- 1Center of Agricultural Biochemistry and Biotechnology (CABB), University of Agriculture, Faisalabad, Pakistan
- 2Department of Molecular and Cellular Biology, Summerlee Science Complex, University of Guelph, Guelph, ON, Canada
A tobacco chloroplast hypothetical open reading frame 4 (YCF4) has been reported as a non-essential assembly factor for photosynthesis based on an incomplete knockout of YCF4, just 93 of 184 amino acids from the N-terminus were knocked out. On the other hand, we removed the complete sequence of YCF4 from tobacco chloroplasts and observed that ΔYCF4 plants were unable to survive photoautotrophically as their growth was hampered in the absence of an external carbon supply, clearly showing that the YCF4 is essential for photosynthesis. Initially, the aadA gene was introduced into the tobacco plastome replacing the complete YCF4 gene through homologous recombination events. The replacement of YCF4 with aadA was confirmed by PCR and Southern blot analysis in ΔYCF4 plants. Homoplasmic ΔYCF4 plants had a light green phenotype, and the leaves became pale yellow as the plants grew older. The structure of chloroplasts of ΔYCF4 mutants of light green phenotype was studied using a transmission electron microscope (TEM), and the micrographs demonstrated structural anomalies in the chloroplasts; including shape, size, and grana stacking compared to the wild-type plants. Further, transcriptome analysis revealed that the expression of PSI, PSII, and ribosomal genes remained unchanged in ∆YCF4 plants. On the other hand, transcriptome levels of rbcL (Ribulose 1,5-bisphosphate carboxylase/oxygenase large subunit), LHC (Light-Harvesting Complex), and ATP Synthase (atpB and atpL) decreased, indicating that the YCF4 has the function(s) in addition to assembling the photosynthetic complex. This was confirmed by in-silico protein-protein interactions of full-length YCF4 as well as 93 and 91 of 184 amino acids from N- and C-termini of the full-length protein, which revealed that the C-terminus (91 aa) of YCF4 is important in interacting with other chloroplast proteins. These findings provide genetic support for the plastid YCF4 gene’s critical role in regulating the plastid gene expression and assembling the photosynthetic complex.
Introduction
Plastid origin can be traced back over a billion years when a fossil red alga was discovered, providing substantial proof for the theory (Embley and Martin, 2006; Song et al., 2018) that plastids arose from endosymbiotic photosynthetic bacteria (Schimper, 1883). Plastids are the defining traits of plants and green algae, yet they maintain many prokaryotic properties. The bacterial genome has reduced dramatically over time, owing to gene loss and large-scale gene transfer to the nuclear genome. Thus, plastid genomes referred to as ‘plastomes’ contain around 120-130 genes, the majority of which encode components of the organelle’s gene expression mechanism and photosynthetic apparatus and are structured in nucleoids. Plastids, on the other hand, have far more proteins than their plastomes can code for. As a result, most plastid proteins are now encoded by the nuclear genome and must be transported into the organelle after translation. Plastids have an extensive membrane system, thylakoids, which are condensed to form, grana, in addition to two envelope membranes. In thylakoid membranes of plants including cyanobacteria and algae two enormous multimeric chlorophyll-binding protein complexes, photosystems I and II are embedded that performs the first step in the oxygenic photosynthesis this process, responsible for converting sunlight into chemical energy (Blankenship, 2010; Kargul and Barber, 2011).
The plastid genome was first sequenced in tobacco followed by hundreds of higher plants that have been sequenced and characterized. This plethora of plastid genome data insights researchers for the functional characterization of plastid-encoded genes. Most of the chloroplast genes have been characterized for their role in the organellar stability or metabolic activities within the chloroplasts, yet few of these are still to be worked out for their function. They are labeled as ycfs (hypothetical chloroplast open reading frames). Few of these have been characterized as non-essential genes whereas others as essentials. The YCF4 gene, one of these ycfs, is a highly conserved protein in cyanobacteria, green algae, and land plants. Previously, it was discovered that the YCF4 gene product is involved in the formation of the PSI complex in Chlamydomonas reinhardtii (Boudreau et al., 1997). In YCF4 mutants, PSI activity was completely lost, resulting in autotrophic growth failure. Inactivation of the YCF4 homolog causes an increase in the PSII-to-PSI ratio in the cyanobacterium Synechocystis sp. PCC 6803. (Wilde et al., 1995). The thylakoid membrane-intrinsic YCF4 was found in complexes with the PSI subunits PsaA through PsaF and the opsin-related eyespot protein COP2 in Chlamydomonas (Ozawa et al., 2009). Because RNA interference did not influence PSI accumulation in the alga, the COP2 protein is unlikely to play a role in PSI biogenesis (Ozawa et al., 2009). YCF4 has also been found as a protein component of the eyespot in Chlamydomonas chloroplasts (Schmidt et al., 2006), suggesting that this has a second function in the eyespot (in conjunction with COP2). Furthermore, utilizing reverse genetics, YCF4 has been knocked out in tobacco, and the mutants have shown autotrophic growth (Krech et al., 2012). Tobacco YCF4 knockout mutants were able to assemble enough PSI to enable modest autotrophic growth. However, the YCF4 mutants did not grow autotrophically in our investigations, and extremely slow growth was detected under controlled settings using sucrose as a carbon source, but not in wild conditions.
In the present studies, we have removed the complete YCF4 gene sequence encoding 184 amino acids and developed homoplasmic ΔYCF4 plants, exhibiting very slow growth on an artificial medium supplemented with varied sucrose levels. These mutants unlike previously reported YCF4 mutants (Krech et al., 2012) where the partial sequence of YCF4 encoding 93 amino acids from the N-terminal region of YCF4 has been removed leaving 91 amino acids of the C-terminal intact, failed to grow autotrophically under normal conditions in peat moss-containing pots. It was observed by in-silico protein-protein interactions of full-length YCF4 as well as 93 and 91 of 184 amino acids from N- and C-termini, respectively of the full-length protein that the C-terminus (91 aa) of YCF4 is interacting with other chloroplast proteins.
Materials and methods
Plant material for targeted knockout of YCF4
Nicotiana tabacum L. var. Petit Havana was grown at 25 ± 1˚C under 16 hrs light (white light: 100 µmol·m-2·s-1) and 8 hrs dark regime in a growth room. The sterilized seeds were cultured on RMOP medium containing MS salts (4.33 g/L), Myoinositol (100 mg/L), BAP (1.0 mg/L), NAA (0.1mg/L) sucrose (3%) solidified with 0.026% Gelrite. Fully mature dark green leaves of 4-6 weeks old plants were used for targeted knockout of the YCF4 gene (Nazir and Khan, 2013).
Development of chloroplast transformation vector and selection of putative knocked out plants
Considering the location of the YCF4 gene where rbcL, accD, and psaI are in the upstream region and ycf10, petA, and psbJ are in the downstream region of the YCF4. To develop chloroplast transformation vector for the targeted inactivation of YCF4, ycf10 sequence was cloned as right border flanking sequence whereas PsaI along with few nucleotides of accD were cloned as left border flanking sequences. FLARE-S cassette having aadA (aminoglycoside 3́-adenyltransferase) and gfp (green fluorescent protein) was cloned in between the plastid flanking sequences (Khan and Maliga, 1999). The resultant plastid transformation vector was coated on 0.6 μm gold particles and were bombarded on tobacco leaves using particle gun (Bio-Rad, USA). The bombarded leaves were chopped into tiny slices and cultured on RMOP medium containing 500 mg/L spectinomycin. Antibiotic resistant shoots were rooted on MS medium containing 30 g/L sucrose.
