- Key Laboratory of Forest Genetics & Biotechnology of Ministry of Education, Co-Innovation Center for Sus-tainable Forestry in Southern China, College of Forestry, Nanjing Forestry University, Nanjing, China
Liriodendron chinense is a relic tree species of the family Magnoliaceae with multiple uses in timber production, landscape decoration, and afforestation. L. chinense often experiences drought stress in arid areas. However, the molecular basis underlying the drought response of L. chinense remains unclear. Many studies have reported that the xyloglucan endotransglucosylase/hydrolase (XTH) family plays an important role in drought stress resistance. Hereby, to explore the drought resistance mechanism of L. chinense, we identify XTH genes on a genome-wide scale in L. chinense. A total of 27 XTH genes were identified in L. chinense, and these genes were classified into three subfamilies. Drought treatment and RT-qPCR analysis revealed that six LcXTH genes significantly responded to drought stress, especially LcXTH21. Hence, we cloned the LcXTH21 gene and overexpressed it in tobacco via gene transfer to analyze its function. The roots of transgenic plants were more developed than those of wild-type plants under different polyethylene glycol (PEG) concentration, and further RT-qPCR analysis showed that LcXTH21 highly expressed in root compared to aboveground organs, indicating that LcXTH21 may play a role in drought resistance through promoting root development. The results of this study provide new insights into the roles of LcXTH genes in the drought stress response. Our findings will also aid future studies of the molecular mechanisms by which LcXTH genes contribute to the drought response.
Introduction
Plants are continuously exposed to various types of abiotic stress because they are sessile, and drought stress has a negative effect on the growth, yield, and cultivation of plants. Given that water scarcity reduces plant performance, improving the drought resistance of plants is a major goal of breeding efforts (Boyer, 1982; Bray et al., 2000; Cushman and Bohnert, 2000; Mittler, 2006). Plant improvement of drought resistance via molecular breeding approaches is a clear trend that further promotes the development of modern agriculture. With the various genome resources available, mining and utilizing genes that provide high resistance to drought stress will promote the development of plant molecular breeding (Parmar et al., 2017; Wai et al., 2020).
In recent years, identification and functional analysis of various abiotic stress-responsive genes and transcription factors and their applications in breeding stress-tolerant plants were favored (Roy et al., 2011; Sharma et al., 2017). For example, overexpression of HhGRAS14 in Arabidopsis thaliana significantly improved the drought tolerance of transgenic plants (Ni et al., 2022). And orphan gene PpARDT was found to be involved in drought tolerance potentially by enhancing ABA response in Physcomitrium patens (Dong et al., 2022). MdFLP enhanced drought tolerance by regulating the expression of MdNAC019 in self-rooted apple stocks (Wang et al., 2022). The bZIP transcription factor ABP9 in maize is involved in the regulation of drought resistance, and overexpression of OsNAC10 in rice increased grain yield under drought stress (Jeong et al., 2010; Wang et al., 2017). The overexpression of a rice OsSalT in tobacco showed increased root growth and resulted in improved drought tolerance (Kaur et al., 2022). In Manihot esculenta, MeRSZ21b was found to be involved in drought tolerance, and plants overexpressed MeRSZ21b gene had longer roots than WT (Chen et al., 2022).
Generally, there are gene families related to abiotic stress in plant genomes. The xyloglucan endotransglucosylase/hydrolase (XTH) family is a typical example (Eklöf and Brumer, 2010). XTH family belongs to the glycoside hydrolase 16 family (GH16), and XTH genes can be divided into four groups: I/II III-A, III-B, and the early diverging group (Campbell and Braam, 1999b). Increasing evidence has revealed that genes in the XTH family play an important role in drought stress. For example, plants overexpressing GmXTH23 had stronger drought tolerance and greater root lengths than wild-type (WT) plants (Long, 2020). Following overexpression of the hot pepper gene CaXTH3 in tomato, half of the stomata of transgenic plants were open under drought stress, and most of the stomata of WT plants were closed, a condition that was suggestive of transgenic plants having a higher drought tolerance than WT plants (Choi et al., 2011). XTH genes have also been shown to be involved in plant growth. For example, GUS staining of A. thaliana has shown that AtXTH genes might be expressed throughout all growth stages (Becnel et al., 2006). The expression levels of DcXTH2 and DcXTH3 increased dramatically during flowering, confirming that XTH genes play a role in petal growth (Harada et al., 2011). In addition, some plants use xyloglucan as a storage polysaccharide for embryonic development (Reid, 1985; Buckeridge et al., 2000). Due to their important roles in drought stress and growth, XTH family members have been identified in various plants, including A. thaliana (33 genes), Oryza sativa (29 genes), Hordeum vulgare (24 genes), Populus spp. (41 genes), Ananas comosus (24 genes), and Schima superba (34 genes) (Yokoyama and Nishitani, 2001; Yokoyama et al., 2004; Geisler-Lee et al., 2006; Fu et al., 2019; Li et al., 2019; Yang et al., 2022).
L. chinense is a relict species in the Magnoliaceae family that has been widely used in timber production, landscape decoration, and afforestation. Drought stress has become a major barrier restricting the cultivation of L. chinense (He and Hao, 1999). As mentioned above, XTH family has been widely reported to be involved in drought response. However, the precise role and molecular mechanisms of the XTH family under drought stress remains unclear in L. chinense. Hereby, in order to have a better insight into the roles of XTH genes in L. chinense, we identified LcXTH genes on a genome-wide scale, uncovered LcXTH genes in relation to drought resistance and characterized their function. Overall, our study provides new insights into the possible roles of XTH genes in L. chinense and will aid future studies of drought resistance mechanisms in woody plants.
Materials and methods
Plant materials and growth conditions
L. chinense material used in this study was obtained from Xiashu Forest Station at Nanjing Forestry University, Jurong, Jiangsu, China. In April 2021, leaves, roots and leaf buds were collected from a 30-year-old L. chinense tree originating form Songyang, Zhejiang Province, and immediately frozen in liquid nitrogen, and then stored at –80°C until further use. L. chinense seeds were soaked in water for 2 days, transplanted to soil, and cultivated in 40 cm × 30 cm × 4 cm trays for 2 months with a 16-h/8-h light/dark photoperiod. All L. chinense seedlings were watered twice a week. To investigate the response of L. chinense to drought stress, seedlings were watered with 10% PEG (100 g/L) solution. Samples of L. chinense were taken at 0, 3, 6, 12, 24, and 48 h and immediately frozen in liquid nitrogen for RT-qPCR analysis. All seedlings were grown under the same conditions, with the exception of plants in the drought stress treatment.
Tobacco (Nicotiana benthamiana) was used for transgenic assays. Before sowing, seeds were sterilized with 75% (v/v) alcohol for 30 s, NaClO (v/v) for 15 min, and double-distilled water four times. Seeds were then sown on 1/2 MS medium (Murashige and Skoog, 2006) and vernalized at 4°C in the dark for 2 days. These seeds were placed in an incubator (SANYO, Japan) under a photoperiod 16-h/8-h light/dark photoperiod at 23°C for 10 days; they were then transplanted into MS medium and cultivated for 20 days. WT tobacco was used as a control, and all seedlings were grown in the same environment. The medium of the 30-day-old transgenic and WT tobacco seedlings was removed, and seedlings were planted in trays. Transgenic and WT tobacco plants were treated with 10% PEG (100 g/L) solution for 5 days, and root length was measured.
Identification and characterization of XTH family members in L. chinense
The two XTH domains (PF00722 and PF06955) were used as queries to search the L. chinense genome with HMMER (v. 3.0). The default settings and cutoff values were set to 0.001 (Potter et al., 2018; El-Gebali et al., 2019). Potential sequences were filtered using the Conserved Domain Search Service website (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) (Marchler-Bauer and Bryant, 2004), and the candidate genes were identified using the SMART database (https://smart.embl.de/) (Letunic et al., 2021). Redundant genes were removed manually. The molecular weight (MW), isoelectric point (pI), and protein length were analyzed using the ExPASy website (https://web.expasy.org/protparam/) (Wilkins et al., 1999). Single peptides and the subcellular localization of LcXTH genes were predicted by SignalP (https://dtu.biolib.com/SignalP-6) and Plant-mPLoc (v. 2.0) (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/), respectively (Chou and Shen, 2010; Teufel et al., 2022). The Blast program was used to identify homologous genes (Chen et al., 2020).
Chromosomal localization, synteny analysis, and tandem repeat analysis
Information on the chromosomal location of LcXTH genes was obtained from the genome GFF file. Ka and Ks values, protein similarity matrices, and collinear gene pairs were analyzed using TBtools (Chen et al., 2020).
Phylogenetic analysis of LcXTH genes
A. thaliana proteins were downloaded from the TAIR website (https://www.A.thaliana.org/). H. vulgare proteins, O. sativa proteins were download from NCBI website (https://www.ncbi.nlm.nih.gov/). The sequences were aligned with ClustalW software (v. 2.1). A phylogenetic tree was constructed by MEGA 7.0 software using the neighbor-joining method with the following parameters: Poisson model, pairwise deletion, and 1000 bootstrap replicates (Kumar et al., 2016). The evolview website (https://www.evolgenius.info/evolview-v2/) was used to modify the phylogenetic tree (Subramanian et al., 2019).
Expression pattern analysis and GO annotation
The expression levels of LcXTH genes were evaluated using fragments per kilobase of transcript per million mapped reads (FPKM) values based on transcriptome data (https://www.ncbi.nlm.nih.gov/, PRJNA559687) from different tissues of L. chinense, and heat maps were constructed using TBtools (Chen et al., 2020). Gene Ontology (GO) analysis was performed using the clusterProfiler 4.0 (Wu et al., 2021).
Analysis of motifs and gene structure
The online software MEME (https://meme-suite.org/meme/doc/meme.html) was used to analyze the conserved motifs of XTH proteins with a maximum of 10 motifs (Bailey et al., 2015). The Gene Structure Display Server (http://gsds.gao-lab.org/) was used to analyze gene structure (Hu et al., 2014). The promoter sequence (2000 bp upstream of the start codon) of each LcXTH gene was extracted and then analyzed using PlantCARE online software (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/); the results were visualized using TBtools. The protein sequences were submitted to ESPript Web server (http://espript.ibcp.fr/ESPript/ESPript/) for secondary structure prediction.
