
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 16 December 2022
Sec. Crop and Product Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1020847
This article is part of the Research Topic Environmental and Endogenous Signals: Crop Yield and Quality Regulation View all 7 articles
Introduction: Plant height and grain length are important agronomic traits in rice, exhibiting a strong effect on plant architecture and grain quality of rice varieties.
Methods: Methods: A novel rice chromosomal segment substitution line (CSSL), i.e., CSSL-Z1357, with significantly increased plant height (PH) and grain length (GL) was identified from CSSLs constructed by using Nipponbare as a receptor and a restorer line Xihui 18 as a donor. Seven agronomic traits of PH, PL, GL, GW, GPP, SPP, and TGW were phenotyped, and REML implemented in HPMIXED of SAS were used to detect the QTL for these traits. Secondary CSSLs were screened out via marker-assisted selection (MAS) to estimate the additive and epistatic effects of detected QTLs, evaluating the potential utilization of pyramiding the target QTLs for yield and quality improvement of rice varieties.
Results and Discussion: Results and Discussion: CSSL-Z1357 carried nine segments from Xihui 18 with an average segment length of 4.13 Mb. The results show that the long grain of CSSL-Z1357 was caused by the increased number of surface cells and the length of the inner glume. Thirteen quantitative trait loci were identified via the F2 population of Nipponbare/CSSL-Z1357, including three each for GL (qGL-3, qGL-6, and qGL-7) and PH (qPH-1, qPH-7, and qPH-12I), among which qGL-3 increased GL by 0.23 mm with synergistic allele from CSSL-Z1357. Additionally, three single (S1 to S3), two double (D1, D2), and one triple segment (T1) substitution lines were developed in F3 via MAS. Results show that pyramiding the segments from Chr.3 (qGL-3 and qPH-3), Chr.6 (qGL-6 and qPH-6), and Chr.7 (Null and qPH-7) tended to result in better phenotype of increased GL and PH and decreased grain width, providing a potential basis for enhancing grain yield and quality in rice breeding.
Grain-related traits, including length, width, thickness, and weight, in rice (Oryza sativa L.) determine yield and quality of rice varieties, making them important for both breeders and scientists (Xu et al., 2004). Up to now, a large number of major quantitative trait loci (QTLs) controlling grain length (GL) and other related traits were detected; some of these QTLs were cloned and functionally profiled, e.g., BG1, PGL1, GL2/3, GS2/3/9, GSE5, and GW2/5/7. In addition, several signaling pathways, including phytohormones, ubiquitin-proteasome, G-protein, mitogen-activated protein kinase (MAPK), transcription factors (TFs), and secreted peptides, that control grain size have been revealed (Fan and Li, 2019; Li et al., 2021). Brassinosteroids (BRs), auxin, and cytokinin (CK) are the phytohormones that play important roles in controlling grain size (Che et al., 2015).
In rice, more than 10 genes are reported to regulate grain size through phytohormone-related pathways. For example, eight genes, i.e., GSE5/GW5 (Weng et al., 2008; Duan et al., 2017; Liu et al., 2017), GSK2 (Tong et al., 2012; Lyu et al., 2020), GSK3 (Gao et al., 2019; Liu et al., 2021), GS5 (Li et al., 2011), GS2 (Duan et al., 2015; Hu et al., 2015), and qGL3/OsPPKL1 (Zhang et al., 2012; Gao et al., 2019), are documented to regulate rice grain size via the BR signaling pathway. In addition, six genes [qTGW3 (Hu et al., 2018), BG1 (Liu et al., 2015), TGW6 (Ishimaru et al., 2013), RBG1 (Lo et al., 2020), and qGL5/OsAUX3 (Qiao et al., 2021)] associated with the auxin pathway, and two genes [OsPUP7 (Qi and Xiong, 2013) and BG3 (Xiao et al., 2019; Yin et al., 2020)] linked to the CK pathway are reported to regulate grain size in rice.
The ubiquitination-proteasome pathway genes, including GW2 (Song et al., 2007; Hao et al., 2021), OsOTUB1/WTG1 (Wang et al., 2017; Huang et al., 2017), LG1/OsUBP15 (Shi et al., 2019), and TUD1(Hu et al., 2013), also play crucial roles in the regulation of grain size. The genes DEP1, GGC2, and GS3 (Fan et al., 2006; Mao et al., 2010; Sun et al., 2018; Liu et al., 2018); D1/RGA1, RGB1 (Zhang et al., 2021a); RGG1 (Tao et al., 2020); RGG2 (Miao et al., 2019); and LGY3 (Liu et al., 2018) belonging to the G-protein pathway are also involved in regulating grain size (Liu et al., 2018; Sun et al., 2018). In addition, the combined MAPK module of OsMKKK10-OsMKK4-OsMAPK6 positively regulates grain size by affecting cell proliferation in rice (Xu et al., 2018; Fan and Li, 2019; Li et al., 2021). The MAPK pathway can promote rice grain development via the activation of OsWRKY53, whereas OsMKP1/GSN1 tends to suppress OsMAPK6 and, thus, depress the functioning of this pathway (Fan and Li, 2019; Li et al., 2021). Several TFs are also involved in the regulation of grain size in rice, such as GLW7/OsSPL13 (Si et al., 2016), OsSPL16/GW8 (Wang et al., 2012), GS9 (Zhao et al., 2018), An-1 (Luo et al., 2013), and GL6/SG6 (Wang et al., 2019; Zhou and Xue, 2020).
GS5 is a major QTL controlling grain width (GW), grain filling, and grain weight in rice (Li et al., 2011). The product of GS5, a putative serine carboxypeptidase, tended to regulate the grain size positively, thus serving as a potential candidate for yield improvement in rice and other cereals (Li et al., 2011). The encoded protein of GS9 had a conserved domain with unknown function that could alter cell division and regulate grain shape in rice (Zhao et al., 2018). In addition to regulating grain shape, the GS9 allele exhibited functions of improving grain appearance (as a marker of quality) in rice (Zhao et al., 2018).
Some other phenotypic traits also contribute to the grain size in rice. For example, grain number per panicle/plant, a typical quantitative trait that determines rice yield, generally correlates negatively with grains size traits (Lu et al., 2022). Downregulation of GW10 that encodes a P450 subfamily protein in rice tends to result in decreased GL and GW but increased grain number (Zhan et al., 2021). A loss of function of gad1 (grain number, grain length and awn development1) caused by a frame-shift insertion increases grain number while decreasing GL in rice (Jin et al., 2016). Plant height (PH) is another typical quantitative trait that determines rice plant architecture and affects the yield of rice varieties. Although weak correlations between PH and GL are documented in rice (Sabesan et al., 2009; Lakshmi et al., 2014), some genes for PH have pleiotropic effects influencing GL in rice. Huang and colleagues reported that the rice mutant ZPDM1 with a new allele of BRD2 (BRASSINOSTEROID DEFICIENT DWARF 2) tended to result in the phenotype of reduced PH and smaller grain size (Huang et al., 2022). On the other hand, a novel rice germplasm with increased grain size but decreased height was developed, confirming the weak correlation between PH and GL or size. Tomita and colleagues pyramided grain size–related gene GW2 and plant height-related gene sd1 in the background of Koshihikari in rice, and the newly developed line Koshihikary-sd1GW2 exhibited significantly increased GL and GW but semi-dwarfed PH (Tomita et al., 2022).
The profiling of major QTLs for grain-related traits enriches our knowledge of grain architecture in rice as well as provides potential genes for molecular breeding in rice variety improvement. However, grain-related traits exhibit typical quantitative characteristics with incompletely dissected molecular mechanisms of inheritance. Hence, new novel grain-related mutants are needed. In our previous work, we introduced chromosomal segments from Xihui 18 rice, an elite restorer line, to the background of Nipponbare, constructing a series of chromosome segment substitution lines (CSSLs). From these CSSLs, we identified a distinctive CSSL-Z1357 with significantly increased GL, using SSR-based marker-assisted selection (MAS). In the present study, we characterized CSSL-Z1357, constructed a segregating population to detect QTLs for GL and other traits of interest, and estimated their effects under independent and pyramiding backgrounds. Related results provide a basis for candidate gene dissection and grain-related trait improvement in rice breeding.
CSSL-Z1357 (Z1357) was screened out from the CSSLs produced from the crossing of Nipponbare (receptor) and Xihui 18 (donor). Z1357 carried nine chromosomal segments from the donor Xihui 18 as asserted via SSR-based MAS. The F2 population was constructed by crossing Z1357 and the receptor Nipponbare for QTL mapping of agronomic traits of interest. In 2017, F2 individuals containing target QTLs were self-pollinated to construct advanced CSSLs, including three lines carrying a single substitution segment (SSSL, S1 to S3), two lines with double substitution segments (DSSL, D1 and D2), and one line with triple-substitution segments (TSSL, T1). In 2020, all six screened lines (S1 to S3, D1, D2, and T1) were used to estimate the additive and epistatic effects of target QTLs.