Genomic analysis of putative transplastomic clones for targeted inactivation of YCF4
Total cellular DNA was isolated from the transformed and untransformed tobacco plants using CTAB (hexadecyltrimethyl ammonium bromide) method with certain modifications. The isolated DNA was subjected to PCR (Polymerase Chain Reaction) to confirm the transgene integration using primers flanking aadA selectable marker gene (A19: 5’-GGC TCC GCA GTG GAT GGC GGC CTG-3’; A20: 5’-GGG CTG ATA CTG GGC CGG CAG G-3’) Further, the homoplasmic status of transplastomics were accessed using S19 and S20 primers flanking psaI and ycf10 genes. Following PCR profile was used for the amplification of the above-mentioned gene(s): 94°C for 1.5 minutes 56°C for 1.0 minutes and 72°C for 3 minutes with a final extension at 72°C for 10 minutes and a total of 35 cycles.
Homoplasmy of the YCF4 knock-out plants was confirmed by Southern blot analysis. A total of 13 μg genomic DNA was digested with the restriction enzyme BamHI restriction endonuclease and was resolved on 1% agarose gel. After washing with depurination, denaturation, and neutralization solutions, the restricted DNA was blotted onto a nitrocellulose membrane. Thereafter, it was hybridized with a biotin-labeled probe followed by detection with a chromogenic detection kit (Thermo scientific, USA).
Transcript analysis of ∆YCF4 plants
Total cellular RNA was isolated from the confirmed purified ∆YCF4 and wild type in vitro growing tobacco plants by RNeasy Plant Mini Kit (Qiagen, USA). The isolated RNA was treated with DNase I (Thermo Fisher Scientifc, USA) to remove genomic DNA following the manufacturer’s instructions. RevertAid First Strand cDNA synthesis kit (Thermo Fisher Scientific, Cat. # K1621) was used for the synthesis of cDNA using 1.0 µg RNA following the manufacturer’s instructions. The resultant cDNA was used for semi-quantitative expression analyses of psaA, psaB, psaC, psaH (PhotosystemI), psbA, psbB, psbC, psbD and psbE (Photosystem II), ATP synthase, rps16, rps2, rrn16, ycf10, rbcL clpP, rpoA, and rpoB.
Electron microscopy to study chloroplast ultrastructural variation in ∆YCF4 plants
Transmission Electron Microscopy (TEM) was carried out following the protocol given by Islam et al., 2008. Leaves from ∆YCF4 and wild-type tobacco plants were cut into 1 to 2 mm2 pieces and fixation was carried out using 0.2 M phosphate buffer (PBS, pH 7.2) containing 4.0% glutaraldehyde (v/v) at 4°C for 6-8 hours. Postfixation of samples was carried out in 1% osmium tetroxide (OsO4) for 1 h at 4°C and then incubated in 0.2M PBS (pH 7.2) for 1-2 hours at room temperature. The dehydration was carried out using a graded series of ethanol and acetone and finally, the leaf pieces were embedded in Spurr’s resin. Ultrathin sections (~70 nm) of embedded leaf samples were prepared on an ultramicrotome (RMC Mt 7000) and mounted on copper grids to be viewed in the TEM (JEOL, Model JEM-1010) at an accelerating voltage of 90.0 kV. Multiple images of each section were recorded by exposing a photographic film and later developed in the darkroom.
Determination of physiological parameters
Various photosynthesis-related physiological parameters were determined using IRGA (Infrared gas analyzer). The physiological parameters included photosynthetic rate, transpiration rate, substomatal conductance, substomatal CO2, and light intensity.
Heterotrophic/autotrophic nature of growth of YCF4 mutants
To assess the impact of carbon starvation on the growth, mutant as well as wild-type tobacco plants were cultured on MS medium augmented with different levels of carbon (0, 0.5, 1.0, 1.5, 2.0, 2.5, and 3.0% sucrose). The plants were grown in normal light. Data were recorded to see the impact of carbon starvation on mutant tobacco plants.
Molecular docking of YCF4 protein with other photosynthesis-related proteins
The molecular docking of full-length and truncated versions of YCF4 with other photosynthesis-responsive proteins was performed using an online web server ClusPro 2.0. It predicts the interaction between the candidate proteins through rigid-body docking based on Fast Fourier Transform, followed by clustering, and minimizing the docked complex to get the highly populated cluster having the least-energy conformation (Kozakov et al., 2017). The first 93 amino acids of the YCF4 were knocked out by Krech et al., 2012 in their study leaving 91 amino acids intact. Whereas, we have used 93 amino acids from the N-terminal and 91 amino acids from the C-terminal side of the YCF4 as well as full-length YCF4 for studying their interaction with YCF10, ribosomal proteins or RNA (rps16, rps2, rrn16), subunits of PS-I (psaA, psaB, psaC, psaH), subunits of PS-II (psbA, psbB, psbC, psbD, psbE), alpha and beta chains of ATP synthase (atpB, atpI) and other photosynthetic proteins (rbcL, clpP, rpoA, rpoB, accD, petA, Light-harvesting complex (LHC) using ClusPro. The docked complexes with the maximum clustering members and minimum energy scores were selected. DIMPLOT program of Ligplot+ v.4.5.3 was used to find the number of hydrogen bonds and the bond lengths between the interacting residues (Wallace et al., 1996).