Extraction of RNA and RT-qPCR analysis
A SteadyPure Plant RNA Extraction Kit (AG21019, Accurate Biotechnology, Hunan, Co., Ltd.) was used for RNA extraction following the user manual. The quality of RNA was assessed using a NanoDrop 2000 spectrophotometer. A260/A280 values ranged from 1.8 to 2.0, and values of A260/A230 ranged from 1.9 to 2.1; a total of 500 ng of RNA was used to synthesize complementary DNA (cDNA).
RT-qPCR was used to analyze the expression profiles of LcXTH genes, and Actin97 was used as the reference housekeeping gene (Tu et al., 2019). The thermal cycling conditions for RT-qPCR were based on instructions provided in the SYBR Green Premix Pro Taq HS qPCR Kit (AG11701, Accurate Biotechnology, Hunan, Co., Ltd.). We used the 2−ΔΔCT method to calculate relative levels of expression (Livak and Schmittgen, 2001). Three biological replicates and technical replicates were conducted to ensure the accuracy of the results. And primers used in this study are listed in Table S1.
Full-length cDNA cloning of LcXTH21 and plant transformation
The coding sequence of LcXTH21 was obtained from the genome of L. chinense, and primers were designed using Oligo software (v. 7). The full-length cDNA of LcXTH21 was amplified, and an 876-bp open reading frame sequence was obtained (File S1). The cDNA of LcXTH21 was then cloned into the modified pBI-121 vector, which was digested with XbaI and BamHI QuickCut enzymes (Takara Biomedical Technology, Dalian, China). The transgene construct was introduced into Agrobacterium tumefaciens strain GV105, which was then transformed into tobacco using a leaf-disc infection method. We cut off the edge of wild-type tobacco leaves that had been cultured for about 30 days, put them in solid MS medium at 25°C for 2 days in the dark. Then we activated the transformed A. tumefaciens with liquid MS medium for 30 min to prepare an infection solution, then immersed the leaves in the infection solution for 10 min. After soaking, the leaves were cultivated continuously in the dark at 25°C for two days, then placed them at 25°C for 16 h in the light and 8 h in the dark. When callus grown on the edge of the leaves, we isolated callus and cultured on MS medium containing kanamycin. After 20 days of culture, PCR was used to determine whether the plants were positive, and positive plants were cultured until they reached maturity. The details can be seen in Figure S1.
Results
Identification and characteristics of LcXTH genes
HMM searches were used to identify XTH genes. We originally obtained 29 putative XTH genes. These 29 candidate genes were then submitted to the SMART database, and incomplete sequences were removed manually. Finally, 27 XTH genes were obtained, which were named LcXTH1–LcXTH27.
The characteristics and subcellular localization of LcXTH proteins were also predicted. The length of LcXTH proteins ranged from 243 to 337 amino acids, LcXTH26 and LcXTH27 were the largest proteins with 337 amino acids, and LcXTH17 was the smallest (243 aa). The theoretical pI values for LcXTH proteins ranged from 4.85 to 9.65, and the MW of these proteins ranged from 27.61 to 38.47 kDa. All LcXTH members were predicted to be localized to the cell wall, and 15 LcXTH members were predicted to be localized to the cytoplasm. Details are provided in Table S2.
Chromosome location and homology analysis
The chromosomal location of genes is determined by prior evolutionary events. We thus investigated the chromosomal locations of LcXTH genes. LcXTH genes were randomly distributed on nine chromosomes (Figure S2). However, because the genome assembly was incomplete, the specific chromosomal locations could not be determined for two genes: LcXTH26 and LcXTH27. Most LcXTH genes were clustered on chromosomes 13, 14, and 17; chromosome 17 had 13 genes; and chromosomes 1, 3, 5, 9, and 16 had only one gene.
Synteny within the LcXTH family was analyzed to clarify the evolutionary relationships among LcXTH genes, and three homologous pairs (LcXTH08-LcXTH12, LcXTH09-LcXTH14, and LcXTH02-LcXTH05) were identified (Figure S2). The identity of LcXTH08-LcXTH12, LcXTH09-LcXTH14, and LcXTH02-LcXTH05 was 81.30%, 67.25%, and 82.8%, respectively. The substitution rates (Ka/Ks) of these three gene pairs were calculated to assess whether LcXTH genes have been subjected to selection (Table S3). The substitution rates ranged from 0.099 to 0.114, which indicated that they have experienced purifying selection. These homologous pairs (LcXTH08-LcXTH12, LcXTH09-LcXTH14, and LcXTH02-LcXTH05) diverged approximately 66.42, 188.19, and 67.38 million years ago, respectively. We also evaluated the density of these genes across the entire genome. The density of these genes was high in regions with related homologous genes.
Tandem duplication is an important mechanism underlying the expansion of gene families, and tandemly duplicated genes often occur in clusters (Kozak et al., 2009). Hence, we calculated the protein similarity matrix to investigate the identity of LcXTH genes in three gene clusters (Figure S3 and Table S4). The identity of the LcXTH genes on chromosome 13 was 60.54%, and the identity of the three LcXTH genes on chromosome 14 ranged between 58.46% and 73.29%. Chromosome 17 contained 13 LcXTH genes, and the identity ranged from 47.16% to 96.59%. We speculate that the high identity of these closely arranged genes indicates that they are products of tandem duplication; generally, tandem duplication might be the major force driving the expansion of LcXTH genes.
Phylogenetic analysis of XTH proteins
To further investigate the evolutionary relationships among LcXTH family members, we constructed a phylogenetic tree using 113 XTH proteins. (Figure 1). Phylogenetic analysis revealed that all XTH proteins were classified into four groups (group I/II, group III-A, group III-B, and the early diverging group). In L. chinense, most LcXTH genes (24) were categorized into group I/II. Only one gene (LcXTH03) was classified in group III-A. The remaining genes (LcXTH26 and LcXTH27) were grouped into III-B. No genes were classified into the early diverging group in L. chinense, which might stem from the incomplete genome annotation or gene loss.
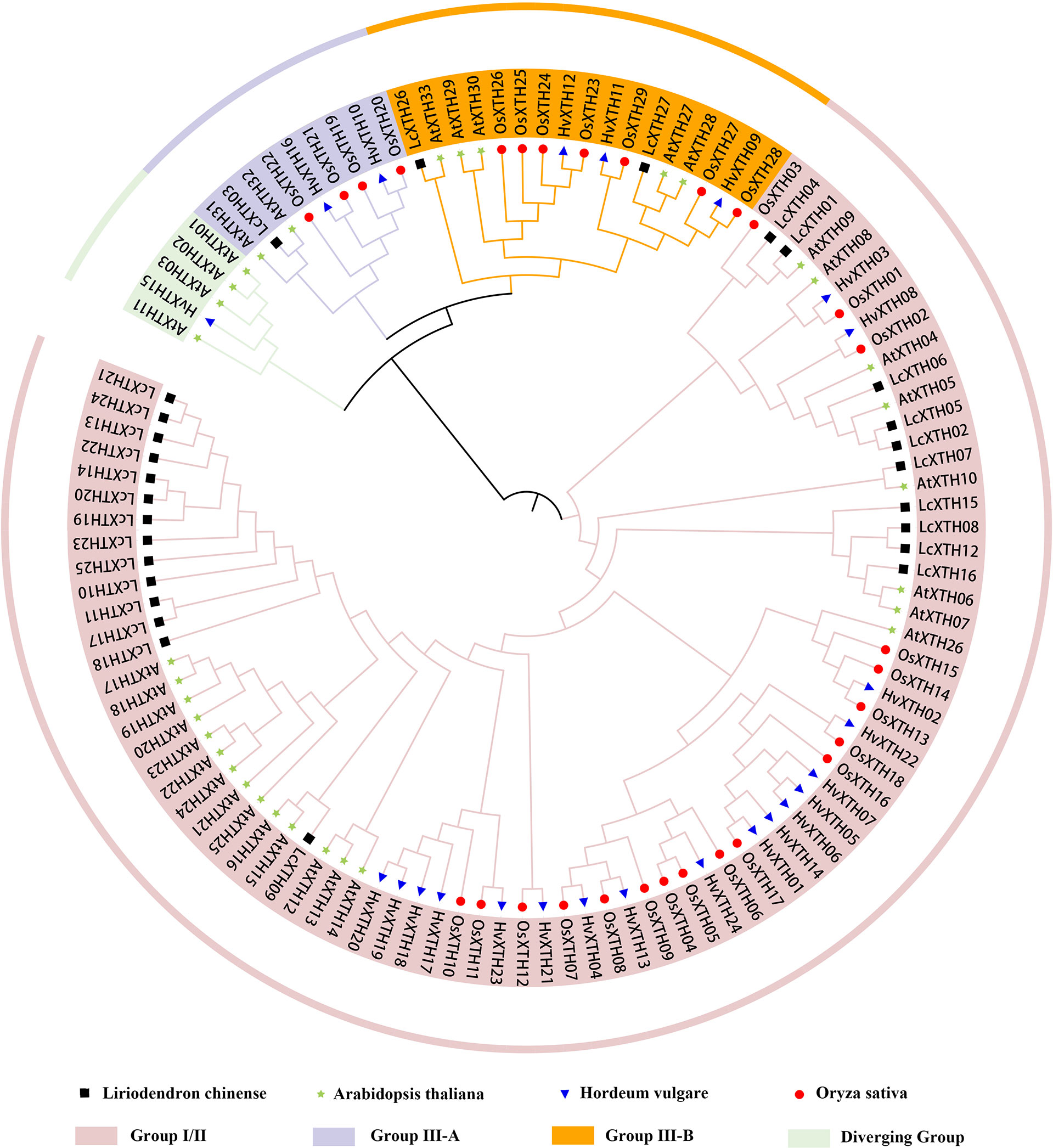
Figure 1 Classification of XTH proteins. The phylogenetic tree was constructed using 27 Liriodendron chinense XTH proteins, 24 Hordeum vulgare XTH proteins, 29 Oryza sativa XTH proteins and 33 Arabidopsis thaliana XTH proteins. Branches ending with Black square, blue triangle, red circle, green star indicate L. chinense, H. vulgare, O. sativa and A. thaliana, respectively. Proteins with a green, purple, orange and pink background indicate the early diverging group, group III-A, group III-B, and group I/II, respectively.
