
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Plant Sci., 31 October 2022
Sec. Plant Development and EvoDevo
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1018559
This article is part of the Research TopicBeyond genetics: modifications of nucleic acid and chromatinView all 6 articles
 Ralf Müller-Xing*
Ralf Müller-Xing* Qian Xing
Qian XingPluripotent stem-cells are slowly dividing cells giving rise to daughter cells that can either differentiate to new tissues and organs, or remain stem-cells. In plants, stem-cells are located in specific niches of the shoot and root apical meristems (SAMs and RAMs). After ablation of stem-cell niches, pluripotent meristematic cells can establish new stem-cells, whereas the removal of the whole meristem destructs the regeneration process. In tissue cultures, after detached plant organs are transferred to rooting or callus induction medium (G5 or CIM), vasculature-associated pluripotent cells (VPCs) immediately start proliferation to form adventitious roots or callus, respectively, while other cell types of the organ explants basically play no part in the process. Hence, in contrast to the widely-held assumption that all plant cells have the ability to reproduce a complete organism, only few cell types are pluripotent in practice, raising the question how pluripotent stem-cells differ from differentiated cells. It is now clear that, in addition to gene regulatory networks of pluripotency factors and phytohormone signaling, epigenetics play a crucial role in initiation, maintenance and determination of plant stem-cells. Although, more and more epigenetic regulators have been shown to control plant stem-cell fate, only a few studies demonstrate how they are recruited and how they change the chromatin structure and transcriptional regulation of pluripotency factors. Here, we highlight recent breakthroughs but also revisited classical studies of epigenetic regulation and chromatin dynamics of plant stem-cells and their pluripotent precursor-cells, and point out open questions and future directions.
Unlike animals, plant growth and organ formation occur post-embryonically, mediated by meristems that are located on the tips of growth axes in shoots and roots (Doerner, 2003). Meristems contain a specialized cellular microenvironment known as stem-cell niche (SCN) that provides the signals and physical support to maintain the pluripotent stem-cells (Sablowski, 2011). The SCN is surrounded by a transitory population of indeterminate cells that give rise to determinate cells and organs. Shoot and root apical meristem (SAM and RAM), which are formed during embryogenesis, only contributes to the main stem and main root, respectively. Branched structures rise post-embryonically from secondary meristems initiated from a few cells that retain meristematic characteristics (Nicolas and Laufs, 2022). In SAMs, the stem-cells, located at the top of the meristematic dome, secret the signal peptide CLAVATA3 (CLV3) that represses the pluripotency gene WUSCHEL (WUS) in cells of the organizing center (OC) underneath the stem-cells (Müller-Xing and Xing, 2021). Along with the pluripotency factor SHOOT MERISTEMLESS (STM), WUS maintains the stem-cells that form with the OC the shoot SCN. RAMs include mitotically less active organizer cells called the quiescent center (QC) and the surrounding initials, which together compose the root SCN. Similar to WUS, the pluripotency factor WOX5 maintains the stemness of the initials (Sarkar et al., 2007). Hence, ‘organizing’ cells maintain the stem-cells and SCNs by continuous short-range signaling. In 2007, Ben Scheres proposed in his landmark review that the SCNs of plant and animal kingdoms are specified by kingdom-specific patterning mechanisms, but that connect to a related core of epigenetic stem-cell factors (Scheres, 2007). Growing evidence endorses that plant stem-cell fate is also determined by epigenetic mechanisms, since the first models for an epigenetic control of plant SCNs were proposed 15 years ago.
High regeneration capacity is a feature of plant development. After loss of the SCN by laser ablation, the pluripotent meristematic cells can establish new stem-cells in SAMs and RAMs (Reinhardt et al., 2003; Xu et al., 2006). The removal of whole meristems leads to destruction of the regeneration process (Sachs, 1994), indicating that meristematic cells are more pluripotent than somatic cells. In the RAM, the stem-cell regeneration competence correlates with the expression gradient of pluripotency factor PLETHORA2 (PLT2) (Figure 1A) (Durgaprasad et al., 2019). There are at least two ways how epigenetic regulators control stem-cell fate: (i) they directly regulate the gene loci of pluripotency factors; or (ii) they regulate genes that are required to maintain the meristem organization that indirectly preserves the SCN, this may include indirect regulation via phytohormone pathways. For instance, altered levels of the repressive histone mark H3K27me3 at PIN gene loci, which encode Auxin efflux-carriers, results in altered auxin gradients, RAM size and lateral root primordia (LRP) numbers (Figure 1C). This perspective focuses on the epigenetic regulation of pluripotency genes that control stem-cell fate in Arabidopsis thaliana.
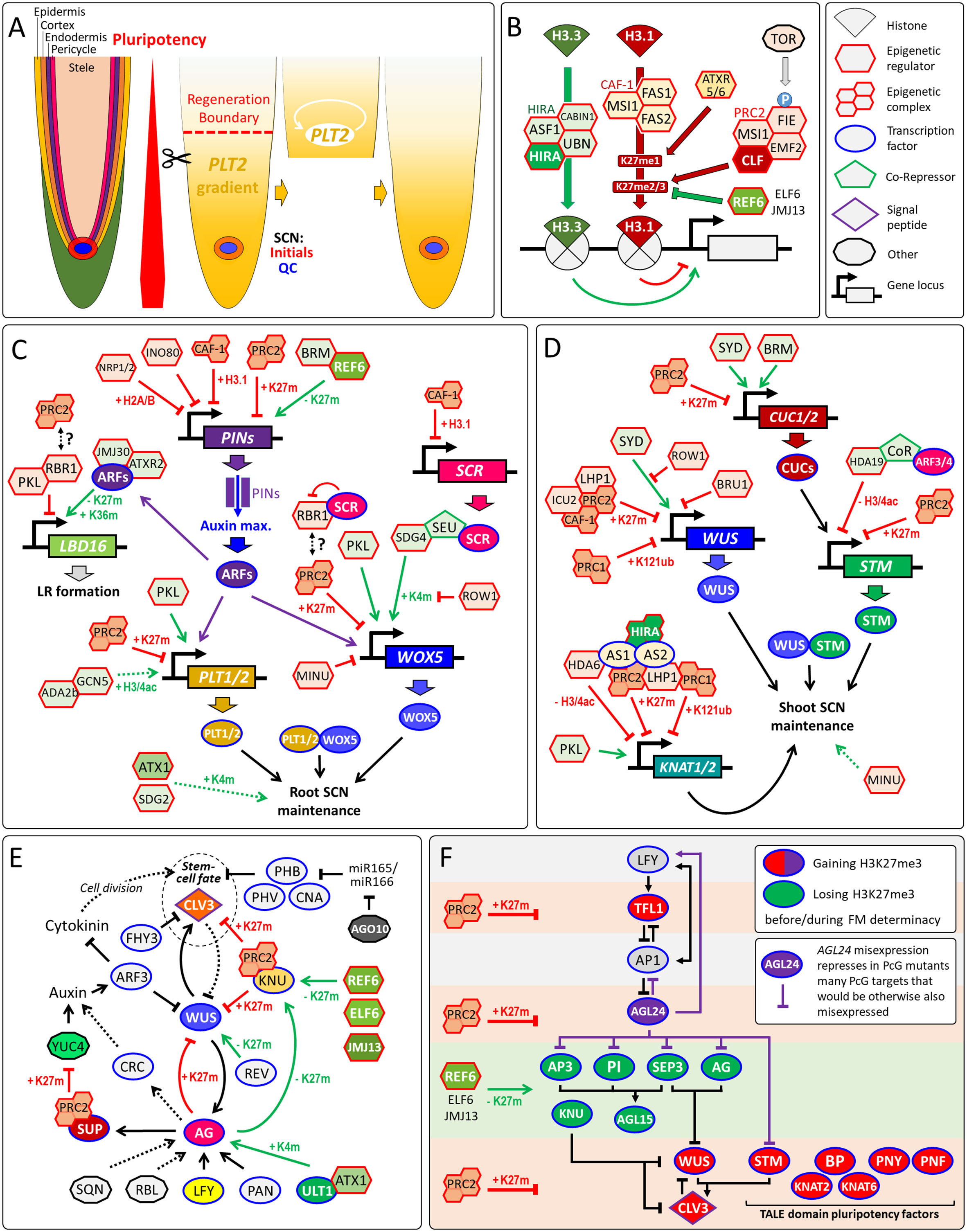
