
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 23 September 2022
Sec. Plant Bioinformatics
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1018029
This article is part of the Research Topic Advances and Applications of Cost-Effective, High-Throughput Genotyping Technologies for Sustainable Agriculture View all 14 articles
High-throughput sequencing technology has been facilitated the development of new methodologies and approaches for studying the origin and evolution of plant genomes and subgenomes, population domestication, and functional genomics. Orchids have tens of thousands of members in nature. Many of them have promising application potential in the extension and conservation of the ecological chain, the horticultural use of ornamental blossoms, and the utilization of botanical medicines. However, a large-scale gene knockout mutant library and a sophisticated genetic transformation system are still lacking in the improvement of orchid germplasm resources. New gene editing tools, such as the favored CRISPR-Cas9 or some base editors, have not yet been widely applied in orchids. In addition to a large variety of orchid cultivars, the high-precision, high-throughput genome sequencing technology is also required for the mining of trait-related functional genes. Nowadays, the focus of orchid genomics research has been directed to the origin and classification of species, genome evolution and deletion, gene duplication and chromosomal polyploidy, and flower morphogenesis-related regulation. Here, the progressing achieved in orchid molecular biology and genomics over the past few decades have been discussed, including the evolution of genome size and polyploidization. The frequent incorporation of LTR retrotransposons play important role in the expansion and structural variation of the orchid genome. The large-scale gene duplication event of the nuclear genome generated plenty of recently tandem duplicated genes, which drove the evolution and functional divergency of new genes. The evolution and loss of the plastid genome, which mostly affected genes related to photosynthesis and autotrophy, demonstrated that orchids have experienced more separate transitions to heterotrophy than any other terrestrial plant. Moreover, large-scale resequencing provide useful SNP markers for constructing genetic maps, which will facilitate the breeding of novel orchid varieties. The significance of high-throughput sequencing and gene editing technologies in the identification and molecular breeding of the trait-related genes in orchids provides us with a representative trait-improving gene as well as some mechanisms worthy of further investigation. In addition, gene editing has promise for the improvement of orchid genetic transformation and the investigation of gene function. This knowledge may provide a scientific reference and theoretical basis for orchid genome studies.
The Orchidaceae family of monocotyledonous plants have the second-largest members after Compositae. This family contains over 750 genera and nearly 28,000 species (Zhang et al., 2017). Conventional orchids could be classified into five subfamilies (Apostasioideae, Vanilloideae, Cypripedioideae, Epidendroideae, and Orchidoideae) by their morphology and anatomy (Lu et al., 2019). The habitat of wild orchids has been gravely affected by natural and manmade factors. Many endangered species are on the edge of extinction due to indiscriminate gathering. The current protection efforts for orchids include the construction of nature reserves and genetic resource nurseries, as well as seed-preservation and in vitro tissue culture (Williams et al., 2018). Although this act ensures a huge number of original germs, the seedlings degenerate and eventually lose their ability to differentiate during the subculture processes, which makes it difficult to maintain the original genetic background. Besides, most orchids are cross-pollinated, and artificial pollination is considered essential in most cases (Suetsugu, 2015). Because of their huge species diversity and significant economic value, orchids have been the focus of study in botany and ecology for many years. China has a long history of cultivating orchids and has bred numerous varieties. So far, 187 genera and 1500 species of wild orchids have been recorded, including some subspecies and varieties (Chase et al., 2015). There are still several ornamental wild orchids to be created, preserved, and exploited in nature. In addition to its high economic and ornamental value, the orchid also has a profound historic origin. In Chinese traditional culture, the orchid referred to be one of the “four gentlemen among the flowers,” the others being the Prunus mume, Chrysanthemum morifolium, and Sasa pygmaea (Li et al., 2021).
Before the emergence of molecular-assisted breeding, distant hybridization was one of the most commonly used methodology for fertilizing orchids. In recent years, high-throughput sequencing technology and gene editing have been widely applied in the molecular biology, genomics, and discovery of trait-related genes in orchids, as well as modern genetic engineering breeding (Paun et al., 2010; Hsiao et al., 2021; Hsu et al., 2022; Li et al., 2022a). Whole genome sequencing of non-model organisms is now common due to the rapid advancement and lower cost of next-generation sequencing. The draft genome of Phalaenopsis equestris, a tropical epiphytic orchid that is normally utilized as a parent species in orchid breeding, was the first real achievement (Cai et al., 2015). Due to the fast development of ultralong sequencing and new assembly algorithms, whole-genome shotgun sequencing and single molecule sequencing have been done on even more orchid species, such as Dendrobium officinale, Dendrobium catenatum, Dendrobium huoshanense, Phalaenopsis ‘KHM190, Phalaenopsis aphrodite, Gastrodia elata, Vanilla planifolia, Apostasia shenzhenica, etc. (Yan et al., 2015; Huang et al., 2016; Zhang et al., 2016; Zhang et al., 2016; Zhang et al., 2017; Chao et al., 2018; Yuan et al., 2018; Hu et al., 2019; Han et al., 2020; Niu et al., 2021). The growing number of orchid species with high-quality genomes and the use of advanced genetic analysis tools make it much easier to study the functional genes, especially those that are of interest for molecular breeding. The new advancement of genome editing technologies, such as the CRISPR/Cas9 system, is beneficial to this continuing endeavor (Wang et al., 2021). Depending on many defined gene transformation systems in orchids, the CRISPR/Cas9 tool has been effectively implemented in P. equestris by having to take tiny insertion/deletion or reversal mutations into target genes or perhaps the establishing kilobase-scale deletions of genes of interest. (Kui et al., 2017; Tong et al., 2020; Li et al., 2022b).
The market for orchids has expanded in size and diversity as a result of economic globalization, driving scientists and biologists to develop new varieties with distinctive looks, improved adaptability, and premium features (Li et al., 2021). Traditional breeding, despite being time- consuming, is always the predominant means of orchid cultivation. Because of the limitations and inefficiencies of the traditional approaches, hybridization and mutagenesis can not be used to get some desirable traits, like the spotted blooms and foliage of a single plant. Agrobacterium-mediated transformation and particle bombardment methods have been routinely used in transgenic molecular breeding, leading to significant progress in horticultural development (Liau et al., 2003; Men et al., 2003; Hsing et al., 2016; Khumkarjorn et al., 2017; Lin et al., 2018; Kayika Febryanti et al., 2020; Setiawati et al., 2020). Our understanding of orchid reproductive biology will undoubtedly change as a result of these efforts to enhance orchid genome-editing tools and the power of large-scale genome sequencing, which will enable us to better understand the inherent roles of orchid genes and changes to genes of interest for desired blooming and floral features (Molla and Yang 2019; Nopitasari et al., 2020; Tong et al., 2020; Guo et al., 2022). Here, we systematically summarized the studies on orchid genomes, including plastid genomes, especially the molecular evolution of orchids based on high-throughput sequencing technology and the identification and functional studies of trait-related genes. In addition, the application of gene editing and genetic transformation technologies in orchids was also discussed in detail.
Ten years ago, only bacterial artificial chromosome (BAC) end sequences were used in genetic investigations of Phalaenopsis orchids. Short sequences can be used as molecular markers to assist in gene mapping and the construction of genetic maps. These sequences contained several repetitive DNA and SSR markers (Hsu et al., 2011). Cytogenetic evidence is only available for few orchid species (Felix and Guerra, 2010). Cattleya, Cymbidium, Dendrobium, Oncidium, Phalaenopsis, Paphiopedilum, Vanilla, and Vanda are examples of commercially significant genera that are valuable in floriculture, medicinal, and food condiments (da Rocha Perini et al., 2016; Vilcherrez-Atoche et al., 2022). Chromosomal counting and nuclear DNA content estimation with flow cytometry (FCM) are the most popular techniques employed for polyploid identification in these orchids (Younis et al., 2013; Mohammadi et al., 2021). Using flow cytometry, the genetic traits and types of endoreplication of 149 orchid species were compared. The variations in genome size and particularly in GC contents were inextricably bound with evolutionary transitions from the conventional mode of endoreplication to partial endoreplication (Trávníček et al., 2019). In eukaryotic species, nuclear genome size is an inherited quantitative feature with both biological and practical relevance. Genome size, karyotype, and nucleobase composition vary significantly across angiosperms, with potential adaptive consequences. A systematic analysis of the major plant families could help us understand the biological significance of the huge differences in genome size within plants. Several studies have assessed C-values in 48 orchid species in order to analyze the distributions of nuclear DNA quantities and identify tissues suited for accurate estimations of nuclear DNA content (Trávníček et al., 2015; Rewers et al., 2021). Additional analysis on the size of the genomes of Pleurothallidinae species showed that those with partial endoreplication (PE) had much bigger genomes and that the number of genomic repeats was closely linked to the size of the non-endoreplicated part of the genome (Chumová et al., 2021). According to previous investigations on the variation of Apostasioideae genome size, the predicted 1C-values vary from 0.38 pg in Apostasia nuda to 5.96 pg in Neuwiedia zollingeri var. javanica, a roughly 16-fold difference. The genome sizes of the two genera did not overlap. Apostasia had much smaller genomes than Neuwiedia, which suggested that smaller genomes were common in the Apostasioideae subfamily (Jersáková et al., 2013). The genome of Apostasia ramifera showed the population size histories of many orchid species, as well as a continual fall in population size in seven orchid genomes (Zhang W. et al., 2021). Some research had shown that the incorporation of LTR retrotransposons Orchid-rt1 and Gypsy1 into Phalaenopsis genomes might be linked to genome size growth (Hsu et al., 2020). Genome size is also linked to cellular and developmental characteristics. The evolutionary connection between genome size, floral lifespan, and labellum epidermal cell size in Paphiopedilum revealed that genome size was connected to floral duration but negatively relevant to labellum epidermal cell size (Zhang and Zhang, 2021).
In addition to flow cytometry, k-mer analysis-based genome survey sequencing is also a common method for estimating genome size. It has the advantages of high-throughput sequencing, high speed, and large amounts of data, which can quickly determine the size and heterozygosity of the genome (Lee et al., 2017). The k-mer depth values are often derived from the curves used to estimate genome size. Through the distribution of the k-mer curve, the genomic characteristics are estimated, and the ratio of the heterozygous peak to the homozygous peak is calculated to obtain the heterozygous rate (Jersáková et al., 2013). For determining the size of orchid genomes, k-mer analysis based on the Illumina Hiseq sequencing platform has been widely applied. The genome of C. ensifolium was evaluated using 17-mer analysis, which indicated the genome size and heterozygozity to be 3.56 Gb and 1.40%, respectively (Ai et al., 2021). The estimated genome size of G. menghaiensis based on k-mers is 0.98 Gb, with 0.1% heterozygosity and high repeats. The 17-mer distribution is Poisson-distributed and is dependent on the properties of the genome (Jiang Y. et al., 2022). Using k-mer distribution analysis, the genome size, heterozygozity, and repetitive ratio of D. officinale were determined. The largest peak of 17 k-mer frequency was seen at a depth of 90, allowing the determination of the genome size, heterozygosity, and repetitive ratio (Niu et al., 2021).