Structure and motif patterns of LcXTH genes
To investigate the structure of LcXTH proteins, we submitted the protein sequences to the SMART database to identify conserved domains. We analyis the pylogenetic analysis of LcXTH proteins and two domains (Glyco_hydro_16 and XET_C) were identified in all LcXTH proteins (Figure 2A and Figure 2B). XET_C is a unique domain among GH16 family members, and the proteins in this family had a common structure known as the β-jellyroll fold (Atkinson et al., 2009; Eklöf and Brumer, 2010; Behar et al., 2018). We also performed conserved motif analysis on LcXTH proteins (Figure 2C) and found that the motif composition of LcXTH family members was similar. As shown in the schematic, the Glyco_hydro_16 domain included motifs 10, 6, 8, 3, 4, 2, and 1, and the XET_C domain included motifs 9, 5, and 7. Motifs 1, 3, 4 (ExDxE), and 5 were conserved in all LcXTH proteins.
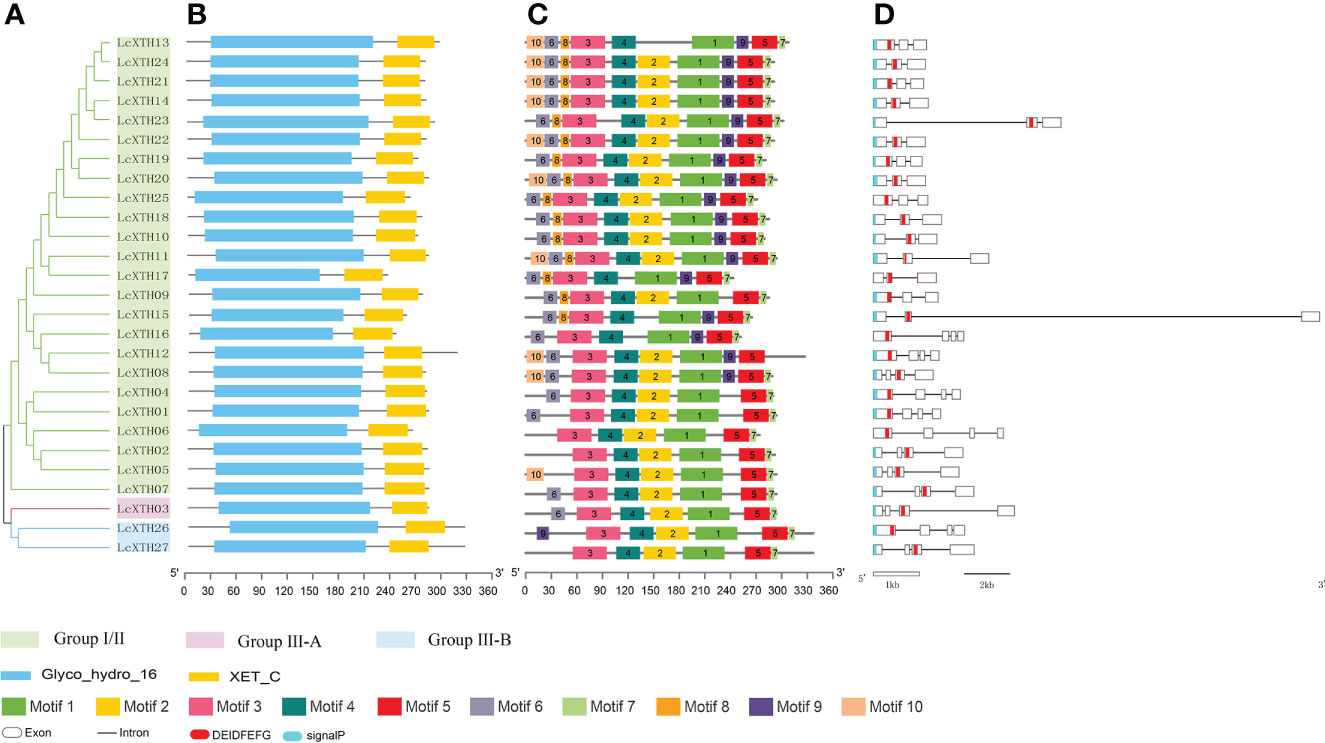
Figure 2 Gene structure and conserved domains of LcXTH proteins. (A) The cluster of LcXTH proteins. The green, pink blue background indicate group I/II group III-A and group III-B, respectively. (B) Conserved domains were predicted using the SMART database. The blue blocks indicate Glyco_hydro_16 domains, and the yellow blocks indicate XET_C domains (C) Conserved motifs in 27 LcXTH proteins were predicted using the MEME tool, and different color blocks correspond to different motifs. (D) The exon–intron distribution was visualized using the GSDS 2.0 server, and white boxes, black lines, red blocks, and blue blocks indicate exons, introns, the conserved domain ExDxE, and signal peptides, respectively.
Previous studies have shown that the exon distribution of AtXTH genes is conserved within each subfamily in A. thaliana (Yokoyama and Nishitani, 2001; Yokoyama and Nishitani, 2008). We used the online tool GSDS 2.0 to analyze the exon–intron organization of the 27 LcXTH genes (Figure 2D). There were three or four exons in LcXTH genes, and the ExDxE domain was randomly distributed in these genes, with exception of the fourth exon. The signal peptides of LcXTH proteins were predicted. A total of 23 LcXTH proteins had signal peptides, and they were all located on the first exon. These short amino acid sequences might be responsible for transmembrane transport and have secretory functions.
Structure-based sequence alignment
To further characterize LcXTH proteins, two fully resolved structures of PttXET16-34 (PDB ID: 1UN1) and TmNXG1 (PDB ID: 2UWA) were used to characterize the secondary structures of XTH proteins with ESPript software. The schematic of the secondary structures shows that all LcXTH proteins contained the conserved ExDxE domain (Figure 3). The first glutamic acid residue (E) acts as a catalyzed nucleophile, which typically initiates enzymatic reactions, and the second E residue acts as a base that activates the entering substrate (Fu et al., 2019). The N-glycosylation domain (NXT/S/Y) is thought to be critical for protein stability and is indicated in the figure. This N-glycosylation site was conserved in all group I/II proteins, but it was missing in nearly all group III-A XTH proteins (Eklöf and Brumer, 2010). However, the N-glycosylation site was observed in all LcXTH members, including LcXTH03 (a member of group III-A). The N-glycosylation sites of LcXTH proteins in group III were shifted by approximately 20 amino acids from the ExDxE domain to the C-terminus.
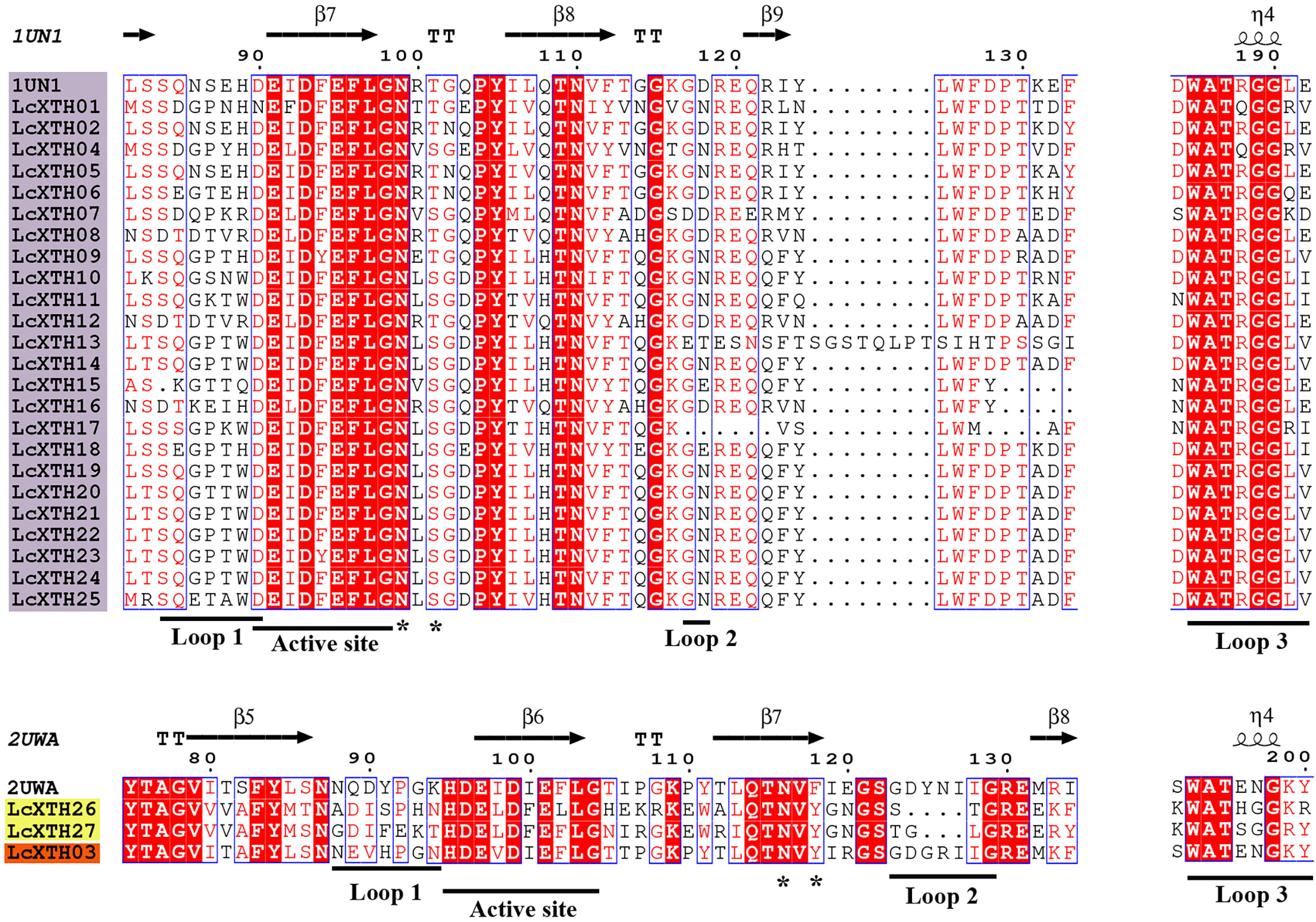
Figure 3 Structure-based sequence alignment of LcXTH proteins. Proteins with a purple background, proteins with a yellow background, and proteins with an orange background indicate Group I/II Group III-B, and Group III-A members, respectively. Blue frames, white letters in red boxes, and red letters in white boxes indicate conserved residues, strict identity, and similarity, respectively. The secondary structures of β sheets (arrows), α-helices (spiral), N-glycosylation site (*), and loops 1, 2, and 3 (lines) are indicated.