Figure 1 Regeneration competence of RAMs and epigenetic regulation of pluripotency genes. (A) Organization of the RAM and dependency of root tip regeneration on the pluripotency factor PLT2. After resection, PLT2 confers regeneration potential to differentiating cells via auto-activation. Notably, resection beyond the regeneration boundary prevents regeneration of the SCN. (B) Epigenetic gene regulation by HIRA, CAF-1, PRC2 and REF6. Beside the eponymous histone chaperone, the HIRA complex contains Anti Silencing Factor 1 (ASF1), Calcineurin Binding protein 1 (CABIN1) and Ubinuclein (UBN) 1/2 (Nie et al., 2014), while FASCIATA1/2 (FAS1/2) and MULTICOPY SUPPRESSOR OF IRA1 (MSI1) are subunits of CAF-1 complex (Kaya et al., 2001). The demethylase REF6 can interact with BRM and bind sequence-specifically its target genes to remove H3K27me3 (Li et al., 2016). (C) Direct and indirect epigenetic control of root pluripotency factors. Removal of H3K27me3 by REF6 is required to maintain PIN1/3/7 expression, while PRC2 represses PIN1 by H3K27me3 (Gu et al., 2014; Wang et al., 2019). Note that altered PIN expression indirectly changes the expression of pluripotency genes via AUXIN RESPONSE FACTORs (ARFs). ARF7/19 recruits H3K27me3-demethylase JMJ30 and H3K36me3-transferase ATXR2 to activate LATERAL ORGAN BOUNDARIES DOMAIN16/29 (LBD16/29) that encode key regulators of lateral root (LR) formation, while PKL recruits RBR1 that represses LBD16 (Lee et al., 2018a; Lee et al., 2018b; Ötvös et al., 2021). ROW1 specifically binds H3K4me3 at the WOX5 promoter to repress its transcription (Zhang et al., 2015). General Control Nonderepressible protein5 (GCN5), and its activator ADA2b promote PLT1/2 expression (Kornet and Scheres, 2009). Note that most pluripotency factors are PcG/H3K27me3-targets (Zhang et al., 2007; Shu et al., 2019). Hence, global changes of H3K27me3 could affect their expression directly and indirectly via changed auxin signaling by altered PIN levels. RBR1 interacts with the PRC2 component FIE (Mosquna et al., 2004), but it remains unclear whether this connection is relevant for the GRN of the root SCN. (D) Maintenance of the shoot SCN by epigenetic regulation of root pluripotency factors. (E) Floral stem-cell determinacy through silencing of WUS by a complex GRN (Reviewed in (Shang et al., 2019). During flower stage 2, LEAFY (LFY) and WUS as well as the TrxG proteins ULT1 and ATX1 activate AG that in turn activate SUPERMAN (SUP) and KNUCKLES (KNU). In flower stage 6, KNU represses CLV3 and WUS (Shang et al., 2021). Many of these transcriptional regulations are supported or driven by changes in H3K4me3 (K4m) or H3K27me3 (K27m), which is also confirmed by the flower indeterminacy phenotypes by loss of H3K27me3 demethylases REF6, ELF6 and JMJ13 or PRC2 (Yan et al., 2018; Müller-Xing et al., 2022). Other genes directly or indirectly involved in WUS silencing: ARGONAUTE1 (AGO1), AUXIN RESPONSE TRANSCRIPTION FACTOR3 (ARF3/ETT), CRABS CLAW (CRC), FAR-RED ELONGATED HYPOCOTYLS3 (FHY3), PERIANTHIA (PAN), REBELOTE (RBL), SQUINT (SQN), YUCCA4 (YUC4) and the HD-ZIP class III transcription factors CORONA (CNA), PHABULOSA (PHB), PHAVOLUTA (PHV) and REVOLUTA (REV). (F) GRN of floral stem-cell determinacy sustained by changes in H3K27me3 levels at gene loci of pluripotency genes and other key regulators (after (Müller-Xing et al., 2022), modified). STM enhances binding of WUS to the CLV3 chromatin through STM-WUS heterodimerization (Su et al., 2020). Arrows, transcriptional activation. Arrows with blunt ends represent repression. Double-sided arrows, protein-protein interaction. Dotted arrows indicate either indirect regulation or possible direct regulation that is not yet verified. Arrows in green indicate positive epigenetic regulation, in red, negative epigenetic regulation, and in black, non-epigenetic regulation. K4m, H3K4me3; K27m, H3K27me3; K36m, H3K36me3; H3/4ac, H3/H4 acetylation.
Epigenetic gene regulation bases on post-translational histone modifications, nucleosome assembly and ATP-dependent chromatin remodeling, which controls the accessibility of chromatin to transcription factors. Although both chaperone complexes target histone H3 variants, Histone Regulator A (HIRA) complex and chromatin assembly factor-1 (CAF-1) complex have opposite effects on epigenetic gene regulation (Figure 1B). The HIRA complex deposits H3.3, which facilitates transcription, in a DNA synthesis-independent manner (Nie et al., 2014), whereas H3.1 deposition, which is essential for maintenance of the repressive H3K27me3 mark through cell division, relies on the heterotrimeric CAF-1 complex (Jiang and Berger, 2017). CAF-1 maintains cellular and functional organization and expression of the pluripotency gene SCARECROW (SCR) in the RAM (Kaya et al., 2001). CAF-1 and the H2A/H2B histone chaperone NAP1-RELATED PROTEIN1/2 (NRP1/2) play synergistic roles in root SCN maintenance by rather maintaining of the auxin gradient maximum at the QC than directly controlling the expression of pluripotency genes such as WOX5 and PLT1 (Ma et al., 2018). Similarly, NRP1/2 and the chromatin-remodeling factor INOSITOL AUXOTROPHY 80 (INO80) synergistically maintain histone H3 levels within the chromatin regions of PIN1, while the simultaneous loss of these three genes results in higher PIN1 protein levels, perturbed auxin gradients and misexpression of the root pluripotency factors WOX5 and PLT1/2 (Kang et al., 2019). On the contrary, the SWI2/SNF2-family chromatin-remodeling factor BRAHMA (BRM) positively regulates the expression of several PINs as well as PLT1/2, but ChIP experiments indicate that only the PIN genes are directly targeted by BRM (Yang et al., 2015). Hence, loss of BRM affects the expression of PLT1/2 rather indirectly through impaired auxin signaling by reduced PIN expression levels. In contrast, the non-canonical SWI2/SNF2-type ATPase MINUSCULE2 (MINU2) directly activates the promoter of WOX5 (Sang et al., 2012).
The repressive Polycomb group (PcG) proteins and the activation-related Trithorax group (TrxG) proteins have been implicated to regulate SCN maintenance and meristem activity (Singh et al., 2020). The Polycomb Repressive Complex 2 (PRC2) deposits the repressive H3K27me3 mark, while PRC1 sets the repressive H2AK121ub mark independently of H3K27me3 (Zhou et al., 2017). TrxG proteins were defined as their antagonists that can range from chromatin-remodelers to histone modifiers that deposit activation-related marks such as H3K4me3 and histone acetylation (Müller-Xing et al., 2014b).
