
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
EDITORIAL article
Front. Plant Sci. , 08 September 2022
Sec. Plant Pathogen Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1014516
This article is part of the Research Topic DNA Virus and Host Plant Interactions from Antagonism to Endogenization View all 7 articles
Editorial on the Research Topic
DNA virus and host plant interactions from antagonism to endogenization
The findings and reviews in the article collection of the Research Topic on “DNA Virus and Host Plant Interactions from Antagonism to Endogenization” investigate the complexity of parasitic, mutual, or commensal virus interactions with the host cell and highlights the evolved diversity of DNA viruses infecting major crop plants, ornamentals as well as weeds.
Even though by current definition viruses are not regarded as living organisms, their regulative force within the ecosystem and their impact on evolution of life mediating horizontal DNA transfer is more and more revealed and recognized (Suttle, 2007; Gilbert and Feschotte, 2018; Loiseau et al., 2021). Significant knowledge has been obtained from studies focusing on viruses of bacteria, animals and humans (Krupovic and Forterre, 2015; Pisano et al., 2020) but less is known for the environment comprising plants and particular for their infecting DNA virus(es). This is surprising since almost 3 decades ago, the literally “breakthrough” of viral sequences into the plant genome had been established for geminiviruses (Bejarano et al., 1996) as well as for some pararetroviruses, taxon Caulimoviridae, neither of them relying on chromosomal integration in their replication cycle (Hohn et al., 2008).
Plant DNA viruses belong either to the families of Geminiviridae or Nanoviridae harboring single stranded (ss)DNA genomes in their capsids or to the family Caulimoviridae that encapsidate double stranded (ds)DNA genomes. Viruses being acellular, parasitic entities occur generally as episomes in their host cell on which their replication depends (Figure 1).
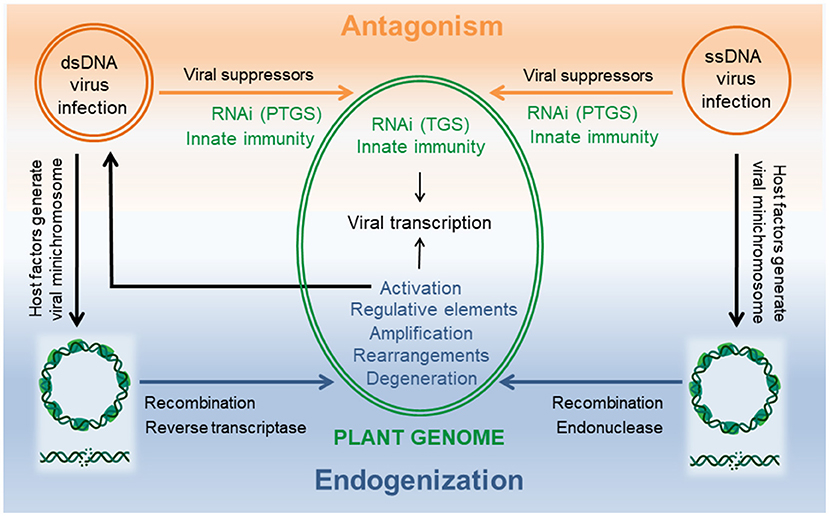
Figure 1. DNA viruses and host plant interactions from antagonism to endogenization. Successful viral infection is established when viral suppressors can overcome the host cellular defense machinery consisting of RNAi and innate immunity. Multiple levels of interactions in the nucleus as well as in the cytoplasm are possible during this arms race. Common to both viruses is a viral minichromosome that is generated using host factors and serves as template for viral transcription in the nucleus. Double strand DNA breaks result in linearized molecules that can promote endogenization by illegitimate recombination and/or interference with reverse trancriptase (dsDNA viruses) or endonuclease (ssDNA viruses). Evolution of the chimeric host-virus genome can create diversity by rearrangements, insertion of viral regulative elements, amplification and/or degeneration of virus sequences. Integrants that allow transcription of a genome length RNA are the source for virus replication and systemic infection. Minichromosomes and dsDNA breaks created with BioRender.com.