At maturity, more than eight randomly selected plants of Z1357 and Nipponbare and all F2 individuals were cut at the ground level; then plant height (PH, cm) and panicle length (PL, cm) were measured. After air-drying, the grain number (filled grains) and spikelet number (total grains of both filled and unfilled grains) per panicle (abbreviated as GPP and SPP, respectively) were counted; 1000 filled grains were randomly selected for measuring of 1000-grain weight (TGW, g). Ten random filled grains were selected for measuring grain length (average grain length of 10 grains, GL, mm) and grain width (average grain width of 10 grais, GW, mm).
During the early heading stage, young panicles of Z1357 and Nipponbare were harvested and placed on ice for scanning electron microscope (SEM) examination. About 5 mm segments of middle glume were excised for observation of inner and outer epidermal cells according to the procedures described by Zhuang et al. (2020). The cell length and cell number of the inner and outer epidermis and outer parenchyma of glume within the same field of view were collected via software Simpal PCI.
DNA of all lines was extracted via the CTAB procedure. All the F2 individuals were genotyped via the polymorphic markers screened by the introgressed segments of Z1357 (Xiang et al., 2015). QTLs for seven agronomic traits of interest, i.e., PH, PL, GPP, SPP, TGW, GL, and GW, were detected by the method of restricted maximum likelihood (REML) implemented in HPMIXED of SAS. The threshold for identifying the association of candidate QTL and the particular trait was set to P <.05. All detected QTLs were named as per the pattern of qPH-1, whereby q referred to a QTL; PH to the target trait, i.e., PH; and the number to the chromosome of QTL location.
For each SSSL, DSSL, and TSSL, according to the genetic model P0 = μ + ϵ for Nipponbare and Pi = μ + ai + ϵ for the SSSLi carrying a specific QTL, Pij = μ + ai + aj + Iij + ϵ for DSSL, and Pijk = μ + ai + aj + ak + Iijk + ϵ for TSSL, P0 and Pi represent the phenotype value of any plant of Nipponbare and the SSSLi carrying the substitution segment i; Pij and Pijk represent the phenotype value of any plant in the DSSLij and TSSLijk; μ represents the mean value of Nipponbare population; ai, aj, and ak represent the additive effect of the QTL in substitution segments i, j, and k, respectively; Iij and Iijk represent the aiaj epistatic effect between QTLs in substitution segments i and j; aiajak denotes the epistatic effect between QTLs in the substitution segments i, j, and k, respectively; and ϵ represents the random error (Liang et al., 2021; Wang et al., 2021). Thus, the additive effect to the target trait was estimated via the formula (SSSLi-Nipponbare)/2 based on S1 to S3. The epistatic effect of each QTL pair (or substitution segment pair) to the control trait was estimated via the formula ((Nipponbare + DSSLij) – (SSSLi + SSSLj))/2 based on both DSSLs (D1 and 2) and SSSLs (S1 to S3). The epistatic effect of three QTLs (or three substitution segments) to the control trait was estimated via ((Nipponbare×2+TSSLijk) – (SSSLi+SSSLj+SSSLk))/2 (Liang et al., 2021; Wang et al., 2021). All calculations were carried out using Microsoft-Excel 2016.
Phenotyping results show that Z1357 exhibited significantly changed plant and panicle/grain architecture compared with the receptor, i.e., Nipponbare (Nipp) (Figures 1A–D). At maturity, seven traits of interest, including PH (cm), PL (cm), SPP, GPP, GL (mm), GW (mm), and TGW (g), were further characterized between Z1357 and Nipp. Both Z1357 and Nipp had similar SPP (104.90 ± 2.96 for Z1347 and 105.10 ± 9.66 for Nipp) and GPP (94.96 ± 5.07 for Z1347 and 98.99 ± 9.40 for Nipp), exhibiting nonsignificant differences (P >.05). The contrasting trends were observed for the remaining five traits (Figures 1E–I). Z1357 had statistically higher PH, PL, GL, and TGW than those of Nipp (Figures 1E–I). However, the average GW of Z1357 was 3.23 mm, significantly lower than that of Nipp (3.42 mm, P <.05, Figure 1H).
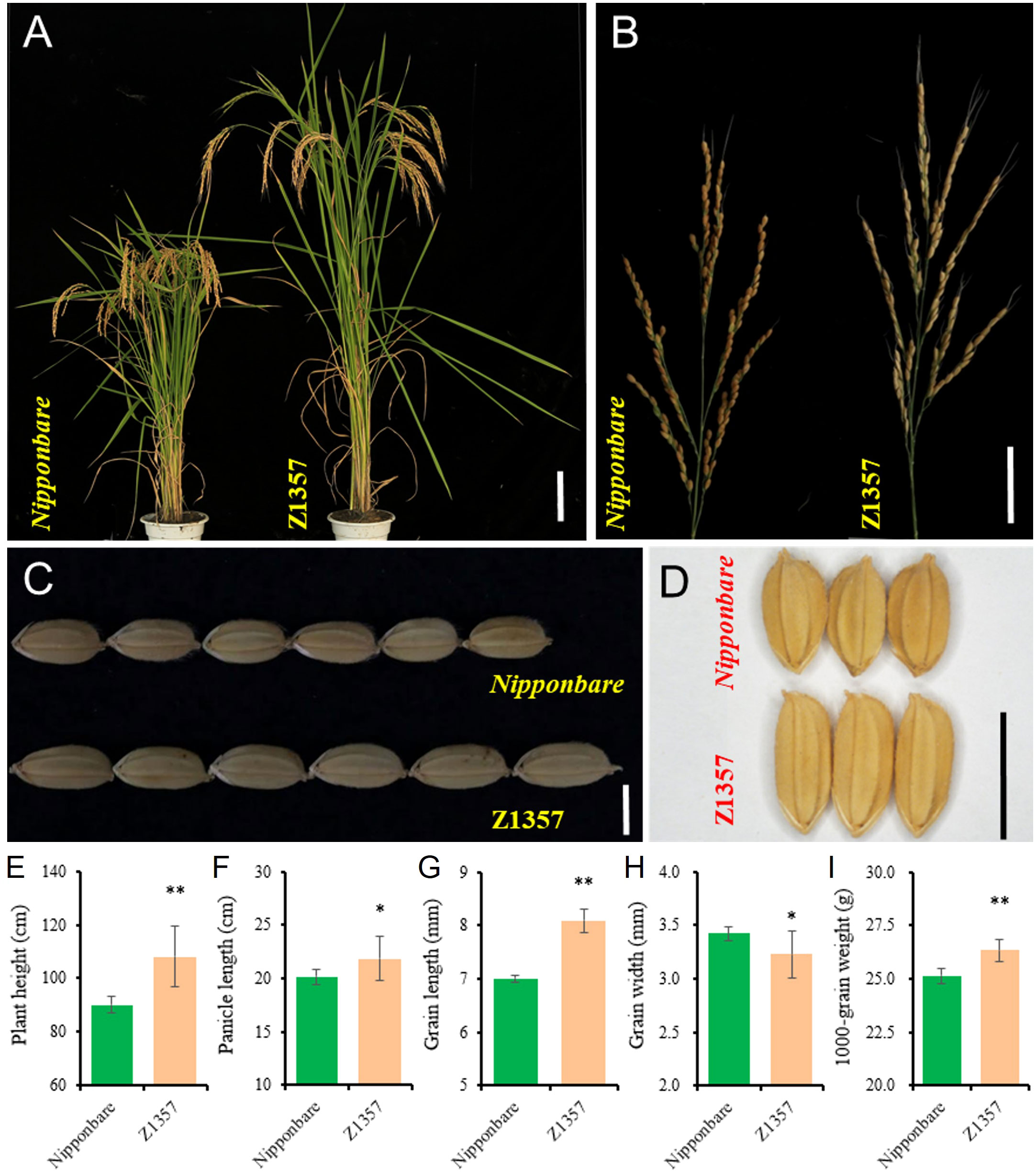
Figure 1 Phenotypes of Nipponbare and Z1357. (A) Plant architecture of Nipponbare and Z1357; (B) Panicles of Nipponbare and Z1357; (C, D) Phenotypes of grain length (C) and width (D) of Nipponbare and Z1357; (E–I) Comparison of PH (E), PL (F), GL (G), GW (H), and TGW (I) between Nipponbare and Z1357. Bar in A refers to 20 cm, in B to 5 cm, in C and D to 1 cm. * and ** refer to the significant differences at P < .05 and P < .01, respectively, by the t-test.
The morphology of glume cells was observed by SEM (Figures 2A–D). The results show that the length of glume inner cells of Z1357 was significantly longer (12.48%) than that of Nipp (Figures 2A, C) (P <.05, t-test, Figure 2E). On the other hand, the cell width of Z1357 glumes (33.55 µm) was statistically smaller than that of Nipp (40.86 µm) (P <.01, Figure 2F), but no significant difference was observed between cell numbers of Z1357 and Nipp (Figure 2G). These results suggest that the increased GL of Z1357 was mainly caused by the increased length of glume cells.