The development of the orchid industry benefits greatly from the ploidy identification of orchid germplasm resources. Chromosomal and cytological investigations revealed that Cymbidium species contained a prevalence of 40 chromosomes along with variations found in C. serratum (41, 43, 60, and 80). From the earliest polyploids recorded at the beginning of the 20th century, it has been feasible to create a number of Cymbidium polyploid cultivars through biological and artificial approaches (Xie et al., 2017). Since then, Cymbidium cultivars have been known to be diploids, triploids, and tetraploids with distinct chromosomal morphology (Younis et al., 2013). About 75.8% of C. hybridum cultivars harbor polyploids, indicating a link between the intentional or unintentional selection of polyploids instead of diploids for superior features (Vilcherrez-Atoche et al., 2022). The majority of Dendrobium species contained 38 chromosomes, with the exception of D. leonis and D. dixanthum, which both have 40 chromosomes (Zheng et al., 2018). The majority of Phalaenopsis species have 38 chromosomes, with the exception of the Aphyllae, which has only 34 or 36 chromosomes (Lee et al., 2017). However, a significant heterogeneity of genome size was detected among species and hybrids within this genus (Chang et al., 2006; Lee et al., 2017). Phalaenopsis cultivars have a wide range of chromosomal numbers (38, 57, and 76 more), indicating polyploidy. Flower gardening traditionally employs Phalaenopsis hybrid cultivars. Only one diploid cultivar has been documented, whereas over 80% of tetraploid cultivars have 76 chromosomes (Lee S. Y. et al., 2020). The domination of commercial tetraploid cultivars demonstrates the relevance of polyploidy in the development of better Phalaenopsis cultivars. These tetraploid species are implemented as parentals to create subgroups of Phalaenopsis cultivars with the goal of achieving desirable colors for commercial purposes (Bolaños-Villegas and Chen, 2007; Li et al., 2012). Vanda, like Dendrobium and Phalaenopsis, has 38 chromosomes and naturally occurs in tetraploid and hexaploid species (Khan et al., 2019; Liu et al., 2020). In Oncidium, it is assumed that x = 7 is the basic number of chromosomes, but unlike other genera, there is a huge chromosomal variation across species, with the majority exhibiting polyploidy (Su et al., 2013).
The continuity and integrity of model plant genomes have also been greatly improved due to the continuous development of genome research and the improvement of sequencing technology. The orchid genome has gone through the draft genome obtained by ordinary next-generation sequencing, to the chromosome-level genome assembled by PacBio or ONT sequencing technology combined with Hi-C, and then to the near complete genome obtained by ONT (N50>50Kb) assembly (Figure 1). By combining ONT ultra-long and PacBio HiFi techniques, those gap-free genomes assembled at telomere to telomere (T2T) level will be a new direction in the future. The whole genome sequencing of the A. shenzhenica helps us better understand the origins and evolution within subfamilies (Zhang et al., 2017). The whole genome duplication (WGD) that has occurred more than once in plant genomes is a noteworthy feature (Clark and Donoghue, 2018). Angiosperm genome sequences provide information regarding polyploidy and genome evolution. By evaluating the prevalence of synonymous substitutions per synonymous site (Ks) throughout all paralogous genes and duplicated genes situated in synteny blocks based on the Phalaenopsis and Dendrobium genome sequences, two WGDs were projected to have evolved in the D. catenatum lineage. (Zhang et al., 2016). The nearest WGD event is shared by Dendrobium, Phalaenopsis, and Apostasia, and it could have occurred near the Cretaceous-Paleogene (K/Pg) boundary. Peaks in older Ks distributions are thought to be an additional ancient WGD event shared by monocot ancestors (Cai et al., 2015; Zhang et al., 2016; Zhang et al., 2017). The draft genome sequencing revealed compelling evidence of a whole-genome duplication that all orchids share and that came right before their divergence (Zhang et al., 2017). The MADS-box family members may govern a wide spectrum of developmental events during orchid evolution. A chromosomal-scale genome and chromosome linkage groups of P. aphrodite were first created, which contributed to the variation in labellum and pollinium morphology and structures (Chao et al., 2018). A chromosome-scale genome assembly of C. goeringii suggested several new gene families, resistance-related homologs and variations within the MADS-box genes may regulate a wide set of developmental processes during adaptive evolution (Chung et al., 2022). A haplotype-resolved genome of Bletilla striata reveals its evolutionary relationship with other orchids, which have experienced an ancient WGD event shared with monocots and a recent WGD event within all orchids. The biochemical machinery of B. striata polysaccharide (BSP) biosynthesis indicated that MYB2 interacted physically with some BSP-regulated genes (Jiang L. et al., 2022). Partial endoreplication has been discovered across all Vanilla species. A chromosome-scaled genome of Vanilla planifolia showed that the genome size discrepancy was driven by the presence of PE (Piet et al., 2022).
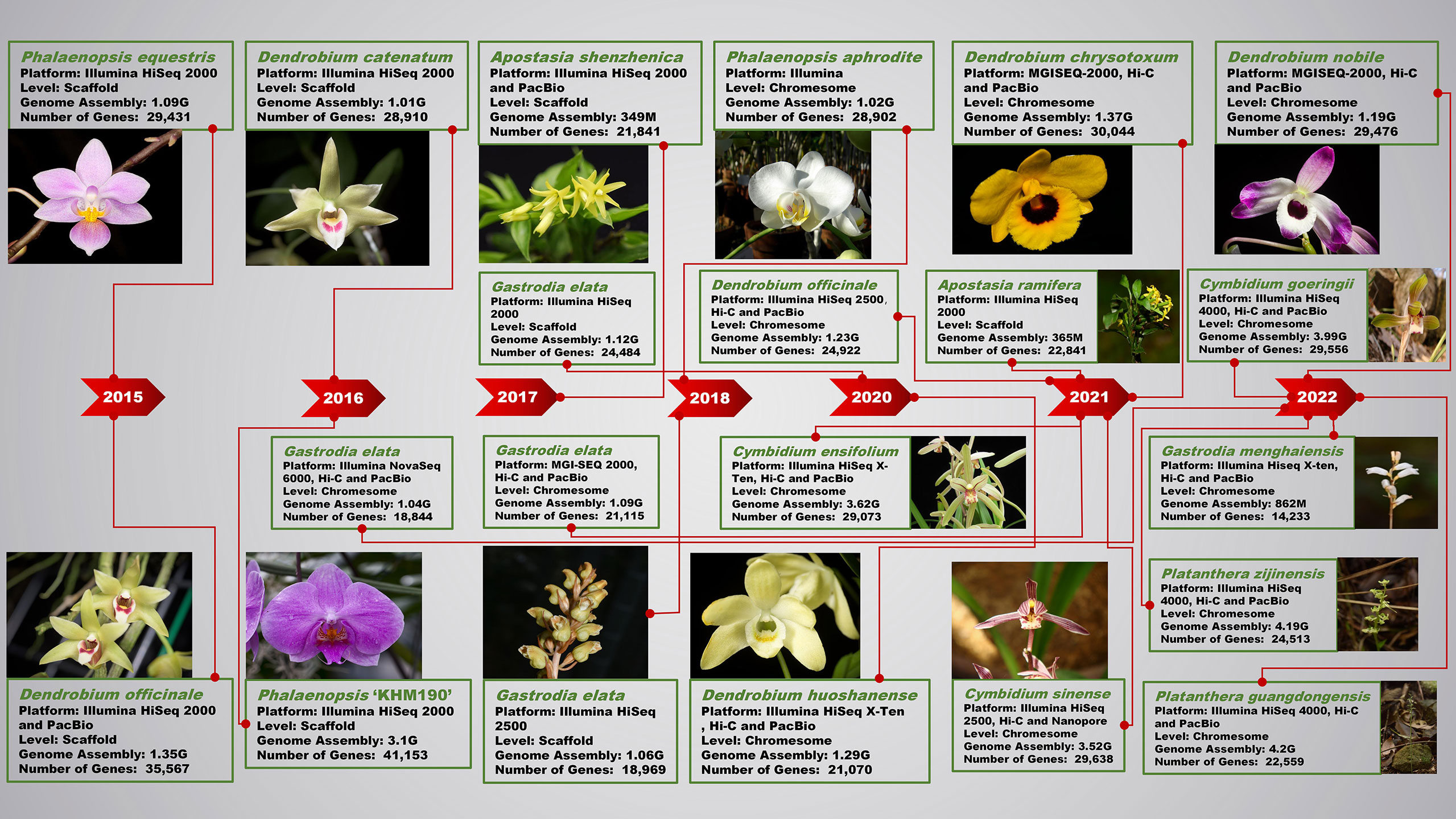
Figure 1 Research progress of next-generation sequencing and third-generation sequencing technology in orchid genomes.
Mycoheterotrophic and parasitic plants get some or all of the nutrients they need from other organisms. Gastrodia fungi are typically perennial, achlorophyllous orchids with a unique evolutionary mechanism for adaptability to a non-photosynthetic lifestyle. The genome of G. elata reveals the genetic basics of most adaptive changes in photosynthesis, leaf development, and plastid division (Chen S. et al., 2020). Comparative genomics studies revealed that G. elata and other completely heterotrophic species dropped nearly 10% of the conserved orthogroups, including those important for autotrophs (Xu et al., 2021). Photosynthesis, circadian clock, flowering control, immunity, food intake, and root and leaf growth are all governed by these orthogroups. Recent assembly of the G. elata genome also showed a strong contraction of genes which involved in multiple biosynthetic processes and cellular components but also an expansion of genes for some metabolic processes and mycorrhizal interactions (Bae et al., 2022). Many genes involved in arbuscular mycorrhizae colonization and biological interaction between Gatrodia and symbiotic microbes were identified in the genome of G. menghaiensis (Jiang Y. et al., 2022). The loss and conservation of symbiotic genes associated with the evolution of unique symbionts in plants were determined by analyzing a broad array of plant genome and transcriptomics data. A shared symbiosis network progressed at the same time as intracellular endosymbioses, from the primitive arbuscular mycorrhiza to the more recent ericoid and orchid mycorrhizae in angiosperms and ericoid-like connections in bryophytes (Radhakrishnan et al., 2020). The comparison of Platanthera zijinensis and Platanthera guangdongensis genomes indicated that mycoheterotrophy is linked to higher rates of gene loss and alternation, and that the deletion of most photoreceptor and auxin transporter genes might explain how fully mycoheterotrophic orchids look so different from other orchids. Some trehalase genes have grown, which makes sense since orchid non-endosperm seeds need carbohydrates from fungi to sprout when they are in the protocorm stage (Li M-H. et al., 2022; Minasiewicz et al., 2022).