The architecture of proteins was conserved within specific groups; for example, other conserved domains adjacent to the ExDxE domain were identified in LcXTH proteins, which were referred to as loop 1, loop 2, and loop 3. Previous studies have demonstrated that the extension of loop 2 plays a key role in determining the activity of XTH proteins (Baumann et al., 2007). In this study, Loop 2 was significantly shorter in groups I/II and III-B than in group III-A, suggesting that the difference in the length of loop 2 among subfamilies of L. chinense might partly account for the differences in the classification of these proteins and their functions. The sequence DWATRGG of loop 3 was present in most group I/II proteins; however, this sequence was replaced by SWATEN in group III-A members.
Tissue expression patterns and GO analysis of LcXTH genes
We analyzed the expression patterns of LcXTH genes across several tissues (including bracts, sepals, petals, stamens, pistils, leaves, and shoots) (Figure 4A). LcXTH genes showed tissue-specific expression patterns. LcXTH04, LcXTH12, LcXTH08, LcXTH25, and LcXTH26 were highly expressed in bracts, and the expression levels of these genes in other tissues were low. LcXTH07, LcXTH16, and LcXTH27 were significantly expressed in leaves. The expression levels of LcXTH03, LcXTH18, and LcXTH10 were high in pistils, suggesting that they are involved in pistil development. The expression levels of almost all LcXTH genes were low in sepals, petals, and stamens, suggesting that LcXTH genes were not expressed during flowering. The expression patterns of the three tandem arrays on chromosomes 13, 14, and 17 were not consistent. LcXTH13, LcXTH19, LcXTH20, LcXTH21, LcXTH22, and LcXTH24 were significantly expressed in shoots and had similar expression patterns, indicating that their functions might be redundant. However, the expression patterns of the other genes on chromosome 17 differed, suggesting that they might have acquired new functions. In addition, LcXTH01, LcXTH02, LcXTH11, and LcXTH17 were not expressed in any of the tissues examined in this study; this indicates that they are not expressed or have specific expression patterns that could not be detected in this study.
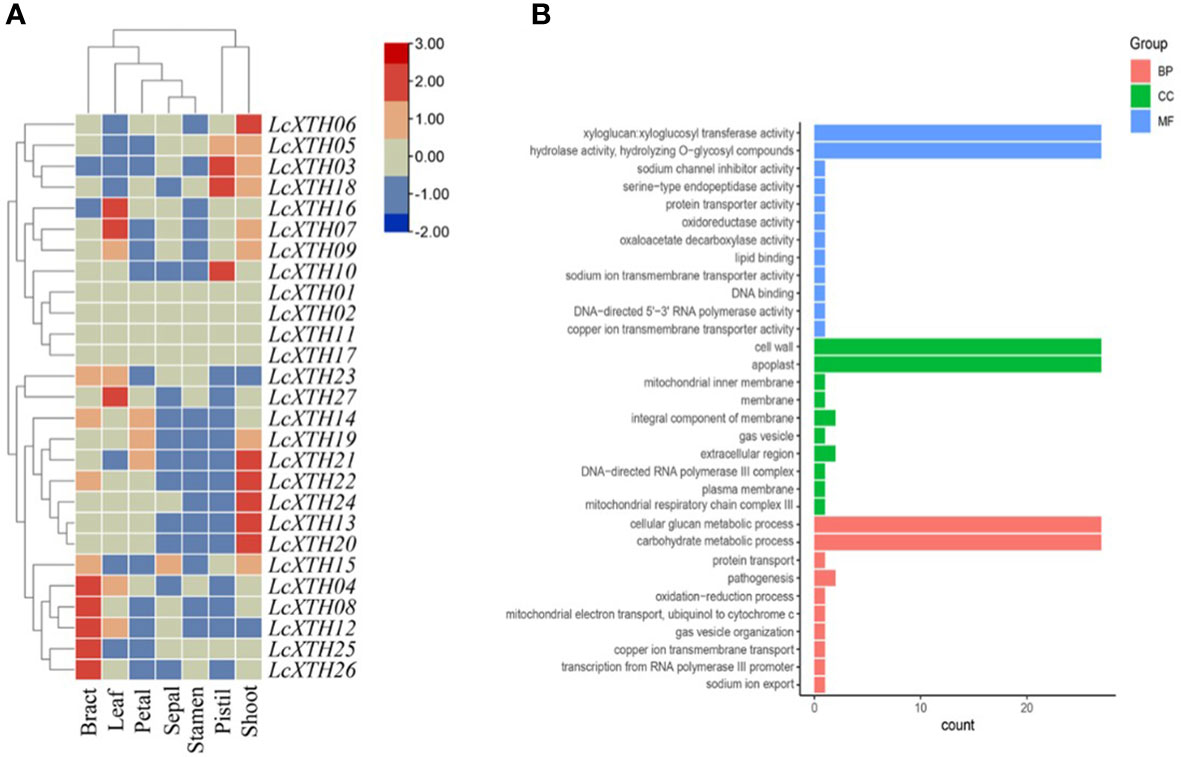
Figure 4 Expression patterns and GO annotations of LcXTH genes. (A) Heat map of the expression levels of LcXTH genes across different tissues in L. chinense based on FPKM values from public transcriptomic data. (B) GO classification of LcXTHs. Details of the GO annotations are provided in Table S5.
GO analysis was performed to clarify the functions of LcXTH genes. All LcXTH genes encoded proteins with xyloglucan xyloglucosyl transferase and hydrolase activity, and they were all localized to the cell wall and the apoplast, which was consistent with the subcellular localization prediction (Figure 4B). All LcXTH genes were predicted to be involved in cellular glucan metabolic process and carbohydrate metabolic process.
Cis-element prediction and RT-qPCR analysis
To determine the expression patterns and regulatory characteristics of LcXTH genes, the 2000-bp upstream sequence of the translation initiation site (ATG) was extracted, and cis-elements were predicted (Figure 5A). A large number of cis-elements were involved in plant growth and development, stress responses, and phytohormone responses (Figures 5B, C). In the first category, the main cis-elements were motif CAT-box (33.64%), motif CCAAT-box (27.1%), motif O2-site (23.36%), and GCN4-motif (11.21%). Cis-elements involved in stress responses mainly included the ARE motif (40.78%), MBS motif (30.1%), LTR motif (14.56%), TC-rich repeats (7.77%), and GC-motif (5.83%). In the third category, the main cis-elements were abscisic acid-responsive element (ABRE, 58.48%), methyl jasmonate-responsiveness element (CGTCA, 14.8%), and auxin-responsive element (TGA-element, 7.97%). The most abundant phytohormone response element is associated with abscisic acid (ABA)-responsiveness, indicating that LcXTH genes might be regulated by ABA.
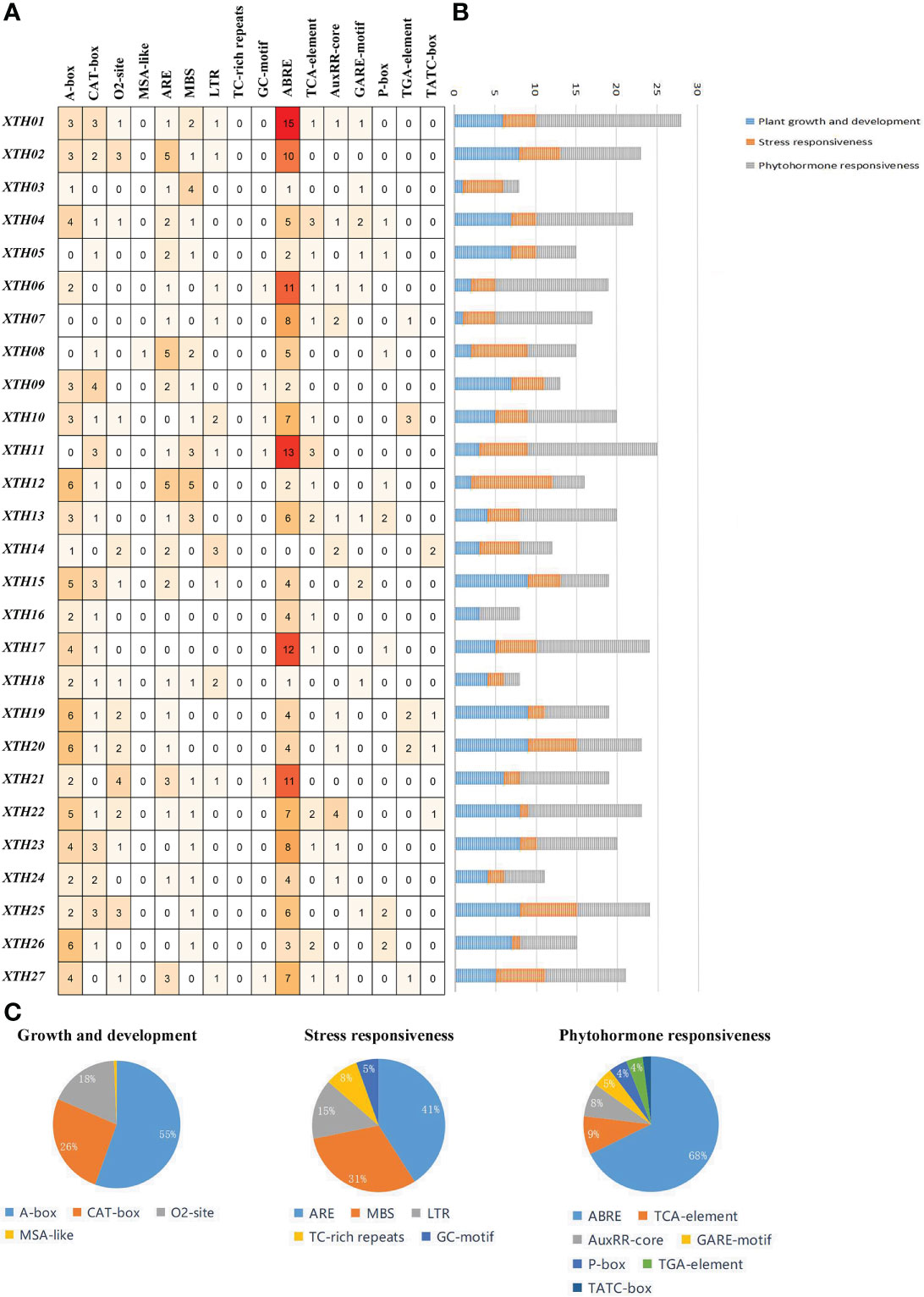
Figure 5 Analysis of cis-elements in LcXTH promoters. (A) Number of distinct cis-elements in each LcXTH. (B) Sum of cis-elements for each class of LcXTH proteins. (C) Proportion of each item in different categories.