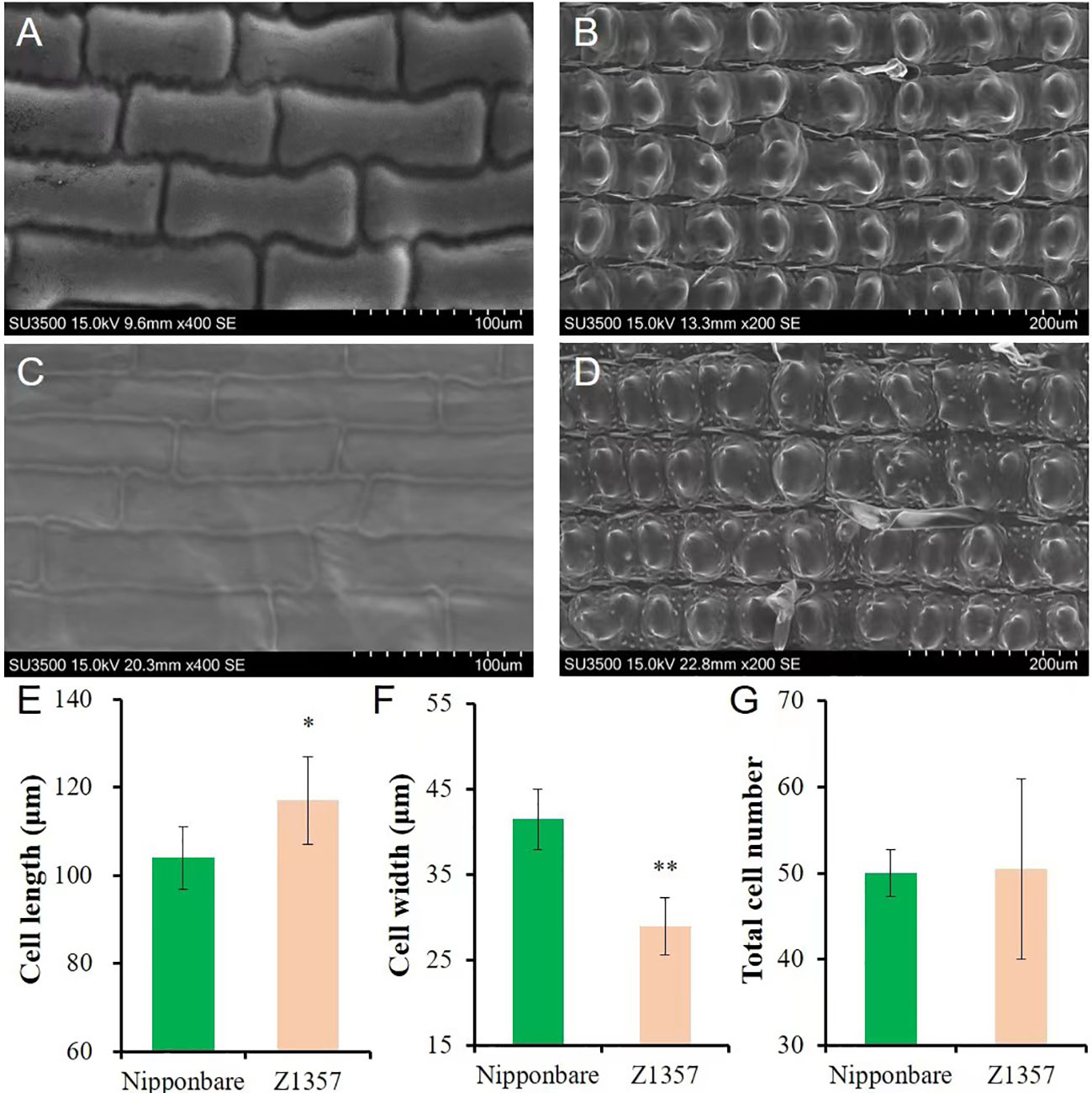
Figure 2 Observation and analysis of glume cells between Nipponbare and Z1357 via scanning electron microscope (SEM). A-B: SEM images of glume inner (A) and surface (B) cells of Nipponbare; (C, D) SEM images of glume inner (C) and surface (D) cells of Z1357; E-F: Comparison of inner glume cell length (E) and width (F) between Nipponbare and Z1357; (G) Comparison of glume surface cells number between Nipponbare and Z1357. * and ** refer to the significant differences at P < .05 and P < .01, respectively, by the t-test.
Nine segments were introduced into the genome of Z1357, located on seven chromosomes (Chr): Chr1, Chr3, Chr4, Chr6 to Chr8, and Chr12. The characterization results of 10 selected molecular markers showed that the introgressed segments carried by Z1357 exhibited the same genotypes to those of the donor line, Xihui18, indicating that the introgressed chromosomal segments from Xihui18 were homologous in the genome of Z1357. Among all seven Chrs carrying the introgressed segments, Chr3 and Chr12 contained two introgressed segments each, and the remaining five chromes had only one segment each (Figure 3).
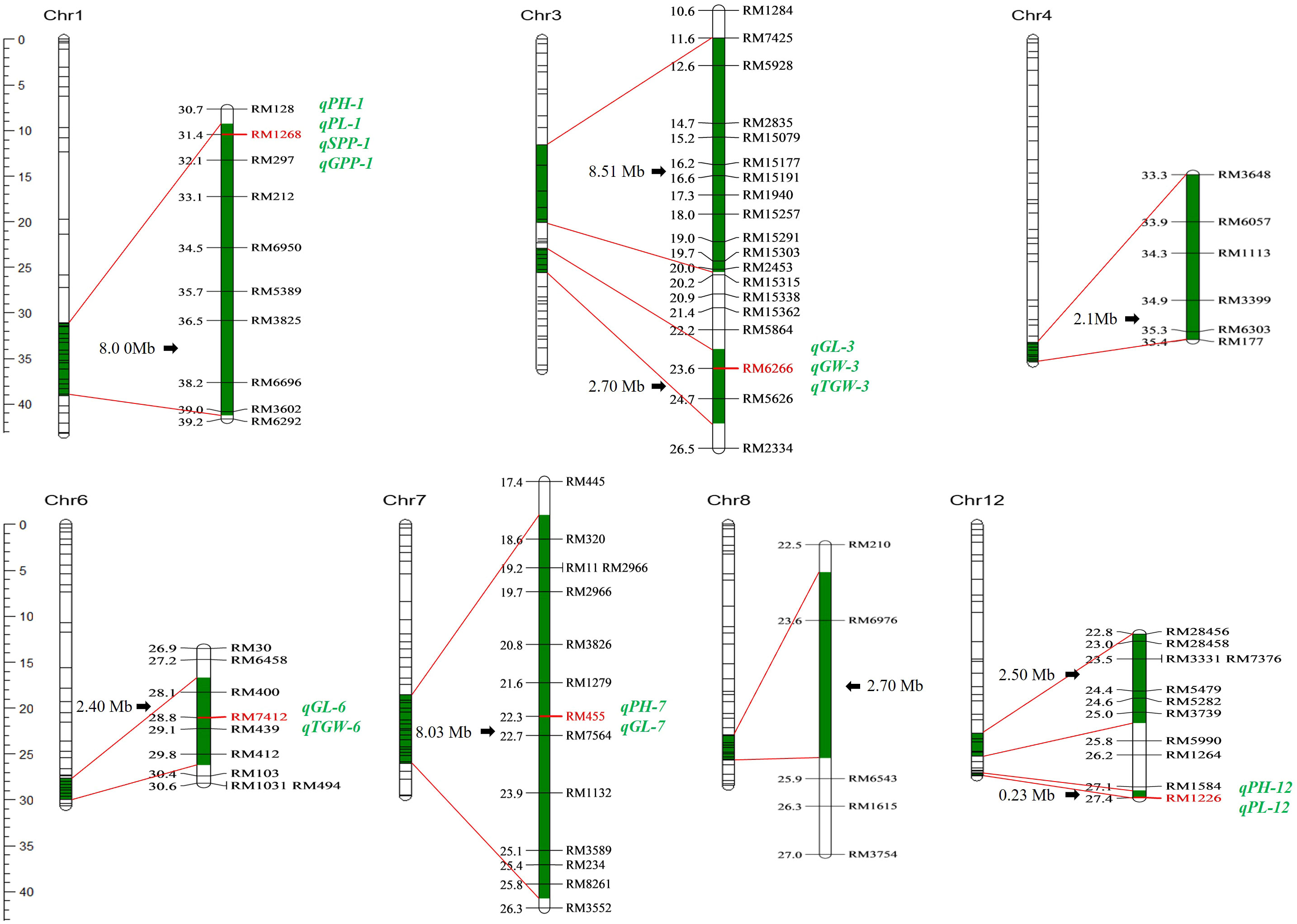
Figure 3 Genome distribution of substitution segments in Z1357. Substitution fragments are labeled in dark green. The left bar refers to the physical position (Mb) of markers (short lines of each Chr. bar) on seven chromosomes. All detected QTLs are listed in italics (green), and the linked marker of each QTL is listed in bold (red).