Dendrobium is the second biggest genus in Orchidaceae. The first genome of a lithophytic orchid, D. catenatum (now recognized as D. officinale), showed wide duplication of genes associated with glucomannan synthase (Yan et al., 2015; Zhang et al., 2016). Recent assembly of the D. officinale genome has brought new insights into the evolution of this Dendrobium spp. (Niu et al., 2021). Our previous study released a chromosome-level assembly of the D. huoshanense genome with PacBio sequencing and Hi-C method (Han et al., 2020). A chromosome-scale reference genome of D. chrysotoxum was also obtained based on PacBio sequencing and Hi-C methods. The phylogeny of the SWEET gene family implied that gene expansion occurred in clade II may associated with fleshy stems rich in polysaccharides (Zhang Y. et al., 2021). Cymbidium is famous for its distinctive leaves, flower morphology, and pleasant aroma (Yang L. et al., 2021). The genome of C. ensifolium has undergone two WGD events, and the abnormal expression of MADS-box genes might be related to flower development and shape mutations (Ai et al., 2021). A chromosome- scale genome of D. nobile showed two polyploidization events occurred. The expression profile of TPS and CYP450 genes suggested that the distinct distribution of TPS-b subclade may contribute to the species-specific alkaloid biosynthesis pathways (Xu et al., 2022). Finally, a phylogenetic tree was constructed based on single-copy genes to better demonstrate the evolutionary relationship between orchid species (Figure S1).
The associated mapping method performed statistical analyses to discover the importance of the relationship between genetic variants and polymorphism in a group of individuals with genetic variations (Ogura and Busch, 2015). Large-scale resequencing has been broadly used for gene mapping of crop quality traits and differential analysis of SNP loci within genes. However, investigations for genome-wide association studies (GWAS) based on genotyping-by-sequencing (GBS) have received less attention in orchids. Through NGS technology, a large number of SNP markers have been found through sequencing to create a high-density genetic map. A total of 691,532 SNP sites were identified to generate a genetic linkage map for marker-assisted selection breeding by resequencing Phalaenopsis pulcherrima and denovo sequencing of Phalaenopsis ‘KHM190’ (Huang et al., 2016). Species-specific markers could help to identify unknown intraspecies and validate the parentage of interspecifc hybrid offspring. Genomics-based diversity analysis of Vanilla species indicated that the value of the GBS approach to interpret diversity in Vanilla collections has been demonstrated to be the paternal parent of hybrids more efficiently than other methods (Hu et al., 2019). The interspecific hybridization of D. nobile and Dendrobium wardianum was used to construct a population with 100 F1 individuals (Li J. et al., 2019). A total of 331,642 SNP markers were obtained, 9645 of which were used to build a high-density genetic map with 19 linkage groups, and three QTLs identified may be associated with stem length and diameter. The genetic diversity and variations among Dendrobium mutants and common Dendrobium cultivars were compared based on SNPs by GBS (Ryu et al., 2019). A total of 517,660 SNPs were identified, 37,721 of which were used to discriminate the differences across Dendrobium genotypes. 129 accessions were collected from 10 wild cultivated populations to explore the genetic diversity and population structure of D. nobile in China (He et al., 2022). Approximately 830,000 SNPs were obtained and used for genetic variation analysis. The recent completion of the chromosome-level assembly of the D. officinale genome provides a reliable data basis for its genetic background and breeding improvement. Niu and his colleagues performed D. officinale resequencing to conduct a GWAS investigation on 38 cultivars and five related species (Niu et al., 2021). A total of 13 GWAS loci were identified to associate with some morphologic traits.
The chloroplast genome (cp) contains more conserved structures than the nuclear and mitochondrial genomes, which is beneficial for systematics and species identification. Studies on the chloroplast genomes of Orchidaceae have remained prominent in recent years (Table 1). The chloroplast genomes of D. officinale and Cypripedium macranthos were compared, and there were parallels in structure as well as gene order and content, but there were differences in the organization of the inverted repeat/small single-copy junction and ndh genes (Luo et al., 2014). Since ndh genes are truncated or excluded in the cp genomes of some autotrophic Epidendroideae orchids, some studies had mentioned that these gene deletion events are independent (Lin et al., 2015). By comparing 53 cp genomes, it was indicated that the expansion of inverted repeats in Paphiopedilum and Vanilla is also associated with a loss of ndh genes (Niu et al., 2017b). Bulbophyllum Thou. is one of the biggest genera with over 2,000 species, found in rainforest regions (Gamisch and Comes, 2019). Long-term geographic isolation exposed Asian and South American Bulbophyllum cp genomes to varying selective pressures (Yang et al., 2022). Besides the Bulbophyllum orchids, plastid genome sequencing has been reported for a large number of Dendrobium species, which are commonly used for phylogenetic studies and variety authentication (Zhang et al., 2018; Wu X.-Y. et al., 2019; Liu et al., 2021). Phalaenopsis orchids are another orchid species that has received significant interest (Chang et al., 2006; Kim et al., 2016; Wang et al., 2019; Xia et al., 2021). Paphiopedilum, also known as slipper orchid, is well-known for its large, specialized lip, as well as its lovely flowers and colors. The cp genome of many Paphiopedilum orchids was investigated to provide the phylogenomic analysis of this species and its relatives (Zhao et al., 2019; Tang F. L. et al., 2020; Hu et al., 2022). Furthermore, the cp genomes of some other orchid genera or subtribes have been published, including Pelatantheria scolopendrifolia, Cymbidium ensifolium, Eulophia flava, Calanthe arcuata, and Coelogyne fimbr (Yun et al., 2018; Bertrand et al., 2019; Jiang et al., 2019; Li H. et al., 2019; Zhong et al., 2019). These results are important for figuring out how chloroplasts have changed over time and how gene structures vary in orchids (Zeng et al., 2007). A phylogenetic tree of 58 representative orchid species was constructed to investigate the relationship of cp genomes within subfamilies or subtribes (Figure S2). The results also revealed that these varieties could be classified into five subfamilies, with the majority of individuals belonging to the Epidendroideae and Orchidoideae.
Orchids have undergone more independent transitions to heterotrophy than any other land plants. Another interesting fact is that some heterotrophic orchids lose photosynthesis and autotrophy-related genes on chloroplasts throughout evolution, which provides an excellent opportunity to explore the effects of shifting selective regimes on genome evolution (Li M.-H. et al., 2022). As a consequence of the relaxation of functional restrictions on photosynthesis, certain heterotrophic plants, such as mycoheterotrophs and parasites, exhibit enormous gene losses. The comparative genomics of 12 tribe Neottieae orchids indicated that genes related to the NAD(P)H dehydrogenase complex, photosystems, and RNA polymerase were functionally lost many times (Feng et al., 2016). A phylogenetic analysis of 26 full plastome sequences from Epidendreae suggested that photosynthesis-related genes such as the atp complex had undergone severe gene loss (Lee S. Y. et al., 2020). Numerous investigation have identified evidence of fast plastome degradation in heterotrophic orchids based on the accumulation of pseudogenes and substantial deletions (Barrett and Kennedy, 2018; Barrett et al., 2019; Kim et al., 2019). Infraspecific analysis of the plastome evolution of leafless Corallorhiza revealed that considerable changes in plastome size and functional gene composition occurred in just a few million years as a consequence of decreasing selection constraints on photosynthesis (Barrett et al., 2018).
Orchid genome sequencing initiatives and other cuttingedge technologies, such as genome editing tools are undoubtedly facilitating molecular genetic studies on orchid reproductive development. The genome sequencing of the tropical epiphytic orchid P. equestris, which provide an important resource for beginning to explore orchid diversity and evolution at the genome level, was a significant step forward in orchid genome study (Cai et al., 2015). It is now possible to identify and compare gene families that might have new functions across the whole genome with the availability of whole genome sequences (Lin et al., 2016; Cao et al., 2019; Chen T. C. et al., 2020; Song et al., 2021). As most orchid plants contain both C4 metabolism and CAM, phosphoenolpyruvate carboxylase (PEPC) plays an important role in photosynthetic performance and CO2 efficiency. For green plants, especially CAM plants, little is known about the evolutionary history of the PEPC gene family. Using high-throughput sequencing and comprehensive phylogenetic analysis, the results indicated that CAM or C4-related PEPC may originate from the PPC-1M1 clade. The WGD event was responsible for the increment of PEPC gene copies (Deng et al., 2016). The plant-specific YABBY TFs regulate leaf polarity. Two DROOPING LEAF/CRABS CLAW (DL/CRC) genes were discovered in P. equestris, where PeDLs have demonstrated conserved function in floral meristem and carpel development (Chen et al., 2021). Protocorm-like bodies (PLBs) are commonly utilized in orchid micropropagation (Ren et al., 2020). According to certain research, SHOOT MERISTEMLESS (STM)-dependent organogenesis is required for PLB formation (Fang et al., 2022). Overexpression of PaSTM improved the regeneration from vegetative tissue-based explants of Phalaenopsis.
Moreover, many studies have demonstrated that MADS-box family genes control flower formation and morphogenesis (Teo et al., 2019). So far, a total of 51, 56, and 63 putative ones have been noticed in P. equestris, P. aphrodite and D. catenatum, respectively (Cai et al., 2015; Zhang et al., 2016; Chao et al., 2018). Despite having fewer MADS-box genes than Arabidopsis (107 genes) and rice (80 genes), orchids have more MADS-box genes involved in floral organ production (Leseberg et al., 2006). This distinction suggests that higher MADS-box gene diversity might be connected with highly specific floral morphological traits in orchids (Cai et al., 2015; Chao et al., 2018). This hypothesis is backed further by the fact that the number of MADS-box genes differs across Apostasioideae and the other orchid subfamilies. A. shenzhenica, a member of the Apostasioideae subfamily, yields solanum-type flowers with undifferentiated lips and somewhat simple gynostemia (Chen et al., 2012). A. shenzhenica contains fewer B-class AP3-clade and E-class MADS-box genes than Dendrobium and Phalaenopsis (Cai et al., 2015; Zhang et al., 2016). Notably, all modern orchids have shared a WGD event, which may be related to their diversification (Zhang et al., 2017; Yuan et al., 2018). The B-class AP3-clade and E-class genes may have increased just after WGD in the common ancestor of all orchids. Nevertheless, their paralogous genes may have been eliminated in Apostasia, culminating in a reversion to an earlier form with the plesiomorphic bloom (Zhang et al., 2017).