Results
Development of YCF4 mutants and their purification to homoplasmic level
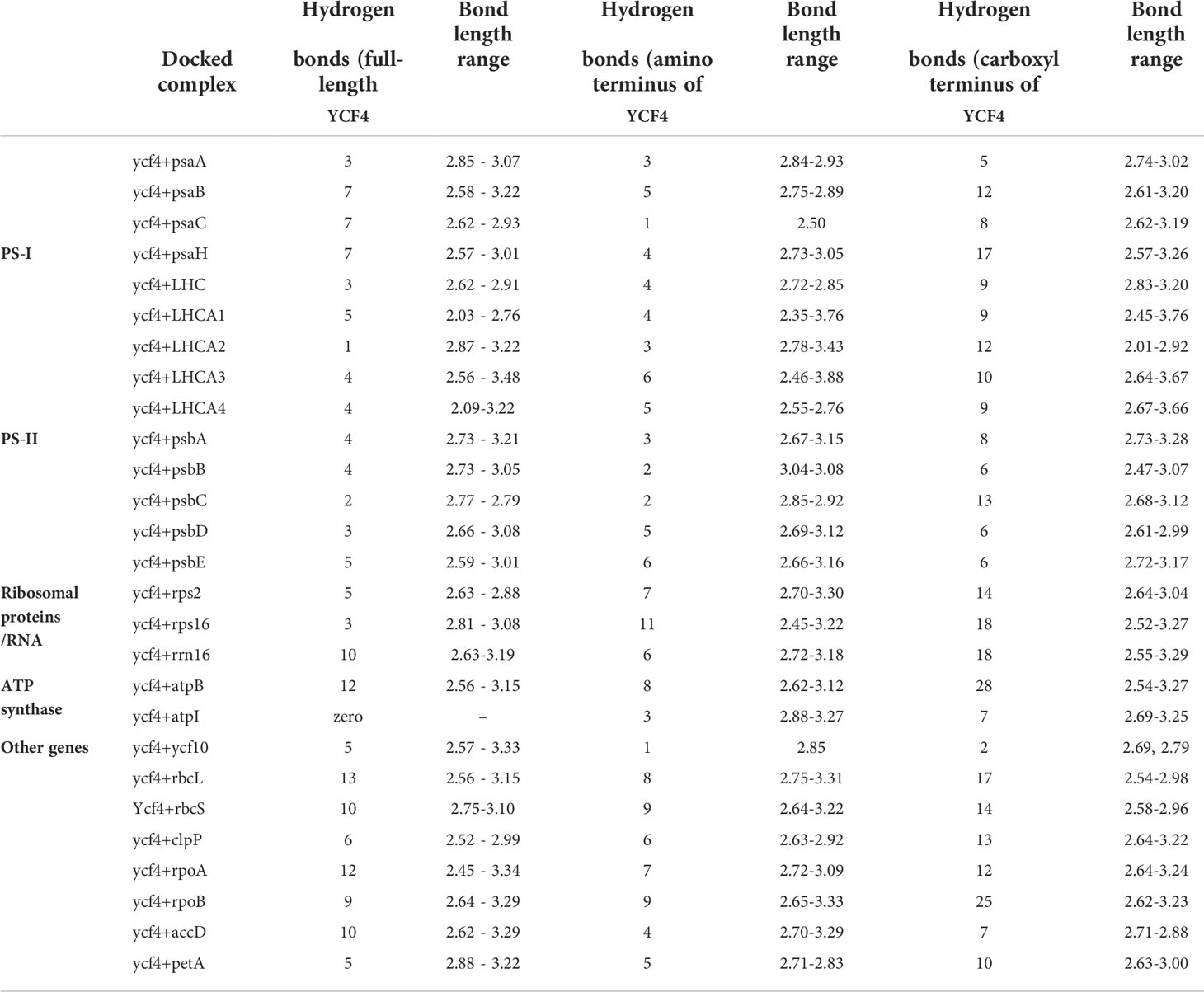
The YCF4 mutants were developed by targeting the FLARE-S (aadA and gfp) cassette with flanking sequences of ycf10 and psaI. Sequential amplification and cloning of ycf10 as right border flanking sequence and PsaI together with a few nucleotides of accD as left border flanking sequence resulted in the development of a plasmid transformation vector. The FLARE-S and its regulatory sequences were cloned in between the plastid flanking sequences. The resulting cassette was inserted into the plastid genome to inactivate YCF4 specifically. Leaf sections were cultivated in an RMOP medium containing a selective agent, spectinomycin (500 mg/L), after particle bombardment. The antibiotic-resistant shoots were putative transformants, but they were indistinguishable from untransformed tobacco plants growing in comparable conditions. For the purification of transplastomic cells, they were exposed to additional rounds of selection and screening. Leaves were chopped into small pieces and cultured on the selective agent for this purpose. All of the plants had a light green to yellow phenotype after several rounds of continuous selection and screening, showing homotransplasmicity (Figure 1). Antibiotic-resistant shoots were validated using various primer sets. To validate the integration of transgene and deletion of YCF4, putative transgenic plants were tested using primers flanking the aadA gene and the deletion cassette (Figure 2A). The incorporation of the aadA into the targeted genome was confirmed by amplification of a 552 kb fragment (Figure 2B). Furthermore, YCF4 knockout was validated using primers flanking the gene with no amplification like wild-type plant samples. Between psaI and ycf10, amplification of a 2.0 kb fragment from wild-type tobacco plants and a 4.0 kb fragment from ΔYCF4 plants demonstrated transgene incorporation into the plastid genome (Figure 2C). The amplification of 2kb fragment in transformants indicates that the transgenic plants are heteroplasmic, and the YCF4 has not been deleted completely from the plastome. The possibility of selecting nuclear transformants, spontaneous mutants (owing to mutations in the 16SrRNA gene), or escapes was eliminated. The plasmy level of the selected transformed shoots was also confirmed by Southern Blot Analysis (Figure 3A). The purified cellular DNA was restricted with BamHI and transferred onto blotting membrane. The presence of a single large-size hybridizing fragment of ~4.0 kb confirmed the homoplasmy of the FLARE-S integration and YCF4 gene deletion (Figure 3B) from the tobacco plants.

Figure 1 Purification of Δycf4 plants on spectinomycin-containing selection and regeneration medium. (A) Wild-type tobacco plants with intact ycf4 showing normal growth with lush green leaves. (B) Heteroplasmic plant showing leaves carrying chimeric tissues with both knocked-out and wild-type cells. (C) Homoplasmic Δycf4 tobacco plant growing under low light. (D) Homoplasmic Δycf4 tobacco plant growing under normal light.
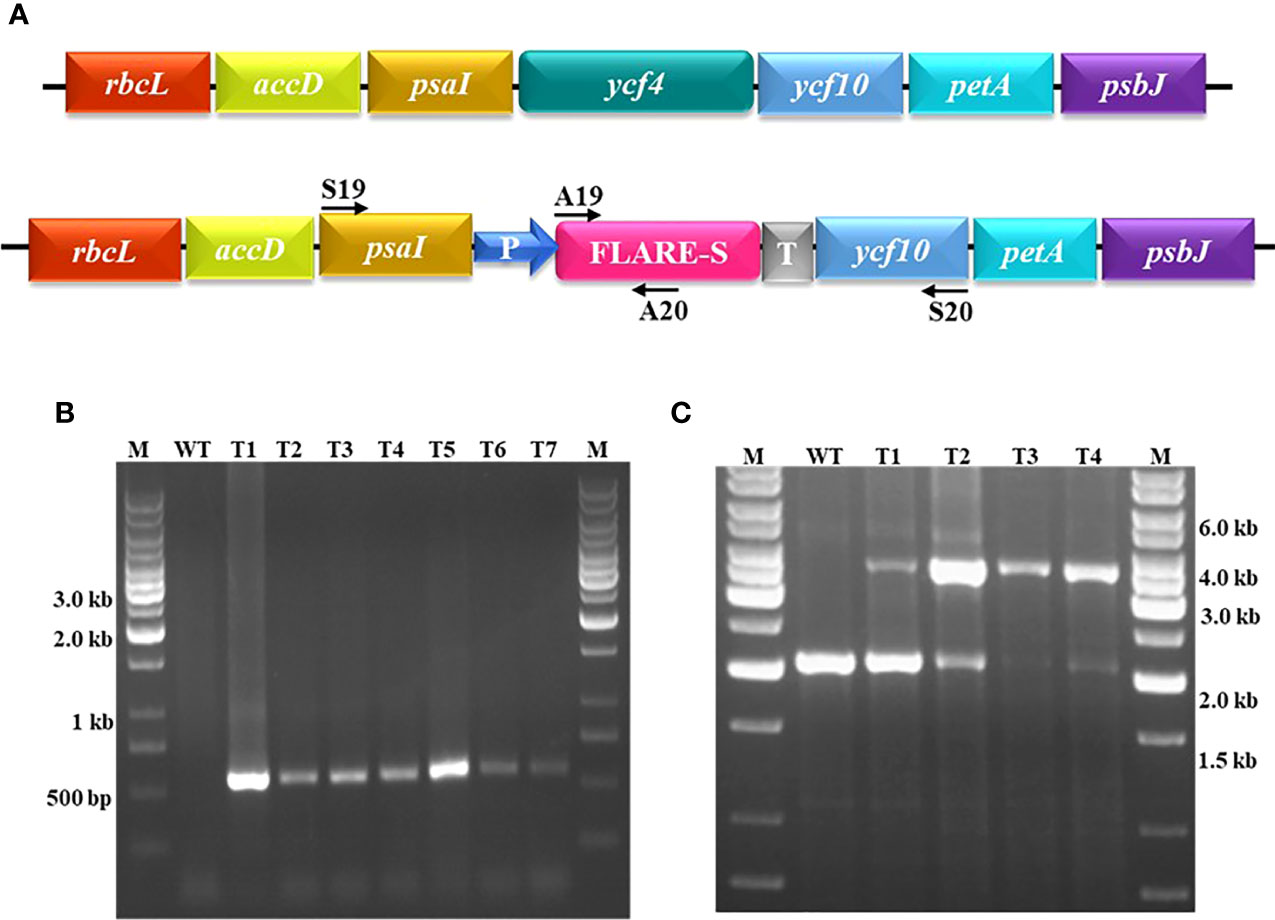
Figure 2 Confirmation of transgene integration into plastid genome and determining the plasmy level by Polymerase Chain Reaction. (A) Physical map showing the positions of the primers used. (B) Entire pool of spectinomycin-resistant plants regenerated on regeneration medium were screened for the presence of marker gene aadA-specific primer set (A19/A20): M is 1 kb marker DNA, WT is the untransformed tobacco, and T1-T7 are putative transplastomic plants. (C) Out of screened plants four as T1-T4 were randomly selected for the determination of plasmy level using S19/S20 primer set that lands on flanking sequences: M represents 1.0 kb DNA ladder, WT represents wild-type tobacco plant whereas T1-T4 represents deletion level of ycf4 from the chloroplast transgenic plants regenerated on spectinomycin-containing regeneration medium. Amplification of a fragment of 4.0 kb indicates transgene integration whereas amplification of a fragment of 2.0 kb represents wild-type plastid DNA.