The total estimated length of all the nine introgressed segments was 37.17 Mb with an average of 4.13 Mb. The largest segment was detected on Chr3 with a length of 8.51 Mb (Figure 3). The shortest segment was detected on Chr12 with a length of 0.23 Mb (Figure 3). The length of the remaining seven segments ranged from 2.1 Mb to 8.03 Mb.
A total of 13 QTLs were detected for six agronomic traits (Table 1; Figure 3). These QTLs were located on five chromosomes, including Chr1 (four QTLs), Chr3 (three QTLs), Chr6 (two QTLs), Chr7 (two QTLs), and Chr12 (two QTLs) (Table 1; Figure 3). No QTLs were detected for any traits of interest on the first introgressed segment in Chr3 (from the top), nor introduced segments in Chr4 and Chr8 (Table 1; Figure 3).
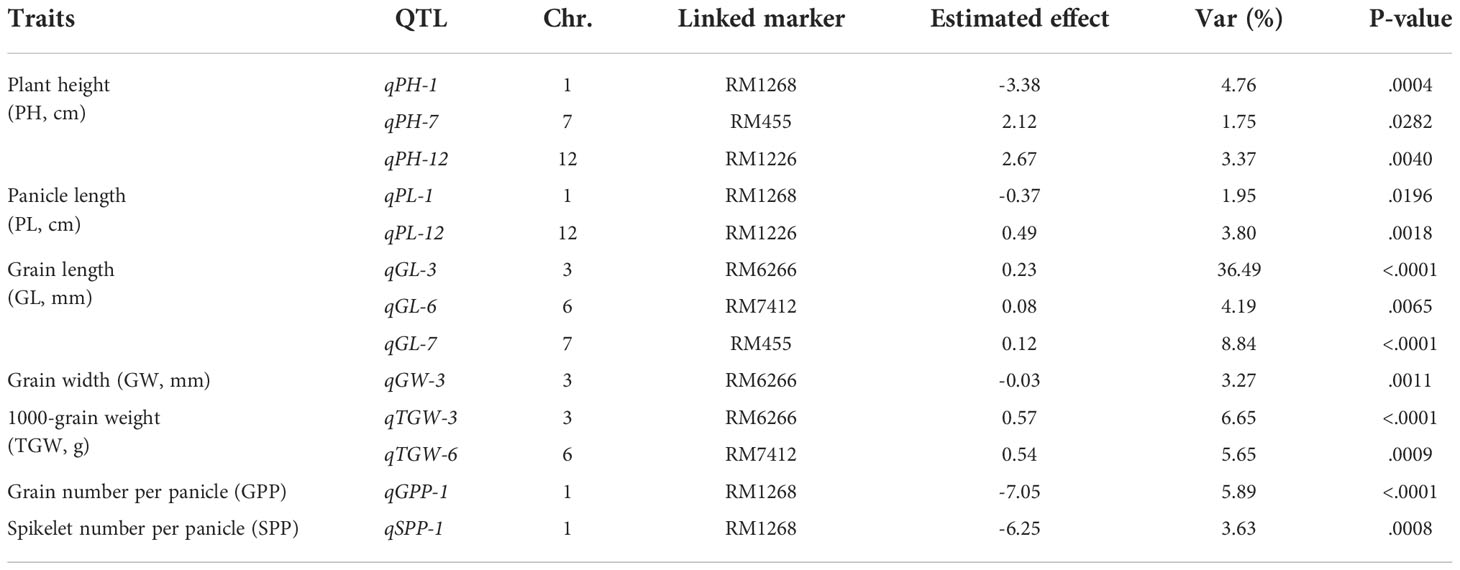
Table 1 Summary of 13 QTLs identified for the six interest traits via the F2 population of Nipponbare/Z1357.
Three QTLs, i.e., qPH-1, qPH-7, and qPH-12, were detected for PH and were located on Chr1, Chr7, and Chr12. Mapping results showed that qPH-1 could decrease PH by 3.38 cm, whereas qPH-7 and qPH-12 increased it by 2.12 and 2.67 cm, respectively, with Var <5% (Table 1). Two QTLs controlling PL, qPL-1 and qPL-12, were located on Chr1 and Chr12 (Table 1; Figure 3). Both qPH-1 and qPL-1 shared the same linking marker (RM1268, Figure 3).
Three QTLs, i.e., qGL-3, qGL-6, and qGL-7, were detected for GL and were located on Chr3, Chr6, and Chr7 (Figure 3). Mapping results showed that qGL-3/6/7 was associated with the additive effects of 0.23, 0.08, and 0.12 mm to GL with Var of 36.49%, 4.19%, and 8.84%, respectively (Table 1). The linking marker of qGL-3 was RM6266 and the two other QTLs, i.e., qGW-3 and qkGW-3, controlling GW and TGW, respectively, also linked with the same marker (Table 1; Figure 3). Additionally, qTGW-3 exhibited relatively higher Var (6.65%) regarding TGW (Table 1).
Only one QTL was detected for both SPP and GPP, i.e., qSPP-1 and qGPP-1 (Figure 3). They shared the same linking marker (RM1268) with qPH-1 and qPL-1, with the corresponding Var values of 3.63% and 5.89% (Table 1).
Based on the QTL mapping results using F2, three SSSLs (S1, S2, S3) were screened out from the F3 generation via MAS. Among these SSSLs, S1 carried four QTLs on Chr3 for GL, GW, PH, and TGW, and they had positive additive effects of 0.45 and 0.07 mm, 9.95 cm, and 1.91 g, respectively, causing statistically higher values of these traits than those of the receptor genotype Nipp (Figure 4). The line S2 also carried four QTLs for GL, GW, PH, and TGW. These QTLs were on Chr6. Two QTL (for GL and PH) showed positive additive effects of 0.28 mm and 9.24 cm, respectively, whereas the other two QTLs (for GW and TGW) exhibited negatively additive effects of -0.07 mm and -0.97 g, respectively, causing significantly increased GL and PH but significantly decreased GW and TGW than those of Nipp (Figure 4). The line S3 contained two QTLs for PH and TGW, presenting additive effects of 10.13 cm and -1.10 g, respectively, causing statistically increased PH but deceased TGW compared with those of Nipp (Figure 4).
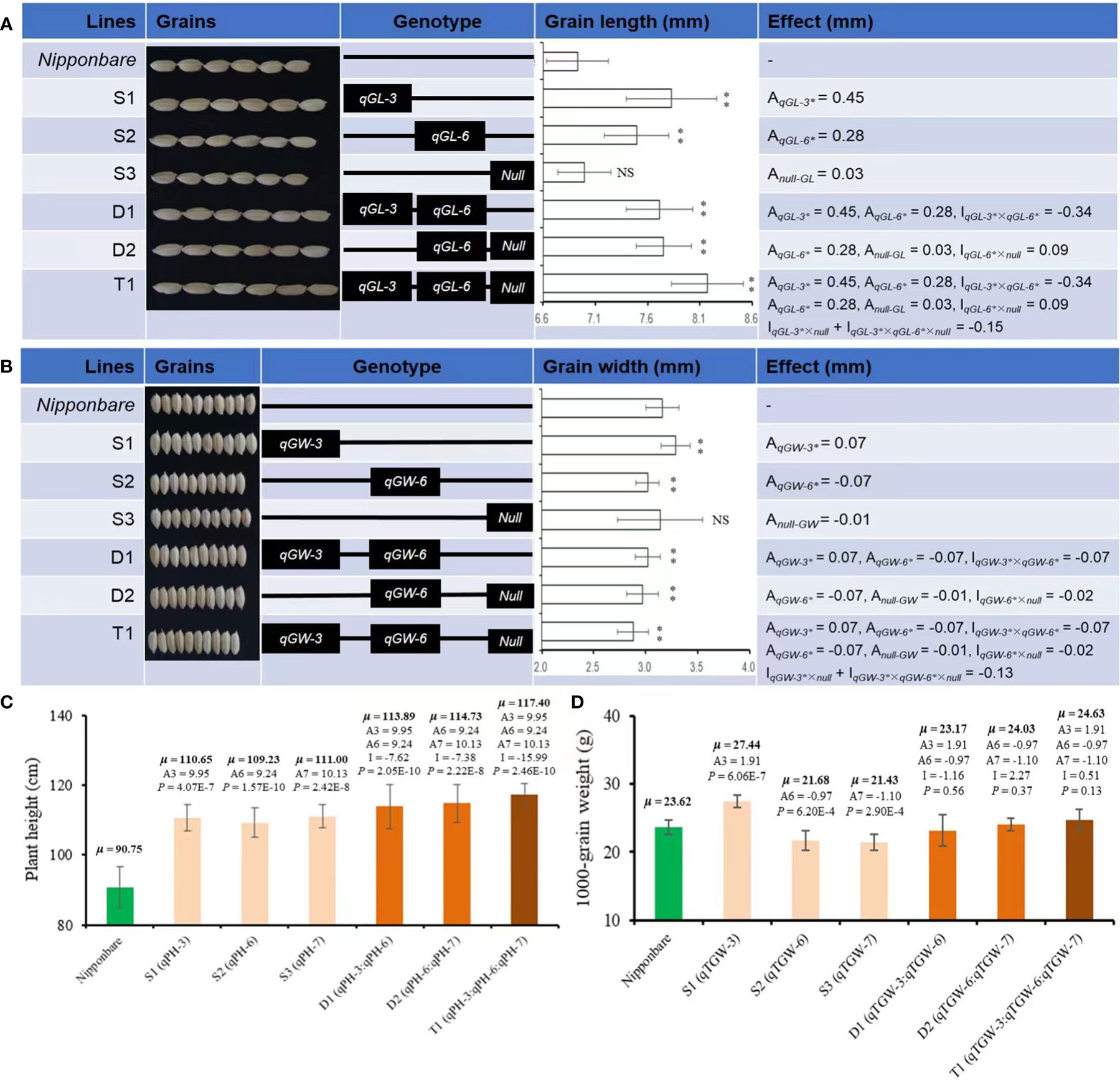
Figure 4 Phenotypes and estimated effects of QTLs for GL, GW, PH, and TGW among Nipponbare and the screened CSSLs. (A) Grain length (mm), (B) Grain width (mm), (C) Plant height (cm), (D) 1000-grain weight (g). μ: the average phenotypic value, a: denotes the additive effect of QTLs, i: denotes the additive × additive epistatic effect among QTLs. **, significant at.01 level (P < .01); NS, not significant. The P-value for a SSSL indicates the probability of a significant difference between the SSSL and Nipponbare.