In the long term, the orchid breeding paradigm has seen the transition from conventional selection to cross-breeding, from molecular-assisted breeding to gene editing breeding (Li et al., 2021). Except for some self-incompatible species, the hybrid progeny preserve the parents’ superior genetic features (Niu et al., 2018). However, the fertility of the hybrid combination and the genetic instability of the embryo after fertilization, the mapping of important agronomic traits and the selection of homozygotes are challenges (Su et al., 2019). Among them, seed germination is closely related to hybridization efficiency. When hybrid seeds are obtained, a proper cultivation technique is required to maintain the population. In vitro cultivation is a common method of seed propagation that has been used in the cultivation of numerous orchid species (Gao et al., 2020). The major goals of in vitro propagation are hybrid gex and a reduced breeding cycle. Mutagenesis breeding is also broadly applied for selecting elite crop and horticultural plant varieties. Many orchid varieties, including Dendrobium, Phalaenopsis, Cymbidium, Oncidium, etc., have successfully undergone polyploid breeding by colchicine induction (Vilcherrez-Atoche et al., 2022). The high heterozygosity of orchids can lead to an increase in the perceived mutation rate and result in a flurry of good mutation types. However, unpredictable mutations can occur throughout the genome, and those negative mutations may occur, with only minor changes frequently achieved (Su et al., 2019). Molecular marker-assisted breeding is fast, efficient, and independent of environmental factors. Techniques such as AFLP, RFLP, SSR, RAPD, etc. are regularly employed to identify trait-related differential sequences (Poczai et al., 2013). These markers, when combined with function annotation given by unigenes, enable the identification of candidates with specific roles. Moreover, the completion of large-scale chromosome-level genomes lays the foundation for gene editing breeding and precise breeding based on features.
Polyploidy is the driving force behind species adaptation, diversity, and genome evolution. Some superior orchid cultivars are produced through chromosomal polyploidy in the domain of horticulture (Vilcherrez-Atoche et al., 2022). Domestication and polyploidy have a close link since polyploid plants are randomly selected for their greater vigor, and consequently, polyploid species are more profitable and attractive for domestication than wild ones. The size of a genome is mostly determined by endoreplication and LTR retrotransposon insertion during expansion (Chumová et al., 2021). Initially, FCM and k-mer analysis was used to calculate the size of these genomes. Large-scale tandem duplication and segmental duplication within the chromosome drive the generation of new genes and species evolution (Clark and Donoghue, 2018). In most cases, orchids underwent WGD more than once, including a historical WGD event and a recent WGD event shared by all orchids. There are both mycoheterotrophic and parasitic orchids, in addition to the vast majority of ornamental orchids. The loss and survival of symbiotic genes related to the evolution of specific symbionts span from the ancestral arbuscular mycorrhiza to the recent ericoid and orchid mycorrhizae (Barrett et al., 2019; Gao et al., 2020). Fully mycoheterotrophic orchids look very different from other orchids. This might be due to the loss of most of their photoreceptor and auxin transporter genes. Large-scale resequencing has been utilized to pinpoint key genes or chromosomal regions linked with some trait characteristics. GWAS based on GBS has sparked a lot of interest in several orchids. Some valuable SNP markers are widely applied to discriminate against orchid varieties (Kumagai et al., 2019). Furthermore, a small single-copy region in the cp genome of Paphiopedilum lost a large number of sequences, implying its significance in adaptive evolution (Trávníček et al., 2015). In this study, a phylogenetic tree of 58 orchid species was constructed to investigate the relationship of cp genomes within five subfamilies. The major sequenced species are those designated as Epidendroideae and Orchidoideae. MADS-box transcriptional factors are one of the most studied gene families in orchids, with evidence that they are involved in the regulation of various developmental processes as well as responses to environmental stimuli (Teo et al., 2019). The biological functions of these MADS-box proteins and the mechanisms by which they contribute to flowering or floral organ development are detailed. The molecular mechanisms underpinning flowering and floral development can be exploited for both traditional orchid breeding and targeted manipulation for desired blooming features.
Despite recent advancements in the field of orchid reproductive development, molecular genetic studies of flowering initiation and development continue to lag behind those in other model plants due to a number of bottlenecks. These included the prolonged vegetative stage, the inefficiency of established genetic transformation systems, and available data on genome sequences (Wang et al., 2017). Consequently, the majority of studies on orchid reproductive development have concentrated on genes that are homologs of other well-known genes in model plants.The duplication of genes in the genomes of some orchids may be beneficial for the inheritance of specific characteristics that contribute to the adaption to various environments. Furthermore, clarifying the inherent roles of the key genes in homologous orchid transgenic systems is critical (Hsing et al., 2016; Zhang et al., 2017). This technique involves the ongoing development of a few orchid-specific technical platforms, such as in vitro tissue culture, gene transformation, and genome editing tools (Hsiao et al., 2011; Li et al., 2022b). Many recent studies on the crop pan-genome have successfully identified core genes, individual-specific genes, and structural variation between many subspecies, providing new insights into the genetic underpinning of intricate biological characteristics (Liao et al., 2004; Li et al., 2021). A pan-genome encompasses more genetic variation within plants than a single reference genome. Therefore, another research hotspot of orchids may be concentrated on pan-genome and next-generation breeding technologies under the genetic background of different species (Tsai et al., 2017). Together, these efforts and the ever-improving use of multi-omics techniques to find specific molecular markers linked with morphological changes in orchid reproductive development will pave the way to figure out the molecular basis of specialized orchid reproductive processes.
CS, FZ, and YC discussed the writing plan. CS, YW and DM wrote the draft manuscript. CS, MM, and PPW edited the manuscript. FZ and CS acquired the funding. All the authors have read and approved the submitted version.
This work was supported by Anhui Province Postdoctoral Fund (2020B454), High-level Talents Research Initiation Fund of West Anhui University (WGKQ2022025), Postdoctoral Fund of West Anhui University (WXBSH2019001), and Anhui Provincial Administration of Traditional Chinese Medicine Project (2020zcyb09).
We apologize to those authors whose excellent work could not be cited because of space restrictions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1018029/full#supplementary-material
Supplementary Figure 1 | The phylogenetic tree of 11 orchid species with publicly available protein sequences based on the identified single-copy genes. A. thaliana was regarded as an outgroup. The tree was visulized by the iTOL online service (https://itol.embl.de/).
Supplementary Figure 2 | The maximum-likelihood (ML) tree of 58 Orchidaceae species based on the chloroplast genomes. Alignments of the cp genomes were performed using MAFFT (v7.505) based on the FFT-NS-2 method (https://mafft.cbrc.jp/alignment/software/). The Archaeopteryx.js tool was used to display the ML tree (https://sites.google.com/site/cmzmasek/home/software/archaeopteryx).
Ai, Y., Li, Z., Sun, W. H., Chen, J., Zhang, D., Ma, L., et al. (2021). The cymbidium genome reveals the evolution of unique morphological traits. Hortic. Res. 8, 1–15. doi: 10.1038/s41438-021-00683-z
Ai, Y., Xie, T. X., Chen, J., Zhou, J., Chen, M. K., Liu, Z. J. (2019a). The complete chloroplast genome of cymbidium floribundum var. pumilum (Orchidaceae). Mitochondrial DNA Part B. Resour. 4, 3648–3649. doi: 10.1080/23802359.2019.1678419
Ai, Y., Xie, T. X., Liu, D. K., Tu, X., Zhou, J., Liu, Z. J. (2019b). Complete chloroplast genome of arundina graminifolia (Orchidaceae). Mitochondrial DNA Part B. Resour. 4, 2898–2899. doi: 10.1080/23802359.2019.1660281
Amiryousefi, A., Hyvonen, J., Poczai, P. (2017). The plastid genome of vanillon (Vanilla pompona, orchidaceae). Mitochondrial DNA Part B. Resour. 2, 689–691. doi: 10.1080/23802359.2017.1383201
Bae, E. K., An, C., Kang, M. J., Lee, S. A., Lee, S. J., Kim, K. T., et al. (2022). Chromosome-level genome assembly of the fully mycoheterotrophic orchid gastrodia elata. G3 Genes. Genomes. Genet. 12, 1–11. doi: 10.1093/g3journal/jkab433
Barrett, C. F., Kennedy, A. H. (2018). Plastid genome degradation in the endangered, mycoheterotrophic, north American orchid hexalectris warnockii. Genome Biol. Evol. 10, 1657–1662. doi: 10.1093/gbe/evy107