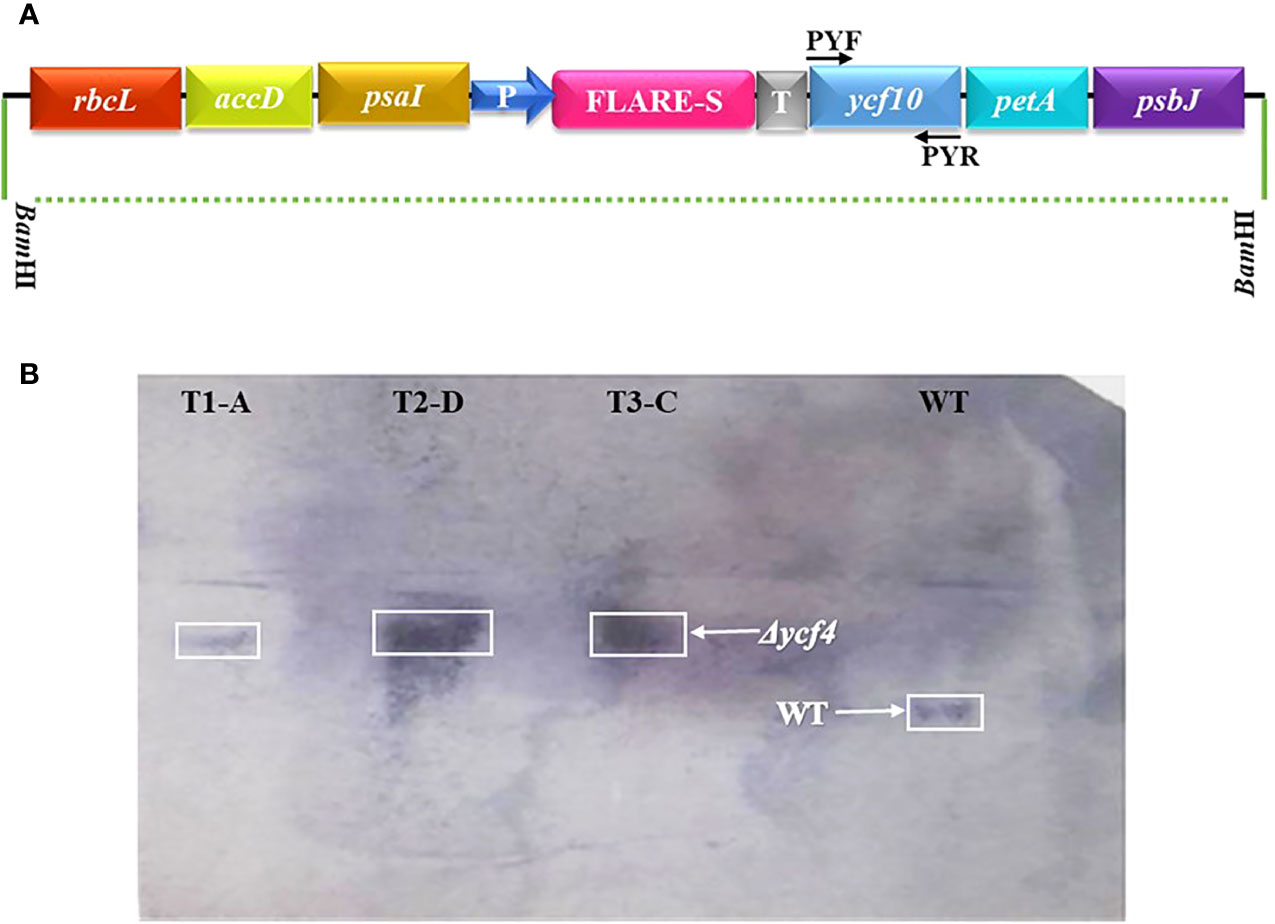
Figure 3 Confirmation of plasmy level of Δycf4 plants through Southern Blot Analysis. (A) Physical map showing location of primers used to amplify the probe. (B) Total cellular DNA of Δycf4 and wild-type tobacco plants was digested with BamH1, transferred onto the nitrocellulose membrane, and hybridized against a biotin-labeled probe prepared from ycf10 that amplifies both transformed and wild-type chloroplast DNA since ycf10 gene-specific probe detects all plastome copies irrespective to their genotype whether transformed or wild-type (untransformed). However, these plants were selected from an already screened pool (T1-T4 as shown in Figure 2C) of transgenic plants.
Impact of YCF4 deletion on phenotype and mode of nutrition
Homoplastic ∆YCF4 plants appeared to have a distinct phenotype. The newly emerged younger leaves were green which gradually bleached out with the attainment of maturity. Lowermost leaves were almost white with minimum chlorophyll whereas top-most leaves were green with comparatively higher content of chlorophyll when plants were grown under normal light. The leaves continued to bleach until the whole plant was discolored and showed stunted growth with an inability to grow autotrophically. To eliminate the possibility that standard light (60 µmol m-2 s-1) may cause photobleaching, the plants were grown under low light (30 µmol m-2 s-1), and a light green phenotype was maintained in the ∆YCF4 plants. Further, to assess the impact of the autotrophic mode of nutrition, plants were grown at different levels of sucrose (0, 0.5, 1.0, 1.5, 2.0, 2.5, and 3.0%). Mutant plants cultured on 0, 0.5, and 1% sucrose were unable to survive and no leaf development was observed. However, plants cultured on 1.5, 2, 2.5, and 3.0% sucrose showed growth, and leaf development was observed with increased sucrose concentration. The ∆YCF4 plants cultured on 3% sucrose were light leaves green in color compared with the wild-type plants which were green in color with normal growth (Figure 4). Further, several mutant plants growing on 3% sucrose were shifted to compost-containing pots for acclimatization, but they were unable to survive. This confirmed that YCF4 mutants are unable to survive autotrophically rather they require an additive carbon source for sustainability and growth.

Figure 4 (A) Homotransplastomic Δycf4 tobacco plants were cultured on different levels of sucrose (0.0%, 0.5%, 1.0%, 1.5%, 2.0%, 2.5% and 3%) to assess autotrophic/heterotrophic nature of Δycf4 plants in comparison with wild-type in vitro growing tobacco plants. (B) Plants at A were taken out of the jars.