Besides SSSLs (S1-S3), two DSSLs (D1 and D2) and one TSSL (T1) were also purposefully screened out from F3 generation via MAS according to the target QTL. D1 carried two chromosomal segments from Chr3 and Chr6 for GL, GW, PH, and TGW, and the broad interactive or epistatic effects (I) were detected for QTLs controlling the corresponding traits (Figure 4). For GL, the estimated I between qGL-3 and qGL-6 was -0.34 mm, and the pyramiding of these two QTLs caused significantly greater GL of D1 than that of Nipp (Figure 4A). A contrasting trend was observed for GW of D1. The estimated I between qGW-3 and qGW-6 was -0.07 mm, and the pyramiding of these two QTLs caused significantly decreased GW of D1 compared with Nipp (Figure 4B). Additionally, the estimated I between qPH-3 and qPH-6 was -7.62 cm. The epistatic effect between qTGW-3 and qTGW-6 was -1.16 g for TGW; the interaction effects caused significantly increased PH but nonsignificant changed TGW in D1 compared with Nipp (Figures 4C, D).
D2 captured two segments from Chr6 and Chr7. Although D2 carried only one QTL for each of GL (qGL-6) and GW (qGW-6), the interaction effects (I) were also observed between the introgressed segments with and without (Null) the target QTL. The estimated I between qGL-6 and Null was 0.09 mm, whereas the estimated epistatic effect between qGW-6 and Null was -0.02 mm (Figures 4A, B). These interaction effects caused significantly increased GL but decreased GW of D2 (P <.01), which was similar to the findings on D1 (Figures 4A, B). The estimated difference between qPH-6 and qPH-7 was -7.38 cm, and that between qTGW-6 and qTGW-7 was 2.27 g (Figures 4C, D). These diverse interaction effects for PH and TGW caused similar phenotypes of D2 and D1 by the pyramiding of the target QTL.
T1 captured all three segments from Chr3, Chr6, and Chr7. The pyramiding of qGL-3, qGL-6, and Null caused the highest GL of T1, and the estimated interaction effect of qGL-3 vs Null and qGL-3 vs qGL-6 vs Null was -0.15 mm (Figure 4A). In contrast to GL, T1 also had the lowest GW by pyramiding qGW-3, qGW-6, and Null, and the estimated interaction effect of qGW-3 vs Null and qGW-3 vs qGW-6 vs Null was -0.13 mm (Figure 4B). Trends similar to those of GL were also observed for PH and TGW of T1. The estimated interaction effect of qPH-3 vs qPH-6 vs qPG-7 was -15.99 cm, and that of qTGW-3 vs qTGW-6 vs qTGW-7 was 0.51 g; hence, pyramiding of these QTLs significantly increased PH (P <0.01) but did not significantly change TGW (P <0.13) of T1 (Figures 4C, D). Additionally, the pyramiding of targets QTLs in T1 also caused improved quality of T1 grains (Figure 5A). For example, the chalkiness rate of T1 grains was 11.86%, statistically lower than that of Nipp (32.00%, P <.01, Figure 5B). The overall chalky grain rate of T1 (41.02%) was also significantly lower than that of Nipp (69.36%, P <.01, Figure 5C).
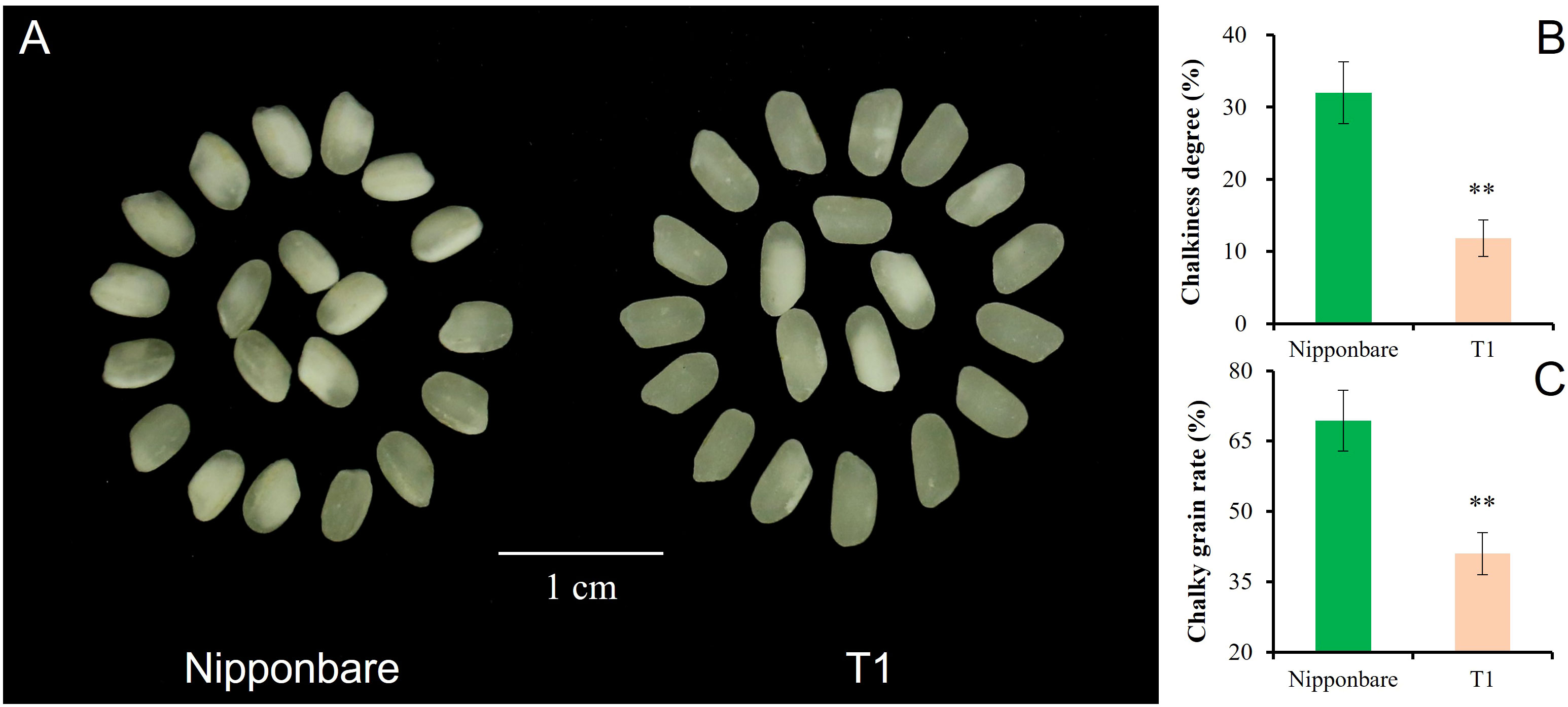
Figure 5 Chalkiness comparison of Nipponbare and T1. (A) Polished grain of Nipponbare and T1; (B) Chalkiness degree (%) of Nipponbare and T1; (C) Chalky grain rate (%) of Nipponbare and T1. **, significant at.01 level (P < .01).