To clarify the roles of LcXTH family members in drought resistance, we used BLASTp to identify stress-related LcXTH genes based on previous studies, and genes containing plant defense and stress elements (TC-rich motifs) were screened out (Xu et al., 2020). A total of eight LcXTH genes (LcXTH07, 15, 18, 19, 20, 21, 25, and 27) were identified, and their expression patterns under drought stress were clarified. The RT-qPCR results revealed that there were significant differences in the expression of these genes at different times under drought stress (Figure 6). LcXTH07 expression was up-regulated at 0 and 6 h, down-regulated at 6 and 24 h, and highest at 48 h. Some genes exhibited similar expression patterns. For example, the expression patterns of LcXTH18, 19, 20, and 25 did not change significantly during 0 and 6 h, but significantly increased at 12 h. LcXTH21 exhibited the most rapid and strongest response to drought stress, and it was the most highly expressed gene. Some genes exhibited opposite expression patterns; the expression levels of LcXTH15 and LcXTH27 changed slightly, indicating that they might not play important roles in the response to drought stress.
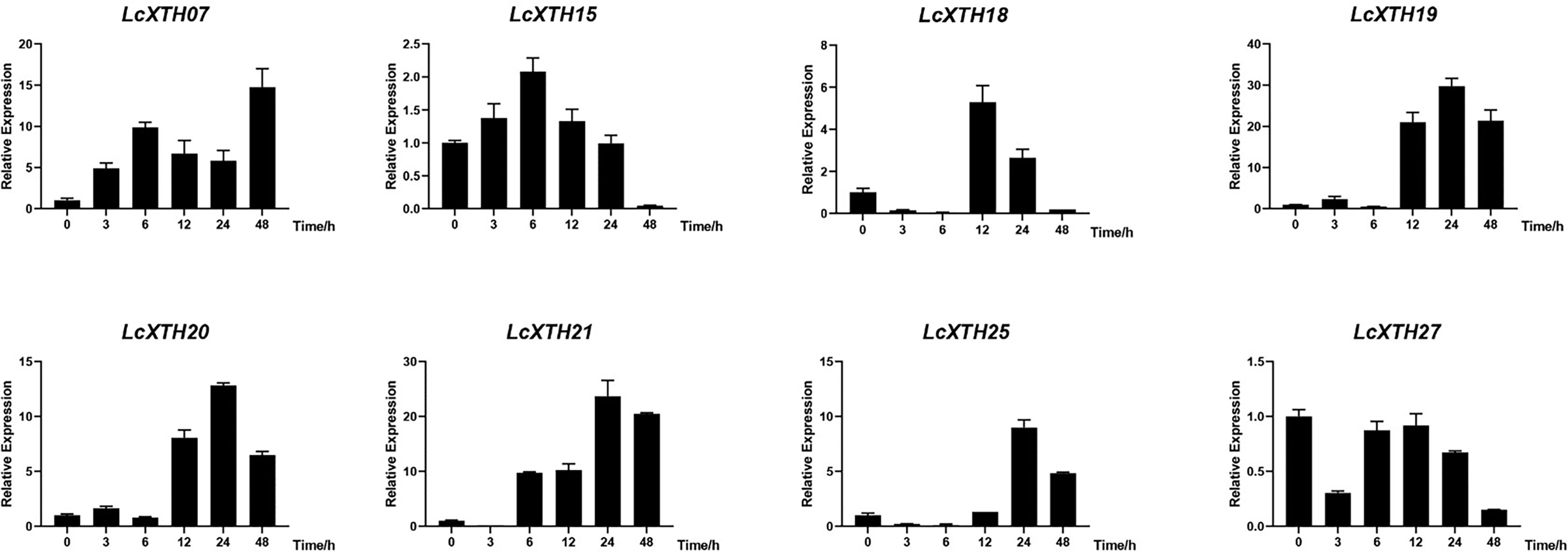
Figure 6 Expression profiles of LcXTH genes in different drought stress periods. Three independent experiments were performed using Actin97 as an internal reference gene. Error bars on the graph indicate the mean standard deviation for each triplicate treatment.
Overexpression of LcXTH21 in tobacco promoted root development under drought stress
In light of the strong response of LcXTH21 under drought stress, we cloned LcXTH21 and overexpressed it in tobacco to analyze its function; the characteristics of transgenic and WT tobacco plants were noted. Then, we cultivated transgenic plants and WT on 1/2 MS medium containing different PEG concentration. With the increase in PEG concentration, the root length decreased to varying degrees. Ten-day-old transgenic plants on 1/2 MS medium had an average root length of 1.26 cm, which was 68% longer than that of WT plants (Figures 7A, B). Under 5% PEG, the average root length in transgenic plants decreased by 52%, however, the WT root length decreased by 79% (Figures 7C, D). When the PEG concentration was increased to 10%, the average root length of transgenic tobacco and WT decreased by 95% and 154%, respectively (Figures 7E, F). Next, we treated thirty-day-old seedlings with 10% PEG for 5 days, and the root length were compared. The average root length was 2.11-fold that of WT plants (Figures 7G, H). To investigate the expression level between root and aboveground, we took samples of root and aboveground for RT-qPCR analysis. The results shown that the expression level of root was 42.06-fold than that of aboveground tissues/organs (Figure 7I). The plant hormone ABA plays a key role in regulating the resistance of plants to drought stress, whereas the NCED enzyme is a key rate-limiting ABA biosynthetic enzyme (Leung and Giraudat, 1998; Zhang et al., 2009; Zhu et al., 2017). Hence, to further study the relationship of the LcXTH21 genes and ABA signaling, we performed RT-qPCR analysis to detect the expression of NCED gene in thirty-day-old WT and transgenic tobacco (Figure 7J). The results showed that the relative expression level of NCED in transgenic plants was about 45-fold higher than that in WT controls, thereby suggesting that LcXTH21 might contribute to drought resistance by promoting ABA biosynthesis.
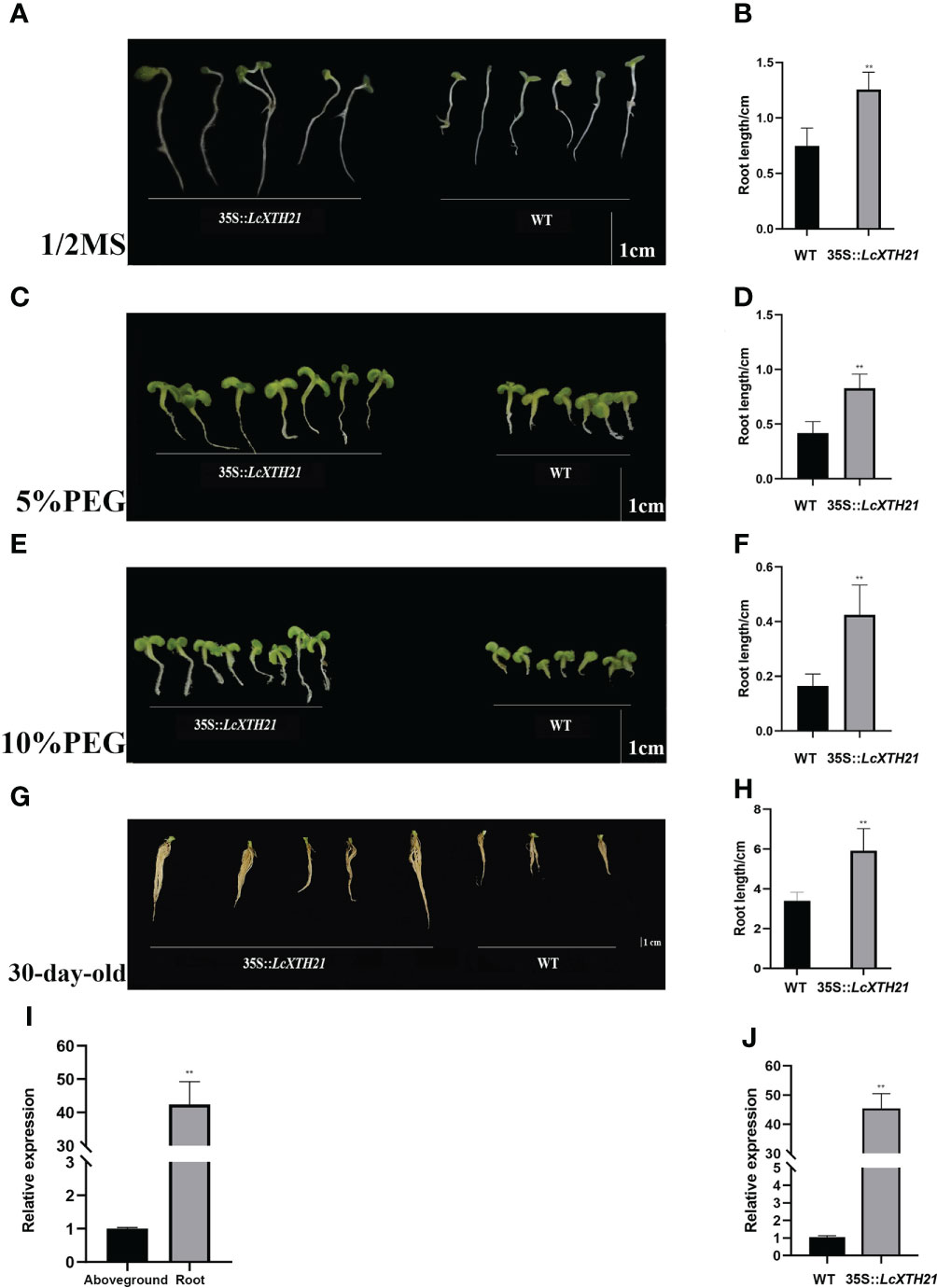
Figure 7 Phenotypic changes caused by LcXTH21 overexpression in tobacco, and histograms of root length in 10-day-old and 30-day-old transgenic tobacco and WT seedlings under different PEG concentration. (A) Comparison of the root length between 10-day-old transgenic tobacco and WT seedlings on 1/2 MS medium. (B) Statistical histograms of root length in 10-day-old transgenic tobacco and WT seedlings on 1/2 MS medium. (C) Comparison of the root length between 10-day-old transgenic tobacco and WT seedlings under 5% PEG. (D) Statistical histogram of root length in 10-day-old transgenic tobacco and WT seedlings under 5% PEG. (E) Comparison of the root length between 10-day-old transgenic tobacco and WT seedlings under 10% PEG. (F) Statistical histogram of seedling height in 10-day-old transgenic tobacco and WT seedlings under 10% PEG. (G) Comparison of the root length between 30-day-old transgenic tobacco and WT seedlings under 10% PEG. (H) Statistical histogram of seedling height in 30-day-old transgenic tobacco and WT seedlings under 10% PEG. (I) Relative expression of LcXTH21 between root and aboveground. Bars = 1 cm. (J) Relative expression of NCED gene between WT and transgenic tobacco. Statistical analyses were performed using t-tests (* P < 0.05, ** P < 0.01).