For survival, DNA viruses that compete with their host for DNA synthesis had to adapt to cellular niches that allow their multiplication without interfering with the host DNA replication in the nucleus. Thus, plant DNA viruses have evolved different strategies. They copy the host's nuclear DNA structures by forming viral minichromosomes and employ for their transcription the host plant DNA-dependent RNA polymerase II (Hull, 2014). Geminiviridae synthesize their genomic ssDNA using rolling-circle replication in the nucleus, whereas Caulimoviridae generate the genomic dsDNA using reverse transcription in the cytoplasm.
When genotoxic agents damage the viral episomes, they change their topology from circular to linearized molecules. These can be recognized by the host DNA repair machinery leading to multiple pathways of genome invasions by illegitimate recombination (Richert-Pöggeler et al.). Additionally, for geminiviruses the multifunctional replication initiator protein (Rep) encoded by ORF AC1 comprising also endonuclease activity is likely to play a key role in promoting host genome accessions of viral sequences (Hanley-Bowdoin et al., 2013; Ruhel and Chakraborty, 2019). It is noteworthy, that in Macademia viral sequences originating from both types of DNA viruses have been reported (Zakeel et al., 2021).
In order to infect their hosts successfully, viruses have evolved various approaches to overcome the RNAi based plant surveillance system in the nucleus as well as in the cytoplasm as illustrated and discussed by Richert-Pöggeler et al. and Zhai et al. In case of Croton yellow vein mosaic virus, a monopartite begomovirus, and its cognate beta-satellite four viral suppressors of RNAi (VSR), namely V2, C2, C4, and βC1 are developed to overcome plant defense mechanism and establish a sustainable infection. Furthermore, the authors reveal distinct functions of the investigated viral suppressors according to their subcellular localization, interactions and roles in symptom induction and intercellular movement.
The importance of miRNAs regulating both host as well as viral gene expression for plant immunity is illustrated by beet curly top virus interactions with its host sugar beet (Majumdar et al.). The observed cross-kingdom RNAi, e.g., plant derived miRNAs targeting the viral capsid protein supports the hypothesis of a chimeric scenario for the origin of viruses, which postulates that replicons existed at an precellular stage and proteins for virion formation derived from the host (Krupovic et al., 2019).
The assembled publications of this Research Topic give examples for the dynamics in autonomous DNA virus evolution as well as in co-evolution with their hosts during endogenization. Thus generated viral diversity and spectrum of virus-host interferences require adapted detection methodology as well as risk assessments (Silva et al.; Umber et al.). This is especially true for collections of germplasm from major food crops like yam and banana. The optimized multiplex PCR-dependent denaturing gradient gel electrophoresis facilitated the screening significantly and allowed comprehensive detection and analysis of endogenous Dioscorea bacilliform viruses (Silva et al.). Umber et al. revealed the dynamics of activation for infectious endogenous pararetroviruses (EPRVs) in banana. The authors paid special attention in their risk studies to cultivation methods comparing tissue culture with field cultivation and indicated the impact of time and altitude respectively for activation on EPRVs in banana.
Design of bioinformatics pipelines are seminal for exploration and functional analyses of integrated viral DNA sequences and related transcriptomes in the plant hosts (Serfraz et al.). Such comprehensive approach identified two previously unreported endogenous badnaviruses in the genus Solanum. In depth analysis of the genomic location of these endogenous badnaviruses found them adjacent or within the late blight resistance gene of their host Solanum melongena. Moreover, the authors located Ty-1 copia mobile elements–also known as Pseudoviridae- in this genomic niche. Future studies addressing the mechanisms that resulted in the co-localization of reverse transcribing elements such as Caulimoviridae and phylogenetically closely related Metaviridae as well as Pseudoviridae are highly desirable.
The ongoing global warming creates selection pressure on evolution of viruses, their associated vectors and hosts resulting in adaptation to the new environment (Amari et al., 2021). Thereby triggered changes in virus epidemiology, host range and pathogenicity can contribute to the emergence of novel viral diseases (Elena et al., 2014).