Barrett, C. F., Sinn, B. T., Kennedy, A. H., Pupko, T. (2019). Unprecedented parallel photosynthetic losses in a heterotrophic orchid genus. Mol. Biol. Evol. 36, 1884–1901. doi: 10.1093/molbev/msz111
Barrett, C. F., Wicke, S., Sass, C. (2018). Dense infraspecific sampling reveals rapid and independent trajectories of plastome degradation in a heterotrophic orchid complex. New Phytol. 218, 1192–1204. doi: 10.1111/nph.15072
Bertrand, J. A. M., Gibert, A., Llauro, C., Panaud, O. (2019). Characterization of the complete plastome of ophrys aveyronensis, a Euro-Mediterranean orchid with an intriguing disjunct geographic distribution. Mitochondrial DNA Part B. Resour. 4, 3256–3257. doi: 10.1080/23802359.2019.1670748
Bolaños-Villegas, P., Chen, F. C. (2007). Cytological identification of chromosomal rearrangements in doritaenopsis and phalaenopsis. J. Int. Coop. 2, 1–11. doi: 10.13140/RG.2.1.1641.8007
Cai, J., Liu, X., Vanneste, K., Proost, S., Tsai, W.-C., Liu, K.-W., et al. (2015). The genome sequence of the orchid phalaenopsis equestris. Nat. Genet. 47, 65–72. doi: 10.1038/ng.3149
Cao, Y., Meng, D., Han, Y., Chen, T., Jiao, C., Chen, Y., et al. (2019). Comparative analysis of b-BOX genes and their expression pattern analysis under various treatments in Dendrobium officinale. BMC Plant Biol. 19, 1–16. doi: 10.1186/s12870-019-1851-6
Chang, C. C., Lin, H. C., Lin, I. P., Chow, T. Y., Chen, H. H., Chen, W. H., et al. (2006). The chloroplast genome of phalaenopsis aphrodite (Orchidaceae): Comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol. Biol. Evol. 23, 279–291. doi: 10.1093/molbev/msj029
Chao, Y. T., Chen, W. C., Chen, C. Y., Ho, H. Y., Yeh, C. H., Kuo, Y. T., et al. (2018). Chromosome-level assembly, genetic and physical mapping of phalaenopsis aphrodite genome provides new insights into species adaptation and resources for orchid breeding. Plant Biotechnol. J. 16, 2027–2041. doi: 10.1111/pbi.12936
Chase, M. W., Cameron, K. M., Freudenstein, J. V., Pridgeon, A. M., Salazar, G., van den Berg, C., et al. (2015). An updated classification of orchidaceae. Bot. J. Linn. Soc 177, 151–174. doi: 10.1111/boj.12234
Chen, J., Chen, M.-K., Zheng, Q.-D., Ma, S.-H., Liu, Z.-J., Ai, Y. (2020). Chloroplast characterization and phylogenetic relationship of cymbidium aloifolium (Orchidaceae). Mitochondrial DNA Part B. 5, 478–479. doi: 10.1080/23802359.2019.1704656
Chen, M. K., Chen, J., Zhou, J., Ma, S. H., Zheng, Q. D., Xie, T. X., et al. (2019). The complete chloroplast genome sequence of habenaria ciliolaris (Orchidaceae). Mitochondrial DNA Part B. Resour. 4, 4132–4133. doi: 10.1080/23802359.2019.1692727
Chen, Y. Y., Hsiao, Y. Y., Li, C. I., Yeh, C. M., Mitsuda, N., Yang, H. X., et al. (2021). The ancestral duplicated DL/CRC orthologs, PeDL1 and PeDL2, function in orchid reproductive organ innovation. J. Exp. Bot. 72, 5442–5461. doi: 10.1093/jxb/erab195
Chen, Y. Y., Lee, P. F., Hsiao, Y. Y., Wu, W. L., Pan, Z. J., Lee, Y. I., et al. (2012). C-and d-class MADS-box genes from phalaenopsis equestris (orchidaceae) display functions in gynostemium and ovule development. Plant Cell Physiol. 53, 1053–1067. doi: 10.1093/pcp/pcs048
Chen, T. C., Liu, Y. C., Wang, X., Wu, C. H., Huang, C. H., Chang, C. C. (2017). Whole plastid transcriptomes reveal abundant RNA editing sites and differential editing status in phalaenopsis aphrodite subsp. formosana. Bot. Stud. 58, 1–14. doi: 10.1186/s40529-017-0193-7
Chen, T. C., Su, Y. Y., Wu, C. H., Liu, Y. C., Huang, C. H., Chang, C. C. (2020). Analysis of mitochondrial genomics and transcriptomics reveal abundant RNA edits and differential editing status in moth orchid, phalaenopsis aphrodite subsp. formosana. Sci. Hortic. (Amsterdam). 267, 1–13. doi: 10.1016/j.scienta.2020.109304
Chen, S., Wang, X., Wang, Y., Zhang, G., Song, W., Dong, X., et al. (2020). Improved de novo assembly of the achlorophyllous orchid gastrodia elata. Front. Genet. 11. doi: 10.3389/fgene.2020.580568
Choi, H., Lyu, J., Lee, H. O., Kim, J. B., Kim, S. H. (2020). Complete chloroplast genome sequence of an orchid hybrid cymbidium sinense (♀) × c. goeringii (♂). Mitochondrial DNA Part B. Resour. 5, 3802–3803. doi: 10.1080/23802359.2020.1839367
Chumová, Z., Záveská, E., Hloušková, P., Ponert, J., Schmidt, P. A., Čertner, M., et al. (2021). Repeat proliferation and partial endoreplication jointly shape the patterns of genome size evolution in orchids. Plant J. 107, 511–524. doi: 10.1111/tpj.15306
Chung, O., Kim, J., Bolser, D., Kim, H. M., Jun, J. H., Choi, J. P., et al. (2022). A chromosome-scale genome assembly and annotation of the spring orchid (Cymbidium goeringii). Mol. Ecol. Resour. 22, 1168–1177. doi: 10.1111/1755-0998.13537
Clark, J. W., Donoghue, P. C. J. (2018). Whole-genome duplication and plant macroevolution. Trends Plant Sci. 23, 933–945. doi: 10.1016/j.tplants.2018.07.006
da Rocha Perini, V., Leles, B., Furtado, C., Prosdocimi, F. (2016). Complete chloroplast genome of the orchid cattleya crispata (Orchidaceae:Laeliinae), a Neotropical rupiculous species. Mitochondrial DNA Part A. DNA Mapping. Seq. Anal. 27, 4075–4077. doi: 10.3109/19401736.2014.1003850
Deng, H., Zhang, L. S., Zhang, G. Q., Zheng, B. Q., Liu, Z. J., Wang, Y. (2016). Evolutionary history of PEPC genes in green plants: Implications for the evolution of CAM in orchids. Mol. Phylogenet. Evol. 94, 559–564. doi: 10.1016/j.ympev.2015.10.007
Du, Z., Yang, X., Tan, G., Chen, Z. (2021). The complete chloroplast genome of cymbidium dayanum (Orchidaceae). Mitochondrial DNA Part B. Resour. 6, 1897–1898. doi: 10.1080/23802359.2021.1934173
Fang, S.-C., Chen, J.-C., Chang, P.-Y., Lin, H.-Y. (2022). Co-Option of the SHOOT MERISTEMLESS network regulates protocorm-like body development in phalaenopsis aphrodite. Plant Physiol. 1–19, 1–19. doi: 10.1093/plphys/kiac100
Fan, J., Huang, M. Y. (2019). Chloroplast genome structure and phylogeny of spiranthes sinensis, an endangered medicinal orchid plant. Mitochondrial DNA Part B. Resour. 4, 2994–2996. doi: 10.1080/23802359.2019.1664345
Fan, Z. F., Yu, D. Y., Ma, C. (2021). The complete chloroplast genome sequence of phalaenopsis wilsonii Rolfe, a vulnerable wild moth orchid species (Orchidaceae). Mitochondrial DNA Part B. Resour. 6, 2903–2905. doi: 10.1080/23802359.2021.1923420
Felix, L. P., Guerra, M. (2010). Variation in chromosome number and the basic number of subfamily epidendroideae (Orchidaceae). Bot. J. Linn. Soc 163, 234–278. doi: 10.1111/j.1095-8339.2010.01059.x
Feng, Y. L., Wicke, S., Li, J. W., Han, Y., Lin, C. S., Li, D. Z., et al. (2016). Lineage-specific reductions of plastid genomes in an orchid tribe with partially and fully mycoheterotrophic species. Genome Biol. Evol. 8, 2164–2175. doi: 10.1093/gbe/evw144
Gamisch, A., Comes, H. P. (2019). Clade-age-dependent diversification under high species turnover shapes species richness disparities among tropical rainforest lineages of bulbophyllum (Orchidaceae). BMC Evol. Biol. 19, 1–16. doi: 10.1186/s12862-019-1416-1
Gao, Y., Zhao, Z., Li, J., Liu, N., Jacquemyn, H., Guo, S., et al. (2020). Do fungal associates of co-occurring orchids promote seed germination of the widespread orchid species gymnadenia conopsea? Mycorrhiza 30, 221–228. doi: 10.1007/s00572-020-00943-1
Ge, L. P., Tang, L., Li, L., Luo, Y. (2020). The complete chloroplast genome of an endangered orchid paphiopedilum spicerianum. Mitochondrial DNA Part B. Resour. 5, 3594–3595. doi: 10.1080/23802359.2020.1830727
Guo, M., Chen, H., Dong, S., Zhang, Z., Luo, H. (2022). CRISPR-cas gene editing technology and its application prospect in medicinal plants. Chin. Med. (United. Kingdom). 17, 1–19. doi: 10.1186/s13020-022-00584-w
Han, B., Jing, Y., Dai, J., Zheng, T., Gu, F., Zhao, Q., et al. (2020). A chromosome-level genome assembly of dendrobium huoshanense using long reads and hi-c data. Genome Biol. Evol. 12, 2486–2490. doi: 10.1093/GBE/EVAA215
He, L. F., Qiang, S. J., Zhang, Y. H. (2021). The complete chloroplast genome of pleione maculata, an orchid with important ornamental value and medicinal value. Mitochondrial DNA Part B. Resour. 6, 2263–2265. 69, 1–16. doi: 10.1080/23802359.2021.1948366
He, T., Ye, C., Zeng, Q., Fan, X., Huang, T. (2022). Genetic diversity and population structure of cultivated dendrobium nobile lindl. in southwest of China based on genotyping-by-sequencing. Genet. Resour. Crop Evol. doi: 10.1007/s10722-022-01401-x
Hsiao, Y. Y., Fu, C. H., Ho, S. Y., Li, C. I., Chen, Y. Y., Wu, W. L., et al. (2021). OrchidBase 4.0: a database for orchid genomics and molecular biology. BMC Plant Biol. 21, 1–11. doi: 10.1186/s12870-021-03140-0
Hsiao, Y. Y., Pan, Z. J., Hsu, C. C., Yang, Y. P., Hsu, Y. C., Chuang, Y. C., et al. (2011). Research on orchid biology and biotechnology. Plant Cell Physiol. 52, 1467–1486. doi: 10.1093/pcp/pcr100
Hsing, H. X., Lin, Y. J., Tong, C. G., Li, M. J., Chen, Y. J., Ko, S. S. (2016). Efficient and heritable transformation of phalaenopsis orchids. Bot. Stud. 57. doi: 10.1186/s40529-016-0146-6
Hsu, C. C., Chen, S. Y., Chiu, S. Y., Lai, C. Y., Lai, P. H., Shehzad, T., et al. (2022). High-density genetic map and genome-wide association studies of aesthetic traits in phalaenopsis orchids. Sci. Rep. 12, 1–15. doi: 10.1038/s41598-022-07318-w