Discussion
The evolution and structure of LcXTH family members
The evolution of the XTH family has received wide research interest. XTH genes were first detected in Zygnematophyceae and non-charophycean taxa; bacterial licheninases have long been considered non-plant ancestors of the XTH family because of their sequence similarities (Barbeyron et al., 1998; Michel et al., 2001; Del Bem and Vincentz, 2010; Behar et al., 2018). The expansion of XTH genes has also received much research interest. The relaxation of substrate specificity, including the broader specificity of group I/IImembers, might have contributed to the expansion of group I/II members (Shinohara and Nishitani, 2021). Tandem duplication is one of the important mechanisms underlying the expansion of XTH family members, and tandem duplications comprise 4.56% of the genome of L. chinense (Chen et al., 2019). We detected a large number of tandemly duplicated LcXTH genes, accounting for 66.7% of all LcXTH genes, indicating that tandem duplication is the main mechanism underlying the expansion of LcXTH family genes. All the tandem repeats identified in this study were group I/II members, indicating that tandem duplication has been a particularly important mechanism underlying the expansion of group I/II members. Previous studies have shown that some gene family members have gained new functions or undergone defunctionalization (Lynch and Conery, 2000). In this study, tandemly duplicated genes on chromosome 17 were of particular interest. The expression patterns of LcXTH13, 19, 20, 21, 22, and 24 were similar, and their functions might have been retained after genome duplication. The expression patterns of LcXTH16, 18, and 25 were inconsistent. The differential expression of these genes indicates that they might have gained new functions. LcXTH01, 02, 11, and 17 were not expressed in any tissues in this study, suggesting that they might have undergone defunctionalization.
Understanding the classification of subfamilies is important for studying the evolution of the XTH gene family. Previous studies of monocotyledonous rice and dicotyledonous A. thaliana have shown that group I and group II members are difficult to distinguish; they have thus been usually classified into one group (group I/II) (Yokoyama et al., 2004). However, both sequence analysis and catalytic measurements have confirmed the divergence of group III (Fanutti et al., 1993; Yokoyama et al., 2004). In this study, a total of 27 LcXTH genes were identified, and they were grouped into three subfamilies. None of these genes were classified in the early diverging group, which might stem from an incomplete genome assembly or gene loss. Domain loop 2 in XTH family members plays an important role in subfamily classification, and previous studies have shown that the length of loop 2 contributes to the difference in the activity between XET and XEH members (Baumann et al., 2007). For example, the extension of loop 2 is thought to be the major structural change responsible for its endohydrolase activity in Fragaria vesca (Opazo et al., 2017). In this study, the extension of loop 2 was only observed in group III-A, and this might be an important factor contributing to the divergence of group III members. Glycosylation as one of key post-translational modifications can affect stability and the molecular weight of the target proteins (Ahmadizadeh et al., 2020; Heidari et al., 2020). In addition, proteins associated with the secretory pathway are firstly glycosylated in the endoplasmic reticulum. The above suggests that XET proteins may be involved in the secretory pathway. In this study, the N-glycosylation domain was observed in all LcXTH members, and previous research has demonstrated that the removal of this region significantly reduces the stability of some XET proteins (Campbell and Braam, 1999a; Kallas et al., 2005).
The roles of LcXTH family members in abiotic stress and root development
Stomatal closure is one of the first events that takes place in plants in response to drought stress to prevent water evaporation, and XTH genes play a crucial role in modifying the cell wall and altering its elongation under drought stress, which improves drought resistance (Kim et al., 2010). For example, overexpression of HvXTH1 in barley resulted in the enlargement of the stomata of transgenic plants relative to those of WT plants under drought treatment, indicating that transgenic plants were more drought tolerant (Fu, 2019). Overexpression of XTH genes from rose (Rosa rugosa) increased the drought tolerance of transgenic China rose plants (Chen et al., 2016). In addition, the presence of xyloglucan in early land plants suggests that XTH gene family members have played a key role in the transition from wetter to drier habitats (Popper, 2008). In our research, the drought treatment and RT-qPCR analysis showed that six LcXTH genes significantly responded to drought stress. The expression patterns of LcXTH18, 19, 20, and 25 were similar, suggesting that they exhibit similar responses to drought stress. ABA is an important signal in plants that mediates the response to drought stress. The most abundant phytohormone response element of LcXTH genes was associated with the ABA response (ABRE motif, 58.48%). Moreover, the augmented NCED expression levels detected in transgenic tobacco indicated that the increased drought stress resistance provided by the LcXTH21 transgene probably involved the promotion of ABA biosynthesis.
The “balanced growth” hypothesis proposes that some plants can stimulate or maintain root growth while reducing shoot growth in response to drought stress (Bloom and Mooney, 1985). In Eucalyptus globulus, a drought-tolerant clone was found to have higher root growth rate than a drought-sensitive one (Costa et al., 2004). Moreover, the development of the root system largely determines the performance of plants under drought conditions. Thus, increased root biomass is one of the primary mechanisms used by plants to avoid, or reduce, drought stress (Kashiwagi et al., 2005). For example, the overexpression of OsNAC5 was found to enhance drought tolerance by increasing root diameter (Jeong et al., 2013). XTH gene family members have been widely studied considering the different roles they are known to play in root development. For example, seven XTH genes from rice were specifically expressed in the roots of seedlings (Yokoyama et al., 2004). AtXTH19 and AtXTH23 were involved in lateral root development via the BES1-dependent pathway, indicating that XTH genes play a role in root development (Xu et al., 2020). Later, the expression levels of AtXTH11, AtXTH29, and AtXTH33 in the roots and aboveground organs were found to differ in A. thaliana plants subjected to high temperature and drought stress, thereby suggesting that these genes might mediate rapid responses to drought stress (De Caroli et al., 2021). In a study performed in grapevine, the transcription levels of VvXTH genes presented the largest changes in roots and leaves under drought and salt stress, indicating that VvXTH genes vigorously respond to abiotic stress in leaves and roots (Qiao et al., 2022).
Molecular breeding is a promising approach and great progress has been made in its use for improving the efficiency of plant breeding programs (Varshney et al., 2009). Combined with molecular marker-assisted selection, greater and faster genetic progress can be achieved. For example, in a study of Triticum aestivum, Iquebal et al. (2019) found that the maximum number of drought responsive quantitative trait loci were detected at the seedling stage and further analyzed the regulatory networks of key candidate genes and their roles in responding to drought stress in order to identify putative markers for breeding applications. Besides, in a recent study, Chauhan et al. (2022) used morphological, biochemical, and molecular markers from Withania somnifera to assess the 25 accessions of Indian ginseng, and concluded that these markers could be used to select superior ginseng genotypes. In addition, XTH genes were also reported to be involved in other abiotic stress, such as salt, heat and cold stress (Han et al., 2017; Hidvégi et al., 2020; De Caroli et al., 2021). Hence, we suggest that LcXTH genes play roles in drought resistance and other abiotic stresses. In this study, we performed functional characterization of LcXTH21, and transgenic tobacco showed higher drought resistance and more developed roots during seedling stage. Therefore, LcXTH genes could be potential functional markers when conducting marker-assisted-selection (MAS) for breeding varieties with high resistance to abiotic stress. As an example, we could select genotypes with high LcXTH21expression levels and these MAS genotypes could be expected to have higher resistance to drought stress at the adult stage. This procedure could represent an alternative way to breed L. chinense varieties with increased stress resistance associated with a highly developed root system in the coming decade.
Conclusion
In this study, 27 LcXTH genes were identified, and they were divided into three subfamilies. Tandem duplication was probably the major contributor to the expansion of the LcXTH family, and six LcXTH genes significantly responded to drought stress. Overexpression of LcXTH21 in tobacco resulted in a more developed root system. In summary, these findings enhance our understanding of the LcXTH gene family and lay the foundation for further exploration on drought resistance mechanisms in L. chinense.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: NCBI, PRJNA559687.
Author contributions
JW performed the experiments, analyzed the data and wrote the manuscript. YZ contributed to the data analysis and preparation of the manuscript. ZT took a role in experimental design and data analysis. ZH analyzed the data. LY, WL and ZC participated in plant sample collection and experimental assay. HL conceived the project, designed the experiments and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was financially supported by the National Natural Science Foundation of China (31770718 and 31470660) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1014339/full#supplementary-material
Supplementary Figure 1 | Different transgenic plant stages.
Supplementary Figure 2 | Distribution and synteny of LcXTH genes in L. chinense chromosomes. Blue lines indicate synteny between LcXTH genes. Different colors indicate different chromosomes and syntenies. Gene densities are shown as red lines in the box. And the LcXTH genes were marked.
Supplementary Figure 3 | The identity heat map of LcXTH genes.
Supplementary Table 1 | The primer used in this study.
Supplementary Table 2 | The characteristics and subcellular localization of LcXTH proteins.
Supplementary Table 3 | The substitution rates (Ka/Ks) and diverging time of LcXTH gene pairs.
Supplementary Table 4 | The protein similarity matrix of LcXTH genes.
Supplementary Table 5 | The GO annotations of LcXTH genes.
File S1 | The protein sequences used in phylogenetic tree constructing, and open reading frame sequence of LcXTH21.