A key role of PRC1 is the repression of several key regulators such as WOX5 and PLT1/2 controlling root SCN specification and cell proliferation (Merini et al., 2017). The CHD3-type chromatin-remodeler PICKLE (PKL) is a TrxG protein and acts antagonistically to the PcG protein and H3K27me3-transferase CURLY LEAF (CLF) controlling RAM activity (Aichinger et al., 2011). Altered meristematic activity in pkl and clf mutants correlates with changed H3K27me3 levels and altered expression of these pluripotency genes (Aichinger et al., 2011). Loss of PKL decreases meristematic activity with an increased H3K27me3 level at WOX5 and PLT1/2, whereas mutation in CLF increases meristematic activity of the root with loss of the H3K27me3 level. In line with the negative effect of H3K27me3 on root pluripotency genes and SCN, the H3K4-histone methyltransferases ARABIDOPSIS HOMOLOG of TRITHORAX1 (ATX1/SDG27) and SET DOMAIN GROUP2 (SDG2) promote the root SCN integrity (Yao et al., 2013; Napsucialy-Mendivil et al., 2014), but it is not yet known whether ATX1 and SDG2 directly bind to the chromatin of pluripotency genes.
Most chromatin modifiers and remodelers do not possess a DNA-binding domain that would allow sequence-specific binding to their target genes. Hence, one of the recruitment strategies of epigenetic regulators such as PRC2 is through intermediary transcription factors, which facilitate recruitment to the target chromatin (Godwin and Farrona, 2022). Notably, pluripotency factors can be such recruiters of epigenetic regulators. WOX5, which is expressed in the QC, functioning as a mobile organizer signal that represses differentiation in neighboring columella stem-cells. There, WOX5 recruits the co-repressors TPL/TPRs that, in turn, recruit histone deacetylase HDA19 to silence the differentiation factor CDF4 via histone deacetylation (Pi et al., 2015). Recently, it has been implied that the EAR-domain of TPL can also recruit PRC2-activity, but the proof of a direct TPL-PRC2 interaction is pending (Baile et al., 2021). WOX5 transcription is also epigenetically regulated. SCR recruits SEUSS (SEU), a homologue of the animal LIM-domain binding (LDB) proteins, to the WOX5 promoter. Subsequently, SEU recruits the methyltransferase SDG4 that deposits H3K4me3 at the WOX5 promoter activating this key pluripotency factor in the RAM (Zhai et al., 2020).
RETINOBLASTOMA-RELATED1 (RBR1) is a master regulator of the cell cycle and root development. In 2007, Ben Scheres suggested RBR1 as one potential link between stem-cell regulation and chromatin modifications in plants (Scheres, 2007). This assumption largely relied on a study in mammalian research revealing that retinoblastoma (RB) protein targets PRC2 to the promoter of cell-cycle control genes (Kotake et al., 2007). Since, it has be shown that RBR1 is required for silencing of late embryonic genes by increasing H3K27me3 levels via PRC2 in plants (Gutzat et al., 2011). Several studies deepened our knowledge about how RBR control RAM development (reviewed in (Desvoyes and Gutierrez, 2020) but elucidation of a potential RBR1-PRC2 pathway involved in controlling the root SCN awaits further research.
In retrospect, the article of (Takeda et al., 2004) about TONSOKU (TSK/BRUSHY1/MGOUN3) can be considered as one of the first studies that addressed the role of epigenetics in maintaining the shoot SCN. The authors showed that loss of TSK results in disorganized SAMs with an abnormal, dispersed WUS expression pattern, and suggested that TSK links DNA-repair and epigenetic gene silencing, but the exact molecular function of TSK remained vague. A most recent study revealed that TSK-mediated DNA-repair, which can rescue broken DNA-replication forks, involves specific interaction of TSK with H3.1 via recognition of alanine 31 (Davarinejad et al., 2022). H3.1 is essential for maintenance of H3K27me3 by PRC2 through cell division and silencing of the TALE class I KNOX and pluripotency genes STM, BP/KNAT1, KNAT2, and KNAT6 (Jiang and Berger, 2017). Hence, the ability of TSK to distinguish H3.1 from other H3 variants might play a role in TSK-dependent epigenetic gene repression and should be addressed in future studies.
SWI2/SNF2-type chromatin-remodelers, such as BRM and its homologue SPLAYED (SYD), play an important role in the accessibility of cis-regulatory DNA regions to transcription factors. In SAMs, SYD is recruited to the WUS promoter and directly activates WUS transcription, which is involved in maintenance of the shoot SCN (Kwon et al., 2005). The recruiter of SYD to the WUS chromatin is not yet known and is a question to be addressed. BRM and SYD are not likely to activate STM directly but via direct activation of CUC genes (Kwon et al., 2006). The PHD-domain protein REPRESSOR OF WUSCHEL1 (ROW1/BARD1) confines WUS expression into the OC by binding to the WUS promoter near the SYD-binding site. Since ROW1 and SYD interact in Co-IP assays, BARD1 may repress WUS transcription via inhibition of SYD-dependent chromatin-remodelling that activates WUS expression (Han et al., 2008; Han and Zhu, 2009). Also two non-canonical SWI2/SNF2-type ATPases MINU1/MINU2 are essential to maintain the shoot SCN but do not bind to the WUS chromatin (Sang et al., 2012).
Strongly depleted PRC2 function results in enlarged SAMs with increased STM expression levels and expanded expression domain of the stem-cell marker CLV3, while WUS expression and domain size were decreased, indicating an uncoupling of stem-cell fate and WUS expression levels (Müller-Xing et al., 2014a; Müller-Xing et al., 2015; Müller-Xing et al., 2022). This downregulation of WUS might be caused by upregulation of WUS repressors such as KNU that is an H3K27me3-target. The mRNAs of STM and KNAT1 are expressed in the SAM but are silent in leaf and early stages of flower primordia (Lincoln et al., 1994; Long et al., 1996). The ASYMMETRIC LEAVES1 (AS1)-AS2 transcription factor complex recruits PRC1, PRC2 and the PcG-associated protein LHP1 to chromatin of KNAT1, KNAT2, and KNAT6 to silence these pluripotency genes (Lodha et al., 2013; Z. Li et al., 2016). Similarly, ARF3/4 silence directly STM via histone-deacetylation in flower primordia, although the co-repressor, which links ARF3/4 with HDA19, still waits to be discovered (Chung et al., 2019). This strong epigenetic repression of pluripotency genes in organ primordia and differentiated tissue leads to the question how the stem-cells of auxiliary (AMs) and flower meristems (FMs) can be de novo initiated.
Shoot branching requires the initiation of auxiliary meristems (AMs) and de novo stem-cell formation from a few cells of the leaf axil that retain some meristematic characteristics, named premeristems (Nicolas and Laufs, 2022). In the premeristemic cells, ATH1-STM heterodimer binds the pluripotency gene STM to maintain its expression at a low level, which endows permissive STM chromatin for subsequent upregulation of STM during AM initiation (Figure 2A). During the premeristemic stage, low STM transcription maintains low levels of repressive H3K27me3 and high levels of the active chromatin marker H3 acetylation, which allows the STM locus to remain epigenetically active (Cao et al., 2020). Hence, pluripotency seems sustained through low and epigenetically maintained expression of pluripotency genes in precursor-cells of stem-cells. It would be certainly interesting to investigate whether this is true throughout the plant life-cycle demonstrating that pluripotency is epigenetically maintained in plant stem-cell lineages.