Electron microscopy revealed ultrastructural variations between normal and ∆YCF4 plants
To investigate the ultrastructural variations in chloroplasts due to the absence of YCF4, leaf tissues from ∆YCF4 mutant and wild-type plants were analyzed using a transmission electron microscope (TEM). Ultrastructural studies revealed that chloroplasts in knockout plants underwent substantial structural changes that may be correlated with the absence of YCF4 protein. Figure 5 revealed distinct variations in chloroplast size and shape between knockout plants and wild-type (normal) plants. The TEM results showed that chloroplasts in wild-type plants were oblong in shape and larger in size than those of mutant knockout plants which were almost rounded. Further, the thylakoid membranes were densely packed in chloroplasts of wild-type plants as compared to those of knockout plants. In knockout plants, the grana thylakoids were less discrete, and their stacks exhibited a loss of their orderly structure. As the thylakoid membranes became less organized some vesicular structures appeared in mutant chloroplasts (Figure 5A).
Figure 5 Transmission electron microscopy of Δycf4 and untransformed tobacco leaves shows the presence of defective plastids in the leaves of bleached plants. (A, B) Chloroplasts in light green leaves of Δycf4 plants (C, D) Chloroplasts in lush green leaves of wild-type plants. The micrographs showed that ycf4 knockout caused structural anomalies including smaller size and rounded shape of chloroplasts in leaves.
Physiological and photosynthetic performance of YCF4 mutants
The YCF4 gene deletion appeared to have profound effects on the photosynthetic and physiological performance of mutant plants (Figure 6A). The mutants were unable to attain normal contents of total chlorophyll as the topmost young leaves of mutant plants accumulated 2.6 mg/g of chlorophyll compared to the wild-type plant leaves (3.1 mg/g). The levels were decreased up to 99.98% in non-photosynthetic cells of mutants as the plant matures, from top to bottom (Figure 6B). Likewise, physiological parameters including photosynthetic rate (A), transpiration rate (E), stomatal conductance (gs), sub-stomatal CO2 (Ci), and photosynthetic photon flux density (q also lux) also revealed that YCF4 plants were physiologically incompetent as compared with normal tobacco plants (Figure 7).
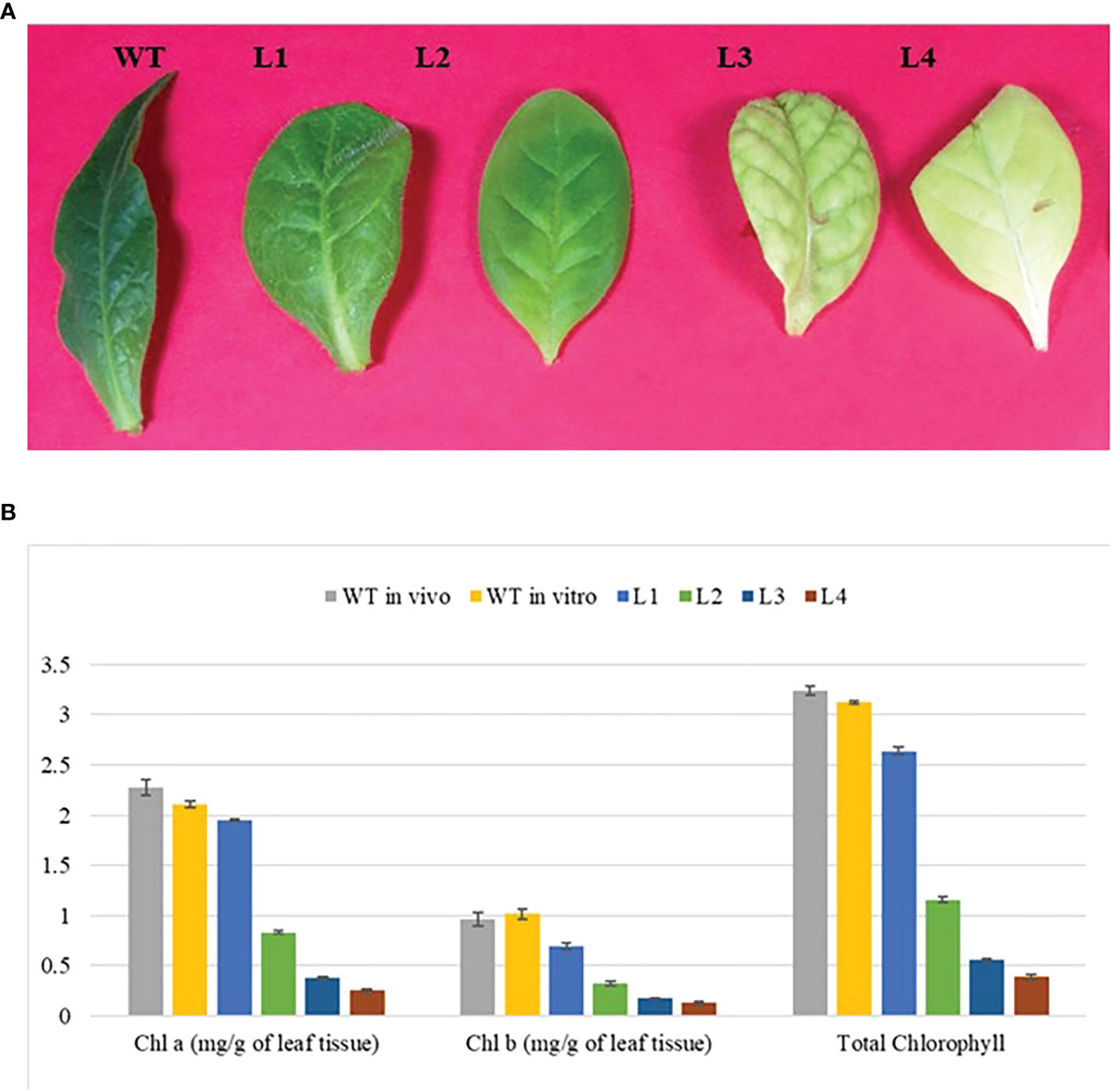
Figure 6 Photosynthetic performance of ∆ycf4 plants. (A) Leaves samples used for chlorophyll estimation (B) Graphical representation showing chlorophyll content in wild-type and transgenic plants. The lower contents of photosynthetic pigments in the ∆ycf4 plants indicate that they were physiologically incompetent as compared with wild-type tobacco plants. WT represents wild-type whereas L1, L2, L3, and L4 represent 1st, 2nd, 3rd and 4th leaf of ∆ ycf4 plants from top to bottom, respectively.