References
Ahmadizadeh, M., Rezaee, S., Heidari, P. (2020). Genome-wide characterization and expression analysis of fatty acid desaturase gene family in Camelina sativa. Gene Rep. 21, 100894. doi: 10.1016/j.genrep.2020.100894
Atkinson, R. G., Johnston, S. L., Yauk, Y.-K., Sharma, N. N., Schröder, R. (2009). Analysis of xyloglucan endotransglucosylase/hydrolase (XTH) gene families in kiwifruit and apple. Postharvest Biol. Tec. 51, 149–157. doi: 10.1016/j.postharvbio.2008.06.014
Bailey, T. L., Johnson, J., Grant, C. E., Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Barbeyron, T., Gerard, A., Potin, P., Henrissat, B., Kloareg, B. (1998). The kappa-carrageenase of the marine bacterium Cytophaga drobachiensis. structural and phylogenetic relationships within family-16 glycoside hydrolases. Mol. Biol. Evol. 15, 528–537. doi: 10.1093/oxfordjournals.molbev.a025952
Baumann, M. J., Eklof, J. M., Michel, G., Kallas, A. M., Teeri, T. T., Czjzek, M., et al. (2007). Structural evidence for the evolution of xyloglucanase activity from xyloglucan endo-transglycosylases: biological implications for cell wall metabolism. Plant Cell 19, 1947–1963. doi: 10.1105/tpc.107.051391
Becnel, J., Natarajan, M., Kipp, A., Braam, J. (2006). Developmental expression patterns of Arabidopsis XTH genes reported by transgenes and genevestigator. Plant Mol. Biol. 61, 451–467. doi: 10.1007/s11103-006-0021-z
Behar, H., Graham, S. W., Brumer, H. (2018). Comprehensive cross-genome survey and phylogeny of glycoside hydrolase family 16 members reveals the evolutionary origin of EG16 and XTH proteins in plant lineages. Plant J. 95, 1114–1128. doi: 10.1111/tpj.14004
Bloom, A. J., Chapin, F. S., Mooney, H. A. (1985). Resource limitation in plants-an economic analogy. Annu. Rev. Ecol. Sys. 16, 363–392. doi: 10.1146/annurev.es.16.110185.002051
Boyer, J. S. (1982). Plant productivity and environment. Science 218, 443–448. doi: 10.1126/science.218.4571.443
Bray, E. A., Bailey-Serres, J., Weretilnyk, E. (2000). “Responses to abiotic stresses,” in Biochemistry and molecular biology of plants. Eds. Buchanan, B. B., Gruissem, W., Jones, R. L. (Rockville, MD: American Society of Plant Physiologists), 1158–1203.
Buckeridge, M. S., Pessoa dos Santos, H., Tiné, M. A. S. (2000). Mobilisation of storage cell wall polysaccharides in seeds. Plant Physiol. Bioch. 38, 141–156. doi: 10.1016/S0981-9428(00)00162-5
Campbell, P., Braam, J. (1999a). In vitro activities of four xyloglucan endotransglycosylases from Arabidopsis. Plant J. 18, 371–382. doi: 10.1046/j.1365-313x.1999.00459.x
Campbell, P., Braam, J. (1999b). Xyloglucan endotransglycosylases: diversity of genes, enzymes and potential wall-modifying functions. Trends Plant Sci. 4, 361–366. doi: 10.1016/s1360-1385(99)01468-5
Chauhan, S., Mandliya, T., Jain, D., Joshi, A., Khatik, C. L., Singh, A., et al. (2022). Early selective strategies for higher yielding bio-economic Indian ginseng based on genotypic study through metabolic and molecular markers. Saudi J. Biol. Sci. 29, 3051–3061. doi: 10.1016/J.SJBS.2022.01.030
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, J.-R., Chen, Y.-B., Ziemiańska, M., Liu, R., Deng, Z.-N., Niedźwiecka-Filipiak, ,. I., et al. (2016). Co-Expression of MtDREB1C and RcXET enhances stress tolerance of transgenic China rose (Rosa chinensis jacq.). J. Plant Growth Regul. 35, 586–599. doi: 10.1007/s00344-015-9564-z
Chen, J., Hao, Z., Guang, X., Zhao, C., Wang, P., Xue, L., et al. (2019). Liriodendron genome sheds light on angiosperm phylogeny and species-pair differentiation. Nat. Plants 5, 18–25. doi: 10.1038/s41477-018-0323-6
Chen, Y., Weng, X., Zhou, X., Gu, J., Hu, Q., Luo, Q., et al. (2022). Overexpression of cassava RSZ21b enhances drought tolerance in Arabidopsis. J. Plant Physiol. 268, 153574. doi: 10.1016/J.JPLPH.2021.153574
Choi, J. Y., Seo, Y. S., Kim, S. J., Kim, W. T., Shin, J. S. (2011). Constitutive expression of CaXTH3, a hot pepper xyloglucan endotransglucosylase/hydrolase, enhanced tolerance to salt and drought stresses without phenotypic defects in tomato plants (Solanum lycopersicum cv. dotaerang). Plant Cell Rep. 30, 867–877. doi: 10.1007/s00299-010-0989-3
Chou, K. C., Shen, H. B. (2010). Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PloS One 5, e11335. doi: 10.1371/journal.pone.0011335
Costa, E. S. F., Shvaleva, A., Maroco, J. P., Almeida, M. H., Chaves, M. M., Pereira, J. S. (2004). Responses to water stress in two Eucalyptus globulus clones differing in drought tolerance. Tree Physiol. 24, 1165–1172. doi: 10.1093/treephys/24.10.1165
Cushman, J. C., Bohnert, H. J. (2000). Genomic approaches to plant stress tolerance. Curr. Opin. Plant Biol. 3, 117–124. doi: 10.1016/s1369-5266(99)00052-7
De Caroli, M., Manno, E., Piro, G., Lenucci, M. S. (2021). Ride to cell wall: Arabidopsis XTH11, XTH29 and XTH33 exhibit different secretion pathways and responses to heat and drought stress. Plant J. 107, 448–466. doi: 10.1111/tpj.15301
Del Bem, L. E., Vincentz, M. G. (2010). Evolution of xyloglucan-related genes in green plants. BMC Evol. Biol. 10, 341. doi: 10.1186/1471-2148-10-341
Dong, X. M., Pu, X. J., Zhou, S. Z., Li, P., Luo, T., Chen, Z. X., et al. (2022). Orphan gene PpARDT positively involved in drought tolerance potentially by enhancing ABA response in Physcomitrium (Physcomitrella) patens. Plant Sci. 319, 111222. doi: 10.1016/j.plantsci.2022.111222
Eklöf, J. M., Brumer, H. (2010). The XTH gene family: an update on enzyme structure, function, and phylogeny in xyloglucan remodeling. Plant Physiol. 153, 456–466. doi: 10.1104/pp.110.156844
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The pfam protein families database in 2019. Nucleic Acids Res. 47, D427–d432. doi: 10.1093/nar/gky995
Fanutti, C., Gidley, M. J., Reid, J. S. (1993). Action of a pure xyloglucan endo-transglycosylase (formerly called xyloglucan-specific endo-(1–>4)-beta-D-glucanase) from the cotyledons of germinated nasturtium seeds. Plant J. 3, 691–700. doi: 10.1046/j.1365-313x.1993.03050691.x
Fu, M. M. (2019). Cloning and functional characterization of HvXTH1 in drought tolerance in Tibetan wild barley Ph.D. dissertation
Fu, M. M., Liu, C., Wu, F. (2019). Genome-wide identification, characterization and expression analysis of xyloglucan endotransglucosylase/hydrolase genes family in barley (Hordeum vulgare). Molecules 24. doi: 10.3390/molecules24101935
Geisler-Lee, J., Geisler, M., Coutinho, P. M., Segerman, B., Nishikubo, N., Takahashi, J., et al. (2006). Poplar carbohydrate-active enzymes. gene identification and expression analyses. Plant Physiol. 140, 946–962. doi: 10.1104/pp.105.072652
Han, Y., Han, S., Ban, Q., He, Y., Jin, M., Rao, J. (2017). Overexpression of persimmon DkXTH1 enhanced tolerance to abiotic stress and delayed fruit softening in transgenic plants. Plant Cell Rep. 36, 583–596. doi: 10.1007/s00299-017-2105-4
Harada, T., Torii, Y., Morita, S., Onodera, R., Hara, Y., Yokoyama, R., et al. (2011). Cloning, characterization, and expression of xyloglucan endotransglucosylase/hydrolase and expansin genes associated with petal growth and development during carnation flower opening. J. Exp. Bot. 62, 815–823. doi: 10.1093/jxb/erq319
He, S. A., Hao, R. M. (1999). Study on the natural population dynamics and the endangering habitat of Liriodendron chinese in China. Acta Phytoecol. Sin. 01, 88–96. doi: CNKI:SUN:ZWZY.0.1995-01-000
Heidari, P., Mazloomi, F., Nussbaumer, T., Barcaccia, G. (2020). Insights into the SAM synthetase gene family and its roles in tomato seedlings under abiotic stresses and hormone treatments. Plants 9, 1–17. doi: 10.3390/plants9050586
Hidvégi, N., Gulyás, A., Dobránszki, J., Teixeira da Silva, J. A. (2020). Mining sequences with similarity to XTH genes in the Solanum tuberosum l. transcriptome: introductory step for identifying homologous XTH genes. Plant Signal. Behav. 15, 1797294. doi: 10.1080/15592324.2020.1797294
Hu, B., Jin, J., Guo, A.-Y., Zhang, H., Luo, J., Gao, G. (2014). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Iquebal, M. A., Sharma, P., Jasrotia, R. S., Jaiswal, S., Kaur, A., Saroha, M., et al. (2019). RNAseq analysis reveals drought-responsive molecular pathways with candidate genes and putative molecular markers in root tissue of wheat. Sci. Rep. 9, 13917. doi: 10.1038/s41598-019-49915-2
Jeong, J. S., Kim, Y. S., Baek, K. H., Jung, H., Ha, S.-H., Do Choi, Y., et al. (2010). Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 153, 185–197. doi: 10.1104/pp.110.154773
Jeong, J. S., Kim, Y. S., Redillas, M. C., Jang, G., Jung, H., Bang, S. W., et al. (2013). OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol. J. 11, 101–114. doi: 10.1111/pbi.12011
Kallas, A. M., Piens, K., Denman, S. E., Henriksson, H., Fäldt, J., Johansson, P., et al. (2005). Enzymatic properties of native and deglycosylated hybrid aspen (Populus tremulaxtremuloides) xyloglucan endotransglycosylase 16A expressed in Pichia pastoris. Biochem. J. 390, 105–113. doi: 10.1042/bj20041749