Hsu, C. C., Chen, S. Y., Lai, P. H., Hsiao, Y. Y., Tsai, W. C., Liu, Z. J., et al. (2020). Identification of high-copy number long terminal repeat retrotransposons and their expansion in phalaenopsis orchids. BMC Genomics 21, 1–13. doi: 10.1186/s12864-020-07221-6
Hsu, C. C., Chung, Y. L., Chen, T. C., Lee, Y. L., Kuo, Y. T., Tsai, W. C., et al. (2011). An overview of the phalaenopsis orchid genome through BAC end sequence analysis. BMC Plant Biol. 11, 1–13. doi: 10.1186/1471-2229-11-3
Huang, J. Z., Lin, C. P., Cheng, T. C., Huang, Y. W., Tsai, Y. J., Cheng, S. Y., et al. (2016). The genome and transcriptome of phalaenopsis yield insights into floral organ development and flowering regulation. PeerJ 2016, 1–17. doi: 10.7717/peerj.2017
Hu, C., Jiang, K., Zeng, X., Huang, W. (2022). The complete chloroplast genome sequence of a critically endangered orchid paphiopedilum gratrixianum (Orchidaceae). Mitochondrial DNA Part B. 7, 609–610. doi: 10.1080/23802359.2021.1891005
Huo, X., Zhao, Y., Qian, Z., Liu, M. (2018). Characterization of the complete chloroplast genome of eulophia zollingeri, an endangered orchid in China. Conserv. Genet. Resour. 10, 817–819. doi: 10.1007/s12686-017-0938-3
Hu, Y., Resende, M. F. R., Bombarely, A., Brym, M., Bassil, E., Chambers, A. H. (2019). Genomics-based diversity analysis of vanilla species using a vanilla planifolia draft genome and genotyping-By-Sequencing. Sci. Rep. 9, 1–16. doi: 10.1038/s41598-019-40144-1
Hu, G., Zhou, H., Zhang, S., Zhao, P. (2020). Characterization of the complete chloroplast genome of orchid family species cymbidium bicolor (Orchidaceae). Mitochondrial DNA Part B. Resour. 5, 323–324. doi: 10.1080/23802359.2019.1703591
Jersáková, J., Trávníček, P., Kubátová, B., Krejčíková, J., Urfus, T., Liu, Z. J., et al. (2013). Genome size variation in orchidaceae subfamily apostasioideae: Filling the phylogenetic gap. Bot. J. Linn. Soc 172, 95–105. doi: 10.1111/boj.12027
Jheng, C. F., Chen, T. C., Lin, J. Y., Chen, T. C., Wu, W. L., Chang, C. C. (2012). The comparative chloroplast genomic analysis of photosynthetic orchids and developing DNA markers to distinguish phalaenopsis orchids. Plant Sci. 190, 62–73. doi: 10.1016/j.plantsci.2012.04.001
Jiang, Y., Hu, X., Yuan, Y., Guo, X., Chase, M. W., Ge, S., et al. (2022). The gastrodia menghaiensis (Orchidaceae) genome provides new insights of orchid mycorrhizal interactions. BMC Plant Biol. 22, 1–14. doi: 10.1186/s12870-022-03573-1
Jiang, Y. T., Lin, R. Q., Liu, B., Zeng, Q. M., Liu, Z. J., Chen, S. P. (2019). Complete chloroplast genome of cymbidium ensifolium(Orchidaceae). Mitochondrial DNA Part B. Resour. 4, 2236–2237. doi: 10.1080/23802359.2019.1624637
Jiang, L., Lin, M., Wang, H., Song, H., Zhang, L., Huang, Q., et al. (2022). Haplotype-resolved genome assembly of bletilla striata ( thunb .) reichb . f . to elucidate medicinal value. Plant J. 111, 1–14. doi: 10.1111/tpj.15892
Kao, H., Zhao, Y., Yang, M., Sun, Y., Cheng, J. (2021). The complete chloroplast genome sequences of an endangered orchid species paphiopedilum parishii (Orchidaceae). Mitochondrial DNA Part B. Resour. 6, 2521–2522. doi: 10.1080/23802359.2021.1959437
Kayika Febryanti, N. L. P., Nurliana, S., Gutierrez-Marcos, J., Semiarti, E. (2020). The expression analysis of AtRKD4 transgene in dendrobium lineale rolfe transgenic orchid carrying 35S::GR::AtRKD4 for micropropagation. AIP. Conf. Proc. 2260, 1–6. doi: 10.1063/5.0015876
Khan, H., Marya, Belwal, T., Mohd Tariq, Atanasov, A. G., Devkota, H. P. (2019). Genus vanda: A review on traditional uses, bioactive chemical constituents and pharmacological activities. J. Ethnopharmacol. 229, 46–53. doi: 10.1016/j.jep.2018.09.031
Khumkarjorn, N., Thanonkeo, S., Yamada, M., Klanrit, P., Thanonkeo, P. (2017). Agrobacterium-mediated transformation of dendrobium orchid with the flavanone 3-hydroxylase gene. Turk. J. Bot. 41, 442–454. doi: 10.3906/bot-1701-13
Kim, Y. K., Jo, S., Cheon, S. H., Joo, M. J., Hong, J. R., Kwak, M. H., et al. (2019). Extensive losses of photosynthesis genes in the plastome of a mycoheterotrophic orchid, cyrtosia septentrionalis (Vanilloideae: Orchidaceae). Genome Biol. Evol. 11, 565–571. doi: 10.1093/gbe/evz024
Kim, G. B., Kwon, Y., Yu, H. J., Lim, K. B., Seo, J. H., Mun, J. H. (2016). The complete chloroplast genome of phalaenopsis “Tiny star”. Mitochondrial DNA 27, 1300–1302. doi: 10.3109/19401736.2014.945566
Konhar, R., Debnath, M., Vishwakarma, S., Bhattacharjee, A., Sundar, D., Tandon, P., et al. (2019). The complete chloroplast genome of dendrobium nobile, an endangered medicinal orchid from north-east India and its comparison with related dendrobium species. PeerJ 2019, 1–28. doi: 10.7717/peerj.7756
Kui, L., Chen, H., Zhang, W., He, S., Xiong, Z., Zhang, Y., et al. (2017). Building a genetic manipulation tool box for orchid biology: Identification of constitutive promoters and application of CRISPR/Cas9 in the orchid, dendrobium officinale. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.02036
Kumagai, M., Nishikawa, D., Kawahara, Y., Wakimoto, H., Itoh, R., Tabei, N., et al. (2019). Tasuke1: A web-based platform for exploring GWAS results and large-scale resequencing data. DNA Res. 26, 445–452. doi: 10.1093/dnares/dsz022
Lee, Y. I., Chung, M. C., Kuo, H. C., Wang, C. N., Lee, Y. C., Lin, C. Y., et al. (2017). The evolution of genome size and distinct distribution patterns of rDNA in phalaenopsis (Orchidaceae). Bot. J. Linn. Soc 185, 65–80. doi: 10.1093/BOTLINNEAN/BOX049
Lee, S. Y., Kaikai, M., Zhou, R., Liao, W. (2020). Severe plastid genome size reduction in a mycoheterotrophic orchid, danxiaorchis singchiana, reveals heavy gene loss and gene relocations. Plants 9, 521. doi: 10.3390/plants9040521
Lee, Y. I., Tseng, Y. F., Lee, Y. C., Chung, M. C. (2020). Chromosome constitution and nuclear DNA content of phalaenopsis hybrids. Sci. Hortic. (Amsterdam). 262, 109089. doi: 10.1016/j.scienta.2019.109089
Leseberg, C. H., Li, A., Kang, H., Duvall, M., Mao, L. (2006). Genome-wide analysis of the MADS-box gene family in populus trichocarpa. Gene 378, 84–94. doi: 10.1016/j.gene.2006.05.022
Liao, L. J., Pan, I. C., Chan, Y. L., Hsu, Y. H., Chen, W. H., Chan, M. T. (2004). Transgene silencing in phalaenopsis expressing the coat protein of cymbidium mosaic virus is a manifestation of RNA-mediated resistance. Mol. Breed. 13, 229–242. doi: 10.1023/B:MOLB.0000022527.68551.30
Liau, C. H., You, S. J., Prasad, V., Hsiao, H. H., Lu, J. C., Yang, N. S., et al. (2003). Agrobacterium tumefaciens-mediated transformation of an oncidium orchid. Plant Cell Rep. 21, 993–998. doi: 10.1007/s00299-003-0614-9
Li, T. Z., Chen, L. J., Wang, M., Chen, J. B., Huang, J. (2019). The complete chloroplast genome of vanilla shenzhenica (Orchidaceae). Mitochondrial DNA Part B. Resour. 4, 2610–2611. doi: 10.1080/23802359.2019.1642165
Li, C., Dong, N., Zhao, Y., Wu, S., Liu, Z., Zhai, J. (2021). A review for the breeding of orchids: Current achievements and prospects. Hortic. Plant J. 7, 380–392. doi: 10.1016/j.hpj.2021.02.006
Li, Y., Li, Z., Hu, Q., Zhai, J., Liu, Z., Wu, S. (2019). Complete plastid genome of apostasia shenzhenica (Orchidaceae). Mitochondrial DNA Part B. Resour. 4, 1388–1389. doi: 10.1080/23802359.2019.1591192
Li, M.-H., Liu, K.-W., Li, Z., Lu, H.-C., Ye, Q.-L., Zhang, D., et al. (2022). Genomes of leafy and leafless platanthera orchids illuminate the evolution of mycoheterotrophy. Nat. Plants 8, 373–388. doi: 10.1038/s41477-022-01127-9
Li, H., Li, R., Zhang, J., Yin, Q., Zhang, Y. (2019). Complete chloroplast genome of ornamental orchids eulophia flava. Mitochondrial DNA Part B. Resour. 4, 2997–2998. doi: 10.1080/23802359.2019.1664946
Li, R., Wang, A., Sun, S., Liang, S., Wang, X., Ye, Q., et al. (2012). Functional characterization of FT and MFT ortholog genes in orchid (Dendrobium nobile Lindl) that regulate the vegetative to reproductive transition in Arabidopsis. Plant Cell. Tissue Organ Cult. 111, 143–151. doi: 10.1007/s11240-012-0178-x
Lin, B. Y., Chang, C. D., Huang, L. L. H., Liu, Y. C., Su, Y. Y., Chen, T. C., et al. (2016). The mitochondrial DNA markers for distinguishing phalaenopsis species and revealing maternal phylogeny. Biol. Plant 60, 68–78. doi: 10.1007/s10535-015-0566-2
Lin, H. Y., Chen, J. C., Fang, S. C. (2018). A protoplast transient expression system to enable molecular, cellular, and functional studies in phalaenopsis orchids. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00843
Lin, C. S., Chen, J. J. W., Huang, Y. T., Chan, M. T., Daniell, H., Chang, W. J., et al. (2015). The location and translocation of ndh genes of chloroplast origin in the orchidaceae family. Sci. Rep. 5, 1–10. doi: 10.1038/srep09040
Liu, J. F. (2020). The complete chloroplast genome sequence of liparis bootanensis (Orchidaceae). Mitochondrial DNA Part B. Resour. 5, 2058–2059. doi: 10.1080/23802359.2020.1763866
Liu, D. K., Tu, X., Zhang, S., Li, M. H. (2020). The complete plastid genome of vanda subconcolor (Orchidaceae, aeridinae). Mitochondrial DNA Part B. Resour. 5, 1712–1713. doi: 10.1080/23802359.2020.1749169
Liu, F., Xiao, X., An, M., Li, Z. (2021). The complete chloroplast genome of dendrobium comatum (Orchidaceae). Mitochondrial DNA Part B. Resour. 6, 3229–3230. doi: 10.1080/23802359.2021.1990152
Li, J., Xu, Y.c., Wang, Z.h. (2019). Construction of a high-density genetic map by RNA sequencing and eQTL analysis for stem length and diameter in dendrobium (Dendrobium nobile × dendrobium wardianum). Ind. Crops Prod. 128, 48–54. doi: 10.1016/j.indcrop.2018.10.073