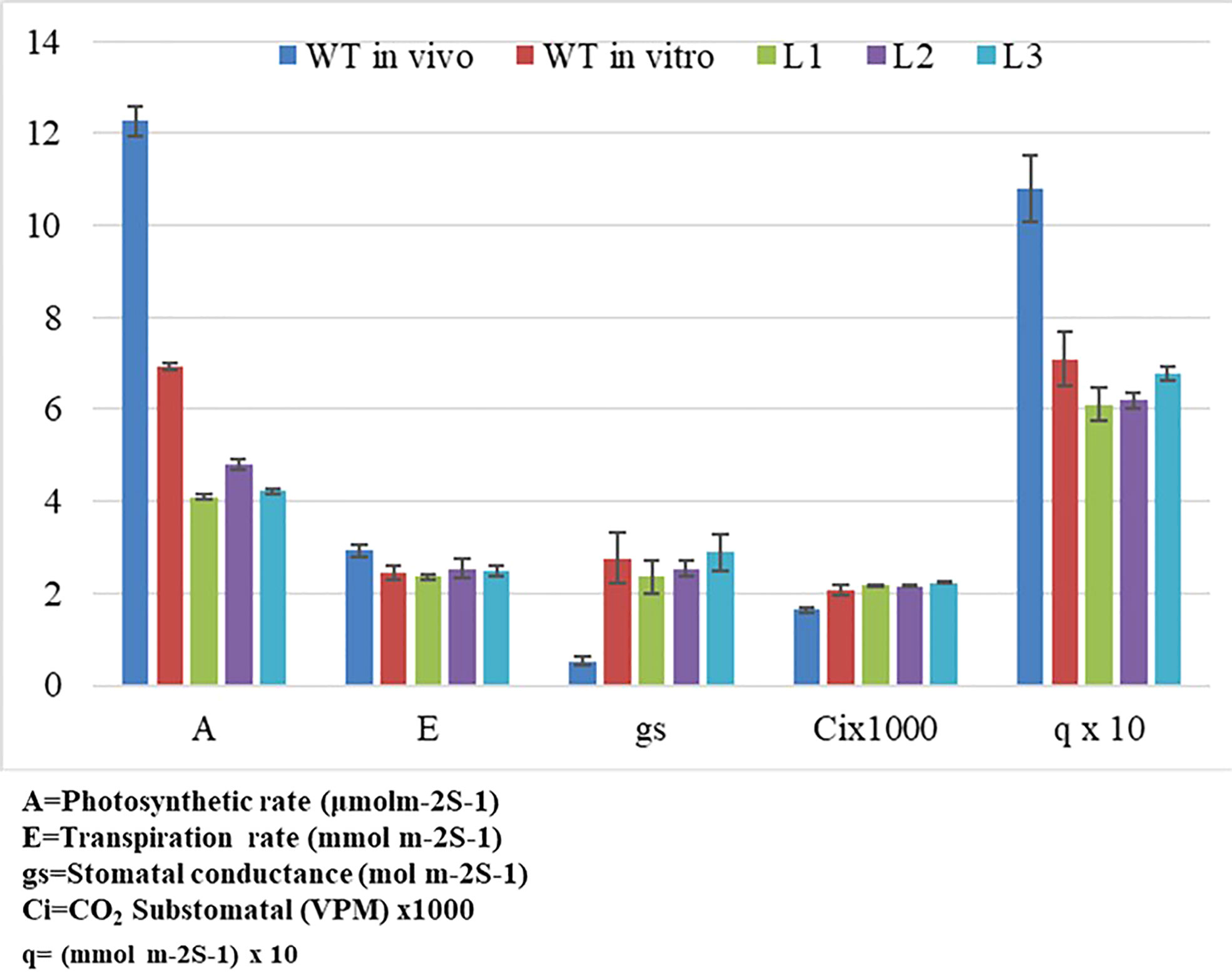
Figure 7 Impact assessment of ycf4 deletion on physiological performance of transplastomic ∆ycf4 and wild-type (WT) tobacco plants. WT represents Wild-type whereas L1, L2, and L3 represent 1st, 2nd, and 3rd leaf of ∆ycf4 plants from top to bottom, respectively.
Transcript analysis of plastid-encoded genes in ∆YCF4 plants
The effect of YCF4 deletion on the expression of plastid-encoded genes was determined by transcript analysis of mutant plants. No transcripts were detected in purified ∆YCF4 plants since the gene has been completely replaced with a marker gene. The transcripts levels of psaA, psaB, psaC and psaH encoding photosystem-I proteins appeared the same in the ∆YCF4 plants and in normal untransformed tobacco plants. Likewise, the accumulation of psbA, psbB, psbC, psbD, psbE transcripts encoding Photosystem-II proteins were not decreased significantly in these mutants suggesting that YCF4 deletion did not have a direct role in the transcriptional regulation of PS-I and PS-II genes. Further, the transcript levels of ribosomal protein-encoding genes rps16, rps2, and rrn16, and of ycf10, rpoA, rpoB, aacD and petA remained unchanged. Interestingly, the transcript levels of rbcL and lhc genes were significantly reduced, suggesting that YCF4 deletion affects the accumulation of RUBISCO and LHC1, eventually the photosynthesis (Figure 8).
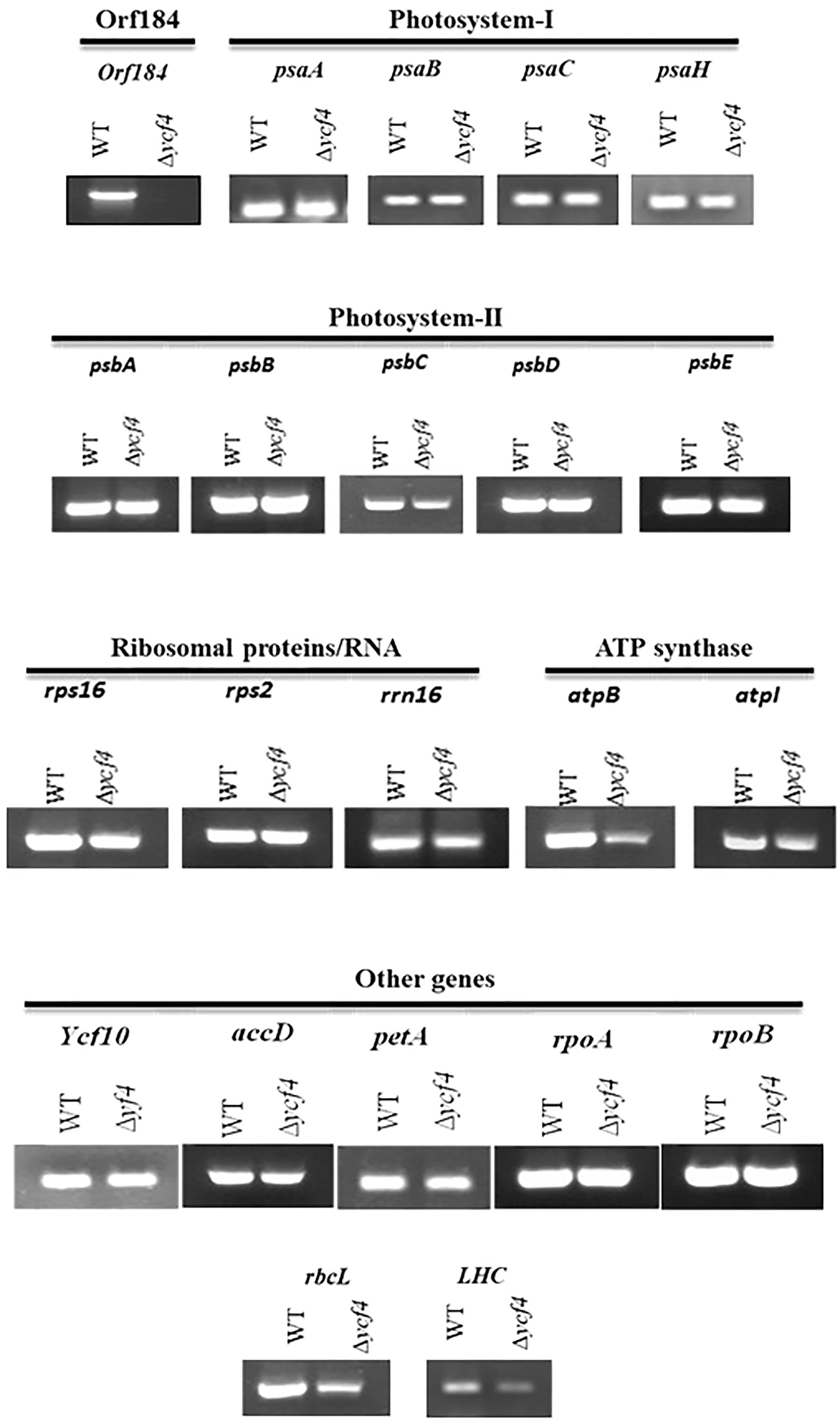
Figure 8 Transcript analysis of ∆ycf4 and wild-type tobacco plants. Transcripts show the expression level of genes in both wild-type and ∆ycf4 plants. The expression of various subunits of photosystem-I (psaA, psaB, psaC, psaH) and photosystem-II (psbA, psbB, psbC, psbD, psbE) remained unaltered. The absence of a transcript in ∆ycf4 indicates the purification of the ∆ycf4 plants to homoplasmy.