Populations consisting of SSSLs are one of the major sources for QTL detection of complex quantitative traits for the distinctive features of eliminating background influences to the mapping procedures (Li et al., 2005; Tian et al., 2017; Chen et al., 2018; Zhang, 2021b). SSSLs have a background nearly the same as the receptor parent except for the introduced segment from the donor (Li et al., 2005; Chen et al., 2018; Tian et al., 2018). In our previous work, a set of CSSLs were developed by crossing Nipponbare (donor parent) and Xihui 18 (donor parent), and a distinctive line, Z1357, was screened out with significantly increased GL and PH but decreased GW (Figure 1). Results from phenotypic characterization also suggest that the introduced chromosomal segments from donor parent into Z1357 showed similar traits of GPP and SPP to those of the receptor parent (P >.05, Figure 1). Although the final yield of Z1357 was significantly lower than that of the receptor, the distinctive characteristics of Z1357 regarding increased GL (15.6%, P <.01) and PH (20%, P <.01), but decreased GW (5.6%, P <.05) showed a potential for utilization in grain quality improvement in rice breeding and research.
SEM showed that the lengthening and narrowing of grains were caused by the increase of cell length (12.48%) and the decrease of cell width (17.89%) in the glumes compared with Nipponbare (Figure 2). Dissection of other traits of interest using Z1357 would be useful in the related future research on these traits.
The QTL mapping results based on the F2 indicated that five out of nine substitution segments contained in Z1357 carried QTLs for six traits of interest, i.e., QTLs for PH and PL on segments of Chr1, Chr7, and Chr12 and QTLs for GL and GW on Chr3 and Chr6, respectively (Table 1). We also found that each introduced fragment contained multiple QTLs for different traits, and for each trait, the detected QTLs were located on different introgression segments, which indicated a complex cross-talk between introgression segments and the traits of interest.
Grain type in rice, especially GL and GW, is one of the most important components for quality improvement of rice varieties. As expected, GL was a typical quantitative trait and was controlled by both the major and minor QTLs. Numerous QTLs associated with GL have been reported across the entire rice genome using populations of F2, F3, and recombinant inbred lines (Zhang et al., 2021). However, the features of those populations limit the thorough dissection of candidate QTLs, such as cloning and functional profiling. In the present study, we identified the major QTLs for GL and GW within the introduced segments of Chr3 and Chr6, respectively; in particular, qGL-3 on Chr3 increased GL by 0.23 mm with Var of 36.5% (Table 1). At the locus of qGL-3, a functional gene of GL3.1 controlling the GL in rice was reported (Qi et al., 2012). The coding product of GL3.1 is Ser/Thr phosphatase that belongs to protein phosphatase kelch-like (PPKL) family (Qi et al., 2012). Qi and colleagues revealed that GL3.1 functioned by influencing the phenotype of grain size and yield of rice via regulating a cell cycle-related protein (cyclin-T1;3) via dephosphorylation (Qi et al., 2012). The downregulation of cyclin-T1;3 by dephosphorylation tended to produce shortened rice grains (Qi et al., 2012).
On Chr6, the reported functional gene, GW6a, was located in the same region as qGL-6 (Song et al., 2015). The product of GW6a is a new-type GNAT-like protein, serving as intrinsic histone acetyltransferase (OsglHAT1) (Song et al., 2015). Overexpression of OsglHAT1 tended to increase the overall acetylation levels of histone H4 and the cell number in grains, resulting in enlarged grain size and enhanced final yield of rice (Song et al., 2015). Another reported gene, GW7/GL7 is located in the same region as qGL-7 (Wang et al., 2015a, b). The product of GW7/GL7 corresponds to longitudinal cell elongation, homologous to the LONGIFOLIA protein of Arabidopsis (Wang et al., 2015). The increased abundance of GW7/GL7 coding product in vitro accelerated the longitudinal cell division and decreased the transverse cell division, resulting in increased GL and improved grain quality in terms of appearance (Wang et al., 2015).
The gene OsBZR1 is responsible for PH in rice and is located in the physical interval of qGL-7 (Qiao et al., 2017). The OsBZR1 serves as the signal molecule downstream of the brassinolide transduction pathway (Qiao et al., 2017). Overexpression of OsBZR1 tended to increase the sugar accumulation in developing anthers and seeds and also enhanced GL, GW, thickness, TGW, and spikelet number (Qiao et al., 2017). In summary, the QTLs reported in the present study (qGL-3, qGL-6, and qGL-7) were located within the same regions as the four genes reported in the literature, i.e., GL3.1, GW6.1, GW7/GL7, and OsBZR1. Further work is needed to reveal whether those three QTLs contain the allelic variations of the reported genes.
Based on the results of QTL mapping, six further substitution fragment lines (S1-S3, D1-D2, T1) were screened out (Figure 4). Characterization of these six lines provides the practical knowledge for molecular or QTL-based improvement of target traits in rice. For example, in rice variety improvement, breeders prefer to select longer grains or grains with higher ratio of length/width (Li et al., 2021). The results of mapping and effect estimation indicated that qGL-3 and qGL-6 could both increase the GL in rice (Figure 4A). If these two QTLs acted together, then the CSSL D1 that carried both QTLs should show increased GL by 1.46 mm (0.45x2+0.28x2) with respect to the receptor parent Nipp. Hence, the GL of D1 should be 8.39 mm (6.93 + 1.46) if qG-3 and qGL6 acted jointly on GL. However, the observed GL of D1 was 7.71 mm (i.e., <8.39 mm), suggesting that qGL3 and qGL6 acted antagonistically to each other regarding GL in rice (Figure 4A). Furthermore, the t-test results showed that the GL of D1 was higher than that of S2 that carried only qGL6 (P <.01), and qGL6 interacted even negatively with qGL3. This result suggests that, in the breeding activities related to GL improvement, we can either utilize qGL3 independently or pyramid qGL-3 and qGL-6 to produce longer grains in rice.
Interestingly, using the F2 population, qGL-7 for GL was detected on the introgressed segment from Chr7, but no QTL for GL was identified via the CSSL S3; even the core marker linked to qGL-7 was fixed within the introgression segment on Chr7 carried by S3 (Figure 4A). However, the phenotypic results showed that the introgressed segment carried by S3 (Null) exhibited a weak increasing effect on GL (0.03 mm, Figure 4A). Such a weak effect caused by Null might have resulted from complex relationships of GL, PH, PL, and other agronomic traits (Zhang et al., 2020). In addition, Null interacted with qGL-6 and exhibited a positive epistatic effect of 0.09 mm to GL (Figure 4A). When qGL-3, qGL-6, and Null were pyramided, i.e., T1, the GL was the longest (8.17 mm, Figure 4A). These results indicated that using the target QTLs or gene(s) for improvement of candidate trait(s) should be done with a strong consideration of the locus or chromosomal regions without target QTLs.
Despite the increasing effect of qGW-3 on GW, both qGW-6 and Null had negative effects to GW, and the exacerbated negative trends were observed after pyramiding qGW-3, qGW-6, and Null (Figure 4B). Phenotyping results showed that the strongest decreasing effects were caused by the epistatic interaction of qGW-3, qGW-6, and Null (T1), then qGW-6 and Null (D2), and then qGW-3 and qGW-6 (D1, Figure 4B). These results suggest that, if one tended to decrease GW in rice slightly, one could utilize qGW-6 alone or combine qGW-3 and qGW-6 together, which would cause approximately a 4% decrease of GW. To achieve a moderate decrease of GW, the integration of qGW-6 and Null would tend to produce approximately a 7% decrease of GW. Finally, the pyramiding qGW-3, qGW-6, and Null in breeding activities might result in about a 10% decrease of GW in rice. In addition to the improved phenotypic performances of GL and GW, the pyramiding of target QTLs in T1 also caused side effects for better quality of polished grains in T1 (Figure 5), implying more potential utilization for yield and quality improvement of rice varieties.
In the present study, an elite CSSL named Z1357, screened from the progeny derived from crossing of Nipponbare as the receptor and Xihui18 as the donor, was characterized by carrying nine substitution segments with the average length of 4.13 Mb. Thirteen QTLs were detected on nine substitution segments for the seven traits of interest. Results via other CSSLs, i.e., S1 to S3, D1, D2, and T1, showed that pyramiding the segments from Chr3 (qGL-3, qPH-3, and qGW-3), Chr6 (qGL-6, qPH-6, and qGW-6), and Chr7 (qPH-7 and qTGW-7) tended to produce increased GL and PH and decreased GW, providing a potential theoretical basis for enhancing grain yield and quality in rice breeding.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
ZM, XD, SX, QC, XM, and MC performed the experiments, ZM and XD drafted this manuscript. FZ and YL designed the experiments, developed genetic populations, and planned the structure of the manuscript. ZY, FZ, and YL participated in the development of Z1357. All authors read and approved the final manuscript.
This work was supported by the Chongqing Outstanding Scientist Foundation (cstc2022ycjh-bgzxm0134) and Undergraduate Innovation and Entrepreneurship Training Program of Southwest University (S202210635248).
