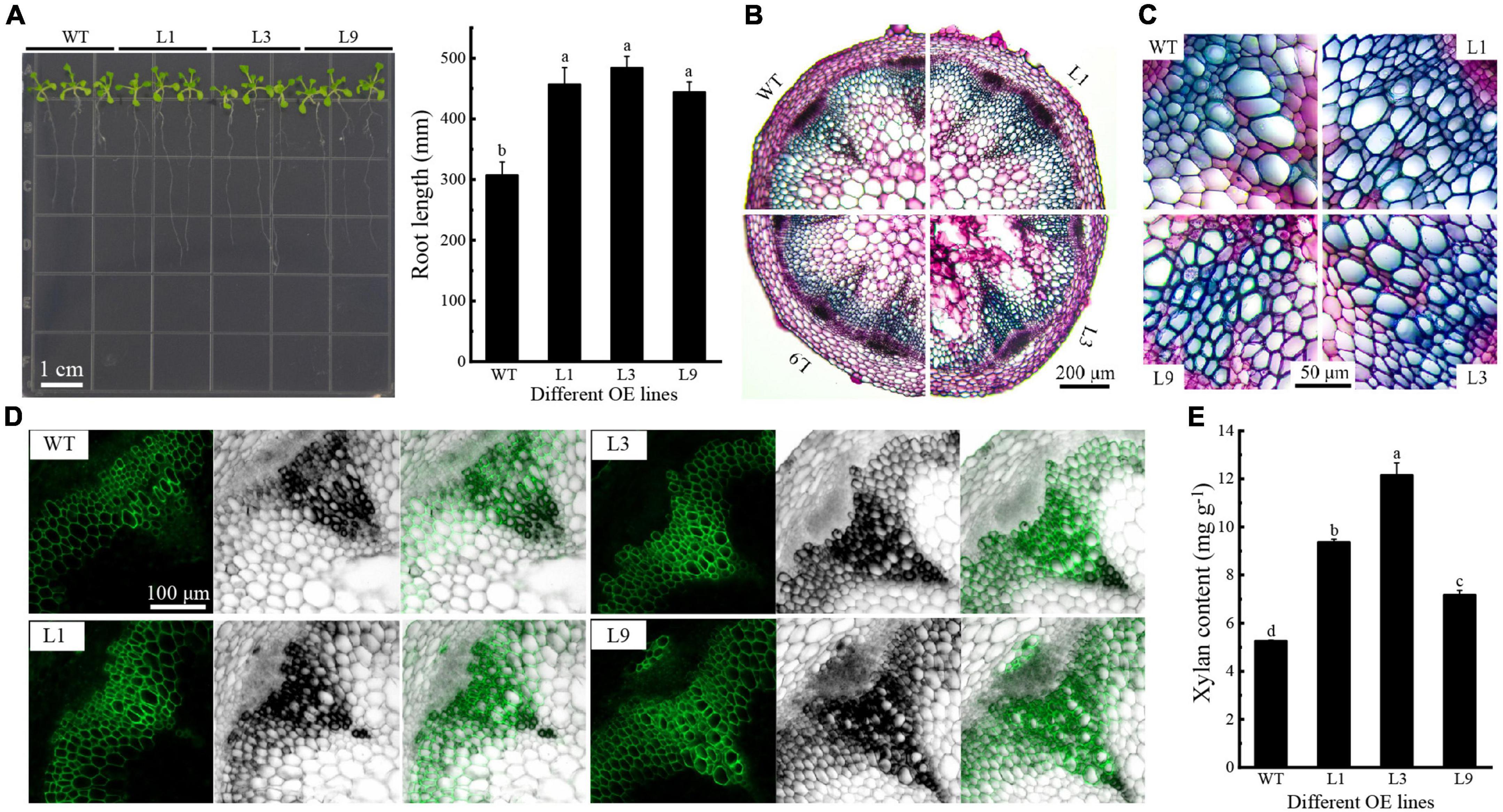
Figure 7. The function analysis of PH02Gene04386 overexpressed in Arabidopsis. (A) Root length of Arabidopsis growing on xylose medium. (B) Histochemical staining of cross section of inflorescence stems. (C) Vascular bundle in cross section of inflorescence stems in (B). (D) Xylan immunofluorescence localization in inflorescence stems. (E) Xylan contents in inflorescence stems. Lowercase letters a, b, c, and d indicate significant differences (P < 0.05).