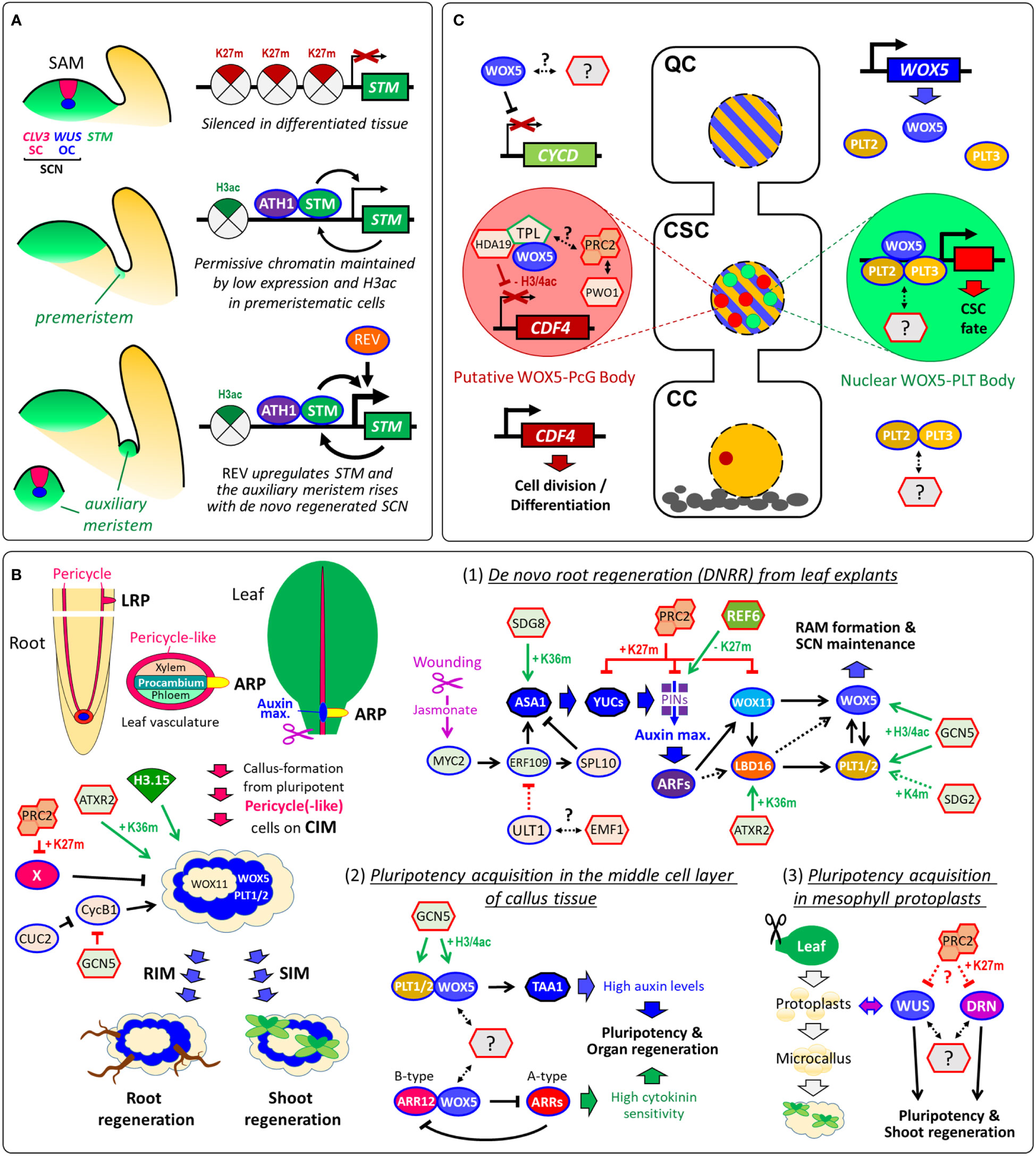
Figure 2 Pluripotency acquisition during de novo stem-cell formation and de novo organogenesis. (A) Organization of the SAM and epigenetic maintenance of premeristem cells for shoot branching. STM is silenced by H3K27me3 in differentiated tissue, while low expression rates of STM, which interacts with ATH1, auto-activates its own transcription in premeristemic cells. REV directly upregulates STM expression in leaf axil meristematic cells (Shi et al., 2016). (B) Vascular-associated pluripotent cells are key for callus formation and de novo organogenesis. Several epigenetic regulators control callus development including PRC2 that promotes callus formation by repressing leaf-regulatory genes (X) by H3K27me3 (He et al., 2012). Although CUC2 and GCN5 promote shoot regeneration, both suppress M/G2-phase marker CYCB1;1 and cell proliferation in callus tissue (Daimon et al., 2003; Kim et al., 2018; Li et al., 2016). (1) Conceptional model of the transcriptional and epigenetic regulation of DNRR (after (Jing et al., 2020); modified): ULT1, which can associate with PcG protein EMF1, suppresses DNRR by negative regulation of ERF109 that is activated by the wounding-induced Jasmonate signaling (Zhang et al., 2019; Tian et al., 2022). ERF109 and ERF111 activate the auxin synthesis gene ASA1 and the transcription factor SPL10 that in turn represses ASA1 (Ye et al., 2020). Auxin synthesis and transport produce an auxin maximum near the wounding site, which triggers the GRN composed of ARFs, WOX11/12, LBD16/18, WOX5/7 and PLT1/2. (2) Pluripotency acquisition in the middle cell layer of callus: WOX5 and PLT1/2 directly interact to activate TAA1 to accelerate endogenous auxin production. WOX5 also interacts with the B-type ARABIDOPSIS RESPONSE REGULATOR12 (ARR12), which represses A-type ARRs breaking the negative feedback loop in cytokinin signaling. Overall, the promotion of auxin biosynthesis and the enhancement of cytokinin sensitivity are both required for pluripotency acquisition for shoot and root regeneration (Zhai and Xu, 2021). (3) Pluripotency acquisition in microcallus: Protoplast isolation from differentiated mesophyll cells induces stochastic gene expression of WUS and DORNRÖSCHEN/ENHANCER OF SHOOT REGENERATION1 (DRN/ESR1), which accelerate microcallus formation and, subsequently, shoot regeneration (Xu et al., 2021). The stochastic gene expression of WUS and DRN in protoplasts and microcalli might be caused by variant levels of epigenetic modifications such as H3K27me3 by PRC2. (C) Models of cell-type-specific transcriptional and epigenetic regulation, interaction and subnuclear localization during distal root SCN maintenance. WOX5 represses QC divisions by repressing CYCLIN D (CYCD) activity within the QC (Forzani et al., 2014). Although WOX5 and PLT2/3 proteins are present in nuclei of QC and columella stem-cells (CSCs), PLT3 recruits WOX5 only in the CSCs into nuclear bodies. The H3K27me3-reader PWWP-DOMAIN INTERACTOR OF POLYCOMBS1 (PWO1) associates with PRC2 and can form nuclear PcG bodies (Hohenstatt et al., 2018; Mikulski et al., 2019). Since TPL can recruit PRC2 activity, WOX5-TPL-HDA19 and PWO1-PRC2 might co-localize in the same nuclear body. In the columella cells (CCs), CDF4 is expressed promoting cell division und differentiation. (B, C) In future studies, it would be interesting to address whether WUS, WOX5 and other pluripotency factors interact with further epigenetic regulators. Arrows, transcriptional activation. Arrows with blunt ends represent repression. Double-sided arrows, protein-protein interaction. Dotted arrows indicate either indirect regulation or possible direct regulation that is not yet verified. Arrows in green indicate positive epigenetic regulation, in red, negative epigenetic regulation, and in black, non-epigenetic regulation. K4m, H3K4me3; K27m, H3K27me3; K36m, H3K36me3; H3ac, H3 acetylation; H3/4ac, H3/H4 acetylation.
Flowers are determinate structures. A feedback loop, built by WUS and AGAMOUS (AG), terminates the stem-cell pool of FMs by silencing WUS during floral stage 6, which is time-buffered via epigenetic regulation by PcG and TrxG proteins (Müller and Goodrich, 2011). In the last two decades, near a dozen pathways were discovered, which also contribute to the epigenetic silencing of WUS by H3K27me3 (Figure 1E). We recently revealed that during FM arrest also several other pluripotency genes, including STM and other TALE KNOX genes, are silenced by H3K27me3 (Figure 1F). This synchronized silencing of several pluripotency genes could accelerate FM determinacy in a way that cannot be achieved by silencing WUS alone (Müller-Xing et al., 2022). Considering the complexity of the gene regulatory network (GRN) controlling H3K27me3-mediated silencing of WUS (Figure 1D), similar complex regulation might wait to be revealed for the other H3K27me3-silenced floral pluripotency genes.