YCF4 interaction with other photosynthesis proteins
The molecular interaction of full-length YCF4 with PS-I subunits indicated strong interaction with psaB, psaC, and psaH, each having seven hydrogen bonds with YCF4. But YCF4+psaC complex showed the most stable interaction among them with bond lengths of 2.62-2.93Å (Supplementary Figure S1). The bond length corresponds to the strength of interaction between interacting molecules and should be <4Å as the greater the bond length, the weaker will be the interaction. Likewise, among the PS-II subunits psaE showed a peculiar binding pattern and formed five hydrogen bonds with YCF4 (Supplementary Figure S2). The ATP synthase consists of two chains namely the alpha chain and beta chain. Both chains are known to interact with YCF4 to carry out photosynthesis (Yamori et al., 2011). Only the beta chain (atpB) revealed an effective docking pattern with YCF4 forming twelve hydrogen bonds with a bond length of 2.56-3.15Å (Supplementary Figure S3). Ribosomal proteins were also investigated as they are reported to play an essential role in the development of plant phenotypes (Horiguchi et al., 2012). rrn16 showed a strong affinity with YCF4 protein forming ten hydrogen bonds within the bond length range of 2.63-3.19Å. Likewise, the interaction of YCF4 with other plastidic proteins was also determined that showed the maximum binding affinity with rbcL among all the candidate proteins.
These photosynthetic proteins were further docked with carboxylic and amino terminus of YCF4 protein to elucidate which portion of the protein is more essential for photosynthesis. Among PS-I proteins, the amino terminus of YCF4 revealed maximum interaction with psaB forming five hydrogen bonds while twelve hydrogen bonds were observed in the carboxyl terminus of the YCF4+psaH complex. The psbE of PS-II bonded with the amino terminus of YCF4 effectively as compared to other proteins but formed only six hydrogen bonds while psbC strongly docked making thirteen hydrogen bonds with the terminus of YCF4. The carboxyl terminus of YCF4 also demonstrated durable interaction with other PS-II proteins but weaker than psbC. The docked complexes of the carboxyl terminus of YCF4 with rps2, rps16 and rrn16 showed 7, 11, and 6 hydrogen bonds respectively, while the amino terminus of YCF4 bonded with these proteins comparatively stronger showing 14, 18, 18 hydrogen bonds with rps2, rps16 and rrn16 (Supplementary Figure S4). Furthermore, the beta chain of ATP synthase is supposed to interact better compared to the alpha chain. The beta chain formed only eight hydrogen bonds with the amino terminus of YCF4 however twenty-eight hydrogen bonds were found in the carboxyl terminus of YCF4+atpB (Supplementary Figure S3). Likewise, assessing other proteins, rpoB found to be well-interacted with the truncated versions of YCF4 than other proteins revealing nine bonds with amino terminus and twenty-five bonds with carboxyl terminus of YCF4 (Supplementary Figure S5).
The YCF4 was also docked with the four core subunits of LHC of PS-I and a nuclear-encoded small subunit (RBCS) of RUBISCO to further validate the findings. Their carboxyl terminus showed stronger interaction with a high number of hydrogen bonds forming between them. The relevance of the carboxyl terminus of YCF4 is once again demonstrating its strong connection with LHCA1, LHCA2, LHCA3, LHCA4, and RRBC (Table 1). The number and length of hydrogen bonds in the docked complexes of full-length and truncated YCF4 variants with many other proteins are shown in Table 1. Thus, these in-silico investigations support the current study’s notion that the carboxylic terminus of YCF4 is more crucial for photosynthesis than the amino terminus.
Discussion
The photosystem I complex requires both plastid and nuclear-encoded proteins for biosynthesis. PS1 assembly requires the association of several redox cofactors, chromophores, and Fe-S clusters. Ycf4 is a crucial auxiliary element in the PS1 assembly process. It has previously been shown to be important in photosystem I (PSI) formation in the unicellular green alga Chlamydomonas reinhardtii, with ycf3- and YCF4-deficient mutants unable to develop photoautotrophically and accumulate PSI (Boudreau et al., 1997). They concluded that Ycf3 and Ycf4 are not essential for PSI subunit synthesis but are most likely involved in PSI complex assembly. Orf184 mutants of Cyanobacterium synechocystis grew normally like wild-type cells, according to Wilde et al. (1995). However, the pigment content of mutant cells (especially the phycocyanin to chlorophyll ratio) differed significantly from that of wild-type cells. Another study found that YCF4 mutants could maintain photoautotrophic growth. They believed the YCF4 gene product was not necessary for photosynthesis. Despite the fact that mutants had lower PSI levels, this was not due to a deficit in plastid gene expression.
They concluded that Ycf3 and Ycf4 are not required for the synthesis of PSI subunits but are most likely involved in the assembly of the PSI complex. However, pigment composition (particularly phycocyanin to chlorophyll ratio) of mutant cells was distinctly different from those of wild-type cells. Another research group also concluded that YCF4 mutants were able to sustain photoautotrophic growth. They were of the view that the YCF4 gene product is not essential for photosynthesis. Though mutants were deficient in PSI contents, this deficiency was not caused by a defect in plastid gene expression. Rather, it was suggested that YCF4 plays a key role at the post-translational stage, resulting in faulty PSI assembly or reducing its stability (Krech et al., 2012).
Contrarily, we establish that tobacco homoplastic YCF4 mutants are heterotrophic and cannot survive in an autotrophic environment. Carbon starvation appeared to inhibit plant growth, as mutants were unable to grow on MS medium containing sucrose up to 10 mg/L. Plants appeared to survive at 15-30 mg/L, although their growth was limited. When switched to compost-containing pots, these plants were photosynthetically incompetent and could not grow photoautotrophically. Our findings contradict that of Krech et al. (2012), who claimed that photoautotrophic growth was possible in YCF4 mutant plants.
Our findings, however, are consistent with those of Boudreau et al. (1997), who knocked out the nearly entire YCF4 gene from C. reinhardtii and concluded that the YCF4 gene product is essential for photosynthesis. The expression of the plastid-encoded genes encoding for the key subunits of photosystem I (psaA, psaB, psaC, and psaH) and Photosystem-II (psbA, psbB, psbC, psbD, psbE) were unaffected in YCF4 plants, indicating that Ycf4 deletion did not appear to have any direct role in photosystem biogenesis. However, YCF4 plants had much lower levels of lhc (light-harvesting complex) and rbcL (large subunit of ribulose-1,5-bisphosphate carboxylase-oxygenase) expression than wild-type normal plants. The LHC is important for the formation of a super complex photosystem (PSI) whereas rbcL is critical for RUBISCO. Decreased expression of LHC and rbcL may affect the conformation of the photosystem and accumulation of functional RUBISCO respectively, resulting in defective photosynthesis.
In the knockout plants, the microscopic studies revealed that chloroplast structure was abnormal, presumably due to the lack of YCF4 protein. Wild-type chloroplasts were substantially larger and oblong in shape, but knockout chloroplasts were much smaller and spherical. The thylakoid membranes appeared to be less organized with certain vesicular structures. Chloroplasts of non-green senescing Broccoli florets have shown similar disorganization and subsequent formation of vesicular structure (Terai and Watada, 2000) that has been attributed to the disorganization and disintegration of thylakoid membranes.