The edible quality decline rapidly during storage, limiting the spread for fresh bamboo shoots in the market. The shoots of moso bamboo and P. prominens showed significant lignification during storage, with lower accumulation rate of lignin and cellulose in moso bamboo shoots (Zhang et al., 2020). PAL and POD catalyze the oxidation of polyphenols, which is presumed to be the cause of severe lignification and browning in P. prominens shoots (Nguyen et al., 2003; Roura et al., 2008). In present study, the degree of browning of moso bamboo shoots is lower than the results in the above study (Figure 1A), probably due to the retention of culm sheaths during storage, thus indicating that storage with culm sheaths is beneficial for preservation of moso bamboo shoots.
The consumption of carbohydrate was an important factor to reduce the edible quality of bamboo shoots. The soluble sugar and hemicellulose contents of the moso bamboo shoots in this study decreased significantly at S2 and continued to decrease during storage (Figures 1C,D), which was similar to the variation of non-structural sugar and hemicellulose in Lei bamboo and green bamboo shoots (Yu et al., 2021). However, the variation of cellulose content was different (Figure 1E; Li et al., 2018; Zhang et al., 2020). It might be explained by the different storage methods and bamboo shoot treatments, by which refrigeration was generally used in the former studies for the shoots with culm sheath removal and bagging in slices, while the shoots with culm sheaths at room temperature used in this study. These results are value for reasonable storage and transportation of bamboo shoots.
The physiological changes of bamboo shoots during storage depend on the spatial and temporal expression of the relevant genes. In this study, we tried to interpret the molecular mechanism of the physiological changes in moso bamboo shoots based on the expression changes of DEGs. Carbohydrate-related DEGs were the most enriched (Figure 2D), which were closely related to the significant changes of carbohydrate content in moso bamboo shoots. Therefore, it could infer that the carbohydrate-related DEGs had important effect on the edible quality of moso bamboo shoots. Exogenous fructose treatment on Lei bamboo shoots could effectively reduce the lignification and prolong the shelf life by regulating the enzymes involved in starch and sucrose metabolism (Zhou et al., 2020). A large number of DEGs enriched in carbohydrate metabolism such as starch, cellulose and pectin were found in blueberry and African Pride (Annona cherimola × A. squamosa) during storage (Chen et al., 2019; Cárcamo de la Concepción et al., 2021). In the similar way, the changing pattern of the synthesis-related DEGs in non-structural sugar metabolism was characterized by a decreasing expression and those hydrolysis-related DEGs had an increasing expression along with the physiological changes in this study.
In addition, a large number of DEGs involved in structural polysaccharide metabolism were also identified, among which DEGs involved in the biosynthesis of mannose, xylan and pectin were significantly down-regulated (Figure 3), while those involved in the biosynthesis of arabinoxylans were either up-regulated or first up-regulated and then down-regulated. Studies have shown that the arabinose side chain of xylan binding to the ferulic acid of lignin could increase the stability of the cell wall, and the added xylan acts as a scaffold for lignin dehydrogenation polymer deposition in polysaccharides matrix (Li et al., 2015). This might explain the up-regulation of arabinose biosynthesis related DEGs in this study.
TFs and miRNAs affect carbohydrate metabolism through transcriptional regulation. GhMYB212 regulated the length of fibers and the accumulation of sucrose simultaneously by controlling the expression of a sucrose transporter gene GhSWEET12 (Sun et al., 2019). Two MYBs of PH02Gene12213 and PH02Gene31429 were suggested to be involved in the biosynthesis of lignin, xylan and cellulose, and lignin deposition, respectively (Yang et al., 2019), both of which had potential regulatory relationships with xylan biosynthesis related DEGs like GT43 and GT47 by bioinformatics and Y1H assay (Figures 5B, 6B). A number of miRNAs involved in carbohydrate metabolism were also identified in this study, such as miRNA167s and miRNA160s (Figure 5B), which had been confirmed to have regulatory roles in starch and sucrose metabolism and structural polysaccharide metabolism in cotton and Acacia (Acacia auriculiformis and A. mangium) (Wong et al., 2011; Wang et al., 2017). In addition, there were several novel miRNAs in the regulatory network, such as miRNA100 and miRNA175, which provided important information for further investigating the function of miRNAs in polysaccharide metabolism.