Similar to shoot branching, de novo organogenesis depends on de novo stem-cell regeneration from pluripotent precursor-cells. De novo root regeneration (DNRR) can occur directly from detached organs such as leaves (Chen et al., 2014) or indirectly, like shoot regeneration, from auxin-induced callus tissue (Skoog and Miller, 1957). Lateral roots (LRs), adventitious roots (ARs) and callus derivate from pluripotent cells that can be collectively termed vasculature-associated pluripotent cells (VPCs): LRs rise from pericycle cells (Malamy and Benfey, 1997), the root-founder cells of ARs emerge through cell-fate-transition from procambium cells or its nearby parenchyma cells (Liu et al., 2014), and callus origins from xylem-pole pericycle and pericycle-like cells (Figure 2B) (Atta et al., 2009; Sugimoto et al., 2010). The formation of LRs, ARs and callus is regulated by several epigenetic regulators including PRC2 (Reviewed in (Jing et al., 2020). Callus does not consist of dedifferentiated cells (Fehér, 2019) and comprise rather features of LRPs including stem-like cells. Treatment on shoot or root induction medium (SIM or RIM) gives raise to de novo organogenesis (Figure 2B). A recent study using single-cell transcriptomics revealed that only the middle cell layer of callus acquires pluripotency, which is required for organ regeneration (Zhai and Xu, 2021). In this QC-like middle layer, pluripotency depends on WOX5 that functions as master regulator activating auxin synthesis while suppressing cytokinin signaling (Figure 2B). The pluripotency acquisition of callus depends also on histone modifications and nucleosome assembly, since overexpression of the atypical histone variant H3.15, which cannot be H3K27me3, promotes callus formation (Yan et al., 2020). During shoot regeneration, WUS is essential for de novo establishment of the shoot SCN. After transferring the callus to SIM, WUS is epigenetically reactivated by a two-step mechanism: (i) the cytokinin-rich environment initially promotes the removal of repressive H3K27me3 at WUS in a cell cycle-dependent manner; subsequently (ii), B-type ARABIDOPSIS RESPONSE REGULATORs (ARRs) spatially activate WUS expression through binding with HD-ZIP III transcription factor REVOLUTA (REV) (Zhang et al., 2017). With extreme treatments, single somatic cells also can reacquire pluripotency and totipotency forming into entire plants (Takebe et al., 1971). While WOX5 provides pluripotency in auxin-induced callus, WUS accelerates microcalli formation and, subsequently, shoot regeneration from mesophyll protoplasts (Xu et al., 2021). It is yet not known whether WUS and WOX5 interact with epigenetic regulators to provide pluripotency during de novo organogenesis (Figure 2B).
In the last 15 years, numerous studies have enhanced our understanding about how epigenetics play a crucial role in initiation, maintenance and determination of plant stem-cells, yet many open questions remain. For example, the HIRA complex deposits the histone variant H3.3 that correlates with active gene transcription (Nie et al., 2014). However, HIRA interacts with AS1 and AS2 to repress the class I KNOX genes (Phelps-Durr et al., 2005). HIRA has at least the potential to act in gene repression since HIRA contributes to nucleosome occupancy also at heterochromatic regions that are silenced (Duc et al., 2015). Similarly, TrxG factors ATX1 and ULTRAPETALA1 (ULT1) and the PcG protein EMF1 interact to prevent synergistically seed gene misexpression in RAMs (Xu et al., 2018). Yet the exact molecular mechanisms remain unclear how epigenetic regulators such as HIRA and ATX1/ULT1, whose main function is transcriptional activation, achieve gene repression. In general, the question remains how the epigenetic pathways corporate with each other and which upstream signals modulate their spatiotemporal specificity. Most recently, it has been shown that glucose-activated TOR kinase controls genome-wide H3K27me3, which limits SAM size (Ye et al., 2022) indicating that glucose signaling controls stem-cell fate also epigenetically.
Some findings of single-gene approaches were recently challenged by whole-genome studies. For example, STM misexpression in PRC1 mutants suggested that STM is a direct target (Xu and Shen, 2008), while recent ChIP-Seq data show that STM is an H3K27me3-target but not of the PRC1 mark H2AK121ub (Zhou et al., 2017). Similarly, manipulation of cell cycles with pharmacological agents suggests that KNU loses H3K27me3 passively by cell division in flower primordia (Sun et al., 2014), while ChIP-Seq data show that demethylases RELATIVE OF EARLY FLOWERING6 (REF6), ELF6 and JMJ13 are required for active H3K27me3-removal at KNU in inflorescences (Yan et al., 2018). It should be emphasized that passive and active H3K27me3-removal could work hand-in-hand at the KNU gene locus.
A recent study showed that the pluripotency factors WOX5, PLT2 and PLT3 form alternative complexes in different cell-types of the RAM (Burkart et al., 2022). It seems reasonable to anticipate that epigenetic complexes and their recruiters are also cell-type-specifically formed (Figure 2C). Although single-cell studies are currently rather used for whole-genome profiling, cell type-specific investigation will also become more common in single-gene/protein approaches in which protein-protein and protein-DNA interactions are investigated. Advanced confocal microscope approaches reached now a new level that allows in vivo FRET-FLIM in single cells of the RAM (Long et al., 2017), which will reveal cell-type-specific protein interactions of epigenetic complexes in the future.
The original contributions presented in the study are included in the article. Further inquiries can be directed to the corresponding author.
RM-X conceived, illustrated, and wrote the manuscript with the help of QX. All authors contributed to the article and approved the submitted version.
This work was kindly funded by the National Natural Science Foundation of China (project nos 31640054 and 31771602), and Starting Grants of the Lushan Botanical Garden, Chinese Academy of Sciences, Jiujiang (project nos 2021ZWZX24 and 2021ZWZX25).