We thank Professor Shizhong Xu from University of California, Riverside, USA, who wrote the program for QTL mapping.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1020847/full#supplementary-material
Chen, J., Wang, J., Chen, W., Sun, W., Peng, M., Yuan, Z., et al. (2018). Metabolome analysis of multi-connected biparental chromosome segment substitution line populations. Plant Physiol. 178 (2), 612–625. doi: 10.1104/pp.18.00490
Che, R., Tong, H., Shi, B., Liu, Y., Fang, S., Liu, D., et al. (2015). Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants 2 (1), 15195. doi: 10.1038/nplants.2015.195
Duan, P., Ni, S., Wang, J., Zhang, B., Xu, R., Wang, Y., et al. (2015). Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat. Plants 2, 15203. doi: 10.1038/nplants.2015.203
Duan, P., Xu, J., Zeng, D., Zhang, B., Geng, M., Zhang, G., et al. (2017). Natural variation in the promoter of GSE5 contributes to grain size diversity in rice. Mol. Plant 10 (5), 685–694. doi: 10.1016/j.molp.2017.03.009
Fan, Y., Li, Y. (2019). Molecular, cellular and yin-yang regulation of grain size and number in rice. Mol. Breed. 39 (12), 163. doi: 10.1007/s11032-019-1078-0
Fan, C., Xing, Y., Mao, H., Lu, T., Han, B., Xu, C., et al. (2006). GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112 (6), 1164–1171. doi: 10.1007/s00122-006-0218-1
Gao, X., Zhang, J., Zhang, X., Zhou, J., Jiang, Z., Huang, P., et al. (2019). Rice qGL3/OsPPKL1 functions with the GSK3/SHAGGY-like kinase OsGSK3 to modulate brassinosteroid signaling. Plant Cell 31 (5), 1077–1093. doi: 10.1105/tpc.18.00836
Hao, J., Wang, D., Wu, Y., Huang, K., Duan, P., Li, N., et al. (2021). The GW2-WG1-OsbZIP47 pathway controls grain size and weight in rice. Mol. Plant 14 (8), 1266–1280. doi: 10.1016/j.molp.2021.04.011
Huang, J., Chen, Z., Lin, J., Chen, J., Wei, M., Liu, L., et al. (2022). Natural variation of the BRAD2 allele affects plant and grain size in rice. Planta 256, 27. doi: 10.1007/s00425-022-03939-7
Huang, K., Wang, D., Duan, P., Zhang, B., Xu, R., Li, N., et al. (2017). WIDE AND THICK GRAIN 1, which encodes an otubain-like protease with deubiquitination activity, influences grain size and shape in rice. Plant J. 91 (5), 849–860. doi: 10.1111/tpj.13613
Hu, Z., Lu, S. J., Wang, M. J., He, H., Sun, L., Wang, H., et al. (2018). Novel QTL qTGW3 encodes the GSK3/SHAGGY-like kinase OsGSK5/OsSK41 that interacts with OsARF4 to negatively regulate grain size and weight in rice. Mol. Plant 11 (5), 736–749. doi: 10.1016/j.molp.2018.03.005
Hu, X., Qian, Q., Xu, T., Zhang, Y., Dong, G., Gao, T., et al. (2013). The U-box E3 ubiquitin ligase TUD1 functions with a heterotrimeric G α subunit to regulate brassinosteroid-mediated growth in rice. PloS Genet. 9 (3), e1003391. doi: 10.1371/journal.pgen.1003391
Hu, J., Wang, Y., Fang, Y., Zeng, L., Xu, J., Yu, H., et al. (2015). A rare allele of GS2 enhances grain size and grain yield in rice. Mol. Plant 8 (10), 1455–1465. doi: 10.1016/j.molp.2015.07.002
Ishimaru, K., Hirotsu, N., Madoka, Y., Murakami, N., Hara, N., Onodera, H., et al. (2013). Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 45 (6), 707–711. doi: 10.1038/ng.2612
Jin, J., Hua, L., Zhu, Z., Tan, L., Zhao, X., Zhang, W., et al. (2016). GAD1 encodes a secreted peptide that regulates grain number, grain length, and awn development in rice domestication. Plant Cell 28, 2453–2463. doi: 10.1105/tpc.16.00379
Lakshmi, M. V., Suneetha, Y., Yugandhar, G., Lakshmi, N. V. (2014). Correlation studies in rice (Oryza stativa l.). Int. J. Genet. Eng. Biotechnol. 5, 121–126. www.ripublication.com/irph/ijgeb-spl/ijgebv5n2_06.pdf
Liang, P., Wang, H., Zhang, Q., Zhou, K., Li, M., Li, R., et al. (2021). Identification and pyramiding of QTLs for rice grain size based on short-wide grain CSSL-Z563 and fine-mapping of qGL3-2. Rice 14 (1), 35. doi: 10.1186/s12284-021-00477-w
Li, Y., Fan, C., Xing, Y., Jiang, Y., Luo, L., Sun, L., et al. (2011). Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43 (12), 1266–1269. doi: 10.1038/ng.977
Li, Z., Fu, B., Gao, Y., Xu, J., Ali, J., Lafitte, H., et al. (2005). Genome-wide introgerssion lines and their use in genetic and molecular dissection of complex phenotypes in rice (Oryza sativa l.). Plant Mol. Biol. 59 (1), 33–52. doi: 10.1007/s11103-005-8519-3
Li, G., Tang, J., Zheng, J., Chu, C. (2021). Exploration of rice yield potential: decoding agronomic and physiological traits. Crop J. 9 (3), 577–589. doi: 10.1016/j.cj.2021.03.014
Liu, J., Chen, J., Zheng, X., Wu, F., Lin, Q., Heng, Y., et al. (2017). GW5 acts in the brassinosteroid signaling pathway to regulate grain width and weight in rice. Nat. Plants 3, 17043. doi: 10.1038/nplants.2017.43
Liu, Q., Han, R., Wu, K., Zhang, J., Ye, Y., Wang, S., et al. (2018). G-Protein βγ subunits determine grain size through interaction with MADS-domain transcription factors in rice. Nat. Commun. 9 (1), 852. doi: 10.1038/s41467-018-03047-9
Liu, L., Tong, H., Xiao, Y., Che, R., Xu, F., Hu, B., et al. (2015). Activation of Big Grain1 significantly improves grain size by regulating auxin transport in rice. Proc. Natl. Acad. Sci. U.S.A. 112 (35), 11102–11107. doi: 10.1073/pnas.1512748112
Liu, D., Yu, Z., Zhang, G., Yin, W., Li, L., Niu, M., et al. (2021). Diversification of plant agronomic traits by genome editing of brassinosteroid signaling family genes in rice. Plant Physiol. 187 (4), 2563–2576. doi: 10.1093/plphys/kiab394
Lo, S., Cheng, M., Hsing, Y., Chen, Y., Lee, K., Hong, Y., et al. (2020). Rice big grain 1 promotes cell division to enhance organ development, stress tolerance and grain yield. Plant Biotechnol. J. 18 (9), 1969–1983. doi: 10.1111/pbi.13357
Lu, Y., Chuan, M., Wang, H., Chen, R., Tao, T., Zhou, Y., et al. (2022). Genetic and molecular factors in determining grain number per panicle of rice. Front. Plant Sci. 13, 964246. doi: 10.3389/fpls.2022.964246
Luo, J., Liu, H., Zhou, T., Gu, B., Huang, X., Shang, G., et al. (2013). An-1 encodes a basic helix-loop-helix protein that regulates awn development, grain size, and grain number in rice. Plant Cell 25 (9), 3360–3376. doi: 10.1105/tpc.113.113589
Lyu, J., Wang, D., Duan, P., Liu, Y., Huang, K., Zeng, D., et al. (2020). Control of grain size and weight by the GSK2-LARGE1/OML4 pathway in rice. Plant Cell 32 (6), 1905–1918. doi: 10.1105/tpc.19.00468
Mao, H., Sun, S., Yao, J., Yao, J., Wang, C., Yu, S., et al. (2010). Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. U.S.A. 107 (45), 19579–19584. doi: 10.1073/pnas.1014419107
Miao, J., Yang, Z., Zhang, D., Wang, Y., Xu, M., Zhou, L., et al. (2019). Mutation of RGG2, which encodes a type b heterotrimeric G protein γ subunit, increases grain size and yield production in rice. Plant Biotechnol. J. 17 (3), 650–664. doi: 10.1111/pbi.13005
Qiao, J., Jiang, H., Lin, Y., Shang, L., Wang, M., Li, D., et al. (2021). A novel miR167a-OsARF6-OsAUX3 module regulates grain length and weight in rice. Mol. Plant 14 (10), 1683–1698. doi: 10.1016/j.molp.2021.06.023
Qiao, S., Sun, S., Wang, L., Wu, Z., Li, C., Li, X., et al. (2017). The RLA1/SMOS1 transcription factor functions with OsBZR1 to regulate brassinosteroid signaling and rice architecture. Plant Cell 29 (2), 292–309. doi: 10.1105/tpc.16.00611
Qi, P., Lin, Y., Song, X., Shen, J., Huang, W., Shan, J., et al. (2012). The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating cyclin-T1; 3. Cell Res. 22, 1666–1680. doi: 10.1038/cr.2012.151