Structural polysaccharides are important components of plant cell walls, which are synthesized by glycosyltransferases like GT43. In this study, overexpression of PH02Gene04386 encoding glycosyltransferase 43 promoted the elongation of OE Arabidopsis seedling roots on the medium with UDP xylose as a carbon source (Figure 7A), suggesting PH02Gene04386 could increase the efficiency of xylose utilization. The change of xylan content and its distribution in PH02Gene04386 OE Arabidopsis in this study (Figures 7D,E) supported the important role of GT43 family members involved in xylan biosynthesis. Similar results were also found in GhGT43Al and GhGT43C1 OE cotton plants (Li et al., 2014), IRX9 and IRX14 in Arabidopsis, rice and cotton (Wu et al., 2010), and GT43A and GT43B in polar (Ratke et al., 2018). A model to elucidate the regulatory mechanism involved in carbohydrate metabolism of moso bamboo shoots during storage was constructed (Figure 8). Overall, these results will not only be useful for understanding the physiological changes and molecular regulatory mechanisms of moso bamboo shoots during storage, but also provide an important potential candidate gene for the breeding related to xylan.
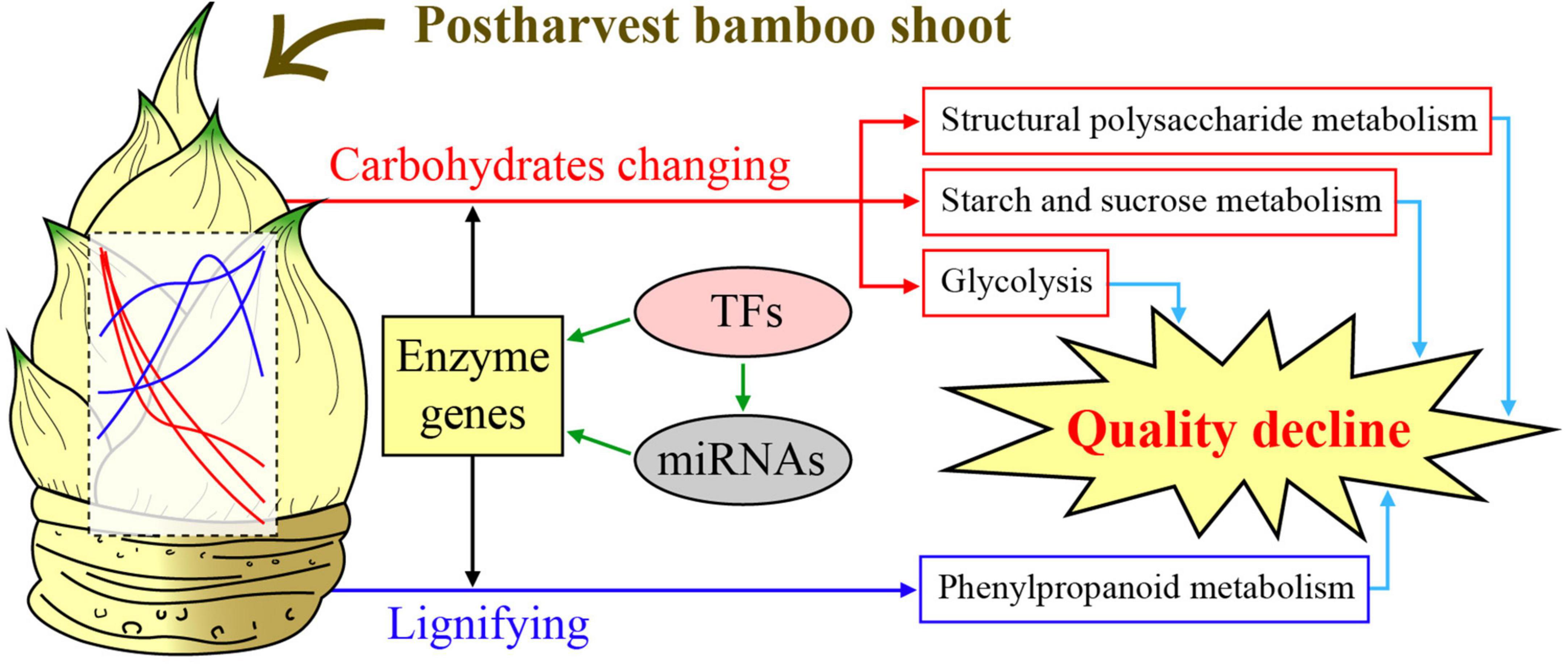
Figure 8. A model to elucidate the regulatory mechanism involved in carbohydrate metabolism of moso bamboo shoots during storage.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA840860 and https://www.ncbi.nlm.nih.gov/, PRJNA840882.