We thank Danyu Kong and Xiaokun Liu for critical reading and comments on the manuscript. We would like to apologize for mostly focusing on histone H3 modifications and H3 variants in Arabidopsis thaliana and not covering many fundamental original studies due to space limitations. For a broader and deeper insight to the field of epigenetic regulation of plant stem-cells, we recommend the review by (Singh et al., 2020) to the readers.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aichinger, E., Villar, C. B. R., Di Mambro, R., Sabatini, S., Köhler, C. (2011). The CHD3 chromatin remodeler PICKLE and polycomb group proteins antagonistically regulate meristem activity in the arabidopsis root. Plant Cell 23, 1047–1060. doi: 10.1105/tpc.111.083352
Atta, R., Laurens, L., Boucheron-Dubuisson, E., Guivarc'h, A., Carnero, E., Giraudat-Pautot, V., et al. (2009). Pluripotency of arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro. Plant J. Cell Mol. Biol. 57, 626–644. doi: 10.1111/j.1365-313X.2008.03715.x
Baile, F., Merini, W., Hidalgo, I., Calonje, M. (2021). EAR domain-containing transcription factors trigger PRC2-mediated chromatin marking in arabidopsis. Plant Cell 33, 2701–2715. doi: 10.1093/plcell/koab139
Burkart, R. C., Strotmann, V. I., Kirschner, G. K., Akinci, A., Czempik, L., Dolata, A., et al. (2022). PLETHORA-WOX5 interaction and subnuclear localization control arabidopsis root stem cell maintenance. EMBO Rep. 23, 1–18. doi: 10.15252/embr.202154105
Cao, X., Wang, J., Xiong, Y., Yang, H., Yang, M., Ye, P., et al. (2020). A self-activation loop maintains meristematic cell fate for branching. Curr. Biol. 30, 1893–1904.e4. doi: 10.1016/j.cub.2020.03.031
Chen, X., Qu, Y., Sheng, L., Liu, J., Huang, H., Xu, L. (2014). A simple method suitable to study de novo root organogenesis. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00208
Chung, Y., Zhu, Y., Wu, M.-F., Simonini, S., Kuhn, A., Armenta-Medina, A., et al. (2019). Auxin response factors promote organogenesis by chromatin-mediated repression of the pluripotency gene SHOOTMERISTEMLESS. Nat. Commun. 10, 886. doi: 10.1038/s41467-019-08861-3
Daimon, Y., Takabe, K., Tasaka, M. (2003). The CUP-SHAPED COTYLEDON genes promote adventitious shoot formation on calli. Plant Cell Physiol. 44, 113–121. doi: 10.1093/pcp/pcg038
Davarinejad, H., Huang, Y.-C., Mermaz, B., LeBlanc, C., Poulet, A., Thomson, G., et al. (2022). The histone H3.1 variant regulates TONSOKU-mediated DNA repair during replication. Science 375, 1281–1286. doi: 10.1126/science.abm5320
Desvoyes, B., Gutierrez, C. (2020). Roles of plant retinoblastoma protein: Cell cycle and beyond. EMBO J. 39, 774109. doi: 10.15252/embj.2020105802
Doerner, P. (2003). Plant meristems: A merry-Go-Round of signals review. Curr. Biol. 13, R368–R374. doi: 10.1016/S0960-9822(03)00280-X
Duc, C., Benoit, M., Le Goff, S., Simon, L., Poulet, A., Cotterell, S., et al. (2015). The histone chaperone complex HIR maintains nucleosome occupancy and counterbalances impaired histone deposition in CAF-1 complex mutants. Plant J. 81, 707–722. doi: 10.1111/tpj.12758
Durgaprasad, K., Roy, M. V., Venugopal M., A., Kareem, A., Raj, K., Willemsen, V., et al. (2019). Gradient expression of transcription factor imposes a boundary on organ regeneration potential in plants. Cell Rep. 29, 453–463.e3. doi: 10.1016/j.celrep.2019.08.099
Fehér, A. (2019). Callus, dedifferentiation, totipotency, somatic embryogenesis: What these terms mean in the era of molecular plant biology? Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00536
Forzani, C., Aichinger, E., Sornay, E., Willemsen, V., Laux, T., Dewitte, W., et al. (2014). WOX5 suppresses CYCLIN d activity to establish quiescence at the center of the root stem cell niche. Curr. Biol. 24, 1939–1944. doi: 10.1016/j.cub.2014.07.019
Godwin, J., Farrona, S. (2022). The importance of networking: Plant polycomb repressive complex 2 and its interactors. Epigenomes 6, 8. doi: 10.3390/epigenomes6010008
Gutzat, R., Borghi, L., Fütterer, J., Bischof, S., Laizet, Y.’H., Hennig, L., et al. (2011). RETINOBLASTOMA-RELATED PROTEIN controls the transition to autotrophic plant development. Development 138, 2977–2986. doi: 10.1242/dev.060830
Gu, X., Xu, T., He, Y. (2014). A histone H3 lysine-27 methyltransferase complex represses lateral root formation in arabidopsis thaliana. Mol. Plant 7, 977–988. doi: 10.1093/mp/ssu035
Han, P., Li, Q., Zhu, Y.-X. (2008). Mutation of arabidopsis BARD1 causes meristem defects by failing to confine WUSCHEL expression to the organizing center. Plant Cell 20, 1482–1493. doi: 10.1105/tpc.108.058867
Han, P., Zhu, Y.-X. (2009). BARD1 may be renamed ROW1 because it functions mainly as a REPRESSOR OF WUSCHEL1. Plant Signaling Behav. 4, 52–54. doi: 10.4161/psb.4.1.7312
He, C., Chen, X., Huang, H., Xu, L. (2012). Reprogramming of H3K27me3 is critical for acquisition of pluripotency from cultured arabidopsis tissues. PloS Genet. 8, e1002911. doi: 10.1371/journal.pgen.1002911
Hohenstatt, M. L., Mikulski, P., Komarynets, O., Klose, C., Kycia, I., Jeltsch, A., et al. (2018). PWWP-DOMAIN INTERACTOR OF POLYCOMBS1 interacts with polycomb-group proteins and histones and regulates arabidopsis flowering and development. Plant Cell 30, 117–133. doi: 10.1105/tpc.17.00117
Jiang, D., Berger, F. (2017). DNA Replication-coupled histone modification maintains polycomb gene silencing in plants. Sci. (New York N.Y.) 357, 1146–1149. doi: 10.1126/science.aan4965
Jing, T., Ardiansyah, R., Xu, Q., Xing, Q., Müller-Xing, R. (2020). Reprogramming of cell fate during root regeneration by transcriptional and epigenetic networks. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00317
Kang, H., Ma, J., Wu, D, Shen, W.-H., Zhu, Y. (2019). Functional coordination of the chromatin-remodeling factor AtINO80 and the histone chaperones NRP1/2 in inflorescence meristem and root apical meristem. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00115
Kaya, H., Shibahara, K.-I., Taoka, K.-I., Iwabuchi, M., Stillman, B., Araki, T. (2001). FASCIATA genes for chromatin assembly factor-1 in arabidopsis maintain the cellular organization of apical meristems. Cell 104, 131–142. doi: 10.1016/S0092-8674(01)00197-0
Kim, J.-Y., Yang, W., Forner, J., Lohmann, J. U., Noh, B., Noh, Y.-S. (2018). Epigenetic reprogramming by histone acetyltransferase HAG1/AtGCN5 is required for pluripotency acquisition in arabidopsis. EMBO J. 37, 1–16. doi: 10.15252/embj.201798726
Kornet, N., Scheres, B. (2009). Members of the GCN5 histone acetyltransferase complex regulate PLETHORA-mediated root stem cell niche maintenance and transit amplifying cell proliferation in arabidopsis. Plant Cell 21, 1070–1079. doi: 10.1105/tpc.108.065300
Kotake, Y., Cao, R., Viatour, P., Sage, J., Zhang, Y., Xiong, Y. (2007). pRB family proteins are required for H3K27 trimethylation and polycomb repression complexes binding to and silencing p16 INK4a tumor suppressor gene. Genes Dev. 21, 49–54. doi: 10.1101/gad.1499407
Kwon, C. S., Chen, C., Wagner, D. (2005). WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in arabidopsis. Genes Dev. 19, 992–1003. doi: 10.1101/gad.1276305
Kwon, C. S., Hibara, K.-I., Pfluger, J., Bezhani, S., Metha, H., Aida, M., et al. (2006). A role for chromatin remodeling in regulation of CUC gene expression in the arabidopsis cotyledon boundary. Development 133, 3223–3230. doi: 10.1242/dev.02508
Lee, K., Park, O.-S., Seo, P. J. (2018a). ATXR2 as a core regulator of de novo root organogenesis. Plant Signaling Behav. 13, e1449543. doi: 10.1080/15592324.2018.1449543