The size of the deleted YCF4 section may be a fundamental difference in the performance of YCF4 tobacco mutants generated by Krech et al. (2012) and mutants reported in the current investigations. The carboxyl terminus spanning 91 amino acids of the YCF4 protein had not been knocked out, and the resultant mutants were able to grow photoautotrophically whereas, we have knocked out the entire open reading frame and found that knocked plants could not survive photoautotrophically and grew stuntedly at 30 g/L sucrose. The results of the in-silico studies supported our findings since the interactions between the photosystem-I subunits psaB, psaC, psaH, and LHC and the carboxyl terminus of the YCF4 were stronger than those between the amino and carboxyl termini. Similar to this, the interaction of YCF4 with the large (encoded by chloroplast) and small (encoded by nuclear) subunits of RuBicCO also supported the significance of the carboxyl terminus. Hence, this study is direct evidence of the fact that deletion of full-length YCF4 has made the plants unable to survive photo-autotrophically as interaction with all the proteins engaged directly or indirectly in photosynthesis has revealed strong binding patterns with the carboxyl terminus of YCF4.
We came to the conclusion that deletion of the entire YCF4 open reading frame prevents tobacco plants from growing autotrophically, even though photosystem biogenesis is a complex process and various co-factors and related proteins still need to be investigated. Under heterotrophic circumstances (30g/L sucrose), plants could thrive, but they grew stuntedly and died when placed in pots with peat moss.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding author.
Author contributions
MK conceived the idea, supervise the students, and wrote the manuscript. MM, RR, KM, and GM performed data analysis. Whereas, FJ handled microscopic studies and helped in the manuscript write-up. All authors contributed to the article and approved the submitted version.
Funding
Funds were provided by the Ministry of Science & Technology, Islamabad Pakistan to MK.
Acknowledgments
The authors are highly thankful to the Ministry of Science & Technology, Islamabad Pakistan.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1014236/full#supplementary-material
Abbreviations
YCF4, Hypothetical chloroplast open reading frame; PSI, Photosystem I; PSII, Photosystem II; Cytb6f, cytochrome b6f complex.
References
Blankenship, R. E. (2010). Early evolution of photosynthesis. Plant Physiol. 154 (2), 6434–438. doi: 10.1104/fpp.110.161687
Boudreau, E., Takahashi, Y., Lemieux, C., Turmel, M., Rochaix, J. D. (1997). The chloroplast ycf3 and YCF4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J. 16, 6095–6104. doi: 10.1093/emboj/16.20.6095
Embley, T. M., Martin, W. (2006). Eukaryotic evolution, changes and challenges. Nature 440 (7084), 623–630. doi: 10.1038/nature04546
Horiguchi, G., Van Lijsebettens, M., Candela, H., Micol, J. L., Tsukaya, H. (2012). Ribosomes and translation in plant developmental control. Plant Sci. 191, 24–34. doi: 10.1016/j.plantsci.2012.04.008
Islam, E., Liu, D., Li, T., Yang, X., Jin, X., Mahmood, Q., et al. (2008). Effect of Pb toxicity on leaf growth, physiology and ultrastructure in the two ecotypes of Elsholtzia argyi. J. Hazard. Mater. 154 (1-3), 914–926. doi: 10.1016/j.jhazmat.2007.10.121
Kargul, J., Barber, J. (2011). Structure and function of photosynthetic reaction centres. Molecular solar fuels Cambridge: Royal Society of Chemistry, 107–142. doi: 10.1039/9781849733038-00107
Khan, M. S., Maliga, P. (1999). Fluorescent antibiotic resistance marker for tracking plastid transformation in higher plants. Nat. Biotechnol. 17 (9), 910–915. doi: 10.1038/12907
Kozakov, D., Hall, D. R., Xia, B., Porter, K. A., Padhorny, D., Yueh, C., et al. (2017). The ClusPro web server for protein–protein docking. Nat. Protoc. 12 (2), 255–278. doi: 10.1038/nprot.2016.169
Krech, K., Ruf, S., Masduki, F. F., Thiele, W., Bednarczyk, D., Albus, C. A., et al. (2012). The plastid genome-encoded Ycf4 protein functions as a nonessential assembly factor for photosystem I in higher plants. Plant Physiol. 159 (2), 579–591. doi: 10.1104/pp.112.196642
Nazir, S., Khan, M. S. (2013). Integration of novel chlorophyll genes from black pine into the chloroplast genome of tobacco. Pak. J. Bot. 45 (S1), 595–600.
Ozawa, S., Nield, J., Terao, A., Stauber, E. J., Hippler, M., Koike, H., et al (2009). Biochemical and structural studies of the large Ycf4-photosystem I assembly complex of the green alga Chlamydomonas reinhardti. Plant Cell 21 (18), 2424–2442. doi: 10.1105/tpc.108.063313
Schimper, A. F. W. (1883). Ueber die entwickelung der chlorophyllkoerner und farbkoerper. Bot. Zeit. 41, 105–113.
Schmidt, M., Gebner, G., Luff, M., Heiland, I., Wagner, V., Kaminski, M., et al. (2006). Proteomic analysis of the eyespot of Chlamydomonas reinhardtii provides novel insights into its components and tactic movements. Plant Cell 18 (8), 1908–1930. doi: 10.1105/tpc.106.041749
Song, M., Kuo, L. Y., Huiet, L., Pryer, K. M., Rothfels, C. J., Li, F. W. (2018). A novel chloroplast gene reported for flagellate plants. Am. J. Bot. 105 (1), 117–121. doi: 10.1002/ajb2.1010
Terai, H., Watada, A. E. (2000). Scanning electron microscopic study of modified chloroplasts in senescing broccoli florets. Hortic. Sci. 35 (1), 99–103. doi: 10.21273/HORTSCI.35.1.99
Wallace, A. C., Laskowski, R. A., Thornton, J. M. (1996). LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 8, 127–134. doi: 10.1093/protein/8.2.127
Wilde, A., Hartel, H., Hubschmann, T., Hoffmann, P., Shestakov, S. V., Borner, T. (1995). Lnactivation of a synechocystis sp strain pcc 6803 gene with homology to conserved chloroplast open reading frame 184 increases the photosystem II-to-photosystem I ratio. Plant Cell 7, 649–658. doi: 10.1105/tpc.7.5.649
Keywords: YCF4, heterotrophic, knockout, transcript analysis, null alleles, homoplasmic
Citation: Khan MS, Riaz R, Majid M, Mehmood K, Mustafa G and Joyia FA (2022) The tobacco chloroplast YCF4 gene is essential for transcriptional gene regulation and plants photoautotrophic growth. Front. Plant Sci. 13:1014236. doi: 10.3389/fpls.2022.1014236
Received: 08 August 2022; Accepted: 10 October 2022;
Published: 24 October 2022.
Edited by:
Niaz Ahmad, National Institute for Biotechnology and Genetic Engineering (NIBGE), PakistanReviewed by:
David Herrin, University of Texas at Austin, United StatesManzar Abbas, Yibin University, China
Copyright © 2022 Khan, Riaz, Majid, Mehmood, Mustafa and Joyia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muhammad Sarwar Khan, c2Fyd2Fya2hhbl80MEBob3RtYWlsLmNvbQ==
 Muhammad Sarwar Khan
Muhammad Sarwar Khan Rimsha Riaz1
Rimsha Riaz1 Ghulam Mustafa
Ghulam Mustafa Faiz Ahmad Joyia
Faiz Ahmad Joyia