ZL designed and performed the experiments and wrote the manuscript. XX constructed the conceptions and worked for the bamboo sample collection. KY and CZ assisted with Y1H assay. YL assisted in completing the histochemical staining. ZG checked the data analysis and revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key Research and Development Program of China (No. 2021YFD2200502) and the National Natural Science Foundation of China (No. 31971736).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer ZZ declared a shared affiliation with the author(s) XX to the handling editor at the time of review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1021161/full#supplementary-material
Supplementary Figure 1 | Gene expression analysis in moso bamboo during storage. (A) Correlation analysis between different samples based on gene expression. (B) Number of DEGs encoding transcript factors. (C) Number of DEGs between different comparison groups. S1, S2, S3, and S4 represent bamboo shoots stored for 0, 3, 6, and 12 d.
Supplementary Figure 2 | GO (A) and KEGG (B) analyses of DEGs in cluster III of Rich Factor represents the value of enrichment factor which is the quotient of foreground value (the number of DEGs), and the larger the value, the more significant enrichment; Coloring indicates P-value with higher in red and lower in blue, and the lower P-value, the more significantly enriched. Point size indicates the number of DEGs.
Supplementary Figure 3 | The expression patterns of DEGs involved in starch and sucrose metabolism. AMY, α-amylase; BMY, β-amylases; GBE, glucan branching enzyme; GH, glycosyl hydrolase; INV, fructosidase; glgA, starch synthase; glgC, glucose-1-phosphate adenylyltransferase; MGAM, maltase-glucoamylase; pgm, phosphoglucomutase; SPS, sucrose phosphate synthase; SUS, sucrose synthase; UGP, glucose-1-phosphate uridylyltransferase. The color bar indicates log2-based fragments per kilobase per million (FPKM) with higher in red and lower in blue.
Supplementary Figure 4 | The expression patterns of DEGs involved in glycolysis. ALDO, fructose-bisphosphate aldolase; ENO, enolase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; gpmI. phosphoglycerate mutase; HK, hexokinase; PFK, phosphofructokinase; PGK, phosphoglycerate kinase; PK, pyruvate kinase. The color bar indicates log2-based fragments per kilobase per million (FPKM) with higher in red and lower in blue.
Supplementary Figure 5 | Length and family classification of miRNA. (A) Length of known miRNA. (B) Length of novel miRNA. (C) Percentage of miRNA members in each family.
Supplementary Figure 6 | Function analysis of miRNA-target pairs.
Supplementary Figure 7 | WGCNA modules based on DEG data. (A) Cluster dendrogram. (B) Module–trait relationships. Each column corresponds to a flavor compound. The color of each cell at the row-column intersection indicates the correlation coefficient between the module and the substance. (C) Expression heat map and expression level of DEGs in the module.
Supplementary Figure 8 | Validation of PH02Gene04386 overexpressed plants. (A) Overexpression vector of PH02Gene04386. (B) Screening of transgenic plants by PCR. (C) Detection of PH02Gene04386 expression in transgenic plants by qPCR. WT: Wild type Arabidopsis. L1, L3, L6, L7, L9: Transgenic lines. Lowercase letters a and b indicate significant differences (P < 0.05).