Lee, K., Park, O.-S., Seo, P. J. (2018b). JMJ30-mediated demethylation of H3K9me3 drives tissue identity changes to promote callus formation in arabidopsis. Plant J. Cell Mol. Biol. 95, 961–975. doi: 10.1111/tpj.14002
Li, C., Gu, L., Gao, L., Chen, C., Wei, C.-Q., Qiu, Q., et al. (2016). Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in arabidopsis. Nat. Genet. 48, 687–693. doi: 10.1038/ng.3555
Li, Z., Li, B., Liu, J., Guo, Z., Liu, Y., Li, Y., et al. (2016). Transcription factors AS1 and AS2 interact with LHP1 to repress KNOX genes in arabidopsis. Integr. Plant Biol. 58, 959–970. doi: 10.1111/jipb.12485
Lincoln, C., Long, J., Yamaguchi, J., Serikawa, K., Hake, S. (1994). A knotted1-like homeobox gene in arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6, 1859–1876. doi: 10.1105/tpc.6.12.1859
Liu, J., Sheng, L., Xu, Y., Li, J., Yang, Z., Huang, H., et al. (2014). WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in arabidopsis. Plant Cell 26, 1081–1093. doi: 10.1105/tpc.114.122887
Li, X., Zheng, Y., Xing, Q., Ardiansyah, R., Zhou, H., Ali, S., et al. (2020). Ectopic expression of the transcription factor CUC2 restricts growth by cell cycle inhibition in arabidopsis leaves. Plant Signaling Behav. 15, 1706024. doi: 10.1080/15592324.2019.1706024
Lodha, M., Marco, C. F., Timmermans, M. C. P. (2013). The ASYMMETRIC LEAVES complex maintains repression of KNOX homeobox genes via direct recruitment of polycomb-repressive complex2. Genes Dev. 27, 596–601. doi: 10.1101/gad.211425.112
Long, J. A., Moan, E. I., Medford, J. I., Barton, M. K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of arabidopsis. Nature 379, 66–69. doi: 10.1038/379066a0
Long, Y., Stahl, Y., Weidtkamp-Peters, S., Postma, M., Zhou, W., Goedhart, J., et al. (2017). In vivo FRET–FLIM reveals cell-type-specific protein interactions in arabidopsis roots. Nature 548, 97–102. doi: 10.1038/nature23317
Malamy, J. E., Benfey, P. N. (1997). Organization and cell differentiation in lateral roots of arabidopsis thaliana. Development 124, 33–44. doi: 10.1242/dev.124.1.33
Ma, J., Liu, Y., Zhou, W., Zhu, Y., Dong, A., Shen, W.-H. (2018). Histone chaperones play crucial roles in maintenance of stem cell niche during plant root development. Plant J. 95, 86–100. doi: 10.1111/tpj.13933
Merini, W., Romero-Campero, F. J., Gomez-Zambrano, A., Zhou, Y., Turck, F., Calonje, M. (2017). The arabidopsis polycomb repressive complex 1 (PRC1) components AtBMI1A, b, and c impact gene networks throughout all stages of plant development. Plant Physiol. 173, 627–641. doi: 10.1104/pp.16.01259
Mikulski, P., Hohenstatt, M. L., Farrona, S., Smaczniak, C., Stahl, Y., Kalyanikrishna, et al. (2019). The chromatin-associated protein PWO1 interacts with plant nuclear lamin-like components to regulate nuclear size. Plant Cell 31, 1141–1154. doi: 10.1105/tpc.18.00663
Mosquna, A., Katz, A., Shochat, S., Grafi, G., Ohad, N. (2004). Interaction of FIE, a polycomb protein, with pRb: A possible mechanism regulating endosperm development. Mol. Genet. Genomics 271, 651–657. doi: 10.1007/s00438-004-1024-6
Müller, R., Goodrich, J (2011). Sweet memories: Epigenetic control in flowering. F1000 Biol. Rep. 3, 13. doi: 10.3410/B3-13
Müller-Xing, R., Ardiansyah, R., Xing, Q., Faivre, L., Tian, J., Wang, G., et al. (2022). Polycomb proteins control floral determinacy by H3K27me3-mediated repression of pluripotency genes in arabidopsis thaliana. J. Exp. Bot. 8, e0127. doi: 10.1093/jxb/erac013
Müller-Xing, R., Clarenz, O., Pokorny, L., Goodrich, J., Schubert, D. (2014a). Polycomb-group proteins and FLOWERING LOCUS T maintain commitment to flowering in arabidopsis thaliana. Plant Cell 26, 2457–2471. doi: 10.1105/tpc.114.123323
Müller-Xing, R., Schubert, D., Goodrich, J. (2015). Non-inductive conditions expose the cryptic bract of flower phytomeres in arabidopsis thaliana. Plant Signaling Behav. 10, e1010868. doi: 10.1080/15592324.2015.1010868
Müller-Xing, R., Xing, Q. (2021). In da club: The cytoplasmic kinase MAZZA joins CLAVATA signaling and dances with CLV1-like receptors. J. Exp. Bot. 72, 4596–4599. doi: 10.1093/jxb/erab203
Müller-Xing, R., Xing, Q., Goodrich, J. (2014b). Footprints of the sun: Memory of UV and light stress in plants. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00474
Napsucialy-Mendivil, S., Alvarez-Venegas, R., Shishkova, S., Dubrovsky, J. G. (2014). Arabidopsis homolog of trithorax1 (ATX1) is required for cell production, patterning, and morphogenesis in root development. J. Exp. Bot. 65, 6373–6384. doi: 10.1093/jxb/eru355
Nicolas, A., Laufs, P. (2022). Meristem initiation and de novo stem cell formation. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.891228
Nie, X., Wang, H., Li, J., Holec, S., Berger, F. (2014). The HIRA complex that deposits the histone H3.3 is conserved in arabidopsis and facilitates transcriptional dynamics. Biol. Open 3, 794–802. doi: 10.1242/bio.20148680
Ötvös, K., Miskolczi, P., Marhavý, P., Cruz-Ramírez, A., Benková, E., Robert, S., et al. (2021). Pickle recruits retinoblastoma related 1 to control lateral root formation in arabidopsis. IJMS 22, 3862. doi: 10.3390/ijms22083862
Phelps-Durr, T. L., Thomas, J., Vahab, P., Timmermans, M. C. P. (2005). Maize rough sheath2 and its arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell 17, 2886–2898. doi: 10.1105/tpc.105.035477
Pi, L., Aichinger, E., van der Graaff, E., Llavata-Peris, C. I., Weijers, D., Hennig, L., et al. (2015). Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Dev. Cell 33, 576–588. doi: 10.1016/j.devcel.2015.04.024
Reinhardt, D., Frenz, M., Mandel, T., Kuhlemeier, C. (2003). Microsurgical and laser ablation analysis of interactions between the zones and layers of the tomato shoot apical meristem. Development 130, 4073–4083. doi: 10.1242/dev.00596
Sablowski, R. (2011). Plant stem cell niches: From signalling to execution. Curr. Opin. Plant Biol. 14, 4–9. doi: 10.1016/j.pbi.2010.08.001
Sang, Y., Silva-Ortega, C. O., Wu, S., Yamaguchi, N., Wu, M.-F., Pfluger, J., et al. (2012). Mutations in two non-canonical arabidopsis SWI2/SNF2 chromatin remodeling ATPases cause embryogenesis and stem cell maintenance defects. Plant J. 72, 1000–1014. doi: 10.1111/tpj.12009
Sarkar, A. K., Luijten, M., Miyashima, S., Lenhard, M., Hashimoto, T., Nakajima, K., et al. (2007). Conserved factors regulate signalling in arabidopsis thaliana shoot and root stem cell organizers. Nature 446, 811–814. doi: 10.1038/nature05703
Scheres, B. (2007). Stem-cell niches: Nursery rhymes across kingdoms. Nat. Rev. Mol. Cell Biol. 8, 345–354. doi: 10.1038/nrm2164
Shang, E., Ito, T., Sun, B. (2019). Control of floral stem cell activity in arabidopsis. Plant Signaling Behav. 14, 1659706. doi: 10.1080/15592324.2019.1659706
Shang, E., Wang, X., Li, T., Guo, F., Ito, T., Sun, B. (2021). Robust control of floral meristem determinacy by position-specific multifunctions of KNUCKLES. Proc. Natl. Acad. Sci. 118, 393. doi: 10.1073/pnas.2102826118
Shi, B., Zhang, C., Tian, C., Wang, J., Wang, Q., Xu, T., et al. (2016). Two-step regulation of a meristematic cell population acting in shoot branching in arabidopsis. PloS Genet. 12, e1006168. doi: 10.1371/journal.pgen.1006168
Shu, J., Chen, C., Thapa, R. K., Bian, S., Nguyen, V., Yu, K., et al. (2019). Genome-wide occupancy of histone H3K27 methyltransferases CURLY LEAF and SWINGER in arabidopsis seedlings. Plant direct 3, e00100. doi: 10.1002/pld3.100
Singh, S., Singh, A., Singh, A., Yadav, S., Bajaj, I., Kumar, S, et al. (2020). Role of chromatin modification and remodeling in stem cell regulation and meristem maintenance in arabidopsis. J. Exp. Bot. 71, 778–792. doi: 10.1093/jxb/erz459
Skoog, F., Miller, C. O. (1957). Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp. Soc. Exp. Biol. 11, 118–130.