Li, Y., Zhang, B., Yu, H. (2022a). Kilobase-scale genomic deletion of DOTFL1 in dendrobium orchids. J. Genet. Genomics 49, 81–84. doi: 10.1016/j.jgg.2021.07.008
Li, Y., Zhang, B., Yu, H. (2022b). Molecular genetic insights into orchid reproductive development. J. Exp. Bot. 73, 1841–1852. doi: 10.1093/jxb/erac016
Li, M., Zhao, Z., He, J., Cheng, J., Xie, L. (2019). The complete chloroplast genome sequences of a highly endangered orchid species paphiopedilum barbigerum (Orchidaceae). Mitochondrial DNA Part B. Resour. 4, 2928–2929. doi: 10.1080/23802359.2019.1660269
Lu, H., Liu, Z., Lan, S. (2019). Genome sequencing reveals the role of MADS-box gene families in the floral morphology evolution of orchids. Hortic. Plant J. 5, 247–254. doi: 10.1016/j.hpj.2019.11.005
Luo, J., Hou, B. W., Niu, Z. T., Liu, W., Xue, Q. Y., Ding, X. Y. (2014). Comparative chloroplast genomes of photosynthetic orchids: Insights into evolution of the orchidaceae and development of molecular markers for phylogenetic applications. PloS One 9, 1–15. doi: 10.1371/journal.pone.0099016
Ma, X., Lin, H., Chen, Y., Lan, S., Ming, R. (2019). The complete chloroplast genome of a gynodioecious deciduous orchid satyrium ciliatum (Orchidaceae) female. Mitochondrial DNA Part B. Resour. 4, 3876–3877. doi: 10.1080/23802359.2019.1687359
Men, S., Ming, X., Wang, Y., Liu, R., Wei, C., Li, Y. (2003). Genetic transformation of two species of orchid by biolistic bombardment. Plant Cell Rep. 21, 592–598. doi: 10.1007/s00299-002-0559-4
Minasiewicz, J., Krawczyk, E., Znaniecka, J., Vincenot, L., Zheleznaya, E., Korybut-Orlowska, J., et al. (2022). Weak population spatial genetic structure and low infraspecific specificity for fungal partners in the rare mycoheterotrophic orchid epipogium aphyllum. J. Plant Res. 135, 275–293. doi: 10.1007/s10265-021-01364-7
Mohammadi, M., Kaviani, B., Sedaghathoor, S. (2021). In vivo polyploidy induction of phalaenopsis amabilis in a bubble bioreactor system using colchicine. Ornam. Hortic. 27, 204–212. doi: 10.1590/2447-536X.V27I2.2275
Molla, K. A., Sretenovic, S., Bansal, K. C., Qi, Y. (2021). Precise plant genome editing using base editors and prime editors. Nat. Plants 7, 1166–1187. doi: 10.1038/s41477-021-00991-1
Molla, K. A., Yang, Y. (2019). CRISPR/Cas-mediated base editing: Technical considerations and practical applications. Trends Biotechnol. 37, 1121–1142. doi: 10.1016/j.tibtech.2019.03.008
Mo, P., Zhou, J., Zhou, F., Chen, Y., Huang, K., Mo, P., et al. (2022). The complete chloroplast genome sequence of gomesa flexuosa. Mitochondrial DNA Part B. 7, 1237–1239. doi: 10.1080/23802359.2022.2093670
Niu, S. C., Huang, J., Xu, Q., Li, P. X., Yang, H. J., Zhang, Y. Q., et al. (2018). Morphological type identification of self-incompatibility in dendrobium and its phylogenetic evolution pattern. Int. J. Mol. Sci. 19, 1–18. doi: 10.3390/ijms19092595
Niu, Z., Pan, J., Zhu, S., Li, L., Xue, Q., Liu, W., et al. (2017a). Comparative analysis of the complete plastomes of apostasia wallichii and neuwiedia singapureana (Apostasioideae) reveals different evolutionary dynamics of IR/SSC boundary among photosynthetic orchi. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01713
Niu, Z., Xue, Q., Zhu, S., Sun, J., Liu, W., Ding, X. (2017b). The complete plastome sequences of four orchid species: Insights into the evolution of the orchidaceae and the utility of plastomic mutational hotspots. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00715
Niu, Z., Zhu, F., Fan, Y., Li, C., Zhang, B., Zhu, S., et al. (2021). The chromosome-level reference genome assembly for dendrobium officinale and its utility of functional genomics research and molecular breeding study. Acta Pharm. Sin. B. 11, 2080–2092. doi: 10.1016/j.apsb.2021.01.019
Nopitasari, S., Setiawati, Y., Lawrie, M. D., Purwantoro, A., Widada, J., Sasongko, A. B., et al. (2020). Development of an agrobacterium-delivered crispr/cas9 for phalaenopsis amabilis (L.) blume genome editing system. AIP. Conf. Proc. 2260, 1–10. doi: 10.1063/5.0015868
Ogura, T., Busch, W. (2015). From phenotypes to causal sequences: Using genome wide association studies to dissect the sequence basis for variation of plant development. Curr. Opin. Plant Biol. 23, 98–108. doi: 10.1016/j.pbi.2014.11.008
Pan, Y. Y., Li, T. Z., Chen, J. B., Huang, J., Rao, W. H. (2019). Complete chloroplast genome of dendrobium thyrsiflorum (Orchidaceae). Mitochondrial DNA Part B. Resour. 4, 3192–3193. doi: 10.1080/23802359.2019.1667918
Paun, O., Bateman, R. M., Fay, M. F., Hedrén, M., Civeyrel, L., Chase, M. W. (2010). Stable epigenetic effects impact adaptation in allopolyploid orchids (Dactylorhiza: Orchidaceae). Mol. Biol. Evol. 27, 2465–2473. doi: 10.1093/molbev/msq150
Peng, X., Ye, H., Liu, H., Zhao, Z., Hu, G., Zhao, P. (2020). Characterization of the complete chloroplast genome of orchid family species cymbidium bicolor (Orchidaceae). Mitochondrial DNA Part B. Resour. 5, 323–324. doi: 10.1080/23802359.2019.1703591
Piet, Q., Droc, G., Marande, W., Sarah, G., Bocs, S., Klopp, C., et al. (2022). A chromosome-level, haplotype-phased vanilla planifolia genome highlights the challenge of partial endoreplication for accurate whole-genome assembly. Plant Commun. 3, 100330. doi: 10.1016/j.xplc.2022.100330
Poczai, P., Varga, I., Laos, M., Cseh, A., Bell, N., Valkonen, J. P. T., et al. (2013). Advances in plant gene-targeted and functional markers: A review. Plant Methods 9, 1–31. doi: 10.1186/1746-4811-9-6
Radhakrishnan, G. V., Keller, J., Rich, M. K., Vernié, T., Mbadinga Mbadinga, D. L., Vigneron, N., et al. (2020). An ancestral signalling pathway is conserved in intracellular symbioses-forming plant lineages. Nat. Plants 6, 280–289. doi: 10.1038/s41477-020-0613-7
Ren, R., Gao, J., Lu, C., Wei, Y., Jin, J., Wong, S. M., et al. (2020). Highly efficient protoplast isolation and transient expression system for functional characterization of flowering related genes in cymbidium orchids. Int. J. Mol. Sci. 21, 1–18. doi: 10.3390/ijms21072264
Rewers, M., Jedrzejczyk, I., Rewicz, A., Jakubska-Busse, A. (2021). Genome size diversity in rare, endangered, and protected orchids in Poland. Genes (Basel). 12, 1–19. doi: 10.3390/genes12040563
Ryu, J., Kim, W. J., Im, J., Kang, K. W., Kim, S. H., Jo, Y. D., et al. (2019). Single nucleotide polymorphism (SNP) discovery through genotyping-by-sequencing (GBS) and genetic characterization of dendrobium mutants and cultivars. Sci. Hortic. (Amsterdam). 244, 225–233. doi: 10.1016/j.scienta.2018.09.053
Setiawati, Y., Nopitasari, S., Lawrie, M. D., Purwantoro, A., Widada, J., Sasongko, A. B., et al. (2020). Agrobacterium-mediated transformation facillitates the CRISPR/Cas9 genome editing system in dendrobium macrophyllum a. rich orchid. AIP. Conf. Proc. 2260, 1Њ9. doi: 10.1063/5.0016200
Shi, Y., Yang, L., Yang, Z., Ji, Y. (2018). The complete chloroplast genome of pleione bulbocodioides (Orchidaceae). Conserv. Genet. Resour. 10, 21–25. doi: 10.1007/s12686-017-0753-x
Song, C., Li, G., Dai, J., Deng, H. (2021). Genome-wide analysis of PEBP genes in dendrobium huoshanense: Unveiling the antagonistic functions of FT/TFL1 in flowering time. Front. Genet. 12. doi: 10.3389/fgene.2021.687689
Su, C. L., Chao, Y. T., Yen, S. H., Chen, C. Y., Chen, W. C., Chang, Y. C. A., et al. (2013). Orchidstra: An integrated orchid functional genomics database. Plant Cell Physiol. 54, 1–19. doi: 10.1093/pcp/pct004
Suetsugu, K. (2015). Autonomous self-pollination in the nectarless orchid pogonia minor. Plant Species Biol. 30, 37–41. doi: 10.1111/1442-1984.12037
Su, J., Jiang, J., Zhang, F., Liu, Y., Ding, L., Chen, S., et al. (2019). Current achievements and future prospects in the genetic breeding of chrysanthemum: a review. Hortic. Res. 6. doi: 10.1038/s41438-019-0193-8
Tang, F. L., Deng, L. L., Qin, H. Z., Shi, Y. C. (2020). Complete chloroplast genome of paphiopedilum emersonii (Orchidaceae). Mitochondrial DNA Part B. Resour. 5, 3518–3519. doi: 10.1080/23802359.2020.1827069
Tang, J. M., Tang, F. L., Shi, Y. C. (2020). The complete chloroplast genome of geodorum densiflorum (Orchidaceae). Mitochondrial DNA Part B. Resour. 5, 2056–2057. doi: 10.1080/23802359.2020.1763865
Teo, Z. W. N., Zhou, W., Shen, L. (2019). Dissecting the function of MADS-box transcription factors in orchid reproductive development. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01474
Tong, C. G., Wu, F. H., Yuan, Y. H., Chen, Y. R., Lin, C. S. (2020). High-efficiency CRISPR/Cas-based editing of phalaenopsis orchid MADS genes. Plant Biotechnol. J. 18, 889–891. doi: 10.1111/pbi.13264
Trávníček, P., Čertner, M., Ponert, J., Chumová, Z., Jersáková, J., Suda, J. (2019). Diversity in genome size and GC content shows adaptive potential in orchids and is closely linked to partial endoreplication, plant life-history traits and climatic conditions. New Phytol. 224, 1642–1656. doi: 10.1111/nph.15996
Trávníček, P., Ponert, J., Urfus, T., Jersáková, J., Vrána, J., Hřibová, E., et al. (2015). Challenges of flow-cytometric estimation of nuclear genome size in orchids, a plant group with both whole-genome and progressively partial endoreplication. Cytom. Part A. 87, 958–966. doi: 10.1002/cyto.a.22681
Tsai, W. C., Dievart, A., Hsu, C. C., Hsiao, Y. Y., Chiou, S. Y., Huang, H., et al. (2017). Post genomics era for orchid research. Bot. Stud. 58, 1–22. doi: 10.1186/s40529-017-0213-7