Cárcamo de la Concepción, M., Sargent, D. J., Šurbanovski, N., Colgan, R. J., and Moretto, M. (2021). De novo sequencing and analysis of the transcriptome of two highbush blueberry (Vaccinium corymbosum L.) cultivars ‘Bluecrop’ and ‘Legacy’ at harvest and following post-harvest storage. PLoS One 16:e0255139. doi: 10.1371/journal.pone.0255139
Chen, C., Xie, F., Hua, Q., Zur, N. T., Zhang, L., Zhang, Z., et al. (2020). Integrated sRNAome and RNA-Seq analysis reveals miRNA effects on betalain biosynthesis in pitaya. BMC Plant Biol. 20:437. doi: 10.1186/s12870-020-02622-x
Chen, J., Duan, Y., Hu, Y., Li, W., Sun, D., Hu, H., et al. (2019). Transcriptome analysis of atemoya pericarp elucidates the role of polysaccharide metabolism in fruit ripening and cracking after harvest. BMC Plant Biol. 19:219. doi: 10.1186/s12870-019-1756-4
Fan, C., Ma, J., Guo, Q., Li, X., Wang, H., and Lu, M. (2013). Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis). PLoS One 8:e56573. doi: 10.1371/journal.pone.0056573
Hou, D., Lu, H., Zhao, Z., Pei, J., Yang, H., Wu, A., et al. (2022). Integrative transcriptomic and metabolomic data provide insights into gene networks associated with lignification in postharvest Lei bamboo shoots under low temperature. Food Chem. 368:130822. doi: 10.1016/j.foodchem.2021.130822
Keppler, B. D., and Showalter, A. M. (2010). IRX14 and IRX14-LIKE, two glycosyl transferases involved in glucuronoxylan biosynthesis and drought tolerance in Arabidopsis. Mol. Plant 3, 834–841. doi: 10.1093/mp/ssq028
Li, C., Suo, J., Xuan, L., Ding, M., Zhang, H., Song, L., et al. (2019). Bamboo shoot-lignification delay by melatonin during low temperature storage. Postharvest Biol. Technol. 156:110933. doi: 10.1016/j.postharvbio.2019.110933
Li, D., Limwachiranon, J., Li, L., Zhang, L., Xu, Y., Fu, M., et al. (2019). Hydrogen peroxide accelerated the lignification process of bamboo shoots by activating the phenylpropanoid pathway and programmed cell death in postharvest storage. Postharvest Biol. Technol. 153, 79–86. doi: 10.1016/j.postharvbio.2019.03.012
Li, X., Xie, L., Zheng, H., Cai, M., Cheng, Z., Bai, Y., et al. (2019). Transcriptome profiling of postharvest shoots identifies PheNAP2- and PheNAP3-promoted shoot senescence. Tree Physiol. 39, 2027–2044. doi: 10.1093/treephys/tpz100
Li, C., Xuan, L., He, Y., Wang, J., Zhang, H., Ying, Y., et al. (2018). Molecular mechanism of xylogenesis in moso bamboo (Phyllostachys edulis) shoots during cold storage. Polymers 11:38. doi: 10.3390/polym11010038
Li, H., Lin, Q., Yan, M., Wang, M., Wang, P., Zhao, H., et al. (2021). Relationship between secondary metabolism and mirna for important flavor compounds in different tissues of tea plant (Camellia sinensis) as revealed by genome-wide mirna analysis. J. Agric. Food Chem. 69, 2001–2012. doi: 10.1021/acs.jafc.0c07440
Li, L., Huang, J., Qin, L., Huang, Y., Zeng, W., Rao, Y., et al. (2014). Two cotton fiber-associated glycosyltransferases, GhGT43A1 and GhGT43C1, function in hemicellulose glucuronoxylan biosynthesis during plant development. Physiol. Plant. 152, 367–379. doi: 10.1111/ppl.12190
Li, Q., Koda, K., Yoshinaga, A., Takabe, K., Shimomura, M., Hirai, Y., et al. (2015). Dehydrogenative polymerization of coniferyl alcohol in artificial polysaccharides matrices: Effects of xylan on the polymerization. J. Agric. Food Chem. 63, 4613–4620. doi: 10.1021/acs.jafc.5b01070
Liu, Z., Li, L., Luo, Z., Zeng, F., Jiang, L., and Tang, K. (2016). Effect of brassinolide on energy status and proline metabolism in postharvest bamboo shoot during chilling stress. Postharvest Biol. Technol. 111, 240–246. doi: 10.1016/j.postharvbio.2015.09.016