It will take the united efforts of virologists covering all taxonomic kingdoms to understand and preserve virosphere as well as to predict virus emergence and to prevent future virus outbreaks.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
YK was supported by the Japan Society for the Promotion of Science program Grants-in-Aid for Scientific Research (JSPS KAKENHI No. 19H00937).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Amari, K., Huang, C., and Heinlein, M. (2021). Potential impact of global warming on virus propagation in infected plants and agricultural productivity. Front. Plant Sci. 12, 649768. doi: 10.3389/fpls.2021.649768
Bejarano, E. R., Khashoggi, A., Witty, M., and Lichtenstein, C. (1996). Integration of multiple repeats of geminiviral DNA into the nuclear genome of tobacco during evolution. Proc. Natl. Acad. Sci. U. S. A. 759–764. doi: 10.1073/pnas.93.2.759
Elena, S. F., Fraile, A., and Garcia-Arenal, F. (2014). Chapter three- evolution and emergence of plant viruses. Adv. Vir. Res. 88, 161–191. doi: 10.1016/B978-0-12-800098-4.00003-9
Gilbert, C., and Feschotte, C. (2018). Horizontal acquisition of transposable elements and viral sequences: patterns and consequences. Curr. Opin. Genet. Dev. 49, 15–24. doi: 10.1016/j.gde.2018.02.007
Hanley-Bowdoin, L., Bejarano, E. R., Robertson, D., and Mansoor, S. (2013). Geminiviruses: masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 11, 777–788. doi: 10.1038/nrmicro3117
Hohn, T., Richert-Pöggeler, K. R., Staginnus, C., Harper, G., Schwarzacher, T., Teo, C. H., et al. (2008). “Evolution of integrated plant viruses,” in Plant Virus Evolution, ed M. J. Roossinck (Berlin, Heidelberg: Springer), 4. doi: 10.1007/978-3-540-75763-4_4
Krupovic, M., Dolja, V. V., and Koonin, E. V. (2019). Origin of viruses: primordial replicators recruiting capsids from hosts. Nat. Rev. Microbiol. 17, 449–458. doi: 10.1038/s41579-019-0205-6
Krupovic, M., and Forterre, P. (2015). Single-stranded DNA viruses employ a variety of mechanisms for integration into host genomes. Ann. N. Y. Acad. Sci. 1341, 41–53. doi: 10.1111/nyas.12675
Loiseau, V., Peccoud, J., Bouzar, C., Guillier, S., Fan, J., Gueli Alletti, G., et al. (2021). Monitoring insect transposable elements in large double-stranded DNA viruses reveals host-to-virus and virus-to-virus transposition. Mol. Biol. Evol. 38, 3512–3530. doi: 10.1093/molbev/msab198
Pisano, M. P., Grandi, N., and Tramontano, E. (2020). High-throughput sequencing is a crucial tool to investigate the contribution of human endogenous retroviruses (HERVs) to human biology and development. Viruses 12, 60633. doi: 10.3390/v12060633
Ruhel, R., and Chakraborty, S. (2019). Multifunctional roles of geminivirus encoded replication initiator protein. Virus Dis. 30, 66–73. doi: 10.1007/s13337-018-0458-0
Suttle, C. A. (2007). Marine viruses–major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812. doi: 10.1038/nrmicro1750
Keywords: plant DNA viruses, virus evolution, virus integration, RNAi, virus ecology
Citation: Richert-Pöggeler KR, Iskra-Caruana M-L and Kishima Y (2022) Editorial: DNA virus and host plant interactions from antagonism to endogenization. Front. Plant Sci. 13:1014516. doi: 10.3389/fpls.2022.1014516
Received: 08 August 2022; Accepted: 24 August 2022;
Published: 08 September 2022.
Edited and reviewed by: Giorgio Gambino, Institute for Sustainable Plant Protection (CNR), Italy
Copyright © 2022 Richert-Pöggeler, Iskra-Caruana and Kishima. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katja R. Richert-Pöggeler, a2F0amEucmljaGVydC1wb2VnZ2VsZXJAanVsaXVzLWt1ZWhuLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.