Vilcherrez-Atoche, J. A., Iiyama, C. M., Cardoso, J. C. (2022). Polyploidization in orchids: From cellular changes to breeding applications. Plants 11, 1–21. doi: 10.3390/plants11040469
Wang, J. Y., Liu, Z. J., Zhang, G. Q., Peng, C. C. (2019). The complete chloroplast genome sequence of phalaenopsis lowii (Orchidaceae). Mitochondrial DNA Part B. Resour. 4, 3569–3570. doi: 10.1080/23802359.2019.1674715
Wang, H. M., Tong, C. G., Jang, S. (2017). Current progress in orchid flowering/flower development research. Plant Signal. Behav. 12, 1–6. doi: 10.1080/15592324.2017.1322245
Wang, M., Wang, Z., Mao, Y., Lu, Y., Yang, R., Tao, X., et al. (2019). Optimizing base editors for improved efficiency and expanded editing scope in rice. Plant Biotechnol. J. 17, 1697–1699. doi: 10.1111/pbi.13124
Wang, T., Zhang, C., Zhang, H., Zhu, H. (2021). CRISPR/Cas9-mediated gene editing revolutionizes the improvement of horticulture food crops. J. Agric. Food Chem. 69, 13260–13269. doi: 10.1021/acs.jafc.1c00104
Wei, Z., Chen, B., Cao, Y., Zheng, Y., Zhang, Y., Zhao, K., et al. (2021). The complete chloroplast genome of cymbidium hookerianum (Orchidaceae): genome structure and basic analysis. Mitochondrial DNA Part B. Resour. 6, 36–37. doi: 10.1080/23802359.2020.1845996
Williams, S. J., Gale, S. W., Hinsley, A., Gao, J., St. John, F. A. V. (2018). Using consumer preferences to characterize the trade of wild-collected ornamental orchids in China. Conserv. Lett. 11, 1–8. doi: 10.1111/conl.12569
Wu, X.-Y., Li, T.-Z., Chen, G.-Z., Xu, Q., Pan, Y.-Y., Chen, L. J. (2019). The complete chloroplast genome of dendrobium longicornu (Orchidaceae). Mitochondrial DNA Part B. 4, 3776–3777. doi: 10.1080/23802359.2019.1666049
Wu, S. S., Shen, L. M., Ling, R., Dai, Z. W., Liu, Z. J., Lan, S. R. (2019). Next-generation sequencing yields the complete chloroplast genome of pleione chunii, a vulnerable orchid in China. Mitochondrial DNA Part B. Resour. 4, 2576–2578. doi: 10.1080/23802359.2019.1640642
Xia, K., Liu, D. K., Wang, J. Y. (2021). The complete chloroplast genome sequence of phalaenopsis wilsoniii (Orchidaceae). Mitochondrial DNA Part B. Resour. 6, 3303–3305. doi: 10.1080/23802359.2021.1994889
Xie, T.-X., Yu, X., Zheng, Q.-D., Ma, S.-H., Liu, Z.-J., Ai, Y. (2020). The complete chloroplast genome of tainia dunnii (Orchidaceae): genome structure and evolution. Mitochondrial DNA Part B. 5, 3–4. doi: 10.1080/23802359.2019.1693935
Xie, L., Zhou, S. S., Wang, M. G., Zeng, R. Z., Guo, H. R., Zhang, Z. S. (2017). Creation and micropropagation of polyploids in cymbidium hybridum. Acta Hortic. 1167, 107–114. doi: 10.17660/ActaHortic.2017.1167.16
Xu, Y., Lei, Y., Su, Z., Zhao, M., Zhang, J., Shen, G., et al. (2021). A chromosome-scale gastrodia elata genome and large-scale comparative genomic analysis indicate convergent evolution by gene loss in mycoheterotrophic and parasitic plants. Plant J. 108, 1609–1623. doi: 10.1111/tpj.15528
Xu, Q., Niu, S.-C., Li, K.-L., Zheng, P.-J., Zhang, X.-J., Jia, Y., et al. (2022). Chromosome-scale assembly of the dendrobium nobile genome provides insights into the molecular mechanism of the biosynthesis of the medicinal active ingredient of dendrobium. Front. Genet. 13. doi: 10.3389/fgene.2022.844622
Yang, F. X., Gao, J., Wei, Y. L., Ren, R., Zhang, G. Q., Lu, C. Q., et al. (2021). The genome of cymbidium sinense revealed the evolution of orchid traits. Plant Biotechnol. J. 19, 2501–2516. doi: 10.1111/pbi.13676
Yang, L., Wu, Q., Yang, M., Zhang, D., Dong, S., Cheng, J. (2021). The complete chloroplast genome sequence of the endemic and rare orchid nothodoritis zhejiangensis (Orchidaceae) in China. Mitochondrial DNA Part B. Resour. 6, 2931–2932. doi: 10.1080/23802359.2021.1972867
Yang, J., Zhang, F., Ge, Y., Yu, W., Xue, Q., Wang, M., et al. (2022). Effects of geographic isolation on the bulbophyllum chloroplast genomes. BMC Plant Biol. 22, 1–14. doi: 10.1186/s12870-022-03592-y
Yan, L., Wang, X., Liu, H., Tian, Y., Lian, J., Yang, R., et al. (2015). The genome of dendrobium officinale illuminates the biology of the important traditional Chinese orchid herb. Mol. Plant 8, 922–934. doi: 10.1016/j.molp.2014.12.011
Younis, A., Ryu, K. B., Co, V. T., Hwang, Y.-J., Jee, S. O., Kim, M.-S., et al. (2013). Analysis of chromosomes and nuclear DNA content in nine genotypes of cymbidium. Korean. Soc Floric. Sci. 21, 158–161. doi: 10.11623/frj.2013.21.4.31
Yuan, Y., Jin, X., Liu, J., Zhao, X., Zhou, J., Wang, X., et al. (2018). The gastrodia elata genome provides insights into plant adaptation to heterotrophy. Nat. Commun. 9, 1–11. doi: 10.1038/s41467-018-03423-5
Yue, Z., Kao, H., Zhang, Y., Wang, T., Dong, S., Cheng, J. (2020). The complete chloroplast genome sequence of a medicinal orchid species coelogyne fimbriata (Orchidaceae). Mitochondrial DNA Part B. Resour. 5, 3458–3460. doi: 10.1080/23802359.2020.1823264
Yu, C. W., Lian, Q., Wu, K. C., Yu, S. H., Xie, L. Y., Wu, Z. J. (2016). The complete chloroplast genome sequence of anoectochilus roxburghii. Mitochondrial DNA 27, 2477–2478. doi: 10.3109/19401736.2015.1033706
Yun, S. A., Son, H. D., Im, H. T., Kim, S. C. (2018). Two complete chloroplast genomes of an endangered orchid species, pelatantheria scolopendrifolia (Orchidaceae), in Korea. Mitochondrial DNA Part B. Resour. 3, 225–226. doi: 10.1080/23802359.2018.1437815
Zeng, W. H., Liao, S. C., Chang, C. C. (2007). Identification of RNA editing sites in chloroplast transcripts of phalaenopsis aphrodite and comparative analysis with those of other seed plants. Plant Cell Physiol. 48, 362–368. doi: 10.1093/pcp/pcl058
Zhang, D., Liu, D. K., Hao, Y., Lan, S. R., Liu, Z. J. (2019). The complete chloroplast genome sequence of liparis vivipara (Orchidaceae). Mitochondrial DNA Part B. Resour. 4, 2223–2224. doi: 10.1080/23802359.2019.1624638
Zhang, G. Q., Liu, Z. J., Lan, S. R. (2019). Complete chloroplast genome of an orchid species cymbidium floribundum lindl. Mitochondrial DNA Part B. Resour. 4, 2940–2941. doi: 10.1080/23802359.2019.1662743
Zhang, G. Q., Liu, K. W., Li, Z., Lohaus, R., Hsiao, Y. Y., Niu, S. C., et al. (2017). The apostasia genome and the evolution of orchids. Nature 549, 379–383. doi: 10.1038/nature23897
Zhang, Y. J., Ma, C., Feng, Y., Cheng, X., Song, J. (2018). The complete chloroplast genome sequence of an endangered orchid species dendrobium bellatulum (Orchidaceae). Mitochondrial DNA Part B. Resour. 3, 233–234. doi: 10.1080/23802359.2018.1437811
Zhang, G. Q., Xu, Q., Bian, C., Tsai, W. C., Yeh, C. M., Liu, K. W., et al. (2016). The dendrobium catenatum lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci. Rep. 6, 1–10. doi: 10.1038/srep19029
Zhang, F. P., Zhang, S. B. (2021). Genome size and labellum epidermal cell size are evolutionarily correlated with floral longevity in paphiopedilum species. Front. Plant Sci. 12 1–14. doi: 10.3389/fpls.2021.793516
Zhang, W., Zhang, G., Zeng, P., Zhang, Y., Hu, H., Liu, Z., et al. (2021). Genome sequence of apostasia ramifera provides insights into the adaptive evolution in orchids. BMC Genomics 22, 1–12. doi: 10.1186/s12864-021-07852-3
Zhang, Y., Zhang, G. Q., Zhang, D., Liu, X. D., Xu, X. Y., Sun, W. H., et al. (2021). Chromosome-scale assembly of the dendrobium chrysotoxum genome enhances the understanding of orchid evolution. Hortic. Res. 8. doi: 10.1038/s41438-021-00621-z
Zhao, Z., Li, M., He, J., Cheng, J., Xie, L. (2019). Complete chloroplast genome sequences of an important horticultural orchid: Paphiopedilum hirsutissimum (Orchidaceae). Mitochondrial DNA Part B. Resour. 4, 2950–2951. doi: 10.1080/23802359.2019.1662752
Zheng, F., Chen, J. B., Liu, W. R., Wang, M. (2021). The complete chloroplast genome of an endangered species apostasia ramifera (Orchidaceae). Mitochondrial DNA Part B. Resour. 6, 470–471. doi: 10.1080/23802359.2021.1872429
Zheng, S.g., Hu, Y.d., Zhao, R., Yan, S., Zhang, X.q., Zhao, T.m., et al. (2018). Genome-wide researches and applications on dendrobium. Planta 248, 769–784. doi: 10.1007/s00425-018-2960-4
Zheng, F., Pan, Y. Y., Li, T. Z., Chen, G. Z., Wu, X. Y., Li, L. Q. (2019). The complete chloroplast genome of an endangered species cymbidium mastersii (Orchidaceae). Mitochondrial DNA Part B. Resour. 4, 3068–3069. doi: 10.1080/23802359.2019.1666047
Zhong, H., Shen, L. M., Liu, H. P., Liu, Z. J., Wu, S. S., Zhai, J. W. (2019). The complete chloroplast genome of calanthe arcuata, an endemic terrestrial orchid in China. Mitochondrial DNA Part B. Resour. 4, 2629–2630. doi: 10.1080/23802359.2019.1639561
Keywords: third-generation sequencing, orchid, genome assembly, polyploidy, functional genomics, molecular breeding
Citation: Song C, Wang Y, Manzoor MA, Mao D, Wei P, Cao Y and Zhu F (2022) In-depth analysis of genomes and functional genomics of orchid using cutting-edge high-throughput sequencing. Front. Plant Sci. 13:1018029. doi: 10.3389/fpls.2022.1018029
Received: 12 August 2022; Accepted: 05 September 2022;
Published: 23 September 2022.
Edited by:
Nisha Singh, Gujarat Biotechnology University, IndiaReviewed by:
Junpeng Shi, Sun Yat-sen University, ChinaCopyright © 2022 Song, Wang, Manzoor, Mao, Wei, Cao and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fucheng Zhu, ZnVjaGVuZzMyM0AxNjMuY29t; Yunpeng Cao, eGZjeXBlbmdAMTI2LmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.