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Nguyen, T., Ketsa, S., and Doorn, W. (2003). Relationship between browning and the activities of polyphenoloxidase and phenylalanine ammonia lyase in banana peel during low temperature storage. Postharvest Biol. Technol. 30, 187–193. doi: 10.1016/S0925-5214(03)00103-0
Rajagopalan, R., Vaucheret, H., Trejo, J., and Bartel, D. P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Gene Dev. 20, 3407–3425. doi: 10.1101/gad.1476406
Ratke, C., Terebieniec, B. K., Winestrand, S., Derba-Maceluch, M., Grahn, T., Schiffthaler, B., et al. (2018). Downregulating aspen xylan biosynthetic GT43 genes in developing wood stimulates growth via reprograming of the transcriptome. New Phytol. 219, 230–245. doi: 10.1111/nph.15160
Roura, S. I., Pereyra, L., and Del Valle, C. E. (2008). Phenylalanine ammonia lyase activity in fresh cut lettuce subjected to the combined action of heat mild shocks and chemical additives. LWT-Food Sci. Technol. 41, 919–924. doi: 10.1016/j.lwt.2007.06.005
Schouten, R. E., Woltering, E. J., and Tijskens, L. M. M. (2016). Sugar and acid interconversion in tomato fruits based on biopsy sampling of locule gel and pericarp tissue. Postharvest Biol. Technol. 111, 83–92. doi: 10.1016/j.postharvbio.2015.07.032
Singhal, P., Bal, L. M., Satya, S., Sudhakar, P., and Naik, S. N. (2013). Bamboo shoots: A novel source of nutrition and medicine. Crit. Rev. Food Sci. 53, 517–534. doi: 10.1080/10408398.2010.531488
Sun, H., Liu, Y., Ma, J., Wang, Y., Song, H., Li, J., et al. (2021). Transcriptome analysis provides strategies for postharvest lotus seeds preservation. Postharvest Biol. Technol. 179:111583. doi: 10.1016/j.postharvbio.2021.111583
Sun, W., Gao, Z., Wang, J., Huang, Y., Chen, Y., Li, J., et al. (2019). Cotton fiber elongation requires the transcription factor GhMYB212 to regulate sucrose transportation into expanding fibers. New Phytol. 222, 864–881. doi: 10.1111/nph.15620
Tanabe, C., Furuta, K., Maeda, M., and Kimura, Y. (2017). Structural feature of N-glycans of bamboo shoot glycoproteins: Useful source of plant antigenic N-glycans. Biosci. Biotechnol. Biochem. 81, 1405–1408. doi: 10.1080/09168451.2017.1320519
Tarazona, S., García-Alcalde, F., Dopazo, J., Ferrer, A., and Conesa, A. (2011). Differential expression in RNA-seq: A matter of depth. Genome Res. 21, 2213–2223. doi: 10.1101/gr.124321.111
Tian, X., Zhu, L., Yang, N., Song, J., Zhao, H., Zhang, J., et al. (2021). Proteomics and metabolomics reveal the regulatory pathways of ripening and quality in post-harvest Kiwifruits. J. Agric. Food Chem. 69, 824–835. doi: 10.1021/acs.jafc.0c05492
Wang, J., Jiang, J., Wang, J., Wang, Z., Yang, X., and Jia, L. (2019). The influence of gamma irradiation on the storage quality of bamboo shoots. Radiat. Phys. Chem. 159, 124–130. doi: 10.1016/j.radphyschem.2019.02.021
Wang, M., Sun, R., Li, C., Wang, Q., and Zhang, B. (2017). MicroRNA expression profiles during cotton (Gossypium hirsutum L) fiber early development. Sci. Rep. 7:44454. doi: 10.1038/srep44454
Wang, Y., Chen, J., Wang, D., Ye, F., He, Y., Hu, Z., et al. (2020). A systematic review on the composition, storage, processing of bamboo shoots: Focusing the nutritional and functional benefits. J. Funct. Foods 71:104015. doi: 10.1016/j.jff.2020.104015
Wen, B., Cheng, Z., Hu, Y., Boon-Ek, Y., Wongs-Aree, C., and Supapanich, S. (2019). Ultraviolet-C treatment maintains physicochemical quality of water bamboo (Zizania latifolia) shoots during postharvest storage. Postharvest Biol. Technol. 152, 65–72. doi: 10.1016/j.postharvbio.2019.02.017