Kashiwagi, J., Krishnamurthy, L., Upadhyaya, H. D., Krishna, H., Chandra, S., Vadez, V., et al. (2005). Genetic variability of drought-avoidance root traits in the mini-core germplasm collection of chickpea (Cicer arietinum l.). Euphytica 146, 213–222. doi: 10.1007/s10681-005-9007-1
Kaur, N., Prashanth, K. H., Bhatti, M. S., Pati, P. K. (2022). OsSalT gene cloned from rice provides evidence of its role in salinity and drought stress tolerance. Plant Sci. 320, 111306. doi: 10.1016/J.plantsci.2022.111306
Kim, T. H., Böhmer, M., Hu, H., Nishimura, N., Schroeder, J. I. (2010). Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 61, 561–591. doi: 10.1146/annurev-arplant-042809-112226
Kozak, K. H., Mendyk, R. W., Wiens, J. J. (2009). Can parallel diversification occur in sympatry? repeated patterns of body-size evolution in coexisting clades of north American salamanders. Evolution 63, 1769–1784. doi: 10.1111/j.1558-5646.2009.00680.x
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Letunic, I., Khedkar, S., Bork, P. (2021). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–d460. doi: 10.1093/nar/gkaa937
Leung, J., Giraudat, J. (1998). Abscisic acid signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 199–222. doi: 10.1146/annurev.arplant.49.1.199
Li, Q., Li, H., Yin, C., Wang, X., Jiang, Q., Zhang, R., et al. (2019). Genome-wide identification and characterization of xyloglucan endotransglycosylase/hydrolase in Ananas comosus during development. Genes 10, 537. doi: 10.3390/genes10070537
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Long, C. (2020). Cloning of GmXTH23 gene and identification of its drought resistance. Acta Agric. Univ. Jiangxiensis 42, 898–905. doi: 10.13836/j.jjau.2020101
Lynch, M., Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. doi: 10.1126/science.290.5494.1151
Marchler-Bauer, A., Bryant, S. H. (2004). CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32, W327–W331. doi: 10.1093/nar/gkh454
Michel, G., Chantalat, L., Duee, E., Barbeyron, T., Henrissat, B., Kloareg, B., et al. (2001). The kappa-carrageenase of P. carrageenovora features a tunnel-shaped active site: a novel insight in the evolution of clan-b glycoside hydrolases. Structure 9, 513–525. doi: 10.1016/s0969-2126(01)00612-8
Mittler, R. (2006). Abiotic stress, the field environment and stress combination. Trends Plant Sci. 11, 15–19. doi: 10.1016/j.tplants.2005.11.002
Murashige, T., Skoog, F. (2006). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Ni, L., Wang, Z., Liu, X., Wu, S., Hua, J., Liu, L., et al. (2022). Genome-wide study of the GRAS gene family in Hibiscus hamabo sieb. et zucc and analysis of HhGRAS14-induced drought and salt stress tolerance in Arabidopsis. Plant Sci. 319, 111260. doi: 10.1016/j.plantsci.2022.111260
Opazo, M. C., Lizana, R., Stappung, Y., Davis, T. M., Herrera, R., Moya-Leon, M. A. (2017). XTHs from Fragaria vesca: genomic structure and transcriptomic analysis in ripening fruit and other tissues. BMC Genomics 18, 852. doi: 10.1186/s12864-017-4255-8
Parmar, N., Singh, K. H., Sharma, D., Singh, L., Kumar, P., Nanjundan, J., et al. (2017). Genetic engineering strategies for biotic and abiotic stress tolerance and quality enhancement in horticultural crops: a comprehensive review. 3 Biotech. 7, 239. doi: 10.1007/s13205-017-0870-y
Popper, Z. A. (2008). Evolution and diversity of green plant cell walls. Curr. Opin. Plant Biol. 11, 286–292. doi: 10.1016/j.pbi.2008.02.012
Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R., Finn, R. D. (2018). HMMER web server: 2018 update. Nucleic Acids Res. 46, W200–W204. doi: 10.1093/nar/gky448
Qiao, T., Zhang, L., Yu, Y., Pang, Y., Tang, X., Wang, X., et al. (2022). Identification and expression analysis of xyloglucan endotransglucosylase/hydrolase (XTH) family in grapevine (Vitis vinifera l.). Peerj 10, e13546. doi: 10.7717/peerj.13546
Reid, J. S. G. (1985). Cell wall storage carbohydrates in seeds–biochemistry of the seed “gums” and “hemicelluloses”. Adv. Bot. Res. 11, 125–155.
Roy, B., Konjengbam, N., Mandal, A., Basu, A. (2011). Genetic engineering for abiotic stress tolerance in agricultural crops. Biotechnology 10, 1–22. doi: 10.3923/biotech.2011.1.22
Sharma, I., Kaur, N., Pati, P. K. (2017). Brassinosteroids: A promising option in deciphering remedial strategies for abiotic stress tolerance in rice. Front. Plant Sci. 8 2151. doi: 10.3389/fpls.2017.02151
Shinohara, N., Nishitani, K. (2021). Cryogenian origin and subsequent diversification of the plant cell-wall enzyme XTH family. Plant Cell Physiol. 62, 1874–1889. doi: 10.1093/pcp/pcab093
Subramanian, B., Gao, S., Lercher, M. J., Hu, S., Chen, W.-H. (2019). Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 47, W270–W275. doi: 10.1093/nar/gkz357
Teufel, F., Almagro Armenteros, J. J., Johansen, A. R., Gíslason, M. H., Pihl, S. I., Tsirigos, K. D., et al. (2022). SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 40, 1023–1025. doi: 10.1038/s41587-021-01156-3
Tu, Z., Hao, Z., Zhong, W., Li, H. (2019). Identification of suitable reference genes for RT-qPCR assays in Liriodendron chinense (Hemsl.) sarg. Forests 10, 441. doi: 10.3390/f10050441
Varshney, R. K., Hoisington, D. A., Nayak, S. N., Graner, A. (2009). Molecular plant breeding: methodology and achievements. Methods Mol. Biol. 513, 283–304. doi: 10.1007/978-1-59745-427-8_15
Wai, A. H., Naing, A. H., Lee, D.-J., Kim, C. K., Chung, M.-Y. (2020). Molecular genetic approaches for enhancing stress tolerance and fruit quality of tomato. Plant Biotechnol. Rep. 14, 515–537. doi: 10.1007/s11816-020-00638-1
Wang, Z., Li, J., Yang, X., Hu, Y., Yin, Y., Shen, X. (2022). MdFLP enhances drought tolerance by regulating MdNAC019 in self-rooted apple stocks. Plant Sci. 321, 111331. doi: 10.1016/j.plantsci.2022.111331
Wang, C., Lu, G., Hao, Y., Guo, H., Guo, Y., Zhao, J., et al. (2017). ABP9, a maize bZIP transcription factor, enhances tolerance to salt and drought in transgenic cotton. Planta 246, 453–469. doi: 10.1007/s00425-017-2704-x
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., Williams, K. L., Appel, R. D., et al. (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552. doi: 10.1385/1-59259-584-7:531
Wu, T., Hu, E., Xu, S., Chen, M., Guo, P., Dai, Z., et al. (2021). clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2, 100141. doi: 10.1016/j.xinn.2021.100141
Xu, P., Fang, S., Chen, H., Cai, W. (2020). The brassinosteroid-responsive xyloglucan endotransglucosylase/hydrolase 19 (XTH19) and XTH23 genes are involved in lateral root development under salt stress in Arabidopsis. Plant J. 104, 59–75. doi: 10.1111/tpj.14905
Yang, Z., Zhang, R., Zhou, Z. (2022). The XTH gene family in Schima superba: Genome-wide identification, expression profiles, and functional interaction network analysis. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.911761
Yokoyama, R., Nishitani, K. (2001). A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant Cell Physiol. 42, 1025–1033. doi: 10.1093/pcp/pce154
Yokoyama, R., Nishitani, K. (2008). Functional diversity of xyloglucan-related proteins and its implications in the cell wall dynamics in plants. Plant Biol. 2, 598–604. doi: 10.1055/s-2000-16643
Yokoyama, R., Rose, J. K., Nishitani, K. (2004). A surprising diversity and abundance of xyloglucan endotransglucosylase/hydrolases in rice classification and expression analysis. Plant Physiol. 134, 1088–1099. doi: 10.1104/pp.103.035261
Zhang, M., Leng, P., Zhang, G., Li, X. (2009). Cloning and functional analysis of 9-cis-epoxycarotenoid dioxygenase (NCED) genes encoding a key enzyme during abscisic acid biosynthesis from peach and grape fruits. J. Plant Physiol. 166, 1241–1252. doi: 10.1016/j.jplph.2009.01.013
Zhu, F., Luo, T., Liu, C., Wang, Y., Yang, H., Yang, W., et al. (2017). An R2R3-MYB transcription factor represses the transformation of α- and β-branch carotenoids by negatively regulating expression of CrBCH2 and CrNCED5 in flavedo of Citrus reticulate. New Phytol. 216, 178–192. doi: 10.1111/nph.14684
Keywords: Liriodendron chinense, XTH family, drought stress response, genome identification, root development
Citation: Wu J, Zong Y, Tu Z, Yang L, li W, Cui Z, Hao Z and Li H (2022) Genome-wide identification of XTH genes in Liriodendron chinense and functional characterization of LcXTH21. Front. Plant Sci. 13:1014339. doi: 10.3389/fpls.2022.1014339
Received: 08 August 2022; Accepted: 11 October 2022;
Published: 27 October 2022.
Edited by:
John Paul Délano-Frier, Instituto Politécnico Nacional de México (CINVESTAV), MexicoReviewed by:
Parviz Heidari, Shahrood University of Technology, IranPeng Zhao, Northwest University, China
Copyright © 2022 Wu, Zong, Tu, Yang, li, Cui, Hao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huogen Li, aGdsaUBuamZ1LmVkdS5jbg==
 Junpeng Wu
Junpeng Wu Yaxian Zong
Yaxian Zong Zhonghua Tu
Zhonghua Tu Lichun Yang
Lichun Yang Ziyuan Hao
Ziyuan Hao Huogen Li
Huogen Li