Qi, Z., Xiong, L. (2013). Characterization of a purine permease family gene OsPUP7 involved in growth and development control in rice. J. Integr. Plant Biol. 55 (11), 1119–1135. doi: 10.1111/jipb.12101
Sabesan, T., Suresh, R., Saravanan, K. (2009). Genetic variability and correlation for yield and grain quality characters of rice grown in coastal saline low land of tamilnadu. Electronic. J. Plant Breed. 1, 56–59. www.cabdirect.org/cabdirect/abstract/20103117225
Shi, C., Ren, Y., Liu, L., Wang, F., Zhang, H., Tian, P., et al. (2019). Ubiquitin specific protease 15 has an important role in regulating grain width and size in rice. Plant Physiol. 180 (1), 381–391. doi: 10.1104/pp.19.00065
Si, L., Chen, J., Huang, X., Gong, H., Luo, J., Hou, Q., et al. (2016). OsSPL13 controls grain size in cultivated rice. Nat. Genet. 48 (4), 447–456. doi: 10.1038/ng.3518
Song, X., Huang, W., Shi, M., Zhu, M., Lin, H. (2007). A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39 (5), 623–630. doi: 10.1038/ng2014
Song, X., Kuroha, T., Ayano, M., Furuta, T., Nagai, K., Komeda, N., et al. (2015). Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice. Proc. Natl. Acad. Sci. U.S.A. 112 (1), 76–81. doi: 10.1073/pnas.1421127112
Sun, S., Wang, L., Mao, H., Shao, L., Li, X., Xiao, J., et al. (2018). A G-protein pathway determines grain size in rice. Nat. Commun. 9, 851. doi: 10.1038/s41467-018-03141-y
Tao, Y., Miao, J., Wang, J., Li, W., Xu, Y., Wang, F., et al. (2020). RGG1, involved in the cytokinin regulatory pathway, controls grain size in rice. Rice 13 (1), 76. doi: 10.1186/s12284-020-00436-x
Tian, X., Li, X., Zhou, W., Ren, Y., Wang, Z., Liu, Z., et al. (2017). Transcription factor OsWRKY53 positively regulates brassinosteroid signaling and plant architecture. Plant Physiol. 175 (3), 1337–1349. doi: 10.1104/pp.17.00946
Tomita, M., Ebata, H., Nakayama, K. (2022). Large-Grain and semidarf isogenic rice koshihikari integrated with GW2 and sd1. Sustainability 14, 11075. doi: 10.3390/su141711075
Tong, H., Liu, L., Jin, Y., Du, L., Yin, Y., Qian, Q., et al. (2012). DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell 24 (6), 2562–2577. doi: 10.1105/tpc.112.097394
Wang, A., Hou, Q., Si, L., Huang, X., Luo, J., Lu, D., et al. (2019). The PLATZ transcription factor GL6 affects grain length and number in rice. Plant Physiol. 180 (4), 2077–2090. doi: 10.1104/pp.18.01574
Wang, S., Li, S., Liu, Q., Wu, K., Zhang, J., Wang, S., et al. (2015a). The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 47 (8), 949–954. doi: 10.1038/ng.3352
Wang, S., Wu, K., Qian, Q., Liu, Q., Li, Q., Pan, Y., et al. (2017). Non-canonical regulation of SPL transcription factors by a human OTUB1-like deubiquitinase defines a new plant type rice associated with higher grain yield. Cell Res. 27 (9), 1142–1156. doi: 10.1038/cr.2017.98
Wang, S., Wu, K., Yuan, Q., Liu, X., Liu, Z., Lin, X., et al. (2012). Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44 (8), 950–954. doi: 10.1038/ng.2327
Wang, Y., Xiong, G., Hu, J., Jiang, L., Yu, H., Xu, J., et al. (2015b). Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 47 (8), 944–948. doi: 10.1038/ng.3346
Wang, D., Zhou, K., Xiang, S., Zhang, Q., Li, R., Li, M., et al. (2021). Identification, pyramid and candidate genes of QTLs for associated traits based on a dense erect panicle rice CSSL-Z749 and five SSSLs, three DSSLs and one TSSL. Rice 14 (1), 55. doi: 10.1186/s12284-021-00496-7
Weng, J., Gu, S., Wan, X., Gao, H., Guo, T., Su, N., et al. (2008). Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 18, 1199–1209. doi: 10.1038/cr.2008.307
Xiang, J., Li, Y., Fan, Y. W., Xu, J. H., Zheng, L. Y., He, G. H., et al. (2015). Identification and morphological analysis of a rice chromosome segment substitution line carrying a major effect gene for late heading date and mapping of Ehd4-2. Agron. Sin. 41(5), 683–691. doi: 10.3724/SP.J.1006.2015.00683
Xiao, Y., Liu, D., Zhang, G., Gao, S., Liu, L., Xu, F., et al. (2019). Big Grain3, encoding a purine permease, regulates grain size via modulating cytokinin transport in rice. J. Integr. Plant Biol. 61 (5), 581–597. doi: 10.1111/jipb.12727
Xu, R., Duan, P., Yu, H., Zhou, Z., Zhang, B., Wang, R., et al. (2018). Control of grain size and weight by the OsMKKK10-OsMKK4-OsMAP K6 signaling pathway in rice. Mol. Plant 11 (6), 860–873. doi: 10.1016/j.molp.2018.04.004
Xu, Z. J., Chen, W. F., Ma, D. R., Lü, Y. N., Zhou, S. Q., Liu, L. X.. (2004). Correlations between rice grain shapes and main qualitative characteristics. Acta. Agron. Sin. 30(9), 894–900. doi: 10.3321/j.issn:0496-3490.2004.09.009
Yin, W., Xiao, Y., Niu, M., Meng, W., Li, L., Zhang, X., et al. (2020). ARGONAUTE2 enhances grain length and salt tolerance by activating BIG GRAIN3 to modulate cytokinin distribution in rice. Plant Cell 32 (7), 2292–2306. doi: 10.1105/tpc.19.00542
Zhang, G. (2021b). Target chromosome-segment substitution: A way to breeding by design in rice. Crop J. 9 (3), 658–668. doi: 10.1016/j.cj.2021.03.001
Zhang, F., Shi, Y., Ali, J., Xu, J., Li, Z. (2021). Breeding by selective introgression: Theory, practices, and lessons learned from rice. Crop J. 9 (3), 646–657. doi: 10.1016/j.cj.2021.03.006
Zhang, X., Wang, J., Huang, J., Lan, H., Wang, C., Yin, C., et al. (2012). Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. U.S.A. 109 (52), 21534–21539. doi: 10.1073/pnas.1219776110
Zhang, T., Wang, S., Sun, S., Zhang, Y., Li, J., You, J., et al. (2020). QTL analysis based on a long-grain CSSL-Z1392 and SSSLs and fine mapping of qGL6. Rice 13, 40. doi: 10.1186/s12284-020-00399-z
Zhang, D., Zhang, M., Liang, J. (2021a). RGB1 regulates grain development and starch accumulation through its effect on OsYUC11-mediated auxin biosynthesis in rice endosperm cells. Front. Plant Sci. 12, 585174. doi: 10.3389/fpls.2021.585174
Zhan, P., Wei, X., Xiao, Z., Wang, X., Ma, S., Lin, S., et al. (2021). GW10, a member of P450 subfamily regulates grain size and grain number in rice. Theor. Appl. Genet. 134, 3941–3950. doi: 10.1007/s00122-021-03939-3
Zhao, D., Li, Q., Zhang, C., Zhang, C., Yang, Q., Pan, L., et al. (2018). GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat. Commun. 9 (1), 1240. doi: 10.1038/s41467-018-03616-y
Zhou, S., Xue, H. (2020). The rice PLATZ protein SHORT GRAIN6 determines grain size by regulating spikelet hull cell division. J. Integr. Plant Biol. 62 (6), 847–864. doi: 10.1111/jipb.12851
Keywords: rice (Oryza sativa L.), chromosome segment substitution lines, grain-related traits, QTL identification, pyramiding
Citation: Mao Z, Di X, Xia S, Chen Q, Ma X, Chen M, Yang Z, Zhao F and Ling Y (2022) Detecting and pyramiding target QTL for plant- and grain-related traits via chromosomal segment substitution line of rice. Front. Plant Sci. 13:1020847. doi: 10.3389/fpls.2022.1020847
Received: 16 August 2022; Accepted: 26 October 2022;
Published: 16 December 2022.
Edited by:
Yufeng Hu, Sichuan Agricultural University, ChinaReviewed by:
Deyong Ren, China National Rice Research Institute (CAAS), ChinaCopyright © 2022 Mao, Di, Xia, Chen, Ma, Chen, Yang, Zhao and Ling. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fangming Zhao, emhhb2ZhbmdtaW5nMjAwNEAxNjMuY29t; Yinghua Ling, bGluZ3loNjVAc3d1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.