Wong, M. M., Cannon, C. H., and Wickneswari, R. (2011). Identification of lignin genes and regulatory sequences involved in secondary cell wall formation in Acacia auriculiformis and Acacia mangium via de novo transcriptome sequencing. BMC Genom. 12:342. doi: 10.1186/1471-2164-12-342
Wu, A. M., Hörnblad, E., Voxeur, A., Gerber, L., Rihouey, C., Lerouge, P., et al. (2010). Analysis of the Arabidopsis IRX9/IRX9-L and IRX14/IRX14-L pairs of glycosyltransferase genes reveals critical contributions to biosynthesis of the hemicellulose glucuronoxylan. Plant Physiol. 153, 542–554. doi: 10.1104/pp.110.154971
Wu, M. X., Zou, Y., Yu, Y. H., Chen, B. X., Zheng, Q. W., Ye, Z. W., et al. (2021). Comparative transcriptome and proteome provide new insights into the regulatory mechanisms of the postharvest deterioration of Pleurotus tuoliensis fruitbodies during storage. Food Res. Int. 147:110540. doi: 10.1016/j.foodres.2021.110540
Xie, C., Mao, X., Huang, J., Ding, Y., Wu, J., Dong, S., et al. (2011). KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucl. Acids Res. 39:W316–W322. doi: 10.1093/nar/gkr483
Yang, K., Li, L., Lou, Y., Zhu, C., Li, X., and Gao, Z. (2021). A regulatory network driving shoot lignification in rapidly growing bamboo. Plant Physiol. 187, 900–916. doi: 10.1093/plphys/kiab289
Yang, K., Li, Y., Wang, S., Xu, X., Sun, H., Zhao, H., et al. (2019). Genome-wide identification and expression analysis of the MYB transcription factor in moso bamboo (Phyllostachys edulis). PeerJ 6:e6242. doi: 10.7717/peerj.6242
Yao, S., Wang, Z., Cao, Q., Xie, J., Wang, X., Zhang, R., et al. (2020). Molecular basis of postharvest granulation in orange fruit revealed by metabolite, transcriptome and methylome profiling. Postharvest Biol. Technol. 166:111205. doi: 10.1016/j.postharvbio.2020.111205
Yu, L., Pei, J., Zhao, Y., and Wang, S. (2021). Physiological changes of bamboo (Fargesia yunnanensis) shoots during storage and the related cold storage mechanisms. Front. Plant Sci. 12:731977. doi: 10.3389/fpls.2021.731977
Zhang, H., Ying, Y. Q., Wang, J., Zhao, X. H., Zeng, W., Beahan, C., et al. (2018). Transcriptome analysis provides insights into xylogenesis formation in Moso bamboo (Phyllostachys edulis) shoot. Sci. Rep. 8:3951. doi: 10.1038/s41598-018-21766-3
Zhang, Z., Li, C., Zhang, H., Ying, Y., Hu, Y., and Song, L. (2020). Comparative analysis of the lignification process of two bamboo shoots stored at room temperature. Plants 9:1399. doi: 10.3390/plants9101399
Zhou, D., Wang, K. T., Kuang, W. L., Lei, C. Y., Qiu, L. L., Li, C. H., et al. (2020). Effects of fructose treatment on quality and lignification of bamboo shoots (Phyllostachys praecox) during cold storage and its regulation mode involved [in Chinese]. Food Ferment. Ind. 46, 175–183. doi: 10.13995/j.cnki.11-1802/ts.023916
Keywords: Phyllostachys edulis shoot, postharvest physiology, carbohydrate metabolism, co-expression network, xylan biosynthesis
Citation: Li Z, Xu X, Yang K, Zhu C, Liu Y and Gao Z (2022) Multifaceted analyses reveal carbohydrate metabolism mainly affecting the quality of postharvest bamboo shoots. Front. Plant Sci. 13:1021161. doi: 10.3389/fpls.2022.1021161
Received: 17 August 2022; Accepted: 31 August 2022;
Published: 21 September 2022.
Edited by:
Guo-Fei Tan, Guizhou Academy of Agricultural Sciences (CAAS), ChinaReviewed by:
Yogesh Kumar Ahlawat, University of Florida, United StatesCopyright © 2022 Li, Xu, Yang, Zhu, Liu and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhimin Gao, Z2FvemhpbWluQGljYnIuYWMuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.