Sugimoto, K., Jiao, Y., Meyerowitz, E. M. (2010). Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev. Cell 18, 463–471. doi: 10.1016/j.devcel.2010.02.004
Sun, B., Looi, L.-S., Guo, S., He, Z., Gan, E.-S., Huang, J., et al. (2014). Timing mechanism dependent on cell division is invoked by polycomb eviction in plant stem cells. Science 343, 1248559. doi: 10.1126/science.1248559
Su, Y. H., Zhou, C., Li, Y. J., Yu, Y., Tang, L. P., Zhang, W. J., et al. (2020). Integration of pluripotency pathways regulates stem cell maintenance in the arabidopsis shoot meristem. Proc. Natl. Acad. Sci. 117, 22561–22571. doi: 10.1073/pnas.2015248117
Takebe, I., Labib, G., Melchers, G. (1971). Regeneration of whole plants from isolated mesophyll protoplasts of tobacco. Naturwissenschaften 58, 318–320. doi: 10.1007/BF00624737
Takeda, S., Tadele, Z., Hofmann, I., Probst, A. V., Angelis, K. J., Kaya, H., et al. (2004). BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in arabidopsis. Genes Dev. 18, 782–793. doi: 10.1101/gad.295404
Tian, J., Xing, Q., Jing, T., Fan, X., Zhang, Q., Müller-Xing, R. (2022). The epigenetic regulator ULTRAPETALA1 suppresses de novo root regeneration from arabidopsis leaf explants. Plant Signaling Behav. 11, 118. doi: 10.1080/15592324.2022.2031784
Wang, X., Gao, J., Gao, S., Li, Z., Kuai, B., Ren, G. (2019). REF6 promotes lateral root formation through de-repression of PIN1/3/7 genes. J. Integr. Plant Biol. 61, 383–387. doi: 10.1111/jipb.12726
Xu, M., Du, Q., Tian, C., Wang, Y., Jiao, Y. (2021). Stochastic gene expression drives mesophyll protoplast regeneration. Sci. Adv. 7. doi: 10.1126/sciadv.abg8466
Xu, J., Hofhuis, H., Heidstra, R., Sauer, M., Friml, J., Scheres, B. (2006). A molecular framework for plant regeneration. Science 311, 385–388. doi: 10.1126/science.1121790
Xu, F., Kuo, T., Rosli, Y., Liu, M.-S., Wu, L., Oliver Chen, L.-F., et al. (2018). Trithorax group proteins act together with a polycomb group protein to maintain chromatin integrity for epigenetic silencing during seed germination in arabidopsis. Mol. Plant 11 (5), 659–677. doi: 10.1016/j.molp.2018.01.010
Xu, L., Shen, W.-H. (2008). Polycomb silencing of KNOX genes confines shoot stem cell niches in arabidopsis. Curr. Biol. 18, 1966–1971. doi: 10.1016/j.cub.2008.11.019
Yan, A., Borg, M., Berger, F., Chen, Z. (2020). The atypical histone variant H3.15 promotes callus formation in arabidopsis thaliana. Development 57, 626. doi: 10.1242/dev.184895
Yan, W., Chen, D., Smaczniak, C., Engelhorn, J., Liu, H., Yang, W., et al. (2018). Dynamic and spatial restriction of polycomb activity by plant histone demethylases. Nat. Plants 4, 681–689. doi: 10.1038/s41477-018-0219-5
Yang, S., Li, C., Zhao, L., Gao, S., Lu, J., Zhao, M., et al. (2015). The arabidopsis SWI2/SNF2 chromatin remodeling ATPase BRAHMA targets directly to PINs and is required for root stem cell niche maintenance. Plant Cell 27, 1670–1680. doi: 10.1105/tpc.15.00091
Yao, X., Feng, H., Yu, Y., Dong, A., Shen, W.-H. (2013). SDG2-mediated H3K4 methylation is required for proper arabidopsis root growth and development. PloS One 8, e56537. doi: 10.1371/journal.pone.0056537
Ye, B.-B., Shang, G.-D., Pan, Y., Xu, Z.-G., Zhou, C.-M., Mao, Y.-B., et al. (2020). AP2/ERF transcription factors integrate age and wound signals for root regeneration. Plant Cell 32, 226–241. doi: 10.1105/tpc.19.00378
Ye, R., Wang, M., Du, H., Chhajed, S., Koh, J., Liu, K.-H., et al. (2022). Glucose-driven TOR–FIE–PRC2 signalling controls plant development. Nature 609, 986–993. doi: 10.1038/s41586-022-05171-5
Zhai, N., Xu, L. (2021). Pluripotency acquisition in the middle cell layer of callus is required for organ regeneration. Nat. Plants 7, 1453–1460. doi: 10.1038/s41477-021-01015-8
Zhai, H., Zhang, X., You, Y., Lin, L., Zhou, W., Li, C. (2020). SEUSS integrates transcriptional and epigenetic control of root stem cell organizer specification. EMBO J. 39, 793. doi: 10.15252/embj.2020105047
Zhang, X., Clarenz, O., Cokus, S., Bernatavichute, Y. V., Pellegrini, M., Goodrich, J., et al. (2007). Whole-genome analysis of histone H3 lysine 27 trimethylation in arabidopsis. PloS Biol. 5, e129. doi: 10.1371/journal.pbio.0050129
Zhang, Y., Jiao, Y., Liu, Z., Zhu, Y.-X. (2015). ROW1 maintains quiescent centre identity by confining WOX5 expression to specific cells. Nat. Commun. 6, 6003. doi: 10.1038/ncomms7003
Zhang, T.-Q., Lian, H., Zhou, C.-M., Xu, L., Jiao, Y., Wang, J.-W. (2017). A two-step model for de Novo activation of WUSCHEL during plant shoot regeneration. Plant Cell 29, 1073–1087. doi: 10.1105/tpc.16.00863
Zhang, G., Zhao, F., Chen, L., Pan, Y., Sun, L., Bao, N., et al. (2019). Jasmonate-mediated wound signalling promotes plant regeneration. Nat. plants 5, 491–497. doi: 10.1038/s41477-019-0408-x
Keywords: stem-cell formation and determinacy, pluripotent stem-cell lineages, callus formation, adventitious shoots and roots, pluripotency factors, epigenetic regulators, chromatin remodeling and modifications
Citation: Müller-Xing R and Xing Q (2022) The plant stem-cell niche and pluripotency: 15 years of an epigenetic perspective. Front. Plant Sci. 13:1018559. doi: 10.3389/fpls.2022.1018559
Received: 13 August 2022; Accepted: 17 October 2022;
Published: 31 October 2022.
Edited by:
Yue Zhou, Peking University, ChinaReviewed by:
Xiaozeng Yang, Beijing Academy of Agricultural and Forestry Sciences, ChinaCopyright © 2022 Müller-Xing and Xing. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ralf Müller-Xing, UmFsZi5NdWVsbGVyQGhodS5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.